SUPPLEMENTARY INFORMATION. A comparative study of outer membrane proteome between a paired colistinsusceptible
|
|
|
- Camron Newton
- 5 years ago
- Views:
Transcription
1 SUPPLEMENTARY INFORMATION A comparative study of outer membrane proteome between a paired colistinsusceptible and extremely colistin-resistant Klebsiella pneumoniae strains Raad Jasim, Mark A. Baker, 2 Yan Zhu, 3 Meiling Han, 3 Elena K. Schneider- Futschik, 4 Maytham Hussein, 4 Daniel Hoyer, 4,5,6 Jian Li, 3* Tony Velkov 4* Drug Development and Innovation, Drug Delivery, Disposition and Dynamics. Monash Institute of Pharmaceutical Sciences, Monash University, 38 Royal Parade, Parkville 3052, Victoria, Australia. 2 Priority Research Centre in Reproductive Science, School of Environmental and Life Sciences, University of Newcastle, Callaghan, NSW, 2308, Australia. 3 Monash Biomedicine Discovery Institute, Department of Microbiology, Monash University, Clayton, Victoria 3800, Australia. 4 Department of Pharmacology and Therapeutics, University of Melbourne, Parkville, Victoria 300, Australia. 5 The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, 30 Royal Parade, Parkville, Victoria 3052, Australia. 6 Department of Molecular Medicine, The Scripps Research Institute, 0550 N. Torrey Pines Road, La Jolla, CA 92037, USA. Correspondence: Tony Velkov, Phone: Fax: E- mail: colistin.polymyxin@gmail.com or Tony.Velkov@unimelb.edu.au * Corresponding authors S
2 Content: -Supplementary Tables: 8 -Supplementary Figures: 3 -Total pages: 0 S2
3 Table S8. OMP proteome differences between paired colistin-susceptible and extremely colistin-resistant K. pneumoniae ATCC 3883 strains. Sensitive K. pneumoniae ATCC 3883 Resistant K. pneumoniae ATCC 3883 Protein name Number of peptides detected Protein name Number of peptides detected Cobyric acid synthase 4-aminobutyraldehyde Phosphoheptose isomerase (EC ) Purine nucleoside phosphorylase (EC ) FIG004088: inner membrane protein YebE Selenocysteine-specific translation elongation factor Sulfur carrier protein adenylyltransferase ThiF Maltose/maltodextrin transport ATPbinding protein MalK (EC ) ATP-dependent protease La (EC ) Type I Glycoprotein-polysaccharide metabolism DNA-binding protein H-NS PTS system, mannose-specific IIA component (EC ) / PTS system, mannose-specific IIB component (EC ) Glycerol-3-phosphate acyltransferase (EC ) LPS-assembly lipoprotein RlpB precursor (Rare lipoprotein B) dehydrogenase (EC.2..9) Galactose/methyl galactoside ABC transport system, permease protein MglC (TC 3.A..2.3) LSU ribosomal protein L3p (L3e) Galactose/methyl galactoside ABC transport system, ATP-binding protein MglA (EC ) SSU ribosomal protein S2p (SAe) Myo-inositol 2-dehydrogenase (EC...8) Glutathione S-transferase (EC ) LSU ribosomal protein L5p (Le) Threonyl-tRNA synthetase (EC 6...3) Purine nucleoside phosphorylase (EC ) Osmotically inducible lipoprotein B precursor Nucleoside permease NupC NADH-ubiquinone oxidoreductase chain M (EC.6.5.3) Cell division protein FtsA LSU ribosomal protein L3p (L3Ae) Adenylate cyclase (EC 4.6..) Serine hydroxymethyltransferase (EC 2..2.) PTS system, N-acetylglucosaminespecific IIA component (EC ) / PTS system, N-acetylglucosaminespecific IIB component (EC ) / PTS system, N-acetylglucosamine- Phage shock protein A S3
4 specific IIC component (EC ) Hypothetical lipoprotein YajG precursor FIG : hypothetical protein Small heat shock protein Alcohol dehydrogenase (EC...) Lysyl-tRNA synthetase (class II) (EC 6...6) Sulfur carrier protein adenylyltransferase ThiF Putative outer membrane protein LSU ribosomal protein L22p (L7e) Putative phosphosugar isomerase Putative lipoprotein Cysteine synthase (EC ) 2 kda hemolysin precursor Threonyl-tRNA synthetase (EC 6...3) Glutamine ABC transporter, periplasmic glutamine-binding protein (TC 3.A..3.2) Transcriptional activator of maltose Universal stress protein E regulon, MalT 4-aminobutyraldehyde dehydrogenase (EC.2..9) Tail-specific protease precursor (EC ) Acetaldehyde dehydrogenase, ethanolamine utilization cluster FIG : hypothetical protein Chromosome partition protein MukB Maltoporin (maltose/maltodextrin high-affinity receptor, phage lambda receptor protein) UDP-N-acetylmuramoylalanyl-Dglutamate--2,6-diaminopimelate ligase (EC ) SSU ribosomal protein Sp Osmotically inducible lipoprotein B precursor 6-phosphofructokinase class II (EC 2.7..) ATP synthase A chain (EC ) LSU ribosomal protein L5p (L27Ae) Alkyl hydroperoxide reductase protein C (EC.6.4.-) Cobalt-precorrin-2 C20- methyltransferase (EC ) Pyruvate formate-lyase (EC ) Lipoprotein Galactose/methyl galactoside ABC transport system, permease protein MglC (TC 3.A..2.3) LSU ribosomal protein Lp (L0Ae) Cell division protein FtsZ (EC ) SSU ribosomal protein S5p (S2e) LSU ribosomal protein L24p (L26e) Pyruvate dehydrogenase E component (EC.2.4.) S-adenosylmethionine synthetase Entericidin B precursor (EC ) Inosine-5'-monophosphate dehydrogenase (EC...205) Maltose/maltodextrin ABC transporter, substrate binding periplasmic protein MalE Phosphoglycerate kinase (EC Asparagine synthetase [glutaminehydrolyzing] ) (EC ) LSU ribosomal protein L22p (L7e) Putative phosphosugar isomerase LSU ribosomal protein L3p (L3e) Chaperone protein HtpG SgrR, sugar-phosphate stress, LSU ribosomal protein L20p S4
5 transcriptional activator of SgrS small RNA LSU ribosomal protein L6p (L9e) Cysteine synthase (EC ) GTP-binding protein TypA/BipA Cytochrome d ubiquinol oxidase subunit II (EC.0.3.-) LSU ribosomal protein L5p (Le) Small heat shock protein DNA-directed RNA polymerase beta' Aspartate--ammonia ligase (EC subunit (EC ) 6.3..) Adenosine deaminase (EC ) Membrane fusion protein of RND family multidrug efflux pump NADP-dependent malic enzyme (EC Septum site-determining protein...40) MinD IncF plasmid conjugative transfer Chaperone protein DnaK surface exclusion protein TraT LSU ribosomal protein L28p LSU ribosomal protein L0p (P0) Entericidin B precursor Di/tripeptide permease DtpB UDP-N-acetylglucosamine - Carbamoyl-phosphate synthase carboxyvinyltransferase (EC ) large chain (EC ) LSU ribosomal protein Lp (L0Ae) ABC transporter, ATP-binding protein Lysine decarboxylase, inducible (EC 4...8) Uncharacterized protein YeaG Formate efflux transporter (TC 2.A.44 Uridine phosphorylase (EC ) family) Maltose-6'-phosphate glucosidase (EC ) Hexuronate transporter Preprotein translocase secy subunit (TC 3.A.5..) Septum site-determining protein MinD Maltoporin (maltose/maltodextrin high-affinity receptor, phage lambda receptor protein) SSU ribosomal protein SSU ribosomal protein S8p, zincindependent Phosphate acetyltransferase (EC ) Osmotically inducible lipoprotein E precursor LSU ribosomal protein L2p Sugar diacid utilization regulator SdaR PTS system, sucrose-specific IIB component (EC ) / PTS system, sucrose-specific IIC component (EC ) 5'-methylthioadenosine nucleosidase (EC ) / S- adenosylhomocysteine nucleosidase (EC ) S5
