Large-Scale Discovery of Enhancers from Human Heart Tissue
|
|
|
- Nelson Grant
- 5 years ago
- Views:
Transcription
1 Large-Scale Discovery of Enhancers from Human Heart Tissue Dalit May, Matthew J. Blow, Tommy Kaplan, David J. McCulley, Brian C. Jensen, Jennifer A. Akiyama, Amy Holt, Ingrid Plajzer-Frick, Malak Shoukry, Crystal Wright, Veena Afzal, Paul C. Simpson, Edward M. Rubin, Brian L. Black, James Bristow, Len A. Pennacchio, Axel Visel - Supplementary Information -
2 May et al: ChIP-seq Discovery of Human Heart Enhancers - 2 Supplementary Note Enrichment of p300/cbp peaks near PolII binding sites To further investigate the association between p300/cbp binding sites in the human heart and transcription of nearby genes, we used ChIP-seq to map genome-wide binding of RNA polymerase II (PolII) in human fetal heart (Methods). Enrichment analysis of this data-set identified 5,540 proximal peaks (less than 2.5kb of transcriptional start sites). There is a significant enrichment of human heart candidate enhancers near proximal PolII peaks, extending up to 150kb away (Supplementary Fig. 4). These results show that p300/cbp peaks are correlated with RNA polymerase II peaks, supporting our findings that the identified p300/cbp peaks are linked to transcription in the human heart. Functional conservation of human candidate enhancers in mice Limited sequence conservation of candidate enhancers is potentially indicative of functional changes in regulatory architecture that have occurred across species 1-3. To further evaluate the functional conservation of human candidate heart enhancers across mammalian species, we performed p300/cbp ChIP-seq on mouse heart tissue at postnatal day 2, a stage at which the developmental progression and gene expression profile of the mouse heart is broadly similar to that of human fetal heart at gestational week 16 4,5. Using identical methods as for human heart tissue, we identified 6,564 candidate enhancers (distal ChIP-seq peaks, Supplementary Table 1) and compared them to the human fetal candidate enhancer sets. We catalogued the human heart candidate enhancers by their p300/cbp binding conservation to mouse into 4 classes: 1) Significant p300/cbp binding at the orthologous genomic site in the mouse heart (21%); 2) Human peaks whose mouse homologous regions were not called as p300/cbp peaks but exceeded the genome-wide background (25%); 3) p300/cbp binding at the orthologous genomic site in the mouse genome does not exceed background levels (41%); 4) Human candidate enhancers that could not be aligned to a unique position in the mouse genome (13%) 6 (Supplementary Fig. 7a, Methods). For each class of peaks, - 2 -
3 May et al: ChIP-seq Discovery of Human Heart Enhancers - 3 we further characterized the degree of sequence constraint across multiple mammalian species using their phastcons score 7 (Supplementary Fig. 7b). These results suggest that while a fraction of the human candidate heart enhancers are functionally conserved between the two species, more than half (54%) of all candidate enhancers identified from human heart tissue did not show any evidence of an orthologous p300/cbp binding site in the mouse heart data set. A considerable proportion (1,296; 26%) of fetal human heart candidate enhancers show neither substantial sequence constraint nor binding conservation to the mouse genome and therefore could not have been identified using sequence-based or binding conservation (asterisks in Supplementary Fig. 7b)
4 May et al: ChIP-seq Discovery of Human Heart Enhancers - 4 Supplementary Figures Supplementary Figure 1: Reproducibility of p300/cbp ChIP-seq. The enrichment of reads from the primary human fetal heart sample analyzed in this study was compared to that from a second human fetal heart sample sequenced at lower coverage (see Methods), using the 2,233 candidate adult human heart enhancers identified in this study as anchoring points. Read coverage was cumulated across all enhancers, and corrected for the different number of reads from each sample. Both the original and validation fetal heart data sets exhibit highly significant enrichment at candidate heart enhancers (*, p < 1e-90, Fisher s Exact test). In contrast, no enrichment was observed near heart enhancers in human input DNA (see Methods). The quantitatively similar degree of enrichment in the two fetal heart data sets supports the reproducibility of p300/cbp ChIP-seq from human heart tissue
5 May et al: ChIP-seq Discovery of Human Heart Enhancers - 5 Supplementary Figure 2: p300/cbp binding conservation in fetal and adult human heart. p300/cbp binding sites in the human genome were compared between fetal and adult human heart. a) Heat map of human ChIP-seq read density in 100kb windows surrounding each of the human adult heart candidate enhancers in the adult human heart (left), compared to ChIP-seq read density at the same sites in the fetal data set (right). b) Heat map of human ChIP-seq read density in 100kb windows surrounding each of the human fetal heart candidate enhancers in the fetal human heart (left), compared to ChIP-seq read density at the same sites in the adult data set (right). Peaks fall into three categories, based on ChIP-seq signal in the respective other data set (from top): 1. significant binding in both data sets, 2. read density in the respective other data set is above background, but below peak significance threshold, 3. read density in the respective other data set not exceeding genome-wide background (stage-specific). c) Overlap of candidate enhancers identified in both fetal and adult heart tissues, with stage-specific and suggestively enriched regions shown in separate colors. Minor differences between total peaks and counts in categories of overlap are due to cases of peaks overlapping more than one peak in the respective other dataset. -5 -
6 May et al: ChIP-seq Discovery of Human Heart Enhancers - 6 Supplementary Figure 3: p300/cbp distal peaks near heart-expressed and non-heart-expressed genes. Frequency of adult human heart candidate enhancers (red) compared to matched random regions (black) near genes highly expressed in adult heart (a) and near genes highly expressed in 10 other tissues (b). The graphs display the frequency of the different elements up to 200kb away from the transcription start site and show significant enrichment of heart p300/cbp candidate enhancers in proximity to heart-expressed genes (starting at 2.5kb away from TSS). Fold enrichment of the most proximal peaks (2.5-10kb from TSS) is 3.6 (p = 1e-11, binomial distribution). The significant enrichment over random peaks and over nonheart genes is maintained up to 100kb (binomial distribution p < 0.002). Errors bars indicate 95% confidence intervals. Supplementary Figure 4: Enrichment of human heart p300/cbp ChIP-seq peaks near human heart RNA polymerase II peaks. Frequency of fetal human heart candidate enhancers near genes occupied by RNA polymerase II in the fetal human heart (red) compared to random sequences (black) showing significant enrichment up to 130kb away from the nearest PolII peak (5,540 peaks). Error bars indicate 95% confidence intervals
7 May et al: ChIP-seq Discovery of Human Heart Enhancers - 7 Supplementary Figure 5: p300/cbp distal peaks are enriched near heart disease-associated genes. Cumulative plot of frequency of fetal (a-b) and adult (c-d) human heart candidate enhancers (red) compared to matched random regions (black) near genes associated with heart diseases (a,c) and control genes not associated with heart diseases (b,d). The graphs display the cumulative frequency of the different elements up to 200kb away from the transcription start and show significant enrichment of heart p300/cbp candidate enhancers in proximity to heart disease-associated genes (starting at 2.5kb away from TSS). The significant enrichment over random peaks and over non-heart genes is maintained up to 200kb (for all bins, binomial distribution p < 0.05). Errors bars indicate 95% confidence intervals
8 May et al: ChIP-seq Discovery of Human Heart Enhancers - 8 Supplementary Figure 6: Sequence constraint properties of human heart candidate enhancers. Direct comparison between the constraint properties of human heart candidate enhancers and previously identified embryonic mouse candidate enhancers. To enable a direct comparison between data sets, 1kb windows centered on the peak maxima of the 500 most significant peaks in each data set were assigned the score of the most highly constrained overlapping vertebrate phastcons 7 element in the relevant genome 8, resulting in a larger proportion of peaks with no evidence of constraint compared to Figure 3. a) Fraction of candidate enhancers that were under extreme evolutionary constraint (defined as a phastcons score of > 600). b) Fraction of candidate enhancers that were under no detectable constraint (no overlapping vertebrate constrained element)
9 May et al: ChIP-seq Discovery of Human Heart Enhancers - 9 Supplementary Figure 7: Cross-species functional conservation. Heart tissue p300/cbp binding sites in the human genome (left) were compared to p300/cbp binding at orthologous positions in the mouse genome (right). a) Heat map of human ChIP-seq read density in 100kb windows surrounding each of the human fetal heart candidate enhancers in the human genome (left), compared to ChIP-seq read density at orthologous sites in the mouse genome (right). Human peaks fall into four categories based on ChIP-seq signal in the mouse (from top): 1. significant binding at orthologous site in the mouse genome, 2. read density in the mouse genome above background, but below peak significance threshold, 3. read density in mouse genome not exceeding genome-wide background, 4. human peak sequence not alignable to mouse genome. b) Proportion of peaks in the different binding conservation categories and their sequence constraint properties (as determined by maximum score of overlapping phastcons elements). Asterisks indicate enhancer populations that are both poorly constrained and absent in the mouse genome, precluding their discovery by comparative genomics or from mouse-derived ChIP-seq data. Highly constrained, phastcons > 450; Moderately constrained, phastcons ; Weakly constrained, phastcons 350; No detectable constraint, no overlapping phastcons element
