Supplemental Table 1 Fasting-response genes (353 genes) WT fast-wt fed Fold Change 1.5, p<0.05 Up-regulated on Fasting (143 genes)
|
|
|
- Elisabeth Armstrong
- 5 years ago
- Views:
Transcription
1 Supplemental Table 1 Fasting-response genes (353 genes) WT fast-wt fed Fold Change 1.5, p<0.05 Up-regulated on Fasting (143 genes) Gene Name UniGene Common Fold Change Description _at Mm Lepr leptin receptor _at Mm.4533 Apoa apolipoprotein A-IV _at Mm Insig insulin induced gene _s_at Mm Mte mitochondrial acyl-coa thioesterase _at Mm.1978 Acot cytosolic acyl-coa thioesterase _at 5.67 BB RIKEN full-length enriched, brain CRL-1443 BC3H1 cdna _at Mm Cyp4a cytochrome P450, family 4, subfamily a, polypeptide _at Mm Cyp8b cytochrome P450, family 8, subfamily b, polypeptide _s_at 4.48 AU Mouse two-cell stage embryo cdna _at Mm Trp53inp transformation related protein 53 inducible nuclear protein _s_at 4.42 BB RIKEN full-length enriched, adult male urinary bladder Mus musculus cdna clone _s_at 4.28 L0288G10-3 NIA Mouse Newborn Ovary cdna Library Mus musculus cdna clone _at Mm.1262 Cyp17a cytochrome P450, family 17, subfamily a, polypeptide _at Mm Crat 3.36 carnitine acetyltransferase _at Mm N10Rik 3.25 Adult male cecum cdna, RIKEN full-length enriched library, clone: h04 product:unknown EST _at Mm Ehhadh 3.25 enoyl-coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase _at 3.18 BB RIKEN full-length enriched, brain CRL-1443 BC3H1 cdna Mus musculus cdna clone _at Mm.3527 Ccng cyclin G _at Mm Pole polymerase (DNA-directed), epsilon 4 (p12 subunit) _at Mm Got glutamate oxaloacetate transaminase 1, soluble _at 2.74 RBM3; Mus musculus RNA-binding motif protein 3 mrna, complete cds _at Mm C05Rik 2.71 RIKEN cdna C05 gene _at Mm AA Transcribed sequences _at Mm H09Rik 2.62 Similar to phospholysine phosphohistidine inorganic pyrophosphate phosphata _at Mm Cxcl chemokine (C-X-C motif) ligand _at Mm Gfra glial cell line derived neurotrophic factor family receptor alpha _at Mm.6994 Hadh hydroxyacyl-coenzyme A dehydrogenase type II _a_at Mm.1566 Atf activating transcription factor _at Mm.381 Adfp 2.46 adipose differentiation related protein _at Mm C330008I15Rik 2.45 RIKEN cdna C330008I15 gene _a_at Mm Hnrpa Transcribed sequence with strong similarity to protein ref:np_ _a_at Mm.8362 Nnmt 2.40 nicotinamide N-methyltransferase _at Mm.1810 Ctgf 2.38 connective tissue growth factor _s_at Mm Rbpms 2.37 RNA binding protein gene with multiple splicing _at Mm Cyp4a cytochrome P450, family 4, subfamily a, polypeptide _at Mm Litaf 2.35 LPS-induced TN factor _at Mm Slc22a solute carrier family 22 (organic cation transporter), member _s_at Mm Actg 2.28 actin, gamma, cytoplasmic _a_at 2.28 AV Mus musculus hippocampus C57BL/6J adult Mus musculus cdna clone _s_at Mm P08Rik 2.28 RIKEN cdna P08 gene _at Mm Cyp2c cytochrome P450, family 2, subfamily c, polypeptide _a_at Mm Il17rb 2.28 interleukin 17 receptor B _at Mm Lrg leucine-rich alpha-2-glycoprotein _at Mm Cltb 2.27 clathrin, light polypeptide (Lcb) _at Mm K07Rik 2.20 RIKEN cdna K07 gene _a_at Mm Slc4a solute carrier family 4 (anion exchanger), member _a_at Mm Pabpc poly A binding protein, cytoplasmic _a_at Mm Nap1l nucleosome assembly protein 1-like _at Mm O18Rik 2.11 RIKEN cdna O18 gene _a_at Mm.4676 Slc7a solute carrier family 7 (cationic amino acid transporter, y+ system), member _a_at Mm Trim tripartite motif protein _s_at Mm Tpbpb 2.08 trophoblast specific protein beta _at Mm Asl 2.07 argininosuccinate lyase _at Mm Arl6ip ADP-ribosylation factor-like 6 interacting protein _at Mm P20Rik 2.06 RIKEN cdna P20 gene _a_at Mm Fh fumarate hydratase _at Mm O06Rik 2.04 RIKEN cdna O06 gene _at Mm Gbif 2.01 globin inducing factor, fetal _a_at Taf1b; Tafi86; mtafi68; RIKEN full-length enriched library, clone: g _a_at Mm Pla2g12a 1.99 phospholipase A2, group XIIA _at Mm F03Rik 1.96 RIKEN cdna F03 gene _at Mm Adk 1.96 adenosine kinase _a_at Mm Eya eyes absent 3 homolog (Drosophila) _at Mm Mtmr myotubularin related protein _at Mm Tbrg transforming growth factor beta regulated gene _at Mm Gata GATA binding protein _at Mm Grpel GrpE-like 2, mitochondrial _at Mm.8137 Chd chromodomain helicase DNA binding protein _at Mm Gosr golgi SNAP receptor complex member _at Mm AA expressed sequence AA _at Mm C01Rik 1.89 RIKEN cdna C01 gene _at Mm B10Rik 1.88 RIKEN cdna B10 gene _at Slc25a solute carrier family 25 (mitochondrial carrier; ornithine transporter), member _at Mm Rbms RNA binding motif, single stranded interacting protein _at Mm D11Ertd730e 1.86 DNA segment, Chr 11, ERATO Doi 730, expressed _at Mm Gpiap GPI-anchored membrane protein _s_at Mm Ccni 1.83 cyclin I
2 _s_at Mm Eps8l EPS8-like _a_at Mm Slc20a solute carrier family 20, member _a_at Mm Snx sorting nexin _at Mm.6720 Kctd potassium channel tetramerisation domain containing _at Mm H23Rik 1.80 RIKEN cdna H23 gene _s_at Mm Ppp2r2d 1.80 protein phosphatase 2, regulatory subunit B, delta isoform _a_at H08Rik; Cyt RIKEN full-length enriched library, clone: h _at Mm Lgals lectin, galactose binding, soluble _at Mm D16H22S680E 1.78 DNA segment, Chr 16, human D22S680E, expressed _at Mm Similar to ubiquitin specific protease 9, X-linked isoform _at Mm D7Ertd156e 1.75 DNA segment, Chr 7, ERATO Doi 156, expressed _x_at Mm Elovl BB RIKEN full-length enriched, Mus musculus cdna clone _at Mm Metap methionyl aminopeptidase _s_at Mm Atp6v1b ATPase, H+ transporting, V1 subunit B, isoform _at Mm AA expressed sequence AA _at Mm Cd CD84 antigen _at Mm Cpt1a 1.71 carnitine palmitoyltransferase 1a, liver _at Mm F11Rik 1.69 AV Mus musculus 18-day embryo C57BL/6J Mus musculus cdna clone _at Mm Dlst 1.69 dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) _s_at Mm AA expressed sequence AA _at Pzf 1.68 Mus musculus Balb/c zinc finger protein PZF (Pzf) mrna, complete cds _at Mm Ap3s adaptor-related protein complex 3, sigma 2 subunit _a_at Mm Atp9a 1.68 ATPase, class II, type 9A _at Mm Atp2a ATPase, Ca++ transporting, cardiac muscle, slow twitch _at Mm.3596 Cops COP9 (constitutive photomorphogenic) homolog, subunit 2 (Arabidopsis thaliana) _at Mm Bcap B-cell receptor-associated protein _at Mm Tomm translocase of outer mitochondrial membrane 20 homolog (yeast) _at Mm N03Rik 1.65 RIKEN cdna N03 gene _at Mm Akp alkaline phosphatase 2, liver _at Mm.3065 Xrn '-3' exoribonuclease _at Mm Transcribed sequence with strong similarity to protein sp:p00722 (E. coli) BGAL_ECOLI Beta-galactosidase _a_at 1.61 Transcribed sequence with moderate similarity to protein ref:np_ (H.sapiens) hypothetical protein _s_at Mm Galnt UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase _a_at Mm Psmd proteasome (prosome, macropain) 26S subunit, non-atpase, _at Mm M01Rik 1.60 RIKEN cdna M01 gene _at Mm.2941 Mttp 1.59 microsomal triglyceride transfer protein _at Mm Senp SUMO/sentrin specific protease _at Mm BC cdna sequence BC _at Mm.5062 Pctp 1.58 phosphatidylcholine transfer protein _at Mm.4428 Prkrir 1.58 protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (P58 repressor) _at Mm Gldc 1.57 glycine decarboxylase _a_at Mm C expressed sequence C _at Mm.3810 Pik3c2a 1.56 phosphatidylinositol 3-kinase, C2 domain containing, alpha polypeptide _a_at Mm Rbl retinoblastoma-like _s_at Mm G22Rik 1.56 RIKEN cdna G22 gene _x_at Mm Hspa heat shock protein _at Mm E08Rik 1.55 Transcribed sequences _at Mm Mat1a 1.55 methionine adenosyltransferase I, alpha _at Mm Tcp t-complex protein _at Mm F13Rik 1.55 RIKEN cdna F13 gene _at Mm M19Rik 1.54 RIKEN cdna M19 gene _at Mm Zfr 1.54 zinc finger RNA binding protein _a_at Mm Igfbp insulin-like growth factor binding protein _at Mm Arhg 1.54 ras homolog gene family, member G _at Mm Abhd abhydrolase domain containing _at Mrps mitochondrial ribosomal protein S _at Mm Ran 1.52 RAN, member RAS oncogene family _at Mm Etf eukaryotic translation termination factor _at Mm C730036B01Rik 1.52 RIKEN cdna C730036B01 gene _x_at 1.52 AV Mus musculus head C57BL/6J 13-day embryo Mus musculus cdna clone _at Mm Abhd abhydrolase domain containing _a_at Mm Amy amylase 1, salivary _a_at Mm Nf neurofibromatosis _at Mm Hspa9a 1.51 heat shock protein, A _at Mm Ap1b adaptor protein complex AP-1, beta 1 subunit _a_at Mm Ctsz 1.50 Cathepsin Z Down-regulated on Fasting (210 genes) Gene Name UniGene Common Fold Change Description _at Mm Thrsp 0.04 thyroid hormone responsive SPOT14 homolog (Rattus) _at Mm Fdps 0.07 farnesyl diphosphate synthetase _a_at Mm.741 Fabp fatty acid binding protein 5, epidermal _at Mm Idi isopentenyl-diphosphate delta isomerase _at Mm Cyp2c cytochrome P450, family 2, subfamily c, polypeptide _a_at Mm Sc4mol 0.09 sterol-c4-methyl oxidase-like _at Mm Aqp aquaporin _s_at Mm Gstm glutathione S-transferase, mu _at Mm Cyp4f14; O15Rik 0.14 Mus musculus cytochrome P450, family 4, subfamily f, polypeptide 14, mrna _at Mm Fasn 0.15 fatty acid synthase _at Stard4; C16Rik 0.15 RIKEN full-length enriched library, clone: c _at Mm Hmgcs hydroxy-3-methylglutaryl-Coenzyme A synthase 1
3 _at Mm BC cdna sequence BC _a_at Mm Lifr 0.17 leukemia inhibitory factor receptor _at Mm Cyp2c Similar to cytochrome P450, subfamily IIC (mephenytoin 4-hydroxylase), polypeptide _at Mm L01Rik 0.18 RIKEN cdna L01 gene _at Mm Ces carboxylesterase _at Mm Abca8a 0.21 ATP-binding cassette, sub-family A (ABC1), member 8a _at Mm Hmgcr hydroxy-3-methylglutaryl-Coenzyme A reductase _at Mm F02Rik 0.22 RIKEN cdna F02 gene _at Mm Rdh retinol dehydrogenase _at Mm C8a 0.23 complement component 8, alpha polypeptide _a_at Mm Nudt nudix (nucleoside diphosphate linked moiety X)-type motif _at Mm Dhcr dehydrocholesterol reductase _s_at 0.26 AV Mus musculus C57BL/6J day embryo Mus musculus cdna clone _x_at Mm Slc13a BB RIKEN full-length enriched, 0 day neonate kidney Mus musculus cdna clone _at Mm Dhcr dehydrocholesterol reductase _at Mm.2131 Ela elastase 1, pancreatic _at Mm Kcnk potassium channel, subfamily K, member _at Mm Gba glucosidase beta _at Mm.2436 Bhlhb basic helix-loop-helix domain containing, class B _x_at Mm Alas aminolevulinic acid synthase _x_at Mm AL expressed sequence AL _a_at Mm Wbscr Williams-Beuren syndrome chromosome region 14 homolog (human) _at Mm Upp uridine phosphorylase _at Mm Bucs butyryl Coenzyme A synthetase _s_at Mm Dia diaphorase 1 (NADH) _a_at Mm Acas acetyl-coenzyme A synthetase 2 (ADP forming) _at Mm F coagulation factor XI _a_at Mm C8b 0.33 complement component 8, beta subunit _at Mm B18Rik 0.33 RIKEN cdna B18 gene _at Mm Pklr 0.34 pyruvate kinase liver and red blood cell _at Strm 0.34 striamin _x_at 0.34 BB RIKEN full-length enriched, adult male pituitary gland Mus musculus cdna clone _at Mm Pstpip proline-serine-threonine phosphatase-interacting protein _at Mm Echdc enoyl Coenzyme A hydratase domain containing _at Mm C complement component _at Papss '-phosphoadenosine 5'-phosphosulfate synthase _at Mm Ly6a 0.37 lymphocyte antigen 6 complex, locus A _at Mm Abcb ATP-binding cassette, sub-family B (MDR/TAP), member _x_at Mm Acly 0.38 BB RIKEN full-length enriched, 10 days neonate cortex Mus musculus cdna clone _at Mm Dock dedicator of cytokinesis _x_at Mm Cetn BB RIKEN full-length enriched, 0 day neonate kidney Mus musculus cdna clone _at Mm Cyp7b cytochrome P450, family 7, subfamily b, polypeptide _at Mm C730032N17Rik 0.39 RIKEN cdna C730032N17 gene _at Mm Hpgd 0.39 hydroxyprostaglandin dehydrogenase 15 (NAD) _at Mm Ptprf 0.39 protein tyrosine phosphatase, receptor type, F _s_at Mm ORF open reading frame _s_at Mm Syne synaptic nuclear envelope _a_at Mm Rgs BB RIKEN full-length enriched, embryonic body between diaphragm region and neck Mus musculus cdna clone _at Mm Ifi interferon activated gene _a_at Mm Klhl kelch-like 13 (Drosophila) _a_at Mm Pdxk 0.41 pyridoxal (pyridoxine, vitamin B6) kinase _at Mm Zfp zinc finger protein _at Mm Nudt nudix (nucleoside diphosphate linked moiety X)-type motif _at Mm Slc3a solute carrier family 3, member _at Mm H04Rik 0.43 RIKEN cdna H04 gene _a_at Mm Mscp 0.44 mitochondrial solute carrier protein _at Mm K01Rik 0.45 RIKEN cdna K01 gene _at Mm.4067 Lisch liver-specific bhlh-zip transcription factor _at Mm.4578 Foxa forkhead box A _at Mm Sh3yl Sh3 domain YSC-like _a_at Mm K13Rik 0.45 RIKEN cdna K13 gene _at Mm Ulk Unc-51 like kinase 2 (C. elegans) _at Mm A230075M04Rik 0.45 RIKEN full-length enriched library, clone:d330042i16 product:unknown EST, full insert sequence _at Mm Aldh1a aldehyde dehydrogenase family 1, subfamily A _at Mm D11Ertd18e 0.46 DNA segment, Chr 11, ERATO Doi 18, expressed _a_at Mm.9394 Nfix 0.47 nuclear factor I/X _s_at Mm AW expressed sequence AW _at Mm.8655 Cfh 0.47 complement component factor h _x_at 0.47 AV RIKEN full-length enriched, ES cells Mus musculus cdna clone I20 3' similar to U _at Mm Rala 0.47 v-ral simian leukemia viral oncogene homolog A (ras related) _at Mm Slc39a solute carrier family 39 (zinc transporter), member _x_at Mm Aco BB RIKEN full-length enriched, 0 day neonate kidney Mus musculus cdna clone _a_at Mm Rdx 0.49 radixin _at Mm Ctsc 0.49 cathepsin C _at Mm Dgat diacylglycerol O-acyltransferase _a_at Mm Smarcd SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member _x_at Mm H21Rik 0.50 RIKEN cdna H21 gene _at Mm Cox7a cytochrome c oxidase, subunit VIIa _at Mm Cklfsf chemokine-like factor super family _at Mm Dexi 0.50 dexamethasone-induced transcript _s_at Mm Gch 0.50 GTP cyclohydrolase _at Mm Gckr 0.50 glucokinase regulatory protein _a_at Mm F18Rik 0.50 RIKEN cdna F18 gene
