b kb
|
|
|
- Abigayle Willis
- 5 years ago
- Views:
Transcription
1 Supplementary Figure S1 COX7RP mrna/gapdh COX activity (OD 45 nm/mg protein) Relative DNA amount COX7RP mrna/gapdh a c pcxn2-flag-cox7rp CAG Flag CMV enhancer Chiken b-actin promoter COX7RP Muscle pa Rabbit b-globin polya signal d b kb BAT.4.4 e.2 KO KO f KO KO * (Days) Supplementary Figure S1. Generation of COX7RP transgenic () mice. a, Schematic representation of the COX7RP transgene. b, Genotyping of COX7RP- mice. Representative litters containing COX7RP transgene were shown. Primers specific for the transgene were used to amplify the allele (6 bp). c and d, Overexpression of COX7RP in COX7RP- mice. mrnas of endogenous Cox7rp and introduced human COX7RP were quantified in quadriceps femoris muscles (c) and BAT (d) simultaneously by qpcr using common primers for mouse and human., P <.1; Dunnett s post hoc test. e, Increase in COX activity in COX7RP- MEFs. COX activity was assessed using MEFs from and COX7RP- mice. Data are presented as means ± s.d. (n = 3). *, P <.5 for versus COX7RP- ; Student s t-test. f, Upregulation of growth in COX7RP- MEFs. Data are presented as means ± s.d. (n = 3). *, P <.5,, P <.1for versus COX7RP- at each indicated time point; Student s t-test.
2 Supplementary Figure S2 Quadriceps femoris muscle /body weight Body weight (g) BAT/body weight a b KO KO * c KO KO * KO KO Supplementary Figure S2. Decreased skeletal muscle weight and increased BAT weight in Cox7rpKO mice. a, Body weight of 4-month-old mice (a) with genotypes, Cox7rpKO (KO, n = 9), COX7RP- (, n = 1), COX7RP-: Cox7rpKO (KO, n = 9), and (n = 7), were measured. b and c, Quadriceps femoris muscle (b) and BAT (c) was dissected and weight of these tissues were adjusted with corresponding body weight and data are presented as means ± s.d. *, P <.5,, P <.1; Dunnett s post hoc test.
3 Supplementary Figure S3 COX activity (O.D. 45 nm/mg protein) 3 2 * * 1 Cox7rp: Muscle: KO KO KO Supplementary Figure S3. Decreased COX activity in muscles of Cox7rpKO mice. The COX activity of quadriceps femoris, gastrocnemius and heart was measured from the tissue homogenates. Data are presented as means ± s.d. (n = 3). *, P <.5,, P <.1 for versus KO in each muscular tissue; Student s t-test.
4 Supplementary Figure S4 After 4 min KO After 7 min KO KO After 4 min KO After 7 min Supplementrary Figure S4. Compromised exercise performance in Cox7rpKO mice. Four-monthold Cox7rpKO (KO), COX7RP- (), COX7RP-:Cox7rpKO (KO) and mice were subjected to a forced treadmill exercise test. Movie captures at 4 and 7 min after the forced treadmill exercise test. Cox7rpKO mice showed exhaustion after 4 min.
5 Supplementary Figure S5 VO 2 (ml/min/g) VCO 2 (ml/min/g) RQ.1 * Cox7rp: KO KO KO KO KO KO Supplementary Figure S5. Decrease in O 2 consumption rate in Cox7rpKO mice in the steady state. CO 2 production, O 2 consumption and respiratory quotient (RQ) were analyzed for 1 h. Data are presented as means ± s.d. (n = 7 or 8). *, P <.5,, P <.1; Dunnett s post hoc test.
6 Supplementary Figure S6 mrna/ GAPDH Pparα Pparγ Pparδ Pgc-1α Pcg-1β Errα Errβ Errγ Nrf1 Tfam.2.15 Cox7rpKO.1.5 Supplementary Figure S6. Downregulation of transcriptional regulatory factors associated with mitochondrial function in Cox7rpKO mice. qpcr analysis was performed using gene-specific primers with total RNAs prepared from BAT of and Cox7rpKO male mice. Data are presented as means ± s.d. (n = 3). *, P <.5,, P <.1; Student s t-test.
7 Supplementary Figure S7 SDS-PAGE SDS-PAGE SDS-PAGE SDS-PAGE a BN-PAGE b BN-PAGE I/III 2 /IV n III 2 /IV n IV 2 IV I/III 2 /IV n III 2 /IV n (%) WB: (%) 46 UQCRC NDUFA9 38 IV 2 IV WB: UQCRC2 NDUFA (%) COX4 COX7RP (%) COX4 COX7RP c BN-PAGE 293T/vec d BN-PAGE 293T/COX7RP I/III 2 /IV n III 2 /IV n (%) (%) WB: UQCRC2 NDUFA9 RISP I/III 2 /IV n III 2 /IV n (%) (%) WB: UQCRC2 NDUFA9 RISP T/vec 293T/COX7RP Supplementary Figure S7. COX7RP stimulates the assembly of mitochondrial respiratory chain supercomplexes. Mitochondrial proteins of 293T cells transfected with empty vector (a and c) and COX7RP (b and d) were solubilized with digitonin in the concentration of 2 g/g protein (a and b) or.5 g/g protein (c and d), and subjected to BN-PAGE followed by second-dimensional SDS- PAGE. Western blot analysis was performed with antibodies for NDUFA9 of complex I, UQCRC2 of complex III, RISP of complex III, COX4 of complex IV, and COX7RP. Positions corresponding to mitochondrial supercomplex species I/III 2 /IV n, I/III 2, and III 2 /IV n, as well as dimerized complex IV (IV 2 ) and monomerized complex IV (IV) are indicated. Percentages of UQCRC2, RISP or COX4 signal intensities in each whole amount are indicated in the images.
8 Supplementary Figure S8 Fig. 1c Fig. 4a Fig. 4b Fig. 4c Fig. 4d Supplementary Figure S8. Full length images of immunoblots.
9 Supplemantary Table S1. Downregulated pathway genes in the muscle of Cox7rpKO mice Term Count P value Genes Muscle protein 1 6.9E-13 Tnnt2, Myl7, Myl2, Tnnc1, Myl3, Myh7, Myh6, Tnni3, Tnni1, Tpm3 Cardiac muscle contraction Dilated cardiomyopathy E-11 Tnnt2, Myl2, Atp2A2, Tnnc1, Myl3, Ryr2, Myh7, Myh6, Cox7A2L, Tnni3, Tpm E-1 Tnnt2, Myl2, Atp2A2, Tnnc1, Myl3, Pln, Ryr2, Myh7, Myh6, Tnni3, Tpm3 Vmn2R43, Vmn1R125, Olfr76, Vmn2R42, Vmn1R148, Olfr331, Vmn1R121, Vmn2R26, Vmn2R-Ps6, Gm769, Olfr728, Vmn1R168, Acvr1C, Adrb3, Olfr36, Receptor 33 2.E-9 Olfr1333, Vmn1R93, Olfr26, Olfr1134, Olfr247, Vmn1R132, Olfr145, Vmn2R51, Vmn2R34, Olfr124, Vmn1R179, Olfr137, Olfr715, Vmn2R37, Olfr1287, Csprs, Vmn2R122, Vmn1R-Ps4, Chrm2, Vmn1R223, Olfr8, V1Rg1, Ryr2, Olfr329, Vmn1R191, Tshr, Olfr121, Vmn1R1 Hypertrophic cardiomyopathy 1 4.7E-9 Tnnt2, Myl2, Atp2A2, Tnnc1, Myl3, Ryr2, Myh7, Myh6, Tnni3, Tpm3 G-protein coupled receptor protein signaling pathway 3 1.7E-8 Vmn2R43, Vmn1R125, Olfr76, Vmn2R42, Olfr331, Vmn1R148, Vmn1R121, Vmn2R26, Vmn2R-Ps6, Olfr728, Vmn1R168, Adrb3, Olfr36, Olfr1333, Vmn1R93, Olfr26, Olfr1134, Olfr247, Vmn1R132, Olfr145, Vmn2R51, Vmn2R34, Olfr124, Vmn1R179, Olfr715, Olfr137, Vmn2R37, Olfr1287, Vmn2R122, Vmn1R-Ps4, Chrm2, Vmn1R223, Olfr8, V1Rg1, Olfr329, Vmn1R191, Tshr, Olfr121, Vmn1R1 G-protein coupled receptor E-6 Tnnt2, Myl7, Myl2, Tnnc1, Myl3, Myh7, Myh6, Tnni3, Tnni1, Tpm3 Cell surface receptor linked signal transduction E-6 Tnnt2, Myl7, Myl2, Tnnc1, Myl3, Myh7, Myh6, Tnni3, Tnni1, Tpm3 Transducer E-6 Tnnt2, Myl7, Myl2, Tnnc1, Myl3, Myh7, Myh6, Tnni3, Tnni1, Tpm3 Heart 4 6.3E-6 Tnnt2, Myl2, Atp2A2, Tnnc1, Myl3, Ryr2, Myh7, Myh6, Cox7A2L, Tnni3, Tpm3
