Supplementary Figures
|
|
|
- Leslie Hancock
- 5 years ago
- Views:
Transcription
1 Supplementary Figures
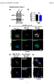
2 Supplementary Figure 1. Single cell gene expression analysis during collective cell migration. (a) Florescence and bright-field images of MCF7 cells transfected with dslna probes targeting β-actin mrna, Dll4 mrna, and random sequence near the leading edge. The model wounds were created by scratching cell monolayers with sterilized 1000 µl pipette tips. Images are merged to correlate the cell morphology with gene expression in leader cells. Images are representatives of four independent experiments. Scale bar, 25 μm. (b-c) Effects of dslna probes on cell migration. Migration speed (b) and distance (c) of cells transfected with Dll4 mrna and random probes. Control represents cells without probe transfection. No statistically significant difference was found between Dll4 and random probes (P > 0.05; paired Student s t- test). Data are expressed as mean ± s.e.m. (n=3).
3 Supplementary Figure 2. Distributions of gene expressions in cells at different locations from the leading edge. (a-d) Intensity distributions of dslna probes targeting β-actin mrna, Dll4 mrna, HO-1 mrna and random sequence in the first four rows from the leading edge. Each row is approximately 25 µm in width. Lines serve as visual guides. Over 200 cells were analyzed for each probe. Data are representatives of three independent experiments.
4 Supplementary Figure 3. Dynamics of leader cell formation during collective cell migration. (a) Time-lapse images illustrating the formation of leader cells after wounding. Examples of leader cells originated from the first row (red arrow) or second row (green arrow) in the boundary. Scale bar, 50 µm. (b) Time-lapse fluorescence images showing the dynamics of Dll4 mrna expression after wounding. A model wound was created in the top half of the image by scratching the monolayer. The cells could decrease (yellow arrows) or increase (blue arrow) their Dll4 expressions after wounding. Representative images of four independent experiments are shown. Scale bar, 50 μm.
5 Supplementary Figure 4. Immunofluorescence images of Dll4 and Notch1 in cell monolayers. (a-c) Effects of DMSO (a), DAPT (b) and Jagged-1 (c) treatments on Dll4 and Notch1 expressions. (d-f) Effects of control sirna (d), Notch1 sirna (e) and Dll4 sirna (f) on Dll4 and Notch1 expressions. Representative images of four independent experiments are shown. Scale bars, 50 μm.
6 Supplementary Figure 5. Immunofluorescence images of Notch1 and Notch1 mrna near the boundary. (a-c) Effects of DMSO (a), DAPT (b) and Jagged-1 (c) treatments on Notch1 and Notch1 mrna expressions. Representative images of three independent experiments are shown. Scale bars, 50 μm.
7 Supplementary Figure 6. Characterization of sirna efficiency. (a-b) Cells were transfected with either scrambled sirna or sirna against Notch1 or Dll4. Notch1 and Dll4 expressions were measured by immunoblotting. The asterisk indicates a nonspecific band. (c) Quantification of sirna efficiency. The sirna efficiency, represented by the knockdown rate, was 76.0±3.5% for Notch1 and 50.1%±5.7 for Dll4. Data
8 represent mean ± S.E. of three independent experiments. (d) Effects of Dll4 sirna and Notch1 sirna on leader cell density. The experiments were performed in the presence or the absence of the Dll4 dslna probe. Data are expressed as mean ± s.e.m. (n=3, * P < 0.05, *** P < 0.001; unpaired Student s t-test)
9 Supplementary Figure 7. Cellular stress distribution near the boundary. (a-b) Representative bright-field image (a) and the corresponding traction force distribution (b) of a model wound. The model wound was created by scratching the left hand side of the monolayer. The traction force distribution was obtained using traction force microscopy plugins in ImageJ. The unit of the scale bar is Pascal. Scale bar, 50 µm. (c) Average normal cell traction force at different positions from the boundary. Data are
10 expressed as mean ± s.d. (n=3). (d) Stress distribution within the monolayer as a result of the cumulative cell traction force. Data are expressed as mean ± s.d. (n=3). (e) A tug-of-war model predicting stress accumulation near the leading edge. Supplementary Figure 8. Bright-field images of cells treated with cellular force modulating agents (Calycuing-A, Nocodazole, Y-27632, and blebbistatin) and Notch signaling modulating agents (DAPT and Jagged-1). Model wounds were created on the top of the monolayer. Images correspond to the fluorescence images in Fig. 4 of the main text. Scale bars, 100 μm.
11 Supplementary Figure 9. Notch1-Dll4 lateral inhibition for cell-fate determination. The cells may initially express the same levels of Notch1 and Dll4. With a small perturbation in the system, the cell with a stronger Dll4 level induces Notch1 activity in the neighboring cell. The neighboring cell increases the Notch activity and decreases the Dll4 level. The Dll4 expressing cell reinforces the expression of Dll4. This results in mutual inactivation of Notch receptor and ligand and cell-fate determination.
12 Supplementary Figure 10. Dynamics of Notch1-Dll4 lateral inhibition and the formation of checker box patterns. Numerical simulation of Notch1-Dll4 lateral inhibition predicted by equations S1-S3 was performed in a homogeneous cell population with small initial fluctuations. The Dll4 expression is shown in the top row and the corresponding Notch1 expression is shown in the bottom row. Parameters in the simulation are shown in Supplementary Table 3. The computational time scale and the gene expression values are in arbitrary units.
13 Supplementary Figure 11. Dynamics of Notch1-Dll4 lateral inhibition. Tracking of Notch1 and Dll4 expressions in representative cells. Cells 1 and 2 are adjacent to each other and cells 3 and 4 are adjacent to each other. Supplementary Figure 12. Numerical simulation of Notch1-Dll4 activity with position dependent expression of Notch1 and Dll4. L=16 and m=3. All other parameters are the same as Supplementary Table 2.