4 _s_at Mm.3758 Bcar BB RIKEN full-length enriched, 7 days neonate cerebellum Mus musculus cdna clone _a_at Mm Enpp ectonucleotide pyrophosphatase/phosphodiesterase _a_at Mm Slc33a solute carrier family 33 (acetyl-coa transporter), member _a_at Mm.1109 Wbp WW domain binding protein _x_at Mm H07Rik 0.51 BB RIKEN full-length enriched, 8 days embryo Mus musculus cdna clone _at Mm E08Rik 0.51 RIKEN cdna E08 gene _s_at Mm Arhgdia 0.51 Rho GDP dissociation inhibitor (GDI) alpha _x_at Mm D09Rik 0.52 BB RIKEN full-length enriched, 2 days neonate sympathetic ganglion Mus musculus cdna clone _a_at Mm Taldo transaldolase _at Mm BC cdna sequence BC _a_at Mm Polr3k 0.52 AV RIKEN full-length enriched, adult male testis (DH10B) Mus musculus cdna clone _a_at Klf13; Bteb3; NSLP1; FK 0.52 RIKEN full-length enriched library, clone: c13 product:kruppel-like factor 13, full insert sequence _x_at Mm H2-K 0.52 histocompatibility 2, K region _s_at Mm LOC esterase 31-like _at Mm H18Rik 0.52 RIKEN cdna H18 gene _x_at Mm Armcx BB RIKEN full-length enriched, 0 day neonate cerebellum Mus musculus cdna clone _at _a_at Mm.1775 Hn hematological and neurological expressed sequence _a_at Mm D22Rik 0.53 RIKEN cdna D22 gene _at Mm Arhgap Rho GTPase activating protein _at Mm Impa inositol (myo)-1(or 4)-monophosphatase _at Mm A17Rik 0.53 RIKEN cdna A17 gene _at Mm Clcn chloride channel _at Mm Lactb lactamase, beta _a_at Mm Ndfip Nedd4 family interacting protein _at Mm E04Rik 0.55 RIKEN cdna E04 gene _x_at Mm E16Rik 0.55 RIKEN cdna E16 gene _at Mm Gprc5c 0.55 G protein-coupled receptor, family C, group 5, member C _at Mm Nr3c nuclear receptor subfamily 3, group C, member _at Mm D3Ertd330e 0.55 DNA segment, Chr 3, ERATO Doi 330, expressed _at Mm B230342M21Rik 0.55 RIKEN cdna B230342M21 gene _x_at Mm Tmlhe 0.55 trimethyllysine hydroxylase, epsilon _x_at Mm Sh3bgrl 0.56 BB RIKEN full-length enriched, 7 days neonate cerebellum Mus musculus cdna clone _at Mm O22Rik 0.56 RIKEN cdna O22 gene _at Mm I11Rik 0.56 RIKEN cdna I11 gene _at Mm Dap 0.56 death-associated protein _s_at Wld 0.56 WldS-specific chimeric protein; Mus musculus UFD2/D4COLE1E fusion protein mrna, complete cds _s_at Mm Mapre microtubule-associated protein, RP/EB family, member _at Mm Abcg ATP-binding cassette, sub-family G (WHITE), member _at Mm Cnot CCR4-NOT transcription complex, subunit _at Mm Slc6a solute carrier family 6 (neurotransmitter transporter, GABA), member _x_at 0.56 BB RIKEN full-length enriched, in vitro fertilized eggs Mus musculus cdna clone _a_at Mm.2026 Birc baculoviral IAP repeat-containing _at Mm Slc7a solute carrier family 7 (cationic amino acid transporter, y+ system), member _a_at Mm Map2k mitogen activated protein kinase kinase _at Pbef pre-b-cell colony-enhancing factor _at Mm Wdr WD repeat domain _at Mm B05Rik 0.58 RIKEN cdna B05 gene _a_at Mm Atp6v1g ATPase, H+ transporting, V1 subunit G isoform _a_at Mm G24Rik 0.58 RIKEN cdna G24 gene _at Mm Manba 0.59 mannosidase, beta A, lysosomal _x_at Mm Sdbcag serologically defined breast cancer antigen _at Mm.6635 Ivd 0.59 isovaleryl coenzyme A dehydrogenase _a_at Mm Sumo AV Mus musculus 18-day embryo C57BL/6J Mus musculus cdna clone _at Mm.4827 F coagulation factor VII _at Mm Slc38a solute carrier family 38, member _at Mm Gmnn 0.60 geminin _a_at 0.60 Mus musculus mrna similar to dual specificity phosphatase 9 (cdna clone MGC:6681 IMAGE: ), complete cds _at Mm Slc12a solute carrier family 12, member _a_at Mm D530020C15Rik 0.60 RIKEN cdna D530020C15 gene _at Mm.1151 Dpp dipeptidylpeptidase _at Mm.1213 Masp mannan-binding lectin serine protease _at Mm Mmaa 0.60 methylmalonic aciduria (cobalamin deficiency) type A _at Mm E01Rik 0.61 RIKEN cdna E01 gene _at Mm Aox aldehyde oxidase _at Mm Gstm glutathione S-transferase, mu _at Mm Xdh 0.61 xanthine dehydrogenase _at Mm Qprt 0.61 quinolinate phosphoribosyltransferase _at Mm.2046 Cdadc cytidine and dcmp deaminase domain containing _x_at Sema4a 0.61 BB RIKEN full-length enriched, adult male urinary bladder Mus musculus cdna clone _a_at Mm D18Rik 0.61 RIKEN cdna D18 gene _at Mm Ap3m adaptor-related protein complex 3, mu 1 subunit _at Mm Sec61b 0.62 Sec61 beta subunit _at Mm Zfp zinc finger protein _at Mm Gstm glutathione S-transferase, mu _at Mm Snrk 0.62 SNF related kinase _a_at Mm Igf insulin-like growth factor _s_at Mm Lman lectin, mannose-binding, _at Mm Pygl 0.62 liver glycogen phosphorylase _a_at Mm Scotin 0.62 BB RIKEN full-length enriched, 0 day neonate lung Mus musculus cdna clone _a_at Mm Cri CREBBP/EP300 inhibitory protein _x_at Mm Rpn ribophorin I _at Mm Cds BB RIKEN full-length enriched, 13 days embryo lung Mus musculus cdna clone
5 _x_at Mm Transcribed sequences _at Mm Krtcap keratinocyte associated protein _at Mm Casp caspase _a_at Mm Zfp zinc finger protein _at Mm Apg10l 0.63 autophagy 10-like (S. cerevisiae) _at Mm Dusp dual specificity phosphatase _at Mm E10Rik 0.63 RIKEN cdna E10 gene _at D4Wsu53e 0.63 DNA segment, Chr 4, Wayne State University 53, expressed _at Mm Spag sperm associated antigen _at Mm Nsbp nucleosome binding protein _a_at Mm L04Rik 0.63 RIKEN cdna L04 gene _a_at Mm Pdcd programmed cell death _at Mm Rga 0.64 recombination activating gene 1 gene activation _at Mm Transcribed sequence with moderate similarity to protein pir:r5hu35 (H.sapiens) R5HU35 ribosomal protein L35a - human _at Mm.3636 Dbt 0.64 dihydrolipoamide branched chain transacylase E _a_at Mm D4Wsu43e 0.64 DNA segment, Chr 4, Wayne State University 43, expressed _at Mm E09Rik 0.64 RIKEN cdna E09 gene _a_at Mm Pacsin protein kinase C and casein kinase substrate in neurons _x_at Mm Ampd AV RIKEN full-length enriched, adult male medulla oblongata Mus musculus cdna clone _at Mm Chn chimerin (chimaerin) _at Mm days embryo head cdna, RIKEN full-length enriched library, clone:c130029f _at Mm Spred sprouty protein with EVH-1 domain 1, related sequence _at Mm O10Rik 0.65 RIKEN cdna O10 gene _a_at Mm Uxs UDP-glucuronate decarboxylase _a_at Mm G12Rik 0.66 RIKEN cdna G12 gene _x_at Agl 0.66 amylo-1,6-glucosidase, 4-alpha-glucanotransferase _at Mm Ankrd ankyrin repeat domain _at Mm Atp2c ATPase, Ca++-sequestering _at Mm Gclm 0.66 glutamate-cysteine ligase, modifier subunit _a_at Mm Risc 0.66 retinoid-inducible serine caroboxypetidase _at Mm Naglu 0.67 alpha-n-acetylglucosaminidase (Sanfilippo disease IIIB) _at Mm Ttrap 0.69 Traf and Tnf receptor associated protein
Supplementary Table SI: Y strain T. cruzi infection results in the upregulation of 381 genes at the site of infection.
