SUPPLEMENTARY INFORMATION
|
|
|
- Dina Hunter
- 5 years ago
- Views:
Transcription
1 FULL METHODS Animals. Male, Bmal1 -/-, Per Luc, and Per Luc ; mice were produced and maintained on a C57BL/6J background at the Northwestern University Center for Comparative Medicine. Bmal1 flx/flx mice (kindly provided by Dr Christopher Bradfield, University of Wisconsin) were crossed with transgenic mice (kindly provided by Dr Douglas Melton, Harvard University) to generate ; Bmal1 flx/flx as well as Bmal1 fxl/flx and littermate controls 1,. Offspring from this cross were on a mixed B6x19xICR background. Ai14 reporter mice (transgenic knock-in mice containing lox-stop-lox-tdtomato in the Rosa6 locus) (N) on C57Bl/6J background were crossed with mice. Mice were maintained on a 1:1 light:dark cycle and allowed free access to regular chow and water. All animal studies were conducted in accordance with regulations of the Committee on Animal Care and Use at Northwestern University. Circadian Expression of PER::LUC. Approximately h before lights off, pancreatic islets were isolated from Per Luc and Per Luc ; mice as described below, and ~15-5 islets per mouse were cultured in ml media containing.1 mm luciferin sodium salt (Biosynth AG). Islets were cultured at 7 C in a Lucite environmentally controlled chamber (Solent Scientific). Bioluminescence from samples was continuously imaged using an XR/MEGA-1Z cooled CCD camera (Stanford Photonics), and signal was integrated over 7-minute intervals. After several days in culture, islets were given 1μM forskolin (Sigma) for 1h and incubated in fresh media. Discrete regions of images were quantified using CellCycle software package 1
2 (Actimetrics). Other tissues were cultured as described in Yoo et al 4. SCN were isolated from μm coronal sections. All tissues were cultured on tissue culture membranes (Millipore). Period and damping rate were calculated using Lumicycle Analysis software (Actimetrics). Period was determined using a modified best-fit sine wave analysis, and damping rate represents the number of days for the sine wave to decay by 1/e. 4h Glucose and Insulin Measurements. Blood glucose and plasma insulin levels in ad lib fed mo and 8-9mo old male and animals, as well as -4mo old ; Bmal1 flx/flx, Bmal1 fxl/flx and mice were assessed at 4h intervals from tail vein bleeds. Insulin was measured by ELISA (Crystal Chem Inc). Results were shown as the average for time points during light and dark periods. Glucose and Insulin Tolerance Tests. Glucose tolerance tests were performed in mice fasted for 14hrs. Blood glucose was measured at the indicated times following IP glucose injection of g/kg body weight. Glucose-stimulated insulin secretion was assessed following IP glucose injection of g/kg body weight. Plasma insulin was measured by ELISA. Insulin tolerance tests were performed after a h fast, followed by IP injection of.75u insulin/kg body weight (Humulin R, Eli Lilly). Tests were performed at ZT and ZT14. Islet Isolation and in Vitro Insulin Secretion Analysis. Pancreatic islets were isolated via bile duct collagenase digestion (Collagenase P, Roche) and Ficoll gradient separation and left to recover overnight at 7 C in RPMI 164 with 1% FBS, 1% L- glutamine and 1% penicillin/streptomycin. For insulin release assays, 5 islets were
3 statically incubated in Krebs-Ringer Buffer and stimulated for 1hr at 7 C with various glucose concentrations, mm KCl, 1nM exendin 4,.5μM forskolin, or 1mM 8-BrcAMP. Supernatant was collected and assayed for insulin content by ELISA. Islets were then sonicated in acid-ethanol solution and solubilized overnight at 4 C before assaying total insulin content by ELISA. Immunohistochemical Analysis. For all immunohistochemical experiments, pancreata from 9-1mo old mice were removed and fixed in 4% paraformaldehyde in.1m phosphate buffer and embedded in paraffin. Six blocks of serial sections (6μm) spaced 5μm apart were mounted on slides. To assess pancreatic islet morphology, slides were double-stained with anti-insulin and anti-glucagon (Dako) antibodies. β-cell mass was assessed by morphometric analysis of insulin immunostained pancreatic sections (Dako; Histomouse TM Plus kit, Invitrogen) using a modified protocol described by Pontoglio et al 5. Two pancreatic sections per block were stained for each animal, and endocrine vs total pancreas area was estimated using ImageJ software. For measurement of β-cell proliferation, one pancreas section per block was stained for the nuclear protein doublet Ki-67 (Dako) using a standard labeled streptavidin-biotin method at the Northwestern University Pathology Core Facility. For measurement of islet apoptosis, one pancreas section per block was stained using DeadEnd Fluorometric TUNEL System (Promega) according to the manufacturer s protocol. Proliferation and apoptosis were assessed by estimating percentage of stained cells from total number of islet cells. All pancreas section images were acquired using an inverted Leica DM IRE Fluorescence Microscope system (Leica Microsystems) and
4 Openlab software. All sections were blinded before quantitation and read by one observer. For all immunohistochemical experiments involving pancreas-specific Bmal1 knockout animals, -4mo old mice were anesthetized with IP injection of Avertin (.5% tribromoethanol) and perfused with PBS followed by 4% paraformaldehyde (PFA) (Electron Microscopy Sciences). Brain and pancreas were removed and post-fixed with 4% PFA for 4 hrs at 4 C. Tissues were then cryoprotected with % sucrose (MPBiomedicals) and frozen in Tissue-Tek O.C.T. (Tissue Tek). 1μm pancreas and 14μm brain sections were collected for various antibody stainings. The following primary antibodies were used: guinea pig anti-insulin (1:1, DAKO), mouse antiglucagon (1:, Sigma), rabbit anti-bmal1 (1:1, AbCam), and rabbit anti-vip (1:1, AbCam). Triple staining was visualized with AMCA, DyLight488 and Cy- conjugated secondary antibodies (1:5, Jackson ImmunoResearch). collected with Zeiss Axiovision 4.6 using a Zeiss Axiovert M. Images were Proliferation measurements were done as described above. Islet Size and Pancreas Insulin Content Measurements. Pancreatic islets were isolated as described above. Bright-field images of isolated islets were obtained using Leica DMRB Microscope (Leica Microsystems) and Openlab software. Islet area was analyzed using ImageJ software. Total insulin content was measured after whole pancreas acid-ethanol extraction, quantified by ELISA, and normalized to pancreas weight 5. 4
5 Intracellular Free Ca + Measurements. Islets were loaded with 5μM Fura- AM (Invitrogen) by a min incubation at 7 C in Krebs-Ringer Buffer with mm glucose. Samples were then placed into a temperature-controlled perfusion chamber (Medical Systems Inc.) mounted on an inverted epifluorescence microscope (TE-U, Nikon Inc.) and perifused at speed of 5ml per minute with Krebs-Ringer Buffer (mm glucose) at 7 C. Islets were subjected to Krebs-Ringer Buffer with 1mM glucose or mm KCl, with a 1min Krebs-Ringer Buffer (mm glucose) rinse between each treatment. Free Ca + concentration changes in Fura- loaded islets were measured via dual-wavelength excitation at 4 and 8 nm and imaged with a Cascade CCD Camera (Roper Scientific). Images were acquired and analyzed with Metafluor imaging software (Universal Imaging). Free cytosolic Ca + concentrations in response to secretagogues were calculated as average percent change in 4/8 ratio. Quantitation of mrna by Real-Time RT-PCR. Pancreatic islets were isolated from 1-11mo old or -4mo old pancreas-specific Bmal1 KO mice as described above. Liver samples and 1-15 islets per mouse were collected from 4 animals per time point at 4h intervals for mice and at 8hr intervals for ; Bmal1 flx/flx animals. Total islet mrna was extracted immediately after islet isolation using Tri Reagent (Molecular Research Center, Inc) according to manufacturer s protocol. Total mrna was extracted from frozen liver samples. cdnas were synthesized from total islet and liver mrna using MultiScribe TM reverse transcriptase (Applied Biosystems). Real-time RT-PCR analysis with SYBR Green Master Mix (Applied Biosystems) was performed and analyzed using an Applied Biosystems 79 Fast Real-Time PCR 5
6 System. Relative expression levels were determined using comparative C T method to normalize target gene mrna to GAPDH. Microarray Analysis. Transcriptional profiling was done on islet-derived RNA from four and three animals using the Illumina whole-genome MouseRef-8 v1.1 Expression BeadChip. The BeadChip targets 5,45 well-annotated transcripts, using -micron beads bearing covalently attached 5-base oligonucleotide probes. Each probe interrogates a single gene, and each bead type is represented with an average -fold redundancy on every array. The array content is derived from the National Center for Biotechnology Information Reference Sequence (NCBI RefSeq) database (Release 5), Stanford/UCSF MEEBO ( and RIKEN FANTOM databases. 1ng of total RNA was used to produce an amplified pool of biotin-labeled RNA corresponding to the polyadenylated (mrna) fraction. The labeled RNA was then hybridized to the array and stained with fluorescently labeled anti-biotin antibody. The BeadChip was scanned on Illumina BeadArray Reader, and images were acquired using Illumina s BeadStudio application. Only genes that were defined as detectable in at least one sample (Illumina Detection p-value <.5) were further analyzed. MRI Measurements. Body composition of live mice was determined by quantitative NMR. Data was analyzed using software by EchoMRI TM technology (Echo Medical Systems). Fat and lean mass was reported as % body weight. 6
