Signal Log Ratio _r_at M12289
|
|
|
- Darlene Cunningham
- 6 years ago
- Views:
Transcription
1 dbm 8wk _r_at M12289 M12289:Myosin heavy chain, skeletal muscle, perinatal /cds=(0,344) /gb=m12289 /gi= /ug=mm.2772 /len=493 /NOTE=replacement for probe set(s) 97212_at on MG-U74A _at X58196 X58196:H19 fetal liver mrna /cds=unknown /gb=x58196 /gi=51131 /ug=mm /len= _r_at AV AV294670:AV Mus musculus cdna, 3 end /clone= j12 /clone_end=3 /gb=av /gi= /ug=mm.3692 /len= _r_at AV AV326267:AV Mus musculus cdna, 3 end /clone= p04 /clone_end=3 /gb=av /gi= /ug=mm /len=254 /NOTE=replacement for probe set(s) 99693_f_at on MG-U74A M80423:Mus castaneus IgK chain gene, C-region, 3 end /cds=(0,322) /gb=m80423 /gi= /ug=mm _f_at M80423 /len=323 C77070:C77070 Mus musculus cdna, 3 end /clone=j0025f02 /clone_end=3 /gb=c77070 /gi= _at C77070 /ug=mm /len=592 U48797:Mus musculus astrotactin mrna, complete cds /cds=(1242,3770) /gb=u48797 /gi= /ug=mm _at U48797 /len= _at AF AF084183:Mus musculus SH2/SH3 adaptor protein (Nck) mrna, complete cds /cds=(0,1133) /gb=af /gi= /ug=mm.8201 /len=1137 /NOTE=replacement for probe set(s) _at on MG-U74A U48716:Mus musculus anti-hiv-1 reverse transcriptase single-chain variable fragment mrna, complete cds _at U48716 /cds=(0,788) /gb=u48716 /gi= /ug=mm /len=789 U10410:Mus musculus recombinant antineuraminidase single chain Ig VH and VL domains mrna, complete cds _r_at U10410 /cds=(0,821) /gb=u10410 /gi= /ug=mm /len=831 AB019029:Mus musculus gene, complete cds, similar to EXLM1 /cds=(369,1781) /gb=ab /gi= _at AB /ug=mm /len=1866 AI314227:uj35g09.x1 Mus musculus cdna, 3 end /clone=image /clone_end=3 /gb=ai _at AI /gi= /ug=mm /len=510 AI853977:UI-M-BH0-aiv-f-01-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh0-aiv-f-01-0-ui /clone_end= _at AI /gb=ai /gi= /ug=mm /len=265 AI120671:ub71d01.r1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=ai _at AI /gi= /ug=mm /len= _at U77967 U77967:Neuronal PAS domain protein 1 /cds=(60,1844) /gb=u77967 /gi= /ug=mm.2525 /len=1965 AI153421:uc53a11.r1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=ai /gi= /ug=mm /len=385 AW121732:UI-M-BH2.3-aoe-b-05-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh2.3-aoe-b-05-0-ui /clone_end=3 /gb=aw /gi= /ug=mm /len=377 AB015200:Mus musculus gene for hippocalcin /cds=(182,763) /gb=ab /gi= /ug=mm /len= _f_at AI _at AW _at AB _at M34815 M34815:Monokine induced by gamma interferon /cds=(94,474) /gb=m34815 /gi= /ug=mm.766 /len= _r_at AV AV242220:AV Mus musculus cdna, 3 end /clone= k05 /clone_end=3 /gb=av /gi= /ug=mm /len=251 /NOTE=replacement for probe set(s) _f_at on MG-U74A _at 98470_at AW AF AW049299:UI-M-BH1-amr-c-07-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh1-amr-c-07-0-ui /clone_end=3 /gb=aw /gi= /ug=mm /len=275 /NOTE=replacement for probe set(s) 95038_at on MG-U74A AF076981:Mus musculus brain mitochondrial carrier protein BMCP1 (Bmcp1) mrna, complete cds /cds=(422,1390) /gb=af /gi= /ug=mm /len= _f_at AA AA048058:mj24a07.r1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=aa /gi= /ug=mm /len=218 /NOTE=replacement for probe set(s) _f_at on MG-U74A _at AI AI836205:UI-M-AI0-aap-f-04-0-UI.s2 Mus musculus cdna, 3 end /clone=ui-m-ai0-aap-f-04-0-ui /clone_end=3 /gb=ai /gi= /ug=mm /len=455 /NOTE=replacement for probe set(s) 99177_at on MG-U74A AW122364:UI-M-BH2.2-aow-a-11-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh2.2-aow-a-11-0-ui 95749_at AW /clone_end=3 /gb=aw /gi= /ug=mm /len= _at U28168 U28168:Adenomatosis polyposis coli /cds=(35,592) /gb=u28168 /gi= /ug=mm.2698 /len= _at AV AV298145:AV Mus musculus cdna, 3 end /clone= o14 /clone_end=3 /gb=av /gi= /ug=mm /len=230 /NOTE=replacement for probe set(s) 98162_f_at on MG-U74A AA216858:mu91d03.r1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=aa _at AA /gi= /ug=mm /len= _at AF AF022371:Interferon activated gene 203 /cds=(187,1413) /gb=af /gi= /ug=mm /len= _i_at AW AW122632:UI-M-BH2.2-aol-c-11-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh2.2-aol-c-11-0-ui /clone_end=3 /gb=aw /gi= /ug=mm /len= _s_at X84014 X84014:Laminin, alpha 3 /cds=(0,7710) /gb=x84014 /gi= /ug=mm /len= _at L16919 L16919:Hydroxysteroid dehydrogenase-4, delta<5>-3-beta /cds=(108,1229) /gb=l16919 /gi= /ug=mm /len= _at AA AA795198:vq95b01.r1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=aa /gi= /ug=mm /len= _at J04967 J04967:CD3 antigen, zeta polypeptide /cds=unknown /gb=j04967 /gi= /ug=mm.1224 /len= _f_at AV AV214912:AV Mus musculus cdna, 3 end /clone= e12 /clone_end=3 /gb=av /gi= /ug=mm /len=248 /NOTE=replacement for probe set(s) 98746_at on MG-U74A 96055_at X59520 X59520:Cholecystokinin /cds=(64,411) /gb=x59520 /gi=50316 /ug=mm.2619 /len= _at AW AW047653:UI-M-BH1-alo-a-12-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh1-alo-a-12-0-ui /clone_end=3 /gb=aw /gi= /ug=mm /len= _r_at AV AV012076:AV Mus musculus cdna /clone= h17 /clone_end=3 /gb=av /gi= /ug=mm /len=257 /NOTE=replacement for probe set(s) _f_at on MG-U74A X83202:M.musculus mrna for 11beta-hydroxysteroid dehydrogenase/carbonyl reductase /cds=(127,1005) 97867_at X83202 /gb=x83202 /gi= /ug=mm /len=1350 AI837530:UI-M-AL0-abs-b-11-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-al0-abs-b-11-0-ui /clone_end= _at AI /gb=ai /gi= /ug=mm /len=218 Z72486:Polymerase (DNA directed), delta 2, regulatory subunit (50kDa) /cds=(83,1492) /gb=z72486 /gi= _r_at Z72486 /ug=mm /len=
2 dbm 8wk _at 92840_at 94906_at U68058 AA M22679 U68058:Secreted frizzled-related sequence protein 3 /cds=(12,983) /gb=u68058 /gi= /ug=mm.3246 /len=2176 AA683712:vm70f08.s1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=aa /gi= /ug=mm /len=492 M22679:Mouse alcohol dehydrogenase class I gene (ADH-A-2) /cds=(0,1127) /gb=m22679 /gi= /ug=mm.2409 /len= _at U90535 U90535:Mus musculus flavin-containing monooxygenase 5 (FMO5) mrna, complete cds /cds=(812,2413) /gb=u90535 /gi= /ug=mm.1668 /len=3168 /NOTE=replacement for probe set(s) 93110_f_at on MG-U74A AI647632:uk36b03.x1 Mus musculus cdna, 3 end /clone=image /clone_end=3 /gb=ai _f_at AI /gi= /ug=mm /len=656 AI854331:UI-M-BG1-aic-b-07-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bg1-aic-b-07-0-ui /clone_end= _at AI /gb=ai /gi= /ug=mm /len= _at AI AI854839:UI-M-BH0-akb-b-08-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh0-akb-b-08-0-ui /clone_end= /gb=ai /gi= /ug=mm /len=457 /NOTE=replacement for probe set(s) 95687_at on MG-U74A AB023401:Mus musculus MEN1 mrna for menin, complete cds /cds=(49,1884) /gb=ab /gi= _at AB /ug=mm /len= _f_at X06358 X06358:UDP-glucuronosyltransferase 2 family, member 5 /cds=(12,1604) /gb=x06358 /gi=55119 /ug=mm /len= _at AW AW125538:UI-M-BH2.2-aqm-h-04-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh2.2-aqm-h-04-0-ui /clone_end=3 /gb=aw /gi= /ug=mm /len= _at AI AI837318:UI-M-AK0-adf-b-11-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-ak0-adf-b-11-0-ui /clone_end=3 /gb=ai /gi= /ug=mm /len= _at U36993 U36993:Cytochrome P450, 7b1 /cds=(80,1603) /gb=u36993 /gi= /ug=mm.4781 /len= _f_at AV AV141027:AV Mus musculus cdna /clone= i11 /clone_end=3 /gb=av /gi= /ug=mm /len=185 /NOTE=replacement for probe set(s) 94609_i_at, 94610_r_at on MG-U74A _at AA AA690863:vt32b05.r1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=aa /gi= /ug=mm /len= _i_at AI AI647632:uk36b03.x1 Mus musculus cdna, 3 end /clone=image /clone_end=3 /gb=ai /gi= /ug=mm /len= _at AA AA726364:vu90e02.r1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=aa /gi= /ug=mm /len= _r_at U62674 U62674:Mus musculus histone H2a (H2a-615), and histone H (H3-615) genes, complete cds /cds=(43,435) /gb=u62674 /gi= /ug=mm /len= _f_at AI AI848699:UI-M-AH1-agv-g-05-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-ah1-agv-g-05-0-ui /clone_end= /gb=ai /gi= /ug=mm /len= _at U35741 U35741:Thiosulfate sulfurtransferase, mitochondrial /cds=(14,907) /gb=u35741 /gi= /ug=mm /len=1016 /NOTE=replacement for probe set(s) 93052_at on MG-U74A _at X81059 X81059:M.musculus tex271 mrna /cds=(55,477) /gb=x81059 /gi= /ug=mm /len= _at _at _at _at 93067_f_at _f_at 92834_at 92496_at 96269_at _at 94916_at _at 95588_at _at AI AI M64068 AI U62674 AI X51528 AF AA M12330 AW AI U89906 AI AI853930:UI-M-BH0-aju-g-05-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh0-aju-g-05-0-ui /clone_end=3 /gb=ai /gi= /ug=mm.3886 /len=442 AI852101:UI-M-BH0-ajb-a-02-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh0-ajb-a-02-0-ui /clone_end=3 /gb=ai /gi= /ug=mm /len=310 M64068:Mouse zinc finger protein (bmi-1) gene, complete cds /cds=(0,749) /gb=m64068 /gi= /ug=mm.7719 /len=750 AI842264:UI-M-AI1-afp-g-06-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-ai1-afp-g-06-0-ui /clone_end=3 /gb=ai /gi= /ug=mm /len=384 U62674:Mus musculus histone H2a (H2a-615), and histone H (H3-615) genes, complete cds /cds=(43,435) /gb=u62674 /gi= /ug=mm /len=515 AI527477:uf12b04.y1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=ai /gi= /ug=mm.732 /len=450 X51528:Tissue specific transplantation antigen P198-7 /cds=(18,629) /gb=x51528 /gi=55049 /ug=mm /len=654 AF035643:Mus musculus VAMP5 mrna, complete cds /cds=(9,317) /gb=af /gi= /ug=mm /len=612 AA716963:vu69f10.r1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=aa /gi= /ug=mm /len=273 M12330:Ornithine decarboxylase, structural /cds=(0,854) /gb=m12330 /gi= /ug=mm /len=1181 /NOTE=replacement for probe set(s) 93049_at on MG-U74A AW122260:UI-M-BH2.2-aov-g-11-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh2.2-aov-g-11-0-ui /clone_end=3 /gb=aw /gi= /ug=mm /len=161 AI846682:UI-M-AN1-afi-e-03-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-an1-afi-e-03-0-ui /clone_end=3 /gb=ai /gi= /ug=mm /len=331 U89906:Mus musculus alpha-methylacyl-coa racemase mrna, complete cds /cds=(25,1107) /gb=u89906 /gi= /ug=mm.2787 /len=1515 AI315647:uj31c11.x1 Mus musculus cdna, 3 end /clone=image /clone_end=3 /gb=ai /gi= /ug=mm /len= _at Z36774 Z36774:M.musculus mrna for alpha-2 antiplasmin /cds=(36,1511) /gb=z36774 /gi= /ug=mm.934 /len= _at AW AW208818:uo64d02.x1 Mus musculus cdna, 3 end /clone=image /clone_end=3 /gb=aw /gi= /ug=mm /len= _at L15193 L15193:Meprin 1 beta /cds=(30,2144) /gb=l15193 /gi= /ug=mm.2682 /len= _at M75886 M75886:Hydroxysteroid dehydrogenase-2, delta<5>-3-beta /cds=unknown /gb=m75886 /gi= /ug=mm /len= _s_at M27938 M27938:Male enhanced antigen 1 /cds=(80,604) /gb=m27938 /gi= /ug=mm.2395 /len= _at AW AW121625:UI-M-BH2.2-aoq-g-12-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh2.2-aoq-g-12-0-ui /clone_end=3 /gb=aw /gi= /ug=mm /len=322 /NOTE=replacement for probe set(s) _at on MG-U74A _at AI AI848235:UI-M-AH1-agu-a-10-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-ah1-agu-a-10-0-ui /clone_end=3 /gb=ai /gi= /ug=mm.9811 /len=569 /NOTE=replacement for probe set(s) _at on MG-U74A AW048712:UI-M-BH1-amy-c-04-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh1-amy-c-04-0-ui 94424_at AW /clone_end=3 /gb=aw /gi= /ug=mm /len=527 AW060791:UI-M-BH1-anu-f-12-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh1-anu-f-12-0-ui /clone_end= 95121_at AW /gb=aw /gi= /ug=mm /len=
3 dbm 8wk 96114_at 95451_at 95028_r_at 94339_at 93539_at 94356_at _r_at AW AA C77009 AI AI AI AA AW122076:UI-M-BH2.2-aoo-f-05-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh2.2-aoo-f-05-0-ui /clone_end=3 /gb=aw /gi= /ug=mm /len=363 AA855382:vw72a04.r1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=aa /gi= /ug=mm /len=314 C77009:C77009 Mus musculus cdna, 3 end /clone=j0024d07 /clone_end=3 /gb=c77009 /gi= /ug=mm /len=496 AI841330:UI-M-AM0-adv-b-11-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-am0-adv-b-11-0-ui /clone_end= /gb=ai /gi= /ug=mm /len=332 AI118854:uc14f05.r1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=ai /gi= /ug=mm.2000 /len=281 AI593074:vv10f06.x1 Mus musculus cdna, 3 end /clone=image /clone_end=3 /gb=ai /gi= /ug=mm /len=604 AA388099:vc88g07.r1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=aa /gi= /ug=mm /len= _s_at AI AI840713:UI-M-AM0-adr-g-11-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-am0-adr-g-11-0-ui /clone_end= /gb=ai /gi= /ug=mm.5324 /len=557 /NOTE=replacement for probe set(s) _at on MG-U74A X57377:Mouse dilute myosin heavy chain gene for novel heavy chain with unique C-terminal region /cds=(40,5601) 98968_at X57377 /gb=x57377 /gi=50714 /ug=mm.3645 /len= _at M93264 M93264:Alpha-2-macroglobulin /cds=(49,4536) /gb=m93264 /gi= /ug=mm /len= _at Y09974 Y09974:M.musculus mrna for TAFI95 /cds=(234,2744) /gb=y09974 /gi= /ug=mm /len= _at AF AF070970:Mus musculus PAR-6 (Par6) mrna, complete cds /cds=(90,1130) /gb=af /gi= /ug=mm /len= _at U79962 U79962:TAR (HIV) RNA binding protein 2 /cds=(91,1188) /gb=u79962 /gi= /ug=mm.176 /len= _f_at AV AV376940:AV Mus musculus cdna, 3 end /clone= d23 /clone_end=3 /gb=av /gi= /ug=mm /len=240 /NOTE=replacement for probe set(s) _at on MG-U74A _at 95694_at 95966_at _at AW X70956 C77609 Y12887 AW122453:UI-M-BH2.2-aou-a-11-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh2.2-aou-a-11-0-ui /clone_end=3 /gb=aw /gi= /ug=mm /len=335 /NOTE=replacement for probe set(s) 95051_at on MG-U74A X70956:M.musculus TOP gene for topoisomerase I, exons /cds=(0,347) /gb=x70956 /gi=57932 /ug=mm /len=348 C77609:C77609 Mus musculus cdna, 3 end /clone=j0034e03 /clone_end=3 /gb=c77609 /gi= /ug=mm /len=636 Y12887:M.musculus mrna for carboxylesterase /cds=(41,1738) /gb=y12887 /gi= /ug=mm /len= _at L34570 L34570:Arachidonate 15-lipoxygenase /cds=(10,2001) /gb=l34570 /gi= /ug=mm.4584 /len= _f_at AJ AJ007909:Mus musculus mrna for erythroid differentiation regulator, partial /cds=(154,687) /gb=aj /gi= /ug=mm /len= _at AI AI447589:mr28a08.x1 Mus musculus cdna, 3 end /clone=image /clone_end=3 /gb=ai /gi= /ug=mm.295 /len= _at L12589 L12589:CD80 antigen /cds=(315,1244) /gb=l12589 /gi= /ug=mm /len= _f_at X61232 X61232:Mouse mrna for carboxypeptidase H /cds=(39,1472) /gb=x61232 /gi=50312 /ug=mm /len= _at L40406 L40406:Heat shock protein, 105 kda /cds=(139,2718) /gb=l40406 /gi= /ug=mm /len= _at M12347 M12347:Actin, alpha 1, skeletal muscle /cds=(70,1203) /gb=m12347 /gi= /ug=mm /len= _at Y11356 Y11356:M.musculus mrna for beta-2-glycoprotein I /cds=unknown /gb=y11356 /gi= /ug=mm.2266 /len= _f_at AF AF061260:Mus musculus immunosuperfamily protein Bl2 mrna, complete cds /cds=(308,1195) /gb=af /gi= /ug=mm /len= _at AJ AJ223361:Mus musculus mrna for myosin heavy chain 2B, partial /cds=(0,93) /gb=aj /gi= /ug=mm /len= _at Z11886 Z11886:Notch gene homolog 1, (Drosophila) /cds=(78,7673) /gb=z11886 /gi= /ug=mm /len= _at M60348 M60348:Mouse multidrug-resistance protein (MDR1) gene, exons 1 and 2 /cds=(153,217) /gb=m60348 /gi= /ug=mm.6404 /len= _at U37119 U37119:Mus musculus arylamine N-acetyltransferase (NAT1) gene, complete cds /cds=(0,872) /gb=u37119 /gi= /ug=mm /len= _at U32330 U32330:Endothelin 3 /cds=(349,993) /gb=u32330 /gi= /ug=mm.9478 /len= _at X78545 X78545:M.musculus MCP-8 mrna for serine protease /cds=(10,753) /gb=x78545 /gi= /ug=mm /len= _r_at AV AV255941:AV Mus musculus cdna, 3 end /clone= c11 /clone_end=3 /gb=av /gi= /ug=mm /len=230 /NOTE=replacement for probe set(s) _f_at on MG-U74A U51123:Mus musculus G-protein coupled inwardly rectifying K+ channel (Girk2A-1) mrna, complete cds 92524_at U51123 /cds=(480,1757) /gb=u51123 /gi= /ug=mm.4276 /len=2514 AF032459:Mus musculus BimEL mrna, complete cds /cds=(0,590) /gb=af /gi= /ug=mm _g_at AF /len=591 AB013850:Mus musculus mrna for peptidylarginine deiminase type IV, complete cds /cds=(16,2016) _at AB /gb=ab /gi= /ug=mm /len=2276 C79663:C79663 Mus musculus cdna, 3 end /clone=j0070b02 /clone_end=3 /gb=c79663 /gi= _at C79663 /ug=mm /len=540 C77756:C77756 Mus musculus cdna, 3 end /clone=j0037b08 /clone_end=3 /gb=c77756 /gi= _at C77756 /ug=mm /len= _f_at AV AV054675:AV Mus musculus cdna /clone= a07 /clone_end=3 /gb=av /gi= /ug=mm /len=181 /NOTE=replacement for probe set(s) 95763_at on MG-U74A AW061307:UI-M-BH1-anw-d-02-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh1-anw-d-02-0-ui 95152_g_at AW /clone_end=3 /gb=aw /gi= /ug=mm /len=438 L26507:Myocyte nuclear factor /cds=(9,1862) /gb=l26507 /gi= /ug=mm /len= _at L26507 /NOTE=replacement for probe set(s) _at on MG-U74A _at U14135 U14135:Integrin alpha V (Cd51) /cds=(220,3354) /gb=u14135 /gi= /ug=mm.4427 /len=
