Article. Actin Filament Severing by Cofilin Dismantles Actin Patches and Produces Mother Filaments for New Patches
|
|
|
- Randell Hancock
- 6 years ago
- Views:
Transcription
1 Current Biology 23, , July 8, 2013 ª2013 Elsevier Ltd All rights reserved Actin Filament Severing by Cofilin Dismantles Actin Patches and Produces Mother Filaments for New Patches Article Qian Chen 1 and Thomas D. Pollard 1,2,3, * 1 Department of Molecular, Cellular, and Developmental Biology 2 Department of Molecular Biophysics and Biochemistry 3 Department of Cell Biology Yale University, P.O. Box , New Haven, CT , USA Summary Background: Yeast cells depend on Arp2/3 complex to assemble actin filaments at sites of endocytosis, but the source of the initial filaments required to activate Arp2/3 complex is not known. Results: We tested the proposal that cofilin severs actin filaments during endocytosis in fission yeast cells using a mutant cofilin defective in severing. We used quantitative fluorescence microscopy to track mgfp-tagged proteins, including early endocytic adaptor proteins, activators of Arp2/3 complex, and actin filaments. Consistent with the hypothesis, actin patches disassembled far more slowly in cells depending on severing-deficient cofilin than in wild-type cells. Even more interesting, actin patches assembled slowly in these cofilin mutant cells. Adaptor proteins End4p and Pan1p accumulated and persisted at endocytic sites more than ten times longer than in wild-type cells, followed by slow but persistent recruitment of activators of Arp2/3 complex, including WASP and myosin-i. Mutations revealed that actin filament binding sites on adaptor proteins Pan1p and End4p contribute to initiating actin polymerization in actin patches. Conclusions: We propose a sever, diffuse, and trigger model for the nucleation of actin filaments at sites of endocytosis, whereby cofilin generates actin filament fragments that diffuse through the cytoplasm, bind adaptor proteins at nascent sites of endocytosis, and serve as mother filaments to initiate the autocatalytic assembly of the branched actin filament network of each new patch. This hypothesis explains the source of the mother filaments that are absolutely required for Arp2/3 complex to nucleate actin polymerization. Introduction Assembly of actin plays a key role in clathrin-mediated endocytosis in yeast by providing both scaffold for the membrane invagination and force for scission of the vesicle against turgor pressure [1 3] (for review see [4, 5]). The sites of endocytosis are called actin patches. Genetics and quantitative fluorescence microscopy provided many details about the molecular composition and assembly pathway of actin patches, including evidence that Arp2/3 complex nucleates the actin filaments [6 8], but questions remain about every stage of the process. Biochemical studies showed that Arp2/3 complex must bind both a nucleation-promoting factor (NPF) and a preexisting filament to *Correspondence: thomas.pollard@yale.edu initiate a new filament [9]. However, the source of these mother filaments is unclear in fungi [7]. An immediately adjacent actin patch can be a source of actin filaments [10], but most patches form in regions of cortex apparently devoid of actin filaments [11]. However, electron microscopy of animal cells suggested that actin filaments may be present at sites of endocytosis earlier than indicated by fluorescence microscopy [12]. Questions also remain about disassembly of the actin coat. Simulations of a mathematical model of fission yeast endocytic patches demonstrated that dissociation of subunits from the ends of actin filaments is too slow to account for disappearance of the filaments in 10 s, unless the filaments are severed into fragments small enough to diffuse out of the patch and disassemble elsewhere [7]. Dissociation of Arp2/3 complex from the branched actin filaments has been proposed as a mechanism [13], and while cofilin stimulates debranching [14], this reaction was not necessary to account for the turnover of filaments in patches [7]. The small protein cofilin severs actin filaments [15 17] and contributes to many processes, including endocytosis, cell motility, and cytokinesis [18 20]. Cofilin concentrates in yeast endocytic actin patches [21 23], and cofilin mutations cause endocytic defects [19, 21, 23]. Here we used a mutant cofilin with a selective defect in severing [18] to study how actin filament severing contributes to endocytosis in fission yeast. Endocytic actin patches disassembled very slowly in mutant cells, as anticipated by studies in budding yeast [21, 23]. Surprisingly, actin patch assembly took longer in cofilin mutant cells, with prolonged accumulation of both membrane adaptor proteins and activators of Arp2/3 complex before assembly of actin. Depletion of the actin monomer pool could not explain these defects in cofilin mutant cells. Instead, we present evidence to support a feedback mechanism where severing by cofilin generates short actin filaments that interact with endocytic adaptor proteins End4p and Pan1p and Arp2/3 complex to initiate actin polymerization at sites of endocytosis. Results Cofilin Concentrates in Actin Patches We studied the localization of cofilin in fission yeast by expressing cofilin tagged on the N terminus with mgfp (Figure 1A; see also Figure S1A available online). The protein is the product of the adf1 + gene, but we use the common name, cofilin or mgfp-cofilin, because adf1, actin-depolymerizing factor 1, is a misleading name for a protein that does not depolymerize actin filaments [15 17]. This fusion protein must be overexpressed from either the adh1 (alcohol dehydrogenase 1) or the strong 3nmt1 promoter to complement a Dadf1 null mutation [18, 22]. The adf1::padh-mgfp-adf1 cells expressed about 10-fold more mgfp-adf1p than Adf1p in the adf1 + cells (Figure S1B). These cells grew normally at both 25 C and 30 C but slightly more slowly at 36 C, compared to wild-type cells (Figure S1C). After correcting for excluded volume [24], the total fluorescence in adf1::padh-mgfp-adf1 cells corresponded to a cytoplasmic concentration of 200 mm mgfp-cofilin.
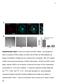
2 Cofilin Contributes to Assembly of Actin Patch 1155 Figure 1. Cofilin Localization in Endocytic Actin Patches of Fission Yeast Cells and Effects of a Cofilin Mutation that Reduces Severing on Actin Filament Accumulation in Patches (A) Time course of mgfp-cofilin in actin patches. Upper panel shows a time series of negative-contrast fluorescence micrographs of an actin patch at 1 s intervals. Images are sums of five median slices of the cell. Lower panel shows a graph of the average number of molecules of mgfp-cofilin per patch (6SD) over the time. (B) Time courses of normalized fluorescence intensities of mgfp-cofilin (green) and Fim1p-mCherry (red) in one actin patch. See also Figure S1. (C and D) Comparisons of actin patches in wild-type adf1 + and cofilin mutant adf1-m2 cells. Maximum-intensity projections of z series of fluorescence micrographs of adf1 + cells and adf1-m2 cells fixed and stained with rhodamine-phalloidin for actin filaments are shown in (C). Histogram of fluorescence intensities of 50 actin patches from adf1 + (open) and adf1-m2 (black) cells are shown in (D). See also Figure S1. We used two-color fluorescence microscopy to compare mgfp-cofilin with either End4p-mCherry, an early endocytic adaptor protein, or Fim1p-mCherry, the fission yeast homolog of the actin filament binding protein fimbrin (Figures 1B and S1D). Actin patches in cells depending on overexpressed mgfp-cofilin assembled and disassembled Fim1p-mCherry normally (Figure S1E), and the lifetime of End4p increased only slightly from 36 s to 39 s (n = 20). mgfp-cofilin appeared at sites of endocytosis 5 s before actin patches began to move from the cell surface (defined as time zero), peaked at 6,000 molecules at time +10 s, and then gradually dissipated over the next 10 s (Figure 1A). The mgfp-cofilin fluorescence peaked after both End4p and Fim1p and persisted after both proteins dissociated from moving patches (Figures 1B and S1D). Endocytic Defects in Cofilin Mutant adf1-m2 Cells To study the role of cofilin during endocytosis, we used cells depending on mutant cofilin-m2, which binds and severs actin filaments far more slowly than the wild-type cofilin [18]. Fission yeast adf1-m2 cells have larger diameters than the wild-type cells, similar to many other endocytic mutants [25]. Calcofluor white stained the cell walls more intensely in these mutant cells than in wild-type cells (data not shown), a defect observed in other endocytic mutants. The average fluorescence intensity of actin patches stained by rhodamine-phalloidin was 2-fold greater in adf1-m2 cells ( , n = 50) than in wild-type cells ( , n = 50) (Figures 1C and 1D). During interphase, these bright actin patches packed more densely at the poles of cofilin mutant cells than in wild-type cells (Figure 1C). We looked for endocytic defects in the adf1-m2 cells using two approaches: (1) localization of the fission yeast homolog of the SNARE protein synaptobrevin Syb1p (Figure S2F) and (2) pulse-chase experiments with the fluorescent lipophilic dye FM4-64 (Figure S2G). After exocytosis, Syb1p recycles from the plasma membrane through the endocytic pathway, a process compromised by many endocytic mutations in both budding and fission yeast [10, 26]. GFP-Syb1p concentrated in numerous cytosolic puncta in both wild-type and adf1-m2 cells but associated mostly with the plasma membrane in adf1-m2 cells (Figure S2F), consistent with defects in endocytosis. Pulse-chase experiments confirmed that adf1- M2 cells internalized FM4-64 very slowly. Wild-type cells took up FM4-64 from the plasma membrane into numerous cytoplasmic puncta in <8 min (Figure S2G), but after 16 min most of the dye remained on the surface of adf1-m2 cells with very few fluorescence puncta in the cytoplasm (Figure S2G). Defects in Actin Patch Assembly and Disassembly in Cofilin Mutant adf1-m2 Cells We used quantitative fluorescence microscopy to compare actin patch dynamics of wild-type and adf1-m2 cells expressing six endocytic proteins tagged with mgfp (Figures 2 and S2A). We aligned time courses of all actin patches based on their initial inward movement (time zero). As in budding yeast [4, 8], fission yeast initiate endocytosis by sequentially recruiting clathrin, adaptor proteins, NPFs for Arp2/3 complex, and finally Arp2/3 complex itself [6]. We tracked two adaptor proteins, End4p/Sla2p (a homolog of Hip1r [27]) and Pan1p (an Eps15 protein [28]). We also tracked NPFs, the homolog of Wiskott-Aldrich syndrome protein (Wsp1p), and myosin-i (Myo1p), which stimulate Arp2/3 complex to initiate the explosive assembly of actin filaments [7, 25, 29, 30] in cooperation with the F-BAR proteins [31]. We tracked actin filaments with either Fim1p-mGFP expressed from its genomic locus or mgfp-act1p (mgfp-actin) expressed at a low level from the leu1 locus under the control of a suppressible 41nmt1 promoter in cells with a wild-type actin gene. Actin patches in adf1-m2 cells accumulated mgfp-actin at the same rate as wild-type cells, but the accumulation phase lasted three times longer (30 s versus 10 s) (Figure 2B), so the peak number of mgfp-actin molecules per actin patch was 3-fold higher in cofilin mutant cells than in wild-type cells (Figure 2B), confirming the more intense staining with rhodamine-phalloidin (Figure 1C). Nevertheless, these actin patches in adf1-m2 cells accumulated the normal amount of the actinbinding protein Fim1p-mGFP but more slowly than wild-type cells (Figure 2A). After time zero, mgfp-actin and Fim1pmGFP took about 10 s to disappear from patches in wildtype cells (Figure 2A), but both proteins persisted far longer in adf1-m2 cells, taking as long as 180 s to completely disappear from some patches (Figure 2A). In many cases, these
