Supplementary Material
|
|
|
- Maude Wheeler
- 6 years ago
- Views:
Transcription
1 Supplementary Material HLA-DM Captures Partially Empty HLA-DR Molecules for Catalyzed Peptide Removal Anne-Kathrin Anders, Melissa J. Call, Monika-Sarah E. D. Schulze, Kevin D. Fowler, David A. Schuert, Nilufer P. Seth, Eric J. Sunderg & Kai W. Wucherpfennig 1
2 a 2 DR2 WT Protein injection Buffer injection 2 DR2 S53D Fc4 DM WT Fc3 DM Mut2 Fc2 SA 1 5 Fc4 DM WT Fc3 DM Mut2 Fc2 SA Supplementary Figure 1. Minimal ackground inding y a HLA-DR mutant compared to the streptavidin surface. In SPR experiments, the sensor detects refractive index changes when only uffer or protein in uffer is injected. Therefore, it is a widely accepted convention in Biacore studies to sutract the response in a control flow cell from the experimental flow cell. Such a control also addresses non-specific inding. We went one step further to rigorously control for non-specific inding: we designed a HLA-DM mutant that closely resemled the iophysical properties of wild-type HLA- DM ut which had no catalytic activity and did not ind to HLA-DR peptide complexes (HLA-DM Mut2). The HLA-DM Mut2 control surface (flow cell 3, Fc3) did not show increased ackground inding compared to the streptavidin surface (flow cell 2, Fc2) in all experiments that were performed. The two graphs compare results from HLA-DR2 WT (a) and the HLA-DR2 S53D mutant () for three surfaces: HLA-DM WT, HLA-DM Mut2 and streptavidin (SA). HLA-DR2 inding curves were very similar when HLA-DM Mut2 and streptavidin surfaces were compared. Data are representative of more than five (a) and of two () independent experiments, respectively. m m 2
3 a μl/min 25 μl/min 5 μl/min Supplementary Figure 2. Asence of significant reinding of HLA-DR to HLA-DM (mass transport). (a) HLA-DR2 CLIP complexes (2 μm) were run through HLA-DM WT and HLA-DM Mut2 flow cells (1 of immoilized protein) for 5 minutes at the indicated flow rates in a citratephosphate uffer with a ph of 5.35 at 3 C. This was followed y injection of uffer (stage 2) and 2 μm CLIP (stage 3). Binding in the HLA-DM Mut2 flow cell was sutracted from measurements in the HLA-DM WT flow cell. Data are representative of two independent experiments as shown and of more than three independent experiments under similar conditions. 3
4 a Antiody injection 2 αdnp (1:2) αdnp (1:5) αdnp (1:1) Control (1:1) c 1 75 Antiody injection 5 25 αdnp Control Supplementary Figure 3. The complex of HLA-DR and HLA-DM in stage 2 is devoid of peptide. (a) Specific inding of the DNP antiody to a DNP-laeled peptide ound to HLA-DR2. HLA- DR2 CLIP (control, no DNP group on peptide) or HLA-DR2 MBP(85-99)-DNP (5 of each protein) were immoilized on a streptavidin chip. DNP or control antiody (at the indicated dilutions) were injected for 2 min (25 C, 15 μl/min), followed y injection of uffer (citratephosphate uffer, ph 6). Binding in the HLA-DR2 CLIP (no DNP) control flow cell was sutracted from inding in the two experimental flow cells. () The stale complex of HLA-DM and HLA-DR in stage 2 is devoid of peptide. HLA-DR2 CLIP(87-11)-DNP (5 μm) was injected for 1 minutes (25 C, 15 μl/min), followed y injection of uffer and a 2 minute injection of antiody (DNP or control antiody at 2 μg/ml) in citrate-phosphate uffer (ph6). The SPR assay was carried out in consecutive flow cells with immoilized HLA-DM WT or HLA-DM Mut2 (5 of each protein) in citrate-phosphate uffer (ph 5.35). Binding in the HLA-DM Mut2 flow cell was sutracted from measurements in the HLA-DM WT flow cell. Data are representative of two 4
5 independent experiments. (c) The DNP antiody detects the DNP group of a covalently linked peptide. HLA-DR1 HA 6 P 2 -P 11 complex (.5 μm) with a C-terminal DNP group on the linked peptide was injected (as in ), followed y injection of DNP or control antiody in stage 2. 5
6 a DR2-CLIP DR2-MBP ΔFP (mp) DR2-CLIP +DM DR2-MBP +DM DR2-CLIP DR2-MBP Time (min) Supplementary Figure 4. Comparison of the kinetics of HLA-DM inding and HLA-DM catalysis for low-affinity versus high-affinity HLA-DR peptide complexes. (a) Comparison of HLA-DM inding y low-affinity HLA-DR2 CLIP and high-affinity HLA-DR2 MBP complexes. HLA-DR2 complexes preloaded with the indicated peptides (1 μm) were injected for 1 minutes (stage 1), followed y injection of uffer (stage 2) and 5 μm MBP (stage 3). The SPR assay was carried out in consecutive flow cells with 5 of immoilized HLA-DM WT and HLA-DM Mut2 in a citrate-phosphate uffer (ph 5.35) at 37 C at a flow rate of 15 μl/min. Measurements from the HLA-DM Mut2 flow cell were sutracted from data otained from the HLA-DM WT flow cell. HLA-DM inding y HLA-DR2 MBP was 8.7-fold slower compared to HLA-DR2 CLIP, ased on measurement of the initial rates. () Comparison of HLA-DM susceptiility of HLA-DR2 CLIP and HLA-DR2 MBP complexes. Binding of 2 nm Alexa488-laeled MBP to HLA-DR2 (2 nm) preloaded with the indicated peptides was measured in the presence or asence of 1 nm HLA-DM in 5 mm citrate-phosphate uffer (ph 5.3) containing 15 mm NaCl. The initial rate of HLA-DM-catalyzed peptide inding was 11.8-fold slower when HLA-DR2 MBP rather than HLA-DR2 CLIP was used as the input protein in the reaction. Data are representative of two independent experiments as shown (a, ) and of more than three under similar conditions (a) (FP assays were performed in triplicates). 6
7 a Elution c DR1-P 6 Cys no peptide HA6 P2-P11 51 kda 39 kda DRα-HA 6 P 2 -P 11 DRα 28 kda DRβ 14 kda Supplementary Figure 5. Production of homogenous disulfide-linked HLA-DR1 peptide complexes. (a-c) Purification of disulfide-linked complexes of HLA-DR1 P 6 Cys and HA 6 P 2 -P 11 peptide. (a) Removal of excess free DNP-laeled peptide following peptide loading and disulfide ond formation. HLA-DR1 P 6 Cys molecules were separated from DNP-laeled HA 6 P 2 -P 11 peptide y gel filtration (Superose 12). The lue trace (28nm) represents protein asorance, the red 7
8 trace (35 nm) DNP asorance. The peak etween 11-15ml contains the HLA-DR peptide complex, the later peak the free peptide. () Affinity purification of disulfide-linked HLA-DR1 HA 6 P 2 -P 11 complexes y DNP affinity chromatography from other HLA-DR species. Protein and peptide are traced as descried in (a). The earlier peak represents the flow-through, the later peak the peptide-loaded complex (elution with CAPS uffer, ph 11.5). (c) Analysis of disulfide linkage of HA 6 P 2 -P 11 peptide to the HLA-DRα chain y non-reducing SDS PAGE. Data are representative of at least four independent experiments. 8
