Genomic complexity and AKT-dependence in serous ovarian cancer
|
|
|
- Madlyn Stanley
- 5 years ago
- Views:
Transcription
1 Genomic complexity and KT-dependence in serous ovarian cancer Hanrahan et al.- Supplementary Text and Figures Corresponding uthor: David B. Solit, Memorial Sloan-Kettering Cancer Center, Human Oncology and Pathogenesis Program, 275 York venue, Box 2, NY, NY 65. Phone: ; Fax: ; SUPPLEMENTRY METHODS Haplotype nalysis of Ovarian Cancer Cell Lines To exclude the potential for prior contamination or misidentification, all cell lines were subject to a MS-based genetic fingerprinting assay designed to screen 42 highly variable single nucleotide polymorphisms, covering all chromosomes as previously described (). The panel of 22 ovarian cancer cells lines were screened against each other and 247 other cell lines and tumors samples of varied lineages to ensure distinct identities. nimal Studies 6-8 week old nu/nu athymic female mice (Harlan Labs) were maintained in ventilated cages. Experiments were performed in accordance with approved ICUC protocols and animals were treated humanely in compliance with institutional guidelines. Tumors were generated by implanting x 7 cancer cells in a : mixture with media and groh factor-reduced Matrigel (BD Biosciences) into the right flank (2uL/mouse). Mice with well-established tumors were selected and randomized approximately 7-34 days postimplantation (size 5-2 mm 3 ), prior to treatment. The first treatment was then given (indicated as day on graphs) and tumor size was subsequently monitored and recorded every 3-4 days as indicated on the graph. KTi/2 was prepared in 25% hydroxypropyl
2 β-cyclodextrin ph 4-5, and administered subcutaneously at mg/kg once per day, M-F as previously determined (2). MK226 was formulated in 3% captisol and given orally at either 24mg/kg or 5mg/kg once per day, M-W-F based on unpublished dose response experiments and previous reports (3). PD3259 was formulated in.5% hydroxypropyl methylcellulose +.2% Tween 8 and administered by oral gavage at 2.5mg/kg, daily, M-F (). Control mice received a vehicle solution based on the formulation of KTi/2 or MK226, depending on which drug was used in the experiment. Tumors were measured by caliper and volume calculated such that mm 3 = π/6 x large diameter x (smaller diameter) 2. Significant p values <.5 were determined by unpaired, two-tailed student t-tests. n= mice per group for IGROV xenograft experiments +/- KTi/2 and MK226 (Supp. Fig. S2). n= 5 mice per group for OVCR5 xenograft combination experiment (Supp. Fig. S4). This OVCR5 xenograft combination study was repeated with analogous results. Tumors were excised and snap frozen in liquid nitrogen, homogenized in 2% SDS lysis buffer and processed for immunoblotting or fixed in 4%PF and processed for immunohistochemical studies. Lentiviral short hairpin infections OVCR-5 cells were infected with lentiviral, short hairpin constructs targeted to KT3 acquired from Dr. Hakim Djaballah and the High Throughput Sequencing Core Facility at MSKCC (shkt3: CCGGGCTGCTCTGTCTTCTTTCTCGGTTGTGCGTGCGCTTTTT; shkt3-6: CCGGGTCTGGCGTGTTTCTCGGTTCTCTTGCCGTTTCTTTTT) or a scrambled sequence (shrn control vector from Sigma-ldrich, SCH2: CCGGCCGTGGGCCCCTCGGTTGGTGCTCTTCTCTTGTTGTTTTT). Stable lines were
3 selected in puromycin for 2 weeks and subsequently tested for KT3 knockdown. OVCR-5 cell lines stably expressing shkt3 constructs or 6 or the scrambled control (OVCR5+ shkt3, +shkt3-6, +shscrmbled) were used for further analysis. SUPPLEMENTRY REFERENCES. Janakiraman M, Vakiani E, Zeng Z, Pratilas C, Taylor BS, Chitale D, et al. Genomic and biological characterization of exon 4 KRS mutations in human cancer. Cancer Res 2;7: She QB, Chandarlapaty S, Ye Q, Lobo J, Haskell KM, Leander KR, et al. Breast tumor cells with PI3K mutation or HER2 amplification are selectively addicted to kt signaling. PLoS One 28;3:e Chandarlapaty S, Sawai, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V, et al. KT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell 2;9: Wolf CR, Hayward IP, Lawrie SS, Buckton K, McIntyre M, dams DJ, et al. Cellular heterogeneity and drug resistance in two ovarian adenocarcinoma cell lines derived from a single patient. Int J Cancer 987;39: Langdon SP, Lawrie SS, Hay FG, Hawkes MM, McDonald, Hayward IP, et al. Characterization and properties of nine human ovarian adenocarcinoma cell lines. Cancer Res 988;48:
4 Supplementary Table S Hanrahan et al.-genomic complexity and KT-dependence in serous ovarian cancer Supplementary Table S. Identity scores for haplotype mapping of ovarian cancer cell lines. 22 ovarian cancer cell lines were subjected to haplotype analysis to distinguish unique cell lines. ll redundant cell lines had a Bonferroni correction of less than 9E signifying that the cell lines are genetically identical with an insignificant probability that they matched by chance (see Supplementary Methods). One pair of cells lines, PEO and PEO-4, were derived from the ascites of a single patient s poorly differentiated ovarian adenocarcinoma before and after acquisition of chemotherapy resistance, respectively (4, 5). This pair serves as an internal control for the analysis. Five additional cell lines mapped as genetically overlapping suggesting either prior cross contamination or mislabeling and were thus excluded from further analysis. The remaining 7 unique cell lines used for further analysis are listed, with genotypes, in Supplementary Table S2.
5 Supplementary Table S2 Hanrahan et al.-genomic complexity and KT-dependence in serous ovarian cancer Cell Line PIK3C KT-3 PTEN ERBB H,N,KRS BRF MEK RB BG E545K SKOV-3 H47R amp OVCR-3 KT2:amp 278 K28- R3del IGROV KT2: V98 Y55C SKOV-6 KT2: V98 R3 R233 loss SKOV-8 KRS: amp OVCR-5 KRS: G2V ES V6E D67N OV-9 N486- P49del SKOV-433 W274G del COV-3 W274G loss OV847 KRS: P2H loss PEO-4 loss SK774 PEO TOV2d Supplementary Table S2. Mutational and copy-number analysis of 7 ovarian cancer cell lines. Mutational and copy-number analyses were performed on 7 unique ovarian cancer cell lines (see methods). Four genetically distinct groups were identified: cells with ) PI3k/KT pathway alterations (red), 2) RS/RF pathway aberrations (blue), 3) RB loss (green), 4) those wild-type for all the preceding genetic changes (black).
6 Supplementary Figure S Hanrahan et al.-genomic complexity and KT-dependence in ovarian cancer 3 ERBB2 amplification in SKOV-3 B KT2 amplification in OVCR-3 Copy number (log2) 2 Copy number (log2) 3 2 ERBB2 chr7: Chromosome 7 position (mb) KT2 chr9: Chromosome 9 position (mb) Supplementary Fig. S. Representative probe-level copy-number data for the ERBB2 and KT2 genes in two ovarian cancer cell lines. -B, Probe-level data (blue dots) and segmented data (red lines) from gilent 244K arrays identifying representative copy-number gains of ERBB2 and KT2 in SKOV-3 and OVCR-3 cells, respectively. The vertical green line crosses the x-axis at the chromosomal position of the gene. Y-axis indicates log2 copy-number expression.
