Macmillan Publishers Limited. All rights reserved
|
|
|
- Anna Newman
- 6 years ago
- Views:
Transcription
1 LETTER doi:1.138/nture11433 Codon-usge-sed inhiition of HIV protein synthesis y humn schlfen 11 Mnqing Li 1, Eline Ko 1, Xi Go 1, Hilry Sndig 1, Kirsten Limmer 1, Mrin Pvon-Eternod 2, Thoms E. Jones 2, Sestien Lndry 3, To Pn 2, Mtthew D. Weitzmn 3 & Michel Dvid 1,4 In mmmls, one of the most pronounced consequences of virl infection is the induction of type I interferons, cytokines with potent ntivirl ctivity. Schlfen (Slfn) genes re suset of interferon-stimulted erly response genes (ISGs) tht re lso induced directly y pthogens vi the interferon regultory fctor 3 (IRF3) pthwy 1. However, mny ISGs re of unknown or incompletely understood function. Here we show tht humn SLFN11 potently nd specificlly rogtes the production of retroviruses such s humn immunodeficiency virus 1 (HIV-1). Our study reveled tht SLFN11 hs no effect on the erly steps of the retrovirl infection cycle, including reverse trnscription, integrtion nd trnscription. Rther, SLFN11 cts t the lte stge of virus production y selectively inhiiting the expression of virl proteins in codon-usge-dependent mnner. We further find tht SLFN11 inds trnsfer RNA, nd countercts chnges in the trna pool elicited y the presence of HIV. Our studies identified novel ntivirl mechnism within the innte immune response, in which SLFN11 selectively inhiits virl protein synthesis in HIV-infected cells y mens of codon-is discrimintion. SLFN genes encode fmily of proteins limited to mmmlin orgnisms. Nine murine nd six humn SLFN genes shre conserved NH2-terminus contining puttive AAA-domin, nd long SLFN genes possess motifs resemling DNA/RNA helicse domins, trit they shre with the nucleic cid sensors RIG-I nd MDA-5 2. Beyond tht, SLFN proteins hrour no sequence similrity to other proteins. In vivo, short nd long murine SLFN proteins inhiit T-cell development 3 5, nd levels of murine SLFN proteins re elevted fter infection with Brucell or Listeri 4. Lipopolyscchride, poly-inosine-cytosine (poly-ic) or interferon (IFN)-/ tretment of mcrophges results in induction of severl murine Slfn genes (our unpulished results). Tretment of humn foreskin firolsts with IFN-, poly-ic or poly-dadt reveled similr induction of SLFN genes (Supplementry Fig. 1), nd humn SLFN5 nd SLFN11 were consistently the most prominent fmily memers (Supplementry Fig. 1). Notly, we oserved striking difference in SLFN levels etween HEK () nd HEKT (T) cells (Supplementry Fig. 1), nd exploited this differentil expression to focus on SLFN11 for further studies. We further used SLFN11-trgeted short hirpin RNA to generte stle cells tht specificlly lck SLFN11 expression (shrna SLFN ) (Supplementry Fig. 1c, d). To test whether lck of SLFN11 in shrna SLFN or T cells lters their ility to sudue virl infections, we infected these cells with vesiculr stomtitis virus (VSV)-G pseudotyped HIV (HIV VSV-G ), or mphotropic murine stem cell virus (MSCV), deno-ssocited virus (AAV), or herpes simplex virus (HSV). HIV VSV-G -infected cells expressed luciferse fter integrtion of the virl complementry DNAs into the host genome. Regrdless of SLFN11 expression, ll cell lines hd comprle luciferse levels fter HIV VSV-G infection (Supplementry Fig. 2). A similr lck of influence of SLFN11 ws oserved when cells were infected with MSCV, AAV or HSV (not shown). HEKT cells re used s pckging cells for production of retroviruses, nd we therefore considered the possiility tht virus production rther thn the response towrds them is fflicted y SLFN11. Indeed, T cells produced mrkedly higher HIV VSV-G (Fig. 1) or MCSV (Supplementry Fig. 3) titres thn cells from the virl vectors pnl4-3.luc.r 1 E 2 or MSCV-IRES-GFP, respectively. Most importntly, this increse in virl titre ws lso clerly evident in shrna SLFN cells, wheres nd shrna Ctl cells produced the sme low levels of virus (Fig. 1 nd Supplementry Fig. 3). Notly, the modultion of virus production is limited to prticulr viruses, s friction of retroviruses (Fig. 1 nd Supplementry Fig. 3), ut not of AAV (Supplementry Fig. 3c), ws ffected y SLFN11. We lso did not oserve ny modultion of ISGs such s ISG15, ISG54 or APOBEC3G (not shown) s consequence of SLFN11 expression, supporting the notion tht SLFN11 does not crete generl virusresistnt phenotype. To corroorte tht the oserved differences re ttriutle to dissimilr SLFN11 expression, we expressed full-length SLFN11 (mino cids 1 91) in T cells nd nlysed their ility to produce HIV VSV-G. Indeed, SLFN11 strongly inhiited HIV VSV-G (Fig. 1) or MSCV (Supplementry Fig. 3) production from T cells, with the inhiitory ctivity residing in the AAA-domin-contining, mino-terminl region (SLFN11-N; mino cids 1 579). No effect of the isolted croxy-terminl region (SLFN11-C; mino cids ) hrouring the puttive helicse sequence ws oserved (Fig. 1 nd Supplementry Fig. 3). Intriguingly, SLFN5 filed to inhiit retrovirus production ut yielded slightly elevted virl titres, illustrting specificity mong SLFN proteins in their ntivirl ctivity. To discern whether SLFN11 reduced the numer or the viility of relesed virus, we mesured p24 cpsid nd virl RNA (vrna) levels in superntnts of pnl4-3.luc.r 1 E 2 -trnsfected, HIV VSV-G - producing cells. Extrcellulr p24 (Fig. 1c nd d) or vrna (Fig. 1e nd f) concentrtion ptterns closely mtched the titre results from infection ssys, demonstrting tht SLFN11 diminishes the numer of virl prticles relesed from the cells. To ssess possile reduction of intrcellulr vrna, we determined its levels in T cells expressing chlormphenicol cetyl trnsferse (CAT), SLFN5, SLFN11, SLFN11-N or SLFN11-C (Fig. 1g), s well s in, shrna Ctl nd shrna SLFN cells (Fig. 1h). In contrst to the pronounced vritions in extrcellulr vrna, only insignificnt differences in intrcellulr vrna were evident mong those cells. We lso nlysed vrna in the cytoplsmic frction specificlly, s nucler export of unspliced vrna is hllmrk of retrovirl RNA processing 6 8, ut gin found no significnt ltertions ttriutle to SLFN11 (Supplementry Fig. 4). Unlike in BST2-expressing cells 9,1, electron-microscopic nlysis filed to document ccumultion of virions inside or on the surfce of virus-producing cells in the presence of SLFN11 (Supplementry Fig. 4). As such, SLFN11 gretly diminishes the formtion of virl prticles inside the cell, despite the fct tht vrna is eqully ville. 1 Section ofmoleculr Biology, Division of Biologicl Sciences, University of Cliforni SnDiego, L Joll, Cliforni 9293, USA. 2 Deprtment of Biochemistry nd Moleculr Biology, University of Chicgo, Chicgo, Illinois 6637, USA. 3 Lortory of Genetics, The Slk Institute for Biologicl Studies, L Joll, Cliforni 9237, USA. 4 Moores Cncer Center, University of Cliforni Sn Diego, L Joll, Cliforni 9293, USA. 212 Mcmilln Pulishers Limited. All rights reserved MONTH 212 VOL NATURE 1
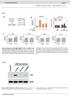
2 RESEARCH LETTER Reltive HIV virus titre c Reltive HIV p24 levels e Reltive HIV RNA levels g Reltive intrcellulr HIV RNA levels T CAT T CAT 1.75 T SLFN T SLFN5 T SLFN T CAT T SLFN T SLFN11-N T CAT T SLFN5 T SLFN11 T SLFN11-N T SLFN11-C.1.23 T SLFN11 T SLFN11-N T SLFN T SLFN11-C When the two cell sets were nlysed for the expression of virl proteins encoded y pnl4-3.luc.r 1 E 2, we noted mrked effect of SLFN11 on p55 Gg nd p24 cpsid proteins 11 (Fig. 2, ). To crete clerer picture of the effect of SLFN11 on p55, we olished expression of the virl protese y introducing stop codon into pro in pnl4-3.luc.r 1 E 2. As nticipted, p24 could no longer e detected; however, modultion of p55 expression y SLFN11 ws clerly present (Supplementry Fig. 5). As with gg-derived proteins, SLFN11 hd notle effect on the protein levels of RT, Vif, Vpu nd Vpr (Fig. 2, ). Intriguingly, we did not oserve ny reduction of enhnced green fluorescent protein (EGFP) derived from co-trnsfected vector, or of (Fig. 2,, ottom), indicting tht the limited virl protein production is not due to glol shutdown of protein synthesis. Even more surprising ws tht luciferse expression, coded in pnl4-3.luc.r 1 E 2 in plce of nef, ws mostly impervious to SLFN11 (Fig. 2c, top), contrsting the SLFN11-medited diminishment of other proteins encoded on this vector. Notly, Nef expression ws inhiited y SLFN11 in the context of pnl4-3-denv-egfp, which hs prt of env replced y EGFP, ut retins nef in its originl position. (Fig. 2c, ottom). We thus conclude tht SLFN11 selectively suppresses virl protein expression vi trnscript-intrinsic properties d T SLFN11-C T SLFN11-N T SLFN11-C Reltive HIV virus titre f h shrna SLFN 5.55 shrna Ctl shrna SLFN shrna SLFN shrna SLFN shrna Ctl shrna Ctl T T 8.17 T shrna Ctl Figure 1 SLFN11 inhiits retrovirus production without ffecting intrcellulr vrna levels. h, T cells were trnsfected with pnl4-3.luc.r 1 E 2 /pcmv-vsv-g together with SLFN5, SLFN11, SLFN11-N, SLFN11- CorCAT(, c, e, g), or, shrna Ctl, shrna SLFN nd T cells were trnsfected with pnl4-3.luc.r 1 E 2 nd pcmv-vsv-g (, d, f, h).,,vsv-gpseudotyped HIV production ws ssyed y titrted infection nd luciferse ssy. c, d, Virl prticle content in superntnts ws nlysed y p24 ELISA. e, f, extrcellulr vrna concentrtion ws nlysed y qpcr of p24. g, h, intrcellulr vrna ws determined y qpcr of p24. (verge 6 s.d.; n 5 3). T c T CAT T SLFN5 WB Gg T SLFN WB RT WB Vif.7.1 WB Vpu. WB Vpr WB GFP 1.2 WB TSlfn11N Vpr TSlfn11C p55 p24 p66 p51 Vif Vpu EGFP shrna Ctl WB Gg shrna SLFN WB RT WB Vif WB Vpu WB Vpr WB GFP WB T p55 p24 p66 p51 Vif Vpu Vpr EGFP rther thn externl fctors or positionl elements. This notion is further corroorted y the fct tht SLFN11 hd no effect on Rev response element (RRE)-medited events such s nucler export of unspliced vrna (Supplementry Fig. 5). Virl genomes hve ised nucleotide compositions different from humn genes Extremely high frequencies of A nucleotides re found in the RNA genomes of lentiviruses nd influenz virus 13,17,19,2. Wild isoltes of HIV-1, prticulrly gg nd pol sequences, re chrcterized y low GC content nd suoptiml codon usge compred to the host cell preference 14, The unusul rre codon is fvours A/U in the third position, which induces riosome pusing nd inefficient trnsltion. As the inhiitory effect of SLFN11 on virl protein expression is intrinsic to the trnscripts, we proposed tht SLFN11 exploits virl codon preferences to specificlly ttenute virl protein synthesis. We therefore generted vectors contining only the open reding frme of HIV-1 gg with either virl codon-is (Gg vir ), or synonymous sustitutions optimizing for humn cell expression (Gg opt ). As shown in Fig. 2d, SLFN11 strongly ffected expression of Gg vir, ut ws without consequence for Gg opt expression. Differences in trnsltion initition re not likely, s oth Gg vir nd Gg opt contin the sme trnsltion strt sequences. This finding strongly indictes tht SLFN11 is exploiting the distinct virl codon is to selectively ttenute the expression of virl proteins. Previous reports indicted chnges in cellulr trna levels fter HIV infection 2, prompting us to investigte whether SLFN11 lters the trna composition in the sence or presence of HIV. Using trna rrys 25,26, we oserved little to no chnges in trna levels s d T CAT T SLFN5 T SLFN11 shrna Ctl pnl4-3.luc.r + E WB Luc WB Luc W: V5 W: V5.9.8 WB pnl4-3-δenv-egfp WB Nef WB Nef.1 WB T CAT T SLFN5 WB V5 T SLFN shrna SLFN WB WB shrna Ctl WB V5 shrna SLFN Luc Nef Gg vir Gg opt Figure 2 SLFN11 selectively inhiits virl protein expression on the sis of codon usge., T cells were trnsfected with pnl4-3.luc.r 1 E 2 nd pcdna5-egfp together with SLFN5, SLFN11 or CAT, nd cell lystes immunolotted for HIV proteins, EGFP nd.,, shrna Ctl nd shrna SLFN cells were trnsfected with pnl4-3.luc.r 1 E 2 nd pcdna5-egfp, nd lystes nlysed s in. c, Top: T cells were cotrnsfected with pnl4-3.luc.r 1 E 2 nd SLFN5, SLFN11 or CAT (left), or, shrna Ctl nd shrna SLFN cells were trnsfected with pnl4-3.luc.r 1 E 2 (right). Lystes were proed for luciferse nd. Bottom: s ove, except pnl4-3-denv-egfp ws used insted of pnl4-3.luc.r 1 E 2, nd lystes were immunolotted for Nef nd ; d, Cells were trnsfected with virl codon-usge-sed gg (Gg vir, top) or synonymous humn codon usge-optimized gg (Gg opt, ottom). Expression of Gg in cell lystes ws determined y nti-v5 immunolotting. 2 NATURE VOL MONTH Mcmilln Pulishers Limited. All rights reserved
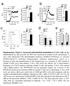
