Genome-wide nucleosome positioning during embryonic stem cell development
|
|
|
- Jocelin Leonard
- 5 years ago
- Views:
Transcription
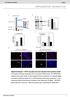
1 Genome-wide nuleosome positioning during emryoni stem ell development Vldimir B Teif 1,2, Yevhen Vinshtein 2,3, Mïwen Cudron-Herger 1,2, Jn-Philipp Mllm 1,2, Croline Mrth 1,2, Thoms Höfer 2,3 & Krsten Rippe 1,2 We determined genome-wide nuleosome oupnies in mouse emryoni stem ells nd their neurl progenitor nd emryoni firolst ounterprts to ssess fetures ssoited with nuleosome positioning during linege ommitment. Cell-type- nd protein-speifi inding preferenes of trnsription ftors to sites with either low (My, Klf4 nd Zfx) or high (Nnog, Ot4 nd Sox2) nuleosome oupny s well s omplex ptterns for CTCF were identified. Nuleosome-depleted regions round trnsription strt nd trnsription termintion sites were rod nd more pronouned for tive genes, with distint ptterns for promoters lssified ording to CpG ontent or histone methyltion mrks. Throughout the genome, nuleosome oupny ws orrelted with ertin histone methyltion or etyltion modifitions. In ddition, the verge nuleosome repet length inresed during differentition y 5 7 se pirs, with lol vritions for speifi regions. Our results revel regultory mehnisms of ell differentition tht involve nuleosome repositioning. Emryoni stem ells nd differentited ells derived from them shre the sme DNA sequene ut hve distint ellulr funtions. Mny of the underlying ell-fte deisions our through hnges to hromtin fetures tht ffet gene expression. The speifi lotion of nuleosomes on the DNA is importnt for ontrolling ess to the DNA 1,2. Binding of protein ftors to the se pirs of DNA wrpped round the histone otmer ore is frequently impeded, wheres the linker DNA etween nuleosomes is more esily essile. Reent dvnements in high-throughput sequening methods llowed for the genome-wide mpping of individul nuleosomes t single se pir resolution 3,4, with yest serving s model system for the initil pioneering studies 5 7. More reently, tissue- nd disese-speifi fetures of nuleosome positions in higher orgnisms were reported These inlude studies of humn ell lines 8 10 nd mouse heptoyte ells 15. We set out to identify fetures of nuleosome positioning t funtionl genomi elements during linege ommitment in mouse emryoni stem ells (s) nd neurl progenitor ells (s) derived from these s, s well s mouse emryoni firolsts (s) from the orresponding mouse strin. By ompring these three ell types, we identified lol nd glol rerrngements of nuleosome oupny tht reveled importnt roles of nuleosome positioning in ell differentition. RESULTS Nuleosome oupny mps of s, s nd s We mpped nuleosome positions y genome-wide pired-end sequening of nuleosoml DNA from mouse s, s nd s fter digesting the linker DNA etween nuleosomes with mirool nulese (MNse) (Online Methods, Supplementry Fig. 1, Supplementry Note). Exmples of the resulting nuleosome overge mps re depited in Figure 1. We lulted these ptterns round trnsription ftor inding sites tht were determined previously y hromtin immunopreipittion nd DNA sequening (ChIP-seq) in mouse s 16 or, in the se of the CCCTC-inding ftor (CTCF), in oth s nd s 17,18. Nuleosome positioning nd trnsription-ftor inding followed omplex reltion (Fig. 1). Some trnsription-ftor sites identified y ChIP-seq in s were nuleosome depleted, with nuleosome oupny redued to 40 80% of tht of the flnking regions in ll ell types. In ontrst, other trnsription ftors were preferentilly ound to nuleosomeenrihed regions or displyed distint ptterns tht hnged during ell differentition, s desried in further detil elow. DNA sequene dependent inding ffinities of the histone otmer lso ontriuted to nuleosome positioning. This is inferred from n exemplry omprison of the experimentl nuleosome oupnies to those predited from the DNA sequenes 19, in whih the omputed peks of nuleosome oupny orrelted well with our experimentl nuleosome positions in s ut displyed lrge differenes reltive to the dt set (Fig. 1). Thus, for the ltter ell type the intrinsi inding preferenes were overwritten y other ftors, for exmple y the presene of CTCF tht inds in s to the region shown nd is flnked y two well-positioned nuleosomes. We found mny promoter regions to e nuleosome depleted t the trnsription strt site (TSS), s shown for the Smr4 promoter in Figure 1. At this lous, one nuleosome ws onstitutively sent downstrem of the TSS in ll three ell lines. An dditionl nuleosome ws lso removed upstrem of the TSS in s, where this gene ws upregulted y out two-fold in omprison to s. 1 Reserh Group Genome Orgniztion nd Funtion, Deutshes Kresforshungszentrum (DKFZ), Heidelerg, Germny. 2 BioQunt, Heidelerg, Germny. 3 Division of Theoretil Systems Biology, Deutshes Kresforshungszentrum (DKFZ), Heidelerg, Germny. Correspondene should e ddressed to V.B.T. (v.teif@dkfz.de) or K.R. (krsten.rippe@dkfz.de). Reeived 10 April; epted 19 Septemer; pulished online 21 Otoer 2012; doi: /nsm.2419 nture struturl & moleulr iology VOLUME 19 NUMBER 11 NOVEMBER
2 Nuleosome oupny Tfp2l1 Klf4 Nnog Sox2 Stt3 Esrr, Klf4, n-my, Ot4, Smd1, Sox2, Tfp2l1, Zfx Position on hromosome 17 E2f1 Zfx Esrr Smd1 Sox2 Tfp2l1 35,640,000 +1,000 +2,000 Nuleosome oupny 0.5 CTCF in 4,407,000 +1,000 Position on hromosome 1 Predited 0.5 TSS Smr4 21,420,000 +1,000 Position on hromosome 9 Figure 1 Nuleosome oupnies of exemplry genomi regions in s, s nd s. () Nuleosome oupnies in s (lk), s (red) nd s (lue) round puttive enhner region residing in gene-reh lous, with the nerest gene eing Pou5f1, whih enodes the Ot4 trnsription ftor. The region shown ontins 11 different developmentl trnsription ftors ound in s t the indited sites, s determined in ref. 16. () Nuleosome oupnies in s (lk) nd s (lue) for n exemplry genomi region ontining CTCF ound in s, s identified in ref. 18. The experimentlly determined nuleosome oupny is ompred to tht predited from the DNA sequene (yellow) ording to previously desried lgorithm 19. () Promoter of the Smr4 gene tht enodes the hromtin remodeler Brg1, whih is involved in development. Color odes for, nd re the sme s in. As desried in the following, speifi fetures of individul nuleosome oupny profiles t funtionl genomi elements like trnsription ftor inding sites or TSSs n e evluted in genome-wide nlysis of the experimentl nuleosome oupny profiles. Nuleosome oupny t trnsription ftor inding sites We lulted verge nuleosome oupny profiles for inding sites tht were experimentlly determined y ChIP-seq in s nd tht omprised 12 developmentlly importnt trnsription ftors 16, p300 histone etyltrnsferse 18, hromtin remodelers Nuleosome oupny -My n-my Zfx Distne from site (p) Stt3 Distne from site (p) p300 Chd7 (ref. 20) nd Brg1 (ref. 21), s well s DNse I hypersensitivity sites 20. Four types of ptterns were oserved (Fig. 2). (i) Some trnsription ftors, suh s -My, n-my, Zfx nd Klf4, were preferentilly ound in nuleosome-depleted regions in s (Fig. 2). These regions retined lrgely redued nuleosome oupny in s nd s. A similr pttern ws oserved for the DNse I hypersensitivity sites, supporting the previous onlusion tht these reflet nuleosome-depleted regions 20. (ii) For nother lss of proteins, suh s Stt3 nd p300, the inding sites resided in nuleosome-depleted regions in s ut eme preferentilly Distne from site (p) E2f1 Klf4 Distne from site (p) Tfp2l1 DNse I Distne from site (p) Essr Nuleosome oupny Distne from site (p) Distne from site (p) Figure 2 Nuleosome oupnies t inding sites of developmentlly importnt trnsription ftors nd enzymes. Nuleosome oupnies in s (lk), s (red) nd s (lue) round protein inding sites re shown. () Trnsription ftors with nuleosome-depleted inding sites in ll three ell types (-My, n-my, Zfx nd Klf4) nd DNse I hypersensitivity sites. () The inding sites of Stt3 nd p300 re nuleosome depleted in s nd s ut eome oupied y nuleosomes in s. () Binding sites of E2f1, Tfp2l1 nd Essr reside in prtilly nuleosomedepleted regions with lol nuleosome enrihment in s nd s tht eomes preferentilly oupied y nuleosome in s. (d) Pluripoteny mster regultors Sox2, Ot4, Nnog nd Smd1 s well s hromtin remodelers Chd7 nd Brg1 preferentilly ind to the DNA regions overed y the nuleosome. d Nuleosome oupny Distne from site (p) Ot4 Distne from site (p) Distne from site (p) Distne from site (p) Distne from site (p) Nnog Sox2 Smd1 Chd7 Brg1 Distne from site (p) 1186 VOLUME 19 NUMBER 11 NOVEMBER 2012 nture struturl & moleulr iology
3 Nuleosome oupny ll CTCF sites only CTCF sites ll CTCF sites only CTCF sites s nd s ommon sites d only 21,000 ommon sites Common only 1, ,000 Distne from ound CTCF (p) 1, ,000 Distne from ound CTCF (p) 1, ,000 Distne from ound CTCF (p) sites (34,000) sites (41,000) Figure 3 Genome-wide verge nuleosome ptterns t CTCF inding sites. ( ) The experimentlly determined CTCF inding sites from ENCODE 18 were split into five groups: (i) CTCF inding sites in s ( ll ), (ii) CTCF inding sites in s ( ll ), (iii) ommon CTCF inding sites found simultneously in oth ell lines ( Common ), (iv) CTCF sites deteted in s ut not found in s ( only ) nd (v) CTCF sites found in s ut not found in s ( only ). Shown re nuleosome oupny in s t ll (lk) nd t only sites (red) (); in s t sites where CTCF ws ound in s (lk) or only in s (red) (); nd in s (lk) nd s (red) t the ommon sites where CTCF ws ound simultneously in oth s nd s (). (d) Venn digrm showing the distriution of CTCF sites etween s nd s. oupied y nuleosomes in differentited ells, possily euse these ftors were no longer ound (Fig. 2). (iii) Some proteins, suh s E2f1, Tfp2l1 nd Essr, showed more omplex pttern with smll nuleosome oupny pek t the inding sites, surrounded y wider regions of redued nuleosome oupny (Fig. 2). This ould reflet regultory role of nuleosome positioning for those trnsription ftors tht remin expressed t different developmentl stges 22. In ddition, inding for this trnsriptionftor group might require tive trnslotion or evition of nuleosome y hromtin remodelers 23,24. (iv) Another trnsription-ftor lss inluding the mster regultors Nnog, Sox2 nd Ot4 hd inding sites in s tht oinided with well-positioned nuleosomes (Fig. 2d). We onlude tht these ftors n effiiently ind while the DNA trget site interts with histone otmer, s hs een postulted for so-lled pioneering ftors tht initite ellulr progrms. Cell type dependent CTCF-direted nuleosome positioning To further explore nuleosome rerrngement round trnsription ftor inding sites, we nlyzed CTCF inding sites tht hve een previously mpped in oth s nd s 17,18 (Fig. 3, Supplementry Fig. 2). CTCF estlishes insultory or oundry elements to demrte repressive nd tive hromtin regions 25 y setting lol oundry orgnizing nuleosomes 26 s well s y ridging distnt hromtin regions 27. CTCF inding sites were reently identified y ChIP-seq y nother study 17 nd y the ENCODE projet 18. Aording to the ENCODE dt set used in our nlysis, the totl numer of CTCF inding sites is 34,000 in s nd 41,000 in s, with only ~30% of sites oiniding in the two ell types (Fig. 3d). Both s nd s displyed nuleosome-depleted region of ~200 se pirs (p) t the enter of the CTCF inding sites, whih ws somewht more pronouned in s (Fig. 3). Two nuleosomes were positioned diretly djent to this site nd flnked y up to nine regulrly sped nuleosomes, similr to the pttern reported Nuleosome oupny Nuleosome oupny Gene expression: 5% lowest 5% highest Other genes Distne from TSS (p) Gene expression: 5% lowest 5% highest Other genes Distne from TTS (p) previously for humn T ells 26. Notly, the suset of CTCF sites tht were oupied only in s ut not in s ws found to lredy e prtly nuleosome depleted in s (Fig. 3). In s, nuleosomes were sustntilly rerrnged round the CTCF sites (Fig. 3). The previous positions of CTCF tht were unique to s ut not ssoited with CTCF in s displyed n ~20% inrese in nuleosome oupny. The sites of ound CTCF in s fell into two lsses with respet to their nuleosome oupny (Fig. 3 nd Supplementry Fig. 2). (i) The onstitutive CTCF sites present oth in s nd s were lso nuleosome depleted in s ut to lesser extent thn in s. This might e relted to n ~27% redution of the rtio of CTCF to ore histone H4 expression (Supplementry Tle 1). Notly, the CTCF sites tht were unique to s displyed n dditionl lol pek of nuleosome oupny (Supplementry Fig. 2). This suggests tht CTCF is le to ind to nuleosoml DNA t these sites, possily through dditionl intertion prtners. In support of this view, n exmple with CTCF ound in the middle of the nuleosome pek is shown in Supplementry Figure 2. (ii) In ddition, suset of CTCF sites loted within enhner elements displyed very different pttern (Supplementry Fig. 2). Distne from TSS (p) Distne from TTS (p) Distne from TSS (p) Distne from TTS (p) Figure 4 Averge nuleosome oupny ptterns round TSSs nd TTSs in s, s nd s. () Nuleosome oupnies t the TSS for the 5% highest- (lk) nd 5% lowest-expressed genes (lue) in omprison to the remining 90% (red). () Sme s in, ut for TTSs. nture struturl & moleulr iology VOLUME 19 NUMBER 11 NOVEMBER
