Temporal Patterning of Neuroblasts Controls Notch-Mediated Cell Survival through Regulation of Hid or Reaper
|
|
|
- Flora Evans
- 5 years ago
- Views:
Transcription
1 Temporal Patterning of Neuroblasts Controls Notch-Mediated Cell Survival through Regulation of Hid or Reaper Claire Bertet, 1 Xin Li, 1 Ted Erclik, 1 Matthieu Cavey, 1 Brent Wells, 1 and Claude Desplan 1,2, * 1 Department of Biology, New York University, 100 Washington Square East, New York, NY 10003, USA 2 CGSB, New York University Abu Dhabi, United Arab Emirates *Correspondence: cd38@nyu.edu SUMMARY Temporal patterning of neural progenitors is one of the core mechanisms generating neuronal diversity in the central nervous system. Here, we show that, in the tips of the outer proliferation center (topc) of the developing Drosophila optic lobes, a unique temporal series of transcription factors not only governs the sequential production of distinct neuronal subtypes but also controls the mode of progenitor division, as well as the selective apoptosis of Notch OFF or Notch ON neurons during binary cell fate decisions. Within a single lineage, intermediate precursors initially do not divide and generate only one neuron; subsequently, precursors divide, but their Notch ON progeny systematically die through Reaper activity, whereas later, their Notch OFF progeny die through Hid activity. These mechanisms dictate how the topc produces neurons for three different optic ganglia. We conclude that temporal patterning generates neuronal diversity by specifying both the identity and survival/death of each unique neuronal subtype. INTRODUCTION A central challenge in developmental neurobiology is to understand how an apparently uniform pool of embryonic progenitors produces the vast diversity of neurons and glial cells found in the adult central nervous system (CNS). Studies in vertebrates and insects have revealed that four primary mechanisms generate neural cell diversity: (1) spatial patterning cues provide a unique lineage identity to each progenitor (Rogulja-Ortmann and Technau, 2008; Technau et al., 2006); (2) temporal progression of transcription factors in progenitors instructs them to orderly produce different neuronal and glial subtypes (Baumgardt et al., 2009; Brody and Odenwald, 2000; Elliott et al., 2008; Isshiki et al., 2001; Lee et al., 1999; Li et al., 2013; Livesey and Cepko, 2001; Zhu et al., 2006); (3) binary cell fate decisions mediated by Notch lead to the formation of two postmitotic cells with different fates during the final mitosis of intermediate precursors (Buescher et al., 1998; Truman et al., 2010), with one of these cells sometimes undergoing apoptosis (Lin et al., 2010; Lundell et al., 2003); (4) progenitors can generate intermediate precursors with different proliferation modes (He et al., 2012). Some intermediate precursors divide once to produce two daughter cells (type 1 neuroblasts) (Buescher et al., 1998), some undergo multiple divisions to amplify the lineage (type 2 neuroblasts) (Bello et al., 2008; Boone and Doe, 2008), or some directly differentiate into a neuron without further cell divisions (Karcavich and Doe, 2005; Ulvklo et al., 2012) (which we name type 0 neuroblasts). Although these mechanisms are well characterized, very little is known about how they interact with each other to specify the unique fate of each neuron. For instance, it has been suggested that Notch integrates spatial signals to determine neuronal survival or apoptosis during binary cell fate decisions (Lin et al., 2010), but these signals and the nature of their interaction with Notch remain enigmatic. The Drosophila visual system is an excellent model for studying complex neurogenesis. Each optic lobe is composed of four ganglia: the lamina, medulla, lobula, and lobula plate (Hofbauer and Campos-Ortega, 1990; Meinertzhagen and Hanson, 1993). The medulla, which has the largest neuropil, is composed of 40,000 neurons comprising more than 70 subtypes (Fischbach and Dittrich, 1989; Morante and Desplan, 2008). It derives from a single-layered crescent-shaped neuroepithelium in the larval brain called the outer proliferation center (OPC). The OPC is progressively converted into progenitors (called neuroblasts) by a wave of expression of the proneural gene lethal of scute that sweeps from the edge toward the center of the crescent over time (Egger et al., 2007; Egger et al., 2010; Ngo et al., 2010; Yasugi et al., 2010) (Figure S1A available online). We and others have recently shown that neuronal diversity in the medulla is generated by a combination of spatial patterning, temporal patterning, and binary cell fate decisions (Li et al., 2013; Suzuki et al., 2013). Indeed, the expression of four genes, Vsx1, optix, decapentaplegic (dpp), and wingless (wg) divides the OPC into four spatial regions along the anteroposterior axis (Kaphingst and Kunes, 1994) (T.E., X.L., C.B., Z. Chen, R. Baumert, J. Ng, Rudy B., A. del Valle Rodriguez, L. Senderowicz, N. Negre, K.P. White, and C.D., unpublished data). Each of these spatially distinct regions produces different neuronal subtypes. In addition, neuroblasts deriving from the main OPC (which includes the regions defined by Vsx1, Optix, and Dpp) all sequentially Cell 158, , August 28, 2014 ª2014 Elsevier Inc. 1173
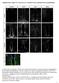
2 Figure 1. The topc Temporal Series Sequentially Produces Four Neuronal Classes Unless specified, all pictures are posterior views of third instar larvae (L3). (A) Model of a larval optic lobe showing that a wave of neurogenesis (orange) converts OPC neuroepithelial cells (NE, purple) into neuroblasts (NBs, yellow). The topc is defined by wg-gal4 expression (green). A, anterior; P, posterior; V, ventral; D, dorsal; L, lateral; and M, medial. (B) The topc region (wg > GFP, green, dashed lines). E-Cad stains the neuroepithelium (purple). (C G) topc neuroblasts sequentially express Dll (magenta in C and D), Ey (cyan in D and E), Slp (red in E and F), and D (yellow in F). White dashed lines show the topc. (legend continued on next page) 1174 Cell 158, , August 28, 2014 ª2014 Elsevier Inc.
3 express five temporal factors Homothorax (Hth), Eyeless (Ey), Sloppy-paired (Slp), Dichaete (D), and Tailless (Tll) as they age (Figures S1A and S1B). These factors and their overlap determine about 12 neuroblast fates that generate distinct neuronal subtypes. Finally, these neuroblasts produce intermediate precursors called ganglion mother cells (GMCs) that divide asymmetrically to generate two distinct neurons, one Notch OFF and one Notch ON neuron expressing Apterous (Ap) (Figure S1B). As a result, the larval medulla cortex is composed of several layers, each composed of Notch ON Ap-positive neurons intermingled with Notch OFF neurons. Here, we focus on the development of the tips of the OPC defined by wingless expression (topc, Figures 1A, 1B, and S1A), which provides a unique opportunity for studying how temporal patterning of progenitors interplays with Notch to control neuronal survival. We show that topc progenitors undergo complex neurogenesis involving two dramatic transitions, one in the mode of intermediate precursor division and the other in systematic apoptosis of one of their neuronal progeny during Notchmediated binary cell fate decisions. We provide evidence that, in addition to specifying distinct neuronal subtypes over time, temporal patterning of topc progenitors also controls the transition in apoptosis by specifying the systematic death of Notch ON neurons in a first phase and of Notch OFF neurons in a second phase through the regulation of Reaper (Rpr) and Head involution defective (Hid), respectively. This complex neurogenesis dictates how the topc produces neurons for three different neuropils of the adult optic lobes. We conclude that temporal patterning of progenitors generates neuronal diversity by controlling multiple aspects of neurogenesis, including neuronal identity and Notch-mediated cell survival. RESULTS topc Neuroblasts Undergo Temporal Patterning In order to understand how medulla neuronal diversity is generated, we conducted an antibody screen to identify transcription factors expressed in the developing optic lobes (Li et al., 2013). The medulla derives from the larval OPC, where the progressive conversion of neuroepithelium into neuroblasts allows us to visualize in one snapshot neuroblasts at different temporal stages. Oldest neuroblasts are found at the medial edge of the OPC near the neuroepithelium, whereas newly specified neuroblasts are found more laterally (Figures 1A and 1B). This screen led us to identify four transcriptions factors, Distalless (Dll), Eyeless (Ey), Sloppy-paired (Slp), and Dichaete (D), expressed in consecutive stripes in topc neuroblasts of increasing ages (Figures 1C 1F), with Dll expressed in the youngest topc neuroblasts (Figures 1C and 1D) and D expressed in the oldest neuroblasts (Figure 1F). Neighboring stripes partially overlap, suggesting a gradual replacement of one transcription factor by the next as neuroblasts age. Therefore, topc neuroblasts express a unique series of transcription factors, Dll, Ey, Slp, and D (Figure 1G), which is different from the main OPC in which neuroblasts sequentially express Hth, Ey, Slp, D, and Tll (Figures S1A and S1B). Next, we examined the progeny of topc neuroblasts. topc neuroblasts divide asymmetrically multiple times to selfrenew and produce a GMC, which, in turn, generates neurons. The progeny of each topc neuroblast therefore forms a chain, with newly specified neurons occupying the most superficial layer and the oldest neurons the deepest layer (Figure 1H). Despite screening over 200 antibodies, we found only four different classes of larval topc neurons based on their expression of transcription factors. More importantly, in contrast to the main OPC, in which different neuronal classes are intermingled, neuronal classes are organized as homogeneous clusters in the topc (Figure 1I), suggesting that this region has a distinct mode of neurogenesis. To decipher the mode of topc neurogenesis, we first determined which temporal windows produce each of the four neuronal clusters, which are localized in different layers that correlate with their birth order. Class 1 neurons are localized in the deepest layer and coexpress Dll, Spalt major (Salm), Runt, and D (the latter only in the ventral topc) (Figures S1D S1F). Young neuroblast clones in which the neuroblasts are at the Dll + stage include Dll + GMCs and neurons (Figure 1J), indicating that class 1 Salm/Runt neurons are produced during the Dll time window. Class 2 neurons are localized in the layer above class 1 neurons and express Seven-up (Svp). Although Ey expression is not transmitted to these neurons, GFP driven by ey-gal4 is expressed in almost all of them due to perdurance of Gal4 and GFP (Figures 1K and S1G). This suggests that class 2 Svp neurons are produced during the Ey time window. Class 3 neurons are localized above class 2 neurons and express Twin-ofeyeless (Toy). Slp is not transmitted to these neurons, but we identified a slp-gal4 line expressed in almost all Slp-expressing neuroblasts (Figure S1H). This Gal4 line drives GFP expression in the majority of Toy neurons (also due to perdurance, Figure 1L), suggesting that class 3 Toy neurons are produced during the Slp time window. Finally, class 4 neurons are localized in the most superficial layer near the neuroblasts and coexpress Toy and D (Figure S1I). Late neuroblast clones in which the neuroblasts are at the D + stage include D + GMCs and neurons, indicating that class 4 Toy/D neurons are produced at the end of topc lineages during the D time window (Figure 1M). In summary, four temporal windows in neuroblasts sequentially produce four classes of neurons in the larval topc (Figure 1N). Initially, Dll is expressed and produces class 1 (H) Model of a cross-section showing neuroblasts (yellow circles), GMCs (yellow), and neurons (cyan). A single neuroblast clone is shown with black thick outlines. (I) Cross-section view showing the clustered organization of three of the four medulla neuronal classes: class 1 (Salm, magenta), class 2 (Svp, cyan), and class 3 (Toy, red). (J) Cross-section view. Early neuroblast clone (GFP, dashed lines) showing that Dll (magenta) is transmitted from neuroblasts (NBs) to neurons (n). (K) Perduring GFP driven by ey-gal4 (dashed lines) is expressed in almost all Svp neurons (cyan). (L) Perduring GFP driven by slp-gal4 (dashed lines) is expressed in all Toy neurons (red). (M) Cross-section view. Late neuroblast clone (dashed lines) showing that D expression (yellow) is transmitted to the neurons. (N) Schematic model of topc neurogenesis. Only one neuroblast/gmc is shown for each stage. See also Figure S1. Cell 158, , August 28, 2014 ª2014 Elsevier Inc. 1175
4 Figure 2. Notch Status of topc Neurons Posterior views of L3 optic lobes. (A and B) Hey (white) is expressed by class 3 Toy and class 4 Toy/D neurons (A), but not by class 1 Salm/Runt (magenta, white arrows) and class 2 Svp (cyan, red arrows) neurons (B). Dashed lines indicate the topc. (C) Class 3 Toy and class 4 Toy/D neurons (red) are lost in su(h) mutant clones (GFP, dashed lines). (D) Control brains showing class 1 Salm/Runt (magenta, arrowheads) and class 2 Svp (cyan, arrows) neurons. (E) Ectopic Notch signaling (insc > N ICD ) leads to the loss of class 2 Svp neurons (cyan, arrows) but has no effect on class 1 Salm/Runt neurons (magenta, arrowheads). Dashed lines indicate the topc. (F) Average number of topc neurons in control and Notch gain of function (n = 7 brains/genotype). Magenta, class 1 Salm/Runt neurons; cyan, class 2 Svp neurons. All data represent mean ± SD. NS, not significant. ***p < (G) Summary. See also Figure S2. Salm/Runt neurons (magenta). These neuroblasts then switch to Ey expression and produce class 2 Svp neurons (cyan), followed by expression of Slp to produce class 3 Toy neurons (red) and, finally, D expression to produce class 4 Toy/D neurons (yellow). Three Groups of topc Neurons Based on Their Notch Status The clustered organization of topc neurons could be due to systematic apoptosis of one of the neuronal progeny of type 1 neuroblasts. Alternatively, it could result from a type 0 mode of division of topc neuroblasts (Figure S1C). To test this, we first determined the Notch status of the four classes of topc neurons. We monitored the expression of the bhlh-o protein Hairy/enhancer-of-split like a Y (Hey), which has been used as a Notch sensor in the Drosophila CNS (Monastirioti et al., 2010). Hey expression is lost when Notch signaling is abolished (suppressor of hairless, su(h) mutant clones, Figure S2A) and is expanded when Notch signaling is constitutively active (overexpression of the Notch intracellular domain, Notch ICD ; Figures S2B and S2C). Therefore, Hey is also a good sensor of Notch activity in optic lobe neurons. In the topc, Hey marks class 3 Toy and class 4 Toy/D neurons, which must therefore be Notch ON (Figure 2A). The absence of Hey in class 1 Salm/Runt and class 2 Svp neurons suggests that they are Notch OFF (Figure 2B). Consistent with this, class 1 Salm/Runt and class 2 Svp neurons are not affected in su(h) mutant clones (Figures S2D and S2E), whereas class 3 Toy and class 4 Toy/D neurons are completely lost (Figure 2C). Conversely, ectopic activation of Notch signaling in the topc decreases the number of class 2 Svp neurons (Figures 2D 2F: control 282 ± 16; 1176 Cell 158, , August 28, 2014 ª2014 Elsevier Inc.
