b PolyA RNA-seq c RNA-seq read distribution FPKM
|
|
|
- Allen Rose
- 5 years ago
- Views:
Transcription
1 a Repliate 2 (FPKM) Poly+ RN-seq R2 = Repliate 1 (FPKM) Repliate 2 (FPKM) Poly RN-seq R2 = Repliate 1 (FPKM) RN-seq rea istriution Intron 12% 14% Intergeni Poly+ 77% Exon Intron Intergeni 27% 36% Poly 38% Exon anonial histones Hist3h3 Hist3h2 Hist3h2a Hist3h2a Hist2h4 Hist2h2e Hist2h2a Hist2h2a Hist2h2aa3 Hist1h4m Hist1h4 Hist1h3 Hist1h2q Hist1h2o Hist1h2l Hist1h2h Hist1h2f Hist1h2 Hist1h2l1 Hist1h2 Hist1h2a Hist1h2ao Hist1h2ak Hist1h2ail1 Hist1h2ail Hist1h2ail Hist1h2ai Hist1h2ah Hist1h2ah Hist1h2af Hist1h2a Hist1h2aa Hist1h1 Hist1h1 Hist1h1a Poly+ Poly FPKM Supplementary Figure 1. Poly+ an Poly RN-seq valiation. a, orrelation etween iniviual iologial repliates for Poly+ RN-seq (linear regression, P < 1)., orrelation etween iniviual iologial repliates for Poly RN-seq (linear regression, P < 1)., Distriution of mappe sequene reas in Poly+ (top) an Poly (ottom) RN-seq. Perentages sum to greater than ue to rea overhang etween features., Expression estimates from Poly+ an Poly RN-seq for anonial histone transripts, whih are known to lak polyaenylation signals. Reas mapping to these transripts were more aunant in the Poly lirary for all histone genes, iniating speifi seletion of Poly transripts in the Poly RN-seq. Data are expresse as mean ± s.e.m.
2 a MBD-seq Extrat genomi DN, fragment Forwar reas m TSS Reverse reas - alignment to annotate genome - ioinformatis - Inuate with MBD2, apture with magneti eas T T - lirary onstrution - next generation sequening - MBD2 Biotin Elute methylate DN Streptaviin Magneti ea ene methylation (RPM) Repliate n ou - n MBD-seq (promoter) R2 = ene methylation (RPM) Repliate 1 BD M MBD-seq (gene oy) 1-1 ap MBD-apture Promoter methylation (RPM) Repliate 2 m DN/non-m DN 1-2 R2 = Promoter methylation (RPM) Repliate 1 e ene Rattus norvegius (Rn5 assemly) hr4: pr19 rel2 kn1 pol1 Dx47 m Repliate 1 Input Repliate 2 2k Supplementary Figure 2. MBD-seq pipeline an valiation. a, MBD-seq workflow., Valiation of methylate DN apture with MBD-IP. Inuation with MBD2 resulte in roust enrihment of methylate DN in IP sample an epletion from unoun fration (n = 4 per group; 2-taile Mann-Whitney test; P = 286). Data are expresse as mean ± s.e.m., orrelation of gene oy methylation levels etween iniviual iologial repliates in MBD-seq (linear regression, P < 1 for eah omparison)., orrelation of promoter methylation levels etween iniviual iologial repliates in MBD-seq. e, Representative genomi lous highlights DN methylation patterns surrouning gene oies an lak of efine peaks in input (non-ip) sample.
3 m - input k p islan Start En Intergeni ll islans Intrageni Promoter TSS +1k ll p Islans Intergeni Intrageni Promoter Overlapping TSS - DN methylation (m - input) m - input Promoter m (-2.5 to 2.5k from TSS) Non-I p < 1 p < 1 I ern quartile m - input Intrageni pi m p < 1 ern quartile Supplementary Figure 3. enome-wie DN methylation y p islan status. a, Meta-feature profile of DN methylation at p islans, stratifie y genomi loation. TSS-spanning p islans are hypomethylate, whereas intergeni p islans are hypermethylate. Data represents average methylation aross ategories, aligne to p islan start an en loations., omparison of p islan methylation y genomi feature (n = per group; one-way NOV, F(4,35696) = 176.2; P < 1, Tukey s post-ho test for iniviual omparisons)., Promoter DN methylation aoring to ern rank, stratifie y presene (re points) or asene (orange points) of p islan (., DN methylation at intrageni p islans, y ern rank of orresponing gene. ern levels are signifiantly orrelate with methylation of intrageni p islans.
4 RNPII inhiitor DRB (4hr treatment) RNPIII inhiitor ML-6218 (4hr treatment) Fos mrn tf4 mrn Fos ern * tf4 ern Fos mrn tf4 mrn * Fos ern tf4 ern RNP3 hip Fos Promoter Kl Supplementary Figure 4. Fos an tf4 ern are ifferentially regulate y RN polymerases. a, Treatment with the RN polymerase II-epenent transriptional inhiitor 5,6-ihloro-1-eta-D-riofuranosylenzimiazole (DRB) loke Fos mrn an ern proution. DRB treatment ha no effet on tf4 ern ut erease tf4 mrn in a ose-epenent manner., Treatment with the RN polymerase III inhiitor ML-6218 ha no effet on Fos or tf4 mrn ut erease Fos an tf4 ern (n = per group for DRB experiments an per group for ML-6218 experiments; one-way NOV, Tukey s post-ho test for multiple omparisons)., fter Kl inution, RN polymerase III ining inreases at the Fos promoter (n = 2 per group; Stuent s t-test versus vehile, P = 396). ll ata are expresse as mean ± s.e.m. Iniviual omparisons, *P < 5, P < 1, *P < 1, an P <1.
5 ern-1 Sequene 5 -y Base omposition :3 :9 :6 :7 /:2 Preite seonary struture 1 proaility omplexes Proe Free proe +DNMT3a-D ern-1 ern-2 Sequene 5 -y Base omposition :5 :6 :7 :7 /:3 Preite seonary struture 1 proaility omplexes Proe Free proe +DNMT3a-D ern-2 Free proe Ig DNMT3a-D ern-1 poly I- omplexes Proe mutant ern-1 Sequene 5 -y Base omposition :3 :9 :6 :7 /:4 Preite seonary struture 1 proaility omplexes Proe Free proe +DNMT3a-D mutant ern-1 mutant ern-2 Sequene 5 -y Base omposition :5 :6 :7 :7 /:3 Preite seonary struture 1 proaility omplexes Proe Free proe +DNMT3a-D mutant ern-2 Free proe Ig DNMT3a-D ern-2 poly I- omplexes Proe Supplementary Figure 5. ern interations with DNMT3a are not regulate y stem-loop seonary struture an are maintaine in the presene of a non-speifi ompetitor. a, Base sequene, omposition, an preite seonary struture for syntheti ern-1 an mutant ern-1 proes. Mutant ern-1 possesses iential ase omposition ut laks seonary struture. DNMT3a-D ining was not impaire y removal of seonary struture., Base sequene, omposition, an preite seonary struture for syntheti ern-2 an mutant ern-2 proes. Mutant ern-2 possesses iential ase omposition ut laks seonary struture. DNMT3a-D ining is not altere in mutant ern-2., ern-1 an ern-2 proes (1nM) o not in to Ig (.2μM). Inreasing amounts of the non-speifi ompetitor poly I- (1nM-nM) oes not aolish Fos ern ining to DNMT3a-D.
