Hs LOC Hs.7100 T Hs YARS Tyrosyl-tRNA synthetase 0.24
|
|
|
- Florence York
- 5 years ago
- Views:
Transcription
1 Supplementary Table S1 Genes differently expressed between LNM35 and N15 UniGene ID Gene symbol Description Ratio Genes up-regulated in LNM35 Hs LOC Hs SLC35B2 Solute carrier family 35, member B Hs.7100 T Genes down-regulated in LNM35 Hs AGR2 Anteriogradient 2 homolog 0.16 Hs YARS Tyrosyl-tRNA synthetase 0.24 Hs EIF2S2 Eukaryotic translation initiation factor 2, subunit 2 beta 0.29
2 Supplementary Table S2 Proteins differently expressed in shcim- vs. VC LNM35 No. Gene ID Gene symbol Description Ratio Genes up-regulated in shcim-lnm35 1 gi CPS1 carbamoyl-phosphate synthetase 1, mitochondrial gi NUP160 nucleoporin 160kDa gi PDIA3 protein disulfide isomerase family A gi WARS tryptophanyl-trna synthetase gi RPLP1 ribosomal protein P gi LGALS3 galectin gi GRP75 heat shock 70kDa protein 9B precursor gi MRLC3 myosin regulatory light chain gi SERPINE2 plasminogen activator inhibitor type gi RPL4 ribosomal protein L gi RPLP2 ribosomal protein P gi RPL6 ribosomal protein L gi RPS5 ribosomal protein S5 2.8
3 14 gi HIST2H4B histone cluster 2, H4b gi HSPB1 heat shock 27kDa protein gi ANXA7 annexin VII gi RPL31 ribosomal protein L gi PD1A6 protein disulfide isomerase-associated gi ATIC 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/imp cyclohydrolase gi CAST calpastatin gi DPP7 dipeptidyl-peptidase gi RPL36 ribosomal protein L gi RPS3A ribosomal protein S3a gi SPTBN1 spectrin, beta, non-erythrocytic 1 isoform gi NUP214 nucleoporin 214kDa gi NUP62 nucleoporin 62kDa gi PSMD1 proteasome 26S non-atpase subunit Genes down-regulated in shcim-lnm35 1 gi SERPINB5 serine (or cysteine) proteinase inhibitor, clade B (ovalbumin), member gi AKR1B10 aldo-keto reductase family 1, member B10 0.5
4 3 gi EIF4A1 eukaryotic translation initiation factor 4A isoform gi AKR1C1 aldo-keto reductase family 1, member C gi RPS13 ribosomal protein S gi RPSA ribosomal protein SA gi RPL5 ribosomal protein L gi HNRNPU heterogeneous nuclear ribonucleoprotein U isoform a gi FLNA filamin A gi SERPINH1 serine (or cysteine) proteinase inhibitor, clade H, member 1 precursor gi STIP1 stress-induced-phosphoprotein 1 (Hsp70/Hsp90-organizing protein) gi GANAB alpha glucosidase II alpha subunit isoform gi HISTH1 histone cluster 1, H1e gi MYH9 myosin, heavy polypeptide 9, non-muscle gi PSMA1 proteasome alpha 1 subunit isoform gi HNRNPL heterogeneous nuclear ribonucleoprotein L isoform a gi RPS14 ribosomal protein S gi PSMA5 proteasome alpha 5 subunit gi ACTB beta actin gi RPL3 ribosomal protein L3 isoform a 0.4
5 21 gi LDHB lactate dehydrogenase B gi PSMB3 proteasome beta 3 subunit gi PA2GA proliferation-associated 2G gi ANXA1 annexin A gi EEF1A1 eukaryotic translation elongation factor 1 alpha gi BiP Immunoglobulin heavy chain-binding protein gi HSPA8 heat shock 70kDa protein 8 isoform gi HNRNPC heterogeneous nuclear ribonucleoprotein C isoform b gi CALM1 calmodulin gi AGR2 anterior gradient 2 homolog gi RPS4X ribosomal protein S4, X-linked X isoform gi RPS16 ribosomal protein S gi FLNB filamin B gi SERPINB2 serine (or cysteine) proteinase inhibitor, clade B (ovalbumin), member gi EEF2 eukaryotic translation elongation factor gi ALDH1A1 aldehyde dehydrogenase 1A gi TPI1 triosephosphate isomerase gi HSP90AB1 heat shock 90kDa protein 1, beta 0.3
6 39 gi HISTH1 histone cluster 1, H1c Hs ALDOA Aldolase A Hs ENO1 enolase Hs LDHA lactate dehydrogenase A Hs HNRPA1 heterogeneous nuclear ribonucleoprotein A1 isoform a Hs ANXA3 annexin A Hs RPL13A ribosomal protein L13a Hs ANXA2 annexin A2 isoform Hs VIM vimentin 0.1
7 Supplementary Table S3 Gene Ontology (GO) terms significantly related to the acquisition of metastatic phenotype GO aspect GO term GO accession P Biological processes translation GO: < cellular macromolecule metabolic process GO: cellular macromolecule biosynthetic process GO: cellular biosynthetic process GO: unfolded protein response GO: protein stimulus response GO: glycolysis GO: cellular protein metabolic process GO: protein metabolic process GO: glucose catabolic process GO: primary metabolic process GO: Molecular functions structural constituent of ribosome GO: structural molecule activity GO: Ca-dependent phospholipid binding GO: phospholipase inhibitor activity GO:
8 lipase inhibitor activity GO: enzyme inhibitor activity GO: Cellular component cytoplasm GO: < ribonucleoprotein complex GO: < macromolecular complex GO: < cytoplasmic part GO: ribosome GO: cytosol GO: non-membrane-bounded organelle GO: intracellular non-membrane-bounded organelle GO: intracellular part GO: organellar ribosome GO: mitochondrial ribosome GO:
9 Supplementary Information Primer sequence for generation of a CIM cdna probe by polymerase chain reaction a sense primer, 5'-TCTGCCCACAACTGGAGTTT an antisense primer, 5'-GGCCAACATATTCCCAAGGT Oligonucleotides sequence for generation of a CIM-specific shrna expression vector si#5; sense 5'-GATCCCC(GTGCAAAGAATCAGATTCA)ttcaagaga(TGAATCTGATTCTTTGCAC)TTTTTG GAAA-3', anti-sense 3'-GGG(CACGTTTCTTAGTCTAAGT)aagttctct(ACTTAGACTAAGAAACGTG)AAAAACCTTT TCGA-5' si#6; sense 5'-GATCCCC(GGACAACCCACATATCCAA)ttcaagaga(TTGGATATGTGGGTTGTCC)TTTTTG GAAA-3' anti-sense 3'-GGG(CCTGTTGGGTGTATATGGTT)aagttctct(AACCTATACACCCAACAGG)AAAAACCTT TTCGA-5') Control; sense, 5 -GATCCCCTTTTTTTTGGAAA antisense, 5 -AGCTTTTCCAAAAAAAAGGG
10 Oligonucleotides sequence for generation of a CIM expression vector CIMS; Sense 5'- AAACTGCAGGATGGAGGAAGGAGGCG CIMAS; anti-sense 5'- CGGGATCCGAGACACAGTGGGACAGAAA Two dimensional HPLC Instrument; DiNa Nano-flow LC system (KYA Technologies, Tokyo, Japan) Column; HiQ Sil SCX, 0.5 mm inside diameter (i.d.) x 35 mm (KYA Technologies) HiQ Sil C18-3 reverse-phase (RP) trap, 0.8 mm i.d. x 3 mm (KYA Technologies) HiQ Sil C18-3 Gradient RP analytical column, 0.15 mm i.d. x 50 mm (KYA Technologies) Elution buffer for a SCX column; 10, 30, 50, 80, 100, 150, 200, 300, and 500 mm of ammonium-formate in ph 3.0, containing 2 % acetonitrile) Condition for RP column; 0 to 50% solvent B in solvent A over 125 minutes (solvent A: 0.1 % formic acid (FA), 2% acetonitrile; solvent D: 0.1% FA, 70% acetonitrile) and % solvent B for 10 minutes at a flow rate 200 nl/minute Mass spectrometry analysis Instrument; Q-STAR XL mass spectrometer (Applied Biosystems Inc.) Analytical mode; Information-Dependent Acquisition (IDA) mode with the scan cycles set to perform a 1-second MS Scan followed by 3 MS/MS scans for 2 seconds each.