7 Supplementary Figure S2 Hanrahan et al.-genomic complexity and KT-dependence in serous ovarian cancer B IGROV (PTEN Y55C) xenograft IGROV xenograft IGROV xenograft V KTi/2 V MK p-kt S473 p-kt T38 KT Vehicle h KTi/2 6h C IGROV (PTEN Y55C) xenograft Vehicle KTi/2 mg/kg 5days/wk 5 H&E IGROV (PTEN Y55C) xenograft Vehicle MK226 24mg/kg MWF p-kt S473 Tumor Volume (mm 3 ) st dose p<. Tumor Volume (mm 3 ) st dose p< Days fter st Treatment Days fter st Treatment Supplementary Fig. S2. KT inhibition suppresses the groh of PTEN-mutant ovarian xenografts., Mice bearing established IGROV (PTEN-mutant) xenografts were treated with KTi/2 (mg/kg daily M-F), MK226 (MK; 24mg/kg 3days/week, M-W- F) or vehicle control (V), then euthanized 6h after the last administration of drug. The expression of phosphorylated KT (S473 and T38) was measured by immunoblot. B, Mice bearing IGROV xenografts were treated with one dose of vehicle (hr) or KTi- /2 (6hr; mg/kg), euthanized, and tumors excised and embedded in paraffin. H&E and immunohistochemical staining for p-kt (Ser473) are shown for representative tumors from vehicle and KTi/2 treated mice. C, Mice with established IGROV xenografts were treated as in (). Data points represent mean tumor volume +/- S.E (asterisks indicates significant tumor suppression vs. vehicle at time of sacrifice).
8 Supplementary Figure S3 Hanrahan et al.- Genomic complexity and KT-dependence in serous ovarian cancer OVCR-5 control no DN sintp sikt sikt2 sikt3 sikt+2 sikt+2+3 KT KT2 KT3 B IGROV % cells in sub-g phase 64h control no DN sintp sikt sikt2 sikt3 sikt+2 sikt+2+3 % cells in G phase 64h control no DN sintp sikt sikt2 sikt3 sikt+2 sikt+2+3 Supplementary Fig. S3. Efficiency of KT3 knockdown in OVCR5 cells and consequence of KT isoform knockdown in IGROV cells., Control sirn (sintp) or sirn against individual KT isoforms, alone or in combination, was transfected into OVCR-5 cells and incubated for 64h as in IGROV cells in Fig. 3D. Cell lysates were subject to immunoblotting with isoform specific KT antibodies. sirn transfection resulted in isoformspecific and efficient (>9%) knockdown of KT isoforms, including KT3. Therefore, although we view sikt3 transfection in IGROV cells in Fig. 3D as a control, we postulate that sikt3 may be functioning to abrogate any residual KT3 protein that may be below our threshold of detection. B, In IGROV cells, sirn was transfected as in Fig. 3D for 64h. Isoform-selective knockdown of KT, or combinations that included sikt, resulted in a cytostatic response as depicted by significant accumulation of cells in G phase (asterisks indicates p <., n > 3; same experiments as in Fig. 3D). No accumulation of cells in sub- G was observed following any single- or combination- of KT isoform knockdown.
9 Supplementary Figure S4 Hanrahan et al.-genomic complexity and KT-dependence in serous ovarian cancer B OVCR-5 (KRS G2V) OVCR-5 (KRS G2V) % cells in sub-g at 72hr % cells in sub-g at 64hr DMSO PD9 5nM MK226 µm ZVD 2µM QVD 2µM PD + MK PD + MK + ZVD PD + MK + QVD Cl. caspase-3 DMSO nodn MK226 µm sintp sikrs sintp + MK sikrs + MK KRS p-erk Cl. PRP ERK p-kt S473 p-kt S473 p-erk KT ctinin p-s6 S24/244 C OVCR-5 (KRS G2V) xenograft Vehicle MK226 5mg/kg MWF PD9 2.5mg/kg M-F MK226 + PD9 7 6 V MK MK + PD PD 6hr p-kt S473 Tumor Volume (mm 3 ) st dose p<.4 p<.43 p-kt T38 KT p-erk ERK Days fter st Treatment Supplementary Fig. S4. Combined inhibition of KT and MEK in KRS-mutant, KT3 expressing OVCR-5 cells and xenografts., Induction of apoptosis in OVCR-5 cells at 72hr following combined treatment with 5nM PD9 and μm MK226 was inhibited by co-treatment with 2μM of the pan-caspase inhibitor ZVD-FMK or QVD-OPH as measured by FCS analysis. Treatment with ZVD/QVD also inhibited PD+MK treatment-induced cleaved PRP and cleaved caspase-3 at 72hr. sterisks indicate p<.28 vs. PD+MK treatment, n=3. B, Control sirn (sintp) or sirn against KRS were transfected into OVCR-5 cells, co-treated with μm MK226 and incubated for 64hr. Cells were collected for FCS analysis or subject to immunoblotting with the indicated antibodies. sterisk indicates significant increase [p=.3] vs. sikrs alone, n=3 each in duplicates). C, Mice bearing established OVCR-5 (KRS G2V) xenografts were treated with MK226 (MK; 5mg/kg, 3day/week, M-W-F), PD9 (2.5mg/kg, daily, M-F), the combination, or vehicle control (V). Data points represent mean tumor volume +/- S.E (asterisks indicate significant tumor suppression vs. vehicle at time of sacrifice). C right panel, Mice from (C) were treated as indicated, then euthanized 6h after the last administration of drug. The expression of phosphorylated KT (S473 and T38) and ERK and total KT/ERK were measured by immunoblot.
10 Supplementary Figure S5 Hanrahan et al.- Genomic complexity and KT-dependence in serous ovarian cancer % cells in sub-g at 72hr OVCR-5 parental OVCR-5 +shscrmbled OVCR-5 +shkt3 OVCR-5 +shkt3-6 DMSO PD9 5nM KTi/2 µm KTi/2 + PD9 Supplementary Fig. S5. Consequences of stable KT3 knockdown in RasG2V mutant OVCR-5 cells. Lentiviral short hairpin constructs targeting KT3 (shkt3) or a scrambled sequence (shscrmbled) were used to generate OVCR-5 stable cells lines with KT3 knockdown. Induction of apoptosis was only observed in OVCR-5 expressing the shkt3 construct, when they were co-treated with KTi/2 (um) and PD9 (5nM) (asterisks indicates significant increase in subg compared to OVCR-5+shSCRMBLED line, p=.442, n=6). Similar results were seen with a second shkt3 construct, shkt3-6. These data recapitulate the increase in cell death observed following combined treatment of OVCR-5 parental cells with MK226 and PD9 (Fig. 4C) and suggest that KT3 plays a role in survival in ovarian cancer cells that express high levels of KT3.
11 Supplementary Figure S6 Hanrahan et al.-genomic complexity and KT-dependence in serous ovarian cancer 2. BRF missense mutation nonsense mutation 2. KT2.5.5 log 2 mrn expression Het loss Diploid Gain Copy-number status p-value =.56e mp Het loss Diploid Gain Copy-number status p-value = 2.693e-42 mp B 7.5mb on chr 7q 6mb on chr 9q 36 ovarian TCG samples BRF focally amplified KT2 focally amplified Supplementary Fig. S6. Copy-number and mrn expression for BRF and KT2 in 36 ovarian tumors from the TCG dataset., log2 mrn expression was assessed as a function of DN copy-number calls (homozygous deletion, hemizygous loss, diploid, gain, high-level amplification) across all samples for the BRF and KT2 genes. P-values were generated using NOV analysis. B, Copy-number heatmaps demonstrating the focality of amplifications for BRF and KT2 loci for all samples in ovarian TCG dataset.
