Insulin-inducible SMILE inhibits hepatic gluconeogenesis
|
|
|
- Lewis Palmer
- 6 years ago
- Views:
Transcription
1 Page 1 of 39 1 Insulin-inducible SMILE inhibits hepatic gluconeogenesis Running title: SMILE inhibits gluconeogenesis Ji-Min Lee 1*, Woo-Young Seo 2*, Hye-Sook Han 2, Kyoung-Jin Oh 2, Yong-Soo Lee 1, Don-Kyu Kim 1, Seri Choi 2, Byeong Hun Choi 2, Robert A. Harris 4, Chul-Ho Lee 3, Seung-Hoi Koo 2, and Hueng-Sik Choi 1 1 National Creative Research Initiatives Center for Nuclear Receptor Signals and Hormone Research Center, School of Biological Sciences and Technology, Chonnam National University, Gwangju , Republic of Korea, 2 Division of Life Sciences, College of Life Sciences & Biotechnology, Korea University, 145 Anam-Ro, Seongbuk-Gu, Seoul , Republic of Korea, 3 Korea Research Institute of Bioscience and Biotechnology, Daejeon , Republic of Korea, and the 4 Richard Roudebush Veterans Affairs Medical Center and the Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, Indianapolis, Indiana * These authors contributed equally to this work. Address correspondence to: Hueng-Sik Choi Hormone Research Center, School of Biological Sciences and Technology, Chonnam National University, Gwangju , Republic of Korea Tel: , Fax: , hsc@chonnam.ac.kr Seung-Hoi Koo Division of Life Sciences, College of Life Sciences & Biotechnology, Korea University, 145 Anam-Ro, Seongbuk-Gu, Seoul , Republic of Korea, Tel: , Fax: , koohoi@korea.ac.kr This manuscript contains 4,071 words, and 8 figures. This is the author's manuscript of the article published in final edited form as: Lee, J.-M., Seo, W.-Y., Han, H.-S., Oh, K.-J., Lee, Y.-S., Kim, D.-K., Choi, H.-S. (2016). Insulin-Inducible SMILE Inhibits Hepatic Gluconeogenesis., 65(1),
2 Page 2 of 39 2 ABSTRACT The role of glucagon/pka inducible coactivator peroxisome proliferator activated receptor γ coactivator-1 α (PGC-1α) signaling pathway is well-characterized in hepatic gluconeogenesis. However, an opposing PKB/Akt inducible corepressor signaling pathway is unknown. A previous report has demonstrated that Small heterodimer partner interacting leucine zipper protein (SMILE) regulates the nuclear receptors and transcriptional factors that control hepatic gluconeogenesis. Here, we show that hepatic SMILE expression was induced by feeding in normal mice, but not in db/db and high fat diet-fed (HFD) mice. Interestingly, SMILE expression was induced by insulin in mouse primary hepatocyte and liver. Hepatic SMILE expression was not altered by refeeding in liver specific insulin receptor knockout (LIRKO) or protein kinase B beta-deficient (PKBβ -/- ) mice. At the molecular level, SMILE inhibited HNF4-mediated transcriptional activity via direct competition with PGC-1α. Moreover, ablation of SMILE augmented gluconeogenesis and increased blood glucose levels in mice. Conversely, overexpression of SMILE reduced hepatic gluconeogenic genes expression and ameliorated hyperglycemia and glucose intolerance in db/db and high fat diet-fed mice. Therefore, SMILE is an insulin-inducible corepressor that suppresses hepatic gluconeogenesis. Small molecules that enhance SMILE expression would have potential for treating hyperglycemia in diabetes.
3 Page 3 of 39 3 INTRODUCTION Insulin induces the insulin receptor tyrosine kinase-mediated activation of the phosphatidylinositol 3-kinase (PI3K) pathway that controls hepatic glucose production. Ablation of insulin signalling leads to the increased gluconeogenesis in type 2 diabetes (1-3). In the fed condition, insulin inhibits hepatic gluconeogenesis by down regulating the expression of phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6- phosphatase (G6Pase). This pathway involves phosphorylation of the forkhead transcription factor FOXO1 and CBP (4-7) and recruitment of coactivators, including peroxisome proliferator activated receptor γ coactivator-1α (PGC-1α) and CREBregulated transcription co-activator 2 (CRTC2) (8, 9). Small heterodimer partner interacting leucine zipper protein (SMILE), including two alternative translation-derived isoforms, SMILE-L (CREBZF; long form of SMILE) and SMILE-S (Zhangfei; short form of SMILE), has been classified as a member of the CREB/ATF family of basic region-leucine zipper (bzip) transcription factors. However, SMILE cannot bind to DNA as a homodimer (10, 11, 12). SMILE has also been reported to function as a coactivator of ATF4 or as a corepressor of HCFbinding transcription factor (13, 14). Previously, we have reported that SMILE is a corepressor of the estrogen receptor-related receptor gamma (ERRgamma), glucocorticoid receptor (GR), constitutive androstane receptor (CAR), hepatocyte nuclear factor 4α (HNF4α), and camp responsive element-binding protein H (CREBH) (15, 16, 17). A recent study demonstrated that SMILE activates tumor suppressor p53 and inhibits the function of BMP-6 by interacting with Smads (18, 19). However, the roles of SMILE in hepatic glucose metabolism still need to be clarified. PGC-1α is a multifunctional transcriptional coactivator involved in diverse
4 Page 4 of 39 4 physiological metabolisms. In the liver, PGC-1α expression is induced in the fasting state and increases the expression of gluconeogenic genes via direct interaction with transcription factors including HNF4α, FOXO1, and GR (8, 20). Overexpression of PGC-1α leads to the increased expression of G6Pase and PEPCK, key enzymes in the hepatic glucose production. Conversely, knockdown or knockout of PGC-1α results in the lower blood glucose levels as a result of reduced gluconeogenesis. In this report, we show that SMILE is tightly regulated by the nutritional status, displaying an opposite expression pattern compared to PGC-1α. Whereas hepatic SMILE expression was elevated in the fed state relative to fasting, this regulation was lost in the absence of insulin signaling. Moreover, SMILE regulates hepatic gluconeogenesis via inhibition of PGC-1α-mediated gluconeogenic genes expression. Collectively, this study indicates that SMILE counteracts the stimulatory effect of PGC- 1α on hepatic gluconeogenesis and plays important role for insulin action on hepatic glucose metabolism. RESEARCH DESIGN AND METHODS Animal experiments Four-weeks-old male C57BL/6 mice (for the DIO model) purchased from Orient (Seongnam, Korea) were fed a high-fat diet (60 kcal% fat diet; D12492 of Research Diets, Inc.) for 12 weeks (12:12 h light-dark cycle). Male 8~12 weeks-old C57BL/6 and db/db mice were provided with a standard rodent diet. Livers of LIRKO and PKBβ / mice were provided by Dr. S. B. Biddinger (Harvard Medical School, Boston, MA, USA) and Dr. B. A. Hemmings (Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland) as described previously (21). Adenoviruses GFP and SMILE were
5 Page 5 of 39 5 delivered by tail vein injection on the 7th day. Seven days later, protein and total RNA were extracted from liver for Western blot and quantitative PCR analyses, respectively. To measure fasting blood glucose levels, animals were fasted for 4 h with free access to water. To activate the insulin signaling pathway, either PBS (for control) or insulin (0.5 unit/kg body wt) was injected intraperitoneally for 1-12 h before the collection of liver for further analyses. All procedures were performed in a specific pathogen-free facility at the Korea University College of Medicine (Seoul, Korea), based on the protocols that were approved by the Korea University Institutional Animal Care & Use Committee. Glucose, insulin and pyruvate tolerance test For the glucose or pyruvate tolerance tests, mice were fasted for 16 h and then injected intraperitoneally with 2 g/kg glucose or g/kg pyruvate (on day 6 post-adenoviral injection). Plasma glucose levels were measured using blood drawn from the tail vein at designated time points using an automatic glucose monitor (One Touch, LifeScan Ltd., Milpitas, CA USA). For the insulin tolerance test, mice were fasted for 4 h and then injected intraperitoneally with unit/kg of insulin on day 6 after adenoviral injection. Insulin measurement Plasma insulin was measured using plasma collected at time of sacrifice by using mouse insulin ELISA kit (Shibayagi Co., Ltd., Ishihara Japan). Glycogen assay Liver tissue 30mg are homogenized in ice cold 0.1M Citrate buffer(ph 4.2)+NaF and glycogen were measured using Enzychrom Glycogen assay kit ((Bioassay Systems, Hayward, CA, USA), according to the manufacturer s instructions.