7 Activity and Feeding Measurements. Locomotor activity was analyzed for month old pancreas-specific Bmal1 KO mice and their littermate controls. All animals were individually housed in standard mouse cages equipped with infrared sensors and allowed free access to food and water. Mice were placed in a 1:1 LD cycle for 14 days, followed by 14 days in constant darkness (DD). Total activity counts were quantified as the total number of infrared sensor beam breaks (Chronobiology Kit, Stanford Software Systems). Activity data was analyzed in 6-minute bouts using ClockLab software (Actimetrics). The free-running period was determined as the duration of time between the major activity periods on consecutive days. Period was calculated using a Chi-square periodogram for days Food consumption was analyzed in - month old pancreas-specific Bmal1 KO mice and their littermate controls. All animals were individually housed with free access to water and regular chow. Day- and night-time food consumption was determined by manual measurement of food at both ZT and ZT1 for 5 consecutive days. Plasma Metabolic Analysis. Plasma samples for analysis of free fatty acid and triglyceride levels were collected from ad lib fed and 14 hour fasted animals at both ZT and ZT14. Free fatty acids (Wako Diagnostics) and triglycerides (GMRD-195, Analox Instruments) were determined according to the manufacturers instructions. Statistical Analysis. Where appropriate, results were expressed as a mean ± S.E.M. Statistical analysis was performed by unpaired two-tailed Student s t-test, 1-way repeated measures analysis of variance (ANOVA), or -way generalized linear models 7
8 ANOVA as described in figure legends (Number Crunchers Statistical Software). Posthoc comparisons, where appropriate, were assessed with Tukey-Kramer Multiple Comparison Test (for time course data of transcripts). Microarray statistical analysis was performed using Partek Genomic Suite where data from detectable genes was log transformed, normalized using the Quantile method 7 and submitted to a Student s t-test. p<.5 was considered to be statistically significant. Functional Analysis of Microarray Data. GeneGo Inc MetaCore Pathway Software Suite was used to determine the most representative ontologic functions (Enrichment Analysis) of the genes which were transcriptionally altered in islets compared to wild type islets in the microarray dataset. Enrichment was conducted across Gene Ontology Processes and GeneGo s Processes Ontology and displayed as percent representation of biological process from the top 5 determined. Key observations are represented on a signaling map created by GeneGo. The interactome (by protein function) tool was also used as an alternative method of analysis to determine the density of interactions for altered gene products in the dataset. The relative connectivity (number of interactions for each gene) can be determined against the data set or the whole human interactome. The statistical relevance of connectivity is calculated according to hypergeometric distribution statistics. 8
9 SUPPORTING DESCRIPTIONS Supporting Description 1: To determine whether this shift in the phase of gene expression in mutant islets reflected a unique disturbance in islets, or rather occurred in a broad range of metabolic tissues, we also examined the expression of metabolic regulatory pathway genes in liver (Fig Suppl ). In contrast to islets, expression of genes in liver exhibited a bimodal peak, with some transcripts rising at ZT6 and others at ZT18, as previously reported 8. Strikingly, at both ZT6 and ZT18, we observed a phase-delay in the expression of some, but not all, of the same oscillating transcripts comparing liver and islet expression patterns. Thus, the phasedelay in gene expression in circadian mutant animals is gene and tissue-specific. The observed difference in timing of gene expression between liver and islet reflects the unique physiological requirements for each tissue at different times in the light-dark cycle. While expression of several metabolic genes in the pancreatic islet, including Glut and Gck, peaks at the beginning of the dark period in mice, genes involved in hepatic gluconeogenesis (Pck) and lipogenesis (Pparα, Srebp-1c, Dbp) rise during the light period when animals are primarily at rest. Both the absolute expression and temporal pattern of these gene oscillations are altered in mutant mice. The reduced expression of glucose sensing and insulin signaling genes around the time of feeding may impede nutrient metabolism and contribute to impaired glucose tolerance at the onset of activity. 9
10 Supporting Description : Because both and mice were fasted prior to the glucose challenge test, the impaired glucose tolerance and diminished insulin secretion in mice is not simply be attributed to differences in the timing of food intake. Supporting Description : Illumina Mouse-Ref8 microbead arrays (Illumina, San Diego) were used for microarray analyses comparing mutant and islets. These analyses revealed significant changes in 57 genes (fold change>1.4, p<.5) in the mutant islets compared to littermate controls, with 7% of these genes down-regulated and % up-regulated (Fig Suppl 6 and Suppl Tables 1-). Supporting Description 4: The GeneGo Ontology analysis is particularly advantageous to identify functional pathways, as opposed to individual genes, that have been significantly altered. The enrichment analysis across Gene Ontology revealed that the most significantly altered processes represented by the affected mutant islets were: 1) protein and vesicle trafficking and localization (including Wnk1, Cdk5r1, Tubb5, Rab9b, Vamp, Syntaxin6, exo7, and Tgoln1), ) cellular metabolism (including Pfk, PIK (p55 subunit), Vipr1, and Slca6), ) post translational modification and protein packaging (Bgalt6, Cln, and Ogt), 4) RNA, chromatin and helicase processes (Phax, Snrnp7, Rpp5), 5) cell cycle (including CyclinD1, CyclinD, Etv1, p1, p57, p16, Neu, Stat, 1
11 and Pak), 6) EGFR signaling (Egfr), and 7) apoptosis and ER stress (S1a6, Atf, and Bik) (Fig Suppl 6d). Supporting Description 5: We further assessed the specificity of the Bmal1 mutation by examining 4 hr expression profiles of circadian genes in liver and pancreatic islets (Fig 4) to confirm that disruption of the transcriptional profile of the circadian clock exists just within pancreatic islets and not within other peripheral tissues such as liver. RevErbα, a downstream target of the CLOCK:BMAL1 heterodimer, was significantly downregulated in islets from ;Bmal1 flx/flx mice. Furthermore, we found a partial compensatory increase of Bmal1 mrna levels in the mutant islets (since the qpcr primers amplify a region outside of the targeted region). Importantly, there was no change in expression of RevErbα and Bmal1 in livers of pancreas-specific Bmal1 mutants compared to controls. Interestingly, we also detected a marked upregulation in expression of the pro-apoptotic gene S1a6 in islets, but not livers, of conditional Bmal1 KO mice, suggesting an increase in islet cell death in the mutants. Taken together, these results provide further evidence for a pancreas-specific perturbation of the clock network in the ;Bmal1 flx/flx animals and argue for the strength of this model to determine the role of the pancreatic clock in whole body glucose homeostasis. 11
12 a Glucose uptake and metabolism c Cell cycle Glut CyclinD1 Relative mrna abundance Gck Gskβ b Insulin signaling d Beta cell growth Ins 1& Hnf4α InsR Hnf1α Irs Pdx1 PiK - p85 NeuroD1 4 Akt 1 4 Time of mrna isolation (ZT) Figure S1: Altered gene expression in 1-1 month old mutant islets. Expression of mrnas encoding selected genes involved in (a) glucose uptake and metabolism, (b) insulin signaling, (c) cell cycle, and (d) β-cell growth and development as assessed by real-time PCR in islets of 1-1 mo old and mice at 4hr intervals across the 1:1 LD cycle (n=4 mice/genotype/timepoint). Fold-change values are displayed as relative abundance (RA; mean ± S.E.M.) after normalization to glyceraldehyde--phosphate dehydrogenase (GAPDH). -way ANOVA was used to determine variance with respect to time and genotype (p<.5). Figure S: Altered gene expression in 1-1 month old mutant livers. Expression of mrnas encoding selected genes involved in (a) circadian rhythmicity, (b) 1
13 Figure S Metabolic gene expression in the liver a Circadian c Nuclear receptors e Glucose and lipid metabolism Bmal1 Rorα Gck Per Pparα Glut Relative mrna abundance 1 RevErbα Lxrα G6p 5 Dbp1 Rxrα Pck Srebp1c b Cell cycle d Insulin signaling CyclinD1 InsR Figure S1: Altered gene expression in 1-1 month old mutant Hnf1α islets. Expression of mrnas encoding selected genes involved in (a) glucose uptake and metabolism, (b) insulin signaling, (c) cell cycle, and (d) β-cell growth and development Gskβ Pik - p85 Hnf4α as assessed by real-time PCR in islets of 1-1 mo old and mice at 4hr intervals across the 1:1 LD cycle (n=4 mice/genotype/timepoint). Fold-change values 1 4 Time of mrna isolation (ZT) are displayed as relative abundance (RA; mean ± S.E.M.) after normalization to Akt glyceraldehyde--phosphate dehydrogenase (GAPDH). -way ANOVA was used to determine variance with respect to time and genotype (p<.5). Figure S: Altered gene expression in 1-1 month old mutant livers. Expression of mrnas encoding selected genes involved in (a) circadian rhythmicity, (b) cell cycle, (c) nuclear receptors, (d) insulin signaling, and (e) glucose and lipid metabolism, as assessed by real-time PCR. Livers from mutant mice and littermate controls were collected at 4hr intervals across the 1:1 LD cycle (n=4 mice/genotype/timepoint). Values are displayed as relative abundance (RA; mean ± S.E.M.) after normalization to glyceraldehyde--phosphate dehydrogenase (GAPDH) expression levels in the same samples. Note that for visual clarity the RA scales vary for the different transcripts. -way ANOVA was used to determine variance with respect to time and genotype (p<.5). 1
