SH3 domain of c-src governs its dynamics at focal adhesions and the cell membrane
|
|
|
- Gwendolyn Casey
- 5 years ago
- Views:
Transcription
1 SH3 domain of c-src governs its dynamics at focal adhesions and the cell membrane Hiroaki Machiyama 1, Tomoyuki Yamaguchi 1, Yasuhiro Sawada 2, Tomonobu M. Watanabe 1,3 and Hideaki Fujita 1,3 1 Immunology Frontier Research Center, Osaka University, Suita, Japan 2 Research Institute, National Rehabilitation Center for Persons with Disabilities, Saitama, Japan 3 Quantitative Biology Center, Riken, Suita, Osaka, Japan Keywords focal adhesion; phosphorylation; SH3 domain; Src; TIRF microscopy Correspondence H. Fujita, Immunology Frontier Research Center, Osaka University, OLABB, Osaka University, Furuedai, Suita, Osaka , Japan Fax: Tel: fujita@ifrec.osaka-u.ac.jp (Received 5 February 2015, revised 4 August 2015, accepted 5 August 2015) doi: /febs We studied the role of the Src SH3 domain in its dynamics at the cell membrane using site-directed mutagenesis and live cell imaging. Physiologically, cell proliferation and migration require the expression of Src family kinases. Hyperactivation of Src molecules has been detected in various cancer cells. Although the activation mechanism of Src has been intensively studied, the dynamics of Src at the cell membrane are still unclear. Although Src molecules also exist at various cellular locations, we found that activated Src molecules are mainly localized at peripheral cell adhesion sites. Src phosphorylation status and subdomain conformations are thought to regulate Src activation and translocation. In this study, we analyzed the single-molecule dynamics of wild-type Src and SH2- and SH3- mutated Src at the cell membrane. Introducing mutations in the SH3 domain resulted in reduced Src motility at the cell membrane, both inside and outside of focal adhesions. Disruption of the actin cytoskeleton resulted in less diffusive Src movement at the cell membrane. We demonstrate that, inside focal adhesions, the SH3 domain enhanced dissociation of Src from the adhesion site and disruption of the SH3 domain altered the distribution of Src at the cell membrane. Inside focal adhesions, kinase activity of Src was essential for the Src mobility reduction by SH3 domain mutation, suggesting that rapid mobility of Src at focal adhesions mediated by the SH3 domain is catalytic-activity-dependent. These findings show that the SH3 domain of Src governs the dynamics of Src at the cell membrane and may be involved in rapid signal transduction in cells. Introduction Src family kinases are major non-receptor tyrosine kinases in normal and cancer cells. Signal transduction via tyrosine phosphorylation is involved in various cellular functions, such as cell proliferation, cytoskeletal rearrangement and cell motility [1 3]. Abnormal hyperphosphorylation leads to many diseases, such as Alzheimer s disease and cancer progression [4,5]. Previous reports have demonstrated that hyperactivation of Src kinases is correlated with cancer proliferation and migration/invasion in many human cancer types [6 8]. Cellular-Src (c-src) is a proto-oncogene tyrosine kinase and its target substrates are ubiquitous, including membrane proteins and nucleoproteins. Controlling these multiple signal transductions via Src might be an effective treatment method for various diseases [9]; indeed, some reports Abbreviations EYFP, enhanced yellow fluorescent protein; MSD, mean-square displacement; PAmCherry, photoactivatable mcherry; SH domain, Srchomology domain; SrcWT, wild-type Src; TIRF, total internal reflection fluorescence. 4034
2 H. Machiyama et al. Roles of SH3 domain in the dynamics of c-src have targeted Src family kinases in cancer or brain damage treatment [10 12]. The roles of Src subdomains on the activation of the kinase have been intensively studied (reviewed in [13]). Individual Src molecules are composed of an N- terminal domain, conserved Src-homology domains (SH2 and SH3), a tyrosine kinase domain and the C terminus [14]. Unless the tyrosine residue 530 (referring to the residue in humans, hereafter) in the C terminus is dephosphorylated, Src remains inactive in the cytoplasm [15]. Myristoylation at the N-terminal domain induces Src translocation to the cell membrane [16]. When tyrosine 530 is dephosphorylated, Src becomes activated via autophosphorylation at tyrosine 419 in the tyrosine kinase domain [17,18]. The SH2 and SH3 domains directly interact with phosphorylated tyrosine residues and proline-rich motifs in the target substrates, respectively [19,20]. By exposing the SH2 and SH3 domains, activated Src at the cell membrane is able to bind to and phosphorylate its substrates, triggering intracellular signal transduction pathways for various cellular functions [21,22]. Mutation of tryptophan 121 to alanine (SrcW121A) in the SH3 domain and mutation of arginine 178 to alanine (SrcR178A) in the SH2 domain are known to decrease the binding of Src to its substrates [23,24]. In addition, the phosphorylation of serine 75 located next to the SH3 domain induces ubiquitination of Src [25], enhancing proteasomal degradation of Src. Thus, Srcmediated signal transduction is regulated by the phosphorylation status of Src, subcellular localization and interaction with the substrate via the SH2 and SH3 domains. Among the many functions of Src-mediated signaling, the interaction between the extracellular matrix receptor and the extracellular matrix induces the accumulation of activated Src molecules at focal adhesions, which is crucial for cell behavior, especially in migrating cells [26]. A number of focal adhesion proteins, such as talin, FAK and paxillin, are known Src substrates [27]. The interaction of Src via its SH2/SH3 domains to substrates at focal adhesions facilitates the phosphorylation of tyrosine residues in the substrates [28]. In addition, focal adhesions are connected by actin cytoskeleton, and the Src kinase-driven signaling pathway regulates their remodeling, especially in the peripheral region of cells [29,30]. Many focal adhesion proteins that serve as Src substrates are transported depending on actin cytoskeletal dynamics [31,32]. These studies indicate that actin cytoskeletal dynamics and proper localization of Src substrates at focal adhesions are essential for the regulation of cellular dynamics by Src-mediated signaling. Therefore, the dynamics of Src molecules at the cell membrane might be crucial for Src signaling. In this study, we analyzed the dynamics of Src molecules at the cell membrane to understand how Src signaling is controlled. Although the importance of Src translocation to the cell membrane in signaling has been described [33], Src molecules do not show specific localization at the cell membrane [34]. The dynamics of Src molecules at the cell membrane have never been directly observed, especially at the singlemolecule level; therefore, how Src molecules are recruited to focal adhesions and how Src molecules dissociate from focal adhesions are still open questions. We revealed the dynamics of wild-type and mutant Src molecules at and around adhesion sites using single-molecule imaging technology. The mutation in the SH3 domain (SrcW121A), but not in the SH2 domain of Src (SrcR178A), decreased movement at the cell membrane. Furthermore, the SrcW121A mutant demonstrated increased residence time of Src molecules at focal adhesions owing to slower dissociation from adhesion sites. The actin cytoskeleton, especially actin polymerization, also facilitated the Src dynamics at the cell membrane, which involved the enhancement of Src targeting to focal adhesions. Furthermore, the SrcW121A mutant demonstrated slower mobility and increased residence time at focal adhesions. Low Src mobility by the SrcW121A mutation was diminished by introducing a kinase activity defective mutation in the SrcW121A mutant. These findings indicate that SH3-domain-mediated rapid Src mobility at focal adhesions is Src catalytic-activity-dependent. Results The distribution of Src and its active form at the cell membrane To examine the Src dynamics at the cell membrane, we first visualized the distribution of Src molecules at the cell membrane in HeLa cells expressing GFPtagged paxillin, a focal adhesion marker, using total internal reflection fluorescence (TIRF) microscopy. TIRF microscopy is suitable for monitoring biological events near the cell membrane, where focal adhesion complexes are located [35]. While we did not detect specific subcellular localization of Src within the cell membrane as previously reported [34], the majority of activated Src molecules were localized at focal adhesions (Fig. 1), indicating that the dynamics of Src within focal adhesions may differ from those of Src in cell membrane regions excluding focal adhesions. 4035
3 Roles of SH3 domain in the dynamics of c-src H. Machiyama et al. Fig. 1. The majority of activated Src molecules are localized at focal adhesions. Immunofluorescence images of Src (upper) and phospho-srcy419 (lower) in HeLa cells expressing paxillin-gfp (green). Src and phosphor-srcy419 were labeled with AlexaFluor 555 (red). Scale bar 10 lm. Therefore, we separately analyzed the dynamics of Src inside and outside focal adhesions. The dynamics of Src at the cell membrane outside focal adhesions Since the Src subdomains SH2 and SH3 play important roles in Src activation and downstream signaling [33,36], we next examined whether they affect Src dynamics at the cell membrane. To observe Src dynamics at the cell membrane, we traced individual Src molecules at the single-molecule level. To achieve this, we constructed wild-type Src (SrcWT) tagged with photoactivatable mcherry (PAmCherry) [37] and Src with mutations in the SH2 and SH3 domains tagged with PAmCherry, named SrcW121A and SrcR178A, respectively (Fig. 2A). According to previous studies, SrcW121A and SrcR178A mutants decrease the binding of Src to its substrates via the SH3 and SH2 domains [38,39]. PAmCherry-tagged Src variants (WT, W121A and R178A) and enhanced yellow fluorescent protein (EYFP) tagged paxillin were co-transfected to HeLa cells. After gentle activation of PAmCherry, the single-molecule fluorescence emitted from Src-PAm- Cherry was traced by TIRF microscopy. Since the myristoylation of the N-terminal domain is known to regulate Src translocation from the cytosol to the cell membrane [16], all Src variants used in this study can translocate to the cell membrane owing to the intact N-terminal domain. We analyzed the individual Src- PAmCherry molecules at the cell membrane outside focal adhesions (Fig. 2B). SrcWT, SrcW121A and SrcR178A moved dynamically for a few seconds at the cell membrane (Fig. 2C). To characterize the movement of Src variants, we analyzed the velocity, duration, total run length and