ARTICLE. Identification of essential genes for cancer immunotherapy
|
|
|
- Eustacia McCormick
- 5 years ago
- Views:
Transcription
1 doi:1.138/nture23477 Identifiction of essentil genes for cncer immunotherpy Shshnk J. Ptel 1,2 *, Neville E. Snjn 3,4 *, Rigel J. Kishton 1, Arsh Eidizdeh 1, Sumn K. Vodnl 1, Mggie Cm 1, Jred J. Grtner 1, Li Ji 1, Seth M. Steinerg 1, Tori N. Ymmoto 1,5, Annd S. Merchnt 1, Gutm U. Meht 1, Ann Chichur 1, Ophir Shlem 6, Eric Trn 1, Roert Eil 1, Mdhusudhnn Sukumr 1, Ev Perez Guijrro 1, Chi-Ping Dy 1, Pul Roins 1, Steve Feldmn 1, Glenn Merlino 1, Feng Zhng 7,8 & Nichols P. Restifo 1,9 Somtic gene muttions cn lter the vulnerility of cncer cells to T-cell-sed immunotherpies. Here we pertured genes in humn melnom cells to mimic loss-of-function muttions involved in resistnce to these therpies, y using genome-scle CRISPR Cs9 lirry tht consisted of round 123, single-guide RNAs, nd profiled genes whose loss in tumour cells impired the effector function of CD8 + T cells. The genes tht were most enriched in the screen hve key roles in ntigen presenttion nd interferon-γ signlling, nd correlte with cytolytic ctivity in ptient tumours from The Cncer Genome Atls. Among the genes vlidted using different cncer cell lines nd ntigens, we identified multiple loss-of-function muttions in, encoding the pelin receptor, in ptient tumours tht were refrctory to immunotherpy. We show tht intercts with JAK1, modulting interferon-γ responses in tumours, nd tht its functionl loss reduces the efficcy of doptive cell trnsfer nd checkpoint lockde immunotherpies in mouse models. Our results link the loss of essentil genes for the effector function of CD8 + T cells with the resistnce or nonresponsiveness of cncer to immunotherpies. Genetic errtions re generted in most cncers s product of their neoplstic evolution 1. Somtic muttions cn give rise to neontigens tht re cple of eliciting potent T cell responses driven y current immunotherpies 2 7. However, muttions cn lso induce resistnce to immunotherpies. For exmple, loss-of-function muttions in β -2-microgloulin (B2M) nd Jnus kinses (JAK1 nd JAK2) hve een reported in ptients unresponsive to immunotherpies 8,9. Nevertheless, the identity of functionlly essentil genes in cncer cells tht fcilitte immune selection y immunotherpies remins unknown. To systemticlly ctlogue genes in tumours whose loss cn enle immune escpe from T-cell-medited cytolysis, we used genomescle CRISPR Cs9 mutgenesis screen in humn melnom cells. CRISPR Cs9 screens hve een used to identify genes tht re criticl for prolifertion 1,11, drug resistnce 12,13, nd metstsis 14 of cncer cells. To identify the genes essentil in tumours for the effector function of T cells (EFT), we developed two cell type (2CT)-CRISPR ssy consisting of humn T cells s effectors nd melnom cells s trgets. We sought to understnd how genetic mnipultions in one cell type cn ffect complex interction with nother cell type. In ddition, the 2CT ssy enled us to perform pooled screens with higher lirry representtion thn cn e chieved in vivo. Here we report severl previously undescried genes nd micrornas tht hve role in fcilitting tumour destruction y T cells. We exmined the correltion of expression of these cndidte genes with nlyses of cytolytic ctivity in round 11, humn tumours from The Cncer Genome Atls (TCGA), nd lso report how the loss of cndidte gene cn medite resistnce to T-cell-sed immunotherpies in humn nd mouse tumours. A 2CT-CRISPR ssy system To identify cell types nd genes relted to the nti-tumour function of immune cells tht might e necessry for immunotherpy, we performed correltion nlysis on gene expression dt from ptients with melnom treted with the nti-ctla4 ntiody ipilimum 3. Consistent with previous reports 15,16, we found tht intrtumorl cytoly tic ctivity (computed s the geometric men of perforin (PRF1) nd grnzyme (GZMA) expression) ws strongly correlted with CD8 + T cell infiltrtion in the tumours (Extended Dt Fig. 1; r =.96, P < ) nd with the expression of genes involved in the MHC clss I ntigen processing/presenttion pthwy (Extended Dt Fig. 1; r >.54, P <.1 for ech gene), ut wekly correlted with interferon-γ (IFNγ ) signlling genes (Extended Dt Fig. 1c). We oserved tht the reduction in the overll survivl of these ptients ws significntly ssocited with loss of expression of B2M nd TAP1 in tumours iopsied efore ipilimum tretment (Fig. 1 c, Extended Dt Fig. 1d g). Given these ssocitions, we chose to use CD8 + T cells nd MHC clss I genes to develop the 2CT-CRISPR ssy system. The 2CT-CRISPR ssy employs gene-engineered CD8 + T cell to specificlly trget n ntigen expressed in n HLA-clss I-restricted mnner (Extended Dt Fig. 2 d). We used primry humn T cells trnsduced with recominnt T cell receptor (TCR) specific for the NY-ESO-1 ntigen (NY-ESO-1: epitope) presented in n HLA-A* 2-restricted mnner (ESO T cells) tht we previously reported to medite tumour regression in ptients with melnoms nd synovil cell srcoms (Fig. 1d, e, Extended Dt Fig. 2 c). We optimized the 2CT ssy to control the selection pressure, llorectivity nd ystnder killing 2 exerted y T cells y modulting the effector to trget (E:T) rtio nd length of co-incution (Extended Dt Fig. 2e i). To test whether the loss of ntigen presenttion genes cn directly compromise T-cell-medited cell lysis of humn cncer cells using our 2CT-CRISPR ssy, we trgeted TAP2 nd B2M with three unique single-guide RNAs (sgrnas) cloned into the lenticrisprv2 lentivirl vector in NY-ESO-1 + Mel624 melnom cells. Fluorescence-ctivted cell sorting (FACS) nlysis confirmed tht B2M-trgeting lentivirl 1 Ntionl Cncer Institute, Ntionl Institutes of Helth (NIH), Bethesd, Mrylnd 2892, USA. 2 NIH-Georgetown University Grdute Prtnership Progrm, Georgetown University Medicl School, Wshington DC 257, USA. 3 New York Genome Center, New York, New York 113, USA. 4 Deprtment of Biology, New York University, New York, New York 112, USA. 5 Immunology Grdute Group, University of Pennsylvni, Phildelphi, Pennsylvni 1914, USA. 6 Children s Hospitl of Phildelphi nd Deprtment of Genetics, University of Pennsylvni, Pennsylvni 1914, USA. 7 Brod Institute of MIT nd Hrvrd, Cmridge, Msschusetts 2142, USA. 8 McGovern Institute for Brin Reserch, Msschusetts Institute of Technology, Cmridge, Msschusetts 2139, USA. 9 Center for Cell-sed Therpy, Center for Cncer Reserch, Ntionl Institutes of Helth (NIH), Bethesd, Mrylnd 2892, USA. *These uthors contriuted eqully to this work. MONTH 217 VO L N A T URE Mcmilln Pulishers Limited, prt of Springer Nture. All rights reserved.
2 RESEARCH ARTICLE Survivl proility B2M c TAP1 1. TAP2 High Low Leukpheresis of primry humn CD8+ T cells HR.36 P =.21 HR.15 P =.18 HR.9 P = Retrovirl trnsduction with NY-ESO-1 TCR 6 T cell Time fter ipilimum tretment (months) Cs9 trgeting SK23 NY-ESO-1 ntigen SK23 Mel *** Ctrl TAP2 B2M CRISPR edited Figure 1 2CT-CRISPR ssy system confirms functionl essentility of ntigen presenttion genes for immunotherpy. c, Kpln Meier survivl plots of ptient overll survivl with the expression of ntigen presenttion genes B2M (), TAP1 () nd TAP2 (c) fter ipilimum immunotherpy. Ptients were ctegorized into high nd low groups ccording to the highest nd the lowest qurtiles of ech individul gene expression (RPKM). Reported P vlues hve een djusted y multiplying the undjusted P vlue y 3 to ccount for the comprison of the two extreme qurtiles. Hzrd rtio ws clculted using Mntel Henszel test; with hzrd rtio rnges: B2M (.2.31), TAP1 (.4.52) nd TAP2 ( ). Dt re derived from 42 ptients with melnom from the Vn Allen et l. cohort3. d, Schemtic of the 2CT-CRISPR ssy to identify essentil genes for EFT. e, NY-ESO-1 ntigen-specific lysis of melnom cells fter 24 h of co-culture of ESO T cells with NY-ESO-1 SK23, NY-ESO-1+ SK23 nd NY-ESO-1+ Mel624 cells (n = 3 iologicl replictes) t E:T rtio of 1. f, Survivl of Mel624 cells modified through lentivirl CRISPR trgeting of MHC clss I ntigen presenttion/processing genes fter introduction of ESO T cells. CRISPR-modified Mel624 cells were co-cultured with ESO T cells t E:T rtio of.5 for 12 h. Live cell survivl (%) ws clculted from control cells unexposed to T cell selection. Dt re from three independent infection replictes. All vlues re men ± s.e.m. ***P <.1 s determined y Student s t-test. CRISPRs chieved round 95% protein knockout (Extended Dt Fig. 3). Significnt resistnce ws detected in the cells trnsduced with B2M sgrnas (72 ± 5%) nd with TAP2 sgrnas (13 ± 2%) ginst the cytolytic ctivity of ESO T cells (Fig. 1f, Extended Dt Fig. 3, c). These results show tht loss of key MHC clss I genes promotes evsion of T-cell-medited tumour killing in the optimized 2CT-CRISPR ssy. Genome-wide 2CT-CRISPR screen for EFT Synthesize CRISPR lirry nd produce lentivirus To identify the tumour-intrinsic genes essentil for EFT on genomescle, we trnsduced Mel624 cells with the genome-scle CRISPR knockout (GeCKOv.2) lirry t multiplicity of infection (MOI) <.3 (Fig. 2). The GeCKOv.2 lirry consists of 123,411 sgrnas tht trget 19,5 protein-coding genes (6 sgrnas per gene) nd 1,864 micrornas (4 sgrnas per microrna), nd lso includes round 1, non-trgeting control sgrnas21. We exposed trnsduced tumour cells to ESO T cells t E:T rtios of.3 nd.5 for 12 h in independent screens tht resulted in round 76% nd round 9% tumour cell lysis, respectively. Using deep sequencing, we exmined the sgrna lirry representtion in tumour cells efore nd fter T cell co-incution (Extended Dt Fig. 4, ). We oserved tht the distriution of the sgrna reds in T-cell-treted smples versus controls ws significntly ltered in screens with the higher numer of T cells, E:T of.5 (Kolmogorov Smirnov test, P = ), nd not with n E:T of.3 (Extended Dt Fig. 4, P =.7), indicting tht the efficiency of this 2CT-CRISPR ssy ws dependent on the selection pressure pplied y T cells. We quntified consistent enrichment of cndidte genes y multiple methods: (1) rnking genes y their second-most-enriched sgrna (Fig. 2); (2) the RNAi Gene Enrichment Rnking (RIGER) metric22 c HLA-A SRP54 2 TAP 1 Trnsduce NY-ESO-1+ Mel624 cells 8 B2M SOX1 mir-11-2 CD Second-est sgrna (norm. score) log1(riger P) Lytic grnules 1 Compre chnge in sgrna counts Control *** 8 6 CD58 HLA-A SRP54 B2M SOX1 TAF3 TAP1 4 JAK1 STAT1 RPL23 COL17A1 2 d e 6 HLA-A Antigen presenttion Plsm memrne B2M Peptide frgments 4 ER TAP1 TAP2 MHC-I 2 PSMB5 RPL1A RNPS1 TAP2 HLA-A COL17A1 TAP1 CRKL mir-548s MAD2L1 SRP54 TAF3 mir-11-2 RPL23 PSMB5 MLANA B2M SOX1 CD58 X f Mel624 surviving (%) X No T cells ESO T cells 2 Cell count ( 15) EFT regultors Recominnt TCR-engineered T cell Top sgrna (norm. score) e Tumour cell Numer of sgrnas in the top 5% most-enriched sgrnas d Tret cells with T cells or control Proteosome Cytoplsm Trnsporter ssocited with ntigen processing (TAP) Figure 2 Genome-wide CRISPR mutgenesis revels essentil genes for the effector function of T cells in trget cell., Design of the genome-wide 2CT-CRISPR ssy to identify loss-of-function genes conferring resistnce to T-cell-medited cytolysis., Sctterplot of the normlized enrichment of the most-enriched sgrna versus the secondmost-enriched sgrnas for ll genes fter T-cell-sed selection (inset). The top 1 genes y second-most-enriched sgrna rnk re displyed in the enlrged region. c, Identifiction of top enriched genes using the RIGER nlysis. d, Consistency of multiple sgrna enrichment for the top 2 rnked genes y second-most-enriched sgrna score. The numer of sgrnas trgeting ech gene tht re found in the top 5% of most-enriched sgrnas overll is plotted. e, Schemtic of MHC clss I ntigen processing pthwy with cndidte genes scoring in the top.1% of ll genes in the lirry highlighted. (Fig. 2c); nd (3) the numer of sgrnas for ech gene enriched in the top 5% of ll sgrnas in the lirry (Fig. 2d). All three methods showed high degree of overlp (Fig. 2 d, Extended Dt Fig. 4c, Supplementry Tle 1). Despite the disprity in the enriched sgrna distriutions etween screens with E:T of.3 nd.5, severl highlyrnked genes nd micrornas re shred etween the screens (Extended Dt Fig. 4d f). On the sis of our initil optimiztion of the 2CT-CRISPR ssy, we expected tht genes directly ssocited with MHC clss I ntigen processing nd presenttion would e enriched in our screens, nd found tht HLA-A, B2M, TAP1, TAP2, nd were mong the most highly-enriched genes in our screen (Fig. 2d, e). In ddition, mny genes without n estlished connection to the EFT were rnked mongst the top 2 enriched genes in this genome-scle nlysis, such s SOX1, CD58, MLANA, PSMB5, RPL23 nd (Fig. 2c, d). Biologicl role of puttive essentil genes for EFT We sought to ssess the clinicl nd iologicl significnce of the most-enriched 2CT-CRISPR cndidtes (554 genes t flse discovery rte (FDR) <.1%) y compring the cndidtes with pulicly ville cncer genome sequencing nd querying ssocition with 2 NAT U R E VO L M O N T H Mcmilln Pulishers Limited, prt of Springer Nture. All rights reserved.
3 RESEARCH Frequency c FDR.1% cut-off sgrna enrichment Numer of genes correlted with CYT in 2CT-CRISPR top-rnked genes top-rnked genes Non-trgeting Gene-trgeting TCGA RNA-seq (36 cncer sutypes) Genes correlted with CYT Induciility y effector cytokines Pthwy enrichment Cncer sutypes Assocition with intrtumorl CYT Muttion frequency in cncer genomes 2CT-CRISPR top-rnked genes CHOL USC KICH MESO DLBC READ SKCM ESCA UCEC LAML THYM CESC ACC UVM OV HNSC SARC LUAD LIHC STAD PCPG COADREAD COAD PAAD LUSC PRAD TGCT KIRP BLCA THCA GBM BRCA LGG KIRC GBMLGG KIPAN Figure 3 Ctegoriztion of cndidte essentil genes for EFT using ville knowledge dtse., Cndidte genes sed on FDR cut-off of.1% (n = 554 genes) were functionlly ctegorized y multiple methods to understnd the iologicl significnce for EFT., Most significnt pthwys y Ingenuity Pthwy Anlysis (P <.5) from top 2CT-CRISPR cndidtes. c, d, Assocition of top 554 cndidte genes with intrtumorl cytolytic ctivity (CYT, expression of perforin nd grnzyme) using TCGA dtsets. c, RNA-sequencing (RNA-seq) dt for 36 humn cncers were d Genes correlted with CYT Pthwy enrichment EIF2 signlling Antigen presenttion Endoplsmic reticulum stress RNA polymerse II complex CTL-medited poptosis Interferon signlling IL-15 production TNFR1 signlling Protein uiquitintion pthwy Deth receptor signlling TCGA cncer sutypes 1 1 Correltion with CYT (Person s r) log 1 (P) TBXAS1 GMIP OTOA LAIR1 CLECL1 GPSM3 TRAF1 JAK2 TABP L ICAM1 LILRA1 LILRA3 STAT1 HLA-A B2M HLA-F TAP1 TAP2 otined from the TCGA dtse nd genes positively correlted with cytolytic ctivity (P <.5, Person s correltion) were intersected with 2CT-CRISPR cndidtes to quntify numer of overlpping genes in ech cncer sutype. d, Hetmp showing the prtitioning of the clusters of genes sed on Person s correltion coefficient vlues of CRISPR screen hits with cytolytic ctivity using pn-cncer TCGA dt. Individul cncer nd pthwy hetmps re ville t pulictions/restifo. known cncer/immune pthwys (Fig. 3). As prt of their effector mechnism, T cells induce trnscriptionl chnges in the tumour microenvironment vi secretion of cytokines such s IFNγ nd tumour necrosis fctor-α (TNFα ) to enhnce recognition nd lysis of cncer cells 15, To ssess whether ny of these genes induced y effector cytokines re likely to hve functionl role in modultion of the EFT, we intersected the gene expression profiles of cytokine- induced genes with the cndidte genes. We found tht 13 IFNγ -induced genes nd 3 TNFα -induced genes were cptured in our 554 top cndidtes (Extended Dt Fig. 5), olstering the functionl relevnce of these cytokine-induced genes in tumour cells for EFT. Using gene ontology nd pthwy nlyses, we found tht in ddition to ntigen presenttion nd IFNγ signlling, 2CT-enriched genes function in EIF2 signlling, endoplsmic reticulum stress, poptosis, ssemly of RNA polymerse II, TNF receptor signlling nd protein uiquitintion pthwys (F-test, Bonferroni corrected P <.5; Fig. 3, Supplementry Tles 2, 3). These dt revel previously unrecognized signlling circuitry in tumours, the loss of which cn dmpen EFT, wrrnting further investigtion. To ssess whether loss of cndidte genes identified in the 2CT-CRISPR screen ws ssocited with decresed cytolytic ctivity in ptient tumours, we otined gene expression profiles of 11,49 humn tumours spnning 36 tumour-types from the TCGA dtse nd mesured the correltion etween cndidte genes nd cytolytic ctivity in these dtsets (Fig. 3c, d, Extended Dt Fig. 5, c). With this pproch, we generted list of genes tht ssocite with cytolytic ctivity for ech TCGA cncer type nd re lso enriched in our 2CT screens (Supplementry Tle 4, we resource cncer.gov/pulictions/restifo). Using hierrchicl clustering, we identified set of 19 genes tht re correlted with cytolytic ctivity cross most of the 36 cncer types (Fig. 3d, Extended Dt Fig. 5d, Supplementry Tles 5, 6). 1 of these 19 genes were inducile y IFNγ, indicting tht these genes my e upregulted in cncers owing to n incresed infiltrtion of T cells. Loss of expression of these 19 genes within tumours could diminish or extinguish the presenttion of tumour ntigens (including HLA-A, HLA-F, B2M, TAP1 nd TAP2); T cell co- stimultion (ICAM1, CLECL1, LILRA1 nd LILRA3); or cytokine production nd signlling (JAK2 nd STAT1) in the tumour microenvironment tht drive infiltrtion nd ctivtion of T cells, nd thus serve s principl mechnism in immune evsion. We interrogted muttion frequency of the top enriched genes in different tumour types in the TCGA dtsets (Extended Dt Fig. 5e, f, Supplementry Fig. 1). We found COL17A1, B2M, HLA-A, TAF3, DEFB134, ABR, QSER1 nd DHX9 ech to e mutted in > 1 ptient tumours cross different mlignncies, demonstrting tht muttions in these cndidte genes nturlly occur in humn cncers. Vlidtion nd generlizility of top cndidtes We elected to independently vlidte 17 genes on the sis of two criteri: the gene should e rnked mong the top 2 cndidtes in either screen, nd the gene should not e previously descried in nti-tumour function of T cells. We included CTAG1B (encoding tumour ntigen, NY-ESO-1) nd (encoding tpsin involved in MHC clss I ntigen processing) s positive controls. We trgeted ech gene in NY-ESO-1 + melnom cells, Mel624 nd A375, with four sgrnas (Supplementry Tle 7) nd individully MONTH 217 VO L N A T URE Mcmilln Pulishers Limited, prt of Springer Nture. All rights reserved.
