Brat Is a Miranda Cargo Protein that Promotes Neuronal Differentiation and Inhibits Neuroblast Self-Renewal
|
|
|
- Jasmin Lawson
- 5 years ago
- Views:
Transcription
1 Developmental Cell 10, , April, 2006 ª2006 Elsevier Inc. DOI /j.devcel Brat Is a Miranda Cargo Protein that Promotes Neuronal Differentiation and Inhibits Neuroblast Self-Renewal Cheng-Yu Lee, 1 Brian D. Wilkinson, 2 Sarah E. Siegrist, 1 Robin P. Wharton, 2, * and Chris Q. Doe 1, * 1 Institutes of Neuroscience and Molecular Biology Howard Hughes Medical Institute University of Oregon Eugene, Oregon Howard Hughes Medical Institute Department of Molecular Genetics and Microbiology Duke University Medical Center Durham, North Carolina Summary An important question in stem cell biology is how a cell decides to self-renew or differentiate. Drosophila neuroblasts divide asymmetrically to self-renew and generate differentiating progeny called GMCs. Here, we report that the Brain tumor (Brat) translation repressor is partitioned into GMCs via direct interaction with the Miranda scaffolding protein. In brat mutants, another Miranda cargo protein (Prospero) is not partitioned into GMCs, GMCs fail to downregulate neuroblast gene expression, and there is a massive increase in neuroblast numbers. Single neuroblast clones lacking Prospero have a similar phenotype. We conclude that Brat suppresses neuroblast stem cell self-renewal and promotes neuronal differentiation. Introduction Asymmetric cell division is important for generating cell diversity during development and for maintaining stem cell identity (Betschinger and Knoblich, 2004; Lechler and Fuchs, 2005; Yamashita et al., 2003). Drosophila neuroblasts are an excellent model system for investigating both asymmetric cell division and stem cell selfrenewal (Betschinger and Knoblich, 2004; Lee et al., 2005). Mitotic neuroblasts have distinct apical and basal cortical domains, and the mitotic spindle aligns with the axis of cortical polarity, resulting in asymmetric cell division. The large apical sibling is a self-renewing neuroblast, and the small basal sibling is a ganglion mother cell (GMC) that typically generates two postmitotic neurons. Apical cortical proteins include atypical protein kinase C (apkc), Bazooka (Baz/Par3), Par6, Inscuteable (Insc), Partner of Inscuteable (Pins), and Gai. These apical proteins regulate neuroblast spindle orientation, spindle asymmetry, sibling cell size asymmetry, and targeting of proteins to the basal cortex (Betschinger and Knoblich, 2004). Basal cortical proteins include Numb, Miranda (coiled-coil scaffold), and two Miranda cargo proteins Prospero (homeodomain transcriptional repressor) and Staufen (RNA binding protein that localizes prospero mrna) (Broadus et al., 1998; Ikeshima- *Correspondence: rwharton@duke.edu (R.P.W.); cdoe@uoregon. edu (C.Q.D.) Kataoka et al., 1997; Knoblich et al., 1995; Rhyu et al., 1994; Schuldt et al., 1998; Spana and Doe, 1995). Miranda basal localization requires the amino-terminal portion of the protein (1 405 aa), and the adjacent coiledcoil central domain of Miranda is required for basal localization of Prospero and Staufen cargo proteins (Fuerstenberg et al., 1998; Ikeshima-Kataoka et al., 1997; Matsuzaki et al., 1998; Schuldt et al., 1998). To date, the only cell fate determinants known to be partitioned into the GMC are Numb and Prospero; Numb has no known function in GMCs (Skeath and Doe, 1998), whereas Prospero is thought to promote GMC exit from the cell cycle (Li and Vaessin, 2000). It is unknown whether there are other cell fate determinants segregated into the GMC, and whether the Miranda scaffolding protein has cargo proteins other than Prospero and Staufen. Drosophila neuroblasts have also emerged as a model system for investigating stem cell self-renewal. Larval neuroblasts can produce hundreds of GMCs without losing cell volume, losing proliferation potential, or differentiating (Lee et al., 2005; Rolls et al., 2003); in contrast, a GMC typically undergoes a reductive division to generate only two postmitotic neurons (Lee and Luo, 2001). Apical proteins, which are partitioned into the neuroblast at cytokinesis, would be excellent candidates for promoting neuroblast self-renewal. Indeed, we have recently shown that apkc mutants have fewer neuroblasts, whereas apkc overexpression can induce ectopic neuroblast renewal and massively increase the population of larval neuroblasts (Lee et al., 2005). Conversely, basal proteins would be excellent candidates for promoting neuronal differentiation and inhibiting neuroblast self-renewal; consistent with this model, it has recently been shown that prospero, miranda, and numb mutant brain tissue can form metastatic tumors when transplanted into wild-type adult hosts (Caussinus and Gonzalez, 2005). However, the cellular phenotype of prospero, miranda, and numb mutants has not been determined, so it remains unclear whether they induce tumor formation as a consequence of genomic instability (Caussinus and Gonzalez, 2005) or by allowing GMCto-neuroblast cell fate transformations. We have been screening for mutants that affect larval neuroblast self-renewal and asymmetric cell division (C.-Y.L. and C.Q.D.), and we identified a mutation in brain tumor (brat) that resulted in ectopic larval brain neuroblasts. Brat is a translational repressor containing an evolutionarily conserved NHL (NCL-1, HT2A, and LIN- 41) domain that recruits Pumilio and Nanos to block hunchback mrna translation during early embryogenesis (Sonoda and Wharton, 2001; Arama et al., 2000), and brat mutants are pupal lethal with a brain overgrowth phenotype (Arama et al., 2000). Here, we report that brat mutants have ectopic neuroblasts and fail to differentiate GMCs. We show that Brat protein is asymmetrically partitioned into GMCs via direct interaction with the Miranda basal scaffolding protein, and that, in brat mutants, the Miranda cargo protein Prospero fails to be partitioned into GMCs. Thus, Brat is a Miranda binding protein that is partitioned into the GMC, where it
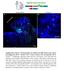
2 Developmental Cell 442 Figure 1. brat Mutants Have Ectopic Neuroblasts (A and B) Images of single brain lobes of wild-type or brat 11 /brat 11 mutants 96 hr after larval hatching (ALH) stained for the neuroblast markers Miranda (Mira) and Deadpan (Dpn) and the neuronal marker Elav and assayed for proliferation by performing a 4 hr BrdU pulse prior to fixation. Each panel shows a single optical section. Representative neuroblasts, arrowheads; GMCs, arrows. (C) Quantification of neuroblast numbers per brain lobe was based on Dpn staining in wild-type (wt; white bars) and brat 11 /brat 11 mutants (brat; black bars). Average neuroblast number and standard deviation (in parentheses): 24 hr ALH wild-type, 47.1 (5.0) and brat, 16.4 (5.1); 48 hr ALH wild-type, 79 (14.2) and brat, 61.2 (8.1); 72 hr ALH wild-type, 100 (5.7) and brat, (27.2); 96 hr ALH wild-type, 103 (4.1) and brat, >500 (n.a.). The lower numbers of neuroblasts in hr ALH brat mutant larvae are likely due to organismal developmental delay, as brat mutants are slower to hatch into larvae and slower to undergo each larval molt (data not shown). inhibits neuroblast self-renewal and promotes neuronal differentiation. Results brat Mutants Have Supernumerary Brain Neuroblasts In order to identify genes that regulate larval neuroblast self-renewal and asymmetric cell division, we screened a collection of transposable P element-induced lethal mutants for an altered number of brain neuroblasts or mislocalized cell polarity markers (C.-Y.L. and C.Q.D.). We identified a mutation in l(2)k06028, a weak loss-offunction allele of brat, that resulted in increased brain neuroblast numbers (Figure S1; see the Supplemental Data available with this article online). To determine the brat null mutant phenotype, we quantified neuroblast numbers in brat 11 homozygous mutant larvae from 24 to 96 hr after larval hatching (ALH). We identified neuroblasts by using molecular and proliferation markers. Neuroblasts express the markers Deadpan (Dpn), Worniu, and Miranda (each is also very transiently present in newborn GMCs); lack the neuronal marker Elav and the glial marker Repo; and are proliferative based on their ability to incorporate BrdU. In wild-type larvae, the number of proliferating neuroblasts gradually increased from 45 neuroblasts at 24 hr ALH to 100 neuroblasts at 72 hr ALH and remained at w100 neuroblasts until the onset of metamorphosis (Figures 1A and 1C) (Datta, 1995; Lee et al., 2005). These 100 neuroblasts generate many thousands of Elav + postmitotic neurons comprising the larval and adult brain (Figure 1A). In brat mutant larvae, we observed three phenotypes. First, we detected a dramatic increase in neuroblast number over 500 by 96 hr ALH and an estimated several thousand by 120 hr ALH (Figures 1B and 1C; data not shown). Second, we observed a corresponding decrease in Elav + neurons (Figure 1B), suggesting that the ectopic neuroblasts may be generated at the expense of neurons. Finally, we noticed that the GMCs adjacent to each neuroblast failed to downregulate neuroblast marker expression (e.g., maintaining Dpn, Miranda, and Worniu staining; Figure 1B and data not shown). The fate of these cells will be addressed in the next section. We conclude that brat mutants have a late-onset increase in proliferative neuroblasts and a corresponding decrease in differentiated neurons. The fact that the neuroblast population expands simultaneously with loss of neurons strongly suggests that at least some GMCs are developing as neuroblasts rather than neurons (see Discussion). brat Mutants Have a Delay in GMC Differentiation To further characterize the fate of the small GMCs that maintain neuroblast marker expression in brat mutant brains, we performed BrdU pulse-chase experiments. Our reasoning was that BrdU incorporation can be used to identify cells that continue to proliferate (they initially incorporate BrdU but rapidly dilute it out during the chase period) and cells that differentiate (they incorporate BrdU during their terminal division and maintain BrdU during the chase period). In wild-type larvae, a 4 hr pulse of BrdU followed by immediate fixation and BrdU staining revealed BrdU incorporation in all neuroblasts and a few adjacent newborn, smaller GMCs (Figures 1A and 2A). After a 24 hr BrdU-free chase, however, all BrdU labeling was restricted to the pool of Elav + neurons born during the BrdU pulse window (Figure 2B). Thus, the proliferative neuroblasts dilute out BrdU incorporation, whereas postmitotic Elav + neurons retain BrdU incorporation (Figures 2A and 2B, schematic). We next assayed brat mutant larvae in these pulsechase experiments. Immediately after the 4 hr BrdU pulse, nearly all neuroblasts were labeled with BrdU as well as adjacent smaller, presumptive GMCs (Figures 1B and 2C). After a 24 hr chase, each Dpn + large neuroblast was surrounded by GMCs that maintained Dpn,