12 Supplementary Figure S7 Hanrahan et al.-genomic complexity and KT-dependence in serous ovarian cancer RB NF log 2 mrn expression missense mutation nonsense mutation frameshift missense mutation nonsense mutation frameshift Homo del Hemi loss Diploid Gain Copy-number status p-value =.969e mp Homo del Hemi loss Diploid Gain Copy-number status p-value = 6.367e mp B.3mb on chr 3q 36 ovarian TCG samples 3mb on chr 7q RB focally deleted NF focally deleted Supplementary Fig. S7. Copy-number and mrn expression for RB and NF in 36 ovarian tumors from the TCG dataset., log2 mrn expression was assessed as a function of DN copy-number calls (homozygous deletion, hemizygous loss, diploid, gain, high-level amplification) across all samples for the RB and NF genes. P-values were generated using NOV analysis. B, Copy-number heatmaps demonstrating the focality of deletions for RB and NF loci for all samples in ovarian TCG dataset.
13 Supplementary Table S3 Hanrahan et al.-genomic complexity and KT-dependence in serous ovarian cancer TCG log2 PTEN PTEN GISTIC score RE score PTEN Other tumor mrn IHC score for PTEN for PTEN genotype genotype identification # expression TCG3-95 TCG3-72 TCG3-8 TCG359 TCG3-758 TCG3-725 TCG3492 TCG3-9 N N N KT amp, RB del PTEN hemizygous KT amp, RB mt PIK3C amp PTEN hemizygous/r233fs22 PTEN hemizygous TCG3497 TCG3-924 TCG3488 TCG3-893 TCG3-89 TCG348 TCG3-724 TCG3-96 N PTEN hemizygous PTEN hemizygous PTEN hemizygous PTEN hemizygous PIK3C amp RB del PIK3C amp RB del BRF amp BRF amp PIK3C amp PIK3C amp TCG345 TCG3-87 TCG3-899 TCG3-75 TCG3-795 TCG3-93 TCG3-923 TCG3-889 TCG3-92 TCG3-885 TCG3-9 TCG347 TCG3-76 TCG3-755 TCG3-77 TCG3-726 TCG3485 TCG355 TCG3494 TCG34 TCG344 TCG3-793 TCG3495 TCG3-92 TCG35 TCG3-883 TCG3-85 TCG3-799 TCG3-727 TCG3-99 TCG3-89 TCG3-894 TCG3-73 TCG3477 TCG349 TCG3-897 N PTEN hemizygous PTEN hemizygous PTEN hemizygous PTEN hemizygous PIK3C amp PTEN hemizygous PTEN hemizygous RB del PTEN hemizygous PIK3C amp PTEN hemizygous KRS amp PTEN hemizygous PTEN hemizygous PTEN hemizygous PIK3C amp KRS amp, KT2 amp RB del KRS amp KRS amp KRS amp KRS amp RB del PTEN gain PTEN gain PTEN gain PTEN gain PTEN gain KRS amp, KT2 amp PTEN gain PTEN gain PTEN gain Supplementary Table S3. PTEN IHC scores, copy-number calls, mrn expression and PTEN/other genotypes for 52 ovarian tumors. Immunohistochemical (IHC) staining for PTEN was performed on 52 ovarian tumors. PTEN IHC was scored as,, or staining. GISTIC and RE scores for PTEN, log2 PTEN mrn expression and PTEN and other genotypes (see Supp. Fig. S8 for a more detailed genomic fingerprint of the status of other genes of interest) were determined as part of TCG project (see Methods). PTEN genotype calls are based on GISTIC scores as categorized in Fig. 6B. Copy-number analysis clearly indicated deletion of PTEN, however this sample was unable to be incorporated into GISTIC, RE or mrn expression analyses.
14 Supplementary Figure S8 Hanrahan et al.-genomic complexity and KT-dependence in serous ovarian cancert 5 of 36 cases with corresponding IHC data Somatic Mutation mplification Homozygous Deletion Up-Regulation Epigenetic Silencing PIK3C KT KT2 KT3 PTEN ERBB KRS HRS NRS NF BRF MP2K RB BRC BRC2 TP53 Supplementary Fig. S8. Genomic fingerprint for 5 ovarian tumors from TCG dataset. Heatmap depicts compiled mutational, copynumber and mrn expression analysis for 5 ovarian tumors subjected to further immunohistochemical analysis of PTEN (see Supplementary Table S3).
15 Supplementary Figure S9 Hanrahan et al.-genomic complexity and KT-dependence in serous ovarian cancer B log2 PTEN PRR score.. p-kt S473 IHC score log2 p-kt S473 RPP score. Homo del Hemi loss Diploid/ Gain PTEN genotype calls by GISTIC. Homo del Hemi loss Diploid/ Gain PTEN genotype calls by GISTIC. Supplementary Fig. S9. PTEN and p-kt S473 RPP expression data from a subset of ovarian TCG samples., log2 PTEN reverse-phase protein array (RPP) scores from 29 of the 52 TCG samples used in the IHC analysis plotted as a function of GISTIC-based copy-number calls at the PTEN locus. Similar to the PTEN IHC data (Fig. 6C, Supp. Table 3), PTEN expression by RPP correlates with PTEN homozygous deletion, while significantly higher levels of PTEN protein were found in the PTEN hemizygous loss and PTEN diploid/gain cohorts. Symbols represent average PTEN RPP score +/- S.E for each group. Green asterisk indicates p=.2 between PTEN hemi loss and diploid gain groups. Black asterisk indicates p=.4 vs. PTEN homozygous deleted group. B, log2 p-kt S473 reverse-phase protein array (RPP) scores from 29 of the 52 TCG samples used in the IHC analysis plotted as a function of GISTIC-based copy-number calls at the PTEN locus. Similar to the p-kt S473 IHC data (in black, left y-axis, as seen in Fig. 6D), high p-kt S473 expression by RPP (in red, right y- axis) correlates with PTEN homozygous deletion, while significantly lower levels of KT phosphorylation were found in the PTEN hemizygous loss and PTEN diploid/gain cohorts. Symbols represent average p-kt IHC or RPP score +/- S.E for each group. Black asterisks indicate p=.477 (hemi loss) and p=.5 (diploid/gain) as compared to PTEN homozygous deleted group. Red asterisks indicate p=.267 (hemi loss) and p=.34 (diploid/gain) as compared to PTEN homozygous deleted group.
