Report. Loss of Individual MicroRNAs Causes Mutant Phenotypes in Sensitized Genetic Backgrounds in C. elegans
|
|
|
- Jeffry Little
- 6 years ago
- Views:
Transcription
1 Current Biology 20, , July 27, 2010 ª2010 Elsevier Ltd All rights reserved DOI /j.cub Loss of Individual MicroRNAs Causes Mutant Phenotypes in Sensitized Genetic Backgrounds in C. elegans Report John L. Brenner, 1 Kristen L. Jasiewicz, 1,2 Alisha F. Fahley, 1,3 Benedict J. Kemp, 1 and Allison L. Abbott 1, * 1 Department of Biological Sciences, Marquette University, Milwaukee, WI 53201, USA Summary MicroRNAs (mirnas) are small, noncoding RNAs that regulate the translation and/or stability of their mrna targets. Previous work showed that for most mirna genes of C. elegans, single-gene knockouts did not result in detectable mutant phenotypes [1]. This may be due, in part, to functional redundancy between mirnas. However, in most cases, worms carrying deletions of all members of a mirna family do not display strong mutant phenotypes [2]. They may function together with unrelated mirnas or with nonmirna genes in regulatory networks, possibly to ensure the robustness of developmental mechanisms. To test this, we examined worms lacking individual mirnas in genetically sensitized backgrounds. These include genetic backgrounds with reduced processing and activity of all mirnas or with reduced activity of a wide array of regulatory pathways [3]. With these two approaches, we identified mutant phenotypes for 25 out of 31 mirnas included in this analysis. Our findings describe biological roles for individual mirnas and suggest that the use of sensitized genetic backgrounds provides an efficient approach for mirna functional analysis. Results Loss of Individual mirnas Can Enhance or Suppress alg-1 Developmental Defects Genetic analysis demonstrates that development of worms, flies, fish, and mice requires micrornas (mirnas) [4 9]. Although it is clear that normal development of the worm requires mirna biogenesis, functions have been described for only a few individual mirnas. In C. elegans, the majority of individual mirnas are not required for viability or for development; most loss-of-function mirna mutants display no obvious developmental abnormalities [1]. To test the hypothesis that the absence of phenotypes in mirna mutant worms is due to functional redundancy with other mirnas, we determined whether loss of specific mirnas in a sensitized genetic background resulted in observable mutant phenotypes. For a sensitized background, we used a loss-of-function allele of alg-1 (argonaute like 1), which is one of two Argonaute-encoding genes that function in the mirna pathway in worms. Compared to wild-type worms, alg-1 mutants have lower levels of mature mirnas [6, 10]. Whereas worms that lack alg-1 and alg-2 activities die during embryogenesis [6], alg-1 *Correspondence: allison.abbott@marquette.edu 2 Present address: Des Moines University, Des Moines, IA 50312, USA 3 Present address: School of Medicine and Public Health, University of Wisconsin- Madison, Madison, WI 53706, USA single-mutant worms are viable, displaying developmental timing defects, molting defects, and early adult-stage lethality [11 14]. A subset of 26 mirna deletion alleles covering 31 mirnaencoding genes were chosen for analysis (Table 1). Because a subset of mirnas are clustered in the genome, some alleles affect multiple mirnas. For example, ndf58 is a deletion of three mirnas: mir-54, mir-55, and mir-56 (mir-54-56). We selected mirnas that show evolutionary conservation of mirna family members on the basis of their seed sequence or a developmentally regulated expression pattern [15 19]. First, all of the strains carrying deletion alleles were outcrossed with wild-type N2 worms (see Table S1 available online). A set of multiply mutant strains was then constructed that carried a single mirna deletion allele and the gk214 allele of alg-1. We performed phenotypic analysis of our collection of 25 alg-1;mir multiply mutant strains. This analysis focused on readily observable, quantifiable, developmental phenotypes: embryonic, larval, and adult lethality, gross morphology and motility, dauer formation, gonad migration, and alae formation. A summary of the phenotypic analysis is shown in Table 1, which includes the identification of novel enhanced or synthetic phenotypes for 19 of the 25 strains analyzed. First, we observed that loss of mir-51, mir-57, mir-59, mir-77, mir-228, mir-240 mir 786, or mir-246 in the alg-1-sensitized background resulted in a significant increase in the percentage of embryonic lethality, from 3% in alg-1 single mutants to 7% 13% in the alg-1;mir multiple mutants (Table 1). Consistent with a role in embryogenesis, embryonic expression has been reported for mir-51, mir-57, mir-59, and mir-228 [17, 20]. However, no embryonic expression has been reported for mir-77, mir-240 mir-786, or mir-246. The point at which these embryos arrested was not determined, and because the increase in embryonic lethality was relatively modest, transgenic rescue was not performed. Recently, it has been shown that worms lacking all six members of the mir-51 mirna family display a penetrant embryonic-lethal phenotype [2], demonstrating an essential role for this mirna family in embryogenesis. Second, we observed that loss of mir-1, mir-59, mir-83, mir- 124, mir-247 mir-797, or mir-259 in the alg-1-sensitized background resulted in a significant increase in the percentage of worms that had defective distal tip cell migration as determined by the gonad morphology in young adult worms (Table 1). In contrast to wild-type animals that execute a single, developmentally regulated reflex of each gonad arm, 8% of alg-1 single mutants displayed abnormal distal tip cell migration, with the distal tip cell executing an extra turn during development. The distal tip cell of the gonad arms migrated normally to the anterior or posterior, made a dorsal turn, and initiated migration back to the midline normally but then executed an extra turn back away from the midline. The extra turn was observed to occur predominantly, but not exclusively, in the posterior gonad arm. In all six alg-1; mir strains, this enhanced gonad migration phenotype was rescued by the extrachromosomal expression of a wild-type genomic fragment containing the mirna stem-loop sequence along
2 Current Biology Vol 20 No Table 1. Phenotypic Characterization of mirna Mutants in alg-1-sensitized Genetic Background Developmental Timing Gonad Migration Embryonic Lethality Adult Lethality Strain Genotype % Incomplete Alae Formation a % Abnormal b % Unhatched c % Dead at 72 hr d N2 wild type 0% 0% 0% 0% RF54 alg-1(gk214) 61% 8% 3% 63% RF70 mir-1(n4102); alg-1(gk214) 57% 25%** 3% 53% RF129 mir-34(n4276) alg-1(gk214) 59% 13% 5% 72% RF420 mir-51(n4473); alg-1(gk214) 31%** 7% 8%* 51% RF411 mir-52(n4114); alg-1(gk214) 3%** 0% 6% 16%** RF398 mir-53(n4113); alg-1(gk214) 60% 17% 1% 57% RF410 mir-54-55(ndf45) alg-1(gk214) 4%** 11% 2% 12%** RF89 mir-54-56(ndf58) alg-1(gk214) 23%** 4% 3% 5%** RF133 mir-57(gk175); alg-1(gk214) 51% 5% 8%** 73% RF137 mir-59(n4604); alg-1(gk214) 69% 23%** 10%** 83%** RF153 mir-72(n4130); alg-1(gk214) 56% 2% 5% 48% RF81 mir-73-74(ndf47) alg-1(gk214) 75% 7% 3% 40%** RF178 mir-77(n4285); alg-1(gk214) 54% 8% 12%** 59% RF65 mir-83(n4638); alg-1(gk214) 51% 25%** 3% 77% RF141 mir-85(n4117); alg-1(gk214) 48% 4% 3% 54% RF77 mir-124(n4255); alg-1(gk214) 69% 18%* 2% 69% RF145 mir-228(n4382); alg-1(gk214) 39%** 5% 13%** 53% RF93 mir-234(n4520); alg-1(gk214) 53% 5% 3% 57% RF182 mir-235(n4504); alg-1(gk214) 56% 7% 0% 80%** RF85 mir-237(n4296) alg-1(gk214) 47% 12% 3% 75% RF163 mir-238(n4112); mir-239a-b(ndf62) 22%** 3% 1% 11%** alg-1(gk214) RF60 mir-240 mir-786(n4541) alg-1(gk214) 50% 13% 7%** 45%* RF186 mir-244(n4367); alg-1(gk214) 13%** 9% 5% 53% RF149 mir-246(n4636); alg-1(gk214) 44% 7% 8%** 71% RF368 mir-247 mir-797(n4505) alg-1(gk214) 41% 25%** 4% 57% RF343 mir-259(n4106); alg-1(gk214) 34%** 28%** 4% 59% *p < 0.05; **p < 0.01 by the chi-square test, as compared to alg-1 single mutants. a Alae were scored at the L4m by DIC microscopy. n > 39 (range: ) worms scored for each strain. b Gonad morphology was scored in young adult worms by DIC microscopy. n > 41 (range: ) worms scored for each strain. c Embryos were transferred to a new plate and scored after hr for the presence of unhatched embryos. n > 78 (range: ) embryos scored for each strain. d Synchronized L1-stage worms were transferred to plates to initiate development. Lethality was scored 72 hr after plating at 20 C; n > 76 (range: ) worms scored for each strain. with flanking sequence (Table S1). The expression patterns of most of these mirnas are consistent with a possible role in the regulation of secreted or membrane-bound guidance molecules: mir-1 is expressed in the body wall muscles [21], mir-59 and mir-83 are expressed in the intestine [20], and mir-247 mir-797 is expressed in the distal tip cell [20]. Proteins from the body wall muscle and intestinal cells are required for guidance cues in the body wall basement membrane, over which the distal tip cells migrate [22, 23]. Third, we observed that loss of mir-59 and mir-235 resulted in an increased level of adult lethality compared to alg-1 single mutants (Table 1). alg-1 single mutants appeared healthy at the L4-to-adult transition, but 63% died as young adults (Table 1). About 62% of alg-1 worms entered a supernumerary molt in the adult stage (Table S2). Nearly all of the adult worms that entered the molt subsequently died with a bag of worms phenotype (Table S2), probably due to a failure to properly complete the molting cycle that compromises the worm s ability to lay embryos. Execution of a supernumerary molt is a heterochronic phenotype also observed in worms missing let-7 family members [24, 25]. This adult lethality was enhanced in mir-59;alg-1 and mir-235;alg-1 worms, with 83% and 80% adult lethality, respectively (Table 1). The increase in adult lethality observed in these strains is primarily due to an increase in the percentage of worms that enter a supernumerary lethargus (Table S2). In contrast to this enhancement of the alg-1 adult lethality phenotype, we found that loss of other mirnas suppressed alg-1 phenotypes. In particular, loss of mir-73-74, mir-238/ mir-239a-b, mir-240 mir-786, or certain mir-51 family members (mir-52, mir-54-55, and mir-54-56) in the alg-1 background resulted in a reduced level of adult lethality when compared to alg-1 single mutants (Table 1). This decrease in adult lethality is primarily due to a decrease in the percentage of worms that enter a supernumerary lethargus (Table S2). Similarly, we observed that loss of mir-228, mir-238/mir-239a-b, mir-244, mir-259, ormir-51 family members (mir-51, mir-52, mir-54-55, and mir-54-56), resulted in a reduced penetrance of developmental timing defects, as assayed by the formation of an adult-specific cuticle structure called alae (Table 1). About 60% of alg-1 single mutants showed incomplete alae formation owing to a partial reiteration of larval stage programs, which is probably due to reduced levels of lin-4 and let-7 family mirnas [6]. Incomplete alae formation indicates an inappropriate larval cell fate for some of the cells in the hypodermis at the L4-to-adult transition. Introduction of a transgene with the mir-54-56, mir-238, mir-244, or mir-259 genomic loci into the respective alg-1;mir strain restored the level of incomplete alae formation to that observed in alg-1 single mutants (Table S1). Four strains, mir-52; alg-1, mir alg-1, mir alg-1, and mir-238; mir-239a-b alg-1 displayed both reduced adult lethality and reduced alae formation defects (Table 1).
