SUPPLEMENTARY FIGURE S1: nlp-22 is expressed in the RIA interneurons and is secreted. (a) An animal expressing both the RIA specific reporter
|
|
|
- Melina Pitts
- 5 years ago
- Views:
Transcription
1 1 SUPPLEMENTARY FIGURE S1: nlp-22 is expressed in the RIA interneurons and is secreted. (a) An animal expressing both the RIA specific reporter Pglr-3:mCherry (red) and Pnlp-22:gfp (green) shows co-localization in the RIA neurons (yellow). Only one of the pair of the RIA neurons is shown in this focal plane. (b) A Translational reporter in which gfp coding sequence replaces the nlp-22 stop codon shows GFP spread to other neurons in the head (arrows, left panel), and in the ventral nerve cord (arrows, right panel). Anterior is to the left. Bright mcherry fluorescence in the pharynx, which was used as a marker for transgenesis, shows a signal in the gfp channel (arrow head). (c) A translational reporter in which gfp coding sequence was placed at the N-terminus of the NLP protein prior to the signal sequence shows GFP concentrated at the perimeter of the RIA neurons, suggesting that the GFP is interfering with secretion, and now marks the RIA plasma membranes. The transgenic animal is also marked with a red pharynx. Scale Bars = 5µM. 1
2 2 SUPPLEMENTARY FIGURE S2: Over-expression of Neuromedin S (17-33) does not induce behavioral quiescence. a) NLP-22 and human NMS-17, which consists of the last 17 amino acids of the full length NMS, are similar at the primary amino acid (top) and structural (bottom) levels. The conserved GR dipeptide and FRP tripeptide motifs are shown in white. b) Overexpression of NMS-17 in the context of the C. elegans nlp-22 preproprotein using an inducible heat-shock promoter does not result in an inhibition of pharyngeal pumping (Student s twotailed t-test; Error bars represent s.e.m, N 20). 2
3 3 Strain Genotype Array Pumps per 20 Sec N N2 WT ± TJ375 WT gpis1[phsp-16.2:gfp] 91.3 ± NQ216 unc-119(ed3) qnex95[phsp-16.2:nlp-22,pmyo-2:mcherry, unc-119(+)] 8.4 ± NQ251 WT qnis142[phsp-16.2:nlp-22;phsp-16.2:gfp; Pmyo-2:mCherry; unc-119(+)] 6.2 ± NQ670 WT qnex95[phsp-16.2:nlp-22;pmyo-2:mcherry; unc-119(+)] 12.8 ± DA509 unc-31(e928)iv ± NQ256 unc-31(e928)iv qnex ± 4.9* 25 KP1873 egl-3(nu349)v ± NQ319 egl-3(nu349)v qnex ± 11.3* 24 KP2018 egl-21(n476)iv ± NQ320 egl-21(n476)iv qnex ± 5.8* 36 MT1074 egl-4(n479)iv ± NQ321 egl-4(n479)iv qnex ± 14.0* 32 NQ668 egl-4(n479)iv qnis ± 10.2* 30 IB16 ceh-17(np1)i ± NQ230 ceh-17(np1)i qnex ± 9.8* 10 KG532 kin-2(ce179)x ± NQ605 kin-2(ce179)x qnis ± 23.1* 25 NQ667 kin-2(ce179)x qnex ± 12.8* 20 RB1288 nmur-1(ok1387)x ± NQ274 nmur-1(ok1387)x qnex ± 12.9* 10 RB2526 nmur-2(ok3502)ii ± NQ285 nmur-2(ok3502)ii qnex ± 19.5* 20 VC1974 nmur-3(ok2295)x ± NQ275 nmur-3(ok2295)x qnex ± 5.4* 10 RB1284 nmur-4(ok1381)i - 76 ± NQ235 nmur-4(ok1381)i qnex ± 6.5* 7 Supplementary Table S1: nlp-22 over-expression analyses. nlp-22 over-expression reduces pharyngeal pumping rates in all genetic backgrounds tested, including those previously implicated in regulating quiescence and those implicated in processing some neuropeptides. Shown are the average pumping rates with standard deviations. *P<.005, 2-tailed Wilcoxon Rank Sum test, comparing the mutant over-expressing nlp-22 to mutant alone. 3
4 4 Strain Genotype Transgene Array Total Peak Fraction L4 lethargus End of L3L to Start Quiescence (min) Quiescence Duration (min) of L4L (hr) N N2 WT ± ± ± ± 0.27^ 10 NQ596 nlp-22(gk509904)x ± 5.4** 0.67 ± 0.04* ± 29.6** ± 0.72^ 10 NQ596 nlp-22(gk509904)x ± ± ± NQ603 nlp-22(gk509904)x qnex311[nlp-22(+);myo:cherry;unc-119(+) ] 81.3 ± 9.0** 0.68 ± ± 16.1** - 11 NQ305 sid-1(pk3321)v qnis137[dpy-7:nlp-22(rnai);myo-2>mcherry;unc-119(+)] ± ± ± NQ376 sid-1(pk3321)v qnis157[glr-3:nlp-22(rnai);myo-2:mcherry;unc-119(+)] 61.5 ± 5.4** 0.67 ± 0.15* ± 34.9* - 11 NQ156 lin-15(n765ts)x qnex48[pins-4:gfp;lin-15(+)] ± ± ± VM1345 lin-15(n765ts)x (2 RIAs DEAD) akex211[pglr-3:gfp;pglr-3:ice;lin-15(+)] 62.4 ± 8.9** 0.66 ± 0.06* ± 17.2* - 9 NQ156 lin-15(n765ts)x qnex48[pins-4:gfp;lin-15(+)] ± ± ± VM1345 lin-15(n765ts)x (1 RIA DEAD) akex211[pglr-3:gfp;pglr-3:ice;lin-15(+)] 96.7 ± 9.7* 0.83 ± ± Supplementary Table S2: Quiescence measurements. (Student s two-tailed t-test. **p<0.005;*p<0.05) Data is from 8 worms only since 2 of the 10 worms were monitored starting only in L4 larval stage. Total quiescence and peak fraction quiescence measurements shown are the average ± standard error of the mean. Lethargus duration and duration between L3 and L4 lethargus measurements are the average ± standard deviation. Measurements were made in a paired fashion. Therefore, there are two rows describing nlp-22(gk509904) data, one paired with simultaneously recorded wild-type data and one paired with simultaneously recorded data from nlp-22(gk509904); qnex311. Data from paired experiments is listed in consecutive rows shaded in the same color (gray or white). The strain NQ156, which was used as a lin-15 transgenic control for VM1345, carries a transgene array with a GFP reporter expressed in a small set of neurons. 4