Supplementary Figure 1
 Supplementary Figure 1 6 HE-50 HE-116 E-1 HE-108 Supplementary Figure 1. Targeted drug response curves of endometrial cancer cells. Endometrial cancer cell lines were incubated with serial dilutions of
Supplementary Figure 1 6 HE-50 HE-116 E-1 HE-108 Supplementary Figure 1. Targeted drug response curves of endometrial cancer cells. Endometrial cancer cell lines were incubated with serial dilutions of
Supplementary Figure 1. Basal level EGFR across a panel of ESCC lines. Immunoblots demonstrate the expression of phosphorylated and total EGFR as
 Supplementary Figure 1. Basal level EGFR across a panel of ESCC lines. Immunoblots demonstrate the expression of phosphorylated and total EGFR as well as their downstream effectors across a panel of ESCC
Supplementary Figure 1. Basal level EGFR across a panel of ESCC lines. Immunoblots demonstrate the expression of phosphorylated and total EGFR as well as their downstream effectors across a panel of ESCC
Expanded View Figures
 Shao-Ming Shen et al Role of I in MT of cancers MO reports xpanded View igures igure V1. nalysis of the expression of I isoforms in cancer cells and their interaction with PTN. RT PR detection of Ish and
Shao-Ming Shen et al Role of I in MT of cancers MO reports xpanded View igures igure V1. nalysis of the expression of I isoforms in cancer cells and their interaction with PTN. RT PR detection of Ish and
Expanded View Figures
 Molecular Systems iology Tumor CNs reflect metabolic selection Nicholas Graham et al Expanded View Figures Human primary tumors CN CN characterization by unsupervised PC Human Signature Human Signature
Molecular Systems iology Tumor CNs reflect metabolic selection Nicholas Graham et al Expanded View Figures Human primary tumors CN CN characterization by unsupervised PC Human Signature Human Signature
Cyclin E amplification, a novel mechanism of resistance to trastuzumab in HER2 amplified breast cancer Sunday, Apr 18, 2010, 2:25 PM - 2:40 PM
 Abstract Print View Page 1 of 1 Presentation Abstract Abstract Number: Presentation Title: Presentation Time: Location: Author Block: 22 Cyclin E amplification, a novel mechanism of resistance to trastuzumab
Abstract Print View Page 1 of 1 Presentation Abstract Abstract Number: Presentation Title: Presentation Time: Location: Author Block: 22 Cyclin E amplification, a novel mechanism of resistance to trastuzumab
Supplementary Information Titles Journal: Nature Medicine
 Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC
 Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH
 EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
Expanded View Figures
 Sarah Kit Leng Lui et al USP26 stabilizes SM7 MO reports xpanded View igures igure V1. USP26 enhances SM2 phosphorylation and T-b-mediated transcription. raph representing relative luciferase values obtained
Sarah Kit Leng Lui et al USP26 stabilizes SM7 MO reports xpanded View igures igure V1. USP26 enhances SM2 phosphorylation and T-b-mediated transcription. raph representing relative luciferase values obtained
(a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable
 Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figure 1: Digitoxin induces apoptosis in primary human melanoma cells but not in normal melanocytes, which express lower levels of the
 Supplementary Figure 1: Digitoxin induces apoptosis in primary human melanoma cells but not in normal melanocytes, which express lower levels of the cardiac glycoside target, ATP1A1. (a) The percentage
Supplementary Figure 1: Digitoxin induces apoptosis in primary human melanoma cells but not in normal melanocytes, which express lower levels of the cardiac glycoside target, ATP1A1. (a) The percentage
Supplementary Figure 1
 Supplementary Figure 1 a γ-h2ax MDC1 RNF8 FK2 BRCA1 U2OS Cells sgrna-1 ** 60 sgrna 40 20 0 % positive Cells (>5 foci per cell) b ** 80 sgrna sgrna γ-h2ax MDC1 γ-h2ax RNF8 FK2 MDC1 BRCA1 RNF8 FK2 BRCA1
Supplementary Figure 1 a γ-h2ax MDC1 RNF8 FK2 BRCA1 U2OS Cells sgrna-1 ** 60 sgrna 40 20 0 % positive Cells (>5 foci per cell) b ** 80 sgrna sgrna γ-h2ax MDC1 γ-h2ax RNF8 FK2 MDC1 BRCA1 RNF8 FK2 BRCA1
B-cell expansion and lymphomagenesis induced by chronic CD40-signaling is
 -cell expansion and lymphomagenesis induced by chronic CD4-signaling is strictly dependent on CD19 Caroline Hojer 1*, Samantha Frankenberger 1*, Lothar J. Strobl 1, Samantha Feicht 1, Kristina Djermanovic
-cell expansion and lymphomagenesis induced by chronic CD4-signaling is strictly dependent on CD19 Caroline Hojer 1*, Samantha Frankenberger 1*, Lothar J. Strobl 1, Samantha Feicht 1, Kristina Djermanovic
Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma
 Supplementary information for: Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma Rintaro Hashizume 1, Noemi Andor 2, Yuichiro Ihara 2, Robin Lerner 2, Haiyun
Supplementary information for: Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma Rintaro Hashizume 1, Noemi Andor 2, Yuichiro Ihara 2, Robin Lerner 2, Haiyun
(A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and a
 Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
Appendix Figure S1 A B C D E F G H
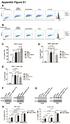 ppendix Figure S1 C D E F G H ppendix Figure S1. RT and chemotherapy alter PD-L1 expression in PDC cells. Flow cytometric analysis of PD-L1 expression in () KPC and () Pan02 cells following treatment with
ppendix Figure S1 C D E F G H ppendix Figure S1. RT and chemotherapy alter PD-L1 expression in PDC cells. Flow cytometric analysis of PD-L1 expression in () KPC and () Pan02 cells following treatment with
Supplementary Figure 1
 A B D Relative TAp73 mrna p73 Supplementary Figure 1 25 2 15 1 5 p63 _-tub. MDA-468 HCC1143 HCC38 SUM149 MDA-468 HCC1143 HCC38 SUM149 HCC-1937 MDA-MB-468 ΔNp63_ TAp73_ TAp73β E C Relative ΔNp63 mrna TAp73
A B D Relative TAp73 mrna p73 Supplementary Figure 1 25 2 15 1 5 p63 _-tub. MDA-468 HCC1143 HCC38 SUM149 MDA-468 HCC1143 HCC38 SUM149 HCC-1937 MDA-MB-468 ΔNp63_ TAp73_ TAp73β E C Relative ΔNp63 mrna TAp73
p.r623c p.p976l p.d2847fs p.t2671 p.d2847fs p.r2922w p.r2370h p.c1201y p.a868v p.s952* RING_C BP PHD Cbp HAT_KAT11
 ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
ARID2 p.r623c KMT2D p.v650fs p.p976l p.r2922w p.l1212r p.d1400h DNA binding RFX DNA binding Zinc finger KMT2C p.a51s p.d372v p.c1103* p.d2847fs p.t2671 p.d2847fs p.r4586h PHD/ RING DHHC/ PHD PHD FYR N
Nature Genetics: doi: /ng Supplementary Figure 1. Somatic coding mutations identified by WES/WGS for 83 ATL cases.
