Single cell tuning of Myc expression by antigen receptor signal strength and interleukin-2 in T lymphocytes
|
|
|
- Joel Poole
- 5 years ago
- Views:
Transcription
1 Article Single cell tuning of expression by antigen receptor signal strength and interleukin-2 in T lymphocytes Gavin C Preston 1, Linda V Sinclair 1, Aneesa Kaskar 1,2, Jens L Hukelmann 1,2, Maria N Navarro 1,3, Isabel Ferrero 4, H Robson MacDonald 4, Victoria H Cowling 2 & Doreen A Cantrell 1,* Abstract controls the metabolic reprogramming that supports effector T cell differentiation. The expression of is regulated by the T cell antigen receptor (TCR) and pro-inflammatory cytokines such as interleukin-2 (). We now show that the TCR is a digital switch for mrna and protein expression that allows the strength of the antigen stimulus to determine the frequency of T cells that express. signalling strength also directs expression but in an analogue process that fine-tunes quantity in individual cells via post-transcriptional control of protein. Fine-tuning matters and is possible as protein has a very short half-life in T cells due to its constant phosphorylation by glycogen synthase kinase 3 (GSK3) and subsequent proteasomal degradation. We show that only accumulates in T cells exhibiting high levels of amino acid uptake allowing T cells to match expression to biosynthetic demands. The combination of digital and analogue processes allows tight control of expression at the population and single cell level during immune responses. Keywords cytokine signals; metabolism; ; T lymphocytes; TCR signals Subject Categories Immunology; Signal Transduction DOI /embj Received 7 October 214 Revised 15 May 215 Accepted 18 May 215 Published online 1 July 215 The EMBO Journal (215) 34: See also: C Chou & T Egawa (August 215) Introduction Immune-activated T lymphocytes undergo rapid clonal expansion and differentiation into effector subpopulations. Activated T cells also strikingly increase nutrient uptake and lipid and protein biosynthesis. Naïve T lymphocytes thus have low rates of amino acid and glucose uptake and use oxidative phosphorylation to efficiently metabolise glucose to generate ATP. In contrast, effector T cells upregulate amino acid and glucose uptake and switch to metabolising glucose through glycolysis (Greiner et al, 1994; Fox et al, 25; Jacobs et al, 28); they also metabolise glutamine to pyruvate and lactate via glutaminolysis (Brand et al, 1984; Newsholme et al, 1985). These changes in T cell metabolism are necessary to support the production of the cytokines and cytolytic effector molecules that are essential for T cell immune responses (Pollizzi & Powell, 214). One key controller of metabolic reprogramming in T cells is (myelocytomatosis oncogene, c-). In the absence of, mature T cells cannot respond to antigen receptor engagement to increase the expression of glucose and glutamine transporters and fail to initiate glycolysis or glutamine catabolism (Wang et al, 211a). loss also has a global impact on the T cell transcriptome. may thus have multiple transcriptional targets or act to amplify expression of active genes in T cells (Nie et al, 212). The importance of in T lymphocytes is also exemplified by its role in T cell development in the thymus (Dose et al, 26, 29; ko et al, 29; Jiang et al, 21) and by its key role in T cell malignancies. For example, elevated expression is observed in most cases of T cell acute lymphoblastic leukaemia (T-ALL), either as a consequence of a chromosomal translocation event (t(8;14)(q24; q11)) that places under the control of the TCR alpha chain promoter (Erikson et al, 1986; Charrin, 1996) or more commonly as a consequence of Notch mutations that lead to transcriptional activation of (Weng et al, 24, 26; Palomero et al, 26). It was recognised many years ago that upregulation of mrna accompanied T cell activation (Reed et al, 1987; Kelly & Siebenlist, 1988; Wang et al, 211a). More recently, the Immunological Genome Project has mapped the transcriptional profiles of multiple T cell populations and reported high levels of mrna in immune-activated effector T cells (Best et al, 213). However, the expression of mrna and protein can be quite discordant because of post-transcriptional control of protein (Vervoorts et al, 26; Junttila & Westermarck, 28; Thomas & Tansey, 211; Ehninger et al, 214). For example, stabilisation of protein by a 1 Department of Cell Signalling & Immunology, College of Life Sciences, University of Dundee, Dundee, UK 2 Centre for Gene Regulation and Expression, College of Life Sciences, University of Dundee, Dundee, UK 3 Instituto Investigación Sanitaria/Hospital Universitario de la Princesa, Universidad Autónoma de Madrid, Madrid, Spain 4 Ludwig Center for Cancer Research of the University of Lausanne, Epalinges, Switzerland *Corresponding author. Tel: ; d.a.cantrell@dundee.ac.uk 28 The EMBO Journal Vol 34 No ª 215 The Authors. Published under the terms of the CC BY 4. license
2 Gavin C Preston et al T cell expression matched to metabolic demand The EMBO Journal T58A mutation has been described in Burkitt s lymphoma (Yano et al, 1993; Gregory & Hann, 2) and inactivating mutations of the E3 ubiquitin ligase Fbw7 that target for proteasomal degradation are frequently found in T-ALL (Welcker et al, 24a; O Neil et al, 27). The relevance of understanding the control of protein levels in T cells was highlighted recently: immune-activated T cells lacking the system L amino acid transporter Slc7a5 upregulate mrna but fail to express protein and hence have global metabolic defects (Sinclair et al, 213). In this context, many experiments have analysed mrna in T cells activated pharmacologically with phorbol esters, calcium ionophores and plant lectins (Kelly et al, 1983; Reed et al, 1987, 1988; Kelly & Siebenlist, 1988), but there is almost no detailed analysis of protein in T cells responding to physiological stimuli. Accordingly, the present study has explored how engagement of the T cell antigen receptor (TCR) with peptide/ major histocompatibility complexes (p/mhc) controls expression. We have also examined how a key pro-inflammatory cytokine interleukin-2 () impacts on expression. We show that the TCR is a digital switch that couples the strength of the antigen stimulus to the frequency of cells within a population that express. However, TCR triggering alone cannot sustain expression of and post-transcriptional control of protein by, a cc cytokine that triggers Janus-associated kinase (JAK)-dependent signalling pathways, fine-tunes levels in individual T cells. We show that is constantly phosphorylated by the serine/threonine kinase GSK3 in activated T cells and targeted for proteasomal degradation. protein thus only accumulates in T cells with high levels of protein synthesis. This study also affords the insight that the cellular concentration of matters in the context of T cell metabolism. Results TCR signal strength determines protein expression in a digital manner The expression of in T cells is controlled by the TCR (Wang et al, 211a). A crucial question is how TCR signalling strength controls the pool size of T cells that differentiate to effector cells. In this respect, although is required for T cell differentiation, the impact of antigen signalling strength on expression has not been explored. To explore the sensitivity of induction to different strengths of TCR ligand, we used the OT1 TCR transgenic model where there are well-characterised peptide ligands with different affinities for the TCR (Daniels et al, 26). These permit exploration of how the affinity of peptide-major histocompatibility complexes (p/mhc), the physiological ligand for TCR complexes, dictates expression. In these experiments, we activated OT1 T cells with TCR agonists of varying affinity: SIIQFEHL (Q4H7: K d nm), SIITFEKL (T4: K d of nm), SIIQFERL (Q4R7: K d nm), SIIQFEKL (Q4: K d nm) and SIINFEKL (N4: K d nm) (Daniels et al, 26). Western blot analysis of OT1 T cells shows that the level of protein within a T cell population is determined by the strength of the TCR ligand (Fig 1A). Cellular levels of mrna are also determined by TCR ligand strength (Fig 1B), revealing that one fundamental way TCR agonists control expression in T cells is by controlling the expression of mrna. Changes in levels in response to TCR ligand strength could reflect that ligand affinity governs the frequency of T cells within a population that express. Alternatively, TCR ligand strength may dictate the amount of expressed by each individual activated T cell. To distinguish these possibilities, we needed to directly visualise protein expression at the single cell level. Accordingly, we used a mouse model in which a fusion protein of and enhanced green fluorescent protein () is expressed from the endogenous locus ( KI ) (Huang et al, 28). We then used OT1 KI T cells to assess whether the strength of the TCR ligand influenced the frequency of T cells expressing within a population. Fig 1C shows the pattern of expression in OT1 T cells activated with TCR agonists of varying affinity for the TCR. is not expressed in naïve CD8 + T cells but is rapidly induced following exposure to TCR peptide agonists (2 h). Following TCR engagement, two distinct cell populations were observed: a population with high expression and a population with no detectable expression (Fig 1C, left panel). It was striking that the frequency of T cells that express is determined by the strength of the TCR ligand (Fig 1C, middle panel). This on/off Figure 1. T cell receptor signalling drives digital expression of. A, B CD8 + T cells were purified from OT1 lymph node cells that had been stimulated for 4 h with 1 ng/ml of peptides SIIQFEHL (Q4H7), SIIQFERL (Q4R7), SIIQFEKL (Q4) or SIINFEKL (N4) or were left unstimulated. (A) Western blot data show and ERK1/2 protein expression, representative of 3 biological replicates. (B) qpcr data show mrna expression relative to unstimulated naïve control (n = 3, mean SEM). C F Flow cytometry data from lymph node cells of OT1 KI mice stimulated through the TCR with peptide or left unstimulated. Data are from at least 3 biological replicates. (C) expression in CD8 + T cells stimulated for 2 h with 1 ng/ml peptides Q4H7,T4, Q4R7, Q4 or N4 (left panel). The middle panel shows the percentage pos cells (mean SEM, ANOVA was used to determine statistical significance, **P <.1). The right panel shows the MFIs of the GFP pos and GFP neg populations (mean SEM). (D) expression in CD8 + T cells stimulated for 2 h with varying concentrations of Q4 (ng/ml) or N4 (1 ng/ml) (left panel). The right panel shows the percentage pos cells (mean SEM, ANOVA was used to determine statistical significance, *P <.5, **P <.1). (E) (top panel) and CD69 (bottom panel) expression on CD8 + T cells stimulated for 2 h with varying concentrations of Q4 and T4 peptides, indicated on the histograms. The right panels show the percentage of pos or CD69 + populations. (F) expression in CD69 + or CD69 CD8 + T cells stimulated with 1.1 ng/ml of T4 peptide for 2 h (left panel). The right panel shows the MFI of the CD69 + populations of cells treated as in (E) (mean SEM). G neg and pos CD8 + T cells were purified from OT1 KI lymph node cells that had been activated for 2 h with N4 peptide (1 ng/ml) or had been left unstimulated. Graph shows the expression of mrna measured by qpcr relative to naïve unstimulated CD8 + T cells (*P <.5, ANOVA was used to determine statistical significance). Data show mean+sem of 3 biological replicates. H, I Flow cytometry data from lymph node cells of WT or KI mice stimulated using CD3 and CD28 antibodies or left unstimulated. Data are from at least 3 biological replicates. (H) expression over time in CD4 + or CD8 + T cells following stimulation with CD3 (1 lg/ml) and CD28 (3 lg/ml) antibodies. (I) The left panel shows the percentage pos CD8 + T cells stimulated with varying concentrations of CD3 (lg/ml as on graph) and CD28 (3 lg/ml) antibodies for 4 h. The right panel shows the MFIs of the pos and neg populations (mean SEM, ****P <.1, ANOVA was used to determine statistical significance). Source data are available online for this figure. ª 215 The Authors The EMBO Journal Vol 34 No
3 The EMBO Journal T cell expression matched to metabolic demand Gavin C Preston et al A 75 5 Erk 1/2 39 Unstim. Q4H7 Q4R7 Q4 N4 C GFP N4 Q4 Q4R7 T4 Q4H7 Unstim % GFP + cells ** Q4H7 T4 Q4R7 Q4 N4 1 3 MFI 1 1 pos neg Q4H7 T4 Q4R7 Q4 N4 B mrna rel.to naive naive. mrna Q4H7 Q4R7 Q4 N4 D GFP peptide (ng/ml) N4 1 Q4 1 Q4 3.3 Q4 1.1 Q4.3 Q4.1 Unstim % GFP + cells * ** N4 1 Q4 peptide (ng/ml) Increasing ligand potency E CD8 + Q4(ng/ml) CD8 + T4(ng/ml) %GFP+ cells Q4 T4 H 2hr KI Unstim WT CD3/CD28 KI CD4 CD8 GFP.1.1 Unstim CD %CD69+ cells Unstim peptide (ng/ml) 4hr 6hr F CD8 + T4 (1.1ng/ml) CD69 + CD GFP MFI 1 1 GFP- neg peptide (ng/ml) Q4 T4 G 5 mrna rel. to naive1 Naive mrna ns * neg pos 24hr 48hr I 5 **** 1 3 pos neg % GFP +ve cells MFI 72hr Unstim CD3 antibody (μg/ml) CD3 antibody (μg/ml) GFP Figure The EMBO Journal Vol 34 No ª 215 The Authors
