Supplemental Data Functional characterization of core components of the Bacillus subtilis c- di-
|
|
|
- Ambrose Sullivan
- 5 years ago
- Views:
Transcription
1 Supplemental Data Functional characterization of core components of the Bacillus subtilis c- di- GMP signaling pathway Xiaohui Gao 1,3, Sampriti Mukherjee 2, Paige M. Matthews 1, Loubna A. Hammad 1, Daniel B. Kearns 2, Charles E. Dann III 1 *, Department of Chemistry 1, Department of Biology 2 and Biochemistry Graduate Program 3 Indiana University, Bloomington, IN
2 FIG. S1. In vivo analysis of c- di- GMP production by putative DGCs in inducible expression strains derived from wild type B. subtilis. All signals obtained from MRM transitions m/z (red), m/z (blue) and m/z (black) via LC- MS/MS are displayed for (A) c- di- GMP standard and bacterial extracts of (B) wild type B. subtilis and strains expressing putative DGCs: (C) YdaK, (D) DgcK (E) DgcP, and (F) DgcW. Transitions m/z (red), m/z (blue) are not unique to c- di- GMP in B. subtilis lysates. Only two transitions are shown in insets to indicate relative abundance of the unique identifier (m/z in black) and the most abundant fragmentation product (m/z in red).
3 FIG. S2. In vivo analysis of c- di- GMP production by putative DGCs in a background lacking all endogenous DGCs. All signals obtained from MRM transitions m/z (red), m/z (blue) and m/z (black) via LC- MS/MS are displayed for (A) c- di- GMP standard and bacterial extracts of (B) ΔydaK dgck dgcw dgcp :: tet B. subtilis - labeled as mutant - and strains expressing putative DGCs: (C) YdaK, (D) DgcK (E) DgcP, and (F) DgcW. Transitions m/z (red), m/z (blue) are not unique to c- di- GMP in B. subtilis lysates. Only two transitions are shown in insets to indicate relative abundance of the unique identifier (m/z in black) and the most abundant fragmentation product (m/z in red).
4 FIG. S3. In vivo detection of c- di- GMP in knockout strains of PdeH, a c- di- GMP phosphodiesterase. LC- MS/MS was used to detect c- di- GMP via the MRM transitions m/z (red), m/z (blue) and m/z (black) for c- di- GMP standard (A) and B. subtilis extracts from the following strains: (B) wild type;(c) ΔpdeH::kan lacking a putative c- di- GMP Phosphodiesterase; (D) Δ pdeh::kan, amye::pdeh showing active phosphodiesterase complementation and (E)Δ ydak dgck dgcw dgcp::tet pdeh::kan lacking a putative c- di- GMP phosphodiesterase and all possible diguanylate cyclases. Transitions m/z (red), m/z (blue) are not unique to c- di- GMP in B. subtilis lysates. Only two transitions are shown in insets to indicate relative abundance of the unique identifier (m/z in black) and the most abundant fragmentation product (m/z in red).
5 FIG. S4. In vitro detection of enzymatic activity of putative B. subtilis DGCs. To assess the activity of putative DGCs in vitro, LC- MS/MS was employed with MRM to detect enzymatic production of c- di- GMP from purified single or dual domain constructs containing GGDEF motifs. All ion peaks indicate production of c- di- GMP in in vitro assays. In total, four GGDEF single domain proteins were analyzed - (A) YdaK, (B) DgcK, (C) DgcP and (E) DgcW - as well as two dual domain constructs containing GGDEF and either GAF or PAS domains (D, F). As c- di- GMP co- purifies with the DgcK GGDEF protein exogenously expressed and purified from E. coli, uniformly labeled 13 C, 15 N- GTP was used as a substrate for the DgcK GGDEF enzymatic reactions. For this isotope, the MRM transitions m/z (red line), m/z (blue line) and m/z (black line) were utilized (B), which
6 corresponds to the m/z (red), m/z (blue) and m/z (black) transitions used in all other cases. FIG. S5. Measurement of binding affinities of c- di- GMP to (A) YdaK and (B) DgrA. At least three titrations were carried out at 25 C in each case with a single representative titration shown. The measured dissociation constant and standard deviations for c- di- GMP binding to YdaK and DgrA were 1.1 ± 0.2 μm and 11.0 ± 0.4 nm, respectively. Buffer conditions: 10 mm sodium citrate phosphate, 200 mm NaCl, ph 7.5.
7 FIG. S6. B. subtilis biofilm formation is unaffected by alterations in c- di- GMP signaling. Strains with deletions or disruptions in any individual or all B. subtilis GGDEF protein(s) show no assembly defect in biofilm or pellicle formation assays, indicating c- di- GMP is not required for biofilm production (top two rows). Likewise, overexpression of each individual GGDEF protein does not alter biofilm or pellicle formation. The epsh::tet mutant is a control to illustrate a context in which these processes are disrupted.
8 FIG. S7. Swarm expansion assays of strains constitutively expressing YdaK or YkuI, potential c- di- GMP receptors, in a background containing elevated c- di- GMP levels (pdeh). Swarming of B. subtilis wild type; DK391, a pdeh deletion; NPS265, a constitutive expression strain of ykui in pdeh background; and NPS266, a constitutive expression strain of ydak in pdeh were shown. YkuI and YdaK do not act as c- di- GMP receptors involved in motility, as each protein does not inhibit motility when expressed in a background containing c- di- GMP (cf. dgra in Fig 9Q). Each point is an average of three replicates.
9 FIG. S8. PCR verification for dgc knockout mutants. Primer pairs for amplification of wild type and mutant sequences are listed on the right.
