Supplementary Information
|
|
|
- Joan Dorsey
- 5 years ago
- Views:
Transcription
1 Supplementary Information Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa Serap Erkek, Mizue Hisano, Ching-Yeu Liang, Mark Gill, Rabih Murr, Jürgen Dieker, Dirk Schübeler, Johan van der Vlag, Michael B. Stadler and Antoine H.F.M. Peters Supplementary Figures Supplementary Figure 1: Supplementary Figure 2: Supplementary Figure 3: Supplementary Figure 4: Supplementary Figure 5: Supplementary Figure 6: Supplementary Figure 7: Genome-wide distribution of nucleosome peaks and motif enrichment analysis for nucleosome peaks. Nucleosome occupancy in Saccharomyces cerevisiae and human sperm. Chromatin states of imprinting control regions (ICRs) in sperm and round spermatids. Specificity of the H3.3 antibody and average profiles of canonical and variant histone occupancy along genes in relation to expression status in embryonic stem cells. Histone proteins and modification patterns in round spermatids, sperm and ESCs. Chromatin states of genes representative of the gene clusters described in Fig. 6c and of pluripotency associated genes in sperm and round spermatids. Reproducibility of ChIP experiments in sperm. Supplementary Tables Supplementary Table 1: GO-term analysis for the gene clusters described in Fig. 6c. Supplementary Note 1
2 a b c Top 20 motifs enriched CGCCGCG CGCGGCG GGCCGCG CGCGGCC CGCGCCG GCCCGCG CGGCGCG GCGGCGG CCGCCGC GCCGCGG CCGCGGC CGCGGGC GCGCGGG CCCGCGC CGCGACG CGGCGGC CGTCGCG CCGCGGG GCCGCCG Supplementary Figure 1: Genome-wide distribution of nucleosome peaks and motif enrichment analysis for nucleosome peaks. The mouse genome is classified into promoter (± 1 kb around TSS), exon, intron, repeat and intergenic (non-repeat) regions. (a) Observed and expected fractions of nucleosome peaks by genomic region. Observed refers to experimentally identified nucleosome peaks and expected is calculated assuming uniform distribution of the same number of peaks in the genome. (b) Barplot showing the data from (a) as log2 enrichments (observed/expected) of nucleosome peaks in different genomic regions. (c) Motif abundance versus enrichment plot showing the enrichment of 7-mer motifs in nucleosome peaks compared to background abundance (see Supplementary Methods for the description of background). Top 20 enriched motifs are shown next to the plot. 2
3 a b c Pearson correlation coefficient Pearson correlation coefficient Pearson correlation coefficient A C G T AA AC AG AT CA CC CG CT GA GC GG GT TA TC TG TT A C G T AA AC AG AT CA CC CG CT GA GC GG GT TA TC TG TT e Average DNA methylation (%) Nucleosome enrichment (log2) d Pearson correlation coefficient f Average DNA methylation (%) Nucleosome enrichment (log2) Supplementary Figure 2: Nucleosome occupancy in Saccharomyces cerevisiae and human sperm. (a) and (b) Correlation of single nucleotide frequencies (left) and dinucleotide frequencies normalized for single nucleotide composition (right) with nucleosome occupancy in Saccharomyces cerevisiae 1 in 1kb windows tiling the yeast genome based on (a) in vitro reconstituted nucleosomes and (b) in vivo determined nucleosomal occupancies (YPD medium). (c) and (d) Correlation of single nucleotide frequencies (left) and dinucleotide frequencies normalized for single nucleotide composition (right) with nucleosome occupancy in human sperm in 1kb windows tiling the human genome. (e) and (f) Correlation of human sperm DNA methylation 2 with nucleosome occupancy in human sperm in 1kb windows tiling the human genome. (c) and (e) display human sperm nucleosome data from Brykczynska et al. 3 while (d) and (f) display data from Hammoud et al. 4. 3
4 a b Round spermatids H3K4me3 H3K4me3 H3.1/H3.2 H3.3 Nucleosome CGI CpG Blastocyst Oocyte DNA methylation Supplementary Figure 3: Chromatin states of imprinting control regions (ICRs) in sperm and round spermatids. From top to bottom, images show the DNA methylation status for spermatozoa, oocytes and blastocyst embryos 5, CpG density, CGI localization, nucleosome, histone variant and histone modification states in sperm and histone modification states in round spermatids. Imprinting control regions 5 for paternally imprinted genes are shown with light blue boxes and imprinting control regions for maternally imprinted genes are shown with light pink boxes. (a) H19. (b) Rasgrf1. (c) Dlk1-Meg3. (d) Kcnq1ot1. (e) Nespas-Gnas. (f) Snrpn. (g) Peg10. DNA methylation states refer to percentage of methylation at individual CpGs as determined by shot gun bisulfite sequencing 5. Chromatin images represent read count per base averaged over a moving 300 bps interval. Averaged reads counts were normalized for total read counts across samples. 4
5 c Round spermatids H3K4me3 H3K4me3 H3.1/H3.2 H3.3 Nucleosome CGI CpG Blastocyst Oocyte DNA methylation 5
6 d e Round spermatids H3K4me3 H3K4me3 H3.1/H3.2 H3.3 Nucleosome CGI CpG Blastocyst Oocyte DNA methylation 6
7 f g Blastocyst Oocyte DNA methylation H3K4me3 H3.1/H3.2 H3.3 Nucleosome CGI CpG Round spermatids H3K4me3 Sgce Peg chr6 (Mbp) 7
8 H3.3-HA over H3.2-HA ratio (log2) 2.0 H3.3-HA enrichment (log2) 1.5 H3.2-HA enrichment (log2) b 3 a TSS TES Position around transcript (kb) Expression in embryonic stem cells: Not detected Low Medium High Supplementary Figure 4: Specificity of the H3.3 antibody and average profiles of canonical and variant histone occupancy along genes in relation to expression status in embryonic stem cells. (a) Western blots showing endogenous and exogenously expressed histone variant H3.3 as detected by H3.3 and tag antibodies. 293 cells were transfected with constructs encoding tagged canonical H3.1, H3.2 and variant H3.3 histones. (b) Average profiles of histone variants H3.2-HA and H3.3-HA in embryonic stem cells (ESC)6. Genes were classified according to expression status in ESC7. Transcripts without any aligned reads were classified as not detected. Remaining transcripts were classified on the basis of increasing expression values into three equally sized groups. In the bottom panel, the ratio between H3.3HA over H3.2-HA is shown. 8
9 a H3K4me3 enrichment in sperm (log2) H3K4me3 enrichment in sperm (log2) H3K4me3 enrichment in sperm (log2) enrichment in sperm (log2) b c H3K4me3 enrichment in RS (log2) H3K4me3 enrichment in RS (log2) H3K4me3 enrichment in RS (log2) H3K4me3 relative enrichment in ESC enrichment in sperm (log2) enrichment in sperm (log2) enrichment in RS (log2) enrichment in RS (log2) enrichment in RS (log2) relative enrichment in ESC Clusters of Fig. 6c Supplementary Figure 5: Histone proteins and modification patterns in round spermatids, sperm and ESCs. (a) Scatter plots showing the comparison of the enrichments of H3K4me3 (left) and (right) in round spermatids (RS) and sperm at TSS (± 1kb); (b) in relation to the five gene clusters as described and color coded in Fig. 6c; (c) in comparison to H3K4me3 8 and 7 levels in mouse ESCs. (d) Scatter plots showing the correlation between percentage of CpGs at TSS (± 1kb) and H3.3 over H3.1/H3.2 ratio in RS in relation to relative enrichment of H3K4me3 in RS (top left), in RS (top right), H3K4me3 in sperm (bottom left) and in sperm (bottom right). (e) H3.3 enrichments (left panel) and H3.1/H3.2 enrichments (right panel) were calculated genome-wide for 1kb windows which do not intersect TSS regions (± 3kb) and CGIs and were modeled as a linear combination of explanatory variables (CpG content and histone measurements in RS). The unique contribution of each variable to observed sperm variation is shown in percentage. Combinatorial effects refer to variation which is explained by combinations of variables used in the analysis. 9
10 d H3K4me3 (RS) (RS) H3.3 over H3.1/H3.2 ratio in RS (log2) H3K4me3 () () Relative enrichment Percentage of CpG (±1kb around TSS) e Explained variation (%) CpG% H3.3 H3.1/H3.2 H3K4me3 Round spermatids Combinatorial effects CpG% H3.3 H3.1/H3.2 H3K4me3 Round spermatids Combinatorial effects 10
11 a b Round spermatids H3K4me3 H3K4me3 H3.1/H3.2 H3.3 Nucleosome CGI CpG Blastocyst Oocyte DNA methylation Supplementary Figure 6: Chromatin states of genes representative of the gene clusters described in Fig. 6c and of pluripotency associated genes in sperm and round spermatids. From top to bottom, images show at various loci the DNA methylation status for spermatozoa, oocytes and blastocyst embryos 5, CpG density, CGI localization, nucleosome, histone variant and histone modification states in sperm and histone modifications in round spermatids. (a, b) examples for cluster 1, Sfrs6 and H3f3a. (c, d) examples for cluster 2, Rps14 and Dnajb1. (e, f) examples for cluster 3, Tsen2 and Hint3. (g, h) examples for cluster 4, T (Brachyury) and Gata2. (i, j) examples for cluster 5, Olfr family and Cts6. (k) Pou5f1 (l) Nanog (m) Sox2 (n) Esrrb (o) Klf4 (p) Klf5. DNA methylation states refer to percentage of methylation at individual CpGs as determined by shot gun bisulfite sequencing 5. Chromatin images represent read count per base averaged over a moving 300 bps interval. Averaged reads counts were normalized for total read counts across samples. 11
12 c d Round spermatids H3K4me3 H3K4me3 H3.1/H3.2 H3.3 Nucleosome CGI CpG Blastocyst Oocyte DNA methylation 12
13 e f Round spermatids H3K4me3 H3K4me3 H3.1/H3.2 H3.3 Nucleosome CGI CpG Blastocyst Oocyte DNA methylation 13