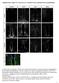
3 Brat Inhibits Neuroblast Self-Renewal 443 Figure 2. brat Mutants Have a Failure in GMC Differentiation (A E) Wild-type or brat 11 /brat 11 mutant larvae 72 hr ALH were given a 4 hr BrdU pulse and either fixed immediately or chased in BrdU-free media for 24 or 72 hr. One brain lobe stained for the indicated markers is shown in each panel, and high-magnification single-label images of the boxed region are shown below each panel. Dpn + large neuroblasts, dotted circles; Dpn 2 BrdU + GMCs, arrows; Dpn + BrdU + GMCs, arrowheads; Elav + neurons, solid outline (in [D] and [E]). Schematic summaries: Dpn, red; BrdU, green; Elav, blue. (A and B) Wild-type. (A) A 4 hr BrdU pulse labels all neuroblasts and newborn GMCs. (B) After a 24 hr chase, BrdU can only be detected in Elav + neurons (the large Dpn + neuroblasts have diluted out BrdU). (C E) brat mutants. (C) A 4 hr BrdU pulse labels all neuroblasts and many small newborn GMCs that fail to downregulate Dpn. (D) After a 24 hr chase, the large neuroblasts have diluted out BrdU, but BrdU is observed in small Dpn + GMCs (arrowhead) and Elav + neurons (solid outline). (E) After a 72 hr chase, virtually all BrdU is restricted to the Elav + neurons (solid outline). did not express Elav, and did not dilute out BrdU (Dpn + BrdU + Elav 2 )(Figure 2D); these are likely to be arrested GMCs that have neither differentiated nor resumed proliferation. We also detected a small number of BrdU + Elav + neurons, indicating that some GMCs are able to differentiate normally (Figure 2D). After a 72 hr chase, there were many fewer BrdU + cells overall, and the remaining BrdU + cells were virtually all Elav + (Figure 2E), showing that some small Dpn + cells seen at 24 hr post- BrdU eventually differentiate into neurons. Based on the steep decline in the number of Dpn + BrdU + small cells during the hr chase interval, and the corresponding increase in neuroblast number during this interval (Figure 1C), we propose that some Dpn + GMCs enlarge into neuroblasts and reenter the cell cycle (Figures 2C 2E, schematic; see Discussion). We conclude that brat neuroblasts generate GMC-sized progeny that are cell cycle delayed and continue to express the neuroblast markers Dpn, Miranda, and Worniu (Figure 2 and data not shown); some of these GMCs differentiate into neurons, but many appear to develop into proliferative neuroblasts. Brat Is Asymmetrically Partitioned into the GMC during Neuroblast Division brat mutants show GMC gene regulation defects, so we next assayed for Brat protein localization in neuroblasts and GMCs of the embryonic and larval CNS. In wild-type
4 Developmental Cell 444 Figure 3. Brat and Miranda Colocalize and Are Partitioned into the GMC during Embryonic Neuroblast Asymmetric Division (A and A 0 ) Wild-type and miranda AB78 /miranda AB78 mutant stage-11 embryos. Wild-type embryos have high Brat protein levels in the GMC layer (arrowhead) compared to the neuroblast layer (arrow), but this difference is abolished in miranda mutants. Apical is up. (B D) Wild-type embryonic neuroblasts in vivo. Brat and Miranda colocalize in a basal crescent (between arrowheads) and are partitioned into the GMC. Apical is up. (E J) Wild-type embryonic neuroblasts isolated in vitro. Isolated neuroblasts undergo normal asymmetric cell division (Broadus and Doe, 1997; Grosskortenhaus et al., 2005; Siegrist and Doe, 2006) and reveal the basal cortex better due to the absence of associated GMCs. Brat and Miranda colocalize throughout the cell cycle and are partitioned specifically into the GMC. Apical is up (defined by the position of the apkc crescent). The scale bar is 5 mm. embryos, Brat was detected at a low level in neuroblasts, and it was highly enriched in the GMC layer (Figure 3A). Examination of mitotic embryonic neuroblasts revealed that Brat was colocalized with the basal scaffolding protein Miranda from prometaphase through telophase, and both proteins were partitioned into the GMC after cytokinesis (95% basal, n = 25; Figures 3B 3D; Table 1). To obtain a clearer view of the Brat protein basal crescent, we cultured embryonic neuroblasts in vitro by using standard methods; these neuroblasts undergo normal asymmetric cell divisions (Broadus and Doe, 1997; Grosskortenhaus et al., 2005; Siegrist and Doe, 2006). Again, we find that Brat and Miranda are precisely colocalized in a basal protein crescent from prometaphase through telophase, with both proteins being specifically partitioned into the newborn GMC (Figures 3E 3J). In addition, larval neuroblasts showed the identical colocalization of Brat and Miranda in basal cortical crescents throughout the cell cycle, resulting in the partitioning of both proteins into the newborn GMC (Figures S2A S2C). In fact, Brat/Miranda colocalization was maintained even in mutant genotypes where Miranda was mislocalized: in apkc mutants, both Miranda and Brat were uniform cortical in metaphase neuroblasts, and in
5 Brat Inhibits Neuroblast Self-Renewal 445 Table 1. Quantification of Neuroblast Cortical Polarity in Wild- Type and Mutant Genotypes Wild-Type a brat Mutant a apkc b Apical (metaphase) 100% (n = 43) 60% (n = 136) Prospero b Basal (metaphase) 91.4% (n = 35) 5.1% (n = 59) Basal (telophase) 88% (n = 25) 7.9% (n = 63) miranda Mutant a Brat b Basal (metaphase) 96% (n = 25) 0% (n = 20) a Genotypes: wild-type, heterozygous siblings (brat/+ or miranda/+); brat mutant, brat 11 homozygotes; miranda mutant, miranda AB78 or miranda L44 homozygotes. b apkc and Prospero protein localization scored in larval central brain neuroblasts at 120 hr ALH; Brat protein localization scored in embryonic neuroblasts. n, number of neuroblasts scored; apical localization, complete restriction to apical quadrant; basal localization, detectable protein at the basal quadrant. lgl mutant neuroblasts, both Miranda and Brat were predominantly cytoplasmic at metaphase (Figures S2D and S2E). We conclude that Brat and Miranda are invariably colocalized in mitotic neuroblasts, and that Brat is partitioned into the GMC during neuroblast asymmetric cell division. Brat Is a Miranda Cargo Protein To test whether asymmetric localization of Brat is mediated by Miranda, we examined Brat protein localization in two miranda mutant genotypes: miranda L44, a genetic null that makes no detectable protein, and miranda AB78, which generates a protein of 405 amino acids that lacks the central Prospero/Staufen binding domain and C- terminal domain (Fuerstenberg et al., 1998). The miranda AB78 homozygous embryos lacked Brat enrichment in the GMC layer, instead showing equalized Brat protein levels between the neuroblast and GMC layers (Figure 3A 0 ). Examination of mitotic neuroblasts in both miranda mutant genotypes revealed that Brat protein was completely delocalized into the cytoplasm (100% delocalized, n = 20; Table 1; Figure 4). We conclude that Brat is a Miranda cargo protein that requires the Miranda coiled-coil cargo binding domain ( aa) for proper basal targeting into the GMC. Independently of our genetic screen, we identified Miranda in a yeast two-hybrid screen for proteins that bind the Brat NHL domain (B.D.W and R.P.W.). All brat alleles exhibiting brain overgrowth, including the genetic null brat 11, that have been sequenced possess molecular lesions in the NHL domain (Sonoda and Wharton, 2001; Arama et al., 2000). We identified six different clones encoding Miranda; each clone contains a portion of the Miranda central cargo binding domain (data not shown). We next tested Miranda binding to a panel of Brat proteins containing single alanine substitutions, which collectively decorate both surfaces of the NHL domain (Edwards et al., 2003). We observed that mutations mapping to the top surface of the NHL domain had no effect on Miranda/Brat association, but that several mutations mapping to the lower surface of the NHL domain prevented Miranda/Brat binding (Figure 5). In particular, the substitution of glycine 774 or tyrosine 829 Figure 4. Miranda Is Required for Brat Basal Asymmetric Localization (A D) (A) Wild-type and (B D) miranda mutant embryonic neuroblasts double labeled for Miranda (Mira) and Brat; panels were acquired