Supplemental Information. Competition for Space Is Controlled. by Apoptosis-Induced Change. of Local Epithelial Topology
|
|
|
- Russell Osborne
- 6 years ago
- Views:
Transcription
1 Current Biology, Volume 28 Supplemental Information Competition for Space Is Controlled by Apoptosis-Induced Change of Local Epithelial Topology Alice Tsuboi, Shizue Ohsawa, Daiki Umetsu, Yukari Sando, Erina Kuranaga, Tatsushi Igaki, and Koichi Fujimoto
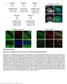
2 A Normalized frequency Rate of division : Simulation (Eq ) Simulation (Eq 2) Experiment (Farhadifar et al., 27) and more Side number n C Rate of division : in simulation Rate of division :3 in simulation simulations, 2988 cells (Eq ) simulations, 38 cells (Eq 2) B Normalized apical surface area <An>/<A> Rate of division : Simulation (Eq ) Simulation (Eq 2) Experiment (Farhadifar et al., 27) simulations, 2988 cells (Eq ) simulations, 38 cells (Eq 2) Side number n D E Cell anisotropy Cell anisotropy simulations simulations..7 Rate of division : Rate of division : Number of cells inside a clone Cell area (a.u.) F G Rate of division : Rate of division :3 4 8 Number of cells inside a clone simulations simulations Ratio Ratio3 Ratio Ratio3 Ratio Ratio3 Ratio Ratio3 Ratio Ratio 3 Ratio Ratio 3 Ratio Ratio 3 Ratio Ratio 3 Normalized cell area G= G= G= G=2 G= G= G= G=2 H I WT WT WT sqh RNAi WT hpo WT hpo 42-47, sqh RNAi WT WT WT Fraction of anisotropic cells Fisher s exact test with WT Fisher s exact test with hpo in vivo P < -2 P < -4 P < -4 P =.3 P = Fisher s exact test with WT Fisher s exact test with wts X in vivo P < -4 P < -4 P =.9 P < -4 P < -4 J Normalized frequency wild-type beside control (3 nota, cells) wild-type beside Yki SAS8AS2A :V (3 nota, 47 cells) apoptotic cells induced byyki SAS8AS2A :V ( nota, 39 cells) wild-type beside Ras V2 (4 nota, 28 cells) WT sqh RNAi hpo hpo 42-47, sqh WT shg RNAi wts X wts X, shg RNAi RNAi discs discs discs 4 discs discs discs 4 discs 4 discs 433 cells 2 cells 49 cells 3 cells 433 cells 7 cells 4 cells 399 cells Cell anisotropy Figure S. Cell deformation due to localized overgrowth robustly appeared and in vivo, Related to Figures, 2, and 4. (A, B) Cell packing topologies under conditions of identical division rate in simulations (Eq. and Eq. 2) account for that of normal epithelial cells in Drosophila [S]. Frequency distribution of side number (i.e., n-side cells) (A). Average area of cells with n-side (A n) normalized to that of all cells (A) (i.e., A n/a where < > denotes the population average) (B). Error bars represent s.d. for simulations. (C G) Additional simulation analysis for Figure using Eq.. (C) Time course of the simulated proliferation process under conditions of identical (upper) or differential (lower) proliferation rate, where the number of clonal cells (green) is (far left), 3,, 7, or 9 (far right). (D, E) Cell anisotropy of non-clonal cells located directly adjacent to the clone boundary (D) and area of clonal cells (E) as a function of number of cells inside of a clone, under conditions of identical (closed circles) and differential (open circles) proliferation rate. Error bars represent s.d. for simulations of each clone at each of the tested times and division rate ratios. (F, G) Robustness of cell deformation under identical and differential proliferation rate (representing Ratio and Ratio 3, respectively) with respect to model mechanical parameters: Line tension parameter for all edges was examined in four different values (G ij =,,, 2 in Eq. ), while the other parameters were kept constant (n H =, l =.). Cell anisotropy of non-clonal cells (white cells in C) directly adjacent to the clone
3 boundary (F). Area of clonal cells (green cells in C) beside the clone boundary, which is normalized by that of non-clonal cells (white in C) beside clone (G). Increased cell anisotropy and decreased area under differential proliferation rate were reproducible on all tested parameters. Error bars represent s.d. for simulations. The number of clonal cells was in the simulation. (H, I) Additional experimental analysis for Figures A and B. Hippo pathway mutant clones induce anisotropy in slower dividing wild-type cells directly adjacent to the clone even with modulation of junction contractility or adhesion in Drosophila wing disc hinge region. Genotypes of clones (green) are shown at top of panels in (H). Images for WT and wts X (H) are identical to those in Figure A. See Methods for the strain preparation. Scale bar, μm. (I) Fraction of cells whose anisotropy values if greater than.38 (see Figure B right for definition). Values for WT and wts X are identical to those in Figure B, right. Statistical test is a Fisher s exact test compared with WT (I, blue), hpo (I left, gray), or wts X (I right, gray). (J) Additional experimental analysis for Figures 2A and 2B. Filled circles: The distribution of cell anisotropy of wild-type cells directly adjacent to clones overexpressing p{tubp-gal8 ts }2 (control, blue), p{tubp-gal8 ts }2; UAS-yki SA,S8A,S2A :V (Yki SA,S8A,S2A :V, Orange), and p{tubp-gal8 ts }2; UAS-Ras V2 (Ras V2, Yellow) in Drosophila pupal notum. Open rectangles: The distribution of cell anisotropy of apoptotic cells 4 minutes before apoptosis completion was measured in a pupa of Video S.
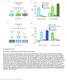
4 Ratio Rate of division : Cell anisotropy of apoptotic cell before death Rate of division : Frequency of boundary intercalation Clone-boundary-oriented cell anisotropy of apoptotic cell before death Corr. Coef =.44, P < -4 Corr. Coef = -.48, P < -4 Rate of division : Corr. Coef = -.3, P < -4 Corr. Coef =.38, P < Rate of division :3 I.2 Corr. Coef = -.2, P < -4 Corr. Coef =.2, P < -4. Cell anisotropy of apoptotic cell before apoptosis Clone (white cells) (green cells).8.4 Non-clone Corr. Coef = -.29, P < -4 (Boundary intercalation) Corr. Coef = -., P =.37 (Non-boundary intercalation) Corr. Coef =.29, P < -4 (Perpendicular intercalation).. H.2 Rate of division :3.8 Clone-boundary-oriented cell anisotropy of apoptotic cell before death G Acquired area after apoptosis (a.u.) Rate of division : Corr. Coef = -.3, P <. (Boundary intercalation) Corr. Coef = -., P <. (Non-boundary intercalation) Corr. Coef =.2, P < -3 (Perpendicular intercalation) Normalized frequency of cell intercalation Acquired area after apoptosis (a.u.) Corr. Coef =.2, P < -4 Corr. Coef = -.3, P <. Clone (white cells) (green cells) Ratio Non-clone Ratio F Rate of division :3 clones, 3 apoptosis.2 E Corr. Coef =.4, P < -4 Corr. Coef = -., P =.7 Rate of division : clones, 337 apoptosis.3 Ratio.2 D simulations Normalized cell area Rate of division :3 C simulations Cell anisotropy Rate of division : B Acquired area after apoptosis (a.u.) A Rate of division : Frequency of non-boundary intercalation Frequency of perpendicular intercalation Figure S2. All the series of events leading to winner-dominated cell expansion robustly occurred in another well-used type of cell-vertex model simulation, Related to Figures, 3, and. (A I) All results in this figure were obtained in Eq. 2. (A C) Cell deformations under identical and differential proliferation rate [representing Ratio and Ratio3, respectively, in (B) and (C)]. Consistent with the results shown in Figures SF and SG (using Eq. ), anisotropy of slower dividing cells directly adjacent to the clone boundary increased (B) and the normalized area of faster dividing cells at the clone boundary decreased (C). See Figures SF and SG for definition of these measures. Error bars represent s.d. of simulations of each clone at each tested division rate. (D I) Apoptosis simulation. (D) Median of averaged acquired area of clonal cells (green) was larger than that of non-clonal cells (white) at division rate ratio = 3 (Wilcoxon signed rank test; P < 2.2 -), whereas it was smaller at division rate ratio [clone (green) / non-clone (white)] = (Wilcoxon signed rank test; P =. -4). (E) The positive correlation between winner-cell expansion and shape anisotropy of apoptotic cells was consistent with the results obtained using Eq. [Figure S3H for (E left), Figure 3D for (E right)]. The y-axis values in (E left) and (E right) are the same. (F) Frequency of boundary (red), non-boundary (orange), and perpendicular (blue) intercalations observed in a single event of apoptosis as a function of shape anisotropy of an apoptotic cell
5 before death were consistent with the results obtained using Eq. [Figure SC for (F left), Figure F for (F right)]. The y-axis values in (F left) and (F right) are the same. (G I) The acquired area averaged over faster dividing cells adjacent to apoptosis was plotted as a function of the frequency of cell intercalation. This tendency was consistent with Figures I, L, and O. See corresponding legends in main figures for method and plot setup. Unless noted (upper panel of A, Ratio in B and C, left panel of D), the division rate ratio [clone (green) / non-clone (white)] = 3 for all panels. The number of clonal cells (green) was 2 for all panels. Corr. coef., Pearson correlation coefficient.