14 g h Round spermatids H3K4me3 H3K4me3 H3.1/H3.2 H3.3 Nucleosome CGI CpG Blastocyst Oocyte DNA methylation 14
15 i j Round spermatids H3K4me3 H3K4me3 H3.1/H3.2 H3.3 Nucleosome CGI CpG Blastocyst Oocyte DNA methylation 15
16 k l Round spermatids H3K4me3 H3K4me3 H3.1/H3.2 H3.3 Nucleosome CGI CpG Blastocyst Oocyte DNA methylation 16
17 m n Round spermatids H3K4me3 H3K4me3 H3.1/H3.2 H3.3 Nucleosome CGI CpG Blastocyst Oocyte DNA methylation 17
18 o p Round spermatids H3K4me3 H3K4me3 H3.1/H3.2 H3.3 Nucleosome CGI CpG Blastocyst Oocyte DNA methylation 18
19 a b ± 1kb TSS region Intersecting a peak Not intersecting a peak c d e f Nucleosome H3.3 H3.1/H3.2 H3K4me3 CpG rep1 rep2 rep3 rep1 rep2 rep1 rep2 rep1 rep2 rep1 rep2 Feature density Supplementary Figure 7: Reproducibility of ChIP experiments in sperm. (a - e) Scatter plots show the correlation of the replicates for nucleosome (a), H3.3 (b), H3.1/H3.2 (c) H3K4me3 (d) and (e) ± 1kb around TSS. TSS regions which intersect a peak are shown in red. For each histone variant / modification sample, peaks were identified in a similar way as described for nucleosomes. (f) Heatmap of genes illustrating CpG density, nucleosome (3 replicates), H3.3 (2 replicates), H3.1/H3.2 (2 replicates), H3K4me3 (2 replicates) and (2 replicates) coverage around TSS (± 3kb) in sperm. Feature density shows the scaled read densities from ChIP-seq experiments genes were randomly selected for visualization. 19
20 Supplementary Table 1: GO-term analysis for the gene clusters described in Fig. 6c. Top 50 terms associated with each cluster are shown. Cluster 1 GO.ID Term Annotated Significant Expected classicfisher Over-representation GO: protein modification by small protein conjugation or removal GO: spermatogenesis GO: male gamete generation GO: protein ubiquitination GO: cellular protein catabolic process GO: ubiquitin-dependent protein catabolic process GO: proteolysis involved in cellular protein catabolic process GO: protein modification by small protein conjugation GO: sexual reproduction GO: cellular macromolecule metabolic process GO: modification-dependent protein catabolic process GO: cellular protein metabolic process GO: modification-dependent macromolecule catabolic process GO: protein catabolic process GO: cellular macromolecule catabolic process GO: proteasomal ubiquitin-dependent protein catabolic process GO: gamete generation GO: proteasomal protein catabolic process GO: cellular metabolic process GO: protein polyubiquitination GO: spermatid development GO: protein folding GO: macromolecule catabolic process GO: spermatid differentiation GO: multicellular organism reproduction GO: multicellular organismal reproductive process GO: primary metabolic process GO: macromolecule metabolic process GO: catabolic process GO: ER-associated protein catabolic process GO: protein K63-linked ubiquitination GO: germ cell development GO: mrna transport GO: cellular catabolic process GO: protein metabolic process GO: protein K48-linked ubiquitination
21 GO: nucleic acid transport GO: RNA transport GO: establishment of RNA localization GO: RNA splicing GO: translational initiation GO: macromolecule modification GO: protein polyglycylation GO: cellular protein modification process GO: protein modification process GO: nucleobase-containing compound transport GO: inactivation of MAPK activity GO: reproduction GO: RNA localization GO: protein monoubiquitination Cluster 2 GO.ID Term Annotated Significant Expected classicfisher Over-representation GO: RNA splicing < 1e GO: mrna processing < 1e GO: mrna metabolic process < 1e GO: RNA processing < 1e GO: nuclear division < 1e GO: mitosis < 1e GO: organelle fission < 1e GO: M phase of mitotic cell cycle < 1e GO: DNA repair < 1e GO: M phase < 1e GO: chromosome organization < 1e GO: cell cycle phase < 1e GO: cell cycle process < 1e GO: mitotic cell cycle < 1e GO: DNA metabolic process < 1e GO: cell cycle < 1e GO: organelle organization < 1e GO: cellular protein metabolic process < 1e cellular component organization or biogenesis at cellular GO: level < 1e GO: nucleic acid metabolic process < 1e GO: cellular macromolecule metabolic process < 1e GO: cellular component organization at cellular level < 1e GO: nucleobase-containing compound metabolic process < 1e GO: gene expression < 1e
22 GO: RNA metabolic process < 1e GO: cellular nitrogen compound metabolic process < 1e GO: nitrogen compound metabolic process < 1e GO: macromolecule metabolic process < 1e GO: cellular metabolic process < 1e GO: primary metabolic process < 1e GO: metabolic process < 1e GO: protein metabolic process E GO: cell division E GO: protein modification by small protein conjugation or removal E GO: response to DNA damage stimulus E GO: ribonucleoprotein complex biogenesis E GO: cellular macromolecule biosynthetic process E GO: macromolecule localization E GO: cellular component organization or biogenesis E GO: chromosome segregation E GO: chromatin modification E GO: intracellular transport E GO: chromatin organization E GO: cellular component biogenesis at cellular level E GO: translation E GO: protein localization E GO: ncrna metabolic process E GO: macromolecule modification E GO: microtubule-based process E GO: macromolecule biosynthetic process E Cluster 3 GO.ID Term Annotated Significant Expected classicfisher Over-representation GO: metabolic process E GO: cellular metabolic process E GO: cofactor metabolic process E GO: small molecule metabolic process E GO: primary metabolic process E GO: cofactor biosynthetic process E GO: coenzyme metabolic process E GO: cellular lipid metabolic process E GO: type I interferon production E GO: cofactor transport E GO: cellular ketone metabolic process E GO: B cell homeostasis E
23 GO: interferon-beta production E GO: B cell differentiation GO: oxidation-reduction process GO: biosynthetic process GO: lipid metabolic process GO: regulation of interferon-beta production GO: alcohol metabolic process GO: glycerolipid biosynthetic process GO: sterol metabolic process GO: coenzyme biosynthetic process GO: carboxylic acid metabolic process GO: oxoacid metabolic process GO: lipid biosynthetic process GO: iron ion homeostasis GO: lymphocyte homeostasis GO: cellular nitrogen compound metabolic process GO: magnesium ion transport GO: cellular catabolic process GO: nitrogen compound metabolic process GO: nucleoside metabolic process GO: sterol biosynthetic process GO: tetrapyrrole biosynthetic process GO: intracellular signal transduction GO: positive regulation of innate immune response GO: organic acid metabolic process GO: GPI anchor biosynthetic process GO: protein lipidation GO: B cell activation GO: carbohydrate derivative metabolic process GO: tetrapyrrole metabolic process GO: heterocycle metabolic process GO: type I interferon biosynthetic process GO: protein homotetramerization GO: glycerophospholipid biosynthetic process GO: regulation of type I interferon production GO: response to oxidative stress GO: nucleoside monophosphate biosynthetic process GO: porphyrin-containing compound metabolic process
24 Cluster 4 GO.ID Term Annotated Significant Expected classicfisher Over-representation GO: cell fate commitment <1e GO: embryonic organ morphogenesis <1e GO: axonogenesis <1e GO: regionalization <1e GO: pattern specification process <1e cell morphogenesis involved in neuron GO: differentiation <1e GO: cell morphogenesis involved in differentiation <1e GO: neuron projection morphogenesis <1e GO: brain development <1e GO: central nervous system development <1e GO: embryonic morphogenesis <1e GO: regulation of nervous system development <1e GO: regulation of cell development <1e GO: organ morphogenesis <1e GO: tissue morphogenesis <1e GO: neuron projection development <1e GO: neuron differentiation <1e GO: neuron development <1e GO: epithelium development <1e GO: generation of neurons <1e GO: neurogenesis <1e GO: cell-cell signaling <1e GO: nervous system development <1e GO: positive regulation of developmental process <1e GO: cell morphogenesis <1e GO: regulation of multicellular organismal development <1e GO: cell development <1e GO: cell migration <1e GO: tissue development <1e GO: embryo development <1e GO: regulation of cell differentiation <1e GO: locomotion <1e GO: anatomical structure morphogenesis <1e GO: regulation of developmental process <1e GO: regulation of multicellular organismal process <1e GO: organ development <1e GO: system development <1e GO: cell differentiation <1e GO: regulation of cell communication <1e GO: regulation of localization <1e
25 GO: cellular developmental process <1e GO: multicellular organismal development <1e GO: anatomical structure development <1e GO: regulation of signaling <1e GO: developmental process <1e GO: positive regulation of cellular process <1e GO: positive regulation of biological process <1e GO: negative regulation of cellular process <1e GO: negative regulation of biological process <1e GO: multicellular organismal process <1e Cluster 5 GO.ID Term Annotated Significant Expected classicfisher Over-representation GO: detection of chemical stimulus involved in sensory perception of smell < 1e GO: detection of chemical stimulus involved in sensory perception < 1e GO: sensory perception of smell < 1e GO: detection of chemical stimulus < 1e GO: sensory perception of chemical stimulus < 1e GO: detection of stimulus involved in sensory perception < 1e GO: detection of stimulus < 1e GO: sensory perception < 1e GO: defense response to bacterium < 1e GO: G-protein coupled receptor signaling pathway < 1e GO: neurological system process < 1e GO: system process < 1e GO: cell surface receptor signaling pathway < 1e GO: response to chemical stimulus < 1e GO: defense response < 1e GO: signal transduction < 1e GO: signaling < 1e GO: cell communication < 1e GO: cellular response to stimulus < 1e GO: response to stimulus < 1e GO: multicellular organismal process < 1e GO: biological regulation < 1e GO: regulation of biological process < 1e GO: regulation of cellular process E GO: response to other organism E GO: response to biotic stimulus E GO: response to bacterium E GO: immune response E