6 Fos gene MeDIP DN methylation (fol versus vehile) Enhaner Promoter ene oy ile mrn SO-2 ern SO-1 * Fos mrn SO * Fos ern SO-1 treatment Fos ern SO SO treatment (ays) * n.s mrn ern Supplementary Figure 6. SO meiate knokown of Fos mrn an ern in ortial ultures. a, Fos ern knokown resulte in inrease enhaner, promoter, an gene oy methylation (n = 4 per group; two-way NOV versus vehile, Tukey s post-ho test for multiple omparisons)., SO meiate ern knokown persiste for up to 7 ays, an resulte in a progressive eline in Fos mrn expression (n = 6 per group, two-way NOV, Siak s post-ho test for multiple omparisons). Data are expresse as mean ± s.e.m. Iniviual omparisons, *P < 5, P < 1, *P < 1, an P <1.
7 1 2 juste 3 ern 3' ern aniate genes (1,771) quantifiation 5' gene X Poly+ RPM 2k 3' Poly- RPM 3' ern laking genes (6,948) Poly- RPM juste 3' ern inex = Poly+ RPM ,719 juste 3 ern rank Poly+ (FPKM) mrn p < 1 m - input juste 3 ern inex Promoter (-2.5 to 2.5k) juste 3 ern quartile (3 ern aniates only) p < 1 e tf4 ria2 Fos Poly- > Poly+ Poly- < Poly+ Promoter m perentile Promoter m - input No 3'eRN 3'eRN Density (a.u.) Min Max juste 3 ern perentile Supplementary Figure 7. lternative analysis onfirms genome-wie relationship etween ern an promoter DN methylation. a, lternative ern quantifiation ajusts ern levels for Poly+ expression for the same lous, removing potential ontamination from overlapping genes or inorret gene annotations., lternative analysis reveale at least 1,771 genes with 3 Poly- RPM greater than 3 Poly+ RPM., This group of genes exhiite less promoter methylation than genes laking 3 ern proution. -e, 3 ern status traks mrn an DN methylation.
100 μm. Axon growth cones. Tubulin (red) + scr (green)
 Supplementary Figures mirorna-9 regulates axon extension an ranhing y targeting Map1 in mouse ortial neurons Dajas-Bailaor, F., Bonev, B., Garez, P., Stanley, P., Guillemot, F., Papalopulu, N. a 2 μm 1
Supplementary Figures mirorna-9 regulates axon extension an ranhing y targeting Map1 in mouse ortial neurons Dajas-Bailaor, F., Bonev, B., Garez, P., Stanley, P., Guillemot, F., Papalopulu, N. a 2 μm 1
Supplementary Material for. Inflammation induced repression of Foxp3-bound chromatin in regulatory T cells
 Supplementary Material for Inflammation inue repression of -oun hromatin in regulatory T ells Aaron Arvey 1,2, Joris van er Veeken 1,2, Roert M. Samstein 1,2, Yongqiang Feng 1,2, John A. Stamatoyannopoulos
Supplementary Material for Inflammation inue repression of -oun hromatin in regulatory T ells Aaron Arvey 1,2, Joris van er Veeken 1,2, Roert M. Samstein 1,2, Yongqiang Feng 1,2, John A. Stamatoyannopoulos
Supplementary Figure 1 - Ling
 HA1 (%) Insulin seretion (SI) e 7. p=....... 7 p=.7 1 7 a-ell granules -ell granules BMI (kg/m ) -ells / (a- + -ells) p=. 1 7 1. p=.1..... 7 Age: -ell ontent: % Age: -ell ontent: % Age: -ell ontent: %
HA1 (%) Insulin seretion (SI) e 7. p=....... 7 p=.7 1 7 a-ell granules -ell granules BMI (kg/m ) -ells / (a- + -ells) p=. 1 7 1. p=.1..... 7 Age: -ell ontent: % Age: -ell ontent: % Age: -ell ontent: %
% of Nestin-EGFP (+) cells
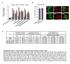 8 7 6 3 Nestin CD % of Nestin-EGFP (+) ells 8 6 Nestin-EGFP Marge Nestin-EGFP Marge Expression levels of Expression levels of Tumorigeniity y stemness markers ifferentiation markers limiting ilution assay
8 7 6 3 Nestin CD % of Nestin-EGFP (+) ells 8 6 Nestin-EGFP Marge Nestin-EGFP Marge Expression levels of Expression levels of Tumorigeniity y stemness markers ifferentiation markers limiting ilution assay
a n a ly s i s Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture
 Proailisti moeling of Hi-C ontat maps eliminates systemati iases to haraterize gloal hromosomal arhiteture Eitan Yaffe & Amos Tanay 2 Nature Ameria, In. All rights reserve. Hi-C experiments measure the
Proailisti moeling of Hi-C ontat maps eliminates systemati iases to haraterize gloal hromosomal arhiteture Eitan Yaffe & Amos Tanay 2 Nature Ameria, In. All rights reserve. Hi-C experiments measure the
Spontaneous persistent ac.vity in entorhinal cortex modulates cor.co- hippocampal
 Spontaneous persistent a.vity in entorhinal ortex moulates or.o- hippoampal intera.ons in vivo Thomas T. G. Hahn, James M. MFarlan, Sven Bererih, Bert Sakmann, Mayank R. Mehta Neoortial persistent Up states
Spontaneous persistent a.vity in entorhinal ortex moulates or.o- hippoampal intera.ons in vivo Thomas T. G. Hahn, James M. MFarlan, Sven Bererih, Bert Sakmann, Mayank R. Mehta Neoortial persistent Up states
Learned spatiotemporal sequence recognition and prediction in primary visual cortex
 Supplementary Materials for Learned spatiotemporal sequene reognition and predition in primary visual ortex Jeffrey P. Gavornik and Mark F. Bear Howard Hughes Medial Institute Piower Institute for Learning
Supplementary Materials for Learned spatiotemporal sequene reognition and predition in primary visual ortex Jeffrey P. Gavornik and Mark F. Bear Howard Hughes Medial Institute Piower Institute for Learning
Table S1. Total and mapped reads produced for each ChIP-seq sample
 Tale S1. Total and mapped reads produced for each ChIP-seq sample Sample Total Reads Mapped Reads Col- H3K27me3 rep1 125662 1334323 (85.76%) Col- H3K27me3 rep2 9176437 7986731 (87.4%) atmi1a//c H3K27m3
Tale S1. Total and mapped reads produced for each ChIP-seq sample Sample Total Reads Mapped Reads Col- H3K27me3 rep1 125662 1334323 (85.76%) Col- H3K27me3 rep2 9176437 7986731 (87.4%) atmi1a//c H3K27m3
