Supplementary Information for. Autoimmunity as a mechanism for hybrid necrosis, a genetic incompatibility syndrome in plants
|
|
|
- Cornelius Cox
- 5 years ago
- Views:
Transcription
1 for Autoimmunity as a mechanism for hybrid necrosis, a genetic incompatibility syndrome in plants Kirsten Bomblies, Janne Lempe, Petra Epple, Norman Warthmann, Christa Lanz, Jeffery L. Dangl, and Detlef Weigel Figure S1 (Alignment of DM1 from Uk-3 with Col-0 alleles)...s2 Figure S2 (Trypan Blue staining of seedlings)...s3 Figure S3 (Graph showing PR gene expression in hybrids)...s4 Table S1A (Significantly over-represented GO terms)...s5 Table S1B (Gene categories in differentially expressed genes)...s6 Table S2 (Accession numbers of lines)...s8 Table S3 (Crosses performed and hybrid phenotypes)...s11 Table S4 (Intercrossability of parents of several incompatibilities)...s21 Table S5 (H. parasitica resistance of hybrids and parents)...s22 S1
2 Figure S1 Alignment of Uk-3 DM1 allele with Col-0 At5g41740 and At5g41750 colorcoded for conservation. S2
3 Figure S2 Trypan Blue staining of seedlings. Untreated, ten day old seedlings of the indicated accessions were grown at 23 C and cotyledons were stained with Trypan Blue to visualize ectopic cell death. S3
4 Figure S3 Expression on microarrays of common pathogen response marker genes. Normalized expression of several common pathogen response marker genes for each parent and hybrid combination analyzed. S4
5 Table S1A Significantly over-represented GO terms (among genes different in Uk-1/Uk- 3 F 1 versus parents, but not between parents). GO process # genes Uk F 1 list # in At genome P-value Systemic acquired resistance (SAR) response to biotic stimulus response to pathogen response to other organism response to pest, pathogen or parasite response to stimulus defense response anthocyanin biosynthesis protein targeting to vacuole flavonoid biosynthesis flavonoid metabolism response to stress defense response, response to pathogen, incompatible interaction defense response to pathogen Notes: Statistically significant over-representation of gene ontology (GO) term categories was calculated in GoStat ( P-values are Fisher s Exact test p- values corrected for multiple testing with Benjamini correction as implemented in GoStat. GO term annotation was based on the latest TAIR release ( S5
6 Table S1B Representation of gene categories in list of genes differentially regulated in at least one hybrid versus its parents. Category on array in list expected foldexcess Total chitinase BON1/copine trypsin inhibitor tropinone reductase harpin-induced family glutamate receptor WRKY transcription factor glutathione S-transferase, putative receptor-like kinase MutT/nudix heavy-metal associated FAD binding domain xyloglucan flavin-containing monooxygenase family protein / FMO family protein peroxidase NAM Oxidoreductase, 2OG-Fe(II) oxygenase family protein pathogenesis-related calcium/calmodulin lipase cytochrome P glycosyl hydrolase S-locus protein kinase disease resistance family plant defensin S6
7 leucine-rich repeat GDSL-motif lipase/hydrolase family protein protein kinase nodulin heat shock Senescence-associated (SAG) glycoside hydrolase LEA glycine-rich protein expressed protein AP2 domain DEAD-box zinc finger protein bhlh glycosyl transferase DnaJ transducin-related F-box ethylene-responsive MADS B3 family transcription factor Note: The gene list consists of 1,080 genes (4.7% of genes on the microarray), which are differentially expressed in at least one of the four hybrids tested with respect to its parents. S7
8 S8 Table S2 Accession numbers of lines used. Name NASC ABRC Aa-0 N935 Ak-1 N939 ANH1 N6607 Ba-1 N952 Bay-0 CS22676 Bch-3 N959 Bch-4 N961 Bd-0 N963 Be-1 N967 BG1 N22341 BG2 N22342 BG5 N22345 BG7 N22347 BG9 N22349 Bl-1 N969 Bla-1 N971 Bla-11 N985 Bla-12 N987 Bla-14 N989 Bla-2 N973 Bla-3 N975 Bla-4 N977 Bla-5 N978 Bla-6 N980 Blh-1 N6645 Blh-2 N6657 Bor-4 CS22677 Br-0 N995 Bs-1 N997 Bs-2 N6628 Bs-5 N1001 Bsch-0 N1003 Bsch-1 Bsch-2 N1005 Bu-0 N1007 Bu-11 N1025 Bu-13 N1027 Bur-0 N1029 C24 CS22680 Ca-0 N1061 Cal-0 N1063 Can-0 N1065 Cen-0 N1067 Cha-0 N1069 Chi-0 N1073 Chi-1 N1075 Chi-2 N1077 CIBC1 N22220 CIBC10 N22229 Cit-0 N1081 Cnt-1 N6921 Co-1 N1085 Co-3 N1089 Co-4 N1091 Col-0 N1093 CSHL1 N22419 Ct-1 N1095 Cvi-0 CS22682 Db-0 N1101 Db-1 N1103 Db-2 N6679 Di-2 N1111 Dr-0 N1115 Dra-0 N1117 Dra-1 N1119 Edi-0 N1123 Ei-2 N1125 Ei-4 N1127 Ei-5 N6691 Ei-6 N1131 Eil-0 N1133 El-0 N1135 Ema-1 N1681 En-1 N1137 En-2 N1139 Enk-T N6176 Ep-0 N1141 Er-0 N1143 Est-0 N1149 Est-1 N1151 Estland N6173 Et-0 N1153 Fe-1 N1155 Fei-0 CS22684 Fi-0 N1157 Fi-1 N1159 FM11 N22392 Fr-2 N1169 Fr-3 N1171 Fr-4 N1173 Fr-5 N1175 Fr-6 N1177 Fr-7 N1179 Ga-0 N1181 Ga-2 N1183
9 S9 Gd-1 N1185 Ge-1 N1189 Ge-2 N1191 Gie-0 N1193 Gö-0 N1195 Gö-2 N1197 GOT1 N22277 GOT10 N22286 GOT7 CS22685 Gr-1 N1199 Gr-2 N1201 Gr-3 N1203 Gr-5 N1207 Gr-6 N6728 Gre-0 N1211 Gü-0 N1213 Gü-1 N1215 Gy-0 N1217 Ha-0 N1219 Hau-0 N1221 Hh-0 N1225 Hi-0 N1227 Hl-0 N1229 Hl-2 N1231 Hl-3 N1233 Hn-0 N1235 Hodja-Obi N6178 HR14 N22213 HR15 N22214 HR5 N22205 HR8 N22208 Hs-0 N1237 HS1 N22351 In-0 N1239 Is-0 N1241 Is-1 N1243 Je-0 N1247 Jl-1 N1249 Jl-2 N1251 Jm-1 N1261 Js-0 N1671 Kb-0 N1269 Kil-0 N1271 Kin-0 N1273 Kl-0 N1275 Kl-1 N1277 Kn-0 N1287 KNO1 N22401 Kondara N6175 Kro-0 N1301 KZ10 N22442 KZ11 N22443 La-1 N1303 Lan-0 N1305 Le-0 N1309 Ler-1 N6928 4nLer N3900 Li-10 N6911 Li-2 N6908 Li2:1 N1315 Lip-0 N1337 Ll-0 N1339 Ll-11 N1651 Ll-2 N1343 Lm-2 N1345 Lö-1 N1347 Lö-2 N1349 Lov-5 CS22695 M7943S N6186 Ma-0 N1357 Mc-0 N1363 Me-0 N1365 Mh-1 N1369 Mir-0 N1379 Mnz-0 N1371 Mr-0 N1373 Mrk-0 N1375 Ms-0 N1377 Mt-0 N1381 Mv-0 N1387 Mz-0 N1383 Na-1 N1385 Nc-1 N1389 Nd-0 N1391 Nd-1 N1680 NFA8 CS22687 NFC10 N22191 NFE1 N22163 NFE10 N22172 Nie-0 N1393 Nok-0 N1399 Nok-1 N1401 Nok-2 N1403 Nok-3 N1405 Np-0 N1397 Nw-0 N1409 Nw-1 N1411 Nw-2 N1413 Nw-3 N1415 Ob-0 N1419
10 Ob-1 N1421 Ob-2 N1423 Ob-3 N1425 Old-1 N1427 Old-2 N1429 Or-0 N1433 Ost-0 N1431 Ove-0 N1435 Oy-1 N6929 Pa-2 N1441 Pa-3 N1443 Per-2 N1449 Per-3 N1451 Pf-0 N1453 Pi-0 N1455 Pi-2 N1457 Pla-0 N1459 Pla-1 N1461 Pla-2 N1463 Po-1 N1473 Pog-0 N1477 Pr-0 N1475 PUZ16 N22451 Ra-0 N1481 Rd-0 N1482 REN11 N22263 Ri-0 N1493 RLD1 N913 Rou-0 N1489 RP1 N22362 RP10 N22371 RRS10 CS22689 RRS7 CS22688 Rsch-0 N1491 Rsch-4 N1495 Sav-0 N1515 Se-0 N1503 Sf-2 N1517 Sg-1 N1519 Sg-2 N1521 Shahdara N6180 CS22690 Shokei N1676 Sp-0 N1531 SQ4 N22243 St-0 N1535 Ste-0 N1537 Stw-0 N1539 Su-0 N1541 Ta-0 N1549 Tamm-2 CS22691 Te-0 N1551 Ts-1 N1553 Ts-5 N6871 Ts-6 N1561 Ts-7 N1563 Tsu-1 CS22693 Tu-1 N1569 Ty-0 N1573 Uk-1 N1575 Uk-10 New collection Uk-11 New collection Uk-12 New collection Uk-13 New collection Uk-14 New collection Uk-2 N1579 Uk-3 N1577 Uk-4 N1581 Uk-5 New collection Uk-6 New collection Uk-7 New collection Uk-8 New collection Uk-9 New collection Van-0 N1585 Wa-1 N1587 Wc-1 N1589 Wc-2 N1591 Wei-0 N6182 Wei-1 N1683 Wil-1 N1595 Wil-3 N1599 Wl-0 N1631 Wt-1 N1605 Wt-4 N1611 Wt-5 N1613 Yo-0 N1622 Zü-0 N1627 ABRC = Arabidopsis Biological Resources Center; NASC = Nottingham Arabidopsis Stock Centre. S10