6 Uridine phosphorylase (EC ) Alkyl hydroperoxide reductase subunit C-like protein Translation initiation factor 2 Rod shape-determining protein MreB UDP-N-acetylglucosamine--Nacetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N- acetylglucosamine transferase (EC ) SSU ribosomal protein S5p (S2e) LSU ribosomal protein L9p 5-keto-2-deoxygluconokinase (EC ) / uncharacterized domain Aspartate aminotransferase (EC 2.6..) Glucose-6-phosphate isomerase (EC ) ATP synthase alpha chain (EC ) FIG : hypothetical protein S6
7 Figure S Figure S. Alignment of the mgrb genes of the colistin-susceptible K. pneumoniae ATCC 3883 (GenBank: JSZI ) and the paired extremely colistin-resistant K. pneumoniae ATCC S7
8 Figure S2 Figure S2. Alignment of the PhoP genes of the colistin-susceptible K. pneumoniae ATCC 3883 (GenBank: JSZI ) and the paired extremely colistin-resistant K. pneumoniae ATCC S8
9 Figure S3 S9
10 Figure S3. Alignment of the PhoQ genes of the colistin-susceptible K. pneumoniae ATCC 3883 (GenBank: JSZI ) and the paired extremely colistin-resistant K. pneumoniae ATCC S0
Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function
 Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSL0024 26223 26621 + LrgA family BPSL0025 26690 27412 + hypothetical
Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSL0024 26223 26621 + LrgA family BPSL0025 26690 27412 + hypothetical
f(x) = x R² = RPKM (M8.MXB) f(x) = x E-014 R² = 1 RPKM (M31.
 14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
Supplemental Information
 Supplemental Information Screening of strong constitutive promoters in the S. albus transcriptome via RNA-seq The total RNA of S. albus J1074 was isolated after 24 hrs and 72 hrs of cultivation at 30 C
Supplemental Information Screening of strong constitutive promoters in the S. albus transcriptome via RNA-seq The total RNA of S. albus J1074 was isolated after 24 hrs and 72 hrs of cultivation at 30 C
Protein Class/Name KEGG Pathways
 Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 4. Proteins Increased in Either Soil medium at 37 o C and 25 o C Table 4A. Proteins
Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 4. Proteins Increased in Either Soil medium at 37 o C and 25 o C Table 4A. Proteins
Protein Class/Name. Pathways. bat00250, bat00280, bat00410, bat00640, bat bat00270, bat00330, bat00410, bat00480
 Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 1. Proteins Increased in Either Soil or Laboratory media Table 1A. Proteins Increased
Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 1. Proteins Increased in Either Soil or Laboratory media Table 1A. Proteins Increased
Biological processes. Mitochondrion Metabolic process Catalytic activity Oxidoreductase
 Full name Glyceraldehyde3 phosphate dehydrogenase Succinatesemialdehyde Glutamate dehydrogenase 1, Alcohol dehydrogenase [NADP+] 2',3'cyclicnucleotide 3' phosphodiesterase Dihydropyrimidinaserelated 2
Full name Glyceraldehyde3 phosphate dehydrogenase Succinatesemialdehyde Glutamate dehydrogenase 1, Alcohol dehydrogenase [NADP+] 2',3'cyclicnucleotide 3' phosphodiesterase Dihydropyrimidinaserelated 2
Figure S1: Abundance of Fe related proteins in oceanic Synechococcus sp. strain WH8102 in. Ferredoxin. Ferredoxin. Fe ABC transporter
 0.004 SW nm 0.04 0.03 nm nm 0.16 0.1 nm nm 0.40 0.3 nm nm 0.80 0.5 nm 1.6 1 nm 16 10 nm 160 100 nm 0 nm 0.04 nm 0.16 nm 0.40 nm 0.80 nm 1.6 nm 16 nm 160 nm Spectral counts 0 nm 0.04 nm 0.16 nm 0.40 nm
0.004 SW nm 0.04 0.03 nm nm 0.16 0.1 nm nm 0.40 0.3 nm nm 0.80 0.5 nm 1.6 1 nm 16 10 nm 160 100 nm 0 nm 0.04 nm 0.16 nm 0.40 nm 0.80 nm 1.6 nm 16 nm 160 nm Spectral counts 0 nm 0.04 nm 0.16 nm 0.40 nm
NBCE Mock Board Questions Biochemistry
 1. Fluid mosaic describes. A. Tertiary structure of proteins B. Ribosomal subunits C. DNA structure D. Plasma membrane structure NBCE Mock Board Questions Biochemistry 2. Where in the cell does beta oxidation
1. Fluid mosaic describes. A. Tertiary structure of proteins B. Ribosomal subunits C. DNA structure D. Plasma membrane structure NBCE Mock Board Questions Biochemistry 2. Where in the cell does beta oxidation
Protein Class/Name. Pathways. bat00250, bat00280, bat bat00640, bat00650 bat00270, bat00330, bat00410, bat00480 bat00270, bat00330,
 Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 2. Proteins Increased in Either Blood or Horizon media Table 2A. Proteins Increased
Electronic Supplementary Material (ESI) for Analyst. This journal is The Royal Society of Chemistry 2017 Supplemental Table 2. Proteins Increased in Either Blood or Horizon media Table 2A. Proteins Increased
Bacterial cellulose synthesis mechanism of facultative
 1 2 3 4 Bacterial cellulose synthesis mechanism of facultative anaerobe Kaihua Ji 1 +, Wei Wang 2, 3 +, Bing Zeng 1, Sibin Chen 1, Qianqian Zhao 4, Yueqing Chen 1, Guoqiang Li 1* and Ting Ma 1* 5 6 7 Figure
1 2 3 4 Bacterial cellulose synthesis mechanism of facultative anaerobe Kaihua Ji 1 +, Wei Wang 2, 3 +, Bing Zeng 1, Sibin Chen 1, Qianqian Zhao 4, Yueqing Chen 1, Guoqiang Li 1* and Ting Ma 1* 5 6 7 Figure
Chapter 10. 이화작용 : 에너지방출과보존 (Catabolism: Energy Release and Conservation)
 Chapter 10 이화작용 : 에너지방출과보존 (Catabolism: Energy Release and Conservation) 1 Fueling Processes Respiration 1 Most respiration involves use of an electron transport chain As electrons pass through the electron
Chapter 10 이화작용 : 에너지방출과보존 (Catabolism: Energy Release and Conservation) 1 Fueling Processes Respiration 1 Most respiration involves use of an electron transport chain As electrons pass through the electron
Electron Transport Chain and Oxidative phosphorylation
 Electron Transport Chain and Oxidative phosphorylation So far we have discussed the catabolism involving oxidation of 6 carbons of glucose to CO 2 via glycolysis and CAC without any oxygen molecule directly
Electron Transport Chain and Oxidative phosphorylation So far we have discussed the catabolism involving oxidation of 6 carbons of glucose to CO 2 via glycolysis and CAC without any oxygen molecule directly
Lecture 11 - Biosynthesis of Amino Acids