Partial Occlusion Modulates Contour-Based Shape Encoding in Primate Area V4
 12 The Journal of Neurosiene, Marh 16, 11 31(11):12 24 Behavioral/Systems/Cognitive Partial Olusion Moulates Contour-Base Shape Enoing in Primate Area V4 Brittany N. Bushnell, Philip J. Haring, Yoshito
12 The Journal of Neurosiene, Marh 16, 11 31(11):12 24 Behavioral/Systems/Cognitive Partial Olusion Moulates Contour-Base Shape Enoing in Primate Area V4 Brittany N. Bushnell, Philip J. Haring, Yoshito
Supplementary Figure 1
 Supplementary Figure 1 how HFD how HFD Epi WT p p Hypothalamus p p Inguinal WT T Liver Lean mouse adipocytes p p p p p p Obese mouse adipocytes Kidney Muscle Spleen Heart p p p p p p p p Extracellular
Supplementary Figure 1 how HFD how HFD Epi WT p p Hypothalamus p p Inguinal WT T Liver Lean mouse adipocytes p p p p p p Obese mouse adipocytes Kidney Muscle Spleen Heart p p p p p p p p Extracellular
Supplementary Information
 Supplementary Information Optial ontrolling reveals time-dependent roles for adult-orn dentate granule ells Yan Gu, Maithe Arruda-Carvalho 3,4, Jia Wang, Stephen Janoshka,, Sheena Josselyn 3-5, Paul Frankland
Supplementary Information Optial ontrolling reveals time-dependent roles for adult-orn dentate granule ells Yan Gu, Maithe Arruda-Carvalho 3,4, Jia Wang, Stephen Janoshka,, Sheena Josselyn 3-5, Paul Frankland
SUPPLEMENTARY INFORMATION
 { OI: 1.138/n31 Srifie n nlyze APs on week 1 s of iet 1 4 6 High-ft iet BrU High-ft iet BrU 4 High-ft iet BrU 6 High-ft iet BrU Lin - Lin - : C34 + : C9 + 1 1 3 1 4 1 5 C45 1 C34 1 1 1 1 3 1 4 1 5 S-1
{ OI: 1.138/n31 Srifie n nlyze APs on week 1 s of iet 1 4 6 High-ft iet BrU High-ft iet BrU 4 High-ft iet BrU 6 High-ft iet BrU Lin - Lin - : C34 + : C9 + 1 1 3 1 4 1 5 C45 1 C34 1 1 1 1 3 1 4 1 5 S-1
Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq
 Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq Philipp Bucher Wednesday January 21, 2009 SIB graduate school course EPFL, Lausanne ChIP-seq against histone variants: Biological
Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq Philipp Bucher Wednesday January 21, 2009 SIB graduate school course EPFL, Lausanne ChIP-seq against histone variants: Biological
Rho-kinase Regulates Energy Balance by Targeting Hypothalamic Leptin Receptor Signaling
 Rho-kinase Regulates Energy Balane y Targeting Hypothalami Leptin Reeptor Signaling Hu Huang 1, Dong Kong 1, Kyung Hee Byun 2, Chianping Ye 1, Shuihi Koda 1, Dae Ho Lee 1, Byung-Chul Oh 2, Sam W Lee 3,
Rho-kinase Regulates Energy Balane y Targeting Hypothalami Leptin Reeptor Signaling Hu Huang 1, Dong Kong 1, Kyung Hee Byun 2, Chianping Ye 1, Shuihi Koda 1, Dae Ho Lee 1, Byung-Chul Oh 2, Sam W Lee 3,
Tethering by lamin A stabilizes and targets the ING1 tumour suppressor
 Tethering y lamin A stailizes an targets the tumour suppressor Xijing Han 1,6, Xiaolan Feng 1,6, Jerome B. Rattner 2, Heather Smith 1, Pinaki Bose 1, Keiko Suzuki 1, Mohame A. Soliman 1,3, Mihelle S. Sott
Tethering y lamin A stailizes an targets the tumour suppressor Xijing Han 1,6, Xiaolan Feng 1,6, Jerome B. Rattner 2, Heather Smith 1, Pinaki Bose 1, Keiko Suzuki 1, Mohame A. Soliman 1,3, Mihelle S. Sott
genomics for systems biology / ISB2020 RNA sequencing (RNA-seq)
 RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
Figure S1, Beyer et al.
 Figure S1, eyer et al. Pax7 Myogenin si sitrl Hoechst T = 72h 14 1.8.6.4.2 12 1 8 6 4 2 24h 48h 96h diff. sitrl siset1 212 72h diff. b1 td r t Se km MyH Vinculin Myogenin β-ctin Vinculin MW b1 ka td r
Figure S1, eyer et al. Pax7 Myogenin si sitrl Hoechst T = 72h 14 1.8.6.4.2 12 1 8 6 4 2 24h 48h 96h diff. sitrl siset1 212 72h diff. b1 td r t Se km MyH Vinculin Myogenin β-ctin Vinculin MW b1 ka td r
ALX4, an epigenetically down regulated tumor suppressor, inhibits breast cancer progression by interfering Wnt/β-catenin pathway
 Yang et al. Journal of Experimental & Clinial Caner Researh (2017) 36:170 DOI 10.1186/s13046-017-0643-9 RESEARCH Open Aess ALX4, an epigenetially own regulate tumor suppressor, inhiits reast aner progression
Yang et al. Journal of Experimental & Clinial Caner Researh (2017) 36:170 DOI 10.1186/s13046-017-0643-9 RESEARCH Open Aess ALX4, an epigenetially own regulate tumor suppressor, inhiits reast aner progression
100 osimertinib. concentration of drugs (nm) Ba/F3 C797S/del19. concentration of drugs (nm) Ba/F3 C797S/T790M/L858R. relative cell vaibility (% of
 Ba/F3 del19 Ba/F3 L858R a afatini afatini EGF-816 EGF-816 ty (% of ontrol) relative ell vaiilit elative ell vaiility (% of ontrol) 5 1 1 5 ty (% of ontrol) relative ell vaiilit 5 1 1 Ba/F3 T79M/L858R Ba/F3
Ba/F3 del19 Ba/F3 L858R a afatini afatini EGF-816 EGF-816 ty (% of ontrol) relative ell vaiilit elative ell vaiility (% of ontrol) 5 1 1 5 ty (% of ontrol) relative ell vaiilit 5 1 1 Ba/F3 T79M/L858R Ba/F3
Supporting information
 Supporting information Evolution of Hollow TiO 2 Nanostrutures via the Kirkendall Effet Driven y Cation Exhange with Enhaned Photoeletrohemial Performane Yanhao Yu, 1 Xin Yin, 1 Alexander Kvit, 1,2 Xudong
Supporting information Evolution of Hollow TiO 2 Nanostrutures via the Kirkendall Effet Driven y Cation Exhange with Enhaned Photoeletrohemial Performane Yanhao Yu, 1 Xin Yin, 1 Alexander Kvit, 1,2 Xudong
Nature Immunology: doi: /ni Supplementary Figure 1. Characteristics of SEs in T reg and T conv cells.
 Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Rice in vivo RNA structurome reveals RNA secondary structure conservation and divergence in plants
 Rice in vivo RN structurome reveals RN secondary structure conservation and divergence in plants Hongjing Deng 1,2,,5, Jitender heema 3, Hang Zhang 2, Hugh Woolfenden 2, Matthew Norris 2, Zhenshan Liu
Rice in vivo RN structurome reveals RN secondary structure conservation and divergence in plants Hongjing Deng 1,2,,5, Jitender heema 3, Hang Zhang 2, Hugh Woolfenden 2, Matthew Norris 2, Zhenshan Liu
Funding: MPIC/Agrochemical Industry USDA/ARS
 Evaluation and omparison of iofungiides and fungiides for the ontrol of post harvest potato tuer diseases. E. Gahango, W. W. Kirk, P. S. Wharton, R. Shafer and P. Tumalam. Department of Plant Pathology,
Evaluation and omparison of iofungiides and fungiides for the ontrol of post harvest potato tuer diseases. E. Gahango, W. W. Kirk, P. S. Wharton, R. Shafer and P. Tumalam. Department of Plant Pathology,
FOXO3 signalling links ATM to the p53 apoptotic pathway following DNA damage
 Reeive 19 Jan 212 Aepte 11 Jul 212 Pulishe 14 Aug 212 DOI: 1.138/nomms28 signalling links to the p apoptoti pathway following DNA amage Young Min Chung 1, *, See-Hyoung Park 1, *, Wen-Bin Tsai 2, Shih-Ya
Reeive 19 Jan 212 Aepte 11 Jul 212 Pulishe 14 Aug 212 DOI: 1.138/nomms28 signalling links to the p apoptoti pathway following DNA amage Young Min Chung 1, *, See-Hyoung Park 1, *, Wen-Bin Tsai 2, Shih-Ya
SUPPLEMENTARY INFORMATION
 % ells with ili (mrke y A-Tu) Reltive Luiferse % ells with ili (mrke y Arl13) % ells with ili DOI: 1.138/n2259 A-Tuulin Hoehst % Cilite Non-ilite -Serum 9% 8% 7% 1 6% % 4% +Serum 1 3% 2% 1% % Serum: -
% ells with ili (mrke y A-Tu) Reltive Luiferse % ells with ili (mrke y Arl13) % ells with ili DOI: 1.138/n2259 A-Tuulin Hoehst % Cilite Non-ilite -Serum 9% 8% 7% 1 6% % 4% +Serum 1 3% 2% 1% % Serum: -
Spleen VAT FMO. Nature Immunology: doi: /ni Ki67 DX5 CD27. CD11b 2B4 KLRG1 CD69 NKG2D CXCR3 CD44 NKG2A/C/E CD62L CD103 CD94 CD48.
 IFN (% of ells) CD7 Marophages Medium NCR LPS PMA/Ionomyin CD IFN (% of ells) T ells Marophages B ells DX CD9 KLRG NKGD B CDL CD CD CXCR CD9 CD NKGA/C/E NCR Ly9h Ly9G Ly9d Ly9i Ly9/i/h FMO CD7 CD9a CD7
IFN (% of ells) CD7 Marophages Medium NCR LPS PMA/Ionomyin CD IFN (% of ells) T ells Marophages B ells DX CD9 KLRG NKGD B CDL CD CD CXCR CD9 CD NKGA/C/E NCR Ly9h Ly9G Ly9d Ly9i Ly9/i/h FMO CD7 CD9a CD7
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Figure 1. Dnmt3b expression in murine and human knee joint cartilage. (A) Representative images
 Figure Legends Figure. expression in murine and human knee joint cartilage. () Representative images showing that Dnmta is not expressed in chondrocytes from mo W articular cartilage [Dnmta expression
Figure Legends Figure. expression in murine and human knee joint cartilage. () Representative images showing that Dnmta is not expressed in chondrocytes from mo W articular cartilage [Dnmta expression
Overview. On the computational aspects of sign language recognition. What is ASL recognition? What makes it hard? Christian Vogler
 On the omputational aspets of sign language reognition Christian Vogler Overview Problem statement Basi probabilisti framework Reognition of multiple hannels Reognition features Disussion Gallaudet Researh
On the omputational aspets of sign language reognition Christian Vogler Overview Problem statement Basi probabilisti framework Reognition of multiple hannels Reognition features Disussion Gallaudet Researh
Supplementary Figure 1. Schematic illustrating major conclusions of this study.
 ORNs GABA A GABA B glomeruli LN PNs Supplementary Figure 1. Shemati illustrating major onlusions of this study. This study represents the most diret evidene to date of inhiitory interations etween olfatory
ORNs GABA A GABA B glomeruli LN PNs Supplementary Figure 1. Shemati illustrating major onlusions of this study. This study represents the most diret evidene to date of inhiitory interations etween olfatory
Supplemental Figures Legends and Supplemental Figures. for. pirna-guided slicing of transposon transcripts enforces their transcriptional
 Supplemental Figures Legends and Supplemental Figures for pirn-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear pirn repertoire Kirsten-ndré
Supplemental Figures Legends and Supplemental Figures for pirn-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear pirn repertoire Kirsten-ndré
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Use of Parents, Sibs, and Unrelated Controls for Detection of Associations between Genetic Markers and Disease
 Am. J. Hum. Genet. 63:492 506, 998 Use of Parents, Sibs, an Unrelate Controls for Detetion of Assoiations between Geneti Markers an Disease Daniel J. Shai,2 an Charles Rowlan Departments of Health Sienes
Am. J. Hum. Genet. 63:492 506, 998 Use of Parents, Sibs, an Unrelate Controls for Detetion of Assoiations between Geneti Markers an Disease Daniel J. Shai,2 an Charles Rowlan Departments of Health Sienes
Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the
 Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the genome-wide methylation microarray data. Mean ± s.d.; Student
Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the genome-wide methylation microarray data. Mean ± s.d.; Student
Confinement-optimized three-dimensional T cell amoeboid motility is modulated via myosin IIA regulated adhesions
 Confinement-optimize three-imensional T ell amoeoi motility is moulate via myosin IIA regulate ahesions Joran Jaoelli 1, Rahel S Frieman 1, Mary Anne Conti 2, Ana-Maria Lennon-Dumenil 3, Matthieu Piel
Confinement-optimize three-imensional T ell amoeoi motility is moulate via myosin IIA regulate ahesions Joran Jaoelli 1, Rahel S Frieman 1, Mary Anne Conti 2, Ana-Maria Lennon-Dumenil 3, Matthieu Piel
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Implication of Long noncoding RNAs in the endothelial cell response to hypoxia revealed y RNAsequencing. Voellenkle C., Garcia-Manteiga J. M., Pedrotti S., Perfetti A., De Toma
SUPPLEMENTARY INFORMATION Implication of Long noncoding RNAs in the endothelial cell response to hypoxia revealed y RNAsequencing. Voellenkle C., Garcia-Manteiga J. M., Pedrotti S., Perfetti A., De Toma
iii Neurones carry information in the form of electrical impulses. Sensory neurones carry impulses from receptors to the central nervous system.