 Supplementary Table SI: Y strain T. cruzi infection results in the upregulation of 381 genes at the site of infection. Genes found to be significantly upregulated (FDR2) in Y strain
Supplementary Table SI: Y strain T. cruzi infection results in the upregulation of 381 genes at the site of infection. Genes found to be significantly upregulated (FDR2) in Y strain
Table S1 Differentially expressed genes showing > 2 fold changes and p <0.01 for 0.01 mm NS-398, 0.1mM ibuprofen and COX-2 RNAi.
 Table S1 Differentially expressed genes showing > 2 fold changes and p
Table S1 Differentially expressed genes showing > 2 fold changes and p
TNFSF13B tumor necrosis factor (ligand) superfamily, member 13b NF-kB pathway cluster, Enrichment Score: 3.57
 Appendix 2. Highly represented clusters of genes in the differential expression of data. Immune Cluster, Enrichment Score: 5.17 GO:0048584 positive regulation of response to stimulus GO:0050778 positive
Appendix 2. Highly represented clusters of genes in the differential expression of data. Immune Cluster, Enrichment Score: 5.17 GO:0048584 positive regulation of response to stimulus GO:0050778 positive
SUPPLEMENTAL TABLE I. Identified Proteins in Bovine Testicular Hyaluronidase Type I-S via LC-MS/MS
 SUPPLEMENTAL TABLE I. Identified Proteins in Bovine Testicular Hyaluronidase Type I-S via LC-MS/MS No. Protein 1 serum albumin precursor gi 30794280 2 annexin A2 gi 27807289 3 Phosphatidylethanolamine-binding
SUPPLEMENTAL TABLE I. Identified Proteins in Bovine Testicular Hyaluronidase Type I-S via LC-MS/MS No. Protein 1 serum albumin precursor gi 30794280 2 annexin A2 gi 27807289 3 Phosphatidylethanolamine-binding
Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function
 Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSL0024 26223 26621 + LrgA family BPSL0025 26690 27412 + hypothetical
Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSL0024 26223 26621 + LrgA family BPSL0025 26690 27412 + hypothetical
Signal Transduction Cascades
 Signal Transduction Cascades Contents of this page: Kinases & phosphatases Protein Kinase A (camp-dependent protein kinase) G-protein signal cascade Structure of G-proteins Small GTP-binding proteins,
Signal Transduction Cascades Contents of this page: Kinases & phosphatases Protein Kinase A (camp-dependent protein kinase) G-protein signal cascade Structure of G-proteins Small GTP-binding proteins,
Mouse Meda-4 : chromosome 5G bp. EST (547bp) _at. 5 -Meda4 inner race (~1.8Kb)
 Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
SUPPLEMENTARY FIG. S2. b-galactosidase staining of
 SUPPLEMENTARY FIG. S1. b-galactosidase staining of senescent cells in 500 mg=dl glucose (10 magnification). SUPPLEMENTARY FIG. S3. Toluidine blue staining of chondrogenic-differentiated adipose-tissue-derived
SUPPLEMENTARY FIG. S1. b-galactosidase staining of senescent cells in 500 mg=dl glucose (10 magnification). SUPPLEMENTARY FIG. S3. Toluidine blue staining of chondrogenic-differentiated adipose-tissue-derived
BIOL 158: BIOLOGICAL CHEMISTRY II
 BIOL 158: BIOLOGICAL CHEMISTRY II Lecture 5: Vitamins and Coenzymes Lecturer: Christopher Larbie, PhD Introduction Cofactors bind to the active site and assist in the reaction mechanism Apoenzyme is an
BIOL 158: BIOLOGICAL CHEMISTRY II Lecture 5: Vitamins and Coenzymes Lecturer: Christopher Larbie, PhD Introduction Cofactors bind to the active site and assist in the reaction mechanism Apoenzyme is an
Enzyme-coupled Receptors. Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors
 Enzyme-coupled Receptors Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors Cell-surface receptors allow a flow of ions across the plasma
Enzyme-coupled Receptors Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors Cell-surface receptors allow a flow of ions across the plasma
Chapter 10. Introduction to Nutrition and Metabolism, 3 rd edition David A Bender Taylor & Francis Ltd, London 2002
 Chapter 10 Introduction to Nutrition and Metabolism, 3 rd edition David A Bender Taylor & Francis Ltd, London 2002 Chapter 10: Integration and Control of Metabolism Press the space bar or click the mouse
Chapter 10 Introduction to Nutrition and Metabolism, 3 rd edition David A Bender Taylor & Francis Ltd, London 2002 Chapter 10: Integration and Control of Metabolism Press the space bar or click the mouse
Supplementary Materials Modeling cholesterol metabolism by gene expression profiling in the hippocampus
 Supplementary Materials Modeling cholesterol metabolism by gene expression profiling in the hippocampus Christopher M. Valdez 1, Clyde F. Phelix 1, Mark A. Smith 3, George Perry 1,2, and Fidel Santamaria
Supplementary Materials Modeling cholesterol metabolism by gene expression profiling in the hippocampus Christopher M. Valdez 1, Clyde F. Phelix 1, Mark A. Smith 3, George Perry 1,2, and Fidel Santamaria
Validated Mouse Quantitative RT-PCR Genes Gene Gene Bank Accession Number Name
 5HT1a NM_008308.4 Serotonin Receptor 1a 5HT1b NM_010482.1 Serotonin Receptor 1b 5HT2a NM_172812.2 Serotonin Receptor 2a ACO-2 NM_080633.2 Aconitase 2 Adora2A NM_009630.02 Adenosine A2a Receptor Aif1 (IbaI)
5HT1a NM_008308.4 Serotonin Receptor 1a 5HT1b NM_010482.1 Serotonin Receptor 1b 5HT2a NM_172812.2 Serotonin Receptor 2a ACO-2 NM_080633.2 Aconitase 2 Adora2A NM_009630.02 Adenosine A2a Receptor Aif1 (IbaI)
Post-translational modifications of proteins in gene regulation under hypoxic conditions
 203 Review Article Post-translational modifications of proteins in gene regulation under hypoxic conditions 1, 2) Olga S. Safronova 1) Department of Cellular Physiological Chemistry, Tokyo Medical and
203 Review Article Post-translational modifications of proteins in gene regulation under hypoxic conditions 1, 2) Olga S. Safronova 1) Department of Cellular Physiological Chemistry, Tokyo Medical and
Biological processes. Mitochondrion Metabolic process Catalytic activity Oxidoreductase
 Full name Glyceraldehyde3 phosphate dehydrogenase Succinatesemialdehyde Glutamate dehydrogenase 1, Alcohol dehydrogenase [NADP+] 2',3'cyclicnucleotide 3' phosphodiesterase Dihydropyrimidinaserelated 2
Full name Glyceraldehyde3 phosphate dehydrogenase Succinatesemialdehyde Glutamate dehydrogenase 1, Alcohol dehydrogenase [NADP+] 2',3'cyclicnucleotide 3' phosphodiesterase Dihydropyrimidinaserelated 2
Fig. S1. Summary of the altered metabolism pathways in alcoholic fatty liver disease using MetPA analysis (panel A).
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Fig. S1. Summary of the altered metabolism pathways in alcoholic fatty liver disease
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Fig. S1. Summary of the altered metabolism pathways in alcoholic fatty liver disease
Table S1. Differentially expressed transcripts between hepatic inkt cells. sorted form WT and plck-hcd1d tg mice. Differentially expressed
 Supplementary Information Table S1. Differentially expressed transcripts between hepatic inkt cells sorted form and plck-hcd1d tg mice. Differentially expressed transcripts identified by gene expression
Supplementary Information Table S1. Differentially expressed transcripts between hepatic inkt cells sorted form and plck-hcd1d tg mice. Differentially expressed transcripts identified by gene expression
Energy storage in cells
 Energy storage in cells Josef Fontana EC - 58 Overview of the lecture Introduction to the storage substances of human body Overview of storage compounds in the body Glycogen metabolism Structure of glycogen
Energy storage in cells Josef Fontana EC - 58 Overview of the lecture Introduction to the storage substances of human body Overview of storage compounds in the body Glycogen metabolism Structure of glycogen
2. What is molecular oxygen directly converted into? a. Carbon Dioxide b. Water c. Glucose d. None of the Above
 Biochem 1 Mock Exam 3 Chapter 11: 1. What is glucose completely oxidized into? a. Carbon Dioxide and Water b. Carbon Dioxide and Oxygen c. Oxygen and Water d. Water and Glycogen 2. What is molecular oxygen
Biochem 1 Mock Exam 3 Chapter 11: 1. What is glucose completely oxidized into? a. Carbon Dioxide and Water b. Carbon Dioxide and Oxygen c. Oxygen and Water d. Water and Glycogen 2. What is molecular oxygen
Supplementary Figure 1. Mitochondrial function in skeletal muscle and plasma parameters of STZ mice A D-F
 Supplementary Figure 1. Mitochondrial function in skeletal muscle and plasma parameters of STZ mice A Expression of representative subunits of the mitochondrial respiratory chain was analyzed by real time
Supplementary Figure 1. Mitochondrial function in skeletal muscle and plasma parameters of STZ mice A Expression of representative subunits of the mitochondrial respiratory chain was analyzed by real time
Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion
 Probe Set ID RefSeq Transcript ID Gene Title Gene Symbol 205980_s_at NM_001017526 /// NM_181334 /// NM_181335 Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion ARHGAP8 /// LOC553158 FC ALK shrna
Probe Set ID RefSeq Transcript ID Gene Title Gene Symbol 205980_s_at NM_001017526 /// NM_181334 /// NM_181335 Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion ARHGAP8 /// LOC553158 FC ALK shrna
Companion to Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism Lecture Notes
 Companion to Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism Lecture Notes The major site of acetoacetate and 3-hydorxybutyrate production is in the liver. 3-hydorxybutyrate is the
Companion to Biosynthesis of Ketones & Cholesterols, Regulation of Lipid Metabolism Lecture Notes The major site of acetoacetate and 3-hydorxybutyrate production is in the liver. 3-hydorxybutyrate is the
Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction. Marker Sequence (5 3 ) Accession No.
 Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Cell Physiology Final Exam Fall 2008
 Cell Physiology Final Exam Fall 2008 Guys, The average on the test was 69.9. Before you start reading the right answers please do me a favor and remember till the end of your life that GLUCOSE TRANSPORT
Cell Physiology Final Exam Fall 2008 Guys, The average on the test was 69.9. Before you start reading the right answers please do me a favor and remember till the end of your life that GLUCOSE TRANSPORT
Chapter 10. 이화작용 : 에너지방출과보존 (Catabolism: Energy Release and Conservation)
 Chapter 10 이화작용 : 에너지방출과보존 (Catabolism: Energy Release and Conservation) 1 Fueling Processes Respiration 1 Most respiration involves use of an electron transport chain As electrons pass through the electron
Chapter 10 이화작용 : 에너지방출과보존 (Catabolism: Energy Release and Conservation) 1 Fueling Processes Respiration 1 Most respiration involves use of an electron transport chain As electrons pass through the electron
University of Guelph Department of Chemistry and Biochemistry Structure and Function In Biochemistry
 University of Guelph Department of Chemistry and Biochemistry 19-356 Structure and Function In Biochemistry Final Exam, April 21, 1997. Time allowed, 120 min. Answer questions 1-30 on the computer scoring
University of Guelph Department of Chemistry and Biochemistry 19-356 Structure and Function In Biochemistry Final Exam, April 21, 1997. Time allowed, 120 min. Answer questions 1-30 on the computer scoring
Cell Signaling part 2
 15 Cell Signaling part 2 Functions of Cell Surface Receptors Other cell surface receptors are directly linked to intracellular enzymes. The largest family of these is the receptor protein tyrosine kinases,
15 Cell Signaling part 2 Functions of Cell Surface Receptors Other cell surface receptors are directly linked to intracellular enzymes. The largest family of these is the receptor protein tyrosine kinases,
Receptor mediated Signal Transduction
 Receptor mediated Signal Transduction G-protein-linked receptors adenylyl cyclase camp PKA Organization of receptor protein-tyrosine kinases From G.M. Cooper, The Cell. A molecular approach, 2004, third
Receptor mediated Signal Transduction G-protein-linked receptors adenylyl cyclase camp PKA Organization of receptor protein-tyrosine kinases From G.M. Cooper, The Cell. A molecular approach, 2004, third
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies
 Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
CO 2 tolerance of Atlantic salmon post-smolts in recirculating aquaculture systems
 CO 2 tolerance of Atlantic salmon post-smolts in recirculating aquaculture systems Vasco Mota*, Tom Nilsen, Elizabeth Ytteborg, Grete Baeverfjord, Aleksei Krasnov, Jelena Kolarevic, Lars Ebbesson, Steven
CO 2 tolerance of Atlantic salmon post-smolts in recirculating aquaculture systems Vasco Mota*, Tom Nilsen, Elizabeth Ytteborg, Grete Baeverfjord, Aleksei Krasnov, Jelena Kolarevic, Lars Ebbesson, Steven
* Kyoto Encyclopedia of Genes and Genomes.
 Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
Supplemental Material Complete gene expression data using Affymetrix 3PRIME IVT ID Chip (54,614 genes) and human immature dendritic cells stimulated with rbmasnrs, IL-8 and control (media) has been deposited
Signaling Through Immune System Receptors (Ch. 7)
 Signaling Through Immune System Receptors (Ch. 7) 1. General principles of signal transduction and propagation. 2. Antigen receptor signaling and lymphocyte activation. 3. Other receptors and signaling
Signaling Through Immune System Receptors (Ch. 7) 1. General principles of signal transduction and propagation. 2. Antigen receptor signaling and lymphocyte activation. 3. Other receptors and signaling
BIOLOGY 103 Spring 2001 MIDTERM LAB SECTION
 BIOLOGY 103 Spring 2001 MIDTERM NAME KEY LAB SECTION ID# (last four digits of SS#) STUDENT PLEASE READ. Do not put yourself at a disadvantage by revealing the content of this exam to your classmates. Your
BIOLOGY 103 Spring 2001 MIDTERM NAME KEY LAB SECTION ID# (last four digits of SS#) STUDENT PLEASE READ. Do not put yourself at a disadvantage by revealing the content of this exam to your classmates. Your
NBCE Mock Board Questions Biochemistry
 1. Fluid mosaic describes. A. Tertiary structure of proteins B. Ribosomal subunits C. DNA structure D. Plasma membrane structure NBCE Mock Board Questions Biochemistry 2. Where in the cell does beta oxidation
1. Fluid mosaic describes. A. Tertiary structure of proteins B. Ribosomal subunits C. DNA structure D. Plasma membrane structure NBCE Mock Board Questions Biochemistry 2. Where in the cell does beta oxidation
Role of metabolism in Drug-Induced Liver Injury (DILI) Drug Metab Rev. 2007;39(1):
 Role of metabolism in Drug-Induced Liver Injury (DILI) Drug Metab Rev. 2007;39(1):159-234 Drug Metab Rev. 2007;39(1):159-234 Drug Metab Rev. 2007;39(1):159-234 A schematic representation of the most relevant
Role of metabolism in Drug-Induced Liver Injury (DILI) Drug Metab Rev. 2007;39(1):159-234 Drug Metab Rev. 2007;39(1):159-234 Drug Metab Rev. 2007;39(1):159-234 A schematic representation of the most relevant
CHE 242 Exam 3 Practice Questions
 CHE 242 Exam 3 Practice Questions Glucose metabolism 1. Below is depicted glucose catabolism. Indicate on the pathways the following: A) which reaction(s) of glycolysis are irreversible B) where energy
CHE 242 Exam 3 Practice Questions Glucose metabolism 1. Below is depicted glucose catabolism. Indicate on the pathways the following: A) which reaction(s) of glycolysis are irreversible B) where energy
Tala Saleh. Razi Kittaneh ... Nayef Karadsheh
 Tala Saleh Razi Kittaneh... Nayef Karadsheh β-oxidation of Fatty Acids The oxidation of fatty acids occurs in 3 steps: Step 1: Activation of the Fatty acid FA + HS-CoA + ATP FA-CoA + AMP + PPi - The fatty
Tala Saleh Razi Kittaneh... Nayef Karadsheh β-oxidation of Fatty Acids The oxidation of fatty acids occurs in 3 steps: Step 1: Activation of the Fatty acid FA + HS-CoA + ATP FA-CoA + AMP + PPi - The fatty
Lehninger 5 th ed. Chapter 17
 Lehninger 5 th ed. Chapter 17 December 26, 2010 Prof. Shimon Schuldiner Email: Shimon.Schuldiner@huji.ac.il Phone: 6585992 CHAPTER 17 Fatty Acid Catabolism Key topics: How fats are digested in animals
Lehninger 5 th ed. Chapter 17 December 26, 2010 Prof. Shimon Schuldiner Email: Shimon.Schuldiner@huji.ac.il Phone: 6585992 CHAPTER 17 Fatty Acid Catabolism Key topics: How fats are digested in animals
ACCESSORY PUBLICATION. Supplementary Table 1. Genes up regulated in the skin of both HR and LR animals. after tick larval challenge e
 ACCESSORY PUBLICATION Supplementary Table 1. Genes up regulated in the skin of both HR and LR animals after tick larval challenge e GenBank Accession a No. of elemen ts b t=0 Signal intensity c t=0 Description
ACCESSORY PUBLICATION Supplementary Table 1. Genes up regulated in the skin of both HR and LR animals after tick larval challenge e GenBank Accession a No. of elemen ts b t=0 Signal intensity c t=0 Description
Cellular Signaling Pathways. Signaling Overview
 Cellular Signaling Pathways Signaling Overview Signaling steps Synthesis and release of signaling molecules (ligands) by the signaling cell. Transport of the signal to the target cell Detection of the
Cellular Signaling Pathways Signaling Overview Signaling steps Synthesis and release of signaling molecules (ligands) by the signaling cell. Transport of the signal to the target cell Detection of the
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at
 Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array.
 Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Enzymes Part III: regulation II. Dr. Mamoun Ahram Summer, 2017
 Enzymes Part III: regulation II Dr. Mamoun Ahram Summer, 2017 Advantage This is a major mechanism for rapid and transient regulation of enzyme activity. A most common mechanism is enzyme phosphorylation
Enzymes Part III: regulation II Dr. Mamoun Ahram Summer, 2017 Advantage This is a major mechanism for rapid and transient regulation of enzyme activity. A most common mechanism is enzyme phosphorylation
Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting
 Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting Question No. 1 of 10 Question 1. Which of the following statements about the nucleus is correct? Question #01 A. The
Molecular Cell Biology Problem Drill 16: Intracellular Compartment and Protein Sorting Question No. 1 of 10 Question 1. Which of the following statements about the nucleus is correct? Question #01 A. The
G-Protein Signaling. Introduction to intracellular signaling. Dr. SARRAY Sameh, Ph.D
 G-Protein Signaling Introduction to intracellular signaling Dr. SARRAY Sameh, Ph.D Cell signaling Cells communicate via extracellular signaling molecules (Hormones, growth factors and neurotransmitters
G-Protein Signaling Introduction to intracellular signaling Dr. SARRAY Sameh, Ph.D Cell signaling Cells communicate via extracellular signaling molecules (Hormones, growth factors and neurotransmitters
Glycolysis. Cellular Respiration
 Glucose is the preferred carbohydrate of cells. In solution, it can change from a linear chain to a ring. Energy is stored in the bonds of the carbohydrates. Breaking these bonds releases that energy.
Glucose is the preferred carbohydrate of cells. In solution, it can change from a linear chain to a ring. Energy is stored in the bonds of the carbohydrates. Breaking these bonds releases that energy.
Lecture #27 Lecturer A. N. Koval
 Lecture #27 Lecturer A. N. Koval Hormones Transduce Signals to Affect Homeostatic Mechanisms Koval A. (C), 2011 2 Lipophilic hormones Classifying hormones into hydrophilic and lipophilic molecules indicates
Lecture #27 Lecturer A. N. Koval Hormones Transduce Signals to Affect Homeostatic Mechanisms Koval A. (C), 2011 2 Lipophilic hormones Classifying hormones into hydrophilic and lipophilic molecules indicates
Mitochondria and ATP Synthesis
 Mitochondria and ATP Synthesis Mitochondria and ATP Synthesis 1. Mitochondria are sites of ATP synthesis in cells. 2. ATP is used to do work; i.e. ATP is an energy source. 3. ATP hydrolysis releases energy
Mitochondria and ATP Synthesis Mitochondria and ATP Synthesis 1. Mitochondria are sites of ATP synthesis in cells. 2. ATP is used to do work; i.e. ATP is an energy source. 3. ATP hydrolysis releases energy
Lecture 10 - Protein Turnover and Amino Acid Catabolism
 Lecture 10 - Protein Turnover and Amino Acid Catabolism Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction 2 Proteins are degraded into amino acids. Protein
Lecture 10 - Protein Turnover and Amino Acid Catabolism Chem 454: Regulatory Mechanisms in Biochemistry University of Wisconsin-Eau Claire 1 Introduction 2 Proteins are degraded into amino acids. Protein
Epigenetics. Lyle Armstrong. UJ Taylor & Francis Group. f'ci Garland Science NEW YORK AND LONDON
 ... Epigenetics Lyle Armstrong f'ci Garland Science UJ Taylor & Francis Group NEW YORK AND LONDON Contents CHAPTER 1 INTRODUCTION TO 3.2 CHROMATIN ARCHITECTURE 21 THE STUDY OF EPIGENETICS 1.1 THE CORE
... Epigenetics Lyle Armstrong f'ci Garland Science UJ Taylor & Francis Group NEW YORK AND LONDON Contents CHAPTER 1 INTRODUCTION TO 3.2 CHROMATIN ARCHITECTURE 21 THE STUDY OF EPIGENETICS 1.1 THE CORE
Under aerobic conditions, pyruvate enters the mitochondria where it is converted into acetyl CoA.
 Under aerobic conditions, pyruvate enters the mitochondria where it is converted into acetyl CoA. Acetyl CoA is the fuel for the citric acid cycle, which processes the two carbon acetyl unit to two molecules
Under aerobic conditions, pyruvate enters the mitochondria where it is converted into acetyl CoA. Acetyl CoA is the fuel for the citric acid cycle, which processes the two carbon acetyl unit to two molecules
SP GPI Found in exosomes
 Accession /Name/Functional group Spectr. counts SP GPI Found in exosomes Description Metabolism and energy production (21) CNAG_06770 fructose-bisphosphate aldolase 0.94 - - + gluconeogenesis and glycolysis
Accession /Name/Functional group Spectr. counts SP GPI Found in exosomes Description Metabolism and energy production (21) CNAG_06770 fructose-bisphosphate aldolase 0.94 - - + gluconeogenesis and glycolysis
Syllabus for BASIC METABOLIC PRINCIPLES
 Syllabus for BASIC METABOLIC PRINCIPLES The video lecture covers basic principles you will need to know for the lectures covering enzymes and metabolism in Principles of Metabolism and elsewhere in the
Syllabus for BASIC METABOLIC PRINCIPLES The video lecture covers basic principles you will need to know for the lectures covering enzymes and metabolism in Principles of Metabolism and elsewhere in the
Comparison of catabolic and anabolic pathways
 Comparison of catabolic and anabolic pathways Three stages of catabolism Glucose Synthesis of compounds e.g. lactose glycolipids Glucose-6-P Pentosephosphate Pathway Glycolysis Glycogenesis Acetyl-CoA
Comparison of catabolic and anabolic pathways Three stages of catabolism Glucose Synthesis of compounds e.g. lactose glycolipids Glucose-6-P Pentosephosphate Pathway Glycolysis Glycogenesis Acetyl-CoA
Lecture 29: Membrane Transport and metabolism
 Chem*3560 Lecture 29: Membrane Transport and metabolism Insulin controls glucose uptake Adipose tissue and muscles contain a passive glucose transporter GluT4 which takes up glucose from blood. (This is
Chem*3560 Lecture 29: Membrane Transport and metabolism Insulin controls glucose uptake Adipose tissue and muscles contain a passive glucose transporter GluT4 which takes up glucose from blood. (This is
MITOCHONDRIA LECTURES OVERVIEW
 1 MITOCHONDRIA LECTURES OVERVIEW A. MITOCHONDRIA LECTURES OVERVIEW Mitochondrial Structure The arrangement of membranes: distinct inner and outer membranes, The location of ATPase, DNA and ribosomes The
1 MITOCHONDRIA LECTURES OVERVIEW A. MITOCHONDRIA LECTURES OVERVIEW Mitochondrial Structure The arrangement of membranes: distinct inner and outer membranes, The location of ATPase, DNA and ribosomes The
SUPPLEMENTARY INFORMATION
 Supplementary Table 1- Differential expression of stress-related genes. Feature Number Probe Name Systematic Name Gene Name Description 30248 A_52_P342860 NM_007954 Es1 Mus musculus esterase 1 (Es1), mrna
Supplementary Table 1- Differential expression of stress-related genes. Feature Number Probe Name Systematic Name Gene Name Description 30248 A_52_P342860 NM_007954 Es1 Mus musculus esterase 1 (Es1), mrna
Lecture 16. Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III
 Lecture 16 Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III The Powertrain of Human Metabolism (verview) CARBHYDRATES PRTEINS
Lecture 16 Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III The Powertrain of Human Metabolism (verview) CARBHYDRATES PRTEINS
Biosynthesis of Fatty Acids
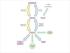 Biosynthesis of Fatty Acids Fatty acid biosynthesis takes place in the cytosol rather than the mitochondria and requires a different activation mechanism and different enzymes and coenzymes than fatty
Biosynthesis of Fatty Acids Fatty acid biosynthesis takes place in the cytosol rather than the mitochondria and requires a different activation mechanism and different enzymes and coenzymes than fatty
LIPID METABOLISM
 LIPID METABOLISM LIPOGENESIS LIPOGENESIS LIPOGENESIS FATTY ACID SYNTHESIS DE NOVO FFA in the blood come from :- (a) Dietary fat (b) Dietary carbohydrate/protein in excess of need FA TAG Site of synthesis:-
LIPID METABOLISM LIPOGENESIS LIPOGENESIS LIPOGENESIS FATTY ACID SYNTHESIS DE NOVO FFA in the blood come from :- (a) Dietary fat (b) Dietary carbohydrate/protein in excess of need FA TAG Site of synthesis:-
Activation of the endocannabinoid system by organophosphorus nerve agents
 1 Supplementary Information Activation of the endocannabinoid system by organophosphorus nerve agents Daniel K. Nomura, 1 Jacqueline L. Blankman, 2 Gabriel M. Simon, 2 Kazutoshi Fujioka, 1 Roger S. Issa,
1 Supplementary Information Activation of the endocannabinoid system by organophosphorus nerve agents Daniel K. Nomura, 1 Jacqueline L. Blankman, 2 Gabriel M. Simon, 2 Kazutoshi Fujioka, 1 Roger S. Issa,
Chapter 9. Cellular Signaling
 Chapter 9 Cellular Signaling Cellular Messaging Page 215 Cells can signal to each other and interpret the signals they receive from other cells and the environment Signals are most often chemicals The
Chapter 9 Cellular Signaling Cellular Messaging Page 215 Cells can signal to each other and interpret the signals they receive from other cells and the environment Signals are most often chemicals The
Protein Name. IFLENVIR,DSVTYTEHAK,TV TALDVVYALK,KTVTALDVV YALK,TVTALDVVYALKR,IF LENVIRDSVTYTEHAK gi Histone H2B
 Table 1. A functional category list of proteins (Lentinula edodes) identified by 1-DGE and nesi-lc-ms/ms. The table lists indicated fraction numbers, matching peptides, scores, accession numbers, protein
Table 1. A functional category list of proteins (Lentinula edodes) identified by 1-DGE and nesi-lc-ms/ms. The table lists indicated fraction numbers, matching peptides, scores, accession numbers, protein
MBB 115:511 and 694:407 Final Exam Niederman/Deis
 MBB 115:511 and 694:407 Final Exam Niederman/Deis Tue. Dec. 19, 2006 Name Row Letter Seat Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is worth two points.