14 8mo of age a Glucose tolerance test - glucose levels (ZT) b Glucose tolerance test - insulin levels (ZT) 4 1. Glucose (mg/dl) 1 Insulin (ng/ml) Time (min) Time (min) c Insulin tolerance test (ZT) d Insulin tolerance test (ZT14) Glucose (% baseline) Glucose (% baseline) Time (min) Time (min) e Average fasting glucose levels f Individual fasting glucose levels Glucose (mg/dl) 15 1 Glucose (mg/dl) 5 15 average median 5 ZT ZT14 5 ZT ZT14 Figure S: Diabetic phenotype in 8 mo old mutant mice. (a,b) Glucose tolerance (n=1) and glucose-responsive insulin release (n=8-9) in mutant animals at the beginning of the rest period (ZT). (c,d) Insulin tolerance at both rest and activity periods in mutant animals (n=1-15). (e,f) Fasting plasma glucose levels in mutant animals (n=19-1). Data was analyzed by 1-way ANOVA (a-d) and by Student s t-test (e). p<.5. Values represent mean ± S.E.M. (a-e) or individual animal measurements (f). 14
15 mo of age a Ad lib glucose b Ad lib insulin Glucose (mg/dl) 15 p=.7 p=.98 p=.5 p=.6 Insulin (ng/ml) p=.11 p=.16 p=. p=.19 1 Light -1 ZT.5 Dark Light Dark Light Dark Light Dark ZT ZT ZT c Glucose tolerance test - glucose levels (ZT) d Glucose tolerance test - glucose gevels (ZT14) Glucose (mg/dl) Glucose (mg/dl) Time (min) Time (min) e Insulin tolerance test (ZT) f Insulin tolerance test (ZT14) 1 1 Glucose (% baseline) 9 6 Glucose (% baseline) Time (min) Time (min) g Insulin release (% content) Islet glucose stimulated insulin release 1 p=.46 Glucose (mm) Figure S4: Primary islet defect in young month old mutant mice. (a,b) Glucose and insulin levels in ad lib fed mutant animals (n=6-9). (c,d) Glucose tolerance tests in young mutant animals during light and dark phase (n=8-9). (e,f) Abnormal insulin tolerance in mutant mice in the light and dark phase (n=5). (g) Glucose-responsive insulin secretion in islets from mutant animals (n=6-7). Data was analyzed by Student s t-test (a,b,g) and 1-way ANOVA (c-f). p<.5. All values represent mean ± S.E.M. 15
16 a Pancreas morphology b Insulin (ng/mg pancreas) 4/8 ratio (% change) 5µm 5µm Pancreas insulin content animals at the beginning of the rest period (ZT). (c,d) Insulin 1 tolerance at both rest and levels in mutant animals (n=19-1). Data was.5analyzed by 1-way ANOVA 4/8 ratio (% change) Cell death (% islet cells) Relative endocrine area (% total) c Islet apoptosis d Islet insulin secretion - Ca + influx - e Islet insulin secretion - Ca + influx - Figure S4: Primary islet defect in young month old mutant mice mm Glu 1mM Glu Time (min) mm Glu mm KCl mm Glu p=.7 Figure S: Diabetic phenotype in 8 mo old p= mutant mice. (a,b) Glucose p=.54 tolerance (n=1) and glucose-responsive insulin release (n=8-9) in mutant activity periods in mutant animals (n=1-15). (e,f) Fasting plasma glucose (a-d) and by Student s t-test (e). p<.5. Values represent mean ± S.E.M. (a-e) or individual animal measurements (f). 1mM Glu (a,b) Glucose and insulin levels in ad lib fed mutant animals (n=6-9). (c,d) Glucose tolerance tests in young mutant animals during light and dark phase (n=8-9). (e,f) Abnormal insulin tolerance in mutant mice in the light and dark phase (n=5). (g) Glucose-responsive insulin secretion in islets from mutant animals (n=6-7). Data was analyzed by Student s t-test (a,b,g) and 1-way ANOVA (c-f). p<.5. All values represent mean ± S.E.M Time (min) mm Glu mm KCl Figure S5: Pancreatic islet defect in 8 month old circadian mutant mice. (a) Morphometric analysis of pancreas slices stained for insulin (brown) and quantitation of endocrine mass in mutant mice (n=5-6 mice/genotype). (b) Total pancreas insulin content in mutants (n=6-9). (c) Islet apoptosis in mice as assessed by TUNEL staining (n=5-6). (d,e) Islet Ca + influx in response to insulin secretagogues (n=15-7 islets from mice per genotype). Data in (a-c) was analyzed by Student s t-test. Values represent mean ± S.E.M. (a-c) or ± S.D. (d,e). 16
17 a Total number of genes changed (microarray) c Microarray data validation by real time PCR Number of genes changed up down Fold change by RT-PCR 4 1 S1a6 Bmal1 CyclinD1 Per1 Per RevErbα Dbp b Altered gene expression (microarray) d Major functional groups changed on microarray (gene ontology analysis) Symbol Fold change Function S1a6.4 apoptosis Bmal1 1.8 circadian p1 1.8 apoptosis Atf 1.7 apoptosis Bik 1.5 apoptosis Tgoln1-1.4 vesicle transport Rab9b -1.4 vesicle transport Pfkfb -1.4 cell cycle Wnk1-1.4 vesicle transport Per -1.6 circadian Per1-1.7 circadian Cyclin D1-1.6 proliferation RevErbα -1.8 circadian Neu -1.9 differentiation Tef -. metabolism Pak -.5 proliferation DBP -. metabolism Cell cycle 6% Cellular metabolism (carbohydrate, amino acids, RNA) 1% ds RNA, chromatin, helicase 14% Protein (vesicle) transport and localization 9% ERBB and EGFR signaling % Post translational modification and protein packaging 17% Figure S6. Altered gene expression networks in 8 month old mutant islets as assessed by microarray analysis. (a) Total number of genes with altered expression in mutant islets (fold change >1.4, p<.5, n=-4, Illumina Mouse Ref8 BeadChip). (b) Expression of genes involved in circadian rhythmicity, proliferation, apoptosis, vesicle transport, and metabolism in mutant islets. (c) Microarray data validation by real-time PCR (n=4-5; Student s t-test, p<.5, p<.1). (d) GeneGo enrichment analysis across Gene ontology determined the top functional groups altered in the mutant islets. 17
18 Syntaxin 4 Syntaxin 1A SNAP- Synaptotagmin VII AP complex 1 VAMP Clathrin Clathrin Heavy Chain Transporter NSF VAMP Docked Insulin Vesicle Insulin Clathrin-dependant endocytosis Generic Binding Protein Protein Kinase WNK 1 MUNC 18- CDK5R1 (p5) Tubulin HSP 9 Regulator Ras Family GTPase PKC-lambda/iota PKC-zeta CDK 5 CSNKA1 Alpha-actinin 4 Protein EXO7 TC1 Syntaxin6 VAMP SNAP-5 Myosin VIIa VAMP MYRIP Filamin A Actin Cofilin Receptor Ligand overexpressed Golgin-97 PIST(CAL) Golgi Rab-7A Rabphilin-A underexpressed Rab-A Insulin Insulin Vesicle Figure S7. Altered vesicle trafficking in mutant islets. Shown is a custom GeneGo Ontology map representation of the insulin secretion pathway, which was identified as a functional pathway that was significantly altered in mutant islets. Microarray data is overlaid on the map with green and red arrows indicating a respective decrease or increase in expression. The results indicate a specific loss of vesicle fusion at the membrane, actin-mediated vesicle trafficking, and a gain in traffic in the Golgi. 18
19 Figure S8 a Brain histology 1 ARC ;Bmal1flx/flx V ARC & DMH ;Bmal1flx/flx DMH V 1 ARC ARC ARC V VMH ARC ARC BMAL1 DAPI b ;Bmal1flx/flx PVN PVN PVN V ARC c expression - islet staining ; Ai14 V VMH PVN DMH PVN V expression - SCN staining ; Ai14 ; Ai14 V V SCN SCN SCN Body weight Body weight (g) 4 e p=.69 Bmal1flx/flx ; Bmal1flx/flx p=.1 1 g h 15 p=.4 p=.4 1 Light Dark p= i p=.58 p=.46 p=. 4 Bmal1flx/flx ; Bmal1flx/flx Fat Lean Fat Lean Fat Lean p=.8 Total daily food intake 6 5 Period Light Dark Bmal1flx/flx ; Bmal1flx/flx 1 5 Bmal1flx/flx ; Bmal1flx/flx j p= f Bmal1flx/flx ; Bmal1flx/flx Activity rhythms Activity (% total) Food intake (% total) Feeding rhythms SCN VIP DAPI Body composition Body composition (% body weight) d Glucagon DAPI Food intake (g) Insulin Bmal1flx/flx ; Bmal1flx/flx.5 4 Period (hr) Actograms Bmal1flx/flx ; Bmal1flx/flx LD LD DD DD DD Days LD Time (hr) 48 Figure S8. Normal activity and feeding behavior in -4 month old pancreasspecific Bmal1 knockout mice. (a) BMAL1 expression in feeding centers in the hypothalamus of ;Bmal1flxflx mice as assessed by fluorescent staining of frozen brain slices. Scale bar represents 5 μm. (b) Expression of in islets of ; Ai14 mice as assessed by tdtomato fluorescent staining (red); insulin (green) and glucagon (green) were used as markers for pancreatic islets. Scale bar represents 5 μm. (c) No expression of (red) in the hypothalamus of ;Ai14 mice; VIP (green) was used as a marker for SCN. Scale bar represents 5 μm. (d,e) Body 19
20 glucagon (green) were used as markers for pancreatic islets. Scale bar represents 5 μm. (c) No expression of (red) in the hypothalamus of ;Ai14 mice; VIP (green) was used as a marker for SCN. Scale bar represents 5 μm. (d,e) Body weight and composition (n=5-8), and (f) total daily food intake (n=5) of pancreasspecific Bmal1 KO mice. (g,h) Feeding and activity rhythms in ;Bmal1 flxflx animals (n=4-5). (i) Period of activity in DD, calculated using Chi-square periodogram for days 18-8 (n=4). (j) Actograms showing locomotor activity over a day period in representative individually housed Bmal1 flx/flx, and ;Bmal1 flx/flx mice. Arrow denotes switch from 1:1 LD to DD. Data was analyzed by Student s t-test (d-i). All values represent mean ± S.E.M.