end-to-end distance (Fig. 2D G). Duration is defined as the time between first appearance of an activated PAmCherry fluorescence in the recording region and its disappearance. We found that the velocity, which is calculated by dividing the total run length by duration, of SrcW121A was significantly lower than that of SrcWT and SrcR178A (Fig. 2D), indicating that SH3 domain disruption decreases the motility of Src at the cell membrane. On the other hand, the duration and total run length were comparable among Src variants (WT, W121A and R178A) (Fig. 2E,F), indicating that cell membrane targeting and the dissociation of Src from the cell membrane are not regulated by the SH2 and SH3 domains and may be stochastic. The SrcW121A mutation but not the SrcR178A mutation significantly reduced the end-to-end distance, defined as the length between the start and end positions (Fig. 2G). Although we could not distinguish between dissociation of Src from focal adhesions and photobleaching of activated PAmCherry at the end point of singlemolecule tracking, a short end-to-end distance still indicates low directional movement or low diffusion because photobleaching occurs at the same rate in both mutants and the fluorescence lifetime (~ 4s) is longer than the observed duration. The persistency of movement was defined as the ratio of the end-to-end distance to the total run length (Fig. 2B). The persistency of SrcW121A was significantly smaller than that of SrcWT and SrcR178A (Fig. 2H). There are two possible reasons for low persistency by SH3 domain disruption: (a) low directional movement of Src molecules at the cell membrane; or (b) confined diffusion of 4036
4 H. Machiyama et al. Roles of SH3 domain in the dynamics of c-src 4037
5 Roles of SH3 domain in the dynamics of c-src H. Machiyama et al. Fig. 2. Mutation in the SH3 domain decreases the dynamics of Src at the cell membrane. (A) Schematic representation of Src and its variants: human wild-type c-src (SrcWT), SrcW121A (mutation in the SH3 domain) and SrcR178A (mutation in the SH2 domain). (B) A merged image of the bright field and Venus fluorescence (green) images in a HeLa cell expressing paxillin-eyfp. The dotted rectangle indicates an example of the outside of the focal adhesion area used in this study ( lm 2 ) and an example trajectory is overlaid. Start and end points are shown by a circle and square, respectively. The value of persistency is calculated by dividing the end-to-end distance by the total run length, labeled 1 and 2, respectively. Scale bar 5 lm. (C) Representative trajectories outside focal adhesions in the cells expressing Src variants (WT, W121A and R178A). Start and end points of trajectories are shown by solid circles and open squares. The length of recording for each trace is shown. The inset shows a trajectory of activated PAmCherry from SrcWT-PAmCherry and paxillin-yfp co-expressed HeLa cells fixed with 4% paraformaldehyde for 30 min at room temperature. Each line indicates the movement of particles for 100 ms. (D) (H) Velocity (D), duration (E), total run length (F), end-to-end distance (G) and persistency value (H) (see Materials and methods) are shown. The dotted line in (H) indicates the mean persistency in fixed HeLa cells expressing SrcWT-PAmCherry. Statistical analyses were performed with one-way ANOVA followed by Tukey Kramer post hoc testing among Src variants; *P < 0.05, n = 153 for WT, n = 82 for W121A and n = 105 for R178A. (I) The mean square displacement r 2 of the obtained trajectories in SrcWT (rectangles), SrcW121A (circles) and SrcR178A (triangles) in HeLa cells and SrcWT in fixed HeLa cells (diamonds) are plotted as a function of the time interval Dt. Dotted lines indicate the results of curve fitting using Eqn (1). the Src mutant. To distinguish between these possibilities, we plotted the mean-square displacement (MSD; r 2 ) as a function of the time interval Dt (Fig. 2I), fitted by hr 2 i¼4dðdtþ c ð1þ where D and c are the diffusion constant and the actual scaling factor, respectively. When calculating the c value, the experimental errors were removed to avoid underestimation, as previously reported [40]. The c value in Src variants (WT, W121A and R178A) was ~ 1 (Table 1), indicating free diffusion at the cell membrane. However, the slope, which is an estimate of the diffusion constant, was lower for SrcW121A than for SrcWT or SrcR178A (Table 1). This result indicates that mutations in the SH3 domain result in less motility at the cell membrane compared with SrcWT or SrcR178A variants. Although the persistency of SrcW121A was low, the c value of SrcW121A was nearly equal to 1, indicating that the SrcW121A mutant freely diffuses outside focal adhesions. Since the molecules would show directional movement when Table 1. Diffusion constants and c values of Src variants at the cell membrane outside focal adhesions in HeLa cells. Src D (lm 2 s 1 ) c Figure WT Figs 2, 4A and 5A W121A Fig. 2 R178A Fig. 2 WT (Fixed) Fig. 2 K298M Fig. 4A W121A/K298M Fig. 4A R178A/K298M Fig. 4A Y530F Fig. 5A W121A/Y530F Fig. 5A R178A/Y530F Fig. 5A the c value is > 1, the latter explanation is a reasonable interpretation of the low persistency of SrcW121A, i.e. the SH3 domain of Src seems to play a role in the acceleration of its movement at the cell membrane, outside focal adhesions. The effect of Src catalytic activity on SH3- domain-regulated acceleration of Src movement outside focal adhesions To assess whether the altered movement of SrcW121A molecules at the cell membrane depends on the Src activation status [17,18], we examined the tyrosine 419 (Y419) phosphorylation level of Src and observed Src behavior outside focal adhesions with different Src kinase activity conditions. Immunoblotting analysis demonstrated that the Y419 phosphorylation levels of both SrcW121A and SrcR178A were higher than that of SrcWT (Fig. 3). According to the Tyr419 phosphorylation level scaled by total Src expression level (Fig. 3), the activation level of SrcW121A- and SrcR178A-PAmCherry was comparable to that of constitutive activation mutant SrcY530F. To remove the effect of Src catalytic activity, we next made kinase dead mutant K298M [41] with SH3 and SH2 domain disruption (W121A/K298M and R178A/ K298M) and performed single-molecule analyses (Fig. 4A). Although the velocity of SrcK298M mutant was comparable with that of SrcWT, SrcW121A/ K298M mutant showed slightly but significantly lower velocity compared to SrcWT and SrcK298M (Fig. 4A). The duration, total run length, end-to-end distance and persistency were not significantly different between SrcWT, SrcK298M, SrcW121A/K298M and SrcR178A/K298M (Fig. 4A). However, the MSD-t plot (Fig. 4A) shows that the diffusion constant of Src outside focal adhesions was lower in the SrcW121A mutation (Table 1). We also treated Src 4038
6 H. Machiyama et al. Roles of SH3 domain in the dynamics of c-src Fig. 3. Mutations in either the SH2 or SH3 domain of Src induce hyperactivation of Src. (A) Expression and phosphorylation of Venus-tagged Src molecules analyzed by anti-phospho-src (Src py419), anti-src (Src) and anti-a-tubulin (Tubulin) immunoblotting. The relative phosphorylation level was scaled by the Src expression level, and the value obtained for SrcWT-Venusexpressing HeLa cells was set at 1. Statistical analyses were performed for comparisons between cells expressing SrcWT (black box) and those expressing another Src variant (n = 4, *P < 0.05 by t tests). Lane 1, Venus; lane 2, SrcWT-Venus; lane 3, SrcW121A- Venus; lane 4, SrcR178A-Venus. (B) Expression and phosphorylation of Src molecules in HeLa cells expressing SrcWT and SrcY530F analyzed by anti-phospho-src (Src py419), anti-src (Src) and anti-gapdh (GAPDH) immunoblotting. Lane 1, vector only; lane 2, SrcWT; lane 3, SrcY530F. family kinase inhibitor CGP77675 to HeLa cells expressing SrcWT- and SrcW121A-PAmCherry and performed single-molecule imaging. Treatment of CGP77675 did not affect any parameters obtained in this study for Src movement at the cell membrane, including velocity, duration, total run length, end-toend distance, persistency, diffusion constant and c value, in both SrcWT and SrcW121A (Fig. 4B and Table 2), indicating that the SH3-domain-regulated Src behavior at the cell membrane does not relate to Src catalytic activity. Taken together, these results indicate that the contribution of Src activation status on the SH3-domain-dependent movement of Src at the cell membrane outside focal adhesions seems to be low. We next tested the effect of constitutive activation of Src on the behavior of Src outside focal adhesions. Since SrcY530F mutant shows constitutive activation by hampering the formation of a closed structure, we made constructs having the Y530F mutation with SH3 and SH2 disruption (W121A/Y530F and R178A/ Y530F). SrcY530F mutation did not affect any parameters in this study except for duration outside focal adhesions. SrcW121A/Y530F and SrcR178A/Y530F mutants also showed a similar velocity to SrcWT (Fig. 5A), while the duration including the SrcY530F mutation (SrcY530F, SrcW121A/Y530F and SrcR178A/Y530F) was significantly longer than that of SrcWT (Fig. 5A). These results indicate that the SrcY530F mutation affects the ability of cell membrane targeting and the dissociation of Src from the cell membrane, and that lowering Src mobility by SH3 domain disruption is eliminated once the ability is lost by the Y530F mutation. In this study, we used overexpression conditions of Src in HeLa cells. To minimize the effect of overexpression of Src, exogenous Src variants (WT, W121A and R178A) were expressed in SYF cells where Src, Yes and Fyn are knocked out [42]. All Src motility parameters of Src variants at cell membranes in SYF cells were comparable to those in HeLa cells (Fig. 5B and Table 3). Therefore we concluded that the results shown in this study are not an artifact of the overexpression system. Actin cytoskeleton regulates the dynamics of Src at the cell membrane We further examined whether the actin cytoskeleton is involved in Src behavior at the cell membrane outside focal adhesions because actin cytoskeleton dynamics are known to affect the mobility of membrane proteins and focal adhesion proteins [31,32], and the actin cytoskeleton was reported to be essential for the targeting of viral Src to focal adhesions via its SH3 domain [43]. Either the myosin II inhibitor blebbistatin (25 lm) or the actin polymerization inhibitor cytochalasin D (10 lm) was added to HeLa cells expressing Src-PAmCherry and single-molecule tracking was performed. Although cell morphology and the focal adhe- 4039
