Dual PI3K/mTOR inhibition shows antileukemic activity in MLL rearranged acute. Supplemental method S1: Confirmation of RAS mutations
|
|
|
- Brittany Terry
- 5 years ago
- Views:
Transcription
1 Supplementary information Dual PI3K/mTOR inhibition shows antileukemic activity in MLL rearranged acute myeloid leukemia Sandhöfer N. et al. Supplemental methods S1-S4: Supplemental method S1: Confirmation of RAS mutations Supplemental method S2: Generation of stable murine MLL-ENL transduced and immortalized cells Supplemental method S3: Microarray analysis Supplemental method S4: quantitative real-time PCR (qrt-pcr) Supplemental tables S1-S4 Supplemental table S1: Characteristics of patient samples used for in vitro treatment experiments (n=18). Supplemental table S2: Genetic background of human cell lines. Supplemental table S3: Characteristics of patient samples used for gene expression analysis. Supplemental table S4: Frequencies of specific codon mutations in cytogenetically normal (CN) AML (n=125) and MLL rearranged AML patients (n=38) Supplemental figures S1-S9 Supplemental figure S1: Pathway inhibition in patient-derived xenografted cells. Supplemental figure S2: Spontaneous apoptosis and Ki67 expression.
2 Supplemental figure S3: Pathway inhibition in murine MLL-ENL immortalized (MLL-ENL ICs) bone marrow progenitor cells. Supplemental figure S4: Inhibition of PI3K/AKT/mTOR signaling in cell lines. Supplemental figure S5: MK-226 and Rapamycin inhibit PI3K/AKT/mTOR signaling but do not induce apoptosis. Supplemental figure S6: Tumor load and body weight during the treatment with BEZ-235 and AC22. Supplemental figure S7: EVI1 and PTEN expression in AML patient samples and cell lines. Supplemental figure S8: Mutational analysis in AML patient samples. Supplemental figure S9: Combination treatment with AZD-6244 in human AML cell lines and PDX cells.
3 Supplemental method S1: Confirmation of RAS mutations KRAS mutations were independently verified using PCR followed by 454 Titanium amplicon chemistry sequencing (Roche Applied Science, Mannheim, Germany), and NRAS mutations by PCR and melting curve analysis. For 454 Titanium amplicon chemistry, a working dilution of 1 6 molecules/l was used as starting concentration for the GS Junior empcr Kit (Lib-A) (Roche Applied Science). Following the empcr amplification, clonally amplified beads were enriched for 454 next-generation sequencing according to the manufacturer s recommendations. The 454 sequencing data were generated on a GS Junior using the GS Junior Titanium Sequencing Kit (Roche Applied Science). Data analysis was performed utilizing JSI Sequence Pilot, SEQNext (JSI medical systems GmbH, Kippenheim, Germany). Supplemental method S2: Generation of stable murine MLL-ENL transduced and immortalized cells The retroviral vectors were purchased from Addgene (pmscv neo empty vector and pmscv neo -MLL-ENL). The production of retroviral supernatants by Phoenix eco cells was conducted as described previously (1). Four to six weeks old C57BL/6 mice were injected intravenously with 15 mg/kg 5-FU in PBS. Two days later bone marrow cells were harvested and cultured overnight in RPMI Glutamax (Life Technologies, Carlsbad, CA, USA) supplemented with 2% fetal calf serum (FCS; Pan Biotech, Aidenbach, Germany), interleukin-3 (IL-3; 1 ng/ml), interleukin-6 (IL-6; 1 ng/ml), stem cell factor (SCF; 1 ng/ml; all from Immunotools, Friesoythe, Germany). The next day cells were infected by spinoculation (2 hour at 25 g at 33 C in 2 ml retroviral supernatant in the presence of 8 µg/ml polybrene) which was
4 repeated on the next day. After the second spinoculation cells were again cultured overnight in RPMI supplemented with 2% FCS, IL-3, IL-6 and SCF. 4, cells were seeded per 1.1 ml of Methocult M3534 (Stem Cell Technologies Inc., Vancouver, Canada) with or without.75 mg/ml G-418 (Life Technologies). Cells transduced with the empty vector could not be replated more than three rounds. After four rounds of serial replating in Methocult3534 MLL-ENL transduced cells were cultured in RPMI Glutamax supplemented with 2% FCS, 1 ng/ml IL-3, 1 ng/ml IL-6 and 5 ng/ml SCF. Supplemental method S3: Microarray data Total RNA was extracted from 562 bone marrow samples as described previously (2) and analyzed using Affymetrix HG-U133 A/B and 2. plus oligonucleotide microarrays (Affymetrix, Santa Clara, CA). Hybridization and image acquisition followed official Affymetrix protocols. No cell sorting was performed. For probes to probe set annotation we used custom chip definition files (CDFs) based on GeneAnnot version 2., synchronized with GeneCards Version 3.4 (available at (3). These CDFs decrease the total number of probe sets (one probe set per gene), and potentially increases the specificity of the analyses by eliminating cross-hybridizing probes (probes are restricted by sequence specificity). Data normalization was performed using the Robust Multichip Average (RMA) method as described by Irizarry (4). Only the probe sets present both on the A, B chips and the 2. plus chips were included in the analysis. Some probe sets on the A, B chips tend to have lower mean signal levels and higher standard deviations than the corresponding probe sets on the Plus 2. chips. To eliminate this
5 batch effect resulting from the different chip designs, we performed a second normalization using an empirical Bayesian method. Supplemental method S4: quantitative real-time PCR (qrt-pcr) Total RNA was extracted from eleven human myeloid cell lines (RNeasy RNA isolation kit (Qiagen,Hilden Germany) and 1 µg RNA was reverse transcribed into copydna (RevertAid First Strand cdna Synthesis Kit; Thermo Scientific, Waltham, MA, USA). The expression levels of the targets (EVI1, PTEN) and a housekeeping gene (GAPDH) were determined using the TaqMan Gene Expression assay kit (Applied Biosystems, Darmstadt, Germany). Amplification and quantification was performed using the 75 Fast Real-Time PCR System (Applied Biosystems). At least three independent experiments were performed with each reaction in duplicates. Supplemental references 1. Polzer H, Janke H, Schmid D, Hiddemann W, Spiekermann K. Casitas B- lineage lymphoma mutants activate AKT to induce transformation in cooperation with class III receptor tyrosine kinases. Experimental hematology. 213;41(3):271-8 e4. 2. Haferlach T, Kern W, Schnittger S, Schoch C. Modern diagnostics in acute leukemias. Critical reviews in oncology/hematology. 25;56(2): Ferrari F, Bortoluzzi S, Coppe A, Sirota A, Safran M, Shmoish M, et al. Novel definition files for human GeneChips based on GeneAnnot. BMC bioinformatics. 27;8:446.
