SUPPLEMENTARY INFORMATION
|
|
|
- Elizabeth McGee
- 5 years ago
- Views:
Transcription
1 Figure S1. The expression of Plgf, Vegfa, Vegfc or Myh7 generates lower correlation coefficients to the mitochondrial cluster, as compared to Vegfb. The intensity of Plgf, Vegfa, Vegfc and the myocyte marker Myh7 expression (black), plotted against the mean expressional intensity of the mitochondrial cluster (red) in 708 microarrays, as described in the Methods Summary section. The Pearson correlation coefficients (r) for each comparison are listed under the respective graphs. 1
2 Figure S2. Vegfb has similar expression pattern as Cycs in an array of mouse tissues. Relative mrna expression of Vegfb and Cycs in 7 wt mouse tissues analyzed by qpcr. Data in is shown as percentage of expression compared to the ribosomal L19 gene (n=3 mice). Primer sequences are listed in Table S3. Figure S3. VEGF-B does not regulate mitochondrial gene expression or copy number. a, Relative mrna expression of Ndufa5 and Cycs in hearts from fed or fasted wt and VEGF-B -/- mice. Data is shown as percentage of expression compared to the ribosomal L19 gene (n=3 mice). c, Mitochondrial DNA copy number in hearts of wt and VEGF-B -/- mice. Total DNA from hearts of wt and VEGF-B -/- mice was phenolchloroform extracted. The final aqueous phase was used to assess mitochondrial DNA (mtdna) content relative to nuclear DNA (ndna) content by qpcr, using 2 ng of total DNA as template and primers for cytochrome c oxidase II (Cox II, mitochondrial genome) and Gapdh (nuclear genome). Primer sequences are listed in Table S3. 2
3 Figure S4. Mouse transformed ECs express combinations of Fatp1, Fatp3 and Fatp4, whereas human primary and transformed ECs express Fatp1, Fatp3, Fatp4 and Fatp5. mrna expression of FATPs, VEGFR1 and NRP1 in different mouse and human endothelial cell lines. mrna was extracted from cultured cells as described in Supplemental Methods, and analyzed by conventional RT-PCR using specific primers listed in Table S3. bend3, mouse brain-capillary derived ECs (polyoma mt immortalized); MS-1, mouse pancreatic ECs (SV40T immortalized); HDMEC, primary human dermal microvascular ECs; HUVEC, primary human umbilical vein ECs; TIME, human dermal telomerase immortalized microvascular ECs. 3
4 Figure S5. VEGF-B treatment induces Fatp1 and Fatp3 expression in human primary EC lines, and Fatp1, Fatp3 and Fatp4 expression in mouse EC lines. a, Cell line specific expression of FATPs in response to VEGF-B treatment. bend3 cells are shown in Figure 2. Data is presented as fold change versus svegfr1 treated cells set to 1 a.u. b, Immunoblot of FATP4 in bend3 ECs after VEGF-B 186 treatment. *P <0.05, **P <0.01, ***P <
5 Figure S6. VEGF-B does not induce Fatp4 expression in cultured NIH3T3 fibroblasts. a, Expression of VEGFR1, VEGFR2 and NRP1 in NIH3T3 cells. b, mrna levels of Fatp4 analyzed by qpcr. NIH3T3 cells were cultured using standard procedures, and treated as described in Supplemental Methods. Data is presented as fold change versus svegfr1 treated cells set to 1 a.u. Figure S7. VEGF-B regulation of FATPs is dependent on PI3K activity mrna expression of Fatp3 and Fatp4 in bend3 cells after co-treatment with VEGF- B and protein kinase inhibitors. Cells were treated and analyzed as described in Supplemental Methods. Data is presented as fold change versus svegfr1-treated cells set to 1 a.u. LY294002, PI3K inhibitor; PD98059, MEK1 inhibitor; U0126 MEK1 and MEK2 inhibitor. 5
6 Figure S8. mrna expression levels of Vegfb, Vegfa, Plgf and Pecam1 after adenoviral transduction a, Mouse and human Vegfb and Vegfa mrna expression levels were analyzed by qpcr using previously published species-specific primers 1. Data is presented as endogenous levels of mvegfb and mvegfa in AdhVEGF-B and AdhVEGF-A transduced mouse hearts, in relation to total Vegfb and Vegfa mrna levels by combining the results obtained with specific mouse and human primers. b, Mouse Plgf mrna expression was determined in AdLacZ and AdmPlGF transduced mouse hearts using primers specific for mplgf transcripts. Primer sequences are found in Table S3. L19 was used as normalization gene, and data is shown as fold change versus endogenous or AdLacZ levels set to 1 a.u. ±SEM. c, The induction of Fatp3 and Fatp4 mrna by AdVEGF-B transduction was not due to proliferation of ECs in mouse hearts, as the mrna levels of Pecam1 were unchanged. Gene expression was analyzed by qpcr as described in Supplemental Methods. Data is presented as fold change versus AdLacZ levels set to 1 a.u. 6
7 Figure S9. mrna expression of FATPs in wt and VEGF-B -/- tissues. a, Expression pattern of FATPs in wt mouse tissues, analyzed by conventional RT- PCR using specific primers listed in Table S3. b, mrna levels of FATPs in VEGF-B -/- mice were determined by qpcr in oxidative skeletal muscle and in ibat as described in Supplemental Methods. Data is shown as fold change versus wt levels set to 1 a.u. ibat, interscapular BAT. **P <0.01. Figure S10. Endothelial enrichment in isolated mouse EC-fractions from heart. Enrichment of isolated EC and non-ec fractions from heart analyzed by conventional RT-PCR. Pecam1 and Vegfr1 were used as marker genes for ECs, and Myh6, Tnnt2 and Vegfb for cardiomyocytes. Total cdna from heart was used as positive controls (Heart); NTC, non-template control. Myh6, cardiac myosin heavy polypeptide 6; Tnnt2, cardiac troponin T2 7
8 Figure S11. The vascular FATPs, Fatp3 and Fatp4, are downregulated in ECs isolated from BAT of VEGF-B -/- animals, but not in ECs isolated from lung. a, Relative expression of FATPs in isolated cell fractions from wt BAT (n=2-3). Data is shown as percentage of expression compared to L19 (n=2-3). b, mrna levels of FATPs in EC fractions isolated from wt and VEGF-B -/- BAT and lung, and in non-ec fractions isolated from wt and VEGF-B -/- BAT (n=3). Data is shown as fold change versus wt levels set to 1 a.u. Figure S12. FATP3 is expressed in the endothelium in oxidative skeletal muscle, whereas FATP4 is also expressed in the myocytes, especially in type I fibers. Immunohistochemical localization of FATP3, FATP4 and type I myosin in serial sections from wt soleus muscles. Asterisks indicate type I fibers. Scale bars, 50 µm (n=2-3 mice). 8
9 Figure S13. FATP3 and FATP4 can induce LCFA-uptake in cultured ECs. BODIPY-FA uptake into bend3 cells after transfection with the indicated plasmids ± a 10x molar excess of OA (lower panel). Quantification is shown in Figure 3. Figure S14. Silencing Fatp3 and Fatp4 expression by sirnas blunts both the transcriptional, and lipid-transporting effects of VEGF-B treatment. a-c, Relative mrna expression of Fatp3 (a), Fatp4 (b) and Gapdh (c) in bend3 cells that were transfected with either scrambled control sirna or the specific targeting sirnas, and treated with VEGF-B 186 or anti-vegf-b mab as described in 9
10 Supplemental Methods. Results are presented as percentage of expression relative to the normalization gene L19. d, Quantification of BODIPY-FA uptake into bend3 cells that were transfected with Fatp3 and/or Fatp4 targeting sirnas, and treated with VEGF-B 186 or anti-vegf-b mab. Representative images are shown in Figure 3. *P <0.05, **P <0.01, ***P < Figure S15. VEGF-B treatment does not increase endothelial permeability. a, Measurements of trans-endothelial resistance (TER) over membranes that were unseeded (No cells), or seeded with bend3 cells (Cells). Cells were subsequently treated with VEGFs, or mock treated, and TER was measured before treatment, as well as after 5 hrs and 30 hrs of treatment. b, Trans-endothelial leakage was assessed by adding the inert carbohydrate 14 C-inulin to bend3 monolayers after treatment with different VEGFs for 30 hrs. c, Trans-endothelial transport of LCFAs across bend3 cell layers. 14 C-labeled OA was added to cell culture inserts unseeded or seeded with bend3 cells. Transport was determined by measuring the radioactivity in the lower compartments in b-c. *P <0.05, **P <0.01, ***P <
