Multiple Roles of Notch Signaling in the Regulation of Epidermal Development
|
|
|
- Louisa Paul
- 5 years ago
- Views:
Transcription
1 Article Multiple Roles of Notch Signaling in the Regulation of Epidermal Development Mariko Moriyama, 1 André-Dante Durham, 2 Hiroyuki Moriyama, 1 Kiyotaka Hasegawa, 1 Shin-Ichi Nishikawa, 3 Freddy Radtke, 2 and Masatake Osawa 1,3, * 1 Cutaneous Biology Research Center, Massachusetts General Hospital and Harvard Medical School, MGH-East, Building 149, 13th Street, Charlestown, MA 02129, USA 2 Swiss Institute for Experimental Cancer Research (ISREC), Ecole Polytechnique Fédérale de Lausanne (EPFL), Chemin des Boveresses 155, 1066 Epalinges, Switzerland 3 Laboratory for Stem Cell Biology, RIKEN Center for Developmental Biology, Minatojima, Minami-machi, Cuo-ku, Kobe, Hyogo , Japan *Correspondence: masatake.osawa@cbrc2.mgh.harvard.edu DOI /j.devcel SUMMARY Recent studies have shown that Notch signaling plays an important role in epidermal development, but the underlying molecular mechanisms remain unclear. Here, by integrating loss- and gain-of-function studies of Notch receptors and Hes1, we describe molecular information about the role of Notch signaling in epidermal development. We show that Notch signaling determines spinous cell fate and induces terminal differentiation by a mechanism independent of Hes1, but Hes1 is required for maintenance of the immature state of spinous cells. Notch signaling induces Ascl2 expression to promote terminal differentiation, while simultaneously repressing Ascl2 through Hes1 to inhibit premature terminal differentiation. Despite the critical role of Hes1 in epidermal development, Hes1 null epidermis transplanted to adult mice showed no obvious defects, suggesting that this role of Hes1 may be restricted to developmental stages. Overall, we conclude that Notch signaling orchestrates the balance between differentiation and immature programs in suprabasal cells during epidermal development. INTRODUCTION The epidermis of the skin is a stratified epithelium. Stratification of the epidermis is a key process in epidermal development, but it is not known how generation of a stratified epithelium from a single-layered ectoderm is coordinated. It has been recently shown, however, that Notch signaling plays a crucial role in the initiation of epidermal development (Blanpain et al., 2006). The Notch signaling pathway is a highly conserved molecular network, but the outcome of Notch signaling differs depending on the cellular and developmental context. Once activated by ligand, the intracellular domain of the Notch receptor (NICD) is cleaved by g-secretase, leading to translocation of the NICD into the nucleus. NICD associates with the transcription factor RBP-J to generate a transactivation complex that initiates transcription of target genes such as the hairy/enhancer of split (Hes) transcriptional repressors (Kageyama et al., 2005). Conditional ablation of Notch signaling in epidermal development results in loss of the spinous and granular layers due to hypoproliferation of the epidermis, indicating that Notch signaling is required for commitment of basal keratinocytes to spinous cell differentiation at early stages of epidermal development (Blanpain et al., 2006). By contrast, postnatal ablation of Notch1 causes hyperproliferation of basal keratinocytes, suggesting that signaling from Notch1 is required for cell cycle withdrawal of the basal keratinocytes to promote terminal differentiation in the postnatal epidermis (Nicolas et al., 2003; Rangarajan et al., 2001). These apparently contradictory functions of Notch signaling in the regulation of keratinocyte proliferation and differentiation may reflect differences in the cell-context-specific functions of Notch signaling between embryonic and postnatal keratinocytes or may be due to differential use of either canonical or noncanonical pathways in the regulation of epidermal keratinocytes. In both cases, however, the molecular mechanisms underlying the regulation of epidermal development by Notch signaling remain largely unknown. In our current study we demonstrate multiple roles of Notch signaling in epidermal development. By combining both lossand gain-of-function studies, we confirmed that Notch signaling promotes spinous cell commitment from basal cells and induces their terminal differentiation into granular cells. Moreover, we reveal a crucial role for the Hes1 transcriptional repressor in determining the outcome of Notch signaling via coordination of the balance between maintenance of the spinous cell fate and the induction of granular cell differentiation. Our present data thus provide new insights into our understanding of how Notch signaling accomplishes apparently contradictory tasks simultaneously, i.e., activating cell fate determination and terminal differentiation programs while also preventing a terminal differentiation. RESULTS Hes1 Is Required for the Generation of Spinous Layers in the Interfollicular Epidermis We found that at E18.5 mouse embryos lacking Hes1 (Hes1 KO) displayed a consistently thinner epidermis on their backs 594 Developmental Cell 14, , April 2008 ª2008 Elsevier Inc.
2 number. By contrast, no overt defects were evident in cornified layers (Figures 1A 1D). A more detailed analysis by immunohistochemical staining of the E18.5 epidermis with the granular cell markers involucrin and loricrin clearly revealed the loss of spinous layers in the Hes1 KO epidermis, whereas at least 2 3 layers of spinous cells, which are negative for these differentiation markers, are normally present (Figures 1E 1J). Hes1 KO mice also exhibit severe neurulation defects and die during the later stages of embryogenesis (Ishibashi et al., 1995). To investigate the role of Hes1 in the postnatal skin, we grafted E18.5 skin fragments from Hes1 KO and WT mice onto nude mice and examined them 26 days later. Despite the clear E18.5 Hes1 KO epidermal phenotype, we could not detect obvious defects in Hes1 KO postnatal skin (see Figure S1 in the Supplemental Data available with this article online). We conclude therefore that Hes1 is critical for the generation and/or maintenance of spinous cells in epidermal development, but its role, if any, in the postnatal skin is less clear. Figure 1. Loss of Spinous Layers in Hes1 KO Epidermis at E18.5 (A and B) Hematoxylin and eosin staining of dorsal skin at E18.5. BL, basal layer; SP, spinous layer; GR, granular layer; SC, stratum corneum (cornified layer). (C and D) Transmission electron micrographs of ultrathin sections. Boxed areas correspond to the higher magnification images in the bottom panels. Granular cells having keratohyalin granules (KG) are present both WT and Hes1 KO epidermis, while spinous cells containing keratin filament bundles (Kf) are absent in Hes1 KO epidermis. BL represents basal layer cell. (E J) Immunofluorescence staining of E18.5 skin sections with antibodies against K10 (green) and K15 (red) (E and F), involucrin (G and H), or loricrin (I and J). Representative results for WT littermates (A, C, E, G, and I) and Hes1 KO mice (B, D, F, H, and J) are shown. The blue signals indicate nuclear staining. The dotted lines indicate the boundary between the epidermis and the dermis. Scale bars, 20 mm. compared to wild-type (WT) mice (Figures 1A and 1B). Histological and ultrastructural analyses of Hes1 KO epidermis revealed that the spinous layer, characterized by suprabasal cells containing dense keratin bundles in their cytoplasm, was dramatically reduced (Figures 1C and 1D). Granular layer cells were present in Hes1 KO epidermis, although slightly decreased in Loss of Hes1 Causes Premature Differentiation of Suprabasal Keratinocytes To investigate how loss of Hes1 affects stratification of the epidermis, we first determined the Hes1 expression profile during epidermal development. We performed immunofluorescent staining in skin and, beginning at E14.5, found that Hes1 protein was readily detectable in the nuclei of the first suprabasal layer (spinous layer) cells. From E15.5, Hes1 expression was confined to the spinous layer cells (Figures 2A 2D). Therefore, consistent with loss of the spinous layer cells in Hes1 KO epidermis, Hes1 expression was restricted to the spinous cells, where it was expected to be functioning. As Hes1 expression was initiated at E14.5, we focused our histological analysis of the epidermis at E14.5 and E15.5 to further characterize the defects in the Hes1 KO mice. The gross appearance of skin sections of Hes1 KO embryos at E14.5 was similar to that of WT littermates (Figures 2F and 2G). However, at E15.5, Hes1 KO embryos displayed a thinner epidermis than agematched WT littermates (Figures 2N and 2O), indicating that Hes1 function is important in the epidermis during these developmental stages. To clarify the role of Hes1, Hes1 KO epidermis at E14.5 and E15.5 was further investigated by immunohistochemical analysis of keratinocyte differentiation and proliferation markers. The general view is that Hes1 negatively regulates terminal differentiation (Kageyama et al., 2005). Therefore, we tested whether Hes1 plays a role in regulation of terminal keratinocyte differentiation during epidermal development. In WT epidermis, at E15.5 both involucrin and loricrin became detectable in the outermost layer, and this is when the earliest terminally differentiated granular cells appear after the initiation of stratification (Figures 2T and 2W). At E14.5, expression of these markers is essentially undetectable (Figures 2J and 2L). In Hes1 KO epidermis, however, these proteins were already present in the spinous layer at E14.5 (Figures 2K and 2M), suggesting premature differentiation of the spinous layer cells. Keratin 5 (K5), a marker for immature basal cells, was occasionally detected in the spinous cells of WT epidermis at E15.5 (Figure 2P), but in Hes1 KO epidermis this expression was strictly confined to the single basal layer (Figure 2Q), suggesting acceleration of differentiation. Thus, these data provide evidence for Developmental Cell 14, , April 2008 ª2008 Elsevier Inc. 595
3 Figure 2. Premature Differentiation of Spinous Cells into Granular Cells in the Hes1 KO Epidermis (A E) Frozen dorsal skin sections at the indicated developmental stages were analyzed by immunofluorescence with an antibody against Hes1. WT dorsal skin is shown in (A) (D). Hes1 KO dorsal skin at E15.5 (E) was used as a negative control. (F and G) Hematoxylin and eosin staining of dorsal skin from WT littermates (F) and Hes1 KO mice (G) at E14.5. (H M) Frozen dorsal skin sections at E14.5 were analyzed by immunofluorescence with polyclonal antibodies against keratin 10 (K10) (H and I), involucrin (J and K), and loricrin (L and M). Representative results for WT littermates (H, J, and L) and Hes1 KO (I, K, and M) are shown. (N and O) Hematoxylin and eosin staining of dorsal skin at E15.5 in WT littermates (N) and Hes1 KO mice (O). (P X) Frozen dorsal skin sections from E15.5 embryos were analyzed by immunofluorescence with antibodies against keratin5 (K5) (P and Q), K10 (R and S), involucrin (T and U), and loricrin (W and X). Skin sections from WT littermates (P, R, T, and W) and Hes1 KO mice (Q, S, U, and X) are shown. The blue signals indicate nuclear staining. The dotted lines indicate the boundary between the epidermis and the dermis. Scale bars, 20 mm. (Y) Expression of the indicated keratinocyte differentiation markers in Hes1 KO epidermis. The results of Q-PCR analyses of K1, K10, involucrin, and loricrin are shown for WT or Hes1 KO epidermis at E14.5. Each expression value was calculated based on DDCt method using Actb as an internal control. The data represent the average of three individual animals. Error bars indicate SD. premature differentiation of spinous layer cells in Hes1 KO epidermis at E14.5 E15.5. In parallel with these immunohistochemical analyses, quantitative PCR (Q-PCR) also revealed that both involucrin and loricrin mrna levels were dramatically elevated in the Hes1 KO epidermis at E14.5 (Figure 2Y), indicating that this premature expression is regulated at the transcriptional level. Recent in vitro studies suggest that canonical Notch signaling via Hes1 triggers the expression of suprabasal genes such as keratin 1 (K1) and keratin 10 (K10) (Blanpain et al., 2006). We thus evaluated whether Hes1 is also involved in fate determination of spinous cells in vivo. We performed immunohistochemical staining for K10 in the Hes1 KO epidermis, as this is one of the earliest markers of spinous cell fate commitment from immature basal cells and is normally expressed throughout the suprabasal epidermal layers (Figures 2H and 2R). We detected strong K10 expression comparable to WT in the Hes1 KO epidermis (Figures 2I and 2S), indicating that suprabasal fate commitment may take place in vivo even in the absence of Hes1. In support of these immunohistochemical results, Q-PCR analysis further showed that K1 and K10 mrna levels in Hes1 KO epidermis were comparable with WT (Figure 2Y), indicating that this expression is not dependant on Hes1 in vivo. Therefore, in contrast to a previous report, our findings suggest that Hes1 is not required for the commitment of spinous cells from basal cells. Taken together, our data indicate that Hes1 KO epidermis undergoes premature differentiation of spinous layer cells during E14.4 E15.5, but the commitment switch for spinous layer cells is initiated in a Hes1-independent manner. Hes1 Maintains the Proliferative Potential of Both Suprabasal and Basal Keratinocytes in Embryonic Epidermis The Hes1 KO skin phenotype, which is characterized by a markedly thinner epidermis at E18.5 (Figure 1), is not likely to be caused solely by aberrant differentiation of suprabasal keratinocytes, because during the early stages of normal epidermal development the spinous and basal layers proliferate continuously, allowing scope for compensatory activity. In addition, immunostaining with an antibody recognizing cleaved caspase Developmental Cell 14, , April 2008 ª2008 Elsevier Inc.
