Supplementary Materials
|
|
|
- Tabitha Harris
- 5 years ago
- Views:
Transcription
1 Supplementary Materials Supplemental figure 1. Absolute cell count of naïve/memory compartment of CD4 and CD8 T cells in early years Absolute cell number of naïve (CD45RA+CCR7+), Stem cell memory T cell (Tscm, CD45RA+CCR7+CD28+CD27+FAS+), central memory (Tcm, CD45RA- CCR7+), effector memory (Tem, CD45RA- CCR7- ), and Temra (effector memory re- expressing CD45RA, CD45RA+CCR7+) T cells respectively (young HC, n=9-10, black circles; young Tx, n=10, gray circles) A) For CD4 T cells, B) CD8 T cells. median shown. Statistical difference is indicated with * for P<0.05, Mann- Whitney U test used.
2 Supplemental figure 2. Thymic regeneration occurs in the majority of children and restores the naïve T cell compartment A) Absolute cell number of CD3+, CD4+ and CD8+ T cells (lymphocytes x 10^9/L) in neonatally thymectomized children, aged 10 years or older, divided by high CD31 expression (gray squares, n=17) and low CD31 expression (open squares, n=7). B) Absolute cell number of naïve (CD45RA+CCR7+), Tscm (Stem cell memory T cell, CD45RA+CCR7+CD28+CD27+FAS+), central memory (Tcm, CD45RA- CCR7+), effector memory (Tem, CD45RA- CCR7- ), and Temra (effector memory re- expressing CD45RA, CD45RA+CCR7+) for CD4+ T cells respectively. C) Absolute numbers of CD8 naïve, Tscm, Tem, Tcm and Temra respectively. D) Percentages of CD8 naïve, Tscm, Tem, Tcm, Temra respectively. (HC>10yr, n=10, black squares; Tx>10yr, n=24, gray squares; Tx>10yr low CD31, open gray squares) Median depicted. Statistical difference is indicated with * for P<0.05, Mann- Whitney U test used.
3 Supplemental figure 3. CD4+ naïve T cells of young thymectomized children show lower calcium flux and IL- 8 intensity, but have similar calcium flux capacity as healthy children. A) Left panel, Calcium flux of HC>10yr (older HC) naïve CD4+ (black line, n=4) and CD4+ memory (gray line, n=4); Right panel, AUC of paired calcium flux of naïve and memory CD4+ T cells. B) Maximum calcium flux after stimulation with ionomycin (HC 1-5yr, n=4; Tx 1-5yr, n=6). Statistical difference is indicated with * for P<0.05, Mann- Whitney U test used.
4 Supplemental figure 4. CD31- naïve CD4+ T cells have increased IFNγ production IFNγ production of CD31+ and CD31- naïve CD4+ T cells in young healthy and thymectomized children. (HC 1-5yr, n=14, black circles; Tx1-5yr, n=15, gray circles). Statistical difference is indicated with * for P<0.05, Mann- Whitney U test used for unpaired data and Wilcoxon signed rank test for paired data
5 Supplemental figure 5. Production of IL- 8 by CD31+ naïve CD4+ is correlated to PTK7 A) Representative histogram of IL- 8 production in PTK7+ (black line) and PTK7- (gray) CD31+ naïve CD4+ T cells. Geo mean of PTK7 expression in IL- 8 negative (gray) and IL- 8 positive (black) CD31+ naïve CD4+ T cells (n=5; HC 1-5yr). B) representative FACSplot of PTK7 expression of each cell division after cytokine stimulation. C) representative FACSplot of IL- 8 expression of each cell division after cytokine stimulation. D) combined data set for corresponding percentages of IL- 8 and PTK7 per patient and division (n=4; HC1-5yr) E) representative FACSplot of CD31 expression after cytokine stimulation per cell division. Right panel, summary of mean fluorescence intensity (MFI) of CD31 per cell division after cytokine stimulation (n=6; HC>10yr). F) left panel, representative FACSplot of IFNγ expression after cytokine stimulation per cell division. Right panel, summary of IFNγ percentage per cell division after cytokine stimulation (n=6; HC>10yr). Statistical difference is indicated with * for P<0.05, Wilcoxon signed rank test for paired data
6 Supplemental figure 6. Representative FACS gating strategy for sorting of CD31+ naive CD4+ T cells. Single cell lymphocytes were gated on CD3+ and CD4+ expression (1 st panel), followed by exclusion of CD25+CD127- (Treg) cells (non- Treg, 2 nd panel), selection of CD31+CD45RA+ expression (3 rd panel), and further exclusion of CD45RO+ and CCR7- T cells to exclude possible Temra or CD45RA+CD45RO+ double positive T cells contamination (non- Temra, 4 th panel)
7 Supplemental figure 7. Pathway enrichment analysis for CD31+ naïve T cells of Tx 1-5yr and HC 1-5yr. A) Description of significant pathways that are differentially expressed between CD31+ naïve CD4+ T cells of Tx1-5yr (n=4) and HC1-5yr (n=3). B) Description of pathways upregulated in CD31+ naïve CD4+ T cells of Tx1-5yr (n=4) in comparison to HC1-5yr (n=3). Significance determined by FDR (B&H) p- value.
8 Supplemental table 1. Differential gene expression between CD31+ naïve CD4+ T cells of thymectomized and healthy children. Gene name Refseq id Base Mean log2foldchange p- value p- value adjusted GBP5 NM_ , , ,31901E- 18 2,94496E- 14 BHLHE40 NM_ , , ,64317E- 18 2,94496E- 14 GBP5 NM_ , , ,37768E- 17 8,14943E- 14 EDA NM_ , , ,69756E- 16 2,3771E- 12 EDA NM_ , , ,69756E- 16 2,3771E- 12 TOX NM_ , , ,22234E- 15 3,61527E- 12 MYO15B NR_ , , ,5154E- 15 6,3769E- 12 PTGER2 NM_ , , ,91551E- 13 1,09038E- 09 PREX1 NM_ , , ,57843E- 13 1,69148E- 09 AFF2 NM_ , , ,09039E- 12 9,03341E- 09 RICS NM_ , , ,92632E- 10 6,33423E- 07 FAM101B NM_ , , ,50437E- 10 1,29742E- 06 ISM1 NM_ , , ,83679E- 10 1,29742E- 06 SUSD4 NM_ , , ,62934E- 09 3,33287E- 06 TIMP1 NM_ , , ,11067E- 09 4,8632E- 06 RICS NM_ , , ,61642E- 09 5,12019E- 06 CACNA1I NM_ , , ,58998E- 08 2,55344E- 05 CACNA1I NM_ , , ,58998E- 08 2,55344E- 05 HES4 NM_ , , ,48062E- 08 3,19725E- 05 HES4 NM_ , , ,60334E- 08 3,19725E- 05 PYHIN1 NM_ , , ,36155E- 08 3,51818E- 05 PYHIN1 NM_ , , ,36155E- 08 3,51818E- 05 PLXDC1 NM_ , , ,56066E- 08 6,60511E- 05 RAB15 NM_ , , ,0371E- 07 7,37388E- 05 SCRN1 NM_ , , ,03881E- 07 7,37388E- 05 TTC28 NM_ , , ,08174E- 07 7,38329E- 05 ENO1 NM_ , , ,70381E- 07 0, HDGFRP3 NM_ , , ,62199E- 07 0, STMN1 NM_ , , ,60945E- 07 0, STMN1 NM_ , , ,60945E- 07 0, STMN1 NM_ , , ,60945E- 07 0,
9 SLC40A1 NM_ , , ,86228E- 07 0, TRIM32 NM_ , , ,59059E- 07 0, TRIM32 NM_ , , ,59059E- 07 0, EDARADD NM_ , , ,41268E- 07 0, NPAS2 NM_ , , ,38705E- 07 0, EDARADD NM_ , , ,41268E- 07 0, GGT1 NM_ , , ,7316E- 07 0, TIMP2 NM_ , , ,78859E- 07 0, GGT1 NM_ , , ,84562E- 07 0, FLNB NM_ , , ,47648E- 07 0, FLNB NM_ , , ,5911E- 07 0, SNHG3 NR_ , , ,80958E- 07 0, FLNB NM_ , , ,47648E- 07 0, FLNB NM_ , , ,5911E- 07 0, GGT1 NM_ , , ,51955E- 07 0, GZMK NM_ , , ,39043E- 07 0, GGT1 NM_ , , ,51955E- 07 0, MT2A NM_ , , ,23163E- 07 0, TCF12 NM_ , , ,93308E- 07 0, TCF12 NM_ , , ,93308E- 07 0, TCF12 NM_ , , ,93308E- 07 0, TCF12 NM_ , , ,93308E- 07 0, FLJ40330 NR_ , , ,61281E- 07 0, STAT4 NM_ , , ,71544E- 07 0, NIN NM_ , , ,93085E- 07 0, CYB561 NM_ , , ,00554E- 06 0, CYB561 NM_ , , ,00554E- 06 0, CYB561 NM_ , , ,00291E- 06 0, ZC3H12D NM_ , , ,13055E- 06 0, NIN NM_ , , ,20456E- 06 0, LSR NM_ , , ,23148E- 06 0, TCF12 NM_ , , ,29194E- 06 0, LSR NM_ , , ,41593E- 06 0, LSR NM_ , , ,42313E- 06 0,