10 Supplementary Table S2. Upregulated pathway genes in the muscle of Cox7rpKO mice Term Count P value Genes Amino-acid biosynthesis 4 1.3E-5 Pycr1, Aldh18A1, Asns, Psat1 Cellular amino acid biosynthetic process Amine biosynthetic process 4 2.2E-4 Pycr1, Aldh18A1, Asns, Psat1 4 1.E-3 Pycr1, Aldh18A1, Asns, Psat1 Oxidoreductase 7 1.6E-3 Mthfd2, Pycr1, Cyb5R1, Aldh18A1, Egln3, Impdh2, Srxn1 Glutamine family amino acid metabolic process Microtubule-associated protein MAP1B Organic acid biosynthetic process Carboxylic acid biosynthetic process L-type amino acid transporter Nitrogen compound biosynthetic process 3 6.1E-3 Pycr1, Aldh18A1, Asns 2 6.4E-3 Mtap1A, Mtap1B 4 6.6E-3 Pycr E-3 Pycr1, Aldh18A1, Asns, Psat E-3 Slc7A5, Slc7A E-3 Pycr1, Aldh18A1, Asns, Psat1, Impdh2
11 Supplementary Table S3. Downregulated pathway genes in the BAT of Cox7rpKO mice Term Count P value Genes Mitochondrion E-17 Ldha, Pdp2, Aldh1L1, Nit2, Ehhadh, Ldhd, Bnip3, Echdc2, Echdc3, Car5B, Acss3, Mccc2, Pecr, Gpx1, Acss1, D5Rik, Slc22A4, Gstz1,Tfb2M, Pxmp2, Prkaca, Slc25A1, Elovl6, Hmgcl, Rtn4Ip1, Hemk1, Brp44, Suox, Aldh6A1, Aco1, Aldh5A1, Lgals12, C3318D2Rik, Immp1L, Hagh, Sccpdh, Glul, Nme3, Ppm1K, Slc25A35, D12Rik, Gm9568, Gpam, Slc27A2, Pcca, Mpst, Mavs, Me1, Ndufb3, Hsd17B1, Apool, Acadsb, Bcat2, Adhfe1, Abcd1, Hk2, Echs1, Cox7A2L, Acat2, Ccbl2, Acat1, Sfxn5, Acat3, Ivd, Fasn, Abcd2, Idh1, Aldh4A1, Mrpl18, Slc25A44, Cabc1, Slc25A42, Acsl5, Pdk1, Gpd2, Gpd1, Gcdh, Maob, Acaca, Acly, Acacb, Ucp1, Ndufa1, Tst, Acsm3, Pcx, Hmgcs2, Slc25A1, Hdhd3, Brp44L, Slc25A19, Acad11, Scp2, Mgst1, Acsm5, D1Jhu81E Oxidation reduction E-14 Sepx1, Steap4, Cyp2D1, Ldha, Aldh1L1, Ltbp2, Ehhadh, Ldhd, Fdft1, Gpx1, Pecr, P4Ha1, Gpx8, Rtn4Ip1, Aldh6A1, Suox, Cyp1A1, Aldh5A1, Aldh3B2, Cyp2E1, Cyp2B1, Cdo1, Dhdh, Dhrs7, C3318D2Rik, Sccpdh, H6Pd, Hsd11B1, Gm9568, Ndufb3, Me1, Hsd17B1, Acadsb, Adhfe1, Cyp2F2, Hsd3B7, Hsd17B12, Egln3, Egln1, Fmo5, Ivd, Fmo1, Fasn, Aldh4A1, Idh1, Aldh1A7, Adh6B, Gpd2, Gcdh, Gpd1, Maob, F5Rik, Cyp4V3, Ndufa1, Vat1L, Dio2, Dpyd, Acad11, Retsat Oxidoreductase E-13 Sepx1, Steap4, Cyp2D1, Ldha, Aldh1L1,Ehhadh, Ldhd, Fdft1, Gpx1, Pecr, P4Ha1, Gpx8, Aldh6A1, Suox, Cyp1A1, Aldh5A1, Cyp2E1, Cyp2B1, Cdo1, Dhdh, Dhrs7, Sccpdh, H6Pd, Hsd11B1, Gm9568, Me1, Hsd17B1, Acadsb, Adhfe1, Cyp2F2, Hsd3B7, Hsd17B12, Egln3, Egln1, Fmo5, Fmo1, Ivd, Fasn, Aldh4A1, Idh1, Aldh1A7, Gpd2, Gcdh, Gpd1, F5Rik, Maob, Cyp4V3, Vat1L, Dio2, Dpyd, Acad11, Retsat Fatty acid metabolic process Lipid metabolism E E-1 Slc27A1, Acadsb, Ehhadh, Abhd5, Prkag2, Echs1, Echdc2, Pecr, Agpat6, Ptges, Elovl3, Acot11, Fasn, Elovl6, Ghr, Acsl5, Aldh5A1, Acaca, Adipor2, Acacb, Adipoq, Acsm3, Aacs, Slc27A2, Gpam, Acsm5, Mgst2 Gcdh, Ces3, Acadsb, Acaca, Echdc1, Echs1, Echdc2, Acacb, Acsm3, Mccc2, Ivd, Bche, D5Rik, Acad11, Acsm5, Pcca, Au18778, Acsl5 Brown fat cell differentiation E-1 Cebpa, Adrb3, Aldh6A1, Rgs2, Slc2A4, Mrap, Lrg1, Nudt7, Bnip3, Ucp1, Adipoq, Arl4A Mitochondrion 56 1.E-9 Fat cell differentiation E-9 Transferase E-9 Mitochondrial part E-8 Ldhd, Echdc2, Bnip3, Echdc3, Car5B, Acss3, Mccc2, Acss1, Tfb2M, Elovl6, Rtn4Ip1, Hmgcl, Aldh6A1, Suox, Aldh5A1, Hagh, Immp1L, Ppm1K, Slc25A35, Gpam, Pcca, Mpst, Ndufb3, Mavs, Hsd17B1, Acadsb, Bcat2, Adhfe1, Echs1, Cox7A2L, Acat1, Sfxn5, Ivd, Mrpl18, Aldh4A1, Slc25A44, Cabc1, Slc25A42, Acsl5, Gpd2, Pdk1, Gcdh, Maob, Ucp1, Ndufa1, Acsm3, Tst, Pcx, Hmgcs2, Slc25A1, Slc25A19, Brp44L, Scp2, Acsm5, Mgst1, D1Jhu81E Cebpa, Aldh6A1, Steap4, Mrap, Bnip3, Ucp1, Adipoq, Gpx1, Retn, Adrb3, Slc2A4, Rgs2, Lrg1, Nudt7, Arl4A Nampt, Sephs2, Gstm6, Pmvk, Itpka, Pnp, Fdft1, Gstm7, Acvr1C, St3Gal1, St6Galnac6, Agpat6, St3Gal5, St3Gal4, Gstz1, Tfb2M, Prkaca, Agphd1, Pcyt2, Hemk1, Sgk2, Pfkl, Chpt1, Pnpla3, Cdkl5, Chst1, Prkd1, Pank3, Nme3, Dgat2, Pank1, Adk, D12Rik, Gpam, Hs3St3B1, Mpst, Bcat2, Taldo1, Fut7, Pfkfb3, Pfkfb1, Abhd5, Mknk2, Mapkapk3, Hk2, Adrbk2, Acat2, Ccbl2, Acat1, Acat3, Gm135, Sbk1, Hnmt, Ptk2B, Fasn, Gys2, Uck1, Gsto2, Cabc1, Mettl7A1, Pdk1, Gsta3, Mogat1, B3Galt2, Acly, Tkt, Inmt, Dolk, Tst, Ntrk3, Hmgcs2, Pygl, Ntrk2, Gpt, Scp2, Mgst1, Nat8L, Mgst2, Nnmt Mavs, Ndufb3, Hsd17B1, Apool, Acadsb, Echs1, Bnip3, Cox7A2L, Acat1, Sfxn5, Mccc2, Acss1, Ivd, Aldh4A1, Slc25A1, Slc25A44, Slc25A42, Hmgcl, Acsl5, Gpd2, Pdk1, Gcdh, Suox, Maob, Ucp1, Ndufa1, Tst, Hagh, Acsm3, Immp1L, Pcx, Hmgcs2, Ppm1K, Slc25A1, Slc25A35, Slc25A19, Brp44L, Gpam, Pcca, Mgst1, Acsm5, Mpst
12 Supplementary Table S4. Upregulated pathway genes in the BAT of Cox7rpKO mice Term Count P value Genes Phosphoprotein E-2 Xrcc5, Naf1, Jub, Tubb2B, Plekhm2, Surf6, Tubb2A, Rusc2, L1Cam, Rpl22L1, Snora65, Kifc3, Lphn2, Actg1, App, Mak16, Rpl12, Ptprj, R3Hcc1, Ctnna1, Myh9, Sirpa, Jup, Ifi22B, Grb1, Pitpnm3, Aaas, Rcc2, Spag5, Jun, Eif2S2, Mybbp1A, Dcbld2, Pltp, Spp1, Pppde2, Rps24, Odc1, Kif3A, Ppfibp1, Gars, Atr, Rps8, Fnip2, Gcfc1, Rgs1, Parp9, Lasp1, Gas2L3, Slc2A1, Trpv2, Nap1L1, Atp1D, Gm8292, Hsf2, Spg2, Synj2, Myc, Arhgef2, Gm5643, Rap2C, Anp32B, Acta2, Arhgef6, Gmeb1, Slc3A2, Hn1, Clic1, Exosc1, Arhgef12, Flnb, Trmt61A, Flna, Arhgef1, Elmo1, Ccnd1, Tmsb4X, Stbd1, Them4, Wdr43, Srgap1, Aw1462, Reps1, Tbc1D9, Gpr65, Eri1, Nufip1, Abi2, Hk1, Rtkn, 9432K1Rik, Il7R, Igsf1, Sorbs3, Kirrel, Anks6, Armcx3, Tcea1, Exoc1, Mid1, Cpt1A, Gla, Plscr2, Phf16, Phldb2, Akna, Ctnnal1, Rbm3, Eif5, Slc7A5, Fntb, Nrcam, Unc5B, Ank2, Mdfic, Snd1, Slc2A1, Ddx21, Sptlc2, Stap1, Stk24, Baiap2, Actn1, Tmsb1, Tmem29, Mrto4, Hspb6, Mtap1A, Hspb8, Mtap1B, Bmp2K, Stmn1, Pam, Nr2C1, Dock2, Smarcb1, Gp49A, Mettl1, Jund, Sh3Tc2, Mrc1, Trp53, Hcn2, Nin, Myo1D, Golim4, Myo1F, Pold3, Plk4, Atf4, Ddx56, Slc7A2, Fabp3, Acox2, Cyb5R2, Fosl2, Atl1, Cldn5, Wwc1, Gabbr2, Acot6, Ptma, Sdc3, Slc1A4, Prmt3, Spry2, Wars, Prmt1, Grwd1, Hmox1, Sntb2, Loxl4, Gm5841, Cdk5Rap2, Lrrfip1, Nfil3, Dlg2, Sgk1, Sars, Stxbp1, Stxbp4, Hltf, Ncl, Slc7A11, Ipo4, Frmd4A, Ipo5, Ern1, Mapre2, Lcp1, Lcp2, Ablim2, Vars, Esyt2, Chd7, Atic, Inpp5J, Naip5, Gpnmb, Ehd4, G7Rik, Slc12A4, Anxa1, Creb5, C3327C9Rik, Anxa4, Tab2, Pwp2, Pwp1, Cyba, Jak1, Rpp38, Dync1Li1, Sncg, G4Rik, Stard3Nl, Ptpn23, Ptpn21, Pdlim1, Got2, Got1, Gab2, Cd44, L9Rik, Stam, Zfp33, Ddx1, Matn2, C5Ar1, Stc2, Aars, Glce, Sfrs3, Baz1A, Pgm1, Lilrb4, Cdca7L, Nup43, Nek6, G22Rik, Aldoc, Akap12, Sfpi1, Igf2Bp2, Chek2, Myo9A, Plaa, Fam65B, Sema5A, Slc11A1, Fam12A, Vrk1, Vrk2, Fcer1G, Emd, Tyrobp, Sash1, Lgals3, Crip2, Osbpl6, Eprs, Ptpn12, Hn1L, Pla2G4A, Atp2A3, Clec7A, Utp14A, Utp2, Chaf1B, Klf4, Nars, Anpep, Cpeb1, 