11 Table S3 Crosses performed and hybrid phenotypes. Female Male F 1 pheno Comment type 2n Ler Blh-1 Normal 4n Ler Bla-5 Normal 4n Ler Wa-1 Normal Aa-0 Ga-0 Normal Aa-0 HR15 Normal Aa-0 Uk-1 Normal Ak-1 Uk-1 Normal Ak-1 Uk-3 Normal ANH1 Gr-1 Normal ANH1 Kro-0 Normal Ba-1 Chi-1 Normal Ba-1 Lov-5 Normal Ba-1 Shokei Normal Bay-0 Br-0 Normal Bay-0 C24 Normal Bay-0 Col-0 Normal Bay-0 Fei-0 Normal Bay-0 Tamm2 Normal Bay-0 Ts-1 Normal Bch-3 BG9 Normal Bch-4 Ei-6 Normal Bch-4 Pr-0 Normal Bd-0 Ep-0 Normal Be-1 Bu-13 Normal Be-1 Di-2 Normal Be-1 Ga-2 Normal Be-1 Hi-0 Normal Be-1 Nie-0 Normal BG1 Rsch-0 Normal BG1 Uk-3 Normal BG2 BG7 Normal BG2 Bsch-0 Normal BG5 Kro-0 Normal BG5 Ra-0 Normal BG7 Po-1 Normal Bla-1 Bla-11 Normal Bla-1 Bla-12 Normal Bla-1 Bla14n Normal Bla-1 Bla-2 Normal Bla-1 Bla-3 Normal Bla-1 Bla-4 Normal Bla-1 Bla-6 Normal Bla-1 Hh-0 Class 2 necrosis Bla-1 KZ10 Normal Bla-1 Mir-0 Normal Bla-1 Pla-0 Normal Bla-1 Pla-2 Normal Bla-1 Ts-5 Normal Bla-1 Ts-7 Normal Bla-1 Uk-1 Normal Bla-1 Uk-3 Normal Bla-1 Sha. Class 2 necrosis Bla-11 Mir-0 Normal Bla-11 Se-0 Normal Bla-12 Bla-11 Normal Bla-12 Bla-14 Normal Bla-12 Bla-2 Normal Bla-12 Bla-3 Normal Bla-12 Hh-0 Normal Bla-12 Mir-0 Normal Bla-12 Se-0 Normal Bla-12 Sha. Normal Bla-12 Ts-6 Normal Bla-14 Bla-11 Normal Bla-14 Bla-2 Normal Bla-14 Bla-3 Normal Bla-14 Bla-4 Normal Bla-14 Ll-0 Normal Bla-14 Mir-0 Normal Bla-14 Mt-0 Normal Bla-14 Pi-2 Normal Bla-14 Pla-0 Normal Bla-14 Se-0 Normal Bla-14 Sha. Normal Bla-14 Ts-7 Normal Bla14n Ep-0 Normal Bla14n Est-0 Normal Bla14n Ga-0 Normal Bla14p Per-3 Normal Bla-2 Bla-11 Normal Bla-2 Bla-3 Normal Bla-2 Bla-6 Normal Bla-2 Ll-11 Normal Bla-2 Mir-0 Normal Bla-2 Se-0 Normal Bla-2 Sha. Class 2 necrosis Bla-2 Ts-7 Normal Bla-2 Uk-1 Normal Bla-2 Uk-3 Normal Bla-3 Bla-11 Normal Bla-3 Bla-4 Normal Bla-3 Bla-6 Normal Bla-3 Hh-0 Normal Bla-3 Ll-11 Normal S11
12 Bla-3 Mir-0 Normal Bla-3 Pla-0 Normal Bla-3 Pla-2 Normal Bla-3 Se-0 Class 1 necrosis Bla-3 Sha. Normal Bla-3 Ts-5 Normal Bla-3 Ts-6 Normal Bla-3 Ts-7 Normal Bla-3 Uk-1 Normal Bla-3 Uk-3 Normal Bla-4 Bla-11 Normal Bla-4 Bla-12 Normal Bla-4 Bla-2 Normal Bla-4 Bla-6 Normal Bla-4 Hh-0 Normal Bla-4 Ll-0 Class 1 necrosis Mild Bla-4 Ll-11 Normal Bla-4 Mir-0 Normal Bla-4 Pla-1 Normal Bla-4 Pla-2 Normal Bla-4 Se-0 Normal Bla-4 Shahdara Normal Bla-5 4nLer Normal RC Bla-5 Bla-14 HSL Ploidy Bla-5 Blh-1 HSL Ploidy Bla-5 Hh-0 Class 2 necrosis (also ploidy) Bla-5 Wa-1 Normal Bla-6 Bla-11 Normal Bla-6 Bla-12 Normal Bla-6 Bla-14 Normal Bla-6 Mir-0 Normal Bla-6 Se-0 Normal Bla-6 Ts-7 Normal Blh-1 2nLer Normal Blh-1 Bla-5 HSL Ploidy;RC Blh-1 Hh-0 Normal Blh-1 Wa-1 HSL Ploidy Blh-2 Bla-5 HSL Ploidy Blh-2 Blh-1 Normal Bor-4 Bay-0 Normal Bor-4 Br-0 Normal Bor-4 C24 Normal Bor-4 Cvi-0 Normal Bor-4 Fei-0 Normal Bor-4 NFA8 Normal Br-0 C24 Normal Br-0 Col-0 Normal Br-0 Fei-0 Normal Br-0 St-0 Normal Br-0 Wil-3 Normal Bs-1 Bs-2 Normal Bs-1 Uk-1 Normal Bs-1 Uk-3 Normal Bs-2 Lö-2 Normal Bs-2 Uk-1 Normal Bs-2 Uk-3 Normal Bs-5 Bs-2 Normal Bs-5 Bs-2.2 Normal Bs-5 Kn-0 Normal Bs-5 Lö-1 Normal Bs-5 Lö-2 Normal Bs-5 Uk-1 Normal Bs-5 Uk-3 Normal Bsch-0 HR5 Normal Bsch-0 HR8 Normal Bsch-0 NFE10 Normal Bsch-1 GOT10 Normal Bu-0 Bsch-2 Normal Bu-0 NFE10 Normal Bu-11 Po-1 Normal Bur-0 Bay-0 Normal Bur-0 Br-0 Normal Bur-0 C24 Normal Bur-0 Col-0 Normal Bur-0 Est-1 Normal Bur-0 Lov-5 Normal Bur-0 Pf-0 Normal Bur-0 Ts-1 Normal Ca-0 Bu-11 Normal Cal-0 Bur-0 Normal Cal-0 Ei-5 Normal Cal-0 Lm-2 Normal Cal-0 Wil-1 Normal Can-0 Bla-1 Normal Can-0 Mt-0 Normal Can-0 St-0 Normal Cen-0 Dr-0 Normal Cen-0 Hl-2 Normal Cen-0 Ost-0 Normal Cen-0 Se-0 Normal Cha-0 Pla-0 Normal Cha-0 Se-0 Normal Chi-0 Ak-1 Normal Chi-0 Bla-1 Normal Chi-0 Uk-1 Normal Chi-0 Uk-3 Normal Chi-2 Chi-0 Normal Chi-2 KZ10 Normal S12
13 CIBC1 Bu-11 Normal CIBC1 Chi-1 Normal CIBC10 Bl-1 Normal CIBC10 Hodja- Normal Obi Cit-0 CIBC10 Normal Cit-0 Ei-6 Normal Cit-0 Gy-0 Normal Cnt-1 Gr-1 Normal Co-1 Co-3 Normal Co-1 Oy-1 Normal Co-3 Te-0 Normal Co-4 Co-3 Normal Co-4 Wil-1 Normal Col-0 Bla-5 HSL Ploidy Col-0 Blh-1 Normal Col-0 C24 Normal Col-0 Chi-2 Normal Col-0 Fei-0 Normal Col-0 GOT7 Normal Col-0 NFA8 Normal Col-0 Nie-0 Normal Col-0 Tamm2 Normal Col-0 Ts-1 Normal Ct-1 St-0 Normal Cvi-0 Bur-0 Normal Cvi-0 RRS7 Normal Cvi-0 Uk-1 Normal Cvi-0 Uk-3 Normal Db-1 Db-0 Normal Di-2 Ra-0 Normal Dra-1 Dra-0 Normal Edi-0 En-2 Normal Ei-2 Lm-2 Normal Ei-2 Na-1 Normal Ei-2 Ove-0 Normal Ei-4 CIBC1 Normal Ei-5 Ei-2 Normal Ei-5 Oy-1 Normal Eil-0 El-0 Normal El-0 Eil-0 Normal RC Ema-1 Estland Normal En-1 Uk-3 Normal En-2 RLD1 Normal Er-0 Ang-1 Normal Er-0 Dra-1 Normal Est-0 Hau-0 Normal Est-0 Wa-1 HSL Ploidy Est-1 Br-0 Normal Est-1 Cvi-0 Normal Est-1 Est-0 Normal Est-1 Fei-0 Normal Est-1 Mt-0 Normal Est-1 RRS10 Normal Est-1 Ts-1 Normal Et-0 Bla-1 Normal Et-0 Fe-1 Normal Fe-1 Ge-2 Normal Fe-1 Uk-1 Normal Fe-1 Uk-3 Normal Fei-0 NFA8 Normal Fei-0 Ts-1 Normal Fi-0 Gr-3 Normal FM11 KZ10 Normal FM11 Pa-2 Normal Fr-3 Hodja- Normal Obi Fr-4 Fr-2 Normal Fr-5 Fr-4 Normal Fr-6 RLD1 Normal Fr-6 Uk-1 Normal Fr-6 Uk-3 Normal Fr-7 ENK-T Normal Fr-7 Uk-1 Normal Fr-7 Uk-3 Normal Gd-1 BG5 Normal Gd-1 Bu-13 Normal Gd-1 NFC10 Normal Gd-1 Ste-0 Normal Ge-1 Li-2:1 Normal Ge-2 Cha-0 Normal Gie-0 Fi-1 Normal Gö-0 Gö-2 Normal GOT1 Chi-2 Normal GOT1 Gr-2 Normal GOT1 Gy-0 Normal GOT1 Nok-3 Normal GOT7 Bur-0 Normal GOT7 C24 Normal GOT7 Cvi-0 Normal GOT7 Est-1 Normal GOT7 Sha. Normal GOT7 Ts-1 Normal Gr-1 M7943S Normal Gr-2 Can-0 Normal Gr-2 GOT1 Normal RC Gr-2 Gr-6 Normal Gr-2 In-0 Normal Gr-3 Fi-0 Normal RC Gr-3 Gr-5 Normal S13