4 Gavin C Preston et al T cell expression matched to metabolic demand The EMBO Journal pattern of expression suggested a bimodal/digital response pattern of regulation. Consistent with a digital response, the mean fluorescence intensity (MFI) of expression in the GFP- pos populations is not significantly different for the T4, Q4R7, Q4 or N4 peptides (Fig 1C, right panel). Hence, increasing the affinity of the TCR ligand does not increase the amount of in individual responding cells. We also examined expression following naïve OT1 T cell activation with increasing concentrations of the p/mhc ligand Q4. Fig 1D shows that the concentration of TCR ligand dictates the frequency of T cells that express. The bimodal, on/off pattern of expression was also maintained during a sustained (2 h) T cell response to p/mhc ligands (Fig 1E). Here we compared the TCR ligand dose response for induction of and the activation marker CD69. Fig 1E (bottom panel) shows that the upregulation of CD69 is more sensitive to the TCR ligand dose than induction (top panel). The expression of CD69 marks T cells that have responded to antigen, and in this respect, it was of note that expression was restricted to the CD69-expressing T cells and was bimodal in the CD69 pos TCRactivated lymphocytes (Fig 1F, left panel). We did however note that at this sustained time point, the level of p/mhc ligand not only directed the frequency of T cells that initiated expression but also influenced the level of per cell (Fig 1F, right panel). One important question was whether the frequency of cells that expressed protein in activated T cells was explained by differences in the frequency of T cells that express mrna. To answer this question, OT1 KI T cells were activated with the N4 TCR agonist for 2 h before pos and neg T cells were purified by FACS and mrna was quantified. Fig 1G shows that N4-stimulated pos OT1 T cells express high levels of mrna compared with neg OT1 T cells. This result is consistent with the hypothesis that the TCR controls the frequency of cells that express because the TCR controls the frequency of cells that express mrna. We next examined whether digital expression is common to both CD4 + and CD8 + T cells. Fig 1H shows that is not expressed in naïve lymph node-derived CD4 + and CD8 + T cells but is rapidly induced in both populations following polyclonal T cell activation with CD3 and CD28 antibodies with a clear digital response pattern of regulation. In these experiments, we used saturating levels of CD3 antibodies, and within the first few hours of the activation response, there was an increase in the percentage of T cells that expressed but no increase in the maximal amount of expressed. Under these potent activation conditions, all T cells expressed at the 24-h time point. This response was sustained over a 72-h period, but it was notable that the level of per cell had decreased at the 48- and 72-h time points (Fig 1H). When T cells were activated polyclonally with different concentrations of CD3 antibody, then the frequency of -expressing cells directly correlated with the amount of CD3 antibody used (Fig 1I), and the digital nature of the response was preserved. In these experiments, the level of the costimulatory antibody CD28 was constant, indicating that it is the level of TCR engagement that determined expression. The digital nature of induction in response to TCR engagement is thus a common feature of the TCR response to both physiological and polyclonal ligands and occurs in both CD4 + and CD8 + T cells. /JAK signalling controls expression in TCR-activated T cells The differentiation of antigen-activated CD8 + T cells is regulated by cc cytokines such as interleukin-2 (). Antigen-primed T cells cultured in thus differentiate to effector cytotoxic T cells (CTL) (Kalia et al, 21; Pipkin et al, 21). It has also been described that controls the expression of (Reed et al, 1988; Lord et al, 2). In this context, we observed that antigen receptor-triggered T cells maintained in culture in the presence of receptor-saturating levels of expressed high levels of (Fig 2A). Moreover, it was striking that sustained signalling is required to maintain the high expression of protein in CTL, with levels decreasing within 1 h of withdrawal from (Fig 2B). We also examined the effect of two other cc cytokines, IL-7 and IL-15, on the expression of in CTL. In this context, expression is essential in vivo Figure 2. cc cytokine signalling supports expression in an analogue fashion. A D CD8 + CTL were generated from splenocytes as described in Materials and Methods and maintained in (2 ng/ml). Data are representative of at least 3 experiments. (A) Western blot data show and ERK1/2 protein expression in CTL switched into the indicated concentration of for 2 h. (B) and GSK3a protein expression in CTL maintained in or deprived of for the times indicated. (C) and SMC1 expression in CTL maintained in or switched into IL-15 (2 ng/ml) or IL-7 (5 ng/ml) for 18 h. Histogram shows densitometry analysis of expression relative to SMC1 expression. (D), py694stat5 and panstat5 protein expression in CTL after 2-h treatment with the JAK inhibitor tofacitinib at the indicated concentrations. E Lymph node cells from KI mice were stimulated using CD3 (1 lg/ml) and CD28 (3 lg/ml) antibodies in the presence or absence of JAK inhibitor tofacitinib (1 nm). Flow cytometry data show expression in CD4 + (left) or CD8 + (right) T cells at the indicated times. Data are representative of at least 3 experiments. F Lymph node cells from KI mice were stimulated using CD3 (1 lg/ml) and CD28 (3 lg/ml) antibodies in the presence of receptor-blocking antibody PC61 (2 lg/ml) or isotype control for 6 or 48 h. Flow cytometry data show expression in CD4 + (left) or CD8 + (right) T cells. Data are representative of 3 biological replicates. G T cells from lymph nodes of KI mice were activated with CD3 antibody (6 ng/ml) for 48 h. The plots show the fluorescence intensity of and CD25 expression from flow cytometry data modelled by linear regression in CD4 + cells (left panel) and CD8 + cells (right panel). Data are representative of 3 experiments. H, I KI KI and WT mice were immunised with attenuated Listeria monocytogenes, and after 24 h, spleens were harvested for analysis. Data are from 2 independent experiments with 2 control and 3 test animals in each experiment. (H) Flow cytometry data showing the gating of activated (CD25 pos and CD44 pos ) and resting (CD25 neg and CD44 neg )CD8 + T cells from control (left panel) and immunised (middle panel) mice. The right panel shows expression of activated and resting CD8 + T cells with the corresponding MFI values. (I) Flow cytometry data showing the gating of CD25 high and CD25 low CD8 + T cells (left panel) and corresponding expression in the KI cells compared to WT cells (right panel). Source data are available online for this figure. ª 215 The Authors The EMBO Journal Vol 34 No
5 The EMBO Journal A T cell expression matched to metabolic demand [IL -2] (ng/ml) B 2 Time (min) Gavin C Preston et al ERK 1/ GSK3α 5 D SMC py694 STAT5 1.5 relative to SMC1 1 5 Tofacitinib (nm) 2 IL IL -7 IL -2 1 C 1 1. pan STAT5.5. IL2 E CD4 IL7 IL15 F CD8 CD3/CD28 CD3/CD28 + Tofacitinib Unstim. control 4hr CD4 CD8 CD3/CD28 + isotype CD3/CD28 + PC61 Unstim. control 6hr 24hr 48hr 48hr CD4 25 CD8 25 r =.599 p <.1 2 r =.6151 p < CD I CD44 Activated KI CD25hi KI Resting KI Activated WT CD25lo KI CD25hi WT CD25 Splenic CD8 T cells Unimmunised Immunised CD25 H 2 CD25 G CD8 Figure The EMBO Journal Vol 34 No ª 215 The Authors
6 Gavin C Preston et al T cell expression matched to metabolic demand The EMBO Journal for signalling by IL-15 (Bianchi et al, 26; Dose et al, 26, 29; ko et al, 29; Jiang et al, 21). Fig 2C shows that, IL-15 and IL-7 are different in their ability to maintain protein expression in effector CTL. has the strongest effect, IL-15 is much less potent at maintaining expression, and there are only very low levels of in effector CTL maintained in IL-7. and IL-15 signal via a receptor complex that includes the common gamma chain (cc) and a b subunit (CD122). Triggering of this receptor complex activates the tyrosine kinases JAK1 and JAK3. is able to sustain a much higher level of signalling in activated T cells than IL-15, even when both cytokines are at the receptorsaturating concentrations (Cornish, 26). The differential effect of and IL-15 on expression suggests that the level of JAK kinase activity might determine the expression of. Recently, inhibitors of JAK kinases have been described, notably tofacitinib (Changelian et al, 23). We could therefore test directly the contribution of JAK signalling to the regulation of expression in - maintained CTL. Fig 2D shows that tofacitinib treatment leads to a rapid loss of protein expression in -maintained CTL. Immune-activated T cells produce and can have autocrine responses to this cytokine (Feau et al, 211; Sa et al, 213). The ability of to control expression in CTL made us question whether autocrine /JAK signalling had any role in controlling expression in TCR-activated naïve T cells. We therefore examined the effect of the JAK inhibitor tofacitinib on induction in TCR- and CD28-activated T cells. Fig 2E shows that there was no effect of tofacitinib on expression in T cells activated with CD3 and CD28 antibodies for 4 h, whereas there was a clear reduction in expression in tofacitinib-treated T cells at the 24- and 48-h time point. It was notable that treatment of activated T cells with the JAK inhibitor did not change the frequency of activated T cells that express but rather reduced the amount of expression per cell. In further experiments, we examined the impact of PC61, a CD25 antibody that blocks binding to its high-affinity receptor (Lowenthal et al, 1985), on expression in T cells activated polyclonally with CD3 and CD28 antibodies. Fig 2F shows that blockade of binding to its receptor reduced expression in the longterm-activated T cells. However, the neutralising antibody had no impact on the digital TCR-induced response seen at 6 h. Inhibition of autocrine did not reduce the frequency of activated T cells that express but could reduce expression level per cell. Hence, control of expression is an analogue response unlike the bimodal response pattern for TCR control of expression. The data above argue that there is no role for autocrine /JAK signalling in the immediate response to TCR engagement, but during the sustained immune activation response, signalling does have an impact on expression. In further experiments to explore this question, we took advantage of the fact that the level of signalling in a T cell is determined by the level of CD25 expression (Cantrell & Smith, 1984). Moreover, CD25 expression in T cells is controlled by a positive feedback mediated by JAK activation of the transcription factor STAT5 (Nakajima et al, 1997). If expression in effector T cells is determined by the strength of signalling, then levels will be highest in cells that express high levels of CD25. Fig 2G shows that the level of protein in activated CD4 + and CD8 + T cells correlates with the expression of CD25. Similarly, when KI mice are immunised with an attenuated strain of Listeria monocytogenes over 24 h, it is possible to identify immune-activated CD25-positive effector T cells (Fig 2H, left panels). These activated CD8 + T cells express, whereas no expression is detected in non-responding naïve CD8 + T cells from the same animal (Fig 2H, right panel). Importantly, expression levels in the activated CD8 + T cells correlate with the level of CD25 expression (Fig 2I). Collectively, these data are consistent with the hypothesis that activation of JAK signalling pathways controls cellular levels of in effector T cells. Transcriptional and post-transcriptional control of expression in T cells T cell antigen receptor control of expression was explained by TCR control of the frequency of cells that express mrna. regulates an analogue response that controls the amount of expressed by each cell. We therefore assessed whether the analogue response reflected the control of mrna levels. Fig 2A shows that although there is a clear dose response for protein expression, there is no equivalent dose response for mrna in CTL (Fig 3A). Similarly, the JAK inhibitor tofacitinib causes CTL to rapidly lose protein but not mrna (Figs 2D and 3B). Moreover, CTL maintained in, IL-15 or IL-7 have very different levels of protein but express equivalent levels of mrna (Figs 2C and 3C). These data argue that the cc cytokines and IL-15 primarily regulate levels via post-transcriptional mechanisms. To rigorously test this model further, we transduced activated T cells with a FLAG- WT -IRES-GFP retroviral construct (encoding a FLAG-tagged wild-type cdna and an IRES-controlled GFP cdna) and then cultured the cells in the presence or absence of. CTL transduced with the -IRES-GFP vector only overexpress ectopic protein if the cells are cultured with (Fig 3D), whereas the IRES-controlled GFP (Fig 3D) and mrna (Fig 3E) were detected in both the -maintained and -deprived T cells. These data show that in the absence of, CTL cannot sustain the expression of from endogenous or ectopically expressed retroviral transcripts. protein has a short half-life in T cells One striking observation was the rapidity with which protein was lost from CTL following withdrawal (Fig 2B). These data argue that has a very short half-life in activated T cells. One previously described mechanism to control the expression of is a phosphorylation-dependent pathway that targets for proteasomal degradation (Gregory & Hann, 2). Phosphorylation of on T58 by glycogen synthase kinase 3b (GSK3b) promotes its interaction with Fbw7, the substrate recognition component of the SCF Fbw7 ubiquitin ligase, and directs ubiquitination and proteasomal degradation (Gregory et al, 23; Welcker et al, 24a,b). We have previously used high-resolution mass spectrometry to map the phosphoproteome of -maintained CTL (Navarro et al, 211). Analysis of this data revealed that is phosphorylated on Ser62 and Thr58 in activated T cells (Fig 4A). In this respect, it has been described that the phosphorylation of Ser62 primes for subsequent Thr58 phosphorylation by GSK3b (Sears et al, 1999, 2). These data argue that is constantly ª 215 The Authors The EMBO Journal Vol 34 No