3 Analysis of mirnas in Sensitized Backgrounds 1323 Table 2. Summary of hub Genes and Synthetic Interactions with Developmental Pathways Gene Description a Enhancement with Developmental Regulatory Components b din-1 Ortholog of human transcriptional corepressor SHARP/SPEN, EGF, Wnt, Notch, ephrin receptor encodes large RNA-binding protein of RRM superfamily dpy-22 Ortholog of the human transcriptional Mediator protein TRAP230 EGF, Wnt, Notch, E2F/synMuvB, ephrin receptor, RAC1/GTPase egl-27 Ortholog of human MTA1, part of nucleosome remodeling EGF, Notch, E2F/synMuvB, ephrin receptor and histone deacetylation (NURD) complex hmg-1.2 DNA binding protein with HMG box EGF, Wnt, Notch, ephrin receptor trr-1 Ortholog of component of the NuA4/Tip60 histone acetyltransferase complexes, atypical protein kinase of TRAAP subfamily EGF, Wnt, Notch, E2F/synMuvB, GTPase a Information from b See [3] for details. Loss of Individual mirnas Confers Enhanced or Synthetic Sterility with Knockdown of a Set of Chromatin Regulator hub Genes We next tested whether mirna genes functionally interact with broadly acting pathways of gene regulation. A small set of genes, termed hub genes, have been identified that show a high level of connectivity to many developmental processes [3]. These hub genes encode chromatin-regulatory proteins and interact with a wide range of signaling pathways that are critical for normal development, including the Wnt, EGF, and Notch pathways [3]. Therefore, we hypothesized that the reduced activity of these hub genes could provide a sensitized genetic background in which to reveal additional relationships between individual mirna mutants and other developmental processes. To address this question directly, we determined whether individual mirna deletion alleles displayed enhanced or synthetic interactions upon knockdown of one of five hub genes by RNAi: egl-27, din-1, hmg-1.2, dpy-22, and trr-1 (Table 2). We performed phenotypic assays on eleven strains carrying mirna deletion alleles to identify embryoniclethal or -sterile phenotypes upon knockdown of hub genes. We found that RNAi knockdown of egl-27, din-1, hmg-1.2, dpy-22, and trr-1 in wild-type worms did not result in a significant number of worms that were sterile as compared to the negative control RNAi (Table 3). However, four mirna mutant strains displayed a synthetic sterile phenotype after knockdown of individual hub genes. When egl-27 or hmg-1.2 activity was knocked down in mir-1 mutant worms, 24% and 29% of worms were sterile, respectively (Table 3). No sterility was observed when mir-1 worms were fed bacteria containing an empty-vector construct. Like mir-1 worms, mir-59 worms showed a synthetic sterile phenotype with knockdown of egl-27 or hmg-1.2 (Table 3). When either egl-27 or trr-1 activity was knocked down in mir-247 mir-797 mutant worms, 29% and 40% of worms were sterile, respectively, compared to 3% in the negative control group (Table 3). The strongest synthetic phenotype was observed in mir-240 mir-786 worms after knockdown of hmg-1.2 or trr-1, with 76% and 83% sterility (Table 3). These worms exhibited germline defects (Figure S1). For all strains that displayed a sterile phenotype upon hub gene knockdown, there was no significant embryonic lethality (Table S3). Few, if any, embryos were found on plates with mir-240 mir-786 worms after knockdown of hmg-1.2 or trr-1 activity (Table S3). These data suggest that mir-1, mir-59, mir-240 mir-786, and mir-247 mir-797 function in regulatory pathways that are essential for germline development. Of these, only mir-240 mir-786 has been shown to be expressed in the gonad by the use of mirna promoter reporter transgenes, with expression observed in the uterus, spermatheca, and gonadal sheath [20]. It is possible that mir-1, mir-59, and mir-247 mir- 797 are expressed in the germline but that this expression is undetectable with reporter transgenes or that these mirnas function to control developmental signals required for fertility from outside of the germline. Discussion The goal of this work was to test the hypothesis that mirnas have overlapping or redundant functions and to identify phenotypes associated with the loss of individual mirnas. We found that loss of individual mirnas resulted in developmental abnormalities in an alg-1 genetic background, which Table 3. Phenotypic Characterization of mirna Mutants after Knockdown of hub Gene Activity by RNAi % Sterile Worms Strain Genotype Empty-Vector RNAi din-1 RNAi dpy-22 RNAi egl-27 RNAi hmg1.2 RNAi trr-1 RNAi N2 wild-type 0% 11% 4% 3% 7% 14% RF71 mir-1(n4102) 0% 16% 4% 24%* 29%* 16% RF124 mir-34(n4276) 3% 2% 0% 7% 3% 0% RF90 mir-54-56(ndf58) 0% 4% 0% 0% 5% 21% RF15 mir-59(n4604) 7% 24% 8% 21%* 26%* 27% RF66 mir-83(n4638) 0% 5% 5% 0% 11% 7% RF78 mir-124(n4255) 0% 4% 0% 0% 14% 24% RF94 mir-234(n4520) 0% 7% 0% 9% 11% 9% RF86 mir-237(n4296) 2% 0% 0% 0% 0% 12% RF61 mir-240 mir-786(n4541) 0% 17% 13% 4% 76%** 83%** RF126 mir-246(n4636) 0% 5% 3% 15% 16% 33% RF24 mir-247 mir-797 (n4505) 3% 11% 9% 29%** 13% 40%* *p < 0.05; **p < 0.01 by the chi-square test as compared to wild-type worms with the corresponding hub gene RNAi.