21 a Insulin (ng/ml) Ad lib insulin p=.97 5 p=.1 4 p=.57 p=.1 1 Light Dark ZT -1 ZT 1-4 Bmal1 flx/flx ; Bmal1 flx/flx b Glucose/Insulin 1 4 Ad lib glucose/insulin p=.6 Light Dark ZT -1 ZT 1-4 Bmal1 flx/flx ; Bmal1 flx/flx c Glucose tolerance test - glucose levels (ZT14) d Glucose tolerance test - insulin levels (ZT14) Glucose (mg/dl) Time (min) Bmal1 flx/flx ; Bmal1 flx/flx Insulin (ng/ml) Time (min) Bmal1 flx/flx ; Bmal1 flx/flx e Islet size f Islet proliferation Bmal1 flx/flx p= p=.1 8 ; Bmal1 flx/flx < > Islet size (1 μm ) Islet size distribution (%) Proliferation (% islet cells) Bmal1 flx/flx ; Bmal1 flx/flx Figure S9. Diabetic phenotype in -4 month old pancreas-specific Bmal1 knockout mice. (a) Insulin and (b) glucose/insulin levels in ad lib fed ;Bmal1 flx/flx mice (n=9-1). Data was collected at 4 hr intervals and presented as average for the timepoints during the light and dark periods. (c,d) Glucose tolerance (n=6-8) and glucose-responsive insulin release (n=9-11) in pancreas-specific Bmal1 knockout animals at the beginning of the activity period (ZT14). (e) Size distribution and (f) proliferation as assessed by Ki67 staining (n=5) in islets isolated from pancreas-specific Bmal1 mutant mice. Data was analyzed by Student s t-test (a,b,e,f) and 1-way ANOVA (c,d). All values represent mean ± S.E.M. 1
22 Table 1 Illumina ID Symbol Definition Mean Clock Mean P-value Fold ChangeAccession Number Synonyms ILMN_6166 Dbp Mus musculus D site albumin promoter binding protein (Dbp), mrna NM_ ILMN_ LOC6677 PREDICTED: Mus musculus similar to interferon-induced protein with tetratricopep XM_ ILMN_11669 Pak Mus musculus p1 (CDKN1A)-activated kinase (Pak), mrna NM_8778. mpak-; Pak65beta; PAK-; Stk4; Pak65alpha ILMN_1991 LOC14747 PREDICTED: Mus musculus similar to thyroid hormone receptor (LOC14747), mrna XM_ ILMN_64481 A94L6Rik Mus musculus RIKEN cdna A94L6 gene (A94L6Rik), mrna NM_ E11N9Rik; AL67; AW1968; RP-1A.1; AI85465 ILMN_ Pdyn Mus musculus prodynorphin (Pdyn), mrna NM_1886. Dyn ILMN_6964 Hnrpdl Mus musculus heterogeneous nuclear ribonucleoprotein D-like (Hnrpdl), mrna NM_1669. hnrnp-dl; AA4741; JKTBP; D5Ertd65e; D5Wsu145e; AA ILMN_1458 Ifit Mus musculus interferon-induced protein with tetratricopeptide repeats (Ifit) NM_151. Ifi49; MGC171 ILMN_15985 Serpina7 Mus musculus serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiprot NM_ TBG; C74N1Rik ILMN_11691 Wipi1 Mus musculus WD repeat domain, phosphoinositide interacting 1 (Wipi1), mrna NM_ MGC6416; AW411817; D11Ertd498e; 495H1Rik ILMN_65858 Rgs17 Mus musculus regulator of G-protein signaling 17 (Rgs17), mrna NM_ Rgsz; 6457P11Rik ILMN_ Ifit Mus musculus interferon-induced protein with tetratricopeptide repeats (Ifit) NM_8. Ifi54; AV8 ILMN_ H14Rik Mus musculus RIKEN cdna 4958H14 gene (4958H14Rik), mrna NM_658. AI19547; Osap ILMN_ Tnfrsf Mus musculus tumor necrosis factor receptor superfamily, member (Tnfrsf) NM_68. Tnfrh; Tnfrsf1al; 818K6Rik; SOBa; mdctrailr; C15G6Rik ILMN_1511 C71H18Rik Mus musculus RIKEN cdna C71H18 gene (C71H18Rik), mrna NM_765. ILMN_667 Rgs11 Mus musculus regulator of G-protein signaling 11 (Rgs11), mrna NM_ C7848 ILMN_68554 Car15 Mus musculus carbonic anhydrase 15 (Car15), mrna NM_558. Cals; AI154 ILMN_14451 Gbp Mus musculus guanylate nucleotide binding protein (Gbp), mrna NM_1874. Gbp4; AW8655 ILMN_117 Zcchc1 Mus musculus zinc finger, CCHC domain containing 1 (Zcchc1), mrna NM_85. AV167; 818A1Rik ILMN_4544 Tnfrsf NM_68 ILMN_ Tef Mus musculus thyrotroph embryonic factor (Tef), transcript variant 1, mrna NM_ DRik ILMN_1799 Gale Mus musculus galactose-4-epimerase, UDP (Gale), mrna NM_ AI96; 1A1Rik ILMN_7586 EG976 Mus musculus predicted gene, EG976 (EG976), mrna NM_ D81H1 ILMN_14589 LOC664 Mus musculus phosphatidylserine decarboxylase pseudogene (LOC664) NR_517.1 ILMN_7699 Gm NM_166 ILMN_ KRik NM_666.1 ILMN_6874 Echdc Mus musculus enoyl Coenzyme A hydratase domain containing (Echdc), mrna NM_ MRik; D4Ertd765e; 117C1Rik ILMN_1441 Slc8a Mus musculus solute carrier family 8, member (Slc8a), mrna NM_ mkiaa18; SAT; 54L14Rik; AI16867; ATA ILMN_6678 Neu Mus musculus neuraminidase (Neu), mrna NM_1575. MTS; MSS; MBS ILMN_64455 Trabd Mus musculus TraB domain containing (Trabd), mrna NM_6485. AL9; 575D15Rik ILMN_ Nr1d1 Mus musculus nuclear receptor subfamily 1, group D, member 1 (Nr1d1), mrna NM_ A57C9Rik; R751 ILMN_14899 Col16a1 Mus musculus procollagen, type XVI, alpha 1 (Col16a1), mrna NM_866.4 A55MRik; 77F1Rik; CA1F; AI8866 ILMN_15657 LOC1888 PREDICTED: Mus musculus hypothetical protein LOC1888 (LOC1888), mrna XM_ ILMN_646 Nptx Mus musculus neuronal pentraxin (Nptx), mrna NM_ Narp; np ILMN_1189 LOC1419 PREDICTED: Mus musculus hypothetical protein LOC1419 (LOC1419), mrna XM_ ILMN_1498 Npy Mus musculus neuropeptide Y (Npy), mrna NM_ A5Rik ILMN_ M1Rik NM_181.1 ILMN_6454 E18DRik Mus musculus RIKEN cdna E18D gene (E18DRik), mrna NM_1771. FLJ116 ILMN_ Ccdc11 Mus musculus coiled-coil domain containing 11 (Ccdc11), mrna. XM_ NM_161. BC596; Psrc ILMN_14967 Pdia4 Mus musculus protein disulfide isomerase associated 4 (Pdia4), mrna NM_9787. Erp7; AI987846; Cai ILMN_64915 Etv1 Mus musculus ets variant gene 1 (Etv1), mrna NM_796. Etsrp81; ER81 ILMN_481 Clk1 Mus musculus CDC-like kinase 1 (Clk1), transcript variant 1, mrna NM_ STY ILMN_6151 Ntnl Mus musculus netrin -like (chicken) (Ntnl), mrna NM_1947. Ntn ILMN_59974 Fndca Mus musculus fibronectin type III domain containing a (Fndca), mrna NM_766. D14Ertd45e; Fndc; 1794E19Rik; sys; F717H4Rik ILMN_1185 Tmem118 Mus musculus transmembrane protein 118 (Tmem118), mrna NM_ AW498; B88P19Rik ILMN_11911 Ccdc84 Mus musculus coiled-coil domain containing 84 (Ccdc84), mrna NM_17. Gm1114; D644F4Rik; MGC1685 ILMN_11615 Gmppa Mus musculus GDP-mannose pyrophosphorylase A (Gmppa), mrna NM_ N1Rik ILMN_1479 Per1 Mus musculus period homolog 1 (Drosophila) (Per1), mrna NM_1165. MGC111; Per; mper1; m-rigui ILMN_ Jam Mus musculus junction adhesion molecule (Jam), mrna NM_844.4 AU1617; 41167M4Rik; JAM-B; VE-JAM; 111NRik; 41G1Rik; JAM- ILMN_688 Oas1g Mus musculus '-5' oligoadenylate synthetase 1G (Oas1g), mrna NM_1185. Oas1a; Oias1; L; AI44956; Oias-1; Mmu-L; Mmu-L ILMN_76547 Luc7l Mus musculus LUC7-like (S. cerevisiae) (Luc7l), mrna NM_ CGI-74; AA51; 49471C18Rik; Cgi74; CGI-59; AU1569 ILMN_197 Inha Mus musculus inhibin alpha (Inha), mrna NM_ AW55578 ILMN_6914 Per Mus musculus period homolog (Drosophila) (Per), mrna NM_1167. mper; 8149O6Rik ILMN_186 Dcx Mus musculus doublecortin (Dcx), mrna NM_15. Dbct ILMN_14855 Samd9l PREDICTED: Mus musculus sterile alpha motif domain containing 9-like, transcript XM_686. ILMN_15689 Rgs9 Mus musculus regulator of G-protein signaling 9 (Rgs9), mrna NM_1168. RGS9-1; MGC1568; MGC1567; Rgs9- ILMN_ H6Rik Mus musculus RIKEN cdna 49546H6 gene (49546H6Rik), mrna NM_95. B6P6Rik ILMN_ Ergic Mus musculus ERGIC and golgi (Ergic), mrna NM_ NY-BR-84; 115B14Rik; DKFZp547A19; CGI-54; DUcla1; Sdbcag84; AV1884 ILMN_1194 Sdfl1 Mus musculus stromal cell-derived factor -like 1 (Sdfl1), mrna NM_4. ILMN_ Alox5ap NM_966 ILMN_1567 Ccnd1 Mus musculus cyclin D1 (Ccnd1), mrna NM_761. AI79; PRAD1; bcl-1; Cyl-1 ILMN_74177 Snurf Mus musculus SNRPN upstream reading frame (Snurf), mrna NM_174. Snrpn; MGC1864; MGC5; 4145I1Rik ILMN_666 Scotin Mus musculus scotin gene (Scotin), transcript variant, mrna NM_ I5Rik; 18D1Rik ILMN_ N4Rik XM_ ILMN_7185 Fhl1 Mus musculus four and a half LIM domains 1 (Fhl1), transcript variant 1, mrna NM_ KyoT ILMN_ A5Rik XM_897 ILMN_1875 Rasd PREDICTED: Mus musculus RASD family, member (Rasd), mrna XM_487.8 ILMN_669 Isgfg Mus musculus interferon dependent positive acting transcription factor gamma NM_894. Irf-9; p48 ILMN_ A7Rik Mus musculus RIKEN cdna 814A7 gene (814A7Rik), mrna NM_8814. AI451678; AI565; A416P18Rik ILMN_ Cbs Mus musculus cystathionine beta-synthase (Cbs), transcript variant 1, mrna NM_ MGC7; MGC18895; AI4754; AI44; MGC18856; HIP4 ILMN_148 Kif1 Mus musculus kinesin family member 1 (Kif1), mrna NM_1616. ILMN_ Tro Mus musculus trophinin (Tro), transcript variant 1, mrna NM_17. Magedl; Maged; Tnn; magphinin; AA4948; Trol ILMN_7566 C65L NM_ ILMN_7416 H1 Mus musculus histocompatibility 1 (H1), mrna NM_176. Spp; H-1; AV44; 4944L17Rik; 16O9Rik; 5144B4Rik; PSL ILMN_181 Dtxl Mus musculus deltex -like (Drosophila) (Dtxl), mrna NM_1171. MGC16; BC741; AU4 ILMN_684 Hes6 Mus musculus hairy and enhancer of split 6 (Drosophila) (Hes6), mrna NM_ AI689 ILMN_1467 Lefty1 Mus musculus left right determination factor 1 (Lefty1), mrna NM_194. lefty; Stra; lefty-1; Leftb; AI455; Tgfb4 ILMN_ Tubb5 Mus musculus tubulin, beta 5 (Tubb5), mrna NM_ AI59618; M(beta)5; AA4857; B1C14Rik ILMN_1556 Fkbp14 Mus musculus FK56 binding protein 14 (Fkbp14), mrna NM_ BC919; MGC1877; FKBP ILMN_77 Shmt Mus musculus serine hydroxymethyltransferase (mitochondrial) (Shmt), mrna NM_8. 