10 May et al: ChIP-seq Discovery of Human Heart Enhancers - 10 Supplementary Figure 8: Intersections with ENCODE datasets. a-c) Percentage of p300/cbp fetal human heart candidate enhancers overlapping ENCODE datasets (red) compared to expected overlap for random distribution (black). a) Histone mark H3K4me1 in 8 human cell lines (fold enrichment , hypergeometric distribution p < 1e-15). b) Histone mark H3K27ac in 8 human cell lines (fold enrichment , hypergeometric distribution p < 1e-15). c) Clusters of DNaseI hypersensitivity regions (DHS) in different cell lines (fold enrichment 12.2, hypergeometric distribution p < 1e-15). d-f) Number of positive in vivo enhancers tested previously 9,10 overlapping ENCODE datasets: d) Histone mark H3K4me1 in 8 human cell lines. e) Histone mark H3K27ac in 8 human cell lines. f) Clusters of DNaseI hypersensitivity regions (DHS) in different cell lines. The vast majority of genome regions identified by the cell linederived marks considered here do not act as in vivo heart enhancers. The ENCODE datasets used are: clusters of DNaseI hypersensitivity sites (DHS), the top 100,000 H3K4me1 peaks in 8 human cell lines and the top 100,000 H3K27ac peaks in 8 human cell lines
11 May et al: ChIP-seq Discovery of Human Heart Enhancers - 11 Supplementary Figure 9: In vivo testing of heart enhancer predictions. Comparison of the frequency of in vivo heart enhancers among tested sequences in different data sets. Human fetal heart p300/cbp ChIP-seq predictions (n=65): the elements tested in this study. Mouse e11.5 heart p300-chip-seq (n=130): mouse data sets 8 showing similar success rate in predicting active heart enhancers. Conserved elements near heart-expressed genes (n=33): a subset of 33 tested elements from previously described data sets 9,10 located within 100kb from the transcript start sites of highly heart-expressed genes. Conserved elements near heart genes (n=17): the testing of 17 moderately conserved elements located near genes implicated in embryonic heart development through genetic or developmental biology studies (Supplementary Table 8). In addition to moderate evolutionary conservation, these sequences contained sequence motifs hypothesized to be cardiac-associated 11. * p < 0.001, chi-square test
12 May et al: ChIP-seq Discovery of Human Heart Enhancers - 12 Supplementary Figure 10: Transcription factor binding site analysis of candidate heart enhancers. The top 500 p300/cbp peaks from each tissue were divided into equally sized bins containing the 250 most conserved and 250 least conserved candidate enhancers, respectively, and searched using the JASPAR database 12 of non-redundant vertebrate transcription factor binding motifs (see Methods). Results are expressed as fold enrichment / depletion of transcription factor binding sites compared to random genomic DNA, and sorted by average fold enrichment across most conserved and least conserved candidate enhancers from P2 mouse heart. Notably, human and mouse p300/cbp peaks, including those with weak evolutionary conservation, tend to be enriched in the same types of transcription factor binding sites. This suggests that while many heart enhancers are poorly conserved between human and mouse, they nonetheless share a similar repertoire of binding sites, providing a possible explanation for the activity of even poorly conserved human heart enhancers in the mouse assay
13 May et al: ChIP-seq Discovery of Human Heart Enhancers - 13 Supplementary Figure 11: Reproducible enhancer activity at e11.5 of adult vs. fetal human candidate heart enhancers. 41 of the 65 tested elements are candidate enhancers in the adult heart, and the proportion of reproducible enhancers is similar to the annotated activity of the complete set of tested sequences (n=65)
14 May et al: ChIP-seq Discovery of Human Heart Enhancers - 14 Supplementary Figure 12: Transgenic results for cardiac enhancers tested in mice at e11.5 and at postnatal day 28. See Fig. 4 for description of panels. The four elements shown in Fig. 4 are also reproduced here for comparison. a) hs1750 The activity of the enhancer is seen throughout the right and left ventricle and the outflow tract with minimal activity in the atria, both at e11.5 and in the P28 heart. b) hs The enhancer is active throughout the chambers (left and right atria and ventricles) with an atria predominance in the P28 heart. c) hs1751 The enhancer is active primarily in the outflow tract in both the embryonic and P28 heart. At e11.5, there is also activity in the right ventricle and minimal activity in the left ventricle. At 4 weeks of age, the pattern is more restricted to the outflow tract, with sporadic activity in the chambers. d) hs At e11.5, the enhancer shows activity in the heart, with the main activity located in the outflow tract myocardium and with sporadic activity in the myocardium of the ventricles. This pattern is maintained at P28. e)/f) hs1760 and hs1763 The enhancers are active throughout the embryonic and P28 heart. g) hs1959 The enhancer is active primarily in the left ventricle with minimal activity in the right ventricle both at e11.5 and in the P28 heart. h) hs1753 The tested element is active mainly in the left and right ventricle both at e11.5 and in the P28 heart. LV-left ventricle; RV-right ventricle; LA-left atrium; RA-right atrium; OFT-outflow tract; PA-pulmonary artery; Ao-Aorta; DA-dorsal aorta
15 May et al: ChIP-seq Discovery of Human Heart Enhancers - 15 Supplementary Tables Human fetal heart p300/cbp Human adult heart p300/cbp Mouse P2 heart p300/cbp Total Reads 27,073, % 26,246, % 27,181, % Reads mapping to reference genome Reads mapping to unique location in the reference genome Non-clonal reads mapping to unique location in the reference genome 13,966, % 16,444, % 12,720, % 11,463, % 13,261, % 9,596, % 10,090, % 9,021, % 8,641, % Total peaks 12, % 5, % 17, % Peaks passing filtering 10, % 4, % 12, % Proximal peaks (<=2.5kb from nearest TSS) Distal peaks (>2.5kb from nearest TSS) 5, % 2, % 6, % 5, % 2, % 6, % Supplementary Table 1: Properties of ChIP-seq data sets. Human fetal heart, 16 week gestational age; Human adult heart, adult failing heart; Mouse P2 heart, CD1 strain hearts from postnatal day
16 May et al: ChIP-seq Discovery of Human Heart Enhancers - 16 Phenotypes Binom FDR Q-value Binom fold enrichment abnormal cardiac muscle morphology 3.32E abnormal vitelline vasculature 1.13E abnormal epicardium morphology 5.23E thin ventricular wall 3.43E abnormal myocardial trabeculae morphology 7.74E decreased cardiac muscle contractility 2.79E abnormal renal glomerulus morphology 3.34E anemia 1.14E pericardial effusion 7.44E abnormal cardiac muscle contractility 4.35E abnormal erythrocyte cell number 2.83E abnormal placenta vasculature 4.35E impaired muscle contractility 6.80E abnormal vascular branching morphogenesis 9.51E abnormal erythrocyte morphology 1.66E abnormal looping morphogenesis 4.90E abnormal myocardial fiber morphology 6.41E poorly developed ventricular trabeculae 7.91E abnormal dorsal aorta morphology 8.31E abnormal liver development 1.33E abnormal liver/biliary system development 3.66E abnormal capillary morphology 1.02E disorganized myocardium 1.32E thin myocardial wall 1.72E abnormal tail development 4.23E abnormal ventricular trabeculae morphology 1.48E cortical renal glomerulopathies 1.49E branchial arch hypoplasia 1.79E abnormal mitochondrial physiology 3.86E abnormal placenta labyrinth morphology 6.09E abnormal outflow tract septation 7.91E decreased heart rate 2.91E dilated left ventricle 7.53E abnormal wound healing 2.03E dilated heart ventricles 2.78E congestive heart failure 6.52E pale yolk sac 6.48E cardiac hypertrophy 7.02E abnormal exercise endurance 7.49E abnormal heart tube morphology 7.45E decreased ventricle muscle contractility 3.60E abnormal trophoblast layer morphology 7.03E
17 May et al: ChIP-seq Discovery of Human Heart Enhancers - 17 abnormal vascular endothelial cell morphology 7.13E abnormal cell differentiation 1.06E hydrops fetalis 1.87E increased stomach ph 3.86E abnormal muscle weight 7.92E failure of looping morphogenesis 8.63E abnormal QRS complex 1.43E abnormal placental labyrinth vasculature morphology 5.25E Supplementary Table 2: Top enriched annotations of putative target genes near candidate human heart enhancers
18 May et al: ChIP-seq Discovery of Human Heart Enhancers - 18 Chr Peak start Peak end Gene symbol SNPs chr LMNA rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr TNNT2 rs , rs , rs763208, rs , rs , rs , rs , rs , rs , rs , rs832153, rs17190, rs , rs , rs , rs , rs , rs , rs832152, rs832151, rs , rs , rs832150, rs chr TNNT2 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs947376, rs947375, rs947374, rs chr AGT rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs868143, rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr AGT rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr ACTN2 rs661219, rs663798, rs507130, rs507841, rs662513, rs535411, rs676377, rs chr ACTN2 rs , rs , rs697660, rs , rs , rs chr ACTN2 rs , rs , rs , rs891337, rs891338, rs , rs , rs , rs , rs , rs , rs707201, rs , rs chr ACTN2 rs , rs , rs , rs , rs , rs , rs chr RYR2 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr RYR2 rs , rs chr RYR2 rs , rs , rs , rs , rs chr VCL rs , rs , rs chr LDB3 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr LDB3 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs ,