1627N9Rik, Ckb, Blzf1, Rrp1B, Mtbp, Tubb5, Zfp238, Tubb6, Fam129B, Dock1, Fam129A, Agap1, Rbbp5, Zcchc11, Mbd2, Rad5, Rab11Fip5, Nol1, Farsb, Ltv1, Bid, Lims2, Slc39A1, Mif, Plcl2, Sqstm1, Bcl2, Aatf, Kif21B, Tes, Fyb, Plek, Aff3, Ilf3, Atp1A1, Npr3, Samd4, Iws1, Svil, Capg, Phgdh, Nop58, Nop56, Zc3Hav1, Wfs1, Ctps, Gabrb1, Mbip, Tlr7, Pus7L, Dysf, Tiam1, Gnl2, Odz2, Gnl3, Cars, Dars, Rrp9, Arntl, Tns3, 49356M7Rik, Lat2, Trim33, Fnbp1L, 4116, Mvp, Plxnc1, Blm, Agfg1, Ssh2, Itgb2, Fxyd5, Srf, Eif3C, Eif3A, Laptm5, Npm1, Stambpl1, Rnf11, Strbp, Entpd1, Tsr1, Brca2, Mafk, Coro1C, Epb4.1L4A, Wdr1, St5, Plekha1, Fam175A, Ppip5K2, Prrx1, Mtss1L, Ddr2, Bzw2, Ccdc41, Evi2A, Pcgf6, Zyx, Msn, Tuba1A, Tuba1B, Ttc39B, Atp8B4, Pilra, Lair1, G3Bp1, Mtap4, Mpp6, Foxj3, Palld, Krtcap2, Arhgap25, Dapk1, Cd84, Prkd2, Ddr1, Tars, Umps, Eya2, Prnp, Ppp1R15A, Cnn3, Tnc, Cyth4, Hat1, Tpm4, Rgs1, Cep57, m2, Lars, Thyn1, Camk2B, Cnn2, Il2Rg, Myof, Dctd, Ptprc, Ampd3, Fam6A, Sh3Bp4, Slc6A8, Sh3Bp2, Nfib Nucleolus E-11 Rpp38, Taf1A, Surf6, Utp15, Eri1, Nufip1, Snora65, Dimt1, G2E3, Mak16, Rrp1B, Mdfic, Grwd1, Hmox1, Npm1, D19Bwg1357E, Aatf, Zfp33, Ddx21, Rpl12, Gnl2, Gnl3, Trp53, Exosc8, Tsr1, Ilf3, Rrp9, Exosc1, Mbd2, Ncl, Pwp2, Mrto4, Dis3, Plk4, Ddx56, Eya2, Rcc2, Nol1, Nol11, Nop58, Nop56, Utp14A, Utp2, Mybbp1A, Wdr43 ncrna metabolic process Intracellular nonmembranebounded organelle Nonmembranebounded organelle AminoacyltRNA synthetase Ribosome biogenesis trna metabolic process Ribonucleoprotein complex biogenesis E E E-11 Rpp38, Naf1, Nars, Utp15, Eri1, Qtrt1, Vars, Pus7L, Iars, Dimt1, Wars, Wdr36, Rrp1B, Trdmt1, Mettl1, Lars, Cars, Yars, Exosc8, Zcchc11, Dars, Sars, Aars, Gars, Eprs, Exosc1, Rrp9, Trmt61A, Dis3, Tars, Ddx56, Pop1, Farsb, Utp14A, Dus4L Ctnnal1, Rpp38, Xrcc5, Dync1Li1, Sncg, Jub, Tubb2B, Surf6, Tubb2A, Pdlim1, Ptpn21, Rpl22L1, Snora65, Kifc3, Actg1, Actg2, App, G2E3, Mak16, Ank2, Mdfic, D19Bwg1357E, Ddx21, Zfp33, Rpl12, Gnl2, Gnl3, Kif5A, Actn1, Tmsb1, Rrp9, Ctnna1, Myh9, Mrto4, Jup, Baz1A, Rcc2, Fnbp1L, Spag5, Mtap1A, Mtap1B, Rps12, Stmn1, Mybbp1A, Nup43, 4116, Blm, Ssh2, Akap12, Myo9A, Itgam, Dock2, Eif3A, Trim66, Smarcb1, Npm1, Strbp, Emd, Rps24, Trp53, Kif3A, Nin, Tsr1, Myo1E, Myo1D, Myo1F, Syce2, Atr, 41163, Rps8, Coro1C, Plk4, Ddx56, Lasp1, Pcna, Epb4.1L4A, Wdr1, Utp14A, Utp2, Taf1A, Utp15, Pot1A, Cpeb1, Prmt3, Slc1A4, Spry2, Des, Gm8292, Rrp1B, Grwd1, Hmox1, Tubb5, Tubb6, Sntb2, Cdk5Rap2, Zyx, Msn, Tuba1A, Myc, Tuba1B, Dlg2, Arhgef2, Exosc8, Acta2, Mtap4, Exosc1, Mbd2, Palld, Ncl, Flnb, Rad5, Flna, Elmo1, Eya2, Sgcg, Frmd4A, Dnahc7B, Nol1, Nol11, Tmsb4X, Mapre2, Wdr43, Lcp1, Ablim2, Eri1, Nufip1, Abi2, Tpm4, Dimt1, Sorbs3, Chd7, Cep57, Aatf, Kif21B, Cep192, Ilf3, Mid1, Smc2, Lrrc49, Pwp2, Dis3, Svil, Nop58, Jak1, Nop56 Ctnnal1, Rpp38, Xrcc5, Dync1Li1, Sncg, Jub, Tubb2B, Surf6, Tubb2A, Pdlim1, Ptpn21, Rpl22L1, Snora65, Kifc3, Actg1, Actg2, App, G2E3, Mak16, Ank2, Mdfic, D19Bwg1357E, Ddx21, Zfp33, Rpl12, Gnl2, Gnl3, Kif5A, Actn1, Tmsb1, Rrp9, Ctnna1, Myh9, Mrto4, Jup, Baz1A, Rcc2, Fnbp1L, Spag5, Mtap1A, Mtap1B, Rps12, Stmn1, Mybbp1A, Nup43, 4116, Blm, Ssh2, Akap12, Myo9A, Itgam, Dock2, Eif3A, Trim66, Smarcb1, Npm1, Strbp, Emd, Rps24, Trp53, Kif3A, Nin, Tsr1, Myo1E, Myo1D, Myo1F, Syce2, Atr, 41163, Rps8, Coro1C, Plk4, Ddx56, Lasp1, Pcna, Epb4.1L4A, Wdr1, Utp14A, Utp2, Taf1A, Utp15, Pot1A, Cpeb1, Prmt3, Slc1A4, Spry2, Des, Gm8292, Rrp1B, Grwd1, Hmox1, Tubb5, Tubb6, Sntb2, Cdk5Rap2, Zyx, Msn, Tuba1A, Myc, Tuba1B, Dlg2, Arhgef2, Exosc8, Acta2, Mtap4, Exosc1, Mbd2, Palld, Ncl, Flnb, Rad5, Flna, Elmo1, Eya2, Sgcg, Frmd4A, Dnahc7B, Nol1, Nol11, Tmsb4X, Mapre2, Wdr43, Lcp1, Ablim2, Eri1, Nufip1, Abi2, Tpm4, Dimt1, Sorbs3, Chd7, Cep57, Aatf, Kif21B, Cep192, Ilf3, Mid1, Smc2, Lrrc49, Pwp2, Dis3, Svil, Nop58, Jak1, Nop E-9 Yars, Cars, Dars, Nars, Sars, Aars, Gars, Eprs, Vars, Iars, Wars, Tars, Lars, Farsb E E E-8 Naf1, Exosc8, Tsr1, Surf6, Utp15, Eri1, Rrp9, Exosc1, Mrto4, Dimt1, Dis3, Ddx56, Wdr36, Rrp1B, Ipo4, Npm1, Nop58, Aatf, Nop56, Gnl2, Utp14A, Znhit6 Rpp38, Yars, Cars, Nars, Dars, Sars, Aars, Gars, Eprs, Qtrt1, Vars, Trmt61A, Iars, Pus7L, Wars, Tars, Trdmt1, Mettl1, Farsb, Lars, Pop1, Dus4L Naf1, Exosc8, Tsr1, Surf6, Utp15, Eri1, Exosc1, Rrp9, Mrto4, Dimt1, Dis3, Ddx56, Eif3A, Wdr36, Prmt7, Rrp1B, Ipo4, Npm1, Nop58, Aatf, Nop56, Gnl2, Utp14A, Znhit6
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature11986 relative IL-6 expression Viable intracellular Bp per well 18 16 1 1 5 5 3 1 6 +DG 8 8 8 Control DG Time (h) hours B. pertussis IL-6 (pg/ml) 15 Control DG
SUPPLEMENTARY INFORMATION doi:1.138/nature11986 relative IL-6 expression Viable intracellular Bp per well 18 16 1 1 5 5 3 1 6 +DG 8 8 8 Control DG Time (h) hours B. pertussis IL-6 (pg/ml) 15 Control DG
Supplemental Figure 1
 Supplemental Figure 1 mir 26a mir 26b Supplemental Figure 1. Expression of mir-26a is higher than mir-26b in the mouse liver. Dot blot analysis of mir-26a/b expression in the livers of wild-type mice fed
Supplemental Figure 1 mir 26a mir 26b Supplemental Figure 1. Expression of mir-26a is higher than mir-26b in the mouse liver. Dot blot analysis of mir-26a/b expression in the livers of wild-type mice fed
Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle.
 Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle. (a) mrna levels of Dj1 measured by quantitative RT-PCR in soleus, gastrocnemius (Gastroc.) and extensor digitorum longus
Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle. (a) mrna levels of Dj1 measured by quantitative RT-PCR in soleus, gastrocnemius (Gastroc.) and extensor digitorum longus
Supplementary Table 1.