6 Page 6 of 39 6 Glucose output assay Glucose production from primary hepatocytes was measured according to the manufacturer's protocol, using a colorimetric glucose oxidase assay (Sigma). Primary hepatocytes were seeded, cultured for 24 h, the cells were washed three times with phosphate-buffered saline and the medium was then replaced with glucose production buffer (glucose-free DMEM, ph 7.4, containing 20 mmol/liter sodium lactate, 1 mmol/liter sodium pyruvate, and 15 mmol/liter HEPES, without phenol red). The glucose assays were performed in triplicate. Chemicals Insulin was purchased from Sigma Aldrich (St. Louis, MO, USA). Forskolin (FSK) was purchased from Calbiochem and dissolved in the recommended solvents. Cell culture and transient transfection assay AML12 cells were maintained in Dulbecco's modified Eagle's medium/f-12 (Invitrogen), supplemented with 10% fetal bovine serum (Cambrex Bio Science Walkersville, Inc., Walkersville, MD) and antibiotics (Invitrogen). Cells were split in 24-well plates ( cells/well) at the day before transfection. Transient transfections were performed using the Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's instruction. Total DNA used in each transfection was adjusted to 0.8 µg/well by adding appropriate amount of empty vector, and CMV-β-galactosidase plasmids were cotransfected as an internal control. The cells were harvested 48 h after the transfection, and luciferase activity was measured and normalized to β-galactosidase activity. Culture of primary hepatocytes
7 Page 7 of 39 7 Primary hepatocytes were prepared from C57BL/6 mice by the collagenase perfusion method as described previously (22). After attachment, cells were infected with adenoviruses for 18 h. Subsequently, cells were maintained in the serum-free Medium 199 media (Mediatech) for overnight and treated with 10 µm forskolin and/or 100 nm insulin. Recombinant adenovirus Adenoviruses expressing unspecific shrna, shsmile, control GFP, SMILE, HNF4α, PGC-1α, SREBP-1c and dn-srebp-1c were described previously (17, 22, 23, 24). All viruses were purified by using CsCl 2 or the Adeno-X Maxi Purification Kit (Clontech, Mountain View, CA USA). Chromatin immunoprecipitation (ChIP) assay The ChIP assay was performed as described previously (17). In brief, mouse livers (40 mg/sample) were fixed with 1% formaldehyde and sonicated. The soluble chromatin was then subjected to immunoprecipitation using anti-ha (#2367, Cell Signaling Technology), anti-flag (#2368, Cell Signaling Technology), anti-pgc-1α (sc-13067, Santa Cruz Biotechnology, Santa Cruz, CA, USA) and anti-smile (sc-49329, Santa Cruz Biotechnology, Santa Cruz, CA, USA) followed by treatment with Protein A- agarose/salmon sperm DNA (Upstate Biotechnology, Upstate, NY, USA). DNA samples were quantified by quantitative real time PCR using two pairs primers of the mouse Pepck promoter follows: forward (5'-GATGTAGACTCTGTCCTG-3') and reverse (5'-GATTGCTCTGCTATGAGT-3') or mouse G6Pase promoter follows: forward (5'-CCTTGCCCCTGTTTTATATGCC-3') and reverse (5'-CGTAAATCACCC TGAACATGTTTG-3') or mouse PGC-1α promoter follows: forward (5'-GCCTATGA
8 Page 8 of 39 8 GCACGAAAGGCT-3') and reverse (5'-GCGCTCTTCAATTGCTTTCT-3'). Western blot analysis Whole-cell extracts were prepared using RIPA buffer (Elpis-Biotech, Korea). Proteins from whole-cell lysates were separated by 10% SDS-PAGE, and then transferred to nitrocellulose membranes. The membranes were probed with indicated antibodies. Immunoreactive proteins were visualized using an ECL kit Amersham ECL kit (GE Healthcare, Piscataway, NJ, USA), according to the manufacturer s instructions. Quantitative PCR Total RNA from primary hepatocytes or liver was extracted using an RNeasy mini-kit (Qiagen). All of the samples were combined for the experiment. cdna was generated by Superscript II enzyme (Invitrogen) and analyzed by Q-PCR using a SYBR green PCR kit and an TP800 Thermal Cycler DICE Real Time system (Takara). All data were normalized to ribosomal L32 expression. Statistical analyses All values are expressed as means ± S.E.M. The significance between mean values was evaluated by a two-tailed unpaired Student s t-test. RESULTS Hepatic SMILE gene expression is elevated by refeeding in normal mice, but not in insulin-resistant mice. To investigate whether SMILE plays a role in glucose homeostasis, we monitored the effect of different nutritional conditions on hepatic SMILE gene expression in mouse liver. Ad libitum feeding increased SMILE expression in liver compared with 6 h fasted mice (Fig. 1A). To further confirm the induction of
9 Page 9 of 39 9 SMILE expression in the refed condition, protein levels of SMILE were measured in the liver from fasted and refed mice. As expected, hepatic SMILE protein levels were increased by refeeding in a time dependent manner while PGC-1α protein levels were decreased (Fig. 1B). These results indicated that SMILE expression may be elevated by a signal induced by feeding. On the basis of SMILE gene expression in different nutritional conditions, we investigated whether insulin signaling is responsible for the modulation of hepatic SMILE gene expression in vivo. To confirm this, we examined whether SMILE expression is also increased by refeeding in insulin resistant mouse models. qpcr analysis showed that the expression of hepatic SMILE was significantly higher in the refed conditions in normal mice but not in db/db mice (Fig. 1C). A similar pattern of SMILE protein levels was observed in response to fasting and refeeding conditions in normal versus db/db mice (Fig. 1D). Furthermore, SMILE mrna (Fig. 1E) and protein levels (Fig. 1F) were decreased in the livers of high fat fed mice compared with those of normal chow diet mice. Collectively, these results demonstrate that hepatic expression of SMILE was tightly regulated by the nutritional condition in normal mice but not in insulin resistant mouse models. The insulin receptor/pkb signaling pathway regulates SMILE gene expression during refeeding. On the basis of different SMILE gene expression in normal mice and insulin resistant mouse models, insulin signaling would be expected to modulate hepatic SMILE gene expression in vivo. To examine this hypothesis, we injected wild type mice with insulin and measured hepatic SMILE gene expression levels. As expected, insulin injection led to a significant induction of hepatic SMILE mrna and protein in wild type mice (Fig. 2A and B). Therefore, we hypothesized that if insulin activates SMILE gene expression during refeeding condition, then liver specific insulin receptor knockout
10 Page 10 of (LIRKO) mice and protein kinase B beta-deficient (PKBβ -/- ) mice should be resistant to these effects. To examine this hypothesis, we measured the expression levels of SMILE after fasting and refeeding in wild type and LIRKO mice. LIRKO and PKBβ -/- mice exhibits elevated levels of plasma insulin, glucose and hepatic gluconeogenic gene expression during refeeding compared to WT mice (21). Although hepatic SMILE gene expression was significantly increased by refeeding in wild type mice, the hepatic SMILE mrna and protein levels were not changed by fasting or refeeding in LIRKO mice (Fig. 2C and D). In contrast, the basal expression of hepatic PGC-1α was higher in LIRKO mice than in wild type mice, but not altered during fasting and refeeding. To further verify whether PKB signal pathway is involved in the induction of hepatic SMILE gene expression in vivo, we measured the expression levels of SMILE after fasting and refeeding in both PKBβ +/+ and PKBβ -/- mice. Consistent with SMILE gene expression pattern in LIRKO mice, the hepatic SMILE mrna and protein levels were not altered by refeeding in PKBβ -/- mice compared with PKBβ +/+ mice (Fig. 2E and F). Taken together, these results demonstrate that insulin signaling via PKB is responsible for the induction of hepatic SMILE gene expression during refeeding. Insulin signaling activates the SMILE expression at the transcriptional level. To verify whether insulin activates SMILE expression at the transcriptional level, we performed transient transfection assay. Insulin treatment significantly activated SMILE promoter activity in a dose dependent manner (Fig. 3A). We also found that insulin triggered induction of SMILE levels in a time dependent manner (Fig. 3B). Consistent with the increased SMILE mrna expression, exposure of mouse primary hepatocytes to insulin also increased the protein levels of SMILE in a time dependent manner (Fig. 3C). Based on the ability of insulin to enhance the hepatic SMILE gene expression, we
11 Page 11 of examined whether PKB increases SMILE gene expression using constitutively active form of PKB (PKBca) adenovirus. Adenoviral overexpression of constitutively active PKB strongly increased SMILE mrna and protein levels (Fig. 3D and E). Because insulin-induced protein modification such as phosphorylation often changes proteins amounts by triggering their nuclear export (5-8), we examined the effects of insulin on subcellular localization of SMILE. Insulin treatment triggered increased accumulation of SMILE in the nucleus in a time dependent manner, whereas the amounts of SMILE in the cytoplasm were not altered (Fig. 3F). Interestingly, forskolin treatment decreased the basal and insulin-stimulated SMILE gene expression (Fig. 3G). Previous report demonstrated that SREBP-1c increased SMILE gene expression in rat insulinoma cells (INS-1) (24). Consistent with that report, adenoviral overexpression of SREBP-1c dose dependently increased SMILE gene expression in mouse primary hepatocytes (Supplementary Fig. 1A). Moreover, insulin-induced SMILE expression was blocked by overexpression of dominant negative SREBP-1c (dnsrebp-1c) (Supplementary Fig. 1B). Collectively, these results indicated that insulin/pkb signal pathway increases hepatic SMILE gene expression at the transcriptional level rather than at the level of protein modification. SMILE decreases HNF4α-induced gluconeogenic gene expression via competing with PGC-1α. PGC-1α is induced by camp/pka signaling in hepatocytes and is strongly increased in the liver by fasting (25). Here, we found that hepatic PGC-1α was expressed in an opposite pattern to SMILE and was significantly elevated in both LIRKO and PKBβ -/- mice. Having seen the opposite expression pattern between SMILE and PGC-1α, we then examined the effect of SMILE on PGC-1α transcriptional activity. SMILE inhibited PGC-1α-mediated G6Pase, and Pepck promoter activity (Fig. 4A).
