Supplemental Information. Alteration of Tumor Metabolism by CD4+ T Cells. Leads to TNF-a-Dependent Intensification
|
|
|
- Mabel Francis
- 6 years ago
- Views:
Transcription
1 Cell Metabolism, Volume 8 Supplemental Information Alteration of Tumor Metabolism by CD4+ T Cells Leads to TNF-a-Dependent Intensification of Oxidative Stress and Tumor Cell Death Tsadik Habtetsion, Zhi-Chun Ding, Wenhu Pi, Tao Li, Chunwan Lu, Tingting Chen, Caixia Xi, Helena Spartz, Kebin Liu, Zhonglin Hao, Nahid Mivechi, Yuqing Huo, Bruce R. Blazar, David H. Munn, and Gang Zhou
2 Tumor size (mm ) Tumor size (mm ) Tumor size (mm ) SUPPLEMENTAL INFORMATION Supplemental Figures A CT6HA or CT6WT CTX HA-CD4 5 B HA-CD4 CT6HA CTX 5 IFN RKO 3 CT6WT (n=4) CT6HA (n=4) Days after treatment 3 Days after treatment C CT6HA HA-CD4 CTX agr mab or clodronate 5 D Tumor rechallenge 5 anti-gr mab (n=5) Clodronate liposomes (n=5) Days after treatment naive cured S. Figure S, related to Figure : The curative effect of CTX+CD4 AT is antigen-driven and dependent on host response to IFN, but does not require the involvement of host myeloid cells (A) Adoptive transfer of HA-specific CD4+ T cells following CTX has no therapeutic effect on wildtype CT6 tumors. The schema depicts the timeline of experimental procedures. Mice were inoculated with either CT6HA or CT6WT tumor cells. Mice with established tumors were treated with CTX followed by adoptive transfer of HA-specific CD4+ T cells. Tumor growth curves of each mouse are shown. (B) CTX+CD4 AT treatment fails to reject tumors in IFN RKO mice. CT6HA
3 tumor cells were subcutaneously injected to IFN RKO mice. Mice with established tumors received CTX+CD4 AT and tumor growth was monitored. (C) Depletion of macrophages or myeloid cells does not affect the curative effect of CTX+CD4 AT. The schema depicts the timeline of experimental procedures. Mice with established CT6HA tumors were treated with CTX +CD4 AT. Some mice received clodronate liposomes to deplete macrophages, other mice were injected with anti-gr mab to deplete myeloid cells. Tumor growth curves of each mouse are shown. (D) Mice cured by CTX+CD4 AT are resistant to CT6HA tumor rechallenge. Mice with established CT6HA tumors were treated with CTX+CD4 AT. A group of mice with complete tumor regression were selected for tumor rechallenge after becoming tumor-free for 3 weeks. x 6 CT6HA tumor cells were subcutaneously injected to these cured mice and a group of naive mice which used as controls. Tumor sizes in mice 3 weeks after tumor inoculation are shown.
4 Scaled intensity Scaled intensity Scaled intensity Glutaminolysis A Glycolysis Glucose G6P F6P FBP DHAP G3P Lactate Pyruvate PEP PG 3PG BPG Acetyl-CoA Citrate Glutamine Oxaloacetate Malate TCA cycle Isocitrate Glutamate A-Ketoglutarate Fumarate Succinyl-CoA Succinate NADH FADH ADP NAD+ FAD ATP Oxidative phosphorylation B Glycolysis Glucose 3 Day 5 Day G6P Day 5 Day F6P DHAP 3PG PEP Lactate Day 5 Day Day 5 Day Day 5 Day Day 5 Day Day 5 Day C Glutaminolysis and TCA cycle Citrate a-ketoglutarate Succinate Fumarate Malate Glutamine Glutamate Day 5 Day Day 5 Day Day 5 Day Day 5 Day Day 5 Day Day 5 Day Day 5 Day D Oxidative phosphorylation NAD+ FAD Day 5 Day Day 5 Day ADP Day 5 Day No Tx CTX CTX+CD4 AT Figure S Figure S, related to Figure : The impact of CTX+CD4 AT treatment on tumor glycolysis, glutaminolysis, TCA cycle and oxidative phosphorylation (A) Simplified schematic presentation of the major metabolic pathways involved in bioenergetics. Tumor sample preparation and metabolic data acquisition are described in Figure. Line plot graphics are provided for detected metabolites involved in glycolysis (B), glutaminolysis and TCA cycle (C), and oxidative phosphorylation (D). 3
5 A 4THA model HA-CD4 CTX 4T-HA mm Day 7 analysis B Thy. C % of Max CD4 AT CTX+CD4 AT.5%.3% FSC 45% no stimul. PMA/ionomycin D SSC CD45. no Tx CD4 AT CTX CTX+CD4 AT DCFDA % ROS tumor cells ** * TNF DCFDA Figure S3. Figure S3, related to Figure 4: CTX+CD4 AT treatment leads to intensified ROS accumulation in 4THA tumors (A) The schema depicts the experimental procedures. x 5 4THA tumor cells were injected to female BALB/c mice in the second mammary fat pad. When tumor sizes reached ~mm, which usually took -5 days, mice were randomized into 4 groups to receive no treatment (no Tx) or one of the following treatments: CD4 AT, CTX and CTX+CD4 AT. 6 days after adoptive transfer of HA-specific CD4+ T cells, tumor tissues were resected and processed into single cell suspensions. The presence of the donor T cells (Thy.+) in tumor was evaluated by FACS. Numbers in dot plots indicate percentage of donor T cells. (C) Tumor-infiltrating donor T cells are capable of TNF production. T cells in tumor samples were stimulated with PMA and ionomycin for 4 hours before TNF ICS. % of TNF + donor CD4+ T cells is given in histogram. (D) ROS levels in tumor cells. Tumor-derived single cell suspension was stained for CD45.. ROS levels in CD45. - tumor cells were evaluated by CM-HDCFDA staining. * indicates p <.5, ** indicates p <.. 4
6 A % of Max 4T MC38 A No Tx TNF mel mel+tnf B % of Max CT6 No Tx mel+ifn mel+il mel+tnf DCFDA DCFDA DCFDA DCFDA C RNA fold-change (relative to no Tx) Cat Gpx Gsr Hmox Nrf Gclc Gclm ** no Tx TNF mel mel+tnf D Annexin V No Tx TNF mel mel+tnf E GSH (um) ** GSH/GSSG ratio ** ** DAPI F 8 G H Figure S4. % DAPI+ cells % DAPI+ cells % of Max Intracellular GSH GSH MFI 5 5 * * no Tx media TNF mel media mel TNF Figure S4, related to Figure 5: The impact of TNF on chemotherapy-experienced tumor cells (A) The combination of chemotherapy and TNF leads to increased ROS accumulation in multiple tumor cell lines. 4T mammary carcinoma, MC38 colon cancer and A lymphoma cells were subjected to the indicated culturing conditions. After overnight culture, cells were evaluated for ROS levels by CM-HDCFDA staining. (B) CT6 cells were treated with the combination of melphalan and TNF, or IFN, or IL overnight. Intracellular ROS levels were measured by CM- HDCFDA staining. (C) Mel+TNF treatment activates antioxidant responses. CT6 cells were subjected to the indicated treatment conditions. After overnight culture, cells were collected for 5
7 RNA extraction. mrna levels of a panel of antioxidant genes were evaluated by quantitative realtime PCR. The target gene transcripts are normalized to -actin, and fold-changes relative to the no treatment sample are shown in bar graphs. Results are shown as mean SEM of triplicate reactions. (D) The combination of chemotherapy and TNF intensifies tumor cell death. CT6 cells were subjected to the indicated conditions. After a -hour culture, cells were harvested and stained for Annexin V and DAPI. Representative dot plots are shown. Numbers indicate percent of cells in the corresponding quadrant. (E) Mel+TNF causes reductions in GSH content and GSH/GSSG ratio in 4T cells. 4T cells were subjected to the indicated culture conditions for 7 hours. Intracellular GSH and GSSG contents were measured using a GSH/GSSG-Glo TM Assay Kit (Promega) and the GSH/GSSG ratio was calculated. Results are summarized in bar graphs and shown as mean SEM of triplicate cultures. (F) Inhibiting the thioredoxin pathway does not worsen cell death induced by mel+tnf. CT6 cells were treated with indicated combination of mel, TNF, sulfasalazine (SSA), and auranofin (AUR). After overnight culture, cell viability was evaluated by DAPI. Results are summarized in bar graphs and shown as mean SEM of triplicate cultures. (G) Chemotherapy sensitizes tumor cells to TNF toxicity. CT6 cells were preincubated with media or mel for 4 hr, washed thoroughly, and cultured in media or TNF for 5 hr before evaluation of cell viability by DAPI. (H) TNF -driven GSH reduction occurs in tumor cells that have been previously exposed to chemotherapy. Cultured cells described in (G) were also assayed for GSH levels by FACS using an Intracellular GSH Assay Kit (Abcam). Gating on live cells (DAPI - ), histograms show overlay of GSH curves under different culturing conditions. The bar graph summarizes GSH MFI shown as mean SEM of triplicate cultures. * indicates p <.5, ** indicates p <., indicates p <., indicates not significant. 6