14 Gr-3 Gr-6 Normal Gr-3 Per-3 Normal Gr-6 Co-4 Normal Gr-6 Gr-3 Normal RC Gr-6 Hl-0 Normal Gr-6 Oy-1 Normal Gr-6 Rsch-4 Normal Gr-6 Wa-1 HSL Ploidy Gre-0 Bla-5 HSL Ploidy Gre-0 Chi-0 Normal Gü-0 Gü-1 Normal Ha-0 Db-2 Normal Hau-0 Est-0 Normal RC Hau-0 Sha. Normal Hau-0 Wa-1 HSL Ploidy Hh-0 Bla-1 Class 2 necrosis; RC Hh-0 Bla-11 Normal Hh-0 Bla-14 Normal Hh-0 Bla-2 Class 2 necrosis Hh-0 Bla-6 Normal Hh-0 Col-0 Normal Hh-0 Mir-0 Normal Hh-0 Nc-1 Normal Hh-0 Se-0 Class 1 necrosis Hh-0 Ts-7 Normal Hh-0 Uk-1 Normal Hh-0 Uk-3 Normal Hi-0 Be-1 Normal RC Hi-0 Pr-0 Normal Hl-0 Bla-1 Normal Hl-0 Gie-0 Normal Hl-0 Pi-0 Normal Hl-2 Et-0 Normal Hl-2 Mt-0 Normal Hl-3 Sha. Normal Hl-3 Wa-1 HSL Ploidy Hn-0 Bch-4 Normal Hn-0 Ra-0 Normal Hodja- Bu-13 Normal Obi Hodja- Mz-0 Normal Obi HR15 Aa-0 Normal RC HR15 Bs-1 Normal HR15 Gr-6 Normal HR15 Te-0 Normal HR5 Bu-13 Normal Hs-0 Hl-0 Normal Hs-1 ENK-T Normal Hs-1 Mz-0 Normal Is-0 Is-1 Normal Is-1 Nd-0 Normal Je-0 Gie-0 Normal Je-0 Pa-3 Normal Jl-1 Eil-0 Normal Jl-1 Sha. Normal Jl-1 Uk-1 Normal Jl-1 Uk-3 Normal Jl-2 Aa-0 Normal Jl-2 Jl-1 Normal Jl-2 Mt-0 Normal Jl-2 Uk-1 Normal Jl-2 Uk-3 Normal Js-0 Bu-13 Normal Kb-0 M7943S Normal Kil-0 An-1 Normal Kl-0 Kl-1 Normal Kn-0 Uk-3 Normal KNO1 M7943S Normal Kondara NFE1 Normal Kondara YGAP1 Normal Kro-0 Sf-2 Normal KZ10 Gie-0 Normal KZ10 Hh-0 Normal KZ10 Mir-0 Normal KZ10 Mrk-0 Class 3 necrosis KZ10 Uk-1 Normal KZ10 Uk-3 Normal KZ11 Ei-4 Normal KZ11 HR14 Normal KZ11 Mrk-0 Class 3 necrosis KZ11 NFC1 Normal KZ11 Ob-0 Normal KZ11 YGAP1 Normal La-1 ENK-T Normal La-1 Fr-6 Normal La-1 YGAP1 Normal Lan-0 Ll-2 Normal Le-0 GOT1 Normal Le-0 Ll-0 Normal Le-0 Ost-0 Normal Le-0 Pf-0 Normal Ler-1 Br-0 Normal Ler-1 Fei-0 Normal Ler-1 Ts-1 Normal Ler-1 Van-0 Normal Li-10 Kin-0 Normal Li-2 Hodja- Normal Obi Li-2 HR5 Normal Li-2:1 Bla-5 HSL Ploidy S14
15 Li-2:1 Chi-2 Normal Li-2:1 Wa-1 HSL Ploidy Li-2:1 Wil-3 Normal Lip-0 Ei-6 Normal Lip-0 Kn-0 Normal Lip-0 Uk-1 Normal Lip-0 Uk-3 Normal Ll-0 Bla-1 Normal Ll-0 Bla-11 Normal Ll-0 Bla-12 Normal Ll-0 Bla-2 Normal Ll-0 Bla-3 Normal Ll-0 Bla-6 Normal Ll-0 Br-0 Normal Ll-0 Fr-2 Normal Ll-0 Hh-0 Normal Ll-0 Ll-11 Normal Ll-0 Ll-2 Normal Ll-0 Mir-0 Normal Ll-0 Se-0 Normal Ll-0 Ts-5 Normal Ll-0 Ts-7 Normal Ll-11 Bla-1 Normal Ll-11 Bla-11 Normal Ll-11 Bla-12 Normal Ll-11 Bla-14 Normal Ll-11 Bla-6 Normal Ll-11 Cha-0 Normal Ll-11 Hh-0 Normal Ll-11 Ll-0 Normal RC Ll-11 Mir-0 Normal Ll-11 Ove-0 Normal Ll-11 Ts-1 Normal Ll-11 Ts-6 Class 2 necrosis (transient) Ll-11 Ts-7 Normal Ll-2 Bla-1 Normal Ll-2 Bla-11 Normal Ll-2 Bla-12 Normal Ll-2 Bla-14 Normal Ll-2 Bla-2 Normal Ll-2 Bla-3 Normal Ll-2 Bla-4 Normal Ll-2 Bla-6 Normal Ll-2 Hh-0 Normal Ll-2 Ll-11 Normal Ll-2 Mir-0 Normal Ll-2 Se-0 Normal Ll-2 Ts-1 Normal Ll-2 Ts-5 Normal Ll-2 Ts-7 Normal Lm-2 Ei-2 Normal RC Lö-1 Bs-1 Normal Lö-1 Bs-2 Normal Lö-1 Ost-0 Normal Lö-1 Uk-1 Normal Lö-1 Uk-3 Normal Lö-1 Uk-4 Normal Lö-2 Bs-1 Normal Lö-2 Lö-1 Normal Lö-2 Shokei Normal Lö-2 Uk-1 Normal Lö-2 Uk-3 Normal Lö-2 Yo-0 Normal Lov-5 Bay-0 Normal Lov-5 Col-0 Normal Lov-5 Est-1 Normal Lov-5 Fei-0 Normal Lov-5 GOT7 Normal Lov-5 Ts-1 Normal Ma-0 Jm-1 Normal Me-0 Hn-0 Normal Me-0 Kro-0 Normal Mh-1 M78845 Normal Mir-0 Hau-0 Normal Mir-0 Se-0 Class 1 necrosis Mir-0 Uk-3 Normal Mir-0 Wil-3 Normal Mnz-0 BG7 Normal Mnz-0 Hl-0 Normal Mrk-0 Bla-1 Normal Mrk-0 Bla-1 Normal Mrk-0 Co-4 Normal Mrk-0 Hh-0 Normal Mrk-0 KZ10 Class 3 necrosis; RC Mrk-0 Mir-0 Normal Mrk-0 Sha. Normal Mrk-0 Uk-1 Normal Mrk-0 Uk-3 Normal Ms-0 Lm-2 Normal Ms-0 Pa-2 Normal Mt-0 Aa-0 Normal Mt-0 Jl-2 Normal RC Mt-0 Nw-2 Normal Mt-0 Pi-2 Normal Mt-0 Uk-1 Normal Mv-0 Br-0 Normal Mv-0 Uk-2 Normal Mz-0 BG5 Normal Mz-0 Bu-13 Normal S15
16 Na-1 Hl-3 Normal Na-1 Ove-0 Normal Na-1 Uk-1 Normal Na-1 Uk-3 Normal Nd-0 Nd-1 Normal Nd-0 Uk-1 Normal NFA8 Br-0 Normal NFA8 Est-1 Normal NFA8 Ts-1 Normal NFA8 Van-0 Normal NFC1 Be-1 Normal NFC1 FM11 Normal NFC1 Hh-0 Normal NFC1 Np-0 Normal NFC10 Kb-0 Normal NFE10 Me-0 Normal Nie-0 BG5 Normal Nie-0 Cen-0 Normal Nie-0 Chi-2 Normal Nie-0 Fr-6 Normal Nie-0 HR5 Normal Nie-0 Li-10 Normal Nie-0 Sp-0 Normal Nok-0 Nok-1 Normal Nok-0 Uk-1 Normal Nok-0 Uk-3 Normal Nok-2 Nok-0 Normal Nok-2 Nok-1 Normal Nok-2 Nok-3 Normal Nok-2 Pla-0 Normal Nok-3 Nok-0 Normal Nok-3 Nok-1 Normal Nw-1 Nw-0 Normal Nw-1 Uk-1 Normal Nw-1 Uk-3 Normal Nw-2 Aa-0 Normal Nw-2 Jl-2 Normal Nw-2 Mt-0 Normal RC Nw-2 Nw-3 Normal Nw-2 Uk-1 Normal Nw-2 Uk-3 Normal Ob-0 BG5 Normal Ob-0 Chi-1 Normal Ob-0 Hodja- Normal Obi Ob-1 En-1 Normal Ob-2 Ob-1 Normal Old-1 Fr-6 Normal Old-2 Db-2 Normal Or-0 Gy-0 Normal Ost-0 Chi-2 Normal Ost-0 Per-2 Normal Ove-0 Lm-2 Normal Oy-1 Nok-0 Normal Oy-1 Pla-2 Normal Oy-1 Uk-1 Normal Oy-1 Uk-3 Normal Pa-2 Pa-3 Normal Per-2 NFC1 Normal Per-2 Uk-1 Normal Per-3 Gr-3 Normal RC Per-3 Pi-2 Normal Per-3 Rd-0 Normal Pf-0 Uk-1 Normal Pf-0 Uk-3 Normal Pi-0 Bur-0 Normal Pla-0 Bla-1 Normal RC Pla-0 Bla-11 Normal Pla-0 Bla-12 Normal Pla-0 Bla-2 Normal Pla-0 Bla-4 Normal Pla-0 Bla-6 Normal Pla-0 Ge-2 Normal Pla-0 Hh-0 Normal Pla-0 Ll-0 Normal Pla-0 Ll-11 Normal Pla-0 Ll-2 Normal Pla-0 Mir-0 Normal Pla-0 Pla-1 Normal Pla-0 Se-0 Normal Pla-0 Sha. Normal Pla-0 Ts-5 Normal Pla-0 Ts-6 Normal Pla-1 Bla-1 Normal Pla-1 Bla-11 Normal Pla-1 Bla-12 Normal Pla-1 Bla-14 Normal Pla-1 Bla-2 Normal Pla-1 Bla-3 Class 1 necrosis Pla-1 Bla-6 Normal Pla-1 Hh-0 Class 1 necrosis Pla-1 Ll-0 Normal Pla-1 Ll-11 Normal Pla-1 Ll-2 Normal Pla-1 Mir-0 Class 1 necrosis Pla-1 Pla-2 Normal Pla-1 Se-0 Normal Pla-1 Ts-7 Normal Pla-2 Bla-11 Normal Pla-2 Bla-12 Normal S16
17 Pla-2 Bla-14 Normal Pla-2 Bla-2 Normal Pla-2 Bla-6 Normal Pla-2 Hh-0 Normal Pla-2 Ll-0 Normal Pla-2 Ll-11 Normal Pla-2 Ll-2 Normal Pla-2 Mir-0 Normal Pla-2 Pla-0 Normal Pla-2 Sha. Normal Pla-2 Ts-7 Normal Po-1 Wei-0 Normal Pog-0 Uk-1 Normal Pog-0 Uk-3 Normal PUZ16 CSHL1 Normal PUZ16 Fr-2 Normal PUZ16 Pog-0 Normal Rd-0 Rou-0 Normal Rd-0 Uk-1 Normal REN11 Wei-1 Class 3 necrosis Ri-0 Rd-0 Normal RLD1 NFC10 Normal RLD2 Me-0 Normal Rou-0 Rd-0 Normal RC Rou-0 Wt-5 Normal RP1 Blh-2 Normal RP1 RP10 Normal RP-1 Mv-0 Normal RP10 Blh-1 Normal RP10 Fe-1 Normal RRS10 Bor-4 Normal RRS10 Br-0 Normal RRS10 Col-0 Normal RRS10 Fei-0 Normal RRS10 NFA8 Normal RRS10 RRS7 Normal RRS10 Ts-1 Normal RRS7 Bur-0 Normal RRS7 Bay-0 Normal RRS7 Col-0 Normal RRS7 Est-1 Normal RRS7 Tamm2 Normal Rsch-0 BG1 Normal RC Rsch-0 Pog-0 Normal Rsch-4 Wl-0 Normal Se-0 Bla-1 Normal Se-0 KZ10 Normal Se-0 Mir-0 Class 1 necrosis; RC Se-0 Mrk-0 Normal Se-0 Uk-3 Normal Sg-1 Bs-1 Normal Sg-1 Sg-2 Normal Sg-1 Uk-1 Normal Sg-1 Uk-3 Normal Sg-2 Lö-2 Normal Sg-2 Sg-1 Normal Sg-2 Uk-1 Normal Sg-2 Uk-3 Normal Sha. Bla-1 Class 2 necrosis; RC Sha. Bla-11 Normal Sha. Bla-6 Normal Sha. Br-0 Normal Sha. C24 Normal Sha. Col-0 Normal Sha. Est-1 Normal Sha. Fei-0 Normal Sha. Hh-0 Normal Sha. KZ10 Normal Sha. Ll-0 Class 2 necrosis; mild Sha. Ll-11 Normal Sha. Ll-2 Normal Sha. Lov-5 Normal Sha. Mir-0 Normal Sha. Pla-1 Normal Sha. RRS7 Normal Sha. Se-0 Normal Sha. Ts-7 Normal Sha. Uk-1 Normal Sha. Uk-3 Normal Sha. Hl-3 Normal RC Sha. Kondara Normal Sp-0 Gr-1 Normal Sp-0 Li-10 Normal SQ4 Uk-2 Normal Ste-0 Bu-0 Normal Ste-0 Sf-2 Normal Stw-0 Jl-1 Normal Su-0 Ak-1 Normal Su-0 Blh-1 Normal Su-0 Chi-0 Normal Su-0 Col-0 Normal Su-0 Hh-0 Normal Su-0 Tsu-1 Normal Ta-0 SQ4 Normal Tamm-2 Bor-4 Normal Tamm-2 Br-0 Normal Tamm-2 Cvi-0 Normal Tamm-2 Est-1 Normal Tamm-2 Fei-0 Normal S17