74D8Rik; AA48; AA9869 ILMN_ JRik XM_ ILMN_1457 Ngef NM_19867 ILMN_ M6Rik PREDICTED: Mus musculus RIKEN cdna 5746M6 gene, transcript variant XM_954. ILMN_64117 Sult1d1 Mus musculus sulfotransferase family 1D, member 1 (Sult1d1), mrna NM_ P1Rik; AI987815; Sultn; ST1d1; SULT-N ILMN_77517 Clk4 Mus musculus CDC like kinase 4 (Clk4), mrna NM_ AI987988; C85119 ILMN_7116 Coq1b Mus musculus coenzyme Q1 homolog B (S. cerevisiae) (Coq1b), transcript variant NM_ I1Rik; AW74641; AV7; 9577A17Rik; 1541JRik ILMN_6641 Srm Mus musculus spermidine synthase (Srm), mrna NM_97.4 SpdSy; AA47669 ILMN_67959 Snrp7 Mus musculus U1 small nuclear ribonucleoprotein polypeptide A (Snrp7), mrna NM_94.4 Rnulp7; R7487; 7kDa; NRik; Srnp7; SNRNP7; 7N1Rik; U1-7 ILMN_ Slca17 Mus musculus solute carrier family (organic cation transporter), member NM_1551. BOIT; Boct; mboct; AW55566; AU4198; 1794CRik ILMN_5179 Impact Mus musculus imprinted and ancient (Impact), mrna NM_878. E416J11Rik ILMN_699 Mcfl Mus musculus mcf. transforming sequence-like (Mcfl), mrna NM_ mkiaa6; Ost; C14GRik; Ost gamma; Dbs ILMN_76765 BC181 Mus musculus cdna sequence BC181 (BC181), mrna NM_1458. MGC95; MGC818; MGC9 ILMN_764 BC79 Mus musculus cdna sequence BC79 (BC79), mrna NM_169. MGC58 ILMN_1745 Aox1 Mus musculus aldehyde oxidase 1 (Aox1), mrna NM_9676. Ro; Ao; MGC1774; AI555; Aox; Moro; AI19651; Aox-1; Aox- ILMN_14717 Tnrc15 Mus musculus trinucleotide repeat containing 15 (Tnrc15), mrna NM_ A88HRik; AW59676; 6116F1Rik; BC685; Gigyf; MGC8689; AI8561 ILMN_ P8Rik Mus musculus RIKEN cdna 1P8 gene (1P8Rik), mrna NM_61.1 Luc7a; RP-44C.4 ILMN_1474 Usp5 Mus musculus ubiquitin specific peptidase 5 (Usp5), mrna NM_199. AW7477; AI4784; 114O4Rik; mkiaa71 ILMN_1149 Scotin Mus musculus scotin gene (Scotin), transcript variant 1, mrna NM_ I5Rik; mshisa5; 18D1Rik ILMN_ Ivns1abp Mus musculus influenza virus NS1A binding protein (Ivns1abp), transcript variant NM_ I16Rik; Nd1-L; 1194M8Rik; Nd1-S; NS1-BP; NS-1; ND1; HSPC68; AA9644 ILMN_ K14Rik XM_ ILMN_665 Lgals4 Mus musculus lectin, galactose binding, soluble 4 (Lgals4), mrna NM_176.1 galectin-4 ILMN_14778 Ogt Mus musculus O-linked N-acetylglucosamine (GlcNAc) transferase NM_ P4Rik; 4814N1Rik; AI11555 ILMN_58866 Txndc11 Mus musculus thioredoxin domain containing 11 (Txndc11), transcript variant NM_ E11Rik; Txdc11; EFP1; AI478 ILMN_58955 Cpeb Mus musculus cytoplasmic polyadenylation element binding protein (Cpeb), mrna NM_198. KIAA94; mkiaa94; O18Rik ILMN_ Arfgap1 Mus musculus ADP-ribosylation factor GTPase activating protein 1 (Arfgap1), mrna NM_ Arf1gap; AI11577 ILMN_ Oasl Mus musculus '-5' oligoadenylate synthetase-like (Oasl), mrna NM_ Mmu-OASL; Oasl; M14 ILMN_68658 Atp6ap Mus musculus ATPase, H+ transporting, lysosomal accessory protein (Atp6ap) NM_749. M8-9; APT6M8-9; 574E6Rik; Atp6ip; ATP6M8-9 ILMN_ NRik Mus musculus RIKEN cdna 1616N gene (1616NRik), mrna NM_85.1 ILMN_6874 Col7a1 Mus musculus procollagen, type XXVII, alpha 1 (Col7a1), mrna NM_ JRik; mkiaa187; AI44966 ILMN_1875 Fkbp Mus musculus FK56 binding protein (Fkbp), mrna NM_8. mfkbp; 1kDa; mfkbp1 ILMN_65668 Fmnl Mus musculus formin-like (Fmnl), mrna NM_1749. Man; 5445K4Rik ILMN_79891 Cldn1 Mus musculus claudin 1 (Cldn1), mrna NM_ AI59671 ILMN_1878 H-T1 Mus musculus histocompatibility, T region locus 1 (H-T1), mrna NM_195.5 H-T1 ILMN_7457 Reln Mus musculus reelin (Reln), mrna NM_1161. reeler; rl ILMN_6461 Parp NM_ ILMN_19648 Tmed4 Mus musculus transmembrane emp4 protein transport domain containing 4 (Tmed4) NM_ L17Rik; RP-198N14.6; AI646 ILMN_7816 LOC67415 PREDICTED: Mus musculus similar to HLA-G protein (LOC67415), misc RNA XR_464. ILMN_16888 Rangap NM_1141 ILMN_ IRik Mus musculus RIKEN cdna 1711I gene (1711IRik), mrna NM_ ERik ILMN_64545 Erbbip Mus musculus Erbb interacting protein (Erbbip), transcript variant 1, mrna NM_ Erbin; 178E5Rik; mkiaa15 ILMN_14171 Safb Mus musculus scaffold attachment factor B (Safb), mrna NM_ AA894; mkiaa18; AI5517 ILMN_4497 Tnrc6a Mus musculus trinucleotide repeat containing 6a (Tnrc6a), mrna NM_ GW18; D1A7Rik; Tnrc6; MGC119; 11I5Rik; CAGH6; 1154G1Rik ILMN_ N11Rik NM_5888. ILMN_19455 Rnf1 Mus musculus ring finger protein 1 (Rnf1), mrna NM_145. D11Ertd759e; 64J1; mkiaa1554 ILMN_79 Gbp6 Mus musculus guanylate binding protein 6 (Gbp6), mrna NM_181.1 Gbp7; 98147J4Rik ILMN_17149 Gtl Mus musculus GTL, imprinted maternally expressed untranslated mrna (Gtl) NR_6.1 Meg; D1Bwg166e; 115O7Rik; AW184; 916C5Rik; 648G6Rik ILMN_469 LOC66 PREDICTED: Mus musculus hypothetical LOC66 (LOC66), mrna XM_ ILMN_ Hnrph1 Mus musculus heterogeneous nuclear ribonucleoprotein H1 (Hnrph1), mrna NM_151. Hnrnph; AI648; E45G16Rik ILMN_ Tspan6 Mus musculus tetraspanin 6 (Tspan6), mrna NM_ AI16786; T45; Tm4sf6; TSPAN-6; 6747L1Rik; MGC1179; Tm4sf ILMN_18649 Rbm1 Mus musculus RNA binding motif protein 1 (Rbm1), mrna NM_ E49K1Rik; mkiaa1; MGC786 ILMN_6557 Ctnnd Mus musculus catenin (cadherin associated protein), delta (Ctnnd), mrna NM_879. Catnd; Nprap; neurojugin ILMN_ HRik Mus musculus RIKEN cdna 1719H gene (1719HRik), mrna NM_ MGC858; BC1957 ILMN_64787 Nav1 Mus musculus neuron navigator 1 (Nav1), mrna NM_ ARik; POMFIL; mkiaa1151; unc5h1; 9589B19; C8M11Rik; steerin-1 ILMN_71717 Ddx58 Mus musculus DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 (Ddx58), mrna NM_ C1E1; 6457DRik; RIG-I ILMN_655 Tmed Mus musculus transmembrane emp4 domain containing (Tmed), mrna NM_56.1 1G1Rik; P4b; AW54667 ILMN_1151 Gsdm1 Mus musculus gasdermin 1 (Gsdm1), mrna NM_147. Gsdm; BB149167; H1E; Gsdma1 ILMN_14 Immpl Mus musculus IMP inner mitochondrial membrane peptidase-like (S. cerevisiae) NM_51. AI8588; IMP ILMN_69967 Lsr Mus musculus lipolysis stimulated lipoprotein receptor (Lsr), mrna NM_ Lisch7 ILMN_ Gpr18 Mus musculus G protein-coupled receptor 18 (Gpr18), mrna NM_84. C791; AA589464; 18115L19Rik ILMN_64891 Car11 Mus musculus carbonic anhydrase 11 (Car11), mrna NM_98.4 ILMN_67888 Eif5a Mus musculus eukaryotic translation initiation factor 5A (Eif5a), mrna NM_ uorf; MGC675; uorf; uorf; D19Wsu54e; AA4158; uorf1; Eif4d ILMN_7156 Pfkfb Mus musculus 6-phosphofructo--kinase/fructose-,6-biphosphatase (Pfkfb), mrna NM_1. ipfk-; E1HRik ILMN_6 Heg1 Mus musculus HEG homolog 1 (zebrafish) (Heg1), transcript variant, mrna NM_ BC8614; 5541IRik; 46417DRik; 955L16Rik; Gm69 ILMN_7698 Ppox Mus musculus protoporphyrinogen oxidase (Ppox), mrna NM_8911. Ppo ILMN_68588 Gdpd Mus musculus glycerophosphodiester phosphodiesterase domain containing (Gdpd) NM_ ERik ILMN_7549 AW1464 Mus musculus expressed sequence AW1464 (AW1464), mrna NM_ ILMN_14678 Safb PREDICTED: Mus musculus scaffold attachment factor B, transcript variant XM_ ILMN_6555 Wnk1 Mus musculus WNK lysine deficient protein kinase 1 (Wnk1), mrna NM_ Prkwnk1; mkiaa44; 6457HRik ILMN_187 Pib5pa Mus musculus phosphatidylinositol (4,5) bisphosphate 5-phosphatase, A (Pib5pa) NM_1749. Pipp ILMN_1554 Dapp1 Mus musculus dual adaptor for phosphotyrosine and -phosphoinositides 1 (Dapp1) NM_119. Bam ILMN_6549 Rab9b Mus musculus RAB9B, member RAS oncogene family (Rab9b), mrna NM_ MGC18; 9195CRik ILMN_6519 Wdr9 PREDICTED: Mus musculus WD repeat domain 9, transcript variant 1 (Wdr9), mrna XM_ ILMN_111 Arl5a Mus musculus ADP-ribosylation factor-like 5A (Arl5a), mrna NM_ T554; AW61751; 4115N4Rik; Arl5; 8141PRik; AA4871 ILMN_14 LOC PREDICTED: Mus musculus similar to synaptotagmin XI (LOC145981), mrna XM_ ILMN_1517 Dad1 Mus musculus defender against cell death 1 (Dad1), mrna NM_115. AI71 ILMN_18119 Itgav Mus musculus integrin alpha V (Itgav), mrna NM_ F14Rik; 618E1Rik; CD51; D44G1Rik ILMN_15671 Fez1 Mus musculus fasciculation and elongation protein zeta 1 (zygin I) (Fez1), mrna NM_ UNC-76; UNC76; MGC199 ILMN_65791 LOC67596 PREDICTED: Mus musculus similar to RIKEN cdna 915E1 gene (LOC67596), mrna XM_ ILMN_ Tmem59l Mus musculus transmembrane protein 59-like (Tmem59l), mrna NM_ G16Rik ILMN_ Rnf6 Mus musculus ring finger protein (CHC type) 6 (Rnf6), mrna NM_8774. AA575; 57419H5Rik; 11I8Rik ILMN_ Tapbpl Mus musculus TAP binding protein-like (Tapbpl), mrna NM_ BC1761; TAPBPL-R; MGC7679 ILMN_18 Parp9 Mus musculus poly (ADP-ribose) polymerase family, member 9 (Parp9), mrna NM_5. MGC7868; Bagl; AW1446; Bal; BC81
23 ILMN_6196 Bfar Mus musculus bifunctional apoptosis regulator (Bfar), mrna NM_5976. BAR; AW17665; 11A7Rik; AI66677; RNF47 ILMN_475 Zch7a Mus musculus zinc finger CCCH type containing 7 A (Zch7a), mrna NM_ AI44794; HSPC55; Zchdc7; A414C18Rik; Zch7; AW55619 ILMN_451 Zfp179 Mus musculus zinc finger protein 179 (Zfp179), mrna NM_9548. bfp; ZNF179; R75574; AI85496 ILMN_ Zfp9 PREDICTED: Mus musculus zinc finger protein 9, transcript variant 4 (Zfp9) XM_ ILMN_ Il15ra Mus musculus interleukin 15 receptor, alpha chain (Il15ra), transcript variant NM_186.1 AA69181 ILMN_77691 Mobkl Mus musculus MOB1, Mps One Binder kinase activator-like (yeast) (Mobkl), mrna NM_ B1Rik; Prei ILMN_ M1Rik Mus musculus RIKEN cdna 57469M1 gene (57469M1Rik), mrna NM_7464. AU48 ILMN_11868 Bicd1 Mus musculus bicaudal D homolog 1 (Drosophila) (Bicd1), mrna NM_975.1 B89D6Rik; mkiaa415; KIAA415 ILMN_148 LOC64746 PREDICTED: Mus musculus similar to tripartite motif protein TRIM4 delta XM_ ILMN_67 Pfkp Mus musculus phosphofructokinase, platelet (Pfkp), mrna NM_197. PFK-C; 915N4Rik; 115HRik ILMN_677 Tceal5 Mus musculus transcription elongation factor A (SII)-like 5 (Tceal5), mrna NM_ A5Rik ILMN_641 Hook1 Mus musculus hook homolog 1 (Drosophila) (Hook1), mrna NM_14. azh; A9L17Rik ILMN_14566 Cep17 Mus musculus centrosomal protein 17 (Cep17), mrna NM_ AI1955; 4946LRik; A4A1Rik ILMN_66 Gnas Mus musculus GNAS (guanine nucleotide binding protein, alpha stimulating) NM_. Gnasxl; P; PHP1A; P1; GPSA; A97G11Rik; P; PHP1B; POH; Oedsml ILMN_77 LOC147 PREDICTED: Mus musculus similar to phosphodiesterase B (LOC147), mrna XM_ ILMN_ F6Rik XM_111.5 ILMN_ B5Rik Mus musculus RIKEN cdna 911B5 gene (911B5Rik), mrna NM_ W91666; 11L8Rik Table S1: Significantly downregulated genes in 8 month old mutant islets based on microarray analysis. Shown are genes detected on the array with fold change > 1.4 and p <.5.
Table S1 Differentially expressed genes showing > 2 fold changes and p <0.01 for 0.01 mm NS-398, 0.1mM ibuprofen and COX-2 RNAi.