8 Figu A TsnfR P E E E3 E4 E5 E6 E7 E8 E9 E B -5 AGGGACTTGTCTGGAAGACACAGATGAGAGTCAGTGGGAGTGACCTTGGGTCAGGACCCAGGCAGGGTAGAAAAGAACATTTGATTTCCCTCCCGGTTTG Screening primer F C Ctl Cas9-4 GGCTGACTGGGTAAACGTCCAAGAAAGTGCGATTAGAGGGAACTCAGAGGGAGGGAGAGCGCGAAGCAAAGGAGTTCTCGGCGGGCGAGCATCAGATATT sgrna# -3 GCCCTCCCCCTAATAACTCCCGCCTGCCGTCCCGTGAGATAGGTGGGCTGGCTTCCTTCCGTGAAATTGACAACAGGCTTAGAGGGGTTAGTCCACCGGT - CTAAGCTCACCCACTTCAAAAGCAAAGGAGTCCGAACTGGAATTCTGTGGACTCTCAAGCTCTTAAGTGGTGTTAAGTGGGTTTGGGGCGCCAAGCTACG GGACCCGGGCTACAAAGCGTTAAAGAACCTTGGCCCTCTCCTCACTCCTCCTAGTTCCCTCCCTCCTCCTCCCTCGCCTCTCCCCAGGCTCTTCCGGTCC + CGCTCTTGCAACACCACCCCCGCCACTCTCCCTTCCCCTCCTACCTTCTCTCTCCCCTCAGCTTAAATTTTCTCCGAGTTTTCCGAACTCTGGCTCATGA + TCGGGCCTACTGGGTGCGAGGTCCTGGAGGACCGTACCCTGATCTCTATCTGCCTCTGACTTTCAGCTTCTCGAACTCGAGGCCCAGGCTGCCATCGCCC + GGGCCACCTGGTCCGATCATCTTACTTCATTCACGAGCGTTGTCAATTGCTGCCCTGTCCCCAGCCCCAATGGGGGAGTGAGAGGCCACTGCCGGCCGGA +3 CATGGGTCTCCCCACCGTGCCTGGCCTGCTGCTGTCACTGGTGAGATGGGAGACTGGAGGGGAGGGTGGGTTGCTAGCGAGGCGCCGGGCAGGAGTGGGC sgrna# +4 TTGTACTAGGGGTAGAGATGGGGGCAGGGGCTTCTGTGCTTTAGTTTGACGCCAGGTTTGCTGTGCCTCACCTGGGGCTGCCCTGAGATGTGGAAGGGAG +5 AAATCTGAGAGGGAGAGAATCTGGCAAGCCAAGCTGGCAAGCCTGGGCCCCTGACAGTCAAGGTCCTGGCCCGTGTTGGGCTGTGGTCAGGCCAGCTGCT +6 CGGCCAGTGGAGTTAGACGGAGAAGCCTTCTTTCTCTGGC Screening primer R D WT 5 -TTCTCGGCGGGCGAGCATCAGATATTGCCCTCCCCCTAAT(69bp)CTAGGGGTAGAGATGGGGGCAGGGGCTTCTGTGCTTTAGTTTGACGCCAGGT-3 Clone : 5 -CGGCGGGCGAGCATCAGATATTGC (deletion) ggggcaggggcttctgtgctttagtttgacgccaggt-3 E CT6HA. TNFR CT6HA Clone 6: 5 -CGGCGGGCGAGCATCAGATATTGCCCTCCCCCTAA--(del)--TAGGGGTAGAGATGGGGGCAGGGGCTTCTGTGCTTTAGTTTGACGCCAGGT-3 Clone 9: 5 -CGGCGGGCGAGCATCAGATATTGCCCTCCCCCTAA--(del)--TAGGGGGCTAGATGGGGACACCAGGTTCGGTGCTTCTCTCCGGGGCCTGGC-3 Clone: 5 -CGGCGGGCGAGCATCAGATATTGC (deletion) GGGGCAGGGGCTTCTGTGCTTTAGTTTGACGCCAGGT-3 CDa (TNFR) F % ROS G % GSH reduction H GSH/GSSG ratio 4 3 ** CT6HA CT6HA. TNFR Figure S5, related to Figure 5: Generation of CT6HA. TNFR cells by CRISPR/Cas9-based gene editing (A) Schematic illustration of guide RNAs targeting the promoter and exon region of the TnfR gene locus. The yellow box denotes the promoter region and the blue boxed indicate exons. (B) TnfR gene fragment showing the sequences of the targeted region. The -nt sgrna target sequences are showed in blue. PAM sequences are shown in red. Primer sequences used to identify gene editing (screening primers) are underlined and exon sequence is showed in box. (C) A representative DNA gel of PCR products using the screening primers. Tumor cells were 7
9 transfected with sgrnas and Cas9. 3 days later, /3 of transfected CT6HA cells were harvested for DNA extraction and PCR analysis, other cells were used for limiting dilution to generate single cell clones. PCR reactions using the screening primers generated a ~kb fragment in untransfected control CT6HA cells (Ctl). A ~33bp PCR product appeared in sgrna/cas 9- transfected CT6HA cells (Cas 9), indicating Cas 9-mediated gene editing. (D) Confirmation of targeted gene editing by sequencing. 4 (#, 6, 9 and ) out of single cell clones were identified with deletion. DNA sequencing results are shown. (E) Confirmation of loss of TNFR expression in CT6HA. TNFR cells. One of the clones (#6) appeared to have the desired deletion in TNFR genome. This clone was designated as CT6HA. TNFR. CT6HA. TNFR cells and the parental CT6HA cells were stained with CDa mab to assess the surface expression of TNFR. (F) CT6HA. TNFR cells have reduced ROS induction after mel+tnf treatment. CT6HA. TNFR and CT6HA cells were treated with mel+tnf overnight before evaluating ROS levels by CM-HDCFDA. (G) Treatment-induced GSH reduction is lessened in CT6HA. TNFR cells. CT6HA. TNFR and CT6HA cells were either untreated or treated with mel+tnf for 7 hours before being harvested for GSH and GSSG measurements using the GSH/GSSG-Glo Assay Kit (Promega). % GSH reduction, calculated using the formula (GSH in untreated cells GSH in treated cells) / GSH in untreated cells. Changes in GSH/GSSG ratio are shown in (H). Data shown in bar graphs represent mean SEM of triplicate cultures. ** indicates p <., indicates p <.. 8
10 Sg# Sg# A Cyba E E E3 E4 E5 E6 B AACGTTCCAACCAGGGTGTAGCACTGGACAGCTATGGTGGAGTTCAGCTTGCTGCGGCTACCTCCCTGAGTGTCCTCTCTGTCCCTGAACCCCTTGCTGC Screening primer F C Ctl Cas9 ACAGTGCTCTGCTCCAGTAGGCAGAGAATTGGAGATGGTCCCAAGCCCGGGGCAAGCTCAAGCTGGCTCACACCTGCAGCAGATGGCACATCCTAACCAC sgrna# Exon CTCTCTCCATCCTTAGTTCTCATCACTGGGGGCATCGTGGCTACTGCTGGACGTTTCACACAGTGGTATTTCGGCGCCTACTCTATGTATCCTTCTACTG GGGACCAGGAAGGTGGGGTGATGGGAGTGCTCTCAGCATGAGCACATCAGTTCACGGAGTGCTACACATAGACTGACCACAGCCTCCAGAGACAGGACCA GCTCCGTCTGCAGCTTTAGAGGCCAAGGCCCAGGGCTGGATGCCCCCTTGCCAAAGGCCACAGACGGGGTTCAGGCAAATGTGGAGCTGAGGCCGAAGGT CCCAAGGCTTGGACTGGCGTCCATTGCCCCTCTGTCCCACCCTGCTGCACAGTAGACTCCTAGGCCAGGGCCTGAGAACAGAAGAGGCAGAGGGCAGCAG Exon3 CCTAGGCCTAGCATGGCTTTCCTTAACCTCACCGCAGCGCTGCAGGTGTGCTCATCTGTCTGCTGGAGTATCCCCGGGGAAAGAGGAAAAAGGGGTCCAC sgrna# CATGGAGCGATGGTAAGTCATTCCTACCCGAGAGGCCTGGGGACCTGCCCCATGCAGCCCACTCGTCCAGAGCTCCCAGCCCAAGGCCTGCCCAGCTGCC AGGCTCATGTGCCCAGGAGGAAAAGGACCCATAACCCTGGAAGGGGACAGATAAGGAGTGACAAGGGCTTCTAGGTGTGGTCGGTGCTGACCCTCTGACG TTTTGTCTTCCCGCTGTCTAGTGGACAGAAGTACCTGACCTCTGTGGTGAAGCTTTTCGGGCCCCTCACCAGGAATTACTACGTCCGGGCTGCCCTCCAC TTCCTGTGAGTCCCTCTGTC Screening primer R D WT: 5 -TCCTAACCACCTCTCTCCATCCTTAGTTC (4bp) AGAGGAAAAAGGGGTCCACCATGGAGCGATGGTAAGTCATTCCTACCCGA-3 E Clone : 5 -ACCACCTCTCTCCATCCT deletion TGGAGCGATGGTAAGTCATTCCTACCCGA-3 Clone 6: 5 -ACCACCTCTTTCCCC deletion AGGGGGTGATGGTAAGTCATTCCTACCCGA-3 p phox Clone 7: 5 -ACCACCTCTCTCCATCCTTAG deletion TGGAGCGATGGTAAGTCATTCCTACCCGA-3 Clone 33: 5 -ACCACCTCTCTCCATCCT deletion AAGTCATTCCTACCCGA-3 Actin Figure S6, related to Figure 6: Generation of CT6HA. Cyba cells by CRISPR/Cas9 gene editing (A) Schematic illustration of guide RNAs targeting the exon and exon 3 regions of the Cyba gene locus. (B) Cyba gene fragment showing the sequences of the targeted region. The -nt Figure S6. sgrna target sequences are showed in blue. PAM sequences are shown in red. Primer sequences used to identify gene editing (screening primers) are underlined and exon sequences are showed in box. (C) A representative DNA gel of PCR products using the screening primers. Tumor cells were transfected with sgrnas and Cas9. 3 days later, /3 of transfected CT6HA cells were harvested for DNA extraction and PCR analysis, other cells were used for limiting dilution to generate single cell clones. PCR reactions using the screening primers generated a ~8bp fragment in untransfected control CT6HA cells (Ctl). A ~36bp PCR product appeared in sgrna/cas 9-transfected CT6HA cells (Cas 9), indicating Cas 9-mediated gene editing. (D) Confirmation of targeted gene editing by sequencing. 4 (#, 6, 7 and 33) out of 8 single cell clones were identified with deletion. DNA sequencing results are shown. (E) Confirmation of loss 9
11 of p phox expression in CT6HA. Cyba cells. One of the clones (#7) appeared to have the desired deletion in Cyba genome. This clone was designated as CT6HA. Cyba. CT6HA. Cyba cells and the parental CT6HA cells were subjected to Western blot to assess the expression of p phox.