4 Current Biology Vol 20 No has lower total mirna activity, and in backgrounds with reduced hub gene activity, in which multiple regulatory pathways are compromised. Our analysis described mutant phenotypes associated with loss of 25 out of the 31 mirnas included in our study. These results identified only a limited number of phenotypes in the alg-1;mir strains. This indicates that only a subset of mirna-regulated pathways are sufficiently sensitized in alg-1 worms, whereas others may require a further reduction in the activity of mirnas to become sensitized. Additional knockdown of alg-2 activity, which is the other Argonauteencoding gene that acts in the mirna pathway, could be used to enhance the sensitivity of this assay. However, complete loss of alg-1 and alg-2 results in embryonic lethality [6]. The synthetic sterility observed with individual mirnas and hub genes indicates that pathways that are essential for germline development are sensitized in these backgrounds. Additional work is needed to identify the specific regulatory pathways and targets that are controlled by individual mirnas. Our assays identified multiple mirnas that interact with either alg-1 or individual hub genes to give similar phenotypes. One parsimonious model for this observation is that these sets of mirnas may regulate shared mrna targets. For example, mir-1, mir-59, and mir-240 mir-786 all interact with hmg-1.2 (RNAi) to give a synthetic sterile phenotype. To examine this hypothesis, we identified predicted mrna targets by using Targetscan [19] or mirwip [26]. Both prediction algorithms identified possible shared targets for pairs of these mirnas (Table S4), with three targets that are possible shared targets for three mirnas (mir-1, mir-59, and mir-786): daf-12, hbl-1, and cfim-2. Interestingly, cfim-2 encodes an mrna cleavage and polyadenylation factor that is required for fertility [27]. This analysis of target predictions provides a platform to identify the biologically relevant targets and to test the hypothesis that multiple mirnas regulate shared targets. Alternatively, the phenotypes we observe may reflect mirna regulation of distinct targets in convergent developmental pathways. Interestingly, loss of a subset of mirnas, mir-228, mir-238; mir-239a/b, mir-244, mir-259, or certain mir-51 family members (mir-51, mir-52, and mir-54-56) resulted in a suppression of alg-1 alae formation defects. This could indicate regulation of specific targets in the developmental timing pathway. In this model, some mirnas could function antagonistically to the lin-4 or let-7 family mirnas, which are largely responsible for the alg-1 developmental timing defects [6]. An alternative explanation for the observed suppression of alg-1 phenotypes by certain mirna gene mutations could be that these mirnas regulate specific targets that control mirisc activity, such that in their absence, mirisc activity is enhanced. For example, the mirisc cofactor, nhl-2 [13], is predicted to be a target of mir-244 and mir-228 by TargetScan [19] and mirwip [26] mirna target-prediction algorithms, suggesting a possible mechanism for suppression. It is also possible that the observed suppression may be due to a more general effect on mirisc activity. If individual mirnas compete for loading into mirisc, then loss of a highly abundant mirna, such as mir-52 [17, 19], may allow for greater activity of remaining mirnas, including the let-7 family. However, this cannot account for the observed suppression in all alg-1;mir strains given that loss of abundant mirnas does not suppress alg-1 defects in all cases. Additionally, strong suppression of alg-1 defects is observed with loss of weakly expressed mirnas, such as mir-238 or mir-259 [19]. Interestingly, Targetscan identifies alg-2 as a candidate target for mir-259 [19]. Few phenotypes have been described for individual mirna mutants (see [28]). In addition, few phenotypes are observed in strains that lack most or all mirna family members, suggesting that the lack of detectable phenotypes for individual mirna mutants is not due primarily to overlapping function with related mirna family members [2]. Our results indicate that mirnas may function together with unrelated mirnas or non-mirna genes, perhaps functioning together to ensure the robustness of developmental mechanisms. The use of sensitized genetic backgrounds is a fruitful approach to identify phenotypes associated with the loss of specific mirnas. Genetic backgrounds can be selected that have compromised activity of specific regulatory pathways, which may render them sensitive to the loss of individual mirnas. It is possible that functions of some mirnas may be revealed only when sufficient genetic or environmental variation is introduced. Experimental Procedures General Methods and Strains C. elegans strains were maintained under standard conditions as previously described [29]. Worms were kept on NGM plates seeded with E. coli strain AMA1004 [30]. The wild-type strain used was var. Bristol N2 [31]. RF54 was used as the alg-1(gk214)x control in phenotypic analysis of mir; alg-1 strains. All strains were kept at 20 C unless otherwise indicated. mirna mutant strains were first outcrossed to wild-type N2 (Table S1). For building multiply mutant strains, presence of the mirna deletion alleles and the alg-1 allele in F2s were identified by performing PCR with primers that amplified the genomic region flanking the deletion mutation. Sequences for primers used for genotyping can be found in Table S1. Fluorescence and differential interference contrast (DIC) microscopy were performed with a Nikon Eclipse 80i equipped with a Photometrics CoolSNAP HQ2 monochrome digital camera and RS Image software (Roper Scientific). RNAi Experiments Four L4-stage worms were placed on RNAi plates (NGM with 0.2% [w/v] lactose and 100 mg/ml ampicillin) seeded with bacteria to knock down egl-27, din-1, hmg-1.2, dpy-22, or trr-1. Empty vector (L4440) was used as the negative control. Bacteria for RNAi experiments were isolated from the Ahringer RNAi library [32]. Worms were placed at 20 C overnight. 48 F1 embryos were transferred to a new RNAi plate, seeded with the same bacteria. The next day, live worms and unhatched embryos were scored. The surviving larval worms were cloned into 96-well culture plates, with each well containing RNAi liquid media (M9 containing 0.2% lactose and 100 mg/ml ampicillin) plus bacteria. The bacteria were from frozen bacterial pellets taken from 5 ml stationary phase overnight cultures of appropriate bacteria strain in LB containing 0.2% lactose and 100 mg/ml ampicillin. Ninety-six-well plates were placed at 20 C for 96 hr, and then wells were scored for the presence of F2 progeny. Wells in which the F1 worm died were not scored. Transgene Rescue Experiments For the creation of transgenic animals, germline transformation was performed as described [33]. Injection mixes contained 5 25 ng/ml of the rescue plasmid, ng/ml of a coinjection marker (myo-2::gfp or myo-2::dsred) containing plasmid, and prs413 plasmid for a final DNA concentration of 150 ng/ml (Table S1). Transgenic animals expressing GFP or dsred in the pharynx were assayed for alae formation or for gonad migration defects. Supplemental Information Supplemental Information includes five tables and one figure and can be found with this article online at doi: /j.cub Acknowledgments We thank Chris Hammell and Victor Ambros (University of Massachusetts Medical School) for helpful discussions and advice on preparation of this manuscript and Molly Hammell (University of Massachusetts Medical School) for sharing additonal target predictions for mir-59. We also thank Rob Shaw and Eric Miska (University of Cambridge) for providing a strain
5 Analysis of mirnas in Sensitized Backgrounds 1325 carrying the mjex160 extrachromosomal array for the mir rescue experiment. Some strains used in this study were obtained from the C. elegans Genetics Center (CGC), which is supported by a grant from the National Institutes of Health (NIH, National Center for Research Resources). This work was supported by a grant from the NIH (National Institute of General Medical Sciences, GM084451). Received: March 25, 2010 Revised: May 10, 2010 Accepted: May 27, 2010 Published online: June 24, 2010 References 1. Miska, E.A., Alvarez-Saavedra, E., Abbott, A.L., Lau, N.C., Hellman, A.B., McGonagle, S.M., Bartel, D.P., Ambros, V.R., and Horvitz, H.R. (2007). Most Caenorhabditis elegans micrornas are individually not essential for development or viability. PLoS Genet. 3, e Alvarez-Saavedra, E., and Horvitz, H.R. (2010). Many families of C. elegans micrornas are not essential for development or viability. Curr. Biol. 20, Lehner, B., Crombie, C., Tischler, J., Fortunato, A., and Fraser, A.G. (2006). Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat. Genet. 38, Bernstein, E., Kim, S.Y., Carmell, M.A., Murchison, E.P., Alcorn, H., Li, M.Z., Mills, A.A., Elledge, S.J., Anderson, K.V., and Hannon, G.J. (2003). Dicer is essential for mouse development. Nat. Genet. 35, Giraldez, A.J., Cinalli, R.M., Glasner, M.E., Enright, A.J., Thomson, J.M., Baskerville, S., Hammond, S.M., Bartel, D.P., and Schier, A.F. (2005). MicroRNAs regulate brain morphogenesis in zebrafish. Science 308, Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. (2001). Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15, Lee, Y.S., Nakahara, K., Pham, J.W., Kim, K., He, Z., Sontheimer, E.J., and Carthew, R.W. (2004). Distinct roles for Drosophila Dicer-1 and Dicer-2 in the sirna/mirna silencing pathways. Cell 117, Wienholds, E., Koudijs, M.J., van Eeden, F.J., Cuppen, E., and Plasterk, R.H. (2003). The microrna-producing enzyme Dicer1 is essential for zebrafish development. Nat. Genet. 35, Lee, R.C., Hammell, C.M., and Ambros, V. (2006). Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA 12, Ding, L., Spencer, A., Morita, K., and Han, M. (2005). The developmental timing regulator AIN-1 interacts with miriscs and may target the Argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell 19, Frand, A.R., Russel, S., and Ruvkun, G. (2005). Functional genomic analysis of C. elegans molting. PLoS Biol. 3, e Hammell, C.M., Lubin, I., Boag, P.R., Blackwell, T.K., and Ambros, V. (2009). nhl-2 Modulates microrna activity in Caenorhabditis elegans. Cell 136, Morita, K., and Han, M. (2006). Multiple mechanisms are involved in regulating the expression of the developmental timing regulator lin-28 in Caenorhabditis elegans. EMBO J. 25, Kato, M., de Lencastre, A., Pincus, Z., and Slack, F.J. (2009). Dynamic expression of small non-coding RNAs, including novel micrornas and pirnas/21u-rnas, during Caenorhabditis elegans development. Genome Biol. 10, R Lee, R.C., and Ambros, V. (2001). An extensive class of small RNAs in Caenorhabditis elegans. Science 294, Lim, L.P., Lau, N.C., Weinstein, E.G., Abdelhakim, A., Yekta, S., Rhoades, M.W., Burge, C.B., and Bartel, D.P. (2003). The micrornas of Caenorhabditis elegans. Genes Dev. 17, Martinez, N.J., Ow, M.C., Barrasa, M.I., Hammell, M., Sequerra, R., Doucette-Stamm, L., Roth, F.P., Ambros, V.R., and Walhout, A.J. (2008). A C. elegans genome-scale microrna network contains composite feedback motifs with high flux capacity. Genes Dev. 22, Ruby, J.G., Jan, C., Player, C., Axtell, M.J., Lee, W., Nusbaum, C., Ge, H., and Bartel, D.P. (2006). Large-scale sequencing reveals 21U-RNAs and additional micrornas and endogenous sirnas in C. elegans. Cell 127, Martinez, N.J., Ow, M.C., Reece-Hoyes, J.S., Barrasa, M.I., Ambros, V.R., and Walhout, A.J. (2008). Genome-scale spatiotemporal analysis of Caenorhabditis elegans microrna promoter activity. Genome Res. 18, Simon, D.J., Madison, J.M., Conery, A.L., Thompson-Peer, K.L., Soskis, M., Ruvkun, G.B., Kaplan, J.M., and Kim, J.K. (2008). The microrna mir-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell 133, Kubota, Y., Kuroki, R., and Nishiwaki, K. (2004). A fibulin-1 homolog interacts with an ADAM protease that controls cell migration in C. elegans. Curr. Biol. 14, Lehmann, R. (2001). Cell migration in invertebrates: Clues from border and distal tip cells. Curr. Opin. Genet. Dev. 11, Abbott, A.L., Alvarez-Saavedra, E., Miska, E.A., Lau, N.C., Bartel, D.P., Horvitz, H.R., and Ambros, V. (2005). The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell 9, Hayes, G.D., Frand, A.R., and Ruvkun, G. (2006). The mir-84 and let-7 paralogous microrna genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development 133, Hammell, M., Long, D., Zhang, L., Lee, A., Carmack, C.S., Han, M., Ding, Y., and Ambros, V. (2008). mirwip: microrna target prediction based on microrna-containing ribonucleoprotein-enriched transcripts. Nat. Methods 5, Simmer, F., Moorman, C., van der Linden, A.M., Kuijk, E., van den Berghe, P.V., Kamath, R.S., Fraser, A.G., Ahringer, J., and Plasterk, R.H. (2003). Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1, E Smibert, P., and Lai, E.C. (2008). Lessons from microrna mutants in worms, flies and mice. Cell Cycle 7, Wood, W.B. (1988). The Nematode Caenorhabditis Elegans (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). 30. Casadaban, M.J., Martinez-Arias, A., Shapira, S.K., and Chou, J. (1983). Beta-galactosidase gene fusions for analyzing gene expression in escherichia coli and yeast. Methods Enzymol. 100, Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, Kamath, R.S., Fraser, A.G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, Mello, C.C., Kramer, J.M., Stinchcomb, D., and Ambros, V. (1991). Efficient gene transfer in C.elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10,
he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003)
 MicroRNAs: Genomics, Biogenesis, Mechanism, and Function (D. Bartel Cell 2004) he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003) Vertebrate MicroRNA Genes (Lim et al. Science
MicroRNAs: Genomics, Biogenesis, Mechanism, and Function (D. Bartel Cell 2004) he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003) Vertebrate MicroRNA Genes (Lim et al. Science
The mir-51 Family of MicroRNAs Functions in Diverse Regulatory Pathways in Caenorhbditis elegans
 Marquette University e-publications@marquette Biological Sciences Faculty Research and Publications Biological Sciences, Department of 5-16-2012 The mir-51 Family of MicroRNAs Functions in Diverse Regulatory
Marquette University e-publications@marquette Biological Sciences Faculty Research and Publications Biological Sciences, Department of 5-16-2012 The mir-51 Family of MicroRNAs Functions in Diverse Regulatory
Phenomena first observed in petunia
 Vectors for RNAi Phenomena first observed in petunia Attempted to overexpress chalone synthase (anthrocyanin pigment gene) in petunia. (trying to darken flower color) Caused the loss of pigment. Bill Douherty
Vectors for RNAi Phenomena first observed in petunia Attempted to overexpress chalone synthase (anthrocyanin pigment gene) in petunia. (trying to darken flower color) Caused the loss of pigment. Bill Douherty
MicroRNA and Male Infertility: A Potential for Diagnosis
 Review Article MicroRNA and Male Infertility: A Potential for Diagnosis * Abstract MicroRNAs (mirnas) are small non-coding single stranded RNA molecules that are physiologically produced in eukaryotic
Review Article MicroRNA and Male Infertility: A Potential for Diagnosis * Abstract MicroRNAs (mirnas) are small non-coding single stranded RNA molecules that are physiologically produced in eukaryotic
BIOL2005 WORKSHEET 2008
 BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment
 Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development
Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
Cross species analysis of genomics data. Computational Prediction of mirnas and their targets
 02-716 Cross species analysis of genomics data Computational Prediction of mirnas and their targets Outline Introduction Brief history mirna Biogenesis Why Computational Methods? Computational Methods
02-716 Cross species analysis of genomics data Computational Prediction of mirnas and their targets Outline Introduction Brief history mirna Biogenesis Why Computational Methods? Computational Methods
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/8/e1701143/dc1 Supplementary Materials for Impaired DNA replication derepresses chromatin and generates a transgenerationally inherited epigenetic memory Adam
advances.sciencemag.org/cgi/content/full/3/8/e1701143/dc1 Supplementary Materials for Impaired DNA replication derepresses chromatin and generates a transgenerationally inherited epigenetic memory Adam
A Genetic Program for Embryonic Development
 Concept 18.4: A program of differential gene expression leads to the different cell types in a multicellular organism During embryonic development, a fertilized egg gives rise to many different cell types
Concept 18.4: A program of differential gene expression leads to the different cell types in a multicellular organism During embryonic development, a fertilized egg gives rise to many different cell types
Circular RNAs (circrnas) act a stable mirna sponges
 Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Programmed Cell Death (apoptosis)
 Programmed Cell Death (apoptosis) Stereotypic death process includes: membrane blebbing nuclear fragmentation chromatin condensation and DNA framentation loss of mitochondrial integrity and release of
Programmed Cell Death (apoptosis) Stereotypic death process includes: membrane blebbing nuclear fragmentation chromatin condensation and DNA framentation loss of mitochondrial integrity and release of
MicroRNAs, RNA Modifications, RNA Editing. Bora E. Baysal MD, PhD Oncology for Scientists Lecture Tue, Oct 17, 2017, 3:30 PM - 5:00 PM
 MicroRNAs, RNA Modifications, RNA Editing Bora E. Baysal MD, PhD Oncology for Scientists Lecture Tue, Oct 17, 2017, 3:30 PM - 5:00 PM Expanding world of RNAs mrna, messenger RNA (~20,000) trna, transfer
MicroRNAs, RNA Modifications, RNA Editing Bora E. Baysal MD, PhD Oncology for Scientists Lecture Tue, Oct 17, 2017, 3:30 PM - 5:00 PM Expanding world of RNAs mrna, messenger RNA (~20,000) trna, transfer
Bi 8 Lecture 17. interference. Ellen Rothenberg 1 March 2016
 Bi 8 Lecture 17 REGulation by RNA interference Ellen Rothenberg 1 March 2016 Protein is not the only regulatory molecule affecting gene expression: RNA itself can be negative regulator RNA does not need
Bi 8 Lecture 17 REGulation by RNA interference Ellen Rothenberg 1 March 2016 Protein is not the only regulatory molecule affecting gene expression: RNA itself can be negative regulator RNA does not need
mir-355 Functions as An Important Link between p38 MAPK Signaling and Insulin Signaling in the Regulation of Innate Immunity
 www.nature.com/scientificreports Received: 7 July 2017 Accepted: 24 October 2017 Published: xx xx xxxx OPEN mir-355 Functions as An Important Link between p38 MAPK Signaling and Insulin Signaling in the
www.nature.com/scientificreports Received: 7 July 2017 Accepted: 24 October 2017 Published: xx xx xxxx OPEN mir-355 Functions as An Important Link between p38 MAPK Signaling and Insulin Signaling in the
mirna Dr. S Hosseini-Asl
 mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
Prokaryotes and eukaryotes alter gene expression in response to their changing environment
 Chapter 18 Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development and is responsible for differences
Chapter 18 Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development and is responsible for differences
MicroRNA expression profiling and functional analysis in prostate cancer. Marco Folini s.c. Ricerca Traslazionale DOSL
 MicroRNA expression profiling and functional analysis in prostate cancer Marco Folini s.c. Ricerca Traslazionale DOSL What are micrornas? For almost three decades, the alteration of protein-coding genes
