KIM-1-/TIM-1-mediated phagocytosis links ATG5-/ULK1-dependent clearance of apoptotic cells to antigen presentation
|
|
|
- Tracey George
- 5 years ago
- Views:
Transcription
1 Article KIM-1-/TIM-1-mediated phagocytosis links ATG5-/ULK1-dependent clearance of apoptotic cells to antigen presentation Craig R Brooks 1,*, Melissa Y Yeung 1,2, Yang S Brooks 3,4, Hui Chen 5, Takaharu Ichimura 1, Joel M Henderson 5 & Joseph V Bonventre 1,6,** Abstract Phagocytosis of apoptotic cells by both professional and semiprofessional phagocytes is required for resolution of organ damage and maintenance of immune tolerance. KIM-1/TIM-1 is a phosphatidylserine receptor that is expressed on epithelial cells and can transform the cells into phagocytes. Here, we demonstrate that KIM-1 phosphorylation and association with p85 results in encapsulation of phagosomes by lipidated LC3 in multi-membrane organelles. KIM-1-mediated phagocytosis is not associated with increased ROS production, and NOX inhibition does not block LC3 lipidation. Autophagy gene expression is required for efficient clearance of apoptotic cells and phagosome maturation. KIM-1- mediated phagocytosis leads to pro-tolerogenic antigen presentation, which suppresses CD4 T-cell proliferation and increases the percentage of regulatory T cells in an autophagy gene-dependent manner. Taken together, these data reveal a novel mechanism of epithelial biology linking phagocytosis, autophagy and antigen presentation to regulation of the inflammatory response. Keywords acute kidney injury; T regs; biomarker; heterophagy; MHC Subject Categories Immunology; Membrane & Intracellular Transport DOI /embj Received 18 August 2014 Revised 24 June 2015 Accepted 1 July 2015 Published online 17 August 2015 The EMBO Journal (2015) 34: See also: C Münz (October 2015) Introduction Tissue remodeling as a result of organ injury, involution or organism development requires bulk clearance of apoptotic and necrotic cells (Erwig & Henson, 2007). Epithelial cells can be responsible for the majority of the uptake and degradation of dead cells from solid organs (Erwig & Henson, 2007; Poon et al, 2014). This clearance process may downregulate the inflammatory response associated with organ injury, but the immunomodulatory pathways involved are poorly defined (Erwig & Henson, 2007). Phagocytosis and clearance of dead cells in the injured kidney are largely carried out by proximal tubule cells (PTCs) expressing kidney injury molecule 1/ T-cell immunoglobulin and mucin domain 1 (KIM-1/TIM-1) (Ichimura et al, 2008; Yang et al, 2015). KIM-1 is a phosphatidylserine (PS) and scavenger receptor and is the only known apoptotic cell phagocytic receptor expressed on PTCs that transforms epithelial cells into phagocytes (Ichimura et al, 2008). KIM-1 has been linked to immunomodulation since polymorphisms in the human gene encoding KIM-1 protein have been associated with various diseases including allergy, asthma, rheumatoid arthritis, atopic dermatitis, and lupus, but this link has been postulated to derive from KIM-1/ TIM-1 function on inflammatory cells where the expression of the protein is order of magnitudes less than that on kidney epithelial cells after injury (Chae et al, 2003, 2005; McIntire et al, 2004; Page et al, 2006; Garcia-Lozano et al, 2010; Lee et al, 2011; Xu et al, 2012). We have defined the cellular pathway linking the uptake of apoptotic debris by epithelial cells to autophagy, antigen presentation, and immune modulation. Phagocytosis and elimination of foreign, non-self material by the lysosomal pathway is referred to as heterophagy, while macroautophagy is a mechanism for the degradation of self components, such as organelles (Oczypok et al, 2013). It remains to be resolved if the canonical (macro)autophagic pathway is involved in heterophagy. The macroautophagic pathway involves activation of a complex containing unc-51-like kinase/ autophagy-related 1 (ULK1/ ATG1), ATG13, and FIP200 (Randall-Demllo et al, 2013). This complex recruits the membrane that becomes autophagosome precursor where microtubule-associated protein 1 light chain 3 (LC3) is inserted, lipidating LC3 I to form LC3 II (Randall-Demllo et al, 1 Department of Medicine, Renal Division, Brigham and Women s Hospital, Harvard Medical School, Boston, MA, USA 2 Transplantation Research Center, Brigham and Women s Hospital, Harvard Medical School, Boston, MA, USA 3 Cutaneous Biology Research Center, Massachusetts General Hospital, Charlestown, MA, USA 4 Department of Dermatology, Harvard Medical School, Boston, MA, USA 5 Department of Pathology and Laboratory Medicine, Boston University School of Medicine, Boston, MA, USA 6 Harvard Stem Cell Institute, Cambridge, MA, USA *Corresponding author. Tel: ; crbrooks@partners.org **Corresponding author. Tel: ; joseph_bonventre@hms.harvard.edu ª 2015 The Authors The EMBO Journal Vol 34 No
2 The EMBO Journal KIM-1 links autophagy factors to MHC presentation Craig R Brooks et al 2013). The autophagosome membrane surrounds the material to be eliminated, creating a double membrane (Randall-Demllo et al, 2013). In addition to heterophagy, recent studies have demonstrated a role of non-canonical autophagy in cell corpse clearance, termed LC3-associated phagocytosis (LAP). In this clearance pathway, LC3 is targeted to the phagosome in an ATG5-, ATG7-, and Beclin1- dependent manner; however, this process does not involve the engagement of upstream autophagy proteins such as ULK1 (Sanjuan et al, 2007; Florey et al, 2011; Martinez et al, 2011; Kim et al, 2013b). For the purposes of this study, processes that involve the canonical macroautophagy pathway will be referred to as autophagy. In the current study, we report that epithelial cell heterophagy of apoptotic cells activates the autophagy pathway. We link the process of KIM-1-mediated phagocytosis to autophagy induction, MHC presentation and maintenance of peripheral T-cell tolerance. LC3 punctae formation and LC3 lipidation are induced by KIM-1- mediated phagocytosis of apoptotic cells. Knockdown of autophagy genes delays clearance of apoptotic cells and prevents LC3 II accumulation. Mechanistically, KIM-1 phosphorylation and interaction with p85 are necessary for localization of LC3 to the phagosome and increased LC3 lipidation, while inhibition of lysosome degradation results in the accumulation of multi-membrane phagosomes. KIM-1- mediated phagocytosis is not associated with increased ROS production and NOX inhibition does not block LC3 lipidation. Following KIM-1-mediated phagocytosis, PTCs present antigens from phagocytosed material on MHC II and cross-present antigens on MHC I. Importantly, antigen presentation reduced T-cell proliferation and increased regulatory T-cell (Tregs) proliferation, in an autophagydependent manner. Thus, KIM-1-mediated MHC presentation regulated T-cell proliferation by inducing Tregs. In this way, KIM-1- mediated phagocytosis modulates the immune response through phagocytosis, autophagy-mediated phagosome maturation and antigen presentation. This provides a mechanism by which epithelial cell self-antigen presentation actively protects against autoimmunity. To our knowledge, this is the first study to illustrate a complete pathway from epithelial cell receptor phagocytosis of apoptotic cells to T-cell response, including the uptake of apoptotic cells, phagosome maturation via autophagy, lysosomal degradation, presentation of antigens, and induction of a pro-tolerogenic T-cell response to presented antigens. Results KIM-1 co-localizes with lipidated LC3 at the phagosome KIM-1 expression was associated with LC3 punctae formation in mouse kidney tissue detected by immunofluorescence. In normal animals, KIM-1 was not detectable and LC3 staining was evenly distributed throughout the PTC cytosol with stronger staining in some interstitial cells (Fig 1A upper panel). As a positive control for LC3 punctae, mice were food deprived. After food withdrawal for 12 h, kidneys showed a dramatic increase in LC3 punctae in the PTCs, without the expression of KIM-1 (Fig 1A middle panel), reflecting the absence of kidney injury. Following injury to the kidney by bilateral ischemia/reperfusion injury (IRI), KIM-1 protein was markedly up-regulated in many proximal tubules, and LC3 punctae were present in KIM-1-expressing proximal tubule cells (Fig 1A lower panel). While KIM-1 expression was localized primarily to the apical aspect of in the PTC, KIM-1 also co-localized with intracellular organelles, which co-labeled with LC3 (Fig 1A lower panel excerpts and arrows). Induction of LC3 punctae post-ischemia was confirmed by immunoblot analysis at 6 and 24 h after bilateral ischemia. During autophagosome formation, the cytosolic form of LC3 (LC3 I) is conjugated to phosphatidylethanolamine, yielding LC3 II which is recruited to the autophagosomal membrane. The lipidation of LC3 makes the protein travel faster by electrophoresis resulting in a lower band (Fujita et al, 2008). At both 6 and 24 h post-iri, there was an increase in LC3 II accompanied by a marked increase in KIM-1 protein expression (Fig 1B). Figure 1. KIM-1 co-localizes with LC3 in vitro and in vivo and induces LC3 lipidation, which is upregulated by phagocytosis. A KIM-1 (green) and LC3 (red) staining in kidney sections from mice which were treated with bi-lateral ischemia (IRI), unfed for 12 h, or untreated. Exerts show higher magnification of areas of LC3 and KIM-1 co-localization and the punctate pattern of LC3 (arrows). Pearson correlation coefficient (PCC) = and and the Mander s overlap coefficient (MOC) = and for the unfed and IRI treatment groups, respectively. n > 10 LC3- positive organelles from two different mice. B Immunoblot analysis of KIM-1 and LC3 from post-ischemic kidney cortex (left panel) and quantification of immunoblots (right panel). *P = , n = 3 mice. C The percentage of GFP-LC3- and KIM-1-transfected cells positive for LC3 punctae was measured by cell counting. *P < 0.001, n = 4. D Immunoblot analysis of LC3 II and ATG5/ATG12 complex formation in LLC-PK1 cells expressing KIM-1 incubated with or without apoptotic cells. Representative blot from three independent experiments. E LLC-PK1 cells were transfected with GFP-LC3 alone or GFP-LC3 and KIM-1. Cells were then untreated or incubated with apoptotic cells and stained for KIM-1 (red). PCC = , , and and MOC = , , and for the LC3 alone, and KIM-1 +LC3 and KIM-1 + LC3 + apoptotic cells treatment groups, respectively. n 6. F Immunoblot of LC3 II levels in KIM-1-transfected cells (left panel) and quantification of LC3 II (right panel). *P = for pcdna vs. KIM-1 and P = for pcdna + apop vs. KIM-1+ apop. n = 3. G Immunoblot of p62 in LLC-PK1 cells expressing pcdna or KIM-1 (left panel) and quantification of p62 levels (right panel). H Uptake of PS-coated, fluorescein-labeled latex beads by pcdna or KIM-1 transfected cells. I Immunoblot analysis of isolated phagosomes for LC3, KIM-1, and organelle markers comparing phagosome (Pha) lysates vs. whole cell lysates (WCL). Representative of five independent experiments. J LC3 and p62 immunoblot analysis of isolated phagosomes from cells treated with Baf (100 nm), Spautin1 (10 lm), or DPI (12.5 lm). K Quantification of ROS production with DCFH-DA in KIM-1-expressing LLC-PK1 cells treated with apoptotic cells with and without DPI. *P = for DMSO vs. DPI and P = for DMSO vs. Apop. DPI. Data information: Data presented as mean SD. Statistical comparisons were calculated by ANOVA followed by Bonferroni s post hoc analysis. Scale bars, 30 lm (A), 10 lm (E), and 50 lm (H). Source data are available online for this figure The EMBO Journal Vol 34 No ª 2015 The Authors
3 Published online: August 17, 2015 Craig R Brooks et al A The EMBO Journal KIM-1 links autophagy factors to MHC presentation B E C D F H J I K G Figure 1. ª 2015 The Authors The EMBO Journal Vol 34 No
