A TSC signaling node at the peroxisome regulates mtorc1 and autophagy in response to ROS
|
|
|
- Randell Wood
- 5 years ago
- Views:
Transcription
1 A TSC signaling node at the peroxisome regulates mtorc1 and autophagy in response to ROS The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters. Citation Published Version Accessed Citable Link Terms of Use Zhang, J., J. Kim, A. Alexander, S. Cai, D. N. Tripathi, R. Dere, A. R. Tee, et al A TSC signaling node at the peroxisome regulates mtorc1 and autophagy in response to ROS. Nature cell biology 15 (10): doi: /ncb doi: /ncb2822 August 21, :30:12 AM EDT This article was downloaded from Harvard University's DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at (Article begins on next page)
2 NIH Public Access Author Manuscript Published in final edited form as: Nat Cell Biol October ; 15(10): doi: /ncb2822. A TSC signaling node at the peroxisome regulates mtorc1 and autophagy in response to ROS Jiangwei Zhang 1,8, Jinhee Kim 2,8, Angela Alexander 1, Shengli Cai 1, Durga Nand Tripathi 1, Ruhee Dere 1, Andrew R. Tee 3, Jacqueline Tait-Mulder 4, Alessia Di Nardo 5, Juliette M. Han 5, Erica Kwiatkowski 5, Elaine A. Dunlop 3, Kayleigh M. Dodd 3, Rebecca D. Folkerth 6, Phyllis L. Faust 7, Michael B. Kastan 4, Mustafa Sahin 5, and Cheryl Lyn Walker 1,9 1 Center for Translational Cancer Research, Institute for Biosciences and Technology, Texas A&M Health Science Center, Houston, TX 77030, USA 2 Korea Institute of Oriental Medicine, Dajeon, , South Korea 3 Institute of Medical Genetics, Cardiff University, Cardiff, Wales, UK 4 Departments of Pediatrics, Pharmacology and Cancer Biology, Duke Cancer Institute, Duke University Medical Center, Durham, NC 27710, USA 5 The F.M. Kirby Neurobiology Center, Department of Neurology, Children s Hospital Boston, Harvard Medical School, Boston, MA 02115, USA 6 Department of Pathology (Neuropathology), Brigham and Women s Hospital, Children s Hospital Boston, Harvard Medical School, Boston, MA 02115, USA 7 Department of Pathology and Cell Biology, Columbia University, New York, NY 10032, USA Abstract Subcellular localization is emerging as an important mechanism for mtorc1 regulation. We report that the tuberous sclerosis complex (TSC) signaling node, TSC1, TSC2 and Rheb, localizes to peroxisomes, where it regulates mtorc1 in response to reactive oxygen species (ROS). TSC1 and TSC2 were bound by PEX19 and PEX5, respectively, and peroxisome-localized TSC functioned as a Rheb GAP to suppress mtorc1 and induce autophagy. Naturally occurring pathogenic mutations in TSC2 decreased PEX5 binding, abrogated peroxisome localization, Rheb GAP activity, and suppression of mtorc1 by ROS. Cells lacking peroxisomes were deficient in mtorc1 repression by ROS and peroxisome-localization deficient TSC2 mutants caused polarity defects and formation of multiple axons in neurons. These data identify a role for TSC in responding to ROS at the peroxisome, and identify the peroxisome as a signaling organelle involved in regulation of mtorc1. Tuberous sclerosis complex (TSC) is a hereditary hamartoma syndrome caused by defects in either the TSC1 or TSC2 genes 1, 2. The TSC tumor suppressor is a heterodimer comprised of tuberin (TSC2), a GTPase activating protein (GAP), and its activation partner hamartin 9 Correspondence should be addressed to C.L.W. (cwalker@ibt.tamhsc.edu), (713) (Phone), (713) (Fax). 8 These authors contributed equally to this work. AUTHOR CONTRIBUTIONS J.Z., J.K. and C.L.W designed research; J.Z., J.K., A.A., S.C., D.N.T., R.D., A.R.T., J.T.-M., A.D.N., J.M.H., E.K., E.A.D. and K.M.D., performed research; J.Z., J.K., A.R.T., R.D.F., P.L.F., M.B.K., M.S. and C.L.W analyzed data; J.Z., J.K. and C.L.W wrote the paper. COMPETING FINANCIAL INTERESTS The authors declare that they have no competing financial interests.
3 Zhang et al. Page 2 RESULTS (TSC1), which localizes the TSC tumor suppressor to endomembranes and protects TSC2 from proteasomal degradation 3, 4. TSC inhibits the activity of the small GTPase Rheb to repress mammalian target of rapamycin complex 1 (mtorc1) signaling, a negative regulator of autophagy mtorc1 is regulated by a variety of cellular stimuli including amino acids, mitogens such as insulin, glucose, and energy stress In the case of amino acids, which do not signal through TSC-Rheb pathway 15, mtorc1 activity is regulated by the Rag GTPases, which form the Ragulator complex that localizes mtorc1 to the late endosome or lysosome compartment of cells We recently reported that TSC functions in a signaling node downstream of ataxia telangiectasia mutated (ATM) to repress mtorc1 in response to reactive oxygen species (ROS) 19. However, identification of the specific subcellular compartment(s) in which the TSC tumor suppressor functions to regulate mtorc1 in response to ROS has heretofore remained elusive. Peroxisomes, carry out key metabolic functions in the cell including β-oxidation of fatty acids, and are a major source of cellular ROS 20, 21. Like mitochondria, peroxisomes are autonomously replicating organelles. Peroxisome biogenesis requires peroxin (PEX) proteins, which are essential for assembly of functional peroxisomes 22. Specific PEX proteins, such as PEX5, function as import receptors that recognize peroxisome targeting signals (PTS) in proteins to target them to the peroxisome 22. Here we show that TSC1 and TSC2 associate with PEX proteins and are localized to peroxisomal membranes, where they regulate mtorc1 signaling in response to ROS. Activation of TSC2 and repression of mtorc1 by peroxisomal ROS induces autophagy, and in cells lacking peroxisomes, TSC2 is unable to repress mtorc1 in response to ROS. Disease-associated point mutations in the TSC2 gene result in loss of PEX5 binding, abrogate peroxisomal localization, TSC2 GAP activity for Rheb, and ROS-induced repression of mtorc1. Our data reveal a previously unknown role for the peroxisome as a signaling organelle involved in regulation of mtorc1 by TSC, and suggest that this TSC signaling node functions as a cellular sensor for ROS to regulate mtorc1 and autophagy. The TSC tumor suppressor localizes to peroxisome In rat liver (FAO) cells, endogenous TSC1, TSC2 and Rheb localized to discrete vesicular structures, the majority of which co-localized with the peroxisomal marker, peroxisomal membrane protein 70 (PMP70) (Fig. 1a). Quantitative analysis of these images revealed a correlation between TSC and PMP70 colocalization when compared with the lysosomal marker LAMP1 (lysosome associated membrane protein=lamp1) (Fig. 1b). As expected, TSC1 and TSC2 also co-localized with each other in discrete vesicular structures (Fig. 1c). Endogenous TSC2 colocalization with Rheb (which also co-localized with PMP70) was also observed (Fig. 1a, bottom panels). Controls demonstrating the absence of co-localization with markers for other cellular organelles including mitochondria (mitochondrial cox 2=MTCO2) and endosomes (early endosome antigen 1=EEA1) (Supplementary Fig. S1a). Specificity for the antibody used to detect endogenous TSC2 was confirmed using sirna knockdown of TSC2 (Fig. 1d f), and TSC2-deficient mouse embryonic fibroblasts (MEFs) (Supplementary Fig. S1b). Cell fractionation confirmed the TSC signaling node (endogenous TSC1, TSC2 and Rheb) in the peroxisome fraction, which contained negligible amounts of other organelles (Fig. 2a,b). Loss of TSC2 (but not TSC1 or Rheb) from the peroxisome fraction was observed in TSC2 / MEFs (Fig. 2c). TBC1D7 (a third subunit of the TSC1-TSC2 complex 23 ) was also found in the peroxisome fraction (Fig. 2a and Supplementary Fig. S1c), whereas AKT was detected in cytosolic (primarily) compartments (Fig. 2a c and Supplementary Fig. S1c,d).
4 Zhang et al. Page 3 Endogenous TSC2 was detected in both the membrane and cytosolic fractions (Fig. 2a,b and Supplementary Fig. S1c,d), consistent with what is known about membrane localization of active, and cytosolic sequestration of inactive, forms of this tumor suppressor, respectively Peroxisome localization of the TSC signaling node was also observed in HEK 293 (Supplementary Fig. S1c) and HeLa (Supplementary Fig. S1d), confirming the generalizability to other cell types. The TSC tumor suppressor is on the cytosolic surface of the peroxisome TSC2 tumor suppressor activity occurs via membrane dissociation and cytosolic binding of this tumor suppressor by proteins, sequestering it away from its activation partner TSC1 and its GAP target Rheb 24. In MCF-7 cells expressing constitutively active myristoylated AKT (myr-akt), AKT phosphorylation inactivates TSC2, with loss of TSC2 from the membrane and peroxisomal fractions and increased TSC2 in the cytosol (Fig. 3a). As predicted, TSC2 was released from peroxisome and membrane compartments into the cytosol with mitogenic stimulation by insulin (Fig. 3b). Furthermore, TSC2 phosphorylated by AKT at S939 was abundant in the cytosolic, but absent or barely detectable in the peroxisomal fraction of cells (Fig. 3c). Correlative data that TSC2 resided at the peroxisomal membrane were supported with direct evidence from a protease protection assay, where peroxisomes were proteinase K treated in the absence or presence of membrane disrupting detergent (Triton X-100). The peroxisomal membrane protein PMP70 and TSC1, TSC2 and Rheb were degraded in both the absence and presence of detergent, indicating that they were membrane-associated (Fig. 3d), while catalase, a matrix protein control was resistant to proteinase K, confirming the TSC signaling node resides in peroxisomal membranes (Fig. 3d). The TSC signaling node functions at the peroxisome to induce autophagy in response to ROS As a major site of ROS generation, we hypothesized TSC at the peroxisome might induce autophagy, which is regulated by both mtorc1 6 8, 11 and ROS 27, 28. Consistent with this hypothesis, overexpression of both Flag-TSC1 and TSC2 increased autophagosome formation relative to overexpression of either Flag-TSC1 or Flag-TSC2 alone (Fig. 4a,b). In these cells, exogenous ROS (H 2 O 2 ), induced rapid (<1 hr) suppression of mtorc1, decreased p62 by 24 hrs, and increased ratio of LC3 II/Actin (Supplementary Fig. S2a,b). The decrease in p62 was significantly inhibited by Bafliomycin A1 (Baf A1), as was the increase in LC3 II (Supplementary Fig. S2c,d). Similarly, as shown in Supplementary Fig. S2e, decreased p62 in response to H 2 O 2 occurred only in autophagy-proficient (Atg5 +/+ MEFs) but not in autophagy-deficient cells (i.e. Atg5 / MEFs 29 ). Finally, increased autophagic flux in response to H 2 O 2 was also demonstrated by accelerated degradation of GFP-LC3 relative to RFP-LC3 9, 10, which is less stable in the acidic ph of the autolysosomes (Fig. 4c,d), confirming the ability of TSC to response to ROS to regulate autophagy. Drugs such as fibrates (e.g., Wy-14643), activate peroxisome proliferator-activated receptor alpha (PPAR-alpha), to induce peroxisome proliferation in rodent liver cells 30, 31, and increase levels of peroxisomal ROS 20. FAO cells were treated with Wy-14643, which induced the expected increase in PPAR-alpha-inducible proteins peroxisomal bifunctional enzyme (EHHADH) and peroxisomal thiolase (ACAA1) (Fig. 4e), and increased superoxide radical production shown by dihydroethidium (DHE) staining (Fig. 4f). Fibrate-induced ROS suppressed mtorc1 (Fig. 4e), to induce autophagy, as confirmed by increased LC3 puncta and electron microscopy (Fig. 4g i).