7 Roles of SH3 domain in the dynamics of c-src H. Machiyama et al. sion distribution dramatically changed after the addition of inhibitors (Fig. 6A), treatment with blebbistatin or cytochalasin D did not alter the velocity, duration, total run length, end-to-end distance or the persistency of the movement (Fig. 6B F). However, when we analyzed the Src movement with an MSD-t plot and fitted with Eqn (1), both the diffusion constant and the c value decreased after treatment with blebbistatin or cytochalasin D (Fig. 6G, Table 2), indicating that diffusion of Src molecules at the cell membrane is reduced by inhibiting actin polymerization and actomyosin contraction. The reduction in Src mobility by cytochalasin D was higher than that by blebbistatin, indicating that actin cytoskeleton, especially actin polymerization, enlarges the diffusion area of Src at the cell membrane outside focal adhesions, possibly to facilitate the targeting of Src to its substrate located at the cell membrane. Single-molecule imaging of Src at focal adhesions We next examined whether the Src subdomain plays a role in Src dynamics at focal adhesions. Although activated Src molecules mainly localized at focal adhesions (Fig. 1), the components of focal adhesions, including many Src substrates, do not stably reside at focal adhesions but dynamically move in and out within seconds [44,45]. Thus, Src molecules may also exhibit associations/dissociations with their substrate at focal adhesions. Again, PAmCherry-tagged Src variants (WT, W121A and R178A) and EYFP-tagged paxillin were co-transfected to HeLa cells and viewed under a TIRF microscope. After identifying the position of focal adhesions using paxillin-eyfp, we traced individual photoactivated Src-PAmCherry molecules (Figs 7A and 8). Similar to the behavior of Src at the cell membrane outside focal adhesions, while SrcWT and SrcR178A dynamically moved inside focal adhesions, the mobility of Src was reduced by introducing a mutation in the SH3 domain (Fig. 7A). The velocity and duration of SrcW121A at focal adhesions was significantly faster and longer, respectively, than those of SrcWT, whereas those of SrcR178A were comparable with those of SrcWT (Fig. 7B,C). Although the total run length was not significantly different among Src variants (Fig. 7D), end-to-end distance and persistency of SrcW121A were significantly lower than those of SrcWT and SrcR178A (Fig. 7E,F). When we analyzed the Src movement with MSD-t plots fitted with Eqn (1), the diffusion constant of SrcW121A was much lower than those of SrcWT and SrcR178A (Fig. 7G and Table 4). We also detected the spot within focal adhesions where the movement of the SrcW121A mutant was severely reduced (Fig. 7A). Since this spot size was comparable with that of SrcWT in fixed HeLa cells (Fig. 7A), SrcW121A molecules appeared to be trapped within focal adhesions, indicating that SrcW121A mutations suppressed the dissociation ability from focal adhesions. Therefore, we propose that the SH3 domain of Src is likely to promote its dynamics within focal adhesions by accelerating the dissociation from focal adhesions. To access the effect of Src catalytic activity on the SH3-domain-regulated movement of Src at focal adhesions, we performed single-molecule analyses using kinase dead mutation K298M (Fig. 9A). Although the velocity, duration, total run length, endto-end distance and persistency did not show significant differences between SrcWT, SrcK298M, SrcW121A/K298M and SrcR178A/K298M, we found that the diffusion constant of SrcW121A/K298M mutant was lower than that of SrcK298M (Fig. 9A and Table 4). The difference in D value between SrcK298M (D = lm 2 /s) and SrcW121A/ K298M (D = lm 2 /s) was much smaller than that between SrcWT (D = lm 2 /s) and SrcW121A (D = lm 2 /s) (Table 4), indicating that the lower mobility of SrcW121A at focal adhesions requires Src catalytic activity. On the other hand, although no parameters on Src mobility at focal adhesions showed statistically significant differences between with and without CGP77675 treatment for both SrcWT and SrcW121A, the D value of SrcW121A was slightly increased by CGP77675 treatment (Fig. 9B and Table 5). This also indicates that kinase activity is required for the reduction of Src mobility by SrcW121A mutations. Taken together, we suggest that the SH3 domain of Src has a role in the regulation of Src movement at focal adhesions which is strongly dependent on Src catalytic activity. We further tested the effect of the Y530F mutation on Src movement at focal adhesions by single-molecule analysis in HeLa cells expressing PAmCherry-tagged Src (Y530F, W121A/Y530F and R178A/Y530F). All parameters used in this study were comparable among Src variants (Fig. 10 and Table 4). Similar to outside focal adhesions, a lowering of Src movement at focal adhesions by SH3 domain disruption was eliminated by the Y530F mutation. Considering that constitutively activated mutant SrcY530F hampers the formation of a closed structure, reversible open close conformational changes of Src may be required for the regulation of Src movement at focal adhesions by the SH3 domain. 4040
8 H. Machiyama et al. Roles of SH3 domain in the dynamics of c-src 4041
9 Roles of SH3 domain in the dynamics of c-src H. Machiyama et al. Fig. 4. The contribution of Src activation status to the SH3-domain-dependent movement of Src at the cell membrane outside focal adhesions. (A) HeLa cells co-expressing Src-PAmCherry (WT, K298M, W121A/K298M or R178A/K298M) and paxillin-eyfp were cultured in a collagen-coated glass-based chamber. Single particles from individually activated PAmCherry molecules were traced outside focal adhesions located at peripheral regions. The velocity (a), duration (b), total run length (c), end-to-end distance (d) and persistency value (e) of the particles and the MSD-t plot (f) are shown. Statistical analyses were performed to compare cells expressing SrcWT and those expressing other Src variants (n > 80, *P < 0.05 by t tests). A significant difference is shown between K298M and W121A/K298M in (a). (B) HeLa cells co-expressing Src- or SrcW121A-PAmCherry and paxillin-eyfp treated with DMSO or 1 lm CGP77675, 1 h before TIRF observation. Single activated PAmCherry molecules were traced outside focal adhesions. The velocity (a), duration (b), total run length (c), end-to-end distance (d) and persistency value (e) of the particles and the MSD-t plot (f) are shown. Statistical analyses were performed between CGP77675 treated and untreated cells (n > 100 by t tests). Non-significant differences are shown between all combinations of groups. Table 2. Diffusion constants and c values of SrcWT and SrcW121A at the cell membrane treated with various drugs in HeLa cells. Src Drug D (lm 2 s 1 ) c Figure WT DMSO Figs 6 and 4B WT Blebbistatin Fig. 6 WT Cytochasin D Fig. 6 WT CGP Fig. 4B W121A DMSO Fig. 4B W121A CGP Fig. 4B Mutations in the SH3 domain alter the distribution of Src at the cell membrane To examine whether the acceleration of Src dissociation from adhesion sites via the SH3 domain affects the distribution of Src at the cell membrane, we constructed SrcWT, SrcW121A and SrcR178A tagged with Venus, a modified yellow fluorescent protein. When we observed HeLa cells co-expressing the Src variants and mcherry-tagged paxillin under a TIRF microscope, colocalization with paxillin-mcherry was not detected for SrcWT-Venus or SrcR178A-Venus (Fig. 11A). However, the distribution of SrcW121A- Venus exhibited a peripheral punctate pattern and was highly correlated with that of paxillin-mcherry (Fig. 11A). To quantify this observation, we assessed the correlation between the distributions of Src and paxillin by calculating the Pearson s coefficient for the two acquired images [46]. SrcW121A exhibited a significantly higher colocalization index than SrcWT and SrcR178A (Fig. 11B), indicating that the mutation in SH3 but not in SH2 altered the Src distribution at the cell membrane. Since the SH3 domain can facilitate the dissociation of Src from adhesion sites, the SH3 domain of Src is crucial for maintaining a uniform distribution of Src at the cell membrane. We also constructed a double mutant in which both the SH2 and the SH3 domains were disrupted (SrcW121A/R178A). The colocalization index of SrcW121A/R178A was slightly but significantly higher than that of SrcWT, and the colocalization index had a larger standard deviation than the other Src variants (Fig. 11B). However, as shown in Fig. 12, the distribution of SrcW121A/R178A-Venus was highly variable: some cells showed a high correlation between the distribution of Src and paxillin, whereas an obvious peripheral punctate pattern was not detected for other cells (Fig. 12). Owing to the variable results for the double mutant with respect to the Src distribution at the cell membrane, we did not use this double mutant for further analysis. Phosphorylation of serine 75 is not highly correlated with Src localization It has been reported that the ubiquitination of Src enhances proteasomal degradation of Src, and the phosphorylation of serine 75 located near the SH3 domain is reported to induce ubiquitination of Src [25], which may contribute to the dissociation of Src from focal adhesions. Therefore, we introduced phosphomimetic (S75E) and phosphodefective (S75A) mutations into SrcWT-Venus (Fig. 13A). These Src mutants were co-transfected with paxillin-mcherry to HeLa cells, observed under a TIRF microscope (Fig. 13B) and quantified using the colocalization index (Fig. 13C,D). While the colocalization index of SrcS75E was comparable with that of SrcWT, the index of SrcS75A was significantly higher than that of SrcWT (Fig. 13C). These results indicate that the phosphodefective mutation at serine 75 facilitates the localization of Src within cell adhesion sites. However, the SrcW121A mutation more drastically altered the distribution of Src at the cell membrane than did the SrcS75A mutation (Figs 11A and 13B). Therefore, the phosphorylation of serine 75 induces dissociation of Src from adhesion sites, but seems to be less effective than mutations in the SH3 domain. 4042
10 H. Machiyama et al. Roles of SH3 domain in the dynamics of c-src 4043