18 Tamm-2 Lov-5 Normal Tamm-2 RRS10 Normal Tamm-2 Ts-1 Normal Te-0 Bla-1 Normal Te-0 Je-0 Normal Te-0 Pf-0 Normal Te-0 Uk-1 Normal Te-0 Uk-3 Normal Te-0 Wil-1 Normal Ts-1 Bla-1 Normal Ts-1 Bla-11 Normal Ts-1 Bla-12 Normal Ts-1 Bla-14 Normal Ts-1 Bla-2 Normal Ts-1 Bla-3 Normal Ts-1 Bla-4 Normal Ts-1 Bla-6 Normal Ts-1 Cvi-0 Normal Ts-1 Hh-0 Normal Ts-1 Ll-0 Normal Ts-1 Mir-0 Normal Ts-1 Pla-0 Normal Ts-1 Pla-1 Normal Ts-1 Pla-2 Normal Ts-1 Se-0 Normal Ts-1 Sha. Class 2 necrosis (transient); RC Ts-1 Ts-7 Normal Ts-1 Uk-1 Normal Ts-1 Uk-3 Normal Ts-5 Bla-11 Normal Ts-5 Bla-12 Normal Ts-5 Bla-14 Normal Ts-5 Bla-2 Normal Ts-5 Bla-4 Normal Ts-5 Bla-6 Normal Ts-5 Hh-0 Normal Ts-5 Ll-0 Normal RC Ts-5 Ll-11 Normal Ts-5 Mir-0 Normal Ts-5 Pla-1 Normal Ts-5 Pla-2 Normal Ts-5 Se-0 Normal Ts-5 Sha. Normal Ts-5 Ts-1 Normal Ts-5 Ts-6 Normal Ts-5 Ts-7 Normal Ts-5 Uk-1 Normal Ts-6 Bla-1 Normal Ts-6 Bla-11 Normal Ts-6 Bla-12 Normal RC Ts-6 Bla-14 Normal Ts-6 Bla-2 Normal Ts-6 Bla-4 Normal Ts-6 Bla-6 Normal Ts-6 Hh-0 Normal Ts-6 Ll-0 Normal Ts-6 Ll-2 Normal Ts-6 Mir-0 Normal Ts-6 Pla-1 Normal Ts-6 Pla-2 Normal Ts-6 Se-0 Normal Ts-6 Sha. Normal Ts-6 Ts-1 Normal Ts-6 Ts-7 Normal Ts-6 Uk-1 Normal Ts-6 Uk-3 Normal Ts-7 Bla-1 Normal RC Ts-7 Bla-12 Normal Ts-7 Bla-4 Normal Ts-7 Hh-0 Normal RC Ts-7 Mir-0 Normal Ts-7 Nw-1 Normal Ts-7 Pa-3 Normal Ts-7 Pla-0 Normal Ts-7 Se-0 Normal Tsu-1 Bay-0 Normal Tsu-1 C24 Normal Tsu-1 Col-0 Normal Tsu-1 Est-1 Normal Tsu-1 RRS10 Normal Tsu-1 Tamm2 Normal Tu-1 Pa-3 Normal Tu-1 Ts-7 Normal Ty-0 Mc-0 Normal Ty-0 NFC1 Normal Ty-0 Tsu-1 Normal Uk-1 Bd-0 Normal Uk-1 BG1 Normal Uk-1 Col-0 Normal Uk-1 En-1 Normal Uk-1 Er-0 Normal Uk-1 Kn-0 Normal Uk-1 Mir-0 Normal Uk-1 Nc-1 Class 3 necrosis Uk-1 Np-0 Normal Uk-1 Se-0 Normal Uk-1 Uk-10 Normal Uk-1 Uk-11 Normal Uk-1 Uk-12 Normal S18
19 Uk-1 Uk-13 Normal Uk-1 Uk-2 Normal Uk-1 Uk-3 Class 3 necrosis Uk-1 Uk-5 Normal Uk-1 Uk-7 Normal Uk-1 Uk-8 Normal Uk-1 Uk-9 Normal Uk-1 Wt-4 Normal Uk-10 Uk-11 Normal Uk-10 Uk-9 Normal Uk-11 Uk-12 Normal Uk-11 Uk-14 Normal Uk-11 Uk-5 Normal Uk-11 Uk-7 Normal Uk-11 Uk-9 Normal Uk-13 Uk-10 Normal Uk-13 Uk-11 Normal Uk-13 Uk-12 Normal Uk-13 Uk-7 Normal Uk-14 Uk-1 Normal Uk-14 Uk-10 Normal Uk-14 Uk-12 Normal Uk-14 Uk-13 Normal Uk-14 Uk-7 Normal Uk-3 Aa-0 Normal Uk-3 Bd-0 Normal Uk-3 Col-0 Normal Uk-3 Er-0 Normal Uk-3 Mt-0 Normal Uk-3 Nc-1 Normal Uk-3 Np-0 Normal Uk-3 Uk-1 Class 3 necrosis; RC Uk-3 Uk-10 Normal Uk-3 Uk-11 Normal Uk-3 Uk-12 Normal Uk-3 Uk-13 Normal Uk-3 Uk-14 Normal Uk-3 Uk-2 Normal Uk-3 Uk-5 Normal Uk-3 Uk-6 Normal Uk-3 Uk-7 Normal Uk-3 Uk-8 Normal Uk-3 Uk-9 Normal Uk-3 Wt-4 Normal Uk-4 Pa-3 Normal Uk-4 Uk-1 Normal Uk-4 Uk-3 Normal Uk-5 Uk-10 Normal Uk-5 Uk-12 Normal Uk-5 Uk-13 Normal Uk-5 Uk-14 Normal Uk-5 Uk-6 Normal Uk-5 Uk-9 Normal Uk-6 Uk-1 Class 3 necrosis Uk-6 Uk-10 Normal Uk-6 Uk-11 Normal Uk-6 Uk-12 Normal Uk-6 Uk-13 Normal Uk-6 Uk-14 Normal Uk-6 Uk-7 Normal Uk-6 Uk-8 Normal Uk-6 Uk-9 Normal Uk-7 Uk-10 Normal Uk-7 Uk-12 Normal Uk-7 Uk-5 Normal Uk-7 Uk-8 Normal Uk-7 Uk-9 Normal Uk-8 Uk-10 Normal Uk-8 Uk-11 Normal Uk-8 Uk-12 Normal Uk-8 Uk-13 Normal Uk-8 Uk-14 Normal Uk-8 Uk-5 Normal Uk-8 Uk-9 Normal Uk-9 Uk-12 Normal Uk-9 Uk-13 Normal Uk-9 Uk-14 Normal Van-0 Bay-0 Normal Van-0 Bor-4 Normal Van-0 Br-0 Normal Van-0 C24 Normal Van-0 Fei-0 Normal Van-0 Ts-1 Normal Wa-1 4nLer Normal Wa-1 Bla-5 Normal Wa-1 Blh-1 HSL Ploidy Wa-1 Ob-3 HSL Ploidy Wc-2 Tu-1 Normal Wc-2 Wc-1 Normal Wei-0 Be-1 Normal Wei-1 REN11 Class 3 necrosis; RC Wil-1 Mir-0 Normal Wil-1 Pla-0 Normal Wil-1 Wil-3 Normal Wil-3 Br-0 Normal RC Wil-3 Col-0 Normal Wl-0 Bur-0 Normal Wt-1 Wt-4 Normal RC Wt-4 Gr-3 Normal Wt-4 Per-3 Normal S19
20 Wt-4 Wt-1 Normal Wt-5 Rou-0 Normal RC Yo-0 Ba-1 Normal Yo-0 Mr-0 Normal Zü-0 Li-2 Normal Zü-0 Sav-0 Normal Zü-1 Be-1 Normal Sha. = Shahdara RC = Reciprocal cross; HSL = high seed lethality; Ploidy = Ploidy difference between parents. S20
21 Table S4 Intercrossability of parents of several incompatibilities. Uk-1 Uk-3 Mir-0 Se-0 Bla-1 Hh-0 Shah dara KZ10 Mrk-0 Uk-1 I (III) C C C C C C C Uk-3 C C C C C C C Mir-0 I (I) C C C C C Se-0 C I (I) C C C Bla-1 I (II) I (II) C C Hh-0 C C C Shahdara C C KZ10 I (III) Mrk-0 C = compatible, I = incompatible ( class in parentheses). S21
22 Table S5 H. parasitica resistance of hybrids and parents. Cala2 Emco5 Noco2 Emwa1 Uk-1 R S nd nd Uk-3 R S nd nd Uk-1/Uk-3 nd R nd nd Bla-1 R R R S Hh-0 R R R S Bla-1/Hh-0 nd nd nd S Mrk-0 S S nd nd KZ10 S R nd nd Mrk-0/KZ10 R nd nd nd Se-0 S R S 1 S Mir-0 R R S 1 R Se-0/Mir-0 nd nd R nd 1 weak sporulator (approx. 2 sporangiophores/cotyledon) S22
GENETICS. Supporting Information
 GENETI upporting Information http://www.genetics.org/cgi/content/full/genetics.110.116392/d1 Natural Diversity in lowering Responses of rabidopsis thaliana ausedbyvariationinatandemgenerray arah Marie
GENETI upporting Information http://www.genetics.org/cgi/content/full/genetics.110.116392/d1 Natural Diversity in lowering Responses of rabidopsis thaliana ausedbyvariationinatandemgenerray arah Marie
The natural genetic variation of the fatty-acyl composition of seed oils in di erent ecotypes of Arabidopsis thaliana
 Phytochemistry 52 (1999) 1029±1033 The natural genetic variation of the fatty-acyl composition of seed oils in di erent ecotypes of Arabidopsis thaliana Anthony A. Millar, Ljerka Kunst* Department of Botany,
Phytochemistry 52 (1999) 1029±1033 The natural genetic variation of the fatty-acyl composition of seed oils in di erent ecotypes of Arabidopsis thaliana Anthony A. Millar, Ljerka Kunst* Department of Botany,
WT siz1-2 siz1-3. WT siz1-2 siz1-3. -Pi, 0.05 M IAA. -Pi, 2.5 M NPA
 A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
A WT siz1-2 siz1-3 B WT siz1-2 siz1-3 -Pi C +Pi WT siz1-2 siz1-3 D WT siz1-2 siz1-3 E +Pi, 0.05 M IAA -Pi, 0.05 M IAA WT siz1-2 siz1-3 WT siz1-2 siz1-3 F +Pi, 2.5 M NPA -Pi, 2.5 M NPA Supplemental Figure
Natural variation in Arabidopsis, a tool to identify genetic bases of nitrogen use efficiency
 Natural variation in rabidopsis, a tool to identify genetic bases of nitrogen use efficiency. Chaillou, F. Chardon, J. Laurette,. Ikram, O. Loudet and F. Daniel-Vedele IN Versailles Plants have different
Natural variation in rabidopsis, a tool to identify genetic bases of nitrogen use efficiency. Chaillou, F. Chardon, J. Laurette,. Ikram, O. Loudet and F. Daniel-Vedele IN Versailles Plants have different
Supplemental Table S1 Up-regulated genes in irjazh plants compared to WT plants by microarray.
