A CRISPR dropout screen identifies genetic vulnerabilities in acute myeloid leukaemia. Kosuke Yusa Wellcome Trust Sanger Institute
|
|
|
- Valentine French
- 5 years ago
- Views:
Transcription
1 A CRISPR dropout screen identifies genetic vulnerabilities in acute myeloid leukaemia Kosuke Yusa Wellcome Trust Sanger Institute
2 CRISPR screens Dec 2013 PLX4720 resistance in BRAF V600E melanoma. Resistance to etoposide in leukaemic cells. Resistance to a toxin in mouse ES cells. How about dropout? Genes essential for cell identity Genes required for tumour cell proliferation Genes whose mutation sensitises to drugs Novel therapeutic targets
3 End point CRISPR screening in drug target discovery Genetic vulnerability screen Pooled lentiviral library Cytotoxic/cytostatic Cas9-expressing cells Increased proliferation Initial population Drug resistance/sensitization screen Pooled lentiviral library Non-treated Cas9-expressing cells Sensitive Treated Resistant
4 Dropout with the v1 Mouse library Dropout in mouse ESCs with v1 library (analysed using MAGeCK, Li et al. 2014) Ribosomal genes Sox2 Pou5f1 Nanog
5 Dropout with the v1 Mouse library Dropout in mouse ESCs with v1 library (analysed using MAGeCK, Li et al. 2014) Ribosomal genes Sox2 Pou5f1 Nanog Particular bases favoured T at the 3 end disfavoured due to poliii termination
6 Higher KO efficiency with the improved grna scaffold
7 V2 library generation Dropout in mouse ESCs with v1 library (analysed using MAGeCK, Li et al. 2014) Ribosomal genes Sox2 Pou5f1 Particular bases favoured Nanog v2 GW-gRNA design pipeline - Avoid T stretch at the 3 end - Not use on-target prediction - Design 5 guides per gene by relaxing cut-off T at the 3 end disfavoured due to poliii termination Backbone vector - Use the improved scaffold
8 Improved gene depletion with the v2 library Dropout in mouse ESCs with v1 library Dropout in mouse ESCs with v2 library Ribosomal genes Sox2 Pou5f1 Pou5f1 Sox2 Ribosomal genes Nanog Nanog
9 Improved library does not impair specificity grna-level scatter plot Gene-level scatter plot
10 BFP-positive [%] BFP CRISPR screening toolkit No virus 1 ul/well 2 ul/well 5 ul/well 10 ul/well 20 ul/well Virus [ul/well]
11 CRISPR screening toolkit Cas9 functional assay Mock Empty ggfp Wt mescs Cas9 mescs
12 Counts More even representation with Twist s oligo pool v # reads per guide Unpublished
13 Counts Counts Counts More even representation with Twist s oligo pool v1 v # reads per guide Human v Human v1.1-twist 95.23% log2 (normalised readcounts) 95.19% log2 (normalised readcounts) Unpublished
14 CRISPR screen in HT-29 colon cancer cell line Human v1 HT-29 colon cancer HT-29C clone 3 x3 7~25 dpi Number of depleted genes
15 CRISPR screen in HT-29 colon cancer cell line Human v1 HT-29 colon cancer HT-29C clone 3 x3 7~25 dpi Biased region
16 CRISPR screen in HT-29 colon cancer cell line Human v1 HT-29 colon cancer HT-29C clone 3 x3 7~25 dpi Chr 8 Chr 20
17 CRISPR screen in HT-29 colon cancer cell line Human v1 HT-29 colon cancer HT-29C clone 3 x3 7~25 dpi Gene depletion in time-course analyses Translation Splicing Cell cycle Mitochondria
18 CRISPR screen in 5 AML cell lines Cell lines MOLM-13 Collaboration with George Vassiliou, Kostas Tzelepis MV4-11 HL-60 OCI-AML2 OCI-AML3 HT-29 Nature Methods 2013 HT-1080 AMLs 492 HT-1080 HT-29
19 Genetic and pharmacological validation of KAT2A Validation by grna KAT2A inhibitor, MB-3
20 KAT2A suppression induces differentiation and apoptosis RNA-seq (MOLM-13, 48h MB-3) ChIP-qPCR (MOLM-13, 24h MB-3)
21 KAT2A inhibition induces differentiation and apoptosis Differentiation Apoptosis
22 KAT2A inhibition is effective against human primary AML cells AML samples CD34+ cells
23 Summary Optimised CRISPR-Cas9 platform for genome-wide screens Genome-wide recessive screen focused on AML Identified a large number of potential therapeutic targets Identified KAT2A as a promising anti-aml target
24 Acknowledgements Wellcome Trust Sanger Institute Team 203 Hiroko Koike-Yusa Swee Hoe Ong Leandro Lo Cascio Meng Li Bahar Mirshekar-Syahkal Sianadh Dunn Jason Yu Yan Zi Au Shuhei Ohnishi Team 154 Yilong Li Team 163 Kostas Tzelepis Etienne De Braekeleer George Vassiliou Team 215 Fiona Behan Mathew Garnett Team 30 Sumana Sharma Shan Chong Gavin Wrights EMBL-EBI Francesco Iorio Julio Saez-Rodriguez University of Cambridge Brian Huntly Cristina Pina Funding Team 155 Vera Grinkevich Steven Williams Ultan McDermott Sequence facility Flow cytometry facility RSF
25
CRISPR/CAS9 based high-throughput screening. Journal club Caihong Zhu
 CRISPR/CAS9 based high-throughput screening Journal club Caihong Zhu 29.04.2014 High Throughput Screening (HTS) HTS is a method for scientific experimentation especially used in drug discovery and relevant
CRISPR/CAS9 based high-throughput screening Journal club Caihong Zhu 29.04.2014 High Throughput Screening (HTS) HTS is a method for scientific experimentation especially used in drug discovery and relevant
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 MSI2 interactors are associated with the riboproteome and are functionally relevant. (a) Coomassie blue staining of FLAG-MSI2 immunoprecipitated complexes. (b) GO analysis of MSI2-interacting
Supplementary Figure 1 MSI2 interactors are associated with the riboproteome and are functionally relevant. (a) Coomassie blue staining of FLAG-MSI2 immunoprecipitated complexes. (b) GO analysis of MSI2-interacting
Complexity of intra- and inter-pathway loops in colon cancer and melanoma: implications for targeted therapies
 Complexity of intra- and inter-pathway loops in colon cancer and melanoma: implications for targeted therapies René Bernards The Netherlands Cancer Institute Amsterdam The Netherlands Molecular versus
Complexity of intra- and inter-pathway loops in colon cancer and melanoma: implications for targeted therapies René Bernards The Netherlands Cancer Institute Amsterdam The Netherlands Molecular versus
7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans.
 Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Nature Genetics: doi: /ng Supplementary Figure 1. Details of sequencing analysis.
 Supplementary Figure 1 Details of sequencing analysis. (a) Flow chart showing which patients fall into each category and were used for analysis. (b) Graph showing the average and median coverage for all
Supplementary Figure 1 Details of sequencing analysis. (a) Flow chart showing which patients fall into each category and were used for analysis. (b) Graph showing the average and median coverage for all
Supplementary Figure 1. a. b. Relative cell viability. Nature Genetics: doi: /ng SCR shyap1-1 shyap
 Supplementary Figure 1. a. b. p-value for depletion in vehicle (DMSO) 1e-05 1e-03 1e-01 1 0 1000 2000 3000 4000 5000 Genes log2 normalized shrna counts in T0 0 2 4 6 8 sh1 shluc 0 2 4 6 8 log2 normalized
Supplementary Figure 1. a. b. p-value for depletion in vehicle (DMSO) 1e-05 1e-03 1e-01 1 0 1000 2000 3000 4000 5000 Genes log2 normalized shrna counts in T0 0 2 4 6 8 sh1 shluc 0 2 4 6 8 log2 normalized
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers
 Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Analysis of Massively Parallel Sequencing Data Application of Illumina Sequencing to the Genetics of Human Cancers Gordon Blackshields Senior Bioinformatician Source BioScience 1 To Cancer Genetics Studies
Large conserved domains of low DNA methylation maintained by Dnmt3a
 Supplementary information Large conserved domains of low DNA methylation maintained by Dnmt3a Mira Jeong# 1, Deqiang Sun # 2, Min Luo# 1, Yun Huang 3, Grant A. Challen %1, Benjamin Rodriguez 2, Xiaotian
Supplementary information Large conserved domains of low DNA methylation maintained by Dnmt3a Mira Jeong# 1, Deqiang Sun # 2, Min Luo# 1, Yun Huang 3, Grant A. Challen %1, Benjamin Rodriguez 2, Xiaotian
Supplementary Information. Supplementary Figure 1
 Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Control shrna#9 shrna#12. shrna#12 CD14-PE CD14-PE
 a Control shrna#9 shrna#12 c Control shrna#9 shrna#12 e Control shrna#9 shrna#12 h 14 12 CFU-E BFU-E GEMM GM b Colony number 7 6 5 4 3 2 1 6 pm A pa pc CFU-E BFU-E GEMM GM pu pgm A p pg B d f CD11b-APC
a Control shrna#9 shrna#12 c Control shrna#9 shrna#12 e Control shrna#9 shrna#12 h 14 12 CFU-E BFU-E GEMM GM b Colony number 7 6 5 4 3 2 1 6 pm A pa pc CFU-E BFU-E GEMM GM pu pgm A p pg B d f CD11b-APC
Whole Genome and Transcriptome Analysis of Anaplastic Meningioma. Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute
 Whole Genome and Transcriptome Analysis of Anaplastic Meningioma Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute Outline Anaplastic meningioma compared to other cancers Whole genomes
Whole Genome and Transcriptome Analysis of Anaplastic Meningioma Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute Outline Anaplastic meningioma compared to other cancers Whole genomes
genomics for systems biology / ISB2020 RNA sequencing (RNA-seq)
 RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
Session 4 Rebecca Poulos
 The Cancer Genome Atlas (TCGA) & International Cancer Genome Consortium (ICGC) Session 4 Rebecca Poulos Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 28
The Cancer Genome Atlas (TCGA) & International Cancer Genome Consortium (ICGC) Session 4 Rebecca Poulos Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 28
User Guide. Association analysis. Input
 User Guide TFEA.ChIP is a tool to estimate transcription factor enrichment in a set of differentially expressed genes using data from ChIP-Seq experiments performed in different tissues and conditions.
User Guide TFEA.ChIP is a tool to estimate transcription factor enrichment in a set of differentially expressed genes using data from ChIP-Seq experiments performed in different tissues and conditions.
