** *** population. (A) Representative FACS plots showing the gating strategy for T N
|
|
|
- Janis Parker
- 6 years ago
- Views:
Transcription
1 Sca-1 (MFI) CD122 (MFI) CD44 A CD8 + gated: resting spleen CD8 + gated: T Mem -derived CD8 + gated: T N -derived, expanded alone B CD62L CD44 low CD62L +, resting CD44 high, T Mem -derived CD44 low CD62L +, T N -derived C CD44 low CD62L +, resting naive CD44 low CD62L +, T Mem -derived CD44 high, T N -derived *** *** 10 3 ** *** Supplemental Figure 1. Precocious differentiation causes attrition of the T SCM population. (A) Representative FACS plots showing the gating strategy for T N /T SCM and T Mem populations. (B) Representative FACS plots and (C) scatter plots showing Sca-1 and CD122 expression on resting T N CD8 + T cells and T N - or T Mem -derived CD8 + T cells expanded alone for 6d with CD3/CD28- specific antibodies and IL-2. Data shown are representative of two independently performed experiments (n = 2-3 independent mice or cultures per condition) with results shown as mean ± SEM. Statistical comparisons performed using an unpaired 2-tailed Student s t test corrected for multiple comparisons by a Bonferroni adjustment (***P<0.001; **P<0.01).
2 CD44 CD44 Fold Expansion A ** 5 0 T N -derived, alone T N -derived, mix CD8 + Ly gated: B Naïve-derived population: 8.6 T N (Ly5.2) alone 71.2 T N (Ly5.2) + T Mem (Ly5.1) OT-1 CD8 + C CD62L Open-repertoire CD8 + CD62L Supplemental Figure 2. Precocious differentiation is not attributable to differences in cell expansion and is not TCR-specific. (A) Expansion of T N -derived cells 6d after priming alone or in the presence of a 1:1 mixture T Mem using CD3/CD28-specific antibodies and IL-2. Results shown as mean ± SEM from n = 3 independently maintained cultures per condition. Statistical comparison performed using an unpaired 2-tailed Student s t-test (**P<0.01). Representative flow cytometry analyses showing the distribution of Ly5.2 + T N -derived CD8 + subsets from either (B) OT-1 TCR transgenic mice or (C) polyclonal WT mice 6d following priming alone or in the presence of a 1:1 mixture with Ly5.1 + T Mem using CD3/CD28-specific antibodies and IL-2. Cells gated on live + CD8 + Ly5.2 + T cells. Numbers represent the percentage of cells in each quadrant. All experiments shown were repeated at least twice with similar results.
3 CD44 A CD8 + Ly5.2-gated: B Stimulation / cytokine conditions: T N (Ly5.2) alone T N (Ly5.2) + T Mem (Ly5.1) T N alone T N + T Mem Anti-CD3/CD28 + IL-2 (standard) Fold increase T EM Anti-CD3/CD28 + IL-2 (high) Fold derease T SCM Stim conditions: IL-2 (standard) IL-2 (high) T N alone T N + T Mem Anti-CD3/CD28 + IL Fold increase T EM anti-cd3 alone + IL-2 (standard) Fold derease T SCM CD62L Stim conditions: IL-15 anti-cd3 alone + IL-2 Supplemental Figure 3. Precocious differentiation occurs independently of cytokine and priming conditions. (A) Representative flow cytometry analyses showing the distribution of Ly5.2 + T N -derived CD8 + subsets 6d following priming alone or in the presence of a 1:3 mixture with Ly5.1 + T Mem using CD3/CD28-specific antibodies or anti-cd3 alone and either standard dose IL-2 (4.4ng/mL), high dose IL-2 (10ng/mL), or IL-15 (10ng/mL). Cells gated on live + CD8 + Ly5.2 + T cells. Numbers represent the percentage of cells in each quadrant. (B) Summary plot showing the fold change in T EM or T SCM populations under different cytokine and stimulation conditions. (n = 3 independent cultures per condition). Experiments shown were repeated twice with similar results.
4 CD44 A T N (Ly5.2) alone CD8 + Ly5.2-gated: T N (Ly5.2) + CD8 + T Mem (Ly5.1) T N (Ly5.2) + Bulk T Mem (Ly5.1) Fold increase T EM B T N alone T N + T Mem 0.5 CD62L Fold derease T SCM Antigen-experienced population : CD8 + T Mem Bulk T Mem Supplemental Figure 4. Evaluation of precocious differentiation using CD8 + T Mem versus a bulk population of T Mem. (A) Representative flow cytometry analyses showing the distribution of Ly5.2 + T N -derived CD8 + subsets 6d following priming alone or in the presence of a 1:3 mixture with CD8+ or bulk Ly5.1 + T Mem using CD3/CD28-specific antibodies and standard dose IL-2 (4.4ng/mL). The bulk T Mem population was ~2:1 CD4 + :CD8 + T cells. Cells gated on live + CD8 + Ly5.2 + T cells. Numbers represent the percentage of cells in each quadrant. (B) Summary plot showing the fold change in T EM or T SCM populations using CD8 + versus bulk T Mem populations (n = 3 independent cultures per condition). Experiments shown were repeated twice with similar results.
5 pmel-1 (cells per spleen) T Mem, alone T Mem, mix Time (d) after treatment Supplemental Figure 5. T Mem expansion is not altered by the co-transfer of T N. In vivo expansion and persistence of 3x10 5 Ly5.1 + CD8 + T Mem injected alone or in combination with 1x10 5 adoptively transferred Thy1.1 + T N cells into Ly5.2 + WT mice bearing 10d established B16 melanomas. All treated mice received 6Gy irradiation, i.v. rvv-gp100 (2x10 7 pfu), and 3d of i.p. IL-2 (10mg/dose twice daily). n = 3 mice per group per time point. Results are displayed as mean ± SEM. Data shown are representative of 2 independently performed experiments.
6 CD44 CD44 A T N alone, no stim T N alone, acd3/acd28 stim CD8 + T N gated: T N mix, acd3/acd28 stim T N mix, no stim T N mix, transwell acd3/acd28 stim CD62L B T N (Ly5.1) + media control CD8 + Ly5.1 gated: T N (Ly5.1) + T Mem supernatant T N (Ly5.1) + T Mem (Ly5.2) Day 0: Day 4: CD62L Supplemental Figure 6. Precocious differentiation is contact-dependent and is not transmissible through transfer of T Mem supernatant. (A) Representative flow cytometry analysis showing the distribution of Ly5.1 + T N -derived CD8 + T cell subsets 6d following priming alone or with Ly5.2 + T Mem either in physical contact or separated by a 0.4 mm semi-permeable membrane. (B) Representative FACS analysis and plots summarizing the distribution of CD8 + T cell subsets on Ly5.1 + T N -derived subsets primed alone or in the presence or absence of Ly5.2 + T Mem cells or supernatant derived from stimulated T Mem cells. For FACS plots, numbers represent the percentage of cells in each quadrant.
7 CD44 T cell subset (%) A T N (Ly5.1) + IgG CD8 + Ly5.1-gated: T N (Ly5.1) + afasl B T SCM T CM ns ns T EM ns CD62L 0 T N -derived, alone + IgG T N -derived, alone + afasl Supplemental Figure 7. Blockade of FasL on T N CD8 + T cells primed in the absence of T Mem not alter the distribution of T-cell subsets. (A) Representative FACS plot and (B) summary bar graph showing the distribution of T cell subsets 6d after Ly5.1 + T N (CD44 low CD62L + ) pmel-1 CD8 + T cells were primed alone with CD3/CD28-specific antibodies and IL-2 in the presence of 5ug/mL of afasl or IgG control antibody. Numbers represent the percentage of cells in each quadrant for FACS plots after gating on live + CD8 + Ly5.1 + cells; graphs represents the mean ± SEM of n = 6 independently maintained cultures per condition from 2 pooled experiments. unpaired 2-tailed Student s t test corrected for multiple comparisons by a Bonferroni adjustment (ns, not significant).
8 Expression relative to Actb Expression relative to Actb Expression relative to Actb Expression relative to Actb (10-3 ) Expression relative to Actb 10-1 Sell 10-2 Il7ra 10-1 Klf2 5 Tfrc 10-1 Gzmb lz-fasl (ng ml -1 ) lz-fasl (ng ml -1 ) lz-fasl (ng ml -1 ) lz-fasl (ng ml -1 ) lz-fasl (ng ml -1 ) Supplemental Figure 8. Agonizing Fas-signaling in T N in the absence of T Mem causes dosedependent changes in differentiation-associated genes. Quantitative PCR analysis of Sell, Il7ra, Klf2, Tfrc, and Gzmb in CD8 + T N -derived cells primed with CD3/CD28-specific antibodies and IL-2 alone or in the presence of titrated concentrations of lz-fasl. Data shown as mean ± SEM of n = 3 independent cultures for each condition.
9 population (%) population (%) population (%) PI A lz-fasl (50 ng ml -1 ): T SCM T CM T EM (-) (+) B Annexin V viable early apoptotic necrotic 100 T SCM lz-fasl: (-) (+) 100 T CM lz-fasl: (-) (+) 100 T EM lz-fasl: (-) (+) Supplemental Figure 9. T EM CD8 + T cells have a higher level of baseline and lz-fasl induced apoptosis relative to T SCM and T CM subsets. (A) Representative FACS plots and (B) summary bar graphs demonstrating annexin/pi staining at baseline or 6h following exposure to 50 ng/ml of lz- FasL (+) or vehicle control (-) in T SCM, T CM, and T EM subsets. Bar graphs represents the mean ± SEM of n = 3 independently maintained cultures. Results from one of two representative experiments is shown.
10 Live + CD8 + -gated: T N Vs. T CM Vs. T EM CD95 Supplemental Figure 10. T CM and T EM CD8 + T cell subsets express similar amounts of surface Fas. Fas expression on naïve (CD44 low CD62L + ), T CM (CD44 high CD62L + ), and T EM (CD44 high CD62L - ) CD8 + T cells subsets. This experiment has been repeated >3 times with similar results.