5 Zhang et al. Page 4 Peroxisome biogenesis disorders (PBDs) are caused by genetic defects in key proteins that participate in this process 21, 32, with Zellweger syndrome being the most severe 32. PBDassociated disease mutations result in an absence of functional peroxisomes. Co-localization with PMP70 for TSC2 and Rheb was lost in peroxisome-deficient human Zellweger cells (Fig. 5a). These cells are deficient in PEX5-mediated protein import but can still assemble peroxisomal membrane proteins, accounting for the occasional PMP-positive puncta, which interestingly also show colocalization with TSC1 (Fig. 5a). Peroxisome-deficient Zellweger (untransformed GM13267) cells were deficient in mtorc1 repression in response to H 2 O 2 relative to control peroxisome-proficient (untransformed GM15871) cells (Fig. 5b). H 2 O 2 activated ATM, AMPK and TSC2 to repress mtorc1 and induce autophagy in control fibroblasts, whereas Zellweger fibroblasts lacking peroxisomes were resistant to mtorc1 repression by H 2 O 2 (Fig. 5b). These data were confirmed in two other control (GM13427) and Zellweger (GM13269) cell cultures (Supplementary Fig. S3a). As predicted 19, in peroxisome-proficient cells, repression of mtorc1 by H 2 O 2 was rescued with N-acetyl cysteine (NAC) (Fig. 5c), and decreased p62 was inhibited by Baf A1 (Supplementary Fig. S3b,c). In contrast to ROS repression of mtorc1, which is TSC2-dependent, activation of mtorc1 by amino acids does not signal through TSC-Rheb 13, 15. This predicts that Zellweger cells would still activate mtorc1 in response to stimulation with amino acids. As shown in Fig. 5d, mtorc1 signaling was activated by amino acids to an equivalent degree in both peroxisome-deficient Zellweger fibroblasts (GM13267) and control peroxisome-proficient fibroblasts (GM15871), which was confirmed in two other control (GM13427) and Zellweger (GM13269) cell (Supplementary Fig. S3d). TSC1 and TSC2 bind peroxins PEX19 and PEX5 PEX proteins mediate localization of peroxisomal proteins and enzymes. We observed by co-immunoprecipitation of both endogenous and Flag-tagged proteins that TSC2 was bound by PEX5 (Fig. 6a,d). PEX5 is known to import proteins into the peroxisomal matrix 22, whereas TSC2 localizes to endomembranes (our data and reference 33 ), therefore additional experiments were performed to confirm that TSC2 was a bona fide target for the PEX5 import receptor. In silico analysis of the TSC2 [ 34 ] identified a region of homology with known PTS1 sequences. This ARL motif 1739 KWIARLRHIKR 1749 was located 63 amino acids from the C-terminal of TSC2 protein (Fig. 6b). Although the majority of PEX5 targets identified to date contain a PTS1 at their extreme C-terminus 22, internal sequences may mediate some peroxisomal targeting To demonstrate this ARL sequence functioned as a PTS1, we substituted the ARL sequence for an SKL PTS1 motif in a DsRed- SKL fusion protein that localizes to the peroxisome. The DsRed-ARL fusion protein colocalized with a GFP fusion protein containing the SKL PTS1, and peroxisome marker PMP70 and catalase (Fig. 6c), whereas deletion of the inserted ARL sequence in this DsRed fusion protein (DsRed-Del-ARL) resulted in loss of cytoplasmic staining (Fig. 6c, top panel), indicating that a C-terminal ARL motif was capable of targeting proteins to the peroxisome in mammalian cells. Importantly, several pathogenic TSC2 mutations were identified in this ARL site (Table 1). To determine whether TSC2 localization to the peroxisome was impaired by these mutations, we introduced three naturally occurring mutations into a wild-type TSC2 (WT) expression construct at amino acid position 1743, R1743Q (RQ), R1743G (RG) and R1743W (RW) (Table 1 and Fig. 6b). All three TSC2 mutants reduced PEX5 import receptor association with TSC2 (Fig. 6d), identifying this ARL sequence as a PEX5 binding sequence (PxBS) in TSC2. Peroxisomal localization of all three PxBS mutants was greatly
6 Zhang et al. Page 5 diminished (Fig. 6e and Supplementary Fig. S4a); the PxBS TSC2 mutants, co-localized with neither lysosome (Supplementary Fig. S4b), nor endoplasmic reticulum (ER) (Calnexin) nor mitochondria (voltage-dependent anion channel=vdac) markers (Supplementary Fig. S4c). Importantly, introduction of nine nucleotides encoding an exogenous SKL PTS1 into the TSC2-RQ mutant, just before the C-terminal stop codon (TSC2-RQ-9NT) (Fig. 6b) restored PEX5 binding to TSC2-RQ-9NT (Fig. 6f) and peroxisome localization of TSC2-RQ-9NT (Fig. 6g). Consistent with the proposed function of TSC1 as the membrane tether for TSC2 26, 38, endogenous TSC1 co-immunoprecipitated with PEX19, which localizes proteins to peroxisomal membranes, but not PEX5 (Fig. 6a), and we identified a potential PEX19 binding site in TSC1 at the amino terminal ( 129 LTTGVLvLIt 138 ) [ peroxisomedb.org 34 ]. While a very small amount of Flag-TSC2 was associated with the PEX19-TSC1 complex (Fig. 6h), this association was abrogated with the TSC2-G294E mutant (a TSC1 binding-deficient mutant 39 ), indicating the little TSC2 detected in this complex was likely associated with TSC1, rather than directly bound to PEX19 (Fig. 6h). As a control we demonstrated that mtor (another large protein), did not coimmunoprecipitated with either PEX5 or PEX19 (Fig. 6a). Rheb did not contain predicted PTS sequences, however, farnesylation of the C-terminal CaaX (Cysteine aliphatic aliphatic any) motif has been previously shown to localize Rheb to endomembranes and be required for Rheb activation of mtorc1 3. We found that in contrast to WT-Rheb, CaaX defective Rheb was not detected in the peroxisomal fraction of cells (Fig. 6i). TSC functions at the peroxisome to suppress mtorc1 Inactivating PEX5 using shrna to inhibit delivery of TSC2 to the peroxisome increased mtorc1 signaling (Fig. 7a), suggesting that localization to the peroxisome played a role in TSC GAP activity for Rheb and suppression of mtorc1 signaling. Functional assays in cells expressing Flag-tagged WT-TSC2 or PxBS TSC2 mutants (RQ, RW, and RG) cotransfected with Flag-TSC1, myc-rheb and either HA-S6K (Fig. 7b, left panel) or HA-4E- BP1 (Fig. 7b, right panel) showed that while WT-TSC2 suppressed Rheb activation of mtorc1 (evidenced by decreased phosphorylation of S6K1 and 4E-BP1 and [ 32 P]- radiolabeled-s6), all three pathogenic PxBS TSC2 mutants were deficient in mtorc1 repression (Fig. 7b). Inability to suppress mtorc1 signaling was not due to inability to bind TSC1, as control experiments revealed equivalent binding to TSC1 of WT-TSC2 and mutants (RQ, RG, and RW) (Supplementary Fig. S4d). To determine directly whether inability to localize to the peroxisome compromised the Rheb GAP activity of PxBS mutants, we compared their GTPase activity with WT-TSC2, using immunoprecipitated TSC heterodimers against GST-Rheb preloaded with [α- 32 P]-GTP. In this in vitro assay, WT-TSC2 promoted the intrinsic GTPase activity of Rheb as observed by accumulation of [α- 32 P]-GDP bound Rheb, while none of the PxBS mutants functioned as a Rheb GAP, similar to what was observed in control experiments with the TSC2 GAP domain mutant (L1624P) protein (Fig. 7c). This suggests that TSC2 GAP activity depends on the delivery of TSC2 to the peroxisome. However, addition of the SKL PTS1 to the TSC2-RQ mutant restored significant repression of mtorc1 signaling (Fig. 7d,e and Supplementary Fig. S4e). This was confirmed in the in vivo Rheb guanine nucleotide binding assay, which demonstrated that TSC2-RQ-9NT had regained significant Rheb GAP activity (Fig. 7f,g), indicating a significant proportion of loss of activity in this mutant was attributable to loss of peroxisome localization, rather than loss of intrinsic GAP activity. Overexpression of TSC1and TSC2 inhibits axon growth, resulting in an increased proportion of neurons with no axon, while loss of TSC1 and TSC2 results in an aberrant neuronal morphology with multiple axons extending out of the cell body, a phenotype