5 insc-gal4 > Notch ICD 54 ± 12; p < 0.001) but does not affect class 3 Toy and class 4 Toy/D neurons (Figure S2F). Surprisingly, although class 1 Salm/Runt neurons appear to be Notch OFF, they are not affected by high levels of Notch signaling, suggesting that these neurons acquire their fate in a Notch-independent manner (Figures 2D 2F: control 211 ± 10; insc-gal4 > Notch ICD 221 ± 10; p = 0.1). Thus, based on their Notch status, the topc neurons can be divided into three groups (Figure 2G): Notch independent neurons (class 1), Notch OFF neurons (class 2), and Notch ON neurons (classes 3 and 4). Origin of the Clustered Organization of topc Neurons The presence of both Notch OFF and Notch ON neurons suggests that most topc neuroblasts are type 1 neuroblasts and that one of the progeny of their GMCs undergoes apoptotic cell death. To test this, we stained topc neurons with the cell death marker cleaved Caspase-3. Cleaved Caspase-3 is expressed in a few neurons of the class 2 cluster that forms during the Ey time window. Indeed, although this cluster is almost exclusively composed of Notch OFF Svp neurons, we detected close to the neuroblast layer a small number of Notch ON neurons that all express cleaved Caspase-3, as well as Toy (white arrows, Figure S3A). This suggests that Ey neuroblasts are type 1 neuroblasts which, at each round of division, produce one Notch OFF Svp neuron that survives and one Notch ON Toy neuron that undergoes apoptosis. Consistent with this, forcing the survival of Notch ON Toy neurons via P35 expression increased their number (Figure 3E: Control 529 ± 51; wg > P ± 36; p < ) and creates an intermingling between these neurons (red) and the Notch OFF class 2 Svp neurons (cyan, compare top parts of Figures 3A, A 0, S3B, and S3C). Similarly, we found evidence that cell death is implicated in the formation of class 3 and class 4 clusters. In these two Notch ON clusters, we detected a small number of Notch OFF neurons expressing cleaved Caspase-3 and Svp (yellow arrows, Figure S3A). Forcing the survival of these neurons via P35 expression strongly increased their number (Figure 3E: control 282 ± 16; wg > p ± 36; p < ). Moreover, P35 expression creates an intermingling between these Notch OFF Svp neurons (cyan) and the Notch ON class 3 Toy neurons (red) in the Slp time window (compare bottom parts of Figures 3A and 3A 0 ). These Notch OFF Svp neurons are also intermingled with Notch ON class 4 Toy/D neurons (yellow) (compare Figures 3B and 3B 0 ) and, because there is no cell migration during topc neurogenesis, they must be produced locally during the D time window. Thus, during the Slp and D time windows, an inversion in the apoptotic pattern causes Notch OFF Svp-expressing neurons to die, leading to the formation of the Notch ON class 3 Toy and class 4 Toy/D clusters. We next examined the mode of division of Dll neuroblasts. These neuroblasts produce class 1 Salm/Runt neurons independently of Notch and show no indication of apoptosis because no new neurons appear in the presence of P35 (Figures 3C, 3C 0, and 3E: control 221 ± 12; wg > p ± 22; p = 0.12). This opened the possibility that Dll neuroblasts undergo a type 0 mode of division and produce GMCs that do not divide but instead directly differentiate into a neuron. To test this, we generated single neuroblast clones and analyzed the progeny of newly specified Dll-expressing neuroblasts (Figure 3D). These clones always contain one GMC (stained with Asense) and only one class 1 Salm/Runt neuron. Thus, during the Dll time window, topc neuroblasts are type 0 neuroblasts that produce a single neuron at each round of division. In conclusion, the clustered organization of topc neurons results from two phenomena (Figure 3F): first, topc neuroblasts are initially specified as type 0 neuroblasts (Dll time window) and produce a single class of neurons. Second, they then switch to type 1 neuroblasts (Ey, Slp, and D time windows), but half of their progeny undergo apoptosis; Notch ON die first in the Ey time window, whereas Notch OFF die later, in the Slp and D time windows. This leads to the production, within the same lineage, of four hemilineages composed of neuronal classes 1, 2, 3, and 4, respectively. Cross-Regulations between topc Temporal Factors We next examined whether, like in the main OPC, cross-regulation among Dll, Ey, Slp, and D contributes to the transition from one factor to the next. The main OPC and topc temporal series begin with different factors, Hth and Dll, respectively, raising the possibility that Dll represses Hth in the topc. However, this is not the case because Hth is not expressed in topc dll mutant clones (Figure S4A). In addition, ey, slp, and D expression is not affected in dll clones (data not shown), indicating that Dll is not required to turn on subsequent factors of the temporal cascade. In ey loss of function (mutants and RNAi, see Experimental Procedures), dll expression remains restricted to the youngest neuroblasts (Figure S4B), whereas slp and D are lost (compare Figures S4C and S4C 0 ). Therefore, Ey is required to turn on the next factor in the series, slp, but is not required to turn off dll. Inslp mutant clones, ey expression expands, whereas D is lost (Figure S4D). Therefore, Slp turns off ey expression and is required to turn on D expression. Finally, in D mutant clones, slp expands, showing that D turns off slp expression (Figure S4E). Thus, like in the main OPC, Ey, Slp, and D are required to turn on the next transcription factor, whereas Slp and D are also required for turning off the preceding transcription factor (Figure S4F). topc Temporal Patterning Controls Notch-Mediated Neuronal Survival We then tested the role of Dll, Ey, Slp, and D during topc neurogenesis. Temporal factors have been shown to specify distinct neuronal identities over time. In addition, in the topc, the transition in apoptosis pattern (from Notch ON to Notch OFF dying) suggests that this process is temporally regulated. We therefore tested whether topc temporal factors specify both the identity and the survival of the four classes of neurons. Newly specified neuroblasts express high levels of Dll and produce class 1 Salm/Runt neurons. However, in dll mutant clones, none of the markers of this neuronal class (Salm, Runt, and D) are lost (Figure S4G and data not shown), suggesting that Dll does not specify the identity of class 1 Salm/Runt neurons. Interestingly, although loss of Dll does not alter Ey expression in neuroblasts, it affects the neurons produced during the Ey time window. Indeed, in dll clones, Notch OFF class 2 Svp neurons are either partially (80% of the cases, n = 17/22) or completely (20% of the cases, n = 5/22) lost, and they do not appear to be Cell 158, , August 28, 2014 ª2014 Elsevier Inc. 1177
6 Figure 3. Origin of the Clustered Organization of topc Neurons Posterior views of L3 optic lobes. (A C) P35 expression in topc neurons. Dashed lines show the topc. (A and B) Control brains showing the NotchOFF Class 2 Svp (cyan in A), NotchON Class 3 Toy (red in A), and NotchON Class 4 Toy/D clusters (yellow in B). In P35-expressing brains, NotchOFF Svp neurons are completely intermingled with NotchON Toy (A0 ) in the Ey and Slp time windows and with NotchON Toy/D (B0 ) neurons in the D time window. White bars in (A) and (A0 ) delimitate the border between the Ey and the Slp time windows. (legend continued on next page) 1178 Cell 158, , August 28, 2014 ª2014 Elsevier Inc.