10 Table S1. B. subtilis strains used in this study Strain Genotype 3610 DK391 DK392 DK393 DK404 DK405 DK406 DK407 DS4856 DS4857 DS4858 DS4859 DS9883 DS9289 DS9305 DS9537 DS9408 DS9544 DS9545 DS9548 DS9884 NPS079 NPS080 NPS081 NPS082 NPS207 NPS208 NPS209 NPS215 NPS216 NPS217 NPS218 NPS219 NPS220 NPS221 NPS224 NPS233 NPS235 NPS236 pdeh::kan ΔydaK ΔdgcK ΔdgcW dgcp::tet pdeh::kan pdeh::kan dgra::erm dgcp::tet pdeh::kan ΔdgcW pdeh::kan ΔydaK pdeh::kan ΔdgcK pdeh::kan amye::physpank- dgcw spec amye::physpank- dgcp spec amye::physpank- ydak spec amye::physpank- dgck spec ΔdgcW ΔydaK ΔdgcK dgcp :: tet ΔydaK ΔdgcK ΔydaK dgcp::tet ΔdgcK dgcp::tet ΔydaK ΔdgcK dgcp::tet ΔydaK ΔdgcK ΔdgcW dgcp::tet amye::physpank- dgcw spec ΔydaK ΔdgcK ΔdgcW dgcp::tet amye::physpank- dgcp spec ΔydaK ΔdgcK ΔdgcW dgcp::tet amye::physpank- ydak spec ΔydaK ΔdgcK ΔdgcW dgcp::tet amye::physpank- dgck spec ΔydaK ΔdgcK ΔdgcW dgcp::tet amye::physpank- dgcw ΔEAL spec, ΔydaK ΔdgcK ΔdgcW dgcp::tet amye::physpank- dgck GGGGF mutant spec, ΔydaK ΔdgcK ΔdgcW dgcp::tet amye::physpank- dgcp GGGGL mutant spec, ΔydaK ΔdgcK ΔdgcW dgcp::tet amye::physpank- dgck GGAAF mutant spec, ΔydaK ΔdgcK ΔdgcW dgcp::tet amye::physpank- dgcp GGAAL mutant spec, ΔydaK ΔdgcK ΔdgcW dgcp::tet dgra :: erm mls dgra :: erm mls, ΔydaK ΔdgcK ΔdgcW dgcp::tet amye::physpank- dgcp spec, dgra :: erm mls ΔydaK ΔdgcK ΔdgcW dgcp::tet amye::physpank- dgck spec, dgra :: erm mls ΔydaK ΔdgcK ΔdgcW dgcp::tet amye::physpank- dgcw ΔEAL spec dgra :: erm mls ΔydaK ΔdgcK ΔdgcW dgcp::te amye::physpank- dgcw PAS- GGAAF ΔEAL spec ΔydaK ΔdgcK ΔdgcW dgcp::tet amye:: pdeh cat, pdeh::kan amye::pconstitutive- dgra spec, pdeh::kan amye::pconstitutive- dgra spec, pdeh::kan, ΔydaK ΔdgcK ΔdgcW dgcp::tet
11 NPS247 NPS252 NPS254 NPS255 NPS265 NPS266 thrc::physpank- dgcp mls, amye::pconstitutive- dgra spec, pdeh::kan, ΔydaK ΔdgcK ΔdgcW dgcp::tet thrc::physpank- Bs_dgcP - dcpa mls,amye::pconstitutive- dgra spec, pdeh::kan, ΔydaK ΔdgcK ΔdgcW dgcp::tet thrc::physpank- Bs_dgcP - cd1420 mls,amye::pconstitutive- dgra spec, pdeh::kan, ΔydaK ΔdgcK ΔdgcW dgcp::tet thrc::physpank- Bs_dgcP - Atu1297 mls,amye::pconstitutive- dgra spec, pdeh::kan, ΔydaK ΔdgcK ΔdgcW dgcp::tet amye::pconstitutive- YkuI spec, pdeh::kan amye::pconstitutive- YdaK spec, pdeh::kan
12 Table S2 Plasmids used in this study Plasmid Genotype/reference pah25 amye::spec amp (gift from Dr. Amy Camp, Mount Holyoke College) pah52 erm amp pdg1515 tet amp (Guérout- Fleury et al., 1995) pdg364 amye cat pdp150 thrc_ Physpank mls pdp391 ΩΔdgcW mls amp pdp392 ΩΔydaK mls amp pdp393 ΩΔdgcK mls amp pdp407 ypfa::erm amp pdr90 amye::phyperspac spec amp (gift from Dr. David Rudner, Harvard) pdr111 amye intergration vector that contains Physpank, spec and amp phis- parallel N- terminal His- tag expression vector amp pkb149 amye_pconstitutive spec pminimad ori BsTs mls (Patrick and Kearns, 2008) pmbp- parallel N- terminal MBP- tag expression vector amp phs001 phis- parallel with pdeh, amp pxg075 phis- parallel with ydak GGDEF domain, amp pxg076 phis- parallel with dgck GGDEF domain, amp pxg061 pmbp- parallel with dgcp GGDEF domain, amp pxg074 phis- parallel with dgcp GAF- GGDEF domain, amp pxg077 pmbp- parallel with dgcw GGDEF domain, amp pxg079 phis- parallel with dgcw PAS- GGDEF domain, amp pxg001 amye::physpank- dgcw, spec pxg002 amye::physpank- dgcp, spec pxg003 amye::physpank- ydak, spec pxg004 amye::physpank- dgck, spec pxg085 phis- parallel with dgra, amp pxg086 amye::physpank- dgcw ΔEAL, spec pxg087 amye::physpank- dgck GGGGF mutant, spec pxg088 amye::physpank- dgcp GGGGL, spec pxg090 amye::physpank- dgck GGAAF, spec pxg091 amye::physpank- dgck GGAAF, spec pxg092 amye::physpank- dgcw GGAAF ΔEAL, spec pxg093 phis- parallel with dgra C4, amp pxg094 amye:: pdeh, cat (complementation) pxg095 amye::pconstitutive- dgra, spec pxg100 thrc::physpank- dgcp, mls pxg101 thrc::physpank- dgcp, mls pxg102 thrc::physpank- dgcp- dcpa, mls pxg104 thrc::physpank- dgcp- CD1420, mls pxg105 thrc::physpank- dgcp- Atu1297, mls
13 pxg124 pxg125 amye::pconstitutive- ykui, spec amye::pconstitutive- ydak, spec
14 Table S3 Primers used in this study Primers for enzymatic activity assays in vitro GXH466 YdaK GGDEF domain(ncoi) ggg ccatgg atcacgatattacagcagag GXH460 YdaK GGDEF domain(ecori) ggg gaattc ttatagttcattcatcatcatctcgttttcc GXH467 DgcK GGDEF domain (NcoI) ggg ccatgg atcatgaaacaaatgcac GXH462 DgcK GGDEF domain(ecori) ggg gaattc tcattcttttttttctgaaaaacacactctg GXH454 DgcP GGDEF (NcoI) ggg ccatgg tgaaaacagatcagctgactg GXH440 DgcP GGDEF(EcoRI) ggg gaattc tcatgagtcatgaatcatcaagcg GXH465 DgcP GAF- GGDEF (NcoI) ggg ccatgg atcaggaaagactctataac GXH458 DgcP GAF- GGDEF(EcoRI) ggg gaattc ttattttattgagtcatgaatcatcaagcgg GXH453 DgcW GGDEF (NcoI) ggg ccatgg ctcattatgattcgcttacagac GXH471 DgcW GGDEF (EcoRI) ggc gaattc ttaagaatagtatctgtatttgcttttg GXH464 DgcW PAS- GGDEF (stui) gggaa aggcct aaatattgatgcaatttttattt GXH468 DgcW PAS- GGDEF (SpeI) ggc actagt ttaagaatagtatctgtatttgcttttg GXH480 PdeH (NcoI) gggg ccatgg gaagggtgtttgttgcaagacag GXH481 PdeH (EcoRI) gggg gaattc tcattttgcgtccataagattatgacac GXH482 DgrA(NcoI) ggg ccatgg gaatagagattggagaaaatgtacttttag GXH483 DgrA (EcoRI) gggg gaattc ttattccattcgggcctttcttc GXH513 DgrA (EcoRI) gggg gaattc ttactttcttctcttatttaactggcgc Primers for overexpression/ constitutive expression in vivo GXH301 dgcwf (SalI) aggaggtcgacgtcaacaagactggaacacacgat GXH302 dgcwr (NheI) ctcctgctagcaaggagtacgattcatgtgtagtaa GXH303 dgcpf(nhei) aggaggctagcgcttcaccgtttatgatataataaac GXH304 dgcpr(sphi) ctcctgcatgcgacgaaattgatgaaaaatacttaact GXH305 ydakf(nhei) aggaggctagcacgcattcatttgacaatctgctg GXH306 ydakr(sphi) ctcctgcatgcaatagaagcatcatgacgcatagc GXH307 dgckf(hindiii) aggagaagcttttccgatgggtcaagtgcgaatt GXH308 dgckr(nhei) ctcctgctagcaagaatatcagggagatcttgtcc GXH516 dgraf (SphI) gggg gcatgcgtgagtatattgaatgcgaaagagtgaag GXH517 dgrar (BamHI) gggg ggatccggaagtgtttgccgatgtttattcc GXH532 dgcp (HindIII) attgtgagcggataacaattaagcttgcttcaccgtttatgatata ataaac GXH533 dgcp (SphI) cgtttccaccgaattagcttgcatgcgacgaaattgatgaaaaat acttaact GXH534 dgcp(- 60~+3) attgtgagcggataacaattaagc GXH535 dgcp (- 60~+3) tttattatatcataaacggtgaagcaagcttaattgttatccgctca ca GXH536 dgcp (- 60~+3) ttgcttcaccgtttatgatataataaacaaaaattagtggtaaactt ttaggtg GXH537 dgcp (- 60~+3) cactagtgctagccatgtttttatcacctaaaagtttaccactaattt ttg GXH538 dgcp (- 60~+3) ataaaaacatggctagcactagtgcatgcaagctaattcggtgga