3 LETTER RESEARCH consequence of SLFN11 expression (Fig. 3, left column). However, wheres HIV triggered sustntil chnges in trna concentrtions in SLFN11-knockdown cells, no chnges were oserved in the presence of SLFN11 (Fig. 3, middle nd right). Thus, SLFN11 countercts HIVinduced chnges in trna composition, presumly initited to promote virl protein synthesis. To test whether SLFN11 intercts directly with trna, we used humn trna s electrophoretic moility shift ssy (EMSA) proe with fst protein liquid chromtogrphy (FPLC)- purified His-conjugted SLFN11-N. As shown in Fig. 3, SLFN11-N shrna Ctl / shrna SLFN11 shrna SLFN11: +/ HIV shrna Ctl: +/ HIV e (E)GFP (protein/mrna rtio) Ile-UAU Leu-UAA Vl-UAC Gly-UCC Ar-UCU Ar-CCG/UCG Glu-UUC Lys-UUU Ser-IGA/UGA Thr-UGU Ile-IAU/GAU Leu-IAG/UAG Leu-CAG Leu-CAA Vl-IAC/CAC Gly-GCC/CCC Ar-ICG Ar-CCU Glu-CUC/UUC Lys-CUU Ser-CGA Ser-GCU Thr-IGU/CGU Thr-CGU Met-i Met-CAU Phe-GAA Trp-CCA Al-IGC Al-IGC/CGC/UGC Cys-GCA Pro-IGG/CGG/UGG Asp-GUC Asn-GUU Gln-CUG/UUG His-GUG Sec-UCA2 Tyr-GUA.9 c 1.8 GFP V5 T CAT No protein No protein EGFP Myc.9 T SLFN5 GFP Anti- S11 NS SLFN11-N T SLFN11 d SLFN11-N T CAT T SLFN5 No protein T SLFN11 GFP V5 SLFN11-N EGFP Myc GFP V5 EGFP Myc.3.7 +trna shrna Ctl shrna SLFN 3.7 +HIV RNA GFP EGFP 1.3 shrna Ctl shrna SLFN Figure 3 SLFN11 inds trnas nd selectively inhiits protein expression sed on codon usge., shrna Ctl nd shrna SLFN cells were trnsfected with pnl4-3.luc.r 1 E 2 or control vector (indicted s 1/2 HIV). Reltive undnces of mture trna species were nlysed y microrry s descried 25,26., Left: incresing mounts of SLFN11-N or GFP were incuted with 32 P-lelled trna nd sujected to EMSA. Right: 2 3 or 1 3 unlelled trna or in-vitro-trnscried vrna corresponding to the gg-pol frmeshifting sequence (12 ses) were dded to the inding rection. c, Asin, except non-specific (NS) or nti-slfn11 (Anti-S11) monoclonl ntiody ws dded to the inding rection. d, T cells were trnsfected with V5- tgged GFP, Myc-tgged EGFP nd pnl4-3.luc.r 1 E 2 together with either CAT, SLFN5 or SLFN11 (left). Lystes were proed for V5 GFP, Myc EGFP nd. Similrly, V5-tgged GFP, Myc-tgged EGFP nd pnl4-3.luc.r 1 E 2 were co-trnsfected into, shrna Ctl nd shrna SLFN cells (right), nd V5 GFP, Myc EGFP nd expression determined y immunolotting. e, V5 GFP nd Myc EGFP protein levels determined from d were quntified nd normlized to V5 GFP nd Myc EGFP mrna levels, respectively (verge 6 s.d.; n 5 4). produced cler shift of the trna in dose-dependent mnner. The shifted nd ws competed with unlelled trna, ut not in-vitrotrnscried vrna (Fig. 3), nd ws disrupted y n ntiody ginst SLFN11 (Fig. 3c). To determine whether SLFN11 hs inding preference for prticulr trnas, we performed EMSAs with incresing mounts of SLFN11-N to otin pproximtely 5% shifting efficiency (Supplementry Fig. 6). When the shifted (S), unshifted (U) nd totl trna (T) nds were isolted nd recovered trnas hyridized to trna rrys, no enrichment of prticulr trnas in the shifted nd ws noticele (Supplementry Fig. 6), indicting lck of discerning inding preference of SLFN11-N. The results shown led to the premise tht ny protein tht uses similr codon usge s HIV would e modulted y HIV nd/or SLFN11. To demonstrte unequivoclly tht codon-is rther thn cryptic regultory elements in gg ccounted for the influence of SLFN11, we tested this hypothesis using non-virl proteins. Nturl green fluorescent protein (GFP) hrours similr codon-is s tht of HIV, wheres EGFP hs een optimized for mmmlin expression y sustituting synonymous codons of highly expressed humn genes throughout the GFP open reding frme 22. As nticipted, co-trnsfection of pnl4-3.luc.r 1 E 2 with V5-tgged GFP nd Myc-tgged EGFP in the sence of SLFN11 resulted in incresed GFP, ut not EGFP, expression (not shown). Most importntly, SLFN11 inhiited the expression of GFP in mnner identicl to virl proteins, wheres EGFP ws lrgely refrctory to suppression y SLFN11 (Fig. 3d, e). As (E)GFP protein levels hve een normlized to their respective messenger RNA levels (Fig. 3e), we conclude tht ny differences in GFP expression re the consequence of ltered protein synthesis rther thn vritions in trnscription or mrna stility. Finlly, to demonstrte tht the nti-retrovirl effects of SLFN11 re not limited to system using HIV VSV-G nd cells, we used CEM cells, T-cell line widely used to ssess HIV repliction kinetics. Humn peripherl lood mono-nucler cells (PBMCs) disply similr levels of SLFN11 fter IFN- stimultion s CEM cells (Fig. 4). Control or SLFN11-directed shrnas were used to generte stle CEM vrints (Fig. 4), nd the resulting CEMshRNA Ctl nd CEMshRNA SLFN cells nd prentl CEM cells were infected with wild-type X4-HIV-1 LAI, nd virl repliction ws ssessed vi p24 enzyme-linked immunosorent ssys (ELISA) of the superntnts. c HIV p24 (ng ml 1 ) 6, 5, 4, 3, 2, 1, WB SLFN11 PBMC 8 h 24 h WB CEM CEM IFN-β SLFN11 CEMshRNA Ctl CEMshRNA SLFN WB SLFN11 CEM CEM shrna Ctl CEM shrna SLFN WB Dys post infection Figure 4 SLFN11 inhiits repliction of wild-type HIV-1 LAI in CEM cells., Humn PBMCs were stimulted with 6, U ml 21 IFN-. SLFN11 expression in the derived lystes nd in CEM cell lystes ws nlysed y immunolotting., SLFN11 expression in CEM, CEMshRNA Ctl nd CEMshRNA SLFN cells s nlysed y immunolotting. c, CEM, CEMshRNA Ctl nd CEMshRNA SLFN cells were infected with HIV-1 LAI t multiplicity of infection (m.o.i.) of.1, nd virl repliction ws ssyed y p24 ELISA of culture superntnts (verge 6 s.d.; n 5 4). 212 Mcmilln Pulishers Limited. All rights reserved MONTH 212 VOL NATURE 3
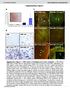
4 RESEARCH LETTER As nticipted, CEM nd CEMshRNA Ctl cells produced comprle virl titres t ll time points. In contrst, CEMshRNA SLFN cells fcilitted significntly enhnced HIV-1 repliction, yielding n exponentilly incresing difference in virl titres (Fig. 4c). A second CEMshRNA SLFN cell line estlished y mens of distinct SLFN11 shrna further corroorted these results (Supplementry Fig. 7). In summry, systemtic nlysis of the HIV repliction cycle reveled tht SLFN11 does not inhiit reverse trnscription, integrtion or production nd nucler export of virl RNA, nor did we oserve lock in udding or relese of virl prticles. However, we discovered selective inhiition in the synthesis of virlly encoded proteins. Intriguingly, specific inhiition of virl protein synthesis in HIVinfected cells in response to interferon ws previously oserved, ut the fctors responsile for the effect were not identified 27. We postulte tht SLFN11 cts t the point of virus protein synthesis y exploiting the unique virl codon is towrds A/T nucleotides. This model supports the notion tht the ntivirl ctivity of SLFN11 extends to other viruses with rre codon is such s influenz, ut pprently not to AAV or HSV. The exct mechnism y which HIV lters trna function in its fvour nd how SLFN11 countercts this process will require considerle further nlysis. Evidently, SLFN11 intercts with ll trnas in vitro. Direct inding of SLFN11 to trna offers the possiility tht SLFN11 either sequesters trnas, prevents their mturtion vi post-trnscriptionl processing or ccelertes their decyltion. In either cse, if lredy rre trnas re further reduced, trna vilility might mnifest s the rte-limiting step in the synthesis of proteins involving those trnas. In contrst, lesser or no impct would e expected on proteins synthesized vi plentiful trnas, s even if frction of those trnas is eliminted, trnsltion initition will likely remin the rte-limiting event. In conclusion, our results estlish SLFN11 s potent, interferon-inducile restriction fctor ginst retroviruses such s HIV, mediting its ntivirl effects on the sis of codon usge discrimintion. METHODS SUMMARY Plsmids nd ntiodies. SLFN5 nd SLFN11 were cloned into pcdna6/v5-his. PCR frgments of SLFN11 were cloned into pcdna6/v5-his to generte SLFN11-N (mino cids 1 579) nd SLFN11-C (mino cids ). The pnl4-3.luc.r 1 E 2 HIV-1 vector hs een descried previously 28. SLFN11 ntiodies were from Sigm nd Amrt. pnl4-3-denv-egfp, pnl-gfp- RRE(SA) nd ntiodies ginst HIV-1 proteins were otined through the NIH AIDS Reserch nd Reference Regent Progrm. Genertion of Gg vir, Gg opt nd (E)GFP vectors is in Methods. plko.1 shrna lentivirus constructs were from Open Biosystems. Virus production nd titre ssys. HIV production ws determined y trnsfection of pnl4-3.luc.r 1 E 2 nd pcmv-vsv-g into cells of interest. Superntnts were used to spin-infect HEKT cells, nd luciferse ws mesured y Bright- Glo Luciferse Assy (Promeg). p24 ELISAs were performed t the Center for AIDS Reserch, UCSD using Allince p24 ELISA kits (Perkin Elmer). HIV repliction kinetics. Wild type X4 strin of HIV-1 LAI ws used to inoculte CEM, CEMshRNA Ctl nd CEMshRNA SLFN cells t multiplicity of infection of.1, nd virl titres were mesured y p24 ELISA. Dt nlysis nd presenttion. Unless indicted otherwise, results presented in grphs re verge 6 s.d. of t lest three independent trnsfections or infections. For immunolots, representtive of t lest three independent trnsfections or infections is shown. Full Methods nd ny ssocited references re ville in the online version of the pper. Received 22 Novemer 211; ccepted 23 July 212. Pulished online 23 Septemer Sohn, W. J. et l. Novel trnscriptionl regultion of the schlfen-2 gene in mcrophges in response to TLR-triggered stimultion. Mol. Immunol. 44, (27). 2. Yoneym, M. et l. Shrednduniquefunctions ofthe DExD/H-ox helicses RIG- I, MDA5, nd LGP2 in ntivirl innte immunity. J. Immunol. 175, (25). 3. Schwrz, D. A., Ktym, C. D. & Hedrick, S. M. Schlfen, new fmily of growth regultory genes tht ffectthymocytedevelopment. Immunity 9, (1998). 4. Geserick, P., Kiser, F., Klemm, U., Kufmnn, S. H. & Zerrhn, J. Modultionof T cell development nd ctivtion y novel memers of the Schlfen (slfn) gene fmily hrouring n RNA helicse-like motif. Int. Immunol. 16, (24). 5. Berger, M. et l. An Slfn2 muttion cuses lymphoid nd myeloid immunodeficiency due to loss of immune cell quiescence. Nture Immunol. 11, (21). 6. Fritz, C. C. & Green, M. R. HIV Rev uses conserved cellulr protein export pthwy for the nucleocytoplsmic trnsport of virl RNAs. Curr. Biol. 6, (1996). 7. Fischer, U., Huer, J., Boelens, W. C., Mttj, I. W. & Luehrmnn, R. The HIV-1 Rev ctivtion domin is nucler export signl tht ccesses n export pthwy used y specific cellulr RNAs. Cell 82, (1995). 8. Mlim, M. H., Huer, J., Le, S. Y., Mizel, J. V. & Cullen, B. R. The HIV-1 rev trnsctivtor cts through structured trget sequence to ctivte nucler export of unspliced virl mrna. Nture 338, (1989). 9. Vn Dmme, N. et l. The interferon-induced protein BST-2 restricts HIV-1 relese nd is downregulted from the cell surfce y the virl Vpu protein. Cell Host Microe 3, (28). 1. Neil, S. J., Zng, T. & Bienisz, P. D. Tetherin inhiits retrovirus relese nd is ntgonized y HIV-1 Vpu. Nture 451, (28). 11. Gnser-Pornillos, B. K., Yeger, M. & Sundquist, W. I. The structurl iology of HIV ssemly. Curr. Opin. Struct. Biol. 18, (28). 12. Colemn, J. R. et l. Virus ttenution y genome-scle chnges in codon pir is. Science 32, (28). 13. Zhou, T., Gu, W., M, J., Sun, X. & Lu, Z. Anlysis of synonymous codon usge in H5N1 virus nd other influenz A viruses. Biosystems 81, (25). 14. Meintjes, P. L. & Rodrigo, A. G. Evolution of reltive synonymous codon usge in humn immunodeficiency virus type-1. J. Bioinform. Comput. Biol. 3, (25). 15. Frelin, L. et l. Codon optimiztion nd mrna mplifiction effectively enhnces the immunogenicity of the heptitis C virus nonstructurl 3/4A gene. Gene Ther. 11, (24). 16. Forserg, R. & Christinsen, F. B. A codon-sed model of host-specificselection in prsites, with n ppliction to the influenz A virus. Mol. Biol. Evol. 2, (23). 17. Plotkin, J. B. & Dushoff, J. Codon is nd frequency-dependent selection on the hemgglutinin epitopes of influenz A virus. Proc. Ntl Acd. Sci. USA 1, (23). 18. Grnthm, P. & Perrin, P. AIDS virus nd HTLV-I differ in codon choices. Nture 319, (1986). 19. Wong, E. H. M., Smith, D. K., Rdn, R., Peiris, M. & Poon, L. L. M. Codon usge is nd the evolution of influenz A viruses. Codon usge ises of influenz virus. BMC Evol. Biol. 1, 253 (21). 2. vn Weringh, A. et l. HIV-1 modultes the trna pool to improve trnsltion efficiency. Mol. Biol. Evol. 28, (211). 21. Kofmn, A. et l. HIV-1 gg expression is quntittively dependent on the rtio of ntive nd optimized codons. Tsitologii 45, (23). 22. Hs, J., Prk, E. C. & Seed, B. Codon usge limittion in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 6, (1996). 23. Berkhout, B. & vn Hemert, F. J. The unusul nucleotide content of the HIV RNA genome results in ised mino cid composition of HIV proteins. Nucleic Acids Res. 22, (1994). 24. Kypr, J. & Mrzek, J. Unusul codon usge of HIV. Nture 327, 2 (1987). 25. Pvon-Eternod, M. et l. trna over-expression in rest cncer nd functionl consequences. Nucleic Acids Res. 37, (29). 26. Dittmr, K. A., Moley, E. M., Rdek, A. J. & Pn, T. Exploring the regultion of trna distriution on the genomic scle. J. Mol. Biol. 337, (24). 27. Cocci, E. M., Krust, B. & Hovnessin, A. G. Specific inhiition of virl protein synthesis in HIV-infected cells in response to interferon tretment. J. Biol. Chem. 269, (1994). 28. Connor, R. I., Chen, B. K., Choe, S. & Lndu, N. R. Vpr is required for efficient repliction of humn immunodeficiency virus type-1 in mononucler phgocytes. Virology 26, (1995). Supplementry Informtion is ville in the online version of the pper. Acknowledgements We thnk D. Smith for help with the HIV repliction studies; M. Wood for performing the electron microscope nlysis; D. Xu nd D.-Y. Song for technicl ssistnce; nd J. Young, J. Gutelli, D. Smith, M. Kul nd S. Chnd for discussion. This work ws supported in prt y NIH AI8119 nd AI74967 to M.D.W., NIH P1AI9935, R1GM11982 nd R21AI8849 to M.D., nd y resources from the UCSD Center for AIDS Reserch, NIH P3AI36214, nd the HINT Progrm, NIH P1AI9935. The uthors declre no competing finncil interests. Author Contriutions M.L., M.D.W., T.P. nd M.D. plnned the experiments; M.L., E.K., X.G., M.P.-E., K.L., H.S., T.E.J. nd S.L. conducted the experiments; M.L., T.P., M.D.W. nd M.D. nlysed the dt; nd M.L. nd M.D. wrote the mnuscript. Author Informtion Reprints nd permissions informtion is ville t The uthors declre no competing finncil interests. Reders re welcome to comment on the online version of the pper. Correspondence nd requests for mterils should e ddressed to M.D. (midvid@ucsd.edu). 4 NATURE VOL MONTH Mcmilln Pulishers Limited. All rights reserved