11 Proliferation Index Cell tracer ef450 A Ratio T Mem : T N T N alone 1:1 3:1 9:1 27:1 T Mem alone Pre stimulation Post stimulation (D+4) B Cell tracer ef HD2 HD1 HD Ratio T Mem : T N Supplemental Figure 11. Gating strategy to assess the phenotype and proliferation of mixed cultures containing human T N and T Mem cells using alternative membrane-associated dyes. Human T N and T Mem CD8 + T cells subsets were obtained by magnetic bead isolation and labeled with cell tracer ef450 or ef670 dyes, respectively. Naïve CD8 + T cells were subsequently stimulated with CD3/CD28-specific antibodies and IL-2 alone or in the presence of indicated ratios with T Mem. (A) Representative FACS plots demonstrating the gating strategy used to distinguish T N -derived from T Mem cells immediately after membrane dye labeling and mixing (top) or 4d after stimulation (bottom). Data is shown after gating on viable CD8 + lymphocytes and is representative of 3 independently performed experiments. (B) Assessment of the proliferation index (PI) of T N -derived cells 4d after expansion alone or in the presence of indicated ratios with T Mem cells. Results shown as mean ± SEM of n = 3 independent cultures for each condition for all three donors.
12 Relative cell yield (%) CD27 A 0 lz-fasl (ng ml -1 ) B C 150 CCR lz-fasl (ng ml -1 ) Supplemental Figure 12. Proliferation, expansion, and division-normalized differentiation of human CD8 + T cells expanded in the presence of titrated amounts of lz-fasl. (A) Representative FACS plots demonstrating the proliferation of human CD8 + T cells labeled with Cell tracer ef450 and expanded for 6d with CD3/CD28-specific antibodies and IL-2 alone or in the presence of indicated concentrations of lz-fasl. Similar results were obtained in 2 independent experiments after gating on live + CD8 + T cell that had undergone at least one cell division. (B) Representative FACS plots demonstrating the division-normalized surface phenotype for CD27 and CCR7 expression of expanded CD8 + T cells that had diluted an equivalent amount of Cell tracer ef450 dye. The arrows indicate the gated cell division analyzed and numbers represent the percentage of cells in each quadrant. (C) Relative overall cell yield of CD8 + T cells expanded in indicated concentrations of lz-fasl. All values normalized to the vehicle only control group. Similar results were obtained in 2 independent experiments.
13 Tumor area (mm 2 ) Survival (%) A 500 No treatment T N -derived + IgG T N -derived + afasl B *** Time (d) after treatment Time (d) after treatment Supplemental Figure 13. Blockade of FasL during ex vivo cell expansion does not influence the in vivo antitumor activity of isolated T N cells. (A) Tumor regression and (B) animal survival of mice bearing 10d established s.c. B16 melanomas left untreated or treated with 2.5x10 5 T N -derived pmel-1 cells primed alone in the presence of afasl or IgG control. All mice received 6Gy irradiation prior to cell infusion in addition to i.v. rvv-gp100 (2x10 7 pfu) and 3d of i.p. IL-2 (10mg/dose twice daily). Data shown are representative of 2 independent experiments with results displayed as mean ± SEM. Statistical comparisons performed using the log-rank test for animal survival (***P<0.01).
14 Supplemental table 1: Differentially expressed genes in naïve-derived T cells primed alone or with memory T cells at 18h. Gene Symbol RefSeq Fold-Change (TN alone 18h vs. TN mix 18h) p-value (TN alone 18h vs. TN mix 18h) Akr1c18 NM_ E-05 Il7r NM_ E-07 Kif21b NM_ E-07 Cd7 NM_ E-06 Slc12a7 NM_ E-07 Pde2a NM_ E-09 Tmem71 NM_ E-08 A430078G23Rik NM_ E-10 Cdc25b NM_ E-07 Fam65b NM_ E-07 H2-Ob NM_ E-08 Fam78a BC E-09 B3gnt5 NM_ E-05 Slc28a2 AF E-06 Rapgef4 NM_ E-07 Itga4 NM_ E-07 Cyp2s1 NM_ E-06 Arhgef18 NM_ E-08 Gm10673 ENSMUST Zfp831 NM_ E-06 Card6 ENSMUST E-07 Rasgrp2 NM_ E-08 Hmha1 NM_ E-07 Slc6a19 NM_ E-09 St6gal1 NM_ E-07 Il6ra NM_ E-08 Acss2 NM_ E-07 Gria3 NM_ E-06 Sepp1 NM_ E-05 A130014H13Rik AK Cmah NM_ E-06 Klrd1 NM_ E-06 Scml4 NM_ E-07 Orai2 NM_ E-07 Cd55 NM_ E-05 Sh2d3c NM_ E-06 Pik3ip1 NM_ E-07 1
15 Dzip1 NM_ E-07 Ctdsp2 NM_ E-07 Fam101b ENSMUST E-06 Zfp652 NM_ E-06 S1pr1 NM_ E-08 St6gal1 NM_ E-05 Itgb7 NM_ E-07 St8sia1 NM_ E-05 Ssh2 ENSMUST Klf13 NM_ E-07 Acss1 NM_ E-07 Lypd6b NM_ E-05 Rasa3 NM_ E-08 Zbtb20 ENSMUST E-05 Apol7e NM_ E-06 Smpdl3a NM_ E E14Rik ENSMUST E-07 Adcy7 NM_ E-07 Acp5 NM_ E-06 Kbtbd11 NM_ E-05 Bcl6 NM_ E-05 Fam102a NM_ E-08 Myo1f NM_ E-07 Dnahc8 NM_ E-06 Apol7b NM_ E E13Rik BC E-05 Thra NM_ E-06 Arrb1 NM_ E-06 Actn1 NM_ E-08 Cdkn1b NM_ Ttyh3 NM_ E-05 C630004H02Rik BC E-05 Slc12a6 NM_ E-07 Klf3 NM_ E-06 Rnasel NM_ E-05 Map4k2 NM_ E-06 Tmem63a NM_ E-06 Gramd1a NM_ E-09 Abcd1 NM_ E-06 Itgae NM_ E-06 2
16 J21Rik NM_ E-05 Sla2 NM_ E-06 Prkd2 NM_ E-06 Tmc6 NM_ E I18Rik AK E-05 Slc28a2 NM_ E-06 Gm10345 AK E-05 Xkrx NM_ E-05 Appl2 NM_ E-05 Nipal3 NM_ E-06 Mkl1 NM_ E-06 Klhl6 NM_ E-07 Tspan32 NM_ E-06 Traf3ip3 NM_ E-06 Hexb NM_ E-05 Cdkn1b NM_ E-06 Zbtb20 NM_ E-06 Mll2 ENSMUST E-05 Kif1b NM_ E-06 Arhgap15 NM_ E-06 Tpcn1 NM_ E-06 Unc84b NM_ E-08 Plaur NM_ E-06 Cks2 NM_ Gm5521 XR_ AI BC Idi2 NM_ E N04Rik NM_ E-05 B3galt6 NM_ E-05 Bhlhe40 NM_ E-05 Rpa3 NM_ Pros1 NM_ E-06 BC BC Gp49a NM_ E-05 Idi2 NM_ E-05 Hexim1 NM_ E-06 Mrpl47 NM_ Ifng NM_ E-06 Ifitm3 NM_ E-05 Mphosph6 NM_
17 Gadd45g NM_ E-06 LOC M E-06 Lad1 NM_ E-08 Gadd45b NM_ E-08 Tnfsf4 NM_ E-06 Rgs1 NM_ E-06 Casp3 NM_ E-06 Eomes NM_ E-07 LOC M E-05 Hsf2 NM_ E-06 Fam129a NM_ E-06 Gzmc NM_ E-07 LOC M E-05 Serpinb6b NM_ E-08 Il2 NM_ E-06 Scd2 NM_ E-07 Serpinb9 NM_ E-07 Cd200 NM_ E-07 Cd83 NM_ E-07 AA ENSMUST E-06 Cd24a NM_ E-05 Il3 NM_ E-07 4
18 Supplemental table 2: Differentially expressed genes in naïve-derived T cells primed alone or with memory T cells at 96h. Gene Symbol RefSeq Fold-Change (TN alone 96h vs. TN mix 96h) p-value (TN alone 96h vs. TN mix 96h) Snord116 NR_ E-06 H2-Aa NM_ E-10 Lyz2 NM_ E-09 Snord116 AF E-06 Gpr83 NM_ E-07 Snord115 AF E-05 Gm3079 XM_ E-05 Ifng NM_ E-10 Gria3 NM_ E-09 Itgae NM_ E-09 A130014H13Rik AK E-08 Xcl1 NM_ E-07 Hba-a1 NM_ E-07 Lgals3 NM_ E-09 Cd74 NM_ E-08 Hba-a2 NM_ E-07 Gm3079 XM_ E-05 Gm8020 ENSMUST Cpa3 NM_ E-05 Serpine2 NM_ E-06 Gm8020 ENSMUST Ccr4 NM_ E-07 Klrb1b NM_ E-08 Ccr9 NM_ Cd80 NM_ E-07 Foxp3 NM_ E-09 Rbpms NM_ E-08 Ifit3 NM_ E-07 Vmn2r96 NM_ Nr4a3 NM_ E-08 Vmn1r100 NM_ Vmn1r158 NM_ Vmn1r101 NM_ Vmn1r117 NM_ Mir186 NR_ Ccl3 NM_ E-07 Gpr56 NM_ E-08 5