7 Zhang et al. Page 6 DISCUSSION dependent on hyperactivation of mtorc1 40. Expression of WT-TSC2 resulted in a significant increase in the number of neurons with no axon and a significant decrease in neurons with multiple axons, compared to controls expressing GFP alone (Fig. 7h). In comparison, all three of the PxBS TSC2 mutants when transfected with TSC1 had the opposite effect, significantly reducing the fraction of cells with no axons and increasing the number of cells with multiple axons (Fig. 7h), providing a functional link between loss of TSC signaling at the peroxisome and the neuronal pathophysiology of TSC. Here we describe an intimate and previously unappreciated, relationship between the TSC signaling node and the peroxisome. We found TSC1 and TSC2 bound to PEX19 and PEX5, respectively, and an ARL sequence required for PEX5 binding and localization to peroxisomal membranes was identified. Missense mutations within the TSC2 ARL sequence are pathogenic 41, 42, and TSC2 s ability to suppress mtorc1 in response to ROS is abrogated by these mutations. Neuronal expression of PxBS mutant TSC2 causes the formation of multiple axons, indicating that peroxisomal TSC2 localization is crucial for proper neuronal morphology and polarity, and providing a link between aberrant TSC signaling and brain pathophysiology. This suggests a model (Supplementary Fig. S5), where the TSC tumor suppressor resides on the exterior of peroxisomal membranes, in proximity to its GAP target Rheb, and where it can be negatively regulated by cytosolic kinases such as AKT and binding, and activated by peroxisomal ROS to repress mtorc1 and induce autophagy. PTS1 sequences located at the extreme C-terminus of cargo proteins, interact with the C- terminal region of PEX5 via the seven tetratricopeptide repeat (TPR) domains 22. The ARL sequence in TSC2 that functions as a PEX5-binding domain is 63 amino acids internal from the C-terminus. While an ARL sequence at the C-terminus can function as a PTS1 to target proteins to the peroxisome in yeast 43, plants 44 and as we demonstrated with a DsRed-ARL fusion protein in mammalian cells, for an internal ARL to act as a true PTS1, the protein loop containing this ARL sequence must be accommodated in the well-characterized ringlike structure formed by the PEX5-TPR domains 36. However, an increasing number of PEX5 dependent targets have been discovered where a PTS1 motif is not essential or even absent 36, 37, and internal sequences that mediate PEX5 binding have been proposed 37. For example, yeast Acyl-CoA oxidase (Pox1p) interacts directly with PEX5 but does not contain any recognizable PTS1, and C-terminal deletions of Pox1p do not affect PEX5 interaction 45. Current dogma dictates that PEX5 cargo is delivered to the matrix of the peroxisome. We demonstrated that TSC2 is part of the PEX5 import complex, but is localized to the peroxisomal membrane facing the cytosol. We considered two possibilities to potentially explain this conundrum: 1) TSC2 docks with TSC1 to be retained at the cytosolic membrane, abrogating import by PEX5; 2) TSC2 is delivered into the matrix and then recycles out to the cytosolic membrane. If the TSC2-TSC1 heterodimer is more stable than TSC2-PEX5 complex, peroxisomal TSC1 may function as an attractor or anchor to capture TSC2 at the cytosolic peroxisomal membrane, abrogating PEX5-mediated import of TSC2 into the matrix. Alternatively, the TSC2-PEX5 complex may enter the peroxisome matrix via PEX14 peroxisomal translocation machinery and then recycle out to interact with peroxisomal membrane-localized TSC1. Such peroxisomal protein recycling is well described for PEX5 22, and also occurs with rotavirus VP4 protein 46, 47. Specific localization of TSC and mtor to several subcellular compartments has been reported, including lysosome or endosome 13 18, 23, cytosol 26, 48, 49, Golgi apparatus 50 and nucleus 48, 51. Interesting, Rheb was recently localized to mitochondrial membranes, where it
8 Zhang et al. Page 7 was shown to regulate mitophagy 52. While shuttling between these compartments is likely to occur, TSC1, TSC2 and mtor resident in these various compartments may respond to specific stimuli. For example, mtor is localized to the late endosome or lysosome compartment by the Ragulator complex in response to amino acids 13, 16 18, and the TSC1- TSC2-TBC1D7 Rhebulator complex has been reported to be regulated by growth factors and energy stress in lysosomes 23. Our finding of a TSC signaling node resident in peroxisome that responds to ROS identifies a subcellular compartment in which TSC functions to repress mtorc1 and induce autophagy in response to oxidative stress. Induction of autophagy is one of many cellular responses to oxidative stress, with superoxide (which is produced by peroxisomes) being the major ROS regulating autophagy 27. Cytoplasmic ATM, which responds to ROS to activate AMPK and TSC2 19, has been previously localized to peroxisomes and a putative PTS1 sequence identified at its C-terminus 53. ATM is directly activated by reactive oxygen species (ROS), with the oxidized form of this kinase forming an active di-sulfide cross-linked dimer 54. Localization of ATM and the TSC signaling node to the peroxisome sets up the intriguing possibility that direct activation of ATM by peroxisomal ROS is the proximal signal that engages TSC to repress mtorc1, indicating characterization of this, and elucidation of other signaling cascades resident at the peroxisome, now merit further study. Supplementary Material Acknowledgments References Refer to Web version on PubMed Central for supplementary material. We thank Drs. Gordon Mills and Yiling Lu (University of Texas M.D. Anderson Cancer Center, Houston, TX) for the MCF7 cell line stably expressing GFP-LC3, and RIKEN BRC for providing the ATG5 +/+ MEFs and ATG5 / MEFs. We are also grateful for the assistance of Mr. Kenneth Dunner in electron microscopy image acquisition and analysis and Tia Berry, Xuefei Tong, and Sean Hensley for technical assistance. This work was supported by National Institutes of Health (NIH) Grant R01 CA to C.L.W., NIH R01CA to M.B.K., NIH R01NS058956, John Merck Fund, Children s Hospital Boston Translational Research Program to M.S. A.R.T. was supported by the Association for International Cancer Research Career Development Fellowship [No /915]. 1. Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006; 355: [PubMed: ] 2. Gomez, MR.; Sampson, JR.; Whittemore, VH. Tuberous sclerosis complex. 3. Oxford University Press; New York; Oxford: Aspuria PJ, Tamanoi F. The Rheb family of GTP-binding proteins. Cell Signal. 2004; 16: [PubMed: ] 4. Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008; 412: [PubMed: ] 5. He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009; 43: [PubMed: ] 6. Hosokawa N, et al. Nutrient-dependent mtorc1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009; 20: [PubMed: ] 7. Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mtor signaling to the autophagy machinery. Mol Biol Cell. 2009; 20: [PubMed: ] 8. Kim J, Kundu M, Viollet B, Guan KL. AMPK and mtor regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011; 13: [PubMed: ] 9. Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012; 8: [PubMed: ]
9 Zhang et al. Page Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010; 140: [PubMed: ] 11. Nazio F, et al. mtor inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013; 15: [PubMed: ] 12. Yu L, et al. Termination of autophagy and reformation of lysosomes regulated by mtor. Nature. 2010; 465: [PubMed: ] 13. Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mtorc1. Science. 2008; 320: [PubMed: ] 14. Sengupta S, Peterson TR, Sabatini DM. Regulation of the mtor complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010; 40: [PubMed: ] 15. Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mtor. Nat Rev Mol Cell Biol. 2013; 14: [PubMed: ] 16. Flinn RJ, Yan Y, Goswami S, Parker PJ, Backer JM. The late endosome is essential for mtorc1 signaling. Mol Biol Cell. 2010; 21: [PubMed: ] 17. Korolchuk VI, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol. 2011; 13: [PubMed: ] 18. Sancak Y, et al. Ragulator-Rag complex targets mtorc1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010; 141: [PubMed: ] 19. Alexander A, et al. ATM signals to TSC2 in the cytoplasm to regulate mtorc1 in response to ROS. Proc Natl Acad Sci U S A. 2010; 107: [PubMed: ] 20. Schrader M, Fahimi HD. Peroxisomes and oxidative stress. Biochim Biophys Acta. 2006; 1763: [PubMed: ] 21. Wanders RJ, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem. 2006; 75: [PubMed: ] 22. Ma C, Agrawal G, Subramani S. Peroxisome assembly: matrix and membrane protein biogenesis. J Cell Biol. 2011; 193:7 16. [PubMed: ] 23. Dibble CC, et al. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mtorc1. Mol Cell. 2012; 47: [PubMed: ] 24. Cai SL, et al. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J Cell Biol. 2006; 173: [PubMed: ] 25. Inoki K, Guan KL. Tuberous sclerosis complex, implication from a rare genetic disease to common cancer treatment. Hum Mol Genet. 2009; 18:R [PubMed: ] 26. Plank TL, Yeung RS, Henske EP. Hamartin, the product of the tuberous sclerosis 1 (TSC1) gene, interacts with tuberin and appears to be localized to cytoplasmic vesicles. Cancer Res. 1998; 58: [PubMed: ] 27. Chen Y, Azad MB, Gibson SB. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009; 16: [PubMed: ] 28. Scherz-Shouval R, et al. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. Embo J. 2007; 26: [PubMed: ] 29. Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004; 432: [PubMed: ] 30. Duclos S, Bride J, Ramirez LC, Bournot P. Peroxisome proliferation and beta-oxidation in Fao and MH1C1 rat hepatoma cells, HepG2 human hepatoblastoma cells and cultured human hepatocytes: effect of ciprofibrate. Eur J Cell Biol. 1997; 72: [PubMed: ] 31. Scotto C, Keller JM, Schohn H, Dauca M. Comparative effects of clofibrate on peroxisomal enzymes of human (Hep EBNA2) and rat (FaO) hepatoma cell lines. Eur J Cell Biol. 1995; 66: [PubMed: ] 32. Weller S, Gould SJ, Valle D. Peroxisome biogenesis disorders. Annu Rev Genomics Hum Genet. 2003; 4: [PubMed: ] 33. Harrington LS, Findlay GM, Lamb RF. Restraining PI3K: mtor signalling goes back to the membrane. Trends Biochem Sci. 2005; 30: [PubMed: ]