 Table S1 Differentially expressed genes showing > 2 fold changes and p
Table S1 Differentially expressed genes showing > 2 fold changes and p
Supplementary Information
 Scientific Reports Supplementary Information Upregulated expression of FGF13/FHF2 mediates resistance to platinum drugs in cervical cancer cells Tomoko Okada, Kazuhiro Murata, Ryoma Hirose, Chie Matsuda,
Scientific Reports Supplementary Information Upregulated expression of FGF13/FHF2 mediates resistance to platinum drugs in cervical cancer cells Tomoko Okada, Kazuhiro Murata, Ryoma Hirose, Chie Matsuda,
Table 1A. Genes enriched as over-expressed in DPM treatment group
 Table 1A. s enriched as over-expressed in DPM treatment group Affy Probeset mrna Accession Description 10538247 Npy NM_023456 Mus musculus neuropeptide Y (Npy), mrna. 2.0024 10598062 --- NC_005089 gi 34538597
Table 1A. s enriched as over-expressed in DPM treatment group Affy Probeset mrna Accession Description 10538247 Npy NM_023456 Mus musculus neuropeptide Y (Npy), mrna. 2.0024 10598062 --- NC_005089 gi 34538597
control kda ATGL ATGLi HSL 82 GAPDH * ** *** WT/cTg WT/cTg ATGLi AKO/cTg AKO/cTg ATGLi WT/cTg WT/cTg ATGLi AKO/cTg AKO/cTg ATGLi iwat gwat ibat
 body weight (g) tissue weights (mg) ATGL protein expression (relative to GAPDH) HSL protein expression (relative to GAPDH) ### # # kda ATGL 55 HSL 82 GAPDH 37 2.5 2. 1.5 1..5 2. 1.5 1..5.. Supplementary
body weight (g) tissue weights (mg) ATGL protein expression (relative to GAPDH) HSL protein expression (relative to GAPDH) ### # # kda ATGL 55 HSL 82 GAPDH 37 2.5 2. 1.5 1..5 2. 1.5 1..5.. Supplementary
Supplementary Table 1. Primer sequences for conventional RT-PCR on mouse islets
 Supplementary Table 1. Primer sequences for conventional RT-PCR on mouse islets Gene 5 Forward 3 5 Reverse 3.T. Product (bp) ( C) mnox1 GTTCTTGGGCTGCCTTGG GCTGGGGCGGCGG 60 300 mnoxa1 GCTTTGCCGCGTGC GGTTCGGGTCCTTTGTGC
Supplementary Table 1. Primer sequences for conventional RT-PCR on mouse islets Gene 5 Forward 3 5 Reverse 3.T. Product (bp) ( C) mnox1 GTTCTTGGGCTGCCTTGG GCTGGGGCGGCGG 60 300 mnoxa1 GCTTTGCCGCGTGC GGTTCGGGTCCTTTGTGC
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
KEY CONCEPT QUESTIONS IN SIGNAL TRANSDUCTION
 Signal Transduction - Part 2 Key Concepts - Receptor tyrosine kinases control cell metabolism and proliferation Growth factor signaling through Ras Mutated cell signaling genes in cancer cells are called
Signal Transduction - Part 2 Key Concepts - Receptor tyrosine kinases control cell metabolism and proliferation Growth factor signaling through Ras Mutated cell signaling genes in cancer cells are called
Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion
 Probe Set ID RefSeq Transcript ID Gene Title Gene Symbol 205980_s_at NM_001017526 /// NM_181334 /// NM_181335 Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion ARHGAP8 /// LOC553158 FC ALK shrna
Probe Set ID RefSeq Transcript ID Gene Title Gene Symbol 205980_s_at NM_001017526 /// NM_181334 /// NM_181335 Rho GTPase activating protein 8 /// PRR5- ARHGAP8 fusion ARHGAP8 /// LOC553158 FC ALK shrna
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Table SI: Y strain T. cruzi infection results in the upregulation of 381 genes at the site of infection.
 Supplementary Table SI: Y strain T. cruzi infection results in the upregulation of 381 genes at the site of infection. Genes found to be significantly upregulated (FDR2) in Y strain
Supplementary Table SI: Y strain T. cruzi infection results in the upregulation of 381 genes at the site of infection. Genes found to be significantly upregulated (FDR2) in Y strain
General Laboratory methods Plasma analysis: Gene Expression Analysis: Immunoblot analysis: Immunohistochemistry:
 General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Figure S1 (A) Comparison of body weight and blood glucose levels in male wild-type (WT) and Cre littermates. Mice were maintained on a normal chow
 Figure S1 (A) Comparison of body weight and blood glucose levels in male wild-type (WT) and Cre littermates. Mice were maintained on a normal chow diet (n = 7 per genotype). Body weight and fed blood glucose
Figure S1 (A) Comparison of body weight and blood glucose levels in male wild-type (WT) and Cre littermates. Mice were maintained on a normal chow diet (n = 7 per genotype). Body weight and fed blood glucose
SUPPLEMENTARY DATA. Supplementary Table 1. Primers used for PCR and qpcr Primer Name
 Supplementary Table. Primers used for PCR and qpcr Primer Name ccession Number Fwd Rev Type of PCR Cre NC_8 GGCGTCTTCCGC GTGCCCCTCGTTTG Standard PCR LoUcp CCGGGCTGTCTCCGCGG GGCTGTTCGCCCGGCC Standard PCR
Supplementary Table. Primers used for PCR and qpcr Primer Name ccession Number Fwd Rev Type of PCR Cre NC_8 GGCGTCTTCCGC GTGCCCCTCGTTTG Standard PCR LoUcp CCGGGCTGTCTCCGCGG GGCTGTTCGCCCGGCC Standard PCR
TNFSF13B tumor necrosis factor (ligand) superfamily, member 13b NF-kB pathway cluster, Enrichment Score: 3.57
 Appendix 2. Highly represented clusters of genes in the differential expression of data. Immune Cluster, Enrichment Score: 5.17 GO:0048584 positive regulation of response to stimulus GO:0050778 positive
Appendix 2. Highly represented clusters of genes in the differential expression of data. Immune Cluster, Enrichment Score: 5.17 GO:0048584 positive regulation of response to stimulus GO:0050778 positive
SUPPLEMENTARY DATA. Supplementary Table 1. Primers used in qpcr
 Supplementary Table 1. Primers used in qpcr Gene forward primer (5'-3') reverse primer (5'-3') β-actin AGAGGGAAATCGTGCGTGAC CAATAGTGATGACCTGGCCGT Hif-p4h-2 CTGGGCAACTACAGGATAAAC GCGTCCCAGTCTTTATTTAGATA
Supplementary Table 1. Primers used in qpcr Gene forward primer (5'-3') reverse primer (5'-3') β-actin AGAGGGAAATCGTGCGTGAC CAATAGTGATGACCTGGCCGT Hif-p4h-2 CTGGGCAACTACAGGATAAAC GCGTCCCAGTCTTTATTTAGATA
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism SUPPLEMENTARY FIGURES, LEGENDS AND METHODS
 A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
U118MG. Supplementary Figure 1 U373MG U118MG 3.5 A CCF-SSTG
 A172 CCF-SSTG1 15 - - - 1 1 1 2 2 3 4 6 7 8 10101112131617192022-1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22 T98G U373MG - - - 1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22-1 1 1 2 2 3 4 6 7
A172 CCF-SSTG1 15 - - - 1 1 1 2 2 3 4 6 7 8 10101112131617192022-1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22 T98G U373MG - - - 1 1 1 2 2 3 4 6 7 8 10 10 11 12 13 16 17 19 20 22-1 1 1 2 2 3 4 6 7
Cell Quality Control. Peter Takizawa Department of Cell Biology
 Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Supplementary Figure 1.
 Supplementary Figure 1. Increased β cell mass and islet diameter in βtsc2 -/- mice up to 35 weeks A: Reconstruction of multiple anti-insulin immunofluorescence images showing differences in β cell mass
Supplementary Figure 1. Increased β cell mass and islet diameter in βtsc2 -/- mice up to 35 weeks A: Reconstruction of multiple anti-insulin immunofluorescence images showing differences in β cell mass
Supplementary Figure 1 Validation of Per2 deletion in neuronal cells in N Per2 -/- mice. (a) Western blot from liver extracts of mice held under ad
 Supplementary Figure 1 Validation of Per2 deletion in neuronal cells in N Per2 -/- mice. (a) Western blot from liver extracts of mice held under ad libitum conditions detecting PER2 protein in brain and
Supplementary Figure 1 Validation of Per2 deletion in neuronal cells in N Per2 -/- mice. (a) Western blot from liver extracts of mice held under ad libitum conditions detecting PER2 protein in brain and
Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction. Marker Sequence (5 3 ) Accession No.
 Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Supplementary Tables Supplementary Table S1. Primers used for quantitative real-time polymerase chain reaction Marker Sequence (5 3 ) Accession No. Angiopoietin 1, ANGPT1 A CCCTCCGGTGAATATTGGCTGG NM_001146.3
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array.
 Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Signal Transduction Cascades
 Signal Transduction Cascades Contents of this page: Kinases & phosphatases Protein Kinase A (camp-dependent protein kinase) G-protein signal cascade Structure of G-proteins Small GTP-binding proteins,
Signal Transduction Cascades Contents of this page: Kinases & phosphatases Protein Kinase A (camp-dependent protein kinase) G-protein signal cascade Structure of G-proteins Small GTP-binding proteins,
Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle.
 Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle. (a) mrna levels of Dj1 measured by quantitative RT-PCR in soleus, gastrocnemius (Gastroc.) and extensor digitorum longus
Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle. (a) mrna levels of Dj1 measured by quantitative RT-PCR in soleus, gastrocnemius (Gastroc.) and extensor digitorum longus
Homework Hanson section MCB Course, Fall 2014
 Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
Homework Hanson section MCB Course, Fall 2014 (1) Antitrypsin, which inhibits certain proteases, is normally secreted into the bloodstream by liver cells. Antitrypsin is absent from the bloodstream of
CIRCADIAN SIGNALING NETWORKS
 Transcription Regulation And Gene Expression in Eukaryotes Cycle G2 (lecture 13709) FS 2014 P. Matthias and RG Clerc Roger G. Clerc 07.05.2014 CIRCADIAN SIGNALING NETWORKS Master pacemaker SCN «Slave clocks»
Transcription Regulation And Gene Expression in Eukaryotes Cycle G2 (lecture 13709) FS 2014 P. Matthias and RG Clerc Roger G. Clerc 07.05.2014 CIRCADIAN SIGNALING NETWORKS Master pacemaker SCN «Slave clocks»
Supplementary Information Titles Journal: Nature Medicine
 Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
GPR120 *** * * Liver BAT iwat ewat mwat Ileum Colon. UCP1 mrna ***
 a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
A Central Role of MG53 in Metabolic Syndrome. and Type-2 Diabetes
 A Central Role of MG53 in Metabolic Syndrome and Type-2 Diabetes Yan Zhang, Chunmei Cao, Rui-Ping Xiao Institute of Molecular Medicine (IMM) Peking University, Beijing, China Accelerated Aging in China
A Central Role of MG53 in Metabolic Syndrome and Type-2 Diabetes Yan Zhang, Chunmei Cao, Rui-Ping Xiao Institute of Molecular Medicine (IMM) Peking University, Beijing, China Accelerated Aging in China
Supplementary Figure 1
 Supplementary Figure 1 a Percent of body weight! (%) 4! 3! 1! Epididymal fat Subcutaneous fat Liver SD Percent of body weight! (%) ** 3! 1! SD Percent of body weight! (%) 6! 4! SD ** b Blood glucose (mg/dl)!
Supplementary Figure 1 a Percent of body weight! (%) 4! 3! 1! Epididymal fat Subcutaneous fat Liver SD Percent of body weight! (%) ** 3! 1! SD Percent of body weight! (%) 6! 4! SD ** b Blood glucose (mg/dl)!