9 Elution Elution a c DR1-HA DR1-HA 6 DR1-HA 6 P 2 -P 11 IP: W4F.5B L F1 W4F.5B L F1 W4F.5B L F1 98 kda 62 kda 49 kda 38 kda 28 kda IB: 17 kda 1 DR antiserum 2 α-rait HRP d ΔFP (mp) DR1-HA P 2 -P 11 DR1-HA 6 + DTT P 2 -P 11 no DTT DR1-CLIP low + DTT DR1-CLIP low no DTT 9
10 Supplementary Figure 6. Biochemical characterization of HLA-DR1 molecules with covalently linked full length or partial peptides. (a,) Affinity purification of disulfide-linked HLA-DR1 HA 6 (a) and HLA-DR1 HA 6 P 2 -P 11 () complexes y DNP affinity chromatography from other HLA-DR species. The lue trace (28nm) represents protein asorance, the red trace (35 nm) DNP asorance. The earlier peak represents the flow through, the later peak the peptide-loaded complex (elution with CAPS uffer, ph 11.5). The scales of 28 nm and 35 nm readings were adjusted ased on prior experiments with affinity purified, fully loaded HLA-DR peptide complexes such that identical peak heights reflected full occupancy of the peptide inding groove y DNP-laeled peptide. Based on these measurements, the peptide inding groove of oth HLA-DR1 HA 6 (a) and HLA- DR1 HA 6 P 2 -P 11 () was fully occupied. (c) Immunoprecipitation of covalent HLA-DR1-peptide complexes y conformation-sensitive antiodies to HLA-DRα and HLA-DRβ chains. HLA-DR1- HA peptide complexes (without covalent linkage) as well as HLA-DR1 HA 6 and HLA-DR1 HA 6 P 2 -P 11 complexes were immunoprecipitated with a control ma (W4F.5B), ma L243 (to HLA-DRα chain) or ma 9.3.F1 (to HLA-DRβ chain). The Western lot was laeled with a polyclonal anti-dr serum, followed y a polyclonal HRP-laeled rait IgG antiserum. (d) Lowaffinity HA 6 P 2 -P 11 peptide exchanges quickly from HLA-DR1 upon reduction of the disulfide ond. Binding of Alexa488-laeled HA (65 nm) to the indicated HLA-DR1 complexes (2 nm) was followed y FP with or without prior reduction of the disulfide ond etween the peptide and the HLA-DRα helix. Equilirium inding data are shown for reactions in which either the HLA-DR1 HA 6 P 2 -P 11 complex or the HLA-DR1 CLIP low complex (positive control) were used as input proteins. Data are representative of at least two independent experiments (FP assay performed in triplicates). 1
11 a C 3 C 25 C 2 C C 3 C 25 C 2 C Supplementary Figure 7. HLA-DM inding to HLA-DR loaded with full-length peptide is highly temperature dependent. (a,) Temperature profiles for HLA-DM inding y HLA-DR molecules loaded with CLIP peptide (HLA-DR2 CLIP, a) or a low-affinity CLIP variant (HLA-DR1 CLIP low, ). (a,) HLA-DR2 CLIP complexes (.5 μm, a) and HLA-DR1 CLIP low complexes (1 μm, ) were run over the HLA-DM WT and HLA-DM Mut2 flow cells (5 of immoilized proteins) at a flow rate of 15 μl/min in a citrate-phosphate uffer with a ph of 5.35 at the indicated temperatures. This was followed y injection of uffer and injection of 2 μm CLIP (a) or 5 μm HA (). Binding in the HLA-DM Mut2 flow cell was sutracted from measurements in the HLA-DM WT flow cell. Data are representative of two independent experiments shown (a, ) and of at least four independent experiments under similar conditions (a). 11
12 a k a (M -1 s -1 ) k d (s -1 ) c Temperature ( C) Temperature ( C) K D (M) Temperature ( C) Supplementary Figure 8. Kinetic parameters for HLA-DM inding to HLA-DR1 HA 6 P 2 -P 11 complex. (a-c) HLA-DR1 HA 6 P 2 -P 11 complexes (.1 to 5 μm) were injected into HLA-DM WT and HLA- DM Mut2 flow cells at temperatures ranging from 2 to 37 C. The datasets were fitted as in Fig. 7c and k a, k d and K d values were plotted against the corresponding temperatures. Data are representative of two independent experiments. 12
13 a ph 4.6 ph 5. ph 5.5 FP (mp) 25 2 ph 5.6 ph 5.11 ph Time (min) Supplementary Figure 9. Slower dissociation of HLA-DM HLA-DR complex at lower ph. (a) Low ph increases HLA-DM inding to HLA-DR CLIP and slows dissociation of HLA-DM HLA-DR complexes. HLA-DR2 CLIP complexes (5 μm) were run over the HLA-DM flow cell (5 of immoilized HLA-DM) and the SA flow cell for 1 minutes at a flow rate of 15 μl/min in a citrate-phosphate uffer with the indicated ph at 3 C, followed y injection of uffer (stage 2) and CLIP at a concentration of 2 μm (stage 3). Binding in the SA flow cell was sutracted from the measurements in the HLA-DM WT flow cell. () ph dependent peptide inding to HLA-DR2 molecules after photolysis of ound peptide. HLA-DR2 MBP complexes preloaded with UV-cleavale MBP peptide were UV irradiated for 1 minutes in 1 mm Tris ph 7. on ice to generate empty HLA-DR2 molecules. Binding of Alexa-488 laeled MBP (3 nm) to empty HLA-DR2 (15 nm) was measured directly following UV-cleavage using FP at the indicated ph. Data are representative of at least two independent experiments (FP assay performed in triplicates). 13
14 Stage 1 Stage 2 Stage 3 peptide DRα DM 5 High affinity peptide inding; DM dissociation high affinity peptide DRβ DR with tightly ound peptide; no DM recognition Partial peptide dissociation DM recognition Stale DM-DR complex low affinity peptide 6 Editing - transient occupancy Supplementary Figure 1. Model of the interaction of HLA-DM with HLA-DR. HLA-DR molecules with a fully ound peptide (1) do not interact with HLA-DM. Partial peptide dissociation at the N-terminus (2) enales HLA-DM inding (3). HLA-DM catalyzes peptide removal and forms a long-lived complex with HLA-DR in the asence of peptide (4). Binding of a high-affinity peptide to this complex will most likely result in its dissociation (5), while the HLA- DM HLA-DR complex is more likely to remain intact upon inding of a low-affinity peptide (6). Low-affinity peptide can e removed y the catalytic action of HLA-DM (editing) and the complex of empty HLA-DR and HLA-DM (4) is then again availale for peptide inding. The conformational change in HLA-DR molecules upon release of the N-terminal peptide segment is indicated here y a change in color, compared to HLA-DR molecules with tightly ound peptides. For all individual molecular steps, the corresponding stages of SPR experiments are indicated (stages 1-3). 14
Lane: 1. Spectra BR protein ladder 2. PFD 3. TERM 4. 3-way connector 5. 2-way connector
 kda 1 2 3 4 5 26 14 1 7 Lane: 1. Spectra BR protein ladder 2. PFD 3. TERM 4. 3-way connector 5. 2-way connector 5 4 35 25 15 1 Supplementary Figure 1. SDS-PAGE of acterially expressed and purified proteins.
kda 1 2 3 4 5 26 14 1 7 Lane: 1. Spectra BR protein ladder 2. PFD 3. TERM 4. 3-way connector 5. 2-way connector 5 4 35 25 15 1 Supplementary Figure 1. SDS-PAGE of acterially expressed and purified proteins.