4 RESEARCH ARTICLE sgrna score Cell survivl (%) 1 5 Genes Select top genes in 2CT-CRISPR screen A375 melnom:eso T Cs9 cut site Design new sgrnas not in GeCKO v.2 lirry c d Control sg SOX1 RPL23 SRP54 PTCD2 MLANA TWF1 CD58 PSMB5 COL17A1 DEFB134 mir-11-2 BBS1 At lest two sgrnas with >1% resistnce to cytolysis SOX1 RPL23 Mel624 BBS1 CD58 COL17A1 NY-ESO-1 DEFB134 MLANA mir-11-2 mesured resistnce ginst ESO T cells (Fig. 4). Fifteen genes showed significnt resistnce to T-cell-medited cytolysis with t lest 1 sgrna in these cells (Fig. 4, Extended Dt Fig. 6, Student s t-test, P <.5 compred to control sgrna, n = 3 iologicl replictes). To mitigte concerns of off-trget ctivity, we considered cndidte to e criticl gene for the EFT if loss-of-function resulted in resistnce phenotype with t lest two distinct sgrnas. Using these criteri, we vlidted nine cndidtes in oth Mel624 nd A375 cells (Fig. 4c). The trnscription fctor SOX1, whose expression is elevted round 16-fold in melnom over other cncers (Supplementry Fig. 2) nd trgets other top 2CT screen hits (for exmple, MITF), ws vlidted only in Mel624 cells (Fig. 4c), which my e owing to its higher expression ( 1.8-fold higher) in these cells compred to A375 cells 26. To test whether these vlidted genes generlize to n unrelted T cell receptor ntigen (MART-1), we introduced sgrnas trgeting severl of these genes into MART-1/MLANA + Mel624 cells nd then co-cultured them with MART-1 TCR-trnsduced primry humn T cells 18 (MART-1 T cells; Extended Dt Figs 2d, 6). MART-1 T cells re highly potent ginst tumour cells owing to their high vidity TCRs nd ystnder destruction of cncer cells 18,2. Tumour cells trnsduced with control non-trgeting sgrnas were completely erdicted upon co-culture with MART-1 T cells, wheres ESO T cells only induced cytolysis of 6% of tumour cells with the sme incution (Supplementry Fig. 3). MART-1 + Mel624 cells with sgrnas trgeting ech of the 9 cndidte genes showed incresed resistnce ginst these T cells (Extended Dt Fig. 6, P <.5). We confirmed on-trget gene disruption efficiency of sgrnas y performing deep sequencing nlysis t the trget Cs9 cut-sites in these melnom cells (Extended Dt Figs 6c e, 7). The genes tht we vlidted cross different melnom cell lines nd different ntigen TCR comintions include cellulr cytoskeleton genes, COL17A1 (collgen type XVII lph 1) nd TWF1 (twinfilin-1), microrna (hs-mir-11-2) nd 6S riosoml suunit (RPL23, Supplementry Discussion). These findings underscore the role of these iologicl processes in tumour cells in modultion of EFT. TCR MHC clss I A375 Different Different Different melnoms cncers ntigens Vlidte T cell Determine whether trgets generlize resistnce in Mel624 cross cncers nd ntigens PSMB5 PTCD2 SRP54 TWF1 Cell survivl (%) 2245R renl cell crcinom:eso T 1 5 Control sg BBS1 CD58 Figure 4 Vlidtion of top cndidte genes cross cncers., Schemtic of the rry vlidtion strtegy for top cndidtes from the pooled 2CT- CRISPR screen to test generliztion cross melnoms, MHC clss I ntigens nd other (non-melnom) cncer., Survivl of HLA-A* 2 + NY-ESO-1 + A375 cells edited with individul sgrnas (2 4 per gene) fter co-culture with ESO T cells t E:T rtio of.5 (n = 3 iologicl replictes per sgrna). c, Shred nd unique genes essentil for EFT vlidted in Mel624 nd A375 melnom cells. d, Survivl of HLA-A* 2 + NY-ESO-1 + renl cell crcinom 2245R cells edited with individul sgrnas (4 per gene) fter co-culture with ESO T cells t E:T rtio of.5 in 2CT ssy (n = 4 iologicl replictes per sgrna). d, Grey dots indicte sgrnas without significnt chnge in cell survivl. Coloured dots (purple or green) indicte P <.5. P vlue clculted for positively enriched gene-trgeting sgrnas compred to control sgrna y Student s t-test. Dt representtive of t lest two independent experiments. Next, we tested whether immune resistnce due to the loss of the cellulr signlling-relted genes, encoding G-protein-coupled pelin receptor, nd BBS1, encoding Brdet Biedl syndrome 1 protein, nd the dhesion-relted gene CD58 cn e generlized to other cncer types. To this end, we overexpressed NY-ESO-1 (CTAG1B) in ptient- derived HLA-A* 2 + renl cell crcinom (2245R) cell line using retrovirl vector nd trgeted ech gene with four sgrnas. For ech gene,, BBS1 nd CD58, we found tht t lest two sgrnas exhiited > 5% resistnce to ESO T-cell-medited lysis (Fig. 4d, P <.5). Thus, we vlidted key genes found in our CRISPR screen using multiple cell lines, trget ntigens nd tumour cell types. To etter understnd the function of gene previously unknown to hve role in EFT, we focused on, which ws vlidted cross different cell lines nd ntigens. In vivo relevnce of is G-protein-coupled receptor tht is mutted in severl cncers 27. To determine whether loss-of-function muttions in re present in ptient tumours treted with T-cell-sed immunotherpies, we mined ville whole-exome sequencing dtsets 3,5,28,29 from ptients with metsttic melnom nd lung cncer treted with checkpoint lockde therpies including nti-ctla4 (ipilimum) nd nti-pd1 (pemrolizum or nivolum) ntiodies. From these dtsets, we identified seven non-synonymous muttions in, one of which is nonsense muttion (W261X), resulting in deletion of the seventh trnsmemrne helix nd cytoplsmic C-terminl til of the receptor (Fig. 5, Extended Dt Fig. 8). The C terminus of contins residues criticl for memrne ssocition, receptor internliztion, receptor dimeriztion, nd interction with cytosolic proteins 3, nd thus its loss is likely to e deleterious to protein function. We lso crried out whole-exome sequencing of metsttic lung lesion resected from ptient (SB-444) with melnom resistnt to oth nti-ctla4 (ipilimum) nd nti-pd1 (nivolum) immunotherpies. We found two non-synonymous muttions, T44S nd C181S, in (Fig. 5, Extended Dt Fig. 8) within this melnom lesion. We selected four non-synonymous muttions (T44S, C181S, P292L, G349E) from the Vn Allen et l. cohort 3 nd ptient SB-444 to determine whether these muttions cn limit EFT. In -knockout A375 cells, we re-introduced mutted or wild-type using lentivirus (Extended Dt Fig. 8). Although re-introduction of wild type, or with C181S or P292L muttions rescued the sensitivity of -knockout cells to T-cell-medited cytolysis, T44S nd G349E muttions only resulted in prtil rescue, indicting tht these muttions in reduce EFT (Fig. 5). The presence of these loss-of-function muttions in non- responding tumour lesions suggests tht the functionl loss of in tumours could e ssocited with immunosuppression in vivo. To investigte potentil mechnisms y which regultes EFT, we nlysed the trnscriptome of -knockout cells using RNA-sequencing. We did not find ny sustntil differences in mrna trnscript levels of genes involved in ntigen presenttion, T cell inhiition or co- stimultion (Supplementry Fig. 4). We thus exmined whether regultes EFT y modulting protein signlling. hs een reported to interct with 96 different intrcellulr proteins (BioGRID interction dtse 31 ), nd in genome-scle CRISPR screens, trgeting multiple genes in the sme pthwy cn result in similr phenotypes. Of these 96 previously reported inding prtners of, JAK1 ws the most enriched in our screen (enriched in the top.5% of ll genes, Fig. 5c). Using immunoprecipittion pull-down, we confirmed tht inds to JAK1 in A375 nd HEK293T cells (Fig. 5d, Extended Dt Fig. 9). Retrovirl overexpression of coincided with n increse in JAK1 protein levels (Extended Dt Fig. 9). In ddition, this overexpression of significntly incresed the sensitivity of tumour cells to EFT (Extended Dt Fig. 9c, increse in cytolysis from 71.7 ± 1% to 81.1 ±.9%). 4 N A T URE VO L M ONTH Mcmilln Pulishers Limited, prt of Springer Nture. All rights reserved.
5 RESEARCH 1 2 AA e sgrna Time fter IFNγ (h) h i Tumour re (mm 2 ) Cytolysis (%) sg1 IFI3 JAK1 TAP1 TAP2 IRF1 4 2 EC 6 3 T44S Non-trg. rescue Min. **** * * NS NS Fold chnge Gene-trgeted CRISPR or RNA interference lentivirus No tretment WT G349E T44S C181S P292L Mx. Trnsduce cells C181S D184N 7-trnsmemrne domin c f CRSIPR screen score β2m induction (%) 1..5 P292L G349E G349W BioGRID score for interction 1 5 Gene-pertured B16 melnom cells W261X R236C 3 38 Pemro Ipi IFNγ -driven phosphoryltion of JAK1 stimultes the JAK STAT signlling cscde to ugment ntigen processing nd presenttion in tumours, which in turn enhnces recognition nd cytolysis y T cells 32. First, we determined whether pelin tretment of tumour cells cn directly induce phosphoryltion of JAK1. We did not oserve ny JAK1 **** **** Ctrl-sg sg1 sg2 Pmel ACT IFNγ (pg ml 1 ) IP: IB 4, 2, E367K Ctrl-sg IgG Ipi, Pemro Ipi, Nivo, ACT JAK1 JAK1 ACTIN ******** **** **** sg1 sg2 sg1 sg2 NY-ESO-1 Control Dys fter ACT Dys fter ACT j Survivl (%) g 1 d CS Input Pmel T cell ACT on dy 1 Sucutneously trnsplnt B16 cells into syngenic mouse Mesure tumour growth nd survivl 5 ** B2m-sg * Aplnr-sg Figure 5 Functionl loss of reduces efficcy of cncer immunotherpy., Non-synonymous muttions detected in in humn tumours refrctory to indicted immunotherpies. Ipi, ipilimum; nivo, nivolum; pemro, pemrolizum; CS, cytoplsmic signlling domin; EC, extrcellulr domin., Muttions encoding G349E nd T44S in resist the restortion of T-cellmedited cytolysis in -pertured tumour cells (n = 3 iologicl replictes). c, Second-most-enriched sgrna score in CRISPR screens for 96 -intercting proteins from the BioGrid dtse 31. d, Western lot proed for fter immunoprecipittion pull-down of JAK1 from A375 cell lystes. Results were similr in n independent replicte experiment (not shown). e, Quntittive PCR nlysis of JAK1 STAT1 pthwy induced genes in wild-type nd -edited cells t, 8 nd 24 h post tretment with 1 µ g ml 1 IFNγ (n = 3 iologicl replictes). f, Induction of surfce expression of β 2M on -edited cells upon co-culture with ESO T cells s mesured y FACS (n = 3 iologicl replictes). g, IFNγ secretion from ESO T cells fter overnight co-culture with CRISPR edited A375 cells determined using ELISA (n = 3 iologicl replictes). h, Schemtic of in vivo experiments to test the response of B2m nd Aplnr knockout tumours to ACT in immunocompetent mice. i, j, Sucutneous tumour growth in mice receiving ACT of Pmel-1 T cells. Tumour re (i) nd overll survivl (j) re shown. Significnce for tumour growth kinetics were clculted y Wilcoxon rnk-sum test. Survivl significnce ws ssessed y log-rnk Mntel Cox test (n = 5 mice per no tretment groups). For Pmel ACT groups, n = 9 mice in control group, n = 1 mice per B2m-sg nd Aplnr-sg groups. All vlues re men ± s.e.m. * * * * P <.1, * * P <.1, * P <.5. Dt re representtive of two independent experiments. chnges in JAK1 phosphoryltion with pelin tretments (Extended Dt Fig. 9d). We lso tested whether ctivtion of using peptide lignds ([Pyr 1 ]pelin 13, pelin 17 nd pelin 36) or chemicl gonist (ML233) on tumour cells lters EFT. We did not oserve ny significnt effect of these tretments, suggesting tht my regulte EFT independent of its cnonicl G-protein-coupled receptor signlling. Next, we tested whether loss of lters the responsiveness of tumour cells to IFNγ tretment. We found tht knockout cells exhiited reduced induction of JAK STAT signlling upon IFNγ tretment, s mesured y the phosphoryltion sttus of JAK1 nd STAT1 (Extended Dt Fig. 9d, e) nd induction of specific IFNγ response gene trnscripts (Fig. 5e, Extended Dt Fig. 9f). Upon co-culture with ESO T cells for 6 h, -knockout cells induced less β 2M expression on the cell surfce compred to unedited cells (Fig. 5f, Extended Dt Fig. 9g). In greement with the reduced sensitivity of -knockout cells to IFNγ -medited upregultion of ntigen processing nd presenttion genes, we noted significnt decrese in the recognition of -knockout tumour cells y ESO T cells (Fig. 5g, Extended Dt Fig. 9h, Supplementry Fig. 5). Tken together, these dt suggest tht ugments EFT y modulting IFNγ signlling in tumour cells. Finlly, to test the relevnce of for immunotherpy in n in vivo setting, we trgeted Aplnr in mouse B16 melnom tumours with multiple sgrnas (Fig. 5h). We lso included B2m-trgeting sgrnas s positive control. For ech trget gene, we performed western lot nlysis on the cells to select the sgrnas with the highest protein reduction (Supplementry Fig. 6). Upon sucutneous implnttion of these cells in immunocompetent C57BL/6 mice, we did not oserve ny significnt chnges in tumour growth kinetics etween untreted control cells nd CRISPR-modified cells (Fig. 5i). However, fter tretment of these tumours with doptive trnsfer of tumourspecific Pmel-1 TCR trnsgenic T cells (which trget melnom ntigen gp1) 33, B2m knockout in these tumours significntly diminished the efficcy of doptive cell trnsfer (ACT) tretment s mesured y tumour growth (Wilcoxon rnk-sum test, P =.2, Fig. 5i) nd host survivl (log-rnk test, P <.1, Fig. 5j). Similrly, for Aplnr-knockout tumour cells, we oserved significnt reduction in the tumour clernce (P =.42, Fig. 5i) nd host survivl (P =.19, Fig. 5j) fter ACT. As n orthogonl method to our genome-edited B16 tumours, we confirmed tht short hirpin RNA (shrna)- medited Aplnr knockdown in B16 using two independent hirpins lso reduced ACT efficcy (Extended Dt Fig. 1). In ddition, we found tht Aplnrknockout in B295 mouse melnom reduced the efficiency of nti- CTLA4 lockde in vivo with out of 1 mice experiencing complete regression with sg-aplnr-trnsduced cells versus 5 out of 1 mice with cells trnsduced with control sgrna (Supplementry Fig. 7). Tken together, these dt demonstrte tht loss reduces the effectiveness of T-cell-sed cncer therpies, including immune checkpoint lockde nd ACT. Discussion To cpture the genes necessry for complex phenomenon such s the EFT in cncer immunotherpy, we developed 2CT-CRISPR screening system nd used it to identify severl genes cple of modulting melnom growth when trgeted y T cells. Given the ssocition of these genes with intrtumorl cytolytic ctivity in pn-cncer TCGA dtsets, this resource provides rich trove of new trgets to investigte resistnce to T-cell-sed therpies. Reverting or ypssing the functionl loss of such genes in tumours using drugs such s epigenetic modultors 34,35 my llow development of new comintoril tretment strtegies to improve clinicl responses with immunotherpies. Our dt revels novel role of in regulting the nti- tumour response of T cells vi modultion of the JAK STAT signlling in trget cells. It hs een shown tht ngiotensin receptor, G-proteincoupled receptor closely relted to, lso directly regultes the JAK STAT pthwy 36. A recent study hs shown tht T cell effector MONTH 217 VO L N A T URE Mcmilln Pulishers Limited, prt of Springer Nture. All rights reserved.