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Supplemental methods. Total RNA was extracted from the stomach, liver, pancreas, pituitary, and
 Supplemental methods Real-time quantitative RT-PCR and Semi-quantitative PCR Total RNA was extracted from the stomach, liver, pancreas, pituitary, and hypothalamus as previously described (). Primers and
Supplemental methods Real-time quantitative RT-PCR and Semi-quantitative PCR Total RNA was extracted from the stomach, liver, pancreas, pituitary, and hypothalamus as previously described (). Primers and
Supporting Information
 Supporting Information Franco et al. 10.1073/pnas.1015557108 SI Materials and Methods Drug Administration. PD352901 was dissolved in 0.5% (wt/vol) hydroxyl-propyl-methylcellulose, 0.2% (vol/vol) Tween
Supporting Information Franco et al. 10.1073/pnas.1015557108 SI Materials and Methods Drug Administration. PD352901 was dissolved in 0.5% (wt/vol) hydroxyl-propyl-methylcellulose, 0.2% (vol/vol) Tween
Enzyme-coupled Receptors. Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors
 Enzyme-coupled Receptors Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors Cell-surface receptors allow a flow of ions across the plasma
Enzyme-coupled Receptors Cell-surface receptors 1. Ion-channel-coupled receptors 2. G-protein-coupled receptors 3. Enzyme-coupled receptors Cell-surface receptors allow a flow of ions across the plasma
Supplementary Table 1 Gene clone ID for ShRNA-mediated gene silencing TNFα downstream signals in in vitro Symbol Gene ID RefSeqID Clone ID
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 Supplementary Table 1 Gene clone ID for ShRNA-mediated gene silencing TNFα downstream
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 Supplementary Table 1 Gene clone ID for ShRNA-mediated gene silencing TNFα downstream
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at
 Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Targeting of the circadian clock via CK1δ/ε to improve glucose homeostasis in obesity
 Targeting of the circadian clock via CK1δ/ε to improve glucose homeostasis in obesity Peter S. Cunningham, Siobhán A. Ahern, Laura C. Smith, Carla S. da Silva Santos, Travis T. Wager and David A. Bechtold
Targeting of the circadian clock via CK1δ/ε to improve glucose homeostasis in obesity Peter S. Cunningham, Siobhán A. Ahern, Laura C. Smith, Carla S. da Silva Santos, Travis T. Wager and David A. Bechtold
SUPPLEMENTARY DATA. Supplementary experimental procedures
 Supplementary experimental procedures Glucose tolerance, insulin secretion and insulin tolerance tests Glucose tolerance test (GTT) and glucose stimulated insulin secretion (GSIS) was measured in over-night
Supplementary experimental procedures Glucose tolerance, insulin secretion and insulin tolerance tests Glucose tolerance test (GTT) and glucose stimulated insulin secretion (GSIS) was measured in over-night
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
G-Protein Signaling. Introduction to intracellular signaling. Dr. SARRAY Sameh, Ph.D
 G-Protein Signaling Introduction to intracellular signaling Dr. SARRAY Sameh, Ph.D Cell signaling Cells communicate via extracellular signaling molecules (Hormones, growth factors and neurotransmitters
G-Protein Signaling Introduction to intracellular signaling Dr. SARRAY Sameh, Ph.D Cell signaling Cells communicate via extracellular signaling molecules (Hormones, growth factors and neurotransmitters
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein
 Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein Lei Wang 1, Tian-Peng Zhang 1, Yuan Zhang 2, Hai-Lian
Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein Lei Wang 1, Tian-Peng Zhang 1, Yuan Zhang 2, Hai-Lian
SUPPLEMENTARY DATA. Assessment of cell survival (MTT) Assessment of early apoptosis and cell death
 Overexpression of a functional calcium-sensing receptor dramatically increases osteolytic potential of MDA-MB-231 cells in a mouse model of bone metastasis through epiregulinmediated osteoprotegerin downregulation
Overexpression of a functional calcium-sensing receptor dramatically increases osteolytic potential of MDA-MB-231 cells in a mouse model of bone metastasis through epiregulinmediated osteoprotegerin downregulation
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system
 Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Cell Biology Lecture 9 Notes Basic Principles of cell signaling and GPCR system Basic Elements of cell signaling: Signal or signaling molecule (ligand, first messenger) o Small molecules (epinephrine,
Supplemental Information. Tissue Myeloid Progenitors Differentiate. into Pericytes through TGF-b Signaling. in Developing Skin Vasculature
 Cell Reports, Volume 18 Supplemental Information Tissue Myeloid Progenitors Differentiate into Pericytes through TGF-b Signaling in Developing Skin Vasculature Tomoko Yamazaki, Ani Nalbandian, Yutaka Uchida,
Cell Reports, Volume 18 Supplemental Information Tissue Myeloid Progenitors Differentiate into Pericytes through TGF-b Signaling in Developing Skin Vasculature Tomoko Yamazaki, Ani Nalbandian, Yutaka Uchida,
Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR
 Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) cadl CTTGGGGGCGCGTCT CTGTTCTTTTGTGCCGTTTCG cyl-coenzyme Dehydrogenase, very
Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) cadl CTTGGGGGCGCGTCT CTGTTCTTTTGTGCCGTTTCG cyl-coenzyme Dehydrogenase, very
Supporting Information
 Supporting Information Pang et al. 10.1073/pnas.1322009111 SI Materials and Methods ELISAs. These assays were performed as previously described (1). ELISA plates (MaxiSorp Nunc; Thermo Fisher Scientific)
Supporting Information Pang et al. 10.1073/pnas.1322009111 SI Materials and Methods ELISAs. These assays were performed as previously described (1). ELISA plates (MaxiSorp Nunc; Thermo Fisher Scientific)
Figure legends Supplemental Fig.1. Glucose-induced insulin secretion and insulin content of islets. Supplemental Fig. 2.
 Figure legends Supplemental Fig.. Glucose-induced insulin secretion and insulin content of islets. Insulin secretory responses to.,., and. mm glucose (A) (n = 7-), and the insulin content in the islets
Figure legends Supplemental Fig.. Glucose-induced insulin secretion and insulin content of islets. Insulin secretory responses to.,., and. mm glucose (A) (n = 7-), and the insulin content in the islets
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Transcription Regulation And Gene Expression in Eukaryotes (Cycle G2 # )
 Transcription Regulation And Gene Expression in Eukaryotes (Cycle G2 #13709-01) CIRCADIAN SIGNALING NETWORKS RG. Clerc May 19. 2010 www.fmi.ch/training/teaching Circadian rythms : most physiological processes
Transcription Regulation And Gene Expression in Eukaryotes (Cycle G2 #13709-01) CIRCADIAN SIGNALING NETWORKS RG. Clerc May 19. 2010 www.fmi.ch/training/teaching Circadian rythms : most physiological processes
p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO
 Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
What would you observe if you fused a G1 cell with a S cell? A. Mitotic and pulverized chromosomes. B. Mitotic and compact G1 chromosomes.
 What would you observe if you fused a G1 cell with a S cell? A. Mitotic and pulverized chromosomes. B. Mitotic and compact G1 chromosomes. C. Mostly non-compact G1 chromosomes. D. Compact G1 and G2 chromosomes.
What would you observe if you fused a G1 cell with a S cell? A. Mitotic and pulverized chromosomes. B. Mitotic and compact G1 chromosomes. C. Mostly non-compact G1 chromosomes. D. Compact G1 and G2 chromosomes.
SUPPLEMENTARY FIG. S2. b-galactosidase staining of
 SUPPLEMENTARY FIG. S1. b-galactosidase staining of senescent cells in 500 mg=dl glucose (10 magnification). SUPPLEMENTARY FIG. S3. Toluidine blue staining of chondrogenic-differentiated adipose-tissue-derived
SUPPLEMENTARY FIG. S1. b-galactosidase staining of senescent cells in 500 mg=dl glucose (10 magnification). SUPPLEMENTARY FIG. S3. Toluidine blue staining of chondrogenic-differentiated adipose-tissue-derived
1. endoplasmic reticulum This is the location where N-linked oligosaccharide is initially synthesized and attached to glycoproteins.
 Biology 4410 Name Spring 2006 Exam 2 A. Multiple Choice, 2 pt each Pick the best choice from the list of choices, and write it in the space provided. Some choices may be used more than once, and other
Biology 4410 Name Spring 2006 Exam 2 A. Multiple Choice, 2 pt each Pick the best choice from the list of choices, and write it in the space provided. Some choices may be used more than once, and other
Requires Signaling though Akt2 Independent of the. Transcription Factors FoxA2, FoxO1, and SREBP1c
 Cell Metabolism, Volume 14 Supplemental Information Postprandial Hepatic Lipid Metabolism Requires Signaling though Akt2 Independent of the Transcription Factors FoxA2, FoxO1, and SREBP1c Min Wan, Karla
Cell Metabolism, Volume 14 Supplemental Information Postprandial Hepatic Lipid Metabolism Requires Signaling though Akt2 Independent of the Transcription Factors FoxA2, FoxO1, and SREBP1c Min Wan, Karla
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
Supplementary Information. Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase
 Journal: Nature Medicine Supplementary Information Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase 1,2 Peng Wang PhD, 1,2 Juan-Carlos Alvarez-Perez
Journal: Nature Medicine Supplementary Information Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase 1,2 Peng Wang PhD, 1,2 Juan-Carlos Alvarez-Perez
Supplementary Information for
 Supplementary Information for Involvement of urinary bladder Connexin43 and the circadian clock in the coordination of diurnal micturition rhythm Hiromitsu Negoro, 1,2 Akihiro Kanematsu, 1,3 Masao Doi,
Supplementary Information for Involvement of urinary bladder Connexin43 and the circadian clock in the coordination of diurnal micturition rhythm Hiromitsu Negoro, 1,2 Akihiro Kanematsu, 1,3 Masao Doi,
Signal Transduction: G-Protein Coupled Receptors
 Signal Transduction: G-Protein Coupled Receptors Federle, M. (2017). Lectures 4-5: Signal Transduction parts 1&2: nuclear receptors and GPCRs. Lecture presented at PHAR 423 Lecture in UIC College of Pharmacy,
Signal Transduction: G-Protein Coupled Receptors Federle, M. (2017). Lectures 4-5: Signal Transduction parts 1&2: nuclear receptors and GPCRs. Lecture presented at PHAR 423 Lecture in UIC College of Pharmacy,
Supplementary Figure 1. Mitochondrial function in skeletal muscle and plasma parameters of STZ mice A D-F
 Supplementary Figure 1. Mitochondrial function in skeletal muscle and plasma parameters of STZ mice A Expression of representative subunits of the mitochondrial respiratory chain was analyzed by real time
Supplementary Figure 1. Mitochondrial function in skeletal muscle and plasma parameters of STZ mice A Expression of representative subunits of the mitochondrial respiratory chain was analyzed by real time
Mouse Meda-4 : chromosome 5G bp. EST (547bp) _at. 5 -Meda4 inner race (~1.8Kb)
 Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
Hormones. Prof. Dr. Volker Haucke Institut für Chemie-Biochemie Takustrasse 6
 Hormones Prof. Dr. Volker Haucke Institut für Chemie-Biochemie Takustrasse 6 Tel. 030-8385-6920 (Sekret.) 030-8385-6922 (direkt) e-mail: vhaucke@chemie.fu-berlin.de http://userpage.chemie.fu-berlin.de/biochemie/aghaucke/teaching.html
Hormones Prof. Dr. Volker Haucke Institut für Chemie-Biochemie Takustrasse 6 Tel. 030-8385-6920 (Sekret.) 030-8385-6922 (direkt) e-mail: vhaucke@chemie.fu-berlin.de http://userpage.chemie.fu-berlin.de/biochemie/aghaucke/teaching.html
Cell Physiology Final Exam Fall 2008