4 The EMBO Journal KIM-1 links autophagy factors to MHC presentation Craig R Brooks et al To evaluate the effects of KIM-1-induced phagocytosis on LC3 punctae formation in vitro, LLC-PK1 cells were transfected with GFP-LC3 plus pcdna or KIM-1. Cells were incubated with apoptotic thymocytes or left untreated. Transfection of KIM-1 was sufficient to induce LC3-positive autophagosome formation in ~33% of cells, as measured by counting GFP-LC3 punctae. Incubation with apoptotic cells increased the number of cells with autophagosomes to ~59% (Fig 1C). Incubation of apoptotic cells with KIM-1-expressing cells resulted in increased levels of lipidated LC3, LC3 II, and a shift of monomeric ATG5 into a complex with ATG12, a complex required for autophagosome formation (Fig 1D). As shown in Fig 1C and E, compared to cells not expressing KIM-1, a greater percentage of KIM-1-positive cells displayed large numbers of LC3 punctae (Fig 1E upper and middle panels). Following the uptake of apoptotic cells, intracellular KIM-1 co-localized with LC3 in phagosomes containing apoptotic cells (Figs 1E lower panel and excerpts and EV1A). KIM-1 also co-localized with LC3 in primary PTCs (Fig EV1B). Thus, KIM-1 expression alone induced an increase in the number of cells displaying large numbers of LC3 punctae, which was further enhanced by phagocytosis. The up-regulation of LC3 punctae by KIM-1 expression was further confirmed by immunoblot analysis of LC3 II formation and p62 levels (Fig 1F and G). KIM-1 led to the reduction of p62 levels, which was reversed upon incubation with the lysosomal inhibitor bafilomycin A1 (Baf) (Fig 1G). Note that these experiments were carried out in full medium with 10% fetal calf serum. Hence, the observed effects on LC3 by KIM-1 expression alone may be related to KIM-1-mediated uptake of media components and/or the uptake of debris of dead cells that are present in cell culture systems. Exogenous KIM-1 expression from the transfection of LLC-PK1 cells was similar to the expression of KIM-1 in primary PTCs (Fig EV1C). To further confirm that LC3 co-localized with KIM-1 in phagosomes, we isolated KIM-1-expressing phagosomes. Latex beads coated with PS were added to pcdna or KIM-1-transfected cells. Coating with PS was necessary for beads to be taken up by KIM- 1-expressing cells. Phagosomes were isolated by homogenization of the cells followed by sucrose gradient centrifugation. As shown in Fig 1H, KIM-1-transfected cells took up the PS-coated latex beads (green), whereas the control pcdna transfected cells did not. Phagosomes isolated from KIM-1-expressing cells were found to contain KIM-1 and LC3 by immunoblot analysis. The phagosome preparation did not react with antibodies specific for other organelles, including mitochondrial (cytochrome c), nuclear (histone H3), or cytosol (S6 kinase) markers (Fig 1I). LC3 and p62 association with the phagosomes was evaluated in the presence of Baf or the autophagy inhibitor Spautin1. We found that p62 and LC3 II levels were markedly increased with Baf and were suppressed with Spautin1 (Fig 1J). LC3 recruitment to phagosomes has been reported to be dependent on NADPH oxidase (NOX)-dependent reactive oxygen species (ROS) production in LAP but not autophagy in macrophages and dendritic cells (Ma et al, 2012; Lam et al, 2013). To test if LC3 accumulation on the phagosomes was dependent on NOX-mediated ROS production, phagosomes were isolated from cells treated with diphenylene iodonium (DPI), a NOX inhibitor. DPI did not inhibit LC3 localization to the phagosome or LC3 lipidation in KIM-1 cells incubated with apoptotic cells (Figs 1J and EV1D). DPI was found to inhibit basal ROS levels; however, contrary to what has been reported in macrophages and dendritic cells, KIM-1-mediated phagocytosis in epithelial cells did not induce ROS production (Figs 1K and EV1E) (Huang & Brumell, 2009; Huang et al, 2009; Ma et al, 2012; Lam et al, 2013; Romao et al, 2013). Thus, KIM-1-mediated phagocytosis induced LC3 punctae formation and increased LC3 II by a NOX-independent process and KIM-1-positive phagosomes co-localized with LC3. KIM-1 and LC3 co-localize following phagocytosis To determine at what point during phagocytosis LC3 localized to the phagosome, primary cultured PTCs expressing KIM-1 (~50% of cells), renal cell carcinoma (769p) cells which endogenously express KIM-1, and LLC-PK1 cells expressing KIM-1-GFP were transfected with RFP-LC3 and phagosome formation was imaged over time. The majority of phagosomes co-localized with LC3 following phagocytosis. As shown in Fig 2A, a KIM-1-GFP-expressing LLC-PK1 cell binds apoptotic cells and KIM-1 is enriched at the binding site (Fig 2A, 52 min; Video EV1). The apoptotic cell is then phagocytosed, but LC3 is not initially localized to the phagosome (Fig 2A, 72 min). LC3 then surrounds the KIM-1-positive phagosome (92 min), and the phagosome becomes further encapsulated by LC3 (Fig 2A, 132 min). At later time points, additional intracellular phagocytosed apoptotic cells become encapsulated with LC3 (Fig 2A, 272 min). In a sub-population of cells, KIM-1 and LC3 colocalized prior to complete phagocytosis. In the example shown, KIM-1 and LC3 first co-localized at the phagocytic cup (Fig 2B middle and right panels and Video EV2). Then, both KIM-1 and LC3 encapsulate the apoptotic cell forming a KIM-1- and LC3-positive phagosome (Fig 2B middle and right panels). KIM-1- and LC3- positive phagosomes could be visualized as early as 10 min after the addition of apoptotic cells (Fig 2B). Most of LC3 localization to the phagosome occurred later when the phagosome moved from the membrane to the cytosol (~80%) with plasma membrane colocalization of KIM-1 and LC3 seen in only a small subset (~16%) of LLC-PK1 cells (Fig 2C). The overall rate of PTC epithelial cell Figure 2. KIM-1 and LC3 co-localize following phagocytosis. A, B Time series of RFP-LC3 and KIM-1-GFP co-localization (arrows) in LLC-PK1 cells incubated with fluorescently labeled apoptotic thymocytes imaged at 20-min intervals (A) or 5-min intervals (B). C Quantification of phagocytosis and LC3 co-localization at plasma membrane or cytosol in LLC-PK1 cells, n = 132. D Co-localization of apoptotic cells and LC3 (arrows) in primary cultured proximal tubule cells were transfected with RFP-LC3, incubated with apoptotic thymocytes, and imaged at 15-min intervals. E Quantification of phagocytosis and LC3 co-localization at plasma membrane or cytosol in primary PTCs, n = 117. F Co-localization of RFP-LC3 and apoptotic cells (arrows) in 769p cells. 769p cells were transfected with LC3, incubated with fluorescently labeled apoptotic cells, and imaged every 10 min. G Quantification of phagocytosis and LC3 co-localization at plasma membrane or cytosol in 769p cells, n = 33. Data information: Scale bars, 10 lm (A), 15 lm (B), 10 lm (D), and 15 lm (F) The EMBO Journal Vol 34 No ª 2015 The Authors
5 Craig R Brooks et al KIM-1 links autophagy factors to MHC presentation The EMBO Journal A B C D E F G Figure 2. ª 2015 The Authors The EMBO Journal Vol 34 No
6 The EMBO Journal KIM-1 links autophagy factors to MHC presentation Craig R Brooks et al Figure 3. KIM-1 co-localizes with LC3 prior to the lysosome and inhibiting KIM-1-mediated phagocytosis blocks LC3 lipidation. A Representative confocal images from 5 independent experiments of ptflc3-expressing LLC-PK1 cells without apoptotic cells (left panel) after apoptotic cell uptake (middle panel) and after phagosome acidification (right panel). B Representative time course from four independent experiments of ptflc3 localization to the phagosome. C Time course of LC3-GFP and LAMP1-RFP localization to the phagosome (upper panels) and line scans of the phagosome (lower panels). Representative of four independent experiments. D Representative time course of LC3-GFP and RFP-Ub colocalization from two independent experiments imagining 20 fields in each experiment. E Confocal micrographs of LLC-PK1 cells co-transfected with LC3-GFP and KIM-1, then incubated with apoptotic cells and stained for ubiquitin. Data information: Data presented as mean SD. Scale bars, 5 lm. phagosome maturation was slower compared to professional phagocytes, such as macrophages and dendritic cells (Fig EV2 and Video EV6). To confirm that LC3 localized with phagosomes after apoptotic cells bound to endogenous KIM-1, LC3-RFP localization to the phagosome was monitored in primary PTCs and 769p cells. In the example shown in Fig 2D, two apoptotic thymocytes are bound to a PTC (Fig 2D arrows and Video EV3). At the 14-min time point, one of the apoptotic cells was phagocytosed, and LC3 did not co-localize with these cells. At 44 and 74 min, however, LC3 surrounded both phagocytosed apoptotic cells (Fig 2D right panels, arrows). LC3 co-localized with the apoptotic cell at the plasma membrane in < 1% of the primary PTCs (Fig 2E). In 769p cells, a renal cell carcinoma cell line that endogenously expresses high levels of KIM-1, apoptotic cell uptake can be seen as early as 81 min. LC3 enrichment to the phagosome was not observed until 181 min (Fig 2F and Video EV4). LC3 accumulation at the plasma membrane was only observed in fewer than 5% of 769p cells (Fig 2G). Thus, in most cases, LC3 localizes to the phagosome after phagocytosis in both primary PTCs and 769p cells. Both phagocytic and autophagic pathways ultimately lead to lysosomal degradation. To confirm that the encapsulation of KIM-1- positive phagosomes with LC3 occurred prior to the lysosome, a tandem fluorescent LC3 (ptflc3) reporter construct was used with RFP and GFP tags on the LC3 protein. The GFP tag is ph sensitive and loses fluorescent signal in the lysosome, while the RFP signal is maintained (Kimura et al, 2007). KIM-1-expressing LLC-PK1 cells, transfected with ptflc3, were incubated with apoptotic cells and imaged for GFP-RFP-LC3. Both the GFP and RFP signals were detected at the phagosome early after phagocytosis (Fig 3A, B at 90 min and Video EV5). At 150 min, the GFP signal was diminished and the RFP signal remained, indicating fusion to the lysosome (Fig 3A, B and Video EV5). We also analyzed LC3-GFP and LAMP1- RFP localization to the phagosome. LC3-GFP was found to localize to the phagosome prior to LAMP1 (Fig 3C). Quantification of the phagosomes in Fig 3C demonstrates that LC3 is enriched around the phagosome prior to LAMP1 localization (Fig 3C lower panels). We also found that RFP-ubiquitin (Ub) localized to phagosomes at similar time or immediately prior to LC3-GFP (Fig 3D and E). These results suggest that phagosomes are ubiquitinated, and LC3 is added prior to fusion of phagosomes to lysosomes. Of note, in some live cell imaging experiments, LC3-positive structures merged with, and dock to, KIM-1 phagosomes, prior to the phagosome becoming surrounded by LC3 in a continuous manner (Fig EV3A). It has been reported that phagocytosed bacteria targeted by autophagy are surrounded by vesicles which fuse to form a double-membrane structure (Rich et al, 2003). Likewise, vesicles and discontinuous double membranes are observed around phagosomes by EM (Fig EV3B). Knockdown of KIM-1 expression or expression of phagocytosisdeficient mutant KIM-1 inhibits phagocytosis and LC3 lipidation in primary cultured mouse PTCs Mouse and human KIM-1 have similar domain structure (Fig 4A). shrna knockdown of KIM-1 inhibited the uptake of fluorescently labeled apoptotic cells by primary cultured PTCs (Fig 4B and C). Thus, phagocytosis of apoptotic cells by primary PTCs was KIM-1 dependent. Knockdown of KIM-1 resulted in less membrane-bound LC3 in the absence or presence of apoptotic cells as reflected by decreased LC3 II levels (Fig 4D). To confirm that KIM-1-mediated induction of LC3 lipidation was phagocytosis dependent, we generated a KIM-1 phagocytosis-deficient mutant in which the third exon of the gene, encoding the mucin domain, was deleted (Xiao et al, 2012). Polymorphisms in the mucin domain in humans are associated with asthma and other diseases (McIntire et al, 2004). KIM-1 Dmucin -expressing proximal tubule cells took up significantly fewer apoptotic cells (Fig 4E and F). The marked decrease of phagocytosis in these cells was associated with a reduction in the levels of LC3 II (Fig 4G). Thus, KIM-1-mediated phagocytosis was associated with enhanced LC3 lipidation in primary cultured PTCs. KIM-1-induced LC3 lipidation is dependent on ligand binding Mutation of the KIM-1-binding domain completely blocked phagocytosis of apoptotic cells and subsequent LC3 lipidation. LLC-PK1 cells, transfected with three targeted mutations in the mouse KIM-1 sequence, amino acids (Fig 4A) in the PS-binding domain (Kobayashi et al, 2007; Santiago et al, 2007) (WF/AA, ND/AA, and WFND/AAAA), these mutant proteins did not bind to or take up apoptotic cells (Fig 4H and I), although each was expressed at levels equivalent to wild-type KIM-1 in kidney epithelial cells which take up apoptotic cells (Fig 4H and K). LC3 lipidation, as measured by an increase in cells displaying large numbers of LC3 punctae and LC3 II levels by Western blot, was not induced in any of the cells carrying one of the three PSbinding domain mutants (Fig 4J L). Incubation of apoptotic cells further increased LC3 punctae formation and LC3 II levels in wildtype but not mutant KIM-1-expressing cells (Fig 4J and K). By contrast, serum starvation induced an increase in cells with LC3 punctae in all transfected cells, indicating that the transfections did not alter the capacity for growth factor deprivation to induce LC3 lipidation (Fig 4J and K) The EMBO Journal Vol 34 No ª 2015 The Authors