MBB 115:511 and 694:407 Final Exam Niederman/Deis Tue. Dec. 19, 2006 Name Row Letter Seat Number This exam consists of two parts. Part I is multiple choice. Each of these 25 questions is worth two points.
Citric acid cycle and respiratory chain. Pavla Balínová
 Citric acid cycle and respiratory chain Pavla Balínová Mitochondria Structure of mitochondria: Outer membrane Inner membrane (folded) Matrix space (mtdna, ribosomes, enzymes of CAC, β-oxidation of FA,
Citric acid cycle and respiratory chain Pavla Balínová Mitochondria Structure of mitochondria: Outer membrane Inner membrane (folded) Matrix space (mtdna, ribosomes, enzymes of CAC, β-oxidation of FA,
March 19 th Batool Aqel
 March 19 th - 2013 6 Batool Aqel Hormones That Bind to Nuclear Receptor Proteins Hormones bind to their receptors.whether the receptor is found in the nucleus or the cytoplasm, at the end they are translocated
March 19 th - 2013 6 Batool Aqel Hormones That Bind to Nuclear Receptor Proteins Hormones bind to their receptors.whether the receptor is found in the nucleus or the cytoplasm, at the end they are translocated
Signal Transduction: G-Protein Coupled Receptors
 Signal Transduction: G-Protein Coupled Receptors Federle, M. (2017). Lectures 4-5: Signal Transduction parts 1&2: nuclear receptors and GPCRs. Lecture presented at PHAR 423 Lecture in UIC College of Pharmacy,
Signal Transduction: G-Protein Coupled Receptors Federle, M. (2017). Lectures 4-5: Signal Transduction parts 1&2: nuclear receptors and GPCRs. Lecture presented at PHAR 423 Lecture in UIC College of Pharmacy,
Transcription and chromatin. General Transcription Factors + Promoter-specific factors + Co-activators
 Transcription and chromatin General Transcription Factors + Promoter-specific factors + Co-activators Cofactor or Coactivator 1. work with DNA specific transcription factors to make them more effective
Transcription and chromatin General Transcription Factors + Promoter-specific factors + Co-activators Cofactor or Coactivator 1. work with DNA specific transcription factors to make them more effective
number Done by Corrected by Doctor Nayef Karadsheh
 number 11 Done by حسام أبو عوض Corrected by Moayyad Al-Shafei Doctor Nayef Karadsheh 1 P a g e General Regulatory Aspects in Metabolism: We can divide all pathways in metabolism to catabolicand anabolic.
number 11 Done by حسام أبو عوض Corrected by Moayyad Al-Shafei Doctor Nayef Karadsheh 1 P a g e General Regulatory Aspects in Metabolism: We can divide all pathways in metabolism to catabolicand anabolic.
Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section B and ONE question from Section C.
 UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2014-15 FUNDAMENTALS OF CELL BIOLOGY AND BIOCHEMISTRY BIO-4004B Time allowed: 2 hours Answer ALL questions in Section
UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2014-15 FUNDAMENTALS OF CELL BIOLOGY AND BIOCHEMISTRY BIO-4004B Time allowed: 2 hours Answer ALL questions in Section
Review of Neurochemistry
 Review of Neurochemistry UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY PBL MBBS YEAR III SEMINAR VJ Temple 1 CEREBRAL METABOLISM What are some
Review of Neurochemistry UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY PBL MBBS YEAR III SEMINAR VJ Temple 1 CEREBRAL METABOLISM What are some
f(x) = x R² = RPKM (M8.MXB) f(x) = x E-014 R² = 1 RPKM (M31.
 14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
Fatty acid breakdown
 Fatty acids contain a long hydrocarbon chain and a terminal carboxylate group. Most contain between 14 and 24 carbon atoms. The chains may be saturated or contain double bonds. The complete oxidation of
Fatty acids contain a long hydrocarbon chain and a terminal carboxylate group. Most contain between 14 and 24 carbon atoms. The chains may be saturated or contain double bonds. The complete oxidation of
Points 1. Following is the overall reaction catalyzed by the Calvin-Benson cycle:
 BCH 4054 February 22, 2002 HOUR TEST 2 NAME_ Points 1. Following is the overall reaction catalyzed by the Calvin-Benson cycle: CO 2 + 3ATP + 2NADPH 1/3 glyceraldehyde-3-p + 3ADP + 2NADP + Give the structures
BCH 4054 February 22, 2002 HOUR TEST 2 NAME_ Points 1. Following is the overall reaction catalyzed by the Calvin-Benson cycle: CO 2 + 3ATP + 2NADPH 1/3 glyceraldehyde-3-p + 3ADP + 2NADP + Give the structures
Table 1A. Genes enriched as over-expressed in DPM treatment group
 Table 1A. s enriched as over-expressed in DPM treatment group Affy Probeset mrna Accession Description 10538247 Npy NM_023456 Mus musculus neuropeptide Y (Npy), mrna. 2.0024 10598062 --- NC_005089 gi 34538597
Table 1A. s enriched as over-expressed in DPM treatment group Affy Probeset mrna Accession Description 10538247 Npy NM_023456 Mus musculus neuropeptide Y (Npy), mrna. 2.0024 10598062 --- NC_005089 gi 34538597
MCB*4010 Midterm Exam / Winter 2008
 MCB*4010 Midterm Exam / Winter 2008 Name: ID: Instructions: Answer all 4 questions. The number of marks for each question indicates how many points you need to provide. Write your answers in point form,
MCB*4010 Midterm Exam / Winter 2008 Name: ID: Instructions: Answer all 4 questions. The number of marks for each question indicates how many points you need to provide. Write your answers in point form,
Endoplasmic Reticulum
 Endoplasmic Reticulum What s ER? How is ER? Why is ER? definition description functions Nissl s bodies neurons Berg s bodies hepatocytes Organelle structure histocytochemical evidences Ergastoplasm pancreatic
Endoplasmic Reticulum What s ER? How is ER? Why is ER? definition description functions Nissl s bodies neurons Berg s bodies hepatocytes Organelle structure histocytochemical evidences Ergastoplasm pancreatic
Principles of cell signaling Lecture 4
 Principles of cell signaling Lecture 4 Johan Lennartsson Molecular Cell Biology (1BG320), 2014 Johan.Lennartsson@licr.uu.se 1 Receptor tyrosine kinase-induced signal transduction Erk MAP kinase pathway
Principles of cell signaling Lecture 4 Johan Lennartsson Molecular Cell Biology (1BG320), 2014 Johan.Lennartsson@licr.uu.se 1 Receptor tyrosine kinase-induced signal transduction Erk MAP kinase pathway
Regulation of P450. b. competitive-inhibitor but not substrate. c. non-competitive inhibition. 2. Covalent binding to heme or protein
 10. Inhibition of P450 Regulation of P450 a. competitive-2 chemicals metabolized by same isoform b. competitive-inhibitor but not substrate c. non-competitive inhibition 11. Induction of P450 1. Non-covalent
10. Inhibition of P450 Regulation of P450 a. competitive-2 chemicals metabolized by same isoform b. competitive-inhibitor but not substrate c. non-competitive inhibition 11. Induction of P450 1. Non-covalent
Cell Communication. Local and Long Distance Signaling
 Cell Communication Cell to cell communication is essential for multicellular organisms Some universal mechanisms of cellular regulation providing more evidence for the evolutionary relatedness of all life
Cell Communication Cell to cell communication is essential for multicellular organisms Some universal mechanisms of cellular regulation providing more evidence for the evolutionary relatedness of all life
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12864 Supplementary Table 1 1 2 3 4 5 6 7 Peak Gene code Screen Function or Read analysis AMP reads camp annotation reads minor Tb927.2.1810 AMP ISWI Confirmed
SUPPLEMENTARY INFORMATION doi:10.1038/nature12864 Supplementary Table 1 1 2 3 4 5 6 7 Peak Gene code Screen Function or Read analysis AMP reads camp annotation reads minor Tb927.2.1810 AMP ISWI Confirmed
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression. Chromatin Array of nuc
 Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Lecture: 26 OXIDATION OF FATTY ACIDS
 Lecture: 26 OXIDATION OF FATTY ACIDS Fatty acids obtained by hydrolysis of fats undergo different oxidative pathways designated as alpha ( ), beta ( ) and omega ( ) pathways. -oxidation -Oxidation of fatty
Lecture: 26 OXIDATION OF FATTY ACIDS Fatty acids obtained by hydrolysis of fats undergo different oxidative pathways designated as alpha ( ), beta ( ) and omega ( ) pathways. -oxidation -Oxidation of fatty
Protein Trafficking in the Secretory and Endocytic Pathways
 Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Protein Trafficking in the Secretory and Endocytic Pathways The compartmentalization of eukaryotic cells has considerable functional advantages for the cell, but requires elaborate mechanisms to ensure
Lecture 16. Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III
 Lecture 16 Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III The Powertrain of Human Metabolism (verview) CARBHYDRATES PRTEINS
Lecture 16 Finish lipid metabolism (Triglycerides, Isoprenoids/Steroids, Glyoxylate cycle) Amino acid metabolism (Urea cycle) Google Man III The Powertrain of Human Metabolism (verview) CARBHYDRATES PRTEINS
1- Which of the following statements is TRUE in regards to eukaryotic and prokaryotic cells?