4 Nuleosome oupny 1, g Trnsript numer 1, Trnsript numer 1 1,723 1Trnsript numer ivlent promoters Distne from TSS (p) ivlent promoters Distne from TSS (p) ivlent promoters Distne from TSS (p) I II III IV V 0.2 1Trnsript numer 5,560 1Trnsript numer h 1,723 Distne from TSS (p) Nuleosome oupnies t TSSs nd trnsription end sites To hrterize the nuleosome oupny t promoters, we ligned the mps of mouse trnsripts t their TSSs nd lustered them ording to their expression levels for eh ell type into tive (top 5% of expression level), intive (ottom 5% of expression level) nd the remining 90% of the trnsripts. We oserved rod nuleosome-depleted region downstrem of the TSS for s, s nd s (Fig. 4). It extended into the gene ody similrly to the pttern reported for mouse heptoytes 15 ut very differently from the typil ptterns oserved in yest nd invertertes 5 7,28,29. The nuleosome oupny profiles in s, s nd s were dependent on the gene expression level. For the most tive genes, the nuleosome-depleted region eme wider nd deeper. Supplementry Figure 3 shows the oupny mps of the 5% of genes with the highest nd lowest expression levels. In generl, the TSS nuleosome oupny displyed signifint ntiorreltion with expression of the orresponding trnsripts (Supplementry Tle 2), inditing tht redued nuleosome oupny t the promoter fvors gene expression. Next, we grouped verge nuleosome profiles t the trnsription termintion sites (TTSs) of the 5% highest, the 5% lowest nd the remining 90% of genes in s, s nd s, ording to gene expression level (Fig. 4). Intive genes were hrterized y reltively smll nuleosome-depleted region round the TTS, 1Trnsript numer HCG promoters Distne from TSS (p) Distne from TSS (p) ivlent promoters I II III IV V e i Trnsript numer 1 1,398 Trnsript numer 1 LCG promoters d HCG promoters f LCG promoters 5,560 1, Nuleosome oupny 0.2 Distne from TSS (p) Distne from TSS (p) Averge oupny luster I Distne from TSS (p) Figure 5 Nuleosome oupny ptterns round TSSs in different lsses of promoters. Bivlent promoters tht rry oth the H3K4me3 nd H3K27me3 histone-modifition mrks in s, HCG promoters nd LCG promoters were nlyzed in terms of their nuleosome oupny. Blk, s; red, s; lue, s. () Averge nuleosome profiles round ivlent promoters. () Cluster plots of nuleosome oupnies t the TSS for ivlent promoters in s (top) nd s (ottom). Eh horizontl line orresponds to the promoter ssoited with given trnsript. The olors indite nuleosome oupny from low (drk lue) to high (red). For s, the sorting ws ording to the similrity of the nuleosome oupny profile, strting with the lowest oupny on top. This order ws mintined in s to visulize ny nuleosome oupny hnges etween s nd s. () Averge nuleosome profiles round HCG promoters. (d) Cluster plots s in, onduted for HCG promoters. (e) Averge nuleosome profiles round LCG promoters. (f) Cluster plots s in, onduted for LCG promoters. (g) Hierrhil luster plot of ivlent promoters in s split into five groups. Cluster I hs nuleosome overing the region from out 500 to 350 p reltive to the TSS. (h) Hierrhil luster plot for s with the sme promoters nd ordering s in g. It is pprent tht the 500 to 350 p region in luster I looses nuleosome. (i) Averge nuleosome oupny of the genes in luster I in s, s nd s. possily due to nuleosome-exluding DNA sequene 30. This nuleosome-depleted region ws lrgely inresed upstrem of the TTS for the highly tive genes in ll ell types (Fig. 4 nd Supplementry Fig. 4) nd ws different from the nuleosome-depleted region downstrem of the TTS reported previously for yest Thus, oth TSSs nd TTSs hve unique nuleosome signtures tht might reflet more omplex regultory mehnisms thn those found in simpler eukryotes. Promoter nuleosome oupny nd gene expression hnges To investigte whether hnges in nuleosome oupny etween s nd s were orrelted with gene expression hnges, we evluted nd identified three different promoter lsses (Fig. 5, Supplementry Fig. 5 nd Supplementry Tles 3 nd 4). The first lss ws defined y the simultneous presene of the trimethyltion modifition of histone H3 t lysine residue 4 (H3K4me3) nd t lysine 27 (H3K27me3) in s nd is referred to s ivlent promoters 34,35 (Fig. 5, nd Supplementry Fig. 5). Two other lsses were distinguished ording to their DNA sequene omposition s high CpG (HCG) (Fig. 5,d) or low CpG (LCG) promoters (Fig. 5e,f) 36. For ivlent nd HCG promoters, the nuleosome oupny profiles were hrterized y strong nuleosome-depleted region (Fig. 5 d) similr to the verge TSS ptterns (Fig. 4) nd with no sustntil differenes etween the three ell types. In ontrst, the nuleosome oupny round the TSS ws high for LCG promoters (Fig. 5e,f) nd for promoters tht rried only the H3K27me3 modifition (Supplementry Fig. 5). We then sorted the TSSs within eh lss ording to their verge nuleosome oupny 1188 VOLUME 19 NUMBER 11 NOVEMBER 2012 nture struturl & moleulr iology
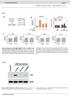
5 Figure 6 Averge nuleosome oupny ptterns round lusters of H3K9me3, H3K9 nd H3K27 histone modifitions. Nuleosome oupnies in s (lk), s (red) nd s (lue) ligned with respet to the luster enters of the indited histone modifitions. ( ) Nuleosome oupny t the lusters of H3K9me3 (), H3K9 () nd H3K27 () found in s. (d) Comprison of nuleosome oupny profiles round the H3K27 modifition in s nd s. H3K27 lusters were split into those tht were present only in s ( only ) or only in s ( only ) nd ommon sites tht were present simultneously in s nd s. in s in the region t 500 to 500 p round the TSS nd ompred these to s while keeping the sme ordering. The overll pttern remined very similr etween s nd s for ivlent (Fig. 5) nd HCG promoters (Fig. 5d) ut hnged for lrge numer of LCG promoters (Fig. 5f). To test whether nuleosome oupny hnges in the region t 500 to 500 p were linked to gene expression hnges etween s nd s, we onduted orreltion nlysis of the orresponding log 2 rtios (Supplementry Tles 3 nd 4). In this nlysis we found no simple orreltion etween the two prmeters exept for LCG promoters. In ddition, for the two smll groups of ivlent promoters tht hd trnsripts deteted y RNA-seq in s ut not in s (36 genes) nd H3K27me3 promoters tht were found to e expressed in s ut not in s (20 genes), the dt were inditive of n inrese of nuleosome oupny t the TSS during silening. We further disseted the reltion etween nuleosome oupny nd gene expression hnges for sugroups of ivlent (H3K4me3 nd H3K27me3) promoters in s. In s these resolve into promoters tht either rry only the H3K4me3 or only the H3K27me3 mrk 34,35. For the verged profiles, we did not oserve sustntil hnges of nuleosome oupny (Supplementry Fig. 5). Clustering of ivlent promoters ording to their nuleosome oupny pttern reveled one group, designted s luster I, whih showed wellpositioned nuleosome oupny pek in the region from 500 to 350 p upstrem of the TSS (Fig. 5g). This nuleosome ws preferentilly removed in oth s nd s (Fig. 5h,i) with onomitnt verge gene expression inrese of 2.2-fold in s nd 4.6-fold in s, similr to the verge gene expression hnge oserved for ll ivlent promoters. Thus, tivtion of luster I genes ould Normlized ounts Nuleosome oupny ,000 1,500 Nuleosome-nuleosome distne (p) 500 1,000 1,500 Distne from ound CTCF (p) Pek position (p) d Distne from CTCF (p) 2,000 1,500 1, ,400 1,200 1, , NRL = (± ) p, NRL = (± ) p, NRL = (± 0.5) p Pek numer CTCF in s NRL = (± 1.5) p CTCF in s NRL = (± 0.7) p Pek numer Nuleosome oupny Nuleosome oupny 1.4 H3K9me3 Distne from H3K9me3 pek (p) 1.4 H3K27 Distne from H3K27 pek (p) 1.4 H3K9 Distne from H3K9 pek (p) d 1.4 nuleosomes t only H3K27 sites nuleosomes t only H3K27 sites nuleosomes t ommon H3K27 sites nuleosomes t ommon H3K27 sites Distne from H3K27 pek (p) involve the omplete removl of nuleosome insted of hnging its ssoited histone modifitions. Within this luster, gene ontology tegories were enrihed tht re ssoited with differentited ell funtion (lood-vessel development, 9 genes, P < ; positive regultion of trnsription nd gene expression, 11 genes, P < ; ell migrtion nd ell motility, 7 genes, P < ; xonogenesis, neuron projetion morphogenesis nd ell morphogenesis, 6 genes, P < ; Supplementry Tle 5). Histone modifition dependent nuleosome oupnies Next, we investigted whether nuleosome oupnies hnged etween trnsriptionlly tive or intive hromtin regions. Histone modifitions were determined y ChIP-seq, nd welldefined peks (P < 10 5 ) were seleted. We identified out 10,000 lusters for eh histone mrk studied. These inluded the on fide repressive trimethyltion modifition of histone H3 t lysine residue 9 (H3K9me3), s well s the permissive etyltion of histone H3 t either lysine residue 9 (H3K9) or t lysine 27 (H3K27) in s nd s. Averge nuleosome oupny ptterns in s, s nd s were lulted round the enters of these lusters (Fig. 6). We found tht H3K9me3 lusters displyed inresed nuleosome oupny (Fig. 6), wheres H3K9 nd H3K27 lusters showed the opposite trend (Fig. 6,). H3K27 sites ommon to s nd s were sustntilly depleted of nuleosomes in oth s nd s (Fig. 6d). However, genomi positions tht were etylted in s ut not in s hd pronouned pek of nuleosome oupny in s. The sme ws true for the -speifi H3K27 Figure 7 Nuleosome repet length in s, s nd s. () Histogrm of distnes etween ll nuleosome strt positions. Peks reflet the distnes etween nerest-neighor nuleosomes, next-nerest neighors, nd so on. The lrgest frequeny pek found for smll distne vlues round zero is not relevnt for the NRL nlysis euse it reflets the experimentl unertinty for determining the oundry of the sme nuleosome. () Plot of the pek position long the x xis versus the pek numer. The slope of the line fitted through the plot yields the NRL for given ell type. () Determintion of the lol NRL for the region round CTCF inding sites in s nd s. The entrlly positioned nuleosomes flnking the CTCF site were exluded from the nlysis, nd the djent 8 nuleosomes on the right side were evluted. (d) Liner fit for NRL determintion in regions of ound CTCF in s nd s for the peks identified in. nture struturl & moleulr iology VOLUME 19 NUMBER 11 NOVEMBER
6 sites: these positions hd inresed nuleosome oupny in s, where they were not etylted. Thus, histone modifitions ould mrk nuleosomes for hnges in their density long the DNA in the orresponding regions. Inrese of nuleosome repet length during differentition An essentil prmeter tht desries the primry hromtin orgniztion is the nuleosome repet length (NRL), whih is the verge distne etween two neighoring nuleosomes. We determined NRLs ording to previously desried method 9. It is sed on lulting the frequeny of nuleosome distnes etween the strts of ll mononuleosoml DNA frgments nd then nlyzing the preferred distnes etween the nerest-neighor nuleosomes, next-nerest neighors, et. The resulting plots yielded well-defined peks for the preferred internuleosome distnes (Fig. 7). From the plot of pek position nd orresponding nuleosome numer, we otined vlues of ± p (s), ± p (s) nd ± 0.5 p (s): tht is, the verge NRL inresed y 5 7 p during differentition (Fig. 7). On the sis of previous findings, the hnges in NRL ould involve hnge in the molr rtio of linker to ore histones 37. We found tht for only the linker histone vrints H nd H1.7, gene expression in reltion to tht of the ore histones ws rised sustntilly in oth s nd s (Supplementry Tle 1). Aordingly, these vrints might e prtiulrly importnt for induing n NRL inrese during differentition. Nuleosome position distnes t speifi genomi loi displyed lrge lol vritions from these verge NRLs, for exmple t the TSS nd TTS, due to the inding of other protein ftors. Moreover, in 4-k region of orderly pked nuleosomes round CTCF inding sites, the NRL ws redued to ± 1.5 p for s nd ± 0.7 p for s: tht is, oth vlues were ~10 p smller thn the orresponding genome-wide NRLs (Fig. 7,d). DISCUSSION Our nlysis of nuleosome positioning in mouse s in omprison to their linege-ommitted nd ounterprts reveled distint profiles t funtionl genomi elements tht re relevnt for ell differentition. The nlysis of nuleosome oupny t trnsription ftor inding sites indited the presene of different hromtin intertion mehnisms (Figs. 1, 2 nd 3); some inding sites were onstitutively depleted of nuleosomes in ll three ell types (Fig. 2). This might e n importnt feture of ertin set of onstitutive sites tht re lwys ompetent for trnsription-ftor inding if the pproprite ftor eomes expressed. In ontrst, for other trnsription ftors the nuleosome oupny t trget sites showed little orreltion with trnsription-ftor inding, whih suggested tht these trnsription ftors n ind to nuleosoml DNA or tht inding ours in only smll frtion of the ells t ny given point in time (Fig. 2d). In third group, trnsriptionftor nd histone-otmer inding ppered to e in ompetitive equilirium, in whih n inrese of the trnsription-ftor onentrtion during development ould e suffiient to disple the nuleosome from position tht interferes with inding to result in oupny profiles like those oserved in Figure 2,. In ddition, hromtin-remodeling omplexes like Chd7 nd Brg1 (Fig. 2d) ould ind to nuleosome nd tively trnslote it long the DNA to new position, s disussed previously 23. Beuse the DNA oupny of developmentl trnsription ftors is highly preditive of gene expression in mouse s 38, ny modultion of this prmeter y nuleosome oupny t given trnsription ftor inding site ould diretly influene the expression of the trget genes. A prtiulrly interesting exmple of the omplex reltionship etween protein inding nd nuleosome positioning ws reveled here for CTCF (Fig. 3 nd Supplementry Fig. 2). Consistent with the previous findings 39, the CTCF inding sites in s were loted in regions with redued nuleosome oupny nd ted s nuleosome oundry element to position djent nuleosomes. Notly, some CTCF inding sites speifi for s pper to e predisposed in s for lter CTCF inding (Fig. 3). In ontrst, CTCF inding sites unique to s eme oupied with nuleosomes in s, whih would e onsistent with the dissoition of CTCF nd possily relted to its deresed expression in s (Supplementry Tle 1). As reported previously, CTCF inding sites in generl hve n intrinsilly high ffinity for the histone otmer, whih ould promote their inorportion into nuleosome if CTCF dissoites 9. Unexpetedly, frtion of CTCF proteins in s ws pprently ssoited with nuleosomes (Supplementry Fig. 2,), nd for CTCF inding sites loted within enhners, no regulr nuleosome positioning pttern ws deteted (Supplementry Fig. 2). Thus, our results suggest tht multiple modes of CTCF intertion with hromtin exist, whih might involve other protein ftors or RNAs tht medite CTCF inding to nuleosoml DNA t ertin sites during differentition. In order to investigte the gene-speifi funtions