6 RESEARCH ARTICLE cytokines IFNγ nd TNFα exert n nti-tumour effect y ltering endothelil cells of lood vessels (where is highly expressed 37 ) to induce ischemi in the tumour strom 38. Given our finding tht intercts with JAK1 to ugment IFNγ response, cn further increse the sensitivity of tumour lood vessels to IFNγ nd thus improve nti-tumour efficcy of T cells. Hence, we speculte tht might hve role eyond direct tumour cell recognition in vivo, nd its expression on tumour lood vessels nd stroml cells should e investigted in future studies. In summry, we hve used two-cell-type CRISPR screen to discover oth well-estlished nd novel genes in cncer cells tht regulte EFT. Our findings hve direct clinicl implictions s these dt my serve s functionl lueprint to study the emergence of tumour resistnce to T-cell-sed cncer therpies. Recent clinicl trils hve rised the question of why most ptients fil to experience complete regressions of their cncers 7. Our study provides comprehensive list of genes tht cn contriute to resistnce of humn tumours to immunotherpy nd hve lso identified loss-of-function muttions shred with those oserved in ptients tht fil to respond to immunotherpy (Supplementry Discussion, Supplementry Fig. 8). A creful evlution nd vlidtion of muttions in these genes on personlized sis in immunotherpy-resistnt ptients my llow identifiction of novel mechnisms of immune escpe nd speed the development of new drugs tht circumvent these escpe mechnisms. Online Content Methods, long with ny dditionl Extended Dt disply items nd Source Dt, re ville in the online version of the pper; references unique to these sections pper only in the online pper. Received 16 Decemer 216; ccepted 13 July 217. Pulished online 7 August Lwrence, M. S. et l. Muttionl heterogeneity in cncer nd the serch for new cncer-ssocited genes. Nture 499, (213). 2. Chn, T. A., Wolchok, J. D. & Snyder, A. Genetic sis for clinicl response to CTLA-4 lockde in melnom. N. Engl. J. Med. 373, 1984 (215). 3. Vn Allen, E. M. et l. Genomic correltes of response to CTLA-4 lockde in metsttic melnom. Science 35, (215). 4. Hugo, W. et l. Genomic nd trnscriptomic fetures of response to nti-pd-1 therpy in metsttic melnom. Cell 165, (216). 5. Rizvi, N. A. et l. Cncer immunology. Muttionl lndscpe determines sensitivity to PD-1 lockde in non-smll cell lung cncer. Science 348, (215). 6. Trn, E. et l. Immunogenicity of somtic muttions in humn gstrointestinl cncers. Science 35, (215). 7. Le, D. T. et l. Mismtch-repir deficiency predicts response of solid tumors to PD-1 lockde. Science n6733 (217). 8. Restifo, N. P. et l. Loss of functionl β 2-microgloulin in metsttic melnoms from five ptients receiving immunotherpy. J. Ntl Cncer Inst. 88, 1 18 (1996). 9. Zretsky, J. M. et l. Muttions ssocited with cquired resistnce to PD-1 lockde in melnom. N. Engl. J. Med. 375, (216). 1. Wng, T. et l. Identifiction nd chrcteriztion of essentil genes in the humn genome. Science 35, (215). 11. Hrt, T. et l. High-resolution CRISPR screens revel fitness genes nd genotype-specific cncer liilities. Cell 163, (215). 12. Shlem, O. et l. Genome-scle CRISPR Cs9 knockout screening in humn cells. Science 343, (214). 13. Wng, T., Wei, J. J., Stini, D. M. & Lnder, E. S. Genetic screens in humn cells using the CRISPR Cs9 system. Science 343, 8 84 (214). 14. Chen, S. et l. Genome-wide CRISPR screen in mouse model of tumor growth nd metstsis. Cell 16, (215). 15. Rooney, M. S., Shukl, S. A., Wu, C. J., Getz, G. & Hcohen, N. Moleculr nd genetic properties of tumors ssocited with locl immune cytolytic ctivity. Cell 16, (215). 16. Kvistorg, P. et l. Anti-CTLA-4 therpy rodens the melnom-rective CD8 + T cell response. Sci. Trnsl. Med. 6, 254r128 (214). 17. Roins, P. F. et l. Single nd dul mino cid sustitutions in TCR CDRs cn enhnce ntigen-specific T cell functions. J. Immunol. 18, (28). 18. Johnson, L. A. et l. Gene trnsfer of tumor-rective TCR confers oth high vidity nd tumor rectivity to nonrective peripherl lood mononucler cells nd tumor-infiltrting lymphocytes. J. Immunol. 177, (26). 19. Roins, P. F. et l. A pilot tril using lymphocytes geneticlly engineered with n NY-ESO-1-rective T-cell receptor: long-term follow-up nd correltes with response. Clin. Cncer Res. 21, (215). 2. Spiotto, M. T., Rowley, D. A. & Schreier, H. Bystnder elimintion of ntigen loss vrints in estlished tumors. Nt. Med. 1, (24). 21. Snjn, N. E., Shlem, O. & Zhng, F. Improved vectors nd genome-wide lirries for CRISPR screening. Nt. Methods 11, (214). 22. Luo, B. et l. Highly prllel identifiction of essentil genes in cncer cells. Proc. Ntl Acd. Sci. USA 15, (28). 23. Indrccolo, S. et l. Identifiction of genes selectively regulted y IFNs in endothelil cells. J. Immunol. 178, (27). 24. Snd, C. et l. Differentil gene induction y type I nd type II interferons nd their comintion. J. Interferon Cytokine Res. 26, (26). 25. Viemnn, D. et l. TNF induces distinct gene expression progrms in microvsculr nd mcrovsculr humn endothelil cells. J. Leukoc. Biol. 8, (26). 26. Klijn, C. et l. A comprehensive trnscriptionl portrit of humn cncer cell lines. Nt. Biotechnol. 33, (215). 27. Kn, Z. et l. Diverse somtic muttion ptterns nd pthwy ltertions in humn cncers. Nture 466, (21). 28. Roh, W. et l. Integrted moleculr nlysis of tumor iopsies on sequentil CTLA-4 nd PD-1 lockde revels mrkers of response nd resistnce. Sci. Trnsl. Med. 9, h356 (217). 29. Nthnson, T. et l. Somtic muttions nd neoepitope homology in melnoms treted with CTLA-4 lockde. Cncer Immunol. Res. 5, (217). 3. O Crroll, A.-M., Lolit, S. J., Hrris, L. E. & Pope, G. R. The pelin receptor APJ: journey from n orphn to multifceted regultor of homeostsis. J. Endocrinol. 219, R13 R35 (213). 31. Strk, C. et l. BioGRID: generl repository for interction dtsets. Nucleic Acids Res. 34, D535 D539 (26). 32. Dunn, G. P., Koeel, C. M. & Schreier, R. D. Interferons, immunity nd cncer immunoediting. Nt. Rev. Immunol. 6, (26). 33. Overwijk, W. W. et l. Tumor regression nd utoimmunity fter reversl of functionlly tolernt stte of self-rective CD8 + T cells. J. Exp. Med. 198, (23). 34. Wng, L.-X. et l. Low dose decitine tretment induces CD8 expression in cncer cells nd stimultes tumor specific cytotoxic T lymphocyte responses. PLoS One 8, e62924 (213). 35. Wrngle, J. et l. Altertions of immune response of non-smll cell lung cncer with Azcytidine. Oncotrget 4, (213). 36. Mrrero, M. B. et l. Direct stimultion of Jk/STAT pthwy y the ngiotensin II AT1 receptor. Nture 375, (1995). 37. Kidoy, H. et l. The pelin/apj system induces mturtion of the tumor vsculture nd improves the efficiency of immune therpy. Oncogene 31, (212). 38. Kmmertoens, T. et l. Tumour ischemi y interferon-γ resemles physiologicl lood vessel regression. Nture 545, (217). Supplementry Informtion is ville in the online version of the pper. Acknowledgements The reserch ws supported y the Intrmurl Reserch Progrm of the NCI, nd y the Cncer Moonshot progrm for the Center for Cell-sed Therpy t the NCI, NIH. The work ws lso supported y the Milstein Fmily Foundtion. We thnk S. A. Rosenerg, K. Hnd, A. Wellstein, C. Hurley nd L. M. Weiner for their vlule discussions nd intellectul input, M. Kruhlk, Z. Yu, C. Surmnim, C. Kriy, A. J. Leonrdi, N. H, H. Xu, M. A. Blck nd H. Chinnsmy for technicl ssistnce in this project. This work used the computtionl resources of the NIH HPC Biowulf cluster ( nih.gov). The results here re in prt sed upon dt generted y the TCGA Reserch Network: This study ws done in prtil fulfilment of PhD in Tumor Biology to S.J.P. N.E.S. is supported y the NIH through NHGRI (R-HG8171) nd Sidney Kimmel Scholr Awrd. Author Contriutions S.J.P., N.E.S., nd N.P.R. designed the study nd wrote the mnuscript. S.J.P. crried out CRISPR screens nd vlidtion experiments. N.E.S., O.S. nd S.J.P. nlysed CRISPR screen dt. S.J.P. nd N.E.S. nlysed humn muttion dtsets from immunotherpy cohorts. T.N.Y., G.U.M., A.C., M.S. nd S.F. ssisted in genertion of TCR-engineered T cells nd CRISPRedited cells. R.E., A.E., T.N.Y., S.K.V., G.U.M., A.C. nd M.S. edited the mnuscript. S.J.P., A.E. nd S.K.V. crried out mouse experiments. G.M., E.P.G. nd C.-P.D. developed B295 mouse model for nti-ctla4 experiments. S.K.V. nd L.J. nlysed RNA-seq dt. M.C. nd A.S.M. nlysed TCGA dtsets. J.J.G. performed indel nlyses. S.M.S. nlysed clinicl dt. R.J.K. performed western lots nd immunoprecipittion experiments. F.Z., E.T. nd P.R. contriuted regents. N.P.R. supervised the study. Author Informtion Reprints nd permissions informtion is ville t The uthors declre no competing finncil interests. Reders re welcome to comment on the online version of the pper. Pulisher s note: Springer Nture remins neutrl with regrd to jurisdictionl clims in pulished mps nd institutionl ffilitions. Correspondence nd requests for mterils should e ddressed to S.J.P. (ptelsj@mil.nih.gov), N.E.S. (nsnjn@nygenome.org) or N.P.R. (restifon@mil.nih.gov). Reviewer Informtion Nture thnks R. Levine nd the other nonymous reviewer(s) for their contriution to the peer review of this work. 6 N A T URE VO L M ONTH Mcmilln Pulishers Limited, prt of Springer Nture. All rights reserved.
7 RESEARCH METHODS Dt reporting. Smple size for the mouse experiments ws determined on the sis of our previous experience with B16 nd B295 tumour models. Tumours were rndomized during tretments. The mesurements for implnted tumours were performed in doule-linded fshion. Humn specimens. Peripherl lood mononucler cells (PBMCs) were isolted from helthy donors nd tumour smples were isolted from ptients with melnom. All humn specimens were collected with informed consent nd procedures pproved y the institutionl review ord (IRB) of the Ntionl Cncer Institute (NCI). Mice. Animl experiments were pproved y the Institutionl Animl Cre nd Use Committees of the NCI nd performed in ccordnce with NIH guidelines. C57BL/6J were purchsed from The Jckson Lortory. Femle mice ged 6 8 weeks were used for tumour implnttion experiments. Cell culture. The melnom cell lines HLA-A* 2 + /MART-1 + /NY-ESO-1 + (Mel624.38, Mel13), HLA-A* 2 (Mel938) nd HLA-A* 2 + /NY-ESO-1 (Mel526) were isolted from surgiclly resected metstses s previously descried 17, nd were cultured in RPMI 164 (Invitrogen) medium supplemented with 1% fetl ovine serum (FBS, Hyclone, Logn, UT). The A375 (HLA-A* 2 + / NY-ESO-1 + ) cell line ws otined from the Americn Type Culture Collection (Mnsss, VA). The SK23 cell line trnsduced with retrovirus contining NY-ESO-1 expressing vector (SK23 NY-ESO-1 + ) nd HLA-A* 2 + renl cell crcinom (2245R) cells ws otined from Ken-ichi Hnd (Surgery Brnch, NCI) nd cultured in RPMI 164 medium supplemented with 1% FBS. A375 cells were cultured in D1 medium contining DMEM supplemented 1% FBS, 2 mm l-glutmine, nd 1% penicillin streptomycin (Life Technologies). Cell lines were confirmed mycoplsm negtive using Mycoplsm detection kit (Biotool #B393). All PBMCs nd lymphocytes used for trnsduction nd s feeder cells were otined from phereses of NCI Surgery Brnch ptients on IRB-pproved protocols. They were cultured in T cell medium, which is: AIM-V medium (Invitrogen) supplemented with 5% humn AB serum (Vlley Biomedicl, Winchester, VA), 1 U ml 1 penicillin nd 1 µ g ml 1 streptomycin, 2 mm l-glutmine nd 12.5 mm HEPES (Life Technologies). Retrovirl vectors nd trnsduction of humn T cells. Retrovirl vectors for TCRs recognizing the HLA-A* 2-restricted melnom ntigens NY-ESO-1 (NY-ESO-1: epitope) nd MART-1 (MART-1:27 35 epitope, DMF5) were generted s previously descried 17,18. Clinicl grde cgmp-qulity retrovirl superntnts were produced y the Ntionl Gene Vector Lortories t Indin University. For trnsduction, peripherl lood lymphocytes (2 1 6 cells per ml) were stimulted with IL-2 (3 IU ml 1 ) nd nti-cd3 ntiody OKT3 (3 IU ml 1 ) on dy. Non-tissue culture treted six-well pltes were coted with 2 ml per well of 1 µ g ml 1 RetroNectin (Tkr Bio) on dy 1 nd stored overnight t 4 C. Vector superntnt (4 ml per well, diluted with D1 medi) were pplied to pltes on dy 2 followed y centrifugtion t 2,g for 2 h t 32 C. Hlf the volume ws spirted nd peripherl lood lymphocytes were dded ( cells per ml, 4 ml per well), centrifuged for 1 min t 1,g, nd incuted t 37 C/5% CO 2. A second trnsduction on dy 3 ws performed s descried ove. Cells were mintined in culture t cells per ml. After collection, cells underwent rpid expnsion protocol in the presence of solule OKT3 (3 IU ml 1 ), IL-2 (6, IU ml 1 ) nd irrdited feeders s previously descried 18. After dy 5 of the rpid expnsion protocol, cells were frozen down for co-culture lter. Lentivirus production nd purifiction. To generte lentivirus, HEK293FT cells (Invitrogen) were cultured in D1 medium. One dy efore trnsfection, HEK293FT cells were seeded in T-225 flsks t 6% confluency. One to two hours efore trnsfection, DMEM medi ws replced with 13 ml of pre-wrmed serumfree OptiMEM medi (Invitrogen). Cells were trnsfected using Lipofectmine 2 nd Plus regent (Invitrogen). For ech flsk, 4 ml of OptiMEM ws mixed with 2 µ l of Plus regent, 2 µ g of lenticrisprv2 plsmid or pooled plsmid humn GeCKOv.2 (Genome-scle CRISPR Knockout) lirry, 15 µ g pspax2 (Addgene) nd 1 µ g pmd2.g (Addgene). 1 µ l Lipofectmine 2 ws diluted with 4 ml of OptiMEM nd ws comined to the mixture of plsmids nd Plus regent. This trnsfection mixture ws incuted for 2 min nd then dded dropwise to the cells. 6 8 h fter trnsfection, the medi ws replced to 2 ml of DMEM supplemented with 1% FBS nd 1% BSA (Sigm). Virus contining medi ws collected 48 h post-trnsfection. Virl titer ws ssyed with Lenti-X GoStix (Clontech). Cell deris were removed y centrifugtion of medi t 3, r.c.f. nd 4 C for 1 min followed y filtrtion of the superntnt through.45 µ m low-protein inding memrne (Millipore Steriflip HV/PVDF). For individul lenticrisprv2 plsmids, virl superntnts were frozen in liquots t 8 C. For pooled lirry plsmids, virl superntnts were concentrted y centrifugtion t 4, r.c.f. nd 4 C for 35 min in Amicon Ultr-15 filters (Millipore Ultrcel- 1K). Concentrted virl superntnts were stored in liquots t 8 C. 2CT T cell nd tumour cell co-culture ssy. NY-ESO-1 nd MART-1 T cells were used for co-culture ssys. Two dys efore co-culture, cells were thwed in T cell medi contining 3 U ml 1 DNse (Genentech Inc.) overnight. Tumour cells were seeded t specific density on this dy in the sme medi s the T cells. T cells were then cultured in T cell medi contining 3 IU ml 1 IL-2 for 24 h. T cells were co-cultured with tumour cells t vrious effector:trget (E:T rtios) for vrying time periods. To reduce T cell killing ctivity nd enrich for resistnt tumour cells during the recovery phse, T cells were removed y creful 2 phosphte-uffered sline (PBS) wshes following the ddition of D1 medi without IL-2. At the end of recovery phse of co-culture, tumour cells were detched using trypsin (Invitrogen) nd wshed twice with PBS. Tumour cells were then stined with fixle Live/Ded dye (Invitrogen) followed y humn nti-cd3 ntiody (clone SK7, BD) in FACS stining uffer (PBS +.2% BSA). Cell counts were mesured using CountBright Asolute Counting Beds (Invitrogen) y FACS. 2CT GeCKOv.2 screens nd genomic DNA extrctions. Cell cell interction genome-wide screens were performed using Mel624 cells trnsduced independently with oth A nd B GeCKOv.2 lirries 21. For one screen, we split two groups of trnsduced Mel624 cells. One group ws co-cultured with ptient-derived NY-ESO-1 T cells (E:T rtio of 1:3 or.3) for ech lirry. A second (control) group were cultured under the sme density nd conditions ut without T cells. The co-culture phse ws mintined for 12 h fter which the T cells were removed s descried ove. The recovery phse ws mintined for nother 48 h. In this initil screen, NY-ESO-1 T cells lysed 76% of tumour cells, nd the surviving cells were frozen to evlute sgrna enrichment lter. For second screen, we similrly trnsduced Mel624 cells with the GeCKOv.2 lirry nd split them into two groups of trnsduced cells. For one group, we incresed the E:T rtio to.5 y co-culture of NY-ESO-1 T cells with trnsduced Mel624 cells for ech lirry while keeping ll other conditions similr. As efore, the second group of Mel624 cells were used s controls, which were cultured under the sme density nd conditions, ut without T cells. By incresing the selection pressure, we oserved tht T cells killed 9% of lirry-trnsduced Mel624 cells. To evlute sgrna enrichment, cells with nd without T cell co-culture (long with n erly time point collected fter puromycin selection) were collected nd frozen. For gdna extrction from collected tumour cells, we used our previously optimized mmonium cette nd lcohol precipittion procedure to isolte gdna 14 ut sustituted AL uffer (Qigen) for the initil cell lysis step. 2CT-CRISPR pooled screen redout nd dt nlysis. To determine sgrna undnce s the redout of lirry screens, two-step PCR mplifiction were performed on gdna using Tkr Ex-Tq polymerse (Clontech). The first PCR step (PCR1) included mplifiction of the region contining sgrna cssette using v2adptor_f nd v2adptor_r primers, nd the second step PCR (PCR2) included mplifiction using uniquely rcoded P7 nd P5 dptor-contining primers to llow multiplexing of smples in single HiSeq run 12. All PCR1 nd PCR2 primer sequences, including full rcodes, re listed on the Addgene wesite ( ddgene.org/pooled-lirry/zhng-humn-gecko-v2/). Assuming 6.6 pg of gdna per cell, 15 µ g of gdna ws used per smple ( 35-fold sgrna representtion), in 15 PCR1 rections. For ech PCR1 rection, 1 µ g gdna were used in 1 µ l rection performed under cycling conditions: 95 C for 5 min, 18 cycles of (95 C for 3 s, 62 C for 3 s, 72 C for 3 s), nd 72 C for 3 min. PCR1 products for ech smple were pooled nd used for mplifiction with rcoded second step PCR primers. For ech smple, we performed 7 PCR2 rections using 5 µ l of the pooled PCR1 product per PCR2 rection. PCR2 products were pooled nd then normlized within ech iologicl smple efore comining uniquely rcoded seprte iologicl smples. The pooled product ws then gel-purified from 2% E-gel EX (Life Technologies) using the QiQuick gel extrction kit (Qigen). The purified, pooled lirry ws then quntified with Tpesttion 42 (Agilent Technologies). Diluted lirries with 5 2% PhiX were sequenced on HiSeq 2. Demultiplexed reds were trimmed y cutdpt using 12 p flnking sequences round the 2 p guide sequence 39. Trimmed reds were ligned using Bowtie 4 to the GeCKOv.2 indexes creted from lirry CSV files downloded from the Addgene wesite ( Red lignment ws performed with prmeters -m 1 -v 1 norc, which llows up to one mismtch nd discrds ny reds tht do not lign in the forwrd orienttion or tht hve multiple possile lignments. Aligned counts of lirry sgrnas were imported into R/RStudio. Counts were normlized y the totl reds for ech smple nd then log-trnsformed. A gene rnking ws computed using the second-most-enriched sgrna for ech gene (second-mostenriched sgrna score) nd the RIGER weighted-sum method 22. Gene pthwy enrichment nlysis. For pthwy nlysis, we constructed set of enriched cndidte genes y tking dvntge of the 1, control (non- trgeting) 217 Mcmilln Pulishers Limited, prt of Springer Nture. All rights reserved.