6 4. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 23;4(2):
7 Table S1: Characteristics of patient samples used for in vitro treatment experiments (n=18). # Age [y] Gender Blast cells FAB MLL FLT3 RAS NPM1 CEBPA Type 1 68 M 8% M1 WT WT NA WT WT BM 2 45 F 83% M4 MLL-PTD ITD NA WT NA BM 3 53 F 88% M WT ITD WT NPM1c WT PB 4 2 F 95% M5a MLL-t WT WT WT NA BM 5 77 F 76% M2 WT WT NRAS G12D NPM1c NA BM 6 6 F 8% M2 WT ITD WT NPM1c NA BM 7 58 F 77% M1 MLL-PTD ITD WT WT WT BM 8 65 F 87% M1 WT TKD NRAS G12C NPM1c WT BM 9 79 F 83% M1 WT WT NRAS Q61R KRAS G12D WT WT BM 1 73 F 8% M5 MLL-t WT WT WT NA BM F 85% NA WT ITD WT WT WT BM M 88% M1 WT TKD NRAS G12S WT NA BM F 84% M4 WT TKD NA NPM1c WT BM F 71% M2 WT ITD WT WT WT PB M 1% M4 MLL-t WT KRAS G13D KRAS Q61H WT WT BM F 8% M1 WT WT WT WT WT BM M 74% M2 MLL-PTD ITD NA WT WT BM M 77% M3 WT ITD NA WT WT BM M: male, F: female, FAB: French-American-British Classification, WT: wildtype, MLL-t: MLL-translocation, MLL-PTD: MLLpartial tandem duplication, FLT3-ITD: internal tandem duplication, FLT3-TKD: tyrosine kinase domain, NA: not analyzed, BM: bone marrow, PB: peripheral blood Table S2: Genetic background of human cell lines. FAB MLL FLT3 CBL NRAS KRAS p53 NPM1 DNMT3A MOLM-13 M5 MLL-AF9 ITD 49del WT WT WT WT WT MV4-11 M5 MLL-AF4 ITD WT WT WT R248W WT WT THP-1 M5 MLL-AF9 WT WT G12D WT R174fs*3 WT WT OCI-AML3 M4 WT WT WT WT WT WT NPM1c R882C K-562 CML WT WT WT WT WT WT WT WT HL-6 M2 WT WT WT Q61L WT 1182del WT WT CML: chronic myeloid leukemia, ITD: internal tandem duplication, WT: wildtype
8 Table S3: Characteristics of patient samples used for gene expression analysis (n=562). Variable Number of patients No. of patients 562 Median age, years (range) 58 (18-85) Female sex, no. (%) 282 (5.2) Bone marrow blasts, %, median(range) 8 (1-1) de novo AML 437 (83.1) taml 27 (5.1) MDS 14 (2.7) saml 48 (9.1) FAB M, no. (%) 22 (3.9) M1, no. (%) 113 (2.1) M2, no. (%) 164 (29.2) M3, no. (%) 15 (2.7) M3v, no. (%) 12 (2.1) M4, no. (%) 121 (21.5) M5, no. (%) 66 (11.7) M6, no. (%) 22 (3.9) M7, no. (%) 3 (.1) NA, no. (%) 24 (4.3) Cytogenetic CN-AML (total), no. (%) 199 (35.4) Complex Cytogenetics, no. (%) 74 (13.2) inv16/t(16;16), no. (%) 38 (6.7) t(8;21), no. (%) 3 (5.3) t(15;17), no. (%) 24 (4.2) (v;11)(v;q23), no. (%) 19 (3.3) t(9;11), no. (%) 19 (3.3) del(7q)/monosomy 7, no. (%) 16 (2.8) inv3/t(3/3), no. (%) 15 (2.6) del(5q)/monosomie 5, no. (%) 6 (1.1) t(6;9), no. (%) 4 (.7)
9 del(9q) soley 7 (1.2) Isolated trisomy 8 14 (2.5) Isolated trisomy 13 9 (1.6) Isolated trisomy 11 7 (1.2) Various (non-complex), no. (%) 51 (9.1) NA, no. (%) 3 (5.3) ELN classification Favorable, no. (%) 141 (25.1) Intermediate-I, no. (%) 111 (19.8) Intermediate-II, no. (%) 17 (19) Adverse, no. (%) 134 (23.8) NA 69 (12.3) CN: cytogentically normal, del: deletion, inv: inversion, MDS: myelodysplastic syndrome, NA: not analyzed, No: number, saml: secondary AML, taml: therapy-related AML, t: translocation Table S4: Frequencies of specific codon mutations in cytogenetically normal (CN) AML (n=125) and MLL rearranged AML patients (n=38) NRAS KRAS codon 12 codon 13 codon 61 others codon 12 codon 13 codon 61 others CN- AML MLL-t AML 43.6% 2.5% 2.5% 15.4% 36.4% 27.3% 9.% 27.3% 7.% 2.% - 1.% 31.6% 26.3% 31.6% 1.5%
10 Supplemental figure S1 untreated BEZ-235 MK-226 Rapamycin isotype PS6rp 62.5 nm 25 nm 1 nm untreated treated 25 nm 1 nm 1 nm counts 1 nm 4 nm 1, nm P-S6rp Supplemental figure S1: Pathway inhibition in patient-derived xenografted cells: Patient-derived xenograft cells were treated with BEZ- 235, MK-226 and Rapamycin, respectively. Phosphorylation of ribosomal protein S6 (S6rp) was measured by FACS staining after three hours. The isotype control is shown in grey, untreated cells are shown in black, treated cells are shown in blue.
11 Supplemental figure S2 A B spontaneous apoptosis [%] spontaneous apoptosis [%] BEZ-235 resistent MLL WT p=.78 BEZ-235 sensitiv p=.64 MLL-t / MLL-PTD C log2 intensities Ki67 expression MLL wt MLL mu Supplemental figure S2: Spontaneous apoptosis and Ki67 expression. Human AML blast cells were freshly isolated and cultured for hours with or without inhibitors. The spontaneous apoptosis of untreated cells was measured by FACS analysis and compared between BEZ-235 sensitive and non-sensitive (A) and between MLL wildtype and MLL mutated samples (B). There was no significant difference. By using microarray analysis Ki67 expression levels were determined and compared between MLL wildtype and MLL rearranged samples (C).
12 Supplemental figure S3 untreated isotype PS6rp BEZ-235 untreated treated counts 125 nm 25 nm 1 nm P-S6rp Supplemental figure S3: Pathway inhibition in murine MLL-ENL immortalized (MLL-ENL ICs) bone marrow progenitor cells Cells were treated with BEZ-235. Phosphorylation of ribosomal protein S6 (S6rp) was measured by FACS staining after three hours. The isotype control is shown in grey, untreated cells are shown in black, treated cells are shown in blue.
13 Supplemental figure S4 A MOLM-13 BEZ-235 MK226 Rapamycin ps6k T389 HL-6 PC -,6, S6K BEZ-235 MK226 Rapamycin PC -,6, ps6k T389 S6K pakt S473 pakt S473 AKT AKT perk T22/Y24 perk T22/Y24 ERK ERK Actin Actin B P-AKT/AKT ratio 1.2 * MDA-MB-453 THP-1 MOLM-13 MV4-11 HL-6 K-562 OCI-AML3.2 Supplemental figure S4: Inhibition of PI3K/AKT/mTOR signaling in cell lines. The human myeloid cell lines were cultured for six hours with or without BEZ-235, MK-226, and Rapamycin. As a positive control (PC) lysates from the human breast cancer cell line MDA-MB-453 were used. Activation of signaling pathways was analyzed by western blot (A) and quantification of P-AKT/AKT ratios was done using Image J software (B; * p<.5). Shown are the mean values +/- standard error of at least three quantifications.
