Late assembly of the Vibrio cholerae cell division machinery postpones
|
|
|
- Hugo Eaton
- 5 years ago
- Views:
Transcription
1 2 Late assembly of the Vibrio cholerae cell division machinery postpones septation to the last % of the cell cycle 3 4 Elisa Galli, Evelyne Paly and François-Xavier Barre,# Institute for Integrative Biology of the Cell (I2BC), Université Paris-Saclay, CEA, CRS, Université Paris Sud, France
2 Supplementary Figure. a. Cells were grown overnight in LB at 3 C. After normalization for cell density, each culture was serially diluted. 5 μl of each dilution was spotted on a LB plate incubated overnight either at 3 C or at 42 C and photographed. b. Phase-contrast images of V. cholerae 696 cells carrying the ftsz ts or ftsi ts allele after h and 3min growth at 42 C. c. Combined phasecontrast and fluorescent image of ftsi ts V. cholerae cells ectopically expressing YGFP-FtsI at 42 C. Scale bar = 2 μm. Supplementary Figure 2. a. Cellular localisation of FtsZ-RFPT, YGFP-FtsA, RFPT-ZapA, FtsK-YGFP, YGFP-FtsL, sfgfp-ftsi, sfgfp-fts, DsbAss-mCherry- SPR and HubP-sfGFP in V. cholerae 696 cells. Scale bar = 2 μm. b. Phasecontrast image and cellular localisation of YGFP-FtsA, RFPT-ZapA, FtsK-YGFP, YGFP-FtsL, sfgfp-ftsi and sfgfp-fts in cephalexin treated cells. Scale bar = 2 μm. c. Percentage of minicells formed at each division in LB as determined from 2 independent time-lapse experiments. hubp-sfgfp is integrated at hubp native locus and expressed from the native promoter. Error bars represent standard deviation. d. Cellular localisation of HubP-YFP ectopically expressed from a PBAD promoter in addition to hubp wild-type copy. Scale bar = 2 μm. e. Demograph of V. cholerae 696 cells ectopically expressing HubP-YFP from a PBAD promoter in addition to hubp wild-type copy. Cells were not oriented. Supplementary Figure 3. Theoretical deduction of the cell cycle advancement of cells using their length. a. V. cholerae cell length increases exponentially with cell cycle. Log scale representation of cell length as a function of cell cycle advancement for individual V. cholerae cells grown on a % (w/v) agarose pad in M9 minimal medium,.2% fructose and μg/ml thiamine. Blue circles: results for 25 complete lineages, i.e. for cells in which both birth and division were 2
3 observed; red line: median log of the length as a function of the cell cycle; black line: linear regression; M: length at division; m: length at birth. b. Scheme depicting the relationship between cell cycle advancement and cell length. At 5% of the cell cycle, cell elongation is smaller than half of the total elongation from birth to division. M; division length; m: birth length; (M+m)/2: half of the total elongation from birth to division. c. Cell length distribution of 33 V. cholerae cells ectopically producing a fluorescently tagged copy of FtsZ. M: largest observed cell length; m: smallest observed cell length. Marks showing the theoretical cell cycle stages are shown as black (.-.4,.6-.9) and red (,.,.5,.,) ticks. Tick positions were determined using the exponential relationship between cell length and cell cycle and assuming that M and m correspond to the division length and the birth length. d. FtsZ-localisation in demograph representation. Marks showing the theoretical cell cycle stages are shown as black (.2-.4,.6,.8) and red (,.,.5,.,) ticks. Each cell cycle stage tick is located at the projection profile of the longest cell whose length is below the theoretical length reached at this particular stage. e. Cell length distribution of 784 V. cholerae cells ectopically producing a fluorescently tagged copy of FtsK. M: largest observed cell length; m: smallest observed cell length. Marks showing the theoretical cell cycle stages are shown as black (.-.4,.6-.9) and red (,.,.5,.,) ticks. Tick positions were determined using the exponential relationship between cell length and cell cycle and assuming that M and m correspond to the division length and the birth length. f. FtsK-localisation in demograph representation. Marks showing the theoretical cell cycle stages are shown as black (.2-.4,.6,.8) and red (,.,.5,.,) ticks. Each cell cycle stage tick is located at the projection profile of the longest cell whose length is 3
4 below the theoretical length reached at this particular stage. Supplementary Figure 4. Examples of time-lapse individual lineages in the and representation., old pole;, new pole. a. FtsZ-RFPT. b. YGFP- FtsA. c. RFPT-ZapA. d. FtsK-YGFP. e. DsbAss-mCherry-SPR. Supplementary Figure 5. a. cell cycle distribution of DsbAss-mCherry- SPR (compilation of 44 single cell cycles). b. Examples (i, ii) of time-lapse images of 696 cells expressing YGFP-FtsK and DsbAss-mCherry-SPR. ne frame was taken every 3 minutes. n the top-right corner of each frame is indicated the time in minutes from the beginning of imaging. White arrows show the arrival of tagged proteins to mid-cell. Scale bar = 2 μm. Supplementary Figure 6. a. cell cycle distribution of HubP-sfGFP (compilation of 88 single cell cycles), YGFP-ParB (compilation of 69 single cell cycles) and oric locus (compilation of 5 single cell cycles). b. cell cycle distribution of YGFP-FtsK (compilation of 73 single cell cycles) in V. cholerae 696 ΔhubP cells. c. Time-lapse images of 696 cells expressing DsbAssmCherry-SPR and HubP-YFP, ectopically expressed from a PBAD promoter in addition to hubp wild-type copy. ne frame was taken every 4 minutes. n the top-right corner of each frame is indicated the time in minutes from the beginning of imaging. White arrows mark proteins arrival at the division sites. Scale bar = 2 μm. Supplementary Figure 7. Examples of time-lapse individual lineages in the and representation., old pole;, new pole. a. HubP-sfGFP. b. YGFP- ParB. c. Movement of oric followed by inserting a lac array in its proximity and expressing a LacI-YGFP fusion. Supplementary Table. List of strains and plasmids used in this study. 4