8 RESEARCH ARTICLE sgrnas emedded in our pooled lirry with t lest one sgrna enriched ove the most-enriched non-trgeting control (Fig. 3, FDR <.1%, second-mostenriched sgrna score >.5). This yielded list of 554 genes. In this list, we looked for gene ctegory over-representtion using Ingenuity Pthwy Anlysis (QIAGEN Redwood City, The nlysis criteri were set s follows: (1) querying for molecules with Ingenuity Knowledge Bse s reference set; (2) restricted to humn species; nd (3) experimentlly oserved findings s confidence level. Fisher s exct test (P <.5) ws used to compute significnce for over-representtion of genes in prticulr pthwy or iologicl process. The Cncer Genome Atls (TCGA) correltion nd muttion nlysis. TCGA RNA-seq dt in form of normlized RNA-seq y Expecttion-Mximiztion (RSEM) vlues from multiple cncer dtsets ws downloded from the Firehose Brod GDAC ( DOI for dt relese: 1.798/ C11GKM9) using the TCGA2STAT pckge for R 41, nd used to find overlp etween TCGA gene expression indictive of cytolytic ctivity nd genes from our pooled screen where loss-of-function confers resistnce to T cell killing. We first identified the genes correlted with previously identified cytolytic ctivity signture (CYT), nmely the geometric men of grnzyme A (GZMA) nd perforin 1 (PRF1) expression 15. To identify these genes in the TCGA dt, we clculted the geometric men of GZMA nd PRF1 in ech dtset nd serched for ny genes with positive correltion to this quntity cross ptients (Person s r >, P <.5). We then exmined the intersection etween genes whose expression ws correlted with cytolytic ctivity (TCGA dtsets) nd the enriched genes found in the CRISPR screen (554 genes). Individul hetmps for the two sets of clustered genes were regenerted for ech cncer type nd cn e found t For the top 2 rnked gene cndidtes from ny of the screens, we otined ptient muttion dt from the TCGA dtse using cbioportl 42,43. For muttion frequency counts, tumours contining likely loss-of-function genetic errtions (defined s homozygous deletion, missense, nonsense, frme-shift, truncted or splice-site muttions) were included in the nlysis. Arryed vlidtion of 2CT-CRISPR screen genes. Individul lenticrisprs were produced s ove except tht virl superntnts were not concentrted. For ech gene, we used 3 4 sgrna guide sequences s listed in Supplementry Tle 7, where 2 sgrnas sequences were designed de novo nd the other 2 sgrnas were from GeCKOv.2 lirry. We cloned these sgrnas into the lenticrisprv2 vector (Addgene) s previously descried 21. To produce virus in high-throughput formt, HEK293FT cells were seeded nd trnsfected in six-well pltes where ech well received different lenticrispr plsmid. The lentivirl production protocol ws the sme s the one descried ove for GeCKOv.2 lirry lentivirus production (scled to 6-well formt). Mel624 nd A375 cells with unique gene perturtions were generted y trnsduction with these virl superntnts. Typiclly, we used 5 µ l of lenticrisprv2 virus per cells for Mel624 nd A375. Puromycin selection (1 µ g ml 1 ) ws pplied to these cells for 5 7 dys. Over this period, untrnsduced cells were killed completely y puromycin. After tking suset of cells for nlysis of insertion deletion (indel) muttions, the reminder were normlized to seed cells per well in 96-well pltes. During the rryed screen, ech cell line ws co-cultured with pproprite T cells (either NY-ESO-1 or MART-1) in 96-well plte formt t n E:T rtio of 1:3 for 12 h in T cell medi. As in the pooled screen, we performed gentle 2 PBS wshes to remove the T cells. Mel624 or A375 cells were collected fter recovery phse culture of 48 h for high-throughput FACS nlysis. Tumour cell counts were mesured using FACS-sed CountBright ed method (Life Technologies). We noticed vriility in prolifertion nd survivl rtes cross cells depending on the sgrna received. To ccount for this vriility, we clculted reltive per cent chnge for ech sgrna: (%, 2CT versus not) in tumour cells co-cultured with T cells (2CT) compred to tumour cell counts without T cells (not). The normlized cell survivl ws clculted y the following formul: Normlized cell survivl(%) = (%,2CT vs not)gene trgeting sgrna 1 1 (%, 2CT vs not)non trgeting sgrna For co-culture with MART-1 T cells, ll prmeters were the sme, except tht the co-culture period durtion ws 24 h with the sme recovery period s with NY-ESO-1 experiments. Indel muttion fetection for rry 2CT vlidtion. Genomic DNA from thwed cell pellets ws purified using the Blood & Cell Culture kit (Qigen). sgrna trget site PCR mplifictions were performed for ech genomic loci using conditions s descried previously 14, pooled together nd then sequenced in single Illumin MiSeq run. All primer sequences for indel detection cn e found in Supplementry Tle 8. To nlyse the dt from the MiSeq run, pired-end reds were trimmed for qulity using trimmomtic with prmeters SLIDINGWINDOW:5: Reds with surviving mte pirs were then ligned to their trgeted mplicon sequence using Bowtie2 45. To determine indel sizes, we clculted the size difference etween oserved reds nd predicted red size sed on the genomic reference sequence. If oserved red size ws equl to the predicted size, these reds were scored s no indels. The size difference ws used to detect insertions or deletions. Flow cytometry nd immunossys. Tumour cells or T cells suspended in FACS stining uffer were stined with fluorochrome-conjugted ntiodies ginst comintions of the following surfce humn ntigens: CD4 (RPA-T4, BD), CD8 (SK1, BD), CD3e (SK7, BD), HLA-A* 2/MART-1:27-35 tetrmer (DMF5, Beckmn Coulter Immunotech); NY-ESO-1 tetrmer (NIH Tetrmer Core Fcility); PD1-L1 (MIH1, ebiosciences); PD1-L2 (24F.1C12, Biolegend); IFNγ ( , BD), CD58 (1C3, BD) nd β 2M (2M2, Biolegend). Cell viility ws determined using propidium iodide exclusion or fixle Live/Ded kit (Invitrogen). Intrcellulr stining ssy (ICS) on ESO T cells fter 5 6 h of co-culture with A375 cells, where T cells were pre-treted with monensin (51292KZ, BD) nd refeldin A (51231KZ, BD), ws performed using mnufcturer s instructions for BD Flow cytometry ICS. Flow cytometric dt were cquired using either FACSCnto II or LSRII Fortess cytometer (BD), nd dt were nlysed with FlowJo version 7.5 softwre (FlowJo LLC). The mount of IFNγ relese y T cells fter co-culture with tumour cells ws mesured y sndwich ELISA ssy using nti-ifnγ (Thermo Scientific M7A) coted 96-well pltes, iotin-lelled nti-ifnγ (M71B), horserdish peroxidse (HRP)-conjugted streptvidin (N1) nd TMB sustrte solution (N31). Quntittive PCR nlysis. Totl RNA ws extrcted from cells using RNesy Plus Mini Kit (Qigen). Gene expression ws quntified using TqMn RNAto-Ct 1-Step Kit (Thermo Fisher) nd Tqmn ssy proes (Invitrogen): Aplnr (Mm442191_s1), Act (Mm67939_s1), APLN (Hs936329), ACTB (Hs16665_g1), IRF1 (Hs971965_m1), IFI3 (Hs173838_m1), TAP1 (Hs388675_m1), TAP2 (Hs2416_m1), (Hs917451_g1) nd JAK1 (Hs126983_m1). Reltive mrna expression ws determined vi the C t method. Immunoprecipittion nd western lot nlysis. For immunoprecipittion, cells were lysed on ice in IP lysis uffer (Thermo Fisher #87787) supplemented with protese nd phosphtse inhiitors (Thermo Fisher #88668) for 15 min. Lystes were clered y centrifugtion t 16,g for 1 min t 4 C nd superntnts were incuted with indicted ntiodies for 24 h t 4 C. Following incution, cells were dded to pre-wshed Dyneds coupled to protein G or protein A (Life Technologies) nd incuted for 3 h t 4 C. Immune complexes were purified on mgnets with extensive wshing with lysis uffer. Purified complexes were mixed with smple uffer nd ssyed y western lot. Antiodies used included: nti-jk1 (BD 61231), nti-jk2 (Cell Signling 323), nti-flg (Cell Signling #8146). In some cses, immune complexes were isolted with sephrose-conjugted ntiodies specific for IgG (Cell Signling 342) or Flg (Cell Signling 575). For immunolots, totl protein ws extrcted with 1 RIPA lysis uffer (Millipore) with 1 protese inhiitor (Roche). Protein concentrtion ws determined using the BCA ssy (Thermo/Pierce). Cell lystes were resolved on 4 2% Tris-Glycine gels (Invitrogen), trnsferred to PVDF memrnes (Millipore), nd incuted overnight t 4 C with the pproprite primry ntiodies: nti-aplnr (Acm ), nti-β 2M (Acm 75853) nd nti-β -ctin (Acm 8227) for mouse tumour cells; nti- (Acm 97452), nti-phospho-jak1 (Cell Signling 3331) nd nti-phospho-stat1 (Cell Signling 7649). Signls were detected using HRP-conjugted secondry ntiodies (Snt Cruz Biotechnology) nd SuperSignl West Femto Chemiluminescent Sustrte (Thermo/Pierce). Imges were cptured using ChemiDoc Touch imging system (Bio-Rd). Whole-exome sequencing nd trnscriptomics nlysis. Whole-exome sequencing of ptient tumours ws performed s previously descried 46. For trnscriptome nlysis on -knockout cells, totl RNA ws extrcted using the RNesy Plus Mini Kit (Qigen). The qulity nd quntity of extrcted totl RNA ws ssessed using TpeSttion 22 (Agilent). All RNA-sequencing (RNA-seq) nlyses were performed using three iologicl replictes. 2 ng of totl RNA ws used to prepre RNA-seq lirry using the TruSeq RNA smple prep kit (Illumin). Pired-end RNA sequencing ws performed on HiSeq 2 (Illumin). Sequenced reds were ligned to the humn genome (hg19) with TopHt (v.2..11) 47 nd the mpped dt were then processed y Cufflinks. Gene expression vlues were normlized to otin RPKM (reds per kilose exon model per million mpped reds) vlues using CuffDiff 48. To define differentilly expressed genes, we used twofold chnge nd P <.5 difference etween groups. Chrcteriztion of ptient muttions using 2CT ssy. ws pertured in A375 cells using LentiCRISPRv2 encoding _sg1 (Supplementry Tle 7). Cells were selected with puromycin for 5 7 dys. For rescue experiments, cells were trnsduced with lentivirl (lsticidin- selectle) 217 Mcmilln Pulishers Limited, prt of Springer Nture. All rights reserved.
9 RESEARCH plsmid encoding codon-shuffled sequences (either wild-type or with specific ptient muttions). sequences were re-coded to ensure tht _sg1 cnnot introduce Cs9-medited reks in the trnsgene. Trnsduced A375 cells were selected with 1 µ g ml 1 lsticidin for 5 dys. Expression of these constructs in cells ws verified y western lot nlysis. Cells were suject to 2CT-ssy with ESO T cells s descried ove. Adoptive cell trnsfer nd tumour immunotherpy. For immunotherpy experiments, we used mouse humn chimeric gp1 ntigen overexpressing B16 melnom (mhgp1-b16) cells tht re responsive to doptive trnsfer of Pmel-1 TCR trnsgenic CD8 + T cells in n in vivo setting. We generted gene-deleted mhgp1-b16 cells using lentiviruses encoding sgrnas trgeting Aplnr (5 -GATCTTGGTGCCATTTTCCG-3 ) or B2m (5 -GAAAT CCAAATGCTGAAGAA-3 ) loci s descried ove. C57BL/6 mice were sucutneously implnted with mhgp1-b16 cells. After 1 dys of tumour implnttion, mice (n = 1 for tretment groups) were su-lethlly irrdited (6 cgy), nd injected intrvenously with Pmel-1 CD8 + T cells nd received intrperitonel injections of IL-2 in PBS (6 1 4 IU per.5 ml) once dily for 3 consecutive dys. All tumour mesurements were linded. Tumour size ws mesured in linded fshion pproximtely every two dys fter trnsfer nd tumour re ws clculted s length width of the tumour. Mice with tumours greter thn 4 mm 2 were euthnized. The products of the perpendiculr tumour dimeters re presented s men ± s.e.m. t the indicted times fter ACT. In nother experimentl setting, we lso used RNA-interference-medited knock-down of Aplnr in mhgp1-b16 cells to perform doptive trnsfer s ove. Lentivirl prticles encoding shrnas (Dhrmcon, shrna1- V2LMM_3664; shrna2- V3LMM_517943) were used to knock-down expression of Aplnr in mhgp1-b16 cells. qpcr nlysis ws performed on these cells to confirm knock-down efficiencies of these constructs. B295 is melnom cell line derived from spontneous tumour induced y UV irrdition of C57BL/6-HGF trnsgenic mice 49. We generted gene-deleted B295 cells using lenticrispr encoding Aplnr sgrna s ove. We implnted tumour cells sucutneously in C57BL/6 mice. Intrperitonel injections of 25 µ g of nti-ctla4 monoclonl ntiody (Bio X cell, BE131) or IgG control ntiody were given on dys 1, 13, 16 nd 19 post implnttions. Tumour size ws mesured in linded fshion pproximtely every 1 dys nd tumour re ws clculted s length width of the tumour. Owing to lrge intr-individul differences in tumour growth rtes (Supplementry Fig. 7), the significnce of resistnce to nti-ctla4 tretment ws determined y Fisher s exct test comprison of control sgrna versus sg-aplnr groups using the numer of progressing tumours nd completely regressed tumours in ech group. Sttisticl nlysis. Dt etween two groups were compred using two-tiled unpired Student s t-test or the Mnn Whitney test s pproprite for the type of dt (depending on normlity of the distriution). Unless otherwise indicted, P vlue less thn or equl to.5 ws considered sttisticlly significnt for ll nlyses, nd not corrected for multiple comprisons. To compre multiple groups, we used n nlysis of vrince (ANOVA) with the Bonferroni correction. All group results re represented s men ± s.e.m., if not stted otherwise. Prism (GrphPd Softwre Inc.) ws used for these nlyses. Dt vilility. Dt tht support the findings of this study re ville from the corresponding uthor upon resonle request. 39. Mrtin, M. Cutdpt removes dpter sequences from high-throughput sequencing reds. EMBnet.journl 17, 1 12 (211). 4. Lngmed, B., Trpnell, C., Pop, M. & Slzerg, S. L. Ultrfst nd memoryefficient lignment of short DNA sequences to the humn genome. Genome Biol. 1, R25 (29). 41. Wn, Y.-W., Allen, G. I. & Liu, Z. TCGA2STAT: simple TCGA dt ccess for integrted sttisticl nlysis in R. Bioinformtics 32, (216). 42. Go, J. et l. Integrtive nlysis of complex cncer genomics nd clinicl profiles using the cbioportl. Sci. Signl. 6, pl1 (213). 43. Cermi, E. et l. The cbio cncer genomics portl: n open pltform for exploring multidimensionl cncer genomics dt. Cncer Discov. 2, (212). 44. Bolger, A. M., Lohse, M. & Usdel, B. Trimmomtic: flexile trimmer for Illumin sequence dt. Bioinformtics 3, (214). 45. Lngmed, B. & Slzerg, S. L. Fst gpped-red lignment with Bowtie 2. Nt. Methods 9, (212). 46. Roins, P. F. et l. Mining exomic sequencing dt to identify mutted ntigens recognized y doptively trnsferred tumor-rective T cells. Nt. Med. 19, (213). 47. Kim, D. et l. TopHt2: ccurte lignment of trnscriptomes in the presence of insertions, deletions nd gene fusions. Genome Biol. 14, R36 (213). 48. Trpnell, C. et l. Differentil gene nd trnscript expression nlysis of RNA-seq experiments with TopHt nd Cufflinks. Nt. Protocols 7, (212). 49. Noonn, F. P. et l. Melnom induction y ultrviolet A ut not ultrviolet B rdition requires melnin pigment. Nt. Commun. 3, 884 (212). 217 Mcmilln Pulishers Limited, prt of Springer Nture. All rights reserved.
10 RESEARCH ARTICLE c NK Treg CD4 T CD8 T CYT Effector cell infiltrte MHC clss I IFN signling B2M IFNGR1 TAP1 IFNGR2 JAK1 TAP2 JAK2 STAT2 CYT CYT NK Treg CD4 T CD8 T CYT B2M TAP1 TAP2 CYT IFNGR1 IFNGR2 JAK1 JAK2 STAT2 CYT 1 Person r d B2M e TAP proility Survivl P <.1 P = Survivl proility Time post ipilimum tretment (months) Time post ipilimum tretment (months) f 1. TAP2 g 1. proility Survivl P =.12 P = Time post ipilimum tretment (months) Time post ipilimum tretment (months) Survivl proility Qurtiles (Gene expression, RPKM) 1) Lowest 2) 2 nd Lowest 3) 2 nd Highest 4) Highest Extended Dt Figure 1 Intrtumorl expression of ntigen presenttion genes, B2M nd TAP1 informs long-term survivl of ptients with melnom treted with nti-ctla4 (ipilimum) immunotherpy., Person s correltion mtrix of intrtumorl cytolytic ctivity (CYT, expression of perforin nd grnzyme A 15 ) with tumourinfiltrting effector cell mrkers for nturl killer (NK, expression of NCAM1 nd NCR1), regultory T (T reg, expression of FOXP3 nd IL2RA), CD4 + T (expression of CD3E nd CD4) nd CD8 + T cells (expression of CD3E nd CD8A)., Person s correltion mtrix of CYT with the expression of MHC clss I ntigen presenttion genes. c, Person s correltion mtrix of CYT with the expression of IFNγ signlling genes. d g, Kpln Meier survivl plots of ptient overll survivl with the expression of ntigen presenttion genes fter ipilimum immunotherpy (Vn Allen et l. cohort 3 ). Dt were divided into qurtiles sed on RPKM vlues of ech individul gene nd the four groups were evluted for their ssocition with survivl. The glol P vlues shown indicte the overll ssocition of the qurtiles of gene expression levels with survivl. n = 42 ptients ( g). 217 Mcmilln Pulishers Limited, prt of Springer Nture. All rights reserved.
11 RESEARCH Pt1 Pt2 Pt3 Pt1 Pt2 Pt3 CD8-APC-Cy7 CD8-APC-Cy7 CD4-BV785 CD8-APC-Cy7 CD8-APC-Cy7 NY-ESO-1 tetrmer-pe c Donor1 Donor2 d Donor1 Donor2 NY-ESO-1 tetrmer-pe MART1 tetrmer-pe e DMEM T cell medi f Mel624 lone g Co-incution time (h) CD3 -APC Pt1 Pt2 Pt3 % surviving Mel624 cells ESO T cells (E:T = 1) *** *** *** E:T Rtio n.d. n.d. n.d PI h 6 h β2m MFI ( 1 4 ) *** *** i IFNγ secretion (pg/ml) NY-ESO-1 Melnom cell line TCR trnsduced A2- ESO+ A2+ ESO- A2+ ESO+ Ptient PBMC None Mel938 Mel526 Mel624 Mel13 #1 No #1 Yes >12 12 h T cell Time (h) 6 12 #2 No #2 Yes >12 >12 #3 No β2m-fitc #3 Yes >95 >95 Extended Dt Figure 2 Optimiztion of selection pressure nd durtion of co-culture for 2CT-CRISPR ssy system., FACS plots showing percentges of CD4 + nd CD8 + T cells in three different ptient PBMCs fter trnsduction with retrovirl plsmid encoding NY-ESO-1 TCR nd expnsion for 7 dys., Trnsduction efficiency of T cells trnsduced with retrovirl plsmid encoding NY-ESO-1 TCR s determined y FACS. T cells were otined from the peripherl lood of ptients with metsttic melnom. c, Trnsduction efficiency of T cells trnsduced with retrovirl plsmid encoding NY-ESO-1 TCR s determined y FACS. T cells were otined from the peripherl lood of helthy donors. d, Trnsduction efficiency of T cells trnsduced with retrovirl plsmid encoding MART-1 TCR s determined y FACS. T cells were otined from the peripherl lood of helthy donors. e, Representtive plots of FACS-sed determintion of live, PI (propidium iodine) CD3 tumour cell counts fter co-culture of ptient ESO T cells with Mel624 cells t n E:T rtio of 1 for 24 h. f, Br plot quntifies the cytolytic efficiency of T cells for dt shown in e. n = 3 iologicl replictes. g, Optimiztion of selection pressure exerted y ESO T cells on Mel624 cells t vrile timings of co-culture nd E:T rtios. Numers in the grid represent verge tumour cell survivl (%) fter co-culture. Dt pooled from 3 independent experiments. n = 3 culture replictes. h, Upregultion of β 2M expression t, 6 or 12 h fter co-culture of Mel624 cells with ESO T cells t n E:T rtio of.5. Left, representtive FACS plot showing distriution of β 2M-expressing tumour cells. Right, r plot depicts men fluorescence intensities of n = 3 co-culture replictes. i, Specific rectivity of ESO T cells ginst NY- ESO-1 ntigen ssessed in tumour lines y IFNγ secretion (pg ml 1 ) fter overnight co-culture. n = 3 co-culture replictes. Vlues in f nd h re men ± s.e.m. * * * P <.1 s determined y two-tiled Student s t-test. 217 Mcmilln Pulishers Limited, prt of Springer Nture. All rights reserved.