of nuleosome positioning, we onduted n nlysis of nuleosome oupny t the TSS (Figs. 4 nd 5). The profile of the nuleosome-depleted region vried for different lsses of promoters tht were seleted either on the sis of the presene of the H3K4me3 nd H3K27me3 histone modifitions or the CpG ontent of the DNA sequene (Fig. 5 nd Supplementry Fig. 5). For the mjority of promoters, nuleosomedepleted region entered round +100 p ws present tht eme more pronouned for highly tive genes. This profile might e relted to the presene of RNA polymerse II tht hs een mpped reently t the promoters of mouse s 40. RNA polymerse II ould e present in either trnsriptionlly engged form or ound in stlled stte tht requires dditionl ftors for initition of trnsription 36. Notly, the trnsript ends displyed derese of nuleosome oupny towrd the gene ody for the most tive genes (Fig. 4). The verge TSS nuleosome pttern determined here ws similr to nuleosome profiles reported previously for mouse heptoytes 15 nd seleted humn promoters 9,10,41 tht were lso hrterized y rther rod nuleosome-depleted region. This pttern is mrkedly different from those previously reported for simpler eukryotes, in whih single nuleosome ws missing upstrem of the TSS nd downstrem of the TTS, nd this ws followed y n osilltory pttern of severl regulrly positioned nuleosomes 5 7,28,29. In ddition, nuleosome-depleted region extending into the gene ody ws found t the TTSs, nd it eme more pronouned with inresed gene expression (Fig. 4). This feture ould e relted to oupling of the TTS with 3 polydenyltion of the trnsript 42. When evluting ll promoters within one ell type, we found tht n inresed nuleosome oupny in the region of 500 p round the TSS ws orrelted with redution of trnsription (Supplementry Tle 2). Although no orresponding ntiorrelted hnges of promoter nuleosome oupny nd gene expression were found etween s nd s for the mjority of genes (Supplementry Tles 3 nd 4), we identified ertin speifi groups of promoters tht displyed suh ehvior. This qulifies them s potentil ndidtes for regultory mehnism tht would operte through nuleosome repositioning during differentition. For exmple, the ivlent promoters in s ontined group of promoters with speifi nuleosome oupny profile. This luster showed orreltion etween the loss 1190 VOLUME 19 NUMBER 11 NOVEMBER 2012 nture struturl & moleulr iology
7 of nuleosome, t position 500 to 350 p round the TSS in oth s nd s, ompnied y n inrese in gene expression (Fig. 5g i). In ddition, other ivlent nd H3K27me3-only promoters s well s LCG promoters displyed ntiorrelted reltions etween nuleosome oupny nd gene expression hnges during differentition (Supplementry Tles 3 nd 4). These hnges might e diretly relted to the ddition or removl of ertin histone mrks, s onluded from our nlysis of exemplry histone modifitions with respet to nuleosome oupny (Fig. 6). H3K9me3 lusters in s were found to e nuleosome enrihed, wheres H3K9 nd H3K27 lusters were nuleosome depleted. H3K9me3 nd H3K9 re prtiulrly notle euse H3K9me3 is highly orrelted with lol muttion rtes in ner ells, wheres H3K9 is strongly ntiorrelted 43. Together with reent findings tht hromtin regions hrterized y different histone modifitions vry in their nuleosome repet length 9, our dt link histone modifitions with importnt struturl funtions with respet to nuleosome positioning nd oupny. As reviewed reently, hromtin-remodeling omplexes reognize vriety of histone modifitions 24. Thus, it is tempting to speulte tht these moleulr mhines re involved in hnges of nuleosome oupny t trnsription ftor inding sites nd promoters fter mrking given nuleosome with speifi histone modifition signls. Finlly, we oserved genome-wide hnges of the primry hromtin struture during ell differentition, s refleted in the inrese of the verge NRL in s of ± p y 7 p (s) nd 5 p (s) (Fig. 7). As reported previously, this NRL hnge is relted to n inrese in the rtio of linker to ore histones 37. Although mouse s hve histone H1/nuleosome rtio of 6 (ref. 37), this prmeter inreses to in vrious differentited mouse tissues 44,45. The upregulted gene expression of linker histone vrints H nd H1.7 in s nd s versus s (Supplementry Tle 1) indites tht these ftors might e prtiulrly importnt for the hnge in the NRL. It is noted tht the 5 7-p differene in NRL oserved etween s nd s nd s ould hve lrge effets on the folding properties of the nuleosome hin euse the helil phsing of the DNA doule helix would relote neighoring nuleosomes y torsionl ngle of out 36 per dditionl se pir. In greement with this view, lrge differenes in the hromtin folding properties s funtion of NRL hve een oserved experimentlly s, for exmple, reported in ref. 46. In ddition, our lultion for seleted short genomi regions reveled tht the NRL shows lol vrition, s for exmple in the 4-k region surrounding CTCF inding sites tht hd n ~10 p smller NRL thn the genome-wide verge vlue. In summry, we identified numer of sustntil rerrngements of nuleosome positions t different funtionl genomi elements like trnsription ftor inding sites nd promoters tht re likely to modulte protein inding to these regions. In ddition, glol hnges of nuleosome density tht our throughout the genome during ell linege ommitment of mouse s ould lso ffet the DNA essiility y hnging the folding of the nuleosome hin. Aordingly, we onlude tht the ell type speifi orgniztion of nuleosomes on identil genomes represents n dditionl regultory lyer tht ontrols DNA ess of protein ftors for seleting tissue-speifi gene expression progrms. Methods Methods nd ny ssoited referenes re ville in the online version of the pper. Aession odes. MNse-seq, ChIP-seq nd RNA-seq dt hve een deposited to the GEO dtse under the ession numer GSE Note: Supplementry informtion is ville in the online version of the pper. Aknowledgments We re grteful to A. Vlouev nd R. Chereji for help with the lgorithms for lultions of NRL nd verge TSS ptterns, respetively; to M. Gerstein for dvie on dt proessing; to G. Längst, G. Wedemnn nd K. Fejes Tóth for disussions; nd to the Deutshes Kresforshungszentrum Sequening Core Fility for onduting the sequening. This work ws funded within projet EpiGenSys y the Germn Federl Ministry of Edution nd Reserh (BMBF) s prtner of the ERASysBio+ inititive in the EU FP7 ERA-NET Plus progrm through grnt A to K.R. Computtionl resoures nd dt storge were provided y grnts from the BMBF (01IG07015G, Servies@ MediGRID) nd the Germn Reserh Foundtion (DFG INST 295/27-1). V.B.T. knowledges the support from the Heidelerg Center for Modeling nd Simultion in the Biosienes nd Deutshes Kresforshungszentrum intrmurl grnt, nd Y.V. ws supported y BMBF MedSys grnt E to T.H. AUTHOR CONTRIBUTIONS V.B.T. nd K.R. designed the reserh. M.C.-H., J.-P.M. nd C.M. performed experiments. V.B.T., Y.V., J.-P.M., T.H. nd K.R. nlyzed dt. V.B.T., T.H. nd K.R. wrote the mnusript. COMPETING FINANCIAL INTERESTS The uthors delre no ompeting finnil interests. Pulished online t Reprints nd permissions informtion is ville online t reprints/index.html. 1. Sdeh, R. & Allis, C.D. Genome-wide re -modeling of nuleosome positions. Cell 147, (2011). 2. Rndo, O.J. & Winston, F. Chromtin nd trnsription in yest. Genetis 190, (2012). 3. Zhng, Z. & Pugh, B.F. High-resolution genome-wide mpping of the primry struture of hromtin. Cell 144, (2011). 4. Cui, K. & Zho, K. Genome-wide pprohes to determining nuleosome oupny in metzons using MNse-Seq. Methods Mol. Biol. 833, (2012). 5. Yun, G.C. et l. Genome-sle identifition of nuleosome positions in S. erevisie. Siene 309, (2005). 6. Ioshikhes, I.P., Alert, I., Znton, S.J. & Pugh, B.F. Nuleosome positions predited through omprtive genomis. Nt. Genet. 38, (2006). 7. Segl, E. et l. A genomi ode for nuleosome positioning. Nture 442, (2006). 8. Hu, G. et l. Regultion of nuleosome lndspe nd trnsription ftor trgeting t tissue-speifi enhners y BRG1. Genome Res. 21, (2011). 9. Vlouev, A. et l. Determinnts of nuleosome orgniztion in primry humn ells. Nture 474, (2011). 10. Ott, C.J. et l. Nuleosome oupny revels regultory elements of the CFTR promoter. Nulei Aids Res. 40, (2012). 11. Shones, D.E. et l. Dynmi regultion of nuleosome positioning in the humn genome. Cell 132, (2008). 12. Zhng, L., M, H. & Pugh, B.F. Stle nd dynmi nuleosome sttes during meioti developmentl proess. Genome Res. 21, (2011). 13. Tirosh, I., Sigl, N. & Brki, N. Widespred remodeling of mid-oding sequene nuleosomes y Isw1. Genome Biol. 11, R49 (2010). 14. Moshkin, Y.M. et l. Remodelers orgnize ellulr hromtin y ounterting intrinsi histone-dna sequene preferenes in lss-speifi mnner. Mol. Cell Biol. 32, (2012). 15. Li, Z., Shug, J., Tutej, G., White, P. & Kestner, K.H. The nuleosome mp of the mmmlin liver. Nt. Strut. Mol. Biol. 18, (2011). 16. Chen, X. et l. Integrtion of externl signling pthwys with the ore trnsriptionl network in emryoni stem ells. Cell 133, (2008). 17. Mrtin, D. et l. Genome-wide CTCF distriution in vertertes defines equivlent sites tht id the identifition of disese-ssoited genes. Nt. Strut. Mol. Biol. 18, (2011). 18. Shen, Y. et l. A mp of the is-regultory sequenes in the mouse genome. Nture 488, (2012). 19. Kpln, N. et l. The DNA-enoded nuleosome orgniztion of eukryoti genome. Nture 458, (2009). 20. Shnetz, M.P. et l. CHD7 trgets tive gene enhner elements to modulte ES ell-speifi gene expression. PLoS Genet. 6, e (2010). 21. Ho, L. et l. An emryoni stem ell hromtin remodeling omplex, esbaf, is n essentil omponent of the ore pluripoteny trnsriptionl network. Pro. Ntl. Ad. Si. USA 106, (2009). nture struturl & moleulr iology VOLUME 19 NUMBER 11 NOVEMBER
8 22. Zielke, N. et l. Control of Drosophil endoyles y E2F nd CRL4(CDT2). Nture 480, (2011). 23. Teif, V.B. & Rippe, K. Prediting nuleosome positions on the DNA: omining intrinsi sequene preferenes nd remodeler tivities. Nulei Aids Res. 37, (2009). 24. Erdel, F., Krug, J., Lngst, G. & Rippe, K. Trgeting hromtin remodelers: signls nd serh mehnisms. Biohim. Biophys. At 1809, (2011). 25. Cuddph, S. et l. Glol nlysis of the insultor inding protein CTCF in hromtin rrier regions revels demrtion of tive nd repressive domins. Genome Res. 19, (2009). 26. Fu, Y., Sinh, M., Peterson, C.L. & Weng, Z. The insultor inding protein CTCF positions 20 nuleosomes round its inding sites ross the humn genome. PLoS Genet. 4, e (2008). 27. Hndoko, L. et l. CTCF-medited funtionl hromtin intertome in pluripotent ells. Nt. Genet. 43, (2011). 28. Mvrih, T.N. et l. Nuleosome orgniztion in the Drosophil genome. Nture 453, (2008). 29. Vlouev, A. et l. A high-resolution, nuleosome position mp of C. elegns revels lk of universl sequene-ditted positioning. Genome Res. 18, (2008). 30. Segl, E. & Widom, J. Poly(dA:dT) trts: mjor determinnts of nuleosome orgniztion. Curr. Opin. Strut. Biol. 19, (2009). 31. Zhng, Y. et l. Intrinsi histone-dna intertions re not the mjor determinnt of nuleosome positions in vivo. Nt. Strut. Mol. Biol. 16, (2009). 32. Weiner, A., Hughes, A., Yssour, M., Rndo, O.J. & Friedmn, N. High-resolution nuleosome mpping revels trnsription-dependent promoter pkging. Genome Res. 20, (2010). 33. Fn, X. et l. Nuleosome depletion t yest termintors is not intrinsi nd n our y trnsriptionl mehnism linked to 3 -end formtion. Pro. Ntl. Ad. Si. USA 107, (2010). 34. Ku, M. et l. Genomewide nlysis of PRC1 nd PRC2 oupny identifies two lsses of ivlent domins. PLoS Genet. 4, e (2008). 35. Mikkelsen, T.S. et l. Genome-wide mps of hromtin stte in pluripotent nd linege-ommitted ells. Nture 448, (2007). 36. Lenhrd, B., Sndelin, A. & Crnini, P. Metzon promoters: emerging hrteristis nd insights into trnsriptionl regultion. Nt. Rev. Genet. 13, (2012). 37. Fn, Y. et l. Histone H1 depletion in mmmls lters glol hromtin struture ut uses speifi hnges in gene regultion. Cell 123, (2005). 38. Cheng, C. & Gerstein, M. Modeling the reltive reltionship of trnsription ftor inding nd histone modifitions to gene expression levels in mouse emryoni stem ells. Nulei Aids Res. 40, (2012). 39. Knduri, M. et l. Multiple nuleosome positioning sites regulte the CTCF-medited insultor funtion of the H19 imprinting ontrol region. Mol. Cell Biol. 22, (2002). 40. Min, I.M. et l. Regulting RNA polymerse pusing nd trnsription elongtion in emryoni stem ells. Genes Dev. 25, (2011). 41. Phm, C.D., Sims, H.I., Arher, T.K. & Shnitzler, G.R. Multiple distint stimuli inrese mesured nuleosome oupny round humn promoters. PLoS ONE 6, e23490 (2011). 42. Zho, J., Hymn, L. & Moore, C. Formtion of mrna 3 ends in eukryotes: mehnism, regultion, nd interreltionships with other steps in mrna synthesis. Miroiol. Mol. Biol. Rev. 63, (1999). 43. Shuster-Bökler, B. & Lehner, B. Chromtin orgniztion is mjor influene on regionl muttion rtes in humn ner ells. Nture 488, (2012). 44. Woodok, C.L., Skoulthi, A.I. & Fn, Y. Role of linker histone in hromtin struture nd funtion: H1 stoihiometry nd nuleosome repet length. Chromosome Res. 14, (2006). 45. Fn, Y. et l. H1 linker histones re essentil for mouse development nd ffet nuleosome sping in vivo. Mol. Cell Biol. 23, (2003). 46. Correll, S.J., Shuert, M.H. & Grigoryev, S.A. Short nuleosome repets impose rottionl modultions on hromtin fire folding. EMBO J. 31, (2012) VOLUME 19 NUMBER 11 NOVEMBER 2012 nture struturl & moleulr iology