12 Page 12 of Moreover, PGC-1α-mediated G6Pase and Pepck expression and hepatic glucose production were significantly decreased by SMILE (Fig. 4B and C). A previous report demonstrated that PGC-1α activates G6Pase and Pepck gene expression by functional interaction with hepatic nuclear factor 4alpha (HNF4α) (8). We have also reported that SMILE interacts with HNF4α and inhibits transcriptional activity of the HNF4α (16). To explore whether PGC-1α-induced HNF4α transcriptional activity is repressed by SMILE, AML-12 cells were co-transfected with HNF4α, PGC-1α and SMILE expression vector together with gluconeogenic genes promoter-reporter constructs. HNF4α-mediated induction of G6Pase and PEPCK promoter activity was significantly decreased by SMILE but was recovered by coexpression of PGC-1α (Fig. 4D). Next, to examine whether SMILE can affect the PGC-1α occupancy on the HNF4α binding region of the G6Pase and Pepck promoter in different nutritional conditions, we performed ChIP assays using fasted or refed mouse liver tissues. We observed increased occupancy of PGC-1α in the HNF4α binding region of the G6Pase and Pepck promoter in the fasted condition compared to the refed condition, whereas the occupancy of SMILE was significantly increased in the refed condition, which is completely opposite to the occupancy of PGC-1α (Fig. 4E). Moreover, overexpression of SMILE significantly decreased PGC-1α occupancy in the HNF4α binding region of the G6Pase and Pepck promoter. In contrast, overexpression of PGC-1α decreased SMILE occupancy in the HNF4α binding region (Fig. 4F). These results suggest that SMILE competes with PGC-1α in forming a complex with HNF4α on G6Pase and Pepck promoter. Moreover, adenoviral overexpression of SMILE markedly inhibited HNF4α-mediated G6Pase, and Pepck gene expression (Fig. 4G) or hepatic glucose production (Fig. 4H). We have also assessed the effect of SMILE on glycolytic gene
13 Page 13 of expression. Overexpression of HNF4α increased the glucokinase (Gck) gene expression, whereas HNF4α-induced Gck gene expression was not affected by SMILE overexpression. Moreover, insulin-induced Gck expression was not altered by knockdown of SMILE (Supplementary Fig. 2A and B). These results indicate that SMILE specifically affects gluconeogenesis in the liver. A previous report showed that activation of FOXO1 by PGC-1a is required for gluconeogenic genes expression (20). We found that SMILE inhibits FOXO1-mediated hepatic gluconeogenic genes promoter activity or expression. Moreover, SMILE directly interacts with FOXO1 and competes with PGC-1α to inhibit FOXO1 transcriptional activity (Supplementary Fig. 3A-D). Taken together, these results demonstrate that SMILE regulates the HNF4α-mediated PEPCK and G6Pase gene expression via competing with PGC-1α and stimulatory effect of PGC-1α on FOXO1 is also inhibited by SMILE. Knockdown of SMILE gene expression causes hyperglycemia in mice. The action of insulin in suppression of gluconeogenesis occurs rapidly via phosphorylation or dephosphorylation of its target molecules, whereas SMILE is regulated by insulin through gene transcription level. We assessed the effect of insulin-mediated SMILE gene expression on hepatic gluconeogenesis in early or delayed response to insulin. In the early response to insulin, knockdown of SMILE did not show any significant effect on insulin-mediated repression, whereas insulin-mediated repression of hepatic glucose output (Fig. 5A) and gluconeogenic gene expression (Fig. 5B) were significantly relieved by knockdown of SMILE in the delayed time. These results indicate that prolonged inhibition of hepatic gluconeogenesis by insulin occurs via induction of SMILE expression in the later time point. Next, we hypothesized that alteration in SMILE gene expression would affect hepatic glucose metabolism in vivo. To explore the
14 Page 14 of action of hepatic SMILE in vivo, we injected mice with an adenovirus expressing shrna against SMILE. As expected, knockdown of endogenous SMILE in the liver elicited a marked increase of fasting blood glucose levels in mice (Fig. 5C). However, plasma insulin levels were unaltered in Ad-shSMILE injected mice compared with Ad- US injected mice (Fig. 5D). Moreover, we confirmed a marked induction of hepatic gluconeogenic gene expression such as Pepck and G6Pase upon SMILE knockdown (Fig. 5E). Next, we further investigated whether ablation of endogenous SMILE affect glucose excursion, insulin sensitivity, and hepatic glucose production. During an intraperitoneal glucose tolerance test (GTT), glucose tolerance was significantly attenuated in Ad-shSMILE injected mice compared with control mice (Fig. 5F). However, ablation of endogenous SMILE did not lead to the significant changes in insulin sensitivity as observed by insulin tolerance test (ITT) in vivo (Fig. 5G). To examine the impact of ablation of endogenous SMILE on hepatic glucose production, we also performed a pyruvate tolerance test (PTT). Ablation of endogenous SMILE elevated the pyruvate-dependent increase of blood glucose levels (Fig. 5H). Overexpression of SMILE in normal mice improves glucose tolerance without markedly affecting fasting blood glucose level. Based on the fact that ablation of SMILE increases blood glucose levels or hepatic gluconeogenesis, we suspected that induction of SMILE would have effects on hepatic glucose metabolism in normal mice. Adenovirus-mediated expression of SMILE did not elicit a significant change in overnight fasting blood glucose levels or liver glycogen contents (Fig. 6A and B). On the contrary, overexpression of SMILE significantly improved glucose tolerance and reduced gluconeogenesis as shown by improved pyruvate tolerance and reduced gluconeogenic gene expression (Fig. 6C-E). In addition, overexpression of SMILE did
15 Page 15 of not cause significant induction of insulin signaling in the liver or in the muscle of normal chow-fed mice (Supplementary Fig. 4A and B). These results indicate that SMILE significantly improved glucose tolerance or pyruvate tolerance without affecting glycogen levels in normal chow diet fed-mice. SMILE decreases gluconeogenic gene expression and improves hyperglycemia in diabetic mice models. We next hypothesized that alteration in SMILE expression would affect hepatic glucose metabolism in diabetic mice. To further assess the functional consequences of SMILE-mediated repression in gluconeogenic gene expression and improvement of blood glucose level, we performed adenoviral overexpression of SMILE in db/db and high fat diet (HFD) mice. The adenovirus GFP (Ad-GFP) and SMILE (Ad-SMILE) injected mice showed similar body weights (Fig. 7A), but overexpression of SMILE significantly reduced fasting blood glucose levels in db/db and HFD mice (Fig. 7B). Moreover, we observed a significant improvement of glucose tolerance and pyruvate tolerance without affecting insulin sensitivity in Ad- SMILE injected db/db or HFD mice compared with Ad-GFP injected db/db or HFD mice (Fig. 7C-E). Concomitantly, there was a significant reduction in mrna levels of hepatic gluconeogenic gene, such as G6Pase, Pepck and Pgc-1α in Ad-SMILE injected db/db or HFD mice compared with Ad-GFP injected db/db or HFD mice (Fig. 7F and G). However, overexpression of SMILE did not lead to significant enhanced insulin signaling either in the liver or in the muscle of HFD mice (Supplementary Fig. 4C and D). Taken together, these results suggest that SMILE directly regulate hepatic glucose metabolism in diabetic mice. Therefore, activation of SMILE could be a new therapeutic approach to treat diabetes by suppressing gluconeogenesis and ameliorating hyperglycemia.
16 Page 16 of DISCUSSION Insulin is the major hormone for the regulation of hepatic glucose metabolism by suppressing the expression of gluconeogenic enzyme genes (7, 9, 20). SMILE expression was increased by refeeding condition, but not in fasting condition or insulin resistant status such as HFD and db/db mice. Thus, it seemed that the insulin/pkb pathway plays a major role in the regulation of SMILE expression. In support of this hypothesis, we found that insulin increases hepatic expression of SMILE both in vitro and in vivo. However, the identity of the transcription factor that mediates activation of SMILE gene expression by insulin signaling is still elusive and requires further exploration. One potential candidate transcription factor for mediating insulin signaling is SREBP-1c, a well known factor in the transcriptional activation of lipogenesis. Indeed, SREBP-1c increased SMILE gene expression in rat insulinoma cells (INS-1) (24). In primary hepatocytes, adenoviral overexpression of SREBP-1c increased SMILE gene expression and insulin-induced SMILE expression was blocked by dominant negative SREBP-1c (dnsrebp-1c). These results suggest that insulin-mediated induction of SREBP-1c expression could increase SMILE gene expression in hepatocytes. Meanwhile, our previous study suggested that AMPK-induced SMILE decreases SREBP-1c gene expression (26). We speculated that the physiological induction of SMILE expression mainly represses gluconeogenesis under feeding conditions, while supraphysiological activation of SMILE expression could also reduce SREBP-1c expression and the subsequent lipogenic program. Alternatively, SMILE might be critical in fine-tuning the levels of fatty acid biosynthesis in the liver under feeding conditions. Reduced expression of SMILE in the livers of insulin resistance models
17 Page 17 of could thus be associated with increased triacylglycerol synthesis in this setting. Taken together, insulin-mediated induction of SREBP-1c increases SMILE gene expression, which could decrease SREBP-1c expression via negative feedback mechanism. Under normal physiological conditions, SREBP-1c-mediated SMILE induction may inhibit the SREBP-1c gene expression to maintain lipid and glucose homeostasis. While SMILE gene expression was significantly increased by refeeding in normal mice, LIRKO and PKBβ -/- mice did not display induction of SMILE gene expression during refeeding. Thus, SMILE could be the nutritional status sensing nuclear corepressor by insulin signaling that represses gluconeogenesis in the fed status. Collectively, we suggest that enhanced expression of SMILE in the fed state is another critical mechanism by which insulin regulates hepatic gluconeogenesis. We have also shown that activation of AMPK by curcumin, a polyphenol compound, increases SMILE gene expression, which leads to the SMILE-mediated repression of CREBH transcriptional activity via competing with PGC-1α (17). These data suggest that pharmacological activation of SMILE expression can be utilized as a new therapeutic strategy to relieve hyperglycemia. Genetically altered animal models of SMILE are also needed to fully understand the physiological and pathological role of SMILE in hepatic gluconeogenesis. Gluconeogenic enzyme gene expression is regulated by both transcription factors and coactivator, such as PGC-1α (8). In this study, SMILE inhibits HNF4α- or FOXO1-mediated hepatic gluconeogenic genes promoter activity or expression via competing with PGC-1α. In addition, previous report revealed that SMILE inhibits glucocorticoid receptor (GR) which plays an important role in regulation of gluconeogenic genes expression such as PEPCK and G6Pase (16). Thus, SMILE could
18 Page 18 of function as a universal transcriptional corepressor for PGC-1α-interacting transcription factors such as HNF4α, FOXO1, and GR. A previous report showed that insulin and PKB inhibit FOXO1 and PGC-1α activity by a phosphorylation-dependent mechanism (5, 20, 27). In the immediate early response of insulin, gluconeogenesis is regulated by classical phosphorylation mechanism. Here, we found the role of the delayed effect of insulin on the regulation of gluconeogenesis via induction of co-repressor SMILE. These results suggest that SMILE has an important role in delayed action of insulin, and the induction of SMILE is another way by which insulin counteracts the action of PGC- 1α in the regulation of hepatic glucose metabolism. In this study, we employed the adenoviral approach to explore the regulatory role of SMILE in glucose homeostasis. Thus, genetically altered animal models of SMILE are also needed to understand the physiological and pathological role of SMILE in hepatic gluconeogenesis. In fasting condition, PKA/PGC-1α pathway increases hepatic gluconeogenesis. In the fed condition, the PKB/SMILE pathway turns off the PGC-1α-induced hepatic gluconeogenesis via a competitive binding mechanism (Fig. 8). Overall, our observations provide insight into a novel mechanism for insulin-mediated regulation in hepatic glucose metabolism that can be utilized as a novel therapeutic approach for the treatment of hyperglycemia in diabetes.