4 dbm 8wk 92663_at 97925_at AW AB AW046257:UI-M-BH1-ala-d-11-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh1-ala-d-11-0-ui /clone_end=3 /gb=aw /gi= /ug=mm.4450 /len=455 AB028241:Mus musculus mrna for casein kinase, complete cds /cds=(88,1338) /gb=ab /gi= /ug=mm /len= _i_at AV AV369921:AV Mus musculus cdna, 3 end /clone= g05 /clone_end=3 /gb=av /gi= /ug=mm /len=231 /NOTE=replacement for probe set(s) 98216_f_at on MG-U74A _r_at AV AV136210:AV Mus musculus cdna /clone= f01 /clone_end=3 /gb=av /gi= /ug=mm /len=289 /NOTE=replacement for probe set(s) 94606_i_at, 94607_r_at on MG-U74A C77679:C77679 Mus musculus cdna, 3 end /clone=j0035h12 /clone_end=3 /gb=c77679 /gi= _at C77679 /ug=mm /len=599 AJ245706:Mus musculus mrna for nicotinic acetylcholine receptor subunit alpha6 (NICA6 gene) /cds=(80,1564) 96996_at AJ /gb=aj /gi= /ug=mm /len= _at AA AA647449:vq82d03.s1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=aa /gi= /ug=mm /len=425 /NOTE=replacement for probe set(s) 97286_at on MG-U74A 98345_at AA AA674593:vm73h01.s1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=aa /gi= /ug=mm.4328 /len= _at AI AI843258:UI-M-AO1-aei-b-08-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-ao1-aei-b-08-0-ui /clone_end=3 /gb=ai /gi= /ug=mm.2875 /len= _at AI AI834849:UI-M-AN0-acm-d-08-0-UI.s2 Mus musculus cdna, 3 end /clone=ui-m-an0-acm-d-08-0-ui /clone_end=3 /gb=ai /gi= /ug=mm.9210 /len= _at AA AA536986:vj86g06.r1 Mus musculus cdna, 3 end /clone=image /clone_end=3 /gb=aa /gi= /ug=mm.6902 /len= _at AF AF052219:Mus musculus MTG8-related protein 1 gene, exon 3 and partial cds /cds=(0,241) /gb=af /gi= /ug=mm /len= _at AI AI844250:UI-M-AL1-ahk-g-10-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-al1-ahk-g-10-0-ui /clone_end=3 /gb=ai /gi= /ug=mm.6667 /len= _at AA AA438122:vd22f11.s1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=aa /gi= /ug=mm.3807 /len= _at AF AF022908:Multiple drug resistance-associated protein /cds=(0,4586) /gb=af /gi= /ug=mm /len= _s_at M63503 M63503:Mouse keratinocyte growth factor receptor mrna, complete cds /cds=(675,2798) /gb=m63503 /gi= /ug=mm /len= _at X75650 X75650:M.musculus mrna for mouse hair acidic 3 /cds=(0,971) /gb=x75650 /gi= /ug=mm /len= _at AI AI842588:UI-M-AQ1-aed-a-08-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-aq1-aed-a-08-0-ui /clone_end=3 /gb=ai /gi= /ug=mm /len= _at U14133 U14133:Silver /cds=(0,1880) /gb=u14133 /gi= /ug=mm /len= _at X00472 X00472:Nerve growth factor, gamma /cds=(0,407) /gb=x00472 /gi=54260 /ug=mm /len= _at U84207 U84207:CTP phosphocholine cytidylyl transferase /cds=(10,1113) /gb=u84207 /gi= /ug=mm.3905 /len= _at AI AI853668:UI-M-BH0-ajq-g-01-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh0-ajq-g-01-0-ui /clone_end=3 /gb=ai /gi= /ug=mm /len= _at AF AF060245:Mus musculus strain C57BL/6 x DBA zinc finger protein 106 (Zfp106) mrna, partial cds /cds=(0,865) /gb=af /gi= /ug=mm.8022 /len= _at AI AI851990:UI-M-BH0-aiq-e-01-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh0-aiq-e-01-0-ui /clone_end=3 /gb=ai /gi= /ug=mm /len= _at AF AF015881:Mus musculus nuclear factor erythroid-related factor 1 (Nrf1) gene, complete cds /cds=(0,941) /gb=af /gi= /ug=mm /len= _at Y08830 Y08830:M.musculus trop2 gene /cds=(0,953) /gb=y08830 /gi= /ug=mm /len= _at AI AI647493:uk42h02.x1 Mus musculus cdna, 3 end /clone=image /clone_end=3 /gb=ai /gi= /ug=mm.2621 /len= _at L03529 L03529:Mus musculus thrombin receptor mrna, complete cds /cds=(14,1306) /gb=l03529 /gi= /ug=mm /len= _at X62669 X62669:M.musculus genes HOX-4.4 and Hox-4.5 /cds=(0,1019) /gb=x62669 /gi=51414 /ug=mm /len=1020 AI850794:UI-M-BG1-aii-e-08-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bg1-aii-e-08-0-ui /clone_end=3 /gb=ai /gi= /ug=mm.2050 /len=377 AW048332:UI-M-BH1-akk-c-05-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh1-akk-c-05-0-ui /clone_end=3 /gb=aw /gi= /ug=mm /len=333 AB005623:Mus musculus mrna for 1-acyl-sn-glycerol-3-phosphate acyltransferase, complete cds /cds=(318,1175) /gb=ab /gi= /ug=mm.8684 /len=1256 X99253:M.musculus mrna for protein expressed at high levels in testis /cds=unknown /gb=x99253 /gi= /ug=mm /len=1393 AA882416:vx44e09.r1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=aa /gi= /ug=mm /len=571 AF032115:Mus musculus cysteine string protein mrna, complete cds /cds=(0,596) /gb=af /gi= /ug=mm /len=597 AA274528:vb02g05.r1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=aa /gi= /ug=mm /len= _at AI _at AW _at AB _at X _at AA _at AF _at AA _f_at AV AV372577:AV Mus musculus cdna, 3 end /clone= l22 /clone_end=3 /gb=av /gi= /ug=mm /len=237 /NOTE=replacement for probe set(s) 99294_f_at on MG-U74A 98994_at AF AF081499:Mus musculus type IIb Na/phosphate-cotransporter (Npt2b) mrna, complete cds /cds=(44,2137) /gb=af /gi= /ug=mm.3786 /len= _at Y11092 Y11092:M.musculus mrna for map kinase interacting kinase, Mnk2 /cds=(92,1330) /gb=y11092 /gi= /ug=mm /len= _at AB AB033887:Mus musculus macs4 variant2 mrna for Acyl-CoA synthetase 4 variant2, complete cds /cds=(317,2452 /gb=ab /gi= /ug=mm /len= _at AA AA794350:vr46a11.s1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=aa /gi= /ug=mm /len= _at AF AF084466:Mus musculus Ras-like GTP-binding protein Rad mrna, complete cds /cds=(6,932) /gb=af /gi= /ug=mm /len= _s_at X76085 X76085:Biliary glycoprotein 2 /cds=(98,916) /gb=x76085 /gi= /ug=mm /len= _at D84655 D84655:Mus musculus mrna for TRAF6, complete cds /cds=(259,1851) /gb=d84655 /gi= /ug=mm.5172 /len=
5 dbm 8wk _at _f_at 96679_at 96606_at 93974_at _s_at AI M26005 AW AB AW AB AI846673:UI-M-AN1-afi-d-03-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-an1-afi-d-03-0-ui /clone_end=3 /gb=ai /gi= /ug=mm /len=382 M26005:Mouse endogenous retrovirus truncated gag protein, complete cds, clone del env /cds=(0,731) /gb=m26005 /gi= /ug=mm /len=732 AW120711:UI-M-BH2.3-anz-e-10-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh2.3-anz-e-10-0-ui /clone_end=3 /gb=aw /gi= /ug=mm /len=497 AB025217:Mus musculus mrna for Sid470p, complete cds /cds=(55,642) /gb=ab /gi= /ug=mm /len=782 AW212475:uo89c05.x1 Mus musculus cdna, 3 end /clone=image /clone_end=3 /gb=aw /gi= /ug=mm /len=230 AB011000:Mus musculus mrna for choline/ethanolamine kinase, complete cds /cds=(299,1483) /gb=ab /gi= /ug=mm /len= _s_at U03715 U03715:Procollagen, type XVIII, alpha 1 /cds=(0,3947) /gb=u03715 /gi= /ug=mm /len= _at AW AW124483:UI-M-BH2.1-ape-h-06-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh2.1-ape-h-06-0-ui /clone_end=3 /gb=aw /gi= /ug=mm /len= _i_at X76850 X76850:M. musculus mrna for MAP kinase-activated protein kinase 2 /cds=(0,1157) /gb=x76850 /gi= /ug=mm /len=1247 /NOTE=replacement for probe set(s) 95720_at on MG-U74A AI881987:ul56h05.x1 Mus musculus cdna, 3 end /clone=image /clone_end=3 /gb=ai _r_at AI /gi= /ug=mm.4609 /len=512 Z36549:M.musculus mrna for zinc finger protein /cds=(822,2105) /gb=z36549 /gi= /ug=mm _at Z36549 /len=2353 /NOTE=replacement for probe set(s) 95055_at on MG-U74A AF077003:Mus musculus SH3 domain-containing adapter protein mrna, complete cds /cds=(6,1931) /gb=af _at AF /gi= /ug=mm /len=2431 AI837006:UI-M-AJ0-aaz-h-01-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-aj0-aaz-h-01-0-ui /clone_end= _at AI /gb=ai /gi= /ug=mm /len=324 AW124083:UI-M-BH2.1-apn-h-01-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh2.1-apn-h-01-0-ui _at AW /clone_end=3 /gb=aw /gi= /ug=mm /len=426 U43205:Mus musculus frizzled-3 protein mrna, complete cds /cds=(312,2312) /gb=u43205 /gi= _at U43205 /ug=mm /len= _at V00722 V00722:Mouse gene for beta-1-globin /cds=(53,496) /gb=v00722 /gi=50155 /ug=mm.2329 /len= _at Y07611 Y07611:M.musculus PKN gene and gene for prostanoid EP1 receptor /cds=(0,852) /gb=y07611 /gi= /ug=mm.4501 /len= _i_at AV AV087000:AV Mus musculus cdna /clone= f03 /clone_end=3 /gb=av /gi= /ug=mm /len=181 /NOTE=replacement for probe set(s) 94575_r_at on MG-U74A _at X84239 X84239:M.musculus mrna for rab5b protein /cds=(50,697) /gb=x84239 /gi= /ug=mm /len= _at AW AW121876:UI-M-BH2.3-aoi-d-06-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh2.3-aoi-d-06-0-ui /clone_end=3 /gb=aw /gi= /ug=mm /len= _at AB AB009275:Mus musculus gene for matrin3, complete cds /cds=(0,920) /gb=ab /gi= /ug=mm /len= _at AW AW122036:UI-M-BH2.2-aoo-b-12-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh2.2-aoo-b-12-0-ui /clone_end=3 /gb=aw /gi= /ug=mm /len= _at D31969 D31969:Vitamin D receptor /cds=(108,1376) /gb=d31969 /gi= /ug=mm /len= _r_at W91600 W91600:MTA.G B Mus musculus cdna, 3 end /clone=mta.g /clone_end=3 /gb=w91600 /gi= /ug=mm /len= _i_at AV AV262491:AV Mus musculus cdna, 3 end /clone= h07 /clone_end=3 /gb=av /gi= /ug=mm /len=271 /NOTE=replacement for probe set(s) 95180_f_at, 95181_r_at on MG-U74A U87619:Mus musculus 129SV encoding transcription factor Spi-B form IA /cds=(97,165) /gb=u87619 /gi= _s_at U87619 /ug=mm.6240 /len=670 AF039839:Mus musculus adriamycin-resistant related protein (Arr) mrna, complete cds /cds=(153,308) _at AF /gb=af /gi= /ug=mm /len= _at AI AI842657:UI-M-AQ1-aed-h-05-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-aq1-aed-h-05-0-ui /clone_end=3 /gb=ai /gi= /ug=mm /len=288 /NOTE=replacement for probe set(s) 97476_at on MG-U74A AW060793:UI-M-BH1-anu-g-02-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh1-anu-g-02-0-ui 92294_at AW /clone_end=3 /gb=aw /gi= /ug=mm.3727 /len=453 U13836:Mus musculus vacuolar adenosine triphosphatase subunit Ac116 mrna, complete cds /cds=(18,2537) _at U13836 /gb=u13836 /gi= /ug=mm /len=2557 AW122517:UI-M-BH2.2-aox-a-10-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh2.2-aox-a-10-0-ui 93463_at AW /clone_end=3 /gb=aw /gi= /ug=mm /len=439 AA683850:vr05f02.r1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=aa _at AA /gi= /ug=mm /len=535 L09192 Mus musculus pyruvate carboxylase mrna, complete cds (_5, _MA, MB, _3 represent transcript regions 5 FX-PyruCarbMur/L091 L09192 prime, MiddleA, MiddleB, and 3 prime respectively) AI853061:UI-M-BH0-ajn-g-12-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-bh0-ajn-g-12-0-ui /clone_end= _at AI /gb=ai /gi= /ug=mm /len= _g_at X95761 X95761:M.musculus mrna for new-rhobin /cds=unknown /gb=x95761 /gi= /ug=mm.7634 /len= _at D87900 D87900:Mus musculus mrna for ARF3, complete cds /cds=(197,742) /gb=d87900 /gi= /ug=mm /len= _at AF AF074266:Homolog of human MLLT2 unidentified gene /cds=(0,3653) /gb=af /gi= /ug=mm.6949 /len= _at AI AI573601:ud46a12.y1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=ai /gi= /ug=mm /len= _at AI AI836034:UI-M-AQ0-aad-f-06-0-UI.s2 Mus musculus cdna, 3 end /clone=ui-m-aq0-aad-f-06-0-ui /clone_end=3 /gb=ai /gi= /ug=mm /len= _at AI AI839906:UI-M-AH0-acw-e-01-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-ah0-acw-e-01-0-ui /clone_end= /gb=ai /gi= /ug=mm /len= _at AI AI842849:UI-M-AN1-afb-b-11-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-an1-afb-b-11-0-ui /clone_end=3 /gb=ai /gi= /ug=mm /len= _at AI AI877157:uc54h09.r1 Mus musculus cdna, 5 end /clone=image /clone_end=5 /gb=ai /gi= /ug=mm /len= _at AI AI842192:UI-M-AI1-afm-d-03-0-UI.s1 Mus musculus cdna, 3 end /clone=ui-m-ai1-afm-d-03-0-ui /clone_end=3 /gb=ai /gi= /ug=mm /len=
UPREGULATED GENES (2.0 fold OR higher):
 RATIO 12.06 9.01 9.00 8.99 8.65 7.57 7.38 7.21 6.76 5.45 5.41 5.26 5.19 4.87 4.73 4.71 4.61 4.58 4.57 UPREGULATED GENES (2.0 fold OR higher): GENE NAME Cluster Incl AI840579:UI-M-AM0-adp-h-12-0-UI.s1 Mus
RATIO 12.06 9.01 9.00 8.99 8.65 7.57 7.38 7.21 6.76 5.45 5.41 5.26 5.19 4.87 4.73 4.71 4.61 4.58 4.57 UPREGULATED GENES (2.0 fold OR higher): GENE NAME Cluster Incl AI840579:UI-M-AM0-adp-h-12-0-UI.s1 Mus
Probe Avg Abs Diff Fold Descriptions Set Diff Call Call Change 99941_at P D ~314.1 Cluster Incl L11333:Mouse carboxyesterase mrna
 Probe Avg Abs Diff Fold Descriptions Set Diff Call Call Change 99941_at 64596.9 P D ~314.1 Cluster Incl L11333:Mouse carboxyesterase mrna /cds=unknown /gb=l11333 /gi=309136 /ug=mm.4350 /len=2006 /STRA=for
Probe Avg Abs Diff Fold Descriptions Set Diff Call Call Change 99941_at 64596.9 P D ~314.1 Cluster Incl L11333:Mouse carboxyesterase mrna /cds=unknown /gb=l11333 /gi=309136 /ug=mm.4350 /len=2006 /STRA=for
-8 J04423 E coli bioc protein (-5 and -3 represent transcript regions 5 prime and 3 prime respectively)
 AFFX-MurIL2_at -51.6 M16762 Mouse interleukin 2 (IL-2) gene, exon 4 AFFX-MurIL10_at 10.8 M37897 Mouse interleukin 10 mrna, complete cds AFFX-MurIL4_at 14.1 M25892 Mus musculus interleukin 4 (Il-4) mrna,
AFFX-MurIL2_at -51.6 M16762 Mouse interleukin 2 (IL-2) gene, exon 4 AFFX-MurIL10_at 10.8 M37897 Mouse interleukin 10 mrna, complete cds AFFX-MurIL4_at 14.1 M25892 Mus musculus interleukin 4 (Il-4) mrna,
201291_s_at Consensus includes gb:au /FEA=EST /DB_XREF=gi: /DB_XREF=est:AU /CLONE=Y79AA /UG=Hs topoisomerase (DNA)
 201291_s_at Consensus includes gb:au159942 /FEA=EST /DB_XREF=gi:11021463 /DB_XREF=est:AU159942 /CLONE=Y79AA1000724 /UG=Hs.156346 topoisomerase (DNA) II alpha (170kD) /FL=gb:J04088.1 gb:nm_001067.1 201664_at
201291_s_at Consensus includes gb:au159942 /FEA=EST /DB_XREF=gi:11021463 /DB_XREF=est:AU159942 /CLONE=Y79AA1000724 /UG=Hs.156346 topoisomerase (DNA) II alpha (170kD) /FL=gb:J04088.1 gb:nm_001067.1 201664_at
Table S1 Differentially expressed genes showing > 2 fold changes and p <0.01 for 0.01 mm NS-398, 0.1mM ibuprofen and COX-2 RNAi.
 Table S1 Differentially expressed genes showing > 2 fold changes and p
Table S1 Differentially expressed genes showing > 2 fold changes and p
List of Genes Expressed in Quiescent Human Mesangial Cells
 List of Genes Expressed in Quiescent Human Mesangial Cells Genbank No Affymetrtix Descriptions Signal- 0h Detection- 0h Detection p- value - 0h U14573 U14573 Alu-Sq subfamily consensus sequence. 45314.4
List of Genes Expressed in Quiescent Human Mesangial Cells Genbank No Affymetrtix Descriptions Signal- 0h Detection- 0h Detection p- value - 0h U14573 U14573 Alu-Sq subfamily consensus sequence. 45314.4
SUPPLEMENTAL TABLE I. Identified Proteins in Bovine Testicular Hyaluronidase Type I-S via LC-MS/MS
 SUPPLEMENTAL TABLE I. Identified Proteins in Bovine Testicular Hyaluronidase Type I-S via LC-MS/MS No. Protein 1 serum albumin precursor gi 30794280 2 annexin A2 gi 27807289 3 Phosphatidylethanolamine-binding
SUPPLEMENTAL TABLE I. Identified Proteins in Bovine Testicular Hyaluronidase Type I-S via LC-MS/MS No. Protein 1 serum albumin precursor gi 30794280 2 annexin A2 gi 27807289 3 Phosphatidylethanolamine-binding
Fig. S1. Summary of the altered metabolism pathways in alcoholic fatty liver disease using MetPA analysis (panel A).