11 Roles of SH3 domain in the dynamics of c-src H. Machiyama et al. Fig. 5. Assessments of constitutive activation and overexpression of Src in the movement of Src outside focal adhesions. (A) HeLa cells coexpressing Src-PAmCherry (WT, Y530F, W121A/Y530F or R178A/Y530F) and paxillin-eyfp were cultured in a collagen-coated glass-based chamber. Single activated PAmCherry molecules were traced outside focal adhesions located at peripheral regions. The velocity (a), duration (b), total run length (c), end-to-end distance (d) and persistency value (e) of the particles and the MSD-t plot (f) are shown. Statistical analyses were performed to compare cells expressing SrcWT and those expressing other Src variants (n > 50, *P < 0.05 by t tests). (B) SYF cells co-expressing Src-PAmCherry (WT, W121A or R178A) and paxillin-eyfp were cultured in a collagen-coated glass-based chamber. Single activated PAmCherry molecules were traced outside focal adhesions located at peripheral regions. The velocity (a), duration (b), total run length (c), end-to-end distance (d) and persistency value (e) of the particles and the MSD-t plot (f) are shown. Statistical analyses were performed with one-way ANOVA followed by Tukey Kramer post hoc testing among Src variants; *P < 0.05, n > 50. Table 3. Diffusion constants and c values of Src variants at the cell membrane outside focal adhesions in SYF cells. Src D (lm 2 s 1 ) c Figure WT Fig. 5B W121A Fig. 5B R178A Fig. 5B To further examine whether the effects of the SH3 domain and the phosphorylation of serine 75 on the dissociation of Src from adhesion sites are parallel or sequential, we introduced phosphomimetic (SrcS75E/ W121A) and phosphodefective (SrcS75A/W121A) mutations to Venus-tagged SrcW121A (Fig. 13A), cotransfected these mutants with paxillin-mcherry to HeLa cells, observed the cells under a TIRF microscope (Fig. 13B) and estimated the colocalization index (Fig. 13C,D). Both SrcS75A/W121A and SrcS75E/W121A were highly colocalized with paxillinmcherry (Fig. 13B,D), indicating that phosphomimetic and phosphodefective mutations in the SrcW121A mutant did not affect the localization of Src molecules. These findings suggest that the prolonged localization of SrcW121A at focal adhesions (Fig. 7B) is not correlated with the phosphorylation of serine 75, which may lead to proteasomal degradation of Src. Discussion Src molecules showed a uniform distribution at the cell membrane, whereas the majority of activated Src molecules mainly localized at focal adhesions (Fig. 1). To achieve this distribution, activated Src molecules must alter their association with adhesion sites, either by increasing the recruitment or by decreasing the dissociation to/from focal adhesions. As shown in Table 1, the diffusion constant of SrcWT at the cell membrane ( lm 2 s 1 ) was much larger than that of transmembrane proteins (e.g. epidermal growth factor receptor, (0.5 5) lm 2 s 1 [47]). Since Src is a peripheral membrane protein, not a transmembrane protein, it is expected that it can move dynamically at the cell membrane. In addition, singlemolecule imaging demonstrated that the SH3 mutation decreased the mobility of Src both inside and outside the focal adhesion (Figs 2 and 7). Decreased mobility of Src SH3 mutant outside focal adhesions is expected to decrease the probability of the molecule colliding with and entering into the focal adhesion. Therefore, it can be speculated that strong focal adhesion localization of the Src SH3 mutant results from decreased dissociation from adhesion sites. The actin cytoskeleton facilitated free diffusion of Src at the cell membrane outside the focal adhesion (Fig. 6). Actin polymerization and actomyosin contraction are involved in centrifugal and centripetal movement of actin binding proteins. In this study, treatment with cytochalasin D, an inhibitor of actin polymerization, showed a stronger effect on Src movement at the cell membrane outside the focal adhesion than did treatment with blebbistatin, an inhibitor of actomyosin contraction (Fig. 6G and Table 2). Therefore, we propose that actin polymerization promotes the centrifugal movement of Src to the cell peripheral region and subsequently enhances the recruitment of Src to adhesion sites. Because a c value < 1 indicates confined diffusion, the movement of Src in actin cytoskeleton disrupted cells seems to be confined. While the actin cytoskeleton is generally considered to form the cage under the cell membrane, there are some reports showing that disruption of the actin cytoskeleton decreased the c value of proteins [48,49]. It can be assumed that actin-filament-driven motility may be used to escape from the cage; thus disruption of the actin cytoskeleton results in a decreased c value of the MSD-t plot. In this case, a cage that impedes the movement may be formed by other cytoskeletons (e.g. microtubules or intermediate filaments). Taken together, the localization of activated Src at the focal adhesion is achieved via the recruitment of Src by enhancing the movement of non-activated Src at the cell membrane and decreasing the dissociation of activated Src from focal adhesions. 4044
12 H. Machiyama et al. Roles of SH3 domain in the dynamics of c-src Fig. 6. Actin cytoskeleton dynamics are required for Src dynamic movement at the cell membrane. HeLa cells co-expressing Src- PAmCherry and paxillin-eyfp treated with DMSO, 25 lm blebbsistatin or 10 lm cytochalasin D, 1 h before TIRF observation. Individual activated PAmCherry molecules were traced outside focal adhesions. (A) Bright field and EYFP fluorescence images of DMSO-, blebbistatinand cytochalasin D treated cells. Scale bar 10 lm. (B) (F) Velocity (B), duration (C), total run length (D), end-to-end distance (E) and persistency value (F) are shown. Statistical analyses were performed with one-way ANOVA followed by Tukey Kramer post hoc testing among Src variants; n = 60 for DMSO, n = 178 for blebbistatin and n = 76 for cytochalasin D. Non-significant differences are shown between all combinations of groups. (G) The mean square displacement r 2 of the obtained trajectories in DMSO (rectangles), blebbistatin (circles) and cytochalasin D (triangles) treated HeLa cells expressing SrcWT-PAmCherry as a function of the time interval Dt. Dotted lines indicate the results of curve fitting using Eqn (1). Although we showed that the disruption of the SH3 domain significantly decreased Src motility both inside and outside focal adhesions (Figs 2 and 7, Tables 1 and 4), Src catalytic activity was required for the reduction of Src motility by the SrcW121A mutation inside but not outside focal adhesions (Fig. 9, Tables 4 and 5). This may indicate that SrcW121A phosphorylates the substrate which slows down the movement of Src at focal adhesions, and that if SrcW121A is unable to phosphorylate the substrate then it moves as quickly as SrcWT. On the other hand, the SrcW121A but not SrcR178A mutation altered the Src distribu- 4045
13 Roles of SH3 domain in the dynamics of c-src H. Machiyama et al. tion at the cell membrane (Fig. 11A,B). Previous studies have demonstrated that phosphorylated substrate proteins are co-immunoprecipitated with Src, but binding is reduced by the SrcW121A or SrcR178A mutation [23,24]. Therefore, the binding ability with Src substrates is likely to be disrupted in SrcW121A and SrcR178A mutants. The different effect depending on which subdomain was disrupted may be attributed to the difference in binding manner with the substrate. The SH3 domain binds with the proline-rich region of the target substrate, whereas the SH2 domain binds with the phosphorylated tyrosine residues [19,20]. Immunoblotting analysis demonstrated that SrcW121A and SrcR178A mutants exhibited hyperactivation (Fig. 3). The behavior of the SrcR178A mutant at focal adhesions can be normal even under hyperphosphorylation of the substrates, because the SH3 domain retained in SrcR178A binds to the target region in a phosphorylation-independent manner. However, since Src hyperactivation involves hyperphosphorylation of Src substrates, the SH2 domain in the SrcW121A mutant is likely to associate strongly with targets within the adhesion site. Indeed, we detected the spot where movement of the SrcW121A mutant stopped within the focal adhesion. Therefore, the Src SH3 domain mediates the dissociation of Src from adhesion sites by enhancing the unbinding of the SH2 domain of Src and the substrate at focal adhesions in a catalytic-activity-dependent manner. Although we showed that the Src SH3 domain facilitated dissociation of Src from focal adhesions, the mechanism is still unclear. Single-molecule analyses demonstrated that the duration of Src at focal adhesions was ~ 1 s, similar to many Src substrates at focal adhesions that exhibit rapid exchanges [44]. In addition, the Src substrates are known to interact with Src at focal adhesions via the Src SH2 and/or Src SH3 domains. Src substrates within focal adhesions may take Src away from focal adhesions through SH2- or SH3-domain-mediated interactions. We further tested whether phosphorylation of serine 75 alters the binding manner to the focal adhesion. Introduction of phosphodefective or phosphomimetic mutations at serine 75 did not result in obvious changes in the dissociation dynamics of SrcW121A (Fig. 13D). This result indicates that proteasomal degradation of Src through the phosphorylation of serine 75 reported previously [25] is not related to the Src SH3-domain-dependent dissociation of Src from focal adhesions. Introduction of the W121A mutation to Src molecules reduces the dissociation from adhesion sites (Figs 7 and 11); in other words, the mutant remains at focal adhesions for longer durations due to lower dissociation ability. Compared to normal cells, v-srctransformed cells demonstrate a higher invasive ability due to strong binding of the SH3 domain to FAK, which is an Src substrate and also a kinase of Src at focal adhesions [50]. The enhanced Src/FAK interaction reinforces the downstream signaling related to cell migration through the rearrangement of the actin cytoskeleton and focal adhesion turnover [28]. By reducing the interaction between FAK and the SH3 domain in v-src, the invasive phenotype significantly diminishes [50]. Although the disruption of the SH3 domain showed hyperactivation of Src molecules (Fig. 3), it has been reported that the mutation does not lead to cell transformation [24]. Therefore, longer residence of Src molecules at focal adhesions may reduce the opportunity for other Src molecules to bind with FAK via the SH3 domain, which can be a trigger for the transforming signal pathway. The biological relevance of the uniform distribution of Src at the cell membrane is still unclear. As discussed above, the uniform distribution of Src at the cell membrane is highly correlated with the enhanced dynamics by the SH3 domain. Since the Src substrates localize at the cell membrane both inside and outside focal adhesions, for Src to serve as a kinase at the cell membrane Src molecules need to continuously bind with and dissociate from their substrates at the cell membrane. Our findings that the SH3 domain of Src enhances its dynamics at the cell membrane indicate that the uniform distribution of Src at the cell membrane may have an advantage for quick signal transduction by the continuous targeting of Src to its substrates. Materials and methods DNA constructs To construct the expression vectors for human c-src tagged with Venus or PAmCherry, the cdnas of SrcWT, Venus and PAmCherry were amplified by PCR and cloned into the pcdna3.1 vector (Thermo Fisher Scientific, Waltham, MA, USA). The cdnas of SrcW121A, SrcR178A, SrcK298M, SrcY530F, SrcW121A/R178A, SrcW121A/ K298M, SrcW121A/Y530F, SrcR178A/K298M and SrcR178A/Y530F were generated by introducing point mutations using the megaprimer method as previously described [51]. Venus or PAmCherry was fused to the C terminus of Src, because fusion of fluorescent proteins to the Src N terminus inhibits membrane targeting due to the lack of myristoylation of the N-terminal region, which is essential for membrane targeting [16]. Since direct fusion of fluorescent proteins to the Src C terminus without any lin- 4046