Hidenobu Kanda, Rebecca Newton, Russell Klein, Yuka Morita, Michael D. Gunn & Steven D. Rosen
 Autotaxin, a lysophosphatidic acid-producing ectoenzyme, promotes lymphocyte entry into secondary lymphoid organs Hidenou Kanda, Reecca Newton, Russell Klein, Yuka Morita, Michael D. Gunn & Steven D. Rosen
Autotaxin, a lysophosphatidic acid-producing ectoenzyme, promotes lymphocyte entry into secondary lymphoid organs Hidenou Kanda, Reecca Newton, Russell Klein, Yuka Morita, Michael D. Gunn & Steven D. Rosen
Surface plasmon resonance (SPR) analysis
 Surface plasmon resonance (SPR) analysis Soluble CD8αα and was manufactured as described previously. 1,2 The C12C heterodimeric TCR was produced using an engineered disulfide bridge between the constant
Surface plasmon resonance (SPR) analysis Soluble CD8αα and was manufactured as described previously. 1,2 The C12C heterodimeric TCR was produced using an engineered disulfide bridge between the constant
SUPPLEMENTAL INFORMATION
 SUPPLEMENTAL INFORMATION EXPERIMENTAL PROCEDURES Tryptic digestion protection experiments - PCSK9 with Ab-3D5 (1:1 molar ratio) in 50 mm Tris, ph 8.0, 150 mm NaCl was incubated overnight at 4 o C. The
SUPPLEMENTAL INFORMATION EXPERIMENTAL PROCEDURES Tryptic digestion protection experiments - PCSK9 with Ab-3D5 (1:1 molar ratio) in 50 mm Tris, ph 8.0, 150 mm NaCl was incubated overnight at 4 o C. The
Crystallization-grade After D After V3 cocktail. Time (s) Time (s) Time (s) Time (s) Time (s) Time (s)
 Ligand Type Name 6 Crystallization-grade After 447-52D After V3 cocktail Receptor CD4 Resonance Units 5 1 5 1 5 1 Broadly neutralizing antibodies 2G12 VRC26.9 Resonance Units Resonance Units 3 1 15 1 5
Ligand Type Name 6 Crystallization-grade After 447-52D After V3 cocktail Receptor CD4 Resonance Units 5 1 5 1 5 1 Broadly neutralizing antibodies 2G12 VRC26.9 Resonance Units Resonance Units 3 1 15 1 5
Supplementary Information CAND1 controls in vivo dynamics of the Cullin 1-RING ubiquitin ligase repertoire
 Supplementary Information CAD1 controls in vivo dynamics of the Cullin 1-RIG uiquitin ligase repertoire Shuangding Wu, Wenhong Zhu, Tina han, Julia I. Toth, Matthew D. Petroski, Dieter A. Wolf 1 c knd1
Supplementary Information CAD1 controls in vivo dynamics of the Cullin 1-RIG uiquitin ligase repertoire Shuangding Wu, Wenhong Zhu, Tina han, Julia I. Toth, Matthew D. Petroski, Dieter A. Wolf 1 c knd1
2. Which of the following amino acids is most likely to be found on the outer surface of a properly folded protein?
 Name: WHITE Student Number: Answer the following questions on the computer scoring sheet. 1 mark each 1. Which of the following amino acids would have the highest relative mobility R f in normal thin layer
Name: WHITE Student Number: Answer the following questions on the computer scoring sheet. 1 mark each 1. Which of the following amino acids would have the highest relative mobility R f in normal thin layer
glpx Rv1100 Probe 4.4 kb 75 kda 50 kda 37 kda 25 kda 20 kda
 PvuI 2.0 k PvuI PvuI Rv1101c Rv1100 glpx fum 4.4 k PvuI PvuI ΔglpX Rv1101c Rv1100 HygR fum ΔglpX 5 k 4 k 3 k c 75 kda 50 kda 37 kda GLPX Δ Δ/C GLPX 2 k 1.5 k 25 kda 20 kda PRCB 1 k 0.5 k Supplementary
PvuI 2.0 k PvuI PvuI Rv1101c Rv1100 glpx fum 4.4 k PvuI PvuI ΔglpX Rv1101c Rv1100 HygR fum ΔglpX 5 k 4 k 3 k c 75 kda 50 kda 37 kda GLPX Δ Δ/C GLPX 2 k 1.5 k 25 kda 20 kda PRCB 1 k 0.5 k Supplementary
Supplementary Information
 Supplementary Information Concerted action of cellular JNK and Pin1 restricts HIV1 genome integration to activated CD4 T lymphocytes Lara Manganaro 1, Marina Lusic 1,#, Maria Ines Gutierrez 1, Anna Cereseto
Supplementary Information Concerted action of cellular JNK and Pin1 restricts HIV1 genome integration to activated CD4 T lymphocytes Lara Manganaro 1, Marina Lusic 1,#, Maria Ines Gutierrez 1, Anna Cereseto
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Nair S, Branagan AR, Liu J, Boddupalli CS, Mistry PK, Dhodapkar
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Nair S, Branagan AR, Liu J, Boddupalli CS, Mistry PK, Dhodapkar
Purification of Glucagon3 Interleukin-2 Fusion Protein Derived from E. coli
 Purification of Glucagon3 Interleukin-2 Fusion Protein Derived from E. coli Hye Soon Won Dept. of Chem. Eng. Chungnam National University INTRODUCTION Human interleukin-2(hil-2) - known as T Cell Growth
Purification of Glucagon3 Interleukin-2 Fusion Protein Derived from E. coli Hye Soon Won Dept. of Chem. Eng. Chungnam National University INTRODUCTION Human interleukin-2(hil-2) - known as T Cell Growth
<Supplemental information>
 The Structural Basis of Endosomal Anchoring of KIF16B Kinesin Nichole R. Blatner, Michael I. Wilson, Cai Lei, Wanjin Hong, Diana Murray, Roger L. Williams, and Wonhwa Cho Protein
The Structural Basis of Endosomal Anchoring of KIF16B Kinesin Nichole R. Blatner, Michael I. Wilson, Cai Lei, Wanjin Hong, Diana Murray, Roger L. Williams, and Wonhwa Cho Protein
a Anti-Dab2 RGD Merge
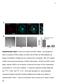 a Anti-Da Merge *** ** Percentage of cells adhered 8 6 4 RGE RAD RGE RAD Supplementary Figure Da are localized at clusters. (a) Endogenous Da is localized at clusters. (), ut not RGE nor RAD peptides can
a Anti-Da Merge *** ** Percentage of cells adhered 8 6 4 RGE RAD RGE RAD Supplementary Figure Da are localized at clusters. (a) Endogenous Da is localized at clusters. (), ut not RGE nor RAD peptides can
Data are contained in multiple tabs in Excel spreadsheets and in CSV files.
 Contents Overview Curves Methods Measuring enzymatic activity (figure 2) Enzyme characterisation (Figure S1, S2) Enzyme kinetics (Table 3) Effect of ph on activity (figure 3B) Effect of metals and inhibitors
Contents Overview Curves Methods Measuring enzymatic activity (figure 2) Enzyme characterisation (Figure S1, S2) Enzyme kinetics (Table 3) Effect of ph on activity (figure 3B) Effect of metals and inhibitors
Structural Characterization of Prion-like Conformational Changes of the Neuronal Isoform of Aplysia CPEB
 Structural Characterization of Prion-like Conformational Changes of the Neuronal Isoform of Aplysia CPEB Bindu L. Raveendra, 1,5 Ansgar B. Siemer, 2,6 Sathyanarayanan V. Puthanveettil, 1,3,7 Wayne A. Hendrickson,
Structural Characterization of Prion-like Conformational Changes of the Neuronal Isoform of Aplysia CPEB Bindu L. Raveendra, 1,5 Ansgar B. Siemer, 2,6 Sathyanarayanan V. Puthanveettil, 1,3,7 Wayne A. Hendrickson,
Analyzing Trace-Level Impurities of a Pharmaceutical Intermediate Using an LCQ Fleet Ion Trap Mass Spectrometer and the Mass Frontier Software Package
 Analyzing Trace-Level Impurities of a Pharmaceutical Intermediate Using an LCQ Fleet Ion Trap Mass Spectrometer and the Mass Frontier Software Package Chromatography and Mass Spectrometry Application Note
Analyzing Trace-Level Impurities of a Pharmaceutical Intermediate Using an LCQ Fleet Ion Trap Mass Spectrometer and the Mass Frontier Software Package Chromatography and Mass Spectrometry Application Note
IP: anti-gfp VPS29-GFP. IP: anti-vps26. IP: anti-gfp - + +
 FAM21 Strump. WASH1 IP: anti- 1 2 3 4 5 6 FAM21 Strump. FKBP IP: anti-gfp VPS29- GFP GFP-FAM21 tail H H/P P H H/P P c FAM21 FKBP Strump. VPS29-GFP IP: anti-gfp 1 2 3 FKBP VPS VPS VPS VPS29 1 = VPS29-GFP
FAM21 Strump. WASH1 IP: anti- 1 2 3 4 5 6 FAM21 Strump. FKBP IP: anti-gfp VPS29- GFP GFP-FAM21 tail H H/P P H H/P P c FAM21 FKBP Strump. VPS29-GFP IP: anti-gfp 1 2 3 FKBP VPS VPS VPS VPS29 1 = VPS29-GFP
The Immunoassay Guide to Successful Mass Spectrometry. Orr Sharpe Robinson Lab SUMS User Meeting October 29, 2013
 The Immunoassay Guide to Successful Mass Spectrometry Orr Sharpe Robinson Lab SUMS User Meeting October 29, 2013 What is it? Hey! Look at that! Something is reacting in here! I just wish I knew what it
The Immunoassay Guide to Successful Mass Spectrometry Orr Sharpe Robinson Lab SUMS User Meeting October 29, 2013 What is it? Hey! Look at that! Something is reacting in here! I just wish I knew what it
The MOLECULES of LIFE
 The MOLECULES of LIFE Physical and Chemical Principles Solutions Manual Prepared by James Fraser and Samuel Leachman Chapter 16 Principles of Enzyme Catalysis Problems True/False and Multiple Choice 1.