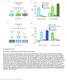
7 transformed into another subtype (Figure 4A). This suggests that Dll promotes survival of the Notch OFF neurons produced during the Ey window (Figure 4A 0 ). To test whether Dll is sufficient to promote the survival of Notch OFF neurons, we ectopically expressed it in the Slp and D later time windows (Figures 4B and 4B 0 ) using slp-gal4. If Dll promotes the survival of these neurons, it should rescue the Svp-expressing neurons (the Notch OFF siblings of class 3 and class 4 Notch ON neurons) that normally die in this part of the lineage. This is indeed the case: although ectopic Dll does not affect the expression of Ey, Slp, and D in neuroblasts (data not shown), like P35 expression, it creates an intermingling between Notch OFF Svp-expressing neurons and Notch ON class 3 and class 4 Toy neurons in the dorsal topc tip (arrow in Figure 4B; Svp neurons: control 142 ± 12; slp > dll 204 ± 16; p < 10 4 ), but not in the control ventral tip in which slp-gal4 is only weakly expressed (arrowhead in Figure 4B). These results thus demonstrate that, during topc neurogenesis, Dll does not specify neuronal identities but instead acts in the Ey temporal window to promote the survival of Notch OFF class 2 Svp neurons (Figure 4H). Neuroblasts expressing high levels of Ey produce Notch OFF class 2 Svp neurons. Removing ey activity does not lead to the loss of these neurons, suggesting that Ey does not specify their identity. However, removing ey activity generates two striking effects. First, it dramatically increases the number of Notch OFF class 2 Svp neurons (Figures 4C 4E: WT 282 ± 16; wg > ey- RNAi 695 ± 41, p < 10 4 ), indicating that Ey induces apoptosis of Notch OFF neurons. Second, it leads to the complete loss of Notch ON class 3 and 4 Toy-expressing neurons that are normally produced during the later Slp and D time windows (compare Figures 4C and 4D). These Notch ON Hey-positive neurons are still generated, but they all express the apoptosis marker Cleaved Caspase-3 and rapidly disappear (Figure 4F). Coexpression of P35 with ey-rnai rescues their death and generates an intermingling between these Notch ON neurons and Notch OFF class 2 Svp neurons (Figures 4G and 4G 0 ). Thus, Ey is required for the survival of Notch ON neurons, and in its absence, the switch in the apoptosis pattern no longer happens (Figure 4F 0 ): Notch OFF neurons survive, whereas Notch ON neurons die throughout neurogenesis, resulting in a massive increase of class 2 Svp neurons. In summary, Ey plays a dual role in the control of neuronal survival it induces apoptosis of Notch OFF neurons, and it promotes survival of Notch ON neurons. Interestingly, when we forced the survival of Hey-positive Notch ON neurons in ey loss of function (ey RNAi + UAS-P35), we noticed that these neurons no longer express Toy (Figure 4G), suggesting that Ey is also required for the Toy identity. Removing ey function affects the neurons produced during the later Slp and D time windows (Figure 4H). Because Ey is required for Slp expression (and thus indirectly for D expression), we next tested whether the ey phenotype could be due to the loss of Slp in neuroblasts. If this hypothesis is correct, removing Slp should produce the same phenotype as ey loss of function i.e., expansion of Notch OFF class 2 Svp neurons and loss of Notch ON class 3 Toy neurons. In slp mutant clones, Notch OFF class 2 Svp neurons do not expand (Figure S4H), indicating that Slp does not induce apoptosis of Notch OFF neurons. However, Notch ON class 3 neurons (marked by Toy and Hey) express cleaved Caspase-3 and systematically undergo apoptosis (Figures 5A, 5A 0, and S4I). Expression of P35 in slp clones rescues their death and generates an intermingling between these Notch ON Toy neurons and Notch OFF class 2 Svp neurons (Figures 5B, 5B 0, and S4J). Thus, Slp promotes survival of Notch ON neurons. Ey induces apoptosis of Notch OFF neurons, whereas Slp promotes survival of Notch ON neurons. We next investigated the potential targets of Ey and Slp. In Drosophila, the cell death pathway is initiated by the expression of the genes reaper (rpr), head involution defective (hid), and grim (Figure 5C), which, by inhibiting DIAP-1, lead to the activation of initiator Caspase-9 (Dronc). Dronc in turn activates Caspase-3, which triggers the apoptotic process (Xu et al., 2009). P35 acts by preventing Caspase-3 from killing the cells. Like with P35 overexpression, removing rpr, hid, and grim (def(3l)h99 clones; Figures 5D and 5E) or Dronc (Figures 5D, 5F, S5A, and S5B) blocks apoptosis and generates an intermingling between Notch OFF and Notch ON neurons in the Ey, Slp, and D time windows. Strikingly, in Dronc mutants in which neurons initiate apoptosis but do not complete it, Hid is only expressed in Notch OFF neurons that normally die during the Slp and D time windows (Figure 5G). This suggests that Hid specifically initiates apoptosis of Notch OFF neurons. Consistent with this, removing Hid activity (Hid hypomorph /def(3l) H99 or wg > hid-rnai) specifically rescues apoptosis of Notch- OFF Svp neurons produced during the Slp and D time windows (Figure 5H, 5H 0, S5C, and S5C 0 ). By contrast, removing rpr specifically rescues apoptosis of Notch ON neurons produced during the Ey time window (Figures 5I and 5I 0 ). Thus, Rpr initiates apoptosis of Notch ON neurons, whereas Hid initiates apoptosis of Notch OFF neurons. Based on these results, we propose that Ey induces apoptosis of Notch OFF neurons by activating hid, whereas Slp promotes survival of Notch ON neurons by repressing rpr (Figure 5J). The Sequential Expression of Dll, Ey, and Slp Governs the Switch in Apoptosis Our data suggest that the default fate of Notch ON neurons is to undergo apoptosis (through rpr) and that Ey promotes their survival indirectly by activating Slp expression in neuroblasts. This explains why Notch ON neurons die during the Ey time window but survive during the later Slp and D time windows (Figure 5J). Although Ey also induces apoptosis of Notch OFF neurons, these (C) Control brain stained with Hey (green) and Salm (magenta). (C 0 ) P35 expression has no effect on the Notch indep Salm/Runt cluster. (D) Single neuroblast clones showing that newly specified neuroblasts generate GMCs (Asense, blue) that directly differentiate into one class 1 neuron (Salm, magenta). Neuroepithelium (NE, arrowhead) is stained with DE-Cad (white). Asterisk indicates a newly formed neuroblast that has not produced any neurons yet. (E) Average number of Salm (magenta), Svp (cyan), Toy (red), and D (yellow) neurons in control and wg > P35 (n = 6 brains/genotype). All data represent mean ± SD. NS, not significant. ***p < (F) topc neurogenesis mode. See also Figure S3. Cell 158, , August 28, 2014 ª2014 Elsevier Inc. 1179
8 Figure 4. Dll and Ey Control Notch-Mediated Neuronal Survival Posterior views of L3 optic lobes. Unless otherwise mentioned, dashed lines show the topc. (A and A0 ) dll mutant clones (green, dashed lines). Removing dll affects class 2 Svp neurons (cyan, arrow). (B and B0 ) Dll overexpression. slp-gal4 is strongly expressed in the dorsal topc but is weakly expressed in the ventral topc that serves as an internal control (B). Expressing Dll (green) in Slp and D neuroblasts rescues the death of class 2 Svp neurons (cyan, compare arrow with arrowhead on control side). (C) Maximal projection of a WT brain showing class 2 (Svp, cyan) and class 3 and 4 (Toy, red) neuronal clusters. (D) Maximal projection of an ey mutant brain showing the disappearance of NotchON class 3 and 4 neurons (red) and the expansion of NotchOFF class 2 Svp neurons (cyan, arrowheads). (E) Average number of class 2 Svp neurons in various genotypes (n = 6 brains/genotype). All data represent mean ± SD. NS, not significant. ***p < (F and F0 ) In wg > ey-rnai, NotchON Hey-positive neurons (white) all express the death marker cleaved Caspase-3 (yellow, arrows). (G and G0 ) Coexpressing P35 with ey-rnai rescues the death of NotchON Hey-positive neurons (white) and generates an intermingling between these neurons and NotchOFF class 2 Svp neurons (cyan). The rescued NotchON neurons do not express Toy (red). (H) Timing of Dll and Ey activities. See also Figure S Cell 158, , August 28, 2014 ª2014 Elsevier Inc.
9 neurons survive in the Ey time window, suggesting that one or several factors repress Ey activity during the Ey time window. Dll could be one of these factors because it is active in the Ey time window and it is sufficient to promote survival of Notch OFF neurons. To test whether we could overcome this repression, we overexpressed Ey in topc neuroblasts with insc-gal4. Ectopic Ey has no effect on neurons produced during the Dll, Slp, and D time windows but leads to a significant decrease in the number of Notch OFF class 2 Svp neurons that form during the Ey time window (control 282 ± 16; insc > UAS-ey 194 ± 17; p < ). Thus, when Ey levels are very high, its repressors (including Dll) are no longer able to prevent it from inducing apoptosis of Notch OFF neurons. Altogether, our data demonstrate that Dll, Ey and Slp control neuronal survival and that their sequential expression determines the switch in the pattern of apoptosis. We propose the following model (Figure 5J): by default, Notch ON neurons undergo Rpr-dependent apoptosis. During the Ey time window, Dll and unknown repressors antagonize Ey activity, whereas Slp is not expressed yet. As a result, Notch OFF neurons survive, whereas Notch ON neurons undergo apoptosis. In the Slp time window, Slp promotes survival of Notch ON neurons, whereas Ey repressors are no longer expressed/active to prevent Eyinduced apoptosis of Notch OFF neurons (through Hid), therefore causing an inversion in the apoptosis pattern. This inversion lasts until the end of neurogenesis, implying that one or several factors relay Ey and Slp activities. D, which defines the last time window, does not control neuronal survival. Indeed, in D mutant clones, Notch OFF neurons keep dying, whereas Notch ON class 4 Toy/D neurons disappear and are replaced by Notch ON class 3 Toy neurons (Figure S4K). This suggests that D does not relay Ey and Slp activities and instead only specifies neuronal fate. We conclude that temporal patterning of progenitors generates neuronal diversity by controlling both neuronal identity and Notch-mediated survival decisions. The topc Produces Neurons for Three Different Optic Ganglia In order to understand the consequences of the complex mode of neurogenesis in the topc, we finally investigated which types of neurons are produced in this region. The adult optic lobes comprise four neuropils, the lamina, medulla, lobula, and lobula plate. The lamina derives from the inner part of the OPC crescent, whereas the main region of the OPC generates the medulla. The inner proliferation center (IPC) and the lobula plug (which derives from the IPC) together generate the lobula and lobula plate (Figure S6A). We tested which neurons the topc specifically produces. Because wg-gal4 expression is not maintained in adult brains, we generated a memory cassette tool to determine the adult neuronal subtypes produced by the topc that we named FLEXAMP for flip-out LexA amplification (Figure 6A, see Experimental Procedures). We crossed FLEXAMP to wg-gal4, which is expressed in the entire topc and a subset of IPC cells, and with slp-gal4, which is specifically expressed in Slp and D-expressing neuroblasts of the topc (due to Gal4 per durance). We generated early clones to label dividing neuroepithelial cells (Figures S6B and S6C). Unexpectedly, these clones revealed that the topc produces distinct neuronal subtypes for three of the four adult neuropils: distal medulla neurons (arrow 1), medulla tangential and proximal medulla neurons (arrows 2), lobula columnar neurons (arrows 3), and lobula complex columnar neurons (arrows 4). To determine during which temporal window each of these neuronal subtypes is produced, we stained the nuclei of adult wg > and slp > FLEXAMP clones with antibodies specific to the four larval neuronal classes (Figure 6). Strikingly, cell bodies of the same neuronal class (i.e., of neurons produced during the same time window) are all localized in the same area of the optic lobes. The identification of these neuronal subtypes is made according to Fischbach and Dittrich (1989), and their detailed morphology is described in the Figure 6 legend. Because Salm and Runt expression is not maintained in adult topc neurons, we could not precisely determine the terminal identity of class 1 neurons. However, in midpupal brains, Salm and Runt are still expressed, and we found that cell bodies of class 1 neurons are localized in the lobula plate and appear to include lobula plate intrinsic neurons (lopi neurons, Figure 6B). Class 2 Svp neurons migrate from their birthplace in the topc, and their cell bodies are found in the distal medulla cortex. These neurons resemble previously described Distal medulla 6 neurons (Dm6 neurons, Figures 6C and 6C 0 ), but because their arborizations are larger, we named them Dm6b. Cell bodies of class 3 Toy neurons are all found in the medulla rim, the region between the medulla and the lobula/lobula plate neuropils (Figure S6A). These neurons can be classified into three groups, depending on where they project. The first group is composed of Medulla tangential neurons Mt8 and Mt4 that connect different layers of the medulla with the central brain (Figures 6D, 6D 0, and S7A). The second group is composed of previously undescribed proximal medulla neurons (Pm) that project into medulla layer M7 (Figures 6E and 6E 0 ). We named these neurons Pm7a, b, and c, depending on the length of their arborizations (Figures S7B S7D). The third group is composed of lobula columnar neurons 6 (LCN6), which connect layers of the lobula with the central brain (Figures 6F and S7D). The presence of three different groups of class 3 Toy neurons (Mt, Pm, and LCN) raises the possibility that unknown temporal genes divide the Slp time window into three smaller windows, each producing one of these groups. Alternatively, this subdivision may be achieved by spatial patterning genes. Finally, cell bodies of class 4 Toy/D neurons are localized in the lobula plate cortex. These neurons resemble lobula complex columnar neurons 2 (Lccn2) that connect the lobula and the lobula plate with the central brain. We could not obtain single neuron clones, but we could identify at least two subtypes of these neurons projecting into different layers of the lobula plate neuropil (Figures 6G and S7E). We named them Lcnn2a and b. As in the Slp time window, the fact that we could identify two subtypes of Toy/D neurons suggests the existence of additional temporal and/or spatial patterning genes. In conclusion, unlike the main OPC, the topc region produces several neuronal cell types that innervate three different neuropils of the adult optic lobes the medulla, lobula, and lobula plate (Figure 6H). Therefore, in addition to leading to the production of distinct neuronal subtypes over time, the four temporal windows defined by Dll, Ey, Slp, and D also determine in which Cell 158, , August 28, 2014 ª2014 Elsevier Inc. 1181
10 Figure 5. topc Temporal Factors and the Cell Death Pathway Posterior views of L3 optic lobes. Unless specified, dashed lines show the topc. (A and A0 ) In slp mutant clones (dashed lines), NotchON Toy-expressing neurons (red) die as shown by expression of cleaved Caspase-3 (yellow, arrows). (B and B0 ) Expressing P35 in slp clones rescues the death of NotchON Toy neurons (red). (C) The cell death pathway. (D) NotchOFF class 2 (Svp, cyan) and NotchON class 3 and 4 clusters (Toy, red) in a WT topc. (E) Removing hid, rpr, and grim (def(3l)h99 clones, dashed lines) rescues the death of NotchON Toy neurons (red) in the Ey time window (arrowhead) and that of NotchOFF Svp neurons (cyan) in the Slp and D time windows (arrow). (legend continued on next page) 1182 Cell 158, , August 28, 2014 ª2014 Elsevier Inc.