14 Supplemental figure S5 A -ps6k -S6K -pakt -AKT 6 h 24 h -ps6k -S6K -pakt -AKT -Aktin -Aktin BEZ-235 [μm] MK-226 [μm] Rapamycin [μm], ,1,1-1 1, ,1,1-1 1 B specific apoptosis [%] BEZ-235 [μm] MK-226 [μm] Rapamycin [μm] viable cells [%] BEZ-235 [μm] MK-226 [μm] Rapamycin [μm] * ** Supplemental figure S5: MK-226 and Rapamycin inhibit PI3K/AKT/mTOR signaling but do not induce apoptosis HL-6 cells were cultured for 6 or 24 hours (h) with or without inhibitors. Activation of signaling pathways was analyzed by Western Blot (A). Similar concentrations were used to culture cells in the absence or presence of those inhibitors. Apoptosis was determined after 72 hours by FACS analysis and is depicted as specific apoptosis in comparison to untreated controls. Shown are mean values +/- standard error of at least three independent experiments. Cells were seeded in the absence or presence of inhibitors and after 72 hours viable cells were counted by Trypan Blue exclusion. Results show the relative cell number compared to untreated controls and represent mean +/- standard error of at least three independent experiments (B). *p<.5; **p<.1
15 Supplemental figure S6 A dpi AC B Vehicle; n=2 BEZ-235; n=2 AC22; n=2 CD45-APC C Body weight [g] GFP 26 Vehicle dpi Body weight [g] BEZ dpi Body weightt [g] AC dpi Supplemental figure S6: Tumor load and body weight during the treatment with BEZ-235 and AC22. Tumor progression of AC22-treated mice. Shown are imaging files of one representative animal of this group at different time points (A). At day 15 two animals out of each group were sacrificed and their tumor load in the bone marrow was determined by measuring human CD45 positive cells in total mouse bone marrow. Shown are the FACS blots of all six animals (B). The body weight of all mice was determined during the treatment period of 21 days (C).
16 Supplemental figure S7 A log2 Intensity EVI1 expression *** *** *** B log2 intensities EVI1 PTEN ns 1st 2nd 3d 4th quartiles CN-AML inv(3) t(3;3) t(8;21) t(15;17) inv(16) del(5q) t(9;11) t(v;11)(v;q23) t(16;16) C EVI1 PTEN MOLM-13 MV4-11 THP-1 MM6 # # MOLM-13 MV4-11 THP-1 MM6 HL-6 OCI-AML3 U937 K562 # HL-6 OCI-AML3 U937 K562 K562 EV K562 MLL-AF9 K562 EV K562 MLL-AF9 HEL HEL Delta Ct-value Delta Ct-value Supplemental figure S7: EVI1 and PTEN expression in in AML patient samples and cell lines. EVI1 expression was analyzed in 35 AML patient samples and compared between different cytogenetic subgroups. MLL-rearranged patient samples are indicated in blue (A). EVI1 expression quartiles were correlated with PTEN expression in 38 MLLrearranged patient samples (B). EVI1 and PTEN expression levels were determined via qpcr in 11 human cell lines. Cell lines expressing an MLL-rearrangement are indicated in blue. Shown are mean delta Ct-values from three independent experiments. +/- standard errors. Gapdh was used as house-keeping gene (C) EV: empty vector, #: not detectable, ns: not significant, ***p<.1.
17 Supplemental figure S8 CN-AML MLL FLT3 NRAS KRAS CBL 37% 19% 9% 5% Supplemental figure S8: Mutational analysis in AML patient samples. 125 cytogenetically normal (CN)-AML patient samples were analyzed for mutations in FLT3, RAS and CBL. Mutations are indicated in red and frequency rates are shown for each gene.
18 Figure for reviewer only specific apoptosis [%] Daunorubicine [.1 μm] n=17 ns; p=.4 48 h 72 h incubation time specific apoptosis [%] BEZ-235 [4 μm] n=16 ns; p=.8 48 h 72 h incubation time Comparison between primary patient samples treated for 48 hours and 72 hours, respectively. Freshly isolated blast cells from AML samples were cultured in the presence of Daunorubicine or BEZ-235 in duplicates. Apoptosis was determined by FACS analysis after 48 hours (white bars) - 72 hours (green bars) and is depicted as specific apoptosis in comparison to untreated controls. The average of two values is shown. There were no significant differences between 48 hours and 72 hours. ns: not significant
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Patel JP, Gönen M, Figueroa ME, et al. Prognostic relevance
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Patel JP, Gönen M, Figueroa ME, et al. Prognostic relevance
Supplementary Materials for
 www.sciencetranslationalmedicine.org/cgi/content/full/8/339/339ra69/dc1 Supplementary Materials for The caspase-8 inhibitor emricasan combines with the SMAC mimetic birinapant to induce necroptosis and
www.sciencetranslationalmedicine.org/cgi/content/full/8/339/339ra69/dc1 Supplementary Materials for The caspase-8 inhibitor emricasan combines with the SMAC mimetic birinapant to induce necroptosis and
Molecular Markers in Acute Leukemia. Dr Muhd Zanapiah Zakaria Hospital Ampang
 Molecular Markers in Acute Leukemia Dr Muhd Zanapiah Zakaria Hospital Ampang Molecular Markers Useful at diagnosis Classify groups and prognosis Development of more specific therapies Application of risk-adjusted
Molecular Markers in Acute Leukemia Dr Muhd Zanapiah Zakaria Hospital Ampang Molecular Markers Useful at diagnosis Classify groups and prognosis Development of more specific therapies Application of risk-adjusted
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung
 Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 MSI2 interactors are associated with the riboproteome and are functionally relevant. (a) Coomassie blue staining of FLAG-MSI2 immunoprecipitated complexes. (b) GO analysis of MSI2-interacting
Supplementary Figure 1 MSI2 interactors are associated with the riboproteome and are functionally relevant. (a) Coomassie blue staining of FLAG-MSI2 immunoprecipitated complexes. (b) GO analysis of MSI2-interacting
Nature Genetics: doi: /ng Supplementary Figure 1. HOX fusions enhance self-renewal capacity.
 Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
All patients with FLT3 mutant AML should receive midostaurin-based induction therapy. Not so fast!
 All patients with FLT3 mutant AML should receive midostaurin-based induction therapy Not so fast! Harry P. Erba, M.D., Ph.D. Professor, Internal Medicine Director, Hematologic Malignancy Program University
All patients with FLT3 mutant AML should receive midostaurin-based induction therapy Not so fast! Harry P. Erba, M.D., Ph.D. Professor, Internal Medicine Director, Hematologic Malignancy Program University
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Test Name Results Units Bio. Ref. Interval. Positive
 LL - LL-ROHINI (NATIONAL REFERENCE 135091534 Age 36 Years Gender Female 1/9/2017 120000AM 1/9/2017 105316AM 2/9/2017 104147AM Ref By Final LEUKEMIA GENETIC ROFILE ANY SIX MARKERS, CR QUALITATIVE AML ETO
LL - LL-ROHINI (NATIONAL REFERENCE 135091534 Age 36 Years Gender Female 1/9/2017 120000AM 1/9/2017 105316AM 2/9/2017 104147AM Ref By Final LEUKEMIA GENETIC ROFILE ANY SIX MARKERS, CR QUALITATIVE AML ETO
Supplemental Material. The new provisional WHO entity RUNX1 mutated AML shows specific genetics without prognostic influence of dysplasia
 Supplemental Material The new provisional WHO entity RUNX1 mutated AML shows specific genetics without prognostic influence of dysplasia Torsten Haferlach, 1 Anna Stengel, 1 Sandra Eckstein, 1 Karolína
Supplemental Material The new provisional WHO entity RUNX1 mutated AML shows specific genetics without prognostic influence of dysplasia Torsten Haferlach, 1 Anna Stengel, 1 Sandra Eckstein, 1 Karolína
MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands)
 Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Nature Immunology: doi: /ni Supplementary Figure 1. Huwe1 has high expression in HSCs and is necessary for quiescence.
 Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Electron micrograph of phosphotungstanic acid-stained exosomes derived from murine
 1 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Physical properties of murine DC-derived exosomes. a, Electron micrograph of phosphotungstanic acid-stained exosomes derived from
1 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Physical properties of murine DC-derived exosomes. a, Electron micrograph of phosphotungstanic acid-stained exosomes derived from
Supplemental Figure S1A Notch1
 Supplemental Figure S1A Notch1 erage) epth of Cove ormalized De Log1(No Notch exons Figure S1: A) Relative coverage of Notch1 and Notch 2 exons in HCC2218, HCC1187, MB157, MDA-MB157 cell lines. Blue color
Supplemental Figure S1A Notch1 erage) epth of Cove ormalized De Log1(No Notch exons Figure S1: A) Relative coverage of Notch1 and Notch 2 exons in HCC2218, HCC1187, MB157, MDA-MB157 cell lines. Blue color
Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3
 Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11095 Supplementary Table 1. Summary of the binding between Angptls and various Igdomain containing receptors as determined by flow cytometry analysis. The results were summarized from
doi:10.1038/nature11095 Supplementary Table 1. Summary of the binding between Angptls and various Igdomain containing receptors as determined by flow cytometry analysis. The results were summarized from
HEMATOLOGIC MALIGNANCIES BIOLOGY
 HEMATOLOGIC MALIGNANCIES BIOLOGY Failure of terminal differentiation Failure of differentiated cells to undergo apoptosis Failure to control growth Neoplastic stem cell FAILURE OF TERMINAL DIFFERENTIATION
HEMATOLOGIC MALIGNANCIES BIOLOGY Failure of terminal differentiation Failure of differentiated cells to undergo apoptosis Failure to control growth Neoplastic stem cell FAILURE OF TERMINAL DIFFERENTIATION
New drugs in Acute Leukemia. Cristina Papayannidis, MD, PhD University of Bologna
 New drugs in Acute Leukemia Cristina Papayannidis, MD, PhD University of Bologna Challenges to targeted therapy in AML Multiple subtypes based upon mutations/cytogenetic aberrations No known uniform genomic
New drugs in Acute Leukemia Cristina Papayannidis, MD, PhD University of Bologna Challenges to targeted therapy in AML Multiple subtypes based upon mutations/cytogenetic aberrations No known uniform genomic
Supplementary Figure 1
 A B D Relative TAp73 mrna p73 Supplementary Figure 1 25 2 15 1 5 p63 _-tub. MDA-468 HCC1143 HCC38 SUM149 MDA-468 HCC1143 HCC38 SUM149 HCC-1937 MDA-MB-468 ΔNp63_ TAp73_ TAp73β E C Relative ΔNp63 mrna TAp73
A B D Relative TAp73 mrna p73 Supplementary Figure 1 25 2 15 1 5 p63 _-tub. MDA-468 HCC1143 HCC38 SUM149 MDA-468 HCC1143 HCC38 SUM149 HCC-1937 MDA-MB-468 ΔNp63_ TAp73_ TAp73β E C Relative ΔNp63 mrna TAp73
Abstract. Introduction
 Activating FLT3 Mutants Show Distinct Gain-of-Function Phenotypes In Vitro and a Characteristic Signaling Pathway Profile Associated with Prognosis in Acute Myeloid Leukemia Hanna Janke 1,2 *, Friederike
Activating FLT3 Mutants Show Distinct Gain-of-Function Phenotypes In Vitro and a Characteristic Signaling Pathway Profile Associated with Prognosis in Acute Myeloid Leukemia Hanna Janke 1,2 *, Friederike
Supplementary Figure 1: Normal vs. Tumor expression of PIM kinases mrna across 17 tissue types
 Supplementary Figure 1: Normal vs. Tumor expression of PIM kinases mrna across 17 tissue types Supplementary Figure 1 Legend: PIM1, PIM2 and PIM3 mrna expression Expression levels in cancer tissues derived
Supplementary Figure 1: Normal vs. Tumor expression of PIM kinases mrna across 17 tissue types Supplementary Figure 1 Legend: PIM1, PIM2 and PIM3 mrna expression Expression levels in cancer tissues derived
a b G75 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server.
 a b G75 2 2 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server. a. Overlay of top 10 models generated by I-TASSER illustrates the potential effect of 7 amino acid insertion
a b G75 2 2 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server. a. Overlay of top 10 models generated by I-TASSER illustrates the potential effect of 7 amino acid insertion
York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
 MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
MATERIALS AND METHODS Study population Blood samples were obtained from 15 patients with AS fulfilling the modified New York criteria, 6 RA patients and 10 age- and gender-matched healthy controls (HCs).
Supplementary Figure 1. Characterization of basophils after reconstitution of SCID mice
 Supplementary figure legends Supplementary Figure 1. Characterization of after reconstitution of SCID mice with CD4 + CD62L + T cells. (A-C) SCID mice (n = 6 / group) were reconstituted with 2 x 1 6 CD4
Supplementary figure legends Supplementary Figure 1. Characterization of after reconstitution of SCID mice with CD4 + CD62L + T cells. (A-C) SCID mice (n = 6 / group) were reconstituted with 2 x 1 6 CD4
Cost-Effective Strategies in the Workup of Hematologic Neoplasm. Karl S. Theil, Claudiu V. Cotta Cleveland Clinic
 Cost-Effective Strategies in the Workup of Hematologic Neoplasm Karl S. Theil, Claudiu V. Cotta Cleveland Clinic In the past 12 months, we have not had a significant financial interest or other relationship
Cost-Effective Strategies in the Workup of Hematologic Neoplasm Karl S. Theil, Claudiu V. Cotta Cleveland Clinic In the past 12 months, we have not had a significant financial interest or other relationship
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Schlenk RF, Döhner K, Krauter J, et al. Mutations and treatment
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Schlenk RF, Döhner K, Krauter J, et al. Mutations and treatment
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 EGFR inhibition activates signaling pathways (a-b) EGFR inhibition activates signaling pathways (a) U251EGFR cells were treated with erlotinib (1µM) for the indicated times followed
Supplementary Figure 1 EGFR inhibition activates signaling pathways (a-b) EGFR inhibition activates signaling pathways (a) U251EGFR cells were treated with erlotinib (1µM) for the indicated times followed
Nature Medicine: doi: /nm.4078
 Supplementary Figure 1. Cetuximab induces ER stress response in DiFi cells. (a) Scheme of SILAC proteome. (b) MS-base read out of SILAC experiment. The histogram of log 2 -transformed normalized H/L ratios
Supplementary Figure 1. Cetuximab induces ER stress response in DiFi cells. (a) Scheme of SILAC proteome. (b) MS-base read out of SILAC experiment. The histogram of log 2 -transformed normalized H/L ratios
Control shrna#9 shrna#12. shrna#12 CD14-PE CD14-PE
 a Control shrna#9 shrna#12 c Control shrna#9 shrna#12 e Control shrna#9 shrna#12 h 14 12 CFU-E BFU-E GEMM GM b Colony number 7 6 5 4 3 2 1 6 pm A pa pc CFU-E BFU-E GEMM GM pu pgm A p pg B d f CD11b-APC
a Control shrna#9 shrna#12 c Control shrna#9 shrna#12 e Control shrna#9 shrna#12 h 14 12 CFU-E BFU-E GEMM GM b Colony number 7 6 5 4 3 2 1 6 pm A pa pc CFU-E BFU-E GEMM GM pu pgm A p pg B d f CD11b-APC
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
Reporting cytogenetics Can it make sense? Daniel Weisdorf MD University of Minnesota
 Reporting cytogenetics Can it make sense? Daniel Weisdorf MD University of Minnesota Reporting cytogenetics What is it? Terminology Clinical value What details are important Diagnostic Tools for Leukemia
Reporting cytogenetics Can it make sense? Daniel Weisdorf MD University of Minnesota Reporting cytogenetics What is it? Terminology Clinical value What details are important Diagnostic Tools for Leukemia
Figure S1. Generation of inducible PTEN deficient mice and the BMMCs (A) B6.129 Pten loxp/loxp mice were mated with B6.