 Lecture 11 - Biosynthesis of Amino Acids Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction Biosynthetic pathways for amino acids, nucleotides and lipids
Lecture 11 - Biosynthesis of Amino Acids Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction Biosynthetic pathways for amino acids, nucleotides and lipids
Coenzymes, vitamins and trace elements 209. Petr Tůma Eva Samcová
 Coenzymes, vitamins and trace elements 209 Petr Tůma Eva Samcová History and nomenclature of enzymes 1810, Gay-Lussac made an experiment with yeats alter saccharide to ethanol and CO 2 Fermentation From
Coenzymes, vitamins and trace elements 209 Petr Tůma Eva Samcová History and nomenclature of enzymes 1810, Gay-Lussac made an experiment with yeats alter saccharide to ethanol and CO 2 Fermentation From
number Done by Corrected by Doctor
 number 35 Done by حسام ا بوعوض Corrected by عبدالرحمن الحنبلي Doctor Diala 1 P age We mentioned at the end of the last lecture that ribonucleotide reductase enzyme can be inhibited preventing the synthesis
number 35 Done by حسام ا بوعوض Corrected by عبدالرحمن الحنبلي Doctor Diala 1 P age We mentioned at the end of the last lecture that ribonucleotide reductase enzyme can be inhibited preventing the synthesis
respiration mitochondria mitochondria metabolic pathways reproduction can fuse or split DRP1 interacts with ER tubules chapter DRP1 ER tubule
 mitochondria respiration chapter 3-4 shape highly variable can fuse or split structure outer membrane inner membrane cristae intermembrane space mitochondrial matrix free ribosomes respiratory enzymes
mitochondria respiration chapter 3-4 shape highly variable can fuse or split structure outer membrane inner membrane cristae intermembrane space mitochondrial matrix free ribosomes respiratory enzymes
Protein Name. IFLENVIR,DSVTYTEHAK,TV TALDVVYALK,KTVTALDVV YALK,TVTALDVVYALKR,IF LENVIRDSVTYTEHAK gi Histone H2B
 Table 1. A functional category list of proteins (Lentinula edodes) identified by 1-DGE and nesi-lc-ms/ms. The table lists indicated fraction numbers, matching peptides, scores, accession numbers, protein
Table 1. A functional category list of proteins (Lentinula edodes) identified by 1-DGE and nesi-lc-ms/ms. The table lists indicated fraction numbers, matching peptides, scores, accession numbers, protein
Six Types of Enzyme Catalysts
 Six Types of Enzyme Catalysts Although a huge number of reactions occur in living systems, these reactions fall into only half a dozen types. The reactions are: 1. Oxidation and reduction. Enzymes that
Six Types of Enzyme Catalysts Although a huge number of reactions occur in living systems, these reactions fall into only half a dozen types. The reactions are: 1. Oxidation and reduction. Enzymes that
2. What is molecular oxygen directly converted into? a. Carbon Dioxide b. Water c. Glucose d. None of the Above
 Biochem 1 Mock Exam 3 Chapter 11: 1. What is glucose completely oxidized into? a. Carbon Dioxide and Water b. Carbon Dioxide and Oxygen c. Oxygen and Water d. Water and Glycogen 2. What is molecular oxygen
Biochem 1 Mock Exam 3 Chapter 11: 1. What is glucose completely oxidized into? a. Carbon Dioxide and Water b. Carbon Dioxide and Oxygen c. Oxygen and Water d. Water and Glycogen 2. What is molecular oxygen
Syllabus for BASIC METABOLIC PRINCIPLES
 Syllabus for BASIC METABOLIC PRINCIPLES The video lecture covers basic principles you will need to know for the lectures covering enzymes and metabolism in Principles of Metabolism and elsewhere in the
Syllabus for BASIC METABOLIC PRINCIPLES The video lecture covers basic principles you will need to know for the lectures covering enzymes and metabolism in Principles of Metabolism and elsewhere in the
Biochemistry: A Short Course
 Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 31 Amino Acid Synthesis 2013 W. H. Freeman and Company Chapter 31 Outline Although the atmosphere is approximately 80% nitrogen,
Tymoczko Berg Stryer Biochemistry: A Short Course Second Edition CHAPTER 31 Amino Acid Synthesis 2013 W. H. Freeman and Company Chapter 31 Outline Although the atmosphere is approximately 80% nitrogen,
BIOLOGY 103 Spring 2001 MIDTERM LAB SECTION
 BIOLOGY 103 Spring 2001 MIDTERM NAME KEY LAB SECTION ID# (last four digits of SS#) STUDENT PLEASE READ. Do not put yourself at a disadvantage by revealing the content of this exam to your classmates. Your
BIOLOGY 103 Spring 2001 MIDTERM NAME KEY LAB SECTION ID# (last four digits of SS#) STUDENT PLEASE READ. Do not put yourself at a disadvantage by revealing the content of this exam to your classmates. Your
Supplementary figure legends
 Supplementary figure legends Fig. S1. Lineweaver-Burk plot of putrescine uptake by YeeF. An overnight culture of SK629 was inoculated in 100-mL LBG medium in 500-mL Erlenmeyer flasks. The medium was supplemented
Supplementary figure legends Fig. S1. Lineweaver-Burk plot of putrescine uptake by YeeF. An overnight culture of SK629 was inoculated in 100-mL LBG medium in 500-mL Erlenmeyer flasks. The medium was supplemented
Biochemistry 2 Recita0on Amino Acid Metabolism
 Biochemistry 2 Recita0on Amino Acid Metabolism 04-20- 2015 Glutamine and Glutamate as key entry points for NH 4 + Amino acid catabolism Glutamine synthetase enables toxic NH 4 + to combine with glutamate
Biochemistry 2 Recita0on Amino Acid Metabolism 04-20- 2015 Glutamine and Glutamate as key entry points for NH 4 + Amino acid catabolism Glutamine synthetase enables toxic NH 4 + to combine with glutamate
Electron transport chain, oxidative phosphorylation, mitochondrial transport systems
 Electron transport chain, oxidative phosphorylation, mitochondrial transport systems JAN ILLNER Respiratory chain & oxidative phosphorylation INTERMEMBRANE SPACE ubiquinone cytochrome c ATPase Production
Electron transport chain, oxidative phosphorylation, mitochondrial transport systems JAN ILLNER Respiratory chain & oxidative phosphorylation INTERMEMBRANE SPACE ubiquinone cytochrome c ATPase Production
Chapter 10. Introduction to Nutrition and Metabolism, 3 rd edition David A Bender Taylor & Francis Ltd, London 2002
 Chapter 10 Introduction to Nutrition and Metabolism, 3 rd edition David A Bender Taylor & Francis Ltd, London 2002 Chapter 10: Integration and Control of Metabolism Press the space bar or click the mouse
Chapter 10 Introduction to Nutrition and Metabolism, 3 rd edition David A Bender Taylor & Francis Ltd, London 2002 Chapter 10: Integration and Control of Metabolism Press the space bar or click the mouse
Fig. S1. Summary of the altered metabolism pathways in alcoholic fatty liver disease using MetPA analysis (panel A).