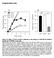
6 Apoptotic cell A C Cell anisotropy G Rate of division :3 G b G c clones B G c /G /.. i ii N.D. iii iv v vi vii viii Rate of division :3 for all panels D i ii iii iv v vi vii viii Rate of division :3 /.. G b /G Acquired area after apoptosis (a.u.) P < -4 P < -4 P < -4 P < -4 P < -4 P < -4 P < -4 P < -4 clones, 22 apoptosis clones, 28 apoptosis clones, 28 apoptosis clones, 2 apoptosis clones, 23 apoptosis clones, 297 apoptosis clones, 28 apoptosis clones, 237 apoptosis.4 E F G Acquired area after apoptosis (a.u.) Normalized frequency of cell intercalation Delamination Type I Delamination Type II Rate of division :3 Delamination Type I i G/G = nh/nh = ii iii iv v vi vii viii P < -4 P < -4 P < -4 P < -4 P < -4 Rate of division :3 T T T Shrinkage of apical surface area T T T G/G =2 G/G =4 nh/nh = n H/nH =. (Tension increase) (Pressure decrease) Delamination Type II Boundary Non-boundary Perpendicular H I J Acquired area after apoptosis (a.u.) Acquired area after apoptosis (a.u.) Acquired area after apoptosis (a.u.) Cell anisotropy of apoptotic cell before death Rate of division :3 Rate of division :3 P < -4 P < -4 Rate of division :3 clones, 2 apoptosis -cell neighbors 2-cell neighbors 3-cell neighbors clones, 2 apoptosis P < -4 P < -4 P < -4 Faster-dividing cells (Corr. Coef =.44, p< -4 ) Slower-dividing cells (Corr. Coef =.7, p< -2 ) -cell neighbors 2-cell neighbors 3-cell neighbors G/G = nh/nh = Delamination Type I G/G =2 G/G =4 nh/nh = n =. H/nH (Tension increase) (Pressure decrease) Delamination Type II Control Stop proliferation Reduced pressure in winner clone
7 Figure S3. Robust expansion of winner cells occurred with different mechanical parameters between clonal and non-clonal cells and in different apoptosis conditions in simulation, Related to Figures, 3, 4,, and 7. (A D) Simulations under different combinations of relative line tension parameters (G b / G and G c / G; Eq. ) with the conditions of differential proliferation rate. (A) G is default tension parameter at edges between non-clonal cells (white) (left); G b is clone boundary tension parameter at edges on clone boundary (middle); and G c is inner clonal tension parameter at edges between clonal cells (green) (right). (B) Close-up images of clone boundary. Note that the model did not work at the parameter indicated by N.D. due to divergence of simulations. (C, D) Cell anisotropy of slower dividing cells (white cells in B) directly adjacent to clone boundary (C) and averaged acquired area after apoptosis of slower and faster dividing cells directly in contact with apoptotic cells (gray and green box plots, respectively) (D). Numerals (i viii) correspond to those in B. Wilcoxon signed rank testing was performed. All simulations were conducted with faster dividing cells (green). The division rate ratio [clone (green) / non-clone (white)] = 3 for all panels. (E G) Simulations with respect to different model settings of apoptosis (Eq. ). (E) Two types of cell-delaminating processes were numerically simulated. The area of the apoptotic cell (magenta) was decreased through junctional loss by repeated cell intercalation (red edges) while () keeping the mechanical parameter values constant (delamination type I; model for the D delamination [S2,S3]), or (2) either increasing the value of the line tension parameter (G ij) or decreasing the value of the pressure parameter (n H) (Eq. ) before repeating the cell intercalation (delamination type II; model for the D2 delamination [S2,S3]). Note that while D2 delamination leads to transitory rosette formation after rapid apical surface shrinkage [S3], our model did not reproduce the intermediate rosette structure due to model limitation on tricellular junctions. (F) Averaged acquired area after apoptosis of slower and faster dividing cells directly in contact with apoptotic cells (gray and green box plots, respectively). G and n H represent parameters of apoptotic cells, whereas G and n H represent those of non-apoptotic cells. Both delaminating processes showed winner-dominated cell expansion. The measurement method and box plot setup are identical to Figures 2H and 2I. Plots of delamination type I (the two leftmost boxplots) are identical with Figure 3C upper. Wilcoxon signed rank testing was performed. (G) Normalized frequency of boundary (red), non-boundary (orange), and perpendicular (blue) intercalations observed during apoptosis. As the anisotropic shape of dying cells was maintained during delamination type II, perpendicular intercalation dominated similarly to that which occurred during delamination type I. All apoptotic simulations were conducted with faster dividing cells (green). The division rate ratio [clone (green) / non-clone (white)] = 3. (H) Winner cells expanded more dominantly with increasing anisotropy (Eq. ) of dying loser cells. The y-axis value is the same as Figure 3D. Corr. coef., Pearson correlation coefficient. (I) Averaged acquired areas after apoptosis of cells directly adjacent to apoptotic cells (right, -cell neighbors; identical to Figure 3C upper) and cells at two or three cells away from the apoptotic cell (right, 2-cell neighbors or 3-cell neighbors). Slower and faster dividing cells are shown in gray and green box plots, respectively. The plot
8 setup is identical to that in Figures 2H and 2I. Wilcoxon signed rank testing was performed. (J) Averaged acquired area after apoptosis of slower and faster dividing cells directly adjacent to apoptotic cells (gray and green box plots, respectively) under simulation with ceasing all cell proliferation during apoptosis (represented by Stop proliferation ), or simulation reducing pressure in the faster dividing clone during apoptosis [represented by Reduced pressure in winner clone ; n Clone Non-clone H / n H =.72, where n Clone H and n Non-clone H represent pressure parameters (Eq. ) of clonal (faster dividing) and non-clonal (slower dividing) cells]. The box plots represented by Control are the same as in Figure 3C upper, which was obtained without additional parameter manipulations. Winner cells still dominated expansion in the absence of the global tissue-scale pushing force generated by winner cells. Wilcoxon signed rank testing was performed.
9 UAS-ykiSAS8AS2A:V (Corr. Coef =.29, p<-2) wild-type (Corr. Coef = -.24, p<.) B in vivo (Yki) 2 nota, 7 apoptosis - C in vivo (Yki) in vivo (Yki) nota, 7 apoptosis Cell anisotropy of apoptotic cell before death D Corr. Coef =.23, P < -2 Corr. Coef =.9, P =. Corr. Coef = -.39, P < E-cad UAS-RasV Cell anisotropy of apoptotic cell before death E-cad UAS-ykiSAS8AS2A :V Acquired area after apoptosis (μm2) 2 Normalized edge length A F Segmentation E-cad Segmentation T (2nd T) & T2 Perpendicular T (st T) G Side number Area H ❶ ❺ ❷ st T Pe rp T Acquired area after apoptosis (μm2) Normalized apical surface area <An>/<A> in vivo (Ras) Corr. Coef =.9, P <. Corr. Coef = -., P <. 3 2 Acquired area after apoptosis (μm2) in vivo (Yki) nota, 7 apoptosis 8 7 Corr. Coef = -.2, P =.8 Corr. Coef =.3, P = in vivo (Yki) nota, apoptosis L 3 2 Corr. Coef = -., P =.9 Corr. Coef =.8, P =.2 in vivo (Yki) nota, apoptosis M ❺ ❶ ❷ ❺ ❷ ❶ ❺ ❷ ❶ ❺.8 Frequency of boundary intercalation in vivo (Yki) Clone-boundary-oriented cell anisotropy of apoptotic cells before death.2 ❶ nota, 7 apoptosis ❶ ❷ ❺ Corr. Coef =.2, P <. Corr. Coef = -.2, P =. Cell anisotropy of apoptotic cell before death side number n K ) / 2 =. - 4 ar WT cells at clone boundary (4 nota, 28 cells) l cu WT cells located two cells or more away from clone boundary (4 nota, 34 cells) di 2 RasV2 cells at clone boundary (4 nota, 82 cells).. 3 RasV2 cells located two cells or more away from clone boundary (4 nota, 479 cells). en J ❺ ❷ 4µm2 E 2. T T2 Perpendicular intercalation = ( + µm2 ry T2 da 9 un ry T 8 nda -bou 7 Non T2 T2 Bo Probability Non-boundary intercalation = ( +. ) / 2 =.2 4 r T ❹ st T 2nd T Total T occurrence 3 icula ❶ ❶ ❷ ❺ ❷ ❺ ❷ ❶ Boundary intercalation = ( +. ) / 2 =.2 more than end Normalized frequency Perp Perpendicular T 2nd T ❺ I ❺ ❹ Corr. Coef =.22, P <. Corr. Coef = -.8, P =.24 in vivo (Yki) nota, apoptosis Frequency of non-boundary intercalation Frequency of perpendicular intercalation Figure S4. Winner-dominated cell expansion in vivo, Related to Figures 2, 4, and. (A C) Acquired areas after apoptosis (A), length distributions of three edge types (B), and normalized
10 frequencies of three intercalation types observed in a single apoptosis event (C) were plotted as a function of anisotropy (Eq. in Methods) of apoptotic cells 4 minutes before complete delamination. The averaged values and error bars on the y-axes are the same as Figure 2K for (A), Figure B for (B), and Figure G for (C). (D) Top-left: Z projection of a live pupal notum (2 hours after Gal4-mediated transcription activation). The fly genotype and image frame are the same as in Figure 2B; the anterior direction is upward. Top-right: Image segmentation for adherens junctions. Green and gray cells represent Ras V2 -overexpressing and wild-type cells at the clone boundary, and light green and white cells represent Ras V2 -overexpressing and wild-type cells located two-cell-distance or more away from the clone boundary. Bottom: Side number (left) and area (right) of each cell are indicated with colors. The white thick line represents the clone boundary. (E) Averaged area of cells with n-sides (A n) normalized by that of all cells for each cell category (A) (i.e. A n/a where < > denotes the population average) 24 2 hours after Gal4-mediated transcription activation. Cells were classified into four categories: green and gray solid lines are Ras V2 -overexpressing and wild-type cells, respectively, at the clone boundary (green and gray cells in D, Top-Right), and green and gray broken lines are Ras V2 -overexpressing and wild-type cells, respectively, located two cells or more away from the clone boundary (light green and white cells in D, Top-Right). Error bars represent s.e. (F I) Inference method of intercalated edges when multiple intercalations (T transition) and/or T2 transition occurred simultaneously. (F, G) An example of cell intercalation upon apoptosis. Pentagonal dying cell (magenta in bottom panels of F) is delaminated by sequential occurrence of two T transitions (intercalations) and one T2 transition. The first T transition is a perpendicular intercalation (i.e., an edge belonging to cell 4 in (F) is lost; G), whereas the type of second T transition cannot be distinguished due to the simultaneous occurrence of T and T2 transitions among the five cells (an apoptotic cell and cells, 2, 3, and ). Fly genotype is the same as in Figure 2A. (H) All geometrically possible combinations of cell rearrangement during the latter half of the delaminating process (right two panels in F), in which the quadrangular dying cell disappears through a second T transition and a T2 transition. Because the quadrangular cell has four edges, there are four possible patterns of rearrangement. Four candidate rearrangements are narrowed down to two (black dashed boxes) that are consistent with cell geometry after delamination complete (rightmost panels in F): boundary or non-boundary intercalation is estimated to occur in the second T transition. Numerical cell labels correspond to those in the bottom panels of F. (I) Method of calculation of normalized frequency for each type of intercalation. Dashed box labeled st T or 2 nd T represents the probability of each type of intercalation for each T event. Probability of perpendicular intercalation at st T is, whereas that of boundary and non-boundary intercalations in the second T is. because the number of candidates is two (H). Dashed box labeled Total T occurrence represents total number of T events ( st and 2 nd ) during apoptosis. (J M) The y-axis values for all panels are the acquired areas after apoptosis without averaging for individual apoptosis events (thin lines in Figure 2G). The x-axis values are the same as Figure S4A for (J left), Figure 2K for (J right), Figure J for (K), Figure M for (L), and Figure P for (M).