 Figure S1. Generation of inducible PTEN deficient mice and the BMMCs (A) B6.129 Pten loxp/loxp mice were mated with B6.129-Gt(ROSA)26Sor tm1(cre/ert2)tyj /J mice. To induce deletion of the Pten locus,
Figure S1. Generation of inducible PTEN deficient mice and the BMMCs (A) B6.129 Pten loxp/loxp mice were mated with B6.129-Gt(ROSA)26Sor tm1(cre/ert2)tyj /J mice. To induce deletion of the Pten locus,
Supplementary Figure 1. Basal level EGFR across a panel of ESCC lines. Immunoblots demonstrate the expression of phosphorylated and total EGFR as
 Supplementary Figure 1. Basal level EGFR across a panel of ESCC lines. Immunoblots demonstrate the expression of phosphorylated and total EGFR as well as their downstream effectors across a panel of ESCC
Supplementary Figure 1. Basal level EGFR across a panel of ESCC lines. Immunoblots demonstrate the expression of phosphorylated and total EGFR as well as their downstream effectors across a panel of ESCC
Examining Genetics and Genomics of Acute Myeloid Leukemia in 2017
 Examining Genetics and Genomics of Acute Myeloid Leukemia in 2017 Elli Papaemmanuil, PhD Memorial Sloan Kettering Cancer Center New York, New York, United States Today s Talk Cancer genome introduction
Examining Genetics and Genomics of Acute Myeloid Leukemia in 2017 Elli Papaemmanuil, PhD Memorial Sloan Kettering Cancer Center New York, New York, United States Today s Talk Cancer genome introduction
Corporate Medical Policy. Policy Effective February 23, 2018
 Corporate Medical Policy Genetic Testing for FLT3, NPM1 and CEBPA Mutations in Acute File Name: Origination: Last CAP Review: Next CAP Review: Last Review: genetic_testing_for_flt3_npm1_and_cebpa_mutations_in_acute_myeloid_leukemia
Corporate Medical Policy Genetic Testing for FLT3, NPM1 and CEBPA Mutations in Acute File Name: Origination: Last CAP Review: Next CAP Review: Last Review: genetic_testing_for_flt3_npm1_and_cebpa_mutations_in_acute_myeloid_leukemia
General Laboratory methods Plasma analysis: Gene Expression Analysis: Immunoblot analysis: Immunohistochemistry:
 General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
Impact of Biomarkers in the Management of Patients with Acute Myeloid Leukemia
 Impact of Biomarkers in the Management of Patients with Acute Myeloid Leukemia Hartmut Döhner Medical Director, Department of Internal Medicine III Director, Comprehensive Cancer Center Ulm Ulm University,
Impact of Biomarkers in the Management of Patients with Acute Myeloid Leukemia Hartmut Döhner Medical Director, Department of Internal Medicine III Director, Comprehensive Cancer Center Ulm Ulm University,
Objectives. Morphology and IHC. Flow and Cyto FISH. Testing for Heme Malignancies 3/20/2013
 Molecular Markers in Hematologic Malignancy: Ways to locate the needle in the haystack. Objectives Review the types of testing for hematologic malignancies Understand rationale for molecular testing Marcie
Molecular Markers in Hematologic Malignancy: Ways to locate the needle in the haystack. Objectives Review the types of testing for hematologic malignancies Understand rationale for molecular testing Marcie
Next Generation Sequencing in Haematological Malignancy: A European Perspective. Wolfgang Kern, Munich Leukemia Laboratory
 Next Generation Sequencing in Haematological Malignancy: A European Perspective Wolfgang Kern, Munich Leukemia Laboratory Diagnostic Methods Cytomorphology Cytogenetics Immunophenotype Histology FISH Molecular
Next Generation Sequencing in Haematological Malignancy: A European Perspective Wolfgang Kern, Munich Leukemia Laboratory Diagnostic Methods Cytomorphology Cytogenetics Immunophenotype Histology FISH Molecular
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis
 Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
Supplemental Figure S1. RANK expression on human lung cancer cells.
 Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
mtor Inhibition Specifically Sensitizes Colorectal Cancers with KRAS or BRAF Mutations to BCL-2/BCL-
 Supplementary Material for mtor Inhibition Specifically Sensitizes Colorectal Cancers with KRAS or BRAF Mutations to BCL-2/BCL- XL Inhibition by Suppressing MCL-1 Anthony C. Faber 1,2 *, Erin M. Coffee
Supplementary Material for mtor Inhibition Specifically Sensitizes Colorectal Cancers with KRAS or BRAF Mutations to BCL-2/BCL- XL Inhibition by Suppressing MCL-1 Anthony C. Faber 1,2 *, Erin M. Coffee
Test Name Results Units Bio. Ref. Interval. Positive
 LL - LL-ROHINI (NATIONAL REFERENCE 135091533 Age 28 Years Gender Male 1/9/2017 120000AM 1/9/2017 105415AM 4/9/2017 23858M Ref By Final LEUKEMIA DIAGNOSTIC COMREHENSIVE ROFILE, ANY 6 MARKERS t (1;19) (q23
LL - LL-ROHINI (NATIONAL REFERENCE 135091533 Age 28 Years Gender Male 1/9/2017 120000AM 1/9/2017 105415AM 4/9/2017 23858M Ref By Final LEUKEMIA DIAGNOSTIC COMREHENSIVE ROFILE, ANY 6 MARKERS t (1;19) (q23
Sunitinib, an orally available receptor tyrosine kinase inhibitor, induces monocytic
 Sunitinib, an orally available receptor tyrosine kinase inhibitor, induces monocytic differentiation of acute myeogenouse leukemia cells that is enhanced by 1,25-dihydroxyviatmin D 3. To the Editor: Sunitinib,
Sunitinib, an orally available receptor tyrosine kinase inhibitor, induces monocytic differentiation of acute myeogenouse leukemia cells that is enhanced by 1,25-dihydroxyviatmin D 3. To the Editor: Sunitinib,
AP VP DLP H&E. p-akt DLP
 A B AP VP DLP H&E AP AP VP DLP p-akt wild-type prostate PTEN-null prostate Supplementary Fig. 1. Targeted deletion of PTEN in prostate epithelium resulted in HG-PIN in all three lobes. (A) The anatomy
A B AP VP DLP H&E AP AP VP DLP p-akt wild-type prostate PTEN-null prostate Supplementary Fig. 1. Targeted deletion of PTEN in prostate epithelium resulted in HG-PIN in all three lobes. (A) The anatomy
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
Supplementary Figure 1. BMS enhances human T cell activation in vitro in a
 Supplementary Figure 1. BMS98662 enhances human T cell activation in vitro in a concentration-dependent manner. Jurkat T cells were activated with anti-cd3 and anti-cd28 antibody in the presence of titrated
Supplementary Figure 1. BMS98662 enhances human T cell activation in vitro in a concentration-dependent manner. Jurkat T cells were activated with anti-cd3 and anti-cd28 antibody in the presence of titrated
The Past, Present, and Future of Acute Myeloid Leukemia
 The Past, Present, and Future of Acute Myeloid Leukemia Carter T. Davis, MD Hematology-Oncology Fellow Duke University Health System September 10, 2016 Overview Overview of Acute Myeloid Leukemia Review
The Past, Present, and Future of Acute Myeloid Leukemia Carter T. Davis, MD Hematology-Oncology Fellow Duke University Health System September 10, 2016 Overview Overview of Acute Myeloid Leukemia Review
Supplementary Information
 Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
GENETIC TESTING FOR FLT3, NPM1 AND CEBPA VARIANTS IN CYTOGENETICALLY NORMAL ACUTE MYELOID LEUKEMIA
 CYTOGENETICALLY NORMAL ACUTE MYELOID LEUKEMIA Non-Discrimination Statement and Multi-Language Interpreter Services information are located at the end of this document. Coverage for services, procedures,
CYTOGENETICALLY NORMAL ACUTE MYELOID LEUKEMIA Non-Discrimination Statement and Multi-Language Interpreter Services information are located at the end of this document. Coverage for services, procedures,
Supplemental Figure 1
 Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplementary Figure 1
 Supplementary Figure 1 6 HE-50 HE-116 E-1 HE-108 Supplementary Figure 1. Targeted drug response curves of endometrial cancer cells. Endometrial cancer cell lines were incubated with serial dilutions of
Supplementary Figure 1 6 HE-50 HE-116 E-1 HE-108 Supplementary Figure 1. Targeted drug response curves of endometrial cancer cells. Endometrial cancer cell lines were incubated with serial dilutions of
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
well for 2 h at rt. Each dot represents an individual mouse and bar is the mean ±
 Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Concomitant WT1 mutations predicted poor prognosis in CEBPA double-mutated acute myeloid leukemia
 Concomitant WT1 mutations predicted poor prognosis in CEBPA double-mutated acute myeloid leukemia Feng-Ming Tien, Hsin-An Hou, Jih-Luh Tang, Yuan-Yeh Kuo, Chien-Yuan Chen, Cheng-Hong Tsai, Ming Yao, Chi-Cheng
Concomitant WT1 mutations predicted poor prognosis in CEBPA double-mutated acute myeloid leukemia Feng-Ming Tien, Hsin-An Hou, Jih-Luh Tang, Yuan-Yeh Kuo, Chien-Yuan Chen, Cheng-Hong Tsai, Ming Yao, Chi-Cheng
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Molecular Markers. Marcie Riches, MD, MS Associate Professor University of North Carolina Scientific Director, Infection and Immune Reconstitution WC
 Molecular Markers Marcie Riches, MD, MS Associate Professor University of North Carolina Scientific Director, Infection and Immune Reconstitution WC Overview Testing methods Rationale for molecular testing
Molecular Markers Marcie Riches, MD, MS Associate Professor University of North Carolina Scientific Director, Infection and Immune Reconstitution WC Overview Testing methods Rationale for molecular testing
Supplementary Figure S1. Flow cytometric analysis of the expression of Thy1 in NH cells. Flow cytometric analysis of the expression of T1/ST2 and
 Supplementary Figure S1. Flow cytometric analysis of the expression of Thy1 in NH cells. Flow cytometric analysis of the expression of T1/ST2 and Thy1 in NH cells derived from the lungs of naïve mice.
Supplementary Figure S1. Flow cytometric analysis of the expression of Thy1 in NH cells. Flow cytometric analysis of the expression of T1/ST2 and Thy1 in NH cells derived from the lungs of naïve mice.
Supplementary Table 1. Characterization of HNSCC PDX models established at MSKCC
 Supplementary Table 1. Characterization of HNSCC PDX models established at MSKCC Supplementary Table 2. Drug content and loading efficiency estimated with F-NMR and UV- Vis Supplementary Table 3. Complete
Supplementary Table 1. Characterization of HNSCC PDX models established at MSKCC Supplementary Table 2. Drug content and loading efficiency estimated with F-NMR and UV- Vis Supplementary Table 3. Complete
B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small cell lung cancer
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Experimental Methods Cell culture B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Experimental Methods Cell culture B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
Acute myeloid leukemia: prognosis and treatment. Dimitri A. Breems, MD, PhD Internist-Hematoloog Ziekenhuis Netwerk Antwerpen Campus Stuivenberg
 Acute myeloid leukemia: prognosis and treatment Dimitri A. Breems, MD, PhD Internist-Hematoloog Ziekenhuis Netwerk Antwerpen Campus Stuivenberg Patient Female, 39 years History: hypothyroidism Present:
Acute myeloid leukemia: prognosis and treatment Dimitri A. Breems, MD, PhD Internist-Hematoloog Ziekenhuis Netwerk Antwerpen Campus Stuivenberg Patient Female, 39 years History: hypothyroidism Present:
Supplementary Information Supplementary Fig. 1. Elevated Usp9x in melanoma and NRAS mutant melanoma cells are dependent on NRAS for 3D growth.
 Supplementary Information Supplementary Fig. 1. Elevated Usp9x in melanoma and NRAS mutant melanoma cells are dependent on NRAS for 3D growth. a. Immunoblot for Usp9x protein in NRAS mutant melanoma cells
Supplementary Information Supplementary Fig. 1. Elevated Usp9x in melanoma and NRAS mutant melanoma cells are dependent on NRAS for 3D growth. a. Immunoblot for Usp9x protein in NRAS mutant melanoma cells
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
SUPPLEMENTARY FIG. S3. Kaplan Meier survival analysis followed with log-rank test of de novo acute myeloid leukemia patients selected by age <60, IA
 Supplementary Data Supplementary Appendix A: Treatment Protocols Treatment protocols of 123 cases patients were treated with the protocols as follows: 110 patients received standard DA (daunorubicin 45
Supplementary Data Supplementary Appendix A: Treatment Protocols Treatment protocols of 123 cases patients were treated with the protocols as follows: 110 patients received standard DA (daunorubicin 45
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
IV Simposio International Sao Paulo Nov Hematologic malignant diseases molecular information, present and future
 IV Simposio International Sao Paulo Nov 7 2012 Hematologic malignant diseases molecular information, present and future Dr. rer. nat. Alexander Kohlmann, MLL Munich Leukemia Laboratory Spectrum of Methods
IV Simposio International Sao Paulo Nov 7 2012 Hematologic malignant diseases molecular information, present and future Dr. rer. nat. Alexander Kohlmann, MLL Munich Leukemia Laboratory Spectrum of Methods
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/430/ra57/dc1 Supplementary Materials for The 4E-BP eif4e axis promotes rapamycinsensitive growth and proliferation in lymphocytes Lomon So, Jongdae Lee, Miguel
www.sciencesignaling.org/cgi/content/full/9/430/ra57/dc1 Supplementary Materials for The 4E-BP eif4e axis promotes rapamycinsensitive growth and proliferation in lymphocytes Lomon So, Jongdae Lee, Miguel
5K ALDEFLUOR-positive/ CXCR1-negative. 5K ALDEFLUOR-positive/ CXCR1-positive BAAA BAAA CXCR1-APC BAAA BAAA CXCR1-APC
 A +DEAB -DEAB K ALDEFLUOR-positive/ CXCR-negative BAAA BAAA CXCR-APC B +DEAB -DEAB K ALDEFLUOR-positive/ CXCR-positive BAAA BAAA CXCR-APC C Supplemental Figure. Tumorigenicity of the ALDEFLUOR-positive/CXCR-positive
A +DEAB -DEAB K ALDEFLUOR-positive/ CXCR-negative BAAA BAAA CXCR-APC B +DEAB -DEAB K ALDEFLUOR-positive/ CXCR-positive BAAA BAAA CXCR-APC C Supplemental Figure. Tumorigenicity of the ALDEFLUOR-positive/CXCR-positive
The Role of CD164 in Metastatic Cancer Aaron M. Havens J. Wang, Y-X. Sun, G. Heresi, R.S. Taichman Mentor: Russell Taichman
 The Role of CD164 in Metastatic Cancer Aaron M. Havens J. Wang, Y-X. Sun, G. Heresi, R.S. Taichman Mentor: Russell Taichman The spread of tumors, a process called metastasis, is a dreaded complication
The Role of CD164 in Metastatic Cancer Aaron M. Havens J. Wang, Y-X. Sun, G. Heresi, R.S. Taichman Mentor: Russell Taichman The spread of tumors, a process called metastasis, is a dreaded complication
sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5
 sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
Please Silence Your Cell Phones. Thank You
 Please Silence Your Cell Phones Thank You Utility of NGS and Comprehensive Genomic Profiling in Hematopathology Practice Maria E. Arcila M.D. Memorial Sloan Kettering Cancer Center New York, NY Disclosure
Please Silence Your Cell Phones Thank You Utility of NGS and Comprehensive Genomic Profiling in Hematopathology Practice Maria E. Arcila M.D. Memorial Sloan Kettering Cancer Center New York, NY Disclosure
