Supplementary Information
|
|
|
- Berenice Harris
- 5 years ago
- Views:
Transcription
1 Supplementary Information Concerted action of cellular JNK and Pin1 restricts HIV1 genome integration to activated CD4 T lymphocytes Lara Manganaro 1, Marina Lusic 1,#, Maria Ines Gutierrez 1, Anna Cereseto 2, Giannino Del Sal 3 and Mauro Giacca 1 1 Molecular Medicine Laoratory, International Centre for Genetic Engineering and Biotechnology (ICGEB), Area Science Park, Padriciano 99, Trieste, Italy 2 Molecular Biology Laoratory, Scuola Normale Superiore, 561 Pisa, Italy 3 Laoratorio Nazionale CIB (LNCIB), Area Science Park, Padriciano 99, Trieste and Dipartimento di Scienze della Vita, Universita' degli Studi di Trieste, Via Giorgieri 1, Trieste, Italy Nature Medicine: doi:1.138/nm.212
2 1 Luciferase activity (% of 48hinduced) Time after PHA/IL2 (h): Suppl. Fig. 1. Primary CD4 T cells are efficiently infected with HIV1 only after prolonged stimulation. Primary CD4 T cells were purified from peripheral lood of three normal donors and either left untreated (>95% resting cells; not shown) or stimulated with PHA/IL2 for 1, 18 or 48 hours prior to infection with VSVGNL4Luc. Luciferase activity of the sample stimulated for 48 hours prior to infection was set as 1% (mean±sem of at least three different experiments). Nature Medicine: doi:1.138/nm.212
3 Numer of copies (% of PHA/IL2 at 7 h) c Numer of copies (% of PHA/IL2 at 7 h) Numer of copies (% of PHA/IL2 at 24 h) Early reverse transcripts Time after infection (h) Late reverse transcripts Time after infection (h) LTR circles Time after infection (h) Not induced PHA/IL2 PHA/IL PHA/IL2SP6125 PHA/IL2AZT Not induced PHA/IL2 PHA/IL PHA/IL2SP6125 PHA/IL2AZT Not induced PHA/IL2 PHA/IL PHA/IL2SP6125 PHA/IL2AZT d e f Integrated HIV DNA (% of activated) VSVGNL4Luc PHA/IL2 SP6125 Integrated HIV DNA (% of infectedactivated) Integrated HIV DNA (% of 48 hinduced) HIV1 BRU PHA/IL2 SP6125 (µm) WB: antijnk WB: antituulin p52 p46 Tuulin 4 Suppl. Fig. 2. JNK activity is required for efficient HIV1 cdna integration in primary lymphocytes. (a, ) JNK inhiition does not impair reverse transcription. PHA/IL2 stimulated PBLs were infected with HIV1BRU with or without treatment with SP6125 or in the presence of AZT. The levels of early (a) and late () reverse transcripts were determined at different time points. Values are mean±sem of infection of three different donors after normalization for the amount of total genomic DNA. (c) JNK inhiition reduces 2LTR circle formation. Time course analysis of 2LTR circles in samples infected with HIV1 BRU with or without treatment with SP6125 or in the presence of AZT (mean±sem of infection of three different donors). (d) JNK inhiition locks integration of pseudotyped HIV1NL4luciferase. PBLs were stimulated or not with PHA/IL2 and treated with or with SP6125 or left untreated for 15 h prior to infection with VSVGNL4Luc; the relative levels of integrated DNA were measured 24 h post infection y real time AluPCR. Each sample was normalized for the amount of total genomic DNA using the Lamin B2 B13 primers and proe (mean±sem of at least three different experiments). (e) JNK inhiition locks HIV1 integration in a dose dependent manner. Primary human CD4 T cells, stimulated with PHA/IL2 as aove, were treated with the indicated concentrations of JNK inhiitor SP6125 for 15 h prior to infection with HIV1BRU. The levels of integrated HIV1 DNA were assayed y real time AluPCR. Each sample was normalized for the amount of total genomic DNA (mean±sem of at least three different experiments). (f) Parallel kinetics of increase of the amount of integrated HIV1 and endogenous JNK levels. CD4 Tlymphocytes were infected with HIV1 BRU either in resting conditions or at different times (1, 18 and 48 h) after activation with PHA/IL2, followed y the measurement of the levels of integrated viral DNA y real time AluPCR after additional 24 h postinfection. Efficient viral integration required 48 h of PHAIL2 stimulation, while it was highly ineffective (>1fold less) upon shorter stimulation. Lysates from the same cells were proed y western lotting with antijnk or antituulin antiodies (lower panel). Of notice, the levels of JNK were arely appreciale in unstimulated cells or in cells stimulated for 1 or 18 h, while the protein was clearly detectale only at 48 h, the same time at which HIV1 integration occurred efficiently. Nature Medicine: doi:1.138/nm.212
4 Pep 49 AMC 65 PPep 49 AMC 65 Peptide (ng) PPep Pep P WB: antip total IgG d Phosphorylated/Total (% of control) PD9859 SP6125 SB2358 c PPep Pep HIV1 BRU λppase WB: antip WB: antiflag IP: antiflag P WB: antip affinitypurified Ratio P: λppase λppase e Total levels of (% of control) PD9859 SP6125 SB2358 Suppl. Fig. 3. A novel antiphospho antiody detects phosphorylated HIV1 in vivo phosphorylation is impaired upon treatment with the JNK inhiitor SP6125. (a) Amino acid sequences of the and phospho peptides used for immunization of raits. Serine 57 is the site of JNK phosphorylation. () IgG polyclonal and affinitypurifed antiphospho antiody (antip) recognizes the phospho peptide. Serial dilutions of phosphos57 and S57, lotted on a Protran BA79 memrane, were proed with anti phospho antiserum purified for IgGs (upper memrane). After an additional purification y affinity chromatography, the phosphorylated peptide was exclusively recognized y the antip antiody (lower memrane). (c) Phosphorylated can e detected in the context of viral infection. 15x16 SupT1 cells were infected with wt HIV1 BRU Flag virus (7.6 µg p24) and the cell lysates were immunoprecipitated with an antiflag antiody, followed y visualization with the same antiody or with our affinitypurified anti P antiody. Prior to western lotting, the precipitate was incuated for 1 hour with PPase at 3 C. Results were quantified densitometrically and presented in the graph on the right. (d) Levels of phosphorylation. The graph shows the percentage of phosphorylated protein in transfected cells after cell treatment with the indicated MAPK inhiitors dissolved in. Values are expressed as a percentage of phosphorylated in cells treated with control, after standardization for the total levels shown in (e); (mean±sd of at least three different experiments). (e) Total levels of. The histogram shows the percentage of total protein in cells treated with MAPK inhiitors as compared to control, treated cells (mean±sd of at least three different experiments). Nature Medicine: doi:1.138/nm.212