26 GO: negative regulation of peptidase activity E GO: multi-organism process E GO: inflammatory response E GO: negative regulation of endopeptidase activity E GO: sensory perception of taste E GO: detection of chemical stimulus involved in sensory perception of bitter taste E GO: response to wounding E GO: detection of chemical stimulus involved in sensory perception of taste E GO: humoral immune response E GO: sensory perception of bitter taste E GO: defense response to Gram-positive bacterium E GO: innate immune response E GO: acute-phase response E GO: interferon-gamma production E GO: negative regulation of hydrolase activity E GO: regulation of interferon-gamma production E GO: acute inflammatory response E GO: cytokine production E GO: defense response to Gram-negative bacterium E GO: regulation of peptidase activity E GO: immune effector process E GO: regulation of immune response E
27 Supplementary Note Mononucleosome-BisSeq (MN-BisSeq) library preparation The protocol was adapted from Illumina Genomic DNA Sample Preparation Guide. Briefly, 2 μg of mononucleosomal fraction DNA were end repaired by incubation at 20 C for 30 minutes with 200µM dntp, 7.5 units of T4 DNA polymerase (NEB #M0203S), 5 units of DNA Polymerase I Large Fragment (Klenow) (NEB #M0210S), 25 units of T4 PNK (NEB #M0201S), 1x T4 DNA ligase buffer containing 10mM ATP (NEB). 3 ends of DNA fragments were adenylated by incubation at 37 C for 30 minutes with 100µM datp, 1xNEB Buffer 2, 10 units Klenow Fragment (3 5 exo ) (NEB # M0212L). Single End adapter sequences were produced based on Illumina adapter sequences (Oligonucleotide sequences Illumina, Inc. All rights reserved). 5 P- GATXGGAAGAGXTXGTATGXXGTXTTXTGXTTG and 5 AXAXTXTTTXXXTAXAXGAXGXTXTTXXGATXT, where X is a methylated cytosine. Adapters were ordered as single stranded oligos (Microsynth AG), resuspended in annealing buffer (10mM Tris ph7.5, 50mM NaCl, 1mM EDTA), annealed by heating at 95 C for 10 minutes and cooling down slowly. Annealed adapters were ligated to the DNA fragments as per manufacturer s instructions for genomic DNA library construction. Adapter-ligated DNA of 250 bp was isolated on 2% agarose gel electrophoresis. Gel purified DNA was then converted with sodium bisulfite using the Imprint DNA Modification Kit (Sigma-Aldrich) following the manufacturer s instructions. Half of the bisulfite-converted, adapter-ligated DNA molecules wasenriched by 7 cycles of PCR with the following reaction composition: 2.5 U of uracilinsensitive PfuTurboCx Hotstart DNA polymerase (Stratagene), 5 μl 10X PfuTurbo reaction buffer, 25 μm dntps, 0.5µM of Illumina single end PCR primers. The thermocycling parameters were: 95 C 2 min, 98 C 30 sec, then 7 cycles of 98 C 15 sec, 65 C 30 sec and 72 C 3 min, ending with one 72 C 5 min elongation step. The reaction products were purified using the MinElute PCR purification kit (Qiagen, Valencia, CA), resolved by 2% agarose gel electrophoresis to separate the library from adapter-adapter ligation products, and purified from the gel using the MinElute gel purification kit (Qiagen, Valencia, CA). Quality of the libraries and template size distribution were assessed by running an aliquot of the library on an Agilent 2100 Bioanalyzer (Agilent Technologies). 27
28 Analysis of bisulfite converted sequencing (BisSeq) data Read filtering and alignment of the BisSeq data from this study (bisulfite converted mononucleosome associated sperm DNA), published sperm whole methylomes (mouse sperm 5 and human sperm 2 ) and published oocyte and blastocyst methylomes 5 were performed as described 7. Peak identification Peak identification for nucleosome data was performed by training a two state hidden semi- Markov model, R-mhsmm package 9 on nucleosome enrichments calculated for 1 kb windows. The two-state model (non-peak and peak states) was initialized using Gaussian emissions (means of 0, 1, and variances of 0.5, 0.5), a gamma sojourn distribution with shape=2 and scale=10, and initial state probabilities of 0.5, 0.5. Parameter estimation of the model was performed by using 1kb window enrichments on chr1 as a training data set and by selecting a maximum of 200 windows in one state. Model parameters were estimated using EM algorithm, and the fitted model with emission distribution means of -0.21, 0.57, variances of 0.071, 0.48, and sojourn distribution parameters shape of 1.32, 0.38, and scale of 52.84, 7.86 was used to predict the maximum likelihood state path for the sequence of all 1kb windows in the genome. Adjacent 1kb windows with identical state labels were fused. Classification of nucleosome peaks Nucleosome enrichments were quantified on the peaks identified by a hidden semi- Markov model. Peaks were classified into three equal sized groups according to enrichment levels, termed weak, intermediate and strong peak groups (Fig. 5a). Genome-wide modeling of nucleosome occupancy Modeling of genome-wide mouse sperm nucleosome occupancy (Fig. 2d) was performed by using enrichment values in 1kb windows. Nucleosome occupancy data was modeled by a linear model with CpG dinucleotide frequency and the average % DNA methylation in 1kb windows as regressors, including only windows with detectable levels of nucleosomes, defined as log2 nucleosome occupancy greater than 0.2. This filtering excludes the majority of 1kb windows 28
29 without nucleosomes from the analysis which is required since the majority of the genome does not contain any nucleosomes in mouse sperm. In the model, DNA methylation data which was obtained by bisulfite converted sequencing of nucleosome associated DNA was used. Average DNA methylation was calculated by taking the ratio of total number of reads for methylated C over total number of reads for all C (methylated or umethylated) per window. Windows with less than 5 total reads for all Cs were excluded from the analysis. Finally, n= (9% of all windows) were used in modeling of nucleosome occupancy in mouse sperm. The same windows were used to analyze the relationship between mouse sperm nucleosome occupancy and DNA methylation / sequence composition (Fig. 2a, 2b). To analyze the relationship between human sperm nucleosome occupancy 3,4 and DNA methylation / sequence composition (Supplementary Fig. 2c - 2f), 1kb windows were processed in a similar way as for mouse sperm. Finally, n= (29% of all windows) of 3 and n= (17% of all windows) of 4 were used in the analysis. Quantifying expression in round spermatids Round spermatid expression data was quantified by summing the total number of reads mapping to Refseq transcripts. Concerning the classification of the expression status, transcripts without any aligned reads were classified as not detected, and the remaining transcripts were classified on the basis of increasing expression values into three equally sized groups termed low, medium and high. Motif finding for histone peaks 7-mer motif frequencies in the foreground and background sets were calculated by using Bioconductor package Biostrings. Foreground refers to histone peaks identified via the hidden semi-markov model approach. As a background, we used CGI that do not intersect any of the peak regions in the foreground set. Motif enrichment (M) and abundance (A) values were calculated as follows: M=log2((fg Z /fg Total min(fg Total, bg Total ))+pscnt)-log2((bg Z /bg Total min(fg Total,bg Total ))+pscnt) A=(log2((fg Z /fg Total min(fg Total,bg Total ))+pscnt)+log2((bg Z /bg Total min(fg Total,bg Total) )+pscnt))0.5 29
30 where fg Z is the number of counts of motif Z in the foreground, fg Total is the total number of all motifs in foreground, bg Z is the number of counts of motif Z in background, bg Total is the total number of all motifs in the background, and pscnt is a constant number (8). Results were visualized in a motif enrichment abundance plot (MA plot) and the top 20 motifs enriched in the foreground set were displayed. Defining expression for oogenesis or early embryogenesis Expression data from 10 was processed as described in 3. Expression states are referred as not expressed, oocyte, 2-8 cell and blastocyst. Embryonic expression was classified according to the first expression stage during development. The distinction between maternal and embryonic expression in 2-cell embryos was made by comparing the expression levels in early embryos treated or untreated with α-amanitin. References 1. Kaplan, N. et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458, (2009). 2. Molaro, A. et al. methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell 146, (2011). 3. Brykczynska, U. et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol 17, (2010). 4. Hammoud, S.S. et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460, (2009). 5. Kobayashi, H. et al. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet 8, e (2012). 6. Goldberg, A.D. et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, (2010). 7. Stadler, M.B. et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480, (2011). 8. Mikkelsen, T.S. et al. Genome-wide maps of chromatin state in pluripotent and lineagecommitted cells. Nature 448, (2007). 9. J., O.C. & S., H. Hidden Semi Markov Models for Multiple Observation Sequences: The mhsmm Package for R. Journal of Statistical Software 39, 1-22 (2011). 10. Zeng, F. & Schultz, R.M. RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Developmental biology 283, (2005). 30
Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq
 Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq Philipp Bucher Wednesday January 21, 2009 SIB graduate school course EPFL, Lausanne ChIP-seq against histone variants: Biological
Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq Philipp Bucher Wednesday January 21, 2009 SIB graduate school course EPFL, Lausanne ChIP-seq against histone variants: Biological
Nature Structural & Molecular Biology: doi: /nsmb.2419
 Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Supplementary information for: Community detection for networks with. unipartite and bipartite structure. Chang Chang 1, 2, Chao Tang 2
 Supplementary information for: Community detection for networks with unipartite and bipartite structure Chang Chang 1, 2, Chao Tang 2 1 School of Life Sciences, Peking University, Beiing 100871, China
Supplementary information for: Community detection for networks with unipartite and bipartite structure Chang Chang 1, 2, Chao Tang 2 1 School of Life Sciences, Peking University, Beiing 100871, China
7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans.
 Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Nature Genetics: doi: /ng Supplementary Figure 1. Assessment of sample purity and quality.
 Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
Supplementary Figure 1 Assessment of sample purity and quality. (a) Hematoxylin and eosin staining of formaldehyde-fixed, paraffin-embedded sections from a human testis biopsy collected concurrently with
An epigenetic approach to understanding (and predicting?) environmental effects on gene expression
 www.collaslab.com An epigenetic approach to understanding (and predicting?) environmental effects on gene expression Philippe Collas University of Oslo Institute of Basic Medical Sciences Stem Cell Epigenetics
www.collaslab.com An epigenetic approach to understanding (and predicting?) environmental effects on gene expression Philippe Collas University of Oslo Institute of Basic Medical Sciences Stem Cell Epigenetics
Regulation of Gene Expression in Eukaryotes
 Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
Ch. 19 Regulation of Gene Expression in Eukaryotes BIOL 222 Differential Gene Expression in Eukaryotes Signal Cells in a multicellular eukaryotic organism genetically identical differential gene expression
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature19360 Supplementary Tables Supplementary Table 1. Number of monoclonal reads in each sample Sample Number of cells Total reads Aligned reads Monoclonal reads
SUPPLEMENTARY INFORMATION doi:10.1038/nature19360 Supplementary Tables Supplementary Table 1. Number of monoclonal reads in each sample Sample Number of cells Total reads Aligned reads Monoclonal reads
Allelic reprogramming of the histone modification H3K4me3 in early mammalian development
 Allelic reprogramming of the histone modification H3K4me3 in early mammalian development 张戈 Method and material STAR ChIP seq (small-scale TELP-assisted rapid ChIP seq) 200 mouse embryonic stem cells PWK/PhJ
Allelic reprogramming of the histone modification H3K4me3 in early mammalian development 张戈 Method and material STAR ChIP seq (small-scale TELP-assisted rapid ChIP seq) 200 mouse embryonic stem cells PWK/PhJ
Cluster Dendrogram. dist(cor(na.omit(tss.exprs.chip[, c(1:10, 24, 27, 30, 48:50, dist(cor(na.omit(tss.exprs.chip[, c(1:99, 103, 104, 109, 110,
 A Transcriptome (RNA-seq) Transcriptome (RNA-seq) 3. 2.5 2..5..5...5..5 2. 2.5 3. 2.5 2..5..5...5..5 2. 2.5 Cluster Dendrogram RS_ES3.2 RS_ES3. RS_SHS5.2 RS_SHS5. PS_SHS5.2 PS_SHS5. RS_LJ3 PS_LJ3..4 _SHS5.2
A Transcriptome (RNA-seq) Transcriptome (RNA-seq) 3. 2.5 2..5..5...5..5 2. 2.5 3. 2.5 2..5..5...5..5 2. 2.5 Cluster Dendrogram RS_ES3.2 RS_ES3. RS_SHS5.2 RS_SHS5. PS_SHS5.2 PS_SHS5. RS_LJ3 PS_LJ3..4 _SHS5.2
Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types.
 Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Stem Cell Epigenetics
 Stem Cell Epigenetics Philippe Collas University of Oslo Institute of Basic Medical Sciences Norwegian Center for Stem Cell Research www.collaslab.com Source of stem cells in the body Somatic ( adult )
Stem Cell Epigenetics Philippe Collas University of Oslo Institute of Basic Medical Sciences Norwegian Center for Stem Cell Research www.collaslab.com Source of stem cells in the body Somatic ( adult )
Table S4A- GOBP functional annotation of celecoxib-modulated genes common between CO
 Table S4A- GOBP functional annotation of celecoxib-modulated genes common between CO Terms ListHits ListTotal PopulationHits cell proliferation 44 228 1156 cell cycle 34 228 806 DNA replication and chromosome
Table S4A- GOBP functional annotation of celecoxib-modulated genes common between CO Terms ListHits ListTotal PopulationHits cell proliferation 44 228 1156 cell cycle 34 228 806 DNA replication and chromosome
Genetics and Genomics in Medicine Chapter 6 Questions
 Genetics and Genomics in Medicine Chapter 6 Questions Multiple Choice Questions Question 6.1 With respect to the interconversion between open and condensed chromatin shown below: Which of the directions
Genetics and Genomics in Medicine Chapter 6 Questions Multiple Choice Questions Question 6.1 With respect to the interconversion between open and condensed chromatin shown below: Which of the directions
Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment
 Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development
Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development
Studying Alternative Splicing
 Studying Alternative Splicing Meelis Kull PhD student in the University of Tartu supervisor: Jaak Vilo CS Theory Days Rõuge 27 Overview Alternative splicing Its biological function Studying splicing Technology
Studying Alternative Splicing Meelis Kull PhD student in the University of Tartu supervisor: Jaak Vilo CS Theory Days Rõuge 27 Overview Alternative splicing Its biological function Studying splicing Technology
Accessing and Using ENCODE Data Dr. Peggy J. Farnham
 1 William M Keck Professor of Biochemistry Keck School of Medicine University of Southern California How many human genes are encoded in our 3x10 9 bp? C. elegans (worm) 959 cells and 1x10 8 bp 20,000
1 William M Keck Professor of Biochemistry Keck School of Medicine University of Southern California How many human genes are encoded in our 3x10 9 bp? C. elegans (worm) 959 cells and 1x10 8 bp 20,000
Results. Abstract. Introduc4on. Conclusions. Methods. Funding
 . expression that plays a role in many cellular processes affecting a variety of traits. In this study DNA methylation was assessed in neuronal tissue from three pigs (frontal lobe) and one great tit (whole
. expression that plays a role in many cellular processes affecting a variety of traits. In this study DNA methylation was assessed in neuronal tissue from three pigs (frontal lobe) and one great tit (whole
Single-cell RNA-Seq profiling of human pre-implantation embryos and embryonic stem cells
 Single-cell RNA-Seq profiling of human pre-implantation embryos and embryonic stem cells Liying Yan,2,5, Mingyu Yang,5, Hongshan Guo, Lu Yang, Jun Wu, Rong Li,2, Ping Liu, Ying Lian, Xiaoying Zheng, Jie
Single-cell RNA-Seq profiling of human pre-implantation embryos and embryonic stem cells Liying Yan,2,5, Mingyu Yang,5, Hongshan Guo, Lu Yang, Jun Wu, Rong Li,2, Ping Liu, Ying Lian, Xiaoying Zheng, Jie
Histones modifications and variants
 Histones modifications and variants Dr. Institute of Molecular Biology, Johannes Gutenberg University, Mainz www.imb.de Lecture Objectives 1. Chromatin structure and function Chromatin and cell state Nucleosome
Histones modifications and variants Dr. Institute of Molecular Biology, Johannes Gutenberg University, Mainz www.imb.de Lecture Objectives 1. Chromatin structure and function Chromatin and cell state Nucleosome
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Not IN Our Genes - A Different Kind of Inheritance.! Christopher Phiel, Ph.D. University of Colorado Denver Mini-STEM School February 4, 2014
 Not IN Our Genes - A Different Kind of Inheritance! Christopher Phiel, Ph.D. University of Colorado Denver Mini-STEM School February 4, 2014 Epigenetics in Mainstream Media Epigenetics *Current definition:
Not IN Our Genes - A Different Kind of Inheritance! Christopher Phiel, Ph.D. University of Colorado Denver Mini-STEM School February 4, 2014 Epigenetics in Mainstream Media Epigenetics *Current definition:
Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63.
 Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
levels of genes were separated by their expression levels; 2,000 high, medium, and low
 Figure S1. Histone modification profiles near transcription start sites. The overall histone modification around transcription start sites (TSSs) was calculated. Histone modification levels of genes were
Figure S1. Histone modification profiles near transcription start sites. The overall histone modification around transcription start sites (TSSs) was calculated. Histone modification levels of genes were
For the 5 GATC-overhang two-oligo adaptors set up the following reactions in 96-well plate format:
 Supplementary Protocol 1. Adaptor preparation: For the 5 GATC-overhang two-oligo adaptors set up the following reactions in 96-well plate format: Per reaction X96 10X NEBuffer 2 10 µl 10 µl x 96 5 -GATC
Supplementary Protocol 1. Adaptor preparation: For the 5 GATC-overhang two-oligo adaptors set up the following reactions in 96-well plate format: Per reaction X96 10X NEBuffer 2 10 µl 10 µl x 96 5 -GATC
Supplementary Information. Deciphering the Venomic Transcriptome of Killer- Wasp Vespa velutina
 Supplementary Information Deciphering the Venomic Transcriptome of Killer- Wasp Vespa velutina Zhirui Liu, Shuanggang Chen, You Zhou, Cuihong Xie, Bifeng Zhu, Huming Zhu, Shupeng Liu, Wei Wang, Hongzhuan
Supplementary Information Deciphering the Venomic Transcriptome of Killer- Wasp Vespa velutina Zhirui Liu, Shuanggang Chen, You Zhou, Cuihong Xie, Bifeng Zhu, Huming Zhu, Shupeng Liu, Wei Wang, Hongzhuan
Methylation reprogramming dynamics and defects in gametogenesis and embryogenesis: implications for reproductive medicine
 Univ.-Prof. Dr. Thomas Haaf Methylation reprogramming dynamics and defects in gametogenesis and embryogenesis: implications for reproductive medicine Epigenetics and DNA methylation Heritable change of
Univ.-Prof. Dr. Thomas Haaf Methylation reprogramming dynamics and defects in gametogenesis and embryogenesis: implications for reproductive medicine Epigenetics and DNA methylation Heritable change of
Mechanisms of alternative splicing regulation
 Mechanisms of alternative splicing regulation The number of mechanisms that are known to be involved in splicing regulation approximates the number of splicing decisions that have been analyzed in detail.
Mechanisms of alternative splicing regulation The number of mechanisms that are known to be involved in splicing regulation approximates the number of splicing decisions that have been analyzed in detail.
Formalin-fixed paraffin-embedded sections of liver from a recipient mouse sacrificed after two rounds
 Supplementary figure legends Supplementary Figure 1 Fah + hepatocytes in a Fah -/- mouse transplanted with sorted cells. Formalin-fixed paraffin-embedded sections of liver from a recipient mouse sacrificed
Supplementary figure legends Supplementary Figure 1 Fah + hepatocytes in a Fah -/- mouse transplanted with sorted cells. Formalin-fixed paraffin-embedded sections of liver from a recipient mouse sacrificed
Nature Immunology: doi: /ni Supplementary Figure 1. Characteristics of SEs in T reg and T conv cells.
 Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Fragile X Syndrome. Genetics, Epigenetics & the Role of Unprogrammed Events in the expression of a Phenotype
 Fragile X Syndrome Genetics, Epigenetics & the Role of Unprogrammed Events in the expression of a Phenotype A loss of function of the FMR-1 gene results in severe learning problems, intellectual disability
Fragile X Syndrome Genetics, Epigenetics & the Role of Unprogrammed Events in the expression of a Phenotype A loss of function of the FMR-1 gene results in severe learning problems, intellectual disability
Heintzman, ND, Stuart, RK, Hon, G, Fu, Y, Ching, CW, Hawkins, RD, Barrera, LO, Van Calcar, S, Qu, C, Ching, KA, Wang, W, Weng, Z, Green, RD,
 Heintzman, ND, Stuart, RK, Hon, G, Fu, Y, Ching, CW, Hawkins, RD, Barrera, LO, Van Calcar, S, Qu, C, Ching, KA, Wang, W, Weng, Z, Green, RD, Crawford, GE, Ren, B (2007) Distinct and predictive chromatin
Heintzman, ND, Stuart, RK, Hon, G, Fu, Y, Ching, CW, Hawkins, RD, Barrera, LO, Van Calcar, S, Qu, C, Ching, KA, Wang, W, Weng, Z, Green, RD, Crawford, GE, Ren, B (2007) Distinct and predictive chromatin
Supplementary Information
 Supplementary Information 5-hydroxymethylcytosine-mediated epigenetic dynamics during postnatal neurodevelopment and aging By Keith E. Szulwach 1,8, Xuekun Li 1,8, Yujing Li 1, Chun-Xiao Song 2, Hao Wu
Supplementary Information 5-hydroxymethylcytosine-mediated epigenetic dynamics during postnatal neurodevelopment and aging By Keith E. Szulwach 1,8, Xuekun Li 1,8, Yujing Li 1, Chun-Xiao Song 2, Hao Wu
Supplemental Figure S1. Tertiles of FKBP5 promoter methylation and internal regulatory region
 Supplemental Figure S1. Tertiles of FKBP5 promoter methylation and internal regulatory region methylation in relation to PSS and fetal coupling. A, PSS values for participants whose placentas showed low,
Supplemental Figure S1. Tertiles of FKBP5 promoter methylation and internal regulatory region methylation in relation to PSS and fetal coupling. A, PSS values for participants whose placentas showed low,
Identification of Tissue-Specific Protein-Coding and Noncoding. Transcripts across 14 Human Tissues Using RNA-seq
 Identification of Tissue-Specific Protein-Coding and Noncoding Transcripts across 14 Human Tissues Using RNA-seq Jinhang Zhu 1, Geng Chen 1, Sibo Zhu 1,2, Suqing Li 3, Zhuo Wen 3, Bin Li 1, Yuanting Zheng
Identification of Tissue-Specific Protein-Coding and Noncoding Transcripts across 14 Human Tissues Using RNA-seq Jinhang Zhu 1, Geng Chen 1, Sibo Zhu 1,2, Suqing Li 3, Zhuo Wen 3, Bin Li 1, Yuanting Zheng
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers
 Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Selective depletion of abundant RNAs to enable transcriptome analysis of lowinput and highly-degraded RNA from FFPE breast cancer samples
 DNA CLONING DNA AMPLIFICATION & PCR EPIGENETICS RNA ANALYSIS Selective depletion of abundant RNAs to enable transcriptome analysis of lowinput and highly-degraded RNA from FFPE breast cancer samples LIBRARY
DNA CLONING DNA AMPLIFICATION & PCR EPIGENETICS RNA ANALYSIS Selective depletion of abundant RNAs to enable transcriptome analysis of lowinput and highly-degraded RNA from FFPE breast cancer samples LIBRARY
ChIP-seq analysis. J. van Helden, M. Defrance, C. Herrmann, D. Puthier, N. Servant, M. Thomas-Chollier, O.Sand
 ChIP-seq analysis J. van Helden, M. Defrance, C. Herrmann, D. Puthier, N. Servant, M. Thomas-Chollier, O.Sand Tuesday : quick introduction to ChIP-seq and peak-calling (Presentation + Practical session)
ChIP-seq analysis J. van Helden, M. Defrance, C. Herrmann, D. Puthier, N. Servant, M. Thomas-Chollier, O.Sand Tuesday : quick introduction to ChIP-seq and peak-calling (Presentation + Practical session)
Peak-calling for ChIP-seq and ATAC-seq
 Peak-calling for ChIP-seq and ATAC-seq Shamith Samarajiwa CRUK Autumn School in Bioinformatics 2017 University of Cambridge Overview Peak-calling: identify enriched (signal) regions in ChIP-seq or ATAC-seq
Peak-calling for ChIP-seq and ATAC-seq Shamith Samarajiwa CRUK Autumn School in Bioinformatics 2017 University of Cambridge Overview Peak-calling: identify enriched (signal) regions in ChIP-seq or ATAC-seq
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression. Chromatin Array of nuc
 Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Eukaryotic Gene Regulation
 Eukaryotic Gene Regulation Chapter 19: Control of Eukaryotic Genome The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different,
Eukaryotic Gene Regulation Chapter 19: Control of Eukaryotic Genome The BIG Questions How are genes turned on & off in eukaryotes? How do cells with the same genes differentiate to perform completely different,
Supplementary Fig.S1. MeDIP RPKM distribution of 5kb windows. Relative expression of DNMT. Percentage of genome. (fold change)
 Supplementary Fig.S1 Percentage of genome 10% 8% 6% 4% 2% 0 MeDIP RPKM distribution of 5kb windows 0 0. 0.5 0.75 >1 MeDIP RPKM value EC Relative expression of DNMT (fold change) 12 6 4 3 2 1 EC DNMT1 DNMT2
Supplementary Fig.S1 Percentage of genome 10% 8% 6% 4% 2% 0 MeDIP RPKM distribution of 5kb windows 0 0. 0.5 0.75 >1 MeDIP RPKM value EC Relative expression of DNMT (fold change) 12 6 4 3 2 1 EC DNMT1 DNMT2
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
Ch. 18 Regulation of Gene Expression
 Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
Ch. 18 Regulation of Gene Expression 1 Human genome has around 23,688 genes (Scientific American 2/2006) Essential Questions: How is transcription regulated? How are genes expressed? 2 Bacteria regulate
RNA-seq Introduction
 RNA-seq Introduction DNA is the same in all cells but which RNAs that is present is different in all cells There is a wide variety of different functional RNAs Which RNAs (and sometimes then translated
RNA-seq Introduction DNA is the same in all cells but which RNAs that is present is different in all cells There is a wide variety of different functional RNAs Which RNAs (and sometimes then translated
Jayanti Tokas 1, Puneet Tokas 2, Shailini Jain 3 and Hariom Yadav 3
 Jayanti Tokas 1, Puneet Tokas 2, Shailini Jain 3 and Hariom Yadav 3 1 Department of Biotechnology, JMIT, Radaur, Haryana, India 2 KITM, Kurukshetra, Haryana, India 3 NIDDK, National Institute of Health,
Jayanti Tokas 1, Puneet Tokas 2, Shailini Jain 3 and Hariom Yadav 3 1 Department of Biotechnology, JMIT, Radaur, Haryana, India 2 KITM, Kurukshetra, Haryana, India 3 NIDDK, National Institute of Health,
he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003)
 MicroRNAs: Genomics, Biogenesis, Mechanism, and Function (D. Bartel Cell 2004) he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003) Vertebrate MicroRNA Genes (Lim et al. Science
MicroRNAs: Genomics, Biogenesis, Mechanism, and Function (D. Bartel Cell 2004) he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003) Vertebrate MicroRNA Genes (Lim et al. Science
Measuring DNA Methylation with the MinION. Winston Timp Department of Biomedical Engineering Johns Hopkins University 12/1/16
 Measuring DNA Methylation with the MinION Winston Timp Department of Biomedical Engineering Johns Hopkins University 12/1/16 Epigenetics: Modern Modern Definition of epigenetics involves heritable changes
Measuring DNA Methylation with the MinION Winston Timp Department of Biomedical Engineering Johns Hopkins University 12/1/16 Epigenetics: Modern Modern Definition of epigenetics involves heritable changes
The Insulator Binding Protein CTCF Positions 20 Nucleosomes around Its Binding Sites across the Human Genome
 The Insulator Binding Protein CTCF Positions 20 Nucleosomes around Its Binding Sites across the Human Genome Yutao Fu 1, Manisha Sinha 2,3, Craig L. Peterson 3, Zhiping Weng 1,4,5 * 1 Bioinformatics Program,
The Insulator Binding Protein CTCF Positions 20 Nucleosomes around Its Binding Sites across the Human Genome Yutao Fu 1, Manisha Sinha 2,3, Craig L. Peterson 3, Zhiping Weng 1,4,5 * 1 Bioinformatics Program,
Integrated analysis of sequencing data
 Integrated analysis of sequencing data How to combine *-seq data M. Defrance, M. Thomas-Chollier, C. Herrmann, D. Puthier, J. van Helden *ChIP-seq, RNA-seq, MeDIP-seq, Transcription factor binding ChIP-seq
Integrated analysis of sequencing data How to combine *-seq data M. Defrance, M. Thomas-Chollier, C. Herrmann, D. Puthier, J. van Helden *ChIP-seq, RNA-seq, MeDIP-seq, Transcription factor binding ChIP-seq
Epigenetics and Chromatin Remodeling
 Epigenetics and Chromatin Remodeling Bradford Coffee, PhD, FACMG Emory University Atlanta, GA Speaker Disclosure Information Grant/Research Support: none Salary/Consultant Fees: none Board/Committee/Advisory
Epigenetics and Chromatin Remodeling Bradford Coffee, PhD, FACMG Emory University Atlanta, GA Speaker Disclosure Information Grant/Research Support: none Salary/Consultant Fees: none Board/Committee/Advisory
Epigenetic Principles and Mechanisms Underlying Nervous System Function in Health and Disease Mark F. Mehler MD, FAAN
 Epigenetic Principles and Mechanisms Underlying Nervous System Function in Health and Disease Mark F. Mehler MD, FAAN Institute for Brain Disorders and Neural Regeneration F.M. Kirby Program in Neural
Epigenetic Principles and Mechanisms Underlying Nervous System Function in Health and Disease Mark F. Mehler MD, FAAN Institute for Brain Disorders and Neural Regeneration F.M. Kirby Program in Neural
MIR retrotransposon sequences provide insulators to the human genome
 Supplementary Information: MIR retrotransposon sequences provide insulators to the human genome Jianrong Wang, Cristina Vicente-García, Davide Seruggia, Eduardo Moltó, Ana Fernandez- Miñán, Ana Neto, Elbert
Supplementary Information: MIR retrotransposon sequences provide insulators to the human genome Jianrong Wang, Cristina Vicente-García, Davide Seruggia, Eduardo Moltó, Ana Fernandez- Miñán, Ana Neto, Elbert
An Unexpected Function of the Prader-Willi Syndrome Imprinting Center in Maternal Imprinting in Mice
 An Unexpected Function of the Prader-Willi Syndrome Imprinting Center in Maternal Imprinting in Mice Mei-Yi Wu 1 *, Ming Jiang 1, Xiaodong Zhai 2, Arthur L. Beaudet 2, Ray-Chang Wu 1 * 1 Department of
An Unexpected Function of the Prader-Willi Syndrome Imprinting Center in Maternal Imprinting in Mice Mei-Yi Wu 1 *, Ming Jiang 1, Xiaodong Zhai 2, Arthur L. Beaudet 2, Ray-Chang Wu 1 * 1 Department of
SUPPLEMENTAL DATA AGING, July 2014, Vol. 6 No. 7
 SUPPLEMENTAL DATA Figure S1. Muscle mass changes in different anatomical regions with age. (A) The TA and gastrocnemius muscle showed a significant loss of weight in aged mice (24 month old) compared to
SUPPLEMENTAL DATA Figure S1. Muscle mass changes in different anatomical regions with age. (A) The TA and gastrocnemius muscle showed a significant loss of weight in aged mice (24 month old) compared to
Prokaryotes and eukaryotes alter gene expression in response to their changing environment
 Chapter 18 Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development and is responsible for differences
Chapter 18 Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development and is responsible for differences
Lecture 27. Epigenetic regulation of gene expression during development
 Lecture 27 Epigenetic regulation of gene expression during development Development of a multicellular organism is not only determined by the DNA sequence but also epigenetically through DNA methylation
Lecture 27 Epigenetic regulation of gene expression during development Development of a multicellular organism is not only determined by the DNA sequence but also epigenetically through DNA methylation
Supplemental Figure 1. Genes showing ectopic H3K9 dimethylation in this study are DNA hypermethylated in Lister et al. study.
 mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
Epigenetics. Jenny van Dongen Vrije Universiteit (VU) Amsterdam Boulder, Friday march 10, 2017
 Epigenetics Jenny van Dongen Vrije Universiteit (VU) Amsterdam j.van.dongen@vu.nl Boulder, Friday march 10, 2017 Epigenetics Epigenetics= The study of molecular mechanisms that influence the activity of
Epigenetics Jenny van Dongen Vrije Universiteit (VU) Amsterdam j.van.dongen@vu.nl Boulder, Friday march 10, 2017 Epigenetics Epigenetics= The study of molecular mechanisms that influence the activity of
Imprinting. Joyce Ohm Cancer Genetics and Genomics CGP-L2-319 x8821
 Imprinting Joyce Ohm Cancer Genetics and Genomics CGP-L2-319 x8821 Learning Objectives 1. To understand the basic concepts of genomic imprinting Genomic imprinting is an epigenetic phenomenon that causes
Imprinting Joyce Ohm Cancer Genetics and Genomics CGP-L2-319 x8821 Learning Objectives 1. To understand the basic concepts of genomic imprinting Genomic imprinting is an epigenetic phenomenon that causes
Patterns of Histone Methylation and Chromatin Organization in Grapevine Leaf. Rachel Schwope EPIGEN May 24-27, 2016
 Patterns of Histone Methylation and Chromatin Organization in Grapevine Leaf Rachel Schwope EPIGEN May 24-27, 2016 What does H3K4 methylation do? Plant of interest: Vitis vinifera Culturally important
Patterns of Histone Methylation and Chromatin Organization in Grapevine Leaf Rachel Schwope EPIGEN May 24-27, 2016 What does H3K4 methylation do? Plant of interest: Vitis vinifera Culturally important
QIAsymphony DSP Circulating DNA Kit
 QIAsymphony DSP Circulating DNA Kit February 2017 Performance Characteristics 937556 Sample to Insight Contents Performance Characteristics... 4 Basic performance... 4 Run precision... 6 Equivalent performance
QIAsymphony DSP Circulating DNA Kit February 2017 Performance Characteristics 937556 Sample to Insight Contents Performance Characteristics... 4 Basic performance... 4 Run precision... 6 Equivalent performance
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Development Supplementary information. Supplementary Figures * * +/+ +/- -/- +/+ +/- -/-
 Development 144: doi:1.1242/dev.1473: Supplementary information Supplementary Figures A (f) FRT LoxP 2 3 4 B All Males Females I Ovary 1 (+) 77 bps (f) 78 bps (-) >13 bps (-) 2 4 (-) 424 bps M +/f +/-
Development 144: doi:1.1242/dev.1473: Supplementary information Supplementary Figures A (f) FRT LoxP 2 3 4 B All Males Females I Ovary 1 (+) 77 bps (f) 78 bps (-) >13 bps (-) 2 4 (-) 424 bps M +/f +/-
SUPPLEMENTARY FIGURE 1: f-i
 SUPPLEMENTARY FIGURE 1: Comparisons of the biological replicates of ChIP-seq, Input and Bisulfite-seq. (a) Density plot of MeCP2 ChIP-seq genome coverage (calculated using tiled 15 bp windows) shows high
SUPPLEMENTARY FIGURE 1: Comparisons of the biological replicates of ChIP-seq, Input and Bisulfite-seq. (a) Density plot of MeCP2 ChIP-seq genome coverage (calculated using tiled 15 bp windows) shows high
Dissecting gene regulation network in human early embryos. at single-cell and single-base resolution
 Dissecting gene regulation network in human early embryos at single-cell and single-base resolution Fuchou Tang BIOPIC, College of Life Sciences Peking University 07/10/2015 Cockburn and Rossant, 2010
Dissecting gene regulation network in human early embryos at single-cell and single-base resolution Fuchou Tang BIOPIC, College of Life Sciences Peking University 07/10/2015 Cockburn and Rossant, 2010
Where Splicing Joins Chromatin And Transcription. 9/11/2012 Dario Balestra
 Where Splicing Joins Chromatin And Transcription 9/11/2012 Dario Balestra Splicing process overview Splicing process overview Sequence context RNA secondary structure Tissue-specific Proteins Development
Where Splicing Joins Chromatin And Transcription 9/11/2012 Dario Balestra Splicing process overview Splicing process overview Sequence context RNA secondary structure Tissue-specific Proteins Development
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Frequency of alternative-cassette-exon engagement with the ribosome is consistent across data from multiple human cell types and from mouse stem cells. Box plots showing AS frequency
Supplementary Figure 1 Frequency of alternative-cassette-exon engagement with the ribosome is consistent across data from multiple human cell types and from mouse stem cells. Box plots showing AS frequency
Supplementary figure legends
 Supplementary figure legends Fig. S1. Lineweaver-Burk plot of putrescine uptake by YeeF. An overnight culture of SK629 was inoculated in 100-mL LBG medium in 500-mL Erlenmeyer flasks. The medium was supplemented
Supplementary figure legends Fig. S1. Lineweaver-Burk plot of putrescine uptake by YeeF. An overnight culture of SK629 was inoculated in 100-mL LBG medium in 500-mL Erlenmeyer flasks. The medium was supplemented
Supplementary data Table S3. GO terms, pathways and networks enriched among the significantly correlating genes using Tox-Profiler
 Supplementary data Table S3. GO terms, pathways and networks enriched among the significantly correlating genes using Tox-Profiler DR CALUX Boys Girls Database Systemic lupus erythematosus 4.4 0.0021 6.7
Supplementary data Table S3. GO terms, pathways and networks enriched among the significantly correlating genes using Tox-Profiler DR CALUX Boys Girls Database Systemic lupus erythematosus 4.4 0.0021 6.7
Genome-Wide Analysis of Histone Modifications in Human Endometrial Stromal Cells
 ORIGINAL RESEARCH Genome-Wide Analysis of Histone Modifications in Human Endometrial Stromal Cells Isao Tamura, Yasuyuki Ohkawa, Tetsuya Sato, Mikita Suyama, Kosuke Jozaki, Maki Okada, Lifa Lee, Ryo Maekawa,
ORIGINAL RESEARCH Genome-Wide Analysis of Histone Modifications in Human Endometrial Stromal Cells Isao Tamura, Yasuyuki Ohkawa, Tetsuya Sato, Mikita Suyama, Kosuke Jozaki, Maki Okada, Lifa Lee, Ryo Maekawa,
Genome-Wide Localization of Protein-DNA Binding and Histone Modification by a Bayesian Change-Point Method with ChIP-seq Data
 Genome-Wide Localization of Protein-DNA Binding and Histone Modification by a Bayesian Change-Point Method with ChIP-seq Data Haipeng Xing, Yifan Mo, Will Liao, Michael Q. Zhang Clayton Davis and Geoffrey
Genome-Wide Localization of Protein-DNA Binding and Histone Modification by a Bayesian Change-Point Method with ChIP-seq Data Haipeng Xing, Yifan Mo, Will Liao, Michael Q. Zhang Clayton Davis and Geoffrey
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Multiplex target enrichment using DNA indexing for ultra-high throughput variant detection
 Multiplex target enrichment using DNA indexing for ultra-high throughput variant detection Dr Elaine Kenny Neuropsychiatric Genetics Research Group Institute of Molecular Medicine Trinity College Dublin
Multiplex target enrichment using DNA indexing for ultra-high throughput variant detection Dr Elaine Kenny Neuropsychiatric Genetics Research Group Institute of Molecular Medicine Trinity College Dublin
Supplementary information
 Supplementary information High fat diet-induced changes of mouse hepatic transcription and enhancer activity can be reversed by subsequent weight loss Majken Siersbæk, Lyuba Varticovski, Shutong Yang,
Supplementary information High fat diet-induced changes of mouse hepatic transcription and enhancer activity can be reversed by subsequent weight loss Majken Siersbæk, Lyuba Varticovski, Shutong Yang,
Supplemental Information. A Highly Sensitive and Robust Method. for Genome-wide 5hmC Profiling. of Rare Cell Populations
 Molecular ell, Volume 63 Supplemental Information Highly Sensitive and Robust Method for enome-wide hm Profiling of Rare ell Populations Dali Han, Xingyu Lu, lan H. Shih, Ji Nie, Qiancheng You, Meng Michelle
Molecular ell, Volume 63 Supplemental Information Highly Sensitive and Robust Method for enome-wide hm Profiling of Rare ell Populations Dali Han, Xingyu Lu, lan H. Shih, Ji Nie, Qiancheng You, Meng Michelle
Measuring DNA Methylation with the MinION
 Measuring DNA Methylation with the MinION Winston Timp Department of Biomedical Engineering Johns Hopkins University Epigenetics: Modern Modern Definition of epigenetics involves heritable changes other
Measuring DNA Methylation with the MinION Winston Timp Department of Biomedical Engineering Johns Hopkins University Epigenetics: Modern Modern Definition of epigenetics involves heritable changes other
Genomic structural variation
 Genomic structural variation Mario Cáceres The new genomic variation DNA sequence differs across individuals much more than researchers had suspected through structural changes A huge amount of structural
Genomic structural variation Mario Cáceres The new genomic variation DNA sequence differs across individuals much more than researchers had suspected through structural changes A huge amount of structural
The Epigenome Tools 2: ChIP-Seq and Data Analysis