19 Page 19 of ACKNOWLEDGEMENTS This work was supported by National Creative Research Initiatives Grant ( ) through the National Research Foundation of Korea (NRF) funded by the Korean government (Ministry of Science, ICT & Future Planning). S-H Koo is supported by the National Research Foundation of Korea (grant nos.: NRF- 2012M3A9B , NRF-2015R1A5A and NRF-2015R1A2A1A ), funded by the Ministry of Science, ICT & Future Planning, Republic of Korea, and Korea University, Seoul, Republic of Korea. No potential conflicts of interest relevant to this article were reported. J.-M.L, W.-Y.S, H.-S.C, S.-H.K designed most of the experiments. J.-M.L, W.-Y.S, K.- J.H, Y.-S.L, D.-K.K, H.-S.H, S.R.C, B.H.C performed the experiments. J.-M.L wrote the manuscript and S.-H.K, H.-S.C, Robert A. Harris and C.-H.L contributed to discussion, review, and editing of the manuscript. Dr. Hueng-Sik Choi and Dr. Seung- Hoi Koo are the guarantors of this work, had full access to all the data in the study, and take responsibility for the integrity of the data and for the accuracy of the data analysis. This paper is dedicated to the memory of the late Dr. Richard W. Hanson (Case Western Reserve University) in recognition of his many great contributions to science.
20 Page 20 of REFERENCES 1. Saltiel AR, Kahn CR: Insulin signalling and the regulation of glucose and lipid metabolism. Nature 2001; 414: Saltiel AR & Pessin JE: Insulin signaling pathways in time and space. Trends Cell Biol 2002; 12, Saltiel AR: New perspectives into the molecular pathogenesis and treatment of type 2 diabetes. Cell 2001; 104: Cho H, Mu J, Kim JK, Thorvaldsen JL, Chu Q, Crenshaw EB 3rd, Kaestner KH, Bartolomei MS, Shulman GI, Birnbaum MJ: Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Insulin resistance an Akt2 (PKB beta). Science 2001;292, Nakae, J., Park, B. C. & Accili, D: Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a Wortmanninsensitive pathway. J. Biol. Chem 1999;274, Taniguchi CM, Kondo T, Sajan M, Luo J, Bronson R, Asano T, Farese R, Cantley LC, Kahn CR: Divergent regulation of hepatic glucose and lipid metabolism by phosphoinositide 3-kinase via Akt and PKClambda/zeta. Cell Metab 2006;3, Zhou XY, Shibusawa N, Naik K, Porras D, Temple K, Ou H, Kaihara K, Roe MW, Brady MJ, Wondisford FE: Insulin regulation of hepatic gluconeogenesis through phosphorylation of CREB-binding protein. Nature Med 2004;10, Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM: Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 2001;413: Renaud Dentin, Yi Liu, Seung-Hoi Koo, Susan Hedrick, Thomas Vargas, Jose Heredia, John Yates III & Marc Montminy: Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature 2007;449, Lu R, Misra V: Zhangfei: a second cellular protein interacts with herpes simplex virus accessory factor HCF in a manner similar to Luman and VP16. Nucleic Acids Res 2000;28(12): Xie Y. B., Lee O. H., Nedumaran B., Seong H. A., Lee K. M., Ha H., Lee I. K., Yun Y., Choi H. S: SMILE, a new orphan nuclear receptor SHP-interacting protein, regulates SHP-repressed estrogen receptor transactivation. Biochem. J 2008;416, Akhova, O., Bainbridge, M., and Misra, V: The neuronal host cell factor-binding protein Zhangfei inhibits herpes simplex virus replication. J. Virol 2005;79, Hogan M. R., Cockram G. P., Lu R: Cooperative interaction of Zhangfei and ATF4 in transactivation of the cyclic AMP response element. FEBS Lett 2006;580, Misra,V., Rapin,N., Akhova,O., Bainbridge,M. and Korchinski,P: Zhangfei is a potent and specific inhibitor of the host cell factor-binding transcription factor Luman. J Biol Chem 2005;280,
21 Page 21 of Xie YB, Park JH, Kim DK, Hwang JH, Oh S, Park SB, Shong M, Lee IK, Choi HS: Transcriptional corepressor SMILE recruits SIRT1 to inhibit nuclear receptor estrogen receptor-related receptor gamma transactivation. J Biol Chem 2009;284(42): Xie Y. B., Nedumaran B., Choi H. S: Molecular characterization of SMILE as a novel corepressor of nuclear receptors. Nucleic Acids Res 2009;37, Misra J, Chanda D, Kim DK, Li T, Koo SH, Back SH, Chiang JY, Choi HS: Curcumin differentially regulates endoplasmic reticulum stress through transcriptional corepressor SMILE (small heterodimer partner-interacting leucine zipper protein)-mediated inhibition of CREBH (camp responsive element-binding protein H). J Biol Chem 2011; 286, López-Mateo I, Villaronga MA, Llanos S, Belandia B: The transcription factor CREBZF is a novel positive regulator of p53. Cell Cycle 2012;11, Lee JH, Lee GT, Kwon SJ, Jeong J, Ha YS, Kim WJ, Kim IY: CREBZF, a novel Smad8-binding protein. Mol Cell Biochem 2012;368, Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, Spiegelman BM: Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature 2003;423(6939): Kim DK, Kim YH, Hynx D, Wang Y, Yang KJ, Ryu D, Kim KS, Yoo EK, Kim JS, Koo SH, Lee IK, Chae HZ, Park J, Lee CH, Biddinger SB, Hemmings BA, Choi HS: PKB/Akt phosphorylation of ERRγ contributes to insulin-mediated inhibition of hepatic gluconeogenesis. Diabetologia 2014; 57(12): Nedumaran B, Hong S, Xie YB, Kim YH, Seo WY, Lee MW, Lee CH, Koo SH, Choi HS: DAX-1 acts as a novel corepressor of orphan nuclear receptor HNF4alpha and negatively regulates gluconeogenic enzyme gene expression. J Biol Chem 2009; 284(40): Kim DK, Kim JR, Koh M, Kim YD, Lee JM, Chanda D, Park SB, Min JJ, Lee CH, Park TS, Choi HS: Estrogen-related receptor γ (ERRγ) is a novel transcriptional regulator of phosphatidic acid phosphatase, LIPIN1, and inhibits hepatic insulin signaling. J Biol Chem 2011; 286(44): Lee KM, Seo YJ, Kim MK, Seo HA, Jeong JY, Choi HS, Lee IK, Park KG: Mediation of glucolipotoxicity in INS-1 rat insulinoma cells by small heterodimer partner interacting leucine zipper protein (SMILE). Biochem Biophys Res Commun 2012; 23;419(4): Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M: CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 2001;413(6852): Lee JM, Gang GT, Kim DK, Kim YD, Koo SH, Lee CH, Choi HS: Ursodeoxycholic acid inhibits liver X receptor α-mediated hepatic lipogenesis via induction of the nuclear corepressor SMILE. J Biol Chem 2014; 10;289(2): Xinghai Li, Bobby Monks, Qingyuan Ge & Morris J. Birnbaum: Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature 2007; 447(7147):1012-6
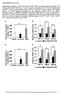
22 Page 22 of FIGURE LEGENDS FIG. 1. Hepatic SMILE expression is induced in feeding condition, but not in insulin-resistant condition. A: Q-PCR analysis was performed using total RNA from mice liver samples after ad libitum fed or 6 h fasting. SMILE, Pgc-1α, Pepck, G6pase and β-actin genes were amplified by qpcr using corresponding primers. B: C57BL6 mice (n=4) fasted for 6 h and refed for 1-6 h. Western blot analysis (left) showing SMILE expression during refed time course. Graphical representation (right). All of the samples were combined for the western blot analysis. C-D: Expression of SMILE and Pgc-1α. Normal and db/db mice were fasted for 6 h and refed for 2 h. Q-PCR analysis was performed to measure the expression of SMILE and Pgc-1α using total RNA from mice liver samples (C) and Western blot analysis was performed using SMILE, PGC-1α and α-tubulin antibodies (D). All of the samples were combined for the western blot analysis. E-F: C57BL6 mice were maintained under normal chow or high fat diet (HFD) for 12 weeks. Q-PCR analysis was performed using total RNA from mice liver of normal and HFD mice to measure SMILE, Pgc-1α and L32 expression (E) and performed Western blot analysis using SMILE, PGC-1α and α-tubulin antibodies (F). Error bars show ± s.e.m. *P<0.05, **P<0.01 by two-tailed Student t-test. FIG. 2. Insulin receptor signalling requires for SMILE gene induction during refeeding. A-B: Expression of hepatic SMILE in mice (n=4) injected with insulin. Q- PCR analysis was performed using total RNA from mice liver injected with insulin. SMILE and β-actin genes were amplified using specific primers for PCR (A). Western blot analysis (left) and graphical representation (right) showing SMILE expression in liver and p-pkb levels were used for positive control of insulin signaling (B). C-D: LIRKO mice exhibits marked reduction of hepatic SMILE gene expression. Normal and