7 The EMBO Journal T cell expression matched to metabolic demand Gavin C Preston et al A Relative mrna expression mrna ns concentration (ng/ml) B Relative mrna expression mrna ns Tofacitinib (nm) C mrna Relative mrna expression ns IL-7 IL-15 D No E -IRES-GFP FLAG- - WT GFP SMC Relative mrna expression mrna ns *** *** No -IRES-GFP neg -IRES-GFP pos Figure 3. Post-transcriptional regulation of protein expression by cc cytokine signalling. A mrna expression in CTL, generated as described in Materials and Methods, switched into decreasing concentrations of for 2 h, shown relative to - deprived CTL (2 h) (n = 3, mean SEM). B mrna expression in -maintained CTL treated with the indicated concentration of tofacitinib for 2 h, shown relative to untreated -maintained CTL (n = 3, mean SEM). C mrna expression in CTL switched into IL-7 (5 ng/ml) or IL-15 (2 ng/ml) or maintained in (2 ng/ml) for 18 h, shown relative to -maintained CTL (n = 3, mean SEM). D, E -IRES-GFP-transduced CD8 + cells were sorted by FACS, and the -IRES-GFP pos and -IRES-GFP neg CTL were maintained in (2 ng/ml) or deprived of for 2 h. Data are representative of 3 experiments. (D) Western blot data show the expression of, GFP and SMC1. (E) qpcr data show mrna expression relative to -maintained control (-IRES-GFP neg ) CTL (mean SEM, ANOVA used to determine statistical significance of multiple comparisons, ***P <.1). Source data are available online for this figure. phosphorylated and targeted for degradation in -maintained CTL. To explore this hypothesis further, we examined the impact of MG132, a well-characterised inhibitor of the 26S proteasome, on the expression of in -maintained and -deprived CTL. These data show that MG132 promoted the expression of in -maintained CTL and could also prevent the loss of expression that normally accompanies withdrawal (Fig 4B). Moreover, if CTL were treated with the GSK3 inhibitor CHIR9921, they were able to sustain expression following withdrawal (Fig 4C). 214 The EMBO Journal Vol 34 No ª 215 The Authors
8 Gavin C Preston et al T cell expression matched to metabolic demand The EMBO Journal B MG No A 51 SMC1 97 C No CHIR9921 (μm) GSK3α/β 5 D Construct evgfp None T58A GSK3α/β 5 5 Figure 4. protein expression is post-transcriptionally regulated in response to signals by GSK3 and proteasome activity in CTL. A Sequence data from phospho-proteomic analysis of -maintained CTL. Phosphorylated in CTL demonstrated by manual sequencing of acquired multistage activation spectra for the peptide KFELLPTPPLSPSRR (amino acid residues 52 66) with Biemann Roepstorff nomenclature; asterisks (y and b ion series) indicate neutral loss of one (*) or two (**) phosphate groups; 2+ indicates double-charged fragment ions. Above: vertical lines indicate a fragmented bond after collisioninduced dissociation; horizontal lines indicate the fragment retaining the charge. Data are representative of three experiments. B, C CTL were cultured in the presence or absence of and inhibitor as indicated for 2 h. Data are representative of 3 independent experiments. (B) Western blot data show and SMC1 expression in CTL treated with or without proteasome inhibitor MG132 (25 lm). (C) Western blot data show and GSK3a/b expression in CTL treated with or without increasing concentration of the GSK3 inhibitor CHIR9921. D evgfp- or T58A -transduced CD8 + CTL were sorted by FACS for GFP expression. evgfp, T58A and non-transduced CTL were then maintained in (2 ng/ml) or deprived of for 18 h. Western blot data show and GSK3a/b expression. Data information: (C, D) Data are from the same lysates but run on parallel gels. Data are representative of 3 experiments. Source data are available online for this figure. These data indicate that the short half-life of in T cells is controlled by GSK3b-mediated phosphorylation of Thr58, which then targets for proteasomal degradation. This predicts that a protein with the Thr58 mutated to a non-phosphorylatable residue such as alanine would be expressed in T cells independent of the presence of cc cytokines. To test this hypothesis, we transduced antigen receptor-activated T cells with a retroviral construct encoding T58A -IRES-eGFP and then cultured the cells in the presence or absence of. The data show that T58A expression is sustained in CTL deprived of (Fig 4D). This is in marked contrast to the inability of -deprived T cells to sustain the expression of endogenous or ectopically expressed retroviral wild-type (Fig 3D). The present results demonstrate that GSK3 constantly phosphorylates on Thr58 in T cells and hence constantly targets for degradation. Moreover, it was striking that the treatment of T cells with the proteasome inhibitor MG132 causes to accumulate in -maintained CTL (Fig 4B). This demonstrates that there is a constant high rate of proteolysis by the proteasome in - maintained T cells. Accordingly, will only be able to accumulate in activated T cells when rates of synthesis exceed the rates of degradation. is very potent at inducing amino acid uptake and protein synthesis in CTL (Cornish, 26; Sinclair et al, 213). High rates of amino acid uptake and consequently high rates of protein synthesis in -maintained CTL could therefore explain why accumulates in -maintained CTL and could also explain why IL-15 is unable to sustain high levels of in CTL: IL-15 is much less potent than in its ability to promote protein synthesis (Fig 5A). ª 215 The Authors The EMBO Journal Vol 34 No
9 The EMBO Journal T cell expression matched to metabolic demand Gavin C Preston et al Figure 5. protein expression is dependent on sustained system L amino acid uptake and protein synthesis in CD8 + T cells. A 3 H methionine incorporation into CTL maintained in IL-15 or or switched from into varying concentrations of (as shown) for 18 h. B Data show qpcr analysis of slc7a5 mrna in CTL maintained in, IL-15 or naïve lymph node cells (left panel). The data are expressed relative to -maintained CTL. The right panel shows flow cytometry data of CD98 expression on CTL maintained in or IL-15, with corresponding MFIs. C 3 H phenylalanine uptake in CTL maintained in, IL-15 or naïve lymph node cells. The data are expressed relative to -maintained cells. D 3 H leucine uptake (left panel) and 3 H methionine incorporation (right panel) measured in -maintained CTL treated with or without JAK inhibitor tofacitinib (1 nm) for 18 h. E Western blot data show and SMC1 expression in CTL maintained in with or without amino acids in culture media for 15 or 3 min. F Lymph node cells from KI mice were stimulated with CD3 (1 lg/ml) and CD28 (3 lg/ml) for 18 h. Flow cytometry data (left panel) show expression in CD8 + T cells stimulated through the TCR (black) or unstimulated (grey) compared to CD8 + T cells stimulated in the absence of amino acids (red, middle panel) and CD8 + T cells stimulated in the presence of the system L transporter inhibitor BCH (5 mm) (red, right panel). MFIs are indicated on each panel. G neg and pos CD8 + T cells were purified from OT1 KI lymph node cells that had been activated for 2 h with N4 peptide (1 ng/ml). Graph shows expression of slc7a5 and CD98 mrna measured by qpcr relative to naïve unstimulated CD8 + T cells. Data information: (A E, G) Data shown are from at least 3 experiments (mean SEM, *P <.5, **P <.1, ***P <.1, Multiple analyses were performed with ANOVA, comparisons between two groups were performed using Student's t-test.). (F) Data are representative of 3 biological replicates. Source data are available online for this figure. Recently, we have shown that expression of system L amino acid transporter Slc7a5 by TCR-activated cells is essential for the expression of protein (Sinclair et al, 213). can sustain high levels of Slc7a5 expression and CD98, the two subunits of the system L amino acid transporter (Fig 5B), and system L-mediated amino acid transport in CTL (Fig 5C). In contrast, IL-15 is much less potent at inducing Slc7a5 and CD98 expression (Fig 5B) and IL-15- maintained cells exhibit much lower rates of system L amino acid transport (Fig 5C). We considered whether the ability of the JAK inhibitor tofacitinib to decrease expression in T cells could be caused by a requirement of JAK activity for system L amino acid uptake and protein synthesis in activated T cells. Fig 5D addresses this question and reveals a striking reduction of system L transport activity and very low rates of protein synthesis in CTL treated with tofacitinib. These results collectively argue that T cells need continual high rates of amino acid uptake to sustain high levels of protein. To test this hypothesis directly, we looked at the impact of amino acid deprivation on expression in activated T cells; indeed, amino acid deprivation caused -maintained CTL to rapidly lose the expression of (Fig 5E). Additionally, blockade of system L transport by the inhibitor 2-amino-2-norbornanecarboxylic acid (BCH) also prevented the upregulation of protein in activated CD8 + T cells (Fig 5F). Moreover, TCR-stimulated pos OT1 T cells have high levels of Slc7a5 and CD98 mrna compared to neg OT1 T cells (Fig 5G). Thus, protein levels in immune-activated T cells are matched to high expression of amino acid transporters and to high rates of amino acid uptake. One consideration in these experiments is that inhibition of amino acid transport will cause loss of mtorc1 activity (Sinclair et al, 213). However, we have shown previously that inhibition of mtorc1 does not prevent expression in TCR- or -activated CD8 + T cells (Finlay et al, 212; Sinclair et al, 213). levels are important for activated T cells The mechanisms whereby TCR signalling strength controls the pool size of T cells that differentiate to effector cells are not fully understood. In the absence of, T cells cannot respond to TCR triggering to proliferate or differentiate to effector cells (Wang et al, 211b). The present data now show that when KI OT1 T cells are activated with p/mhc ligands, it is the pos cells that express the effector cytokine interferon gamma (IFNc) mrna, whereas neg cells from the same cultures do not (Fig 6A). Moreover, in the sustained responses to p/mhc ligands, it is the -expressing OT1 T cells that produce high levels of IFNc and not the neg T cells (Fig 6B). We also observed that the level of IFNc in an individual cell correlates with the level of expression of (Fig 6C). T cell antigen receptor engagement triggers a growth response in T cells causing them to increase in cell size (blastogenesis) as judged by flow cytometric analysis of the forward and side scatter properties of activated T cells. We thus used flow cytometry to analyse the forward and side scatter properties of KI OT1 T cells activated with the p/mhc ligands Q4 and T4. The expression of in the CD69 + TCR-activated cells had a bimodal response as described in Fig 1. Fig 6D shows that it is the -expressing cells that have undergone blastogenesis. We also analysed blastogenesis of KI T cell populations activated polyclonally with CD3 and CD28 antibodies. As described above, when T cells are activated polyclonally with different concentrations of CD3 antibody, then the frequency of -expressing cells directly correlated with the dose of TCR ligand used. Fig 6E shows and CD69 expression in CD8 + T cells stimulated with a low concentration of the CD3 antibody. A large percentage of the cells responded to TCR ligation to induce CD69 expression, but only a subset of the CD69 + cells express ; that is, induction is bimodal in the TCR-activated CD69 + T cells. Fig 6F shows that CD69 + CD8 + T cells that do not express have increased cell size in comparison with the CD69 quiescent CD8 T cells. However, the -expressing CD69 + CD8 + T cells are much larger than the -negative CD69 + CD8 + T cells. Importantly, T cells with both alleles deleted do not blast in response to TCR and CD28 activation (Fig 6G). How important is it that the amount of protein expressed in a single, activated T cell can be fine-tuned? Strategies to address this issue require the ability to quantitatively analyse a -regulated response in single T cells. In this regard, there is strong evidence that regulates transferrin receptor expression (O Donnell et al, 26) (Holland et al, 212). Moreover, there are good tools for single cell analysis of this pathway that would allow a direct analysis of the importance of protein levels for transferrin uptake. We first addressed whether was indeed important for transferrin receptor expression in T cells. Naïve T cells do not express the 216 The EMBO Journal Vol 34 No ª 215 The Authors