19 May et al: ChIP-seq Discovery of Human Heart Enhancers - 19 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr LDB3 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr LDB3 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr ANKRD1 rs , rs chr ANKRD1 rs , rs , rs , rs , rs , rs chr KCNQ1 rs179436, rs , rs , rs179437, rs , rs757086, rs , rs chr KCNQ1 rs , rs , rs , rs , rs81205, rs163149, rs163150, rs , rs , rs , rs , rs , rs81204, rs , rs163148, rs163147, rs , rs , rs , rs , rs , rs189261, rs , rs , rs , rs , rs163146, rs233434, rs , rs , rs , rs , rs163145, rs , rs , rs234886, rs chr KCNQ1 rs , rs149145, rs , rs , rs , rs , rs , rs , rs233439, rs151317, rs , rs149373, rs , rs234875, rs , rs , rs , rs , rs , rs chr KCNQ1 rs , rs , rs chr KCNQ1 rs , rs , rs , rs , rs , rs515774, rs , rs , rs chr KCNQ1 rs371972, rs , rs441613, rs400643, rs , rs , rs439926, rs445108, rs444956, rs446396, rs384037, rs , rs , rs , rs , rs367003, rs429573, rs , rs chr CSRP3 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr CSRP3 rs , rs , rs , rs chr CSRP3 rs , rs , rs , rs , rs793280, rs , rs , rs , rs793279, rs , rs , rs , rs , rs chr CSRP3 rs , rs , rs924692, rs924691, rs998263, rs , rs , rs , rs chr MYL2 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr TBX5 rs , rs , rs , rs , rs
20 May et al: ChIP-seq Discovery of Human Heart Enhancers - 20 chr TBX5 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr TBX5 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr TPM1 rs , rs , rs , rs , rs , rs , rs , rs , rs chr TPM1 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr TCAP rs , rs , rs907087, rs , rs , rs , rs , rs903502, rs , rs903504, rs , rs903503, rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs907088, rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs732083, rs , rs , rs907089, rs907090, rs732084, rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr TCAP rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr JUP rs , rs , rs , rs chr JUP rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs753663, rs , rs , rs , rs , rs , rs , rs873150, rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr SGCA rs , rs , rs , rs , rs , rs , rs , rs chr SGCA rs , rs , rs , rs , rs , rs , rs , rs chr TNNI3 rs , rs604216, rs , rs , rs682884, rs , rs , rs , rs , rs , rs , rs , rs , rs , rs575144, rs , rs , rs , rs667451, rs , rs , rs , rs , rs , rs chr DYSF rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr DYSF rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr DYSF rs934068, rs , rs , rs , rs , rs , rs , rs , rs , rs934066, rs , rs , rs , rs , rs , rs , rs934067, rs750355, rs , rs , rs , rs751473, rs750356, rs751474, rs , rs , rs , rs , rs751475, rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr DYSF rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs ,
21 May et al: ChIP-seq Discovery of Human Heart Enhancers - 21 rs , rs , rs , rs , rs934069, rs749314, rs chr DES rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr KCNE2 rs , rs , rs , rs chr KCNE2 rs , rs , rs , rs , rs , rs , rs , rs chr KCNE1 rs724386, rs , rs , rs chr KCNE1 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr SCN5A rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr SCN5A rs , rs , rs9932, rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr SCN5A rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr SCN5A rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr TNNC1 rs613519, rs , rs chr NKX2-5 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr NKX2-5 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr NKX2-5 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs869236, rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr NKX2-5 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs
22 May et al: ChIP-seq Discovery of Human Heart Enhancers - 22 chr NKX2-5 rs , rs , rs , rs876580, rs , rs876579, rs , rs chr NKX2-5 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr NKX2-5 rs , rs , rs , rs , rs , rs , rs chr NKX2-5 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr DSP rs , rs , rs , rs , rs , rs , rs chr PRKAG2 rs , rs , rs , rs , rs , rs chr PRKAG2 rs , rs , rs , rs , rs , rs , rs , rs , rs chr PRKAG2 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr PRKAG2 rs , rs , rs , rs , rs , rs , rs chr PRKAG2 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr PRKAG2 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr PRKAG2 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr PRKAG2 rs , rs , rs , rs , rs , rs chr PRKAG2 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr PRKAG2 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr GATA4 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr GATA4 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs ,
23 May et al: ChIP-seq Discovery of Human Heart Enhancers - 23 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chr GATA4 rs , rs , rs , rs , rs , rs , rs904006, rs , rs , rs , rs chr NOTCH1 rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chrx FLNA rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs , rs chrx TAZ rs , rs , rs , rs , rs , rs Supplementary Table 3: Candidate enhancers near genes implicated in heart diseases and their corresponding SNPs. Coordinates are in human genome version hg18. SNP Candidate enhancer location (hg18) Gene rs chr1: MTHFR rs chr10: ZMIZ1 rs chr14: DHRS4 rs chr7: GIMAP4 rs chr19: DUS3L rs chr15: ANKDD1A rs chr10: ASCC1 rs chr11: PRDM11 rs chr22: PACSIN2 rs chr9: NUDT2 rs chr12: RPS26 Supplementary Table 4: SNPs within candidate heart enhancers that are eqtls for gene expression in primary human cells
24 May et al: ChIP-seq Discovery of Human Heart Enhancers - 24 No detectable constraint Weakly constrained Moderately constrained Highly constrained Maximum scoring phastcons element overlapping core 1kb centered on peak summit Fetal human heart candidate enhancers (n=5,047) Tested elements (n=65) Maximum scoring phastcons element overlapping region Fetal human heart candidate enhancers (n=5,047) Tested peaks (n=65) Tested element (cloned region) (n=65) 1931 (38%) 22 (34%) 793 (16%) 5 (8%) 1 (2%) 1284 (26%) 20 (31%) 1204 (24%) 18 (28%) 20 (31%) 752 (15%) 11 (17%) 1004 (20%) 16 (25%) 18 (28%) 1080 (21%) 12 (18%) 2046 (40%) 26 (40%) 26 (40%) Supplementary Table 5: Sequence constraint properties of human heart candidate enhancers and tested elements. The distribution of tested elements is overall similar to the genome-wide set of ChIP-seq peaks. A moderate bias towards constrained elements among the tested regions is in part due to the frequent inclusion of additional weakly or moderately constrained sequences flanking the peak region in the tested constructs
25 May et al: ChIP-seq Discovery of Human Heart Enhancers - 25 Candidate enhancers Top 100 Top 500 Top 1000 All (n=5,047) Tested peaks (n=65) Significant in mouse 52 (52%) 217 (43%) 361 (36%) 1047 (21%) 24 (37%) Sub-significant in mouse 31 (31%) 183 (37%) 369 (37%) 1285 (25%) 25 (38%) Human-specific 12 (12%) 64 (13%) 185 (18.5%) 2079 (41%) 9 (14%) Non-alignable 5 (5%) 36 (7%) 85 (8.5%) 635 (13%) 7 (11%) Supplementary Table 6: Functional conservation properties of human heart candidate enhancers and in vivo tested elements. Candidate human fetal heart enhancers were ranked by the statistical significance of ChIP-seq enrichment at the respective peaks, and the top 100, 500, and 1000 peaks were examined for functional conservation in the mouse genome. See Supplementary Fig. 7 for definitions of functional conservation classes. Overall, more significant binding is positively correlated with increased likelihood of functional conservation of peaks between human and mouse. All categories of functional conservation are represented among the in vivo tested candidate enhancers
26 May et al: ChIP-seq Discovery of Human Heart Enhancers - 26 ID Annotation Whole-mount 1747 Tested peak: chr Reproducibility: 7/12 Flanking genes: HEMK1-CISH (intergenic) 1667 Tested peak: chr Reproducibility: 5/6 Flanking genes: CTAGE1-RBBP8 (intergenic) 1748 Tested peak: chr Reproducibility: 3/4 Flanking genes: GDPD5-SERPINH1 (intergenic) 1750 Tested peak: chr Reproducibility: 7/7 Flanking genes: OTOS-GPC1 (intergenic) 1751 Tested peak: chr Reproducibility: 8/8 Flanking genes: CIDEA-TUBB6 (intergenic) 1752 Tested peak: chr Reproducibility: 5/6 Flanking genes: IL17B-CSNK1A1 (intergenic) 1753 Tested peak: chr Reproducibility: 14/18 Flanking genes: VIPR2 (intronic) 1754 Tested peak: chr Reproducibility: 9/11 Flanking genes: LDLR-SPC24 (intergenic) 1881 Tested peak: chr Reproducibility: 6/14 Flanking genes: LOC CASC4 (intergenic) 1759 Tested peak: chr Reproducibility: 8/13 Flanking genes: NCRNA00084-MALAT1 (intergenic)
27 May et al: ChIP-seq Discovery of Human Heart Enhancers Tested peak: chr Reproducibility: 14/14 Flanking genes: PDP2-CDH16 (intergenic) 1763 Tested peak: chr Reproducibility: 4/5 Flanking genes: INPP5A (intronic) 1764 Tested peak: chr Reproducibility: 7/9 Flanking genes: TNS1 (intronic) 1766 Tested peak: chr Reproducibility: 12/12 Flanking genes: C1orf198 (intronic) 1767 Tested peak: chr Reproducibility: 7/12 Flanking genes: ZMIZ1 (intronic) 1769 Tested peak: chr Reproducibility: 6/7 Flanking genes: PGAP3 (intronic) 1882 Tested peak: chr Reproducibility: 8/8 Flanking genes: GPR20-PTP4A3 (intergenic) 1886 Tested peak: chr Reproducibility: 5/8 Flanking genes: C2orf57-PTMA (intergenic) 1887 Tested peak: chr Reproducibility: 4/5 Flanking genes: TEAD3 (intronic) 1891 Tested peak: chr Reproducibility: 7/7 Flanking genes: BCAN-NES (intergenic) 1909 Tested peak: chr Reproducibility: 9/9 Flanking genes: HAAO-ZFP36L2 (intergenic)