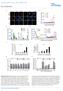
5 LETTER RESEARCH METHODS Cell culture. HEK, HEKT, NIH3T3, HeL, HFFs (humn foreskin firolsts) nd their derivtive cells were mintined in high glucose DMEM. CEM nd their derivtive cells were mintined in RPMI 164 medium. Both medi were supplemented with 1% het-inctivted fetl ovine serum, 1 U ml 21 penicillin, 1 mgml 21 streptomycin, 2 mm L-glutmine, MEM non-essentil mino cids, 1 mm sodium pyruvte nd 55 mm 2-mercptoethnol. The Phoenix mphotropic retrovirus pckging cell line (T) origintes from the lortory of G. Noln. Humn PBMCs were otined from the Sn Diego Blood Bnk. Plsmids. The templte vectors encoding SLFN5 nd SLFN11 sequences were cquired from Open Biosystems (Humn MGC Verified FL cdna (IRAU), clone ID nd ). The coding sequences were mplified y PCR with PfuUltr (Strtgene) nd sucloned into the pcdna6/v5-his vector (Invitrogen) for mmmlin cell expression. Prtils of the humn SLFN11 coding sequence (GenBnk ccession no. NM_114587) were mplified y PCR nd sucloned into the pcdna6/v5-his vector to generte the SLFN11-N (mino cids 1 579) nd SLFN11-C (mino cids ) trunction mutnts. The pcdna6/v5-his CAT (chlormphenicol cetyl trnsferse) vector ws generted the sme wy using pcdna5/frt/to CAT (Invitrogen) s the PCR templte for the CAT sequence. The following plsmids were otined from the Addgene plsmid repository: MSCV-IRES-GFP (Addgene plsmid 2672, source D. Bltimore); pcl-eco retrovirus pckging vector (Addgene plsmid 12371, source I. Verm); pspax2 lentivirus pckging vector (Addgene plsmid 1226, source D. Trono); pmd2.g helper vector (Addgene plsmid 12259, source D. Trono) nd pcmv-vsv-g vector (Addgene plsmid 8454, source R. M. Weinerg). The following plsmids were otined through the NIH AIDS Reserch nd Reference Regent Progrm, Division of AIDS, NIAID, NIH: pnl-gfp- RRE(SA) (ctlogue no ) from J. Mrsh nd Y. Wu, pnl4-3-delte- EGFP (ctlogue no. 111) from H. Zhng, Y. Zhou nd R. Silicino. The pnl4-3.luc.r 1 E 2 HIV-1 vector hs een descried previously 28. The pnl4-3.luc.r 1 E 2 PRstop construct ws creted y deletion of thymidine 2478 in pnl4-3.luc.r 1 E 2 HIV-1 to put the following TAGTAG into frme such tht no pol-encoded virl enzyme proteins would e expressed. Gg vir nd Gg opt constructs: gg sequences were cloned into pcdna6/v5/his etween AflII nd XhoI sites. Nine se pirs of the wild-type virl sequence preceding the initition codon were present in oth finl constructs to ssure identicl trnsltion initition sites. Gg vir ws generted through PCR mplifiction of the gg sequence from pgg-egfp (NIH AIDS Reserch & Reference Regent Progrm, no ) in which only the inhiitory RNA sequence (INS) ws eliminted through introduction of silent muttions while otherwise retining the originl virl codon usge. Gg opt ws generted through PCR mplifiction of the gg sequence from p96zm651gg-opt (NIH AIDS Reserch & Reference Regent Progrm, no. 8675) in which the entire gg sequence ws converted to use codon usge in line with highly expressed humn genes. GFP nd EGFP sequences were cloned into pcdna6.2/v5 using the pcdna6.2/gw/d-topo kit (Invitrogen). The GFP sequence ws tgged with V5 epitope wheres the EGFP sequence ws tgged with Myc tg (59-GAGCAGAAGCTGATCA GCGAGGAGGACCTG-39). The GFP sequence used retins most of the originl wild-type protein sequence. The EGFP sequence contins more thn 19 silent se chnges following humn genes codon-usge preferences. Sequences flnking oth GFP nd EGFP trnsltion initition site hve een converted to Kozk consensus trnsltion initition site (59-CACCATGGTGAGC ) to ensure identicl trnsltion initition efficiency in eukryotic cells. shrna constructs. The plko.1 shrna lentivirus constructs (TRCN14899, TRCN15223, TRCN155578, TRCN157747, TRCN15257, TRCN155564) ginst SLFN11 were otined from The RNAi Consortium vi Open Biosystems. Construct TRCN14899 (59-CCGGGCTCAGAA TTTCCGTACTGAACTCGAGTTCAGTACGGAAATTCTGAGCTTTTTTG-39) produced mximum SLFN11 knockdown t the protein level nd ws thus designted s SLFN11 shrna in this study. plko.1-blsticidin construct ws creted y replcing puromycin resistnce gene of the originl plko.1 construct with lsticidin resistnce gene using BmHI nd KpnI sites. The control shrna construct in the sme plko.1 vector system (59-CCGGTGAAGAACTAA CCCGGGACTTCTCGAGAAGTCCCGGGTTAGTTCTTCATTTTTG-39) ws lso otined from The RNAi Consortium vi Open Biosystems nd does not mtch ny humn genes. Antiodies. Anti-SLFN11 ntiody for immunolotting ws purchsed from Sigm Life Sciences (Atls Antiodies). Monoclonl nti-slfn11 ntiody for supershift experiments ws custom-generted y Amrt. Murine nti-v5 ntiody (E1) ntiodies were from ebiosciences nd Snt Cruz Biotechnology, respectively. Got polyclonl nti-luciferse ntiody ws otined from Promeg, rit nti-gfp (D5.1) nd rit nti- monoclonl ntiodies (14C1) were cquired from Cell Signling Technology. The following ntiodies were otined through the AIDS Reserch nd Reference Regent Progrm, Division of AIDS, NIAID, NIH: monoclonl ntiody ginst HIV-1 Gg/p24 (AG3.) from J. Alln; nti-hiv-1 RT monoclonl ntiody (8C4) from D. E. Hellnd; nti-hiv-1 Vif monoclonl ntiody (no. 319) from M. H. Mlim; nti-hiv-1 Vpr (1-5) ntiody from J. Kopp; rit nti-hiv-1 NL4-3 Vpu ntiserum from K. Streel, nd nti-hiv-1 Nef (Ag11) monoclonl ntiody from J. Hoxie. Quntittive PCR nd primers. Totl cellulr RNA ws extrcted with QIAGEN RNesy Mini kits, nd virion RNA in culture superntnts ws extrcted with QIAGEN QIAmp Virl RNA Mini kits. Cytoplsmic RNA ws prepred with QIAGEN RNesy Mini kits following the mnufcturer s protocol. Briefly, freshly hrvested cells were gently lysed on ice for 5 min in uffer RLN (5 mm Tris?Cl, ph 8., 14 mm NCl, 1.5 mm MgCl 2,.5% (v/v) IGEPAL CA-63, 1, U ml 21 RNsin Plus RNse Inhiitor (Promeg) nd 1 mm DTT). Intct cell nuclei were removed y centrifugtion t 4 uc for 2 min t 3g, nd cytoplsmic RNA ws extrcted from the remining superntnt. RNA ws clened using Amion DNAfree kits nd reverse trnscried with Applied Biosystems High Cpcity cdna Reverse Trnscription kit. qpcr ws performed on Applied Biosystems 7 Sequence Detection System or Applied Biosystems StepOnePlus Rel-Time PCR System using Power SYBR Green PCR Mster Mix. Reltive RNA level were clculted fter normliztion to 18S rrna unless specified otherwise. The following primer sequences were used in these ssys: SLFN5 forwrd 59-CAAGCCTGTGTGCATTCATAA-39, reverse 59-TCTGGAGTATATACCA CTCTGTCTGAA-39; SLFN11 forwrd 59-AAGGCCTGGAACATAAAAAGG-39, reverse 59-GGAGTATATCGCAAATATCCTGGT-39; SLFN12 forwrd 59-CTT TGTTCAACACGCCAAGA-39, reverse 59-ATGCAGTGTCCAAGCAGAAA-39; SLFN13 forwrd 59-GAGAAAATGATGGACGCAGAT-39, reverse 59-AGACTC AAAGGCCTCAGCAA-39; SLFN12L forwrd 59-GAAAGTCAGTTTCTGAGG AATTTCA-39, reverse 59-CCAGCTCAGCATAGTTTGTGTC-39; SLFN14 forwrd 59-GGTGGTCATGATGCTGGATA-39, reverse 59-TGATGAAATCA GGCAAGAGTTG-39; ISG54 forwrd 59-TGGTGGCAGAAGAGGAAGAT-39, reverse 59-CCAAGGAATTCTTATTGTTCTCACT-39; TBP forwrd 59-GCTGG CCCATAGTGATCTTT-39, reverse 59-CTTCACACGCCAAGAAACAGT-39; 18S rrna forwrd 59-GGATGCGTGCATTTATCAGA-39,reverse 59-GTTGATA GGGCAGACGTTCG-39;GFPforwrd59-CTGGAGTTGTCCCAATTCTTG-39, reverse 59-TCACCCTCTCCACTGACAGA-39; EGFP forwrd 59-CAGCAGAA CACCCCCATC-39, reverse 59-TGGGTGCTCAGGTAGTGGTT-39; luciferse forwrd 59-AGGTCTTCCCGACGATGA-39, reverse 59-GTCTTTCCGTGCTCC AAAAC-39; HIV p24 forwrd 59-TGCATGGGTAAAAGTAGTAGAAGAGA-39, reverse 59-TGATAATGCTGAAAACATGGGTA-39. Genertion of stle SLFN11 knockdown HEK nd CEM cell lines. HEKT cells were trnsfected with the SLFN11 shrna or control lentivirus vector nd the lentivirus pckging vectors pspax2 nd pmd2.g using Lipofectmine 2 (Invitrogen) ccording to the mnufcturer s protocol. The cell culture medium ws collected nd replced with fresh medium 48, 72 nd 96 h fter the trnsfection. The comined superntnts were clered y centrifugtion t 1,g t 4 uc for 15 min. To generte stle SLFN11 knockdown HEK cell lines, clered superntnts were dded to pre-plted HEK cells in the presence of 8 mgml 21 polyrene nd centrifuged t 6g for 9 min t room temperture. After 48 h, infected cells were suject to resistnce selection with 2 mgml 21 puromycin. The efficiency of SLFN11 knockdown in the selected cells ws ssyed y qpcr nd immunolotting. To generte stle SLFN11 knockdown CEM cell lines, cells were first infected nd selected s outlined ove. Although the sme shrna construct ws used, knockdown in CEM cells ws less efficient when compred to HEK cells. To improve the SLFN11 knockdown in CEM cells, puromycin-selected CEM cells were re-infected with the plko.1-blsticidin construct crrying the sme shrna nd suject to resistnce selection with 15 mgml 21 lsticidin nd 2 mgml 21 puromycin. Such doule-selected cells displyed. 9% knockdown of SLFN11 (see Fig. 4). Assy for MSCV, HIV nd AAV production. To nlyse MSCV retrovirus production, MSCV-IRES-GFP (MIG) plsmid nd pcl Eco pckging vector were trnsfected into the cells of interest using Lipofectmine 2 (Invitrogen) ccording to the mnufcturer s instructions. After 24 h the cells were moved to 32 uc, nd superntnts were collected fter 24 h nd clered y centrifugtion. Trnsfection efficiency ws determined y nlysis of GFP expression in the cells nd ws susequently incorported into the virus titre clcultions. The mount of (infectious) virus prticles in the superntnts ws determined y oth infection ssys nd qpcr nlysis of virl RNA. For infection ssys, pre-plted NIH3T3 cells were spin-infected with tenfold serilly diluted superntnts in the presence of 8 mgml 21 polyrene t 6g for 9 min t room temperture. 48 h fter the spin infection, the NIH3T3 cells were exmined for GFP expression y flow cytometry, nd the numer of GFP 1 cells ws used to clculte reltive virl titres. For virl RNA qpcr ssys, virl RNA ws extrcted from the virus superntnt nd 212 Mcmilln Pulishers Limited. All rights reserved