12 RNA fold-change (relative to no Tx) RNA fold-change (relative to no Tx) Hour: Hour: Nox Nox 3. Nrf Nqo Hmox 4 Cat TNF mel mel+tnf Figure S7, related to Figure 6: Kinetics of antioxidant gene expression in tumor cells after treatment CT6 cells were subjected to the indicated treatment conditions. hours and 9 hours after treatment, cells were collected for RNA extraction. mrna levels of a panel of antioxidant genes were evaluated by quantitative real-time PCR. The target gene transcripts are normalized to - actin, and fold-changes relative to the no treatment sample are shown in bar graphs.
13 Table S, related to STAR Methods (RNA extraction and quantitative real-time PCR): Primer sequences and sources GENE PRIMER SEQUENCES SOURCE IDENTIFIER NAME Ifnɣ F TCAAGTGGCATAGATGTGGAAGAA Overbergh et al., 3 N/A IFNɣ R TGGCTCTGCAGGATTTTCATG Overbergh et al., 3 N/A Tsp F CCAAAGCCTGCAAGAAAGAC Overbergh et al., 3 N/A Tsp R CCTGCTTGTTGCAAACTTGA Overbergh et al., 3 N/A Il-6 F GAGGATACCACTCCCAACAGACC Overbergh et al., 3 N/A Il-6 R AAGTGCATCATCGTTGTTCATACA Overbergh et al., 3 N/A Ccl3 F GTGGAATCTTCCGGCTGTAG Overbergh et al., 3 N/A Ccl3 R ACCATGACACTCTGCAACCA Overbergh et al., 3 N/A Ccl5 F CCCACTTCTTCTCTGGGTTG Overbergh et al., 3 N/A Ccl5 R GTGCCCACGTCAAGGAGTAT Overbergh et al., 3 N/A Tnfα F CATCTTCTCAAAATTCGAGTGACAA Overbergh et al., 3 N/A Tnfα R TGGGAGTAGACAAGGTACAACCC Overbergh et al., 3 N/A IL-β F CAACCAACAAGTGATATTCTCCAT Overbergh et al., 3 N/A IL-β R GATCCACACTCTCCAGCTGCA Overbergh et al., 3 N/A Ifnβ F CCATCCAAGAGATGCTCCAG Overbergh et al., 3 N/A Ifnβ R GTGGAGAGCAGTTGAGGACA Overbergh et al., 3 N/A Gmcsf F GCCATCAAAGAAGCCCTGAA Overbergh et al., 3 N/A Gmcsf R GCGGGTCTGCACACATGTTA Overbergh et al., 3 N/A Nrf F CGAGATATACGCAGGAGAGGTAAGA Jian et al., 8 N/A Nrf R GCTCGACAATGTTCTCCAGCTT Jian et al., 8 N/A Gpx F GAAGAACTTGGGCCATTTGG Jian et al., 8 N/A Gpx R TCTCGCCTGGCTCCTGTTT Jian et al., 8 N/A Sod F GTGATTGGGATTGCGCAGTA Jian et al., 8 N/A Sod R TGGTTTGAGGGTAGCAGATGAGT Jian et al., 8 N/A Cat F TGAGAAGCCTAAGAACGCAATTC Jian et al., 8 N/A Cat R TGAGAAGCCTAAGAACGCAATTC Jian et al., 8 N/A Ho F CACGCCAGCCACACAGCACTA Laura et al., 3 N/A Ho R GGCTGTCGATGTTCGGGAAGG Laura et al., 3 N/A Gclc F GCACGGCATCCTCCAGTTCCT Laura et al., 3 N/A Gclc R TCGGATGGTTGGGGTTTGTCC Laura et al., 3 N/A Gclm F GGCTTCGCCTCCGATTGAAGA Laura et al., 3 N/A Gclm R TCACACAGCAGGAGGCCAGGT Laura et al., 3 N/A Nox F ATGCCCAGGATCG GGT Ki Soon Kim et al., N/A Nox R GATGGAAGCAAAGGGAGTGA Ki Soon Kim et al., N/A Nox F CCCTTTGGTACAGCCAGTGAAGAT Ki Soon Kim et al., N/A Nox R CAATCCCGGCTCCCACTAACATCA Ki Soon Kim et al., NA
14 Table S, related to STAR Methods (Generation of CT6HA. TNFR and CT6HA. Cyba cell lines by CRSIPR/Cas 9): In vitro sgrna synthsis oligos and screening primer sequences OLIGO NAME SEQUENCES (5 3 ) SOURCE IDENTIFIER TNFR- sgrna# Forward Reverse TNFR- sgrna# Forward Reverse TNFR Screening Forward Reverse Cyba sgrna# Forward Reverse Cyba sgrna# Forward Reverse Cyba Screening Forward Reverse ACGGCAGGCGGGAGTTATTA TAATAACTCCCGCCTGCCGT CAGGAGTGGGCTTGTACTAG CTAGTACAAGCCCACTCCTG TTTGGGCTGACTGGGTAAAC TCTCCGTCTAACTCCACTGG GCCCCCAGTGATGAGAACTA TAGTTCTCATCACTGGGGGC GACTTACCATCGCTCCATGG CCATGGAGCGATGGTAAGTC CCAGTAGGCAGAGAATTGGA AGCTTCACCACAGAGGTCAG This paper This paper This paper This paper This paper This paper N/A N/A N/A N/A N/A N/A 3
Respiration. Energy is everything!
 Respiration Energy is everything! Tesla was incredible Everyone was intrigued by Tesla Tesla showed that energy does not need to be feared So what does this have to do with twinkies? Everything! Cellular
Respiration Energy is everything! Tesla was incredible Everyone was intrigued by Tesla Tesla showed that energy does not need to be feared So what does this have to do with twinkies? Everything! Cellular
Respiration. Energy is everything!
 Respiration Energy is everything! Tesla was incredible Everyone was intrigued by Tesla Tesla showed that energy does not need to be feared So what does this have to do with twinkies? Everything! Cellular
Respiration Energy is everything! Tesla was incredible Everyone was intrigued by Tesla Tesla showed that energy does not need to be feared So what does this have to do with twinkies? Everything! Cellular
CELLULAR RESPIRATION SUMMARY EQUATION. C 6 H 12 O 6 + O 2 6CO2 + 6H 2 O + energy (ATP) STEPWISE REDOX REACTION
 CELLULAR RESPIRATION SUMMARY EQUATION C 6 H 12 O 6 + O 2 6CO2 + 6H 2 O + energy (ATP) STEPWISE REDOX REACTION Oxidation: partial or complete loss of electrons Reduction: partial or complete gain of electrons
CELLULAR RESPIRATION SUMMARY EQUATION C 6 H 12 O 6 + O 2 6CO2 + 6H 2 O + energy (ATP) STEPWISE REDOX REACTION Oxidation: partial or complete loss of electrons Reduction: partial or complete gain of electrons
Notes CELLULAR RESPIRATION SUMMARY EQUATION C 6 H 12 O 6 + O 2. 6CO 2 + 6H 2 O + energy (ATP) STEPWISE REDOX REACTION
 AP BIOLOGY CELLULAR ENERGETICS ACTIVITY #2 Notes NAME DATE HOUR SUMMARY EQUATION CELLULAR RESPIRATION C 6 H 12 O 6 + O 2 6CO 2 + 6H 2 O + energy (ATP) STEPWISE REDOX REACTION Oxidation: partial or complete
AP BIOLOGY CELLULAR ENERGETICS ACTIVITY #2 Notes NAME DATE HOUR SUMMARY EQUATION CELLULAR RESPIRATION C 6 H 12 O 6 + O 2 6CO 2 + 6H 2 O + energy (ATP) STEPWISE REDOX REACTION Oxidation: partial or complete
(A) Urea cycle (B) TCA cycle (C) Glycolysis (D) Pyruvate oxidation (E) Respiratory chain
 Biochemistry - Problem Drill 15: Citric Acid Cycle No. 1 of 10 1. Which of the following statements is not a metabolic pathway involved in carbohydrate catabolism and ATP production. (A) Urea cycle (B)
Biochemistry - Problem Drill 15: Citric Acid Cycle No. 1 of 10 1. Which of the following statements is not a metabolic pathway involved in carbohydrate catabolism and ATP production. (A) Urea cycle (B)
Supplementary Figure 1
 Supplementary Figure 1 Expression of apoptosis-related genes in tumor T reg cells. (a) Identification of FOXP3 T reg cells by FACS. CD45 + cells were gated as enriched lymphoid cell populations with low-granularity.