10 Zhang et al. Page Schluter A, Real-Chicharro A, Gabaldon T, Sanchez-Jimenez F, Pujol A. PeroxisomeDB 2.0: an integrative view of the global peroxisomal metabolome. Nucleic Acids Res. 2010; 38:D [PubMed: ] 35. Freitas MO, et al. PEX5 protein binds monomeric catalase blocking its tetramerization and releases it upon binding the N-terminal domain of PEX14. J Biol Chem. 2011; 286: [PubMed: ] 36. Stanley WA, Wilmanns M. Dynamic architecture of the peroxisomal import receptor Pex5p. Biochim Biophys Acta. 2006; 1763: [PubMed: ] 37. van der Klei IJ, Veenhuis M. PTS1-independent sorting of peroxisomal matrix proteins by Pex5p. Biochim Biophys Acta. 2006; 1763: [PubMed: ] 38. Yamamoto Y, Jones KA, Mak BC, Muehlenbachs A, Yeung RS. Multicompartmental distribution of the tuberous sclerosis gene products, hamartin and tuberin. Arch Biochem Biophys. 2002; 404: [PubMed: ] 39. Hodges AK, et al. Pathological mutations in TSC1 and TSC2 disrupt the interaction between hamartin and tuberin. Hum Mol Genet. 2001; 10: [PubMed: ] 40. Choi YJ, et al. Tuberous sclerosis complex proteins control axon formation. Genes Dev. 2008; 22: [PubMed: ] 41. Coevoets R, et al. A reliable cell-based assay for testing unclassified TSC2 gene variants. Eur J Hum Genet. 2009; 17: [PubMed: ] 42. Jones AC, et al. Comprehensive mutation analysis of TSC1 and TSC2-and phenotypic correlations in 150 families with tuberous sclerosis. Am J Hum Genet. 1999; 64: [PubMed: ] 43. Szewczyk E, Andrianopoulos A, Davis MA, Hynes MJ. A single gene produces mitochondrial, cytoplasmic, and peroxisomal NADP-dependent isocitrate dehydrogenase in Aspergillus nidulans. J Biol Chem. 2001; 276: [PubMed: ] 44. Tilbrook K, Gnanasambandam A, Schenk PM, Brumbley SM. Efficient targeting of polyhydroxybutyrate biosynthetic enzymes to plant peroxisomes requires more than three amino acids in the carboxyl-terminal signal. J Plant Physiol. 2010; 167: [PubMed: ] 45. Klein AT, van den Berg M, Bottger G, Tabak HF, Distel B. Saccharomyces cerevisiae acyl-coa oxidase follows a novel, non-pts1, import pathway into peroxisomes that is dependent on Pex5p. J Biol Chem. 2002; 277: [PubMed: ] 46. Lazarow PB. Viruses exploiting peroxisomes. Curr Opin Microbiol. 2011; 14: [PubMed: ] 47. Mohan KV, Som I, Atreya CD. Identification of a type 1 peroxisomal targeting signal in a viral protein and demonstration of its targeting to the organelle. J Virol. 2002; 76: [PubMed: ] 48. Rosner M, Freilinger A, Hengstschlager M. Akt regulates nuclear/cytoplasmic localization of tuberin. Oncogene. 2007; 26: [PubMed: ] 49. van Slegtenhorst M, et al. Interaction between hamartin and tuberin, the TSC1 and TSC2 gene products. Hum Mol Genet. 1998; 7: [PubMed: ] 50. Wienecke R, et al. Co-localization of the TSC2 product tuberin with its target Rap1 in the Golgi apparatus. Oncogene. 1996; 13: [PubMed: ] 51. Zhang X, Shu L, Hosoi H, Murti KG, Houghton PJ. Predominant nuclear localization of mammalian target of rapamycin in normal and malignant cells in culture. J Biol Chem. 2002; 277: [PubMed: ] 52. Melser S, et al. Rheb regulates mitophagy induced by mitochondrial energetic status. Cell Metab. 2013; 17: [PubMed: ] 53. Watters D, et al. Localization of a portion of extranuclear ATM to peroxisomes. J Biol Chem. 1999; 274: [PubMed: ] 54. Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010; 330: [PubMed: ]
11 Zhang et al. Page 10 Figure 1. TSC1 and TSC2 localization to peroxisomes. (a) Representative images of FAO cells immunostained with TSC2, TSC1 or Rheb (green) and PMP70 (peroxisome marker) or LAMP1 (lysosome marker) (red) antibodies. (Scale bar - 10μm). (b) Pearson s Correlation Coefficient for TSC1 or TSC2 co-localization with PMP70 or LAMP1 calculated using Imaris software. Quantification was performed on 8 12 cells from each of the 4 independent experiment giving rise to a total of 40 cells. All error bars represent s.e.m., *** p < (c) Representative images using HeLa cells transfected with Myc-TSC1 and Flag-TSC2 wild type (WT) and stained with anti-flag (green) and anti-myc (red) antobodies. (Scale bar - 10μm). (d) Representative images of HepG2 cells transfected with control (sicontrol) or TSC2 (sitsc2) sirna immunostained for TSC2 (green) and PMP70 (red). Individual boundaries were shown to identify cells with TSC2 knockdown (white) versus cells that retain TSC2 (yellow). (Scale bar - 10μm). (e) Corresponding immunoblots for HepG2 cells transfected with control (sicontrol) or TSC2 (sitsc2) sirna showing extent of knockdown (average over population of cells). (f) HepG2 cells from Fig. 1d were analyzed for Pearson s Correlation Coefficient of TSC2 co-localization with PMP70 using Imaris software.
12 Zhang et al. Page 11 Quantification was performed on 8 12 cells from each of the 4 independent experiment giving rise to a total of 40 cells. All error bars represent s.e.m., *** p < Uncropped images of western blots are shown in Supplementary Fig. S6. Source data of statistical analysis are shown in Supplementary Table S1.
13 Zhang et al. Page 12 Figure 2. Cell fractionation demonstrating the TSC signaling node at the peroxisome. (a and b) Subcellular fractionation of FAO (a) or HepG2 (b) cells demonstrated the localization of TSC2, TSC1, Rheb, TBC1D7 (FAO, Fig. 2a) and AKT in various subcellular compartments. Catalase, PMP70 and Lamin A/C were used as subcellular markers for the peroxisome (P) and nuclear (N) fractions, respectively. EEA1, LAMP1 and VDAC were used as markers for endosomes, lysosomes, and mitochondria, respectively. WCE whole cell extracts, M- membrane, C-cytosol. (c) Subcellular fractionation of TSC2 +/+ and TSC2 / MEFs demonstrating the localization of TSC2, TSC1, Rheb and AKT. PMP70, catalase (peroxisome fraction - P), LDH (cytosolic fraction - C), β-integrin (membrane fraction - M) and lamin A/C (nuclear fraction - N) were used as subcellular markers. WCE whole cell extracts. Uncropped images of western blots are shown in Supplementary Fig. S6.
14 Zhang et al. Page 13 Figure 3. Active TSC signaling node resident at peroxisome membrane. (a) Western analysis of whole cell extracts (WCE), membrane (M), cytosolic (C) and peroxisome (P) fractions from parental and MCF-7 cells stably expressing constitutively active myristoylated AKT (myr- AKT) immunoblotted for TSC2, LDH, catalase and PMP70. Light and dark represent short and long autoradiographic exposures, respectively. (b) Western analysis of whole cell extracts (WCE), membrane (M), cytosolic (C) and peroxisome (P) fractions from MCF-7 cells stimulated with insulin (200 nm) for 30 min after 1h of serum starvation and immunoblotted for TSC2, LDH, catalase and PMP70. Light and dark represent short and long autoradiographic exposures, respectively. (c) Representative western analysis of HEK 293 whole cell extracts (WCE), cytosolic (C), and peroxisome (P) fractions, immunoblotted for phospho-tsc2 (S939) (inactive), TSC2, and phospho-akt (S473 and T308) (activated). LDH and catalase were used as markers for the cytosolic (C) and peroxisome (P) fractions, respectively. (d) Proteinase K protection assays performed in the presence or absence of Triton X-100 to disrupt peroxisomess on equal masses of peroxisome (P) fractions (HEK 293 cells) collected at the indicated time points. The lysates were immunoblotted for TSC2, TSC1, Rheb, catalase and PMP70. WCE whole cell extract. Uncropped images of western blots are shown in Supplementary Fig. S6.
15 Zhang et al. Page 14 Figure 4. The TSC signaling node at the peroxisome induces autophagy in response to ROS. (a) Representative merged images using GFP-LC3 MCF-7 cells expressing Flag-TSC1, Flag TSC2 or both Flag-TSC1 and Flag-TSC2 showing GFP-LC3 (green) puncta. (Scale bar - 10μm). (b) Quantification of GFP-LC3 puncta was performed and the results are represented as the average puncta fluorescence per cell (±s.e.m., n = 3 independent experiments) from 100 cells per experiment as shown in Fig. 4a. *** p < 0.001, NS, not significant. (c) Representative immunocytochemistry images using MCF-7 cells transfected with an mrfp- GFP-LC3 construct. Cells treated with 0.4 mm H 2 O 2 were analyzed at 0hr (Control), 1hr, 3hr and 6hr. (Scale bar - 10μm). (d) Quantification of autophagosomes (AP, GFP-LC3) and autolysosomes (AL, RFP-LC3) per cell in different conditions as shown in Fig. 4c. The results are represented as the average puncta fluorescence per cell (±s.e.m., n = 3 independent experiments) from 100 cells per experiment. ** p < 0.01, *** p < (e) Western analysis of FAO cells treated with 50 μm WY (WY) or vehicle (DMSO) for PPAR-alpha-inducible proteins (EHHADH and ACAA1), mtorc1 signaling proteins [(ps6 (S235/236), S6, ps6k (T389) and S6K)] and autophagy markers (LC3 and p62). (f)
16 Zhang et al. Page 15 Representative images of FAO cells treated with vehicle (DMSO) or 50 μm WY (WY) for 1hr, with superoxide production detected using dihydroethidium (DHE). (Scale bar - 30μm). (g) Representative images of LC3 puncta (green) in FAO cells following treatment of 50 μm WY (WY) or vehicle (DMSO) for indicated time period. (Scale bar - 10μm). (h) Quantification of LC3 puncta per cell in response to WY (WY) or vehicle (DMSO). The results are represented as the average LC3 puncta fluorescence per cell (±s.e.m., n = 3 independent experiments) from 100 cells per experiment as shown in Fig. 4a. * p < 0.05, ** p < (i) Transmission electron microscopy of FAO cells treated with vehicle (DMSO) or 50 μm WY (WY) for 24, 48 and 72 hr. Peroxisomes and autophagosomes are indicated with red and yellow arrows, respectively. (Scale bar - 500nm). Uncropped images of western blots are shown in Supplementary Fig. S6. Source data of statistical analysis are shown in Supplementary Table S1.