10 SNHG5 NR_ , , ,57707E- 06 0, ADAMTSL5 NM_ , , ,65242E- 06 0, PYHIN1 NM_ , , ,88629E- 06 0, PYHIN1 NM_ , , ,88629E- 06 0, FXYD5 NR_ , , ,99153E- 06 0, FXYD5 NM_ , , ,04134E- 06 0, SCRN1 NM_ , , ,05653E- 06 0, FXYD5 NM_ , , ,11089E- 06 0, FXYD5 NM_ , , ,1153E- 06 0, IGF2R NM_ , , ,33385E- 06 0, SCRN1 NM_ , , ,29672E- 06 0, SCRN1 NM_ , , ,31514E- 06 0, IFITM1 NM_ , , ,94191E- 06 0, FAM195A NM_ , , ,37359E- 06 0, NIN NM_ , , ,58213E- 06 0, LETMD1 NM_ , , ,98676E- 06 0, LETMD1 NM_ , , ,27019E- 06 0, AOAH NM_ , , ,32227E- 06 0, CAPN2 NM_ , , ,52588E- 06 0, CAPN2 NM_ , , ,97574E- 06 0, SH3PXD2A NM_ , , ,24195E- 06 0, SERTAD2 NM_ , , ,79765E- 06 0, DYNLT1 NM_ , , ,01473E- 06 0, APP NM_ , , ,65354E- 06 0, GRIP1 NM_ , , ,0021E- 06 0, HERC1 NM_ , , ,62094E- 06 0, OBSCN NM_ , , ,9033E- 06 0, SLFN13 NM_ , , ,1636E- 06 0, CCDC59 NM_ , , ,66053E- 06 0, SOCS2 NM_ , , ,02186E- 05 0, CA6 NM_ , , ,23101E- 05 0, TOB1 NM_ , , ,29345E- 05 0, APP NM_ , , ,47894E- 05 0, GSTK1 NM_ , , ,53021E- 05 0,
11 TMPRSS3 NR_ , , ,5221E- 05 0, MRC2 NM_ , , ,90555E- 05 0, EPHB6 NM_ , , ,96894E- 05 0, ACCN2 NM_ , , ,03631E- 05 0, ACCN2 NM_ , , ,03631E- 05 0, RNF157 NM_ , , ,00869E- 05 0, TET1 NM_ , , ,17071E- 05 0, APP NM_ , , ,30355E- 05 0, AK1 NM_ , , ,44242E- 05 0, AKAP11 NM_ , , ,47539E- 05 0, TBK1 NM_ , , ,57832E- 05 0, FAM113B NM_ , , ,88917E- 05 0, GAPDH NM_ , , ,9891E- 05 0, HIGD2A NM_ , , ,97574E- 05 0, C8orf59 NM_ , , ,23065E- 05 0, GZMA NM_ , , ,57034E- 05 0, GALNT6 NM_ , , ,55162E- 05 0, C8orf59 NM_ , , ,5128E- 05 0, CAMSAP1L1 NM_ , , ,74657E- 05 0, LY86 NM_ , , ,05315E- 05 0, C5orf13 NM_ , , ,33403E- 05 0, C5orf13 NM_ , , ,245E- 05 0, ANKRD50 NM_ , , ,90519E- 05 0, C5orf13 NM_ , , ,245E- 05 0, C5orf13 NM_ , , ,245E- 05 0, GSTK1 NM_ , , ,33091E- 05 0, TYMP NM_ , , ,88121E- 05 0, C5orf13 NM_ , , ,245E- 05 0, C5orf13 NM_ , , ,245E- 05 0, C8orf59 NM_ , , ,2249E- 05 0, TYMP NM_ , , ,86509E- 05 0, C5orf13 NM_ , , ,245E- 05 0, FOSL2 NM_ , , ,20691E- 05 0, C5orf13 NM_ , , ,33403E- 05 0,
12 C5orf13 NM_ , , ,245E- 05 0, C5orf13 NM_ , , ,07552E- 05 0, C5orf13 NM_ , , ,245E- 05 0, C8orf59 NM_ , , ,47794E- 05 0, SLC35F2 NM_ , , ,49722E- 05 0, TYMP NM_ , , ,45817E- 05 0, APP NM_ , , ,58782E- 05 0, ETS1 NM_ , , ,84558E- 05 0, APP NM_ , , ,96493E- 05 0, ZBTB34 NM_ , , ,12508E- 05 0, RELB NM_ , , ,14297E- 05 0, NHEDC2 NM_ , , ,23906E- 05 0, PSME2 NM_ , , ,31564E- 05 0, GSTK1 NM_ , , ,32027E- 05 0, SNHG1 NR_ , , ,35931E- 05 0, SUSD4 NM_ , , ,84287E- 05 0, ETS1 NM_ , , ,8841E- 05 0, IRS1 NM_ , , ,96524E- 05 0, RCN3 NM_ , , ,21042E- 05 0, SARDH NM_ , , ,44734E- 05 0, SARDH NM_ , , ,44734E- 05 0, VGLL4 NM_ , , ,90658E- 05 0, ACTN1 NM_ , , ,98053E- 05 0, GSTK1 NM_ , , ,33883E- 05 0, HMBOX1 NM_ , , ,3357E- 05 0, ETS1 NM_ , , ,51918E- 05 0, IFITM2 NM_ , , ,59871E- 05 0, YARS NM_ , , ,67056E- 05 0, S100A4 NM_ , , ,96106E- 05 0, PASK NM_ , , ,89663E- 05 0, S100A4 NM_ , , ,96106E- 05 0, ACTN1 NM_ , , ,94999E- 05 0, CERKL NM_ , , ,36694E- 05 0, CERKL NM_ , , ,36694E- 05 0,
13 CERKL NR_ , , ,36694E- 05 0, CERKL NM_ , , ,36694E- 05 0, CERKL NM_ , , ,36694E- 05 0, CERKL NM_ , , ,36694E- 05 0, CERKL NR_ , , ,36694E- 05 0, LDHA NM_ , , ,59064E- 05 0, REST NM_ , , ,11163E- 05 0, ACTN1 NM_ , , ,27091E- 05 0, RPLP1 NM_ , , ,328E- 05 0, OXA1L NM_ , , ,42157E- 05 0, RPLP1 NM_ , , ,53683E- 05 0, ACVR1 NM_ , , ,6995E- 05 0, ACVR1 NM_ , , ,67864E- 05 0, SLU7 NM_ , , ,8286E- 05 0, LDHA NR_ , , ,86006E- 05 0, OBSCN NM_ , , , , SH3BGRL3 NM_ , , , , BCOR NM_ , , , , SRGN NM_ , , , , BCOR NM_ , , , , MTHFD2 NR_ , , , , HMBOX1 NM_ , , , , BCOR NM_ , , , , BCOR NM_ , , , , MTHFD2 NM_ , , , , RGS12 NM_ , , , , NUAK2 NM_ , , , , EIF2C3 NM_ , , , , MDGA1 NM_ , , , , NFKBIZ NM_ , , , , IL16 NM_ , , , , RAPGEF6 NM_ , , , , CHMP7 NM_ , , , , SLCO3A1 NM_ , , , ,
14 IL16 NM_ , , , , FKBP11 NM_ , , , , NFKBIZ NM_ , , , , MDFIC NM_ , , , , MDFIC NM_ , , , , STXBP1 NM_ , , , , STXBP1 NM_ , , , , LDHA NM_ , , , , ISG15 NM_ , , , , LDHA NM_ , , , , CHD9 NM_ , , , , CBL NM_ , , , , RNF41 NM_ , , , , RNF41 NM_ , , , , ODC1 NM_ , , , , FLJ43663 NR_ , , , , B2M NM_ , , , , RNF41 NM_ , , , , LGALS9 NM_ , , , , LDHA NM_ , , , , C17orf49 NM_ , , , , ZC3H12A NM_ , , , , IL21R NM_ , , , , BCL9 NM_ , , , , S100A10 NM_ , , , , C17orf49 NM_ , , , , MARCKSL1 NM_ , , , , NR3C1 NM_ , , , , RWDD1 NM_ , , , , TGFBR2 NM_ , , , , CD40LG NM_ , , , , RWDD1 NM_ , , , , LGALS9 NR_ , , , , OASL NM_ , , , ,
15 SNX6 NM_ , , , , SNX6 NM_ , , , , UPP1 NM_ , , , , CREM NM_ , , , , NBL1 NM_ , , , , CREM NM_ , , , , NBL1 NM_ , , , , CREM NM_ , , , , CREM NM_ , , , , CREM NM_ , , , , SBDS NM_ , , , , RWDD1 NM_ , , , , YBX1 NM_ ,7143 0, , , CHST2 NM_ , , , , TLR2 NM_ , , , , SULT1B1 NM_ , , , , FRY NM_ , , , , TJP3 NM_ , , , , NHP2 NM_ , , , , CLEC2B NM_ , , , , FBXO11 NM_ , , , , TGFBR2 NM_ , , , , BCL11A NM_ , , , , LGALS9 NM_ , , , , SYN1 NM_ , , , , SON NM_ , , , , LSM7 NM_ , , , , SYN1 NM_ , , , , NME2P1 NR_ , , , , FLJ43663 NR_ , , , , PDE7A NM_ , , , , KLF11 NM_ , , , , NCRNA00164 NR_ , , , , UPP1 NM_ , , , ,
16 WHAMM NM_ , , , , APOL3 NR_ , , , , DEGS1 NM_ , , , , ATF4 NM_ , , , , APOL3 NR_ , , , , DCHS1 NM_ , , , , MRPS21 NM_ , , , , VGLL4 NM_ , , , , SON NM_ , , , , CREM NM_ , , , , C7orf50 NM_ , , , , C7orf50 NM_ , , , , C7orf50 NM_ , , , , ZBP1 NM_ , , , , PRDX6 NM_ , , , , SIPA1L3 NM_ , , , , MRPS21 NM_ , , , , FOXO1 NM_ , , , , RPLP0 NM_ , , , , ODZ1 NM_ , , , , RPLP0 NM_ , , , , ODZ1 NM_ , , , , ODZ1 NM_ , , , , LZTFL1 NM_ , , , , ACVR2B NM_ , , , , ZNF696 NM_ , , , , MYBL1 NM_ , , , , CD97 NM_ , , , , MYBL1 NM_ , , , , IL21R NM_ , , , , OASL NM_ , , , , STRN NM_ , , , , NFATC1 NM_ , , , , CD97 NM_ , , , ,
17 DDB2 NM_ , , , , CCDC85B NM_ , , , , EIF5A NM_ , , , , EIF5A NM_ , , , , EIF5A NM_ , , , , ARHGAP26 NM_ , , , , ARHGAP26 NM_ , , , , C8orf45 NM_ , , , , EIF5A NM_ , , , , TSC22D2 NM_ , , , , APOL3 NM_ , , , , APOL3 NM_ , , , , EIF2C3 NM_ , , , , APOL3 NM_ , , , , CD97 NM_ , , , , STMN1 NM_ , , , , SNRPG NM_ , , , , HMBS NM_ , , , , NME2 NM_ , , , , ADAMTS10 NM_ , , , , TNF NM_ , , , , HPCAL4 NM_ , , , , APPL1 NM_ , , , , NME2 NM_ , , , , NME2 NM_ , , , , ABHD2 NM_ , , , , PELI1 NM_ , , , , ABHD2 NM_ , , , , NME2 NM_ , , , , ATF4 NM_ , , , , SLCO3A1 NM_ , , , , GPR132 NM_ , , , , SCAI NM_ , , , , APOL3 NR_ , , , ,