14 H. Machiyama et al. Roles of SH3 domain in the dynamics of c-src 4047
15 Roles of SH3 domain in the dynamics of c-src H. Machiyama et al. Fig. 7. The SH3 domain accelerates Src movement and regulates the residency of Src at focal adhesions. HeLa cells co-expressing Src- PAmCherry (WT, W121A or R178A) and paxillin-yfp were cultured in a collagen-coated glass-based chamber. Single particles from individually activated PAmCherry molecules were traced within focal adhesions located at peripheral regions. (A) Representative trajectories within focal adhesions in the cells expressing Src variants (WT, W121A and R178A). Start and end points of obtained trajectories are shown by solid circles and open squares, respectively. The point-to-point interval is 16 ms. The inset shows a trajectory of activated PAmCherry inside a focal adhesion from SrcWT-PAmCherry and paxillin-yfp co-expressed HeLa cells fixed with 4% paraformaldehyde for 30 min at room temperature. (B) (F) Velocity (B), duration (C), total run length (D), end-to-end distance (E) and persistency value (F) of the particles are shown. (G) The mean square displacements r 2 of the obtained trajectories in SrcWT (rectangles), SrcW121A (circles) and SrcR178A (triangles) in HeLa cells are plotted as a function of the time interval Dt. Dotted lines indicate the results of curve fitting using Eqn (1). Statistical analyses were performed with one-way ANOVA followed by Tukey Kramer post hoc testing among Src variants; *P < 0.05, n = 140 for WT, n = 228 for W121A and n = 109 for R178A. Table 4. Diffusion constants and c values of Src variants at the cell membrane inside focal adhesions in HeLa cells. Src D (lm 2 s 1 ) c Figure WT Figs 7, 9A and 10 W121A Fig. 7 R178A Fig. 7 K298M Fig. 9A W121A/K298M Fig. 9A R178A/K298M Fig. 9A Y530F Fig. 10 W121A/Y530F Fig. 10 R178A/Y530F Fig. 10 ker sequences involves constitutive activation of Src due to the lack of negative regulation by tyrosine 530 phosphorylation [52], we attached a flexible (GGGGS) 3 linker [53] between Src and fluorescent proteins. The expression vectors for GFP-, EYFP-, and mcherry-tagged paxillin were constructed by PCR using the pcdna3.1 vector as a parent vector. Cell culture, transfection and drug treatment The cells used in this study were cultured in Dulbecco s modified Eagle s medium (DMEM) (Sigma-Aldrich, St Louis, MO, USA) containing 10% fetal bovine serum (Thermo Fisher Scientific) supplemented with 1% penicillin/streptomycin (Sigma-Aldrich). Transfection of plasmid DNA was carried out by FuGENE6 HD (Promega, Madison, WI, USA). The transfected cells were transferred to collagen-coated glass dishes (AGC Techno Glass, Shizuoka, Japan) 24 h before microscope observation. CGP77675 (Sigma-Aldrich), cytochalasin D (Wako Pure Chemicals, Osaka, Japan) and blebbistatin (Sigma-Aldrich) were applied 1 h before image acquisition. Immunoblotting Cells were lysed in cell lysis buffer [50 mm Tris/HCl ph 8.0, 150 mm NaCl, 10 mm ethylenediaminetetraacetic acid, 0.5% Triton X-100, 100 mm NaF, 1 mm Na 3 VO 4, 1 mm dithiothreitol and protease inhibitor cocktail (Roche Diagnostics, Basel, Switzerland)] and the lysates were centrifuged at g for 15 min. The supernatants were placed in SDS sample buffer (62.5 mm Tris/HCl ph 6.8, 2.3% SDS, 10% glycerol, 0.37 mm bromophenol blue and 2.5% b-mercaptoethanol) and were boiled for 5 min. Equivalent amounts of the proteins from each sample were electrophoresed on an SDS/PAGE gel (4 15% gradient gel; Bio-Rad, Hercules, CA, USA) and transferred to polyvinylidene difluoride membranes (Bio-Rad). For immunoblotting analysis, the membranes were probed with the appropriate dilution of primary antibodies against Src (1 : 1000; Merck Millipore, Billerica, MA, USA), phospho-src-y419 (1 : 1000; Cell Signaling Technology, Beverly, MA, USA), a-tubulin (1 : 2000; Sigma-Aldrich) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1 : 2000; Abcam, Cambridge, UK). Microscope settings A TIRF analysis system was set up on a conventional inverted microscope (Ti-U; Nikon, Tokyo, Japan) equipped with an EMCCD camera with a pixel chip (ixon3; Andor Technology, Belfast, UK) and 473 and 561 nm lasers to excite GFP or Venus and mcherry/pam- Cherry, respectively. PAmCherry was activated using a 405 nm laser. NIS ELEMENTS software (Nikon) was used for image acquisition and IMAGEJ software (National Institutes of Health, Bethesda, MD, USA) was used for image processing and final figure preparation. Immunofluorescence microscopy HeLa cells expressing GFP-tagged paxillin were cultured overnight in a collagen-coated glass dish. Cells were fixed with 4% paraformaldehyde for 30 min at room temperature, permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature, blocked with 2% BSA in PBS for 1 h at room temperature, and incubated with a polyclonal antibody against Src (Cell Signaling Technology) 4048
16 H. Machiyama et al. Roles of SH3 domain in the dynamics of c-src Fig. 8. Trajectories of Src molecules obtained from focal adhesions. A merged image of the bright field and EYFP fluorescence (green) images in a HeLa cell co-expressing paxillin-eyfp and PAmCherry-tagged Src variants (WT, W121A and R178A) (left). Representative trajectories of activated PAmCherry in a focal adhesion indicated by the arrowhead in the left column are shown (right). and phospho-src-y419 (psrc-419) (Cell Signaling Technology) as a primary antibody in PBS containing 2% BSA overnight at 4 C. Lastly, the cells were incubated with an Alexa594-conjugated goat anti-rabbit IgG (Thermo Fisher Scientific) as a secondary antibody for 1 h at 4 C to fluorescently label Src and psrc-419. Fig. 9. The contribution of Src activation status on the SH3-domain-dependent movement of Src at the cell membrane at focal adhesions. (A) HeLa cells co-expressing Src-PAmCherry (WT, K298M, W121A/K298M or R178A/K298M) and paxillin-eyfp were cultured in a collagencoated glass-based chamber. Single particles from individually activated PAmCherry molecules were traced at focal adhesions. The velocity (a), duration (b), total run length (c), end-to-end distance (d) and persistency value (e) of the particles and the MSD-t plot (f) are shown. Statistical analyses were performed to compare cells expressing SrcWT and those expressing other Src variants (n > 100 by t tests). (B) HeLa cells co-expressing Src- or SrcW121A-PAmCherry and paxillin-eyfp treated with DMSO or 1 lm CGP77675, 1 h before TIRF observation. Single activated PAmCherry molecules were traced at focal adhesions. The velocity (a), duration (b), total run length (c), end-toend distance (d) and persistency value (e) of the particles and the MSD-t plot (f) are shown. Statistical analyses were performed between CGP77675 treated and untreated cells (n > 100 by t tests). Non-significant differences are shown between all combinations of groups in both (A) and (B). 4049
17 Roles of SH3 domain in the dynamics of c-src H. Machiyama et al. 4050
18 H. Machiyama et al. Roles of SH3 domain in the dynamics of c-src Table 5. Diffusion constants and c values of SrcWT and SrcW121A at focal adhesions treated with various drugs in HeLa cells. Src Drug D (lm 2 s 1 ) c Figures WT DMSO Fig. 9B WT CGP Fig. 9B W121A DMSO Fig. 9B W121A CGP Fig. 9B Tracking of single fluorescent particles HeLa and SYF cells co-expressing EYFP-tagged paxillin and PAmCherry-tagged Src variants were cultured on a collagen-coated glass dish. PAmCherry was activated with a gentle pulse of a 405 nm laser to isolate individual fluorescent particles emitted from single activated-pamcherry molecules. To track Src molecule dynamics within focal adhesions, images were obtained every 16 ms and individual analyses were conducted on lm 2 areas including Fig. 10. Assessments of the constitutive activation of Src in the movement of Src at focal adhesions. (A) HeLa cells co-expressing Src- PAmCherry (WT, Y530F, W121A/Y530F or R178A/Y530F) and paxillin-eyfp were cultured in a collagen-coated glass-based chamber. Single particles from individually activated PAmCherry molecules were traced at focal adhesions. The velocity (a), duration (b), total run length (c), end-to-end distance (d) and persistency value (e) of the particles and the MSD-t plot (f) are shown. Statistical analyses were performed to compare cells expressing SrcWT and those expressing other Src variants (n > 100 by t tests). Non-significant differences are shown between all combinations of groups. 4051
19 H. Machiyama et al. Roles of SH3 domain in the dynamics of c-src Fig. 11. Mutations in the SH3 domain of Src alter the distribution of Src at the cell membrane. (A) HeLa cells co-expressing Src-Venus (WT, W121A, R178A or W121A/R178A) and paxillin-mcherry cultured in a collagen-coated glass-based chamber and viewed by TIRF microscopy. TIRF images of Venus (first row) and mcherry (second row) fluorescence are shown. The third row shows the merged TIRF images of Venus and mcherry. Insets in row 3 show the expanded images indicated by white rectangles. Scale bar 10 lm. (B) Box plots of the colocalization index of Venus-tagged Src variants and mcherry-paxillin in HeLa cells. Statistical analyses were performed to compare cells expressing SrcWT (black box) and those expressing other Src variants (n > 27, **P < 0.01 by t tests). Fig. 12. Localization of the SrcW121A/R178A mutant is ambiguous. HeLa cells co-expressing SrcW121A/R178A-Venus and paxillin-mcherry were viewed by TIRF microscopy. (A), (B) Venus (left), mcherry (center) and merged (right) TIRF images in which the cell indicated a high (A) and low (B) colocalization index. Magnified images indicated by white rectangles are shown below. Scale bar 10 lm. single focal adhesions at the cell s peripheral region. To track Src molecule dynamics outside focal adhesions, images were obtained every 100 ms and individual analyses were conducted on lm2 areas at the peripheral region of cells (Fig. 2B). The spatial x-y position of individual fluorescent particles was determined by two-dimensional Gaussian fitting [54] Acknowledgements We thank Kazuki Matsuda and Miwa Fukami for technical assistance. This work was supported by a grant from the Japanese Ministry of Education, Culture, Sports, Science and Technology to HF (# ).