6 RESEARCH LETTER quntified s descried ove with EGFP-specific primers. The production of HIV ws tested similrly fter pnl4-3.luc.r 1 E 2 HIV vector, pcmv-vsv-g vector nd pcdna5 GFP vector were trnsfected into cells of interest using Lipofectmine 2. The trnsfection efficiency ws estlished y flow cytometric determintion of the numer of GFP 1 cells nd used to djust reltive virl titres. Virl titres were determined vi infection ssys, qpcr of virl RNA nd HIV p24 ELISA. The infection ssy ws similr to the MSCV virus infection ssy except HEKT cells were used for nlysis. Expression of luciferse ws mesured y using the Bright-Glo Luciferse Assy System (Promeg) nd microplte luminometer (LUMIstr, BMG-LABTECH) 24 h fter the spin infection. HIV virl RNA ws quntified y qpcr using luciferse- or HIV-p24-specific primers. The HIV p24 ELISA ws performed y the Virl Pthogenesis Core t the UCSD Center for AIDS Reserch using Allince HIV1 p24 ELISA kits (Perkin Elmer). For recominnt AAV production ssys, cells were trnsfected with pxx6 (.6 mg), pxx2 (.2 mg) nd paclaluc (.2 mg) using lipofectmine 2 (Invitrogen), nd collected 72 h fter trnsfection. raavluc-contining lystes were generted y freeze/thw cycles followed y centrifugtion nd were used to infect HEKT cells. Luciferse ctivity ws quntified 48 h post-trnsduction using Stedy-Glo luciferse sustrte regent (Promeg) in 96-well Lumipltes (Greiner Bio-One) in TopCount NXT scintilltion nd luminescence counter (PerkinElmer). Assys for MSCV nd HIV infection. Amphotropic MSCV-IRES-GFP (MIG) retrovirus ws prepred y trnsfecting Phoenix mphotropic retrovirus pckging cells with MSCV-IRES-GFP (MIG) vector using Lipofectmine 2. The cell culture medium ws collected nd replced with fresh medium 48, 72 nd 96 h fter the trnsfection. The comined superntnts were clered y centrifugtion t 1,g t 4 uc for 15 min. Cells to e tested were spin-infected with the indicted dilutions in the presence of 8 mgml 21 polyrene t 6g for 9 min t room temperture. 48 h fter the spin infection, cells were nlysed for GFP expression y flow cytometry. Infection efficiency ws estlished y oth the numer of GFP 1 cells nd the level of GFP expression. VSV-G pseudotyped HIV ws prepred y trnsfecting HEKT cells with pnl4-3.luc.r 1 E 2 HIV nd pcmv-vsv-g vectors using Lipofectmine 2. The cell culture medium ws collected nd clered s descried ove. Cells to e tested were spin-infected with the indicted dilutions in the presence of 8 mgml 21 polyrene t 6g for 9 min t room temperture. 24 h fter the spin infection, luciferse expression ws mesured to determine the reltive infection efficiencies. Trnsmission electron microscopy. HEKT cells were trnsfected with pnl4-3.luc.r 1 E 2 HIV vector together with either pcdna6/v5-his CAT or pcdna6/ V5-His SLFN11 vectors using Lipofectmine h fter the trnsfection, cells were fixed nd processed y the Microscopy Core t the Scripps Reserch Institute. Imges were cquired with Philips CM1 trnsmission electron microscope. Whole-cell lysis nd western lotting. Cells were lysed directly in 1 3 NuPAGE LDS Smple Buffer (Invitrogen) contining 2.5% 2-mercptoethnol nd heted t 9 uc for 5 min. Smples were homogenized y QIAshredder (Qigen), totl protein resolved y NuPAGE (Invitrogen) nd trnsferred to PVDF (polyvinylidene difluoride) memrnes followed y western lotting with the specified ntiodies. Corresponding horserdish peroxidse (HRP)-conjugted secondry ntiodies nd Amershm ECL Plus Western Blotting Detection Regent (GE Helthcre) were used to visulize the signls followed y quntifiction using densitometer nd NIH ImgeJ64 softwre. Alterntively, fluorochromeconjugted secondry ntiodies were used nd signls were cquired nd quntified using Typhoon Trio Vrile Mode Imger nd ImgeQunt softwre. Wild-type HIV virus repliction ssy in CEM cells. HIV-1 LAI ws used to infect the CEM stle cell lines (CEM, CEMshRNA Ctl nd CEMshRNA SLFN ) t m.o.i. of.1 in 5 ml of culture t cells per ml using RPMI/poly medium contining 3 mgml 21 polyrene, 1% glutmine, 1 U ml 21 penicillin, 1 mg ml 21 streptomycin nd 1% FBS. The cultures were smpled t the indicted time points nd the virl titres were mesured y HIV p24 ELISA. trna rrys. mirvn mirna Isoltion kit (Amion) ws used to prepre totl RNA from freshly collected cells following the totl RNA isoltion protocol. RNA smples were lelled nd the reltive undnces of individul trnas were mesured using microrrys s previously descried 25,26. trna electrophoretic moility shift ssy (EMSA). The prtil coding sequence of humn SLFN11 (mino cids 1 579; SLFN11-N) nd the coding sequence of GFP were cloned into the cteril expression vector pet11/d-topo (Invitrogen) with 63His epitope tg. 63His-tgged SLFN11-N nd GFP proteins were expressed in Origmi B Escherichi coli cells nd purified over Ni-NTA nd FPLC columns. The purity of the proteins ws.95% s estimted y Coomssie lue stining. Totl humn trna ws extrcted with the mirvn mirna Isoltion kit (Amion) following the smll RNA isoltion protocol, nd further purified on 15% denturing polycrylmide TBE-ure gel (Invitrogen). To generte EMSA proes, purified humn trna ws decylted to remove chrging mino cid, dephosphorylted nd 59-lelled using [ 32 P]c-ATP. Totl yest trna ws purchsed from Invitrogen. To prepre the HIV virl RNA, the sequence corresponding to the gg-pol frme-shifting region ( , pnl4-3.luc.r 1.E 2 vector) ws mplified y PCR with T7 promoter-contining primers nd in vitro trnscried using MEGAscript T7 kit (Amion). Purified proteins, unlelled competitor RNA or nti-slfn11 ntiody (Amrt), nd proe were comined s specified nd incuted on ice for 3 min in EMSA uffer (5% glycerol, 1 mm ph 7.4 Tris-HCl, 5 mm MgCl 2, 1 mm KCl, 1% Triton X-1, 1 mm DTT nd 1Uml 21 RNsin Plus RNse Inhiitor). After smples were resolved on 6% DNA retrdtion gel (Invitrogen), the gel ws dried nd visulized with Typhoon Trio Vrile Mode Imger (storge phosphor mode). Dt nlysis nd presenttion. Unless indicted otherwise, results re presented in grphs s verge 6 s.d. of t lest three independent trnsfections or infections. For western lots, representtive of t lest three independent trnsfections or infections is shown. For the experiments shown in Fig. 3d, e, GFP nd EGFP protein nd mrna levels were quntified nd djusted to the corresponding protein or mrna concentrtions, respectively. Expression levels of the proteins were then normlized to their corresponding stndrdized mrna concentrtions to compenste for minor vritions in mrna concentrtions etween the smples. 212 Mcmilln Pulishers Limited. All rights reserved
7 SUPPLEMENTARY INFORMATION doi:1.138/nture11433 Ct (TBP-Trget Gene) Unstimulted IFN pdadt pic SLFN5 7.59± ± ± ±.5 SLFN ± ± ± ±.1 SLFN ±.5 8.2± ± ±.2 SLFN ± ± ±.26.67±.46 SLFN12L U.D. 2.2± ± ±.2 SLFN14 U.D. U.D. U.D. U.D. ISG ± ± ± ±.8 Ct (TBP-Trget Gene) HEK HEKT Hel CEM Jurkt hslfn5 2.4±.4 -.4±.2 5.1± ± ±.7 hslfn ±.4 U.D. U.D. 4.58± ±.6 hslfn12 2.1±.8 U.D. 5.±.2-6.7± ±.1 hslfn13-2.4± ± ± ± ±.12 hslfn12l U.D. U.D. U.D. U.D. -3.6±.49 hslfn14-8.2± ±.3 U.D ± ±.51 c Reltive Slfn11 mrna Levels shrna Ctl shrna Slfn T d W: Slfn11 shrnactl W: shrnaslfn T Slfn11 Supplementry Figure 1 Reltive expression nd induciility of huslfns. ) Humn HFF cells were stimulted with IFNβ (5 U/ml), pdadt (1 μg/ml) or pic (1 μg/ml) for 6 hours, nd levels of the indicted mrnas nd TBP were determined y qpcr. The expression of the Slfn nd ISG54 mrnas re presented s ΔCt etween TBP nd the Slfn mrna of interest. ) Totl cellulr RNA ws prepred from the indicted cell lines, nd mrna levels of the huslfns nd of TBP were quntitted y qpcr nd presented s in ). c) huslfn11 mrna levels in, shrnactl, shrnaslfn nd T cells s determined y qpcr; d) Sme s c), except huslfn11 protein ws visulized y immunolotting of whole cell lystes. 1
8 RESEARCH SUPPLEMENTARY INFORMATION Reltive Luciferse Activity shrna Ctl shrna Slfn 1x Dilution 1x Dilution 1x Dilution Supplementry Figure 2 - huslfn11 does not inhiit erly virl infection steps., shrnactl, shrnaslfn cells were infected with pseudotyped HIV-1VSV-G virus t the indicted dilutions, nd infection efficiency ws determined y the expression levels of the trnsduced luciferse in the infected cells. 2
9 SUPPLEMENTARY INFORMATION RESEARCH Reltive MSCV Virus Titer shrna Slfn shrna Ctl T Reltive MSCVVirus Titer T CAT 1.97 T Slfn T Slfn11 T Slfn11N T Slfn11C c Reltive AAV Virus Titer shrna Slfn shrna Ctl T Supplementry Figure 3 huslfn11 inhiits production of MSCV ut not AAV. ), shrnactl, shrnaslfn nd T cells were trnsfected with MSCV-IRES-GFP retrovirus vector nd pcl-eco ecotropic retrovirus pckging vector. Virl titers in the superntnts were determined y infection nd susequent flow-cytometric nlysis of GFP expression of NIH3T3 cells. ) T cells were trnsfected with MSCV-IRES-GFP retrovirus vector/pcl-eco pckging vector nd plsmids encoding either huslfn5, huslfn11, huslfn11n, huslfn11c or CAT s indicted. Virus production ws ssyed y titrted infection of NIH3T3 cells s in ). c), shrnactl, shrnaslfn nd T cells were trnsfected with pxx6, pxx2 nd paclaluc, nd raavluc production determined y luciferse ssy following titrted infection. 3
10 RESEARCH SUPPLEMENTARY INFORMATION Cytoplsmic HIV RNA Levels HIVp24 Luc CAT SLFN5 SLFN11 T CAT T Slfn11 Supplementry Figure 4 huslfn11 does not lter cytoplsmic vrna levels or cuse ccumultion of virl prticles. ) T cells were trnsfected with pnl4-3.luc.r+e- together with either huslfn5, huslfn11 or CAT. After cellulr frctiontion, cytoplsmic vrna concentrtion ws nlyzed y qpcr; ) T cells were trnsfected with HIV-1 pnl4-3.luc.r+e- vector together with pcdna6-cat or pcdna6-slfn hours lter, the cells were fixed nd imges were cquired y electronic microscopy. 4
11 SUPPLEMENTARY INFORMATION RESEARCH pnl4-3.luc.r+e-prstop T CAT T Slfn5 T Slfn11 shrna Ctl shrna Slfn W: Gg W: Gg p55 W: W: pspax2 & pnl-gfp-rre(sa) CAT Slfn5 Slfn11 Slfn11-N Slfn11-C W: GFP GFP W: Gg p24 W: Supplementry Figure 5 huslfn11 inhiition is independent of virl enzymes or nucler export of unspliced RNA. ) T cells were co-trnsfected with HIV-1 pnl4-3.luc.r+e-prstop vector nd either huslfn5, huslfn11 or CAT s indicted (left pnels), or, shrnactl nd shrnaslfn cells were trnsfected with pnl4-3.luc.r+e-prstop vector (right pnels). Whole cell lystes derived 48 hours fter trnsfection were immunolotted with nti-gg nd nti- ntiodies. ) T cells were trnsfected with pspax2 (encoding Gg, Pol, Tt & Rev without ny LTR elements) nd pnl-gfp-rre(sa) (providing splicle RNA yielding GFP only if exported s unspliced RNA vi Rev/RRE interction) together with plsmids encoding either huslfn5, huslfn11, huslfn11n, huslfn11c or CAT s indicted. Whole cell lystes were immunolotted with nti-gfp, nti-gg nd nti- ntiodies. 5
12 RESEARCH SUPPLEMENTARY INFORMATION No Protein Slfn11N Totl (T) Unshifted (U) Shifted (S) S T U Supplementry Figure 6 Non-discriminting trna-inding of huslfn11. Gel purified 32P-leled totl trna ws incuted with incresing mounts of huslfn11n to otin pproximte 5% shifting on ntive gel. 32P-leled trna from shifted nd unshifted nds ws recovered nd smples hyridized on microrrys contining 96 proes (proes re repeted 8 times ech nd re complementry to humn/mouse nucler-encoded nd mitochondril-encoded trnas). 6
13 SUPPLEMENTARY INFORMATION RESEARCH 45 CEM HIV p24 (ng/ml) 3 15 CEMshRNA Ctl CEMshRNA Slfn Dys post infection Supplementry Figure 7 huslfn11 inhiits HIV repliction. The plko.1 shrna vector ginst Slfn11 (TRCN15257, CCGGCAGTCTTTGAGAGAGCTTATTCT- CGAGAATAAGCTCTCTCAAAGACTGTTTTTTG) ws used to independently crete second huslfn11-deficient CEM cell line (CEMshRNASlfn in this figure). CEM, CEMshRNACtl nd the new CEMshRNASlfn cells were infected with HIV-1LAI t n MOI of.1. The extent of virl repliction ws determined y HIV p24 ELISA of culture superntnts t indicted time points. 7
Supplementary figure 1
 Supplementry figure 1 Dy 8 post LCMV infection Vsculr Assoc. Prenchym Dy 3 post LCMV infection 1 5 6.7.29 1 4 1 3 1 2 88.9 4.16 1 2 1 3 1 4 1 5 1 5 1.59 5.97 1 4 1 3 1 2 21.4 71 1 2 1 3 1 4 1 5 1 5.59.22
Supplementry figure 1 Dy 8 post LCMV infection Vsculr Assoc. Prenchym Dy 3 post LCMV infection 1 5 6.7.29 1 4 1 3 1 2 88.9 4.16 1 2 1 3 1 4 1 5 1 5 1.59 5.97 1 4 1 3 1 2 21.4 71 1 2 1 3 1 4 1 5 1 5.59.22
SUPPLEMENTARY INFORMATION
 doi:0.08/nture078 RNse VifHA VifHA βctin 6 Cell lyste IP: ntiha MG VifHA VifHA β ctin 6 7 Cell lyste IP: ntiha Supplementry Figure. Effect of RNse nd MG tretment on the Vif interction., RNse tretment does
doi:0.08/nture078 RNse VifHA VifHA βctin 6 Cell lyste IP: ntiha MG VifHA VifHA β ctin 6 7 Cell lyste IP: ntiha Supplementry Figure. Effect of RNse nd MG tretment on the Vif interction., RNse tretment does
SUPPLEMENTARY INFORMATION
 DOI: 1.138/nc286 Figure S1 e f Medium DMSO AktVIII PP242 Rp S6K1-I Gr1 + + + + + + Strvtion + + + + + IB: Akt-pT38 IB: Akt K-pT389 K IB: Rptor Gr1 shs6k1-a shs6k1-b shs6k1-c shrictor shrptor Gr1 c IB:
DOI: 1.138/nc286 Figure S1 e f Medium DMSO AktVIII PP242 Rp S6K1-I Gr1 + + + + + + Strvtion + + + + + IB: Akt-pT38 IB: Akt K-pT389 K IB: Rptor Gr1 shs6k1-a shs6k1-b shs6k1-c shrictor shrptor Gr1 c IB:
TNF-α (pg/ml) IL-6 (ng/ml)
 Xio, et l., Supplementry Figure 1 IL-6 (ng/ml) TNF-α (pg/ml) 16 12 8 4 1,4 1,2 1, 8 6 4 2 med Cl / Pm3CSK4 zymosn curdln Poly (I:C) LPS flgelin MALP-2 imiquimod R848 CpG TNF-α (pg/ml) IL-6 (ng/ml) 2 1.6
Xio, et l., Supplementry Figure 1 IL-6 (ng/ml) TNF-α (pg/ml) 16 12 8 4 1,4 1,2 1, 8 6 4 2 med Cl / Pm3CSK4 zymosn curdln Poly (I:C) LPS flgelin MALP-2 imiquimod R848 CpG TNF-α (pg/ml) IL-6 (ng/ml) 2 1.6
Ulk λ PPase. 32 P-Ulk1 32 P-GST-TSC2. Ulk1 GST (TSC2) : Ha-Ulk1 : AMPK. WB: Ha (Ulk1) : Glu. h CON - Glu - A.A WB: LC3 AMPK-WT AMPK-DKO
 DOI: 10.1038/ncb2152 C.C + - + - : Glu b Ulk1 - - + λ PPse c AMPK + - + + : ATP P-GST-TSC2 WB: Flg (Ulk1) WB Ulk1 WB: H (Ulk1) GST (TSC2) C.C d e WT K46R - + - + : H-Ulk1 : AMPK - + - + + + AMPK H-Ulk1
DOI: 10.1038/ncb2152 C.C + - + - : Glu b Ulk1 - - + λ PPse c AMPK + - + + : ATP P-GST-TSC2 WB: Flg (Ulk1) WB Ulk1 WB: H (Ulk1) GST (TSC2) C.C d e WT K46R - + - + : H-Ulk1 : AMPK - + - + + + AMPK H-Ulk1
SUPPLEMENTARY INFORMATION
 doi:1.138/nture1794 BR EPFs BRI1? ERECTA TMM BSKs YDA PP2A BSU1 BIN2 pbzr1/2 BZR1/2 MKK4/5/7/9 MPK3/6 SPCH Cell growth Stomtl production Supplementry Figure 1. The model of BR nd stomtl signling pthwys.
doi:1.138/nture1794 BR EPFs BRI1? ERECTA TMM BSKs YDA PP2A BSU1 BIN2 pbzr1/2 BZR1/2 MKK4/5/7/9 MPK3/6 SPCH Cell growth Stomtl production Supplementry Figure 1. The model of BR nd stomtl signling pthwys.