 Supplementary Table 1. Expression of genes involved in brown fat differentiation in WAT of db/db mice treated with HDAC inhibitors. Data are expressed as fold change (FC) versus control. symbol FC SAHA
Supplementary Table 1. Expression of genes involved in brown fat differentiation in WAT of db/db mice treated with HDAC inhibitors. Data are expressed as fold change (FC) versus control. symbol FC SAHA
SUPPLEMENTARY INFORMATION
 DOI:.3/ncb7 Hematopoiesis Expression (Protein) Competition PRC integration Polycomb-mediated gene repression Cell fate HSC PRC SELF-RENEWAL Cbx Cbx4 PRC progenitor genes PROG Cbx PRC Cbx4 DIFFERENTIATION
DOI:.3/ncb7 Hematopoiesis Expression (Protein) Competition PRC integration Polycomb-mediated gene repression Cell fate HSC PRC SELF-RENEWAL Cbx Cbx4 PRC progenitor genes PROG Cbx PRC Cbx4 DIFFERENTIATION
Fig. S1. Validation of ChIP-seq binding sites by single gene ChIP-PCR Fig. S2. Transactivation potential of PPAR
 Fig. S1. Validation of ChIP-seq binding sites by single gene ChIP-PCR ChIP-PCR was performed on PPARγ and RXR-enriched chromatin harvested during adipocyte differentiation at day and day 6 as described
Fig. S1. Validation of ChIP-seq binding sites by single gene ChIP-PCR ChIP-PCR was performed on PPARγ and RXR-enriched chromatin harvested during adipocyte differentiation at day and day 6 as described
Supplemental Table 1. List of genes and mature shrna sequences identified from the shrna screen in BRCA2-mutant PEO1 cells.
 Guillemette_Supplemental Table 1 Gene Mature Sequence Gene Mature Sequence ATTCATAGGATGTCAGCAG LOC157193 TATGCGTAAGTAATAAATTGCT TTAGTTCTGTCTTGGAGG LOC152225 CGCATATATGTTCTGACTT ALDH2 CACTTCAGTGTATGCCT
Guillemette_Supplemental Table 1 Gene Mature Sequence Gene Mature Sequence ATTCATAGGATGTCAGCAG LOC157193 TATGCGTAAGTAATAAATTGCT TTAGTTCTGTCTTGGAGG LOC152225 CGCATATATGTTCTGACTT ALDH2 CACTTCAGTGTATGCCT
Baf60c drives glycolytic muscle formation and improves glucose homeostasis through Deptor-mediated Akt activation
 Baf6c drives glycolytic muscle formation and improves glucose homeostasis through Deptor-mediated Akt activation Zhuo-Xian Meng,2, Siming Li,2, Lin Wang,2, Hwi Jin Ko 3, Yongjin Lee 3, Dae Young Jung 3,
Baf6c drives glycolytic muscle formation and improves glucose homeostasis through Deptor-mediated Akt activation Zhuo-Xian Meng,2, Siming Li,2, Lin Wang,2, Hwi Jin Ko 3, Yongjin Lee 3, Dae Young Jung 3,
Histone demethylase JMJD1A coordinates acute and chronic adaptation to cold stress via thermogenic phospho-switch. Abe et al.
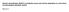 Histone demethylase JMJD1A coordinates acute and chronic adaptation to cold stress via thermogenic phospho-switch Abe et al. a BAT 1 kb Ucp1 H3Kme3 H3K7ac (., ) (., ) -13 kb -. kb -. kb b scwat Chronic
Histone demethylase JMJD1A coordinates acute and chronic adaptation to cold stress via thermogenic phospho-switch Abe et al. a BAT 1 kb Ucp1 H3Kme3 H3K7ac (., ) (., ) -13 kb -. kb -. kb b scwat Chronic
Subject ID: Date Test Started: Specimen Type: Peripheral Blood Date of Report: Date Specimen Obtained:
 TEST PERFORMED Whole exome sequencing (WES) with Focused Diagnostic Panel(s): Neuropathy, Leukodystrophy, Myopathy See below for a complete list of genes analyzed. RESULTS 1 sequence variant with potential
TEST PERFORMED Whole exome sequencing (WES) with Focused Diagnostic Panel(s): Neuropathy, Leukodystrophy, Myopathy See below for a complete list of genes analyzed. RESULTS 1 sequence variant with potential
Supplementary Table 1. Information on the 174 single nucleotide variants identified by whole-exome sequencing
 Supplementary Table 1. Information on the 174 single nucleotide s identified by whole-exome sequencing Chr Position Type AF exomes Database 1 10163148 A/G UBE4B missense 0.000326 1534 2 98 1 4.44 Damaging
Supplementary Table 1. Information on the 174 single nucleotide s identified by whole-exome sequencing Chr Position Type AF exomes Database 1 10163148 A/G UBE4B missense 0.000326 1534 2 98 1 4.44 Damaging
SUPPLEMENTARY,INFORMATIONS,,,, mtorc1,is,required,for,brown,adipose,tissue,recruitment,and, METABOLIC,ADAPTATION,TO,COLD,!! Sébastien!M.!
 SUPPLEMENTARY,INFORMATIONS,,,, mtorc,is,required,for,brown,adipose,tissue,recruitment,and, METABOLIC,ADAPTATION,TO,COLD, SébastienM.Labbé,,MathildeMouchiroud,,AlexandreCaron,,BlandineSecco,, Elizaveta
SUPPLEMENTARY,INFORMATIONS,,,, mtorc,is,required,for,brown,adipose,tissue,recruitment,and, METABOLIC,ADAPTATION,TO,COLD, SébastienM.Labbé,,MathildeMouchiroud,,AlexandreCaron,,BlandineSecco,, Elizaveta
devseek (Sequence(Analysis(Panel(for(Neurodevelopmental(Disorders
 ACSL4 Mental/retardation,/XBlinked/63 XLBR ADSL Adenylosuccinase/Deficiency AR AFF2 Mental/retardation,/XBlinked,/FRAXE/type XLBR ALG6 Congenital/disorder/of/glycosylation/type/Ic AR ANK3 Mental/retardation,/autosomal/recessive/37
ACSL4 Mental/retardation,/XBlinked/63 XLBR ADSL Adenylosuccinase/Deficiency AR AFF2 Mental/retardation,/XBlinked,/FRAXE/type XLBR ALG6 Congenital/disorder/of/glycosylation/type/Ic AR ANK3 Mental/retardation,/autosomal/recessive/37
8 th NUGO week Wageningen. #nugo. Michael Müller Conclusions What is health?
 8 th NUGO week Wageningen #nugo Michael Müller Conclusions What is health? 1 1 You are what you eat What's healthy? 4 4 What is health? WHO 1946:..a state of complete physical, mental, and social well-being
8 th NUGO week Wageningen #nugo Michael Müller Conclusions What is health? 1 1 You are what you eat What's healthy? 4 4 What is health? WHO 1946:..a state of complete physical, mental, and social well-being
Supplementary Information
 1 Supplementary Information Human TSC2 Null Fibroblast-Like Cells Induce Hair Follicle Neogenesis and Hamartoma Morphogenesis Shaowei Li 1, Rajesh L. Thangapazham 1, Ji-an Wang 1, Sangeetha Rajesh 1, Tzu-Cheg
1 Supplementary Information Human TSC2 Null Fibroblast-Like Cells Induce Hair Follicle Neogenesis and Hamartoma Morphogenesis Shaowei Li 1, Rajesh L. Thangapazham 1, Ji-an Wang 1, Sangeetha Rajesh 1, Tzu-Cheg
Supplemental Figure 1. Characterization of CNS lysosomal pathology in 2-month-old IDS-deficient mice. (A) Measurement of IDS activity in brain
 Supplemental Figure 1. Characterization of CNS lysosomal pathology in 2-month-old IDS-deficient mice. (A) Measurement of IDS activity in brain extracts from wild-type (WT) and IDS-deficient males. IDS
Supplemental Figure 1. Characterization of CNS lysosomal pathology in 2-month-old IDS-deficient mice. (A) Measurement of IDS activity in brain extracts from wild-type (WT) and IDS-deficient males. IDS
CFSE Thy1.2 + CD4 + cells
 Time after immunization (d) IFN-g Spleen Lymph nodes Rbpj +/+ ;OT-II Rbpj -/- ;OT-II Rbpj +/+ ;OT-II Rbpj -/- ;OT-II 2.3.2 22.9 5.6 3 23.3 3. 21.3 32.7 8.7 18.5 11.6 13.5 7 84.2 74.2 85. 83.8 CFSE Thy1.2
Time after immunization (d) IFN-g Spleen Lymph nodes Rbpj +/+ ;OT-II Rbpj -/- ;OT-II Rbpj +/+ ;OT-II Rbpj -/- ;OT-II 2.3.2 22.9 5.6 3 23.3 3. 21.3 32.7 8.7 18.5 11.6 13.5 7 84.2 74.2 85. 83.8 CFSE Thy1.2
Supplementary Table 1. Candidate loci that were associated with diabetes- and/or obesity-related traits.
 Supplementary Table 1. Candidate loci that were associated with diabetes- and/or obesity-related traits. All With Fst in the With long With SNP(s) at the 5' top 5% bracket haplotypes regulatory region
Supplementary Table 1. Candidate loci that were associated with diabetes- and/or obesity-related traits. All With Fst in the With long With SNP(s) at the 5' top 5% bracket haplotypes regulatory region
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature7221 Brown fat selective genes 12 1 Control Q-RT-PCR (% of Control) 8 6 4 2 Ntrk3 Cox7a1 Cox8b Cox5b ATPase b2 ATPase f1a1 Sirt3 ERRα Elovl3/Cig3 PPARα Zic1 Supplementary Figure S1. stimulates
doi: 1.138/nature7221 Brown fat selective genes 12 1 Control Q-RT-PCR (% of Control) 8 6 4 2 Ntrk3 Cox7a1 Cox8b Cox5b ATPase b2 ATPase f1a1 Sirt3 ERRα Elovl3/Cig3 PPARα Zic1 Supplementary Figure S1. stimulates
Supplementary Figure 1
 VO (ml kg - min - ) VCO (ml kg - min - ) Respiratory exchange ratio Energy expenditure (cal kg - min - ) Locomotor activity (x count) Body temperature ( C) Relative mrna expression TA Sol EDL PT Heart
VO (ml kg - min - ) VCO (ml kg - min - ) Respiratory exchange ratio Energy expenditure (cal kg - min - ) Locomotor activity (x count) Body temperature ( C) Relative mrna expression TA Sol EDL PT Heart
Supplementary Figures and Tables. An inflammatory gene signature distinguishes neurofibroma Schwann cells and
 Supplementary Figures and Tables An inflammatory gene signature distinguishes neurofibroma Schwann cells and macrophages from cells in the normal peripheral nervous system Kwangmin Choi 1, Kakajan Komurov
Supplementary Figures and Tables An inflammatory gene signature distinguishes neurofibroma Schwann cells and macrophages from cells in the normal peripheral nervous system Kwangmin Choi 1, Kakajan Komurov
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Nature Immunology: doi: /ni Supplementary Figure 1 33,312. Aire rep 1. Aire rep 2 # 44,325 # 44,055. Aire rep 1. Aire rep 2.