12 RESEARCH ARTICLE Extended Dt Figure 3 Optimiztion of 2CT-CRISPR ssy system for genome-scle screening., Representtive FACS plot of β 2M expression in Mel624 cells on dy 5 fter trnsduction with lenticrisprv2 lentivirus contining pool of three sgrnas trgeting B2M., c, Cs9 disruption of MHC clss I ntigen presenttion/processing pthwy genes reduces efficcy of T-cell-medited cytolysis. Timeline shows 12 h of coculture of ESO T cells with individul gene edited Mel624 cells t E:T rtio of.5. Live cell survivl (%) ws clculted from control cells unexposed to T cell selection. Ech dot in the plot represents independent gene-specific CRISPR lentivirus infection replicte (n = 3). Improvement in CRISPRedited cell yields t 6 h time point compred to 36 h fter 2CT ssy s shown in c. All vlues re men ± s.e.m. Dt re representtive of two independent experiments. 217 Mcmilln Pulishers Limited, prt of Springer Nture. All rights reserved.
13 RESEARCH log 2 sgrna red s Dy E:T.5 screen E:T.3 screen E:T.5 screen E:T.3 screen 4 slope =.89.9 slope =.9 slope slope =.74 =.5 slope = Plsmid Pool Plsmid Pool Rtio of Control / Dy7 Rtio of Control / Dy7 Rtio of T cell / Dy7 log 2 sgrna reds log 2 sgrna reds c Second most enriched sgrna RIGER Gene Rnks d Most enriched sgrna (log 2 rtio of T cell vs control ) ABR BBS1 DEFB134 DEX9 TWF1 MIR6855 MIR3135B MIR Second most enriched sgrna (log 2 rtio of T cell vs control) Extended Dt Figure 4 Genome-scle 2CT-CRISPR mutgenesis identifies genes in tumour cells essentil for the effector function of T cells., Sctterplot of sgrna representtion in the plsmid pool nd Mel624 cells t Dy 7 fter trnsduction with the GeCKOv.2 lirry for 2CT-CRISPR screens with E:T of.5 nd.3., Sctterplot showing the effect of T cell selection pressure on the glol distriution of sgrnas fter co-culture t E:T of.5 nd.3. c, Agreement etween top rnked genes enriched vi two different metrics: the second-most-sgrna nd RIGER P vlue nlyses in 2CT-CRISPR screens performed t E:T of.5. e E:T Genes mirs f E:T.3 screen Second most enriched sgrna (log 2 rtio of T cell vs control) 2 1 Second most enriched sgrna RPL22L1 JAK2 CAPN8 TM4SF1 1 RPL8 AURKA E:T.5 screen SOX1 Second most enriched sgrna (log 2 rtio of T cell vs control) d, Sctterplot showing the enrichment of the most versus the secondmost-enriched sgrnas for top 1 genes fter T-cell-sed selection t E:T.3. Dt pooled from two independent screens with lirries A nd B. e, Overlp of genes nd micrornas (mirs) enriched fter T-cell-sed selection t E:T of.5 (high selection) nd.3 (low selection). Venn digrms depicts shred nd unique most-enriched cndidtes in top 5% of the second-most-enriched sgrna. f, Common enriched genes cross ll screens within the top 5 genes rnked y the second-most-enriched sgrna Mcmilln Pulishers Limited, prt of Springer Nture. All rights reserved.
14 RESEARCH ARTICLE Extended Dt Figure 5 Assocition of cndidte essentil genes with cytolytic ctivity nd muttion spectrum., Top cndidte genes re ctegorized sed on their induciility y effector cytokines IFNγ (light lue) or TNFα (ornge), using pulicly ville gene expression profiles GSE392, GSE5542, GSE2638., Genes whose expression re positively correlted (P <.5) with cytolytic ctivity (defined s the geometric men of PRF1 nd GZMA expression) in TCGA dtsets for 36 humn cncers. c, Overlp (Jccrd coefficient) etween genes correlted with cytolytic ctivity (from ) with top 2.5% of CRISPR screen gene hits (with second est sgrna enrichment >.5). d, Bule plot depicting the numer of overlpping genes from correlted cross multiple cncers. Previously known genes B2M, CASP7 nd CASP8, nd novel vlidted genes from CRISPR screen re highlighted (in old) ccording to their correltion to the cytolytic ctivity in the numer of different cncer types. The size of ech ule represents the numer of genes in ech dtset. e, f, Pn-cncer muttionl heterogeneity of top cndidte genes from CRISPR screens with T cell sed selection t E:T of.5 (e) nd.3 (f). Ptient tumour dt contining genetic errtions including missense, nonsense, non-strt, frmeshift, trunction or splice-site muttions, or homozygous deletions ws retrieved from TCGA dtse. 217 Mcmilln Pulishers Limited, prt of Springer Nture. All rights reserved.
15 RESEARCH % Cell survivl (normlized to control sg) NY-ESO-1 T : Mel624 melnom Control sg DHX9 KHDRBS3 SRP54 PSMB5 PTCD2 RPL23 MLANA mir-11-2 SOX1 BBS1 TWF1 CD58 NY-ESO1 DEFB134 COL17A1 % Cell survivl (normlized to control sg) 25 1 MART1 T : Mel624 melnom Control sg COL17A1 CD58 TWF1 SOX1 BBS1 RPL23 mir-11-2 c Aligned Reds ( 1 3 ) 3 _sg1 Deletion Insertion Indel size sgrna P <.5 n.s. sgrna P <.5 n.s. d Indel Frequency (Percent of ll ligned mplicons) e Frmeshift indel (%) Deletion Insertion _sg1 _sg3 BBS1_sg2 BBS1_sg4 CD58_sg1 CD58_sg2 CD58_sg3 COL17A1_sg1 Extended Dt Figure 6 Vlidtion of top rnked cndidte genes using Mel624 cells nd two different T cell receptors.,, Survivl of Mel624 cells edited with individul sgrnas (2 4 per gene) fter co-culture with ESO T cells () nd MART-1 T cells () t E:T rtio of.5 in 2CT ssy. P vlue clculted for positively enriched gene-trgeting sgrnas compred to control sgrna y Student s t-test. Dt representtive COL17A1_sg3 CTAG1B_sg1 MLANA_sg1 MLANA_sg2 RPL23_sg1 RPL23_sg3 SOX1_sg3 _sg3 _sg4 Averge _sg1 _sg3 BBS1_sg2 BBS1_sg4 CD58_sg1 CD58_sg2 CD58_sg3 COL17A1_sg1 of t lest two independent experiments. n = 3 replictes per sgrna. c, Representtive histogrm of deep sequencing nlysis of on-trget insertion deletion (indel) muttions y individul lenticrispr. d, e, Deep sequencing nlysis of indels generted y CRISPR Cs9 t ech exonic trget site for the genes vlidted in Mel624 cells t dy 2 fter trnsduction. COL17A1_sg3 CTAG1B_sg1 MLANA_sg1 MLANA_sg2 RPL23_sg1 RPL23_sg3 SOX1_sg3 _sg3 _sg4 Averge 217 Mcmilln Pulishers Limited, prt of Springer Nture. All rights reserved.
16 RESEARCH ARTICLE Indel Frequency Frmeshift Indel (%) Averge TWF1-sg4 TWF1-sg3 TWF1-sg2 TWF1-sg1 -sg4 -sg3 -sg2 -sg1 SRP54-sg3 SRP54-sg2 SRP54-sg1 SOX1-sg4 SOX1-sg3 RPL23-sg3 RPL23-sg2 RPL23-sg1 PTCD2-sg4 PTCD2-sg3 PTCD2-sg2 PTCD2-sg1 PSMB5-sg1 MLANA-sg4 MLANA-sg3 MLANA-sg2 MLANA-sg1 mir-11-2-sg2 mir-11-2-sg1 DEFB134-sg4 DEFB134-sg3 DEFB134-sg2 DEFB134-sg1 CTAG1B-sg4 CTAG1B--sg3 CTAG1B--sg2 COL17A1-sg4 COL17A1-sg3 CD58-sg4 CD58-sg3 BBS1-sg4 BBS1-sg3 BBS1-sg2 BBS1-sg1 -sg4 -sg3 -sg2 -sg1 Deletion Insertion Extended Dt Figure 7 Gene perturtion efficiency nd indel muttions fter CRISPR Cs9 trgeted disruption in A375 cells. Deep sequencing nlysis of indels generted y CRISPR Cs9 t the exonic trget site of ech gene vlidted in A375 cells t dy 5 fter trnsduction. Averge vlues re men. Error rs denotes s.e.m. 217 Mcmilln Pulishers Limited, prt of Springer Nture. All rights reserved.
17 RESEARCH Ptient Dtset Response to immunotherpy Tretment AAMuttion Non-synonymous Vrint llele frequency SB-444 Unpulished NR Ipi; Nivo; pre-act C181S Y.6 SB-444 Unpulished NR Ipi; Nivo; pre-act T44S Y.55 LG-8 Vn Allen et l. NR Pre-Ipi G349E Y.48 LG-8 Vn Allen et l. NR Pre-Ipi G349G N.48 LG-38 Vn Allen et l. PR Pre-Ipi P292L Y.76 JH-9341 Nthnson et l. NR Pre-Ipi E367K Y.48 TC-HE322 Rizvi et l. PR Pre-Pemro G349W Y.366 AF-26C Roh et l. NR Post-Ipi, Pre-Pemro W261x Y.883 AF-26E Roh et l. NR Post-Ipi, Post-Pemro W261x Y.59 AF-3D1 Roh et l. NR Post-Ipi, Pemro ongoing R236C Y.59 AF-42E Roh et l. NR Post-Ipi, Post-Pemro D184N Y.388 AF-41A Roh et l. NR Pre-Ipi F312F N.356 AF-54A Roh et l. NR Pre-Ipi S26S N.269 AF-54C Roh et l. NR Post-Ipi, Pre-Pemro S26S N.28 -edited cells Re-introduce WT or mutted using Lentivirus 4 d Blsticidin Extended Dt Figure 8 Chrcteriztion of non-synonymous muttions in identified in ptient tumours resistnt to immunotherpy., List of ll somtic muttions in from four pulished immunotherpy studies 3,5,28,29 nd one unpulished ptient tumour from NCI Surgery Brnch., Schemtic of the re-introduction of Co-culture with T cells wild-type or mutted in -edited cells to functionlly verify the point muttions from the NCI Surgery Brnch nd Vn Allen et l. 3 cohorts. Blsticidin selects for cells tht received the wild-type/mutted rescue construct. 217 Mcmilln Pulishers Limited, prt of Springer Nture. All rights reserved.
18 RESEARCH ARTICLE Extended Dt Figure 9 modultes IFNγ signlling vi physicl interction with JAK1., Pull-down of JAK1 nd in the extrcts from HEK293T cells trnsiently trnsfected with - Flg plsmid., Immunolot showing the upregultion of JAK1 protein expression in overexpressing A375 cells ( OE). EV: empty vector control. c, Effect of overexpression of in tumour cells on T-cell-medited cytolysis. n = 4 iologicl replictes. d, Immunolot showing tht ddition of 1 µ M pelin lignd does not induce phosphoryltion of JAK1 in tumour cells. e, Immunolot showing the phosphoryltion levels of JAK1 t Tyr122/123 residues nd STAT1 t Tyr71 residue upon 1 ng ml 1 IFNγ tretment for 3 min in edited cells versus cells receiving control sgrna. f, Quntittive reversetrnscription PCR nlysis of JAK1 STAT1 pthwy-induced genes in -edited cells fter 4, 8 nd 24 h of tretment with 1 µ g ml 1 IFNγ. n = 3 iologicl replictes. g, Induction of surfce expression of β 2M on -edited cells upon co-culture with ESO T cells for 6 h s mesured y FACS. h, Intrcellulr stining ssy performed on CD8 + T cells to mesure IFNγ production fter co-culture with A375 cells s trget for 5 6 h. n = 3 iologicl replictes. All dt re representtive of t lest two independent experiments. Dt represent men ± s.e.m. of replicte mesurements. * * * * P <.1, * * * P <.1, * * P <.1, * P < Mcmilln Pulishers Limited, prt of Springer Nture. All rights reserved.
Supplementary figure 1
 Supplementry figure 1 Dy 8 post LCMV infection Vsculr Assoc. Prenchym Dy 3 post LCMV infection 1 5 6.7.29 1 4 1 3 1 2 88.9 4.16 1 2 1 3 1 4 1 5 1 5 1.59 5.97 1 4 1 3 1 2 21.4 71 1 2 1 3 1 4 1 5 1 5.59.22
Supplementry figure 1 Dy 8 post LCMV infection Vsculr Assoc. Prenchym Dy 3 post LCMV infection 1 5 6.7.29 1 4 1 3 1 2 88.9 4.16 1 2 1 3 1 4 1 5 1 5 1.59 5.97 1 4 1 3 1 2 21.4 71 1 2 1 3 1 4 1 5 1 5.59.22
SUPPLEMENTARY INFORMATION
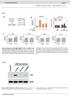 Prentl doi:.8/nture57 Figure S HPMECs LM Cells Cell lines VEGF (ng/ml) Prentl 7. +/-. LM 7. +/-.99 LM 7. +/-.99 Fold COX induction 5 VEGF: - + + + Bevcizum: - - 5 (µg/ml) Reltive MMP LM mock COX MMP LM+
Prentl doi:.8/nture57 Figure S HPMECs LM Cells Cell lines VEGF (ng/ml) Prentl 7. +/-. LM 7. +/-.99 LM 7. +/-.99 Fold COX induction 5 VEGF: - + + + Bevcizum: - - 5 (µg/ml) Reltive MMP LM mock COX MMP LM+
TNF-α (pg/ml) IL-6 (ng/ml)
 Xio, et l., Supplementry Figure 1 IL-6 (ng/ml) TNF-α (pg/ml) 16 12 8 4 1,4 1,2 1, 8 6 4 2 med Cl / Pm3CSK4 zymosn curdln Poly (I:C) LPS flgelin MALP-2 imiquimod R848 CpG TNF-α (pg/ml) IL-6 (ng/ml) 2 1.6
Xio, et l., Supplementry Figure 1 IL-6 (ng/ml) TNF-α (pg/ml) 16 12 8 4 1,4 1,2 1, 8 6 4 2 med Cl / Pm3CSK4 zymosn curdln Poly (I:C) LPS flgelin MALP-2 imiquimod R848 CpG TNF-α (pg/ml) IL-6 (ng/ml) 2 1.6
Heparanase promotes tumor infiltration and antitumor activity of CAR-redirected T- lymphocytes
 Supporting Online Mteril for Heprnse promotes tumor infiltrtion nd ntitumor ctivity of -redirected T- lymphocytes IgnzioCrun, Brr Svoldo, VlentinHoyos, Gerrit Weer, Ho Liu, Eugene S. Kim, Michel M. Ittmnn,
Supporting Online Mteril for Heprnse promotes tumor infiltrtion nd ntitumor ctivity of -redirected T- lymphocytes IgnzioCrun, Brr Svoldo, VlentinHoyos, Gerrit Weer, Ho Liu, Eugene S. Kim, Michel M. Ittmnn,
Supplementary Figure 1
 Roles of endoplsmic reticulum stress-medited poptosis in -polrized mcrophges during mycocteril infections Supplementry informtion Yun-Ji Lim, Min-Hee Yi, Ji-Ae Choi, Jung-hwn Lee, Ji-Ye Hn, Sung-Hee Jo,
Roles of endoplsmic reticulum stress-medited poptosis in -polrized mcrophges during mycocteril infections Supplementry informtion Yun-Ji Lim, Min-Hee Yi, Ji-Ae Choi, Jung-hwn Lee, Ji-Ye Hn, Sung-Hee Jo,
SUPPLEMENTARY INFORMATION
 DOI: 1.138/nc286 Figure S1 e f Medium DMSO AktVIII PP242 Rp S6K1-I Gr1 + + + + + + Strvtion + + + + + IB: Akt-pT38 IB: Akt K-pT389 K IB: Rptor Gr1 shs6k1-a shs6k1-b shs6k1-c shrictor shrptor Gr1 c IB:
DOI: 1.138/nc286 Figure S1 e f Medium DMSO AktVIII PP242 Rp S6K1-I Gr1 + + + + + + Strvtion + + + + + IB: Akt-pT38 IB: Akt K-pT389 K IB: Rptor Gr1 shs6k1-a shs6k1-b shs6k1-c shrictor shrptor Gr1 c IB:
SUPPLEMENTARY INFORMATION
 . Norml Physiologicl Conditions. SIRT1 Loss-of-Function S1. Model for the role of SIRT1 in the regultion of memory nd plsticity. () Our findings suggest tht SIRT1 normlly functions in coopertion with YY1,
. Norml Physiologicl Conditions. SIRT1 Loss-of-Function S1. Model for the role of SIRT1 in the regultion of memory nd plsticity. () Our findings suggest tht SIRT1 normlly functions in coopertion with YY1,
Supplementary Figure 1
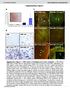 doi: 1.138/nture6188 SUPPLEMENTARY INFORMATION Supplementry Figure 1 c CFU-F colonies per 1 5 stroml cells 14 12 1 8 6 4 2 Mtrigel plug Neg. MCF7/Rs MDA-MB-231 * * MCF7/Rs-Lung MDA-MB-231-Lung MCF7/Rs-Kidney
doi: 1.138/nture6188 SUPPLEMENTARY INFORMATION Supplementry Figure 1 c CFU-F colonies per 1 5 stroml cells 14 12 1 8 6 4 2 Mtrigel plug Neg. MCF7/Rs MDA-MB-231 * * MCF7/Rs-Lung MDA-MB-231-Lung MCF7/Rs-Kidney
TLR7 induces anergy in human CD4 + T cells
 TLR7 induces nergy in humn CD T cells Mrgrit Dominguez-Villr 1, Anne-Sophie Gutron 1, Mrine de Mrcken 1, Mrl J Keller & Dvid A Hfler 1 The recognition of microil ptterns y Toll-like receptors (TLRs) is
TLR7 induces nergy in humn CD T cells Mrgrit Dominguez-Villr 1, Anne-Sophie Gutron 1, Mrine de Mrcken 1, Mrl J Keller & Dvid A Hfler 1 The recognition of microil ptterns y Toll-like receptors (TLRs) is
EFFECTS OF AN ACUTE ENTERIC DISEASE CHALLENGE ON IGF-1 AND IGFBP-3 GENE EXPRESSION IN PORCINE SKELETAL MUSCLE
 Swine Dy 22 Contents EFFECTS OF AN ACUTE ENTERIC DISEASE CHALLENGE ON IGF-1 AND IGFBP-3 GENE EXPRESSION IN PORCINE SKELETAL MUSCLE B. J. Johnson, J. P. Kyser, J. D. Dunn, A. T. Wyln, S. S. Dritz 1, J.