Discovering Novel Cancer Genetic Vulnerabilities with CRISPR Screens
 Discovering Novel Cancer Genetic Vulnerabilities with CRISPR Screens Traver Hart, Ph.D. Assistant Professor Department of Bioinformatics and Computational Biology Genetic screens in cancer cells A brief
Discovering Novel Cancer Genetic Vulnerabilities with CRISPR Screens Traver Hart, Ph.D. Assistant Professor Department of Bioinformatics and Computational Biology Genetic screens in cancer cells A brief
Simple, rapid, and reliable RNA sequencing
 Simple, rapid, and reliable RNA sequencing RNA sequencing applications RNA sequencing provides fundamental insights into how genomes are organized and regulated, giving us valuable information about the
Simple, rapid, and reliable RNA sequencing RNA sequencing applications RNA sequencing provides fundamental insights into how genomes are organized and regulated, giving us valuable information about the
Nature Medicine: doi: /nm.4078
 Supplementary Figure 1. Cetuximab induces ER stress response in DiFi cells. (a) Scheme of SILAC proteome. (b) MS-base read out of SILAC experiment. The histogram of log 2 -transformed normalized H/L ratios
Supplementary Figure 1. Cetuximab induces ER stress response in DiFi cells. (a) Scheme of SILAC proteome. (b) MS-base read out of SILAC experiment. The histogram of log 2 -transformed normalized H/L ratios
Bowel Disease Research Foundation
 Research Project Annual Report Lead investigator Dr Ricky Sharma Institution Department of Oncology, University of Oxford Project Title Making radiotherapy for rectal cancer more effective by identifying
Research Project Annual Report Lead investigator Dr Ricky Sharma Institution Department of Oncology, University of Oxford Project Title Making radiotherapy for rectal cancer more effective by identifying
RNA-Seq guided gene therapy for vision loss. Michael H. Farkas
 RNA-Seq guided gene therapy for vision loss Michael H. Farkas The retina is a complex tissue Many cell types Neural retina vs. RPE Each highly dependent on the other Graw, Nature Reviews Genetics, 2003
RNA-Seq guided gene therapy for vision loss Michael H. Farkas The retina is a complex tissue Many cell types Neural retina vs. RPE Each highly dependent on the other Graw, Nature Reviews Genetics, 2003
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Binding capacity of DNA-barcoded MHC multimers and recovery of antigen specificity
 Supplementary Figure 1 Binding capacity of DNA-barcoded MHC multimers and recovery of antigen specificity (a, b) Fluorescent-based determination of the binding capacity of DNA-barcoded MHC multimers (+barcode)
Supplementary Figure 1 Binding capacity of DNA-barcoded MHC multimers and recovery of antigen specificity (a, b) Fluorescent-based determination of the binding capacity of DNA-barcoded MHC multimers (+barcode)
Host Double Strand Break Repair Generates HIV-1 Strains Resistant to CRISPR/Cas9
 Host Double Strand Break Repair Generates HIV-1 Strains Resistant to CRISPR/Cas9 Kristine E. Yoder, a * and Ralf Bundschuh b a Department of Molecular Virology, Immunology and Medical Genetics, Center
Host Double Strand Break Repair Generates HIV-1 Strains Resistant to CRISPR/Cas9 Kristine E. Yoder, a * and Ralf Bundschuh b a Department of Molecular Virology, Immunology and Medical Genetics, Center
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature19360 Supplementary Tables Supplementary Table 1. Number of monoclonal reads in each sample Sample Number of cells Total reads Aligned reads Monoclonal reads
SUPPLEMENTARY INFORMATION doi:10.1038/nature19360 Supplementary Tables Supplementary Table 1. Number of monoclonal reads in each sample Sample Number of cells Total reads Aligned reads Monoclonal reads
School of Pathology and Laboratory Medicine: Current and New Research Interests
 School of Pathology and Laboratory Medicine: Current and New Research Interests W/Professor Wendy Erber Current Research Interests Viral immunology and immunogenetics Bone pathology and cell signalling
School of Pathology and Laboratory Medicine: Current and New Research Interests W/Professor Wendy Erber Current Research Interests Viral immunology and immunogenetics Bone pathology and cell signalling
Session 4 Rebecca Poulos
 The Cancer Genome Atlas (TCGA) & International Cancer Genome Consortium (ICGC) Session 4 Rebecca Poulos Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 20
The Cancer Genome Atlas (TCGA) & International Cancer Genome Consortium (ICGC) Session 4 Rebecca Poulos Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 20
Sirt1 Hmg20b Gm (0.17) 24 (17.3) 877 (857)
 3 (0.17) 24 (17.3) Sirt1 Hmg20 Gm4763 877 (857) c d Suppl. Figure 1. Screen validation for top candidate antagonists of Dot1L (a) Numer of genes with one (gray), two (cyan) or three (red) shrna scored
3 (0.17) 24 (17.3) Sirt1 Hmg20 Gm4763 877 (857) c d Suppl. Figure 1. Screen validation for top candidate antagonists of Dot1L (a) Numer of genes with one (gray), two (cyan) or three (red) shrna scored
Selective depletion of abundant RNAs to enable transcriptome analysis of lowinput and highly-degraded RNA from FFPE breast cancer samples
 DNA CLONING DNA AMPLIFICATION & PCR EPIGENETICS RNA ANALYSIS Selective depletion of abundant RNAs to enable transcriptome analysis of lowinput and highly-degraded RNA from FFPE breast cancer samples LIBRARY
DNA CLONING DNA AMPLIFICATION & PCR EPIGENETICS RNA ANALYSIS Selective depletion of abundant RNAs to enable transcriptome analysis of lowinput and highly-degraded RNA from FFPE breast cancer samples LIBRARY
Nature Structural & Molecular Biology: doi: /nsmb.2419
 Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Supplementary Figure 1 Mapped sequence reads and nucleosome occupancies. (a) Distribution of sequencing reads on the mouse reference genome for chromosome 14 as an example. The number of reads in a 1 Mb
Supplementary Figure 1
 Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Introduction to Systems Biology of Cancer Lecture 2
 Introduction to Systems Biology of Cancer Lecture 2 Gustavo Stolovitzky IBM Research Icahn School of Medicine at Mt Sinai DREAM Challenges High throughput measurements: The age of omics Systems Biology
Introduction to Systems Biology of Cancer Lecture 2 Gustavo Stolovitzky IBM Research Icahn School of Medicine at Mt Sinai DREAM Challenges High throughput measurements: The age of omics Systems Biology