MicroRNA expression profiling and functional analysis in prostate cancer Marco Folini s.c. Ricerca Traslazionale DOSL What are micrornas? For almost three decades, the alteration of protein-coding genes
Chapter 2. Investigation into mir-346 Regulation of the nachr α5 Subunit
 15 Chapter 2 Investigation into mir-346 Regulation of the nachr α5 Subunit MicroRNA s (mirnas) are small (< 25 base pairs), single stranded, non-coding RNAs that regulate gene expression at the post transcriptional
15 Chapter 2 Investigation into mir-346 Regulation of the nachr α5 Subunit MicroRNA s (mirnas) are small (< 25 base pairs), single stranded, non-coding RNAs that regulate gene expression at the post transcriptional
Mutations in the Caenorhabditis elegans U2AF Large Subunit UAF-1 Al= of a 3' Splice Site In Vivo
 Mutations in the Caenorhabditis elegans U2AF Large Subunit UAF-1 Al= of a 3' Splice Site In Vivo The MIT Faculty has made this article openly available. Please share how this access benefits you. Your
Mutations in the Caenorhabditis elegans U2AF Large Subunit UAF-1 Al= of a 3' Splice Site In Vivo The MIT Faculty has made this article openly available. Please share how this access benefits you. Your
Improved annotation of C. elegans micrornas by deep sequencing reveals structures associated with processing by Drosha and Dicer
 BIOINFORMATICS Improved annotation of C. elegans micrornas by deep sequencing reveals structures associated with processing by Drosha and Dicer M. BRYAN WARF, 1 W. EVAN JOHNSON, 2 and BRENDA L. BASS 1
BIOINFORMATICS Improved annotation of C. elegans micrornas by deep sequencing reveals structures associated with processing by Drosha and Dicer M. BRYAN WARF, 1 W. EVAN JOHNSON, 2 and BRENDA L. BASS 1
MicroRNAs Modulate Hematopoietic Lineage Differentiation
 MicroRNAs Modulate Hematopoietic Lineage Differentiation Harvey F. Lodish, Chang-Zheng Chen, David P. Bartel Whitehead Institute for Biomedical Research, Department of Biology, Massachusetts Institute
MicroRNAs Modulate Hematopoietic Lineage Differentiation Harvey F. Lodish, Chang-Zheng Chen, David P. Bartel Whitehead Institute for Biomedical Research, Department of Biology, Massachusetts Institute
Most Caenorhabditis elegans micrornas Are Individually Not Essential for Development or Viability
 Most Caenorhabditis elegans micrornas Are Individually Not Essential for Development or Viability Eric A. Miska 1,2 a[, Ezequiel Alvarez-Saavedra 1,2[, Allison L. Abbott 3[ b, Nelson C. Lau 4 c, Andrew
Most Caenorhabditis elegans micrornas Are Individually Not Essential for Development or Viability Eric A. Miska 1,2 a[, Ezequiel Alvarez-Saavedra 1,2[, Allison L. Abbott 3[ b, Nelson C. Lau 4 c, Andrew
Correspondence: mirna regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration
 Correspondence: mirna regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration Alison A. Staton, Holger Knaut, and Antonio J. Giraldez Supplementary Note Materials and
Correspondence: mirna regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration Alison A. Staton, Holger Knaut, and Antonio J. Giraldez Supplementary Note Materials and
Supplementary Information
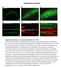 Supplementary Information Supplementary Figure 1: Luminal localization of CCM-3. (a) The CCM-3::GFP fusion protein localizes along the apical (luminal) surface of the pharynx (b) as well as the lumen of
Supplementary Information Supplementary Figure 1: Luminal localization of CCM-3. (a) The CCM-3::GFP fusion protein localizes along the apical (luminal) surface of the pharynx (b) as well as the lumen of
Nature Genetics: doi: /ng Supplementary Figure 1. Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms.
 Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Identification of target genes of micrornas in retinoic acid-induced neuronal differentiation*
 Pure Appl. Chem., Vol. 77, No. 1, pp. 313 318, 2005. DOI: 10.1351/pac200577010313 2005 IUPAC Identification of target genes of micrornas in retinoic acid-induced neuronal differentiation* Hiroaki Kawasaki
Pure Appl. Chem., Vol. 77, No. 1, pp. 313 318, 2005. DOI: 10.1351/pac200577010313 2005 IUPAC Identification of target genes of micrornas in retinoic acid-induced neuronal differentiation* Hiroaki Kawasaki
klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach
![klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach](/thumbs/91/104639484.jpg) DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
Chapter 11. Robert C. Johnsen and Denise V. Clark. Department of Biological Sciences, Simon Fraser University, Burnaby, B.C.
 Chapter 11 Mapping Genes in C. elegans Robert C. Johnsen and Denise V. Clark Department of Biological Sciences, Simon Fraser University, Burnaby, B.C., Canada, V5A 1S6 Robert C. Johnsen received his B.Sc.
Chapter 11 Mapping Genes in C. elegans Robert C. Johnsen and Denise V. Clark Department of Biological Sciences, Simon Fraser University, Burnaby, B.C., Canada, V5A 1S6 Robert C. Johnsen received his B.Sc.
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression. Chromatin Array of nuc
 Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
(,, ) microrna(mirna) 19~25 nt RNA, RNA mirna mirna,, ;, mirna mirna. : microrna (mirna); ; ; ; : R321.1 : A : X(2015)
 35 1 Vol.35 No.1 2015 1 Jan. 2015 Reproduction & Contraception doi: 10.7669/j.issn.0253-357X.2015.01.0037 E-mail: randc_journal@163.com microrna ( 450052) microrna() 19~25 nt RNA RNA ; : microrna (); ;
35 1 Vol.35 No.1 2015 1 Jan. 2015 Reproduction & Contraception doi: 10.7669/j.issn.0253-357X.2015.01.0037 E-mail: randc_journal@163.com microrna ( 450052) microrna() 19~25 nt RNA RNA ; : microrna (); ;
Biology Developmental Biology Spring Quarter Midterm 1 Version A
 Biology 411 - Developmental Biology Spring Quarter 2013 Midterm 1 Version A 75 Total Points Open Book Choose 15 out the 20 questions to answer (5 pts each). Only the first 15 questions that are answered
Biology 411 - Developmental Biology Spring Quarter 2013 Midterm 1 Version A 75 Total Points Open Book Choose 15 out the 20 questions to answer (5 pts each). Only the first 15 questions that are answered
SUPPLEMENTARY FIGURE S1: nlp-22 is expressed in the RIA interneurons and is secreted. (a) An animal expressing both the RIA specific reporter
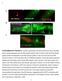 1 SUPPLEMENTARY FIGURE S1: nlp-22 is expressed in the RIA interneurons and is secreted. (a) An animal expressing both the RIA specific reporter Pglr-3:mCherry (red) and Pnlp-22:gfp (green) shows co-localization
1 SUPPLEMENTARY FIGURE S1: nlp-22 is expressed in the RIA interneurons and is secreted. (a) An animal expressing both the RIA specific reporter Pglr-3:mCherry (red) and Pnlp-22:gfp (green) shows co-localization
Utility of Circulating micrornas in Cardiovascular Disease
 Utility of Circulating micrornas in Cardiovascular Disease Pil-Ki Min, MD, PhD Cardiology Division, Gangnam Severance Hospital, Yonsei University College of Medicine Introduction Biology of micrornas Circulating
Utility of Circulating micrornas in Cardiovascular Disease Pil-Ki Min, MD, PhD Cardiology Division, Gangnam Severance Hospital, Yonsei University College of Medicine Introduction Biology of micrornas Circulating
The Biology and Genetics of Cells and Organisms The Biology of Cancer
 The Biology and Genetics of Cells and Organisms The Biology of Cancer Mendel and Genetics How many distinct genes are present in the genomes of mammals? - 21,000 for human. - Genetic information is carried
The Biology and Genetics of Cells and Organisms The Biology of Cancer Mendel and Genetics How many distinct genes are present in the genomes of mammals? - 21,000 for human. - Genetic information is carried
Noncoding or non-messenger RNAs are
 The functions of animal micrornas Victor Ambros Dartmouth Medical School, Department of Genetics, Hanover, New Hampshire 03755, USA (e-mail: vra@dartmouth.edu) MicroRNAs (mirnas) are small RNAs that regulate
The functions of animal micrornas Victor Ambros Dartmouth Medical School, Department of Genetics, Hanover, New Hampshire 03755, USA (e-mail: vra@dartmouth.edu) MicroRNAs (mirnas) are small RNAs that regulate
Regulation of Gene Expression
 Chapter 18 Regulation of Gene Expression PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from
Chapter 18 Regulation of Gene Expression PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from
Sexual Reproduction. For most diploid eukaryotes, sexual reproduction is the only mechanism resulting in new members of a species.
 Sex Determination Sexual Reproduction For most diploid eukaryotes, sexual reproduction is the only mechanism resulting in new members of a species. Meiosis in the sexual organs of parents produces haploid
Sex Determination Sexual Reproduction For most diploid eukaryotes, sexual reproduction is the only mechanism resulting in new members of a species. Meiosis in the sexual organs of parents produces haploid
micrornas (mirna) and Biomarkers
 micrornas (mirna) and Biomarkers Small RNAs Make Big Splash mirnas & Genome Function Biomarkers in Cancer Future Prospects Javed Khan M.D. National Cancer Institute EORTC-NCI-ASCO November 2007 The Human
micrornas (mirna) and Biomarkers Small RNAs Make Big Splash mirnas & Genome Function Biomarkers in Cancer Future Prospects Javed Khan M.D. National Cancer Institute EORTC-NCI-ASCO November 2007 The Human
C. elegans Embryonic Development
 Autonomous Specification in Tunicate development Autonomous & Conditional Specification in C. elegans Embryonic Development Figure 8.36 Bilateral Symmetry in the Egg of the Tunicate Styela partita Fig.
Autonomous Specification in Tunicate development Autonomous & Conditional Specification in C. elegans Embryonic Development Figure 8.36 Bilateral Symmetry in the Egg of the Tunicate Styela partita Fig.