 a 33,312 b rep 1 rep 1 # 44,325 rep 2 # 44,055 [0-84] rep 2 [0-84] 1810043G02Rik Pfkl Dnmt3l Icosl rep 1 [0-165] rep 2 [0-165] Rps14 Cd74 Mir5107 Tcof1 rep 1 [0-69] rep 2 [0-68] Id3 E2f2 Asap3 rep 1 [0-141]
a 33,312 b rep 1 rep 1 # 44,325 rep 2 # 44,055 [0-84] rep 2 [0-84] 1810043G02Rik Pfkl Dnmt3l Icosl rep 1 [0-165] rep 2 [0-165] Rps14 Cd74 Mir5107 Tcof1 rep 1 [0-69] rep 2 [0-68] Id3 E2f2 Asap3 rep 1 [0-141]
1.5 ASK1KO fed. fasted 16 hrs w/o water. Fed. 4th. 4th WT ASK1KO N=29, 11(WT), ,5(ASK1KO) ASK1KO ASK1KO **** Time [h]
![1.5 ASK1KO fed. fasted 16 hrs w/o water. Fed. 4th. 4th WT ASK1KO N=29, 11(WT), ,5(ASK1KO) ASK1KO ASK1KO **** Time [h] 1.5 ASK1KO fed. fasted 16 hrs w/o water. Fed. 4th. 4th WT ASK1KO N=29, 11(WT), ,5(ASK1KO) ASK1KO ASK1KO **** Time [h]](/thumbs/87/97258189.jpg) 7: 13: 19: 1: 7: 151117 a 151117 4th 4th b c RQ.95 KO.9.85.8.75.7 light dark light dark.65 7: 19: 7: 19: 7: Means ± SEM, N=6 RQ 1..9.8.7.6.6 KO CL (-) CL (+) ibat weight ratio (/body weight) [%].5.4.3.2.1
7: 13: 19: 1: 7: 151117 a 151117 4th 4th b c RQ.95 KO.9.85.8.75.7 light dark light dark.65 7: 19: 7: 19: 7: Means ± SEM, N=6 RQ 1..9.8.7.6.6 KO CL (-) CL (+) ibat weight ratio (/body weight) [%].5.4.3.2.1
LMM / emerge III Network Reference Sequences October 2016 LMM / emerge III Network Consensus Actionable Gene List *ACMG56 gene
 LMM / emerge III Network Consensus Actionable Gene List *ACMG56 gene ACTA2* Exon 02-09 NM_001613.2 DSG2* Exon 01-15 NM_001943.3 ACTC1* Exon 01-06 NM_005159.4 DSP* Exon 01-24 NM_004415.2 APC* Exon 01-15
LMM / emerge III Network Consensus Actionable Gene List *ACMG56 gene ACTA2* Exon 02-09 NM_001613.2 DSG2* Exon 01-15 NM_001943.3 ACTC1* Exon 01-06 NM_005159.4 DSP* Exon 01-24 NM_004415.2 APC* Exon 01-15
Figure 6: TERT regulates MYC half-life and ubiquitination.
 TERT or IgG as indicated. For the western blots, representative images of n= independent experiments are shown. Student s t-test was used, and * indicates p
TERT or IgG as indicated. For the western blots, representative images of n= independent experiments are shown. Student s t-test was used, and * indicates p
SUPPLEMENTARY INFORMATION
 -. -. SUPPLEMENTARY INFORMATION DOI: 1.1/ncb86 a WAT-1 WAT- BAT-1 BAT- sk-muscle-1 sk-muscle- mir-133b mir-133a mir-6 mir-378 mir-1 mir-85 mir-378 mir-6a mir-18 mir-133a mir- mir- mir-341 mir-196a mir-17
-. -. SUPPLEMENTARY INFORMATION DOI: 1.1/ncb86 a WAT-1 WAT- BAT-1 BAT- sk-muscle-1 sk-muscle- mir-133b mir-133a mir-6 mir-378 mir-1 mir-85 mir-378 mir-6a mir-18 mir-133a mir- mir- mir-341 mir-196a mir-17
7 Glyceraldehyde 3-Phosphate GAPDH % SYM+G+e NM_ BC JU JU JU %% 45 /
 02234546789:;;;?6@A9587BA6;A6;C4D46E4C;B6;7FBC;C7G:;B6E3GB6H;H464;6854I;H464;8E9A6:5I;7A783;65=49;A@;=8C4;28B9C;J=2KI;7F4;65=49;A@;289CB5A6:;6B6@A9587BL4;2ACB7BA6C;J#M?KI;7F4; 65=49;A@;289CB5A6:;B6@A9587BL4;2ACB7BA6C;J#?KI;5AG43;C4G;B6;8683:CBC;A@;B6GBLBG83;3AEB;JNAG43KI;8EE4CCBA6;65=49C;A@;S.
02234546789:;;;?6@A9587BA6;A6;C4D46E4C;B6;7FBC;C7G:;B6E3GB6H;H464;6854I;H464;8E9A6:5I;7A783;65=49;A@;=8C4;28B9C;J=2KI;7F4;65=49;A@;289CB5A6:;6B6@A9587BL4;2ACB7BA6C;J#M?KI;7F4; 65=49;A@;289CB5A6:;B6@A9587BL4;2ACB7BA6C;J#?KI;5AG43;C4G;B6;8683:CBC;A@;B6GBLBG83;3AEB;JNAG43KI;8EE4CCBA6;65=49C;A@;S.
Supplementary Information
 Scientific Reports Supplementary Information Upregulated expression of FGF13/FHF2 mediates resistance to platinum drugs in cervical cancer cells Tomoko Okada, Kazuhiro Murata, Ryoma Hirose, Chie Matsuda,
Scientific Reports Supplementary Information Upregulated expression of FGF13/FHF2 mediates resistance to platinum drugs in cervical cancer cells Tomoko Okada, Kazuhiro Murata, Ryoma Hirose, Chie Matsuda,
SUPPLEMENTARY DATA. Supplementary Table 1. Primers used in qpcr
 Supplementary Table 1. Primers used in qpcr Gene forward primer (5'-3') reverse primer (5'-3') β-actin AGAGGGAAATCGTGCGTGAC CAATAGTGATGACCTGGCCGT Hif-p4h-2 CTGGGCAACTACAGGATAAAC GCGTCCCAGTCTTTATTTAGATA
Supplementary Table 1. Primers used in qpcr Gene forward primer (5'-3') reverse primer (5'-3') β-actin AGAGGGAAATCGTGCGTGAC CAATAGTGATGACCTGGCCGT Hif-p4h-2 CTGGGCAACTACAGGATAAAC GCGTCCCAGTCTTTATTTAGATA
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10860 Supplementary Discussion It remains unclear why H3K9 demethylation appeared to be more sensitive to suppression than at least some other histone methylation marks as a result of
doi:10.1038/nature10860 Supplementary Discussion It remains unclear why H3K9 demethylation appeared to be more sensitive to suppression than at least some other histone methylation marks as a result of
FH- FH+ DM. 52 Volunteers. Oral & IV Glucose Tolerance Test Hyperinsulinemic Euglycemic Clamp in Non-DM Subjects ACADSB MYSM1. Mouse Skeletal Muscle
 A 52 Volunteers B 6 5 4 3 2 FH- FH+ DM 1 Oral & IV Glucose Tolerance Test Hyperinsulinemic Euglycemic Clamp in Non-DM Subjects ZYX EGR2 NR4A1 SRF target TPM1 ACADSB MYSM1 Non SRF target FH- FH+ DM2 C SRF
A 52 Volunteers B 6 5 4 3 2 FH- FH+ DM 1 Oral & IV Glucose Tolerance Test Hyperinsulinemic Euglycemic Clamp in Non-DM Subjects ZYX EGR2 NR4A1 SRF target TPM1 ACADSB MYSM1 Non SRF target FH- FH+ DM2 C SRF
Supplementary Table 2. Plasma lipid profiles in wild type and mutant female mice submitted to a HFD for 12 weeks wt ERα -/- AF-1 0 AF-2 0
 Supplementary Table 1. List of specific primers used for gene expression analysis. Genes Primer forward Primer reverse Hprt GCAGTACAGCCCCAAAATGG AACAAAGTCTGGCCTGTATCCA Srebp-1c GGAAGCTGTCGGGGTAGCGTC CATGTCTTCAAATGTGCAATCCAT
Supplementary Table 1. List of specific primers used for gene expression analysis. Genes Primer forward Primer reverse Hprt GCAGTACAGCCCCAAAATGG AACAAAGTCTGGCCTGTATCCA Srebp-1c GGAAGCTGTCGGGGTAGCGTC CATGTCTTCAAATGTGCAATCCAT
A synergistic anti-obesity effect by a combination of capsinoids and cold temperature through the promotion of beige adipocyte biogenesis
 A synergistic anti-obesity effect by a combination of capsinoids and cold temperature through the promotion of beige adipocyte biogenesis Kana Ohyama, 1,2 Yoshihito Nogusa, 1 Kosaku Shinoda, 2 Katsuya
A synergistic anti-obesity effect by a combination of capsinoids and cold temperature through the promotion of beige adipocyte biogenesis Kana Ohyama, 1,2 Yoshihito Nogusa, 1 Kosaku Shinoda, 2 Katsuya
ALT (U/L) (Relative expression) HDL (mm) (Relative expression) ALT (U/L) (Relative expression)
 a DMT mrna () 8 6 r =.96 P =. DMT mrna () 8 6 r =. P =.6 DMT mrna () 8 6 r =.99 P =.6 DMT mrna () 8 6 r =. P =.9 DMT mrna () BMI (kg/m ) 8 6 r =.7 P =.966 DMT mrna () 8 ALT (U/L) 8 6 r = -.66 P =.76 DMT
a DMT mrna () 8 6 r =.96 P =. DMT mrna () 8 6 r =. P =.6 DMT mrna () 8 6 r =.99 P =.6 DMT mrna () 8 6 r =. P =.9 DMT mrna () BMI (kg/m ) 8 6 r =.7 P =.966 DMT mrna () 8 ALT (U/L) 8 6 r = -.66 P =.76 DMT