CRISPR and RNAi Platforms for Genome- Wide Loss-of-Function Screens AACR April 21, 2015
 CRISPR and RNAi Platforms for Genome- Wide Loss-of-Function Screens AACR April 21, 2015 Paul Diehl, Ph.D. Director of Business Development Cellecta, Inc. Mountain View, CA Cellecta Overview Founded: April
CRISPR and RNAi Platforms for Genome- Wide Loss-of-Function Screens AACR April 21, 2015 Paul Diehl, Ph.D. Director of Business Development Cellecta, Inc. Mountain View, CA Cellecta Overview Founded: April
Supplementary Information. Novel lentiviral vectors with mutated reverse transcriptase for mrna delivery of TALE nucleases
 Supplementary Information Novel lentiviral vectors with mutated reverse transcriptase for mrna delivery of TALE nucleases Ulrike Mock 1, Kristoffer Riecken 1, Belinda Berdien 1, Waseem Qasim 2, Emma Chan
Supplementary Information Novel lentiviral vectors with mutated reverse transcriptase for mrna delivery of TALE nucleases Ulrike Mock 1, Kristoffer Riecken 1, Belinda Berdien 1, Waseem Qasim 2, Emma Chan
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression. Chromatin Array of nuc
 Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Transcriptional control in Eukaryotes: (chapter 13 pp276) Chromatin structure affects gene expression Chromatin Array of nuc 1 Transcriptional control in Eukaryotes: Chromatin undergoes structural changes
Supplementary Figure 1
 Supplementary Figure 1 # nonsynonymous mutations (tngs, 1561 genes) 600 p=0.056 500 250 200 150 100 50 0 HLA-DR (-) HLA-DR (+) Supplementary Figure 1: HLA-DR(+) melanoma cell lines are associated with
Supplementary Figure 1 # nonsynonymous mutations (tngs, 1561 genes) 600 p=0.056 500 250 200 150 100 50 0 HLA-DR (-) HLA-DR (+) Supplementary Figure 1: HLA-DR(+) melanoma cell lines are associated with
Protein SD Units (P-value) Cluster order
 SUPPLEMENTAL TABLE AND FIGURES Table S1. Signature Phosphoproteome of CD22 E12 Transgenic Mouse BPL Cells. T-test vs. Other Protein SD Units (P-value) Cluster order ATPase (Ab-16) 1.41 0.000880 1 mtor
SUPPLEMENTAL TABLE AND FIGURES Table S1. Signature Phosphoproteome of CD22 E12 Transgenic Mouse BPL Cells. T-test vs. Other Protein SD Units (P-value) Cluster order ATPase (Ab-16) 1.41 0.000880 1 mtor
Nature Immunology: doi: /ni Supplementary Figure 1. Characteristics of SEs in T reg and T conv cells.
 Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Implementation of the DDD/ClinGen OGT (CytoSure v3) Microarray
 Implementation of the DDD/ClinGen OGT (CytoSure v3) Microarray OGT UGM Birmingham 08/09/2016 Dom McMullan Birmingham Women's NHS Trust WM chromosomal microarray (CMA) testing Population of ~6 million (10%)
Implementation of the DDD/ClinGen OGT (CytoSure v3) Microarray OGT UGM Birmingham 08/09/2016 Dom McMullan Birmingham Women's NHS Trust WM chromosomal microarray (CMA) testing Population of ~6 million (10%)
Deploying the full transcriptome using RNA sequencing. Jo Vandesompele, CSO and co-founder The Non-Coding Genome May 12, 2016, Leuven
 Deploying the full transcriptome using RNA sequencing Jo Vandesompele, CSO and co-founder The Non-Coding Genome May 12, 2016, Leuven Roadmap Biogazelle the power of RNA reasons to study non-coding RNA
Deploying the full transcriptome using RNA sequencing Jo Vandesompele, CSO and co-founder The Non-Coding Genome May 12, 2016, Leuven Roadmap Biogazelle the power of RNA reasons to study non-coding RNA
A Slfn2 mutation causes lymphoid and myeloid immunodeficiency due to loss of immune cell quiescence
 Supplementary Information A Slfn mutation causes lymphoid and myeloid immunodeficiency due to loss of immune cell quiescence Michael Berger, Philippe Kres, Karine Crozat, Xiaohong Li, Ben A. Croker, Owen
Supplementary Information A Slfn mutation causes lymphoid and myeloid immunodeficiency due to loss of immune cell quiescence Michael Berger, Philippe Kres, Karine Crozat, Xiaohong Li, Ben A. Croker, Owen
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
Introduction. Introduction
 Introduction We are leveraging genome sequencing data from The Cancer Genome Atlas (TCGA) to more accurately define mutated and stable genes and dysregulated metabolic pathways in solid tumors. These efforts
Introduction We are leveraging genome sequencing data from The Cancer Genome Atlas (TCGA) to more accurately define mutated and stable genes and dysregulated metabolic pathways in solid tumors. These efforts
sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5
 sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
Founded in 2016 to develop life-changing therapies against debilitating aggressive cancers that have limited treatment options
 Founded in 2016 to develop life-changing therapies against debilitating aggressive cancers that have limited treatment options Integrated platform technology for discovery of novel molecular targets and
Founded in 2016 to develop life-changing therapies against debilitating aggressive cancers that have limited treatment options Integrated platform technology for discovery of novel molecular targets and
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
EPIGENOMICS PROFILING SERVICES
 EPIGENOMICS PROFILING SERVICES Chromatin analysis DNA methylation analysis RNA-seq analysis Diagenode helps you uncover the mysteries of epigenetics PAGE 3 Integrative epigenomics analysis DNA methylation
EPIGENOMICS PROFILING SERVICES Chromatin analysis DNA methylation analysis RNA-seq analysis Diagenode helps you uncover the mysteries of epigenetics PAGE 3 Integrative epigenomics analysis DNA methylation
The Cancer Genome Atlas & International Cancer Genome Consortium
 The Cancer Genome Atlas & International Cancer Genome Consortium Session 3 Dr Jason Wong Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 31 st July 2014 1
The Cancer Genome Atlas & International Cancer Genome Consortium Session 3 Dr Jason Wong Prince of Wales Clinical School Introductory bioinformatics for human genomics workshop, UNSW 31 st July 2014 1
Introduction to Cancer Biology
 Introduction to Cancer Biology Robin Hesketh Multiple choice questions (choose the one correct answer from the five choices) Which ONE of the following is a tumour suppressor? a. AKT b. APC c. BCL2 d.