Ch. 18 Regulation of Gene Expression
 Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
Src-INACTIVE / Src-INACTIVE
 Biology 169 -- Exam 1 February 2003 Answer each question, noting carefully the instructions for each. Repeat- Read the instructions for each question before answering!!! Be as specific as possible in each
Biology 169 -- Exam 1 February 2003 Answer each question, noting carefully the instructions for each. Repeat- Read the instructions for each question before answering!!! Be as specific as possible in each
Problem Set 5 KEY
 2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
Regulation of Gene Expression in Eukaryotes
 Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Alternative splicing. Biosciences 741: Genomics Fall, 2013 Week 6
 Alternative splicing Biosciences 741: Genomics Fall, 2013 Week 6 Function(s) of RNA splicing Splicing of introns must be completed before nuclear RNAs can be exported to the cytoplasm. This led to early
Alternative splicing Biosciences 741: Genomics Fall, 2013 Week 6 Function(s) of RNA splicing Splicing of introns must be completed before nuclear RNAs can be exported to the cytoplasm. This led to early
Chapter 11 How Genes Are Controlled
 Chapter 11 How Genes Are Controlled PowerPoint Lectures for Biology: Concepts & Connections, Sixth Edition Campbell, Reece, Taylor, Simon, and Dickey Copyright 2009 Pearson Education, Inc. Lecture by Mary
Chapter 11 How Genes Are Controlled PowerPoint Lectures for Biology: Concepts & Connections, Sixth Edition Campbell, Reece, Taylor, Simon, and Dickey Copyright 2009 Pearson Education, Inc. Lecture by Mary
Report. Many Families of C. elegans MicroRNAs Are Not Essential for Development or Viability. Ezequiel Alvarez-Saavedra 1 and H. Robert Horvitz 1, * 1
 Current Biology 20, 367 373, Feruary 23, 2010 ª2010 Elsevier Ltd All rights reserved DOI 10.1016/j.cu.2009.12.051 Many Families of C. elegans MicroRNAs Are Not Essential for Development or Viaility Report
Current Biology 20, 367 373, Feruary 23, 2010 ª2010 Elsevier Ltd All rights reserved DOI 10.1016/j.cu.2009.12.051 Many Families of C. elegans MicroRNAs Are Not Essential for Development or Viaility Report
NOVEL FUNCTION OF mirnas IN REGULATING GENE EXPRESSION. Ana M. Martinez
 NOVEL FUNCTION OF mirnas IN REGULATING GENE EXPRESSION Ana M. Martinez Switching from Repression to Activation: MicroRNAs can Up-Regulate Translation. Shoba Vasudevan, Yingchun Tong, Joan A. Steitz AU-rich
NOVEL FUNCTION OF mirnas IN REGULATING GENE EXPRESSION Ana M. Martinez Switching from Repression to Activation: MicroRNAs can Up-Regulate Translation. Shoba Vasudevan, Yingchun Tong, Joan A. Steitz AU-rich
Cellular MicroRNA and P Bodies Modulate Host-HIV-1 Interactions. 指導教授 : 張麗冠博士 演講者 : 黃柄翰 Date: 2009/10/19
 Cellular MicroRNA and P Bodies Modulate Host-HIV-1 Interactions 指導教授 : 張麗冠博士 演講者 : 黃柄翰 Date: 2009/10/19 1 MicroRNA biogenesis 1. Pri mirna: primary mirna 2. Drosha: RNaseIII 3. DCR 1: Dicer 1 RNaseIII
Cellular MicroRNA and P Bodies Modulate Host-HIV-1 Interactions 指導教授 : 張麗冠博士 演講者 : 黃柄翰 Date: 2009/10/19 1 MicroRNA biogenesis 1. Pri mirna: primary mirna 2. Drosha: RNaseIII 3. DCR 1: Dicer 1 RNaseIII
Novel RNAs along the Pathway of Gene Expression. (or, The Expanding Universe of Small RNAs)
 Novel RNAs along the Pathway of Gene Expression (or, The Expanding Universe of Small RNAs) Central Dogma DNA RNA Protein replication transcription translation Central Dogma DNA RNA Spliced RNA Protein
Novel RNAs along the Pathway of Gene Expression (or, The Expanding Universe of Small RNAs) Central Dogma DNA RNA Protein replication transcription translation Central Dogma DNA RNA Spliced RNA Protein
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Chapter 11. How Genes Are Controlled. Lectures by Edward J. Zalisko
 Chapter 11 How Genes Are Controlled PowerPoint Lectures for Campbell Essential Biology, Fifth Edition, and Campbell Essential Biology with Physiology, Fourth Edition Eric J. Simon, Jean L. Dickey, and
Chapter 11 How Genes Are Controlled PowerPoint Lectures for Campbell Essential Biology, Fifth Edition, and Campbell Essential Biology with Physiology, Fourth Edition Eric J. Simon, Jean L. Dickey, and
Appendix: Subcellular localization of MIG-14::GFP in C. elegans body wall muscle cells. Pei-Tzu Yang and Hendrik C. Korswagen
 Appendix: Subcellular localization of MIG-14::GFP in C. elegans body wall muscle cells Pei-Tzu Yang and Hendrik C. Korswagen 129 SUBCELLULAR LOCALIZATION OF MIG-14 Abstract MIG-14/Wls is a Wnt binding
Appendix: Subcellular localization of MIG-14::GFP in C. elegans body wall muscle cells Pei-Tzu Yang and Hendrik C. Korswagen 129 SUBCELLULAR LOCALIZATION OF MIG-14 Abstract MIG-14/Wls is a Wnt binding
micrornas Under the Microscope
 DEVELOPMENTAL DYNAMICS 235:846 853, 2006 WHAT S COOL IN DEV BIO micrornas Under the Microscope Julie C. Kiefer* Once regarded as a biological anomaly, micrornas (mirnas) have since been recognized as a
DEVELOPMENTAL DYNAMICS 235:846 853, 2006 WHAT S COOL IN DEV BIO micrornas Under the Microscope Julie C. Kiefer* Once regarded as a biological anomaly, micrornas (mirnas) have since been recognized as a
CONTRACTING ORGANIZATION: Cold Spring Harbor Laboratory Cold Spring Harbor, NY 11724
 AD Award Number: W81XWH-05-1-0256 TITLE: Dicer in Mammary Tumor Stem Cell Maintenance PRINCIPAL INVESTIGATOR: Elizabeth P. Murchison CONTRACTING ORGANIZATION: Cold Spring Harbor Laboratory Cold Spring
AD Award Number: W81XWH-05-1-0256 TITLE: Dicer in Mammary Tumor Stem Cell Maintenance PRINCIPAL INVESTIGATOR: Elizabeth P. Murchison CONTRACTING ORGANIZATION: Cold Spring Harbor Laboratory Cold Spring
MicroRNA function in animal development
 FEBS 29869 FEBS Letters xxx (2005) xxx xxx Minireview MicroRNA function in animal development Erno Wienholds, Ronald H.A. Plasterk * Hubrecht Laboratory, Uppsalalaan 8, 3584 CT Utrecht, The Netherlands
FEBS 29869 FEBS Letters xxx (2005) xxx xxx Minireview MicroRNA function in animal development Erno Wienholds, Ronald H.A. Plasterk * Hubrecht Laboratory, Uppsalalaan 8, 3584 CT Utrecht, The Netherlands
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10495 WWW.NATURE.COM/NATURE 1 2 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 3 4 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 5 6 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 7 8 WWW.NATURE.COM/NATURE
doi:10.1038/nature10495 WWW.NATURE.COM/NATURE 1 2 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 3 4 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 5 6 WWW.NATURE.COM/NATURE WWW.NATURE.COM/NATURE 7 8 WWW.NATURE.COM/NATURE
I) Development: tissue differentiation and timing II) Whole Chromosome Regulation
 Epigenesis: Gene Regulation Epigenesis : Gene Regulation I) Development: tissue differentiation and timing II) Whole Chromosome Regulation (X chromosome inactivation or Lyonization) III) Regulation during
Epigenesis: Gene Regulation Epigenesis : Gene Regulation I) Development: tissue differentiation and timing II) Whole Chromosome Regulation (X chromosome inactivation or Lyonization) III) Regulation during
Prediction of micrornas and their targets
 Prediction of micrornas and their targets Introduction Brief history mirna Biogenesis Computational Methods Mature and precursor mirna prediction mirna target gene prediction Summary micrornas? RNA can
Prediction of micrornas and their targets Introduction Brief history mirna Biogenesis Computational Methods Mature and precursor mirna prediction mirna target gene prediction Summary micrornas? RNA can
Marta Puerto Plasencia. microrna sponges
 Marta Puerto Plasencia microrna sponges Introduction microrna CircularRNA Publications Conclusions The most well-studied regions in the human genome belong to proteincoding genes. Coding exons are 1.5%
Marta Puerto Plasencia microrna sponges Introduction microrna CircularRNA Publications Conclusions The most well-studied regions in the human genome belong to proteincoding genes. Coding exons are 1.5%
Strategies to determine the biological function of micrornas
 Strategies to determine the biological function of micrornas Jan Krützfeldt, Matthew N Poy & Markus Stoffel MicroRNAs (mirnas) are regulators of gene expression that control many biological processes in
Strategies to determine the biological function of micrornas Jan Krützfeldt, Matthew N Poy & Markus Stoffel MicroRNAs (mirnas) are regulators of gene expression that control many biological processes in
SUPPLEMENTARY FIGURE LEGENDS
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Negative correlation between mir-375 and its predicted target genes, as demonstrated by gene set enrichment analysis (GSEA). 1 The correlation between
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Negative correlation between mir-375 and its predicted target genes, as demonstrated by gene set enrichment analysis (GSEA). 1 The correlation between
Identification of mirnas in Eucalyptus globulus Plant by Computational Methods
 International Journal of Pharmaceutical Science Invention ISSN (Online): 2319 6718, ISSN (Print): 2319 670X Volume 2 Issue 5 May 2013 PP.70-74 Identification of mirnas in Eucalyptus globulus Plant by Computational
International Journal of Pharmaceutical Science Invention ISSN (Online): 2319 6718, ISSN (Print): 2319 670X Volume 2 Issue 5 May 2013 PP.70-74 Identification of mirnas in Eucalyptus globulus Plant by Computational
Chapter 5 A Dose Dependent Screen for Modifiers of Kek5
 Chapter 5 A Dose Dependent Screen for Modifiers of Kek5 "#$ ABSTRACT Modifier screens in Drosophila have proven to be a powerful tool for uncovering gene interaction and elucidating molecular pathways.