7 Published online: August 17, 2015 Craig R Brooks et al KIM-1 links autophagy factors to MHC presentation The EMBO Journal A B C D E Figure 3. ª 2015 The Authors The EMBO Journal Vol 34 No
8 The EMBO Journal KIM-1 links autophagy factors to MHC presentation Craig R Brooks et al KIM-1-mediated LC3 lipidation requires KIM-1 phosphorylation and interaction with p85 The dependence of KIM-1-induced LC3 lipidation on ligand binding suggests that it may involve KIM-1 signaling. The KIM-1 extracellular domain, containing the ligand-binding site, can be cleaved and shed by metalloproteases (Bailly et al, 2002). The resulting remnant peptide, consisting of the transmembrane domain and cytosolic domain, contains two tyrosine phosphorylation sites (Bailly et al, 2002). These phosphorylation sites have been shown to play a role in KIM-1 signaling (de Souza et al, 2008). Interestingly, the KIM-1 remnant peptide has different subcellular localization compared to full-length KIM-1 (Fig 5A and B). KIM-1 is localized primarily to the apical membrane, while the remnant peptide localizes to intracellular vesicles (Fig 5A and B). The ligand binding mutant, WFND/ AAAA, primarily localizes to the apical membrane, and the remnant peptide staining is greatly reduced (Fig 5B). Wild-type KIM-1 also colocalized with components of the endocytotic/exocytotic pathways, such as vesicle-associated membrane proteins (VAMP) (Fig 5C). In order to determine if KIM-1 signaling was involved in LC3 lipidation, we examined the effect of deleting different KIM-1 domains on phagocytosis and LC3 lipidation. Deletion of the KIM-1 extracellular domain, while retaining the cytosolic domain and transmembrane domain (cytodomain), ablated LC3 lipidation and phagocytosis (Fig 5D F). Deletion of the cytosolic domain, while retaining the ectodomain and transmembrane domain (ectodomain + TM), did not significantly alter phagocytic response compared to full-length KIM-1 (Fig 5D F). The ectodomain + TM and cytodomain mutants were expressed at similar levels as wild-type KIM-1 (Fig 5F). KIM-1 ectodomain + TM in the absence of the cytodomain, however, was deficient in LC3 lipidation especially when cells were incubated with bafilomycin to prevent lysosomal degradation (Figs 5G and H, and EV4). Therefore, the cytosolic tail of KIM-1 was dispensable for phagocytosis but necessary for LC3 lipidation. We next examined if KIM-1 phosphorylation played a role in KIM-1-induced LC3 lipidation and phagocytosis. Mutation of Y314 or Y350 to phenylalanine reduced KIM-1-mediated phagocytosis (Fig 5I and J). The KIM-1 Y314F mutation more closely correlated with reduced lipidated LC3 levels (Fig 5J and K). Mutation to the human KIM-1 mucin domain in a region similar to the mouse KIM-1 Δmucin mutant, KIM-1Δ , reduced phagocytosis (Fig 5I). Mutation to the juxtamembrane region, KIM-1Δ , which we previously reported to prevent KIM-1 shedding (Bailly et al, 2002; Zhang et al, 2007), did not have an effect on phagocytosis (Fig 5I). These data suggest that KIM-1 phosphorylation regulates phagocytosis and LC3 lipidation. To test whether KIM-1 ligand-binding mutants have an altered phosphorylation response, we examined the phosphorylation status of wild-type mouse KIM-1 and KIM-1 WFND/AAAA. We found that wild-type KIM-1 was tyrosine phosphorylated and the WFND/AAAA mutant had greatly reduced phosphorylation, despite the fact it was expressed at higher levels (Fig 5L). We examined whether KIM-1 co-localized with p85, a phosphoinositide 3-kinase regulatory subunit previously reported to be involved in KIM-1 signaling (de Souza et al, 2008). KIM-1 and p85 co-localized at apoptotic cells that were phagocytosed or in the process of being phagocytosed (Fig 5M). p85 co-immunoprecipitated with wild-type human KIM-1 (Fig 5N). Deletion of the mucin domain (KIM-1 Dmucin ) resulted in a marked reduction in the interaction with p85 (Fig 5O). To determine if p85 plays a role in KIM-1-mediated LC3 lipidation, we examined the effect of KIM-1 overexpression on wt or p85 a/b double knockout (DKO) mouse embryonic fibroblasts (MEFs). Wt cells showed an increase in lipidated LC3 with KIM-1 overexpression, while p85 DKO cells did not (Fig 5P). Taken together, these data support the model that KIM-1 ligand binding increases KIM-1 phosphorylation and interaction with p85 is necessary for KIM-1-induced LC3 lipidation. KIM-1 phagosomes localize to multi-membrane structures To evaluate KIM-1 phagosome processing, the ultrastructure of KIM-1 phagosomes was analyzed by electron microscopy (EM) to detect the presence of multi-membrane-bound phagosomes (Codogno et al, 2011; Randall-Demllo et al, 2013). Phagosomes were imaged at different stages of maturation. Initially, the Figure 4. Mutations to KIM-1 PS-binding domain blocks phagocytosis and autophagy induction. A Diagram of mouse and human KIM-1 protein domains indicating the PS-binding domain, WFND, which was targeted for mutation. B Representative images of apoptotic cell uptake (green) in primary mouse PTCs transfected with shgfp or shkim-1. C Quantification of phagocytosis of CellTracker green-labeled apoptotic cells by PTCs expressing shgfp or shkim-1. *P < 0.01, n = 3. D Immunoblot analysis of KIM-1 and LC3 in PTCs transduced with shgfp or shkim-1, and incubated with apoptotic cells (representative of four experiments). E Representative images of apoptotic cell uptake by cells expressing wt mouse KIM-1 or KIM-1 Dmucin. F Quantification of uptake of fluorescently labeled apoptotic cells by primary PTCs expressing wt KIM-1 or KIM-1 Dmucin.*P < 0.001, n = 4. G Immunoblot analysis of LC3 II formation in wt KIM-1- or KIM-1 Dmucin -expressing PTCs. H Representative images of LLC-PK1 cells transfected with wild-type mouse KIM-1 or mouse KIM-1 PS-binding mutants (red) incubated with apoptotic cells (green, arrows). I Quantification of uptake of apoptotic cells in LLC-PK1 cells transfected with pcdna wt KIM-1 or KIM-1-binding mutants. *P < 0.001, n = 3. J LLC-PK1 cells were transfected with wt KIM-1 or PS-binding mutants and were incubated with medium (**P < 0.01), apoptotic cells (***P < 0.001), and serum-free medium (*P < 0.05, # significantly different from media control, P < 0.001). Autophagy was quantified by scoring cells positive for GFP-LC3 punctae. Lower panel: quantification of immunoblot in (K). # P < 0.05 compared to pcdna control. n = 4. K Immunoblot analysis of the expression of KIM-1 PS-binding domain mutants and LC3 in pcdna, wt KIM-1, WF/AA, ND/AA, and WFND/AAAA mutants with and without incubation with apoptotic cells or serum starvation (Note, the antibody used for immunoblots preferentially recognizes the LC3 II band). L Representative images from five independent experiments of LC3 punctae formation (arrows) in cells transfected with wt KIM-1, WF/AA, ND/AA, and WFND/AAAA. N = nucleus. Data information: Data presented as mean SD. Statistical comparisons were calculated by Student s t-test (C) and ANOVA followed by Bonferroni s post hoc analysis (F, I and J). Scale bars, 100 lm (B), 20 lm (E and H) and 10 lm (I). Source data are available online for this figure The EMBO Journal Vol 34 No ª 2015 The Authors
9 Craig R Brooks et al KIM-1 links autophagy factors to MHC presentation The EMBO Journal A B C D E F G H I J K L Figure 4. ª 2015 The Authors The EMBO Journal Vol 34 No
10 The EMBO Journal KIM-1 links autophagy factors to MHC presentation Craig R Brooks et al Figure 5. KIM-1 phosphorylation and interaction with p85 are required for LC3 lipidation. A, B Immunofluorescence staining of KIM-1 using antibodies specific for the extracellular domain (KIM-1 E Ab, R&D clone ) and cytosolic domain (R9 KIM-1 C Ab, Bonventre Lab) (Ichimura et al, 1998, 2008; Bailly et al, 2002) in mouse kidney tissue following ischemic injury (A) or LLC-PK1 cells transfected with full-length mouse KIM-1 or the WFND/AAAA mutant (B). (Note, 5B was taken at the apical surface of the cell, which consists of numerous microvilli. Representative of four independent experiments). C Representative images from three independent experiments of wild-type KIM-1 co-localization with intracellular VAMP8 and VAMP5. D Representative images of apoptotic cell uptake in LLC-PK1 cells expressing full-length KIM-1 or KIM-1 ectodomain or cytodomain mutants. E Quantification of fluorescently labeled apoptotic cell uptake in LLC-PK1 cells expressing full-length KIM-1 or KIM-1 mutants. *P < 0.05, n = 3. F Immunoblot analysis of KIM-1 or the domain deletion mutants expression. G, H Immunoblot analysis of LC3 lipidation in LLC-PK1 cells expressing full-length KIM-1 or KIM-1 mutants. Note that in the blot shown, the samples were treated with 100 nm bafilomycin. *P = n = 3. I Quantification of fluorescently labeled apoptotic cell uptake in LLC-PK1 cells expressing full-length KIM-1 or KIM-1 mutants. *P < 0.05, n = 3. J, K Immunoblot analysis of the expression of KIM-1 mutants in LLC-PK1 cells and the level of LC3 lipidation. n = 3, *P = L Mouse KIM-1 or KIM-1 WFND/AAAA phosphorylation was examined by immunoprecipitating KIM-1 and probing for p-tyr. M Representative images from 3 independent experiments of co-localization of KIM-1 and p85. N KIM-1 co-immunoprecipitation analysis of the interaction of p85 with human KIM-1. O KIM-1 co-immunoprecipitation analysis of the interaction of p85 with mouse KIM-1 and KIM-1 Dmucin. P Immunoblot analysis of LC3 lipidation in wild-type of p85a/b double knockout MEFs. Right panel: quantification of immunoblots represented in 5P. *P = for WT control vs. WT apop. and for WT apop. vs. DKO apop.. Data information: Data presented as mean SD. Statistical comparisons were calculated by ANOVA followed by Bonferroni s post hoc analysis. Scale bars, 7 lm (A), 5 lm (B), 50 lm (D), and 10 lm (M). Source data are available online for this figure. phagocytic cup formed around the apoptotic cell debris (Fig 6A). Importantly, at the initial phagocytic cup stage, the apoptotic cell membrane and phagocytic cell membrane were indistinguishable at > 150,000 by EM (Fig 6A and data not shown). In subsequent early phagososomal stages, the phagosomes were also found to have only a single membrane (Fig 6B left panels). At later stages following phagocytosis, phagocytosed apoptotic cell debris were surrounded by two membranes (Fig 6B center panels and 6F) or multiple membranes (Fig 6B right panels and 6F). We hypothesized that the multi-membrane structures were the last step before lysosomal fusion and were not more prominent due to rapid degradation of these structures. Multi-membrane autophagosomes have been shown to form due to repeated sequestration but are generally rarely observed because they are rapidly degraded (Corcelle et al, 2007; Li et al, 2013). To test if the number of multi-membrane structures could be amplified, lysosomal degradation was inhibited with bafilomycin (Corcelle et al, 2007). Incubation with bafilomycin, which blocks lysosome acidification and fusion of autophagosomes to lysosomes (Yamamoto et al, 1998; Jahreiss et al, 2008; Klionsky et al, 2008), increased the number of multi-membrane phagosomes as well as phagosomes surrounded by distinct two membrane structures (Fig 6C and F). Cells expressing a KIM-1 PS-binding domain mutant deficient in phagocytosis, WFND/AAAA, did not take up apoptotic cells and did not have phagosomes (Fig 6D). To extend these results in vivo, immunogold staining for KIM-1 was performed in post-ischemic kidneys. KIM-1 localized to the brush border of PTCs (Fig 6G second panel) as well as to intracellular membranebound vesicles (Fig 6G third and fourth panels) (Kabeya et al, 2000, 2004). Co-immunogold labeling revealed that LC3 co-stained the same membrane-bound organelles as KIM-1 (Fig 6H). (Tissues shown in Fig 6G and H were processed using methods that preserve the antigen sites for immunogold staining. As a result of processing steps associated with Lowicryl embedding, cell membranes are difficult to discern, in contrast to standard TEM.). Lysosomal inhibition in vivo (1-time 1 mg/kg i.p. injection of bafilomycin 1 h before induction of ischemia) resulted in large multi-membrane bodies in wild-type KIM-1 proximal tubules (Fig 6I left panels). In the KIM-1 Dmucin mice, there were fewer and smaller multi-membrane bodies (Fig 6I right panels, J and K). Autophagy components are necessary for efficient phagosome maturation and LC3 lipidation To determine if autophagy genes were necessary for phagosome maturation, we analyzed acidification of the phagosomes utilizing phrodo red, a ph-sensitive fluorescent tag, in cells where autophagy genes were depleted by sirna. KIM-1-expressing LLC-PK1 cells were transfected with sirna against the autophagy genes encoding ATG5, Beclin1, ULK1, or scrambled sirna (siscram). Cells were then incubated with apoptotic cells labeled with phrodo red for 2 h. Unattached apoptotic cells were washed off and replaced with fresh media. In KIM-1-expressing cells, transfected with scrambled sirna, internalized apoptotic cells showed increased red fluorescence, indicating that phagosomes fused with the lysosome, a process that peaked at 8 h. By contrast, cells transfected with sirna against ATG5, Beclin1, or ULK1 had fewer fluorescent apoptotic cells (Fig 7A) and reduced LC3 lipidation (Figs 7B and EV5) at the same time point. The effect of autophagy gene knockdown on LC3 II levels was also evaluated in KIM- 1-expressing TCMK mouse kidney tubular epithelial cells using different sirnas against ATG5 or ULK1 (Fig 7C). sirnas against autophagy genes did not alter the uptake of apoptotic cells, indicating autophagy genes are not required for KIM-1-mediated phagocytosis (Fig 7D). The effect of dominant negative (DN) ATG5 (RFP-ATG5-K130R) or DN ULK1 (GFP-ULK1-K46N) on LC3 lipidation was also tested. Compared to control cells not exposed to apoptotic cells, the addition of apoptotic cells to KIM-1-expressing cells resulted in increased LC3 II levels in the empty vectortransfected cells (Fig 7E). Cells expressing either DN ATG5 or DN ULK1 did not have increased levels of LC3 II upon stimulation with apoptotic cells (Fig 7E). Thus, KIM-1-induced LC3 lipidation was dependent on autophagy genes and KIM-1-regulated LC3 lipidation and autophagy are necessary for efficient phagosome maturation in kidney tubule cells The EMBO Journal Vol 34 No ª 2015 The Authors