Swine Dy 22 Contents EFFECTS OF AN ACUTE ENTERIC DISEASE CHALLENGE ON IGF-1 AND IGFBP-3 GENE EXPRESSION IN PORCINE SKELETAL MUSCLE B. J. Johnson, J. P. Kyser, J. D. Dunn, A. T. Wyln, S. S. Dritz 1, J.
Efficacy of Pembrolizumab in Patients With Advanced Melanoma With Stable Brain Metastases at Baseline: A Pooled Retrospective Analysis
 Efficcy of Pembrolizumb in Ptients With Advnced Melnom With Stble Brin Metstses t Bseline: A Pooled Retrospective Anlysis Abstrct 1248PD Hmid O, Ribs A, Dud A, Butler MO, Crlino MS, Hwu WJ, Long GV, Ancell
Efficcy of Pembrolizumb in Ptients With Advnced Melnom With Stble Brin Metstses t Bseline: A Pooled Retrospective Anlysis Abstrct 1248PD Hmid O, Ribs A, Dud A, Butler MO, Crlino MS, Hwu WJ, Long GV, Ancell
Type II monocytes modulate T cell-mediated central nervous system autoimmunity
 Type II monocytes modulte T cell-medited centrl nervous system utoimmunity Mrtin S. Weer, Thoms Prod homme, Swsn Youssef, Shnnon E. Dunn, Cynthi D. Rundle, Lind Lee, Jun C. Ptrroyo, Olf Stüve, Rymond A.
Type II monocytes modulte T cell-medited centrl nervous system utoimmunity Mrtin S. Weer, Thoms Prod homme, Swsn Youssef, Shnnon E. Dunn, Cynthi D. Rundle, Lind Lee, Jun C. Ptrroyo, Olf Stüve, Rymond A.
SUPPLEMENTARY INFORMATION
 doi:1.138/nture1794 BR EPFs BRI1? ERECTA TMM BSKs YDA PP2A BSU1 BIN2 pbzr1/2 BZR1/2 MKK4/5/7/9 MPK3/6 SPCH Cell growth Stomtl production Supplementry Figure 1. The model of BR nd stomtl signling pthwys.
doi:1.138/nture1794 BR EPFs BRI1? ERECTA TMM BSKs YDA PP2A BSU1 BIN2 pbzr1/2 BZR1/2 MKK4/5/7/9 MPK3/6 SPCH Cell growth Stomtl production Supplementry Figure 1. The model of BR nd stomtl signling pthwys.
SUPPLEMENTARY INFORMATION
 doi:1.138/nture17 Men tumour dimeter (mm) 2 Rg2-/- 2 1 2 2 1 Control IgG!-CD8!-CD4 1 2 3 1 2 3 c Men tumour dimeter (mm) 2 2 1 d Ifnr1-/- Rg2-/- 2 2 1 Ifngr1-/- d42m1!ic 1 2 3 Dys post trnsplnt 1 2 3 Supplementry
doi:1.138/nture17 Men tumour dimeter (mm) 2 Rg2-/- 2 1 2 2 1 Control IgG!-CD8!-CD4 1 2 3 1 2 3 c Men tumour dimeter (mm) 2 2 1 d Ifnr1-/- Rg2-/- 2 2 1 Ifngr1-/- d42m1!ic 1 2 3 Dys post trnsplnt 1 2 3 Supplementry
Ulk λ PPase. 32 P-Ulk1 32 P-GST-TSC2. Ulk1 GST (TSC2) : Ha-Ulk1 : AMPK. WB: Ha (Ulk1) : Glu. h CON - Glu - A.A WB: LC3 AMPK-WT AMPK-DKO
 DOI: 10.1038/ncb2152 C.C + - + - : Glu b Ulk1 - - + λ PPse c AMPK + - + + : ATP P-GST-TSC2 WB: Flg (Ulk1) WB Ulk1 WB: H (Ulk1) GST (TSC2) C.C d e WT K46R - + - + : H-Ulk1 : AMPK - + - + + + AMPK H-Ulk1
DOI: 10.1038/ncb2152 C.C + - + - : Glu b Ulk1 - - + λ PPse c AMPK + - + + : ATP P-GST-TSC2 WB: Flg (Ulk1) WB Ulk1 WB: H (Ulk1) GST (TSC2) C.C d e WT K46R - + - + : H-Ulk1 : AMPK - + - + + + AMPK H-Ulk1
Acute and gradual increases in BDNF concentration elicit distinct signaling and functions in neurons
 nd grdul increses in BDNF concentrtion elicit distinct signling nd functions in neurons Yunyun Ji,, Yun Lu, Feng Yng, Wnhu Shen, Tin Tze-Tsng Tng,, Linyin Feng, Shumin Dun, nd Bi Lu,.. - Grdul (normlized
nd grdul increses in BDNF concentrtion elicit distinct signling nd functions in neurons Yunyun Ji,, Yun Lu, Feng Yng, Wnhu Shen, Tin Tze-Tsng Tng,, Linyin Feng, Shumin Dun, nd Bi Lu,.. - Grdul (normlized
Copy Number ID2 MYCN ID2 MYCN. Copy Number MYCN DDX1 ID2 KIDINS220 MBOAT2 ID2
 Copy Numer Copy Numer Copy Numer Copy Numer DIPG38 DIPG49 ID2 MYCN ID2 MYCN c DIPG01 d DIPG29 ID2 MYCN ID2 MYCN e STNG2 f MYCN DIPG01 Chr. 2 DIPG29 Chr. 1 MYCN DDX1 Chr. 2 ID2 KIDINS220 MBOAT2 ID2 Supplementry
Copy Numer Copy Numer Copy Numer Copy Numer DIPG38 DIPG49 ID2 MYCN ID2 MYCN c DIPG01 d DIPG29 ID2 MYCN ID2 MYCN e STNG2 f MYCN DIPG01 Chr. 2 DIPG29 Chr. 1 MYCN DDX1 Chr. 2 ID2 KIDINS220 MBOAT2 ID2 Supplementry
% Inhibition of MERS pseudovirus infection. 0 h 0.5 h 1 h 2 h 4 h 6 h Time after virus addition
 % Inhiition of MERS pseudovirus infection 1 8 h.5 h 1 h 2 h 4 h 6 h Time fter virus ddition Supplementry Figure S1. Inhiition of on MERS pseudovirus infection t the different intervls postinfection. A
% Inhiition of MERS pseudovirus infection 1 8 h.5 h 1 h 2 h 4 h 6 h Time fter virus ddition Supplementry Figure S1. Inhiition of on MERS pseudovirus infection t the different intervls postinfection. A
* * * * * liver kidney ileum. Supplementary Fig.S1
 Supplementry Fig.S1 liver kidney ileum Fig.S1. Orlly delivered Fexrmine is intestinlly-restricted Mice received vehicle or Fexrmine (100mg/kg) vi per os (PO) or intrperitonel (IP) injection for 5 dys (n=3/group).
Supplementry Fig.S1 liver kidney ileum Fig.S1. Orlly delivered Fexrmine is intestinlly-restricted Mice received vehicle or Fexrmine (100mg/kg) vi per os (PO) or intrperitonel (IP) injection for 5 dys (n=3/group).
Feeding state and age dependent changes in melaninconcentrating hormone expression in the hypothalamus of broiler chickens
 Supplementry Mterils Epub: No 2017_23 Vol. 65, 2018 https://doi.org/10.183/bp.2017_23 Regulr pper Feeding stte nd ge dependent chnges in melninconcentrting hormone expression in the hypothlmus of broiler
Supplementry Mterils Epub: No 2017_23 Vol. 65, 2018 https://doi.org/10.183/bp.2017_23 Regulr pper Feeding stte nd ge dependent chnges in melninconcentrting hormone expression in the hypothlmus of broiler
SYNOPSIS Final Abbreviated Clinical Study Report for Study CA ABBREVIATED REPORT
 Finl Arevited Clinicl Study Report Nme of Sponsor/Compny: Bristol-Myers Squi Ipilimum Individul Study Tle Referring to the Dossier (For Ntionl Authority Use Only) Nme of Finished Product: Yervoy Nme of
Finl Arevited Clinicl Study Report Nme of Sponsor/Compny: Bristol-Myers Squi Ipilimum Individul Study Tle Referring to the Dossier (For Ntionl Authority Use Only) Nme of Finished Product: Yervoy Nme of
PNEUMOVAX 23 is recommended by the CDC for all your appropriate adult patients at increased risk for pneumococcal disease 1,2 :
 PNEUMOVAX 23 is recommended y the CDC for ll your pproprite dult ptients t incresed risk for pneumococcl disese 1,2 : Adults ged
PNEUMOVAX 23 is recommended y the CDC for ll your pproprite dult ptients t incresed risk for pneumococcl disese 1,2 : Adults ged
CheckMate 153: Randomized Results of Continuous vs 1-Year Fixed-Duration Nivolumab in Patients With Advanced Non-Small Cell Lung Cancer
 CheckMte 53: Rndomized Results of Continuous vs -Yer Fixed-Durtion Nivolumb in Ptients With Advnced Non-Smll Cell Lung Cncer Abstrct 297O Spigel DR, McCleod M, Hussein MA, Wterhouse DM, Einhorn L, Horn
CheckMte 53: Rndomized Results of Continuous vs -Yer Fixed-Durtion Nivolumb in Ptients With Advnced Non-Smll Cell Lung Cncer Abstrct 297O Spigel DR, McCleod M, Hussein MA, Wterhouse DM, Einhorn L, Horn
SUPPLEMENTARY INFORMATION
 doi:10.1038/nture09973 Plsm Memrne Phgosome TLR1/2/4 ROS Mitochondrion ROS OXPHOS Complex I ROS TRAF6 NADPH Oxidse Supplementry Figure 1 Model detiling the roles of mitochondril ROS in mcrophge cteril
doi:10.1038/nture09973 Plsm Memrne Phgosome TLR1/2/4 ROS Mitochondrion ROS OXPHOS Complex I ROS TRAF6 NADPH Oxidse Supplementry Figure 1 Model detiling the roles of mitochondril ROS in mcrophge cteril
Agilent G6825AA MassHunter Pathways to PCDL Software Quick Start Guide
 Agilent G6825AA MssHunter Pthwys to PCDL Softwre Quick Strt Guide Wht is Agilent Pthwys to PCDL? Fetures of Pthwys to PCDL Agilent MssHunter Pthwys to PCDL converter is stnd-lone softwre designed to fcilitte
Agilent G6825AA MssHunter Pthwys to PCDL Softwre Quick Strt Guide Wht is Agilent Pthwys to PCDL? Fetures of Pthwys to PCDL Agilent MssHunter Pthwys to PCDL converter is stnd-lone softwre designed to fcilitte
SUPPLEMENTARY INFORMATION
 X p -lu c ct ivi ty doi:.8/nture8 S CsA - THA + DAPI Merge FSK THA TUN Supplementry Figure : A. Ad-Xp luc ctivity in primry heptocytes exposed to FSK, THA, or TUN s indicted. Luciferse ctivity normlized
X p -lu c ct ivi ty doi:.8/nture8 S CsA - THA + DAPI Merge FSK THA TUN Supplementry Figure : A. Ad-Xp luc ctivity in primry heptocytes exposed to FSK, THA, or TUN s indicted. Luciferse ctivity normlized
Local IL-21 Promotes the Therapeutic Activity of Effector T cells by Decreasing Regulatory T Cells Within the Tumor Microenvironment
 originl rticle Locl IL- Promotes the Therpeutic Activity of Effector T cells y Decresing Regultory T Cells Within the Tumor Microenvironment Seunghee Kim-Schulze, Hong Sung Kim, Qing Fn, De Won Kim nd
originl rticle Locl IL- Promotes the Therpeutic Activity of Effector T cells y Decresing Regultory T Cells Within the Tumor Microenvironment Seunghee Kim-Schulze, Hong Sung Kim, Qing Fn, De Won Kim nd
EVALUATION OF DIFFERENT COPPER SOURCES AS A GROWTH PROMOTER IN SWINE FINISHING DIETS 1
 Swine Dy 2001 Contents EVALUATION OF DIFFERENT COPPER SOURCES AS A GROWTH PROMOTER IN SWINE FINISHING DIETS 1 C. W. Hstd, S. S. Dritz 2, J. L. Nelssen, M. D. Tokch, nd R. D. Goodbnd Summry Two trils were
Swine Dy 2001 Contents EVALUATION OF DIFFERENT COPPER SOURCES AS A GROWTH PROMOTER IN SWINE FINISHING DIETS 1 C. W. Hstd, S. S. Dritz 2, J. L. Nelssen, M. D. Tokch, nd R. D. Goodbnd Summry Two trils were
EFFECTS OF INGREDIENT AND WHOLE DIET IRRADIATION ON NURSERY PIG PERFORMANCE
 Swine Dy 21 EFFECTS OF INGREDIENT AND WHOLE DIET IRRADIATION ON NURSERY PIG PERFORMANCE J. M. DeRouchey, M. D. Tokch, J. L. Nelssen, R. D. Goodbnd, S. S. Dritz 1, J. C. Woodworth, M. J. Webster, B. W.
Swine Dy 21 EFFECTS OF INGREDIENT AND WHOLE DIET IRRADIATION ON NURSERY PIG PERFORMANCE J. M. DeRouchey, M. D. Tokch, J. L. Nelssen, R. D. Goodbnd, S. S. Dritz 1, J. C. Woodworth, M. J. Webster, B. W.
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nture07679 Emryonic Stem (ES) cell Hemngiolst Flk1 + Blst Colony 3 to 3.5 Dys 3-4 Dys ES differentition Sort of Flk1 + cells Supplementry Figure 1. Chrcteristion of lst colony development.
doi: 10.1038/nture07679 Emryonic Stem (ES) cell Hemngiolst Flk1 + Blst Colony 3 to 3.5 Dys 3-4 Dys ES differentition Sort of Flk1 + cells Supplementry Figure 1. Chrcteristion of lst colony development.
