Supplementary Material
|
|
|
- Jemima Goodwin
- 6 years ago
- Views:
Transcription
1 Supplementary Material Original article in Gastric Cancer A novel splice variant of XIAP-associated factor 1 (XAF1) is expressed in peripheral blood containing gastric cancer-derived circulating tumor cells Keiichi Hatakeyama 1, Yushi Yamakawa 2,3, Yorikane Fukuda 1,4, Keiichi Ohshima 1, Kanako Wakabayashi-Nakao 1, Naoki Sakura 1, Yutaka Tanizawa 2, Yusuke Kinugasa 3, Ken Yamaguchi 5, Masanori Terashima 2* and Tohru Mochizuki 1 1 Medical Genetics Division, Shizuoka Cancer Center Research Institute. 2 Division of Gastric Surgery, Shizuoka Cancer Center Hospital. 3 Division of Colon and Rectal Surgery Shizuoka Cancer Center Hospital. 4 Present address: G&G Science. 5 Shizuoka Cancer Center Hospital and Research Institute Equal contributor. * Corresponding author. m.terashima@scchr.jp. 1
2 Methods Cell cultures Cell lines used in this study are listed in Table S1 in the Supplementary Method. MKN45P cell lines [1] were kindly provided by Dr. Yutaka Yonemura and Dr. Yoshio Endo. These cell lines were cultured and maintained in RPMI 1640, DMEM/F12, or DMEM medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 5 to 10% foetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA), glutamine (0.3 mg/ml), penicillin (100 unit/ml), and streptomycin (0.1 mg/ml), in a humidified 5% CO 2 incubator. mrna analyses were performed using cells found to have 70 90% confluency and greater than 95% viability, as determined using trypan blue staining. RNA sample preparations Total RNA extraction of cell pellet and peripheral blood stabilized with Paxgene were performed using the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) and PAXgene Blood RNA Kit (Qiagen), respectively. The extracted RNA concentration was determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA) and total RNA quality was then confirmed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). The purified total RNA from cancer cell lines and peripheral blood were then reverse-transcribed using ThermoScript Reverse Transcriptase (Invitrogen) and oligo(dt) 20 primers in accordance with the manufacturer s instructions. 2
3 RT-PCR The PCR was designed to detect the splice variants and known XAF1 transcripts (XAF1A and XAF1C). The synthesized cdnas were amplified using Ex Taq polymerase (Takara Bio, Shiga, Japan) for 36 PCR cycles of 95 C for 60 s, 59 C for 30 s, and 68 C for 60 s. Primer sets listed in Table S2 in the Supplementary Method. DNA sequencing analysis For DNA sequencing analysis, the PCR products were analyzed on 1 2% agarose gels by electrophoresis following by gel staining with SYBR Safe (Invitrogen). The bands visualized under ultraviolet light were isolated and purified using the QIAquick Gel Extraction Kit (Qiagen). The purified samples were then cloned into a pcr 2.1-TOPO vector (Invitrogen) using the TOPO TA Cloning Kit for Sequencing (Invitrogen). Positive transformants were sequenced using the Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) and an ABI 3130xl Genetic Analyzer (Applied Biosystems). Quantitative real-time PCR The target fragment was amplified using specific primers (see Table S2) according to the following protocol: preheating at 95 C for 20 s, 40 cycles at 95 C for 1 s and 60 C for 30 s, and then a dissociation curve performed at 95 C for 15 s, 60 C for 15 s, and 95 C for 15 s. The threshold cycle (Ct) values were converted into transcript expression levels using a standard curve prepared using 3
4 purified PCR amplicons that were generated from plasmids containing the sequence of the target transcripts. Nonspecific amplification in qrt-pcr was evaluated using gel electrophoresis, which helped determine the amplicon size. NMD-related genes (ATF3, GADD45B, MAFF, and UPF1) were quantified using the 2 -ΔΔCt method. [2] The fold change was normalized by ACTB expression. 4
5 Results Validation of XAF1F expression levels The novel splice variant XAF1F was detected in 20 cancer cell lines. To properly estimate the expression level of this transcript, we performed a qrt-pcr assay. The specificity of the qrt-pcr was first verified by gel electrophoresis and a dissociation curve analysis (Fig. S1A and S1B in Supplementary Material). As shown in Fig. 2, the XAF1F transcript appeared to be highly expressed in MKN45P (gastric), KP-3L (pancreatic), LS123 (colorectal), and Hs578T (breast) cancer cells. The specificity of qrt-pcr was thus validated using these cells. Amplicons were observed in a single band in electrophoresis, and the size of the bands was identical to the expected size of 116 bp. The firstderivative curves of fluorescence decay showed a single peak of MKN45P in the in qrt-pcr assays. In the other three cells, a single peak was also observed at the same temperature (data not shown). These results indicated that nonspecific PCR products were not amplified in qrt-pcr. Standard curves were next generated from 10-fold serial dilutions of template DNA amplified from plasmids containing the sequences of the target transcripts (Fig. S1C in Supplementary Material). The amplification efficiency (AE) of all transcripts was 1.00 ± 0.05 (R 2 = 0.988, p = ). These results indicate that the target-specific primers and the constructed template DNA provided accurate amplification in our qrt- PCR assay. We finally quantified the XAF1F transcripts by using this validated assay (Fig. S1D in Supplementary Material). As shown in Fig. 2, MKN45, LoVo, PANC-1, and MDA-MB-175-VII cells expressed XAF1A/C but barely expressed XAF1F. qrt-pcr analysis was performed using these cell lines and the four XAF1F-expressing cell lines indicated in Fig. S1A. The mrna expression levels of 5
6 XAF1F determined by qrt-pcr were fairly consistent with the electrophoresis results of the RT-PCR amplicons, indicating that XAF1F in cancer cells was amplified by qrt-pcr without artifacts from the other XAF1 transcripts. Therefore, we conclude that quantification of XAF1F transcript was successfully achieved using our qrt-pcr analysis. 6