5 WB: antijnk WB: antiflag FlagLuc Flag p52 p46 Input (1/5) GST GSTNterm GSTCore GSTCterm GST Luc 35 SJNK IP: antiflag WLC Suppl. Fig. 4. HIV1 inds cellular JNK protein in vitro and in vivo. (a) inds endogenous JNK in vivo. Let panel: extracts from HEK 293T cells transfected with Flag or FlagLuciferase, as a control, were immunoprecipitated with antiflag antiody and the immunoprecipitate was analysed for the presence of endogenous JNK with an antijnk1 antiody; the levels of Flagtagged, immunoprecipitated proteins were controlled y immunolotting with an antiflag antiody. Right panel: western lotting using antijnk and antiflag antiodies, as indicated, on whole cell lysates (WLC) efore ummunoprecipitation, to verify the levels of protein expression. () The core domain is required for interaction with JNK1. In vitro translated, radiolaeled JNK1 p46 was incuated with GST fusion proteins corresponding to full length or to domains 15 (Nterminus), (core) and (Cterminus); ound was resolved y gel electrophoresis. The autoradiograpy strip in the middle shows the levels of recovered at the end of the GST pulldown procedure in a representative experiment; for the same experiment, the Coomassie luestained gel on top shows recominant proteins efore exposure. The histogram at the ottom shows the levels of ound (mean±sd) in three independent experiments, expressed as JNK ound (% of input) Nature Medicine: doi:1.138/nm.212
6 Flag Flag(S57A) FlagLuciferase WB: antip3 p3 c (K3R) (S57A) WB: antiflag Luciferase IgH WB: antip IP: antiflag P IP: antiflag WB: antiflag WB: antip3 p3 WCL (K3R) (S57A) p3 WCL d His pmol His(S57A) WB: antiac Ac P IP: antiflag WB: antiflag WCL S Suppl. Fig. 5. HIV1 S57 phosphorylation is independent from Nterminus acetylation and does not affect enzymatic activity. (a) The (S57A) mutant inds endogenous p3 in vivo similar to wt. Extracts from HEK 293T cells transfected with Flag, Flag(S57A) or Flagluciferase, as indicated, were immunoprecipitated with an antiflag antiody and immunolotted with antip3 (top panel) or antiflag (middle panel) antiodies. Whole cell lysates (WCL) was immunolotted with either antip3 or antiflag antiody (ottom panel) to visualize the levels of endogenous p3. () (S57A) is acetylated on lysines at positions 264, 266 and 273 similar to wt (ref. 8). Lysates of HEK 293T cells transfected with Flag, Flag(K3R) in which lysines 264, 266 and 273 are mutated to arginines, or Flag(S57A), with or without p3, were immunoprecipitated with antiflag antiody and immunolotted with an antiacetylated antiody (antiac). The same lysates were immunolotted with the antiflag antiody to verify protein expression levels (ottom gel). (c) mutant defective for acetylation can still e phosphorylated. Extracts of HEK293T cells transfected with Flag, Flag (K3R) or Flag(S57A) were sujected to immunoprecipitation with antiflag antiody and immunolotting with antip. Protein expression levels were verified y westernlot on total cell lysates with antiflag antiody. (d) enzymatic activity is not affected y the S57A mutation. In vitro strand transfer activity of scalar amounts of wt His or His(S57A) (.2, 1 and 5 pmol). In the first lane, the sustrate without His was incuated with the strand transfer uffer alone as a control. S: DNA sustrate; P: catalytic products. Nature Medicine: doi:1.138/nm.212
7 HIV1 BRU PHA/IL2 IgH IgL WB: antiflag IP: antiflag Suppl. Fig. 6. Detection of upon HIV1 infection of activated ut not resting peripheral lood lymphocytes. 15x1 6 primary PBLs, either stimulated or not with PHA/IL2, were infected with wt HIV1BRUFlag virus (7.6 µg p24). Five hours postinfection, was immunoprecipiated and its total levels were determined y immunolotting with an antiflag antiody. Nature Medicine: doi:1.138/nm.212
8 Reverse transcripts (% of infectednot treated) HIV1 BRU Pi Early RT Late RT Suppl. Fig. 7. Levels of early and late reverse transcripts are not altered y Pin1 inhiition or upon infection with the HIV1 BRU (S57A) virus. (a) Pin1 inhiition does not affect reverse transcription. SupT1 cells were infected with HIV1 BRU after treatment with Pin1 inhiitor dissolved in and analyzed for the levels of early and late reverse transcripts (mean±sem of at least three indipendent experiments). () Mutation of Ser 57 does not impair virion production. Westernlot analysis of HIV1BRU and HIV1 BRU (S57A) viral stocks with antiflag (upper panel) or antip17 (matrix) (lower panel) antiodies is shown. (c) Serine 57 mutation does not affect reverse transcription. SupT1 cells were infected with HIV1BRU or HIV1 BRU (S57A). mutant and analyzed for the levels of early and late reverse transcripts (mean±sem of at least three independent experiments). c 12 HIV1 BRU HIV1 BRU (S57A) WB: antiflag WB: antip17 Virion lysate p17 Reverse transcripts (% of wt HIV1infected) Early RT Late RT HIV1 BRU HIV1 BRU (S57A) Nature Medicine: doi:1.138/nm.212
9 HGQVDCGPGIWQLD HGQVNSDLGTWQMD HGQVDCSPGIWQVD HGQVDASPGVWQMD HGQVNAELGTWQMD HGQVNAELGTWQMD AGCVMRSPNHWQAD HIV1 Sutype A HIV1 Sutype B HIV1 Sutype C HIV1 Sutype D HIV1 Sutype F HIV1 Sutype G HIV1 Sutype H HIV1 Sutype K HIV1 Sutype 1 HIV1 Sutype 2 HIV2 SIV(CPZ.CD.9.ANT) SIV(VER.DE.x.AGM3) SIV(MAC.US.x.251_1A11) SIV(SMM.US.x.PGM3) EIAV GGQLKIGPGIWQMD 2. FIV 3. Luciferase activity (normalized) Luciferase activity (normalized) VSVGpNL4Luc SP6125 VSVGpRodLuc SP6125 Nature Medicine: doi:1.138/nm.212 Suppl. Fig. 8. Conservation of the SP motif and effects of JNK inhiition on HIV2 infection. (a) Conservation of the serineproline sequence (position 5758 in HIV1, in red) in the proteins of different HIV1 sutypes and other Lentiviruses. () HIV2 is insensitive to JNK inhiition. HOS CCR5 cells treated with SP6125 dissolved in were infected with VSVGNL4Luciferase virus or with VSVGRODLuciferase. Luciferase activity was assessed 24 hours after infection and normalized for the levels of ATP present in the cells and the amount of virus used for infection. (mean±sd of at least three independent experiments).