9 ONLINE METHODS Isoltion of nuleosomes. s from 129P2/Ol mie 47 were ultured in ESGRO omplete medium (Millipore). Differentition of s into neuronl preursors ws indued y formtion of emryoid odies in emryoid ody formtion medium (Millipore) nd tretment with 5 µm retinoi id for 4 d. Neuronl emryoid odies were dissoited nd seeded on Mtrigel (BD Biosienes) in neuronl stem ell medium (PAN) for 4 d. s were generted from pregnnt 129P2/Ol E13.5 mie nd ultured in DMEM supplemented with 10% FCS nd glutmine for up to pssge 5. For MNse digestion, ells were hrvested nd resuspended in low-slt uffer (10 mm HEPES, ph 8, 10 mm KCl, 0.5 mm DTT) t 4 C. After disruption of the ells with doune homogenizer, the nulei were olleted y entrifugtion nd wshed one with the MNse Buffer (10 mm Tris-HCl, ph 7.5, 10 mm CCl 2 ), resuspended in the MNse Buffer nd digested with 0.5 units MNse (Ferments) per miroliter nd inuted for 6 11 min t 37 C. The MNse digestion ws stopped y putting the smples on ie nd dding EDTA to onentrtion of 10 mm. After digestion with 0.1 µg µl 1 RNse A (Ferments) nd removl of protein y phenol-hloroform extrtion, the DNA ws ethnol preipitted, nd the resulting DNA pellet ws dissolved in H 2 O. DNA frgments orresponding to mononuleosomes or dinuleosomes were seprted on 2% grose gel y using n E-Gel eletrophoresis system (Life Tehnologies). The lirries for sequening were prepred ording to the stndrd protool for the Illumin HiSeq2000 sequening pltform. Deep sequening of nuleosoml DNA. High-throughput pired-end sequening of t lest 50-p red length ws performed on the Illumin HiSeq2000 pltform t the DKFZ sequening ore fility in Heidelerg, Germny. We mpped out 150 million nuleosome positions per sequening retion nd used in the finl nlysis three iologil-replite experiments for s nd two replite experiments for eh of s nd s, yielding totl of 300 million 450 million nuleosome positions per ell type. In line with the previous studies 48,49, we oserved tht the hromosome-wide nuleosome density ws dependent on the verge GC ontent, whih ws ntiorrelted with the MNse preferenes found with purified genomi mouse DNA (Supplementry Fig. 1, Supplementry Note). For mpping the position of individul nuleosomes, MNse sequene preferenes were found to e negligile, in greement with reent study 50. Following previous findings desried in refs. 32 nd 51, we heked the dependene of the nuleosome mps on the level of MNse digestion. Using slightly different levels of MNse digestions in four replite experiments in s, we otined verge mononuleosome frgment lengths round 150 p, 155 p, 160 p nd 180 p, with ~150 million mpped reds in eh retion. The hnges of the integrl prmeters suh s the nuleosome repet length during the ell differentition were found to e independent of the degree of MNse digestion. However, in line with previous studies 31,48,50, different levels of MNse digestion ffeted nuleosome distriutions, with individul nuleosome peks sometimes missing or ppering, without ler inditions tht one of the smples with 150-p, 155-p or 160-p verge length ws etter representtion of the sitution in vivo. For MNse digestion with 180-p verge frgment length, lrge frtion of the linker remined undigested, leding to lrgely redued overge of individul nuleosome positions. Aordingly, the nuleosome oupny mps used here were generted from omining only smples with MNse digestions tht hd n verge mononuleosome frgment length etween p. Dt nlysis of nuleosome oupnies. DNA reds were ligned on the mm9 ssemly version of the mouse genome, with Bowtie 52 reporting unique hits with up to two mismthes. The nuleosome oupny mps were lulted with ustom-mde Perl sripts y ounting how mny reds overed given DNA se pir (Supplementry Fig. 1). Sites with rtifiilly high overge were onsidered s rtifts nd exluded from the nlysis. No further pek lling or smoothing ws onduted. In ddition, no ssumptions on the length of the nuleosoml DNA hd to e mde to derive the nuleosome oupny mps, s nuleosome oundries were determined on oth sides of the nuleosome y pired-end sequening. The nuleosome signtures t trnsription ftor inding sites nd TSSs or TTSs, respetively, were lulted s the sum of nuleosome oupnies in window of 2,000 to 2,000 p round given site. For eh gene, the sum of reds ws normlized to 1. Then the verged nuleosome profile ws normlized to yield the nuleosome oupny equl to 1 t position 2,000 p 53. For nuleosome lignment round CTCF inding sites, we nlyzed the dt set from previous study 17 downloded from the GEO rhive (GSE27944) nd the dt sets from ENCODE 18 downloded from the UCSC Genome Browser (ession odes: wgenodeem nd wgenodeem001698), whih resulted in qulittively similr ptterns. Only the ptterns otined with ENCODE dt re reported here. For nuleosome lignment round p300 sites, we used the ENCODE dt set wgenodelirtfses- 4P300ME0C57l6StdPk. For nuleosome lignment round inding sites of 12 developmentl trnsription ftors, the nlysis ws onduted ording to the dt set from ref. 16, whih ws initilly mpped to the mm8 genome uild nd onverted to mm9 y using the liftover tool of the UCSC Genome Browser. The histone-modifition dt from refs. 34 nd 35 were lso onverted from mm8 to mm9 efore the nlysis. Brg1 ChIP-seq dt from ref. 21 (GEO rhive GSE14344) were relustered with MACS 54 using P = 10 5 utoff for pek detetion. Chd7 ChIP-seq dt were from ref. 20 (GEO rhive GSM558674). DNse I hypersensitivity rw dt from the ltter study were provided y the uthors nd mpped nd lustered s desried ove. For nuleosome lignment round the TSS nd TTS, we used the Eldordo gene nnottion provided in the Genomtix Genome Anlyzer softwre (Genomtix) 55. Alignments with the RefSeq gene nnottion resulted in similr ptterns. Nuleosome-oupny luster plots for visulizing multiple trnsripts were generted in Mtl (Mthworks). These profiles were sed on the verge oupny t the TSS in the region from position 500 to +500 p. Hierrhil lustering ws done ording to the Wrd s minimum vrine method implemented in Mtl, whih omputes mutully exlusive groups of oupny profiles with minimum within-luster vrine 56. The resulting lusters were nlyzed with the DAVID gene nnottion lustering tool 57. The nuleosome repet length ws lulted essentilly s desried previously 9, with the following modifitions: A histogrm of the numer of ourrenes of nuleosome distnes from 1 3,000 p etween ll nuleosomes ws omputed nd smoothed with 50-p window. The NRL ws then determined from liner fit of the deteted pek positions versus the nuleosome numer. Up to 12 peks ould e identified in this nlysis. To mke lultions more roust, threshold of 20 reds ws set to remove rtifiilly enrihed nuleosome frgments strting t given genomi position. These nuleosomes were given sttistil weight equl to 20. Less-undnt nuleosome frgments entered the lultion with the weights equl to the numer of their ourrenes in the high-throughput sequening. Expression profiling y RNA sequening. Totl RNA ws purified nd prepred for sequening s desried previously 58. Sequening ws performed on Illumin pltforms. RNA reds were ligned with TopHt 59. Further expression nlysis ws performed with the Genomtix softwre using the most reent Eldordo gene nnottion 55. For eh trnsript, normlized expression vlue ws lulted from the red distriution tht ounts for the length differenes nd the mount of mpped reds. The progrm DESeq 60 ws used for the nlysis of differentil expression (Supplementry Tles 6 nd 7). Chromtin immunopreipittion. For eh smple, ells were rosslinked with 1% PFA, nd ell nulei were prepred y using swelling uffer (25 mm HEPES, ph 7.8, 1 mm MgCl 2, 10 mm KCl, 0.1% NP-40, 1 mm DTT). Chromtin ws shered to mononuleosoml frgments. After IgG prelerne the shered hromtin ws inuted overnight with 4 µg of ntiodies ginst either H3K9 (Am, 4441), H3K27 (Am, 4729) or H3K9me3 (Am 8898). After wshes with sonition uffer (10 mm Tris-HCl, ph 8.0, 200 mm NCl, 1 mm EDTA, 0.5% N-luroylsrosine, 0.1% N-deoxyholte), high-slt uffer (50 mm HEPES, ph 7.9, 500 mm NCl, 1mM EDTA, 1% Triton X-100, 0.1% N-deoxyholte, 0.1% SDS), lithium uffer (20 mm Tris-HCl, ph 8.0, 1 mm EDTA, 250 mm LiCl, 0.5% NP-40, 0.5% N-deoxyholte) nd 10 mm Tris-HCl, hromtin ws eluted from the protein G mgneti eds nd the rosslink ws reversed overnight. After RNse A nd proteinse K digestion, DNA ws purified nd loned in roded sequening lirry for the Illumin HiSeq2000 sequening pltform. Single reds of 50-p length were mpped with Bowtie nd lustered with MACS 54, using P-vlue utoff of Mllm, J.P., Tshpe, J.A., Hik, M., Filippov, M.A. & Muller, U.C. Genertion of onditionl null lleles for APP nd APLP2. Genesis 48, (2010). doi: /nsm.2419 nture struturl & moleulr iology
Nucleosome positioning as a determinant of exon recognition
 Nuleosome positioning s determinnt of exon reognition Hgen Tilgner 1,3, Christoforos Nikolou 1,3, Sonj Althmmer 1, Mihel Smmeth 1, Miguel Beto 1, Jun Vlárel 1,2 & Roderi Guigó 1 200 Nture Ameri, In. All
Nuleosome positioning s determinnt of exon reognition Hgen Tilgner 1,3, Christoforos Nikolou 1,3, Sonj Althmmer 1, Mihel Smmeth 1, Miguel Beto 1, Jun Vlárel 1,2 & Roderi Guigó 1 200 Nture Ameri, In. All
EFFECT OF DIETARY ENZYME ON PERFORMANCE OF WEANLING PIGS
 EFFECT OF DIETARY ENZYME ON PERFORMANCE OF WEANLING PIGS Finl report sumitted to Dniso Animl Nutrition E. vn Heugten nd B. Frederik North Crolin Stte University, Deprtment of Animl Siene Summry The urrent
EFFECT OF DIETARY ENZYME ON PERFORMANCE OF WEANLING PIGS Finl report sumitted to Dniso Animl Nutrition E. vn Heugten nd B. Frederik North Crolin Stte University, Deprtment of Animl Siene Summry The urrent
P AND K IN POTATOES. Donald A Horneck Oregon State University Extension Service
 P AND K IN POTATOES Donld A Hornek Oregon Stte University Extension Servie INTRODUCTION Phosphorous nd potssium re importnt to grow high yielding nd qulity pottoes. Muh of the northwest hs hd trditionlly
P AND K IN POTATOES Donld A Hornek Oregon Stte University Extension Servie INTRODUCTION Phosphorous nd potssium re importnt to grow high yielding nd qulity pottoes. Muh of the northwest hs hd trditionlly
Alimonti_Supplementary Figure 1. Pten +/- Pten + Pten. Pten hy. β-actin. Pten - wt hy/+ +/- wt hy/+ +/- Pten. Pten. Relative Protein level (% )
 Alimonti_Supplementry Figure 1 hy 3 4 5 3 Neo 4 5 5 Proe 5 Proe hy/ hy/ /- - 3 6 Neo β-tin d Reltive Protein level (% ) 15 1 5 hy/ /- Reltive Gene Expr. (% ) 15 1 5 hy/ /- Supplementry Figure 1 Chrteriztion
Alimonti_Supplementry Figure 1 hy 3 4 5 3 Neo 4 5 5 Proe 5 Proe hy/ hy/ /- - 3 6 Neo β-tin d Reltive Protein level (% ) 15 1 5 hy/ /- Reltive Gene Expr. (% ) 15 1 5 hy/ /- Supplementry Figure 1 Chrteriztion
SUPPLEMENTARY INFORMATION
 DOI: 1.138/n358 TLR2 nd MyD88 expression in murine mmmry epithelil supopultions. CD24 min plus MRU Myo-epithelil Luminl progenitor (CD61 pos ) Mture luminl (CD61 neg ) CD49f CD61 Reltive expression Krt5
DOI: 1.138/n358 TLR2 nd MyD88 expression in murine mmmry epithelil supopultions. CD24 min plus MRU Myo-epithelil Luminl progenitor (CD61 pos ) Mture luminl (CD61 neg ) CD49f CD61 Reltive expression Krt5
Lesions of prefrontal cortex reduce attentional modulation of neuronal responses. and synchrony in V4
 Lesions of prefrontl ortex reue ttentionl moultion of neuronl responses n synhrony in V4 Georgi G. Gregoriou,, Anrew F. Rossi, 3 Leslie G Ungerleier, 4 Roert Desimone 5 Deprtment of Bsi Sienes, Fulty of
Lesions of prefrontl ortex reue ttentionl moultion of neuronl responses n synhrony in V4 Georgi G. Gregoriou,, Anrew F. Rossi, 3 Leslie G Ungerleier, 4 Roert Desimone 5 Deprtment of Bsi Sienes, Fulty of
SUPPLEMENTARY INFORMATION
 doi: 1.138/nture862 humn hr. 21q MRPL39 murine Chr.16 Mrpl39 Dyrk1A Runx1 murine Chr. 17 ZNF295 Ets2 Znf295 murine Chr. 1 COL18A1 -/- lot: nti-dscr1 IgG hevy hin DSCR1 DSCR1 expression reltive to hevy
doi: 1.138/nture862 humn hr. 21q MRPL39 murine Chr.16 Mrpl39 Dyrk1A Runx1 murine Chr. 17 ZNF295 Ets2 Znf295 murine Chr. 1 COL18A1 -/- lot: nti-dscr1 IgG hevy hin DSCR1 DSCR1 expression reltive to hevy
LHb VTA. VTA-projecting RMTg-projecting overlay. Supplemental Figure 2. Retrograde labeling of LHb neurons. a. VTA-projecting LHb
 SUPPLEMENTARY INFORMATION Supplementl Figure 1 doi:10.1038/nture09742 Lterl 1.0 mm from midline mpfc BNST mpfc BNST Lterl 2.1 mm from midline LHA LHA Lterl 2.7 mm from midline SUPPLEMENTAL INFORMATION
SUPPLEMENTARY INFORMATION Supplementl Figure 1 doi:10.1038/nture09742 Lterl 1.0 mm from midline mpfc BNST mpfc BNST Lterl 2.1 mm from midline LHA LHA Lterl 2.7 mm from midline SUPPLEMENTAL INFORMATION
Cos7 (3TP) (K): TGFβ1(h): (K)
 IP#2: IP#1: Totl Lystes luiferse tivity (K): 6-4 - (K): luiferse tivity luiferse tivity (K): 2 1 RL-: - + + + + + Sm4-3F: + - + + + + MYC-Sm3: - - - - + + TβRI-HA(T204D): - - - + - + α-ha Luiferse Ativity
IP#2: IP#1: Totl Lystes luiferse tivity (K): 6-4 - (K): luiferse tivity luiferse tivity (K): 2 1 RL-: - + + + + + Sm4-3F: + - + + + + MYC-Sm3: - - - - + + TβRI-HA(T204D): - - - + - + α-ha Luiferse Ativity
SUPPLEMENTARY INFORMATION
 DOI: 1.13/n7 Reltive Pprg mrna 3 1 1 Time (weeks) Interspulr Inguinl Epididyml Reltive undne..1.5. - 5 5-51 51-1 1-7 7 - - 1 1-1 Lipid droplet size ( m ) 1-3 3 - - - 1 1-1 1-1 1-175 175-3 3-31 31-5 >5
DOI: 1.13/n7 Reltive Pprg mrna 3 1 1 Time (weeks) Interspulr Inguinl Epididyml Reltive undne..1.5. - 5 5-51 51-1 1-7 7 - - 1 1-1 Lipid droplet size ( m ) 1-3 3 - - - 1 1-1 1-1 1-175 175-3 3-31 31-5 >5
Other Uses for Cluster Sampling
 Other Uses for Cluster Smpling Mesure hnges in the level of n ttriute Hypothesis testing versus intervl estimtion Type I n 2 errors Power of the test Mesuring ttriute t sme time in ifferent sites Exmple:
Other Uses for Cluster Smpling Mesure hnges in the level of n ttriute Hypothesis testing versus intervl estimtion Type I n 2 errors Power of the test Mesuring ttriute t sme time in ifferent sites Exmple:
Poultry No The replacement value of betaine for DL-methionine and Choline in broiler diets
 Poultry No. 1573 The replement vlue of etine for DL-methionine nd Choline in roiler diets Key Informtion In roiler diets defiient in sulfur mino ids ut dequtely supplemented with methyl groups vi dded
Poultry No. 1573 The replement vlue of etine for DL-methionine nd Choline in roiler diets Key Informtion In roiler diets defiient in sulfur mino ids ut dequtely supplemented with methyl groups vi dded
Supplementary Figure 1. Scheme of unilateral pyramidotomy used for detecting compensatory sprouting of intact CST axons.