by using identical confocal settings. The Miranda AB78 protein forms a basal crescent but lacks the central cargo binding domain, whereas the Miranda L44 protein is undetectable. All neuroblasts are in metaphase, except for the neuroblast in (C), which is in telophase; all neuroblasts are at stage 10, except for the neuroblast in (B), which is at stage 13 and is thus smaller. Miranda protein domains present in each genotype are diagrammed at the top (ALD, asymmetric localization domain; cargo, Prospero/Staufen cargo binding domain). Apical is up. The arrow or bracket indicates basal Miranda protein. completely abolished Miranda binding to the NHL domain (Figure 5); both mutants also abolish Brat/Pumilio association (Edwards et al., 2003; Sonoda and Wharton, 2001), suggesting that Miranda and Pumilio have overlapping binding surfaces on the Brat NHL domain. We conclude that the Brat NHL domain interacts with the Miranda cargo domain, confirming our genetic evidence that Brat is a Miranda cargo protein. brat Mutants Have Defects in Prospero and apkc Asymmetric Localization Brat protein is partitioned into the newborn GMC, and brat mutants have defects in GMC development. However, it is possible that the brat phenotype arises in the neuroblast and not the GMC, based on fact that mislocalization of apkc beyond its normal apical cortical domain during neuroblast cell division is sufficient to generate ectopic larval brain neuroblasts (Lee et al., 2005). Here, we test whether the Brat protein, which forms a basal cortical crescent in mitotic neuroblasts, has a role in establishing or maintaining neuroblast cortical polarity. In wild-type neuroblasts, apkc, Baz, and Pins were all localized to the apical cortex at metaphase (Figure 6A; Table 1; data not shown). In brat mutants, Baz and Pins showed normal localization, but apkc was occasionally delocalized to the basal cortex (40%, n = 136; Figure 6B; Table 1). Ectopic apkc was not sufficient to cause the brat mutant phenotype, however, because brat apkc double mutants still developed ectopic neuroblasts, and misexpression of Lgl3A (which is a potent suppressor of apkc-induced ectopic neuroblasts [Lee et al., 2005]) was unable to suppress the ectopic neuroblasts in brat mutants (Figure S3). Examination of the basal cortical markers Miranda and Prospero in brat mutants revealed a more striking
6 Developmental Cell 446 Figure 5. Miranda Directly Interacts with the Brat NHL Domain (A) Diagram of the Brat NHL domain (turquoise). The Brat NHL domain interacts with both Pumilio (Pum; red/yellow; Edwards et al., 2003) and Miranda (Mira) on the lower surface of the NHL domain. The numbers 1 7 represent the following Brat mutations used in the yeast two-hybrid experiments: (1) Y859A, (2) D1012A, (3) E970A, (4) G774A, (5) Y829A, (6) R847A, (7) R875A (Edwards et al., 2003). (B) Yeast two-hybrid experiments were performed as described previously (Edwards et al., 2003); interaction between Brat and Mira is required for growth in the absence of histidine (top row). Substitutions on the lower surface of the NHL domain block interaction both with Pum and with Mira (mutants 4 and 5) or just Pum (mutants 6 and 7); substitutions on the top surface (mutants 1 3) have no effect on Mira or Pum binding. The smallest portion of Mira common to the 6 different clones isolated in the screen consists of amino acids ; exclusion of a single clone that encodes a weakly interacting Mira fragment changes the minimal interaction domain to residues phenotype: although Miranda localization was normal, Prospero was almost always absent from the basal cortex (metaphase 95% absent, n = 59; telophase 92% absent, n = 63; Figure 6B; Table 1). In addition, we observed a significant decrease in overall Prospero levels in brat mutant brains (Figure 6, right-most panels; Figure S1). We conclude that Brat promotes Prospero localization into the GMC and normal Prospero levels in the brain. prospero Mutant Neuroblast Clones Maintain Neuroblast Gene Expression and Fail to Differentiate GMCs To test whether the absence of Prospero was sufficient to mimic the brat mutant phenotype, we removed Prospero function from single neuroblast lineages by using the MARCM technique (Lee and Luo, 2001). We generated a low frequency of prospero 17 null mutant clones in the thoracic ganglia, central brain, and optic lobes of the larval CNS. In every case, the prospero mutant clone was nearly exclusively composed of Dpn + Elav 2 cells, and it was almost completely devoid of Elav + neurons (Figures 7A 7C). We conclude that Prospero is required to promote GMC differentiation, and thus loss of Prospero is sufficient to account for this aspect of the brat mutant phenotype. Discussion Brat Is a Miranda Cargo Protein Here, we show that the Brat translational repressor directly interacts with the Miranda central domain and is a Miranda cargo protein specifically partitioned into the GMC daughter cell during neuroblast asymmetric cell division. To our knowledge, Brat is the first Miranda cargo protein identified since the original finding that Prospero and Staufen were shown to be Miranda cargo proteins over 8 years ago (Fuerstenberg et al., 1998; Ikeshima- Kataoka et al., 1997; Shen et al., 1998). Prospero is a homeodomain transcriptional repressor, and Staufen is an RNA binding protein that interacts with prospero mrna. It is unknown whether Miranda has other cargo proteins in addition to Brat, Prospero, and Staufen, and it is unclear whether all three known cargo proteins can associate with a single Miranda protein. Figure 6. brat Mutants Fail to Localize Prospero Protein into GMCs and Have Reduced Prospero Levels Third instar larval neuroblasts in wild-type and brat 11 /brat 11 mutants stained for the indicated proteins. (A) In wild-type, Bazooka (Baz), Partner of Inscuteable (Pins), and apkc are localized to the apical cortex, and Miranda (Mira) and Prospero (Pros) are localized to the basal cortex. Right panel: Prospero protein staining in a single brain lobe. (B) In brat mutants, Baz, Pins, and Mira show normal localization, while apkc shows partial extension into the basal cortical domain (bracket); Pros is undetectable at the basal cortex. Right panel: Prospero protein levels are reduced in brat brain lobes. Dotted lines mark the apical cortex in telophase neuroblasts. Apical is up.
7 Brat Inhibits Neuroblast Self-Renewal 447 Figure 7. Single Neuroblast prospero Mutant Clones Have Ectopic Neuroblast Self-Renewal at the Expense of GMC Differentiation (A C) A prospero 17 null mutant clone induced in the (A) thoracic ganglia, (B) central brain, and (C) optic lobe. The mutant clone is marked with GFP (green), the Dpn neuroblast marker (red), and the Elav GMC/neuron marker (blue). One optical section is shown; anterior is up. brat Mutant Neuroblasts Have Defects in Prospero and apkc Localization It is unknown how Brat promotes Prospero basal localization. We favor the model that Brat protein stabilizes Prospero/Miranda interactions, so that Prospero protein is cytoplasmic in the absence of Brat. We don t see an obviously elevated level of cytoplasmic Prospero in brat mutant neuroblasts, but delocalization of Prospero protein from the basal crescent might not be visible over background. Alternatively, brat mutant neuroblasts may fail to transcribe or translate prospero in neuroblasts. This would most likely be an indirect effect, as Brat has only been shown to have translational repressor function (Sonoda and Wharton, 2001). We have been unable to detect prospero mrna in wild-type larval neuroblasts, despite robust levels in GMCs (unpublished data), so we have been unable to test this possibility. We also observed that some brat mutant neuroblasts show expanded apkc cortical crescents, in some cases reaching the basal cortex. This phenotype appears specific for apkc, because other apical cortical proteins (e.g., Baz, Pins) are unaffected. Brat might repress apkc translation, leading to increased apkc protein levels in brat mutants. Alternatively, the absence of Prospero or other basal cortical proteins may indirectly affect apkc localization. brat Mutants Show Ectopic Neuroblast Self-Renewal and Fail to Differentiate GMCs brat mutant brains show a dramatic increase in the number of large, proliferating Dpn + neuroblasts between 48 and 96 hr ALH. Where do these hundreds of extra neuroblasts come from? They are unlikely to come from outside the brain, or from dedifferentiation of neurons or glia, although we can t formally rule out these models. They are likely to derive from the pool of Dpn + neuroblasts in the brain, because these are the primary pool of proliferating cells in the larval central brain, and thus the best candidates to generate the thousands of extra cells found in the hypertrophied brat mutant brains. We favor a model in which a subset of brat mutant GMCs enlarge into proliferative neuroblasts (Figure 8). Figure 8. Brat Is a Miranda Cargo Protein Required for Inhibiting Neuroblast Self-Renewal and Promoting GMC Differentiation Top, wild-type. Neuroblasts partition Miranda (Mira), Brat, and Prospero (Pros) into GMCs. In GMCs, Mira is degraded, Brat is cytoplasmic, Pros is nuclear, and neuroblast genes are downregulated. GMCs differentiate into two postmitotic neurons. Bottom, brat mutant. Neuroblasts partition Miranda (Mira), but not Brat or Pros, into GMCs. GMCs maintain neuroblast gene expression and show delayed differentiation; some ultimately form neurons, while others appear to enlarge into proliferative neuroblasts. NB gene expression indicates Dpn, Mira, and Worniu.