Supplementary Information CAND1 controls in vivo dynamics of the Cullin 1-RING ubiquitin ligase repertoire
 Supplementary Information CAD1 controls in vivo dynamics of the Cullin 1-RIG uiquitin ligase repertoire Shuangding Wu, Wenhong Zhu, Tina han, Julia I. Toth, Matthew D. Petroski, Dieter A. Wolf 1 c knd1
Supplementary Information CAD1 controls in vivo dynamics of the Cullin 1-RIG uiquitin ligase repertoire Shuangding Wu, Wenhong Zhu, Tina han, Julia I. Toth, Matthew D. Petroski, Dieter A. Wolf 1 c knd1
Supplementary information. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins
 Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Tel: ; Fax: ;
 Tel.: +98 216 696 9291; Fax: +98 216 696 9291; E-mail: mrasadeghi@pasteur.ac.ir Tel: +98 916 113 7679; Fax: +98 613 333 6380; E-mail: abakhshi_e@ajums.ac.ir A Soluble Chromatin-bound MOI 0 1 5 0 1 5 HDAC2
Tel.: +98 216 696 9291; Fax: +98 216 696 9291; E-mail: mrasadeghi@pasteur.ac.ir Tel: +98 916 113 7679; Fax: +98 613 333 6380; E-mail: abakhshi_e@ajums.ac.ir A Soluble Chromatin-bound MOI 0 1 5 0 1 5 HDAC2
RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh-
 1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
* * A3027. A4623 e A3507 A3507 A3507
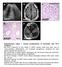 a c L A327 d e A37 A37 A37 Supplementary Figure 1. Clinical manifestations of individuals with mutations. (a) Renal ultrasound of right kidney in A327 reveals small renal cysts, loss of corticomedullary
a c L A327 d e A37 A37 A37 Supplementary Figure 1. Clinical manifestations of individuals with mutations. (a) Renal ultrasound of right kidney in A327 reveals small renal cysts, loss of corticomedullary
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis
 Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Material
 Supplementary Material HLA-DM Captures Partially Empty HLA-DR Molecules for Catalyzed Peptide Removal Anne-Kathrin Anders, Melissa J. Call, Monika-Sarah E. D. Schulze, Kevin D. Fowler, David A. Schuert,
Supplementary Material HLA-DM Captures Partially Empty HLA-DR Molecules for Catalyzed Peptide Removal Anne-Kathrin Anders, Melissa J. Call, Monika-Sarah E. D. Schulze, Kevin D. Fowler, David A. Schuert,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10962 Supplementary Figure 1. Expression of AvrAC-FLAG in protoplasts. Total protein extracted from protoplasts described in Fig. 1a was subjected to anti-flag immunoblot to detect AvrAC-FLAG
doi:10.1038/nature10962 Supplementary Figure 1. Expression of AvrAC-FLAG in protoplasts. Total protein extracted from protoplasts described in Fig. 1a was subjected to anti-flag immunoblot to detect AvrAC-FLAG
Supplementary Figure 1. MAT IIα is Acetylated at Lysine 81.
 IP: Flag a Mascot PTM Modified Mass Error Position Gene Names Score Score Sequence m/z [ppm] 81 MAT2A;AMS2;MATA2 35.6 137.28 _AAVDYQK(ac)VVR_ 595.83-2.28 b Pre-immu After-immu Flag- WT K81R WT K81R / Flag
IP: Flag a Mascot PTM Modified Mass Error Position Gene Names Score Score Sequence m/z [ppm] 81 MAT2A;AMS2;MATA2 35.6 137.28 _AAVDYQK(ac)VVR_ 595.83-2.28 b Pre-immu After-immu Flag- WT K81R WT K81R / Flag
Predictive PP1Ca binding region in BIG3 : 1,228 1,232aa (-KAVSF-) HEK293T cells *** *** *** KPL-3C cells - E E2 treatment time (h)
 Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
Supplementary Figure S1 Stereotypic socio-sexual actions when sexually active male quail are paired in a relatively small cage.
 c d e Supplementary Figure S1 Stereotypic socio-sexual actions when sexually active male quail are paired in a relatively small cage. (a) Struts, () Pecks, (c) Gras, (d) Mounts, (e) Cloacal Contact (CC)-like
c d e Supplementary Figure S1 Stereotypic socio-sexual actions when sexually active male quail are paired in a relatively small cage. (a) Struts, () Pecks, (c) Gras, (d) Mounts, (e) Cloacal Contact (CC)-like
Supplemental information
 Supplemental information PI(3)K p11δ controls the sucellular compartmentalization of TLR4 signaling and protects from endotoxic shock Ezra Aksoy, Salma Taoui, David Torres, Sandrine Delauve, Aderrahman
Supplemental information PI(3)K p11δ controls the sucellular compartmentalization of TLR4 signaling and protects from endotoxic shock Ezra Aksoy, Salma Taoui, David Torres, Sandrine Delauve, Aderrahman
Hidenobu Kanda, Rebecca Newton, Russell Klein, Yuka Morita, Michael D. Gunn & Steven D. Rosen
 Autotaxin, a lysophosphatidic acid-producing ectoenzyme, promotes lymphocyte entry into secondary lymphoid organs Hidenou Kanda, Reecca Newton, Russell Klein, Yuka Morita, Michael D. Gunn & Steven D. Rosen
Autotaxin, a lysophosphatidic acid-producing ectoenzyme, promotes lymphocyte entry into secondary lymphoid organs Hidenou Kanda, Reecca Newton, Russell Klein, Yuka Morita, Michael D. Gunn & Steven D. Rosen
Supplementary Figure S1 Supplementary Figure S2
 Supplementary Figure S A) The blots shown in Figure B were qualified by using Gel-Pro analyzer software (Rockville, MD, USA). The ratio of LC3II/LC3I to actin was then calculated. The data are represented
Supplementary Figure S A) The blots shown in Figure B were qualified by using Gel-Pro analyzer software (Rockville, MD, USA). The ratio of LC3II/LC3I to actin was then calculated. The data are represented
TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer
 Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression
 Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Supplementary Figure 1
 Supplementary Figure 1 a γ-h2ax MDC1 RNF8 FK2 BRCA1 U2OS Cells sgrna-1 ** 60 sgrna 40 20 0 % positive Cells (>5 foci per cell) b ** 80 sgrna sgrna γ-h2ax MDC1 γ-h2ax RNF8 FK2 MDC1 BRCA1 RNF8 FK2 BRCA1
Supplementary Figure 1 a γ-h2ax MDC1 RNF8 FK2 BRCA1 U2OS Cells sgrna-1 ** 60 sgrna 40 20 0 % positive Cells (>5 foci per cell) b ** 80 sgrna sgrna γ-h2ax MDC1 γ-h2ax RNF8 FK2 MDC1 BRCA1 RNF8 FK2 BRCA1
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure S1: Endogenous interaction between RNF2 and H2AX: Whole cell extracts from 293T were subjected to immunoprecipitation with anti-rnf2 or anti-γ-h2ax antibodies
SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure S1: Endogenous interaction between RNF2 and H2AX: Whole cell extracts from 293T were subjected to immunoprecipitation with anti-rnf2 or anti-γ-h2ax antibodies
SUPPLEMENTARY INFORMATION
 Supplementary Table 1. Cell sphingolipids and S1P bound to endogenous TRAF2. Sphingolipid Cell pmol/mg TRAF2 immunoprecipitate pmol/mg Sphingomyelin 4200 ± 250 Not detected Monohexosylceramide 311 ± 18
Supplementary Table 1. Cell sphingolipids and S1P bound to endogenous TRAF2. Sphingolipid Cell pmol/mg TRAF2 immunoprecipitate pmol/mg Sphingomyelin 4200 ± 250 Not detected Monohexosylceramide 311 ± 18
condition. Left panel, the HCT-116 cells were lysed with RIPA buffer containing 0.1%
 FIGURE LEGENDS Supplementary Fig 1 (A) sumoylation pattern detected under denaturing condition. Left panel, the HCT-116 cells were lysed with RIPA buffer containing 0.1% SDS in the presence and absence
FIGURE LEGENDS Supplementary Fig 1 (A) sumoylation pattern detected under denaturing condition. Left panel, the HCT-116 cells were lysed with RIPA buffer containing 0.1% SDS in the presence and absence
Supplemental information
 Carcinoemryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation y Chiang et al. Supplemental
Carcinoemryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation y Chiang et al. Supplemental
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
WT Xid (h) poly(da:dt) Alum. Poly(dA:dT) 0.05
 IL1 (ng/ml) (1 x 1 3 cells) DMSO LFMA13 R46 IL1 (ng/ml) Control shrna TNFa (ng/ml) Alum Nigericin ATP IL1 (ng/ml) IL1 (ng/ml) IL1 (ng/ml) a c f 3 2 1 1 8 6 4 2 Sup 1 8 6 4 DMSO Alum/DMSO Alum/LFMA13 ***
IL1 (ng/ml) (1 x 1 3 cells) DMSO LFMA13 R46 IL1 (ng/ml) Control shrna TNFa (ng/ml) Alum Nigericin ATP IL1 (ng/ml) IL1 (ng/ml) IL1 (ng/ml) a c f 3 2 1 1 8 6 4 2 Sup 1 8 6 4 DMSO Alum/DMSO Alum/LFMA13 ***
Supplemental Materials and Methods Plasmids and viruses Quantitative Reverse Transcription PCR Generation of molecular standard for quantitative PCR
 Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. Neither the activation nor suppression of the MAPK pathway affects the ASK1/Vif interaction. (a, b) HEK293 cells were cotransfected with plasmids encoding
Supplementary Figure 1 Supplementary Figure 1. Neither the activation nor suppression of the MAPK pathway affects the ASK1/Vif interaction. (a, b) HEK293 cells were cotransfected with plasmids encoding
supplementary information
 DOI: 1.138/ncb1 Control Atg7 / NAC 1 1 1 1 (mm) Control Atg7 / NAC 1 1 1 1 (mm) Lamin B Gstm1 Figure S1 Neither the translocation of into the nucleus nor the induction of antioxidant proteins in autophagydeficient
DOI: 1.138/ncb1 Control Atg7 / NAC 1 1 1 1 (mm) Control Atg7 / NAC 1 1 1 1 (mm) Lamin B Gstm1 Figure S1 Neither the translocation of into the nucleus nor the induction of antioxidant proteins in autophagydeficient
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature05732 SUPPLEMENTARY INFORMATION Supplemental Data Supplement Figure Legends Figure S1. RIG-I 2CARD undergo robust ubiquitination a, (top) At 48 h posttransfection with a GST, GST-RIG-I-2CARD
doi: 10.1038/nature05732 SUPPLEMENTARY INFORMATION Supplemental Data Supplement Figure Legends Figure S1. RIG-I 2CARD undergo robust ubiquitination a, (top) At 48 h posttransfection with a GST, GST-RIG-I-2CARD
Neocortex Zbtb20 / NFIA / Sox9
 Neocortex / NFIA / Sox9 Supplementary Figure 1. Expression of, NFIA, and Sox9 in the mouse neocortex at. The lower panels are higher magnification views of the oxed area. Arrowheads indicate triple-positive
Neocortex / NFIA / Sox9 Supplementary Figure 1. Expression of, NFIA, and Sox9 in the mouse neocortex at. The lower panels are higher magnification views of the oxed area. Arrowheads indicate triple-positive
a Anti-Dab2 RGD Merge
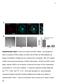 a Anti-Da Merge *** ** Percentage of cells adhered 8 6 4 RGE RAD RGE RAD Supplementary Figure Da are localized at clusters. (a) Endogenous Da is localized at clusters. (), ut not RGE nor RAD peptides can
a Anti-Da Merge *** ** Percentage of cells adhered 8 6 4 RGE RAD RGE RAD Supplementary Figure Da are localized at clusters. (a) Endogenous Da is localized at clusters. (), ut not RGE nor RAD peptides can
Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid.