11 The acquisition method; 1 repetition at any m/z followed by a dynamic exclusion for a period of 60 seconds Data acquisition and analysis software; Analyst 1.1, ProQUANT 1.1 and ProGroup (Applied Biosystems Inc.) Search parameters; tolerance 0.3 Da for the MS and 0.15 Da for the MS/MS
12 Supplementary Figure legends Supplementary Fig. S1. Detection of CIM transcripts by Northern blot analysis in NCI-H460-LNM35 and -N15 clones knocked down for CIM. Supplementary Fig. S2. Detection of CIM transcripts by Northern blot analysis in various adult tissues. MTN TM blots containing 2 µg each of poly(a)+ RNA (Clontech) were used for the analysis of CIM expression. Significant abundant expressions of CIM transcripts 2.5 kb in size were present in testis, skeletal muscle, pancreas, and prostate tissues. CIM splice variant transcripts 2.0 kb in size are also abundantly observed in pancreas and testis. Supplementary Fig. S3. Significant up-regulation of CIM in the highly metastatic LNM35 subline as well as primary lung cancers. (A) Significantly more abundant CIM expression was found in highly metastatic LNM35 in comparison with that in its representative parental clone, N15. (B) Increased expression of CIM analyzed by northern blot in primary lung cancers in vivo. NL, normal lung; SCLC, small cell lung cancer; AD, adenocarcinomas; SQ, squamous cell carcinoma; LA, large cell carcinoma. (C) Increased expression of CIM analyzed by microarray in primary lung cancers in vivo. AD, adenocarcinomas; SQ, squamous cell carcinoma; LA, large cell carcinoma; LCNEC, large cell neuroendocrine carcinoma. Supplementary Fig. S4. Pathway analysis was performed using KeyMolnet according to the differential expression profiles of proteins between shcim- and VC-LNM35. A molecular interaction search was utilized, with the parameter set to 2 path. Closed circles, proteins in the differential expression profile. Open circles, selected by pathway analysis with KeyMolnet.
13 Supplementary Fig. S5. Detection of CIM transcripts by real-time RT-PCR analysis in LNM35 cells transiently transfected with sirna-control or -targeting CIM. Supplementary Fig. S6. (A) The expression of HIF-1α at the primary tumor sites of the xenografts of the shcim-lnm35 clones was reduced, as compared with the VC-LNM35 clones. (B) As compared to the primary tumor sites, the expression level of HIF-1α in metastatic nodules in the lungs of the shcim-lnm35 clones was considerably closer to that in those of the VC-LNM35 clones. Western blotting data presented here were first quantitated, then the relative expression ratio of HIF-1α to α-tubulin was calculated. The mean value for the VC-LNM35 clones was set at 1.0. Supplementary Fig. S7. Interaction of CIM with unfolded protein response proteins. Western blot analysis of total and phosphorylated-p38 (p38 and p-p38, respectively) expressions in shcim and VC clones cultured in the presence or absence of tunicamycin. α-tubulin was used as a loading control. Supplementary Fig. S8. CIM/ERLEC1/XTP3-B has multi-facetted roles, including modulation of unfolded protein response via binding to BiP as well as positive regulation of HIF-1α expression via binding to OS-9. The involvement of CIM/ERLEC1/XTP3-B in ER-associated protein degradation was recently reported (1-3). BiP, immunoglobin heavy chain-binding protein; HIF-1α, hypoxia inducible factor 1α; ER, endoplasmic reticulum; UPR, unfolded protein response; ERAD, ER-associated protein degradation; OS-9, osteosarcoma 9; PHDs, prolyl hydroxylases; SEL1L, Sel-1 suppressor of lin-12-like; Hrd1, Synovial apoptosis inhibitor 1; Ub, ubiquitin; HR, homologous regions (HR) between CIM and OS-9.
14 References 1. Cruciat CM, Hassler C, Niehrs C. The MRH protein Erlectin is a member of the endoplasmic reticulum synexpression group and functions in N-glycan recognition. J Biol Chem 2006; 281: Hosokawa N, Wada I, Nagasawa K, Moriyama T, Okawa K, Nagata K. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J Biol Chem 2008; 283: Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol 2008; 10:
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/2/e1602038/dc1 Supplementary Materials for Mitochondrial metabolic regulation by GRP78 Manoj Prasad, Kevin J. Pawlak, William E. Burak, Elizabeth E. Perry, Brendan
advances.sciencemag.org/cgi/content/full/3/2/e1602038/dc1 Supplementary Materials for Mitochondrial metabolic regulation by GRP78 Manoj Prasad, Kevin J. Pawlak, William E. Burak, Elizabeth E. Perry, Brendan
Proteome analysis of human amniotic mesenchymal stem cells (h A-MSCs) reveals impaired
 Supplementary Information for Proteome analysis of human amniotic mesenchymal stem cells (h A-MSCs) reveals impaired antioxidant ability, cytoskeleton and metabolic functionality in maternal obesity Valentina
Supplementary Information for Proteome analysis of human amniotic mesenchymal stem cells (h A-MSCs) reveals impaired antioxidant ability, cytoskeleton and metabolic functionality in maternal obesity Valentina
SUPPLEMENTAL TABLE I. Identified Proteins in Bovine Testicular Hyaluronidase Type I-S via LC-MS/MS
 SUPPLEMENTAL TABLE I. Identified Proteins in Bovine Testicular Hyaluronidase Type I-S via LC-MS/MS No. Protein 1 serum albumin precursor gi 30794280 2 annexin A2 gi 27807289 3 Phosphatidylethanolamine-binding
SUPPLEMENTAL TABLE I. Identified Proteins in Bovine Testicular Hyaluronidase Type I-S via LC-MS/MS No. Protein 1 serum albumin precursor gi 30794280 2 annexin A2 gi 27807289 3 Phosphatidylethanolamine-binding
Supplemental Table S1. Table of 131 NudC-interacting proteins, classified in functional groups.
