High-Throughput Flow Cytometry Screening Reveals a Role for Junctional Adhesion Molecule A as a Cancer Stem Cell Maintenance Factor
|
|
|
- Leo Harrell
- 5 years ago
- Views:
Transcription
1 Cell Reports Article High-Throughput Flow Cytometry Screening Reveals a Role for Junctional Adhesion Molecule A as a Cancer Stem Cell Maintenance Factor Justin D. Lathia, 1,2,3,4, * Meizhang Li, 1,2,13 Maksim Sinyuk, 1 Alvaro G. Alvarado, 1,3 William A. Flavahan, 2,3 Kevin Stoltz, 1 Ann Mari Rosager, 5 James Hale, 1 Masahiro Hitomi, 1,3 Joseph Gallagher, 2 Qiulian Wu, 2 Jody Martin, 6 Jason G. Vidal, 6 Ichiro Nakano, 7 Rikke H. Dahlrot, 8 Steinbjørn Hansen, 8 Roger E. McLendon, 9 Andrew E. Sloan, 4,10,11 Shideng Bao, 2,3,4 Anita B. Hjelmeland, 2,3,4 Christian T. Carson, 6 Ulhas P. Naik, 12 Bjarne Kristensen, 5 and Jeremy N. Rich 2,3,4, * 1 Department of Cellular and Molecular Medicine, Cleveland Clinic Lerner Research Institute, Cleveland, OH 44195, USA 2 Department of Stem Cell Biology and Regenerative Medicine, Cleveland Clinic Lerner Research Institute, Cleveland, OH 44195, USA 3 Department of Molecular Medicine, Cleveland Clinic Lerner College of Medicine of Case Western Reserve University, Cleveland, OH 44195, USA 4 Case Comprehensive Cancer Center, Cleveland, OH 44106, USA 5 Department of Pathology, Odense University Hospital, Institute of Clinical Research, University of Southern Denmark, Odense 5000, Denmark 6 BD Biosciences, La Jolla, CA 92037, USA 7 Department of Neurological Surgery, James Comprehensive Cancer Center, The Ohio State University, Columbus, OH 43210, USA 8 Department of Oncology, Odense University Hospital, Institute of Clinical Research, University of Southern Denmark, Odense 5000, Denmark 9 Department of Pathology, Duke University Medical Center, Durham, NC 27710, USA 10 Seidman Cancer Center, University Hospital-Case Medical Center, School of Medicine, Case Western Reserve University, Cleveland, OH 44106, USA 11 Departments of Neurological Surgery, Pathology, and Translational Neuroscience, University Hospital-Case Medical Center, School of Medicine, Case Western Reserve University, Cleveland, OH 44106, USA 12 Department of Biology, University of Delaware, Newark, DE 19716, USA 13 Laboratory of Biochemistry and Molecular Biology, School of Life Sciences, Yunnan University, Kunming , China *Correspondence: lathiaj@ccf.org (J.D.L.), drjeremyrich@gmail.com (J.N.R.) This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited. SUMMARY Stem cells reside in niches that regulate the balance between self-renewal and differentiation. The identity of a stem cell is linked with the ability to interact with its niche through adhesion mechanisms. To identify targets that disrupt cancer stem cell (CSC) adhesion, we performed a flow cytometry screen on patient-derived glioblastoma (GBM) cells and identified junctional adhesion molecule A (JAM-A) as a CSC adhesion mechanism essential for self-renewal and tumor growth. JAM-A was dispensable for normal neural stem/progenitor cell (NPC) function, and JAM-A expression was reduced in normal brain versus GBM. Targeting JAM-A compromised the self-renewal of CSCs. JAM-A expression negatively correlated to GBM patient prognosis. Our results demonstrate that GBM-targeting strategies can be identified through screening adhesion receptors and JAM-A represents a mechanism for niche-driven CSC maintenance. INTRODUCTION Stem cells are essential for the growth and homeostasis of organs and, more broadly, an organism. Whereas stem cells have an unlimited proliferation capacity and can differentiate into any cell type in a given organ, they reside in specific anatomical locations or niches that ensure the correct balance of self-renewal and differentiation. Stem cells must be simultaneously maintained for long-term tissue preservation yet constrained to avoid oncogenesis. Stem cells that exit a niche undergo differentiation and fate determination. Niche signaling via secreted factors, cell-to-cell, and cell-to-extracellular-matrix (ECM) interactions represent a powerful control mechanism for stem cell regulation. Many niche factors have also been implicated in the progression of advanced cancers in the context of maintaining a self-renewing, tumorigenic cancer stem cell (CSC) population at the top of a cellular hierarchy. Like normal stem cells, CSCs also reside in distinct anatomical locations and rely on niche interactions to regulate the balance between self-renewal and differentiation, and these interactions provide prosurvival and therapeutic resistance mechanisms (Gilbertson and Rich, 2007; Spradling et al., 2001). Whereas the CSC hypothesis remains controversial, CSCs have been well documented in many advanced cancers, including glioblastoma (GBM; World Health Organization Cell Reports 6, , January 16, 2014 ª2014 The Authors 117
2 118 Cell Reports 6, , January 16, 2014 ª2014 The Authors (legend on next page)
3 grade IV glioma). Current therapeutic approaches for GBM remain only palliative, and the standard care of surgical resection followed by aggressive radiation and chemotherapy has extended median survival to between 12 and 15 months whereas the 5-year survival remains approximately 2% (Stupp et al., 2009). GBM is refractory to the current standard therapies partly due to invasion into the normal brain and cellular heterogeneity (Bonavia et al., 2011). The identification of CSCs in GBM (Galli et al., 2004; Hemmati et al., 2003; Ignatova et al., 2002; Singh et al., 2003, 2004) has generated enthusiasm for the integration of CSCs into models of cancer (Visvader and Lindeman, 2012) and the development of anti-csc therapies. CSCs in GBM are contained within hypoxic (Li et al., 2009) and perivascular niches (Calabrese et al., 2007). The perivascular niche is readily identifiable in vivo, and efforts are underway to characterize the components regulating this niche which include local mitogens, cell-cell communication mechanisms, and unique structural components, such as ECM proteins (Hjelmeland et al., 2011). We previously demonstrated that integrin a6, an ECM receptor, can be used to enrich and target CSCs (Lathia et al., 2010). Additionally, interaction with the specialized ECM of the perivascular niche, which is provided by niche components, also promotes CSC growth (Lathia et al., 2012). These studies provide a paradigm for CSC maintenance whereby adhesion status is a determining factor for the position of a cell within the tumor hierarchy, with CSCs displaying an enhanced adhesion capacity as compared to their differentiated progeny. Whereas similarities exist between CSCs and nonneoplastic stem cells, specific targets for CSCs have been generated by comparing the molecular machinery between the cell types and have been validated in preclinical models (Eyler et al., 2011; Guryanova et al., 2011). However, niche adhesion targets have yet to be developed. The interaction between cell-adhesion mechanisms, including integrins, generates diverse signaling responses based on cell type, location, and the cluster of receptors present in a signaling complex. Defining adhesion-specific programs unique to the CSC niche compartment is difficult as many of these programs exist both in the normal and neoplastic niches (Hale et al., 2012). For example, integrin expression and function in organs, such as the brain and the breast, are similar in the normal and neoplastic context, with both normal stem cells and CSCs being enriched on the basis of integrin a6 (Ali et al., 2011; Hall et al., 2006; Lathia et al., 2010; Shackleton et al., 2006; Stingl et al., 2006), which plays a key role in regulating their growth (Kazanis et al., 2010; Lathia et al., 2010; Loulier et al., 2009). Given the importance of niche adhesion to stem cell maintenance (Niola et al., 2012, 2013), identification of unique CSC niche mechanisms is an immediate priority to facilitate the development of more effective GBM therapies. To identify CSC niche adhesion mechanisms, we performed an unbiased flow cytometry screen of cell adhesion receptors expressed in patient-derived GBM cells with a link to patient survival. Combining additional biochemical, molecular, and biological analyses, we identified junctional adhesion molecule A (JAM-A) as a critical maintenance factor for CSCs. RESULTS Flow Cytometry Screen of Adhesion Receptors Reveals that GBM Cells Express JAM-A Unbiased screening approaches present an opportunity to understand the complexity of a system as well as identify new regulatory nodes that can be then further developed for targeting purposes. In the context of GBM, a variety of genetic (RNAi) and pharmacological (small molecule) screens have been performed that have identified targets for further investigation (Hubert et al., 2013; Visnyei et al., 2011; Wurdak et al., 2010). Whereas these approaches have revealed the complexity of GBM and provided targets, they have provided limited information with respect to niche interaction. Flow cytometry enables the prospective enrichment of CSCs and can be utilized to study intact cells as well as specific cell-surface interactions. To identify cell-adhesion receptors expressed on the surface of GBM cells, we performed a high-throughput flow cytometry screen, which informed the identification of cell-surface antigens differentially expressed during neural differentiation (Yuan et al., 2011). To perform the screen, we utilized six different GBM specimens that had validated differences in self-renewal both in vitro (Figure S1A) and in vivo based on previously reported cell surface markers (Eyler et al., 2011; Guryanova et al., 2011; Lathia et al., 2010; Singh et al., 2003). Each specimen was uniquely identified using a bar-coding approach, and then all six xenografts were pooled together for the screening procedure (Figure 1A). Along with interrogating the cell-surface expression of adhesion receptors, we combined the analysis with a bioinformatics evaluation of patient survival to identify adhesion receptors that were expressed on GBM cells and informative for patient prognosis (Figure 1A). Screening was performed on cell-adhesion molecules for which antibodies were commercially available, and any that were expressed above a level of 5% for three of the six GBM specimens were subsequently evaluated for a negative correlation to glioma patient survival based on Figure 1. High-Throughput Screening on GBM Specimens with a Multiplex Flow Cytometry Identifies Cell-Adhesion Receptors Negatively Correlated to GBM Patient Survival (A) Schematics depicting the work flow of the high-throughput flow cytometry screening method based on bar-coding multiple GBM specimens and screening cell surface marker expression to identify cell-adhesion receptors linked to GBM patient survival. (B) Representative flow cytometry plots for adhesion receptors from GBM xenograft T4121, antigen of interest shown in blue, isotype control shown in red, and percentage of positive cells indicated on each individual plot. (C) Representative Kaplan-Meier survival curves for candidate adhesion molecular generated from the NCI REpository for Molecular BRAin Neoplasia DaTa (REMBRANDT) bioinformatics database ( for the 2-fold upregulated (red) and intermediate (black) groups. Log rank p value for survival curves between upregulated and intermediate groups provided for each adhesion molecule. (D) Representative immunohistochemical data generated for CD321 and CD63 from the human protein atlas ( for the cortex, subventricular zone (SVZ), and glioblastoma (GBM) tumor. Age and sex of each individual indicated directly on the micrographs. See also Table S1 and Figure S1. Cell Reports 6, , January 16, 2014 ª2014 The Authors 119