 Page 4 Coordination 1 a Brain (1); spinal ord (1) i Motor (1) i Neurones arry information in the form of eletrial impulses. Sensory neurones arry impulses from reeptors to the entral nervous system. (3)
Page 4 Coordination 1 a Brain (1); spinal ord (1) i Motor (1) i Neurones arry information in the form of eletrial impulses. Sensory neurones arry impulses from reeptors to the entral nervous system. (3)
Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes
 Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes Kaifu Chen 1,2,3,4,5,10, Zhong Chen 6,10, Dayong Wu 6, Lili Zhang 7, Xueqiu Lin 1,2,8,
Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes Kaifu Chen 1,2,3,4,5,10, Zhong Chen 6,10, Dayong Wu 6, Lili Zhang 7, Xueqiu Lin 1,2,8,
LETTER. Journal Club. Pathogen-induced human T H 17 cells produce IFN-c or IL-10 and are regulated by IL-1b. Markus Geuking April 27 th, 2012
 Journal Clu LETTER oi:1.138/nature1957 Pathogen-inue human T H 17 ells proue IFN- or IL-1 an are regulate y IL-1 Christina E. Zielinski 1, Feerio Mele 1, Dominik Ashenrenner 1, Davi Jarrossay 1, Franesa
Journal Clu LETTER oi:1.138/nature1957 Pathogen-inue human T H 17 ells proue IFN- or IL-1 an are regulate y IL-1 Christina E. Zielinski 1, Feerio Mele 1, Dominik Ashenrenner 1, Davi Jarrossay 1, Franesa
The metabolic checkpoint kinase mtor is essential for IL-15 signaling during the development and activation of NK cells
 The metaoli hekpoint kinase mtor is essential for IL- signaling uring the evelopment an ativation of NK ells Antoine Marçais, Julien Cherfils-Viini 6,, Charlotte Viant 7,, Sophie Degouve, Séastien Viel,8,
The metaoli hekpoint kinase mtor is essential for IL- signaling uring the evelopment an ativation of NK ells Antoine Marçais, Julien Cherfils-Viini 6,, Charlotte Viant 7,, Sophie Degouve, Séastien Viel,8,
Supplementary Fig.S1. MeDIP RPKM distribution of 5kb windows. Relative expression of DNMT. Percentage of genome. (fold change)
 Supplementary Fig.S1 Percentage of genome 10% 8% 6% 4% 2% 0 MeDIP RPKM distribution of 5kb windows 0 0. 0.5 0.75 >1 MeDIP RPKM value EC Relative expression of DNMT (fold change) 12 6 4 3 2 1 EC DNMT1 DNMT2
Supplementary Fig.S1 Percentage of genome 10% 8% 6% 4% 2% 0 MeDIP RPKM distribution of 5kb windows 0 0. 0.5 0.75 >1 MeDIP RPKM value EC Relative expression of DNMT (fold change) 12 6 4 3 2 1 EC DNMT1 DNMT2
Cos7 (3TP) (K): TGFβ1(h): (K)
 IP#2: IP#1: Totl Lystes luiferse tivity (K): 6-4 - (K): luiferse tivity luiferse tivity (K): 2 1 RL-: - + + + + + Sm4-3F: + - + + + + MYC-Sm3: - - - - + + TβRI-HA(T204D): - - - + - + α-ha Luiferse Ativity
IP#2: IP#1: Totl Lystes luiferse tivity (K): 6-4 - (K): luiferse tivity luiferse tivity (K): 2 1 RL-: - + + + + + Sm4-3F: + - + + + + MYC-Sm3: - - - - + + TβRI-HA(T204D): - - - + - + α-ha Luiferse Ativity
Supplementary Figure 1. Implants derived from human embryoid body preparations contain non-cardiac structures. In early studies, infarcted hearts
 a Supplementary Figure 1. Implants derived from human emryoid ody preparations ontain non-ardia strutures. In early studies, infarted hearts reeived ell preparations of low ardia purity (
a Supplementary Figure 1. Implants derived from human emryoid ody preparations ontain non-ardia strutures. In early studies, infarted hearts reeived ell preparations of low ardia purity (
Sequence Analysis using Logic Regression
 Geneti Epidemiology (Suppl ): S66 S6 (00) Sequene Analysis using Logi Regression Charles Kooperberg Ingo Ruzinski, Mihael L. LeBlan, and Li Hsu Division of Publi Health Sienes, Fred Huthinson Caner Researh
Geneti Epidemiology (Suppl ): S66 S6 (00) Sequene Analysis using Logi Regression Charles Kooperberg Ingo Ruzinski, Mihael L. LeBlan, and Li Hsu Division of Publi Health Sienes, Fred Huthinson Caner Researh
Computer mouse use predicts acute pain but not prolonged or chronic pain in the neck and shoulder
 Computer mouse use predits aute pain but not prolonged or hroni pain in the nek and shoulder J H Andersen, 1 M Harhoff, 2 S Grimstrup, 2 I Vilstrup, 1 C F Lassen, 3 L P A Brandt, 4 A I Kryger, 3,5 E Overgaard,
Computer mouse use predits aute pain but not prolonged or hroni pain in the nek and shoulder J H Andersen, 1 M Harhoff, 2 S Grimstrup, 2 I Vilstrup, 1 C F Lassen, 3 L P A Brandt, 4 A I Kryger, 3,5 E Overgaard,
Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types.
 Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
SUPPLEMENTARY FIGURE 1: f-i
 SUPPLEMENTARY FIGURE 1: Comparisons of the biological replicates of ChIP-seq, Input and Bisulfite-seq. (a) Density plot of MeCP2 ChIP-seq genome coverage (calculated using tiled 15 bp windows) shows high
SUPPLEMENTARY FIGURE 1: Comparisons of the biological replicates of ChIP-seq, Input and Bisulfite-seq. (a) Density plot of MeCP2 ChIP-seq genome coverage (calculated using tiled 15 bp windows) shows high
Supplemental Information. A Highly Sensitive and Robust Method. for Genome-wide 5hmC Profiling. of Rare Cell Populations
 Molecular ell, Volume 63 Supplemental Information Highly Sensitive and Robust Method for enome-wide hm Profiling of Rare ell Populations Dali Han, Xingyu Lu, lan H. Shih, Ji Nie, Qiancheng You, Meng Michelle
Molecular ell, Volume 63 Supplemental Information Highly Sensitive and Robust Method for enome-wide hm Profiling of Rare ell Populations Dali Han, Xingyu Lu, lan H. Shih, Ji Nie, Qiancheng You, Meng Michelle
EPIGENOMICS PROFILING SERVICES
 EPIGENOMICS PROFILING SERVICES Chromatin analysis DNA methylation analysis RNA-seq analysis Diagenode helps you uncover the mysteries of epigenetics PAGE 3 Integrative epigenomics analysis DNA methylation
EPIGENOMICS PROFILING SERVICES Chromatin analysis DNA methylation analysis RNA-seq analysis Diagenode helps you uncover the mysteries of epigenetics PAGE 3 Integrative epigenomics analysis DNA methylation
The relative roles of vision and chemosensation in mate recognition of Drosophila melanogaster
 214. Pulished y The Company of Biologists Ltd (214) 217, 2796-28 doi:1.1242/je.1817 RESEARCH ARTICLE The relative roles of vision and hemosensation in mate reognition of Drosophila melanogaster Sweta Agrawal*,
214. Pulished y The Company of Biologists Ltd (214) 217, 2796-28 doi:1.1242/je.1817 RESEARCH ARTICLE The relative roles of vision and hemosensation in mate reognition of Drosophila melanogaster Sweta Agrawal*,
IP: anti-gfp VPS29-GFP. IP: anti-vps26. IP: anti-gfp - + +
 FAM21 Strump. WASH1 IP: anti- 1 2 3 4 5 6 FAM21 Strump. FKBP IP: anti-gfp VPS29- GFP GFP-FAM21 tail H H/P P H H/P P c FAM21 FKBP Strump. VPS29-GFP IP: anti-gfp 1 2 3 FKBP VPS VPS VPS VPS29 1 = VPS29-GFP
FAM21 Strump. WASH1 IP: anti- 1 2 3 4 5 6 FAM21 Strump. FKBP IP: anti-gfp VPS29- GFP GFP-FAM21 tail H H/P P H H/P P c FAM21 FKBP Strump. VPS29-GFP IP: anti-gfp 1 2 3 FKBP VPS VPS VPS VPS29 1 = VPS29-GFP
MODULE 3: TRANSCRIPTION PART II
 MODULE 3: TRANSCRIPTION PART II Lesson Plan: Title S. CATHERINE SILVER KEY, CHIYEDZA SMALL Transcription Part II: What happens to the initial (premrna) transcript made by RNA pol II? Objectives Explain
MODULE 3: TRANSCRIPTION PART II Lesson Plan: Title S. CATHERINE SILVER KEY, CHIYEDZA SMALL Transcription Part II: What happens to the initial (premrna) transcript made by RNA pol II? Objectives Explain
Sirt1 Hmg20b Gm (0.17) 24 (17.3) 877 (857)
 3 (0.17) 24 (17.3) Sirt1 Hmg20 Gm4763 877 (857) c d Suppl. Figure 1. Screen validation for top candidate antagonists of Dot1L (a) Numer of genes with one (gray), two (cyan) or three (red) shrna scored
3 (0.17) 24 (17.3) Sirt1 Hmg20 Gm4763 877 (857) c d Suppl. Figure 1. Screen validation for top candidate antagonists of Dot1L (a) Numer of genes with one (gray), two (cyan) or three (red) shrna scored
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table.
 Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tale. Type of file: VI Title of file for HTML: Supplementary Movie 1 Description:
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tale. Type of file: VI Title of file for HTML: Supplementary Movie 1 Description:
Mitosis. Single Nano Micro Milli Macro. Primary. PCNA expression
 a b c DAPI YFP CC3 DAPI YFP PCNA DAPI YFP ph3 DAPI YFP KI67 e 6 Mitosis f 1 PCNA expression %ph3 + /YFP + n= 63 87 61 3 13 8 n= 15 3 9 1 5 %PCNA+/YFP+ 8 6 Supplementary Figure 1. Proliferation/apoptosis
a b c DAPI YFP CC3 DAPI YFP PCNA DAPI YFP ph3 DAPI YFP KI67 e 6 Mitosis f 1 PCNA expression %ph3 + /YFP + n= 63 87 61 3 13 8 n= 15 3 9 1 5 %PCNA+/YFP+ 8 6 Supplementary Figure 1. Proliferation/apoptosis
Pharmaceutical care of stroke patients. Course activities
 Pharmaeutial are of stroke patients Course ativities Pharmaeutial are of stroke patients Course ativities page 2 Case Stuy 1 4 Case Stuy 2 6 Case Stuy 3 8 Multiple Choie Questionnaire Case Stuy 1 Mr Ferguson
Pharmaeutial are of stroke patients Course ativities Pharmaeutial are of stroke patients Course ativities page 2 Case Stuy 1 4 Case Stuy 2 6 Case Stuy 3 8 Multiple Choie Questionnaire Case Stuy 1 Mr Ferguson
Accessing and Using ENCODE Data Dr. Peggy J. Farnham
 1 William M Keck Professor of Biochemistry Keck School of Medicine University of Southern California How many human genes are encoded in our 3x10 9 bp? C. elegans (worm) 959 cells and 1x10 8 bp 20,000
1 William M Keck Professor of Biochemistry Keck School of Medicine University of Southern California How many human genes are encoded in our 3x10 9 bp? C. elegans (worm) 959 cells and 1x10 8 bp 20,000
ChIP-seq analysis. J. van Helden, M. Defrance, C. Herrmann, D. Puthier, N. Servant, M. Thomas-Chollier, O.Sand
 ChIP-seq analysis J. van Helden, M. Defrance, C. Herrmann, D. Puthier, N. Servant, M. Thomas-Chollier, O.Sand Tuesday : quick introduction to ChIP-seq and peak-calling (Presentation + Practical session)
ChIP-seq analysis J. van Helden, M. Defrance, C. Herrmann, D. Puthier, N. Servant, M. Thomas-Chollier, O.Sand Tuesday : quick introduction to ChIP-seq and peak-calling (Presentation + Practical session)
WHITE PAPER. Increasing Ligation Efficiency and Discovery of mirnas for Small RNA NGS Sequencing Library Prep with Plant Samples
 WHITE PPER Increasing Ligation Efficiency and iscovery of mirns for Small RN NGS Sequencing Library Prep with Plant Samples ioo Scientific White Paper May 2017 bstract MicroRNs (mirns) are 18-22 nucleotide
WHITE PPER Increasing Ligation Efficiency and iscovery of mirns for Small RN NGS Sequencing Library Prep with Plant Samples ioo Scientific White Paper May 2017 bstract MicroRNs (mirns) are 18-22 nucleotide
Molecular basis for the immunosuppressive action of
 Immunology 199 7 379-384 Moleular basis for the immunosuppressive ation of steari aid on T ells P. W. TEBBEY & T. M. BUTTKE Department of Mirobiology and Immunology, East Carolina University Shool of Mediine,
Immunology 199 7 379-384 Moleular basis for the immunosuppressive ation of steari aid on T ells P. W. TEBBEY & T. M. BUTTKE Department of Mirobiology and Immunology, East Carolina University Shool of Mediine,
SUPPLEMENTARY FIGURES: Supplementary Figure 1
 SUPPLEMENTARY FIGURES: Supplementary Figure 1 Supplementary Figure 1. Glioblastoma 5hmC quantified by paired BS and oxbs treated DNA hybridized to Infinium DNA methylation arrays. Workflow depicts analytic
SUPPLEMENTARY FIGURES: Supplementary Figure 1 Supplementary Figure 1. Glioblastoma 5hmC quantified by paired BS and oxbs treated DNA hybridized to Infinium DNA methylation arrays. Workflow depicts analytic
Lecture 8 Understanding Transcription RNA-seq analysis. Foundations of Computational Systems Biology David K. Gifford
 Lecture 8 Understanding Transcription RNA-seq analysis Foundations of Computational Systems Biology David K. Gifford 1 Lecture 8 RNA-seq Analysis RNA-seq principles How can we characterize mrna isoform
Lecture 8 Understanding Transcription RNA-seq analysis Foundations of Computational Systems Biology David K. Gifford 1 Lecture 8 RNA-seq Analysis RNA-seq principles How can we characterize mrna isoform
Computational aspects of ChIP-seq. John Marioni Research Group Leader European Bioinformatics Institute European Molecular Biology Laboratory
 Computational aspects of ChIP-seq John Marioni Research Group Leader European Bioinformatics Institute European Molecular Biology Laboratory ChIP-seq Using highthroughput sequencing to investigate DNA
Computational aspects of ChIP-seq John Marioni Research Group Leader European Bioinformatics Institute European Molecular Biology Laboratory ChIP-seq Using highthroughput sequencing to investigate DNA
MicroRNA-494 promotes cancer progression and targets adenomatous polyposis coli in colorectal cancer
 Zhang et al. Moleular Caner (8) 7: DOI.86/s94-7-75- RESEARCH Open Aess MiroRNA-494 promotes aner progression and targets adenomatous polyposis oli in oloretal aner Ying Zhang, Lu Guo,, Yuhuan Li,, Gui-Hai
Zhang et al. Moleular Caner (8) 7: DOI.86/s94-7-75- RESEARCH Open Aess MiroRNA-494 promotes aner progression and targets adenomatous polyposis oli in oloretal aner Ying Zhang, Lu Guo,, Yuhuan Li,, Gui-Hai
PREPARED FOR: U.S. Army Medical Research and Materiel Command Fort Detrick, Maryland
 AD Award Number: W81XWH-12-1-0298 TITLE: MTHFR Functional Polymorphism C677T and Genomic Instability in the Etiology of Idiopathic Autism in Simplex Families PRINCIPAL INVESTIGATOR: Xudong Liu, PhD CONTRACTING
AD Award Number: W81XWH-12-1-0298 TITLE: MTHFR Functional Polymorphism C677T and Genomic Instability in the Etiology of Idiopathic Autism in Simplex Families PRINCIPAL INVESTIGATOR: Xudong Liu, PhD CONTRACTING
Nature Structural & Molecular Biology: doi: /nsmb.2419
 Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers
 Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
describing DNA reassociation* (renaturation/nucleation inhibition/single strand ends)
 Pro. Nat. Aad. Si. USA Vol. 73, No. 2, pp. 415-419, February 1976 Biohemistry Studies on nulei aid reassoiation kinetis: Empirial equations desribing DNA reassoiation* (renaturation/nuleation inhibition/single
Pro. Nat. Aad. Si. USA Vol. 73, No. 2, pp. 415-419, February 1976 Biohemistry Studies on nulei aid reassoiation kinetis: Empirial equations desribing DNA reassoiation* (renaturation/nuleation inhibition/single
Assignment 5: Integrative epigenomics analysis
 Assignment 5: Integrative epigenomics analysis Due date: Friday, 2/24 10am. Note: no late assignments will be accepted. Introduction CpG islands (CGIs) are important regulatory regions in the genome. What
Assignment 5: Integrative epigenomics analysis Due date: Friday, 2/24 10am. Note: no late assignments will be accepted. Introduction CpG islands (CGIs) are important regulatory regions in the genome. What
Novel PRD-like homeodomain transcription factors and retrotransposon elements in early human
 Novel PRD-like homeodomain transcription factors and retrotransposon elements in early human development Virpi öhönen, Shintaro Katayama, Liselotte Vesterlund, Eeva-Mari Jouhilahti, Mona Sheikhi, Elo Madissoon,
Novel PRD-like homeodomain transcription factors and retrotransposon elements in early human development Virpi öhönen, Shintaro Katayama, Liselotte Vesterlund, Eeva-Mari Jouhilahti, Mona Sheikhi, Elo Madissoon,
Acute transcriptional up-regulation specific to osteoblasts/osteoclasts in medaka fish
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 Aute transriptional up-regulation speifi to osteoblasts/osteolasts in medaka fish immediately after exposure to mirogravity Masahiro Chatani, 1, Hiroya Morimoto,
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 Aute transriptional up-regulation speifi to osteoblasts/osteolasts in medaka fish immediately after exposure to mirogravity Masahiro Chatani, 1, Hiroya Morimoto,
a b Pml F/F Prrx1-cre f 1.4 n.s.