8 Developmental Cell 448 This model is supported by several lines of evidence. (1) brat mutant GMCs maintain neuroblast-specific gene expression (Dpn, Miranda, Worniu); (2) brat mutants show an inverse relationship between increasing neuroblast number and decreasing neuronal number over time, consistent with GMCs forming neuroblasts instead of neurons; (3) brat mutant GMCs can be labeled by a BrdU pulse at their birth, yet most lose BrdU incorporation during the chase interval, showing that they either reenter the cell cycle or undergo cell death, and that cell death is not consistent with the brain overgrowth phenotype; (4) brat mutant telophase profiles show that all GMCs are born as small Miranda + cells, ruling out physically or molecularly symmetric neuroblast divisions as a mechanism for increasing the neuroblast population; and (5) brat mutants show cell enlargement in other tissues, and a similar cell growth phenotype has been observed in mutants in the C. elegans brat ortholog (Frank et al., 2002). What is the cellular origin of the brat mutant phenotype? brat mutant GMCs are compromised in three ways: they lack Brat translational repression activity, lack Prospero, and some may have ectopic apkc. Loss of Brat translational repression activity could well play a role in the ectopic neuroblast self-renewal phenotype, because all brat mutants disrupting the NHL translational repression domain exhibit a brain tumor phenotype, and Brat has been previously shown to negatively regulate cell growth (Frank et al., 2002). Loss of Prospero also plays a role in the brat phenotype: prospero mutant GMCs have a failure to downregulate neuroblast gene expression and a failure in neuronal differentiation, similar to brat mutants. prospero null mutant embryos also show a slight delay in neuronal differentiation (Broadie and Bate, 1993), although they appear to undergo normal neuroblast self-renewal (Doe et al., 1991; Vaessin et al., 1991). Finally, ectopic apkc can also mimic aspects of the brat phenotype, including formation of supernumerary large Dpn + neuroblasts (Lee et al., 2005). Interestingly, the mammalian paralogs of Drosophila apkc (apkcl/z) are expressed in neural progenitors of the ventricular zone (Manabe et al., 2002), and the mammalian Prospero ortholog Prox1 is expressed in differentiating neurons of the subventricular zone (Oliver et al., 1993). Thus, identifying Prospero transcriptional targets and apkc phosphorylation targets may provide further insight into the molecular mechanism of neural stem cell self-renewal in both Drosophila and mammals. Experimental Procedures Fly Stocks and Genetics All mutant chromosomes were balanced over CyO, actin::gfp or TM3, ftz-lacz, Ser, e. We used Oregon R as wild-type, and the following mutant chromosomes: mira L44 and mira AB78 (Fuerstenberg et al., 1998; Ikeshima- Kataoka et al., 1997) brat 11 and brat l(2)k06028 (Arama et al., 2000) lgl 334 (Lee et al., 2005) apkc k06403 (Rolls et al., 2003) worniu-gal4 (Lee et al., 2005) UAS-lgl3A (Betschinger et al., 2003; Lee et al., 2005) MARCM experiments were performed by using published methods (Lee and Luo, 2001) by crossing elav-gal4(c155), UASmCD8:GFP, hsp70-flp; FRT82B tubulin-gal80 to FRT82B prospero 17 and giving the larval progeny a 1 hr 37ºC heat shock at 24 hr ALH followed by clone analysis at 96 hr ALH. Antibody Staining and Neuroblast Culture Larvae were dissected in Schneider s medium (Sigma), fixed in 100 mm PIPES (ph 6.9), 1 mm EGTA, 0.3% Triton X-100, 1 mm MgSO 4 containing 4% formaldehyde for 25 min, and blocked for 1 hr in 13 PBS containing 1% BSA and 0.3% Triton X-100 (PBS-BT). After blocking, specimens were washed in PBS-BT for 1 hr and incubated with primary antibodies in PBS-BT overnight at 4ºC. Embryos were stained as described previously (Spana and Doe, 1995). We used rat anti-brat (1:100), guinea pig anti-dpn (1:1000; J. Skeath), guinea pig anti-miranda (1:400), rat anti-miranda (1:500), rat anti-pins (1:1000), mouse anti-elav (1:50), mouse anti-brdu (1:50; Sigma), mouse anti-prospero bioreactor supernatant (1:100), rabbit antibgal (1:1000), rat anti-elav (1:10; Developmental Study Hybridoma Bank), mouse anti-a-tubulin (1:2000; Sigma), rabbit anti-apkcz (1:1000; Santa Cruz Biotech.), rabbit anti-phosphohistoneh3 (1:1000; Upstate), rat anti-par6 (1:50), guinea pig anti-baz (1:400), rabbit anti-scribble (1:2500), and secondary antibodies from Molecular Probes (details are available upon request). Embryonic neuroblasts (4 5 hr) were isolated in vitro, cultured for 30 min, and antibody stained by using published methods (Grosskortenhaus et al., 2005). Yeast Two-Hybrid Experiments We used the Brat NHL domain as a bait and screened a library of plasmids encoding activation domain fusions to cdnas from 0 4 hr embryos (Dahanukar et al., 1999). Experiments were done as described previously (Edwards et al., 2003). BrdU Pulse-Chase Experiments Larvae were fed BrdU (1 mg/ml; Roche, Indianapolis, IN) in media for 4 hr, and then one pool was processed for BrdU staining (pulse experiments; Figure 1) and a second pool was grown without BrdU for 24 or 72 hr before fixation and BrdU staining (pulse-chase experiments; Figure 2). Larval brains were dissected, processed, and antibody stained as described above with the exception that larval brains were treated in 2N HCl for 30 min prior to primary antibody staining; BrdU detection was performed by following the manufacturer s protocol (details are available upon request). Neuroblast Counting and Brain Orientation A larval brain lobe consists of the medially localized central brain and the laterally localized optic lobe. Neuroblasts can be unambiguously identified by expression of Worniu, Dpn, and Miranda and the absence of the neuronal/glial differentiation markers Elav and Repo (Albertson and Doe, 2003; Lee et al., 2005). Central brain neuroblasts (the focus of this paper) can be distinguished from optic lobe neuroblasts due to their medial/superficial location in the brain, larger size, and dispersed pattern (optic lobe neuroblasts laterally positioned in the brain and spaced very close to each other, forming a ribbon that flanks and encircles the highly stereotypical epithelial shape optic lobe cells [Lee et al., 2005]). All images of neuroblasts shown were collected from central brain, and all brains were mounted with dorsal surface up and ventral surface down; the midline is to the left in all panels. Supplemental Data Supplemental Data including three figures are available at Acknowledgments We thank Allan Shearn, Tory Herman, and the Bloomington stock center for fly stocks and/or antibody reagents; Bruce Bowerman, Karsten Siller, Jason Boone, and Chiswili Chabu for comments on the manuscript; and Chiswili Chabu for helpful discussion. C.-Y.L. is supported by a Damon Runynon postdoctoral fellowship, S.E.S. was supported by a National Institutes of Health (NIH) Developmental Biology Training Grant, and B.D.W. and R.P.W. were supported by NIH grant GM R.P.W. and C.Q.D. are Investigators of the Howard Hughes Medical Institute.
9 Brat Inhibits Neuroblast Self-Renewal 449 Received: November 8, 2005 Revised: January 6, 2006 Accepted: January 26, 2006 Published online: March 22, 2006 References Albertson, R., and Doe, C.Q. (2003). Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat. Cell Biol. 5, Arama, E., Dickman, D., Kimchie, Z., Shearn, A., and Lev, Z. (2000). Mutations in the b-propeller domain of the Drosophila brain tumor (brat) protein induce neoplasm in the larval brain. Oncogene 19, Betschinger, J., and Knoblich, J.A. (2004). Dare to be different: asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr. Biol. 14, R674 R685. Betschinger, J., Mechtler, K., and Knoblich, J.A. (2003). The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature 422, Broadie, K., and Bate, M. (1993). Muscle development is independent of innervation during Drosophila embryogenesis. Development 119, Broadus, J., and Doe, C.Q. (1997). Extrinsic cues, intrinsic cues and microfilaments regulate asymmetric protein localization in Drosophila neuroblasts. Curr. Biol. 7, Broadus, J., Fuerstenberg, S., and Doe, C.Q. (1998). Staufen-dependent localization of prospero mrna contributes to neuroblast daughter-cell fate. Nature 391, Caussinus, E., and Gonzalez, C. (2005). Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat. Genet. 37, Dahanukar, A., Walker, J.A., and Wharton, R.P. (1999). Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol. Cell 4, Datta, S. (1995). Control of proliferation activation in quiescent neuroblasts of the Drosophila central nervous system. Development 121, Doe, C.Q., Chu-LaGraff, Q., Wright, D.M., and Scott, M.P. (1991). The prospero gene specifies cell fates in the Drosophila central nervous system. Cell 65, Edwards, T.A., Wilkinson, B.D., Wharton, R.P., and Aggarwal, A.K. (2003). Model of the brain tumor-pumilio translation repressor complex. Genes Dev. 17, Frank, D.J., Edgar, B.A., and Roth, M.B. (2002). The Drosophila melanogaster gene brain tumor negatively regulates cell growth and ribosomal RNA synthesis. Development 129, Fuerstenberg, S., Peng, C.Y., Alvarez-Ortiz, P., Hor, T., and Doe, C.Q. (1998). Identification of Miranda protein domains regulating asymmetric cortical localization, cargo binding, and cortical release. Mol. Cell. Neurosci. 12, Grosskortenhaus, R., Pearson, B.J., Marusich, A., and Doe, C.Q. (2005). Regulation of temporal identity transitions in Drosophila neuroblasts. Dev. Cell 8, Ikeshima-Kataoka, H., Skeath, J.B., Nabeshima, Y., Doe, C.Q., and Matsuzaki, F. (1997). Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature 390, Knoblich, J.A., Jan, L.Y., and Jan, Y.N. (1995). Asymmetric segregation of Numb and Prospero during cell division. Nature 377, Lechler, T., and Fuchs, E. (2005). Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 437, Lee, C.Y., Robinson, K., and Doe, C.Q. (2005). Lgl, Pins and apkc regulate neuroblast self-renewal versus differentiation. Nature 439, Lee, T., and Luo, L. (2001). Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24, Li, L., and Vaessin, H. (2000). Pan-neural Prospero terminates cell proliferation during Drosophila neurogenesis. Genes Dev. 14, Manabe, N., Hirai, S., Imai, F., Nakanishi, H., Takai, Y., and Ohno, S. (2002). Association of ASIP/mPAR-3 with adherens junctions of mouse neuroepithelial cells. Dev. Dyn. 225, Matsuzaki, F., Ohshiro, T., Ikeshima-Kataoka, H., and Izumi, H. (1998). miranda localizes staufen and prospero asymmetrically in mitotic neuroblasts and epithelial cells in early Drosophila embryogenesis. Development 125, Oliver, G., Sosa-Pineda, B., Geisendorf, S., Spana, E.P., Doe, C.Q., and Gruss, P. (1993). Prox 1, a prospero-related homeobox gene expressed during mouse development. Mech. Dev. 44, Rhyu, M.S., Jan, L.Y., and Jan, Y.N. (1994). Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 76, Rolls, M.M., Albertson, R., Shih, H.P., Lee, C.Y., and Doe, C.Q. (2003). Drosophila apkc regulates cell polarity and cell proliferation in neuroblasts and epithelia. J. Cell Biol. 163, Schuldt, A.J., Adams, J.H., Davidson, C.M., Micklem, D.R., Haseloff, J., St Johnston, D., and Brand, A.H. (1998). Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev. 12, Shen, C.P., Knoblich, J.A., Chan, Y.M., Jiang, M.M., Jan, L.Y., and Jan, Y.N. (1998). Miranda as a multidomain adapter linking apically localized Inscuteable and basally localized Staufen and Prospero during asymmetric cell division in Drosophila. Genes Dev. 12, Siegrist, S.E., and Doe, C.Q. (2006). Extrinsic cues orient the cell division axis in Drosophila embryonic neuroblasts. Development 133, Skeath, J.B., and Doe, C.Q. (1998). Sanpodo and Notch act in opposition to Numb to distinguish sibling neuron fates in the Drosophila CNS. Development 125, Sonoda, J., and Wharton, R.P. (2001). Drosophila Brain Tumor is a translational repressor. Genes Dev. 15, Spana, E.P., and Doe, C.Q. (1995). The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development 121, Vaessin, H., Grell, E., Wolff, E., Bier, E., Jan, L.Y., and Jan, Y.N. (1991). prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell 67, Yamashita, Y.M., Jones, D.L., and Fuller, M.T. (2003). Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 301,
The Tumor Suppressors Brat and Numb Regulate Transit-Amplifying Neuroblast Lineages in Drosophila
 Article The Tumor Suppressors Brat and Numb Regulate Transit-Amplifying Neuroblast Lineages in Drosophila Sarah K. Bowman, 1 Vivien Rolland, 1 Joerg Betschinger, 1,3 Kaolin A. Kinsey, 2 Gregory Emery,
Article The Tumor Suppressors Brat and Numb Regulate Transit-Amplifying Neuroblast Lineages in Drosophila Sarah K. Bowman, 1 Vivien Rolland, 1 Joerg Betschinger, 1,3 Kaolin A. Kinsey, 2 Gregory Emery,
glial cells missing and gcm2 Cell-autonomously Regulate Both Glial and Neuronal
 glial cells missing and gcm2 Cell-autonomously Regulate Both Glial and Neuronal Development in the Visual System of Drosophila Carole Chotard, Wendy Leung and Iris Salecker Supplemental Data Supplemental
glial cells missing and gcm2 Cell-autonomously Regulate Both Glial and Neuronal Development in the Visual System of Drosophila Carole Chotard, Wendy Leung and Iris Salecker Supplemental Data Supplemental
Cell Birth and Death. Chapter Three
 Cell Birth and Death Chapter Three Neurogenesis All neurons and glial cells begin in the neural tube Differentiated into neurons rather than ectoderm based on factors we have already discussed If these
Cell Birth and Death Chapter Three Neurogenesis All neurons and glial cells begin in the neural tube Differentiated into neurons rather than ectoderm based on factors we have already discussed If these
Supplemental Data SUPPLEMENTAL EXPERIMENTAL PROCEDURES
 Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cédric Maurange, Louise Cheng and Alex P. Gould Supplemental Data SUPPLEMENTAL EXPERIMENTAL PROCEDURES
Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cédric Maurange, Louise Cheng and Alex P. Gould Supplemental Data SUPPLEMENTAL EXPERIMENTAL PROCEDURES
Postembryonic Development of Amplifying Neuroblast Lineages in the Drosophila Brain: Proliferation, Differentiation and Projection Patterns.