15 GXH539 dgcp (- 60~+3) gacctcgtttccaccgaattagcttgcat GXH540 dcpa (NheI) tggtaaacttttaggtgataaaaacatggctagcattagatattca actcgtcccatcct GXH521 dcpa (SphI) cgtttccaccgaattagcttgcatgctggtcgtgaggaagcccaaa aaggtgaag GXH542 CD1420 (NheI) tggtaaacttttaggtgataaaaacatggctagctttaaagaaatt tttttaagaacttttccagga GXH543 CD1420 (SphI) gacctcgtttccaccgaattagcttgcatgcatgtacaagtttaaaa aaagaaagataattttaatt GXH544 Atu1297 (NheI) tggtaaacttttaggtgataaaaacatggctagcacggcgagagt tctggttgttgac GXH531 Atu1297 (SphI) cgtttccaccgaattagcttgcatgcgagaggctgcgcaaacagtc gtggttaa GXH583 ykui (SphI) ggg gcatgc gtgcgtataatcagacaaagatagagaagc GXH584 ykui (NheI) ggg gctagc aaacggcctttcagccgttaaatc GXH585 ydak(sphi) ggg gcatgc cacgcattcatttgacaatctgctg GXH586 ydak(nhei) ggg gctagc aatagaagcatcatgacgcatagc Primers for gene knockouts and complementation in vivo 2006 cgccattcgccagggctgcag 2007 ctcctgcatgcacacacattatgccacaccttgtag 2928 dgcwuf (EcoRI) aggaggaattcatcctgccaaaacaacgcca 2929 dgcwur (XhoI) ctcctctcgagatattccatcggcatttccat 2930 dgcwdf (XhoI) aggagctcgaggtgattgccgaaggtgtgg 2931 dgcwdr (BamHI) ctcctggatcccagggtataggccttcgt 2932 ydakuf (EcoRI) aggaggaattcgtctttgaagacatacaatatg 2933 ydakur (XhoI) ctcctctcgagcagagcaccttctgattgct 2934 ydakdf (XhoI) aggagctcgagggaaagtatgtgtcgcttacg 2935 ydakdr (BamHI) ctcctggatccctctgagctgtttcgccac 2936 dgckuf(ecori) aggaggaattctcgttgaagtcggtctagctc 2937 dgckur (XhoI) ctcctctcgagtactagcctttctttaaacagg 2938 dgck DF (XhoI) aggagctcgagcttgccgatcaaatgctttac 2939 dgck DR (BamHI) ctcctggatcctcgggattgctgagtctgac 3037 dgcp UP F 3 (BamHI) aggagggatccgtctgctctattcgaccatg 3038 dgcp UP R 3 (PstI) ctcctctgcaggcacgcaagatggaaacgc 3039 dgcp DN F 3 (EcoRI) aggaggaattcacgtacggccatcagactg 3040 dgcp DN R 3 (XhoI) ctcctctcgagcagaaattgtgcttccgattc GXH489 dgra UPF (EagI) aggagcggccggaacaggaagcaaaacaaattg GXH490 dgra UPR (BamHI) ctcctggatccgtacattttctccaatctctatc GXH491 dgra DNF (XhoI) aggagctcgagcaggctttgcttcaatactgc GXH492 dgra DNR (KpnI) ctcctggtacctctgcatttggcagcacatg 3428 pdeh_kan ITA UP F ggaaagcgctttaatatcccg
16 3429 pdeh_kan ITA UP R caattcgccctatagtgagtcgtacaaacaccctcattgatttatc 3430 pdeh_kan ITA DN F ccagcttttgttccctttagtgagatggacgcaaaatgattgcgg 3431 pdeh_kan ITA DN R ggacatttaaaggacagcagg GXH514 pdeh complementation F tgataagctgtcaaacatgagaattcgaaagaaagtctaccgca ttattaatcagtcc GXH515 pdeh complementation R atggtagcgaccggcgctcaggatcctttgcggtggtcagagga ccgc Primers for DGC mutants in vivo GXH484 dgcw in vivo truncation cctc gctagc ttaagaatagtatctgtatttgcttttg GXH485 dgck GGGGF mutant gatcctgccgctcggatcggcggtggcggttttgccgtgctcctg ccgaactg GXH486 dgck GGGGF mutant cagttcggcaggagcacggcaaaaccgccaccgccgatccga gcggcaggatc GXH487 dgcp GGGGL mutant cggtgcccgctggggaggaggaggactggcgatttatttgccaa atgtgccg GXH488 dgcp GGGGL mutant cggcacatttggcaaataaatcgccagtcctcctcctccccagcg ggcaccg GXH495 dgck GGAAF mutant gatcctgccgctcggatcggcggtgccgcttttgccgtgctcctg ccgaactg GXH496 dgck GGAAF mutant cagttcggcaggagcacggcaaaagcggcaccgccgatccga gcggcaggatc GXH497 dgcp GGAAL mutant cggtgcccgctggggaggagccgccctggcgatttatttgccaa atgtgccg GXH498 dgcp GGAAL mutant cggcacatttggcaaataaatcgccagggcggctcctccccagc gggcaccg GXH499 dgcw GGAAF ΔEAL ccgtcttggcggtgctgcatttattattatattaac GXH500 dgcw GGAAF ΔEAL gttaatataataataaatgcagcaccgccaagacgg
Intercellular Nanotubes Mediate Bacterial Communication
 Supplemental Information Cell, Volume 144 Intercellular Nanotubes Mediate Bacterial Communication Gyanendra P. Dubey and Sigal Ben-Yehuda 1 Table S1. List of B. subtilis Mutants Examined in This Study,
Supplemental Information Cell, Volume 144 Intercellular Nanotubes Mediate Bacterial Communication Gyanendra P. Dubey and Sigal Ben-Yehuda 1 Table S1. List of B. subtilis Mutants Examined in This Study,
Ec DOS: Escherichia coli Direct Oxygen Sensor
 Ec DS: Escherichia coli Direct xygen Sensor 1 Heme-based gas sensor protein Signal (2,, C etc.) or redox state FixL, HemAT, sgc CooA, Ec DS Sensor domain (Heme) Functional domain Protein structural change
Ec DS: Escherichia coli Direct xygen Sensor 1 Heme-based gas sensor protein Signal (2,, C etc.) or redox state FixL, HemAT, sgc CooA, Ec DS Sensor domain (Heme) Functional domain Protein structural change
Culture Density (OD600) 0.1. Culture Density (OD600) Culture Density (OD600) Culture Density (OD600) Culture Density (OD600)
 A. B. C. D. E. PA JSRI JSRI 2 PA DSAM DSAM 2 DSAM 3 PA LNAP LNAP 2 LNAP 3 PAO Fcor Fcor 2 Fcor 3 PAO Wtho Wtho 2 Wtho 3 Wtho 4 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB
A. B. C. D. E. PA JSRI JSRI 2 PA DSAM DSAM 2 DSAM 3 PA LNAP LNAP 2 LNAP 3 PAO Fcor Fcor 2 Fcor 3 PAO Wtho Wtho 2 Wtho 3 Wtho 4 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB Low Iron 2 4 6 8 2 4 6 8 2 22 DTSB
Raffinose, a plant galactoside, inhibits Pseudomonas aeruginosa
 Raffinose, a plant galactoside, inhibits Pseudomonas aeruginosa biofilm formation via binding to LecA and decreasing cellular cyclic diguanylate levels Han-Shin Kim 1, Eunji Cha 1, YunHye Kim 2, Young
Raffinose, a plant galactoside, inhibits Pseudomonas aeruginosa biofilm formation via binding to LecA and decreasing cellular cyclic diguanylate levels Han-Shin Kim 1, Eunji Cha 1, YunHye Kim 2, Young
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the
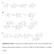 Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
Supplementary Figure 1. The proposed biosynthetic pathways for the A-ring transformation of the kinamycin and lomaiviticin antibiotics. A, by Steven J. Gould; B, by Emily P. Balskus; C, by Bradley S. Moore.