18 NRGN NM_ , , , , VIPR1 NM_ , , , , NRGN NM_ , , , , PLSCR3 NM_ , , , , LOC NM_ , , , , RARRES3 NM_ , , , , IL21R NM_ , , , , CTSW NM_ , , , , EMP3 NM_ , , , , TTC16 NM_ , , , , SCAI NM_ , , , , GADD45B NM_ , , , , PLEKHG2 NM_ , , , , ANKRD44 NM_ , , , , LY6E NM_ , , , , LY6E NM_ , , , , KLF13 NM_ , , , , HMBS NM_ , , , , RGS12 NM_ , , , , LDOC1L NM_ , , , , ZNF792 NM_ , , , , FKBP11 NM_ , , , , DRAP1 NM_ , , , , FAM13A NM_ , , , , MAN1A2 NM_ , , , , BAT2L NM_ , , , , NME1- NME2 NM_ , , , , NDUFV2 NM_ , , , , FAM13A NM_ , , , , CD109 NM_ , , , , GUK1 NM_ , , , , C17orf49 NM_ , , , , TAF4B NM_ , , , , EPSTI1 NM_ , , , ,
19 EPSTI1 NM_ , , , , ZNF394 NM_ , , , , DPP4 NM_ , , , , VGLL4 NM_ , , , , UQCRFS1 NM_ , , , , IDI1 NM_ , , , , GUK1 NM_ , , , , TTC39C NR_ , , , , NT5C3 NM_ , , , , CBR3 NM_ , , , , MAN2C1 NM_ , , , , TP53TG1 NR_ , , , , CTDSP2 NM_ , , , , SEL1L3 NM_ , , , , LBH NM_ , , , , NT5C3 NR_ , , , , GBP2 NM_ , , , , ZBP1 NM_ , , , , CSTB NM_ , , , , GTF3A NM_ , , , , FTH1 NM_ , , , , B4GALT3 NM_ , , , , IFITM3 NM_ , , , , MAPKAPK3 NM_ , , , , NT5C3 NM_ , , , , SLC39A8 NM_ , , , , H1FX NM_ , , , , RAB25 NM_ , , , , CD109 NM_ , , , , MSH2 NM_ , , , , NT5C3 NM_ , , , , NR3C1 NM_ , , , , NR3C1 NM_ , , , , CSDAP1 NR_ , , , ,
20 C19orf70 NM_ , , , , LTA NM_ , , , , VGLL4 NM_ , , , , FNBP1 NM_ , , , , TNFRSF10B NR_ , , , , PVT1 NR_ , , , , TNFRSF1B NM_ , , , , CLIC1 NM_ , , , , SYF2 NM_ , , , , ZBP1 NM_ , , , , NR3C1 NM_ , , , , EMR4P NR_ , , , , PPA2 NM_ , , , , HINT1 NR_ , , , , NR3C1 NM_ , , , , RPL39 NM_ , , , , EPPK1 NM_ , , , , NR3C1 NM_ , , , , MAP1LC3A NM_ , , , , LDHA NM_ ,4828 0, , , NR3C1 NM_ , , , , GPRC5B NM_ , , , , LBXCOR1 NM_ , , , , LOC NR_ , , , , BTLA NM_ , , , , CREM NM_ , , , , GUK1 NM_ , , , , CCNL1 NM_ , , , , OSGIN1 NM_ , , , , OSGIN1 NM_ , , , , PIK3CG NM_ , , , , IL17RA NM_ , , , , C10orf35 NM_ , , , , RAG1AP1 NM_ , , , ,
21 TXNL1 NR_ , , , , TMPRSS3 NM_ , , , , ADCY9 NM_ , , , , TNFRSF10B NM_ , , , , UHMK1 NM_ , , , , PELI2 NM_ , , , , CREM NM_ , , , , NDUFA7 NM_ , , , , ARID1A NM_ , , , , ANKK1 NM_ , , , , RAB4B NM_ , , , , GOLGA7B NM_ , , , , TXNL1 NM_ , , , , ACVR1C NM_ , , , , NHP2 NM_ , , , , SLC39A8 NM_ , , , , LOC NR_ , , , , MAN2A1 NM_ , , , , CDGAP NM_ , , , , BOLA2 NM_ , , , , ZC3HAV1 NM_ , , , , BOLA2B NM_ , , , , RPS29 NM_ , , , , P2RY11 NM_ , , , , SEMA4D NM_ , , , , CREM NM_ , , , , KIAA1432 NM_ , , , , ERC1 NR_ , , , , ARID1A NM_ , , , , RPL28 NM_ , , , , RPL28 NM_ ,6698 0, , , ZBTB44 NM_ , , , , USP10 NM_ , , , , SYNE2 NM_ , , , ,
22 BCL3 NM_ , , , , LAMP3 NM_ , , , , RPL41 NM_ , , , , ZNF445 NM_ , , , , RAG1AP1 NM_ , , , , ODF3B NM_ , , , , HCST NM_ , , , , BTLA NM_ , , , , HCST NM_ , , , , ACVR1C NM_ , , , , COX7A2 NR_ , , , , SSNA1 NM_ , , , , TP53INP2 NM_ , , , , NFKBIA NM_ , , , , ACVR1C NM_ , , , , LTA NM_ , , , , RPL41 NM_ , , , , CUX1 NM_ , , , , RAG1AP1 NM_ , , , , ERC1 NR_ , , , , THOC4 NM_ , , , , OSGIN1 NM_ , , , , RABGGTB NM_ , , , , ERC1 NM_ , , , , MAP3K7IP2 NM_ , , , ,
23 Supplemental table 2. Differential gene expression between CD31+ and CD31- naïve CD4+ T cells of thymectomized children Gene name Refseq id Base Mean log2foldchange p- value p- value adjusted PECAM1 NM_ , , ,32149E- 17 7,66832 E- 13 MAF NM_ , , ,9188E- 09 9,14107 E- 05 FAM129C NM_ , , ,52509E- 08 0, MAF NM_ , , ,05523E- 06 0,
24 Supplemental table 3. Differential gene expression between CD31+ and CD31- naïve CD4+ T cells of healthy children Gene name Refseq id Base Mean log2foldchange p- value p- value adjusted PECAM1 NM_ ,3161 2, ,59E- 30 4,83E- 26 PYHIN1 NM_ ,9948-1, ,62E- 09 3,56E- 05 PYHIN1 NM_ ,9948-1, ,62E- 09 3,56E- 05 PASK NM_ ,527-1, ,65E- 09 3,56E- 05 MAF NM_ ,5755-1, ,07E- 08 4,01E- 05 TOX2 NM_ ,0959 1, ,42E- 08 5,02E- 05 TOX2 NM_ ,5652 1, ,41E- 08 5,02E- 05 TOX2 NM_ ,0959 1, ,42E- 08 5,02E- 05 TOX2 NM_ ,0959 1, ,42E- 08 5,02E- 05 PLXND1 NM_ ,7822 1, ,3E- 08 5,45E- 05 SYK NM_ , , ,97E- 08 5,45E- 05 SYK NR_ , , ,97E- 08 5,45E- 05 SYK NM_ , , ,09E- 08 5,45E- 05 SYK NR_ , , ,09E- 08 5,45E- 05 FLJ40330 NR_ ,1594 1, ,21E- 08 0, FAM101B NM_ ,485 0,7369 1,04E- 07 0, GBP5 NM_ ,3008-1, ,75E- 07 0, PREX1 NM_ ,745-0, ,02E- 07 0, GBP5 NM_ ,047-1, ,78E- 07 0, PYHIN1 NM_ ,2576-1, ,29E- 06 0, PYHIN1 NM_ ,2576-1, ,29E- 06 0, GBP2 NM_ ,085-0, ,78E- 06 0, IKZF2 NM_ ,5382 0, ,65E- 06 0, CD8B NM_ , , ,43E- 06 0, CD8B NM_ , , ,43E- 06 0, MIAT NR_ ,0661-1, ,84E- 06 0, CD8B NM_ , , ,92E- 05 0, FRY NM_ ,3923 0, E- 05 0, FXYD5 NR_ ,535-0, ,7E- 05 0, CD8B NM_ , , ,54E- 05 0, FXYD5 NM_ ,313-0, ,58E- 05 0, UHMK1 NM_ ,1406 0, ,31E- 05 0,014848
25 FXYD5 NM_ ,345-0, ,59E- 05 0, FXYD5 NM_ ,839-0,6223 2,64E- 05 0, LRRN3 NM_ ,823 0, ,86E- 05 0, LRRN3 NM_ ,583 0, ,02E- 05 0, CD8B NM_ , , ,68E- 05 0, CAMK4 NM_ ,337 0, ,63E- 05 0, FCRL2 NM_ ,6392 0, ,86E- 05 0, LRRN3 NM_ ,041 0, ,87E- 05 0, F5 NM_ ,4492-0, ,54E- 05 0, LSR NM_ ,2851-0, ,8E- 05 0, LSR NM_ ,5957-0, ,63E- 05 0, LSR NM_ ,34-0, ,89E- 05 0, KCNA3 NM_ ,57 0, ,67E- 05 0, RHOH NM_ ,377 0, ,57E- 05 0,038839
An integrated transcriptomic and functional genomic approach identifies a. type I interferon signature as a host defense mechanism against Candida
 Supplementary information for: An integrated transcriptomic and functional genomic approach identifies a type I interferon signature as a host defense mechanism against Candida albicans in humans Sanne
Supplementary information for: An integrated transcriptomic and functional genomic approach identifies a type I interferon signature as a host defense mechanism against Candida albicans in humans Sanne
Deconvolution of Heterogeneous Wound Tissue Samples into Relative Macrophage Phenotype Composition via Models based on Gene Expression
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2017 Supplementary Information for: Deconvolution of Heterogeneous Wound Tissue Samples into
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2017 Supplementary Information for: Deconvolution of Heterogeneous Wound Tissue Samples into
Supplementary Figure 1. Microarray data mining and validation
 Supplementary Figure 1. Microarray data mining and validation Microarray (>47,000 probes) Raw data Background subtraction NanoString (124 genes) Set cut-off and threshold values Raw data Normalized to
Supplementary Figure 1. Microarray data mining and validation Microarray (>47,000 probes) Raw data Background subtraction NanoString (124 genes) Set cut-off and threshold values Raw data Normalized to
Nature Immunology: doi: /ni.3745
 Supplementary Figure 1 Experimental settings. (a) Experimental setup and bioinformatic data analysis of transcriptomic changes induced in neonatal and adult monocytes by stimulation with 10 ng/ml LPS for
Supplementary Figure 1 Experimental settings. (a) Experimental setup and bioinformatic data analysis of transcriptomic changes induced in neonatal and adult monocytes by stimulation with 10 ng/ml LPS for
HTG EdgeSeq Immuno-Oncology Assay Gene List
 A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
Supplementary Figures. Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated. for each lesion type.
