Partial carotid ligation and flow pattern validation by high resolution ultrasound
|
|
|
- Ernest Griffin
- 5 years ago
- Views:
Transcription
1 Partial carotid ligation and flow pattern validation by high resolution ultrasound All animal studies were performed with Male C57Bl/6 mice according to the approved IACUC protocol by Emory University. Mice (Jackson Laboratories) were partially ligated between 6 to 8 weeks of age as we recently described 1. Briefly, three of four caudal branches of left common carotid artery (LCA) - left external carotid, internal carotid, and occipital artery - were ligated with 6-0 silk suture while the superior thyroid artery was left intact in anesthetized mice. Six hours post-surgery, each animal was examined by VEVO 770 High-resolution in vivo micro-imaging ultrasound system whether the ligation induced low and oscillatory shear stress in LCA with the contralateral RCA as a control 1. Intimal RNA isolation from carotid arteries Total RNA from intima were separately obtained from LCA and RCA at 12, 24 and 48 hr post-ligation as we described previously 1. Briefly, LCA and RCA were quickly flushed (few seconds) with 150 µl of QIAzol lysis reagent (QIAGEN) using 29G insulin syringe into a microfuge tube. The eluate was then used for total intimal RNA isolation using mirneasy mini kit (QIAGEN). Microarray Procedures Total intimal RNAs were obtained from LCA and RCA at 12hr and 48hr postligation. Intimal RNAs from three LCAs or RCAs were pooled to obtain ~30 ng total RNA. All RNA samples used for the microarray study passed a quality control test using Agilent BioAnalyze NanoChip. Each sample was linearly amplified by WT-Ovation RNA amplification system (NuGEN) and used for the microarray study using MouseWG-6 v2 Expression BeadChip array with 45,281 probes (Illumina) at the Emory Biomarker Service Center. After hybridization, BeadChips are scanned on the Illumina BeadArray Reader to determine the probe fluorescence intensity. The raw probe intensities were then normalized by the quantile normalization algorithm 2 using the GenomeStudio software from Illumina. Microarray Data Analysis and Bioinformatics The microarray data was statistically analyzed by Significance Analysis of Microarrays software (SAM) 3. The differentially expressed genes between LCA and RCA were identified for those which showed more than 1.5 fold-changes at <10% false discovery rate. The lists of differentially expressed genes were interrogated for statistically significant overrepresented cellular functions and
2 disorders using DAVID analysis and Ingenuity Pathway (IPA) Analysis (Ingenuity Systems). Quantitative real time PCR (qpcr) validation Total RNA of each sample was reverse transcribed into cdna using SuperScript III and random primers (Invitrogen) as we described 1. Briefly, qpcr was performed on selected genes using Brilliant II SYBR Green QPCR Master Mix (Stratagene) with custom designed primers on a Real-Time PCR System (ABI StepOne Plus). Predesigned TaqMan Gene Expression Assay probes (Applied Biosystems) were also used for some selected genes. All qpcr results were normalized based on 18S RNA expression in each sample. Fold changes between LCA and RCA were determined using the Ct method 4. Immunohistochemical staining Paraffin section immunostaining Mice were euthanized by CO 2 inhalation and then were pressure-perfused at 100 mmhg with normal saline followed by pressure fixation with a 10% formalin solution. LCA and RCA were collected en block with the trachea and esophagus. Paraffin sections (5 µm) were then microwaved for 20 min in citrate buffer (0.1 M, ph 6.0) for BMP4 and LMO4 staining or in Tris buffer (0.1 M, ph 9.0) for Angpt2 and Jam2 staining. Sections were blocked with 10% donkey serum for 1 hour at room temperature and incubated with primary antibodies specific to BMP4 (5 µg/ml, Biovision), Lmo4 (5 µg/ml, 5-7 ), Jam2 (2 µg/ml, R&D System), and Angpt2 (0.4 µg/ml, Santa Cruz) overnight at 4 C in a humidified chamber 8. To visualize primary antibodies, rhodamine-conjugated secondary antibodies (donkey anti-goat, anti-rat IgG, Jackson) were used for one hour at room temperature. Nuclei were counter stained with Hoechst # All photographs were taken using a Zeiss epifluorescent microscope. Paraffin sections of human coronary arteries from patients undergoing heart transplants were obtained with the patients consent according to the IRB protocol approved at Emory as described previously 8. The same staining method used for mouse carotids as described above was used for Lmo4 staining. En Face staining - Mice were euthanized by CO 2 inhalation and the aortas were pressure-perfused at 100 mmhg with normal saline followed by pressure fixation with a 10% formalin solution. The aortas were carefully dissected in situ and the aortic arches and thoracic aortas were dissected and stained with Lmo4 antibody 5-7, followed by rhodamine-conjugated secondary antibodies for 2 hours at room temperature. The aortas were then mounted on glass slides using Vectashield containing DAPI (Vector Laboratories). They were then opened and lesser curvature and the greater curvature of the arch were separated. En face images were obtained using a Zeiss LSM 510 META confocal microscope. Ex vivo tissue culture
3 Mice were euthanized by CO 2 inhalation and then pressure-perfused with heparinized normal saline. Under sterile conditions, common carotid arteries were harvested and carefully cleaned of perivascular fat. Carotid artery rings (~3 mm) and incubated for 3 to 5 days at 37 C and 5% CO2 in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 100 U/mL of penicillin and 100 µg/ml of streptomycin and 10% of heat-inactivated fetal bovine serum. Statistical analysis Data are presented as mean±sem. Paired Student s t-test was carried out for all qpcr results of each gene to compare LCA vs. RCA and p<0.05 (n=3-5) was considered statistically significant. REFERENCES 1. Nam D, Ni CW, Rezvan A, et al. Partial carotid ligation is a model of acutely induced disturbed flow, leading to rapid endothelial dysfunction and atherosclerosis. Am J Physiol Heart Circ Physiol. 2009;297:H Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19: Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98: Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3: Sum EY, Shackleton M, Hahm K, et al. Loss of the LIM domain protein Lmo4 in the mammary gland during pregnancy impedes lobuloalveolar development. Oncogene. 2005;24: Sum EY, Segara D, Duscio B, et al. Overexpression of LMO4 induces mammary hyperplasia, promotes cell invasion, and is a predictor of poor outcome in breast cancer. Proc Natl Acad Sci U S A. 2005;102: Visvader JE, Venter D, Hahm K, et al. The LIM domain gene LMO4 inhibits differentiation of mammary epithelial cells in vitro and is overexpressed in breast cancer. Proc Natl Acad Sci U S A. 2001;98: Chang K, Weiss D, Suo J, et al. Bone morphogenic protein antagonists are coexpressed with bone morphogenic protein 4 in endothelial cells exposed to unstable flow in vitro in mouse aortas and in human coronary arteries: role of bone morphogenic protein antagonists in inflammation and atherosclerosis. Circulation. 2007;116:
4 Table S.1 The list of gene probes identified as significant expressed in microarray 12hr Down regulated gene probes (LCA/RCA) (38) 12hr Up regulated gene probes (LCA/RCA) (29) Gene ID Gene Name Score(d) Numerator(r) Denominator(s+s0) Fold Change q value(%) Gene ID Gene Name Score(d) Numerator(r) Denominator(s+s0) Fold Change q value(%) ILMN_ Timp ILMN_ Bmp ILMN_ Klf ILMN_ Got ILMN_ Gnaq ILMN_ Ctgf ILMN_ Eln ILMN_ Ctps ILMN_ Capn ILMN_ Galntl ILMN_ Dhh ILMN_ LOC ILMN_ Id ILMN_ Galntl ILMN_ Pdlim ILMN_ LOC ILMN_ Ptprj ILMN_ Ulk ILMN_ Pdgfa ILMN_ Gabarapl ILMN_ Cdkn1a ILMN_ Lmo ILMN_ Icam ILMN_ H16Rik ILMN_ BC ILMN_ Phactr ILMN_ Tek ILMN_ A17Rik ILMN_ E030024M20Rik ILMN_ Birc ILMN_ Dab2ip ILMN_ Snn ILMN_ Klf ILMN_ Aldh6a ILMN_ K01Rik ILMN_ Slc1a ILMN_ Tinagl ILMN_ Dapk ILMN_ Kras ILMN_ Gpm6a ILMN_ Rhpn ILMN_ Ctgf ILMN_ Rab11fip ILMN_ AI ILMN_ C230009C22Rik ILMN_ Angpt ILMN_ F2rl ILMN_ Ier ILMN_ P4ha ILMN_ Fosl ILMN_ Lims ILMN_ Scn3b ILMN_ Inmt ILMN_ Fn3k ILMN_ Capn ILMN_ Mrpl ILMN_ Slc9a3r ILMN_ E030033D05Rik ILMN_ Sertad ILMN_ Rapgef ILMN_ Hspa12b ILMN_ Lims ILMN_ Lsr ILMN_ Klk ILMN_ Col6a ILMN_ Cmklr ILMN_ Kras hr Down regulated gene probes (LCA/RCA) (338) 48hr Up regulated gene probes (LCA/RCA )(250) Gene ID Gene Name Score(d) Numerator(r) Denominator(s+s0) Fold Change q value(%) Gene ID Gene Name Score(d) Numerator(r) Denominator(s+s0) Fold Change q value(%) ILMN_ Id ILMN_ Ncf ILMN_ S ILMN_ Lyzs ILMN_ Atp2b ILMN_ Lyz ILMN_ Pthlh ILMN_ Tyrobp ILMN_ Sdpr ILMN_ G22Rik ILMN_ Bcam ILMN_ Ifitm ILMN_ Kctd ILMN_ Ifi ILMN_ Rbms ILMN_ Smap ILMN_ Gstm ILMN_ scl _ ILMN_ AW ILMN_ Ccl ILMN_ Dhh ILMN_ Fosl ILMN_ Exdl ILMN_ Sell ILMN_ Syp ILMN_ Chi3l ILMN_ G03Rik ILMN_ Sf3b ILMN_ Fbln ILMN_ Edn ILMN_ Phlda ILMN_ Lyz ILMN_ C030034J23Rik ILMN_ J19Rik ILMN_ MGC ILMN_ Apob48r ILMN_ Aldh3a ILMN_ Pira ILMN_ C230096K16Rik ILMN_ Slc9a3r ILMN_ Eln ILMN_ BC ILMN_ Cyb5r ILMN_ Chi3l ILMN_ Fzd ILMN_ Coro1a ILMN_ Ahnak ILMN_ Dnajc ILMN_ Rras ILMN_ H06Rik ILMN_ Clstn ILMN_ Nfkbid ILMN_ F2rl ILMN_ Rgs ILMN_ Pcolce ILMN_ C230067O06Rik ILMN_ Arl4a ILMN_ Laptm ILMN_ D530030K12Rik ILMN_ EG ILMN_ Emp ILMN_ Myo1f ILMN_ Sgcd ILMN_ Cd ILMN_ Plek ILMN_ Cd ILMN_ Kctd ILMN_ Hist1h2bj ILMN_ Cyb5r ILMN_ E20Rik ILMN_ Aoc ILMN_ Prep ILMN_ Rbms ILMN_ Thbs ILMN_ Gja ILMN_ Gp49a ILMN_ Ankrd ILMN_ M18Rik ILMN_ Nkx ILMN_ H17Rik ILMN_ Nucb ILMN_ Hdc ILMN_ Plec ILMN_ K15Rik ILMN_ Fchsd ILMN_ Asprv ILMN_ Tek ILMN_ Amica ILMN_ Hyal ILMN_ Pfkfb ILMN_ Sult1a ILMN_ Fhod ILMN_ Trrp ILMN_ B130015M16Rik ILMN_ Mxd ILMN_ H2 DMb ILMN_ Gja ILMN_ Lrrc ILMN_ Snhg ILMN_ Obfc2a ILMN_ Plec ILMN_ Fmnl
5 ILMN_ Ern ILMN_ Rassf ILMN_ Adam ILMN_ Lst ILMN_ Serpina1b ILMN_ H2 Ab ILMN_ Dhrs ILMN_ Fcgr2b ILMN_ K21Rik ILMN_ Bop ILMN_ Nos ILMN_ Lyzs ILMN_ Ddef ILMN_ Bzw ILMN_ Kctd ILMN_ Lmnb ILMN_ Sec14l ILMN_ Pscd ILMN_ Stxbp3a ILMN_ B17Rik ILMN_ Cygb ILMN_ Mpeg ILMN_ Tmem ILMN_ Ero1lb ILMN_ Foxk ILMN_ Csf3r ILMN_ Arhgef ILMN_ Cd300lf ILMN_ C4a ILMN_ Hmha ILMN_ Atp2b ILMN_ Csk ILMN_ Cytl ILMN_ Wdr ILMN_ LOC ILMN_ Tkt ILMN_ Slc9a3r ILMN_ Rassf ILMN_ E130014J05Rik ILMN_ Ctgf ILMN_ Lims ILMN_ Cxcl ILMN_ Hs3st ILMN_ Gas ILMN_ I09Rik ILMN_ Ctgf ILMN_ Wwp ILMN_ Ccr ILMN_ Lims ILMN_ LOC ILMN_ Taf9b ILMN_ Nipsnap ILMN_ Klk ILMN_ E2f ILMN_ B01Rik ILMN_ Cd ILMN_ Ttc ILMN_ Gpatch ILMN_ St6galnac ILMN_ Rad ILMN_ Adcy ILMN_ A230055O06Rik ILMN_ Nme ILMN_ Fzr ILMN_ Plekha ILMN_ LOC ILMN_ Col4a ILMN_ Lgals ILMN_ Ptn ILMN_ Fcer1g ILMN_ LOC ILMN_ LOC ILMN_ Smarcd ILMN_ Lsg ILMN_ Klf ILMN_ Rnd ILMN_ LOC ILMN_ Coro1a ILMN_ Pi ILMN_ Napsa ILMN_ Tmod ILMN_ B20Rik ILMN_ Klf ILMN_ Map4k ILMN_ Marveld ILMN_ BC ILMN_ Arhgef ILMN_ Cd300a ILMN_ Lrrc3b ILMN_ Itgb ILMN_ BC ILMN_ Dock ILMN_ Ubxd ILMN_ Wdr ILMN_ Lsr ILMN_ Slc38a ILMN_ Klf ILMN_ Ctps ILMN_ Aatk ILMN_ Gmfg ILMN_ K01Rik ILMN_ Lbh ILMN_ Ramp ILMN_ Sema6b ILMN_ C03Rik ILMN_ Lmo ILMN_ D330008E13Rik ILMN_ Rps ILMN_ Klk ILMN_ Ptpn ILMN_ Rhpn ILMN_ Pstpip ILMN_ Sema3f ILMN_ Ivns1abp ILMN_ Elmo ILMN_ Spp ILMN_ Adamts ILMN_ Aif ILMN_ Tbc1d9b ILMN_ G05Rik ILMN_ Jam ILMN_ Akna ILMN_ M22Rik ILMN_ Znhit ILMN_ Dusp ILMN_ Ccr ILMN_ Tmem ILMN_ Sh3bp ILMN_ B130005I07Rik ILMN_ Dnahc ILMN_ Trspap ILMN_ Fcgr2b ILMN_ Amigo ILMN_ Cdh ILMN_ Rdm ILMN_ Ptprc ILMN_ P4ha ILMN_ F08Rik ILMN_ Nos ILMN_ Slfn ILMN_ Cgnl ILMN_ Cd ILMN_ Nagk ILMN_ D16Rik ILMN_ Nos ILMN_ Narg ILMN_ Lama ILMN_ Clec4d ILMN_ Sema3f ILMN_ Ttll ILMN_ Fbxo ILMN_ Bloc1s ILMN_ Mcfd ILMN_ Sema6b ILMN_ Klf ILMN_ Cfp ILMN_ Fbln ILMN_ Cxcl ILMN_ Clip ILMN_ As3mt ILMN_ Mterfd ILMN_ Agpat ILMN_ Ptprr ILMN_ Ctsc ILMN_ Myst ILMN_ Il17ra ILMN_ Rbms ILMN_ Ets ILMN_ Megf ILMN_ Hist1h2bm ILMN_ Sod ILMN_ Cd ILMN_ Add ILMN_ Fcgr ILMN_ Ly6c ILMN_ Pde4b ILMN_ Mal ILMN_ I17Rik ILMN_ Klf ILMN_ Rcc ILMN_ Ltbp ILMN_ H2 Eb ILMN_ Dcn ILMN_ Pira ILMN_ Scamp ILMN_ Coro1a ILMN_ Btbd ILMN_ H2 Ab ILMN_ E330020K23Rik ILMN_ Clec7a ILMN_ Arhgap ILMN_ Tbc1d10c ILMN_ Pdlim ILMN_ Cd300a ILMN_ Ppap2a ILMN_ Cep
6 ILMN_ Prickle ILMN_ Samhd ILMN_ Pbx ILMN_ Syncrip ILMN_ Icam ILMN_ G12Rik ILMN_ Adcy ILMN_ Gemin ILMN_ Pkhd1l ILMN_ Gadd45a ILMN_ Slc9a3r ILMN_ K01Rik ILMN_ C4b ILMN_ Bcl2l ILMN_ Ly6a ILMN_ Frzb ILMN_ Ppm1a ILMN_ LOC ILMN_ LOC ILMN_ Mgl ILMN_ Ccnd ILMN_ G22Rik ILMN_ B20Rik ILMN_ Psmd ILMN_ Cd ILMN_ Gbl ILMN_ Prdm ILMN_ Serhl ILMN_ Map3k ILMN_ Lass ILMN_ Cachd ILMN_ Acot ILMN_ Spna ILMN_ AI ILMN_ Pak ILMN_ LOC ILMN_ A03Rik ILMN_ Got ILMN_ Mknk ILMN_ Pdgfb ILMN_ LOC ILMN_ Zfpn1a ILMN_ Sfxn ILMN_ Mgea ILMN_ AW ILMN_ E2f ILMN_ Itm2b ILMN_ H2 Aa ILMN_ Zmym ILMN_ Hist1h2be ILMN_ Mmrn ILMN_ Myo ILMN_ P4ha ILMN_ Smox ILMN_ Tnrc6c ILMN_ Tmem132a ILMN_ Exdl ILMN_ Cd ILMN_ Elmo ILMN_ Ube2t ILMN_ Tns ILMN_ LOC ILMN_ Pnpla ILMN_ Tgfbi ILMN_ Efcab4a ILMN_ scl _ ILMN_ Rw1 pending ILMN_ Ifitm ILMN_ Ptgs ILMN_ Vcam ILMN_ Pps ILMN_ Klra ILMN_ Ptprj ILMN_ Plcb ILMN_ Hspa12b ILMN_ Vcan ILMN_ Ecm ILMN_ Myo1g ILMN_ She ILMN_ Vcan ILMN_ Wipf ILMN_ Lcp ILMN_ Pde6d ILMN_ Cotl ILMN_ Mgst ILMN_ Thbs ILMN_ Palmd ILMN_ Sgpl ILMN_ Myst ILMN_ Selpl ILMN_ Cdc42ep ILMN_ Itgb ILMN_ Cldn ILMN_ Il1rn ILMN_ Numb ILMN_ Fcgr2b ILMN_ Rbms ILMN_ Rnmtl ILMN_ LOC ILMN_ Sla ILMN_ Ahnak ILMN_ Noc4l ILMN_ Tspo ILMN_ Hspa ILMN_ Gja ILMN_ K02Rik ILMN_ Epas ILMN_ Gmfg ILMN_ Jam ILMN_ Fpr ILMN_ C630004H02Rik ILMN_ Mrps ILMN_ Wfdc ILMN_ Cd ILMN_ Cd ILMN_ Plek ILMN_ Bace ILMN_ Evi2a ILMN_ Muted ILMN_ Hcst ILMN_ Serpina1e ILMN_ Csf3r ILMN_ Kirrel ILMN_ Pygl ILMN_ Rarres ILMN_ Ruvbl ILMN_ Tnrc6c ILMN_ Rcc ILMN_ Krt ILMN_ D030029G14Rik ILMN_ Trib ILMN_ Tnfrsf1b ILMN_ Cbr ILMN_ Arhgap ILMN_ Nfia ILMN_ scl _ ILMN_ C920007D24Rik ILMN_ Prmt ILMN_ Zdhhc ILMN_ Ctage ILMN_ Ccl ILMN_ Rasa ILMN_ Jag ILMN_ LOC ILMN_ AI ILMN_ Fkbp ILMN_ Rab11fip ILMN_ Zfp ILMN_ Mif4gd ILMN_ S100a ILMN_ Pde6d ILMN_ Cugbp ILMN_ Kcnn ILMN_ OTTMUSG ILMN_ N15Rik ILMN_ Ebna1bp ILMN_ Lrrc ILMN_ B05Rik ILMN_ I02Rik ILMN_ LOC ILMN_ Sfxn ILMN_ H2afz ILMN_ Unc13b ILMN_ Rpo ILMN_ Snx ILMN_ Eftud ILMN_ Serping ILMN_ Abcb1b ILMN_ Epas ILMN_ Ptpn ILMN_ Inmt ILMN_ Prkcd ILMN_ C330008K14Rik ILMN_ Eif5a ILMN_ Wasf ILMN_ Ptpre ILMN_ Ncam ILMN_ C330023M02Rik ILMN_ Ctsh ILMN_ Homer ILMN_ Cytl ILMN_ Pgd ILMN_ BC ILMN_ Txndc ILMN_ Serpina1d ILMN_ Trpv ILMN_ Ell ILMN_ Nek ILMN_ Scn3b ILMN_ Selplg ILMN_ Gja ILMN_ Tes ILMN_ Tsc22d ILMN_ Dock ILMN_ Ermp ILMN_ Sirpb
7 ILMN_ D130063P19Rik ILMN_ Golt1b ILMN_ Ldb ILMN_ BC ILMN_ Pkp ILMN_ Ddx ILMN_ Tpcn ILMN_ Klc ILMN_ Traf3ip ILMN_ Trpv ILMN_ D02Rik ILMN_ Ndrg ILMN_ J03Rik ILMN_ Cutc ILMN_ Wscd ILMN_ Stbd ILMN_ Atp2a ILMN_ Kras ILMN_ Plekha ILMN_ Jam ILMN_ Hoxa ILMN_ Myo1c ILMN_ Stmn ILMN_ G02Rik ILMN_ Phlda ILMN_ Rom ILMN_ Bcl9l ILMN_ L10Rik ILMN_ Dync1li ILMN_ Cutc ILMN_ Adamts ILMN_ Mgll ILMN_ C03Rik ILMN_ K01Rik ILMN_ Sp ILMN_ Palmd ILMN_ Igf ILMN_ Nkx ILMN_ Arfl ILMN_ Pqlc ILMN_ Klf ILMN_ B230107H12Rik ILMN_ O14Rik ILMN_ Them ILMN_ Ddit ILMN_ Ndrl ILMN_ Nos ILMN_ Setmar ILMN_ Itgb ILMN_ Casp ILMN_ Pwwp2b ILMN_ Mgll ILMN_ Timp ILMN_ Zbtb7c ILMN_ Peg ILMN_ Chrd ILMN_ K14Rik ILMN_ Ap1g ILMN_ E030024M20Rik ILMN_ Sort ILMN_ B3gnt ILMN_ /9/ ILMN_ Pmp ILMN_ Gcap ILMN_ Fmo ILMN_ Ubxd ILMN_ Itgb ILMN_ Bcorl ILMN_ Pdlim ILMN_ Nr1d ILMN_ N16Rik ILMN_ K03Rik ILMN_ D430023I21Rik ILMN_ Agrn ILMN_ Arl6ip ILMN_ Dab2ip ILMN_ Mgst ILMN_ Cd59a ILMN_ Sp ILMN_ Ephx ILMN_ E230020D15Rik ILMN_ Gm ILMN_ Brd ILMN_ Fgd ILMN_ Thap ILMN_ Pkn ILMN_ Ppp2r3a ILMN_ L14Rik ILMN_ Six ILMN_ Nme ILMN_ Rbbp ILMN_ Zdhhc ILMN_ Scara ILMN_ Mfap
8 Table S2. Common mechanosensitive genes between 12hr and 48hr Up-regulated (LCA/RCA) Ctgf Ctps Fosl2 Got2 Lmo4 Down-regulated (LCA/RCA) K01Rik BC Dab2ip Dhh E030024M20Rik Eln F2rl1 Icam2 Id1 Inmt Klf2 Klf4 Klk10 Kras Lims2 Lsr P4ha2 Pdlim2 Ptprj Rab11fip5 Rhpn2 Slc9a3r2 Tek Timp3
9 Table S3. Taqman qpcr probes Assay Type Gene Name Assay ID Angpt2,mCG1200 angiopoietin 2 Mm _m1 Bcam,mCG4887 basal cell adhesion molecule Mm _m1 Bmp4,mCG4634 bone morphogenetic protein 4 Mm _m1 Cd300a,mCG13614 CD300A antigen Mm _m1 Col4a3,mCG collagen, type IV, alpha 3 Mm _m1 Ctgf,mCG6745 connective tissue growth factor Mm _g1 Cxcl12,mCG chemokine (C-X-C motif) ligand 12 Mm _m1 Cxcl16,mCG21161 chemokine (C-X-C motif) ligand 16 Mm _m1 Dhh desert hedgehog Mm _s1 Dusp8,mCG dual specificity phosphatase 8 Mm _m1 Emp2,mCG epithelial membrane protein 2 Mm _m1 Epas1,mCG20417 endothelial PAS domain protein 1 Mm _m1 Hdc,mCG2100 histidine decarboxylase Mm _m1 Icam1,mCG14043 intercellular adhesion molecule 1 Mm _m1 Igf2,mCG11082 insulin-like growth factor 2 Mm _g1 Jam2,mCG junction adhesion molecule 2 Mm _m1 Klf2,mCG18931 Kruppel-like factor 2 (lung) Mm _g1 Klk10,mCG22144 kallikrein related-peptidase 10 Mm _m1 Lat2,mCG16701 linker for activation of T cells family, member 2 Mm _m1 Mif, mcg3124 macrophage migration inhibitory factor,macrophage migration inhibitory factor-like Mm _gH Nos3,mCG16477 nitric oxide synthase 3, endothelial cell Mm _m1 Pprc1,mCG10300 peroxisome proliferative activated receptor, gamma, coactivator-related 1 Mm _m1 Pthlh,mCG7104 parathyroid hormone-like peptide Mm _m1 Ramp2,mCG20228 receptor (calcitonin) activity modifying protein 2 Mm _g1 Rhpn2,mCG rhophilin, Rho GTPase binding protein 2 Mm _m1 Tgfb1,mCG7649 transforming growth factor, beta 1 Mm _m1 Tyrobp,mCG22805 TYRO protein tyrosine kinase binding protein Mm _m1 Vcam1,mCG19764 vascular cell adhesion molecule 1 Mm _m1
10 Table S4. SYBR Green qpcr probes Gene Symble Gene ID Gene Name Forward primer 5'-3' Reverse primer 5'-3' Ankrd25 Arhgef15 Ctps Cyb5r3 ICAM2 KLF4 Lims2 Lmo4 Mfap5 Pak4 Plec1 Plek2 Ptprj Rab11fip5 Sgcd Slc9a3r2 Tek Timp3 NM_ NM_ NM_ NM_ NM_ NM_ NM_ NM_ NM_ NM_ NM_ NM_ NM_ NM_ NM_ NM_ NM_ NM_ Mus musculus KN motif and ankyrin repeat domains 2 CCAGGTCCTGCATGTGCCCG TCCAGGTCCAGGCGGTAGCC Mus musculus Rho guanine nucleotide exchange factor GGCCCAGCAGGTTCCTGACC ACCTGGGGTGGGGAAGGCTC (GEF) 15 Mus musculus cytidine 5'- triphosphate synthase TCCCTGGGGTGCCAGGACTC ATGGCGAGGGCAACCACAGC Mus musculus cytochrome b5 reductase 3 GTGCGTGAGGCCACCGTCTC GGTGATGGCCGGTGTGGAGC Mus musculus intercellular adhesion molecule 2 CACGGTGTCCCCTGTGCAGC CGTGGCTGTGGCCTCTTGGG Mus musculus Kruppel-like factor 4 (gut GCAGGTGCCCCGACTAACCG CTGCACCAGCTCCGCCACTC Mus musculus LIM and senescent cell antigen like ACGCCAACTGGCATCCTGGC TTGTGGCAAGGCCGGCAGAG domains 2 Mus musculus LIM domain only 4 (Lmo4) GCCGGCTCCCTCTCCTGGAA GGACGTGCCAATGTCGCCCA Mus musculus microfibrillar associated protein 5 GGCCACCGGCAGACAGATCG CCGCGTTGACCACTGACCCC Mus musculus p21 protein (Cdc42/Rac)-activated CCAGGAGGACCCCAGGAGGC GTGGTCCGTGTCAGCCCGTG kinase 4 Mus musculus plectin 1 (Plec1), transcript variant 11 TCAGAGCCTCCGAGGGCAAGA GGTTGTGGCCATCACGGAGGTC Mus musculus pleckstrin 2 (Plek2) ACGGCGTGCTCAAGGAAGGC CCTTGGGCGGGGTTACTCGC Mus musculus protein tyrosine phosphatase, TGCCCCACAGTCCCCTTCCC CTTCCTCCCCACCCCCACCC receptor type, J Mus musculus RAB11 family interacting protein 5 AGTGGGATCCTGGCCCCTGC TCTCCCTGGGGCTCTGTCGC (class I) Mus musculus sarcoglycan, delta (dystrophin-associated CTGCGTCTGCGCCAATGGGA TGCTGCCGGCAATTGTCCACT glycoprotein) Mus musculus solute carrier family 9 (sodium/hydrogen exchanger), member 3 GGGCGAGACGCATCACCAGG AGTGCAGGTCAGTTGCCGCC regulator 2 Mus musculus endothelialspecific receptor tyrosine ACTTGCCGCATGCTCAGCCC TCGGGCCCCCACTTCTGAGC kinase Mus musculus tissue inhibitor of GCTGGAGCCTTGGGCACTGG AGGGCCCCTCCTTCACCAGC metalloproteinase 3