11 Published online: August 17, 2015 Craig R Brooks et al A B C G D H E I K N The EMBO Journal KIM-1 links autophagy factors to MHC presentation J L O F M P Figure 5. ª 2015 The Authors The EMBO Journal Vol 34 No
12 The EMBO Journal KIM-1 links autophagy factors to MHC presentation Craig R Brooks et al Figure 6. KIM-1 phagosomes are enveloped by multi-membrane structures. LLC-PK1 cells were transfected with wild-type KIM-1 or KIM-1 WFND/AAAA and incubated with apoptotic cells. Cells were then analyzed by EM. A Electron micrographs of wild-type KIM-1-expressing LLC-PK1 cell phagocytosis of an apoptotic cell. B EM micrographs of phagosomes within wild-type mkim-1-expressing cells. C EM micrographs of KIM-1-expressing LLC-PK1 cells incubated with apoptotic cells in the presence of 100 nm bafilomycin. D EM micrographs of a cell expressing the phagocytosis-deficient KIM-1 WFND/AAAA mutant incubated with apoptotic cells. E, F Quantification of the number of cells with phagosomes and the number of phagosomes encapsulated by 1, 2, or> 2 membranes. n 24 cells per condition. G Representative immunogold staining (from four experiments) of KIM-1 in post-ischemic kidney tissue (Note: Fig 6G and H reflect processing by Lowicryl embedding in order to preserve the antigen sites for immunogold staining. With Lowicryl embedding, cell membranes are difficult to discern, relative to standard TEM). H Co-immunogold staining of KIM-1 (12 nm beads) and LC3 (18 nm beads) in post-ischemic kidney tissue. Representative of three mice. I EM micrographs of wild-type (left) or KIM-1 Dmucin kidneys from mice injected with Baf and subjected to ischemic injury. J, K Quantification of the number of multi-membrane bodies per cell (J) and the distribution of the size of the multi-membrane bodies (K) from wild-type or KIM-1 Dmucin kidneys from mice injected with Baf and subjected to 25 min bilateral ischemia. Tissue was harvested at 24 h. Statistical comparisons were calculated by Student s t-test. *P < 0.05, n = 3 mice with at least 30 proximal tubule cells analyzed per mouse. Data information: Scale bars, 2 lm (A D, G and H) and 10 lm (I). KIM-1 expression activates ULK1/ATG1 and dominant negative ATG5 or ULK1 blocks LC3 accumulation at the phagosome Activation of ULK1 by phosphorylation at serine 555 is known to occur upon autophagy induction (Hamacher-Brady et al, 2007; Florey et al, 2011; Xiao et al, 2012). pulk1-s555 was increased in KIM-1-expressing LLC-PK1 cells treated with apoptotic cells compared to cells expressing the pcdna empty vector (Fig 7F). AMPK is known to phosphorylate ULK1 at S555 (Zou et al, 2014). AMPK was also phosphorylated with KIM-1 expression (Fig 7F). The AMPK inhibitor, compound C, which is known to block AMPK-induced ULK1 phosphorylation (Zou et al, 2014), reduces the levels of phospho AMPK and ULK1, as well as total ULK1 (Fig 7F). Compound C also reduced phagosome maturation (Fig 7G and H). The autophagy inhibitor Spautin1 also inhibited phagosome maturation (Fig 7G and H). ULK1 also localized to phagosomes (Fig EV6A). To determine if autophagy is required for LC3 accumulation at the autophagosome, human embryonic kidney cells (HEK293) co-expressing KIM-1 plus DN ATG5 and GFP-LC3 or DN ULK1 and RFP-LC3 were exposed to apoptotic cells. As overexpression of wild-type ATG5 or ULK1 can up- or down-modulate autophagy, respectively, cells expressing RFP or GFP plus GFP-LC3 or RFP-LC3, respectively, were used as controls (Hamacher-Brady et al, 2007; Hara et al, 2008). KIM-1-mediated phagocytosis was not affected by the expression of DN ATG5 (ATG5-K130R) or DN ULK1 (ULK1-K46N), indicating that the phagocytic function of KIM-1 does not require ATG5 or ULK1 (Fig 7I L). In cells expressing RFP or GFP, LC3 localized to phagocytosed apoptotic cells (Fig 7I, J (upper panels), K and L). In cells expressing ATG5-K130R or ULK1-K46N, however, LC3 co-localization with phagocytosed apoptotic cells was greatly decreased (Fig 7I, J (lower panels), K and L). Thus, LC3 localization to the phagosome requires functional ATG5 and ULK1. KIM-1-dependent phagocytosis and induction of autophagy leads to antigen presentation Approximately 80% of PTCs isolated from mice express MHC II by flow cytometry, raising the possibility that KIM-1-mediated phagocytosis and autophagy induction can regulate MHC antigen presentation (Fig 8A). The majority of primary PTCs positive for MHC II were also positive for KIM-1 (Fig EV6B). To evaluate the role of KIM-1-mediated autophagy in antigen presentation, we first examined KIM-1 co-localization with the antigen processing machinery. DM acts as a peptide editor and is essential for the processing of antigens (Schulze & Wucherpfennig, 2012). DM co-localizes with KIM-1 in distinct subcellular structures (Fig EV6C). To analyze the ability of PTCs to present antigens, PTCs were incubated with OVA are Ea peptide. Cells were then treated with antibodies that recognized MHC I bound to OVA peptide or MHC II bound to Ea peptide. These antibodies only react with OVA or Ea when these peptides have been presented on MHC I or II, respectively. As controls, cells incubated with the MHC I OVA peptide were stained for MHC II presentation and cells incubated with MHC II Ea peptide were stained for MHC I presentation. Flow cytometric analysis revealed that MHC I cross-presentation and MHC II presentation occurred in KIM-1 wt primary cultured PTCs, and the presentation was greatly reduced in KIM-1 Dmucin cells (Figs 8B and C, and EV6D). To analyze the effect of PTC MHC presentation on T cells, naïve CD4 + T cells were isolated from OT-II transgenic mice, which respond to OVA presented on MHC II. Boston University mouse proximal tubule cells (BUMPT) (Sinha et al, 2005) were incubated with PS liposomes containing OVA for 2 h, washed extensively, and then incubated with CD4 + T cells pre-labeled with CellTrace Violet dye (Invitrogen) to allow cellular proliferation to be tracked through successive generations. After 48 h of incubation with wild-type KIM-1-expressing PTCs fed OVA liposomes, a greater percentage of CD4 + T cells display a naïve CD44 low CD62L high phenotype ( % in the wild-type vs % in the KIM-1 Dmucin cells and %, P < 0.05 in pcdh control PTCs). Furthermore, CD4 + T cells co-incubated with KIM-1-expressing PTCs proliferated less (Fig 8D) and were enriched in FoxP3.GFP + regulatory T cells (Tregs) by percentage (Fig 8E) and absolute number (Fig 8G). Removal of FoxP3 + Tregs from the T-cell population by flow sorting abolished the anti-proliferative effects of KIM-1-mediated MHC presentation (Fig 8F). These data indicate that KIM-1-mediated MHC presentation maintains T cells in their naïve phenotype and reduces T-cell proliferation in a manner dependent on Tregs. To evaluate the role of autophagy on the T-cell response, KIM-1- expressing BUMPT cells were transfected with sirnas against ATG5 and ULK1. Compared to scrambled sirna, knockdown of ATG5 or ULK1 led to greater CD4 + T-cell proliferation and a reduction in Tregs (Fig 8H and I). Thus, KIM-1-induced phagocytosis leads to antigen presentation, but the recognition of antigen by T cells 2452 The EMBO Journal Vol 34 No ª 2015 The Authors
13 Published online: August 17, 2015 Craig R Brooks et al A D The EMBO Journal KIM-1 links autophagy factors to MHC presentation B C E G H I F J K Figure 6. ª 2015 The Authors The EMBO Journal Vol 34 No
14 The EMBO Journal KIM-1 links autophagy factors to MHC presentation Craig R Brooks et al Figure 7. sirna against autophagy genes inhibits phagosome maturation and LC3 lipidation and DN ATG5 or DN ULK1 prevents LC3 lipidation and localization with the phagosome. A KIM-1-expressing LLC-PK1 cells were transfected with sirnas against a scrambled sequence (siscram), Beclin1, ATG5, orulk1, incubated with apoptotic cells fluorescently labeled with ph-sensitive phrodo red for 2 h, and then washed and incubated for 8 h. Quantification of phrodo fluorescence intensity over the time series is shown in the right panel. n = 520 fields from a representative experiment repeated three times. B, C Immunoblot analysis of LC3 lipidation in KIM-1-expressing LLC-PK1 cells (B) or TCMK cells (C) transfected with sirna against autophagy genes and exposed to apoptotic (apop) cells or untreated (Co). D Quantification of phagocytosis of CellTracker green-labeled apoptotic cells by BUMPT cells transfected with siscram, siatg5, or siulk1. E Immunoblot analysis of LC3 lipidation in HEK293 cells expressing KIM-1 and DN ATG5 or DN ULK1, with or without apoptotic cell incubation. F Immunoblot analysis of pulk1 and pampk in LLC-PK1 cells transfected with empty vector or wild-type KIM-1 and incubated with control media or apoptotic cells, with or without compound C (10 lm). Right panel: Quantification of pulk1 levels. *P = for pcdna vs. KIM-1 and P = for pcdna apop vs. KIM-1 apop. n = 3. G, H Representative images (G) and fluorescent quantification (H) of phrodo-stained apoptotic cells incubated with KIM-1-expressing LLC-PK1 cells in the presence of DMSO, Spautin1 (10 lm), or compound C (10 lm) and imaged over time. n = 10 fields from an representative experiment repeated three times. I Co-localization of GFP-LC3 with phagocytosed apoptotic cells in HEK293 cells expressing either RFP or RFP-DN-ATG5. Arrows indicate LC3 accumulation on an phagocytosed apoptotic cell. J Co-localization of RFP-LC3 with phagocytosed apoptotic cells in HEK293 cells expressing either GFP or GFP-DN-ULK1. Arrows indicate LC3 accumulation on an phagocytosed apoptotic cell. Arrowheads indicate areas of DN-ULK1 accumulation on apoptotic cell containing phagosomes. K, L Quantification of the percentage of DN ATG5- or DN ULK1-positive cells with phagosomes and the percentage of DN ATG5- or DN ULK1-positive cells with LC3-positive phagosomes, n 180 cells. Data information: Scale bars, 30 lm (A) and 10 lm (I and J). Data presented as mean SD. Statistical comparisons were calculated by ANOVA followed by Bonferroni s post hoc analysis. Source data are available online for this figure. suppresses CD4 + T-cell activation and promotes expansion of tolerogenic Tregs, in an autophagy-dependent manner. To determine if functional KIM-1 could regulate Tregs in vivo, we analyzed T cells from post-ischemic kidney tissue from wild-type and KIM-1 Dmucin mice. Mice expressing KIM-1 Dmucin have fewer Foxp3 + Tregs and greater infiltration of CD4 + and CD8 + effector T cells (CD44 high and CD62 low ) and lower Foxp3 + Tregs compared to wildtype mice, following ischemic injury (Fig 8J and K). Discussion Epithelial cells, as semi-professional phagocytes, are often involved in clearance of apoptotic cells following tissue injury or remodeling, such as after kidney injury or during mammary involution (Erwig & Henson, 2007). Epithelial cell phagocytosis of neighboring cells reduces inflammation by removal of dead cells prior to recognition by lymphocytes, thus limiting immune cell infiltration, and through anti-inflammatory signaling induced by the uptake of apoptotic cells (Erwig & Henson, 2007). KIM-1/TIM-1 is a phosphatidylserine phagocytosis and scavenger receptor expressed by kidney