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Fig. S1. Summary of the altered metabolism pathways in alcoholic fatty liver disease
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Fig. S1. Summary of the altered metabolism pathways in alcoholic fatty liver disease
Figure S3 Differentially expressed fungal and plant genes organised by catalytic activity ontology Organisation of E. festucae (A) and L.
 sak WT Number of branches 3 WT sak Figure S1 Vasculature of plants infected with the E. festucae sak mutant. Light micrographs of perennial ryegrass blade tissue showing branching between the vasculature
sak WT Number of branches 3 WT sak Figure S1 Vasculature of plants infected with the E. festucae sak mutant. Light micrographs of perennial ryegrass blade tissue showing branching between the vasculature
Carbohydrate. Metabolism
 Carbohydrate Metabolism Dietary carbohydrates (starch, glycogen, sucrose, lactose Mouth salivary amylase Summary of Carbohydrate Utilization Utilization for energy (glycolysis) ligosaccharides and disaccharides
Carbohydrate Metabolism Dietary carbohydrates (starch, glycogen, sucrose, lactose Mouth salivary amylase Summary of Carbohydrate Utilization Utilization for energy (glycolysis) ligosaccharides and disaccharides
Lecture 13 (10/13/17)
 Lecture 13 (10/13/17) Reading: Ch6; 187-189, 204-205 Problems: Ch4 (text); 2, 3 NXT (after xam 2) Reading: Ch6; 190-191, 194-195, 197-198 Problems: Ch6 (text); 5, 6, 7, 24 OUTLIN NZYMS: Binding & Catalysis
Lecture 13 (10/13/17) Reading: Ch6; 187-189, 204-205 Problems: Ch4 (text); 2, 3 NXT (after xam 2) Reading: Ch6; 190-191, 194-195, 197-198 Problems: Ch6 (text); 5, 6, 7, 24 OUTLIN NZYMS: Binding & Catalysis
Physiological Adaptation. Microbial Physiology Module 4
 Physiological Adaptation Microbial Physiology Module 4 Topics Coordination of Metabolic Reactions Regulation of Enzyme Activity Regulation of Gene Expression Global Control, Signal Transduction and Twocomponent
Physiological Adaptation Microbial Physiology Module 4 Topics Coordination of Metabolic Reactions Regulation of Enzyme Activity Regulation of Gene Expression Global Control, Signal Transduction and Twocomponent
E.coli Core Model: Metabolic Core
 1 E.coli Core Model: Metabolic Core 2 LEARNING OBJECTIVES Each student should be able to: Describe the glycolysis pathway in the core model. Describe the TCA cycle in the core model. Explain gluconeogenesis.
1 E.coli Core Model: Metabolic Core 2 LEARNING OBJECTIVES Each student should be able to: Describe the glycolysis pathway in the core model. Describe the TCA cycle in the core model. Explain gluconeogenesis.
ENHANCED IDENTIFICATION OF 2D-GEL ISOLATED PROTEINS FROM ESCHERICHIA COLI USING PSD MX TM
 ENHANCED IDENTIFICATION OF 2D-GEL ISOLATED PROTEINS FROM ESCHERICHIA COLI USING PSD MX TM E. Claude 1, M. Snel 1, T.Franz 2, A. Bathke 2, T. McKenna 1 and J. Langridge 1 1 Waters Corporation, MS Technologies
ENHANCED IDENTIFICATION OF 2D-GEL ISOLATED PROTEINS FROM ESCHERICHIA COLI USING PSD MX TM E. Claude 1, M. Snel 1, T.Franz 2, A. Bathke 2, T. McKenna 1 and J. Langridge 1 1 Waters Corporation, MS Technologies
0.5. Normalized 95% gray value interval h
 Normalized 95% gray value interval.5.4.3.2.1 h Supplemental Figure 1: Symptom score of root samples used in the proteomics study. For each time point, the normalized 95% gray value interval is an averaged
Normalized 95% gray value interval.5.4.3.2.1 h Supplemental Figure 1: Symptom score of root samples used in the proteomics study. For each time point, the normalized 95% gray value interval is an averaged
Cellular Respiration Harvesting Chemical Energy ATP
 Cellular Respiration Harvesting Chemical Energy ATP 2006-2007 What s the point? The point is to make ATP! ATP 2006-2007 Harvesting stored energy Energy is stored in organic molecules carbohydrates, fats,
Cellular Respiration Harvesting Chemical Energy ATP 2006-2007 What s the point? The point is to make ATP! ATP 2006-2007 Harvesting stored energy Energy is stored in organic molecules carbohydrates, fats,
Biochemistry 2000 Sample Question Transcription, Translation and Lipids. (1) Give brief definitions or unique descriptions of the following terms:
 (1) Give brief definitions or unique descriptions of the following terms: (a) exon (b) holoenzyme (c) anticodon (d) trans fatty acid (e) poly A tail (f) open complex (g) Fluid Mosaic Model (h) embedded
(1) Give brief definitions or unique descriptions of the following terms: (a) exon (b) holoenzyme (c) anticodon (d) trans fatty acid (e) poly A tail (f) open complex (g) Fluid Mosaic Model (h) embedded
number Done by Corrected by Doctor Nayef Karadsheh
 number 11 Done by حسام أبو عوض Corrected by Moayyad Al-Shafei Doctor Nayef Karadsheh 1 P a g e General Regulatory Aspects in Metabolism: We can divide all pathways in metabolism to catabolicand anabolic.
number 11 Done by حسام أبو عوض Corrected by Moayyad Al-Shafei Doctor Nayef Karadsheh 1 P a g e General Regulatory Aspects in Metabolism: We can divide all pathways in metabolism to catabolicand anabolic.
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the
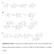 Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
SUPPLEMENTAL TABLE I. Identified Proteins in Bovine Testicular Hyaluronidase Type I-S via LC-MS/MS
 SUPPLEMENTAL TABLE I. Identified Proteins in Bovine Testicular Hyaluronidase Type I-S via LC-MS/MS No. Protein 1 serum albumin precursor gi 30794280 2 annexin A2 gi 27807289 3 Phosphatidylethanolamine-binding
SUPPLEMENTAL TABLE I. Identified Proteins in Bovine Testicular Hyaluronidase Type I-S via LC-MS/MS No. Protein 1 serum albumin precursor gi 30794280 2 annexin A2 gi 27807289 3 Phosphatidylethanolamine-binding
Mitochondria and ATP Synthesis
 Mitochondria and ATP Synthesis Mitochondria and ATP Synthesis 1. Mitochondria are sites of ATP synthesis in cells. 2. ATP is used to do work; i.e. ATP is an energy source. 3. ATP hydrolysis releases energy
Mitochondria and ATP Synthesis Mitochondria and ATP Synthesis 1. Mitochondria are sites of ATP synthesis in cells. 2. ATP is used to do work; i.e. ATP is an energy source. 3. ATP hydrolysis releases energy
CELL BIOLOGY - CLUTCH CH AEROBIC RESPIRATION.