 Postembryonic Development of Amplifying Neuroblast Lineages in the Drosophila Brain: Proliferation, Differentiation and Projection Patterns. Inauguraldissertation zur Erlangung der Würde eines Doktors
Postembryonic Development of Amplifying Neuroblast Lineages in the Drosophila Brain: Proliferation, Differentiation and Projection Patterns. Inauguraldissertation zur Erlangung der Würde eines Doktors
NIH Public Access Author Manuscript Dev Biol. Author manuscript; available in PMC 2007 April 30.
 NIH Public Access Author Manuscript Published in final edited form as: Dev Biol. 2007 January 1; 301(1): 287 297. Drosophila brain tumor metastases express both neuronal and glial cell type markers Michelle
NIH Public Access Author Manuscript Published in final edited form as: Dev Biol. 2007 January 1; 301(1): 287 297. Drosophila brain tumor metastases express both neuronal and glial cell type markers Michelle
Review Article Neural Stem Cells in Drosophila: Molecular Genetic Mechanisms Underlying Normal Neural Proliferation and Abnormal Brain Tumor Formation
 Stem Cells International Volume 2012, Article ID 486169, 10 pages doi:10.1155/2012/486169 Review Article Neural Stem Cells in Drosophila: Molecular Genetic Mechanisms Underlying Normal Neural Proliferation
Stem Cells International Volume 2012, Article ID 486169, 10 pages doi:10.1155/2012/486169 Review Article Neural Stem Cells in Drosophila: Molecular Genetic Mechanisms Underlying Normal Neural Proliferation
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
Bidirectional Notch signaling regulates Drosophila intestinal stem cell multipotency
 RESEARCH RESEARCH ARTICLE SUMMARY STEM CELL REGULATION Bidirectional Notch signaling regulates Drosophila intestinal stem cell multipotency Zheng Guo and Benjamin Ohlstein* INTRODUCTION: In the Drosophila
RESEARCH RESEARCH ARTICLE SUMMARY STEM CELL REGULATION Bidirectional Notch signaling regulates Drosophila intestinal stem cell multipotency Zheng Guo and Benjamin Ohlstein* INTRODUCTION: In the Drosophila
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Expression of escargot (esg) and genetic approach for achieving IPC-specific knockdown. (a) esg MH766 -Gal4 UAS-cd8GFP (green) and esg-lacz B7-2-22 (red) show similar expression
Supplementary Figure 1 Expression of escargot (esg) and genetic approach for achieving IPC-specific knockdown. (a) esg MH766 -Gal4 UAS-cd8GFP (green) and esg-lacz B7-2-22 (red) show similar expression
Supplementary Figure 1
 Supplementary Figure 1 Kif1a RNAi effect on basal progenitor differentiation Related to Figure 2. Representative confocal images of the VZ and SVZ of rat cortices transfected at E16 with scrambled or Kif1a
Supplementary Figure 1 Kif1a RNAi effect on basal progenitor differentiation Related to Figure 2. Representative confocal images of the VZ and SVZ of rat cortices transfected at E16 with scrambled or Kif1a
Problem Set 5 KEY
 2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1. Ras V12 expression in the entire eye-antennal disc does not cause invasive tumours. a, Eye-antennal discs expressing Ras V12 in all cells (marked with GFP, green) overgrow moderately
Supplementary Figure 1. Ras V12 expression in the entire eye-antennal disc does not cause invasive tumours. a, Eye-antennal discs expressing Ras V12 in all cells (marked with GFP, green) overgrow moderately
OSVZ progenitors of human and ferret neocortex are epithelial-like and
 OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling Simone A Fietz, Iva Kelava, Johannes Vogt, Michaela Wilsch-Bräuninger, Denise Stenzel, Jennifer L Fish,
OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling Simone A Fietz, Iva Kelava, Johannes Vogt, Michaela Wilsch-Bräuninger, Denise Stenzel, Jennifer L Fish,
Cell Type Nervous System I. Developmental Readout. Foundations. Stem cells. Organ formation. Human issues.
 7.72 10.11.06 Cell Type Nervous System I Human issues Organ formation Stem cells Developmental Readout Axes Cell type Axon guidance 3D structure Analysis Model + + organisms Foundations Principles 1 What
7.72 10.11.06 Cell Type Nervous System I Human issues Organ formation Stem cells Developmental Readout Axes Cell type Axon guidance 3D structure Analysis Model + + organisms Foundations Principles 1 What
Extrinsic cues orient the cell division axis in Drosophila embryonic neuroblasts
 RESEARCH ARTICLE 529 Development 133, 529-536 doi:10.1242/dev.02211 Extrinsic cues orient the cell division axis in Drosophila embryonic neuroblasts Sarah E. Siegrist and Chris Q. Doe* Cell polarity must
RESEARCH ARTICLE 529 Development 133, 529-536 doi:10.1242/dev.02211 Extrinsic cues orient the cell division axis in Drosophila embryonic neuroblasts Sarah E. Siegrist and Chris Q. Doe* Cell polarity must
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2419 Figure S1 NiGFP localization in Dl mutant dividing SOPs. a-c) time-lapse analysis of NiGFP (green) in Dl mutant SOPs (H2B-RFP, red; clones were identified by the loss of nlsgfp) showing
DOI: 10.1038/ncb2419 Figure S1 NiGFP localization in Dl mutant dividing SOPs. a-c) time-lapse analysis of NiGFP (green) in Dl mutant SOPs (H2B-RFP, red; clones were identified by the loss of nlsgfp) showing
A Genetic Program for Embryonic Development
 Concept 18.4: A program of differential gene expression leads to the different cell types in a multicellular organism During embryonic development, a fertilized egg gives rise to many different cell types
Concept 18.4: A program of differential gene expression leads to the different cell types in a multicellular organism During embryonic development, a fertilized egg gives rise to many different cell types
Regulators of Cell Cycle Progression
 Regulators of Cell Cycle Progression Studies of Cdk s and cyclins in genetically modified mice reveal a high level of plasticity, allowing different cyclins and Cdk s to compensate for the loss of one
Regulators of Cell Cycle Progression Studies of Cdk s and cyclins in genetically modified mice reveal a high level of plasticity, allowing different cyclins and Cdk s to compensate for the loss of one
Viktorin, G. and Riebli, N. and Popkova, A. and Giangrande, A. and Reichert, H.
 Institutional Repository of the University of Basel University Library Schoenbeinstrasse 18-20 CH-4056 Basel, Switzerland http://edoc.unibas.ch/ Year: 2011 Multipotent neural stem cells generate glial
Institutional Repository of the University of Basel University Library Schoenbeinstrasse 18-20 CH-4056 Basel, Switzerland http://edoc.unibas.ch/ Year: 2011 Multipotent neural stem cells generate glial
Supplemental Information. Otic Mesenchyme Cells Regulate. Spiral Ganglion Axon Fasciculation. through a Pou3f4/EphA4 Signaling Pathway
 Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Programmed Cell Death (apoptosis)
 Programmed Cell Death (apoptosis) Stereotypic death process includes: membrane blebbing nuclear fragmentation chromatin condensation and DNA framentation loss of mitochondrial integrity and release of
Programmed Cell Death (apoptosis) Stereotypic death process includes: membrane blebbing nuclear fragmentation chromatin condensation and DNA framentation loss of mitochondrial integrity and release of
Supplementary Figures
 J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
Supplementary Figure S1: TIPF reporter validation in the wing disc.