19 Vmn1r122 NM_ Fcer1g NM_ E-07 Airn NR_ Vmn1r125 NM_ Gm14085 NM_ E-06 Vmn1r103 NM_ Mcpt8 NM_ Gm10375 NM_ Gm5891 NM_ Vmn1r93 NM_ Gm3079 XM_ Vmn1r151 NM_ Vmn1r121 NM_ O19Rik ENSMUST E-05 Vmn1r114 NM_ Timp2 NM_ E-06 Vmn1r158 NM_ Gm2799 NM_ Furin NM_ E-07 Kit NM_ E-08 Rps3a NM_ Cybb NM_ E-09 Fes NM_ E-06 D18Ertd653e BC E-08 H2-Ab1 NM_ E-06 Folr4 NM_ E-06 Vmn1r-ps79 NR_ Gm10688 ENSMUST Vmn1r-ps79 NR_ Eng NM_ E-07 Igfbp4 NM_ E-07 Clec12a NM_ E-08 Il6ra NM_ E-06 Il7r NM_ E-07 Mir15a NR_ Gm3994 ENSMUST E-05 Tgfb3 NM_ E-06 Slc12a7 NM_ E-09 Gm5929 XR_ Cd80 BC E-05 6
20 Gm5111 NM_ E I15Rik ENSMUST E-07 Tnfsf4 NM_ E-05 Snora74a NR_ E-07 Snora69 NR_ S1pr1 NM_ E-07 Speer4e NM_ Gm3994 ENSMUST E-05 Hsd11b1 NM_ E E14Rik BC E-05 Cpd NM_ E-06 Rps13 NM_ E-05 Boll NM_ E-05 Gm2799 NM_ Rps13 NM_ E-05 Ahr NM_ E-06 Gm10406 NM_ Gm3002 NR_ Cd7 NM_ E-06 Csprs NM_ Pgpep1l NM_ E-06 H2-Eb1 NM_ E-07 Epcam NM_ E-05 Pisd-ps1 NR_ E-06 Napsa NM_ E-06 Gm9282 XR_ Gm9282 XR_ Cd200r1 NM_ E-05 Tlr1 NM_ E-06 Gm2799 NM_ Adamts6 NM_ Gm10375 NM_ Lpar6 NM_ E-05 Speer4d NM_ Snord52 NR_ Gm10673 ENSMUST Pla2g7 NM_ E-05 Nr4a1 NM_ E-08 Gm3002 NR_ E-05 Ikzf2 NM_
21 O03Rik BC E-07 Mir181b-1 NR_ Zbtb20 ENSMUST M02Rik NM_ EG NM_ Psrc1 NM_ E-07 Sytl2 NM_ E-05 P4ha2 NM_ E-07 Chd7 NM_ E-05 Prelid2 NM_ E-08 Il10 NM_ Myo1f NM_ E-08 Dsc1 ENSMUST E-08 Fgl2 NM_ E-06 Gls2 NM_ E-07 Rnf157 NM_ E-06 Cercam NM_ E-07 Fads1 NM_ E-07 Vcl NM_ E-07 Fam64a NM_ E-06 Endod1 NM_ E-05 Tmem67 NM_ E-05 Selp NM_ E-07 Slc1a4 NM_ E-07 Zfp280b NM_ E-06 Gm129 BC E-06 Crmp1 NM_ E-08 Lipg NM_ E-06 Ero1l NM_ E-06 Sepw1 NM_ E-07 Pfkl NM_ E-07 Chsy1 NM_ E-08 Mboat2 NM_ Bmpr1a NM_ E-06 Smpd1 NM_ E-07 Lss NM_ E-07 Sqle NM_ E-06 Arsb NM_ E-07 Fbxo44 NM_ E-07 Abcb1a NM_ E-06 8
22 Gys1 NM_ E-07 St3gal6 NM_ E-08 Cxcr3 NM_ E-08 Arsb NM_ E-07 Dgat1 NM_ E-08 Pld4 NM_ E-06 Arrb1 NM_ E-07 Bex6 NM_ E-07 Acsl3 NM_ E-05 Gmfg NM_ E-09 Chac1 NM_ E-06 Serpina3f NM_ E-06 Rgs11 NM_ E-07 Grtp1 NM_ E-07 Slc7a3 NM_ E-06 Ppap2c NM_ E-07 Hpse NM_ E K17Rik BC E-07 Ttc39c NM_ E-05 Hemgn NM_ E-06 P4ha1 NM_ E-07 Entpd1 NM_ E-07 Pgm2 NM_ E-09 Gzmk NM_ E-07 Cysltr2 NM_ Grhpr NM_ E-08 Tgm2 NM_ E-06 Cyp51 NM_ E-06 Ndrg1 NM_ E-09 Mctp2 NM_ E-07 Slc6a9 NM_ E-08 Ctla2b NM_ Gpnmb NM_ E-06 Ifitm2 NM_ E-05 Rundc3b NM_ E-06 E430029J22Rik NM_ E-06 Fosb NM_ E-08 Ly86 NM_ E-06 Hmgcs1 NM_ E-07 Gpt2 NM_ E-06 9
23 Ldlr NM_ E-10 Dennd5a NM_ E-07 Fasl NM_ Scpep1 NM_ E-07 Ctla2a NM_ Fam183b NM_ E-07 Ccr5 NM_ E-06 Ccr5 NM_ E-06 Nrn1 NM_ E-06 Dkkl1 NM_ E-06 Ryk NM_ E-07 Tirap NM_ E-06 Slc2a1 NM_ E-09 AY AY E-05 ATP6 ENSMUST Sla NM_ E-09 Gm129 BC E-09 Ifitm2 NM_ E-07 Plek NM_ E-07 Insig1 NM_ E-08 Ndrg2 NM_ E-08 Plod2 NM_ E-05 Sc4mol NM_ E-05 Mapk11 NM_ E-07 Gp49a NM_ E-06 Klrk1 NM_ E-06 Nov NM_ E-07 Atf6 NM_ E-09 Vaultrc5 NR_ E-05 Serpinb6a NM_ Serpinb6b NM_ E-07 Ifitm2 NM_ E-08 Atf3 NM_ E-10 H1f0 NM_ E-04 Anxa2 NM_ E-07 Pik3ap1 NM_ E-10 Itih5 NM_ E-08 Scd2 NM_ E H14Rik NR_ E-06 Dusp6 NM_ E-09 10
24 Slc2a3 NM_ E-10 Kdm5d NM_ E-07 Idh1 NM_ E-05 Ak3l1 NM_ E-07 Fads2 NM_ E-12 Slc16a3 NM_ E-08 Bnip3 NM_ E-09 Egln3 NM_ E-12 Ccr2 NM_ E-07 Mt1 NM_ E-11 Ddx3y NM_ E-05 Uty NM_ E-07 I830127L07Rik ENSMUST E-09 Aldoc NM_ E-09 Zdhhc2 NM_ E-06 Asns NM_ E-11 Bnip3 NM_ E-10 Mt2 NM_ E-08 Eif2s3y NM_ E-08 Ak3l1 NM_ E-09 Scd1 NM_ E-11 11
25 Supplemental table 3: Differentially expressed genes in memory-derived T cells expanded alone or with naïve T cells at 96h. Gene Symbol RefSeq Fold-Change (TMem alone Vs. TMem mix, 96h) p-value (TMem alone Vs. TMem mix, 96h) Snord14d NR_ E-06 Gm11277 NM_ Mela D E-05 Aldoc NM_ E-06 Nrn1 NM_ E-05 Dapl1 NM_ E-06 Ifit3 NM_ E H14Rik NR_ E-05 Art2b NM_ E-07 Ctla4 NM_ E-06 12
26 Supplemental table 4: Comparative expression of TNF superfamily member ligands in memory-derived versus naïve-derived T cells 18h after stimulation. Gene Symbol RefSeq Soluble only Known expression in T cells P-value Fc (TMem:TN, 18h) Lta (aka Tnfsf1) NM_ yes yes No expression detected No expression detected Tnf (aka Tnfsf2) NM_ no yes No expression detected No expression detected Ltb (aka tumor necrosis factor (ligand) superfamily, member 3) NM_ no yes Tnfsf4 (aka Ox40l) NM_ no yes No expression detected No expression detected Tnfsf5 (aka Cd40l) NM_ no yes No expression detected No expression detected Tnfsf6 (aka Fasl) NM_ no yes Tnfsf7 (aka Cd70) NM_ no yes No expression detected No expression detected Tnfsf8 (aka Cd30l) NM_ no yes No expression detected No expression detected Tnfsf9 (aka 41bbl) NM_ no yes Tnfsf10 (aka Trail) NM_ no yes Tnfsf11 (aka Rankl) NM_ no yes Tnfsf12 (aka Dr3l, Tweak) NM_ no no No expression detected No expression detected Tnfsf13 (aka Tall2, April) NM_ no no No expression detected No expression detected Tnfsf13b (aka Tall1) NM_ no no No expression detected No expression detected Tnfsf14 (aka Hveml) NM_ no yes No expression detected No expression detected Tnfsf15 (aka Tl1a) NM_ no yes No expression detected No expression detected Tnfsf18 (aka Gitr) NM_ no yes No expression Eda NM_ no no detected No expression detected Btla NM_ no yes
CPM (x 10-3 ) Tregs +Teffs. Tregs alone ICOS CLTA-4
 A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
A 2,5 B 4 Number of cells (x 1-6 ) 2, 1,5 1, 5 CPM (x 1-3 ) 3 2 1 5 1 15 2 25 3 Days of culture 1/1 1/2 1/4 1/8 1/16 1/32 Treg/Teff ratio C alone alone alone alone CD25 FoxP3 GITR CD44 ICOS CLTA-4 CD127
Memory T cell driven differentiation of naive cells impairs adoptive immunotherapy
 Memory T cell driven differentiation of naive impairs adoptive immunotherapy Christopher A. Klebanoff,, Richard M. Siegel, Nicholas P. Restifo J Clin Invest. 2016;126(1):318-334. https://doi.org/10.1172/jci81217.
Memory T cell driven differentiation of naive impairs adoptive immunotherapy Christopher A. Klebanoff,, Richard M. Siegel, Nicholas P. Restifo J Clin Invest. 2016;126(1):318-334. https://doi.org/10.1172/jci81217.