10 Gavin C Preston et al T cell expression matched to metabolic demand The EMBO Journal A 3 H methionine cpm *** ** ** * IL-15 (ng/ml) B relative mrna expression *** slc7a5 IL-15 *** naive LN CD IL-15 C E relative uptake Time (min) Amino acids 75 5 Phenylalanine uptake ** + IL-15 naive LN H leucine cpm F D Leucine uptake Methionine incorporation Control Tofacitinib 3 H methionine cpm ** * 8 4 Control Tofacitinib SMC G mrna expression relative to naive naive neg pos Figure 5. slc7a5 CD98 transferrin receptor (CD71), but expression of this receptor, and the cellular uptake of transferrin, is strongly induced in antigen receptor-activated T cells (Fig 7A). T cells with both alleles deleted cannot upregulate CD71 or transferrin uptake in response to TCR engagement (Fig 7B). Is the level of expression per cell important for transferrin receptor expression? To explore this, we first used the KI mouse model and examined CD71 expression versus expression in TCR-activated CD4 + and CD8 + T cells. The data show a clear correlation between expression of CD71 and (Fig 7C). There was also a strong correlation between cellular levels of and expression of transferrin receptors in CTL cultured in the ª 215 The Authors The EMBO Journal Vol 34 No
11 The EMBO Journal T cell expression matched to metabolic demand Gavin C Preston et al A mrna rel. to naive naive IFNγ mrna GFP- neg GFP- pos B Q4 (5ng/ml) T4 (5ng/ml) IFNγ % IFNγ positive Q4 T4 neg pos C R=.2728 p= <.1 D Q4 (3.3ng/ml) CD69 + neg T4 (3.3ng/ml) CD69 + neg E CD IFNγ CD69 + pos CD69 + pos CD Side Scatter Forward Scatter F CD69 - neg CD69 + neg CD69 + pos G Unstim CD4Cre - fl/fl CD3/CD28 CD4Cre + fl/fl CD4 + Side Scatter Forward Scatter CD8 + Side Scatter Forward Scatter Figure 6. protein expression correlates with effector functions. A Lymph node CD8 + T cells were activated with N4 peptide (1 ng/ml) for 2 h, and neg and pos populations were FACS-sorted. Expression of IFNc mrna measured by qpcr relative to naïve unstimulated CD8 + T cells is shown. Data show mean + SEM of 3 biological replicates. B Flow cytometry data show and IFNc expression from spleen OT1 KI CD8 + T cells stimulated with 5 ng/ml Q4 (left panel) or T4 (middle panel) peptides for 24 h. Quadrant percentages are shown. The right panel shows the percentage of pos and neg CD8 + T cells expressing IFNc protein. Data are from 3 biological replicates. C The fluorescence intensity of and IFNc expression from flow cytometry data modelled by linear regression in OT1 KI CD8 + T cells treated with 5 ng/ml Q4 for 24 h. Data are representative of 3 biological replicates. D Forward scatter and side scatter plots of CD69 + neg (upper panels) and CD69 + pos (lower panel) OT1 KI CD8 + T cells stimulated with 3.3 ng/ml Q4 or N4 peptides for 24 h. Data are representative of 2 biological replicates E, F KI CD8 + T cells were stimulated with CD3 antibody (6 ng/ml) for 24 h. Data are representative of 3 biological replicates. (E) Flow cytometry data show CD69 and expression. (F) Forward and side scatter plots of CD69 neg (left panel), CD69 + neg (middle panel) and CD69 + pos (right panel) populations. G CD4Cre fl/fl and CD4cre + fl/fl lymph node cells were activated with CD3 (1 lg/ml) and CD28 (3 lg/ml) antibodies for 24 h. The data show forward and side scatter plots of CD4 + (upper panels) and CD8 + (lower panels) T cells, compared to unstimulated CD4Cre fl/fl cells. Data are representative of at least 3 biological replicates. 218 The EMBO Journal Vol 34 No ª 215 The Authors
12 Gavin C Preston et al T cell expression matched to metabolic demand The EMBO Journal A CD4 CD8 Unstim CD3/CD28 B CD4 CD8 Unstim CD4Cre + fl/fl CD3/CD28 CD4Cre - fl/fl CD71 CD71 CD4Cre + fl/fl CD4Cre - fl/fl CD3/CD28 Transferrin uptake Transferrin uptake C CD CD4 r =.6597 p <.1 F no cytokine IL-15 evgfp T58A 1 CD CD r =.594 p <.1 CD71 Transferrin D no cytokine IL-15 E CD71 MFI no inhibitor CHIR9921 (2μM) CD71 Transferrin (ng/ml) Figure 7. Transferrin receptor (CD71) expression and transferrin uptake are dependent on the level of expression in CD8 + T cells. A, B Lymph node cells were stimulated using CD3 (1 lg/ml) and CD28 (3 lg/ml) antibodies for 48 h, and unstimulated controls were maintained in IL-7 (5 ng/ml). (A) Histograms show CD71 expression (upper panels) and transferrin uptake (lower panels) in CD4 + and CD8 + T cells. (B) Histograms show CD71 expression (upper panels) and transferrin uptake (lower panels) in CD4 + and CD8 + T cells from CD4Cre + fl/fl and CD4Cre fl/fl control mice. C T cells from lymph nodes of KI mice were activated with CD3 antibody (6 ng/ml) for 48 h. The plots show the fluorescence intensity of and CD71 expression from flow cytometry data modelled by linear regression in CD4 + cells (upper panel) and CD8 + cells (lower panel). D Flow cytometry data showing CD71 expression and transferrin uptake (bottom panel) in CTL maintained in (2 ng/ml) or IL-15 (2 ng/ml) or withdrawn from for 18 h (no cytokine). E CD71 MFI data (mean SEM) in CTL treated with decreasing concentrations of in the presence or absence of the GSK3 inhibitor CHIR9921 (2 lm) for 18 h. F evgfp- or T58A -transduced CD8 + CTL were cultured in (2 ng/ml), IL-15 (2 ng/ml) or withdrawn from for final 18 h (no cytokine). Histograms show CD71 expression (upper panels) and transferrin uptake (lower panels) as measured by flow cytometry. Data information: (A D, F) Data are representative of at least 3 biological replicates. (E) Data are quantified from 3 experiments. ª 215 The Authors The EMBO Journal Vol 34 No
13 The EMBO Journal T cell expression matched to metabolic demand Gavin C Preston et al cytokines and IL-15. Fig 7D thus shows that -maintained CTL express high levels of CD71 and uptake high amounts of transferrin compared to T cells deprived of cytokines, with expression of CD71 and transferrin uptake in activated T cells cultured in IL-15 being intermediate. This correlates with the ability of to maintain high levels of protein compared to IL-15 (Fig 2C). These data are correlative; hence, to test whether differences in expression levels were causal for differences in CD71 expression, we examined the impact of increasing levels in T cells cultured under these different conditions. In these experiments, we looked at the impact of expressing the T58A mutant on CD71 expression and transferrin uptake because the use of this mutant bypasses the GSK3-mediated pathway that rapidly targets for degradation and allows overexpression of even when cells are cytokinedeprived. In this respect, Fig 7E shows that GSK3 inhibition using CHIR9921, which prevents degradation, could increase CD71 expression in -maintained CTL. There was also a strong effect of T58A expression on CD71 expression and the ability of cells to take up transferrin in cytokine-maintained CTL. Hence, ectopic expression of T58A increased expression of CD71 and transferrin uptake in both IL-15- and even -cultured CTL (Fig 7F). Ectopic expression of T58A was also able to sustain expression of CD71 and transferrin uptake in -deprived CTL. In addition, increasing levels increased transferrin uptake in IL-15-cultured CTL to the level seen in the control -maintained cells. Hence, the limiting factor for transferrin receptor expression and transferrin uptake in IL-15-maintained CTL was the level of expression. Moreover, the fact that expression of T58A, which increased cellular concentrations of, increases rates of transferrin uptake in - maintained cells (Fig 7G) shows that the concentration of levels per cell dictates transferrin receptor expression and rates of transferrin uptake by immune-activated T cells. There is thus a clear biological consequence of fine-tuning expression in single, activated T cells. Discussion The expression of in CD8 + T cells is regulated by both antigen receptor signals and the cc cytokine. The present study now shows that the TCR and use distinct mechanisms to control levels in T cells. The TCR controls expression in a digital response that determines the frequency of the T cell population that switches on mrna and protein expression. In contrast, the cytokine regulates expression in an analogue response that fine-tunes protein levels per cell. In the immediate response to TCR engagement, the strength of TCR signal controls how many cells express, and at these time points there is no apparent role for /JAK signalling. expression in T cells thus appears to be a two-stage process; expression is digitally induced by the strength of TCR signalling, but the maintenance of high expression then becomes dependent upon other external stimuli such as. is essential for the metabolic reprogramming that controls effector T cell differentiation (Wang et al, 211a). The fact that the strength of the TCR ligand determines the frequency of T cells that express gives new insight about the mechanisms that allow TCR signalling strength to control the pool size of T cells that metabolically reprogramme and differentiate to effector cells. One striking observation from the present study was the discordance between protein and mrna levels in immune-activated T cells. This latter observation is relevant because many previous studies have interrogated mrna expression in T lymphocyte subpopulations (Best et al, 213). The data herein show that T cells can express high levels of RNA but that high levels of protein can only be sustained in T cells that have high rates of amino acid uptake. expression in an antigen receptorstimulated T cell will thus be transient unless the T cell can sustain high rates of biosynthesis of protein. A key mechanism that controls protein expression in activated T cells is mediated by the serine/threonine kinase GSK3. Hence, in activated T cells is phosphorylated by GSK3 on Thr58, which then constantly targets the protein for proteasomal degradation (Gregory et al, 23). It is this constant high rate of degradation in activated T cells that confines high levels of protein expression to T cells with high rates of amino acid uptake and protein synthesis. protein expression thus rapidly declines in situations where T cell protein synthesis and amino acid uptake are limited. Recent studies have shown that there is very tight control of amino acid transport and hence protein synthetic capacity during the immune activation of T cells (Hayashi et al, 213; Sinclair et al, 213). The critical signals that are known to be able to induce and sustain high-level expression of amino acid transporters in T cells are antigen and pro-inflammatory cytokines such as (Wang et al, 211a; Sinclair et al, 213). This means that protein expression will be restricted to T cells responding to immune activation or to T cells exposed to pro-inflammatory cytokines that can maintain amino acid uptake. One of the cytokines that can sustain high levels of protein in T cells is. This molecule is a member of the cc family of cytokines that signal by activating the JAK tyrosine kinases. The present experiments interrogated the role of JAKs regulating expression using the JAK inhibitor tofacitinib: a drug showing clinical promise for the treatment of autoimmune diseases (Hsu & Armstrong, 214; Lee et al, 214). Our data afford the insight that immune-activated T cells can express high levels of mrna and yet be dependent on sustained JAK activity to express protein. Moreover, another novel insight from the present experiments is that part of the tofacitinib mechanism of action as an immunosuppressant may be to prevent T cell reprogramming of -controlled metabolic responses. In the context of cc cytokines and JAKs, the strength of signalling during an immune response determines effector versus memory cell fate of immune-activated CD8 + T cells (Kalia et al, 21). Herein we show that has the ability to sustain high rates of expression compared to other cc cytokines such as IL-7 and IL-15, and this could contribute to the unique role for this cytokine as a regulator of peripheral T cell metabolism and differentiation. What is the mechanistic explanation for the differential effect of, IL-7 and IL-15? They all signal through the cc, and and IL-15 share the same beta receptor subunit. Recent studies have shown that it is predominantly the abundance of the specific alpha chain for and IL-15 that determines the strength of signal mediated by that cytokine (Ring et al, 212). Furthermore, in T cells that highly express CD25, the alpha subunit for the high-affinity receptor, signalling through IL-15 and IL-7 in particular is attenuated because of the sequestration of cc subunits in complete receptors (Cotari et al, 213). Therefore, even at saturating concentrations, IL-7 and IL-15 are less potent than in our system. 22 The EMBO Journal Vol 34 No ª 215 The Authors