 Supplementary Figure S1: TIPF reporter validation in the wing disc. a,b, Test of put RNAi. a, In wildtype discs the Dpp target gene Sal (red) is expressed in a broad stripe in the centre of the ventral
Supplementary Figure S1: TIPF reporter validation in the wing disc. a,b, Test of put RNAi. a, In wildtype discs the Dpp target gene Sal (red) is expressed in a broad stripe in the centre of the ventral
Regulation of Self-renewal and Differentiation in the Drosophila Nervous System
 Regulation of Self-renewal and Differentiation in the Drosophila Nervous System T.D. SOUTHALL, B. EGGER, K.S. GOLD, AND A.H. BRAND The Gurdon Institute and Department of hysiology, Development, and Neuroscience,
Regulation of Self-renewal and Differentiation in the Drosophila Nervous System T.D. SOUTHALL, B. EGGER, K.S. GOLD, AND A.H. BRAND The Gurdon Institute and Department of hysiology, Development, and Neuroscience,
Neuroepithelial Cells and Neural Differentiation
 Neuroepithelial Cells and Neural Differentiation Neurulation The cells of the neural tube are NEUROEPITHELIAL CELLS Neural crest cells migrate out of neural tube Neuroepithelial cells are embryonic stem
Neuroepithelial Cells and Neural Differentiation Neurulation The cells of the neural tube are NEUROEPITHELIAL CELLS Neural crest cells migrate out of neural tube Neuroepithelial cells are embryonic stem
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3443 In the format provided by the authors and unedited. Supplementary Figure 1 TC and SC behaviour during ISV sprouting. (a) Predicted outcome of TC division and competitive Dll4-Notch-mediated
DOI: 10.1038/ncb3443 In the format provided by the authors and unedited. Supplementary Figure 1 TC and SC behaviour during ISV sprouting. (a) Predicted outcome of TC division and competitive Dll4-Notch-mediated
BIOL2005 WORKSHEET 2008
 BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
Control of proliferation activation in quiescent neuroblasts of the Drosophila central nervous system
 Development 121, 1173-1182 (1995) Printed in Great Britain The Company of Biologists Limited 1995 1173 Control of proliferation activation in quiescent neuroblasts of the Drosophila central nervous system
Development 121, 1173-1182 (1995) Printed in Great Britain The Company of Biologists Limited 1995 1173 Control of proliferation activation in quiescent neuroblasts of the Drosophila central nervous system
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature07173 SUPPLEMENTARY INFORMATION Supplementary Figure Legends: Supplementary Figure 1: Model of SSC and CPC divisions a, Somatic stem cells (SSC) reside adjacent to the hub (red), self-renew
doi: 10.1038/nature07173 SUPPLEMENTARY INFORMATION Supplementary Figure Legends: Supplementary Figure 1: Model of SSC and CPC divisions a, Somatic stem cells (SSC) reside adjacent to the hub (red), self-renew
Axis Formation and Mesoderm Induction
 Developmental Biology Biology 4361 Axis Formation and Mesoderm Induction October 27, 2005 Amphibian anteroposterior specification polarized eggs animal/vegetal pigment yolk v. clear cytoplasm mitochondrial
Developmental Biology Biology 4361 Axis Formation and Mesoderm Induction October 27, 2005 Amphibian anteroposterior specification polarized eggs animal/vegetal pigment yolk v. clear cytoplasm mitochondrial
Cancer Stem Cells & Glioblastoma
 Cancer Stem Cells & Glioblastoma JP Hugnot «Brain plasticity, Neural stem cells and Glial tumors» INSERM U1051-UM2 Institut des Neurosciences de Montpellier Montpellier 1-Stem cells and Brain Stem Cells
Cancer Stem Cells & Glioblastoma JP Hugnot «Brain plasticity, Neural stem cells and Glial tumors» INSERM U1051-UM2 Institut des Neurosciences de Montpellier Montpellier 1-Stem cells and Brain Stem Cells
UNC-Duke Biology Course for Residents Fall Cell Cycle Effects of Radiation
 UNC-Duke Biology Course for Residents Fall 2018 1 Cell Cycle: Sequence of changes in a cell starting with the moment the cell is created by cell division, continuing through the doubling of the DNA and
UNC-Duke Biology Course for Residents Fall 2018 1 Cell Cycle: Sequence of changes in a cell starting with the moment the cell is created by cell division, continuing through the doubling of the DNA and
Cell Cycle, Mitosis, and Microtubules. LS1A Final Exam Review Friday 1/12/07. Processes occurring during cell cycle
 Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
Cell Cycle, Mitosis, and Microtubules LS1A Final Exam Review Friday 1/12/07 Processes occurring during cell cycle Replicate chromosomes Segregate chromosomes Cell divides Cell grows Cell Growth 1 The standard
Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates
 Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates L. Cheng, A. Arata, R. Mizuguchi, Y. Qian, A. Karunaratne, P.A. Gray, S. Arata, S. Shirasawa, M. Bouchard,
Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates L. Cheng, A. Arata, R. Mizuguchi, Y. Qian, A. Karunaratne, P.A. Gray, S. Arata, S. Shirasawa, M. Bouchard,
klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach
![klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach](/thumbs/91/104639484.jpg) DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
marker. DAPI labels nuclei. Flies were 20 days old. Scale bar is 5 µm. Ctrl is
 Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Supplementary Figure 1. Electroporation of a stable form of β-catenin causes masses protruding into the IV ventricle. HH12 chicken embryos were
 Supplementary Figure 1. Electroporation of a stable form of β-catenin causes masses protruding into the IV ventricle. HH12 chicken embryos were electroporated with β- Catenin S33Y in PiggyBac expression
Supplementary Figure 1. Electroporation of a stable form of β-catenin causes masses protruding into the IV ventricle. HH12 chicken embryos were electroporated with β- Catenin S33Y in PiggyBac expression
The Biology and Genetics of Cells and Organisms The Biology of Cancer
 The Biology and Genetics of Cells and Organisms The Biology of Cancer Mendel and Genetics How many distinct genes are present in the genomes of mammals? - 21,000 for human. - Genetic information is carried
The Biology and Genetics of Cells and Organisms The Biology of Cancer Mendel and Genetics How many distinct genes are present in the genomes of mammals? - 21,000 for human. - Genetic information is carried
The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein
 THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
EMBO REPORT SUPPLEMENTARY SECTION. Quantitation of mitotic cells after perturbation of Notch signalling.
 EMBO REPORT SUPPLEMENTARY SECTION Quantitation of mitotic cells after perturbation of Notch signalling. Notch activation suppresses the cell cycle indistinguishably both within and outside the neural plate
EMBO REPORT SUPPLEMENTARY SECTION Quantitation of mitotic cells after perturbation of Notch signalling. Notch activation suppresses the cell cycle indistinguishably both within and outside the neural plate
Early cell death (FGF) B No RunX transcription factor produced Yes No differentiation
 Solution Key - Practice Questions Question 1 a) A recent publication has shown that the fat stem cells (FSC) can act as bone stem cells to repair cavities in the skull, when transplanted into immuno-compromised
Solution Key - Practice Questions Question 1 a) A recent publication has shown that the fat stem cells (FSC) can act as bone stem cells to repair cavities in the skull, when transplanted into immuno-compromised
Alpha thalassemia mental retardation X-linked. Acquired alpha-thalassemia myelodysplastic syndrome
 Alpha thalassemia mental retardation X-linked Acquired alpha-thalassemia myelodysplastic syndrome (Alpha thalassemia mental retardation X-linked) Acquired alpha-thalassemia myelodysplastic syndrome Schematic
Alpha thalassemia mental retardation X-linked Acquired alpha-thalassemia myelodysplastic syndrome (Alpha thalassemia mental retardation X-linked) Acquired alpha-thalassemia myelodysplastic syndrome Schematic
A Cxcl12-Cxcr4 Chemokine Signaling Pathway Defines
 Supplemental Data A Cxcl12-Cxcr4 Chemokine Signaling Pathway Defines the Initial Trajectory of Mammalian Motor Axons Ivo Lieberam, Dritan Agalliu, Takashi Nagasawa, Johan Ericson, and Thomas M. Jessell
Supplemental Data A Cxcl12-Cxcr4 Chemokine Signaling Pathway Defines the Initial Trajectory of Mammalian Motor Axons Ivo Lieberam, Dritan Agalliu, Takashi Nagasawa, Johan Ericson, and Thomas M. Jessell
BCHM3972 Human Molecular Cell Biology (Advanced) 2013 Course University of Sydney
 BCHM3972 Human Molecular Cell Biology (Advanced) 2013 Course University of Sydney Page 2: Immune Mechanisms & Molecular Biology of Host Defence (Prof Campbell) Page 45: Infection and Implications for Cell
BCHM3972 Human Molecular Cell Biology (Advanced) 2013 Course University of Sydney Page 2: Immune Mechanisms & Molecular Biology of Host Defence (Prof Campbell) Page 45: Infection and Implications for Cell
Polarity and Segmentation. Chapter Two
 Polarity and Segmentation Chapter Two Polarization Entire body plan is polarized One end is different than the other Head vs. Tail Anterior vs. Posterior Front vs. Back Ventral vs. Dorsal Majority of neural
Polarity and Segmentation Chapter Two Polarization Entire body plan is polarized One end is different than the other Head vs. Tail Anterior vs. Posterior Front vs. Back Ventral vs. Dorsal Majority of neural
Analysis of neural stem cell self-renewal and differentiation by transgenic RNAi in Drosophila
 Institutional Repository of the University of Basel University Library Schoenbeinstrasse 18-20 CH-4056 Basel, Switzerland http://edoc.unibas.ch/ Year: 2013 Analysis of neural stem cell self-renewal and
Institutional Repository of the University of Basel University Library Schoenbeinstrasse 18-20 CH-4056 Basel, Switzerland http://edoc.unibas.ch/ Year: 2013 Analysis of neural stem cell self-renewal and
7.012 Quiz 3 Answers
 MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
MIT Biology Department 7.012: Introductory Biology - Fall 2004 Instructors: Professor Eric Lander, Professor Robert A. Weinberg, Dr. Claudette Gardel Friday 11/12/04 7.012 Quiz 3 Answers A > 85 B 72-84
effects on organ development. a-f, Eye and wing discs with clones of ε j2b10 show no
 Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
a 0,8 Figure S1 8 h 12 h y = 0,036x + 0,2115 y = 0,0366x + 0,206 Labeling index Labeling index ctrl shrna Time (h) Time (h) ctrl shrna S G2 M G1
 (GFP+ BrdU+)/GFP+ Labeling index Labeling index Figure S a, b, y =,x +, y =,x +,,,,,,,, Time (h) - - Time (h) c d S G M G h M G S G M G S G h Time of BrdU injection after electroporation (h) M G S G M
(GFP+ BrdU+)/GFP+ Labeling index Labeling index Figure S a, b, y =,x +, y =,x +,,,,,,,, Time (h) - - Time (h) c d S G M G h M G S G M G S G h Time of BrdU injection after electroporation (h) M G S G M
Nature Neuroscience: doi: /nn Supplementary Figure 1. MADM labeling of thalamic clones.