4 revealed no evidence of apoptosis in the Hes1 KO epidermis (Figure S2), excluding the possibility that this may cause the thinner phenotype. Hence, to explain the Hes1 KO epidermal phenotype, we hypothesized that the proliferative potential of the epidermis may also be affected. To investigate this possibility, we examined Hes1 KO skin samples from E embryos for expression of the proliferation marker Ki67. At E14.5, the Hes1 KO and WT epidermis both showed Ki67-positive nuclear staining in the spinous and basal layers (Figures 3A and 3B). However, a dramatic reduction in Ki67 expression was evident in these layers in the E15.5 Hes1 KO epidermis, while most of the basal cells in the WT epidermis retained Ki67 expression at this stage (Figures 3C and 3D). In addition, transient 5-bromo-2 0 -deoxyuridine (BrdU) incorporation revealed a marked reduction in proliferating cells in the basal layer in KO animals (Figures 3E 3G). These data thus demonstrate that the proliferative potential of spinous and basal cells is reduced in the Hes1 KO epidermis. As it has been previously demonstrated that p63 plays a crucial role in maintenance of the proliferative potential of basal keratinocytes (Senoo et al., 2007), we next investigated expression of p63 in the Hes1 KO epidermis. By immunohistochemical staining, p63 was abundantly expressed in both the spinous and basal cells at E14.5 in WT and Hes1 KO epidermis (Figures 3H and 3I). However, at E15.5, p63 was considerably downregulated in both cell types in Hes1 KO epidermis (Figures 3J and 3K). Consistent with this observation, the level of p63 mrna was also significantly reduced in the E15.5 Hes1 KO epidermis (Figures 3L and 3M). Thus, the epidermal phenotype in the E18.5 Hes1 KO epidermis, where spinous layers are dramatically reduced in the interfollicular epidermis (Figure 1), may result from a combination of premature differentiation of suprabasal cells and reduced proliferation of basal cells. Figure 3. Reduced Proliferative Capacity of the Suprabasal and Basal Cells in the Hes1 KO Epidermis (A D) Frozen dorsal skin sections at the indicated developmental stages were analyzed by immunofluorescence using antibody against the proliferation marker Ki67. Typical results for WT littermates (A and C) and Hes1 KO mice (B and D) are shown. (E G) Staining (E and F) and quantification (G) of BrdU incorporation. WT (E) or Hes1 KO (F) skin sections were stained with antibodies against BrdU and K15. Frequency of BrdU-positive cells in the basal layer of either the WT or Hes1KO epidermis was plotted (G). The data represent the average of three independent experiments. Error bars indicate SD. Notch Signaling Is Required for Both Spinous and Granular Cell Differentiation The phenotype that we observed in Hes1 KO epidermis differs significantly from that seen in previous studies in which Notch signaling was conditionally ablated in either embryonic or postnatal epidermis (Blanpain et al., 2006; Rangarajan et al., 2001). The contrasting results may be due to the differential use of Hes1-dependent and Hes1-independent pathways downstream of Notch signaling. To test this possibility, we performed a series of loss- and gain-of-function studies of Notch in the epidermis to dissect the roles of its different downstream pathways during epidermal development. First, we conditionally downregulated Notch signaling in the embryonic epidermis by generating K14-Cre;Notch1 lox/lox ; Notch2 lox/lox (N1N2 dcko) mice. Consistent with a prior report in which RBP-J was conditionally ablated in the epidermis (RBP-J cko) (Blanpain et al., 2006), the N1N2 dcko epidermis exhibited a considerably thinner epidermis due to a reduction in both the spinous and granular layers (Figure S3). (H M) Frozen dorsal skin sections at the indicated stages were analyzed by immunofluorescence with an antibody against p63 (H K). Frozen dorsal skin sections at E15.5 were analyzed by in situ hybridization using a p63 antisense probe (L and M). Results from WT littermates (H, J, and L) and Hes1 KO mice (I, K, and M) are shown. The blue signals correspond to nuclear staining. The dotted lines indicate the boundary between the epidermis and the dermis. Scale bars, 20 mm. Developmental Cell 14, , April 2008 ª2008 Elsevier Inc. 597
5 Figure 4. Hes1-Independent Notch Signaling Functions in Spinous Cell Fate Determination, whereas Hes1-Dependent Notch Signaling Functions in Granular Cell Fate Determination (A D) Frozen dorsal skin sections from N1 lox/lox N2 lox/lox mice (A and C) and K14Cre; N1 lox/lox N2 lox/lox mice (B and D) were subjected to immunofluorescent staining with antibodies against K10 (A and B) or loricrin (Lor) (C and D). Arrowheads in (B) indicate cells lacking K10 expression in the second layer of the epidermis, where K10-expressing cells normally exist. (E) Q-PCR analysis of K1 and K10 mrna expression in MPEK cells derived from either WT or Hes1 KO (KO) mice, infected with adenoviruses expressing GFP (cont), intracellular domain of Notch1 (NICD), or a constitutively active form of RBP-J (RBP-CA). Each expression value was calculated with the DDCt method using Actb as an internal control. Relative value indicates the fold increases over the WT control. (F) Q-PCR analysis of K1 and K10 mrna expression in MPEK cells infected with adenoviruses expressing either GFP (cont) or Hes1. (G Q) Frozen dorsal skin sections from WT (G, K, and L), K14Hes1 (H and J), K14NICD (I), K1NICD (M and N), or K1Hes1 (O Q) transgenic mice were analyzed by immunofluorescent staining with antibodies against Hes1 (G, H, N, and P), K10 (green) and K15 (red) (I K, and O), and loricrin (Lor) (L, M, and Q). The blue signals indicate nuclear staining. The dotted lines indicate the boundary between the epidermis and the dermis. Scale bars, 20 mm. To further characterize the defects in the N1N2 dcko epidermis, we analyzed immunohistological markers for the basal, suprabasal, and granular layers. Consistent with the thinner epidermal phenotype, K10-positive suprabasal layer cells were selectively reduced in the N1N2 dcko epidermis (Figures 4A and 4B), indicating that Notch signaling is required for the generation and/or maintenance of suprabasal cells during epidermal development. K15, a marker for basal layer cells at this stage of epidermal development, was exclusively expressed in basal layer cells in the control epidermis (Figure 4A). By contrast, a pronounced number of K15-positive/K10-negative cells were also evident in the lowermost suprabasal layer of the N1N2 dcko epidermis, suggesting retardation of spinous cell differentiation (Figure 4B). Furthermore, a significant reduction in loricrin expression was observed in the N1N2 dcko epidermis (Figures 4C and 4D), indicating an important role of Notch signaling in granular cell differentiation. Taken together, our findings suggest that Notch signaling plays a crucial role in promotion of both spinous and granular layer differentiation during epidermal development. Notch Signaling Downregulates Basal Fate and Promotes Spinous Cell Differentiation via a Hes1- Independent Mechanism We next conducted gain-of-function studies to further elucidate the role of Notch signaling during epidermal development. For these experiments, we utilized a transgenic approach with a constitutively active form of the intracellular domain of Notch1 (NICD1) and the Hes1 gene. First, to test a role of Notch signaling in spinous-fate determination, we generated transgenic mice in which NICD1 was expressed under the control of the K14 promoter (K14NICD1) to ectopically activate Notch signaling in basal layer cells. In contrast to the N1N2 dcko epidermis, which exhibited a slight expansion of the K15-positive layers (Figure 4B), constitutive activation of Notch signaling in basal layer cells resulted in a dramatic reduction of K15-expressing cells in the basal layer (Figure 4I), suggesting that Notch signaling plays a suppressive role in maintenance of basal cell fate. Therefore, consistent with prior studies in which Notch signaling was constitutively activated in the entire epidermis (Blanpain et al., 2006), these data demonstrate a crucial role of Notch signaling in suppression of basal fate during epidermal development. Previously, it has been suggested that Hes1 promotes spinous cell fate by stimulating spinous gene expression (Blanpain et al., 598 Developmental Cell 14, , April 2008 ª2008 Elsevier Inc.