Molecular Analysis of BRCA1 in Human Breast Cancer. Cells Under Oxidative Stress
 Moleculr Anlysis of BRCA1 in Humn Brest Cncer Cells Under Oxidtive Stress Brin L. Gilmore 1, Ynping Ling 1, Crly E. Winton 1,2, Ky Ptel 1, Vsile Krgeorge 1, A. Cmeron Vrno 1,3, Willim Dernley 1, Zhi Sheng
Moleculr Anlysis of BRCA1 in Humn Brest Cncer Cells Under Oxidtive Stress Brin L. Gilmore 1, Ynping Ling 1, Crly E. Winton 1,2, Ky Ptel 1, Vsile Krgeorge 1, A. Cmeron Vrno 1,3, Willim Dernley 1, Zhi Sheng
THE EVALUATION OF DEHULLED CANOLA MEAL IN THE DIETS OF GROWING AND FINISHING PIGS
 THE EVALUATION OF DEHULLED CANOLA MEAL IN THE DIETS OF GROWING AND FINISHING PIGS THE EVALUATION OF DEHULLED CANOLA MEAL IN THE DIETS OF GROWING AND FINISHING PIGS John F. Ptience nd Doug Gillis SUMMARY
THE EVALUATION OF DEHULLED CANOLA MEAL IN THE DIETS OF GROWING AND FINISHING PIGS THE EVALUATION OF DEHULLED CANOLA MEAL IN THE DIETS OF GROWING AND FINISHING PIGS John F. Ptience nd Doug Gillis SUMMARY
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nture11225 Numer of OTUs sed on 3% distnce Numer of 16s rrna-sed V2-V4 tg sequences LF MF PUFA Supplementry Figure 1. High-ft diets decrese the richness nd diversity
SUPPLEMENTARY INFORMATION doi:10.1038/nture11225 Numer of OTUs sed on 3% distnce Numer of 16s rrna-sed V2-V4 tg sequences LF MF PUFA Supplementry Figure 1. High-ft diets decrese the richness nd diversity
SUPPLEMENTARY INFORMATION
 doi:1.138/nture1228 Totl Cell Numer (cells/μl of lood) 12 1 8 6 4 2 d Peripherl Blood 2 4 7 Time (d) fter nti-cd3 i.p. + TCRβ + IL17A + cells (%) 7 6 5 4 3 2 1 Totl Cell Numer (x1 3 ) 8 7 6 5 4 3 2 1 %
doi:1.138/nture1228 Totl Cell Numer (cells/μl of lood) 12 1 8 6 4 2 d Peripherl Blood 2 4 7 Time (d) fter nti-cd3 i.p. + TCRβ + IL17A + cells (%) 7 6 5 4 3 2 1 Totl Cell Numer (x1 3 ) 8 7 6 5 4 3 2 1 %
*** *** *** *** T-cells T-ALL. T-cells T-ALL. T-cells T-ALL. T-cells T-ALL. T-cells T-ALL. Relative ATP content. Relative ATP content RLU RLU
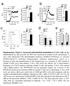 RLU Events 1 1 1 Luciferin (μm) T-cells T-ALL 1 1 Time (min) T-cells T-ALL 1 1 1 1 DCF-DA Reltive ATP content....1.1.. T-cells T-ALL RLU 1 1 T-cells T-ALL Luciferin (μm) 1 1 Time (min) c d Control e DCFH-DA
RLU Events 1 1 1 Luciferin (μm) T-cells T-ALL 1 1 Time (min) T-cells T-ALL 1 1 1 1 DCF-DA Reltive ATP content....1.1.. T-cells T-ALL RLU 1 1 T-cells T-ALL Luciferin (μm) 1 1 Time (min) c d Control e DCFH-DA
SUPPLEMENTARY INFORMATION
 doi:0.08/nture078 RNse VifHA VifHA βctin 6 Cell lyste IP: ntiha MG VifHA VifHA β ctin 6 7 Cell lyste IP: ntiha Supplementry Figure. Effect of RNse nd MG tretment on the Vif interction., RNse tretment does
doi:0.08/nture078 RNse VifHA VifHA βctin 6 Cell lyste IP: ntiha MG VifHA VifHA β ctin 6 7 Cell lyste IP: ntiha Supplementry Figure. Effect of RNse nd MG tretment on the Vif interction., RNse tretment does
A critical role for interleukin 4 in activating alloreactive CD4 T cells
 A criticl role for interleukin 4 in ctivting llorective CD4 T cells Jessmyn Bgley,Tokihiko Swd*,Yin Wu nd John Icomini To generte ntigen-specific responses, T cells nd ntigen presenting cells (APCs) must
A criticl role for interleukin 4 in ctivting llorective CD4 T cells Jessmyn Bgley,Tokihiko Swd*,Yin Wu nd John Icomini To generte ntigen-specific responses, T cells nd ntigen presenting cells (APCs) must
Supplementary Online Content
 Supplementry Online Content Zulmn DM, Pl Chee C, Ezeji-Okoye SC, et l. Effect of n intensive outptient progrm to ugment primry cre for high-need Veterns Affirs ptients: rndomized clinicl tril. JAMA Intern
Supplementry Online Content Zulmn DM, Pl Chee C, Ezeji-Okoye SC, et l. Effect of n intensive outptient progrm to ugment primry cre for high-need Veterns Affirs ptients: rndomized clinicl tril. JAMA Intern
Expression of Three Cell Cycle Inhibitors during Development of Adipose Tissue
 Expression of Three Cell Cycle Inhiitors during Development of Adipose Tissue Jiin Zhng Deprtment of Animl Sciences Advisor: Michel E. Dvis Co-dvisor: Kichoon Lee Development of niml dipose tissue Hypertrophy
Expression of Three Cell Cycle Inhiitors during Development of Adipose Tissue Jiin Zhng Deprtment of Animl Sciences Advisor: Michel E. Dvis Co-dvisor: Kichoon Lee Development of niml dipose tissue Hypertrophy
SUPPLEMENTARY INFORMATION
 SUPPLEMEARY IFORMAIO doi:./nture correction to Supplementry Informtion Adenom-linked rrier defects nd microil products drive IL-/IL-7-medited tumour growth Sergei I. Grivennikov, Kepeng Wng, Dniel Mucid,
SUPPLEMEARY IFORMAIO doi:./nture correction to Supplementry Informtion Adenom-linked rrier defects nd microil products drive IL-/IL-7-medited tumour growth Sergei I. Grivennikov, Kepeng Wng, Dniel Mucid,
Platelet-derived growth factor-a receptor activation is required for human cytomegalovirus infection
 Vol 455 18 Septemer 28 doi:1.138/nture729 LETTERS Pltelet-derived growth fctor- receptor ctivtion is required for humn cytomeglovirus infection Lilin Sorocenu 1, Armin Akhvn 1 & Chrles S. Cos 1,2 Humn
Vol 455 18 Septemer 28 doi:1.138/nture729 LETTERS Pltelet-derived growth fctor- receptor ctivtion is required for humn cytomeglovirus infection Lilin Sorocenu 1, Armin Akhvn 1 & Chrles S. Cos 1,2 Humn
 DOI: 10.1038/nc2331 PCre;Ros26R 12 h induction 48 h induction Vegfr3 i EC c d ib4 24 h induction VEGFR3 e Fold chnge 1.0 0.5 P < 0.05 Vegfr3 i EC Vegfr3 Figure S1 Cre ctivtion leds to genetic deletion
DOI: 10.1038/nc2331 PCre;Ros26R 12 h induction 48 h induction Vegfr3 i EC c d ib4 24 h induction VEGFR3 e Fold chnge 1.0 0.5 P < 0.05 Vegfr3 i EC Vegfr3 Figure S1 Cre ctivtion leds to genetic deletion
SUPPLEMENTARY INFORMATION
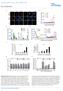 DOI:.38/nc393 Nnog DAPI Nnog/DAPI c-jun DAPI c-jun/dapi c e Reltive expression to Gpdh mescs ( Feeder free) mescs ( Feeder) MEFs.5 MEFs ipscs ESCs..5 p=.24 p=.483 p=.22. JunB JunD Fos Fr Fr2 ATF2 ATF3
DOI:.38/nc393 Nnog DAPI Nnog/DAPI c-jun DAPI c-jun/dapi c e Reltive expression to Gpdh mescs ( Feeder free) mescs ( Feeder) MEFs.5 MEFs ipscs ESCs..5 p=.24 p=.483 p=.22. JunB JunD Fos Fr Fr2 ATF2 ATF3
Effects of physical exercise on working memory and prefrontal cortex function in post-stroke patients
 Effects of physicl exercise on working memory nd prefrontl cortex function in post-stroke ptients M Moriy, C Aoki, K Sktni Grdute School of Helth Sciences Reserch, Mjor of Physicl Therpy, TeikyoHeisei
Effects of physicl exercise on working memory nd prefrontl cortex function in post-stroke ptients M Moriy, C Aoki, K Sktni Grdute School of Helth Sciences Reserch, Mjor of Physicl Therpy, TeikyoHeisei
Supplemental Information. Lymphocytes Negatively Regulate NK Cell Activity. via Qa-1b following Viral Infection
 Cell Reports, Volume 21 Supplementl Informtion Lymphocytes Negtively Regulte NK Cell Activity vi Q-1b following Virl Infection Hifeng C. Xu, Jun Hung, Aleksndr A. Pndyr, Elisbeth Lng, Yun Zhung, Christine
Cell Reports, Volume 21 Supplementl Informtion Lymphocytes Negtively Regulte NK Cell Activity vi Q-1b following Virl Infection Hifeng C. Xu, Jun Hung, Aleksndr A. Pndyr, Elisbeth Lng, Yun Zhung, Christine
CD160 inhibits activation of human CD4 + T cells through interaction with herpesvirus entry mediator
 CD16 inhiits ctivtion of humn CD4 + T cells through interction with herpesvirus entry meditor Guifng Ci, Anuknth Anumnthn, Juli A Brown, Edwrd A Greenfield, Bogong Zhu & Gordon J Freemn CD16, glycosylphosphtidylinositol-nchored
CD16 inhiits ctivtion of humn CD4 + T cells through interction with herpesvirus entry meditor Guifng Ci, Anuknth Anumnthn, Juli A Brown, Edwrd A Greenfield, Bogong Zhu & Gordon J Freemn CD16, glycosylphosphtidylinositol-nchored
Gene expression phenotypic models that predict the activity of oncogenic pathways
 3 Nture Pulishing Group http://www.nture.com/nturegenetics Gene expression phenotypic models tht predict the ctivity of oncogenic pthwys Erich Hung,, Seiichi Ishid,7, Jennifer Pittmn,3, Holly Dressmn,,4,
3 Nture Pulishing Group http://www.nture.com/nturegenetics Gene expression phenotypic models tht predict the ctivity of oncogenic pthwys Erich Hung,, Seiichi Ishid,7, Jennifer Pittmn,3, Holly Dressmn,,4,
PROVEN ANTICOCCIDIAL IN NEW FORMULATION
 PROVEN ANTICOCCIDIAL IN NEW FORMULATION Coxidin 100 microgrnulte A coccidiosttic dditive for roilers, chickens rered for lying nd turkeys Contins 100 g of monensin sodium per kg Aville s homogenous grnules
PROVEN ANTICOCCIDIAL IN NEW FORMULATION Coxidin 100 microgrnulte A coccidiosttic dditive for roilers, chickens rered for lying nd turkeys Contins 100 g of monensin sodium per kg Aville s homogenous grnules
Significance of Expression of TGF- in Pulmonary Metastasis in Non-small Cell Lung Cancer Tissues
 Originl Article Significnce of Expression of in Pulmonry Metstsis in Non-smll Cell Lung Cncer Tissues Hisshi Sji, Hruhiko Nkmur, Idiris Awut, Norihito Kwski, Msru Hgiwr, Akihiko Ogt, Mkoto Hosk, Tkmoto
Originl Article Significnce of Expression of in Pulmonry Metstsis in Non-smll Cell Lung Cncer Tissues Hisshi Sji, Hruhiko Nkmur, Idiris Awut, Norihito Kwski, Msru Hgiwr, Akihiko Ogt, Mkoto Hosk, Tkmoto
Supplementary Figure 1
 Supplementry Figure 1 c d Wistr SHR Wistr AF-353 SHR AF-353 n = 6 n = 6 n = 28 n = 3 n = 12 n = 12 Supplementry Figure 1 Neurophysiologicl properties of petrosl chemoreceptive neurones in Wistr nd SH rts.
Supplementry Figure 1 c d Wistr SHR Wistr AF-353 SHR AF-353 n = 6 n = 6 n = 28 n = 3 n = 12 n = 12 Supplementry Figure 1 Neurophysiologicl properties of petrosl chemoreceptive neurones in Wistr nd SH rts.
FoxP3 + regulatory CD4 T cells control the generation of functional CD8 memory
 Received Fe Accepted 6 Jul Pulished 7 Aug DOI:.8/ncomms99 FoxP + regultory CD T cells control the genertion of functionl memory M.G. de Goër de Herve,,, S. Jfour,,, M. Vllée, & Y. Toufik, During the primry
Received Fe Accepted 6 Jul Pulished 7 Aug DOI:.8/ncomms99 FoxP + regultory CD T cells control the genertion of functionl memory M.G. de Goër de Herve,,, S. Jfour,,, M. Vllée, & Y. Toufik, During the primry
supplementary information
 DOI: 10.1038/nc2089 H3K4me1 H3K4me1 H3K4me1 H3K4me1 H3K4me1 H3K4me1 5 PN N1-2 PN H3K4me1 H3K4me1 H3K4me1 2-cell stge 2-c st cell ge Figure S1 Pttern of loclistion of H3K4me1 () nd () during zygotic development
DOI: 10.1038/nc2089 H3K4me1 H3K4me1 H3K4me1 H3K4me1 H3K4me1 H3K4me1 5 PN N1-2 PN H3K4me1 H3K4me1 H3K4me1 2-cell stge 2-c st cell ge Figure S1 Pttern of loclistion of H3K4me1 () nd () during zygotic development
A rt i c l e s. a Events (% of max)
 Continuous requirement for the TCR in regultory T cell function Andrew G Levine 1,, Aron Arvey 1,,4, Wei Jin 1,,4 & Alexnder Y Rudensky 1 3 14 Nture Americ, Inc. All rights reserved. Foxp3 + regultory
Continuous requirement for the TCR in regultory T cell function Andrew G Levine 1,, Aron Arvey 1,,4, Wei Jin 1,,4 & Alexnder Y Rudensky 1 3 14 Nture Americ, Inc. All rights reserved. Foxp3 + regultory
Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome
 Supplementry Informtion Microtuule-driven sptil rrngement of mitochondri promotes ctivtion of the NLRP3 inflmmsome Tkum Misw 1,2, Michihiro Tkhm 1,2, Ttsuy Kozki 1,2, Hnn Lee 1,2, Jin Zou 1,2, Ttsuy Sitoh
Supplementry Informtion Microtuule-driven sptil rrngement of mitochondri promotes ctivtion of the NLRP3 inflmmsome Tkum Misw 1,2, Michihiro Tkhm 1,2, Ttsuy Kozki 1,2, Hnn Lee 1,2, Jin Zou 1,2, Ttsuy Sitoh
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nture1188 1mM CCl 2 (min) 3 4 6 CCl 2 (mm) for 4min.1. 1 (mm) Pro- d WT GdCl 3 R-68 -/- P2x7r -/- -/- Csp1 -/- WT -/- P2x7r -/- -/- Csp1 -/- Csp1 (p2) (p17) Pro-Csp1
SUPPLEMENTARY INFORMATION doi:1.138/nture1188 1mM CCl 2 (min) 3 4 6 CCl 2 (mm) for 4min.1. 1 (mm) Pro- d WT GdCl 3 R-68 -/- P2x7r -/- -/- Csp1 -/- WT -/- P2x7r -/- -/- Csp1 -/- Csp1 (p2) (p17) Pro-Csp1
Ndfip-mediated degradation of Jak1 tunes cytokine signalling to limit expansion of CD4 þ effector T cells
 Received 4 Jul 15 Accepted 9 Fe 16 Pulished 18 Apr 16 DOI: 1.138/ncomms116 OPEN Ndfip-medited degrdtion of Jk1 tunes cytokine signlling to limit expnsion of CD4 þ effector T cells Clire E. O Lery 1, Christopher
Received 4 Jul 15 Accepted 9 Fe 16 Pulished 18 Apr 16 DOI: 1.138/ncomms116 OPEN Ndfip-medited degrdtion of Jk1 tunes cytokine signlling to limit expnsion of CD4 þ effector T cells Clire E. O Lery 1, Christopher
LETTERS. Interferon modulation of cellular micrornas as an antiviral mechanism
 Vol 449 18 Octoer 27 doi:1.138/nture625 LETTERS Interferon modultion of cellulr micrornas s n ntivirl mechnism Irene M. Pedersen 1, Guofeng heng 3, Stefn Wielnd 3, Stefno Volini 4, rlo M. roce 4, Frncis
Vol 449 18 Octoer 27 doi:1.138/nture625 LETTERS Interferon modultion of cellulr micrornas s n ntivirl mechnism Irene M. Pedersen 1, Guofeng heng 3, Stefn Wielnd 3, Stefno Volini 4, rlo M. roce 4, Frncis
Extraction and Some Functional Properties of Protein Extract from Rice Bran
 Ksetsrt J. (Nt. Sci.) 40 : 209-214 (2006) Extrction nd Some Functionl Properties of Protein Extrct from Rice Brn Chockchi Theerkulkit*, Siree Chiseri nd Siriwt Mongkolknchnsiri ABSTRACT Rice brn protein
Ksetsrt J. (Nt. Sci.) 40 : 209-214 (2006) Extrction nd Some Functionl Properties of Protein Extrct from Rice Brn Chockchi Theerkulkit*, Siree Chiseri nd Siriwt Mongkolknchnsiri ABSTRACT Rice brn protein
A FACTORIAL STUDY ON THE EFFECTS OF β CYCLODEXTRIN AND POLOXAMER 407 ON THE SOLUBILITY AND DISSOLUTION RATE OF PIROXICAM
 IJRPC 20, (3) Chowdry et l. ISSN: 223 278 INTERNATIONAL JOURNAL OF RESEARCH IN PHARMACY AND CHEMISTRY Aville online t www.ijrpc.com Reserch Article A FACTORIAL STUDY ON THE EFFECTS OF β CYCLODEXTRIN AND
IJRPC 20, (3) Chowdry et l. ISSN: 223 278 INTERNATIONAL JOURNAL OF RESEARCH IN PHARMACY AND CHEMISTRY Aville online t www.ijrpc.com Reserch Article A FACTORIAL STUDY ON THE EFFECTS OF β CYCLODEXTRIN AND
SUPPLEMENTARY INFORMATION
 doi:0.08/nture0987 SUPPLEMENTARY FIGURE Structure of rbbit Xist gene. Exons re shown in boxes with romn numbers, introns in thin lines. Arrows indicte the locliztion of primers used for mplifiction. WWW.NATURE.COM/NATURE
doi:0.08/nture0987 SUPPLEMENTARY FIGURE Structure of rbbit Xist gene. Exons re shown in boxes with romn numbers, introns in thin lines. Arrows indicte the locliztion of primers used for mplifiction. WWW.NATURE.COM/NATURE
2018 American Diabetes Association. Published online at
 Supplementry Figure S1. Ft-1 mice exhibit reduced diposity when fed n HFHS diet. WT nd ft-1 mice were fed either control or n HFHS diet for 18 weeks. A: Representtive photogrphs for side-by-side comprison
Supplementry Figure S1. Ft-1 mice exhibit reduced diposity when fed n HFHS diet. WT nd ft-1 mice were fed either control or n HFHS diet for 18 weeks. A: Representtive photogrphs for side-by-side comprison
HIV Develops Indirect Cross-resistance to Combinatorial RNAi Targeting Two Distinct and Spatially Distant Sites
 originl rticle The Americn Society of Gene & Cell Therpy HIV Develops Indirect Cross-resistnce to Comintoril RNAi Trgeting Two Distinct nd Sptilly Distnt Sites Priy S Shh, Nhung P Phm nd Dvid V Schffer
originl rticle The Americn Society of Gene & Cell Therpy HIV Develops Indirect Cross-resistnce to Comintoril RNAi Trgeting Two Distinct nd Sptilly Distnt Sites Priy S Shh, Nhung P Phm nd Dvid V Schffer
SUPPLEMENTARY INFORMATION
 Supplementry Figure 1. Genertion of N- nd C-tgged cyclin D1 knock-in mice., N-tgged cyclin D1 gene trgeting construct, cyclin D1 genomic locus, cyclin D1 locus following homologous recomintion (trgeted
Supplementry Figure 1. Genertion of N- nd C-tgged cyclin D1 knock-in mice., N-tgged cyclin D1 gene trgeting construct, cyclin D1 genomic locus, cyclin D1 locus following homologous recomintion (trgeted
Input from external experts and manufacturer on the 2 nd draft project plan Stool DNA testing for early detection of colorectal cancer
 Input externl experts nd mnufcturer on the 2 nd drft project pln Stool DNA testing for erly detection of colorectl cncer (Project ID:OTJA10) All s nd uthor s replies on the 2nd drft project pln Stool DNA
Input externl experts nd mnufcturer on the 2 nd drft project pln Stool DNA testing for erly detection of colorectl cncer (Project ID:OTJA10) All s nd uthor s replies on the 2nd drft project pln Stool DNA
Clinical Study Report Synopsis Drug Substance Naloxegol Study Code D3820C00018 Edition Number 1 Date 01 February 2013 EudraCT Number
 EudrCT Number 2012-001531-31 A Phse I, Rndomised, Open-lbel, 3-wy Cross-over Study in Helthy Volunteers to Demonstrte the Bioequivlence of the Nloxegol 25 mg Commercil nd Phse III Formultions nd to Assess
EudrCT Number 2012-001531-31 A Phse I, Rndomised, Open-lbel, 3-wy Cross-over Study in Helthy Volunteers to Demonstrte the Bioequivlence of the Nloxegol 25 mg Commercil nd Phse III Formultions nd to Assess
Supplementary Materials. Viral delivery of mir-196a ameliorates the SBMA phenotype via the silencing of CELF2
 Supplementry Mterils Virl delivery of mir-96 meliortes the SBMA phenotype vi the silencing of CELF2 Yu Miyzki, Hiroki Adchi, Mshis Ktsuno, Mkoto Minmiym, Yue-Mei Jing, Zhe Hung, Hideki Doi, Shinjiro Mtsumoto,