Supplementary Figure S1. PTPN2 levels are not altered in proliferating CD8+ T cells. Lymph node (LN) CD8+ T cells from C57BL/6 mice were stained with
 Supplementary Figure S1. PTPN2 levels are not altered in proliferating CD8+ T cells. Lymph node (LN) CD8+ T cells from C57BL/6 mice were stained with CFSE and stimulated with plate-bound α-cd3ε (10µg/ml)
Supplementary Figure S1. PTPN2 levels are not altered in proliferating CD8+ T cells. Lymph node (LN) CD8+ T cells from C57BL/6 mice were stained with CFSE and stimulated with plate-bound α-cd3ε (10µg/ml)
The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep
 SUPPLEMENTARY INFORMATION The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness Jinyi Zhang, Naima
SUPPLEMENTARY INFORMATION The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness Jinyi Zhang, Naima
% of live splenocytes. STAT5 deletion. (open shapes) % ROSA + % floxed
 Supp. Figure 1. a 14 1 1 8 6 spleen cells (x1 6 ) 16 % of live splenocytes 5 4 3 1 % of live splenocytes 8 6 4 b 1 1 c % of CD11c + splenocytes (closed shapes) 8 6 4 8 6 4 % ROSA + (open shapes) % floxed
Supp. Figure 1. a 14 1 1 8 6 spleen cells (x1 6 ) 16 % of live splenocytes 5 4 3 1 % of live splenocytes 8 6 4 b 1 1 c % of CD11c + splenocytes (closed shapes) 8 6 4 8 6 4 % ROSA + (open shapes) % floxed
SUPPLEMENTARY INFORMATION. Supp. Fig. 1. Autoimmunity. Tolerance APC APC. T cell. T cell. doi: /nature06253 ICOS ICOS TCR CD28 TCR CD28
 Supp. Fig. 1 a APC b APC ICOS ICOS TCR CD28 mir P TCR CD28 P T cell Tolerance Roquin WT SG Icos mrna T cell Autoimmunity Roquin M199R SG Icos mrna www.nature.com/nature 1 Supp. Fig. 2 CD4 + CD44 low CD4
Supp. Fig. 1 a APC b APC ICOS ICOS TCR CD28 mir P TCR CD28 P T cell Tolerance Roquin WT SG Icos mrna T cell Autoimmunity Roquin M199R SG Icos mrna www.nature.com/nature 1 Supp. Fig. 2 CD4 + CD44 low CD4
Supplemental Figure 1
 Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
T-cell activation T cells migrate to secondary lymphoid tissues where they interact with antigen, antigen-presenting cells, and other lymphocytes:
 Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
T-cell activation T cells migrate to secondary lymphoid tissues where they interact with antigen, antigen-presenting cells, and other lymphocytes:
 Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
Interactions between innate immunity & adaptive immunity What happens to T cells after they leave the thymus? Naïve T cells exit the thymus and enter the bloodstream. If they remain in the bloodstream,
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. BMS enhances human T cell activation in vitro in a
 Supplementary Figure 1. BMS98662 enhances human T cell activation in vitro in a concentration-dependent manner. Jurkat T cells were activated with anti-cd3 and anti-cd28 antibody in the presence of titrated
Supplementary Figure 1. BMS98662 enhances human T cell activation in vitro in a concentration-dependent manner. Jurkat T cells were activated with anti-cd3 and anti-cd28 antibody in the presence of titrated
NKTR-255: Accessing The Immunotherapeutic Potential Of IL-15 for NK Cell Therapies
 NKTR-255: Accessing The Immunotherapeutic Potential Of IL-15 for NK Cell Therapies Saul Kivimäe Senior Scientist, Research Biology Nektar Therapeutics NK Cell-Based Cancer Immunotherapy, September 26-27,
NKTR-255: Accessing The Immunotherapeutic Potential Of IL-15 for NK Cell Therapies Saul Kivimäe Senior Scientist, Research Biology Nektar Therapeutics NK Cell-Based Cancer Immunotherapy, September 26-27,
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/430/ra57/dc1 Supplementary Materials for The 4E-BP eif4e axis promotes rapamycinsensitive growth and proliferation in lymphocytes Lomon So, Jongdae Lee, Miguel
www.sciencesignaling.org/cgi/content/full/9/430/ra57/dc1 Supplementary Materials for The 4E-BP eif4e axis promotes rapamycinsensitive growth and proliferation in lymphocytes Lomon So, Jongdae Lee, Miguel
Supplementary Figure 1. Using DNA barcode-labeled MHC multimers to generate TCR fingerprints
 Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
MATERIALS AND METHODS. Neutralizing antibodies specific to mouse Dll1, Dll4, J1 and J2 were prepared as described. 1,2 All
 MATERIALS AND METHODS Antibodies (Abs), flow cytometry analysis and cell lines Neutralizing antibodies specific to mouse Dll1, Dll4, J1 and J2 were prepared as described. 1,2 All other antibodies used
MATERIALS AND METHODS Antibodies (Abs), flow cytometry analysis and cell lines Neutralizing antibodies specific to mouse Dll1, Dll4, J1 and J2 were prepared as described. 1,2 All other antibodies used
Supplemental Table I.
 Supplemental Table I Male / Mean ± SEM n Mean ± SEM n Body weight, g 29.2±0.4 17 29.7±0.5 17 Total cholesterol, mg/dl 534.0±30.8 17 561.6±26.1 17 HDL-cholesterol, mg/dl 9.6±0.8 17 10.1±0.7 17 Triglycerides,
Supplemental Table I Male / Mean ± SEM n Mean ± SEM n Body weight, g 29.2±0.4 17 29.7±0.5 17 Total cholesterol, mg/dl 534.0±30.8 17 561.6±26.1 17 HDL-cholesterol, mg/dl 9.6±0.8 17 10.1±0.7 17 Triglycerides,
T cell maturation. T-cell Maturation. What allows T cell maturation?
 T-cell Maturation What allows T cell maturation? Direct contact with thymic epithelial cells Influence of thymic hormones Growth factors (cytokines, CSF) T cell maturation T cell progenitor DN DP SP 2ry
T-cell Maturation What allows T cell maturation? Direct contact with thymic epithelial cells Influence of thymic hormones Growth factors (cytokines, CSF) T cell maturation T cell progenitor DN DP SP 2ry
SUPPLEMENTARY INFORMATION
 Supplemental Figure 1. Furin is efficiently deleted in CD4 + and CD8 + T cells. a, Western blot for furin and actin proteins in CD4cre-fur f/f and fur f/f Th1 cells. Wild-type and furin-deficient CD4 +
Supplemental Figure 1. Furin is efficiently deleted in CD4 + and CD8 + T cells. a, Western blot for furin and actin proteins in CD4cre-fur f/f and fur f/f Th1 cells. Wild-type and furin-deficient CD4 +
ECM1 controls T H 2 cell egress from lymph nodes through re-expression of S1P 1
 ZH, Li et al, page 1 ECM1 controls T H 2 cell egress from lymph nodes through re-expression of S1P 1 Zhenhu Li 1,4,Yuan Zhang 1,4, Zhiduo Liu 1, Xiaodong Wu 1, Yuhan Zheng 1, Zhiyun Tao 1, Kairui Mao 1,
ZH, Li et al, page 1 ECM1 controls T H 2 cell egress from lymph nodes through re-expression of S1P 1 Zhenhu Li 1,4,Yuan Zhang 1,4, Zhiduo Liu 1, Xiaodong Wu 1, Yuhan Zheng 1, Zhiyun Tao 1, Kairui Mao 1,
Intracellular MHC class II molecules promote TLR-triggered innate. immune responses by maintaining Btk activation
 Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining Btk activation Xingguang Liu, Zhenzhen Zhan, Dong Li, Li Xu, Feng Ma, Peng Zhang, Hangping Yao and Xuetao
Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining Btk activation Xingguang Liu, Zhenzhen Zhan, Dong Li, Li Xu, Feng Ma, Peng Zhang, Hangping Yao and Xuetao
NKTR-255: Accessing IL-15 Therapeutic Potential through Robust and Sustained Engagement of Innate and Adaptive Immunity
 NKTR-255: Accessing IL-15 Therapeutic Potential through Robust and Sustained Engagement of Innate and Adaptive Immunity Peiwen Kuo Scientist, Research Biology Nektar Therapeutics August 31 st, 2018 Emerging
NKTR-255: Accessing IL-15 Therapeutic Potential through Robust and Sustained Engagement of Innate and Adaptive Immunity Peiwen Kuo Scientist, Research Biology Nektar Therapeutics August 31 st, 2018 Emerging
Nature Immunology doi: /ni.3268
 Supplementary Figure 1 Loss of Mst1 and Mst2 increases susceptibility to bacterial sepsis. (a) H&E staining of colon and kidney sections from wild type and Mst1 -/- Mst2 fl/fl Vav-Cre mice. Scale bar,
Supplementary Figure 1 Loss of Mst1 and Mst2 increases susceptibility to bacterial sepsis. (a) H&E staining of colon and kidney sections from wild type and Mst1 -/- Mst2 fl/fl Vav-Cre mice. Scale bar,
T Cell Activation. Patricia Fitzgerald-Bocarsly March 18, 2009
 T Cell Activation Patricia Fitzgerald-Bocarsly March 18, 2009 Phases of Adaptive Immune Responses Phases of T cell responses IL-2 acts as an autocrine growth factor Fig. 11-11 Clonal Expansion of T cells
T Cell Activation Patricia Fitzgerald-Bocarsly March 18, 2009 Phases of Adaptive Immune Responses Phases of T cell responses IL-2 acts as an autocrine growth factor Fig. 11-11 Clonal Expansion of T cells
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/3/114/ra23/dc1 Supplementary Materials for Regulation of Zap70 Expression During Thymocyte Development Enables Temporal Separation of CD4 and CD8 Repertoire Selection
www.sciencesignaling.org/cgi/content/full/3/114/ra23/dc1 Supplementary Materials for Regulation of Zap70 Expression During Thymocyte Development Enables Temporal Separation of CD4 and CD8 Repertoire Selection
Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were
 Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were isolated from wild type (PKC-θ- WT) or PKC-θ null (PKC-θ-KO)
Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were isolated from wild type (PKC-θ- WT) or PKC-θ null (PKC-θ-KO)
Out-of-sequence signal 3 as a mechanism for virusinduced immune suppression of CD8 T cell responses
 University of Massachusetts Medical School escholarship@umms Open Access Articles Open Access Publications by UMMS Authors 9-25-2014 Out-of-sequence signal 3 as a mechanism for virusinduced immune suppression
University of Massachusetts Medical School escholarship@umms Open Access Articles Open Access Publications by UMMS Authors 9-25-2014 Out-of-sequence signal 3 as a mechanism for virusinduced immune suppression
Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured
 Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured under Th0, Th1, Th2, Th17, and Treg conditions. mrna
Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured under Th0, Th1, Th2, Th17, and Treg conditions. mrna
Adaptive immune responses: T cell-mediated immunity
 MICR2209 Adaptive immune responses: T cell-mediated immunity Dr Allison Imrie allison.imrie@uwa.edu.au 1 Synopsis: In this lecture we will discuss the T-cell mediated immune response, how it is activated,
MICR2209 Adaptive immune responses: T cell-mediated immunity Dr Allison Imrie allison.imrie@uwa.edu.au 1 Synopsis: In this lecture we will discuss the T-cell mediated immune response, how it is activated,
Examples of questions for Cellular Immunology/Cellular Biology and Immunology
 Examples of questions for Cellular Immunology/Cellular Biology and Immunology Each student gets a set of 6 questions, so that each set contains different types of questions and that the set of questions
Examples of questions for Cellular Immunology/Cellular Biology and Immunology Each student gets a set of 6 questions, so that each set contains different types of questions and that the set of questions
Supplementary Fig. 1 p38 MAPK negatively regulates DC differentiation. (a) Western blot analysis of p38 isoform expression in BM cells, immature DCs
 Supplementary Fig. 1 p38 MAPK negatively regulates DC differentiation. (a) Western blot analysis of p38 isoform expression in BM cells, immature DCs (idcs) and mature DCs (mdcs). A myeloma cell line expressing
Supplementary Fig. 1 p38 MAPK negatively regulates DC differentiation. (a) Western blot analysis of p38 isoform expression in BM cells, immature DCs (idcs) and mature DCs (mdcs). A myeloma cell line expressing
Determinants of Immunogenicity and Tolerance. Abul K. Abbas, MD Department of Pathology University of California San Francisco
 Determinants of Immunogenicity and Tolerance Abul K. Abbas, MD Department of Pathology University of California San Francisco EIP Symposium Feb 2016 Why do some people respond to therapeutic proteins?