SUPPLEMENTARY INFORMATION
 Prentl doi:.8/nture57 Figure S HPMECs LM Cells Cell lines VEGF (ng/ml) Prentl 7. +/-. LM 7. +/-.99 LM 7. +/-.99 Fold COX induction 5 VEGF: - + + + Bevcizum: - - 5 (µg/ml) Reltive MMP LM mock COX MMP LM+
Prentl doi:.8/nture57 Figure S HPMECs LM Cells Cell lines VEGF (ng/ml) Prentl 7. +/-. LM 7. +/-.99 LM 7. +/-.99 Fold COX induction 5 VEGF: - + + + Bevcizum: - - 5 (µg/ml) Reltive MMP LM mock COX MMP LM+
SUPPLEMENTARY INFORMATION
 X p -lu c ct ivi ty doi:.8/nture8 S CsA - THA + DAPI Merge FSK THA TUN Supplementry Figure : A. Ad-Xp luc ctivity in primry heptocytes exposed to FSK, THA, or TUN s indicted. Luciferse ctivity normlized
X p -lu c ct ivi ty doi:.8/nture8 S CsA - THA + DAPI Merge FSK THA TUN Supplementry Figure : A. Ad-Xp luc ctivity in primry heptocytes exposed to FSK, THA, or TUN s indicted. Luciferse ctivity normlized
SUPPLEMENTARY INFORMATION
 doi:10.1038/nture09973 Plsm Memrne Phgosome TLR1/2/4 ROS Mitochondrion ROS OXPHOS Complex I ROS TRAF6 NADPH Oxidse Supplementry Figure 1 Model detiling the roles of mitochondril ROS in mcrophge cteril
doi:10.1038/nture09973 Plsm Memrne Phgosome TLR1/2/4 ROS Mitochondrion ROS OXPHOS Complex I ROS TRAF6 NADPH Oxidse Supplementry Figure 1 Model detiling the roles of mitochondril ROS in mcrophge cteril
Supplementary Information. SAMHD1 Restricts HIV-1 Infection in Resting CD4 + T Cells
 Supplementry Informtion SAMHD Restricts HIV- Infection in Resting CD T Cells Hnn-Mri Blduf,2,, Xioyu Pn,, Elin Erikson,2, Srh Schmidt, Wqo Dddch 3, Mnj Burggrf, Kristin Schenkov, In Amiel,2, Guido Wnitz
Supplementry Informtion SAMHD Restricts HIV- Infection in Resting CD T Cells Hnn-Mri Blduf,2,, Xioyu Pn,, Elin Erikson,2, Srh Schmidt, Wqo Dddch 3, Mnj Burggrf, Kristin Schenkov, In Amiel,2, Guido Wnitz
EFFECTS OF AN ACUTE ENTERIC DISEASE CHALLENGE ON IGF-1 AND IGFBP-3 GENE EXPRESSION IN PORCINE SKELETAL MUSCLE
 Swine Dy 22 Contents EFFECTS OF AN ACUTE ENTERIC DISEASE CHALLENGE ON IGF-1 AND IGFBP-3 GENE EXPRESSION IN PORCINE SKELETAL MUSCLE B. J. Johnson, J. P. Kyser, J. D. Dunn, A. T. Wyln, S. S. Dritz 1, J.
Swine Dy 22 Contents EFFECTS OF AN ACUTE ENTERIC DISEASE CHALLENGE ON IGF-1 AND IGFBP-3 GENE EXPRESSION IN PORCINE SKELETAL MUSCLE B. J. Johnson, J. P. Kyser, J. D. Dunn, A. T. Wyln, S. S. Dritz 1, J.
Heparanase promotes tumor infiltration and antitumor activity of CAR-redirected T- lymphocytes
 Supporting Online Mteril for Heprnse promotes tumor infiltrtion nd ntitumor ctivity of -redirected T- lymphocytes IgnzioCrun, Brr Svoldo, VlentinHoyos, Gerrit Weer, Ho Liu, Eugene S. Kim, Michel M. Ittmnn,
Supporting Online Mteril for Heprnse promotes tumor infiltrtion nd ntitumor ctivity of -redirected T- lymphocytes IgnzioCrun, Brr Svoldo, VlentinHoyos, Gerrit Weer, Ho Liu, Eugene S. Kim, Michel M. Ittmnn,
Acute and gradual increases in BDNF concentration elicit distinct signaling and functions in neurons
 nd grdul increses in BDNF concentrtion elicit distinct signling nd functions in neurons Yunyun Ji,, Yun Lu, Feng Yng, Wnhu Shen, Tin Tze-Tsng Tng,, Linyin Feng, Shumin Dun, nd Bi Lu,.. - Grdul (normlized
nd grdul increses in BDNF concentrtion elicit distinct signling nd functions in neurons Yunyun Ji,, Yun Lu, Feng Yng, Wnhu Shen, Tin Tze-Tsng Tng,, Linyin Feng, Shumin Dun, nd Bi Lu,.. - Grdul (normlized
supplementary information
 DOI: 10.1038/nc2089 H3K4me1 H3K4me1 H3K4me1 H3K4me1 H3K4me1 H3K4me1 5 PN N1-2 PN H3K4me1 H3K4me1 H3K4me1 2-cell stge 2-c st cell ge Figure S1 Pttern of loclistion of H3K4me1 () nd () during zygotic development
DOI: 10.1038/nc2089 H3K4me1 H3K4me1 H3K4me1 H3K4me1 H3K4me1 H3K4me1 5 PN N1-2 PN H3K4me1 H3K4me1 H3K4me1 2-cell stge 2-c st cell ge Figure S1 Pttern of loclistion of H3K4me1 () nd () during zygotic development
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nture1188 1mM CCl 2 (min) 3 4 6 CCl 2 (mm) for 4min.1. 1 (mm) Pro- d WT GdCl 3 R-68 -/- P2x7r -/- -/- Csp1 -/- WT -/- P2x7r -/- -/- Csp1 -/- Csp1 (p2) (p17) Pro-Csp1
SUPPLEMENTARY INFORMATION doi:1.138/nture1188 1mM CCl 2 (min) 3 4 6 CCl 2 (mm) for 4min.1. 1 (mm) Pro- d WT GdCl 3 R-68 -/- P2x7r -/- -/- Csp1 -/- WT -/- P2x7r -/- -/- Csp1 -/- Csp1 (p2) (p17) Pro-Csp1
SUPPLEMENTARY INFORMATION
 . Norml Physiologicl Conditions. SIRT1 Loss-of-Function S1. Model for the role of SIRT1 in the regultion of memory nd plsticity. () Our findings suggest tht SIRT1 normlly functions in coopertion with YY1,
. Norml Physiologicl Conditions. SIRT1 Loss-of-Function S1. Model for the role of SIRT1 in the regultion of memory nd plsticity. () Our findings suggest tht SIRT1 normlly functions in coopertion with YY1,
LETTERS. Interferon modulation of cellular micrornas as an antiviral mechanism
 Vol 449 18 Octoer 27 doi:1.138/nture625 LETTERS Interferon modultion of cellulr micrornas s n ntivirl mechnism Irene M. Pedersen 1, Guofeng heng 3, Stefn Wielnd 3, Stefno Volini 4, rlo M. roce 4, Frncis
Vol 449 18 Octoer 27 doi:1.138/nture625 LETTERS Interferon modultion of cellulr micrornas s n ntivirl mechnism Irene M. Pedersen 1, Guofeng heng 3, Stefn Wielnd 3, Stefno Volini 4, rlo M. roce 4, Frncis
% Inhibition of MERS pseudovirus infection. 0 h 0.5 h 1 h 2 h 4 h 6 h Time after virus addition
 % Inhiition of MERS pseudovirus infection 1 8 h.5 h 1 h 2 h 4 h 6 h Time fter virus ddition Supplementry Figure S1. Inhiition of on MERS pseudovirus infection t the different intervls postinfection. A
% Inhiition of MERS pseudovirus infection 1 8 h.5 h 1 h 2 h 4 h 6 h Time fter virus ddition Supplementry Figure S1. Inhiition of on MERS pseudovirus infection t the different intervls postinfection. A
Copy Number ID2 MYCN ID2 MYCN. Copy Number MYCN DDX1 ID2 KIDINS220 MBOAT2 ID2
 Copy Numer Copy Numer Copy Numer Copy Numer DIPG38 DIPG49 ID2 MYCN ID2 MYCN c DIPG01 d DIPG29 ID2 MYCN ID2 MYCN e STNG2 f MYCN DIPG01 Chr. 2 DIPG29 Chr. 1 MYCN DDX1 Chr. 2 ID2 KIDINS220 MBOAT2 ID2 Supplementry
Copy Numer Copy Numer Copy Numer Copy Numer DIPG38 DIPG49 ID2 MYCN ID2 MYCN c DIPG01 d DIPG29 ID2 MYCN ID2 MYCN e STNG2 f MYCN DIPG01 Chr. 2 DIPG29 Chr. 1 MYCN DDX1 Chr. 2 ID2 KIDINS220 MBOAT2 ID2 Supplementry
NappHS. rrna. transcript abundance. NappHS relative con W+W 0.8. nicotine [µg mg -1 FM]
![NappHS. rrna. transcript abundance. NappHS relative con W+W 0.8. nicotine [µg mg -1 FM] NappHS. rrna. transcript abundance. NappHS relative con W+W 0.8. nicotine [µg mg -1 FM]](/thumbs/92/110177253.jpg) (A) W+OS 3 min 6 min con L S L S RNA loding control NppHS rrna (B) (C) 8 1 k NppHS reltive trnscript undnce 6 4.5 *** *** *** *** 3 k. + + + line 1 line (D) nicotine [µg mg -1 FM] 1..8.4. con W+W Supplementl
(A) W+OS 3 min 6 min con L S L S RNA loding control NppHS rrna (B) (C) 8 1 k NppHS reltive trnscript undnce 6 4.5 *** *** *** *** 3 k. + + + line 1 line (D) nicotine [µg mg -1 FM] 1..8.4. con W+W Supplementl
SUPPLEMENTARY INFORMATION
 Supplementry Figure 1. Genertion of N- nd C-tgged cyclin D1 knock-in mice., N-tgged cyclin D1 gene trgeting construct, cyclin D1 genomic locus, cyclin D1 locus following homologous recomintion (trgeted
Supplementry Figure 1. Genertion of N- nd C-tgged cyclin D1 knock-in mice., N-tgged cyclin D1 gene trgeting construct, cyclin D1 genomic locus, cyclin D1 locus following homologous recomintion (trgeted
*** *** *** *** T-cells T-ALL. T-cells T-ALL. T-cells T-ALL. T-cells T-ALL. T-cells T-ALL. Relative ATP content. Relative ATP content RLU RLU
 RLU Events 1 1 1 Luciferin (μm) T-cells T-ALL 1 1 Time (min) T-cells T-ALL 1 1 1 1 DCF-DA Reltive ATP content....1.1.. T-cells T-ALL RLU 1 1 T-cells T-ALL Luciferin (μm) 1 1 Time (min) c d Control e DCFH-DA
RLU Events 1 1 1 Luciferin (μm) T-cells T-ALL 1 1 Time (min) T-cells T-ALL 1 1 1 1 DCF-DA Reltive ATP content....1.1.. T-cells T-ALL RLU 1 1 T-cells T-ALL Luciferin (μm) 1 1 Time (min) c d Control e DCFH-DA
Supplementary Figure 1
 Roles of endoplsmic reticulum stress-medited poptosis in -polrized mcrophges during mycocteril infections Supplementry informtion Yun-Ji Lim, Min-Hee Yi, Ji-Ae Choi, Jung-hwn Lee, Ji-Ye Hn, Sung-Hee Jo,
Roles of endoplsmic reticulum stress-medited poptosis in -polrized mcrophges during mycocteril infections Supplementry informtion Yun-Ji Lim, Min-Hee Yi, Ji-Ae Choi, Jung-hwn Lee, Ji-Ye Hn, Sung-Hee Jo,
Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome
 Supplementry Informtion Microtuule-driven sptil rrngement of mitochondri promotes ctivtion of the NLRP3 inflmmsome Tkum Misw 1,2, Michihiro Tkhm 1,2, Ttsuy Kozki 1,2, Hnn Lee 1,2, Jin Zou 1,2, Ttsuy Sitoh
Supplementry Informtion Microtuule-driven sptil rrngement of mitochondri promotes ctivtion of the NLRP3 inflmmsome Tkum Misw 1,2, Michihiro Tkhm 1,2, Ttsuy Kozki 1,2, Hnn Lee 1,2, Jin Zou 1,2, Ttsuy Sitoh
SUPPLEMENTARY INFORMATION
 doi:1.138/nture1228 Totl Cell Numer (cells/μl of lood) 12 1 8 6 4 2 d Peripherl Blood 2 4 7 Time (d) fter nti-cd3 i.p. + TCRβ + IL17A + cells (%) 7 6 5 4 3 2 1 Totl Cell Numer (x1 3 ) 8 7 6 5 4 3 2 1 %
doi:1.138/nture1228 Totl Cell Numer (cells/μl of lood) 12 1 8 6 4 2 d Peripherl Blood 2 4 7 Time (d) fter nti-cd3 i.p. + TCRβ + IL17A + cells (%) 7 6 5 4 3 2 1 Totl Cell Numer (x1 3 ) 8 7 6 5 4 3 2 1 %
Supplementary Figure 1
 doi: 1.138/nture6188 SUPPLEMENTARY INFORMATION Supplementry Figure 1 c CFU-F colonies per 1 5 stroml cells 14 12 1 8 6 4 2 Mtrigel plug Neg. MCF7/Rs MDA-MB-231 * * MCF7/Rs-Lung MDA-MB-231-Lung MCF7/Rs-Kidney
doi: 1.138/nture6188 SUPPLEMENTARY INFORMATION Supplementry Figure 1 c CFU-F colonies per 1 5 stroml cells 14 12 1 8 6 4 2 Mtrigel plug Neg. MCF7/Rs MDA-MB-231 * * MCF7/Rs-Lung MDA-MB-231-Lung MCF7/Rs-Kidney
Using Paclobutrazol to Suppress Inflorescence Height of Potted Phalaenopsis Orchids
 Using Pcloutrzol to Suppress Inflorescence Height of Potted Phlenopsis Orchids A REPORT SUBMITTED TO FINE AMERICAS Linsey Newton nd Erik Runkle Deprtment of Horticulture Spring 28 Using Pcloutrzol to Suppress
Using Pcloutrzol to Suppress Inflorescence Height of Potted Phlenopsis Orchids A REPORT SUBMITTED TO FINE AMERICAS Linsey Newton nd Erik Runkle Deprtment of Horticulture Spring 28 Using Pcloutrzol to Suppress
TLR7 induces anergy in human CD4 + T cells
 TLR7 induces nergy in humn CD T cells Mrgrit Dominguez-Villr 1, Anne-Sophie Gutron 1, Mrine de Mrcken 1, Mrl J Keller & Dvid A Hfler 1 The recognition of microil ptterns y Toll-like receptors (TLRs) is
TLR7 induces nergy in humn CD T cells Mrgrit Dominguez-Villr 1, Anne-Sophie Gutron 1, Mrine de Mrcken 1, Mrl J Keller & Dvid A Hfler 1 The recognition of microil ptterns y Toll-like receptors (TLRs) is
SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef
 doi:1.138/nture154 SERINC3 nd SERINC5 restrict HIV-1 infectivity nd re countercted y Nef Yoshiko Usmi 1, Yunfei Wu 1 & Heinrich G. Göttlinger 1 HIV-1 Nef nd the unrelted mouse leukemi virus glycosylted
doi:1.138/nture154 SERINC3 nd SERINC5 restrict HIV-1 infectivity nd re countercted y Nef Yoshiko Usmi 1, Yunfei Wu 1 & Heinrich G. Göttlinger 1 HIV-1 Nef nd the unrelted mouse leukemi virus glycosylted
 DOI: 10.1038/nc2331 PCre;Ros26R 12 h induction 48 h induction Vegfr3 i EC c d ib4 24 h induction VEGFR3 e Fold chnge 1.0 0.5 P < 0.05 Vegfr3 i EC Vegfr3 Figure S1 Cre ctivtion leds to genetic deletion
DOI: 10.1038/nc2331 PCre;Ros26R 12 h induction 48 h induction Vegfr3 i EC c d ib4 24 h induction VEGFR3 e Fold chnge 1.0 0.5 P < 0.05 Vegfr3 i EC Vegfr3 Figure S1 Cre ctivtion leds to genetic deletion
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nture07679 Emryonic Stem (ES) cell Hemngiolst Flk1 + Blst Colony 3 to 3.5 Dys 3-4 Dys ES differentition Sort of Flk1 + cells Supplementry Figure 1. Chrcteristion of lst colony development.