** *** population. (A) Representative FACS plots showing the gating strategy for T N
 Sca-1 (MFI) CD122 (MFI) CD44 A CD8 + gated: resting spleen CD8 + gated: T Mem -derived CD8 + gated: T N -derived, expanded alone B CD62L CD44 low CD62L +, resting CD44 high, T Mem -derived CD44 low CD62L
Sca-1 (MFI) CD122 (MFI) CD44 A CD8 + gated: resting spleen CD8 + gated: T Mem -derived CD8 + gated: T N -derived, expanded alone B CD62L CD44 low CD62L +, resting CD44 high, T Mem -derived CD44 low CD62L
COMPLETE A FORM FOR EACH SAMPLE SUBMITTED ETHNIC BACKGROUND REPORTING INFORMATION
 DATE SAMPLE DRAWN: SAMPLE TYPE: BLOOD OTHER (SPECIFY): COMPLETE A FORM FOR EACH SAMPLE SUBMITTED PATIENT FIRST NAME: LAST NAME: DOB: SEX: M F UNKNOWN ADDRESS: HOME PHONE: ( ) WORK: ( ) EUROPEAN CAUCASIAN
DATE SAMPLE DRAWN: SAMPLE TYPE: BLOOD OTHER (SPECIFY): COMPLETE A FORM FOR EACH SAMPLE SUBMITTED PATIENT FIRST NAME: LAST NAME: DOB: SEX: M F UNKNOWN ADDRESS: HOME PHONE: ( ) WORK: ( ) EUROPEAN CAUCASIAN
Assessed Molecular Mechanism of Action of Milagro de la Selva
 Assessed Molecular Mechanism of Action of Milagro de la Selva Results A report prepared by Connexios Life Sciences Bangalore 1. Muscle - C2C12 cells. 1.1. Glucose uptake, Glycolysis and Tricarboxylic acid
Assessed Molecular Mechanism of Action of Milagro de la Selva Results A report prepared by Connexios Life Sciences Bangalore 1. Muscle - C2C12 cells. 1.1. Glucose uptake, Glycolysis and Tricarboxylic acid
Tissue-specific pathways and networks underlying sexual dimorphism in nonalcoholic
 Kurt et al. Biology of Sex Differences (2018) 9:46 https://doi.org/10.1186/s13293-018-0205-7 RESEARCH Tissue-specific pathways and networks underlying sexual dimorphism in nonalcoholic fatty liver disease
Kurt et al. Biology of Sex Differences (2018) 9:46 https://doi.org/10.1186/s13293-018-0205-7 RESEARCH Tissue-specific pathways and networks underlying sexual dimorphism in nonalcoholic fatty liver disease
SUPPLEMENTARY DATA. Supplementary Table 1. Primer sequences for qrt-pcr
 Supplementary Table 1. Primer sequences for qrt-pcr Gene PRDM16 UCP1 PGC1α Dio2 Elovl3 Cidea Cox8b PPARγ AP2 mttfam CyCs Nampt NRF1 16s-rRNA Hexokinase 2, intron 9 β-actin Primer Sequences 5'-CCA CCA GCG
Supplementary Table 1. Primer sequences for qrt-pcr Gene PRDM16 UCP1 PGC1α Dio2 Elovl3 Cidea Cox8b PPARγ AP2 mttfam CyCs Nampt NRF1 16s-rRNA Hexokinase 2, intron 9 β-actin Primer Sequences 5'-CCA CCA GCG
Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases
 Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
Large-Scale Discovery of Enhancers from Human Heart Tissue
 Large-Scale Discovery of Enhancers from Human Heart Tissue Dalit May, Matthew J. Blow, Tommy Kaplan, David J. McCulley, Brian C. Jensen, Jennifer A. Akiyama, Amy Holt, Ingrid Plajzer-Frick, Malak Shoukry,
Large-Scale Discovery of Enhancers from Human Heart Tissue Dalit May, Matthew J. Blow, Tommy Kaplan, David J. McCulley, Brian C. Jensen, Jennifer A. Akiyama, Amy Holt, Ingrid Plajzer-Frick, Malak Shoukry,
Supplement Results, Figures and Tables
 Supplement Results, Figures and Tables Results PET imaging The SUV was significantly higher in tumours of the parental HSC-3 cell line than tumours of the EV and the MMP-8 overexpressing cell lines (Figure
Supplement Results, Figures and Tables Results PET imaging The SUV was significantly higher in tumours of the parental HSC-3 cell line than tumours of the EV and the MMP-8 overexpressing cell lines (Figure
WERNER_FIBRO_DN LEI_MYB_REGULATED_GENES BRCA1_OVEREXP_DN SUBCLASSES_DIFF P21_P53_MIDDLE_DN
 Supplemental Table S1. Full List of Up-Regulated Conserved Biological Modules Between Zebrafish Liver Tumor with Human Liver, Lung, Gastric, and Prostate Tumors LvCMs (ZF liver cancer vs. human liver cancers
Supplemental Table S1. Full List of Up-Regulated Conserved Biological Modules Between Zebrafish Liver Tumor with Human Liver, Lung, Gastric, and Prostate Tumors LvCMs (ZF liver cancer vs. human liver cancers
[U- 13 C5] glutamine. Glutamate. Acetyl-coA. Citrate. Citrate. Malate. Malate. Isocitrate OXIDATIVE METABOLISM. Oxaloacetate CO2.
![[U- 13 C5] glutamine. Glutamate. Acetyl-coA. Citrate. Citrate. Malate. Malate. Isocitrate OXIDATIVE METABOLISM. Oxaloacetate CO2. [U- 13 C5] glutamine. Glutamate. Acetyl-coA. Citrate. Citrate. Malate. Malate. Isocitrate OXIDATIVE METABOLISM. Oxaloacetate CO2.](/thumbs/76/74112154.jpg) Supplementary Figures a. Relative mrna levels Supplementary Figure 1 (Christofk) 3.0 2.5 2.0 1.5 1.0 0.5 0.0 LAT1 Fumarate Succinate Palmitate Acetyl-coA Oxaloacetate OXIDATIVE METABOLISM α-ketoglutarate
Supplementary Figures a. Relative mrna levels Supplementary Figure 1 (Christofk) 3.0 2.5 2.0 1.5 1.0 0.5 0.0 LAT1 Fumarate Succinate Palmitate Acetyl-coA Oxaloacetate OXIDATIVE METABOLISM α-ketoglutarate
GPR120 *** * * Liver BAT iwat ewat mwat Ileum Colon. UCP1 mrna ***
 a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
Targeted Genes and Methodology Details for Epilepsy/Seizure Genetic Panels
 Targeted s and Methodology Details for Epilepsy/Seizure tic Panels Reference transcripts based on build GRCh37 (hg19) interrogated by Epilepsy/Seizure tic Panels Epilepsy Expanded Panel Epilepsy Expanded
Targeted s and Methodology Details for Epilepsy/Seizure tic Panels Reference transcripts based on build GRCh37 (hg19) interrogated by Epilepsy/Seizure tic Panels Epilepsy Expanded Panel Epilepsy Expanded
Supplementary Materials
 Supplementary Materials Supplemental figure 1. Absolute cell count of naïve/memory compartment of CD4 and CD8 T cells in early years Absolute cell number of naïve (CD45RA+CCR7+), Stem cell memory T cell
Supplementary Materials Supplemental figure 1. Absolute cell count of naïve/memory compartment of CD4 and CD8 T cells in early years Absolute cell number of naïve (CD45RA+CCR7+), Stem cell memory T cell
Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR
 Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) cadl CTTGGGGGCGCGTCT CTGTTCTTTTGTGCCGTTTCG cyl-coenzyme Dehydrogenase, very
Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) cadl CTTGGGGGCGCGTCT CTGTTCTTTTGTGCCGTTTCG cyl-coenzyme Dehydrogenase, very
Relative Rates. SUM159 CB- 839-Resistant *** n.s Intracellular % Labeled by U- 13 C-Asn 0.
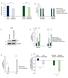 A Relative Growth Rates 1.2 1.8.6.4.2 B Relative Rates 1.6 1.4 1.2 1.8.6.4.2 LPS2 Parental LPS2 Q-Independent SUM159 Parental SUM159 CB-839-Resistant LPS2 Parental LPS2 Q- Independent SUM159 Parental SUM159
A Relative Growth Rates 1.2 1.8.6.4.2 B Relative Rates 1.6 1.4 1.2 1.8.6.4.2 LPS2 Parental LPS2 Q-Independent SUM159 Parental SUM159 CB-839-Resistant LPS2 Parental LPS2 Q- Independent SUM159 Parental SUM159
Nature Immunology doi: /ni Supplementary Figure 1. Raf-1 inhibition does not affect TLR4-induced type I IFN responses.