23 Page 23 of LIRKO mice were fasted for 6 h and refed for 2 h. SMILE and Pgc-1α gene expression were analyzed using Q-PCR analysis (C) and Western (D). E-F: Hepatic expression of SMILE in PKBβ -/- mice. Wild type and PKBβ -/- mice were fasted for 6 h and refed for 2 h. SMILE and Pgc-1α gene expression were analyzed using quantitative PCR analysis (E) and Western blot analysis (F). All of the samples were combined for the western blot analysis. Error bars show ± s.e.m. n.s., not significant. *P<0.05, **P<0.01 by twotailed Student t-test. FIG. 3. Insulin increases SMILE gene expression at a transcriptional level. A: AML-12 cells were co-transfected with SMILE promoter and treated with vehicle or insulin. B-C: Insulin effects on SMILE gene expression in primary hepatocytes. Primary hepatocytes were cultured for 12 h under serum starvation. The cells were then treated with insulin (100 nm) for h. SMILE gene expression were analyzed using Q-PCR analysis (B). Western blot analysis (left) and graphical representation (right) showing hepatic SMILE expression and p-pkb levels were used for positive control of insulin signaling (C). D-E: Overexpression of PKB results in the induction of SMILE gene expression. Adenovirus GFP and PKB ca (constitutive active form) were infected in primary hepatocytes. SMILE gene expression was analyzed using Q-PCR analysis (D). Western blot analysis (left) and graphical representation (right) showing SMILE expression levels (E). F-G: Nuclear expression of SMILE by insulin. Immunoblot of SMILE in cytosolic (C) and nuclear (N) fraction form AML-12 cells exposed to insulin (100 nm) and forskolin (10 µm). Cytoplasmic (C) and nuclear (N) fraction were indicated. LaminA indicates a nuclear protein loading control and α-tubulin indicates a cytoplasmic protein loading control. Error bars show ± s.e.m. *P<0.05, **P<0.01 by two-tailed Student t-test.
24 Page 24 of FIG. 4. SMILE inhibits HNF4α-mediated hepatic gluconeogenesis via competing with PGC-1α. A: SMILE decreases PGC-1α-induced PEPCK and G6Pase promoter activity. AML-12 cells were co-transfected with PEPCK-Luc, G6Pase-Luc, PGC-1α and SMILE expression vector. B: Primary hepatocytes were infected with Ad-GFP, Ad- PGC-1α and Ad-SMILE for 24 h before cell harvest. Expression of Pepck and G6pase were analyzed by quantitative PCR. C: SMILE decreases PGC-1α-induced hepatic glucose production. Glucose output assay was performed using Ad-PGC-1α and Ad- SMILE infected primary hepatocytes. D: SMILE competes with PGC-1α to inhibit HNF4α transcriptional activity. AML-12 cells were co-transfected with PEPCK-Luc, G6Pase-Luc, HNF4α, PGC-1α and SMILE expression vector. E: C57BL6 mice (n=3) were fasted or fed for 12 h and chromatin Immunoprecipitation (ChIP) assay were performed using mice liver sample. F: AML-12 cells were co-transfected with HA- PGC-1α and FLAG-SMILE and ChIP assay were performed. G: Overexpression of SMILE results in the decrease of HNF4α-induced gluconeogenic gene expression. Primary hepatocytes were infected with Ad-GFP, Ad-HNF4α, Ad-PGC-1α and Ad- SMILE for 24 h and gluconeogenic gene expression was analyzed by quantitative PCR. H: SMILE decreases HNF4α-induced hepatic glucose production. Primary hepatocytes were infected with Ad-HNF4α and Ad-SMILE for 24 h and glucose output assay was performed using glucose-free media supplemented with gluconeogenic substrate sodium lactate (20 mm) and sodium pyruvate (1 mm). Error bars show ± s.e.m. *P<0.05 by two-tailed Student t-test. FIG. 5. Ablation of SMILE leads to hyperglycemia in mice. A-B: Knockdown of SMILE rescues insulin-mediated repression hepatic glucose production or
25 Page 25 of gluconeogenic gene expression. Primary hepatocytes were infected with Ad-US or AdshSMILE for 24 h and treated insulin for 2-12 h with glucagon. Glucose output assay was performed using glucose-free media supplemented with gluconeogenic substrate sodium lactate (20 mm) and sodium pyruvate (1 mm) (A). Gluconeogenic gene expression was analyzed by quantitative PCR (B). C-H: Knockdown of SMILE induces hepatic gluconeogenic genes and fasting blood glucose levels. Ad-US or Ad-shSMILE were injected via the tail-vein into mice (n=5-9). Blood glucose levels measured at day 4 after Ad-US or Ad-shSMILE injection (C). Insulin levels measured at day 4 after Ad- US or Ad-shSMILE injection (D). Q-PCR analysis of gluconeogenic gene expression using total RNA isolated from mouse liver after Ad-US or Ad-shSMILE injection (E). Mice were injected intraperitoneally with 2 g/kg glucose, 1 g/kg pyruvate or 1 unit/kg of insulin on day 6 after adenoviral injection for glucose tolerance test (F), insulin tolerance test (G) and pyruvate tolerance test (H). Plasma glucose or insulin was measured on blood drawn from the tail vein at designated time points. Error bars show ± s.e.m. n.s., not significant. *P<0.05, **P<0.01, ***P<0.005 by two-tailed Student t-test. FIG. 6. Minimal effect of SMILE overexpression in normal mice. A-F: C57BL6-8weeks old mice (n=6-8) injected with adenovirus GFP or SMILE. Overnight fasting blood glucose levels (A) and glucagon levels (B) were measured in adenovirus GFP or SMILE injected normal mice. Mice were injected intraperitoneally with 2 g/kg glucose or 2 g/kg pyruvate on day 6 after adenoviral injection for glucose tolerance test (C), pyruvate tolerance test (D). Expression of Pepck and G6pase were analyzed by realtime quantitative RT-PCR in adenovirus GFP or SMILE injected mice (E). Error bars show ± s.e.m. n.s., not significant. *P<0.05, **P<0.01 by two-tailed Student t-test.
26 Page 26 of FIG. 7. SMILE ameliorates hyperglycemia in db/db and HFD mice. A-G: Recombinant adenovirus GFP or SMILE were delivered by tail-vein injection to db/db and HFD mice (n=4-7). Mice were sacrificed at day 7 after adenovirus GFP or SMILE injection. Body weight (A) or blood glucose levels (B) were measured in adenovirus GFP or SMILE injected mice. Adenovirus GFP or SMILE injected mice were fasted for 4 h and injected intraperitoneally with 2 g/kg glucose, 1 unit/kg of insulin or 1.5 g/kg pyruvate for glucose tolerance test (C), pyruvate tolerance test (D) or insulin tolerance test (E). Expression of Pepck, G6pase, Pgc-1α and SMILE were analyzed by real-time quantitative RT-PCR in adenovirus GFP or SMILE injected db/db (F) or HFD mice (G). All data were normalized to ribosomal L32 expression. Error bars show ± s.e.m. *P<0.05, **P<0.01, ***P<0.005 by two-tailed Student t-test. FIG. 8. Schematic diagram of insulin-induced SMILE role in regulation of hepatic gluconeogenesis. PKA/PGC-1α pathway increases hepatic gluconeogenesis during fasting. However, PKB/SMILE pathway turns off PGC-1α-induced hepatic gluconeogenesis during refeeding by a competitive binding mechanism to prevent excess of glucose production.