 Supplemental Table S1. Table of 131 NudC-interacting proteins, classified in functional groups. IPI accession number Swiss-Prot accession number Gene Symbol Protein name Signal Transduction Protein kinases
Supplemental Table S1. Table of 131 NudC-interacting proteins, classified in functional groups. IPI accession number Swiss-Prot accession number Gene Symbol Protein name Signal Transduction Protein kinases
supplementary information
 DOI: 10.1038/ncb1875 Figure S1 (a) The 79 surgical specimens from NSCLC patients were analysed by immunohistochemistry with an anti-p53 antibody and control serum (data not shown). The normal bronchi served
DOI: 10.1038/ncb1875 Figure S1 (a) The 79 surgical specimens from NSCLC patients were analysed by immunohistochemistry with an anti-p53 antibody and control serum (data not shown). The normal bronchi served
Protein sorting (endoplasmic reticulum) Dr. Diala Abu-Hsasan School of Medicine
 Protein sorting (endoplasmic reticulum) Dr. Diala Abu-Hsasan School of Medicine dr.abuhassand@gmail.com An overview of cellular components Endoplasmic reticulum (ER) It is a network of membrane-enclosed
Protein sorting (endoplasmic reticulum) Dr. Diala Abu-Hsasan School of Medicine dr.abuhassand@gmail.com An overview of cellular components Endoplasmic reticulum (ER) It is a network of membrane-enclosed
Antibodies for Unfolded Protein Response
 Novus-lu-2945 Antibodies for Unfolded rotein Response Unfolded roteins ER lumen GR78 IRE-1 GR78 ERK Cytosol GR78 TRAF2 ASK1 JNK Activator Intron RIDD elf2α Degraded mrna XB1 mrna Translation XB1-S (p50)
Novus-lu-2945 Antibodies for Unfolded rotein Response Unfolded roteins ER lumen GR78 IRE-1 GR78 ERK Cytosol GR78 TRAF2 ASK1 JNK Activator Intron RIDD elf2α Degraded mrna XB1 mrna Translation XB1-S (p50)
Supplementary data Table S3. GO terms, pathways and networks enriched among the significantly correlating genes using Tox-Profiler
 Supplementary data Table S3. GO terms, pathways and networks enriched among the significantly correlating genes using Tox-Profiler DR CALUX Boys Girls Database Systemic lupus erythematosus 4.4 0.0021 6.7
Supplementary data Table S3. GO terms, pathways and networks enriched among the significantly correlating genes using Tox-Profiler DR CALUX Boys Girls Database Systemic lupus erythematosus 4.4 0.0021 6.7
Mass is the key. Maartje van den Biggelaar. British Society for Proteome Research meeting CIDA September 29 th Sanquin Plasma Proteins
 Mass is the key Maartje van den Biggelaar British Society for Proteome Research meeting 2013 CIDA September 29 th 2016 Maartje van den Biggelaar Sanquin Plasma Proteins Mass spectrometry approaches to
Mass is the key Maartje van den Biggelaar British Society for Proteome Research meeting 2013 CIDA September 29 th 2016 Maartje van den Biggelaar Sanquin Plasma Proteins Mass spectrometry approaches to
Biological processes. Mitochondrion Metabolic process Catalytic activity Oxidoreductase
 Full name Glyceraldehyde3 phosphate dehydrogenase Succinatesemialdehyde Glutamate dehydrogenase 1, Alcohol dehydrogenase [NADP+] 2',3'cyclicnucleotide 3' phosphodiesterase Dihydropyrimidinaserelated 2
Full name Glyceraldehyde3 phosphate dehydrogenase Succinatesemialdehyde Glutamate dehydrogenase 1, Alcohol dehydrogenase [NADP+] 2',3'cyclicnucleotide 3' phosphodiesterase Dihydropyrimidinaserelated 2
Protein Name. IFLENVIR,DSVTYTEHAK,TV TALDVVYALK,KTVTALDVV YALK,TVTALDVVYALKR,IF LENVIRDSVTYTEHAK gi Histone H2B
 Table 1. A functional category list of proteins (Lentinula edodes) identified by 1-DGE and nesi-lc-ms/ms. The table lists indicated fraction numbers, matching peptides, scores, accession numbers, protein
Table 1. A functional category list of proteins (Lentinula edodes) identified by 1-DGE and nesi-lc-ms/ms. The table lists indicated fraction numbers, matching peptides, scores, accession numbers, protein
1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled
 Protein Targeting Objectives 1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled As a protein is being synthesized, decisions
Protein Targeting Objectives 1. to understand how proteins find their destination in prokaryotic and eukaryotic cells 2. to know how proteins are bio-recycled As a protein is being synthesized, decisions
U118MG. Supplementary Figure 1 U373MG U118MG 3.5 A CCF-SSTG
 A172 CCF-SSTG1 15 - - - 1 1 1 2 2 3 4 6 7 8 10101112131617192022-1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22 T98G U373MG - - - 1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22-1 1 1 2 2 3 4 6 7
A172 CCF-SSTG1 15 - - - 1 1 1 2 2 3 4 6 7 8 10101112131617192022-1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22 T98G U373MG - - - 1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22-1 1 1 2 2 3 4 6 7
7 Glyceraldehyde 3-Phosphate GAPDH % SYM+G+e NM_ BC JU JU JU %% 45 /
 02234546789:;;;?6@A9587BA6;A6;C4D46E4C;B6;7FBC;C7G:;B6E3GB6H;H464;6854I;H464;8E9A6:5I;7A783;65=49;A@;=8C4;28B9C;J=2KI;7F4;65=49;A@;289CB5A6:;6B6@A9587BL4;2ACB7BA6C;J#M?KI;7F4; 65=49;A@;289CB5A6:;B6@A9587BL4;2ACB7BA6C;J#?KI;5AG43;C4G;B6;8683:CBC;A@;B6GBLBG83;3AEB;JNAG43KI;8EE4CCBA6;65=49C;A@;S.