6 2006). We tested this possibility using transgenic mice in which the Hes1 gene was expressed in basal cells under the control of the K14 promoter (K14Hes1). Unlike in the K14NICD1 epidermis, and despite strong expression of Hes1 protein in the basal layer (Figure 4H), K15-positive/K10-negative cells remained in the basal layer of the K14Hes1 epidermis (Figure 4J). Hence, unlike the effect of NICD1, constitutive expression of Hes1 in the basal layer did not suppress basal fate or induce spinous cell differentiation. Taken together, these results suggest that, although activation of Notch signaling in basal layer cells suppresses basal fate, Hes1 is not capable of doing so. However, we cannot completely exclude the possibility that ectopic overexpression of Hes1 causes unexpected effects in the basal cells. In addition to our in vivo analysis of the K14NICD1 and K14Hes1 epidermis, adenoviral overexpression of NICD1 in cultured primary keratinocytes resulted in a dramatic induction of the spinous layer genes K1 and K10 in both WT and Hes1 KO keratinocytes (Figure 4E), indicating a strong spinous cell-promoting activity of Notch signaling by a mechanism that is not dependent on Hes1. Furthermore, overexpression of Hes1 in primary keratinocytes did not induce K1/K10 expression and, in fact, suppressed the expression of both markers (Figure 4F). From integrating the results of these in vivo and in vitro studies, it appears that Notch signaling downregulates basal fate and promotes spinous cell differentiation in a Hes1-independent manner. Activation of Notch Signaling in Spinous Cells Induces Granular Cell Differentiation, but also Maintains Spinous Fate via a Hes1-Dependent Mechanism Our phenotypic analysis of the N1N2 dcko epidermis suggested that Notch signaling was also involved in the induction of granular cell differentiation. To investigate this possibility, we conducted gain-of-function studies of Notch signaling in spinous layers. We generated transgenic mice expressing NICD1 under the control of the K1 promoter (K1NICD1). This promoter was chosen as it has specific transcriptional activity in the lowermost suprabasal cells (spinous cells) from E14.5 (Greenhalgh et al., 1993) (Figure S4). In contrast to WT epidermis, immunofluorescent staining of K1NICD1 epidermis revealed a pronounced expansion of loricrin-positive cell layers (Figures 4L and 4M). Hence, and consistent with previous findings (Uyttendaele et al., 2004), constitutive activation of Notch signaling in spinous cells accelerates granular cell differentiation. In addition, enhanced expression of Hes1 protein was also observed in spinous layer cells of K1NICD1 epidermis, providing clear evidence that Notch activation in spinous cells triggers Hes1 expression (Figure 4N). Despite strong expression of Hes1 in the spinous layers, the effect of NICD1 expression in these layers was quite limited. As endogenous Hes1 function is tightly regulated by both transcriptional and posttranscriptional mechanisms (Hirata et al., 2002), we reasoned that this might be a consequence of negative feedback regulation. If so, the effect of upregulated endogenous Hes1 in the K1NICD1 epidermis might be compensated for by an unknown mechanism. Therefore, to demonstrate the function of Hes1 in the maintenance of spinous layer cells, we generated transgenic mice in which the Hes1 gene was artificially expressed in the spinous layers under the control of the K1 promoter (K1Hes1). In contrast to Hes1 KO epidermis, in which the spinous layers were lost (Figure 1), gain of Hes1 function in the K1Hes1 epidermis resulted in a massive expansion of K10- positive/loricrin-negative spinous layer cells (Figure 4O). Furthermore, in parallel with the expansion of spinous cells, immunofluorescent staining revealed extensive Hes1 expression in the suprabasal cells (Figure 4P), demonstrating that constitutive expression of Hes1 causes an accumulation of spinous layer cells. Collectively, these data demonstrate that Hes1 plays a crucial role in the maintenance of spinous layer cells. The existence of loricrin-positive cells in the outer layers of K1Hes1 epidermis (Figure 4Q) suggested that the expanded spinous cells are capable of differentiating into granular cells. Hes1 expression should be downregulated in the outer suprabasal layers where K1 promoter activity becomes diminished. In addition, a comparison of the expression patterns of loricrin and Hes1 proteins in both the K1NICD1 and K1Hes1 epidermis indicated that their expression in the suprabasal cells is mutually exclusive. This raises the intriguing possibility that Hes1 downregulation in the suprabasal cells may promote granular cell differentiation. Notch Signaling Represses Ascl2 Expression in the Spinous Layers via a Hes1 Repressor, while Acting Directly to Promote the Expression of Ascl2 Our loss- and gain-of-function studies of Notch signaling revealed that it had seemingly contradictory roles in the spinous layers, where it both promotes and prevents granular differentiation. Therefore, an obvious question was how a single signaling pathway could have such contradictory effects in one location. To address this question, we investigated the molecular mechanism of Notch activity in spinous cells. In light of the critical role of Hes1 in the maintenance of spinous cells, we first explored the molecular targets of Hes1 in spinous layer cells. As Hes1 is thought to be a transcriptional repressor, loss of Hes1 is expected to cause aberrant upregulation of genes that are normally repressed in spinous layer cells. To identify such genes, we conducted comparative global transcript analysis by microarray. Among the genes that were preferentially overexpressed in Hes1 KO, we focused on Ascl2, as it is an Achaete/Scute-related basic helix-loop-helix (bhlh) transcriptional activator that has been implicated in trophoblast differentiation (Guillemot et al., 1994). The upregulation of Ascl2 in the Hes1 KO epidermis was confirmed by both quantitative PCR (Q-PCR) and in situ hybridization. Q-PCR analysis revealed a greater than 7-fold increase in the levels of Ascl2 in the Hes1 KO epidermis relative to WT (Figure 5A). In situ hybridization also revealed strong Ascl2 expression in the spinous layer of the Hes1 KO epidermis at E15.5, whereas in the WT epidermis only low levels of Ascl2 were observed in the outer suprabasal epidermis (Figures 5B 5E). As expression of Hes1 is restricted to the spinous layer cells, these observations suggested that Hes1 might play a direct role in repressing Ascl2 expression in this layer. To determine whether Hes1 directly regulates Ascl2 expression, the region 1 kb upstream of the transcription initiation site of the mouse Ascl2 gene was screened for Hes1-binding motifs. We identified at least two Hes1 consensus binding sites in the Ascl2 promoter. A chromatin immunoprecipitation (ChIP) assay Developmental Cell 14, , April 2008 ª2008 Elsevier Inc. 599
7 Figure 5. Dynamic Transcriptional Regulation of Ascl2 by Notch Signaling (A) Q-PCR analysis of Ascl2 expression in freshly isolated E14.5 dorsal skin epidermal sections. The data represent the average of three independent experiments. Error bars indicate SD. (B E) Frozen dorsal skin sections at E15.5 were analyzed by in situ hybridization using an Ascl2 antisense (B and C) and sense probe (D and E). Skin sections from WT littermates (B and D) and Hes1 KO mice (C and E) are shown. K5 staining is indicated by the green signals; nuclear staining is highlighted by the blue signal. Scale bar, 20 mm. (F) Specific binding of Hes1 to the Ascl2 promoter. MPEK cells were infected with adenoviral constructs expressing HA-tagged Hes1, and processed for chromatin immunoprecipitation (ChIP) with an anti-ha antibody and IgG as a nonimmune control. Q-PCR amplification of the region of the Ascl2 gene described in the indicated map (lower panel; nucleotides 6929 to 6808 [1]; nucleotides 247 to +3 [2] containing two Hes1 consensus binding sites) was also performed. The amount of precipitated DNA was calculated relative to the total input chromatin. (G) Q-PCR analysis of Ascl2 expression in MPEK cells derived from WT or Hes1 KO mice infected with adenoviruses expressing GFP control (cont), intracellular domain of Notch1 (NICD), or a constitutively active form of RBP-J (RBP-CA). (H) Specific binding of RBP-J to the Ascl2 promoter. MPEK cells were infected with adenovirus expressing RBP-J, and processed for ChIP with an anti-rbp-j antibody and IgG as a nonimmune control. Q-PCR amplification of the region of the Ascl2 gene indicated in the map (lower panel; nucleotides 6929 to 6808 [1]; nucleotides 1008 to 908 [2] containing a RBP-J consensus binding site; nucleotides 322 to 222 [3] containing a RBP-J consensus binding site) was performed. The amount of precipitated DNA was again calculated relative to the total input chromatin. The data represent the average of three independent experiments. Error bars indicate SD. and subsequent Q-PCR analysis revealed that a DNA fragment containing these sites was strongly amplified from crosslinked chromatin isolated by Hes1 immunoprecipitation (Figure 5F). These data clearly show that Hes1 specifically binds to the promoter region of Ascl2 at its consensus binding sites. To demonstrate the repressible role of Hes1 in the regulation of Ascl2 expression, primary keratinocytes obtained from either WT or Hes1 KO epidermis were infected with recombinant adenovirus expressing NICD1 or a constitutively active form of RBP-J, RBP-CA (Kuroda et al., 1999). Ascl2 mrna was then quantified by Q-PCR. We found that both NICD1 and RBP-CA induced a dramatic increase of Ascl2 expression in Hes1 KO keratinocytes but not in WT keratinocytes (Figure 5G), demonstrating that Hes1 is involved in repression of Acsl2. Intriguingly, these findings also suggested that activation of canonical Notch signaling can promote Ascl2 expression in the absence of Hes1 (Figure 5G). Consistent with this, ChIP analysis revealed specific binding of RBP-J protein to consensus binding sites in the Ascl2 promoter, thus providing evidence that canonical Notch signaling can directly promote Ascl2 expression (Figure 5H). Ascl2 Overexpression in the Suprabasal Keratinocytes Mimics the Epidermal Phenotype of the Hes1 KO Mouse Given the key role of the bhlh transcriptional activators in cell fate specification during animal development, we reasoned that the misexpression of Ascl2 in the Hes1 KO epidermis might be a primary cause of its aberrant differentiation. We investigated this possibility using transgenic mice expressing Ascl2 in spinous layer cells under the control of the K1 promoter (K1Ascl2IresGFP). As expected, K1Ascl2IresGFP mice showed a similar phenotype to Hes1 KO mice in which the epidermis was thinner than that of their WT littermates, K5 expression was restricted to the basal layer, and the downregulation of Ki67 and p63 was also evident in the epidermis (Figure 6). These results indicated that ectopic overexpression of Ascl2 was sufficient to induce aberrant differentiation of the spinous layer cells and reduce the proliferative capacity of the basal and spinous cells, as observed in the Hes1 KO epidermis. Our current data thus demonstrate that Hes1 plays a crucial role in fate specification of spinous and granular cells, in which 600 Developmental Cell 14, , April 2008 ª2008 Elsevier Inc.