 () BDA 2 weeks fter Py () AAVs Cre or GFP t P1 BDA 2 weeks fter Py CSMN CST () Py t P7 or 2 months () Py t 2 months Supplementry Figure 1. Sheme of unilterl pyrmidotomy used for deteting ompenstory sprouting
() BDA 2 weeks fter Py () AAVs Cre or GFP t P1 BDA 2 weeks fter Py CSMN CST () Py t P7 or 2 months () Py t 2 months Supplementry Figure 1. Sheme of unilterl pyrmidotomy used for deteting ompenstory sprouting
Whangarei District Council Class 4 Gambling Venue Policy
 Whngrei Distrit Counil Clss 4 Gmling Venue Poliy April 2013 Whngrei Distrit Counil Clss 4 Gmling Venue Poliy Tle of ontents Introdution... 3 1 Ojetives of the poliy in so fr s promoted y the Gmling At
Whngrei Distrit Counil Clss 4 Gmling Venue Poliy April 2013 Whngrei Distrit Counil Clss 4 Gmling Venue Poliy Tle of ontents Introdution... 3 1 Ojetives of the poliy in so fr s promoted y the Gmling At
TNFa Signaling Exposes Latent Estrogen Receptor Binding Sites to Alter the Breast Cancer Cell Transcriptome
 Artile TNF Signling Exposes Ltent Estrogen Reeptor Binding Sites to Alter the Brest Cner Cell Trnsriptome Grphil Astrt Authors Hetor L. Frno, Anush Ngri, W. Lee Krus Correspondene lee.krus@utsouthwestern.edu
Artile TNF Signling Exposes Ltent Estrogen Reeptor Binding Sites to Alter the Brest Cner Cell Trnsriptome Grphil Astrt Authors Hetor L. Frno, Anush Ngri, W. Lee Krus Correspondene lee.krus@utsouthwestern.edu
Supplementary Figure S1
 Supplementry Figure S1 - UTR m - 3HA - 2-1 hgh - 1 Uiquitin *! *! lk distl promoter m K3R/ K121R-3HA UTR hgh founder lines - HA - - founder lines TG- E1 L A2 B1 F9 G6 H4 H6 B C D2 G1 H3 J2 L - 7 IP: lk
Supplementry Figure S1 - UTR m - 3HA - 2-1 hgh - 1 Uiquitin *! *! lk distl promoter m K3R/ K121R-3HA UTR hgh founder lines - HA - - founder lines TG- E1 L A2 B1 F9 G6 H4 H6 B C D2 G1 H3 J2 L - 7 IP: lk
SUPPLEMENTARY INFORMATION
 doi:.38/nture277 d 25 25 2 Time from sound onset (ms) 25 25 2 Time from sound onset (ms) Firing rte (spikes/s) Firing rte (spikes/s).8.6..2 e f g h.8.6..2 Frtion of neurons Frtion of neurons N = 53 2 2
doi:.38/nture277 d 25 25 2 Time from sound onset (ms) 25 25 2 Time from sound onset (ms) Firing rte (spikes/s) Firing rte (spikes/s).8.6..2 e f g h.8.6..2 Frtion of neurons Frtion of neurons N = 53 2 2
Title of Experiment: Author, Institute and address:
 Title of Experiment: Trsfetion of murine mrophge RAW264.7 ells with METAFECTENE PRO. Author, Institute n ress: Ptrizi Pellegtti n Frneso Di Virgilio. Deprtment of Experimentl n Dignosti Meiine, Setion
Title of Experiment: Trsfetion of murine mrophge RAW264.7 ells with METAFECTENE PRO. Author, Institute n ress: Ptrizi Pellegtti n Frneso Di Virgilio. Deprtment of Experimentl n Dignosti Meiine, Setion
SUPPLEMENTARY INFORMATION
 % ells with ili (mrke y A-Tu) Reltive Luiferse % ells with ili (mrke y Arl13) % ells with ili DOI: 1.138/n2259 A-Tuulin Hoehst % Cilite Non-ilite -Serum 9% 8% 7% 1 6% % 4% +Serum 1 3% 2% 1% % Serum: -
% ells with ili (mrke y A-Tu) Reltive Luiferse % ells with ili (mrke y Arl13) % ells with ili DOI: 1.138/n2259 A-Tuulin Hoehst % Cilite Non-ilite -Serum 9% 8% 7% 1 6% % 4% +Serum 1 3% 2% 1% % Serum: -
PTSE RATES IN PNNI NETWORKS
 PTSE RATES IN PNNI NETWORKS Norert MERSCH 1 Siemens AG, Hofmnnstr. 51, D-81359 Münhen, Germny Peter JOCHER 2 LKN, Tehnishe Universität Münhen, Arisstr. 21, D-80290 Münhen, Germny Lrs BURGSTAHLER 3 IND,
PTSE RATES IN PNNI NETWORKS Norert MERSCH 1 Siemens AG, Hofmnnstr. 51, D-81359 Münhen, Germny Peter JOCHER 2 LKN, Tehnishe Universität Münhen, Arisstr. 21, D-80290 Münhen, Germny Lrs BURGSTAHLER 3 IND,
The microrna mir-31 inhibits CD8 + T cell function in chronic viral infection
 A rt i l e s The mirorna mir-3 inhiits CD8 + T ell funtion in hroni virl infetion Howell F Moffett, Adm N R Crtwright, Hye-Jung Kim, Jernej Gode, Json Pyrdol, Trmo Äijö 3, Gustvo J Mrtinez,6, Anjn Ro,
A rt i l e s The mirorna mir-3 inhiits CD8 + T ell funtion in hroni virl infetion Howell F Moffett, Adm N R Crtwright, Hye-Jung Kim, Jernej Gode, Json Pyrdol, Trmo Äijö 3, Gustvo J Mrtinez,6, Anjn Ro,
SUPPLEMENTARY INFORMATION
 { OI: 1.138/n31 Srifie n nlyze APs on week 1 s of iet 1 4 6 High-ft iet BrU High-ft iet BrU 4 High-ft iet BrU 6 High-ft iet BrU Lin - Lin - : C34 + : C9 + 1 1 3 1 4 1 5 C45 1 C34 1 1 1 1 3 1 4 1 5 S-1
{ OI: 1.138/n31 Srifie n nlyze APs on week 1 s of iet 1 4 6 High-ft iet BrU High-ft iet BrU 4 High-ft iet BrU 6 High-ft iet BrU Lin - Lin - : C34 + : C9 + 1 1 3 1 4 1 5 C45 1 C34 1 1 1 1 3 1 4 1 5 S-1
SUPPLEMENTARY INFORMATION
 DOI: 1.138/n2977 Numer of ells per field 6 4 2 P =.1 Orthotopi eum Normlized ventrl photon flux 1E7 1E6 1E5 1E4 1E3 1E2 n=8 n=9 1 2 3 4 5 6 Dys Dy54 1.5E5 2.4E7 d Mie with lymph node metstsis (%) 1 8 6
DOI: 1.138/n2977 Numer of ells per field 6 4 2 P =.1 Orthotopi eum Normlized ventrl photon flux 1E7 1E6 1E5 1E4 1E3 1E2 n=8 n=9 1 2 3 4 5 6 Dys Dy54 1.5E5 2.4E7 d Mie with lymph node metstsis (%) 1 8 6
Distribution, recognition and regulation of non-cpg methylation in the adult mammalian brain
 Distriution, reognition nd regultion of non-cpg methyltion in the dult mmmlin rin Junjie U Guo,9,, Yijing Su,,, Joo Heon Shin, Jehoon Shin,5, Hongd Li 6, Bin Xie, Chun Zhong,, Shohui Hu 7, Thu Le 8, Guoping
Distriution, reognition nd regultion of non-cpg methyltion in the dult mmmlin rin Junjie U Guo,9,, Yijing Su,,, Joo Heon Shin, Jehoon Shin,5, Hongd Li 6, Bin Xie, Chun Zhong,, Shohui Hu 7, Thu Le 8, Guoping
The GCN5-CITED2-PKA signalling module controls hepatic glucose metabolism through a camp-induced substrate switch
 Reeived 6 Apr 216 Aepted 8 Sep 216 Pulished 22 Nov 216 DOI: 1.138/nomms13147 OPEN The GCN5-CITED2-PKA signlling module ontrols hepti gluose metolism through AMP-indued sustrte swith Mshito Ski 1, Tomoko
Reeived 6 Apr 216 Aepted 8 Sep 216 Pulished 22 Nov 216 DOI: 1.138/nomms13147 OPEN The GCN5-CITED2-PKA signlling module ontrols hepti gluose metolism through AMP-indued sustrte swith Mshito Ski 1, Tomoko
Learning to see: experience and attention in primary visual cortex
 2 Nture Pulishing Group http://neurosi.nture.om rtiles Lerning to see: experiene nd ttention in primry visul ortex 2 Nture Pulishing Group http://neurosi.nture.om Roy E. Crist, Wu Li nd Chrles D. Gilert
2 Nture Pulishing Group http://neurosi.nture.om rtiles Lerning to see: experiene nd ttention in primry visul ortex 2 Nture Pulishing Group http://neurosi.nture.om Roy E. Crist, Wu Li nd Chrles D. Gilert
Interplay of LRRK2 with chaperone-mediated autophagy
 Interply of with hperone-medited utophgy Smnth J Orenstein,, Sheng-Hn Kuo,, Inmuld Tsset,,, Espernz Aris,, Hiroshi Kog,, Irene Fernndez-Crs, Etty Cortes,5, Lwrene S Honig,5, Willim Duer 6, Antonell Consiglio,7,
Interply of with hperone-medited utophgy Smnth J Orenstein,, Sheng-Hn Kuo,, Inmuld Tsset,,, Espernz Aris,, Hiroshi Kog,, Irene Fernndez-Crs, Etty Cortes,5, Lwrene S Honig,5, Willim Duer 6, Antonell Consiglio,7,
Copy Number ID2 MYCN ID2 MYCN. Copy Number MYCN DDX1 ID2 KIDINS220 MBOAT2 ID2
 Copy Numer Copy Numer Copy Numer Copy Numer DIPG38 DIPG49 ID2 MYCN ID2 MYCN c DIPG01 d DIPG29 ID2 MYCN ID2 MYCN e STNG2 f MYCN DIPG01 Chr. 2 DIPG29 Chr. 1 MYCN DDX1 Chr. 2 ID2 KIDINS220 MBOAT2 ID2 Supplementry
Copy Numer Copy Numer Copy Numer Copy Numer DIPG38 DIPG49 ID2 MYCN ID2 MYCN c DIPG01 d DIPG29 ID2 MYCN ID2 MYCN e STNG2 f MYCN DIPG01 Chr. 2 DIPG29 Chr. 1 MYCN DDX1 Chr. 2 ID2 KIDINS220 MBOAT2 ID2 Supplementry
Agilent G6825AA MassHunter Pathways to PCDL Software Quick Start Guide
 Agilent G6825AA MssHunter Pthwys to PCDL Softwre Quick Strt Guide Wht is Agilent Pthwys to PCDL? Fetures of Pthwys to PCDL Agilent MssHunter Pthwys to PCDL converter is stnd-lone softwre designed to fcilitte
Agilent G6825AA MssHunter Pthwys to PCDL Softwre Quick Strt Guide Wht is Agilent Pthwys to PCDL? Fetures of Pthwys to PCDL Agilent MssHunter Pthwys to PCDL converter is stnd-lone softwre designed to fcilitte
Fates-shifted is an F box protein that targets Bicoid for degradation and regulates developmental fate determination in Drosophila embryos
 ARTICLES Ftes-shifted is n F ox protein tht trgets Bioid for degrdtion nd regultes developmentl fte determintion in Drosophil emryos Juno Liu 1 nd Jun M 1,2,3 Bioid (Bd) is morphogeneti protein tht instruts
ARTICLES Ftes-shifted is n F ox protein tht trgets Bioid for degrdtion nd regultes developmentl fte determintion in Drosophil emryos Juno Liu 1 nd Jun M 1,2,3 Bioid (Bd) is morphogeneti protein tht instruts
Receptive field structure varies with layer in the primary visual cortex
 2 Nture Pulishing Group http://www.nture.om/ntureneurosiene Reeptive field struture vries with lyer in the primry visul ortex Luis M Mrtinez, Qingo Wng 2, R Cly Reid 3, Cinthi Pilli 2, José-Mñuel Alonso
2 Nture Pulishing Group http://www.nture.om/ntureneurosiene Reeptive field struture vries with lyer in the primry visul ortex Luis M Mrtinez, Qingo Wng 2, R Cly Reid 3, Cinthi Pilli 2, José-Mñuel Alonso
Maintenance of protein synthesis reading frame by EF-P and m 1 G37-tRNA
 Reeived 23 Nov 214 Aepted 2 Apr 215 Pulished 26 My 215 DOI: 1.138/nomms8226 Mintenne of protein synthesis reding frme y EF-P nd m 1 G37-tRNA Howrd B. Gmper 1, *, Iso Msud 1, *, Miln Frenkel-Morgenstern
Reeived 23 Nov 214 Aepted 2 Apr 215 Pulished 26 My 215 DOI: 1.138/nomms8226 Mintenne of protein synthesis reding frme y EF-P nd m 1 G37-tRNA Howrd B. Gmper 1, *, Iso Msud 1, *, Miln Frenkel-Morgenstern
SUPPLEMENTARY INFORMATION
 doi:.8/nture98 : hr NEMO :5 hr IKK IKK NF-κB p65 p5 p65/-rel NF-κB p65 p5 p65/-rel Cytoplsm Cytoplsm p65/p5 Nuleus Nuleus NEMO IKK IKK d : hr > : hr p65/-rel NF- p65 p5 Cytoplsm Cytoplsm p65/p5 p65/-rel
doi:.8/nture98 : hr NEMO :5 hr IKK IKK NF-κB p65 p5 p65/-rel NF-κB p65 p5 p65/-rel Cytoplsm Cytoplsm p65/p5 Nuleus Nuleus NEMO IKK IKK d : hr > : hr p65/-rel NF- p65 p5 Cytoplsm Cytoplsm p65/p5 p65/-rel