11 C clones, 27 apoptosis Normalized frequency of cell intercalation Edge length (a.u.) Rate of division : clones, 27 apoptosis Rate of division : Cell anisotropy of apoptotic cell before apoptosis D Corr. Coef = -.4, P <. Corr. Coef = -.2, P =.8 Corr. Coef =., P <..2.4 E 2 2 Rate of division :3 clones, 27 apoptosis 2. Corr. Coef = -.32, P < -4 Corr. Coef =.9, P < No Bo un d Cell anisotropy of apoptotic cell before apoptosis da ry.3..8 Rate of division :3 un.4.4. Bo...2 Normalized frequency of perpendicular intercalation No. Rate of division :. ry.7 ula r.8 H da.9.8 nd ic clones, 2232 apoptosis. Normalized frequency of non-boundary intercalation Boundary Non-boundary Perpendicular Rate of division :.4 rpe G.2 Pe.8 ary. un.4 Normalized frequency of boundary intercalation Edge length (a.u.) Corr. Coef =.43, P < -4 Corr. Coef = -.44, P < F 2. Corr. Coef = -.23, P < -3 Corr. Coef =.33, P < -4 Normalized frequency of cell intercalation Side number change after apoptosis 2.. Cell anisotropy of apoptotic cell before apoptosis un da ry Pe rpe nd icu lar Corr. Coef =., P < -4 Corr. Coef =.24, P < -4 Corr. Coef = -.2, P < -4 n-b o B Rate of division :3 n-b o A Rate of division : I Boundary intercalation Averaged side number change 7 Clone : -. Non-clone : +.4 Rate of division : 8 7 Boundary T Non-boundary intercalation Averaged side number change 7 Boundary T Non-boundary T Clone : -. Non-clone : Non-boundary T Non-boundary T Non-boundary T Averaged side number change 7 7 Clone : + Non-clone : -.2 Perpendicular T Perpendicular T Perpendicular T 8 Boundary T Perpendicular intercalation 8 Non-boundary T 8 Perpendicular T Figure S. In silico examination of the contribution of biased cell intercalations upon apoptosis, Related to Figures 3,, and. (A) Time course of apoptosis under identical and differential proliferation rates shown in Figure 3C, where the figures were rotated 9 degrees. (B, C) Length distributions of three edge types (B), and normalized frequency of three intercalation types observed in a single apoptosis event (C) was plotted as a function of cell anisotropy (Eq. in Methods) of apoptotic cells immediately before cell death (normalized time [/loser cell cycle] = 8.3-3). The y-axis values are the same as in Figure A for (B) and Figure F for (C). (D F)
12 Change in side number of winner cells (green) after apoptosis was positively correlated with the normalized frequency of perpendicular intercalation (F), but not with that of boundary and non-boundary intercalation (D, E). The correlation of loser cells (gray) was generally opposite that of winner cells. The x-axis values are the same as in Figure I, L, and O. The division rate ratio [clone (green) / non-clone (white)] = 3 for all panels. (G) The length distribution of three types of edges as a function of cell anisotropy of an apoptotic cell immediately before death (normalized time [/loser cell cycle] = ) under conditions of identical division rate is shown. The distributions were nearly identical for the three types of edges. (H) Normalized frequency of boundary (red), non-boundary (orange), and perpendicular (blue) intercalations when apoptosis was independently introduced into all non-clonal cells directly adjacent to clones with clonal cells. Data for identical (left) and differential (right) proliferation rate are shown. Normalized frequency of the three types of intercalation was nearly identical under identical division rate (left). (I) Apoptotic simulations with a predetermined order of directionally biased cell intercalations with identical division rates. Time course of apoptosis with biased occurrences of boundary (upper), non-boundary (middle) and perpendicular (lower) intercalation shown in Figure B. Arrowheads indicate the shortest edge in each edge type that is lost via cell intercalations in the next frame. The numerals within cells indicate the numbers of cell sides. Under perpendicular intercalation, one of the two clonal cells decreased the side number (from 7 to ; i.e. -), but the other cell increased (from to 8; i.e. +3). Thus, the clonal cells increase their side number on average [i.e. + (= (-+3) / 2)]. Likewise, the averaged side number change (left end) was calculated for non-clones and clones for each intercalation type. Note that with a bias favoring boundary intercalation, dying cells have only non-boundary edges after experiencing sequential induction of boundary intercalations; therefore, only non-boundary intercalations take place after that (upper, right three panels). Under a bias favoring non-boundary intercalation, dying cells only have boundary and perpendicular edges after experiencing sequential induction of non-boundary intercalations; therefore, boundary and/or perpendicular intercalations take place after that (middle, right two panels).
13 Figure S. Anisotropic distortion of loser cells at the clone boundary is maintained despite repeated apoptosis, Related to Figure 7. (A E) Differential proliferation-induced cell anisotropy was maintained in the presence of apoptosis. (A, B) To analyze the effects of apoptosis on the emergence of anisotropic cells, we compared the anisotropy of slower dividing cells in contact with the clone boundary at time points before (A left; normalized time [/loser cell cycle] = ) and after (A right; normalized time [/loser cell cycle] = ) apoptosis. The cell originally in contact with the clone boundary before apoptosis (apoptotic cell; magenta) and the slower dividing cells newly in contact with the clone boundary after apoptosis (gray) are shown (B). Anisotropy immediately after apoptosis (gray) was slightly reduced compared with that before apoptosis (magenta) (B, left two plots), but still higher than that under conditions of identical division rate (B, right two plots). The number of clonal cells was. (C) The average anisotropy of slower dividing cells newly in contact with the clone boundary after apoptosis (gray cells in (A)) increased over time up to that of the apoptotic cell immediately before death (magenta line). Division rate ratio [clone (green) / non-clone (white)] = 3. Error bars represent s.d. of 2 independent apoptosis simulations. P-values are based on Welch s t-test at the indicated times compared with the apoptotic cell immediately before death (normalized time [/loser cell cycle] = ; magenta line); n.s. (not significant), P >.; **P < -3 ; ***P < -4. (D) Robustness of cell anisotropy as a function of apoptosis rate (number of apoptosis events/cell cycle/cell). Note that apoptosis was not induced at apoptosis rate =. Apoptosis was simulated by incorporation of cell delamination in
14 slower dividing cells (white cells in E) directly adjacent to the clone boundary under conditions of differential proliferation rates (division rate ratio [clone (green) / non-clone (white)] = 3), while the number of faster dividing winner cells increased from to 3 cells. Apoptotic cell delamination was simulated by repeating cell intercalation from the shortest edge in an apoptotic cell. When the apoptosis rate is lower than cell division rate (black broken line), anisotropy of slower dividing cells directly adjacent to the clone boundary (black circles with solid line) was 2-fold higher than that of the control (brown line, which represents anisotropy under identical division rate without apoptosis). Error bars represent s.d. of simulations of each clone at each tested apoptosis rate. Anisotropy was measured when the number of clonal cells (green) = 3 in simulations. (E) Close-up snapshots of clone boundary under different apoptosis rates in simulations, where the numerals indicate apoptosis rate given in D. Division rate ratio [clone (green) / non-clone (white)] = 3.