Supplementary Figure 1 Expression of apoptosis-related genes in tumor T reg cells. (a) Identification of FOXP3 T reg cells by FACS. CD45 + cells were gated as enriched lymphoid cell populations with low-granularity.
Supplementary Figure 1.
 Supplementary Figure 1. Female Pro-ins2 -/- mice at 5-6 weeks of age were either inoculated i.p. with a single dose of CVB4 (1x10 5 PFU/mouse) or PBS and treated with αgalcer or control vehicle. On day
Supplementary Figure 1. Female Pro-ins2 -/- mice at 5-6 weeks of age were either inoculated i.p. with a single dose of CVB4 (1x10 5 PFU/mouse) or PBS and treated with αgalcer or control vehicle. On day
Notes CELLULAR RESPIRATION SUMMARY EQUATION C 6 H 12 O 6 + O 2. 6CO 2 + 6H 2 O + energy (ATP) STEPWISE REDOX REACTION
 AP BIOLOGY CELLULAR ENERGETICS ACTIVITY #2 Notes NAME DATE HOUR SUMMARY EQUATION CELLULAR RESPIRATION C 6 H 12 O 6 + O 2 6CO 2 + 6H 2 O + energy (ATP) STEPWISE REDOX REACTION Oxidation: partial or complete
AP BIOLOGY CELLULAR ENERGETICS ACTIVITY #2 Notes NAME DATE HOUR SUMMARY EQUATION CELLULAR RESPIRATION C 6 H 12 O 6 + O 2 6CO 2 + 6H 2 O + energy (ATP) STEPWISE REDOX REACTION Oxidation: partial or complete
Supplemental Figure 1
 Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
New tools bring greater understanding to cellular metabolism research
 New tools bring greater understanding to cellular metabolism research Mourad Ferhat, Ph.D, 7 Juin 2017 FDSS Users Meeting, Hamamatsu mourad.ferhat@promega.com Today s talk : focus on new cell-based assays
New tools bring greater understanding to cellular metabolism research Mourad Ferhat, Ph.D, 7 Juin 2017 FDSS Users Meeting, Hamamatsu mourad.ferhat@promega.com Today s talk : focus on new cell-based assays
MicroRNAs Modulate the Noncanonical NF- B Pathway by Regulating IKK Expression During Macrophage Differentiation
 MicroRNAs Modulate the Noncanonical NF- B Pathway by Regulating IKK Expression During Macrophage Differentiation Tao Li 1 *, Michael J. Morgan 1 *, Swati Choksi 1, Yan Zhang 1, You-Sun Kim 2#, Zheng-gang
MicroRNAs Modulate the Noncanonical NF- B Pathway by Regulating IKK Expression During Macrophage Differentiation Tao Li 1 *, Michael J. Morgan 1 *, Swati Choksi 1, Yan Zhang 1, You-Sun Kim 2#, Zheng-gang
Supplemental Table 1. Primer sequences for transcript analysis
 Supplemental Table 1. Primer sequences for transcript analysis Primer Sequence (5 3 ) Primer Sequence (5 3 ) Mmp2 Forward CCCGTGTGGCCCTC Mmp15 Forward CGGGGCTGGCT Reverse GCTCTCCCGGTTTC Reverse CCTGGTGTGCCTGCTC
Supplemental Table 1. Primer sequences for transcript analysis Primer Sequence (5 3 ) Primer Sequence (5 3 ) Mmp2 Forward CCCGTGTGGCCCTC Mmp15 Forward CGGGGCTGGCT Reverse GCTCTCCCGGTTTC Reverse CCTGGTGTGCCTGCTC
Supplementary figure 1. Systemic delivery of anti-cd47 antibody controls tumor growth in
 T u m o r v o lu m e (m m 3 ) P e rc e n t s u rv iv a l P e rc e n t s u rv iv a l Supplementary data a 1 8 6 4 2 5 1 1 5 2 2 5 3 3 5 4 T im e a fte r tu m o r in o c u la tio n (d ) b c 1 5 1 1 5 * *
T u m o r v o lu m e (m m 3 ) P e rc e n t s u rv iv a l P e rc e n t s u rv iv a l Supplementary data a 1 8 6 4 2 5 1 1 5 2 2 5 3 3 5 4 T im e a fte r tu m o r in o c u la tio n (d ) b c 1 5 1 1 5 * *
III. 6. Test. Respiració cel lular
 III. 6. Test. Respiració cel lular Chapter Questions 1) What is the term for metabolic pathways that release stored energy by breaking down complex molecules? A) anabolic pathways B) catabolic pathways
III. 6. Test. Respiració cel lular Chapter Questions 1) What is the term for metabolic pathways that release stored energy by breaking down complex molecules? A) anabolic pathways B) catabolic pathways
Eosinophils are required. for the maintenance of plasma cells in the bone marrow
 Eosinophils are required for the maintenance of plasma cells in the bone marrow Van Trung Chu, Anja Fröhlich, Gudrun Steinhauser, Tobias Scheel, Toralf Roch, Simon Fillatreau, James J. Lee, Max Löhning
Eosinophils are required for the maintenance of plasma cells in the bone marrow Van Trung Chu, Anja Fröhlich, Gudrun Steinhauser, Tobias Scheel, Toralf Roch, Simon Fillatreau, James J. Lee, Max Löhning
Chapter 13 Carbohydrate Metabolism
 Chapter 13 Carbohydrate Metabolism Metabolism of Foods Food is broken down into carbohydrates, lipids, and proteins and sent through catabolic pathways to produce energy. Glycolysis glucose 2 P i 2 ADP
Chapter 13 Carbohydrate Metabolism Metabolism of Foods Food is broken down into carbohydrates, lipids, and proteins and sent through catabolic pathways to produce energy. Glycolysis glucose 2 P i 2 ADP
Nature Medicine doi: /nm.3957
 Supplementary Fig. 1. p38 alternative activation, IL-21 expression, and T helper cell transcription factors in PDAC tissue. (a) Tissue microarrays of pancreatic tissue from 192 patients with pancreatic
Supplementary Fig. 1. p38 alternative activation, IL-21 expression, and T helper cell transcription factors in PDAC tissue. (a) Tissue microarrays of pancreatic tissue from 192 patients with pancreatic
Supplementary Information. Supplementary Figure 1
 Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Supplementary Material. Contents include:
 Supplementary Material Contents include: 1. Supplementary Figures (p. 2-7) 2. Supplementary Figure Legends (p. 8-9) 3. Supplementary Tables (p. 10-12) 4. Supplementary Table Legends (p. 13) 1 Wellen_FigS1
Supplementary Material Contents include: 1. Supplementary Figures (p. 2-7) 2. Supplementary Figure Legends (p. 8-9) 3. Supplementary Tables (p. 10-12) 4. Supplementary Table Legends (p. 13) 1 Wellen_FigS1
Information transmission
 1-3-3 Case studies in Systems Biology Goutham Vemuri goutham@chalmers.se Information transmission Fluxome Metabolome flux 1 flux flux 3 Proteome metabolite1 metabolite metabolite3 protein 1 protein protein
1-3-3 Case studies in Systems Biology Goutham Vemuri goutham@chalmers.se Information transmission Fluxome Metabolome flux 1 flux flux 3 Proteome metabolite1 metabolite metabolite3 protein 1 protein protein
Supplementary Figure 1. ETBF activate Stat3 in B6 and Min mice colons
 Supplementary Figure 1 ETBF activate Stat3 in B6 and Min mice colons a pstat3 controls Pos Neg ETBF 1 2 3 4 b pstat1 pstat2 pstat3 pstat4 pstat5 pstat6 Actin Figure Legend: (a) ETBF induce predominantly
Supplementary Figure 1 ETBF activate Stat3 in B6 and Min mice colons a pstat3 controls Pos Neg ETBF 1 2 3 4 b pstat1 pstat2 pstat3 pstat4 pstat5 pstat6 Actin Figure Legend: (a) ETBF induce predominantly
Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle.
 Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle. (a) mrna levels of Dj1 measured by quantitative RT-PCR in soleus, gastrocnemius (Gastroc.) and extensor digitorum longus
Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle. (a) mrna levels of Dj1 measured by quantitative RT-PCR in soleus, gastrocnemius (Gastroc.) and extensor digitorum longus
Nature Medicine: doi: /nm.3922
 Title: Glucocorticoid-induced tumor necrosis factor receptor-related protein co-stimulation facilitates tumor regression by inducing IL-9-producing helper T cells Authors: Il-Kyu Kim, Byung-Seok Kim, Choong-Hyun
Title: Glucocorticoid-induced tumor necrosis factor receptor-related protein co-stimulation facilitates tumor regression by inducing IL-9-producing helper T cells Authors: Il-Kyu Kim, Byung-Seok Kim, Choong-Hyun
well for 2 h at rt. Each dot represents an individual mouse and bar is the mean ±
 Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Tutorial 27: Metabolism, Krebs Cycle and the Electron Transport Chain
 Tutorial 27: Metabolism, Krebs Cycle and the Electron Transport Chain Goals: To be able to describe the overall catabolic pathways for food molecules. To understand what bonds are hydrolyzed in the digestion
Tutorial 27: Metabolism, Krebs Cycle and the Electron Transport Chain Goals: To be able to describe the overall catabolic pathways for food molecules. To understand what bonds are hydrolyzed in the digestion
Cellular Respiration Stage 2 & 3. Glycolysis is only the start. Cellular respiration. Oxidation of Pyruvate Krebs Cycle.
 Cellular Respiration Stage 2 & 3 Oxidation of Pyruvate Krebs Cycle AP 2006-2007 Biology Glycolysis is only the start Glycolysis glucose pyruvate 6C 2x 3C Pyruvate has more energy to yield 3 more C to strip
Cellular Respiration Stage 2 & 3 Oxidation of Pyruvate Krebs Cycle AP 2006-2007 Biology Glycolysis is only the start Glycolysis glucose pyruvate 6C 2x 3C Pyruvate has more energy to yield 3 more C to strip
Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured
 Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured under Th0, Th1, Th2, Th17, and Treg conditions. mrna
Supplemental Figure 1. Signature gene expression in in vitro differentiated Th0, Th1, Th2, Th17 and Treg cells. (A) Naïve CD4 + T cells were cultured under Th0, Th1, Th2, Th17, and Treg conditions. mrna
Supplementary Figure 1. BMS enhances human T cell activation in vitro in a
 Supplementary Figure 1. BMS98662 enhances human T cell activation in vitro in a concentration-dependent manner. Jurkat T cells were activated with anti-cd3 and anti-cd28 antibody in the presence of titrated
Supplementary Figure 1. BMS98662 enhances human T cell activation in vitro in a concentration-dependent manner. Jurkat T cells were activated with anti-cd3 and anti-cd28 antibody in the presence of titrated
Biology 30 Structure & Function of Cells (Part 2) Bioenergetics: Energy: Potential energy: Examples: Kinetic energy. Examples:
 Biology 30 Structure & Function of Cells (Part 2) Bioenergetics: Energy: Potential energy: Examples: Kinetic energy Examples: Energy can be transformed: Thermodynamics: First law of Thermodynamics: Second
Biology 30 Structure & Function of Cells (Part 2) Bioenergetics: Energy: Potential energy: Examples: Kinetic energy Examples: Energy can be transformed: Thermodynamics: First law of Thermodynamics: Second
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Figure S1, related to Figure 1. Escaper p38a-expressing cancer cells repopulate the tumors (A) Scheme of the mt/mg reporter that expresses a
 Cancer Cell, Volume 33 Supplemental Information Targeting p38a Increases DNA Damage, Chromosome Instability, and the Anti-tumoral Response to Taxanes in Breast Cancer Cells Begoña Cánovas, Ana Igea, Alessandro
Cancer Cell, Volume 33 Supplemental Information Targeting p38a Increases DNA Damage, Chromosome Instability, and the Anti-tumoral Response to Taxanes in Breast Cancer Cells Begoña Cánovas, Ana Igea, Alessandro
Aerobic Respiration. The four stages in the breakdown of glucose
 Aerobic Respiration The four stages in the breakdown of glucose 1 I. Aerobic Respiration Why can t we break down Glucose in one step? (Flaming Gummy Bear) Enzymes gently lower the potential energy until
Aerobic Respiration The four stages in the breakdown of glucose 1 I. Aerobic Respiration Why can t we break down Glucose in one step? (Flaming Gummy Bear) Enzymes gently lower the potential energy until
sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5
 sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
Pyruvate Alanine 0.15 *** ** ***
 SUPPLEMENTARY FIGURES Glucose ΔµM from fresh media / mg protein -1-2 -3 - -.1 -.3 -.5 Lactate Alanine Formate ΔµM from fresh media / mg protein 5 3 2 1.15.1.5.6..2.. NS-3 WT-NS G93A-NS Supplementary Figure
SUPPLEMENTARY FIGURES Glucose ΔµM from fresh media / mg protein -1-2 -3 - -.1 -.3 -.5 Lactate Alanine Formate ΔµM from fresh media / mg protein 5 3 2 1.15.1.5.6..2.. NS-3 WT-NS G93A-NS Supplementary Figure
3.7 CELLULAR RESPIRATION. How are these two images related?
 3.7 CELLULAR RESPIRATION How are these two images related? CELLULAR RESPIRATION Cellular respiration is the process whereby the body converts the energy that we get from food (glucose) into an energy form
3.7 CELLULAR RESPIRATION How are these two images related? CELLULAR RESPIRATION Cellular respiration is the process whereby the body converts the energy that we get from food (glucose) into an energy form
Cellular Respiration. The Details
 Cellular Respiration The Details LEARNING GOALS By the end of the lesson you will be able to; 1. Describe in detail the 4 stages of cellular respira;on 2. Track the crea;on of ATP and energy carrier molecules
Cellular Respiration The Details LEARNING GOALS By the end of the lesson you will be able to; 1. Describe in detail the 4 stages of cellular respira;on 2. Track the crea;on of ATP and energy carrier molecules
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Data Table of Contents:
 Supplementary Data Table of Contents: - Supplementary Methods - Supplementary Figures S1(A-B) - Supplementary Figures S2 (A-B) - Supplementary Figures S3 - Supplementary Figures S4(A-B) - Supplementary
Supplementary Data Table of Contents: - Supplementary Methods - Supplementary Figures S1(A-B) - Supplementary Figures S2 (A-B) - Supplementary Figures S3 - Supplementary Figures S4(A-B) - Supplementary
Supplementary Table; Supplementary Figures and legends S1-S21; Supplementary Materials and Methods
 Silva et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability Supplementary Table; Supplementary Figures and legends S1-S21; Supplementary
Silva et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability Supplementary Table; Supplementary Figures and legends S1-S21; Supplementary
FREE ENERGY Reactions involving free energy: 1. Exergonic 2. Endergonic
 BIOENERGETICS FREE ENERGY It is the portion of the total energy change in a system that is available for doing work at constant temperature and pressure; it is represented as ΔG. Reactions involving free
BIOENERGETICS FREE ENERGY It is the portion of the total energy change in a system that is available for doing work at constant temperature and pressure; it is represented as ΔG. Reactions involving free
BY: RASAQ NURUDEEN OLAJIDE
 BY: RASAQ NURUDEEN OLAJIDE LECTURE CONTENT INTRODUCTION CITRIC ACID CYCLE (T.C.A) PRODUCTION OF ACETYL CoA REACTIONS OF THE CITIRC ACID CYCLE THE AMPHIBOLIC NATURE OF THE T.C.A CYCLE THE GLYOXYLATE CYCLE
BY: RASAQ NURUDEEN OLAJIDE LECTURE CONTENT INTRODUCTION CITRIC ACID CYCLE (T.C.A) PRODUCTION OF ACETYL CoA REACTIONS OF THE CITIRC ACID CYCLE THE AMPHIBOLIC NATURE OF THE T.C.A CYCLE THE GLYOXYLATE CYCLE
4. Which step shows a split of one molecule into two smaller molecules? a. 2. d. 5
 1. Which of the following statements about NAD + is false? a. NAD + is reduced to NADH during both glycolysis and the citric acid cycle. b. NAD + has more chemical energy than NADH. c. NAD + is reduced
1. Which of the following statements about NAD + is false? a. NAD + is reduced to NADH during both glycolysis and the citric acid cycle. b. NAD + has more chemical energy than NADH. c. NAD + is reduced
Chem 109 C. Fall Armen Zakarian Office: Chemistry Bldn 2217
 Chem 109 C Fall 2014 Armen Zakarian ffice: Chemistry Bldn 2217 Chapter 25 o Glycolysis : fates of pyruvate NADH, H + H + C 2 NADH, H + H - (S)-lactic acid lactate dehydrogenase; anaerobic conditions -
Chem 109 C Fall 2014 Armen Zakarian ffice: Chemistry Bldn 2217 Chapter 25 o Glycolysis : fates of pyruvate NADH, H + H + C 2 NADH, H + H - (S)-lactic acid lactate dehydrogenase; anaerobic conditions -
Yield of energy from glucose
 Paper : Module : 05 Yield of Energy from Glucose Principal Investigator, Paper Coordinator and Content Writer Prof. Ramesh Kothari, Professor Dept. of Biosciences, Saurashtra University, Rajkot - 360005
Paper : Module : 05 Yield of Energy from Glucose Principal Investigator, Paper Coordinator and Content Writer Prof. Ramesh Kothari, Professor Dept. of Biosciences, Saurashtra University, Rajkot - 360005
CELL BIOLOGY - CLUTCH CH AEROBIC RESPIRATION.