 Supplementary Figure 1 Raf-1 inhibition does not affect TLR4-induced type I IFN responses. Real-time PCR analyses of IFNB, ISG15, TRIM5, TRIM22 and APOBEC3G mrna in modcs 6 h after stimulation with TLR4
Supplementary Figure 1 Raf-1 inhibition does not affect TLR4-induced type I IFN responses. Real-time PCR analyses of IFNB, ISG15, TRIM5, TRIM22 and APOBEC3G mrna in modcs 6 h after stimulation with TLR4
Supplemental Fig. 1. Relative mrna Expression. Relative mrna Expression WT KO WT KO RT 4 0 C
 Supplemental Fig. 1 A 1.5 1..5 Hdac11 (ibat) n=4 n=4 n=4 n=4 n=4 n=4 n=4 n=4 WT KO WT KO WT KO WT KO RT 4 C RT 4 C Supplemental Figure 1. Hdac11 mrna is undetectable in KO adipose tissue. Quantitative
Supplemental Fig. 1 A 1.5 1..5 Hdac11 (ibat) n=4 n=4 n=4 n=4 n=4 n=4 n=4 n=4 WT KO WT KO WT KO WT KO RT 4 C RT 4 C Supplemental Figure 1. Hdac11 mrna is undetectable in KO adipose tissue. Quantitative
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/264/rs4/dc1 Supplementary Materials for A Systems Approach for Decoding Mitochondrial Retrograde Signaling Pathways Sehyun Chae, Byung Yong Ahn, Kyunghee Byun,
www.sciencesignaling.org/cgi/content/full/6/264/rs4/dc1 Supplementary Materials for A Systems Approach for Decoding Mitochondrial Retrograde Signaling Pathways Sehyun Chae, Byung Yong Ahn, Kyunghee Byun,
control kda ATGL ATGLi HSL 82 GAPDH * ** *** WT/cTg WT/cTg ATGLi AKO/cTg AKO/cTg ATGLi WT/cTg WT/cTg ATGLi AKO/cTg AKO/cTg ATGLi iwat gwat ibat
 body weight (g) tissue weights (mg) ATGL protein expression (relative to GAPDH) HSL protein expression (relative to GAPDH) ### # # kda ATGL 55 HSL 82 GAPDH 37 2.5 2. 1.5 1..5 2. 1.5 1..5.. Supplementary
body weight (g) tissue weights (mg) ATGL protein expression (relative to GAPDH) HSL protein expression (relative to GAPDH) ### # # kda ATGL 55 HSL 82 GAPDH 37 2.5 2. 1.5 1..5 2. 1.5 1..5.. Supplementary
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies
 Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Nature Genetics: doi: /ng.3731
 Supplementary Figure 1 Circadian profiles of Adarb1 transcript and ADARB1 protein in mouse tissues. (a) Overlap of rhythmic transcripts identified in the previous transcriptome analyses. The mouse liver
Supplementary Figure 1 Circadian profiles of Adarb1 transcript and ADARB1 protein in mouse tissues. (a) Overlap of rhythmic transcripts identified in the previous transcriptome analyses. The mouse liver
Inferring the Mammalian Cardiogenic Gene Regulatory Network. Jason Bazil 10/18/13 Cardiac Physiome Workshop 2013
 Inferring the Mammalian Cardiogenic Gene Regulatory Network Jason Bazil 1/18/13 Cardiac Physiome Workshop 213 Reverse Engineering Gene Regulatory Networks Two main questions need addressed: 1) What s connected
Inferring the Mammalian Cardiogenic Gene Regulatory Network Jason Bazil 1/18/13 Cardiac Physiome Workshop 213 Reverse Engineering Gene Regulatory Networks Two main questions need addressed: 1) What s connected
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
SUPPLEMENTARY FIGURES
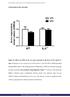 SUPPLEMENTARY FIGURES Figure S1. Effect of a HFD on the Acox gene expression in the livers of WT and IL-6 -/- mice. Expression of Acox in the livers of WT and IL-6 -/- mice fed STD or HFD determined through
SUPPLEMENTARY FIGURES Figure S1. Effect of a HFD on the Acox gene expression in the livers of WT and IL-6 -/- mice. Expression of Acox in the livers of WT and IL-6 -/- mice fed STD or HFD determined through
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
Use of Molecular Assays to Assess Cellular Therapies
 Use of Molecular Assays to Assess Cellular Therapies D. Stroncek, P. Jin, M. Sabatino, J. Ren, L. Castiello, E. Wang, and F.M. Marincola. Cellular Therapies Section Clinical Center, NIH Methods for the
Use of Molecular Assays to Assess Cellular Therapies D. Stroncek, P. Jin, M. Sabatino, J. Ren, L. Castiello, E. Wang, and F.M. Marincola. Cellular Therapies Section Clinical Center, NIH Methods for the
Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna
 Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna targeted for hamster PAQR3. At 24 h after the transfection,
Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna targeted for hamster PAQR3. At 24 h after the transfection,
Novel Insights into Breast Cancer Genetic Variance through RNA Sequencing
 Insights into Breast Cancer Genetic Variance through RNA Sequencing SUPPLEMENTARY DATA Anelia Horvath, 1,2* Suresh Babu Pakala, 2* Prakriti Mudvari, 1* Sirigiri Divijendra Natha Reddy, 2 Kazufumi Ohshiro,
Insights into Breast Cancer Genetic Variance through RNA Sequencing SUPPLEMENTARY DATA Anelia Horvath, 1,2* Suresh Babu Pakala, 2* Prakriti Mudvari, 1* Sirigiri Divijendra Natha Reddy, 2 Kazufumi Ohshiro,
103.5 Membrane metalloendopeptidase TGA GGG GTC ACG ATT TTA GG ATG ATG GTG AGG AGC AGG AC
 Supplementary Table SA Primers used for real-time RT-PCR gene regulation and their confirmation Gene Name Accession no Size Region Forward Primer Reverse Primer Efficiency% AKT v-akt oncogene homolog NM_00563
Supplementary Table SA Primers used for real-time RT-PCR gene regulation and their confirmation Gene Name Accession no Size Region Forward Primer Reverse Primer Efficiency% AKT v-akt oncogene homolog NM_00563
Table SІ. List of genes upregulated by IL-4/IL-13-treatment of NHEK
 Supplemental legends Table SІ. List of genes upregulated by IL-4/IL-13-treatment of NHEK Table SП. List of genes in cluster 1 identified using two-dimensional hierarchical clustering analysis Table SШ.
Supplemental legends Table SІ. List of genes upregulated by IL-4/IL-13-treatment of NHEK Table SП. List of genes in cluster 1 identified using two-dimensional hierarchical clustering analysis Table SШ.
SUPPLEMENTAL TABLE AND FIGURES
 SUPPLEMENTAL TABLE AND FIGURES Zhang et al. (29) - Enzymes in the NAD + Salvage Pathway Regulate SIRT1 Activity at Target Gene Promoters This document contains supplemental data (1 table and 6 figures)
SUPPLEMENTAL TABLE AND FIGURES Zhang et al. (29) - Enzymes in the NAD + Salvage Pathway Regulate SIRT1 Activity at Target Gene Promoters This document contains supplemental data (1 table and 6 figures)
Supplementary Material. Table S1. Summary of mapping results
 Supplementary Material Table S1. Summary of mapping results Sample Total reads Mapped (%) HNE0-1 99687516 80786083 (81.0%) HNE0-2 94318720 77047792 (81.7%) HNE0-3 104033900 84348102 (81.1%) HNE15-1 94426598
Supplementary Material Table S1. Summary of mapping results Sample Total reads Mapped (%) HNE0-1 99687516 80786083 (81.0%) HNE0-2 94318720 77047792 (81.7%) HNE0-3 104033900 84348102 (81.1%) HNE15-1 94426598
Quantitative Real Time PCR (RT-qPCR) Differentiation of human preadipocytes to adipocytes
 Quantitative Real Time PCR (RT-qPCR) First strand cdna was synthesized and RT-qPCR was performed using RT2 first strand kits (Cat#330401) and RT2 qpcr Master Mix (SABioscience, Frederick, MD). Experiments
Quantitative Real Time PCR (RT-qPCR) First strand cdna was synthesized and RT-qPCR was performed using RT2 first strand kits (Cat#330401) and RT2 qpcr Master Mix (SABioscience, Frederick, MD). Experiments
Supplementary Information
 Supplementary Information Notch deficiency decreases hepatic lipid accumulation by induction of fatty acid oxidation No-Joon Song,#, Ui Jeong Yun,#, Sunghee Yang, Chunyan Wu, Cho-Rong Seo, A-Ryeong Gwon,,
Supplementary Information Notch deficiency decreases hepatic lipid accumulation by induction of fatty acid oxidation No-Joon Song,#, Ui Jeong Yun,#, Sunghee Yang, Chunyan Wu, Cho-Rong Seo, A-Ryeong Gwon,,
Research Article Integrated Bioinformatics Analysis of Hub Genes and Pathways in Anaplastic Thyroid Carcinomas
 International Journal of Endocrinology Volume 219, Article ID 965138, 9 pages https://doi.org/.1155/219/965138 Research Article Integrated Bioinformatics Analysis of Hub Genes and Pathways in Anaplastic
International Journal of Endocrinology Volume 219, Article ID 965138, 9 pages https://doi.org/.1155/219/965138 Research Article Integrated Bioinformatics Analysis of Hub Genes and Pathways in Anaplastic
Supplementary Information
 Supplementary Information Title: Role of Cytochrome P450 (CYP)1A in Hyperoxic Lung Injury: Analysis of the Transcriptome and Proteome Authors: Krithika Lingappan a, Suman Maity b, Weiwu Jiang a, Lihua
Supplementary Information Title: Role of Cytochrome P450 (CYP)1A in Hyperoxic Lung Injury: Analysis of the Transcriptome and Proteome Authors: Krithika Lingappan a, Suman Maity b, Weiwu Jiang a, Lihua
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
Integrative Radiation Biology
 Dr. Kristian Unger Integrative Biology Group Research Unit of Radiation Cytogenetics Department of Radiation Sciences Helmholtz-Zentrum München 1 Key Questions Molecular mechanisms of radiation-induced
Dr. Kristian Unger Integrative Biology Group Research Unit of Radiation Cytogenetics Department of Radiation Sciences Helmholtz-Zentrum München 1 Key Questions Molecular mechanisms of radiation-induced
Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3
 Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus
 Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplemental Table 1. 3 types of signals that are involved in T cell activation
 Supplemental Table 1. 3 types of signals that are involved in T cell activation Syn-T cells Allo-T cells CD3/28- T cells First signal MHC-TCR-CD3 pathway Not engaged MHC-alloantigen- TCR-CD3 Engaged CD3-Engaged
Supplemental Table 1. 3 types of signals that are involved in T cell activation Syn-T cells Allo-T cells CD3/28- T cells First signal MHC-TCR-CD3 pathway Not engaged MHC-alloantigen- TCR-CD3 Engaged CD3-Engaged
Supplementary Materials
 Supplementary Materials Using Multi objective Optimization to Identify Dynamical Network Biomarkers as Early warning Signals of Complex Diseases Fatemeh Vafaee 1, 2 1 Charles Perkins Centre, University
Supplementary Materials Using Multi objective Optimization to Identify Dynamical Network Biomarkers as Early warning Signals of Complex Diseases Fatemeh Vafaee 1, 2 1 Charles Perkins Centre, University
Supplementary Files to Töröcsik et al. Factor XIII-A is involved in the regulation of
 Supplementary Files to Töröcsik et al. Factor XIII-A is involved in the regulation of gene expression in alternatively activated human macrophages (Thromb Haemost 2010; 104.3) A Protein synthesis Cell
Supplementary Files to Töröcsik et al. Factor XIII-A is involved in the regulation of gene expression in alternatively activated human macrophages (Thromb Haemost 2010; 104.3) A Protein synthesis Cell
Glia in Alzheimer's disease and aging: Molecular mechanisms underlying astrocyte and microglia reactivity Orre, A.M.