5 5 Oligo Name Description onq366 onq367 onq368 onq372 onq373 For- nlp-22 - Adds Heat Shock Promoter tail Rev - Past 3 UTR of nlp-22 Nested to onq367 For - HSPr from Fire Vector ppd49.83 Nested to onq372 onq374 Rev- HSPr from Fire Vector ppd49.83 onq416 onq417 onq470 onq471 onq472 onq473 onq541 onq646 onq647 onq711 onq712 onq713 onq714 onq715 onq716 onq717 onq718 0NQ556 onq557 onq558 onq559 onq533 onq534 onq535 onq536 onq615 onq616 onq641 onq642 For - nlp-22 promoter Nested to onq416 Rev - Before stop codon - Adds GFP tail For - GFP in ppd Adds nlp-22 tail Rev - In unc-54 3 UTR in ppd95.75 Nested to onq472 Sequence Phsp-16.2:nlp-22 CTTCGAAAATCCTCATCGGATCCATGCGTTCCATAATCGTC GTGAAGTAGCGCCAGTG AAGACATGGTTATGGCAC GCCAAGCTTGCATGCCTG CAGGTCGACTCTAGAGG GGATCCGATGAGGATTTTC Pnlp-22:nlp-22::gfp TATCAGTCGTCAAGGATTCCG TCAATGCCATTTGCAGAGAG AAAAGTTCTTCTCCTTTACTCATTACTCCGATTGGGAATCCAGTT AACTGGATTCCCAATCGGAGTAATGAGTAAAGGAGAAGAACTTTT AAAAGAAGCTAAAAAACAAAGAAATTA GAGAAGTTTTTTGATAATAACAAAAATAGG Mutated versions of Phsp-16.2:nlp-22 (Each pair ( ) was used with onq366 and onq367, respectively) For- Used with onq367 Rev - Mutates KR to AA For - Mutates KR to AA For - FRPG to MRPG Rev - FRPG to MRPG For - FRPG to FEPG Rev - FRPG to FEPG For - FRPG to FREG Rev - FRPG to FREG For - FRPG to FRPE Rev - FRPG to FRPE For - GFP in ppd Used with onq559 Rev - nlp-22 promoter - Used with onq416 For - nlp-22 - Used with onq367 Rev - GFP (before stop) For - nlp-22 coding sequence for RNAi Rev - nlp-22 coding sequence for RNAi Rev - Nested to onq534 for sense - with 616 Rev - Nested to onq534 for anti - with 616 For - glr-3 promoter Nested to 615 Rev - glr-3 promoter - Adds sense tail Rev - glr-3 promoter - Adds antisense tail AAATCTTCAAACTATAATCATGCAGACCTCAGCTCTTGGGCTTCAG CGTTGTTGCTGCCCCTGGACGGAATCCGGCTCG CAGGGGCAGCAACAACGGACGAACTAACTGGATT ATGCGTCCAGGGAAACGAACAACG TTGTTCGTTTCCCTGGACGCATTCCGGCTCGCCCAATCGCAATC TCGAACCAGGGAAACGAACAACGGAC GTTTCCCTGGTTCGAATCCGGCTCGCCCAATCGCA CGTGAAGGGAAACGAACAACGGACGAA GTTGTTCGTTTCCCTTCACGGAATCCGGCTCGCCCAAT TCCAGAAAAACGAACAACGGACGAACTA CGTTGTTCGTTTTTCTGGACGGAATCCGGCTCGCCC Pnlp-22:gfp::nlp-22 TTTCCCAACTCGGAAATGAGTAAAGGAGAAGAACTT AAAAGTTCTTCTCCTTTACTCATTTCCGAGTTGGGAAAGTTCGAG ATGGCATGGATGAACTATACAAAATGCGTTCCATAATCGTCTTC ATGAAGACGATTATGGAACGCATTTTGTATAGTTCATCCATGCC Pglr-3:nlp-22(RNAi) TTAGCTTTACACAATGTTAAAGGAAAA AACAATTAACATCAGAAAATTCATACT ATTAACATCAGAAAATTCATACTCCG GCTTTACACAATGTTAAAGGAAAAG CTTCAATCTTCAAAAAAGGGCATT CTTCAAAAAAGGGCATTAAAAACAGT TTTTCCTTTAACATTGTGTAAAGCTAAAATCCAGAACATATGTTAATAGCAAA AGTATGAATTTTCTGATGTTAATTGTTAATCCAGAACATATGTTAATAGCAAA Pglr-3:mCherry 5
6 6 onq856 Rev - glr-3 promoter - used with 615 onq857 For - mcherry from pcfj90 - used with 472 onq567 For - dpy-7 promoter onq568 Nested to 567 onq569 Rev - dpy-7 promoter - Adds sense tail onq570 Rev - dpy-7 promoter - Adds antisense tail onq648 Rev - GFP - Adds nlp-22 intron tail - with 557 onq649 For - Intron nlp-22 -Adds gfp tail - with 650 onq650 Rev - intron nlp-22 -Adds gfp tail onq651 For - GFP - Adds nlp-22 intron tail - with 472 onq617 Rev - glr-3 promoter ChR2 tail - with 615 onq618 For - ChR2 glr-3 tail - with 472 onq552 Forward onq534 Reverse onq1167 Rev - engineers NMS(17-33) in nlp-22 - with 366 onq1168 For - engineers NMS(17-33) in nlp22 - with 367 Probe Primer 1 Primer 2 GAAAAGTTCTTCTCCTTTACTCATAATCGCAATCGACTTTTTCATGAT CTTTTTTGTACAAACTTGTCATGAACATATGTTAATAGCAAATATT Pdpy-7:nlp-22(RNAi) CAGCAGCGGTAACGGAGACAT GGTAACGGAGACATCAGTGACCT TTTTCCTTTAACATTGTGTAAAGCTAATTATCTGGAACAAAATGTAAGAATA AGTATGAATTTTCTGATGTTAATTGTTTTATCTGGAACAAAATGTAAGAATA Pnlp-22(w/Intron):gfp TTGTGTAAAGCTAACGTGTCTTGTAGTTCCCGTCAT GGGAACTACAAGACACGTTAGCTTTACACAATGTTA ACCTGAAATTACGGTGCTTGTTG ACCGTAATTTCAGGTGCTGAAGTCAAGTTTGAAG Pglr-3:ChannelRhodopsin-2 CAGGGCGCCTCCATAATCCATAATCCAGAACATATGTTAATAGCAAA AATATTTGCTATTAACATATGTTCTGGATTATGGATTATGGAGGCGCCCTG Sequencing gk GGATTGACGATCTTCGCGTTG AACAATTAACATCAGAAAATTCATACT Phsp-16.2:Human NMS(17-33) AAGGAAGAATGGGCGTCCCCAGGTGGCGGTGTGGTCCTTTTTCATGATTGGAAGGGTTCG ACCTGGGGACGCCCATTCTTCCTTTTCCGCCCACGTAACGGGAAACGAACAACGGACGAA nlp-22 qpcr /56-FAM/AAGCGCCCC/ZEN/TCACGAGTGT/3IABkFQ/ ATCCTTCACCGAGCAAACTAC ATGATTGGAAGGGTTCGGAG Supplementary Table S3: Oligonucleotides used in this study. DNA constructs were made using overlapextension PCR 48. Mutated versions of nlp-22 were made by PCR and then TOPO cloned into a pcr 2.1- TOPO TA Vector (Invitrogen TM ). They were sequenced to verify that the intended mutation was successfully introduced and that no additional mutations were introduced. 6