4 the National Cancer Institute REpository for Molecular BRAin Neoplastic Data (NCI REMBRANDT) database (Figures 1B and 1C). Our screen identified nine cell-adhesion receptors that met the selection criteria and were ranked based on negative correlation to glioma patient survival (Table S1) with varying correlation between individual targets (Figure S1B). The screen revealed the following cell-surface adhesion receptors: CD321 (JAM-A), CD63 (a Tetraspanin family member), CD87 (urokinase receptor), CD49e (integrin a5), CD26 (dipeptidyl peptidase-4), CD54 (ICAM1), CD29 (integrin b1), CD44 (a hyaluronic acid receptor), and CD49f (integrin a6). Among the list were three previously published CSC markers, CD49f and CD29 (Lathia et al., 2010) as well as CD44 (Anido et al., 2010; Jijiwa et al., 2011), providing evidence that our approach was capable of identifying functionally relevant adhesion receptors. CD26, a regulator of colon CSC metastasis that interacts with integrin b1 (Pang et al., 2010), was also identified, and CD87, which is linked to GBM survival (Hu et al., 2011) and invasion (Raghu et al., 2011), was likewise identified. The top two targets, CD321 and CD63, were further validated with respect to tumor-specific expression using the human protein atlas (Figure 1D). JAM-A, which had a median survival of 13.4 months, was of interest based on its reported role in cell-to-cell adhesion and facilitation of integrin signaling (McSherry et al., 2011; Severson et al., 2009). These functions have not been previously tested in GBM and are likely to be an important mechanism by which cells communicate with niche s ECM. In the nervous system, integrin a6 predominantly uses integrin b1 as a binding partner to initiate signaling events upon activation (Lathia et al., 2007; Loulier et al., 2009). We previously demonstrated that integrin a6 expression can enrich for CSCs (Lathia et al., 2010). Further evaluation of the activation state of integrin b1 revealed that it was also informative for sphere-formation potential and indeed blocking integrin b1 attenuated sphere formation (Figures S1C S1E). Integrin b1 is regulated by a variety of coreceptors that are cell type and context dependent. In the mammary epithelium, where integrins a6 and b1 are also elevated in normal and neoplastic stem cells (Ali et al., 2011; Shackleton et al., 2006; Stingl et al., 2006), JAM-A interacts with integrin b1 and correlates with poor patient prognosis (McSherry et al., 2011; Murakami et al., 2011). Given the relationship between integrin b1 and JAM-A in the mammary gland, we wanted to determine the role of JAM-A in GBM and its role in regulating CSC maintenance. JAM-A Is Elevated in CSCs To confirm that JAM-A was expressed on GBM cells and exclude the contribution of nonneoplastic cells, we performed flow cytometry analyses on xenografted (T4121 and T387) specimens and a primary-patient-derived specimen (CCF2418) in combination with known endothelial cell markers (CD105 and CD31 [data not shown]). JAM-A was expressed in xenografted and primary GBM specimens on both endothelial and nonendothelial cells (Figure S2A). To verify that JAM-A-positive/endothelial-cell-marker-negative cells were tumor cells, we isolated these cells from GBM xenografts and confirmed their tumor cell origin by fluorescence in situ hybridization (FISH) (Figures S2B and S2C). Taken together, these results confirm that JAM-A is expressed in GBM cells. To validate our flow cytometry screen findings, we evaluated JAM-A expression by immunofluorescence analysis on cryosections from intracranial GBM xenografts. JAM-A was expressed in a fraction of tumor cells (T387: 28.8%; T3691: 28.1%; T4121: 32.7%; data not shown), and colabeling revealed that a large fraction of JAM-A-positive cells expressed CSC markers Sox2 or integrin a6 (Figures 2A and 2B; T387: 97.0%; T3691: 92.6%; T4121: 97.3%; data not shown). The coexpression of JAM-A with integrin a6 was further confirmed by flow cytometry analysis of xenografted patient specimens (T4121 and T387) as well as primary GBM specimens (1217 and 1221) and found to be at least 70% or higher (data not shown). These data suggest that JAM-A may be elevated in CSCs. To directly evaluate this possibility, we evaluated JAM-A expression by immunoblotting, flow cytometry and immunostaining analysis. Immunoblotting analysis of matched CSCs and non-cscs from a GBM patient xenograft enriched on the basis of a cell-surface marker previously shown to distinguish tumor formation potential (Eyler et al., 2011; Guryanova et al., 2011; Lathia et al., 2010; Li et al., 2009) revealed that JAM-A was elevated in the CSC-marker-positive population as compared with the matched CSC-marker-negative population from the same GBM xenograft (Figure 2C). The differential expression of JAM-A between CSCs and non-cscs was also confirmed by flow cytometry analysis comparing parental xenografted GBM cells to CSCs enriched from the resulting xenografts and propagated as CSC-enriched sphere cultures (Figure 2D). The coexpression between JAM-A and integrin a6 in CSC-enriched sphere cultures was confirmed by immunofluorescence analysis (Figure 2E). To evaluate if JAM-A expression changed as a result of loss of stemness, we placed CSC-enriched sphere cultures under a 7-day serum-induced differentiation paradigm. During differentiation, JAM-A expression was reduced as was the expression of CSC markers Sox2 and Olig2 whereas the differentiation markers Map2 and glial fibrillary acidic protein (GFAP) were increased (Figure 2F). These data verify that the expression of JAM-A is enriched in CSCs as compared to their non-csc counterparts and is suppressed during differentiation. JAM-A Is Not Expressed by NPCs and Is Not Required for NPC Proliferation Our results suggest that JAM-A is expressed on a fraction of GBM cells positive for CSC markers and encourage the evaluation of the expression profile of JAM-A in normal tissue in order to facilitate therapeutic approaches. To evaluate the expression and importance of JAM-A in normal neural function, we evaluated both human and mouse normal brain tissues for JAM-A expression. To evaluate the expression of JAM-A in human brain, we utilized both cultured fetal neural progenitor cells (NPCs) as well as an immunohistochemistry approach of adult brain tissue. Immunoblotting of NPCs and GBM cells (matched CSCs and non-cscs) confirmed that JAM-A expression was elevated in CSCs and low in NPCs (Figure S2D). Using an immunohistochemistry analysis, we observed limited staining in the cortex or white matter areas of human adult brain (Figures S2E and S2F). JAM-A staining in the subependymal/subventricular zone (SEZ/SVZ) was limited to the ependymal cell layer (Figure S2G). To determine 120 Cell Reports 6, , January 16, 2014 ª2014 The Authors
5 Figure 2. JAM-A Is Elevated in CSCs (A and B) Confocal micrographs show that JAM-A is coexpressed with CSC markers Sox2 (A) and integrin a6 (B) on GBM cells within an intracranial xenograft (T4121) using antibodies against JAM-A (green) and costained with Sox2 (red) or integrin a6 (red). Areas of interest are indicated by yellow arrow and shown at higher power, and the scale bar represents 50 mm. (C) JAM-A is elevated in CSCs (enriched by cell surface expression of CD133 from GBM xenograft tumor T4121) as compared with matched non- CSCs as shown in representative immunoblots that show separation between populations with respect to Sox2 and Olig2. (D) Bar graphs summarizing flow cytometry analyses between parental cells and CSCs demonstrate that CSCs have significantly higher expression of JAM-A as compared to parental unenriched tumor cells from GBM xenografts (T3691, T387, T4121, and T4302). (E) CSCs grown in sphere culture (from tumor T4121) coexpress JAM-A (green) and integrin a6 (red) as shown in confocal micrographs. An area of interest indicated by white box is shown with enlarged images below, nuclei were counterstained with Hoechst 33342, and scale bar represents 50 mm. (F) Upon CSC differentiation (enriched from GBM xenograft tumor T4121) using a 10% fetal bovine serum (FBS) differentiation paradigm over a 7-day period, JAM-A expression is suppressed with concomitant loss in CSC markers Sox2 and Olig2 and an increase in differentiation markers GFAP and Map2 as shown in representative immunoblots. Data for bar graphs displayed as mean values ± SD, blue value represents fold change, and ***p < as assessed by one-way ANOVA. See also Figure S2. relative levels of JAM-A expression, we also utilized grade II astrocytoma and GBM tissue and observed low but detectable staining in grade II tumors and strong staining in GBM tissue (Figures S2H S2J). JAM-A knockout mice are viable, although the birth ratios are not Mendelian (1 [+/+]: 1.7 [+/ ]: 0.6 [ / ]), and their endothelial cells display a fibroblast growth factor 2-induced angiogenesis defect (Cooke et al., 2006). However, the impact of JAM-A deletion in the brains of these mice is yet to be determined. To determine the role of JAM-A in NPCs, we evaluated proliferation in a key neurogenic niche (Alvarez-Buylla and Lim, 2004) in the brains of wild-type and JAM-A / mice, the SVZ adjacent to the lateral ventricles. Analysis of SVZ proliferation using two molecular markers (proliferating cell nuclear antigen [PCNA] and phosphohistone H3 [PH3]) revealed no differences between adult wild-type (WT) and JAM-A / mice (Figures 3A 3C) suggesting that JAM-A is not essential for NPC proliferation. JAM-A has previously been reported to be expressed in the mouse corpus callosum on NG2-expressing cells (Stelzer et al., 2010), likely to be glial progenitors and pericytes. However, the expression in the neurogenic niches or NPCs is yet to be determined. The JAM-A antibody used only stained JAM-A expression in WT mice, but not in JAM-A / mice confirming the specificity of the JAM-A antibody (Figure 3D). JAM-A expression did not overlap with NPCs and astrocytes (marked by GFAP; Figure 3E), NPCs (marked by Sox2; Figure 3F), or neuroblasts (marked by doublecortin, Dcx; Figure 3G). JAM-A expression, however, was detected in the SVZ and was found to overlap with Iba1-positive cells, reported to be putative microglia and macrophages (Figure 3H). Iba1-positive cells were detected in both WT and JAM-A / mice, suggesting that the genesis of microglia and macrophages is not dependent on JAM-A (Figures S3A and S3B). JAM-A was not detected on additional SVZ components (ependymal cells, blood vessels, and glial Cell Reports 6, , January 16, 2014 ª2014 The Authors 121