 Supplementary Figure 1 Somatic coding mutations identified by WES/WGS for 83 ATL cases. (a) The percentage of targeted bases covered by at least 2, 10, 20 and 30 sequencing reads (top) and average read
Supplementary Figure 1 Somatic coding mutations identified by WES/WGS for 83 ATL cases. (a) The percentage of targeted bases covered by at least 2, 10, 20 and 30 sequencing reads (top) and average read
Whole Genome and Transcriptome Analysis of Anaplastic Meningioma. Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute
 Whole Genome and Transcriptome Analysis of Anaplastic Meningioma Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute Outline Anaplastic meningioma compared to other cancers Whole genomes
Whole Genome and Transcriptome Analysis of Anaplastic Meningioma Patrick Tarpey Cancer Genome Project Wellcome Trust Sanger Institute Outline Anaplastic meningioma compared to other cancers Whole genomes
Supplemental Figure S1A Notch1
 Supplemental Figure S1A Notch1 erage) epth of Cove ormalized De Log1(No Notch exons Figure S1: A) Relative coverage of Notch1 and Notch 2 exons in HCC2218, HCC1187, MB157, MDA-MB157 cell lines. Blue color
Supplemental Figure S1A Notch1 erage) epth of Cove ormalized De Log1(No Notch exons Figure S1: A) Relative coverage of Notch1 and Notch 2 exons in HCC2218, HCC1187, MB157, MDA-MB157 cell lines. Blue color
Supplementary Information
 Supplementary Information - chimeric fusion transcript in human gastric cancer promotes tumorigenesis through activation of PI3K/AKT signaling Sun Mi Yun, Kwiyeom Yoon, Sunghoon Lee, Eunjeong Kim, Seong-Ho
Supplementary Information - chimeric fusion transcript in human gastric cancer promotes tumorigenesis through activation of PI3K/AKT signaling Sun Mi Yun, Kwiyeom Yoon, Sunghoon Lee, Eunjeong Kim, Seong-Ho
injected subcutaneously into flanks of 6-8 week old athymic male nude mice (LNCaP SQ) and body
 SUPPLEMENTAL FIGURE LEGENDS Figure S1: Generation of ENZR Xenografts and Cell Lines: (A) 1x10 6 LNCaP cells in matrigel were injected subcutaneously into flanks of 6-8 week old athymic male nude mice (LNCaP
SUPPLEMENTAL FIGURE LEGENDS Figure S1: Generation of ENZR Xenografts and Cell Lines: (A) 1x10 6 LNCaP cells in matrigel were injected subcutaneously into flanks of 6-8 week old athymic male nude mice (LNCaP
hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This
 SUPPLEMENTAL FIGURE LEGEND Fig. S1. Generation and characterization of. (A) Coomassie staining of soluble hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This protein was expressed
SUPPLEMENTAL FIGURE LEGEND Fig. S1. Generation and characterization of. (A) Coomassie staining of soluble hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This protein was expressed
underlying metastasis and recurrence in HNSCC, we analyzed two groups of patients. The
 Supplementary Figures Figure S1. Patient cohorts and study design. To define and interrogate the genetic alterations underlying metastasis and recurrence in HNSCC, we analyzed two groups of patients. The
Supplementary Figures Figure S1. Patient cohorts and study design. To define and interrogate the genetic alterations underlying metastasis and recurrence in HNSCC, we analyzed two groups of patients. The
Supplementary Figure 1. IHC and proliferation analysis of pten-deficient mammary tumors
 Wang et al LEGENDS TO SUPPLEMENTARY INFORMATION Supplementary Figure 1. IHC and proliferation analysis of pten-deficient mammary tumors A. Induced expression of estrogen receptor α (ERα) in AME vs PDA
Wang et al LEGENDS TO SUPPLEMENTARY INFORMATION Supplementary Figure 1. IHC and proliferation analysis of pten-deficient mammary tumors A. Induced expression of estrogen receptor α (ERα) in AME vs PDA
Supplementary Materials and Methods
 DD2 suppresses tumorigenicity of ovarian cancer cells by limiting cancer stem cell population Chunhua Han et al. Supplementary Materials and Methods Analysis of publicly available datasets: To analyze
DD2 suppresses tumorigenicity of ovarian cancer cells by limiting cancer stem cell population Chunhua Han et al. Supplementary Materials and Methods Analysis of publicly available datasets: To analyze
Supplementary Figure 1
 Supplementary Figure 1 14 12 SEM4C PLXN2 8 SEM4C C 3 Cancer Cell Non Cancer Cell Expression 1 8 6 6 4 log2 ratio Expression 2 1 4 2 2 p value.1 D Supplementary Figure 1. Expression of Sema4C and Plexin2
Supplementary Figure 1 14 12 SEM4C PLXN2 8 SEM4C C 3 Cancer Cell Non Cancer Cell Expression 1 8 6 6 4 log2 ratio Expression 2 1 4 2 2 p value.1 D Supplementary Figure 1. Expression of Sema4C and Plexin2
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1
 Supplementary Figure 1 how HFD how HFD Epi WT p p Hypothalamus p p Inguinal WT T Liver Lean mouse adipocytes p p p p p p Obese mouse adipocytes Kidney Muscle Spleen Heart p p p p p p p p Extracellular
Supplementary Figure 1 how HFD how HFD Epi WT p p Hypothalamus p p Inguinal WT T Liver Lean mouse adipocytes p p p p p p Obese mouse adipocytes Kidney Muscle Spleen Heart p p p p p p p p Extracellular
RESISTANCE RELATIONSHIPS BETWEEN PLATINUM AND PARP-INHIBITORS IN OVARIAN CANCER.
 RESISTANCE RELATIONSHIPS BETWEEN PLATINUM AND PARP-INHIBITORS IN OVARIAN CANCER. Britta Stordal 1, Yidong Chen 2, Angela Farrelly 3, Danielle Gallagher 3, Steven Busschots 1, Mattia Cremona 3, Mark Carey
RESISTANCE RELATIONSHIPS BETWEEN PLATINUM AND PARP-INHIBITORS IN OVARIAN CANCER. Britta Stordal 1, Yidong Chen 2, Angela Farrelly 3, Danielle Gallagher 3, Steven Busschots 1, Mattia Cremona 3, Mark Carey
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis
 Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols.
 Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1: Quality assessment of formalin-fixed paraffin-embedded (FFPE)-derived DNA and nuclei. (a) Multiplex PCR analysis of unrepaired and repaired bulk FFPE gdna from
Supplementary Figure 1 Supplementary Fig. 1: Quality assessment of formalin-fixed paraffin-embedded (FFPE)-derived DNA and nuclei. (a) Multiplex PCR analysis of unrepaired and repaired bulk FFPE gdna from
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
mtor Inhibition Specifically Sensitizes Colorectal Cancers with KRAS or BRAF Mutations to BCL-2/BCL-
 Supplementary Material for mtor Inhibition Specifically Sensitizes Colorectal Cancers with KRAS or BRAF Mutations to BCL-2/BCL- XL Inhibition by Suppressing MCL-1 Anthony C. Faber 1,2 *, Erin M. Coffee
Supplementary Material for mtor Inhibition Specifically Sensitizes Colorectal Cancers with KRAS or BRAF Mutations to BCL-2/BCL- XL Inhibition by Suppressing MCL-1 Anthony C. Faber 1,2 *, Erin M. Coffee
A. Generation and characterization of Ras-expressing autophagycompetent
 Supplemental Material Supplemental Figure Legends Fig. S1 A. Generation and characterization of Ras-expressing autophagycompetent and -deficient cell lines. HA-tagged H-ras V12 was stably expressed in
Supplemental Material Supplemental Figure Legends Fig. S1 A. Generation and characterization of Ras-expressing autophagycompetent and -deficient cell lines. HA-tagged H-ras V12 was stably expressed in
Supplementary Figure 1: Comparison of acgh-based and expression-based CNA analysis of tumors from breast cancer GEMMs.
 Supplementary Figure 1: Comparison of acgh-based and expression-based CNA analysis of tumors from breast cancer GEMMs. (a) CNA analysis of expression microarray data obtained from 15 tumors in the SV40Tag
Supplementary Figure 1: Comparison of acgh-based and expression-based CNA analysis of tumors from breast cancer GEMMs. (a) CNA analysis of expression microarray data obtained from 15 tumors in the SV40Tag
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
Plasma-Seq conducted with blood from male individuals without cancer.
 Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
Supplementary Figures Supplementary Figure 1 Plasma-Seq conducted with blood from male individuals without cancer. Copy number patterns established from plasma samples of male individuals without cancer
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figure 1. Copy Number Alterations TP53 Mutation Type. C-class TP53 WT. TP53 mut. Nature Genetics: doi: /ng.