Supplementary Figure 1. Gene schematics of hyls-1, gasr-8 and k10g6.4, and TEM analysis of TFs in WT and hyls-1 cilia. (a) Gene structure of hyls-1,
 Supplementary Figure 1. Gene schematics of hyls-1, gasr-8 and k10g6.4, and TEM analysis of TFs in WT and hyls-1 cilia. (a) Gene structure of hyls-1, gasr-8 and k10g6.4 based on WormBase (http://wormbase.org),
Supplementary Figure 1. Gene schematics of hyls-1, gasr-8 and k10g6.4, and TEM analysis of TFs in WT and hyls-1 cilia. (a) Gene structure of hyls-1, gasr-8 and k10g6.4 based on WormBase (http://wormbase.org),
Supplementary Information
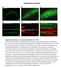 Supplementary Information Supplementary Figure 1: Luminal localization of CCM-3. (a) The CCM-3::GFP fusion protein localizes along the apical (luminal) surface of the pharynx (b) as well as the lumen of
Supplementary Information Supplementary Figure 1: Luminal localization of CCM-3. (a) The CCM-3::GFP fusion protein localizes along the apical (luminal) surface of the pharynx (b) as well as the lumen of
EGFR shrna A: CCGGCGCAAGTGTAAGAAGTGCGAACTCGAGTTCGCACTTCTTACACTTGCG TTTTTG. EGFR shrna B: CCGGAGAATGTGGAATACCTAAGGCTCGAGCCTTAGGTATTCCACATTCTCTT TTTG
 Supplementary Methods Sequence of oligonucleotides used for shrna targeting EGFR EGFR shrna were obtained from the Harvard RNAi consortium. The following oligonucleotides (forward primer) were used to
Supplementary Methods Sequence of oligonucleotides used for shrna targeting EGFR EGFR shrna were obtained from the Harvard RNAi consortium. The following oligonucleotides (forward primer) were used to
ns ns hp761(lf); daf-28(lf) daf-28(gf) Figure S1
 A ns ns B 100 100 80 80 60 60 40 40 20 20 0 0% 0 0% hp761(lf); daf-28(lf) daf-28(gf) Figure S1 A 100 80 15 o C 22 o C 25 o C % Dauers 60 40 20 0 0% 0% * 0% 0% 0% Class I alleles Class II alleles daf-2(lf;ts)
A ns ns B 100 100 80 80 60 60 40 40 20 20 0 0% 0 0% hp761(lf); daf-28(lf) daf-28(gf) Figure S1 A 100 80 15 o C 22 o C 25 o C % Dauers 60 40 20 0 0% 0% * 0% 0% 0% Class I alleles Class II alleles daf-2(lf;ts)
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/8/e1701143/dc1 Supplementary Materials for Impaired DNA replication derepresses chromatin and generates a transgenerationally inherited epigenetic memory Adam
advances.sciencemag.org/cgi/content/full/3/8/e1701143/dc1 Supplementary Materials for Impaired DNA replication derepresses chromatin and generates a transgenerationally inherited epigenetic memory Adam
Concentration memory-dependent synaptic plasticity of a taste circuit regulates salt chemotaxis in Caenorhabditis elegans
 Supplementary Information for: Concentration memory-dependent synaptic plasticity of a taste circuit regulates salt chemotaxis in Caenorhabditis elegans Hirofumi Kunitomo, Hirofumi Sato, Ryo Iwata, Yohsuke
Supplementary Information for: Concentration memory-dependent synaptic plasticity of a taste circuit regulates salt chemotaxis in Caenorhabditis elegans Hirofumi Kunitomo, Hirofumi Sato, Ryo Iwata, Yohsuke
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated
 Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Phenomena first observed in petunia
 Vectors for RNAi Phenomena first observed in petunia Attempted to overexpress chalone synthase (anthrocyanin pigment gene) in petunia. (trying to darken flower color) Caused the loss of pigment. Bill Douherty
Vectors for RNAi Phenomena first observed in petunia Attempted to overexpress chalone synthase (anthrocyanin pigment gene) in petunia. (trying to darken flower color) Caused the loss of pigment. Bill Douherty
Normal sleep bouts are not essential for C. elegans survival and FoxO is important for compensatory changes in sleep
 Bennett et al. BMC Neurosci (218) 19:1 https://doi.org/1.1186/s12868-18-48-1 BMC Neuroscience RESEARCH ARTICLE Open Access Normal sleep bouts are not essential for C. elegans survival and FoxO is important
Bennett et al. BMC Neurosci (218) 19:1 https://doi.org/1.1186/s12868-18-48-1 BMC Neuroscience RESEARCH ARTICLE Open Access Normal sleep bouts are not essential for C. elegans survival and FoxO is important
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid.
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
Supplementary Table 1. List of primers used in this study
 Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
Biology Open (2014) 000, 1 10 doi: /bio
 (2014) 000, 1 10 doi:10.1242/bio.201410041 Supplementary Material Michael Brauchle et al. doi: 10.1242/bio.201410041 Fig. S1. Alignment of GFP, sfgfp, egfp, eyfp, mcherry and mruby2. Sequence-based alignment
(2014) 000, 1 10 doi:10.1242/bio.201410041 Supplementary Material Michael Brauchle et al. doi: 10.1242/bio.201410041 Fig. S1. Alignment of GFP, sfgfp, egfp, eyfp, mcherry and mruby2. Sequence-based alignment
Nature Neuroscience: doi: /nn Supplementary Figure 1. Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites.
 Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
100 mm Sucrose. +Berberine +Quinine
 8 mm Sucrose Probability (%) 7 6 5 4 3 Wild-type Gr32a / 2 +Caffeine +Berberine +Quinine +Denatonium Supplementary Figure 1: Detection of sucrose and bitter compounds is not affected in Gr32a / flies.
8 mm Sucrose Probability (%) 7 6 5 4 3 Wild-type Gr32a / 2 +Caffeine +Berberine +Quinine +Denatonium Supplementary Figure 1: Detection of sucrose and bitter compounds is not affected in Gr32a / flies.
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11306 Supplementary Figures Supplementary Figure 1. Basic characterization of GFP+ RGLs in the dentate gyrus of adult nestin-gfp mice. a, Sample confocal images
SUPPLEMENTARY INFORMATION doi:10.1038/nature11306 Supplementary Figures Supplementary Figure 1. Basic characterization of GFP+ RGLs in the dentate gyrus of adult nestin-gfp mice. a, Sample confocal images
Supplementary Figure 1) GABAergic enhancement by leptin hyperpolarizes POMC neurons A) Representative recording samples showing the membrane
 Supplementary Figure 1) GABAergic enhancement by leptin hyperpolarizes POMC neurons A) Representative recording samples showing the membrane potential recorded from POMC neurons following treatment with
Supplementary Figure 1) GABAergic enhancement by leptin hyperpolarizes POMC neurons A) Representative recording samples showing the membrane potential recorded from POMC neurons following treatment with
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
Nature Genetics: doi: /ng Supplementary Figure 1. Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms.
 Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Nature Neuroscience: doi: /nn Supplementary Figure 1. Lick response during the delayed Go versus No-Go task.
 Supplementary Figure 1 Lick response during the delayed Go versus No-Go task. Trial-averaged lick rate was averaged across all mice used for pyramidal cell imaging (n = 9). Different colors denote different
Supplementary Figure 1 Lick response during the delayed Go versus No-Go task. Trial-averaged lick rate was averaged across all mice used for pyramidal cell imaging (n = 9). Different colors denote different
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus
 Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons
 Supplemental Material mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons Antoine de Chevigny, Nathalie Coré, Philipp Follert, Marion Gaudin, Pascal
Supplemental Material mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons Antoine de Chevigny, Nathalie Coré, Philipp Follert, Marion Gaudin, Pascal
Supplementary Figure 1 hlrrk2 promotes CAP dependent protein translation.
 ` Supplementary Figure 1 hlrrk2 promotes CAP dependent protein translation. (a) Overexpression of hlrrk2 in HeLa cells enhances total protein synthesis in [35S] methionine/cysteine incorporation assays.
` Supplementary Figure 1 hlrrk2 promotes CAP dependent protein translation. (a) Overexpression of hlrrk2 in HeLa cells enhances total protein synthesis in [35S] methionine/cysteine incorporation assays.
293T cells were transfected with indicated expression vectors and the whole-cell extracts were subjected
 SUPPLEMENTARY INFORMATION Supplementary Figure 1. Formation of a complex between Slo1 and CRL4A CRBN E3 ligase. (a) HEK 293T cells were transfected with indicated expression vectors and the whole-cell
SUPPLEMENTARY INFORMATION Supplementary Figure 1. Formation of a complex between Slo1 and CRL4A CRBN E3 ligase. (a) HEK 293T cells were transfected with indicated expression vectors and the whole-cell
Supplementary Information
 Supplementary Information Supplementary s Supplementary 1 All three types of foods suppress subsequent feeding in both sexes when the same food is used in the pre-feeding test feeding. (a) Adjusted pre-feeding
Supplementary Information Supplementary s Supplementary 1 All three types of foods suppress subsequent feeding in both sexes when the same food is used in the pre-feeding test feeding. (a) Adjusted pre-feeding
Supplementary Figure 1
 Count Count Supplementary Figure 1 Coverage per amplicon for error-corrected sequencing experiments. Errorcorrected consensus sequence (ECCS) coverage was calculated for each of the 568 amplicons in the
Count Count Supplementary Figure 1 Coverage per amplicon for error-corrected sequencing experiments. Errorcorrected consensus sequence (ECCS) coverage was calculated for each of the 568 amplicons in the
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Differential expression of mirnas from the pri-mir-17-92a locus.
 Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
Supplementary figure 1: LII/III GIN-cells show morphological characteristics of MC
 1 2 1 3 Supplementary figure 1: LII/III GIN-cells show morphological characteristics of MC 4 5 6 7 (a) Reconstructions of LII/III GIN-cells with somato-dendritic compartments in orange and axonal arborizations
1 2 1 3 Supplementary figure 1: LII/III GIN-cells show morphological characteristics of MC 4 5 6 7 (a) Reconstructions of LII/III GIN-cells with somato-dendritic compartments in orange and axonal arborizations
Supplementary Fig. 1 V-ATPase depletion induces unique and robust phenotype in Drosophila fat body cells.
 Supplementary Fig. 1 V-ATPase depletion induces unique and robust phenotype in Drosophila fat body cells. a. Schematic of the V-ATPase proton pump macro-complex structure. The V1 complex is composed of
Supplementary Fig. 1 V-ATPase depletion induces unique and robust phenotype in Drosophila fat body cells. a. Schematic of the V-ATPase proton pump macro-complex structure. The V1 complex is composed of
Supplemental Figure 1
 1 Supplemental Figure 1 Effects of DATE shortening on HGF promoter activity. The HGF promoter region (-1037 to +56) containing wild-type (30As) or truncated DATE (26As, 27As, 28A, 29As) from breast cancer
1 Supplemental Figure 1 Effects of DATE shortening on HGF promoter activity. The HGF promoter region (-1037 to +56) containing wild-type (30As) or truncated DATE (26As, 27As, 28A, 29As) from breast cancer
Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent Protein.