 !! www.clutchprep.com CONCEPT: OVERVIEW OF AEROBIC RESPIRATION Cellular respiration is a series of reactions involving electron transfers to breakdown molecules for (ATP) 1. Glycolytic pathway: Glycolysis
!! www.clutchprep.com CONCEPT: OVERVIEW OF AEROBIC RESPIRATION Cellular respiration is a series of reactions involving electron transfers to breakdown molecules for (ATP) 1. Glycolytic pathway: Glycolysis
This is an example outline of 3 lectures in BSC (Thanks to Dr. Ellington for sharing this information.)
 This is an example outline of 3 lectures in BSC 2010. (Thanks to Dr. Ellington for sharing this information.) Topic 10: CELLULAR RESPIRATION (lectures 14-16) OBJECTIVES: 1. Know the basic reactions that
This is an example outline of 3 lectures in BSC 2010. (Thanks to Dr. Ellington for sharing this information.) Topic 10: CELLULAR RESPIRATION (lectures 14-16) OBJECTIVES: 1. Know the basic reactions that
Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic
 Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic B cells from WT Nfat2 +/+, TCL1 Nfat2 +/+ and TCL1 Nfat2
Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic B cells from WT Nfat2 +/+, TCL1 Nfat2 +/+ and TCL1 Nfat2
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the T H 17 cell it reg cell balance Rajatava Basu 1,5, Sarah K.
SUPPLEMENTARY MATERIAL IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the T H 17 cell it reg cell balance Rajatava Basu 1,5, Sarah K.
Biochemistry: A Short Course
 Tymoczko Berg Stryer Biochemistry: A Short Course First Edition CHAPTER 19 Harvesting Electrons from the Cycle 2013 W. H. Freeman and Company Chapter 19 Outline The citric acid cycle oxidizes the acetyl
Tymoczko Berg Stryer Biochemistry: A Short Course First Edition CHAPTER 19 Harvesting Electrons from the Cycle 2013 W. H. Freeman and Company Chapter 19 Outline The citric acid cycle oxidizes the acetyl
Nature Immunology: doi: /ni Supplementary Figure 1. Cellularity of leukocytes and their progenitors in naive wild-type and Spp1 / mice.
 Supplementary Figure 1 Cellularity of leukocytes and their progenitors in naive wild-type and Spp1 / mice. (a, b) Gating strategies for differentiated cells including PMN (CD11b + Ly6G hi and CD11b + Ly6G
Supplementary Figure 1 Cellularity of leukocytes and their progenitors in naive wild-type and Spp1 / mice. (a, b) Gating strategies for differentiated cells including PMN (CD11b + Ly6G hi and CD11b + Ly6G
Cellular Metabolism 6/20/2015. Metabolism. Summary of Cellular Respiration. Consists of all the chemical reactions that take place in a cell!
 Cellular Metabolism Biology 105 Lecture 6 Chapter 3 (pages 56-61) Metabolism Consists of all the chemical reactions that take place in a cell! Cellular metabolism: Aerobic cellular respiration requires
Cellular Metabolism Biology 105 Lecture 6 Chapter 3 (pages 56-61) Metabolism Consists of all the chemical reactions that take place in a cell! Cellular metabolism: Aerobic cellular respiration requires
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
L1 on PyMT tumor cells but Py117 cells are more responsive to IFN-γ. (A) Flow
 A MHCI B PD-L1 Fold expression 8 6 4 2 Fold expression 3 2 1 No tx 1Gy 2Gy IFN Py117 Py117 Supplementary Figure 1. Radiation and IFN-γ enhance MHCI expression and PD- L1 on PyMT tumor cells but Py117 cells
A MHCI B PD-L1 Fold expression 8 6 4 2 Fold expression 3 2 1 No tx 1Gy 2Gy IFN Py117 Py117 Supplementary Figure 1. Radiation and IFN-γ enhance MHCI expression and PD- L1 on PyMT tumor cells but Py117 cells
Suppl Video: Tumor cells (green) and monocytes (white) are seeded on a confluent endothelial
 Supplementary Information Häuselmann et al. Monocyte induction of E-selectin-mediated endothelial activation releases VE-cadherin junctions to promote tumor cell extravasation in the metastasis cascade
Supplementary Information Häuselmann et al. Monocyte induction of E-selectin-mediated endothelial activation releases VE-cadherin junctions to promote tumor cell extravasation in the metastasis cascade
Marah Bitar. Faisal Nimri ... Nafeth Abu Tarboosh
 8 Marah Bitar Faisal Nimri... Nafeth Abu Tarboosh Summary of the 8 steps of citric acid cycle Step 1. Acetyl CoA joins with a four-carbon molecule, oxaloacetate, releasing the CoA group and forming a six-carbon
8 Marah Bitar Faisal Nimri... Nafeth Abu Tarboosh Summary of the 8 steps of citric acid cycle Step 1. Acetyl CoA joins with a four-carbon molecule, oxaloacetate, releasing the CoA group and forming a six-carbon
Supplementary Table 1 Clinicopathological characteristics of 35 patients with CRCs
 Supplementary Table Clinicopathological characteristics of 35 patients with CRCs Characteristics Type-A CRC Type-B CRC P value Sex Male / Female 9 / / 8.5 Age (years) Median (range) 6. (9 86) 6.5 (9 76).95
Supplementary Table Clinicopathological characteristics of 35 patients with CRCs Characteristics Type-A CRC Type-B CRC P value Sex Male / Female 9 / / 8.5 Age (years) Median (range) 6. (9 86) 6.5 (9 76).95
Cellular Metabolism 9/24/2013. Metabolism. Cellular Metabolism. Consists of all the chemical reactions that take place in a cell!
 Cellular Metabolism Biology 105 Lecture 6 Chapter 3 (pages 56-61) Metabolism Consists of all the chemical reactions that take place in a cell! Cellular Metabolism Aerobic cellular respiration requires
Cellular Metabolism Biology 105 Lecture 6 Chapter 3 (pages 56-61) Metabolism Consists of all the chemical reactions that take place in a cell! Cellular Metabolism Aerobic cellular respiration requires
Photosynthesis in chloroplasts CO2 + H2O. Cellular respiration in mitochondria ATP. powers most cellular work. Heat energy
 Figure 9-01 LE 9-2 Light energy ECOSYSTEM Photosynthesis in chloroplasts CO2 + H2O Cellular respiration in mitochondria Organic + O molecules 2 powers most cellular work Heat energy LE 9-UN161a becomes
Figure 9-01 LE 9-2 Light energy ECOSYSTEM Photosynthesis in chloroplasts CO2 + H2O Cellular respiration in mitochondria Organic + O molecules 2 powers most cellular work Heat energy LE 9-UN161a becomes
Biological oxidation II. The Cytric acid cycle
 Biological oxidation II The Cytric acid cycle Outline The Cytric acid cycle (TCA tricarboxylic acid) Central role of Acetyl-CoA Regulation of the TCA cycle Anaplerotic reactions The Glyoxylate cycle Localization
Biological oxidation II The Cytric acid cycle Outline The Cytric acid cycle (TCA tricarboxylic acid) Central role of Acetyl-CoA Regulation of the TCA cycle Anaplerotic reactions The Glyoxylate cycle Localization
Supplementary Figure 1. Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Nature Immunology: doi: /ni.
 Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Chapter 9 Cellular Respiration Overview: Life Is Work Living cells require energy from outside sources
 Chapter 9 Cellular Respiration Overview: Life Is Work Living cells require energy from outside sources Some animals, such as the giant panda, obtain energy by eating plants, and some animals feed on other
Chapter 9 Cellular Respiration Overview: Life Is Work Living cells require energy from outside sources Some animals, such as the giant panda, obtain energy by eating plants, and some animals feed on other
Integrative Metabolism: Significance
 Integrative Metabolism: Significance Energy Containing Nutrients Carbohydrates Fats Proteins Catabolism Energy Depleted End Products H 2 O NH 3 ADP + Pi NAD + NADP + FAD + Pi NADH+H + NADPH+H + FADH2 Cell
Integrative Metabolism: Significance Energy Containing Nutrients Carbohydrates Fats Proteins Catabolism Energy Depleted End Products H 2 O NH 3 ADP + Pi NAD + NADP + FAD + Pi NADH+H + NADPH+H + FADH2 Cell
Cellular Metabolism. Biology 105 Lecture 6 Chapter 3 (pages 56-61)
 Cellular Metabolism Biology 105 Lecture 6 Chapter 3 (pages 56-61) Metabolism Consists of all the chemical reactions that take place in a cell! Cellular Metabolism Aerobic cellular respiration requires
Cellular Metabolism Biology 105 Lecture 6 Chapter 3 (pages 56-61) Metabolism Consists of all the chemical reactions that take place in a cell! Cellular Metabolism Aerobic cellular respiration requires
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was
 Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was
 Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was sorted by FACS. Surface markers for sorting were CD11c +
Supplementary Figure 1. mrna expression of chitinase and chitinase-like protein in splenic immune cells. Each splenic immune cell population was sorted by FACS. Surface markers for sorting were CD11c +
Cellular Metabolism. Biol 105 Lecture 6 Read Chapter 3 (pages 63 69)
 Cellular Metabolism Biol 105 Lecture 6 Read Chapter 3 (pages 63 69) Metabolism Consists of all of the chemical reactions that take place in a cell Metabolism Animation Breaking Down Glucose For Energy
Cellular Metabolism Biol 105 Lecture 6 Read Chapter 3 (pages 63 69) Metabolism Consists of all of the chemical reactions that take place in a cell Metabolism Animation Breaking Down Glucose For Energy
Photosynthesis in chloroplasts. Cellular respiration in mitochondria ATP. ATP powers most cellular work
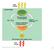 Light energy ECOSYSTEM CO + H O Photosynthesis in chloroplasts Cellular respiration in mitochondria Organic molecules + O powers most cellular work Heat energy 1 becomes oxidized (loses electron) becomes
Light energy ECOSYSTEM CO + H O Photosynthesis in chloroplasts Cellular respiration in mitochondria Organic molecules + O powers most cellular work Heat energy 1 becomes oxidized (loses electron) becomes
Supplemental Materials for. Effects of sphingosine-1-phosphate receptor 1 phosphorylation in response to. FTY720 during neuroinflammation
 Supplemental Materials for Effects of sphingosine-1-phosphate receptor 1 phosphorylation in response to FTY7 during neuroinflammation This file includes: Supplemental Table 1. EAE clinical parameters of
Supplemental Materials for Effects of sphingosine-1-phosphate receptor 1 phosphorylation in response to FTY7 during neuroinflammation This file includes: Supplemental Table 1. EAE clinical parameters of
Supplementary Figure 1. Double-staining immunofluorescence analysis of invasive colon and breast cancers. Specimens from invasive ductal breast
 Supplementary Figure 1. Double-staining immunofluorescence analysis of invasive colon and breast cancers. Specimens from invasive ductal breast carcinoma (a) and colon adenocarcinoma (b) were staining
Supplementary Figure 1. Double-staining immunofluorescence analysis of invasive colon and breast cancers. Specimens from invasive ductal breast carcinoma (a) and colon adenocarcinoma (b) were staining
The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep
 SUPPLEMENTARY INFORMATION The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness Jinyi Zhang, Naima
SUPPLEMENTARY INFORMATION The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness Jinyi Zhang, Naima
Citric Acid Cycle: Central Role in Catabolism. Entry of Pyruvate into the TCA cycle
 Citric Acid Cycle: Central Role in Catabolism Stage II of catabolism involves the conversion of carbohydrates, fats and aminoacids into acetylcoa In aerobic organisms, citric acid cycle makes up the final
Citric Acid Cycle: Central Role in Catabolism Stage II of catabolism involves the conversion of carbohydrates, fats and aminoacids into acetylcoa In aerobic organisms, citric acid cycle makes up the final
BIOCHEMICHISTRY OF EYE TISSUE. Sri Widia A Jusman Department of Biochemistry & Molecular Biology FMUI
 BIOCHEMICHISTRY OF EYE TISSUE Sri Widia A Jusman Department of Biochemistry & Molecular Biology FMUI 1 METABOLIC PATHWAYS IN EYE TISSUE Glycolysis ( aerobic & anaerobic) HMP shunt Poliol pathway TCA cycle
BIOCHEMICHISTRY OF EYE TISSUE Sri Widia A Jusman Department of Biochemistry & Molecular Biology FMUI 1 METABOLIC PATHWAYS IN EYE TISSUE Glycolysis ( aerobic & anaerobic) HMP shunt Poliol pathway TCA cycle
doi: /nature10642
 doi:10.1038/nature10642 Supplementary Fig. 1. Citric acid cycle (CAC) metabolism in WT 143B and CYTB 143B cells. a, Proliferation of WT 143B and CYTB 143B cells. Doubling times were 28±1 and 33±2 hrs for
doi:10.1038/nature10642 Supplementary Fig. 1. Citric acid cycle (CAC) metabolism in WT 143B and CYTB 143B cells. a, Proliferation of WT 143B and CYTB 143B cells. Doubling times were 28±1 and 33±2 hrs for
MULTIPLE CHOICE QUESTIONS
 MULTIPLE CHOICE QUESTIONS 1. Which of the following statements concerning anabolic reactions is FALSE? A. They are generally endergonic. B. They usually require ATP. C. They are part of metabolism. D.
MULTIPLE CHOICE QUESTIONS 1. Which of the following statements concerning anabolic reactions is FALSE? A. They are generally endergonic. B. They usually require ATP. C. They are part of metabolism. D.
Pathway overview. Glucose + 2NAD + + 2ADP +2Pi 2NADH + 2pyruvate + 2ATP + 2H 2 O + 4H +
 Glycolysis Glycolysis The conversion of glucose to pyruvate to yield 2ATP molecules 10 enzymatic steps Chemical interconversion steps Mechanisms of enzyme conversion and intermediates Energetics of conversions
Glycolysis Glycolysis The conversion of glucose to pyruvate to yield 2ATP molecules 10 enzymatic steps Chemical interconversion steps Mechanisms of enzyme conversion and intermediates Energetics of conversions
2/4/17. Cellular Metabolism. Metabolism. Cellular Metabolism. Consists of all of the chemical reactions that take place in a cell.
 Metabolism Cellular Metabolism Consists of all of the chemical reactions that take place in a cell. Can be reactions that break things down. (Catabolism) Or reactions that build things up. (Anabolism)
Metabolism Cellular Metabolism Consists of all of the chemical reactions that take place in a cell. Can be reactions that break things down. (Catabolism) Or reactions that build things up. (Anabolism)
Glycolysis. Intracellular location Rate limiting steps
 Glycolysis Definition Fx Fate Site Intracellular location Rate limiting steps Regulation Consume ATP Subs level phosphoryla tion Key reactions control points Nb Oxidation of glucose to give pyruvate (
Glycolysis Definition Fx Fate Site Intracellular location Rate limiting steps Regulation Consume ATP Subs level phosphoryla tion Key reactions control points Nb Oxidation of glucose to give pyruvate (
Chapter 9. Cellular Respiration: Harvesting Chemical Energy
 Chapter 9 Cellular Respiration: Harvesting Chemical Energy Living cells require energy from outside sources Energy flows into an ecosystem as sunlight and leaves as heat Photosynthesis generates O 2 and
Chapter 9 Cellular Respiration: Harvesting Chemical Energy Living cells require energy from outside sources Energy flows into an ecosystem as sunlight and leaves as heat Photosynthesis generates O 2 and
Citric acid cycle. Tomáš Kučera.
 itric acid cycle Tomáš Kučera tomas.kucera@lfmotol.cuni.cz Department of Medical hemistry and linical Biochemistry 2nd Faculty of Medicine, harles University in Prague and Motol University Hospital 2017
itric acid cycle Tomáš Kučera tomas.kucera@lfmotol.cuni.cz Department of Medical hemistry and linical Biochemistry 2nd Faculty of Medicine, harles University in Prague and Motol University Hospital 2017
TCA CYCLE (Citric Acid Cycle)
 TCA CYCLE (Citric Acid Cycle) TCA CYCLE The Citric Acid Cycle is also known as: Kreb s cycle Sir Hans Krebs Nobel prize, 1953 TCA (tricarboxylic acid) cycle The citric acid cycle requires aerobic conditions!!!!
TCA CYCLE (Citric Acid Cycle) TCA CYCLE The Citric Acid Cycle is also known as: Kreb s cycle Sir Hans Krebs Nobel prize, 1953 TCA (tricarboxylic acid) cycle The citric acid cycle requires aerobic conditions!!!!