 Cell Physiology Final Exam Fall 2008 Guys, The average on the test was 69.9. Before you start reading the right answers please do me a favor and remember till the end of your life that GLUCOSE TRANSPORT
Cell Physiology Final Exam Fall 2008 Guys, The average on the test was 69.9. Before you start reading the right answers please do me a favor and remember till the end of your life that GLUCOSE TRANSPORT
Lecture: CHAPTER 13 Signal Transduction Pathways
 Lecture: 10 17 2016 CHAPTER 13 Signal Transduction Pathways Chapter 13 Outline Signal transduction cascades have many components in common: 1. Release of a primary message as a response to a physiological
Lecture: 10 17 2016 CHAPTER 13 Signal Transduction Pathways Chapter 13 Outline Signal transduction cascades have many components in common: 1. Release of a primary message as a response to a physiological
Supplemental Information. Otic Mesenchyme Cells Regulate. Spiral Ganglion Axon Fasciculation. through a Pou3f4/EphA4 Signaling Pathway
 Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Endocannabinoid-activated Nlrp3 inflammasome in infiltrating macrophages mediates β- cell loss in type 2 diabetes
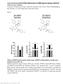 Endocannabinoid-activated Nlrp3 inflammasome in infiltrating macrophages mediates β- cell loss in type 2 diabetes T Jourdan, G Godlewski, R Cinar, A Bertola, G Szanda, J Liu, J Tam, T Han, B Mukhopadhyay,
Endocannabinoid-activated Nlrp3 inflammasome in infiltrating macrophages mediates β- cell loss in type 2 diabetes T Jourdan, G Godlewski, R Cinar, A Bertola, G Szanda, J Liu, J Tam, T Han, B Mukhopadhyay,
Supporting Information
 Supporting Information Soltani et al. 10.1073/pnas.1102715108 SI Experimental Procedures Evaluation of Mice and Drug Administration. IPGTT, insulin tolerance test, and glucagon tolerance test were performed
Supporting Information Soltani et al. 10.1073/pnas.1102715108 SI Experimental Procedures Evaluation of Mice and Drug Administration. IPGTT, insulin tolerance test, and glucagon tolerance test were performed
ENERGY FROM INGESTED NUTREINTS MAY BE USED IMMEDIATELY OR STORED
 QUIZ/TEST REVIEW NOTES SECTION 1 SHORT TERM METABOLISM [METABOLISM] Learning Objectives: Identify primary energy stores of the body Differentiate the metabolic processes of the fed and fasted states Explain
QUIZ/TEST REVIEW NOTES SECTION 1 SHORT TERM METABOLISM [METABOLISM] Learning Objectives: Identify primary energy stores of the body Differentiate the metabolic processes of the fed and fasted states Explain
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus
 Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Receptor mediated Signal Transduction
 Receptor mediated Signal Transduction G-protein-linked receptors adenylyl cyclase camp PKA Organization of receptor protein-tyrosine kinases From G.M. Cooper, The Cell. A molecular approach, 2004, third
Receptor mediated Signal Transduction G-protein-linked receptors adenylyl cyclase camp PKA Organization of receptor protein-tyrosine kinases From G.M. Cooper, The Cell. A molecular approach, 2004, third
hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This
 SUPPLEMENTAL FIGURE LEGEND Fig. S1. Generation and characterization of. (A) Coomassie staining of soluble hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This protein was expressed
SUPPLEMENTAL FIGURE LEGEND Fig. S1. Generation and characterization of. (A) Coomassie staining of soluble hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This protein was expressed
AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination
 AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination Supplementary Figure 1. Generation of the adult-onset, liver-specific GH receptor knock-down (alivghrkd, Kd) mouse
AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination Supplementary Figure 1. Generation of the adult-onset, liver-specific GH receptor knock-down (alivghrkd, Kd) mouse
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.
 Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
ACC ELOVL MCAD. CPT1α 1.5 *** 0.5. Reverbα *** *** 0.5. Fasted. Refed
 Supplementary Figure A 8 SREBPc 6 5 FASN ELOVL6.5.5.5 ACC.5.5 CLOCK.5.5 CRY.5.5 PPARα.5.5 ACSL CPTα.5.5.5.5 MCAD.5.5 PEPCK.5.5 G6Pase 5.5.5.5 BMAL.5.5 Reverbα.5.5 Reverbβ.5.5 PER.5.5 PER B Fasted Refed
Supplementary Figure A 8 SREBPc 6 5 FASN ELOVL6.5.5.5 ACC.5.5 CLOCK.5.5 CRY.5.5 PPARα.5.5 ACSL CPTα.5.5.5.5 MCAD.5.5 PEPCK.5.5 G6Pase 5.5.5.5 BMAL.5.5 Reverbα.5.5 Reverbβ.5.5 PER.5.5 PER B Fasted Refed
CELLS. Cells. Basic unit of life (except virus)
 Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
Basic unit of life (except virus) CELLS Prokaryotic, w/o nucleus, bacteria Eukaryotic, w/ nucleus Various cell types specialized for particular function. Differentiation. Over 200 human cell types 56%
Supplementary Figure 1
 VO (ml kg - min - ) VCO (ml kg - min - ) Respiratory exchange ratio Energy expenditure (cal kg - min - ) Locomotor activity (x count) Body temperature ( C) Relative mrna expression TA Sol EDL PT Heart
VO (ml kg - min - ) VCO (ml kg - min - ) Respiratory exchange ratio Energy expenditure (cal kg - min - ) Locomotor activity (x count) Body temperature ( C) Relative mrna expression TA Sol EDL PT Heart
Summary of Endomembrane-system
 Summary of Endomembrane-system 1. Endomembrane System: The structural and functional relationship organelles including ER,Golgi complex, lysosome, endosomes, secretory vesicles. 2. Membrane-bound structures
Summary of Endomembrane-system 1. Endomembrane System: The structural and functional relationship organelles including ER,Golgi complex, lysosome, endosomes, secretory vesicles. 2. Membrane-bound structures
Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 )
 770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
Metabolic ER stress and inflammation in white adipose tissue (WAT) of mice with dietary obesity.
 Supplementary Figure 1 Metabolic ER stress and inflammation in white adipose tissue (WAT) of mice with dietary obesity. Male C57BL/6J mice were fed a normal chow (NC, 10% fat) or a high-fat diet (HFD,
Supplementary Figure 1 Metabolic ER stress and inflammation in white adipose tissue (WAT) of mice with dietary obesity. Male C57BL/6J mice were fed a normal chow (NC, 10% fat) or a high-fat diet (HFD,
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/439/ra78/dc1 Supplementary Materials for Small heterodimer partner mediates liver X receptor (LXR) dependent suppression of inflammatory signaling by promoting
www.sciencesignaling.org/cgi/content/full/9/439/ra78/dc1 Supplementary Materials for Small heterodimer partner mediates liver X receptor (LXR) dependent suppression of inflammatory signaling by promoting
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Figure S1. Body composition, energy homeostasis and substrate utilization in LRH-1 hep+/+ (white bars) and LRH-1 hep-/- (black bars) mice.
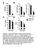 Figure S1. Body composition, energy homeostasis and substrate utilization in LRH-1 hep+/+ (white bars) and LRH-1 hep-/- (black bars) mice. (A) Lean and fat masses, determined by EchoMRI. (B) Food and water
Figure S1. Body composition, energy homeostasis and substrate utilization in LRH-1 hep+/+ (white bars) and LRH-1 hep-/- (black bars) mice. (A) Lean and fat masses, determined by EchoMRI. (B) Food and water
Week 3, Lecture 5a. Pathophysiology of Diabetes. Simin Liu, MD, ScD
 Week 3, Lecture 5a Pathophysiology of Diabetes Simin Liu, MD, ScD General Model of Peptide Hormone Action Hormone Plasma Membrane Activated Nucleus Cellular Trafficking Enzymes Inhibited Receptor Effector
Week 3, Lecture 5a Pathophysiology of Diabetes Simin Liu, MD, ScD General Model of Peptide Hormone Action Hormone Plasma Membrane Activated Nucleus Cellular Trafficking Enzymes Inhibited Receptor Effector
Table S1. Differentially expressed transcripts between hepatic inkt cells. sorted form WT and plck-hcd1d tg mice. Differentially expressed
 Supplementary Information Table S1. Differentially expressed transcripts between hepatic inkt cells sorted form and plck-hcd1d tg mice. Differentially expressed transcripts identified by gene expression
Supplementary Information Table S1. Differentially expressed transcripts between hepatic inkt cells sorted form and plck-hcd1d tg mice. Differentially expressed transcripts identified by gene expression
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3
 Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
marker. DAPI labels nuclei. Flies were 20 days old. Scale bar is 5 µm. Ctrl is
 Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Week 3 The Pancreas: Pancreatic ph buffering:
 Week 3 The Pancreas: A gland with both endocrine (secretion of substances into the bloodstream) & exocrine (secretion of substances to the outside of the body or another surface within the body) functions
Week 3 The Pancreas: A gland with both endocrine (secretion of substances into the bloodstream) & exocrine (secretion of substances to the outside of the body or another surface within the body) functions
Supplementary Information
 Supplementary Information Overexpression of Fto leads to increased food intake and results in obesity Chris Church, Lee Moir, Fiona McMurray, Christophe Girard, Gareth T Banks, Lydia Teboul, Sara Wells,
Supplementary Information Overexpression of Fto leads to increased food intake and results in obesity Chris Church, Lee Moir, Fiona McMurray, Christophe Girard, Gareth T Banks, Lydia Teboul, Sara Wells,
Insulin mrna to Protein Kit
 Insulin mrna to Protein Kit A 3DMD Paper BioInformatics and Mini-Toober Folding Activity Student Handout www.3dmoleculardesigns.com Insulin mrna to Protein Kit Contents Becoming Familiar with the Data...
Insulin mrna to Protein Kit A 3DMD Paper BioInformatics and Mini-Toober Folding Activity Student Handout www.3dmoleculardesigns.com Insulin mrna to Protein Kit Contents Becoming Familiar with the Data...
Growth and Differentiation Phosphorylation Sampler Kit
 Growth and Differentiation Phosphorylation Sampler Kit E 0 5 1 0 1 4 Kits Includes Cat. Quantity Application Reactivity Source Akt (Phospho-Ser473) E011054-1 50μg/50μl IHC, WB Human, Mouse, Rat Rabbit
Growth and Differentiation Phosphorylation Sampler Kit E 0 5 1 0 1 4 Kits Includes Cat. Quantity Application Reactivity Source Akt (Phospho-Ser473) E011054-1 50μg/50μl IHC, WB Human, Mouse, Rat Rabbit
PHSI3009 Frontiers in Cellular Physiology 2017
 Overview of PHSI3009 L2 Cell membrane and Principles of cell communication L3 Signalling via G protein-coupled receptor L4 Calcium Signalling L5 Signalling via Growth Factors L6 Signalling via small G-protein
Overview of PHSI3009 L2 Cell membrane and Principles of cell communication L3 Signalling via G protein-coupled receptor L4 Calcium Signalling L5 Signalling via Growth Factors L6 Signalling via small G-protein
Autonomic regulation of islet hormone secretion
 Autonomic regulation of islet hormone secretion Implications for health and disease Billy & Bree Paper 1: Autonomic regulation of islet hormone secretion : Implications for health and disease By Team BBB
Autonomic regulation of islet hormone secretion Implications for health and disease Billy & Bree Paper 1: Autonomic regulation of islet hormone secretion : Implications for health and disease By Team BBB
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11464 Supplemental Figure S1. The expression of Vegfb is increased in obese and diabetic mice as compared to lean mice. a-b, Body weight and postprandial blood
SUPPLEMENTARY INFORMATION doi:10.1038/nature11464 Supplemental Figure S1. The expression of Vegfb is increased in obese and diabetic mice as compared to lean mice. a-b, Body weight and postprandial blood