3 Current Biology Vol 23 No Figure 2. Accumulation and Disappearance of Fluorescent Proteins in Actin Patches Time courses of the accumulation and disappearance of fluorescent proteins in actin patches of wild-type adf1 + cells (blue lines) and cofilin mutant adf1-m2 cells (red lines). Graphs show the average number of molecules at each point in time. Data from individual patches were aligned at the time they started to move (time zero, green line). Inserts are kymographs of fluorescence micrographs of individual actin patches in wild-type cells (blue outline) or adf1-m2 cells (red outline). Inserts in (E) and (F) are expanded timescales for data in dashed boxes. Shown are Fim1p-mGFP (A), mgfp-act1p (B), End4p-mGFP (C), Pan1p-mGFP (D), mgfp-wsp1p (E), and mgfp-myo1p (F). See also Figure S2. persisting actin patches in adf1-m2 cells moved from their initial locations, and occasionally they appeared to merge with nearby actin patches. Quantitative microscopy revealed that the early stages of actin patch assembly were prolonged in adf1-m2 cells (Figures 2C and 2D). Two adaptor proteins, End4p-mGFP and Pan1p-mGFP, appeared at actin patches around time 230 s in wild-type cells, but both appeared before 2100 s in adf1- M2 cells and accumulated to higher levels than in wild-type cells (Figures 2C and 2D). Prolonged accumulation of End4p and Pan1p resulted in w1.5-fold more of these adaptors in actin patches (Figure 3A) at the time NPFs arrived prior to actin polymerization. Imaging actin patches in adf1-m2 and wild-type cells side by side confirmed the presence of more End4p-mGFP in the long-lived actin patches of the mutant cells (Figure 3B), as observed by quantitative fluorescence imaging. The NPFs Wsp1p and Myo1p also accumulated more slowly in the actin patches of adf1-m2 cells than in wild-type cells (Figures 2E and 2F), but the cofilin mutation did not alter their peak numbers (Figure 3A). Both membrane adaptor proteins dissociated from actin patches in 20 s in adf1-m2 cells, slightly more slowly than in wild-type cells (10 s) (Figures 2C and 2D). The movements of patches away from the cell surface were indistinguishable in adf1- M2 and wild-type cells (Figure S2B). Similarly, both NPFs disappeared slightly more slowly than normal from patches in adf1-m2 cells. We used two-color fluorescence microscopy of live cells expressing a tagged adaptor protein and either Myo1p or Fim1p to confirm the timing of these events (Figures 3C 3F). Thus, defective actin filament severing in adf1-m2 cells not only slowed disassembly of actin patches (as expected) but also prolonged the accumulation of adaptor proteins and NPFs, delaying the initiation of actin filament assembly in actin patches, a defect that we confirmed in cells depending on the mutant cofilin-m3 with a slightly milder defect in severing actin filaments (Figures S2C and S2D) [18]. We considered two mechanisms that might explain these assembly defects: (1) depletion of the cytoplasmic pool of actin monomers and (2) a deficiency of actin filament fragments to stimulate Arp2/3 complex in adjacent actin patches. Depletion of Actin Monomers Does Not Mimic the Defects in Actin Patch Assembly of Cofilin Mutant Cells We investigated whether slow severing and depolymerization of actin filaments in adf1-m2 cells might deplete the cytoplasmic pool of actin monomers, leading secondarily to defects in actin patch assembly. A shortage of actin monomers would not only slow nucleation and elongation of actin filaments but also might compromise binding of Arp2/3 complex to mother filaments [32]. Quantitative fluorescence microscopy of cells expressing mgfp-actin from the 41nmt1 promoter in the leu1 locus showed that the cytoplasmic mgfp-actin fluorescence was 10% lower in adf1-m2 cells than in wild-type cells (Figure S2E). To test whether this slightly lower pool of cytoplasmic actin might cause endocytic defects in adf1-m2 cells, we studied actin patches in wild-type cells treated with latrunculin A (LatA), a small molecule that binds actin monomers with high affinity and inhibits polymerization [33 35]. At 8 mm, LatA increased the cytoplasmic fluorescence of mgfp-actin w30% (Figure S2F). We interpret this change to be sequestered mgfp-actin bound to LatA. This effect of LatA on the pool of unpolymerized actin available to assemble actin patches should be sufficient to mimic the 10% smaller amount of cytoplasmic actin in adf1-m2 cells.
4 Cofilin Contributes to Assembly of Actin Patch 1157 Figure 3. Accumulation of Endocytic Adaptor Proteins in Actin Patches of Wild-Type and Cofilin Mutant Cells before Actin Assembly (A) Histogram of peak numbers of proteins per actin patch in wild-type adf1 + cells (open bars) and cofilin mutant adf1-m2 cells (shaded bars). Mean numbers 6 SD are normalized against wild-type actin patches. p values are from two-tailed Student s t tests. (B) Side-by-side comparison of actin patches in wild-type (asterisk) and adf1-m2 cells expressing End4p-mGFP. Top panel shows a fluorescence micrograph (sum of five middle z slices of the cells) of a mixed population of cells. Wild-type cells were identified by expression of Fim1p-mCherry. Data for the kymograph were collected along the white line. Bottom panel shows a kymograph of pseudocolored fluorescence micrographs of actin patches in a pair of neighboring cells showing three actin patches (arrows) in the wild-type cell top and one prolonged actin patch in the adf-m2 cell bottom. (C and D) Kymographs of fluorescence micrographs of actin patches in wild-type adf1 + (left) and adf1-m2 (right) cells expressing pairs of fluorescent proteins, either End4p-mYFP and mcfp-myo1p (C) or End4p-mGFP and Fim1p-mCherry (D). Bottom panels are merged images with End4 (in green) and Myo1p/Fim1p (in red). Arrows designate the interval between the appearance of End4p and Myo1p/Fim1p in the patches. (E and F) Time courses of normalized fluorescence intensities of End4p-mGFP (green) and Fim1p-mCherry (red) in actin patches of wild-type adf1 + (E) and adf1-m2 cells (F). See also Figure S2. Low concentrations of LatA (0 8 mm) prolonged the accumulation of both End4p-mYFP and mcfp-myo1p in patches (Figures S2F S2H). However, LatA only led to a small delay in the interval between the appearance of End4p and Myo1p in actin patches of wild-type cells (Figures S2F S2H), as observed in adf1-m2 cells (Figure 3). Furthermore, low concentrations of LatA inhibited the inward movement of actin patches, a defect not observed in cofilin mutant cells. For example, most actin patches (nine out of ten) failed to move inward in cells treated with 8 mm LatA. Therefore, sequestering a fraction of the pool of monomeric actin with LatA did not replicate the phenotype of cofilin mutant cells. Roles of End4p and Pan1p in Initiating Actin Polymerization in Actin Patches A second hypothesis to explain the defects in actin patch assembly in cofilin mutant cells is that the process depends on short actin filaments normally severed by cofilin from endocytic patches to serve as mother filaments to activate Arp2/3 complex. We also considered the possibility that proteins recruited early in the endocytic pathway might tether these filaments and enhance their impact. Among the proteins that join clathrin early in the endocytic pathway [6, 8], Pan1p and End4p are known to bind actin filaments [36, 37], making them candidates to retain diffusing actin filaments at endocytic sites before the arrival of NPFs and Arp2/3 complex. Eps15 homolog Pan1p [28] is essential in fission yeast (Figure S3A). End4p is a nonessential homolog of budding yeast Sla2p and animal Hip1r [27, 38]. Actin patches in Dend4 cells formed elongated tails as they moved from the cell surface (Figure S3D), similarly to Dsla2 deletion mutations in budding yeast [8]. Biochemical experiments showed that fission yeast Pan1p binds actin filaments like budding yeast Pan1p [39]. Pan1p has a conserved WH2 (W) motif, located at the very C terminus
5 Current Biology Vol 23 No Figure 4. Analysis of Pan1p Binding to Actin Filaments and Effects of Deleting the Actin-Binding Region of Pan1p on Actin Patches (A) Domain structures of budding yeast and fission yeast Pan1p (numbers are total amino acid residues) and Pan1p truncation mutants. (B) Graph of the dependence of the fraction of Pan pelleted on the concentration of actin filaments. Samples of 2 mm Pan with 0 20 mm actin filaments were incubated at room temperature for 30 min in KMEI buffer before pelleting at 212,300 3 g for 40 min. A bimolecular binding isotherm fit the data well with a K d of 2.9 mm. Only 40% of GST-Pan (APW) was depleted from the supernatant with 20 mm actin, indicating that much of the protein was inactive. (C) Viability of strains with the pan1dw truncation mutation, adf1-m2 mutation, or both mutations, measured by the growth of the dilution series for 2 days at 30 C. (D) Time course of average number of molecules of Pan1p-mGFP (n = 10) or Pan1pDW-mGFP (n = 12) per actin patch. (E and F) Kymographs of fluorescence micrographs of actin patches from cells expressing wild-type or mutant Pan1p. The cells expressed Pan1pDAPWtdTomato and mgfp-myo1p (E) or Pan1pDAPW-mGFP and Fim1p-mCherry (F). See also Figure S3. after acidic (A) and polyproline (PP) motifs (Figures 4A and S3B). A purified C-terminal fragment of Pan1p (Pan1p ), including the A, PP, and W domains, bound muscle actin filaments with a K d of about 3 mm (Figure 4B), although only 40% of the GST-tagged protein was active. We tested the effects on actin patch dynamics of two Pan1p truncation mutants designed to compromise binding to actin filaments. We deleted either the W domain alone in pan1d (pan1dw) or the A, PP, and W domains altogether in pan1d (pan1dapw) (Figure 4A). Cells expressed the pan1 truncation mutant proteins at levels similar to full-length Pan1p (Figure S3C). Both mutations had negative