5 Movie. Examples of FtsZ-RFPT time-lapses in V. cholerae cells. ne frame was taken every 3 minutes. Cells were grown in M9 minimal medium supplemented with.2% fructose and μg/ml thiamine at 3 C. Movies start from the beginning of imaging. Movie 2. Time-lapse of FtsZ-RFPT and FtsK-YGFP localisation in V. cholerae cells. ne frame was taken every 4 minutes. Cells were grown in M9 minimal medium supplemented with.2% fructose and μg/ml thiamine at 3 C. μg/ml cephalexin was added to the agarose slide. Movie 3. Time-lapse of FtsK-YGFP and DsbAss-mCherry-SPR localisation in V. cholerae cells. ne frame was taken every 3 minutes. Cells were grown in M9 minimal medium supplemented with.2% fructose and μg/ml thiamine at 3 C. μg/ml cephalexin was added to the agarose slide. Movie 4. Time-lapse of FtsK-YGFP and HubP-RFPT localisation in V. cholerae cells. ne frame was taken every 3 minutes. Cells were grown in M9 minimal medium supplemented with.2% fructose and μg/ml thiamine at 3 C. μg/ml cephalexin was added to the agarose slide. Movie 5. Time-lapse of DsbAss-mCherry-SPR and HubP-sfGFP localisation in V. cholerae cells. ne frame was taken every 3 minutes. Cells were grown in M9 minimal medium supplemented with.2% fructose and μg/ml thiamine at 3 C. μg/ml cephalexin was added to the agarose slide. Movie 6. Time-lapse of DsbAss-mCherry-SPR and HubP-YFP localisation in V. cholerae cells. HubP-YFP was ectopically expressed from a PBAD promoter in addition to hubp wild-type copy. ne frame was taken every 4 minutes. Cells were grown in M9 minimal medium supplemented with.2% fructose and μg/ml thiamine at 3 C. μg/ml cephalexin was added to the agarose slide. 5
6 a b dilutions ftsz ts 42 C wt 3 C ftsz ts ftsi ts ftsi ts 42 C wt ftsz ts ftsi ts c 42 C ftsi ts + YGFP-FtsI 42 C Supplementary Figure
7 a FtsZ FtsA ZapA FtsK FtsL FtsI Fts SPR HubP A b Cephalexin-treated V. cholerae cells FtsA FtsK Phase contrast ZapA FtsL d c e 5 5 minicell per divisions (%) Pbad HubP-YFP FtsI Cell length Supplementary Figure 2 Fts Pbad HubP-YFP
8 a M b M Cell Length (log) Cell Length m+m 2 m..5. Cell Cycle m..5. Cell Cycle c 8 FtsZ d..3 FtsZ number of cells Cell Cycle.4.5 m Cell Length M Cell Cycle..8 e f FtsK 2 FtsK..3 number of cells Cell Cycle.4.5 m Cell Length M..5. Cell Cycle.6..8 Supplementary Figure 3
9 a FtsZ b FtsA c ZapA d FtsK e SPR Cell cycle progression (%) Cell cycle progression (%) Cell cycle progression (%) Cell cycle progression (%) Cell cycle progression (%) Supplementary Figure 4
10 a SPR, IStime Cell length 5 Cell cycle SPR FtsK b i SPR FtsK ii Supplementary Figure
11 a HubP, IStime ParB, IStime Cell length oric, IStime Cell cycle..5.. Cell cycle..5 Cell cycle FtsK in hubp, IScycle b Cell length. c..5 Cell cycle Pbad:: HubP SPR. Supplementary Figure
12 a HubP b ParB c oric Cell cycle progression (%) Cell cycle progression (%) Cell cycle progression (%) Cell cycle progression (%) Cell cycle progression (%) Supplementary Figure 7
13 Supplementary Table. List of bacterial strains and plasmids Strains ame Relevant genotype or features Reference EGV 696 ChapR ΔlacZ::(P BAD ::ftsz-rfpt-sh ble) zeo R gm R () EGV 696 ChapR ΔlacZ::(P BAD ::RFPT-zapA-Sh ble) zeo R gm R This study EGV ΔlacZ::(P BAD ::ftsz-rfpt-sh ble) ΔhapR::(P lac ::laci- YGFP-cat) + lac array-aph inserted on chr at position.5 Mb (53355 bp) (oric) zeo R gm R cml R kan R This study EGV ChapR ΔlacZ P ftsk ::ftsk-ygfp-sh ble zeo R gm R () EGV ΔlacZ P ftsk ::ftsk-ygfp-sh ble ΔhapR::(P BAD ::ftsz-rfptcat) () R zeo R gm R cml EGV ChapR ΔlacZ mincd::sh ble zeo R gm R () EGV ΔlacZ::(P BAD ::YGFP-ftsA-Sh ble) zeo R gm R This study EGV ChapR ΔlacZ::(P lac ::YGFP-parB-Sh ble) zeo R gm R This study EGV ChapR ΔlacZ::(P BAD ::YGFP-ftsL-Sh ble) zeo R gm R This study EGV 696 ChapR ΔlacZ ΔhubP mincd::sh ble zeo R gm R () EGV ChapR ΔlacZ::(P BAD ::dsba ss -mcherry-spr-sh ble) This study SPR domain of E. coli Fts (Fts[243-39]) (signal sequence from DsbA) zeo R gm R EGV ChapR ΔlacZ::(P BAD ::hubp-yfp-sh ble) zeo R gm R This study EGV ChapR ΔlacZ::(P BAD ::RFPT-zapA-Sh ble) carrying the ftsz This study ts chromosomal mutation (G6S) zeo R gm R EGV ChapR ΔlacZ P ftsk ::ftsk-ygfp-sh ble carrying the ftsz ts This study chromosomal mutation (G6S) zeo R rif R gm R EGV ChapR ΔlacZ::(P BAD ::YGFP-ftsA-Sh ble) carrying the ftsz This study ts chromosomal mutation (G6S) zeo R gm R EGV ChapR ΔlacZ::(P BAD ::YGFP-ftsL-Sh ble) carrying the ftsz This study ts chromosomal mutation (G6S) zeo R gm R EGV ChapR ΔlacZ::(P BAD ::YGFP-ftsI-Sh ble) carrying the ftsz This study ts chromosomal mutation (G6S) zeo R gm R EGV ChapR ΔlacZ::(P BAD ::dsba ss -mcherry-spr-sh ble) carrying the ftsz ts chromosomal mutation (G6S) SPR domain of E. coli Fts (Fts[243-39]) (signal sequence from DsbA) zeo R gm R This study EGV ΔlacZ P ftsk ::ftsk-ygfp-sh ble ΔhapR::(P BAD ::dsba ss - mcherry-spr-cat) SPR domain of E. coli Fts (Fts[243-39]) (signal sequence from DsbA) zeo R gm R cml R EGV ΔlacZ::(P BAD ::hubp-yfp-sh ble) ΔhapR::(P BAD ::dsba ss - mcherry-spr-cat) SPR domain of E. coli Fts (Fts[243-39]) (signal sequence from DsbA) zeo R gm R cml R This study This study EGV ChapR ΔlacZ P ftsk ::ftsk-ygfp-sh ble P hubp ::hubp- This study RFPT-aadA zeo R spec R gm R EGV ChapR ΔlacZ P hubp ::hubp-rfpt-aada carrying the ftsz ts This study chromosomal mutation (G6S) spec R rif R gm R EGV ChapR ΔlacZ::(P lac ::sfgfp-fts-sh ble) carrying the ftsz This study ts chromosomal mutation (G6S) zeo R gm R EGV ChapR ΔlacZ ΔhubP P ftsk ::ftsk-ygfp-sh ble zeo R gm R This study EPV5 696 ChapR ΔlacZ gm R (2) EPV ChapR ΔlacZ carrying the ftsz ts chromosomal mutation () (G6S) rif R gm R EPV ChapR ΔlacZ::(P lac ::sfgfp-fts-sh ble) zeo R gm R This study EPV432 EPV437 EPV ChapR ΔlacZ carrying the ftsi ts chromosomal mutation (Y388D) rif R gm R 696 ChapR ΔlacZ::(P BAD ::ftsz-rfpt-sh ble) carrying the ftsi ts chromosomal mutation (Y388D) zeo R gm R 696 ChapR ΔlacZ::(P BAD ::dsba ss -mcherry-spr-sh ble) carrying the ftsi ts chromosomal mutation (Y388D) SPR domain This study This study This study