Supporting Information
 Supporting Information Keller et al. 10.1073/pnas.1222607110 MB1 Ori puc18 Ori AscI (8713) pdad downstream flank pdad PpdaD SphI (6970) Msed1993 ApaI (10721) Apr ApaLI (9115) NcoI (8) PmeI (8796) PvuI
Supporting Information Keller et al. 10.1073/pnas.1222607110 MB1 Ori puc18 Ori AscI (8713) pdad downstream flank pdad PpdaD SphI (6970) Msed1993 ApaI (10721) Apr ApaLI (9115) NcoI (8) PmeI (8796) PvuI
Supplementary Figure 1. Structure models of the c4 variant proteins based on the structures of maltose binding protein (MBP) in red and TEM-1
 Supplementary Figure 1. Structure models of the c4 variant proteins based on the structures of maltose binding protein (MBP) in red and TEM-1 β-lactamase (BLA) in blue. (a) c4 model and the close-up view
Supplementary Figure 1. Structure models of the c4 variant proteins based on the structures of maltose binding protein (MBP) in red and TEM-1 β-lactamase (BLA) in blue. (a) c4 model and the close-up view
Supplementary Figure 1. Chemical structures of activity-based probes (ABPs) and of click reagents used in this study.
 Supplementary Figure 1. Chemical structures of activity-based probes (ABPs) and of click reagents used in this study. In this study, one fluorophosphonate (FP, 1), three nitrophenol phosphonate probes
Supplementary Figure 1. Chemical structures of activity-based probes (ABPs) and of click reagents used in this study. In this study, one fluorophosphonate (FP, 1), three nitrophenol phosphonate probes
Controlling Macromolecular Topology with Genetically Encoded. SpyTag-SpyCatcher Chemistry
 Electronic Supporting Information for Controlling Macromolecular Topology with Genetically Encoded SpyTag-SpyCatcher Chemistry Wen-Bin Zhang, Fei Sun, David A. Tirrell,* and Frances H. Arnold* Division
Electronic Supporting Information for Controlling Macromolecular Topology with Genetically Encoded SpyTag-SpyCatcher Chemistry Wen-Bin Zhang, Fei Sun, David A. Tirrell,* and Frances H. Arnold* Division
Enhanced pinocembrin production in Escherichia coli by regulating cinnamic acid metabolism
 1 Enhanced pinocembrin production in Escherichia coli by regulating cinnamic acid metabolism 2 Weijia Cao 1,2, Weichao Ma 1,2,3, Xin wang 1,2, Bowen Zhang 1,2, Xun Cao 1,2, Kequan Chen 1,2*, Yan Li 1,2
1 Enhanced pinocembrin production in Escherichia coli by regulating cinnamic acid metabolism 2 Weijia Cao 1,2, Weichao Ma 1,2,3, Xin wang 1,2, Bowen Zhang 1,2, Xun Cao 1,2, Kequan Chen 1,2*, Yan Li 1,2
Luminescent platforms for monitoring changes in the solubility of amylin and huntingtin in living cells
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
glpx Rv1100 Probe 4.4 kb 75 kda 50 kda 37 kda 25 kda 20 kda
 PvuI 2.0 k PvuI PvuI Rv1101c Rv1100 glpx fum 4.4 k PvuI PvuI ΔglpX Rv1101c Rv1100 HygR fum ΔglpX 5 k 4 k 3 k c 75 kda 50 kda 37 kda GLPX Δ Δ/C GLPX 2 k 1.5 k 25 kda 20 kda PRCB 1 k 0.5 k Supplementary
PvuI 2.0 k PvuI PvuI Rv1101c Rv1100 glpx fum 4.4 k PvuI PvuI ΔglpX Rv1101c Rv1100 HygR fum ΔglpX 5 k 4 k 3 k c 75 kda 50 kda 37 kda GLPX Δ Δ/C GLPX 2 k 1.5 k 25 kda 20 kda PRCB 1 k 0.5 k Supplementary
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
Supplemental Data. Deinlein et al. Plant Cell. (2012) /tpc
 µm Zn 2+ 15 µm Zn 2+ Growth (% of control) empty vector NS1 NS2 NS3 NS4 S. pombe zhfδ Supplemental Figure 1. Functional characterization of. halleri NS genes in Zn 2+ hypersensitive S. pombe Δzhf mutant
µm Zn 2+ 15 µm Zn 2+ Growth (% of control) empty vector NS1 NS2 NS3 NS4 S. pombe zhfδ Supplemental Figure 1. Functional characterization of. halleri NS genes in Zn 2+ hypersensitive S. pombe Δzhf mutant
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis
 Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Information
 Supplementary Information Supplementary Figure 1. EBV-gB 23-431 mainly exists as trimer in HEK 293FT cells. (a) Western blotting analysis for DSS crosslinked FLAG-gB 23-431. HEK 293FT cells transfected
Supplementary Information Supplementary Figure 1. EBV-gB 23-431 mainly exists as trimer in HEK 293FT cells. (a) Western blotting analysis for DSS crosslinked FLAG-gB 23-431. HEK 293FT cells transfected
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10962 Supplementary Figure 1. Expression of AvrAC-FLAG in protoplasts. Total protein extracted from protoplasts described in Fig. 1a was subjected to anti-flag immunoblot to detect AvrAC-FLAG
doi:10.1038/nature10962 Supplementary Figure 1. Expression of AvrAC-FLAG in protoplasts. Total protein extracted from protoplasts described in Fig. 1a was subjected to anti-flag immunoblot to detect AvrAC-FLAG
Supplementary Information CAND1 controls in vivo dynamics of the Cullin 1-RING ubiquitin ligase repertoire
 Supplementary Information CAD1 controls in vivo dynamics of the Cullin 1-RIG uiquitin ligase repertoire Shuangding Wu, Wenhong Zhu, Tina han, Julia I. Toth, Matthew D. Petroski, Dieter A. Wolf 1 c knd1
Supplementary Information CAD1 controls in vivo dynamics of the Cullin 1-RIG uiquitin ligase repertoire Shuangding Wu, Wenhong Zhu, Tina han, Julia I. Toth, Matthew D. Petroski, Dieter A. Wolf 1 c knd1
Supplementary Figure 1
 Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplemental Information for: Localization of anionic phospholipids in Escherichia coli cells
 Supplemental Information for: Localization of anionic phospholipids in Escherichia coli cells Piercen M. Oliver, a John A. Crooks, a Mathias Leidl, b Earl J. Yoon, a Alan Saghatelian, b and Douglas B.
Supplemental Information for: Localization of anionic phospholipids in Escherichia coli cells Piercen M. Oliver, a John A. Crooks, a Mathias Leidl, b Earl J. Yoon, a Alan Saghatelian, b and Douglas B.