proximal tubule epithelial cells and, at a much lower level, in a subset of immune cells (Ichimura & Mou, 2008; Ichimura et al, 2012). The phagocytic function of KIM-1 is best characterized in kidney tubular cells (Ichimura et al, 2008). In the injured kidney, KIM-1 is upregulated more than a thousand fold and aids in the recovery of organ injury through phagocytosis of apoptotic cells and cellular debris (Ichimura et al, 2008; Yang et al, 2015). Kidney injury in mice carrying a non-functional mutant KIM-1 results in a worsening of the injury with the accumulation of unphagocytosed apoptotic cells and increased inflammation (Yang et al, 2015). The current study provides mechanistic insight into how the phagocytic capability of epithelial cells that express KIM-1 can trigger a cascade of events that result in modulation of the immune response. KIM-1 expression and phagocytosis of apoptotic cells leads to increased LC3 lipidation. Lipidated LC3 encompasses the phagosome in a manner dependent on ATG5, Beclin1, and ULK1. Mutations to the KIM-1 sequence that prevent phagocytosis also block KIM-1 induction of LC3 lipidation. Material taken up by KIM-1 is processed and presented on MHC. Presented antigens result in a suppression of T-cell proliferation. These data indicate that phagocytosis mediated by KIM-1 expression in epithelial cells can limit inflammation by both removal of dead cells and induction of anti-inflammatory Treg signaling. To our knowledge, KIM-1 is the first epithelial receptor shown to mediate phagocytosis, enhance autophagy, and induce pro-tolerogenic antigen presentation. We first characterized KIM-1 as an epithelial cell PS and scavenger receptor capable of transforming epithelial cells into phagocytes (Ichimura et al, 2008). KIM-1 protein is a type one transmembrane protein that contains an IgG domain, mucin domain, and cytosolic domain with two tyrosine phosphorylation sites. While KIM-1 s phagocytic function is known, the role of the different protein domains have not previously been characterized. Through mutagenesis studies, we have now defined the role of the major KIM-1 protein domains. First, we demonstrated in primary cultured proximal tubule epithelial cells that KIM-1 is the dominant apoptotic cell phagocytosis receptor by knocking down KIM-1 in these cells and quantifying apoptotic cell uptake. Mutation to the ligand-binding site, WFND, in the IgG domain or deletion of the majority of the mucin domain blocks KIM-1-mediated phagocytosis and LC3 lipidation. We next demonstrated that the ectodomain plus the transmembrane domain was sufficient to mediate phagocytosis but not LC3 lipidation. The cytodomain plus transmembrane alone domain did not regulate phagocytosis or LC3 lipidation. These data demonstrate that the extracellular + transmembrane domains of KIM-1 were sufficient for phagocytosis, while the full-length protein was necessary for processing of the phagosome via autophagy proteins. PI3K regulatory subunit p85 is involved in KIM-1 phosphorylationmediated signaling through interaction with the cystosolic domain (de Souza et al, 2008). p85 binding to p110 inhibits PI3K activity and promotes autophagy, and p85 depletion reduces autophagy in HEK293 cells (Kuchay et al, 2013). p85 is also known to regulate starvation-induced autophagy through IKK. In this pathway, phosphorylation of p85 in the SH2 domain leads to inhibition of 2454 The EMBO Journal Vol 34 No ª 2015 The Authors
15 Published online: August 17, 2015 Craig R Brooks et al The EMBO Journal KIM-1 links autophagy factors to MHC presentation A C F B D E G I J K L H Figure 7. ª 2015 The Authors The EMBO Journal Vol 34 No
16 The EMBO Journal KIM-1 links autophagy factors to MHC presentation Craig R Brooks et al Figure 8. KIM-1 phagocytosis promotes MHC I cross-presentation and MHC II presentation in an autophagy-dependent manner. A Flow cytometric analysis of primary PTCs stained for MHC II (representative of three experiments). B, C Flow cytometric analysis of OVA-MHC I cross-presentation and Ea-MHC II presentation in primary cultured PTCs from wild-type KIM-1 or KIM-1 Dmucin (representative of three independent experiments). D, E OT-II CD4 + (7AAD-) T cells were incubated with BUMPT PTCs expressing empty vector, wild-type mouse KIM-1 (mkim-1), or mutant KIM-1 Dmucin and proliferation as well as expansion of FoxP3.GFP Tregs were analyzed by flow cytometry. F OT-II CD4 + (7AAD-) T cells were flow sorted to remove the Foxp3.GFP + cells and then incubated with BUMPT PTCs expressing wild-type or mutant KIM-1. G Absolute numbers of FoxP3 + CD4 + regulatory T cells from CD4 + cells incubated with pcdh, mkim-1, or KIM-1 Dmucin. H, I Analysis of T-cell proliferation and Treg expansion from T cells incubated with KIM-1-expressing PTCs transfected with sirna targeting autophagy genes. J, K Analysis of Foxp3 +,CD4 +,orcd8 + infiltrating cell in post-ischemic kidneys from wild-type mkim-1 or KIM-1 Dmucin (mut) mice. Data information: Statistical comparisons were calculated by Student s t-test (K) and ANOVA followed by Bonferroni s post hoc analysis (G J). *P < 0.05, **P < 0.01, n = 3. Data presented as mean SD. KIM-1 Dmucin was cloned from the KIM-1 Dmucin mouse, experiments with KIM-1 Dmucin expression vectors utilized mouse KIM-1 as a wildtype control. PI3K and induction of autophagy (Comb et al, 2012). We explored the role of KIM-1 phosphorylation and p85 signaling in KIM-1- mediated phagocytosis and LC3 lipidation. The cytosolic KIM-1 phosphorylation sites were required for phagocytosis and LC3 lipidation. We also found that KIM-1 ligand-binding mutants have reduced phosphorylation, which correlated with decreased interaction with p85. Knockout of p85 prevented KIM-1-induced LC3 lipidation. These data provide the first evidence of a role of KIM-1 phosphorylation and interaction with p85 in KIM-1-mediated phagocytosis and autophagy induction. Recent studies have demonstrated a role of non-canonical autophagy in cell corpse clearance, termed LC3-associated phagocytosis (LAP) (Sanjuan et al, 2007). In this clearance pathway, LC3 is targeted to the phagosome in an ATG5-, ATG7-, and Beclin1- dependent manner; however, this process does not involve the engagement of upstream autophagy proteins such as ULK1 (Sanjuan et al, 2007; Florey et al, 2011; Martinez et al, 2011; Kim et al, 2013b). The requirement for LAP, and not autophagy, for corpse degradation is not universal, however (Cadwell & Philips, 2013; Cheng et al, 2013; Parihar et al, 2013). In particular, a recent study in C. elegans demonstrated that many autophagy genes are required for corpse degradation, including ULK1, which was not implicated in LAP (Cheng et al, 2013). In our studies, EM immunogold staining revealed KIM-1-positive organelles characteristic of autophagy. The ROS inhibitor DPI which has been shown to block LC3 lipidation in LAP, but not autophagy, did not inhibit LC3 lipidation in our system. The process and timing of multi-membrane formation in our model is similar to what has been reported in autophagic clearance of intracellular bacteria, such as Listeria monocytogenes (Rich et al, 2003). sirna against ULK1, as well as dominant negative ULK1, inhibited apoptotic cell clearance, LC3 lipidation, and LC3 localization to the phagosome induced by KIM-1 phagocytosis, implicating autophagy and not LAP. Also, unlike in LAP, KIM-1 expression induced phosphorylation and activation of ULK1/ATG1, which is essential for autophagosome formation. KIM-1 phagosomes are targeted by ubiquitin in a similar time course as LC3. The current study highlights some fundamental differences between the uptake of apoptotic cells by semi-professional phagocytes compared to professional phagocytes. One notable difference between epithelial KIM-1 action and scavenger function in immune cells is the rate of maturation of the phagocyte. As measured by a decrease in ph of the phagosome, as it fuses with the lysosome, phagosome maturation occurs much faster in professional phagocytes compared to epithelial cells (Fig EV2 compared to time courses in Fig 7). In our study, using live cell imaging, LC3-positive vesicles were sometimes observed to localize around the phagosome before becoming a continuous ring (Figs 2A and D, and EV3A). By EM, we observe discontinuous double membranes around phagosomes (Fig EV3B). It has been reported in macrophages that bacteria are targeted by multiple vesicles that fuse to form a double-membrane autophagic vesicles (Rich et al, 2003). It is likely that epithelial cells employ a similar mechanism to surround the phagosome with a double membrane. This could also argue against the involvement of LAP in KIM-1-mediated phagocytosis. The theory behind LAP is that LC3 is recruited and directly inserted in the phagosomal membrane; on the other hand, we observe LC3- positive structures interacting with the phagosome. The finding that KIM-1-mediated phagocytosis is not associated with upregulated ROS production is novel and could explain many of the differences we observe in epithelial cell phagocytosis vs. that of professional phagocytes. For instance, maturation of phagosomes derived from KIM-1-mediated phagocytosis is slower than that in macrophages or dendritic cells. In professional phagocytes, NOX enzymes form an electron transport system on the phagosomal membranes to transfer electrons into the phagosome, generating ROS in the lumen (Cross & Segal, 2004). The ROS flux begins the degradation process and lowers the phagosomal ph, also quickening the degradation process (Cross & Segal, 2004). Phagosomal ROS production is necessary for LC3 recruitment in LAP (Huang & Brumell, 2009; Huang et al, 2009; Ma et al, 2012; Lam et al, 2013; Romao et al, 2013). Without the upregulation of ROS, epithelial cells likely need to utilize more of the autophagy machinery to degrade the phagocytosed cargo. The biochemical assays performed to differentiate canonical autophagy from LAP suggest that canonical autophagy is responsible for LC3 localization to the phagosomes in PTCs; however, multimembrane phagosomes only account for ~10% of the phagosomes at a given time point. Under conditions of lysosomal inhibition, multi-membrane phagosomes become more prominent (Figs 6C, F and I, and 9). Importantly, multi-membrane lamellar bodies have been shown to require autophagy for their formation (Baba et al, 1994; Hariri et al, 2000; Lajoie et al, 2005; Corcelle et al, 2007; Li et al, 2013) and form due to repeated sequestration of autophagosome membranes but are generally rarely observed because they are rapidly degraded (Corcelle et al, 2007; Li et al, 2013). These data suggest three possible hypotheses: (i) KIM-1 phagosomes are degraded by autophagy; (ii) KIM-1 phagosomes are degraded by LAP, but epithelial cells require more autophagy factors, such as ULK1, to induce LAP compared to professional phagocytes; (iii) 2456 The EMBO Journal Vol 34 No ª 2015 The Authors
17 Craig R Brooks et al KIM-1 links autophagy factors to MHC presentation The EMBO Journal A B C D E F G H I J K Figure 8. ª 2015 The Authors The EMBO Journal Vol 34 No