7 Table S1 Human cancer cell lines used in this study Tissue Cell line Histology Supplier Catalogue No. Note about metastasis Gstric MKN45 carcinoma JCRB 1) JCRB MKN45P subcloned from MKN45* - - Highly metastatic cell derived from xenograft of MKN45 MKN1 adenosquamous carcinoma JCRB JCRB MKN74 adenocarcinoma JCRB JCRB0255 Stomach cancer metastasized to liver KATOIII signet ring cell carcinoma JCRB JCRB0611 Stomach cancer, signet ring cell carcinoma SNU-1 carcinoma ATCC 2) CRL-5971 Stomach; derived from metastatic site: ascites SNU-16 carcinoma ATCC CRL-5974 Stomach; derived from metastatic site: ascites AGS adenocarcinoma ECACC 3) EC Hs746T carcinoma ATCC HTB NCI-N87 carcinoma ATCC CRL-5822 Stomach; derived from metastatic site: liver. The tumor was passaged as a xenograft in athymic nude mice for three passages before the cell line was established. HGC-27 carcinoma ECACC EC Pancreatic ACBRI 515 normal pancreatic CEL 4) RI KP-3 adenosquamous carcinoma JCRB JCRB KP-3L adenosquamous carcinoma JCRB JCRB Cells were established from the resulting tumor nodules in the liver of mouse, into the spleen where KP-3 cells had been injected. AsPC-1 adenocarcinoma ATCC CRL-1682 Pancreas; derived from metastatic site: ascites BxPC-3 adenocarcinoma ATCC CRL Capan-1 adenocarcinoma ATCC HTB-79 Pancreas; derived from metastatic site: liver Capan-2 adenocarcinoma ATCC HTB-80 - CFPAC-1 ductal adenocarcinoma ATCC CRL-1918 Pancreas; derived from metastatic site: liver HPAF-II adenocarcinoma ATCC CRL-1997 HPAF-II is a human pancreatic adenocarcinoma cell line derived from peritoneal ascitic fluid of a 44 year old Caucasian male with primary pancreatic adenocarcinoma and metastases to the liver, diaphragm and lymph nodes. KP-2 adenocarcinoma JCRB JCRB KP-4 pancreatic duct carcinoma RCB 5) RCB MIAPaca-2 carcinoma ATCC CRL PANC-1 epithelioid carcinoma ATCC CRL
8 Continued Colonrectal LoVo colorectal carcinoma ATCC CCL-229 Colon; derived from metastatic site: left supraclavicular region. DLD-1 adenocarcinoma ECACC EC Colo-205 colorectal carcinoma ATCC CCL-222 Colon; derived from metastatic site: ascites SW948 adenocarcinoma ATCC CCL SK-CO-1 adenocarcinoma ATCC HTB-39 Colon; derived from metastatic site: ascites HCC-56 5) adenocarcinoma JCRB JCRB LS123 adenocarcinoma ECACC EC C170 carcinoma ECACC EC Colo741 carcinoma ECACC EC Derived from a pelvic wall metastasis of a 69 year old female with colon carcinoma SNU-C1 adenocarcinoma ATCC CRL-5972 Colon; derived from metastatic site: peritoneum SW480 adenocarcinoma ECACC EC SW620 adenocarcinoma ATCC CCL-227 Colon; derived from metastatic site: lymph node Breast MDA-MB-175-VII epithelial ductal carcinoma ATCC HTB-25 Mammary gland; breast/duct; derived from metastatic site: pleural effusion MDA-MB-361 adenocarcinoma ATCC HTB-27 Mammary gland/breast; derived from metastatic site:brain HCC38 primary ductal carcinoma ATCC CRL-2314 HCC38 was initiated from a 50-year-old white female with a prior history of leiomyosarcoma; her mother died of breast cancer. T-47D ductal carcinoma ATCC HTB-133 Mammary gland; derived from metastatic site: pleural effusion Hs 578T carcinoma ECACC EC Tumorigenic in immunosuppressed mice and form colonies in semisolid medium MFM 223 carcinoma ECACC EC HCC70 primary ductal carcinoma ATCC CRL UACC-812 ductal carcinoma ATCC CRL UACC-893 primary ductal carcinoma ATCC CRL ) Japanese Collection of Research Bioresources 2) American Type Culture Collection 3) European Collection of Cell Cultures 4) Cell Systems Corporation 5) Riken Bioresource Center Cell Bank 6) The individual-specific DNA profile of HCC-56 is identical to that of other cell line, HSC-57 * Iinuma H, et al: Intracellular targeting therapy of cisplatin-encapsulated transferrin-polyethylene glycol liposome on peritoneal dissemination of gastric cancer. Int. J. Cancer 2002, 99(1):
9 Table S2 DNA sequences of RT-PCR and quantitative RT-PCR primers Assay Gene Primer name Sequence (5 to 3 ) Orientation RT-PCR and qrt-pcr XAF1 qfs TGGACCCACATCTGGTGTGT Sense qfas GGCACTCATTGGCCTTATGAA Antisense RT-PCR XAF1 S5* AGTGTGAGGAGCCTGTCC Sense AS* CAGAAGTCCTCGCTGGAGTTTC Sense AAS* TTCAGGAGCTGAAATTCTTTCC Antisense CAS* TTATGGCCACAGATGTGCACT Antisense DAS* AGTCTGGACAACATTTACCCTTTC Antisense ACTB AT1 ATTCCTATGTGGGCGACGAGGC Sense AT2 TGGATAGCAACGTACATGGCTGG Antisense qrt-pcr ATF3 ATF-S GATGTCCTCTGCGCTGGAAT Sense ATF-A CTCGTCGCCTCTTTTTCCTTT Antisense GADD45B GAD-S GCCAGGATCGCCTCACAGT Sense GAD-A GATTTGCAGGGCGATGTCAT Antisense MAFF MAF-S GAGGGCACCTTCTGCAAACA Sense MAF-A CGCAGATGCCGGTTCAG Antisense UPF1 UPF-S AGATCACGGCACAGCAGAT Sense UPF-A TGGCAGAAGGGTTTTCCTT Antisense BIRC5 qsv1 TCAAGGACCACCGCATCTCT Sense qsv2 GCCAAGTCTGGCTCGTTCTC Antisense ACTB qat1 TGGCACCCAGCACAATGA Sense qat2 CCGATCCACACGGAGTACTTG Antisense * These primers were designed based on previous report [3] 9
10 Table S3 Relationship between age and expression of transcripts Gene symbol Spearman's rank correlation coefficient (p value 1) ) Healthy volunteer Patient whole samples 2) BIRC (0.352) (0.402) 0.18 (0.204) XAF1F 0.22 (0.318) (0.696) 0.07 (0.399) XAF1A 0.14 (0.528) (0.943) 0.14 (0.129) XAF1C 0.08 (0.731) (0.770) 0.11 (0.249) 1) Using the t distribution. 2) Healthy volunteer + Patient 10
11 Figure S1 (A) (B) Derivative MKN45P KP-3L LS123 MKN45P Hs578T 116 bp Temperature ( C) (C) Ct value (-) R 2 = p = 1.88 x AE = 1.00 ± Input of XAF1F log 10 (n) (D) XAF1F expression level MKN45 MKN45P PANC-1 KP-3L 0 gastric pancreatic LoVo colorectal LS123 MDA-MB-175-VII breast Hs578T Figure S1. Validation of qrt-pcr assay for XAF1F transcript. (A) Specificity verification of qrt- PCR primers using agarose gel electrophoresis of amplicons. The amplicon size of XAF1F obtained by qrt-pcr using primers qfs and qfas was 116 bp. (B) Dissociation curve analysis of qrt-pcr amplicons obtained from MKN45P cells. First-derivative curves of the fluorescence decay derived from dissociation of the intercalator and DNA were plotted versus temperature. (C) Standard curve constructed from 10-fold serial dilutions of purified PCR amplicons that were generated from plasmids containing the sequences of the target transcripts. Amplification efficiency (AE) was calculated from the slope of the regression line according to the equation AE = 10-1/slope -1 and is represented as the mean value ± SD obtained from pentaplicate experiments. (D) Quantification of XAF1F levels in cancer cell lines. For demonstration of qrt-pcr, XAF1F-expressing cells and XAF1A-expressing cells without XAF1F expression were selected from the experiment depicted in Fig
12 Figure S Relative expression level XAF1A XAF1C XAF1F Relative expression level XAF1A XAF1C XAF1F 0 MKN45 MKN45P 0 KP-3 KP-3L gastric pancreatic Figure S2. Expression analysis of XAF1 transcripts in pairs of cell lines used as a metastatic model. The pairs of gastric cells (MKN45 vs. MKN45P) and pancreatic cells (KP-3 vs. KP-3L) are shown in the left and right panels, respectively. Expression level of XAF1F was evaluated using qrt-pcr, and that of other XAF1 transcripts was estimated by densitometric scaning of RT-PCR amplicons based on previous report. [3] The expression levels of XAF1 transcripts in each parental cell (MKN45 or KP-3) are shown as