genetic interactions with the adf1-m2 mutation, with double mutants growing more slowly than the wild-type cells at 30 C(Figure 4C). Pan1pDAPW-mGFP took twice as long as full-length Pan1p-mGFP to accumulate to normal levels in actin patches but dissociated from actin patches over the same time course as full-length Pan1p-mGFP (Figure 4D). The behavior of Pan1pDW-mGFP was similar (data not shown). Strains depending on these pan1 truncation mutants took w40 s longer than wild-type cells to recruit Myo1p to actin patches (Figure 4E), so actin polymerization was delayed (Figure 4F). Similarly the interval between the appearances of End4p and Myo1p was longer in actin patches of pan1 truncation mutants than in wild-type cells (data not shown). Therefore, the WH2 domain of Pan1p contributes to the initiation of actin patch assembly but is not essential. We tested the effects on actin patch dynamics of two End4p truncation mutants designed to compromise binding to actin filaments. The C-terminal talin-like (TL) domain (also called an I/LWEQ domain) of budding yeast End4p can bind actin filaments [36, 40], but removing the TL domain had no detectable effect on the assembly of actin patches [40]. Using homology-based structural prediction ( Protein Homology/analogY Recognition Engine, we found that the End4p sequence between the central coiled-coil and C-terminal TL domain has a high degree of sequence homology to the actin filament binding domain of talin (Figure 5A). This previously unrecognized TL domain may have compensated for deletion of the C-terminal TL domain in previous work. To compromise actin filament binding of End4p, we deleted either the C-terminal TL domain
6 Cofilin Contributes to Assembly of Actin Patch 1159 Figure 5. Actin Filament Binding Domains of End4p and Pan1p Cooperate to Initiate Actin Polymerization in Endocytic Patches (A and B) Deletion of one or both talin-like domains from End4p. Domain structure of End4p and the two truncation mutants are shown in (A). Kymographs of fluorescence micrographs of actin patches in cells expressing either End4p or End4p truncation mutants tagged with mgfp and Fim1p-mCherry are shown in (B). (C) Viability of strains with the pan1dw mutation, end4dtld mutation, and both mutations (pan1dw end4dtld), measured by growth of dilution series for 2 days at 30 C. (D) Box plot of the time interval between the appearances of Ent1p and Myo1p in actin patches of wild-type, end4dtld, pan1dw, and end4dtld pan1dw cells. Horizontal lines in the boxes are average values. The boxes include 625% from the average values. Vertical lines mark minimum and maximum values, except for outliners. (E) Kymographs of fluorescence micrographs of actin patches in the wild-type, end4dtld, pan1dw, end4dtld pan1dw double mutant, and adf1-m2 cells expressing both Ent1p-Tdtomato (top panels) and mgfp-myo1p (bottom panels). The bottom panels are merged kymographs with Ent1p (in red) and Myo1p (in green). (F) Kymographs of fluorescence micrographs of actin patches in end4dtld pan1dw cells expressing End4pDTLD- mgfp (top panels) and Fim1p-Tdtomato (bottom panels). Bottom panels are merged kymographs with End4pDTLD (in green) and Fim1p (in red). Recruitment of Fim1p (left) was delayed in 83% of patches and failed in 17% of patches (right). See also Figures S3 and S4. (end4d ) or both TL domains (end4d / end4dtld) (Figure 5A). Both end4 TL domain deletion mutations had negative, temperature-sensitive interactions with the cofilin mutation adf1- M2 (Figure S3E). mgfp-end4p lacking one or both TL domains was expressed at the same level as the full-length protein (Figure S3F), and it accumulated in and disappeared from actin patches normally (Figures S3G and S3H). Both end4 mutant strains initiated actin assembly without delay (Figure 5B). The end4 mutants lacking TL domains had strong negative genetic interactions with pan1 mutants lacking the WH2 motif (Figure 5C). The double mutant pan1dw end4dtld was temperature sensitive, growing very slowly at 30 C(Figure 5C) and dead at 36 C. On the other hand, the function of the newly identified actin filament binding motif of the fission yeast homolog of Epsin (Ent1p) [41] did not overlap with those of Pan1p (Figure S4 and Supplemental Results). The pan1dw end4dtld cells took up FM4-64 much more slowly than wildtype cells, even at the permissive temperature of 25 C. Wildtype cells took up most of the cell surface FM4-64 in less than 9 min, but most of the dye remained on the cell surface of double-mutant cells after 15 min (Figure S3I). We observed endocytic adaptor protein Ent1p-tdTomato and mgfp-myo1p to confirm that serious defects in actin patch assembly compromise endocytosis in pan1dw end4dtld double-mutant cells (Figures 5D and 5E). Ent1p normally
7 Current Biology Vol 23 No Figure 6. Sever, Diffuse, and Trigger Model for Actin Filament Turnover in Actin Patches The seven steps (numbers next to the arrows) are: (1) Clathrin-coated pits bind adaptor proteins End4p and Pan1p. (2) Short, diffusing actin filaments bind to End4p and Pan1p associated with coated pits. (3) Arp2/3 complex interacts with these mother filaments and nucleation-promoting factors to (4) initiate branching nucleation of actin filaments that promote elongation of the endocytic tubule. (5) After abscission of the vesicle, (6) cofilin severs actin filaments to (7) disassemble actin patches and generate a pool of short, diffusing actin filaments, some of which return to the cycle at step 2. See also Figure S5. appears in actin patches w30 s before initiation of actin assembly. The interval between the appearances of Ent1p and Myo1p in actin patches was s (n = 20) in wild-type cells, s (n = 30) in end4dtld cells, s (n = 30) in pan1dw cells, and s (n = 31) in end4dtld pan1dw doublemutant cells at the permissive temperature (Figure 5D). Actin patches in adf1-m2 cells also recruited Myo1p slowly (Figure 2F). Actin assembly followed with Fim1p-mCherry was severely compromised in the double-mutant cells, with 17% (n = 30) of actin patches failing to recruit Fim1p-mCherry and initiate actin polymerization (Figure 5F). The patches in the double-mutant cells that polymerized actin did so with an average of s (n = 25) between the appearance of End4p and Fim1p compared with just 25 s in wild-type cells (Figure 5F). Although we did not detect small actin filaments diffusing from patches in cells expressing mgfp-lifeact (explained in Supplemental Results), we found that the depletion of actin filaments in cells by LatA inhibited recruitment of Arp2/3 complex and Myo1p at sites of endocytosis without preventing the accumulation of both End4p and Wsp1p (Figure S5 and Supplemental Results). Discussion Our experiments with a cofilin mutant defective in severing actin filaments confirmed that actin filament severing not only is essential for disassembling actin filaments in endocytic patches but also contributes to the assembly of actin patches. We attribute the endocytic defects of adf1-m2 cells to slow severing by the mutant cofilin. Although the affinity of cofilin-m2 for actin filaments is 5-fold lower than wild-type cofilin, cooperative binding allows 2 mm cofilin-m2 to saturate 1 mm actin filaments [18], so cofilin-m2 will bind actin filaments but sever them slowly in the mutant cells. The effects of truncation mutations of two endocytic adaptor proteins support the proposal that cofilin produces short actin filaments that bind adaptor proteins and serve as primers to initiate actin polymerization at the site of endocytosis. Actin Filament Severing by Cofilin Disassembles Endocytic Actin Patches Disassembly of endocytic actin patches depends on cofilin in budding yeast [8, 21, 23], and our experiments demonstrate that the serving activity of cofilin is required (step 6, Figure 6). Slow binding of the mutant cofilin to actin filaments should also slow dissociation of branches [14] and may contribute to the slow turnover of patches. Since the cofilin mutation had little effect on the movement of actin patches (Figure S2B), neither scission of endocytic vesicles from the plasma membrane (step 5) nor vesicle movements appear to depend on disassembly of actin filaments. Membrane adaptor proteins and NPFs dissociated normally from actin patches (step 7), so these events are also independent of actin filament severing. A Sever, Diffuse, and Trigger Hypothesis Explains the Cofilin Requirement for Actin Patch Assembly Our most important finding is that the early steps in the assembly of actin patches depend on both actin filament severing by cofilin and actin filament binding by early endocytic adaptor proteins. These two seemingly disparate features suggest a sever, diffuse, and trigger hypothesis (Figure 6): cofilin generates actin filament fragments that diffuse through the cytoplasm (step 6), bind adaptor proteins at nascent sites of endocytosis (step 2), and serve as mother filaments (step 3) to initiate the autocatalytic assembly of the branched actin filament network of each new patch (step 4). This hypothesis offers one feasible explanation for the source of mother filaments that are absolutely required for Arp2/3 complex to nucleate polymerization [9] but appear to be absent from the cortex of yeast [11, 42]. We recognize other potential sources of mother filaments, including the possibility that Wsp1p/ Las17p may nucleate actin filaments in patches [43]. Our hypothesis is an extension of a proposal that actin patches can provide mother filaments to initiate the assembly of an adjacent new patch through a touch and trigger mechanism [10]. Actin patches were observed to trigger actin filament assembly at a nearby site of endocytosis by direct contact. Although touch and trigger events were rare in wild-type cells, they were frequent in the dip1 mutant, which has defects in nucleation of actin filaments in patches [10]. The sever, diffuse, and trigger mechanism can act over longer distances to explain actin assembly at sites of endocytosis physically separated from other actin patches. Our hypothesis does not preclude the possibility that the actin filament binding domains of adaptor proteins have separate roles at a later stage, after the assembly of actin patches, as suggested by studies in budding yeast [1, 41]. Indeed, our studies suggest that the TLD domain of End4p could complement the