 Supplementary Figure a Copy Number Alterations in M-class b TP53 Mutation Type Recurrent Copy Number Alterations 8 6 4 2 TP53 WT TP53 mut TP53-mutated samples (%) 7 6 5 4 3 2 Missense Truncating M-class
Supplementary Figure a Copy Number Alterations in M-class b TP53 Mutation Type Recurrent Copy Number Alterations 8 6 4 2 TP53 WT TP53 mut TP53-mutated samples (%) 7 6 5 4 3 2 Missense Truncating M-class
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
SUPPLEMENTARY FIGURES AND TABLES
 SUPPLEMENTARY FIGURES AND TABLES Supplementary Figure S1: CaSR expression in neuroblastoma models. A. Proteins were isolated from three neuroblastoma cell lines and from the liver metastasis of a MYCN-non
SUPPLEMENTARY FIGURES AND TABLES Supplementary Figure S1: CaSR expression in neuroblastoma models. A. Proteins were isolated from three neuroblastoma cell lines and from the liver metastasis of a MYCN-non
BRaf V600E cooperates with Pten silencing to elicit metastatic melanoma (Nature Genetics Supplementary Information)
 BRaf V600E cooperates with Pten silencing to elicit metastatic melanoma (Nature Genetics Supplementary Information) David Dankort, David P. Curley, Robert A. Cartlidge, Betsy Nelson, Anthony N. Karnezis,
BRaf V600E cooperates with Pten silencing to elicit metastatic melanoma (Nature Genetics Supplementary Information) David Dankort, David P. Curley, Robert A. Cartlidge, Betsy Nelson, Anthony N. Karnezis,
Supplementary Information. Induction of p53-independent apoptosis by ectopic expression of HOXA5
 Supplementary Information Induction of p53-independent apoptosis by ectopic expression of in human liposarcomas Dhong Hyun Lee 1, *, Charles Forscher 1, Dolores Di Vizio 2, 3, and H. Phillip Koeffler 1,
Supplementary Information Induction of p53-independent apoptosis by ectopic expression of in human liposarcomas Dhong Hyun Lee 1, *, Charles Forscher 1, Dolores Di Vizio 2, 3, and H. Phillip Koeffler 1,
Figure S1. Generation of inducible PTEN deficient mice and the BMMCs (A) B6.129 Pten loxp/loxp mice were mated with B6.
 Figure S1. Generation of inducible PTEN deficient mice and the BMMCs (A) B6.129 Pten loxp/loxp mice were mated with B6.129-Gt(ROSA)26Sor tm1(cre/ert2)tyj /J mice. To induce deletion of the Pten locus,
Figure S1. Generation of inducible PTEN deficient mice and the BMMCs (A) B6.129 Pten loxp/loxp mice were mated with B6.129-Gt(ROSA)26Sor tm1(cre/ert2)tyj /J mice. To induce deletion of the Pten locus,
m 6 A mrna methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer
 SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3
 Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
AACR 101st Annual Meeting 2010, Washington D.C. Experimental and Molecular Therapeutics Section 29; Abstract #3855
 Investigation of the Growth Inhibitory Activity of the MEK Inhibitor ARRY-162 in Combination with Everolimus in a Variety of KRas and PI3K Pathway Mutant Cancers Brian Tunquist, Tyler Risom, Debbie Anderson,
Investigation of the Growth Inhibitory Activity of the MEK Inhibitor ARRY-162 in Combination with Everolimus in a Variety of KRas and PI3K Pathway Mutant Cancers Brian Tunquist, Tyler Risom, Debbie Anderson,
Effec<ve Use of PI3K and MEK Inhibitors to Treat Mutant K Ras G12D and PIK3CA H1047R Murine Lung Cancers
 Effec
Effec
Supplementary Figure 1. a. b. Relative cell viability. Nature Genetics: doi: /ng SCR shyap1-1 shyap
 Supplementary Figure 1. a. b. p-value for depletion in vehicle (DMSO) 1e-05 1e-03 1e-01 1 0 1000 2000 3000 4000 5000 Genes log2 normalized shrna counts in T0 0 2 4 6 8 sh1 shluc 0 2 4 6 8 log2 normalized
Supplementary Figure 1. a. b. p-value for depletion in vehicle (DMSO) 1e-05 1e-03 1e-01 1 0 1000 2000 3000 4000 5000 Genes log2 normalized shrna counts in T0 0 2 4 6 8 sh1 shluc 0 2 4 6 8 log2 normalized
Supplementary note: Comparison of deletion variants identified in this study and four earlier studies
 Supplementary note: Comparison of deletion variants identified in this study and four earlier studies Here we compare the results of this study to potentially overlapping results from four earlier studies
Supplementary note: Comparison of deletion variants identified in this study and four earlier studies Here we compare the results of this study to potentially overlapping results from four earlier studies
Infect MCF-7 cells carrying dcas9-vp64 + psm2-p65-hsf1 with SAM library or vector. Introduce AKT reporter
 Infect MCF-7 cells carrying dcas9-vp64 + psm2-p65-hsf1 with SM library or vector Introduce reporter Grow cells in presence of puromycin for 5 days Vector control SM library fewer surviving cells More surviving
Infect MCF-7 cells carrying dcas9-vp64 + psm2-p65-hsf1 with SM library or vector Introduce reporter Grow cells in presence of puromycin for 5 days Vector control SM library fewer surviving cells More surviving
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/439/ra78/dc1 Supplementary Materials for Small heterodimer partner mediates liver X receptor (LXR) dependent suppression of inflammatory signaling by promoting
www.sciencesignaling.org/cgi/content/full/9/439/ra78/dc1 Supplementary Materials for Small heterodimer partner mediates liver X receptor (LXR) dependent suppression of inflammatory signaling by promoting
Supplementary Materials
 Supplementary Materials Figure S1. MTT Cell viability assay. To measure the cytotoxic potential of the oxidative treatment, the MTT [3-(4,5-dimethylthiazol- 2-yl)-2,5-diphenyl tetrazolium bromide] assay
Supplementary Materials Figure S1. MTT Cell viability assay. To measure the cytotoxic potential of the oxidative treatment, the MTT [3-(4,5-dimethylthiazol- 2-yl)-2,5-diphenyl tetrazolium bromide] assay
Supplemental Table 1. Biochemical and Cellular Potency and Selectivity of PF
 Supplemental Table 1. Biochemical and Cellular Potency and Selectivity of PF- 02341066 Assay IC 50 nm Selectivity Ratio d Biochemical Activity In Vitro c-met/hgfr enzyme (Ki, nm) a 4 NA Cellular Activity
Supplemental Table 1. Biochemical and Cellular Potency and Selectivity of PF- 02341066 Assay IC 50 nm Selectivity Ratio d Biochemical Activity In Vitro c-met/hgfr enzyme (Ki, nm) a 4 NA Cellular Activity
Targeting the PI3K-Akt-mTOR pathway with GDC-0068, a novel selective ATP competitive Akt inhibitor
 Targeting the PI3K-Akt-mTOR pathway with GDC-0068, a novel selective ATP competitive Akt inhibitor Josep Tabernero, Andrés Cervantes, Cristina Saura, Desamparados Roda, Yibing Yan, Kui Lin, Sumati Murli,
Targeting the PI3K-Akt-mTOR pathway with GDC-0068, a novel selective ATP competitive Akt inhibitor Josep Tabernero, Andrés Cervantes, Cristina Saura, Desamparados Roda, Yibing Yan, Kui Lin, Sumati Murli,
Characterisation of structural variation in breast. cancer genomes using paired-end sequencing on. the Illumina Genome Analyser
 Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
Characterisation of structural variation in breast cancer genomes using paired-end sequencing on the Illumina Genome Analyser Phil Stephens Cancer Genome Project Why is it important to study cancer? Why
a b G75 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server.