13 Figure S3 colorectal breast Colo205 LS123 Hs578T Figure S3. Expression levels of XAF1 transcripts in NMD-inhibited cancer cells. NMD was inhibited by caffeine and actd treatment (+caffeine). Treatment with actd alone (-caffeine) was used to observe RNA stability after blockade of transcription. Cancer cells that were not treated with reagents are presented as the control. For verification of upregulation of XAF1F through NMD inhibition, XAF1F-expressing cells were selected from the experiment depicted in Fig
14 Figure S4 (A) MKN45P patient 1 patient 2 patient bp (B) MKN45P (C) R 2 = p = 3.14 x AE = 0.93 ± 0.04 Derivative Ct value (-) Temperature ( C) Input of XAF1F log 10 (n) Figure S4. Validation of quntitative real-time (qrt)-pcr assay for BIRC5 transcript. (A) Specificity verification of qrt-pcr primers using agarose gel electrophoresis of amplicons. The amplicon size of BIRC5 obtained by qrt-pcr using primers qsv1 and qsv2 (detailed in Table S2) was 124 bp. (B) Dissociation curve analysis of qrt-pcr amplicons obtained from MKN45P cells. First-derivative curves of the fluorescence decay derived from dissociation of the intercalator and DNA were plotted versus temperature. (C) Standard curve constructed from 10-fold serial dilutions of purified PCR amplicons that were generated from plasmids containing the sequences of BIRC5 transcript. Amplification efficiency (AE) was calculated from the slope of the regression line according to the equation AE = 10-1/slope -1 and is represented as the mean values ± S.D. obtained from pentaplicate experiments. 14
15 Figure S5 100 XAF1C 100 XAF1F Sensitivity (%) Cut-off: sensitivity: 51.04% specificity: 86.36% Sensitivity (%) Cut-off: sensitivity: 58.33% specificity: 72.73% 20 AUC, (95%CI, ) Specificity (%) 20 0 AUC, (95%CI, ) Specificity (%) Figure S5. ROC curve of XAF1C and XAF1F expressions in peripheral blood. The black solid line indicates the curve of XAF1C or XAF1F in all peripheral blood samples (n = 118). The optimal threshold is represented by the black arrow. 15
16 References 1. Iinuma H, Maruyama K, Okinaga K, et al. Intracellular targeting therapy of cisplatin-encapsulated transferrinpolyethylene glycol liposome on peritoneal dissemination of gastric cancer. Int J Cancer. 2002;99(1): Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(- Delta Delta C(T)) Method. Methods. 2001;25(4): Chung SK, Lee MG, Ryu BK, et al. Frequent alteration of XAF1 in human colorectal cancers: implication for tumor cell resistance to apoptotic stresses. Gastroenterology. 2007;132(7):
RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using
 Supplementary Information Materials and Methods RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using Trizol reagent (Invitrogen,Carlsbad, CA) according to the manufacturer's instructions.
Supplementary Information Materials and Methods RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using Trizol reagent (Invitrogen,Carlsbad, CA) according to the manufacturer's instructions.
America, Hershey, PA Australia, Melbourne, VIC Europe, Munich
 America, Hershey, PA Australia, Melbourne, VIC Europe, Munich info@vivopharm.com www.vivopharm.com Bladder MB-9-luc 2 Mouse urinary bladder carcinoma yes yes yes C57BL/6 2 Bone UMR-106 Rat osteosarcoma
America, Hershey, PA Australia, Melbourne, VIC Europe, Munich info@vivopharm.com www.vivopharm.com Bladder MB-9-luc 2 Mouse urinary bladder carcinoma yes yes yes C57BL/6 2 Bone UMR-106 Rat osteosarcoma
Species Tumor Type Comment for in vivo work Lead Time for in vivo studies [weeks] MB-49-luc-2 Mouse urinary bladder carcinoma C57BL/6 2
![Species Tumor Type Comment for in vivo work Lead Time for in vivo studies [weeks] MB-49-luc-2 Mouse urinary bladder carcinoma C57BL/6 2 Species Tumor Type Comment for in vivo work Lead Time for in vivo studies [weeks] MB-49-luc-2 Mouse urinary bladder carcinoma C57BL/6 2](/thumbs/88/116309357.jpg) America, Hershey, PA Australia, Melbourne, VIC Europe, Munich info@vivopharm.com www.vivopharm.com Tissue Bladder Species Tumor Type Comment for in vivo work Lead Time for in vivo studies [weeks] MB-49-luc-
America, Hershey, PA Australia, Melbourne, VIC Europe, Munich info@vivopharm.com www.vivopharm.com Tissue Bladder Species Tumor Type Comment for in vivo work Lead Time for in vivo studies [weeks] MB-49-luc-
Supplementary Materials and Methods
 Supplementary Materials and Methods Immunoblotting Immunoblot analysis was performed as described previously (1). Due to high-molecular weight of MUC4 (~ 950 kda) and MUC1 (~ 250 kda) proteins, electrophoresis
Supplementary Materials and Methods Immunoblotting Immunoblot analysis was performed as described previously (1). Due to high-molecular weight of MUC4 (~ 950 kda) and MUC1 (~ 250 kda) proteins, electrophoresis
Online Data Supplement. Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2
 Online Data Supplement Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2 Yi Lin and Zhongjie Sun Department of physiology, college of
Online Data Supplement Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2 Yi Lin and Zhongjie Sun Department of physiology, college of
The mir-199a/brm/egr1 axis is a determinant of anchorage-independent growth in epithelial tumor cell lines
 Supplementary information Supplementary Figure -9 Supplementary Table -4 The mir-99a/brm/egr axis is a determinant of anchorage-independent growth in epithelial tumor cell lines Kazuyoshi Kobayashi, Kouhei
Supplementary information Supplementary Figure -9 Supplementary Table -4 The mir-99a/brm/egr axis is a determinant of anchorage-independent growth in epithelial tumor cell lines Kazuyoshi Kobayashi, Kouhei
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplemental Materials and Methods Plasmids and viruses Quantitative Reverse Transcription PCR Generation of molecular standard for quantitative PCR
 Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
Online Appendix Material and Methods: Pancreatic RNA isolation and quantitative real-time (q)rt-pcr. Mice were fasted overnight and killed 1 hour (h)
 Online Appendix Material and Methods: Pancreatic RNA isolation and quantitative real-time (q)rt-pcr. Mice were fasted overnight and killed 1 hour (h) after feeding. A small slice (~5-1 mm 3 ) was taken
Online Appendix Material and Methods: Pancreatic RNA isolation and quantitative real-time (q)rt-pcr. Mice were fasted overnight and killed 1 hour (h) after feeding. A small slice (~5-1 mm 3 ) was taken
Title page. Title: MicroRNA-155 Controls Exosome Synthesis and Promotes Gemcitabine Resistance in
 Title page Title: MicroRNA- Controls Synthesis and Promotes Gemcitabine Resistance in Pancreatic Ductal Adenocarcinoma Authors Manabu Mikamori, Daisaku Yamada, Hidetoshi Eguchi, Shinichiro Hasegawa, Tomoya
Title page Title: MicroRNA- Controls Synthesis and Promotes Gemcitabine Resistance in Pancreatic Ductal Adenocarcinoma Authors Manabu Mikamori, Daisaku Yamada, Hidetoshi Eguchi, Shinichiro Hasegawa, Tomoya