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
Brd4 Coactivates Transcriptional Activation of NF- B via Specific Binding to Acetylated RelA
 MOLECULAR AND CELLULAR BIOLOGY, Mar. 2009, p. 1375 1387 Vol. 29, No. 5 0270-7306/09/$08.00 0 doi:10.1128/mcb.01365-08 Copyright 2009, American Society for Microbiology. All Rights Reserved. Brd4 Coactivates
MOLECULAR AND CELLULAR BIOLOGY, Mar. 2009, p. 1375 1387 Vol. 29, No. 5 0270-7306/09/$08.00 0 doi:10.1128/mcb.01365-08 Copyright 2009, American Society for Microbiology. All Rights Reserved. Brd4 Coactivates
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/10/471/eaah5085/dc1 Supplementary Materials for Phosphorylation of the exocyst protein Exo84 by TBK1 promotes insulin-stimulated GLUT4 trafficking Maeran Uhm,
www.sciencesignaling.org/cgi/content/full/10/471/eaah5085/dc1 Supplementary Materials for Phosphorylation of the exocyst protein Exo84 by TBK1 promotes insulin-stimulated GLUT4 trafficking Maeran Uhm,
William C. Comb, Jessica E. Hutti, Patricia Cogswell, Lewis C. Cantley, and Albert S. Baldwin
 Molecular Cell, Volume 45 Supplemental Information p85 SH2 Domain Phosphorylation by IKK Promotes Feedback Inhibition of PI3K and Akt in Response to Cellular Starvation William C. Comb, Jessica E. Hutti,
Molecular Cell, Volume 45 Supplemental Information p85 SH2 Domain Phosphorylation by IKK Promotes Feedback Inhibition of PI3K and Akt in Response to Cellular Starvation William C. Comb, Jessica E. Hutti,
supplementary information
 DOI: 10.1038/ncb1875 Figure S1 (a) The 79 surgical specimens from NSCLC patients were analysed by immunohistochemistry with an anti-p53 antibody and control serum (data not shown). The normal bronchi served
DOI: 10.1038/ncb1875 Figure S1 (a) The 79 surgical specimens from NSCLC patients were analysed by immunohistochemistry with an anti-p53 antibody and control serum (data not shown). The normal bronchi served
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/439/ra78/dc1 Supplementary Materials for Small heterodimer partner mediates liver X receptor (LXR) dependent suppression of inflammatory signaling by promoting
www.sciencesignaling.org/cgi/content/full/9/439/ra78/dc1 Supplementary Materials for Small heterodimer partner mediates liver X receptor (LXR) dependent suppression of inflammatory signaling by promoting
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Schwarz et al. Activity-Dependent Ubiquitination of GluA1 Mediates a Distinct AMPAR Endocytosis
 Schwarz et al Activity-Dependent Ubiquitination of GluA1 Mediates a Distinct AMPAR Endocytosis and Sorting Pathway Supplemental Data Supplemental Fie 1: AMPARs undergo activity-mediated ubiquitination
Schwarz et al Activity-Dependent Ubiquitination of GluA1 Mediates a Distinct AMPAR Endocytosis and Sorting Pathway Supplemental Data Supplemental Fie 1: AMPARs undergo activity-mediated ubiquitination
(a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable
 Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table.
 Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tale. Type of file: VI Title of file for HTML: Supplementary Movie 1 Description:
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Tale. Type of file: VI Title of file for HTML: Supplementary Movie 1 Description:
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
glpx Rv1100 Probe 4.4 kb 75 kda 50 kda 37 kda 25 kda 20 kda
 PvuI 2.0 k PvuI PvuI Rv1101c Rv1100 glpx fum 4.4 k PvuI PvuI ΔglpX Rv1101c Rv1100 HygR fum ΔglpX 5 k 4 k 3 k c 75 kda 50 kda 37 kda GLPX Δ Δ/C GLPX 2 k 1.5 k 25 kda 20 kda PRCB 1 k 0.5 k Supplementary
PvuI 2.0 k PvuI PvuI Rv1101c Rv1100 glpx fum 4.4 k PvuI PvuI ΔglpX Rv1101c Rv1100 HygR fum ΔglpX 5 k 4 k 3 k c 75 kda 50 kda 37 kda GLPX Δ Δ/C GLPX 2 k 1.5 k 25 kda 20 kda PRCB 1 k 0.5 k Supplementary
Ctrl CCT7 CCT2 GAPDH * * ** ** * Ctrl Size of LC3 dots (a.u.) mrfp GFP mrfp-gfp-lc3 CCT5 KD CCT7 KD
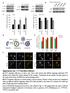 CCT7 Size of dots (a.u.) CCT7 CCT7 /GAPH deitometry (a.u.) /GAPH deitometry (a.u.) a CCT7 Primary cortical neuro 55 CCT5 55 CCT7 55 CCT7 55 55 55 c 3 25 2 DMSO 2 8 Bafilomycin A 5 6 4 5 2 2 3 2 3 d mrfpgfp
CCT7 Size of dots (a.u.) CCT7 CCT7 /GAPH deitometry (a.u.) /GAPH deitometry (a.u.) a CCT7 Primary cortical neuro 55 CCT5 55 CCT7 55 CCT7 55 55 55 c 3 25 2 DMSO 2 8 Bafilomycin A 5 6 4 5 2 2 3 2 3 d mrfpgfp
Recruitment of HBO1 Histone Acetylase and Blocks
 Molecular Cell, Volume 44 Supplemental Information JNK1 Phosphorylation of Cdt1 Inhibits Recruitment of HO1 Histone cetylase and locks Replication Licensing in Response to Stress enoit Miotto and Kevin
Molecular Cell, Volume 44 Supplemental Information JNK1 Phosphorylation of Cdt1 Inhibits Recruitment of HO1 Histone cetylase and locks Replication Licensing in Response to Stress enoit Miotto and Kevin
tom tom 24hpf tom tom 48hpf tom 60hpf tom tom 72hpf tom
 a 24hpf c 48hpf d e 60hpf f g 72hpf h i j k ISV ISV Figure 1. Vascular integrity defects and endothelial regression in mutant emryos. (a,c,e,g,i) Bright-field and (,d,f,h,j) corresponding fluorescent micrographs
a 24hpf c 48hpf d e 60hpf f g 72hpf h i j k ISV ISV Figure 1. Vascular integrity defects and endothelial regression in mutant emryos. (a,c,e,g,i) Bright-field and (,d,f,h,j) corresponding fluorescent micrographs
Supplementary information
 Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
Sirt1 Hmg20b Gm (0.17) 24 (17.3) 877 (857)
 3 (0.17) 24 (17.3) Sirt1 Hmg20 Gm4763 877 (857) c d Suppl. Figure 1. Screen validation for top candidate antagonists of Dot1L (a) Numer of genes with one (gray), two (cyan) or three (red) shrna scored
3 (0.17) 24 (17.3) Sirt1 Hmg20 Gm4763 877 (857) c d Suppl. Figure 1. Screen validation for top candidate antagonists of Dot1L (a) Numer of genes with one (gray), two (cyan) or three (red) shrna scored
Nature Immunology: doi: /ni Supplementary Figure 1. IC261 inhibits a virus-induced type I interferon response.
 Supplementary Figure 1 IC261 inhibits a virus-induced type I interferon response. (a) HEK293T cells were cultured in 384 wells and transiently transfected with 50 ng of the IFN-β promoter-luc construct
Supplementary Figure 1 IC261 inhibits a virus-induced type I interferon response. (a) HEK293T cells were cultured in 384 wells and transiently transfected with 50 ng of the IFN-β promoter-luc construct
p = formed with HCI-001 p = Relative # of blood vessels that formed with HCI-002 Control Bevacizumab + 17AAG Bevacizumab 17AAG
 A.. Relative # of ECs associated with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 ol b p < 0.001 Relative # of blood vessels that formed with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 l b p = 0.002 Control IHC:
A.. Relative # of ECs associated with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 ol b p < 0.001 Relative # of blood vessels that formed with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 l b p = 0.002 Control IHC:
Incorporation of photo-caged lysine (pc-lys) at K273 of human LCK allows specific control of the enzyme activity.