 a b G75 2 2 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server. a. Overlay of top 10 models generated by I-TASSER illustrates the potential effect of 7 amino acid insertion
a b G75 2 2 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server. a. Overlay of top 10 models generated by I-TASSER illustrates the potential effect of 7 amino acid insertion
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
Genetic alterations of histone lysine methyltransferases and their significance in breast cancer
 Genetic alterations of histone lysine methyltransferases and their significance in breast cancer Supplementary Materials and Methods Phylogenetic tree of the HMT superfamily The phylogeny outlined in the
Genetic alterations of histone lysine methyltransferases and their significance in breast cancer Supplementary Materials and Methods Phylogenetic tree of the HMT superfamily The phylogeny outlined in the
Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier
 Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier curves for progression-free survival (PFS) and overall survival (OS) in a cohort of patients (N=52) with stage III primary ovarian
Supplementary Figure S1 Expression of mir-181b in EOC (A) Kaplan-Meier curves for progression-free survival (PFS) and overall survival (OS) in a cohort of patients (N=52) with stage III primary ovarian
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06994 A phosphatase cascade by which rewarding stimuli control nucleosomal response A. Stipanovich*, E. Valjent*, M. Matamales*, A. Nishi, J.H. Ahn, M. Maroteaux, J. Bertran-Gonzalez,
doi: 10.1038/nature06994 A phosphatase cascade by which rewarding stimuli control nucleosomal response A. Stipanovich*, E. Valjent*, M. Matamales*, A. Nishi, J.H. Ahn, M. Maroteaux, J. Bertran-Gonzalez,
MEDICAL GENOMICS LABORATORY. Peripheral Nerve Sheath Tumor Panel by Next-Gen Sequencing (PNT-NG)
 Peripheral Nerve Sheath Tumor Panel by Next-Gen Sequencing (PNT-NG) Ordering Information Acceptable specimen types: Blood (3-6ml EDTA; no time limitations associated with receipt) Saliva (OGR-575 DNA Genotek;
Peripheral Nerve Sheath Tumor Panel by Next-Gen Sequencing (PNT-NG) Ordering Information Acceptable specimen types: Blood (3-6ml EDTA; no time limitations associated with receipt) Saliva (OGR-575 DNA Genotek;
EGFR shrna A: CCGGCGCAAGTGTAAGAAGTGCGAACTCGAGTTCGCACTTCTTACACTTGCG TTTTTG. EGFR shrna B: CCGGAGAATGTGGAATACCTAAGGCTCGAGCCTTAGGTATTCCACATTCTCTT TTTG
 Supplementary Methods Sequence of oligonucleotides used for shrna targeting EGFR EGFR shrna were obtained from the Harvard RNAi consortium. The following oligonucleotides (forward primer) were used to
Supplementary Methods Sequence of oligonucleotides used for shrna targeting EGFR EGFR shrna were obtained from the Harvard RNAi consortium. The following oligonucleotides (forward primer) were used to
Supplemental Data. TGF-β-mediated mir-181a expression promotes breast cancer metastasis by targeting Bim.
 Supplemental Data TGF-β-mediated mir-181a expression promotes breast cancer metastasis by targeting Bim. Molly A. Taylor 1, Khalid Sossey-Alaoui 2, Cheryl L. Thompson 3, David Danielpour 4, and William
Supplemental Data TGF-β-mediated mir-181a expression promotes breast cancer metastasis by targeting Bim. Molly A. Taylor 1, Khalid Sossey-Alaoui 2, Cheryl L. Thompson 3, David Danielpour 4, and William
Supplementary Information. Detection and delineation of oral cancer with a PARP1 targeted optical imaging agent
 Supplementary Information Detection and delineation of oral cancer with a PARP1 targeted optical imaging agent Authors: Susanne Kossatz a, Christian Brand a, Stanley Gutiontov b, Jonathan T.C. Liu c, Nancy
Supplementary Information Detection and delineation of oral cancer with a PARP1 targeted optical imaging agent Authors: Susanne Kossatz a, Christian Brand a, Stanley Gutiontov b, Jonathan T.C. Liu c, Nancy
Supplementary Material
 Supplementary Material accompanying the manuscript Interleukin 37 is a fundamental inhibitor of innate immunity Marcel F Nold, Claudia A Nold-Petry, Jarod A Zepp, Brent E Palmer, Philip Bufler & Charles
Supplementary Material accompanying the manuscript Interleukin 37 is a fundamental inhibitor of innate immunity Marcel F Nold, Claudia A Nold-Petry, Jarod A Zepp, Brent E Palmer, Philip Bufler & Charles
Nature Genetics: doi: /ng Supplementary Figure 1. Mutational signatures in BCC compared to melanoma.
 Supplementary Figure 1 Mutational signatures in BCC compared to melanoma. (a) The effect of transcription-coupled repair as a function of gene expression in BCC. Tumor type specific gene expression levels
Supplementary Figure 1 Mutational signatures in BCC compared to melanoma. (a) The effect of transcription-coupled repair as a function of gene expression in BCC. Tumor type specific gene expression levels
MSI positive MSI negative
 Pritchard et al. 2014 Supplementary Figure 1 MSI positive MSI negative Hypermutated Median: 673 Average: 659.2 Non-Hypermutated Median: 37.5 Average: 43.6 Supplementary Figure 1: Somatic Mutation Burden
Pritchard et al. 2014 Supplementary Figure 1 MSI positive MSI negative Hypermutated Median: 673 Average: 659.2 Non-Hypermutated Median: 37.5 Average: 43.6 Supplementary Figure 1: Somatic Mutation Burden
AP VP DLP H&E. p-akt DLP
 A B AP VP DLP H&E AP AP VP DLP p-akt wild-type prostate PTEN-null prostate Supplementary Fig. 1. Targeted deletion of PTEN in prostate epithelium resulted in HG-PIN in all three lobes. (A) The anatomy
A B AP VP DLP H&E AP AP VP DLP p-akt wild-type prostate PTEN-null prostate Supplementary Fig. 1. Targeted deletion of PTEN in prostate epithelium resulted in HG-PIN in all three lobes. (A) The anatomy
ERK1/2/MAPK pathway-dependent regulation of the telomeric factor TRF2
 ERK1/2/MAPK pathway-dependent regulation of the telomeric factor TRF2 SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Conservation of the D domain throughout evolution. Alignment of TRF2 sequences
ERK1/2/MAPK pathway-dependent regulation of the telomeric factor TRF2 SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Conservation of the D domain throughout evolution. Alignment of TRF2 sequences
S1a S1b S1c. S1d. S1f S1g S1h SUPPLEMENTARY FIGURE 1. - si sc Il17rd Il17ra bp. rig/s IL-17RD (ng) -100 IL-17RD
 SUPPLEMENTARY FIGURE 1 0 20 50 80 100 IL-17RD (ng) S1a S1b S1c IL-17RD β-actin kda S1d - si sc Il17rd Il17ra rig/s15-574 - 458-361 bp S1f S1g S1h S1i S1j Supplementary Figure 1. Knockdown of IL-17RD enhances
SUPPLEMENTARY FIGURE 1 0 20 50 80 100 IL-17RD (ng) S1a S1b S1c IL-17RD β-actin kda S1d - si sc Il17rd Il17ra rig/s15-574 - 458-361 bp S1f S1g S1h S1i S1j Supplementary Figure 1. Knockdown of IL-17RD enhances
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
Introduction to LOH and Allele Specific Copy Number User Forum
 Introduction to LOH and Allele Specific Copy Number User Forum Jonathan Gerstenhaber Introduction to LOH and ASCN User Forum Contents 1. Loss of heterozygosity Analysis procedure Types of baselines 2.
Introduction to LOH and Allele Specific Copy Number User Forum Jonathan Gerstenhaber Introduction to LOH and ASCN User Forum Contents 1. Loss of heterozygosity Analysis procedure Types of baselines 2.
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus
 Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
A Comprehensive Study of TP53 Mutations in Chronic Lymphocytic Leukemia: Analysis of 1,287 Diagnostic CLL Samples
 A Comprehensive Study of TP53 Mutations in Chronic Lymphocytic Leukemia: Analysis of 1,287 Diagnostic CLL Samples Sona Pekova, MD., PhD. Chambon Ltd., Laboratory for molecular diagnostics, Prague, Czech
A Comprehensive Study of TP53 Mutations in Chronic Lymphocytic Leukemia: Analysis of 1,287 Diagnostic CLL Samples Sona Pekova, MD., PhD. Chambon Ltd., Laboratory for molecular diagnostics, Prague, Czech
Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients.