Supplemental Table 1. Primers used for RT-PCR analysis of inflammatory cytokines Gene Primer Sequence
 Supplemental Table 1. Primers used for RT-PCR analysis of inflammatory cytokines Gene Primer Sequence IL-1α Forward primer 5 -CAAGATGGCCAAAGTTCGTGAC-3' Reverse primer 5 -GTCTCATGAAGTGAGCCATAGC-3 IL-1β
Supplemental Table 1. Primers used for RT-PCR analysis of inflammatory cytokines Gene Primer Sequence IL-1α Forward primer 5 -CAAGATGGCCAAAGTTCGTGAC-3' Reverse primer 5 -GTCTCATGAAGTGAGCCATAGC-3 IL-1β
fl/+ KRas;Atg5 fl/+ KRas;Atg5 fl/fl KRas;Atg5 fl/fl KRas;Atg5 Supplementary Figure 1. Gene set enrichment analyses. (a) (b)
 KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
Supplementary information. The proton-sensing G protein-coupled receptor T-cell death-associated gene 8
 1 Supplementary information 2 3 The proton-sensing G protein-coupled receptor T-cell death-associated gene 8 4 (TDAG8) shows cardioprotective effects against myocardial infarction 5 Akiomi Nagasaka 1+,
1 Supplementary information 2 3 The proton-sensing G protein-coupled receptor T-cell death-associated gene 8 4 (TDAG8) shows cardioprotective effects against myocardial infarction 5 Akiomi Nagasaka 1+,
CITRIC ACID CYCLE ERT106 BIOCHEMISTRY SEM /19 BY: MOHAMAD FAHRURRAZI TOMPANG
 CITRIC ACID CYCLE ERT106 BIOCHEMISTRY SEM 1 2018/19 BY: MOHAMAD FAHRURRAZI TOMPANG Chapter Outline (19-1) The central role of the citric acid cycle in metabolism (19-2) The overall pathway of the citric
CITRIC ACID CYCLE ERT106 BIOCHEMISTRY SEM 1 2018/19 BY: MOHAMAD FAHRURRAZI TOMPANG Chapter Outline (19-1) The central role of the citric acid cycle in metabolism (19-2) The overall pathway of the citric
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/310/ra11/dc1 Supplementary Materials for STAT3 Induction of mir-146b Forms a Feedback Loop to Inhibit the NF-κB to IL-6 Signaling Axis and STAT3-Driven Cancer
www.sciencesignaling.org/cgi/content/full/7/310/ra11/dc1 Supplementary Materials for STAT3 Induction of mir-146b Forms a Feedback Loop to Inhibit the NF-κB to IL-6 Signaling Axis and STAT3-Driven Cancer
Supporting Information Table of Contents
 Supporting Information Table of Contents Supporting Information Figure 1 Page 2 Supporting Information Figure 2 Page 4 Supporting Information Figure 3 Page 5 Supporting Information Figure 4 Page 6 Supporting
Supporting Information Table of Contents Supporting Information Figure 1 Page 2 Supporting Information Figure 2 Page 4 Supporting Information Figure 3 Page 5 Supporting Information Figure 4 Page 6 Supporting
CD14 + S100A9 + Monocytic Myeloid-Derived Suppressor Cells and Their Clinical Relevance in Non-Small Cell Lung Cancer
 CD14 + S1A9 + Monocytic Myeloid-Derived Suppressor Cells and Their Clinical Relevance in Non-Small Cell Lung Cancer Po-Hao, Feng M.D., Kang-Yun, Lee, M.D. Ph.D., Ya-Ling Chang, Yao-Fei Chan, Lu- Wei, Kuo,Ting-Yu
CD14 + S1A9 + Monocytic Myeloid-Derived Suppressor Cells and Their Clinical Relevance in Non-Small Cell Lung Cancer Po-Hao, Feng M.D., Kang-Yun, Lee, M.D. Ph.D., Ya-Ling Chang, Yao-Fei Chan, Lu- Wei, Kuo,Ting-Yu
Supplementary Figure 1. Generation of knockin mice expressing L-selectinN138G. (a) Schematics of the Sellg allele (top), the targeting vector, the
 Supplementary Figure 1. Generation of knockin mice expressing L-selectinN138G. (a) Schematics of the Sellg allele (top), the targeting vector, the targeted allele in ES cells, and the mutant allele in
Supplementary Figure 1. Generation of knockin mice expressing L-selectinN138G. (a) Schematics of the Sellg allele (top), the targeting vector, the targeted allele in ES cells, and the mutant allele in
Krebs Cycle. Color Index: Original slides. Important. 436 Notes 438 notes. Extra information Biochemistry team 438. Red boxes are IMPORTANT!
 Red boxes are IMPORTANT! Krebs Cycle Color Index: Original slides. Important. 436 Notes 438 notes : ل ی د ع ت ل ا ط ب ا ر https://docs.google.com/document/d/1wvdec1atp7j- ZKWOUSukSLsEcosjZ0AqV4z2VcH2TA0/edit?usp=sharing
Red boxes are IMPORTANT! Krebs Cycle Color Index: Original slides. Important. 436 Notes 438 notes : ل ی د ع ت ل ا ط ب ا ر https://docs.google.com/document/d/1wvdec1atp7j- ZKWOUSukSLsEcosjZ0AqV4z2VcH2TA0/edit?usp=sharing
Glycolysis Part 2. BCH 340 lecture 4
 Glycolysis Part 2 BCH 340 lecture 4 Regulation of Glycolysis There are three steps in glycolysis that have enzymes which regulate the flux of glycolysis These enzymes catalyzes irreversible reactions of
Glycolysis Part 2 BCH 340 lecture 4 Regulation of Glycolysis There are three steps in glycolysis that have enzymes which regulate the flux of glycolysis These enzymes catalyzes irreversible reactions of
a b c d e C 3 ]Aspartate [ 20 minutes C 3 ]Hexose-P * * *
![a b c d e C 3 ]Aspartate [ 20 minutes C 3 ]Hexose-P * * * a b c d e C 3 ]Aspartate [ 20 minutes C 3 ]Hexose-P * * *](/thumbs/92/110334219.jpg) Supplemental Figure 1 Metabolic flux with [U- C]Lactate - [ 12 C]Glutamine in primary hepatocytes a b c d e [ C 3 ]Pyruvate [ C 3 ]Malate [ C 3 ]Aspartate [ C 3 ]Citrate [ C 2 ] -Ketoglutarate f g h [
Supplemental Figure 1 Metabolic flux with [U- C]Lactate - [ 12 C]Glutamine in primary hepatocytes a b c d e [ C 3 ]Pyruvate [ C 3 ]Malate [ C 3 ]Aspartate [ C 3 ]Citrate [ C 2 ] -Ketoglutarate f g h [
Figure S1A. Blood glucose levels in mice after glucose injection
 ## Figure S1A. Blood glucose levels in mice after glucose injection Blood glucose (mm/l) 25 2 15 1 5 # 15 3 6 3+3 Time after glucose injection (min) # Figure S1B. α-kg levels in mouse livers after glucose
## Figure S1A. Blood glucose levels in mice after glucose injection Blood glucose (mm/l) 25 2 15 1 5 # 15 3 6 3+3 Time after glucose injection (min) # Figure S1B. α-kg levels in mouse livers after glucose
Cellular Respiration and Fermentation
 CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 7 Cellular Respiration and Fermentation Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION
CAMPBELL BIOLOGY IN FOCUS URRY CAIN WASSERMAN MINORSKY REECE 7 Cellular Respiration and Fermentation Lecture Presentations by Kathleen Fitzpatrick and Nicole Tunbridge, Simon Fraser University SECOND EDITION
Metabolism Gluconeogenesis/Citric Acid Cycle
 Metabolism Gluconeogenesis/Citric Acid Cycle BIOB111 CHEMISTRY & BIOCHEMISTRY Session 21 Session Plan Gluconeogenesis Cori Cycle Common Metabolic Pathway The Citric Acid Cycle Stoker 2014, p859 Gluconeogenesis
Metabolism Gluconeogenesis/Citric Acid Cycle BIOB111 CHEMISTRY & BIOCHEMISTRY Session 21 Session Plan Gluconeogenesis Cori Cycle Common Metabolic Pathway The Citric Acid Cycle Stoker 2014, p859 Gluconeogenesis
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Increased ABHD5 expression in human colon cancer associated macrophages. (a) Murine peritoneal macrophages were treated with regular culture medium (Ctrl) or
Supplementary Figures Supplementary Figure 1 Increased ABHD5 expression in human colon cancer associated macrophages. (a) Murine peritoneal macrophages were treated with regular culture medium (Ctrl) or
I tried to put as many questions as possible, but unfortunately only answers were found without the questions.
 I tried to put as many questions as possible, but unfortunately only answers were found without the questions. These are some questions from doctor2015 med exam : 1. One of them isn t acute phase protein
I tried to put as many questions as possible, but unfortunately only answers were found without the questions. These are some questions from doctor2015 med exam : 1. One of them isn t acute phase protein
I tried to put as many questions as possible, but unfortunately only answers were found without the questions.
 I tried to put as many questions as possible, but unfortunately only answers were found without the questions. These are some questions from doctor2015 med exam : 1. One of them isn t acute phase protein
I tried to put as many questions as possible, but unfortunately only answers were found without the questions. These are some questions from doctor2015 med exam : 1. One of them isn t acute phase protein
METABOLISM Sri Widia A Jusman Department of Biochemistry & Molecular Biology FMUI
 METABOLISM Sri Widia A Jusman Department of Biochemistry & Molecular Biology FMUI 1 METABOLISM Process of how cells acquire, transform, store and use energy Study of the chemistry, regulation and energetics
METABOLISM Sri Widia A Jusman Department of Biochemistry & Molecular Biology FMUI 1 METABOLISM Process of how cells acquire, transform, store and use energy Study of the chemistry, regulation and energetics
Chapter 9. Cellular Respiration and Fermentation
 Chapter 9 Cellular Respiration and Fermentation Energy flows into an ecosystem as sunlight and leaves as heat Photosynthesis generates O 2 and organic molecules, which are used in cellular respiration
Chapter 9 Cellular Respiration and Fermentation Energy flows into an ecosystem as sunlight and leaves as heat Photosynthesis generates O 2 and organic molecules, which are used in cellular respiration