Determinants of Immunogenicity and Tolerance Abul K. Abbas, MD Department of Pathology University of California San Francisco EIP Symposium Feb 2016 Why do some people respond to therapeutic proteins?
SUPPLEMENTARY INFORMATION
 Complete but curtailed T-cell response to very-low-affinity antigen Dietmar Zehn, Sarah Y. Lee & Michael J. Bevan Supp. Fig. 1: TCR chain usage among endogenous K b /Ova reactive T cells. C57BL/6 mice
Complete but curtailed T-cell response to very-low-affinity antigen Dietmar Zehn, Sarah Y. Lee & Michael J. Bevan Supp. Fig. 1: TCR chain usage among endogenous K b /Ova reactive T cells. C57BL/6 mice
Tbk1-TKO! DN cells (%)! 15! 10!
 a! T Cells! TKO! B Cells! TKO! b! CD4! 8.9 85.2 3.4 2.88 CD8! Tbk1-TKO! 1.1 84.8 2.51 2.54 c! DN cells (%)! 4 3 2 1 DP cells (%)! 9 8 7 6 CD4 + SP cells (%)! 5 4 3 2 1 5 TKO! TKO! TKO! TKO! 15 1 5 CD8
a! T Cells! TKO! B Cells! TKO! b! CD4! 8.9 85.2 3.4 2.88 CD8! Tbk1-TKO! 1.1 84.8 2.51 2.54 c! DN cells (%)! 4 3 2 1 DP cells (%)! 9 8 7 6 CD4 + SP cells (%)! 5 4 3 2 1 5 TKO! TKO! TKO! TKO! 15 1 5 CD8
Insulin Resistance. Biol 405 Molecular Medicine
 Insulin Resistance Biol 405 Molecular Medicine Insulin resistance: a subnormal biological response to insulin. Defects of either insulin secretion or insulin action can cause diabetes mellitus. Insulin-dependent
Insulin Resistance Biol 405 Molecular Medicine Insulin resistance: a subnormal biological response to insulin. Defects of either insulin secretion or insulin action can cause diabetes mellitus. Insulin-dependent
Supporting Information Table of Contents
 Supporting Information Table of Contents Supporting Information Figure 1 Page 2 Supporting Information Figure 2 Page 4 Supporting Information Figure 3 Page 5 Supporting Information Figure 4 Page 6 Supporting
Supporting Information Table of Contents Supporting Information Figure 1 Page 2 Supporting Information Figure 2 Page 4 Supporting Information Figure 3 Page 5 Supporting Information Figure 4 Page 6 Supporting
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis
 Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Appendix Figure S1 A B C D E F G H
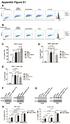 ppendix Figure S1 C D E F G H ppendix Figure S1. RT and chemotherapy alter PD-L1 expression in PDC cells. Flow cytometric analysis of PD-L1 expression in () KPC and () Pan02 cells following treatment with
ppendix Figure S1 C D E F G H ppendix Figure S1. RT and chemotherapy alter PD-L1 expression in PDC cells. Flow cytometric analysis of PD-L1 expression in () KPC and () Pan02 cells following treatment with
Follicular Lymphoma. ced3 APOPTOSIS. *In the nematode Caenorhabditis elegans 131 of the organism's 1031 cells die during development.
 Harvard-MIT Division of Health Sciences and Technology HST.176: Cellular and Molecular Immunology Course Director: Dr. Shiv Pillai Follicular Lymphoma 1. Characterized by t(14:18) translocation 2. Ig heavy
Harvard-MIT Division of Health Sciences and Technology HST.176: Cellular and Molecular Immunology Course Director: Dr. Shiv Pillai Follicular Lymphoma 1. Characterized by t(14:18) translocation 2. Ig heavy
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
Supplementary Fig. 1 No relative growth advantage of Foxp3 negative cells.
 Supplementary Fig. 1 Supplementary Figure S1: No relative growth advantage of Foxp3 negative cells. itreg were induced from WT (A) or FIR (B) CD4 + T cells. FIR itregs were then removed from the TCR signal
Supplementary Fig. 1 Supplementary Figure S1: No relative growth advantage of Foxp3 negative cells. itreg were induced from WT (A) or FIR (B) CD4 + T cells. FIR itregs were then removed from the TCR signal
TCR, MHC and coreceptors
 Cooperation In Immune Responses Antigen processing how peptides get into MHC Antigen processing involves the intracellular proteolytic generation of MHC binding proteins Protein antigens may be processed
Cooperation In Immune Responses Antigen processing how peptides get into MHC Antigen processing involves the intracellular proteolytic generation of MHC binding proteins Protein antigens may be processed
Nature Medicine doi: /nm.3957
 Supplementary Fig. 1. p38 alternative activation, IL-21 expression, and T helper cell transcription factors in PDAC tissue. (a) Tissue microarrays of pancreatic tissue from 192 patients with pancreatic
Supplementary Fig. 1. p38 alternative activation, IL-21 expression, and T helper cell transcription factors in PDAC tissue. (a) Tissue microarrays of pancreatic tissue from 192 patients with pancreatic
Chapter 11. B cell generation, Activation, and Differentiation. Pro-B cells. - B cells mature in the bone marrow.
 Chapter B cell generation, Activation, and Differentiation - B cells mature in the bone marrow. - B cells proceed through a number of distinct maturational stages: ) Pro-B cell ) Pre-B cell ) Immature
Chapter B cell generation, Activation, and Differentiation - B cells mature in the bone marrow. - B cells proceed through a number of distinct maturational stages: ) Pro-B cell ) Pre-B cell ) Immature
Generation of the Immune Response
 Generation of the Immune Response Sheet 18 immunity I only added extra notes that were explained in the lecture, refer back to the slides. SLIDE 3: In the generation of Immune response whether by B or
Generation of the Immune Response Sheet 18 immunity I only added extra notes that were explained in the lecture, refer back to the slides. SLIDE 3: In the generation of Immune response whether by B or
T Cell Activation, Costimulation and Regulation
 1 T Cell Activation, Costimulation and Regulation Abul K. Abbas, MD University of California San Francisco 2 Lecture outline T cell antigen recognition and activation Costimulation, the B7:CD28 family
1 T Cell Activation, Costimulation and Regulation Abul K. Abbas, MD University of California San Francisco 2 Lecture outline T cell antigen recognition and activation Costimulation, the B7:CD28 family
Nature Structural & Molecular Biology: doi: /nsmb.3218
 Supplementary Figure 1 Endogenous EGFR trafficking and responses depend on biased ligands. (a) Lysates from HeLa cells stimulated for 2 min. with increasing concentration of ligands were immunoblotted
Supplementary Figure 1 Endogenous EGFR trafficking and responses depend on biased ligands. (a) Lysates from HeLa cells stimulated for 2 min. with increasing concentration of ligands were immunoblotted
The T cell receptor for MHC-associated peptide antigens
 1 The T cell receptor for MHC-associated peptide antigens T lymphocytes have a dual specificity: they recognize polymporphic residues of self MHC molecules, and they also recognize residues of peptide
1 The T cell receptor for MHC-associated peptide antigens T lymphocytes have a dual specificity: they recognize polymporphic residues of self MHC molecules, and they also recognize residues of peptide
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature775 4 O.D. (595-655) 3 1 -ζ no antibody isotype ctrl Plated Soluble 1F6 397 7H11 Supplementary Figure 1 Soluble and plated anti- Abs induce -! signalling. B3Z cells stably expressing!
doi: 1.138/nature775 4 O.D. (595-655) 3 1 -ζ no antibody isotype ctrl Plated Soluble 1F6 397 7H11 Supplementary Figure 1 Soluble and plated anti- Abs induce -! signalling. B3Z cells stably expressing!
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original
 Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original phenotypic screening was n=40. For specific tests, the
Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original phenotypic screening was n=40. For specific tests, the
Nature Immunology: doi: /ni Supplementary Figure 1
 Supplementary Figure 1 Fatty acid oxidation is emphasized in 1 macrophages compared with that in macrophages. Gene expression of mitochondrial OXPHOS (Atp5j, Cox4i1, Uqcrc1/2, Ndufs1, Sdhb) and β-oxidation
Supplementary Figure 1 Fatty acid oxidation is emphasized in 1 macrophages compared with that in macrophages. Gene expression of mitochondrial OXPHOS (Atp5j, Cox4i1, Uqcrc1/2, Ndufs1, Sdhb) and β-oxidation
Supplemental Materials for. Effects of sphingosine-1-phosphate receptor 1 phosphorylation in response to. FTY720 during neuroinflammation
 Supplemental Materials for Effects of sphingosine-1-phosphate receptor 1 phosphorylation in response to FTY7 during neuroinflammation This file includes: Supplemental Table 1. EAE clinical parameters of
Supplemental Materials for Effects of sphingosine-1-phosphate receptor 1 phosphorylation in response to FTY7 during neuroinflammation This file includes: Supplemental Table 1. EAE clinical parameters of
Supplementary Figures
 Supplementary Figures Supplementary Fig. 1. Surface thiol groups and reduction of activated T cells. (a) Activated CD8 + T-cells have high expression levels of free thiol groups on cell surface proteins.