The MOLECULES of LIFE Physical and Chemical Principles Solutions Manual Prepared by James Fraser and Samuel Leachman Chapter 16 Principles of Enzyme Catalysis Problems True/False and Multiple Choice 1.
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL Divergent effects of intrinsically active MEK variants on developmental Ras signaling Yogesh Goyal,2,3,4, Granton A. Jindal,2,3,4, José L. Pelliccia 3, Kei Yamaya 2,3, Eyan Yeung
SUPPLEMENTARY MATERIAL Divergent effects of intrinsically active MEK variants on developmental Ras signaling Yogesh Goyal,2,3,4, Granton A. Jindal,2,3,4, José L. Pelliccia 3, Kei Yamaya 2,3, Eyan Yeung
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
االمتحان النهائي لعام 1122
 االمتحان النهائي لعام 1122 Amino Acids : 1- which of the following amino acid is unlikely to be found in an alpha-helix due to its cyclic structure : -phenylalanine -tryptophan -proline -lysine 2- : assuming
االمتحان النهائي لعام 1122 Amino Acids : 1- which of the following amino acid is unlikely to be found in an alpha-helix due to its cyclic structure : -phenylalanine -tryptophan -proline -lysine 2- : assuming
Time after injection (hours) ns ns
 Platelet life span (% iotinylated platelets) 1 8 6 4 2 4 24 48 72 96 Time after injection (hours) 6 4 2 IgG GPIα GPIβ GPII GPVI Receptor expression (GeoMean, fluorescence inteity) Supplementary figure
Platelet life span (% iotinylated platelets) 1 8 6 4 2 4 24 48 72 96 Time after injection (hours) 6 4 2 IgG GPIα GPIβ GPII GPVI Receptor expression (GeoMean, fluorescence inteity) Supplementary figure
A mechanism for glycoconjugate vaccine activation of the adaptive immune system and its implications for vaccine design
 A mechanism for glycoconjugate vaccine activation of the adaptive immune system and its implications for vaccine design Fikri Y. Avci 1,2, Xiangming Li 3, Moriya Tsuji 3, Dennis L. Kasper 1,2* Supplementary
A mechanism for glycoconjugate vaccine activation of the adaptive immune system and its implications for vaccine design Fikri Y. Avci 1,2, Xiangming Li 3, Moriya Tsuji 3, Dennis L. Kasper 1,2* Supplementary
a surface permeabilized
 a surface permeabilized RAW 64.7 P388D1 J774 b CD11b + Ly-6G - Blood Monocytes WT Supplementary Figure 1. Cell surface expression on macrophages and DCs. (a) RAW64.7, P388D1, and J774 cells were subjected
a surface permeabilized RAW 64.7 P388D1 J774 b CD11b + Ly-6G - Blood Monocytes WT Supplementary Figure 1. Cell surface expression on macrophages and DCs. (a) RAW64.7, P388D1, and J774 cells were subjected
بسم هللا الرحمن الرحيم
 بسم هللا الرحمن الرحيم Q1: the overall folding of a single protein subunit is called : -tertiary structure -primary structure -secondary structure -quaternary structure -all of the above Q2 : disulfide
بسم هللا الرحمن الرحيم Q1: the overall folding of a single protein subunit is called : -tertiary structure -primary structure -secondary structure -quaternary structure -all of the above Q2 : disulfide
NC bp. b 1481 bp
 Kcna3 NC 11 178 p 1481 p 346 p c *** *** d relative expression..4.3.2.1 CD4 T cells Kcna1 Kcna2 Kcna3 Kcna4 Kcna Kcna6 Kcna7 Kcnn4 relative expression..4.3.2.1 CD8 T cells Kcna1 Kcna2 Kcna3 Kcna4 Kcna
Kcna3 NC 11 178 p 1481 p 346 p c *** *** d relative expression..4.3.2.1 CD4 T cells Kcna1 Kcna2 Kcna3 Kcna4 Kcna Kcna6 Kcna7 Kcnn4 relative expression..4.3.2.1 CD8 T cells Kcna1 Kcna2 Kcna3 Kcna4 Kcna
Table S1. Total and mapped reads produced for each ChIP-seq sample
 Tale S1. Total and mapped reads produced for each ChIP-seq sample Sample Total Reads Mapped Reads Col- H3K27me3 rep1 125662 1334323 (85.76%) Col- H3K27me3 rep2 9176437 7986731 (87.4%) atmi1a//c H3K27m3
Tale S1. Total and mapped reads produced for each ChIP-seq sample Sample Total Reads Mapped Reads Col- H3K27me3 rep1 125662 1334323 (85.76%) Col- H3K27me3 rep2 9176437 7986731 (87.4%) atmi1a//c H3K27m3
Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random
 S1 Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random Conical Tilt (RCT) reconstruction (left: -50,right:
S1 Supplementary Figure 1 (previous page). EM analysis of full-length GCGR. (a) Exemplary tilt pair images of the GCGR mab23 complex acquired for Random Conical Tilt (RCT) reconstruction (left: -50,right:
RAPID SAMPLE PREPARATION METHODS FOR THE ANALYSIS OF N-LINKED GLYCANS
 RAPID SAMPLE PREPARATION METHODS FOR THE ANALYSIS OF N-LINKED GLYCANS Zoltan Szabo, András Guttman, Tomas Rejtar and Barry L. Karger Barnett Institute, Boston, MA, USA PCT Workshop,Boston, 21 May, 2010.
RAPID SAMPLE PREPARATION METHODS FOR THE ANALYSIS OF N-LINKED GLYCANS Zoltan Szabo, András Guttman, Tomas Rejtar and Barry L. Karger Barnett Institute, Boston, MA, USA PCT Workshop,Boston, 21 May, 2010.
Biology 2180 Laboratory #3. Enzyme Kinetics and Quantitative Analysis
 Biology 2180 Laboratory #3 Name Introduction Enzyme Kinetics and Quantitative Analysis Catalysts are agents that speed up chemical processes and the catalysts produced by living cells are called enzymes.
Biology 2180 Laboratory #3 Name Introduction Enzyme Kinetics and Quantitative Analysis Catalysts are agents that speed up chemical processes and the catalysts produced by living cells are called enzymes.