18 The EMBO Journal KIM-1 links autophagy factors to MHC presentation Craig R Brooks et al Figure 9. Schematic representation of KIM-1-mediated phagocytosis. KIM-1 coats the apical surface of PTCs and binds tightly to apoptotic cells. Following phagocytosis, the phagosome is then targeted by ULK1- and LC3- positive membranes, leading to lysosomal degradation. Antigens derived from apoptotic cells targeted for degradation via autophagy machinery are presented on MHC I and MHC II. Autophagosomes fuse together at the phagosome, coating the phagosome with LC3. We also observe that smaller autophagosomes tend to fuse at the phagosome to create an enveloped phagosome (Figs 2A and D, and EV3A and B). Although a similar phenomenon has been proposed in macrophages for phagocytosis of bacteria (Rich et al, 2003), this would represent a novel mechanism by which autophagosomes generated in an ULK1-dependent manner target phagosomes for degradation in epithelial cells. We believe the data suggest that KIM-1 phagosomes are degraded by autophagy; however, we cannot completely rule out LAP, due to the fact that lysosomal inhibition is required to readily observe multi-membrane phagosome. Either hypothesis suggests that semi-professional phagocytes, such as epithelial cells, require more of the autophagy machinery to process phagocytosed material, likely due to the fact that epithelial cells have reduced ROS production in response to phagocytosis, limiting the clearance capacity of epithelial cells when compared to professional phagocytes. The links drawn by our results between phagocytosis, autophagy, and antigen presentation have important implications for immune responses associated with epithelial cell injury. Presentation of apoptotic cell self-antigens by macrophages and dendritic cells has been shown to be tolerogenic in steady-state conditions and crucial to the maintenance of peripheral T-cell tolerance. For instance, the presentation of apoptotic cell antigens have been shown to result in abortive T-cell proliferation and deletion (Kurts et al, 1997). Blockage of phagocytosis through genetic deletion or inhibition of key phagocytic proteins, such as milk fatty acid globule EGF factor 8, results in increased auto-antigen production (Nakayama et al, 2009; Albacker et al, 2010; Peng & Elkon, 2011; Baghdadi et al, 2013). Failure to take up apoptotic cells also leads to autoantibody production and autoimmune disease (Duffield et al, 2001; Nakayama et al, 2009). Our data demonstrate that, following phagocytosis in epithelial cells, phagosomes are targeted for degradation via the autophagy pathway, and material processed through autophagy is presented on MHC I and MHC II. While KIM- 1-mediated uptake of apoptotic cells leads to enhanced MHC presentation, the resultant adaptive immune response is one of tolerance. We found that there was a decrease in T-cell proliferation in response to antigens processed in this manner and an associated increase in the number and percentage of Tregs. Knockdown of autophagy genes or removal of the Treg population reversed the anti-inflammatory phenotype, leading to greater T-cell proliferation. These data could help explain evidence that KIM-1 polymorphisms have been associated with autoimmune disease (Lee et al, 2011). Mice expressing a mutant KIM-1 protein spontaneously developed autoimmune disease and lupus-associated autoantigens (Lee et al, 2011; Xiao et al, 2012). KIM-1 recognition of apoptotic cells has been linked to experimental models of asthma (Kim et al, 2013a). Our findings, combined with these recent studies, strongly suggest that KIM-1-mediated apoptotic cell phagocytosis and antigen presentation help maintain self-tolerance. In conclusion, we have shown that KIM-1-induced phagocytosis enhances autophagy and KIM-1 phagosomes are targeted to the autophagosome. KIM-1 phosphorylation and interaction with p85 are necessary for autophagy induction. KIM-1 induction of autophagy is dependent on ligand binding, involves phosphorylation of ULK1/ATG1, and leads to presentation of antigens on both MHC I and MHC II on the proximal tubule cell. Presentation of antigens derived from KIM-1 phagosomes is pro-tolerogenic, limiting T-cell proliferation by inducing Tregs. These data provide a hitherto unidentified link between epithelial injury, elimination of dead cells by autophagy, and regulation of the immune response via antigen presentation. Materials and Methods Materials LLC-PK1, 769p, TCMK, HEK293 and 293T cells were purchased from ATCC. Boston University mouse proximal tubular (BUMPT) cells were originally from Dr. Steve Borkan (Sinha et al, 2005). Cell lines were routinely tested for mycoplasma (Mycoalert Mycoplasma Detection Kit, Lonza, Cat#LT07-318) and, in the case of kidney cells, the presence of species-specific/cell-type markers as well as growth and morphological characteristics of kidney tubular epithelial cells. DMEM, RPMI 1640, and DMEM/F12 were from Cellgro, and FBS was from Gibco. Anti-KIM-1 antibodies to the human and rodent cytosolic domain (1400 and R9) and human ectodomain (AKG7) were generated and profiled as described previously (Bonventre lab) (Ichimura et al, 1998, 2008; Bailly et al, 2002; Zhang et al, 2007; Humphreys et al, 2013), and secondary antibodies were from Jackson ImmunoReasearch Laboratories Inc. Anti-KIM-1 goat polyclonal antibody which reacted with mouse KIM-1 ectodomain was purchased from R&D Systems (Cat# AF1817). Anti-Myc antibody was purchased from the Brigham and Women s Hospital antibody core facility. Anti-LC3 (for immunoblot of LLC-PK1 or human cell lysates) (clone D11), anti-pulk1 (clone D1H4 (Ser555)), total ULK1 (D8H5), AMPK (Cat# 2532), pampk (Cat# 2458 The EMBO Journal Vol 34 No ª 2015 The Authors
Supplemental Figures:
 Supplemental Figures: Figure 1: Intracellular distribution of VWF by electron microscopy in human endothelial cells. a) Immunogold labeling of LC3 demonstrating an LC3-positive autophagosome (white arrow)
Supplemental Figures: Figure 1: Intracellular distribution of VWF by electron microscopy in human endothelial cells. a) Immunogold labeling of LC3 demonstrating an LC3-positive autophagosome (white arrow)
SUPPLEMENTARY INFORMATION
 Figure S1 Induction of non-apoptotic death of SV40-transformed and primary DKO MEFs, and DKO thymocytes. (A-F) STS-induced non-apoptotic death of DKO MEF. (A, B) Reduced viability of DKO MEFs after exposure
Figure S1 Induction of non-apoptotic death of SV40-transformed and primary DKO MEFs, and DKO thymocytes. (A-F) STS-induced non-apoptotic death of DKO MEF. (A, B) Reduced viability of DKO MEFs after exposure
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
A. Generation and characterization of Ras-expressing autophagycompetent
 Supplemental Material Supplemental Figure Legends Fig. S1 A. Generation and characterization of Ras-expressing autophagycompetent and -deficient cell lines. HA-tagged H-ras V12 was stably expressed in
Supplemental Material Supplemental Figure Legends Fig. S1 A. Generation and characterization of Ras-expressing autophagycompetent and -deficient cell lines. HA-tagged H-ras V12 was stably expressed in
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting. protein3) regulate autophagy and mitophagy in renal tubular cells in. acute kidney injury
 Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
Kim-1/Tim-1 and Immune Cells: Shifting Sands
 Kim-1/Tim-1 and Immune Cells: Shifting Sands The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters. Citation Published Version Accessed
Kim-1/Tim-1 and Immune Cells: Shifting Sands The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters. Citation Published Version Accessed
Expanded View Figures
 PEX13 functions in selective autophagy Ming Y Lee et al Expanded View Figures Figure EV1. PEX13 is required for Sindbis virophagy. A, B Quantification of mcherry-capsid puncta per cell (A) and GFP-LC3
PEX13 functions in selective autophagy Ming Y Lee et al Expanded View Figures Figure EV1. PEX13 is required for Sindbis virophagy. A, B Quantification of mcherry-capsid puncta per cell (A) and GFP-LC3
Supplemental Figure 1. Western blot analysis indicated that MIF was detected in the fractions of
 Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were
 Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were isolated from wild type (PKC-θ- WT) or PKC-θ null (PKC-θ-KO)
Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were isolated from wild type (PKC-θ- WT) or PKC-θ null (PKC-θ-KO)
Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained
 Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained with MitoTracker (red), then were immunostained with
Supplementary Figure 1. Confocal immunofluorescence showing mitochondrial translocation of Drp1. Cardiomyocytes treated with H 2 O 2 were prestained with MitoTracker (red), then were immunostained with
Supplementary Figure 1
 Supplementary Figure 1 a γ-h2ax MDC1 RNF8 FK2 BRCA1 U2OS Cells sgrna-1 ** 60 sgrna 40 20 0 % positive Cells (>5 foci per cell) b ** 80 sgrna sgrna γ-h2ax MDC1 γ-h2ax RNF8 FK2 MDC1 BRCA1 RNF8 FK2 BRCA1
Supplementary Figure 1 a γ-h2ax MDC1 RNF8 FK2 BRCA1 U2OS Cells sgrna-1 ** 60 sgrna 40 20 0 % positive Cells (>5 foci per cell) b ** 80 sgrna sgrna γ-h2ax MDC1 γ-h2ax RNF8 FK2 MDC1 BRCA1 RNF8 FK2 BRCA1
Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson
 Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson General principles Properties of lysosomes Delivery of enzymes to lysosomes Endocytic uptake clathrin, others Endocytic pathways recycling vs.
Lysosomes and endocytic pathways 9/27/2012 Phyllis Hanson General principles Properties of lysosomes Delivery of enzymes to lysosomes Endocytic uptake clathrin, others Endocytic pathways recycling vs.
6. TNF-α regulates oxidative stress, mitochondrial function and autophagy in neuronal cells
 6. TNF-α regulates oxidative stress, mitochondrial function and autophagy in neuronal cells 6.1 TNF-α induces mitochondrial oxidative stress in SH-SY5Y cells. The dysregulation of mitochondria and oxidative
6. TNF-α regulates oxidative stress, mitochondrial function and autophagy in neuronal cells 6.1 TNF-α induces mitochondrial oxidative stress in SH-SY5Y cells. The dysregulation of mitochondria and oxidative
Figure S1. Western blot analysis of clathrin RNA interference in human DCs Human immature DCs were transfected with 100 nm Clathrin SMARTpool or
 Figure S1. Western blot analysis of clathrin RNA interference in human DCs Human immature DCs were transfected with 100 nm Clathrin SMARTpool or control nontargeting sirnas. At 90 hr after transfection,
Figure S1. Western blot analysis of clathrin RNA interference in human DCs Human immature DCs were transfected with 100 nm Clathrin SMARTpool or control nontargeting sirnas. At 90 hr after transfection,
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous
 Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Peli1 negatively regulates T-cell activation and prevents autoimmunity
 Peli1 negatively regulates T-cell activation and prevents autoimmunity Mikyoung Chang 1,*, Wei Jin 1,5,*, Jae-Hoon Chang 1, Yi-chuan Xiao 1, George Brittain 1, Jiayi Yu 1, Xiaofei Zhou 1, Yi-Hong Wang
Peli1 negatively regulates T-cell activation and prevents autoimmunity Mikyoung Chang 1,*, Wei Jin 1,5,*, Jae-Hoon Chang 1, Yi-chuan Xiao 1, George Brittain 1, Jiayi Yu 1, Xiaofei Zhou 1, Yi-Hong Wang
Tumor suppressor Spred2 interaction with LC3 promotes autophagosome maturation and induces autophagy-dependent cell death
 www.impactjournals.com/oncotarget/ Oncotarget, Supplementary Materials 2016 Tumor suppressor Spred2 interaction with LC3 promotes autophagosome maturation and induces autophagy-dependent cell death Supplementary
www.impactjournals.com/oncotarget/ Oncotarget, Supplementary Materials 2016 Tumor suppressor Spred2 interaction with LC3 promotes autophagosome maturation and induces autophagy-dependent cell death Supplementary
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis
 Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Nature Immunology doi: /ni.3268
 Supplementary Figure 1 Loss of Mst1 and Mst2 increases susceptibility to bacterial sepsis. (a) H&E staining of colon and kidney sections from wild type and Mst1 -/- Mst2 fl/fl Vav-Cre mice. Scale bar,
Supplementary Figure 1 Loss of Mst1 and Mst2 increases susceptibility to bacterial sepsis. (a) H&E staining of colon and kidney sections from wild type and Mst1 -/- Mst2 fl/fl Vav-Cre mice. Scale bar,
Supplementary Fig. 1 V-ATPase depletion induces unique and robust phenotype in Drosophila fat body cells.
 Supplementary Fig. 1 V-ATPase depletion induces unique and robust phenotype in Drosophila fat body cells. a. Schematic of the V-ATPase proton pump macro-complex structure. The V1 complex is composed of
Supplementary Fig. 1 V-ATPase depletion induces unique and robust phenotype in Drosophila fat body cells. a. Schematic of the V-ATPase proton pump macro-complex structure. The V1 complex is composed of
(a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable
 Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
William C. Comb, Jessica E. Hutti, Patricia Cogswell, Lewis C. Cantley, and Albert S. Baldwin
 Molecular Cell, Volume 45 Supplemental Information p85 SH2 Domain Phosphorylation by IKK Promotes Feedback Inhibition of PI3K and Akt in Response to Cellular Starvation William C. Comb, Jessica E. Hutti,
Molecular Cell, Volume 45 Supplemental Information p85 SH2 Domain Phosphorylation by IKK Promotes Feedback Inhibition of PI3K and Akt in Response to Cellular Starvation William C. Comb, Jessica E. Hutti,
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified
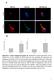 Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified for RFP-LC3 puncta (red dots) representing both autolysosomes
Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified for RFP-LC3 puncta (red dots) representing both autolysosomes
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11700 Figure 1: RIP3 as a potential Sirt2 interacting protein. Transfected Flag-tagged Sirt2 was immunoprecipitated from cells and eluted from the Sepharose beads using Flag peptide.
doi:10.1038/nature11700 Figure 1: RIP3 as a potential Sirt2 interacting protein. Transfected Flag-tagged Sirt2 was immunoprecipitated from cells and eluted from the Sepharose beads using Flag peptide.