11 Figure S16. Normal expression of fatty acid handling proteins in the small intestine of VEGF-B -/- animals. Expression of the fatty acid handling proteins Fatp4 and Cd36 in the small intestine of wt and VEGF-B -/- mice. Male wt and VEGF-B -/- mice were sacrificed and the proximal half of the small intestine was dissected and rinsed with PBS. RNA was isolated using RNeasy kit (Qiagen) as described in Supplemental Methods. The expression of Fatp4 and Cd36 was analyzed by qpcr and the results are presented as fold change versus the level in wt mice, which was set to 1 a.u. (n=3-6 mice). Figure S17. VEGF-B -/- mice have normal basal plasma levels of triglycerides (TGs) and non-esterified fatty acids (NEFAs). Blood was collected from hearts of Avertin-anesthetized, normally fed and agematched 8-12 week old male mice using EDTA syringes, and centrifuged for 1 min at 16,000 g. TG and NEFA content in plasma was determined using the Serum Triglyceride Determination Kit (Sigma), and the Wako NEFA C test kit (Wako Diagnostics), respectively, according to the manufactures instructions. 11
12 Figure S18. Accumulation of the 14 C-OA tracer in different WAT depots. Relative radioactivity in retroperitoneal (rwat), inguinal (iwat) and subcutaneous WAT (scwat) in VEGF-B -/- mice as compared to controls 24 hrs after oral gavage, preformed as described in Supplemental Methods (n=4 mice). ewat is shown in Figure 4. *P <0.05, **P <0.01. Figure S19. VEGF-B -/- mice accumulate lipids in WAT a, Weight of epididymal (ewat), retroperitoneal (rwat), inguinal (iwat) and subcutaneous WAT (scwat) fat pads from wt and VEGF-B -/- mice normalized to body weight (n=5-7). b, Quantification of total body fat from MRI scans performed as described in Supplemental Methods. *P <0.05, **P <
13 Figure S20. Quantitation of ORO staining in heart and muscle, and mean lipid vacuole size in BAT. Quantifications of ORO- and HE-staining of sections from wt and VEGF-B -/- mice (a), and from NRP1-EC-WT and NRP1-EC-KO mice (b). Representative images are shown in Figure 4. *P <0.05, **P <0.01, ***P < Figure S21. hvegf-b content in plasma 5 days after adenoviral administration. Wt and VEGF-B -/- mice were treated, and plasma was collected and analyzed by ELISA as described in Supplemental methods (n=4-5 mice). Recombinant hvegf-b protein was used for the standard curve. 13
14 Figure S22. Administration of AdVEGF-B rescues the phenotype of the VEGFB - /- mice but not the NRP-EC-KO mice. a, Cardiac Fatp4 mrna levels after adenoviral administration. Data is presented as fold change versus wt+adcmv or NRP1-EC-WT+AdCMV hearts set to 1 a.u. b, Quantification of ORO staining of heart sections from wt and VEGF-B -/- hearts (n=4-5) and from NRP1-EC-WT and NRP1-EC-KO hearts (n=3-4) after adenoviral administration. c, Representative ORO images from NRP1-EC-WT and NRP1-EC- KO mice after adenoviral delivery. *P <0.05, **P <
15 Figure S23. VEGF-B -/- mice have normal capacity for lipid β-oxidation, and VEGF-B per se does not induce oxidation of fatty acids in cultured myotubes. a, Ex vivo β-oxidation in isolated organs from wt and VEGF-B -/- mice. Dissected organs were weighted and incubated with 14 C-OA, after which formed 14 CO 2 was collected onto alkalinized membranes and analyzed by liquid scintillation (n=5). b, Level of oxidized 14 C-OA in cultured C2C12 myotubes in response to treatment with VEGF-B 186, globular adiponectin (gadiponectin, positive control) or mock treatment. Formed 14 CO 2 was collected onto alkalinized membranes and analyzed by liquid scintillation (n=3 wells per treatment). Both methods were essentially performed as previously described 2,3. Figure S24. Direct measurements of radioactivity in dissected hearts confirm the results from PET image analysis. Dissected mouse hearts from VEGF-B -/- mice show higher cardiac radioactivity, as compared to wt hearts, 60 min after [ 18 F]FDG i.v. injection. Data shows mean radioactivity as percentage of injected [ 18 F]FDG dose (n= 3-4 mice). *P <
16 Table S1. Vegfb is included in the mitochondrial co-expression cluster. The clustering of co-expressed genes (r>0.80) was preformed as described in Supplemental Methods. Vegfb is emphasized in bold. Sdhd succinate dehydrogenase complex, subunit D, integral membrane protein Atp5c1 ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 Atp5c1 ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 Ndufa8 NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 Ndufs1 NADH dehydrogenase (ubiquinone) Fe-S protein 1 Sdhc succinate dehydrogenase complex, subunit C, integral membrane protein Ndufab1 NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, Mus musculus cdna clone IMAGE: , partial cds Atp5o ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit Ndufb10 NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 Ndufv3 NADH dehydrogenase (ubiquinone) flavoprotein 3 Ndufs3 NADH dehydrogenase (ubiquinone) Fe-S protein 3 Ogdh oxoglutarate dehydrogenase (lipoamide) Ndufa9 NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9 Sdhb succinate dehydrogenase complex, subunit B, iron sulfur (Ip) Ndufa3 NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3 Uqcr ubiquinol-cytochrome c reductase (6.4kD) subunit Ndufc1 NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1 Ndufv1 NADH dehydrogenase (ubiquinone) flavoprotein 1 Ndufs2 NADH dehydrogenase (ubiquinone) Fe-S protein 2 Uqcrc1 ubiquinol-cytochrome c reductase core protein 1 Coq9 coenzyme Q9 homolog (yeast) Ndufb7 NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 Ndufb2 NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 Ndufb9 NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9 Np15 nuclear protein 15.6 Ndufb2 NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 Atp5e ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit --- Mus musculus cdna clone IMAGE: , with apparent retained intron Atp5l ATP synthase, H+ transporting, mitochondrial F0 complex, subunit g Uqcrh ubiquinol-cytochrome c reductase hinge protein Cox5b cytochrome c oxidase, subunit Vb Ndufb8 NADH dehydrogenase (ubiquinone) 1 beta subcomplex 8 Ndufv2 NADH dehydrogenase (ubiquinone) flavoprotein 2 Ndufs5 NADH dehydrogenase (ubiquinone) Fe-S protein 5 Fh1 fumarate hydratase E04Rik RIKEN cdna E04 gene Ech1 enoyl coenzyme A hydratase 1, peroxisomal Etfb electron transferring flavoprotein, beta polypeptide Dci dodecenoyl-coenzyme A delta isomerase (3,2 trans-enoyl-coenyme A isomerase) Hadhb hydroxyacyl-coenzyme A dehydrogenase/3-ketoacyl-coenzyme A thiolase/enoyl- Coenzyme A hydratase (trifunctional protein), beta subunit Hsdl2 hydroxysteroid dehydrogenase like 2 Etfdh electron transferring flavoprotein, dehydrogenase Cpt2 carnitine palmitoyltransferase 2 Pdha1 pyruvate dehydrogenase E1 alpha 1 Mrps36 mitochondrial ribosomal protein S36 Cox7a1 cytochrome c oxidase, subunit VIIa 1 Idh3g isocitrate dehydrogenase 3 (NAD+), gamma Cs citrate synthase Chchd10 coiled-coil-helix-coiled-coil-helix domain containing 10 AI expressed sequence AI Vegfb vascular endothelial growth factor B Chchd3 coiled-coil-helix-coiled-coil-helix domain containing 3 Uqcrb ubiquinol-cytochrome c reductase binding protein Mcee methylmalonyl CoA epimerase Mor1 malate dehydrogenase, mitochondrial 16