Lipid Composition of Egg Yolk and Serum in Laying Hens Fed Diets Containing Black Cumin (Nigella sativa)
 Interntionl Journl of Poultry Siene 5 (6): 574-578, 2006 ISSN 682-8356 Asin Network for Sientifi Informtion, 2006 Lipid Composition of Egg Yolk nd Serum in Lying Hens Fed Diets Contining Blk Cumin (Nigell
Interntionl Journl of Poultry Siene 5 (6): 574-578, 2006 ISSN 682-8356 Asin Network for Sientifi Informtion, 2006 Lipid Composition of Egg Yolk nd Serum in Lying Hens Fed Diets Contining Blk Cumin (Nigell
Chloride Nutrition Regulates Water Balance in Plants
 XII Portuguese-Spnish Symposium on Plnt Wter Reltions Chloride Nutrition Regultes Wter Blne in Plnts Frno-Nvrro JD 1, Brumós J, Rosles MA 1, Vázquez-Rodríguez A 1, Sñudo BJ 1, Díz- Rued P 1, Rivero C 1,
XII Portuguese-Spnish Symposium on Plnt Wter Reltions Chloride Nutrition Regultes Wter Blne in Plnts Frno-Nvrro JD 1, Brumós J, Rosles MA 1, Vázquez-Rodríguez A 1, Sñudo BJ 1, Díz- Rued P 1, Rivero C 1,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:.8/nture89 4 4 Ilr -/- Ilr -/- Ilr -/- Cspse- -/- As -/- Nlrp -/- Il8 -/- Ilr -/- Supplementl figure. Inresed severity of NASH in inflmmsome-defiient mie, ut not in Ilr-defiient
SUPPLEMENTARY INFORMATION doi:.8/nture89 4 4 Ilr -/- Ilr -/- Ilr -/- Cspse- -/- As -/- Nlrp -/- Il8 -/- Ilr -/- Supplementl figure. Inresed severity of NASH in inflmmsome-defiient mie, ut not in Ilr-defiient
SUPPLEMENTARY INFORMATION
 doi:10.1038/nture10754 Supplementry note 1 To ompre our dt with previous studies, we mesured the width of spikes from identified dopminergi neurons nd unidentified neurons from DATCre mie. Previous studies
doi:10.1038/nture10754 Supplementry note 1 To ompre our dt with previous studies, we mesured the width of spikes from identified dopminergi neurons nd unidentified neurons from DATCre mie. Previous studies
Association between haloacetic acid degradation and heterotrophic bacteria in water distribution systems
 wter reserh 43 (29) 971 978 Aville t www.sienediret.om journl homepge: www.elsevier.om/lote/wtres Assoition etween hloeti id degrdtion nd heterotrophi teri in wter distriution systems Hsin-hsin Tung, *,
wter reserh 43 (29) 971 978 Aville t www.sienediret.om journl homepge: www.elsevier.om/lote/wtres Assoition etween hloeti id degrdtion nd heterotrophi teri in wter distriution systems Hsin-hsin Tung, *,
Supplementary Information
 Supplementry Informtion A new lss of plnt lipid is essentil for protetion ginst phosphorus depletion Yozo Okzki 1, Hitomi Otsuki 1, Tomoko Nrisw 1, Mkoto Koyshi 1, Storu Swi 2, Yukiko Kmide 1, Miyko Kusno
Supplementry Informtion A new lss of plnt lipid is essentil for protetion ginst phosphorus depletion Yozo Okzki 1, Hitomi Otsuki 1, Tomoko Nrisw 1, Mkoto Koyshi 1, Storu Swi 2, Yukiko Kmide 1, Miyko Kusno
Operating Systems Principles. Page Replacement Algorithms
 Operting Systems Priniples Pge Replement Algorithms Steve Gor gor@se.unl.eu http://www.se.unl.eu/~gor/courses/csce45 Virtul Memory Mngement Funmentl issues Plement strtegy Replement strtegies Lo ontrol
Operting Systems Priniples Pge Replement Algorithms Steve Gor gor@se.unl.eu http://www.se.unl.eu/~gor/courses/csce45 Virtul Memory Mngement Funmentl issues Plement strtegy Replement strtegies Lo ontrol
Reward expectation differentially modulates attentional behavior and activity in visual area V4
 Rewrd expettion differentilly modultes ttentionl ehvior nd tivity in visul re V4 Jll K Bruni 1,6, Brin Lu 1,5,6 & C Dniel Slzmn 1 4 npg 215 Nture Ameri, In. All rights reserved. Neurl tivity in visul re
Rewrd expettion differentilly modultes ttentionl ehvior nd tivity in visul re V4 Jll K Bruni 1,6, Brin Lu 1,5,6 & C Dniel Slzmn 1 4 npg 215 Nture Ameri, In. All rights reserved. Neurl tivity in visul re
Using Paclobutrazol to Suppress Inflorescence Height of Potted Phalaenopsis Orchids
 Using Pcloutrzol to Suppress Inflorescence Height of Potted Phlenopsis Orchids A REPORT SUBMITTED TO FINE AMERICAS Linsey Newton nd Erik Runkle Deprtment of Horticulture Spring 28 Using Pcloutrzol to Suppress
Using Pcloutrzol to Suppress Inflorescence Height of Potted Phlenopsis Orchids A REPORT SUBMITTED TO FINE AMERICAS Linsey Newton nd Erik Runkle Deprtment of Horticulture Spring 28 Using Pcloutrzol to Suppress
Neural population coding of sound level adapts to stimulus statistics
 COMPUTATION AND SYSTEMS 25 Nture Pulishing Group http://www.nture.om/ntureneurosiene Neurl popultion oding of sound level dpts to stimulus sttistis Isel Den 1, Niol S Hrper 1,2 & Dvid MAlpine 1 Mmmls n
COMPUTATION AND SYSTEMS 25 Nture Pulishing Group http://www.nture.om/ntureneurosiene Neurl popultion oding of sound level dpts to stimulus sttistis Isel Den 1, Niol S Hrper 1,2 & Dvid MAlpine 1 Mmmls n
Open Access RESEARCH ARTICLE. Genetics Selection Evolution
 DOI 10.1186/s12711-016-0222-0 Genetis Seletion Evolution RESEARCH ARTICLE Open Aess Comprison of host geneti ftors influening pig response to infetion with two North Amerin isoltes of porine reprodutive
DOI 10.1186/s12711-016-0222-0 Genetis Seletion Evolution RESEARCH ARTICLE Open Aess Comprison of host geneti ftors influening pig response to infetion with two North Amerin isoltes of porine reprodutive
SUPPLEMENTARY INFORMATION
 Prentl doi:.8/nture57 Figure S HPMECs LM Cells Cell lines VEGF (ng/ml) Prentl 7. +/-. LM 7. +/-.99 LM 7. +/-.99 Fold COX induction 5 VEGF: - + + + Bevcizum: - - 5 (µg/ml) Reltive MMP LM mock COX MMP LM+
Prentl doi:.8/nture57 Figure S HPMECs LM Cells Cell lines VEGF (ng/ml) Prentl 7. +/-. LM 7. +/-.99 LM 7. +/-.99 Fold COX induction 5 VEGF: - + + + Bevcizum: - - 5 (µg/ml) Reltive MMP LM mock COX MMP LM+
Plant Physiology Preview. Published on February 21, 2017, as DOI: /pp
 Plnt Physiology Preview. Pulished on Ferury 21, 217, s DOI:1.114/pp.16.1928 1 2 3 4 5 6 7 8 9 1 11 12 13 14 15 16 17 18 Running title: Spliing regultor STA1 in het stress dpttion Corresponding Author:
Plnt Physiology Preview. Pulished on Ferury 21, 217, s DOI:1.114/pp.16.1928 1 2 3 4 5 6 7 8 9 1 11 12 13 14 15 16 17 18 Running title: Spliing regultor STA1 in het stress dpttion Corresponding Author:
Adaptive echolocation behavior in bats for the analysis of auditory scenes
 9 The Journl of Experimentl Biology, 9- Pulished y The Compny of Biologists 9 doi:./je.7 Adptive eholotion ehvior in ts for the nlysis of uditory senes Chen Chiu*, Wei Xin nd Cynthi F. Moss Deprtment of
9 The Journl of Experimentl Biology, 9- Pulished y The Compny of Biologists 9 doi:./je.7 Adptive eholotion ehvior in ts for the nlysis of uditory senes Chen Chiu*, Wei Xin nd Cynthi F. Moss Deprtment of
Shear behaviour of regular and irregular rock joints under cyclic conditions
 Pper No. 69 ISMS 2016 Sher ehviour of regulr n irregulr rok joints uner yli onitions S. M. Mhi Niktr, *, K. Seshgiri Ro, Amit Kumr Shrivstv Deprtment of Civil Engineering, Inin Institute of Tehnology Delhi,
Pper No. 69 ISMS 2016 Sher ehviour of regulr n irregulr rok joints uner yli onitions S. M. Mhi Niktr, *, K. Seshgiri Ro, Amit Kumr Shrivstv Deprtment of Civil Engineering, Inin Institute of Tehnology Delhi,
EFFECTS OF AN ACUTE ENTERIC DISEASE CHALLENGE ON IGF-1 AND IGFBP-3 GENE EXPRESSION IN PORCINE SKELETAL MUSCLE
 Swine Dy 22 Contents EFFECTS OF AN ACUTE ENTERIC DISEASE CHALLENGE ON IGF-1 AND IGFBP-3 GENE EXPRESSION IN PORCINE SKELETAL MUSCLE B. J. Johnson, J. P. Kyser, J. D. Dunn, A. T. Wyln, S. S. Dritz 1, J.
Swine Dy 22 Contents EFFECTS OF AN ACUTE ENTERIC DISEASE CHALLENGE ON IGF-1 AND IGFBP-3 GENE EXPRESSION IN PORCINE SKELETAL MUSCLE B. J. Johnson, J. P. Kyser, J. D. Dunn, A. T. Wyln, S. S. Dritz 1, J.
SUPPLEMENTARY INFORMATION
 . Norml Physiologicl Conditions. SIRT1 Loss-of-Function S1. Model for the role of SIRT1 in the regultion of memory nd plsticity. () Our findings suggest tht SIRT1 normlly functions in coopertion with YY1,
. Norml Physiologicl Conditions. SIRT1 Loss-of-Function S1. Model for the role of SIRT1 in the regultion of memory nd plsticity. () Our findings suggest tht SIRT1 normlly functions in coopertion with YY1,
Mechanisms underlying cross-orientation suppression in cat visual cortex
 Mehnisms underlying ross-orienttion suppression in t visul ortex 6 Nture Pulishing Group http://www.nture.om/ntureneurosiene Nihols J Priee & Dvid Ferster In simple ells of the t primry visul ortex, null-oriented
Mehnisms underlying ross-orienttion suppression in t visul ortex 6 Nture Pulishing Group http://www.nture.om/ntureneurosiene Nihols J Priee & Dvid Ferster In simple ells of the t primry visul ortex, null-oriented
Nature Structural & Molecular Biology: doi: /nsmb.2419
 Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Differential expression of cyclin G2, cyclin dependent kinase inhibitor 2C and peripheral myelin protein 22 genes during adipogenesis..
 The Ohio Stte University From the SeletedWorks of Jiin Zhng Summer My 5, 214 Differentil expression of ylin G2, ylin dependent kinse inhiitor 2C nd peripherl myelin protein 22 genes during dipogenesis..pdf
The Ohio Stte University From the SeletedWorks of Jiin Zhng Summer My 5, 214 Differentil expression of ylin G2, ylin dependent kinse inhiitor 2C nd peripherl myelin protein 22 genes during dipogenesis..pdf
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION CD169 + MACROPHAGES PRESENT LIPID ANTIGENS TO MEDIATE EARLY ACTIVATION OF INVARIANT NKT CELLS IN LYMPH NODES Ptrii Brrl, Polo Polzell, Andres Brukuer, Nio vn Rooijen, Gurdyl S.
SUPPLEMENTARY INFORMATION CD169 + MACROPHAGES PRESENT LIPID ANTIGENS TO MEDIATE EARLY ACTIVATION OF INVARIANT NKT CELLS IN LYMPH NODES Ptrii Brrl, Polo Polzell, Andres Brukuer, Nio vn Rooijen, Gurdyl S.