02234546789:;;;?6@A9587BA6;A6;C4D46E4C;B6;7FBC;C7G:;B6E3GB6H;H464;6854I;H464;8E9A6:5I;7A783;65=49;A@;=8C4;28B9C;J=2KI;7F4;65=49;A@;289CB5A6:;6B6@A9587BL4;2ACB7BA6C;J#M?KI;7F4; 65=49;A@;289CB5A6:;B6@A9587BL4;2ACB7BA6C;J#?KI;5AG43;C4G;B6;8683:CBC;A@;B6GBLBG83;3AEB;JNAG43KI;8EE4CCBA6;65=49C;A@;S.
Table S1 Differentially expressed genes showing > 2 fold changes and p <0.01 for 0.01 mm NS-398, 0.1mM ibuprofen and COX-2 RNAi.
 Table S1 Differentially expressed genes showing > 2 fold changes and p
Table S1 Differentially expressed genes showing > 2 fold changes and p
PTM Discovery Method for Automated Identification and Sequencing of Phosphopeptides Using the Q TRAP LC/MS/MS System
 Application Note LC/MS PTM Discovery Method for Automated Identification and Sequencing of Phosphopeptides Using the Q TRAP LC/MS/MS System Purpose This application note describes an automated workflow
Application Note LC/MS PTM Discovery Method for Automated Identification and Sequencing of Phosphopeptides Using the Q TRAP LC/MS/MS System Purpose This application note describes an automated workflow
Supporting Information: Protein Corona Analysis of Silver Nanoparticles Exposed to Fish Plasma
 Supporting Information: Protein Corona Analysis of Silver Nanoparticles Exposed to Fish Plasma Jiejun Gao 1, Lu Lin 2, Alexander Wei 2,*, and Maria S. Sepúlveda 1,* 1 Department of Forestry and Natural
Supporting Information: Protein Corona Analysis of Silver Nanoparticles Exposed to Fish Plasma Jiejun Gao 1, Lu Lin 2, Alexander Wei 2,*, and Maria S. Sepúlveda 1,* 1 Department of Forestry and Natural
Practice Exam 2 MCBII
 1. Which feature is true for signal sequences and for stop transfer transmembrane domains (4 pts)? A. They are both 20 hydrophobic amino acids long. B. They are both found at the N-terminus of the protein.
1. Which feature is true for signal sequences and for stop transfer transmembrane domains (4 pts)? A. They are both 20 hydrophobic amino acids long. B. They are both found at the N-terminus of the protein.
Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting
 Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting Question No. 1 of 10 Question 1. Which of the following statements about the nucleus is correct? Question #01 A. The
Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting Question No. 1 of 10 Question 1. Which of the following statements about the nucleus is correct? Question #01 A. The
2013 John Wiley & Sons, Inc. All rights reserved. PROTEIN SORTING. Lecture 10 BIOL 266/ Biology Department Concordia University. Dr. S.
 PROTEIN SORTING Lecture 10 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University Introduction Membranes divide the cytoplasm of eukaryotic cells into distinct compartments. The endomembrane
PROTEIN SORTING Lecture 10 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University Introduction Membranes divide the cytoplasm of eukaryotic cells into distinct compartments. The endomembrane
!"#$%!&%##&'()*+,-.&/01*-0)*,(&%,2345()
 !"#$%!&%##&'()*+,-.&/01*-0)*,(&%,2345() Date of Submission Name: Email: Lab Antibody Name: Target: Company/ Source: Catalog Number, database ID, laboratory Lot Number Antibody Description: Target Description:
!"#$%!&%##&'()*+,-.&/01*-0)*,(&%,2345() Date of Submission Name: Email: Lab Antibody Name: Target: Company/ Source: Catalog Number, database ID, laboratory Lot Number Antibody Description: Target Description:
for the Identification of Phosphorylated Peptides
 Application of a Data Dependent Neutral-Loss Experiment on the Finnigan LTQ for the Identification of Phosphorylated Peptides Gargi Choudhary Diane Cho Thermo Electron, San Jose, CA Abstracted from posters
Application of a Data Dependent Neutral-Loss Experiment on the Finnigan LTQ for the Identification of Phosphorylated Peptides Gargi Choudhary Diane Cho Thermo Electron, San Jose, CA Abstracted from posters
Sequence Coverage (%) Profilin-1 P UD 2
 Protein Name Accession Number (Swissprot) Sequence Coverage (%) No. of MS/MS Queries Mascot Score 1 Reference Cytoskeletal proteins Beta-actin P60709 37 14 298 Alpha-actin P68032 33 10 141 20 Beta-actin-like
Protein Name Accession Number (Swissprot) Sequence Coverage (%) No. of MS/MS Queries Mascot Score 1 Reference Cytoskeletal proteins Beta-actin P60709 37 14 298 Alpha-actin P68032 33 10 141 20 Beta-actin-like
Intracellular Compartments and Protein Sorting
 Intracellular Compartments and Protein Sorting Intracellular Compartments A eukaryotic cell is elaborately subdivided into functionally distinct, membrane-enclosed compartments. Each compartment, or organelle,
Intracellular Compartments and Protein Sorting Intracellular Compartments A eukaryotic cell is elaborately subdivided into functionally distinct, membrane-enclosed compartments. Each compartment, or organelle,
CELLS. Cells. Basic unit of life (except virus)
 Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein
 Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
REGULATION OF ENZYME ACTIVITY. Medical Biochemistry, Lecture 25
 REGULATION OF ENZYME ACTIVITY Medical Biochemistry, Lecture 25 Lecture 25, Outline General properties of enzyme regulation Regulation of enzyme concentrations Allosteric enzymes and feedback inhibition
REGULATION OF ENZYME ACTIVITY Medical Biochemistry, Lecture 25 Lecture 25, Outline General properties of enzyme regulation Regulation of enzyme concentrations Allosteric enzymes and feedback inhibition
BIO360 Fall 2013 Quiz 1
 BIO360 Fall 2013 Quiz 1 1. Examine the diagram below. There are two homologous copies of chromosome one and the allele of YFG carried on the light gray chromosome has undergone a loss-of-function mutation.
BIO360 Fall 2013 Quiz 1 1. Examine the diagram below. There are two homologous copies of chromosome one and the allele of YFG carried on the light gray chromosome has undergone a loss-of-function mutation.
Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis
 Supplementary information Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis Yasuyuki Yamada, Fumihiko Sato
Supplementary information Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis Yasuyuki Yamada, Fumihiko Sato
EGFR shrna A: CCGGCGCAAGTGTAAGAAGTGCGAACTCGAGTTCGCACTTCTTACACTTGCG TTTTTG. EGFR shrna B: CCGGAGAATGTGGAATACCTAAGGCTCGAGCCTTAGGTATTCCACATTCTCTT TTTG
 Supplementary Methods Sequence of oligonucleotides used for shrna targeting EGFR EGFR shrna were obtained from the Harvard RNAi consortium. The following oligonucleotides (forward primer) were used to
Supplementary Methods Sequence of oligonucleotides used for shrna targeting EGFR EGFR shrna were obtained from the Harvard RNAi consortium. The following oligonucleotides (forward primer) were used to
Office number.
 The University of Jordan Faculty: Pharmacy Department: Biopharmaceutics and Clinical Pharmacy Program: Pharmacy Academic Year/ Fall Semester: 2014/15 BIOCHEMISTRY 2 [1203253] Credit hours 3 Level 2 nd
The University of Jordan Faculty: Pharmacy Department: Biopharmaceutics and Clinical Pharmacy Program: Pharmacy Academic Year/ Fall Semester: 2014/15 BIOCHEMISTRY 2 [1203253] Credit hours 3 Level 2 nd
Cell Quality Control. Peter Takizawa Department of Cell Biology
 Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Supporting Information
 Supporting Information Palmisano et al. 10.1073/pnas.1202174109 Fig. S1. Expression of different transgenes, driven by either viral or human promoters, is up-regulated by amino acid starvation. (A) Quantification
Supporting Information Palmisano et al. 10.1073/pnas.1202174109 Fig. S1. Expression of different transgenes, driven by either viral or human promoters, is up-regulated by amino acid starvation. (A) Quantification
Validated Mouse Quantitative RT-PCR Genes Gene Gene Bank Accession Number Name
 5HT1a NM_008308.4 Serotonin Receptor 1a 5HT1b NM_010482.1 Serotonin Receptor 1b 5HT2a NM_172812.2 Serotonin Receptor 2a ACO-2 NM_080633.2 Aconitase 2 Adora2A NM_009630.02 Adenosine A2a Receptor Aif1 (IbaI)
5HT1a NM_008308.4 Serotonin Receptor 1a 5HT1b NM_010482.1 Serotonin Receptor 1b 5HT2a NM_172812.2 Serotonin Receptor 2a ACO-2 NM_080633.2 Aconitase 2 Adora2A NM_009630.02 Adenosine A2a Receptor Aif1 (IbaI)
Supplementary Data A
 Supplementary Data A Proteomic data and MS/MS fragmentation spectra from 2-DE and GeLC-MS/MS analysis of the bovine teat canal lining and the outer teat skin epithelium Table A1: 2-DE proteins spots identified
Supplementary Data A Proteomic data and MS/MS fragmentation spectra from 2-DE and GeLC-MS/MS analysis of the bovine teat canal lining and the outer teat skin epithelium Table A1: 2-DE proteins spots identified
Proteasomes. When Death Comes a Knock n. Warren Gallagher Chem412, Spring 2001
 Proteasomes When Death Comes a Knock n Warren Gallagher Chem412, Spring 2001 I. Introduction Introduction The central dogma Genetic information is used to make proteins. DNA RNA Proteins Proteins are the
Proteasomes When Death Comes a Knock n Warren Gallagher Chem412, Spring 2001 I. Introduction Introduction The central dogma Genetic information is used to make proteins. DNA RNA Proteins Proteins are the
Fig. S1. Summary of the altered metabolism pathways in alcoholic fatty liver disease using MetPA analysis (panel A).
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Fig. S1. Summary of the altered metabolism pathways in alcoholic fatty liver disease
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Fig. S1. Summary of the altered metabolism pathways in alcoholic fatty liver disease
Human recombinat MIF protein (hrmif), MW: Da. m/z. hrmif ( Da) + 4-IPP (282 Da) MWtot ~ Da. m/z.
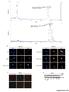 Intensity % Intensity % A Human recombinat MIF protein (hrmif), MW: 12428.31 Da m/z hrmif (12428.31 Da) + 4-IPP (282 Da) MWtot ~ 12715.21 Da m/z B HTC/C3 DAPI phistone-h3 Merge HTC/C3 DAPI phistone-h3
Intensity % Intensity % A Human recombinat MIF protein (hrmif), MW: 12428.31 Da m/z hrmif (12428.31 Da) + 4-IPP (282 Da) MWtot ~ 12715.21 Da m/z B HTC/C3 DAPI phistone-h3 Merge HTC/C3 DAPI phistone-h3
Point total. Page # Exam Total (out of 90) The number next to each intermediate represents the total # of C-C and C-H bonds in that molecule.
 This exam is worth 90 points. Pages 2- have questions. Page 1 is for your reference only. Honor Code Agreement - Signature: Date: (You agree to not accept or provide assistance to anyone else during this
This exam is worth 90 points. Pages 2- have questions. Page 1 is for your reference only. Honor Code Agreement - Signature: Date: (You agree to not accept or provide assistance to anyone else during this
Human Anatomy & Physiology
 PowerPoint Lecture Slides prepared by Barbara Heard, Atlantic Cape Community College Ninth Edition Human Anatomy & Physiology C H A P T E R 3 Annie Leibovitz/Contact Press Images 2013 Pearson Education,
PowerPoint Lecture Slides prepared by Barbara Heard, Atlantic Cape Community College Ninth Edition Human Anatomy & Physiology C H A P T E R 3 Annie Leibovitz/Contact Press Images 2013 Pearson Education,
Zool 3200: Cell Biology Exam 4 Part I 2/3/15
 Name: Key Trask Zool 3200: Cell Biology Exam 4 Part I 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
Name: Key Trask Zool 3200: Cell Biology Exam 4 Part I 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
Endoplasmic Reticulum
 Endoplasmic Reticulum What s ER? How is ER? Why is ER? definition description functions Nissl s bodies neurons Berg s bodies hepatocytes Organelle structure histocytochemical evidences Ergastoplasm pancreatic
Endoplasmic Reticulum What s ER? How is ER? Why is ER? definition description functions Nissl s bodies neurons Berg s bodies hepatocytes Organelle structure histocytochemical evidences Ergastoplasm pancreatic
BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001
 BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) The
BIOL 4374/BCHS 4313 Cell Biology Exam #1 February 13, 2001 SS# Name This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses. Good luck! 1. (2) The
BIO 5099: Molecular Biology for Computer Scientists (et al)
 BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
Comparison of nanoparticle protein corona profiles resulting from changes in nanoparticle environment and engineered properties.