Chapter 5 A Dose Dependent Screen for Modifiers of Kek5 "#$ ABSTRACT Modifier screens in Drosophila have proven to be a powerful tool for uncovering gene interaction and elucidating molecular pathways.
The genetics of heterochromatin. in metazoa. mutations by means of X-ray irradiation" "for the discovery of the production of
 The genetics of heterochromatin in metazoa 1 Hermann Joseph Muller 1946 Nobel Prize in Medicine: "for the discovery of the production of mutations by means of X-ray irradiation" 3 4 The true meaning of
The genetics of heterochromatin in metazoa 1 Hermann Joseph Muller 1946 Nobel Prize in Medicine: "for the discovery of the production of mutations by means of X-ray irradiation" 3 4 The true meaning of
Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles
 Research Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles Jianbin Wang, 1 Benjamin Czech, 2 Amanda Crunk, 1 Adam Wallace,
Research Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles Jianbin Wang, 1 Benjamin Czech, 2 Amanda Crunk, 1 Adam Wallace,
BIOLOGY. Regulation of Gene Expression CAMPBELL. Reece Urry Cain Wasserman Minorsky Jackson
 CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 18 Regulation of Gene Expression Lecture Presentation by Nicole Tunbridge and Kathleen Fitzpatrick 2014 Pearson Education, Inc.
CAMPBELL BIOLOGY TENTH EDITION Reece Urry Cain Wasserman Minorsky Jackson 18 Regulation of Gene Expression Lecture Presentation by Nicole Tunbridge and Kathleen Fitzpatrick 2014 Pearson Education, Inc.
Vertebrate Limb Patterning
 Vertebrate Limb Patterning What makes limb patterning an interesting/useful developmental system How limbs develop Key events in limb development positioning and specification initiation of outgrowth establishment
Vertebrate Limb Patterning What makes limb patterning an interesting/useful developmental system How limbs develop Key events in limb development positioning and specification initiation of outgrowth establishment
Genetic suppressors and enhancers provide clues to gene regulation and genetic pathways
 Genetic suppressors and enhancers provide clues to gene regulation and genetic pathways Suppressor mutation: a second mutation results in a less severe phenotype than the original mutation Suppressor mutations
Genetic suppressors and enhancers provide clues to gene regulation and genetic pathways Suppressor mutation: a second mutation results in a less severe phenotype than the original mutation Suppressor mutations
Neurotrophic factor GDNF and camp suppress glucocorticoid-inducible PNMT expression in a mouse pheochromocytoma model.
 161 Neurotrophic factor GDNF and camp suppress glucocorticoid-inducible PNMT expression in a mouse pheochromocytoma model. Marian J. Evinger a, James F. Powers b and Arthur S. Tischler b a. Department
161 Neurotrophic factor GDNF and camp suppress glucocorticoid-inducible PNMT expression in a mouse pheochromocytoma model. Marian J. Evinger a, James F. Powers b and Arthur S. Tischler b a. Department
doi: /j.bbrc
 doi: 10.1016/j.bbrc.2009.11.006 Fatty Acid Metabolism is Involved in Stress Resistance Mechanisms of Caenorhabditis elegans Makoto Horikawa a, b, Kazuichi Sakamoto a, 1 a Graduate School of Life and Environmental
doi: 10.1016/j.bbrc.2009.11.006 Fatty Acid Metabolism is Involved in Stress Resistance Mechanisms of Caenorhabditis elegans Makoto Horikawa a, b, Kazuichi Sakamoto a, 1 a Graduate School of Life and Environmental
FATTY ACID METABOLISM IN CAENORHABDITIS ELEGANS: CHARACTERIZATION OF THE DELTA 9 FATTY ACID DESATURASES AND IDENTIFICATION OF A KEY REGULATOR, NHR-80
 FATTY ACID METABOLISM IN CAENORHABDITIS ELEGANS: CHARACTERIZATION OF THE DELTA 9 FATTY ACID DESATURASES AND IDENTIFICATION OF A KEY REGULATOR, NHR-80 By TRISHA JANE BROCK A dissertation submitted in partial
FATTY ACID METABOLISM IN CAENORHABDITIS ELEGANS: CHARACTERIZATION OF THE DELTA 9 FATTY ACID DESATURASES AND IDENTIFICATION OF A KEY REGULATOR, NHR-80 By TRISHA JANE BROCK A dissertation submitted in partial
Qualifying Fat Storage Phenotypes Using Gas Chromatography in Caenorhabditis elegans
 University of Colorado, Boulder CU Scholar Undergraduate Honors Theses Honors Program Spring 2014 Qualifying Fat Storage Phenotypes Using Gas Chromatography in Caenorhabditis elegans Sabrina Fechtner University
University of Colorado, Boulder CU Scholar Undergraduate Honors Theses Honors Program Spring 2014 Qualifying Fat Storage Phenotypes Using Gas Chromatography in Caenorhabditis elegans Sabrina Fechtner University
MicroRNA in Cancer Karen Dybkær 2013
 MicroRNA in Cancer Karen Dybkær RNA Ribonucleic acid Types -Coding: messenger RNA (mrna) coding for proteins -Non-coding regulating protein formation Ribosomal RNA (rrna) Transfer RNA (trna) Small nuclear
MicroRNA in Cancer Karen Dybkær RNA Ribonucleic acid Types -Coding: messenger RNA (mrna) coding for proteins -Non-coding regulating protein formation Ribosomal RNA (rrna) Transfer RNA (trna) Small nuclear
Chapter 18. Regulation of Gene Expression. Lecture Outline. Overview: Conducting the Genetic Orchestra
 Chapter 18 Regulation of Gene Expression Lecture Outline Overview: Conducting the Genetic Orchestra Both prokaryotes and eukaryotes alter their patterns of gene expression in response to changes in environmental
Chapter 18 Regulation of Gene Expression Lecture Outline Overview: Conducting the Genetic Orchestra Both prokaryotes and eukaryotes alter their patterns of gene expression in response to changes in environmental
Alternative RNA processing: Two examples of complex eukaryotic transcription units and the effect of mutations on expression of the encoded proteins.
 Alternative RNA processing: Two examples of complex eukaryotic transcription units and the effect of mutations on expression of the encoded proteins. The RNA transcribed from a complex transcription unit
Alternative RNA processing: Two examples of complex eukaryotic transcription units and the effect of mutations on expression of the encoded proteins. The RNA transcribed from a complex transcription unit
General Biology 1004 Chapter 11 Lecture Handout, Summer 2005 Dr. Frisby
 Slide 1 CHAPTER 11 Gene Regulation PowerPoint Lecture Slides for Essential Biology, Second Edition & Essential Biology with Physiology Presentation prepared by Chris C. Romero Neil Campbell, Jane Reece,
Slide 1 CHAPTER 11 Gene Regulation PowerPoint Lecture Slides for Essential Biology, Second Edition & Essential Biology with Physiology Presentation prepared by Chris C. Romero Neil Campbell, Jane Reece,
Supplementary information for: Human micrornas co-silence in well-separated groups and have different essentialities
 Supplementary information for: Human micrornas co-silence in well-separated groups and have different essentialities Gábor Boross,2, Katalin Orosz,2 and Illés J. Farkas 2, Department of Biological Physics,
Supplementary information for: Human micrornas co-silence in well-separated groups and have different essentialities Gábor Boross,2, Katalin Orosz,2 and Illés J. Farkas 2, Department of Biological Physics,
A. Incorrect! All the cells have the same set of genes. (D)Because different types of cells have different types of transcriptional factors.