 !! www.clutchprep.com CONCEPT: OVERVIEW OF AEROBIC RESPIRATION Cellular respiration is a series of reactions involving electron transfers to breakdown molecules for (ATP) 1. Glycolytic pathway: Glycolysis
!! www.clutchprep.com CONCEPT: OVERVIEW OF AEROBIC RESPIRATION Cellular respiration is a series of reactions involving electron transfers to breakdown molecules for (ATP) 1. Glycolytic pathway: Glycolysis
Dental Students Biochemistry Exam V Questions ( Note: In all cases, the only correct answer is the best answer)
 Dental Students Biochemistry Exam V Questions - 2006 ( Note: In all cases, the only correct answer is the best answer) 1. Essential fatty acids are: A. precursors of biotin B. precursors of tyrosine C.
Dental Students Biochemistry Exam V Questions - 2006 ( Note: In all cases, the only correct answer is the best answer) 1. Essential fatty acids are: A. precursors of biotin B. precursors of tyrosine C.
Genome-wide association studies for understanding pathogen evolution. Samuel K. Sheppard
 Genome-wide association studies for understanding pathogen evolution Samuel K. Sheppard Pathogenic bacteria Population genomics and evolution Sheppard et al. Science 2008; 11: 237-239. Sheppard et al.
Genome-wide association studies for understanding pathogen evolution Samuel K. Sheppard Pathogenic bacteria Population genomics and evolution Sheppard et al. Science 2008; 11: 237-239. Sheppard et al.
A) Choose the correct answer: 1) Reduction of a substance can mostly occur in the living cells by:
 Code: 1 1) Reduction of a substance can mostly occur in the living cells by: (a) Addition of oxygen (b) Removal of electrons (c) Addition of electrons (d) Addition of hydrogen 2) Starting with succinate
Code: 1 1) Reduction of a substance can mostly occur in the living cells by: (a) Addition of oxygen (b) Removal of electrons (c) Addition of electrons (d) Addition of hydrogen 2) Starting with succinate
III. Metabolism The Citric Acid Cycle
 Department of Chemistry and Biochemistry University of Lethbridge III. Metabolism The Citric Acid Cycle Slide 1 The Eight Steps of the Citric Acid Cycle Enzymes: 4 dehydrogenases (2 decarboxylation) 3
Department of Chemistry and Biochemistry University of Lethbridge III. Metabolism The Citric Acid Cycle Slide 1 The Eight Steps of the Citric Acid Cycle Enzymes: 4 dehydrogenases (2 decarboxylation) 3
Electron Transport and Oxidative. Phosphorylation
 Electron Transport and Oxidative Phosphorylation Electron-transport chain electron- Definition: The set of proteins and small molecules involved in the orderly sequence of transfer to oxygen within the
Electron Transport and Oxidative Phosphorylation Electron-transport chain electron- Definition: The set of proteins and small molecules involved in the orderly sequence of transfer to oxygen within the
7 Pathways That Harvest Chemical Energy
 7 Pathways That Harvest Chemical Energy Pathways That Harvest Chemical Energy How Does Glucose Oxidation Release Chemical Energy? What Are the Aerobic Pathways of Glucose Metabolism? How Is Energy Harvested
7 Pathways That Harvest Chemical Energy Pathways That Harvest Chemical Energy How Does Glucose Oxidation Release Chemical Energy? What Are the Aerobic Pathways of Glucose Metabolism? How Is Energy Harvested
Oxidative Phosphorylation
 Electron Transport Chain (overview) The NADH and FADH 2, formed during glycolysis, β- oxidation and the TCA cycle, give up their electrons to reduce molecular O 2 to H 2 O. Electron transfer occurs through
Electron Transport Chain (overview) The NADH and FADH 2, formed during glycolysis, β- oxidation and the TCA cycle, give up their electrons to reduce molecular O 2 to H 2 O. Electron transfer occurs through
CH395G FINAL (3 rd ) EXAM Kitto/Hackert - Fall 2003
 CH395G FINAL (3 rd ) EXAM Kitto/Hackert - Fall 2003 1. A cell in an active, catabolic state has a. a high (ATP/ADP) and a high (NADH/NAD + ) ratio b. a high (ATP/ADP) and a low (NADH/NAD + ) ratio c. a
CH395G FINAL (3 rd ) EXAM Kitto/Hackert - Fall 2003 1. A cell in an active, catabolic state has a. a high (ATP/ADP) and a high (NADH/NAD + ) ratio b. a high (ATP/ADP) and a low (NADH/NAD + ) ratio c. a
Chemistry 107 Exam 4 Study Guide
 Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
WT siz1-2 siz1-3. WT siz1-2 siz1-3. -Pi, 0.05 M IAA. -Pi, 2.5 M NPA
 A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
If you ate a clown, would it taste funny? Oh, wait, that s cannibalism . Anabolism
 If you ate a clown, would it taste funny? Oh, wait, that s cannibalism. Anabolism is about putting things together. Anabolism: The Use of Energy in Biosynthesis Anabolism energy from catabolism is used
If you ate a clown, would it taste funny? Oh, wait, that s cannibalism. Anabolism is about putting things together. Anabolism: The Use of Energy in Biosynthesis Anabolism energy from catabolism is used
University of Palestine. Final Exam 2016/2017 Total Grade:
 Part 1 : Multiple Choice Questions (MCQs) 1)Which of the following statements about Michaelis-Menten kinetics is correct? a)k m, the Michaelis constant, is defined as the concentration of substrate required
Part 1 : Multiple Choice Questions (MCQs) 1)Which of the following statements about Michaelis-Menten kinetics is correct? a)k m, the Michaelis constant, is defined as the concentration of substrate required
Biochemistry 423 Final Examination NAME:
 Biochemistry 423 Final Examination NAME: 1 Circle the single BEST answer (3 points each) 1. At equilibrium the free energy of a reaction G A. depends only on the temperature B. is positive C. is 0 D. is
Biochemistry 423 Final Examination NAME: 1 Circle the single BEST answer (3 points each) 1. At equilibrium the free energy of a reaction G A. depends only on the temperature B. is positive C. is 0 D. is
SP GPI Found in exosomes
 Accession /Name/Functional group Spectr. counts SP GPI Found in exosomes Description Metabolism and energy production (21) CNAG_06770 fructose-bisphosphate aldolase 0.94 - - + gluconeogenesis and glycolysis
Accession /Name/Functional group Spectr. counts SP GPI Found in exosomes Description Metabolism and energy production (21) CNAG_06770 fructose-bisphosphate aldolase 0.94 - - + gluconeogenesis and glycolysis
Photosynthesis in chloroplasts CO2 + H2O. Cellular respiration in mitochondria ATP. powers most cellular work. Heat energy
 Figure 9-01 LE 9-2 Light energy ECOSYSTEM Photosynthesis in chloroplasts CO2 + H2O Cellular respiration in mitochondria Organic + O molecules 2 powers most cellular work Heat energy LE 9-UN161a becomes
Figure 9-01 LE 9-2 Light energy ECOSYSTEM Photosynthesis in chloroplasts CO2 + H2O Cellular respiration in mitochondria Organic + O molecules 2 powers most cellular work Heat energy LE 9-UN161a becomes
Find this material useful? You can help our team to keep this site up and bring you even more content consider donating via the link on our site.