17 Table S2. Transcriptional analyses of β-oxidation related genes revealed no major changes in VEGF-B -/- organs. mrna levels of genes in lipid metabolism determined by qpcr in tissues from VEGF-B -/- mice, normalized by the mean value of wt mice set to 1 unit (n=3 mice). *P <0.05, **P <0.01. ns= non significant. Cpt1/2, carnitine palmitoyltransferase, Slc25a20, solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20; Pgc1a, PPAR gamma coactivator 1α; Acsl, acyl-coa synthetase long-chain; Acadm, acyl-coenzyme A dehydrogenase medium chain; Acox1, acyl-coenzyme A oxidase; Ucp1/3, Uncoupling protein; Srebp, sterol regulatory element binding transcription factor; Acc, acetyl-coa carboxylase; Glut4, facilitated glucose transporter 4. Gene/Organ Heart Muscle ibat Cpt1a/b 0.57±0.07 * 0.61±0.15 * 0.72±0.04 * Cpt2 0.76±0.06 ns 0.75±0.05 * 0.75±0.11 ns Slc25a ±0.13 ns 0.71±0.12 * 0.63±0.07 ** Pgc1a 0.98±0.12 ns 0.92±0.07 ns 1.08±0.1 ns Ppara 1.13±0.08 ns 0.81±0.08 ns - Pparg ±0.12 ns Acsl 0.98±0.12 ns 0.95±0.15 ns 0.85±0.07 ns Mcad (Acadm) 1.01±0.08 ns 0.93±0.02 ns 1.14±0.12 ns Acox1 0.86±0.11 ns 0.78±0.09 ns 1.00±0.11 ns Ucp ±0.16 ns Ucp ±0.37 ns Srebp1c 1.27±0.06 ns 1.02±0.08 ns 2.13±0.20 ** Srebp2 1.44±0.03 * 1.21±0.05 * 1.28±0.09 * Acc 1.58±0.09 * 1.65±0.12 * 1.68±0.13 ** Glut4 1.79±0.12 ** 1.12±0.7 ns 1.42±0.07 ** 17
18 Table S3. Primer sequences. All sequences are written 5 3 Fwd primer Rev primer mvegfb TCTGAGCATGGAACTCATGG TCTGCATTCACATTGGCTGT mndufa5 ATCACCTTCGAGAAGCTGGA ACTTCACCACCCTGAAGCAA mcycs CCAAATCTCCACGGTCTGTT CCAGGTGATGCCTTTGTTCT mplgf CCCACACCCAGCTCACGTATTTA TCCCCTCTACATGCCTTCAATGC mvegfa CAGGCTGCTGTAACGATGAA TATGTGCTGGCTTTGGTGAT ml19 GGTGACCTGGATGAGAAGGA TTCAGCTTGTGGATGTGCTC mbeta actin ACTCTTCCAGCCTTCCTTC ATCTCCTTCTGCATCCTGTC mb2m CTGACCGGCCTGTATGCTAT CCGTTCTTCAGCATTTGGAT mcpt1a CATGTCAAGCCAGAGGAAGA TGGTAGGAGAGCAGCACCTT mcpt1b CCCATGTGCTCCTACCAGAT CCTTGAAGAAGCGACCTTTG mcpt2 CAGCATATGATGGCTGAGTG GTGGTTTATCCGCTGGTATG mslc25a20 TTTGCAGGGATCTTCAACTG CCCTTTGTACAAGGAGGTGA mpgc1a GGAGCCGTGACCACTGACA TGGTTTGCTGCATGGTTCTG mppara GACAAGGCCTCAGGGTACCA GCCGAATAGTTCGCCGAAA mpparg CCATTCTGGCCCACCAAC AATGCGAGTGGTCTTCCATCA macsl ATATCTACCTGCGGAGTGAAG CCTTCCCAAGTTTCAACAAGTC macadm TCGGAGGCTATGGATTCAAC CAGCCTCTGAATTTGTGCAG macox1 ACTTGTTTGAGTGGGCCAAG AGAGATTCGGCCTCTCTGTG mhsd17b4 CGGTTTTGAGAAGCCCATATT ATTCTGTTTCCTTCCTTCCACA mhmgcs1 CAGTACTCACCTCAGCAGTTG CACACAAGTTCTCGAGTCAAG mucp1 CTGCCAGGACAGTACCCAAG GCCACAAACCCTTTGAAAAA mucp2 GCCACAAACCCTTTGAAAAA TACAAGGGGTTCATGCCTTC mucp3 AGCCCTCTGCACTGTATGCT CAGAAAGGAGGGCACAAATC msrebp1c* GGCACTAAGTGCCCTCAACCT GCCACATAGATCTCTGCCAGTGT msrebp2** GGATCATCCAGCAGCCTTTGA ACCGGGACCTGCACCTGT macc* CCCAGCAGAATAAAGCTACTTTGG TCCTTTTGTGCAACTAGGAACGT mglut4 ACTCTTGCCACACAGGCTCT CCTTGCCCTGTCAGGTATGT mfatp1 TCAATGTACCAGGAATTACAGAAGG GAGTGAGAAGTCGCCTGCAC mfatp2 CATGAGCTAAACCACCAGGG TTCCTGAGGATACAAGATACCATTG mfatp3 CGCAGGCTCTGAACCTGG TCGAAGGTCTCCAGACAGGAG or CCTCGGTTTCTCAGGCTCCA CTGTACCGGGCAGGTGTGA mfatp4 GCAAGTCCCATCAGCAACTG GGGGGAAATCACAGCTTCTC mfatp5 GTGGTCAGAGATTCCAGGTTCC GCTATACCAGCATGTCCGCTC mfatp6 TACAACCAAGTGGTGACATCTCTG AATCTCTTCGGTCAATGGGAC mcd36 GATGAGCATAGGACATACTTAGATGTG CACCACTCCAATCCCAAGTAAG mfabp1 CCAGAAAGGGAAGGACATCA GTCTCCAGTTCGCACTCCTC mfabp3 TTCAGCTGGGAATAGAGTTCG CTGCACATGGATGAGTTTGC mfabp4 GATGGTGACAAGCTGGTGGT AATTTCCATCCAGGCCTCTT mfabp5 GGAAGATGGCGCCTGGTGGA CCGAGTACAGGTGACATTGT mpecam1 AGAGACGGTCTTGTCGCAGT TACTGGGCTTCGAGAGCATT mvegfr1 GGAGGAGTACAACACCACGG TTGAGGAGCTTTCACCGAAC mmyh6 AGCTGGAGAATGAGCTGGAG GCAGCCGCATTAAGTTCTTC mtnnt2 CGTGAGGAGGAGGAGAACAG TCCTCTCTGCCAGGATCTTC mvegfr2 AGCACCTCTCTCGTGATTTCC AGTAAAAGCAGGGAGTCTGTGG mnrp1 GGAGCTACTGGGCTGTGAAG CCTCCTGTGAGCTGGAAGTC CoxII*** TCTCCCCTCTCTACGCATTCTA ACGGATTGGAAGTTCTATTGGC Gapdh TGCGACTTCAACAGCAACTC GCCTCTCTTGCTCAGTGTCC hl19 GGCACATGGGCATAGGTAAG CCATGAGAATCCGCTTGTTT hvegfr1 CTTCACCTGGACTGACAGCAA CTCAGCGTGGTCGTAGGT hnrp1 CGCTACCAGAAGCCAGAGGA CATCCACAGCAATCCCACCAA hfatp-1 TCTATGGGGTGGCTGTTCCA TCAAACCCTCTCGCTGCA hfatp-2 AGGGGAAAATGTGGCCACCA TTAGAAACCGGGGCCTTGCA hfatp-3 CAGAGGTCTTCGAGGCCCTA AAGGTCTCTGTGGTGGCCAA hfatp-4 CTTTTCCAGCCGCTTCCACA TGGCTGGCAGGGAATGCA hfatp-5 TGTGCGTGCCAGGTTGTGA ATCCCCACATTGAAGCCCTCA hfatp-6 GCTGGGCCTTATAAGCACACA CAACCTCAGTGGTTGCGACA The following primer sequneces were taken from these publications: * Primer sequence copied from reference 4 ** Primer sequence copied from reference 5 *** Primer sequence copied from reference
19 References 1. Thijssen, V.L., Brandwijk, R.J., Dings, R.P. & Griffioen, A.W. Angiogenesis gene expression profiling in xenograft models to study cellular interactions. Exp Cell Res 299, (2004). 2. Fruebis, J., et al. Proteolytic cleavage product of 30-kDa adipocyte complement-related protein increases fatty acid oxidation in muscle and causes weight loss in mice. Proc Natl Acad Sci U S A 98, (2001). 3. Mao, X., et al. APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function. Nature cell biology 8, (2006). 4. Jiang, T., et al. Diet-induced obesity in C57BL/6J mice causes increased renal lipid accumulation and glomerulosclerosis via a sterol regulatory elementbinding protein-1c-dependent pathway. J Biol Chem 280, (2005). 5. Zhou, R.H., et al. Vascular endothelial growth factor activation of sterol regulatory element binding protein: a potential role in angiogenesis. Circ Res 95, (2004). 6. Villena, J.A., et al. Orphan nuclear receptor estrogen-related receptor alpha is essential for adaptive thermogenesis. Proc Natl Acad Sci U S A 104, (2007). 19
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11464 Supplemental Figure S1. The expression of Vegfb is increased in obese and diabetic mice as compared to lean mice. a-b, Body weight and postprandial blood
SUPPLEMENTARY INFORMATION doi:10.1038/nature11464 Supplemental Figure S1. The expression of Vegfb is increased in obese and diabetic mice as compared to lean mice. a-b, Body weight and postprandial blood
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at
 Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Fig. S1. Dose-response effects of acute administration of the β3 adrenoceptor agonists CL316243, BRL37344, ICI215,001, ZD7114, ZD2079 and CGP12177 at doses of 0.1, 0.5 and 1 mg/kg on cumulative food intake
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies
 Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR
 Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) cadl CTTGGGGGCGCGTCT CTGTTCTTTTGTGCCGTTTCG cyl-coenzyme Dehydrogenase, very
Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) cadl CTTGGGGGCGCGTCT CTGTTCTTTTGTGCCGTTTCG cyl-coenzyme Dehydrogenase, very
Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle.
 Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle. (a) mrna levels of Dj1 measured by quantitative RT-PCR in soleus, gastrocnemius (Gastroc.) and extensor digitorum longus
Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle. (a) mrna levels of Dj1 measured by quantitative RT-PCR in soleus, gastrocnemius (Gastroc.) and extensor digitorum longus
Supplementary Figure 1. Mitochondrial function in skeletal muscle and plasma parameters of STZ mice A D-F
 Supplementary Figure 1. Mitochondrial function in skeletal muscle and plasma parameters of STZ mice A Expression of representative subunits of the mitochondrial respiratory chain was analyzed by real time
Supplementary Figure 1. Mitochondrial function in skeletal muscle and plasma parameters of STZ mice A Expression of representative subunits of the mitochondrial respiratory chain was analyzed by real time
GPR120 *** * * Liver BAT iwat ewat mwat Ileum Colon. UCP1 mrna ***
 a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
SUPPLEMENTARY,INFORMATIONS,,,, mtorc1,is,required,for,brown,adipose,tissue,recruitment,and, METABOLIC,ADAPTATION,TO,COLD,!! Sébastien!M.!