MATERIALS AND METHODS. Neutralizing antibodies specific to mouse Dll1, Dll4, J1 and J2 were prepared as described. 1,2 All
 MATERIALS AND METHODS Antibodies (Abs), flow cytometry analysis and cell lines Neutralizing antibodies specific to mouse Dll1, Dll4, J1 and J2 were prepared as described. 1,2 All other antibodies used
MATERIALS AND METHODS Antibodies (Abs), flow cytometry analysis and cell lines Neutralizing antibodies specific to mouse Dll1, Dll4, J1 and J2 were prepared as described. 1,2 All other antibodies used
Supplementary Figure Legends. group) and analyzed for Siglec-G expression utilizing a monoclonal antibody to Siglec-G (clone SH2.1).
 Supplementary Figure Legends Supplemental Figure : Naïve T cells express Siglec-G. Splenocytes were isolated from WT B or Siglec-G -/- animals that have not been transplanted (n= per group) and analyzed
Supplementary Figure Legends Supplemental Figure : Naïve T cells express Siglec-G. Splenocytes were isolated from WT B or Siglec-G -/- animals that have not been transplanted (n= per group) and analyzed
Supplementary Figure 1. Characterization of basophils after reconstitution of SCID mice
 Supplementary figure legends Supplementary Figure 1. Characterization of after reconstitution of SCID mice with CD4 + CD62L + T cells. (A-C) SCID mice (n = 6 / group) were reconstituted with 2 x 1 6 CD4
Supplementary figure legends Supplementary Figure 1. Characterization of after reconstitution of SCID mice with CD4 + CD62L + T cells. (A-C) SCID mice (n = 6 / group) were reconstituted with 2 x 1 6 CD4
Supplementary Figures
 Supplementary Figures Supplementary Fig. 1. Surface thiol groups and reduction of activated T cells. (a) Activated CD8 + T-cells have high expression levels of free thiol groups on cell surface proteins.
Supplementary Figures Supplementary Fig. 1. Surface thiol groups and reduction of activated T cells. (a) Activated CD8 + T-cells have high expression levels of free thiol groups on cell surface proteins.
Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was
 Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was sorted by FACS. Surface markers for sorting were CD11c +
Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was sorted by FACS. Surface markers for sorting were CD11c +
Supplemental Figure 1. Activated splenocytes upregulate Serpina3g and Serpina3f expression.
 Relative Serpin expression 25 2 15 1 5 Serpina3f 1 2 3 4 5 6 8 6 4 2 Serpina3g 1 2 3 4 5 6 C57BL/6 DBA/2 Supplemental Figure 1. Activated splenocytes upregulate Serpina3g and Serpina3f expression. Splenocytes
Relative Serpin expression 25 2 15 1 5 Serpina3f 1 2 3 4 5 6 8 6 4 2 Serpina3g 1 2 3 4 5 6 C57BL/6 DBA/2 Supplemental Figure 1. Activated splenocytes upregulate Serpina3g and Serpina3f expression. Splenocytes
Supplemental Table 1: Enriched GO categories per cluster
 Supplemental Table 1: Enriched GO categories per cluster Cluster Enriched with #genes Raw p-value Corrected p-value Frequency in set (%) Cluster 1 immune response - GO:0006955 32 2.36E-14 0.001 13.5 Cluster
Supplemental Table 1: Enriched GO categories per cluster Cluster Enriched with #genes Raw p-value Corrected p-value Frequency in set (%) Cluster 1 immune response - GO:0006955 32 2.36E-14 0.001 13.5 Cluster
Eosinophils! 40! 30! 20! 10! 0! NS!
 A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
and follicular helper T cells is Egr2-dependent. (a) Diagrammatic representation of the
 Supplementary Figure 1. LAG3 + Treg-mediated regulation of germinal center B cells and follicular helper T cells is Egr2-dependent. (a) Diagrammatic representation of the experimental protocol for the
Supplementary Figure 1. LAG3 + Treg-mediated regulation of germinal center B cells and follicular helper T cells is Egr2-dependent. (a) Diagrammatic representation of the experimental protocol for the
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the T H 17 cell it reg cell balance Rajatava Basu 1,5, Sarah K.
SUPPLEMENTARY MATERIAL IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the T H 17 cell it reg cell balance Rajatava Basu 1,5, Sarah K.
Nature Medicine: doi: /nm.3922
 Title: Glucocorticoid-induced tumor necrosis factor receptor-related protein co-stimulation facilitates tumor regression by inducing IL-9-producing helper T cells Authors: Il-Kyu Kim, Byung-Seok Kim, Choong-Hyun
Title: Glucocorticoid-induced tumor necrosis factor receptor-related protein co-stimulation facilitates tumor regression by inducing IL-9-producing helper T cells Authors: Il-Kyu Kim, Byung-Seok Kim, Choong-Hyun
Supplementary Figure 1. Immune profiles of untreated and PD-1 blockade resistant EGFR and Kras mouse lung tumors (a) Total lung weight of untreated
 1 Supplementary Figure 1. Immune profiles of untreated and PD-1 blockade resistant EGFR and Kras mouse lung tumors (a) Total lung weight of untreated (U) EGFR TL mice (n=7), Kras mice (n=7), PD-1 blockade
1 Supplementary Figure 1. Immune profiles of untreated and PD-1 blockade resistant EGFR and Kras mouse lung tumors (a) Total lung weight of untreated (U) EGFR TL mice (n=7), Kras mice (n=7), PD-1 blockade
HTG EdgeSeq Immuno-Oncology Assay Gene List
 A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
A2M ABCB1 ABCB11 ABCC2 ABCG2 ABL1 ABL2 ACTB ADA ADAM17 ADGRE5 ADORA2A AICDA AKT3 ALCAM ALO5 ANA1 APAF1 APP ATF1 ATF2 ATG12 ATG16L1 ATG5 ATG7 ATM ATP5F1 AL BATF BA BCL10 BCL2 BCL2L1 BCL6 BID BIRC5 BLNK
Supplementary Figure S1. PTPN2 levels are not altered in proliferating CD8+ T cells. Lymph node (LN) CD8+ T cells from C57BL/6 mice were stained with
 Supplementary Figure S1. PTPN2 levels are not altered in proliferating CD8+ T cells. Lymph node (LN) CD8+ T cells from C57BL/6 mice were stained with CFSE and stimulated with plate-bound α-cd3ε (10µg/ml)
Supplementary Figure S1. PTPN2 levels are not altered in proliferating CD8+ T cells. Lymph node (LN) CD8+ T cells from C57BL/6 mice were stained with CFSE and stimulated with plate-bound α-cd3ε (10µg/ml)
Engineered T cells: Next-generation cancer immunotherapy
 Engineered T cells: Next-generation cancer immunotherapy Rahul Purwar, Assistant Professor Department of Biosciences & Bioengineering Indian Institute of Technology Bombay (IIT Bombay) Mumbai, India, 400076
Engineered T cells: Next-generation cancer immunotherapy Rahul Purwar, Assistant Professor Department of Biosciences & Bioengineering Indian Institute of Technology Bombay (IIT Bombay) Mumbai, India, 400076
SUPPLEMENTARY INFORMATION
 Complete but curtailed T-cell response to very-low-affinity antigen Dietmar Zehn, Sarah Y. Lee & Michael J. Bevan Supp. Fig. 1: TCR chain usage among endogenous K b /Ova reactive T cells. C57BL/6 mice
Complete but curtailed T-cell response to very-low-affinity antigen Dietmar Zehn, Sarah Y. Lee & Michael J. Bevan Supp. Fig. 1: TCR chain usage among endogenous K b /Ova reactive T cells. C57BL/6 mice
Supplemental Table I.