 Find this material useful? You can help our team to keep this site up and bring you even more content consider donating via the link on our site. Still having trouble understanding the material? Check
Find this material useful? You can help our team to keep this site up and bring you even more content consider donating via the link on our site. Still having trouble understanding the material? Check
MEMBRANE-BOUND ELECTRON TRANSFER AND ATP SYNTHESIS (taken from Chapter 18 of Stryer)
 MEMBRANE-BOUND ELECTRON TRANSFER AND ATP SYNTHESIS (taken from Chapter 18 of Stryer) FREE ENERGY MOST USEFUL THERMODYNAMIC CONCEPT IN BIOCHEMISTRY Living things require an input of free energy for 3 major
MEMBRANE-BOUND ELECTRON TRANSFER AND ATP SYNTHESIS (taken from Chapter 18 of Stryer) FREE ENERGY MOST USEFUL THERMODYNAMIC CONCEPT IN BIOCHEMISTRY Living things require an input of free energy for 3 major
University of Guelph Department of Chemistry and Biochemistry Structure and Function In Biochemistry
 University of Guelph Department of Chemistry and Biochemistry 19-356 Structure and Function In Biochemistry Final Exam, April 21, 1997. Time allowed, 120 min. Answer questions 1-30 on the computer scoring
University of Guelph Department of Chemistry and Biochemistry 19-356 Structure and Function In Biochemistry Final Exam, April 21, 1997. Time allowed, 120 min. Answer questions 1-30 on the computer scoring
BIOCHEMISTRY - CLUTCH REVIEW 6.
 !! www.clutchprep.com CONCEPT: AMINO ACID OXIDATION Urea cycle occurs in liver, removes amino groups from amino acids so they may enter the citric acid cycle 2 nitrogen enter the cycle to ultimately leave
!! www.clutchprep.com CONCEPT: AMINO ACID OXIDATION Urea cycle occurs in liver, removes amino groups from amino acids so they may enter the citric acid cycle 2 nitrogen enter the cycle to ultimately leave
Metabolic Biochemistry / BIBC 102 Midterm Exam / Spring 2005
 Metabolic Biochemistry / BIBC 102 Midterm Exam / Spring 2005 I. (20 points) Fill in all of the enzyme catalyzed reactions which convert glycogen to lactate. Draw the correct structure for each intermediate
Metabolic Biochemistry / BIBC 102 Midterm Exam / Spring 2005 I. (20 points) Fill in all of the enzyme catalyzed reactions which convert glycogen to lactate. Draw the correct structure for each intermediate
Answer three from questions 5, 6, 7, 8, and 9.
 BCH 4053 May 1, 2003 FINAL EXAM NAME There are 9 pages and 9 questions on the exam. nly five are to be answered, each worth 20 points. Answer two from questions 1, 2, 3, and 4 Answer three from questions
BCH 4053 May 1, 2003 FINAL EXAM NAME There are 9 pages and 9 questions on the exam. nly five are to be answered, each worth 20 points. Answer two from questions 1, 2, 3, and 4 Answer three from questions
Microbiology AN INTRODUCTION
 TORTORA FUNKE CASE Microbiology AN INTRODUCTION EIGHTH EDITION B.E Pruitt & Jane J. Stein Chapter 5, part A Microbial Metabolism PowerPoint Lecture Slide Presentation prepared by Christine L. Case Microbial
TORTORA FUNKE CASE Microbiology AN INTRODUCTION EIGHTH EDITION B.E Pruitt & Jane J. Stein Chapter 5, part A Microbial Metabolism PowerPoint Lecture Slide Presentation prepared by Christine L. Case Microbial
Biology 638 Biochemistry II Exam-2
 Biology 638 Biochemistry II Exam-2 Biol 638, Exam-2 (Code-1) 1. Assume that 16 glucose molecules enter into a liver cell and are attached to a liner glycogen one by one. Later, this glycogen is broken-down
Biology 638 Biochemistry II Exam-2 Biol 638, Exam-2 (Code-1) 1. Assume that 16 glucose molecules enter into a liver cell and are attached to a liner glycogen one by one. Later, this glycogen is broken-down
Points 1. Following is the overall reaction catalyzed by the Calvin-Benson cycle:
 BCH 4054 February 22, 2002 HOUR TEST 2 NAME_ Points 1. Following is the overall reaction catalyzed by the Calvin-Benson cycle: CO 2 + 3ATP + 2NADPH 1/3 glyceraldehyde-3-p + 3ADP + 2NADP + Give the structures
BCH 4054 February 22, 2002 HOUR TEST 2 NAME_ Points 1. Following is the overall reaction catalyzed by the Calvin-Benson cycle: CO 2 + 3ATP + 2NADPH 1/3 glyceraldehyde-3-p + 3ADP + 2NADP + Give the structures
Metabolism. Metabolic pathways. BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 11: Metabolic Pathways
 BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 11: Metabolic Pathways http://compbio.uchsc.edu/hunter/bio5099 Larry.Hunter@uchsc.edu Metabolism Metabolism is the chemical change of
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 11: Metabolic Pathways http://compbio.uchsc.edu/hunter/bio5099 Larry.Hunter@uchsc.edu Metabolism Metabolism is the chemical change of
MITOCHONDRIA LECTURES OVERVIEW
 1 MITOCHONDRIA LECTURES OVERVIEW A. MITOCHONDRIA LECTURES OVERVIEW Mitochondrial Structure The arrangement of membranes: distinct inner and outer membranes, The location of ATPase, DNA and ribosomes The
1 MITOCHONDRIA LECTURES OVERVIEW A. MITOCHONDRIA LECTURES OVERVIEW Mitochondrial Structure The arrangement of membranes: distinct inner and outer membranes, The location of ATPase, DNA and ribosomes The
Midterm 2 Results. Standard Deviation:
 Midterm 2 Results High: Low: Mean: Standard Deviation: 97.5% 16% 58% 16.3 Lecture 17 Amino Acid Metabolism Urea Cycle N and S assimilation Last cofactors: THF and SAM Dietary (Exogenous) Proteins Hydrolyzed
Midterm 2 Results High: Low: Mean: Standard Deviation: 97.5% 16% 58% 16.3 Lecture 17 Amino Acid Metabolism Urea Cycle N and S assimilation Last cofactors: THF and SAM Dietary (Exogenous) Proteins Hydrolyzed
Type III CRISPR complexes from Thermus thermophilus
 Regular paper Supplementary Material, Vol. 63, 2016 377 386 http://dx.doi.org/10.18388/abp.2016_1261 Type III CRISPR complexes from Thermus thermophilus Marta Szychowska 1,2,a,#, Wojciech Siwek 1,b,#,
Regular paper Supplementary Material, Vol. 63, 2016 377 386 http://dx.doi.org/10.18388/abp.2016_1261 Type III CRISPR complexes from Thermus thermophilus Marta Szychowska 1,2,a,#, Wojciech Siwek 1,b,#,
9/10/2012. The electron transfer system in the inner membrane of mitochondria in plants
 LECT 6. RESPIRATION COMPETENCIES Students, after mastering the materials of Plant Physiology course, should be able to: 1. To explain the process of respiration (the oxidation of substrates particularly
LECT 6. RESPIRATION COMPETENCIES Students, after mastering the materials of Plant Physiology course, should be able to: 1. To explain the process of respiration (the oxidation of substrates particularly
BIOLOGY - CLUTCH CH.9 - RESPIRATION.