Supplementary Figures Supplementary Fig. 1. Surface thiol groups and reduction of activated T cells. (a) Activated CD8 + T-cells have high expression levels of free thiol groups on cell surface proteins.
The Adaptive Immune Responses
 The Adaptive Immune Responses The two arms of the immune responses are; 1) the cell mediated, and 2) the humoral responses. In this chapter we will discuss the two responses in detail and we will start
The Adaptive Immune Responses The two arms of the immune responses are; 1) the cell mediated, and 2) the humoral responses. In this chapter we will discuss the two responses in detail and we will start
Supplementary Figure 1. Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Nature Immunology: doi: /ni.
 Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Nature Immunology: doi: /ni.3631
 Supplementary Figure 1 SPT analyses of Zap70 at the T cell plasma membrane. (a) Total internal reflection fluorescent (TIRF) excitation at 64-68 degrees limits single molecule detection to 100-150 nm above
Supplementary Figure 1 SPT analyses of Zap70 at the T cell plasma membrane. (a) Total internal reflection fluorescent (TIRF) excitation at 64-68 degrees limits single molecule detection to 100-150 nm above
Haematopoietic stem cells
 Haematopoietic stem cells Neil P. Rodrigues, DPhil NIH Centre for Biomedical Research Excellence in Stem Cell Biology Boston University School of Medicine neil.rodrigues@imm.ox.ac.uk Haematopoiesis: An
Haematopoietic stem cells Neil P. Rodrigues, DPhil NIH Centre for Biomedical Research Excellence in Stem Cell Biology Boston University School of Medicine neil.rodrigues@imm.ox.ac.uk Haematopoiesis: An
Supplementary Figure 1. Characterization of basophils after reconstitution of SCID mice
 Supplementary figure legends Supplementary Figure 1. Characterization of after reconstitution of SCID mice with CD4 + CD62L + T cells. (A-C) SCID mice (n = 6 / group) were reconstituted with 2 x 1 6 CD4
Supplementary figure legends Supplementary Figure 1. Characterization of after reconstitution of SCID mice with CD4 + CD62L + T cells. (A-C) SCID mice (n = 6 / group) were reconstituted with 2 x 1 6 CD4
COURSE: Medical Microbiology, MBIM 650/720 - Fall TOPIC: Antigen Processing, MHC Restriction, & Role of Thymus Lecture 12
 COURSE: Medical Microbiology, MBIM 650/720 - Fall 2008 TOPIC: Antigen Processing, MHC Restriction, & Role of Thymus Lecture 12 FACULTY: Dr. Mayer Office: Bldg. #1, Rm B32 Phone: 733-3281 Email: MAYER@MED.SC.EDU
COURSE: Medical Microbiology, MBIM 650/720 - Fall 2008 TOPIC: Antigen Processing, MHC Restriction, & Role of Thymus Lecture 12 FACULTY: Dr. Mayer Office: Bldg. #1, Rm B32 Phone: 733-3281 Email: MAYER@MED.SC.EDU
Principles of Adaptive Immunity
 Principles of Adaptive Immunity Chapter 3 Parham Hans de Haard 17 th of May 2010 Agenda Recognition molecules of adaptive immune system Features adaptive immune system Immunoglobulins and T-cell receptors
Principles of Adaptive Immunity Chapter 3 Parham Hans de Haard 17 th of May 2010 Agenda Recognition molecules of adaptive immune system Features adaptive immune system Immunoglobulins and T-cell receptors
Supplementary Figure 1
 d f a IL7 b IL GATA RORγt h HDM IL IL7 PBS Ilra R7 PBS HDM Ilra R7 HDM Foxp Foxp Ilra R7 HDM HDM Ilra R7 HDM. 9..79. CD + FOXP + T reg cell CD + FOXP T conv cell PBS Ilra R7 PBS HDM Ilra R7 HDM CD + FOXP
d f a IL7 b IL GATA RORγt h HDM IL IL7 PBS Ilra R7 PBS HDM Ilra R7 HDM Foxp Foxp Ilra R7 HDM HDM Ilra R7 HDM. 9..79. CD + FOXP + T reg cell CD + FOXP T conv cell PBS Ilra R7 PBS HDM Ilra R7 HDM CD + FOXP
Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic
 Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic B cells from WT Nfat2 +/+, TCL1 Nfat2 +/+ and TCL1 Nfat2
Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic B cells from WT Nfat2 +/+, TCL1 Nfat2 +/+ and TCL1 Nfat2
Antigen Presentation and T Lymphocyte Activation. Abul K. Abbas UCSF. FOCiS
 1 Antigen Presentation and T Lymphocyte Activation Abul K. Abbas UCSF FOCiS 2 Lecture outline Dendritic cells and antigen presentation The role of the MHC T cell activation Costimulation, the B7:CD28 family
1 Antigen Presentation and T Lymphocyte Activation Abul K. Abbas UCSF FOCiS 2 Lecture outline Dendritic cells and antigen presentation The role of the MHC T cell activation Costimulation, the B7:CD28 family
Nature Immunology: doi: /ni Supplementary Figure 1. Id2 and Id3 define polyclonal T H 1 and T FH cell subsets.
 Supplementary Figure 1 Id2 and Id3 define polyclonal T H 1 and T FH cell subsets. Id2 YFP/+ (a) or Id3 GFP/+ (b) mice were analyzed 7 days after LCMV infection. T H 1 (SLAM + CXCR5 or CXCR5 PD-1 ), T FH
Supplementary Figure 1 Id2 and Id3 define polyclonal T H 1 and T FH cell subsets. Id2 YFP/+ (a) or Id3 GFP/+ (b) mice were analyzed 7 days after LCMV infection. T H 1 (SLAM + CXCR5 or CXCR5 PD-1 ), T FH
Chapter 11. B cell generation, Activation, and Differentiation. Pro-B cells. - B cells mature in the bone marrow.
 Chapter B cell generation, Activation, and Differentiation - B cells mature in the bone marrow. - B cells proceed through a number of distinct maturational stages: ) Pro-B cell ) Pre-B cell ) Immature
Chapter B cell generation, Activation, and Differentiation - B cells mature in the bone marrow. - B cells proceed through a number of distinct maturational stages: ) Pro-B cell ) Pre-B cell ) Immature
The Major Histocompatibility Complex (MHC)
 The Major Histocompatibility Complex (MHC) An introduction to adaptive immune system before we discuss MHC B cells The main cells of adaptive immune system are: -B cells -T cells B cells: Recognize antigens
The Major Histocompatibility Complex (MHC) An introduction to adaptive immune system before we discuss MHC B cells The main cells of adaptive immune system are: -B cells -T cells B cells: Recognize antigens
Cells communicate with each other via signaling ligands which interact with receptors located on the surface or inside the target cell.
 BENG 100 Frontiers of Biomedical Engineering Professor Mark Saltzman Chapter 6 SUMMARY In this chapter, cell signaling was presented within the context of three physiological systems that utilize communication
BENG 100 Frontiers of Biomedical Engineering Professor Mark Saltzman Chapter 6 SUMMARY In this chapter, cell signaling was presented within the context of three physiological systems that utilize communication
Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was
 Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was sorted by FACS. Surface markers for sorting were CD11c +
Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was sorted by FACS. Surface markers for sorting were CD11c +
Medical Virology Immunology. Dr. Sameer Naji, MB, BCh, PhD (UK) Head of Basic Medical Sciences Dept. Faculty of Medicine The Hashemite University
 Medical Virology Immunology Dr. Sameer Naji, MB, BCh, PhD (UK) Head of Basic Medical Sciences Dept. Faculty of Medicine The Hashemite University Human blood cells Phases of immune responses Microbe Naïve
Medical Virology Immunology Dr. Sameer Naji, MB, BCh, PhD (UK) Head of Basic Medical Sciences Dept. Faculty of Medicine The Hashemite University Human blood cells Phases of immune responses Microbe Naïve
D CD8 T cell number (x10 6 )
 IFNγ Supplemental Figure 1. CD T cell number (x1 6 ) 18 15 1 9 6 3 CD CD T cells CD6L C CD5 CD T cells CD6L D CD8 T cell number (x1 6 ) 1 8 6 E CD CD8 T cells CD6L F Log(1)CFU/g Feces 1 8 6 p
IFNγ Supplemental Figure 1. CD T cell number (x1 6 ) 18 15 1 9 6 3 CD CD T cells CD6L C CD5 CD T cells CD6L D CD8 T cell number (x1 6 ) 1 8 6 E CD CD8 T cells CD6L F Log(1)CFU/g Feces 1 8 6 p
RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh-
 1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
Nature Immunology: doi: /ni Supplementary Figure 1. Characteristics of SEs in T reg and T conv cells.
 Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system
 Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Can we classify cancer using cell signaling?
 Can we classify cancer using cell signaling? Central hypotheses (big ideas) Alterations to signaling genes would cause leukemic cells to react in an inappropriate or sensitized manner to environmental
Can we classify cancer using cell signaling? Central hypotheses (big ideas) Alterations to signaling genes would cause leukemic cells to react in an inappropriate or sensitized manner to environmental
Peli1 negatively regulates T-cell activation and prevents autoimmunity
 Peli1 negatively regulates T-cell activation and prevents autoimmunity Mikyoung Chang 1,*, Wei Jin 1,5,*, Jae-Hoon Chang 1, Yi-chuan Xiao 1, George Brittain 1, Jiayi Yu 1, Xiaofei Zhou 1, Yi-Hong Wang
Peli1 negatively regulates T-cell activation and prevents autoimmunity Mikyoung Chang 1,*, Wei Jin 1,5,*, Jae-Hoon Chang 1, Yi-chuan Xiao 1, George Brittain 1, Jiayi Yu 1, Xiaofei Zhou 1, Yi-Hong Wang
Evidence for an Alternative Glycolytic Pathway in Rapidly Proliferating Cells. Matthew G. Vander Heiden, et al. Science 2010
 Evidence for an Alternative Glycolytic Pathway in Rapidly Proliferating Cells Matthew G. Vander Heiden, et al. Science 2010 Introduction The Warburg Effect Cancer cells metabolize glucose differently Primarily
Evidence for an Alternative Glycolytic Pathway in Rapidly Proliferating Cells Matthew G. Vander Heiden, et al. Science 2010 Introduction The Warburg Effect Cancer cells metabolize glucose differently Primarily
REGULATION OF ENZYME ACTIVITY. Medical Biochemistry, Lecture 25
 REGULATION OF ENZYME ACTIVITY Medical Biochemistry, Lecture 25 Lecture 25, Outline General properties of enzyme regulation Regulation of enzyme concentrations Allosteric enzymes and feedback inhibition
REGULATION OF ENZYME ACTIVITY Medical Biochemistry, Lecture 25 Lecture 25, Outline General properties of enzyme regulation Regulation of enzyme concentrations Allosteric enzymes and feedback inhibition
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Supplementary Figure 1 Cytokine receptors on developing thymocytes that can potentially signal Runx3d expression.