doi: 10.1038/nture07679 Emryonic Stem (ES) cell Hemngiolst Flk1 + Blst Colony 3 to 3.5 Dys 3-4 Dys ES differentition Sort of Flk1 + cells Supplementry Figure 1. Chrcteristion of lst colony development.
SUPPLEMENTARY INFORMATION
 TM TM tip link horizontl top connectors 1 leucine-rich (21 %) otoncorin-like 1809 ntigenic peptides B D signl peptide hydrophoic segment proline/threonine-rich (79 %) Supplementry Figure 1. () The outer
TM TM tip link horizontl top connectors 1 leucine-rich (21 %) otoncorin-like 1809 ntigenic peptides B D signl peptide hydrophoic segment proline/threonine-rich (79 %) Supplementry Figure 1. () The outer
Supplementary Figure 1
 Supplementry Figure 1 c d Wistr SHR Wistr AF-353 SHR AF-353 n = 6 n = 6 n = 28 n = 3 n = 12 n = 12 Supplementry Figure 1 Neurophysiologicl properties of petrosl chemoreceptive neurones in Wistr nd SH rts.
Supplementry Figure 1 c d Wistr SHR Wistr AF-353 SHR AF-353 n = 6 n = 6 n = 28 n = 3 n = 12 n = 12 Supplementry Figure 1 Neurophysiologicl properties of petrosl chemoreceptive neurones in Wistr nd SH rts.
SUPPLEMENTARY INFORMATION
 doi:1.138/nture17 Men tumour dimeter (mm) 2 Rg2-/- 2 1 2 2 1 Control IgG!-CD8!-CD4 1 2 3 1 2 3 c Men tumour dimeter (mm) 2 2 1 d Ifnr1-/- Rg2-/- 2 2 1 Ifngr1-/- d42m1!ic 1 2 3 Dys post trnsplnt 1 2 3 Supplementry
doi:1.138/nture17 Men tumour dimeter (mm) 2 Rg2-/- 2 1 2 2 1 Control IgG!-CD8!-CD4 1 2 3 1 2 3 c Men tumour dimeter (mm) 2 2 1 d Ifnr1-/- Rg2-/- 2 2 1 Ifngr1-/- d42m1!ic 1 2 3 Dys post trnsplnt 1 2 3 Supplementry
Ras enhances TGF-β signaling by decreasing cellular protein levels of its type II receptor negative regulator SPSB1
 Liu et l. Cell Communiction nd Signling (2018) 16:10 https://doi.org/10.1186/s12964-018-0223-4 RESEARCH Open Access Rs enhnces TGF-β signling y decresing cellulr protein levels of its type II receptor
Liu et l. Cell Communiction nd Signling (2018) 16:10 https://doi.org/10.1186/s12964-018-0223-4 RESEARCH Open Access Rs enhnces TGF-β signling y decresing cellulr protein levels of its type II receptor
Supplementary Materials. Viral delivery of mir-196a ameliorates the SBMA phenotype via the silencing of CELF2
 Supplementry Mterils Virl delivery of mir-96 meliortes the SBMA phenotype vi the silencing of CELF2 Yu Miyzki, Hiroki Adchi, Mshis Ktsuno, Mkoto Minmiym, Yue-Mei Jing, Zhe Hung, Hideki Doi, Shinjiro Mtsumoto,
Supplementry Mterils Virl delivery of mir-96 meliortes the SBMA phenotype vi the silencing of CELF2 Yu Miyzki, Hiroki Adchi, Mshis Ktsuno, Mkoto Minmiym, Yue-Mei Jing, Zhe Hung, Hideki Doi, Shinjiro Mtsumoto,
Agilent G6825AA MassHunter Pathways to PCDL Software Quick Start Guide
 Agilent G6825AA MssHunter Pthwys to PCDL Softwre Quick Strt Guide Wht is Agilent Pthwys to PCDL? Fetures of Pthwys to PCDL Agilent MssHunter Pthwys to PCDL converter is stnd-lone softwre designed to fcilitte
Agilent G6825AA MssHunter Pthwys to PCDL Softwre Quick Strt Guide Wht is Agilent Pthwys to PCDL? Fetures of Pthwys to PCDL Agilent MssHunter Pthwys to PCDL converter is stnd-lone softwre designed to fcilitte
SUPPLEMENTARY INFORMATION
 DOI:.38/nc393 Nnog DAPI Nnog/DAPI c-jun DAPI c-jun/dapi c e Reltive expression to Gpdh mescs ( Feeder free) mescs ( Feeder) MEFs.5 MEFs ipscs ESCs..5 p=.24 p=.483 p=.22. JunB JunD Fos Fr Fr2 ATF2 ATF3
DOI:.38/nc393 Nnog DAPI Nnog/DAPI c-jun DAPI c-jun/dapi c e Reltive expression to Gpdh mescs ( Feeder free) mescs ( Feeder) MEFs.5 MEFs ipscs ESCs..5 p=.24 p=.483 p=.22. JunB JunD Fos Fr Fr2 ATF2 ATF3
Microarrays of lentiviruses for gene function screens in immortalized and primary cells
 26 Nture Pulishing Group http://www.nture.com/nturemethods Microrrys of lentiviruses for gene function screens in immortlized nd primry cells Steve N Biley, Sirj M Ali, Anne E Crpenter, Citlin O Higgins
26 Nture Pulishing Group http://www.nture.com/nturemethods Microrrys of lentiviruses for gene function screens in immortlized nd primry cells Steve N Biley, Sirj M Ali, Anne E Crpenter, Citlin O Higgins
Supplementary Fig. 1. Aortic micrornas are differentially expressed in PFM v. GFM.
 Reltive expression 5 1 15 2-5 5 (n = 3) (n = 3) GFM nd PFM PFM GFM mmu-mir-145 mmu-mir-143 mmu-mir-72 mmu-mir-22 mmu-mir-27 mmu-mir-125-5p mmu-mir-23 mmu-mir-29 mmu-mir-126-3p mmu-let-7d mmu-let-7c mmu-mir-199-3p
Reltive expression 5 1 15 2-5 5 (n = 3) (n = 3) GFM nd PFM PFM GFM mmu-mir-145 mmu-mir-143 mmu-mir-72 mmu-mir-22 mmu-mir-27 mmu-mir-125-5p mmu-mir-23 mmu-mir-29 mmu-mir-126-3p mmu-let-7d mmu-let-7c mmu-mir-199-3p
SUPPLEMENTARY INFORMATION
 doi:0.08/nture0987 SUPPLEMENTARY FIGURE Structure of rbbit Xist gene. Exons re shown in boxes with romn numbers, introns in thin lines. Arrows indicte the locliztion of primers used for mplifiction. WWW.NATURE.COM/NATURE
doi:0.08/nture0987 SUPPLEMENTARY FIGURE Structure of rbbit Xist gene. Exons re shown in boxes with romn numbers, introns in thin lines. Arrows indicte the locliztion of primers used for mplifiction. WWW.NATURE.COM/NATURE
Supplementary Figure S1
 Supplementry Figure S Connexin4 TroponinI Merge Plsm memrne Met Intrcellulr Met Supplementry Figure S H9c rt crdiomyolsts cell line. () Immunofluorescence of crdic mrkers: Connexin4 (green) nd TroponinI
Supplementry Figure S Connexin4 TroponinI Merge Plsm memrne Met Intrcellulr Met Supplementry Figure S H9c rt crdiomyolsts cell line. () Immunofluorescence of crdic mrkers: Connexin4 (green) nd TroponinI
Identification of a tripartite interaction between the N terminus of HIV 1 Vif and CBFβ that is critical for Vif function
 DOI 1.1186/s12977-17-346-5 Retrovirology RESEARCH Open Access Identifiction of triprtite interction etween the N terminus of HIV 1 nd CBFβ tht is criticl for function Belete A. Desimmie 1, Jessic L. Smith
DOI 1.1186/s12977-17-346-5 Retrovirology RESEARCH Open Access Identifiction of triprtite interction etween the N terminus of HIV 1 nd CBFβ tht is criticl for function Belete A. Desimmie 1, Jessic L. Smith
SUPPLEMENTARY INFORMATION
 doi:10.1038/nture09663 Scrmle shnlrp3 shcsp1 IL-1β (p17) IL-1β (pg/ml) 2000 1500 1000 500 Wt Nlrp3-/- Ipf-/- 0 APDC IL-1β (p17) Supplementl Figure 1. Mitochondril ROS cn trigger NLRP3 inflmmsome ctivtion,
doi:10.1038/nture09663 Scrmle shnlrp3 shcsp1 IL-1β (p17) IL-1β (pg/ml) 2000 1500 1000 500 Wt Nlrp3-/- Ipf-/- 0 APDC IL-1β (p17) Supplementl Figure 1. Mitochondril ROS cn trigger NLRP3 inflmmsome ctivtion,
Gene expression phenotypic models that predict the activity of oncogenic pathways
 3 Nture Pulishing Group http://www.nture.com/nturegenetics Gene expression phenotypic models tht predict the ctivity of oncogenic pthwys Erich Hung,, Seiichi Ishid,7, Jennifer Pittmn,3, Holly Dressmn,,4,
3 Nture Pulishing Group http://www.nture.com/nturegenetics Gene expression phenotypic models tht predict the ctivity of oncogenic pthwys Erich Hung,, Seiichi Ishid,7, Jennifer Pittmn,3, Holly Dressmn,,4,
AMPK maintains energy homeostasis and survival in cancer cells via. regulating p38/pgc-1α-mediated mitochondrial biogenesis
 SUPPLEMENTARY INFORMATION AMPK mintins energy homeostsis nd survivl in cncer cells vi regulting p38/pgc-1α-medited mitochondril iogenesis Blkrishn Chue 1, Prmnnd Mlvi 1, Shivendr Vikrm Singh 1, Noshd Mohmmd
SUPPLEMENTARY INFORMATION AMPK mintins energy homeostsis nd survivl in cncer cells vi regulting p38/pgc-1α-medited mitochondril iogenesis Blkrishn Chue 1, Prmnnd Mlvi 1, Shivendr Vikrm Singh 1, Noshd Mohmmd
A rt i c l e s. a Events (% of max)
 Continuous requirement for the TCR in regultory T cell function Andrew G Levine 1,, Aron Arvey 1,,4, Wei Jin 1,,4 & Alexnder Y Rudensky 1 3 14 Nture Americ, Inc. All rights reserved. Foxp3 + regultory
Continuous requirement for the TCR in regultory T cell function Andrew G Levine 1,, Aron Arvey 1,,4, Wei Jin 1,,4 & Alexnder Y Rudensky 1 3 14 Nture Americ, Inc. All rights reserved. Foxp3 + regultory
PROVEN ANTICOCCIDIAL IN NEW FORMULATION
 PROVEN ANTICOCCIDIAL IN NEW FORMULATION Coxidin 100 microgrnulte A coccidiosttic dditive for roilers, chickens rered for lying nd turkeys Contins 100 g of monensin sodium per kg Aville s homogenous grnules
PROVEN ANTICOCCIDIAL IN NEW FORMULATION Coxidin 100 microgrnulte A coccidiosttic dditive for roilers, chickens rered for lying nd turkeys Contins 100 g of monensin sodium per kg Aville s homogenous grnules
Effect of Aqueous Extract of Carica papaya Dry Root Powder on Lactation of Albino Rats
 Effect of Aqueous Extrct of Cric ppy Dry Root Powder on Lcttion of Alino Rts G. Tosswnchuntr nd S. Aritjt Deprtment of Biology Fculty of Science Ching Mi University Ching Mi 50200 Thilnd Keywords: mmmry
Effect of Aqueous Extrct of Cric ppy Dry Root Powder on Lcttion of Alino Rts G. Tosswnchuntr nd S. Aritjt Deprtment of Biology Fculty of Science Ching Mi University Ching Mi 50200 Thilnd Keywords: mmmry
MicroRNA-dependent localization of targeted mrnas to mammalian P-bodies
 MicroRNA-dependent locliztion of trgeted mrnas to mmmlin P-odies Jidong Liu 1, Mrco Antonio Vlenci-Snchez 2, Gregory J. Hnnon 1,3 nd Roy Prker 2,3 Smll RNAs, including smll interfering RNAs (sirnas) nd
MicroRNA-dependent locliztion of trgeted mrnas to mmmlin P-odies Jidong Liu 1, Mrco Antonio Vlenci-Snchez 2, Gregory J. Hnnon 1,3 nd Roy Prker 2,3 Smll RNAs, including smll interfering RNAs (sirnas) nd
SUPPLEMENTARY INFORMATION
 doi:.38/nture73 Glucose SSP THF Methyl- THF Purines 3- PG PHGDH PSAT PSPH Glycine SHMT GSH Lctte Pyruvte ROS TCA Cycle Glucose p53 p Purines 3- PG PHGDH PSAT PSPH Glycine SHMT GSH Lctte Pyruvte ROS TCA
doi:.38/nture73 Glucose SSP THF Methyl- THF Purines 3- PG PHGDH PSAT PSPH Glycine SHMT GSH Lctte Pyruvte ROS TCA Cycle Glucose p53 p Purines 3- PG PHGDH PSAT PSPH Glycine SHMT GSH Lctte Pyruvte ROS TCA
Supplementary Information
 Supplementry Informtion Cutneous immuno-surveillnce nd regultion of inflmmtion y group 2 innte lymphoid cells Ben Roediger, Ryn Kyle, Kwok Ho Yip, Nitl Sumri, Thoms V. Guy, Brin S. Kim, Andrew J. Mitchell,
Supplementry Informtion Cutneous immuno-surveillnce nd regultion of inflmmtion y group 2 innte lymphoid cells Ben Roediger, Ryn Kyle, Kwok Ho Yip, Nitl Sumri, Thoms V. Guy, Brin S. Kim, Andrew J. Mitchell,
Ndfip-mediated degradation of Jak1 tunes cytokine signalling to limit expansion of CD4 þ effector T cells
 Received 4 Jul 15 Accepted 9 Fe 16 Pulished 18 Apr 16 DOI: 1.138/ncomms116 OPEN Ndfip-medited degrdtion of Jk1 tunes cytokine signlling to limit expnsion of CD4 þ effector T cells Clire E. O Lery 1, Christopher
Received 4 Jul 15 Accepted 9 Fe 16 Pulished 18 Apr 16 DOI: 1.138/ncomms116 OPEN Ndfip-medited degrdtion of Jk1 tunes cytokine signlling to limit expnsion of CD4 þ effector T cells Clire E. O Lery 1, Christopher
Feeding state and age dependent changes in melaninconcentrating hormone expression in the hypothalamus of broiler chickens
 Supplementry Mterils Epub: No 2017_23 Vol. 65, 2018 https://doi.org/10.183/bp.2017_23 Regulr pper Feeding stte nd ge dependent chnges in melninconcentrting hormone expression in the hypothlmus of broiler
Supplementry Mterils Epub: No 2017_23 Vol. 65, 2018 https://doi.org/10.183/bp.2017_23 Regulr pper Feeding stte nd ge dependent chnges in melninconcentrting hormone expression in the hypothlmus of broiler
* * * * * liver kidney ileum. Supplementary Fig.S1
 Supplementry Fig.S1 liver kidney ileum Fig.S1. Orlly delivered Fexrmine is intestinlly-restricted Mice received vehicle or Fexrmine (100mg/kg) vi per os (PO) or intrperitonel (IP) injection for 5 dys (n=3/group).