8 Cofilin Contributes to Assembly of Actin Patch 1161 ABD domain of Ent1p in a function separate from initiating the assembly of actin patches (Figure S4), perhaps coupling the actin filament network to the membrane of endocytic vesicles (step 5). Simulations of actin patch dynamics [7] estimated that severing releases short actin filaments at a prodigious rate of w30 per second from each >100 actin patches. The cytoplasmic concentration of actin in such fragments would rapidly reach the micromolar range, even if further severing and polymerization turn over the fragments in 10 s. A filament of 30 subunits has a translational diffusion coefficient of 5.2 mm 2 /s (see Experimental Procedures) and shorter fragments move even faster, so they diffuse rapidly throughout the cytoplasm. These short, rapidly diffusing filaments [7, 44] are difficult to distinguish from cytoplasmic actin monomers in cells with a small fraction of the total actin tagged with mgfp. Severing of filaments in actin cables nucleated by formin For3p may provide a second source of actin filament fragments [45]. Animal cells may also have small diffusing actin filaments that contribute to endocytosis. Fluorescence correlation spectroscopy (FCS) detected short (w80 subunits) actin filaments diffusing in the cortex of tissue culture cells [46], and electron microscopy of HeLa cells showed actin filaments associated with early endocytic invaginations that were not detected by correlated fluorescence microscopy [12]. Cofilin may generate these filaments, perhaps explaining how cofilin contributes to actin polymerization at the leading edge of animal cells [20]. Actin filament binding sites on adaptor proteins Pan1p and End4p are important to initiate actin nucleation in patches. The affinities of the APW motif of Pan1p and the TLD domain of End4p for actin filaments (K d = w3 mm) are higher than alpha-actinins (K d =20mM), which bind actin filaments in physiological settings. Given 240 molecules of End4p and Pan1p on the tiny surface of a coated pit (12,000 nm 2, assuming a diameter of 50 nm [47]), the density of actin filament binding sites is 1/50 nm 2. This high density of actin filament binding sites on the cytoplasmic surface of a coated pit may favor multivalent binding of short filaments. Why Are Actin Filament Severing and Actin Filament Binding by Adaptor Proteins Required to Terminate the Accumulation of Adaptor Proteins and Recruit NPFs? Slow severing of actin filaments by cofilin and deletion of actinbinding sites from the adaptor proteins End4p and Pan1p cause similar defects, prolonged recruitment of both adaptors (step 3, Figure 6) at sites of endocytosis and delayed recruitment of the NPFs Wsp1p and Myo1p. Our experiments show that the actin filament binding sites on End4p and Pan1p are complementary, since deletion of both sites causes more severe defects than deletion of the individual sites. Interestingly, a budding yeast pan1 mutant is synthetic lethal with a clathrin light chain (clc1) truncation mutant that is missing its sla2/end4-binding motif [48]. These features suggest that recruitment of actin filaments by adaptor proteins is part of a negative feedback loop that limits the accumulation of the adaptor proteins, as well as a positive signal for binding NFPs. Actin filaments are not required for adaptor proteins to concentrate at nascent sites of endocytosis, since both accumulate in the presence of LatA. However, nothing is known about how binding of actin filaments to these adaptor proteins provides negative feedback to limit their accumulation. Similarly, the LatA experiments showed that actin filaments are not required for Wsp1p to concentrate with the adaptor proteins at sites of endocytosis. Nevertheless, when severing is compromised or the adaptor proteins lack actin filament binding sites, Wsp1p binds only after the adaptor proteins have accumulated to much higher levels. Thus actin filaments must directly or indirectly favor Wsp1p binding. On the other hand, LatA reduced the recruitment of both Myo1p and Arp2/3 complex and eliminated actin filament polymerization. Verprolin (Vrp1p) may recruit a small number of Myo1p molecules to patches by linking to Wsp1p [25], but direct interactions with actin filaments are likely to recruit most Myo1p to actin patches [25, 29]. Since Myo1p appears in patches prior to Arp2/3 complex [6, 24], the initial binding sites for Myo1p may be actin filament fragments bound to End4p and Pan1p. Experimental Procedures Fission Yeast Strains, Cell Culture, and Microscopy Table S1 lists the strains used in this study. Standard yeast culture techniques were used. Cells were imaged with an Olympus IX71 microscope equipped with a confocal spinning-disk unit. The Supplemental Experimental Procedures provide details. Construction of the adf1::gfp-adf1 Strain We amplified the strong ADH promoter (Padh) sequence from fission yeast genomic DNA by PCR and subcloned it into the pfa6a-kanmx6-41nmt1- mgfp vector [49] in place of the 41nmt1 promoter. We amplified the KanMX6-Padh-mGFP fragment for targeting to the 5 0 end of the endogenous adf1 coding sequence by homologous recombination [49]. Tracking and Counting the Numbers of Fluorescent Fusion Proteins in Actin Patches We tracked actin patches semimanually with ImageJ plug-ins. We selected actin patches in time-lapse fluorescence micrographs with maximum fluorescence intensities within the middle three slices of a five-slice z series. We measured the fluorescence intensity of each actin patch as the sum of the fluorescence in three slices within a circle with a diameter of 0.67 mm (five pixels), after subtracting the background cytosolic fluorescence, based on the fluorescence surrounding the patch. Final values of the fluorescence intensities of the patches were corrected for exposure time, camera noise, uneven illumination, and photobleaching [6, 31]. We converted the fluorescence intensities of actin patches into the numbers of mgfp fusion proteins with a standard curve based on the total cellular intensities of seven standard proteins [6, 24]. Protein Purification and Binding Assay GST-Pan was expressed in E. coli and purified by passing through affinity, gel filtration, and ion exchange columns. GST-Pan was bound to actin filaments at room temperature and centrifuged at 213,300 3 g for 40 min. For details, please see Supplemental Experimental Procedures. Supplemental Information Supplemental Information includes five figures, one table, Supplemental Experimental Procedures, and Supplemental Results and can be found with this article online at Acknowledgments This work was supported by NIH research grant GM to T.D.P. The authors thank Julien Berro for providing ImageJ plug-ins and Rajesh Arasada for providing strains and for helpful discussions. The authors also thank Jian-Qiu Wu and Masaki Edamatsu for providing strains. Received: December 14, 2012 Revised: April 2, 2013 Accepted: May 2, 2013 Published: May 30, 2013
9 Current Biology Vol 23 No References 1. Kaksonen, M., Toret, C.P., and Drubin, D.G. (2005). A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell 123, Newpher, T.M., Smith, R.P., Lemmon, V., and Lemmon, S.K. (2005). In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev. Cell 9, Aghamohammadzadeh, S., and Ayscough, K.R. (2009). Differential requirements for actin during yeast and mammalian endocytosis. Nat. Cell Biol. 11, Weinberg, J., and Drubin, D.G. (2012). Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 22, Mooren, O.L., Galletta, B.J., and Cooper, J.A. (2012). Roles for actin assembly in endocytosis. Annu. Rev. Biochem. 81, Sirotkin, V., Berro, J., Macmillan, K., Zhao, L., and Pollard, T.D. (2010). Quantitative analysis of the mechanism of endocytic actin patch assembly and disassembly in fission yeast. Mol. Biol. Cell 21, Berro, J., Sirotkin, V., and Pollard, T.D. (2010). Mathematical modeling of endocytic actin patch kinetics in fission yeast: disassembly requires release of actin filament fragments. Mol. Biol. Cell 21, Kaksonen, M., Sun, Y., and Drubin, D.G. (2003). A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115, Mullins, R.D., Heuser, J.A., and Pollard, T.D. (1998). The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA 95, Basu, R., and Chang, F. (2011). Characterization of dip1p reveals a switch in Arp2/3-dependent actin assembly for fission yeast endocytosis. Curr. Biol. 21, Mulholland, J., Preuss, D., Moon, A., Wong, A., Drubin, D., and Botstein, D. (1994). Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J. Cell Biol. 125, Collins, A., Warrington, A., Taylor, K.A., and Svitkina, T. (2011). Structural organization of the actin cytoskeleton at sites of clathrinmediated endocytosis. Curr. Biol. 21, Martin, A.C., Welch, M.D., and Drubin, D.G. (2006). Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation. Nat. Cell Biol. 8, Chan, C., Beltzner, C.C., andpollard, T.D. (2009). Cofilin dissociates Arp2/ 3 complex and branches from actin filaments. Curr. Biol. 19, Andrianantoandro, E., and Pollard, T.D. (2006). Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24, Pavlov, D., Muhlrad, A., Cooper, J., Wear, M., and Reisler, E. (2007). Actin filament severing by cofilin. J. Mol. Biol. 365, Maciver, S.K., Zot, H.G., and Pollard, T.D. (1991). Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J. Cell Biol. 115, Chen, Q., and Pollard, T.D. (2011). Actin filament severing by cofilin is more important for assembly than constriction of the cytokinetic contractile ring. J. Cell Biol. 195, Lappalainen, P., and Drubin, D.G. (1997). Cofilin promotes rapid actin filament turnover in vivo. Nature 388, Ghosh, M., Song, X., Mouneimne, G., Sidani, M., Lawrence, D.S., and Condeelis, J.S. (2004). Cofilin promotes actin polymerization and defines the direction of cell motility. Science 304, Okreglak, V., and Drubin, D.G. (2007). Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J. Cell Biol. 178, Nakano, K., and Mabuchi, I. (2006). Actin-depolymerizing protein Adf1 is required for formation and maintenance of the contractile ring during cytokinesis in fission yeast. Mol. Biol. Cell 17, Lin, M.C., Galletta, B.J., Sept, D., and Cooper, J.A. (2010). Overlapping and distinct functions for cofilin, coronin and Aip1 in actin dynamics in vivo. J. Cell Sci. 123, Wu, J.Q., and Pollard, T.D. (2005). Counting cytokinesis proteins globally and locally in fission yeast. Science 310, Sirotkin, V., Beltzner, C.C., Marchand, J.B., and Pollard, T.D. (2005). Interactions of WASp, myosin-i, and verprolin with Arp2/3 complex during actin patch assembly in fission yeast. J. Cell Biol. 170, Lewis, M.J., Nichols, B.J., Prescianotto-Baschong, C., Riezman, H., and Pelham, H.R. (2000). Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell 11, Engqvist-Goldstein, A.E., Kessels, M.M., Chopra, V.S., Hayden, M.R., and Drubin, D.G. (1999). An actin-binding protein of the Sla2/ Huntingtin interacting protein 1 family is a novel component of clathrin-coated pits and vesicles. J. Cell Biol. 147, Wendland, B., and Emr, S.D. (1998). Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J. Cell Biol. 141, Lee, W.L., Bezanilla, M., and Pollard, T.D. (2000). Fission yeast myosin-i, Myo1p, stimulates actin assembly by Arp2/3 complex and shares functions with WASp. J. Cell Biol. 151, Lechler, T., Shevchenko, A., and Li, R. (2000). Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J. Cell Biol. 148, Arasada, R., and Pollard, T.D. (2011). Distinct roles for F-BAR proteins Cdc15p and Bzz1p in actin polymerization at sites of endocytosis in fission yeast. Curr. Biol. 21, Ti, S.C., Jurgenson, C.T., Nolen, B.J., and Pollard, T.D. (2011). Structural and biochemical characterization of two binding sites for nucleationpromoting factor WASp-VCA on Arp2/3 complex. Proc. Natl. Acad. Sci. USA 108, E463 E Coué, M., Brenner, S.L., Spector, I., and Korn, E.D. (1987). Inhibition of actin polymerization by latrunculin A. FEBS Lett. 213, Ayscough, K.R., Stryker, J., Pokala, N., Sanders, M., Crews, P., and Drubin, D.G. (1997). High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-a. J. Cell Biol. 137, Morton, W.M., Ayscough, K.R., and McLaughlin, P.J. (2000). Latrunculin alters the actin-monomer subunit interface to prevent polymerization. Nat. Cell Biol. 2, McCann, R.O., and Craig, S.W. (1997). The I/LWEQ module: a conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc. Natl. Acad. Sci. USA 94, Toshima, J., Toshima, J.Y., Martin, A.C., and Drubin, D.G. (2005). Phosphoregulation of Arp2/3-dependent actin assembly during receptor-mediated endocytosis. Nat. Cell Biol. 7, Iwaki, T., Tanaka, N., Takagi, H., Giga-Hama, Y., and Takegawa, K. (2004). Characterization of end4+, a gene required for endocytosis in Schizosaccharomyces pombe. Yeast 21, Duncan, M.C., Cope, M.J., Goode, B.L., Wendland, B., and Drubin, D.G. (2001). Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat. Cell Biol. 3, Wesp, A., Hicke, L., Palecek, J., Lombardi, R., Aust, T., Munn, A.L., and Riezman, H. (1997). End4p/Sla2p interacts with actin-associated proteins for endocytosisinsaccharomyces cerevisiae. Mol. Biol. Cell8, Skruzny, M., Brach, T., Ciuffa, R., Rybina, S., Wachsmuth, M., and Kaksonen, M. (2012). Molecular basis for coupling the plasma membrane to the actin cytoskeleton during clathrin-mediated endocytosis. Proc. Natl. Acad. Sci. USA 109, E2533 E Rodal, A.A., Kozubowski, L., Goode, B.L., Drubin, D.G., and Hartwig, J.H. (2005). Actin and septin ultrastructures at the budding yeast cell cortex. Mol. Biol. Cell 16, Urbanek, A.N., Smith, A.P., Allwood, E.G., Booth, W.I., and Ayscough, K.R. (2013). A novel actin-binding motif in Las17/WASP nucleates actin filaments independently of Arp2/3. Curr. Biol. 23, Young, M.E., Cooper, J.A., and Bridgman, P.C. (2004). Yeast actin patches are networks of branched actin filaments. J. Cell Biol. 166, Martin, S.G., and Chang, F. (2006). Dynamics of the formin for3p in actin cable assembly. Curr. Biol. 16, Gowrishankar, K., Ghosh, S., Saha, S., C, R., Mayor, S., and Rao, M. (2012). Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell 149, Kukulski, W., Schorb, M., Kaksonen, M., and Briggs, J.A. (2012). Plasma membrane reshaping during endocytosis is revealed by time-resolved electron tomography. Cell 150, Boettner, D.R., Friesen, H., Andrews, B., and Lemmon, S.K. (2011). Clathrin light chain directs endocytosis by influencing the binding of the yeast Hip1R homologue, Sla2, to F-actin. Mol. Biol. Cell 22, Bähler, J., Wu, J.Q., Longtine, M.S., Shah, N.G., McKenzie, A., 3rd, Steever, A.B., Wach, A., Philippsen, P., and Pringle, J.R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14,
Plasma Membrane Reshaping during Endocytosis Is Revealed by Time-Resolved Electron Tomography
 Plasma Membrane Reshaping during Endocytosis Is Revealed by Time-Resolved Electron Tomography Wanda Kukulski, 1,2 Martin Schorb, 1 Marko Kaksonen, 1,2, * and John A.G. Briggs 1,2, * 1 Structural and Computational
Plasma Membrane Reshaping during Endocytosis Is Revealed by Time-Resolved Electron Tomography Wanda Kukulski, 1,2 Martin Schorb, 1 Marko Kaksonen, 1,2, * and John A.G. Briggs 1,2, * 1 Structural and Computational
Mechanisms of Clathrin-Mediated Endocytosis in Budding Yeast. Akemi Marian Kunibe. A dissertation submitted in partial satisfaction of the
 Mechanisms of Clathrin-Mediated Endocytosis in Budding Yeast by Akemi Marian Kunibe A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Molecular
Mechanisms of Clathrin-Mediated Endocytosis in Budding Yeast by Akemi Marian Kunibe A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Molecular
MCB Topic 19 Regulation of Actin Assembly- Prof. David Rivier
 MCB 252 -Topic 19 Regulation of Actin Assembly- Prof. David Rivier MCB 252 Spring 2017 MCB 252 Cell Biology Topic 19 Regulation of Actin Assembly Reading: Lodish 17.2-17.3, 17.7 MCB 252 Actin Cytoskeleton
MCB 252 -Topic 19 Regulation of Actin Assembly- Prof. David Rivier MCB 252 Spring 2017 MCB 252 Cell Biology Topic 19 Regulation of Actin Assembly Reading: Lodish 17.2-17.3, 17.7 MCB 252 Actin Cytoskeleton
Cell Cycle, Mitosis, and Microtubules. LS1A Final Exam Review Friday 1/12/07. Processes occurring during cell cycle
 Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson
 Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson General principles Properties of lysosomes Delivery of enzymes to lysosomes Endocytic uptake clathrin, others Endocytic pathways recycling vs.
Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson General principles Properties of lysosomes Delivery of enzymes to lysosomes Endocytic uptake clathrin, others Endocytic pathways recycling vs.
Regulation of Clathrin-Mediated Endocytosis by Dynamic Ubiquitination/Deubiquitination. of Ede1. Jasper Stanley Weinberg
 Regulation of Clathrin-Mediated Endocytosis by Dynamic Ubiquitination/Deubiquitination of Ede1 by Jasper Stanley Weinberg A dissertation submitted in partial satisfaction of the requirement for the degree
Regulation of Clathrin-Mediated Endocytosis by Dynamic Ubiquitination/Deubiquitination of Ede1 by Jasper Stanley Weinberg A dissertation submitted in partial satisfaction of the requirement for the degree
04_polarity. The formation of synaptic vesicles
 Brefeldin prevents assembly of the coats required for budding Nocodazole disrupts microtubules Constitutive: coatomer-coated Selected: clathrin-coated The formation of synaptic vesicles Nerve cells (and
Brefeldin prevents assembly of the coats required for budding Nocodazole disrupts microtubules Constitutive: coatomer-coated Selected: clathrin-coated The formation of synaptic vesicles Nerve cells (and
Introduction to Cofilin and its Regulation of Actin Dynamics
 Article ID: ISSN 2046-1690 Introduction to Cofilin and its Regulation of Actin Dynamics Author(s):Mr. Kuen Yeow Chin Corresponding Author: Mr. Kuen Yeow Chin, MSc, Department of Cell and Developmental
Article ID: ISSN 2046-1690 Introduction to Cofilin and its Regulation of Actin Dynamics Author(s):Mr. Kuen Yeow Chin Corresponding Author: Mr. Kuen Yeow Chin, MSc, Department of Cell and Developmental
Homework Hanson section MCB Course, Fall 2014
 Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
Cytoskeleton and cell communication
 Cytoskeleton and cell communication Actin monomer has subdomains 1-4. A simplified cartoon is at right. ATP binds, along with Mg ++, within a deep cleft between subdomains 2 & 4. G-actin (globular actin),
Cytoskeleton and cell communication Actin monomer has subdomains 1-4. A simplified cartoon is at right. ATP binds, along with Mg ++, within a deep cleft between subdomains 2 & 4. G-actin (globular actin),
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
Distinct Dynamics of Endocytic Clathrin-Coated Pits and Coated Plaques
 Distinct Dynamics of Endocytic Clathrin-Coated Pits and Coated Plaques Saveez Saffarian, Emanuele Cocucci, Tomas Kirchhausen* Department of Cell Biology, Harvard Medical School, Children s Hospital and
Distinct Dynamics of Endocytic Clathrin-Coated Pits and Coated Plaques Saveez Saffarian, Emanuele Cocucci, Tomas Kirchhausen* Department of Cell Biology, Harvard Medical School, Children s Hospital and
El Azzouzi et al., http ://www.jcb.org /cgi /content /full /jcb /DC1
 Supplemental material JCB El Azzouzi et al., http ://www.jcb.org /cgi /content /full /jcb.201510043 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Acquisition of -phluorin correlates negatively with podosome
Supplemental material JCB El Azzouzi et al., http ://www.jcb.org /cgi /content /full /jcb.201510043 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Acquisition of -phluorin correlates negatively with podosome
CMB621: Cytoskeleton. Also known as How the cell plays with LEGOs to ensure order, not chaos, is temporally and spatially achieved
 CMB621: Cytoskeleton Also known as How the cell plays with LEGOs to ensure order, not chaos, is temporally and spatially achieved Lecture(s) Overview Lecture 1: What is the cytoskeleton? Membrane interaction
CMB621: Cytoskeleton Also known as How the cell plays with LEGOs to ensure order, not chaos, is temporally and spatially achieved Lecture(s) Overview Lecture 1: What is the cytoskeleton? Membrane interaction
Nature Methods: doi: /nmeth.4257
 Supplementary Figure 1 Screen for polypeptides that affect cellular actin filaments. (a) Table summarizing results from all polypeptides tested. Source shows organism, gene, and amino acid numbers used.
Supplementary Figure 1 Screen for polypeptides that affect cellular actin filaments. (a) Table summarizing results from all polypeptides tested. Source shows organism, gene, and amino acid numbers used.
Las17p Vrp1p but not Las17p Arp2/3 interaction is important for actin patch polarization in yeast
 Las17p Vrp1p but not Las17p Arp2/3 interaction is important for actin patch polarization in yeast Author Rajmohan, Rajamuthiah, Wong, Ming Hwa, Meng, Lei, Munn, Alan, Thanabalu, Thirumaran Published 2009
Las17p Vrp1p but not Las17p Arp2/3 interaction is important for actin patch polarization in yeast Author Rajmohan, Rajamuthiah, Wong, Ming Hwa, Meng, Lei, Munn, Alan, Thanabalu, Thirumaran Published 2009
Selective protection of an ARF1-GTP signaling axis by a bacterial scaffold induces bidirectional trafficking arrest.
 Selective protection of an ARF1-GTP signaling axis by a bacterial scaffold induces bidirectional trafficking arrest. Andrey S. Selyunin, L. Evan Reddick, Bethany A. Weigele, and Neal M. Alto Supplemental
Selective protection of an ARF1-GTP signaling axis by a bacterial scaffold induces bidirectional trafficking arrest. Andrey S. Selyunin, L. Evan Reddick, Bethany A. Weigele, and Neal M. Alto Supplemental
Zool 3200: Cell Biology Exam 4 Part II 2/3/15
 Name:Key Trask Zool 3200: Cell Biology Exam 4 Part II 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
Name:Key Trask Zool 3200: Cell Biology Exam 4 Part II 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Courtheoux et al., http://www.jcb.org/cgi/content/full/jcb.200902093/dc1 S1 Figure S2. Visualization of multiple merotelic attachments.
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Courtheoux et al., http://www.jcb.org/cgi/content/full/jcb.200902093/dc1 S1 Figure S2. Visualization of multiple merotelic attachments.
Nature Neuroscience: doi: /nn Supplementary Figure 1. Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites.
 Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
Supplemental Data. Hao et al. (2014). Plant Cell /tpc
 Supplemental Figure 1. Confocal Images and VA-TIRFM Analysis of GFP-RbohD in Arabidopsis Seedlings. (A) RbohD expression in whole Arabidopsis seedlings. RbohD was expressed in the leaves, hypocotyl, and
Supplemental Figure 1. Confocal Images and VA-TIRFM Analysis of GFP-RbohD in Arabidopsis Seedlings. (A) RbohD expression in whole Arabidopsis seedlings. RbohD was expressed in the leaves, hypocotyl, and
Supplemental information contains 7 movies and 4 supplemental Figures
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplemental information contains 7 movies and 4 supplemental Figures Movies: Movie 1. Single virus tracking of A4-mCherry-WR MV
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplemental information contains 7 movies and 4 supplemental Figures Movies: Movie 1. Single virus tracking of A4-mCherry-WR MV
The clathrin adaptor Numb regulates intestinal cholesterol. absorption through dynamic interaction with NPC1L1
 The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Lu et al., http://www.jcb.org/cgi/content/full/jcb.201012063/dc1 Figure S1. Kinetics of nuclear envelope assembly, recruitment of Nup133
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Lu et al., http://www.jcb.org/cgi/content/full/jcb.201012063/dc1 Figure S1. Kinetics of nuclear envelope assembly, recruitment of Nup133
Nature Immunology: doi: /ni.3631
 Supplementary Figure 1 SPT analyses of Zap70 at the T cell plasma membrane. (a) Total internal reflection fluorescent (TIRF) excitation at 64-68 degrees limits single molecule detection to 100-150 nm above
Supplementary Figure 1 SPT analyses of Zap70 at the T cell plasma membrane. (a) Total internal reflection fluorescent (TIRF) excitation at 64-68 degrees limits single molecule detection to 100-150 nm above
F-actin VWF Vinculin. F-actin. Vinculin VWF
 a F-actin VWF Vinculin b F-actin VWF Vinculin Supplementary Fig. 1. WPBs in HUVECs are located along stress fibers and at focal adhesions. (a) Immunofluorescence images of f-actin (cyan), VWF (yellow),
a F-actin VWF Vinculin b F-actin VWF Vinculin Supplementary Fig. 1. WPBs in HUVECs are located along stress fibers and at focal adhesions. (a) Immunofluorescence images of f-actin (cyan), VWF (yellow),
Molecular Cell Biology 5068 In Class Exam 1 September 29, Please print your name:
 Molecular Cell Biology 5068 In Class Exam 1 September 29, 2015 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your
Molecular Cell Biology 5068 In Class Exam 1 September 29, 2015 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your
Genomic analysis of essentiality within protein networks
 Genomic analysis of essentiality within protein networks Haiyuan Yu, Dov Greenbaum, Hao Xin Lu, Xiaowei Zhu and Mark Gerstein Department of Molecular Biophysics and Biochemistry, 266 Whitney Avenue, Yale
Genomic analysis of essentiality within protein networks Haiyuan Yu, Dov Greenbaum, Hao Xin Lu, Xiaowei Zhu and Mark Gerstein Department of Molecular Biophysics and Biochemistry, 266 Whitney Avenue, Yale
The Molecular Mechanism of Intracellular Membrane Fusion. Richard H. Scheller
 The Molecular Mechanism of Intracellular Membrane Fusion Richard H. Scheller The human brain contains approximately 10 15 connections between nerve cells (Figure 2). The specific formation and modulation
The Molecular Mechanism of Intracellular Membrane Fusion Richard H. Scheller The human brain contains approximately 10 15 connections between nerve cells (Figure 2). The specific formation and modulation
1. endoplasmic reticulum This is the location where N-linked oligosaccharide is initially synthesized and attached to glycoproteins.
 Biology 4410 Name Spring 2006 Exam 2 A. Multiple Choice, 2 pt each Pick the best choice from the list of choices, and write it in the space provided. Some choices may be used more than once, and other
Biology 4410 Name Spring 2006 Exam 2 A. Multiple Choice, 2 pt each Pick the best choice from the list of choices, and write it in the space provided. Some choices may be used more than once, and other
Vesicle Transport. Vesicle pathway: many compartments, interconnected by trafficking routes 3/17/14
 Vesicle Transport Vesicle Formation Curvature (Self Assembly of Coat complex) Sorting (Sorting Complex formation) Regulation (Sar1/Arf1 GTPases) Fission () Membrane Fusion SNARE combinations Tethers Regulation
Vesicle Transport Vesicle Formation Curvature (Self Assembly of Coat complex) Sorting (Sorting Complex formation) Regulation (Sar1/Arf1 GTPases) Fission () Membrane Fusion SNARE combinations Tethers Regulation
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Beck et al., http://www.jcb.org/cgi/content/full/jcb.201011027/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Membrane binding of His-tagged proteins to Ni-liposomes.
Supplemental material Beck et al., http://www.jcb.org/cgi/content/full/jcb.201011027/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Membrane binding of His-tagged proteins to Ni-liposomes.
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
Life Sciences 1A Midterm Exam 2. November 13, 2006
 Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
Name: TF: Section Time Life Sciences 1A Midterm Exam 2 November 13, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use
UNIVERSITY OF YORK BSc Stage 2 Degree Examinations Department: BIOLOGY. Title of Exam: Cell Biology
 Examination Candidate Number: Desk Number: UNIVERSITY OF YORK BSc Stage 2 Degree Examinations 2017-18 Department: BIOLOGY Title of Exam: Cell Biology Time allowed: 1 hour and 30 minutes Total marks available
Examination Candidate Number: Desk Number: UNIVERSITY OF YORK BSc Stage 2 Degree Examinations 2017-18 Department: BIOLOGY Title of Exam: Cell Biology Time allowed: 1 hour and 30 minutes Total marks available
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Edens and Levy, http://www.jcb.org/cgi/content/full/jcb.201406004/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nuclear shrinking does not depend on the cytoskeleton
Supplemental material Edens and Levy, http://www.jcb.org/cgi/content/full/jcb.201406004/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nuclear shrinking does not depend on the cytoskeleton
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division The Cell Cycle: Cell Growth, Cell Division 2007-2008 2007-2008 Getting from there to here Going from egg to baby. the original
Biology is the only subject in which multiplication is the same thing as division The Cell Cycle: Cell Growth, Cell Division 2007-2008 2007-2008 Getting from there to here Going from egg to baby. the original
endomembrane system internal membranes origins transport of proteins chapter 15 endomembrane system
 endo system chapter 15 internal s endo system functions as a coordinated unit divide cytoplasm into distinct compartments controls exocytosis and endocytosis movement of molecules which cannot pass through
endo system chapter 15 internal s endo system functions as a coordinated unit divide cytoplasm into distinct compartments controls exocytosis and endocytosis movement of molecules which cannot pass through
Supplemental Materials Molecular Biology of the Cell
 Supplemental Materials Molecular Biology of the Cell Gilberti et al. SUPPLEMENTAL FIGURE LEGENDS: Figure S1: The effect of pharmacological inhibitors on particle uptake. The data presented in Figure 1
Supplemental Materials Molecular Biology of the Cell Gilberti et al. SUPPLEMENTAL FIGURE LEGENDS: Figure S1: The effect of pharmacological inhibitors on particle uptake. The data presented in Figure 1
The Cell Cycle and How Cells Divide
 The Cell Cycle and How Cells Divide 1 Phases of the Cell Cycle The cell cycle consists of Interphase normal cell activity The mitotic phase cell divsion INTERPHASE Growth G 1 (DNA synthesis) Growth G 2
The Cell Cycle and How Cells Divide 1 Phases of the Cell Cycle The cell cycle consists of Interphase normal cell activity The mitotic phase cell divsion INTERPHASE Growth G 1 (DNA synthesis) Growth G 2
Report. Rapid Glucose Depletion Immobilizes Active Myosin V on Stabilized Actin Cables. Li Xu 1 and Anthony Bretscher 1, * 1
 Current Biology 24, 2471 2479, October 20, 2014 ª2014 Elsevier Ltd All rights reserved http://dx.doi.org/10.1016/j.cub.2014.09.017 Rapid Glucose Depletion Immobilizes Active Myosin V on Stabilized Actin
Current Biology 24, 2471 2479, October 20, 2014 ª2014 Elsevier Ltd All rights reserved http://dx.doi.org/10.1016/j.cub.2014.09.017 Rapid Glucose Depletion Immobilizes Active Myosin V on Stabilized Actin
Overview of clathrin-mediated endocytosis
 Overview of clathrin-mediated endocytosis Accessory and adaptor proteins promote clathrin nucleation on the plasma membrane and some help deform membrane. Clathrin assembly into lattices stabilize the
Overview of clathrin-mediated endocytosis Accessory and adaptor proteins promote clathrin nucleation on the plasma membrane and some help deform membrane. Clathrin assembly into lattices stabilize the
Requirements of Slm proteins for proper eisosome organization, endocytic trafficking and recycling in the yeast Saccharomyces cerevisiae
 Requirements of Slm proteins for proper eisosome organization, endocytic trafficking and recycling in the yeast Saccharomyces cerevisiae CHITRA KAMBLE, SANDHYA JAIN, ERIN MURPHY and KYOUNGTAE KIM* Department
Requirements of Slm proteins for proper eisosome organization, endocytic trafficking and recycling in the yeast Saccharomyces cerevisiae CHITRA KAMBLE, SANDHYA JAIN, ERIN MURPHY and KYOUNGTAE KIM* Department
Reactive Oxygen Species Regulate Protrusion Efficiency by Controlling Actin Dynamics
 Reactive Oxygen Species Regulate Protrusion Efficiency by Controlling Actin Dynamics Nicolas Taulet, Violaine D. Delorme-Walker, Céline DerMardirossian* Department of Immunology and Microbial Science,
Reactive Oxygen Species Regulate Protrusion Efficiency by Controlling Actin Dynamics Nicolas Taulet, Violaine D. Delorme-Walker, Céline DerMardirossian* Department of Immunology and Microbial Science,
APGRU4L1 Chap 12 Extra Reading Cell Cycle and Mitosis
 APGRU4L1 Chap 12 Extra Reading Cell Cycle and Mitosis Dr. Ramesh Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008
APGRU4L1 Chap 12 Extra Reading Cell Cycle and Mitosis Dr. Ramesh Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Stelter et al., http://www.jcb.org/cgi/content/full/jcb.201105042/dc1 S1 Figure S1. Dyn2 recruitment to edid-labeled FG domain Nups.
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Stelter et al., http://www.jcb.org/cgi/content/full/jcb.201105042/dc1 S1 Figure S1. Dyn2 recruitment to edid-labeled FG domain Nups.
Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) (b) (c) (d) (e) (f) (g) .
 Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) The inhibitory effects of new antibodies (Mab17 and Mab18). They were investigated in in vitro fertilization
Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) The inhibitory effects of new antibodies (Mab17 and Mab18). They were investigated in in vitro fertilization
Module 3 Lecture 7 Endocytosis and Exocytosis
 Module 3 Lecture 7 Endocytosis and Exocytosis Endocytosis: Endocytosis is the process by which cells absorb larger molecules and particles from the surrounding by engulfing them. It is used by most of
Module 3 Lecture 7 Endocytosis and Exocytosis Endocytosis: Endocytosis is the process by which cells absorb larger molecules and particles from the surrounding by engulfing them. It is used by most of
MOLECULAR CELL BIOLOGY
 1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
Xenopus Actin-interacting Protein 1 (XAip1) Enhances Cofilin Fragmentation of Filaments by Capping Filament Ends*
 THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 277, No. 45, Issue of November 8, pp. 43011 43016, 2002 2002 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Xenopus Actin-interacting
THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 277, No. 45, Issue of November 8, pp. 43011 43016, 2002 2002 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Xenopus Actin-interacting
Nature Biotechnology: doi: /nbt.3828
 Supplementary Figure 1 Development of a FRET-based MCS. (a) Linker and MA2 modification are indicated by single letter amino acid code. indicates deletion of amino acids and N or C indicate the terminus
Supplementary Figure 1 Development of a FRET-based MCS. (a) Linker and MA2 modification are indicated by single letter amino acid code. indicates deletion of amino acids and N or C indicate the terminus
genome edited transient transfection, CMV promoter
 Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
Figure S1. (A) SDS-PAGE separation of GST-fusion proteins purified from E.coli BL21 strain is shown. An equal amount of GST-tag control, LRRK2 LRR
 Figure S1. (A) SDS-PAGE separation of GST-fusion proteins purified from E.coli BL21 strain is shown. An equal amount of GST-tag control, LRRK2 LRR and LRRK2 WD40 GST fusion proteins (5 µg) were loaded
Figure S1. (A) SDS-PAGE separation of GST-fusion proteins purified from E.coli BL21 strain is shown. An equal amount of GST-tag control, LRRK2 LRR and LRRK2 WD40 GST fusion proteins (5 µg) were loaded
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a
Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a
a Anti-Dab2 RGD Merge
 a Anti-Da Merge *** ** Percentage of cells adhered 8 6 4 RGE RAD RGE RAD Supplementary Figure Da are localized at clusters. (a) Endogenous Da is localized at clusters. (), ut not RGE nor RAD peptides can
a Anti-Da Merge *** ** Percentage of cells adhered 8 6 4 RGE RAD RGE RAD Supplementary Figure Da are localized at clusters. (a) Endogenous Da is localized at clusters. (), ut not RGE nor RAD peptides can
Problem-solving Test: The Mechanism of Protein Synthesis
 Q 2009 by The International Union of Biochemistry and Molecular Biology BIOCHEMISTRY AND MOLECULAR BIOLOGY EDUCATION Vol. 37, No. 1, pp. 58 62, 2009 Problem-based Learning Problem-solving Test: The Mechanism
Q 2009 by The International Union of Biochemistry and Molecular Biology BIOCHEMISTRY AND MOLECULAR BIOLOGY EDUCATION Vol. 37, No. 1, pp. 58 62, 2009 Problem-based Learning Problem-solving Test: The Mechanism
Molecular Cell Biology 5068 In class Exam 1 October 2, Please print your name: Instructions:
 Molecular Cell Biology 5068 In class Exam 1 October 2, 2012 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your number
Molecular Cell Biology 5068 In class Exam 1 October 2, 2012 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your number
Cofilin is one crucial mediator of actin cytoskeletal dynamics
 Chronophin coordinates cell leading edge dynamics by controlling active cofilin levels Violaine Delorme-Walker a,b, Ji-Yeon Seo a,b, Antje Gohla c, Bruce Fowler a,b, Ben Bohl a,b, and Céline DerMardirossian
Chronophin coordinates cell leading edge dynamics by controlling active cofilin levels Violaine Delorme-Walker a,b, Ji-Yeon Seo a,b, Antje Gohla c, Bruce Fowler a,b, Ben Bohl a,b, and Céline DerMardirossian
Practice Exam 2 MCBII
 1. Which feature is true for signal sequences and for stop transfer transmembrane domains (4 pts)? A. They are both 20 hydrophobic amino acids long. B. They are both found at the N-terminus of the protein.
1. Which feature is true for signal sequences and for stop transfer transmembrane domains (4 pts)? A. They are both 20 hydrophobic amino acids long. B. They are both found at the N-terminus of the protein.
SUPPLEMENTARY INFORMATION
 Figure S1. Loss of Ena/VASP proteins inhibits filopodia and neuritogenesis. (a) Bar graph of filopodia number per stage 1 control and mmvvee (Mena/ VASP/EVL-null) neurons at 40hrs in culture. Loss of all
Figure S1. Loss of Ena/VASP proteins inhibits filopodia and neuritogenesis. (a) Bar graph of filopodia number per stage 1 control and mmvvee (Mena/ VASP/EVL-null) neurons at 40hrs in culture. Loss of all
Name: Multiple choice questions. Pick the BEST answer (2 pts ea)
 Exam 1 202 Oct. 5, 1999 Multiple choice questions. Pick the BEST answer (2 pts ea) 1. The lipids of a red blood cell membrane are all a. phospholipids b. amphipathic c. glycolipids d. unsaturated 2. The
Exam 1 202 Oct. 5, 1999 Multiple choice questions. Pick the BEST answer (2 pts ea) 1. The lipids of a red blood cell membrane are all a. phospholipids b. amphipathic c. glycolipids d. unsaturated 2. The
Endocytosis involves a succession of plasma membrane
 Determinants of endocytic membrane geometry, stability, and scission Takuma Kishimoto, Yidi Sun, Christopher user, Jian Liu 1, Alphée Michelot, and David G. Drubin 2 Department of Molecular and Cell iology,
Determinants of endocytic membrane geometry, stability, and scission Takuma Kishimoto, Yidi Sun, Christopher user, Jian Liu 1, Alphée Michelot, and David G. Drubin 2 Department of Molecular and Cell iology,
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a
Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a
Biology Open (2014) 000, 1 10 doi: /bio
 (2014) 000, 1 10 doi:10.1242/bio.201410041 Supplementary Material Michael Brauchle et al. doi: 10.1242/bio.201410041 Fig. S1. Alignment of GFP, sfgfp, egfp, eyfp, mcherry and mruby2. Sequence-based alignment
(2014) 000, 1 10 doi:10.1242/bio.201410041 Supplementary Material Michael Brauchle et al. doi: 10.1242/bio.201410041 Fig. S1. Alignment of GFP, sfgfp, egfp, eyfp, mcherry and mruby2. Sequence-based alignment
Molecular Cell Biology 5068 In Class Exam 1 October 3, 2013
 Molecular Cell Biology 5068 In Class Exam 1 October 3, 2013 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your number
Molecular Cell Biology 5068 In Class Exam 1 October 3, 2013 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your number
Chapter 12 The Cell Cycle: Cell Growth, Cell Division
 Chapter 12 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a period at the end of a sentence And now look at you How did you get from there to
Chapter 12 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Where it all began You started as a cell smaller than a period at the end of a sentence And now look at you How did you get from there to
Appendix: Subcellular localization of MIG-14::GFP in C. elegans body wall muscle cells. Pei-Tzu Yang and Hendrik C. Korswagen
 Appendix: Subcellular localization of MIG-14::GFP in C. elegans body wall muscle cells Pei-Tzu Yang and Hendrik C. Korswagen 129 SUBCELLULAR LOCALIZATION OF MIG-14 Abstract MIG-14/Wls is a Wnt binding
Appendix: Subcellular localization of MIG-14::GFP in C. elegans body wall muscle cells Pei-Tzu Yang and Hendrik C. Korswagen 129 SUBCELLULAR LOCALIZATION OF MIG-14 Abstract MIG-14/Wls is a Wnt binding
Supplementary Materials. High affinity binding of phosphatidylinositol-4-phosphate. by Legionella pneumophila DrrA
 Supplementary Materials High affinity binding of phosphatidylinositol-4-phosphate by Legionella pneumophila DrrA Running title: Molecular basis of PtdIns(4)P-binding by DrrA Stefan Schoebel, Wulf Blankenfeldt,
Supplementary Materials High affinity binding of phosphatidylinositol-4-phosphate by Legionella pneumophila DrrA Running title: Molecular basis of PtdIns(4)P-binding by DrrA Stefan Schoebel, Wulf Blankenfeldt,
AP Biology Summer Assignment Chapter 3 Quiz
 AP Biology Summer Assignment Chapter 3 Quiz 2016-17 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. All of the following are found in a DNA nucleotide
AP Biology Summer Assignment Chapter 3 Quiz 2016-17 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. All of the following are found in a DNA nucleotide
Chapter 7: Membranes
 Chapter 7: Membranes Roles of Biological Membranes The Lipid Bilayer and the Fluid Mosaic Model Transport and Transfer Across Cell Membranes Specialized contacts (junctions) between cells What are the
Chapter 7: Membranes Roles of Biological Membranes The Lipid Bilayer and the Fluid Mosaic Model Transport and Transfer Across Cell Membranes Specialized contacts (junctions) between cells What are the
Drs2p-Dependent Formation of Exocytic Clathrin-Coated Vesicles In Vivo
 Current Biology, Vol. 12, 1623 1627, September 17, 2002, 2002 Elsevier Science Ltd. All rights reserved. PII S0960-9822(02)01148-X Drs2p-Dependent Formation of Exocytic Clathrin-Coated Vesicles In Vivo
Current Biology, Vol. 12, 1623 1627, September 17, 2002, 2002 Elsevier Science Ltd. All rights reserved. PII S0960-9822(02)01148-X Drs2p-Dependent Formation of Exocytic Clathrin-Coated Vesicles In Vivo
Supplemental Data. Wu et al. (2010). Plant Cell /tpc
 Supplemental Figure 1. FIM5 is preferentially expressed in stamen and mature pollen. The expression data of FIM5 was extracted from Arabidopsis efp browser (http://www.bar.utoronto.ca/efp/development/),
Supplemental Figure 1. FIM5 is preferentially expressed in stamen and mature pollen. The expression data of FIM5 was extracted from Arabidopsis efp browser (http://www.bar.utoronto.ca/efp/development/),
Section 6. Junaid Malek, M.D.