 SUPPLEMENTARY,INFORMATIONS,,,, mtorc,is,required,for,brown,adipose,tissue,recruitment,and, METABOLIC,ADAPTATION,TO,COLD, SébastienM.Labbé,,MathildeMouchiroud,,AlexandreCaron,,BlandineSecco,, Elizaveta
SUPPLEMENTARY,INFORMATIONS,,,, mtorc,is,required,for,brown,adipose,tissue,recruitment,and, METABOLIC,ADAPTATION,TO,COLD, SébastienM.Labbé,,MathildeMouchiroud,,AlexandreCaron,,BlandineSecco,, Elizaveta
control kda ATGL ATGLi HSL 82 GAPDH * ** *** WT/cTg WT/cTg ATGLi AKO/cTg AKO/cTg ATGLi WT/cTg WT/cTg ATGLi AKO/cTg AKO/cTg ATGLi iwat gwat ibat
 body weight (g) tissue weights (mg) ATGL protein expression (relative to GAPDH) HSL protein expression (relative to GAPDH) ### # # kda ATGL 55 HSL 82 GAPDH 37 2.5 2. 1.5 1..5 2. 1.5 1..5.. Supplementary
body weight (g) tissue weights (mg) ATGL protein expression (relative to GAPDH) HSL protein expression (relative to GAPDH) ### # # kda ATGL 55 HSL 82 GAPDH 37 2.5 2. 1.5 1..5 2. 1.5 1..5.. Supplementary
SUPPLEMENTARY DATA. Supplementary Table 1. Primers used in qpcr
 Supplementary Table 1. Primers used in qpcr Gene forward primer (5'-3') reverse primer (5'-3') β-actin AGAGGGAAATCGTGCGTGAC CAATAGTGATGACCTGGCCGT Hif-p4h-2 CTGGGCAACTACAGGATAAAC GCGTCCCAGTCTTTATTTAGATA
Supplementary Table 1. Primers used in qpcr Gene forward primer (5'-3') reverse primer (5'-3') β-actin AGAGGGAAATCGTGCGTGAC CAATAGTGATGACCTGGCCGT Hif-p4h-2 CTGGGCAACTACAGGATAAAC GCGTCCCAGTCTTTATTTAGATA
Metabolism of cardiac muscle. Dr. Mamoun Ahram Cardiovascular system, 2013
 Metabolism of cardiac muscle Dr. Mamoun Ahram Cardiovascular system, 2013 References This lecture Mark s Basic Medical Biochemistry, 4 th ed., p. 890-891 Hand-out Why is this topic important? Heart failure
Metabolism of cardiac muscle Dr. Mamoun Ahram Cardiovascular system, 2013 References This lecture Mark s Basic Medical Biochemistry, 4 th ed., p. 890-891 Hand-out Why is this topic important? Heart failure
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature7221 Brown fat selective genes 12 1 Control Q-RT-PCR (% of Control) 8 6 4 2 Ntrk3 Cox7a1 Cox8b Cox5b ATPase b2 ATPase f1a1 Sirt3 ERRα Elovl3/Cig3 PPARα Zic1 Supplementary Figure S1. stimulates
doi: 1.138/nature7221 Brown fat selective genes 12 1 Control Q-RT-PCR (% of Control) 8 6 4 2 Ntrk3 Cox7a1 Cox8b Cox5b ATPase b2 ATPase f1a1 Sirt3 ERRα Elovl3/Cig3 PPARα Zic1 Supplementary Figure S1. stimulates
1.5 ASK1KO fed. fasted 16 hrs w/o water. Fed. 4th. 4th WT ASK1KO N=29, 11(WT), ,5(ASK1KO) ASK1KO ASK1KO **** Time [h]
![1.5 ASK1KO fed. fasted 16 hrs w/o water. Fed. 4th. 4th WT ASK1KO N=29, 11(WT), ,5(ASK1KO) ASK1KO ASK1KO **** Time [h] 1.5 ASK1KO fed. fasted 16 hrs w/o water. Fed. 4th. 4th WT ASK1KO N=29, 11(WT), ,5(ASK1KO) ASK1KO ASK1KO **** Time [h]](/thumbs/87/97258189.jpg) 7: 13: 19: 1: 7: 151117 a 151117 4th 4th b c RQ.95 KO.9.85.8.75.7 light dark light dark.65 7: 19: 7: 19: 7: Means ± SEM, N=6 RQ 1..9.8.7.6.6 KO CL (-) CL (+) ibat weight ratio (/body weight) [%].5.4.3.2.1
7: 13: 19: 1: 7: 151117 a 151117 4th 4th b c RQ.95 KO.9.85.8.75.7 light dark light dark.65 7: 19: 7: 19: 7: Means ± SEM, N=6 RQ 1..9.8.7.6.6 KO CL (-) CL (+) ibat weight ratio (/body weight) [%].5.4.3.2.1
Supplemental Information Supplementary Table 1. Tph1+/+ Tph1 / Analyte Supplementary Table 2. Tissue Vehicle LP value
 Supplemental Information Supplementary Table. Urinary and adipose tissue catecholamines in Tph +/+ and Tph / mice fed a high fat diet for weeks. Tph +/+ Tph / Analyte ewat ibat ewat ibat Urine (ng/ml)
Supplemental Information Supplementary Table. Urinary and adipose tissue catecholamines in Tph +/+ and Tph / mice fed a high fat diet for weeks. Tph +/+ Tph / Analyte ewat ibat ewat ibat Urine (ng/ml)
MITOCHONDRIA LECTURES OVERVIEW
 1 MITOCHONDRIA LECTURES OVERVIEW A. MITOCHONDRIA LECTURES OVERVIEW Mitochondrial Structure The arrangement of membranes: distinct inner and outer membranes, The location of ATPase, DNA and ribosomes The
1 MITOCHONDRIA LECTURES OVERVIEW A. MITOCHONDRIA LECTURES OVERVIEW Mitochondrial Structure The arrangement of membranes: distinct inner and outer membranes, The location of ATPase, DNA and ribosomes The
doi: /nature10642
 doi:10.1038/nature10642 Supplementary Fig. 1. Citric acid cycle (CAC) metabolism in WT 143B and CYTB 143B cells. a, Proliferation of WT 143B and CYTB 143B cells. Doubling times were 28±1 and 33±2 hrs for
doi:10.1038/nature10642 Supplementary Fig. 1. Citric acid cycle (CAC) metabolism in WT 143B and CYTB 143B cells. a, Proliferation of WT 143B and CYTB 143B cells. Doubling times were 28±1 and 33±2 hrs for
A microrna-34a/fgf21 Regulatory Axis and Browning of White Fat
 A microrna-34a/fgf21 Regulatory Axis and Browning of White Fat Jongsook Kim Kemper, Ph.D Department of Molecular and Integrative Physiology, University of Illinois at Urbana-Champaign, USA 213 International
A microrna-34a/fgf21 Regulatory Axis and Browning of White Fat Jongsook Kim Kemper, Ph.D Department of Molecular and Integrative Physiology, University of Illinois at Urbana-Champaign, USA 213 International
Mouse Meda-4 : chromosome 5G bp. EST (547bp) _at. 5 -Meda4 inner race (~1.8Kb)
 Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
Supporting Information Table of content
 Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
Males- Western Diet WT KO Age (wks) Females- Western Diet WT KO Age (wks)
 Relative Arv1 mrna Adrenal 33.48 +/- 6.2 Skeletal Muscle 22.4 +/- 4.93 Liver 6.41 +/- 1.48 Heart 5.1 +/- 2.3 Brain 4.98 +/- 2.11 Ovary 4.68 +/- 2.21 Kidney 3.98 +/-.39 Lung 2.15 +/-.6 Inguinal Subcutaneous
Relative Arv1 mrna Adrenal 33.48 +/- 6.2 Skeletal Muscle 22.4 +/- 4.93 Liver 6.41 +/- 1.48 Heart 5.1 +/- 2.3 Brain 4.98 +/- 2.11 Ovary 4.68 +/- 2.21 Kidney 3.98 +/-.39 Lung 2.15 +/-.6 Inguinal Subcutaneous
SUPPLEMENTARY INFORMATION
 -. -. SUPPLEMENTARY INFORMATION DOI: 1.1/ncb86 a WAT-1 WAT- BAT-1 BAT- sk-muscle-1 sk-muscle- mir-133b mir-133a mir-6 mir-378 mir-1 mir-85 mir-378 mir-6a mir-18 mir-133a mir- mir- mir-341 mir-196a mir-17
-. -. SUPPLEMENTARY INFORMATION DOI: 1.1/ncb86 a WAT-1 WAT- BAT-1 BAT- sk-muscle-1 sk-muscle- mir-133b mir-133a mir-6 mir-378 mir-1 mir-85 mir-378 mir-6a mir-18 mir-133a mir- mir- mir-341 mir-196a mir-17
General Laboratory methods Plasma analysis: Gene Expression Analysis: Immunoblot analysis: Immunohistochemistry:
 General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
HSP72 HSP90. Quadriceps Muscle. MEF2c MyoD1 MyoG Myf5 Hsf1 Hsp GLUT4/GAPDH (AU)
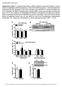 Supplementary Figure 1. Impaired insulin action in HSP72 deficient muscle and myotubes in culture cannot be explained by altered myogenesis or reduced total GLUT4 expression. Genes associated with myogenesis
Supplementary Figure 1. Impaired insulin action in HSP72 deficient muscle and myotubes in culture cannot be explained by altered myogenesis or reduced total GLUT4 expression. Genes associated with myogenesis
Supplementary Table 1 Gene clone ID for ShRNA-mediated gene silencing TNFα downstream signals in in vitro Symbol Gene ID RefSeqID Clone ID
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 Supplementary Table 1 Gene clone ID for ShRNA-mediated gene silencing TNFα downstream
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 Supplementary Table 1 Gene clone ID for ShRNA-mediated gene silencing TNFα downstream
Lehninger 5 th ed. Chapter 17
 Lehninger 5 th ed. Chapter 17 December 26, 2010 Prof. Shimon Schuldiner Email: Shimon.Schuldiner@huji.ac.il Phone: 6585992 CHAPTER 17 Fatty Acid Catabolism Key topics: How fats are digested in animals
Lehninger 5 th ed. Chapter 17 December 26, 2010 Prof. Shimon Schuldiner Email: Shimon.Schuldiner@huji.ac.il Phone: 6585992 CHAPTER 17 Fatty Acid Catabolism Key topics: How fats are digested in animals
Supplementary Table 1.