11 Table S5. Overrepresented Gene Ontology categories regulated by flow-disturbance in mouse carotid endothelium 12hr post-ligation No. of Genes 48hr post-ligation No. of Genes Diseases and Disorders Developmental Disorder 10 Cancer 9 Immunological Disease 4 Cardiovascular Disease 6 Respiratory Disease 4 Diseases and Disorders Immunological Disease Inflammatory Response Connective Tissue Disorders Inflammatory Disease Skeletal and Muscular Disorders Molecular and Cellular Functions Molecular and Cellular Functions Cellular Growth and Proliferation 12 Cellular Movement 70 Cellular Development 20 Cell-To-Cell Signaling and Interaction 76 Cell Morphology 12 Antigen Presentation 30 Cellular Function and Maintenance 6 Cellular Function and Maintenance 44 Cell Cycle 4 Cellular Growth and Proliferation 73
12 Table S6. Comparison of flow-sensitive genes found in vivo mouse carotid endothelium to cultured HUVEC Gene Symbol Downregulated (LCA/RCA) Gene Name Carotid 48hr (LCA/RCA) microarray Carotid 48hr (LCA/RCA) qpcr HUVEC 24hr (OS/LS) microarray Congruency KLK10 kallikrein related-peptidase ND N Col4a3 collagen, type IV, alpha ND N Bcam basal cell adhesion molecule Y Pthlh parathyroid hormone-like peptide Y Rhpn2 rhophilin, Rho GTPase binding protein N KLF2 kruppel-like factor Y Dusp8 dual specificity phosphatase Y KLF4 kruppel-like factor Y IGF2 insulin-like growth factor Y Slc9a3r2 solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator Y Ptprj protein tyrosine phosphatase, receptor type, J ND N Mfap5 microfibrillar associated protein N Dhh desert hedgehog Y Emp2 epithelial membrane protein N Lims2 LIM and senescent cell antigen like domains Y Jam2 junction adhesion molecule Y Pak4 p21 (CDKN1A)-activated kinase Y Ramp2 receptor (calcitonin) activity modifying protein Y NOS3 nitric oxide synthase Y ICAM2 intercellular adhesion molecule N Plek2 pleckstrin Y Epas1 endothelial PAS domain protein Y Ankrd25 ankyrin repeat domain ND N Sgcd sarcoglycan, delta N Tek endothelial-specific receptor tyrosine kinase Y Timp3 tissue inhibitor of metalloproteinase N Arhgef15 Rho guanine nucleotide exchange factor (GEF) N Cyb5r3 cytochrome b5 reductase Y Plec1 plectin Y Rab11fip5 RAB11 family interacting protein 5 (class I) (Rab11fip5), transcript variant Y Upregulated (LCA/RCA) Angpt2 angiopoietin Y Ctgf connective tissue growth factor Y Cd300a CD300A antigen ND N
13 Tyrobp TYRO protein tyrosine kinase binding protein ND N Cxcl12 chemokine (C-X-C motif) ligand N Cxcl16 chemokine (C-X-C motif) ligand N Ctps cytidine 5'-triphosphate synthase Y Lmo4 LIM domain only N VCAM1 vascular cell adhesion molecule N BMP4 Bone morphogenetic protein Y 23/42
14 Table S7. Comparison of gene expressions between different microarray studies Gene Symbol Gene Name Mouse Carotid 48hr (LCA/RCA) microarray HUVEC 24hr (OS/LS) microarray HUVEC 24hr (RF/HSS) microarray Pig Aorta (DF/UF) microarray Category * Downregulated (LCA/RCA) KLK10 kallikrein related-peptidase ND ND NA - Col4a3 collagen, type IV, alpha ND ND NA - Bcam basal cell adhesion molecule NA b Pthlh parathyroid hormone-like peptide NA b,c Rhpn2 rhophilin, Rho GTPase binding protein NA - KLF2 kruppel-like factor NA b,c Dusp8 dual specificity phosphatase NA b,c KLF4 kruppel-like factor a,b,c IGF2 insulin-like growth factor b,c Slc9a3r2 solute carrier family 9 (sodium/hydrogen exchanger), member NA b,c regulator 2 Ptprj protein tyrosine phosphatase, receptor type, J 0.33 ND 0.98 NA - Mfap5 microfibrillar associated protein ND NA - Dhh desert hedgehog NA b Emp2 epithelial membrane protein NA c Lims2 LIM and senescent cell antigen like domains NA b,c Jam2 junction adhesion molecule NA b,c Pak4 p21 (CDKN1A)-activated kinase NA b Ramp2 receptor (calcitonin) activity modifying protein NA b,c NOS3 nitric oxide synthase b,c ICAM2 intercellular adhesion molecule NA c, Plek2 pleckstrin NA b,c Epas1 endothelial PAS domain protein NA b,c Ankrd25 ankyrin repeat domain ND 0.58 NA c Sgcd sarcoglycan, delta ND NA - Tek endothelial-specific receptor tyrosine kinase NA b,c Timp3 tissue inhibitor of metalloproteinase ND NA - Arhgef15 Rho guanine nucleotide exchange factor (GEF) NA - Cyb5r3 cytochrome b5 reductase NA b
15 Plec1 plectin NA b Rab11fip5 RAB11 family interacting protein 5 (class I) (Rab11fip5), transcript NA variant 1 b,c Upregulated (LCA/RCA) Angpt2 angiopoietin b,c Ctgf connective tissue growth factor b Cd300a CD300A antigen 4.31 ND 2.02 NA c Tyrobp TYRO protein tyrosine kinase binding protein 3.64 ND 2.04 NA c Cxcl12 chemokine (C-X-C motif) ligand NA - Cxcl16 chemokine (C-X-C motif) ligand NA c Ctps cytidine 5'-triphosphate synthase NA b Lmo4 LIM domain only NA - VCAM1 vascular cell adhesion molecule ND NA - BMP4 Bone morphogenetic protein NA b * a : gene expression is consistent between mouse and pig (2) b : gene expression is consistent between mouse and HUVEC (OS/LS) (23) c : gene expression is consistent between mouse and HUVEC (RF/HF) (21)
16 Copy numbers / 10E6 18S * * * * * * Day 0 Day 3 Day 5 KLF2 Dhh KLK10 Lmo4 Fig. S1 Endothelial expression of KLF2, Dhh, and KLK10, but not Lmo4, decreased during ex vivo tissue culture. Mouse carotid rings were incubated ex vivo in a growth medium. Intimal RNAs were collected after 0, 3, and 5 days during culture. qpcr analyses were carried out to examine the mrna levels of KLF2, Dhh, KLK10 and Lmo4. mrna copy numbers were normalized against 18S and were shown as mean± SEM (n=3), * p<0.05 (vs. Day 0).
17 Gene expression (OS/LS, Log 2) Pthlh KLF2 Dhh * * * Ramp2 NOS3 ICAM1 IGF2 * * * Bcam Jam2 * Cxcl16 Epas1 Hdc ND Tyrobp ND KLK10 ND Col4a3 Emp2 Tgfb1 Pprc1 Dusp8 Cxcl12 * * Mif Rhpn2 Coro1a Cd300a VCAM1 * Ctgf * BMP4 * Angpt2 Fig. S2 Validation of shear-sensitive mrnas in HUVEC by qpcr. Total RNAs were collected from HUVECs exposed to OS or LS for 24hr. qpcr analysis was then performed using SYBR green with custom design primers. mrna copy numbers were normalized against 18S and are shown as mean± SEM (n=4). p*< 0.05 (OS vs, LS). ND is not detectable in qpcr.
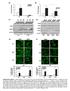
18 A B C Fig. S3. The raw images of the composite figure shown in Fig 4B. Paraffin sections of human left anterior descending coronary artery were stained for Lmo4 protein expression. A) Fluorescence photomicroscopic images were taken from six different but overlapping regions of the stained section. B) The six images were then overlapped together to make a composite image in order to show an overall staining pattern of the entire coronary artery section. C) White dashed lines indicate the edges of each image. C. White dashed lines were removed for a cleaner view and the final picture is shown in Figure 4B.
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array.
 Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction. Marker Sequence (5 3 ) Accession No.
 Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Table S1. Differentially expressed transcripts between hepatic inkt cells. sorted form WT and plck-hcd1d tg mice. Differentially expressed
 Supplementary Information Table S1. Differentially expressed transcripts between hepatic inkt cells sorted form and plck-hcd1d tg mice. Differentially expressed transcripts identified by gene expression
Supplementary Information Table S1. Differentially expressed transcripts between hepatic inkt cells sorted form and plck-hcd1d tg mice. Differentially expressed transcripts identified by gene expression
Supplemental Figure 1. Characterization of CNS lysosomal pathology in 2-month-old IDS-deficient mice. (A) Measurement of IDS activity in brain
 Supplemental Figure 1. Characterization of CNS lysosomal pathology in 2-month-old IDS-deficient mice. (A) Measurement of IDS activity in brain extracts from wild-type (WT) and IDS-deficient males. IDS
Supplemental Figure 1. Characterization of CNS lysosomal pathology in 2-month-old IDS-deficient mice. (A) Measurement of IDS activity in brain extracts from wild-type (WT) and IDS-deficient males. IDS
Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3
 Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
Table 1A. Genes enriched as over-expressed in DPM treatment group
 Table 1A. s enriched as over-expressed in DPM treatment group Affy Probeset mrna Accession Description 10538247 Npy NM_023456 Mus musculus neuropeptide Y (Npy), mrna. 2.0024 10598062 --- NC_005089 gi 34538597
Table 1A. s enriched as over-expressed in DPM treatment group Affy Probeset mrna Accession Description 10538247 Npy NM_023456 Mus musculus neuropeptide Y (Npy), mrna. 2.0024 10598062 --- NC_005089 gi 34538597
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature11986 relative IL-6 expression Viable intracellular Bp per well 18 16 1 1 5 5 3 1 6 +DG 8 8 8 Control DG Time (h) hours B. pertussis IL-6 (pg/ml) 15 Control DG
SUPPLEMENTARY INFORMATION doi:1.138/nature11986 relative IL-6 expression Viable intracellular Bp per well 18 16 1 1 5 5 3 1 6 +DG 8 8 8 Control DG Time (h) hours B. pertussis IL-6 (pg/ml) 15 Control DG
B16F1 B16F10 Supplemental Figure S1
 B16F1 B16F1 Supplemental Figure S1 Representative microangiography images of B16F1 and B16F1 tumors grown in the cranial windows. FITC-dextran (2 million MW) was injected systemically to visualize blood
B16F1 B16F1 Supplemental Figure S1 Representative microangiography images of B16F1 and B16F1 tumors grown in the cranial windows. FITC-dextran (2 million MW) was injected systemically to visualize blood
Supplementary Information Titles Journal: Nature Medicine
 Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Table S1 Differentially expressed genes showing > 2 fold changes and p <0.01 for 0.01 mm NS-398, 0.1mM ibuprofen and COX-2 RNAi.
 Table S1 Differentially expressed genes showing > 2 fold changes and p
Table S1 Differentially expressed genes showing > 2 fold changes and p
Erzsebet Kokovay, Susan Goderie, Yue Wang, Steve Lotz, Gang Lin, Yu Sun, Badrinath Roysam, Qin Shen,
 Cell Stem Cell, Volume 7 Supplemental Information Adult SVZ Lineage Cells Home to and Leave the Vascular Niche via Differential Responses to SDF1/CXCR4 Signaling Erzsebet Kokovay, Susan Goderie, Yue Wang,
Cell Stem Cell, Volume 7 Supplemental Information Adult SVZ Lineage Cells Home to and Leave the Vascular Niche via Differential Responses to SDF1/CXCR4 Signaling Erzsebet Kokovay, Susan Goderie, Yue Wang,
Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression
 Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
CHANGES IN GENE EXPRESSION RELATED TO DECLINE OF THE AGING IMMUNE SYSTEM
 CHANGES IN GENE EXPRESSION RELATED TO DECLINE OF THE AGING IMMUNE SYSTEM HHMI Summer 2012 Oregon State University Student: Jenny Tran Mentor: Adrian F. Gombart, Ph.D. My project involves Studying age-
CHANGES IN GENE EXPRESSION RELATED TO DECLINE OF THE AGING IMMUNE SYSTEM HHMI Summer 2012 Oregon State University Student: Jenny Tran Mentor: Adrian F. Gombart, Ph.D. My project involves Studying age-
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Figure 1. Lkb1-deficient lung ADC progressively transdifferentiates into SCC. (a) A scheme showing the progression pattern of atypical
 Supplementary Figure 1. Lkb1-deficient lung ADC progressively transdifferentiates into SCC. (a) A scheme showing the progression pattern of atypical adenomatous hyperplasia/epithelial hyperplasia (AAH/EH),
Supplementary Figure 1. Lkb1-deficient lung ADC progressively transdifferentiates into SCC. (a) A scheme showing the progression pattern of atypical adenomatous hyperplasia/epithelial hyperplasia (AAH/EH),
Supplementary Information
 Scientific Reports Supplementary Information Upregulated expression of FGF13/FHF2 mediates resistance to platinum drugs in cervical cancer cells Tomoko Okada, Kazuhiro Murata, Ryoma Hirose, Chie Matsuda,
Scientific Reports Supplementary Information Upregulated expression of FGF13/FHF2 mediates resistance to platinum drugs in cervical cancer cells Tomoko Okada, Kazuhiro Murata, Ryoma Hirose, Chie Matsuda,
SUPPLEMENTARY DATA. Assessment of cell survival (MTT) Assessment of early apoptosis and cell death
 Overexpression of a functional calcium-sensing receptor dramatically increases osteolytic potential of MDA-MB-231 cells in a mouse model of bone metastasis through epiregulinmediated osteoprotegerin downregulation
Overexpression of a functional calcium-sensing receptor dramatically increases osteolytic potential of MDA-MB-231 cells in a mouse model of bone metastasis through epiregulinmediated osteoprotegerin downregulation
Suppl Video: Tumor cells (green) and monocytes (white) are seeded on a confluent endothelial
 Supplementary Information Häuselmann et al. Monocyte induction of E-selectin-mediated endothelial activation releases VE-cadherin junctions to promote tumor cell extravasation in the metastasis cascade
Supplementary Information Häuselmann et al. Monocyte induction of E-selectin-mediated endothelial activation releases VE-cadherin junctions to promote tumor cell extravasation in the metastasis cascade
In vivo bromodeoxyuridine (BrdU) incorporation was performed to analyze cell
 Supplementary Methods BrdU incorporation in vivo In vivo bromodeoxyuridine (BrdU) incorporation was performed to analyze cell proliferation in the heart. Mice were subjected to LI-TAC, and 5 days later
Supplementary Methods BrdU incorporation in vivo In vivo bromodeoxyuridine (BrdU) incorporation was performed to analyze cell proliferation in the heart. Mice were subjected to LI-TAC, and 5 days later
Supplementary Materials. for Garmy-Susini, et al, Integrin 4 1 signaling is required for lymphangiogenesis and tumor metastasis
 Supplementary Materials for Garmy-Susini, et al, Integrin 4 1 signaling is required for lymphangiogenesis and tumor metastasis 1 Supplementary Figure Legends Supplementary Figure 1: Integrin expression
Supplementary Materials for Garmy-Susini, et al, Integrin 4 1 signaling is required for lymphangiogenesis and tumor metastasis 1 Supplementary Figure Legends Supplementary Figure 1: Integrin expression
General Laboratory methods Plasma analysis: Gene Expression Analysis: Immunoblot analysis: Immunohistochemistry:
 General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
Pair-fed % inkt cells 0.5. EtOH 0.0
 MATERIALS AND METHODS Histopathological analysis Liver tissue was collected 9 h post-gavage, and the tissue samples were fixed in 1% formalin and paraffin-embedded following a standard procedure. The embedded
MATERIALS AND METHODS Histopathological analysis Liver tissue was collected 9 h post-gavage, and the tissue samples were fixed in 1% formalin and paraffin-embedded following a standard procedure. The embedded
SUPPLEMENTARY METHODS
 SUPPLEMENTARY METHODS Histological analysis. Colonic tissues were collected from 5 parts of the middle colon on day 7 after the start of DSS treatment, and then were cut into segments, fixed with 4% paraformaldehyde,
SUPPLEMENTARY METHODS Histological analysis. Colonic tissues were collected from 5 parts of the middle colon on day 7 after the start of DSS treatment, and then were cut into segments, fixed with 4% paraformaldehyde,
Table SІ. List of genes upregulated by IL-4/IL-13-treatment of NHEK
 Supplemental legends Table SІ. List of genes upregulated by IL-4/IL-13-treatment of NHEK Table SП. List of genes in cluster 1 identified using two-dimensional hierarchical clustering analysis Table SШ.
Supplemental legends Table SІ. List of genes upregulated by IL-4/IL-13-treatment of NHEK Table SП. List of genes in cluster 1 identified using two-dimensional hierarchical clustering analysis Table SШ.