 The Epigenome Tools 2: ChIP-Seq and Data Analysis Chongzhi Zang zang@virginia.edu http://zanglab.com PHS5705: Public Health Genomics March 20, 2017 1 Outline Epigenome: basics review ChIP-seq overview
The Epigenome Tools 2: ChIP-Seq and Data Analysis Chongzhi Zang zang@virginia.edu http://zanglab.com PHS5705: Public Health Genomics March 20, 2017 1 Outline Epigenome: basics review ChIP-seq overview
Mapping by recurrence and modelling the mutation rate
 Mapping by recurrence and modelling the mutation rate Shamil Sunyaev Broad Institute of M.I.T. and Harvard Current knowledge is from Comparative genomics Experimental systems: yeast reporter assays Potential
Mapping by recurrence and modelling the mutation rate Shamil Sunyaev Broad Institute of M.I.T. and Harvard Current knowledge is from Comparative genomics Experimental systems: yeast reporter assays Potential
Assignment 5: Integrative epigenomics analysis
 Assignment 5: Integrative epigenomics analysis Due date: Friday, 2/24 10am. Note: no late assignments will be accepted. Introduction CpG islands (CGIs) are important regulatory regions in the genome. What
Assignment 5: Integrative epigenomics analysis Due date: Friday, 2/24 10am. Note: no late assignments will be accepted. Introduction CpG islands (CGIs) are important regulatory regions in the genome. What
Cellular Reproduction, Part 1: Mitosis Lecture 10 Fall 2008
 Cell Theory 1 Cellular Reproduction, Part 1: Mitosis Lecture 10 Fall 2008 Cell theory: All organisms are made of cells All cells arise from preexisting cells How do new cells arise? Cell division the reproduction
Cell Theory 1 Cellular Reproduction, Part 1: Mitosis Lecture 10 Fall 2008 Cell theory: All organisms are made of cells All cells arise from preexisting cells How do new cells arise? Cell division the reproduction
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis
 APPLICATION NOTE Cell-Free DNA Isolation Kit A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis Abstract Circulating cell-free DNA (cfdna) has been shown
APPLICATION NOTE Cell-Free DNA Isolation Kit A complete next-generation sequencing workfl ow for circulating cell-free DNA isolation and analysis Abstract Circulating cell-free DNA (cfdna) has been shown
Epigenetics. Lyle Armstrong. UJ Taylor & Francis Group. f'ci Garland Science NEW YORK AND LONDON
 ... Epigenetics Lyle Armstrong f'ci Garland Science UJ Taylor & Francis Group NEW YORK AND LONDON Contents CHAPTER 1 INTRODUCTION TO 3.2 CHROMATIN ARCHITECTURE 21 THE STUDY OF EPIGENETICS 1.1 THE CORE
... Epigenetics Lyle Armstrong f'ci Garland Science UJ Taylor & Francis Group NEW YORK AND LONDON Contents CHAPTER 1 INTRODUCTION TO 3.2 CHROMATIN ARCHITECTURE 21 THE STUDY OF EPIGENETICS 1.1 THE CORE
Systems Analysis Of Chromatin-Related Protein Complexes In Cancer READ ONLINE
 Systems Analysis Of Chromatin-Related Protein Complexes In Cancer READ ONLINE If looking for the book Systems Analysis of Chromatin-Related Protein Complexes in Cancer in pdf format, then you have come
Systems Analysis Of Chromatin-Related Protein Complexes In Cancer READ ONLINE If looking for the book Systems Analysis of Chromatin-Related Protein Complexes in Cancer in pdf format, then you have come
Session 6: Integration of epigenetic data. Peter J Park Department of Biomedical Informatics Harvard Medical School July 18-19, 2016
 Session 6: Integration of epigenetic data Peter J Park Department of Biomedical Informatics Harvard Medical School July 18-19, 2016 Utilizing complimentary datasets Frequent mutations in chromatin regulators
Session 6: Integration of epigenetic data Peter J Park Department of Biomedical Informatics Harvard Medical School July 18-19, 2016 Utilizing complimentary datasets Frequent mutations in chromatin regulators
oocytes were pooled for RT-PCR analysis. The number of PCR cycles was 35. Two
 Supplementary Fig. 1 a Kdm3a Kdm4b β-actin Oocyte Testis Oocyte Testis Oocyte Testis b 1.8 Relative expression.6.4.2 Kdm3a Kdm4b RT-PCR analysis of Kdm3a and Kdm4b expression in oocytes and testes. Twenty
Supplementary Fig. 1 a Kdm3a Kdm4b β-actin Oocyte Testis Oocyte Testis Oocyte Testis b 1.8 Relative expression.6.4.2 Kdm3a Kdm4b RT-PCR analysis of Kdm3a and Kdm4b expression in oocytes and testes. Twenty
Circular RNAs (circrnas) act a stable mirna sponges
 Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Breast cancer. Risk factors you cannot change include: Treatment Plan Selection. Inferring Transcriptional Module from Breast Cancer Profile Data
 Breast cancer Inferring Transcriptional Module from Breast Cancer Profile Data Breast Cancer and Targeted Therapy Microarray Profile Data Inferring Transcriptional Module Methods CSC 177 Data Warehousing
Breast cancer Inferring Transcriptional Module from Breast Cancer Profile Data Breast Cancer and Targeted Therapy Microarray Profile Data Inferring Transcriptional Module Methods CSC 177 Data Warehousing
Epigenetics DNA methylation. Biosciences 741: Genomics Fall, 2013 Week 13. DNA Methylation
 Epigenetics DNA methylation Biosciences 741: Genomics Fall, 2013 Week 13 DNA Methylation Most methylated cytosines are found in the dinucleotide sequence CG, denoted mcpg. The restriction enzyme HpaII
Epigenetics DNA methylation Biosciences 741: Genomics Fall, 2013 Week 13 DNA Methylation Most methylated cytosines are found in the dinucleotide sequence CG, denoted mcpg. The restriction enzyme HpaII
Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research
 Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research Application Note Authors John McGuigan, Megan Manion,
Mutation Detection and CNV Analysis for Illumina Sequencing data from HaloPlex Target Enrichment Panels using NextGENe Software for Clinical Research Application Note Authors John McGuigan, Megan Manion,
Iso-Seq Method Updates and Target Enrichment Without Amplification for SMRT Sequencing
 Iso-Seq Method Updates and Target Enrichment Without Amplification for SMRT Sequencing PacBio Americas User Group Meeting Sample Prep Workshop June.27.2017 Tyson Clark, Ph.D. For Research Use Only. Not
Iso-Seq Method Updates and Target Enrichment Without Amplification for SMRT Sequencing PacBio Americas User Group Meeting Sample Prep Workshop June.27.2017 Tyson Clark, Ph.D. For Research Use Only. Not
Chapter 11 How Genes Are Controlled
 Chapter 11 How Genes Are Controlled PowerPoint Lectures for Biology: Concepts & Connections, Sixth Edition Campbell, Reece, Taylor, Simon, and Dickey Copyright 2009 Pearson Education, Inc. Lecture by Mary
Chapter 11 How Genes Are Controlled PowerPoint Lectures for Biology: Concepts & Connections, Sixth Edition Campbell, Reece, Taylor, Simon, and Dickey Copyright 2009 Pearson Education, Inc. Lecture by Mary
Gene Regulation. Bacteria. Chapter 18: Regulation of Gene Expression
 Chapter 18: Regulation of Gene Expression A Biology 2013 1 Gene Regulation rokaryotes and eukaryotes alter their gene expression in response to their changing environment In multicellular eukaryotes, gene
Chapter 18: Regulation of Gene Expression A Biology 2013 1 Gene Regulation rokaryotes and eukaryotes alter their gene expression in response to their changing environment In multicellular eukaryotes, gene
EPIGENOMICS PROFILING SERVICES
 EPIGENOMICS PROFILING SERVICES Chromatin analysis DNA methylation analysis RNA-seq analysis Diagenode helps you uncover the mysteries of epigenetics PAGE 3 Integrative epigenomics analysis DNA methylation
EPIGENOMICS PROFILING SERVICES Chromatin analysis DNA methylation analysis RNA-seq analysis Diagenode helps you uncover the mysteries of epigenetics PAGE 3 Integrative epigenomics analysis DNA methylation
Chapter 18 Regulation of Gene Expression
 Chapter 18 Regulation of Gene Expression Differential Expression of Genes Prokaryotes and eukaryotes precisely regulate gene expression in response to environmental conditions In multicellular eukaryotes,
Chapter 18 Regulation of Gene Expression Differential Expression of Genes Prokaryotes and eukaryotes precisely regulate gene expression in response to environmental conditions In multicellular eukaryotes,
Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified
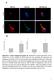 Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified for RFP-LC3 puncta (red dots) representing both autolysosomes
Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified for RFP-LC3 puncta (red dots) representing both autolysosomes
Back to the Basics: Methyl-Seq 101
 Back to the Basics: Methyl-Seq 101 Presented By: Alex Siebold, Ph.D. October 9, 2013 Field Applications Scientist Agilent Technologies Life Sciences & Diagnostics Group Life Sciences & Diagnostics Group
Back to the Basics: Methyl-Seq 101 Presented By: Alex Siebold, Ph.D. October 9, 2013 Field Applications Scientist Agilent Technologies Life Sciences & Diagnostics Group Life Sciences & Diagnostics Group