 Supplementary Table 1. Expression of genes involved in brown fat differentiation in WAT of db/db mice treated with HDAC inhibitors. Data are expressed as fold change (FC) versus control. symbol FC SAHA
Supplementary Table 1. Expression of genes involved in brown fat differentiation in WAT of db/db mice treated with HDAC inhibitors. Data are expressed as fold change (FC) versus control. symbol FC SAHA
Citric acid cycle and respiratory chain. Pavla Balínová
 Citric acid cycle and respiratory chain Pavla Balínová Mitochondria Structure of mitochondria: Outer membrane Inner membrane (folded) Matrix space (mtdna, ribosomes, enzymes of CAC, β-oxidation of FA,
Citric acid cycle and respiratory chain Pavla Balínová Mitochondria Structure of mitochondria: Outer membrane Inner membrane (folded) Matrix space (mtdna, ribosomes, enzymes of CAC, β-oxidation of FA,
Supplementary Figure 1. DNA methylation of the adiponectin promoter R1, Pparg2, and Tnfa promoter in adipocytes is not affected by obesity.
 Supplementary Figure 1. DNA methylation of the adiponectin promoter R1, Pparg2, and Tnfa promoter in adipocytes is not affected by obesity. (a) Relative amounts of adiponectin, Ppar 2, C/ebp, and Tnf mrna
Supplementary Figure 1. DNA methylation of the adiponectin promoter R1, Pparg2, and Tnfa promoter in adipocytes is not affected by obesity. (a) Relative amounts of adiponectin, Ppar 2, C/ebp, and Tnf mrna
number Done by Corrected by Doctor Faisal Al-Khatibe
 number 24 Done by Mohammed tarabieh Corrected by Doctor Faisal Al-Khatibe 1 P a g e *Please look over the previous sheet about fatty acid synthesis **Oxidation(degradation) of fatty acids, occurs in the
number 24 Done by Mohammed tarabieh Corrected by Doctor Faisal Al-Khatibe 1 P a g e *Please look over the previous sheet about fatty acid synthesis **Oxidation(degradation) of fatty acids, occurs in the
Immuno_BeadChip_ (Illumina Inc., San Diego, CA, USA) according to the
 Supplementary Methods Genotyping of the Milwaukee cohort DNA samples were genotyped for 196,524 markers using the Human Immuno_BeadChip_1149691 (Illumina Inc., San Diego, CA, USA) according to the manufacture's
Supplementary Methods Genotyping of the Milwaukee cohort DNA samples were genotyped for 196,524 markers using the Human Immuno_BeadChip_1149691 (Illumina Inc., San Diego, CA, USA) according to the manufacture's
CELL BIOLOGY - CLUTCH CH AEROBIC RESPIRATION.
 !! www.clutchprep.com CONCEPT: OVERVIEW OF AEROBIC RESPIRATION Cellular respiration is a series of reactions involving electron transfers to breakdown molecules for (ATP) 1. Glycolytic pathway: Glycolysis
!! www.clutchprep.com CONCEPT: OVERVIEW OF AEROBIC RESPIRATION Cellular respiration is a series of reactions involving electron transfers to breakdown molecules for (ATP) 1. Glycolytic pathway: Glycolysis
Mitochondrial Fatty Acid Oxidation Deficiencies Prof. Niels Gregersen
 Mitochondrial Fatty Acid Oxidation Deficiencies Research Unit for Molecular Medicine Clinical Institute Aarhus University Hospital and Faculty of Health Sciences Aarhus University, Aarhus, Denmark 1 Menu
Mitochondrial Fatty Acid Oxidation Deficiencies Research Unit for Molecular Medicine Clinical Institute Aarhus University Hospital and Faculty of Health Sciences Aarhus University, Aarhus, Denmark 1 Menu
SUPPLEMENTARY DATA. Nature Medicine: doi: /nm.4171
 SUPPLEMENTARY DATA Supplementary Figure 1 a b c PF %Change - -4-6 Body weight Lean mass Body fat Tissue weight (g).4.3.2.1. PF GC iwat awat BAT PF d e f g week 2 week 3 NEFA (mmol/l) 1..5. PF phsl (Ser565)
SUPPLEMENTARY DATA Supplementary Figure 1 a b c PF %Change - -4-6 Body weight Lean mass Body fat Tissue weight (g).4.3.2.1. PF GC iwat awat BAT PF d e f g week 2 week 3 NEFA (mmol/l) 1..5. PF phsl (Ser565)
2-more complex molecules (fatty acyl esters) as triacylglycerols.
 ** Fatty acids exist in two forms:- 1-free fatty acids (unesterified) 2-more complex molecules (fatty acyl esters) as triacylglycerols. ** most tissues might use fatty acids as source of energy during
** Fatty acids exist in two forms:- 1-free fatty acids (unesterified) 2-more complex molecules (fatty acyl esters) as triacylglycerols. ** most tissues might use fatty acids as source of energy during
Implications of mitochondrial skeletal muscle metabolism on diabetes and obesity before and after weight loss
 GG2 Implications of mitochondrial skeletal muscle metabolism on diabetes and obesity before and after weight loss Dr Giacomo Gastaldi CHRU Montpellier Folie 1 GG2 19.10.2009 GG_PC; 12.10.2009 Plan Introduction
GG2 Implications of mitochondrial skeletal muscle metabolism on diabetes and obesity before and after weight loss Dr Giacomo Gastaldi CHRU Montpellier Folie 1 GG2 19.10.2009 GG_PC; 12.10.2009 Plan Introduction
Tala Saleh. Razi Kittaneh ... Nayef Karadsheh
 Tala Saleh Razi Kittaneh... Nayef Karadsheh β-oxidation of Fatty Acids The oxidation of fatty acids occurs in 3 steps: Step 1: Activation of the Fatty acid FA + HS-CoA + ATP FA-CoA + AMP + PPi - The fatty
Tala Saleh Razi Kittaneh... Nayef Karadsheh β-oxidation of Fatty Acids The oxidation of fatty acids occurs in 3 steps: Step 1: Activation of the Fatty acid FA + HS-CoA + ATP FA-CoA + AMP + PPi - The fatty
Can physical exercise and exercise mimetics improve metabolic health in humans?
 Can physical exercise and exercise mimetics improve metabolic health in humans? Patrick Schrauwen, PhD NUTRIM school for Nutrition and Translational Research in Metabolism Department of Human Biology,
Can physical exercise and exercise mimetics improve metabolic health in humans? Patrick Schrauwen, PhD NUTRIM school for Nutrition and Translational Research in Metabolism Department of Human Biology,
Nature Immunology: doi: /ni Supplementary Figure 1
 Supplementary Figure 1 Fatty acid oxidation is emphasized in 1 macrophages compared with that in macrophages. Gene expression of mitochondrial OXPHOS (Atp5j, Cox4i1, Uqcrc1/2, Ndufs1, Sdhb) and β-oxidation
Supplementary Figure 1 Fatty acid oxidation is emphasized in 1 macrophages compared with that in macrophages. Gene expression of mitochondrial OXPHOS (Atp5j, Cox4i1, Uqcrc1/2, Ndufs1, Sdhb) and β-oxidation
Supplemental Table 1. Plasma NEFA and liver triglyceride levels in ap2-hif1ako and ap2-hif2ako mice under control and high fat diets.
 Supplemental Table 1. Plasma NEFA and liver triglyceride levels in Hif1aKO and Hif2aKO mice under control and high fat diets. Hif1a (n=6) Hif1aK O (n=6) Hif2a Hif2aK O Hif1a (n=5) Hif1aKO (n=5) Hif2a Hif2aK
Supplemental Table 1. Plasma NEFA and liver triglyceride levels in Hif1aKO and Hif2aKO mice under control and high fat diets. Hif1a (n=6) Hif1aK O (n=6) Hif2a Hif2aK O Hif1a (n=5) Hif1aKO (n=5) Hif2a Hif2aK
Figure 1. Effects of FGF21 on adipose tissue. (A) Representative histological. findings of epididymal adipose tissue (B) mrna expression of
 SUPPLEMENTAL MATERIAL EN-12-2276 Figure 1. Effects of FGF21 on adipose tissue. (A) Representative histological findings of epididymal adipose tissue (B) mrna expression of adipocytokines in adipose tissue.
SUPPLEMENTAL MATERIAL EN-12-2276 Figure 1. Effects of FGF21 on adipose tissue. (A) Representative histological findings of epididymal adipose tissue (B) mrna expression of adipocytokines in adipose tissue.