 Comparison of nanoparticle protein corona profiles resulting from changes in nanoparticle environment and engineered properties. Korin Wheeler Dept. of Chemistry & Biochemistry Santa Clara University at
Comparison of nanoparticle protein corona profiles resulting from changes in nanoparticle environment and engineered properties. Korin Wheeler Dept. of Chemistry & Biochemistry Santa Clara University at
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being A Eukaryote. Eukaryotic Cells. Basic eukaryotes have:
 BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 15: Being a Eukaryote: From DNA to Protein, A Tour of the Eukaryotic Cell. Christiaan van Woudenberg Being A Eukaryote Basic eukaryotes
Moving Proteins into Membranes and Organelles
 13 Moving Proteins into Membranes and Organelles Review the Concepts 1. In eukaryotes, protein translocation across the endoplasmic reticulum (ER) membrane is most commonly cotranslational; it can also
13 Moving Proteins into Membranes and Organelles Review the Concepts 1. In eukaryotes, protein translocation across the endoplasmic reticulum (ER) membrane is most commonly cotranslational; it can also
Summary of Endomembrane-system
 Summary of Endomembrane-system 1. Endomembrane System: The structural and functional relationship organelles including ER,Golgi complex, lysosome, endosomes, secretory vesicles. 2. Membrane-bound structures
Summary of Endomembrane-system 1. Endomembrane System: The structural and functional relationship organelles including ER,Golgi complex, lysosome, endosomes, secretory vesicles. 2. Membrane-bound structures
What is the Warburg Effect
 What is the Warburg Effect Roles nutrients play in the biochemistry of a cell Thus, proliferating cells must acquire more nutrients, convert them into biosynthetic building blocks, and coordinate the reactions
What is the Warburg Effect Roles nutrients play in the biochemistry of a cell Thus, proliferating cells must acquire more nutrients, convert them into biosynthetic building blocks, and coordinate the reactions
Molecular Biology (BIOL 4320) Exam #2 May 3, 2004
 Molecular Biology (BIOL 4320) Exam #2 May 3, 2004 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
Molecular Biology (BIOL 4320) Exam #2 May 3, 2004 Name SS# This exam is worth a total of 100 points. The number of points each question is worth is shown in parentheses after the question number. Good
Bio 401 Sp2014. Multiple Choice (Circle the letter corresponding to the correct answer)
 MIDTERM EXAM KEY (60 pts) You should have 5 pages. Please put your name on every page. You will have 70 minutes to complete the exam. You may begin working as soon as you receive the exam. NOTE: the RED
MIDTERM EXAM KEY (60 pts) You should have 5 pages. Please put your name on every page. You will have 70 minutes to complete the exam. You may begin working as soon as you receive the exam. NOTE: the RED
Aberrant Promoter CpG Methylation is a Mechanism for Lack of Hypoxic Induction of
 Aberrant Promoter CpG Methylation is a Mechanism for Lack of Hypoxic Induction of PHD3 in a Diverse Set of Malignant Cells Abstract The prolyl-hydroxylase domain family of enzymes (PHD1-3) plays an important
Aberrant Promoter CpG Methylation is a Mechanism for Lack of Hypoxic Induction of PHD3 in a Diverse Set of Malignant Cells Abstract The prolyl-hydroxylase domain family of enzymes (PHD1-3) plays an important
Chapter 3 Part 2! Pages (10 th and 11 th eds.)! The Cellular Level of Organization! Cellular Organelles and Protein Synthesis!
 Chapter 3 Part 2! Pages 65 89 (10 th and 11 th eds.)! The Cellular Level of Organization! Cellular Organelles and Protein Synthesis! The Cell Theory! Living organisms are composed of one or more cells.!
Chapter 3 Part 2! Pages 65 89 (10 th and 11 th eds.)! The Cellular Level of Organization! Cellular Organelles and Protein Synthesis! The Cell Theory! Living organisms are composed of one or more cells.!
The endoplasmic reticulum is a network of folded membranes that form channels through the cytoplasm and sacs called cisternae.
 Endoplasmic reticulum (ER) The endoplasmic reticulum is a network of folded membranes that form channels through the cytoplasm and sacs called cisternae. Cisternae serve as channels for the transport of
Endoplasmic reticulum (ER) The endoplasmic reticulum is a network of folded membranes that form channels through the cytoplasm and sacs called cisternae. Cisternae serve as channels for the transport of
5/12/2015. Cell Size. Relative Rate of Reaction
 Cell Makeup Chapter 4 The Cell: The Fundamental Unit of Life We previously talked about the cell membrane The cytoplasm is everything inside the membrane, except the nucleus Includes Cytosol = liquid portion
Cell Makeup Chapter 4 The Cell: The Fundamental Unit of Life We previously talked about the cell membrane The cytoplasm is everything inside the membrane, except the nucleus Includes Cytosol = liquid portion
Predictive PP1Ca binding region in BIG3 : 1,228 1,232aa (-KAVSF-) HEK293T cells *** *** *** KPL-3C cells - E E2 treatment time (h)
 Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3311 A B TSC2 -/- MEFs C Rapa Hours WCL 0 6 12 24 36 pakt.s473 AKT ps6k S6K CM IGF-1 Recipient WCL - + - + - + pigf-1r IGF-1R pakt ps6 AKT D 1 st SILAC 2 nd SILAC E GAPDH FGF21 ALKPGVIQILGVK
DOI: 10.1038/ncb3311 A B TSC2 -/- MEFs C Rapa Hours WCL 0 6 12 24 36 pakt.s473 AKT ps6k S6K CM IGF-1 Recipient WCL - + - + - + pigf-1r IGF-1R pakt ps6 AKT D 1 st SILAC 2 nd SILAC E GAPDH FGF21 ALKPGVIQILGVK
microrna-200b and microrna-200c promote colorectal cancer cell proliferation via
 Supplementary Materials microrna-200b and microrna-200c promote colorectal cancer cell proliferation via targeting the reversion-inducing cysteine-rich protein with Kazal motifs Supplementary Table 1.
Supplementary Materials microrna-200b and microrna-200c promote colorectal cancer cell proliferation via targeting the reversion-inducing cysteine-rich protein with Kazal motifs Supplementary Table 1.
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/439/ra78/dc1 Supplementary Materials for Small heterodimer partner mediates liver X receptor (LXR) dependent suppression of inflammatory signaling by promoting
www.sciencesignaling.org/cgi/content/full/9/439/ra78/dc1 Supplementary Materials for Small heterodimer partner mediates liver X receptor (LXR) dependent suppression of inflammatory signaling by promoting
1- Which of the following statements is TRUE in regards to eukaryotic and prokaryotic cells?