6 Figure 3. JAM-A Is Not Required for Neurogenesis (A C) No detectable difference was observed in SEZ/SVZ proliferation between wild-type (WT) and JAM-A / mice as shown in summary bar graphs (A) and confocal micrographs of proliferation markers PCNA (B) and PH3 (C). (D) Confocal micrographs show specific staining for JAM-A antibody in the SEZ/SVZ of WT mice but no staining in JAM-A / mice. (E H) Analysis of the mouse SEZ/SVZ reveals that JAM-A (green) is present in the SEZ/SVZ and not on NSCs/astrocytes (stained with an antibody against GFAP, red [E]), NSC/NPCs (stained with an antibody against Sox2, red [F]), or neuroblasts (stained with an antibody against Dcx, red [G]) but on microglia and macrophages (stained with an antibody against Iba1, red [H]) as shown in confocal micrographs. Areas of interest are indicated by yellow arrow and areas shown as insets. In (F), another example of JAM-A-positive, Sox2-negative cell is indicated by a white arrow. Nuclei were counterstained with Hoechst and the scale bar represents 25 mm. Data for bar graphs are displayed as mean values ± SD. See also Figure S3. progenitors) or proliferating cells (Figures S3C S3F). JAM-A was also not detected on additional cell types in another neurogenic niche, the subgranular zone (SGZ) of the dentate gyrus in the hippocampus, with the exception of microglia and macrophages (Figures S3G S3J). Our analysis of the neurogenic regions in the mouse brain revealed that in the SVZ and SGZ, JAM-A was only coexpressed with Iba1- positive cells (Figures S3K and S3L) and that JAM-A / does not affect lba1+ cell genesis. Taken together, our human and mouse analysis suggests that JAM-A is not highly expressed on NPCs and not likely to be essential for their function. 122 Cell Reports 6, , January 16, 2014 ª2014 The Authors
7 Figure 4. Targeting JAM-A by Blocking Antibodies Decreases Growth, Self-Renewal, and Adhesion and Increases Apoptosis of CSCs but Does Not Impact Survival of Nonstem Tumor Cells or NPCs JAM-A-blocking antibody (red line/red bar) was used to treat CSCs (enriched from GBM xenografts T4121 and T387) and matched non-cscs at 10 mg/ml as compared to a control antibody (black line/black bar). (A D) A strong decrease was seen in CSC growth over time (A and C) but was not observed in non- CSCs (B and D). Ab, antibody. (E and F) An attenuation in self-renewal as assessed by sphere formation was observed in CSCs in the presence of the JAM-A-blocking antibody. (G and H) A significant reduction in CSC adhesion onto a tumor ECM (Geltrex) was also observed with the JAM-A-blocking antibody as well as an integrin-a6-blocking antibody. (I K) Apoptosis induced by the JAM-A-blocking antibody in CSCs (I and J) was not observed in NPCs (K). Data for all graphs displayed as mean values ± SD; *p < 0.05, **p < 0.01, and ***p < as assessed by one-way ANOVA. See also Figure S4. Targeting JAM-A by Blocking Antibody Attenuates CSC Growth, Self-Renewal, and Adhesion but Does Not Affect Non-CSCs or NPCs Our analysis confirmed the expression of JAM-A on CSCs and the very low or nondetectable levels of expression in matched non-cscs or NPCs, supporting the hypothesis that JAM-A is primarily utilized by CSCs. To evaluate the impact of disrupting JAM-A on CSC growth, we utilized a blocking antibody. CSCs treated with the JAM-A-blocking antibody at previously reported concentrations of 1 and 10 mg/ml (Severson et al., 2009) showed significant reduction in growth over a 7-day time course (Figures 4A, 4C, S4A, and S4C), which was not observed to the same extent in the non-csc population (Figures 4B, 4D, S4B, and S4D). A longer time course (days 10 14) did reveal a significant reduction in non-csc growth at the 10 mg/ml, but not the 1 mg/ml antibody concentration (data not shown). Sphere formation, a surrogate of self-renewal, was also significantly attenuated in the presence of the JAM- A-blocking antibody at both 10 mg/ml (Figures 4E and 4F) and 1 mg/ml (data not shown). To determine if the effects of the JAM-A-blocking antibody on CSCs are due, in part, to apoptosis, we evaluated caspase 3/caspase 7 activity and found a significant increase in CSCs at both concentrations of the JAM-A blocking-antibody (Figures S4E S4H). For equivalent conditions tested for non- CSCs, only a significant induction of caspase activity was observed from GBM specimen T4121 at 10 mg/ml (Figure S4F), but this was 4.8-fold lower when compared to CSCs from the same specimen. Unlike non-cscs from GBM specimen T4121 over a longer time course, caspase 3/caspase 7 activity was not induced by the JAM-A blocking antibody at both concentrations in non-cscs from T387 (Figures S4G and S4H). A key function of JAM-A is to stabilize integrins and thereby facilitate adhesion, and to evaluate if blocking JAM-A compromised this function, we performed short-term adhesion assays. Using a representative tumor ECM enriched in laminins (Geltrex), we found that the JAM-A-blocking antibody significantly inhibited cell adhesion to the matrix (Figures 4G and 4H), which was phenocopied with an integrin-a6-blocking antibody (Figures 4G and 4H). These differences in adhesion were not observed when assays were performed on fibronectin, another ECM protein, suggesting that JAM-A-mediated adhesion is laminin-specific. Finally, to determine if the JAM-A-blocking antibody induced apoptosis in NPCs, we evaluated CSCs and NPCs treated with Cell Reports 6, , January 16, 2014 ª2014 The Authors 123
8 Figure 5. Targeting JAM-A by RNAi Decreases Stem Cell Expression, Growth, Self-Renewal, and Tumor Formation Capacity of CSCs (A) Two independent shrna constructs (knockdown [KD] 1 and 2) were generated against JAM- A, and reduced JAM-A expression as compared to nontargeting controls was verified in CSCs from GBM xenograft T4121 by flow cytometry. (B) Attenuation of JAM-A in CSCs (enriched from GBM xenograft T4121) results in a decrease in integrin b1 expression as well as other core CSC markers (Sox2, Olig2) as shown by immunoblots. (C and D) Reduction of JAM-A in CSCs (enriched from GBM xenograft T4121) also resulted in decreased self-renewal (C) and CSC growth (D). (E and F) Slower tumor growth after grafting CSCs (enriched from GBM xenograft T4121) was also observed when JAM-A expression was attenuated by shrna (E), and overall tumor size was reduced as shown in bar graphs and images from brain sections 30 days after transplantation of 1,000 CSCs (F). Data for all graphs displayed as mean values ± SD; *p < 0.05, **p < 0.01, and ***p < as assessed by one-way ANOVA. For multiple groups, #p < 0.05, ##p < 0.01, and ###p < NT versus KD1; *p < 0.05, **p < 0.01, and ***p < NT versus KD2; and p < 0.05 KD1 versus KD2 as assessed by one-way ANOVA. 10 mg/ml of the JAM-A blocking antibody and observed a significant induction of cleaved caspase 3 by flow cytometry only in CSCs, but not in NPCs (Figures 4I 4K) measured after 3 days. Taken together, these data suggest that blockade of JAM-A in CSCs attenuates growth, self-renewal, and adhesion and induces cell death while leaving non-cscs and NPCs intact. Genetic Disruption of JAM-A Results in Decreased CSC Growth, Self-Renewal, and Tumor Formation Acute disruption of JAM-A by a blocking antibody compromised CSC adhesion, growth, and self-renewal. To confirm the role of JAM-A in CSC maintenance, we also utilized a genetic approach to attenuate JAM-A expression by RNAi. We developed two independent, nonoverlapping small hairpin RNA (shrna) constructs to knockdown JAM-A (knockdown 1 [KD1] and KD2). When compared to a nontargeting (NT) control construct, both JAM-A knockdown constructs reduced JAM-A expression levels as evaluated by quantitative PCR (qpcr) (data not shown), flow cytometry (Figure 5A), and immunoblotting (Figure 5B). JAM-A plays a major role in integrin stabilization (Severson et al., 2009), and knockdown of JAM-A resulted in reduced expression of integrin b1(figure 5B). In addition, flow cytometry analysis revealed a reduction in integrin a6 cell surface levels after JAM-A knockdown (data not shown), further supporting a role for JAM-A in integrin stabilization. CSCs with attenuated JAM-A also displayed deficiencies in sphere formation (Figure 5C) and proliferation (Figure 5D). To evaluate if JAM-A was required for tumor formation, the key functional criteria for a CSC, we transplanted NT or KD CSCs into the brain of immunocompromised mice and observed significantly less tumor formation with JAM-A-deficient CSCs (Figure 5E). Additionally, analysis of mice 30 days after CSC transplantation revealed significantly smaller tumor area in the mouse brains receiving JAM-A knockdown CSCs than in the ones receiving CSCs with NT construct (Figure 5F). These results indicate that JAM-A is critical for integrin stabilization and that attenuation of JAM-A results in a deficiency of CSC phenotypes, including growth, self-renewal, and tumor formation. JAM-A Expression Is Informative for Tumor Grade and Patient Prognosis The molecular genetics of GBM have been extensively investigated permitting the association between gene profiles and patient outcome (Brennan et al., 2013; Cancer Genome Atlas Research Network, 2008; Frattini et al., 2013; Phillips et al., 124 Cell Reports 6, , January 16, 2014 ª2014 The Authors