28 May et al: ChIP-seq Discovery of Human Heart Enhancers Tested peak: chr Reproducibility: 4/4 Flanking genes: PRDM16 (intronic) 1915 Tested peak: chr Reproducibility: 4/9 Flanking genes: PVRL1-TRIM29 (intergenic) 1919 Tested peak: chr Reproducibility: 8/13 Flanking genes: TMEM127 (intronic) 1920 Tested peak: chr Reproducibility: 9/10 Flanking genes: MYEOV-CCND1 (intergenic) 1927 Tested peak: chr Reproducibility: 5/6 Flanking genes: PEBP4-RHOBTB2 (intergenic) 1931 Tested peak: chr Reproducibility: 11/11 Flanking genes: CCDC85C (intronic) 1932 Tested peak: chr Reproducibility: 6/14 Flanking genes: IGF1R (intronic) 1933 Tested peak: chr Reproducibility: 5/9 Flanking genes: TBC1D8 (intronic) 1937 Tested peak: chr Reproducibility: 8/9 Flanking genes: OTOS-GPC1 (intergenic) 1945 Tested peak: chr Reproducibility: 4/9 Flanking genes: RAB17-LRRFIP1 (intergenic) 1948 Tested peak: chr Reproducibility: 7/10 Flanking genes: B3GNT8-C19orf69 (intergenic)
29 May et al: ChIP-seq Discovery of Human Heart Enhancers Tested peak: chr Reproducibility: 4/4 Flanking genes: HECTD2-PPP1R3C (intergenic) 1951 Tested peak: chr Reproducibility: 11/18 Flanking genes: COPS8-COL6A3 (intergenic) 1955 Tested peak: chr Reproducibility: 5/8 Flanking genes: QKI-C6orf118 (intergenic) 1958 Tested peak: chr Reproducibility: 4/4 Flanking genes: ADARB2-PFKP (intergenic) 1959 Tested peak: chr Reproducibility: 7/7 Flanking genes: CDH13 (intronic) 1962 Tested peak: chr Reproducibility: 15/17 Flanking genes: DCTN6-RBPMS (intergenic) 1963 Tested peak: chr Reproducibility: 12/12 Flanking genes: NUP214-FAM78A (intergenic) 1967 Tested peak: chr Reproducibility: 8/9 Flanking genes: TTC7A (intronic) 1971 Tested peak: chr Reproducibility: 10/10 Flanking genes: RBM38-HMGB1L1 (intergenic) 1974 Tested peak: chr Reproducibility: 4/5 Flanking genes: RPS24-LOC (intergenic) 1466 Tested peak: chr Reproducibility: 4/8 Flanking genes: C14orf166B-C14orf4 (intergenic)
30 May et al: ChIP-seq Discovery of Human Heart Enhancers - 30 Supplementary Table 7: Summary of in vivo heart enhancers. Representative images of whole embryos of all 43 positive in vivo heart enhancers identified in this study. ID: identifier of the tested element in the Vista enhancer browser ( Tested peak: coordinates of the injected DNA sequence in human genome hg18; Reproducibility: fraction of stained embryos with reproducible staining in the exact same sub-region shown in the representative whole-mount; Flanking Genes: nearest UCSC Known Genes upstream and downstream the tested element. ID Region (hg18) phastcons Nearest heart gene In vivo testing results Distance 494 chr4: HAND2 negative chr10: ANKRD1 other (hindbrain, midbrain, nose) 497 chr22: TBX1 negative chr12: TBX5 negative chr4: PITX2 negative chr18: GATA6 heart chr5: MEF2C heart chr2: DES negative chr5: NKX2-5 negative chr4: PITX2 negative chr8: GATA4 heart chr5: ISL1 negative chr10: FGF8 other (tail) chr10: FGF8 other (dorsal root ganglion, cranial nerve) 514 chr5: HAND1 negative chr22: TBX1 other (limb) chr5: FGF10 other (limb) Supplementary Table 8: Summary of the testing of 17 moderately conserved elements located near genes implicated in embryonic heart development through genetic or developmental biology studies. In addition to moderate evolutionary conservation, these sequences contained sequence motifs hypothesized to be cardiac-associated 11. We observed that 3 of the 17 sequences (18%) were reproducible heart enhancers
31 May et al: ChIP-seq Discovery of Human Heart Enhancers - 31 Human fetal heart p300/cbp-chip-seq predictions e11.5 mouse heart p300 ChIP-seq predictions Total tested elements Elements with reproducible cardiac patterns 43 (66%) 81 (60%) Average positive embryos per element Reproducible LacZ staining in the heart Average reproducibility 7.2/9.3 (77%) 5.8/7.7 (75%) Average peak size (STD) (2326.6) (292.9) Average element size (STD) 3719 (808) 1648 (868) Supplementary Table 9: Reproducibility of positive heart enhancers in the present study and comparison with mouse heart enhancer predictions 8. While average peak size in the human data set was substantially larger than in the mouse data set, elements tested in transgenic mice typically included only the most significantly covered subregions of longer peaks (see example in Fig. 1), resulting in a more moderate difference in the average size of tested elements
32 May et al: ChIP-seq Discovery of Human Heart Enhancers - 32 mcs/cs264 Mouse ortholog (mm9) Human ortholog (hg18) ID mm123 hs 1652 Tested region chr6:72,165,180-72,168,822 chr2:85,850,978-85,854,516 Reproducibility - heart - other human mouse lift-over Flanking genes Whole-mount heart 5/11 Limb 7, neural tube 9, nose % of bases, 100.0% of span intronic Atoh8 heart 6/8 Other 6 P300/CBP coverage (called yes/no) 55 (yes) mcs/cs271 Mouse ortholog (mm9) Human ortholog (hg18) ID mm130 hs1659 Tested region chr9:24,691,516-24,694,512 chr7:35,379,250-35,382,874 Reproducibility - heart - other human mouse lift-over Flanking genes Whole mount Heart 7/10 other % of bases, 99.4% of span Tbx20 / Herpud2 heart 9/9 P300/CBP coverage (called yes/no) 27 (yes) mcs/cs272 Mouse ortholog (mm9) Human ortholog (hg18) ID mm131 hs1660 Tested region chr7:69,037,006-69,038,618 chr5:132,704, ,706,219 Reproducibility - heart - other human mouse lift-over Flanking genes Whole-mount Heart 6/7 ear % of bases, 99.9% of span Intronic Auts2 -- Ear 5/8 P300/CBP coverage (called yes/no) 5 (no)
33 May et al: ChIP-seq Discovery of Human Heart Enhancers - 33 mcs/cs273 Mouse ortholog (mm9) Human ortholog (hg18) ID mm132 hs1661 Tested region chr13:46,666,689-46,669,718 chr5:153,556, ,558,157 Reproducibility - heart - other human mouse lift-over Flanking genes Whole mount heart 7/7 branchial arch 5, eye 6, midbrain 6, somite % of bases, 100.0% of span Intronic CAP P300/CBP coverage (called yes/no) -- mcs/cs277 Mouse ortholog (mm9) Human ortholog (hg18) ID mm136 hs1665 Tested region chr8:28,216,799-28,218,903 chr8:37,802,248-37,804,505 Reproducibility - heart - other human mouse lift-over Flanking genes Whole mount Heart 7/10, blood vessels 4 other % of bases, 100.0% of span Intronic GPR124 Heart 5/7, blood vessels 7 P300/CBP coverage (called yes/no) 14 (no) mcs/cs279 Mouse ortholog (mm9) Human ortholog (hg18) ID mm138 hs1667 Tested region chr18:11,371,312-11,374,453 chr18:18,303,642-18,306,902 Reproducibility - heart - other human mouse lift-over heart 8/8 heart 5/6 87.3% of bases, 100.0% of span Flanking genes Gata6 Rbbp8 CTAGE1 - Rbbp8 Whole mount P300/CBP coverage (called yes/no) 87 (yes)
34 May et al: ChIP-seq Discovery of Human Heart Enhancers - 34 mcs/cs284 Mouse ortholog (mm9) Human ortholog (hg18) ID mm143 hs1672 Tested region chr3:104,400, ,404,522 chr1:113,341, ,343,543 Reproducibility - heart - other human mouse lift-over Flanking genes Whole mount heart 3/3 heart 7/7 87.0% of bases, 99.9% of span Slc16A1 LRIG2 P300/CBP coverage (called yes/no) 17 (no) mcs/cs287 Mouse ortholog (mm9) Human ortholog (hg18) ID mm146 hs1675 Tested region chr11:77,235,971-77,237,943 chr17:25,018,828-25,021,000 Reproducibility - heart heart 9/9 heart 8/10 - other human mouse lift-over 55.2% of bases, 100.0% of span Flanking genes Intron SSH2 Whole mount P300/CBP coverage (called yes/no) 23 (yes) Supplementary Table 10: Summary of mouse human orthologous elements tested in vivo. To establish that the transgenic mouse enhancer assay correctly reports the activity of human heart enhancers, we tested the human orthologous sequences of eight mouse heart enhancers originally identified through ChIP-seq from mouse heart tissue 8. Six of eight (75%) human sequences showed positive heart enhancer activity at e11.5. While the reporter staining in the heart appeared in several cases fainter with the human heart enhancer, we observed overall no significant difference in the reproducibility of the positive enhancers in mouse vs. human (t-test, P=0.71). These results suggest that, with few exceptions, this transgenic assay accurately reports the in vivo activity of human heart enhancers
ChIP-seq Identification of Weakly Conserved Heart Enhancers
 ChIP-seq Identification of Weakly Conserved Heart Enhancers Matthew J. Blow, David J. McCulley, Zirong Li, Tao Zhang, Jennifer A. Akiyama, Amy Holt, Ingrid Plajzer-Frick, Malak Shoukry, Crystal Wright,
ChIP-seq Identification of Weakly Conserved Heart Enhancers Matthew J. Blow, David J. McCulley, Zirong Li, Tao Zhang, Jennifer A. Akiyama, Amy Holt, Ingrid Plajzer-Frick, Malak Shoukry, Crystal Wright,
Nature Genetics: doi: /ng Supplementary Figure 1. Assessment of sample purity and quality.
 Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63.
 Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans.
 Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Accessing and Using ENCODE Data Dr. Peggy J. Farnham
 1 William M Keck Professor of Biochemistry Keck School of Medicine University of Southern California How many human genes are encoded in our 3x10 9 bp? C. elegans (worm) 959 cells and 1x10 8 bp 20,000
1 William M Keck Professor of Biochemistry Keck School of Medicine University of Southern California How many human genes are encoded in our 3x10 9 bp? C. elegans (worm) 959 cells and 1x10 8 bp 20,000
Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types.
 Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplementary Figure 1. Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Nature Immunology: doi: /ni.
 Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Title: Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease
 1 Supplemental Materials 2 3 Title: Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease 4 5 6 Authors: Madhumita Basu, 1 Jun-Yi Zhu, 2 Stephanie LaHaye 1,3, Uddalak
1 Supplemental Materials 2 3 Title: Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease 4 5 6 Authors: Madhumita Basu, 1 Jun-Yi Zhu, 2 Stephanie LaHaye 1,3, Uddalak
Heart Development and Congenital Heart Disease
 Heart Development and Congenital Heart Disease Sally Dunwoodie s.dunwoodie@victorchang.edu.au Developmental and Stem Cell Biology Division Victor Chang Cardiac Research Institute for the heart of Australia...