 Name: NetID: Exam 3 - Version 1 October 23, 2017 Dr. A. Pimentel Each question has a value of 4 points and there are a total of 160 points in the exam. However, the maximum score of this exam will be capped
Name: NetID: Exam 3 - Version 1 October 23, 2017 Dr. A. Pimentel Each question has a value of 4 points and there are a total of 160 points in the exam. However, the maximum score of this exam will be capped
Biochemistry 673 Lecture 2 Jason Kahn, UMCP Introduction to steroid hormone receptor (nuclear receptor) signalling
 Biochemistry 673 Lecture 2 Jason Kahn, UMCP Introduction to steroid hormone receptor (nuclear receptor) signalling Resources: Latchman Lodish chapter 10, 20 Helmreich, chapter 11 http://www.nursa.org,
Biochemistry 673 Lecture 2 Jason Kahn, UMCP Introduction to steroid hormone receptor (nuclear receptor) signalling Resources: Latchman Lodish chapter 10, 20 Helmreich, chapter 11 http://www.nursa.org,
FATTY ACID SYNTHESIS
 FATTY ACID SYNTHESIS Malonyl- CoA inhibits Carni1ne Palmitoyl Transferase I. Malonyl- CoA is a precursor for fa=y acid synthesis. Malonyl- CoA is produced from acetyl- CoA by the enzyme Acetyl- CoA Carboxylase.
FATTY ACID SYNTHESIS Malonyl- CoA inhibits Carni1ne Palmitoyl Transferase I. Malonyl- CoA is a precursor for fa=y acid synthesis. Malonyl- CoA is produced from acetyl- CoA by the enzyme Acetyl- CoA Carboxylase.
INTERACTION DRUG BODY
 INTERACTION DRUG BODY What the drug does to the body What the body does to the drug Receptors - intracellular receptors - membrane receptors - Channel receptors - G protein-coupled receptors - Tyrosine-kinase
INTERACTION DRUG BODY What the drug does to the body What the body does to the drug Receptors - intracellular receptors - membrane receptors - Channel receptors - G protein-coupled receptors - Tyrosine-kinase
Cornstarch
 Electronic Supplementary Material (ESI) for Food & Function. This journal is The Royal Society of Chemistry 2018 Supplementary data : Supplementary Table 1: Diet composition (g/kg) on the basis of the
Electronic Supplementary Material (ESI) for Food & Function. This journal is The Royal Society of Chemistry 2018 Supplementary data : Supplementary Table 1: Diet composition (g/kg) on the basis of the
Cell Communication. Chapter 11. Biology Eighth Edition Neil Campbell and Jane Reece. PowerPoint Lecture Presentations for
 Chapter 11 Cell Communication PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp
Chapter 11 Cell Communication PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from Joan Sharp
KEY CONCEPT QUESTIONS IN SIGNAL TRANSDUCTION
 Signal Transduction - Part 2 Key Concepts - Receptor tyrosine kinases control cell metabolism and proliferation Growth factor signaling through Ras Mutated cell signaling genes in cancer cells are called
Signal Transduction - Part 2 Key Concepts - Receptor tyrosine kinases control cell metabolism and proliferation Growth factor signaling through Ras Mutated cell signaling genes in cancer cells are called
Chemistry 107 Exam 4 Study Guide
 Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
Chemistry 107 Exam 4 Study Guide Chapter 10 10.1 Recognize that enzyme catalyze reactions by lowering activation energies. Know the definition of a catalyst. Differentiate between absolute, relative and
Molecular Cell Biology - Problem Drill 10: Gene Expression in Eukaryotes
 Molecular Cell Biology - Problem Drill 10: Gene Expression in Eukaryotes Question No. 1 of 10 1. Which of the following statements about gene expression control in eukaryotes is correct? Question #1 (A)
Molecular Cell Biology - Problem Drill 10: Gene Expression in Eukaryotes Question No. 1 of 10 1. Which of the following statements about gene expression control in eukaryotes is correct? Question #1 (A)
Supplementary Information
 Scientific Reports Supplementary Information Upregulated expression of FGF13/FHF2 mediates resistance to platinum drugs in cervical cancer cells Tomoko Okada, Kazuhiro Murata, Ryoma Hirose, Chie Matsuda,
Scientific Reports Supplementary Information Upregulated expression of FGF13/FHF2 mediates resistance to platinum drugs in cervical cancer cells Tomoko Okada, Kazuhiro Murata, Ryoma Hirose, Chie Matsuda,
Lecture 15. Signal Transduction Pathways - Introduction
 Lecture 15 Signal Transduction Pathways - Introduction So far.. Regulation of mrna synthesis Regulation of rrna synthesis Regulation of trna & 5S rrna synthesis Regulation of gene expression by signals
Lecture 15 Signal Transduction Pathways - Introduction So far.. Regulation of mrna synthesis Regulation of rrna synthesis Regulation of trna & 5S rrna synthesis Regulation of gene expression by signals
Oxidation of Long Chain Fatty Acids
 Oxidation of Long Chain Fatty Acids Dr NC Bird Oxidation of long chain fatty acids is the primary source of energy supply in man and animals. Hibernating animals utilise fat stores to maintain body heat,
Oxidation of Long Chain Fatty Acids Dr NC Bird Oxidation of long chain fatty acids is the primary source of energy supply in man and animals. Hibernating animals utilise fat stores to maintain body heat,
HORMONES (Biomedical Importance)
 hormones HORMONES (Biomedical Importance) Hormones are the chemical messengers of the body. They are defined as organic substances secreted into blood stream to control the metabolic and biological activities.
hormones HORMONES (Biomedical Importance) Hormones are the chemical messengers of the body. They are defined as organic substances secreted into blood stream to control the metabolic and biological activities.
Regulation of Enzymatic Activity. Lesson 4
 Regulation of Enzymatic Activity Lesson 4 Regulation of Enzymatic Activity no real regulation: - regulation of enzyme expression and turnover - control of enzyme trafficking - supply of cofactors real
Regulation of Enzymatic Activity Lesson 4 Regulation of Enzymatic Activity no real regulation: - regulation of enzyme expression and turnover - control of enzyme trafficking - supply of cofactors real
Oxidative Phosphorylation
 Electron Transport Chain (overview) The NADH and FADH 2, formed during glycolysis, β- oxidation and the TCA cycle, give up their electrons to reduce molecular O 2 to H 2 O. Electron transfer occurs through
Electron Transport Chain (overview) The NADH and FADH 2, formed during glycolysis, β- oxidation and the TCA cycle, give up their electrons to reduce molecular O 2 to H 2 O. Electron transfer occurs through