 Supplementary Figure 1 Incorporation of photo-caged lysine (pc-lys) at K273 of human LCK allows specific control of the enzyme activity. (a) Modeling of the kinase domain of LCK with ATP (left) or pc-lys
Supplementary Figure 1 Incorporation of photo-caged lysine (pc-lys) at K273 of human LCK allows specific control of the enzyme activity. (a) Modeling of the kinase domain of LCK with ATP (left) or pc-lys
SUPPLEMENTARY INFORMATION. Supplementary Figures S1-S9. Supplementary Methods
 SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
Time after injection (hours) ns ns
 Platelet life span (% iotinylated platelets) 1 8 6 4 2 4 24 48 72 96 Time after injection (hours) 6 4 2 IgG GPIα GPIβ GPII GPVI Receptor expression (GeoMean, fluorescence inteity) Supplementary figure
Platelet life span (% iotinylated platelets) 1 8 6 4 2 4 24 48 72 96 Time after injection (hours) 6 4 2 IgG GPIα GPIβ GPII GPVI Receptor expression (GeoMean, fluorescence inteity) Supplementary figure
IP: anti-gfp VPS29-GFP. IP: anti-vps26. IP: anti-gfp - + +
 FAM21 Strump. WASH1 IP: anti- 1 2 3 4 5 6 FAM21 Strump. FKBP IP: anti-gfp VPS29- GFP GFP-FAM21 tail H H/P P H H/P P c FAM21 FKBP Strump. VPS29-GFP IP: anti-gfp 1 2 3 FKBP VPS VPS VPS VPS29 1 = VPS29-GFP
FAM21 Strump. WASH1 IP: anti- 1 2 3 4 5 6 FAM21 Strump. FKBP IP: anti-gfp VPS29- GFP GFP-FAM21 tail H H/P P H H/P P c FAM21 FKBP Strump. VPS29-GFP IP: anti-gfp 1 2 3 FKBP VPS VPS VPS VPS29 1 = VPS29-GFP
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Chairoungdua et al., http://www.jcb.org/cgi/content/full/jcb.201002049/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Expression of CD9 and CD82 inhibits Wnt/ -catenin
Supplemental material Chairoungdua et al., http://www.jcb.org/cgi/content/full/jcb.201002049/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Expression of CD9 and CD82 inhibits Wnt/ -catenin
a b G75 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server.
 a b G75 2 2 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server. a. Overlay of top 10 models generated by I-TASSER illustrates the potential effect of 7 amino acid insertion
a b G75 2 2 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server. a. Overlay of top 10 models generated by I-TASSER illustrates the potential effect of 7 amino acid insertion
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
S1a S1b S1c. S1d. S1f S1g S1h SUPPLEMENTARY FIGURE 1. - si sc Il17rd Il17ra bp. rig/s IL-17RD (ng) -100 IL-17RD
 SUPPLEMENTARY FIGURE 1 0 20 50 80 100 IL-17RD (ng) S1a S1b S1c IL-17RD β-actin kda S1d - si sc Il17rd Il17ra rig/s15-574 - 458-361 bp S1f S1g S1h S1i S1j Supplementary Figure 1. Knockdown of IL-17RD enhances
SUPPLEMENTARY FIGURE 1 0 20 50 80 100 IL-17RD (ng) S1a S1b S1c IL-17RD β-actin kda S1d - si sc Il17rd Il17ra rig/s15-574 - 458-361 bp S1f S1g S1h S1i S1j Supplementary Figure 1. Knockdown of IL-17RD enhances
INI1/hSNF5-interaction defective HIV-1 IN mutants exhibit impaired particle morphology, reverse transcription and integration in vivo
 Mathew et al. Retrovirology 213, 1:66 RESEARCH Open Access INI1/hSNF5-interaction defective HIV-1 IN mutants exhibit impaired particle morphology, reverse transcription and integration in vivo Sheeba Mathew
Mathew et al. Retrovirology 213, 1:66 RESEARCH Open Access INI1/hSNF5-interaction defective HIV-1 IN mutants exhibit impaired particle morphology, reverse transcription and integration in vivo Sheeba Mathew
Supplementary Figure 1. Ex vivo IFNγ production by Tregs. Nature Medicine doi: /nm % CD127. Empty SSC 98.79% CD25 CD45RA.
 SSC CD25 1.8% CD127 Empty 98.79% FSC CD45RA CD45RA Foxp3 %IFNγ + cells 4 3 2 1 + IL-12 P =.3 IFNγ p=.9 %IL-4+ cells 3 2 1 IL-4 P =.4 c %IL-1 + cells IFNγ 4 3 2 1 Control Foxp3 IL-1 P =.41.64 4.76 MS 2.96
SSC CD25 1.8% CD127 Empty 98.79% FSC CD45RA CD45RA Foxp3 %IFNγ + cells 4 3 2 1 + IL-12 P =.3 IFNγ p=.9 %IL-4+ cells 3 2 1 IL-4 P =.4 c %IL-1 + cells IFNγ 4 3 2 1 Control Foxp3 IL-1 P =.41.64 4.76 MS 2.96
Recombinant Protein Expression Retroviral system
 Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
Tbk1-TKO! DN cells (%)! 15! 10!