15 A Rate of division : Rate of division : Total of area change (a.u.) (per layer) E up to -cell distance up to 3-cell distance exp(μ2%) =.3 clones Rate of division : 3 2. exp(μ2%) = 2.2 P < -4 Inside Border.... Normalized time ( -3/WT cell cycle) G H clones clones 3 y=x Normalized time ( -3/WT cell cycle) y = x/2 Net increase of clone area by apoptosis (a.u.) 32 Clone area (a.u.) exp(μ2%) =.3. All slower dividing cells exp(μ2%) =. P < up to -cell distance with apoptosis (Regular intercalation) μ2% =.29 μ2% =.38 μ2% =.27 μ2% =.79 Target region of apoptosis (Rate of division : 3) without apoptosis Rate of division : 3 Border 2 Rate of division : 3 clones Inside clones Rate of division : 3 P < -4 2 Area final / Area initial (log scale) (per cell) P =.9 4 with apoptosis (Regular intercalation) 2 Total of side number change (per layer) clones Randomized intercalation Total of area change (a.u.) (per layer) Total of side number change (per layer) Regular intercalation without apoptosis Rate of division : 3 Rate of division : 3 Normalized time ( -3/WT cell cycle) F D clones with apoptosis C Side no. final - Side no. initial (per cell) clones Total of area change (a.u.) (per layer) without apoptosis B Total of side number change (per layer) Total area of apoptosis (a.u.) Figure S7. Robust expansion of winner cells occurred under repetitive apoptosis during long-term clone development, Related to Figure 7. (A E) Apoptosis simulations, with the setting described in Figure 7. (A C) Sum of changes in side number (B) and cell area (C) per indicated layers, which are classified according to the location in relation to clone boundary (A), with layer (cells located immediately adjacent to the clone boundary), layer 2 (cells next to layer ), and layer 3 (cells next to layer 2). (B, C) Green and gray lines represent faster and slower dividing cells and different markers represent different layers defined in (A). Layers are identified at every time point. Error bars represent s.d. of five simulations of each clone at each simulation setup. (D, E) Difference between
16 the final and initial side number per cell (D) and fold change in cell area per cell (E; log scale) in the presence and absence of apoptosis. See the definition of the criteria and simulation setups for Figure 7B. Box plot setup is identical to that in Figure 2H and 2I. μ 2% is average of the values ranked in the top 2% for each box plot in E. Wilcoxon rank sum testing was performed. (F H) Winner-clone expansion varied depending on the spatial distribution of apoptosis. The division rate ratio (clonal/non-clonal = 3) and total numbers of apoptosis events (about 4 cells) were the same for all panels. Simulation setups other than the target regions of apoptosis were the same as in Figure 7. (F) Slower dividing cells for targeting apoptosis are marked in purple (directly adjacent to clone boundary), blue (up to 3-cell distance away from clone boundary), light-blue (up to -cell distance away from clone boundary), and orange (all slower dividing cells). Apoptotic cells were randomly selected from these regions. All faster dividing cells are marked in green. (G) Temporal change in clone area. The line colors (purple, blue, light-blue, orange) are as in (F). The black line is the case without apoptosis, which is the same as in Figure 7C. Black and purple lines are the same as in Figure 7C. (H) The net increase in clone area due to apoptosis was calculated by subtracting the clone area from that value in the absence of apoptosis as a function of total area of apoptosis. Dashed/dotted lines indicate cases in which winner clones capture %/% of the territory made available by apoptosis. Purple line is the same as in Figure 7D.
17 Supplemental References S. Farhadifar, R., Röper, J.-C., Aigouy, B., Eaton, S., and Jülicher, F. (27). The influence of cell mechanics, cell-cell interactions, and proliferation on epithelial packing. Curr. Biol. 7, S2. Levayer, R., Dupont, C., and Moreno, E. (2). Tissue Crowding Induces Caspase-Dependent Competition for Space. Curr. Biol. 2, S3. Marinari, E., Mehonic, A., Curran, S., Gale, J., Duke, T., and Baum, B. (22). Live-cell delamination counterbalances epithelial growth to limit tissue overcrowding. Nature 484, 42 4.
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1. Ras V12 expression in the entire eye-antennal disc does not cause invasive tumours. a, Eye-antennal discs expressing Ras V12 in all cells (marked with GFP, green) overgrow moderately
Supplementary Figure 1. Ras V12 expression in the entire eye-antennal disc does not cause invasive tumours. a, Eye-antennal discs expressing Ras V12 in all cells (marked with GFP, green) overgrow moderately
Supporting Information
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 Supporting Information Variances and biases of absolute distributions were larger in the 2-line
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 Supporting Information Variances and biases of absolute distributions were larger in the 2-line
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2419 Figure S1 NiGFP localization in Dl mutant dividing SOPs. a-c) time-lapse analysis of NiGFP (green) in Dl mutant SOPs (H2B-RFP, red; clones were identified by the loss of nlsgfp) showing
DOI: 10.1038/ncb2419 Figure S1 NiGFP localization in Dl mutant dividing SOPs. a-c) time-lapse analysis of NiGFP (green) in Dl mutant SOPs (H2B-RFP, red; clones were identified by the loss of nlsgfp) showing
Transient β-hairpin Formation in α-synuclein Monomer Revealed by Coarse-grained Molecular Dynamics Simulation
 Transient β-hairpin Formation in α-synuclein Monomer Revealed by Coarse-grained Molecular Dynamics Simulation Hang Yu, 1, 2, a) Wei Han, 1, 3, b) Wen Ma, 1, 2 1, 2, 3, c) and Klaus Schulten 1) Beckman
Transient β-hairpin Formation in α-synuclein Monomer Revealed by Coarse-grained Molecular Dynamics Simulation Hang Yu, 1, 2, a) Wei Han, 1, 3, b) Wen Ma, 1, 2 1, 2, 3, c) and Klaus Schulten 1) Beckman
Supplementary Figure 1. Nature Neuroscience: doi: /nn.4547
 Supplementary Figure 1 Characterization of the Microfetti mouse model. (a) Gating strategy for 8-color flow analysis of peripheral Ly-6C + monocytes from Microfetti mice 5-7 days after TAM treatment. Living
Supplementary Figure 1 Characterization of the Microfetti mouse model. (a) Gating strategy for 8-color flow analysis of peripheral Ly-6C + monocytes from Microfetti mice 5-7 days after TAM treatment. Living
Nature Neuroscience doi: /nn Supplementary Figure 1. Characterization of viral injections.
 Supplementary Figure 1 Characterization of viral injections. (a) Dorsal view of a mouse brain (dashed white outline) after receiving a large, unilateral thalamic injection (~100 nl); demonstrating that
Supplementary Figure 1 Characterization of viral injections. (a) Dorsal view of a mouse brain (dashed white outline) after receiving a large, unilateral thalamic injection (~100 nl); demonstrating that
Changing expectations about speed alters perceived motion direction
 Current Biology, in press Supplemental Information: Changing expectations about speed alters perceived motion direction Grigorios Sotiropoulos, Aaron R. Seitz, and Peggy Seriès Supplemental Data Detailed
Current Biology, in press Supplemental Information: Changing expectations about speed alters perceived motion direction Grigorios Sotiropoulos, Aaron R. Seitz, and Peggy Seriès Supplemental Data Detailed
Nature Neuroscience: doi: /nn Supplementary Figure 1. Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites.
 Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
Nature Neuroscience: doi: /nn Supplementary Figure 1. Behavioral training.
 Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
Supplementary Figure 1 Behavioral training. a, Mazes used for behavioral training. Asterisks indicate reward location. Only some example mazes are shown (for example, right choice and not left choice maze
Stretching Cardiac Myocytes: A Finite Element Model of Cardiac Tissue
 Megan McCain ES240 FEM Final Project December 19, 2006 Stretching Cardiac Myocytes: A Finite Element Model of Cardiac Tissue Cardiac myocytes are the cells that constitute the working muscle of the heart.
Megan McCain ES240 FEM Final Project December 19, 2006 Stretching Cardiac Myocytes: A Finite Element Model of Cardiac Tissue Cardiac myocytes are the cells that constitute the working muscle of the heart.
John Nguyen, Nozomi Nishimura, Robert Fetcho, Costantino Iadecola, Chris B. Schaffer
 Supplemental figures and text for Occlusion of cortical ascending venules causes blood flow decreases, reversals in flow direction, and vessel dilation in upstream capillaries John Nguyen, Nozomi Nishimura,
Supplemental figures and text for Occlusion of cortical ascending venules causes blood flow decreases, reversals in flow direction, and vessel dilation in upstream capillaries John Nguyen, Nozomi Nishimura,
Supplementary Figures
 J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
Nature Neuroscience: doi: /nn Supplementary Figure 1. MADM labeling of thalamic clones.
 Supplementary Figure 1 MADM labeling of thalamic clones. (a) Confocal images of an E12 Nestin-CreERT2;Ai9-tdTomato brain treated with TM at E10 and stained for BLBP (green), a radial glial progenitor-specific
Supplementary Figure 1 MADM labeling of thalamic clones. (a) Confocal images of an E12 Nestin-CreERT2;Ai9-tdTomato brain treated with TM at E10 and stained for BLBP (green), a radial glial progenitor-specific
Theta sequences are essential for internally generated hippocampal firing fields.