Stewart et al. CD36 ligands promote sterile inflammation through assembly of a TLR 4 and 6 heterodimer
 NFκB (fold induction) Stewart et al. ligands promote sterile inflammation through assembly of a TLR 4 and 6 heterodimer a. mrna (fold induction) 5 4 3 2 1 LDL oxldl Gro1a MIP-2 RANTES mrna (fold induction)
NFκB (fold induction) Stewart et al. ligands promote sterile inflammation through assembly of a TLR 4 and 6 heterodimer a. mrna (fold induction) 5 4 3 2 1 LDL oxldl Gro1a MIP-2 RANTES mrna (fold induction)
Supplement Results, Figures and Tables
 Supplement Results, Figures and Tables Results PET imaging The SUV was significantly higher in tumours of the parental HSC-3 cell line than tumours of the EV and the MMP-8 overexpressing cell lines (Figure
Supplement Results, Figures and Tables Results PET imaging The SUV was significantly higher in tumours of the parental HSC-3 cell line than tumours of the EV and the MMP-8 overexpressing cell lines (Figure
qpcr-array Analysis Service
 qpcr-array Analysis Service Customer Name Institute Telephone Address E-mail PO Number Service Code Report Date Service Laboratory Department Phalanx Biotech Group, Inc 6 Floor, No.6, Technology Road 5,
qpcr-array Analysis Service Customer Name Institute Telephone Address E-mail PO Number Service Code Report Date Service Laboratory Department Phalanx Biotech Group, Inc 6 Floor, No.6, Technology Road 5,
VEGFR2-Mediated Vascular Dilation as a Mechanism of VEGF-Induced Anemia and Bone Marrow Cell Mobilization
 Cell Reports, Volume 9 Supplemental Information VEGFR2-Mediated Vascular Dilation as a Mechanism of VEGF-Induced Anemia and Bone Marrow Cell Mobilization Sharon Lim, Yin Zhang, Danfang Zhang, Fang Chen,
Cell Reports, Volume 9 Supplemental Information VEGFR2-Mediated Vascular Dilation as a Mechanism of VEGF-Induced Anemia and Bone Marrow Cell Mobilization Sharon Lim, Yin Zhang, Danfang Zhang, Fang Chen,
Alternatively Activated Macrophages Determine the Repair of the Infarcted
 Alternatively Activated Macrophages Determine the Repair of the Infarcted Adult Murine Heart (Shiraishi et al.) List of Supplemental Materials Supplemental Methods Supplemental Figure 1. Cardiac CD206
Alternatively Activated Macrophages Determine the Repair of the Infarcted Adult Murine Heart (Shiraishi et al.) List of Supplemental Materials Supplemental Methods Supplemental Figure 1. Cardiac CD206
Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion
 Probe Set ID RefSeq Transcript ID Gene Title Gene Symbol 205980_s_at NM_001017526 /// NM_181334 /// NM_181335 Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion ARHGAP8 /// LOC553158 FC ALK shrna
Probe Set ID RefSeq Transcript ID Gene Title Gene Symbol 205980_s_at NM_001017526 /// NM_181334 /// NM_181335 Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion ARHGAP8 /// LOC553158 FC ALK shrna
SUPPLEMENTAL INFORMATIONS
 1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
Eosinophils! 40! 30! 20! 10! 0! NS!
 A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
A Macrophages Lymphocytes Eosinophils Neutrophils Percentage (%) 1 ** 4 * 1 1 MMA SA B C Baseline FEV1, % predicted 15 p = 1.11 X 10-9 5 CD4:CD8 ratio 1 Supplemental Figure 1. Cellular infiltrate in the
Table 1 Demographic data of PD-patients. Data are represented as counts or median and full range.
 Table 1 Demographic data of PD-patients. Data are represented as counts or median and full range. Parameter Number Patients 5 Diabetes mellitus 0 Previous NTX 0 Steroids 0 Patients with residual renal
Table 1 Demographic data of PD-patients. Data are represented as counts or median and full range. Parameter Number Patients 5 Diabetes mellitus 0 Previous NTX 0 Steroids 0 Patients with residual renal
Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases
 Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
Supplementary Tables: Supplementary Table 1: List of the 242 hypoxia/reoxygenation marker genes collected from literature and databases Supplementary Table 2: Contingency tables used in the fisher exact
CD80 and PD-L2 define functionally distinct memory B cell subsets that are. Griselda V Zuccarino-Catania, Saheli Sadanand, Florian J Weisel, Mary M
 Supplementary Figures CD8 and PD-L define functionally distinct memory B cell subsets that are independent of antibody isotype Running title: Memory B Cell Subset Function Griselda V Zuccarino-Catania,
Supplementary Figures CD8 and PD-L define functionally distinct memory B cell subsets that are independent of antibody isotype Running title: Memory B Cell Subset Function Griselda V Zuccarino-Catania,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
SUPPLEMENTARY FIG. S2. b-galactosidase staining of
 SUPPLEMENTARY FIG. S1. b-galactosidase staining of senescent cells in 500 mg=dl glucose (10 magnification). SUPPLEMENTARY FIG. S3. Toluidine blue staining of chondrogenic-differentiated adipose-tissue-derived
SUPPLEMENTARY FIG. S1. b-galactosidase staining of senescent cells in 500 mg=dl glucose (10 magnification). SUPPLEMENTARY FIG. S3. Toluidine blue staining of chondrogenic-differentiated adipose-tissue-derived
Deconvolution of Heterogeneous Wound Tissue Samples into Relative Macrophage Phenotype Composition via Models based on Gene Expression
 Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2017 Supplementary Information for: Deconvolution of Heterogeneous Wound Tissue Samples into
Electronic Supplementary Material (ESI) for Integrative Biology. This journal is The Royal Society of Chemistry 2017 Supplementary Information for: Deconvolution of Heterogeneous Wound Tissue Samples into
Supplementary data Supplementary Figure 1 Supplementary Figure 2
 Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
TNFSF13B tumor necrosis factor (ligand) superfamily, member 13b NF-kB pathway cluster, Enrichment Score: 3.57
 Appendix 2. Highly represented clusters of genes in the differential expression of data. Immune Cluster, Enrichment Score: 5.17 GO:0048584 positive regulation of response to stimulus GO:0050778 positive
Appendix 2. Highly represented clusters of genes in the differential expression of data. Immune Cluster, Enrichment Score: 5.17 GO:0048584 positive regulation of response to stimulus GO:0050778 positive
Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein
 Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein Lei Wang 1, Tian-Peng Zhang 1, Yuan Zhang 2, Hai-Lian
Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein Lei Wang 1, Tian-Peng Zhang 1, Yuan Zhang 2, Hai-Lian
Cardiovascular Division, Brigham and Women s Hospital, Harvard Medical School
 Low Endothelial Shear Stress Upregulates Atherogenic and Inflammatory Genes Extremely Early in the Natural History of Coronary Artery Disease in Diabetic Hyperlipidemic Juvenile Swine Michail I. Papafaklis,
Low Endothelial Shear Stress Upregulates Atherogenic and Inflammatory Genes Extremely Early in the Natural History of Coronary Artery Disease in Diabetic Hyperlipidemic Juvenile Swine Michail I. Papafaklis,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH
 EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
IL-13 Augments Compressive Stress-induced Tissue Factor Expression in Human Airway Epithelial Cells
 IL-13 Augments Compressive Stress-induced Tissue Factor Expression in Human Airway Epithelial Cells Jennifer A. Mitchel, Silvio Antoniak, Joo-Hyeon Lee, Sae-Hoon Kim, Maureen McGill, David I. Kasahara,
IL-13 Augments Compressive Stress-induced Tissue Factor Expression in Human Airway Epithelial Cells Jennifer A. Mitchel, Silvio Antoniak, Joo-Hyeon Lee, Sae-Hoon Kim, Maureen McGill, David I. Kasahara,
well for 2 h at rt. Each dot represents an individual mouse and bar is the mean ±
 Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Supplementary information. Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls
 Supplementary information Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls angiogenesis in breast cancer Ali Flores-Pérez, Laurence A. Marchat, Sergio Rodríguez-Cuevas, Verónica Bautista-Piña,
Supplementary information Dual targeting of ANGPT1 and TGFBR2 genes by mir-204 controls angiogenesis in breast cancer Ali Flores-Pérez, Laurence A. Marchat, Sergio Rodríguez-Cuevas, Verónica Bautista-Piña,
Islet viability assay and Glucose Stimulated Insulin Secretion assay RT-PCR and Western Blot
 Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Profiles of gene expression & diagnosis/prognosis of cancer. MCs in Advanced Genetics Ainoa Planas Riverola
 Profiles of gene expression & diagnosis/prognosis of cancer MCs in Advanced Genetics Ainoa Planas Riverola Gene expression profiles Gene expression profiling Used in molecular biology, it measures the
Profiles of gene expression & diagnosis/prognosis of cancer MCs in Advanced Genetics Ainoa Planas Riverola Gene expression profiles Gene expression profiling Used in molecular biology, it measures the
Epithelial interleukin-25 is a key mediator in Th2-high, corticosteroid-responsive
 Online Data Supplement: Epithelial interleukin-25 is a key mediator in Th2-high, corticosteroid-responsive asthma Dan Cheng, Zheng Xue, Lingling Yi, Huimin Shi, Kan Zhang, Xiaorong Huo, Luke R. Bonser,
Online Data Supplement: Epithelial interleukin-25 is a key mediator in Th2-high, corticosteroid-responsive asthma Dan Cheng, Zheng Xue, Lingling Yi, Huimin Shi, Kan Zhang, Xiaorong Huo, Luke R. Bonser,
Proteomic profiling of small-molecule inhibitors reveals dispensability of MTH1 for cancer cell survival
 Supplementary Information for Proteomic profiling of small-molecule inhibitors reveals dispensability of MTH1 for cancer cell survival Tatsuro Kawamura 1, Makoto Kawatani 1, Makoto Muroi, Yasumitsu Kondoh,
Supplementary Information for Proteomic profiling of small-molecule inhibitors reveals dispensability of MTH1 for cancer cell survival Tatsuro Kawamura 1, Makoto Kawatani 1, Makoto Muroi, Yasumitsu Kondoh,
Supplemental Table 1. Primer sequences for transcript analysis
 Supplemental Table 1. Primer sequences for transcript analysis Primer Sequence (5 3 ) Primer Sequence (5 3 ) Mmp2 Forward CCCGTGTGGCCCTC Mmp15 Forward CGGGGCTGGCT Reverse GCTCTCCCGGTTTC Reverse CCTGGTGTGCCTGCTC
Supplemental Table 1. Primer sequences for transcript analysis Primer Sequence (5 3 ) Primer Sequence (5 3 ) Mmp2 Forward CCCGTGTGGCCCTC Mmp15 Forward CGGGGCTGGCT Reverse GCTCTCCCGGTTTC Reverse CCTGGTGTGCCTGCTC
Supplementary Figure 1
 Supplementary Figure 1 a Percent of body weight! (%) 4! 3! 1! Epididymal fat Subcutaneous fat Liver SD Percent of body weight! (%) ** 3! 1! SD Percent of body weight! (%) 6! 4! SD ** b Blood glucose (mg/dl)!