17 Zhang et al. Page 16 Figure 5. TSC s ability to suppress mtorc1 is abrogated in peroxisome-deficient Zellweger cells. (a) Representative images of Zellweger cells (GM13267) showing endogenous TSC2, TSC1 and Rheb (green) co-localization with PMP70 (red). (Scale bar - 15μm). (b) Western analysis of human fibroblasts obtained from Zellweger (GM13267) or corresponding control patient with [Ehlers-Danlos syndrome (GM15871)] treated with indicated doses of H 2 O 2 for 1hr. mtorc1 signaling was assessed by western analysis for ps6k (T389), S6K, ps6 (S235/236), S6, p4ebp1 (T37/46), 4EBP1, patm (S1981), ATM, pampk (T172), AMPK, p62 and LC3. (c) Western analysis of human fibroblast (GM13427) cells pre-incubated with 3 mm NAC (ROS scavenger) for 1hr before treated with 0.4 mm H 2 O 2 for 1hr using antips6k (T389), S6K, p62 and LC3 antibodies. (d) Representative western analysis using cell extracts from human fibroblasts obtained from a Zellweger patient (GM13267) or control
18 Zhang et al. Page 17 fibroblasts (GM15871) treated with amino acid free media for 60 min, and stimulated with mixture of amino acid for 10 min. mtor signaling was monitored using anti-ps6k (T389), S6K, ps6 (S235/236), S6, p4ebp1(t37/46), and 4EBP1 antibodies. Uncropped images of western blots are shown in Supplementary Fig. S6.
19 Zhang et al. Page 18 Figure 6. TSC1 and TSC2 interact with PEX19 and PEX5. (a) Immunoprecipitation was performed with anti-pex5 (left panel), anti-pex19 (right panel) in HEK 293 cells and immunoblotted for endogenous TSC1, TSC2, mtor (negative control) and PMP70, and PEX1 (PEX19 cargo) or catalase (PEX5 cargo). (b) Schematic of TSC2 C-terminus (aa1517 aa1807) showing the GAP and PEX5 binding sequence (PxBS), naturally occurring mutations and the re-introduced SKL sequence. (c) Representative images of FAO cells transfected with DsRed-Del-ARL (deleted ARL sequence) (red) and GFP-PTS1 (green) (top panel). FAO cells were also transfected with DsRed-ARL (red) and stained for PMP70 or catalase (green) as indicated. (Scale bar - 10μm). (d) HEK 293 cells expressing Flag-TSC2 wild type (WT) or Flag-TSC2 mutants (RQ, RW, and RG) were co-immunoprecipitated using anti-flag or anti-pex5, and blotted for PEX5 and Flag-TSC2. (e) Representative experiment showing subcellular fractionation of HEK 293 cells overexpressing Flag-TSC2 wild type (WT) and
20 Zhang et al. Page 19 Flag-TSC2 mutants (RQ, RW and RG). Lysates from peroxisome (P) or membrane (M) fractions and whole cell extracts (WCE) were immunoblotted with Flag or PMP70 antibodies. (f) Co-immunoprecipitation of HEK 293 cells overexpressing Flag-TSC2 wild type (WT), or Flag-TSC2 mutants (RQ) or Flag-TSC2 rescue mutant (RQ-9NT) using anti- Flag antibody or control IgG and immunoblotted with anti-flag and anti-pex5 antibodies. (g) Subcellular fractionation of TSC2 / MEFs transfected with Flag-TSC2 wild type (WT), or Flag-TSC2 mutants (RQ or RQ-9NT). β-integrin and catalase were used as subcellular markers for membrane (M) and peroxisome (P) fractions, respectively. WCE whole cell extract. The arrow indicates the position of overexpressed Flag-TSC2. (h) HEK 293 cells co-transfected with Flag-TSC1 and Flag-TSC2 wild type (WT) or Flag-TSC2 G294E mutant (TSC2 mutant that cannot bind TSC1). Lysates were immunoprecipitated using anti-pex19 and blotted for PEX19 and Flag. (i) Representative blots from subcellular fractionation of HEK 293 cells overexpressing wild type (WT) or mutant (CaaX mutant) Flag-Rheb. LDH, Lamin A/C and catalase were used as subcellular markers for the cytoplasmic (Cp), nuclear (N), and peroxisome (P) fractions, respectively. WCE whole cell extracts. Uncropped images of western blots are shown in Supplementary Fig. S6.
21 Zhang et al. Page 20 Figure 7. TSC2 functions at the peroxisome to repress mtorc1. (a) HEK 293 cells were transfected with either control shrna or a PEX5 targeting shrna, and lysates were blotted using ps6k (T389), S6K, ps6 (S235/236) and S6 antibodies. (b) TSC2 functional assay was performed in HEK 293 cells co-expressing myc-rheb, Flag-TSC1 and Flag-TSC2 wild type (WT), or Flag-TSC2 mutants (RG, RQ and RW) and HA-S6K (left panel) or HA-4EBP1 (right panel). Cells transfected with empty vector (EV) with or without myc-rheb were used as controls. Lysates were further analyzed for phospho-s6k (T389), S6K, phospho-4ebp1 (S65), 4EBP1 and 32 P-incorporation into S6. (c) Rheb GTPase activity assays was performed by co-immunoprecipitating TSC heterodimers from HEK 293 cells expressing Flag-TSC1 and Flag-TSC2 wild type (WT), Flag-TSC2 mutants (RG, RQ and RW), or Flag-TSC2 GAPmutant (L1624P) performed for the indicated time. (d) TSC2 functional assay was
22 Zhang et al. Page 21 performed using TSC2 / MEFs co-transfected with Flag-TSC1, HA-S6K, and Flag-TSC2 wild type (WT) or Flag-TSC2 mutants (RQ or RQ-9NT), with mock transfected cells as controls. Arrows denote the positions of Flag-TSC1 and Flag-TSC2. (e) Quantitation of the ratio of phospho-s6k to total HA-S6K from Fig 7d. (±s.e.m., n = 3 independent experiments). *p < 0.05, **p < (f) In vivo guanine nucleotide loading assays of Rheb was measured using HEK 293 cells overexpressing Flag-TSC1 and Flag-TSC2 wild type (WT), or Flag-TSC2 mutants (RQ and RQ-9NT) with or without Rheb as indicated. (g) Graph shows quantitation of the percentage of Rheb bound to GTP (indicative of Rheb GTPase activity, ±s.e.m., n = 4 independent experiments). *p < 0.05, ***p < (h) Quantification of axon number in hippocampal neurons co-transfected with GFP, Flag-TSC1 and Flag-TSC2 WT or Flag-TSC2 mutants (RG, RQ, and RW). No axons = yellow, one axon = green, and multiple axons = red. Quantification was performed on neurons from each of the 3 independent experiment and the results are represented as polarity (%). All error bars represent s.e.m., *p < 0.05, ** p < 0.01 compared to GFP only transfected control. Uncropped images of western blots are shown in Supplementary Fig. S6. Source data of statistical analysis are shown in Supplementary Table S1.