Supplementary Table 1. Properties of lysates of E. coli strains expressing CcLpxI point mutants
 Supplementary Table 1. Properties of lysates of E. coli strains expressing CcLpxI point mutants Species UDP-2,3- diacylglucosamine hydrolase specific activity (nmol min -1 mg -1 ) Fold vectorcontrol specific
Supplementary Table 1. Properties of lysates of E. coli strains expressing CcLpxI point mutants Species UDP-2,3- diacylglucosamine hydrolase specific activity (nmol min -1 mg -1 ) Fold vectorcontrol specific
Supporting Information
 Supporting Information Dauvillée et al. 10.1073/pnas.0907424106 Fig. S1. Iodine screening of the C. cohnii mutant bank. Each single colony was grown on rich-medium agar plates then vaporized with iodine.
Supporting Information Dauvillée et al. 10.1073/pnas.0907424106 Fig. S1. Iodine screening of the C. cohnii mutant bank. Each single colony was grown on rich-medium agar plates then vaporized with iodine.
Supplementary Data 1. Alanine substitutions and position variants of APNCYGNIPL. Applied in
 Supplementary Data 1. Alanine substitutions and position variants of APNCYGNIPL. Applied in Supplementary Fig. 2 Substitution Sequence Position variant Sequence original APNCYGNIPL original APNCYGNIPL
Supplementary Data 1. Alanine substitutions and position variants of APNCYGNIPL. Applied in Supplementary Fig. 2 Substitution Sequence Position variant Sequence original APNCYGNIPL original APNCYGNIPL
Robert E. Murphy, Arvind Kinhikar, Joselyn Del Rosario, Ryan Preston, Mike Shields, and Nancy Levin
 Combined use of Immunoassay and Two-Dimensional Liquid Chromatography Mass Spectrometry for the Detection and Identification of Metabolites from Biotherapeutic Pharmacokinetic Samples Robert E. Murphy,
Combined use of Immunoassay and Two-Dimensional Liquid Chromatography Mass Spectrometry for the Detection and Identification of Metabolites from Biotherapeutic Pharmacokinetic Samples Robert E. Murphy,
Supplemental information
 Carcinoemryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation y Chiang et al. Supplemental
Carcinoemryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation y Chiang et al. Supplemental
Supplementary Materials. High affinity binding of phosphatidylinositol-4-phosphate. by Legionella pneumophila DrrA
 Supplementary Materials High affinity binding of phosphatidylinositol-4-phosphate by Legionella pneumophila DrrA Running title: Molecular basis of PtdIns(4)P-binding by DrrA Stefan Schoebel, Wulf Blankenfeldt,
Supplementary Materials High affinity binding of phosphatidylinositol-4-phosphate by Legionella pneumophila DrrA Running title: Molecular basis of PtdIns(4)P-binding by DrrA Stefan Schoebel, Wulf Blankenfeldt,
Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers
 Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers Cheryl L. Day, 1,2 Nilufer P. Seth, 3 Michaela Lucas, 4 Heiner Appel, 3 Laurent Gauthier, 3 Georg
Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers Cheryl L. Day, 1,2 Nilufer P. Seth, 3 Michaela Lucas, 4 Heiner Appel, 3 Laurent Gauthier, 3 Georg
Supplemental information
 Supplemental information PI(3)K p11δ controls the sucellular compartmentalization of TLR4 signaling and protects from endotoxic shock Ezra Aksoy, Salma Taoui, David Torres, Sandrine Delauve, Aderrahman
Supplemental information PI(3)K p11δ controls the sucellular compartmentalization of TLR4 signaling and protects from endotoxic shock Ezra Aksoy, Salma Taoui, David Torres, Sandrine Delauve, Aderrahman
TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer
 Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Supplementary Figures
 Supplementary Figures a b c d PDI activity in % ERp72 activity in % 4 3 2 1 1 1 ERp activity in % e ΔRFU min -1 1 1 ERp7 activity in % 1 1 Supplementary Figure 1. Selectivity of the bepristat-mediated
Supplementary Figures a b c d PDI activity in % ERp72 activity in % 4 3 2 1 1 1 ERp activity in % e ΔRFU min -1 1 1 ERp7 activity in % 1 1 Supplementary Figure 1. Selectivity of the bepristat-mediated
UV Tracer TM Maleimide NHS ester
 UV Tracer TM Maleimide HS ester Product o.: 1020 Product ame: UV-Tracer TM Maleimide-HS ester Chemical Structure: Chemical Composition: C 41 H 67 5 18 Molecular Weight: 1014.08 Appearance: Storage: Yellow
UV Tracer TM Maleimide HS ester Product o.: 1020 Product ame: UV-Tracer TM Maleimide-HS ester Chemical Structure: Chemical Composition: C 41 H 67 5 18 Molecular Weight: 1014.08 Appearance: Storage: Yellow
Supplementary Material
 Supplementary Material Materials and methods Enzyme assay The enzymatic activity of -glucosidase toward salicin was measured with the Miller method (Miller, 1959) using glucose as the standard. A total
Supplementary Material Materials and methods Enzyme assay The enzymatic activity of -glucosidase toward salicin was measured with the Miller method (Miller, 1959) using glucose as the standard. A total
Aminoglycoside activity observed on single pre-translocation ribosome complexes
 correction notice Nat. Chem. Biol. 6, 54 62 (2010) Aminoglycoside activity observed on single pre-translocation ribosome complexes Michael B Feldman, Daniel S Terry, Roger B Altman & Scott C Blanchard
correction notice Nat. Chem. Biol. 6, 54 62 (2010) Aminoglycoside activity observed on single pre-translocation ribosome complexes Michael B Feldman, Daniel S Terry, Roger B Altman & Scott C Blanchard
Manja Henze, Dorothee Merker and Lothar Elling. 1. Characteristics of the Recombinant β-glycosidase from Pyrococcus
 S1 of S17 Supplementary Materials: Microwave-Assisted Synthesis of Glycoconjugates by Transgalactosylation with Recombinant Thermostable β-glycosidase from Pyrococcus Manja Henze, Dorothee Merker and Lothar
S1 of S17 Supplementary Materials: Microwave-Assisted Synthesis of Glycoconjugates by Transgalactosylation with Recombinant Thermostable β-glycosidase from Pyrococcus Manja Henze, Dorothee Merker and Lothar
Mitochondrial Trifunctional Protein (TFP) Protein Quantity Microplate Assay Kit
 PROTOCOL Mitochondrial Trifunctional Protein (TFP) Protein Quantity Microplate Assay Kit DESCRIPTION Mitochondrial Trifunctional Protein (TFP) Protein Quantity Microplate Assay Kit Sufficient materials
PROTOCOL Mitochondrial Trifunctional Protein (TFP) Protein Quantity Microplate Assay Kit DESCRIPTION Mitochondrial Trifunctional Protein (TFP) Protein Quantity Microplate Assay Kit Sufficient materials
Simultaneous targeting of two ligand-binding sites on VEGFR2 using biparatopic Affibody molecules results in dramatically improved affinity
 Simultaneous targeting of two ligand-binding sites on VEGFR2 using biparatopic Affibody molecules results in dramatically improved affinity Filippa Fleetwood 1, Susanne Klint 2, Martin Hanze 1, Elin Gunneriusson
Simultaneous targeting of two ligand-binding sites on VEGFR2 using biparatopic Affibody molecules results in dramatically improved affinity Filippa Fleetwood 1, Susanne Klint 2, Martin Hanze 1, Elin Gunneriusson
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table.
 Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tale. Type of file: VI Title of file for HTML: Supplementary Movie 1 Description:
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tale. Type of file: VI Title of file for HTML: Supplementary Movie 1 Description:
Supplementary Information
 Supplementary Information Targeted Disruption of the EZH2/EED Complex Inhibits EZH2- dependent Cancer Woojin Kim 1,2,3, Gregory H. Bird 2,3,4, Tobias Neff 5, Guoji Guo 1,2,3, Marc A. Kerenyi 1,2,3, Loren
Supplementary Information Targeted Disruption of the EZH2/EED Complex Inhibits EZH2- dependent Cancer Woojin Kim 1,2,3, Gregory H. Bird 2,3,4, Tobias Neff 5, Guoji Guo 1,2,3, Marc A. Kerenyi 1,2,3, Loren
Study of different types of ubiquitination
 Study of different types of ubiquitination Rudi Beyaert (rudi.beyaert@irc.vib-ugent.be) VIB UGent Center for Inflammation Research Ghent, Belgium VIB Training Novel Proteomics Tools: Identifying PTMs October
Study of different types of ubiquitination Rudi Beyaert (rudi.beyaert@irc.vib-ugent.be) VIB UGent Center for Inflammation Research Ghent, Belgium VIB Training Novel Proteomics Tools: Identifying PTMs October
Chapter 8. Interaction between the phosphatidylinositol 3- kinase SH3 domain and a photocleavable cyclic peptide
 Interaction between the phosphatidylinositol 3- kinase SH3 domain and a photocleavable cyclic peptide 129 Abstract The interaction of the PI3K SH3 domain with a cyclic photocleavable peptide and the linear
Interaction between the phosphatidylinositol 3- kinase SH3 domain and a photocleavable cyclic peptide 129 Abstract The interaction of the PI3K SH3 domain with a cyclic photocleavable peptide and the linear
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/439/ra78/dc1 Supplementary Materials for Small heterodimer partner mediates liver X receptor (LXR) dependent suppression of inflammatory signaling by promoting
www.sciencesignaling.org/cgi/content/full/9/439/ra78/dc1 Supplementary Materials for Small heterodimer partner mediates liver X receptor (LXR) dependent suppression of inflammatory signaling by promoting
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
Biological Sciences 4087 Exam I 9/20/11
 Name: Biological Sciences 4087 Exam I 9/20/11 Total: 100 points Be sure to include units where appropriate. Show all calculations. There are 5 pages and 11 questions. 1.(20pts)A. If ph = 4.6, [H + ] =
Name: Biological Sciences 4087 Exam I 9/20/11 Total: 100 points Be sure to include units where appropriate. Show all calculations. There are 5 pages and 11 questions. 1.(20pts)A. If ph = 4.6, [H + ] =
Tivadar Orban, Beata Jastrzebska, Sayan Gupta, Benlian Wang, Masaru Miyagi, Mark R. Chance, and Krzysztof Palczewski
 Structure, Volume Supplemental Information Conformational Dynamics of Activation for the Pentameric Complex of Dimeric G Protein-Coupled Receptor and Heterotrimeric G Protein Tivadar Orban, Beata Jastrzebska,
Structure, Volume Supplemental Information Conformational Dynamics of Activation for the Pentameric Complex of Dimeric G Protein-Coupled Receptor and Heterotrimeric G Protein Tivadar Orban, Beata Jastrzebska,
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. (a) Uncropped version of Fig. 2a. RM indicates that the translation was done in the absence of rough mcirosomes. (b) LepB construct containing the GGPG-L6RL6-
Supplementary Figures Supplementary Figure 1. (a) Uncropped version of Fig. 2a. RM indicates that the translation was done in the absence of rough mcirosomes. (b) LepB construct containing the GGPG-L6RL6-
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Beck et al., http://www.jcb.org/cgi/content/full/jcb.201011027/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Membrane binding of His-tagged proteins to Ni-liposomes.
Supplemental material Beck et al., http://www.jcb.org/cgi/content/full/jcb.201011027/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Membrane binding of His-tagged proteins to Ni-liposomes.
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 The UBL and RING1 interface remains associated in the complex structures of Parkin and pub. a) Asymmetric Unit of crystal structure of UBLR0RBR and pub complex showing UBL (green),
Supplementary Figure 1 The UBL and RING1 interface remains associated in the complex structures of Parkin and pub. a) Asymmetric Unit of crystal structure of UBLR0RBR and pub complex showing UBL (green),
Supplementary Figure 1. Ent inhibits LPO activity in a dose- and time-dependent fashion.
 Supplementary Information Supplementary Figure 1. Ent inhibits LPO activity in a dose- and time-dependent fashion. Various concentrations of Ent, DHBA or ABAH were pre-incubated for 10 min with LPO (50
Supplementary Information Supplementary Figure 1. Ent inhibits LPO activity in a dose- and time-dependent fashion. Various concentrations of Ent, DHBA or ABAH were pre-incubated for 10 min with LPO (50
Europium Labeling Kit
 Europium Labeling Kit Catalog Number KA2096 100ug *1 Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle of the Assay...
Europium Labeling Kit Catalog Number KA2096 100ug *1 Version: 03 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Intended Use... 3 Background... 3 Principle of the Assay...
Review of Biochemistry
 Review of Biochemistry Chemical bond Functional Groups Amino Acid Protein Structure and Function Proteins are polymers of amino acids. Each amino acids in a protein contains a amino group, - NH 2,
Review of Biochemistry Chemical bond Functional Groups Amino Acid Protein Structure and Function Proteins are polymers of amino acids. Each amino acids in a protein contains a amino group, - NH 2,
Page 1 of 5 Biochemistry I Fall 2017 Practice for Exam2 Dr. Stone Name
 Page 1 of 5 Biochemistry I Fall 2017 Practice for Exam2 Dr. Stone ame o answers will be provided. ere are some constants and equations that may be useful: K a = [+][A-]/[A] p = pka + log [A-]/[A] K a for
Page 1 of 5 Biochemistry I Fall 2017 Practice for Exam2 Dr. Stone ame o answers will be provided. ere are some constants and equations that may be useful: K a = [+][A-]/[A] p = pka + log [A-]/[A] K a for
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/2/4/e1500980/dc1 Supplementary Materials for The crystal structure of human dopamine -hydroxylase at 2.9 Å resolution Trine V. Vendelboe, Pernille Harris, Yuguang
advances.sciencemag.org/cgi/content/full/2/4/e1500980/dc1 Supplementary Materials for The crystal structure of human dopamine -hydroxylase at 2.9 Å resolution Trine V. Vendelboe, Pernille Harris, Yuguang
Supplementary Figure 1
 Supplementary Figure 1 6 HE-50 HE-116 E-1 HE-108 Supplementary Figure 1. Targeted drug response curves of endometrial cancer cells. Endometrial cancer cell lines were incubated with serial dilutions of
Supplementary Figure 1 6 HE-50 HE-116 E-1 HE-108 Supplementary Figure 1. Targeted drug response curves of endometrial cancer cells. Endometrial cancer cell lines were incubated with serial dilutions of
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11095 Supplementary Table 1. Summary of the binding between Angptls and various Igdomain containing receptors as determined by flow cytometry analysis. The results were summarized from
doi:10.1038/nature11095 Supplementary Table 1. Summary of the binding between Angptls and various Igdomain containing receptors as determined by flow cytometry analysis. The results were summarized from
Nature Methods: doi: /nmeth Supplementary Figure 1. Salipro lipid particles.