14 Supplementary Tables Supplementary Table 1. Sequences of dslna probes with alternating DNA/LNA monomers for single cell gene expression analysis. Bold italic letters represent LNA monomers. Probe Name Sequence & Label Length (base) β-actin Dll4 Notch1 HO-1 Random Donor (D) Quencher (Q) Donor (D) Quencher (Q) Donor (D) Quencher (Q) Donor (D) Quencher (Q) Donor (D) Quencher (Q) 5'-/FAM- 6/AGGAAGGAAGGCTGGAAGAG/ /CTTCCTTCCT/Iowa Black FQ/-3' 10 5'-/FAM- 6/AAGGGCAGTTGGAGAGGGTT/-3' /AACTGCCCTT/Iowa Black FQ/-3' 10 5'- /FAM6/TGCGGTCTGTCTGGTTGTGC/-3' ACAGACCGCA/Iowa Black FQ/-3' 10 5'-/FAM- 6/AAGACTGGGCTCTCCTTGTT/ /GCCCAGTCTT/Iowa Black FQ/-3' 10 5'-/FAM- 6/ACGCGACAAGCGCACCGATA/-3' /CTTGTCGCGT/Iowa Black FQ/-3' 10
15 Supplementary Table 2. Sequences of sirna in this study. sirna Name Sequence & Label Length (base) Notch1 Dll4 Sense 5 -GGGAC AUCAC GGAUC AUAUT T-3 21 Antisense 5 -AUAUG AUCCG UGAUG UCCCG G- 3 Sense 5 -GAGUC UAGUA UUUCA AUAAT T-3 21 Antisense 5 -UUAUU GAAAU ACUAG ACUCC A Supplementary Table 3. Parameters in the computational model of Notch1-Dll4 lateral inhibition. Parameters Symbols Values Notch1 expression rate R N 0.01 Dll4 expression rate R D 0.01 Hill coefficient for Notch1 H 3 Hill coefficient for Dll4 k 3 Dissociation constant for Notch1 a 0.01 Dissociation constant for Dll4 b 100 Notch receptor degradation rate µ Notch ligand degradation rate Ρ Cell contact coefficient α 0.16 Cis inhibition coefficient k c 10
16 Supplementary Note 1 Notch lateral inhibition. Notch is an evolutionarily conserved intercellular signaling pathway that regulates numerous cell-fate specification events 1, 2, 3. Notch receptors and ligands are typically transmembrane proteins and Notch signaling is activated via direct cell-cell contact. One of the key functions of Notch signaling is lateral inhibition, which controls binary cell-fate decision in physically adjacent cells. In Notch lateral inhibition, the expressions of Notch ligands (e.g., Dll4) and receptors (e.g., Notch1) may be similar among the cells initially. With a small difference in Dll4, the cell with a stronger level of Dll4 inhibits the expression of Dll4 in the neighboring cell via Notch1 signaling. Transcriptional feedback amplifies and consolidates the differences between Notch1 and Dll4 expressions, resulting in Notch ligand and receptor expressing cells (Supplementary Fig. 9). The cells may also be under the influence of intrinsic and extrinsic signals in the microenvironment. The mutual inactivation of Notch receptor and ligand can create spatial patterns of cells and controls the proper ratio of specific cell types to regulate the tissue architecture and morphology 4, 5, 6. Additional information on Notch signaling can be found from recent review articles. Computational model of Notch lateral inhibition. Several numerical models have been developed for studying Notch receptor and ligand lateral inhibition. These models have been applied to study numerous developmental events, such as bristle patterning, boundary formation, and angiogenesis sprouting, in both invertebrate and vertebrate species 4, 5, 6, 7, 8. We developed a computational model by modifying an established
17 model of Notch lateral inhibition 8 to study the formation of leader cell near the boundary. Equations 1-3 show the basic model, which considers the expression of Notch1, N, and Dll4, D. In equation S1, the first term shows the rate of change in Notch1 which is modeled by the Hill equation. This trans-inhibition term depends on the Dll4 level of the neighboring cells, D in. The second term of equation S1 describes the first order degradation of the Notch1. The third term shows the cis-inhibition representing the mutual inactivation of Notch receptor and ligand in the same cell. The cis-inhibition is shown to accelerate patterning dynamics and improve robustness to perturbation 4. The activity of the Dll4 is modeled similarly by equation S2 except that the expression rate depends on the level of Notch1 in the same cell, effectively implementing the mutual inactivation of Notch1 and Dll4. In equation S3, the immediate and second layers of cells are considered to be in contact to a cell to represent the dynamic filopodial activity of the cells 8. (S1) (S2) (S3) To study the behaviors of this system, an agent based approach was implemented 9. In particular, a rectangular array of cells was created to represent the cell monolayer. In each cell, the Notch1 and Dll4 levels, N(i,j,t) and D(i,,j,t), were described by equations S1-3, where i and j are position coordinates and t is the time step in the simulation. The equations were discretized in space and time using the finite difference approach and
18 implemented in MATLAB 10. In the simulation, 16 by 16 elements were considered. For initial conditions, low levels of Dll4 and Notch1 were assigned with small random fluctuations. The model parameters are described in Table N1. Following the original model 8, the expressions and time scale are expressed in arbitrary units. In the simulation, Notch1-Dll4 lateral inhibition was found to robustly drive the spatial pattern. The system parameters have little effects on the spatial patterns observed. A periodic boundary condition was applied to the monolayer. For wounding, the periodic boundary condition at top edge of the cells was removed to effectively create the boundary. Notch1-Dll4 lateral inhibition in homogeneous monolayer. The behaviors of the basic model was first studied (Supplementary Fig. 10 and Supplementary Movie 2). Upon the initiation of the simulation, some cells rapidly increased their Dll4 levels while reducing their Notch levels. These Dll4 expressing cells also inhibited the Dll4 levels of neighboring cells. Occasionally, cells with intermediate levels of Dll4 were adjacent to each other, resulting in transient competition of the Dll4 level. Examples of such transient Dll4 competition are shown in Supplementary Fig. 10 (T = 1250, light blue and orange cells). However, the transient competition was not stable. Only one cell won the competition and emerged to be the Dll4 expressing cell. The neighboring cells eventually reduced the Dll4 level and expressed a high level of Notch1. The inhibition of Dll4 activity in neighboring cells resulted in the checkbox pattern with approximately equal spacing between the Dll4 expressing cells. Examining the activities of individual cells further illustrates the dynamic behaviors of Notch1-Dll4 lateral inhibition. Supplementary Fig. 11 shows typical behaviors observed
19 in the numerical simulation. In particular, most cells rapidly emerged to either high levels of Dll4 or Notch1 (cells 1 and 2 in Supplementary Fig. 11). Cells with intermediate activities were observed to complete for the Dll4 activity (cells 3 and 4 in Supplementary Fig. 11). These behaviors are consistent with our experimental observations (see Fig. 2). Spatial distribution of Notch1-Dll4 activity near the boundary. The Notch1-Dll4 lateral inhibition model correctly predicted the regular spacing between the leader cells and the dynamic competition of Dll4 activities in adjacent cells. Nevertheless, the checker box pattern observed in the basic model with homogeneous cell distribution did not fully describe the experimental observations. As shown by the dslna probe and immunostaining, leader cells were only initiated near the leading edge and the Notch1 expression was upregulated in a region ~300 μm near the boundary. The creation of a boundary by removing the periodic boundary condition only induced a small number of leader cells at random positions near the boundary (data not shown). Therefore, additional regulatory signals are likely involved in controlling the formation of leader cells. Specifically, it is known that cells exhibit a gradient in response to the model wound and express various genes in a spatiotemporal manner 11. We therefore modify the basic model by incorporating position dependence in Notch1-Dll4 interaction near the boundary. Mathematically, the rate constants, R N and R D, were replaced by and to implement the position dependence. In the rate equations, x is the position of the cell from the leading edge and L is the characteristic length. The exponent, m, represents the slope of the gradient in response. Supplementary Fig. 12 shows the behavior of this spatial model with m = 3
20 and L = 16. In this model, the formation of Dll4 expressing leader cells was only observed near the leading edge (i.e., the top boundary). The model also correctly describes the spatial distribution of Notch1 near the boundary. To study the effects of traction force on Dll4 activity, m was chosen from 1, 3, and 5, which effectively modulates the force distribution. To study the effects of Notch signaling, the cell contact coefficient was adjusted between 0.04, 0.16, and Supplementary References 1. Fortini ME. Notch Signaling: The Core Pathway and Its Posttranslational Regulation. Dev Cell 16, (2009). 2. Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science 284, (1999). 3. Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol 7, (2006). 4. Sprinzak D, et al. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature 465, 86-U95 (2010). 5. Sprinzak D, Lakhanpal A, LeBon L, Garcia-Ojalvo J, Elowitz MB. Mutual Inactivation of Notch Receptors and Ligands Facilitates Developmental Patterning. Plos Comput Biol 7, (2011).