 Supplementary Figures Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated for each lesion type. a b Supplementary Figure 2. Non-negative matrix factorization-derived
Supplementary Figures Supplementary Figure 1. Dinucleotide variant proportions. These are described and quantitated for each lesion type. a b Supplementary Figure 2. Non-negative matrix factorization-derived
Low Avidity CMV + T Cells accumulate in Old Humans
 Supplementary Figure Legends Supplementary Figure 1. CD45RA expressing CMVpp65-specific T cell populations accumulate within HLA-A*0201 and HLA-B*0701 individuals Pooled data showing the size of the NLV/HLA-A*0201-specific
Supplementary Figure Legends Supplementary Figure 1. CD45RA expressing CMVpp65-specific T cell populations accumulate within HLA-A*0201 and HLA-B*0701 individuals Pooled data showing the size of the NLV/HLA-A*0201-specific
Supplemental Table 1: Enriched GO categories per cluster
 Supplemental Table 1: Enriched GO categories per cluster Cluster Enriched with #genes Raw p-value Corrected p-value Frequency in set (%) Cluster 1 immune response - GO:0006955 32 2.36E-14 0.001 13.5 Cluster
Supplemental Table 1: Enriched GO categories per cluster Cluster Enriched with #genes Raw p-value Corrected p-value Frequency in set (%) Cluster 1 immune response - GO:0006955 32 2.36E-14 0.001 13.5 Cluster
Transduction of lentivirus to human primary CD4+ T cells
 Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10860 Supplementary Discussion It remains unclear why H3K9 demethylation appeared to be more sensitive to suppression than at least some other histone methylation marks as a result of
doi:10.1038/nature10860 Supplementary Discussion It remains unclear why H3K9 demethylation appeared to be more sensitive to suppression than at least some other histone methylation marks as a result of
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature11986 relative IL-6 expression Viable intracellular Bp per well 18 16 1 1 5 5 3 1 6 +DG 8 8 8 Control DG Time (h) hours B. pertussis IL-6 (pg/ml) 15 Control DG
SUPPLEMENTARY INFORMATION doi:1.138/nature11986 relative IL-6 expression Viable intracellular Bp per well 18 16 1 1 5 5 3 1 6 +DG 8 8 8 Control DG Time (h) hours B. pertussis IL-6 (pg/ml) 15 Control DG
Supplemental Table 1. List of genes and mature shrna sequences identified from the shrna screen in BRCA2-mutant PEO1 cells.
 Guillemette_Supplemental Table 1 Gene Mature Sequence Gene Mature Sequence ATTCATAGGATGTCAGCAG LOC157193 TATGCGTAAGTAATAAATTGCT TTAGTTCTGTCTTGGAGG LOC152225 CGCATATATGTTCTGACTT ALDH2 CACTTCAGTGTATGCCT
Guillemette_Supplemental Table 1 Gene Mature Sequence Gene Mature Sequence ATTCATAGGATGTCAGCAG LOC157193 TATGCGTAAGTAATAAATTGCT TTAGTTCTGTCTTGGAGG LOC152225 CGCATATATGTTCTGACTT ALDH2 CACTTCAGTGTATGCCT
pplementary Figur Supplementary Figure 1. a.
 pplementary Figur Supplementary Figure 1. a. Quantification by RT-qPCR of YFV-17D and YFV-17D pol- (+) RNA in the supernatant of cultured Huh7.5 cells following viral RNA electroporation of respective
pplementary Figur Supplementary Figure 1. a. Quantification by RT-qPCR of YFV-17D and YFV-17D pol- (+) RNA in the supernatant of cultured Huh7.5 cells following viral RNA electroporation of respective
HD1 (FLU) HD2 (EBV) HD2 (FLU)
 ramer staining + anti-pe beads ramer staining a HD1 (FLU) HD2 (EBV) HD2 (FLU).73.11.56.46.24 1.12 b CD127 + c CD127 + d CD127 - e CD127 - PD1 - PD1 + PD1 + PD1-1 1 1 1 %CD127 + PD1-8 6 4 2 + anti-pe %CD127
ramer staining + anti-pe beads ramer staining a HD1 (FLU) HD2 (EBV) HD2 (FLU).73.11.56.46.24 1.12 b CD127 + c CD127 + d CD127 - e CD127 - PD1 - PD1 + PD1 + PD1-1 1 1 1 %CD127 + PD1-8 6 4 2 + anti-pe %CD127
x Lymphocyte count /µl CD8+ count/µl 800 Calculated
 % Lymphocyte in CBC A. 50 40 30 20 10 Lymphocyte count /µl B. x10 3 2.5 1.5 C. 50 D. 1000 % CD3+CD8+ Cells 40 30 20 Calculated CD8+ count/µl 800 600 400 200 10 0 #61 #63 #64 #65 #68 #71 #72 #75 Figure
% Lymphocyte in CBC A. 50 40 30 20 10 Lymphocyte count /µl B. x10 3 2.5 1.5 C. 50 D. 1000 % CD3+CD8+ Cells 40 30 20 Calculated CD8+ count/µl 800 600 400 200 10 0 #61 #63 #64 #65 #68 #71 #72 #75 Figure
A genome-wide association study identifies vitiligo
 A genome-wide association study identifies vitiligo susceptibility loci at MHC and 6q27 Supplementary Materials Index Supplementary Figure 1 The principal components analysis (PCA) of 2,546 GWAS samples
A genome-wide association study identifies vitiligo susceptibility loci at MHC and 6q27 Supplementary Materials Index Supplementary Figure 1 The principal components analysis (PCA) of 2,546 GWAS samples
Fetal-maternal alignment of regulatory T cells correlates with Interleukin 10 mediated Bcl-
 Fetal-maternal alignment of regulatory T cells correlates with Interleukin 10 mediated Bcl- 2 upregulation in pregnancy B. Santner-Nanan, K. Straubinger, P. Hsu, G. Parnell, B. Tang, B. Xu, A. Makris,
Fetal-maternal alignment of regulatory T cells correlates with Interleukin 10 mediated Bcl- 2 upregulation in pregnancy B. Santner-Nanan, K. Straubinger, P. Hsu, G. Parnell, B. Tang, B. Xu, A. Makris,
Table SІ. List of genes upregulated by IL-4/IL-13-treatment of NHEK
 Supplemental legends Table SІ. List of genes upregulated by IL-4/IL-13-treatment of NHEK Table SП. List of genes in cluster 1 identified using two-dimensional hierarchical clustering analysis Table SШ.
Supplemental legends Table SІ. List of genes upregulated by IL-4/IL-13-treatment of NHEK Table SП. List of genes in cluster 1 identified using two-dimensional hierarchical clustering analysis Table SШ.
Supplementary Table S1: Gene expression targets
 Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion
 Probe Set ID RefSeq Transcript ID Gene Title Gene Symbol 205980_s_at NM_001017526 /// NM_181334 /// NM_181335 Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion ARHGAP8 /// LOC553158 FC ALK shrna
Probe Set ID RefSeq Transcript ID Gene Title Gene Symbol 205980_s_at NM_001017526 /// NM_181334 /// NM_181335 Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion ARHGAP8 /// LOC553158 FC ALK shrna
Supporting Information
 Supporting Information Lin et al. 10.1073/pnas.1408759111 Fig. S1. Safety and potential toxicity evaluation of M1 in immunocompetent mice. Immunocompetent BALB/c mice received two doses of i.v. infusion
Supporting Information Lin et al. 10.1073/pnas.1408759111 Fig. S1. Safety and potential toxicity evaluation of M1 in immunocompetent mice. Immunocompetent BALB/c mice received two doses of i.v. infusion
SDC (Supplemental Digital Content) Figure S1. Percentages of Granzyme B expressing CD8 + T lymphocytes
 Figure S1. Percentages of Granzyme expressing D8 + T lymphocytes 13.5% 1.4%.6% 77% 21% 24% 76% 8 8 Granzyme + (%) 6 4 2 Granzyme + (%) 6 4 2 D8 + Tnaive Tcm Tem Temra D8 + D8 + D28 + D8 + D28 null We analyzed
Figure S1. Percentages of Granzyme expressing D8 + T lymphocytes 13.5% 1.4%.6% 77% 21% 24% 76% 8 8 Granzyme + (%) 6 4 2 Granzyme + (%) 6 4 2 D8 + Tnaive Tcm Tem Temra D8 + D8 + D28 + D8 + D28 null We analyzed
Moyamoya disease susceptibility gene RNF213 links inflammatory and angiogenic
 Supplementary Information for Moyamoya disease susceptibility gene RNF213 links inflammatory and angiogenic signals in endothelial cells Kazuhiro Ohkubo 1,, Yasunari Sakai 1,*,, Hirosuke Inoue 1, Satoshi
Supplementary Information for Moyamoya disease susceptibility gene RNF213 links inflammatory and angiogenic signals in endothelial cells Kazuhiro Ohkubo 1,, Yasunari Sakai 1,*,, Hirosuke Inoue 1, Satoshi
Supplementary Fig. 1: Ex vivo tetramer enrichment with anti-c-myc beads
 Supplementary Fig. 1: Ex vivo tetramer enrichment with anti-c-myc beads Representative example of comparative ex vivo tetramer enrichment performed in three independent experiments with either conventional
Supplementary Fig. 1: Ex vivo tetramer enrichment with anti-c-myc beads Representative example of comparative ex vivo tetramer enrichment performed in three independent experiments with either conventional
The CAPRI-T was one of two (along with the CAPRI-NK, see reference. [29]) basic immunological studies nested within the CAMELIA trial.