9 Figure 6. JAM-A Correlates with Glioma Malignancy and Is Informative for Patient Survival (A and B) Box and whisker plots of expression level for JAM-A using Oncomine ( oncomine.org/) demonstrates significantly higher expression (p < as assessed by one-way ANOVA) of JAM-A in glioblastoma tissue than in neural progenitor cells (A) and significantly higher expression (p < as assessed by one-way ANOVA) of JAM-A in glioblastoma tissue as compared with oligodendroglia (Oligo) tumors, low-grade astrocytoma, or mixed glioma and astrocytoma (B). (C) JAM-A is also informative for GBM patient survival as demonstrated by Kaplan-Meier survival plots. The red line indicates the high expression group (ten patients) with greater than one SD above the mean of all patients. The blue line indicates the low expression group (nine patients) with greater than one SD below the mean of all patients. The black line indicates the group (57 patients) within one SD of mean. (D and E) Kaplan-Meier survival plots for glioma (D) and GBM (E) patients with differential tumor JAM-A expression using the NCI REMBRANDT database at a level of 2-fold upregulation indicates that JAM-A levels inversely correlates with patient survival (red line indicates 2-fold upregulation [137 glioma patients, 70 GBM patients], blue line indicates 2-fold downregulation [eight glioma patients], and black line represents intermediate expression [198 glioma patients, 110 GBM patients]). (F and G) Analysis of JAM-A using histochemical staining in a patient data set (Odense data set, Odense University Hospital Denmark) indicates that JAM-A is elevated in GBM as compared to normal brain controls (F) and that high-jam-a expression correlates with poor patient prognosis ([G]; 102 patients in the JAM-A-high group and 83 patients in the JAM-A-low group). Data for all graphs displayed as mean values ± SD; **p < 0.01 and ***p < as assessed by oneway ANOVA. For survival analysis, the log rank p value for significance of difference between groups is indicated on each individual graph. See also Figure S ; Verhaak et al., 2010). To evaluate the relationship between JAM-A and GBM patient survival and determine if JAM-A was informative for tumor grade and patient prognosis, we interrogated brain tumor bioinformatics databases. To determine if elevated JAM-A expression was restricted to GBM or also present in other neural cell types, such as NPCs, we evaluated the Lee data set (Lee et al., 2006) from Oncomine and observed a significantly higher level of JAM-A in GBM as compared to NPCs (Figure 6A). Evaluation of JAM-A expression in relation to tumor grade from the Freije data set (Freije et al., 2004) in the Oncomine database indicated a significant elevation of JAM-A expression in GBM as compared to lower-grade tumors (oligodendroglioma, mixed glioma, grade III astrocytoma; Figure 6B). Kaplan-Meier analysis also indicated that high-jam-a expression (at least one SD above the mean) correlated with poor survival, whereas low-jam-a expression (at least one SD below the mean) correlated with better survival in the Phillips data set (Phillips et al., 2006) from Oncomine (data not shown). Likewise, JAM-A informed poor patient prognosis in GBM from the same data set (Figure 6C). We further confirmed the association between JAM-A expression and glioma patient survival using the NCI REMBRANDT database and found that high- JAM-A expression (>2-fold) correlated to poor glioma patient survival whereas low-jam-a expression (<2-fold) correlated to increased patient survival (Figure 6D). The correlation between elevated JAM-A expression and poor patient prognosis was observed in GBM patients as well (Figure 6E). To determine if these differences in survival were specific to JAM-A or shared by the other JAM family members, JAM-B and JAM-C, we evaluated the REMBRANDT database and Phillips data set in Cell Reports 6, , January 16, 2014 ª2014 The Authors 125
10 Oncomine and found neither isoform was informative for glioma or GBM patient prognosis (Figure S5). Taken together, our bioinformatics analysis reveals that JAM-A is associated with increased malignancy and poor patient prognosis. The ability for a biomarker to provide informative molecular diagnosis is useful and is further enhanced by its capacity to inform survival based on protein expression. Using immunohistochemical approaches, we observed that JAM-A expression increased with malignancy and was elevated in GBM tissue as compared to nonneoplastic cortical tissue (Figures 1D, 6F, and S2E S2J). We examined the capacity of JAM-A protein expression to predict GBM patient outcome in a clinically annotated data set (Dahlrot et al., 2013). High-JAM-A protein level also significantly correlated to poor GBM patient prognosis as compared with low-jam-a expression (Figure 6G). Taken together, our findings demonstrate that JAM-A is increased with malignancy and is a negative indicator of patient prognosis. DISCUSSION The ability for a stem cell to interact with its niche through adhesion mechanisms enables it to maintain a physical location as well as receive specialized signals from the niche (Spradling et al., 2001). Adhesion receptors are essential transducers of this process and facilitate niche adhesion as well as stem-cellmaintenance signaling. Adhesion receptor expression has been used to prospectively enrich them from normal and neoplastic tissues, including the breast (Ali et al., 2011; Shackleton et al., 2006; Stingl et al., 2006; Vaillant et al., 2008) and brain (Hall et al., 2006; Lathia et al., 2010). Stem cell niches, themselves, also contain unique ECM expression, which for the brain includes elevated laminin expression (Lathia et al., 2007, 2012). The close association of adhesion to stem cell maintenance is further supported by functional studies targeting niche adhesion that results in stem cell detachment and subsequent differentiation and /or death (Georges-Labouesse et al., 1998; Graus-Porta et al., 2001; Loulier et al., 2009; Niola et al., 2012). Whereas the majority of niche adhesion studies involve adhesion receptors, such as integrins or CD44. Here, we provide evidence that JAM-A promotes CSC maintenance in GBM, and our results pose several outstanding questions. Why is JAM-A elevated in GBM? Why might JAM-A be useful to CSCs rather than other adhesion molecules? Possible explanations include the necessity to form cell-cell contacts within a GBM to promote tumor progression and the need for CSCs to directly interact with other cells within the niche. JAM-A is considered to be an epithelial marker as it is expressed at tight junctions and regulates cell-cell adhesion (Severson and Parkos, 2009). The JAM family members play a variety of roles in normal and neoplastic processes, including angiogenesis (Naik et al., 2008), leukocyte migration (Wojcikiewicz et al., 2009), platelet activation (Naik et al., 2012), and the regulation of cell morphology and polarity (Rehder et al., 2006). JAMs can execute these programs by several methods, including adjacent cell JAM-to-JAM interaction, stabilization of integrins on the same cell, or the direct interaction of integrins on an adjacent cell (Severson and Parkos, 2009). There are conflicting reports as to the role of JAM-A in breast cancer progression, with evidence to suggest that loss of JAM-A increases migration and invasion (Naik and Naik, 2008), whereas others suggest that JAM-A is increased during tumor cell migration and adhesion (McSherry et al., 2011). Our work suggests that in CSCs, JAM-A is instrumental in regulating stabilization of integrins on the same cell, although interaction of JAM-A dimers between adjacent CSCs as well as JAM-A-integrin interactions with adjacent cells may also be possible and may contribute to the regulation of CSC maintenance by JAM-A. Future studies expanding the signaling network of JAM-A regulation in CSCs is likely to connect it to other key GBM pathways as well as other stem cell processes, such as epithelial-to-mesenchymal transition. In addition, future studies focusing on the transcriptome, cell cycle, and cell division mode changes as a result of JAM-A targeting are likely to identify additional mechanisms governing JAM-A function in CSCs. Further interrogation of JAM-A function in CSCs from other advanced cancer may also demonstrate a conserved role for JAM-A in CSC maintenance. Given the importance of adhesion in stem cell maintenance, the question arises whether this process can be targeted in CSCs. This therapeutic paradigm is not without complexity and challenges as many adhesion mechanisms, including integrins, are present in both normal and neoplastic stem cells (Hale et al., 2012). Alternative strategies to develop CSC antiadhesion therapies include the identification of coreceptors and/or other signaling pathway members unique to the CSC population. With this goal in mind, our studies demonstrate that JAM-A is specifically used by CSCs to maintain integrin b1 activation and promote stem cell maintenance. Moreover, targeting of JAM-A compromises CSC growth but does not adversely impact non- CSCs or NPCs, suggesting that only tumor cells at the top of the cellular hierarchy are sensitive to JAM-A inhibition. These data provide a rationale for further evaluation of JAM-A in a clinical setting. Taken together, our data demonstrate that CSCs possess specific cell-adhesion mechanisms to regulate their self-renewal and can be targeted with limited disruption to the normal tissue, representing a promising anti-gbm therapeutic paradigm. EXPERIMENTAL PROCEDURES CSC Derivation, Culture, and Analysis Human GBM cells were derived under written informed consent and approved institutional review board protocols from Cleveland Clinic (CCF2352 and CCF2418), University Hospitals-Seidman Cancer Center at Case Western Reserve University (1217 and 1221) for experimental analysis, and from Duke University to establish xenografts in BalbC nu/nu (athymic nude) mice for maintenance of the tumor hierarchy as previously published (Bao et al., 2006; Eyler et al., 2011; Guryanova et al., 2011; Lathia et al., 2010; Li et al., 2009). All procedures involving mice were done under an approved Cleveland Clinic Institutional Animal Care and Use Committee protocol. GBM xenografts for four tumors (T3691, T387, T4121, and T4302) were used for experimental studies. Only low (<5) passage cells were used for analysis to minimize cellular drift, and the majority of cells were used immediately after dissociation. Dissociation and culture of cells was done as previously described (Bao et al., 2006; Eyler et al., 2011; Guryanova et al., 2011; Lathia et al., 2010; Li et al., 2009) and detailed in the Supplemental Information. Flow Cytometry Screening Flow cytometry screening was done as previously described with the BD Lyoplate system (Yuan et al., 2011), and additional parameters are provided in the Supplemental Information. 126 Cell Reports 6, , January 16, 2014 ª2014 The Authors