Supplementary Information. Inhibition of USP10 induces degradation of oncogenic FLT3
 Supplementary Information Inhibition of USP10 induces degradation of oncogenic FLT3 Ellen L. Weisberg 1, *, Nathan Schauer 2 *, Jing Yang 2 *, Ilaria Lamberto 2 *, Laura Doherty 2, Shruti Bhatt 1, Atsushi
Supplementary Information Inhibition of USP10 induces degradation of oncogenic FLT3 Ellen L. Weisberg 1, *, Nathan Schauer 2 *, Jing Yang 2 *, Ilaria Lamberto 2 *, Laura Doherty 2, Shruti Bhatt 1, Atsushi
Supplementary data Supplementary Figure 1 Supplementary Figure 2
 Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Supplementary Figure 1
 Supplementary Figure 1 Constitutive EGFR signaling does not activate canonical EGFR signals (a) U251EGFRInd cells with or without tetracycline exposure (24h, 1µg/ml) were treated with EGF for 15 minutes
Supplementary Figure 1 Constitutive EGFR signaling does not activate canonical EGFR signals (a) U251EGFRInd cells with or without tetracycline exposure (24h, 1µg/ml) were treated with EGF for 15 minutes
Significance of Chromosome Changes in Hematological Disorders and Solid Tumors
 Significance of Chromosome Changes in Hematological Disorders and Solid Tumors Size of Components of Human Genome Size of haploid genome 3.3 X 10 9 DNA basepairs Estimated genetic constitution 30,000
Significance of Chromosome Changes in Hematological Disorders and Solid Tumors Size of Components of Human Genome Size of haploid genome 3.3 X 10 9 DNA basepairs Estimated genetic constitution 30,000
Significance of Chromosome Changes in Hematological Disorders and Solid Tumors
 Significance of Chromosome Changes in Hematological Disorders and Solid Tumors Size of Components of Human Genome Size of haploid genome! Estimated genetic constitution! Size of average chromosome
Significance of Chromosome Changes in Hematological Disorders and Solid Tumors Size of Components of Human Genome Size of haploid genome! Estimated genetic constitution! Size of average chromosome
Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein
 Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein Lei Wang 1, Tian-Peng Zhang 1, Yuan Zhang 2, Hai-Lian
Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein Lei Wang 1, Tian-Peng Zhang 1, Yuan Zhang 2, Hai-Lian
Acute myeloid leukemia. M. Kaźmierczak 2016
 Acute myeloid leukemia M. Kaźmierczak 2016 Acute myeloid leukemia Malignant clonal disorder of immature hematopoietic cells characterized by clonal proliferation of abnormal blast cells and impaired production
Acute myeloid leukemia M. Kaźmierczak 2016 Acute myeloid leukemia Malignant clonal disorder of immature hematopoietic cells characterized by clonal proliferation of abnormal blast cells and impaired production
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Information Titles Journal: Nature Medicine
 Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
(A) Dose response curves of HMLE_shGFP (blue circle), HMLE_shEcad (red square),
 Supplementary Figures and Tables Figure S1. Validation of EMT-selective small molecules (A) Dose response curves of HMLE_shGFP (blue circle), HMLE_shEcad (red square), and HMLE_Twist (black diamond) cells
Supplementary Figures and Tables Figure S1. Validation of EMT-selective small molecules (A) Dose response curves of HMLE_shGFP (blue circle), HMLE_shEcad (red square), and HMLE_Twist (black diamond) cells
The MILE study: Expression microarray analysis for diagnosis of leukaemia. Ken Mills CCRCB Queen s University Belfast
 The MILE study: Expression microarray analysis for diagnosis of leukaemia Ken Mills CCRCB Queen s University Belfast Outline MILE Prephase Stage I Stage II Implementation MDS Diagnosis into Prognosis Leukaemia
The MILE study: Expression microarray analysis for diagnosis of leukaemia Ken Mills CCRCB Queen s University Belfast Outline MILE Prephase Stage I Stage II Implementation MDS Diagnosis into Prognosis Leukaemia
Supplementary Figure 1. Characterization of ALDH-positive cell population in MCF-7 cells. (a) Expression level of stem cell markers in MCF-7 cells or
 Supplementary Figure 1. Characterization of ALDH-positive cell population in MCF-7 cells. (a) Expression level of stem cell markers in MCF-7 cells or ALDH-positive cell population by qpcr. Data represent
Supplementary Figure 1. Characterization of ALDH-positive cell population in MCF-7 cells. (a) Expression level of stem cell markers in MCF-7 cells or ALDH-positive cell population by qpcr. Data represent
Supplementary Figure 1. Supernatants electrophoresis from CD14+ and dendritic cells. Supernatants were resolved by SDS-PAGE and stained with
 Supplementary Figure 1. Supernatants electrophoresis from CD14+ and dendritic cells. Supernatants were resolved by SDS-PAGE and stained with Coomassie brilliant blue. One µg/ml recombinant human (rh) apo-e
Supplementary Figure 1. Supernatants electrophoresis from CD14+ and dendritic cells. Supernatants were resolved by SDS-PAGE and stained with Coomassie brilliant blue. One µg/ml recombinant human (rh) apo-e
Mixed Phenotype Acute Leukemias
 Mixed Phenotype Acute Leukemias CHEN GAO; AMY M. SANDS; JIANLAN SUN NORTH AMERICAN JOURNAL OF MEDICINE AND SCIENCE APR 2012 VOL 5 NO.2 INTRODUCTION Most cases of acute leukemia can be classified based
Mixed Phenotype Acute Leukemias CHEN GAO; AMY M. SANDS; JIANLAN SUN NORTH AMERICAN JOURNAL OF MEDICINE AND SCIENCE APR 2012 VOL 5 NO.2 INTRODUCTION Most cases of acute leukemia can be classified based
Supplementary Figure 1
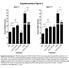 Supplementary Figure 1 60 HRAS G12V 40 * 55 35 Percent BRDU Positive 50 45 40 30 20 NS Percent BRDU Positive 30 25 20 15 10 NS 10 5 0 -dox dox doxkt5720 -doxkt5720 Treatment TSHKT5720 0 -dox dox doxkt5720
Supplementary Figure 1 60 HRAS G12V 40 * 55 35 Percent BRDU Positive 50 45 40 30 20 NS Percent BRDU Positive 30 25 20 15 10 NS 10 5 0 -dox dox doxkt5720 -doxkt5720 Treatment TSHKT5720 0 -dox dox doxkt5720
WHO Classification of Myeloid Neoplasms with Defined Molecular Abnormalities
 WHO Classification of Myeloid Neoplasms with Defined Molecular Abnormalities Robert W. McKenna, M.D. 1/2009 WHO Classification of Myeloid Neoplasms (4th Edition)--2008 Incorporates new information that
WHO Classification of Myeloid Neoplasms with Defined Molecular Abnormalities Robert W. McKenna, M.D. 1/2009 WHO Classification of Myeloid Neoplasms (4th Edition)--2008 Incorporates new information that
BCR-ABL - LSK BCR-ABL + LKS - (%)
 Marker Clone BCR-ABL + LSK (%) BCR-ABL + LKS - (%) BCR-ABL - LSK (%) P value vs. BCR-ABL + LKS - P value vs. BCR-ABL - LSK CD2 RM2-5 12.9 ± 3.6 36.7 ± 6.5 19.3 ± 2.4 0.01 0.10 CD5 53-7.3 13.9 ± 3.2 20.8
Marker Clone BCR-ABL + LSK (%) BCR-ABL + LKS - (%) BCR-ABL - LSK (%) P value vs. BCR-ABL + LKS - P value vs. BCR-ABL - LSK CD2 RM2-5 12.9 ± 3.6 36.7 ± 6.5 19.3 ± 2.4 0.01 0.10 CD5 53-7.3 13.9 ± 3.2 20.8
Pearson r = P (one-tailed) = n = 9
 8F4-Specific Lysis, % 1 UPN1 UPN3 8 UPN7 6 Pearson r =.69 UPN2 UPN5 P (one-tailed) =.192 4 UPN8 n = 9 2 UPN9 UPN4 UPN6 5 1 15 2 25 8 8F4, % Max MFI Supplementary Figure S1. AML samples UPN1-UPN9 show variable
8F4-Specific Lysis, % 1 UPN1 UPN3 8 UPN7 6 Pearson r =.69 UPN2 UPN5 P (one-tailed) =.192 4 UPN8 n = 9 2 UPN9 UPN4 UPN6 5 1 15 2 25 8 8F4, % Max MFI Supplementary Figure S1. AML samples UPN1-UPN9 show variable
Tbk1-TKO! DN cells (%)! 15! 10!