![The CAPRI-T was one of two (along with the CAPRI-NK, see reference. [29]) basic immunological studies nested within the CAMELIA trial. The CAPRI-T was one of two (along with the CAPRI-NK, see reference. [29]) basic immunological studies nested within the CAMELIA trial.](/thumbs/88/117921481.jpg) 1 SUPPLEMENTARY INFORMATION Patient description and recruitment The CAPRI-T was one of two (along with the CAPRI-NK, see reference [29]) basic immunological studies nested within the CAMELIA trial. Patients
1 SUPPLEMENTARY INFORMATION Patient description and recruitment The CAPRI-T was one of two (along with the CAPRI-NK, see reference [29]) basic immunological studies nested within the CAMELIA trial. Patients
Supplementary appendix
 Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Kennedy GA, Varelias A, Vuckovic S, et al.
Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Kennedy GA, Varelias A, Vuckovic S, et al.
Supplementary Table 1: List of parameters used in this study. Parameter Unit Parameter Unit Clinical parameters CD4 T cells Microbial translocation
 Supplementary Table 1: List of parameters used in this study. Left table shows parameters available for all subjects and right table shows parameters available for a smaller subset of subjects. Parameters
Supplementary Table 1: List of parameters used in this study. Left table shows parameters available for all subjects and right table shows parameters available for a smaller subset of subjects. Parameters
Nature Neuroscience: doi: /nn.4295
 Supplementary Figure 1 Gene expression profiling of EGFRvIII-expressing human brain tumor stem cells (BTSCs) and mouse astrocytes (a) The protein lysates of human BTSCs were analyzed by immunoblotting
Supplementary Figure 1 Gene expression profiling of EGFRvIII-expressing human brain tumor stem cells (BTSCs) and mouse astrocytes (a) The protein lysates of human BTSCs were analyzed by immunoblotting
Supplementary Figure 1. Immune profiles of untreated and PD-1 blockade resistant EGFR and Kras mouse lung tumors (a) Total lung weight of untreated
 1 Supplementary Figure 1. Immune profiles of untreated and PD-1 blockade resistant EGFR and Kras mouse lung tumors (a) Total lung weight of untreated (U) EGFR TL mice (n=7), Kras mice (n=7), PD-1 blockade
1 Supplementary Figure 1. Immune profiles of untreated and PD-1 blockade resistant EGFR and Kras mouse lung tumors (a) Total lung weight of untreated (U) EGFR TL mice (n=7), Kras mice (n=7), PD-1 blockade
Eosinophils! 40! 30! 20! 10! 0! NS!
 A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
Supplemental Information. Checkpoint Blockade Immunotherapy. Induces Dynamic Changes. in PD-1 CD8 + Tumor-Infiltrating T Cells
 Immunity, Volume 50 Supplemental Information Checkpoint Blockade Immunotherapy Induces Dynamic Changes in PD-1 CD8 + Tumor-Infiltrating T Cells Sema Kurtulus, Asaf Madi, Giulia Escobar, Max Klapholz, Jackson
Immunity, Volume 50 Supplemental Information Checkpoint Blockade Immunotherapy Induces Dynamic Changes in PD-1 CD8 + Tumor-Infiltrating T Cells Sema Kurtulus, Asaf Madi, Giulia Escobar, Max Klapholz, Jackson
Nature Immunology: doi: /ni.3494
 Supplementary Figure 1 High-throughput screen for novel inducer of T FH cell differentiation. (a) Schematic of primary screen. Purified human naïve CD4 T cells were stimulated by anti-cd3/cd28 beads on
Supplementary Figure 1 High-throughput screen for novel inducer of T FH cell differentiation. (a) Schematic of primary screen. Purified human naïve CD4 T cells were stimulated by anti-cd3/cd28 beads on
Supplementary Fig. 1 p38 MAPK negatively regulates DC differentiation. (a) Western blot analysis of p38 isoform expression in BM cells, immature DCs
 Supplementary Fig. 1 p38 MAPK negatively regulates DC differentiation. (a) Western blot analysis of p38 isoform expression in BM cells, immature DCs (idcs) and mature DCs (mdcs). A myeloma cell line expressing
Supplementary Fig. 1 p38 MAPK negatively regulates DC differentiation. (a) Western blot analysis of p38 isoform expression in BM cells, immature DCs (idcs) and mature DCs (mdcs). A myeloma cell line expressing
Supplement Results, Figures and Tables
 Supplement Results, Figures and Tables Results PET imaging The SUV was significantly higher in tumours of the parental HSC-3 cell line than tumours of the EV and the MMP-8 overexpressing cell lines (Figure
Supplement Results, Figures and Tables Results PET imaging The SUV was significantly higher in tumours of the parental HSC-3 cell line than tumours of the EV and the MMP-8 overexpressing cell lines (Figure
The epigenetic landscape of T cell subsets in SLE identifies known and potential novel drivers of the autoimmune response
 Abstract # 319030 Poster # F.9 The epigenetic landscape of T cell subsets in SLE identifies known and potential novel drivers of the autoimmune response Jozsef Karman, Brian Johnston, Sofija Miljovska,
Abstract # 319030 Poster # F.9 The epigenetic landscape of T cell subsets in SLE identifies known and potential novel drivers of the autoimmune response Jozsef Karman, Brian Johnston, Sofija Miljovska,
** *** population. (A) Representative FACS plots showing the gating strategy for T N
 Sca-1 (MFI) CD122 (MFI) CD44 A CD8 + gated: resting spleen CD8 + gated: T Mem -derived CD8 + gated: T N -derived, expanded alone B CD62L CD44 low CD62L +, resting CD44 high, T Mem -derived CD44 low CD62L
Sca-1 (MFI) CD122 (MFI) CD44 A CD8 + gated: resting spleen CD8 + gated: T Mem -derived CD8 + gated: T N -derived, expanded alone B CD62L CD44 low CD62L +, resting CD44 high, T Mem -derived CD44 low CD62L
Transcription factor Foxp3 and its protein partners form a complex regulatory network
 Supplementary figures Resource Paper Transcription factor Foxp3 and its protein partners form a complex regulatory network Dipayan Rudra 1, Paul deroos 1, Ashutosh Chaudhry 1, Rachel Niec 1, Aaron Arvey
Supplementary figures Resource Paper Transcription factor Foxp3 and its protein partners form a complex regulatory network Dipayan Rudra 1, Paul deroos 1, Ashutosh Chaudhry 1, Rachel Niec 1, Aaron Arvey
Supplementary Information
 1 Supplementary Information Human TSC2 Null Fibroblast-Like Cells Induce Hair Follicle Neogenesis and Hamartoma Morphogenesis Shaowei Li 1, Rajesh L. Thangapazham 1, Ji-an Wang 1, Sangeetha Rajesh 1, Tzu-Cheg
1 Supplementary Information Human TSC2 Null Fibroblast-Like Cells Induce Hair Follicle Neogenesis and Hamartoma Morphogenesis Shaowei Li 1, Rajesh L. Thangapazham 1, Ji-an Wang 1, Sangeetha Rajesh 1, Tzu-Cheg
Supplementary Figure 1
 Supplementary Figure 1 Identification of IFN-γ-producing CD8 + and CD4 + T cells with naive phenotype by alternative gating and sample-processing strategies. a. Contour 5% probability plots show definition
Supplementary Figure 1 Identification of IFN-γ-producing CD8 + and CD4 + T cells with naive phenotype by alternative gating and sample-processing strategies. a. Contour 5% probability plots show definition
Fluorochrome Panel 1 Panel 2 Panel 3 Panel 4 Panel 5 CTLA-4 CTLA-4 CD15 CD3 FITC. Bio) PD-1 (MIH4, BD) ICOS (C398.4A, Biolegend) PD-L1 (MIH1, BD)
 Additional file : Table S. Antibodies used for panel stain to identify peripheral immune cell subsets. Panel : PD- signaling; Panel : CD + T cells, CD + T cells, B cells; Panel : Tregs; Panel :, -T, cdc,
Additional file : Table S. Antibodies used for panel stain to identify peripheral immune cell subsets. Panel : PD- signaling; Panel : CD + T cells, CD + T cells, B cells; Panel : Tregs; Panel :, -T, cdc,
BD Rhapsody Immune Response Panel Hs
 Doc ID: 55665 Rev. 2.0 PRODUCT INSERT BD Rhapsody Immune Response Panel Hs BD Biosciences Kit Part Number: 633750 Purpose The BD Rhapsody Immune Response Panel Hs is a set of ready-to-use primer pools
Doc ID: 55665 Rev. 2.0 PRODUCT INSERT BD Rhapsody Immune Response Panel Hs BD Biosciences Kit Part Number: 633750 Purpose The BD Rhapsody Immune Response Panel Hs is a set of ready-to-use primer pools
Nature Immunology: doi: /ni Supplementary Figure 1. Transcriptional program of the TE and MP CD8 + T cell subsets.
 Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
 Table S1. Metadata for transcriptome interaction network and pathway analysis of 5448 intracelluarly infected TEpi cells in comparison to mock TEpi cells. Gene symbol Full name Log2FC gene expression in
Table S1. Metadata for transcriptome interaction network and pathway analysis of 5448 intracelluarly infected TEpi cells in comparison to mock TEpi cells. Gene symbol Full name Log2FC gene expression in
Supplementary Files to Töröcsik et al. Factor XIII-A is involved in the regulation of
 Supplementary Files to Töröcsik et al. Factor XIII-A is involved in the regulation of gene expression in alternatively activated human macrophages (Thromb Haemost 2010; 104.3) A Protein synthesis Cell
Supplementary Files to Töröcsik et al. Factor XIII-A is involved in the regulation of gene expression in alternatively activated human macrophages (Thromb Haemost 2010; 104.3) A Protein synthesis Cell
WT-TP53 DLBCL WT-TP53 GCB-DLBCL
 SUPPLEMENTAL TABLES AND FIGURES Supplemental Table S1. Summary of gene expression profiling analysis results listing counts of significant differentially expressed transcripts between defined two groups
SUPPLEMENTAL TABLES AND FIGURES Supplemental Table S1. Summary of gene expression profiling analysis results listing counts of significant differentially expressed transcripts between defined two groups
SUPPLEMENTARY MATERIAL
 Supplementary Material The Open AIDS Journal, 2012, Volume 6 i SUPPLEMENTARY MATERIAL Immune Reconstitution During the First Year of Antiretroviral Therapy of HIV-1-Infected Adults in Rural Burkina Faso
Supplementary Material The Open AIDS Journal, 2012, Volume 6 i SUPPLEMENTARY MATERIAL Immune Reconstitution During the First Year of Antiretroviral Therapy of HIV-1-Infected Adults in Rural Burkina Faso
Statistical Issues in Translational Cancer Research
 Statistical Issues in Translational Cancer Research Martin Bøgsted Department of Haematology Aalborg University Hospital and Department of Clinical Medicine Aalborg University The only useful function
Statistical Issues in Translational Cancer Research Martin Bøgsted Department of Haematology Aalborg University Hospital and Department of Clinical Medicine Aalborg University The only useful function
The broadest portfolio for your T cell research
 quick guide The broadest portfolio for your T cell research Sample preparation Cell separation Cell culture Flow cytometry Mouse T cells Overview CD4 + T cell subsets Polarization reagents Naive CD4 +
quick guide The broadest portfolio for your T cell research Sample preparation Cell separation Cell culture Flow cytometry Mouse T cells Overview CD4 + T cell subsets Polarization reagents Naive CD4 +
W/T Itgam -/- F4/80 CD115. F4/80 hi CD115 + F4/80 + CD115 +
 F4/8 % in the peritoneal lavage 6 4 2 p=.15 n.s p=.76 CD115 F4/8 hi CD115 + F4/8 + CD115 + F4/8 hi CD115 + F4/8 + CD115 + MHCII MHCII Supplementary Figure S1. CD11b deficiency affects the cellular responses
F4/8 % in the peritoneal lavage 6 4 2 p=.15 n.s p=.76 CD115 F4/8 hi CD115 + F4/8 + CD115 + F4/8 hi CD115 + F4/8 + CD115 + MHCII MHCII Supplementary Figure S1. CD11b deficiency affects the cellular responses
ankylosing spondylitis Department of Clinical Immunology, Xijing Hospital, The Fourth Military
 Functional defects in CD4 + CD25 high FoxP3 + regulatory cells in ankylosing spondylitis Huifang Guo 1, 2, 3, Ming Zheng 1, 2, 3, Kui Zhang 1, 3, Fengfan Yang 1, 3, Xin Zhang 1, 3, Qing Han 1, 3, Zhi-Nan
Functional defects in CD4 + CD25 high FoxP3 + regulatory cells in ankylosing spondylitis Huifang Guo 1, 2, 3, Ming Zheng 1, 2, 3, Kui Zhang 1, 3, Fengfan Yang 1, 3, Xin Zhang 1, 3, Qing Han 1, 3, Zhi-Nan
Appendix Figure S1 A B C D E F G H
 ppendix Figure S1 C D E F G H ppendix Figure S1. RT and chemotherapy alter PD-L1 expression in PDC cells. Flow cytometric analysis of PD-L1 expression in () KPC and () Pan02 cells following treatment with
ppendix Figure S1 C D E F G H ppendix Figure S1. RT and chemotherapy alter PD-L1 expression in PDC cells. Flow cytometric analysis of PD-L1 expression in () KPC and () Pan02 cells following treatment with
Bronchoalveolar lavage fluid analysis showed increased eosinophil in 30% of subjects with IrIa BAL
 Bronchoalveolar lavage fluid analysis showed increased eosinophil in 30% of subjects with IrIa BAL UBC Chan-Yeung Centre for Occupational and Environmental Respiratory Disease Understanding exposure effects
Bronchoalveolar lavage fluid analysis showed increased eosinophil in 30% of subjects with IrIa BAL UBC Chan-Yeung Centre for Occupational and Environmental Respiratory Disease Understanding exposure effects
CPM (x 10-3 ) Tregs +Teffs. Tregs alone ICOS CLTA-4
 A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
Analysis of gene expression in blood before diagnosis of ovarian cancer
 Analysis of gene expression in blood before diagnosis of ovarian cancer Different statistical methods Note no. Authors SAMBA/10/16 Marit Holden and Lars Holden Date March 2016 Norsk Regnesentral Norsk
Analysis of gene expression in blood before diagnosis of ovarian cancer Different statistical methods Note no. Authors SAMBA/10/16 Marit Holden and Lars Holden Date March 2016 Norsk Regnesentral Norsk
Supplemental Figures/Tables. Methylation forward GTCGGGGCGTATTTAGTTC. Methylation reverse AACGACGTAAACGAAAATATCG
 Supplemental Figures/Tables Supplementary Tables Table S1. Primer sequences MSP Primers SPARC2 Unmethylation forward TTTTTTAGATTGTTTGGAGAGTG Unmethylation reverse AACTAACAACATAAACAAAAATATC Methylation
Supplemental Figures/Tables Supplementary Tables Table S1. Primer sequences MSP Primers SPARC2 Unmethylation forward TTTTTTAGATTGTTTGGAGAGTG Unmethylation reverse AACTAACAACATAAACAAAAATATC Methylation
Nature Immunology: doi: /ni Supplementary Figure 1 33,312. Aire rep 1. Aire rep 2 # 44,325 # 44,055. Aire rep 1. Aire rep 2.
 a 33,312 b rep 1 rep 1 # 44,325 rep 2 # 44,055 [0-84] rep 2 [0-84] 1810043G02Rik Pfkl Dnmt3l Icosl rep 1 [0-165] rep 2 [0-165] Rps14 Cd74 Mir5107 Tcof1 rep 1 [0-69] rep 2 [0-68] Id3 E2f2 Asap3 rep 1 [0-141]
a 33,312 b rep 1 rep 1 # 44,325 rep 2 # 44,055 [0-84] rep 2 [0-84] 1810043G02Rik Pfkl Dnmt3l Icosl rep 1 [0-165] rep 2 [0-165] Rps14 Cd74 Mir5107 Tcof1 rep 1 [0-69] rep 2 [0-68] Id3 E2f2 Asap3 rep 1 [0-141]
Supplementary. presence of the. (c) mrna expression. Error. in naive or
 Figure 1. (a) Naive CD4 + T cells were activated in the presence of the indicated cytokines for 3 days. Enpp2 mrna expression was measured by qrt-pcrhr, infected with (b, c) Naive CD4 + T cells were activated
Figure 1. (a) Naive CD4 + T cells were activated in the presence of the indicated cytokines for 3 days. Enpp2 mrna expression was measured by qrt-pcrhr, infected with (b, c) Naive CD4 + T cells were activated
Multi-class cancer outlier differential gene expression detection Supplementary Information
 Multi-class cancer outlier differential gene expression detection Supplementary Information Fang Liu, Baolin Wu 1 Breast cancer microarray data analysis The breast cancer microarray data (West et al.,
Multi-class cancer outlier differential gene expression detection Supplementary Information Fang Liu, Baolin Wu 1 Breast cancer microarray data analysis The breast cancer microarray data (West et al.,
Supplementary Figure 1 a
 Supplementary Figure 1 a b d c Supplementary Figure 1. Poor sensitivity for mutation detection using established single-cell RNA-sequencing methods. (a) Representative bioanalyzer trace showing expected
Supplementary Figure 1 a b d c Supplementary Figure 1. Poor sensitivity for mutation detection using established single-cell RNA-sequencing methods. (a) Representative bioanalyzer trace showing expected
Supplementary Fig.S1 Microarray quality control chart. Supplementary Fig.S2 Display of the overview of pathway network from Reactome online.