 a b Pml F/F Prrx1-re X Prrx1-re-Pml F/F Relative CFU-F numbers 2. Prrx1-Cre-PML +/+ n.s. d early passages e f 1.4 n.s. Adsorbane Adsorbane.8.6.4.2.8.6.4.2 1 2 3 4 days in ulture late passages 2 4 6 8 1
a b Pml F/F Prrx1-re X Prrx1-re-Pml F/F Relative CFU-F numbers 2. Prrx1-Cre-PML +/+ n.s. d early passages e f 1.4 n.s. Adsorbane Adsorbane.8.6.4.2.8.6.4.2 1 2 3 4 days in ulture late passages 2 4 6 8 1
Mineral Oil Application Experiments: Reducing Current Season PVY
 Mineral Oil Appliation Experiments: Reduing Current Season PVY Prinipal Investigator(s). Russell L. Groves, Assistant Professor and Entomology Extension Speialist, Department of Entomology, 537 Russell
Mineral Oil Appliation Experiments: Reduing Current Season PVY Prinipal Investigator(s). Russell L. Groves, Assistant Professor and Entomology Extension Speialist, Department of Entomology, 537 Russell
Measurement of Dose Rate Dependence of Radiation Induced Damage to the Current Gain in Bipolar Transistors 1
 Measurement of Dose Rate Dependene of Radiation Indued Damage to the Current Gain in Bipolar Transistors 1 D. Dorfan, T. Dubbs, A. A. Grillo, W. Rowe, H. F.-W. Sadrozinski, A. Seiden, E. Spener, S. Stromberg,
Measurement of Dose Rate Dependene of Radiation Indued Damage to the Current Gain in Bipolar Transistors 1 D. Dorfan, T. Dubbs, A. A. Grillo, W. Rowe, H. F.-W. Sadrozinski, A. Seiden, E. Spener, S. Stromberg,
SUPPLEMENTAL INFORMATION
 SUPPLEMENTAL INFORMATION GO term analysis of differentially methylated SUMIs. GO term analysis of the 458 SUMIs with the largest differential methylation between human and chimp shows that they are more
SUPPLEMENTAL INFORMATION GO term analysis of differentially methylated SUMIs. GO term analysis of the 458 SUMIs with the largest differential methylation between human and chimp shows that they are more
Large Virchow-Robin Spaces:
 929 Large Virhow-Robin Spaes: MR-Ciinial Correlation Linda A. Heier 1 Cristel J. Bauer 1 Larry Shwartz 1 Robert D. Zimmerman 1 Susan Morgello 2 Mihael D. F. Dek 1 High-field MR sans frequently show Virhow-Robin
929 Large Virhow-Robin Spaes: MR-Ciinial Correlation Linda A. Heier 1 Cristel J. Bauer 1 Larry Shwartz 1 Robert D. Zimmerman 1 Susan Morgello 2 Mihael D. F. Dek 1 High-field MR sans frequently show Virhow-Robin
Nature Genetics: doi: /ng.2995
 Supplementary Figure 1 Kaplan-Meier survival curves of patients with brainstem tumors. (a) Comparison of patients with PPM1D mutation versus wild-type PPM1D. (b) Comparison of patients with PPM1D mutation
Supplementary Figure 1 Kaplan-Meier survival curves of patients with brainstem tumors. (a) Comparison of patients with PPM1D mutation versus wild-type PPM1D. (b) Comparison of patients with PPM1D mutation
7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans.
 Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Role of b3-adrenergic receptors in the action of a tumour lipid mobilizing factor
 British Journal of Caner (22) 86, 424 428 ª 22 The Caner Researh Campaign All rights reserve 7 92/2 $25. www.bjaner.om Role of b3-arenergi reeptors in the ation of a tumour lipi mobilizing fator ST Russell
British Journal of Caner (22) 86, 424 428 ª 22 The Caner Researh Campaign All rights reserve 7 92/2 $25. www.bjaner.om Role of b3-arenergi reeptors in the ation of a tumour lipi mobilizing fator ST Russell
Introduction to epigenetics: chromatin modifications, DNA methylation and the CpG Island landscape. Héctor Corrada Bravo CMSC702 Spring 2014
 Introduction to epigenetics: chromatin modifications, DN methylation and the p Island landscape Héctor orrada Bravo MS702 Spring 2014 enetics: the alphabet of life Letters of DN sequence carry information
Introduction to epigenetics: chromatin modifications, DN methylation and the p Island landscape Héctor orrada Bravo MS702 Spring 2014 enetics: the alphabet of life Letters of DN sequence carry information
Supplemental Table 1. Primers used for real-time quantitative RT-PCR assay. Nucleotide sequence
 Supplemental Table 1. Primers used for real-time quantitative RT-PCR assay Name FoxO1 forward FoxO1 reverse GK forward GK reverse INS-1 forward INS-1 reverse INS-2 forward INS-2 reverse Glut2 forward Glut2
Supplemental Table 1. Primers used for real-time quantitative RT-PCR assay Name FoxO1 forward FoxO1 reverse GK forward GK reverse INS-1 forward INS-1 reverse INS-2 forward INS-2 reverse Glut2 forward Glut2
SUPPLEMENTARY INFORMATION
 DOI: 1.138/n358 TLR2 nd MyD88 expression in murine mmmry epithelil supopultions. CD24 min plus MRU Myo-epithelil Luminl progenitor (CD61 pos ) Mture luminl (CD61 neg ) CD49f CD61 Reltive expression Krt5
DOI: 1.138/n358 TLR2 nd MyD88 expression in murine mmmry epithelil supopultions. CD24 min plus MRU Myo-epithelil Luminl progenitor (CD61 pos ) Mture luminl (CD61 neg ) CD49f CD61 Reltive expression Krt5
Survivin withdrawal by nuclear export failure as a physiological switch to commit cells to apoptosis
 Citation: (2010) 1, e57; doi:10.1038/ddis.2010.34 & 2010 Mamillan Pulishers Limited All rights reserved 2041-4889/10 www.nature.om/ddis withdrawal y nulear export failure as a physiologial swith to ommit
Citation: (2010) 1, e57; doi:10.1038/ddis.2010.34 & 2010 Mamillan Pulishers Limited All rights reserved 2041-4889/10 www.nature.om/ddis withdrawal y nulear export failure as a physiologial swith to ommit
Supplemental Figure 1. Genes showing ectopic H3K9 dimethylation in this study are DNA hypermethylated in Lister et al. study.
 mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
Supplementary Figure 1. Verification of drug infusions into the IPN. a. Representative
 Supplementary Figure 1. Verifiation of drug infusions into the IPN. a. Representative neutral red-stained oronal setion from a mouse with a guide annula targeting the IPN. The guide annula sar is irled
Supplementary Figure 1. Verifiation of drug infusions into the IPN. a. Representative neutral red-stained oronal setion from a mouse with a guide annula targeting the IPN. The guide annula sar is irled
articles Published online: 11 March 2002, DOI: /nn823
 Mutant SOD1 auses motor neuron disease independent of opper haperone mediated opper loading Jamuna R. Suramaniam 1, W. Ernest Lyons 1, Jian Liu 2, Thomas B. Bartnikas 3, Jeffrey Rothstein 4,5, Donald L.
Mutant SOD1 auses motor neuron disease independent of opper haperone mediated opper loading Jamuna R. Suramaniam 1, W. Ernest Lyons 1, Jian Liu 2, Thomas B. Bartnikas 3, Jeffrey Rothstein 4,5, Donald L.
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression. Chromatin Array of nuc
 Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Nature Medicine: doi: /nm.4322
 1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
Nature Neuroscience: doi: /nn Supplementary Figure 1. Confirmation that optogenetic inhibition of dopaminergic neurons affects choice
 Supplementary Figure 1 Confirmation that optogenetic inhibition of dopaminergic neurons affects choice (a) Sample behavioral trace as in Figure 1d, but with NpHR stimulation trials depicted as green blocks
Supplementary Figure 1 Confirmation that optogenetic inhibition of dopaminergic neurons affects choice (a) Sample behavioral trace as in Figure 1d, but with NpHR stimulation trials depicted as green blocks
Variation in Broccoli Cultivar Phytochemical Content under Organic and Conventional Management Systems: Implications in Breeding for Nutrition
 Variation in Brooli Cultivar Phytohemial Content under Organi and Conventional Management Systems: Impliations in Breeding for Nutrition Renaud ENC, Lammerts van Bueren ET, Myers JR, Paulo MJ, van Eeuwijk
Variation in Brooli Cultivar Phytohemial Content under Organi and Conventional Management Systems: Impliations in Breeding for Nutrition Renaud ENC, Lammerts van Bueren ET, Myers JR, Paulo MJ, van Eeuwijk
Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition
 In the format provided y the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 217.15 Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition Chloé
In the format provided y the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 3 ARTICLE NUMBER: 217.15 Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition Chloé
Keywords: congested heart failure,cardiomyopathy-targeted areas, Beck Depression Inventory, psychological distress. INTRODUCTION:
 International Journal of Medial Siene and Eduation An offiial Publiation of Assoiation for Sientifi and Medial Eduation (ASME) Original Researh Artile ASSOCIATION BETWEEN QUALITY OF LIFE AND ANXIETY, DEPRESSION,
International Journal of Medial Siene and Eduation An offiial Publiation of Assoiation for Sientifi and Medial Eduation (ASME) Original Researh Artile ASSOCIATION BETWEEN QUALITY OF LIFE AND ANXIETY, DEPRESSION,
Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first
 Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first intron IGLL5 mutation depicting biallelic mutations. Red arrows highlight the presence of out of phase
Supplementary Figure 1: Features of IGLL5 Mutations in CLL: a) Representative IGV screenshot of first intron IGLL5 mutation depicting biallelic mutations. Red arrows highlight the presence of out of phase
Lesions of prefrontal cortex reduce attentional modulation of neuronal responses. and synchrony in V4
 Lesions of prefrontl ortex reue ttentionl moultion of neuronl responses n synhrony in V4 Georgi G. Gregoriou,, Anrew F. Rossi, 3 Leslie G Ungerleier, 4 Roert Desimone 5 Deprtment of Bsi Sienes, Fulty of
Lesions of prefrontl ortex reue ttentionl moultion of neuronl responses n synhrony in V4 Georgi G. Gregoriou,, Anrew F. Rossi, 3 Leslie G Ungerleier, 4 Roert Desimone 5 Deprtment of Bsi Sienes, Fulty of
A post-translational modification switch controls coactivator function of histone methyltransferases G9a and GLP
 rticle post-translational modification switch controls coactivator function of histone methyltransferases G9a and GLP oralie Poulard, anielle ittencourt, ai-ying Wu Michael R Stallcup *, Yixin Hu, aniel
rticle post-translational modification switch controls coactivator function of histone methyltransferases G9a and GLP oralie Poulard, anielle ittencourt, ai-ying Wu Michael R Stallcup *, Yixin Hu, aniel
Ambient temperature regulated flowering time
 Ambient temperature regulated flowering time Applications of RNAseq RNA- seq course: The power of RNA-seq June 7 th, 2013; Richard Immink Overview Introduction: Biological research question/hypothesis
Ambient temperature regulated flowering time Applications of RNAseq RNA- seq course: The power of RNA-seq June 7 th, 2013; Richard Immink Overview Introduction: Biological research question/hypothesis
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
Supplementary Figures Supplementary Figure 1. Pan-cancer analysis of global and local DNA methylation variation a) Variations in global DNA methylation are shown as measured by averaging the genome-wide
phosphatidylcholine by high performance liquid chromatography: a partial resolution of molecular species
 A large-sale purifiation of phosphatidylethanolamine, lysophosphatidylethanolamine, and phosphatidylholine by high performane liquid hromatography: a partial resolution of moleular speies R. S. Fager,
A large-sale purifiation of phosphatidylethanolamine, lysophosphatidylethanolamine, and phosphatidylholine by high performane liquid hromatography: a partial resolution of moleular speies R. S. Fager,
Chemotherapy triggers HIF-1 dependent glutathione synthesis and copper chelation that induces the breast cancer stem cell phenotype
 hemotherapy triggers HIF-1 dependent glutathione synthesis and opper helation that indues the breast aner stem ell phenotype Haiquan Lu a, ebangshu Samanta a, Lisha Xiang a, Huimin Zhang a, Hongxia Hu
hemotherapy triggers HIF-1 dependent glutathione synthesis and opper helation that indues the breast aner stem ell phenotype Haiquan Lu a, ebangshu Samanta a, Lisha Xiang a, Huimin Zhang a, Hongxia Hu