Electron Transport Chain and Oxidative phosphorylation
 Electron Transport Chain and Oxidative phosphorylation So far we have discussed the catabolism involving oxidation of 6 carbons of glucose to CO 2 via glycolysis and CAC without any oxygen molecule directly
Electron Transport Chain and Oxidative phosphorylation So far we have discussed the catabolism involving oxidation of 6 carbons of glucose to CO 2 via glycolysis and CAC without any oxygen molecule directly
Unit of Muscular and Neurodegenerative Diseases; G. Gaslini Institute; Genova, Italy
 Research paper Cell Cycle 10:23, 4047-4064; December 1, 2011; 2011 Landes Bioscience Hyperactivation of oxidative mitochondrial metabolism in epithelial cancer cells in situ Visualizing the therapeutic
Research paper Cell Cycle 10:23, 4047-4064; December 1, 2011; 2011 Landes Bioscience Hyperactivation of oxidative mitochondrial metabolism in epithelial cancer cells in situ Visualizing the therapeutic
Chapter 14. Energy conversion: Energy & Behavior
 Chapter 14 Energy conversion: Energy & Behavior Why do you Eat and Breath? To generate ATP Foods, Oxygen, and Mitochodria Cells Obtain Energy by the Oxidation of Organic Molecules Food making ATP making
Chapter 14 Energy conversion: Energy & Behavior Why do you Eat and Breath? To generate ATP Foods, Oxygen, and Mitochodria Cells Obtain Energy by the Oxidation of Organic Molecules Food making ATP making
BCH 4054 Spring 2001 Chapter 24 Lecture Notes
 BCH 4054 Spring 2001 Chapter 24 Lecture Notes 1 Chapter 24 Fatty Acid Catabolism 2 Fatty Acids as Energy Source Triglycerides yield 37 kj/g dry weight Protein 17 kj/g Glycogen 16 kj/g (even less wet weight)
BCH 4054 Spring 2001 Chapter 24 Lecture Notes 1 Chapter 24 Fatty Acid Catabolism 2 Fatty Acids as Energy Source Triglycerides yield 37 kj/g dry weight Protein 17 kj/g Glycogen 16 kj/g (even less wet weight)
LIPID METABOLISM
 LIPID METABOLISM LIPOGENESIS LIPOGENESIS LIPOGENESIS FATTY ACID SYNTHESIS DE NOVO FFA in the blood come from :- (a) Dietary fat (b) Dietary carbohydrate/protein in excess of need FA TAG Site of synthesis:-
LIPID METABOLISM LIPOGENESIS LIPOGENESIS LIPOGENESIS FATTY ACID SYNTHESIS DE NOVO FFA in the blood come from :- (a) Dietary fat (b) Dietary carbohydrate/protein in excess of need FA TAG Site of synthesis:-
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
Synthesis and degradation of fatty acids Martina Srbová
 Synthesis and degradation of fatty acids Martina Srbová martina.srbova@lfmotol.cuni.cz Fatty acids (FA) mostly an even number of carbon atoms and linear chain in esterified form as component of lipids
Synthesis and degradation of fatty acids Martina Srbová martina.srbova@lfmotol.cuni.cz Fatty acids (FA) mostly an even number of carbon atoms and linear chain in esterified form as component of lipids
Supplemental Information. Metabolic Maturation during Muscle Stem Cell. Differentiation Is Achieved by mir-1/133a-mediated
 Cell Metabolism, Volume 27 Supplemental Information Metabolic Maturation during Muscle Stem Cell Differentiation Is Achieved by mir-1/133a-mediated Inhibition of the Dlk1-Dio3 Mega Gene Cluster Stas Wüst,
Cell Metabolism, Volume 27 Supplemental Information Metabolic Maturation during Muscle Stem Cell Differentiation Is Achieved by mir-1/133a-mediated Inhibition of the Dlk1-Dio3 Mega Gene Cluster Stas Wüst,
Supplemental Information. Intermittent Fasting Promotes. White Adipose Browning and Decreases Obesity. by Shaping the Gut Microbiota
 Cell Metabolism, Volume 26 Supplemental Information Intermittent Fasting Promotes White Adipose Browning and Decreases Obesity by Shaping the Gut Microbiota Guolin Li, Cen Xie, Siyu Lu, Robert G. Nichols,
Cell Metabolism, Volume 26 Supplemental Information Intermittent Fasting Promotes White Adipose Browning and Decreases Obesity by Shaping the Gut Microbiota Guolin Li, Cen Xie, Siyu Lu, Robert G. Nichols,
Supplementary Figure 1.
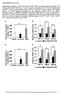 Supplementary Figure 1. FGF21 does not exert direct effects on hepatic glucose production. The liver explants from C57BL/6J mice (A, B) or primary rat hepatocytes (C, D) were incubated with rmfgf21 (2
Supplementary Figure 1. FGF21 does not exert direct effects on hepatic glucose production. The liver explants from C57BL/6J mice (A, B) or primary rat hepatocytes (C, D) were incubated with rmfgf21 (2
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/264/rs4/dc1 Supplementary Materials for A Systems Approach for Decoding Mitochondrial Retrograde Signaling Pathways Sehyun Chae, Byung Yong Ahn, Kyunghee Byun,
www.sciencesignaling.org/cgi/content/full/6/264/rs4/dc1 Supplementary Materials for A Systems Approach for Decoding Mitochondrial Retrograde Signaling Pathways Sehyun Chae, Byung Yong Ahn, Kyunghee Byun,
f(x) = x R² = RPKM (M8.MXB) f(x) = x E-014 R² = 1 RPKM (M31.
 14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
14 12 f(x) = 1.633186874x - 21.46732234 R² =.995616541 RPKM (M8.MXA) 1 8 6 4 2 2 4 6 8 1 12 14 RPKM (M8.MXB) 14 12 f(x) =.821767782x - 4.192595677497E-14 R² = 1 RPKM (M31.XA) 1 8 6 4 2 2 4 6 8 1 12 14
OVERVIEW M ET AB OL IS M OF FR EE FA TT Y AC ID S
 LIPOLYSIS LIPOLYSIS OVERVIEW CATABOLISM OF FREE FATTY ACIDS Nonesterified fatty acids Source:- (a) breakdown of TAG in adipose tissue (b) action of Lipoprotein lipase on plasma TAG Combined with Albumin
LIPOLYSIS LIPOLYSIS OVERVIEW CATABOLISM OF FREE FATTY ACIDS Nonesterified fatty acids Source:- (a) breakdown of TAG in adipose tissue (b) action of Lipoprotein lipase on plasma TAG Combined with Albumin
Dietary Lipid Utilization by Haddock (Melanogrammus aeglefinus)
 Dietary Lipid Utilization by Haddock (Melanogrammus aeglefinus) Santosh P. Lall & Dominic A. Nanton National Research Council of Canada Halifax, Canada vis, California ne 23, 2003 Body Components of Wild
Dietary Lipid Utilization by Haddock (Melanogrammus aeglefinus) Santosh P. Lall & Dominic A. Nanton National Research Council of Canada Halifax, Canada vis, California ne 23, 2003 Body Components of Wild
Expanded View Figures
 Expanded View Figures A B C D E F G H I J K L Figure EV1. The dysregulated lipid metabolic phenotype of mouse models of metabolic dysfunction is most pronounced in the fasted state. A L Male 12-weeks-old
Expanded View Figures A B C D E F G H I J K L Figure EV1. The dysregulated lipid metabolic phenotype of mouse models of metabolic dysfunction is most pronounced in the fasted state. A L Male 12-weeks-old
Lipid metabolism. Degradation and biosynthesis of fatty acids Ketone bodies
 Lipid metabolism Degradation and biosynthesis of fatty acids Ketone bodies Fatty acids (FA) primary fuel molecules in the fat category main use is for long-term energy storage high level of energy storage:
Lipid metabolism Degradation and biosynthesis of fatty acids Ketone bodies Fatty acids (FA) primary fuel molecules in the fat category main use is for long-term energy storage high level of energy storage:
Lipid Metabolism. Catabolism Overview
 Lipid Metabolism Pratt & Cornely, Chapter 17 Catabolism Overview Lipids as a fuel source from diet Beta oxidation Mechanism ATP production Ketone bodies as fuel 1 High energy More reduced Little water
Lipid Metabolism Pratt & Cornely, Chapter 17 Catabolism Overview Lipids as a fuel source from diet Beta oxidation Mechanism ATP production Ketone bodies as fuel 1 High energy More reduced Little water
PHM142 Energy Production + The Mitochondria
 PHM142 Energy Production + The Mitochondria 1 The Endosymbiont Theory of Mitochondiral Evolution 1970: Lynn Margulis Origin of Eukaryotic Cells Endosymbiant Theory: the mitochondria evolved from free-living
PHM142 Energy Production + The Mitochondria 1 The Endosymbiont Theory of Mitochondiral Evolution 1970: Lynn Margulis Origin of Eukaryotic Cells Endosymbiant Theory: the mitochondria evolved from free-living
3-Thia Fatty Acids A New Generation of Functional Lipids?
 Conference on Food Structure and Food Quality 3-Thia Fatty Acids A New Generation of Functional Lipids? Rolf K. Berge rolf.berge@med.uib.no Fatty acids- Essential cellular metabolites Concentrations must
Conference on Food Structure and Food Quality 3-Thia Fatty Acids A New Generation of Functional Lipids? Rolf K. Berge rolf.berge@med.uib.no Fatty acids- Essential cellular metabolites Concentrations must
Integrative Metabolism: Significance
 Integrative Metabolism: Significance Energy Containing Nutrients Carbohydrates Fats Proteins Catabolism Energy Depleted End Products H 2 O NH 3 ADP + Pi NAD + NADP + FAD + Pi NADH+H + NADPH+H + FADH2 Cell
Integrative Metabolism: Significance Energy Containing Nutrients Carbohydrates Fats Proteins Catabolism Energy Depleted End Products H 2 O NH 3 ADP + Pi NAD + NADP + FAD + Pi NADH+H + NADPH+H + FADH2 Cell
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
Fuel the Failing Heart: glucose or fatty acids? Rong Tian, MD, PhD Mitochondria and Metabolism Center University of Washington, Seattle
 Fuel the Failing Heart: glucose or fatty acids? Rong Tian, MD, PhD Mitochondria and Metabolism Center University of Washington, Seattle Metabolic Remodeling: Fatty Acids Carbohydrates PCr/ATP Glucose vs.