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
(A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC,
 Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
Human cell line list Production of Exosome Standards - Cell Lysates
 Human cell line list 2016 - Production of Exosome Standards - Cell Lysates 380 peripheral blood leukemia, pre-b cell 1301 blood leukemia, acute lymphoblastic, T cell 5637 bladder carcinoma 8305C thyroid
Human cell line list 2016 - Production of Exosome Standards - Cell Lysates 380 peripheral blood leukemia, pre-b cell 1301 blood leukemia, acute lymphoblastic, T cell 5637 bladder carcinoma 8305C thyroid
Department of Pharmaceutical Sciences, School of Pharmacy, Northeastern University, Boston, MA 02115, USA 2
 Pancreatic Cancer Cell Exosome-Mediated Macrophage Reprogramming and the Role of MicroRNAs 155 and 125b2 Transfection using Nanoparticle Delivery Systems Mei-Ju Su 1, Hibah Aldawsari 2, and Mansoor Amiji
Pancreatic Cancer Cell Exosome-Mediated Macrophage Reprogramming and the Role of MicroRNAs 155 and 125b2 Transfection using Nanoparticle Delivery Systems Mei-Ju Su 1, Hibah Aldawsari 2, and Mansoor Amiji
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
Islet viability assay and Glucose Stimulated Insulin Secretion assay RT-PCR and Western Blot
 Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Supplementary Information
 Supplementary Information - chimeric fusion transcript in human gastric cancer promotes tumorigenesis through activation of PI3K/AKT signaling Sun Mi Yun, Kwiyeom Yoon, Sunghoon Lee, Eunjeong Kim, Seong-Ho
Supplementary Information - chimeric fusion transcript in human gastric cancer promotes tumorigenesis through activation of PI3K/AKT signaling Sun Mi Yun, Kwiyeom Yoon, Sunghoon Lee, Eunjeong Kim, Seong-Ho
Epithelial interleukin-25 is a key mediator in Th2-high, corticosteroid-responsive
 Online Data Supplement: Epithelial interleukin-25 is a key mediator in Th2-high, corticosteroid-responsive asthma Dan Cheng, Zheng Xue, Lingling Yi, Huimin Shi, Kan Zhang, Xiaorong Huo, Luke R. Bonser,
Online Data Supplement: Epithelial interleukin-25 is a key mediator in Th2-high, corticosteroid-responsive asthma Dan Cheng, Zheng Xue, Lingling Yi, Huimin Shi, Kan Zhang, Xiaorong Huo, Luke R. Bonser,
Figure S1 Time-dependent down-modulation of HER3 by EZN No Treatment. EZN-3920, 2 μm. Time, h
 Figure S1 Time-dependent down-modulation of HER3 by EZN-392 HE ER3 mrna A, %Contr rol 12 No Treatment EZN-392, 2 μm 1 8 6 4 2 2 8 24 Time, h Figure S2. Specific target down-modulation by HER3 (EZN-392)
Figure S1 Time-dependent down-modulation of HER3 by EZN-392 HE ER3 mrna A, %Contr rol 12 No Treatment EZN-392, 2 μm 1 8 6 4 2 2 8 24 Time, h Figure S2. Specific target down-modulation by HER3 (EZN-392)
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
(A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and a
 Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation
 Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
Phosphate buffered saline (PBS) for washing the cells TE buffer (nuclease-free) ph 7.5 for use with the PrimePCR Reverse Transcription Control Assay
 Catalog # Description 172-5080 SingleShot Cell Lysis Kit, 100 x 50 µl reactions 172-5081 SingleShot Cell Lysis Kit, 500 x 50 µl reactions For research purposes only. Introduction The SingleShot Cell Lysis
Catalog # Description 172-5080 SingleShot Cell Lysis Kit, 100 x 50 µl reactions 172-5081 SingleShot Cell Lysis Kit, 500 x 50 µl reactions For research purposes only. Introduction The SingleShot Cell Lysis
p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO
 Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1: Cryopreservation alters CD62L expression by CD4 T cells. Freshly isolated (left) or cryopreserved PBMCs (right) were stained with the mix of antibodies described
Supplementary Figure 1 Supplementary Figure 1: Cryopreservation alters CD62L expression by CD4 T cells. Freshly isolated (left) or cryopreserved PBMCs (right) were stained with the mix of antibodies described
A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis
 APPLICATION NOTE Cell-Free DNA Isolation Kit A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis Abstract Circulating cell-free DNA (cfdna) has been shown
APPLICATION NOTE Cell-Free DNA Isolation Kit A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis Abstract Circulating cell-free DNA (cfdna) has been shown
Supplementary Figure 1. mrna targets were found in exosomes and absent in free-floating supernatant. Serum exosomes and exosome-free supernatant were
 Supplementary Figure 1. mrna targets were found in exosomes and absent in free-floating supernatant. Serum exosomes and exosome-free supernatant were separated via ultracentrifugation and lysed to analyze
Supplementary Figure 1. mrna targets were found in exosomes and absent in free-floating supernatant. Serum exosomes and exosome-free supernatant were separated via ultracentrifugation and lysed to analyze
Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3
 Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
Supporting Information
 Supporting Information Palmisano et al. 10.1073/pnas.1202174109 Fig. S1. Expression of different transgenes, driven by either viral or human promoters, is up-regulated by amino acid starvation. (A) Quantification
Supporting Information Palmisano et al. 10.1073/pnas.1202174109 Fig. S1. Expression of different transgenes, driven by either viral or human promoters, is up-regulated by amino acid starvation. (A) Quantification
Genetic Alteration Panels
 TM Genetic Alteration Panels GENETIC ALTERATION PANELS Table of Contents AKT Genetic Alteration Panel (ATCC No. TCP-1029 )...1 BRAF Genetic Alteration Panel (ATCC No. TCP-1032 )...5 EGFR Genetic Alteration
TM Genetic Alteration Panels GENETIC ALTERATION PANELS Table of Contents AKT Genetic Alteration Panel (ATCC No. TCP-1029 )...1 BRAF Genetic Alteration Panel (ATCC No. TCP-1032 )...5 EGFR Genetic Alteration
CHAPTER 4 RESULTS. showed that all three replicates had similar growth trends (Figure 4.1) (p<0.05; p=0.0000)
 CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
CHAPTER 4 RESULTS 4.1 Growth Characterization of C. vulgaris 4.1.1 Optical Density Growth study of Chlorella vulgaris based on optical density at 620 nm (OD 620 ) showed that all three replicates had similar
CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer
 European Review for Medical and Pharmacological Sciences 2018; 22: 3713-3718 CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer N. LIU 1, J. ZHANG 1, L.-Y. ZHANG 1, L.
European Review for Medical and Pharmacological Sciences 2018; 22: 3713-3718 CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer N. LIU 1, J. ZHANG 1, L.-Y. ZHANG 1, L.