21 6. Shaya O, Sprinzak D. From Notch signaling to fine-grained patterning: Modeling meets experiments. Curr Opin Genet Dev 21, (2011). 7. Jakobsson L, et al. Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nat Cell Biol 12, (2010). 8. Cohen M, Georgiou M, Stevenson NL, Miodownik M, Baum B. Dynamic Filopodia Transmit Intermittent Delta-Notch Signaling to Drive Pattern Refinement during Lateral Inhibition. Dev Cell 19, (2010). 9. Long J, Junkin M, Wong PK, Hoying J, Deymier P. Calcium Wave Propagation in Networks of Endothelial Cells: Model-based Theoretical and Experimental Study. Plos Comput Biol 8, e (2012). 10. LeVeque RJ. Finite difference methods for ordinary and partial differential equations : steady-state and time-dependent problems. Society for Industrial and Applied Mathematics (2007). 11. Riahi R, et al. Single cell gene expression analysis in injury-induced collective cell migration. Integrative biology : quantitative biosciences from nano to macro 6, (2014).
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3443 In the format provided by the authors and unedited. Supplementary Figure 1 TC and SC behaviour during ISV sprouting. (a) Predicted outcome of TC division and competitive Dll4-Notch-mediated
DOI: 10.1038/ncb3443 In the format provided by the authors and unedited. Supplementary Figure 1 TC and SC behaviour during ISV sprouting. (a) Predicted outcome of TC division and competitive Dll4-Notch-mediated
Supplementary Data Dll4-containing exosomes induce capillary sprout retraction ina 3D microenvironment
 Supplementary Data Dll4-containing exosomes induce capillary sprout retraction ina 3D microenvironment Soheila Sharghi-Namini 1, Evan Tan 1,2, Lee-Ling Sharon Ong 1, Ruowen Ge 2 * and H. Harry Asada 1,3
Supplementary Data Dll4-containing exosomes induce capillary sprout retraction ina 3D microenvironment Soheila Sharghi-Namini 1, Evan Tan 1,2, Lee-Ling Sharon Ong 1, Ruowen Ge 2 * and H. Harry Asada 1,3
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Díaz et al., http://www.jcb.org/cgi/content/full/jcb.201209151/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Hypoxia induces invadopodia formation in different epithelial
Supplemental material Díaz et al., http://www.jcb.org/cgi/content/full/jcb.201209151/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Hypoxia induces invadopodia formation in different epithelial
s u p p l e m e n ta ry i n f o r m at i o n
 Figure S1 Characterization of tet-off inducible cell lines expressing GFPprogerin and GFP-wt lamin A. a, Western blot analysis of GFP-progerin- or GFP-wt lamin A- expressing cells before induction (0d)
Figure S1 Characterization of tet-off inducible cell lines expressing GFPprogerin and GFP-wt lamin A. a, Western blot analysis of GFP-progerin- or GFP-wt lamin A- expressing cells before induction (0d)
Tanimoto et al., http ://www.jcb.org /cgi /content /full /jcb /DC1
 Supplemental material JCB Tanimoto et al., http ://www.jcb.org /cgi /content /full /jcb.201510064 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Method for aster 3D tracking, extended characterization of
Supplemental material JCB Tanimoto et al., http ://www.jcb.org /cgi /content /full /jcb.201510064 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Method for aster 3D tracking, extended characterization of
Supplementary Figure 1. Rab27a-KD inhibits speed and persistence of HEp3 cells migrating in the chick CAM. (a) Western blot analysis of Rab27a
 Supplementary Figure 1. Rab27a-KD inhibits speed and persistence of HEp3 cells migrating in the chick CAM. (a) Western blot analysis of Rab27a expression in GFP-expressing HEp3 cells. (b) Representative
Supplementary Figure 1. Rab27a-KD inhibits speed and persistence of HEp3 cells migrating in the chick CAM. (a) Western blot analysis of Rab27a expression in GFP-expressing HEp3 cells. (b) Representative
Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at
 Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at E10.5 were double-stained for TUNEL (red) and PECAM-1 (green).
Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at E10.5 were double-stained for TUNEL (red) and PECAM-1 (green).
Boucher et al NCOMMS B
 1 Supplementary Figure 1 (linked to Figure 1). mvegfr1 constitutively internalizes in endothelial cells. (a) Immunoblot of mflt1 from undifferentiated mouse embryonic stem (ES) cells with indicated genotypes;
1 Supplementary Figure 1 (linked to Figure 1). mvegfr1 constitutively internalizes in endothelial cells. (a) Immunoblot of mflt1 from undifferentiated mouse embryonic stem (ES) cells with indicated genotypes;
Interleukin-6 promotes pancreatic cancer cell migration by rapidly activating the small GTPase CDC42
 Interleukin-6 promotes pancreatic cancer cell migration by rapidly activating the small GTPase CDC42 Gina L. Razidlo, Kevin M. Burton, and Mark A. McNiven SUPPORTING INFORMATION Figure S1. IL-6 promotes
Interleukin-6 promotes pancreatic cancer cell migration by rapidly activating the small GTPase CDC42 Gina L. Razidlo, Kevin M. Burton, and Mark A. McNiven SUPPORTING INFORMATION Figure S1. IL-6 promotes
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
supplementary information
 DOI: 10.1038/ncb1875 Figure S1 (a) The 79 surgical specimens from NSCLC patients were analysed by immunohistochemistry with an anti-p53 antibody and control serum (data not shown). The normal bronchi served
DOI: 10.1038/ncb1875 Figure S1 (a) The 79 surgical specimens from NSCLC patients were analysed by immunohistochemistry with an anti-p53 antibody and control serum (data not shown). The normal bronchi served
SUPPLEMENTARY LEGENDS...