Fuel the Failing Heart: glucose or fatty acids? Rong Tian, MD, PhD Mitochondria and Metabolism Center University of Washington, Seattle Metabolic Remodeling: Fatty Acids Carbohydrates PCr/ATP Glucose vs.
FSP27 contributes to efficient energy storage in murine white adipocytes by promoting the formation of unilocular lipid droplets
 Research article Related Commentary, page 2693 FSP27 contributes to efficient energy storage in murine white adipocytes by promoting the formation of unilocular lipid droplets Naonobu Nishino, 1 Yoshikazu
Research article Related Commentary, page 2693 FSP27 contributes to efficient energy storage in murine white adipocytes by promoting the formation of unilocular lipid droplets Naonobu Nishino, 1 Yoshikazu
Hmgcoar AGCTTGCCCGAATTGTATGTG TCTGTTGTAACCATGTGACTTC. Cyp7α GGGATTGCTGTGGTAGTGAGC GGTATGGAATCAACCCGTTGTC
 Supplement Table I: primers for Real Time RT-PCR Gene Foward Reverse Hmgcoar AGCTTGCCCGAATTGTATGTG TCTGTTGTAACCATGTGACTTC Cyp7α GGGATTGCTGTGGTAGTGAGC GGTATGGAATCAACCCGTTGTC Cyp27a1 GTGGTCTTATTGGGTACTTGC
Supplement Table I: primers for Real Time RT-PCR Gene Foward Reverse Hmgcoar AGCTTGCCCGAATTGTATGTG TCTGTTGTAACCATGTGACTTC Cyp7α GGGATTGCTGTGGTAGTGAGC GGTATGGAATCAACCCGTTGTC Cyp27a1 GTGGTCTTATTGGGTACTTGC
A Central Role of MG53 in Metabolic Syndrome. and Type-2 Diabetes
 A Central Role of MG53 in Metabolic Syndrome and Type-2 Diabetes Yan Zhang, Chunmei Cao, Rui-Ping Xiao Institute of Molecular Medicine (IMM) Peking University, Beijing, China Accelerated Aging in China
A Central Role of MG53 in Metabolic Syndrome and Type-2 Diabetes Yan Zhang, Chunmei Cao, Rui-Ping Xiao Institute of Molecular Medicine (IMM) Peking University, Beijing, China Accelerated Aging in China
Fatty acid breakdown
 Fatty acids contain a long hydrocarbon chain and a terminal carboxylate group. Most contain between 14 and 24 carbon atoms. The chains may be saturated or contain double bonds. The complete oxidation of
Fatty acids contain a long hydrocarbon chain and a terminal carboxylate group. Most contain between 14 and 24 carbon atoms. The chains may be saturated or contain double bonds. The complete oxidation of
Voet Biochemistry 3e John Wiley & Sons, Inc.
 * * Voet Biochemistry 3e Lipid Metabolism Part I: (Chap. 25, sec.1-3) Glucose C 6 H 12 O 6 + 6 O 2 6 CO 2 + 6 H 2 O G o = -2823 kj/mol Fats (palmitic acid) C 16 H 32 O 2 + 23 O 2 16 CO 2 + 16 H 2 O G o
* * Voet Biochemistry 3e Lipid Metabolism Part I: (Chap. 25, sec.1-3) Glucose C 6 H 12 O 6 + 6 O 2 6 CO 2 + 6 H 2 O G o = -2823 kj/mol Fats (palmitic acid) C 16 H 32 O 2 + 23 O 2 16 CO 2 + 16 H 2 O G o
Biology 638 Biochemistry II Exam-2
 Biology 638 Biochemistry II Exam-2 Biol 638, Exam-2 (Code-1) 1. Assume that 16 glucose molecules enter into a liver cell and are attached to a liner glycogen one by one. Later, this glycogen is broken-down
Biology 638 Biochemistry II Exam-2 Biol 638, Exam-2 (Code-1) 1. Assume that 16 glucose molecules enter into a liver cell and are attached to a liner glycogen one by one. Later, this glycogen is broken-down
X P. Supplementary Figure 1. Nature Medicine: doi: /nm Nilotinib LSK LT-HSC. Cytoplasm. Cytoplasm. Nucleus. Nucleus
 a b c Supplementary Figure 1 c-kit-apc-eflu780 Lin-FITC Flt3-Linc-Kit-APC-eflu780 LSK Sca-1-PE-Cy7 d e f CD48-APC LT-HSC CD150-PerCP-cy5.5 g h i j Cytoplasm RCC1 X Exp 5 mir 126 SPRED1 SPRED1 RAN P SPRED1
a b c Supplementary Figure 1 c-kit-apc-eflu780 Lin-FITC Flt3-Linc-Kit-APC-eflu780 LSK Sca-1-PE-Cy7 d e f CD48-APC LT-HSC CD150-PerCP-cy5.5 g h i j Cytoplasm RCC1 X Exp 5 mir 126 SPRED1 SPRED1 RAN P SPRED1
Supplemental Fig. 1. Relative mrna Expression. Relative mrna Expression WT KO WT KO RT 4 0 C
 Supplemental Fig. 1 A 1.5 1..5 Hdac11 (ibat) n=4 n=4 n=4 n=4 n=4 n=4 n=4 n=4 WT KO WT KO WT KO WT KO RT 4 C RT 4 C Supplemental Figure 1. Hdac11 mrna is undetectable in KO adipose tissue. Quantitative
Supplemental Fig. 1 A 1.5 1..5 Hdac11 (ibat) n=4 n=4 n=4 n=4 n=4 n=4 n=4 n=4 WT KO WT KO WT KO WT KO RT 4 C RT 4 C Supplemental Figure 1. Hdac11 mrna is undetectable in KO adipose tissue. Quantitative
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
Fatty Acid Degradation. Catabolism Overview. TAG and FA 11/11/2015. Chapter 27, Stryer Short Course. Lipids as a fuel source diet Beta oxidation
 Fatty Acid Degradation Chapter 27, Stryer Short Course Catabolism verview Lipids as a fuel source diet Beta oxidation saturated Unsaturated dd chain Ketone bodies as fuel Physiology High energy More reduced
Fatty Acid Degradation Chapter 27, Stryer Short Course Catabolism verview Lipids as a fuel source diet Beta oxidation saturated Unsaturated dd chain Ketone bodies as fuel Physiology High energy More reduced
Fatty acids synthesis
 Fatty acids synthesis The synthesis start from Acetyl COA the first step requires ATP + reducing power NADPH! even though the oxidation and synthesis are different pathways but from chemical part of view
Fatty acids synthesis The synthesis start from Acetyl COA the first step requires ATP + reducing power NADPH! even though the oxidation and synthesis are different pathways but from chemical part of view
Diosgenin, antagonism of LXRs 36 DNA microarray, sesame seed lignan regulation of liver fatty acid metabolism gene expression 12 18, 22, 23
 Subject Index ACC, see Acyl-CoA carboxylase 1 -Acetoxychavicol acetate, inducible nitric oxide synthase suppression in cancer chemoprevention 199, 200 Acetylene carotenoids, anti-inflammatory activity
Subject Index ACC, see Acyl-CoA carboxylase 1 -Acetoxychavicol acetate, inducible nitric oxide synthase suppression in cancer chemoprevention 199, 200 Acetylene carotenoids, anti-inflammatory activity
Biochemistry - I SPRING Mondays and Wednesdays 9:30-10:45 AM (MR-1307) Lectures Based on Profs. Kevin Gardner & Reza Khayat
 Biochemistry - I Mondays and Wednesdays 9:30-10:45 AM (MR-1307) SPRING 2017 Lectures 21-22 Based on Profs. Kevin Gardner & Reza Khayat 1 Outline Vertebrate processing of dietary lipids Mobilization of
Biochemistry - I Mondays and Wednesdays 9:30-10:45 AM (MR-1307) SPRING 2017 Lectures 21-22 Based on Profs. Kevin Gardner & Reza Khayat 1 Outline Vertebrate processing of dietary lipids Mobilization of
Gene Polymorphisms and Carbohydrate Diets. James M. Ntambi Ph.D
 Gene Polymorphisms and Carbohydrate Diets James M. Ntambi Ph.D Fatty Acids that Flux into Tissue Lipids are from Dietary Sources or are Made De novo from Glucose or Fructose Glucose Fructose Acetyl-CoA
Gene Polymorphisms and Carbohydrate Diets James M. Ntambi Ph.D Fatty Acids that Flux into Tissue Lipids are from Dietary Sources or are Made De novo from Glucose or Fructose Glucose Fructose Acetyl-CoA
Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna
 Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna targeted for hamster PAQR3. At 24 h after the transfection,
Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna targeted for hamster PAQR3. At 24 h after the transfection,
HIV VPR alters fat metabolism. Dorothy E Lewis PhD/Ashok Balasubramanyam MD
 HIV VPR alters fat metabolism Dorothy E Lewis PhD/Ashok Balasubramanyam MD Old Dogma for HIV associated lipodystrophy Differentiation Block (PI) Lipoatrophy Apoptosis (NRTI) Stem cell Preadipocyte Adipocyte
HIV VPR alters fat metabolism Dorothy E Lewis PhD/Ashok Balasubramanyam MD Old Dogma for HIV associated lipodystrophy Differentiation Block (PI) Lipoatrophy Apoptosis (NRTI) Stem cell Preadipocyte Adipocyte
SUPPLEMENTARY DATA. Supplementary Table 1. Primer sequences for qrt-pcr
 Supplementary Table 1. Primer sequences for qrt-pcr Gene PRDM16 UCP1 PGC1α Dio2 Elovl3 Cidea Cox8b PPARγ AP2 mttfam CyCs Nampt NRF1 16s-rRNA Hexokinase 2, intron 9 β-actin Primer Sequences 5'-CCA CCA GCG