 Supplementary Fig.S1 Microarray quality control chart. Data after normalization. X-axis: sample; Y-axis: log2 expression value. Box plot showed the distribution of value data for the samples we selected
Supplementary Fig.S1 Microarray quality control chart. Data after normalization. X-axis: sample; Y-axis: log2 expression value. Box plot showed the distribution of value data for the samples we selected
SCIENCE CHINA Life Sciences
 SCIENCE CHINA Life Sciences RESEARCH PAPER April 2013 Vol.56 No.4: 1 7 doi: 10.1007/s11427-013-4460-x Table S1 Human paired-end RNA-Seq data sets from the Sequence Read Archive (SRA) database Experiment
SCIENCE CHINA Life Sciences RESEARCH PAPER April 2013 Vol.56 No.4: 1 7 doi: 10.1007/s11427-013-4460-x Table S1 Human paired-end RNA-Seq data sets from the Sequence Read Archive (SRA) database Experiment
Table S1. Differentially expressed transcripts between hepatic inkt cells. sorted form WT and plck-hcd1d tg mice. Differentially expressed
 Supplementary Information Table S1. Differentially expressed transcripts between hepatic inkt cells sorted form and plck-hcd1d tg mice. Differentially expressed transcripts identified by gene expression
Supplementary Information Table S1. Differentially expressed transcripts between hepatic inkt cells sorted form and plck-hcd1d tg mice. Differentially expressed transcripts identified by gene expression
supplemental Figure 1
 supplemental Figure 1 A T cell T1 anti-ny-eso-117-16/hla-a*:1 CDζ CH/CH scfv B T cell T1 anti-ny-eso-117-16/hla-a*:1 CDζ CH/CH scfv C T cell BW1/6 anti-cea CDζ CH/CH scfv supplemental Figure 1.79.9.87
supplemental Figure 1 A T cell T1 anti-ny-eso-117-16/hla-a*:1 CDζ CH/CH scfv B T cell T1 anti-ny-eso-117-16/hla-a*:1 CDζ CH/CH scfv C T cell BW1/6 anti-cea CDζ CH/CH scfv supplemental Figure 1.79.9.87
Supplementary information. The proton-sensing G protein-coupled receptor T-cell death-associated gene 8
 1 Supplementary information 2 3 The proton-sensing G protein-coupled receptor T-cell death-associated gene 8 4 (TDAG8) shows cardioprotective effects against myocardial infarction 5 Akiomi Nagasaka 1+,
1 Supplementary information 2 3 The proton-sensing G protein-coupled receptor T-cell death-associated gene 8 4 (TDAG8) shows cardioprotective effects against myocardial infarction 5 Akiomi Nagasaka 1+,
Table 1 Demographic data of PD-patients. Data are represented as counts or median and full range.
 Table 1 Demographic data of PD-patients. Data are represented as counts or median and full range. Parameter Number Patients 5 Diabetes mellitus 0 Previous NTX 0 Steroids 0 Patients with residual renal
Table 1 Demographic data of PD-patients. Data are represented as counts or median and full range. Parameter Number Patients 5 Diabetes mellitus 0 Previous NTX 0 Steroids 0 Patients with residual renal
Supplementary Figure 1
 Count Count Supplementary Figure 1 Coverage per amplicon for error-corrected sequencing experiments. Errorcorrected consensus sequence (ECCS) coverage was calculated for each of the 568 amplicons in the
Count Count Supplementary Figure 1 Coverage per amplicon for error-corrected sequencing experiments. Errorcorrected consensus sequence (ECCS) coverage was calculated for each of the 568 amplicons in the
Supplementary Information
 Scientific Reports Supplementary Information Upregulated expression of FGF13/FHF2 mediates resistance to platinum drugs in cervical cancer cells Tomoko Okada, Kazuhiro Murata, Ryoma Hirose, Chie Matsuda,
Scientific Reports Supplementary Information Upregulated expression of FGF13/FHF2 mediates resistance to platinum drugs in cervical cancer cells Tomoko Okada, Kazuhiro Murata, Ryoma Hirose, Chie Matsuda,
Schrader et al. Cell Communication and Signaling 2012, 10:43
 Schrader et al. Cell Communication and Signaling 2012, 10:43 RESEARCH Open Access Global gene expression changes of in vitro stimulated human transformed germinal centre B cells as surrogate for oncogenic
Schrader et al. Cell Communication and Signaling 2012, 10:43 RESEARCH Open Access Global gene expression changes of in vitro stimulated human transformed germinal centre B cells as surrogate for oncogenic
Supplementary Figure 1. BMS enhances human T cell activation in vitro in a
 Supplementary Figure 1. BMS98662 enhances human T cell activation in vitro in a concentration-dependent manner. Jurkat T cells were activated with anti-cd3 and anti-cd28 antibody in the presence of titrated
Supplementary Figure 1. BMS98662 enhances human T cell activation in vitro in a concentration-dependent manner. Jurkat T cells were activated with anti-cd3 and anti-cd28 antibody in the presence of titrated
Supplementary Table 1. mirna modulated in D HF and ND HF patients. Table shows in a linear scale the same values of Fig. 2.
 Supplementary Table 1. mirna modulated in D HF and ND HF patients. Table shows in a linear scale the same values of Fig. 2. mir D HF VS CTR ND HF VS CTR D HF VS ND HF FOLD CHANGE p FOLD CHANGE p FOLD CHANGE
Supplementary Table 1. mirna modulated in D HF and ND HF patients. Table shows in a linear scale the same values of Fig. 2. mir D HF VS CTR ND HF VS CTR D HF VS ND HF FOLD CHANGE p FOLD CHANGE p FOLD CHANGE
Blocking antibodies and peptides. Rat anti-mouse PD-1 (29F.1A12, rat IgG2a, k), PD-
 Supplementary Methods Blocking antibodies and peptides. Rat anti-mouse PD-1 (29F.1A12, rat IgG2a, k), PD- L1 (10F.9G2, rat IgG2b, k), and PD-L2 (3.2, mouse IgG1) have been described (24). Anti-CTLA-4 (clone
Supplementary Methods Blocking antibodies and peptides. Rat anti-mouse PD-1 (29F.1A12, rat IgG2a, k), PD- L1 (10F.9G2, rat IgG2b, k), and PD-L2 (3.2, mouse IgG1) have been described (24). Anti-CTLA-4 (clone
Supplementary Figure S1. PTPN2 levels are not altered in proliferating CD8+ T cells. Lymph node (LN) CD8+ T cells from C57BL/6 mice were stained with
 Supplementary Figure S1. PTPN2 levels are not altered in proliferating CD8+ T cells. Lymph node (LN) CD8+ T cells from C57BL/6 mice were stained with CFSE and stimulated with plate-bound α-cd3ε (10µg/ml)
Supplementary Figure S1. PTPN2 levels are not altered in proliferating CD8+ T cells. Lymph node (LN) CD8+ T cells from C57BL/6 mice were stained with CFSE and stimulated with plate-bound α-cd3ε (10µg/ml)
Supplemental Information. lncrna Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncrna that Interacts with MYC
 Cancer Cell, Volume 33 Supplemental Information lncrna Epigenetic Landscape Analysis Identifies as an Oncogenic lncrna that Interacts with and Promotes Cell-Cycle Progression in Cancer Zehua Wang, Bo Yang,
Cancer Cell, Volume 33 Supplemental Information lncrna Epigenetic Landscape Analysis Identifies as an Oncogenic lncrna that Interacts with and Promotes Cell-Cycle Progression in Cancer Zehua Wang, Bo Yang,
IgG3 regulates tissue-like memory B cells in HIV-infected individuals
 SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41590-018-0180-5 In the format provided by the authors and unedited. IgG3 regulates tissue-like memory B cells in HIV-infected individuals Lela
SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41590-018-0180-5 In the format provided by the authors and unedited. IgG3 regulates tissue-like memory B cells in HIV-infected individuals Lela
CD80 and PD-L2 define functionally distinct memory B cell subsets that are. Griselda V Zuccarino-Catania, Saheli Sadanand, Florian J Weisel, Mary M
 Supplementary Figures CD8 and PD-L define functionally distinct memory B cell subsets that are independent of antibody isotype Running title: Memory B Cell Subset Function Griselda V Zuccarino-Catania,
Supplementary Figures CD8 and PD-L define functionally distinct memory B cell subsets that are independent of antibody isotype Running title: Memory B Cell Subset Function Griselda V Zuccarino-Catania,
Supplementary Material. Table S1. Summary of mapping results
 Supplementary Material Table S1. Summary of mapping results Sample Total reads Mapped (%) HNE0-1 99687516 80786083 (81.0%) HNE0-2 94318720 77047792 (81.7%) HNE0-3 104033900 84348102 (81.1%) HNE15-1 94426598
Supplementary Material Table S1. Summary of mapping results Sample Total reads Mapped (%) HNE0-1 99687516 80786083 (81.0%) HNE0-2 94318720 77047792 (81.7%) HNE0-3 104033900 84348102 (81.1%) HNE15-1 94426598
Äijö et al. BMC Genomics 2012, 13:572
 Äijö et al. BMC Genomics 2012, 13:572 RESEARCH ARTICLE Open Access An integrative computational systems biology approach identifies differentially regulated dynamic transcriptome signatures which drive
Äijö et al. BMC Genomics 2012, 13:572 RESEARCH ARTICLE Open Access An integrative computational systems biology approach identifies differentially regulated dynamic transcriptome signatures which drive
Epigenetic Regulation by Chromatin Activation Mark H3K4me3 in Primate Progenitor
 Epigenetic Regulation by Chromatin Activation Mark H3K4me3 in Primate Progenitor Cells within Adult Neurogenic Niche Richard S. Sandstrom 1, Michael R. Foret 2, Douglas A. Grow 2,3, Eric Haugen 1, Christopher
Epigenetic Regulation by Chromatin Activation Mark H3K4me3 in Primate Progenitor Cells within Adult Neurogenic Niche Richard S. Sandstrom 1, Michael R. Foret 2, Douglas A. Grow 2,3, Eric Haugen 1, Christopher
Supplementary Online Content
 Supplementary Online Content Lyall DM, Celis-Morales C, Ward J, et al. Association of body mass index with cardiometabolic disease in the UK Biobank: a mendelian randomization study. JAMA Cardiol. Published
Supplementary Online Content Lyall DM, Celis-Morales C, Ward J, et al. Association of body mass index with cardiometabolic disease in the UK Biobank: a mendelian randomization study. JAMA Cardiol. Published
Supplementary Figures and Tables. An inflammatory gene signature distinguishes neurofibroma Schwann cells and
 Supplementary Figures and Tables An inflammatory gene signature distinguishes neurofibroma Schwann cells and macrophages from cells in the normal peripheral nervous system Kwangmin Choi 1, Kakajan Komurov
Supplementary Figures and Tables An inflammatory gene signature distinguishes neurofibroma Schwann cells and macrophages from cells in the normal peripheral nervous system Kwangmin Choi 1, Kakajan Komurov
p5+mir30: AATGATACGGCGACCACCGACTAAAGTAGCCCCTTGAATTC
 SUPPORTING INFORMATION METHODS Massively parallel sequencing and shrna screen data analysis pipeline Genomic DNA extraction and purification from surviving MCF7 cells in the genome wide screen was carried
SUPPORTING INFORMATION METHODS Massively parallel sequencing and shrna screen data analysis pipeline Genomic DNA extraction and purification from surviving MCF7 cells in the genome wide screen was carried
Supplementary appendix
 Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Ellingford JM, Sergouniotis PI, Lennon R, et
Supplementary appendix This appendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Ellingford JM, Sergouniotis PI, Lennon R, et
F-CD8 +/+ 313F-CD8 -/ ShcA-CD8 +/+ ShcA-CD8 -/- * * 313F-IFN +/+ 313F-IFN -/- 2F-IFN -/- % Tumor Free.