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Fig. S1. Summary of the altered metabolism pathways in alcoholic fatty liver disease
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Fig. S1. Summary of the altered metabolism pathways in alcoholic fatty liver disease
SUPPLEMENTARY INFORMATION
 Supplementary Table 1- Differential expression of stress-related genes. Feature Number Probe Name Systematic Name Gene Name Description 30248 A_52_P342860 NM_007954 Es1 Mus musculus esterase 1 (Es1), mrna
Supplementary Table 1- Differential expression of stress-related genes. Feature Number Probe Name Systematic Name Gene Name Description 30248 A_52_P342860 NM_007954 Es1 Mus musculus esterase 1 (Es1), mrna
Supplementary Information
 Scientific Reports Supplementary Information Upregulated expression of FGF13/FHF2 mediates resistance to platinum drugs in cervical cancer cells Tomoko Okada, Kazuhiro Murata, Ryoma Hirose, Chie Matsuda,
Scientific Reports Supplementary Information Upregulated expression of FGF13/FHF2 mediates resistance to platinum drugs in cervical cancer cells Tomoko Okada, Kazuhiro Murata, Ryoma Hirose, Chie Matsuda,
Mouse Meda-4 : chromosome 5G bp. EST (547bp) _at. 5 -Meda4 inner race (~1.8Kb)
 Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
Supplemental Figure Legends
 Supplemental Figure Legends Supplemental Figure 1. Transcriptional changes are dependent upon Fpn and occur in peritoneal macrophages. A. Mouse bone marrow macrophages were transfected with either WT Fpn-GFP
Supplemental Figure Legends Supplemental Figure 1. Transcriptional changes are dependent upon Fpn and occur in peritoneal macrophages. A. Mouse bone marrow macrophages were transfected with either WT Fpn-GFP
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at
 Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Table 1A. Genes enriched as over-expressed in DPM treatment group
 Table 1A. s enriched as over-expressed in DPM treatment group Affy Probeset mrna Accession Description 10538247 Npy NM_023456 Mus musculus neuropeptide Y (Npy), mrna. 2.0024 10598062 --- NC_005089 gi 34538597
Table 1A. s enriched as over-expressed in DPM treatment group Affy Probeset mrna Accession Description 10538247 Npy NM_023456 Mus musculus neuropeptide Y (Npy), mrna. 2.0024 10598062 --- NC_005089 gi 34538597
Supplemental Information. Host-Microbiota Interactions in the Pathogenesis. of Antibiotic-Associated Diseases
 Cell Reports, Volume Supplemental Information Host-Microbiota Interactions in the Pathogenesis of -Associated Diseases Joshua S. Lichtman, Jessica A. Ferreyra, Katharine M. Ng, Samuel A. Smits, Justin
Cell Reports, Volume Supplemental Information Host-Microbiota Interactions in the Pathogenesis of -Associated Diseases Joshua S. Lichtman, Jessica A. Ferreyra, Katharine M. Ng, Samuel A. Smits, Justin
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies
 Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function
 Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSL0024 26223 26621 + LrgA family BPSL0025 26690 27412 + hypothetical
Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSL0024 26223 26621 + LrgA family BPSL0025 26690 27412 + hypothetical
Validated Mouse Quantitative RT-PCR Genes Gene Gene Bank Accession Number Name
 5HT1a NM_008308.4 Serotonin Receptor 1a 5HT1b NM_010482.1 Serotonin Receptor 1b 5HT2a NM_172812.2 Serotonin Receptor 2a ACO-2 NM_080633.2 Aconitase 2 Adora2A NM_009630.02 Adenosine A2a Receptor Aif1 (IbaI)
5HT1a NM_008308.4 Serotonin Receptor 1a 5HT1b NM_010482.1 Serotonin Receptor 1b 5HT2a NM_172812.2 Serotonin Receptor 2a ACO-2 NM_080633.2 Aconitase 2 Adora2A NM_009630.02 Adenosine A2a Receptor Aif1 (IbaI)
f(x) = x R² = RPKM (M8.MXB) f(x) = x E-014 R² = 1 RPKM (M31.
 14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
Under the Radar Screen: How Bugs Trick Our Immune Defenses
 Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 7: Cytokines Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research HHV-8 Discovered in the 1980 s at the
Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 7: Cytokines Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research HHV-8 Discovered in the 1980 s at the
Supporting Information: Protein Corona Analysis of Silver Nanoparticles Exposed to Fish Plasma
 Supporting Information: Protein Corona Analysis of Silver Nanoparticles Exposed to Fish Plasma Jiejun Gao 1, Lu Lin 2, Alexander Wei 2,*, and Maria S. Sepúlveda 1,* 1 Department of Forestry and Natural
Supporting Information: Protein Corona Analysis of Silver Nanoparticles Exposed to Fish Plasma Jiejun Gao 1, Lu Lin 2, Alexander Wei 2,*, and Maria S. Sepúlveda 1,* 1 Department of Forestry and Natural
Chemistry 107 Exam 4 Study Guide
 Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction. Marker Sequence (5 3 ) Accession No.
 Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR
 Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) cadl CTTGGGGGCGCGTCT CTGTTCTTTTGTGCCGTTTCG cyl-coenzyme Dehydrogenase, very
Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) cadl CTTGGGGGCGCGTCT CTGTTCTTTTGTGCCGTTTCG cyl-coenzyme Dehydrogenase, very
BIOL212 Biochemistry of Disease. Metabolic Disorders - Obesity
 BIOL212 Biochemistry of Disease Metabolic Disorders - Obesity Obesity Approx. 23% of adults are obese in the U.K. The number of obese children has tripled in 20 years. 10% of six year olds are obese, rising
BIOL212 Biochemistry of Disease Metabolic Disorders - Obesity Obesity Approx. 23% of adults are obese in the U.K. The number of obese children has tripled in 20 years. 10% of six year olds are obese, rising
Electron Transport Chain and Oxidative phosphorylation
 Electron Transport Chain and Oxidative phosphorylation So far we have discussed the catabolism involving oxidation of 6 carbons of glucose to CO 2 via glycolysis and CAC without any oxygen molecule directly
Electron Transport Chain and Oxidative phosphorylation So far we have discussed the catabolism involving oxidation of 6 carbons of glucose to CO 2 via glycolysis and CAC without any oxygen molecule directly
Supporting Information
 Comparative Proteomic Study of Fatty Acid-treated Myoblasts Reveals Role of Cox-2 in Palmitate-induced Insulin Resistance Supporting Information Xiulan Chen 1#, Shimeng Xu 2,3#, Shasha Wei 1,3, Yaqin Deng
Comparative Proteomic Study of Fatty Acid-treated Myoblasts Reveals Role of Cox-2 in Palmitate-induced Insulin Resistance Supporting Information Xiulan Chen 1#, Shimeng Xu 2,3#, Shasha Wei 1,3, Yaqin Deng
Significance of the MHC
 CHAPTER 7 Major Histocompatibility Complex (MHC) What is is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) Significance of the MHC role in immune response role in organ
CHAPTER 7 Major Histocompatibility Complex (MHC) What is is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) Significance of the MHC role in immune response role in organ
Processing of RNA II Biochemistry 302. February 14, 2005 Bob Kelm
 Processing of RNA II Biochemistry 302 February 14, 2005 Bob Kelm What s an intron? Transcribed sequence removed during the process of mrna maturation (non proteincoding sequence) Discovered by P. Sharp
Processing of RNA II Biochemistry 302 February 14, 2005 Bob Kelm What s an intron? Transcribed sequence removed during the process of mrna maturation (non proteincoding sequence) Discovered by P. Sharp
ACCESSORY PUBLICATION. Supplementary Table 1. Genes up regulated in the skin of both HR and LR animals. after tick larval challenge e
 ACCESSORY PUBLICATION Supplementary Table 1. Genes up regulated in the skin of both HR and LR animals after tick larval challenge e GenBank Accession a No. of elemen ts b t=0 Signal intensity c t=0 Description
ACCESSORY PUBLICATION Supplementary Table 1. Genes up regulated in the skin of both HR and LR animals after tick larval challenge e GenBank Accession a No. of elemen ts b t=0 Signal intensity c t=0 Description
Minjia Tan, Ph.D Shanghai Institute of Materia Medica, Chinese Academy of Sciences
 Lysine Glutarylation Is a Protein Posttranslational Modification Regulated by SIRT5 Minjia Tan, Ph.D Shanghai Institute of Materia Medica, Chinese Academy of Sciences Lysine: the most frequently modified
Lysine Glutarylation Is a Protein Posttranslational Modification Regulated by SIRT5 Minjia Tan, Ph.D Shanghai Institute of Materia Medica, Chinese Academy of Sciences Lysine: the most frequently modified
Receptor mediated Signal Transduction
 Receptor mediated Signal Transduction G-protein-linked receptors adenylyl cyclase camp PKA Organization of receptor protein-tyrosine kinases From G.M. Cooper, The Cell. A molecular approach, 2004, third
Receptor mediated Signal Transduction G-protein-linked receptors adenylyl cyclase camp PKA Organization of receptor protein-tyrosine kinases From G.M. Cooper, The Cell. A molecular approach, 2004, third
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array.
 Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
* Kyoto Encyclopedia of Genes and Genomes.
 Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
Processing of RNA II Biochemistry 302. February 13, 2006
 Processing of RNA II Biochemistry 302 February 13, 2006 Precursor mrna: introns and exons Intron: Transcribed RNA sequence removed from precursor RNA during the process of maturation (for class II genes:
Processing of RNA II Biochemistry 302 February 13, 2006 Precursor mrna: introns and exons Intron: Transcribed RNA sequence removed from precursor RNA during the process of maturation (for class II genes:
Pre-mRNA has introns The splicing complex recognizes semiconserved sequences
 Adding a 5 cap Lecture 4 mrna splicing and protein synthesis Another day in the life of a gene. Pre-mRNA has introns The splicing complex recognizes semiconserved sequences Introns are removed by a process
Adding a 5 cap Lecture 4 mrna splicing and protein synthesis Another day in the life of a gene. Pre-mRNA has introns The splicing complex recognizes semiconserved sequences Introns are removed by a process
VIRUSES AND CANCER Michael Lea
 VIRUSES AND CANCER 2010 Michael Lea VIRAL ONCOLOGY - LECTURE OUTLINE 1. Historical Review 2. Viruses Associated with Cancer 3. RNA Tumor Viruses 4. DNA Tumor Viruses HISTORICAL REVIEW Historical Review
VIRUSES AND CANCER 2010 Michael Lea VIRAL ONCOLOGY - LECTURE OUTLINE 1. Historical Review 2. Viruses Associated with Cancer 3. RNA Tumor Viruses 4. DNA Tumor Viruses HISTORICAL REVIEW Historical Review
Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion
 Probe Set ID RefSeq Transcript ID Gene Title Gene Symbol 205980_s_at NM_001017526 /// NM_181334 /// NM_181335 Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion ARHGAP8 /// LOC553158 FC ALK shrna
Probe Set ID RefSeq Transcript ID Gene Title Gene Symbol 205980_s_at NM_001017526 /// NM_181334 /// NM_181335 Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion ARHGAP8 /// LOC553158 FC ALK shrna
Revision. camp pathway
 االله الرحمن الرحيم بسم Revision camp pathway camp pathway Revision camp pathway Adenylate cyclase Adenylate Cyclase enzyme Adenylate cyclase catalyses the formation of camp from ATP. Stimulation or inhibition
االله الرحمن الرحيم بسم Revision camp pathway camp pathway Revision camp pathway Adenylate cyclase Adenylate Cyclase enzyme Adenylate cyclase catalyses the formation of camp from ATP. Stimulation or inhibition
LIPID METABOLISM
 LIPID METABOLISM LIPOGENESIS LIPOGENESIS LIPOGENESIS FATTY ACID SYNTHESIS DE NOVO FFA in the blood come from :- (a) Dietary fat (b) Dietary carbohydrate/protein in excess of need FA TAG Site of synthesis:-
LIPID METABOLISM LIPOGENESIS LIPOGENESIS LIPOGENESIS FATTY ACID SYNTHESIS DE NOVO FFA in the blood come from :- (a) Dietary fat (b) Dietary carbohydrate/protein in excess of need FA TAG Site of synthesis:-
Recombinant Protein Expression Retroviral system
 Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
HORMONES AND CELL SIGNALLING
 HORMONES AND CELL SIGNALLING TYPES OF CELL JUNCTIONS CHEMICAL SIGNALS AND MODES OF ACTION Endocrine system produces chemical messages = hormones that are transported from endocrine gland to target cell
HORMONES AND CELL SIGNALLING TYPES OF CELL JUNCTIONS CHEMICAL SIGNALS AND MODES OF ACTION Endocrine system produces chemical messages = hormones that are transported from endocrine gland to target cell
Antibodies and T Cell Receptor Genetics Generation of Antigen Receptor Diversity
 Antibodies and T Cell Receptor Genetics 2008 Peter Burrows 4-6529 peterb@uab.edu Generation of Antigen Receptor Diversity Survival requires B and T cell receptor diversity to respond to the diversity of
Antibodies and T Cell Receptor Genetics 2008 Peter Burrows 4-6529 peterb@uab.edu Generation of Antigen Receptor Diversity Survival requires B and T cell receptor diversity to respond to the diversity of
Biosignals, Chapter 8, rearranged, Part I
 Biosignals, Chapter 8, rearranged, Part I Nicotinic Acetylcholine Receptor: A Ligand-Binding Ion Channel Classes of Receptor Proteins in Eukaryotes, Heterotrimeric G Proteins Signaling View the Heterotrimeric
Biosignals, Chapter 8, rearranged, Part I Nicotinic Acetylcholine Receptor: A Ligand-Binding Ion Channel Classes of Receptor Proteins in Eukaryotes, Heterotrimeric G Proteins Signaling View the Heterotrimeric
Supplementary Material Correlation matrices on FP and FN profiles
 Supplementary Material Correlation matrices on FP and FN profiles The following two tables give the correlation coefficients for the FP profiles and the FN profiles of a single tagging solutions against
Supplementary Material Correlation matrices on FP and FN profiles The following two tables give the correlation coefficients for the FP profiles and the FN profiles of a single tagging solutions against
Mohammad Husain Department of Biotechnology, Jamia Millia Islamia New Delhi
 Role of Vitamin D receptor (VDR) in HIV induced tubular injury Mohammad Husain Department of Biotechnology, Jamia Millia Islamia New Delhi 07/10/2015 INTRODUCTION Vitamin D is technically not a Vitamin;
Role of Vitamin D receptor (VDR) in HIV induced tubular injury Mohammad Husain Department of Biotechnology, Jamia Millia Islamia New Delhi 07/10/2015 INTRODUCTION Vitamin D is technically not a Vitamin;
Immune suppression in Atlantic salmon: smoltification, sea water transfer and breeding
 Immune suppression in Atlantic salmon: smoltification, sea water transfer and breeding Aleksei Krasnov, Nofima FHFs fiskehelsesamling 1.-2. september 2015 Smoltifiction and breeding suppress immunity in
Immune suppression in Atlantic salmon: smoltification, sea water transfer and breeding Aleksei Krasnov, Nofima FHFs fiskehelsesamling 1.-2. september 2015 Smoltifiction and breeding suppress immunity in
Signal Transduction: G-Protein Coupled Receptors
 Signal Transduction: G-Protein Coupled Receptors Federle, M. (2017). Lectures 4-5: Signal Transduction parts 1&2: nuclear receptors and GPCRs. Lecture presented at PHAR 423 Lecture in UIC College of Pharmacy,
Signal Transduction: G-Protein Coupled Receptors Federle, M. (2017). Lectures 4-5: Signal Transduction parts 1&2: nuclear receptors and GPCRs. Lecture presented at PHAR 423 Lecture in UIC College of Pharmacy,
Syllabus for BASIC METABOLIC PRINCIPLES
 Syllabus for BASIC METABOLIC PRINCIPLES The video lecture covers basic principles you will need to know for the lectures covering enzymes and metabolism in Principles of Metabolism and elsewhere in the
Syllabus for BASIC METABOLIC PRINCIPLES The video lecture covers basic principles you will need to know for the lectures covering enzymes and metabolism in Principles of Metabolism and elsewhere in the
Supplemental Table 1 Age and gender-specific cut-points used for MHO.
 Supplemental Table 1 Age and gender-specific cut-points used for MHO. Age SBP (mmhg) DBP (mmhg) HDL-C (mmol/l) TG (mmol/l) FG (mmol/l) Boys 6-11 90th * 90th * 1.03 1.24 5.6 12 121 76 1.13 1.44 5.6 13 123
Supplemental Table 1 Age and gender-specific cut-points used for MHO. Age SBP (mmhg) DBP (mmhg) HDL-C (mmol/l) TG (mmol/l) FG (mmol/l) Boys 6-11 90th * 90th * 1.03 1.24 5.6 12 121 76 1.13 1.44 5.6 13 123
the nature and importance of biomacromolecules in the chemistry of the cell: synthesis of biomacromolecules through the condensation reaction lipids
 the nature and importance of biomacromolecules in the chemistry of the cell: synthesis of biomacromolecules through the condensation reaction lipids and their sub-units; the role of lipids in the plasma
the nature and importance of biomacromolecules in the chemistry of the cell: synthesis of biomacromolecules through the condensation reaction lipids and their sub-units; the role of lipids in the plasma
Regulation of P450. b. competitive-inhibitor but not substrate. c. non-competitive inhibition. 2. Covalent binding to heme or protein
 10. Inhibition of P450 Regulation of P450 a. competitive-2 chemicals metabolized by same isoform b. competitive-inhibitor but not substrate c. non-competitive inhibition 11. Induction of P450 1. Non-covalent
10. Inhibition of P450 Regulation of P450 a. competitive-2 chemicals metabolized by same isoform b. competitive-inhibitor but not substrate c. non-competitive inhibition 11. Induction of P450 1. Non-covalent
Oncogenomics to Target the Tumor Cell in its Microenvironment
 Oncogenomics to Target the Tumor Cell in its Microenvironment Kenneth C. Anderson, M.D. Jerome Lipper Multiple Myeloma Center Dana-Farber Cancer Institute Harvard Medical School Tumor-Promoting Influence
Oncogenomics to Target the Tumor Cell in its Microenvironment Kenneth C. Anderson, M.D. Jerome Lipper Multiple Myeloma Center Dana-Farber Cancer Institute Harvard Medical School Tumor-Promoting Influence
Supplementary Table SI: Y strain T. cruzi infection results in the upregulation of 381 genes at the site of infection.