Late assembly of the Vibrio cholerae cell division machinery postpones
 2 Late assembly of the Vibrio cholerae cell division machinery postpones septation to the last % of the cell cycle 3 4 Elisa Galli, Evelyne Paly and François-Xavier Barre,# 5 6 7 Institute for Integrative
2 Late assembly of the Vibrio cholerae cell division machinery postpones septation to the last % of the cell cycle 3 4 Elisa Galli, Evelyne Paly and François-Xavier Barre,# 5 6 7 Institute for Integrative
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/2/5/e1501632/dc1 Supplementary Materials for Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms
advances.sciencemag.org/cgi/content/full/2/5/e1501632/dc1 Supplementary Materials for Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms
JBC Papers in Press. Published on January 14, 2002 as Manuscript M
 JBC Papers in Press. Published on January 14, 2002 as Manuscript M109976200 Pig Liver Carnitine Palmitoyltransferase: Chimera Studies Show That Both the N- and C-Terminal Regions of the Enzyme Are Important
JBC Papers in Press. Published on January 14, 2002 as Manuscript M109976200 Pig Liver Carnitine Palmitoyltransferase: Chimera Studies Show That Both the N- and C-Terminal Regions of the Enzyme Are Important
SUPPLEMENTARY DATA. Materials and Methods
 SUPPLEMENTARY DATA Materials and Methods HPLC-UV of phospholipid classes and HETE isomer determination. Fractionation of platelet lipid classes was undertaken on a Spherisorb S5W 150 x 4.6 mm column (Waters
SUPPLEMENTARY DATA Materials and Methods HPLC-UV of phospholipid classes and HETE isomer determination. Fractionation of platelet lipid classes was undertaken on a Spherisorb S5W 150 x 4.6 mm column (Waters
Supplementary Material
 Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
Supplementary Table 1. Properties of lysates of E. coli strains expressing CcLpxI point mutants
 Supplementary Table 1. Properties of lysates of E. coli strains expressing CcLpxI point mutants Species UDP-2,3- diacylglucosamine hydrolase specific activity (nmol min -1 mg -1 ) Fold vectorcontrol specific
Supplementary Table 1. Properties of lysates of E. coli strains expressing CcLpxI point mutants Species UDP-2,3- diacylglucosamine hydrolase specific activity (nmol min -1 mg -1 ) Fold vectorcontrol specific
igem2013 Microbiology BMB SDU
 igem2013 Microbiology BMB SDU Project type: Creation of Biobrick with Prenyltransferase Project title: Biobrick_Prenyltransferase Sub project: Creation date: 13.08.09 Written by: SIS Performed by: PRA,
igem2013 Microbiology BMB SDU Project type: Creation of Biobrick with Prenyltransferase Project title: Biobrick_Prenyltransferase Sub project: Creation date: 13.08.09 Written by: SIS Performed by: PRA,
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Expression constructs
 Gene expressed in bebe3 ZmBEa Expression constructs 35S ZmBEa Pnos:Hygromycin r 35S Pnos:Hygromycin r 35S ctp YFP Pnos:Hygromycin r B -1 Chl YFP- Merge Supplemental Figure S1: Constructs Used for the Expression
Gene expressed in bebe3 ZmBEa Expression constructs 35S ZmBEa Pnos:Hygromycin r 35S Pnos:Hygromycin r 35S ctp YFP Pnos:Hygromycin r B -1 Chl YFP- Merge Supplemental Figure S1: Constructs Used for the Expression
Identification, characterization of a flavin-containing monooxygenase MoA and its function in a
 1 Applied Microbiology and Biotechnology 2 Supplementary Material 3 4 Identification, characterization of a flavin-containing monooxygenase MoA and its function in a specific sophorolipid molecule metabolism
1 Applied Microbiology and Biotechnology 2 Supplementary Material 3 4 Identification, characterization of a flavin-containing monooxygenase MoA and its function in a specific sophorolipid molecule metabolism
Transient Ribosomal Attenuation Coordinates Protein Synthesis and Co-translational Folding
 SUPPLEMENTARY INFORMATION: Transient Ribosomal Attenuation Coordinates Protein Synthesis and Co-translational Folding Gong Zhang 1,2, Magdalena Hubalewska 1 & Zoya Ignatova 1,2 1 Department of Cellular
SUPPLEMENTARY INFORMATION: Transient Ribosomal Attenuation Coordinates Protein Synthesis and Co-translational Folding Gong Zhang 1,2, Magdalena Hubalewska 1 & Zoya Ignatova 1,2 1 Department of Cellular
SUPPLEMENTARY INFORMATION
 In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION DOI: 10.1038/NCHEM.2919 Direct observation of the influence of cardiolipin and antibiotics on lipid II binding to MurJ Jani
In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION DOI: 10.1038/NCHEM.2919 Direct observation of the influence of cardiolipin and antibiotics on lipid II binding to MurJ Jani
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS
 Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
7.012 Problem Set 4 October 28, 2005
 Name TA Sec 7.012 Problem Set 4 October 28, 2005 Answers to this problem set must be inserted into the box outside 68-120 by 4:30 am, Thursday, November 3 rd. Problem sets will not be accepted late. Solutions
Name TA Sec 7.012 Problem Set 4 October 28, 2005 Answers to this problem set must be inserted into the box outside 68-120 by 4:30 am, Thursday, November 3 rd. Problem sets will not be accepted late. Solutions
Table S1. Strains, Plasmids and Phage
 Table S1. Strains, Plasmids and Phage Strain, Plasmid, Phage Genotype or Description Reference S. enterica 14028 wild type ATCC strain (1) HK132 yfin; 14028 with deletion of yfin This study QW262 yhjh
Table S1. Strains, Plasmids and Phage Strain, Plasmid, Phage Genotype or Description Reference S. enterica 14028 wild type ATCC strain (1) HK132 yfin; 14028 with deletion of yfin This study QW262 yhjh
UNDERSTANDING THE ROLE OF ATP HYDROLYSIS IN THE SPLICEOSOME
 UNDERSTANDING THE ROLE OF ATP HYDROLYSIS IN THE SPLICEOSOME RNA Splicing Lecture 2, Biological Regulatory Mechanisms, H. Madhani Dept. of Biochemistry and Biophysics MAJOR MESSAGES Eight essential ATPases
UNDERSTANDING THE ROLE OF ATP HYDROLYSIS IN THE SPLICEOSOME RNA Splicing Lecture 2, Biological Regulatory Mechanisms, H. Madhani Dept. of Biochemistry and Biophysics MAJOR MESSAGES Eight essential ATPases
Figure S1. Analysis of genomic and cdna sequences of the targeted regions in WT-KI and
 Figure S1. Analysis of genomic and sequences of the targeted regions in and indicated mutant KI cells, with WT and corresponding mutant sequences underlined. (A) cells; (B) K21E-KI cells; (C) D33A-KI cells;
Figure S1. Analysis of genomic and sequences of the targeted regions in and indicated mutant KI cells, with WT and corresponding mutant sequences underlined. (A) cells; (B) K21E-KI cells; (C) D33A-KI cells;
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
Characterization of the DNA-mediated Oxidation of Dps, a Bacterial Ferritin
 SUPPORTING INFORMATION Characterization of the DNA-mediated Oxidation of Dps, a Bacterial Ferritin Anna R. Arnold, Andy Zhou, and Jacqueline K. Barton Division of Chemistry and Chemical Engineering, California
SUPPORTING INFORMATION Characterization of the DNA-mediated Oxidation of Dps, a Bacterial Ferritin Anna R. Arnold, Andy Zhou, and Jacqueline K. Barton Division of Chemistry and Chemical Engineering, California
Supplementary Information
 Supplementary Information Supplementary s Supplementary 1 All three types of foods suppress subsequent feeding in both sexes when the same food is used in the pre-feeding test feeding. (a) Adjusted pre-feeding
Supplementary Information Supplementary s Supplementary 1 All three types of foods suppress subsequent feeding in both sexes when the same food is used in the pre-feeding test feeding. (a) Adjusted pre-feeding
Cell Signaling part 2
 15 Cell Signaling part 2 Functions of Cell Surface Receptors Other cell surface receptors are directly linked to intracellular enzymes. The largest family of these is the receptor protein tyrosine kinases,
15 Cell Signaling part 2 Functions of Cell Surface Receptors Other cell surface receptors are directly linked to intracellular enzymes. The largest family of these is the receptor protein tyrosine kinases,
Proteins? Protein function. Protein folding. Protein folding diseases. Protein interactions. Macromolecular assemblies. The end product of Genes
 Proteins? Protein function Protein folding Protein folding diseases Protein interactions Macromolecular assemblies The end product of Genes Protein Unfolding DOD Acid Catalysis DOD HDOD + N H N D C N C
Proteins? Protein function Protein folding Protein folding diseases Protein interactions Macromolecular assemblies The end product of Genes Protein Unfolding DOD Acid Catalysis DOD HDOD + N H N D C N C
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Stelter et al., http://www.jcb.org/cgi/content/full/jcb.201105042/dc1 S1 Figure S1. Dyn2 recruitment to edid-labeled FG domain Nups.
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Stelter et al., http://www.jcb.org/cgi/content/full/jcb.201105042/dc1 S1 Figure S1. Dyn2 recruitment to edid-labeled FG domain Nups.
Supporting Information Table of content
 Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
Supplemental Data. Wu et al. (2010). Plant Cell /tpc
 Supplemental Figure 1. FIM5 is preferentially expressed in stamen and mature pollen. The expression data of FIM5 was extracted from Arabidopsis efp browser (http://www.bar.utoronto.ca/efp/development/),
Supplemental Figure 1. FIM5 is preferentially expressed in stamen and mature pollen. The expression data of FIM5 was extracted from Arabidopsis efp browser (http://www.bar.utoronto.ca/efp/development/),
CA Tel.: ; Fax: ; ucsd.edu.
 THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 283, NO. 10, pp. 6300 6311, March 7, 2008 2008 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. The Phosphatase PHLPP
THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 283, NO. 10, pp. 6300 6311, March 7, 2008 2008 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. The Phosphatase PHLPP
Nature Immunology doi: /ni.3268
 Supplementary Figure 1 Loss of Mst1 and Mst2 increases susceptibility to bacterial sepsis. (a) H&E staining of colon and kidney sections from wild type and Mst1 -/- Mst2 fl/fl Vav-Cre mice. Scale bar,
Supplementary Figure 1 Loss of Mst1 and Mst2 increases susceptibility to bacterial sepsis. (a) H&E staining of colon and kidney sections from wild type and Mst1 -/- Mst2 fl/fl Vav-Cre mice. Scale bar,
Supplementary Figure 1. The CagA-dependent wound healing or transwell migration of gastric cancer cell. AGS cells transfected with vector control or
 Supplementary Figure 1. The CagA-dependent wound healing or transwell migration of gastric cancer cell. AGS cells transfected with vector control or 3xflag-CagA expression vector were wounded using a pipette
Supplementary Figure 1. The CagA-dependent wound healing or transwell migration of gastric cancer cell. AGS cells transfected with vector control or 3xflag-CagA expression vector were wounded using a pipette
EFFECTORS IMPLICATED IN THE AC1 INHIBITORY EFFECT ON CELL MIGRATION IN PANCREATIC CANCER CELLS
 EFFECTORS IMPLICATED IN THE AC1 INHIBITORY EFFECT ON CELL MIGRATION IN PANCREATIC CANCER CELLS VIDYA MEDEPALLI ADVISOR: Maria Eugenia Sabbatini, PhD PHI KAPPA PHI RESEARCH CONFERENCE MARCH 8 th, 2017 Pancreatic
EFFECTORS IMPLICATED IN THE AC1 INHIBITORY EFFECT ON CELL MIGRATION IN PANCREATIC CANCER CELLS VIDYA MEDEPALLI ADVISOR: Maria Eugenia Sabbatini, PhD PHI KAPPA PHI RESEARCH CONFERENCE MARCH 8 th, 2017 Pancreatic
m 6 A mrna methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer
 SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
Supporting Information
 Supporting Information Mass Spectrometry Imaging Shows Cocaine and Methylphenidate have Opposite Effects on Major Lipids in Drosophila Brain Mai H. Philipsen *, Nhu T. N. Phan *, John S. Fletcher *, Per
Supporting Information Mass Spectrometry Imaging Shows Cocaine and Methylphenidate have Opposite Effects on Major Lipids in Drosophila Brain Mai H. Philipsen *, Nhu T. N. Phan *, John S. Fletcher *, Per
The mechanism of patellamide macrocyclization revealed by study of the Prochloron sp PatG macrocyclase domain
 The mechanism of patellamide macrocyclization revealed by study of the Prochloron sp PatG macrocyclase domain Jesko Koehnke 1,2, Andrew Bent 1,2, Wael E. Houssen 2,3,4, David Zollman 1, Falk Morawitz 1,
The mechanism of patellamide macrocyclization revealed by study of the Prochloron sp PatG macrocyclase domain Jesko Koehnke 1,2, Andrew Bent 1,2, Wael E. Houssen 2,3,4, David Zollman 1, Falk Morawitz 1,
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Use of a camp BRET Sensor to Characterize a Novel Regulation of camp by the Sphingosine-1-phosphate/G 13 Pathway
 Use of a camp BRET Sensor to Characterize a Novel Regulation of camp by the Sphingosine-1-phosphate/G 13 Pathway SUPPLEMENTAL DATA Characterization of the CAMYEL sensor and calculation of intracellular
Use of a camp BRET Sensor to Characterize a Novel Regulation of camp by the Sphingosine-1-phosphate/G 13 Pathway SUPPLEMENTAL DATA Characterization of the CAMYEL sensor and calculation of intracellular
Supporting Information. Lysine Propionylation to Boost Proteome Sequence. Coverage and Enable a Silent SILAC Strategy for
 Supporting Information Lysine Propionylation to Boost Proteome Sequence Coverage and Enable a Silent SILAC Strategy for Relative Protein Quantification Christoph U. Schräder 1, Shaun Moore 1,2, Aaron A.
Supporting Information Lysine Propionylation to Boost Proteome Sequence Coverage and Enable a Silent SILAC Strategy for Relative Protein Quantification Christoph U. Schräder 1, Shaun Moore 1,2, Aaron A.
Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted. residues are colored red. Numbers indicate the position of each residue.
 Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted residues are colored red. Numbers indicate the position of each residue. Supplementary Figure 2. SDS-PAGE analysis of purified recombinant
Supplementary Figure 1. Amino acid sequences of GodA and GodA*. Inserted residues are colored red. Numbers indicate the position of each residue. Supplementary Figure 2. SDS-PAGE analysis of purified recombinant
Formation of the Peroxisome Lumen Is Abolished by Loss of Pichia pastoris Pas7p, a Zinc-Binding Integral Membrane Protein of the Peroxisome
 MOLECULAR AND CELLULAR BIOLOGY, Nov. 1995, p. 6406 6419 Vol. 15, No. 11 0270-7306/95/$04.00 0 Copyright 1995, American Society for Microbiology Formation of the Peroxisome Lumen Is Abolished by Loss of
MOLECULAR AND CELLULAR BIOLOGY, Nov. 1995, p. 6406 6419 Vol. 15, No. 11 0270-7306/95/$04.00 0 Copyright 1995, American Society for Microbiology Formation of the Peroxisome Lumen Is Abolished by Loss of
Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene
 Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene YUELIN ZHANG, WEIHUA FAN, MARK KINKEMA, XIN LI, AND
Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene YUELIN ZHANG, WEIHUA FAN, MARK KINKEMA, XIN LI, AND
Supporting information
 Supporting information Figure legends Supplementary Table 1. Specific product ions obtained from fragmentation of lithium adducts in the positive ion mode comparing the different positional isomers of
Supporting information Figure legends Supplementary Table 1. Specific product ions obtained from fragmentation of lithium adducts in the positive ion mode comparing the different positional isomers of
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation
 Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
Construction of a hepatocellular carcinoma cell line that stably expresses stathmin with a Ser25 phosphorylation site mutation J. Du 1, Z.H. Tao 2, J. Li 2, Y.K. Liu 3 and L. Gan 2 1 Department of Chemistry,
Supplemental Data. Shin et al. Plant Cell. (2012) /tpc YFP N
 MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
Chapter 3. Expression of α5-megfp in Mouse Cortical Neurons. on the β subunit. Signal sequences in the M3-M4 loop of β nachrs bind protein factors to
 22 Chapter 3 Expression of α5-megfp in Mouse Cortical Neurons Subcellular localization of the neuronal nachr subtypes α4β2 and α4β4 depends on the β subunit. Signal sequences in the M3-M4 loop of β nachrs
22 Chapter 3 Expression of α5-megfp in Mouse Cortical Neurons Subcellular localization of the neuronal nachr subtypes α4β2 and α4β4 depends on the β subunit. Signal sequences in the M3-M4 loop of β nachrs
The MOLECULES of LIFE
 The MOLECULES of LIFE Physical and Chemical Principles Solutions Manual Prepared by James Fraser and Samuel Leachman Chapter 16 Principles of Enzyme Catalysis Problems True/False and Multiple Choice 1.
The MOLECULES of LIFE Physical and Chemical Principles Solutions Manual Prepared by James Fraser and Samuel Leachman Chapter 16 Principles of Enzyme Catalysis Problems True/False and Multiple Choice 1.