11 Database Mining and Processing To further refine the flow cytometry screen and interrogate the association of JAM-A to GBM patient survival, the following databases were used: NCI REpository for Molecular BRAin Neoplasia DaTa bioinformatics database ( REMBRANDT), human protein atlas ( The Cancer Genome Atlas (TCGA) data (accessed at and Oncomine (Compendia Bioscience; Relevant analytical parameters are provided in the Supplemental Information. Flow Cytometry Analysis and FISH For flow cytometry analysis, GBM cells were dissociated from patient specimens or xenografts, recovered overnight in supplemented neurobasal media, and sorted on a BD FACS Aria II as previously described (Lathia et al., 2010) using the following antibodies: JAM-A-PE (BD Biosciences), CD31-FITC (BD Biosciences), and CD105-FITC (BD Biosciences). Appropriate isotype control antibodies were used to set gates and live/dead determination was based on 7AAD exclusion (BD Biosciences). Additional flow cytometry analysis was done with JAM-A and an integrin a6-fitc antibody (BD Biosciences). JAM-A-positive/CD105-negative cells were collected for fluorescence in situ hybridization of chromosome 7 and epidermal growth factor (EGFR) using a Vysis EGFR/CEP 7 FISH Probe Kit (Abbott Molecular) as per the manufacturers protocol. Images were taken with a 633 oil immersion objective using a Leica widefield microscope and images were processed and assembled in Photoshop (Adobe). Immunoblotting Analysis Protein analysis was done by immunoblotting as previously described (Eyler et al., 2011; Guryanova et al., 2011; Li et al., 2009). Briefly, 10 mg of total protein was loaded per condition and probed using the following antibodies: JAM-A (Santa Cruz Biotechnology, 1:1,000), Sox2 (Sigma-Aldrich, 1:1,000), integrin a6 (Cell Signaling Technologies, 1:1,000), integrin b1 (Millipore, 1:1,000), Olig2 (Millipore, 1:1,000), GFAP (Sigma, 1:1,000), Map2 (Covance, 1:1,000), Actin (Santa Cruz Biotechnology, 1:2,500), and a-tubulin antibody (Sigma- Aldrich, 1:500) was used as a loading control. Species-specific horseradish peroxidase-conjugated secondary antibodies were used for detection (Invitrogen, 1:2,000). Immunostaining and Immunohistochemical Analysis Immunostaining analysis on xenografted GBM specimens (generated by intracranial injection of GBM specimen T4121) or tumorspheres was done as previously described (Lathia et al., 2010) using 10 mm frozen sections obtained from the Duke University Brain Tumor Center Tissue Bank and histologically confirmed by a neuropathologist (R.E.M.) using antibodies against integrin a6 (Millipore, 1:100), integrin b1 (Millipore, 1:100), Sox2 (R&D Systems, 1:200), and JAM-A (Santa Cruz Biotechnology, 1:50) and species-appropriate secondary antibodies (Alexa 488, 568, or 633; Invitrogen, 1:200). Nuclei were counterstained with Hoechst (1:1,000 dilution from a 5 mg/ml stock solution; Invitrogen). All confocal imaging was done using a Leica SP-5 confocal microscope as described previously (Lathia et al., 2010), and images were processed and assembled in Photoshop (Adobe). Immunohistochemical analysis was done using standard procedures (Li et al., 2009), and details are provided in the Supplemental Information. JAM-A Null Mice and Mouse Histological Analysis JAM-A null ( / ) and wild-type (WT) control mice were obtained from the University of Delaware and generated as previously described (Cooke et al., 2006). For histological analysis of PCNA and PH3, six age-matched JAM-A / and WT were evaluated at three anatomical depths per brain for the SVZ and dentate gyrus. For expression analysis, the following antibodies were used: JAM-A (Santa Cruz Biotechnology, 1:100), PCNA (Abcam, 1:200), PH3 (Millipore 1:500), Doublecortin (Dcx; Cell Signaling, 1:100), Sox2 (R&D Systems, 1:200), GFAP (Dako, 1:200), Iba1 (Millipore, 1:200), S100b (Dako, 1:200), collagen IV (Millipore, 1:500), NG2 (Millipore, 1:100), and NeuN (Millipore, 1:200). Nuclei were counterstained with Hoechst (1:1,000 dilution from a 5 mg/ml stock solution; Invitrogen). All imaging was done using a Leica SP-5 confocal microscope as described previously (Lathia et al., 2010), and images were processed and assembled in Photoshop (Adobe). ECM Adhesion Assays To evaluate adhesion, CSCs were trypsinized and allowed to recover for a minimum of 5 hr. Subsequently, CSCs were plated at 100,000 cells per well in a 24-well plate and treated overnight with either 10 mg/ml immunoglobulin G1 (IgG1) control antibody (Millipore), 10 mg/ml JAM-A-blocking antibody (Santa Cruz Biotechnology), or 10 mg/ml GoH3 integrin-a6-blocking antibody (Millipore). Cells were transferred to a 6-well plate coated with Geltrex (24 mg/ml) or fibronectin (10 mg/ml), as per manufacturer s instructions, and allowed to adhere for 30 min at 37 C. Following adhesion, wells were washed with neurobasal medium and cells were fixed with 4% paraformaldehyde for 15 min at room temperature. After two washes with 13 PBS, nuclei were counterstained with DAPI or propidium iodide (1:1,000). Cells were imaged using the well plate acquisition tool on a Hamamatsu C9100 imaging system. The number of cells adhered to each well were quantified using ImageJ software. Values are expressed in comparison to the nonimmune IgG1-treated samples, which were set to a relative value of 1. JAM-A Lentiviral shrna and Overexpression Construct Production Lentiviral shrna constructs were prepared as we previously reported (Eyler et al., 2011; Guryanova et al., 2011; Lathia et al., 2010; Li et al., 2009). In short, using Lipofectamine 2000 (Invitrogen), 293FT cells were cotransfected with packaging vectors pspax2 and pci-vsvg (Addgene) and lentiviral vectors directing expression of shrna (Sigma) specific to JAM-A (TRCN [KD1] and TRCN [KD2]) or a nontargeting control (NT) shrna (SHC002) to produce virus. Media of the 293FT cell cultures were changed 18 hr after transfection, and viral supernatants were collected 24 and 48 hr later and filtered for immediate use or concentrated with polyethylene glycol precipitation and stored at 80 C for future use. Cell Proliferation, Caspase Activity, and Tumorsphere Formation Assays Assessments were made as previously described (Bao et al., 2006; Eyler et al., 2011; Guryanova et al., 2011; Lathia et al., 2010; Li et al., 2009) and outlined in the Supplemental Information. qpcr qpcr analysis was performed as previously described (Wang et al., 2008) using procedures outlined in the Supplemental Information. In Vivo Tumor Formation Assays For in vivo tumor formation evaluation, CSCs were transplanted into the frontal lobe of BalbC Nu/Nu mice at 100, 1,000, or 5,000 cells per mouse and evaluated as previously described (Bao et al., 2006; Eyler et al., 2011; Guryanova et al., 2011; Lathia et al., 2010; Li et al., 2009). Additional information is provided in the Supplemental Information. Statistical Analysis Reported values are mean values ± SD from studies done in at least triplicate. Unless otherwise stated, one-way ANOVA was used to calculate statistical significance with p values detailed in the text and figure legends. SUPPLEMENTAL INFORMATION Supplemental Information includes Supplemental Experimental Procedures, five figures, and one table and can be found with this article online at dx.doi.org/ /j.celrep ACKNOWLEDGMENTS We thank the members of the Lathia and Rich labs for critical comments on the manuscript. We thank C. Shemo, M. Morgan, P. Barrett, and S. O Bryant for flow cytometry assistance. We thank the Cleveland Clinic Foundation Tissue Procurement Service and S. Staugatis, R. Weil, and M. McGraw. We also thank Cell Reports 6, , January 16, 2014 ª2014 The Authors 127
Supplemental Data. Integrin Alpha 6 Regulates Glioblastoma Stem Cells. Supplemental Data Inventory Supplemental Data
 Cell Stem Cell, Volume 6 Supplemental Data Integrin Alpha 6 Regulates Glioblastoma Stem Cells Justin D. Lathia, Joseph Gallagher, John M. Heddleston, Jialiang Wang, Christine E. Eyler, Jennifer MacSwords,
Cell Stem Cell, Volume 6 Supplemental Data Integrin Alpha 6 Regulates Glioblastoma Stem Cells Justin D. Lathia, Joseph Gallagher, John M. Heddleston, Jialiang Wang, Christine E. Eyler, Jennifer MacSwords,
Differential Connexin Function Enhances Self-Renewal in Glioblastoma
 CSU ID: 2558414 Name: Sonal Patel Differential Connexin Function Enhances Self-Renewal in Glioblastoma Cancer is one of the most common disease in the US with more than a million people affected every
CSU ID: 2558414 Name: Sonal Patel Differential Connexin Function Enhances Self-Renewal in Glioblastoma Cancer is one of the most common disease in the US with more than a million people affected every
Supplemental Figure 1. Intracranial transduction of a modified ptomo lentiviral vector in the mouse
 Supplemental figure legends Supplemental Figure 1. Intracranial transduction of a modified ptomo lentiviral vector in the mouse hippocampus targets GFAP-positive but not NeuN-positive cells. (A) Stereotaxic
Supplemental figure legends Supplemental Figure 1. Intracranial transduction of a modified ptomo lentiviral vector in the mouse hippocampus targets GFAP-positive but not NeuN-positive cells. (A) Stereotaxic
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
Sema3C Promotes the Survival and Tumorigenicity of Glioma Stem Cells through Rac1 Activation
 Cell Reports, Volume 9 Supplemental Information Sema3C Promotes the Survival and Tumorigenicity of Glioma Stem Cells through Rac1 Activation Jianghong Man, Jocelyn Shoemake, Wenchao Zhou, Xiaoguang Fang,
Cell Reports, Volume 9 Supplemental Information Sema3C Promotes the Survival and Tumorigenicity of Glioma Stem Cells through Rac1 Activation Jianghong Man, Jocelyn Shoemake, Wenchao Zhou, Xiaoguang Fang,
CRIPTO-1 A POSSIBLE NEW BIOMARKER IN GLIOBLASTOMA MULTIFORME PIA OLESEN, MD, PHD STUDENT
 CRIPTO-1 A POSSIBLE NEW BIOMARKER IN GLIOBLASTOMA MULTIFORME PIA OLESEN, MD, PHD STUDENT Glioblastoma WHO Grade IV Glioma Heterogenic Undiffenrentiated phenotype 50% of all Gliomas Around 600 patients
CRIPTO-1 A POSSIBLE NEW BIOMARKER IN GLIOBLASTOMA MULTIFORME PIA OLESEN, MD, PHD STUDENT Glioblastoma WHO Grade IV Glioma Heterogenic Undiffenrentiated phenotype 50% of all Gliomas Around 600 patients
Supplementary Figure 1. Validation of astrocytes. Primary astrocytes were
 Supplementary Figure 1. Validation of astrocytes. Primary astrocytes were separated from the glial cultures using a mild trypsinization protocol. Anti-glial fibrillary acidic protein (GFAP) immunofluorescent
Supplementary Figure 1. Validation of astrocytes. Primary astrocytes were separated from the glial cultures using a mild trypsinization protocol. Anti-glial fibrillary acidic protein (GFAP) immunofluorescent
Erzsebet Kokovay, Susan Goderie, Yue Wang, Steve Lotz, Gang Lin, Yu Sun, Badrinath Roysam, Qin Shen,
 Cell Stem Cell, Volume 7 Supplemental Information Adult SVZ Lineage Cells Home to and Leave the Vascular Niche via Differential Responses to SDF1/CXCR4 Signaling Erzsebet Kokovay, Susan Goderie, Yue Wang,
Cell Stem Cell, Volume 7 Supplemental Information Adult SVZ Lineage Cells Home to and Leave the Vascular Niche via Differential Responses to SDF1/CXCR4 Signaling Erzsebet Kokovay, Susan Goderie, Yue Wang,
Gene co-expression networks in the mouse, monkey, and human brain July 16, Jeremy Miller Scientist I
 Gene co-expression networks in the mouse, monkey, and human brain July 16, 2013 Jeremy Miller Scientist I jeremym@alleninstitute.org Outline 1. Brief introduction to previous WGCNA studies in brain 2.
Gene co-expression networks in the mouse, monkey, and human brain July 16, 2013 Jeremy Miller Scientist I jeremym@alleninstitute.org Outline 1. Brief introduction to previous WGCNA studies in brain 2.