 Supplemental Table S1 Up-regulated genes in irjazh plants compared to WT plants by microarray. Classification Fold Change Bincode (TAIR) Annotation Probe number ACC number Cell wall. Degradation 3.29 10.6.2
Supplemental Table S1 Up-regulated genes in irjazh plants compared to WT plants by microarray. Classification Fold Change Bincode (TAIR) Annotation Probe number ACC number Cell wall. Degradation 3.29 10.6.2
A Sound Track to Reading
 A Sound Track to Reading Blending Flashcards Prepared by Donald L Potter June 1, 2018 Mr. Potter prepared these cards to be used with Sister Monica Foltzer s advanced intensive phonics program and reader,
A Sound Track to Reading Blending Flashcards Prepared by Donald L Potter June 1, 2018 Mr. Potter prepared these cards to be used with Sister Monica Foltzer s advanced intensive phonics program and reader,
MOLECULAR BIOLOGY AND GENETICS OF PROGRAMMED CELL DEATH IN ARABIDOPSIS THALIANA
 PROCEEDINGS OF THE BALKAN SCIENTIFIC CONFERENCE OF BIOLOGY IN PLOVDIV (BULGARIA) FROM 19 TH TILL 21 ST OF MAY 2005 (EDS B. GRUEV, M. NIKOLOVA AND A. DONEV), 2005 (P. 157 161) MOLECULAR BIOLOGY AND GENETICS
PROCEEDINGS OF THE BALKAN SCIENTIFIC CONFERENCE OF BIOLOGY IN PLOVDIV (BULGARIA) FROM 19 TH TILL 21 ST OF MAY 2005 (EDS B. GRUEV, M. NIKOLOVA AND A. DONEV), 2005 (P. 157 161) MOLECULAR BIOLOGY AND GENETICS
A Comprehensive Study of TP53 Mutations in Chronic Lymphocytic Leukemia: Analysis of 1,287 Diagnostic CLL Samples
 A Comprehensive Study of TP53 Mutations in Chronic Lymphocytic Leukemia: Analysis of 1,287 Diagnostic CLL Samples Sona Pekova, MD., PhD. Chambon Ltd., Laboratory for molecular diagnostics, Prague, Czech
A Comprehensive Study of TP53 Mutations in Chronic Lymphocytic Leukemia: Analysis of 1,287 Diagnostic CLL Samples Sona Pekova, MD., PhD. Chambon Ltd., Laboratory for molecular diagnostics, Prague, Czech
Lecture 3-4. Identification of Positive Regulators downstream of SA
 Lecture 3-4 Identification of Positive Regulators downstream of SA Positive regulators between SA and resistance/pr Systemic resistance PR gene expression? SA Local infection Necrosis SA What could they
Lecture 3-4 Identification of Positive Regulators downstream of SA Positive regulators between SA and resistance/pr Systemic resistance PR gene expression? SA Local infection Necrosis SA What could they
SUPPLEMENTARY INFORMATION
 Supplementary igure 1. The expression level of PP4 (At4g16860) is diminished in the rpp4 mutant (SK017569). The mna was extracted to analyze the expression level of PP4 using quantitative PC (qpc). Gene
Supplementary igure 1. The expression level of PP4 (At4g16860) is diminished in the rpp4 mutant (SK017569). The mna was extracted to analyze the expression level of PP4 using quantitative PC (qpc). Gene
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated
 Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Supplementary Figure 1 Transcription assay of nine ABA-responsive PP2C genes. Transcription assay of nine ABA-responsive PP2C genes. Total RNA was isolated from 7 day-old seedlings treated with or without
Testing the ABC floral-organ identity model: expression of A and C function genes
 Objectives: Testing the ABC floral-organ identity model: expression of A and C function genes To test the validity of the ABC model for floral organ identity we will: 1. Use the model to make predictions
Objectives: Testing the ABC floral-organ identity model: expression of A and C function genes To test the validity of the ABC model for floral organ identity we will: 1. Use the model to make predictions
EOPS PROBATION STUDENT LIST
 EOPS PROBATION STUDENT LIST Fall 2018 EOPS The following students have been placed on EOPS Probation due to their not satisfying the semester g.p.a. and/or semester units completed requirements of the
EOPS PROBATION STUDENT LIST Fall 2018 EOPS The following students have been placed on EOPS Probation due to their not satisfying the semester g.p.a. and/or semester units completed requirements of the
Supplementary Tables 1
 Supplementary Tables 1 Supplementary Table 1: The 11 genes in the geneset for Case Study 1 At1g62570 At1g09350 At1g60470 At2g47180 At4g17090 At5g20830 At2g16890 At3g51240 At3g55120 At4g27560 At5g08640
Supplementary Tables 1 Supplementary Table 1: The 11 genes in the geneset for Case Study 1 At1g62570 At1g09350 At1g60470 At2g47180 At4g17090 At5g20830 At2g16890 At3g51240 At3g55120 At4g27560 At5g08640
Figure S3 Differentially expressed fungal and plant genes organised by catalytic activity ontology Organisation of E. festucae (A) and L.
 sak WT Number of branches 3 WT sak Figure S1 Vasculature of plants infected with the E. festucae sak mutant. Light micrographs of perennial ryegrass blade tissue showing branching between the vasculature
sak WT Number of branches 3 WT sak Figure S1 Vasculature of plants infected with the E. festucae sak mutant. Light micrographs of perennial ryegrass blade tissue showing branching between the vasculature
Plasmodium and host glutathione reductase: molecular function and biological process
 Plasmodium and host glutathione reductase: molecular function and biological process Viroj Wiwanitkit Department of Laboratory Medicine, Faculty of Medicine, Chulalongkorn University, Bangkok Thailand
Plasmodium and host glutathione reductase: molecular function and biological process Viroj Wiwanitkit Department of Laboratory Medicine, Faculty of Medicine, Chulalongkorn University, Bangkok Thailand
Supplemental Figure 1. Genes showing ectopic H3K9 dimethylation in this study are DNA hypermethylated in Lister et al. study.
 mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
How sarcoma/gist research & expert care are organized in Italy
 How sarcoma/gist research & expert care are organized in Italy Organization of care Historical migration for medical care PR VA LC VI TV BG LU GR PG CN AL MI Desi o LO PV Alzano Lombardo CO Casalpusterlen
How sarcoma/gist research & expert care are organized in Italy Organization of care Historical migration for medical care PR VA LC VI TV BG LU GR PG CN AL MI Desi o LO PV Alzano Lombardo CO Casalpusterlen
The N-end rule pathway regulates pathogen responses. in plants
 SUPPLEMENTARY INFORMATION The N-end rule pathway regulates pathogen responses in plants Rémi de Marchi 1,2, Maud Sorel 1, Brian Mooney 1, Isabelle Fudal 3, Kevin Goslin 1, Kamila Kwaśniewska 4, Patrick
SUPPLEMENTARY INFORMATION The N-end rule pathway regulates pathogen responses in plants Rémi de Marchi 1,2, Maud Sorel 1, Brian Mooney 1, Isabelle Fudal 3, Kevin Goslin 1, Kamila Kwaśniewska 4, Patrick
HIV-1 resistance testing from proviral DNA
 HIV-1 resistance testing from proviral DNA Alexander Thielen AREVIR 2018 Resistance testing from proviral DNA? amount of samples with low viral loads increasing desire to switch under successful therapy
HIV-1 resistance testing from proviral DNA Alexander Thielen AREVIR 2018 Resistance testing from proviral DNA? amount of samples with low viral loads increasing desire to switch under successful therapy
Genomic Analysis of QTLs and Genes Altering Natural Variation in Stochastic Noise
 Genomic Analysis of QTLs and Genes Altering Natural Variation in Stochastic Noise Jose M. Jimenez-Gomez 1., Jason A. Corwin 2., Bindu Joseph 2., Julin N. Maloof 1, Daniel J. Kliebenstein 2 * 1 Department
Genomic Analysis of QTLs and Genes Altering Natural Variation in Stochastic Noise Jose M. Jimenez-Gomez 1., Jason A. Corwin 2., Bindu Joseph 2., Julin N. Maloof 1, Daniel J. Kliebenstein 2 * 1 Department
DCP Networks
 www.bps.org.uk/dcp DCP Networks To support and promote clinical psychology, working collaboratively with people who use our services and wider communities to improve wellbeing and to promote the unique
www.bps.org.uk/dcp DCP Networks To support and promote clinical psychology, working collaboratively with people who use our services and wider communities to improve wellbeing and to promote the unique
33VASTVNGATSANNHGEPPS51PADARPR58
 Pro-rich region Trans-membrane region 214 246 359 381 UL50 1 397 211SSRTAS216PPPPPR222 NLS CR1 CR2 CR3 CR4 UL53 1 376 11RERRS15ALRS19LLRKRRR25 33VASTVNGATSANNHGEPPS51PADARPR58 FIG S1. UL97 phosphorylation
Pro-rich region Trans-membrane region 214 246 359 381 UL50 1 397 211SSRTAS216PPPPPR222 NLS CR1 CR2 CR3 CR4 UL53 1 376 11RERRS15ALRS19LLRKRRR25 33VASTVNGATSANNHGEPPS51PADARPR58 FIG S1. UL97 phosphorylation
Regulation of Floral Organ Identity. Dr. Chloe Diamond Mara
 Regulation of Floral Organ Identity Dr. Chloe Diamond Mara Flower Development Angiosperms (flowering plants) are the most widespread group of land plants Flowers are the reproductive organs that consist
Regulation of Floral Organ Identity Dr. Chloe Diamond Mara Flower Development Angiosperms (flowering plants) are the most widespread group of land plants Flowers are the reproductive organs that consist
Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene
 Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene YUELIN ZHANG, WEIHUA FAN, MARK KINKEMA, XIN LI, AND
Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene YUELIN ZHANG, WEIHUA FAN, MARK KINKEMA, XIN LI, AND
7th International Congress of the Spanish Society of Obesity Surgery. Valladolid Spain May, 2004.