Introduction to Cancer Biology Robin Hesketh Multiple choice questions (choose the one correct answer from the five choices) Which ONE of the following is a tumour suppressor? a. AKT b. APC c. BCL2 d.
RESEARCHER S NAME: Làszlò Tora RESEARCHER S ORGANISATION: Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC)
 Thursday 5 November EU-India PARTNERING EVENT Theme: Health RESEARCHER S NAME: Làszlò Tora RESEARCHER S ORGANISATION: Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC) CNRS, INSERM,
Thursday 5 November EU-India PARTNERING EVENT Theme: Health RESEARCHER S NAME: Làszlò Tora RESEARCHER S ORGANISATION: Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC) CNRS, INSERM,
CRISPR-mediated Editing of Hematopoietic Stem Cells for the Treatment of β-hemoglobinopathies
 CRISPR-mediated Editing of Hematopoietic Stem Cells for the Treatment of β-hemoglobinopathies Jennifer Gori American Society of Gene & Cell Therapy May 11, 2017 editasmedicine.com 1 Highlights Developed
CRISPR-mediated Editing of Hematopoietic Stem Cells for the Treatment of β-hemoglobinopathies Jennifer Gori American Society of Gene & Cell Therapy May 11, 2017 editasmedicine.com 1 Highlights Developed
Supplementary Figure 1. Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Nature Immunology: doi: /ni.
 Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Dual PI3K/mTOR inhibition shows antileukemic activity in MLL rearranged acute. Supplemental method S1: Confirmation of RAS mutations
 Supplementary information Dual PI3K/mTOR inhibition shows antileukemic activity in MLL rearranged acute myeloid leukemia Sandhöfer N. et al. Supplemental methods S1-S4: Supplemental method S1: Confirmation
Supplementary information Dual PI3K/mTOR inhibition shows antileukemic activity in MLL rearranged acute myeloid leukemia Sandhöfer N. et al. Supplemental methods S1-S4: Supplemental method S1: Confirmation
Supporting Information
 Supporting Information Horwitz et al. 73/pnas.35295 A Copies ml - C 3NC7 7 697 698 7 7 73 76-2 2 Days Gp2 residue G458D G459D T278A 7/36 N28 K D 28 459 A28T ID# 697 ID# 698 ID# 7 ID# 7 ID# 73 ID# 76 ID#
Supporting Information Horwitz et al. 73/pnas.35295 A Copies ml - C 3NC7 7 697 698 7 7 73 76-2 2 Days Gp2 residue G458D G459D T278A 7/36 N28 K D 28 459 A28T ID# 697 ID# 698 ID# 7 ID# 7 ID# 73 ID# 76 ID#
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Nature Genetics: doi: /ng Supplementary Figure 1. HOX fusions enhance self-renewal capacity.
 Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Differential expression of mirnas from the pri-mir-17-92a locus.
 Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
Molecular mechanisms of the T cellinflamed tumor microenvironment: Implications for cancer immunotherapy
 Molecular mechanisms of the T cellinflamed tumor microenvironment: Implications for cancer immunotherapy Thomas F. Gajewski, M.D., Ph.D. Professor, Departments of Pathology and Medicine Program Leader,
Molecular mechanisms of the T cellinflamed tumor microenvironment: Implications for cancer immunotherapy Thomas F. Gajewski, M.D., Ph.D. Professor, Departments of Pathology and Medicine Program Leader,
HOW TO FIND THE PROJECT. Different ways Reference from your Mentor National Societies ESMO pathway Try to find your way
 HOW TO FIND THE PROJECT Different ways Reference from your Mentor National Societies ESMO pathway Try to find your way Educational Fellowships Translational Research Unit Visit Clinical Unit Visits Palliative
HOW TO FIND THE PROJECT Different ways Reference from your Mentor National Societies ESMO pathway Try to find your way Educational Fellowships Translational Research Unit Visit Clinical Unit Visits Palliative
The role of DNMT3A and HOXA9 hypomethylation in acute myeloid leukemia (AML)
 Campbell Drohan BIOL 463 December 2016 The role of DNMT3A and HOXA9 hypomethylation in acute myeloid leukemia (AML) Introduction Epigenetic modifications of DNA and histones are key to gene regulation
Campbell Drohan BIOL 463 December 2016 The role of DNMT3A and HOXA9 hypomethylation in acute myeloid leukemia (AML) Introduction Epigenetic modifications of DNA and histones are key to gene regulation
Illumina Trusight Myeloid Panel validation A R FHAN R A FIQ
 Illumina Trusight Myeloid Panel validation A R FHAN R A FIQ G E NETIC T E CHNOLOGIST MEDICAL G E NETICS, CARDIFF To Cover Background to the project Choice of panel Validation process Genes on panel, Protocol
Illumina Trusight Myeloid Panel validation A R FHAN R A FIQ G E NETIC T E CHNOLOGIST MEDICAL G E NETICS, CARDIFF To Cover Background to the project Choice of panel Validation process Genes on panel, Protocol
Enterprise Interest Thermo Fisher Scientific / Employee
 Enterprise Interest Thermo Fisher Scientific / Employee A next-generation sequencing assay to estimate tumor mutation load from FFPE research samples Fiona Hyland. Director of R&D, Bioinformatics Clinical
Enterprise Interest Thermo Fisher Scientific / Employee A next-generation sequencing assay to estimate tumor mutation load from FFPE research samples Fiona Hyland. Director of R&D, Bioinformatics Clinical
ESCs were lysed with Trizol reagent (Life technologies) and RNA was extracted according to
 Supplemental Methods RNA-seq ESCs were lysed with Trizol reagent (Life technologies) and RNA was extracted according to the manufacturer's instructions. RNAse free DNaseI (Sigma) was used to eliminate