 a 864 5372 6738 ShcA-CD8 +/+ ShcA-CD8 -/- 2F-CD8 +/+ 2F-CD8 -/- b 4788 5376 623 313F-CD8 +/+ 313F-CD8 -/- 1 1 1 75 75 75 5 5 5 25 25 25 2 4 6 Time after Inoculation (d) 2 4 6 Time after Inoculation (d)
a 864 5372 6738 ShcA-CD8 +/+ ShcA-CD8 -/- 2F-CD8 +/+ 2F-CD8 -/- b 4788 5376 623 313F-CD8 +/+ 313F-CD8 -/- 1 1 1 75 75 75 5 5 5 25 25 25 2 4 6 Time after Inoculation (d) 2 4 6 Time after Inoculation (d)
Supplementary Figure 1. Antibiotic partially rescues mice from sepsis. (ab) BALB/c mice under CLP were treated with antibiotic or PBS.
 1 Supplementary Figure 1. Antibiotic partially rescues mice from sepsis. (ab) BALB/c mice under CLP were treated with antibiotic or PBS. (a) Survival curves. WT Sham (n=5), WT CLP or WT CLP antibiotic
1 Supplementary Figure 1. Antibiotic partially rescues mice from sepsis. (ab) BALB/c mice under CLP were treated with antibiotic or PBS. (a) Survival curves. WT Sham (n=5), WT CLP or WT CLP antibiotic
Kerdiles et al - Figure S1
 Kerdiles et al - Figure S1 a b Homo sapiens T B ce ce l ls c l M ls ac r PM oph N ag es Mus musculus Foxo1 PLCγ Supplementary Figure 1 Foxo1 expression pattern is conserved between mouse and human. (a)
Kerdiles et al - Figure S1 a b Homo sapiens T B ce ce l ls c l M ls ac r PM oph N ag es Mus musculus Foxo1 PLCγ Supplementary Figure 1 Foxo1 expression pattern is conserved between mouse and human. (a)
Neonatal and adult recent thymic emigrants produce IL-8 and express complement receptors CR1 and CR2
 Neonatal and adult recent thymic emigrants produce IL-8 and express complement receptors CR1 and CR2 Marcin L. Pekalski,, John A. Todd, Linda S. Wicker JCI Insight. 2017;2(16):e93739.. Research Article
Neonatal and adult recent thymic emigrants produce IL-8 and express complement receptors CR1 and CR2 Marcin L. Pekalski,, John A. Todd, Linda S. Wicker JCI Insight. 2017;2(16):e93739.. Research Article
Supplementary Table 1. Information on the 174 single nucleotide variants identified by whole-exome sequencing
 Supplementary Table 1. Information on the 174 single nucleotide s identified by whole-exome sequencing Chr Position Type AF exomes Database 1 10163148 A/G UBE4B missense 0.000326 1534 2 98 1 4.44 Damaging
Supplementary Table 1. Information on the 174 single nucleotide s identified by whole-exome sequencing Chr Position Type AF exomes Database 1 10163148 A/G UBE4B missense 0.000326 1534 2 98 1 4.44 Damaging
From the Diagnostic Immunology Laboratories
 Immunology Laboratories ISSUE 9 SPRING 2012 PAGE 1 Spring Meetings Please stop by and say hello in May at this year s American Society of Pediatric Hematology/Oncology (ASPHO) Annual Meeting in New Orleans
Immunology Laboratories ISSUE 9 SPRING 2012 PAGE 1 Spring Meetings Please stop by and say hello in May at this year s American Society of Pediatric Hematology/Oncology (ASPHO) Annual Meeting in New Orleans
Proteome Analysis of Human Perilymph using an Intraoperative Sampling Method
 Supporting Information Article Title Proteome Analysis of Human Perilymph using an Intraoperative Sampling Method Authors Heike A. Schmitt 1,3, Andreas Pich 2, Anke Schröder 2, Verena Scheper 1,3, Giorgio
Supporting Information Article Title Proteome Analysis of Human Perilymph using an Intraoperative Sampling Method Authors Heike A. Schmitt 1,3, Andreas Pich 2, Anke Schröder 2, Verena Scheper 1,3, Giorgio
g. RA Bardot et al., 2016 Supplementary Figure 1 RV LV E15.5 E6.5 E18.5 E7.5 VE RV LV E8.5 RV LV E10.5 Brightfield Foxa2Cre:YFP Brightfield
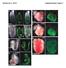 Bardot et al., 216 Supplementary Figure 1 a. VE e. RA LA LA RV LV E15.5 E6.5 b. E7.5 VE f. RA E18.5 LA c. E8.5 ML N CM g. RA RV LV LA P7 PHT RV LV d. E1.5 Brightfield Foxa2Cre:YFP H Brightfield Foxa2Cre:YFP
Bardot et al., 216 Supplementary Figure 1 a. VE e. RA LA LA RV LV E15.5 E6.5 b. E7.5 VE f. RA E18.5 LA c. E8.5 ML N CM g. RA RV LV LA P7 PHT RV LV d. E1.5 Brightfield Foxa2Cre:YFP H Brightfield Foxa2Cre:YFP
15. Supplementary Figure 9. Predicted gene module expression changes at 24hpi during HIV
 Supplementary Information Table of content 1. Supplementary Table 1. Summary of RNAseq data and mapping statistics 2. Supplementary Table 2. Biological functions enriched in 12 hpi DE genes, derived from
Supplementary Information Table of content 1. Supplementary Table 1. Summary of RNAseq data and mapping statistics 2. Supplementary Table 2. Biological functions enriched in 12 hpi DE genes, derived from
Optimized between-group classification: a new jackknife-based gene selection procedure for genome-wide expression data Supplementary information
 Optimized between-group classification: a new jackknife-based gene selection procedure for genome-wide expression data Supplementary information Florent Baty 1 florent.baty@unibas.ch Michel P Bihl 1 michel.bihl@unibas.ch
Optimized between-group classification: a new jackknife-based gene selection procedure for genome-wide expression data Supplementary information Florent Baty 1 florent.baty@unibas.ch Michel P Bihl 1 michel.bihl@unibas.ch
Use of Molecular Assays to Assess Cellular Therapies
 Use of Molecular Assays to Assess Cellular Therapies D. Stroncek, P. Jin, M. Sabatino, J. Ren, L. Castiello, E. Wang, and F.M. Marincola. Cellular Therapies Section Clinical Center, NIH Methods for the
Use of Molecular Assays to Assess Cellular Therapies D. Stroncek, P. Jin, M. Sabatino, J. Ren, L. Castiello, E. Wang, and F.M. Marincola. Cellular Therapies Section Clinical Center, NIH Methods for the
Supplementary information: Forstner, Hofmann et al.
 Supplementary information: Forstner, Hofmann et al., Genome-wide analysis implicates micrornas and their target genes in the development of bipolar disorder Table of contents Supplementary Figure 1: Regional
Supplementary information: Forstner, Hofmann et al., Genome-wide analysis implicates micrornas and their target genes in the development of bipolar disorder Table of contents Supplementary Figure 1: Regional
Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured
 Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured under Th0, Th1, Th2, Th17, and Treg conditions. mrna
Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured under Th0, Th1, Th2, Th17, and Treg conditions. mrna
Allele specific FKBP5 DNA demethylation: a molecular mediator of gene childhood
 Allele specific FKBP5 DNA demethylation: a molecular mediator of gene childhood trauma interactions Torsten Klengel 1, Divya Mehta 1, Christoph Anacker 2, Monika Rex Haffner 1, Jens C. Pruessner 3, Carmine
Allele specific FKBP5 DNA demethylation: a molecular mediator of gene childhood trauma interactions Torsten Klengel 1, Divya Mehta 1, Christoph Anacker 2, Monika Rex Haffner 1, Jens C. Pruessner 3, Carmine
Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was
 Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was sorted by FACS. Surface markers for sorting were CD11c +
Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was sorted by FACS. Surface markers for sorting were CD11c +
Supplementary Information
 Supplementary Information Supplementary Figure 1! a! b! Nfatc1!! Nfatc1"! P1! P2! pa1! pa2! ex1! ex2! exons 3-9! ex1! ex11!!" #" Nfatc1A!!" Nfatc1B! #"!" Nfatc1C! #" DN1! DN2! DN1!!A! #A!!B! #B!!C! #C!!A!
Supplementary Information Supplementary Figure 1! a! b! Nfatc1!! Nfatc1"! P1! P2! pa1! pa2! ex1! ex2! exons 3-9! ex1! ex11!!" #" Nfatc1A!!" Nfatc1B! #"!" Nfatc1C! #" DN1! DN2! DN1!!A! #A!!B! #B!!C! #C!!A!
Integrative Radiation Biology
 Dr. Kristian Unger Integrative Biology Group Research Unit of Radiation Cytogenetics Department of Radiation Sciences Helmholtz-Zentrum München 1 Key Questions Molecular mechanisms of radiation-induced
Dr. Kristian Unger Integrative Biology Group Research Unit of Radiation Cytogenetics Department of Radiation Sciences Helmholtz-Zentrum München 1 Key Questions Molecular mechanisms of radiation-induced