End of the Note Book
 End of the Note Book 16 September 2016 Colonies PCR Mix (25 µl total volume reaction): + clones - 12.5 µl DreamTaq PCR Mastermix 2X (ThermoScientific) - 2.5 µl 10X DreamTaq Green Buffer (ThermoScientific)
End of the Note Book 16 September 2016 Colonies PCR Mix (25 µl total volume reaction): + clones - 12.5 µl DreamTaq PCR Mastermix 2X (ThermoScientific) - 2.5 µl 10X DreamTaq Green Buffer (ThermoScientific)
In Vivo Action of the HRD Ubiquitin Ligase Complex: Mechanisms of Endoplasmic Reticulum Quality Control and Sterol Regulation
 MOLECULAR AND CELLULAR BIOLOGY, July 2001, p. 4276 4291 Vol. 21, No. 13 0270-7306/01/$04.00 0 DOI: 10.1128/MCB.21.13.4276 4291.2001 Copyright 2001, American Society for Microbiology. All Rights Reserved.
MOLECULAR AND CELLULAR BIOLOGY, July 2001, p. 4276 4291 Vol. 21, No. 13 0270-7306/01/$04.00 0 DOI: 10.1128/MCB.21.13.4276 4291.2001 Copyright 2001, American Society for Microbiology. All Rights Reserved.
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein
 Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Figure S1! CrFK! DNA Synthesis! Copies viral DNA/ 100 ng DNA (Log 10 ) % Infected (GFP+) cells! 100! 80! 60! 40! 20! 0! 1! 10! 100! 1000!
 Figure S1! A! B! % Infected (GFP+) cells! 100! 80! 60! 40! 20! 0! 1! 10! 100! 1000! Dose μl! CrFK! B-MLV! N-MLV! Template:! Water! Ube2V2! Ube2V1! Primers:! V2! V1! V2! V1! V2! V1! bp! 500! 300! 200! 150!
Figure S1! A! B! % Infected (GFP+) cells! 100! 80! 60! 40! 20! 0! 1! 10! 100! 1000! Dose μl! CrFK! B-MLV! N-MLV! Template:! Water! Ube2V2! Ube2V1! Primers:! V2! V1! V2! V1! V2! V1! bp! 500! 300! 200! 150!
Supplementary Information
 Supplementary Information Using the pimeloyl-coa synthetase adenylation fold to synthesise fatty acid thioesters Menglu Wang 1a ; Lucile Moynié 2a, Peter J. Harrison 1, Van Kelly 1, Andrew Piper 1, James
Supplementary Information Using the pimeloyl-coa synthetase adenylation fold to synthesise fatty acid thioesters Menglu Wang 1a ; Lucile Moynié 2a, Peter J. Harrison 1, Van Kelly 1, Andrew Piper 1, James
A smart acid nanosystem for ultrasensitive. live cell mrna imaging by the target-triggered intracellular self-assembly
 Electronic Supplementary Material (ESI) for Chemical Science. This journal is The Royal Society of Chemistry 2017 A smart ZnO@polydopamine-nucleic acid nanosystem for ultrasensitive live cell mrna imaging
Electronic Supplementary Material (ESI) for Chemical Science. This journal is The Royal Society of Chemistry 2017 A smart ZnO@polydopamine-nucleic acid nanosystem for ultrasensitive live cell mrna imaging
Supplementary Fig. S1. Schematic diagram of minigenome segments.
 open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
Supplemental Table 1. Plasmid Construct backbone. Restriction sites. PCR template. HLA DRα pcdna3.1 Zeo+
 SupplementalTable1 Plasmid Construct backbone HLA DRα Zeo+ Restriction sites PCR template PCRCloningstrategy Primerpairs(F/R)andpairsofprimersforchimericconstructs(N terminalandc terminal codingregion)weregiven
SupplementalTable1 Plasmid Construct backbone HLA DRα Zeo+ Restriction sites PCR template PCRCloningstrategy Primerpairs(F/R)andpairsofprimersforchimericconstructs(N terminalandc terminal codingregion)weregiven
Chapter 11: Enzyme Catalysis
 Chapter 11: Enzyme Catalysis Matching A) high B) deprotonated C) protonated D) least resistance E) motion F) rate-determining G) leaving group H) short peptides I) amino acid J) low K) coenzymes L) concerted
Chapter 11: Enzyme Catalysis Matching A) high B) deprotonated C) protonated D) least resistance E) motion F) rate-determining G) leaving group H) short peptides I) amino acid J) low K) coenzymes L) concerted
supplementary information
 DOI: 1.138/ncb1 Control Atg7 / NAC 1 1 1 1 (mm) Control Atg7 / NAC 1 1 1 1 (mm) Lamin B Gstm1 Figure S1 Neither the translocation of into the nucleus nor the induction of antioxidant proteins in autophagydeficient
DOI: 1.138/ncb1 Control Atg7 / NAC 1 1 1 1 (mm) Control Atg7 / NAC 1 1 1 1 (mm) Lamin B Gstm1 Figure S1 Neither the translocation of into the nucleus nor the induction of antioxidant proteins in autophagydeficient
Nature Biotechnology: doi: /nbt Supplementary Figure 1. RNAseq expression profiling of selected glycosyltransferase genes in CHO.
 Supplementary Figure 1 RNAseq expression profiling of selected glycosyltransferase genes in CHO. RNAseq analysis was performed on two common CHO lines (CHO-K1, CHO-GS) and two independent CHO-GS triple
Supplementary Figure 1 RNAseq expression profiling of selected glycosyltransferase genes in CHO. RNAseq analysis was performed on two common CHO lines (CHO-K1, CHO-GS) and two independent CHO-GS triple
Trim29 gene-targeting strategy. (a) Genotyping of wildtype mice (+/+), Trim29 heterozygous mice (+/ ) and homozygous mice ( / ).