14 of E. coli Fts (Fts[243-39]) (signal sequence from DsbA) zeo R gm R EPV ChapR ΔlacZ P hubp ::hubp-rfpt-aada carrying the ftsi ts This study chromosomal mutation (Y388D) spec R rif R gm R EPV ChapR ΔlacZ::(P lac ::sfgfp-ftsi-sh ble) zeo R gm R This study EPV ChapR ΔlacZ::(P lac ::sfgfp-ftsi-sh ble) carrying the FtsI ts This study chromosomal mutation (Y388D) zeo R gm R EPV ChapR ΔlacZ P hubp ::hubp-sfgfp gm R This study EPV ChapR P hubp ::hubp-sfgfp ΔlacZ::(P BAD ::dsba ss -mcherry- This study SPR-Sh ble) SPR domain of E. coli Fts (Fts[243-39]) (signal sequence from DsbA) zeo R gm R EPV ChapR ΔlacZ P hubp ::hubp-sfgfp mincd::sh ble zeo R gm R This study Plasmids (2) () () ame Relevant genotype or features Reference pad2 integration-excision vector; Tet -lac array-aph - Tet; sacb; ori R6K; cml R, kan R peg233 P BAD ::ftsz-rfpt-sh ble flanked by the upstream and downstream peg234 P BAD ::RFPT-zapA-Sh ble flanked by the upstream and downstream peg242 P BAD ::YGFP-ftsI-Sh ble flanked by the upstream and downstream This study peg245 P lac ::laci-ygfp-cat flanked by the upstream and downstream () regions of hapr; ori puc; amp R, cml R peg248 ftsk-ygfp-sh ble flanked by the upstream and downstream regions () of ftsk; ori psc; amp R, zeo R peg2 Sh ble flanked by the upstream and downstream regions of mincd; () ori puc; amp R, zeo R peg266 Sh ble flanked by the upstream and downstream regions of hubp; This study ori puc; amp R, zeo R peg27 P BAD ::YGFP-ftsA-Sh ble flanked by the upstream and downstream This study peg282 P lac ::YGFP-parB-Sh ble flanked by the upstream and downstream This study peg287 P BAD ::YGFP-ftsL-Sh ble flanked by the upstream and downstream This study peg323 P BAD ::dsba ss -mcherry-spr-sh ble flanked by the upstream and This study downstream regions of lacz; SPR domain of E. coli Fts (Fts[243-39]) (signal sequence from DsbA); ori puc; amp R, zeo R peg324 P BAD ::hubp-yfp-sh ble flanked by the upstream and downstream This study peg392 P BAD ::dsba ss -mcherry-spr-cat flanked by the upstream and This study downstream regions of hapr; SPR domain of E. coli Fts (Fts[243-39]) (signal sequence from DsbA); ori puc; amp R, cml R peg396 P lac ::sfgfp-fts-sh ble flanked by the upstream and downstream This study peg398 P lac ::sfgfp-ftsi-sh ble flanked by the upstream and downstream This study peg4 integration-excision vector; hubp-rfpt-aada at native locus; This study sacb; ori R6K; amp R, spec R peyy24 integration-excision vector; hubp-sfgfp at native locus; sacb; ori R6K; amp R Y. Yamaichi gift
15 References. Galli E, Poidevin M, Le Bars R, Desfontaines J-M, Muresan L, Paly E, Yamaichi Y, Barre F-X. 26. Cell division licensing in the multichromosomal Vibrio cholerae bacterium. at Microbiol : David A, Demarre G, Muresan L, Paly E, Barre F-X, Possoz C. 24. The two Cis-acting sites, pars and oric, contribute to the longitudinal organisation of Vibrio cholerae chromosome I. PLoS Genet :e4448.
Table S1. Strains, Plasmids and Phage
 Table S1. Strains, Plasmids and Phage Strain, Plasmid, Phage Genotype or Description Reference S. enterica 14028 wild type ATCC strain (1) HK132 yfin; 14028 with deletion of yfin This study QW262 yhjh
Table S1. Strains, Plasmids and Phage Strain, Plasmid, Phage Genotype or Description Reference S. enterica 14028 wild type ATCC strain (1) HK132 yfin; 14028 with deletion of yfin This study QW262 yhjh
klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach
![klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach](/thumbs/91/104639484.jpg) DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
Nature Methods: doi: /nmeth.4257
 Supplementary Figure 1 Screen for polypeptides that affect cellular actin filaments. (a) Table summarizing results from all polypeptides tested. Source shows organism, gene, and amino acid numbers used.
Supplementary Figure 1 Screen for polypeptides that affect cellular actin filaments. (a) Table summarizing results from all polypeptides tested. Source shows organism, gene, and amino acid numbers used.
Real-time acquisition of drug resistance by DNA transfer in live cells
 eal-time acquisition of drug resistance by DNA transfer in live cells Christian LESELIN (INSEM-CNS) Cell-to-cell DNA transfer team (AIP-Avenir FINOVI) MMSB - Molecular Microbiology and Structural Biochemistry
eal-time acquisition of drug resistance by DNA transfer in live cells Christian LESELIN (INSEM-CNS) Cell-to-cell DNA transfer team (AIP-Avenir FINOVI) MMSB - Molecular Microbiology and Structural Biochemistry
Nature Genetics: doi: /ng Supplementary Figure 1. HOX fusions enhance self-renewal capacity.
 Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Gene expression scaled by distance to the genome replication site. Ying et al. Supporting Information. Contents. Supplementary note I p.
 Gene expression scaled by distance to the genome replication site Ying et al Supporting Information Contents Supplementary note I p. 2 Supplementary note II p. 3-5 Supplementary note III p. 5-7 Supplementary
Gene expression scaled by distance to the genome replication site Ying et al Supporting Information Contents Supplementary note I p. 2 Supplementary note II p. 3-5 Supplementary note III p. 5-7 Supplementary
Supporting Information
 Supporting Information Keller et al. 10.1073/pnas.1222607110 MB1 Ori puc18 Ori AscI (8713) pdad downstream flank pdad PpdaD SphI (6970) Msed1993 ApaI (10721) Apr ApaLI (9115) NcoI (8) PmeI (8796) PvuI
Supporting Information Keller et al. 10.1073/pnas.1222607110 MB1 Ori puc18 Ori AscI (8713) pdad downstream flank pdad PpdaD SphI (6970) Msed1993 ApaI (10721) Apr ApaLI (9115) NcoI (8) PmeI (8796) PvuI
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human
 Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
Supplementary Figure 1. Ent inhibits LPO activity in a dose- and time-dependent fashion.
 Supplementary Information Supplementary Figure 1. Ent inhibits LPO activity in a dose- and time-dependent fashion. Various concentrations of Ent, DHBA or ABAH were pre-incubated for 10 min with LPO (50
Supplementary Information Supplementary Figure 1. Ent inhibits LPO activity in a dose- and time-dependent fashion. Various concentrations of Ent, DHBA or ABAH were pre-incubated for 10 min with LPO (50
Supplemental Material for. Figure S1. Identification of TetR responsive promoters in F. novicida and E. coli.