 7th International Congress of the Spanish Society of Obesity Surgery. Valladolid Spain May, 2004. DIMINISHING POSTOPERATIVE RISKS OF GASTRIC BYPASS Stenosis Stenosis Leak Leak Bleeding Bleeding Stenosis
7th International Congress of the Spanish Society of Obesity Surgery. Valladolid Spain May, 2004. DIMINISHING POSTOPERATIVE RISKS OF GASTRIC BYPASS Stenosis Stenosis Leak Leak Bleeding Bleeding Stenosis
Supporting Information for
 Supporting Information for CuO Nanoparticle Interaction with Arabidopsis: Toxicity, Parent-Progeny Transfer and Gene Expression Zhenyu Wang,, Lina Xu, Jian Zhao,,, Xiangke Wang, Jason C. White, and Baoshan
Supporting Information for CuO Nanoparticle Interaction with Arabidopsis: Toxicity, Parent-Progeny Transfer and Gene Expression Zhenyu Wang,, Lina Xu, Jian Zhao,,, Xiangke Wang, Jason C. White, and Baoshan
Supplemental Data. Di Giorgio et al. (2016). Plant Cell /tpc
 Supplemental Figure 1. Synteny analysis of NIP4;1 and NIP4;2. Examination of the NIP4;1-NIP4;2 region in rabidopsis thaliana and equivalent evolutionary regions in other dicot genomes are shown on the
Supplemental Figure 1. Synteny analysis of NIP4;1 and NIP4;2. Examination of the NIP4;1-NIP4;2 region in rabidopsis thaliana and equivalent evolutionary regions in other dicot genomes are shown on the
Supplemental Data. Beck et al. (2010). Plant Cell /tpc
 Supplemental Figure 1. Phenotypic comparison of the rosette leaves of four-week-old mpk4 and Col-0 plants. A mpk4 vs Col-0 plants grown in soil. Note the extreme dwarfism of the mpk4 plants (white arrows)
Supplemental Figure 1. Phenotypic comparison of the rosette leaves of four-week-old mpk4 and Col-0 plants. A mpk4 vs Col-0 plants grown in soil. Note the extreme dwarfism of the mpk4 plants (white arrows)
Supplementary Figures
 Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
Supplementary Figures a miel1-2 (SALK_41369).1kb miel1-1 (SALK_978) b TUB MIEL1 Supplementary Figure 1. MIEL1 expression in miel1 mutant and S:MIEL1-MYC transgenic plants. (a) Mapping of the T-DNA insertion
BASIC COMPATIBILITY. Pl Path 604
 BASIC COMPATIBILITY Pl Path 604 Plant Reactions to Pathogens Interaction Phenotype; appearance of host infected by pathogen at a given point of time and growth stage of host omplete esistance Partial resistance
BASIC COMPATIBILITY Pl Path 604 Plant Reactions to Pathogens Interaction Phenotype; appearance of host infected by pathogen at a given point of time and growth stage of host omplete esistance Partial resistance
Supplemental Figure S1. The number of hydathodes is reduced in the as2-1 rev-1
 Supplemental Data Supplemental Figure S1. The number of hydathodes is reduced in the as2-1 rev-1 and kan1-11 kan2-5 double mutants. A, The numbers of hydathodes in different leaves of Col-0, as2-1 rev-1,
Supplemental Data Supplemental Figure S1. The number of hydathodes is reduced in the as2-1 rev-1 and kan1-11 kan2-5 double mutants. A, The numbers of hydathodes in different leaves of Col-0, as2-1 rev-1,
MG132. GRXS17:V5-His MG132 GRXS17:HA. RuBisCo. Supplemental Data. Walton et al. (2015). Plant Cell /tpc
 MG132 GRXS17:V5-His MG132 GRXS17:HA RuBisCo Supplemental Figure 1. Degradation of GRXS17 by the 26S proteasome. (A) Cell-free degradation assay. Recombinant protein GRXS17:V5-HIS was incubated with total
MG132 GRXS17:V5-His MG132 GRXS17:HA RuBisCo Supplemental Figure 1. Degradation of GRXS17 by the 26S proteasome. (A) Cell-free degradation assay. Recombinant protein GRXS17:V5-HIS was incubated with total
Recent Minima of 161 Eclipsing Binary Stars
 Samolyk, JAAVSO Volume 38, 2010 85 Recent Minima of 161 Eclipsing Binary Stars Gerard Samolyk P.O. Box 20677; Greenfield, WI 53220; gsamolyk@wi.rr.com Received September 23, 2009; accepted September 25,
Samolyk, JAAVSO Volume 38, 2010 85 Recent Minima of 161 Eclipsing Binary Stars Gerard Samolyk P.O. Box 20677; Greenfield, WI 53220; gsamolyk@wi.rr.com Received September 23, 2009; accepted September 25,
(43) Publication date: 04 September 2014 ( ) (22) Filing Date: 27 February 2014 ( )
 (54) Title (EN): INCOHERENT TYPE-III MATERIALS FOR CHARGE CARRIERS CONTROL DEVICES (54) Title (FR): MATÉRIAUX DE TYPE III INCOHÉRENTS POUR DISPOSITIFS DE RÉGULATION DE PORTEURS DE CHARGES (72) Inventor(s):
(54) Title (EN): INCOHERENT TYPE-III MATERIALS FOR CHARGE CARRIERS CONTROL DEVICES (54) Title (FR): MATÉRIAUX DE TYPE III INCOHÉRENTS POUR DISPOSITIFS DE RÉGULATION DE PORTEURS DE CHARGES (72) Inventor(s):
Genetic Analysis of Allosteric Signaling in RhaR from Escherichia coli and Characterization of the VirF Protein from Shigella flexneri
 Genetic Analysis of Allosteric Signaling in RhaR from Escherichia coli and Characterization of the VirF Protein from Shigella flexneri By Bria Collette Kettle Submitted to the graduate degree program in
Genetic Analysis of Allosteric Signaling in RhaR from Escherichia coli and Characterization of the VirF Protein from Shigella flexneri By Bria Collette Kettle Submitted to the graduate degree program in
silent epidemic,. (WHO),
 Tel: 02-740-8686; E-mail: hhbkim@snu.ac.kr silent epidemic,. (WHO),. 5 3, 1. 50 70. 50%, 25%, 20% (12~35%). 2.8% 0.7% 4. ( ). bone remodeling (osteoblast), (osteoclast),.. 3~4.. 70% (osteocyte) (bone lining
Tel: 02-740-8686; E-mail: hhbkim@snu.ac.kr silent epidemic,. (WHO),. 5 3, 1. 50 70. 50%, 25%, 20% (12~35%). 2.8% 0.7% 4. ( ). bone remodeling (osteoblast), (osteoclast),.. 3~4.. 70% (osteocyte) (bone lining
4-1 Dyspnea (Chuan, 喘 )
 4-1 Dyspnea (Chuan, 喘 ) Concept Breathing with difficulty (open wide mouth, raise shoulders) Etiology and pathogenesis Climatic factors Phlegm fluid Emotion Chronic diseases Exertion Over sex Diagnosis
4-1 Dyspnea (Chuan, 喘 ) Concept Breathing with difficulty (open wide mouth, raise shoulders) Etiology and pathogenesis Climatic factors Phlegm fluid Emotion Chronic diseases Exertion Over sex Diagnosis
Phosphorus Deficiency in Potatoes. Helen Bowen, Rory Hayden & Will Spracklen
 A Molecular Diagnostic for Phosphorus Deficiency in Potatoes Philip J. White John P. Hammond, Martin R. Broadley, Helen Bowen, Rory Hayden & Will Spracklen (Warwick, Nottingham, SCRI) IPNC XVI, Sacramento,
A Molecular Diagnostic for Phosphorus Deficiency in Potatoes Philip J. White John P. Hammond, Martin R. Broadley, Helen Bowen, Rory Hayden & Will Spracklen (Warwick, Nottingham, SCRI) IPNC XVI, Sacramento,
Table S1 Differentially expressed genes showing > 2 fold changes and p <0.01 for 0.01 mm NS-398, 0.1mM ibuprofen and COX-2 RNAi.
 Table S1 Differentially expressed genes showing > 2 fold changes and p
Table S1 Differentially expressed genes showing > 2 fold changes and p
Natural variation of root exudates in Arabidopsis thaliana - linking metabolomic and genomic data
 Natural variation of root exudates in Arabidopsis thaliana - linking metabolomic and genomic data Susann Mönchgesang +, Nadine Strehmel +, Stephan Schmidt +, Lore Westphal, Franziska Taruttis, Erik Müller,
Natural variation of root exudates in Arabidopsis thaliana - linking metabolomic and genomic data Susann Mönchgesang +, Nadine Strehmel +, Stephan Schmidt +, Lore Westphal, Franziska Taruttis, Erik Müller,
J. A. Mayfield et al. FIGURE S1. Methionine Salvage. Methylthioadenosine. Methionine. AdoMet. Folate Biosynthesis. Methylation SAH.
 FIGURE S1 Methionine Salvage Methionine Methylthioadenosine AdoMet Folate Biosynthesis Methylation SAH Homocysteine Homocystine CBS Cystathionine Cysteine Glutathione Figure S1 Biochemical pathway of relevant
FIGURE S1 Methionine Salvage Methionine Methylthioadenosine AdoMet Folate Biosynthesis Methylation SAH Homocysteine Homocystine CBS Cystathionine Cysteine Glutathione Figure S1 Biochemical pathway of relevant
Spherical Bearings Heavy Duty Equipments
 Spherical Bearings Heavy Duty Equipments Highlights Quality Service Price Wbf Replacement Parts adaptableto > Caterpillar >Komatsu >Volvo 1 WBF SPHERICAL BEARINGS adaptable to Caterpillar Part No. Description
Spherical Bearings Heavy Duty Equipments Highlights Quality Service Price Wbf Replacement Parts adaptableto > Caterpillar >Komatsu >Volvo 1 WBF SPHERICAL BEARINGS adaptable to Caterpillar Part No. Description
Supplemental data. Uppalapati et al. (2012) Plant Cell /tpc
 PAL Control P. emaculata 0 8 24 48 8 24 48 OPR Control P. emaculata 0 8 24 48 8 24 48 CHS PR3 CHR PR5 CHI PR10 IFS IFR Supplemental Figure 1. Expression profiles for selected genes in phenylpropanoid pathway
PAL Control P. emaculata 0 8 24 48 8 24 48 OPR Control P. emaculata 0 8 24 48 8 24 48 CHS PR3 CHR PR5 CHI PR10 IFS IFR Supplemental Figure 1. Expression profiles for selected genes in phenylpropanoid pathway
Arabidopsis Transcriptome Analysis Reveals Key Roles of Melatonin in Plant Defense Systems
 Virginia Commonwealth University VCU Scholars Compass Study of Biological Complexity Publications Center for the Study of Biological Complexity 2014 Arabidopsis Transcriptome Analysis Reveals Key Roles
Virginia Commonwealth University VCU Scholars Compass Study of Biological Complexity Publications Center for the Study of Biological Complexity 2014 Arabidopsis Transcriptome Analysis Reveals Key Roles
Supplemental Figure S1. Sequence feature and phylogenetic analysis of GmZF351. (A) Amino acid sequence alignment of GmZF351, AtTZF1, SOMNUS, AtTZF5,
 Supplemental Figure S. Sequence feature and phylogenetic analysis of GmZF35. (A) Amino acid sequence alignment of GmZF35, AtTZF, SOMNUS, AtTZF5, and OsTZF. Characters with black background indicate conserved
Supplemental Figure S. Sequence feature and phylogenetic analysis of GmZF35. (A) Amino acid sequence alignment of GmZF35, AtTZF, SOMNUS, AtTZF5, and OsTZF. Characters with black background indicate conserved
Supporting Information
 Supporting Information Supplemental Figure Legends Supplementary Figure 1. Experimental Design. A. To benchmark the performance of software parameter sets proteins were extracted from a pool of Arabidopsis
Supporting Information Supplemental Figure Legends Supplementary Figure 1. Experimental Design. A. To benchmark the performance of software parameter sets proteins were extracted from a pool of Arabidopsis
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
GENE EXPRESSION ANALYSIS OF PROSTATE TUMORS FROM AFRICAN-AMERICAN AND EUROPEAN-AMERICAN MEN
 GENE EXPRESSION ANALYSIS OF PROSTATE TUMORS FROM AFRICAN-AMERICAN AND EUROPEAN-AMERICAN MEN Tiffany A. Wallace Laboratory of Human Carcinogenesis Center for Cancer Research National Cancer Institute Health
GENE EXPRESSION ANALYSIS OF PROSTATE TUMORS FROM AFRICAN-AMERICAN AND EUROPEAN-AMERICAN MEN Tiffany A. Wallace Laboratory of Human Carcinogenesis Center for Cancer Research National Cancer Institute Health
Front Plant Sci Mar 14
 Case Study Front Plant Sci. 2016 Mar 14 30 33 Study Background Cryptochromes (CRY) are blue-light photoreceptors that mediate various light responses in plants and animals. It is known that CRY1 and CRY2
Case Study Front Plant Sci. 2016 Mar 14 30 33 Study Background Cryptochromes (CRY) are blue-light photoreceptors that mediate various light responses in plants and animals. It is known that CRY1 and CRY2
Nature Immunology: doi: /ni Supplementary Figure 1. Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice.
 Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
Arabidopsis: Flower Development and Patterning
 Arabidopsis: Flower Development and Patterning John L Bowman, University of California, Davis, California, USA The development of flowers and floral organs is directed by genetic programmes likely to be
Arabidopsis: Flower Development and Patterning John L Bowman, University of California, Davis, California, USA The development of flowers and floral organs is directed by genetic programmes likely to be
Nicotine biosynthesis pathway: beyond tobacco
 Nicotine biosynthesis pathway: beyond tobacco Masataka Kajikawa, Nicolas Sierro, Haruhiko Kawaguchi, Nicolas Bakaher, Nikolai V Ivanov, Takashi Hashimoto, Tsubasa Shoji Nicotiana tabacum Cultivated as
Nicotine biosynthesis pathway: beyond tobacco Masataka Kajikawa, Nicolas Sierro, Haruhiko Kawaguchi, Nicolas Bakaher, Nikolai V Ivanov, Takashi Hashimoto, Tsubasa Shoji Nicotiana tabacum Cultivated as
Genetic Specification of floral organ identity. Initiating floral development. Deciding when to initiate flowering - induced mutations -in Nature
 Genetic Specification of floral organ identity Initiating floral development Deciding when to initiate flowering - induced mutations -in Nature Flower structure of rabidopsis Stamens arpels Petals Sepals
Genetic Specification of floral organ identity Initiating floral development Deciding when to initiate flowering - induced mutations -in Nature Flower structure of rabidopsis Stamens arpels Petals Sepals
BIOLOGY OF POSTHARVEST DISEASE AND THE ROLE OF PLANT CELL WALLS
 Postharvest Technology of Horticultural Crops 17 June 2010, UC Davis BIOLOGY OF POSTHARVEST DISEASE AND THE ROLE OF PLANT CELL WALLS Ann Powell, Plant Sciences Department, Univ. of California, Davis Dario
Postharvest Technology of Horticultural Crops 17 June 2010, UC Davis BIOLOGY OF POSTHARVEST DISEASE AND THE ROLE OF PLANT CELL WALLS Ann Powell, Plant Sciences Department, Univ. of California, Davis Dario
Appendix A: International Classification of Diseases, 10th Revision, Clinical Modification Codes (ICD-10) Utilized for VTE Events
 Online Appendices to Mahan et al. External validation of a risk assessment model for venous thromboembolism in the hospitalised acutely-ill medical patient (VTE-VALOURR) (Thromb Haemost 2014; 112.4) Appendix
Online Appendices to Mahan et al. External validation of a risk assessment model for venous thromboembolism in the hospitalised acutely-ill medical patient (VTE-VALOURR) (Thromb Haemost 2014; 112.4) Appendix
Supplementary Figures
 Supplementary Figures 9 10 11 Supplementary Figure 1. Old plants are more resistant to insect herbivores than young plants. (a) Image of young (1-day-old, 1D) and old (-day-old, D) plants of Arabidopsis
Supplementary Figures 9 10 11 Supplementary Figure 1. Old plants are more resistant to insect herbivores than young plants. (a) Image of young (1-day-old, 1D) and old (-day-old, D) plants of Arabidopsis
Single Herbs III / Quiz I
 Single Herbs III / Quiz I 1. What herb is good to generate fluids? A. Ren Shen C. Xi Yang Shen B. Tai Zi Shen D. All the Shens 2. What herb is best for Qi collapse? A. Huang Qi C. Dang Shen B. Ren Shen
Single Herbs III / Quiz I 1. What herb is good to generate fluids? A. Ren Shen C. Xi Yang Shen B. Tai Zi Shen D. All the Shens 2. What herb is best for Qi collapse? A. Huang Qi C. Dang Shen B. Ren Shen
Supplementary Figure 1 Weight and body temperature of ferrets inoculated with
 Supplementary Figure 1 Weight and body temperature of ferrets inoculated with A/Anhui/1/2013 (H7N9) influenza virus. (a) Body temperature and (b) weight change of ferrets after intranasal inoculation with
Supplementary Figure 1 Weight and body temperature of ferrets inoculated with A/Anhui/1/2013 (H7N9) influenza virus. (a) Body temperature and (b) weight change of ferrets after intranasal inoculation with
EMPEROR'S COLLEGE MTOM COURSE SYLLABUS HERB FORMULAE II
 COURSE DESCRIPTION The second of three courses in the Herb Formulae series. Categories covered in Formulae II include the Tonify Qi and Blood, Regulate Qi, Invigorate the Blood, Stop Bleeding, Stabilize
COURSE DESCRIPTION The second of three courses in the Herb Formulae series. Categories covered in Formulae II include the Tonify Qi and Blood, Regulate Qi, Invigorate the Blood, Stop Bleeding, Stabilize
Supporting Information
 Supporting Information Hu et al..73/pnas.3489 SI Materials and Methods Histone Extraction and Western Blot Analysis. Rice histone proteins were extracted from -d-old seedlings. After being washed with
Supporting Information Hu et al..73/pnas.3489 SI Materials and Methods Histone Extraction and Western Blot Analysis. Rice histone proteins were extracted from -d-old seedlings. After being washed with
The Arabidopsis mir396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens
 The Arabidopsis mir396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens Mauricio Soto-Suárez, Patricia Baldrich, Detlef Weigel, Ignacio Rubio-Somoza and
The Arabidopsis mir396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens Mauricio Soto-Suárez, Patricia Baldrich, Detlef Weigel, Ignacio Rubio-Somoza and
Supplemental Data. Deinlein et al. Plant Cell. (2012) /tpc
 µm Zn 2+ 15 µm Zn 2+ Growth (% of control) empty vector NS1 NS2 NS3 NS4 S. pombe zhfδ Supplemental Figure 1. Functional characterization of. halleri NS genes in Zn 2+ hypersensitive S. pombe Δzhf mutant
µm Zn 2+ 15 µm Zn 2+ Growth (% of control) empty vector NS1 NS2 NS3 NS4 S. pombe zhfδ Supplemental Figure 1. Functional characterization of. halleri NS genes in Zn 2+ hypersensitive S. pombe Δzhf mutant
Table S1. Total and mapped reads produced for each ChIP-seq sample
 Tale S1. Total and mapped reads produced for each ChIP-seq sample Sample Total Reads Mapped Reads Col- H3K27me3 rep1 125662 1334323 (85.76%) Col- H3K27me3 rep2 9176437 7986731 (87.4%) atmi1a//c H3K27m3
Tale S1. Total and mapped reads produced for each ChIP-seq sample Sample Total Reads Mapped Reads Col- H3K27me3 rep1 125662 1334323 (85.76%) Col- H3K27me3 rep2 9176437 7986731 (87.4%) atmi1a//c H3K27m3
Supplementary. presence of the. (c) mrna expression. Error. in naive or
 Figure 1. (a) Naive CD4 + T cells were activated in the presence of the indicated cytokines for 3 days. Enpp2 mrna expression was measured by qrt-pcrhr, infected with (b, c) Naive CD4 + T cells were activated
Figure 1. (a) Naive CD4 + T cells were activated in the presence of the indicated cytokines for 3 days. Enpp2 mrna expression was measured by qrt-pcrhr, infected with (b, c) Naive CD4 + T cells were activated
A -GLS Arabidopsis Cuscuta gronovii
 -GLS Arabidopsis Cuscuta gronovii internal standard 4MS Wildtype Arabidopsis Cuscuta gronovii base Cuscuta gronovii apex 3MSP internal standard MSP 4MT 8MSO 4MO 1MO Figure S1. Representative HPLC-DAD chromatograms
-GLS Arabidopsis Cuscuta gronovii internal standard 4MS Wildtype Arabidopsis Cuscuta gronovii base Cuscuta gronovii apex 3MSP internal standard MSP 4MT 8MSO 4MO 1MO Figure S1. Representative HPLC-DAD chromatograms
A bts-1 (SALK_016526)
 Electronic Supplementary Material (ESI) for Metallomics. This journal is The Royal Society of Chemistry 217 A ts-1 (SALK_16526) BTS (At3g1829) ATG tsl1 (SALK_1554) 2 p BTSL1 (At1g7477) ATG tsl2 (SAIL_615_HO1)
Electronic Supplementary Material (ESI) for Metallomics. This journal is The Royal Society of Chemistry 217 A ts-1 (SALK_16526) BTS (At3g1829) ATG tsl1 (SALK_1554) 2 p BTSL1 (At1g7477) ATG tsl2 (SAIL_615_HO1)
Supplemental Data. Hiruma et al. Plant Cell. (2010) /tpc Col-0. pen2-1
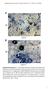 A Ch B Col-0 Cg pen2-1 Supplemental Figure 1. Trypan Blue Staining of Leaves Inoculated with Adapted and Nonadapted Colletotrichum Species.(A) Conidial suspensions of C. higginsianum MAFF305635 (Ch) were
A Ch B Col-0 Cg pen2-1 Supplemental Figure 1. Trypan Blue Staining of Leaves Inoculated with Adapted and Nonadapted Colletotrichum Species.(A) Conidial suspensions of C. higginsianum MAFF305635 (Ch) were
Supplemental Data. Candat et al. Plant Cell (2014) /tpc Cytosol. Nucleus. Mitochondria. Plastid. Peroxisome. Endomembrane system
 Cytosol Nucleus 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 PSORT MultiLoc YLoc SubLoc BaCelLo WoLF PSORT
Cytosol Nucleus 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 PSORT MultiLoc YLoc SubLoc BaCelLo WoLF PSORT
General Detoxification and Stress Responses Are Mediated by Oxidized Lipids through TGA Transcription Factors in Arabidopsis W
 The Plant Cell, Vol. 20: 768 785, March 2008, www.plantcell.org ª 2008 American Society of Plant Biologists General Detoxification and Stress Responses Are Mediated by Oxidized Lipids through TGA Transcription
The Plant Cell, Vol. 20: 768 785, March 2008, www.plantcell.org ª 2008 American Society of Plant Biologists General Detoxification and Stress Responses Are Mediated by Oxidized Lipids through TGA Transcription
Host-parasite interactions: Evolutionary genetics of the House Finch- Mycoplasma epizootic
 Host-parasite interactions: Evolutionary genetics of the House Finch- Mycoplasma epizootic Scott V. Edwards Department of Organismic and Evolutionary Biology Harvard University Cambridge, MA USA http://www.oeb.harvard.edu/faculty/edwards
Host-parasite interactions: Evolutionary genetics of the House Finch- Mycoplasma epizootic Scott V. Edwards Department of Organismic and Evolutionary Biology Harvard University Cambridge, MA USA http://www.oeb.harvard.edu/faculty/edwards
EMPEROR'S COLLEGE MTOM COURSE SYLLABUS HERB FORMULAE I
 COURSE DESCRIPTION The first of three courses in the Herb Formulae series. These courses can be taken in any order. The Herb Formulae series analyzes the functions, ingredients, and properties of approximately
COURSE DESCRIPTION The first of three courses in the Herb Formulae series. These courses can be taken in any order. The Herb Formulae series analyzes the functions, ingredients, and properties of approximately
Figure legends of supplementary figures
 Figure legends of supplementary figures Figure 1. Phenotypic analysis of rice early flowering1 () plants and enhanced expression of floral identity genes in.. Leaf emergence of,, and plants with complementary
Figure legends of supplementary figures Figure 1. Phenotypic analysis of rice early flowering1 () plants and enhanced expression of floral identity genes in.. Leaf emergence of,, and plants with complementary
Nature Immunology: doi: /ni Supplementary Figure 1. Huwe1 has high expression in HSCs and is necessary for quiescence.
 Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Figure S1: Abundance of Fe related proteins in oceanic Synechococcus sp. strain WH8102 in. Ferredoxin. Ferredoxin. Fe ABC transporter
 0.004 SW nm 0.04 0.03 nm nm 0.16 0.1 nm nm 0.40 0.3 nm nm 0.80 0.5 nm 1.6 1 nm 16 10 nm 160 100 nm 0 nm 0.04 nm 0.16 nm 0.40 nm 0.80 nm 1.6 nm 16 nm 160 nm Spectral counts 0 nm 0.04 nm 0.16 nm 0.40 nm
0.004 SW nm 0.04 0.03 nm nm 0.16 0.1 nm nm 0.40 0.3 nm nm 0.80 0.5 nm 1.6 1 nm 16 10 nm 160 100 nm 0 nm 0.04 nm 0.16 nm 0.40 nm 0.80 nm 1.6 nm 16 nm 160 nm Spectral counts 0 nm 0.04 nm 0.16 nm 0.40 nm
Supplemental Figure 1: Characterization of Toc159 co-suppression lines nts
 Supplemental Data. ischof et al. Plant Cell (211)..1/tpc.111.92882 Supplemental Figure 1: Characterization of Toc19 co-suppression lines nts () Phenotype of rabiopsis plants co-suppressing attoc19. Images
Supplemental Data. ischof et al. Plant Cell (211)..1/tpc.111.92882 Supplemental Figure 1: Characterization of Toc19 co-suppression lines nts () Phenotype of rabiopsis plants co-suppressing attoc19. Images
Chapter 5 A Dose Dependent Screen for Modifiers of Kek5
 Chapter 5 A Dose Dependent Screen for Modifiers of Kek5 "#$ ABSTRACT Modifier screens in Drosophila have proven to be a powerful tool for uncovering gene interaction and elucidating molecular pathways.