 TABLE OF CONTENTS SUPPLEMENTARY LEGENDS... 2 11 MOVIE S1... 2 FIGURE S1 LEGEND... 3 FIGURE S2 LEGEND... 4 FIGURE S3 LEGEND... 5 FIGURE S4 LEGEND... 6 FIGURE S5 LEGEND... 7 FIGURE S6 LEGEND... 8 FIGURE
TABLE OF CONTENTS SUPPLEMENTARY LEGENDS... 2 11 MOVIE S1... 2 FIGURE S1 LEGEND... 3 FIGURE S2 LEGEND... 4 FIGURE S3 LEGEND... 5 FIGURE S4 LEGEND... 6 FIGURE S5 LEGEND... 7 FIGURE S6 LEGEND... 8 FIGURE
Chapter 2. Investigation into mir-346 Regulation of the nachr α5 Subunit
 15 Chapter 2 Investigation into mir-346 Regulation of the nachr α5 Subunit MicroRNA s (mirnas) are small (< 25 base pairs), single stranded, non-coding RNAs that regulate gene expression at the post transcriptional
15 Chapter 2 Investigation into mir-346 Regulation of the nachr α5 Subunit MicroRNA s (mirnas) are small (< 25 base pairs), single stranded, non-coding RNAs that regulate gene expression at the post transcriptional
Supplementary Figure 1: Uncropped western blots for Figure 1B. Uncropped blots shown in Figure 1B, showing that NOTCH intracellular domain (NICD) is
 Supplementary Figure 1: Uncropped western blots for Figure 1B. Uncropped blots shown in Figure 1B, showing that NOTCH intracellular domain (NICD) is increased with exposure of HUVEC to 1h FSS, and that
Supplementary Figure 1: Uncropped western blots for Figure 1B. Uncropped blots shown in Figure 1B, showing that NOTCH intracellular domain (NICD) is increased with exposure of HUVEC to 1h FSS, and that
Supplemental Table S1
 Supplemental Table S. Tumorigenicity and metastatic potential of 44SQ cell subpopulations a Tumorigenicity b Average tumor volume (mm ) c Lung metastasis d CD high /4 8. 8/ CD low /4 6./ a Mice were injected
Supplemental Table S. Tumorigenicity and metastatic potential of 44SQ cell subpopulations a Tumorigenicity b Average tumor volume (mm ) c Lung metastasis d CD high /4 8. 8/ CD low /4 6./ a Mice were injected
Supplementary Figures for
 mirns regulate s Supplementary igures for MicroRNs Reprogram Normal ibroblasts into Cancer ssociated ibroblasts in Ovarian Cancer nirban K. Mitra, Marion Zillhardt, Youjia Hua, Payal iwari, ndrea E. Murmann,
mirns regulate s Supplementary igures for MicroRNs Reprogram Normal ibroblasts into Cancer ssociated ibroblasts in Ovarian Cancer nirban K. Mitra, Marion Zillhardt, Youjia Hua, Payal iwari, ndrea E. Murmann,
Fig. S1. Upregulation of K18 and K14 mrna levels during ectoderm specification of hescs. Quantitative real-time PCR analysis of mrna levels of OCT4
 Fig. S1. Upregulation of K18 and K14 mrna levels during ectoderm specification of hescs. Quantitative real-time PCR analysis of mrna levels of OCT4 (n=3 independent differentiation experiments for each
Fig. S1. Upregulation of K18 and K14 mrna levels during ectoderm specification of hescs. Quantitative real-time PCR analysis of mrna levels of OCT4 (n=3 independent differentiation experiments for each
Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis
 Nature Methods Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis Prabuddha Sengupta, Tijana Jovanovic-Talisman, Dunja Skoko 1, Malte Renz, Sarah L Veatch & Jennifer
Nature Methods Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis Prabuddha Sengupta, Tijana Jovanovic-Talisman, Dunja Skoko 1, Malte Renz, Sarah L Veatch & Jennifer
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1. Formation of the AA5x. a, Camera lucida drawing of embryo at 48 hours post fertilization (hpf, modified from Kimmel et al. Dev Dyn. 1995 203:253-310). b, Confocal microangiogram
Supplementary Figure 1. Formation of the AA5x. a, Camera lucida drawing of embryo at 48 hours post fertilization (hpf, modified from Kimmel et al. Dev Dyn. 1995 203:253-310). b, Confocal microangiogram
Figure S1: Effects on haptotaxis are independent of effects on cell velocity A)
 Supplemental Figures Figure S1: Effects on haptotaxis are independent of effects on cell velocity A) Velocity of MV D7 fibroblasts expressing different GFP-tagged Ena/VASP family proteins in the haptotaxis
Supplemental Figures Figure S1: Effects on haptotaxis are independent of effects on cell velocity A) Velocity of MV D7 fibroblasts expressing different GFP-tagged Ena/VASP family proteins in the haptotaxis
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation
 Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Nature Neuroscience: doi: /nn Supplementary Figure 1. Large-scale calcium imaging in vivo.
 Supplementary Figure 1 Large-scale calcium imaging in vivo. (a) Schematic illustration of the in vivo camera imaging set-up for large-scale calcium imaging. (b) High-magnification two-photon image from
Supplementary Figure 1 Large-scale calcium imaging in vivo. (a) Schematic illustration of the in vivo camera imaging set-up for large-scale calcium imaging. (b) High-magnification two-photon image from
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary information. The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic. Spindle Orientation
 Supplementary information The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic Spindle Orientation Running title: Dynein LICs distribute mitotic functions. Sagar Mahale a, d, *, Megha
Supplementary information The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic Spindle Orientation Running title: Dynein LICs distribute mitotic functions. Sagar Mahale a, d, *, Megha
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Edens and Levy, http://www.jcb.org/cgi/content/full/jcb.201406004/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nuclear shrinking does not depend on the cytoskeleton
Supplemental material Edens and Levy, http://www.jcb.org/cgi/content/full/jcb.201406004/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Nuclear shrinking does not depend on the cytoskeleton
SUPPLEMENTARY INFORMATION
 In the format provided by the authors and unedited. 2 3 4 DOI: 10.1038/NMAT4893 EGFR and HER2 activate rigidity sensing only on rigid matrices Mayur Saxena 1,*, Shuaimin Liu 2,*, Bo Yang 3, Cynthia Hajal
In the format provided by the authors and unedited. 2 3 4 DOI: 10.1038/NMAT4893 EGFR and HER2 activate rigidity sensing only on rigid matrices Mayur Saxena 1,*, Shuaimin Liu 2,*, Bo Yang 3, Cynthia Hajal
MII. Supplement Figure 1. CapZ β2. Merge. 250ng. 500ng DIC. Merge. Journal of Cell Science Supplementary Material. GFP-CapZ β2 DNA
 A GV GVBD MI DNA CapZ β2 CapZ β2 Merge B DIC GFP-CapZ β2 Merge CapZ β2-gfp 250ng 500ng Supplement Figure 1. MII A early MI late MI Control RNAi CapZαβ DNA Actin Tubulin B Phalloidin Intensity(A.U.) n=10
A GV GVBD MI DNA CapZ β2 CapZ β2 Merge B DIC GFP-CapZ β2 Merge CapZ β2-gfp 250ng 500ng Supplement Figure 1. MII A early MI late MI Control RNAi CapZαβ DNA Actin Tubulin B Phalloidin Intensity(A.U.) n=10
CD3 coated cover slips indicating stimulatory contact site, F-actin polymerization and
 SUPPLEMENTAL FIGURES FIGURE S1. Detection of MCs. A, Schematic representation of T cells stimulated on anti- CD3 coated cover slips indicating stimulatory contact site, F-actin polymerization and microclusters.
SUPPLEMENTAL FIGURES FIGURE S1. Detection of MCs. A, Schematic representation of T cells stimulated on anti- CD3 coated cover slips indicating stimulatory contact site, F-actin polymerization and microclusters.
Simulation of Chemotractant Gradients in Microfluidic Channels to Study Cell Migration Mechanism in silico
 Simulation of Chemotractant Gradients in Microfluidic Channels to Study Cell Migration Mechanism in silico P. Wallin 1*, E. Bernson 1, and J. Gold 1 1 Chalmers University of Technology, Applied Physics,
Simulation of Chemotractant Gradients in Microfluidic Channels to Study Cell Migration Mechanism in silico P. Wallin 1*, E. Bernson 1, and J. Gold 1 1 Chalmers University of Technology, Applied Physics,
Supplementary Figure 1. Deletion of Smad3 prevents B16F10 melanoma invasion and metastasis in a mouse s.c. tumor model.
 A B16F1 s.c. Lung LN Distant lymph nodes Colon B B16F1 s.c. Supplementary Figure 1. Deletion of Smad3 prevents B16F1 melanoma invasion and metastasis in a mouse s.c. tumor model. Highly invasive growth
A B16F1 s.c. Lung LN Distant lymph nodes Colon B B16F1 s.c. Supplementary Figure 1. Deletion of Smad3 prevents B16F1 melanoma invasion and metastasis in a mouse s.c. tumor model. Highly invasive growth
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1 SNARE Probes for FRET/2pFLIM Analysis Used in the Present Study. mturquoise (mtq) and Venus (Ven) are in blue and yellow, respectively. The soluble N-ethylmaleimide-sensitive
Supplementary Figure 1 Supplementary Figure 1 SNARE Probes for FRET/2pFLIM Analysis Used in the Present Study. mturquoise (mtq) and Venus (Ven) are in blue and yellow, respectively. The soluble N-ethylmaleimide-sensitive
microrna-200b and microrna-200c promote colorectal cancer cell proliferation via
 Supplementary Materials microrna-200b and microrna-200c promote colorectal cancer cell proliferation via targeting the reversion-inducing cysteine-rich protein with Kazal motifs Supplementary Table 1.