8 Figure 6. Similar Epidermal Phenotypes of Ascl2-Overexpressing Transgenic Mice and Hes1 KO Mice Frozen dorsal skin sections from K1Ascl2IresGFP transgenic mice (B, D, F, H, and J) and WT littermates (A, C, E, G, and I) at E15.5 were analyzed by immunofluorescence with antibodies against K5 (A and B), K10 (C and D), p63 (E and F), Ki67 (G and H), and involucrin (Inv) (I and J). Anti-GFP staining (green) indicates specificity transgene expression. The blue signals indicate nuclear staining. The dotted line indicates the boundary between the epidermis and the dermis. Scale bar, 20 mm. Overexpression of Ascl2 mrna or Ascl2 protein was confirmed by RT-PCR and Q-PCR or western blotting analysis (Figure S5). DISCUSSION Notch signaling through Hes1 represses Ascl2 to maintain the spinous cell fate. In the absence of Hes1, however, Notch signaling directly activates Ascl2 expression to promote granular cell fate. The overall model that we propose, based upon our findings, is shown in Figure 7. Figure 7. Model of the Role of Notch Signaling in Epidermal Keratinocyte Differentiation Schematic diagram illustrating the multiple roles of Notch signaling in epidermal development. Notch signaling promotes spinous cell commitment by a mechanism independent on Hes1, while Hes1 plays an essential role for the maintenance of spinous cells. Sustained Notch activation in spinous cells may result in the accumulation of Hes1 protein, leading to the downregulation of Hes1 by the autoregulatory feedback loop. This may subsequently release Ascl2 promoter from Hes1-mediated repression, and promote Ascl2 transcription by Notch/RBP-J to induce granular differentiation. In the absence of Hes1, Notch signaling directly promotes Ascl2 expression to induce premature granular differentiation, resulting in loss of spinous cells. Furthermore, we found loss of proliferating potential of the basal cells in Hes1 KO epidermis, indicating a possibility that non-cell-autonomous signaling from suprabasal cells may regulate proliferation of basal cells, as has been suggested previously (Feng et al., 1997). Multiple Roles of Notch Signaling during Epidermal Development By conducting both loss- and gain-offunction studies of Notch receptors and Hes1, we have obtained strong physiological evidence that Notch signaling plays multiple roles in the coordination of proper stratification during epidermal development. During epidermal development, spinous cells have to carry out at least three distinct tasks simultaneously: upregulation of specific genes required for suprabasal cell differentiation and repression of genes specifically expressed in the basal layer cells; maintenance of their immature and proliferative status to prevent premature terminal differentiation; and eventual initiation of the terminal differentiation program to differentiate into granular cells. Our in vivo studies demonstrate that Notch signaling regulates all of these three tasks in spinous layer cells in either a Hes1-dependent or a Hes1-independent manner. It has previously been suggested that Notch signaling plays a crucial role in determination of the spinous cell fate from basal cells (Blanpain et al., 2006). These data were essentially confirmed in our current study, as we observed retardation of spinous cell differentiation in the N1N2 dcko epidermis and found that constitutive activation of Notch signaling in the basal layer cells in K14NICD1 transgenic mice suppressed basal fate. However, our conclusions regarding the role of Hes1 in this process differs from previous studies in which it has been suggested from in vitro experiments that Hes1 induces the expression of spinous genes such as K1/K10. We observed in our in vivo genetic studies that there was no significant downregulation of K1 or K10 expression in Hes1 KO epidermis either at the mrna or protein level. Moreover, forced expression of Hes1 in basal cells in the K14Hes1 epidermis did not result in upregulation of K1/K10 expression or promote a cell fate conversion from basal cells to spinous cells, as was seen in the K14NICD1 epidermis. In addition, and consistent with these in vivo results, we found from our in vitro analyses that overexpression of Hes1 in primary keratinocytes did not induce K1 or K10, whereas forced expression of NICD1 in Hes1 null keratinocytes did so strongly. These data provide evidence that induction of K1/K10 expression by NICD1 does not require Developmental Cell 14, , April 2008 ª2008 Elsevier Inc. 601
9 Hes1. Taken together, our data thus suggest that Notch signaling in fact promotes spinous cell fate determination via a Hes1-independent mechanism. We did not find a requirement for Hes1 in the induction of suprabasal gene expression, but we did find that Notch signaling acting through Hes1 plays a critical role in the maintenance of spinous cells. Loss of Hes1 function caused premature differentiation of spinous cells into granular cells, resulting in a lack of spinous layers during epidermal development. Conversely, forced expression of Hes1 in spinous layer cells in K1Hes1 epidermis resulted in massive expansion of K1-positive/loricrin-negative spinous layer cells. Moreover, we found marked upregulation of Hes1 in spinous layer cells when NICD1 was overexpressed in the suprabasal cells of the epidermis, indicating that this Hes1 expression was mediated by Notch signaling. Taken together, these data provide physiological evidence that Notch signaling, acting through Hes1, is required for the maintenance of spinous cells at early stages of epidermal development by preventing premature granular differentiation. Therefore, our findings suggest that Hes1 serves as a key gatekeeper that protects the spinous layer cells from inappropriate differentiation during epidermal development. Previously it has been shown that a transgenic expression of NICD1 under the control of the involucrin promoter, i.e., gainof-function Notch signaling, resulted in acceleration of granular cell differentiation (Uyttendaele et al., 2004). Consistent with this, we found that the N1N2 dcko epidermis displays a significant reduction of loricrin, whereas forced expression of NICD1 in the spinous cells of K1NICD1 transgenic mice resulted in accumulation of loricrin-positive cells in the developing epidermis. Thus, from these loss- and gain-of-function studies, we conclude that Notch signaling plays an essential role in promoting granular cell differentiation. Based on the presence of granular layers in the Hes1 KO epidermis, it seems likely that Hes1 is not required for the promotion of granular cell differentiation. Our findings thus reveal a triple function of Notch signaling in spinous cells: it induces spinous-fate commitment, promotes further differentiation into granular cells via a Hes1-independent mechanism, and at the same time maintains spinous cell fate and prevents premature differentiation in a Hes1-dependent manner. Hence, our in vivo studies have elucidated multiple roles of the Notch signaling pathway in spinous cells during the early stages of epidermal development. Intriguingly, and despite the presence of clear defects during epidermal development, we did not observe any obvious abnormality in postnatal Hes1 KO epidermis in our skin graft experiments. These data might suggest that Hes1 function in the epidermis is restricted to epidermal development, although we cannot at this stage rule out the possibility that Hes1 functions in a small portion of the postnatal epidermis, where defects in the Hes1 KO epidermis escaped detection. Indeed, the latter possibility is supported in our immunohistochemical analysis in which Hes1 expression is detectable only a small population in the adult epidermis (Figure S6A). In any case, further studies are required to define the exact role of Hes1 in the postnatal epidermis. To this end, analysis of postnatal epidermis in which the Hes1 gene is conditionally ablated in an epidermis-specific manner is currently underway in our laboratory. In addition to these multiple functions of Notch signaling in spinous cells at early stages of epidermal development, our observations also indicate that loss of Hes1 severely affects p63 expression and proliferation in basal cells. As Hes1 expression is restricted to spinous layer cells during epidermal development, the phenotype of basal cells in Hes1 KO epidermis is probably due to a secondary effect from prematurely differentiated suprabasal cells. Consistent with this idea, we showed that the basal cell phenotype in Hes1 KO epidermis was recapitulated in the K1Ascl2IresGFP epidermis, in which Ascl2 expression was targeted to suprabasal cells, indicating the possibility of non-cell-autonomous regulation of basal cell proliferation, as has been suggested previously (Feng et al., 1997). Furthermore, we observed that suprabasal cell proliferation and p63 expression in Hes1 KO epidermis were comparable to WT at E14.5, when the earliest Hes1 expression is detectable in suprabasal cells, arguing against the possibility that Hes1 acts cell-autonomously to regulate p63 expression and proliferation. Taken together, these data suggest that Hes1 plays an important role in maintaining the p63 expression status and proliferative potential of basal cells through a non-cell-autonomous mechanism. In this context, the underlying molecular mechanism may be different from the recent proposal that Notch signaling and p63 can counteract each other in a cell-autonomous manner (Nguyen et al., 2006). As p63 plays an essential role in maintaining the proliferative potential of basal cells (Senoo et al., 2007), it seems plausible that downregulation of p63 expression contributes to the reduced proliferative potential of basal cells in Hes1 KO epidermis. Future studies will be necessary to clarify a molecular link between Notch/Hes1 signaling and p63 expression in the regulation of basal cell proliferation. Molecular Mechanisms Underlying the Regulation of Spinous Layer Cells by Notch Signaling By integrating our current findings and previous data regarding Notch signaling in epidermal development, we propose a working model to explain the contradictory roles of Notch in the spinous layer cells (Figure 7). First, as shown in previous reports (Blanpain et al., 2006), activation of Notch signaling in the spinous cells promotes their fate determination by initiating genetic programs that are required for suprabasal differentiation, in which Notch signaling induces suprabasal-specific genes such as K1 and K10 and represses basal genes such as K15 and the b1 and a6 integrins. In contrast to previously reported (Blanpain et al., 2006) in vitro data in which Hes1 was shown to promote K1/K10 expression, our current in vivo data indicate that the expression of these markers is mediated through a mechanism that is independent of Hes1. We have shown instead that Hes1 is in fact required for the maintenance of the immature and proliferative status of the spinous cells. Based upon the ability of Notch/RBP-J to bind to the promoter region of Ascl2, we speculate that activation of Notch signaling in spinous cells is capable of promoting the expression of both the Hes1 and Ascl2 genes, whereas subsequent Hes1 binding to the Acsl2 promoter interferes with Notch/RBP-J-induced Ascl2 expression in spinous layer cells, resulting in inhibition of Ascl2. Under such circumstances, the spinous cells can maintain 602 Developmental Cell 14, , April 2008 ª2008 Elsevier Inc.