Supplemental Methods RNA-seq ESCs were lysed with Trizol reagent (Life technologies) and RNA was extracted according to the manufacturer's instructions. RNAse free DNaseI (Sigma) was used to eliminate
Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion
 Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Investigating rare diseases with Agilent NGS solutions
 Investigating rare diseases with Agilent NGS solutions Chitra Kotwaliwale, Ph.D. 1 Rare diseases affect 350 million people worldwide 7,000 rare diseases 80% are genetic 60 million affected in the US, Europe
Investigating rare diseases with Agilent NGS solutions Chitra Kotwaliwale, Ph.D. 1 Rare diseases affect 350 million people worldwide 7,000 rare diseases 80% are genetic 60 million affected in the US, Europe
Supplementary Material
 Supplementary Material Fig. S1. Gpa33 -/- mice do not express Gpa33 mrna or GPA33 protein. (A) The Gpa33 null allele contains a premature translational stop codon (STOP) in exon 2, and almost all of the
Supplementary Material Fig. S1. Gpa33 -/- mice do not express Gpa33 mrna or GPA33 protein. (A) The Gpa33 null allele contains a premature translational stop codon (STOP) in exon 2, and almost all of the
Functional genomics reveal that the serine synthesis pathway is essential in breast cancer
 Functional genomics reveal that the serine synthesis pathway is essential in breast cancer Results Presented by Stacey Lin Lloyd Lab http://www.amsbio.com/expression-ready-lentiviral-particles.aspx Overview
Functional genomics reveal that the serine synthesis pathway is essential in breast cancer Results Presented by Stacey Lin Lloyd Lab http://www.amsbio.com/expression-ready-lentiviral-particles.aspx Overview
The 100,000 Genomes Project
 The 100,000 Genomes Project Dr Matina Prapa, Scientific co ordinator Genomics England Clinical Interpretation Partnership William Harvey Research Institute Queen Mary University of London Genomics England
The 100,000 Genomes Project Dr Matina Prapa, Scientific co ordinator Genomics England Clinical Interpretation Partnership William Harvey Research Institute Queen Mary University of London Genomics England
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
Genome. Institute. GenomeVIP: A Genomics Analysis Pipeline for Cloud Computing with Germline and Somatic Calling on Amazon s Cloud. R. Jay Mashl.
 GenomeVIP: the Genome Institute at Washington University A Genomics Analysis Pipeline for Cloud Computing with Germline and Somatic Calling on Amazon s Cloud R. Jay Mashl October 20, 2014 Turnkey Variant
GenomeVIP: the Genome Institute at Washington University A Genomics Analysis Pipeline for Cloud Computing with Germline and Somatic Calling on Amazon s Cloud R. Jay Mashl October 20, 2014 Turnkey Variant
Tbk1-TKO! DN cells (%)! 15! 10!
 a! T Cells! TKO! B Cells! TKO! b! CD4! 8.9 85.2 3.4 2.88 CD8! Tbk1-TKO! 1.1 84.8 2.51 2.54 c! DN cells (%)! 4 3 2 1 DP cells (%)! 9 8 7 6 CD4 + SP cells (%)! 5 4 3 2 1 5 TKO! TKO! TKO! TKO! 15 1 5 CD8
a! T Cells! TKO! B Cells! TKO! b! CD4! 8.9 85.2 3.4 2.88 CD8! Tbk1-TKO! 1.1 84.8 2.51 2.54 c! DN cells (%)! 4 3 2 1 DP cells (%)! 9 8 7 6 CD4 + SP cells (%)! 5 4 3 2 1 5 TKO! TKO! TKO! TKO! 15 1 5 CD8
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Frequency of alternative-cassette-exon engagement with the ribosome is consistent across data from multiple human cell types and from mouse stem cells. Box plots showing AS frequency
Supplementary Figure 1 Frequency of alternative-cassette-exon engagement with the ribosome is consistent across data from multiple human cell types and from mouse stem cells. Box plots showing AS frequency
Integrated analysis of sequencing data
 Integrated analysis of sequencing data How to combine *-seq data M. Defrance, M. Thomas-Chollier, C. Herrmann, D. Puthier, J. van Helden *ChIP-seq, RNA-seq, MeDIP-seq, Transcription factor binding ChIP-seq
Integrated analysis of sequencing data How to combine *-seq data M. Defrance, M. Thomas-Chollier, C. Herrmann, D. Puthier, J. van Helden *ChIP-seq, RNA-seq, MeDIP-seq, Transcription factor binding ChIP-seq
The Avatar System TM Yields Biologically Relevant Results
 Application Note The Avatar System TM Yields Biologically Relevant Results Liquid biopsies stand to revolutionize the cancer field, enabling early detection and noninvasive monitoring of tumors. In the
Application Note The Avatar System TM Yields Biologically Relevant Results Liquid biopsies stand to revolutionize the cancer field, enabling early detection and noninvasive monitoring of tumors. In the
Joint Department of Biomedical Engineering
 Electronic Supplementary Material (ESI) for Biomaterials Science. This journal is The Royal Society of Chemistry 2018 Supplementary Data Systemic Delivery of CRISPR/Cas9 with PEG-PLGA Nanoparticles for
Electronic Supplementary Material (ESI) for Biomaterials Science. This journal is The Royal Society of Chemistry 2018 Supplementary Data Systemic Delivery of CRISPR/Cas9 with PEG-PLGA Nanoparticles for
mtor Inhibition Specifically Sensitizes Colorectal Cancers with KRAS or BRAF Mutations to BCL-2/BCL-
 Supplementary Material for mtor Inhibition Specifically Sensitizes Colorectal Cancers with KRAS or BRAF Mutations to BCL-2/BCL- XL Inhibition by Suppressing MCL-1 Anthony C. Faber 1,2 *, Erin M. Coffee
Supplementary Material for mtor Inhibition Specifically Sensitizes Colorectal Cancers with KRAS or BRAF Mutations to BCL-2/BCL- XL Inhibition by Suppressing MCL-1 Anthony C. Faber 1,2 *, Erin M. Coffee