 prfp-vector RFP Exon1 Intron RFP Exon2 prfp-mir-124 mir-93/124 RFP Exon1 Intron RFP Exon2 Untransfected prfp-vector prfp-mir-93 prfp-mir-124 Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent
prfp-vector RFP Exon1 Intron RFP Exon2 prfp-mir-124 mir-93/124 RFP Exon1 Intron RFP Exon2 Untransfected prfp-vector prfp-mir-93 prfp-mir-124 Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent
Diabetic pdx1-mutant zebrafish show conserved responses to nutrient overload and anti-glycemic treatment
 Supplementary Information Diabetic pdx1-mutant zebrafish show conserved responses to nutrient overload and anti-glycemic treatment Robin A. Kimmel, Stefan Dobler, Nicole Schmitner, Tanja Walsen, Julia
Supplementary Information Diabetic pdx1-mutant zebrafish show conserved responses to nutrient overload and anti-glycemic treatment Robin A. Kimmel, Stefan Dobler, Nicole Schmitner, Tanja Walsen, Julia
Relative activity (%) SC35M
 a 125 Bat (H17N) b 125 A/WSN (H1N1) Relative activity (%) 0 75 50 25 Relative activity (%) 0 75 50 25 0 Pos. Neg. PA PB1 Pos. Neg. NP PA PB1 PB2 0 Pos. Neg. NP PA PB1 PB2 SC35M Bat Supplementary Figure
a 125 Bat (H17N) b 125 A/WSN (H1N1) Relative activity (%) 0 75 50 25 Relative activity (%) 0 75 50 25 0 Pos. Neg. PA PB1 Pos. Neg. NP PA PB1 PB2 0 Pos. Neg. NP PA PB1 PB2 SC35M Bat Supplementary Figure
P. Mathijs Voorhoeve, Carlos le Sage, Mariette Schrier, Ad J.M. Gillis, Hans Stoop,
 Supplemental Data A Genetic Screen Implicates mirna-372 and mirna-373 As Oncogenes in Testicular Germ Cell Tumors P. Mathijs Voorhoeve, Carlos le Sage, Mariette Schrier, Ad J.M. Gillis, Hans Stoop, Remco
Supplemental Data A Genetic Screen Implicates mirna-372 and mirna-373 As Oncogenes in Testicular Germ Cell Tumors P. Mathijs Voorhoeve, Carlos le Sage, Mariette Schrier, Ad J.M. Gillis, Hans Stoop, Remco
Nature Medicine: doi: /nm.4322
 1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
Supplemental Information. Figures. Figure S1
 Supplemental Information Figures Figure S1 Identification of JAGGER T-DNA insertions. A. Positions of T-DNA and Ds insertions in JAGGER are indicated by inverted triangles, the grey box represents the
Supplemental Information Figures Figure S1 Identification of JAGGER T-DNA insertions. A. Positions of T-DNA and Ds insertions in JAGGER are indicated by inverted triangles, the grey box represents the
Supplemental Materials and Methods Plasmids and viruses Quantitative Reverse Transcription PCR Generation of molecular standard for quantitative PCR
 Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
A conserved neuronal DAF-16/FoxO plays an important role in conveying
 SREP-17-10421.R2 Supplementary Information (clean version) Scientific Reports A conserved neuronal DAF-16/FoxO plays an important role in conveying pheromone signals to elicit repulsion behavior in Caenorhabditis
SREP-17-10421.R2 Supplementary Information (clean version) Scientific Reports A conserved neuronal DAF-16/FoxO plays an important role in conveying pheromone signals to elicit repulsion behavior in Caenorhabditis
Supplementary Figure 1 Information on transgenic mouse models and their recording and optogenetic equipment. (a) 108 (b-c) (d) (e) (f) (g)
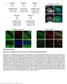 Supplementary Figure 1 Information on transgenic mouse models and their recording and optogenetic equipment. (a) In four mice, cre-dependent expression of the hyperpolarizing opsin Arch in pyramidal cells
Supplementary Figure 1 Information on transgenic mouse models and their recording and optogenetic equipment. (a) In four mice, cre-dependent expression of the hyperpolarizing opsin Arch in pyramidal cells
MII. Supplement Figure 1. CapZ β2. Merge. 250ng. 500ng DIC. Merge. Journal of Cell Science Supplementary Material. GFP-CapZ β2 DNA
 A GV GVBD MI DNA CapZ β2 CapZ β2 Merge B DIC GFP-CapZ β2 Merge CapZ β2-gfp 250ng 500ng Supplement Figure 1. MII A early MI late MI Control RNAi CapZαβ DNA Actin Tubulin B Phalloidin Intensity(A.U.) n=10
A GV GVBD MI DNA CapZ β2 CapZ β2 Merge B DIC GFP-CapZ β2 Merge CapZ β2-gfp 250ng 500ng Supplement Figure 1. MII A early MI late MI Control RNAi CapZαβ DNA Actin Tubulin B Phalloidin Intensity(A.U.) n=10
GAP-FREE NEURAL CIRCUITS CLASS #3: C. elegans touch-induced locomotion
 GAP-FREE NEURAL CIRCUITS CLASS #3: C. elegans touch-induced locomotion OUTLINE: C. elegans background o General background o Neural signaling: no action potentials Excitatory circuits: Touch-induced locomotion
GAP-FREE NEURAL CIRCUITS CLASS #3: C. elegans touch-induced locomotion OUTLINE: C. elegans background o General background o Neural signaling: no action potentials Excitatory circuits: Touch-induced locomotion
Nature Neuroscience: doi: /nn Supplementary Figure 1. Large-scale calcium imaging in vivo.
 Supplementary Figure 1 Large-scale calcium imaging in vivo. (a) Schematic illustration of the in vivo camera imaging set-up for large-scale calcium imaging. (b) High-magnification two-photon image from
Supplementary Figure 1 Large-scale calcium imaging in vivo. (a) Schematic illustration of the in vivo camera imaging set-up for large-scale calcium imaging. (b) High-magnification two-photon image from
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
Nature Genetics: doi: /ng Supplementary Figure 1. HOX fusions enhance self-renewal capacity.
 Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Genome-editing via Oviductal Nucleic Acids Delivery (GONAD) system: a novel microinjection-independent genome engineering method in mice
 Supplementary Information Genome-editing via Oviductal Nucleic Acids Delivery (GONAD) system: a novel microinjection-independent genome engineering method in mice Gou Takahashi, Channabasavaiah B Gurumurthy,
Supplementary Information Genome-editing via Oviductal Nucleic Acids Delivery (GONAD) system: a novel microinjection-independent genome engineering method in mice Gou Takahashi, Channabasavaiah B Gurumurthy,
Supplementary Figure 1
 Supplementary Figure 1 Miniature microdrive, spike sorting and sleep stage detection. a, A movable recording probe with 8-tetrodes (32-channels). It weighs ~1g. b, A mouse implanted with 8 tetrodes in
Supplementary Figure 1 Miniature microdrive, spike sorting and sleep stage detection. a, A movable recording probe with 8-tetrodes (32-channels). It weighs ~1g. b, A mouse implanted with 8 tetrodes in
Postn MCM Smad2 fl/fl Postn MCM Smad3 fl/fl Postn MCM Smad2/3 fl/fl. Postn MCM. Tgfbr1/2 fl/fl TAC
 A Smad2 fl/fl Smad3 fl/fl Smad2/3 fl/fl Tgfbr1/2 fl/fl 1. mm B Tcf21 MCM Tcf21 MCM Smad3 fl/fl Tcf21 MCM Smad2/3 fl/fl Tcf21 MCM Tgfbr1/2 fl/fl αmhc MCM C 1. mm 1. mm D Smad2 fl/fl Smad3 fl/fl Smad2/3
A Smad2 fl/fl Smad3 fl/fl Smad2/3 fl/fl Tgfbr1/2 fl/fl 1. mm B Tcf21 MCM Tcf21 MCM Smad3 fl/fl Tcf21 MCM Smad2/3 fl/fl Tcf21 MCM Tgfbr1/2 fl/fl αmhc MCM C 1. mm 1. mm D Smad2 fl/fl Smad3 fl/fl Smad2/3
Nature Neuroscience: doi: /nn Supplementary Figure 1. Splenic atrophy and leucopenia caused by T3 SCI.
 Supplementary Figure 1 Splenic atrophy and leucopenia caused by T3 SCI. (a) Gross anatomy of representative spleens from control and T3 SCI mice at 28 days post-injury. (b and c) Hematoxylin and eosin
Supplementary Figure 1 Splenic atrophy and leucopenia caused by T3 SCI. (a) Gross anatomy of representative spleens from control and T3 SCI mice at 28 days post-injury. (b and c) Hematoxylin and eosin
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1 SNARE Probes for FRET/2pFLIM Analysis Used in the Present Study. mturquoise (mtq) and Venus (Ven) are in blue and yellow, respectively. The soluble N-ethylmaleimide-sensitive
Supplementary Figure 1 Supplementary Figure 1 SNARE Probes for FRET/2pFLIM Analysis Used in the Present Study. mturquoise (mtq) and Venus (Ven) are in blue and yellow, respectively. The soluble N-ethylmaleimide-sensitive
Nature Immunology: doi: /ni.3631
 Supplementary Figure 1 SPT analyses of Zap70 at the T cell plasma membrane. (a) Total internal reflection fluorescent (TIRF) excitation at 64-68 degrees limits single molecule detection to 100-150 nm above
Supplementary Figure 1 SPT analyses of Zap70 at the T cell plasma membrane. (a) Total internal reflection fluorescent (TIRF) excitation at 64-68 degrees limits single molecule detection to 100-150 nm above
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Nature Neuroscience: doi: /nn Supplementary Figure 1. Behavioral training.
 Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
Supplementary Figure 1
 Supplementary Figure 1 how HFD how HFD Epi WT p p Hypothalamus p p Inguinal WT T Liver Lean mouse adipocytes p p p p p p Obese mouse adipocytes Kidney Muscle Spleen Heart p p p p p p p p Extracellular
Supplementary Figure 1 how HFD how HFD Epi WT p p Hypothalamus p p Inguinal WT T Liver Lean mouse adipocytes p p p p p p Obese mouse adipocytes Kidney Muscle Spleen Heart p p p p p p p p Extracellular
effects on organ development. a-f, Eye and wing discs with clones of ε j2b10 show no
 Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Subcellular segregation of VGluT2-IR and TH-IR within the same VGluT2-TH axon (wild type rats). (a-e) Serial sections of a dual VGluT2-TH labeled axon. This axon (blue outline) has
Supplementary Figure 1 Subcellular segregation of VGluT2-IR and TH-IR within the same VGluT2-TH axon (wild type rats). (a-e) Serial sections of a dual VGluT2-TH labeled axon. This axon (blue outline) has
A conserved dopamine-cholecystokinin signaling pathway shapes context-dependent Caenorhabditis elegans behavior
 University of Massachusetts Medical School escholarship@umms Open Access Articles Open Access Publications by UMMS Authors 8-28-2014 A conserved dopamine-cholecystokinin signaling pathway shapes context-dependent
University of Massachusetts Medical School escholarship@umms Open Access Articles Open Access Publications by UMMS Authors 8-28-2014 A conserved dopamine-cholecystokinin signaling pathway shapes context-dependent
Nature Neuroscience: doi: /nn Supplementary Figure 1. Distribution of starter cells for RV-mediated retrograde tracing.
 Supplementary Figure 1 Distribution of starter cells for RV-mediated retrograde tracing. Parcellation of cortical areas is based on Allen Mouse Brain Atlas and drawn to scale. Thick white curves, outlines
Supplementary Figure 1 Distribution of starter cells for RV-mediated retrograde tracing. Parcellation of cortical areas is based on Allen Mouse Brain Atlas and drawn to scale. Thick white curves, outlines
Supplementary Materials for VAMP4 directs synaptic vesicles to a pool that selectively maintains asynchronous neurotransmission
 Supplementary Materials for VAMP4 directs synaptic vesicles to a pool that selectively maintains asynchronous neurotransmission Jesica Raingo, Mikhail Khvotchev, Pei Liu, Frederic Darios, Ying C. Li, Denise
Supplementary Materials for VAMP4 directs synaptic vesicles to a pool that selectively maintains asynchronous neurotransmission Jesica Raingo, Mikhail Khvotchev, Pei Liu, Frederic Darios, Ying C. Li, Denise
Supplementary Figure 1
 Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Kidney. Heart. Lung. Sirt1. Gapdh. Mouse IgG DAPI. Rabbit IgG DAPI
 a e Na V 1.5 Ad-LacZ Ad- 110KD b Scn5a/ (relative to Ad-LacZ) f 150 100 50 0 p = 0.65 Ad-LacZ Ad- c Heart Lung Kidney Spleen 110KD d fl/fl c -/- DAPI 20 µm Na v 1.5 250KD fl/fl Rabbit IgG DAPI fl/fl Mouse
a e Na V 1.5 Ad-LacZ Ad- 110KD b Scn5a/ (relative to Ad-LacZ) f 150 100 50 0 p = 0.65 Ad-LacZ Ad- c Heart Lung Kidney Spleen 110KD d fl/fl c -/- DAPI 20 µm Na v 1.5 250KD fl/fl Rabbit IgG DAPI fl/fl Mouse
Karen L.P. McNally, Amy S. Fabritius, Marina L. Ellefson, Jonathan R. Flynn, Jennifer A. Milan, and Francis J. McNally
 Developmental Cell, Volume 22 Supplemental Information Kinesin-1 Prevents Capture of the Oocyte Meiotic Spindle by the Sperm Aster Karen L.P. McNally, Amy S. Fabritius, Marina L. Ellefson, Jonathan R.