Supplementary Table 1. Primer sequences for qrt-pcr Gene PRDM16 UCP1 PGC1α Dio2 Elovl3 Cidea Cox8b PPARγ AP2 mttfam CyCs Nampt NRF1 16s-rRNA Hexokinase 2, intron 9 β-actin Primer Sequences 5'-CCA CCA GCG
Supplementary Table 2. Plasma lipid profiles in wild type and mutant female mice submitted to a HFD for 12 weeks wt ERα -/- AF-1 0 AF-2 0
 Supplementary Table 1. List of specific primers used for gene expression analysis. Genes Primer forward Primer reverse Hprt GCAGTACAGCCCCAAAATGG AACAAAGTCTGGCCTGTATCCA Srebp-1c GGAAGCTGTCGGGGTAGCGTC CATGTCTTCAAATGTGCAATCCAT
Supplementary Table 1. List of specific primers used for gene expression analysis. Genes Primer forward Primer reverse Hprt GCAGTACAGCCCCAAAATGG AACAAAGTCTGGCCTGTATCCA Srebp-1c GGAAGCTGTCGGGGTAGCGTC CATGTCTTCAAATGTGCAATCCAT
Nafith Abu Tarboush DDS, MSc, PhD
 Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush OMM: permeable to small molecules (MW
Nafith Abu Tarboush DDS, MSc, PhD natarboush@ju.edu.jo www.facebook.com/natarboush OMM: permeable to small molecules (MW
AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination
 AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination Supplementary Figure 1. Generation of the adult-onset, liver-specific GH receptor knock-down (alivghrkd, Kd) mouse
AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination Supplementary Figure 1. Generation of the adult-onset, liver-specific GH receptor knock-down (alivghrkd, Kd) mouse
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was
 Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Lecture 29: Membrane Transport and metabolism
 Chem*3560 Lecture 29: Membrane Transport and metabolism Insulin controls glucose uptake Adipose tissue and muscles contain a passive glucose transporter GluT4 which takes up glucose from blood. (This is
Chem*3560 Lecture 29: Membrane Transport and metabolism Insulin controls glucose uptake Adipose tissue and muscles contain a passive glucose transporter GluT4 which takes up glucose from blood. (This is
Mitochondria and ATP Synthesis
 Mitochondria and ATP Synthesis Mitochondria and ATP Synthesis 1. Mitochondria are sites of ATP synthesis in cells. 2. ATP is used to do work; i.e. ATP is an energy source. 3. ATP hydrolysis releases energy
Mitochondria and ATP Synthesis Mitochondria and ATP Synthesis 1. Mitochondria are sites of ATP synthesis in cells. 2. ATP is used to do work; i.e. ATP is an energy source. 3. ATP hydrolysis releases energy
Control 7 d cold 7 d CL
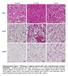 Control 7 d cold 7 d ibat iwat gwat Supplementary Figure 1. Histology of adipose tissues after cold or 3-adrenergic receptor stimulation. C57BL/6J wild-type mice were housed at 4 C or injected daily with
Control 7 d cold 7 d ibat iwat gwat Supplementary Figure 1. Histology of adipose tissues after cold or 3-adrenergic receptor stimulation. C57BL/6J wild-type mice were housed at 4 C or injected daily with
SUPPLEMENTARY DATA Supplementary Figure 1. Body weight and fat mass of AdicerKO mice.
 SUPPLEMENTARY DATA Supplementary Figure 1. Body weight and fat mass of AdicerKO mice. Twelve week old mice were subjected to ad libitum (AL) or dietary restriction (DR) regimens for three months. (A) Body
SUPPLEMENTARY DATA Supplementary Figure 1. Body weight and fat mass of AdicerKO mice. Twelve week old mice were subjected to ad libitum (AL) or dietary restriction (DR) regimens for three months. (A) Body
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/2/e1602038/dc1 Supplementary Materials for Mitochondrial metabolic regulation by GRP78 Manoj Prasad, Kevin J. Pawlak, William E. Burak, Elizabeth E. Perry, Brendan
advances.sciencemag.org/cgi/content/full/3/2/e1602038/dc1 Supplementary Materials for Mitochondrial metabolic regulation by GRP78 Manoj Prasad, Kevin J. Pawlak, William E. Burak, Elizabeth E. Perry, Brendan
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Increased ABHD5 expression in human colon cancer associated macrophages. (a) Murine peritoneal macrophages were treated with regular culture medium (Ctrl) or
Supplementary Figures Supplementary Figure 1 Increased ABHD5 expression in human colon cancer associated macrophages. (a) Murine peritoneal macrophages were treated with regular culture medium (Ctrl) or
Electron transport chain, oxidative phosphorylation, mitochondrial transport systems
 Electron transport chain, oxidative phosphorylation, mitochondrial transport systems JAN ILLNER Respiratory chain & oxidative phosphorylation INTERMEMBRANE SPACE ubiquinone cytochrome c ATPase Production
Electron transport chain, oxidative phosphorylation, mitochondrial transport systems JAN ILLNER Respiratory chain & oxidative phosphorylation INTERMEMBRANE SPACE ubiquinone cytochrome c ATPase Production
Cornstarch
 Electronic Supplementary Material (ESI) for Food & Function. This journal is The Royal Society of Chemistry 2018 Supplementary data : Supplementary Table 1: Diet composition (g/kg) on the basis of the
Electronic Supplementary Material (ESI) for Food & Function. This journal is The Royal Society of Chemistry 2018 Supplementary data : Supplementary Table 1: Diet composition (g/kg) on the basis of the
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
incorporation into fructose-6-phosphate and glucose-6-phosphate (F6P/G6P); 2-
 Relative abundance (%) Relative abundance (%) Relative abundance (%) Relative abundance (%) 100% 80% 60% 40% F6P / G6P M+0 M+1 M+2 M+3 M+4 M+5 M+6 100% 80% 60% 40% 2PG / 3PG M+0 M+1 M+2 M+3 20% 20% 0%
Relative abundance (%) Relative abundance (%) Relative abundance (%) Relative abundance (%) 100% 80% 60% 40% F6P / G6P M+0 M+1 M+2 M+3 M+4 M+5 M+6 100% 80% 60% 40% 2PG / 3PG M+0 M+1 M+2 M+3 20% 20% 0%
Supplementary Information. Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase
 Journal: Nature Medicine Supplementary Information Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase 1,2 Peng Wang PhD, 1,2 Juan-Carlos Alvarez-Perez
Journal: Nature Medicine Supplementary Information Induction of human pancreatic beta cell replication by inhibitors of dual specificity tyrosine regulated kinase 1,2 Peng Wang PhD, 1,2 Juan-Carlos Alvarez-Perez
ANSC/NUTR 618 Lipids & Lipid Metabolism
 I. Overall concepts A. Definitions ANC/NUTR 618 Lipids & Lipid Metabolism 1. De novo synthesis = synthesis from non-fatty acid precursors a. Carbohydrate precursors (glucose, lactate, and pyruvate) b.
I. Overall concepts A. Definitions ANC/NUTR 618 Lipids & Lipid Metabolism 1. De novo synthesis = synthesis from non-fatty acid precursors a. Carbohydrate precursors (glucose, lactate, and pyruvate) b.
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
BIOL2171 ANU TCA CYCLE
 TCA CYCLE IMPORTANCE: Oxidation of 2C Acetyl Co-A 2CO 2 + 3NADH + FADH 2 (8e-s donated to O 2 in the ETC) + GTP (energy) + Heat OVERVIEW: Occurs In the mitochondrion matrix. 1. the acetyl portion of acetyl-coa
TCA CYCLE IMPORTANCE: Oxidation of 2C Acetyl Co-A 2CO 2 + 3NADH + FADH 2 (8e-s donated to O 2 in the ETC) + GTP (energy) + Heat OVERVIEW: Occurs In the mitochondrion matrix. 1. the acetyl portion of acetyl-coa
Total RNA was isolated and reverse transcribed with AffinityScript QPCR cdna Synthesis Kit
 SUPPLEMENTAL MATERIAL RNA analyses Total RNA was isolated and reverse transcribed with AffinityScript QPCR cdna Synthesis Kit (Agilent Technologies). PCR reactions were performed in triplicate in a 96-well
SUPPLEMENTAL MATERIAL RNA analyses Total RNA was isolated and reverse transcribed with AffinityScript QPCR cdna Synthesis Kit (Agilent Technologies). PCR reactions were performed in triplicate in a 96-well