Supplementary Figure 1: Digitoxin induces apoptosis in primary human melanoma cells but not in normal melanocytes, which express lower levels of the
 Supplementary Figure 1: Digitoxin induces apoptosis in primary human melanoma cells but not in normal melanocytes, which express lower levels of the cardiac glycoside target, ATP1A1. (a) The percentage
Supplementary Figure 1: Digitoxin induces apoptosis in primary human melanoma cells but not in normal melanocytes, which express lower levels of the cardiac glycoside target, ATP1A1. (a) The percentage
Developing Best in Class BET Inhibitors for Oncology & AI: from Discovery to the Clinic. Kevin G. McLure, PhD EpiCongress July 2014
 Developing Best in Class BET Inhibitors for Oncology & AI: from Discovery to the Clinic Kevin G. McLure, PhD EpiCongress July 2014 Zenith Epigenetics Overview Formed from Resverlogix as an independent
Developing Best in Class BET Inhibitors for Oncology & AI: from Discovery to the Clinic Kevin G. McLure, PhD EpiCongress July 2014 Zenith Epigenetics Overview Formed from Resverlogix as an independent
Supplemental Figure 1
 Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
The development and function of immune cells
 The development and function of immune cells Our research focusses on the development and function of immune cells. For cells to develop and carry out their jobs properly, instructions must be read and
The development and function of immune cells Our research focusses on the development and function of immune cells. For cells to develop and carry out their jobs properly, instructions must be read and
injected subcutaneously into flanks of 6-8 week old athymic male nude mice (LNCaP SQ) and body
 SUPPLEMENTAL FIGURE LEGENDS Figure S1: Generation of ENZR Xenografts and Cell Lines: (A) 1x10 6 LNCaP cells in matrigel were injected subcutaneously into flanks of 6-8 week old athymic male nude mice (LNCaP
SUPPLEMENTAL FIGURE LEGENDS Figure S1: Generation of ENZR Xenografts and Cell Lines: (A) 1x10 6 LNCaP cells in matrigel were injected subcutaneously into flanks of 6-8 week old athymic male nude mice (LNCaP
Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines.
 Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines. The scores summarize the global expression of the tissue
Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines. The scores summarize the global expression of the tissue
The Wistar Institute is an international leader in biomedical
 A LEADER IN RESEARCH The Wistar Institute is an international leader in biomedical research with special expertise in cancer, immunology and infectious disease research. The Institute works actively to
A LEADER IN RESEARCH The Wistar Institute is an international leader in biomedical research with special expertise in cancer, immunology and infectious disease research. The Institute works actively to
Implementation of nation-wide molecular testing in oncology in the French Health care system : quality assurance issues & challenges
 Implementation of nation-wide molecular testing in oncology in the French Health care system : quality assurance issues & challenges Frédérique Nowak - 21 october 2015 "Putting Science into Standards event:
Implementation of nation-wide molecular testing in oncology in the French Health care system : quality assurance issues & challenges Frédérique Nowak - 21 october 2015 "Putting Science into Standards event:
Genotype analysis by Southern blots of nine independent recombinated ES cell clones by
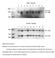 Supplemental Figure 1 Selected ES cell clones show a correctly recombined conditional Ngn3 allele Genotype analysis by Southern blots of nine independent recombinated ES cell clones by hybridization with
Supplemental Figure 1 Selected ES cell clones show a correctly recombined conditional Ngn3 allele Genotype analysis by Southern blots of nine independent recombinated ES cell clones by hybridization with
PREPARED FOR: U.S. Army Medical Research and Materiel Command Fort Detrick, MD
 AD Award Number: W81XWH-11-1-0126 TITLE: Chemical strategy to translate genetic/epigenetic mechanisms to breast cancer therapeutics PRINCIPAL INVESTIGATOR: Xiang-Dong Fu, PhD CONTRACTING ORGANIZATION:
AD Award Number: W81XWH-11-1-0126 TITLE: Chemical strategy to translate genetic/epigenetic mechanisms to breast cancer therapeutics PRINCIPAL INVESTIGATOR: Xiang-Dong Fu, PhD CONTRACTING ORGANIZATION:
MODELLING PRDM14 EXPRESSION IN HUMAN ACUTE MYELOID LEUKEMIA
 MODELLING PRDM14 EXPRESSION IN HUMAN ACUTE MYELOID LEUKEMIA by Yasser Abouelkheer A thesis submitted in conformity with the requirements for the degree of Master of Science The Department of Molecular
MODELLING PRDM14 EXPRESSION IN HUMAN ACUTE MYELOID LEUKEMIA by Yasser Abouelkheer A thesis submitted in conformity with the requirements for the degree of Master of Science The Department of Molecular
Cellecta Overview. Started Operations in 2007 Headquarters: Mountain View, CA
 Cellecta Overview Started Operations in 2007 Headquarters: Mountain View, CA Focus: Development of flexible, scalable, and broadly parallel genetic screening assays to expedite the discovery and characterization
Cellecta Overview Started Operations in 2007 Headquarters: Mountain View, CA Focus: Development of flexible, scalable, and broadly parallel genetic screening assays to expedite the discovery and characterization
VIRAL TITER COUNTS. The best methods of measuring infectious lentiviral titer