 UvA-DARE (Digital Academic Repository) Glia in Alzheimer's disease and aging: Molecular mechanisms underlying astrocyte and microglia reactivity Orre, A.M. Link to publication Citation for published version
UvA-DARE (Digital Academic Repository) Glia in Alzheimer's disease and aging: Molecular mechanisms underlying astrocyte and microglia reactivity Orre, A.M. Link to publication Citation for published version
Table S1. Primer sequences used for qrt-pcr. CACCATTGGCAATGAGCGGTTC AGGTCTTTGCGGATGTCCACGT ACTB AAGTCCATGTGCTGGCAGCACT ATCACCACTCCGAAGTCCGTCT LCOR
 Table S1. Primer sequences used for qrt-pcr. ACTB LCOR KLF6 CTBP1 CDKN1A CDH1 ATF3 PLAU MMP9 TFPI2 CACCATTGGCAATGAGCGGTTC AGGTCTTTGCGGATGTCCACGT AAGTCCATGTGCTGGCAGCACT ATCACCACTCCGAAGTCCGTCT CGGCTGCAGGAAAGTTTACA
Table S1. Primer sequences used for qrt-pcr. ACTB LCOR KLF6 CTBP1 CDKN1A CDH1 ATF3 PLAU MMP9 TFPI2 CACCATTGGCAATGAGCGGTTC AGGTCTTTGCGGATGTCCACGT AAGTCCATGTGCTGGCAGCACT ATCACCACTCCGAAGTCCGTCT CGGCTGCAGGAAAGTTTACA
Regulation of eif4 and p70s6k signaling. Chondroitin and dermatan biosynthesis Interferon signaling
 Supplemental Information Table S1: Differentially regulated pathways in cortical versus cerebellar human astrocytes following treatment with PBS or IFN-β Pathway Regulation of eif4 and p70s6k signaling
Supplemental Information Table S1: Differentially regulated pathways in cortical versus cerebellar human astrocytes following treatment with PBS or IFN-β Pathway Regulation of eif4 and p70s6k signaling
The pluripotency factor NANOG promotes the formation of squamous cell carcinomas
 SUPPLEMENTAL DATA The pluripotency factor NANOG promotes the formation of squamous cell carcinomas Adelaida R. Palla, Daniela Piazzolla, Noelia Alcazar, Marta Cañamero, Osvaldo Graña, Gonzalo Gómez-López,
SUPPLEMENTAL DATA The pluripotency factor NANOG promotes the formation of squamous cell carcinomas Adelaida R. Palla, Daniela Piazzolla, Noelia Alcazar, Marta Cañamero, Osvaldo Graña, Gonzalo Gómez-López,
COMPREHENSIVE DIAGNOSIS FOR MITOCHONDRIAL DISORDERS
 COMPREHENSIVE DIAGNOSIS FOR MITOCHONDRIAL DISORDERS MITOCHONDRIAL DISORDERS NGS PANELS MITOCHONDRIAL DISORDERS NGS PANELS Name Test code Gene Name Mitome200-Nuclear 2086 (164 nuclear genes) AARS2, ACACA,
COMPREHENSIVE DIAGNOSIS FOR MITOCHONDRIAL DISORDERS MITOCHONDRIAL DISORDERS NGS PANELS MITOCHONDRIAL DISORDERS NGS PANELS Name Test code Gene Name Mitome200-Nuclear 2086 (164 nuclear genes) AARS2, ACACA,
A role for Peroxisome Proliferator-Activated Receptor Beta in T cell development
 SUPPLEMENTARY INFORMATION A role for Peroxisome Proliferator-Activated Receptor Beta in T cell development Isabelle Mothe-Satney, Joseph Murdaca, Brigitte Sibille, Anne-Sophie Rousseau, Raphaëlle Squillace,
SUPPLEMENTARY INFORMATION A role for Peroxisome Proliferator-Activated Receptor Beta in T cell development Isabelle Mothe-Satney, Joseph Murdaca, Brigitte Sibille, Anne-Sophie Rousseau, Raphaëlle Squillace,
Breast cancer cells produce tenascin-c as a metastatic niche component to colonize the lungs
 Supplementary figures for: Breast cancer cells produce tenascin-c as a metastatic niche component to colonize the lungs Thordur Oskarsson, Swarnali Acharyya, Xiang H.-F. Zhang, Sakari Vanharanta, Sohail
Supplementary figures for: Breast cancer cells produce tenascin-c as a metastatic niche component to colonize the lungs Thordur Oskarsson, Swarnali Acharyya, Xiang H.-F. Zhang, Sakari Vanharanta, Sohail
Supplementary information. Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls
 Supplementary information Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls angiogenesis in breast cancer Ali Flores-Pérez, Laurence A. Marchat, Sergio Rodríguez-Cuevas, Verónica Bautista-Piña,
Supplementary information Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls angiogenesis in breast cancer Ali Flores-Pérez, Laurence A. Marchat, Sergio Rodríguez-Cuevas, Verónica Bautista-Piña,
Illumina Clinical Services Laboratory
 Illumina Clinical Services Laboratory CLIA Certificate No.: 05D1092911 Illumina Clinical Services Laboratory TruGenome Predisposition Screen Requisition Form The TruGenome Predisposition Screen is intended
Illumina Clinical Services Laboratory CLIA Certificate No.: 05D1092911 Illumina Clinical Services Laboratory TruGenome Predisposition Screen Requisition Form The TruGenome Predisposition Screen is intended
Transduction of lentivirus to human primary CD4+ T cells
 Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
Supplementary information: Forstner, Hofmann et al.
 Supplementary information: Forstner, Hofmann et al., Genome-wide analysis implicates micrornas and their target genes in the development of bipolar disorder Table of contents Supplementary Figure 1: Regional
Supplementary information: Forstner, Hofmann et al., Genome-wide analysis implicates micrornas and their target genes in the development of bipolar disorder Table of contents Supplementary Figure 1: Regional
What Do We Know About Individual Variability and Its
 What Do We Know About Individual Variability and Its Contribution to Disease? Nathaniel Rothman, MD, MPH, MHS Occupational and Environmental Epidemiology Branch, Division of Cancer Epidemiology and Genetics,
What Do We Know About Individual Variability and Its Contribution to Disease? Nathaniel Rothman, MD, MPH, MHS Occupational and Environmental Epidemiology Branch, Division of Cancer Epidemiology and Genetics,
Enabling Informed Clinical Decisions with Deep Insights. Routine Multi-gene Testing for Inherited Neuromuscular Disorders
 Enabling Informed Clinical Decisions with Deep Insights Routine Multi-gene Testing for Inherited Neuromuscular Disorders Introduction Multi-gene Testing for Inherited Neuromuscular Disorders Inherited
Enabling Informed Clinical Decisions with Deep Insights Routine Multi-gene Testing for Inherited Neuromuscular Disorders Introduction Multi-gene Testing for Inherited Neuromuscular Disorders Inherited
Peroxisome proliferator-activated receptor alpha target genes
 University of Wollongong Research Online Illawarra Health and Medical Research Institute Faculty of Science, Medicine and Health 2010 Peroxisome proliferator-activated receptor alpha target genes Maryam
University of Wollongong Research Online Illawarra Health and Medical Research Institute Faculty of Science, Medicine and Health 2010 Peroxisome proliferator-activated receptor alpha target genes Maryam
a Supplementary Figure 1 Celastrol Withaferin A Individual drugs
 Supplementary Figure 1 a 17 27 HSPA1A SLC7A11 HMOX1 GSTA1 DUSP4 GML CHAC1 CDKN1A GSTA4 CA6 BHLHE41 NR1D1 HSPB1 PTX3 HP NFKBIA VDR MVD HAS2 ANGPT1 WDR6 TGFB3 IDI1 VCAM1 H1F HMGCS1 CXCL5 STEAP4 NOS2 b Enrichment
Supplementary Figure 1 a 17 27 HSPA1A SLC7A11 HMOX1 GSTA1 DUSP4 GML CHAC1 CDKN1A GSTA4 CA6 BHLHE41 NR1D1 HSPB1 PTX3 HP NFKBIA VDR MVD HAS2 ANGPT1 WDR6 TGFB3 IDI1 VCAM1 H1F HMGCS1 CXCL5 STEAP4 NOS2 b Enrichment
NGS data processing and alignment: Raw reads generated from the Illumina HiSeq2500
 Supplemental aterials ethods NGS data processing and alignment: Raw reads generated from the Illumina HiSeq25 sequencer were demultiplexed using configurebcl2fastq.pl version 1.8.4. Quality filtering and
Supplemental aterials ethods NGS data processing and alignment: Raw reads generated from the Illumina HiSeq25 sequencer were demultiplexed using configurebcl2fastq.pl version 1.8.4. Quality filtering and
Delineation of Diverse Macrophage Activation Programs in Response to Intracellular Parasites and Cytokines
 Delineation of Diverse Macrophage Activation Programs in Response to Intracellular Parasites and Cytokines Shuyi Zhang 1,2, Charles C. Kim 3, Sajeev Batra 3, James H. McKerrow 1,2, P ng Loke 4 * 1 Department
Delineation of Diverse Macrophage Activation Programs in Response to Intracellular Parasites and Cytokines Shuyi Zhang 1,2, Charles C. Kim 3, Sajeev Batra 3, James H. McKerrow 1,2, P ng Loke 4 * 1 Department
Supporting Information
 Supporting Information Lin et al. 10.1073/pnas.1408759111 Fig. S1. Safety and potential toxicity evaluation of M1 in immunocompetent mice. Immunocompetent BALB/c mice received two doses of i.v. infusion
Supporting Information Lin et al. 10.1073/pnas.1408759111 Fig. S1. Safety and potential toxicity evaluation of M1 in immunocompetent mice. Immunocompetent BALB/c mice received two doses of i.v. infusion
Table S1. List of the genes regulated by thyroid hormones in intestinal crypt cells
 Table S1. List of the genes regulated by thyroid hormones in intestinal crypt cells Probeset ID Gene symbol Cell adhesion/extracellular matrix Gene title Fold change a pvalue a WT THvsPTU T-Test TRa0/0
Table S1. List of the genes regulated by thyroid hormones in intestinal crypt cells Probeset ID Gene symbol Cell adhesion/extracellular matrix Gene title Fold change a pvalue a WT THvsPTU T-Test TRa0/0
Moyamoya disease susceptibility gene RNF213 links inflammatory and angiogenic
 Supplementary Information for Moyamoya disease susceptibility gene RNF213 links inflammatory and angiogenic signals in endothelial cells Kazuhiro Ohkubo 1,, Yasunari Sakai 1,*,, Hirosuke Inoue 1, Satoshi
Supplementary Information for Moyamoya disease susceptibility gene RNF213 links inflammatory and angiogenic signals in endothelial cells Kazuhiro Ohkubo 1,, Yasunari Sakai 1,*,, Hirosuke Inoue 1, Satoshi