 Supplementary Figure 1 MADM labeling of thalamic clones. (a) Confocal images of an E12 Nestin-CreERT2;Ai9-tdTomato brain treated with TM at E10 and stained for BLBP (green), a radial glial progenitor-specific
Supplementary Figure 1 MADM labeling of thalamic clones. (a) Confocal images of an E12 Nestin-CreERT2;Ai9-tdTomato brain treated with TM at E10 and stained for BLBP (green), a radial glial progenitor-specific
SUPPLEMENTARY FIG. S2. Representative counting fields used in quantification of the in vitro neural differentiation of pattern of dnscs.
 Supplementary Data SUPPLEMENTARY FIG. S1. Representative counting fields used in quantification of the in vitro neural differentiation of pattern of anpcs. A panel of lineage-specific markers were used
Supplementary Data SUPPLEMENTARY FIG. S1. Representative counting fields used in quantification of the in vitro neural differentiation of pattern of anpcs. A panel of lineage-specific markers were used
Supplementary Fig. 1 Blocking shh function at the protein level confirms its role as a guidance cue for postcommissural axons.
 Supplementary Fig. 1 Blocking shh function at the protein level confirms its role as a guidance cue for postcommissural axons. As an alternative method to demonstrate the role of shh as a guidance cue
Supplementary Fig. 1 Blocking shh function at the protein level confirms its role as a guidance cue for postcommissural axons. As an alternative method to demonstrate the role of shh as a guidance cue
Control of neural stem cell self-renewal and differentiation in Drosophila
 Institutional Repository of the University of Basel University Library Schoenbeinstrasse 18-20 CH-4056 Basel, Switzerland http://edoc.unibas.ch/ Year: 2014 Control of neural stem cell self-renewal and
Institutional Repository of the University of Basel University Library Schoenbeinstrasse 18-20 CH-4056 Basel, Switzerland http://edoc.unibas.ch/ Year: 2014 Control of neural stem cell self-renewal and
Mitosis and the Cell Cycle
 Mitosis and the Cell Cycle Chapter 12 The Cell Cycle: Cell Growth & Cell Division Where it all began You started as a cell smaller than a period at the end of a sentence Getting from there to here Cell
Mitosis and the Cell Cycle Chapter 12 The Cell Cycle: Cell Growth & Cell Division Where it all began You started as a cell smaller than a period at the end of a sentence Getting from there to here Cell
Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment
 Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development
Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons
 Supplemental Material mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons Antoine de Chevigny, Nathalie Coré, Philipp Follert, Marion Gaudin, Pascal
Supplemental Material mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons Antoine de Chevigny, Nathalie Coré, Philipp Follert, Marion Gaudin, Pascal
MCB140: Second Midterm Spring 2010
 MCB140: Second Midterm Spring 2010 Before you start, print your name and student identification number (S.I.D) at the top of each page. There are 11 pages including this page. You will have 150 minutes
MCB140: Second Midterm Spring 2010 Before you start, print your name and student identification number (S.I.D) at the top of each page. There are 11 pages including this page. You will have 150 minutes
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
JCB. Myosin and the PAR proteins polarize microfilament-dependent forces that shape and position mitotic spindles in Caenorhabditis elegans
 JCB Published Online: 14 April, 2003 Supp Info: http://doi.org/10.1083/jcb.200210171 Downloaded from jcb.rupress.org on January 15, 2019 Report Myosin and the PAR proteins polarize microfilament-dependent
JCB Published Online: 14 April, 2003 Supp Info: http://doi.org/10.1083/jcb.200210171 Downloaded from jcb.rupress.org on January 15, 2019 Report Myosin and the PAR proteins polarize microfilament-dependent
Supplemental Information. Induction of Expansion and Folding. in Human Cerebral Organoids
 Cell Stem Cell, Volume 20 Supplemental Information Induction of Expansion and Folding in Human Cerebral Organoids Yun Li, Julien Muffat, Attya Omer, Irene Bosch, Madeline A. Lancaster, Mriganka Sur, Lee
Cell Stem Cell, Volume 20 Supplemental Information Induction of Expansion and Folding in Human Cerebral Organoids Yun Li, Julien Muffat, Attya Omer, Irene Bosch, Madeline A. Lancaster, Mriganka Sur, Lee
Chapter 2. Mitosis and Meiosis
 Chapter 2. Mitosis and Meiosis Chromosome Theory of Heredity What structures within cells correspond to genes? The development of genetics took a major step forward by accepting the notion that the genes
Chapter 2. Mitosis and Meiosis Chromosome Theory of Heredity What structures within cells correspond to genes? The development of genetics took a major step forward by accepting the notion that the genes
Supplementary Figure S1: Tanycytes are restricted to the central/posterior hypothalamus
 Supplementary Figure S1: Tanycytes are restricted to the central/posterior hypothalamus a: Expression of Vimentin, GFAP, Sox2 and Nestin in anterior, central and posterior hypothalamus. In the anterior
Supplementary Figure S1: Tanycytes are restricted to the central/posterior hypothalamus a: Expression of Vimentin, GFAP, Sox2 and Nestin in anterior, central and posterior hypothalamus. In the anterior
The subcortical maternal complex controls symmetric division of mouse zygotes by
 The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics Xing-Jiang Yu 1,2, Zhaohong Yi 1, Zheng Gao 1,2, Dan-dan Qin 1,2, Yanhua Zhai 1, Xue Chen 1,
The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics Xing-Jiang Yu 1,2, Zhaohong Yi 1, Zheng Gao 1,2, Dan-dan Qin 1,2, Yanhua Zhai 1, Xue Chen 1,
Nature Medicine doi: /nm.2860
 Supplemental Figure Legends Supplemental Figure 1: Hypomorphic expression of IFT88 results in olfactory signaling proteins no longer localizing to the ciliary layer. (a) ACIII localizes to the cilia and
Supplemental Figure Legends Supplemental Figure 1: Hypomorphic expression of IFT88 results in olfactory signaling proteins no longer localizing to the ciliary layer. (a) ACIII localizes to the cilia and
Biology Developmental Biology Spring Quarter Midterm 1 Version A
 Biology 411 - Developmental Biology Spring Quarter 2013 Midterm 1 Version A 75 Total Points Open Book Choose 15 out the 20 questions to answer (5 pts each). Only the first 15 questions that are answered
Biology 411 - Developmental Biology Spring Quarter 2013 Midterm 1 Version A 75 Total Points Open Book Choose 15 out the 20 questions to answer (5 pts each). Only the first 15 questions that are answered
Chapter 5 A Dose Dependent Screen for Modifiers of Kek5
 Chapter 5 A Dose Dependent Screen for Modifiers of Kek5 "#$ ABSTRACT Modifier screens in Drosophila have proven to be a powerful tool for uncovering gene interaction and elucidating molecular pathways.
Chapter 5 A Dose Dependent Screen for Modifiers of Kek5 "#$ ABSTRACT Modifier screens in Drosophila have proven to be a powerful tool for uncovering gene interaction and elucidating molecular pathways.