FRAMEstar. 2-Component PCR Plates
 FRAMEstr -Component Pltes FrmeStr two-omponent tehnology redues evportion from pltes, improving results nd llowing for volume redutions to sve on expensive regents. FrmeStr pltes mximise therml stility
FRAMEstr -Component Pltes FrmeStr two-omponent tehnology redues evportion from pltes, improving results nd llowing for volume redutions to sve on expensive regents. FrmeStr pltes mximise therml stility
Laminar sources of synaptic input to cortical inhibitory interneurons and pyramidal neurons
 rtiles Lminr soures of synpti input to ortil inhiitory interneurons nd pyrmidl neurons J. L. Dntzker nd E. M. Cllwy Systems Neuroiology Lortories, The Slk Institute for Biologil Studies, N. Torrey Pines
rtiles Lminr soures of synpti input to ortil inhiitory interneurons nd pyrmidl neurons J. L. Dntzker nd E. M. Cllwy Systems Neuroiology Lortories, The Slk Institute for Biologil Studies, N. Torrey Pines
Chow KD CR HFD. Fed Fast Refed
 Supplementry Figure 1 Control d/d Chow KD CR Fed Fst Refed Supplementry Figure 1: Liver expression in diet nd disese models. () expression in the livers of ontrol nd d/d mie. () expression in the livers
Supplementry Figure 1 Control d/d Chow KD CR Fed Fst Refed Supplementry Figure 1: Liver expression in diet nd disese models. () expression in the livers of ontrol nd d/d mie. () expression in the livers
(% of adherent cells) *** PBL firm adhesion. Frequency (% ) 4 1 L 2 CXCR3 DP-2
 Chemotxis (% of dded ells) PBL totl dhesion (N ells/mm 2 /1.1 6 PBL) Frequeny (% ) PBL firm dhesion Supplementry Figure 1 4 4 3 3 2 2 1.1-4 1-3 1.1.2. 1 1 8 6 4 2 Adiponetin ( g/ml) - + Adiponetin ( g/ml)
Chemotxis (% of dded ells) PBL totl dhesion (N ells/mm 2 /1.1 6 PBL) Frequeny (% ) PBL firm dhesion Supplementry Figure 1 4 4 3 3 2 2 1.1-4 1-3 1.1.2. 1 1 8 6 4 2 Adiponetin ( g/ml) - + Adiponetin ( g/ml)
Once small always small? To what extent morphometric characteristics and postweaning starter regime affect pig lifetime growth performance
 Huting et l. Porine Helth Mngement (2018) 4:21 https://doi.org/10.1186/s40813-018-0098-1 RESEARCH Open Aess One smll lwys smll? To wht extent morphometri hrteristis nd postwening strter regime ffet pig
Huting et l. Porine Helth Mngement (2018) 4:21 https://doi.org/10.1186/s40813-018-0098-1 RESEARCH Open Aess One smll lwys smll? To wht extent morphometri hrteristis nd postwening strter regime ffet pig
Insulin regulation of heart function in aging fruit flies
 4 Nture Pulishing Group http://www.nture.om/nturegenetis Insulin regultion of hert funtion in ging fruit flies Roert J Wessells, Erin Fitzgerld, Jmes R Cypser, Mr Ttr & Rolf Bodmer Insulin-IGF reeptor
4 Nture Pulishing Group http://www.nture.om/nturegenetis Insulin regultion of hert funtion in ging fruit flies Roert J Wessells, Erin Fitzgerld, Jmes R Cypser, Mr Ttr & Rolf Bodmer Insulin-IGF reeptor
The Role of Background Statistics in Face Adaptation
 The Journl of Neurosiene, Septemer 3, 29 29(39):235 244 235 Behviorl/Systems/Cognitive The Role of Bkground Sttistis in Fe Adpttion Jinhu Wu, * Hong Xu, * Peter Dyn, 2 nd Ning Qin Deprtments of Neurosiene
The Journl of Neurosiene, Septemer 3, 29 29(39):235 244 235 Behviorl/Systems/Cognitive The Role of Bkground Sttistis in Fe Adpttion Jinhu Wu, * Hong Xu, * Peter Dyn, 2 nd Ning Qin Deprtments of Neurosiene
RESEARCH ARTICLE. Supplemental Figure 5
 11.5 2 2 11. RESEARCH ARTICLE RBC ( 1 12 /L) 1.5 1. 9.5 PLT ( 1 9 /L) 1 16 14 HGB (g/l) 19 1 17 16 9. 12 4 4 46 Cellulr & Moleulr Immunology dvne online pulition, PCV (%) 44 MCV (fl) 46 44 ; doi:1.13/mi.214.16
11.5 2 2 11. RESEARCH ARTICLE RBC ( 1 12 /L) 1.5 1. 9.5 PLT ( 1 9 /L) 1 16 14 HGB (g/l) 19 1 17 16 9. 12 4 4 46 Cellulr & Moleulr Immunology dvne online pulition, PCV (%) 44 MCV (fl) 46 44 ; doi:1.13/mi.214.16
Minimum effective dose of chenic acid for gallstone patients: reduction with bedtime administration and
 Gut, 1982, 23, 28-284 Minimum effetive dose of heni id for gllstone ptients: redution with bedtime dministrtion nd low holesterol diet D P MUDGL, R M KUPFER, ND T C NORTHFIELD* From the Normn Tnner Gstroenterology
Gut, 1982, 23, 28-284 Minimum effetive dose of heni id for gllstone ptients: redution with bedtime dministrtion nd low holesterol diet D P MUDGL, R M KUPFER, ND T C NORTHFIELD* From the Normn Tnner Gstroenterology
A maternal junk food diet in pregnancy and lactation promotes an exacerbated taste for junk food and a greater propensity for obesity in rat offspring
 British Journl of Nutrition (27), pge 1 of 9 q The uthors 27 doi: 1.117/S7114781237 mternl junk food diet in pregnny nd lttion promotes n exerted tste for junk food nd greter propensity for oesity in rt
British Journl of Nutrition (27), pge 1 of 9 q The uthors 27 doi: 1.117/S7114781237 mternl junk food diet in pregnny nd lttion promotes n exerted tste for junk food nd greter propensity for oesity in rt
A AOAC Official Method Fat (Total, Saturated, Unsaturated, and Monounsaturated) in Cereal Products
 32.2.02A AOAC Offiil Method 996.01 Ft (Totl, Sturted, Unsturted, nd Monounsturted) in Cerel Produts Aid Hydrolysis Cpillry Gs Chromtogrphi Method First Ation 1996 (Applile for determintion of ft in erel
32.2.02A AOAC Offiil Method 996.01 Ft (Totl, Sturted, Unsturted, nd Monounsturted) in Cerel Produts Aid Hydrolysis Cpillry Gs Chromtogrphi Method First Ation 1996 (Applile for determintion of ft in erel
Introduction to Study Designs II
 Introdution to Study Designs II Commonly used study designs in publi helth & epidemiologi reserh Benjmin Rihrd H. Muthmbi, DrPH, MPH Stte HIV Epidemiologist HIV Epidemiology Investigtion Setion PA Deprtment
Introdution to Study Designs II Commonly used study designs in publi helth & epidemiologi reserh Benjmin Rihrd H. Muthmbi, DrPH, MPH Stte HIV Epidemiologist HIV Epidemiology Investigtion Setion PA Deprtment
CAUSES OF DIARRHEA, PNEUMONIA, AND ABORTION IN 1991 CATTLE SUBMISSIONS TO THE KSU VETERINARY DIAGNOSTIC LABORATORY
 CAUSES OF DIARRHEA, PNEUMONIA, AND ABORTION IN 1991 CATTLE SUBMISSIONS TO THE KSU VETERINARY DIAGNOSTIC LABORATORY 1 1 2 R. K. Frnk, M. W. Vorhies, nd M. M. Chengpp Summry Cuses of dirrhe, pneumoni, nd
CAUSES OF DIARRHEA, PNEUMONIA, AND ABORTION IN 1991 CATTLE SUBMISSIONS TO THE KSU VETERINARY DIAGNOSTIC LABORATORY 1 1 2 R. K. Frnk, M. W. Vorhies, nd M. M. Chengpp Summry Cuses of dirrhe, pneumoni, nd
Single-Molecule Studies of Unlabelled Full-Length p53 Protein Binding to DNA
 Single-Molecule Studies of Unlbelled Full-Length p53 Protein Binding to DNA Philipp Nuttll, 1 Kidn Lee, 2 Pietro Ciccrell, 3 Mrco Crminti, 3 Giorgio Ferrri, 3 Ki- Bum Kim, 2 Tim Albrecht 1* 1 Imperil College
Single-Molecule Studies of Unlbelled Full-Length p53 Protein Binding to DNA Philipp Nuttll, 1 Kidn Lee, 2 Pietro Ciccrell, 3 Mrco Crminti, 3 Giorgio Ferrri, 3 Ki- Bum Kim, 2 Tim Albrecht 1* 1 Imperil College
larvi 2013 Epigenetic regulation of muscle development and growth in Senegalese sole larvae Catarina Campos
 lrvi 213 6th fish & shellfish lrviulture symposium Epigeneti regultion of musle development nd growth in Seneglese sole lrve Ctrin Cmpos ghent university, elgium, 2-5 septemer 213 EPIGENETIC REGULATION
lrvi 213 6th fish & shellfish lrviulture symposium Epigeneti regultion of musle development nd growth in Seneglese sole lrve Ctrin Cmpos ghent university, elgium, 2-5 septemer 213 EPIGENETIC REGULATION
Sonic Hedgehog promotes proliferation of Notch-dependent monociliated choroid plexus tumour cells
 Soni Hedgehog promotes prolifertion of Noth-dependent monoilited horoid plexus tumour ells Li Li, Ktie B. Grusm,2, Jun Wng 3, Melody P. Lun 4,5, Jsmin Ohli 6, Hrt G. W. Lidov 4, Moni L. Clihio 4, Erling
Soni Hedgehog promotes prolifertion of Noth-dependent monoilited horoid plexus tumour ells Li Li, Ktie B. Grusm,2, Jun Wng 3, Melody P. Lun 4,5, Jsmin Ohli 6, Hrt G. W. Lidov 4, Moni L. Clihio 4, Erling
SYNOPSIS Final Abbreviated Clinical Study Report for Study CA ABBREVIATED REPORT
 Finl Arevited Clinicl Study Report Nme of Sponsor/Compny: Bristol-Myers Squi Ipilimum Individul Study Tle Referring to the Dossier (For Ntionl Authority Use Only) Nme of Finished Product: Yervoy Nme of
Finl Arevited Clinicl Study Report Nme of Sponsor/Compny: Bristol-Myers Squi Ipilimum Individul Study Tle Referring to the Dossier (For Ntionl Authority Use Only) Nme of Finished Product: Yervoy Nme of
Check your understanding 3
 1 Wht is the difference etween pssive trnsport nd ctive trnsport? Pssive trnsport is the movement of prticles not requiring energy. Movement of prticles in ctive trnsport uses energy. 2 A gs tp in the
1 Wht is the difference etween pssive trnsport nd ctive trnsport? Pssive trnsport is the movement of prticles not requiring energy. Movement of prticles in ctive trnsport uses energy. 2 A gs tp in the
Toxicity effects of seven Cu compounds/nps in Lettuce (Lactuca sativa) and Alfalfa (Medicago sativa)
 Toxiity effets of seven Cu ompounds/nps in Lettue (Ltu stiv) nd Alflf (Medigo stiv) Jie Hong, Lijun Zho, Cyren Rio, Jose R Perlt-Vide, Jorge Grde-Torresdey The University of Texs t El Pso UC-CEIN Theme
Toxiity effets of seven Cu ompounds/nps in Lettue (Ltu stiv) nd Alflf (Medigo stiv) Jie Hong, Lijun Zho, Cyren Rio, Jose R Perlt-Vide, Jorge Grde-Torresdey The University of Texs t El Pso UC-CEIN Theme
Optimisation of diets for Atlantic cod (Gadus morhua) broodstock: effect of arachidonic acid on egg & larval quality
 Optimistion of diets for Atlntic cod (Gdus morhu) roodstock: effect of rchidonic cid on egg & lrvl qulity Dr Gordon Bell, Ms. An Blnco, Dr Bill Roy, Dr Derek Roertson, Dr Jim Henderson nd Mr Richrd Prickett,
Optimistion of diets for Atlntic cod (Gdus morhu) roodstock: effect of rchidonic cid on egg & lrvl qulity Dr Gordon Bell, Ms. An Blnco, Dr Bill Roy, Dr Derek Roertson, Dr Jim Henderson nd Mr Richrd Prickett,
Imaging analysis of clock neurons reveals light buffers the wake-promoting effect of dopamine
 Imging nlysis of lok neurons revels light uffers the wke-promoting effet of dopmine Yuhu Shng 1,2, Pul Hynes 2, Niolás Pírez 2, Kyle I Hrrington 3, Fng Guo 1,2, Jordn Pollk 3, Pengyu Hong 3, Leslie C Griffith
Imging nlysis of lok neurons revels light uffers the wke-promoting effet of dopmine Yuhu Shng 1,2, Pul Hynes 2, Niolás Pírez 2, Kyle I Hrrington 3, Fng Guo 1,2, Jordn Pollk 3, Pengyu Hong 3, Leslie C Griffith
Olfactory behavior and physiology are disrupted in prion protein knockout mice
 Olftory ehvior nd physiology re disrupted in prion protein knokout mie Clire E Le Pihon 1, Mtthew T Vlley 1, Mgdlini Polymenidou 2,3, Alexnder T Chesler 1, Botir T Sgdullev 1,3, Adrino Aguzzi 2 & Sturt
Olftory ehvior nd physiology re disrupted in prion protein knokout mie Clire E Le Pihon 1, Mtthew T Vlley 1, Mgdlini Polymenidou 2,3, Alexnder T Chesler 1, Botir T Sgdullev 1,3, Adrino Aguzzi 2 & Sturt
CSE 5311 Notes 2: Binary Search Trees
 S Notes : inry Ser Trees (Lst upte /7/ 8:7 M) ROTTIONS Single left rottion t (K rotting ege ) Single rigt rottion t (K rotting ege ) F oule rigt rottion t F G F G Wt two single rottions re equivlent? (OTTOM-UP)
S Notes : inry Ser Trees (Lst upte /7/ 8:7 M) ROTTIONS Single left rottion t (K rotting ege ) Single rigt rottion t (K rotting ege ) F oule rigt rottion t F G F G Wt two single rottions re equivlent? (OTTOM-UP)