 Theta sequences are essential for internally generated hippocampal firing fields. Yingxue Wang, Sandro Romani, Brian Lustig, Anthony Leonardo, Eva Pastalkova Supplementary Materials Supplementary Modeling
Theta sequences are essential for internally generated hippocampal firing fields. Yingxue Wang, Sandro Romani, Brian Lustig, Anthony Leonardo, Eva Pastalkova Supplementary Materials Supplementary Modeling
SDS-Assisted Protein Transport Through Solid-State Nanopores
 Supplementary Information for: SDS-Assisted Protein Transport Through Solid-State Nanopores Laura Restrepo-Pérez 1, Shalini John 2, Aleksei Aksimentiev 2 *, Chirlmin Joo 1 *, Cees Dekker 1 * 1 Department
Supplementary Information for: SDS-Assisted Protein Transport Through Solid-State Nanopores Laura Restrepo-Pérez 1, Shalini John 2, Aleksei Aksimentiev 2 *, Chirlmin Joo 1 *, Cees Dekker 1 * 1 Department
effects on organ development. a-f, Eye and wing discs with clones of ε j2b10 show no
 Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
Supplementary Figure 1 Madm is not required in GSCs and hub cells. (a,b) Act-Gal4-UAS-GFP (a), Act-Gal4-UAS- GFP.nls (b,c) is ubiquitously expressed
 Supplementary Figure 1 Madm is not required in GSCs and hub cells. (a,b) Act-Gal4-UAS-GFP (a), Act-Gal4-UAS- GFP.nls (b,c) is ubiquitously expressed in the testes. The testes were immunostained with GFP
Supplementary Figure 1 Madm is not required in GSCs and hub cells. (a,b) Act-Gal4-UAS-GFP (a), Act-Gal4-UAS- GFP.nls (b,c) is ubiquitously expressed in the testes. The testes were immunostained with GFP
Linkage Mapping in Drosophila Melanogaster
 Linkage Mapping in Drosophila Melanogaster Genetics: Fall 2012 Joshua Hanau Introduction: An experiment was performed in order to determine the presence and degree of gene linkage in Drosophila Melanogaster.
Linkage Mapping in Drosophila Melanogaster Genetics: Fall 2012 Joshua Hanau Introduction: An experiment was performed in order to determine the presence and degree of gene linkage in Drosophila Melanogaster.
Muscle-Tendon Mechanics Dr. Ted Milner (KIN 416)
 Muscle-Tendon Mechanics Dr. Ted Milner (KIN 416) Muscle Fiber Geometry Muscle fibers are linked together by collagenous connective tissue. Endomysium surrounds individual fibers, perimysium collects bundles
Muscle-Tendon Mechanics Dr. Ted Milner (KIN 416) Muscle Fiber Geometry Muscle fibers are linked together by collagenous connective tissue. Endomysium surrounds individual fibers, perimysium collects bundles
Supplementary Materials for
 www.sciencetranslationalmedicine.org/cgi/content/full/7/283/283ra54/dc1 Supplementary Materials for Clonal status of actionable driver events and the timing of mutational processes in cancer evolution
www.sciencetranslationalmedicine.org/cgi/content/full/7/283/283ra54/dc1 Supplementary Materials for Clonal status of actionable driver events and the timing of mutational processes in cancer evolution
Supplementary Figure 1 Information on transgenic mouse models and their recording and optogenetic equipment. (a) 108 (b-c) (d) (e) (f) (g)
 Supplementary Figure 1 Information on transgenic mouse models and their recording and optogenetic equipment. (a) In four mice, cre-dependent expression of the hyperpolarizing opsin Arch in pyramidal cells
Supplementary Figure 1 Information on transgenic mouse models and their recording and optogenetic equipment. (a) In four mice, cre-dependent expression of the hyperpolarizing opsin Arch in pyramidal cells
SUPPLEMENTAL MATERIAL
 1 SUPPLEMENTAL MATERIAL Response time and signal detection time distributions SM Fig. 1. Correct response time (thick solid green curve) and error response time densities (dashed red curve), averaged across
1 SUPPLEMENTAL MATERIAL Response time and signal detection time distributions SM Fig. 1. Correct response time (thick solid green curve) and error response time densities (dashed red curve), averaged across
SUPPLEMENTARY INFORMATION
 doi:1.138/nature1139 a d Whisker angle (deg) Whisking repeatability Control Muscimol.4.3.2.1 -.1 8 4-4 1 2 3 4 Performance (d') Pole 8 4-4 1 2 3 4 5 Time (s) b Mean protraction angle (deg) e Hit rate (p
doi:1.138/nature1139 a d Whisker angle (deg) Whisking repeatability Control Muscimol.4.3.2.1 -.1 8 4-4 1 2 3 4 Performance (d') Pole 8 4-4 1 2 3 4 5 Time (s) b Mean protraction angle (deg) e Hit rate (p
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Illustration of the working of network-based SVM to confidently predict a new (and now confirmed) ASD gene. Gene CTNND2 s brain network neighborhood that enabled its prediction by
Supplementary Figure 1 Illustration of the working of network-based SVM to confidently predict a new (and now confirmed) ASD gene. Gene CTNND2 s brain network neighborhood that enabled its prediction by
Supplementary Information. Supplementary Figures
 Supplementary Information Supplementary Figures.8 57 essential gene density 2 1.5 LTR insert frequency diversity DEL.5 DUP.5 INV.5 TRA 1 2 3 4 5 1 2 3 4 1 2 Supplementary Figure 1. Locations and minor
Supplementary Information Supplementary Figures.8 57 essential gene density 2 1.5 LTR insert frequency diversity DEL.5 DUP.5 INV.5 TRA 1 2 3 4 5 1 2 3 4 1 2 Supplementary Figure 1. Locations and minor
Nature Neuroscience: doi: /nn Supplementary Figure 1. Trial structure for go/no-go behavior
 Supplementary Figure 1 Trial structure for go/no-go behavior a, Overall timeline of experiments. Day 1: A1 mapping, injection of AAV1-SYN-GCAMP6s, cranial window and headpost implantation. Water restriction
Supplementary Figure 1 Trial structure for go/no-go behavior a, Overall timeline of experiments. Day 1: A1 mapping, injection of AAV1-SYN-GCAMP6s, cranial window and headpost implantation. Water restriction
Supplementary Materials
 Supplementary Materials Supplementary Figure S1: Data of all 106 subjects in Experiment 1, with each rectangle corresponding to one subject. Data from each of the two identical sub-sessions are shown separately.
Supplementary Materials Supplementary Figure S1: Data of all 106 subjects in Experiment 1, with each rectangle corresponding to one subject. Data from each of the two identical sub-sessions are shown separately.
Nature Neuroscience: doi: /nn Supplementary Figure 1. Lick response during the delayed Go versus No-Go task.
 Supplementary Figure 1 Lick response during the delayed Go versus No-Go task. Trial-averaged lick rate was averaged across all mice used for pyramidal cell imaging (n = 9). Different colors denote different
Supplementary Figure 1 Lick response during the delayed Go versus No-Go task. Trial-averaged lick rate was averaged across all mice used for pyramidal cell imaging (n = 9). Different colors denote different
Unit 1 Exploring and Understanding Data
 Unit 1 Exploring and Understanding Data Area Principle Bar Chart Boxplot Conditional Distribution Dotplot Empirical Rule Five Number Summary Frequency Distribution Frequency Polygon Histogram Interquartile
Unit 1 Exploring and Understanding Data Area Principle Bar Chart Boxplot Conditional Distribution Dotplot Empirical Rule Five Number Summary Frequency Distribution Frequency Polygon Histogram Interquartile
Supplementary Figures
 Supplementary Figures Spatial arrangement Variation in the morphology of central NCs (shape x size) x Variation in the morphology of satellite NCs (shape x size) x Variations in the spatial arrangement
Supplementary Figures Spatial arrangement Variation in the morphology of central NCs (shape x size) x Variation in the morphology of satellite NCs (shape x size) x Variations in the spatial arrangement
Nature Neuroscience: doi: /nn Supplementary Figure 1. Large-scale calcium imaging in vivo.
 Supplementary Figure 1 Large-scale calcium imaging in vivo. (a) Schematic illustration of the in vivo camera imaging set-up for large-scale calcium imaging. (b) High-magnification two-photon image from
Supplementary Figure 1 Large-scale calcium imaging in vivo. (a) Schematic illustration of the in vivo camera imaging set-up for large-scale calcium imaging. (b) High-magnification two-photon image from
Supplementary Figure S1: TIPF reporter validation in the wing disc.
 Supplementary Figure S1: TIPF reporter validation in the wing disc. a,b, Test of put RNAi. a, In wildtype discs the Dpp target gene Sal (red) is expressed in a broad stripe in the centre of the ventral
Supplementary Figure S1: TIPF reporter validation in the wing disc. a,b, Test of put RNAi. a, In wildtype discs the Dpp target gene Sal (red) is expressed in a broad stripe in the centre of the ventral
Santulli G. et al. A microrna-based strategy to suppress restenosis while preserving endothelial function
 ONLINE DATA SUPPLEMENTS Santulli G. et al. A microrna-based strategy to suppress restenosis while preserving endothelial function Supplementary Figures Figure S1 Effect of Ad-p27-126TS on the expression
ONLINE DATA SUPPLEMENTS Santulli G. et al. A microrna-based strategy to suppress restenosis while preserving endothelial function Supplementary Figures Figure S1 Effect of Ad-p27-126TS on the expression
Table S1: Kinetic parameters of drug and substrate binding to wild type and HIV-1 protease variants. Data adapted from Ref. 6 in main text.
 Dynamical Network of HIV-1 Protease Mutants Reveals the Mechanism of Drug Resistance and Unhindered Activity Rajeswari Appadurai and Sanjib Senapati* BJM School of Biosciences and Department of Biotechnology,
Dynamical Network of HIV-1 Protease Mutants Reveals the Mechanism of Drug Resistance and Unhindered Activity Rajeswari Appadurai and Sanjib Senapati* BJM School of Biosciences and Department of Biotechnology,
Early Learning vs Early Variability 1.5 r = p = Early Learning r = p = e 005. Early Learning 0.