20 H. Machiyama et al. Roles of SH3 domain in the dynamics of c-src Fig. 13. Manipulation of phosphorylation status at serine 75 does not affect the distribution of SrcW121A mutant. (A) Schematic representation of Src mutants: SrcS75A (phosphodefective at serine 75), SrcS75E (phosphomimetic at serine 75), SrcS75A/W121A (phosphodefective at serine 75 and a mutated SH3 domain) and SrcS75E/W121A (phosphomimetic at serine 75 and a mutated SH3 domain). (B) TIRF images of HeLa cells co-expressing Venus-tagged Src mutants (S75A, S75E, S75A/W121A or S75E/W121A) (first column) and mcherry-tagged paxillin (second column). The third column shows the merged TIRF images of Venus and mcherry. (C), (D) Box plots of the colocalization index of Venus-tagged Src variants and mcherry-paxillin in HeLa cells. The data for WT and W121A in (C) and (D), respectively, are the same as those shown in Fig. 11B. All statistical analyses compared cells expressing SrcWT or SrcW121A in (C) or (D) (black box), respectively, and those expressing another Src variant (n > 27 in C, n > 33 in D, **P < 0.01 by t tests). Conflicts of interest We declare that we have no conflicts of interest. Author contributions HM, TY and HF designed the experiments. HM performed the experiments. HM and HF analyzed data. HM, YS, TMW and HF wrote the paper. References 1 Chackalaparampil I & Shalloway D (1988) Altered phosphorylation and activation of pp60c-src during fibroblast mitosis. Cell 52, Schwartzberg PL, Xing L, Hoffmann O, Lowell CA, Garrett L, Boyce BF & Varmus HE (1997) Rescue of osteoclast function by transgenic expression of kinasedeficient Src in src-/- mutant mice. Genes Dev 11,
Nature Immunology: doi: /ni.3631
 Supplementary Figure 1 SPT analyses of Zap70 at the T cell plasma membrane. (a) Total internal reflection fluorescent (TIRF) excitation at 64-68 degrees limits single molecule detection to 100-150 nm above
Supplementary Figure 1 SPT analyses of Zap70 at the T cell plasma membrane. (a) Total internal reflection fluorescent (TIRF) excitation at 64-68 degrees limits single molecule detection to 100-150 nm above
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
El Azzouzi et al., http ://www.jcb.org /cgi /content /full /jcb /DC1
 Supplemental material JCB El Azzouzi et al., http ://www.jcb.org /cgi /content /full /jcb.201510043 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Acquisition of -phluorin correlates negatively with podosome
Supplemental material JCB El Azzouzi et al., http ://www.jcb.org /cgi /content /full /jcb.201510043 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Acquisition of -phluorin correlates negatively with podosome
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplemental Information. Autophagy in Oncogenic K-Ras. Promotes Basal Extrusion. of Epithelial Cells by Degrading S1P. Current Biology, Volume 24
 Current Biology, Volume 24 Supplemental Information Autophagy in Oncogenic K-Ras Promotes Basal Extrusion of Epithelial Cells by Degrading S1P Gloria Slattum, Yapeng Gu, Roger Sabbadini, and Jody Rosenblatt
Current Biology, Volume 24 Supplemental Information Autophagy in Oncogenic K-Ras Promotes Basal Extrusion of Epithelial Cells by Degrading S1P Gloria Slattum, Yapeng Gu, Roger Sabbadini, and Jody Rosenblatt
TFEB-mediated increase in peripheral lysosomes regulates. Store Operated Calcium Entry
 TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
Supplementary information. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins
 Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
crossmark Ca V subunits interact with the voltage-gated calcium channel
 crossmark THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 291, NO. 39, pp. 20402 20416, September 23, 2016 Author s Choice 2016 by The American Society for Biochemistry and Molecular Biology, Inc. Published in
crossmark THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 291, NO. 39, pp. 20402 20416, September 23, 2016 Author s Choice 2016 by The American Society for Biochemistry and Molecular Biology, Inc. Published in
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Jewell et al., http://www.jcb.org/cgi/content/full/jcb.201007176/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. IR Munc18c association is independent of IRS-1. (A and
Supplemental material Jewell et al., http://www.jcb.org/cgi/content/full/jcb.201007176/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. IR Munc18c association is independent of IRS-1. (A and
SUPPLEMENTARY INFORMATION
 In the format provided by the authors and unedited. 2 3 4 DOI: 10.1038/NMAT4893 EGFR and HER2 activate rigidity sensing only on rigid matrices Mayur Saxena 1,*, Shuaimin Liu 2,*, Bo Yang 3, Cynthia Hajal
In the format provided by the authors and unedited. 2 3 4 DOI: 10.1038/NMAT4893 EGFR and HER2 activate rigidity sensing only on rigid matrices Mayur Saxena 1,*, Shuaimin Liu 2,*, Bo Yang 3, Cynthia Hajal
04_polarity. The formation of synaptic vesicles
 Brefeldin prevents assembly of the coats required for budding Nocodazole disrupts microtubules Constitutive: coatomer-coated Selected: clathrin-coated The formation of synaptic vesicles Nerve cells (and
Brefeldin prevents assembly of the coats required for budding Nocodazole disrupts microtubules Constitutive: coatomer-coated Selected: clathrin-coated The formation of synaptic vesicles Nerve cells (and
Supplements. Figure S1. B Phalloidin Alexa488
 Supplements A, DMSO, PP2, PP3 Crk-myc Figure S1. (A) Src kinase activity is necessary for recruitment of Crk to Nephrin cytoplasmic domain. Human podocytes expressing /7-NephrinCD () were treated with
Supplements A, DMSO, PP2, PP3 Crk-myc Figure S1. (A) Src kinase activity is necessary for recruitment of Crk to Nephrin cytoplasmic domain. Human podocytes expressing /7-NephrinCD () were treated with
Bioluminescence Resonance Energy Transfer (BRET)-based studies of receptor dynamics in living cells with Berthold s Mithras
 Bioluminescence Resonance Energy Transfer (BRET)-based studies of receptor dynamics in living cells with Berthold s Mithras Tarik Issad, Ralf Jockers and Stefano Marullo 1 Because they play a pivotal role
Bioluminescence Resonance Energy Transfer (BRET)-based studies of receptor dynamics in living cells with Berthold s Mithras Tarik Issad, Ralf Jockers and Stefano Marullo 1 Because they play a pivotal role
Src-INACTIVE / Src-INACTIVE
 Biology 169 -- Exam 1 February 2003 Answer each question, noting carefully the instructions for each. Repeat- Read the instructions for each question before answering!!! Be as specific as possible in each
Biology 169 -- Exam 1 February 2003 Answer each question, noting carefully the instructions for each. Repeat- Read the instructions for each question before answering!!! Be as specific as possible in each
The role of the scaffolding protein Tks5 in EGF signaling
 The role of the scaffolding protein Tks5 in EGF signaling PhD Thesis Anna Fekete Doctoral School in Biology Head of the School: Dr. Anna Erdei Structural Biochemistry Doctoral Program Head of the Program:
The role of the scaffolding protein Tks5 in EGF signaling PhD Thesis Anna Fekete Doctoral School in Biology Head of the School: Dr. Anna Erdei Structural Biochemistry Doctoral Program Head of the Program:
The unique N-terminal region of SRMS regulates enzymatic activity and phosphorylation of its novel substrate docking protein 1
 The unique N-terminal region of SRMS regulates enzymatic activity and phosphorylation of its novel substrate docking protein 1 Raghuveera K. Goel, Sayem Miah, Kristin Black, Natasha Kalra, Chenlu Dai and
The unique N-terminal region of SRMS regulates enzymatic activity and phosphorylation of its novel substrate docking protein 1 Raghuveera K. Goel, Sayem Miah, Kristin Black, Natasha Kalra, Chenlu Dai and
The pi-value distribution of single-pass membrane proteins at the plasma membrane in immune cells and in total cells.
 Supplementary Figure 1 The pi-value distribution of single-pass membrane proteins at the plasma membrane in immune cells and in total cells. The PI values were measured for the first 10 amino acids in
Supplementary Figure 1 The pi-value distribution of single-pass membrane proteins at the plasma membrane in immune cells and in total cells. The PI values were measured for the first 10 amino acids in
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
SUPPLEMENTARY INFORMATION. Supplementary Figures S1-S9. Supplementary Methods
 SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
293T cells were transfected with indicated expression vectors and the whole-cell extracts were subjected
 SUPPLEMENTARY INFORMATION Supplementary Figure 1. Formation of a complex between Slo1 and CRL4A CRBN E3 ligase. (a) HEK 293T cells were transfected with indicated expression vectors and the whole-cell
SUPPLEMENTARY INFORMATION Supplementary Figure 1. Formation of a complex between Slo1 and CRL4A CRBN E3 ligase. (a) HEK 293T cells were transfected with indicated expression vectors and the whole-cell
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Nature Methods: doi: /nmeth.4257
 Supplementary Figure 1 Screen for polypeptides that affect cellular actin filaments. (a) Table summarizing results from all polypeptides tested. Source shows organism, gene, and amino acid numbers used.
Supplementary Figure 1 Screen for polypeptides that affect cellular actin filaments. (a) Table summarizing results from all polypeptides tested. Source shows organism, gene, and amino acid numbers used.
Nature Biotechnology: doi: /nbt.3828
 Supplementary Figure 1 Development of a FRET-based MCS. (a) Linker and MA2 modification are indicated by single letter amino acid code. indicates deletion of amino acids and N or C indicate the terminus
Supplementary Figure 1 Development of a FRET-based MCS. (a) Linker and MA2 modification are indicated by single letter amino acid code. indicates deletion of amino acids and N or C indicate the terminus
HCC1937 is the HCC1937-pcDNA3 cell line, which was derived from a breast cancer with a mutation
 SUPPLEMENTARY INFORMATION Materials and Methods Human cell lines and culture conditions HCC1937 is the HCC1937-pcDNA3 cell line, which was derived from a breast cancer with a mutation in exon 20 of BRCA1
SUPPLEMENTARY INFORMATION Materials and Methods Human cell lines and culture conditions HCC1937 is the HCC1937-pcDNA3 cell line, which was derived from a breast cancer with a mutation in exon 20 of BRCA1
SUPPLEMENT. Materials and methods
 SUPPLEMENT Materials and methods Cell culture and reagents Cell media and reagents were from Invitrogen unless otherwise indicated. Antibiotics and Tet-certified serum were from Clontech. In experiments
SUPPLEMENT Materials and methods Cell culture and reagents Cell media and reagents were from Invitrogen unless otherwise indicated. Antibiotics and Tet-certified serum were from Clontech. In experiments
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
Supplementary Figure S1
 Supplementary Figure S1 Supplementary Figure S1. PARP localization patterns using GFP-PARP and PARP-specific antibody libraries GFP-PARP localization in non-fixed (A) and formaldehyde fixed (B) GFP-PARPx
Supplementary Figure S1 Supplementary Figure S1. PARP localization patterns using GFP-PARP and PARP-specific antibody libraries GFP-PARP localization in non-fixed (A) and formaldehyde fixed (B) GFP-PARPx
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis
 Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Chapter 3. Expression of α5-megfp in Mouse Cortical Neurons. on the β subunit. Signal sequences in the M3-M4 loop of β nachrs bind protein factors to
 22 Chapter 3 Expression of α5-megfp in Mouse Cortical Neurons Subcellular localization of the neuronal nachr subtypes α4β2 and α4β4 depends on the β subunit. Signal sequences in the M3-M4 loop of β nachrs
22 Chapter 3 Expression of α5-megfp in Mouse Cortical Neurons Subcellular localization of the neuronal nachr subtypes α4β2 and α4β4 depends on the β subunit. Signal sequences in the M3-M4 loop of β nachrs
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system
 Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures
 Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
G-Protein Signaling. Introduction to intracellular signaling. Dr. SARRAY Sameh, Ph.D
 G-Protein Signaling Introduction to intracellular signaling Dr. SARRAY Sameh, Ph.D Cell signaling Cells communicate via extracellular signaling molecules (Hormones, growth factors and neurotransmitters
G-Protein Signaling Introduction to intracellular signaling Dr. SARRAY Sameh, Ph.D Cell signaling Cells communicate via extracellular signaling molecules (Hormones, growth factors and neurotransmitters
Schwarz et al. Activity-Dependent Ubiquitination of GluA1 Mediates a Distinct AMPAR Endocytosis
 Schwarz et al Activity-Dependent Ubiquitination of GluA1 Mediates a Distinct AMPAR Endocytosis and Sorting Pathway Supplemental Data Supplemental Fie 1: AMPARs undergo activity-mediated ubiquitination
Schwarz et al Activity-Dependent Ubiquitination of GluA1 Mediates a Distinct AMPAR Endocytosis and Sorting Pathway Supplemental Data Supplemental Fie 1: AMPARs undergo activity-mediated ubiquitination
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Chen et al., http://www.jcb.org/cgi/content/full/jcb.201210119/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Lack of fast reversibility of UVR8 dissociation. (A) HEK293T
Supplemental material Chen et al., http://www.jcb.org/cgi/content/full/jcb.201210119/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Lack of fast reversibility of UVR8 dissociation. (A) HEK293T