Supplementary Figure 1 a Percent of body weight! (%) 4! 3! 1! Epididymal fat Subcutaneous fat Liver SD Percent of body weight! (%) ** 3! 1! SD Percent of body weight! (%) 6! 4! SD ** b Blood glucose (mg/dl)!
Single Cell Quantitative Polymer Chain Reaction (sc-qpcr)
 Single Cell Quantitative Polymer Chain Reaction (sc-qpcr) Analyzing gene expression profiles from a bulk population of cells provides an average profile which may obscure important biological differences
Single Cell Quantitative Polymer Chain Reaction (sc-qpcr) Analyzing gene expression profiles from a bulk population of cells provides an average profile which may obscure important biological differences
Flow-dependent epigenetic DNA methylation regulates endothelial gene expression and atherosclerosis
 Flow-dependent epigenetic DNA methylation regulates endothelial gene expression and atherosclerosis Jessilyn Dunn, Georgia Institute of Technology Haiwei Qiu, Georgia Institute of Technology Soyeon Kim,
Flow-dependent epigenetic DNA methylation regulates endothelial gene expression and atherosclerosis Jessilyn Dunn, Georgia Institute of Technology Haiwei Qiu, Georgia Institute of Technology Soyeon Kim,
Supplemental Methods RNA sequencing experiment
 Supplemental Methods RNA sequencing experiment Mice were euthanized as described in the Methods and the right lung was removed, placed in a sterile eppendorf tube, and snap frozen in liquid nitrogen. RNA
Supplemental Methods RNA sequencing experiment Mice were euthanized as described in the Methods and the right lung was removed, placed in a sterile eppendorf tube, and snap frozen in liquid nitrogen. RNA
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting. protein3) regulate autophagy and mitophagy in renal tubular cells in. acute kidney injury
 Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small cell lung cancer
 Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Experimental Methods Cell culture B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small
Electronic Supplementary Material (ESI) for ChemComm. This journal is The Royal Society of Chemistry 2017 Experimental Methods Cell culture B16-F10 (Mus musculus skin melanoma), NCI-H460 (human non-small
AmoyDx TM BRAF V600E Mutation Detection Kit
 AmoyDx TM BRAF V600E Mutation Detection Kit Detection of V600E mutation in the BRAF oncogene Instructions For Use Instructions Version: B3.1 Date of Revision: April 2012 Store at -20±2 o C 1/5 Background
AmoyDx TM BRAF V600E Mutation Detection Kit Detection of V600E mutation in the BRAF oncogene Instructions For Use Instructions Version: B3.1 Date of Revision: April 2012 Store at -20±2 o C 1/5 Background
Supplementary Figure 1:
 Supplementary Figure 1: (A) Whole aortic cross-sections stained with Hematoxylin and Eosin (H&E), 7 days after porcine-pancreatic-elastase (PPE)-induced AAA compared to untreated, healthy control aortas
Supplementary Figure 1: (A) Whole aortic cross-sections stained with Hematoxylin and Eosin (H&E), 7 days after porcine-pancreatic-elastase (PPE)-induced AAA compared to untreated, healthy control aortas
Supplementary Figures and Tables. An inflammatory gene signature distinguishes neurofibroma Schwann cells and
 Supplementary Figures and Tables An inflammatory gene signature distinguishes neurofibroma Schwann cells and macrophages from cells in the normal peripheral nervous system Kwangmin Choi 1, Kakajan Komurov
Supplementary Figures and Tables An inflammatory gene signature distinguishes neurofibroma Schwann cells and macrophages from cells in the normal peripheral nervous system Kwangmin Choi 1, Kakajan Komurov
To compare the relative amount of of selected gene expression between sham and
 Supplementary Materials and Methods Gene Expression Analysis To compare the relative amount of of selected gene expression between sham and mice given renal ischemia-reperfusion injury (IRI), ncounter
Supplementary Materials and Methods Gene Expression Analysis To compare the relative amount of of selected gene expression between sham and mice given renal ischemia-reperfusion injury (IRI), ncounter
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at
 Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Fig 1 CD163. CD11b S100A9. Sirius Red. 100μm ** ** CD163. CD11b S100A9 ** Sirius Red (PL) Sirius Red SUM Mo.
 T47D T47D + o SU-59 Fig SU-59 + o IHC score (-3) IHC score (-2) CD63 3 2 IHC score (-3) CD63 3 ** 2 CDb CDb * * SA9 SA9 ** * 2 IHC score (-4) αsa αsa 4 ** ** 2 Sirius Red μm IHC score (%) Sirius Red 8
T47D T47D + o SU-59 Fig SU-59 + o IHC score (-3) IHC score (-2) CD63 3 2 IHC score (-3) CD63 3 ** 2 CDb CDb * * SA9 SA9 ** * 2 IHC score (-4) αsa αsa 4 ** ** 2 Sirius Red μm IHC score (%) Sirius Red 8
Acute myocardial infarction (MI) on initial presentation was diagnosed if there was 20 minutes
 SUPPLEMENTAL MATERIAL Supplemental Methods Diagnosis for acute myocardial infarction Acute myocardial infarction (MI) on initial presentation was diagnosed if there was 20 minutes or more of chest pain
SUPPLEMENTAL MATERIAL Supplemental Methods Diagnosis for acute myocardial infarction Acute myocardial infarction (MI) on initial presentation was diagnosed if there was 20 minutes or more of chest pain
Irf1 fold changes (D) 24h 48h. p-p65. t-p65. p-irf3. t-irf3. β-actin SKO TKO 100% 80% 60% 40% 20%
 Irf7 Fold changes 3 1 Irf1 fold changes 3 1 8h h 8h 8h h 8h p-p6 p-p6 t-p6 p-irf3 β-actin p-irf3 t-irf3 β-actin TKO TKO STKO (E) (F) TKO TKO % of p6 nuclear translocation % % 1% 1% % % p6 TKO % of IRF3
Irf7 Fold changes 3 1 Irf1 fold changes 3 1 8h h 8h 8h h 8h p-p6 p-p6 t-p6 p-irf3 β-actin p-irf3 t-irf3 β-actin TKO TKO STKO (E) (F) TKO TKO % of p6 nuclear translocation % % 1% 1% % % p6 TKO % of IRF3
Supplementary Information and Figure legends
 Supplementary Information and Figure legends Table S1. Primers for quantitative RT-PCR Target Sequence (5 -> 3 ) Target Sequence (5 -> 3 ) DAB2IP F:TGGACGATGTGCTCTATGCC R:GGATGGTGATGGTTTGGTAG Snail F:CCTCCCTGTCAGATGAGGAC
Supplementary Information and Figure legends Table S1. Primers for quantitative RT-PCR Target Sequence (5 -> 3 ) Target Sequence (5 -> 3 ) DAB2IP F:TGGACGATGTGCTCTATGCC R:GGATGGTGATGGTTTGGTAG Snail F:CCTCCCTGTCAGATGAGGAC
Phosphate buffered saline (PBS) for washing the cells TE buffer (nuclease-free) ph 7.5 for use with the PrimePCR Reverse Transcription Control Assay
 Catalog # Description 172-5080 SingleShot Cell Lysis Kit, 100 x 50 µl reactions 172-5081 SingleShot Cell Lysis Kit, 500 x 50 µl reactions For research purposes only. Introduction The SingleShot Cell Lysis
Catalog # Description 172-5080 SingleShot Cell Lysis Kit, 100 x 50 µl reactions 172-5081 SingleShot Cell Lysis Kit, 500 x 50 µl reactions For research purposes only. Introduction The SingleShot Cell Lysis
Supporting Information
 Supporting Information Franco et al. 10.1073/pnas.1015557108 SI Materials and Methods Drug Administration. PD352901 was dissolved in 0.5% (wt/vol) hydroxyl-propyl-methylcellulose, 0.2% (vol/vol) Tween
Supporting Information Franco et al. 10.1073/pnas.1015557108 SI Materials and Methods Drug Administration. PD352901 was dissolved in 0.5% (wt/vol) hydroxyl-propyl-methylcellulose, 0.2% (vol/vol) Tween
RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using
 Supplementary Information Materials and Methods RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using Trizol reagent (Invitrogen,Carlsbad, CA) according to the manufacturer's instructions.
Supplementary Information Materials and Methods RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using Trizol reagent (Invitrogen,Carlsbad, CA) according to the manufacturer's instructions.
!! INL!!!! ONL!! IS/OS!!