Supplementary Materials microrna-200b and microrna-200c promote colorectal cancer cell proliferation via targeting the reversion-inducing cysteine-rich protein with Kazal motifs Supplementary Table 1.
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous
 Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Supporting Information
 Supporting Information Palmisano et al. 10.1073/pnas.1202174109 Fig. S1. Expression of different transgenes, driven by either viral or human promoters, is up-regulated by amino acid starvation. (A) Quantification
Supporting Information Palmisano et al. 10.1073/pnas.1202174109 Fig. S1. Expression of different transgenes, driven by either viral or human promoters, is up-regulated by amino acid starvation. (A) Quantification
Identification of influential proteins in the classical retinoic acid signaling pathway
 Ghaffari and Petzold Theoretical Biology and Medical Modelling (2018) 15:16 https://doi.org/10.1186/s12976-018-0088-7 RESEARCH Open Access Identification of influential proteins in the classical retinoic
Ghaffari and Petzold Theoretical Biology and Medical Modelling (2018) 15:16 https://doi.org/10.1186/s12976-018-0088-7 RESEARCH Open Access Identification of influential proteins in the classical retinoic
Supplementary Figure 1. Mother centrioles can reduplicate while in the close association
 C1-GFP distance (nm) C1-GFP distance (nm) a arrested HeLa cell expressing C1-GFP and Plk1TD-RFP -3 s 1 2 3 4 5 6 7 8 9 11 12 13 14 16 17 18 19 2 21 22 23 24 26 27 28 29 3 b 9 8 7 6 5 4 3 2 arrested HeLa
C1-GFP distance (nm) C1-GFP distance (nm) a arrested HeLa cell expressing C1-GFP and Plk1TD-RFP -3 s 1 2 3 4 5 6 7 8 9 11 12 13 14 16 17 18 19 2 21 22 23 24 26 27 28 29 3 b 9 8 7 6 5 4 3 2 arrested HeLa
Journal Club. 03/04/2012 Lama Nazzal
 Journal Club 03/04/2012 Lama Nazzal NOTCH and the kidneys Is an evolutionarily conserved cell cell communication mechanism. Is a regulator of cell specification, differentiation, and tissue patterning.
Journal Club 03/04/2012 Lama Nazzal NOTCH and the kidneys Is an evolutionarily conserved cell cell communication mechanism. Is a regulator of cell specification, differentiation, and tissue patterning.
Supplementary Figure 1. Nature Neuroscience: doi: /nn.4547
 Supplementary Figure 1 Characterization of the Microfetti mouse model. (a) Gating strategy for 8-color flow analysis of peripheral Ly-6C + monocytes from Microfetti mice 5-7 days after TAM treatment. Living
Supplementary Figure 1 Characterization of the Microfetti mouse model. (a) Gating strategy for 8-color flow analysis of peripheral Ly-6C + monocytes from Microfetti mice 5-7 days after TAM treatment. Living
1. Microfluidic device characteristics and cell compartmentalization
 Supplementary figures: 1. Microfluidic device characteristics and cell compartmentalization Microfluidic channels were molded in PDMS (Fig. S1 A and B) over silicon masters, which were structured with
Supplementary figures: 1. Microfluidic device characteristics and cell compartmentalization Microfluidic channels were molded in PDMS (Fig. S1 A and B) over silicon masters, which were structured with
Supplementary Figure 1. Expression of the inducible tper2 is proportional to Dox/Tet concentration in Rosa-DTG/Per2 Per2-luc/wt MEFs.
 Supplementary Figure 1. Expression of the inducible tper2 is proportional to Dox/Tet concentration in Rosa-DTG/Per2 Per2-luc/wt MEFs. (a) Dose-responsive expression of tper2 by Dox. Note that there are
Supplementary Figure 1. Expression of the inducible tper2 is proportional to Dox/Tet concentration in Rosa-DTG/Per2 Per2-luc/wt MEFs. (a) Dose-responsive expression of tper2 by Dox. Note that there are
d e f Spatiotemporal quantification of subcellular ATP levels in a single HeLa cell during changes in morphology Supplementary Information
 Ca 2+ level (a. u.) Area (a. u.) Normalized distance Normalized distance Center Edge Center Edge Relative ATP level Relative ATP level Supplementary Information Spatiotemporal quantification of subcellular
Ca 2+ level (a. u.) Area (a. u.) Normalized distance Normalized distance Center Edge Center Edge Relative ATP level Relative ATP level Supplementary Information Spatiotemporal quantification of subcellular
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells
 Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells Immunofluorescence staining of H3K9me3 and 53BP1 in PH and HGADFN003 (HG003) cells at 24 h after γ-irradiation. Scale bar,
Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells Immunofluorescence staining of H3K9me3 and 53BP1 in PH and HGADFN003 (HG003) cells at 24 h after γ-irradiation. Scale bar,
Supplemental Figures:
 Supplemental Figures: Figure 1: Intracellular distribution of VWF by electron microscopy in human endothelial cells. a) Immunogold labeling of LC3 demonstrating an LC3-positive autophagosome (white arrow)
Supplemental Figures: Figure 1: Intracellular distribution of VWF by electron microscopy in human endothelial cells. a) Immunogold labeling of LC3 demonstrating an LC3-positive autophagosome (white arrow)
Nature Methods: doi: /nmeth.4257
 Supplementary Figure 1 Screen for polypeptides that affect cellular actin filaments. (a) Table summarizing results from all polypeptides tested. Source shows organism, gene, and amino acid numbers used.
Supplementary Figure 1 Screen for polypeptides that affect cellular actin filaments. (a) Table summarizing results from all polypeptides tested. Source shows organism, gene, and amino acid numbers used.