 Supplementary Figure 1 Cytokine receptors on developing thymocytes that can potentially signal Runx3d expression. (a) Characterization of c-independent SP8 cells. Stainings for maturation markers (top)
Supplementary Figure 1 Cytokine receptors on developing thymocytes that can potentially signal Runx3d expression. (a) Characterization of c-independent SP8 cells. Stainings for maturation markers (top)
Supplemental Information. Checkpoint Blockade Immunotherapy. Induces Dynamic Changes. in PD-1 CD8 + Tumor-Infiltrating T Cells
 Immunity, Volume 50 Supplemental Information Checkpoint Blockade Immunotherapy Induces Dynamic Changes in PD-1 CD8 + Tumor-Infiltrating T Cells Sema Kurtulus, Asaf Madi, Giulia Escobar, Max Klapholz, Jackson
Immunity, Volume 50 Supplemental Information Checkpoint Blockade Immunotherapy Induces Dynamic Changes in PD-1 CD8 + Tumor-Infiltrating T Cells Sema Kurtulus, Asaf Madi, Giulia Escobar, Max Klapholz, Jackson
Tolerance, autoimmunity and the pathogenesis of immunemediated inflammatory diseases. Abul K. Abbas UCSF
 Tolerance, autoimmunity and the pathogenesis of immunemediated inflammatory diseases Abul K. Abbas UCSF Balancing lymphocyte activation and control Activation Effector T cells Tolerance Regulatory T cells
Tolerance, autoimmunity and the pathogenesis of immunemediated inflammatory diseases Abul K. Abbas UCSF Balancing lymphocyte activation and control Activation Effector T cells Tolerance Regulatory T cells
CYTOKINE RECEPTORS AND SIGNAL TRANSDUCTION
 CYTOKINE RECEPTORS AND SIGNAL TRANSDUCTION What is Cytokine? Secreted popypeptide (protein) involved in cell-to-cell signaling. Acts in paracrine or autocrine fashion through specific cellular receptors.
CYTOKINE RECEPTORS AND SIGNAL TRANSDUCTION What is Cytokine? Secreted popypeptide (protein) involved in cell-to-cell signaling. Acts in paracrine or autocrine fashion through specific cellular receptors.
Akt and mtor pathways differentially regulate the development of natural and inducible. T H 17 cells
 Akt and mtor pathways differentially regulate the development of natural and inducible T H 17 cells Jiyeon S Kim, Tammarah Sklarz, Lauren Banks, Mercy Gohil, Adam T Waickman, Nicolas Skuli, Bryan L Krock,
Akt and mtor pathways differentially regulate the development of natural and inducible T H 17 cells Jiyeon S Kim, Tammarah Sklarz, Lauren Banks, Mercy Gohil, Adam T Waickman, Nicolas Skuli, Bryan L Krock,
Proteasome activity regulates CD8 + T lymphocyte metabolism and fate specification
 Proteasome activity regulates CD8 + T lymphocyte metabolism and fate specification Christella E. Widjaja,, Huib Ovaa, John T. Chang J Clin Invest. 2017;127(10):3609-3623. https://doi.org/10.1172/jci90895.
Proteasome activity regulates CD8 + T lymphocyte metabolism and fate specification Christella E. Widjaja,, Huib Ovaa, John T. Chang J Clin Invest. 2017;127(10):3609-3623. https://doi.org/10.1172/jci90895.
Mouse Clec9a ORF sequence
 Mouse Clec9a gene LOCUS NC_72 13843 bp DNA linear CON 1-JUL-27 DEFINITION Mus musculus chromosome 6, reference assembly (C57BL/6J). ACCESSION NC_72 REGION: 129358881-129372723 Mouse Clec9a ORF sequence
Mouse Clec9a gene LOCUS NC_72 13843 bp DNA linear CON 1-JUL-27 DEFINITION Mus musculus chromosome 6, reference assembly (C57BL/6J). ACCESSION NC_72 REGION: 129358881-129372723 Mouse Clec9a ORF sequence
ACTIVATION AND EFFECTOR FUNCTIONS OF CELL-MEDIATED IMMUNITY AND NK CELLS. Choompone Sakonwasun, MD (Hons), FRCPT
 ACTIVATION AND EFFECTOR FUNCTIONS OF CELL-MEDIATED IMMUNITY AND NK CELLS Choompone Sakonwasun, MD (Hons), FRCPT Types of Adaptive Immunity Types of T Cell-mediated Immune Reactions CTLs = cytotoxic T lymphocytes
ACTIVATION AND EFFECTOR FUNCTIONS OF CELL-MEDIATED IMMUNITY AND NK CELLS Choompone Sakonwasun, MD (Hons), FRCPT Types of Adaptive Immunity Types of T Cell-mediated Immune Reactions CTLs = cytotoxic T lymphocytes
Supplementary Figure 1
 Supplementary Figure 1 Expression of apoptosis-related genes in tumor T reg cells. (a) Identification of FOXP3 T reg cells by FACS. CD45 + cells were gated as enriched lymphoid cell populations with low-granularity.
Supplementary Figure 1 Expression of apoptosis-related genes in tumor T reg cells. (a) Identification of FOXP3 T reg cells by FACS. CD45 + cells were gated as enriched lymphoid cell populations with low-granularity.
Nature Immunology: doi: /ni Supplementary Figure 1. Huwe1 has high expression in HSCs and is necessary for quiescence.
 Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
2? If there is a suboptimum stimulus, the potency of which can relate to ligand affinity
 1 Confirmation that T cell receptor activation is digital, not analog. Hence individual T cell cytolytic capacity is independent of initial stimulation strength Abstract. Whether a sub-optimum lymphocyte
1 Confirmation that T cell receptor activation is digital, not analog. Hence individual T cell cytolytic capacity is independent of initial stimulation strength Abstract. Whether a sub-optimum lymphocyte
W/T Itgam -/- F4/80 CD115. F4/80 hi CD115 + F4/80 + CD115 +
 F4/8 % in the peritoneal lavage 6 4 2 p=.15 n.s p=.76 CD115 F4/8 hi CD115 + F4/8 + CD115 + F4/8 hi CD115 + F4/8 + CD115 + MHCII MHCII Supplementary Figure S1. CD11b deficiency affects the cellular responses
F4/8 % in the peritoneal lavage 6 4 2 p=.15 n.s p=.76 CD115 F4/8 hi CD115 + F4/8 + CD115 + F4/8 hi CD115 + F4/8 + CD115 + MHCII MHCII Supplementary Figure S1. CD11b deficiency affects the cellular responses
KEY CONCEPT QUESTIONS IN SIGNAL TRANSDUCTION
 Signal Transduction - Part 2 Key Concepts - Receptor tyrosine kinases control cell metabolism and proliferation Growth factor signaling through Ras Mutated cell signaling genes in cancer cells are called
Signal Transduction - Part 2 Key Concepts - Receptor tyrosine kinases control cell metabolism and proliferation Growth factor signaling through Ras Mutated cell signaling genes in cancer cells are called
T cell development October 28, Dan Stetson
 T cell development October 28, 2016 Dan Stetson stetson@uw.edu 441 Lecture #13 Slide 1 of 29 Three lectures on T cells (Chapters 8, 9) Part 1 (Today): T cell development in the thymus Chapter 8, pages
T cell development October 28, 2016 Dan Stetson stetson@uw.edu 441 Lecture #13 Slide 1 of 29 Three lectures on T cells (Chapters 8, 9) Part 1 (Today): T cell development in the thymus Chapter 8, pages
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.10/nature10195 NCBI gene: Tagged Subunit(s: HA-Vpx; FLAG-Cul4 HA-DCAF1 FLAG-Cul4 HA-FLAG-Vpx Mock Vpx (SIVmac 100 (a ; 0.159 (b ; 0.05 DCAF1 DDB1 DDA1 Cul4A 1; 0.024591
SUPPLEMENTARY INFORMATION doi:10.10/nature10195 NCBI gene: Tagged Subunit(s: HA-Vpx; FLAG-Cul4 HA-DCAF1 FLAG-Cul4 HA-FLAG-Vpx Mock Vpx (SIVmac 100 (a ; 0.159 (b ; 0.05 DCAF1 DDB1 DDA1 Cul4A 1; 0.024591
T Lymphocyte Activation and Costimulation. FOCiS. Lecture outline
 1 T Lymphocyte Activation and Costimulation Abul K. Abbas, MD UCSF FOCiS 2 Lecture outline T cell activation Costimulation, the B7:CD28 family Inhibitory receptors of T cells Targeting costimulators for
1 T Lymphocyte Activation and Costimulation Abul K. Abbas, MD UCSF FOCiS 2 Lecture outline T cell activation Costimulation, the B7:CD28 family Inhibitory receptors of T cells Targeting costimulators for
Nature Immunology: doi: /ni Supplementary Figure 1. Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice.
 Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
Nature Medicine: doi: /nm.3922
 Title: Glucocorticoid-induced tumor necrosis factor receptor-related protein co-stimulation facilitates tumor regression by inducing IL-9-producing helper T cells Authors: Il-Kyu Kim, Byung-Seok Kim, Choong-Hyun
Title: Glucocorticoid-induced tumor necrosis factor receptor-related protein co-stimulation facilitates tumor regression by inducing IL-9-producing helper T cells Authors: Il-Kyu Kim, Byung-Seok Kim, Choong-Hyun
CD4 + T cells recovered in Rag2 / recipient ( 10 5 ) Heart Lung Pancreas
 a CD4 + T cells recovered in Rag2 / recipient ( 1 5 ) Heart Lung Pancreas.5 1 2 4 6 2 4 6 Ctla4 +/+ Ctla4 / Ctla4 / Lung Ctla4 / Pancreas b Heart Lung Pancreas Ctla4 +/+ Ctla4 / Ctla4 / Lung Ctla4 / Pancreas
a CD4 + T cells recovered in Rag2 / recipient ( 1 5 ) Heart Lung Pancreas.5 1 2 4 6 2 4 6 Ctla4 +/+ Ctla4 / Ctla4 / Lung Ctla4 / Pancreas b Heart Lung Pancreas Ctla4 +/+ Ctla4 / Ctla4 / Lung Ctla4 / Pancreas
Effector T Cells and
 1 Effector T Cells and Cytokines Andrew Lichtman, MD PhD Brigham and Women's Hospital Harvard Medical School 2 Lecture outline Cytokines Subsets of CD4+ T cells: definitions, functions, development New
1 Effector T Cells and Cytokines Andrew Lichtman, MD PhD Brigham and Women's Hospital Harvard Medical School 2 Lecture outline Cytokines Subsets of CD4+ T cells: definitions, functions, development New
Nuclear Export of Histone Deacetylase 7 During Thymic Selection is required for Immune Self-tolerance
 The EMBO Journal Peer Review Process File - EMBO-2012-80891 Manuscript EMBO-2012-80891 Nuclear Export of Histone Deacetylase 7 During Thymic Selection is required for Immune Self-tolerance Herbert G Kasler,
The EMBO Journal Peer Review Process File - EMBO-2012-80891 Manuscript EMBO-2012-80891 Nuclear Export of Histone Deacetylase 7 During Thymic Selection is required for Immune Self-tolerance Herbert G Kasler,
T Cell Development II: Positive and Negative Selection
 T Cell Development II: Positive and Negative Selection 8 88 The two phases of thymic development: - production of T cell receptors for antigen, by rearrangement of the TCR genes CD4 - selection of T cells
T Cell Development II: Positive and Negative Selection 8 88 The two phases of thymic development: - production of T cell receptors for antigen, by rearrangement of the TCR genes CD4 - selection of T cells
well for 2 h at rt. Each dot represents an individual mouse and bar is the mean ±
 Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Transcription factor Foxp3 and its protein partners form a complex regulatory network
 Supplementary figures Resource Paper Transcription factor Foxp3 and its protein partners form a complex regulatory network Dipayan Rudra 1, Paul deroos 1, Ashutosh Chaudhry 1, Rachel Niec 1, Aaron Arvey
Supplementary figures Resource Paper Transcription factor Foxp3 and its protein partners form a complex regulatory network Dipayan Rudra 1, Paul deroos 1, Ashutosh Chaudhry 1, Rachel Niec 1, Aaron Arvey