Heart Development and Congenital Heart Disease Sally Dunwoodie s.dunwoodie@victorchang.edu.au Developmental and Stem Cell Biology Division Victor Chang Cardiac Research Institute for the heart of Australia...
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Frequency of alternative-cassette-exon engagement with the ribosome is consistent across data from multiple human cell types and from mouse stem cells. Box plots showing AS frequency
Supplementary Figure 1 Frequency of alternative-cassette-exon engagement with the ribosome is consistent across data from multiple human cell types and from mouse stem cells. Box plots showing AS frequency
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
Development of the Heart
 Development of the Heart Thomas A. Marino, Ph.D. Temple University School of Medicine Stages of Development of the Heart 1. The horseshoe-shaped pericardial cavity. 2. The formation of the single heart
Development of the Heart Thomas A. Marino, Ph.D. Temple University School of Medicine Stages of Development of the Heart 1. The horseshoe-shaped pericardial cavity. 2. The formation of the single heart
cis-regulatory enrichment analysis in human, mouse and fly
 cis-regulatory enrichment analysis in human, mouse and fly Zeynep Kalender Atak, PhD Laboratory of Computational Biology VIB-KU Leuven Center for Brain & Disease Research Laboratory of Computational Biology
cis-regulatory enrichment analysis in human, mouse and fly Zeynep Kalender Atak, PhD Laboratory of Computational Biology VIB-KU Leuven Center for Brain & Disease Research Laboratory of Computational Biology
Introduction to Fetal Medicine: Genetics and Embryology
 Introduction to Fetal Medicine: Genetics and Embryology Question: What do cancer biology, molecular biology, neurobiology, cell biology developmental biology and reproductive biology, all have in common?
Introduction to Fetal Medicine: Genetics and Embryology Question: What do cancer biology, molecular biology, neurobiology, cell biology developmental biology and reproductive biology, all have in common?
Heart Development. Origins of congenital heart defects Properties of cardiac progenitor cells. Robert G. Kelly
 ESC CBCS Summer School on Cardiovascular Sciences Heart Development 19th June 2013 Origins of congenital heart defects Properties of cardiac progenitor cells Robert G. Kelly Animal models of heart development
ESC CBCS Summer School on Cardiovascular Sciences Heart Development 19th June 2013 Origins of congenital heart defects Properties of cardiac progenitor cells Robert G. Kelly Animal models of heart development
Introduction to Fetal Medicine: Genetics and Embryology
 Introduction to Fetal Medicine: Genetics and Embryology Question: What do cancer biology, molecular biology, neurobiology, cell biology developmental biology and reproductive biology, all have in common?
Introduction to Fetal Medicine: Genetics and Embryology Question: What do cancer biology, molecular biology, neurobiology, cell biology developmental biology and reproductive biology, all have in common?
Supplementary Figure 1. A microarray screen of organizers compared to non-organizer tissue reveals a putative organizer gene set.
 Supplementary Figure 1. A microarray screen of organizers compared to non-organizer tissue reveals a putative organizer gene set. (a, b) Venn diagrams of 31 enriched (a) and 17 depleted (b) genes significantly
Supplementary Figure 1. A microarray screen of organizers compared to non-organizer tissue reveals a putative organizer gene set. (a, b) Venn diagrams of 31 enriched (a) and 17 depleted (b) genes significantly
Heart Development. Robert G. Kelly Developmental Biology Institute of Marseilles - Luminy
 ESC CBCS Summer School on Cardiovascular Sciences 15th June 2011 Heart Development Robert G. Kelly Developmental Biology Institute of Marseilles - Luminy Animal models of heart development Tinman/Nkx2.5
ESC CBCS Summer School on Cardiovascular Sciences 15th June 2011 Heart Development Robert G. Kelly Developmental Biology Institute of Marseilles - Luminy Animal models of heart development Tinman/Nkx2.5
Supplemental Figure S1. Tertiles of FKBP5 promoter methylation and internal regulatory region
 Supplemental Figure S1. Tertiles of FKBP5 promoter methylation and internal regulatory region methylation in relation to PSS and fetal coupling. A, PSS values for participants whose placentas showed low,
Supplemental Figure S1. Tertiles of FKBP5 promoter methylation and internal regulatory region methylation in relation to PSS and fetal coupling. A, PSS values for participants whose placentas showed low,
Processing, integrating and analysing chromatin immunoprecipitation followed by sequencing (ChIP-seq) data
 Processing, integrating and analysing chromatin immunoprecipitation followed by sequencing (ChIP-seq) data Bioinformatics methods, models and applications to disease Alex Essebier ChIP-seq experiment To
Processing, integrating and analysing chromatin immunoprecipitation followed by sequencing (ChIP-seq) data Bioinformatics methods, models and applications to disease Alex Essebier ChIP-seq experiment To
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
SUPPLEMENTAL INFORMATION
 SUPPLEMENTAL INFORMATION GO term analysis of differentially methylated SUMIs. GO term analysis of the 458 SUMIs with the largest differential methylation between human and chimp shows that they are more
SUPPLEMENTAL INFORMATION GO term analysis of differentially methylated SUMIs. GO term analysis of the 458 SUMIs with the largest differential methylation between human and chimp shows that they are more
ChIP-seq analysis. J. van Helden, M. Defrance, C. Herrmann, D. Puthier, N. Servant, M. Thomas-Chollier, O.Sand
 ChIP-seq analysis J. van Helden, M. Defrance, C. Herrmann, D. Puthier, N. Servant, M. Thomas-Chollier, O.Sand Tuesday : quick introduction to ChIP-seq and peak-calling (Presentation + Practical session)
ChIP-seq analysis J. van Helden, M. Defrance, C. Herrmann, D. Puthier, N. Servant, M. Thomas-Chollier, O.Sand Tuesday : quick introduction to ChIP-seq and peak-calling (Presentation + Practical session)
ChIP-seq data analysis
 ChIP-seq data analysis Harri Lähdesmäki Department of Computer Science Aalto University November 24, 2017 Contents Background ChIP-seq protocol ChIP-seq data analysis Transcriptional regulation Transcriptional
ChIP-seq data analysis Harri Lähdesmäki Department of Computer Science Aalto University November 24, 2017 Contents Background ChIP-seq protocol ChIP-seq data analysis Transcriptional regulation Transcriptional
Computational aspects of ChIP-seq. John Marioni Research Group Leader European Bioinformatics Institute European Molecular Biology Laboratory
 Computational aspects of ChIP-seq John Marioni Research Group Leader European Bioinformatics Institute European Molecular Biology Laboratory ChIP-seq Using highthroughput sequencing to investigate DNA
Computational aspects of ChIP-seq John Marioni Research Group Leader European Bioinformatics Institute European Molecular Biology Laboratory ChIP-seq Using highthroughput sequencing to investigate DNA
MIR retrotransposon sequences provide insulators to the human genome
 Supplementary Information: MIR retrotransposon sequences provide insulators to the human genome Jianrong Wang, Cristina Vicente-García, Davide Seruggia, Eduardo Moltó, Ana Fernandez- Miñán, Ana Neto, Elbert
Supplementary Information: MIR retrotransposon sequences provide insulators to the human genome Jianrong Wang, Cristina Vicente-García, Davide Seruggia, Eduardo Moltó, Ana Fernandez- Miñán, Ana Neto, Elbert
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
Heintzman, ND, Stuart, RK, Hon, G, Fu, Y, Ching, CW, Hawkins, RD, Barrera, LO, Van Calcar, S, Qu, C, Ching, KA, Wang, W, Weng, Z, Green, RD,
 Heintzman, ND, Stuart, RK, Hon, G, Fu, Y, Ching, CW, Hawkins, RD, Barrera, LO, Van Calcar, S, Qu, C, Ching, KA, Wang, W, Weng, Z, Green, RD, Crawford, GE, Ren, B (2007) Distinct and predictive chromatin
Heintzman, ND, Stuart, RK, Hon, G, Fu, Y, Ching, CW, Hawkins, RD, Barrera, LO, Van Calcar, S, Qu, C, Ching, KA, Wang, W, Weng, Z, Green, RD, Crawford, GE, Ren, B (2007) Distinct and predictive chromatin
Molecular Diagnostic Laboratory 18 Sequencing St, Gene Town, ZY Tel: Fax:
 Molecular Diagnostic Laboratory 18 Sequencing St, Gene Town, ZY 01234 Tel: 555-920-3333 Fax: 555-920-3334 www.moldxlaboratory.com Patient Name: Jane Doe Specimen type: Blood, peripheral DOB: 04/05/1990
Molecular Diagnostic Laboratory 18 Sequencing St, Gene Town, ZY 01234 Tel: 555-920-3333 Fax: 555-920-3334 www.moldxlaboratory.com Patient Name: Jane Doe Specimen type: Blood, peripheral DOB: 04/05/1990
The Insulator Binding Protein CTCF Positions 20 Nucleosomes around Its Binding Sites across the Human Genome
 The Insulator Binding Protein CTCF Positions 20 Nucleosomes around Its Binding Sites across the Human Genome Yutao Fu 1, Manisha Sinha 2,3, Craig L. Peterson 3, Zhiping Weng 1,4,5 * 1 Bioinformatics Program,
The Insulator Binding Protein CTCF Positions 20 Nucleosomes around Its Binding Sites across the Human Genome Yutao Fu 1, Manisha Sinha 2,3, Craig L. Peterson 3, Zhiping Weng 1,4,5 * 1 Bioinformatics Program,
Nature Structural & Molecular Biology: doi: /nsmb.2419
 Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Discovery of Novel Human Gene Regulatory Modules from Gene Co-expression and
 Discovery of Novel Human Gene Regulatory Modules from Gene Co-expression and Promoter Motif Analysis Shisong Ma 1,2*, Michael Snyder 3, and Savithramma P Dinesh-Kumar 2* 1 School of Life Sciences, University
Discovery of Novel Human Gene Regulatory Modules from Gene Co-expression and Promoter Motif Analysis Shisong Ma 1,2*, Michael Snyder 3, and Savithramma P Dinesh-Kumar 2* 1 School of Life Sciences, University
Nature Immunology: doi: /ni Supplementary Figure 1. Characteristics of SEs in T reg and T conv cells.
 Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Nature Immunology: doi: /ni Supplementary Figure 1. Transcriptional program of the TE and MP CD8 + T cell subsets.
 Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Dynamic GATA4 enhancers shape the chromatin landscape central to heart development and disease
 ARTICLE Received Jan Accepted Aug Published Sep DOI:.8/ncomms97 Dynamic GATA enhancers shape the chromatin landscape central to heart development and disease Aibin He,,, Fei Gu,, Yong Hu, Qing Ma, Lillian
ARTICLE Received Jan Accepted Aug Published Sep DOI:.8/ncomms97 Dynamic GATA enhancers shape the chromatin landscape central to heart development and disease Aibin He,,, Fei Gu,, Yong Hu, Qing Ma, Lillian
Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes
 Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes Kaifu Chen 1,2,3,4,5,10, Zhong Chen 6,10, Dayong Wu 6, Lili Zhang 7, Xueqiu Lin 1,2,8,
Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes Kaifu Chen 1,2,3,4,5,10, Zhong Chen 6,10, Dayong Wu 6, Lili Zhang 7, Xueqiu Lin 1,2,8,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature19360 Supplementary Tables Supplementary Table 1. Number of monoclonal reads in each sample Sample Number of cells Total reads Aligned reads Monoclonal reads
SUPPLEMENTARY INFORMATION doi:10.1038/nature19360 Supplementary Tables Supplementary Table 1. Number of monoclonal reads in each sample Sample Number of cells Total reads Aligned reads Monoclonal reads
Allelic reprogramming of the histone modification H3K4me3 in early mammalian development
 Allelic reprogramming of the histone modification H3K4me3 in early mammalian development 张戈 Method and material STAR ChIP seq (small-scale TELP-assisted rapid ChIP seq) 200 mouse embryonic stem cells PWK/PhJ
Allelic reprogramming of the histone modification H3K4me3 in early mammalian development 张戈 Method and material STAR ChIP seq (small-scale TELP-assisted rapid ChIP seq) 200 mouse embryonic stem cells PWK/PhJ
CARDIAC DEVELOPMENT CARDIAC DEVELOPMENT
 CARDIAC DEVELOPMENT CARDIAC DEVELOPMENT Diane E. Spicer, BS, PA(ASCP) University of Florida Dept. of Pediatric Cardiology Curator Van Mierop Cardiac Archive This lecture is given with special thanks to
CARDIAC DEVELOPMENT CARDIAC DEVELOPMENT Diane E. Spicer, BS, PA(ASCP) University of Florida Dept. of Pediatric Cardiology Curator Van Mierop Cardiac Archive This lecture is given with special thanks to
Results. Abstract. Introduc4on. Conclusions. Methods. Funding
 . expression that plays a role in many cellular processes affecting a variety of traits. In this study DNA methylation was assessed in neuronal tissue from three pigs (frontal lobe) and one great tit (whole
. expression that plays a role in many cellular processes affecting a variety of traits. In this study DNA methylation was assessed in neuronal tissue from three pigs (frontal lobe) and one great tit (whole
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes.
 Supplementary Figure 1 Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes. (a,b) Values of coefficients associated with genomic features, separately
Supplementary Figure 1 Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes. (a,b) Values of coefficients associated with genomic features, separately
Peak-calling for ChIP-seq and ATAC-seq
 Peak-calling for ChIP-seq and ATAC-seq Shamith Samarajiwa CRUK Autumn School in Bioinformatics 2017 University of Cambridge Overview Peak-calling: identify enriched (signal) regions in ChIP-seq or ATAC-seq
Peak-calling for ChIP-seq and ATAC-seq Shamith Samarajiwa CRUK Autumn School in Bioinformatics 2017 University of Cambridge Overview Peak-calling: identify enriched (signal) regions in ChIP-seq or ATAC-seq
The Epigenome Tools 2: ChIP-Seq and Data Analysis
 The Epigenome Tools 2: ChIP-Seq and Data Analysis Chongzhi Zang zang@virginia.edu http://zanglab.com PHS5705: Public Health Genomics March 20, 2017 1 Outline Epigenome: basics review ChIP-seq overview
The Epigenome Tools 2: ChIP-Seq and Data Analysis Chongzhi Zang zang@virginia.edu http://zanglab.com PHS5705: Public Health Genomics March 20, 2017 1 Outline Epigenome: basics review ChIP-seq overview
Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation
 Published online: June 1, 15 Article Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation Evelyn N Kouwenhoven 1,, Martin Oti, Hanna Niehues 3, Simon J van
Published online: June 1, 15 Article Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation Evelyn N Kouwenhoven 1,, Martin Oti, Hanna Niehues 3, Simon J van
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 Expression deviation of the genes mapped to gene-wise recurrent mutations in the TCGA breast cancer cohort (top) and the TCGA lung cancer cohort (bottom). For each gene (each pair
Supplementary Figure 1 Expression deviation of the genes mapped to gene-wise recurrent mutations in the TCGA breast cancer cohort (top) and the TCGA lung cancer cohort (bottom). For each gene (each pair
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
When you see this diagram, remember that you are looking at the embryo from above, through the amniotic cavity, where the epiblast appears as an oval
 When you see this diagram, remember that you are looking at the embryo from above, through the amniotic cavity, where the epiblast appears as an oval disc 2 Why the embryo needs the vascular system? When
When you see this diagram, remember that you are looking at the embryo from above, through the amniotic cavity, where the epiblast appears as an oval disc 2 Why the embryo needs the vascular system? When
STAT1 regulates microrna transcription in interferon γ stimulated HeLa cells
 CAMDA 2009 October 5, 2009 STAT1 regulates microrna transcription in interferon γ stimulated HeLa cells Guohua Wang 1, Yadong Wang 1, Denan Zhang 1, Mingxiang Teng 1,2, Lang Li 2, and Yunlong Liu 2 Harbin
CAMDA 2009 October 5, 2009 STAT1 regulates microrna transcription in interferon γ stimulated HeLa cells Guohua Wang 1, Yadong Wang 1, Denan Zhang 1, Mingxiang Teng 1,2, Lang Li 2, and Yunlong Liu 2 Harbin
A Practical Guide to Integrative Genomics by RNA-seq and ChIP-seq Analysis
 A Practical Guide to Integrative Genomics by RNA-seq and ChIP-seq Analysis Jian Xu, Ph.D. Children s Research Institute, UTSW Introduction Outline Overview of genomic and next-gen sequencing technologies
A Practical Guide to Integrative Genomics by RNA-seq and ChIP-seq Analysis Jian Xu, Ph.D. Children s Research Institute, UTSW Introduction Outline Overview of genomic and next-gen sequencing technologies
The epigenetic landscape of T cell subsets in SLE identifies known and potential novel drivers of the autoimmune response
 Abstract # 319030 Poster # F.9 The epigenetic landscape of T cell subsets in SLE identifies known and potential novel drivers of the autoimmune response Jozsef Karman, Brian Johnston, Sofija Miljovska,
Abstract # 319030 Poster # F.9 The epigenetic landscape of T cell subsets in SLE identifies known and potential novel drivers of the autoimmune response Jozsef Karman, Brian Johnston, Sofija Miljovska,
"Lecture Index. 1) Heart Progenitors. 2) Cardiac Tube Formation. 3) Valvulogenesis and Chamber Formation. 4) Epicardium Development.
 "Lecture Index 1) Heart Progenitors. 2) Cardiac Tube Formation. 3) Valvulogenesis and Chamber Formation. 4) Epicardium Development. 5) Septation and Maturation. 6) Changes in Blood Flow during Development.
"Lecture Index 1) Heart Progenitors. 2) Cardiac Tube Formation. 3) Valvulogenesis and Chamber Formation. 4) Epicardium Development. 5) Septation and Maturation. 6) Changes in Blood Flow during Development.
9/16/2009. Fast and slow twitch fibres. Properties of Muscle Fiber Types Fast fibers Slow fibers
 Muscles, muscle fibres and myofibrils Fast and slow twitch fibres Rat hindlimb muscle ATPase staining at different ph and NADH Muscle fibre shortening velocity lengths/second Properties of Muscle Fiber
Muscles, muscle fibres and myofibrils Fast and slow twitch fibres Rat hindlimb muscle ATPase staining at different ph and NADH Muscle fibre shortening velocity lengths/second Properties of Muscle Fiber
Supplemental Information For: The genetics of splicing in neuroblastoma
 Supplemental Information For: The genetics of splicing in neuroblastoma Justin Chen, Christopher S. Hackett, Shile Zhang, Young K. Song, Robert J.A. Bell, Annette M. Molinaro, David A. Quigley, Allan Balmain,
Supplemental Information For: The genetics of splicing in neuroblastoma Justin Chen, Christopher S. Hackett, Shile Zhang, Young K. Song, Robert J.A. Bell, Annette M. Molinaro, David A. Quigley, Allan Balmain,
Lung Met 1 Lung Met 2 Lung Met Lung Met H3K4me1. Lung Met H3K27ac Primary H3K4me1
 a Gained Met-VELs 1.5 1.5 -.5 Lung Met 1 Lung Met Lung Met 3 1. Lung Met H3K4me1 Lung Met H3K4me1 1 Lung Met H3K4me1 Lung Met H3K7ac 1.5 Lung Met H3K7ac Lung Met H3K7ac.8 Primary H3K4me1 Primary H3K7ac
a Gained Met-VELs 1.5 1.5 -.5 Lung Met 1 Lung Met Lung Met 3 1. Lung Met H3K4me1 Lung Met H3K4me1 1 Lung Met H3K4me1 Lung Met H3K7ac 1.5 Lung Met H3K7ac Lung Met H3K7ac.8 Primary H3K4me1 Primary H3K7ac
Hypertrophic Cardiomyopathy
 Hypertrophic Cardiomyopathy From Genetics to ECHO Alexandra A Frogoudaki Second Cardiology Department ATTIKON University Hospital Athens University Athens, Greece EUROECHO 2010, Copenhagen, 11/12/2010
Hypertrophic Cardiomyopathy From Genetics to ECHO Alexandra A Frogoudaki Second Cardiology Department ATTIKON University Hospital Athens University Athens, Greece EUROECHO 2010, Copenhagen, 11/12/2010
Organogenesis Part 2. V. Lateral Plate Mesoderm VI. Endoderm VII. Development of the Tetrapod Limb VIII. Sex Determination. V. Lateral Plate Mesoderm
 Organogenesis Part 2 V. Lateral Plate Mesoderm VI. Endoderm VII. Development of the Tetrapod Limb VIII. Sex Determination V. Lateral Plate Mesoderm chordamesoderm paraxial mesoderm intermediate mesoderm
Organogenesis Part 2 V. Lateral Plate Mesoderm VI. Endoderm VII. Development of the Tetrapod Limb VIII. Sex Determination V. Lateral Plate Mesoderm chordamesoderm paraxial mesoderm intermediate mesoderm
Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at
 Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at E10.5 were double-stained for TUNEL (red) and PECAM-1 (green).
Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at E10.5 were double-stained for TUNEL (red) and PECAM-1 (green).
Genome-wide Association Studies (GWAS) Pasieka, Science Photo Library
 Lecture 5 Genome-wide Association Studies (GWAS) Pasieka, Science Photo Library Chi-squared test to evaluate whether the odds ratio is different from 1. Corrected for multiple testing Source: wikipedia.org
Lecture 5 Genome-wide Association Studies (GWAS) Pasieka, Science Photo Library Chi-squared test to evaluate whether the odds ratio is different from 1. Corrected for multiple testing Source: wikipedia.org
Supplementary Fig.S1. MeDIP RPKM distribution of 5kb windows. Relative expression of DNMT. Percentage of genome. (fold change)
 Supplementary Fig.S1 Percentage of genome 10% 8% 6% 4% 2% 0 MeDIP RPKM distribution of 5kb windows 0 0. 0.5 0.75 >1 MeDIP RPKM value EC Relative expression of DNMT (fold change) 12 6 4 3 2 1 EC DNMT1 DNMT2
Supplementary Fig.S1 Percentage of genome 10% 8% 6% 4% 2% 0 MeDIP RPKM distribution of 5kb windows 0 0. 0.5 0.75 >1 MeDIP RPKM value EC Relative expression of DNMT (fold change) 12 6 4 3 2 1 EC DNMT1 DNMT2
EXPression ANalyzer and DisplayER
 EXPression ANalyzer and DisplayER Tom Hait Aviv Steiner Igor Ulitsky Chaim Linhart Amos Tanay Seagull Shavit Rani Elkon Adi Maron-Katz Dorit Sagir Eyal David Roded Sharan Israel Steinfeld Yossi Shiloh
EXPression ANalyzer and DisplayER Tom Hait Aviv Steiner Igor Ulitsky Chaim Linhart Amos Tanay Seagull Shavit Rani Elkon Adi Maron-Katz Dorit Sagir Eyal David Roded Sharan Israel Steinfeld Yossi Shiloh
EPIGENOMICS PROFILING SERVICES
 EPIGENOMICS PROFILING SERVICES Chromatin analysis DNA methylation analysis RNA-seq analysis Diagenode helps you uncover the mysteries of epigenetics PAGE 3 Integrative epigenomics analysis DNA methylation
EPIGENOMICS PROFILING SERVICES Chromatin analysis DNA methylation analysis RNA-seq analysis Diagenode helps you uncover the mysteries of epigenetics PAGE 3 Integrative epigenomics analysis DNA methylation
Chromosome-Wide Analysis of Parental Allele-Specific Chromatin and DNA Methylation
 MOLECULAR AND CELLULAR BIOLOGY, Apr. 2011, p. 1757 1770 Vol. 31, No. 8 0270-7306/11/$12.00 doi:10.1128/mcb.00961-10 Copyright 2011, American Society for Microbiology. All Rights Reserved. Chromosome-Wide
MOLECULAR AND CELLULAR BIOLOGY, Apr. 2011, p. 1757 1770 Vol. 31, No. 8 0270-7306/11/$12.00 doi:10.1128/mcb.00961-10 Copyright 2011, American Society for Microbiology. All Rights Reserved. Chromosome-Wide
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers
 Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
COMPREHENSIVE EVALUATION OF FETAL HEART R. GOWDAMARAJAN MD
 COMPREHENSIVE EVALUATION OF FETAL HEART R. GOWDAMARAJAN MD Disclosure No Relevant Financial Relationships with Commercial Interests Fetal Echo: How to do it? Timing of Study -optimally between 22-24 weeks
COMPREHENSIVE EVALUATION OF FETAL HEART R. GOWDAMARAJAN MD Disclosure No Relevant Financial Relationships with Commercial Interests Fetal Echo: How to do it? Timing of Study -optimally between 22-24 weeks
Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq
 Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq Philipp Bucher Wednesday January 21, 2009 SIB graduate school course EPFL, Lausanne ChIP-seq against histone variants: Biological
Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq Philipp Bucher Wednesday January 21, 2009 SIB graduate school course EPFL, Lausanne ChIP-seq against histone variants: Biological
Supplementary information
 Supplementary information High fat diet-induced changes of mouse hepatic transcription and enhancer activity can be reversed by subsequent weight loss Majken Siersbæk, Lyuba Varticovski, Shutong Yang,
Supplementary information High fat diet-induced changes of mouse hepatic transcription and enhancer activity can be reversed by subsequent weight loss Majken Siersbæk, Lyuba Varticovski, Shutong Yang,
Patterns of Histone Methylation and Chromatin Organization in Grapevine Leaf. Rachel Schwope EPIGEN May 24-27, 2016
 Patterns of Histone Methylation and Chromatin Organization in Grapevine Leaf Rachel Schwope EPIGEN May 24-27, 2016 What does H3K4 methylation do? Plant of interest: Vitis vinifera Culturally important
Patterns of Histone Methylation and Chromatin Organization in Grapevine Leaf Rachel Schwope EPIGEN May 24-27, 2016 What does H3K4 methylation do? Plant of interest: Vitis vinifera Culturally important
SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer.
 Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Nature Immunology: doi: /ni Supplementary Figure 1 33,312. Aire rep 1. Aire rep 2 # 44,325 # 44,055. Aire rep 1. Aire rep 2.
 a 33,312 b rep 1 rep 1 # 44,325 rep 2 # 44,055 [0-84] rep 2 [0-84] 1810043G02Rik Pfkl Dnmt3l Icosl rep 1 [0-165] rep 2 [0-165] Rps14 Cd74 Mir5107 Tcof1 rep 1 [0-69] rep 2 [0-68] Id3 E2f2 Asap3 rep 1 [0-141]
a 33,312 b rep 1 rep 1 # 44,325 rep 2 # 44,055 [0-84] rep 2 [0-84] 1810043G02Rik Pfkl Dnmt3l Icosl rep 1 [0-165] rep 2 [0-165] Rps14 Cd74 Mir5107 Tcof1 rep 1 [0-69] rep 2 [0-68] Id3 E2f2 Asap3 rep 1 [0-141]
SUPPLEMENTARY INFORMATION
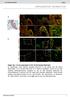 Suppl. Fig. 1 in vivo expression of ISL1 in the human fetal heart. a, Hematoxylin eosin staining showing structures of left atrium and left atrium appendage (*) of a human fetal heart at 11 weeks of gestation.
Suppl. Fig. 1 in vivo expression of ISL1 in the human fetal heart. a, Hematoxylin eosin staining showing structures of left atrium and left atrium appendage (*) of a human fetal heart at 11 weeks of gestation.
CTCF-Mediated Functional Chromatin Interactome in Pluripotent Cells
 SUPPLEMENTARY INFORMATION CTCF-Mediated Functional Chromatin Interactome in Pluripotent Cells Lusy Handoko 1,*, Han Xu 1,*, Guoliang Li 1,*, Chew Yee Ngan 1, Elaine Chew 1, Marie Schnapp 1, Charlie Wah
SUPPLEMENTARY INFORMATION CTCF-Mediated Functional Chromatin Interactome in Pluripotent Cells Lusy Handoko 1,*, Han Xu 1,*, Guoliang Li 1,*, Chew Yee Ngan 1, Elaine Chew 1, Marie Schnapp 1, Charlie Wah
RNA-seq Introduction
 RNA-seq Introduction DNA is the same in all cells but which RNAs that is present is different in all cells There is a wide variety of different functional RNAs Which RNAs (and sometimes then translated
RNA-seq Introduction DNA is the same in all cells but which RNAs that is present is different in all cells There is a wide variety of different functional RNAs Which RNAs (and sometimes then translated
Nature Genetics: doi: /ng Supplementary Figure 1. Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms.
 Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10188 Supplementary Figure 1. Embryonic epicardial genes are down-regulated from midgestation stages and barely detectable post-natally. Real time qrt-pcr revealed a significant down-regulation
doi:10.1038/nature10188 Supplementary Figure 1. Embryonic epicardial genes are down-regulated from midgestation stages and barely detectable post-natally. Real time qrt-pcr revealed a significant down-regulation
SUPPLEMENTARY INFORMATION
 Supplementary text Collectively, we were able to detect ~14,000 expressed genes with RPKM (reads per kilobase per million) > 1 or ~16,000 with RPKM > 0.1 in at least one cell type from oocyte to the morula
Supplementary text Collectively, we were able to detect ~14,000 expressed genes with RPKM (reads per kilobase per million) > 1 or ~16,000 with RPKM > 0.1 in at least one cell type from oocyte to the morula
SUPPLEMENTAL MATERIAL ADDITIONAL TABLES
 SUPPLEMENTAL MATERIAL ADDITIONAL TABLES Table S1. List of distinct rare variants detected by high-throughput sequencing in our cohort. The number of patients with each variant and the reference for the
SUPPLEMENTAL MATERIAL ADDITIONAL TABLES Table S1. List of distinct rare variants detected by high-throughput sequencing in our cohort. The number of patients with each variant and the reference for the
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
Data mining with Ensembl Biomart. Stéphanie Le Gras
 Data mining with Ensembl Biomart Stéphanie Le Gras (slegras@igbmc.fr) Guidelines Genome data Genome browsers Getting access to genomic data: Ensembl/BioMart 2 Genome Sequencing Example: Human genome 2000:
Data mining with Ensembl Biomart Stéphanie Le Gras (slegras@igbmc.fr) Guidelines Genome data Genome browsers Getting access to genomic data: Ensembl/BioMart 2 Genome Sequencing Example: Human genome 2000:
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
Figure 10.1A Transparency Master 79
 Brain Carotid arteries Jugular vein Right front leg Lungs (inflated) Cranial Right atrium To left front leg Left subclavian Bronchus capillaries Brachiocephalic vein Left atrium Dorsal aorta Right ventricle
Brain Carotid arteries Jugular vein Right front leg Lungs (inflated) Cranial Right atrium To left front leg Left subclavian Bronchus capillaries Brachiocephalic vein Left atrium Dorsal aorta Right ventricle
Cardiovascular System
 Cardiovascular System The Heart Cardiovascular System The Heart Overview What does the heart do? By timed muscular contractions creates pressure gradients blood moves then from high pressure to low pressure
Cardiovascular System The Heart Cardiovascular System The Heart Overview What does the heart do? By timed muscular contractions creates pressure gradients blood moves then from high pressure to low pressure
Supplemental Information. Genomic Characterization of Murine. Monocytes Reveals C/EBPb Transcription. Factor Dependence of Ly6C Cells
 Immunity, Volume 46 Supplemental Information Genomic Characterization of Murine Monocytes Reveals C/EBPb Transcription Factor Dependence of Ly6C Cells Alexander Mildner, Jörg Schönheit, Amir Giladi, Eyal
Immunity, Volume 46 Supplemental Information Genomic Characterization of Murine Monocytes Reveals C/EBPb Transcription Factor Dependence of Ly6C Cells Alexander Mildner, Jörg Schönheit, Amir Giladi, Eyal
Histone Modifications Are Associated with Transcript Isoform Diversity in Normal and Cancer Cells
 Histone Modifications Are Associated with Transcript Isoform Diversity in Normal and Cancer Cells Ondrej Podlaha 1, Subhajyoti De 2,3,4, Mithat Gonen 5, Franziska Michor 1 * 1 Department of Biostatistics
Histone Modifications Are Associated with Transcript Isoform Diversity in Normal and Cancer Cells Ondrej Podlaha 1, Subhajyoti De 2,3,4, Mithat Gonen 5, Franziska Michor 1 * 1 Department of Biostatistics
Supplementary Figure 1. Nature Genetics: doi: /ng.3736
 Supplementary Figure 1 Genetic correlations of five personality traits between 23andMe discovery and GPC samples. (a) The values in the colored squares are genetic correlations (r g ); (b) P values of
Supplementary Figure 1 Genetic correlations of five personality traits between 23andMe discovery and GPC samples. (a) The values in the colored squares are genetic correlations (r g ); (b) P values of
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
CARDIOVASCULAR SYSTEM
 CARDIOVASCULAR SYSTEM Overview Heart and Vessels 2 Major Divisions Pulmonary Circuit Systemic Circuit Closed and Continuous Loop Location Aorta Superior vena cava Right lung Pulmonary trunk Base of heart
CARDIOVASCULAR SYSTEM Overview Heart and Vessels 2 Major Divisions Pulmonary Circuit Systemic Circuit Closed and Continuous Loop Location Aorta Superior vena cava Right lung Pulmonary trunk Base of heart
Gene Ontology and Functional Enrichment. Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein
 Gene Ontology and Functional Enrichment Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein The parsimony principle: A quick review Find the tree that requires the fewest
Gene Ontology and Functional Enrichment Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein The parsimony principle: A quick review Find the tree that requires the fewest
TITLE: - Whole Genome Sequencing of High-Risk Families to Identify New Mutational Mechanisms of Breast Cancer Predisposition
 AD Award Number: W81XWH-13-1-0336 TITLE: - Whole Genome Sequencing of High-Risk Families to Identify New Mutational Mechanisms of Breast Cancer Predisposition PRINCIPAL INVESTIGATOR: Mary-Claire King,
AD Award Number: W81XWH-13-1-0336 TITLE: - Whole Genome Sequencing of High-Risk Families to Identify New Mutational Mechanisms of Breast Cancer Predisposition PRINCIPAL INVESTIGATOR: Mary-Claire King,
Supplementary Figure S1 Enlarged coronary artery branches in Edn1-knockout mice. a-d, Coronary angiography by ink injection in wild-type (a, b) and
 Supplementary Figure S1 Enlarged coronary artery branches in Edn1-knockout mice. a-d, Coronary angiography by ink injection in wild-type (a, b) and Edn1-knockout (Edn1-KO) (c, d) hearts. The boxed areas
Supplementary Figure S1 Enlarged coronary artery branches in Edn1-knockout mice. a-d, Coronary angiography by ink injection in wild-type (a, b) and Edn1-knockout (Edn1-KO) (c, d) hearts. The boxed areas
Supplementary Figure S1. Generation of LSL-EZH2 conditional transgenic mice.
 Downstream Col1A locus S P P P EP Genotyping with P1, P2 frt PGKneopA + frt hygro-pa Targeting vector Genotyping with P3, P4 P1 pcag-flpe P2 P3 P4 frt SApA CAG LSL PGKATG frt hygro-pa C. D. E. ormal KRAS
Downstream Col1A locus S P P P EP Genotyping with P1, P2 frt PGKneopA + frt hygro-pa Targeting vector Genotyping with P3, P4 P1 pcag-flpe P2 P3 P4 frt SApA CAG LSL PGKATG frt hygro-pa C. D. E. ormal KRAS
Nature Neuroscience: doi: /nn Supplementary Figure 1. Behavioral training.
 Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
Nuclear RNA Sequencing of the Mouse Erythroid Cell Transcriptome
 Nuclear RNA Sequencing of the Mouse Erythroid Cell Transcriptome Jennifer A. Mitchell 1,2 *., Ieuan Clay 2., David Umlauf 2, Chih-yu Chen 1, Catherine A. Moir 2, Christopher H. Eskiw 2,3, Stefan Schoenfelder
Nuclear RNA Sequencing of the Mouse Erythroid Cell Transcriptome Jennifer A. Mitchell 1,2 *., Ieuan Clay 2., David Umlauf 2, Chih-yu Chen 1, Catherine A. Moir 2, Christopher H. Eskiw 2,3, Stefan Schoenfelder
Assignment 5: Integrative epigenomics analysis
 Assignment 5: Integrative epigenomics analysis Due date: Friday, 2/24 10am. Note: no late assignments will be accepted. Introduction CpG islands (CGIs) are important regulatory regions in the genome. What
Assignment 5: Integrative epigenomics analysis Due date: Friday, 2/24 10am. Note: no late assignments will be accepted. Introduction CpG islands (CGIs) are important regulatory regions in the genome. What
Nature Immunology: doi: /ni Supplementary Figure 1. Huwe1 has high expression in HSCs and is necessary for quiescence.
 Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Genome Control in Cell Identity and Disease! Development and cell identity Loss of cell identity and disease New diagnostics and therapeutics
 Genome Control in Cell Identity and Disease! Development and cell identity Loss of cell identity and disease New diagnostics and therapeutics Development and cell identity! 30,000,000,000 cells!! Control
Genome Control in Cell Identity and Disease! Development and cell identity Loss of cell identity and disease New diagnostics and therapeutics Development and cell identity! 30,000,000,000 cells!! Control
Variant Classification. Author: Mike Thiesen, Golden Helix, Inc.
 Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
Variant Classification Author: Mike Thiesen, Golden Helix, Inc. Overview Sequencing pipelines are able to identify rare variants not found in catalogs such as dbsnp. As a result, variants in these datasets
THE HEART OBJECTIVES: LOCATION OF THE HEART IN THE THORACIC CAVITY CARDIOVASCULAR SYSTEM
 BIOLOGY II CARDIOVASCULAR SYSTEM ACTIVITY #3 NAME DATE HOUR THE HEART OBJECTIVES: Describe the anatomy of the heart and identify and give the functions of all parts. (pp. 356 363) Trace the flow of blood
BIOLOGY II CARDIOVASCULAR SYSTEM ACTIVITY #3 NAME DATE HOUR THE HEART OBJECTIVES: Describe the anatomy of the heart and identify and give the functions of all parts. (pp. 356 363) Trace the flow of blood
the Cardiovascular System I
 the Cardiovascular System I By: Dr. Nabil A Khouri MD, MsC, Ph.D MEDIASTINUM 1. Superior Mediastinum 2. inferior Mediastinum Anterior mediastinum. Middle mediastinum. Posterior mediastinum Anatomy of
the Cardiovascular System I By: Dr. Nabil A Khouri MD, MsC, Ph.D MEDIASTINUM 1. Superior Mediastinum 2. inferior Mediastinum Anterior mediastinum. Middle mediastinum. Posterior mediastinum Anatomy of
ULTRASOUND OF THE FETAL HEART
 ULTRASOUND OF THE FETAL HEART Cameron A. Manbeian, MD Disclosure Statement Today s faculty: Cameron Manbeian, MD does not have any relevant financial relationships with commercial interests or affiliations
ULTRASOUND OF THE FETAL HEART Cameron A. Manbeian, MD Disclosure Statement Today s faculty: Cameron Manbeian, MD does not have any relevant financial relationships with commercial interests or affiliations