 Supplementary Materials Supplementary Figures Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients. Figure S2. Expression level of podocyte
Supplementary Materials Supplementary Figures Figure S1. Reduction in glomerular mir-146a levels correlate with progression to higher albuminuria in diabetic patients. Figure S2. Expression level of podocyte
Statistical Analysis of Single Nucleotide Polymorphism Microarrays in Cancer Studies
 Statistical Analysis of Single Nucleotide Polymorphism Microarrays in Cancer Studies Stanford Biostatistics Workshop Pierre Neuvial with Henrik Bengtsson and Terry Speed Department of Statistics, UC Berkeley
Statistical Analysis of Single Nucleotide Polymorphism Microarrays in Cancer Studies Stanford Biostatistics Workshop Pierre Neuvial with Henrik Bengtsson and Terry Speed Department of Statistics, UC Berkeley
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/4/199/ra75/dc1 Supplementary Materials for Signaling by the Matrix Proteoglycan Decorin Controls Inflammation and Cancer Through PDCD4 and MicroRNA-21 Rosetta
www.sciencesignaling.org/cgi/content/full/4/199/ra75/dc1 Supplementary Materials for Signaling by the Matrix Proteoglycan Decorin Controls Inflammation and Cancer Through PDCD4 and MicroRNA-21 Rosetta
Complexity of intra- and inter-pathway loops in colon cancer and melanoma: implications for targeted therapies
 Complexity of intra- and inter-pathway loops in colon cancer and melanoma: implications for targeted therapies René Bernards The Netherlands Cancer Institute Amsterdam The Netherlands Molecular versus
Complexity of intra- and inter-pathway loops in colon cancer and melanoma: implications for targeted therapies René Bernards The Netherlands Cancer Institute Amsterdam The Netherlands Molecular versus
Supplemental Table 1. Cell Lines used in the CRISPR pooled screen.
 Supplemental Table 1. Cell Lines used in the CRISPR pooled screen. 5637_URINARY_TRACT HEC1B_ENDOMETRIUM NCIH1975_LUNG RCCGS_KIDNEY 639V_URINARY_TRACT HPAFII_PANCREAS NCIH1993_LUNG RCCJW_KIDNEY 647V_URINARY_TRACT
Supplemental Table 1. Cell Lines used in the CRISPR pooled screen. 5637_URINARY_TRACT HEC1B_ENDOMETRIUM NCIH1975_LUNG RCCGS_KIDNEY 639V_URINARY_TRACT HPAFII_PANCREAS NCIH1993_LUNG RCCJW_KIDNEY 647V_URINARY_TRACT
Nature Medicine: doi: /nm.3967
 Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Supplementary Figure 1. Network clustering. (a) Clustering performance as a function of inflation factor. The grey curve shows the median weighted Silhouette widths for varying inflation factors (f [1.6,
Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes.
 Supplementary Figure 1 Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes. (a,b) Values of coefficients associated with genomic features, separately
Supplementary Figure 1 Relationship between genomic features and distributions of RS1 and RS3 rearrangements in breast cancer genomes. (a,b) Values of coefficients associated with genomic features, separately
Functional Genomics : PTPRF-Mediated Contact Inhibition in HCC development
 Functional Genomics : PTPRF-Mediated Contact Inhibition in HCC development Sen-Yung Hsieh, MD, PhD Liver Research Unit Chang Gung Memorial Hospital Taoyuan, Taiwan Aims To identify genes controlling tumor
Functional Genomics : PTPRF-Mediated Contact Inhibition in HCC development Sen-Yung Hsieh, MD, PhD Liver Research Unit Chang Gung Memorial Hospital Taoyuan, Taiwan Aims To identify genes controlling tumor
Computational Systems Biology: Biology X
 Bud Mishra Room 1002, 715 Broadway, Courant Institute, NYU, New York, USA L#4:(October-0-4-2010) Cancer and Signals 1 2 1 2 Evidence in Favor Somatic mutations, Aneuploidy, Copy-number changes and LOH
Bud Mishra Room 1002, 715 Broadway, Courant Institute, NYU, New York, USA L#4:(October-0-4-2010) Cancer and Signals 1 2 1 2 Evidence in Favor Somatic mutations, Aneuploidy, Copy-number changes and LOH
Supplementary methods:
 Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
of TERT, MLL4, CCNE1, SENP5, and ROCK1 on tumor development were discussed.
 Supplementary Note The potential association and implications of HBV integration at known and putative cancer genes of TERT, MLL4, CCNE1, SENP5, and ROCK1 on tumor development were discussed. Human telomerase
Supplementary Note The potential association and implications of HBV integration at known and putative cancer genes of TERT, MLL4, CCNE1, SENP5, and ROCK1 on tumor development were discussed. Human telomerase
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung
 Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines.
 Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines. The scores summarize the global expression of the tissue
Supplementary Figure 1: Tissue of Origin analysis on 152 cell lines. (a) Heatmap representation of the 30 Tissue scores for the 152 cell lines. The scores summarize the global expression of the tissue
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Targeted mass spectrometry (LC/MS/MS) for Olaparib pharmacokinetics. For LC/MS/MS of Olaparib pharmacokinetics metabolites were extracted from
 Supplementary Methods: Targeted mass spectrometry (LC/MS/MS) for Olaparib pharmacokinetics For LC/MS/MS of Olaparib pharmacokinetics metabolites were extracted from mouse tumor samples and analyzed as
Supplementary Methods: Targeted mass spectrometry (LC/MS/MS) for Olaparib pharmacokinetics For LC/MS/MS of Olaparib pharmacokinetics metabolites were extracted from mouse tumor samples and analyzed as
SUPPLEMENTARY FIGURES AND TABLE
 SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
(A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939 (5 µm)
 Supplementary Figure Legends Figure S1. Tankyrase inhibition suppresses cell proliferation in an axin/β-catenin independent manner. (A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939
Supplementary Figure Legends Figure S1. Tankyrase inhibition suppresses cell proliferation in an axin/β-catenin independent manner. (A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939
MEDICAL GENOMICS LABORATORY. Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG)
 Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG) Ordering Information Acceptable specimen types: Fresh blood sample (3-6 ml EDTA; no time limitations associated with receipt)
Next-Gen Sequencing and Deletion/Duplication Analysis of NF1 Only (NF1-NG) Ordering Information Acceptable specimen types: Fresh blood sample (3-6 ml EDTA; no time limitations associated with receipt)
Supplementary Figure S1. Generation of LSL-EZH2 conditional transgenic mice.
 Downstream Col1A locus S P P P EP Genotyping with P1, P2 frt PGKneopA + frt hygro-pa Targeting vector Genotyping with P3, P4 P1 pcag-flpe P2 P3 P4 frt SApA CAG LSL PGKATG frt hygro-pa C. D. E. ormal KRAS
Downstream Col1A locus S P P P EP Genotyping with P1, P2 frt PGKneopA + frt hygro-pa Targeting vector Genotyping with P3, P4 P1 pcag-flpe P2 P3 P4 frt SApA CAG LSL PGKATG frt hygro-pa C. D. E. ormal KRAS
Leucine Deprivation Reveals a Targetable Liability
 Cancer Cell, 19 Supplemental Information Defective Regulation of Autophagy upon Leucine Deprivation Reveals a Targetable Liability of Human Melanoma Cells In Vitro and In Vivo Joon-Ho Sheen, Roberto Zoncu,
Cancer Cell, 19 Supplemental Information Defective Regulation of Autophagy upon Leucine Deprivation Reveals a Targetable Liability of Human Melanoma Cells In Vitro and In Vivo Joon-Ho Sheen, Roberto Zoncu,