Supplementary methods:
 Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
SUPPLEMENTARY INFORMATION
 doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
Supplemental Figure 1
 1 Supplemental Figure 1 Effects of DATE shortening on HGF promoter activity. The HGF promoter region (-1037 to +56) containing wild-type (30As) or truncated DATE (26As, 27As, 28A, 29As) from breast cancer
1 Supplemental Figure 1 Effects of DATE shortening on HGF promoter activity. The HGF promoter region (-1037 to +56) containing wild-type (30As) or truncated DATE (26As, 27As, 28A, 29As) from breast cancer
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary webappendix
 Supplementary webappendix This webappendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Kratz JR, He J, Van Den Eeden SK, et
Supplementary webappendix This webappendix formed part of the original submission and has been peer reviewed. We post it as supplied by the authors. Supplement to: Kratz JR, He J, Van Den Eeden SK, et
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
Oncolytic Adenovirus Complexes Coated with Lipids and Calcium Phosphate for Cancer Gene Therapy
 Oncolytic Adenovirus Complexes Coated with Lipids and Calcium Phosphate for Cancer Gene Therapy Jianhua Chen, Pei Gao, Sujing Yuan, Rongxin Li, Aimin Ni, Liang Chu, Li Ding, Ying Sun, Xin-Yuan Liu, Yourong
Oncolytic Adenovirus Complexes Coated with Lipids and Calcium Phosphate for Cancer Gene Therapy Jianhua Chen, Pei Gao, Sujing Yuan, Rongxin Li, Aimin Ni, Liang Chu, Li Ding, Ying Sun, Xin-Yuan Liu, Yourong
MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands)
 Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
TITLE: CYP1B1 Polymorphism as a Risk Factor for Race-Related Prostate Cancer
 AD Award Number: W81XWH-04-1-0579 TITLE: CYP1B1 Polymorphism as a Risk Factor for Race-Related Prostate Cancer PRINCIPAL INVESTIGATOR: Yuichiro Tanaka, Ph.D. CONTRACTING ORGANIZATION: Northern California
AD Award Number: W81XWH-04-1-0579 TITLE: CYP1B1 Polymorphism as a Risk Factor for Race-Related Prostate Cancer PRINCIPAL INVESTIGATOR: Yuichiro Tanaka, Ph.D. CONTRACTING ORGANIZATION: Northern California
Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing)
 Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing) platform with laser manipulation to efficiently purify lung
Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing) platform with laser manipulation to efficiently purify lung
RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers
 Xu et al. Molecular Cancer (2019) 18:8 https://doi.org/10.1186/s12943-018-0932-8 LETTER TO THE EDITOR RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers
Xu et al. Molecular Cancer (2019) 18:8 https://doi.org/10.1186/s12943-018-0932-8 LETTER TO THE EDITOR RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Establishment of in Vivo Metastasis Model of Human Adenoid Cystic Carcinoma: Detection of Metastasis by PCR with Human β -Globin Gene
 Kobe J. Med. Sci., Vol. 48, No. 5, pp. 145-152, 2002 Establishment of in Vivo Metastasis Model of Human Adenoid Cystic Carcinoma: Detection of Metastasis by PCR with Human β -Globin Gene HIDEKI KOMATSUBARA,
Kobe J. Med. Sci., Vol. 48, No. 5, pp. 145-152, 2002 Establishment of in Vivo Metastasis Model of Human Adenoid Cystic Carcinoma: Detection of Metastasis by PCR with Human β -Globin Gene HIDEKI KOMATSUBARA,
Supporting Information
 Supporting Information Bertolini et al. 10.1073/pnas.0905653106 SI Text Lung Tissue Disaggregation. Solid tissues were finely minced by razorblade, washed in DMEM/F12 (Lonza), and then incubated with Accumax
Supporting Information Bertolini et al. 10.1073/pnas.0905653106 SI Text Lung Tissue Disaggregation. Solid tissues were finely minced by razorblade, washed in DMEM/F12 (Lonza), and then incubated with Accumax
Supplementary Fig. 1: ATM is phosphorylated in HER2 breast cancer cell lines. (A) ATM is phosphorylated in SKBR3 cells depending on ATM and HER2
 Supplementary Fig. 1: ATM is phosphorylated in HER2 breast cancer cell lines. (A) ATM is phosphorylated in SKBR3 cells depending on ATM and HER2 activity. Upper panel: Representative histograms for FACS
Supplementary Fig. 1: ATM is phosphorylated in HER2 breast cancer cell lines. (A) ATM is phosphorylated in SKBR3 cells depending on ATM and HER2 activity. Upper panel: Representative histograms for FACS
A263 A352 A204. Pan CK. pstat STAT3 pstat3 STAT3 pstat3. Columns Columns 1-6 Positive control. Omentum. Rectosigmoid A195.
 pstat3 75 Pan CK A A263 A352 A24 B Columns 1-6 Positive control A195 A22 A24 A183 Rectal Nodule STAT3 pstat3 STAT3 pstat3 Columns 7-12 Omentum Rectosigmoid Left Ovary Right Ovary Omentum Uterus Uterus
pstat3 75 Pan CK A A263 A352 A24 B Columns 1-6 Positive control A195 A22 A24 A183 Rectal Nodule STAT3 pstat3 STAT3 pstat3 Columns 7-12 Omentum Rectosigmoid Left Ovary Right Ovary Omentum Uterus Uterus
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Supplemental Data. TGF-β-mediated mir-181a expression promotes breast cancer metastasis by targeting Bim.
 Supplemental Data TGF-β-mediated mir-181a expression promotes breast cancer metastasis by targeting Bim. Molly A. Taylor 1, Khalid Sossey-Alaoui 2, Cheryl L. Thompson 3, David Danielpour 4, and William
Supplemental Data TGF-β-mediated mir-181a expression promotes breast cancer metastasis by targeting Bim. Molly A. Taylor 1, Khalid Sossey-Alaoui 2, Cheryl L. Thompson 3, David Danielpour 4, and William
Supplementary Information Titles Journal: Nature Medicine
 Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Fig. 1. Identification of acetylation of K68 of SOD2
 Supplementary Fig. 1. Identification of acetylation of K68 of SOD2 A B H. sapiens 54 KHHAAYVNNLNVTEEKYQEALAK 75 M. musculus 54 KHHAAYVNNLNATEEKYHEALAK 75 X. laevis 55 KHHATYVNNLNITEEKYAEALAK 77 D. rerio
Supplementary Fig. 1. Identification of acetylation of K68 of SOD2 A B H. sapiens 54 KHHAAYVNNLNVTEEKYQEALAK 75 M. musculus 54 KHHAAYVNNLNATEEKYHEALAK 75 X. laevis 55 KHHATYVNNLNITEEKYAEALAK 77 D. rerio
Table S1. Primers and PCR protocols for mutation screening of MN1, NF2, KREMEN1 and ZNRF3.
 Table S1. Primers and PCR protocols for mutation screening of MN1, NF2, KREMEN1 and ZNRF3. MN1 (Accession No. NM_002430) MN1-1514F 5 -GGCTGTCATGCCCTATTGAT Exon 1 MN1-1882R 5 -CTGGTGGGGATGATGACTTC Exon
Table S1. Primers and PCR protocols for mutation screening of MN1, NF2, KREMEN1 and ZNRF3. MN1 (Accession No. NM_002430) MN1-1514F 5 -GGCTGTCATGCCCTATTGAT Exon 1 MN1-1882R 5 -CTGGTGGGGATGATGACTTC Exon
Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma.
 Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
Expression of mir-1294 is downregulated and predicts a poor prognosis in gastric cancer
 European Review for Medical and Pharmacological Sciences 2018; 22: 5525-5530 Expression of mir-1294 is downregulated and predicts a poor prognosis in gastric cancer Y.-X. SHI, B.-L. YE, B.-R. HU, X.-J.
European Review for Medical and Pharmacological Sciences 2018; 22: 5525-5530 Expression of mir-1294 is downregulated and predicts a poor prognosis in gastric cancer Y.-X. SHI, B.-L. YE, B.-R. HU, X.-J.
Pair-fed % inkt cells 0.5. EtOH 0.0
 MATERIALS AND METHODS Histopathological analysis Liver tissue was collected 9 h post-gavage, and the tissue samples were fixed in 1% formalin and paraffin-embedded following a standard procedure. The embedded
MATERIALS AND METHODS Histopathological analysis Liver tissue was collected 9 h post-gavage, and the tissue samples were fixed in 1% formalin and paraffin-embedded following a standard procedure. The embedded
Supplementary Information
 Supplementary Information Detection and differential diagnosis of colon cancer by a cumulative analysis of promoter methylation Qiong Yang 1,3*, Ying Dong 2,3, Wei Wu 1, Chunlei Zhu 1, Hui Chong 1, Jiangyang
Supplementary Information Detection and differential diagnosis of colon cancer by a cumulative analysis of promoter methylation Qiong Yang 1,3*, Ying Dong 2,3, Wei Wu 1, Chunlei Zhu 1, Hui Chong 1, Jiangyang
For in vitro Veterinary Diagnostics only. Kylt Rotavirus A. Real-Time RT-PCR Detection.
 For in vitro Veterinary Diagnostics only. Kylt Rotavirus A Real-Time RT-PCR Detection www.kylt.eu DIRECTION FOR USE Kylt Rotavirus A Real-Time RT-PCR Detection A. General Kylt Rotavirus A products are
For in vitro Veterinary Diagnostics only. Kylt Rotavirus A Real-Time RT-PCR Detection www.kylt.eu DIRECTION FOR USE Kylt Rotavirus A Real-Time RT-PCR Detection A. General Kylt Rotavirus A products are
Supplemental File. TRAF6 is an amplified oncogene bridging the Ras and nuclear factor-κb cascade in human lung cancer
 Supplemental File TRAF6 is an amplified oncogene bridging the Ras and nuclear factor-κb cascade in human lung cancer Daniel T. Starczynowski, William W. Lockwood, Sophie Delehouzee, Raj Chari, Joanna Wegrzyn,
Supplemental File TRAF6 is an amplified oncogene bridging the Ras and nuclear factor-κb cascade in human lung cancer Daniel T. Starczynowski, William W. Lockwood, Sophie Delehouzee, Raj Chari, Joanna Wegrzyn,
Products for cfdna and mirna isolation. Subhead Circulating Cover nucleic acids from plasma
 MACHEREY-NAGEL Products for cfdna and mirna isolation Bioanalysis Subhead Circulating Cover nucleic acids from plasma n Flexible solutions for small and large blood plasma volumes n Highly efficient recovery
MACHEREY-NAGEL Products for cfdna and mirna isolation Bioanalysis Subhead Circulating Cover nucleic acids from plasma n Flexible solutions for small and large blood plasma volumes n Highly efficient recovery
microrna-200b and microrna-200c promote colorectal cancer cell proliferation via
 Supplementary Materials microrna-200b and microrna-200c promote colorectal cancer cell proliferation via targeting the reversion-inducing cysteine-rich protein with Kazal motifs Supplementary Table 1.
Supplementary Materials microrna-200b and microrna-200c promote colorectal cancer cell proliferation via targeting the reversion-inducing cysteine-rich protein with Kazal motifs Supplementary Table 1.
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1: Quality assessment of formalin-fixed paraffin-embedded (FFPE)-derived DNA and nuclei. (a) Multiplex PCR analysis of unrepaired and repaired bulk FFPE gdna from
Supplementary Figure 1 Supplementary Fig. 1: Quality assessment of formalin-fixed paraffin-embedded (FFPE)-derived DNA and nuclei. (a) Multiplex PCR analysis of unrepaired and repaired bulk FFPE gdna from
Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures
 Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures Supplementary Figure 1 mir-128-3p is highly expressed in chemoresistant, metastatic
Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures Supplementary Figure 1 mir-128-3p is highly expressed in chemoresistant, metastatic
RD-100i OSNA the new generation of sentinel lymph node analysis in breast cancer
 RD-1i OSNA the new generation of sentinel lymph node analysis in breast cancer www.sysmex-europe.com RD-1i OSNA the new generation of sentinel lymph node analysis in breast cancer Sentinel node biopsy
RD-1i OSNA the new generation of sentinel lymph node analysis in breast cancer www.sysmex-europe.com RD-1i OSNA the new generation of sentinel lymph node analysis in breast cancer Sentinel node biopsy
Retro-X qrt-pcr Titration Kit User Manual
 Takara Bio USA Retro-X qrt-pcr Titration Kit User Manual Cat. No. 631453 PT3952-1 (030218) 1290 Terra Bella Avenue, Mountain View, CA 94043, USA U.S. Technical Support: techus@takarabio.com United States/Canada
Takara Bio USA Retro-X qrt-pcr Titration Kit User Manual Cat. No. 631453 PT3952-1 (030218) 1290 Terra Bella Avenue, Mountain View, CA 94043, USA U.S. Technical Support: techus@takarabio.com United States/Canada
Supplementary Materials for
 immunology.sciencemag.org/cgi/content/full/2/16/eaan6049/dc1 Supplementary Materials for Enzymatic synthesis of core 2 O-glycans governs the tissue-trafficking potential of memory CD8 + T cells Jossef
immunology.sciencemag.org/cgi/content/full/2/16/eaan6049/dc1 Supplementary Materials for Enzymatic synthesis of core 2 O-glycans governs the tissue-trafficking potential of memory CD8 + T cells Jossef
Supplementary Figure 1: Experimental design. DISCOVERY PHASE VALIDATION PHASE (N = 88) (N = 20) Healthy = 20. Healthy = 6. Endometriosis = 33
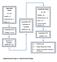 DISCOVERY PHASE (N = 20) Healthy = 6 Endometriosis = 7 EAOC = 7 Quantitative PCR (mirnas = 1113) a Quantitative PCR Verification of Candidate mirnas (N = 24) VALIDATION PHASE (N = 88) Healthy = 20 Endometriosis
DISCOVERY PHASE (N = 20) Healthy = 6 Endometriosis = 7 EAOC = 7 Quantitative PCR (mirnas = 1113) a Quantitative PCR Verification of Candidate mirnas (N = 24) VALIDATION PHASE (N = 88) Healthy = 20 Endometriosis
SUPPLEMENTARY METHODS
 SUPPLEMENTARY METHODS Histological analysis. Colonic tissues were collected from 5 parts of the middle colon on day 7 after the start of DSS treatment, and then were cut into segments, fixed with 4% paraformaldehyde,
SUPPLEMENTARY METHODS Histological analysis. Colonic tissues were collected from 5 parts of the middle colon on day 7 after the start of DSS treatment, and then were cut into segments, fixed with 4% paraformaldehyde,
ANGPTL2 increases bone metastasis of breast cancer cells through. Tetsuro Masuda, Motoyoshi Endo, Yutaka Yamamoto, Haruki Odagiri, Tsuyoshi