 VIRAL TITER COUNTS The best methods of measuring infectious lentiviral titer FLUORESCENCE CYCLES qpcr of Viral RNA SUMMARY Viral vectors are now routinely used for gene transduction in a wide variety of
VIRAL TITER COUNTS The best methods of measuring infectious lentiviral titer FLUORESCENCE CYCLES qpcr of Viral RNA SUMMARY Viral vectors are now routinely used for gene transduction in a wide variety of
Nature Genetics: doi: /ng.3731
 Supplementary Figure 1 Circadian profiles of Adarb1 transcript and ADARB1 protein in mouse tissues. (a) Overlap of rhythmic transcripts identified in the previous transcriptome analyses. The mouse liver
Supplementary Figure 1 Circadian profiles of Adarb1 transcript and ADARB1 protein in mouse tissues. (a) Overlap of rhythmic transcripts identified in the previous transcriptome analyses. The mouse liver
Genomic Methods in Cancer Epigenetic Dysregulation
 Genomic Methods in Cancer Epigenetic Dysregulation Clara, Lyon 2018 Jacek Majewski, Associate Professor Department of Human Genetics, McGill University Montreal, Canada A few words about my lab Genomics
Genomic Methods in Cancer Epigenetic Dysregulation Clara, Lyon 2018 Jacek Majewski, Associate Professor Department of Human Genetics, McGill University Montreal, Canada A few words about my lab Genomics
Biomarkers: An approach to targeting SMARCB1-deficient sarcomas
 Biomarkers: An approach to targeting SMARCB1-deficient sarcomas Description The basis for dual targeting SMARCB1-deficient sarcomas with inhibitors towards PDGFRalpha and FGFR1. Key publications Wong et
Biomarkers: An approach to targeting SMARCB1-deficient sarcomas Description The basis for dual targeting SMARCB1-deficient sarcomas with inhibitors towards PDGFRalpha and FGFR1. Key publications Wong et
Supplementary Information
 Supplementary Information Supplementary Figure 1! a! b! Nfatc1!! Nfatc1"! P1! P2! pa1! pa2! ex1! ex2! exons 3-9! ex1! ex11!!" #" Nfatc1A!!" Nfatc1B! #"!" Nfatc1C! #" DN1! DN2! DN1!!A! #A!!B! #B!!C! #C!!A!
Supplementary Information Supplementary Figure 1! a! b! Nfatc1!! Nfatc1"! P1! P2! pa1! pa2! ex1! ex2! exons 3-9! ex1! ex11!!" #" Nfatc1A!!" Nfatc1B! #"!" Nfatc1C! #" DN1! DN2! DN1!!A! #A!!B! #B!!C! #C!!A!
Living Immunotherapies. Corporate Presentation OCTOBER 2018
 Living Immunotherapies Corporate Presentation OCTOBER 2018 All of the information herein has been prepared by the Company solely for use in this presentation. The information contained in this presentation
Living Immunotherapies Corporate Presentation OCTOBER 2018 All of the information herein has been prepared by the Company solely for use in this presentation. The information contained in this presentation
The common colorectal cancer predisposition SNP rs at chromosome 8q24 confers potential to enhanced Wnt signaling
 SUPPLEMENTARY INFORMATION The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling Sari Tuupanen 1, Mikko Turunen 2, Rainer Lehtonen 1, Outi
SUPPLEMENTARY INFORMATION The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling Sari Tuupanen 1, Mikko Turunen 2, Rainer Lehtonen 1, Outi
Cancer and Gene Alterations - 1
 Cancer and Gene Alterations - 1 Cancer and Gene Alteration As we know, cancer is a disease of unregulated cell growth. Although we looked at some of the features of cancer when we discussed mitosis checkpoints,
Cancer and Gene Alterations - 1 Cancer and Gene Alteration As we know, cancer is a disease of unregulated cell growth. Although we looked at some of the features of cancer when we discussed mitosis checkpoints,
Nature Immunology: doi: /ni Supplementary Figure 1. DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells.
 Supplementary Figure 1 DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells. (a) Scheme for the retroviral shrna screen. (b) Histogram showing CD4 expression (MFI) in WT cytotoxic
Supplementary Figure 1 DNA-methylation machinery is essential for silencing of Cd4 in cytotoxic T cells. (a) Scheme for the retroviral shrna screen. (b) Histogram showing CD4 expression (MFI) in WT cytotoxic
Should novel molecular therapies replace old knowledge of clinical tumor biology?
 Should novel molecular therapies replace old knowledge of clinical tumor biology? Danai Daliani, M.D. Director, 1 st Oncology Clinic Euroclinic of Athens Cancer Treatments Localized disease Surgery XRT
Should novel molecular therapies replace old knowledge of clinical tumor biology? Danai Daliani, M.D. Director, 1 st Oncology Clinic Euroclinic of Athens Cancer Treatments Localized disease Surgery XRT
IDENTIFICATION OF BIOMARKERS FOR EARLY DIAGNOSIS OF BREAST CANCER"
 IDENTIFICATION OF BIOMARKERS FOR EARLY DIAGNOSIS OF BREAST CANCER" Edmond Marzbani, MD December 2, 2008 Early diagnosis of cancer Many solid tumors are potentially curable if diagnosed at an early stage
IDENTIFICATION OF BIOMARKERS FOR EARLY DIAGNOSIS OF BREAST CANCER" Edmond Marzbani, MD December 2, 2008 Early diagnosis of cancer Many solid tumors are potentially curable if diagnosed at an early stage
Supplementary Information
 Supplementary Information Supplementary Figure 1. Effect of mir mimics and anti-mirs on DTPs a, Representative fluorescence microscopy images of GFP vector control or mir mimicexpressing parental and DTP
Supplementary Information Supplementary Figure 1. Effect of mir mimics and anti-mirs on DTPs a, Representative fluorescence microscopy images of GFP vector control or mir mimicexpressing parental and DTP