Supplemental Figure 1. Isolation and characterization of CD133+ neurosphere-like
 SUPPLEMENTL FIGURE LEGENDS Supplemental Figure 1. Isolation and characterization of CD133+ neurosphere-like spheroids from a human brain tumor sample or glioma xenograft. () CD133+ tumor cells isolated
SUPPLEMENTL FIGURE LEGENDS Supplemental Figure 1. Isolation and characterization of CD133+ neurosphere-like spheroids from a human brain tumor sample or glioma xenograft. () CD133+ tumor cells isolated
Supplementary Figure 1
 Supplementary Figure 1 The average sigmoid parametric curves of capillary dilation time courses and average time to 50% peak capillary diameter dilation computed from individual capillary responses averaged
Supplementary Figure 1 The average sigmoid parametric curves of capillary dilation time courses and average time to 50% peak capillary diameter dilation computed from individual capillary responses averaged
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
TEB. Id4 p63 DAPI Merge. Id4 CK8 DAPI Merge
 a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis
 Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the
 Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the genome-wide methylation microarray data. Mean ± s.d.; Student
Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the genome-wide methylation microarray data. Mean ± s.d.; Student
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplemental Information. Induction of Expansion and Folding. in Human Cerebral Organoids
 Cell Stem Cell, Volume 20 Supplemental Information Induction of Expansion and Folding in Human Cerebral Organoids Yun Li, Julien Muffat, Attya Omer, Irene Bosch, Madeline A. Lancaster, Mriganka Sur, Lee
Cell Stem Cell, Volume 20 Supplemental Information Induction of Expansion and Folding in Human Cerebral Organoids Yun Li, Julien Muffat, Attya Omer, Irene Bosch, Madeline A. Lancaster, Mriganka Sur, Lee
Figure S1: Effects on haptotaxis are independent of effects on cell velocity A)
 Supplemental Figures Figure S1: Effects on haptotaxis are independent of effects on cell velocity A) Velocity of MV D7 fibroblasts expressing different GFP-tagged Ena/VASP family proteins in the haptotaxis
Supplemental Figures Figure S1: Effects on haptotaxis are independent of effects on cell velocity A) Velocity of MV D7 fibroblasts expressing different GFP-tagged Ena/VASP family proteins in the haptotaxis
Supplementary Figure 1
 Supplementary Figure 1 Kif1a RNAi effect on basal progenitor differentiation Related to Figure 2. Representative confocal images of the VZ and SVZ of rat cortices transfected at E16 with scrambled or Kif1a
Supplementary Figure 1 Kif1a RNAi effect on basal progenitor differentiation Related to Figure 2. Representative confocal images of the VZ and SVZ of rat cortices transfected at E16 with scrambled or Kif1a
Supplemental Information. Otic Mesenchyme Cells Regulate. Spiral Ganglion Axon Fasciculation. through a Pou3f4/EphA4 Signaling Pathway
 Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
marker. DAPI labels nuclei. Flies were 20 days old. Scale bar is 5 µm. Ctrl is
 Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
CANCER STEM CELLS. Cancer Stem Cell-Specific Scavenger Receptor CD36 Drives Glioblastoma Progression ABSTRACT INTRODUCTION
 CANCER STEM CELLS Cancer Stem Cell-Specific Scavenger Receptor CD36 Drives Glioblastoma Progression JAMES S. HALE, a BALINT OTVOS, a MAKSIM SINYUK, a ALVARO G. ALVARADO, a,b MASAHIRO HITOMI, a,b KEVIN
CANCER STEM CELLS Cancer Stem Cell-Specific Scavenger Receptor CD36 Drives Glioblastoma Progression JAMES S. HALE, a BALINT OTVOS, a MAKSIM SINYUK, a ALVARO G. ALVARADO, a,b MASAHIRO HITOMI, a,b KEVIN
Supplementary Materials. for Garmy-Susini, et al, Integrin 4 1 signaling is required for lymphangiogenesis and tumor metastasis
 Supplementary Materials for Garmy-Susini, et al, Integrin 4 1 signaling is required for lymphangiogenesis and tumor metastasis 1 Supplementary Figure Legends Supplementary Figure 1: Integrin expression
Supplementary Materials for Garmy-Susini, et al, Integrin 4 1 signaling is required for lymphangiogenesis and tumor metastasis 1 Supplementary Figure Legends Supplementary Figure 1: Integrin expression
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
Supplementary Information
 Supplementary Information Title Degeneration and impaired regeneration of gray matter oligodendrocytes in amyotrophic lateral sclerosis Authors Shin H. Kang, Ying Li, Masahiro Fukaya, Ileana Lorenzini,
Supplementary Information Title Degeneration and impaired regeneration of gray matter oligodendrocytes in amyotrophic lateral sclerosis Authors Shin H. Kang, Ying Li, Masahiro Fukaya, Ileana Lorenzini,
Targeting Glioma Stem Cell-Derived Pericytes Disrupts the Blood-Tumor Barrier and Improves Chemotherapeutic Efficacy
 Article Targeting Glioma Stem Cell-Derived Pericytes Disrupts the Blood- Barrier and Improves Chemotherapeutic Efficacy Graphical Abstract Authors Wenchao Zhou, Cong Chen, Yu Shi,..., Jennifer S. Yu, Shideng
Article Targeting Glioma Stem Cell-Derived Pericytes Disrupts the Blood- Barrier and Improves Chemotherapeutic Efficacy Graphical Abstract Authors Wenchao Zhou, Cong Chen, Yu Shi,..., Jennifer S. Yu, Shideng
(A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and a
 Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplemental figure 1. PDGFRα is expressed dominantly by stromal cells surrounding mammary ducts and alveoli. A) IHC staining of PDGFRα in
 Supplemental figure 1. PDGFRα is expressed dominantly by stromal cells surrounding mammary ducts and alveoli. A) IHC staining of PDGFRα in nulliparous (left panel) and InvD6 mouse mammary glands (right
Supplemental figure 1. PDGFRα is expressed dominantly by stromal cells surrounding mammary ducts and alveoli. A) IHC staining of PDGFRα in nulliparous (left panel) and InvD6 mouse mammary glands (right
Award Number: W81XWH TITLE: Characterizing an EMT Signature in Breast Cancer. PRINCIPAL INVESTIGATOR: Melanie C.
 AD Award Number: W81XWH-08-1-0306 TITLE: Characterizing an EMT Signature in Breast Cancer PRINCIPAL INVESTIGATOR: Melanie C. Bocanegra CONTRACTING ORGANIZATION: Leland Stanford Junior University Stanford,
AD Award Number: W81XWH-08-1-0306 TITLE: Characterizing an EMT Signature in Breast Cancer PRINCIPAL INVESTIGATOR: Melanie C. Bocanegra CONTRACTING ORGANIZATION: Leland Stanford Junior University Stanford,
Nature Neuroscience: doi: /nn.2275
 Supplementary Figure S1. The presence of MeCP2 in enriched primary glial cultures from rat or mouse brains is not neuronal. Western blot analysis of protein extracts from (a) rat glial and neuronal cultures.
Supplementary Figure S1. The presence of MeCP2 in enriched primary glial cultures from rat or mouse brains is not neuronal. Western blot analysis of protein extracts from (a) rat glial and neuronal cultures.
Citation for published version (APA): Martina-Mamber, C. E. (2014). GFAP as an understudy in adult neurogenesis. 's-hertogenbosch: Boxpress.
 UvA-DARE (Digital Academic Repository) GFAP as an understudy in adult neurogenesis Mamber, C.E. Link to publication Citation for published version (APA): Martina-Mamber, C. E. (2014). GFAP as an understudy
UvA-DARE (Digital Academic Repository) GFAP as an understudy in adult neurogenesis Mamber, C.E. Link to publication Citation for published version (APA): Martina-Mamber, C. E. (2014). GFAP as an understudy
Glioblastoma Multiforme
 Glioblastoma Multiforme Highly malignant, invasive, difficult-to-treat primary brain tumor" " Frequency: 9,000 cases/year (peak age, 55 65 years)" " Recurrence: rapid growth; size may double every 10 days"
Glioblastoma Multiforme Highly malignant, invasive, difficult-to-treat primary brain tumor" " Frequency: 9,000 cases/year (peak age, 55 65 years)" " Recurrence: rapid growth; size may double every 10 days"
Nature Genetics: doi: /ng Supplementary Figure 1. Phenotypic characterization of MES- and ADRN-type cells.
 Supplementary Figure 1 Phenotypic characterization of MES- and ADRN-type cells. (a) Bright-field images showing cellular morphology of MES-type (691-MES, 700-MES, 717-MES) and ADRN-type (691-ADRN, 700-
Supplementary Figure 1 Phenotypic characterization of MES- and ADRN-type cells. (a) Bright-field images showing cellular morphology of MES-type (691-MES, 700-MES, 717-MES) and ADRN-type (691-ADRN, 700-
supplementary information
 DOI: 10.1038/ncb2153 Figure S1 Ectopic expression of HAUSP up-regulates REST protein. (a) Immunoblotting showed that ectopic expression of HAUSP increased REST protein levels in ENStemA NPCs. (b) Immunofluorescent
DOI: 10.1038/ncb2153 Figure S1 Ectopic expression of HAUSP up-regulates REST protein. (a) Immunoblotting showed that ectopic expression of HAUSP increased REST protein levels in ENStemA NPCs. (b) Immunofluorescent
Supplementary Information and Figure legends
 Supplementary Information and Figure legends Table S1. Primers for quantitative RT-PCR Target Sequence (5 -> 3 ) Target Sequence (5 -> 3 ) DAB2IP F:TGGACGATGTGCTCTATGCC R:GGATGGTGATGGTTTGGTAG Snail F:CCTCCCTGTCAGATGAGGAC
Supplementary Information and Figure legends Table S1. Primers for quantitative RT-PCR Target Sequence (5 -> 3 ) Target Sequence (5 -> 3 ) DAB2IP F:TGGACGATGTGCTCTATGCC R:GGATGGTGATGGTTTGGTAG Snail F:CCTCCCTGTCAGATGAGGAC
SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer.
 Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Neocortex Zbtb20 / NFIA / Sox9
 Neocortex / NFIA / Sox9 Supplementary Figure 1. Expression of, NFIA, and Sox9 in the mouse neocortex at. The lower panels are higher magnification views of the oxed area. Arrowheads indicate triple-positive
Neocortex / NFIA / Sox9 Supplementary Figure 1. Expression of, NFIA, and Sox9 in the mouse neocortex at. The lower panels are higher magnification views of the oxed area. Arrowheads indicate triple-positive
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung
 Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
A new role for meninges as a niche for stem/precursor cells with neural differentiation potential during development up to adulthood
 A new role for meninges as a niche for stem/precursor cells with neural differentiation potential during development up to adulthood Francesco Bifari, MD, PhD Mauro Krampera, MD, PhD Ilaria Decimo, PhD
A new role for meninges as a niche for stem/precursor cells with neural differentiation potential during development up to adulthood Francesco Bifari, MD, PhD Mauro Krampera, MD, PhD Ilaria Decimo, PhD
stem cell products Basement Membrane Matrix Products Rat Mesenchymal Stem Cell Growth and Differentiation Products
 stem cell products Basement Membrane Matrix Products Rat Mesenchymal Stem Cell Growth and Differentiation Products Stem Cell Qualified Extracellular Matrix Proteins Stem cell research requires the finest
stem cell products Basement Membrane Matrix Products Rat Mesenchymal Stem Cell Growth and Differentiation Products Stem Cell Qualified Extracellular Matrix Proteins Stem cell research requires the finest
Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the
 Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the location of the transmembrane (TM), FRM binding (FB)
Supplementary Figure 1 Expression of Crb3 in mouse sciatic nerve: biochemical analysis (a) Schematic of Crb3 isoforms, ERLI and CLPI, indicating the location of the transmembrane (TM), FRM binding (FB)
Nature Genetics: doi: /ng Supplementary Figure 1
 Supplementary Figure 1 MSI2 interactors are associated with the riboproteome and are functionally relevant. (a) Coomassie blue staining of FLAG-MSI2 immunoprecipitated complexes. (b) GO analysis of MSI2-interacting
Supplementary Figure 1 MSI2 interactors are associated with the riboproteome and are functionally relevant. (a) Coomassie blue staining of FLAG-MSI2 immunoprecipitated complexes. (b) GO analysis of MSI2-interacting
Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures
 Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures Supplementary Figure 1 mir-128-3p is highly expressed in chemoresistant, metastatic
Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures Supplementary Figure 1 mir-128-3p is highly expressed in chemoresistant, metastatic
Material and Methods. Flow Cytometry Analyses:
 Material and Methods Flow Cytometry Analyses: Immunostaining of breast cancer cells for HER2 was performed by incubating cells with anti- HER2/neu APC (Biosciences, Cat# 340554), anti-her2/neu PE (Biosciences,
Material and Methods Flow Cytometry Analyses: Immunostaining of breast cancer cells for HER2 was performed by incubating cells with anti- HER2/neu APC (Biosciences, Cat# 340554), anti-her2/neu PE (Biosciences,
SUPPLEMENTARY FIG. S2. Representative counting fields used in quantification of the in vitro neural differentiation of pattern of dnscs.