10 their immature and proliferative status without undergoing further differentiation triggered by Ascl2. Although Hes1 expression is essential for the maintenance of spinous layer cells, this repressor must eventually be downregulated to initiate granular differentiation. The spinous cells would otherwise be aberrantly expanded, as occurred in the K1Hes1 epidermis. The tightly regulated expression of Hes1 in the spinous layers suggests that a specific negative feedback mechanism may operate. Consistent with this idea, we showed that the artificial overexpression of Hes1 via the K1 promoter caused a dramatic expansion of Hes1-expressing spinous layer cells, whereas stimulation of the endogenous Hes1 promoter by NICD1 in the K1NICD1 epidermis caused quite restricted Hes1 expression and limited expansion of spinous cells. This suggests that the expression of Hes1 from its endogenous promoter is tightly regulated by a negative feedback mechanism. In fact, it has already been demonstrated that Hes1 is regulated by an autoregulatory feedback loop, in which the Hes1 protein represses its own gene promoter activity by directly binding to a consensus element therein (Hirata et al., 2002). Accumulation of Hes1 protein in spinous cells could lead to a downregulation of the Hes1 gene by this negative feedback mechanism, which could in turn release the Ascl2 promoter from Hes1-mediated repression and allow activation of Ascl2 transcription via Notch signaling to initiate granular cell differentiation. Therefore, we contend that our proposed model provides a good explanation of how a single signaling entity such as Notch can accomplish multiple tasks in the same spinous layer cells, where it determines spinous cell fate and promotes further differentiation to granular cells, while simultaneously preventing terminal differentiation to maintain the immature state of spinous cells. Despite the crucial role of bhlh transcription factors in orchestrating organogenesis, prior to our study, there was little evidence for a role of activator-type bhlh transcription factors in epidermal development. Therefore, our implication of Ascl2 might provide a new insight into understanding the molecular mechanisms of epidermal development. However, loss-of-function analyses are still required to demonstrate that Ascl2 does in fact play a key role in this context. In preliminary studies, we found that Ascl2 is detectable in suprabasal layers of the adult epidermis (Figure S6B), suggesting that Ascl2 also plays a role in the adult epidermis. Intriguingly, and despite profound upregulation of Ascl2 in the embryonic Hes1 KO epidermis (Figures 5B and 5C), its expression in the adult Hes1 KO epidermis is comparable to that of wild-type epidermis (Figure S6C). Although these data indicate a possibility that Ascl2 expression in the adult epidermis might be regulated by a mechanism independent on Hes1, it is also possible that Hes1 functions to repress Ascl2 expression only in a small population of epidermis, where the aberrant Ascl2 expression in the Hes1 KO epidermis escaped detection. In any case, future studies are necessary to clarify the exact molecular function of Ascl2 in the epidermis. Concluding Remarks Our current findings uncover multiple roles of Notch signaling in the coordination of proper stratification during epidermal development. We found that Notch signaling promotes spinous cell fate specification from basal layer cells and further induces granular differentiation, but simultaneously prevents further differentiation to maintain an immature spinous fate. Our model provides new insights into the understanding of a complex developmental role of Notch signaling in which it both inhibits and promotes differentiation depending on the precise cellular circumstances in which it is acting. EXPERIMENTAL PROCEDURES Mice Hes1 KO mice (Ishibashi et al., 1995) were a generous gift from R. Kageyama, Kyoto, Japan. Details of transgenic mice are included in the Supplemental Data. All animal experiments were performed in accordance with the guidelines of the RIKEN Center for Developmental Biology for animal and recombinant DNA experiments. Histology, In Situ Hybridization, and Immunohistochemical Analysis Skin samples and embryos were fixed in 4% paraformaldehyde, embedded in OCT, frozen, and sectioned at 10 mm. Sections were then either subjected to hematoxylin and eosin staining, in situ hybridization, or immunohistochemical analysis, as previously described (Moriyama et al., 2006). The probes and antibodies used in this study are described in the Supplemental Data. Cell Culture and Adenovirus infection Mouse primary epidermal keratinocyte (MPEK) cells were harvested from the E18.5 WT or Hes1 KO mice and maintained in CnT-07 (CELLnTEC) culture medium according to the manufacturer s protocol. Adenoviruses expressing Hes1, NICD1, and RBP-CA were constructed using the ViraPower adenoviral expression system (Invitrogen) according to the manufacturer s protocol. ChIP Assay The ChIP assay was performed using the ChIP-IT Enzymatic Shearing Kit (Active Motif) according to the manufacturer s instructions. Details are included in the Supplemental Data. Microarray Analysis and Q-PCR Detail of methods for microarray analysis and Q-PCR are provided in the Supplemental Experimental Procedures. SUPPLEMENTAL DATA Supplemental Data include six figures and Supplemental Experimental Procedures and can be found at full/14/4/594/dc1/. ACKNOWLEDGMENTS We thank R. Kageyama for providing the Hes1 knockout mice, D. Roop for providing the hkeratin1 promoter, and N. Brown for supplying the anti-hes1 antibody. We are also very grateful to the Laboratory for Animal Resources and Genetic Engineering in RIKEN CDB for generating the transgenic mice used in this study. This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas ( to M.O.) and a grant for Regenerative Medicine Realization Projects from Ministry of Education, Culture, Sports, Science and Technology of Japan. The authors have no conflicting interests. Received: November 26, 2006 Revised: October 4, 2007 Accepted: January 24, 2008 Published: April 14, 2008 REFERENCES Blanpain, C., Lowry, W.E., Pasolli, H.A., and Fuchs, E. (2006). Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 20, Feng, X., Peng, Z.H., Di, W., Li, X.Y., Rochette-Egly, C., Chambon, P., Voorhees, J.J., and Xiao, J.H. (1997). Suprabasal expression of Developmental Cell 14, , April 2008 ª2008 Elsevier Inc. 603
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Amelio et al., http://www.jcb.org/cgi/content/full/jcb.201203134/dc1 Figure S1. mir-24 regulates proliferation and by itself induces
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Amelio et al., http://www.jcb.org/cgi/content/full/jcb.201203134/dc1 Figure S1. mir-24 regulates proliferation and by itself induces
The Beauty of the Skin
 The Beauty of the Skin Rose-Anne Romano, Ph.D Assistant Professor Department of Oral Biology School of Dental Medicine State University of New York at Buffalo The Big Question How do approximately 50 trillion
The Beauty of the Skin Rose-Anne Romano, Ph.D Assistant Professor Department of Oral Biology School of Dental Medicine State University of New York at Buffalo The Big Question How do approximately 50 trillion
s u p p l e m e n ta ry i n f o r m at i o n
 Figure S1 Characterization of tet-off inducible cell lines expressing GFPprogerin and GFP-wt lamin A. a, Western blot analysis of GFP-progerin- or GFP-wt lamin A- expressing cells before induction (0d)
Figure S1 Characterization of tet-off inducible cell lines expressing GFPprogerin and GFP-wt lamin A. a, Western blot analysis of GFP-progerin- or GFP-wt lamin A- expressing cells before induction (0d)
Fig. S1. Upregulation of K18 and K14 mrna levels during ectoderm specification of hescs. Quantitative real-time PCR analysis of mrna levels of OCT4
 Fig. S1. Upregulation of K18 and K14 mrna levels during ectoderm specification of hescs. Quantitative real-time PCR analysis of mrna levels of OCT4 (n=3 independent differentiation experiments for each
Fig. S1. Upregulation of K18 and K14 mrna levels during ectoderm specification of hescs. Quantitative real-time PCR analysis of mrna levels of OCT4 (n=3 independent differentiation experiments for each
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was
 Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Supplementary Table 1. List of primers used in this study
 Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
Journal Club. 03/04/2012 Lama Nazzal
 Journal Club 03/04/2012 Lama Nazzal NOTCH and the kidneys Is an evolutionarily conserved cell cell communication mechanism. Is a regulator of cell specification, differentiation, and tissue patterning.
Journal Club 03/04/2012 Lama Nazzal NOTCH and the kidneys Is an evolutionarily conserved cell cell communication mechanism. Is a regulator of cell specification, differentiation, and tissue patterning.
Supplemental Information. Otic Mesenchyme Cells Regulate. Spiral Ganglion Axon Fasciculation. through a Pou3f4/EphA4 Signaling Pathway
 Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Loss of RhoA promotes skin tumor formation. Supplementary Figure 1. Loss of RhoA does not impair F-actin organization.