 Supplementary Table SI: Y strain T. cruzi infection results in the upregulation of 381 genes at the site of infection. Genes found to be significantly upregulated (FDR2) in Y strain
Supplementary Table SI: Y strain T. cruzi infection results in the upregulation of 381 genes at the site of infection. Genes found to be significantly upregulated (FDR2) in Y strain
A bioinformatics driven cancer study
 The 11th China-Japan-Korea Bioinformatics Training Course & Symposium, June 16-18, Suzhou, China A bioinformatics driven cancer study Chao Li and Yixue Li yxli@sibs.ac.cn Shanghai Institutes for Bio-Medicine
The 11th China-Japan-Korea Bioinformatics Training Course & Symposium, June 16-18, Suzhou, China A bioinformatics driven cancer study Chao Li and Yixue Li yxli@sibs.ac.cn Shanghai Institutes for Bio-Medicine
Supporting Information Table of content
 Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
Supplementary information, Table S2. Genes belonging to groups in Supplementary information, Figure S8
 Supplementary information, Table S2. Genes belonging to groups in Supplementary information, Figure S8 Genbank NM_012054 NM_153508 NM_011305 NM_011884 NM_013892 NM_008595 NM_008860 NM_212457 NM_009514
Supplementary information, Table S2. Genes belonging to groups in Supplementary information, Figure S8 Genbank NM_012054 NM_153508 NM_011305 NM_011884 NM_013892 NM_008595 NM_008860 NM_212457 NM_009514
BIOL 158: BIOLOGICAL CHEMISTRY II
 BIOL 158: BIOLOGICAL CHEMISTRY II Lecture 5: Vitamins and Coenzymes Lecturer: Christopher Larbie, PhD Introduction Cofactors bind to the active site and assist in the reaction mechanism Apoenzyme is an
BIOL 158: BIOLOGICAL CHEMISTRY II Lecture 5: Vitamins and Coenzymes Lecturer: Christopher Larbie, PhD Introduction Cofactors bind to the active site and assist in the reaction mechanism Apoenzyme is an
PHRM20001 NOTES PART 1 Lecture 1 History of Pharmacology- Key Principles
 PHRM20001 NOTES PART 1 Lecture 1 History of Pharmacology- Key Principles Hippocrates (5 th century BCE):... benefit my patients according to my greatest ability and judgment, and I will do no harm or injustice
PHRM20001 NOTES PART 1 Lecture 1 History of Pharmacology- Key Principles Hippocrates (5 th century BCE):... benefit my patients according to my greatest ability and judgment, and I will do no harm or injustice
FREE ENERGY Reactions involving free energy: 1. Exergonic 2. Endergonic
 BIOENERGETICS FREE ENERGY It is the portion of the total energy change in a system that is available for doing work at constant temperature and pressure; it is represented as ΔG. Reactions involving free
BIOENERGETICS FREE ENERGY It is the portion of the total energy change in a system that is available for doing work at constant temperature and pressure; it is represented as ΔG. Reactions involving free
University of Guelph Department of Chemistry and Biochemistry Structure and Function In Biochemistry
 University of Guelph Department of Chemistry and Biochemistry 19-356 Structure and Function In Biochemistry Final Exam, April 21, 1997. Time allowed, 120 min. Answer questions 1-30 on the computer scoring
University of Guelph Department of Chemistry and Biochemistry 19-356 Structure and Function In Biochemistry Final Exam, April 21, 1997. Time allowed, 120 min. Answer questions 1-30 on the computer scoring
Biological processes. Mitochondrion Metabolic process Catalytic activity Oxidoreductase
 Full name Glyceraldehyde3 phosphate dehydrogenase Succinatesemialdehyde Glutamate dehydrogenase 1, Alcohol dehydrogenase [NADP+] 2',3'cyclicnucleotide 3' phosphodiesterase Dihydropyrimidinaserelated 2
Full name Glyceraldehyde3 phosphate dehydrogenase Succinatesemialdehyde Glutamate dehydrogenase 1, Alcohol dehydrogenase [NADP+] 2',3'cyclicnucleotide 3' phosphodiesterase Dihydropyrimidinaserelated 2
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/2/e1602038/dc1 Supplementary Materials for Mitochondrial metabolic regulation by GRP78 Manoj Prasad, Kevin J. Pawlak, William E. Burak, Elizabeth E. Perry, Brendan
advances.sciencemag.org/cgi/content/full/3/2/e1602038/dc1 Supplementary Materials for Mitochondrial metabolic regulation by GRP78 Manoj Prasad, Kevin J. Pawlak, William E. Burak, Elizabeth E. Perry, Brendan
An#body structure & func#on
 An#body structure & func#on Objec&ves You should be able to: ü Define an&body ü Describe an&body structure ü Compare between different types or classes of an&body and their func&on An#bodies = Immunoglobulin
An#body structure & func#on Objec&ves You should be able to: ü Define an&body ü Describe an&body structure ü Compare between different types or classes of an&body and their func&on An#bodies = Immunoglobulin
Mechanistic Toxicology
 SECOND EDITION Mechanistic Toxicology The Molecular Basis of How Chemicals Disrupt Biological Targets URS A. BOELSTERLI CRC Press Tavlor & France Croup CRC Press is an imp^t o* :H Taylor H Francn C'r,,jpi
SECOND EDITION Mechanistic Toxicology The Molecular Basis of How Chemicals Disrupt Biological Targets URS A. BOELSTERLI CRC Press Tavlor & France Croup CRC Press is an imp^t o* :H Taylor H Francn C'r,,jpi
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system
 Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Energy storage in cells
 Energy storage in cells Josef Fontana EC - 58 Overview of the lecture Introduction to the storage substances of human body Overview of storage compounds in the body Glycogen metabolism Structure of glycogen
Energy storage in cells Josef Fontana EC - 58 Overview of the lecture Introduction to the storage substances of human body Overview of storage compounds in the body Glycogen metabolism Structure of glycogen
Moh Tarek. Razi Kittaneh. Jaqen H ghar
 14 Moh Tarek Razi Kittaneh Jaqen H ghar Naif Karadsheh Gluconeogenesis is making glucose from non-carbohydrates precursors. Although Gluconeogenesis looks like Glycolysis in many steps, it is not the simple
14 Moh Tarek Razi Kittaneh Jaqen H ghar Naif Karadsheh Gluconeogenesis is making glucose from non-carbohydrates precursors. Although Gluconeogenesis looks like Glycolysis in many steps, it is not the simple
There are approximately 30,000 proteasomes in a typical human cell Each proteasome is approximately 700 kda in size The proteasome is made up of 3
 Proteasomes Proteasomes Proteasomes are responsible for degrading proteins that have been damaged, assembled improperly, or that are of no profitable use to the cell. The unwanted protein is literally
Proteasomes Proteasomes Proteasomes are responsible for degrading proteins that have been damaged, assembled improperly, or that are of no profitable use to the cell. The unwanted protein is literally
TNFSF13B tumor necrosis factor (ligand) superfamily, member 13b NF-kB pathway cluster, Enrichment Score: 3.57
 Appendix 2. Highly represented clusters of genes in the differential expression of data. Immune Cluster, Enrichment Score: 5.17 GO:0048584 positive regulation of response to stimulus GO:0050778 positive
Appendix 2. Highly represented clusters of genes in the differential expression of data. Immune Cluster, Enrichment Score: 5.17 GO:0048584 positive regulation of response to stimulus GO:0050778 positive
Cornstarch
 Electronic Supplementary Material (ESI) for Food & Function. This journal is The Royal Society of Chemistry 2018 Supplementary data : Supplementary Table 1: Diet composition (g/kg) on the basis of the
Electronic Supplementary Material (ESI) for Food & Function. This journal is The Royal Society of Chemistry 2018 Supplementary data : Supplementary Table 1: Diet composition (g/kg) on the basis of the
Polyomaviridae. Spring
 Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
Polyomaviridae Spring 2002 331 Antibody Prevalence for BK & JC Viruses Spring 2002 332 Polyoma Viruses General characteristics Papovaviridae: PA - papilloma; PO - polyoma; VA - vacuolating agent a. 45nm
HSP72 HSP90. Quadriceps Muscle. MEF2c MyoD1 MyoG Myf5 Hsf1 Hsp GLUT4/GAPDH (AU)
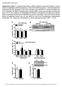 Supplementary Figure 1. Impaired insulin action in HSP72 deficient muscle and myotubes in culture cannot be explained by altered myogenesis or reduced total GLUT4 expression. Genes associated with myogenesis
Supplementary Figure 1. Impaired insulin action in HSP72 deficient muscle and myotubes in culture cannot be explained by altered myogenesis or reduced total GLUT4 expression. Genes associated with myogenesis
Electron transport chain chapter 6 (page 73) BCH 340 lecture 6
 Electron transport chain chapter 6 (page 73) BCH 340 lecture 6 The Metabolic Pathway of Cellular Respiration All of the reactions involved in cellular respiration can be grouped into three main stages
Electron transport chain chapter 6 (page 73) BCH 340 lecture 6 The Metabolic Pathway of Cellular Respiration All of the reactions involved in cellular respiration can be grouped into three main stages
Supplementary Figure 1. Mitochondrial function in skeletal muscle and plasma parameters of STZ mice A D-F
 Supplementary Figure 1. Mitochondrial function in skeletal muscle and plasma parameters of STZ mice A Expression of representative subunits of the mitochondrial respiratory chain was analyzed by real time
Supplementary Figure 1. Mitochondrial function in skeletal muscle and plasma parameters of STZ mice A Expression of representative subunits of the mitochondrial respiratory chain was analyzed by real time
Significance of the MHC
 CHAPTER 8 Major Histocompatibility Complex (MHC) What is is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) Significance of the MHC role in immune response role in organ
CHAPTER 8 Major Histocompatibility Complex (MHC) What is is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) Significance of the MHC role in immune response role in organ
Tumour growth environment modulates Chk1 signalling pathways and sensitivity to Chk1 inhibition
 Tumour growth environment modulates Chk1 signalling pathways and sensitivity to Chk1 inhibition Andrew J Massey Supplementary Information Supplementary Figure S1. Related to Fig. 1. (a) HT29 or U2OS cells
Tumour growth environment modulates Chk1 signalling pathways and sensitivity to Chk1 inhibition Andrew J Massey Supplementary Information Supplementary Figure S1. Related to Fig. 1. (a) HT29 or U2OS cells
Regulation of Glucose Metabolism by Intracellular Compounds
 Regulation of Metabolism by Intracellular ompounds Hexokinase ( ) -6- H ribose-5- or fructose-6- -6-phosphate () ( ) H -6-phosphate GI GM UD-glucose pyrophosphorylase 1-phosphate UD- hosphorylase synthase
Regulation of Metabolism by Intracellular ompounds Hexokinase ( ) -6- H ribose-5- or fructose-6- -6-phosphate () ( ) H -6-phosphate GI GM UD-glucose pyrophosphorylase 1-phosphate UD- hosphorylase synthase
LUMC: LOPAC screen analysis
 LUMC: LOPAC screen analysis Preliminary results November 9, 2012 This document briefly presents the results from the LOPAC compounds screen performed at LUMC. It contains the following information: The
LUMC: LOPAC screen analysis Preliminary results November 9, 2012 This document briefly presents the results from the LOPAC compounds screen performed at LUMC. It contains the following information: The
Alpha thalassemia mental retardation X-linked. Acquired alpha-thalassemia myelodysplastic syndrome
 Alpha thalassemia mental retardation X-linked Acquired alpha-thalassemia myelodysplastic syndrome (Alpha thalassemia mental retardation X-linked) Acquired alpha-thalassemia myelodysplastic syndrome Schematic
Alpha thalassemia mental retardation X-linked Acquired alpha-thalassemia myelodysplastic syndrome (Alpha thalassemia mental retardation X-linked) Acquired alpha-thalassemia myelodysplastic syndrome Schematic
INTERACTION DRUG BODY
 INTERACTION DRUG BODY What the drug does to the body What the body does to the drug Receptors - intracellular receptors - membrane receptors - Channel receptors - G protein-coupled receptors - Tyrosine-kinase
INTERACTION DRUG BODY What the drug does to the body What the body does to the drug Receptors - intracellular receptors - membrane receptors - Channel receptors - G protein-coupled receptors - Tyrosine-kinase
FATTY ACID SYNTHESIS
 FATTY ACID SYNTHESIS Malonyl- CoA inhibits Carni1ne Palmitoyl Transferase I. Malonyl- CoA is a precursor for fa=y acid synthesis. Malonyl- CoA is produced from acetyl- CoA by the enzyme Acetyl- CoA Carboxylase.