11 area of the optic lobes the neuronal cell bodies are localized (Figure 7): the Dll time window gives rise to neurons whose cell bodies are in the lobula plate cortex; the Ey time window produces neurons with cell bodies in the medulla cortex; the Slp time window produces neurons with cell bodies in the medulla rim; and the D time window produces neurons with cell bodies in the lobula plate cortex. DISCUSSION Although apoptosis is a common feature of neurogenesis in both vertebrates and Drosophila, the mechanisms controlling this process are still poorly understood. For instance, several studies in Drosophila have shown that, depending on the context, Notch can either induce neurons to die or allow them to survive during binary cell fate decisions. This is the case in the antennal lobes where Notch induces apoptosis in the antero-dorsal projecting neurons lineage (adpn), whereas it promotes survival in the ventral projecting neurons lineage (vpn) (Lin et al., 2010). In both of these cases, the entire lineage makes the same decision whether the Notch ON or Notch OFF cells survive or die. This suggests that, in this system, Notch integrates spatial signals to specify neuronal survival or apoptosis. Here, we show that, during topc neurogenesis, neuronal survival is determined by the interplay between Notch and temporal patterning of progenitors. Indeed, within the same lineage, Notch signaling leads to two different fates: it first induces neurons to die, whereas later, it allows them to survive. We show that this switch is due to the sequential expression of three highly conserved transcription factors Dll/Dlx, Ey/Pax-6, and Slp/ Fkh in neural progenitors. These three factors have distinct functions, with Dll promoting survival of Notch OFF neurons, Ey inducing apoptosis of Notch OFF neurons, and Slp promoting survival of Notch ON neurons. Our data suggest that Ey induces death of Notch OFF neurons by activating the proapoptotic factor hid. Thus, Dll probably antagonizes Ey activity by preventing Ey from activating hid. Our data also suggest that Notch signaling induces neuronal death by activating the proapoptotic gene rpr. Thus, Slp might promote survival of Notch ON neurons by directly repressing rpr expression or by preventing Notch from activating it. In both cases, the interplay between Notch and Slp modifies the default fate of Notch ON neurons, allowing them to survive. Further investigations will test these hypotheses and determine how Dll, Ey, Slp, and Notch differentially activate/ repress hid and rpr. Although the topc and the main OPC have related temporal sequences, their neurogenesis is very different. This difference is in part due to the fact that newly specified topc neuroblasts express Dll, which controls neuronal survival, instead of Hth. Why do topc neuroblasts express Dll? The topc, which is defined by Wg expression in the neuroepithelium, is flanked by a region expressing Dpp (Kaphingst and Kunes, 1994). Previous studies have shown that high levels of Wg and Dpp activate Dll expression in the distal cells of the Drosophila leg disc (Estella et al., 2008). Wg and Dpp could therefore also activate Dll in the neuroepithelium and at the beginning of the temporal series in topc progenitors. Another difference between the main OPC and topc neurogenesis is that Ey and Slp have completely different functions in these regions. Indeed, unlike in the main OPC, Ey and Slp control the survival of topc neurons. This suggests that autonomous and/or nonautonomous signals interact with these temporal factors and modify their function in the topc. Finally, topc neuroblasts produce neurons for three different neuropils of the adult visual system, the medulla, the lobula, and the lobula plate. This ability could be due to the particular location of this region in the larval optic lobes. Indeed, the topc is very close to the two larval structures giving rise to the lobula and lobula plate neuropils Dll-expressing neuroblasts are located next to the lobula plug, whereas D-expressing neuroblasts are close to the IPC. Interestingly, Dll and D neuroblasts specifically produce lobula plate neurons. This raises the possibility that these neuroblasts and/or the neurons produced by these neuroblasts receive signals from the lobula plug and the IPC, which instruct them to specifically produce lobula plate neurons. These nonautonomous signals could also modify the function of Ey and Slp in the topc. In summary, this study demonstrates that temporal patterning of progenitors, a well-conserved mechanism from Drosophila to vertebrates, generates neural cell diversity by controlling multiple aspects of neurogenesis, including neuronal identity, Notch-mediated cell survival decisions, and the mode of intermediate precursor division. In the topc temporal series, some factors control two of these aspects (Ey), whereas others have a specialized function (Dll, Slp, and D). This suggests that temporal patterning does not consist of a unique series of transcription factors controlling all aspects of neurogenesis but instead consists of multiple superimposed series, each with distinct functions. EXPERIMENTAL PROCEDURES Antibodies and Immunostaining Standard methods were used for antibody staining (see Extended Experimental Procedures). The following antibodies were used: rabbit anti-hth (1:500) (R. Mann), guinea pig anti-dll (1:500) (R. Mann), mouse anti-ey (1:10) (DSHB), rabbit anti-slp (1:500) (segmentation antibodies), guinea-pig anti- Runt (1:500) (segmentation antibodies), rabbit anti-d (1:200) (ModENCODE), guinea-pig anti-d (1:50) (J. Nambu), rabbit anti-salm (1:500) (T. Cook), mouse (F) Removing Dronc generates an intermingling between Notch OFF (Svp, cyan) and Notch ON neurons (Toy, red) in the Ey, Slp, and D time windows. White bars delimitate the border between the Ey and Slp time windows. (G) In Dronc mutants, Hid is only expressed in the Notch OFF Svp neurons that normally die in the Slp and D time windows (arrows). (H and H 0 ) Decreasing Hid activity (hid hypomorph /def(3l)h99) has no effect on the neurons produced during the Ey time window (H). However, this rescues the death of Notch OFF Svp neurons (cyan) in the Slp and D time windows (H 0 ). This phenotype is more pronounced in the ventral side of the topc (arrow). (I and I 0 ) Removing rpr rescues the death of Notch ON Toy neurons in the Ey time window (red, I) but has no effect on the neurons produced during the Slp and D time windows (I 0 ). (J) Interactions between topc temporal factors and cell death genes. See also Figure S5. Cell 158, , August 28, 2014 ª2014 Elsevier Inc. 1183
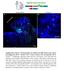
12 Figure 6. Neuronal Subtypes Produced in the topc (A) The FLEXAMP memory cassette. (B G) FLEXAMP memory clones in midpupal (B) and adult brains (C G). In all panels, dashed lines show the neuropils stained by N-Cad (magenta). Arrows point to the neuronal cell bodies, and arrowheads point to the projections. (B and B0 ) wg > FLEXAMP clones (green) suggest that class 1 Salm/Runt neurons (white) are Lopi neurons. (legend continued on next page) 1184 Cell 158, , August 28, 2014 ª2014 Elsevier Inc.
13 crossed to wg-gal4 or slp-gal4 combined with tub-gal80 ts. The progeny of these flies was raised at 18 C, incubated several hours at 29 C (to inactivate Gal80), and put back at 18 C until adult stage. The incubation time to obtain an optimal and analyzable number of GFP-expressing neurons in adults varied depending on the gal4 line (4 hr with wg-gal4 and 24 hr with slp-gal4). Quantifications For each staining, positive cells were counted manually on each z plane using custom-written scripts in Igor Pro (WaveMetrics). Each graph bar represents the average number of neurons for a given genotype. Error bars represent SD, and t tests are used to determine whether the samples are significantly different. SUPPLEMENTAL INFORMATION Supplemental Information includes Extended Experimental Procedures and seven figures and can be found with this article online at /j.cell AUTHOR CONTRIBUTIONS Figure 7. Summary of topc Neurogenesis The Dll, Ey, Slp, and D time windows produce distinct neuronal subtypes, each localized to a specific area of the optic lobes. anti-svp (1:500) (Y. Hiromi), guinea-pig anti-toy (1:500) (U. Walldorf), rabbit anti-ey/toy (1/500) (J. Clements), guinea-pig anti-hey (1:1,000) (C. Delidakis), guinea pig anti-dpn (1:500) (J. Skeath), rabbit anti-ase (1:100) (Y. Jan), rabbit anti-cleaved-caspase-3 (1:100) (Cell Signaling Technology), guinea pig anti- Hid (D. Ryoo), mouse anti-24b10 (1:20), Rat anti-de-cadherin (1:20), and Rat anti-dn-cadherin (1:20) (all from DSHB), sheep anti-gfp (1:500, AbD Serotec). Secondary antibodies are from Jackson or Invitrogen. Genetics and Fly Strains The following Gal4 lines were used: wg-gal4 (ND382, K. Basler); ey OK107 -gal4 (JB. Connolly); slp-gal4 (R35A08, Janelia Gal4 collection), and insc-gal4 (Li et al., 2013). The following mutant stocks were used: y,w;;; ey J5.71 /Miunc- 13 MI00468 (y + ); Dronc I24 /TM3Sb (D. Ryoo); def(3l)h99,knip/tm3sb (Bloomington 1576); hid A22 /TM6c (L. Johnston); and y,w,hsflp;;def(3l)rpr XR38 /TM6b (L. Johnston). Wild-type and mutant clones were generated by 37 C heat shocks at early larval stages. Wandering third instar larvae were analyzed. The progeny of overexpression and RNAi experiments (all involving tubgal80 ts ) was raised at 18 C, heat shocked 2 days at 29 C from early larval stages and dissected right after heat shock. Genotypes of clonal, RNAi, and overexpression experiments are simplified throughout the text and figures. See Extended Experimental Procedures for complete genotypes and crosses. FLEXAMP Memory Cassette FLEXAMP genotype: y,w,uas-flp; If/CyO; act > y[+] > LHV2-86Fb,13Xlex- Aop2-myr::GFP/TM6B. The act > y[+] > LHV2-86Fb stop cassette (K. Basler) contains an optimized version of lex VP16 that has a reduced toxicity and cannot be inhibited by Gal80. The lexaop13x-myr::gfp reporter (B. Pfeiffer) is an optimized version of lexaop, leading to strong expression levels of myr::gfp without being leaky in the optic lobes. To generate clones, FLEXAMP was C.B. designed and performed all experiments. C.B., X.L., and T.E performed the initial antibody screen. X.L. identified the slp-gal4 line. M.C. generated the Igor procedures for quantifications. B.W. contributed to the design of cell death experiments. C.D. contributed to the design of experiments. The manuscript was written by C.B. and C.D., and all authors commented on it. ACKNOWLEDGMENTS We thank the fly community and the modencode team for gifts of antibodies and fly stocks. We especially thank Don Ryoo for his valuable help and advice regarding cell death experiments. We thank the Desplan laboratory members for discussions and support, as well as M. Slaidina, L. Christiaen, C. Maurange, N. Vogt, J. Bos, C. Doe, T. Lee, and R. Jonhston for critically reading the manuscript. This work was supported by a grant from NIH to C.D. (R01 EY017916). Support was also provided by NYU Abu Dhabi grant G-1205C to C.D.; fellowships from EMBO (ALTF ), HFSPO (LT000077/2010-L), and Philippe Foundation to C.B.; The Robert Leet and Clara Guthrie Patterson Trust to X.L.; The Canadian Institutes of Health and Research (CIHR) to T.E.; EMBO (ALTF ) and The Revson Foundation to M.C; and NIH (T32EY ) to B.W. Received: October 30, 2013 Revised: June 11, 2014 Accepted: July 7, 2014 Published: August 28, 2014 REFERENCES Baumgardt, M., Karlsson, D., Terriente, J., Díaz-Benjumea, F.J., and Thor, S. (2009). Neuronal subtype specification within a lineage by opposing temporal feed-forward loops. Cell 139, Bello, B.C., Izergina, N., Caussinus, E., and Reichert, H. (2008). Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 3, 5. (C and C 0 ) (C) wg > FLEXAMP clones (green). Class 2 Svp neurons (cyan) are Dm6b neurons projecting in medulla layers M2 and M3. (C 0 ) Dm6b projections seem to contact R8 photoreceptors axons (Chaoptin, magenta). (D F) slp > FLEXAMP clones (green) showing the three types of class 3 Toy neurons (red). (D and D 0 ) Mt8 neurons project in M6, M7 (white arrowheads), and the central brain (blue arrowhead). (D 0 ) Mt8 projections seem to contact R7 photoreceptors terminations (Chaoptin, magenta). (E) Two Pm7b neurons (medium-size projections) identified with white and yellow arrows, projecting into M7 and contacting R7 projections (Chaoptin, magenta in E 0 ). (F) LCN6 neurons arborize in lobula layers Lo3, 4, and 5 (white arrowheads) and project toward the central brain (blue arrowhead,). (G) slp > FLEXAMP clones (green). Class 4 Toy/D neurons (yellow) are Lccn2a and Lccn2b neurons that connect the lobula plate (white arrowheads) with the lobula (red arrowheads) and the central brain (blue arrowhead). (H) Neuronal subtypes produced by the topc. M, medulla; Lo, lobula; Lop, lobula plate; and CB, central brain. See also Figure S7. Cell 158, , August 28, 2014 ª2014 Elsevier Inc. 1185
glial cells missing and gcm2 Cell-autonomously Regulate Both Glial and Neuronal
 glial cells missing and gcm2 Cell-autonomously Regulate Both Glial and Neuronal Development in the Visual System of Drosophila Carole Chotard, Wendy Leung and Iris Salecker Supplemental Data Supplemental
glial cells missing and gcm2 Cell-autonomously Regulate Both Glial and Neuronal Development in the Visual System of Drosophila Carole Chotard, Wendy Leung and Iris Salecker Supplemental Data Supplemental
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Expression of escargot (esg) and genetic approach for achieving IPC-specific knockdown. (a) esg MH766 -Gal4 UAS-cd8GFP (green) and esg-lacz B7-2-22 (red) show similar expression
Supplementary Figure 1 Expression of escargot (esg) and genetic approach for achieving IPC-specific knockdown. (a) esg MH766 -Gal4 UAS-cd8GFP (green) and esg-lacz B7-2-22 (red) show similar expression
Neurogenesis and neuronal circuit formation in the Drosophila visual center
 The Japanese Society of Developmental Biologists Develop. Growth Differ. (2014) 56, 491 498 doi: 10.1111/dgd.12151 Review Article Neurogenesis and neuronal circuit formation in the Drosophila visual center
The Japanese Society of Developmental Biologists Develop. Growth Differ. (2014) 56, 491 498 doi: 10.1111/dgd.12151 Review Article Neurogenesis and neuronal circuit formation in the Drosophila visual center
Cell Birth and Death. Chapter Three
 Cell Birth and Death Chapter Three Neurogenesis All neurons and glial cells begin in the neural tube Differentiated into neurons rather than ectoderm based on factors we have already discussed If these
Cell Birth and Death Chapter Three Neurogenesis All neurons and glial cells begin in the neural tube Differentiated into neurons rather than ectoderm based on factors we have already discussed If these
Neuroepithelial Cells and Neural Differentiation
 Neuroepithelial Cells and Neural Differentiation Neurulation The cells of the neural tube are NEUROEPITHELIAL CELLS Neural crest cells migrate out of neural tube Neuroepithelial cells are embryonic stem
Neuroepithelial Cells and Neural Differentiation Neurulation The cells of the neural tube are NEUROEPITHELIAL CELLS Neural crest cells migrate out of neural tube Neuroepithelial cells are embryonic stem
Cell migration in Drosophila optic lobe neurons is controlled by eyeless/pax6
 687 Development 138, 687-693 (2011) doi:10.1242/dev.056069 2011. Published by The Company of Biologists Ltd Cell migration in Drosophila optic lobe neurons is controlled by eyeless/pax6 Javier Morante
687 Development 138, 687-693 (2011) doi:10.1242/dev.056069 2011. Published by The Company of Biologists Ltd Cell migration in Drosophila optic lobe neurons is controlled by eyeless/pax6 Javier Morante
Supplementary Figure 1 Madm is not required in GSCs and hub cells. (a,b) Act-Gal4-UAS-GFP (a), Act-Gal4-UAS- GFP.nls (b,c) is ubiquitously expressed
 Supplementary Figure 1 Madm is not required in GSCs and hub cells. (a,b) Act-Gal4-UAS-GFP (a), Act-Gal4-UAS- GFP.nls (b,c) is ubiquitously expressed in the testes. The testes were immunostained with GFP
Supplementary Figure 1 Madm is not required in GSCs and hub cells. (a,b) Act-Gal4-UAS-GFP (a), Act-Gal4-UAS- GFP.nls (b,c) is ubiquitously expressed in the testes. The testes were immunostained with GFP
Article. The Color-Vision Circuit in the Medulla of Drosophila. Javier Morante 1 and Claude Desplan 1, * 1
 Current Biology 18, 1 13, April 22, 2008 ª2008 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2008.02.075 The Color-Vision Circuit in the Medulla of Drosophila Article Javier Morante 1 and Claude Desplan
Current Biology 18, 1 13, April 22, 2008 ª2008 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2008.02.075 The Color-Vision Circuit in the Medulla of Drosophila Article Javier Morante 1 and Claude Desplan
Supplementary Figure S1: TIPF reporter validation in the wing disc.
 Supplementary Figure S1: TIPF reporter validation in the wing disc. a,b, Test of put RNAi. a, In wildtype discs the Dpp target gene Sal (red) is expressed in a broad stripe in the centre of the ventral
Supplementary Figure S1: TIPF reporter validation in the wing disc. a,b, Test of put RNAi. a, In wildtype discs the Dpp target gene Sal (red) is expressed in a broad stripe in the centre of the ventral
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1. Ras V12 expression in the entire eye-antennal disc does not cause invasive tumours. a, Eye-antennal discs expressing Ras V12 in all cells (marked with GFP, green) overgrow moderately
Supplementary Figure 1. Ras V12 expression in the entire eye-antennal disc does not cause invasive tumours. a, Eye-antennal discs expressing Ras V12 in all cells (marked with GFP, green) overgrow moderately
effects on organ development. a-f, Eye and wing discs with clones of ε j2b10 show no
 Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
Supplemental Information. Proprioceptive Opsin Functions. in Drosophila Larval Locomotion
 Neuron, Volume 98 Supplemental Information Proprioceptive Opsin Functions in Drosophila Larval Locomotion Damiano Zanini, Diego Giraldo, Ben Warren, Radoslaw Katana, Marta Andrés, Suneel Reddy, Stephanie
Neuron, Volume 98 Supplemental Information Proprioceptive Opsin Functions in Drosophila Larval Locomotion Damiano Zanini, Diego Giraldo, Ben Warren, Radoslaw Katana, Marta Andrés, Suneel Reddy, Stephanie
Supplemental Data SUPPLEMENTAL EXPERIMENTAL PROCEDURES
 Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cédric Maurange, Louise Cheng and Alex P. Gould Supplemental Data SUPPLEMENTAL EXPERIMENTAL PROCEDURES
Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cédric Maurange, Louise Cheng and Alex P. Gould Supplemental Data SUPPLEMENTAL EXPERIMENTAL PROCEDURES
Concentric zones, cell migration and neuronal circuits in the Drosophila visual center
 983 Development 138, 983-993 (2011) doi:10.1242/dev.058370 2011. Published by The Company of Biologists Ltd Concentric zones, cell migration and neuronal circuits in the Drosophila visual center Eri Hasegawa
983 Development 138, 983-993 (2011) doi:10.1242/dev.058370 2011. Published by The Company of Biologists Ltd Concentric zones, cell migration and neuronal circuits in the Drosophila visual center Eri Hasegawa
Nature Neuroscience: doi: /nn Supplementary Figure 1. MADM labeling of thalamic clones.
 Supplementary Figure 1 MADM labeling of thalamic clones. (a) Confocal images of an E12 Nestin-CreERT2;Ai9-tdTomato brain treated with TM at E10 and stained for BLBP (green), a radial glial progenitor-specific
Supplementary Figure 1 MADM labeling of thalamic clones. (a) Confocal images of an E12 Nestin-CreERT2;Ai9-tdTomato brain treated with TM at E10 and stained for BLBP (green), a radial glial progenitor-specific
Neurodevelopment II Structure Formation. Reading: BCP Chapter 23
 Neurodevelopment II Structure Formation Reading: BCP Chapter 23 Phases of Development Ovum + Sperm = Zygote Cell division (multiplication) Neurogenesis Induction of the neural plate Neural proliferation
Neurodevelopment II Structure Formation Reading: BCP Chapter 23 Phases of Development Ovum + Sperm = Zygote Cell division (multiplication) Neurogenesis Induction of the neural plate Neural proliferation
marker. DAPI labels nuclei. Flies were 20 days old. Scale bar is 5 µm. Ctrl is
 Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
The Tumor Suppressors Brat and Numb Regulate Transit-Amplifying Neuroblast Lineages in Drosophila
 Article The Tumor Suppressors Brat and Numb Regulate Transit-Amplifying Neuroblast Lineages in Drosophila Sarah K. Bowman, 1 Vivien Rolland, 1 Joerg Betschinger, 1,3 Kaolin A. Kinsey, 2 Gregory Emery,
Article The Tumor Suppressors Brat and Numb Regulate Transit-Amplifying Neuroblast Lineages in Drosophila Sarah K. Bowman, 1 Vivien Rolland, 1 Joerg Betschinger, 1,3 Kaolin A. Kinsey, 2 Gregory Emery,
Nature Neuroscience: doi: /nn Supplementary Figure 1. Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites.
 Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
Supplementary Figure 1. Nature Neuroscience: doi: /nn.4547
 Supplementary Figure 1 Characterization of the Microfetti mouse model. (a) Gating strategy for 8-color flow analysis of peripheral Ly-6C + monocytes from Microfetti mice 5-7 days after TAM treatment. Living
Supplementary Figure 1 Characterization of the Microfetti mouse model. (a) Gating strategy for 8-color flow analysis of peripheral Ly-6C + monocytes from Microfetti mice 5-7 days after TAM treatment. Living
Supporting Information
 Supporting Information Fig. S1. Overexpression of Rpr causes progenitor cell death. (A) TUNEL assay of control intestines. No progenitor cell death could be observed, except that some ECs are undergoing
Supporting Information Fig. S1. Overexpression of Rpr causes progenitor cell death. (A) TUNEL assay of control intestines. No progenitor cell death could be observed, except that some ECs are undergoing
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
The Hunchback temporal transcription factor establishes, but is not required to maintain, early-born neuronal identity
 Hirono et al. Neural Development (2017) 12:1 DOI 10.1186/s13064-017-0078-1 RESEARCH ARTICLE Open Access The Hunchback temporal transcription factor establishes, but is not required to maintain, early-born
Hirono et al. Neural Development (2017) 12:1 DOI 10.1186/s13064-017-0078-1 RESEARCH ARTICLE Open Access The Hunchback temporal transcription factor establishes, but is not required to maintain, early-born
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang and Page-McCaw, http://www.jcb.org/cgi/content/full/jcb.201403084/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Extracellular anti-wg staining is specific. Note
Supplemental material Wang and Page-McCaw, http://www.jcb.org/cgi/content/full/jcb.201403084/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Extracellular anti-wg staining is specific. Note
Supplementary Figure 1
 Supplementary Figure 1 Kif1a RNAi effect on basal progenitor differentiation Related to Figure 2. Representative confocal images of the VZ and SVZ of rat cortices transfected at E16 with scrambled or Kif1a
Supplementary Figure 1 Kif1a RNAi effect on basal progenitor differentiation Related to Figure 2. Representative confocal images of the VZ and SVZ of rat cortices transfected at E16 with scrambled or Kif1a
Supplemental Information. Competition for Space Is Controlled. by Apoptosis-Induced Change. of Local Epithelial Topology
 Current Biology, Volume 28 Supplemental Information Competition for Space Is Controlled by Apoptosis-Induced Change of Local Epithelial Topology Alice Tsuboi, Shizue Ohsawa, Daiki Umetsu, Yukari Sando,
Current Biology, Volume 28 Supplemental Information Competition for Space Is Controlled by Apoptosis-Induced Change of Local Epithelial Topology Alice Tsuboi, Shizue Ohsawa, Daiki Umetsu, Yukari Sando,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2419 Figure S1 NiGFP localization in Dl mutant dividing SOPs. a-c) time-lapse analysis of NiGFP (green) in Dl mutant SOPs (H2B-RFP, red; clones were identified by the loss of nlsgfp) showing
DOI: 10.1038/ncb2419 Figure S1 NiGFP localization in Dl mutant dividing SOPs. a-c) time-lapse analysis of NiGFP (green) in Dl mutant SOPs (H2B-RFP, red; clones were identified by the loss of nlsgfp) showing
Supplementary Figures
 J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
A region-specific neurogenesis mode requires migratory progenitors in the Drosophila visual system
 Europe PMC Funders Group Author Manuscript Published in final edited form as: Nat Neurosci. 2015 January ; 18(1): 46 55. doi:10.1038/nn.3896. A region-specific neurogenesis mode requires migratory progenitors
Europe PMC Funders Group Author Manuscript Published in final edited form as: Nat Neurosci. 2015 January ; 18(1): 46 55. doi:10.1038/nn.3896. A region-specific neurogenesis mode requires migratory progenitors
Control of neural stem cell self-renewal and differentiation in Drosophila
 Institutional Repository of the University of Basel University Library Schoenbeinstrasse 18-20 CH-4056 Basel, Switzerland http://edoc.unibas.ch/ Year: 2014 Control of neural stem cell self-renewal and
Institutional Repository of the University of Basel University Library Schoenbeinstrasse 18-20 CH-4056 Basel, Switzerland http://edoc.unibas.ch/ Year: 2014 Control of neural stem cell self-renewal and
Supplementary Table 1. List of primers used in this study
 Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
Viktorin, G. and Riebli, N. and Popkova, A. and Giangrande, A. and Reichert, H.
 Institutional Repository of the University of Basel University Library Schoenbeinstrasse 18-20 CH-4056 Basel, Switzerland http://edoc.unibas.ch/ Year: 2011 Multipotent neural stem cells generate glial
Institutional Repository of the University of Basel University Library Schoenbeinstrasse 18-20 CH-4056 Basel, Switzerland http://edoc.unibas.ch/ Year: 2011 Multipotent neural stem cells generate glial
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature07173 SUPPLEMENTARY INFORMATION Supplementary Figure Legends: Supplementary Figure 1: Model of SSC and CPC divisions a, Somatic stem cells (SSC) reside adjacent to the hub (red), self-renew
doi: 10.1038/nature07173 SUPPLEMENTARY INFORMATION Supplementary Figure Legends: Supplementary Figure 1: Model of SSC and CPC divisions a, Somatic stem cells (SSC) reside adjacent to the hub (red), self-renew
Developmental Biology
 Developmental iology 356 (2011) 553 565 ontents lists available at ScienceDirect Developmental iology journal homepage: www.elsevier.com/developmentalbiology Multipotent neural stem cells generate glial
Developmental iology 356 (2011) 553 565 ontents lists available at ScienceDirect Developmental iology journal homepage: www.elsevier.com/developmentalbiology Multipotent neural stem cells generate glial
Supplementary Figure S1: Tanycytes are restricted to the central/posterior hypothalamus
 Supplementary Figure S1: Tanycytes are restricted to the central/posterior hypothalamus a: Expression of Vimentin, GFAP, Sox2 and Nestin in anterior, central and posterior hypothalamus. In the anterior
Supplementary Figure S1: Tanycytes are restricted to the central/posterior hypothalamus a: Expression of Vimentin, GFAP, Sox2 and Nestin in anterior, central and posterior hypothalamus. In the anterior
Supplemental Information. Otic Mesenchyme Cells Regulate. Spiral Ganglion Axon Fasciculation. through a Pou3f4/EphA4 Signaling Pathway
 Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Developmental Biology
 Developmental Biology 327 (2009) 288 300 Contents lists available at ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology The HLH protein Extramacrochaetae is required
Developmental Biology 327 (2009) 288 300 Contents lists available at ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology The HLH protein Extramacrochaetae is required
HHS Public Access Author manuscript Curr Opin Neurobiol. Author manuscript; available in PMC 2018 February 01.
 Timing temporal transitions during brain development Anthony M. Rossi 1,*, Vilaiwan M. Fernandes 1,*, and Claude Desplan 1,2, 1 Department of Biology, New York University, New York, New York 10003, USA
Timing temporal transitions during brain development Anthony M. Rossi 1,*, Vilaiwan M. Fernandes 1,*, and Claude Desplan 1,2, 1 Department of Biology, New York University, New York, New York 10003, USA
A Drosophila model for alcohol reward. Supplemental Material
 Kaun et al. Supplemental Material 1 A Drosophila model for alcohol reward Supplemental Material Kaun, K.R. *, Azanchi, R. *, Maung, Z. *, Hirsh, J. and Heberlein, U. * * Department of Anatomy, University
Kaun et al. Supplemental Material 1 A Drosophila model for alcohol reward Supplemental Material Kaun, K.R. *, Azanchi, R. *, Maung, Z. *, Hirsh, J. and Heberlein, U. * * Department of Anatomy, University
100 mm Sucrose. +Berberine +Quinine
 8 mm Sucrose Probability (%) 7 6 5 4 3 Wild-type Gr32a / 2 +Caffeine +Berberine +Quinine +Denatonium Supplementary Figure 1: Detection of sucrose and bitter compounds is not affected in Gr32a / flies.
8 mm Sucrose Probability (%) 7 6 5 4 3 Wild-type Gr32a / 2 +Caffeine +Berberine +Quinine +Denatonium Supplementary Figure 1: Detection of sucrose and bitter compounds is not affected in Gr32a / flies.
Dissecting and Staining Drosophila Optic Lobes PAGE PROOFS
 Ch10_DNB:Drosophila Neurobiology Manual 2/19/10 9:11 AM Page 1 10 Dissecting and Staining Drosophila Optic Lobes ABSTRACT The Drosophila visual system is composed of Optic Lobes at Different Stages of
Ch10_DNB:Drosophila Neurobiology Manual 2/19/10 9:11 AM Page 1 10 Dissecting and Staining Drosophila Optic Lobes ABSTRACT The Drosophila visual system is composed of Optic Lobes at Different Stages of
Postembryonic Development of Amplifying Neuroblast Lineages in the Drosophila Brain: Proliferation, Differentiation and Projection Patterns.
 Postembryonic Development of Amplifying Neuroblast Lineages in the Drosophila Brain: Proliferation, Differentiation and Projection Patterns. Inauguraldissertation zur Erlangung der Würde eines Doktors
Postembryonic Development of Amplifying Neuroblast Lineages in the Drosophila Brain: Proliferation, Differentiation and Projection Patterns. Inauguraldissertation zur Erlangung der Würde eines Doktors
Role of Notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster
 RESEARCH ARTICLE 53 Development 137, 53-61 (2010) doi:10.1242/dev.041749 Role of Notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster James W. Truman*,, Wanda
RESEARCH ARTICLE 53 Development 137, 53-61 (2010) doi:10.1242/dev.041749 Role of Notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster James W. Truman*,, Wanda
Supplementary Figure 1. Procedures to independently control fly hunger and thirst states.
 Supplementary Figure 1 Procedures to independently control fly hunger and thirst states. (a) Protocol to produce exclusively hungry or thirsty flies for 6 h water memory retrieval. (b) Consumption assays
Supplementary Figure 1 Procedures to independently control fly hunger and thirst states. (a) Protocol to produce exclusively hungry or thirsty flies for 6 h water memory retrieval. (b) Consumption assays
Cancer Stem Cells & Glioblastoma
 Cancer Stem Cells & Glioblastoma JP Hugnot «Brain plasticity, Neural stem cells and Glial tumors» INSERM U1051-UM2 Institut des Neurosciences de Montpellier Montpellier 1-Stem cells and Brain Stem Cells
Cancer Stem Cells & Glioblastoma JP Hugnot «Brain plasticity, Neural stem cells and Glial tumors» INSERM U1051-UM2 Institut des Neurosciences de Montpellier Montpellier 1-Stem cells and Brain Stem Cells
Control of neuronal cell fate and number by integration of distinct daughter cell proliferation modes with temporal progression
 Control of neuronal cell fate and number by integration of distinct daughter cell proliferation modes with temporal progression Carina Ulvklo, Ryan MacDonald, Caroline Bivik, Magnus Baumgardt, Daniel Karlsson
Control of neuronal cell fate and number by integration of distinct daughter cell proliferation modes with temporal progression Carina Ulvklo, Ryan MacDonald, Caroline Bivik, Magnus Baumgardt, Daniel Karlsson
Cell Type Nervous System I. Developmental Readout. Foundations. Stem cells. Organ formation. Human issues.