 Supplementary Figure 1 Salipro lipid particles. (a) Gel filtration analysis of Saposin A after incubation with the indicated detergent solubilised lipid solutions. The generation of Saposin A-lipid complexes
Supplementary Figure 1 Salipro lipid particles. (a) Gel filtration analysis of Saposin A after incubation with the indicated detergent solubilised lipid solutions. The generation of Saposin A-lipid complexes
Supplementary Figure S1 Stereotypic socio-sexual actions when sexually active male quail are paired in a relatively small cage.
 c d e Supplementary Figure S1 Stereotypic socio-sexual actions when sexually active male quail are paired in a relatively small cage. (a) Struts, () Pecks, (c) Gras, (d) Mounts, (e) Cloacal Contact (CC)-like
c d e Supplementary Figure S1 Stereotypic socio-sexual actions when sexually active male quail are paired in a relatively small cage. (a) Struts, () Pecks, (c) Gras, (d) Mounts, (e) Cloacal Contact (CC)-like
PhosFree TM Phosphate Assay Biochem Kit
 PhosFree TM Phosphate Assay Biochem Kit (Cat. # BK050) ORDERING INFORMATION To order by phone: (303) - 322-2254 To order by Fax: (303) - 322-2257 To order by e-mail: cservice@cytoskeleton.com Technical
PhosFree TM Phosphate Assay Biochem Kit (Cat. # BK050) ORDERING INFORMATION To order by phone: (303) - 322-2254 To order by Fax: (303) - 322-2257 To order by e-mail: cservice@cytoskeleton.com Technical
Supplemental Figure 1
 Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
1. For the following reaction, at equilibrium [S] = 5 mm, [P] = 0.5 mm, and k f = 10 s -1. k f
![1. For the following reaction, at equilibrium [S] = 5 mm, [P] = 0.5 mm, and k f = 10 s -1. k f 1. For the following reaction, at equilibrium [S] = 5 mm, [P] = 0.5 mm, and k f = 10 s -1. k f](/thumbs/77/75734355.jpg) 1. For the following reaction, at equilibrium [S] = 5 mm, [P] = 0.5 mm, and k f = 10 s -1. S k f k r P a) Calculate K eq (the equilibrium constant) and k r. b) A catalyst increases k f by a factor of 10
1. For the following reaction, at equilibrium [S] = 5 mm, [P] = 0.5 mm, and k f = 10 s -1. S k f k r P a) Calculate K eq (the equilibrium constant) and k r. b) A catalyst increases k f by a factor of 10
Predictive PP1Ca binding region in BIG3 : 1,228 1,232aa (-KAVSF-) HEK293T cells *** *** *** KPL-3C cells - E E2 treatment time (h)
 Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
Prerequisites Protein purification techniques and protein analytical methods. Basic enzyme kinetics.
 Case 19 Purification of Rat Kidney Sphingosine Kinase Focus concept The purification and kinetic analysis of an enzyme that produces a product important in cell survival is the focus of this study. Prerequisites
Case 19 Purification of Rat Kidney Sphingosine Kinase Focus concept The purification and kinetic analysis of an enzyme that produces a product important in cell survival is the focus of this study. Prerequisites
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION. Bacterial strains and growth conditions. Streptococcus pneumoniae strain R36A was
 SUPPLEMENTARY INFORMATION Bacterial strains and growth conditions. Streptococcus pneumoniae strain R36A was grown in a casein-based semisynthetic medium (C+Y) supplemented with yeast extract (1 mg/ml of
SUPPLEMENTARY INFORMATION Bacterial strains and growth conditions. Streptococcus pneumoniae strain R36A was grown in a casein-based semisynthetic medium (C+Y) supplemented with yeast extract (1 mg/ml of
a b G75 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server.
 a b G75 2 2 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server. a. Overlay of top 10 models generated by I-TASSER illustrates the potential effect of 7 amino acid insertion
a b G75 2 2 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server. a. Overlay of top 10 models generated by I-TASSER illustrates the potential effect of 7 amino acid insertion
(B D) Three views of the final refined 2Fo-Fc electron density map of the Vpr (red)-ung2 (green) interacting region, contoured at 1.4σ.
 Supplementary Figure 1 Overall structure of the DDB1 DCAF1 Vpr UNG2 complex. (A) The final refined 2Fo-Fc electron density map, contoured at 1.4σ of Vpr, illustrating well-defined side chains. (B D) Three
Supplementary Figure 1 Overall structure of the DDB1 DCAF1 Vpr UNG2 complex. (A) The final refined 2Fo-Fc electron density map, contoured at 1.4σ of Vpr, illustrating well-defined side chains. (B D) Three
Supplementary Figure 1. Using DNA barcode-labeled MHC multimers to generate TCR fingerprints
 Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
G = Energy of activation
 Biochemistry I, CHEM 4400 Exam 2 10:00 Fall 2015 Dr. Stone Name rate forward = k forward [reactants] rate reverse = k reverse [products] K eq = [products]/[reactants] ΔG = ΔH - TΔS ΔG = ΔG + RT ln Q Q
Biochemistry I, CHEM 4400 Exam 2 10:00 Fall 2015 Dr. Stone Name rate forward = k forward [reactants] rate reverse = k reverse [products] K eq = [products]/[reactants] ΔG = ΔH - TΔS ΔG = ΔG + RT ln Q Q
Supplementary Figure S1 Supplementary Figure S2
 Supplementary Figure S A) The blots shown in Figure B were qualified by using Gel-Pro analyzer software (Rockville, MD, USA). The ratio of LC3II/LC3I to actin was then calculated. The data are represented
Supplementary Figure S A) The blots shown in Figure B were qualified by using Gel-Pro analyzer software (Rockville, MD, USA). The ratio of LC3II/LC3I to actin was then calculated. The data are represented
Short, 2 point questions. Be brief, but not vague. Specfic details are needed.
 Biochemistry Exam II Fall 2012 Dr. Stone Name There are 11 short answer/ multiple choice questions worth 2 points each. There are six long answer questions worth a total of 68 points. Short, 2 point questions.
Biochemistry Exam II Fall 2012 Dr. Stone Name There are 11 short answer/ multiple choice questions worth 2 points each. There are six long answer questions worth a total of 68 points. Short, 2 point questions.
Supplementary Figure 1. MAT IIα is Acetylated at Lysine 81.