 Masuda et al. Supplementary information for ANGPTL2 increases bone metastasis of breast cancer cells through enhancing CXCR4 signaling Tetsuro Masuda, Motoyoshi Endo, Yutaka Yamamoto, Haruki Odagiri, Tsuyoshi
Masuda et al. Supplementary information for ANGPTL2 increases bone metastasis of breast cancer cells through enhancing CXCR4 signaling Tetsuro Masuda, Motoyoshi Endo, Yutaka Yamamoto, Haruki Odagiri, Tsuyoshi
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
The Role of CD164 in Metastatic Cancer Aaron M. Havens J. Wang, Y-X. Sun, G. Heresi, R.S. Taichman Mentor: Russell Taichman
 The Role of CD164 in Metastatic Cancer Aaron M. Havens J. Wang, Y-X. Sun, G. Heresi, R.S. Taichman Mentor: Russell Taichman The spread of tumors, a process called metastasis, is a dreaded complication
The Role of CD164 in Metastatic Cancer Aaron M. Havens J. Wang, Y-X. Sun, G. Heresi, R.S. Taichman Mentor: Russell Taichman The spread of tumors, a process called metastasis, is a dreaded complication
B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small cell lung cancer
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Experimental Methods Cell culture B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Experimental Methods Cell culture B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small
Award Number: W81XWH TITLE: CYP1B1 Polymorphism as a Risk Factor for Race-Related Prostate Cancer
 AD Award Number: W81XWH-04-1-0579 TITLE: CYP1B1 Polymorphism as a Risk Factor for Race-Related Prostate Cancer PRINCIPAL INVESTIGATOR: Yuichiro Tanaka, Ph.D. Rajvir Dahiya, Ph.D. CONTRACTING ORGANIZATION:
AD Award Number: W81XWH-04-1-0579 TITLE: CYP1B1 Polymorphism as a Risk Factor for Race-Related Prostate Cancer PRINCIPAL INVESTIGATOR: Yuichiro Tanaka, Ph.D. Rajvir Dahiya, Ph.D. CONTRACTING ORGANIZATION:
Establishment of Human Cancer Cell Clones with Different Characteristics: A Model for Screening Chemopreventive Agents
 Establishment of Human Cancer Cell Clones with Different Characteristics: A Model for Screening Chemopreventive Agents JEFFREY H. WARE 1, ZHAOZONG ZHOU 1, JUN GUAN 1, ANN R. KENNEDY 1 and LEVY KOPELOVICH
Establishment of Human Cancer Cell Clones with Different Characteristics: A Model for Screening Chemopreventive Agents JEFFREY H. WARE 1, ZHAOZONG ZHOU 1, JUN GUAN 1, ANN R. KENNEDY 1 and LEVY KOPELOVICH
The Association of DCC mrna Alternative Splicing with Colorectal Cancer
 University of Colorado, Boulder CU Scholar Undergraduate Honors Theses Honors Program Spring 2017 The Association of DCC mrna Alternative Splicing with Colorectal Cancer Natalie Graham Natalie.Graham@Colorado.EDU
University of Colorado, Boulder CU Scholar Undergraduate Honors Theses Honors Program Spring 2017 The Association of DCC mrna Alternative Splicing with Colorectal Cancer Natalie Graham Natalie.Graham@Colorado.EDU
Effect of Survivin-siRNA on Drug Sensitivity of Osteosarcoma Cell Line MG-63
 68 Chin J Cancer Res 22(1):68-72, 2010 www.springerlink.com Original Article Effect of Survivin-siRNA on Drug Sensitivity of Osteosarcoma Cell Line MG-63 Jing-Wei Wang 1, Yi Liu 2, Hai-mei Tian 2, Wei
68 Chin J Cancer Res 22(1):68-72, 2010 www.springerlink.com Original Article Effect of Survivin-siRNA on Drug Sensitivity of Osteosarcoma Cell Line MG-63 Jing-Wei Wang 1, Yi Liu 2, Hai-mei Tian 2, Wei
Protocol for A-549 VIM RFP (ATCC CCL-185EMT) TGFβ1 EMT Induction and Drug Screening
 Protocol for A-549 VIM RFP (ATCC CCL-185EMT) TGFβ1 EMT Induction and Drug Screening Introduction: Vimentin (VIM) intermediate filament (IF) proteins are associated with EMT in lung cancer and its metastatic
Protocol for A-549 VIM RFP (ATCC CCL-185EMT) TGFβ1 EMT Induction and Drug Screening Introduction: Vimentin (VIM) intermediate filament (IF) proteins are associated with EMT in lung cancer and its metastatic
Nature Medicine doi: /nm.3957
 Supplementary Fig. 1. p38 alternative activation, IL-21 expression, and T helper cell transcription factors in PDAC tissue. (a) Tissue microarrays of pancreatic tissue from 192 patients with pancreatic
Supplementary Fig. 1. p38 alternative activation, IL-21 expression, and T helper cell transcription factors in PDAC tissue. (a) Tissue microarrays of pancreatic tissue from 192 patients with pancreatic
Circular RNA_LARP4 is lower expressed and serves as a potential biomarker of ovarian cancer prognosis
 European Review for Medical and Pharmacological Sciences 2018; 22: 7178-7182 Circular RNA_LARP4 is lower expressed and serves as a potential biomarker of ovarian cancer prognosis T. ZOU, P.-L. WANG, Y.
European Review for Medical and Pharmacological Sciences 2018; 22: 7178-7182 Circular RNA_LARP4 is lower expressed and serves as a potential biomarker of ovarian cancer prognosis T. ZOU, P.-L. WANG, Y.
SUPPLEMENTARY INFORMATION. Divergent TLR7/9 signaling and type I interferon production distinguish
 SUPPLEMENTARY INFOATION Divergent TLR7/9 signaling and type I interferon production distinguish pathogenic and non-pathogenic AIDS-virus infections Judith N. Mandl, Ashley P. Barry, Thomas H. Vanderford,
SUPPLEMENTARY INFOATION Divergent TLR7/9 signaling and type I interferon production distinguish pathogenic and non-pathogenic AIDS-virus infections Judith N. Mandl, Ashley P. Barry, Thomas H. Vanderford,
York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
 MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
SUPPLEMENTAL MATERIAL. Supplementary Methods
 SUPPLEMENTAL MATERIAL Supplementary Methods Culture of cardiomyocytes, fibroblasts and cardiac microvascular endothelial cells The isolation and culturing of neonatal rat ventricular cardiomyocytes was
SUPPLEMENTAL MATERIAL Supplementary Methods Culture of cardiomyocytes, fibroblasts and cardiac microvascular endothelial cells The isolation and culturing of neonatal rat ventricular cardiomyocytes was
Supplementary Figure 1. Validation of astrocytes. Primary astrocytes were
 Supplementary Figure 1. Validation of astrocytes. Primary astrocytes were separated from the glial cultures using a mild trypsinization protocol. Anti-glial fibrillary acidic protein (GFAP) immunofluorescent
Supplementary Figure 1. Validation of astrocytes. Primary astrocytes were separated from the glial cultures using a mild trypsinization protocol. Anti-glial fibrillary acidic protein (GFAP) immunofluorescent
The Relationship Between Bone Metastasis from Human Breast Cancer and Integrin v 3 Expression
 The Relationship Between Bone Metastasis from Human Breast Cancer and Integrin v 3 Expression SHIN TAKAYAMA 1, SEIICHIRO ISHII 2, TADASHI IKEDA 1, SHIGERU MASAMURA 1, MASAKAZU DOI 1 and MASAKI KITAJIMA
The Relationship Between Bone Metastasis from Human Breast Cancer and Integrin v 3 Expression SHIN TAKAYAMA 1, SEIICHIRO ISHII 2, TADASHI IKEDA 1, SHIGERU MASAMURA 1, MASAKAZU DOI 1 and MASAKI KITAJIMA
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A.
 Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
Supplementary Figure 1. Genotyping strategies for Mcm3 +/+, Mcm3 +/Lox and Mcm3 +/- mice and luciferase activity in Mcm3 +/Lox mice. A. Upper part, three-primer PCR strategy at the Mcm3 locus yielding
RD-100i OSNA the new generation of sentinel lymph node analysis in breast cancer
 RD-1i OSNA the new generation of sentinel lymph node analysis in breast cancer RD-1i OSNA the new generation of sentinel lymph node analysis in breast cancer Sentinel node biopsy has rapidly emerged as
RD-1i OSNA the new generation of sentinel lymph node analysis in breast cancer RD-1i OSNA the new generation of sentinel lymph node analysis in breast cancer Sentinel node biopsy has rapidly emerged as
Gastric Carcinoma with Lymphoid Stroma: Association with Epstein Virus Genome demonstrated by PCR
 Gastric Carcinoma with Lymphoid Stroma: Association with Epstein Virus Genome demonstrated by PCR Pages with reference to book, From 305 To 307 Irshad N. Soomro,Samina Noorali,Syed Abdul Aziz,Suhail Muzaffar,Shahid
Gastric Carcinoma with Lymphoid Stroma: Association with Epstein Virus Genome demonstrated by PCR Pages with reference to book, From 305 To 307 Irshad N. Soomro,Samina Noorali,Syed Abdul Aziz,Suhail Muzaffar,Shahid
Supplementary Data Supplementary Table S1. IC 50 values of ipatasertib on cell viability in cell lines with
 Supplementary Data Supplementary Table S1. IC 50 values of ipatasertib on cell viability in cell lines with or without alterations in PTEN or PIK3CA. Cell Line Tissue Type Ipatasertib IC 50 (μmol/l) a
Supplementary Data Supplementary Table S1. IC 50 values of ipatasertib on cell viability in cell lines with or without alterations in PTEN or PIK3CA. Cell Line Tissue Type Ipatasertib IC 50 (μmol/l) a
Supplemental Table S1
 Supplemental Table S. Tumorigenicity and metastatic potential of 44SQ cell subpopulations a Tumorigenicity b Average tumor volume (mm ) c Lung metastasis d CD high /4 8. 8/ CD low /4 6./ a Mice were injected
Supplemental Table S. Tumorigenicity and metastatic potential of 44SQ cell subpopulations a Tumorigenicity b Average tumor volume (mm ) c Lung metastasis d CD high /4 8. 8/ CD low /4 6./ a Mice were injected
SUPPORTING MATREALS. Methods and Materials
 SUPPORTING MATREALS Methods and Materials Cell Culture MC3T3-E1 (subclone 4) cells were maintained in -MEM with 10% FBS, 1% Pen/Strep at 37ºC in a humidified incubator with 5% CO2. MC3T3 cell differentiation
SUPPORTING MATREALS Methods and Materials Cell Culture MC3T3-E1 (subclone 4) cells were maintained in -MEM with 10% FBS, 1% Pen/Strep at 37ºC in a humidified incubator with 5% CO2. MC3T3 cell differentiation
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 EGFR inhibition activates signaling pathways (a-b) EGFR inhibition activates signaling pathways (a) U251EGFR cells were treated with erlotinib (1µM) for the indicated times followed
Supplementary Figure 1 EGFR inhibition activates signaling pathways (a-b) EGFR inhibition activates signaling pathways (a) U251EGFR cells were treated with erlotinib (1µM) for the indicated times followed
Detection of let-7a MicroRNA by Real-time PCR in Colorectal Cancer: a Single-centre Experience from China
 The Journal of International Medical Research 2007; 35: 716 723 Detection of let-7a MicroRNA by Real-time PCR in Colorectal Cancer: a Single-centre Experience from China W-J FANG 1, C-Z LIN 2, H-H ZHANG
The Journal of International Medical Research 2007; 35: 716 723 Detection of let-7a MicroRNA by Real-time PCR in Colorectal Cancer: a Single-centre Experience from China W-J FANG 1, C-Z LIN 2, H-H ZHANG
Electronic Supplementary Information
 Electronic Supplementary Material (ESI) for Journal of Materials Chemistry B. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information Single mammalian cell encapsulation
Electronic Supplementary Material (ESI) for Journal of Materials Chemistry B. This journal is The Royal Society of Chemistry 2016 Electronic Supplementary Information Single mammalian cell encapsulation
Identification of Novel Variant of EML4-ALK Fusion Gene in NSCLC: Potential Benefits of the RT-PCR Method
 International journal of Biomedical science ORIGINAL ARTICLE Identification of Novel Variant of EML4-ALK Fusion Gene in NSCLC: Potential Benefits of the RT-PCR Method Martin K. H. Maus 1, 2, Craig Stephens
International journal of Biomedical science ORIGINAL ARTICLE Identification of Novel Variant of EML4-ALK Fusion Gene in NSCLC: Potential Benefits of the RT-PCR Method Martin K. H. Maus 1, 2, Craig Stephens
Influenza A viruses Detection with real time RT-PCR reagents
 Influenza A viruses Detection with real time RT-PCR reagents Overview:... 1 Products... 2 Influenza A matix FAM-BHQ1 PP500 0.055ml... 2 Influenza A Plasmid 200 pg/ml PLAS500 0.25ml... 2 Detection Influenza
Influenza A viruses Detection with real time RT-PCR reagents Overview:... 1 Products... 2 Influenza A matix FAM-BHQ1 PP500 0.055ml... 2 Influenza A Plasmid 200 pg/ml PLAS500 0.25ml... 2 Detection Influenza
Supplementary Figure (OH) 22 nanoparticles did not affect cell viability and apoposis. MDA-MB-231, MCF-7, MCF-10A and BT549 cells were
 Supplementary Figure 1. Gd@C 82 (OH) 22 nanoparticles did not affect cell viability and apoposis. MDA-MB-231, MCF-7, MCF-10A and BT549 cells were treated with PBS, Gd@C 82 (OH) 22, C 60 (OH) 22 or GdCl
Supplementary Figure 1. Gd@C 82 (OH) 22 nanoparticles did not affect cell viability and apoposis. MDA-MB-231, MCF-7, MCF-10A and BT549 cells were treated with PBS, Gd@C 82 (OH) 22, C 60 (OH) 22 or GdCl