 7.72 10.11.06 Cell Type Nervous System I Human issues Organ formation Stem cells Developmental Readout Axes Cell type Axon guidance 3D structure Analysis Model + + organisms Foundations Principles 1 What
7.72 10.11.06 Cell Type Nervous System I Human issues Organ formation Stem cells Developmental Readout Axes Cell type Axon guidance 3D structure Analysis Model + + organisms Foundations Principles 1 What
Supplementary Information
 Supplementary Information Supplementary s Supplementary 1 All three types of foods suppress subsequent feeding in both sexes when the same food is used in the pre-feeding test feeding. (a) Adjusted pre-feeding
Supplementary Information Supplementary s Supplementary 1 All three types of foods suppress subsequent feeding in both sexes when the same food is used in the pre-feeding test feeding. (a) Adjusted pre-feeding
Supplementary Information
 Supplementary Information Title Degeneration and impaired regeneration of gray matter oligodendrocytes in amyotrophic lateral sclerosis Authors Shin H. Kang, Ying Li, Masahiro Fukaya, Ileana Lorenzini,
Supplementary Information Title Degeneration and impaired regeneration of gray matter oligodendrocytes in amyotrophic lateral sclerosis Authors Shin H. Kang, Ying Li, Masahiro Fukaya, Ileana Lorenzini,
Negative regulation of atonal in proneural cluster formation of Drosophila R8 photoreceptors
 Proc. Natl. Acad. Sci. USA Vol. 96, pp. 5055 5060, April 1999 Developmental Biology Negative regulation of atonal in proneural cluster formation of Drosophila R8 photoreceptors CHIEN-KUO CHEN AND CHENG-TING
Proc. Natl. Acad. Sci. USA Vol. 96, pp. 5055 5060, April 1999 Developmental Biology Negative regulation of atonal in proneural cluster formation of Drosophila R8 photoreceptors CHIEN-KUO CHEN AND CHENG-TING
doi: /nature09554
 SUPPLEMENTARY INFORMATION doi:10.1038/nature09554 Supplementary Figure 1: Optical Tracing with New Photoactivatable GFP Variants Reveals Enhanced Labeling of Neuronal Processes We qualitatively compare
SUPPLEMENTARY INFORMATION doi:10.1038/nature09554 Supplementary Figure 1: Optical Tracing with New Photoactivatable GFP Variants Reveals Enhanced Labeling of Neuronal Processes We qualitatively compare
Supplementary Figure 1. Electroporation of a stable form of β-catenin causes masses protruding into the IV ventricle. HH12 chicken embryos were
 Supplementary Figure 1. Electroporation of a stable form of β-catenin causes masses protruding into the IV ventricle. HH12 chicken embryos were electroporated with β- Catenin S33Y in PiggyBac expression
Supplementary Figure 1. Electroporation of a stable form of β-catenin causes masses protruding into the IV ventricle. HH12 chicken embryos were electroporated with β- Catenin S33Y in PiggyBac expression
Supplementary Figure 1. Flies form water-reward memory only in the thirsty state
 1 2 3 4 5 6 7 Supplementary Figure 1. Flies form water-reward memory only in the thirsty state Thirsty but not sated wild-type flies form robust 3 min memory. For the thirsty group, the flies were water-deprived
1 2 3 4 5 6 7 Supplementary Figure 1. Flies form water-reward memory only in the thirsty state Thirsty but not sated wild-type flies form robust 3 min memory. For the thirsty group, the flies were water-deprived
ErbB4 migrazione II parte
 ErbB4 migrazione II parte Control SVZ cells prefer to migrate on the NRG1 type III substrate the substrate preference of the neuroblasts migrating out of the SVZ explant was evaluated SVZ cells had a strong
ErbB4 migrazione II parte Control SVZ cells prefer to migrate on the NRG1 type III substrate the substrate preference of the neuroblasts migrating out of the SVZ explant was evaluated SVZ cells had a strong
Ahtiainen et al., http :// /cgi /content /full /jcb /DC1
 Supplemental material JCB Ahtiainen et al., http ://www.jcb.org /cgi /content /full /jcb.201512074 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Distinct distribution of different cell cycle phases in the
Supplemental material JCB Ahtiainen et al., http ://www.jcb.org /cgi /content /full /jcb.201512074 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. Distinct distribution of different cell cycle phases in the
Plasticity of Cerebral Cortex in Development
 Plasticity of Cerebral Cortex in Development Jessica R. Newton and Mriganka Sur Department of Brain & Cognitive Sciences Picower Center for Learning & Memory Massachusetts Institute of Technology Cambridge,
Plasticity of Cerebral Cortex in Development Jessica R. Newton and Mriganka Sur Department of Brain & Cognitive Sciences Picower Center for Learning & Memory Massachusetts Institute of Technology Cambridge,
Segmentation. Serial Homology. Hox genes
 Segmentation Hox genes Serial Homology William Bateson 1861-1926 Homeosis: a variation in which something has been changed into the likeness of something else Calvin Bridges at Columbia bithorax 1923 1
Segmentation Hox genes Serial Homology William Bateson 1861-1926 Homeosis: a variation in which something has been changed into the likeness of something else Calvin Bridges at Columbia bithorax 1923 1
atonal Regulates Neurite Arborization but Does Not Act as a Proneural Gene in the Drosophila Brain
 Neuron, Vol. 25, 549 561, March, 2000, Copyright 2000 by Cell Press atonal Regulates Neurite Arborization but Does Not Act as a Proneural Gene in the Drosophila Brain Bassem A. Hassan,* # Nessan A. Bermingham,*
Neuron, Vol. 25, 549 561, March, 2000, Copyright 2000 by Cell Press atonal Regulates Neurite Arborization but Does Not Act as a Proneural Gene in the Drosophila Brain Bassem A. Hassan,* # Nessan A. Bermingham,*
Supplemental Information. Tissue Myeloid Progenitors Differentiate. into Pericytes through TGF-b Signaling. in Developing Skin Vasculature
 Cell Reports, Volume 18 Supplemental Information Tissue Myeloid Progenitors Differentiate into Pericytes through TGF-b Signaling in Developing Skin Vasculature Tomoko Yamazaki, Ani Nalbandian, Yutaka Uchida,
Cell Reports, Volume 18 Supplemental Information Tissue Myeloid Progenitors Differentiate into Pericytes through TGF-b Signaling in Developing Skin Vasculature Tomoko Yamazaki, Ani Nalbandian, Yutaka Uchida,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3443 In the format provided by the authors and unedited. Supplementary Figure 1 TC and SC behaviour during ISV sprouting. (a) Predicted outcome of TC division and competitive Dll4-Notch-mediated
DOI: 10.1038/ncb3443 In the format provided by the authors and unedited. Supplementary Figure 1 TC and SC behaviour during ISV sprouting. (a) Predicted outcome of TC division and competitive Dll4-Notch-mediated
G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila
 nature methods G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila Cory J Evans, John M Olson, Kathy T Ngo, Eunha Kim, Noemi E Lee, Edward Kuoy, Alexander N Patananan, Daniel Sitz, PhuongThao
nature methods G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila Cory J Evans, John M Olson, Kathy T Ngo, Eunha Kim, Noemi E Lee, Edward Kuoy, Alexander N Patananan, Daniel Sitz, PhuongThao
Quantification of early stage lesions for loss of p53 should be shown in the main figures.
 Reviewer #1 (Remarks to the Author): Expert in prostate cancer The manuscript "Clonal dynamics following p53 loss of heterozygosity in Kras-driven cancers" uses a number of novel genetically engineered
Reviewer #1 (Remarks to the Author): Expert in prostate cancer The manuscript "Clonal dynamics following p53 loss of heterozygosity in Kras-driven cancers" uses a number of novel genetically engineered
A Genetic Program for Embryonic Development
 Concept 18.4: A program of differential gene expression leads to the different cell types in a multicellular organism During embryonic development, a fertilized egg gives rise to many different cell types
Concept 18.4: A program of differential gene expression leads to the different cell types in a multicellular organism During embryonic development, a fertilized egg gives rise to many different cell types
Transgenic Expression of the Helicobacter pylori Virulence Factor CagA Promotes Apoptosis or Tumorigenesis through JNK Activation in Drosophila
 Transgenic Expression of the Helicobacter pylori Virulence Factor CagA Promotes Apoptosis or Tumorigenesis through JNK Activation in Drosophila Anica M. Wandler, Karen Guillemin* Institute of Molecular
Transgenic Expression of the Helicobacter pylori Virulence Factor CagA Promotes Apoptosis or Tumorigenesis through JNK Activation in Drosophila Anica M. Wandler, Karen Guillemin* Institute of Molecular
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
T cell maturation. T-cell Maturation. What allows T cell maturation?
 T-cell Maturation What allows T cell maturation? Direct contact with thymic epithelial cells Influence of thymic hormones Growth factors (cytokines, CSF) T cell maturation T cell progenitor DN DP SP 2ry
T-cell Maturation What allows T cell maturation? Direct contact with thymic epithelial cells Influence of thymic hormones Growth factors (cytokines, CSF) T cell maturation T cell progenitor DN DP SP 2ry
File name: Supplementary Information Description: Supplementary Figures, Supplementary Table and Supplementary References
 File name: Supplementary Information Description: Supplementary Figures, Supplementary Table and Supplementary References File name: Supplementary Data 1 Description: Summary datasheets showing the spatial
File name: Supplementary Information Description: Supplementary Figures, Supplementary Table and Supplementary References File name: Supplementary Data 1 Description: Summary datasheets showing the spatial
Control of neuronal cell fate and number by integration of distinct daughter cell proliferation modes with temporal progression
 678 AND STEM CELLS Development 139, 678-689 (2012) doi:10.1242/dev.074500 2012. Published by The Company of Biologists Ltd Control of neuronal cell fate and number by integration of distinct daughter cell
678 AND STEM CELLS Development 139, 678-689 (2012) doi:10.1242/dev.074500 2012. Published by The Company of Biologists Ltd Control of neuronal cell fate and number by integration of distinct daughter cell
Neurogenesis in the insect brain: cellular identification and molecular characterization of brain neuroblasts in the grasshopper embryo
 Development 118, 941-955 (1993) Printed in Great Britain The Company of Biologists Limited 1993 941 Neurogenesis in the insect brain: cellular identification and molecular characterization of brain neuroblasts
Development 118, 941-955 (1993) Printed in Great Britain The Company of Biologists Limited 1993 941 Neurogenesis in the insect brain: cellular identification and molecular characterization of brain neuroblasts
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
Polarity and Segmentation. Chapter Two
 Polarity and Segmentation Chapter Two Polarization Entire body plan is polarized One end is different than the other Head vs. Tail Anterior vs. Posterior Front vs. Back Ventral vs. Dorsal Majority of neural
Polarity and Segmentation Chapter Two Polarization Entire body plan is polarized One end is different than the other Head vs. Tail Anterior vs. Posterior Front vs. Back Ventral vs. Dorsal Majority of neural
Regionalization of the nervous system. Paul Garrity 7.68J/9.013J February 25, 2004
 Regionalization of the nervous system Paul Garrity 7.68J/9.013J February 25, 2004 Patterning along: Rostral/Caudal (AP) axis Dorsal/Ventral (DV) axis Start with DV axial patterning in Spinal Cord Dorsal/Ventral
Regionalization of the nervous system Paul Garrity 7.68J/9.013J February 25, 2004 Patterning along: Rostral/Caudal (AP) axis Dorsal/Ventral (DV) axis Start with DV axial patterning in Spinal Cord Dorsal/Ventral
Nature Neuroscience doi: /nn Supplementary Figure 1. Characterization of viral injections.
 Supplementary Figure 1 Characterization of viral injections. (a) Dorsal view of a mouse brain (dashed white outline) after receiving a large, unilateral thalamic injection (~100 nl); demonstrating that
Supplementary Figure 1 Characterization of viral injections. (a) Dorsal view of a mouse brain (dashed white outline) after receiving a large, unilateral thalamic injection (~100 nl); demonstrating that
Supplemental Data. Wnt/β-Catenin Signaling in Mesenchymal Progenitors. Controls Osteoblast and Chondrocyte
 Supplemental Data Wnt/β-Catenin Signaling in Mesenchymal Progenitors Controls Osteoblast and Chondrocyte Differentiation during Vertebrate Skeletogenesis Timothy F. Day, Xizhi Guo, Lisa Garrett-Beal, and
Supplemental Data Wnt/β-Catenin Signaling in Mesenchymal Progenitors Controls Osteoblast and Chondrocyte Differentiation during Vertebrate Skeletogenesis Timothy F. Day, Xizhi Guo, Lisa Garrett-Beal, and
50mM D-Glucose. Percentage PI. L-Glucose
 Monica Dus, Minrong Ai, Greg S.B Suh. Taste-independent nutrient selection is mediated by a brain-specific Na+/solute cotransporter in Drosophila. Control + Phlorizin mm D-Glucose 1 2mM 1 L-Glucose Gr5a;Gr64a
Monica Dus, Minrong Ai, Greg S.B Suh. Taste-independent nutrient selection is mediated by a brain-specific Na+/solute cotransporter in Drosophila. Control + Phlorizin mm D-Glucose 1 2mM 1 L-Glucose Gr5a;Gr64a
SUPPLEMENTARY INFORMATION
 DOI: 0.038/ncb33 a b c 0 min 6 min 7 min (fixed) DIC -GFP, CenpF 3 µm Nocodazole Single optical plane -GFP, CenpF Max. intensity projection d µm -GFP, CenpF, -GFP CenpF 3-D rendering e f 0 min 4 min 0
DOI: 0.038/ncb33 a b c 0 min 6 min 7 min (fixed) DIC -GFP, CenpF 3 µm Nocodazole Single optical plane -GFP, CenpF Max. intensity projection d µm -GFP, CenpF, -GFP CenpF 3-D rendering e f 0 min 4 min 0
EMBO REPORT SUPPLEMENTARY SECTION. Quantitation of mitotic cells after perturbation of Notch signalling.