 Supplementary Figure Legends Supplementary Figure 1. Loss of RhoA does not impair F-actin organization. a. Representative IF images of F-actin staining of big and small control (left) and RhoA ko tumors
Supplementary Figure Legends Supplementary Figure 1. Loss of RhoA does not impair F-actin organization. a. Representative IF images of F-actin staining of big and small control (left) and RhoA ko tumors
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2535 Figure S1 SOX10 is expressed in human giant congenital nevi and its expression in human melanoma samples suggests that SOX10 functions in a MITF-independent manner. a, b, Representative
DOI: 10.1038/ncb2535 Figure S1 SOX10 is expressed in human giant congenital nevi and its expression in human melanoma samples suggests that SOX10 functions in a MITF-independent manner. a, b, Representative
Molecular Mechanism of MicroRNA-203 Mediated Epidermal Stem Cell Differentiation
 University of Colorado, Boulder CU Scholar Undergraduate Honors Theses Honors Program Spring 2011 Molecular Mechanism of MicroRNA-203 Mediated Epidermal Stem Cell Differentiation Sarah Jackson University
University of Colorado, Boulder CU Scholar Undergraduate Honors Theses Honors Program Spring 2011 Molecular Mechanism of MicroRNA-203 Mediated Epidermal Stem Cell Differentiation Sarah Jackson University
Supplementary Figure 1. Baf60c and baf180 are induced during cardiac regeneration in zebrafish. RNA in situ hybridization was performed on paraffin
 Supplementary Figure 1. Baf60c and baf180 are induced during cardiac regeneration in zebrafish. RNA in situ hybridization was performed on paraffin sections from sham-operated adult hearts (a and i) and
Supplementary Figure 1. Baf60c and baf180 are induced during cardiac regeneration in zebrafish. RNA in situ hybridization was performed on paraffin sections from sham-operated adult hearts (a and i) and
AP VP DLP H&E. p-akt DLP
 A B AP VP DLP H&E AP AP VP DLP p-akt wild-type prostate PTEN-null prostate Supplementary Fig. 1. Targeted deletion of PTEN in prostate epithelium resulted in HG-PIN in all three lobes. (A) The anatomy
A B AP VP DLP H&E AP AP VP DLP p-akt wild-type prostate PTEN-null prostate Supplementary Fig. 1. Targeted deletion of PTEN in prostate epithelium resulted in HG-PIN in all three lobes. (A) The anatomy
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Supplemental Figure 1. Intracranial transduction of a modified ptomo lentiviral vector in the mouse
 Supplemental figure legends Supplemental Figure 1. Intracranial transduction of a modified ptomo lentiviral vector in the mouse hippocampus targets GFAP-positive but not NeuN-positive cells. (A) Stereotaxic
Supplemental figure legends Supplemental Figure 1. Intracranial transduction of a modified ptomo lentiviral vector in the mouse hippocampus targets GFAP-positive but not NeuN-positive cells. (A) Stereotaxic
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
SUPPLEMENTARY INFORMATION
 Figure S1 Treatment with both Sema6D and Plexin-A1 sirnas induces the phenotype essentially identical to that induced by treatment with Sema6D sirna alone or Plexin-A1 sirna alone. (a,b) The cardiac tube
Figure S1 Treatment with both Sema6D and Plexin-A1 sirnas induces the phenotype essentially identical to that induced by treatment with Sema6D sirna alone or Plexin-A1 sirna alone. (a,b) The cardiac tube
Expression of acid base transporters in the kidney collecting duct in Slc2a7 -/-
 Supplemental Material Results. Expression of acid base transporters in the kidney collecting duct in Slc2a7 -/- and Slc2a7 -/- mice. The expression of AE1 in the kidney was examined in Slc26a7 KO mice.
Supplemental Material Results. Expression of acid base transporters in the kidney collecting duct in Slc2a7 -/- and Slc2a7 -/- mice. The expression of AE1 in the kidney was examined in Slc26a7 KO mice.
Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the
 Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the location of the transmembrane (TM), FRM binding (FB)
Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the location of the transmembrane (TM), FRM binding (FB)
Supplementary Information. Preferential associations between co-regulated genes reveal a. transcriptional interactome in erythroid cells
 Supplementary Information Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells Stefan Schoenfelder, * Tom Sexton, * Lyubomira Chakalova, * Nathan
Supplementary Information Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells Stefan Schoenfelder, * Tom Sexton, * Lyubomira Chakalova, * Nathan
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Generation and validation of mtef4-knockout mice.
 Supplementary Figure 1 Generation and validation of mtef4-knockout mice. (a) Alignment of EF4 (E. coli) with mouse, yeast and human EF4. (b) Domain structures of mouse mtef4 compared to those of EF4 (E.
Supplementary Figure 1 Generation and validation of mtef4-knockout mice. (a) Alignment of EF4 (E. coli) with mouse, yeast and human EF4. (b) Domain structures of mouse mtef4 compared to those of EF4 (E.
EMBO REPORT SUPPLEMENTARY SECTION. Quantitation of mitotic cells after perturbation of Notch signalling.
 EMBO REPORT SUPPLEMENTARY SECTION Quantitation of mitotic cells after perturbation of Notch signalling. Notch activation suppresses the cell cycle indistinguishably both within and outside the neural plate
EMBO REPORT SUPPLEMENTARY SECTION Quantitation of mitotic cells after perturbation of Notch signalling. Notch activation suppresses the cell cycle indistinguishably both within and outside the neural plate
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting. protein3) regulate autophagy and mitophagy in renal tubular cells in. acute kidney injury
 Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
Genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1.
 Genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Youngsoo Lee, Sachin Katyal, Yang Li, Sherif F. El-Khamisy, Helen R. Russell, Keith W. Caldecott and Peter J. McKinnon.
Genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Youngsoo Lee, Sachin Katyal, Yang Li, Sherif F. El-Khamisy, Helen R. Russell, Keith W. Caldecott and Peter J. McKinnon.
GFP/Iba1/GFAP. Brain. Liver. Kidney. Lung. Hoechst/Iba1/TLR9!
 Supplementary information a +KA Relative expression d! Tlr9 5!! 5! NSC Neuron Astrocyte Microglia! 5! Tlr7!!!! NSC Neuron Astrocyte! GFP/Sβ/! Iba/Hoechst Microglia e Hoechst/Iba/TLR9! GFP/Iba/GFAP f Brain
Supplementary information a +KA Relative expression d! Tlr9 5!! 5! NSC Neuron Astrocyte Microglia! 5! Tlr7!!!! NSC Neuron Astrocyte! GFP/Sβ/! Iba/Hoechst Microglia e Hoechst/Iba/TLR9! GFP/Iba/GFAP f Brain
Quantification of early stage lesions for loss of p53 should be shown in the main figures.
 Reviewer #1 (Remarks to the Author): Expert in prostate cancer The manuscript "Clonal dynamics following p53 loss of heterozygosity in Kras-driven cancers" uses a number of novel genetically engineered
Reviewer #1 (Remarks to the Author): Expert in prostate cancer The manuscript "Clonal dynamics following p53 loss of heterozygosity in Kras-driven cancers" uses a number of novel genetically engineered
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
Nature Immunology: doi: /ni Supplementary Figure 1. Huwe1 has high expression in HSCs and is necessary for quiescence.
 Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplemental Figure 1. Western blot analysis indicated that MIF was detected in the fractions of
 Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at
 Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at E10.5 were double-stained for TUNEL (red) and PECAM-1 (green).
Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at E10.5 were double-stained for TUNEL (red) and PECAM-1 (green).
MicroRNA and Male Infertility: A Potential for Diagnosis
 Review Article MicroRNA and Male Infertility: A Potential for Diagnosis * Abstract MicroRNAs (mirnas) are small non-coding single stranded RNA molecules that are physiologically produced in eukaryotic
Review Article MicroRNA and Male Infertility: A Potential for Diagnosis * Abstract MicroRNAs (mirnas) are small non-coding single stranded RNA molecules that are physiologically produced in eukaryotic
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Correlation between LKB1 and YAP expression in human lung cancer samples. (a) Representative photos showing LKB1 and YAP immunohistochemical staining in human
Supplementary Figures Supplementary Figure 1 Correlation between LKB1 and YAP expression in human lung cancer samples. (a) Representative photos showing LKB1 and YAP immunohistochemical staining in human
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
CHAPTER 6 SUMMARIZING DISCUSSION
 CHAPTER 6 SUMMARIZING DISCUSSION More than 20 years ago the founding member of the Wnt gene family, Wnt-1/Int1, was discovered as a proto-oncogene activated in mammary gland tumors by the mouse mammary
CHAPTER 6 SUMMARIZING DISCUSSION More than 20 years ago the founding member of the Wnt gene family, Wnt-1/Int1, was discovered as a proto-oncogene activated in mammary gland tumors by the mouse mammary
Early cell death (FGF) B No RunX transcription factor produced Yes No differentiation
 Solution Key - Practice Questions Question 1 a) A recent publication has shown that the fat stem cells (FSC) can act as bone stem cells to repair cavities in the skull, when transplanted into immuno-compromised
Solution Key - Practice Questions Question 1 a) A recent publication has shown that the fat stem cells (FSC) can act as bone stem cells to repair cavities in the skull, when transplanted into immuno-compromised
Supplementary Figure 1: Signaling centers contain few proliferating cells, express p21, and
 Supplementary Figure 1: Signaling centers contain few proliferating cells, express p21, and exclude YAP from the nucleus. (a) Schematic diagram of an E10.5 mouse embryo. (b,c) Sections at B and C in (a)
Supplementary Figure 1: Signaling centers contain few proliferating cells, express p21, and exclude YAP from the nucleus. (a) Schematic diagram of an E10.5 mouse embryo. (b,c) Sections at B and C in (a)
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Figure 1. Quantification of proliferation in thyroid of WT, Ctns -/- and grafted
 Supplemental Figure 1. Quantification of proliferation in thyroid of WT, Ctns -/- and grafted Ctns -/- mice. Cells immunolabeled for the proliferation marker (Ki-67) were counted in sections (n=3 WT, n=4
Supplemental Figure 1. Quantification of proliferation in thyroid of WT, Ctns -/- and grafted Ctns -/- mice. Cells immunolabeled for the proliferation marker (Ki-67) were counted in sections (n=3 WT, n=4
Supporting Information
 Supporting Information Plikus et al. 10.1073/pnas.1215935110 SI Text Movies S1, S2, S3, and S4 are time-lapse recordings from individually cultured Period2 Luc vibrissa follicles show that circadian cycles
Supporting Information Plikus et al. 10.1073/pnas.1215935110 SI Text Movies S1, S2, S3, and S4 are time-lapse recordings from individually cultured Period2 Luc vibrissa follicles show that circadian cycles
Foxm1 Transcription Factor is Required for Lung Fibrosis and Epithelial to Mesenchymal Transition.