 Supplementary Figure 1 Trim29 gene-targeting strategy. (a) Genotyping of wildtype mice (+/+), Trim29 heterozygous mice (+/ ) and homozygous mice ( / ). (b) Immunoblot analysis of TRIM29 in lung primary
Supplementary Figure 1 Trim29 gene-targeting strategy. (a) Genotyping of wildtype mice (+/+), Trim29 heterozygous mice (+/ ) and homozygous mice ( / ). (b) Immunoblot analysis of TRIM29 in lung primary
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Chairoungdua et al., http://www.jcb.org/cgi/content/full/jcb.201002049/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Expression of CD9 and CD82 inhibits Wnt/ -catenin
Supplemental material Chairoungdua et al., http://www.jcb.org/cgi/content/full/jcb.201002049/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Expression of CD9 and CD82 inhibits Wnt/ -catenin
SUPPLEMENTARY INFORMATION. An orthogonal ribosome-trnas pair by the engineering of
 SUPPLEMENTARY INFORMATION An orthogonal ribosome-trnas pair by the engineering of peptidyl transferase center Naohiro Terasaka 1 *, Gosuke Hayashi 1 *, Takayuki Katoh 1, and Hiroaki Suga 1,2 1 Department
SUPPLEMENTARY INFORMATION An orthogonal ribosome-trnas pair by the engineering of peptidyl transferase center Naohiro Terasaka 1 *, Gosuke Hayashi 1 *, Takayuki Katoh 1, and Hiroaki Suga 1,2 1 Department
Double charge of 33kD peak A1 A2 B1 B2 M2+ M/z. ABRF Proteomics Research Group - Qualitative Proteomics Study Identifier Number 14146
 Abstract The 2008 ABRF Proteomics Research Group Study offers participants the chance to participate in an anonymous study to identify qualitative differences between two protein preparations. We used
Abstract The 2008 ABRF Proteomics Research Group Study offers participants the chance to participate in an anonymous study to identify qualitative differences between two protein preparations. We used
The Comparison of High Resolution MS with Triple Quadrupole MS for the Analysis of Oligonucleotides
 The Comparison of High Resolution MS with Triple Quadrupole MS for the Analysis of Oligonucleotides Mohammed Abrar Unilabs York Bioanalytical Solutions Outline Introduction Why LC-MS/MS? Limitations of
The Comparison of High Resolution MS with Triple Quadrupole MS for the Analysis of Oligonucleotides Mohammed Abrar Unilabs York Bioanalytical Solutions Outline Introduction Why LC-MS/MS? Limitations of
EXPANDING THE SET OF KNOWN SUBSTRATES OF NUCLEAR RNASE P. Daniel J Coughlin
 EXPANDING THE SET OF KNOWN SUBSTRATES OF NUCLEAR RNASE P by Daniel J Coughlin A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Biological Chemistry)
EXPANDING THE SET OF KNOWN SUBSTRATES OF NUCLEAR RNASE P by Daniel J Coughlin A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Biological Chemistry)
Measuring Lipid Composition LC-MS/MS
 Project: Measuring Lipid Composition LC-MS/MS Verification of expected lipid composition in nanomedical controlled release systems by liquid chromatography tandem mass spectrometry AUTHORED BY: DATE: Sven
Project: Measuring Lipid Composition LC-MS/MS Verification of expected lipid composition in nanomedical controlled release systems by liquid chromatography tandem mass spectrometry AUTHORED BY: DATE: Sven
SUPPLEMENTARY INFORMATION Glucosylceramide synthase (GlcT-1) in the fat body controls energy metabolism in Drosophila
 SUPPLEMENTARY INFORMATION Glucosylceramide synthase (GlcT-1) in the fat body controls energy metabolism in Drosophila Ayako Kohyama-Koganeya, 1,2 Takuji Nabetani, 1 Masayuki Miura, 2,3 Yoshio Hirabayashi
SUPPLEMENTARY INFORMATION Glucosylceramide synthase (GlcT-1) in the fat body controls energy metabolism in Drosophila Ayako Kohyama-Koganeya, 1,2 Takuji Nabetani, 1 Masayuki Miura, 2,3 Yoshio Hirabayashi
Overview of the Expressway Cell-Free Expression Systems. Expressway Mini Cell-Free Expression System
 Overview of the Expressway Cell-Free Expression Systems The Expressway Cell-Free Expression Systems use an efficient coupled transcription and translation reaction to produce up to milligram quantities
Overview of the Expressway Cell-Free Expression Systems The Expressway Cell-Free Expression Systems use an efficient coupled transcription and translation reaction to produce up to milligram quantities
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
The Impact of a Transposon Insertion in phzf2 on the Specialized Metabolite. Production and Interkingdom Interactions of Pseudomonas aeruginosa
 1 2 3 Supplemental Material The Impact of a Transposon Insertion in phzf2 on the Specialized Metabolite Production and Interkingdom Interactions of Pseudomonas aeruginosa 4 5 6 7 Vanessa V. Phelan a, Wilna
1 2 3 Supplemental Material The Impact of a Transposon Insertion in phzf2 on the Specialized Metabolite Production and Interkingdom Interactions of Pseudomonas aeruginosa 4 5 6 7 Vanessa V. Phelan a, Wilna
Receptor mediated Signal Transduction
 Receptor mediated Signal Transduction G-protein-linked receptors adenylyl cyclase camp PKA Organization of receptor protein-tyrosine kinases From G.M. Cooper, The Cell. A molecular approach, 2004, third
Receptor mediated Signal Transduction G-protein-linked receptors adenylyl cyclase camp PKA Organization of receptor protein-tyrosine kinases From G.M. Cooper, The Cell. A molecular approach, 2004, third
A Yersinia-Secreted Effector Protein Promotes Virulence by Preventing Inflammasome Recognition of the Type III Secretion System
 Cell Host & Microbe, Volume 7 Supplemental Information A Yersinia-Secreted Effector Protein Promotes Virulence by Preventing Inflammasome Recognition of the Type III Secretion System Igor E. Brodsky, Noah
Cell Host & Microbe, Volume 7 Supplemental Information A Yersinia-Secreted Effector Protein Promotes Virulence by Preventing Inflammasome Recognition of the Type III Secretion System Igor E. Brodsky, Noah
Nucleotide degradation and ribose salvage in yeast
 Nucleotide degradation and ribose salvage in yeast Yi-Fan Xu 1,2, Fabien Létisse 4, Farnaz Absalan 3, Wenyun Lu 1, Ekaterina Kuznetsova 5, Greg Brown 5, Amy A. Caudy 6, Alexander F. Yakunin 5, James R.
Nucleotide degradation and ribose salvage in yeast Yi-Fan Xu 1,2, Fabien Létisse 4, Farnaz Absalan 3, Wenyun Lu 1, Ekaterina Kuznetsova 5, Greg Brown 5, Amy A. Caudy 6, Alexander F. Yakunin 5, James R.
PhosFree TM Phosphate Assay Biochem Kit
 PhosFree TM Phosphate Assay Biochem Kit (Cat. # BK050) ORDERING INFORMATION To order by phone: (303) - 322-2254 To order by Fax: (303) - 322-2257 To order by e-mail: cservice@cytoskeleton.com Technical
PhosFree TM Phosphate Assay Biochem Kit (Cat. # BK050) ORDERING INFORMATION To order by phone: (303) - 322-2254 To order by Fax: (303) - 322-2257 To order by e-mail: cservice@cytoskeleton.com Technical
Effec<ve Use of PI3K and MEK Inhibitors to Treat Mutant K Ras G12D and PIK3CA H1047R Murine Lung Cancers
 Effec
Effec
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
Supporting information (protein purification, kinetic characterization, product isolation, and characterization by NMR and mass spectrometry):
 Supporting Information Mechanistic studies of a novel C-S lyase in ergothioneine biosynthesis: the involvement of a sulfenic acid intermediate Heng Song, 1 Wen Hu, 1,2 Nathchar Naowarojna, 1 Ampon Sae
Supporting Information Mechanistic studies of a novel C-S lyase in ergothioneine biosynthesis: the involvement of a sulfenic acid intermediate Heng Song, 1 Wen Hu, 1,2 Nathchar Naowarojna, 1 Ampon Sae
Supplemental Data Macrophage Migration Inhibitory Factor MIF Interferes with the Rb-E2F Pathway
 Supplemental Data Macrophage Migration Inhibitory Factor MIF Interferes with the Rb-E2F Pathway S1 Oleksi Petrenko and Ute M. Moll Figure S1. MIF-Deficient Cells Have Reduced Transforming Ability (A) Soft
Supplemental Data Macrophage Migration Inhibitory Factor MIF Interferes with the Rb-E2F Pathway S1 Oleksi Petrenko and Ute M. Moll Figure S1. MIF-Deficient Cells Have Reduced Transforming Ability (A) Soft
Tandem mass spectrometry analysis of prostaglandins and isoprostanes
 Tandem mass spectrometry analysis of prostaglandins and isoprostanes Jeevan Prasain jprasain@uab.edu 6-2612 verview Introduction to PGs and their synthesis Mass spectrometry characterization of PGs and
Tandem mass spectrometry analysis of prostaglandins and isoprostanes Jeevan Prasain jprasain@uab.edu 6-2612 verview Introduction to PGs and their synthesis Mass spectrometry characterization of PGs and
ARV Mode of Action. Mode of Action. Mode of Action NRTI. Immunopaedia.org.za
 ARV Mode of Action Mode of Action Mode of Action - NRTI Mode of Action - NNRTI Mode of Action - Protease Inhibitors Mode of Action - Integrase inhibitor Mode of Action - Entry Inhibitors Mode of Action
ARV Mode of Action Mode of Action Mode of Action - NRTI Mode of Action - NNRTI Mode of Action - Protease Inhibitors Mode of Action - Integrase inhibitor Mode of Action - Entry Inhibitors Mode of Action
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
Supporting Information
 Supporting Information For Heterologous production of thiostrepton A and biosynthetic engineering of thiostrepton analogs Chaoxuan Li, Feifei Zhang, and Wendy L. Kelly* Table of Contents Figure S1. PCR
Supporting Information For Heterologous production of thiostrepton A and biosynthetic engineering of thiostrepton analogs Chaoxuan Li, Feifei Zhang, and Wendy L. Kelly* Table of Contents Figure S1. PCR