27 Page 27 of x156mm (150 x 150 DPI)
28 Page 28 of x162mm (150 x 150 DPI)
29 Page 29 of x117mm (150 x 150 DPI)
30 Page 30 of x211mm (150 x 150 DPI)
31 Page 31 of x169mm (150 x 150 DPI)
32 Page 32 of x123mm (150 x 150 DPI)
33 Page 33 of x227mm (150 x 150 DPI)
34 Page 34 of x147mm (150 x 150 DPI)
35 Page 35 of 39 Supplementary Informations Insulin-inducible SMILE inhibits hepatic gluconeogenesis Running title: SMILE inhibits gluconeogenesis Ji-Min Lee 1*, Woo-Young Seo 2*, Hye-Sook Han 2, Kyoung-Jin Oh 2, Yong-Soo Lee 1, Don-Kyu Kim 1, Seri Choi 2, Byeong Hun Choi 2, Robert A. Harris 4, Chul-Ho Lee 3, Seung-Hoi Koo 2, and Hueng-Sik Choi 1 1 National Creative Research Initiatives Center for Nuclear Receptor Signals and Hormone Research Center, School of Biological Sciences and Technology, Chonnam National University, Gwangju , Republic of Korea, 2 Division of Life Sciences, College of Life Sciences & Biotechnology, Korea University, 145 Anam-Ro, Seongbuk-Gu, Seoul , Republic of Korea, 3 Korea Research Institute of Bioscience and Biotechnology, Daejeon , Republic of Korea, and the 4 Richard Roudebush Veterans Affairs Medical Center and the Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, Indianapolis, Indiana * These authors contributed equally to this work. CONTENTS Supplementary Figure 1. SREBP-1c increases SMILE gene expression. Supplementary Figure 2. SMILE does not alter glucokinase gene expression. Supplementary Figure 3. SMILE inhibits FOXO1-induced gluconeogenic gene expression. Supplementary Figure 4. SMILE does not change insulin sensitivity in liver and muscle.
36 Page 36 of 39 Supplementary Figure 1. SREBP-1c increases SMILE gene expression. A: Primary hepatocytes were infected with Ad-GFP, Ad-SREBP-1c for 24 h and expression of SMILE was analyzed by quantitative PCR. B: Primary hepatocytes were infected with Ad-GFP, AddnSREBP-1c for 12 h and treated with insulin (100nM) for 9 h. SMILE gene expression were analyzed using Q-PCR analysis.
37 Page 37 of 39 Supplementary Figure 2. SMILE does not alter glucokinase gene expression. A: Primary hepatocytes were infected with Ad-GFP, Ad-HNF4α and Ad-SMILE for 24 h and expression of glucokinase was analyzed by quantitative PCR. B: Primary hepatocytes were infected with Ad-US or Ad-shSMILE for 24 h and treated insulin for 6 h. Glucokinase gene expression was analyzed by quantitative PCR. Error bars show ± s.e.m. n.s., not significant. *P<0.05, ***P<0.005 by two-tailed Student t-test.
Regulation of Hepatic Gluconeogenesis by an ER-Bound Transcription Factor, CREBH
 Cell Metabolism, Volume 11 Supplemental Information Regulation of Hepatic Gluconeogenesis by an ER-Bound Transcription Factor, CREBH Min-Woo Lee, Dipanjan Chanda, Jianqi Yang, Hyunhee Oh, Su Sung Kim,
Cell Metabolism, Volume 11 Supplemental Information Regulation of Hepatic Gluconeogenesis by an ER-Bound Transcription Factor, CREBH Min-Woo Lee, Dipanjan Chanda, Jianqi Yang, Hyunhee Oh, Su Sung Kim,
Effect of BI-1 on insulin resistance through regulation of CYP2E1
 Effect of BI-1 on insulin resistance through regulation of CYP2E1 Geum-Hwa Lee 1, Kyoung-Jin Oh 2, 3, Hyung-Ryong Kim 4, Hye-Sook Han 2, Hwa-Young Lee 1, Keun-Gyu Park 5, Ki-Hoan Nam 6, Seung-Hoi Koo 2
Effect of BI-1 on insulin resistance through regulation of CYP2E1 Geum-Hwa Lee 1, Kyoung-Jin Oh 2, 3, Hyung-Ryong Kim 4, Hye-Sook Han 2, Hwa-Young Lee 1, Keun-Gyu Park 5, Ki-Hoan Nam 6, Seung-Hoi Koo 2
Requires Signaling though Akt2 Independent of the. Transcription Factors FoxA2, FoxO1, and SREBP1c
 Cell Metabolism, Volume 14 Supplemental Information Postprandial Hepatic Lipid Metabolism Requires Signaling though Akt2 Independent of the Transcription Factors FoxA2, FoxO1, and SREBP1c Min Wan, Karla
Cell Metabolism, Volume 14 Supplemental Information Postprandial Hepatic Lipid Metabolism Requires Signaling though Akt2 Independent of the Transcription Factors FoxA2, FoxO1, and SREBP1c Min Wan, Karla
Over-expression of MKP-3 and knockdown of MKP-3 and FOXO1 in primary rat. day, cells were transduced with adenoviruses expressing GFP, MKP-3 or shgfp,
 SUPPLEMENTAL METHODS Over-expression of MKP-3 and knockdown of MKP-3 and FOXO1 in primary rat hepatocytes Primary rat hepatocytes were seeded as described in experimental procedures. The next day, cells
SUPPLEMENTAL METHODS Over-expression of MKP-3 and knockdown of MKP-3 and FOXO1 in primary rat hepatocytes Primary rat hepatocytes were seeded as described in experimental procedures. The next day, cells
ACC ELOVL MCAD. CPT1α 1.5 *** 0.5. Reverbα *** *** 0.5. Fasted. Refed
 Supplementary Figure A 8 SREBPc 6 5 FASN ELOVL6.5.5.5 ACC.5.5 CLOCK.5.5 CRY.5.5 PPARα.5.5 ACSL CPTα.5.5.5.5 MCAD.5.5 PEPCK.5.5 G6Pase 5.5.5.5 BMAL.5.5 Reverbα.5.5 Reverbβ.5.5 PER.5.5 PER B Fasted Refed
Supplementary Figure A 8 SREBPc 6 5 FASN ELOVL6.5.5.5 ACC.5.5 CLOCK.5.5 CRY.5.5 PPARα.5.5 ACSL CPTα.5.5.5.5 MCAD.5.5 PEPCK.5.5 G6Pase 5.5.5.5 BMAL.5.5 Reverbα.5.5 Reverbβ.5.5 PER.5.5 PER B Fasted Refed
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
General Laboratory methods Plasma analysis: Gene Expression Analysis: Immunoblot analysis: Immunohistochemistry:
 General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
Supplementary Table 1. Metabolic parameters in GFP and OGT-treated mice
 Supplementary Table 1. Metabolic parameters in GFP and OGT-treated mice Fasted Refed GFP OGT GFP OGT Liver G6P (mmol/g) 0.03±0.01 0.04±0.02 0.60±0.04 0.42±0.10 A TGs (mg/g of liver) 20.08±5.17 16.29±0.8
Supplementary Table 1. Metabolic parameters in GFP and OGT-treated mice Fasted Refed GFP OGT GFP OGT Liver G6P (mmol/g) 0.03±0.01 0.04±0.02 0.60±0.04 0.42±0.10 A TGs (mg/g of liver) 20.08±5.17 16.29±0.8
Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice
 Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice Julien Bricambert,, Catherine Postic, Renaud Dentin J Clin
Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice Julien Bricambert,, Catherine Postic, Renaud Dentin J Clin
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism SUPPLEMENTARY FIGURES, LEGENDS AND METHODS
 A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
Defective Hepatic Autophagy in Obesity Promotes ER Stress and Causes Insulin Resistance
 Cell Metabolism, Volume 11 Supplemental Information Defective Hepatic Autophagy in Obesity Promotes ER Stress and Causes Insulin Resistance Ling Yang, Ping Li, Suneng Fu, Ediz S. Calay, and Gökhan S. Hotamisligil
Cell Metabolism, Volume 11 Supplemental Information Defective Hepatic Autophagy in Obesity Promotes ER Stress and Causes Insulin Resistance Ling Yang, Ping Li, Suneng Fu, Ediz S. Calay, and Gökhan S. Hotamisligil
Supplementary Information
 Supplementary Information Akt regulates hepatic metabolism by suppressing a Foxo1 dependent global inhibition of adaptation to nutrient intake Mingjian Lu 1, Min Wan 1, Karla F. Leavens 1, Qingwei Chu
Supplementary Information Akt regulates hepatic metabolism by suppressing a Foxo1 dependent global inhibition of adaptation to nutrient intake Mingjian Lu 1, Min Wan 1, Karla F. Leavens 1, Qingwei Chu
The antiparasitic drug ivermectin is a novel FXR ligand that regulates metabolism