 Manuscript EMBO-2012-82682 Foxm1 Transcription Factor is Required for Lung Fibrosis and Epithelial to Mesenchymal Transition. David Balli, Vladimir Ustiyan, Yufang Zhang, I-Ching Wang, Alex J. Masino,
Manuscript EMBO-2012-82682 Foxm1 Transcription Factor is Required for Lung Fibrosis and Epithelial to Mesenchymal Transition. David Balli, Vladimir Ustiyan, Yufang Zhang, I-Ching Wang, Alex J. Masino,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
Title: Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease
 1 Supplemental Materials 2 3 Title: Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease 4 5 6 Authors: Madhumita Basu, 1 Jun-Yi Zhu, 2 Stephanie LaHaye 1,3, Uddalak
1 Supplemental Materials 2 3 Title: Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease 4 5 6 Authors: Madhumita Basu, 1 Jun-Yi Zhu, 2 Stephanie LaHaye 1,3, Uddalak
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figures. T Cell Factor-1 initiates T helper 2 fate by inducing GATA-3 and repressing Interferon-γ
 Supplementary Figures T Cell Factor-1 initiates T helper 2 fate by inducing GATA-3 and repressing Interferon-γ Qing Yu, Archna Sharma, Sun Young Oh, Hyung-Geun Moon, M. Zulfiquer Hossain, Theresa M. Salay,
Supplementary Figures T Cell Factor-1 initiates T helper 2 fate by inducing GATA-3 and repressing Interferon-γ Qing Yu, Archna Sharma, Sun Young Oh, Hyung-Geun Moon, M. Zulfiquer Hossain, Theresa M. Salay,
Supplemental Table 1. Primers used for RT-PCR analysis of inflammatory cytokines Gene Primer Sequence
 Supplemental Table 1. Primers used for RT-PCR analysis of inflammatory cytokines Gene Primer Sequence IL-1α Forward primer 5 -CAAGATGGCCAAAGTTCGTGAC-3' Reverse primer 5 -GTCTCATGAAGTGAGCCATAGC-3 IL-1β
Supplemental Table 1. Primers used for RT-PCR analysis of inflammatory cytokines Gene Primer Sequence IL-1α Forward primer 5 -CAAGATGGCCAAAGTTCGTGAC-3' Reverse primer 5 -GTCTCATGAAGTGAGCCATAGC-3 IL-1β
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Figure 1. The expression of ephrin-b2 H2BGFP persists in the post-hearingonset organ of Corti and is specifically restricted to supporting cells. Sox2 immunolabeling
SUPPLEMENTARY INFORMATION Supplementary Figure 1. The expression of ephrin-b2 H2BGFP persists in the post-hearingonset organ of Corti and is specifically restricted to supporting cells. Sox2 immunolabeling
Neocortex Zbtb20 / NFIA / Sox9
 Neocortex / NFIA / Sox9 Supplementary Figure 1. Expression of, NFIA, and Sox9 in the mouse neocortex at. The lower panels are higher magnification views of the oxed area. Arrowheads indicate triple-positive
Neocortex / NFIA / Sox9 Supplementary Figure 1. Expression of, NFIA, and Sox9 in the mouse neocortex at. The lower panels are higher magnification views of the oxed area. Arrowheads indicate triple-positive
Zhu et al, page 1. Supplementary Figures
 Zhu et al, page 1 Supplementary Figures Supplementary Figure 1: Visual behavior and avoidance behavioral response in EPM trials. (a) Measures of visual behavior that performed the light avoidance behavior
Zhu et al, page 1 Supplementary Figures Supplementary Figure 1: Visual behavior and avoidance behavioral response in EPM trials. (a) Measures of visual behavior that performed the light avoidance behavior
Programmed Cell Death (apoptosis)
 Programmed Cell Death (apoptosis) Stereotypic death process includes: membrane blebbing nuclear fragmentation chromatin condensation and DNA framentation loss of mitochondrial integrity and release of
Programmed Cell Death (apoptosis) Stereotypic death process includes: membrane blebbing nuclear fragmentation chromatin condensation and DNA framentation loss of mitochondrial integrity and release of
marker. DAPI labels nuclei. Flies were 20 days old. Scale bar is 5 µm. Ctrl is
 Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature11463 %Sox17(+) 9 8 7 6 5 4 3 2 1 %Sox17(+) #Sox17(+) d2 d4 d6 d8 d1 d12 d14 d18 25 2 15 1 5 Number of Sox17(+) cells X 1 Supplementary Figure 1: Expression of
SUPPLEMENTARY INFORMATION doi:1.138/nature11463 %Sox17(+) 9 8 7 6 5 4 3 2 1 %Sox17(+) #Sox17(+) d2 d4 d6 d8 d1 d12 d14 d18 25 2 15 1 5 Number of Sox17(+) cells X 1 Supplementary Figure 1: Expression of
Vertebrate Limb Patterning
 Vertebrate Limb Patterning What makes limb patterning an interesting/useful developmental system How limbs develop Key events in limb development positioning and specification initiation of outgrowth establishment
Vertebrate Limb Patterning What makes limb patterning an interesting/useful developmental system How limbs develop Key events in limb development positioning and specification initiation of outgrowth establishment
Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells
 Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells Immunofluorescence staining of H3K9me3 and 53BP1 in PH and HGADFN003 (HG003) cells at 24 h after γ-irradiation. Scale bar,
Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells Immunofluorescence staining of H3K9me3 and 53BP1 in PH and HGADFN003 (HG003) cells at 24 h after γ-irradiation. Scale bar,
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Endothelial PGC 1 - α 1 mediates vascular dysfunction in diabetes
 Endothelial PGC-1α mediates vascular dysfunction in diabetes Reporter: Yaqi Zhou Date: 04/14/2014 Outline I. Introduction II. Research route & Results III. Summary Diabetes the Epidemic of the 21st Century
Endothelial PGC-1α mediates vascular dysfunction in diabetes Reporter: Yaqi Zhou Date: 04/14/2014 Outline I. Introduction II. Research route & Results III. Summary Diabetes the Epidemic of the 21st Century
SUPPLEMENTARY INFORMATION
 Supplementary Discussion The cell cycle machinery and the DNA damage response network are highly interconnected and co-regulated in assuring faithful duplication and partition of genetic materials into
Supplementary Discussion The cell cycle machinery and the DNA damage response network are highly interconnected and co-regulated in assuring faithful duplication and partition of genetic materials into
INTRODUCTION. Human Molecular Genetics, 2006, Vol. 15, No doi: /hmg/ddi470 Advance Access published on January 6, 2006
 Skin lesion development in a mouse model of incontinentia pigmenti is triggered by NEMO deficiency in epidermal keratinocytes and requires TNF signaling Arianna Nenci 1, Marion Huth 1, Alfred Funteh 2,
Skin lesion development in a mouse model of incontinentia pigmenti is triggered by NEMO deficiency in epidermal keratinocytes and requires TNF signaling Arianna Nenci 1, Marion Huth 1, Alfred Funteh 2,
Supplementary Figure 1 (Related with Figure 4). Molecular consequences of Eed deletion. (a) ChIP analysis identifies 3925 genes that are associated
 Supplementary Figure 1 (Related with Figure 4). Molecular consequences of Eed deletion. (a) ChIP analysis identifies 3925 genes that are associated with the H3K27me3 mark in chondrocytes (see Table S1,
Supplementary Figure 1 (Related with Figure 4). Molecular consequences of Eed deletion. (a) ChIP analysis identifies 3925 genes that are associated with the H3K27me3 mark in chondrocytes (see Table S1,
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein
 Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
mirna Dr. S Hosseini-Asl
 mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
Nature Medicine doi: /nm.2860
 Supplemental Figure Legends Supplemental Figure 1: Hypomorphic expression of IFT88 results in olfactory signaling proteins no longer localizing to the ciliary layer. (a) ACIII localizes to the cilia and
Supplemental Figure Legends Supplemental Figure 1: Hypomorphic expression of IFT88 results in olfactory signaling proteins no longer localizing to the ciliary layer. (a) ACIII localizes to the cilia and
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons
 Supplemental Material mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons Antoine de Chevigny, Nathalie Coré, Philipp Follert, Marion Gaudin, Pascal
Supplemental Material mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons Antoine de Chevigny, Nathalie Coré, Philipp Follert, Marion Gaudin, Pascal
Biology Developmental Biology Spring Quarter Midterm 1 Version A
 Biology 411 - Developmental Biology Spring Quarter 2013 Midterm 1 Version A 75 Total Points Open Book Choose 15 out the 20 questions to answer (5 pts each). Only the first 15 questions that are answered
Biology 411 - Developmental Biology Spring Quarter 2013 Midterm 1 Version A 75 Total Points Open Book Choose 15 out the 20 questions to answer (5 pts each). Only the first 15 questions that are answered
Supplemental Information. Myocardial Polyploidization Creates a Barrier. to Heart Regeneration in Zebrafish
 Developmental Cell, Volume 44 Supplemental Information Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish Juan Manuel González-Rosa, Michka Sharpe, Dorothy Field, Mark H.
Developmental Cell, Volume 44 Supplemental Information Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish Juan Manuel González-Rosa, Michka Sharpe, Dorothy Field, Mark H.