 Genetics - Problem Drill 21: Cytogenetics and Chromosomal Mutation No. 1 of 10 1. Why do some cells express one set of genes while other cells express a different set of genes during development? (A) Because
Genetics - Problem Drill 21: Cytogenetics and Chromosomal Mutation No. 1 of 10 1. Why do some cells express one set of genes while other cells express a different set of genes during development? (A) Because
Gene Regulation. Bacteria. Chapter 18: Regulation of Gene Expression
 Chapter 18: Regulation of Gene Expression A Biology 2013 1 Gene Regulation rokaryotes and eukaryotes alter their gene expression in response to their changing environment In multicellular eukaryotes, gene
Chapter 18: Regulation of Gene Expression A Biology 2013 1 Gene Regulation rokaryotes and eukaryotes alter their gene expression in response to their changing environment In multicellular eukaryotes, gene
Epigenetics: The Future of Psychology & Neuroscience. Richard E. Brown Psychology Department Dalhousie University Halifax, NS, B3H 4J1
 Epigenetics: The Future of Psychology & Neuroscience Richard E. Brown Psychology Department Dalhousie University Halifax, NS, B3H 4J1 Nature versus Nurture Despite the belief that the Nature vs. Nurture
Epigenetics: The Future of Psychology & Neuroscience Richard E. Brown Psychology Department Dalhousie University Halifax, NS, B3H 4J1 Nature versus Nurture Despite the belief that the Nature vs. Nurture
Supplementary Figures
 J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
7.013 Problem Set 5 FRIDAY April 2nd, 2004
 MIT Department of Biology 7.013: Introductory Biology - Spring 2004 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel 7.013 Problem Set 5 FRIDAY April 2nd, 2004 Problem sets
MIT Department of Biology 7.013: Introductory Biology - Spring 2004 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel 7.013 Problem Set 5 FRIDAY April 2nd, 2004 Problem sets
Alternative Splicing Regulation During C. elegans Development: Splicing Factors as Regulated Targets
 Alternative Splicing Regulation During C. elegans Development: Splicing Factors as Regulated Targets Sergio Barberan-Soler 1,2, Alan M. Zahler 1,2 * 1 Department of MCD Biology, University of California
Alternative Splicing Regulation During C. elegans Development: Splicing Factors as Regulated Targets Sergio Barberan-Soler 1,2, Alan M. Zahler 1,2 * 1 Department of MCD Biology, University of California
Report The Egg Surface LDL Receptor Repeat-Containing Proteins EGG-1 and EGG-2 Are Required for Fertilization in Caenorhabditis elegans
 Current Biology, Vol. 15, 2222 2229, December 20, 2005, ª2005 Elsevier Ltd All rights reserved. DOI 10.1016/j.cub.2005.10.043 Report The Egg Surface LDL Receptor Repeat-Containing Proteins EGG-1 and EGG-2
Current Biology, Vol. 15, 2222 2229, December 20, 2005, ª2005 Elsevier Ltd All rights reserved. DOI 10.1016/j.cub.2005.10.043 Report The Egg Surface LDL Receptor Repeat-Containing Proteins EGG-1 and EGG-2
MicroRNA-mediated incoherent feedforward motifs are robust
 The Second International Symposium on Optimization and Systems Biology (OSB 8) Lijiang, China, October 31 November 3, 8 Copyright 8 ORSC & APORC, pp. 62 67 MicroRNA-mediated incoherent feedforward motifs
The Second International Symposium on Optimization and Systems Biology (OSB 8) Lijiang, China, October 31 November 3, 8 Copyright 8 ORSC & APORC, pp. 62 67 MicroRNA-mediated incoherent feedforward motifs
Chapter 18 Regulation of Gene Expression
 Chapter 18 Regulation of Gene Expression Differential Expression of Genes Prokaryotes and eukaryotes precisely regulate gene expression in response to environmental conditions In multicellular eukaryotes,
Chapter 18 Regulation of Gene Expression Differential Expression of Genes Prokaryotes and eukaryotes precisely regulate gene expression in response to environmental conditions In multicellular eukaryotes,
The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein
 THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
V16: involvement of micrornas in GRNs
 What are micrornas? V16: involvement of micrornas in GRNs How can one identify micrornas? What is the function of micrornas? Elisa Izaurralde, MPI Tübingen Huntzinger, Izaurralde, Nat. Rev. Genet. 12,
What are micrornas? V16: involvement of micrornas in GRNs How can one identify micrornas? What is the function of micrornas? Elisa Izaurralde, MPI Tübingen Huntzinger, Izaurralde, Nat. Rev. Genet. 12,
Gene Regulation Part 2
 Michael Cummings Chapter 9 Gene Regulation Part 2 David Reisman University of South Carolina Other topics in Chp 9 Part 2 Protein folding diseases Most diseases are caused by mutations in the DNA that
Michael Cummings Chapter 9 Gene Regulation Part 2 David Reisman University of South Carolina Other topics in Chp 9 Part 2 Protein folding diseases Most diseases are caused by mutations in the DNA that
Paternal RNA contributions in the C. elegans zygote
 Manuscript EMBO-2014-88117 Paternal RNA contributions in the C. elegans zygote Marlon Stoeckius, Dominic Grün and Nikolaus Rajewsky Corresponding author: Nikolaus Rajewsky, Max Delbrueck Centrum fuer Molekulare
Manuscript EMBO-2014-88117 Paternal RNA contributions in the C. elegans zygote Marlon Stoeckius, Dominic Grün and Nikolaus Rajewsky Corresponding author: Nikolaus Rajewsky, Max Delbrueck Centrum fuer Molekulare
collection micrornas Unlocking the Secrets of Sponsored by This booklet brought to you by the AAAS/Science Business Office
 collection Unlocking the Secrets of micrornas Sponsored by This booklet brought to you by the AAAS/Science Business Office We re pushing the frontiers of microrna analysis, so you can accelerate discoveries.
collection Unlocking the Secrets of micrornas Sponsored by This booklet brought to you by the AAAS/Science Business Office We re pushing the frontiers of microrna analysis, so you can accelerate discoveries.
Figure S1. Molecular confirmation of the precise insertion of the AsMCRkh2 cargo into the kh w locus.
 Supporting Information Appendix Table S1. Larval and adult phenotypes of G 2 progeny of lines 10.1 and 10.2 G 1 outcrosses to wild-type mosquitoes. Table S2. List of oligonucleotide primers. Table S3.
Supporting Information Appendix Table S1. Larval and adult phenotypes of G 2 progeny of lines 10.1 and 10.2 G 1 outcrosses to wild-type mosquitoes. Table S2. List of oligonucleotide primers. Table S3.
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
RNAi Restricts Virus Infections
 RNAi Restricts Virus Infections RNA silencing is a new immunity mechanism that protects fruit flies, mosquitoes, and nematodes as well as plants against viral infections Shou-Wei Ding Shou-Wei Ding is
RNAi Restricts Virus Infections RNA silencing is a new immunity mechanism that protects fruit flies, mosquitoes, and nematodes as well as plants against viral infections Shou-Wei Ding Shou-Wei Ding is
ns ns hp761(lf); daf-28(lf) daf-28(gf) Figure S1
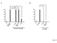 A ns ns B 100 100 80 80 60 60 40 40 20 20 0 0% 0 0% hp761(lf); daf-28(lf) daf-28(gf) Figure S1 A 100 80 15 o C 22 o C 25 o C % Dauers 60 40 20 0 0% 0% * 0% 0% 0% Class I alleles Class II alleles daf-2(lf;ts)
A ns ns B 100 100 80 80 60 60 40 40 20 20 0 0% 0 0% hp761(lf); daf-28(lf) daf-28(gf) Figure S1 A 100 80 15 o C 22 o C 25 o C % Dauers 60 40 20 0 0% 0% * 0% 0% 0% Class I alleles Class II alleles daf-2(lf;ts)
CONTRACTING ORGANIZATION: Cold Spring Harbor Laboratory Cold Spring Harbor, NY 11724
 AD Award Number: W81XWH-06-1-0249 TITLE: Detection of genes modifying sensitivity to proteasome inhibitors using a shrna Library in Breast Cancer PRINCIPAL INVESTIGATOR: Gregory J. Hannon, Ph.D. CONTRACTING
AD Award Number: W81XWH-06-1-0249 TITLE: Detection of genes modifying sensitivity to proteasome inhibitors using a shrna Library in Breast Cancer PRINCIPAL INVESTIGATOR: Gregory J. Hannon, Ph.D. CONTRACTING
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature19357 Figure 1a Chd8 +/+ Chd8 +/ΔSL Chd8 +/+ Chd8 +/ΔL E10.5_Whole brain E10.5_Whole brain E10.5_Whole brain E14.5_Whole brain E14.5_Whole brain E14.5_Whole
SUPPLEMENTARY INFORMATION doi:10.1038/nature19357 Figure 1a Chd8 +/+ Chd8 +/ΔSL Chd8 +/+ Chd8 +/ΔL E10.5_Whole brain E10.5_Whole brain E10.5_Whole brain E14.5_Whole brain E14.5_Whole brain E14.5_Whole
CRS4 Seminar series. Inferring the functional role of micrornas from gene expression data CRS4. Biomedicine. Bioinformatics. Paolo Uva July 11, 2012
 CRS4 Seminar series Inferring the functional role of micrornas from gene expression data CRS4 Biomedicine Bioinformatics Paolo Uva July 11, 2012 Partners Pharmaceutical company Fondazione San Raffaele,
CRS4 Seminar series Inferring the functional role of micrornas from gene expression data CRS4 Biomedicine Bioinformatics Paolo Uva July 11, 2012 Partners Pharmaceutical company Fondazione San Raffaele,
Bio 401 Sp2014. Multiple Choice (Circle the letter corresponding to the correct answer)
 MIDTERM EXAM KEY (60 pts) You should have 5 pages. Please put your name on every page. You will have 70 minutes to complete the exam. You may begin working as soon as you receive the exam. NOTE: the RED
MIDTERM EXAM KEY (60 pts) You should have 5 pages. Please put your name on every page. You will have 70 minutes to complete the exam. You may begin working as soon as you receive the exam. NOTE: the RED