 The temporal structure of motor variability is dynamically regulated and predicts individual differences in motor learning ability Howard Wu *, Yohsuke Miyamoto *, Luis Nicolas Gonzales-Castro, Bence P.
The temporal structure of motor variability is dynamically regulated and predicts individual differences in motor learning ability Howard Wu *, Yohsuke Miyamoto *, Luis Nicolas Gonzales-Castro, Bence P.
Supplemental Information. Proprioceptive Opsin Functions. in Drosophila Larval Locomotion
 Neuron, Volume 98 Supplemental Information Proprioceptive Opsin Functions in Drosophila Larval Locomotion Damiano Zanini, Diego Giraldo, Ben Warren, Radoslaw Katana, Marta Andrés, Suneel Reddy, Stephanie
Neuron, Volume 98 Supplemental Information Proprioceptive Opsin Functions in Drosophila Larval Locomotion Damiano Zanini, Diego Giraldo, Ben Warren, Radoslaw Katana, Marta Andrés, Suneel Reddy, Stephanie
d e f Spatiotemporal quantification of subcellular ATP levels in a single HeLa cell during changes in morphology Supplementary Information
 Ca 2+ level (a. u.) Area (a. u.) Normalized distance Normalized distance Center Edge Center Edge Relative ATP level Relative ATP level Supplementary Information Spatiotemporal quantification of subcellular
Ca 2+ level (a. u.) Area (a. u.) Normalized distance Normalized distance Center Edge Center Edge Relative ATP level Relative ATP level Supplementary Information Spatiotemporal quantification of subcellular
SUPPLEMENTARY INFORMATION
 In the format provided by the authors and unedited. 2 3 4 DOI: 10.1038/NMAT4893 EGFR and HER2 activate rigidity sensing only on rigid matrices Mayur Saxena 1,*, Shuaimin Liu 2,*, Bo Yang 3, Cynthia Hajal
In the format provided by the authors and unedited. 2 3 4 DOI: 10.1038/NMAT4893 EGFR and HER2 activate rigidity sensing only on rigid matrices Mayur Saxena 1,*, Shuaimin Liu 2,*, Bo Yang 3, Cynthia Hajal
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature07173 SUPPLEMENTARY INFORMATION Supplementary Figure Legends: Supplementary Figure 1: Model of SSC and CPC divisions a, Somatic stem cells (SSC) reside adjacent to the hub (red), self-renew
doi: 10.1038/nature07173 SUPPLEMENTARY INFORMATION Supplementary Figure Legends: Supplementary Figure 1: Model of SSC and CPC divisions a, Somatic stem cells (SSC) reside adjacent to the hub (red), self-renew
Supplementary materials for: Executive control processes underlying multi- item working memory
 Supplementary materials for: Executive control processes underlying multi- item working memory Antonio H. Lara & Jonathan D. Wallis Supplementary Figure 1 Supplementary Figure 1. Behavioral measures of
Supplementary materials for: Executive control processes underlying multi- item working memory Antonio H. Lara & Jonathan D. Wallis Supplementary Figure 1 Supplementary Figure 1. Behavioral measures of
marker. DAPI labels nuclei. Flies were 20 days old. Scale bar is 5 µm. Ctrl is
 Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Impact of Response Variability on Pareto Front Optimization
 Impact of Response Variability on Pareto Front Optimization Jessica L. Chapman, 1 Lu Lu 2 and Christine M. Anderson-Cook 3 1 Department of Mathematics, Computer Science, and Statistics, St. Lawrence University,
Impact of Response Variability on Pareto Front Optimization Jessica L. Chapman, 1 Lu Lu 2 and Christine M. Anderson-Cook 3 1 Department of Mathematics, Computer Science, and Statistics, St. Lawrence University,
Learning Utility for Behavior Acquisition and Intention Inference of Other Agent
 Learning Utility for Behavior Acquisition and Intention Inference of Other Agent Yasutake Takahashi, Teruyasu Kawamata, and Minoru Asada* Dept. of Adaptive Machine Systems, Graduate School of Engineering,
Learning Utility for Behavior Acquisition and Intention Inference of Other Agent Yasutake Takahashi, Teruyasu Kawamata, and Minoru Asada* Dept. of Adaptive Machine Systems, Graduate School of Engineering,
Main Study: Summer Methods. Design
 Main Study: Summer 2000 Methods Design The experimental design is within-subject each participant experiences five different trials for each of the ten levels of Display Condition and for each of the three
Main Study: Summer 2000 Methods Design The experimental design is within-subject each participant experiences five different trials for each of the ten levels of Display Condition and for each of the three
Supplementary Figure 1. Procedures to independently control fly hunger and thirst states.
 Supplementary Figure 1 Procedures to independently control fly hunger and thirst states. (a) Protocol to produce exclusively hungry or thirsty flies for 6 h water memory retrieval. (b) Consumption assays
Supplementary Figure 1 Procedures to independently control fly hunger and thirst states. (a) Protocol to produce exclusively hungry or thirsty flies for 6 h water memory retrieval. (b) Consumption assays
RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays
 Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
Supplementary Materials RASA: Robust Alternative Splicing Analysis for Human Transcriptome Arrays Junhee Seok 1*, Weihong Xu 2, Ronald W. Davis 2, Wenzhong Xiao 2,3* 1 School of Electrical Engineering,
Simulation of the power transmission of bone-conducted sound in a finite-element model of the human head
 Biomechanics and Modeling in Mechanobiology (2018) 17:1741 1755 https://doi.org/10.1007/s10237-018-1053-4 ORIGINAL PAPER Simulation of the power transmission of bone-conducted sound in a finite-element
Biomechanics and Modeling in Mechanobiology (2018) 17:1741 1755 https://doi.org/10.1007/s10237-018-1053-4 ORIGINAL PAPER Simulation of the power transmission of bone-conducted sound in a finite-element
Irina V. Ionova, Vsevolod A. Livshits, and Derek Marsh
 Phase Diagram of Ternary Cholesterol/Palmitoylsphingomyelin/Palmitoyloleoyl- Phosphatidylcholine Mixtures: Spin-abel EPR Study of ipid-raft Formation Irina V. Ionova, Vsevolod A. ivshits, and Derek Marsh
Phase Diagram of Ternary Cholesterol/Palmitoylsphingomyelin/Palmitoyloleoyl- Phosphatidylcholine Mixtures: Spin-abel EPR Study of ipid-raft Formation Irina V. Ionova, Vsevolod A. ivshits, and Derek Marsh
Nature Getetics: doi: /ng.3471
 Supplementary Figure 1 Summary of exome sequencing data. ( a ) Exome tumor normal sample sizes for bladder cancer (BLCA), breast cancer (BRCA), carcinoid (CARC), chronic lymphocytic leukemia (CLLX), colorectal
Supplementary Figure 1 Summary of exome sequencing data. ( a ) Exome tumor normal sample sizes for bladder cancer (BLCA), breast cancer (BRCA), carcinoid (CARC), chronic lymphocytic leukemia (CLLX), colorectal
File name: Supplementary Information Description: Supplementary Figures, Supplementary Table and Supplementary References
 File name: Supplementary Information Description: Supplementary Figures, Supplementary Table and Supplementary References File name: Supplementary Data 1 Description: Summary datasheets showing the spatial
File name: Supplementary Information Description: Supplementary Figures, Supplementary Table and Supplementary References File name: Supplementary Data 1 Description: Summary datasheets showing the spatial
Nature Methods: doi: /nmeth Supplementary Figure 1. Activity in turtle dorsal cortex is sparse.
 Supplementary Figure 1 Activity in turtle dorsal cortex is sparse. a. Probability distribution of firing rates across the population (notice log scale) in our data. The range of firing rates is wide but
Supplementary Figure 1 Activity in turtle dorsal cortex is sparse. a. Probability distribution of firing rates across the population (notice log scale) in our data. The range of firing rates is wide but
Supporting Information
 Supporting Information Fig. S1. Overexpression of Rpr causes progenitor cell death. (A) TUNEL assay of control intestines. No progenitor cell death could be observed, except that some ECs are undergoing
Supporting Information Fig. S1. Overexpression of Rpr causes progenitor cell death. (A) TUNEL assay of control intestines. No progenitor cell death could be observed, except that some ECs are undergoing
Supplementary Information A Hydrophobic Barrier Deep Within the Inner Pore of the TWIK-1 K2P Potassium Channel Aryal et al.