 Supplemental Material for Synthetic promoters functional in Francisella novicida and Escherichia coli Ralph L. McWhinnie and Francis E. Nano Department of Biochemistry and Microbiology, University of Victoria,
Supplemental Material for Synthetic promoters functional in Francisella novicida and Escherichia coli Ralph L. McWhinnie and Francis E. Nano Department of Biochemistry and Microbiology, University of Victoria,
Supplementary table 1
 Supplementary table 1 S. pombe strain list Fig. 1A JX38 h + ade6-m216 nda3-km311 PX476 PW775 PX545 PX546 h- ade6-m216 sgo2::ura4 + nda3-km311 h 9 mad2::ura4 + nda3-km311 h + ade6-m21 nda3-km311 rad21 +
Supplementary table 1 S. pombe strain list Fig. 1A JX38 h + ade6-m216 nda3-km311 PX476 PW775 PX545 PX546 h- ade6-m216 sgo2::ura4 + nda3-km311 h 9 mad2::ura4 + nda3-km311 h + ade6-m21 nda3-km311 rad21 +
Nature Immunology: doi: /ni Supplementary Figure 1. DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells.
 Supplementary Figure 1 DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells. (a) Scheme for the retroviral shrna screen. (b) Histogram showing CD4 expression (MFI) in WT cytotoxic
Supplementary Figure 1 DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells. (a) Scheme for the retroviral shrna screen. (b) Histogram showing CD4 expression (MFI) in WT cytotoxic
Global variation in copy number in the human genome
 Global variation in copy number in the human genome Redon et. al. Nature 444:444-454 (2006) 12.03.2007 Tarmo Puurand Study 270 individuals (HapMap collection) Affymetrix 500K Whole Genome TilePath (WGTP)
Global variation in copy number in the human genome Redon et. al. Nature 444:444-454 (2006) 12.03.2007 Tarmo Puurand Study 270 individuals (HapMap collection) Affymetrix 500K Whole Genome TilePath (WGTP)
genome edited transient transfection, CMV promoter
 Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
TetR repressor-based bioreporters for the detection of doxycycline using Escherichia
 Supplementary materials TetR repressor-based bioreporters for the detection of doxycycline using Escherichia coli and Acinetobacter oleivorans Hyerim Hong and Woojun Park * Department of Environmental
Supplementary materials TetR repressor-based bioreporters for the detection of doxycycline using Escherichia coli and Acinetobacter oleivorans Hyerim Hong and Woojun Park * Department of Environmental
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
Supplementary information. The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic. Spindle Orientation
 Supplementary information The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic Spindle Orientation Running title: Dynein LICs distribute mitotic functions. Sagar Mahale a, d, *, Megha
Supplementary information The Light Intermediate Chain 2 Subpopulation of Dynein Regulates Mitotic Spindle Orientation Running title: Dynein LICs distribute mitotic functions. Sagar Mahale a, d, *, Megha
Overview of the Expressway Cell-Free Expression Systems. Expressway Mini Cell-Free Expression System
 Overview of the Expressway Cell-Free Expression Systems The Expressway Cell-Free Expression Systems use an efficient coupled transcription and translation reaction to produce up to milligram quantities
Overview of the Expressway Cell-Free Expression Systems The Expressway Cell-Free Expression Systems use an efficient coupled transcription and translation reaction to produce up to milligram quantities
Use of double- stranded DNA mini- circles to characterize the covalent topoisomerase- DNA complex
 SUPPLEMENTARY DATA Use of double- stranded DNA mini- circles to characterize the covalent topoisomerase- DNA complex Armêl Millet 1, François Strauss 1 and Emmanuelle Delagoutte 1 1 Structure et Instabilité
SUPPLEMENTARY DATA Use of double- stranded DNA mini- circles to characterize the covalent topoisomerase- DNA complex Armêl Millet 1, François Strauss 1 and Emmanuelle Delagoutte 1 1 Structure et Instabilité
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Lu et al., http://www.jcb.org/cgi/content/full/jcb.201012063/dc1 Figure S1. Kinetics of nuclear envelope assembly, recruitment of Nup133
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Lu et al., http://www.jcb.org/cgi/content/full/jcb.201012063/dc1 Figure S1. Kinetics of nuclear envelope assembly, recruitment of Nup133
Nature Genetics: doi: /ng Supplementary Figure 1. Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms.
 Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Chen et al., http://www.jcb.org/cgi/content/full/jcb.201210119/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Lack of fast reversibility of UVR8 dissociation. (A) HEK293T
Supplemental material Chen et al., http://www.jcb.org/cgi/content/full/jcb.201210119/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Lack of fast reversibility of UVR8 dissociation. (A) HEK293T
Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal
 Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal hydrocephalus Fadel TISSIR, Yibo QU, Mireille MONTCOUQUIOL, Libing ZHOU, Kouji KOMATSU, Dongbo SHI, Toshihiko FUJIMORI,
Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal hydrocephalus Fadel TISSIR, Yibo QU, Mireille MONTCOUQUIOL, Libing ZHOU, Kouji KOMATSU, Dongbo SHI, Toshihiko FUJIMORI,
p.r623c p.p976l p.d2847fs p.t2671 p.d2847fs p.r2922w p.r2370h p.c1201y p.a868v p.s952* RING_C BP PHD Cbp HAT_KAT11
 ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
Problem 2 (10 Points) The wild type sequence of the coding region of an mrna is shown below.
 Problem 2 (10 Points) The wild type sequence of the coding region of an mrna is shown below. 5 AUG ACC UGG AAU AAA UGA 3 Use that sequence to answer each of the questions below. a) What is the sequence
Problem 2 (10 Points) The wild type sequence of the coding region of an mrna is shown below. 5 AUG ACC UGG AAU AAA UGA 3 Use that sequence to answer each of the questions below. a) What is the sequence
Supplementary Figure S1 (a) (b)
 Supplementary Figure S1: IC87114 does not affect basal Ca 2+ level nor nicotineinduced Ca 2+ influx. (a) Bovine chromaffin cells were loaded with Fluo-4AM (1 μm) in buffer A containing 0.02% of pluronic
Supplementary Figure S1: IC87114 does not affect basal Ca 2+ level nor nicotineinduced Ca 2+ influx. (a) Bovine chromaffin cells were loaded with Fluo-4AM (1 μm) in buffer A containing 0.02% of pluronic
J. A. Mayfield et al. FIGURE S1. Methionine Salvage. Methylthioadenosine. Methionine. AdoMet. Folate Biosynthesis. Methylation SAH.
 FIGURE S1 Methionine Salvage Methionine Methylthioadenosine AdoMet Folate Biosynthesis Methylation SAH Homocysteine Homocystine CBS Cystathionine Cysteine Glutathione Figure S1 Biochemical pathway of relevant
FIGURE S1 Methionine Salvage Methionine Methylthioadenosine AdoMet Folate Biosynthesis Methylation SAH Homocysteine Homocystine CBS Cystathionine Cysteine Glutathione Figure S1 Biochemical pathway of relevant
Predicted membrane topologies of members of the TMBIM family. N and C terminally tagged alleles localise at the Golgi.