Supplementary methods:
 Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Today. Genomic Imprinting & X-Inactivation
 Today 1. Quiz (~12 min) 2. Genomic imprinting in mammals 3. X-chromosome inactivation in mammals Note that readings on Dosage Compensation and Genomic Imprinting in Mammals are on our web site. Genomic
Today 1. Quiz (~12 min) 2. Genomic imprinting in mammals 3. X-chromosome inactivation in mammals Note that readings on Dosage Compensation and Genomic Imprinting in Mammals are on our web site. Genomic
Yap Tunes Airway Epithelial Size and Architecture by Regulating the Identity, Maintenance, and Self-Renewal of Stem Cells
 Article Yap Tunes Airway Epithelial Size and Architecture by Regulating the Identity, Maintenance, and Self-Renewal of Stem Cells Rui Zhao, 1,2,3,4 Timothy R. Fallon, 1 Srinivas Vinod Saladi, 5 Ana Pardo-Saganta,
Article Yap Tunes Airway Epithelial Size and Architecture by Regulating the Identity, Maintenance, and Self-Renewal of Stem Cells Rui Zhao, 1,2,3,4 Timothy R. Fallon, 1 Srinivas Vinod Saladi, 5 Ana Pardo-Saganta,
Receptor-interacting Protein Kinases Mediate Necroptosis In Neural Tissue Damage After Spinal Cord Injury
 Receptor-interacting Protein Kinases Mediate Necroptosis In Neural Tissue Damage After Spinal Cord Injury Haruo Kanno, M.D., Ph.D., Hiroshi Ozawa, M.D., Ph.D., Satoshi Tateda, M.D., Kenichiro Yahata, M.D.,
Receptor-interacting Protein Kinases Mediate Necroptosis In Neural Tissue Damage After Spinal Cord Injury Haruo Kanno, M.D., Ph.D., Hiroshi Ozawa, M.D., Ph.D., Satoshi Tateda, M.D., Kenichiro Yahata, M.D.,
Project report October 2012 March 2013 CRF fellow: Principal Investigator: Project title:
 Project report October 2012 March 2013 CRF fellow: Gennaro Napolitano Principal Investigator: Sergio Daniel Catz Project title: Small molecule regulators of vesicular trafficking to enhance lysosomal exocytosis
Project report October 2012 March 2013 CRF fellow: Gennaro Napolitano Principal Investigator: Sergio Daniel Catz Project title: Small molecule regulators of vesicular trafficking to enhance lysosomal exocytosis
An HMGA2-IGF2BP2 Axis Regulates Myoblast Proliferation and Myogenesis
 Please cite this article in press as: Li et al., An HMGA2-IGF2BP2 Axis Regulates Myoblast Proliferation and Myogenesis, Developmental Cell (2012), http://dx.doi.org/10.1016/j.devcel.2012.10.019 Developmental
Please cite this article in press as: Li et al., An HMGA2-IGF2BP2 Axis Regulates Myoblast Proliferation and Myogenesis, Developmental Cell (2012), http://dx.doi.org/10.1016/j.devcel.2012.10.019 Developmental
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous
 Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic
 Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic B cells from WT Nfat2 +/+, TCL1 Nfat2 +/+ and TCL1 Nfat2
Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic B cells from WT Nfat2 +/+, TCL1 Nfat2 +/+ and TCL1 Nfat2
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11095 Supplementary Table 1. Summary of the binding between Angptls and various Igdomain containing receptors as determined by flow cytometry analysis. The results were summarized from
doi:10.1038/nature11095 Supplementary Table 1. Summary of the binding between Angptls and various Igdomain containing receptors as determined by flow cytometry analysis. The results were summarized from
Cell Type Nervous System I. Developmental Readout. Foundations. Stem cells. Organ formation. Human issues.
 7.72 10.11.06 Cell Type Nervous System I Human issues Organ formation Stem cells Developmental Readout Axes Cell type Axon guidance 3D structure Analysis Model + + organisms Foundations Principles 1 What
7.72 10.11.06 Cell Type Nervous System I Human issues Organ formation Stem cells Developmental Readout Axes Cell type Axon guidance 3D structure Analysis Model + + organisms Foundations Principles 1 What
Supplementary Information Titles Journal: Nature Medicine
 Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Distinct mechanisms of TGF-b1 mediated epithelial-to-mesenchymal transition and metastasis during skin carcinogenesis
 Distinct mechanisms of TGF-b1 mediated epithelial-to-mesenchymal transition and metastasis during skin carcinogenesis Gangwen Han,, Molly Kulesz-Martin, Xiao-Jing Wang J Clin Invest. 2005;115(7):1714-1723.
Distinct mechanisms of TGF-b1 mediated epithelial-to-mesenchymal transition and metastasis during skin carcinogenesis Gangwen Han,, Molly Kulesz-Martin, Xiao-Jing Wang J Clin Invest. 2005;115(7):1714-1723.
(A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939 (5 µm)
 Supplementary Figure Legends Figure S1. Tankyrase inhibition suppresses cell proliferation in an axin/β-catenin independent manner. (A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939
Supplementary Figure Legends Figure S1. Tankyrase inhibition suppresses cell proliferation in an axin/β-catenin independent manner. (A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939
Fig. S1. RT-PCR analyses of the expression and distribution of Xdscr6 transcripts during early development.
 Fig. S1. RT-PCR analyses of the expression and distribution of Xdscr6 transcripts during early development. (A) Temporal expression of Xdscr6 at various stages (numbers on the top) and its distribution
Fig. S1. RT-PCR analyses of the expression and distribution of Xdscr6 transcripts during early development. (A) Temporal expression of Xdscr6 at various stages (numbers on the top) and its distribution
NFIL3/E4BP4 controls Type 2 T helper cell cytokine expression
 Manuscript EMBO-2010-75657 NFIL3/E4BP4 controls Type 2 T helper cell cytokine expression Masaki Kashiwada, Suzanne L. Cassel, John D. Colgan and Paul B. Rothman Corresponding author: Paul Rothman, University
Manuscript EMBO-2010-75657 NFIL3/E4BP4 controls Type 2 T helper cell cytokine expression Masaki Kashiwada, Suzanne L. Cassel, John D. Colgan and Paul B. Rothman Corresponding author: Paul Rothman, University
Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained
 Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained with MitoTracker (red), then were immunostained with
Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained with MitoTracker (red), then were immunostained with
Hunk is required for HER2/neu-induced mammary tumorigenesis
 Research article Hunk is required for HER2/neu-induced mammary tumorigenesis Elizabeth S. Yeh, 1 Thomas W. Yang, 1 Jason J. Jung, 1 Heather P. Gardner, 1 Robert D. Cardiff, 2 and Lewis A. Chodosh 1 1 Department
Research article Hunk is required for HER2/neu-induced mammary tumorigenesis Elizabeth S. Yeh, 1 Thomas W. Yang, 1 Jason J. Jung, 1 Heather P. Gardner, 1 Robert D. Cardiff, 2 and Lewis A. Chodosh 1 1 Department
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates
 Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates L. Cheng, A. Arata, R. Mizuguchi, Y. Qian, A. Karunaratne, P.A. Gray, S. Arata, S. Shirasawa, M. Bouchard,
Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates L. Cheng, A. Arata, R. Mizuguchi, Y. Qian, A. Karunaratne, P.A. Gray, S. Arata, S. Shirasawa, M. Bouchard,
Akemi Ishida-Yamamoto
 Journal of Dermatological Science (2003) 31(1):3-8. Loricrin keratoderma: a novel disease entity characterized by nuclear accumulation of mutant loricrin Akemi Ishida-Yamamoto Loricrin Keratoderma. A novel
Journal of Dermatological Science (2003) 31(1):3-8. Loricrin keratoderma: a novel disease entity characterized by nuclear accumulation of mutant loricrin Akemi Ishida-Yamamoto Loricrin Keratoderma. A novel
TGF-β Signaling Regulates Neuronal C1q Expression and Developmental Synaptic Refinement
 Supplementary Information Title: TGF-β Signaling Regulates Neuronal C1q Expression and Developmental Synaptic Refinement Authors: Allison R. Bialas and Beth Stevens Supplemental Figure 1. In vitro characterization
Supplementary Information Title: TGF-β Signaling Regulates Neuronal C1q Expression and Developmental Synaptic Refinement Authors: Allison R. Bialas and Beth Stevens Supplemental Figure 1. In vitro characterization
Supplementary Figure 1. Lkb1-deficient lung ADC progressively transdifferentiates into SCC. (a) A scheme showing the progression pattern of atypical
 Supplementary Figure 1. Lkb1-deficient lung ADC progressively transdifferentiates into SCC. (a) A scheme showing the progression pattern of atypical adenomatous hyperplasia/epithelial hyperplasia (AAH/EH),
Supplementary Figure 1. Lkb1-deficient lung ADC progressively transdifferentiates into SCC. (a) A scheme showing the progression pattern of atypical adenomatous hyperplasia/epithelial hyperplasia (AAH/EH),
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Differential expression of mirnas from the pri-mir-17-92a locus.
 Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
Primary Cilia Can Both Mediate and Suppress Hedgehog Pathway- Dependent Tumorigenesis (Supplementary Figures and Materials)
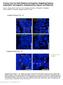 Primary Cilia Can Both Mediate and Suppress Hedgehog Pathway- Dependent Tumorigenesis (Supplementary Figures and Materials) Sunny Y. Wong, Allen D. Seol, Po-Lin So, Alexandre N. Ermilov, Christopher K.
Primary Cilia Can Both Mediate and Suppress Hedgehog Pathway- Dependent Tumorigenesis (Supplementary Figures and Materials) Sunny Y. Wong, Allen D. Seol, Po-Lin So, Alexandre N. Ermilov, Christopher K.
Mass Histology Service
 Mass Histology Service A complete anatomical pathology laboratory www.masshistology.com Telephone: (877) 286-6004 Report on Pathology A Time Course Study of the Local Effects of Intramuscular XXXXXXX Injection
Mass Histology Service A complete anatomical pathology laboratory www.masshistology.com Telephone: (877) 286-6004 Report on Pathology A Time Course Study of the Local Effects of Intramuscular XXXXXXX Injection
(a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable
 Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figures
 Supplementary Figures Supplementary Fig. 1. Galectin-3 is present within tumors. (A) mrna expression levels of Lgals3 (galectin-3) and Lgals8 (galectin-8) in the four classes of cell lines as determined
Supplementary Figures Supplementary Fig. 1. Galectin-3 is present within tumors. (A) mrna expression levels of Lgals3 (galectin-3) and Lgals8 (galectin-8) in the four classes of cell lines as determined
Cesarini et al., http ://www.jcb.org /cgi /content /full /jcb /DC1
 Supplemental material JCB Cesarini et al., http ://www.jcb.org /cgi /content /full /jcb.201504035 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. Lamin A/C depletion generates two distinct phenotypes in
Supplemental material JCB Cesarini et al., http ://www.jcb.org /cgi /content /full /jcb.201504035 /DC1 THE JOU RNAL OF CELL BIO LOGY Figure S1. Lamin A/C depletion generates two distinct phenotypes in
Supplementary Figures
 J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