 Supplementary Information A Hydrophobic Barrier Deep Within the Inner Pore of the TWIK-1 K2P Potassium Channel Aryal et al. Supplementary Figure 1 TWIK-1 stability during MD simulations in a phospholipid
Supplementary Information A Hydrophobic Barrier Deep Within the Inner Pore of the TWIK-1 K2P Potassium Channel Aryal et al. Supplementary Figure 1 TWIK-1 stability during MD simulations in a phospholipid
PROPORTION GAUGE T-BAR TIP
 Proportion PROPORTION GAUGE The Proportion Gauge has the T-bar tip on one end and an In-line tip on the opposite end. The T-bar tip is used on teeth with normal alignment and the In-line tip is used for
Proportion PROPORTION GAUGE The Proportion Gauge has the T-bar tip on one end and an In-line tip on the opposite end. The T-bar tip is used on teeth with normal alignment and the In-line tip is used for
Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes
 Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes Kaifu Chen 1,2,3,4,5,10, Zhong Chen 6,10, Dayong Wu 6, Lili Zhang 7, Xueqiu Lin 1,2,8,
Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor suppressor genes Kaifu Chen 1,2,3,4,5,10, Zhong Chen 6,10, Dayong Wu 6, Lili Zhang 7, Xueqiu Lin 1,2,8,
Power-Based Connectivity. JL Sanguinetti
 Power-Based Connectivity JL Sanguinetti Power-based connectivity Correlating time-frequency power between two electrodes across time or over trials Gives you flexibility for analysis: Test specific hypotheses
Power-Based Connectivity JL Sanguinetti Power-based connectivity Correlating time-frequency power between two electrodes across time or over trials Gives you flexibility for analysis: Test specific hypotheses
Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded
 Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded from TCGA and differentially expressed metabolic genes
Supplementary Figure 1. Metabolic landscape of cancer discovery pipeline. RNAseq raw counts data of cancer and healthy tissue samples were downloaded from TCGA and differentially expressed metabolic genes
Tracking X-ray microscopy for alveolar dynamics in live intact mice
 SUPPLEMENTARY INFORMATION Tracking X-ray microscopy for alveolar dynamics in live intact mice Soeun Chang, Namseop Kwon, Byung Mook Weon, Jinkyung Kim, Chin Kook Rhee, Han Sung Choi, Yoshiki Kohmura, Masaki
SUPPLEMENTARY INFORMATION Tracking X-ray microscopy for alveolar dynamics in live intact mice Soeun Chang, Namseop Kwon, Byung Mook Weon, Jinkyung Kim, Chin Kook Rhee, Han Sung Choi, Yoshiki Kohmura, Masaki
Transport and Concentration of Charged Molecules in a Lipid Membrane
 Transport and Concentration of Charged Molecules in a Lipid Membrane Johannes S. Roth 1, Matthew R. Cheetham 1,2, and Stephen D. Evans* 1 1 University of Leeds, 2 Current Address: University of Oxford
Transport and Concentration of Charged Molecules in a Lipid Membrane Johannes S. Roth 1, Matthew R. Cheetham 1,2, and Stephen D. Evans* 1 1 University of Leeds, 2 Current Address: University of Oxford
Supplementary Figure 1
 Supplementary Figure 1 5 microns C7 B6 unclassified H19 C7 signal H19 guide signal H19 B6 signal C7 SNP spots H19 RNA spots B6 SNP spots colocalization H19 RNA classification Supplementary Figure 1. Allele-specific
Supplementary Figure 1 5 microns C7 B6 unclassified H19 C7 signal H19 guide signal H19 B6 signal C7 SNP spots H19 RNA spots B6 SNP spots colocalization H19 RNA classification Supplementary Figure 1. Allele-specific
Supplemental Data Figure S1 Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells.
 Supplemental Data Figure S1. Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells. (A) Specific lysis of IFN-γ-treated SKBR-3 cells in the absence
Supplemental Data Figure S1. Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells. (A) Specific lysis of IFN-γ-treated SKBR-3 cells in the absence
Supplementary Information on TMS/hd-EEG recordings: acquisition and preprocessing
 Supplementary Information on TMS/hd-EEG recordings: acquisition and preprocessing Stability of the coil position was assured by using a software aiming device allowing the stimulation only when the deviation
Supplementary Information on TMS/hd-EEG recordings: acquisition and preprocessing Stability of the coil position was assured by using a software aiming device allowing the stimulation only when the deviation
Supplementary Information
 Supplementary Information Archaeal Elp3 catalyzes trna wobble uridine modification at C5 via a radical mechanism Kiruthika Selvadurai, Pei Wang, Joseph Seimetz & Raven H Huang* Department of Biochemistry,
Supplementary Information Archaeal Elp3 catalyzes trna wobble uridine modification at C5 via a radical mechanism Kiruthika Selvadurai, Pei Wang, Joseph Seimetz & Raven H Huang* Department of Biochemistry,
Supplementary Information
 Supplementary Information Title Degeneration and impaired regeneration of gray matter oligodendrocytes in amyotrophic lateral sclerosis Authors Shin H. Kang, Ying Li, Masahiro Fukaya, Ileana Lorenzini,
Supplementary Information Title Degeneration and impaired regeneration of gray matter oligodendrocytes in amyotrophic lateral sclerosis Authors Shin H. Kang, Ying Li, Masahiro Fukaya, Ileana Lorenzini,
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL Divergent effects of intrinsically active MEK variants on developmental Ras signaling Yogesh Goyal,2,3,4, Granton A. Jindal,2,3,4, José L. Pelliccia 3, Kei Yamaya 2,3, Eyan Yeung
SUPPLEMENTARY MATERIAL Divergent effects of intrinsically active MEK variants on developmental Ras signaling Yogesh Goyal,2,3,4, Granton A. Jindal,2,3,4, José L. Pelliccia 3, Kei Yamaya 2,3, Eyan Yeung
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
doi: /
 Yiting Xie ; Matthew D. Cham ; Claudia Henschke ; David Yankelevitz ; Anthony P. Reeves; Automated coronary artery calcification detection on low-dose chest CT images. Proc. SPIE 9035, Medical Imaging
Yiting Xie ; Matthew D. Cham ; Claudia Henschke ; David Yankelevitz ; Anthony P. Reeves; Automated coronary artery calcification detection on low-dose chest CT images. Proc. SPIE 9035, Medical Imaging
Supplementary Figure 1: Signaling centers contain few proliferating cells, express p21, and
 Supplementary Figure 1: Signaling centers contain few proliferating cells, express p21, and exclude YAP from the nucleus. (a) Schematic diagram of an E10.5 mouse embryo. (b,c) Sections at B and C in (a)
Supplementary Figure 1: Signaling centers contain few proliferating cells, express p21, and exclude YAP from the nucleus. (a) Schematic diagram of an E10.5 mouse embryo. (b,c) Sections at B and C in (a)
Musculoskeletal System. Terms. Origin (Proximal Attachment) Insertion (Distal Attachment)
 Musculoskeletal System Terms Origin (Proximal Attachment) Insertion (Distal Attachment) Agonist- prime mover Antagonist- provides a braking force Synergist- assists indirectly in the movement Musculoskeletal
Musculoskeletal System Terms Origin (Proximal Attachment) Insertion (Distal Attachment) Agonist- prime mover Antagonist- provides a braking force Synergist- assists indirectly in the movement Musculoskeletal
7 Grip aperture and target shape
 7 Grip aperture and target shape Based on: Verheij R, Brenner E, Smeets JBJ. The influence of target object shape on maximum grip aperture in human grasping movements. Exp Brain Res, In revision 103 Introduction
7 Grip aperture and target shape Based on: Verheij R, Brenner E, Smeets JBJ. The influence of target object shape on maximum grip aperture in human grasping movements. Exp Brain Res, In revision 103 Introduction
Sum of Neurally Distinct Stimulus- and Task-Related Components.
 SUPPLEMENTARY MATERIAL for Cardoso et al. 22 The Neuroimaging Signal is a Linear Sum of Neurally Distinct Stimulus- and Task-Related Components. : Appendix: Homogeneous Linear ( Null ) and Modified Linear
SUPPLEMENTARY MATERIAL for Cardoso et al. 22 The Neuroimaging Signal is a Linear Sum of Neurally Distinct Stimulus- and Task-Related Components. : Appendix: Homogeneous Linear ( Null ) and Modified Linear
Supplementary Figure 1
 Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Auditory Scene Analysis
 1 Auditory Scene Analysis Albert S. Bregman Department of Psychology McGill University 1205 Docteur Penfield Avenue Montreal, QC Canada H3A 1B1 E-mail: bregman@hebb.psych.mcgill.ca To appear in N.J. Smelzer
1 Auditory Scene Analysis Albert S. Bregman Department of Psychology McGill University 1205 Docteur Penfield Avenue Montreal, QC Canada H3A 1B1 E-mail: bregman@hebb.psych.mcgill.ca To appear in N.J. Smelzer
50mM D-Glucose. Percentage PI. L-Glucose
 Monica Dus, Minrong Ai, Greg S.B Suh. Taste-independent nutrient selection is mediated by a brain-specific Na+/solute cotransporter in Drosophila. Control + Phlorizin mm D-Glucose 1 2mM 1 L-Glucose Gr5a;Gr64a
Monica Dus, Minrong Ai, Greg S.B Suh. Taste-independent nutrient selection is mediated by a brain-specific Na+/solute cotransporter in Drosophila. Control + Phlorizin mm D-Glucose 1 2mM 1 L-Glucose Gr5a;Gr64a
Supplementary Figure 1. Using DNA barcode-labeled MHC multimers to generate TCR fingerprints
 Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTARY INFORMATION
 DOI: 0.038/ncb33 a b c 0 min 6 min 7 min (fixed) DIC -GFP, CenpF 3 µm Nocodazole Single optical plane -GFP, CenpF Max. intensity projection d µm -GFP, CenpF, -GFP CenpF 3-D rendering e f 0 min 4 min 0
DOI: 0.038/ncb33 a b c 0 min 6 min 7 min (fixed) DIC -GFP, CenpF 3 µm Nocodazole Single optical plane -GFP, CenpF Max. intensity projection d µm -GFP, CenpF, -GFP CenpF 3-D rendering e f 0 min 4 min 0
Table of content. -Supplementary methods. -Figure S1. -Figure S2. -Figure S3. -Table legend
 Table of content -Supplementary methods -Figure S1 -Figure S2 -Figure S3 -Table legend Supplementary methods Yeast two-hybrid bait basal transactivation test Because bait constructs sometimes self-transactivate