 !! www.clutchprep.com CONCEPT: REDOX REACTIONS Redox reaction a chemical reaction that involves the transfer of electrons from one atom to another Oxidation loss of electrons Reduction gain of electrons
!! www.clutchprep.com CONCEPT: REDOX REACTIONS Redox reaction a chemical reaction that involves the transfer of electrons from one atom to another Oxidation loss of electrons Reduction gain of electrons
Energy storage in cells
 Energy storage in cells Josef Fontana EC - 58 Overview of the lecture Introduction to the storage substances of human body Overview of storage compounds in the body Glycogen metabolism Structure of glycogen
Energy storage in cells Josef Fontana EC - 58 Overview of the lecture Introduction to the storage substances of human body Overview of storage compounds in the body Glycogen metabolism Structure of glycogen
Midterm 2. Low: 14 Mean: 61.3 High: 98. Standard Deviation: 17.7
 Midterm 2 Low: 14 Mean: 61.3 High: 98 Standard Deviation: 17.7 Lecture 17 Amino Acid Metabolism Review of Urea Cycle N and S assimilation Last cofactors: THF and SAM Synthesis of few amino acids Dietary
Midterm 2 Low: 14 Mean: 61.3 High: 98 Standard Deviation: 17.7 Lecture 17 Amino Acid Metabolism Review of Urea Cycle N and S assimilation Last cofactors: THF and SAM Synthesis of few amino acids Dietary
Citric acid cycle and respiratory chain. Pavla Balínová
 Citric acid cycle and respiratory chain Pavla Balínová Mitochondria Structure of mitochondria: Outer membrane Inner membrane (folded) Matrix space (mtdna, ribosomes, enzymes of CAC, β-oxidation of FA,
Citric acid cycle and respiratory chain Pavla Balínová Mitochondria Structure of mitochondria: Outer membrane Inner membrane (folded) Matrix space (mtdna, ribosomes, enzymes of CAC, β-oxidation of FA,
Lecture 10 - Protein Turnover and Amino Acid Catabolism
 Lecture 10 - Protein Turnover and Amino Acid Catabolism Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction 2 Proteins are degraded into amino acids. Protein
Lecture 10 - Protein Turnover and Amino Acid Catabolism Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction 2 Proteins are degraded into amino acids. Protein
Characterization of the use of wastewater from hydrothermal liquefaction as a nitrogen source by the potential algal biofuel strain Picochlorum sp.
 Characterization of the use of wastewater from hydrothermal liquefaction as a nitrogen source by the potential algal biofuel strain Picochlorum sp. Shuyi Wang, Xinguo Shi, Fatima Foflonker, Debashish Bhattacharya,
Characterization of the use of wastewater from hydrothermal liquefaction as a nitrogen source by the potential algal biofuel strain Picochlorum sp. Shuyi Wang, Xinguo Shi, Fatima Foflonker, Debashish Bhattacharya,
1. Investigate the structure of the trna Synthase in complex with a trna molecule. (pdb ID 1ASY).
 Problem Set 11 (Due Nov 25 th ) 1. Investigate the structure of the trna Synthase in complex with a trna molecule. (pdb ID 1ASY). a. Why don t trna molecules contain a 5 triphosphate like other RNA molecules
Problem Set 11 (Due Nov 25 th ) 1. Investigate the structure of the trna Synthase in complex with a trna molecule. (pdb ID 1ASY). a. Why don t trna molecules contain a 5 triphosphate like other RNA molecules
7/5/2014. Microbial. Metabolism. Basic Chemical Reactions Underlying. Metabolism. Metabolism: Overview
 PowerPoint Lecture Presentations prepared by Mindy Miller-Kittrell, North Carolina State University Basic Chemical Reactions Underlying Metabolism Metabolism C H A P T E R 5 Microbial Metabolism Collection
PowerPoint Lecture Presentations prepared by Mindy Miller-Kittrell, North Carolina State University Basic Chemical Reactions Underlying Metabolism Metabolism C H A P T E R 5 Microbial Metabolism Collection
Electron transport chain chapter 6 (page 73) BCH 340 lecture 6
 Electron transport chain chapter 6 (page 73) BCH 340 lecture 6 The Metabolic Pathway of Cellular Respiration All of the reactions involved in cellular respiration can be grouped into three main stages
Electron transport chain chapter 6 (page 73) BCH 340 lecture 6 The Metabolic Pathway of Cellular Respiration All of the reactions involved in cellular respiration can be grouped into three main stages
Cellular Respiration
 Cellular Respiration C 6 H 12 O 6 + 6O 2 -----> 6CO 2 + 6H 2 0 + energy (heat and ATP) 1. Energy Capacity to move or change matter Forms of energy are important to life include Chemical, radiant (heat
Cellular Respiration C 6 H 12 O 6 + 6O 2 -----> 6CO 2 + 6H 2 0 + energy (heat and ATP) 1. Energy Capacity to move or change matter Forms of energy are important to life include Chemical, radiant (heat
CHEM121. Unit 6: Enzymes. Lecture 10. At the end of the lecture, students should be able to:
 CHEM121 Unit 6: Enzymes Lecture 10 At the end of the lecture, students should be able to: Define the term enzyme Name and classify enzymes according to the: type of reaction catalyzed type of specificity
CHEM121 Unit 6: Enzymes Lecture 10 At the end of the lecture, students should be able to: Define the term enzyme Name and classify enzymes according to the: type of reaction catalyzed type of specificity
Metabolism III. Aim: understand gluconeogenesis, pentose phosphate pathway, photosynthesis and amino acid synthesis
 Metabolism III Aim: understand gluconeogenesis, pentose phosphate pathway, photosynthesis and amino acid synthesis Anabolism From a carbon source and inorganic molecules, microbes synthesize new organelles
Metabolism III Aim: understand gluconeogenesis, pentose phosphate pathway, photosynthesis and amino acid synthesis Anabolism From a carbon source and inorganic molecules, microbes synthesize new organelles
Point total. Page # Exam Total (out of 90) The number next to each intermediate represents the total # of C-C and C-H bonds in that molecule.