 Supplementary fig. 1. Predicted membrane topologies of members of the TMBIM family. Multiple amino acid sequence alignment of members of the TMBIM family generated with Clustal W, with transmembrane regions
Supplementary fig. 1. Predicted membrane topologies of members of the TMBIM family. Multiple amino acid sequence alignment of members of the TMBIM family generated with Clustal W, with transmembrane regions
Supplementary Information. Supplementary Figure 1
 Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11306 Supplementary Figures Supplementary Figure 1. Basic characterization of GFP+ RGLs in the dentate gyrus of adult nestin-gfp mice. a, Sample confocal images
SUPPLEMENTARY INFORMATION doi:10.1038/nature11306 Supplementary Figures Supplementary Figure 1. Basic characterization of GFP+ RGLs in the dentate gyrus of adult nestin-gfp mice. a, Sample confocal images
Supplementary Information
 Supplementary Information Overexpression of Fto leads to increased food intake and results in obesity Chris Church, Lee Moir, Fiona McMurray, Christophe Girard, Gareth T Banks, Lydia Teboul, Sara Wells,
Supplementary Information Overexpression of Fto leads to increased food intake and results in obesity Chris Church, Lee Moir, Fiona McMurray, Christophe Girard, Gareth T Banks, Lydia Teboul, Sara Wells,
Supplementary Figure 1. Establishment of prostacyclin-secreting hmscs. (a) PCR showed the integration of the COX-1-10aa-PGIS transgene into the
 Supplementary Figure 1. Establishment of prostacyclin-secreting hmscs. (a) PCR showed the integration of the COX-1-10aa-PGIS transgene into the genomic DNA of hmscs (PGI2- hmscs). Native hmscs and plasmid
Supplementary Figure 1. Establishment of prostacyclin-secreting hmscs. (a) PCR showed the integration of the COX-1-10aa-PGIS transgene into the genomic DNA of hmscs (PGI2- hmscs). Native hmscs and plasmid
ERK1/2/MAPK pathway-dependent regulation of the telomeric factor TRF2
 ERK1/2/MAPK pathway-dependent regulation of the telomeric factor TRF2 SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Conservation of the D domain throughout evolution. Alignment of TRF2 sequences
ERK1/2/MAPK pathway-dependent regulation of the telomeric factor TRF2 SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Conservation of the D domain throughout evolution. Alignment of TRF2 sequences
Supplemental Materials Molecular Biology of the Cell
 Supplemental Materials Molecular Biology of the Cell Garcia-Alvarez et al. Supplementary Figure Legends Figure S1.Expression and RNAi-mediated silencing of STIM1 in hippocampal neurons (DIV, days in vitro).
Supplemental Materials Molecular Biology of the Cell Garcia-Alvarez et al. Supplementary Figure Legends Figure S1.Expression and RNAi-mediated silencing of STIM1 in hippocampal neurons (DIV, days in vitro).
Expression constructs
 Gene expressed in bebe3 ZmBEa Expression constructs 35S ZmBEa Pnos:Hygromycin r 35S Pnos:Hygromycin r 35S ctp YFP Pnos:Hygromycin r B -1 Chl YFP- Merge Supplemental Figure S1: Constructs Used for the Expression
Gene expressed in bebe3 ZmBEa Expression constructs 35S ZmBEa Pnos:Hygromycin r 35S Pnos:Hygromycin r 35S ctp YFP Pnos:Hygromycin r B -1 Chl YFP- Merge Supplemental Figure S1: Constructs Used for the Expression
Near-infrared fluorescent proteins
 nature methods Near-infrared fluorescent proteins Dmitry Shcherbo, Irina I Shemiakina, Anastasiya V Ryabova, Kathryn E Luker, Bradley T Schmidt, Ekaterina A Souslova, Tatiana V Gorodnicheva, Lydia Strukova,
nature methods Near-infrared fluorescent proteins Dmitry Shcherbo, Irina I Shemiakina, Anastasiya V Ryabova, Kathryn E Luker, Bradley T Schmidt, Ekaterina A Souslova, Tatiana V Gorodnicheva, Lydia Strukova,
Supplemental Data Figure S1 Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells.
 Supplemental Data Figure S1. Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells. (A) Specific lysis of IFN-γ-treated SKBR-3 cells in the absence
Supplemental Data Figure S1. Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells. (A) Specific lysis of IFN-γ-treated SKBR-3 cells in the absence
A programmable NOR-based device for transcription profile analyses
 A programmable NOR-based device for transcription profile analyses Tom Ran 1,2, Yehonatan Douek 1, Lilach Milo 1 & Ehud Shapiro 1,2 1 Department of Computer Science and Applied Mathematics 2 Department
A programmable NOR-based device for transcription profile analyses Tom Ran 1,2, Yehonatan Douek 1, Lilach Milo 1 & Ehud Shapiro 1,2 1 Department of Computer Science and Applied Mathematics 2 Department
Figure S1. Generation of inducible PTEN deficient mice and the BMMCs (A) B6.129 Pten loxp/loxp mice were mated with B6.
 Figure S1. Generation of inducible PTEN deficient mice and the BMMCs (A) B6.129 Pten loxp/loxp mice were mated with B6.129-Gt(ROSA)26Sor tm1(cre/ert2)tyj /J mice. To induce deletion of the Pten locus,
Figure S1. Generation of inducible PTEN deficient mice and the BMMCs (A) B6.129 Pten loxp/loxp mice were mated with B6.129-Gt(ROSA)26Sor tm1(cre/ert2)tyj /J mice. To induce deletion of the Pten locus,
Nature Medicine: doi: /nm.4322
 1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
Figure S1: Effects on haptotaxis are independent of effects on cell velocity A)
 Supplemental Figures Figure S1: Effects on haptotaxis are independent of effects on cell velocity A) Velocity of MV D7 fibroblasts expressing different GFP-tagged Ena/VASP family proteins in the haptotaxis
Supplemental Figures Figure S1: Effects on haptotaxis are independent of effects on cell velocity A) Velocity of MV D7 fibroblasts expressing different GFP-tagged Ena/VASP family proteins in the haptotaxis
Figure S1 Expression of AHL gene family members in diploid (Ler Col) and triploid (Ler
 Supplemental material Supplemental figure legends Figure S Expression of AHL gene family members in diploid (Ler ) and triploid (Ler osd) seeds. AHLs from clade B are labelled with (I), and AHLs from clade
Supplemental material Supplemental figure legends Figure S Expression of AHL gene family members in diploid (Ler ) and triploid (Ler osd) seeds. AHLs from clade B are labelled with (I), and AHLs from clade
Title: Authors: Affiliation(s): * Correspondence:
 Title: Bifidobacterium breve UCC2003 metabolises the human milk oligosaccharides lacto-ntetraose and lacto-n-neo-tetraose through overlapping, yet distinct pathways. Authors: Kieran James 1,2, Mary O Connell
Title: Bifidobacterium breve UCC2003 metabolises the human milk oligosaccharides lacto-ntetraose and lacto-n-neo-tetraose through overlapping, yet distinct pathways. Authors: Kieran James 1,2, Mary O Connell
SUPPLEMENTARY INFORMATION. Supp. Fig. 1. Autoimmunity. Tolerance APC APC. T cell. T cell. doi: /nature06253 ICOS ICOS TCR CD28 TCR CD28
 Supp. Fig. 1 a APC b APC ICOS ICOS TCR CD28 mir P TCR CD28 P T cell Tolerance Roquin WT SG Icos mrna T cell Autoimmunity Roquin M199R SG Icos mrna www.nature.com/nature 1 Supp. Fig. 2 CD4 + CD44 low CD4