 Section 6 Junaid Malek, M.D. The Golgi and gp160 gp160 transported from ER to the Golgi in coated vesicles These coated vesicles fuse to the cis portion of the Golgi and deposit their cargo in the cisternae
Section 6 Junaid Malek, M.D. The Golgi and gp160 gp160 transported from ER to the Golgi in coated vesicles These coated vesicles fuse to the cis portion of the Golgi and deposit their cargo in the cisternae
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2294 Figure S1 Localization and function of cell wall polysaccharides in root hair cells. (a) Spinning-disk confocal sections of seven day-old A. thaliana seedlings stained with 0.1% S4B
DOI: 10.1038/ncb2294 Figure S1 Localization and function of cell wall polysaccharides in root hair cells. (a) Spinning-disk confocal sections of seven day-old A. thaliana seedlings stained with 0.1% S4B
Mechanism of Vesicular Transport
 Mechanism of Vesicular Transport Transport vesicles play a central role in the traffic of molecules between different membrane-enclosed enclosed compartments. The selectivity of such transport is therefore
Mechanism of Vesicular Transport Transport vesicles play a central role in the traffic of molecules between different membrane-enclosed enclosed compartments. The selectivity of such transport is therefore
Supplementary Information
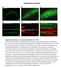 Supplementary Information Supplementary Figure 1: Luminal localization of CCM-3. (a) The CCM-3::GFP fusion protein localizes along the apical (luminal) surface of the pharynx (b) as well as the lumen of
Supplementary Information Supplementary Figure 1: Luminal localization of CCM-3. (a) The CCM-3::GFP fusion protein localizes along the apical (luminal) surface of the pharynx (b) as well as the lumen of
Chapter 8 Cells and Their Environment
 Chapter Outline Chapter 8 Cells and Their Environment Section 1: Cell Membrane KEY IDEAS > How does the cell membrane help a cell maintain homeostasis? > How does the cell membrane restrict the exchange
Chapter Outline Chapter 8 Cells and Their Environment Section 1: Cell Membrane KEY IDEAS > How does the cell membrane help a cell maintain homeostasis? > How does the cell membrane restrict the exchange
Mechanisms of alternative splicing regulation
 Mechanisms of alternative splicing regulation The number of mechanisms that are known to be involved in splicing regulation approximates the number of splicing decisions that have been analyzed in detail.
Mechanisms of alternative splicing regulation The number of mechanisms that are known to be involved in splicing regulation approximates the number of splicing decisions that have been analyzed in detail.
MEMBRANE STRUCTURE. Lecture 8. Biology Department Concordia University. Dr. S. Azam BIOL 266/
 1 MEMBRANE STRUCTURE Lecture 8 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University Plasma Membrane 2 Plasma membrane: The outer boundary of the cell that separates it from the world
1 MEMBRANE STRUCTURE Lecture 8 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University Plasma Membrane 2 Plasma membrane: The outer boundary of the cell that separates it from the world
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supporting Information
 Supporting Information Harries et al. 1.173/pnas.9923916 A Fig. S1. Disruption of microfilaments within epidermal cells after treatment with 5 M Lat. Images of N. benthamiana cells are from plants expressing
Supporting Information Harries et al. 1.173/pnas.9923916 A Fig. S1. Disruption of microfilaments within epidermal cells after treatment with 5 M Lat. Images of N. benthamiana cells are from plants expressing
Supplemental Information. Otic Mesenchyme Cells Regulate. Spiral Ganglion Axon Fasciculation. through a Pou3f4/EphA4 Signaling Pathway
 Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Chapter 3 subtitles Action potentials
 CELLULAR NEUROPHYSIOLOGY CONSTANCE HAMMOND Chapter 3 subtitles Action potentials Introduction (3:15) This third chapter explains the calcium current triggered by the arrival of the action potential in
CELLULAR NEUROPHYSIOLOGY CONSTANCE HAMMOND Chapter 3 subtitles Action potentials Introduction (3:15) This third chapter explains the calcium current triggered by the arrival of the action potential in
Supplementary Figure 1. Overview of steps in the construction of photosynthetic protocellular systems
 Supplementary Figure 1 Overview of steps in the construction of photosynthetic protocellular systems (a) The small unilamellar vesicles were made with phospholipids. (b) Three types of small proteoliposomes
Supplementary Figure 1 Overview of steps in the construction of photosynthetic protocellular systems (a) The small unilamellar vesicles were made with phospholipids. (b) Three types of small proteoliposomes
Hui Chen, Barbara W. Bernstein and James R. Bamburg
 IBS 25 JANUARY 2000 sequence-specific NA binding by acetylation of the p53 C-terminal domain. Cell 90, 595 606 41 Liu, L. et al. (1999) p53 sites acetylated in vitro by PCAF and p300 are acetylated in
IBS 25 JANUARY 2000 sequence-specific NA binding by acetylation of the p53 C-terminal domain. Cell 90, 595 606 41 Liu, L. et al. (1999) p53 sites acetylated in vitro by PCAF and p300 are acetylated in
Supplementary Figure 1
 Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
cell movement and neuronal migration
 cell movement and neuronal migration Paul Letourneau letou001@umn.edu Chapter 16; The Cytoskeleton; Molecular Biology of the Cell, Alberts et al. 1 Cell migration Cell migration in 3 steps; protrusion,
cell movement and neuronal migration Paul Letourneau letou001@umn.edu Chapter 16; The Cytoskeleton; Molecular Biology of the Cell, Alberts et al. 1 Cell migration Cell migration in 3 steps; protrusion,
The dynamic actin cytoskeleton drives membrane shape changes
 Molecular basis for coupling the plasma membrane to the actin cytoskeleton during clathrinmediated endocytosis Michal Skruzny, Thorsten rach, Rodolfo iuffa, Sofia Rybina, Malte Wachsmuth, and Marko Kaksonen
Molecular basis for coupling the plasma membrane to the actin cytoskeleton during clathrinmediated endocytosis Michal Skruzny, Thorsten rach, Rodolfo iuffa, Sofia Rybina, Malte Wachsmuth, and Marko Kaksonen
Biology is the only subject in which multiplication is the same thing as division
 Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Getting from there to here Going from egg to baby. the original
Biology is the only subject in which multiplication is the same thing as division 2007-2008 The Cell Cycle: Cell Growth, Cell Division 2007-2008 Getting from there to here Going from egg to baby. the original
Regulation of Actin Assembly Associated With Protrusion and Adhesion in Cell Migration
 Physiol Rev 88: 489 513, 2008; doi:10.1152/physrev.00021.2007. Regulation of Actin Assembly Associated With Protrusion and Adhesion in Cell Migration CHRISTOPHE LE CLAINCHE AND MARIE-FRANCE CARLIER Laboratoire
Physiol Rev 88: 489 513, 2008; doi:10.1152/physrev.00021.2007. Regulation of Actin Assembly Associated With Protrusion and Adhesion in Cell Migration CHRISTOPHE LE CLAINCHE AND MARIE-FRANCE CARLIER Laboratoire
Chapter 3. Expression of α5-megfp in Mouse Cortical Neurons. on the β subunit. Signal sequences in the M3-M4 loop of β nachrs bind protein factors to
 22 Chapter 3 Expression of α5-megfp in Mouse Cortical Neurons Subcellular localization of the neuronal nachr subtypes α4β2 and α4β4 depends on the β subunit. Signal sequences in the M3-M4 loop of β nachrs
22 Chapter 3 Expression of α5-megfp in Mouse Cortical Neurons Subcellular localization of the neuronal nachr subtypes α4β2 and α4β4 depends on the β subunit. Signal sequences in the M3-M4 loop of β nachrs
Phospholipids. Extracellular fluid. Polar hydrophilic heads. Nonpolar hydrophobic tails. Polar hydrophilic heads. Intracellular fluid (cytosol)
 Module 2C Membranes and Cell Transport All cells are surrounded by a plasma membrane. Eukaryotic cells also contain internal membranes and membrane- bound organelles. In this module, we will examine the
Module 2C Membranes and Cell Transport All cells are surrounded by a plasma membrane. Eukaryotic cells also contain internal membranes and membrane- bound organelles. In this module, we will examine the
Chapter 1: Vesicular traffic. Biochimica cellulare parte B 2017/18
 Chapter 1: Vesicular traffic Biochimica cellulare parte B 2017/18 Major Protein-sorting pathways in eukaryotic cells Secretory and endocytic pathways Unifying principle governs all protein trafficking
Chapter 1: Vesicular traffic Biochimica cellulare parte B 2017/18 Major Protein-sorting pathways in eukaryotic cells Secretory and endocytic pathways Unifying principle governs all protein trafficking
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Molecular Cell Biology - Problem Drill 22: The Mechanics of Cell Division
 Molecular Cell Biology - Problem Drill 22: The Mechanics of Cell Division Question No. 1 of 10 1. Which of the following statements about mitosis is correct? Question #1 (A) Mitosis involves the dividing
Molecular Cell Biology - Problem Drill 22: The Mechanics of Cell Division Question No. 1 of 10 1. Which of the following statements about mitosis is correct? Question #1 (A) Mitosis involves the dividing
2013 John Wiley & Sons, Inc. All rights reserved. PROTEIN SORTING. Lecture 10 BIOL 266/ Biology Department Concordia University. Dr. S.
 PROTEIN SORTING Lecture 10 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University Introduction Membranes divide the cytoplasm of eukaryotic cells into distinct compartments. The endomembrane
PROTEIN SORTING Lecture 10 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University Introduction Membranes divide the cytoplasm of eukaryotic cells into distinct compartments. The endomembrane
A genetically targeted optical sensor to monitor calcium signals in astrocyte processes
 A genetically targeted optical sensor to monitor calcium signals in astrocyte processes 1 Eiji Shigetomi, 1 Sebastian Kracun, 2 Michael V. Sofroniew & 1,2 *Baljit S. Khakh Ψ 1 Departments of Physiology
A genetically targeted optical sensor to monitor calcium signals in astrocyte processes 1 Eiji Shigetomi, 1 Sebastian Kracun, 2 Michael V. Sofroniew & 1,2 *Baljit S. Khakh Ψ 1 Departments of Physiology
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
10.2 The Cell Cycle *
 OpenStax-CNX module: m52672 1 10.2 The Cell Cycle * Shannon McDermott Based on The Cell Cycle by OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License
OpenStax-CNX module: m52672 1 10.2 The Cell Cycle * Shannon McDermott Based on The Cell Cycle by OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License
The subcortical maternal complex controls symmetric division of mouse zygotes by
 The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics Xing-Jiang Yu 1,2, Zhaohong Yi 1, Zheng Gao 1,2, Dan-dan Qin 1,2, Yanhua Zhai 1, Xue Chen 1,
The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics Xing-Jiang Yu 1,2, Zhaohong Yi 1, Zheng Gao 1,2, Dan-dan Qin 1,2, Yanhua Zhai 1, Xue Chen 1,