 This exam is worth 90 points. Pages 2- have questions. Page 1 is for your reference only. Honor Code Agreement - Signature: Date: (You agree to not accept or provide assistance to anyone else during this
This exam is worth 90 points. Pages 2- have questions. Page 1 is for your reference only. Honor Code Agreement - Signature: Date: (You agree to not accept or provide assistance to anyone else during this
Chapter 9. Cellular Respiration and Fermentation
 Chapter 9 Cellular Respiration and Fermentation Energy flows into an ecosystem as sunlight and leaves as heat Photosynthesis generates O 2 and organic molecules, which are used in cellular respiration
Chapter 9 Cellular Respiration and Fermentation Energy flows into an ecosystem as sunlight and leaves as heat Photosynthesis generates O 2 and organic molecules, which are used in cellular respiration
BIOL 158: BIOLOGICAL CHEMISTRY II
 BIOL 158: BIOLOGICAL CHEMISTRY II Lecture 5: Vitamins and Coenzymes Lecturer: Christopher Larbie, PhD Introduction Cofactors bind to the active site and assist in the reaction mechanism Apoenzyme is an
BIOL 158: BIOLOGICAL CHEMISTRY II Lecture 5: Vitamins and Coenzymes Lecturer: Christopher Larbie, PhD Introduction Cofactors bind to the active site and assist in the reaction mechanism Apoenzyme is an
Ch. 9 Cell Respiration. Title: Oct 15 3:24 PM (1 of 53)
 Ch. 9 Cell Respiration Title: Oct 15 3:24 PM (1 of 53) Essential question: How do cells use stored chemical energy in organic molecules and to generate ATP? Title: Oct 15 3:28 PM (2 of 53) Title: Oct 19
Ch. 9 Cell Respiration Title: Oct 15 3:24 PM (1 of 53) Essential question: How do cells use stored chemical energy in organic molecules and to generate ATP? Title: Oct 15 3:28 PM (2 of 53) Title: Oct 19
MDSC 1102/VM1102 Cardiovascular and Renal. Purine nucleotide metabolism
 MDSC 1102/VM1102 Cardiovascular and Renal Purine nucleotide metabolism Dr. J. Foster Biochemistry Unit, Dept. Preclinical Sciences Faculty of Medical Sciences, U.W.I. Learning Objectives Discuss purineand
MDSC 1102/VM1102 Cardiovascular and Renal Purine nucleotide metabolism Dr. J. Foster Biochemistry Unit, Dept. Preclinical Sciences Faculty of Medical Sciences, U.W.I. Learning Objectives Discuss purineand
Electron Transport and oxidative phosphorylation (ATP Synthesis) Dr. Howaida Nounou Biochemistry department Sciences college
 Electron Transport and oxidative phosphorylation (ATP Synthesis) Dr. Howaida Nounou Biochemistry department Sciences college The Metabolic Pathway of Cellular Respiration All of the reactions involved
Electron Transport and oxidative phosphorylation (ATP Synthesis) Dr. Howaida Nounou Biochemistry department Sciences college The Metabolic Pathway of Cellular Respiration All of the reactions involved
Chemistry 5.07 Problem Set
 Chemistry 5.07 Problem Set 8 2013 Problem 1. All oxidation steps in the pathway from glucose to CO 2 result in the production of NADH, except the succinate dehydrogenase (SDH) step in the TCA cycle, which
Chemistry 5.07 Problem Set 8 2013 Problem 1. All oxidation steps in the pathway from glucose to CO 2 result in the production of NADH, except the succinate dehydrogenase (SDH) step in the TCA cycle, which
Chapter 14 - Electron Transport and Oxidative Phosphorylation
 Chapter 14 - Electron Transport and Oxidative Phosphorylation The cheetah, whose capacity for aerobic metabolism makes it one of the fastest animals Prentice Hall c2002 Chapter 14 1 14.4 Oxidative Phosphorylation
Chapter 14 - Electron Transport and Oxidative Phosphorylation The cheetah, whose capacity for aerobic metabolism makes it one of the fastest animals Prentice Hall c2002 Chapter 14 1 14.4 Oxidative Phosphorylation
Supplementary Figure 1. High-affinity methane oxidation (HAMO) dynamics of soils with added methane at ppmv for 1 time and 10 times.
 Supplementary Figure 1. High-affinity methane oxidation () dynamics of soils with added methane at 10000 ppmv for 1 time and 10 times. After the complete consumption of 10000 ppmv methane, the measurement
Supplementary Figure 1. High-affinity methane oxidation () dynamics of soils with added methane at 10000 ppmv for 1 time and 10 times. After the complete consumption of 10000 ppmv methane, the measurement
Synthesis of ATP, the energy currency in metabolism
 Synthesis of ATP, the energy currency in metabolism Note that these are simplified summaries to support lecture material Either Substrate-level phosphorylation (SLP) Or Electron transport phosphorylation
Synthesis of ATP, the energy currency in metabolism Note that these are simplified summaries to support lecture material Either Substrate-level phosphorylation (SLP) Or Electron transport phosphorylation
Photosynthesis in chloroplasts. Cellular respiration in mitochondria ATP. ATP powers most cellular work
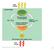 Light energy ECOSYSTEM CO + H O Photosynthesis in chloroplasts Cellular respiration in mitochondria Organic molecules + O powers most cellular work Heat energy 1 becomes oxidized (loses electron) becomes
Light energy ECOSYSTEM CO + H O Photosynthesis in chloroplasts Cellular respiration in mitochondria Organic molecules + O powers most cellular work Heat energy 1 becomes oxidized (loses electron) becomes
Oxidative Phosphorylation
 Oxidative Phosphorylation Energy from Reduced Fuels Is Used to Synthesize ATP in Animals Carbohydrates, lipids, and amino acids are the main reduced fuels for the cell. Electrons from reduced fuels are
Oxidative Phosphorylation Energy from Reduced Fuels Is Used to Synthesize ATP in Animals Carbohydrates, lipids, and amino acids are the main reduced fuels for the cell. Electrons from reduced fuels are
Microbial Metabolism. PowerPoint Lecture Presentations prepared by Bradley W. Christian, McLennan Community College C H A P T E R
 PowerPoint Lecture Presentations prepared by Bradley W. Christian, McLennan Community College C H A P T E R 5 Microbial Metabolism Big Picture: Metabolism Metabolism is the buildup and breakdown of nutrients
PowerPoint Lecture Presentations prepared by Bradley W. Christian, McLennan Community College C H A P T E R 5 Microbial Metabolism Big Picture: Metabolism Metabolism is the buildup and breakdown of nutrients
Amino acid Catabolism
 Enzymatic digestion of dietary proteins in gastrointestinal-tract. Amino acid Catabolism Amino acids: 1. There are 20 different amino acid, they are monomeric constituents of proteins 2. They act as precursors
Enzymatic digestion of dietary proteins in gastrointestinal-tract. Amino acid Catabolism Amino acids: 1. There are 20 different amino acid, they are monomeric constituents of proteins 2. They act as precursors
Chapter 10 Lecture Notes: Microbial Metabolism The Use of Energy in Biosynthesis
 Chapter 10 Lecture Notes: Microbial Metabolism The Use of Energy in Biosynthesis I. Principles governing biosynthesis A. Construct larger macromolecules from smaller subunits to conserve genetic storage
Chapter 10 Lecture Notes: Microbial Metabolism The Use of Energy in Biosynthesis I. Principles governing biosynthesis A. Construct larger macromolecules from smaller subunits to conserve genetic storage
Objective: You will be able to explain how the subcomponents of
 Objective: You will be able to explain how the subcomponents of nucleic acids determine the properties of that polymer. Do Now: Read the first two paragraphs from enduring understanding 4.A Essential knowledge:
Objective: You will be able to explain how the subcomponents of nucleic acids determine the properties of that polymer. Do Now: Read the first two paragraphs from enduring understanding 4.A Essential knowledge:
Physiological Chemistry II Exam IV Dr. Melissa Kelley April 13, 2004
 Name Write your name on the back of the exam Physiological Chemistry II Exam IV Dr. Melissa Kelley April 13, 2004 This examination consists of forty-four questions, each having 2 points. The remaining
Name Write your name on the back of the exam Physiological Chemistry II Exam IV Dr. Melissa Kelley April 13, 2004 This examination consists of forty-four questions, each having 2 points. The remaining
We must be able to make glucose
 Biosynthesis of Carbohydrates Synthesis of glucose (gluconeogenesis) Glycogen Formation of pentoses and NADPH Photosynthesis We must be able to make glucose Compulsory need for glucose (above all the brain)
Biosynthesis of Carbohydrates Synthesis of glucose (gluconeogenesis) Glycogen Formation of pentoses and NADPH Photosynthesis We must be able to make glucose Compulsory need for glucose (above all the brain)