Supp. Fig. 1 a APC b APC ICOS ICOS TCR CD28 mir P TCR CD28 P T cell Tolerance Roquin WT SG Icos mrna T cell Autoimmunity Roquin M199R SG Icos mrna www.nature.com/nature 1 Supp. Fig. 2 CD4 + CD44 low CD4
Nature Medicine doi: /nm.2860
 Supplemental Figure Legends Supplemental Figure 1: Hypomorphic expression of IFT88 results in olfactory signaling proteins no longer localizing to the ciliary layer. (a) ACIII localizes to the cilia and
Supplemental Figure Legends Supplemental Figure 1: Hypomorphic expression of IFT88 results in olfactory signaling proteins no longer localizing to the ciliary layer. (a) ACIII localizes to the cilia and
SUPPLEMENTARY INFORMATION. Supplementary Figures S1-S9. Supplementary Methods
 SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
Identification of vaccine candidate antigens for a Babesia bovis transmission blocking vaccine
 Identification of vaccine candidate antigens for a Babesia bovis transmission blocking vaccine Carlos E. Suarez Animal Disease Research Unit ARS-USDA Department of Veterinary Microbiology and Pathology
Identification of vaccine candidate antigens for a Babesia bovis transmission blocking vaccine Carlos E. Suarez Animal Disease Research Unit ARS-USDA Department of Veterinary Microbiology and Pathology
SelenoMet Dream TM Media Kit (MD12-506)
 SelenoMet Dream TM Media Kit (MD12-506) For the Overnight Expression of Recombinant Proteins About the Kit: MD12-506 contains: (*each packet contains enough to make up 1L of media) Description For the
SelenoMet Dream TM Media Kit (MD12-506) For the Overnight Expression of Recombinant Proteins About the Kit: MD12-506 contains: (*each packet contains enough to make up 1L of media) Description For the
Supplemental information contains 7 movies and 4 supplemental Figures
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplemental information contains 7 movies and 4 supplemental Figures Movies: Movie 1. Single virus tracking of A4-mCherry-WR MV
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplemental information contains 7 movies and 4 supplemental Figures Movies: Movie 1. Single virus tracking of A4-mCherry-WR MV
Characterization of the DNA-mediated Oxidation of Dps, a Bacterial Ferritin
 SUPPORTING INFORMATION Characterization of the DNA-mediated Oxidation of Dps, a Bacterial Ferritin Anna R. Arnold, Andy Zhou, and Jacqueline K. Barton Division of Chemistry and Chemical Engineering, California
SUPPORTING INFORMATION Characterization of the DNA-mediated Oxidation of Dps, a Bacterial Ferritin Anna R. Arnold, Andy Zhou, and Jacqueline K. Barton Division of Chemistry and Chemical Engineering, California
Bioluminescence Resonance Energy Transfer (BRET)-based studies of receptor dynamics in living cells with Berthold s Mithras
 Bioluminescence Resonance Energy Transfer (BRET)-based studies of receptor dynamics in living cells with Berthold s Mithras Tarik Issad, Ralf Jockers and Stefano Marullo 1 Because they play a pivotal role
Bioluminescence Resonance Energy Transfer (BRET)-based studies of receptor dynamics in living cells with Berthold s Mithras Tarik Issad, Ralf Jockers and Stefano Marullo 1 Because they play a pivotal role
TKB1 Competent Cells. Instruction Manual. Research Use Only. Not for Use in Diagnostic Procedures. Catalog # Revision B
 TKB1 Competent Cells Instruction Manual Catalog #200134 Revision B Research Use Only. Not for Use in Diagnostic Procedures. 200134-12 LIMITED PRODUCT WARRANTY This warranty limits our liability to replacement
TKB1 Competent Cells Instruction Manual Catalog #200134 Revision B Research Use Only. Not for Use in Diagnostic Procedures. 200134-12 LIMITED PRODUCT WARRANTY This warranty limits our liability to replacement
Luminescent platforms for monitoring changes in the solubility of amylin and huntingtin in living cells
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) (b) (c) (d) (e) (f) (g) .
 Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) The inhibitory effects of new antibodies (Mab17 and Mab18). They were investigated in in vitro fertilization
Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) The inhibitory effects of new antibodies (Mab17 and Mab18). They were investigated in in vitro fertilization
BA, BSc, and MSc Degree Examinations
 Examination Candidate Number: Desk Number: BA, BSc, and MSc Degree Examinations 2017-8 Department : BIOLOGY Title of Exam: Microbiology Time Allowed: 1 hour and 30 minutes Marking Scheme: Total marks available
Examination Candidate Number: Desk Number: BA, BSc, and MSc Degree Examinations 2017-8 Department : BIOLOGY Title of Exam: Microbiology Time Allowed: 1 hour and 30 minutes Marking Scheme: Total marks available
Rob Ross, MD. Infinity Pharmaceuticals March 9 th, 2011
 Heat Shock Protein 90 (Hsp90) Inhibition as a Potential Novel Approach to the Treatment of Patients with ALK Mutated Non-small Cell Lung Cancer (NSCLC) Rob Ross, MD Infinity Pharmaceuticals March 9 th,
Heat Shock Protein 90 (Hsp90) Inhibition as a Potential Novel Approach to the Treatment of Patients with ALK Mutated Non-small Cell Lung Cancer (NSCLC) Rob Ross, MD Infinity Pharmaceuticals March 9 th,
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation
 Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
Culture Density (OD600) 0.1. Culture Density (OD600) Culture Density (OD600) Culture Density (OD600) Culture Density (OD600)
 A. B. C. D. E. PA JSRI JSRI 2 PA DSAM DSAM 2 DSAM 3 PA LNAP LNAP 2 LNAP 3 PAO Fcor Fcor 2 Fcor 3 PAO Wtho Wtho 2 Wtho 3 Wtho 4 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB
A. B. C. D. E. PA JSRI JSRI 2 PA DSAM DSAM 2 DSAM 3 PA LNAP LNAP 2 LNAP 3 PAO Fcor Fcor 2 Fcor 3 PAO Wtho Wtho 2 Wtho 3 Wtho 4 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB
Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion
 Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical
 Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical representation using all SNPs (n= 13,515,798) including the region on chromosome 1 including SORT1 which was previously
Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical representation using all SNPs (n= 13,515,798) including the region on chromosome 1 including SORT1 which was previously
Supplementary Information Titles Journal: Nature Medicine
 Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Figure 1
 Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Nature Genetics: doi: /ng Supplementary Figure 1. SEER data for male and female cancer incidence from
 Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
CD3 coated cover slips indicating stimulatory contact site, F-actin polymerization and
 SUPPLEMENTAL FIGURES FIGURE S1. Detection of MCs. A, Schematic representation of T cells stimulated on anti- CD3 coated cover slips indicating stimulatory contact site, F-actin polymerization and microclusters.
SUPPLEMENTAL FIGURES FIGURE S1. Detection of MCs. A, Schematic representation of T cells stimulated on anti- CD3 coated cover slips indicating stimulatory contact site, F-actin polymerization and microclusters.