Developmental Cell, Volume 22 Supplemental Information Kinesin-1 Prevents Capture of the Oocyte Meiotic Spindle by the Sperm Aster Karen L.P. McNally, Amy S. Fabritius, Marina L. Ellefson, Jonathan R.
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature07173 SUPPLEMENTARY INFORMATION Supplementary Figure Legends: Supplementary Figure 1: Model of SSC and CPC divisions a, Somatic stem cells (SSC) reside adjacent to the hub (red), self-renew
doi: 10.1038/nature07173 SUPPLEMENTARY INFORMATION Supplementary Figure Legends: Supplementary Figure 1: Model of SSC and CPC divisions a, Somatic stem cells (SSC) reside adjacent to the hub (red), self-renew
Nature Methods: doi: /nmeth.4257
 Supplementary Figure 1 Screen for polypeptides that affect cellular actin filaments. (a) Table summarizing results from all polypeptides tested. Source shows organism, gene, and amino acid numbers used.
Supplementary Figure 1 Screen for polypeptides that affect cellular actin filaments. (a) Table summarizing results from all polypeptides tested. Source shows organism, gene, and amino acid numbers used.
Nature Methods: doi: /nmeth Supplementary Figure 1. Activity in turtle dorsal cortex is sparse.
 Supplementary Figure 1 Activity in turtle dorsal cortex is sparse. a. Probability distribution of firing rates across the population (notice log scale) in our data. The range of firing rates is wide but
Supplementary Figure 1 Activity in turtle dorsal cortex is sparse. a. Probability distribution of firing rates across the population (notice log scale) in our data. The range of firing rates is wide but
Supplementary Figure 1
 Supplementary Figure 1 a γ-h2ax MDC1 RNF8 FK2 BRCA1 U2OS Cells sgrna-1 ** 60 sgrna 40 20 0 % positive Cells (>5 foci per cell) b ** 80 sgrna sgrna γ-h2ax MDC1 γ-h2ax RNF8 FK2 MDC1 BRCA1 RNF8 FK2 BRCA1
Supplementary Figure 1 a γ-h2ax MDC1 RNF8 FK2 BRCA1 U2OS Cells sgrna-1 ** 60 sgrna 40 20 0 % positive Cells (>5 foci per cell) b ** 80 sgrna sgrna γ-h2ax MDC1 γ-h2ax RNF8 FK2 MDC1 BRCA1 RNF8 FK2 BRCA1
SUPPLEMENTARY INFORMATION
 doi:1.138/nature1139 a d Whisker angle (deg) Whisking repeatability Control Muscimol.4.3.2.1 -.1 8 4-4 1 2 3 4 Performance (d') Pole 8 4-4 1 2 3 4 5 Time (s) b Mean protraction angle (deg) e Hit rate (p
doi:1.138/nature1139 a d Whisker angle (deg) Whisking repeatability Control Muscimol.4.3.2.1 -.1 8 4-4 1 2 3 4 Performance (d') Pole 8 4-4 1 2 3 4 5 Time (s) b Mean protraction angle (deg) e Hit rate (p
Nature Neuroscience: doi: /nn Supplementary Figure 1. Trial structure for go/no-go behavior
 Supplementary Figure 1 Trial structure for go/no-go behavior a, Overall timeline of experiments. Day 1: A1 mapping, injection of AAV1-SYN-GCAMP6s, cranial window and headpost implantation. Water restriction
Supplementary Figure 1 Trial structure for go/no-go behavior a, Overall timeline of experiments. Day 1: A1 mapping, injection of AAV1-SYN-GCAMP6s, cranial window and headpost implantation. Water restriction
Supplementary Figure 1. Spitzoid Melanoma with PPFIBP1-MET fusion. (a) Histopathology (4x) shows a domed papule with melanocytes extending into the
 Supplementary Figure 1. Spitzoid Melanoma with PPFIBP1-MET fusion. (a) Histopathology (4x) shows a domed papule with melanocytes extending into the deep dermis. (b) The melanocytes demonstrate abundant
Supplementary Figure 1. Spitzoid Melanoma with PPFIBP1-MET fusion. (a) Histopathology (4x) shows a domed papule with melanocytes extending into the deep dermis. (b) The melanocytes demonstrate abundant
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Cellular Neurobiology BIPN140. 1st Midterm Exam October 18 th, Tuesday Material covered: Lectures 1-6 & Reading
 Cellular Neurobiology BIPN140 1st Midterm Exam October 18 th, Tuesday Material covered: Lectures 1-6 & Reading Review session October 17 th 3500 Pacitic Hall, 6-8 pm (access code is 127895) Come with questions!
Cellular Neurobiology BIPN140 1st Midterm Exam October 18 th, Tuesday Material covered: Lectures 1-6 & Reading Review session October 17 th 3500 Pacitic Hall, 6-8 pm (access code is 127895) Come with questions!