Transgenic Expression of the Helicobacter pylori Virulence Factor CagA Promotes Apoptosis or Tumorigenesis through JNK Activation in Drosophila
 Transgenic Expression of the Helicobacter pylori Virulence Factor CagA Promotes Apoptosis or Tumorigenesis through JNK Activation in Drosophila Anica M. Wandler, Karen Guillemin* Institute of Molecular
Transgenic Expression of the Helicobacter pylori Virulence Factor CagA Promotes Apoptosis or Tumorigenesis through JNK Activation in Drosophila Anica M. Wandler, Karen Guillemin* Institute of Molecular
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
Chapter 12. living /non-living? growth repair renew. Reproduction. Reproduction. living /non-living. fertilized egg (zygote) next chapter
 Chapter 12 How cells divide Reproduction living /non-living? growth repair renew based on cell division first mitosis - distributes identical sets of chromosomes cell cycle (life) Cell Division in Bacteria
Chapter 12 How cells divide Reproduction living /non-living? growth repair renew based on cell division first mitosis - distributes identical sets of chromosomes cell cycle (life) Cell Division in Bacteria
ErbB4 migrazione II parte
 ErbB4 migrazione II parte Control SVZ cells prefer to migrate on the NRG1 type III substrate the substrate preference of the neuroblasts migrating out of the SVZ explant was evaluated SVZ cells had a strong
ErbB4 migrazione II parte Control SVZ cells prefer to migrate on the NRG1 type III substrate the substrate preference of the neuroblasts migrating out of the SVZ explant was evaluated SVZ cells had a strong
WDR62 is associated with the spindle pole and mutated in human microcephaly
 WDR62 is associated with the spindle pole and mutated in human microcephaly Adeline K. Nicholas, Maryam Khurshid, Julie Désir, Ofélia P. Carvalho, James J. Cox, Gemma Thornton, Rizwana Kausar, Muhammad
WDR62 is associated with the spindle pole and mutated in human microcephaly Adeline K. Nicholas, Maryam Khurshid, Julie Désir, Ofélia P. Carvalho, James J. Cox, Gemma Thornton, Rizwana Kausar, Muhammad
Supporting Information
 Supporting Information Fig. S1. Overexpression of Rpr causes progenitor cell death. (A) TUNEL assay of control intestines. No progenitor cell death could be observed, except that some ECs are undergoing
Supporting Information Fig. S1. Overexpression of Rpr causes progenitor cell death. (A) TUNEL assay of control intestines. No progenitor cell death could be observed, except that some ECs are undergoing
Role of Notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster
 RESEARCH ARTICLE 53 Development 137, 53-61 (2010) doi:10.1242/dev.041749 Role of Notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster James W. Truman*,, Wanda
RESEARCH ARTICLE 53 Development 137, 53-61 (2010) doi:10.1242/dev.041749 Role of Notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster James W. Truman*,, Wanda
Neurodevelopment II Structure Formation. Reading: BCP Chapter 23
 Neurodevelopment II Structure Formation Reading: BCP Chapter 23 Phases of Development Ovum + Sperm = Zygote Cell division (multiplication) Neurogenesis Induction of the neural plate Neural proliferation
Neurodevelopment II Structure Formation Reading: BCP Chapter 23 Phases of Development Ovum + Sperm = Zygote Cell division (multiplication) Neurogenesis Induction of the neural plate Neural proliferation
Control of neuronal cell fate and number by integration of distinct daughter cell proliferation modes with temporal progression
 Control of neuronal cell fate and number by integration of distinct daughter cell proliferation modes with temporal progression Carina Ulvklo, Ryan MacDonald, Caroline Bivik, Magnus Baumgardt, Daniel Karlsson
Control of neuronal cell fate and number by integration of distinct daughter cell proliferation modes with temporal progression Carina Ulvklo, Ryan MacDonald, Caroline Bivik, Magnus Baumgardt, Daniel Karlsson
Plasticity of Cerebral Cortex in Development
 Plasticity of Cerebral Cortex in Development Jessica R. Newton and Mriganka Sur Department of Brain & Cognitive Sciences Picower Center for Learning & Memory Massachusetts Institute of Technology Cambridge,
Plasticity of Cerebral Cortex in Development Jessica R. Newton and Mriganka Sur Department of Brain & Cognitive Sciences Picower Center for Learning & Memory Massachusetts Institute of Technology Cambridge,
Title: Chapter 5 Recorded Lecture. Speaker: Amit Dhingra Created by: (remove if same as speaker) online.wsu.edu
 Title: Chapter 5 Recorded Lecture Speaker: Title: What Anthony is the title Berger/Angela of this lecture? Williams Speaker: Amit Dhingra Created by: (remove if same as speaker) online.wsu.edu Chapter
Title: Chapter 5 Recorded Lecture Speaker: Title: What Anthony is the title Berger/Angela of this lecture? Williams Speaker: Amit Dhingra Created by: (remove if same as speaker) online.wsu.edu Chapter
A. Incorrect! All the cells have the same set of genes. (D)Because different types of cells have different types of transcriptional factors.
 Genetics - Problem Drill 21: Cytogenetics and Chromosomal Mutation No. 1 of 10 1. Why do some cells express one set of genes while other cells express a different set of genes during development? (A) Because
Genetics - Problem Drill 21: Cytogenetics and Chromosomal Mutation No. 1 of 10 1. Why do some cells express one set of genes while other cells express a different set of genes during development? (A) Because
Nature Genetics: doi: /ng Supplementary Figure 1. Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms.
 Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Chapter 11. How Genes Are Controlled. Lectures by Edward J. Zalisko
 Chapter 11 How Genes Are Controlled PowerPoint Lectures for Campbell Essential Biology, Fifth Edition, and Campbell Essential Biology with Physiology, Fourth Edition Eric J. Simon, Jean L. Dickey, and
Chapter 11 How Genes Are Controlled PowerPoint Lectures for Campbell Essential Biology, Fifth Edition, and Campbell Essential Biology with Physiology, Fourth Edition Eric J. Simon, Jean L. Dickey, and
Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the
 Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the location of the transmembrane (TM), FRM binding (FB)
Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the location of the transmembrane (TM), FRM binding (FB)
Control of neuronal cell fate and number by integration of distinct daughter cell proliferation modes with temporal progression
 678 AND STEM CELLS Development 139, 678-689 (2012) doi:10.1242/dev.074500 2012. Published by The Company of Biologists Ltd Control of neuronal cell fate and number by integration of distinct daughter cell
678 AND STEM CELLS Development 139, 678-689 (2012) doi:10.1242/dev.074500 2012. Published by The Company of Biologists Ltd Control of neuronal cell fate and number by integration of distinct daughter cell
Supplemental Information. Myocardial Polyploidization Creates a Barrier. to Heart Regeneration in Zebrafish
 Developmental Cell, Volume 44 Supplemental Information Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish Juan Manuel González-Rosa, Michka Sharpe, Dorothy Field, Mark H.
Developmental Cell, Volume 44 Supplemental Information Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish Juan Manuel González-Rosa, Michka Sharpe, Dorothy Field, Mark H.
Developmental Biology
 Developmental Biology 362 (2012) 141 153 Contents lists available at SciVerse ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology Dual function of suppressor of
Developmental Biology 362 (2012) 141 153 Contents lists available at SciVerse ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology Dual function of suppressor of
Nature Neuroscience: doi: /nn Supplementary Figure 1. Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites.
 Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
Supplementary Figure 1 Neuron class-specific arrangements of Khc::nod::lacZ label in dendrites. Staining with fluorescence antibodies to detect GFP (Green), β-galactosidase (magenta/white). (a, b) Class
Emx2 patterns the neocortex by regulating FGF positional signaling
 Emx2 patterns the neocortex by regulating FGF positional signaling Tomomi Fukuchi-Shimogori and Elizabeth A Grove Presented by Sally Kwok Background Cerebral cortex has anatomically and functionally distinct
Emx2 patterns the neocortex by regulating FGF positional signaling Tomomi Fukuchi-Shimogori and Elizabeth A Grove Presented by Sally Kwok Background Cerebral cortex has anatomically and functionally distinct
Zhu et al, page 1. Supplementary Figures
 Zhu et al, page 1 Supplementary Figures Supplementary Figure 1: Visual behavior and avoidance behavioral response in EPM trials. (a) Measures of visual behavior that performed the light avoidance behavior
Zhu et al, page 1 Supplementary Figures Supplementary Figure 1: Visual behavior and avoidance behavioral response in EPM trials. (a) Measures of visual behavior that performed the light avoidance behavior
AP VP DLP H&E. p-akt DLP
 A B AP VP DLP H&E AP AP VP DLP p-akt wild-type prostate PTEN-null prostate Supplementary Fig. 1. Targeted deletion of PTEN in prostate epithelium resulted in HG-PIN in all three lobes. (A) The anatomy
A B AP VP DLP H&E AP AP VP DLP p-akt wild-type prostate PTEN-null prostate Supplementary Fig. 1. Targeted deletion of PTEN in prostate epithelium resulted in HG-PIN in all three lobes. (A) The anatomy
Developmental Biology
 Developmental iology 356 (2011) 553 565 ontents lists available at ScienceDirect Developmental iology journal homepage: www.elsevier.com/developmentalbiology Multipotent neural stem cells generate glial
Developmental iology 356 (2011) 553 565 ontents lists available at ScienceDirect Developmental iology journal homepage: www.elsevier.com/developmentalbiology Multipotent neural stem cells generate glial
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
Impact of Sox9 Dosage and Hes1-mediated Notch Signaling in Controlling the Plasticity of Adult Pancreatic Duct Cells in Mice
 Impact of Sox9 Dosage and Hes1-mediated Notch Signaling in Controlling the Plasticity of Adult Pancreatic Duct Cells in Mice Shinichi Hosokawa 1,3,Kenichiro Furuyama 1,3, Masashi Horiguchi 1,3,Yoshiki
Impact of Sox9 Dosage and Hes1-mediated Notch Signaling in Controlling the Plasticity of Adult Pancreatic Duct Cells in Mice Shinichi Hosokawa 1,3,Kenichiro Furuyama 1,3, Masashi Horiguchi 1,3,Yoshiki
Cancer. October is National Breast Cancer Awareness Month
 Cancer October is National Breast Cancer Awareness Month Objectives 1: Gene regulation Explain how cells in all the different parts of your body develop such different characteristics and functions. Contrast
Cancer October is National Breast Cancer Awareness Month Objectives 1: Gene regulation Explain how cells in all the different parts of your body develop such different characteristics and functions. Contrast
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
Midterm 1. Number of students Score. Mean: 73 Median: 75 Top Score: 98
 Midterm 1 14 12 Number of students 10 8 6 4 2 0 35-40 41-45 Mean: 73 Median: 75 Top Score: 98 46-50 51-55 56-60 61-65 66-70 71-75 Score 76-80 81-85 86-90 91-95 96-100 Write your name and student ID# on
Midterm 1 14 12 Number of students 10 8 6 4 2 0 35-40 41-45 Mean: 73 Median: 75 Top Score: 98 46-50 51-55 56-60 61-65 66-70 71-75 Score 76-80 81-85 86-90 91-95 96-100 Write your name and student ID# on
Supplementary Materials for
 www.sciencetranslationalmedicine.org/cgi/content/full/4/117/117ra8/dc1 Supplementary Materials for Notch4 Normalization Reduces Blood Vessel Size in Arteriovenous Malformations Patrick A. Murphy, Tyson
www.sciencetranslationalmedicine.org/cgi/content/full/4/117/117ra8/dc1 Supplementary Materials for Notch4 Normalization Reduces Blood Vessel Size in Arteriovenous Malformations Patrick A. Murphy, Tyson
Early Embryonic Development
 Early Embryonic Development Maternal effect gene products set the stage by controlling the expression of the first embryonic genes. 1. Transcription factors 2. Receptors 3. Regulatory proteins Maternal
Early Embryonic Development Maternal effect gene products set the stage by controlling the expression of the first embryonic genes. 1. Transcription factors 2. Receptors 3. Regulatory proteins Maternal
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003)
 MicroRNAs: Genomics, Biogenesis, Mechanism, and Function (D. Bartel Cell 2004) he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003) Vertebrate MicroRNA Genes (Lim et al. Science
MicroRNAs: Genomics, Biogenesis, Mechanism, and Function (D. Bartel Cell 2004) he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003) Vertebrate MicroRNA Genes (Lim et al. Science