Crl and RpoS May Not Be Involved in Cross Protection against Tetracycline in Escherichia coli in Response to Heat Stress
 rl and RpoS May Not Be Involved in ross Protection against Tetracycline in Escherichia coli in Response to Heat Stress Irina han, hi Wing heng, Jasmine hin, and Veronica how Department of Microbiology
rl and RpoS May Not Be Involved in ross Protection against Tetracycline in Escherichia coli in Response to Heat Stress Irina han, hi Wing heng, Jasmine hin, and Veronica how Department of Microbiology
Supplemental Data Functional characterization of core components of the Bacillus subtilis c- di-
 Supplemental Data Functional characterization of core components of the Bacillus subtilis c- di- GMP signaling pathway Xiaohui Gao 1,3, Sampriti Mukherjee 2, Paige M. Matthews 1, Loubna A. Hammad 1, Daniel
Supplemental Data Functional characterization of core components of the Bacillus subtilis c- di- GMP signaling pathway Xiaohui Gao 1,3, Sampriti Mukherjee 2, Paige M. Matthews 1, Loubna A. Hammad 1, Daniel
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Courtheoux et al., http://www.jcb.org/cgi/content/full/jcb.200902093/dc1 S1 Figure S2. Visualization of multiple merotelic attachments.
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Courtheoux et al., http://www.jcb.org/cgi/content/full/jcb.200902093/dc1 S1 Figure S2. Visualization of multiple merotelic attachments.
(A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC,
 Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
Supplemental Information For: The genetics of splicing in neuroblastoma
 Supplemental Information For: The genetics of splicing in neuroblastoma Justin Chen, Christopher S. Hackett, Shile Zhang, Young K. Song, Robert J.A. Bell, Annette M. Molinaro, David A. Quigley, Allan Balmain,
Supplemental Information For: The genetics of splicing in neuroblastoma Justin Chen, Christopher S. Hackett, Shile Zhang, Young K. Song, Robert J.A. Bell, Annette M. Molinaro, David A. Quigley, Allan Balmain,
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Afferent lymph-derived T cells and dendritic cells use different CCR7-dependent routes for lymph node entry and intranodal migration
 Braun et al. Supplementary Information 1 Supplementary Information Afferent lymph-derived T cells and dendritic cells use different CCR7-dependent routes for lymph node entry and intranodal migration Asolina
Braun et al. Supplementary Information 1 Supplementary Information Afferent lymph-derived T cells and dendritic cells use different CCR7-dependent routes for lymph node entry and intranodal migration Asolina
Supplementary Information
 Supplementary Information Structural basis of improved second generation 3-nitro-tyrosine trna synthetases Richard B. Cooley, Jessica L. Feldman, Camden M. Driggers, Taylor Bundy, Audrey L. Stokes, P.
Supplementary Information Structural basis of improved second generation 3-nitro-tyrosine trna synthetases Richard B. Cooley, Jessica L. Feldman, Camden M. Driggers, Taylor Bundy, Audrey L. Stokes, P.
Acetyl-CoA or BC acyl-coa. Int1-ACP. FabH. FabF. Anti-FabF Platensimycin ACP. Fak
 Initiation Acetyl-CoA AccBCDA FabZ Int-ACP Elongation [N+] FabG Int1-ACP Acetyl-CoA or BC acyl-coa FabH FabD CoA Malonyl-ACP Malonyl CoA ACP Anti-FabI Triclosan AFN-15 FabI acyl-acp PlsX PlsY Pls C Lipoic
Initiation Acetyl-CoA AccBCDA FabZ Int-ACP Elongation [N+] FabG Int1-ACP Acetyl-CoA or BC acyl-coa FabH FabD CoA Malonyl-ACP Malonyl CoA ACP Anti-FabI Triclosan AFN-15 FabI acyl-acp PlsX PlsY Pls C Lipoic
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Analysis of hair bundle morphology in Ush1c c.216g>a mice at P18 by SEM.
 Supplementary Figure 1 Analysis of hair bundle morphology in Ush1c c.216g>a mice at P18 by SEM. (a-c) Heterozygous c.216ga mice displayed normal hair bundle morphology at P18. (d-i) Disorganized hair bundles
Supplementary Figure 1 Analysis of hair bundle morphology in Ush1c c.216g>a mice at P18 by SEM. (a-c) Heterozygous c.216ga mice displayed normal hair bundle morphology at P18. (d-i) Disorganized hair bundles
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
a) SSR with core motif > 2 and repeats number >3. b) MNR with repeats number>5.
 1 2 APPENDIX Legends to figures 3 4 5 Figure A1: Distribution of perfect SSR along chromosome 1 of V. cholerae (El-Tor N191). a) SSR with core motif > 2 and repeats number >3. b) MNR with repeats number>5.
1 2 APPENDIX Legends to figures 3 4 5 Figure A1: Distribution of perfect SSR along chromosome 1 of V. cholerae (El-Tor N191). a) SSR with core motif > 2 and repeats number >3. b) MNR with repeats number>5.
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures
 Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Subcellular segregation of VGluT2-IR and TH-IR within the same VGluT2-TH axon (wild type rats). (a-e) Serial sections of a dual VGluT2-TH labeled axon. This axon (blue outline) has
Supplementary Figure 1 Subcellular segregation of VGluT2-IR and TH-IR within the same VGluT2-TH axon (wild type rats). (a-e) Serial sections of a dual VGluT2-TH labeled axon. This axon (blue outline) has
Supplementary Information for. Shi and King, Chromosome Nondisjunction Yields Tetraploid Rather than Aneuploid Cells in Human Cell Lines.
 Supplementary Information for Shi and King, Chromosome Nondisjunction Yields Tetraploid Rather than Aneuploid Cells in Human Cell Lines Contains Supplementary Methods Supplementary Figures 1-7 Supplementary
Supplementary Information for Shi and King, Chromosome Nondisjunction Yields Tetraploid Rather than Aneuploid Cells in Human Cell Lines Contains Supplementary Methods Supplementary Figures 1-7 Supplementary
Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma.
 Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
Biology Open (2014) 000, 1 10 doi: /bio
 (2014) 000, 1 10 doi:10.1242/bio.201410041 Supplementary Material Michael Brauchle et al. doi: 10.1242/bio.201410041 Fig. S1. Alignment of GFP, sfgfp, egfp, eyfp, mcherry and mruby2. Sequence-based alignment
(2014) 000, 1 10 doi:10.1242/bio.201410041 Supplementary Material Michael Brauchle et al. doi: 10.1242/bio.201410041 Fig. S1. Alignment of GFP, sfgfp, egfp, eyfp, mcherry and mruby2. Sequence-based alignment
SUPPLEMENTARY LEGENDS...