 Supplemental Table I Male / Mean ± SEM n Mean ± SEM n Body weight, g 29.2±0.4 17 29.7±0.5 17 Total cholesterol, mg/dl 534.0±30.8 17 561.6±26.1 17 HDL-cholesterol, mg/dl 9.6±0.8 17 10.1±0.7 17 Triglycerides,
Supplemental Table I Male / Mean ± SEM n Mean ± SEM n Body weight, g 29.2±0.4 17 29.7±0.5 17 Total cholesterol, mg/dl 534.0±30.8 17 561.6±26.1 17 HDL-cholesterol, mg/dl 9.6±0.8 17 10.1±0.7 17 Triglycerides,
Supplementary Data. Treg phenotype
 Supplementary Data Additional Experiment An additional experiment was performed using cryopreserved peripheral blood mononuclear cells (PBMC) derived from five renal cell carcinoma (RCC) patients [see
Supplementary Data Additional Experiment An additional experiment was performed using cryopreserved peripheral blood mononuclear cells (PBMC) derived from five renal cell carcinoma (RCC) patients [see
Supplemental Figure 1
 Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Blocking antibodies and peptides. Rat anti-mouse PD-1 (29F.1A12, rat IgG2a, k), PD-
 Supplementary Methods Blocking antibodies and peptides. Rat anti-mouse PD-1 (29F.1A12, rat IgG2a, k), PD- L1 (10F.9G2, rat IgG2b, k), and PD-L2 (3.2, mouse IgG1) have been described (24). Anti-CTLA-4 (clone
Supplementary Methods Blocking antibodies and peptides. Rat anti-mouse PD-1 (29F.1A12, rat IgG2a, k), PD- L1 (10F.9G2, rat IgG2b, k), and PD-L2 (3.2, mouse IgG1) have been described (24). Anti-CTLA-4 (clone
Supplementary Figure 1
 Supplementary Figure 1 Identification of IFN-γ-producing CD8 + and CD4 + T cells with naive phenotype by alternative gating and sample-processing strategies. a. Contour 5% probability plots show definition
Supplementary Figure 1 Identification of IFN-γ-producing CD8 + and CD4 + T cells with naive phenotype by alternative gating and sample-processing strategies. a. Contour 5% probability plots show definition
Supplemental Materials for. Effects of sphingosine-1-phosphate receptor 1 phosphorylation in response to. FTY720 during neuroinflammation
 Supplemental Materials for Effects of sphingosine-1-phosphate receptor 1 phosphorylation in response to FTY7 during neuroinflammation This file includes: Supplemental Table 1. EAE clinical parameters of
Supplemental Materials for Effects of sphingosine-1-phosphate receptor 1 phosphorylation in response to FTY7 during neuroinflammation This file includes: Supplemental Table 1. EAE clinical parameters of
Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases
 Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
Supplementary Information:
 Supplementary Information: Follicular regulatory T cells with Bcl6 expression suppress germinal center reactions by Yeonseok Chung, Shinya Tanaka, Fuliang Chu, Roza Nurieva, Gustavo J. Martinez, Seema
Supplementary Information: Follicular regulatory T cells with Bcl6 expression suppress germinal center reactions by Yeonseok Chung, Shinya Tanaka, Fuliang Chu, Roza Nurieva, Gustavo J. Martinez, Seema
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature11986 relative IL-6 expression Viable intracellular Bp per well 18 16 1 1 5 5 3 1 6 +DG 8 8 8 Control DG Time (h) hours B. pertussis IL-6 (pg/ml) 15 Control DG
SUPPLEMENTARY INFORMATION doi:1.138/nature11986 relative IL-6 expression Viable intracellular Bp per well 18 16 1 1 5 5 3 1 6 +DG 8 8 8 Control DG Time (h) hours B. pertussis IL-6 (pg/ml) 15 Control DG
SUPPLEMENTARY INFORMATION
 doi:1.138/nature1554 a TNF-α + in CD4 + cells [%] 1 GF SPF 6 b IL-1 + in CD4 + cells [%] 5 4 3 2 1 Supplementary Figure 1. Effect of microbiota on cytokine profiles of T cells in GALT. Frequencies of TNF-α
doi:1.138/nature1554 a TNF-α + in CD4 + cells [%] 1 GF SPF 6 b IL-1 + in CD4 + cells [%] 5 4 3 2 1 Supplementary Figure 1. Effect of microbiota on cytokine profiles of T cells in GALT. Frequencies of TNF-α
CD Marker Antibodies. atlasantibodies.com
 CD Marker Antibodies atlasantibodies.com CD Marker Antibodies Product Name Product Number Validated Applications Anti-ACE HPA029298 IHC,WB Anti-ACKR1 HPA016421 IHC Anti-ACKR1 HPA017672 IHC Anti-ADAM17
CD Marker Antibodies atlasantibodies.com CD Marker Antibodies Product Name Product Number Validated Applications Anti-ACE HPA029298 IHC,WB Anti-ACKR1 HPA016421 IHC Anti-ACKR1 HPA017672 IHC Anti-ADAM17
Supplementary Figure 1. Example of gating strategy
 Supplementary Figure 1. Example of gating strategy Legend Supplementary Figure 1: First, gating is performed to include only single cells (singlets) (A) and CD3+ cells (B). After gating on the lymphocyte
Supplementary Figure 1. Example of gating strategy Legend Supplementary Figure 1: First, gating is performed to include only single cells (singlets) (A) and CD3+ cells (B). After gating on the lymphocyte
Supplementary. presence of the. (c) mrna expression. Error. in naive or
 Figure 1. (a) Naive CD4 + T cells were activated in the presence of the indicated cytokines for 3 days. Enpp2 mrna expression was measured by qrt-pcrhr, infected with (b, c) Naive CD4 + T cells were activated
Figure 1. (a) Naive CD4 + T cells were activated in the presence of the indicated cytokines for 3 days. Enpp2 mrna expression was measured by qrt-pcrhr, infected with (b, c) Naive CD4 + T cells were activated
% of Cells A B C. Proliferation Index. T cell count (10 6 ) Division Index. % of Max CFSE. %Ki67+ cells. Supplementary Figure 1.
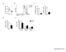 A B C T cell count (1 6 ) 3. 2. 1.. * % of Max CFSE ormoxia ypoxia o Stim. Proliferation Index 2.5 2. 1.5 1. * Division Index 2. 1.5 1..5. D E %Ki67+ cells 1 8 6 4 2 % of Cells 8 ormoxia 6 ypoxia 4 2 *
A B C T cell count (1 6 ) 3. 2. 1.. * % of Max CFSE ormoxia ypoxia o Stim. Proliferation Index 2.5 2. 1.5 1. * Division Index 2. 1.5 1..5. D E %Ki67+ cells 1 8 6 4 2 % of Cells 8 ormoxia 6 ypoxia 4 2 *
D CD8 T cell number (x10 6 )
 IFNγ Supplemental Figure 1. CD T cell number (x1 6 ) 18 15 1 9 6 3 CD CD T cells CD6L C CD5 CD T cells CD6L D CD8 T cell number (x1 6 ) 1 8 6 E CD CD8 T cells CD6L F Log(1)CFU/g Feces 1 8 6 p
IFNγ Supplemental Figure 1. CD T cell number (x1 6 ) 18 15 1 9 6 3 CD CD T cells CD6L C CD5 CD T cells CD6L D CD8 T cell number (x1 6 ) 1 8 6 E CD CD8 T cells CD6L F Log(1)CFU/g Feces 1 8 6 p
Nature Medicine doi: /nm.3957
 Supplementary Fig. 1. p38 alternative activation, IL-21 expression, and T helper cell transcription factors in PDAC tissue. (a) Tissue microarrays of pancreatic tissue from 192 patients with pancreatic
Supplementary Fig. 1. p38 alternative activation, IL-21 expression, and T helper cell transcription factors in PDAC tissue. (a) Tissue microarrays of pancreatic tissue from 192 patients with pancreatic
pro-b large pre-b small pre-b CCCP (µm) Rag1 -/- ;33.C9HCki
 a TMRM FI (Median) b TMRM FI (Median) c 20 15 10 5 0 8 6 4 2 0 pro-b large pre-b small pre-b 0 10 20 30 40 50 60 70 80 90 100 TMRM (nm) pro-b large pre-b small pre-b 0 1 2 4 8 16 32 64 128 256 CCCP (mm)
a TMRM FI (Median) b TMRM FI (Median) c 20 15 10 5 0 8 6 4 2 0 pro-b large pre-b small pre-b 0 10 20 30 40 50 60 70 80 90 100 TMRM (nm) pro-b large pre-b small pre-b 0 1 2 4 8 16 32 64 128 256 CCCP (mm)
Nature Immunology: doi: /ni Supplementary Figure 1. Cytokine pattern in skin in response to urushiol.
 Supplementary Figure 1 Cytokine pattern in skin in response to urushiol. Wild-type (WT) and CD1a-tg mice (n = 3 per group) were sensitized and challenged with urushiol (uru) or vehicle (veh). Quantitative
Supplementary Figure 1 Cytokine pattern in skin in response to urushiol. Wild-type (WT) and CD1a-tg mice (n = 3 per group) were sensitized and challenged with urushiol (uru) or vehicle (veh). Quantitative
Transduction of lentivirus to human primary CD4+ T cells
 Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
Transduction of lentivirus to human primary CD4 + T cells Human primary CD4 T cells were stimulated with anti-cd3/cd28 antibodies (10 µl/2 5 10^6 cells of Dynabeads CD3/CD28 T cell expander, Invitrogen)
Nature Immunology: doi: /ni Supplementary Figure 1. RNA-Seq analysis of CD8 + TILs and N-TILs.
 Supplementary Figure 1 RNA-Seq analysis of CD8 + TILs and N-TILs. (a) Schematic representation of the tumor and cell types used for the study. HNSCC, head and neck squamous cell cancer; NSCLC, non-small
Supplementary Figure 1 RNA-Seq analysis of CD8 + TILs and N-TILs. (a) Schematic representation of the tumor and cell types used for the study. HNSCC, head and neck squamous cell cancer; NSCLC, non-small
Eosinophils are required. for the maintenance of plasma cells in the bone marrow
 Eosinophils are required for the maintenance of plasma cells in the bone marrow Van Trung Chu, Anja Fröhlich, Gudrun Steinhauser, Tobias Scheel, Toralf Roch, Simon Fillatreau, James J. Lee, Max Löhning
Eosinophils are required for the maintenance of plasma cells in the bone marrow Van Trung Chu, Anja Fröhlich, Gudrun Steinhauser, Tobias Scheel, Toralf Roch, Simon Fillatreau, James J. Lee, Max Löhning
Supplemental Materials
 Supplemental Materials Programmed death one homolog maintains the pool size of regulatory T cells by promoting their differentiation and stability Qi Wang 1, Jianwei He 1, Dallas B. Flies 2, Liqun Luo
Supplemental Materials Programmed death one homolog maintains the pool size of regulatory T cells by promoting their differentiation and stability Qi Wang 1, Jianwei He 1, Dallas B. Flies 2, Liqun Luo
Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured
 Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured under Th0, Th1, Th2, Th17, and Treg conditions. mrna
Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured under Th0, Th1, Th2, Th17, and Treg conditions. mrna
CD25-PE (BD Biosciences) and labeled with anti-pe-microbeads (Miltenyi Biotec) for depletion of CD25 +
 Supplements Supplemental Materials and Methods Depletion of CD25 + T-cells from PBMC. Fresh or HD precultured PBMC were stained with the conjugate CD25-PE (BD Biosciences) and labeled with anti-pe-microbeads
Supplements Supplemental Materials and Methods Depletion of CD25 + T-cells from PBMC. Fresh or HD precultured PBMC were stained with the conjugate CD25-PE (BD Biosciences) and labeled with anti-pe-microbeads
Low Avidity CMV + T Cells accumulate in Old Humans
 Supplementary Figure Legends Supplementary Figure 1. CD45RA expressing CMVpp65-specific T cell populations accumulate within HLA-A*0201 and HLA-B*0701 individuals Pooled data showing the size of the NLV/HLA-A*0201-specific
Supplementary Figure Legends Supplementary Figure 1. CD45RA expressing CMVpp65-specific T cell populations accumulate within HLA-A*0201 and HLA-B*0701 individuals Pooled data showing the size of the NLV/HLA-A*0201-specific
Bezzi et al., Supplementary Figure 1 *** Nature Medicine: doi: /nm Pten pc-/- ;Zbtb7a pc-/- Pten pc-/- ;Pml pc-/- Pten pc-/- ;Trp53 pc-/-
 Gr-1 Gr-1 Gr-1 Bezzi et al., Supplementary Figure 1 a Gr1-CD11b 3 months Spleen T cells 3 months Spleen B cells 3 months Spleen Macrophages 3 months Spleen 15 4 8 6 c CD11b+/Gr1+ cells [%] 1 5 b T cells
Gr-1 Gr-1 Gr-1 Bezzi et al., Supplementary Figure 1 a Gr1-CD11b 3 months Spleen T cells 3 months Spleen B cells 3 months Spleen Macrophages 3 months Spleen 15 4 8 6 c CD11b+/Gr1+ cells [%] 1 5 b T cells
Supplementary Figure 1
 d f a IL7 b IL GATA RORγt h HDM IL IL7 PBS Ilra R7 PBS HDM Ilra R7 HDM Foxp Foxp Ilra R7 HDM HDM Ilra R7 HDM. 9..79. CD + FOXP + T reg cell CD + FOXP T conv cell PBS Ilra R7 PBS HDM Ilra R7 HDM CD + FOXP
d f a IL7 b IL GATA RORγt h HDM IL IL7 PBS Ilra R7 PBS HDM Ilra R7 HDM Foxp Foxp Ilra R7 HDM HDM Ilra R7 HDM. 9..79. CD + FOXP + T reg cell CD + FOXP T conv cell PBS Ilra R7 PBS HDM Ilra R7 HDM CD + FOXP
Supplementary Figure 1. Antibiotic partially rescues mice from sepsis. (ab) BALB/c mice under CLP were treated with antibiotic or PBS.