How Viruses Spread Among Computers and People
 Institution: COLUMBIA UNIVERSITY Sign In as Individual FAQ Access Rights Join AAAS Summary of this Article debates: Submit a response to this article Download to Citation Manager Alert me when: new articles
Institution: COLUMBIA UNIVERSITY Sign In as Individual FAQ Access Rights Join AAAS Summary of this Article debates: Submit a response to this article Download to Citation Manager Alert me when: new articles
Supplemental information contains 7 movies and 4 supplemental Figures
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplemental information contains 7 movies and 4 supplemental Figures Movies: Movie 1. Single virus tracking of A4-mCherry-WR MV
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplemental information contains 7 movies and 4 supplemental Figures Movies: Movie 1. Single virus tracking of A4-mCherry-WR MV
Bystander cells enhance NK cytotoxic efficiency by reducing search time
 Bystander cells enhance NK cytotoxic efficiency by reducing search time Xiao Zhou 1, Renping Zhao 1, Karsten Schwarz 2, Matthieu Mangeat 2,4, Eva C. Schwarz 1, Mohamed Hamed 3,5, Ivan Bogeski 1, Volkhard
Bystander cells enhance NK cytotoxic efficiency by reducing search time Xiao Zhou 1, Renping Zhao 1, Karsten Schwarz 2, Matthieu Mangeat 2,4, Eva C. Schwarz 1, Mohamed Hamed 3,5, Ivan Bogeski 1, Volkhard
Cell Migration and Invasion Assays INCUCYTE LIVE-CELL ANALYSIS SYSTEM. Real-time automated measurements of cell motility inside your incubator
 Cell Migration and Invasion Assays INCUCYTE LIVE-CELL ANALYSIS SYSTEM Real-time automated measurements of cell motility inside your incubator See the whole story Real-time cell motility visualization and
Cell Migration and Invasion Assays INCUCYTE LIVE-CELL ANALYSIS SYSTEM Real-time automated measurements of cell motility inside your incubator See the whole story Real-time cell motility visualization and
TITLE: Notch in Pathological Angiogenesis and Lymphangiogenesis
 Award Number: W81XWH-10-1-0304 TITLE: Notch in Pathological Angiogenesis and Lymphangiogenesis PRINCIPAL INVESTIGATOR: Minji Kim CONTRACTING ORGANIZATION: Columbia University New York, NY 10032 REPORT
Award Number: W81XWH-10-1-0304 TITLE: Notch in Pathological Angiogenesis and Lymphangiogenesis PRINCIPAL INVESTIGATOR: Minji Kim CONTRACTING ORGANIZATION: Columbia University New York, NY 10032 REPORT
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Posch et al., http://www.jcb.org/cgi/content/full/jcb.200912046/dc1 Figure S1. Biochemical characterization of the interaction between
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Posch et al., http://www.jcb.org/cgi/content/full/jcb.200912046/dc1 Figure S1. Biochemical characterization of the interaction between
Supplementary Materials for
 www.advances.sciencemag.org/cgi/content/full/1/3/e1400244/dc1 Supplementary Materials for PlGF-induced VEGFR1-dependent vascular remodeling determines opposing antitumor effects and drug resistance to
www.advances.sciencemag.org/cgi/content/full/1/3/e1400244/dc1 Supplementary Materials for PlGF-induced VEGFR1-dependent vascular remodeling determines opposing antitumor effects and drug resistance to
Supplementary Materials for
 www.sciencetranslationalmedicine.org/cgi/content/full/4/117/117ra8/dc1 Supplementary Materials for Notch4 Normalization Reduces Blood Vessel Size in Arteriovenous Malformations Patrick A. Murphy, Tyson
www.sciencetranslationalmedicine.org/cgi/content/full/4/117/117ra8/dc1 Supplementary Materials for Notch4 Normalization Reduces Blood Vessel Size in Arteriovenous Malformations Patrick A. Murphy, Tyson
El Azzouzi et al., http ://www.jcb.org /cgi /content /full /jcb /DC1
 Supplemental material JCB El Azzouzi et al., http ://www.jcb.org /cgi /content /full /jcb.201510043 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Acquisition of -phluorin correlates negatively with podosome
Supplemental material JCB El Azzouzi et al., http ://www.jcb.org /cgi /content /full /jcb.201510043 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Acquisition of -phluorin correlates negatively with podosome
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
Supplementary Material
 Supplementary Material accompanying the manuscript Interleukin 37 is a fundamental inhibitor of innate immunity Marcel F Nold, Claudia A Nold-Petry, Jarod A Zepp, Brent E Palmer, Philip Bufler & Charles
Supplementary Material accompanying the manuscript Interleukin 37 is a fundamental inhibitor of innate immunity Marcel F Nold, Claudia A Nold-Petry, Jarod A Zepp, Brent E Palmer, Philip Bufler & Charles
Culturing embryonic tissues in the computer
 Culturing embryonic tissues in the computer Blood vessel development Roeland Merks Biomodeling & Biosystems Analysis CWI, Life Sciences and Netherlands Institute for Systems Biology Biological development
Culturing embryonic tissues in the computer Blood vessel development Roeland Merks Biomodeling & Biosystems Analysis CWI, Life Sciences and Netherlands Institute for Systems Biology Biological development
Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion
 Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
nature methods Organelle-specific, rapid induction of molecular activities and membrane tethering
 nature methods Organelle-specific, rapid induction of molecular activities and membrane tethering Toru Komatsu, Igor Kukelyansky, J Michael McCaffery, Tasuku Ueno, Lidenys C Varela & Takanari Inoue Supplementary
nature methods Organelle-specific, rapid induction of molecular activities and membrane tethering Toru Komatsu, Igor Kukelyansky, J Michael McCaffery, Tasuku Ueno, Lidenys C Varela & Takanari Inoue Supplementary
supplementary information
 DOI: 10.1038/ncb2133 Figure S1 Actomyosin organisation in human squamous cell carcinoma. (a) Three examples of actomyosin organisation around the edges of squamous cell carcinoma biopsies are shown. Myosin
DOI: 10.1038/ncb2133 Figure S1 Actomyosin organisation in human squamous cell carcinoma. (a) Three examples of actomyosin organisation around the edges of squamous cell carcinoma biopsies are shown. Myosin
Nature Neuroscience doi: /nn Supplementary Figure 1. Characterization of viral injections.
 Supplementary Figure 1 Characterization of viral injections. (a) Dorsal view of a mouse brain (dashed white outline) after receiving a large, unilateral thalamic injection (~100 nl); demonstrating that
Supplementary Figure 1 Characterization of viral injections. (a) Dorsal view of a mouse brain (dashed white outline) after receiving a large, unilateral thalamic injection (~100 nl); demonstrating that
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Subcellular segregation of VGluT2-IR and TH-IR within the same VGluT2-TH axon (wild type rats). (a-e) Serial sections of a dual VGluT2-TH labeled axon. This axon (blue outline) has
Supplementary Figure 1 Subcellular segregation of VGluT2-IR and TH-IR within the same VGluT2-TH axon (wild type rats). (a-e) Serial sections of a dual VGluT2-TH labeled axon. This axon (blue outline) has
SUPPLEMENTARY INFORMATION
 Figure S1. Loss of Ena/VASP proteins inhibits filopodia and neuritogenesis. (a) Bar graph of filopodia number per stage 1 control and mmvvee (Mena/ VASP/EVL-null) neurons at 40hrs in culture. Loss of all