 a! T Cells! TKO! B Cells! TKO! b! CD4! 8.9 85.2 3.4 2.88 CD8! Tbk1-TKO! 1.1 84.8 2.51 2.54 c! DN cells (%)! 4 3 2 1 DP cells (%)! 9 8 7 6 CD4 + SP cells (%)! 5 4 3 2 1 5 TKO! TKO! TKO! TKO! 15 1 5 CD8
a! T Cells! TKO! B Cells! TKO! b! CD4! 8.9 85.2 3.4 2.88 CD8! Tbk1-TKO! 1.1 84.8 2.51 2.54 c! DN cells (%)! 4 3 2 1 DP cells (%)! 9 8 7 6 CD4 + SP cells (%)! 5 4 3 2 1 5 TKO! TKO! TKO! TKO! 15 1 5 CD8
Keywords: Acute Myeloid Leukemia, FLT3-ITD Mutation, FAB Subgroups, Cytogenetic Risk Groups
 Original Articleeee fms Like Tyrosine kinase3- Internal Tandem Duplication (FLT3-ITD) in Acute Myeloid Leukemia, Mutation Frequency and its Relation with Complete Remission, 2007-2008 Emami AH, 1 Shekarriz
Original Articleeee fms Like Tyrosine kinase3- Internal Tandem Duplication (FLT3-ITD) in Acute Myeloid Leukemia, Mutation Frequency and its Relation with Complete Remission, 2007-2008 Emami AH, 1 Shekarriz
X P. Supplementary Figure 1. Nature Medicine: doi: /nm Nilotinib LSK LT-HSC. Cytoplasm. Cytoplasm. Nucleus. Nucleus
 a b c Supplementary Figure 1 c-kit-apc-eflu780 Lin-FITC Flt3-Linc-Kit-APC-eflu780 LSK Sca-1-PE-Cy7 d e f CD48-APC LT-HSC CD150-PerCP-cy5.5 g h i j Cytoplasm RCC1 X Exp 5 mir 126 SPRED1 SPRED1 RAN P SPRED1
a b c Supplementary Figure 1 c-kit-apc-eflu780 Lin-FITC Flt3-Linc-Kit-APC-eflu780 LSK Sca-1-PE-Cy7 d e f CD48-APC LT-HSC CD150-PerCP-cy5.5 g h i j Cytoplasm RCC1 X Exp 5 mir 126 SPRED1 SPRED1 RAN P SPRED1
Signaling changes in the stem cell factor AKT-S6 pathway in diagnostic AML samples are associated with disease relapse
 LETTER TO THE EDITOR Citation: (2011) 1, e3; doi:10.1038/bcj.2010.2 & 2011 Macmillan Publishers Limited All rights reserved 2044-5385/11 www.nature.com/bcj Signaling changes in the stem cell factor AKT-S6
LETTER TO THE EDITOR Citation: (2011) 1, e3; doi:10.1038/bcj.2010.2 & 2011 Macmillan Publishers Limited All rights reserved 2044-5385/11 www.nature.com/bcj Signaling changes in the stem cell factor AKT-S6
Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic
 Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic B cells from WT Nfat2 +/+, TCL1 Nfat2 +/+ and TCL1 Nfat2
Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic B cells from WT Nfat2 +/+, TCL1 Nfat2 +/+ and TCL1 Nfat2
Supplementary Figure 1. Successful excision of genes from WBM lysates and
 Supplementary Information: Supplementary Figure 1. Successful excision of genes from WBM lysates and survival of mice with different genotypes. (a) The proper excision of Pten, p110α, p110α and p110δ was
Supplementary Information: Supplementary Figure 1. Successful excision of genes from WBM lysates and survival of mice with different genotypes. (a) The proper excision of Pten, p110α, p110α and p110δ was
Supporting Information
 Supporting Information van der Windt et al. 10.1073/pnas.1221740110 SI Materials and Methods Mice and Reagents. C57BL/6 and major histocompatibility complex class I-restricted OVA-specific T-cell receptor
Supporting Information van der Windt et al. 10.1073/pnas.1221740110 SI Materials and Methods Mice and Reagents. C57BL/6 and major histocompatibility complex class I-restricted OVA-specific T-cell receptor
Effective Targeting of Quiescent Chronic Myelogenous
 Cancer Cell, Volume 7 Supplemental Information Effective Targeting of Quiescent Chronic Myelogenous Leukemia Stem Cells by Histone Deacetylase Inhibitors in Combination with Imatinib Mesylate Bin Zhang,
Cancer Cell, Volume 7 Supplemental Information Effective Targeting of Quiescent Chronic Myelogenous Leukemia Stem Cells by Histone Deacetylase Inhibitors in Combination with Imatinib Mesylate Bin Zhang,
RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using
 Supplementary Information Materials and Methods RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using Trizol reagent (Invitrogen,Carlsbad, CA) according to the manufacturer's instructions.
Supplementary Information Materials and Methods RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using Trizol reagent (Invitrogen,Carlsbad, CA) according to the manufacturer's instructions.
mtor plays critical roles in pancreatic cancer stem cells through specific and stemness-related functions
 Supplementary Information mtor plays critical roles in pancreatic cancer stem cells through specific and stemness-related functions Shyuichiro Matsubara 1, Qiang Ding 1, Yumi Miyazaki 1, Taisaku Kuwahata
Supplementary Information mtor plays critical roles in pancreatic cancer stem cells through specific and stemness-related functions Shyuichiro Matsubara 1, Qiang Ding 1, Yumi Miyazaki 1, Taisaku Kuwahata