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Bidirectional optogenetic modulation of the tonic activity of CEA PKCδ + neurons in vitro. a, Top, Cell-attached voltage recording illustrating the blue light-induced increase in
Supplementary Figure 1 Bidirectional optogenetic modulation of the tonic activity of CEA PKCδ + neurons in vitro. a, Top, Cell-attached voltage recording illustrating the blue light-induced increase in
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1: Chemokine receptor expression profiles of CCR6 + and CCR6 - CD4 + IL-17A +/ex and Treg cells. Quantitative PCR analysis of chemokine receptor transcript abundance
SUPPLEMENTARY FIGURES Supplementary Figure 1: Chemokine receptor expression profiles of CCR6 + and CCR6 - CD4 + IL-17A +/ex and Treg cells. Quantitative PCR analysis of chemokine receptor transcript abundance
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
Supplementary information
 Supplementary information 1 Supplementary Figure 1. CALM regulates autophagy. (a). Quantification of LC3 levels in the experiment described in Figure 1A. Data are mean +/- SD (n > 3 experiments for each
Supplementary information 1 Supplementary Figure 1. CALM regulates autophagy. (a). Quantification of LC3 levels in the experiment described in Figure 1A. Data are mean +/- SD (n > 3 experiments for each
Supplementary Figure 1. FACS analysis of cells infected with TY93/H5N1 GFP-627E,
 Supplementary Figure 1. FACS analysis of cells infected with TY93/H5N1 GFP-627E, TY93/H5N1 GFP-627K, or the TY93/H5N1 PB2(588-759) virus library. To establish our GFP- FACS screening platform, we compared
Supplementary Figure 1. FACS analysis of cells infected with TY93/H5N1 GFP-627E, TY93/H5N1 GFP-627K, or the TY93/H5N1 PB2(588-759) virus library. To establish our GFP- FACS screening platform, we compared
doi: /nature10632
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10632 Supplementary Figure 1 Lyn mediates neutrophil wound responses as a redox sensor. a, A schematic model. Wounded epithelial cells release H 2 O 2 by an
SUPPLEMENTARY INFORMATION doi:10.1038/nature10632 Supplementary Figure 1 Lyn mediates neutrophil wound responses as a redox sensor. a, A schematic model. Wounded epithelial cells release H 2 O 2 by an
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
Predictive PP1Ca binding region in BIG3 : 1,228 1,232aa (-KAVSF-) HEK293T cells *** *** *** KPL-3C cells - E E2 treatment time (h)
 Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
Supplementary Figure 1 Validation of Per2 deletion in neuronal cells in N Per2 -/- mice. (a) Western blot from liver extracts of mice held under ad
 Supplementary Figure 1 Validation of Per2 deletion in neuronal cells in N Per2 -/- mice. (a) Western blot from liver extracts of mice held under ad libitum conditions detecting PER2 protein in brain and
Supplementary Figure 1 Validation of Per2 deletion in neuronal cells in N Per2 -/- mice. (a) Western blot from liver extracts of mice held under ad libitum conditions detecting PER2 protein in brain and
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplemental Information. Myocardial Polyploidization Creates a Barrier. to Heart Regeneration in Zebrafish
 Developmental Cell, Volume 44 Supplemental Information Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish Juan Manuel González-Rosa, Michka Sharpe, Dorothy Field, Mark H.
Developmental Cell, Volume 44 Supplemental Information Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish Juan Manuel González-Rosa, Michka Sharpe, Dorothy Field, Mark H.
Supplemental Information. Otic Mesenchyme Cells Regulate. Spiral Ganglion Axon Fasciculation. through a Pou3f4/EphA4 Signaling Pathway
 Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
Supplementary Figure 1
 Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplementary Information. Induction of p53-independent apoptosis by ectopic expression of HOXA5
 Supplementary Information Induction of p53-independent apoptosis by ectopic expression of in human liposarcomas Dhong Hyun Lee 1, *, Charles Forscher 1, Dolores Di Vizio 2, 3, and H. Phillip Koeffler 1,
Supplementary Information Induction of p53-independent apoptosis by ectopic expression of in human liposarcomas Dhong Hyun Lee 1, *, Charles Forscher 1, Dolores Di Vizio 2, 3, and H. Phillip Koeffler 1,
Zhu et al, page 1. Supplementary Figures
 Zhu et al, page 1 Supplementary Figures Supplementary Figure 1: Visual behavior and avoidance behavioral response in EPM trials. (a) Measures of visual behavior that performed the light avoidance behavior
Zhu et al, page 1 Supplementary Figures Supplementary Figure 1: Visual behavior and avoidance behavioral response in EPM trials. (a) Measures of visual behavior that performed the light avoidance behavior
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06994 A phosphatase cascade by which rewarding stimuli control nucleosomal response A. Stipanovich*, E. Valjent*, M. Matamales*, A. Nishi, J.H. Ahn, M. Maroteaux, J. Bertran-Gonzalez,
doi: 10.1038/nature06994 A phosphatase cascade by which rewarding stimuli control nucleosomal response A. Stipanovich*, E. Valjent*, M. Matamales*, A. Nishi, J.H. Ahn, M. Maroteaux, J. Bertran-Gonzalez,
genome edited transient transfection, CMV promoter
 Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
Nature Biotechnology: doi: /nbt Supplementary Figure 1. PL gene expression in tomato fruit.
 Supplementary Figure 1 PL gene expression in tomato fruit. Relative expression of five PL-coding genes measured in at least three fruit of each genotype (cv. Alisa Craig) at four stages of development,
Supplementary Figure 1 PL gene expression in tomato fruit. Relative expression of five PL-coding genes measured in at least three fruit of each genotype (cv. Alisa Craig) at four stages of development,
Appendix. Table of Contents
 Appendix Table of Contents Appendix Figures Figure S1: Gp78 is not required for the degradation of mcherry-cl1 in Hela Cells. Figure S2: Indel formation in the MARCH6 sgrna targeted HeLa clones. Figure
Appendix Table of Contents Appendix Figures Figure S1: Gp78 is not required for the degradation of mcherry-cl1 in Hela Cells. Figure S2: Indel formation in the MARCH6 sgrna targeted HeLa clones. Figure
Nature Immunology: doi: /ni Supplementary Figure 1. Cytokine pattern in skin in response to urushiol.
 Supplementary Figure 1 Cytokine pattern in skin in response to urushiol. Wild-type (WT) and CD1a-tg mice (n = 3 per group) were sensitized and challenged with urushiol (uru) or vehicle (veh). Quantitative
Supplementary Figure 1 Cytokine pattern in skin in response to urushiol. Wild-type (WT) and CD1a-tg mice (n = 3 per group) were sensitized and challenged with urushiol (uru) or vehicle (veh). Quantitative
klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach
![klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach](/thumbs/91/104639484.jpg) DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
Supplemental Information. A Visual-Cue-Dependent Memory Circuit. for Place Navigation
 Neuron, Volume 99 Supplemental Information A Visual-Cue-Dependent Memory Circuit for Place Navigation Han Qin, Ling Fu, Bo Hu, Xiang Liao, Jian Lu, Wenjing He, Shanshan Liang, Kuan Zhang, Ruijie Li, Jiwei
Neuron, Volume 99 Supplemental Information A Visual-Cue-Dependent Memory Circuit for Place Navigation Han Qin, Ling Fu, Bo Hu, Xiang Liao, Jian Lu, Wenjing He, Shanshan Liang, Kuan Zhang, Ruijie Li, Jiwei
Nature Genetics: doi: /ng Supplementary Figure 1. SEER data for male and female cancer incidence from
 Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