 Supplementary Information The antiparasitic drug ivermectin is a novel FXR ligand that regulates metabolism Address correspondence to Yong Li (yongli@xmu.edu.cn, Tel: 86-592-218151) GW464 CDCA Supplementary
Supplementary Information The antiparasitic drug ivermectin is a novel FXR ligand that regulates metabolism Address correspondence to Yong Li (yongli@xmu.edu.cn, Tel: 86-592-218151) GW464 CDCA Supplementary
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
Excessive intake of diets rich in fat results in the. Endoplasmic Reticulum Stress Promotes LIPIN2-Dependent Hepatic Insulin Resistance
 ORIGINAL ARTICLE Endoplasmic Reticulum Stress Promotes LIPIN2-Dependent Hepatic Insulin Resistance Dongryeol Ryu, 1 Woo-Young Seo, 1 Young-Sil Yoon, 1 Yo-Na Kim, 2 Su Sung Kim, 2 Hye-Jin Kim, 2 Tae-Sik
ORIGINAL ARTICLE Endoplasmic Reticulum Stress Promotes LIPIN2-Dependent Hepatic Insulin Resistance Dongryeol Ryu, 1 Woo-Young Seo, 1 Young-Sil Yoon, 1 Yo-Na Kim, 2 Su Sung Kim, 2 Hye-Jin Kim, 2 Tae-Sik
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state
 Related Commentary, page 2267 Research article Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state Marc Foretz, 1,2 Sophie Hébrard,
Related Commentary, page 2267 Research article Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state Marc Foretz, 1,2 Sophie Hébrard,
Supplementary Information
 Supplementary Information Notch deficiency decreases hepatic lipid accumulation by induction of fatty acid oxidation No-Joon Song,#, Ui Jeong Yun,#, Sunghee Yang, Chunyan Wu, Cho-Rong Seo, A-Ryeong Gwon,,
Supplementary Information Notch deficiency decreases hepatic lipid accumulation by induction of fatty acid oxidation No-Joon Song,#, Ui Jeong Yun,#, Sunghee Yang, Chunyan Wu, Cho-Rong Seo, A-Ryeong Gwon,,
FOR REVIEW. BMB Reports - Manuscript Submission. Manuscript Draft. Manuscript Number: BMB
 BMB Reports - Manuscript Submission Manuscript Draft Manuscript Number: BMB-18-095 Title: Insulin Receptor Substrate 2:A Bridge between Hippo and AKT Pathways Article Type: Perspective (Invited Only) Keywords:
BMB Reports - Manuscript Submission Manuscript Draft Manuscript Number: BMB-18-095 Title: Insulin Receptor Substrate 2:A Bridge between Hippo and AKT Pathways Article Type: Perspective (Invited Only) Keywords:
MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands)
 Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
TFEB-mediated increase in peripheral lysosomes regulates. Store Operated Calcium Entry
 TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
18s AAACGGCTACCACATCCAAG CCTCCAATGGATCCTCGTTA. 36b4 GTTCTTGCCCATCAGCACC AGATGCAGCAGATCCGCAT. Acc1 AGCAGATCCGCAGCTTG ACCTCTGCTCGCTGAGTGC
 Supplementary Table 1. Quantitative PCR primer sequences Gene symbol Sequences (5 to 3 ) Forward Reverse 18s AAACGGCTACCACATCCAAG CCTCCAATGGATCCTCGTTA 36b4 GTTCTTGCCCATCAGCACC AGATGCAGCAGATCCGCAT Acc1
Supplementary Table 1. Quantitative PCR primer sequences Gene symbol Sequences (5 to 3 ) Forward Reverse 18s AAACGGCTACCACATCCAAG CCTCCAATGGATCCTCGTTA 36b4 GTTCTTGCCCATCAGCACC AGATGCAGCAGATCCGCAT Acc1
AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination
 AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination Supplementary Figure 1. Generation of the adult-onset, liver-specific GH receptor knock-down (alivghrkd, Kd) mouse
AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination Supplementary Figure 1. Generation of the adult-onset, liver-specific GH receptor knock-down (alivghrkd, Kd) mouse
Supplementary Information. Cryptochrome Mediates Circadian Regulation of camp. Signalling and Hepatic Gluconeogenesis
 Supplementary Information Cryptochrome Mediates Circadian Regulation of camp Signalling and Hepatic Gluconeogenesis Eric E. Zhang 1,2*, Yi Liu 3,5*, Renaud Dentin 3, Pagkapol Y. Pongsawakul 1, Andrew C.
Supplementary Information Cryptochrome Mediates Circadian Regulation of camp Signalling and Hepatic Gluconeogenesis Eric E. Zhang 1,2*, Yi Liu 3,5*, Renaud Dentin 3, Pagkapol Y. Pongsawakul 1, Andrew C.
Online Data Supplement. Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2
 Online Data Supplement Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2 Yi Lin and Zhongjie Sun Department of physiology, college of
Online Data Supplement Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2 Yi Lin and Zhongjie Sun Department of physiology, college of
GPR120 *** * * Liver BAT iwat ewat mwat Ileum Colon. UCP1 mrna ***
 a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies
 Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Metformin and Insulin Suppress Hepatic Gluconeogenesis through Phosphorylation of CREB Binding Protein
 Metformin and Insulin Suppress Hepatic Gluconeogenesis through Phosphorylation of CREB Binding Protein Ling He, 1 Amin Sabet, 1 Stephen Djedjos, 2 Ryan Miller, 1 Xiaojian Sun, 3 Mehboob A. Hussain, 1 Sally
Metformin and Insulin Suppress Hepatic Gluconeogenesis through Phosphorylation of CREB Binding Protein Ling He, 1 Amin Sabet, 1 Stephen Djedjos, 2 Ryan Miller, 1 Xiaojian Sun, 3 Mehboob A. Hussain, 1 Sally
Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original
 Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original phenotypic screening was n=40. For specific tests, the
Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original phenotypic screening was n=40. For specific tests, the
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting. protein3) regulate autophagy and mitophagy in renal tubular cells in. acute kidney injury
 Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
Sestrin2 and BNIP3 (Bcl-2/adenovirus E1B 19kDa-interacting protein3) regulate autophagy and mitophagy in renal tubular cells in acute kidney injury by Masayuki Ishihara 1, Madoka Urushido 2, Kazu Hamada
Jang, H; Lee, GY; Selby, CP; Lee, G; Jeon, YG; Lee, JH; Cheng, KY; Titchenell, P; Birnbaum, MJ; Xu, A; Sancar, A; Kim, JB
 Title SREBPc- signalling represses hepatic glucose production by promoting FOXO degradation during refeeding Author(s) Jang, H; Lee, GY; Selby, CP; Lee, G; Jeon, YG; Lee, JH; Cheng, KY; Titchenell, P;
Title SREBPc- signalling represses hepatic glucose production by promoting FOXO degradation during refeeding Author(s) Jang, H; Lee, GY; Selby, CP; Lee, G; Jeon, YG; Lee, JH; Cheng, KY; Titchenell, P;
Supplementary Figure S1. Effect of Glucose on Energy Balance in WT and KHK A/C KO
 Supplementary Figure S1. Effect of Glucose on Energy Balance in WT and KHK A/C KO Mice. WT mice and KHK-A/C KO mice were provided drinking water containing 10% glucose or tap water with normal chow ad
Supplementary Figure S1. Effect of Glucose on Energy Balance in WT and KHK A/C KO Mice. WT mice and KHK-A/C KO mice were provided drinking water containing 10% glucose or tap water with normal chow ad
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature7221 Brown fat selective genes 12 1 Control Q-RT-PCR (% of Control) 8 6 4 2 Ntrk3 Cox7a1 Cox8b Cox5b ATPase b2 ATPase f1a1 Sirt3 ERRα Elovl3/Cig3 PPARα Zic1 Supplementary Figure S1. stimulates
doi: 1.138/nature7221 Brown fat selective genes 12 1 Control Q-RT-PCR (% of Control) 8 6 4 2 Ntrk3 Cox7a1 Cox8b Cox5b ATPase b2 ATPase f1a1 Sirt3 ERRα Elovl3/Cig3 PPARα Zic1 Supplementary Figure S1. stimulates
Supplementary Figure 1.