Table of content -Supplementary methods -Figure S1 -Figure S2 -Figure S3 -Table legend Supplementary methods Yeast two-hybrid bait basal transactivation test Because bait constructs sometimes self-transactivate
To test the possible source of the HBV infection outside the study family, we searched the Genbank
 Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
Supplementary Discussion The source of hepatitis B virus infection To test the possible source of the HBV infection outside the study family, we searched the Genbank and HBV Database (http://hbvdb.ibcp.fr),
Supplementary Material for
 Supplementary Material for Selective neuronal lapses precede human cognitive lapses following sleep deprivation Supplementary Table 1. Data acquisition details Session Patient Brain regions monitored Time
Supplementary Material for Selective neuronal lapses precede human cognitive lapses following sleep deprivation Supplementary Table 1. Data acquisition details Session Patient Brain regions monitored Time
Supplemental Figures. Figure S1: 2-component Gaussian mixture model of Bourdeau et al. s fold-change distribution
 Supplemental Figures Figure S1: 2-component Gaussian mixture model of Bourdeau et al. s fold-change distribution 1 All CTCF Unreponsive RefSeq Frequency 0 2000 4000 6000 0 500 1000 1500 2000 Block Size
Supplemental Figures Figure S1: 2-component Gaussian mixture model of Bourdeau et al. s fold-change distribution 1 All CTCF Unreponsive RefSeq Frequency 0 2000 4000 6000 0 500 1000 1500 2000 Block Size
Afferent lymph-derived T cells and dendritic cells use different CCR7-dependent routes for lymph node entry and intranodal migration
 Braun et al. Supplementary Information 1 Supplementary Information Afferent lymph-derived T cells and dendritic cells use different CCR7-dependent routes for lymph node entry and intranodal migration Asolina
Braun et al. Supplementary Information 1 Supplementary Information Afferent lymph-derived T cells and dendritic cells use different CCR7-dependent routes for lymph node entry and intranodal migration Asolina
Crystallization-grade After D After V3 cocktail. Time (s) Time (s) Time (s) Time (s) Time (s) Time (s)
 Ligand Type Name 6 Crystallization-grade After 447-52D After V3 cocktail Receptor CD4 Resonance Units 5 1 5 1 5 1 Broadly neutralizing antibodies 2G12 VRC26.9 Resonance Units Resonance Units 3 1 15 1 5
Ligand Type Name 6 Crystallization-grade After 447-52D After V3 cocktail Receptor CD4 Resonance Units 5 1 5 1 5 1 Broadly neutralizing antibodies 2G12 VRC26.9 Resonance Units Resonance Units 3 1 15 1 5
mtor Inhibition Specifically Sensitizes Colorectal Cancers with KRAS or BRAF Mutations to BCL-2/BCL-
 Supplementary Material for mtor Inhibition Specifically Sensitizes Colorectal Cancers with KRAS or BRAF Mutations to BCL-2/BCL- XL Inhibition by Suppressing MCL-1 Anthony C. Faber 1,2 *, Erin M. Coffee
Supplementary Material for mtor Inhibition Specifically Sensitizes Colorectal Cancers with KRAS or BRAF Mutations to BCL-2/BCL- XL Inhibition by Suppressing MCL-1 Anthony C. Faber 1,2 *, Erin M. Coffee
B220 CD4 CD8. Figure 1. Confocal Image of Sensitized HLN. Representative image of a sensitized HLN
 B220 CD4 CD8 Natarajan et al., unpublished data Figure 1. Confocal Image of Sensitized HLN. Representative image of a sensitized HLN showing B cell follicles and T cell areas. 20 µm thick. Image of magnification
B220 CD4 CD8 Natarajan et al., unpublished data Figure 1. Confocal Image of Sensitized HLN. Representative image of a sensitized HLN showing B cell follicles and T cell areas. 20 µm thick. Image of magnification
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
Supplementary Material
 Supplementary Material Supplementary Text Text S1. Time distributions of the high FRET efficiency at different concentrations of EF-G.GTP From Fig. 1, the model of ribosomal translocation at non-saturating
Supplementary Material Supplementary Text Text S1. Time distributions of the high FRET efficiency at different concentrations of EF-G.GTP From Fig. 1, the model of ribosomal translocation at non-saturating
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Reward rate affects the decision to begin work. (a) Latency distributions are bimodal, and depend on reward rate. Very short latencies (early peak) preferentially occur when a greater
Supplementary Figure 1 Reward rate affects the decision to begin work. (a) Latency distributions are bimodal, and depend on reward rate. Very short latencies (early peak) preferentially occur when a greater
Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier
 Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier curves for progression-free survival (PFS) and overall survival (OS) in a cohort of patients (N=52) with stage III primary ovarian
Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier curves for progression-free survival (PFS) and overall survival (OS) in a cohort of patients (N=52) with stage III primary ovarian
A Race Model of Perceptual Forced Choice Reaction Time
 A Race Model of Perceptual Forced Choice Reaction Time David E. Huber (dhuber@psyc.umd.edu) Department of Psychology, 1147 Biology/Psychology Building College Park, MD 2742 USA Denis Cousineau (Denis.Cousineau@UMontreal.CA)
A Race Model of Perceptual Forced Choice Reaction Time David E. Huber (dhuber@psyc.umd.edu) Department of Psychology, 1147 Biology/Psychology Building College Park, MD 2742 USA Denis Cousineau (Denis.Cousineau@UMontreal.CA)
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Relative expression of K IR2.1 transcript to enos was reduced 29-fold in capillaries from knockout animals. Relative expression of K IR2.1 transcript to enos was reduced 29-fold
Supplementary Figure 1 Relative expression of K IR2.1 transcript to enos was reduced 29-fold in capillaries from knockout animals. Relative expression of K IR2.1 transcript to enos was reduced 29-fold
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
SUPPLEMENTARY INFORMATION Perceptual learning in a non-human primate model of artificial vision
 SUPPLEMENTARY INFORMATION Perceptual learning in a non-human primate model of artificial vision Nathaniel J. Killian 1,2, Milena Vurro 1,2, Sarah B. Keith 1, Margee J. Kyada 1, John S. Pezaris 1,2 1 Department
SUPPLEMENTARY INFORMATION Perceptual learning in a non-human primate model of artificial vision Nathaniel J. Killian 1,2, Milena Vurro 1,2, Sarah B. Keith 1, Margee J. Kyada 1, John S. Pezaris 1,2 1 Department
Summary of behavioral performances for mice in imaging experiments.
 Supplementary Figure 1 Summary of behavioral performances for mice in imaging experiments. (a) Task performance for mice during M2 imaging experiments. Open triangles, individual experiments. Filled triangles,
Supplementary Figure 1 Summary of behavioral performances for mice in imaging experiments. (a) Task performance for mice during M2 imaging experiments. Open triangles, individual experiments. Filled triangles,
Measuring the User Experience
 Measuring the User Experience Collecting, Analyzing, and Presenting Usability Metrics Chapter 2 Background Tom Tullis and Bill Albert Morgan Kaufmann, 2008 ISBN 978-0123735584 Introduction Purpose Provide
Measuring the User Experience Collecting, Analyzing, and Presenting Usability Metrics Chapter 2 Background Tom Tullis and Bill Albert Morgan Kaufmann, 2008 ISBN 978-0123735584 Introduction Purpose Provide
Mechanisms of Regulating Cell Topology in Proliferating Epithelia: Impact of Division Plane, Mechanical Forces, and Cell Memory
 1 Mechanisms of Regulating Cell Topology in Proliferating Epithelia: Impact of Division Plane, Mechanical Forces, and Cell Memory Yingzi Li 1,2, Hammad Naveed 2, Sema Kachalo 2, Lisa X. Xu 1,3, Jie Liang
1 Mechanisms of Regulating Cell Topology in Proliferating Epithelia: Impact of Division Plane, Mechanical Forces, and Cell Memory Yingzi Li 1,2, Hammad Naveed 2, Sema Kachalo 2, Lisa X. Xu 1,3, Jie Liang
Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines.
 Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines. The scores summarize the global expression of the tissue
Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines. The scores summarize the global expression of the tissue
A genetically targeted optical sensor to monitor calcium signals in astrocyte processes
 A genetically targeted optical sensor to monitor calcium signals in astrocyte processes 1 Eiji Shigetomi, 1 Sebastian Kracun, 2 Michael V. Sofroniew & 1,2 *Baljit S. Khakh Ψ 1 Departments of Physiology
A genetically targeted optical sensor to monitor calcium signals in astrocyte processes 1 Eiji Shigetomi, 1 Sebastian Kracun, 2 Michael V. Sofroniew & 1,2 *Baljit S. Khakh Ψ 1 Departments of Physiology
A Race Model of Perceptual Forced Choice Reaction Time
 A Race Model of Perceptual Forced Choice Reaction Time David E. Huber (dhuber@psych.colorado.edu) Department of Psychology, 1147 Biology/Psychology Building College Park, MD 2742 USA Denis Cousineau (Denis.Cousineau@UMontreal.CA)
A Race Model of Perceptual Forced Choice Reaction Time David E. Huber (dhuber@psych.colorado.edu) Department of Psychology, 1147 Biology/Psychology Building College Park, MD 2742 USA Denis Cousineau (Denis.Cousineau@UMontreal.CA)
Discrimination and Generalization in Pattern Categorization: A Case for Elemental Associative Learning
 Discrimination and Generalization in Pattern Categorization: A Case for Elemental Associative Learning E. J. Livesey (el253@cam.ac.uk) P. J. C. Broadhurst (pjcb3@cam.ac.uk) I. P. L. McLaren (iplm2@cam.ac.uk)
Discrimination and Generalization in Pattern Categorization: A Case for Elemental Associative Learning E. J. Livesey (el253@cam.ac.uk) P. J. C. Broadhurst (pjcb3@cam.ac.uk) I. P. L. McLaren (iplm2@cam.ac.uk)