23 Zhang et al. Page 22 Table 1 Disease causing mutations of TSC2 PEX5 binding sequence (PxBS) disrupts TSC2 s peroxisome localization Exon/Intron Mutation # Reported Comments Exon c>g, 1743 R>G 1 Probably pathogenic Exon c>t, 1743 R>W 28 Pathogenic Exon g>a, 1743 R>Q 18 Pathogenic Exon g>c, 1743 R>P 1 Pathogenic Exon g>t, 1743 R>L 1 Probably pathogenic Based on (
SUPPLEMENTARY INFORMATION
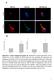
 DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
PREPARED FOR: U.S. Army Medical Research and Materiel Command Fort Detrick, Maryland
 AD Award Number: W81XWH-09-1-0279 TITLE: Regulation of mtor by Nutrients PRINCIPAL INVESTIGATOR: Kun-Liang Guan CONTRACTING ORGANIZATION: University of San Diego La Jolla, CA 92093 REPORT DATE: July 2010
AD Award Number: W81XWH-09-1-0279 TITLE: Regulation of mtor by Nutrients PRINCIPAL INVESTIGATOR: Kun-Liang Guan CONTRACTING ORGANIZATION: University of San Diego La Jolla, CA 92093 REPORT DATE: July 2010
THE ROLE OF ALTERED CALCIUM AND mtor SIGNALING IN THE PATHOGENESIS OF CYSTINOSIS
 Research Foundation, 18 month progress report THE ROLE OF ALTERED CALCIUM AND mtor SIGNALING IN THE PATHOGENESIS OF CYSTINOSIS Ekaterina Ivanova, doctoral student Elena Levtchenko, MD, PhD, PI Antonella
Research Foundation, 18 month progress report THE ROLE OF ALTERED CALCIUM AND mtor SIGNALING IN THE PATHOGENESIS OF CYSTINOSIS Ekaterina Ivanova, doctoral student Elena Levtchenko, MD, PhD, PI Antonella
TITLE: A Genetic Approach to Define the Importance of Rheb in Tuberous Sclerosis
 AD Award Number: W81XWH-05-1-0164 TITLE: A Genetic Approach to Define the Importance of Rheb in Tuberous Sclerosis PRINCIPAL INVESTIGATOR: Fuyuhiko Tamanoi, Ph.D. CONTRACTING ORGANIZATION: The University
AD Award Number: W81XWH-05-1-0164 TITLE: A Genetic Approach to Define the Importance of Rheb in Tuberous Sclerosis PRINCIPAL INVESTIGATOR: Fuyuhiko Tamanoi, Ph.D. CONTRACTING ORGANIZATION: The University
The clathrin adaptor Numb regulates intestinal cholesterol. absorption through dynamic interaction with NPC1L1
 The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
SUPPLEMENTARY INFORMATION
 doi:.38/nature234 Note 1. LINC00961 encodes a lysosomal Type I transmembrane polypeptide In order to determine if the polypeptide encoded by LINC00961 was an integral membrane protein or simply associated
doi:.38/nature234 Note 1. LINC00961 encodes a lysosomal Type I transmembrane polypeptide In order to determine if the polypeptide encoded by LINC00961 was an integral membrane protein or simply associated
Expanded View Figures
 PEX13 functions in selective autophagy Ming Y Lee et al Expanded View Figures Figure EV1. PEX13 is required for Sindbis virophagy. A, B Quantification of mcherry-capsid puncta per cell (A) and GFP-LC3
PEX13 functions in selective autophagy Ming Y Lee et al Expanded View Figures Figure EV1. PEX13 is required for Sindbis virophagy. A, B Quantification of mcherry-capsid puncta per cell (A) and GFP-LC3
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
A particular set of insults induces apoptosis (part 1), which, if inhibited, can switch to autophagy. At least in some cellular settings, autophagy se
 A particular set of insults induces apoptosis (part 1), which, if inhibited, can switch to autophagy. At least in some cellular settings, autophagy serves as a defence mechanism that prevents or retards
A particular set of insults induces apoptosis (part 1), which, if inhibited, can switch to autophagy. At least in some cellular settings, autophagy serves as a defence mechanism that prevents or retards
6. TNF-α regulates oxidative stress, mitochondrial function and autophagy in neuronal cells
 6. TNF-α regulates oxidative stress, mitochondrial function and autophagy in neuronal cells 6.1 TNF-α induces mitochondrial oxidative stress in SH-SY5Y cells. The dysregulation of mitochondria and oxidative
6. TNF-α regulates oxidative stress, mitochondrial function and autophagy in neuronal cells 6.1 TNF-α induces mitochondrial oxidative stress in SH-SY5Y cells. The dysregulation of mitochondria and oxidative
nature methods Organelle-specific, rapid induction of molecular activities and membrane tethering
 nature methods Organelle-specific, rapid induction of molecular activities and membrane tethering Toru Komatsu, Igor Kukelyansky, J Michael McCaffery, Tasuku Ueno, Lidenys C Varela & Takanari Inoue Supplementary
nature methods Organelle-specific, rapid induction of molecular activities and membrane tethering Toru Komatsu, Igor Kukelyansky, J Michael McCaffery, Tasuku Ueno, Lidenys C Varela & Takanari Inoue Supplementary
(a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable
 Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
MOLECULAR CELL BIOLOGY
 1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
A. Generation and characterization of Ras-expressing autophagycompetent
 Supplemental Material Supplemental Figure Legends Fig. S1 A. Generation and characterization of Ras-expressing autophagycompetent and -deficient cell lines. HA-tagged H-ras V12 was stably expressed in
Supplemental Material Supplemental Figure Legends Fig. S1 A. Generation and characterization of Ras-expressing autophagycompetent and -deficient cell lines. HA-tagged H-ras V12 was stably expressed in
IP: anti-gfp VPS29-GFP. IP: anti-vps26. IP: anti-gfp - + +
 FAM21 Strump. WASH1 IP: anti- 1 2 3 4 5 6 FAM21 Strump. FKBP IP: anti-gfp VPS29- GFP GFP-FAM21 tail H H/P P H H/P P c FAM21 FKBP Strump. VPS29-GFP IP: anti-gfp 1 2 3 FKBP VPS VPS VPS VPS29 1 = VPS29-GFP
FAM21 Strump. WASH1 IP: anti- 1 2 3 4 5 6 FAM21 Strump. FKBP IP: anti-gfp VPS29- GFP GFP-FAM21 tail H H/P P H H/P P c FAM21 FKBP Strump. VPS29-GFP IP: anti-gfp 1 2 3 FKBP VPS VPS VPS VPS29 1 = VPS29-GFP
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
SUPPLEMENTARY FIGURE LEGENDS
 SUPPLEMENTARY FIGURE LEGENDS Supplemental FIG. 1. Localization of myosin Vb in cultured neurons varies with maturation stage. A and B, localization of myosin Vb in cultured hippocampal neurons. A, in DIV
SUPPLEMENTARY FIGURE LEGENDS Supplemental FIG. 1. Localization of myosin Vb in cultured neurons varies with maturation stage. A and B, localization of myosin Vb in cultured hippocampal neurons. A, in DIV
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous
 Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Supplementary Materials
 Supplementary Materials Supplementary Figure S1 Regulation of Ubl4A stability by its assembly partner A, The translation rate of Ubl4A is not affected in the absence of Bag6. Control, Bag6 and Ubl4A CRISPR
Supplementary Materials Supplementary Figure S1 Regulation of Ubl4A stability by its assembly partner A, The translation rate of Ubl4A is not affected in the absence of Bag6. Control, Bag6 and Ubl4A CRISPR
293T cells were transfected with indicated expression vectors and the whole-cell extracts were subjected
 SUPPLEMENTARY INFORMATION Supplementary Figure 1. Formation of a complex between Slo1 and CRL4A CRBN E3 ligase. (a) HEK 293T cells were transfected with indicated expression vectors and the whole-cell
SUPPLEMENTARY INFORMATION Supplementary Figure 1. Formation of a complex between Slo1 and CRL4A CRBN E3 ligase. (a) HEK 293T cells were transfected with indicated expression vectors and the whole-cell
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Chairoungdua et al., http://www.jcb.org/cgi/content/full/jcb.201002049/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Expression of CD9 and CD82 inhibits Wnt/ -catenin
Supplemental material Chairoungdua et al., http://www.jcb.org/cgi/content/full/jcb.201002049/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Expression of CD9 and CD82 inhibits Wnt/ -catenin
HHS Public Access Author manuscript Nat Cell Biol. Author manuscript; available in PMC 2011 September 01.
 Inactivation of Rheb by PRAK-mediated phosphorylation is essential for energy depletion-induced suppression of mtorc1 Min Zheng 1, Yan-Hai Wang 1, Xiao-Nan Wu 1, Su-Qin Wu 1, Bao-Ju Lu 2, Meng-Qiu Dong
Inactivation of Rheb by PRAK-mediated phosphorylation is essential for energy depletion-induced suppression of mtorc1 Min Zheng 1, Yan-Hai Wang 1, Xiao-Nan Wu 1, Su-Qin Wu 1, Bao-Ju Lu 2, Meng-Qiu Dong
Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified
 Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified for RFP-LC3 puncta (red dots) representing both autolysosomes
Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified for RFP-LC3 puncta (red dots) representing both autolysosomes
RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh-
 1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
LPS LPS P6 - + Supplementary Fig. 1.
 P6 LPS - - - + + + - LPS + + - - P6 + Supplementary Fig. 1. Pharmacological inhibition of the JAK/STAT blocks LPS-induced HMGB1 nuclear translocation. RAW 267.4 cells were stimulated with LPS in the absence
P6 LPS - - - + + + - LPS + + - - P6 + Supplementary Fig. 1. Pharmacological inhibition of the JAK/STAT blocks LPS-induced HMGB1 nuclear translocation. RAW 267.4 cells were stimulated with LPS in the absence
Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were
 Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were isolated from wild type (PKC-θ- WT) or PKC-θ null (PKC-θ-KO)
Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were isolated from wild type (PKC-θ- WT) or PKC-θ null (PKC-θ-KO)
RAS Genes. The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes.
 ۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
۱ RAS Genes The ras superfamily of genes encodes small GTP binding proteins that are responsible for the regulation of many cellular processes. Oncogenic ras genes in human cells include H ras, N ras,
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Chapter 3. Expression of α5-megfp in Mouse Cortical Neurons. on the β subunit. Signal sequences in the M3-M4 loop of β nachrs bind protein factors to
 22 Chapter 3 Expression of α5-megfp in Mouse Cortical Neurons Subcellular localization of the neuronal nachr subtypes α4β2 and α4β4 depends on the β subunit. Signal sequences in the M3-M4 loop of β nachrs
22 Chapter 3 Expression of α5-megfp in Mouse Cortical Neurons Subcellular localization of the neuronal nachr subtypes α4β2 and α4β4 depends on the β subunit. Signal sequences in the M3-M4 loop of β nachrs
Trehalose, sucrose and raffinose are novel activators of autophagy in human. keratinocytes through an mtor-independent pathway
 Title page Trehalose, sucrose and raffinose are novel activators of autophagy in human keratinocytes through an mtor-independent pathway Xu Chen 1*, Min Li 1*, Li Li 1, Song Xu 1, Dan Huang 1, Mei Ju 1,
Title page Trehalose, sucrose and raffinose are novel activators of autophagy in human keratinocytes through an mtor-independent pathway Xu Chen 1*, Min Li 1*, Li Li 1, Song Xu 1, Dan Huang 1, Mei Ju 1,
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure S1: Endogenous interaction between RNF2 and H2AX: Whole cell extracts from 293T were subjected to immunoprecipitation with anti-rnf2 or anti-γ-h2ax antibodies
SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure S1: Endogenous interaction between RNF2 and H2AX: Whole cell extracts from 293T were subjected to immunoprecipitation with anti-rnf2 or anti-γ-h2ax antibodies
Tumor suppressor Spred2 interaction with LC3 promotes autophagosome maturation and induces autophagy-dependent cell death
 www.impactjournals.com/oncotarget/ Oncotarget, Supplementary Materials 2016 Tumor suppressor Spred2 interaction with LC3 promotes autophagosome maturation and induces autophagy-dependent cell death Supplementary
www.impactjournals.com/oncotarget/ Oncotarget, Supplementary Materials 2016 Tumor suppressor Spred2 interaction with LC3 promotes autophagosome maturation and induces autophagy-dependent cell death Supplementary
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/2/e1602038/dc1 Supplementary Materials for Mitochondrial metabolic regulation by GRP78 Manoj Prasad, Kevin J. Pawlak, William E. Burak, Elizabeth E. Perry, Brendan
advances.sciencemag.org/cgi/content/full/3/2/e1602038/dc1 Supplementary Materials for Mitochondrial metabolic regulation by GRP78 Manoj Prasad, Kevin J. Pawlak, William E. Burak, Elizabeth E. Perry, Brendan
Rapid parallel measurements of macroautophagy and mitophagy in
 Supplemental Figures Rapid parallel measurements of macroautophagy and mitophagy in mammalian cells using a single fluorescent biosensor Sargsyan A, Cai J, Fandino LB, Labasky ME, Forostyan T, Colosimo
Supplemental Figures Rapid parallel measurements of macroautophagy and mitophagy in mammalian cells using a single fluorescent biosensor Sargsyan A, Cai J, Fandino LB, Labasky ME, Forostyan T, Colosimo
SUPPLEMENTARY INFORMATION
 Supplementary Discussion The cell cycle machinery and the DNA damage response network are highly interconnected and co-regulated in assuring faithful duplication and partition of genetic materials into
Supplementary Discussion The cell cycle machinery and the DNA damage response network are highly interconnected and co-regulated in assuring faithful duplication and partition of genetic materials into
Nature Immunology doi: /ni.3268
 Supplementary Figure 1 Loss of Mst1 and Mst2 increases susceptibility to bacterial sepsis. (a) H&E staining of colon and kidney sections from wild type and Mst1 -/- Mst2 fl/fl Vav-Cre mice. Scale bar,
Supplementary Figure 1 Loss of Mst1 and Mst2 increases susceptibility to bacterial sepsis. (a) H&E staining of colon and kidney sections from wild type and Mst1 -/- Mst2 fl/fl Vav-Cre mice. Scale bar,
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human
 Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
Homework Hanson section MCB Course, Fall 2014
 Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3311 A B TSC2 -/- MEFs C Rapa Hours WCL 0 6 12 24 36 pakt.s473 AKT ps6k S6K CM IGF-1 Recipient WCL - + - + - + pigf-1r IGF-1R pakt ps6 AKT D 1 st SILAC 2 nd SILAC E GAPDH FGF21 ALKPGVIQILGVK
DOI: 10.1038/ncb3311 A B TSC2 -/- MEFs C Rapa Hours WCL 0 6 12 24 36 pakt.s473 AKT ps6k S6K CM IGF-1 Recipient WCL - + - + - + pigf-1r IGF-1R pakt ps6 AKT D 1 st SILAC 2 nd SILAC E GAPDH FGF21 ALKPGVIQILGVK
Cell Quality Control. Peter Takizawa Department of Cell Biology
 Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
The PI3K/AKT axis. Dr. Lucio Crinò Medical Oncology Division Azienda Ospedaliera-Perugia. Introduction
 The PI3K/AKT axis Dr. Lucio Crinò Medical Oncology Division Azienda Ospedaliera-Perugia Introduction Phosphoinositide 3-kinase (PI3K) pathway are a family of lipid kinases discovered in 1980s. They have
The PI3K/AKT axis Dr. Lucio Crinò Medical Oncology Division Azienda Ospedaliera-Perugia Introduction Phosphoinositide 3-kinase (PI3K) pathway are a family of lipid kinases discovered in 1980s. They have
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
doi: /nature10642
 doi:10.1038/nature10642 Supplementary Fig. 1. Citric acid cycle (CAC) metabolism in WT 143B and CYTB 143B cells. a, Proliferation of WT 143B and CYTB 143B cells. Doubling times were 28±1 and 33±2 hrs for
doi:10.1038/nature10642 Supplementary Fig. 1. Citric acid cycle (CAC) metabolism in WT 143B and CYTB 143B cells. a, Proliferation of WT 143B and CYTB 143B cells. Doubling times were 28±1 and 33±2 hrs for
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. Neither the activation nor suppression of the MAPK pathway affects the ASK1/Vif interaction. (a, b) HEK293 cells were cotransfected with plasmids encoding
Supplementary Figure 1 Supplementary Figure 1. Neither the activation nor suppression of the MAPK pathway affects the ASK1/Vif interaction. (a, b) HEK293 cells were cotransfected with plasmids encoding
Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson
 Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson General principles Properties of lysosomes Delivery of enzymes to lysosomes Endocytic uptake clathrin, others Endocytic pathways recycling vs.
Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson General principles Properties of lysosomes Delivery of enzymes to lysosomes Endocytic uptake clathrin, others Endocytic pathways recycling vs.
7.06 Cell Biology EXAM #3 April 24, 2003
 7.06 Spring 2003 Exam 3 Name 1 of 8 7.06 Cell Biology EXAM #3 April 24, 2003 This is an open book exam, and you are allowed access to books and notes. Please write your answers to the questions in the
7.06 Spring 2003 Exam 3 Name 1 of 8 7.06 Cell Biology EXAM #3 April 24, 2003 This is an open book exam, and you are allowed access to books and notes. Please write your answers to the questions in the
Supplementary Information
 Supplementary Information Supplementary Figure 1: cholesterol manipulation alters the positioning of autophagosomes in cells, related to figure 1. (a) HeLa cells were treated for 24h under conditions reducing
Supplementary Information Supplementary Figure 1: cholesterol manipulation alters the positioning of autophagosomes in cells, related to figure 1. (a) HeLa cells were treated for 24h under conditions reducing
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
Supplemental Materials. STK16 regulates actin dynamics to control Golgi organization and cell cycle
 Supplemental Materials STK16 regulates actin dynamics to control Golgi organization and cell cycle Juanjuan Liu 1,2,3, Xingxing Yang 1,3, Binhua Li 1, Junjun Wang 1,2, Wenchao Wang 1, Jing Liu 1, Qingsong
Supplemental Materials STK16 regulates actin dynamics to control Golgi organization and cell cycle Juanjuan Liu 1,2,3, Xingxing Yang 1,3, Binhua Li 1, Junjun Wang 1,2, Wenchao Wang 1, Jing Liu 1, Qingsong
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus
 Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Biol403 MAP kinase signalling
 Biol403 MAP kinase signalling The mitogen activated protein kinase (MAPK) pathway is a signalling cascade activated by a diverse range of effectors. The cascade regulates many cellular activities including
Biol403 MAP kinase signalling The mitogen activated protein kinase (MAPK) pathway is a signalling cascade activated by a diverse range of effectors. The cascade regulates many cellular activities including
Zool 3200: Cell Biology Exam 4 Part II 2/3/15
 Name:Key Trask Zool 3200: Cell Biology Exam 4 Part II 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
Name:Key Trask Zool 3200: Cell Biology Exam 4 Part II 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
Supplementary Fig. 1 V-ATPase depletion induces unique and robust phenotype in Drosophila fat body cells.
 Supplementary Fig. 1 V-ATPase depletion induces unique and robust phenotype in Drosophila fat body cells. a. Schematic of the V-ATPase proton pump macro-complex structure. The V1 complex is composed of
Supplementary Fig. 1 V-ATPase depletion induces unique and robust phenotype in Drosophila fat body cells. a. Schematic of the V-ATPase proton pump macro-complex structure. The V1 complex is composed of
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis
 Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
SUPPLEMENTARY INFORMATION
 Supplementary Table 1. Cell sphingolipids and S1P bound to endogenous TRAF2. Sphingolipid Cell pmol/mg TRAF2 immunoprecipitate pmol/mg Sphingomyelin 4200 ± 250 Not detected Monohexosylceramide 311 ± 18
Supplementary Table 1. Cell sphingolipids and S1P bound to endogenous TRAF2. Sphingolipid Cell pmol/mg TRAF2 immunoprecipitate pmol/mg Sphingomyelin 4200 ± 250 Not detected Monohexosylceramide 311 ± 18
Supplemental Figures:
 Supplemental Figures: Figure 1: Intracellular distribution of VWF by electron microscopy in human endothelial cells. a) Immunogold labeling of LC3 demonstrating an LC3-positive autophagosome (white arrow)
Supplemental Figures: Figure 1: Intracellular distribution of VWF by electron microscopy in human endothelial cells. a) Immunogold labeling of LC3 demonstrating an LC3-positive autophagosome (white arrow)
SUPPLEMENTARY INFORMATION
 DOI: 1.13/ncb22 a starvation protocol 1, 5 min HBSS aa recovery,5min complete deprivation,5min 1μm colocalisation / colocalisation, % b 12 1 6 2 c complete deprivation,5min aa recovery,5min complete deprivation,5min
DOI: 1.13/ncb22 a starvation protocol 1, 5 min HBSS aa recovery,5min complete deprivation,5min 1μm colocalisation / colocalisation, % b 12 1 6 2 c complete deprivation,5min aa recovery,5min complete deprivation,5min
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/393/rs9/dc1 Supplementary Materials for Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts
www.sciencesignaling.org/cgi/content/full/8/393/rs9/dc1 Supplementary Materials for Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts
SUPPLEMENTARY INFORMATION In format provided by JAATTELA (NOVEMBER 2005)
 Box S1: Methods for studying lysosomal function and integrity Volume and distribution of the acidic compartment. Acridine orange is a metachromatic fluorochrome and a weak base that accumulates in the
Box S1: Methods for studying lysosomal function and integrity Volume and distribution of the acidic compartment. Acridine orange is a metachromatic fluorochrome and a weak base that accumulates in the
Protein Trafficking in the Secretory and Endocytic Pathways
 Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures
 Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
(A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939 (5 µm)
 Supplementary Figure Legends Figure S1. Tankyrase inhibition suppresses cell proliferation in an axin/β-catenin independent manner. (A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939
Supplementary Figure Legends Figure S1. Tankyrase inhibition suppresses cell proliferation in an axin/β-catenin independent manner. (A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939
A Self-Propelled Biological Process Plk1-Dependent Product- Activated, Feed-Forward Mechanism
 A Self-Propelled Biological Process Plk1-Dependent Product- Activated, Feed-Forward Mechanism The Harvard community has made this article openly available. Please share how this access benefits you. Your
A Self-Propelled Biological Process Plk1-Dependent Product- Activated, Feed-Forward Mechanism The Harvard community has made this article openly available. Please share how this access benefits you. Your
Supplementary information. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins
 Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting. protein3) regulate autophagy and mitophagy in renal tubular cells in. acute kidney injury
 Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
Name Animal source Vendor Cat # Dilutions
 Supplementary data Table S1. Primary and Secondary antibody sources Devi et al, TXNIP in mitophagy A. Primary Antibodies Name Animal source Vendor Cat # Dilutions 1. TXNIP mouse MBL KO205-2 1:2000 (WB)
Supplementary data Table S1. Primary and Secondary antibody sources Devi et al, TXNIP in mitophagy A. Primary Antibodies Name Animal source Vendor Cat # Dilutions 1. TXNIP mouse MBL KO205-2 1:2000 (WB)
Antigen presenting cells
 Antigen recognition by T and B cells - T and B cells exhibit fundamental differences in antigen recognition - B cells recognize antigen free in solution (native antigen). - T cells recognize antigen after
Antigen recognition by T and B cells - T and B cells exhibit fundamental differences in antigen recognition - B cells recognize antigen free in solution (native antigen). - T cells recognize antigen after
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Name: Multiple choice questions. Pick the BEST answer (2 pts ea)
 Exam 1 202 Oct. 5, 1999 Multiple choice questions. Pick the BEST answer (2 pts ea) 1. The lipids of a red blood cell membrane are all a. phospholipids b. amphipathic c. glycolipids d. unsaturated 2. The
Exam 1 202 Oct. 5, 1999 Multiple choice questions. Pick the BEST answer (2 pts ea) 1. The lipids of a red blood cell membrane are all a. phospholipids b. amphipathic c. glycolipids d. unsaturated 2. The
Bioluminescence Resonance Energy Transfer (BRET)-based studies of receptor dynamics in living cells with Berthold s Mithras
 Bioluminescence Resonance Energy Transfer (BRET)-based studies of receptor dynamics in living cells with Berthold s Mithras Tarik Issad, Ralf Jockers and Stefano Marullo 1 Because they play a pivotal role
Bioluminescence Resonance Energy Transfer (BRET)-based studies of receptor dynamics in living cells with Berthold s Mithras Tarik Issad, Ralf Jockers and Stefano Marullo 1 Because they play a pivotal role
Additional methods appearing in the supplement are described in the Experimental Procedures section of the manuscript.