 SUPPLEMENTARY FIGURES GCL INL ONL IS/OS RPE Choroid GR MR A B C HSD2 Cc RPE CV D Cc RPE CV E RPE Cc CV F RPE Cc CV Figure S1 Figure S1: Immunohistochemistry eidence for glucocorticoid receptor (GR), mineralocorticoid
SUPPLEMENTARY FIGURES GCL INL ONL IS/OS RPE Choroid GR MR A B C HSD2 Cc RPE CV D Cc RPE CV E RPE Cc CV F RPE Cc CV Figure S1 Figure S1: Immunohistochemistry eidence for glucocorticoid receptor (GR), mineralocorticoid
Endogenous TNFα orchestrates the trafficking of neutrophils into and within lymphatic vessels during acute inflammation
 SUPPLEMENTARY INFORMATION Endogenous TNFα orchestrates the trafficking of neutrophils into and within lymphatic vessels during acute inflammation Samantha Arokiasamy 1,2, Christian Zakian 1, Jessica Dilliway
SUPPLEMENTARY INFORMATION Endogenous TNFα orchestrates the trafficking of neutrophils into and within lymphatic vessels during acute inflammation Samantha Arokiasamy 1,2, Christian Zakian 1, Jessica Dilliway
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature7221 Brown fat selective genes 12 1 Control Q-RT-PCR (% of Control) 8 6 4 2 Ntrk3 Cox7a1 Cox8b Cox5b ATPase b2 ATPase f1a1 Sirt3 ERRα Elovl3/Cig3 PPARα Zic1 Supplementary Figure S1. stimulates
doi: 1.138/nature7221 Brown fat selective genes 12 1 Control Q-RT-PCR (% of Control) 8 6 4 2 Ntrk3 Cox7a1 Cox8b Cox5b ATPase b2 ATPase f1a1 Sirt3 ERRα Elovl3/Cig3 PPARα Zic1 Supplementary Figure S1. stimulates
Supplementary Figure 1. SA-β-Gal positive senescent cells in various cancer tissues. Representative frozen sections of breast, thyroid, colon and
 Supplementary Figure 1. SA-β-Gal positive senescent cells in various cancer tissues. Representative frozen sections of breast, thyroid, colon and stomach cancer were stained with SA-β-Gal and nuclear fast
Supplementary Figure 1. SA-β-Gal positive senescent cells in various cancer tissues. Representative frozen sections of breast, thyroid, colon and stomach cancer were stained with SA-β-Gal and nuclear fast
Immunostaining was performed on tumor biopsy samples arranged in a tissue-microarray format or on
 Supplemental Methods Immunohistochemical Analyses Immunostaining was performed on tumor biopsy samples arranged in a tissue-microarray format or on prostatectomy sections obtained post-study. Briefly,
Supplemental Methods Immunohistochemical Analyses Immunostaining was performed on tumor biopsy samples arranged in a tissue-microarray format or on prostatectomy sections obtained post-study. Briefly,
Supporting Information
 Supporting Information M1 macrophage-derived nanovesicles potentiate the anticancer efficacy of immune checkpoint inhibitors Yeon Woong Choo, 1, Mikyung Kang, 2, Han Young Kim, 1 Jin Han, 1 Seokyung Kang,
Supporting Information M1 macrophage-derived nanovesicles potentiate the anticancer efficacy of immune checkpoint inhibitors Yeon Woong Choo, 1, Mikyung Kang, 2, Han Young Kim, 1 Jin Han, 1 Seokyung Kang,
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis
 Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
SUPPLEMENTARY INFORMATION
 DOI:.3/ncb7 Hematopoiesis Expression (Protein) Competition PRC integration Polycomb-mediated gene repression Cell fate HSC PRC SELF-RENEWAL Cbx Cbx4 PRC progenitor genes PROG Cbx PRC Cbx4 DIFFERENTIATION
DOI:.3/ncb7 Hematopoiesis Expression (Protein) Competition PRC integration Polycomb-mediated gene repression Cell fate HSC PRC SELF-RENEWAL Cbx Cbx4 PRC progenitor genes PROG Cbx PRC Cbx4 DIFFERENTIATION
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/389/ra79/dc1 Supplementary Materials for HDL-bound sphingosine 1-phosphate acts as a biased agonist for the endothelial cell receptor S1P 1 to limit vascular
www.sciencesignaling.org/cgi/content/full/8/389/ra79/dc1 Supplementary Materials for HDL-bound sphingosine 1-phosphate acts as a biased agonist for the endothelial cell receptor S1P 1 to limit vascular
TEB. Id4 p63 DAPI Merge. Id4 CK8 DAPI Merge
 a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
Supplementary Table S1: Gene expression targets
 Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
Supplementary Table S1: Gene expression targets Probe ID Gene ID Gene Descriptions Cell type Antigen-dependent activation MmugDNA.32422.1.S1_at CD180 CD180 molecule B 1 MmugDNA.18254.1.S1_at CD19 CD19
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Table 1. Primer sequences for conventional RT-PCR on mouse islets
 Supplementary Table 1. Primer sequences for conventional RT-PCR on mouse islets Gene 5 Forward 3 5 Reverse 3.T. Product (bp) ( C) mnox1 GTTCTTGGGCTGCCTTGG GCTGGGGCGGCGG 60 300 mnoxa1 GCTTTGCCGCGTGC GGTTCGGGTCCTTTGTGC
Supplementary Table 1. Primer sequences for conventional RT-PCR on mouse islets Gene 5 Forward 3 5 Reverse 3.T. Product (bp) ( C) mnox1 GTTCTTGGGCTGCCTTGG GCTGGGGCGGCGG 60 300 mnoxa1 GCTTTGCCGCGTGC GGTTCGGGTCCTTTGTGC
FH- FH+ DM. 52 Volunteers. Oral & IV Glucose Tolerance Test Hyperinsulinemic Euglycemic Clamp in Non-DM Subjects ACADSB MYSM1. Mouse Skeletal Muscle
 A 52 Volunteers B 6 5 4 3 2 FH- FH+ DM 1 Oral & IV Glucose Tolerance Test Hyperinsulinemic Euglycemic Clamp in Non-DM Subjects ZYX EGR2 NR4A1 SRF target TPM1 ACADSB MYSM1 Non SRF target FH- FH+ DM2 C SRF
A 52 Volunteers B 6 5 4 3 2 FH- FH+ DM 1 Oral & IV Glucose Tolerance Test Hyperinsulinemic Euglycemic Clamp in Non-DM Subjects ZYX EGR2 NR4A1 SRF target TPM1 ACADSB MYSM1 Non SRF target FH- FH+ DM2 C SRF
FUS-regulated region- and cell-type-specific transcriptome is associated with cell
 FUS-regulated region- and cell-type-specific transcriptome is associated with cell selectivity in ALS/FTLD Yusuke Fujioka, Shinsuke Ishigaki, Akio Masuda, Yohei Iguchi, Tsuyoshi Udagawa, Hirohisa Watanabe,
FUS-regulated region- and cell-type-specific transcriptome is associated with cell selectivity in ALS/FTLD Yusuke Fujioka, Shinsuke Ishigaki, Akio Masuda, Yohei Iguchi, Tsuyoshi Udagawa, Hirohisa Watanabe,
Online Data Supplement. Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2
 Online Data Supplement Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2 Yi Lin and Zhongjie Sun Department of physiology, college of
Online Data Supplement Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2 Yi Lin and Zhongjie Sun Department of physiology, college of
** *** population. (A) Representative FACS plots showing the gating strategy for T N
 Sca-1 (MFI) CD122 (MFI) CD44 A CD8 + gated: resting spleen CD8 + gated: T Mem -derived CD8 + gated: T N -derived, expanded alone B CD62L CD44 low CD62L +, resting CD44 high, T Mem -derived CD44 low CD62L
Sca-1 (MFI) CD122 (MFI) CD44 A CD8 + gated: resting spleen CD8 + gated: T Mem -derived CD8 + gated: T N -derived, expanded alone B CD62L CD44 low CD62L +, resting CD44 high, T Mem -derived CD44 low CD62L
SUPPLEMENTAL MATERIAL. Supplementary Methods
 SUPPLEMENTAL MATERIAL Supplementary Methods Culture of cardiomyocytes, fibroblasts and cardiac microvascular endothelial cells The isolation and culturing of neonatal rat ventricular cardiomyocytes was
SUPPLEMENTAL MATERIAL Supplementary Methods Culture of cardiomyocytes, fibroblasts and cardiac microvascular endothelial cells The isolation and culturing of neonatal rat ventricular cardiomyocytes was
Supplementary Materials and Methods
 Supplementary Materials and Methods Hepatocyte toxicity assay. Freshly isolated hepatocytes were incubated for overnight with varying concentrations (-25 µm) of sodium glycochenodeoxycholate (GCDC) or
Supplementary Materials and Methods Hepatocyte toxicity assay. Freshly isolated hepatocytes were incubated for overnight with varying concentrations (-25 µm) of sodium glycochenodeoxycholate (GCDC) or
Supplemental Data. NKp46 Identifies an NKT Cell Subset Susceptible to Leukemic Transformation in Mouse and Man
 Supplemental Data NKp46 Identifies an NKT Cell Subset Susceptible to Leukemic Transformation in Mouse and Man Jianhua Yu 1,2*, Takeki Mitsui 1,3*, Min Wei 1, Hsiaoyin Mao 1, Jonathan P Butchar 4, Mithun
Supplemental Data NKp46 Identifies an NKT Cell Subset Susceptible to Leukemic Transformation in Mouse and Man Jianhua Yu 1,2*, Takeki Mitsui 1,3*, Min Wei 1, Hsiaoyin Mao 1, Jonathan P Butchar 4, Mithun
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
III. Results and Discussion
 III. Results and Discussion 1. Histological findings in the coronary artery Twenty-four swine had surgical treatments performed in two of the coronary arteries, LAD as well as either the LCX or RCA. A
III. Results and Discussion 1. Histological findings in the coronary artery Twenty-four swine had surgical treatments performed in two of the coronary arteries, LAD as well as either the LCX or RCA. A
Supplementary Figure 1. NAFL enhanced immunity of other vaccines (a) An over-the-counter, hand-held non-ablative fractional laser (NAFL).
 Supplementary Figure 1. NAFL enhanced immunity of other vaccines (a) An over-the-counter, hand-held non-ablative fractional laser (NAFL). (b) Depiction of a MTZ array generated by NAFL. (c-e) IgG production
Supplementary Figure 1. NAFL enhanced immunity of other vaccines (a) An over-the-counter, hand-held non-ablative fractional laser (NAFL). (b) Depiction of a MTZ array generated by NAFL. (c-e) IgG production
SUPPORTING MATREALS. Methods and Materials
 SUPPORTING MATREALS Methods and Materials Cell Culture MC3T3-E1 (subclone 4) cells were maintained in -MEM with 10% FBS, 1% Pen/Strep at 37ºC in a humidified incubator with 5% CO2. MC3T3 cell differentiation
SUPPORTING MATREALS Methods and Materials Cell Culture MC3T3-E1 (subclone 4) cells were maintained in -MEM with 10% FBS, 1% Pen/Strep at 37ºC in a humidified incubator with 5% CO2. MC3T3 cell differentiation
Supplementary Information
 Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands)
 Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