 IP: Flag a Mascot PTM Modified Mass Error Position Gene Names Score Score Sequence m/z [ppm] 81 MAT2A;AMS2;MATA2 35.6 137.28 _AAVDYQK(ac)VVR_ 595.83-2.28 b Pre-immu After-immu Flag- WT K81R WT K81R / Flag
IP: Flag a Mascot PTM Modified Mass Error Position Gene Names Score Score Sequence m/z [ppm] 81 MAT2A;AMS2;MATA2 35.6 137.28 _AAVDYQK(ac)VVR_ 595.83-2.28 b Pre-immu After-immu Flag- WT K81R WT K81R / Flag
SYNOPSIS STUDIES ON THE PREPARATION AND CHARACTERISATION OF PROTEIN HYDROLYSATES FROM GROUNDNUT AND SOYBEAN ISOLATES
 1 SYNOPSIS STUDIES ON THE PREPARATION AND CHARACTERISATION OF PROTEIN HYDROLYSATES FROM GROUNDNUT AND SOYBEAN ISOLATES Proteins are important in food processing and food product development, as they are
1 SYNOPSIS STUDIES ON THE PREPARATION AND CHARACTERISATION OF PROTEIN HYDROLYSATES FROM GROUNDNUT AND SOYBEAN ISOLATES Proteins are important in food processing and food product development, as they are
Chapter 6. Antigen Presentation to T lymphocytes
 Chapter 6 Antigen Presentation to T lymphocytes Generation of T-cell Receptor Ligands T cells only recognize Ags displayed on cell surfaces These Ags may be derived from pathogens that replicate within
Chapter 6 Antigen Presentation to T lymphocytes Generation of T-cell Receptor Ligands T cells only recognize Ags displayed on cell surfaces These Ags may be derived from pathogens that replicate within
SUPPLEMENTARY INFORMATION
 Complete but curtailed T-cell response to very-low-affinity antigen Dietmar Zehn, Sarah Y. Lee & Michael J. Bevan Supp. Fig. 1: TCR chain usage among endogenous K b /Ova reactive T cells. C57BL/6 mice
Complete but curtailed T-cell response to very-low-affinity antigen Dietmar Zehn, Sarah Y. Lee & Michael J. Bevan Supp. Fig. 1: TCR chain usage among endogenous K b /Ova reactive T cells. C57BL/6 mice
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus
 Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
The effects of ph on Type VII-NA Bovine Intestinal Mucosal Alkaline Phosphatase Activity
 The effects of ph on Type VII-NA Bovine Intestinal Mucosal Alkaline Phosphatase Activity ANDREW FLYNN, DYLAN JONES, ERIC MAN, STEPHEN SHIPMAN, AND SHERMAN TUNG Department of Microbiology and Immunology,
The effects of ph on Type VII-NA Bovine Intestinal Mucosal Alkaline Phosphatase Activity ANDREW FLYNN, DYLAN JONES, ERIC MAN, STEPHEN SHIPMAN, AND SHERMAN TUNG Department of Microbiology and Immunology,
Rescue of mutant rhodopsin traffic by metformin-induced AMPK activation accelerates photoreceptor degeneration Athanasiou et al
 Supplementary Material Rescue of mutant rhodopsin traffic by metformin-induced AMPK activation accelerates photoreceptor degeneration Athanasiou et al Supplementary Figure 1. AICAR improves P23H rod opsin
Supplementary Material Rescue of mutant rhodopsin traffic by metformin-induced AMPK activation accelerates photoreceptor degeneration Athanasiou et al Supplementary Figure 1. AICAR improves P23H rod opsin
Supplementary Figure-1. SDS PAGE analysis of purified designed carbonic anhydrase enzymes. M1-M4 shown in lanes 1-4, respectively, with molecular
 Supplementary Figure-1. SDS PAGE analysis of purified designed carbonic anhydrase enzymes. M1-M4 shown in lanes 1-4, respectively, with molecular weight markers (M). Supplementary Figure-2. Overlay of
Supplementary Figure-1. SDS PAGE analysis of purified designed carbonic anhydrase enzymes. M1-M4 shown in lanes 1-4, respectively, with molecular weight markers (M). Supplementary Figure-2. Overlay of
Supplementary Figure S1. TRAIL-induced necroptosis at acidic phe is dependent on
 Online supplementary information Supplementary Figure S1. TRAIL-induced necroptos at acidic phe is dependent on RIPK1 and RIPK3. (a) HT29 cells were tranently transfected with RIPK1, RIPK3, RIPK1/ RIPK3
Online supplementary information Supplementary Figure S1. TRAIL-induced necroptos at acidic phe is dependent on RIPK1 and RIPK3. (a) HT29 cells were tranently transfected with RIPK1, RIPK3, RIPK1/ RIPK3
Effects of Second Messengers
 Effects of Second Messengers Inositol trisphosphate Diacylglycerol Opens Calcium Channels Binding to IP 3 -gated Channel Cooperative binding Activates Protein Kinase C is required Phosphorylation of many
Effects of Second Messengers Inositol trisphosphate Diacylglycerol Opens Calcium Channels Binding to IP 3 -gated Channel Cooperative binding Activates Protein Kinase C is required Phosphorylation of many
Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) (b) (c) (d) (e) (f) (g) .
 Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) The inhibitory effects of new antibodies (Mab17 and Mab18). They were investigated in in vitro fertilization
Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) The inhibitory effects of new antibodies (Mab17 and Mab18). They were investigated in in vitro fertilization
Nature Immunology: doi: /ni.3631
 Supplementary Figure 1 SPT analyses of Zap70 at the T cell plasma membrane. (a) Total internal reflection fluorescent (TIRF) excitation at 64-68 degrees limits single molecule detection to 100-150 nm above
Supplementary Figure 1 SPT analyses of Zap70 at the T cell plasma membrane. (a) Total internal reflection fluorescent (TIRF) excitation at 64-68 degrees limits single molecule detection to 100-150 nm above
Recombinant Protein Expression Retroviral system
 Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
Influenza B Hemagglutinin / HA ELISA Pair Set
 Influenza B Hemagglutinin / HA ELISA Pair Set Catalog Number : SEK11053 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized
Influenza B Hemagglutinin / HA ELISA Pair Set Catalog Number : SEK11053 To achieve the best assay results, this manual must be read carefully before using this product and the assay is run as summarized
Transient Ribosomal Attenuation Coordinates Protein Synthesis and Co-translational Folding
 SUPPLEMENTARY INFORMATION: Transient Ribosomal Attenuation Coordinates Protein Synthesis and Co-translational Folding Gong Zhang 1,2, Magdalena Hubalewska 1 & Zoya Ignatova 1,2 1 Department of Cellular
SUPPLEMENTARY INFORMATION: Transient Ribosomal Attenuation Coordinates Protein Synthesis and Co-translational Folding Gong Zhang 1,2, Magdalena Hubalewska 1 & Zoya Ignatova 1,2 1 Department of Cellular
FIRST BIOCHEMISTRY EXAM Tuesday 25/10/ MCQs. Location : 102, 105, 106, 301, 302
 FIRST BIOCHEMISTRY EXAM Tuesday 25/10/2016 10-11 40 MCQs. Location : 102, 105, 106, 301, 302 The Behavior of Proteins: Enzymes, Mechanisms, and Control General theory of enzyme action, by Leonor Michaelis
FIRST BIOCHEMISTRY EXAM Tuesday 25/10/2016 10-11 40 MCQs. Location : 102, 105, 106, 301, 302 The Behavior of Proteins: Enzymes, Mechanisms, and Control General theory of enzyme action, by Leonor Michaelis
Mouse Cathepsin B ELISA Kit
 GenWay Biotech, Inc. 6777 Nancy Ridge Drive San Diego, CA 92121 Phone: 858.458.0866 Fax: 858.458.0833 Email: techline@genwaybio.com http://www.genwaybio.com Mouse Cathepsin B ELISA Kit Catalog No. GWB-ZZD154
GenWay Biotech, Inc. 6777 Nancy Ridge Drive San Diego, CA 92121 Phone: 858.458.0866 Fax: 858.458.0833 Email: techline@genwaybio.com http://www.genwaybio.com Mouse Cathepsin B ELISA Kit Catalog No. GWB-ZZD154
Mouse TrkB ELISA Kit
 Mouse TrkB ELISA Kit CATALOG NO: IRKTAH5472 LOT NO: SAMPLE INTENDED USE For quantitative detection of mouse TrkB in cell culture supernates, cell lysates and tissue homogenates. BACKGROUND TrkB receptor
Mouse TrkB ELISA Kit CATALOG NO: IRKTAH5472 LOT NO: SAMPLE INTENDED USE For quantitative detection of mouse TrkB in cell culture supernates, cell lysates and tissue homogenates. BACKGROUND TrkB receptor
Biology Open (2014) 000, 1 10 doi: /bio
 (2014) 000, 1 10 doi:10.1242/bio.201410041 Supplementary Material Michael Brauchle et al. doi: 10.1242/bio.201410041 Fig. S1. Alignment of GFP, sfgfp, egfp, eyfp, mcherry and mruby2. Sequence-based alignment
(2014) 000, 1 10 doi:10.1242/bio.201410041 Supplementary Material Michael Brauchle et al. doi: 10.1242/bio.201410041 Fig. S1. Alignment of GFP, sfgfp, egfp, eyfp, mcherry and mruby2. Sequence-based alignment
Insulin (Human) ELISA Kit Protocol. (Cat. No.:EK )
 Insulin (Human) ELISA Kit Protocol (Cat. No.:EK-035-06) 2 PHOENIX PHARMACEUTICALS, INC. PHOENIX PHARMACEUTICALS, INC. 3 TALE OF CONTENT Introduction and Protocol Overview 4 List of Components 6 Materials
Insulin (Human) ELISA Kit Protocol (Cat. No.:EK-035-06) 2 PHOENIX PHARMACEUTICALS, INC. PHOENIX PHARMACEUTICALS, INC. 3 TALE OF CONTENT Introduction and Protocol Overview 4 List of Components 6 Materials