 Supplementary Data SUPPLEMENTARY FIG. S1. Representative counting fields used in quantification of the in vitro neural differentiation of pattern of anpcs. A panel of lineage-specific markers were used
Supplementary Data SUPPLEMENTARY FIG. S1. Representative counting fields used in quantification of the in vitro neural differentiation of pattern of anpcs. A panel of lineage-specific markers were used
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 DOT1L regulates the expression of epithelial and mesenchymal markers. (a) The expression levels and cellular localizations of EMT markers were confirmed by
Supplementary Figures Supplementary Figure 1 DOT1L regulates the expression of epithelial and mesenchymal markers. (a) The expression levels and cellular localizations of EMT markers were confirmed by
Bone marrow-derived mesenchymal stem cells improve diabetes-induced cognitive impairment by
 Nakano et al. Supplementary information 1. Supplementary Figure 2. Methods 3. References Bone marrow-derived mesenchymal stem cells improve diabetes-induced cognitive impairment by exosome transfer into
Nakano et al. Supplementary information 1. Supplementary Figure 2. Methods 3. References Bone marrow-derived mesenchymal stem cells improve diabetes-induced cognitive impairment by exosome transfer into
Supporting Information
 Supporting Information Chan et al. 1.173/pnas.9654916 A Patient B Xenograft C * remaining feature of normal lymph node * * * D lymphocytes Infiltrating transitional carcinoma cells E Enlarged axillary
Supporting Information Chan et al. 1.173/pnas.9654916 A Patient B Xenograft C * remaining feature of normal lymph node * * * D lymphocytes Infiltrating transitional carcinoma cells E Enlarged axillary
Supplemental Figure 1
 Supplemental Figure 1 A S100A4: SFLGKRTDEAAFQKLMSNLDSNRDNEVDFQEYCVFLSCIAMMCNEFFEGFPDK Overlap: SF G DE KLM LD N D VDFQEY VFL I M N FF G PD S100A2: SFVGEKVDEEGLKKLMGSLDENSDQQVDFQEYAVFLALITVMCNDFFQGCPDR
Supplemental Figure 1 A S100A4: SFLGKRTDEAAFQKLMSNLDSNRDNEVDFQEYCVFLSCIAMMCNEFFEGFPDK Overlap: SF G DE KLM LD N D VDFQEY VFL I M N FF G PD S100A2: SFVGEKVDEEGLKKLMGSLDENSDQQVDFQEYAVFLALITVMCNDFFQGCPDR
Predictive Biomarkers in GBM
 Predictive Biomarkers in GBM C. David James, Ph.D. Professor & Associate Director, Brain Tumor Research Center Dept. Neurological Surgery and Helen Diller Comprehensive Cancer Center, University of California
Predictive Biomarkers in GBM C. David James, Ph.D. Professor & Associate Director, Brain Tumor Research Center Dept. Neurological Surgery and Helen Diller Comprehensive Cancer Center, University of California
Supplemental Information. Tissue Myeloid Progenitors Differentiate. into Pericytes through TGF-b Signaling. in Developing Skin Vasculature
 Cell Reports, Volume 18 Supplemental Information Tissue Myeloid Progenitors Differentiate into Pericytes through TGF-b Signaling in Developing Skin Vasculature Tomoko Yamazaki, Ani Nalbandian, Yutaka Uchida,
Cell Reports, Volume 18 Supplemental Information Tissue Myeloid Progenitors Differentiate into Pericytes through TGF-b Signaling in Developing Skin Vasculature Tomoko Yamazaki, Ani Nalbandian, Yutaka Uchida,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2535 Figure S1 SOX10 is expressed in human giant congenital nevi and its expression in human melanoma samples suggests that SOX10 functions in a MITF-independent manner. a, b, Representative
DOI: 10.1038/ncb2535 Figure S1 SOX10 is expressed in human giant congenital nevi and its expression in human melanoma samples suggests that SOX10 functions in a MITF-independent manner. a, b, Representative
(A) Dose response curves of HMLE_shGFP (blue circle), HMLE_shEcad (red square),
 Supplementary Figures and Tables Figure S1. Validation of EMT-selective small molecules (A) Dose response curves of HMLE_shGFP (blue circle), HMLE_shEcad (red square), and HMLE_Twist (black diamond) cells
Supplementary Figures and Tables Figure S1. Validation of EMT-selective small molecules (A) Dose response curves of HMLE_shGFP (blue circle), HMLE_shEcad (red square), and HMLE_Twist (black diamond) cells
TMA-VARESE COHORT-1 TMA-BERN COHORT-2
 Supplementary Figure 1 TMA-VARESE COHORT-1 TOTAL SAMPLES #5 GLEASON SCORE Number Percentage 6 16 32% = 7 17 34% >7 17 34% TUMOR STAGE T2C 28 56% T3A- 21 42% T3C-T4 1 2% NODE STATUS N 42 84% N1 8 16% PSA
Supplementary Figure 1 TMA-VARESE COHORT-1 TOTAL SAMPLES #5 GLEASON SCORE Number Percentage 6 16 32% = 7 17 34% >7 17 34% TUMOR STAGE T2C 28 56% T3A- 21 42% T3C-T4 1 2% NODE STATUS N 42 84% N1 8 16% PSA
Genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1.
 Genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Youngsoo Lee, Sachin Katyal, Yang Li, Sherif F. El-Khamisy, Helen R. Russell, Keith W. Caldecott and Peter J. McKinnon.
Genesis of cerebellar interneurons and the prevention of neural DNA damage require XRCC1. Youngsoo Lee, Sachin Katyal, Yang Li, Sherif F. El-Khamisy, Helen R. Russell, Keith W. Caldecott and Peter J. McKinnon.
Layered-IHC (L-IHC): A novel and robust approach to multiplexed immunohistochemistry So many markers and so little tissue
 Page 1 The need for multiplex detection of tissue biomarkers. There is a constant and growing demand for increased biomarker analysis in human tissue specimens. Analysis of tissue biomarkers is key to
Page 1 The need for multiplex detection of tissue biomarkers. There is a constant and growing demand for increased biomarker analysis in human tissue specimens. Analysis of tissue biomarkers is key to
Supplemental Data. Wnt/β-Catenin Signaling in Mesenchymal Progenitors. Controls Osteoblast and Chondrocyte
 Supplemental Data Wnt/β-Catenin Signaling in Mesenchymal Progenitors Controls Osteoblast and Chondrocyte Differentiation during Vertebrate Skeletogenesis Timothy F. Day, Xizhi Guo, Lisa Garrett-Beal, and
Supplemental Data Wnt/β-Catenin Signaling in Mesenchymal Progenitors Controls Osteoblast and Chondrocyte Differentiation during Vertebrate Skeletogenesis Timothy F. Day, Xizhi Guo, Lisa Garrett-Beal, and
Correlation between expression and significance of δ-catenin, CD31, and VEGF of non-small cell lung cancer
 Correlation between expression and significance of δ-catenin, CD31, and VEGF of non-small cell lung cancer X.L. Liu 1, L.D. Liu 2, S.G. Zhang 1, S.D. Dai 3, W.Y. Li 1 and L. Zhang 1 1 Thoracic Surgery,
Correlation between expression and significance of δ-catenin, CD31, and VEGF of non-small cell lung cancer X.L. Liu 1, L.D. Liu 2, S.G. Zhang 1, S.D. Dai 3, W.Y. Li 1 and L. Zhang 1 1 Thoracic Surgery,
Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION
 Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION X. Shawn Liu 1, 3, Bing Song 2, 3, Bennett D. Elzey 3, 4, Timothy L. Ratliff 3, 4, Stephen F. Konieczny
Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION X. Shawn Liu 1, 3, Bing Song 2, 3, Bennett D. Elzey 3, 4, Timothy L. Ratliff 3, 4, Stephen F. Konieczny
Supplemental Figure S1. RANK expression on human lung cancer cells.
 Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
Supplementary Table; Supplementary Figures and legends S1-S21; Supplementary Materials and Methods
 Silva et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability Supplementary Table; Supplementary Figures and legends S1-S21; Supplementary
Silva et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability Supplementary Table; Supplementary Figures and legends S1-S21; Supplementary
Supplementary Fig. 1: ATM is phosphorylated in HER2 breast cancer cell lines. (A) ATM is phosphorylated in SKBR3 cells depending on ATM and HER2
 Supplementary Fig. 1: ATM is phosphorylated in HER2 breast cancer cell lines. (A) ATM is phosphorylated in SKBR3 cells depending on ATM and HER2 activity. Upper panel: Representative histograms for FACS
Supplementary Fig. 1: ATM is phosphorylated in HER2 breast cancer cell lines. (A) ATM is phosphorylated in SKBR3 cells depending on ATM and HER2 activity. Upper panel: Representative histograms for FACS
SHREE ET AL, SUPPLEMENTAL MATERIALS. (A) Workflow for tumor cell line derivation and orthotopic implantation.