Supplementry Fig.S1 liver kidney ileum Fig.S1. Orlly delivered Fexrmine is intestinlly-restricted Mice received vehicle or Fexrmine (100mg/kg) vi per os (PO) or intrperitonel (IP) injection for 5 dys (n=3/group).
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nture11225 Numer of OTUs sed on 3% distnce Numer of 16s rrna-sed V2-V4 tg sequences LF MF PUFA Supplementry Figure 1. High-ft diets decrese the richness nd diversity
SUPPLEMENTARY INFORMATION doi:10.1038/nture11225 Numer of OTUs sed on 3% distnce Numer of 16s rrna-sed V2-V4 tg sequences LF MF PUFA Supplementry Figure 1. High-ft diets decrese the richness nd diversity
DR. MARC PAGÈS Project Manager R&D Biologicals - Coccidia Projects, HIPRA
 DR. MARC PAGÈS Project Mnger R&D Biologicls - Coccidi Projects, HIPRA Dr. Mrc Pgès Bosch otined Microiology nd Genetics degree t the University of Brcelon in 1998. He otined his PhD working on the synptoneml
DR. MARC PAGÈS Project Mnger R&D Biologicls - Coccidi Projects, HIPRA Dr. Mrc Pgès Bosch otined Microiology nd Genetics degree t the University of Brcelon in 1998. He otined his PhD working on the synptoneml
Platelet-derived growth factor-a receptor activation is required for human cytomegalovirus infection
 Vol 455 18 Septemer 28 doi:1.138/nture729 LETTERS Pltelet-derived growth fctor- receptor ctivtion is required for humn cytomeglovirus infection Lilin Sorocenu 1, Armin Akhvn 1 & Chrles S. Cos 1,2 Humn
Vol 455 18 Septemer 28 doi:1.138/nture729 LETTERS Pltelet-derived growth fctor- receptor ctivtion is required for humn cytomeglovirus infection Lilin Sorocenu 1, Armin Akhvn 1 & Chrles S. Cos 1,2 Humn
Abstract ABSTRACT #69. Abstract. Introduction & Methods. Methods & Results. Results. Results & Conclusions
 Effects of dietry β-glucn on Growth Performnce, Dirrhe, nd Gut Permeility of Wening Pigs Experimentlly Infected with Pthogenic E. coli Kwngwook Kim, Amy Ehrlich, Vivin Perng, Jennifer Chse, Helen Ryould,
Effects of dietry β-glucn on Growth Performnce, Dirrhe, nd Gut Permeility of Wening Pigs Experimentlly Infected with Pthogenic E. coli Kwngwook Kim, Amy Ehrlich, Vivin Perng, Jennifer Chse, Helen Ryould,
SUPPLEMENTARY INFORMATION
 doi:.3/nture93 d 5 Rttlesnke DRG (reds) Rttlesnke TG (reds) c 3 TRPV1 other TRPs 1 1 3 Non-pit snke TG (reds) SFig. 1 5 5 3 other TRPs TRPV1 1 1 3 Non-pit snke DRG (reds) 5 Antomy of the pit orgn nd comprison
doi:.3/nture93 d 5 Rttlesnke DRG (reds) Rttlesnke TG (reds) c 3 TRPV1 other TRPs 1 1 3 Non-pit snke TG (reds) SFig. 1 5 5 3 other TRPs TRPV1 1 1 3 Non-pit snke DRG (reds) 5 Antomy of the pit orgn nd comprison
ESM Table 1. Characterisation of the human non-diabetic cohort used for MRIbased assessment of pancreatic fat and insulin secretion via OGTT.
 ESM Tle 1. Chrcteristion of the humn non-dietic cohort used for MRIsed ssessment of pncretic ft nd insulin secretion vi OGTT. Trit sex Medin (IQR) 86 femles, 5 mles ge (yers) 4.4 (.5-5.57) BMI (kg/m²).62
ESM Tle 1. Chrcteristion of the humn non-dietic cohort used for MRIsed ssessment of pncretic ft nd insulin secretion vi OGTT. Trit sex Medin (IQR) 86 femles, 5 mles ge (yers) 4.4 (.5-5.57) BMI (kg/m²).62
HIV Develops Indirect Cross-resistance to Combinatorial RNAi Targeting Two Distinct and Spatially Distant Sites
 originl rticle The Americn Society of Gene & Cell Therpy HIV Develops Indirect Cross-resistnce to Comintoril RNAi Trgeting Two Distinct nd Sptilly Distnt Sites Priy S Shh, Nhung P Phm nd Dvid V Schffer
originl rticle The Americn Society of Gene & Cell Therpy HIV Develops Indirect Cross-resistnce to Comintoril RNAi Trgeting Two Distinct nd Sptilly Distnt Sites Priy S Shh, Nhung P Phm nd Dvid V Schffer
Overcoming EGFR T790M-based Tyrosine Kinase Inhibitor Resistance with an Allele-specific DNAzyme
 Cittion: Moleculr Therpy Nucleic Acids (214) 3, e15; doi:1.138/mtn.214.3 214 The Americn Society of Gene & Cell Therpy All rights reserved 2162-2531/14 www.nture.com/mtn Overcoming T79M-sed Tyrosine Kinse
Cittion: Moleculr Therpy Nucleic Acids (214) 3, e15; doi:1.138/mtn.214.3 214 The Americn Society of Gene & Cell Therpy All rights reserved 2162-2531/14 www.nture.com/mtn Overcoming T79M-sed Tyrosine Kinse
SUPPLEMENTARY INFORMATION
 Supplementry Informtion Supplementry Figure legends Supplementry tle. List of the fctors identified from the GST-RAR (LBD)-ffinity purifiction. The hrored domins, their puttive cytologicl functions, nd
Supplementry Informtion Supplementry Figure legends Supplementry tle. List of the fctors identified from the GST-RAR (LBD)-ffinity purifiction. The hrored domins, their puttive cytologicl functions, nd
CD160 inhibits activation of human CD4 + T cells through interaction with herpesvirus entry mediator
 CD16 inhiits ctivtion of humn CD4 + T cells through interction with herpesvirus entry meditor Guifng Ci, Anuknth Anumnthn, Juli A Brown, Edwrd A Greenfield, Bogong Zhu & Gordon J Freemn CD16, glycosylphosphtidylinositol-nchored
CD16 inhiits ctivtion of humn CD4 + T cells through interction with herpesvirus entry meditor Guifng Ci, Anuknth Anumnthn, Juli A Brown, Edwrd A Greenfield, Bogong Zhu & Gordon J Freemn CD16, glycosylphosphtidylinositol-nchored
Electronic Supplementary Information for:
 Electronic Supplementry Mteril (ESI) for ChemComm. This journl is The Royl Society of Chemistry 214 Electronic Supplementry Informtion for: Gold nnoprticles functionlized with cresyl violet nd porphyrin
Electronic Supplementry Mteril (ESI) for ChemComm. This journl is The Royl Society of Chemistry 214 Electronic Supplementry Informtion for: Gold nnoprticles functionlized with cresyl violet nd porphyrin
FoxP3 + regulatory CD4 T cells control the generation of functional CD8 memory
 Received Fe Accepted 6 Jul Pulished 7 Aug DOI:.8/ncomms99 FoxP + regultory CD T cells control the genertion of functionl memory M.G. de Goër de Herve,,, S. Jfour,,, M. Vllée, & Y. Toufik, During the primry
Received Fe Accepted 6 Jul Pulished 7 Aug DOI:.8/ncomms99 FoxP + regultory CD T cells control the genertion of functionl memory M.G. de Goër de Herve,,, S. Jfour,,, M. Vllée, & Y. Toufik, During the primry
Hormonal networks involved in phosphate deficiencyinduced cluster root formation of Lupinus albus L.
 Institute of Crop Science (34h) Hormonl networks involved in phosphte deficiencyinduced cluster root formtion of Lupinus lus L. For PSP5 in Montpellier, 214 Zhengrui Wng, A.B.M. Moshiur Rhmn, Guoying Wng,
Institute of Crop Science (34h) Hormonl networks involved in phosphte deficiencyinduced cluster root formtion of Lupinus lus L. For PSP5 in Montpellier, 214 Zhengrui Wng, A.B.M. Moshiur Rhmn, Guoying Wng,
Role of Cell Surface Spikes in Alphavirus Budding
 JOURNAL OF VIROLOGY, Dec. 1992, p. 7089-7095 0022-538X/92/127089-07$02.00/0 Copyright 1992, Americn Society for Microiology Vol. 66, No. 12 Role of Cell Surfce Spikes in Alphvirus Budding HONGXING ZHAO
JOURNAL OF VIROLOGY, Dec. 1992, p. 7089-7095 0022-538X/92/127089-07$02.00/0 Copyright 1992, Americn Society for Microiology Vol. 66, No. 12 Role of Cell Surfce Spikes in Alphvirus Budding HONGXING ZHAO
Innate immune response to a H3N2 subtype swine influenza virus in newborn porcine trachea cells, alveolar macrophages, and precision-cut lung slices
 Delgdo-Orteg et l. Veterinry Reserch 214, 45:42 VETERINARY RESEARCH RESEARCH Open Access Innte immune response to H3N2 sutype swine influenz virus in neworn porcine trche cells, lveolr mcrophges, nd precision-cut
Delgdo-Orteg et l. Veterinry Reserch 214, 45:42 VETERINARY RESEARCH RESEARCH Open Access Innte immune response to H3N2 sutype swine influenz virus in neworn porcine trche cells, lveolr mcrophges, nd precision-cut
Butyrate inhibits pro-proliferative mir-92a by diminishing c-myc-induced mir-17-92a cluster transcription in human colon cancer cells
 Hu et l. Moleculr Cncer (25) 4:8 DOI.86/s2943-5-45-x RESEARCH Open Access inhiits pro-prolifertive mir-92 y diminishing -induced mir-7-92 cluster trnscription in humn colon cncer cells Shien Hu, Ln Liu
Hu et l. Moleculr Cncer (25) 4:8 DOI.86/s2943-5-45-x RESEARCH Open Access inhiits pro-prolifertive mir-92 y diminishing -induced mir-7-92 cluster trnscription in humn colon cncer cells Shien Hu, Ln Liu
PDGF-BB secreted by preosteoclasts induces angiogenesis during coupling with osteogenesis
 Supplementry Informtion PDGF-BB secreted y preosteoclsts induces ngiogenesis during coupling with osteogenesis Hui Xie, Zhung Cui, Long Wng, Zhuying Xi, Yin Hu, Lingling Xin, Chngjun Li, Ling Xie, Jnet
Supplementry Informtion PDGF-BB secreted y preosteoclsts induces ngiogenesis during coupling with osteogenesis Hui Xie, Zhung Cui, Long Wng, Zhuying Xi, Yin Hu, Lingling Xin, Chngjun Li, Ling Xie, Jnet
Supplementary Information Titles
 Supplementry Informtion Titles Journl: Nture Medicine Article Title: Corresponding Author: Modelling colorectl cncer using CRISPR-Cs9-medited engineering of humn intestinl orgnoids Toshiro Sto Supplementry
Supplementry Informtion Titles Journl: Nture Medicine Article Title: Corresponding Author: Modelling colorectl cncer using CRISPR-Cs9-medited engineering of humn intestinl orgnoids Toshiro Sto Supplementry
Supplementary Figure S1
 Supplementry Figure S1 d MAP2 GFAP e MAP2 GFAP GFAP c f Clindin GFAP Supplementry Figure S1. Neuronl deth nd ltered strocytes in the rin of n ffected child. Neuron specific MAP2 ntiody stining in the hippocmpus
Supplementry Figure S1 d MAP2 GFAP e MAP2 GFAP GFAP c f Clindin GFAP Supplementry Figure S1. Neuronl deth nd ltered strocytes in the rin of n ffected child. Neuron specific MAP2 ntiody stining in the hippocmpus
SUPPLEMENTARY INFORMATION
 SUPPLEMEARY IFORMAIO doi:./nture correction to Supplementry Informtion Adenom-linked rrier defects nd microil products drive IL-/IL-7-medited tumour growth Sergei I. Grivennikov, Kepeng Wng, Dniel Mucid,
SUPPLEMEARY IFORMAIO doi:./nture correction to Supplementry Informtion Adenom-linked rrier defects nd microil products drive IL-/IL-7-medited tumour growth Sergei I. Grivennikov, Kepeng Wng, Dniel Mucid,
Clec4A4 is a regulatory receptor for dendritic cells that impairs inflammation and T-cell immunity
 Received 1 Oct 215 Accepted 8 Mr 216 Pulished 12 Apr 216 DOI: 1.138/ncomms11273 OPEN Clec4A4 is regultory receptor for dendritic cells tht impirs inflmmtion nd T-cell immunity Tomofumi Uto 1,, Tomohiro
Received 1 Oct 215 Accepted 8 Mr 216 Pulished 12 Apr 216 DOI: 1.138/ncomms11273 OPEN Clec4A4 is regultory receptor for dendritic cells tht impirs inflmmtion nd T-cell immunity Tomofumi Uto 1,, Tomohiro
The effect of dietary α-linolenic acid levels on regulation of omega-3 lipid synthesis in rat
 The effect of dietry α-linolenic cid levels on regultion of omeg-3 lipid synthesis in rt Wei-Chun Tu School of Agriculture Food nd Wine The University of Adelide Conversion of PUFA to LCPUFA PUFA LCPUFA
The effect of dietry α-linolenic cid levels on regultion of omeg-3 lipid synthesis in rt Wei-Chun Tu School of Agriculture Food nd Wine The University of Adelide Conversion of PUFA to LCPUFA PUFA LCPUFA
SUPPLEMENTARY INFORMATION
 DOI: 1.138/n358 TLR2 nd MyD88 expression in murine mmmry epithelil supopultions. CD24 min plus MRU Myo-epithelil Luminl progenitor (CD61 pos ) Mture luminl (CD61 neg ) CD49f CD61 Reltive expression Krt5