Chapter 5 A Dose Dependent Screen for Modifiers of Kek5 "#$ ABSTRACT Modifier screens in Drosophila have proven to be a powerful tool for uncovering gene interaction and elucidating molecular pathways.
Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid.
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
ABA-dependant signalling of PR genes and potential involvement in the defence of lentil to Ascochyta lentis
 ABA-dependant signalling of PR genes and potential involvement in the defence of lentil to Ascochyta lentis Barkat Mustafa, David Tan, Paul WJ Taylor and Rebecca Ford Melbourne School of Land and Environment
ABA-dependant signalling of PR genes and potential involvement in the defence of lentil to Ascochyta lentis Barkat Mustafa, David Tan, Paul WJ Taylor and Rebecca Ford Melbourne School of Land and Environment
Supplemental Information. A Highly Sensitive and Robust Method. for Genome-wide 5hmC Profiling. of Rare Cell Populations
 Molecular ell, Volume 63 Supplemental Information Highly Sensitive and Robust Method for enome-wide hm Profiling of Rare ell Populations Dali Han, Xingyu Lu, lan H. Shih, Ji Nie, Qiancheng You, Meng Michelle
Molecular ell, Volume 63 Supplemental Information Highly Sensitive and Robust Method for enome-wide hm Profiling of Rare ell Populations Dali Han, Xingyu Lu, lan H. Shih, Ji Nie, Qiancheng You, Meng Michelle
Fatty Acid Desaturation
 Fatty Acid Desaturation Objectives: 1. Isolation of desaturase mutants 2. Substrates for fatty acid desaturation 3. ellular localization of desaturases References: Buchanan et al. 2000. Biochemistry and
Fatty Acid Desaturation Objectives: 1. Isolation of desaturase mutants 2. Substrates for fatty acid desaturation 3. ellular localization of desaturases References: Buchanan et al. 2000. Biochemistry and
Theme song for the 2018 Los Angeles Religious Education Congress Rise Up! Levántate! A/E. j œ. œ œ. Œ œ œ. Dios te. œ œ œ œ
 Theme sog for the 2018 Los Ageles Religious ducatio Cogress Up! vátate! ditio 0140629 Based o zekiel 7:1214; Romas 8:8 11; oh 11:1 45 stela GarcíaLópez ad Rodolfo López & & & & & & INTRO (q = ca 124) (o)
Theme sog for the 2018 Los Ageles Religious ducatio Cogress Up! vátate! ditio 0140629 Based o zekiel 7:1214; Romas 8:8 11; oh 11:1 45 stela GarcíaLópez ad Rodolfo López & & & & & & INTRO (q = ca 124) (o)
SUPPLEMENTARY FIGURES: Supplementary Figure 1
 SUPPLEMENTARY FIGURES: Supplementary Figure 1 Supplementary Figure 1. Glioblastoma 5hmC quantified by paired BS and oxbs treated DNA hybridized to Infinium DNA methylation arrays. Workflow depicts analytic
SUPPLEMENTARY FIGURES: Supplementary Figure 1 Supplementary Figure 1. Glioblastoma 5hmC quantified by paired BS and oxbs treated DNA hybridized to Infinium DNA methylation arrays. Workflow depicts analytic
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature07422 SUPPLEMENTARY INFRMATIN K S(P) R S I M Q(L4) R M 7 6 Sp Q I K R 5 4 3 2 1 L(L0) L L S E 0 +1 +2 +3 Figure S1a Difference electron density (mfo DFc) for the peptide (Qpeptide),
doi: 10.1038/nature07422 SUPPLEMENTARY INFRMATIN K S(P) R S I M Q(L4) R M 7 6 Sp Q I K R 5 4 3 2 1 L(L0) L L S E 0 +1 +2 +3 Figure S1a Difference electron density (mfo DFc) for the peptide (Qpeptide),
BIOL212 Biochemistry of Disease. Metabolic Disorders - Obesity
 BIOL212 Biochemistry of Disease Metabolic Disorders - Obesity Obesity Approx. 23% of adults are obese in the U.K. The number of obese children has tripled in 20 years. 10% of six year olds are obese, rising
BIOL212 Biochemistry of Disease Metabolic Disorders - Obesity Obesity Approx. 23% of adults are obese in the U.K. The number of obese children has tripled in 20 years. 10% of six year olds are obese, rising
Identification and expression of the genes of Metal-Responsive Transcription Factor-1 and glutathione peroxidases of common carp
 Identification and expression of the genes of Metal-Responsive Transcription Factor-1 and glutathione peroxidases of common carp Summery of Ph.D. Thesis Ágnes Ferencz Supervisor: Dr. Edit Hermesz Ph.D.
Identification and expression of the genes of Metal-Responsive Transcription Factor-1 and glutathione peroxidases of common carp Summery of Ph.D. Thesis Ágnes Ferencz Supervisor: Dr. Edit Hermesz Ph.D.
Relative activity (%) SC35M
 a 125 Bat (H17N) b 125 A/WSN (H1N1) Relative activity (%) 0 75 50 25 Relative activity (%) 0 75 50 25 0 Pos. Neg. PA PB1 Pos. Neg. NP PA PB1 PB2 0 Pos. Neg. NP PA PB1 PB2 SC35M Bat Supplementary Figure
a 125 Bat (H17N) b 125 A/WSN (H1N1) Relative activity (%) 0 75 50 25 Relative activity (%) 0 75 50 25 0 Pos. Neg. PA PB1 Pos. Neg. NP PA PB1 PB2 0 Pos. Neg. NP PA PB1 PB2 SC35M Bat Supplementary Figure
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
Gamma-aminobutyric acid (GABA) treatment blocks inflammatory pathways and promotes survival and proliferation of pancreatic beta cells
 Gamma-aminobutyric acid (GABA) treatment blocks inflammatory pathways and promotes survival and proliferation of pancreatic beta cells Gérald J. Prud homme, MD, FRCPC Keenan Research Centre for Biomedical
Gamma-aminobutyric acid (GABA) treatment blocks inflammatory pathways and promotes survival and proliferation of pancreatic beta cells Gérald J. Prud homme, MD, FRCPC Keenan Research Centre for Biomedical
Regulation of Floral-Organ- Type by SUPERMAN
 Regulation of Floral-Organ- Type by SUPERMAN 1. Need for regulators of the organ-identity genes. 2. The Superman mutant phenotype-predicting the role of SUPERMAN. 3. Testing our hypothesis of the role
Regulation of Floral-Organ- Type by SUPERMAN 1. Need for regulators of the organ-identity genes. 2. The Superman mutant phenotype-predicting the role of SUPERMAN. 3. Testing our hypothesis of the role
Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function
 Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSL0024 26223 26621 + LrgA family BPSL0025 26690 27412 + hypothetical
Table S9A: List of taurine regulated genes in Bp K96243 Chr 1 (up regulated >=2 fold) Cluster no GENE ID Start Stop Strand Function 1 BPSL0024 26223 26621 + LrgA family BPSL0025 26690 27412 + hypothetical
Overexpression of the brassinosteroid biosynthetic gene DWF4 in Brassica napus simultaneously increases seed yield and stress tolerance
 Overexpression of the brassinosteroid biosynthetic gene DWF4 in Brassica napus simultaneously increases seed yield and stress tolerance Authors Sangita Sahni 1+, Bishun D. Prasad 1+, Qing Liu 2, Vojislava
Overexpression of the brassinosteroid biosynthetic gene DWF4 in Brassica napus simultaneously increases seed yield and stress tolerance Authors Sangita Sahni 1+, Bishun D. Prasad 1+, Qing Liu 2, Vojislava
Role of GABA Shunt Pathway in Plant Abiotic Stress Tolerance
 Role of GABA Shunt Pathway in Plant Abiotic Stress Tolerance Dr. Nisreen A. AL-Quraan Jordan University of Science and Technology JORDAN What is GABA? GABA : Gamma-aminobutyric Acid Four carbon non-protein
Role of GABA Shunt Pathway in Plant Abiotic Stress Tolerance Dr. Nisreen A. AL-Quraan Jordan University of Science and Technology JORDAN What is GABA? GABA : Gamma-aminobutyric Acid Four carbon non-protein
Co-expressed Cyclin D variants cooperate to regulate proliferation of germline nuclei in. Accession number
 SUPPLEMENTARY MATERIAL Co-expressed Cyclin D variants cooperate to regulate proliferation of germline nuclei in a syncytium. Subramaniam et al. Table S1. Cyclin accession numbers Gene Human Cyclin D1a
SUPPLEMENTARY MATERIAL Co-expressed Cyclin D variants cooperate to regulate proliferation of germline nuclei in a syncytium. Subramaniam et al. Table S1. Cyclin accession numbers Gene Human Cyclin D1a
Laser Lipo Ltd. Strawberry and Strawberry & Cream. Clinical Trial Protocol
 Laser Lipo Ltd K130341 510(k) Strawberry and Strawberry & Cream Clinical Trial Protocol Conducted at: The Heath House Clinic, Crockham Hill, Edenbridge, Kent, UK. Laser Lipo Ltd. K130341 510(k) Page No.
Laser Lipo Ltd K130341 510(k) Strawberry and Strawberry & Cream Clinical Trial Protocol Conducted at: The Heath House Clinic, Crockham Hill, Edenbridge, Kent, UK. Laser Lipo Ltd. K130341 510(k) Page No.
FREE ENERGY Reactions involving free energy: 1. Exergonic 2. Endergonic
 BIOENERGETICS FREE ENERGY It is the portion of the total energy change in a system that is available for doing work at constant temperature and pressure; it is represented as ΔG. Reactions involving free
BIOENERGETICS FREE ENERGY It is the portion of the total energy change in a system that is available for doing work at constant temperature and pressure; it is represented as ΔG. Reactions involving free
Supporting Information
 Supporting Information Lee et al. 10.1073/pnas.0910950106 Fig. S1. Fe (A), Zn (B), Cu (C), and Mn (D) concentrations in flag leaves from WT, osnas3-1, and OsNAS3-antisense (AN-2) plants. Each measurement
Supporting Information Lee et al. 10.1073/pnas.0910950106 Fig. S1. Fe (A), Zn (B), Cu (C), and Mn (D) concentrations in flag leaves from WT, osnas3-1, and OsNAS3-antisense (AN-2) plants. Each measurement
Supplemental Data. Wang et al. (2013). Plant Cell /tpc
 Supplemental Data. Wang et al. (2013). Plant Cell 10.1105/tpc.112.108993 Supplemental Figure 1. 3-MA Treatment Reduces the Growth of Seedlings. Two-week-old Nicotiana benthamiana seedlings germinated on
Supplemental Data. Wang et al. (2013). Plant Cell 10.1105/tpc.112.108993 Supplemental Figure 1. 3-MA Treatment Reduces the Growth of Seedlings. Two-week-old Nicotiana benthamiana seedlings germinated on
injected subcutaneously into flanks of 6-8 week old athymic male nude mice (LNCaP SQ) and body
 SUPPLEMENTAL FIGURE LEGENDS Figure S1: Generation of ENZR Xenografts and Cell Lines: (A) 1x10 6 LNCaP cells in matrigel were injected subcutaneously into flanks of 6-8 week old athymic male nude mice (LNCaP
SUPPLEMENTAL FIGURE LEGENDS Figure S1: Generation of ENZR Xenografts and Cell Lines: (A) 1x10 6 LNCaP cells in matrigel were injected subcutaneously into flanks of 6-8 week old athymic male nude mice (LNCaP
Københavns Universitet
 university of copenhagen Københavns Universitet Linking metabolic QTLs with network and cis-eqtls controlling biosynthetic pathways Wentzell, Adam M.; Rowe, Heather C.; Hansen, Bjarne Gram; Ticconi, Carla;
university of copenhagen Københavns Universitet Linking metabolic QTLs with network and cis-eqtls controlling biosynthetic pathways Wentzell, Adam M.; Rowe, Heather C.; Hansen, Bjarne Gram; Ticconi, Carla;