 1 Supplementary Figure 1. Antibiotic partially rescues mice from sepsis. (ab) BALB/c mice under CLP were treated with antibiotic or PBS. (a) Survival curves. WT Sham (n=5), WT CLP or WT CLP antibiotic
1 Supplementary Figure 1. Antibiotic partially rescues mice from sepsis. (ab) BALB/c mice under CLP were treated with antibiotic or PBS. (a) Survival curves. WT Sham (n=5), WT CLP or WT CLP antibiotic
CD80 and PD-L2 define functionally distinct memory B cell subsets that are. Griselda V Zuccarino-Catania, Saheli Sadanand, Florian J Weisel, Mary M
 Supplementary Figures CD8 and PD-L define functionally distinct memory B cell subsets that are independent of antibody isotype Running title: Memory B Cell Subset Function Griselda V Zuccarino-Catania,
Supplementary Figures CD8 and PD-L define functionally distinct memory B cell subsets that are independent of antibody isotype Running title: Memory B Cell Subset Function Griselda V Zuccarino-Catania,
NK cell flow cytometric assay In vivo DC viability and migration assay
 NK cell flow cytometric assay 6 NK cells were purified, by negative selection with the NK Cell Isolation Kit (Miltenyi iotec), from spleen and lymph nodes of 6 RAG1KO mice, injected the day before with
NK cell flow cytometric assay 6 NK cells were purified, by negative selection with the NK Cell Isolation Kit (Miltenyi iotec), from spleen and lymph nodes of 6 RAG1KO mice, injected the day before with
Supplementary Fig. 1 p38 MAPK negatively regulates DC differentiation. (a) Western blot analysis of p38 isoform expression in BM cells, immature DCs
 Supplementary Fig. 1 p38 MAPK negatively regulates DC differentiation. (a) Western blot analysis of p38 isoform expression in BM cells, immature DCs (idcs) and mature DCs (mdcs). A myeloma cell line expressing
Supplementary Fig. 1 p38 MAPK negatively regulates DC differentiation. (a) Western blot analysis of p38 isoform expression in BM cells, immature DCs (idcs) and mature DCs (mdcs). A myeloma cell line expressing
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. NKT ligand-loaded tumour antigen-presenting B cell- and monocyte-based vaccine induces NKT, NK and CD8 T cell responses. (A) The cytokine profiles of liver
Supplementary Figures Supplementary Figure 1. NKT ligand-loaded tumour antigen-presenting B cell- and monocyte-based vaccine induces NKT, NK and CD8 T cell responses. (A) The cytokine profiles of liver
L-selectin Is Essential for Delivery of Activated CD8 + T Cells to Virus-Infected Organs for Protective Immunity
 Cell Reports Supplemental Information L-selectin Is Essential for Delivery of Activated CD8 + T Cells to Virus-Infected Organs for Protective Immunity Rebar N. Mohammed, H. Angharad Watson, Miriam Vigar,
Cell Reports Supplemental Information L-selectin Is Essential for Delivery of Activated CD8 + T Cells to Virus-Infected Organs for Protective Immunity Rebar N. Mohammed, H. Angharad Watson, Miriam Vigar,
Supplementary Figure 1
 Supplementary Figure 1 Expression of apoptosis-related genes in tumor T reg cells. (a) Identification of FOXP3 T reg cells by FACS. CD45 + cells were gated as enriched lymphoid cell populations with low-granularity.
Supplementary Figure 1 Expression of apoptosis-related genes in tumor T reg cells. (a) Identification of FOXP3 T reg cells by FACS. CD45 + cells were gated as enriched lymphoid cell populations with low-granularity.
NKTR-255: Accessing The Immunotherapeutic Potential Of IL-15 for NK Cell Therapies
 NKTR-255: Accessing The Immunotherapeutic Potential Of IL-15 for NK Cell Therapies Saul Kivimäe Senior Scientist, Research Biology Nektar Therapeutics NK Cell-Based Cancer Immunotherapy, September 26-27,
NKTR-255: Accessing The Immunotherapeutic Potential Of IL-15 for NK Cell Therapies Saul Kivimäe Senior Scientist, Research Biology Nektar Therapeutics NK Cell-Based Cancer Immunotherapy, September 26-27,
Mouse Clec9a ORF sequence
 Mouse Clec9a gene LOCUS NC_72 13843 bp DNA linear CON 1-JUL-27 DEFINITION Mus musculus chromosome 6, reference assembly (C57BL/6J). ACCESSION NC_72 REGION: 129358881-129372723 Mouse Clec9a ORF sequence
Mouse Clec9a gene LOCUS NC_72 13843 bp DNA linear CON 1-JUL-27 DEFINITION Mus musculus chromosome 6, reference assembly (C57BL/6J). ACCESSION NC_72 REGION: 129358881-129372723 Mouse Clec9a ORF sequence
Supplementary Materials for
 immunology.sciencemag.org/cgi/content/full/2/16/eaan6049/dc1 Supplementary Materials for Enzymatic synthesis of core 2 O-glycans governs the tissue-trafficking potential of memory CD8 + T cells Jossef
immunology.sciencemag.org/cgi/content/full/2/16/eaan6049/dc1 Supplementary Materials for Enzymatic synthesis of core 2 O-glycans governs the tissue-trafficking potential of memory CD8 + T cells Jossef
SUPPLEMENTARY INFORMATION
 Supplemental Figure 1. Furin is efficiently deleted in CD4 + and CD8 + T cells. a, Western blot for furin and actin proteins in CD4cre-fur f/f and fur f/f Th1 cells. Wild-type and furin-deficient CD4 +
Supplemental Figure 1. Furin is efficiently deleted in CD4 + and CD8 + T cells. a, Western blot for furin and actin proteins in CD4cre-fur f/f and fur f/f Th1 cells. Wild-type and furin-deficient CD4 +
Supplementary Figure S1. Flow cytometric analysis of the expression of Thy1 in NH cells. Flow cytometric analysis of the expression of T1/ST2 and
 Supplementary Figure S1. Flow cytometric analysis of the expression of Thy1 in NH cells. Flow cytometric analysis of the expression of T1/ST2 and Thy1 in NH cells derived from the lungs of naïve mice.
Supplementary Figure S1. Flow cytometric analysis of the expression of Thy1 in NH cells. Flow cytometric analysis of the expression of T1/ST2 and Thy1 in NH cells derived from the lungs of naïve mice.
Supplemental Figure Legends
 Supplemental Figure Legends Supplemental Figure 1. SemaB / mice have normal immune cell populations. Cells were prepared from the spleens of WT and SemaB / mice, stained with various antibodies and then
Supplemental Figure Legends Supplemental Figure 1. SemaB / mice have normal immune cell populations. Cells were prepared from the spleens of WT and SemaB / mice, stained with various antibodies and then
The encephalitogenicity of TH17 cells is dependent on IL-1- and IL-23- induced production of the cytokine GM-CSF
 CORRECTION NOTICE Nat.Immunol. 12, 568 575 (2011) The encephalitogenicity of TH17 cells is dependent on IL-1- and IL-23- induced production of the cytokine GM-CSF Mohamed El-Behi, Bogoljub Ciric, Hong
CORRECTION NOTICE Nat.Immunol. 12, 568 575 (2011) The encephalitogenicity of TH17 cells is dependent on IL-1- and IL-23- induced production of the cytokine GM-CSF Mohamed El-Behi, Bogoljub Ciric, Hong
Supplementary Figure 1. IL-12 serum levels and frequency of subsets in FL patients. (A) IL-12
 1 Supplementary Data Figure legends Supplementary Figure 1. IL-12 serum levels and frequency of subsets in FL patients. (A) IL-12 serum levels measured by multiplex ELISA (Luminex) in FL patients before
1 Supplementary Data Figure legends Supplementary Figure 1. IL-12 serum levels and frequency of subsets in FL patients. (A) IL-12 serum levels measured by multiplex ELISA (Luminex) in FL patients before
activation with anti-cd3/cd28 beads and 3d following transduction. Supplemental Figure 2 shows
 Supplemental Data Supplemental Figure 1 compares CXCR4 expression in untreated CD8 + T cells, following activation with anti-cd3/cd28 beads and 3d following transduction. Supplemental Figure 2 shows the
Supplemental Data Supplemental Figure 1 compares CXCR4 expression in untreated CD8 + T cells, following activation with anti-cd3/cd28 beads and 3d following transduction. Supplemental Figure 2 shows the
B220 CD4 CD8. Figure 1. Confocal Image of Sensitized HLN. Representative image of a sensitized HLN
 B220 CD4 CD8 Natarajan et al., unpublished data Figure 1. Confocal Image of Sensitized HLN. Representative image of a sensitized HLN showing B cell follicles and T cell areas. 20 µm thick. Image of magnification
B220 CD4 CD8 Natarajan et al., unpublished data Figure 1. Confocal Image of Sensitized HLN. Representative image of a sensitized HLN showing B cell follicles and T cell areas. 20 µm thick. Image of magnification
W/T Itgam -/- F4/80 CD115. F4/80 hi CD115 + F4/80 + CD115 +
 F4/8 % in the peritoneal lavage 6 4 2 p=.15 n.s p=.76 CD115 F4/8 hi CD115 + F4/8 + CD115 + F4/8 hi CD115 + F4/8 + CD115 + MHCII MHCII Supplementary Figure S1. CD11b deficiency affects the cellular responses
F4/8 % in the peritoneal lavage 6 4 2 p=.15 n.s p=.76 CD115 F4/8 hi CD115 + F4/8 + CD115 + F4/8 hi CD115 + F4/8 + CD115 + MHCII MHCII Supplementary Figure S1. CD11b deficiency affects the cellular responses
Nature Immunology: doi: /ni Supplementary Figure 1. Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice.
 Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
HD1 (FLU) HD2 (EBV) HD2 (FLU)
 ramer staining + anti-pe beads ramer staining a HD1 (FLU) HD2 (EBV) HD2 (FLU).73.11.56.46.24 1.12 b CD127 + c CD127 + d CD127 - e CD127 - PD1 - PD1 + PD1 + PD1-1 1 1 1 %CD127 + PD1-8 6 4 2 + anti-pe %CD127
ramer staining + anti-pe beads ramer staining a HD1 (FLU) HD2 (EBV) HD2 (FLU).73.11.56.46.24 1.12 b CD127 + c CD127 + d CD127 - e CD127 - PD1 - PD1 + PD1 + PD1-1 1 1 1 %CD127 + PD1-8 6 4 2 + anti-pe %CD127
Signal 3 requirement for memory CD8 1 T-cell activation is determined by the infectious pathogen
 3176 DOI 1.12/eji.21141537 Eur. J. Immunol. 211. 41: 3176 3186 Signal 3 requirement for memory CD8 1 T-cell activation is determined by the infectious pathogen Selina J. Keppler 1,2 and Peter Aichele 1
3176 DOI 1.12/eji.21141537 Eur. J. Immunol. 211. 41: 3176 3186 Signal 3 requirement for memory CD8 1 T-cell activation is determined by the infectious pathogen Selina J. Keppler 1,2 and Peter Aichele 1
Out-of-sequence signal 3 as a mechanism for virusinduced immune suppression of CD8 T cell responses
 University of Massachusetts Medical School escholarship@umms Open Access Articles Open Access Publications by UMMS Authors 9-25-2014 Out-of-sequence signal 3 as a mechanism for virusinduced immune suppression
University of Massachusetts Medical School escholarship@umms Open Access Articles Open Access Publications by UMMS Authors 9-25-2014 Out-of-sequence signal 3 as a mechanism for virusinduced immune suppression
Supplementary Figure 1. Efficient DC depletion in CD11c.DOG transgenic mice
 Supplementary Figure 1. Efficient DC depletion in CD11c.DOG transgenic mice (a) CD11c.DOG transgenic mice (tg) were treated with 8 ng/g body weight (b.w.) diphtheria toxin (DT) i.p. on day -1 and every
Supplementary Figure 1. Efficient DC depletion in CD11c.DOG transgenic mice (a) CD11c.DOG transgenic mice (tg) were treated with 8 ng/g body weight (b.w.) diphtheria toxin (DT) i.p. on day -1 and every
Manuscript: OX40 signaling is involved in the autoactivation of CD4 + CD28 - T cells and contributes to pathogenesis of autoimmune arthritis
 Manuscript: OX40 signaling is involved in the autoactivation of CD4 + CD28 - T cells and contributes to pathogenesis of autoimmune arthritis Vincent Laufer Rheumatology JC October 17 Disclosures NIH 2
Manuscript: OX40 signaling is involved in the autoactivation of CD4 + CD28 - T cells and contributes to pathogenesis of autoimmune arthritis Vincent Laufer Rheumatology JC October 17 Disclosures NIH 2
1,000 in silico simulated alpha, beta, gamma and delta TCR repertoires were created.
 938 939 940 941 942 Figure S1 Schematic of the in silico TCRminer and MiXCR validation. 1,000 in silico simulated alpha, beta, gamma and delta TCR repertoires were created. Then, 100,000 simulated 80 bp
938 939 940 941 942 Figure S1 Schematic of the in silico TCRminer and MiXCR validation. 1,000 in silico simulated alpha, beta, gamma and delta TCR repertoires were created. Then, 100,000 simulated 80 bp
Appendix Figure S1 A B C D E F G H
 ppendix Figure S1 C D E F G H ppendix Figure S1. RT and chemotherapy alter PD-L1 expression in PDC cells. Flow cytometric analysis of PD-L1 expression in () KPC and () Pan02 cells following treatment with
ppendix Figure S1 C D E F G H ppendix Figure S1. RT and chemotherapy alter PD-L1 expression in PDC cells. Flow cytometric analysis of PD-L1 expression in () KPC and () Pan02 cells following treatment with
Supplementary Materials for
 www.sciencetranslationalmedicine.org/cgi/content/full/8/352/352ra110/dc1 Supplementary Materials for Spatially selective depletion of tumor-associated regulatory T cells with near-infrared photoimmunotherapy
www.sciencetranslationalmedicine.org/cgi/content/full/8/352/352ra110/dc1 Supplementary Materials for Spatially selective depletion of tumor-associated regulatory T cells with near-infrared photoimmunotherapy
Supplemental Figure 1. Characterization of CNS lysosomal pathology in 2-month-old IDS-deficient mice. (A) Measurement of IDS activity in brain
 Supplemental Figure 1. Characterization of CNS lysosomal pathology in 2-month-old IDS-deficient mice. (A) Measurement of IDS activity in brain extracts from wild-type (WT) and IDS-deficient males. IDS
Supplemental Figure 1. Characterization of CNS lysosomal pathology in 2-month-old IDS-deficient mice. (A) Measurement of IDS activity in brain extracts from wild-type (WT) and IDS-deficient males. IDS
An integrated transcriptomic and functional genomic approach identifies a. type I interferon signature as a host defense mechanism against Candida
 Supplementary information for: An integrated transcriptomic and functional genomic approach identifies a type I interferon signature as a host defense mechanism against Candida albicans in humans Sanne
Supplementary information for: An integrated transcriptomic and functional genomic approach identifies a type I interferon signature as a host defense mechanism against Candida albicans in humans Sanne
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
SUPPLEMENT Supplementary Figure 1: (A) (B)
 SUPPLEMENT Supplementary Figure 1: CD4 + naïve effector T cells (CD4 effector) were labeled with CFSE, stimulated with α-cd2/cd3/cd28 coated beads (at 2 beads/cell) and cultured alone or cocultured with
SUPPLEMENT Supplementary Figure 1: CD4 + naïve effector T cells (CD4 effector) were labeled with CFSE, stimulated with α-cd2/cd3/cd28 coated beads (at 2 beads/cell) and cultured alone or cocultured with
Supplemental Figure 1. Immune phenotype of mcd19 targeted CAR T and dose titration of in vivo
 Supplemental Materials Supplemental Methods Supplemental Figure 1. Immune phenotype of mcd19 targeted CAR T and dose titration of in vivo efficacy. Supplemental Figure 2. Gene expression of fluorescent-protein
Supplemental Materials Supplemental Methods Supplemental Figure 1. Immune phenotype of mcd19 targeted CAR T and dose titration of in vivo efficacy. Supplemental Figure 2. Gene expression of fluorescent-protein
Nature Immunology: doi: /ni Supplementary Figure 1. Cellularity of leukocytes and their progenitors in naive wild-type and Spp1 / mice.