FATTY ACID SYNTHESIS Malonyl- CoA inhibits Carni1ne Palmitoyl Transferase I. Malonyl- CoA is a precursor for fa=y acid synthesis. Malonyl- CoA is produced from acetyl- CoA by the enzyme Acetyl- CoA Carboxylase.
WT siz1-2 siz1-3. WT siz1-2 siz1-3. -Pi, 0.05 M IAA. -Pi, 2.5 M NPA
 A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
T cell Receptor. Chapter 9. Comparison of TCR αβ T cells
 Chapter 9 The αβ TCR is similar in size and structure to an antibody Fab fragment T cell Receptor Kuby Figure 9-3 The αβ T cell receptor - Two chains - α and β - Two domains per chain - constant (C) domain
Chapter 9 The αβ TCR is similar in size and structure to an antibody Fab fragment T cell Receptor Kuby Figure 9-3 The αβ T cell receptor - Two chains - α and β - Two domains per chain - constant (C) domain
Chapter 8 Mitochondria and Cellular Respiration
 Chapter 8 Mitochondria and Cellular Respiration Cellular respiration is the process of oxidizing food molecules, like glucose, to carbon dioxide and water. The energy released is trapped in the form of
Chapter 8 Mitochondria and Cellular Respiration Cellular respiration is the process of oxidizing food molecules, like glucose, to carbon dioxide and water. The energy released is trapped in the form of
For all of the following, you will have to use this website to determine the answers:
 For all of the following, you will have to use this website to determine the answers: http://blast.ncbi.nlm.nih.gov/blast.cgi We are going to be using the programs under this heading: Answer the following
For all of the following, you will have to use this website to determine the answers: http://blast.ncbi.nlm.nih.gov/blast.cgi We are going to be using the programs under this heading: Answer the following
MOLECULAR WEIGHT OF DIFFERENT PROTEINS PRESENT IN ZEBRAFISH EMBRYO DURING GASTRULATION PERIOD
 MOLECULAR WEIGHT OF DIFFERENT PROTEINS PRESENT IN ZEBRAFISH EMBRYO DURING GASTRULATION PERIOD 1) 37% molecular weight of about 97KDa 2) 14.6% molecular weight of about 45 KDa 3+4) 27.4% molecular weight
MOLECULAR WEIGHT OF DIFFERENT PROTEINS PRESENT IN ZEBRAFISH EMBRYO DURING GASTRULATION PERIOD 1) 37% molecular weight of about 97KDa 2) 14.6% molecular weight of about 45 KDa 3+4) 27.4% molecular weight
Proteasomes. When Death Comes a Knock n. Warren Gallagher Chem412, Spring 2001
 Proteasomes When Death Comes a Knock n Warren Gallagher Chem412, Spring 2001 I. Introduction Introduction The central dogma Genetic information is used to make proteins. DNA RNA Proteins Proteins are the
Proteasomes When Death Comes a Knock n Warren Gallagher Chem412, Spring 2001 I. Introduction Introduction The central dogma Genetic information is used to make proteins. DNA RNA Proteins Proteins are the
Herpesviruses. Virion. Genome. Genes and proteins. Viruses and hosts. Diseases. Distinctive characteristics
 Herpesviruses Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Virion Enveloped icosahedral capsid (T=16), diameter 125 nm Diameter of enveloped virion 200 nm Capsid
Herpesviruses Virion Genome Genes and proteins Viruses and hosts Diseases Distinctive characteristics Virion Enveloped icosahedral capsid (T=16), diameter 125 nm Diameter of enveloped virion 200 nm Capsid
Development of B and T lymphocytes
 Development of B and T lymphocytes What will we discuss today? B-cell development T-cell development B- cell development overview Stem cell In periphery Pro-B cell Pre-B cell Immature B cell Mature B cell
Development of B and T lymphocytes What will we discuss today? B-cell development T-cell development B- cell development overview Stem cell In periphery Pro-B cell Pre-B cell Immature B cell Mature B cell
GLUTATHIONE TRANSFERASES. Ralf Morgenstern Institute of Environmental Medicine Karolinska Institutet
 GLUTATHIONE TRANSFERASES Ralf Morgenstern Institute of Environmental Medicine Karolinska Institutet GSTs How many enzymes Structures THREE SUPERFAMILIES SOLUBLE GLUTATHIONE-TRANSFERASES (25 kda, dimers)
GLUTATHIONE TRANSFERASES Ralf Morgenstern Institute of Environmental Medicine Karolinska Institutet GSTs How many enzymes Structures THREE SUPERFAMILIES SOLUBLE GLUTATHIONE-TRANSFERASES (25 kda, dimers)
Significance of the MHC
 CHAPTER 8 Major Histocompatibility Complex (MHC) What is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) - MHC molecules were initially discovered during studies aimed
CHAPTER 8 Major Histocompatibility Complex (MHC) What is MHC? HLA H-2 Minor histocompatibility antigens Peter Gorer & George Sneell (1940) - MHC molecules were initially discovered during studies aimed
number Done by Corrected by Doctor Nayef Karadsheh
 number 11 Done by حسام أبو عوض Corrected by Moayyad Al-Shafei Doctor Nayef Karadsheh 1 P a g e General Regulatory Aspects in Metabolism: We can divide all pathways in metabolism to catabolicand anabolic.
number 11 Done by حسام أبو عوض Corrected by Moayyad Al-Shafei Doctor Nayef Karadsheh 1 P a g e General Regulatory Aspects in Metabolism: We can divide all pathways in metabolism to catabolicand anabolic.
MITOCHONDRIA LECTURES OVERVIEW
 1 MITOCHONDRIA LECTURES OVERVIEW A. MITOCHONDRIA LECTURES OVERVIEW Mitochondrial Structure The arrangement of membranes: distinct inner and outer membranes, The location of ATPase, DNA and ribosomes The
1 MITOCHONDRIA LECTURES OVERVIEW A. MITOCHONDRIA LECTURES OVERVIEW Mitochondrial Structure The arrangement of membranes: distinct inner and outer membranes, The location of ATPase, DNA and ribosomes The
The recruitment of leukocytes and plasma proteins from the blood to sites of infection and tissue injury is called inflammation
 The migration of a particular type of leukocyte into a restricted type of tissue, or a tissue with an ongoing infection or injury, is often called leukocyte homing, and the general process of leukocyte
The migration of a particular type of leukocyte into a restricted type of tissue, or a tissue with an ongoing infection or injury, is often called leukocyte homing, and the general process of leukocyte
Mitochondria and ATP Synthesis
 Mitochondria and ATP Synthesis Mitochondria and ATP Synthesis 1. Mitochondria are sites of ATP synthesis in cells. 2. ATP is used to do work; i.e. ATP is an energy source. 3. ATP hydrolysis releases energy
Mitochondria and ATP Synthesis Mitochondria and ATP Synthesis 1. Mitochondria are sites of ATP synthesis in cells. 2. ATP is used to do work; i.e. ATP is an energy source. 3. ATP hydrolysis releases energy
Alternative splicing. Biosciences 741: Genomics Fall, 2013 Week 6
 Alternative splicing Biosciences 741: Genomics Fall, 2013 Week 6 Function(s) of RNA splicing Splicing of introns must be completed before nuclear RNAs can be exported to the cytoplasm. This led to early
Alternative splicing Biosciences 741: Genomics Fall, 2013 Week 6 Function(s) of RNA splicing Splicing of introns must be completed before nuclear RNAs can be exported to the cytoplasm. This led to early
L I F E S C I E N C E S
 1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 OCTOBER 31, 2006
1a L I F E S C I E N C E S 5 -UUA AUA UUC GAA AGC UGC AUC GAA AAC UGU GAA UCA-3 5 -TTA ATA TTC GAA AGC TGC ATC GAA AAC TGT GAA TCA-3 3 -AAT TAT AAG CTT TCG ACG TAG CTT TTG ACA CTT AGT-5 OCTOBER 31, 2006
Lecture: 26 OXIDATION OF FATTY ACIDS
 Lecture: 26 OXIDATION OF FATTY ACIDS Fatty acids obtained by hydrolysis of fats undergo different oxidative pathways designated as alpha ( ), beta ( ) and omega ( ) pathways. -oxidation -Oxidation of fatty
Lecture: 26 OXIDATION OF FATTY ACIDS Fatty acids obtained by hydrolysis of fats undergo different oxidative pathways designated as alpha ( ), beta ( ) and omega ( ) pathways. -oxidation -Oxidation of fatty
Biochemistry 423 Final Examination NAME:
 Biochemistry 423 Final Examination NAME: 1 Circle the single BEST answer (3 points each) 1. At equilibrium the free energy of a reaction G A. depends only on the temperature B. is positive C. is 0 D. is
Biochemistry 423 Final Examination NAME: 1 Circle the single BEST answer (3 points each) 1. At equilibrium the free energy of a reaction G A. depends only on the temperature B. is positive C. is 0 D. is