 a! T Cells! TKO! B Cells! TKO! b! CD4! 8.9 85.2 3.4 2.88 CD8! Tbk1-TKO! 1.1 84.8 2.51 2.54 c! DN cells (%)! 4 3 2 1 DP cells (%)! 9 8 7 6 CD4 + SP cells (%)! 5 4 3 2 1 5 TKO! TKO! TKO! TKO! 15 1 5 CD8
a! T Cells! TKO! B Cells! TKO! b! CD4! 8.9 85.2 3.4 2.88 CD8! Tbk1-TKO! 1.1 84.8 2.51 2.54 c! DN cells (%)! 4 3 2 1 DP cells (%)! 9 8 7 6 CD4 + SP cells (%)! 5 4 3 2 1 5 TKO! TKO! TKO! TKO! 15 1 5 CD8
SUPPLEMENTARY FIGURES AND TABLE
 SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
Supplementary Figure 1: Characterisation of phospho-fgfr-y463 antibody. (A)
 Supplementary Figure 1: Characterisation of phospho-fgfr-y463 antibody. (A) Cells over-expressing hfgfr1-pcdna3 (+) or pcdna3 (-) were stimulated for 10 minutes with 50ng/ml FGF2 and lysates immunoblotted
Supplementary Figure 1: Characterisation of phospho-fgfr-y463 antibody. (A) Cells over-expressing hfgfr1-pcdna3 (+) or pcdna3 (-) were stimulated for 10 minutes with 50ng/ml FGF2 and lysates immunoblotted
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
A Slfn2 mutation causes lymphoid and myeloid immunodeficiency due to loss of immune cell quiescence
 Supplementary Information A Slfn mutation causes lymphoid and myeloid immunodeficiency due to loss of immune cell quiescence Michael Berger, Philippe Kres, Karine Crozat, Xiaohong Li, Ben A. Croker, Owen
Supplementary Information A Slfn mutation causes lymphoid and myeloid immunodeficiency due to loss of immune cell quiescence Michael Berger, Philippe Kres, Karine Crozat, Xiaohong Li, Ben A. Croker, Owen
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Nature Methods: doi: /nmeth Supplementary Figure 1
 Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
Supplementary Figure 1 Subtiligase-catalyzed ligations with ubiquitin thioesters and 10-mer biotinylated peptides. (a) General scheme for ligations between ubiquitin thioesters and 10-mer, biotinylated
+ + + IP: Anti-Flag. Catalytic domain. Regulatory domain. Myr SH3 SH2 Kinase domain PP2. Flag-HK1. c-src ΔSH3. HK1 N-half: C-half:
 a d h ΔSH2 Δ(SH3SH2) Myr SH3 SH2 Kinase domain 85 5 249 55 5 csrc ΔSH3 FlagHK HAcSrc HAcSrcΔSH2 HAcSrcΔSH3 HAcSrcΔ(SH3SH2) WB: FlagHK HAcSrc HAcSrcKD HAcSrcY529F WB: HisHK2HK2 Input GSTcSrc GST : : GST
a d h ΔSH2 Δ(SH3SH2) Myr SH3 SH2 Kinase domain 85 5 249 55 5 csrc ΔSH3 FlagHK HAcSrc HAcSrcΔSH2 HAcSrcΔSH3 HAcSrcΔ(SH3SH2) WB: FlagHK HAcSrc HAcSrcKD HAcSrcY529F WB: HisHK2HK2 Input GSTcSrc GST : : GST
Supplementary Figure 1. Procedures for p38 activity imaging in living cells. (a) Schematic model of the p38 activity reporter. The reporter consists
 Supplementary Figure 1. Procedures for p38 activity imaging in living cells. (a) Schematic model of the p38 activity reporter. The reporter consists of: (i) the YPet domain (an enhanced YFP); (ii) the
Supplementary Figure 1. Procedures for p38 activity imaging in living cells. (a) Schematic model of the p38 activity reporter. The reporter consists of: (i) the YPet domain (an enhanced YFP); (ii) the
Supplemental Information
 Supplemental Information Tobacco-specific Carcinogen Induces DNA Methyltransferases 1 Accumulation through AKT/GSK3β/βTrCP/hnRNP-U in Mice and Lung Cancer patients Ruo-Kai Lin, 1 Yi-Shuan Hsieh, 2 Pinpin
Supplemental Information Tobacco-specific Carcinogen Induces DNA Methyltransferases 1 Accumulation through AKT/GSK3β/βTrCP/hnRNP-U in Mice and Lung Cancer patients Ruo-Kai Lin, 1 Yi-Shuan Hsieh, 2 Pinpin
Identification of Mutation(s) in. Associated with Neutralization Resistance. Miah Blomquist
 Identification of Mutation(s) in the HIV 1 gp41 Subunit Associated with Neutralization Resistance Miah Blomquist What is HIV 1? HIV-1 is an epidemic that affects over 34 million people worldwide. HIV-1
Identification of Mutation(s) in the HIV 1 gp41 Subunit Associated with Neutralization Resistance Miah Blomquist What is HIV 1? HIV-1 is an epidemic that affects over 34 million people worldwide. HIV-1
Supplementary Information. Supplementary Figure 1
 Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Phospho-AKT Sampler Kit
 Phospho-AKT Sampler Kit E 0 5 1 0 0 3 Kits Includes Cat. Quantity Application Reactivity Source Akt (Ab-473) Antibody E021054-1 50μg/50μl IHC, WB Human, Mouse, Rat Rabbit Akt (Phospho-Ser473) Antibody
Phospho-AKT Sampler Kit E 0 5 1 0 0 3 Kits Includes Cat. Quantity Application Reactivity Source Akt (Ab-473) Antibody E021054-1 50μg/50μl IHC, WB Human, Mouse, Rat Rabbit Akt (Phospho-Ser473) Antibody
Nature Medicine: doi: /nm.4322
 1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
7.012 Quiz 3 Answers
 MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
Supplementary Information
 Supplementary Information HBV maintains electrostatic homeostasis by modulating negative charges from phosphoserine and encapsidated nucleic acids Authors: Pei-Yi Su 1,2,3, Ching-Jen Yang 2, Tien-Hua Chu
Supplementary Information HBV maintains electrostatic homeostasis by modulating negative charges from phosphoserine and encapsidated nucleic acids Authors: Pei-Yi Su 1,2,3, Ching-Jen Yang 2, Tien-Hua Chu
SUPPLEMENTARY INFORMATION
 Figure S1. Silver staining and immunoblotting of the purified TAK1 kinase complex. The TAK1 kinase complex was purified through tandem affinity methods (Protein A and FLAG), and aliquots of the purified
Figure S1. Silver staining and immunoblotting of the purified TAK1 kinase complex. The TAK1 kinase complex was purified through tandem affinity methods (Protein A and FLAG), and aliquots of the purified
NC bp. b 1481 bp
 Kcna3 NC 11 178 p 1481 p 346 p c *** *** d relative expression..4.3.2.1 CD4 T cells Kcna1 Kcna2 Kcna3 Kcna4 Kcna Kcna6 Kcna7 Kcnn4 relative expression..4.3.2.1 CD8 T cells Kcna1 Kcna2 Kcna3 Kcna4 Kcna
Kcna3 NC 11 178 p 1481 p 346 p c *** *** d relative expression..4.3.2.1 CD4 T cells Kcna1 Kcna2 Kcna3 Kcna4 Kcna Kcna6 Kcna7 Kcnn4 relative expression..4.3.2.1 CD8 T cells Kcna1 Kcna2 Kcna3 Kcna4 Kcna
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis
 Supplementary information Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis Yasuyuki Yamada, Fumihiko Sato
Supplementary information Tyrosine phosphorylation and protein degradation control the transcriptional activity of WRKY involved in benzylisoquinoline alkaloid biosynthesis Yasuyuki Yamada, Fumihiko Sato