 Supplementary Figure 1 Cellularity of leukocytes and their progenitors in naive wild-type and Spp1 / mice. (a, b) Gating strategies for differentiated cells including PMN (CD11b + Ly6G hi and CD11b + Ly6G
Supplementary Figure 1 Cellularity of leukocytes and their progenitors in naive wild-type and Spp1 / mice. (a, b) Gating strategies for differentiated cells including PMN (CD11b + Ly6G hi and CD11b + Ly6G
Fluorochrome Panel 1 Panel 2 Panel 3 Panel 4 Panel 5 CTLA-4 CTLA-4 CD15 CD3 FITC. Bio) PD-1 (MIH4, BD) ICOS (C398.4A, Biolegend) PD-L1 (MIH1, BD)
 Additional file : Table S. Antibodies used for panel stain to identify peripheral immune cell subsets. Panel : PD- signaling; Panel : CD + T cells, CD + T cells, B cells; Panel : Tregs; Panel :, -T, cdc,
Additional file : Table S. Antibodies used for panel stain to identify peripheral immune cell subsets. Panel : PD- signaling; Panel : CD + T cells, CD + T cells, B cells; Panel : Tregs; Panel :, -T, cdc,
NKTR-255: Accessing IL-15 Therapeutic Potential through Robust and Sustained Engagement of Innate and Adaptive Immunity
 NKTR-255: Accessing IL-15 Therapeutic Potential through Robust and Sustained Engagement of Innate and Adaptive Immunity Peiwen Kuo Scientist, Research Biology Nektar Therapeutics August 31 st, 2018 Emerging
NKTR-255: Accessing IL-15 Therapeutic Potential through Robust and Sustained Engagement of Innate and Adaptive Immunity Peiwen Kuo Scientist, Research Biology Nektar Therapeutics August 31 st, 2018 Emerging
Reviewers' comments: Reviewer #1 (Remarks to the Author):
 Reviewers' comments: Reviewer #1 (Remarks to the Author): In this interesting article, Kondo et al demonstrate that Notch signaling can induce Tscm-like fate in activated T cells. This work extends prior
Reviewers' comments: Reviewer #1 (Remarks to the Author): In this interesting article, Kondo et al demonstrate that Notch signaling can induce Tscm-like fate in activated T cells. This work extends prior
Belatacept: An Opportunity to Personalize Immunosuppression? Andrew Adams MD/PhD Emory Transplant Center
 Belatacept: An Opportunity to Personalize Immunosuppression? Andrew Adams MD/PhD Emory Transplant Center Disclosure Research Funding from BMS. Learning Objectives -Define belatacept-resistant rejection
Belatacept: An Opportunity to Personalize Immunosuppression? Andrew Adams MD/PhD Emory Transplant Center Disclosure Research Funding from BMS. Learning Objectives -Define belatacept-resistant rejection
well for 2 h at rt. Each dot represents an individual mouse and bar is the mean ±
 Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Figure S1. Gating strategy used in NK cells and γδ T lymphocytes coculture An example of flow cytometry analysis shows the gating of NK cells and γδ
 Figure S1. Gating strategy used in NK cells and γδ T lymphocytes coculture An example of flow cytometry analysis shows the gating of NK cells and γδ T lymphocytes used in all NK activation and cytotoxicity
Figure S1. Gating strategy used in NK cells and γδ T lymphocytes coculture An example of flow cytometry analysis shows the gating of NK cells and γδ T lymphocytes used in all NK activation and cytotoxicity
Nature Protocols: doi: /nprot Supplementary Figure 1
 Supplementary Figure 1 Traditional electronic gating strategy for analysing cell death based on A5-FITC and 7-AAD. a, Flow cytometry analysis showing the traditional two-stage electronic gating strategy
Supplementary Figure 1 Traditional electronic gating strategy for analysing cell death based on A5-FITC and 7-AAD. a, Flow cytometry analysis showing the traditional two-stage electronic gating strategy
Programmed death-1 (PD-1) defines a transient and dysfunctional oligoclonal T cell population in acute homeostatic proliferation
 University of Massachusetts Medical School escholarship@umms Open Access Articles Open Access Publications by UMMS Authors 9-6-2007 Programmed death-1 (PD-1) defines a transient and dysfunctional oligoclonal
University of Massachusetts Medical School escholarship@umms Open Access Articles Open Access Publications by UMMS Authors 9-6-2007 Programmed death-1 (PD-1) defines a transient and dysfunctional oligoclonal
Supplemental Information. Aryl Hydrocarbon Receptor Controls. Monocyte Differentiation. into Dendritic Cells versus Macrophages
 Immunity, Volume 47 Supplemental Information Aryl Hydrocarbon Receptor Controls Monocyte Differentiation into Dendritic Cells versus Macrophages Christel Goudot, Alice Coillard, Alexandra-Chloé Villani,
Immunity, Volume 47 Supplemental Information Aryl Hydrocarbon Receptor Controls Monocyte Differentiation into Dendritic Cells versus Macrophages Christel Goudot, Alice Coillard, Alexandra-Chloé Villani,
Nature Immunology: doi: /ni Supplementary Figure 1. Id2 and Id3 define polyclonal T H 1 and T FH cell subsets.
 Supplementary Figure 1 Id2 and Id3 define polyclonal T H 1 and T FH cell subsets. Id2 YFP/+ (a) or Id3 GFP/+ (b) mice were analyzed 7 days after LCMV infection. T H 1 (SLAM + CXCR5 or CXCR5 PD-1 ), T FH
Supplementary Figure 1 Id2 and Id3 define polyclonal T H 1 and T FH cell subsets. Id2 YFP/+ (a) or Id3 GFP/+ (b) mice were analyzed 7 days after LCMV infection. T H 1 (SLAM + CXCR5 or CXCR5 PD-1 ), T FH
Supplementary Materials for
 www.sciencemag.org/content/348/6241/aaa825/suppl/dc1 Supplementary Materials for A mucosal vaccine against Chlamydia trachomatis generates two waves of protective memory T cells Georg Stary,* Andrew Olive,
www.sciencemag.org/content/348/6241/aaa825/suppl/dc1 Supplementary Materials for A mucosal vaccine against Chlamydia trachomatis generates two waves of protective memory T cells Georg Stary,* Andrew Olive,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Pleiotrophin Regulates the Expansion and Regeneration of Hematopoietic Stem Cells Heather A Himburg 1, Garrett G Muramoto 1 *, Pamela Daher 1*, Sarah K Meadows 1, J. Lauren Russell
SUPPLEMENTARY INFORMATION Pleiotrophin Regulates the Expansion and Regeneration of Hematopoietic Stem Cells Heather A Himburg 1, Garrett G Muramoto 1 *, Pamela Daher 1*, Sarah K Meadows 1, J. Lauren Russell
Supplementary Information
 Supplementary Information Supplementary Figure 1! a! b! Nfatc1!! Nfatc1"! P1! P2! pa1! pa2! ex1! ex2! exons 3-9! ex1! ex11!!" #" Nfatc1A!!" Nfatc1B! #"!" Nfatc1C! #" DN1! DN2! DN1!!A! #A!!B! #B!!C! #C!!A!
Supplementary Information Supplementary Figure 1! a! b! Nfatc1!! Nfatc1"! P1! P2! pa1! pa2! ex1! ex2! exons 3-9! ex1! ex11!!" #" Nfatc1A!!" Nfatc1B! #"!" Nfatc1C! #" DN1! DN2! DN1!!A! #A!!B! #B!!C! #C!!A!
Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion
 Probe Set ID RefSeq Transcript ID Gene Title Gene Symbol 205980_s_at NM_001017526 /// NM_181334 /// NM_181335 Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion ARHGAP8 /// LOC553158 FC ALK shrna
Probe Set ID RefSeq Transcript ID Gene Title Gene Symbol 205980_s_at NM_001017526 /// NM_181334 /// NM_181335 Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion ARHGAP8 /// LOC553158 FC ALK shrna
Supplementary Figure 1. Using DNA barcode-labeled MHC multimers to generate TCR fingerprints
 Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
Supplementary Figure 1 Lymphocytes can be tracked for at least 4 weeks after
 Supplementary Figure 1 Lymphocytes can be tracked for at least 4 weeks after photoconversion by using H2B-Dendra2. 4-5 PPs of H2B-Dendra2 BM chimeras were photoconverted and analyzed 7 days (upper panel)
Supplementary Figure 1 Lymphocytes can be tracked for at least 4 weeks after photoconversion by using H2B-Dendra2. 4-5 PPs of H2B-Dendra2 BM chimeras were photoconverted and analyzed 7 days (upper panel)
Therapeutic PD L1 and LAG 3 blockade rapidly clears established blood stage Plasmodium infection
 Supplementary Information Therapeutic PD L1 and LAG 3 blockade rapidly clears established blood stage Plasmodium infection Noah S. Butler, Jacqueline Moebius, Lecia L. Pewe, Boubacar Traore, Ogobara K.
Supplementary Information Therapeutic PD L1 and LAG 3 blockade rapidly clears established blood stage Plasmodium infection Noah S. Butler, Jacqueline Moebius, Lecia L. Pewe, Boubacar Traore, Ogobara K.
Regulating the Regulators for Cancer Immunotherapy: LAG-3 Finally Catches Up. Drew Pardoll Sidney Kimmel Cancer Center Johns Hopkins
 Regulating the Regulators for Cancer Immunotherapy: LAG-3 Finally Catches Up Drew Pardoll Sidney Kimmel Cancer Center Johns Hopkins The hostile immune microenvironment within a tumor Stat3 Stat3 NK Lytic
Regulating the Regulators for Cancer Immunotherapy: LAG-3 Finally Catches Up Drew Pardoll Sidney Kimmel Cancer Center Johns Hopkins The hostile immune microenvironment within a tumor Stat3 Stat3 NK Lytic
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplemental Information. Checkpoint Blockade Immunotherapy. Induces Dynamic Changes. in PD-1 CD8 + Tumor-Infiltrating T Cells
 Immunity, Volume 50 Supplemental Information Checkpoint Blockade Immunotherapy Induces Dynamic Changes in PD-1 CD8 + Tumor-Infiltrating T Cells Sema Kurtulus, Asaf Madi, Giulia Escobar, Max Klapholz, Jackson
Immunity, Volume 50 Supplemental Information Checkpoint Blockade Immunotherapy Induces Dynamic Changes in PD-1 CD8 + Tumor-Infiltrating T Cells Sema Kurtulus, Asaf Madi, Giulia Escobar, Max Klapholz, Jackson
TITLE: MODULATION OF T CELL TOLERANCE IN A MURINE MODEL FOR IMMUNOTHERAPY OF PROSTATIC ADENOCARCINOMA
 AD Award Number: DAMD17-01-1-0085 TITLE: MODULATION OF T CELL TOLERANCE IN A MURINE MODEL FOR IMMUNOTHERAPY OF PROSTATIC ADENOCARCINOMA PRINCIPAL INVESTIGATOR: ARTHUR A HURWITZ, Ph.d. CONTRACTING ORGANIZATION:
AD Award Number: DAMD17-01-1-0085 TITLE: MODULATION OF T CELL TOLERANCE IN A MURINE MODEL FOR IMMUNOTHERAPY OF PROSTATIC ADENOCARCINOMA PRINCIPAL INVESTIGATOR: ARTHUR A HURWITZ, Ph.d. CONTRACTING ORGANIZATION:
Supplementary Figure 1. Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Nature Immunology: doi: /ni.
 Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