Figure S1. Loss of Ena/VASP proteins inhibits filopodia and neuritogenesis. (a) Bar graph of filopodia number per stage 1 control and mmvvee (Mena/ VASP/EVL-null) neurons at 40hrs in culture. Loss of all
Rescue of mutant rhodopsin traffic by metformin-induced AMPK activation accelerates photoreceptor degeneration Athanasiou et al
 Supplementary Material Rescue of mutant rhodopsin traffic by metformin-induced AMPK activation accelerates photoreceptor degeneration Athanasiou et al Supplementary Figure 1. AICAR improves P23H rod opsin
Supplementary Material Rescue of mutant rhodopsin traffic by metformin-induced AMPK activation accelerates photoreceptor degeneration Athanasiou et al Supplementary Figure 1. AICAR improves P23H rod opsin
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 DOT1L regulates the expression of epithelial and mesenchymal markers. (a) The expression levels and cellular localizations of EMT markers were confirmed by
Supplementary Figures Supplementary Figure 1 DOT1L regulates the expression of epithelial and mesenchymal markers. (a) The expression levels and cellular localizations of EMT markers were confirmed by
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
File Name: Supplementary Information Description: Supplementary Figures and Supplementary Table. File Name: Peer Review File Description:
 File Name: Supplementary Information Description: Supplementary Figures and Supplementary Table File Name: Peer Review File Description: Supplementary Table 1 Primers and taqman probes used were the following:
File Name: Supplementary Information Description: Supplementary Figures and Supplementary Table File Name: Peer Review File Description: Supplementary Table 1 Primers and taqman probes used were the following:
Cell-based modeling of angiogenic blood vessel sprouting
 Cell-based modeling of angiogenic blood vessel sprouting Roeland Merks Biomodeling & Biosystems Analysis Centrum Wiskunde & Informatica - Life Sciences Netherlands Institute for Systems Biology Netherlands
Cell-based modeling of angiogenic blood vessel sprouting Roeland Merks Biomodeling & Biosystems Analysis Centrum Wiskunde & Informatica - Life Sciences Netherlands Institute for Systems Biology Netherlands
Astrocyte signaling controls spike timing-dependent depression at neocortical synapses
 Supplementary Information Astrocyte signaling controls spike timing-dependent depression at neocortical synapses Rogier Min and Thomas Nevian Department of Physiology, University of Berne, Bern, Switzerland
Supplementary Information Astrocyte signaling controls spike timing-dependent depression at neocortical synapses Rogier Min and Thomas Nevian Department of Physiology, University of Berne, Bern, Switzerland
Supplementary Figure 1. Procedures for p38 activity imaging in living cells. (a) Schematic model of the p38 activity reporter. The reporter consists
 Supplementary Figure 1. Procedures for p38 activity imaging in living cells. (a) Schematic model of the p38 activity reporter. The reporter consists of: (i) the YPet domain (an enhanced YFP); (ii) the
Supplementary Figure 1. Procedures for p38 activity imaging in living cells. (a) Schematic model of the p38 activity reporter. The reporter consists of: (i) the YPet domain (an enhanced YFP); (ii) the
Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the
 Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the location of the transmembrane (TM), FRM binding (FB)
Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the location of the transmembrane (TM), FRM binding (FB)
Supplementary Materials
 Supplementary Materials Supplementary Figure S1 Regulation of Ubl4A stability by its assembly partner A, The translation rate of Ubl4A is not affected in the absence of Bag6. Control, Bag6 and Ubl4A CRISPR
Supplementary Materials Supplementary Figure S1 Regulation of Ubl4A stability by its assembly partner A, The translation rate of Ubl4A is not affected in the absence of Bag6. Control, Bag6 and Ubl4A CRISPR
Supplementary Figure 1. Characterization of human carotid plaques. (a) Flash-frozen human plaques were separated into vulnerable (V) and stable (S),
 Supplementary Figure 1. Characterization of human carotid plaques. (a) Flash-frozen human plaques were separated into vulnerable (V) and stable (S), regions which were then quantified for mean fluorescence
Supplementary Figure 1. Characterization of human carotid plaques. (a) Flash-frozen human plaques were separated into vulnerable (V) and stable (S), regions which were then quantified for mean fluorescence
Phosphoinositides Regulate Ciliary Protein Trafficking to Modulate Hedgehog Signaling
 Developmental Cell Supplemental Information Phosphoinositides Regulate Ciliary Protein Trafficking to Modulate Hedgehog Signaling Francesc R. Garcia-Gonzalo, Siew Cheng Phua, Elle C. Roberson, Galo Garcia
Developmental Cell Supplemental Information Phosphoinositides Regulate Ciliary Protein Trafficking to Modulate Hedgehog Signaling Francesc R. Garcia-Gonzalo, Siew Cheng Phua, Elle C. Roberson, Galo Garcia
HIF-inducible mir-191 promotes migration in breast cancer through complex regulation of TGFβ-signaling in hypoxic microenvironment.
 HIF-inducible mir-9 promotes migration in breast cancer through complex regulation of TGFβ-signaling in hypoxic microenvironment. Neha Nagpal, Hafiz M Ahmad, Shibu Chameettachal3, Durai Sundar, Sourabh
HIF-inducible mir-9 promotes migration in breast cancer through complex regulation of TGFβ-signaling in hypoxic microenvironment. Neha Nagpal, Hafiz M Ahmad, Shibu Chameettachal3, Durai Sundar, Sourabh
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/305/ra106/dc1 Supplementary Materials for Controlling Long-Term Signaling: Receptor Dynamics Determine Attenuation and Refractory Behavior of the TGF-β Pathway
www.sciencesignaling.org/cgi/content/full/6/305/ra106/dc1 Supplementary Materials for Controlling Long-Term Signaling: Receptor Dynamics Determine Attenuation and Refractory Behavior of the TGF-β Pathway
Influenza virus exploits tunneling nanotubes for cell-to-cell spread
 Supplementary Information Influenza virus exploits tunneling nanotubes for cell-to-cell spread Amrita Kumar 1, Jin Hyang Kim 1, Priya Ranjan 1, Maureen G. Metcalfe 2, Weiping Cao 1, Margarita Mishina 1,
Supplementary Information Influenza virus exploits tunneling nanotubes for cell-to-cell spread Amrita Kumar 1, Jin Hyang Kim 1, Priya Ranjan 1, Maureen G. Metcalfe 2, Weiping Cao 1, Margarita Mishina 1,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Analysis of hair bundle morphology in Ush1c c.216g>a mice at P18 by SEM.
 Supplementary Figure 1 Analysis of hair bundle morphology in Ush1c c.216g>a mice at P18 by SEM. (a-c) Heterozygous c.216ga mice displayed normal hair bundle morphology at P18. (d-i) Disorganized hair bundles
Supplementary Figure 1 Analysis of hair bundle morphology in Ush1c c.216g>a mice at P18 by SEM. (a-c) Heterozygous c.216ga mice displayed normal hair bundle morphology at P18. (d-i) Disorganized hair bundles
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
Supplemental Data Figure S1 Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells.