 Name: NetID: Exam 3 - Version 1 October 23, 2017 Dr. A. Pimentel Each question has a value of 4 points and there are a total of 160 points in the exam. However, the maximum score of this exam will be capped
Name: NetID: Exam 3 - Version 1 October 23, 2017 Dr. A. Pimentel Each question has a value of 4 points and there are a total of 160 points in the exam. However, the maximum score of this exam will be capped
Structures in Cells. Cytoplasm. Lecture 5, EH1008: Biology for Public Health, Biomolecules
 Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression. Chromatin Array of nuc
 Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Zool 3200: Cell Biology Exam 1 1/30/15
 Name: TRASK Zool 3200: Cell Biology Exam 1 1/30/15 Answer each of the following short- and long- answer questions in the space provided; circle the BEST answer or answers for each of the multiple choice
Name: TRASK Zool 3200: Cell Biology Exam 1 1/30/15 Answer each of the following short- and long- answer questions in the space provided; circle the BEST answer or answers for each of the multiple choice
Cellular functions of protein degradation
 Protein Degradation Cellular functions of protein degradation 1. Elimination of misfolded and damaged proteins: Environmental toxins, translation errors and genetic mutations can damage proteins. Misfolded
Protein Degradation Cellular functions of protein degradation 1. Elimination of misfolded and damaged proteins: Environmental toxins, translation errors and genetic mutations can damage proteins. Misfolded
Molecular Cell Biology 5068 In class Exam 1 October 2, Please print your name: Instructions:
 Molecular Cell Biology 5068 In class Exam 1 October 2, 2012 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your number
Molecular Cell Biology 5068 In class Exam 1 October 2, 2012 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your number
RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh-
 1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
Prokaryotes and eukaryotes alter gene expression in response to their changing environment
 Chapter 18 Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development and is responsible for differences
Chapter 18 Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development and is responsible for differences
Tumour growth environment modulates Chk1 signalling pathways and sensitivity to Chk1 inhibition
 Tumour growth environment modulates Chk1 signalling pathways and sensitivity to Chk1 inhibition Andrew J Massey Supplementary Information Supplementary Figure S1. Related to Fig. 1. (a) HT29 or U2OS cells
Tumour growth environment modulates Chk1 signalling pathways and sensitivity to Chk1 inhibition Andrew J Massey Supplementary Information Supplementary Figure S1. Related to Fig. 1. (a) HT29 or U2OS cells
respiration mitochondria mitochondria metabolic pathways reproduction can fuse or split DRP1 interacts with ER tubules chapter DRP1 ER tubule
 mitochondria respiration chapter 3-4 shape highly variable can fuse or split structure outer membrane inner membrane cristae intermembrane space mitochondrial matrix free ribosomes respiratory enzymes
mitochondria respiration chapter 3-4 shape highly variable can fuse or split structure outer membrane inner membrane cristae intermembrane space mitochondrial matrix free ribosomes respiratory enzymes
Homeostatic Control Systems
 Homeostatic Control Systems In order to maintain homeostasis, control system must be able to Detect deviations from normal in the internal environment that need to be held within narrow limits Integrate
Homeostatic Control Systems In order to maintain homeostasis, control system must be able to Detect deviations from normal in the internal environment that need to be held within narrow limits Integrate
Signal Transduction Cascades
 Signal Transduction Cascades Contents of this page: Kinases & phosphatases Protein Kinase A (camp-dependent protein kinase) G-protein signal cascade Structure of G-proteins Small GTP-binding proteins,
Signal Transduction Cascades Contents of this page: Kinases & phosphatases Protein Kinase A (camp-dependent protein kinase) G-protein signal cascade Structure of G-proteins Small GTP-binding proteins,
Eukaryotic Cell Structure
 5 Eukaryotic Cell Structure 1 5.1 A typical eukaryotic cell 1. Compare and contrast eukaryotic, bacterial, and archaeal cells in terms of their use of membranes, size, morphological diversity, and organelles.
5 Eukaryotic Cell Structure 1 5.1 A typical eukaryotic cell 1. Compare and contrast eukaryotic, bacterial, and archaeal cells in terms of their use of membranes, size, morphological diversity, and organelles.
ANSWERS Problem Set 8
 ANSWERS Problem Set 8 Problem 1. All oxidation steps in the pathway from glucose to CO 2 result in the production of NADH, except the succinate dehydrogenase (SDH) step in the TCA cycle, which yields FADH2.
ANSWERS Problem Set 8 Problem 1. All oxidation steps in the pathway from glucose to CO 2 result in the production of NADH, except the succinate dehydrogenase (SDH) step in the TCA cycle, which yields FADH2.
Electron Transport Chain and Oxidative phosphorylation
 Electron Transport Chain and Oxidative phosphorylation So far we have discussed the catabolism involving oxidation of 6 carbons of glucose to CO 2 via glycolysis and CAC without any oxygen molecule directly
Electron Transport Chain and Oxidative phosphorylation So far we have discussed the catabolism involving oxidation of 6 carbons of glucose to CO 2 via glycolysis and CAC without any oxygen molecule directly
SOPten flox/flox (KO) Pten flox/flox (WT) flox allele 6.0 kb. Pten. Actin. ! allele 2.3 kb. Supplementary Figure S1. Yanagi, et al.
 s1 A Pten flox/flox () SOPten flox/flox () flox allele 6. kb B Pten flox/flox () SOPten flox/flox () Pten Actin! allele 2.3 kb Supplementary Figure S1. Yanagi, et al. A B BrdU BrdU positive cells ( ) 3
s1 A Pten flox/flox () SOPten flox/flox () flox allele 6. kb B Pten flox/flox () SOPten flox/flox () Pten Actin! allele 2.3 kb Supplementary Figure S1. Yanagi, et al. A B BrdU BrdU positive cells ( ) 3
Structures in Cells. Lecture 5, EH1008: Biology for Public Health, Biomolecules.
 Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
Structures in Cells Lecture 5, EH1008: Biology for Public Health, Biomolecules Limian.zheng@ucc.ie 1 Cytoplasm Nucleus Centrioles Cytoskeleton Cilia Microvilli 2 Cytoplasm Cellular material outside nucleus
Translation Activity Guide
 Translation Activity Guide Student Handout β-globin Translation Translation occurs in the cytoplasm of the cell and is defined as the synthesis of a protein (polypeptide) using information encoded in an
Translation Activity Guide Student Handout β-globin Translation Translation occurs in the cytoplasm of the cell and is defined as the synthesis of a protein (polypeptide) using information encoded in an
Amino acids. Side chain. -Carbon atom. Carboxyl group. Amino group
 PROTEINS Amino acids Side chain -Carbon atom Amino group Carboxyl group Amino acids Primary structure Amino acid monomers Peptide bond Peptide bond Amino group Carboxyl group Peptide bond N-terminal (
PROTEINS Amino acids Side chain -Carbon atom Amino group Carboxyl group Amino acids Primary structure Amino acid monomers Peptide bond Peptide bond Amino group Carboxyl group Peptide bond N-terminal (
Supplementary Figures
 Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
5 Identification of Binding Partners of the Annexin A2 / P11 Complex by Chemical Cross-Linking
 5 Identification of Binding Partners of the Annexin A2 / P11 Complex by Chemical Cross-Linking In the quest of the omics sciences for holistic schemes, the identification of binding partners of proteins
5 Identification of Binding Partners of the Annexin A2 / P11 Complex by Chemical Cross-Linking In the quest of the omics sciences for holistic schemes, the identification of binding partners of proteins
Protein Trafficking in the Secretory and Endocytic Pathways
 Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Objectives: Prof.Dr. H.D.El-Yassin
 Protein Synthesis and drugs that inhibit protein synthesis Objectives: 1. To understand the steps involved in the translation process that leads to protein synthesis 2. To understand and know about all
Protein Synthesis and drugs that inhibit protein synthesis Objectives: 1. To understand the steps involved in the translation process that leads to protein synthesis 2. To understand and know about all
Cells. Variation and Function of Cells
 Cells Variation and Function of Cells Cell Theory states that: 1. All living things are made of cells 2. Cells are the basic unit of structure and function in living things 3. New cells are produced from
Cells Variation and Function of Cells Cell Theory states that: 1. All living things are made of cells 2. Cells are the basic unit of structure and function in living things 3. New cells are produced from
Intrinsically Disordered Proteins. Alex Cioffi June 22 nd 2013
 Intrinsically Disordered Proteins Alex Cioffi June 22 nd 2013 Implications in Human Disease Adenovirus Early Region 1A (E1A) and Cancer α-synuclein and Parkinson s Disease Amyloid β and Alzheimer s Disease
Intrinsically Disordered Proteins Alex Cioffi June 22 nd 2013 Implications in Human Disease Adenovirus Early Region 1A (E1A) and Cancer α-synuclein and Parkinson s Disease Amyloid β and Alzheimer s Disease
Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion
 Probe Set ID RefSeq Transcript ID Gene Title Gene Symbol 205980_s_at NM_001017526 /// NM_181334 /// NM_181335 Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion ARHGAP8 /// LOC553158 FC ALK shrna
Probe Set ID RefSeq Transcript ID Gene Title Gene Symbol 205980_s_at NM_001017526 /// NM_181334 /// NM_181335 Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion ARHGAP8 /// LOC553158 FC ALK shrna
PCB 3023 Exam 4 - Form A First and Last Name
 PCB 3023 Exam 4 - Form A First and Last Name Student ID # (U Number) A Before beginning this exam, please complete the following instructions: 1) Write your name and U number on the first page of this
PCB 3023 Exam 4 - Form A First and Last Name Student ID # (U Number) A Before beginning this exam, please complete the following instructions: 1) Write your name and U number on the first page of this
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation
 Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
levels of genes were separated by their expression levels; 2,000 high, medium, and low
 Figure S1. Histone modification profiles near transcription start sites. The overall histone modification around transcription start sites (TSSs) was calculated. Histone modification levels of genes were
Figure S1. Histone modification profiles near transcription start sites. The overall histone modification around transcription start sites (TSSs) was calculated. Histone modification levels of genes were
TRANSPORT PROCESSES. 1b. moving proteins into membranes and organelles
 1b. moving proteins into membranes and organelles SLIDE 1 A typical mammalian cell contains up to 10,000 different kinds of proteins. The vast majority of these proteins are synthesized by cytosolic ribosomes,
1b. moving proteins into membranes and organelles SLIDE 1 A typical mammalian cell contains up to 10,000 different kinds of proteins. The vast majority of these proteins are synthesized by cytosolic ribosomes,
Supplementary figures legends
 Supplementary Information to Nijmegen Breakage Syndrome fibroblasts and ipscs: cellular models for uncovering diseaseassociated signaling pathways and establishing a screening platform for anti-oxidants
Supplementary Information to Nijmegen Breakage Syndrome fibroblasts and ipscs: cellular models for uncovering diseaseassociated signaling pathways and establishing a screening platform for anti-oxidants
Proteomic analysis of human spermatozoa proteins with oxidative stress
 Sharma et al. Reproductive Biology and Endocrinology 2013, 11:48 RESEARCH Proteomic analysis of human spermatozoa proteins with oxidative stress Rakesh Sharma 1, Ashok Agarwal 1*, Gayatri Mohanty 1,6,
Sharma et al. Reproductive Biology and Endocrinology 2013, 11:48 RESEARCH Proteomic analysis of human spermatozoa proteins with oxidative stress Rakesh Sharma 1, Ashok Agarwal 1*, Gayatri Mohanty 1,6,
AP Biology
 Tour of the Cell (1) 2007-2008 Types of cells Prokaryote bacteria cells - no organelles - organelles Eukaryote animal cells Eukaryote plant cells Cell Size Why organelles? Specialized structures - specialized
Tour of the Cell (1) 2007-2008 Types of cells Prokaryote bacteria cells - no organelles - organelles Eukaryote animal cells Eukaryote plant cells Cell Size Why organelles? Specialized structures - specialized
Cell Injury MECHANISMS OF CELL INJURY
 Cell Injury MECHANISMS OF CELL INJURY The cellular response to injurious stimuli depends on the following factors: Type of injury, Its duration, and Its severity. Thus, low doses of toxins or a brief duration
Cell Injury MECHANISMS OF CELL INJURY The cellular response to injurious stimuli depends on the following factors: Type of injury, Its duration, and Its severity. Thus, low doses of toxins or a brief duration
Figure 1 Original Advantages of biological reactions being catalyzed by enzymes:
 Enzyme basic concepts, Enzyme Regulation I III Carmen Sato Bigbee, Ph.D. Objectives: 1) To understand the bases of enzyme catalysis and the mechanisms of enzyme regulation. 2) To understand the role of
Enzyme basic concepts, Enzyme Regulation I III Carmen Sato Bigbee, Ph.D. Objectives: 1) To understand the bases of enzyme catalysis and the mechanisms of enzyme regulation. 2) To understand the role of
The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein
 THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting. protein3) regulate autophagy and mitophagy in renal tubular cells in. acute kidney injury
 Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