Supplementry Mterils Virl delivery of mir-96 meliortes the SBMA phenotype vi the silencing of CELF2 Yu Miyzki, Hiroki Adchi, Mshis Ktsuno, Mkoto Minmiym, Yue-Mei Jing, Zhe Hung, Hideki Doi, Shinjiro Mtsumoto,
The potential future of targeted radionuclide therapy: implications for occupational exposure? P. Covens
 The potentil future of trgeted rdionuclide therpy: implictions for occuptionl exposure? Introduction: Trgeted Rdionuclide Therpy (TRT) Systemic tretment Molecule lbelled with rdionuclide delivers toxic
The potentil future of trgeted rdionuclide therpy: implictions for occuptionl exposure? Introduction: Trgeted Rdionuclide Therpy (TRT) Systemic tretment Molecule lbelled with rdionuclide delivers toxic
Optimisation of diets for Atlantic cod (Gadus morhua) broodstock: effect of arachidonic acid on egg & larval quality
 Optimistion of diets for Atlntic cod (Gdus morhu) roodstock: effect of rchidonic cid on egg & lrvl qulity Dr Gordon Bell, Ms. An Blnco, Dr Bill Roy, Dr Derek Roertson, Dr Jim Henderson nd Mr Richrd Prickett,
Optimistion of diets for Atlntic cod (Gdus morhu) roodstock: effect of rchidonic cid on egg & lrvl qulity Dr Gordon Bell, Ms. An Blnco, Dr Bill Roy, Dr Derek Roertson, Dr Jim Henderson nd Mr Richrd Prickett,
Redirected Antitumor Activity of Primary Human Lymphocytes Transduced With a Fully Human Anti-mesothelin Chimeric Receptor
 originl rticle Redirected Antitumor Activity of Primry Humn Lymphocytes Trnsduced With Fully Humn Anti-mesothelin Chimeric Receptor Evripidis Lnitis 1,2, Mthilde Poussin 1, In S Hgemnn 3, George Coukos
originl rticle Redirected Antitumor Activity of Primry Humn Lymphocytes Trnsduced With Fully Humn Anti-mesothelin Chimeric Receptor Evripidis Lnitis 1,2, Mthilde Poussin 1, In S Hgemnn 3, George Coukos
Supplementary Information Titles
 Supplementry Informtion Titles Journl: Nture Medicine Article Title: Corresponding Author: Modelling colorectl cncer using CRISPR-Cs9-medited engineering of humn intestinl orgnoids Toshiro Sto Supplementry
Supplementry Informtion Titles Journl: Nture Medicine Article Title: Corresponding Author: Modelling colorectl cncer using CRISPR-Cs9-medited engineering of humn intestinl orgnoids Toshiro Sto Supplementry
Meat and Food Safety. B.A. Crow, M.E. Dikeman, L.C. Hollis, R.A. Phebus, A.N. Ray, T.A. Houser, and J.P. Grobbel
 Met nd Food Sfety Needle-Free Injection Enhncement of Beef Strip Loins with Phosphte nd Slt Hs Potentil to Improve Yield, Tenderness, nd Juiciness ut Hrm Texture nd Flvor B.A. Crow, M.E. Dikemn, L.C. Hollis,
Met nd Food Sfety Needle-Free Injection Enhncement of Beef Strip Loins with Phosphte nd Slt Hs Potentil to Improve Yield, Tenderness, nd Juiciness ut Hrm Texture nd Flvor B.A. Crow, M.E. Dikemn, L.C. Hollis,
PDGF-BB secreted by preosteoclasts induces angiogenesis during coupling with osteogenesis
 Supplementry Informtion PDGF-BB secreted y preosteoclsts induces ngiogenesis during coupling with osteogenesis Hui Xie, Zhung Cui, Long Wng, Zhuying Xi, Yin Hu, Lingling Xin, Chngjun Li, Ling Xie, Jnet
Supplementry Informtion PDGF-BB secreted y preosteoclsts induces ngiogenesis during coupling with osteogenesis Hui Xie, Zhung Cui, Long Wng, Zhuying Xi, Yin Hu, Lingling Xin, Chngjun Li, Ling Xie, Jnet
Dr. Javier Polo Vice President Research & Development
 Efecto de l suplementción con concentrdo de inmunoglobulins prtir del suero bovino en l Microbiot y su contribución l slud intestinl y l sistem nervioso centrl Dr. Jvier Polo Vice President Reserch & Development
Efecto de l suplementción con concentrdo de inmunoglobulins prtir del suero bovino en l Microbiot y su contribución l slud intestinl y l sistem nervioso centrl Dr. Jvier Polo Vice President Reserch & Development
Polymer-Coated Metal-Oxide Nanoparticles Inhibit IgE Receptor Binding, Cellular Signaling, and Degranulation in a Mast Cell-like Cell Line
 www.mterilsviews.com Polymer-Coted Metl-Oxide Nnoprticles Inhiit IgE Receptor inding, Cellulr Signling, nd Degrnultion in Mst Cell-like Cell Line Vn. Orteg, Jmes D. Ede, Dvid oyle, Jmes L. Stfford, nd
www.mterilsviews.com Polymer-Coted Metl-Oxide Nnoprticles Inhiit IgE Receptor inding, Cellulr Signling, nd Degrnultion in Mst Cell-like Cell Line Vn. Orteg, Jmes D. Ede, Dvid oyle, Jmes L. Stfford, nd
D14 D 10 D 7. Untreated CYTOXAN 5-FU
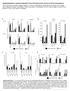 Dipeptidylpeptidse Negtively Regultes Colony Stimulting Fctor Activity nd Stress Hemtopoiesis. Hl E. Broxmeyer, Jonthn Hoggtt, Hether A. O Lery, Chrlie Mntel, Brhmnnd R. Chitteti, Scott Cooper, Steven
Dipeptidylpeptidse Negtively Regultes Colony Stimulting Fctor Activity nd Stress Hemtopoiesis. Hl E. Broxmeyer, Jonthn Hoggtt, Hether A. O Lery, Chrlie Mntel, Brhmnnd R. Chitteti, Scott Cooper, Steven
Consumer perceptions of meat quality and shelf-life in commercially raised broilers compared to organic free range broilers
 Consumer perceptions of met qulity nd shelf-life in commercilly rised roilers compred to orgnic free rnge roilers C.Z. ALVARADO 1 *, E. WENGER 2 nd S. F. O KEEFE 3 1 Texs Tech University, Box 42141 Luock,
Consumer perceptions of met qulity nd shelf-life in commercilly rised roilers compred to orgnic free rnge roilers C.Z. ALVARADO 1 *, E. WENGER 2 nd S. F. O KEEFE 3 1 Texs Tech University, Box 42141 Luock,
Using Paclobutrazol to Suppress Inflorescence Height of Potted Phalaenopsis Orchids
 Using Pcloutrzol to Suppress Inflorescence Height of Potted Phlenopsis Orchids A REPORT SUBMITTED TO FINE AMERICAS Linsey Newton nd Erik Runkle Deprtment of Horticulture Spring 28 Using Pcloutrzol to Suppress
Using Pcloutrzol to Suppress Inflorescence Height of Potted Phlenopsis Orchids A REPORT SUBMITTED TO FINE AMERICAS Linsey Newton nd Erik Runkle Deprtment of Horticulture Spring 28 Using Pcloutrzol to Suppress
Overcoming EGFR T790M-based Tyrosine Kinase Inhibitor Resistance with an Allele-specific DNAzyme
 Cittion: Moleculr Therpy Nucleic Acids (214) 3, e15; doi:1.138/mtn.214.3 214 The Americn Society of Gene & Cell Therpy All rights reserved 2162-2531/14 www.nture.com/mtn Overcoming T79M-sed Tyrosine Kinse
Cittion: Moleculr Therpy Nucleic Acids (214) 3, e15; doi:1.138/mtn.214.3 214 The Americn Society of Gene & Cell Therpy All rights reserved 2162-2531/14 www.nture.com/mtn Overcoming T79M-sed Tyrosine Kinse
SUPPLEMENTARY INFORMATION
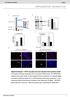 doi: 1.138/nture862 humn hr. 21q MRPL39 murine Chr.16 Mrpl39 Dyrk1A Runx1 murine Chr. 17 ZNF295 Ets2 Znf295 murine Chr. 1 COL18A1 -/- lot: nti-dscr1 IgG hevy hin DSCR1 DSCR1 expression reltive to hevy
doi: 1.138/nture862 humn hr. 21q MRPL39 murine Chr.16 Mrpl39 Dyrk1A Runx1 murine Chr. 17 ZNF295 Ets2 Znf295 murine Chr. 1 COL18A1 -/- lot: nti-dscr1 IgG hevy hin DSCR1 DSCR1 expression reltive to hevy
The effect of dietary α-linolenic acid levels on regulation of omega-3 lipid synthesis in rat
 The effect of dietry α-linolenic cid levels on regultion of omeg-3 lipid synthesis in rt Wei-Chun Tu School of Agriculture Food nd Wine The University of Adelide Conversion of PUFA to LCPUFA PUFA LCPUFA
The effect of dietry α-linolenic cid levels on regultion of omeg-3 lipid synthesis in rt Wei-Chun Tu School of Agriculture Food nd Wine The University of Adelide Conversion of PUFA to LCPUFA PUFA LCPUFA
Supplementary Figure 1 a
 % DAPI + Are Supplementry Figure 1 MOI 1 MOI.5 MOI.25 5 4 3 MOPC MOPC, LCMV reltive VL4 expression 2 1 d1 d2 d3 MOI.125 MOI.625 untreted Supplementry Figure 1: LCMV replictes in MOPC without ffecting cell
% DAPI + Are Supplementry Figure 1 MOI 1 MOI.5 MOI.25 5 4 3 MOPC MOPC, LCMV reltive VL4 expression 2 1 d1 d2 d3 MOI.125 MOI.625 untreted Supplementry Figure 1: LCMV replictes in MOPC without ffecting cell
SUPPLEMENTARY INFORMATION
 SUPPEMENTARY INFORMATION DOI: 1.138/ncb956 Norml CIS Invsive crcinom 4 months months b Bldder #1 Bldder # Bldder #3 6 months (Invsive crcinom) Supplementry Figure 1 Mouse model of bldder cncer. () Schemtic
SUPPEMENTARY INFORMATION DOI: 1.138/ncb956 Norml CIS Invsive crcinom 4 months months b Bldder #1 Bldder # Bldder #3 6 months (Invsive crcinom) Supplementry Figure 1 Mouse model of bldder cncer. () Schemtic
The effect of encapsulated butyric acid and zinc on performance, gut integrity and meat quality in male broiler chickens 1
 The effect of encpsulted utyric cid nd zinc on performnce, gut integrity nd met qulity in mle roiler chickens 1 Astrct This study evluted the impct of encpsulted utyric cid nd zinc (ButiPEARL Z) on performnce
The effect of encpsulted utyric cid nd zinc on performnce, gut integrity nd met qulity in mle roiler chickens 1 Astrct This study evluted the impct of encpsulted utyric cid nd zinc (ButiPEARL Z) on performnce
Identification of a tripartite interaction between the N terminus of HIV 1 Vif and CBFβ that is critical for Vif function
 DOI 1.1186/s12977-17-346-5 Retrovirology RESEARCH Open Access Identifiction of triprtite interction etween the N terminus of HIV 1 nd CBFβ tht is criticl for function Belete A. Desimmie 1, Jessic L. Smith
DOI 1.1186/s12977-17-346-5 Retrovirology RESEARCH Open Access Identifiction of triprtite interction etween the N terminus of HIV 1 nd CBFβ tht is criticl for function Belete A. Desimmie 1, Jessic L. Smith
Supplementary Figure S1
 Supplementry Figure S Connexin4 TroponinI Merge Plsm memrne Met Intrcellulr Met Supplementry Figure S H9c rt crdiomyolsts cell line. () Immunofluorescence of crdic mrkers: Connexin4 (green) nd TroponinI
Supplementry Figure S Connexin4 TroponinI Merge Plsm memrne Met Intrcellulr Met Supplementry Figure S H9c rt crdiomyolsts cell line. () Immunofluorescence of crdic mrkers: Connexin4 (green) nd TroponinI
Identification and selective expansion of functionally superior T cells expressing chimeric antigen receptors
 Identifiction nd selective expnsion of functionlly superior T cells expressing chimeric ntigen receptors The Hrvrd community hs mde this rticle openly vilble. Plese shre how this ccess benefits you. Your
Identifiction nd selective expnsion of functionlly superior T cells expressing chimeric ntigen receptors The Hrvrd community hs mde this rticle openly vilble. Plese shre how this ccess benefits you. Your
Supporting Information. In Situ Supramolecular Assembly and Modular Modification of Hyaluronic Acid Hydrogels for 3D Cellular Engineering
 Supporting Informtion In Situ Suprmoleculr Assemly nd Modulr Modifiction of Hyluronic Acid Hydrogels for 3D Cellulr Engineering Kyeng Min Prk,, Jeong-A Yng,, Hyunte Jung, c Junseok Yeom, Ji Sun Prk, d
Supporting Informtion In Situ Suprmoleculr Assemly nd Modulr Modifiction of Hyluronic Acid Hydrogels for 3D Cellulr Engineering Kyeng Min Prk,, Jeong-A Yng,, Hyunte Jung, c Junseok Yeom, Ji Sun Prk, d
Effect of fungicide timing and wheat varietal resistance on Mycosphaerella graminicola and its sterol 14 α-demethylation-inhibitorresistant
 Effect of fungicide timing nd whet vrietl resistnce on Mycospherell grminicol nd its sterol 14 α-demethyltion-inhiitorresistnt genotypes Didierlurent L., Roisin-Fichter C., Snssené J., Selim S. Pltform
Effect of fungicide timing nd whet vrietl resistnce on Mycospherell grminicol nd its sterol 14 α-demethyltion-inhiitorresistnt genotypes Didierlurent L., Roisin-Fichter C., Snssené J., Selim S. Pltform
SUPPLEMENTARY INFORMATION
 DOI:.3/nc37 This Supplementry Informtion file ws updted with new reference (ref. 3) on Decemer WWW.NATURE.COM/NATURECELLBIOLOGY Mcmilln Pulishers Limited. All rights reserved logfc SUPPLEMENTARY INFORMATION
DOI:.3/nc37 This Supplementry Informtion file ws updted with new reference (ref. 3) on Decemer WWW.NATURE.COM/NATURECELLBIOLOGY Mcmilln Pulishers Limited. All rights reserved logfc SUPPLEMENTARY INFORMATION
Thebiotutor.com A2 Biology OCR Unit F215: Control, genomes and environment Module 1.2 Meiosis and variation Answers
 Theiotutor.com A2 Biology OCR Unit F215: Control, genomes nd environment Module 1.2 Meiosis nd vrition Answers Andy Todd 1 1. () (i) gene length of DNA; codes for (specific), polypeptide / protein / RNA;
Theiotutor.com A2 Biology OCR Unit F215: Control, genomes nd environment Module 1.2 Meiosis nd vrition Answers Andy Todd 1 1. () (i) gene length of DNA; codes for (specific), polypeptide / protein / RNA;
Wnt signaling enhances the activation and survival of human hepatic stellate cells
 FEBS Letters 581 (2007) 2954 2958 Wnt signling enhnces the ctivtion nd survivl of humn heptic stellte cells Sun Jung Myung, Jung-Hwn Yoon, *, Geum-Youn Gwk, Won Kim, Jeong-Hoon Lee, Kng Mo Kim, Chn Soo
FEBS Letters 581 (2007) 2954 2958 Wnt signling enhnces the ctivtion nd survivl of humn heptic stellte cells Sun Jung Myung, Jung-Hwn Yoon, *, Geum-Youn Gwk, Won Kim, Jeong-Hoon Lee, Kng Mo Kim, Chn Soo
Adenovirus Expressing Both Thymidine Kinase and Soluble PD1 Enhances Antitumor Immunity by Strengthening CD8 T-cell Response
 originl rticle The Americn Society of Gene & Cell Therpy Adenovirus Expressing Both Thymidine Kinse nd Solule PD1 Enhnces Antitumor Immunity y Strengthening CD8 T-cell Response Seung-Pil Shin 1,2, Hye-Hyun
originl rticle The Americn Society of Gene & Cell Therpy Adenovirus Expressing Both Thymidine Kinse nd Solule PD1 Enhnces Antitumor Immunity y Strengthening CD8 T-cell Response Seung-Pil Shin 1,2, Hye-Hyun
Supplementary Information
 Supplementry Informtion Cutneous immuno-surveillnce nd regultion of inflmmtion y group 2 innte lymphoid cells Ben Roediger, Ryn Kyle, Kwok Ho Yip, Nitl Sumri, Thoms V. Guy, Brin S. Kim, Andrew J. Mitchell,
Supplementry Informtion Cutneous immuno-surveillnce nd regultion of inflmmtion y group 2 innte lymphoid cells Ben Roediger, Ryn Kyle, Kwok Ho Yip, Nitl Sumri, Thoms V. Guy, Brin S. Kim, Andrew J. Mitchell,
Supplementary Information. SAMHD1 Restricts HIV-1 Infection in Resting CD4 + T Cells
 Supplementry Informtion SAMHD Restricts HIV- Infection in Resting CD T Cells Hnn-Mri Blduf,2,, Xioyu Pn,, Elin Erikson,2, Srh Schmidt, Wqo Dddch 3, Mnj Burggrf, Kristin Schenkov, In Amiel,2, Guido Wnitz
Supplementry Informtion SAMHD Restricts HIV- Infection in Resting CD T Cells Hnn-Mri Blduf,2,, Xioyu Pn,, Elin Erikson,2, Srh Schmidt, Wqo Dddch 3, Mnj Burggrf, Kristin Schenkov, In Amiel,2, Guido Wnitz
Effect of Aqueous Extract of Carica papaya Dry Root Powder on Lactation of Albino Rats
 Effect of Aqueous Extrct of Cric ppy Dry Root Powder on Lcttion of Alino Rts G. Tosswnchuntr nd S. Aritjt Deprtment of Biology Fculty of Science Ching Mi University Ching Mi 50200 Thilnd Keywords: mmmry
Effect of Aqueous Extrct of Cric ppy Dry Root Powder on Lcttion of Alino Rts G. Tosswnchuntr nd S. Aritjt Deprtment of Biology Fculty of Science Ching Mi University Ching Mi 50200 Thilnd Keywords: mmmry
NappHS. rrna. transcript abundance. NappHS relative con W+W 0.8. nicotine [µg mg -1 FM]
![NappHS. rrna. transcript abundance. NappHS relative con W+W 0.8. nicotine [µg mg -1 FM] NappHS. rrna. transcript abundance. NappHS relative con W+W 0.8. nicotine [µg mg -1 FM]](/thumbs/92/110177253.jpg) (A) W+OS 3 min 6 min con L S L S RNA loding control NppHS rrna (B) (C) 8 1 k NppHS reltive trnscript undnce 6 4.5 *** *** *** *** 3 k. + + + line 1 line (D) nicotine [µg mg -1 FM] 1..8.4. con W+W Supplementl
(A) W+OS 3 min 6 min con L S L S RNA loding control NppHS rrna (B) (C) 8 1 k NppHS reltive trnscript undnce 6 4.5 *** *** *** *** 3 k. + + + line 1 line (D) nicotine [µg mg -1 FM] 1..8.4. con W+W Supplementl
Effects of Recombinant Bovine Somatotropin Administration at Breeding on the Cow, Conceptus and Subsequent Offspring Performance of Beef Cattle
 Effects of Recominnt Bovine Somtotropin Administrtion t Breeding on the Cow, Conceptus nd Susequent Offspring Performnce of Beef Cttle V. R. G. Mercdnte 1, F. M. Cirico 1, D. D. Henry 1, P. L. P. Fontes
Effects of Recominnt Bovine Somtotropin Administrtion t Breeding on the Cow, Conceptus nd Susequent Offspring Performnce of Beef Cttle V. R. G. Mercdnte 1, F. M. Cirico 1, D. D. Henry 1, P. L. P. Fontes
SUPPLEMENTARY INFORMATION
 DOI: 1.138/n358 TLR2 nd MyD88 expression in murine mmmry epithelil supopultions. CD24 min plus MRU Myo-epithelil Luminl progenitor (CD61 pos ) Mture luminl (CD61 neg ) CD49f CD61 Reltive expression Krt5
DOI: 1.138/n358 TLR2 nd MyD88 expression in murine mmmry epithelil supopultions. CD24 min plus MRU Myo-epithelil Luminl progenitor (CD61 pos ) Mture luminl (CD61 neg ) CD49f CD61 Reltive expression Krt5
DR. MARC PAGÈS Project Manager R&D Biologicals - Coccidia Projects, HIPRA
 DR. MARC PAGÈS Project Mnger R&D Biologicls - Coccidi Projects, HIPRA Dr. Mrc Pgès Bosch otined Microiology nd Genetics degree t the University of Brcelon in 1998. He otined his PhD working on the synptoneml
DR. MARC PAGÈS Project Mnger R&D Biologicls - Coccidi Projects, HIPRA Dr. Mrc Pgès Bosch otined Microiology nd Genetics degree t the University of Brcelon in 1998. He otined his PhD working on the synptoneml
Invasive Pneumococcal Disease Quarterly Report July September 2018
 Invsive Pneumococcl Disese Qurterly Report July Septemer Introduction Since 17 Octoer 2008, invsive pneumococcl disese (IPD) hs een notifile to the locl Medicl Officer of Helth under the Helth Act 1956.
Invsive Pneumococcl Disese Qurterly Report July Septemer Introduction Since 17 Octoer 2008, invsive pneumococcl disese (IPD) hs een notifile to the locl Medicl Officer of Helth under the Helth Act 1956.