 TABLE OF CONTENTS SUPPLEMENTARY LEGENDS... 2 11 MOVIE S1... 2 FIGURE S1 LEGEND... 3 FIGURE S2 LEGEND... 4 FIGURE S3 LEGEND... 5 FIGURE S4 LEGEND... 6 FIGURE S5 LEGEND... 7 FIGURE S6 LEGEND... 8 FIGURE
TABLE OF CONTENTS SUPPLEMENTARY LEGENDS... 2 11 MOVIE S1... 2 FIGURE S1 LEGEND... 3 FIGURE S2 LEGEND... 4 FIGURE S3 LEGEND... 5 FIGURE S4 LEGEND... 6 FIGURE S5 LEGEND... 7 FIGURE S6 LEGEND... 8 FIGURE
293T cells were transfected with indicated expression vectors and the whole-cell extracts were subjected
 SUPPLEMENTARY INFORMATION Supplementary Figure 1. Formation of a complex between Slo1 and CRL4A CRBN E3 ligase. (a) HEK 293T cells were transfected with indicated expression vectors and the whole-cell
SUPPLEMENTARY INFORMATION Supplementary Figure 1. Formation of a complex between Slo1 and CRL4A CRBN E3 ligase. (a) HEK 293T cells were transfected with indicated expression vectors and the whole-cell
The power of bacterial genetics is the potential for studying rare events
 The types of experiments that a geneticist can do are dictated to a great extent by the model organism that s/he is working with The power of bacterial genetics is the potential for studying rare events
The types of experiments that a geneticist can do are dictated to a great extent by the model organism that s/he is working with The power of bacterial genetics is the potential for studying rare events
Nature Genetics: doi: /ng Supplementary Figure 1. Susceptibility of MTase-deficient E. coli K12 to sublethal ampicillin treatment.
 Supplementary Figure 1 Susceptibility of MTase-deficient E. coli K12 to sublethal ampicillin treatment. (a,b) Wild-type (wt) or MTase-deficient E. coli BW25113 (a) or MG1655 (b) were grown in LB to an
Supplementary Figure 1 Susceptibility of MTase-deficient E. coli K12 to sublethal ampicillin treatment. (a,b) Wild-type (wt) or MTase-deficient E. coli BW25113 (a) or MG1655 (b) were grown in LB to an
Supplementary Figure 1. Experimental paradigm. A combination of genome and exome sequencing coupled with array-comparative genome hybridization was
 Supplementary Figure 1. Experimental paradigm. A combination of genome and exome sequencing coupled with array-comparative genome hybridization was performed on a total of 85 SS patients. Data filtration
Supplementary Figure 1. Experimental paradigm. A combination of genome and exome sequencing coupled with array-comparative genome hybridization was performed on a total of 85 SS patients. Data filtration
iplex genotyping IDH1 and IDH2 assays utilized the following primer sets (forward and reverse primers along with extension primers).
 Supplementary Materials Supplementary Methods iplex genotyping IDH1 and IDH2 assays utilized the following primer sets (forward and reverse primers along with extension primers). IDH1 R132H and R132L Forward:
Supplementary Materials Supplementary Methods iplex genotyping IDH1 and IDH2 assays utilized the following primer sets (forward and reverse primers along with extension primers). IDH1 R132H and R132L Forward:
The unconstrained evolution of fast and efficient antibiotic-resistant bacterial genomes
 In the format provided by the authors and unedited. VOLUME: ARTICLE NUMBER: 5 The unconstrained evolution of fast and efficient antibiotic-resistant bacterial genomes Carlos Reding-Roman, Mark Hewlett,
In the format provided by the authors and unedited. VOLUME: ARTICLE NUMBER: 5 The unconstrained evolution of fast and efficient antibiotic-resistant bacterial genomes Carlos Reding-Roman, Mark Hewlett,
a b G75 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server.
 a b G75 2 2 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server. a. Overlay of top 10 models generated by I-TASSER illustrates the potential effect of 7 amino acid insertion
a b G75 2 2 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server. a. Overlay of top 10 models generated by I-TASSER illustrates the potential effect of 7 amino acid insertion
Auxin-Inducible Degron (AID) System Total Set
 Catalog No. BRS-APC011A Auxin-Inducible Degron (AID) System Total Set This AID System Charactoristics Principle of Auxin-Inducible Degron (AID ) System AID plasmid protein 1. Transfection of AID plasmid
Catalog No. BRS-APC011A Auxin-Inducible Degron (AID) System Total Set This AID System Charactoristics Principle of Auxin-Inducible Degron (AID ) System AID plasmid protein 1. Transfection of AID plasmid
Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.
 Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.32 PCOS locus after conditioning for the lead SNP rs10993397;
Supplementary Figure 1: Attenuation of association signals after conditioning for the lead SNP. a) attenuation of association signal at the 9p22.32 PCOS locus after conditioning for the lead SNP rs10993397;
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Primer DNA sequence (5 to 3 )
 Supplemental Table 1 Oligonucleotide primers used in the study Primer DNA sequence (5 to 3 ) Plasmodium falciparum 18s forward Plasmodium falciparum 18s reverse Plasmodium falciparum MSP1 forward Plasmodium
Supplemental Table 1 Oligonucleotide primers used in the study Primer DNA sequence (5 to 3 ) Plasmodium falciparum 18s forward Plasmodium falciparum 18s reverse Plasmodium falciparum MSP1 forward Plasmodium
Supplementary Figure S1. Generation of LSL-EZH2 conditional transgenic mice.
 Downstream Col1A locus S P P P EP Genotyping with P1, P2 frt PGKneopA + frt hygro-pa Targeting vector Genotyping with P3, P4 P1 pcag-flpe P2 P3 P4 frt SApA CAG LSL PGKATG frt hygro-pa C. D. E. ormal KRAS
Downstream Col1A locus S P P P EP Genotyping with P1, P2 frt PGKneopA + frt hygro-pa Targeting vector Genotyping with P3, P4 P1 pcag-flpe P2 P3 P4 frt SApA CAG LSL PGKATG frt hygro-pa C. D. E. ormal KRAS
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2480 a ~250 kda ~130 ~95 ~72 ~250 kda anti ypk1δ ypk2δ D239A S641A T659A tor2δ YPK2 ypk2δ CherryYPK1 unspecific crossreaction Cherry c b D239A S641A T659A YPK2 D239A S641A T659A tor2δ YPK2
DOI: 10.1038/ncb2480 a ~250 kda ~130 ~95 ~72 ~250 kda anti ypk1δ ypk2δ D239A S641A T659A tor2δ YPK2 ypk2δ CherryYPK1 unspecific crossreaction Cherry c b D239A S641A T659A YPK2 D239A S641A T659A tor2δ YPK2
d e f Spatiotemporal quantification of subcellular ATP levels in a single HeLa cell during changes in morphology Supplementary Information
 Ca 2+ level (a. u.) Area (a. u.) Normalized distance Normalized distance Center Edge Center Edge Relative ATP level Relative ATP level Supplementary Information Spatiotemporal quantification of subcellular
Ca 2+ level (a. u.) Area (a. u.) Normalized distance Normalized distance Center Edge Center Edge Relative ATP level Relative ATP level Supplementary Information Spatiotemporal quantification of subcellular
Supplementary Materials
 Supplementary Materials Supplementary figure 1. Taxonomic representation summarized at genus level. Fecal microbiota from a separate set of Jackson and Harlan mice prior to irradiation. A taxon was included
Supplementary Materials Supplementary figure 1. Taxonomic representation summarized at genus level. Fecal microbiota from a separate set of Jackson and Harlan mice prior to irradiation. A taxon was included
SUPPLEMENTARY INFORMATION
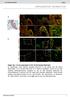 Suppl. Fig. 1 in vivo expression of ISL1 in the human fetal heart. a, Hematoxylin eosin staining showing structures of left atrium and left atrium appendage (*) of a human fetal heart at 11 weeks of gestation.
Suppl. Fig. 1 in vivo expression of ISL1 in the human fetal heart. a, Hematoxylin eosin staining showing structures of left atrium and left atrium appendage (*) of a human fetal heart at 11 weeks of gestation.