DOI: 1.138/n358 TLR2 nd MyD88 expression in murine mmmry epithelil supopultions. CD24 min plus MRU Myo-epithelil Luminl progenitor (CD61 pos ) Mture luminl (CD61 neg ) CD49f CD61 Reltive expression Krt5
Arachidonic acid induces ERK activation via Src SH2 domain association with the epidermal growth factor receptor
 http://www.kidney-interntionl.org & 6 Interntionl Society of Nephrology originl rticle Archidonic cid induces ERK ctivtion vi Src SH2 domin ssocition with the epiderml growth fctor receptor LD Alexnder
http://www.kidney-interntionl.org & 6 Interntionl Society of Nephrology originl rticle Archidonic cid induces ERK ctivtion vi Src SH2 domin ssocition with the epiderml growth fctor receptor LD Alexnder
SUPPLEMENTARY INFORMATION
 doi: 1.138/nture862 humn hr. 21q MRPL39 murine Chr.16 Mrpl39 Dyrk1A Runx1 murine Chr. 17 ZNF295 Ets2 Znf295 murine Chr. 1 COL18A1 -/- lot: nti-dscr1 IgG hevy hin DSCR1 DSCR1 expression reltive to hevy
doi: 1.138/nture862 humn hr. 21q MRPL39 murine Chr.16 Mrpl39 Dyrk1A Runx1 murine Chr. 17 ZNF295 Ets2 Znf295 murine Chr. 1 COL18A1 -/- lot: nti-dscr1 IgG hevy hin DSCR1 DSCR1 expression reltive to hevy
Supporting Information. In Situ Supramolecular Assembly and Modular Modification of Hyaluronic Acid Hydrogels for 3D Cellular Engineering
 Supporting Informtion In Situ Suprmoleculr Assemly nd Modulr Modifiction of Hyluronic Acid Hydrogels for 3D Cellulr Engineering Kyeng Min Prk,, Jeong-A Yng,, Hyunte Jung, c Junseok Yeom, Ji Sun Prk, d
Supporting Informtion In Situ Suprmoleculr Assemly nd Modulr Modifiction of Hyluronic Acid Hydrogels for 3D Cellulr Engineering Kyeng Min Prk,, Jeong-A Yng,, Hyunte Jung, c Junseok Yeom, Ji Sun Prk, d
Molecular Analysis of BRCA1 in Human Breast Cancer. Cells Under Oxidative Stress
 Moleculr Anlysis of BRCA1 in Humn Brest Cncer Cells Under Oxidtive Stress Brin L. Gilmore 1, Ynping Ling 1, Crly E. Winton 1,2, Ky Ptel 1, Vsile Krgeorge 1, A. Cmeron Vrno 1,3, Willim Dernley 1, Zhi Sheng
Moleculr Anlysis of BRCA1 in Humn Brest Cncer Cells Under Oxidtive Stress Brin L. Gilmore 1, Ynping Ling 1, Crly E. Winton 1,2, Ky Ptel 1, Vsile Krgeorge 1, A. Cmeron Vrno 1,3, Willim Dernley 1, Zhi Sheng
Reuven Agami and Yosef Shaul
 The kinse ctivity of ut not v-al is potentited y direct interction with RFXI, protein tht inds the enhncers of severl viruses nd cell-cycle regulted genes Reuven Agmi nd Yosef Shul Oncogene (1998) 16,
The kinse ctivity of ut not v-al is potentited y direct interction with RFXI, protein tht inds the enhncers of severl viruses nd cell-cycle regulted genes Reuven Agmi nd Yosef Shul Oncogene (1998) 16,
Supplementary information for: Low bone mass and changes in the osteocyte network in mice lacking autophagy in the osteoblast lineage
 Supplementry informtion for: Low one mss nd chnges in the osteocyte network in mice lcking utophgy in the osteolst linege Mrilin Piemontese, Meld Onl, Jinhu Xiong, Li Hn, Jeff D. Thostenson, Mri Almeid,
Supplementry informtion for: Low one mss nd chnges in the osteocyte network in mice lcking utophgy in the osteolst linege Mrilin Piemontese, Meld Onl, Jinhu Xiong, Li Hn, Jeff D. Thostenson, Mri Almeid,
THE EVALUATION OF DEHULLED CANOLA MEAL IN THE DIETS OF GROWING AND FINISHING PIGS
 THE EVALUATION OF DEHULLED CANOLA MEAL IN THE DIETS OF GROWING AND FINISHING PIGS THE EVALUATION OF DEHULLED CANOLA MEAL IN THE DIETS OF GROWING AND FINISHING PIGS John F. Ptience nd Doug Gillis SUMMARY
THE EVALUATION OF DEHULLED CANOLA MEAL IN THE DIETS OF GROWING AND FINISHING PIGS THE EVALUATION OF DEHULLED CANOLA MEAL IN THE DIETS OF GROWING AND FINISHING PIGS John F. Ptience nd Doug Gillis SUMMARY
3 Results RESULTS Cell growth and segregation in recombinant bioprocesses
 RESULTS 3 3 Results In this thesis, yest α-glucosidse produced in E. coli ws used s model system in order to otin more comprehensive knowledge out cell physiology in response to overexpression of heterologous
RESULTS 3 3 Results In this thesis, yest α-glucosidse produced in E. coli ws used s model system in order to otin more comprehensive knowledge out cell physiology in response to overexpression of heterologous
Supplemental Materials
 Supplementl Mterils Cellulose deficiency of shv3svl1 is enhnced y hyper ccumultion of exogenous sucrose vi the plsm memrne sucrose/h symporter SUC1 Trevor H. Yets, Hgit Sorek, Dvid E. Wemmer, Chris R.
Supplementl Mterils Cellulose deficiency of shv3svl1 is enhnced y hyper ccumultion of exogenous sucrose vi the plsm memrne sucrose/h symporter SUC1 Trevor H. Yets, Hgit Sorek, Dvid E. Wemmer, Chris R.
Effect of fungicide timing and wheat varietal resistance on Mycosphaerella graminicola and its sterol 14 α-demethylation-inhibitorresistant
 Effect of fungicide timing nd whet vrietl resistnce on Mycospherell grminicol nd its sterol 14 α-demethyltion-inhiitorresistnt genotypes Didierlurent L., Roisin-Fichter C., Snssené J., Selim S. Pltform
Effect of fungicide timing nd whet vrietl resistnce on Mycospherell grminicol nd its sterol 14 α-demethyltion-inhiitorresistnt genotypes Didierlurent L., Roisin-Fichter C., Snssené J., Selim S. Pltform
Human PRP4 kinase is required for stable tri-snrnp association during spliceosomal B complex formation
 Humn PRP4 kinse is required for stle tri-snrnp ssocition during spliceosoml formtion Mrc Schneider 1, He-Hsun Hsio 2, Cindy L Will 1, Régis Giet 3, Henning Urlu 2 & Reinhrd Lührmnn 1 Reversile protein
Humn PRP4 kinse is required for stle tri-snrnp ssocition during spliceosoml formtion Mrc Schneider 1, He-Hsun Hsio 2, Cindy L Will 1, Régis Giet 3, Henning Urlu 2 & Reinhrd Lührmnn 1 Reversile protein
Wnt signaling enhances the activation and survival of human hepatic stellate cells
 FEBS Letters 581 (2007) 2954 2958 Wnt signling enhnces the ctivtion nd survivl of humn heptic stellte cells Sun Jung Myung, Jung-Hwn Yoon, *, Geum-Youn Gwk, Won Kim, Jeong-Hoon Lee, Kng Mo Kim, Chn Soo
FEBS Letters 581 (2007) 2954 2958 Wnt signling enhnces the ctivtion nd survivl of humn heptic stellte cells Sun Jung Myung, Jung-Hwn Yoon, *, Geum-Youn Gwk, Won Kim, Jeong-Hoon Lee, Kng Mo Kim, Chn Soo
Suppressor of cytokine signaling 1 regulates the immune response to infection by a unique inhibition of type I interferon activity
 5 Nture Pulishing Group http://www.nture.com/ntureimmunology Suppressor of cytokine signling 1 regultes the immune response to infection y unique inhiition of type I interferon ctivity Jennifer E Fenner
5 Nture Pulishing Group http://www.nture.com/ntureimmunology Suppressor of cytokine signling 1 regultes the immune response to infection y unique inhiition of type I interferon ctivity Jennifer E Fenner
MUTATIONS. Mutagens. Point mutations substitutions. Mutations. Sickle-cell disease. Point mutations insertions & deletions
 MUTTIONS Mutgens Mutgens re physicl or chemicl gents tht give rise to muttions. High energy rdition UV, X, gmm. se nlogues, intercltors, chemicl chnge inducers. ffect DN structure, se piring, etc. 2004
MUTTIONS Mutgens Mutgens re physicl or chemicl gents tht give rise to muttions. High energy rdition UV, X, gmm. se nlogues, intercltors, chemicl chnge inducers. ffect DN structure, se piring, etc. 2004
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/nc2824 Hcn4 Tx5 Mlc2 c Hcn4- ISH d Tx5- ISH e Mlc2-ISH Hcn4-ISH f e Tx5-ISH f -ISH Figure S1 Section in situ hyridistion nlysis of crescent stge mouse emryos (E7.5). () More nterior section
DOI: 10.1038/nc2824 Hcn4 Tx5 Mlc2 c Hcn4- ISH d Tx5- ISH e Mlc2-ISH Hcn4-ISH f e Tx5-ISH f -ISH Figure S1 Section in situ hyridistion nlysis of crescent stge mouse emryos (E7.5). () More nterior section
The β-globin nuclear compartment in development and erythroid differentiation
 The β-gloin nucler comprtment in development nd erythroid differentition Roert-Jn Plstr 1,2, Bs Tolhuis 1,2, Erik Splinter 1, Rin Nijmeijer 1, Frnk Grosveld 1 & Wouter de Lt 1 Efficient trnscription of
The β-gloin nucler comprtment in development nd erythroid differentition Roert-Jn Plstr 1,2, Bs Tolhuis 1,2, Erik Splinter 1, Rin Nijmeijer 1, Frnk Grosveld 1 & Wouter de Lt 1 Efficient trnscription of
The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages
 The endoplsmic reticulum is the site of cholesterol-induced cytotoxicity in mcrophges Bo Feng 1, Pin Mei Yo 1, Ynkun Li 1, Cecili M. Devlin 1, Djun Zhng 1, Hether P. Hrding 2, Michele Sweeney 3, Jmes X.
The endoplsmic reticulum is the site of cholesterol-induced cytotoxicity in mcrophges Bo Feng 1, Pin Mei Yo 1, Ynkun Li 1, Cecili M. Devlin 1, Djun Zhng 1, Hether P. Hrding 2, Michele Sweeney 3, Jmes X.
Bioactive milk components to secure growth and gut development in preterm pigs ESTER ARÉVALO SUREDA PIGUTNET FA1401 STSM
 Bioctive milk components to secure growth nd gut development in preterm pigs ESTER ARÉVALO SUREDA PIGUTNET FA1401 STSM STSM Pigutnet FA1401 STSM 03/Septemer 30/Novemer/2017 (3 months) Host: Home: Thoms
Bioctive milk components to secure growth nd gut development in preterm pigs ESTER ARÉVALO SUREDA PIGUTNET FA1401 STSM STSM Pigutnet FA1401 STSM 03/Septemer 30/Novemer/2017 (3 months) Host: Home: Thoms
Glucose metabolism inhibits apoptosis in neurons and cancer cells by redox inactivation of cytochrome c
 letters Glucose metolism inhiits poptosis in neurons nd cncer cells y redox inctivtion of cytochrome c Allyson E. Vughn 1 nd Mohnish Deshmukh 1,2,3 Neurons nd cncer cells use glucose extensively, yet the
letters Glucose metolism inhiits poptosis in neurons nd cncer cells y redox inctivtion of cytochrome c Allyson E. Vughn 1 nd Mohnish Deshmukh 1,2,3 Neurons nd cncer cells use glucose extensively, yet the
Gene regulation mediated by calcium signals in T lymphocytes
 Gene regultion medited y clcium signls in T lymphocytes Stefn Feske 1, Jen Giltnne 2, Ricrdo Dolmetsch 3, Louis M. Studt 2 nd Anjn Ro 1 Modultion of mny signling pthwys in ntigen-stimulted T nd B cells
Gene regultion medited y clcium signls in T lymphocytes Stefn Feske 1, Jen Giltnne 2, Ricrdo Dolmetsch 3, Louis M. Studt 2 nd Anjn Ro 1 Modultion of mny signling pthwys in ntigen-stimulted T nd B cells
Control vector. HA-Elfn2. HA-Elfn1
 Control vector c HA-Elfn2 HA- Control vector HA-Elfn2 HA- nti- nti-ha Hippocmpus (CA3) PV DAPI SOM DAPI Hippocmpus (dentte gyrus) PV DAPI SOM DAPI Supplementry Figure 1. Specificity of the nti- ntiody
Control vector c HA-Elfn2 HA- Control vector HA-Elfn2 HA- nti- nti-ha Hippocmpus (CA3) PV DAPI SOM DAPI Hippocmpus (dentte gyrus) PV DAPI SOM DAPI Supplementry Figure 1. Specificity of the nti- ntiody
Unique roles of the unfolded protein response pathway in fungal. development and differentiation. Kwang Woo Jung, Yee Seul So, & Yong Sun Bahn *
 Supplementry Informtion Unique roles of the unfolded protein response pthwy in fungl development nd differentition Kwng Woo Jung, Yee Seul So, & Yong Sun Bhn * Contents Supplementry Figure S1 Supplementry
Supplementry Informtion Unique roles of the unfolded protein response pthwy in fungl development nd differentition Kwng Woo Jung, Yee Seul So, & Yong Sun Bhn * Contents Supplementry Figure S1 Supplementry