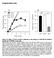 Supplemental Data Figure S1. Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells. (A) Specific lysis of IFN-γ-treated SKBR-3 cells in the absence
Supplemental Data Figure S1. Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells. (A) Specific lysis of IFN-γ-treated SKBR-3 cells in the absence
F-actin VWF Vinculin. F-actin. Vinculin VWF
 a F-actin VWF Vinculin b F-actin VWF Vinculin Supplementary Fig. 1. WPBs in HUVECs are located along stress fibers and at focal adhesions. (a) Immunofluorescence images of f-actin (cyan), VWF (yellow),
a F-actin VWF Vinculin b F-actin VWF Vinculin Supplementary Fig. 1. WPBs in HUVECs are located along stress fibers and at focal adhesions. (a) Immunofluorescence images of f-actin (cyan), VWF (yellow),
Supplementary. limb. bars
 Figure 1. CD163 -/- mice exhibit a similar phenotype ass WT mice in the absence of ischemic injury. a, Laser Doppler analysiss with perfusion quantitation at baseline (n= =10 per group). b, Immunostaining
Figure 1. CD163 -/- mice exhibit a similar phenotype ass WT mice in the absence of ischemic injury. a, Laser Doppler analysiss with perfusion quantitation at baseline (n= =10 per group). b, Immunostaining
SUPPLEMENTARY INFORMATION
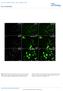 DOI: 10.1038/ncb2610 Figure S1 FSMCs derived from MSLN CLN transgenic mice express smooth muscle-specific proteins. Beta-galactosidase is ubiquitously expressed within cultured FSMCs derived from MSLN
DOI: 10.1038/ncb2610 Figure S1 FSMCs derived from MSLN CLN transgenic mice express smooth muscle-specific proteins. Beta-galactosidase is ubiquitously expressed within cultured FSMCs derived from MSLN
Supplementary Figure 1 Information on transgenic mouse models and their recording and optogenetic equipment. (a) 108 (b-c) (d) (e) (f) (g)
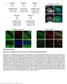 Supplementary Figure 1 Information on transgenic mouse models and their recording and optogenetic equipment. (a) In four mice, cre-dependent expression of the hyperpolarizing opsin Arch in pyramidal cells
Supplementary Figure 1 Information on transgenic mouse models and their recording and optogenetic equipment. (a) In four mice, cre-dependent expression of the hyperpolarizing opsin Arch in pyramidal cells
Supplementary figure 1: LII/III GIN-cells show morphological characteristics of MC
 1 2 1 3 Supplementary figure 1: LII/III GIN-cells show morphological characteristics of MC 4 5 6 7 (a) Reconstructions of LII/III GIN-cells with somato-dendritic compartments in orange and axonal arborizations
1 2 1 3 Supplementary figure 1: LII/III GIN-cells show morphological characteristics of MC 4 5 6 7 (a) Reconstructions of LII/III GIN-cells with somato-dendritic compartments in orange and axonal arborizations
Supplementary Figure 1
 Supplementary Figure 1 14 12 SEM4C PLXN2 8 SEM4C C 3 Cancer Cell Non Cancer Cell Expression 1 8 6 6 4 log2 ratio Expression 2 1 4 2 2 p value.1 D Supplementary Figure 1. Expression of Sema4C and Plexin2
Supplementary Figure 1 14 12 SEM4C PLXN2 8 SEM4C C 3 Cancer Cell Non Cancer Cell Expression 1 8 6 6 4 log2 ratio Expression 2 1 4 2 2 p value.1 D Supplementary Figure 1. Expression of Sema4C and Plexin2
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
Supplementary Information
 Supplementary Information Supplementary Figure 1. EBV-gB 23-431 mainly exists as trimer in HEK 293FT cells. (a) Western blotting analysis for DSS crosslinked FLAG-gB 23-431. HEK 293FT cells transfected
Supplementary Information Supplementary Figure 1. EBV-gB 23-431 mainly exists as trimer in HEK 293FT cells. (a) Western blotting analysis for DSS crosslinked FLAG-gB 23-431. HEK 293FT cells transfected
SUPPLEMENTARY INFORMATION
 SUPPLEENTRY INFORTION DOI: 1.138/ncb2577 Early Telophase Late Telophase B icrotubules within the ICB (percent of total cells in telophase) D G ultinucleate cells (% total) 8 6 4 2 2 15 1 5 T without gaps
SUPPLEENTRY INFORTION DOI: 1.138/ncb2577 Early Telophase Late Telophase B icrotubules within the ICB (percent of total cells in telophase) D G ultinucleate cells (% total) 8 6 4 2 2 15 1 5 T without gaps
SPATIAL DISTRIBUTION OF PROTEIN CONCENTRATION CAN CONTROL CELLULAR PATHWAY FLUXES
 SPATIAL DISTRIBUTION OF PROTEIN CONCENTRATION CAN CONTROL CELLULAR PATHWAY FLUXES BENG 221 October 21, 2011 INTRODUCTION One of the goals of systems biology is to create models for intracellular pathway
SPATIAL DISTRIBUTION OF PROTEIN CONCENTRATION CAN CONTROL CELLULAR PATHWAY FLUXES BENG 221 October 21, 2011 INTRODUCTION One of the goals of systems biology is to create models for intracellular pathway
In vitro scratch assay: method for analysis of cell migration in vitro labeled fluorodeoxyglucose (FDG)
 In vitro scratch assay: method for analysis of cell migration in vitro labeled fluorodeoxyglucose (FDG) 1 Dr Saeb Aliwaini 13/11/2015 Migration in vivo Primary tumors are responsible for only about 10%
In vitro scratch assay: method for analysis of cell migration in vitro labeled fluorodeoxyglucose (FDG) 1 Dr Saeb Aliwaini 13/11/2015 Migration in vivo Primary tumors are responsible for only about 10%
File name: Supplementary Information Description: Supplementary Figures, Supplementary Table and Supplementary References
 File name: Supplementary Information Description: Supplementary Figures, Supplementary Table and Supplementary References File name: Supplementary Data 1 Description: Summary datasheets showing the spatial
File name: Supplementary Information Description: Supplementary Figures, Supplementary Table and Supplementary References File name: Supplementary Data 1 Description: Summary datasheets showing the spatial
Supplementary Figure 1. Characterization of ALDH-positive cell population in MCF-7 cells. (a) Expression level of stem cell markers in MCF-7 cells or
 Supplementary Figure 1. Characterization of ALDH-positive cell population in MCF-7 cells. (a) Expression level of stem cell markers in MCF-7 cells or ALDH-positive cell population by qpcr. Data represent
Supplementary Figure 1. Characterization of ALDH-positive cell population in MCF-7 cells. (a) Expression level of stem cell markers in MCF-7 cells or ALDH-positive cell population by qpcr. Data represent
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Amelio et al., http://www.jcb.org/cgi/content/full/jcb.201203134/dc1 Figure S1. mir-24 regulates proliferation and by itself induces
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Amelio et al., http://www.jcb.org/cgi/content/full/jcb.201203134/dc1 Figure S1. mir-24 regulates proliferation and by itself induces
Transport and Concentration of Charged Molecules in a Lipid Membrane
 Transport and Concentration of Charged Molecules in a Lipid Membrane Johannes S. Roth 1, Matthew R. Cheetham 1,2, and Stephen D. Evans* 1 1 University of Leeds, 2 Current Address: University of Oxford
Transport and Concentration of Charged Molecules in a Lipid Membrane Johannes S. Roth 1, Matthew R. Cheetham 1,2, and Stephen D. Evans* 1 1 University of Leeds, 2 Current Address: University of Oxford
Supplemental Information. Menin Deficiency Leads to Depressive-like. Behaviors in Mice by Modulating. Astrocyte-Mediated Neuroinflammation
 Neuron, Volume 100 Supplemental Information Menin Deficiency Leads to Depressive-like Behaviors in Mice by Modulating Astrocyte-Mediated Neuroinflammation Lige Leng, Kai Zhuang, Zeyue Liu, Changquan Huang,
Neuron, Volume 100 Supplemental Information Menin Deficiency Leads to Depressive-like Behaviors in Mice by Modulating Astrocyte-Mediated Neuroinflammation Lige Leng, Kai Zhuang, Zeyue Liu, Changquan Huang,
microrna as a Potential Vector for the Propagation of Robustness in Protein Expression and Oscillatory Dynamics within a cerna Network
 microrna as a Potential Vector for the Propagation of Robustness in Protein Expression and Oscillatory Dynamics within a cerna Network Claude Gérard*,Béla Novák Oxford Centre for Integrative Systems Biology,
microrna as a Potential Vector for the Propagation of Robustness in Protein Expression and Oscillatory Dynamics within a cerna Network Claude Gérard*,Béla Novák Oxford Centre for Integrative Systems Biology,
p = formed with HCI-001 p = Relative # of blood vessels that formed with HCI-002 Control Bevacizumab + 17AAG Bevacizumab 17AAG
 A.. Relative # of ECs associated with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 ol b p < 0.001 Relative # of blood vessels that formed with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 l b p = 0.002 Control IHC:
A.. Relative # of ECs associated with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 ol b p < 0.001 Relative # of blood vessels that formed with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 l b p = 0.002 Control IHC:
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/6/e1700338/dc1 Supplementary Materials for HIV virions sense plasma membrane heterogeneity for cell entry Sung-Tae Yang, Alex J. B. Kreutzberger, Volker Kiessling,
advances.sciencemag.org/cgi/content/full/3/6/e1700338/dc1 Supplementary Materials for HIV virions sense plasma membrane heterogeneity for cell entry Sung-Tae Yang, Alex J. B. Kreutzberger, Volker Kiessling,