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
293T cells were transfected with indicated expression vectors and the whole-cell extracts were subjected
 SUPPLEMENTARY INFORMATION Supplementary Figure 1. Formation of a complex between Slo1 and CRL4A CRBN E3 ligase. (a) HEK 293T cells were transfected with indicated expression vectors and the whole-cell
SUPPLEMENTARY INFORMATION Supplementary Figure 1. Formation of a complex between Slo1 and CRL4A CRBN E3 ligase. (a) HEK 293T cells were transfected with indicated expression vectors and the whole-cell
SUPPLEMENTARY INFORMATION In format provided by JAATTELA (NOVEMBER 2005)
 Box S1: Methods for studying lysosomal function and integrity Volume and distribution of the acidic compartment. Acridine orange is a metachromatic fluorochrome and a weak base that accumulates in the
Box S1: Methods for studying lysosomal function and integrity Volume and distribution of the acidic compartment. Acridine orange is a metachromatic fluorochrome and a weak base that accumulates in the
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
The clathrin adaptor Numb regulates intestinal cholesterol. absorption through dynamic interaction with NPC1L1
 The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
IP: anti-gfp VPS29-GFP. IP: anti-vps26. IP: anti-gfp - + +
 FAM21 Strump. WASH1 IP: anti- 1 2 3 4 5 6 FAM21 Strump. FKBP IP: anti-gfp VPS29- GFP GFP-FAM21 tail H H/P P H H/P P c FAM21 FKBP Strump. VPS29-GFP IP: anti-gfp 1 2 3 FKBP VPS VPS VPS VPS29 1 = VPS29-GFP
FAM21 Strump. WASH1 IP: anti- 1 2 3 4 5 6 FAM21 Strump. FKBP IP: anti-gfp VPS29- GFP GFP-FAM21 tail H H/P P H H/P P c FAM21 FKBP Strump. VPS29-GFP IP: anti-gfp 1 2 3 FKBP VPS VPS VPS VPS29 1 = VPS29-GFP
SUPPLEMENTARY FIGURE LEGENDS
 SUPPLEMENTARY FIGURE LEGENDS Supplemental FIG. 1. Localization of myosin Vb in cultured neurons varies with maturation stage. A and B, localization of myosin Vb in cultured hippocampal neurons. A, in DIV
SUPPLEMENTARY FIGURE LEGENDS Supplemental FIG. 1. Localization of myosin Vb in cultured neurons varies with maturation stage. A and B, localization of myosin Vb in cultured hippocampal neurons. A, in DIV
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature775 4 O.D. (595-655) 3 1 -ζ no antibody isotype ctrl Plated Soluble 1F6 397 7H11 Supplementary Figure 1 Soluble and plated anti- Abs induce -! signalling. B3Z cells stably expressing!
doi: 1.138/nature775 4 O.D. (595-655) 3 1 -ζ no antibody isotype ctrl Plated Soluble 1F6 397 7H11 Supplementary Figure 1 Soluble and plated anti- Abs induce -! signalling. B3Z cells stably expressing!
T cell maturation. T-cell Maturation. What allows T cell maturation?
 T-cell Maturation What allows T cell maturation? Direct contact with thymic epithelial cells Influence of thymic hormones Growth factors (cytokines, CSF) T cell maturation T cell progenitor DN DP SP 2ry
T-cell Maturation What allows T cell maturation? Direct contact with thymic epithelial cells Influence of thymic hormones Growth factors (cytokines, CSF) T cell maturation T cell progenitor DN DP SP 2ry
Supplementary Figure 1
 Supplementary Figure 1 AAV-GFP injection in the MEC of the mouse brain C57Bl/6 mice at 4 months of age were injected with AAV-GFP into the MEC and sacrificed at 7 days post injection (dpi). (a) Brains
Supplementary Figure 1 AAV-GFP injection in the MEC of the mouse brain C57Bl/6 mice at 4 months of age were injected with AAV-GFP into the MEC and sacrificed at 7 days post injection (dpi). (a) Brains
Principles of Adaptive Immunity
 Principles of Adaptive Immunity Chapter 3 Parham Hans de Haard 17 th of May 2010 Agenda Recognition molecules of adaptive immune system Features adaptive immune system Immunoglobulins and T-cell receptors
Principles of Adaptive Immunity Chapter 3 Parham Hans de Haard 17 th of May 2010 Agenda Recognition molecules of adaptive immune system Features adaptive immune system Immunoglobulins and T-cell receptors
Project report October 2012 March 2013 CRF fellow: Principal Investigator: Project title:
 Project report October 2012 March 2013 CRF fellow: Gennaro Napolitano Principal Investigator: Sergio Daniel Catz Project title: Small molecule regulators of vesicular trafficking to enhance lysosomal exocytosis
Project report October 2012 March 2013 CRF fellow: Gennaro Napolitano Principal Investigator: Sergio Daniel Catz Project title: Small molecule regulators of vesicular trafficking to enhance lysosomal exocytosis
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Intracellular MHC class II molecules promote TLR-triggered innate. immune responses by maintaining Btk activation
 Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining Btk activation Xingguang Liu, Zhenzhen Zhan, Dong Li, Li Xu, Feng Ma, Peng Zhang, Hangping Yao and Xuetao
Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining Btk activation Xingguang Liu, Zhenzhen Zhan, Dong Li, Li Xu, Feng Ma, Peng Zhang, Hangping Yao and Xuetao
McWilliams et al., http :// /cgi /content /full /jcb /DC1
 Supplemental material JCB McWilliams et al., http ://www.jcb.org /cgi /content /full /jcb.201603039 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. In vitro characterization of mito-qc. (A and B) Analysis
Supplemental material JCB McWilliams et al., http ://www.jcb.org /cgi /content /full /jcb.201603039 /DC1 THE JOURNAL OF CELL BIOLOGY Figure S1. In vitro characterization of mito-qc. (A and B) Analysis
T Cell Development. Xuefang Cao, MD, PhD. November 3, 2015
 T Cell Development Xuefang Cao, MD, PhD November 3, 2015 Thymocytes in the cortex of the thymus Early thymocytes development Positive and negative selection Lineage commitment Exit from the thymus and
T Cell Development Xuefang Cao, MD, PhD November 3, 2015 Thymocytes in the cortex of the thymus Early thymocytes development Positive and negative selection Lineage commitment Exit from the thymus and
Myelin suppresses axon regeneration by PIR-B/SHPmediated inhibition of Trk activity
 Manuscript EMBO-2010-76298 Myelin suppresses axon regeneration by PIR-B/SHPmediated inhibition of Trk activity Yuki Fujita, Shota Endo, Toshiyuki Takai and Toshihide Yamashita Corresponding author: Toshihide
Manuscript EMBO-2010-76298 Myelin suppresses axon regeneration by PIR-B/SHPmediated inhibition of Trk activity Yuki Fujita, Shota Endo, Toshiyuki Takai and Toshihide Yamashita Corresponding author: Toshihide
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table
 Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table Title of file for HTML: Peer Review File Description: Innate Scavenger Receptor-A regulates
Title of file for HTML: Supplementary Information Description: Supplementary Figures and Supplementary Table Title of file for HTML: Peer Review File Description: Innate Scavenger Receptor-A regulates
Supplementary Figure 1
 Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
A particular set of insults induces apoptosis (part 1), which, if inhibited, can switch to autophagy. At least in some cellular settings, autophagy se
 A particular set of insults induces apoptosis (part 1), which, if inhibited, can switch to autophagy. At least in some cellular settings, autophagy serves as a defence mechanism that prevents or retards
A particular set of insults induces apoptosis (part 1), which, if inhibited, can switch to autophagy. At least in some cellular settings, autophagy serves as a defence mechanism that prevents or retards
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Chairoungdua et al., http://www.jcb.org/cgi/content/full/jcb.201002049/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Expression of CD9 and CD82 inhibits Wnt/ -catenin
Supplemental material Chairoungdua et al., http://www.jcb.org/cgi/content/full/jcb.201002049/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Expression of CD9 and CD82 inhibits Wnt/ -catenin
THE ROLE OF ALTERED CALCIUM AND mtor SIGNALING IN THE PATHOGENESIS OF CYSTINOSIS
 Research Foundation, 18 month progress report THE ROLE OF ALTERED CALCIUM AND mtor SIGNALING IN THE PATHOGENESIS OF CYSTINOSIS Ekaterina Ivanova, doctoral student Elena Levtchenko, MD, PhD, PI Antonella
Research Foundation, 18 month progress report THE ROLE OF ALTERED CALCIUM AND mtor SIGNALING IN THE PATHOGENESIS OF CYSTINOSIS Ekaterina Ivanova, doctoral student Elena Levtchenko, MD, PhD, PI Antonella
genome edited transient transfection, CMV promoter
 Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
Supplementary Figure 1. In the absence of new protein translation, overexpressed caveolin-1-gfp is degraded faster than caveolin-1-gfp expressed from the endogenous caveolin 1 locus % loss of total caveolin-1-gfp
Central tolerance. Mechanisms of Immune Tolerance. Regulation of the T cell response
 Immunoregulation: A balance between activation and suppression that achieves an efficient immune response without damaging the host. Mechanisms of Immune Tolerance ACTIVATION (immunity) SUPPRESSION (tolerance)
Immunoregulation: A balance between activation and suppression that achieves an efficient immune response without damaging the host. Mechanisms of Immune Tolerance ACTIVATION (immunity) SUPPRESSION (tolerance)
Mechanisms of Immune Tolerance
 Immunoregulation: A balance between activation and suppression that achieves an efficient immune response without damaging the host. ACTIVATION (immunity) SUPPRESSION (tolerance) Autoimmunity Immunodeficiency
Immunoregulation: A balance between activation and suppression that achieves an efficient immune response without damaging the host. ACTIVATION (immunity) SUPPRESSION (tolerance) Autoimmunity Immunodeficiency
Combined Rho-kinase inhibition and immunogenic cell death triggers and propagates immunity against cancer
 Supplementary Information Combined Rho-kinase inhibition and immunogenic cell death triggers and propagates immunity against cancer Gi-Hoon Nam, Eun-Jung Lee, Yoon Kyoung Kim, Yeonsun Hong, Yoonjeong Choi,
Supplementary Information Combined Rho-kinase inhibition and immunogenic cell death triggers and propagates immunity against cancer Gi-Hoon Nam, Eun-Jung Lee, Yoon Kyoung Kim, Yeonsun Hong, Yoonjeong Choi,
CELL BIOLOGY - CLUTCH CH THE IMMUNE SYSTEM.
 !! www.clutchprep.com CONCEPT: OVERVIEW OF HOST DEFENSES The human body contains three lines of against infectious agents (pathogens) 1. Mechanical and chemical boundaries (part of the innate immune system)
!! www.clutchprep.com CONCEPT: OVERVIEW OF HOST DEFENSES The human body contains three lines of against infectious agents (pathogens) 1. Mechanical and chemical boundaries (part of the innate immune system)
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
Immunological Tolerance
 Immunological Tolerance Introduction Definition: Unresponsiveness to an antigen that is induced by exposure to that antigen Tolerogen = tolerogenic antigen = antigen that induces tolerance Important for
Immunological Tolerance Introduction Definition: Unresponsiveness to an antigen that is induced by exposure to that antigen Tolerogen = tolerogenic antigen = antigen that induces tolerance Important for
Part-4. Cell cycle regulatory protein 5 (Cdk5) A novel target of ERK in Carb induced cell death
 Part-4 Cell cycle regulatory protein 5 (Cdk5) A novel target of ERK in Carb induced cell death 95 1. Introduction The process of replicating DNA and dividing cells can be described as a series of coordinated
Part-4 Cell cycle regulatory protein 5 (Cdk5) A novel target of ERK in Carb induced cell death 95 1. Introduction The process of replicating DNA and dividing cells can be described as a series of coordinated
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure S1: Endogenous interaction between RNF2 and H2AX: Whole cell extracts from 293T were subjected to immunoprecipitation with anti-rnf2 or anti-γ-h2ax antibodies
SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure S1: Endogenous interaction between RNF2 and H2AX: Whole cell extracts from 293T were subjected to immunoprecipitation with anti-rnf2 or anti-γ-h2ax antibodies
RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh-
 1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
Supplemental Figure 1
 Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplementary Information
 Supplementary Information Supplementary Figure 1: cholesterol manipulation alters the positioning of autophagosomes in cells, related to figure 1. (a) HeLa cells were treated for 24h under conditions reducing
Supplementary Information Supplementary Figure 1: cholesterol manipulation alters the positioning of autophagosomes in cells, related to figure 1. (a) HeLa cells were treated for 24h under conditions reducing
c Ischemia (30 min) Reperfusion (8 w) Supplementary Figure bp 300 bp Ischemia (30 min) Reperfusion (4 h) Dox 20 mg/kg i.p.