 EMBO REPORT SUPPLEMENTARY SECTION Quantitation of mitotic cells after perturbation of Notch signalling. Notch activation suppresses the cell cycle indistinguishably both within and outside the neural plate
EMBO REPORT SUPPLEMENTARY SECTION Quantitation of mitotic cells after perturbation of Notch signalling. Notch activation suppresses the cell cycle indistinguishably both within and outside the neural plate
Supporting Information
 Supporting Information Plikus et al. 10.1073/pnas.1215935110 SI Text Movies S1, S2, S3, and S4 are time-lapse recordings from individually cultured Period2 Luc vibrissa follicles show that circadian cycles
Supporting Information Plikus et al. 10.1073/pnas.1215935110 SI Text Movies S1, S2, S3, and S4 are time-lapse recordings from individually cultured Period2 Luc vibrissa follicles show that circadian cycles
Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at
 Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at E10.5 were double-stained for TUNEL (red) and PECAM-1 (green).
Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at E10.5 were double-stained for TUNEL (red) and PECAM-1 (green).
mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons
 Supplemental Material mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons Antoine de Chevigny, Nathalie Coré, Philipp Follert, Marion Gaudin, Pascal
Supplemental Material mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons Antoine de Chevigny, Nathalie Coré, Philipp Follert, Marion Gaudin, Pascal
Supplemental Information. Induction of Expansion and Folding. in Human Cerebral Organoids
 Cell Stem Cell, Volume 20 Supplemental Information Induction of Expansion and Folding in Human Cerebral Organoids Yun Li, Julien Muffat, Attya Omer, Irene Bosch, Madeline A. Lancaster, Mriganka Sur, Lee
Cell Stem Cell, Volume 20 Supplemental Information Induction of Expansion and Folding in Human Cerebral Organoids Yun Li, Julien Muffat, Attya Omer, Irene Bosch, Madeline A. Lancaster, Mriganka Sur, Lee
Inner ear development Nervous system development
 Upcoming Sessions April 22: Nervous System Development Lecture April 24: Reviews of Axonal Pathfinding in Sensory Systems April 29: Inner Ear Development Lecture May 1: May 6: May 8: Auditory System Pathfinding
Upcoming Sessions April 22: Nervous System Development Lecture April 24: Reviews of Axonal Pathfinding in Sensory Systems April 29: Inner Ear Development Lecture May 1: May 6: May 8: Auditory System Pathfinding
Bio 3411 Midterm Review:
 Midterm Review: Structure/Development/Systems/ Plastics/Talents/Diseases/Genes Structure General Overview Wednesday 1( 2( THE BRAIN ATLAS 3 rd ed, p. 8! THE BRAIN ATLAS 3 rd ed, p. 9! Mid-line (sagittal)
Midterm Review: Structure/Development/Systems/ Plastics/Talents/Diseases/Genes Structure General Overview Wednesday 1( 2( THE BRAIN ATLAS 3 rd ed, p. 8! THE BRAIN ATLAS 3 rd ed, p. 9! Mid-line (sagittal)
Patterning the Embryo
 Patterning the Embryo Anteroposterior axis Regional Identity in the Vertebrate Neural Tube Fig. 2.2 1 Brain and Segmental Ganglia in Drosophila Fig. 2.1 Genes that create positional and segment identity
Patterning the Embryo Anteroposterior axis Regional Identity in the Vertebrate Neural Tube Fig. 2.2 1 Brain and Segmental Ganglia in Drosophila Fig. 2.1 Genes that create positional and segment identity
Zhu et al, page 1. Supplementary Figures
 Zhu et al, page 1 Supplementary Figures Supplementary Figure 1: Visual behavior and avoidance behavioral response in EPM trials. (a) Measures of visual behavior that performed the light avoidance behavior
Zhu et al, page 1 Supplementary Figures Supplementary Figure 1: Visual behavior and avoidance behavioral response in EPM trials. (a) Measures of visual behavior that performed the light avoidance behavior
Chapter 5 A Dose Dependent Screen for Modifiers of Kek5
 Chapter 5 A Dose Dependent Screen for Modifiers of Kek5 "#$ ABSTRACT Modifier screens in Drosophila have proven to be a powerful tool for uncovering gene interaction and elucidating molecular pathways.
Chapter 5 A Dose Dependent Screen for Modifiers of Kek5 "#$ ABSTRACT Modifier screens in Drosophila have proven to be a powerful tool for uncovering gene interaction and elucidating molecular pathways.
SUPPLEMENTARY FIG. S2. Representative counting fields used in quantification of the in vitro neural differentiation of pattern of dnscs.
 Supplementary Data SUPPLEMENTARY FIG. S1. Representative counting fields used in quantification of the in vitro neural differentiation of pattern of anpcs. A panel of lineage-specific markers were used
Supplementary Data SUPPLEMENTARY FIG. S1. Representative counting fields used in quantification of the in vitro neural differentiation of pattern of anpcs. A panel of lineage-specific markers were used
Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the
 Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the location of the transmembrane (TM), FRM binding (FB)
Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the location of the transmembrane (TM), FRM binding (FB)
Bio 3411 Midterm Review:
 Bio 3411 Midterm Review: Structure/Development/Systems/ Plastics/Talents/Diseases/Genes Structure General Overview Wednesday October 26, 2011 1 2 THE BRAIN ATLAS 3 rd ed, p. 8! THE BRAIN ATLAS 3 rd ed,
Bio 3411 Midterm Review: Structure/Development/Systems/ Plastics/Talents/Diseases/Genes Structure General Overview Wednesday October 26, 2011 1 2 THE BRAIN ATLAS 3 rd ed, p. 8! THE BRAIN ATLAS 3 rd ed,
Nature Neuroscience: doi: /nn Supplementary Figure 1. Splenic atrophy and leucopenia caused by T3 SCI.
 Supplementary Figure 1 Splenic atrophy and leucopenia caused by T3 SCI. (a) Gross anatomy of representative spleens from control and T3 SCI mice at 28 days post-injury. (b and c) Hematoxylin and eosin
Supplementary Figure 1 Splenic atrophy and leucopenia caused by T3 SCI. (a) Gross anatomy of representative spleens from control and T3 SCI mice at 28 days post-injury. (b and c) Hematoxylin and eosin
Chapter 5. Summary and Future directions
 95 Chapter 5 Summary and Future directions 96 Much of our knowledge about glial development in the vertebrate CNS comes from studies of purified oligodendrocyte precursor cells (Raff 1989; Pfeiffer et
95 Chapter 5 Summary and Future directions 96 Much of our knowledge about glial development in the vertebrate CNS comes from studies of purified oligodendrocyte precursor cells (Raff 1989; Pfeiffer et
How strong is it? What is it? Where is it? What must sensory systems encode? 9/8/2010. Spatial Coding: Receptive Fields and Tactile Discrimination
 Spatial Coding: Receptive Fields and Tactile Discrimination What must sensory systems encode? How strong is it? What is it? Where is it? When the brain wants to keep certain types of information distinct,
Spatial Coding: Receptive Fields and Tactile Discrimination What must sensory systems encode? How strong is it? What is it? Where is it? When the brain wants to keep certain types of information distinct,
Spatial Coding: Receptive Fields and Tactile Discrimination
 Spatial Coding: Receptive Fields and Tactile Discrimination What must sensory systems encode? How strong is it? What is it? Where is it? When the brain wants to keep certain types of information distinct,
Spatial Coding: Receptive Fields and Tactile Discrimination What must sensory systems encode? How strong is it? What is it? Where is it? When the brain wants to keep certain types of information distinct,
Biological Bases of Behavior. 3: Structure of the Nervous System
 Biological Bases of Behavior 3: Structure of the Nervous System Neuroanatomy Terms The neuraxis is an imaginary line drawn through the spinal cord up to the front of the brain Anatomical directions are
Biological Bases of Behavior 3: Structure of the Nervous System Neuroanatomy Terms The neuraxis is an imaginary line drawn through the spinal cord up to the front of the brain Anatomical directions are
M Cells. Why parallel pathways? P Cells. Where from the retina? Cortical visual processing. Announcements. Main visual pathway from retina to V1
 Announcements exam 1 this Thursday! review session: Wednesday, 5:00-6:30pm, Meliora 203 Bryce s office hours: Wednesday, 3:30-5:30pm, Gleason https://www.youtube.com/watch?v=zdw7pvgz0um M Cells M cells
Announcements exam 1 this Thursday! review session: Wednesday, 5:00-6:30pm, Meliora 203 Bryce s office hours: Wednesday, 3:30-5:30pm, Gleason https://www.youtube.com/watch?v=zdw7pvgz0um M Cells M cells
SUPPLEMENTARY INFORMATION
 b 350 300 250 200 150 100 50 0 E0 E10 E50 E0 E10 E50 E0 E10 E50 E0 E10 E50 Number of organoids per well 350 300 250 200 150 100 50 0 R0 R50 R100 R500 1st 2nd 3rd Noggin 100 ng/ml Noggin 10 ng/ml Noggin
b 350 300 250 200 150 100 50 0 E0 E10 E50 E0 E10 E50 E0 E10 E50 E0 E10 E50 Number of organoids per well 350 300 250 200 150 100 50 0 R0 R50 R100 R500 1st 2nd 3rd Noggin 100 ng/ml Noggin 10 ng/ml Noggin
Notch Maintains Drosophila Type II Neuroblasts by Suppressing the. Expression of the Fez Transcription Factor Earmuff
 First posted online on 5 May 2016 as 10.1242/dev.136184 Access the most recent version at http://dev.biologists.org/lookup/doi/10.1242/dev.136184 Notch Maintains Drosophila Type II Neuroblasts by Suppressing
First posted online on 5 May 2016 as 10.1242/dev.136184 Access the most recent version at http://dev.biologists.org/lookup/doi/10.1242/dev.136184 Notch Maintains Drosophila Type II Neuroblasts by Suppressing
Diabetic pdx1-mutant zebrafish show conserved responses to nutrient overload and anti-glycemic treatment
 Supplementary Information Diabetic pdx1-mutant zebrafish show conserved responses to nutrient overload and anti-glycemic treatment Robin A. Kimmel, Stefan Dobler, Nicole Schmitner, Tanja Walsen, Julia
Supplementary Information Diabetic pdx1-mutant zebrafish show conserved responses to nutrient overload and anti-glycemic treatment Robin A. Kimmel, Stefan Dobler, Nicole Schmitner, Tanja Walsen, Julia
Organization of The Nervous System PROF. SAEED ABUEL MAKAREM
 Organization of The Nervous System PROF. SAEED ABUEL MAKAREM Objectives By the end of the lecture, you should be able to: List the parts of the nervous system. List the function of the nervous system.
Organization of The Nervous System PROF. SAEED ABUEL MAKAREM Objectives By the end of the lecture, you should be able to: List the parts of the nervous system. List the function of the nervous system.
Emx2 patterns the neocortex by regulating FGF positional signaling
 Emx2 patterns the neocortex by regulating FGF positional signaling Tomomi Fukuchi-Shimogori and Elizabeth A Grove Presented by Sally Kwok Background Cerebral cortex has anatomically and functionally distinct
Emx2 patterns the neocortex by regulating FGF positional signaling Tomomi Fukuchi-Shimogori and Elizabeth A Grove Presented by Sally Kwok Background Cerebral cortex has anatomically and functionally distinct