 SHREE ET AL, SUPPLEMENTAL MATERIALS SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure 1. Derivation and characterization of TS1-TGL and TS2-TGL PyMT cell lines and development of an orthotopic
SHREE ET AL, SUPPLEMENTAL MATERIALS SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure 1. Derivation and characterization of TS1-TGL and TS2-TGL PyMT cell lines and development of an orthotopic
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
Bmi-1 regulates stem cell-like properties of gastric cancer cells via modulating mirnas
 Wang et al. Journal of Hematology & Oncology (2016) 9:90 DOI 10.1186/s13045-016-0323-9 RESEARCH Bmi-1 regulates stem cell-like properties of gastric cancer cells via modulating mirnas Open Access Xiaofeng
Wang et al. Journal of Hematology & Oncology (2016) 9:90 DOI 10.1186/s13045-016-0323-9 RESEARCH Bmi-1 regulates stem cell-like properties of gastric cancer cells via modulating mirnas Open Access Xiaofeng
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Bredel M, Scholtens DM, Yadav AK, et al. NFKBIA deletion in
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Bredel M, Scholtens DM, Yadav AK, et al. NFKBIA deletion in
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
injected subcutaneously into flanks of 6-8 week old athymic male nude mice (LNCaP SQ) and body
 SUPPLEMENTAL FIGURE LEGENDS Figure S1: Generation of ENZR Xenografts and Cell Lines: (A) 1x10 6 LNCaP cells in matrigel were injected subcutaneously into flanks of 6-8 week old athymic male nude mice (LNCaP
SUPPLEMENTAL FIGURE LEGENDS Figure S1: Generation of ENZR Xenografts and Cell Lines: (A) 1x10 6 LNCaP cells in matrigel were injected subcutaneously into flanks of 6-8 week old athymic male nude mice (LNCaP
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
SUPPLEMENTARY INFORMATION
 Supplemental Figure 1. Furin is efficiently deleted in CD4 + and CD8 + T cells. a, Western blot for furin and actin proteins in CD4cre-fur f/f and fur f/f Th1 cells. Wild-type and furin-deficient CD4 +
Supplemental Figure 1. Furin is efficiently deleted in CD4 + and CD8 + T cells. a, Western blot for furin and actin proteins in CD4cre-fur f/f and fur f/f Th1 cells. Wild-type and furin-deficient CD4 +
Supplemental Information. Menin Deficiency Leads to Depressive-like. Behaviors in Mice by Modulating. Astrocyte-Mediated Neuroinflammation
 Neuron, Volume 100 Supplemental Information Menin Deficiency Leads to Depressive-like Behaviors in Mice by Modulating Astrocyte-Mediated Neuroinflammation Lige Leng, Kai Zhuang, Zeyue Liu, Changquan Huang,
Neuron, Volume 100 Supplemental Information Menin Deficiency Leads to Depressive-like Behaviors in Mice by Modulating Astrocyte-Mediated Neuroinflammation Lige Leng, Kai Zhuang, Zeyue Liu, Changquan Huang,
Mitosis. Single Nano Micro Milli Macro. Primary. PCNA expression
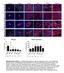 a b c DAPI YFP CC3 DAPI YFP PCNA DAPI YFP ph3 DAPI YFP KI67 e 6 Mitosis f 1 PCNA expression %ph3 + /YFP + n= 63 87 61 3 13 8 n= 15 3 9 1 5 %PCNA+/YFP+ 8 6 Supplementary Figure 1. Proliferation/apoptosis
a b c DAPI YFP CC3 DAPI YFP PCNA DAPI YFP ph3 DAPI YFP KI67 e 6 Mitosis f 1 PCNA expression %ph3 + /YFP + n= 63 87 61 3 13 8 n= 15 3 9 1 5 %PCNA+/YFP+ 8 6 Supplementary Figure 1. Proliferation/apoptosis
Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma
 Supplementary information for: Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma Rintaro Hashizume 1, Noemi Andor 2, Yuichiro Ihara 2, Robin Lerner 2, Haiyun
Supplementary information for: Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma Rintaro Hashizume 1, Noemi Andor 2, Yuichiro Ihara 2, Robin Lerner 2, Haiyun
LGR5, a novel functional glioma stem cell marker, promotes EMT by activating the Wnt/β-catenin pathway and predicts poor survival of glioma patients
 Zhang et al. Journal of Experimental & Clinical Cancer Research (2018) 37:225 https://doi.org/10.1186/s13046-018-0864-6 RESEARCH Open Access LGR5, a novel functional glioma stem cell marker, promotes EMT
Zhang et al. Journal of Experimental & Clinical Cancer Research (2018) 37:225 https://doi.org/10.1186/s13046-018-0864-6 RESEARCH Open Access LGR5, a novel functional glioma stem cell marker, promotes EMT
GFP/Iba1/GFAP. Brain. Liver. Kidney. Lung. Hoechst/Iba1/TLR9!
 Supplementary information a +KA Relative expression d! Tlr9 5!! 5! NSC Neuron Astrocyte Microglia! 5! Tlr7!!!! NSC Neuron Astrocyte! GFP/Sβ/! Iba/Hoechst Microglia e Hoechst/Iba/TLR9! GFP/Iba/GFAP f Brain
Supplementary information a +KA Relative expression d! Tlr9 5!! 5! NSC Neuron Astrocyte Microglia! 5! Tlr7!!!! NSC Neuron Astrocyte! GFP/Sβ/! Iba/Hoechst Microglia e Hoechst/Iba/TLR9! GFP/Iba/GFAP f Brain
Nicholas Szerlip, MD Department of Neurosurgery Wayne State University Karmanos Cancer Institute
 Nicholas Szerlip, MD Department of Neurosurgery Wayne State University Karmanos Cancer Institute Many different names (Heregulin, Aria) Found when investigating ligands that activated HER2 (it actually
Nicholas Szerlip, MD Department of Neurosurgery Wayne State University Karmanos Cancer Institute Many different names (Heregulin, Aria) Found when investigating ligands that activated HER2 (it actually
Conserved properties of dentate gyrus neurogenesis across postnatal development revealed by single-cell RNA sequencing
 SUPPLEMENTARY INFORMATION Resource https://doi.org/10.1038/s41593-017-0056-2 In the format provided by the authors and unedited. Conserved properties of dentate gyrus neurogenesis across postnatal development
SUPPLEMENTARY INFORMATION Resource https://doi.org/10.1038/s41593-017-0056-2 In the format provided by the authors and unedited. Conserved properties of dentate gyrus neurogenesis across postnatal development
Supplementary Figure (OH) 22 nanoparticles did not affect cell viability and apoposis. MDA-MB-231, MCF-7, MCF-10A and BT549 cells were
 Supplementary Figure 1. Gd@C 82 (OH) 22 nanoparticles did not affect cell viability and apoposis. MDA-MB-231, MCF-7, MCF-10A and BT549 cells were treated with PBS, Gd@C 82 (OH) 22, C 60 (OH) 22 or GdCl
Supplementary Figure 1. Gd@C 82 (OH) 22 nanoparticles did not affect cell viability and apoposis. MDA-MB-231, MCF-7, MCF-10A and BT549 cells were treated with PBS, Gd@C 82 (OH) 22, C 60 (OH) 22 or GdCl
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation
 Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
supplementary information
 DOI: 10.1038/ncb2133 Figure S1 Actomyosin organisation in human squamous cell carcinoma. (a) Three examples of actomyosin organisation around the edges of squamous cell carcinoma biopsies are shown. Myosin
DOI: 10.1038/ncb2133 Figure S1 Actomyosin organisation in human squamous cell carcinoma. (a) Three examples of actomyosin organisation around the edges of squamous cell carcinoma biopsies are shown. Myosin
Figure S1. The PDE5 inhibitor sildenafil interacts with celecoxib to kill cancer cell lines. (A) Hepatoma
 Figure S1. The PDE5 inhibitor sildenafil interacts with celecoxib to kill cancer cell lines. (A) Hepatoma cells were treated with celecoxib ( 5.0 M) and/or sildenafil (, 2.0 M). ls were isolated after
Figure S1. The PDE5 inhibitor sildenafil interacts with celecoxib to kill cancer cell lines. (A) Hepatoma cells were treated with celecoxib ( 5.0 M) and/or sildenafil (, 2.0 M). ls were isolated after
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 EGFR inhibition activates signaling pathways (a-b) EGFR inhibition activates signaling pathways (a) U251EGFR cells were treated with erlotinib (1µM) for the indicated times followed
Supplementary Figure 1 EGFR inhibition activates signaling pathways (a-b) EGFR inhibition activates signaling pathways (a) U251EGFR cells were treated with erlotinib (1µM) for the indicated times followed
(A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC,
 Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH
 EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
Supplemental information
 Carcinoemryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation y Chiang et al. Supplemental
Carcinoemryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation y Chiang et al. Supplemental
High expression of fibroblast activation protein is an adverse prognosticator in gastric cancer.
 Biomedical Research 2017; 28 (18): 7779-7783 ISSN 0970-938X www.biomedres.info High expression of fibroblast activation protein is an adverse prognosticator in gastric cancer. Hu Song 1, Qi-yu Liu 2, Zhi-wei
Biomedical Research 2017; 28 (18): 7779-7783 ISSN 0970-938X www.biomedres.info High expression of fibroblast activation protein is an adverse prognosticator in gastric cancer. Hu Song 1, Qi-yu Liu 2, Zhi-wei
Supplementary figure legends
 Supplementary figure legends Supplementary Figure 1. Exposure of CRT occurs independently from the apoptosisassociated loss of the mitochondrial membrane potential (MMP). (A) HeLa cells treated with MTX
Supplementary figure legends Supplementary Figure 1. Exposure of CRT occurs independently from the apoptosisassociated loss of the mitochondrial membrane potential (MMP). (A) HeLa cells treated with MTX
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC
 Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
ECM1 controls T H 2 cell egress from lymph nodes through re-expression of S1P 1
 ZH, Li et al, page 1 ECM1 controls T H 2 cell egress from lymph nodes through re-expression of S1P 1 Zhenhu Li 1,4,Yuan Zhang 1,4, Zhiduo Liu 1, Xiaodong Wu 1, Yuhan Zheng 1, Zhiyun Tao 1, Kairui Mao 1,
ZH, Li et al, page 1 ECM1 controls T H 2 cell egress from lymph nodes through re-expression of S1P 1 Zhenhu Li 1,4,Yuan Zhang 1,4, Zhiduo Liu 1, Xiaodong Wu 1, Yuhan Zheng 1, Zhiyun Tao 1, Kairui Mao 1,
Ophthalmology, Radiation Oncology,
 Supporting Online Material Journal: Nature Neuroscience Article Title: Corresponding Author: All Authors: Affiliations: Tanycytes of the Hypothalamic Median Eminence Form a Diet- Responsive Neurogenic
Supporting Online Material Journal: Nature Neuroscience Article Title: Corresponding Author: All Authors: Affiliations: Tanycytes of the Hypothalamic Median Eminence Form a Diet- Responsive Neurogenic
Supplementary Figure S1: Tanycytes are restricted to the central/posterior hypothalamus
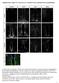 Supplementary Figure S1: Tanycytes are restricted to the central/posterior hypothalamus a: Expression of Vimentin, GFAP, Sox2 and Nestin in anterior, central and posterior hypothalamus. In the anterior
Supplementary Figure S1: Tanycytes are restricted to the central/posterior hypothalamus a: Expression of Vimentin, GFAP, Sox2 and Nestin in anterior, central and posterior hypothalamus. In the anterior
SUPPLEMENTARY INFORMATION. Supplementary Figures S1-S9. Supplementary Methods
 SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
5K ALDEFLUOR-positive/ CXCR1-negative. 5K ALDEFLUOR-positive/ CXCR1-positive BAAA BAAA CXCR1-APC BAAA BAAA CXCR1-APC
 A +DEAB -DEAB K ALDEFLUOR-positive/ CXCR-negative BAAA BAAA CXCR-APC B +DEAB -DEAB K ALDEFLUOR-positive/ CXCR-positive BAAA BAAA CXCR-APC C Supplemental Figure. Tumorigenicity of the ALDEFLUOR-positive/CXCR-positive
A +DEAB -DEAB K ALDEFLUOR-positive/ CXCR-negative BAAA BAAA CXCR-APC B +DEAB -DEAB K ALDEFLUOR-positive/ CXCR-positive BAAA BAAA CXCR-APC C Supplemental Figure. Tumorigenicity of the ALDEFLUOR-positive/CXCR-positive
Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at
 Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at E10.5 were double-stained for TUNEL (red) and PECAM-1 (green).
Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at E10.5 were double-stained for TUNEL (red) and PECAM-1 (green).
Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing)
 Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing) platform with laser manipulation to efficiently purify lung
Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing) platform with laser manipulation to efficiently purify lung