Table S1. Total and mapped reads produced for each ChIP-seq sample
 Tale S1. Total and mapped reads produced for each ChIP-seq sample Sample Total Reads Mapped Reads Col- H3K27me3 rep1 125662 1334323 (85.76%) Col- H3K27me3 rep2 9176437 7986731 (87.4%) atmi1a//c H3K27m3
Tale S1. Total and mapped reads produced for each ChIP-seq sample Sample Total Reads Mapped Reads Col- H3K27me3 rep1 125662 1334323 (85.76%) Col- H3K27me3 rep2 9176437 7986731 (87.4%) atmi1a//c H3K27m3
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/398/rs12/dc1 Supplementary Materials for Quantitative phosphoproteomics reveals new roles for the protein phosphatase PP6 in mitotic cells Scott F. Rusin, Kate
www.sciencesignaling.org/cgi/content/full/8/398/rs12/dc1 Supplementary Materials for Quantitative phosphoproteomics reveals new roles for the protein phosphatase PP6 in mitotic cells Scott F. Rusin, Kate
33VASTVNGATSANNHGEPPS51PADARPR58
 Pro-rich region Trans-membrane region 214 246 359 381 UL50 1 397 211SSRTAS216PPPPPR222 NLS CR1 CR2 CR3 CR4 UL53 1 376 11RERRS15ALRS19LLRKRRR25 33VASTVNGATSANNHGEPPS51PADARPR58 FIG S1. UL97 phosphorylation
Pro-rich region Trans-membrane region 214 246 359 381 UL50 1 397 211SSRTAS216PPPPPR222 NLS CR1 CR2 CR3 CR4 UL53 1 376 11RERRS15ALRS19LLRKRRR25 33VASTVNGATSANNHGEPPS51PADARPR58 FIG S1. UL97 phosphorylation
Endothelial TWIK-related potassium channel-1 is a critical regulator of immune cell trafficking into the CNS
 Endothelial TWIK-related potassium channel- is a critical regulator of immune cell trafficking into the C Stefan Bittner, Toias Ruck, Michael K Schuhmann, Alexander M Herrmann, Hamid Moha ou Maati, Nicole
Endothelial TWIK-related potassium channel- is a critical regulator of immune cell trafficking into the C Stefan Bittner, Toias Ruck, Michael K Schuhmann, Alexander M Herrmann, Hamid Moha ou Maati, Nicole
Study of different types of ubiquitination
 Study of different types of ubiquitination Rudi Beyaert (rudi.beyaert@irc.vib-ugent.be) VIB UGent Center for Inflammation Research Ghent, Belgium VIB Training Novel Proteomics Tools: Identifying PTMs October
Study of different types of ubiquitination Rudi Beyaert (rudi.beyaert@irc.vib-ugent.be) VIB UGent Center for Inflammation Research Ghent, Belgium VIB Training Novel Proteomics Tools: Identifying PTMs October
Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication
 Manuscript EMBO-2010-74756 Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication Tsai-Ling Liao, Chung-Yi Wu, Wen-Chi Su, King-Song Jeng and Michael Lai Corresponding
Manuscript EMBO-2010-74756 Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication Tsai-Ling Liao, Chung-Yi Wu, Wen-Chi Su, King-Song Jeng and Michael Lai Corresponding
Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients.
 Supplementary Materials Supplementary Figures Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients. Figure S2. Expression level of podocyte
Supplementary Materials Supplementary Figures Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients. Figure S2. Expression level of podocyte
Figure S1. (A) SDS-PAGE separation of GST-fusion proteins purified from E.coli BL21 strain is shown. An equal amount of GST-tag control, LRRK2 LRR
 Figure S1. (A) SDS-PAGE separation of GST-fusion proteins purified from E.coli BL21 strain is shown. An equal amount of GST-tag control, LRRK2 LRR and LRRK2 WD40 GST fusion proteins (5 µg) were loaded
Figure S1. (A) SDS-PAGE separation of GST-fusion proteins purified from E.coli BL21 strain is shown. An equal amount of GST-tag control, LRRK2 LRR and LRRK2 WD40 GST fusion proteins (5 µg) were loaded
Supporting Information. FADD regulates NF-кB activation and promotes ubiquitination of cflip L to induce. apoptosis
 1 2 Supporting Information 3 4 5 FADD regulates NF-кB activation and promotes ubiquitination of cflip L to induce apoptosis 6 7 Kishu Ranjan and Chandramani Pathak* 8 9 Department of Cell Biology, School
1 2 Supporting Information 3 4 5 FADD regulates NF-кB activation and promotes ubiquitination of cflip L to induce apoptosis 6 7 Kishu Ranjan and Chandramani Pathak* 8 9 Department of Cell Biology, School
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Jewell et al., http://www.jcb.org/cgi/content/full/jcb.201007176/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. IR Munc18c association is independent of IRS-1. (A and
Supplemental material Jewell et al., http://www.jcb.org/cgi/content/full/jcb.201007176/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. IR Munc18c association is independent of IRS-1. (A and
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11700 Figure 1: RIP3 as a potential Sirt2 interacting protein. Transfected Flag-tagged Sirt2 was immunoprecipitated from cells and eluted from the Sepharose beads using Flag peptide.
doi:10.1038/nature11700 Figure 1: RIP3 as a potential Sirt2 interacting protein. Transfected Flag-tagged Sirt2 was immunoprecipitated from cells and eluted from the Sepharose beads using Flag peptide.
Sequences in the 5 and 3 R Elements of Human Immunodeficiency Virus Type 1 Critical for Efficient Reverse Transcription
 JOURNAL OF VIROLOGY, Sept. 2000, p. 8324 8334 Vol. 74, No. 18 0022-538X/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. Sequences in the 5 and 3 R Elements of Human
JOURNAL OF VIROLOGY, Sept. 2000, p. 8324 8334 Vol. 74, No. 18 0022-538X/00/$04.00 0 Copyright 2000, American Society for Microbiology. All Rights Reserved. Sequences in the 5 and 3 R Elements of Human
COMPARISON OF HIV DRUG-RESISTANT MUTANT DETECTION BY NGS WITH AND WITHOUT UNIQUE MOLECULAR IDENTIFIERS (UMI)
 COMPARISON OF HIV DRUG-RESISTANT MUTANT DETECTION BY NGS WITH AND WITHOUT UNIQUE MOLECULAR IDENTIFIERS (UMI) Kevin D McCormick; Kerri J Penrose; Rahil Sethi; Jacob Waldman; William Schwarzmann; Uma Chandran;
COMPARISON OF HIV DRUG-RESISTANT MUTANT DETECTION BY NGS WITH AND WITHOUT UNIQUE MOLECULAR IDENTIFIERS (UMI) Kevin D McCormick; Kerri J Penrose; Rahil Sethi; Jacob Waldman; William Schwarzmann; Uma Chandran;

 Supplementary Figure 1 a OD c. 1. 1... Time [min] 1 p
Supplementary Figure 1 a OD c. 1. 1... Time [min] 1 p