THE EVALUATION OF DEHULLED CANOLA MEAL IN THE DIETS OF GROWING AND FINISHING PIGS
 THE EVALUATION OF DEHULLED CANOLA MEAL IN THE DIETS OF GROWING AND FINISHING PIGS THE EVALUATION OF DEHULLED CANOLA MEAL IN THE DIETS OF GROWING AND FINISHING PIGS John F. Ptience nd Doug Gillis SUMMARY
THE EVALUATION OF DEHULLED CANOLA MEAL IN THE DIETS OF GROWING AND FINISHING PIGS THE EVALUATION OF DEHULLED CANOLA MEAL IN THE DIETS OF GROWING AND FINISHING PIGS John F. Ptience nd Doug Gillis SUMMARY
REVIEW Study of the Formation of trans Fatty Acids in Model Oils (triacylglycerols) and Edible Oils during the Heating Process
 JARQ 46 (3), 215 220 (2012) http://www.jirs.ffr.go.jp REVIEW Study of the Formtion of trns Ftty Aids in Model Oils (triylglyerols) nd Edible Oils during the Heting Proess Wkko TSUZUKI* Food Resoure Division,
JARQ 46 (3), 215 220 (2012) http://www.jirs.ffr.go.jp REVIEW Study of the Formtion of trns Ftty Aids in Model Oils (triylglyerols) nd Edible Oils during the Heting Proess Wkko TSUZUKI* Food Resoure Division,
 nestin ironetin p75 s1 CNS SKPs Dermo-1 +ve SKPs CNS H2O SCGs Skin Di. SKPs TH SHOX2 GAPDH NCAM D H Figure S1, Immunoytohemil nlysis o SKP spheres ulture rom neontl mouse (nestin, ironetin, S-1) or rt
nestin ironetin p75 s1 CNS SKPs Dermo-1 +ve SKPs CNS H2O SCGs Skin Di. SKPs TH SHOX2 GAPDH NCAM D H Figure S1, Immunoytohemil nlysis o SKP spheres ulture rom neontl mouse (nestin, ironetin, S-1) or rt
SETD1A modulates cell cycle progression through a mirna network that regulates p53 target genes
 SETDA modultes ell yle progression through mirna network tht regultes p5 trget genes The Hrvrd ommunity hs mde this rtile openly vilble. Plese shre how this ess benefits you. Your story mtters Cittion
SETDA modultes ell yle progression through mirna network tht regultes p5 trget genes The Hrvrd ommunity hs mde this rtile openly vilble. Plese shre how this ess benefits you. Your story mtters Cittion
Adiabatic CMOS Circuit Design: Principles and Examples
 Aditi CMOS Ciruit Design: Priniples nd Exmples X.Wu,G.Hng,ndM.Pedrm Astrt: In view of hnging the type of energy onversion in CMOS iruits nd therey hieving ultr-low-power design, this pper investigtes diti
Aditi CMOS Ciruit Design: Priniples nd Exmples X.Wu,G.Hng,ndM.Pedrm Astrt: In view of hnging the type of energy onversion in CMOS iruits nd therey hieving ultr-low-power design, this pper investigtes diti
Mediating Multi-Party Negotiation Through Marker-Based Tracking of Mobile Phones
 Mediting Multi-Prty Negotition Through Mrker-sed Trking of Moile Phones Mihel Rohs Deutshe Telekom Lortories TU erlin, Germny mihel.rohs@telekom.de hristin Kry Informtis Reserh Institute Newstle University,
Mediting Multi-Prty Negotition Through Mrker-sed Trking of Moile Phones Mihel Rohs Deutshe Telekom Lortories TU erlin, Germny mihel.rohs@telekom.de hristin Kry Informtis Reserh Institute Newstle University,
Provide a Buffet and Carvery Service
 CU926 Provide Buffet nd Crvery Servie Unit summry This unit is out prepring the rvery or uffet disply y rrnging items suh s rokery, utlery nd npkins. It lso overs serving ustomers t the rvery or uffet
CU926 Provide Buffet nd Crvery Servie Unit summry This unit is out prepring the rvery or uffet disply y rrnging items suh s rokery, utlery nd npkins. It lso overs serving ustomers t the rvery or uffet
Model for processive movement of myosin V and myosin VI
 Model for proessive movement of myosin V nd myosin VI Ping Xie, Shuo-Xing Dou, nd Peng-Ye Wng Lortory of Soft Mtter Physis, Institute of Physis, Chinese Ademy of Sienes, P.O. ox, 603, eijing 100080, Chin
Model for proessive movement of myosin V nd myosin VI Ping Xie, Shuo-Xing Dou, nd Peng-Ye Wng Lortory of Soft Mtter Physis, Institute of Physis, Chinese Ademy of Sienes, P.O. ox, 603, eijing 100080, Chin
supplementary information
 DOI: 10.1038/nc2089 H3K4me1 H3K4me1 H3K4me1 H3K4me1 H3K4me1 H3K4me1 5 PN N1-2 PN H3K4me1 H3K4me1 H3K4me1 2-cell stge 2-c st cell ge Figure S1 Pttern of loclistion of H3K4me1 () nd () during zygotic development
DOI: 10.1038/nc2089 H3K4me1 H3K4me1 H3K4me1 H3K4me1 H3K4me1 H3K4me1 5 PN N1-2 PN H3K4me1 H3K4me1 H3K4me1 2-cell stge 2-c st cell ge Figure S1 Pttern of loclistion of H3K4me1 () nd () during zygotic development
EFFECTS OF DIETARY CALCIUM LEVELS ON GROWTH-PERFORMANCE AND DIGESTIVE FUNCTION IN CATTLE FED A HIGH-FAT FINISHING DIET
 EFFECTS OF DIETARY CALCIUM LEVELS ON GROWTH-PERFORMANCE AND DIGESTIVE FUNCTION IN CATTLE FED A HIGH-FAT FINISHING DIET R. A. Zinn, Y. Shen, R. Brjs, M. Montño, E. Alvrez, nd E. Rmirez Desert Reserh nd
EFFECTS OF DIETARY CALCIUM LEVELS ON GROWTH-PERFORMANCE AND DIGESTIVE FUNCTION IN CATTLE FED A HIGH-FAT FINISHING DIET R. A. Zinn, Y. Shen, R. Brjs, M. Montño, E. Alvrez, nd E. Rmirez Desert Reserh nd
SUPPLEMENTARY INFORMATION
 doi:.3/nture93 d 5 Rttlesnke DRG (reds) Rttlesnke TG (reds) c 3 TRPV1 other TRPs 1 1 3 Non-pit snke TG (reds) SFig. 1 5 5 3 other TRPs TRPV1 1 1 3 Non-pit snke DRG (reds) 5 Antomy of the pit orgn nd comprison
doi:.3/nture93 d 5 Rttlesnke DRG (reds) Rttlesnke TG (reds) c 3 TRPV1 other TRPs 1 1 3 Non-pit snke TG (reds) SFig. 1 5 5 3 other TRPs TRPV1 1 1 3 Non-pit snke DRG (reds) 5 Antomy of the pit orgn nd comprison
Tbp. Per Relative mrna levels Circadian Time. Liver weight/ body weight (%) n.s. Pernull
 Liver weight/ ody weight (%) Dy Body weight (g) Reltive mrna levels Reltive mrna levels Reltive mrna levels Reltive mrna levels Dy Per1 Per2 Per3 Tp 8 2 8 2. 6 2 8 12162 Cirdin Time 3 2 1 2 1 1 8 12162
Liver weight/ ody weight (%) Dy Body weight (g) Reltive mrna levels Reltive mrna levels Reltive mrna levels Reltive mrna levels Dy Per1 Per2 Per3 Tp 8 2 8 2. 6 2 8 12162 Cirdin Time 3 2 1 2 1 1 8 12162
Input from external experts and manufacturer on the 2 nd draft project plan Stool DNA testing for early detection of colorectal cancer
 Input externl experts nd mnufcturer on the 2 nd drft project pln Stool DNA testing for erly detection of colorectl cncer (Project ID:OTJA10) All s nd uthor s replies on the 2nd drft project pln Stool DNA
Input externl experts nd mnufcturer on the 2 nd drft project pln Stool DNA testing for erly detection of colorectl cncer (Project ID:OTJA10) All s nd uthor s replies on the 2nd drft project pln Stool DNA
Progestin effects on cell proliferation pathways in the postmenopausal mammary gland
 Wood et l. Brest Cner Reserh 213, 15:R62 http://rest-ner-reserh.om/ontent/15/4/r62 RESEARCH ARTICLE Open Aess Progestin effets on ell prolifertion pthwys in the postmenopusl mmmry glnd Chrles E Wood 1,
Wood et l. Brest Cner Reserh 213, 15:R62 http://rest-ner-reserh.om/ontent/15/4/r62 RESEARCH ARTICLE Open Aess Progestin effets on ell prolifertion pthwys in the postmenopusl mmmry glnd Chrles E Wood 1,
The Hippo/YAP pathway interacts with EGFR signaling and HPV oncoproteins to regulate cervical cancer progression
 Reserh Artile The Hippo/ pthwy interts with EGFR signling nd HPV onoproteins to regulte ervil ner progression Chuno He 1,, Dgn Mo 1,3, Guohu Hu 1,, Xingmin Lv 1, Xingheng Chen, Peter C Angeletti 5, Jixin
Reserh Artile The Hippo/ pthwy interts with EGFR signling nd HPV onoproteins to regulte ervil ner progression Chuno He 1,, Dgn Mo 1,3, Guohu Hu 1,, Xingmin Lv 1, Xingheng Chen, Peter C Angeletti 5, Jixin
LETTER. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2
 doi:.8/nture98 Impired hydroxyltion of -methylytosine in myeloid ners with mutnt TET Myunggon Ko {, Yun Hung {, Ann M. Jnkowsk, Utz J. Ppe,, Mmt Thilini, Hozef S. Bndukwl, Jungeun An {, Edwrd D. Lmperti,
doi:.8/nture98 Impired hydroxyltion of -methylytosine in myeloid ners with mutnt TET Myunggon Ko {, Yun Hung {, Ann M. Jnkowsk, Utz J. Ppe,, Mmt Thilini, Hozef S. Bndukwl, Jungeun An {, Edwrd D. Lmperti,
Meat and Food Safety. B.A. Crow, M.E. Dikeman, L.C. Hollis, R.A. Phebus, A.N. Ray, T.A. Houser, and J.P. Grobbel
 Met nd Food Sfety Needle-Free Injection Enhncement of Beef Strip Loins with Phosphte nd Slt Hs Potentil to Improve Yield, Tenderness, nd Juiciness ut Hrm Texture nd Flvor B.A. Crow, M.E. Dikemn, L.C. Hollis,
Met nd Food Sfety Needle-Free Injection Enhncement of Beef Strip Loins with Phosphte nd Slt Hs Potentil to Improve Yield, Tenderness, nd Juiciness ut Hrm Texture nd Flvor B.A. Crow, M.E. Dikemn, L.C. Hollis,
N6-methyladenosine (m6a) is the most prevalent messenger
 https://oi.org/8/s556-8-7- m 6 A mrna methyltion regultes tivity to promote the prolifertion n tumorigeniity of enometril ner Jun Liu,,, Mrk A. Ekert,, Bryn T. Hr,,, Song-Mei Liu,, Zhike Lu,, Kngkng Yu,,5,
https://oi.org/8/s556-8-7- m 6 A mrna methyltion regultes tivity to promote the prolifertion n tumorigeniity of enometril ner Jun Liu,,, Mrk A. Ekert,, Bryn T. Hr,,, Song-Mei Liu,, Zhike Lu,, Kngkng Yu,,5,
GSK-3 is a master regulator of neural progenitor homeostasis
 GSK-3 is mster regultor of neurl progenitor homeostsis Woo-Yng Kim 1, Xinshuo Wng 1, Yohong Wu 1, Brdley W Dole 2, Stish Ptel 3, Jmes R Woodgett 3 & Willim D Snider 1 The development of the rin requires
GSK-3 is mster regultor of neurl progenitor homeostsis Woo-Yng Kim 1, Xinshuo Wng 1, Yohong Wu 1, Brdley W Dole 2, Stish Ptel 3, Jmes R Woodgett 3 & Willim D Snider 1 The development of the rin requires
Role of HCP5-miR-139-RUNX1 Feedback Loop in Regulating Malignant Behavior of Glioma Cells
 originl rtile The Amerin Soiety of Gene & Cell Therpy Role of HCP-miR-19-RUNX1 Feedk Loop in Regulting Mlignnt Behvior of Gliom Cells Ho Teng 1,2, Ping Wng, Yixue Xue, Xioi Liu 1,2, Jun M, Heng Ci 1,2,
originl rtile The Amerin Soiety of Gene & Cell Therpy Role of HCP-miR-19-RUNX1 Feedk Loop in Regulting Mlignnt Behvior of Gliom Cells Ho Teng 1,2, Ping Wng, Yixue Xue, Xioi Liu 1,2, Jun M, Heng Ci 1,2,
TNF-a Downregulates Filaggrin and Loricrin through c-jun N-terminal Kinase: Role for TNF-a Antagonists to Improve Skin Barrier
 ORIGINAL ARTICLE TNF- Downregultes Filggrin nd Loririn through -Jun N-terminl Kinse: Role for TNF- Antgonists to Improve Skin Brrier Byung Eui Kim, Mihel D. Howell,, Emm Guttmn,, Ptrii M. Gilleudeu, Irm
ORIGINAL ARTICLE TNF- Downregultes Filggrin nd Loririn through -Jun N-terminl Kinse: Role for TNF- Antgonists to Improve Skin Brrier Byung Eui Kim, Mihel D. Howell,, Emm Guttmn,, Ptrii M. Gilleudeu, Irm
Prediction of the Wrist Joint Position During a Postural Tremor Using Neural Oscillators and an Adaptive Controller
 Short Communition www.jmss.mui..ir Predition of the Wrist Joint Position During Posturl Tremor Using Neurl Osilltors nd n Adptive Controller Hmid Rez Korvi, Sr Hemmti Ali 1, Msood Vtndoust, Rsoul Mrvi
Short Communition www.jmss.mui..ir Predition of the Wrist Joint Position During Posturl Tremor Using Neurl Osilltors nd n Adptive Controller Hmid Rez Korvi, Sr Hemmti Ali 1, Msood Vtndoust, Rsoul Mrvi
The soy isoflavone genistein promotes apoptosis in mammary epithelial cells by inducing the tumor suppressor PTEN
 Crinogenesis vol. no.1 pp.1793 183, 5 doi:1.193/rin/gi131 dvne ess pulition My 19, 5 The soy isoflvone genistein promotes poptosis in mmmry epithelil ells y induing the tumor suppressor huvnesh Dve 1,,
Crinogenesis vol. no.1 pp.1793 183, 5 doi:1.193/rin/gi131 dvne ess pulition My 19, 5 The soy isoflvone genistein promotes poptosis in mmmry epithelil ells y induing the tumor suppressor huvnesh Dve 1,,