 a Marker Ripk3 +/ 5 bp 3 bp b Ischemia (3 min) Reperfusion (4 h) d 2 mg/kg i.p. 1 w 5 w Sacrifice for IF size A subset for echocardiography and morphological analysis c Ischemia (3 min) Reperfusion (8
a Marker Ripk3 +/ 5 bp 3 bp b Ischemia (3 min) Reperfusion (4 h) d 2 mg/kg i.p. 1 w 5 w Sacrifice for IF size A subset for echocardiography and morphological analysis c Ischemia (3 min) Reperfusion (8
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06994 A phosphatase cascade by which rewarding stimuli control nucleosomal response A. Stipanovich*, E. Valjent*, M. Matamales*, A. Nishi, J.H. Ahn, M. Maroteaux, J. Bertran-Gonzalez,
doi: 10.1038/nature06994 A phosphatase cascade by which rewarding stimuli control nucleosomal response A. Stipanovich*, E. Valjent*, M. Matamales*, A. Nishi, J.H. Ahn, M. Maroteaux, J. Bertran-Gonzalez,
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
GFP-LC3 +/+ CLU -/- kda CLU GFP. Actin. GFP-LC3 +/+ CLU -/- kda CLU GFP. Actin
 Supplementary Fig. 1 a CQ treatment ScrB OGX11 MG132 I II AZD5363 I II b GFP / / GFP / / GFP / / GFP / / GFP GFP Actin Actin ctrl CQ GFP / / GFP / / GFP / / GFP / / GFP GFP Actin Actin rapamycin rapamycincq
Supplementary Fig. 1 a CQ treatment ScrB OGX11 MG132 I II AZD5363 I II b GFP / / GFP / / GFP / / GFP / / GFP GFP Actin Actin ctrl CQ GFP / / GFP / / GFP / / GFP / / GFP GFP Actin Actin rapamycin rapamycincq
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
M.Sc. III Semester Biotechnology End Semester Examination, 2013 Model Answer LBTM: 302 Advanced Immunology
 Code : AS-2246 M.Sc. III Semester Biotechnology End Semester Examination, 2013 Model Answer LBTM: 302 Advanced Immunology A. Select one correct option for each of the following questions:- 2X10=10 1. (b)
Code : AS-2246 M.Sc. III Semester Biotechnology End Semester Examination, 2013 Model Answer LBTM: 302 Advanced Immunology A. Select one correct option for each of the following questions:- 2X10=10 1. (b)
The Immune System: Innate and Adaptive Body Defenses Outline PART 1: INNATE DEFENSES 21.1 Surface barriers act as the first line of defense to keep
 The Immune System: Innate and Adaptive Body Defenses Outline PART 1: INNATE DEFENSES 21.1 Surface barriers act as the first line of defense to keep invaders out of the body (pp. 772 773; Fig. 21.1; Table
The Immune System: Innate and Adaptive Body Defenses Outline PART 1: INNATE DEFENSES 21.1 Surface barriers act as the first line of defense to keep invaders out of the body (pp. 772 773; Fig. 21.1; Table
Supplementary information
 Supplementary information 1 Supplementary Figure 1. CALM regulates autophagy. (a). Quantification of LC3 levels in the experiment described in Figure 1A. Data are mean +/- SD (n > 3 experiments for each
Supplementary information 1 Supplementary Figure 1. CALM regulates autophagy. (a). Quantification of LC3 levels in the experiment described in Figure 1A. Data are mean +/- SD (n > 3 experiments for each
Nature Immunology: doi: /ni.3631
 Supplementary Figure 1 SPT analyses of Zap70 at the T cell plasma membrane. (a) Total internal reflection fluorescent (TIRF) excitation at 64-68 degrees limits single molecule detection to 100-150 nm above
Supplementary Figure 1 SPT analyses of Zap70 at the T cell plasma membrane. (a) Total internal reflection fluorescent (TIRF) excitation at 64-68 degrees limits single molecule detection to 100-150 nm above
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures
 Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
The Adaptive Immune Response: T lymphocytes and Their Functional Types *
 OpenStax-CNX module: m46560 1 The Adaptive Immune Response: T lymphocytes and Their Functional Types * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution
OpenStax-CNX module: m46560 1 The Adaptive Immune Response: T lymphocytes and Their Functional Types * OpenStax This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution
Production of Exosomes in a Hollow Fiber Bioreactor
 Production of Exosomes in a Hollow Fiber Bioreactor John J S Cadwell, President and CEO, FiberCell Systems Inc INTRODUCTION Exosomes are small lipid membrane vesicles (80-120 nm) of endocytic origin generated
Production of Exosomes in a Hollow Fiber Bioreactor John J S Cadwell, President and CEO, FiberCell Systems Inc INTRODUCTION Exosomes are small lipid membrane vesicles (80-120 nm) of endocytic origin generated
Phagocytosis: An Evolutionarily Conserved Mechanism to Remove Apoptotic Bodies and Microbial Pathogens
 Phagocytosis of IgG-coated Targets by s Phagocytosis: An Evolutionarily Conserved Mechanism to Remove Apoptotic Bodies and Microbial s 3 min 10 min Mast Cells Can Phagocytose Too! Extension of an F-actin-rich
Phagocytosis of IgG-coated Targets by s Phagocytosis: An Evolutionarily Conserved Mechanism to Remove Apoptotic Bodies and Microbial s 3 min 10 min Mast Cells Can Phagocytose Too! Extension of an F-actin-rich
Trehalose, sucrose and raffinose are novel activators of autophagy in human. keratinocytes through an mtor-independent pathway
 Title page Trehalose, sucrose and raffinose are novel activators of autophagy in human keratinocytes through an mtor-independent pathway Xu Chen 1*, Min Li 1*, Li Li 1, Song Xu 1, Dan Huang 1, Mei Ju 1,
Title page Trehalose, sucrose and raffinose are novel activators of autophagy in human keratinocytes through an mtor-independent pathway Xu Chen 1*, Min Li 1*, Li Li 1, Song Xu 1, Dan Huang 1, Mei Ju 1,
Hunk is required for HER2/neu-induced mammary tumorigenesis
 Research article Hunk is required for HER2/neu-induced mammary tumorigenesis Elizabeth S. Yeh, 1 Thomas W. Yang, 1 Jason J. Jung, 1 Heather P. Gardner, 1 Robert D. Cardiff, 2 and Lewis A. Chodosh 1 1 Department
Research article Hunk is required for HER2/neu-induced mammary tumorigenesis Elizabeth S. Yeh, 1 Thomas W. Yang, 1 Jason J. Jung, 1 Heather P. Gardner, 1 Robert D. Cardiff, 2 and Lewis A. Chodosh 1 1 Department
Cell Quality Control. Peter Takizawa Department of Cell Biology
 Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Cell Quality Control Peter Takizawa Department of Cell Biology Cellular quality control reduces production of defective proteins. Cells have many quality control systems to ensure that cell does not build
Supplementary Figure 1
 Supplementary Figure 1 Expression of apoptosis-related genes in tumor T reg cells. (a) Identification of FOXP3 T reg cells by FACS. CD45 + cells were gated as enriched lymphoid cell populations with low-granularity.
Supplementary Figure 1 Expression of apoptosis-related genes in tumor T reg cells. (a) Identification of FOXP3 T reg cells by FACS. CD45 + cells were gated as enriched lymphoid cell populations with low-granularity.
Leucine Deprivation Reveals a Targetable Liability
 Cancer Cell, 19 Supplemental Information Defective Regulation of Autophagy upon Leucine Deprivation Reveals a Targetable Liability of Human Melanoma Cells In Vitro and In Vivo Joon-Ho Sheen, Roberto Zoncu,
Cancer Cell, 19 Supplemental Information Defective Regulation of Autophagy upon Leucine Deprivation Reveals a Targetable Liability of Human Melanoma Cells In Vitro and In Vivo Joon-Ho Sheen, Roberto Zoncu,
7.06 Cell Biology EXAM #3 April 24, 2003
 7.06 Spring 2003 Exam 3 Name 1 of 8 7.06 Cell Biology EXAM #3 April 24, 2003 This is an open book exam, and you are allowed access to books and notes. Please write your answers to the questions in the
7.06 Spring 2003 Exam 3 Name 1 of 8 7.06 Cell Biology EXAM #3 April 24, 2003 This is an open book exam, and you are allowed access to books and notes. Please write your answers to the questions in the
Supplementary Table; Supplementary Figures and legends S1-S21; Supplementary Materials and Methods
 Silva et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability Supplementary Table; Supplementary Figures and legends S1-S21; Supplementary
Silva et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability Supplementary Table; Supplementary Figures and legends S1-S21; Supplementary
TITLE: Novel Role of Merlin Tumor Suppressor in Autophagy and Its Implication in Treating NF2-Associated Tumors
 AWARD NUMBER: W81XWH 11 1 0269 TITLE: Novel Role of Merlin Tumor Suppressor in Autophagy and Its Implication in Treating NF2-Associated Tumors PRINCIPAL INVESTIGATOR: Toshifumi Tomoda, M.D., Ph.D CONTRACTING
AWARD NUMBER: W81XWH 11 1 0269 TITLE: Novel Role of Merlin Tumor Suppressor in Autophagy and Its Implication in Treating NF2-Associated Tumors PRINCIPAL INVESTIGATOR: Toshifumi Tomoda, M.D., Ph.D CONTRACTING
Antigen Presentation to T lymphocytes
 Antigen Presentation to T lymphocytes Immunology 441 Lectures 6 & 7 Chapter 6 October 10 & 12, 2016 Jessica Hamerman jhamerman@benaroyaresearch.org Office hours by arrangement Antibodies and T cell receptors
Antigen Presentation to T lymphocytes Immunology 441 Lectures 6 & 7 Chapter 6 October 10 & 12, 2016 Jessica Hamerman jhamerman@benaroyaresearch.org Office hours by arrangement Antibodies and T cell receptors
SUPPLEMENTARY INFORMATION. Supplementary Figures S1-S9. Supplementary Methods
 SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This
 SUPPLEMENTAL FIGURE LEGEND Fig. S1. Generation and characterization of. (A) Coomassie staining of soluble hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This protein was expressed
SUPPLEMENTAL FIGURE LEGEND Fig. S1. Generation and characterization of. (A) Coomassie staining of soluble hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This protein was expressed
LPS LPS P6 - + Supplementary Fig. 1.
 P6 LPS - - - + + + - LPS + + - - P6 + Supplementary Fig. 1. Pharmacological inhibition of the JAK/STAT blocks LPS-induced HMGB1 nuclear translocation. RAW 267.4 cells were stimulated with LPS in the absence
P6 LPS - - - + + + - LPS + + - - P6 + Supplementary Fig. 1. Pharmacological inhibition of the JAK/STAT blocks LPS-induced HMGB1 nuclear translocation. RAW 267.4 cells were stimulated with LPS in the absence
supplementary information
 DOI: 10.1038/ncb1875 Figure S1 (a) The 79 surgical specimens from NSCLC patients were analysed by immunohistochemistry with an anti-p53 antibody and control serum (data not shown). The normal bronchi served
DOI: 10.1038/ncb1875 Figure S1 (a) The 79 surgical specimens from NSCLC patients were analysed by immunohistochemistry with an anti-p53 antibody and control serum (data not shown). The normal bronchi served
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC
 Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
Supplementary Figure 1. Deletion of Smad3 prevents B16F10 melanoma invasion and metastasis in a mouse s.c. tumor model.
 A B16F1 s.c. Lung LN Distant lymph nodes Colon B B16F1 s.c. Supplementary Figure 1. Deletion of Smad3 prevents B16F1 melanoma invasion and metastasis in a mouse s.c. tumor model. Highly invasive growth
A B16F1 s.c. Lung LN Distant lymph nodes Colon B B16F1 s.c. Supplementary Figure 1. Deletion of Smad3 prevents B16F1 melanoma invasion and metastasis in a mouse s.c. tumor model. Highly invasive growth
Immune Regulation and Tolerance
 Immune Regulation and Tolerance Immunoregulation: A balance between activation and suppression of effector cells to achieve an efficient immune response without damaging the host. Activation (immunity)
Immune Regulation and Tolerance Immunoregulation: A balance between activation and suppression of effector cells to achieve an efficient immune response without damaging the host. Activation (immunity)
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
General information. Cell mediated immunity. 455 LSA, Tuesday 11 to noon. Anytime after class.
 General information Cell mediated immunity 455 LSA, Tuesday 11 to noon Anytime after class T-cell precursors Thymus Naive T-cells (CD8 or CD4) email: lcoscoy@berkeley.edu edu Use MCB150 as subject line
General information Cell mediated immunity 455 LSA, Tuesday 11 to noon Anytime after class T-cell precursors Thymus Naive T-cells (CD8 or CD4) email: lcoscoy@berkeley.edu edu Use MCB150 as subject line
TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer
 Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Cigarette Smoke Exposure and HIV-Related Neurologic Disease Progression Basic Mechanisms and Clinical Consequences
 Cigarette Smoke Exposure and HIV-Related Neurologic Disease Progression Basic Mechanisms and Clinical Consequences Walter Royal, III, MD Professor of Neurology University of Maryland School of Medicine
Cigarette Smoke Exposure and HIV-Related Neurologic Disease Progression Basic Mechanisms and Clinical Consequences Walter Royal, III, MD Professor of Neurology University of Maryland School of Medicine
Appendix Table of Contents. 1. Appendix Figure legends S1-S13 and Appendix Table S1 and S2. 2. Appendix Figures S1-S13
 Appendix Table of Contents. Appendix Figure legends S-S3 and Appendix Table S and S. Appendix Figures S-S3 . Appendix Figure legends S-S3 and Appendix Table S and S Appendix Figure S. Western blot analysis
Appendix Table of Contents. Appendix Figure legends S-S3 and Appendix Table S and S. Appendix Figures S-S3 . Appendix Figure legends S-S3 and Appendix Table S and S Appendix Figure S. Western blot analysis
Supplementary Figure S1 Supplementary Figure S2
 Supplementary Figure S A) The blots shown in Figure B were qualified by using Gel-Pro analyzer software (Rockville, MD, USA). The ratio of LC3II/LC3I to actin was then calculated. The data are represented
Supplementary Figure S A) The blots shown in Figure B were qualified by using Gel-Pro analyzer software (Rockville, MD, USA). The ratio of LC3II/LC3I to actin was then calculated. The data are represented