Influenza A virus hemagglutinin and neuraminidase act as novel motile machinery. Tatsuya Sakai, Shin I. Nishimura, Tadasuke Naito, and Mineki Saito
 Supplementary Information for: Influenza A virus hemagglutinin and neuraminidase act as novel motile machinery Tatsuya Sakai, Shin I. Nishimura, Tadasuke Naito, and Mineki Saito Supplementary Methods Supplementary
Supplementary Information for: Influenza A virus hemagglutinin and neuraminidase act as novel motile machinery Tatsuya Sakai, Shin I. Nishimura, Tadasuke Naito, and Mineki Saito Supplementary Methods Supplementary
Cbl ubiquitin ligase: Lord of the RINGs
 Cbl ubiquitin ligase: Lord of the RINGs Not just quite interesting - really interesting! A cell must be able to degrade proteins when their activity is no longer required. Many eukaryotic proteins are
Cbl ubiquitin ligase: Lord of the RINGs Not just quite interesting - really interesting! A cell must be able to degrade proteins when their activity is no longer required. Many eukaryotic proteins are
Data Sheet TIGIT / NFAT Reporter - Jurkat Cell Line Catalog #60538
 Data Sheet TIGIT / NFAT Reporter - Jurkat Cell Line Catalog #60538 Background: TIGIT is a co-inhibitory receptor that is highly expressed in Natural Killer (NK) cells, activated CD4+, CD8+ and regulatory
Data Sheet TIGIT / NFAT Reporter - Jurkat Cell Line Catalog #60538 Background: TIGIT is a co-inhibitory receptor that is highly expressed in Natural Killer (NK) cells, activated CD4+, CD8+ and regulatory
F-actin VWF Vinculin. F-actin. Vinculin VWF
 a F-actin VWF Vinculin b F-actin VWF Vinculin Supplementary Fig. 1. WPBs in HUVECs are located along stress fibers and at focal adhesions. (a) Immunofluorescence images of f-actin (cyan), VWF (yellow),
a F-actin VWF Vinculin b F-actin VWF Vinculin Supplementary Fig. 1. WPBs in HUVECs are located along stress fibers and at focal adhesions. (a) Immunofluorescence images of f-actin (cyan), VWF (yellow),
Islet viability assay and Glucose Stimulated Insulin Secretion assay RT-PCR and Western Blot
 Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Supplementary Information
 Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO
 Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Effects of Second Messengers
 Effects of Second Messengers Inositol trisphosphate Diacylglycerol Opens Calcium Channels Binding to IP 3 -gated Channel Cooperative binding Activates Protein Kinase C is required Phosphorylation of many
Effects of Second Messengers Inositol trisphosphate Diacylglycerol Opens Calcium Channels Binding to IP 3 -gated Channel Cooperative binding Activates Protein Kinase C is required Phosphorylation of many
supplementary information
 DOI: 10.1038/ncb2133 Figure S1 Actomyosin organisation in human squamous cell carcinoma. (a) Three examples of actomyosin organisation around the edges of squamous cell carcinoma biopsies are shown. Myosin
DOI: 10.1038/ncb2133 Figure S1 Actomyosin organisation in human squamous cell carcinoma. (a) Three examples of actomyosin organisation around the edges of squamous cell carcinoma biopsies are shown. Myosin
MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands)
 Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Supplemental Information
 Supplemental Information Tobacco-specific Carcinogen Induces DNA Methyltransferases 1 Accumulation through AKT/GSK3β/βTrCP/hnRNP-U in Mice and Lung Cancer patients Ruo-Kai Lin, 1 Yi-Shuan Hsieh, 2 Pinpin
Supplemental Information Tobacco-specific Carcinogen Induces DNA Methyltransferases 1 Accumulation through AKT/GSK3β/βTrCP/hnRNP-U in Mice and Lung Cancer patients Ruo-Kai Lin, 1 Yi-Shuan Hsieh, 2 Pinpin
Supplementary Data Table of Contents:
 Supplementary Data Table of Contents: - Supplementary Methods - Supplementary Figures S1(A-B) - Supplementary Figures S2 (A-B) - Supplementary Figures S3 - Supplementary Figures S4(A-B) - Supplementary
Supplementary Data Table of Contents: - Supplementary Methods - Supplementary Figures S1(A-B) - Supplementary Figures S2 (A-B) - Supplementary Figures S3 - Supplementary Figures S4(A-B) - Supplementary
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Lu et al., http://www.jcb.org/cgi/content/full/jcb.201012063/dc1 Figure S1. Kinetics of nuclear envelope assembly, recruitment of Nup133
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Lu et al., http://www.jcb.org/cgi/content/full/jcb.201012063/dc1 Figure S1. Kinetics of nuclear envelope assembly, recruitment of Nup133
HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates
 HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates Department of Microbiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, USA
HIV-1 Virus-like Particle Budding Assay Nathan H Vande Burgt, Luis J Cocka * and Paul Bates Department of Microbiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, USA
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Beck et al., http://www.jcb.org/cgi/content/full/jcb.201011027/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Membrane binding of His-tagged proteins to Ni-liposomes.
Supplemental material Beck et al., http://www.jcb.org/cgi/content/full/jcb.201011027/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Membrane binding of His-tagged proteins to Ni-liposomes.
Supplemental information contains 7 movies and 4 supplemental Figures
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplemental information contains 7 movies and 4 supplemental Figures Movies: Movie 1. Single virus tracking of A4-mCherry-WR MV
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplemental information contains 7 movies and 4 supplemental Figures Movies: Movie 1. Single virus tracking of A4-mCherry-WR MV
Protocol for A-549 VIM RFP (ATCC CCL-185EMT) TGFβ1 EMT Induction and Drug Screening
 Protocol for A-549 VIM RFP (ATCC CCL-185EMT) TGFβ1 EMT Induction and Drug Screening Introduction: Vimentin (VIM) intermediate filament (IF) proteins are associated with EMT in lung cancer and its metastatic
Protocol for A-549 VIM RFP (ATCC CCL-185EMT) TGFβ1 EMT Induction and Drug Screening Introduction: Vimentin (VIM) intermediate filament (IF) proteins are associated with EMT in lung cancer and its metastatic
Appendix. Table of Contents
 Appendix Table of Contents Appendix Figures Figure S1: Gp78 is not required for the degradation of mcherry-cl1 in Hela Cells. Figure S2: Indel formation in the MARCH6 sgrna targeted HeLa clones. Figure
Appendix Table of Contents Appendix Figures Figure S1: Gp78 is not required for the degradation of mcherry-cl1 in Hela Cells. Figure S2: Indel formation in the MARCH6 sgrna targeted HeLa clones. Figure
Supplementary Information. Conformational states of Lck regulate clustering in early T cell signaling
 Supplementary Information Conformational states of Lck regulate clustering in early T cell signaling Jérémie Rossy, Dylan M. Owen, David J. Williamson, Zhengmin Yang and Katharina Gaus Centre for Vascular
Supplementary Information Conformational states of Lck regulate clustering in early T cell signaling Jérémie Rossy, Dylan M. Owen, David J. Williamson, Zhengmin Yang and Katharina Gaus Centre for Vascular
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer
 Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Zool 3200: Cell Biology Exam 4 Part II 2/3/15
 Name:Key Trask Zool 3200: Cell Biology Exam 4 Part II 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
Name:Key Trask Zool 3200: Cell Biology Exam 4 Part II 2/3/15 Answer each of the following questions in the space provided, explaining your answers when asked to do so; circle the correct answer or answers
Life Science 1A Final Exam. January 19, 2006
 ame: TF: Section Time Life Science 1A Final Exam January 19, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use non-erasable
ame: TF: Section Time Life Science 1A Final Exam January 19, 2006 Please write legibly in the space provided below each question. You may not use calculators on this exam. We prefer that you use non-erasable
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
a Anti-Dab2 RGD Merge
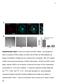 a Anti-Da Merge *** ** Percentage of cells adhered 8 6 4 RGE RAD RGE RAD Supplementary Figure Da are localized at clusters. (a) Endogenous Da is localized at clusters. (), ut not RGE nor RAD peptides can
a Anti-Da Merge *** ** Percentage of cells adhered 8 6 4 RGE RAD RGE RAD Supplementary Figure Da are localized at clusters. (a) Endogenous Da is localized at clusters. (), ut not RGE nor RAD peptides can
ORIGINAL PAPER DISRUPTION OF CELL SPREADING BY THE ACTIVATION OF MEK/ERK PATHWAY IS DEPENDENT ON AP-1 ACTIVITY
 Nagoya J. Med. Sci. 72. 139 ~ 144, 1 ORIGINAL PAPER DISRUPTION OF CELL SPREADING BY THE ACTIVATION OF MEK/ERK PATHWAY IS DEPENDENT ON AP-1 ACTIVITY FENG XU, SATOKO ITO, MICHINARI HAMAGUCHI and TAKESHI
Nagoya J. Med. Sci. 72. 139 ~ 144, 1 ORIGINAL PAPER DISRUPTION OF CELL SPREADING BY THE ACTIVATION OF MEK/ERK PATHWAY IS DEPENDENT ON AP-1 ACTIVITY FENG XU, SATOKO ITO, MICHINARI HAMAGUCHI and TAKESHI
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
d e f Spatiotemporal quantification of subcellular ATP levels in a single HeLa cell during changes in morphology Supplementary Information
 Ca 2+ level (a. u.) Area (a. u.) Normalized distance Normalized distance Center Edge Center Edge Relative ATP level Relative ATP level Supplementary Information Spatiotemporal quantification of subcellular
Ca 2+ level (a. u.) Area (a. u.) Normalized distance Normalized distance Center Edge Center Edge Relative ATP level Relative ATP level Supplementary Information Spatiotemporal quantification of subcellular
Gladstone Institutes, University of California (UCSF), San Francisco, USA
 Fluorescence-linked Antigen Quantification (FLAQ) Assay for Fast Quantification of HIV-1 p24 Gag Marianne Gesner, Mekhala Maiti, Robert Grant and Marielle Cavrois * Gladstone Institutes, University of
Fluorescence-linked Antigen Quantification (FLAQ) Assay for Fast Quantification of HIV-1 p24 Gag Marianne Gesner, Mekhala Maiti, Robert Grant and Marielle Cavrois * Gladstone Institutes, University of
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation
 Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the
 Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the genome-wide methylation microarray data. Mean ± s.d.; Student
Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the genome-wide methylation microarray data. Mean ± s.d.; Student
A Reciprocal Interdependence between Nck and PI(4,5)P 2 Promotes Localized N-WASp-Mediated Actin Polymerization in Living Cells
 Molecular Cell, Volume 36 Supplemental Data A Reciprocal Interdependence between Nck and PI(4,5)P 2 Promotes Localized N-WASp-Mediated Actin Polymerization in Living Cells Gonzalo M. Rivera, Dan Vasilescu,
Molecular Cell, Volume 36 Supplemental Data A Reciprocal Interdependence between Nck and PI(4,5)P 2 Promotes Localized N-WASp-Mediated Actin Polymerization in Living Cells Gonzalo M. Rivera, Dan Vasilescu,
William C. Comb, Jessica E. Hutti, Patricia Cogswell, Lewis C. Cantley, and Albert S. Baldwin
 Molecular Cell, Volume 45 Supplemental Information p85 SH2 Domain Phosphorylation by IKK Promotes Feedback Inhibition of PI3K and Akt in Response to Cellular Starvation William C. Comb, Jessica E. Hutti,
Molecular Cell, Volume 45 Supplemental Information p85 SH2 Domain Phosphorylation by IKK Promotes Feedback Inhibition of PI3K and Akt in Response to Cellular Starvation William C. Comb, Jessica E. Hutti,
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human
 Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
Supplementary Table; Supplementary Figures and legends S1-S21; Supplementary Materials and Methods
 Silva et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability Supplementary Table; Supplementary Figures and legends S1-S21; Supplementary
Silva et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability Supplementary Table; Supplementary Figures and legends S1-S21; Supplementary
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
CHAPTER 3: EGFR ACTIVATION IMPACTS ON FAK PROTEIN EXPRESSION AND PHOSPHORYLATION STATUS IN HOSCC CELL LINES
 CHAPTER 3: EGFR ACTIVATION IMPACTS ON FAK PROTEIN EXPRESSION AND PHOSPHORYLATION STATUS IN HOSCC CELL LINES 3.1 Introduction Developmental processes such as cell migration depend on signals from both the
CHAPTER 3: EGFR ACTIVATION IMPACTS ON FAK PROTEIN EXPRESSION AND PHOSPHORYLATION STATUS IN HOSCC CELL LINES 3.1 Introduction Developmental processes such as cell migration depend on signals from both the
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Proteomic profiling of small-molecule inhibitors reveals dispensability of MTH1 for cancer cell survival
 Supplementary Information for Proteomic profiling of small-molecule inhibitors reveals dispensability of MTH1 for cancer cell survival Tatsuro Kawamura 1, Makoto Kawatani 1, Makoto Muroi, Yasumitsu Kondoh,
Supplementary Information for Proteomic profiling of small-molecule inhibitors reveals dispensability of MTH1 for cancer cell survival Tatsuro Kawamura 1, Makoto Kawatani 1, Makoto Muroi, Yasumitsu Kondoh,
CD3 coated cover slips indicating stimulatory contact site, F-actin polymerization and
 SUPPLEMENTAL FIGURES FIGURE S1. Detection of MCs. A, Schematic representation of T cells stimulated on anti- CD3 coated cover slips indicating stimulatory contact site, F-actin polymerization and microclusters.