 Supplemental Materials: I. Supplemental Methods II. Supplemental Figure Legends III. Supplemental Figures Supplemental Methods Cell Culture and Transfections for Wild Type and JNK1-/-,JNK2-/- MEFs: The
Supplemental Materials: I. Supplemental Methods II. Supplemental Figure Legends III. Supplemental Figures Supplemental Methods Cell Culture and Transfections for Wild Type and JNK1-/-,JNK2-/- MEFs: The
supplementary information
 Figure S1 Nucleotide binding status of RagA mutants. Wild type and mutant forms of MycRagA was transfected into HEK293 cells and the transfected cells were labeled with 32 Pphosphate. MycRagA was immunoprecipitated
Figure S1 Nucleotide binding status of RagA mutants. Wild type and mutant forms of MycRagA was transfected into HEK293 cells and the transfected cells were labeled with 32 Pphosphate. MycRagA was immunoprecipitated
a b G75 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server.
 a b G75 2 2 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server. a. Overlay of top 10 models generated by I-TASSER illustrates the potential effect of 7 amino acid insertion
a b G75 2 2 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server. a. Overlay of top 10 models generated by I-TASSER illustrates the potential effect of 7 amino acid insertion
effects on organ development. a-f, Eye and wing discs with clones of ε j2b10 show no
 Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
McWilliams et al., http :// /cgi /content /full /jcb /DC1
 Supplemental material JCB McWilliams et al., http ://www.jcb.org /cgi /content /full /jcb.201603039 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. In vitro characterization of mito-qc. (A and B) Analysis
Supplemental material JCB McWilliams et al., http ://www.jcb.org /cgi /content /full /jcb.201603039 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. In vitro characterization of mito-qc. (A and B) Analysis
supplementary information
 DOI: 10.1038/ncb1875 Figure S1 (a) The 79 surgical specimens from NSCLC patients were analysed by immunohistochemistry with an anti-p53 antibody and control serum (data not shown). The normal bronchi served
DOI: 10.1038/ncb1875 Figure S1 (a) The 79 surgical specimens from NSCLC patients were analysed by immunohistochemistry with an anti-p53 antibody and control serum (data not shown). The normal bronchi served
SUPPLEMENTARY INFORMATION
 Figure S1 Treatment with both Sema6D and Plexin-A1 sirnas induces the phenotype essentially identical to that induced by treatment with Sema6D sirna alone or Plexin-A1 sirna alone. (a,b) The cardiac tube
Figure S1 Treatment with both Sema6D and Plexin-A1 sirnas induces the phenotype essentially identical to that induced by treatment with Sema6D sirna alone or Plexin-A1 sirna alone. (a,b) The cardiac tube
m 6 A mrna methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer
 SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
Supplementary material Legends to Supplementary Figures Figure S1. Figure S2. Figure S3.
 Supplementary material Legends to Supplementary Figures. Figure S1. Expression of BICD-N-MTS fusion does not affect the distribution of the Golgi and endosomes. HeLa cells were transfected with GFP-BICD-N-MTS
Supplementary material Legends to Supplementary Figures. Figure S1. Expression of BICD-N-MTS fusion does not affect the distribution of the Golgi and endosomes. HeLa cells were transfected with GFP-BICD-N-MTS
Rescue of mutant rhodopsin traffic by metformin-induced AMPK activation accelerates photoreceptor degeneration Athanasiou et al
 Supplementary Material Rescue of mutant rhodopsin traffic by metformin-induced AMPK activation accelerates photoreceptor degeneration Athanasiou et al Supplementary Figure 1. AICAR improves P23H rod opsin
Supplementary Material Rescue of mutant rhodopsin traffic by metformin-induced AMPK activation accelerates photoreceptor degeneration Athanasiou et al Supplementary Figure 1. AICAR improves P23H rod opsin
Supplementary Figure 1
 Supplementary Figure 1 a γ-h2ax MDC1 RNF8 FK2 BRCA1 U2OS Cells sgrna-1 ** 60 sgrna 40 20 0 % positive Cells (>5 foci per cell) b ** 80 sgrna sgrna γ-h2ax MDC1 γ-h2ax RNF8 FK2 MDC1 BRCA1 RNF8 FK2 BRCA1
Supplementary Figure 1 a γ-h2ax MDC1 RNF8 FK2 BRCA1 U2OS Cells sgrna-1 ** 60 sgrna 40 20 0 % positive Cells (>5 foci per cell) b ** 80 sgrna sgrna γ-h2ax MDC1 γ-h2ax RNF8 FK2 MDC1 BRCA1 RNF8 FK2 BRCA1
TFEB-mediated increase in peripheral lysosomes regulates. Store Operated Calcium Entry
 TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Spatial Control of the TSC Complex Integrates Insulin and Nutrient Regulation of mtorc1 at the Lysosome
 Spatial Control of the TSC Complex Integrates Insulin and Nutrient Regulation of mtorc1 at the Lysosome Suchithra Menon, 1,4 Christian C. Dibble, 2,4, * George Talbott, 1 Gerta Hoxhaj, 1 Alexander J. Valvezan,
Spatial Control of the TSC Complex Integrates Insulin and Nutrient Regulation of mtorc1 at the Lysosome Suchithra Menon, 1,4 Christian C. Dibble, 2,4, * George Talbott, 1 Gerta Hoxhaj, 1 Alexander J. Valvezan,
Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells
 Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells Immunofluorescence staining of H3K9me3 and 53BP1 in PH and HGADFN003 (HG003) cells at 24 h after γ-irradiation. Scale bar,
Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells Immunofluorescence staining of H3K9me3 and 53BP1 in PH and HGADFN003 (HG003) cells at 24 h after γ-irradiation. Scale bar,
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/308/ra4/dc1 Supplementary Materials for Antipsychotics Activate mtorc1-dependent Translation to Enhance Neuronal Morphological Complexity Heather Bowling, Guoan
www.sciencesignaling.org/cgi/content/full/7/308/ra4/dc1 Supplementary Materials for Antipsychotics Activate mtorc1-dependent Translation to Enhance Neuronal Morphological Complexity Heather Bowling, Guoan
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
SUPPLEMENTARY INFORMATION. Supplementary Figures S1-S9. Supplementary Methods
 SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting
 Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting Question No. 1 of 10 Question 1. Which of the following statements about the nucleus is correct? Question #01 A. The
Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting Question No. 1 of 10 Question 1. Which of the following statements about the nucleus is correct? Question #01 A. The
Nature Neuroscience: doi: /nn Supplementary Figure 1. PICALM expression in brain capillary endothelium in human brain and in mouse brain.
 Supplementary Figure 1 PICALM expression in brain capillary endothelium in human brain and in mouse brain. a, Double immunostaining for PICALM (red, left) and lectin positive endothelial profiles (blue,
Supplementary Figure 1 PICALM expression in brain capillary endothelium in human brain and in mouse brain. a, Double immunostaining for PICALM (red, left) and lectin positive endothelial profiles (blue,
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06994 A phosphatase cascade by which rewarding stimuli control nucleosomal response A. Stipanovich*, E. Valjent*, M. Matamales*, A. Nishi, J.H. Ahn, M. Maroteaux, J. Bertran-Gonzalez,
doi: 10.1038/nature06994 A phosphatase cascade by which rewarding stimuli control nucleosomal response A. Stipanovich*, E. Valjent*, M. Matamales*, A. Nishi, J.H. Ahn, M. Maroteaux, J. Bertran-Gonzalez,
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS
 Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Chapter 6. Antigen Presentation to T lymphocytes
 Chapter 6 Antigen Presentation to T lymphocytes Generation of T-cell Receptor Ligands T cells only recognize Ags displayed on cell surfaces These Ags may be derived from pathogens that replicate within
Chapter 6 Antigen Presentation to T lymphocytes Generation of T-cell Receptor Ligands T cells only recognize Ags displayed on cell surfaces These Ags may be derived from pathogens that replicate within