SUPPLEMENTAL FIGURES FIGURE S1. Detection of MCs. A, Schematic representation of T cells stimulated on anti- CD3 coated cover slips indicating stimulatory contact site, F-actin polymerization and microclusters.
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. Neither the activation nor suppression of the MAPK pathway affects the ASK1/Vif interaction. (a, b) HEK293 cells were cotransfected with plasmids encoding
Supplementary Figure 1 Supplementary Figure 1. Neither the activation nor suppression of the MAPK pathway affects the ASK1/Vif interaction. (a, b) HEK293 cells were cotransfected with plasmids encoding
Animal Tissue Culture SQG 3242 Biology of Cultured Cells. Dr. Siti Pauliena Mohd Bohari
 Animal Tissue Culture SQG 3242 Biology of Cultured Cells Dr. Siti Pauliena Mohd Bohari The Culture Environment Changes of Cell s microenvironment needed that favor the spreading, migration, and proliferation
Animal Tissue Culture SQG 3242 Biology of Cultured Cells Dr. Siti Pauliena Mohd Bohari The Culture Environment Changes of Cell s microenvironment needed that favor the spreading, migration, and proliferation
Biology Open (2014) 000, 1 10 doi: /bio
 (2014) 000, 1 10 doi:10.1242/bio.201410041 Supplementary Material Michael Brauchle et al. doi: 10.1242/bio.201410041 Fig. S1. Alignment of GFP, sfgfp, egfp, eyfp, mcherry and mruby2. Sequence-based alignment
(2014) 000, 1 10 doi:10.1242/bio.201410041 Supplementary Material Michael Brauchle et al. doi: 10.1242/bio.201410041 Fig. S1. Alignment of GFP, sfgfp, egfp, eyfp, mcherry and mruby2. Sequence-based alignment
UIM domain-dependent recruitment of the endocytic adaptor protein Eps15 to ubiquitin-enriched endosomes
 UIM domain-dependent recruitment of the endocytic adaptor protein Eps15 to ubiquitin-enriched endosomes Gucwa and Brown Gucwa and Brown BMC Cell Biology 2014, 15:34 Gucwa and Brown BMC Cell Biology 2014,
UIM domain-dependent recruitment of the endocytic adaptor protein Eps15 to ubiquitin-enriched endosomes Gucwa and Brown Gucwa and Brown BMC Cell Biology 2014, 15:34 Gucwa and Brown BMC Cell Biology 2014,
Supplemental Data Figure S1 Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells.
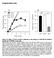 Supplemental Data Figure S1. Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells. (A) Specific lysis of IFN-γ-treated SKBR-3 cells in the absence
Supplemental Data Figure S1. Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells. (A) Specific lysis of IFN-γ-treated SKBR-3 cells in the absence
Electronic Supporting Information
 Modulation of raft domains in a lipid bilayer by boundary-active curcumin Manami Tsukamoto a, Kenichi Kuroda* b, Ayyalusamy Ramamoorthy* c, Kazuma Yasuhara* a Electronic Supporting Information Contents
Modulation of raft domains in a lipid bilayer by boundary-active curcumin Manami Tsukamoto a, Kenichi Kuroda* b, Ayyalusamy Ramamoorthy* c, Kazuma Yasuhara* a Electronic Supporting Information Contents
Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression
 Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous
 Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Bio 401 Sp2014. Multiple Choice (Circle the letter corresponding to the correct answer)
 MIDTERM EXAM KEY (60 pts) You should have 5 pages. Please put your name on every page. You will have 70 minutes to complete the exam. You may begin working as soon as you receive the exam. NOTE: the RED
MIDTERM EXAM KEY (60 pts) You should have 5 pages. Please put your name on every page. You will have 70 minutes to complete the exam. You may begin working as soon as you receive the exam. NOTE: the RED
Human SH-SY5Y neuroblastoma cells (A.T.C.C., Manassas, VA) were cultured in DMEM, F-12
 SUPPLEMENTARY METHODS Cell cultures Human SH-SY5Y neuroblastoma cells (A.T.C.C., Manassas, VA) were cultured in DMEM, F-12 Ham with 25 mm HEPES and NaHCO 3 (1:1) and supplemented with 10% (v/v) FBS, 1.0
SUPPLEMENTARY METHODS Cell cultures Human SH-SY5Y neuroblastoma cells (A.T.C.C., Manassas, VA) were cultured in DMEM, F-12 Ham with 25 mm HEPES and NaHCO 3 (1:1) and supplemented with 10% (v/v) FBS, 1.0
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/6/e1700338/dc1 Supplementary Materials for HIV virions sense plasma membrane heterogeneity for cell entry Sung-Tae Yang, Alex J. B. Kreutzberger, Volker Kiessling,
advances.sciencemag.org/cgi/content/full/3/6/e1700338/dc1 Supplementary Materials for HIV virions sense plasma membrane heterogeneity for cell entry Sung-Tae Yang, Alex J. B. Kreutzberger, Volker Kiessling,
Many G protein-coupled receptors (GPCRs) 2 are rapidly endocytosed after agonist binding, but the pathway of postendocytic
 THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 282, NO. 40, pp. 29646 29657, October 5, 2007 2007 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. Hepatocyte Growth
THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 282, NO. 40, pp. 29646 29657, October 5, 2007 2007 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. Hepatocyte Growth
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
(a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable
 Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Chairoungdua et al., http://www.jcb.org/cgi/content/full/jcb.201002049/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Expression of CD9 and CD82 inhibits Wnt/ -catenin
Supplemental material Chairoungdua et al., http://www.jcb.org/cgi/content/full/jcb.201002049/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Expression of CD9 and CD82 inhibits Wnt/ -catenin
Signaling. Dr. Sujata Persad Katz Group Centre for Pharmacy & Health research
 Signaling Dr. Sujata Persad 3-020 Katz Group Centre for Pharmacy & Health research E-mail:sujata.persad@ualberta.ca 1 Growth Factor Receptors and Other Signaling Pathways What we will cover today: How
Signaling Dr. Sujata Persad 3-020 Katz Group Centre for Pharmacy & Health research E-mail:sujata.persad@ualberta.ca 1 Growth Factor Receptors and Other Signaling Pathways What we will cover today: How
Rabbit Polyclonal antibody to NFkB p65 (v-rel reticuloendotheliosis viral oncogene homolog A (avian))
 Datasheet GeneTex, Inc : Toll Free 1-877-GeneTex (1-877-436-3839) Fax:1-949-309-2888 info@genetex.com GeneTex International Corporation : Tel:886-3-6208988 Fax:886-3-6208989 infoasia@genetex.com Date :
Datasheet GeneTex, Inc : Toll Free 1-877-GeneTex (1-877-436-3839) Fax:1-949-309-2888 info@genetex.com GeneTex International Corporation : Tel:886-3-6208988 Fax:886-3-6208989 infoasia@genetex.com Date :
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC
 Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Validation & Assay Performance Summary
 Validation & Assay Performance Summary LanthaScreen IGF-1R GripTite Cells Cat. no. K1834 Modification Detected: Phosphorylation of Multiple Tyr Residues on IGF-1R LanthaScreen Cellular Assay Validation
Validation & Assay Performance Summary LanthaScreen IGF-1R GripTite Cells Cat. no. K1834 Modification Detected: Phosphorylation of Multiple Tyr Residues on IGF-1R LanthaScreen Cellular Assay Validation
Predictive PP1Ca binding region in BIG3 : 1,228 1,232aa (-KAVSF-) HEK293T cells *** *** *** KPL-3C cells - E E2 treatment time (h)
 Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION
 Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION X. Shawn Liu 1, 3, Bing Song 2, 3, Bennett D. Elzey 3, 4, Timothy L. Ratliff 3, 4, Stephen F. Konieczny
Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION X. Shawn Liu 1, 3, Bing Song 2, 3, Bennett D. Elzey 3, 4, Timothy L. Ratliff 3, 4, Stephen F. Konieczny
ab E3 Ligase Auto- Ubiquitilylation Assay Kit
 ab139469 E3 Ligase Auto- Ubiquitilylation Assay Kit Instructions for Use For testing ubiquitin E3 ligase activity through assessment of their ability to undergo auto-ubiquitinylation This product is for
ab139469 E3 Ligase Auto- Ubiquitilylation Assay Kit Instructions for Use For testing ubiquitin E3 ligase activity through assessment of their ability to undergo auto-ubiquitinylation This product is for
SUPPLEMENTARY INFORMATION
 Figure S1 Treatment with both Sema6D and Plexin-A1 sirnas induces the phenotype essentially identical to that induced by treatment with Sema6D sirna alone or Plexin-A1 sirna alone. (a,b) The cardiac tube
Figure S1 Treatment with both Sema6D and Plexin-A1 sirnas induces the phenotype essentially identical to that induced by treatment with Sema6D sirna alone or Plexin-A1 sirna alone. (a,b) The cardiac tube