 Supplementary Figure 1. FGF21 does not exert direct effects on hepatic glucose production. The liver explants from C57BL/6J mice (A, B) or primary rat hepatocytes (C, D) were incubated with rmfgf21 (2
Supplementary Figure 1. FGF21 does not exert direct effects on hepatic glucose production. The liver explants from C57BL/6J mice (A, B) or primary rat hepatocytes (C, D) were incubated with rmfgf21 (2
SREBPs suppress IRS-2-mediated insulin signalling in the liver
 SREBPs suppress -mediated insulin signalling in the liver Tomohiro Ide 1,Hitoshi Shimano 2,,Naoya Yahagi 2,Takashi Matsuzaka 1,Masanori Nakakuki 1, Takashi Yamamoto 1,Yoshimi Nakagawa 2,Akimitsu Takahashi
SREBPs suppress -mediated insulin signalling in the liver Tomohiro Ide 1,Hitoshi Shimano 2,,Naoya Yahagi 2,Takashi Matsuzaka 1,Masanori Nakakuki 1, Takashi Yamamoto 1,Yoshimi Nakagawa 2,Akimitsu Takahashi
Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna
 Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna targeted for hamster PAQR3. At 24 h after the transfection,
Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna targeted for hamster PAQR3. At 24 h after the transfection,
A Tryptophan Hydroxylase Inhibitor Increases Hepatic FGF21 Production and Decreases Hepatic Gluconeogenesis Independently of Insulin in db/db Mice
 A Tryptophan Hydroxylase Inhibitor Increases Hepatic FGF21 Production and Decreases Hepatic Gluconeogenesis Independently of Insulin in db/db Mice Katsunori Nonogaki, Takao Kaji, Mari Murakami ABSTRACT
A Tryptophan Hydroxylase Inhibitor Increases Hepatic FGF21 Production and Decreases Hepatic Gluconeogenesis Independently of Insulin in db/db Mice Katsunori Nonogaki, Takao Kaji, Mari Murakami ABSTRACT
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supporting Information Table of content
 Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
control kda ATGL ATGLi HSL 82 GAPDH * ** *** WT/cTg WT/cTg ATGLi AKO/cTg AKO/cTg ATGLi WT/cTg WT/cTg ATGLi AKO/cTg AKO/cTg ATGLi iwat gwat ibat
 body weight (g) tissue weights (mg) ATGL protein expression (relative to GAPDH) HSL protein expression (relative to GAPDH) ### # # kda ATGL 55 HSL 82 GAPDH 37 2.5 2. 1.5 1..5 2. 1.5 1..5.. Supplementary
body weight (g) tissue weights (mg) ATGL protein expression (relative to GAPDH) HSL protein expression (relative to GAPDH) ### # # kda ATGL 55 HSL 82 GAPDH 37 2.5 2. 1.5 1..5 2. 1.5 1..5.. Supplementary
Hepatic gluconeogenesis is essential for maintenance
 ORIGINAL ARTICLE Yin Yang 1 Promotes Hepatic Gluconeogenesis Through Upregulation of Glucocorticoid Receptor Yan Lu, 1 Xuelian Xiong, 1 Xiaolin Wang, 1 Zhijian Zhang, 1 Jin Li, 1 Guojun Shi, 1 Jian Yang,
ORIGINAL ARTICLE Yin Yang 1 Promotes Hepatic Gluconeogenesis Through Upregulation of Glucocorticoid Receptor Yan Lu, 1 Xuelian Xiong, 1 Xiaolin Wang, 1 Zhijian Zhang, 1 Jin Li, 1 Guojun Shi, 1 Jian Yang,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Supplementary Figure 1
 VO (ml kg - min - ) VCO (ml kg - min - ) Respiratory exchange ratio Energy expenditure (cal kg - min - ) Locomotor activity (x count) Body temperature ( C) Relative mrna expression TA Sol EDL PT Heart
VO (ml kg - min - ) VCO (ml kg - min - ) Respiratory exchange ratio Energy expenditure (cal kg - min - ) Locomotor activity (x count) Body temperature ( C) Relative mrna expression TA Sol EDL PT Heart
Figure S1A. Blood glucose levels in mice after glucose injection
 ## Figure S1A. Blood glucose levels in mice after glucose injection Blood glucose (mm/l) 25 2 15 1 5 # 15 3 6 3+3 Time after glucose injection (min) # Figure S1B. α-kg levels in mouse livers after glucose
## Figure S1A. Blood glucose levels in mice after glucose injection Blood glucose (mm/l) 25 2 15 1 5 # 15 3 6 3+3 Time after glucose injection (min) # Figure S1B. α-kg levels in mouse livers after glucose
Supplementary data Supplementary Figure 1 Supplementary Figure 2
 Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
SUPPLEMENTARY DATA. Supplementary Table 1. Primers used in qpcr
 Supplementary Table 1. Primers used in qpcr Gene forward primer (5'-3') reverse primer (5'-3') β-actin AGAGGGAAATCGTGCGTGAC CAATAGTGATGACCTGGCCGT Hif-p4h-2 CTGGGCAACTACAGGATAAAC GCGTCCCAGTCTTTATTTAGATA
Supplementary Table 1. Primers used in qpcr Gene forward primer (5'-3') reverse primer (5'-3') β-actin AGAGGGAAATCGTGCGTGAC CAATAGTGATGACCTGGCCGT Hif-p4h-2 CTGGGCAACTACAGGATAAAC GCGTCCCAGTCTTTATTTAGATA
Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle.
 Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle. (a) mrna levels of Dj1 measured by quantitative RT-PCR in soleus, gastrocnemius (Gastroc.) and extensor digitorum longus
Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle. (a) mrna levels of Dj1 measured by quantitative RT-PCR in soleus, gastrocnemius (Gastroc.) and extensor digitorum longus
2.5. AMPK activity
 Supplement Fig. A 3 B phos-ampk 2.5 * Control AICAR AMPK AMPK activity (Absorbance at 45 nm) 2.5.5 Control AICAR Supplement Fig. Effects of AICAR on AMPK activation in macrophages. J774. macrophages were
Supplement Fig. A 3 B phos-ampk 2.5 * Control AICAR AMPK AMPK activity (Absorbance at 45 nm) 2.5.5 Control AICAR Supplement Fig. Effects of AICAR on AMPK activation in macrophages. J774. macrophages were
Dual role of the coactivator TORC2 in modulating hepatic glucose output and insulin signaling
 Dual role of the coactivator TORC2 in modulating hepatic glucose output and insulin signaling Gianluca Canettieri, 1,3,4 Seung-Hoi Koo, 1,2,4 Rebecca Berdeaux, 1 Jose Heredia, 1 Susan Hedrick, 1 Xinmin
Dual role of the coactivator TORC2 in modulating hepatic glucose output and insulin signaling Gianluca Canettieri, 1,3,4 Seung-Hoi Koo, 1,2,4 Rebecca Berdeaux, 1 Jose Heredia, 1 Susan Hedrick, 1 Xinmin
Curriculum Vitae. Dissertation Title M.S. Thesis: Regulation of Notch1 signaling by Runx2 during osteoblast differentiation.
 Curriculum Vitae Name: Eun-Jung Ann Education Mar.2009 : Ph.D. in Graduate School of Biological Sciences and Technology, Chonnam National University Mar.2007 Feb. 2009: M.S. in Graduate School of Biological
Curriculum Vitae Name: Eun-Jung Ann Education Mar.2009 : Ph.D. in Graduate School of Biological Sciences and Technology, Chonnam National University Mar.2007 Feb. 2009: M.S. in Graduate School of Biological
Gallic acid prevents isoproterenol-induced cardiac hypertrophy and fibrosis through regulation of JNK2 signaling and Smad3 binding activity
 Gallic acid prevents isoproterenol-induced cardiac hypertrophy and fibrosis through regulation of JNK2 signaling and Smad3 binding activity Yuhee Ryu 1,+, Li Jin 1,2+, Hae Jin Kee 1,, Zhe Hao Piao 3, Jae
Gallic acid prevents isoproterenol-induced cardiac hypertrophy and fibrosis through regulation of JNK2 signaling and Smad3 binding activity Yuhee Ryu 1,+, Li Jin 1,2+, Hae Jin Kee 1,, Zhe Hao Piao 3, Jae
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplementary Figure 1. Blood glucose and insulin levels in mice during 4-day infusion.
 Supplementary Figure 1. Blood glucose and insulin levels in mice during 4-day infusion. (A-B) WT and HT mice infused with saline or glucose had overlapping achieved blood glucose and insulin levels, necessitating
Supplementary Figure 1. Blood glucose and insulin levels in mice during 4-day infusion. (A-B) WT and HT mice infused with saline or glucose had overlapping achieved blood glucose and insulin levels, necessitating
p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO
 Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression
 Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Obesity frequently leads to insulin-resistance that, in turn, produces
 Bifurcation of insulin signaling pathway in rat liver: mtorc1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis Shijie Li, Michael S. Brown 1, and Joseph L. Goldstein 1 Department
Bifurcation of insulin signaling pathway in rat liver: mtorc1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis Shijie Li, Michael S. Brown 1, and Joseph L. Goldstein 1 Department
SUPPLEMENTARY INFORMATION
 Glucose is a ligand for the oxysterol receptor LXR Supplementary Figure 1 Schematic representation of pathways influencing glucose fate in the liver. Glucose induces insulin secretion, suppressing hepatic
Glucose is a ligand for the oxysterol receptor LXR Supplementary Figure 1 Schematic representation of pathways influencing glucose fate in the liver. Glucose induces insulin secretion, suppressing hepatic
Islet viability assay and Glucose Stimulated Insulin Secretion assay RT-PCR and Western Blot
 Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
Pancreatic insulin is a major anabolic hormone to
 Protein Arginine Methyltransferase 1 Regulates Hepatic Glucose Production in a FoxO1-Dependent Manner Dahee Choi,* Kyoung-Jin Oh,* Hye-Sook Han, Young-Sil Yoon, Chang-Yun Jung, Seong-Tae Kim, Seung-Hoi
Protein Arginine Methyltransferase 1 Regulates Hepatic Glucose Production in a FoxO1-Dependent Manner Dahee Choi,* Kyoung-Jin Oh,* Hye-Sook Han, Young-Sil Yoon, Chang-Yun Jung, Seong-Tae Kim, Seung-Hoi
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11464 Supplemental Figure S1. The expression of Vegfb is increased in obese and diabetic mice as compared to lean mice. a-b, Body weight and postprandial blood
SUPPLEMENTARY INFORMATION doi:10.1038/nature11464 Supplemental Figure S1. The expression of Vegfb is increased in obese and diabetic mice as compared to lean mice. a-b, Body weight and postprandial blood
Figure S1. Body composition, energy homeostasis and substrate utilization in LRH-1 hep+/+ (white bars) and LRH-1 hep-/- (black bars) mice.
 Figure S1. Body composition, energy homeostasis and substrate utilization in LRH-1 hep+/+ (white bars) and LRH-1 hep-/- (black bars) mice. (A) Lean and fat masses, determined by EchoMRI. (B) Food and water
Figure S1. Body composition, energy homeostasis and substrate utilization in LRH-1 hep+/+ (white bars) and LRH-1 hep-/- (black bars) mice. (A) Lean and fat masses, determined by EchoMRI. (B) Food and water
Table S1. Primer sequences used for qrt-pcr. CACCATTGGCAATGAGCGGTTC AGGTCTTTGCGGATGTCCACGT ACTB AAGTCCATGTGCTGGCAGCACT ATCACCACTCCGAAGTCCGTCT LCOR
 Table S1. Primer sequences used for qrt-pcr. ACTB LCOR KLF6 CTBP1 CDKN1A CDH1 ATF3 PLAU MMP9 TFPI2 CACCATTGGCAATGAGCGGTTC AGGTCTTTGCGGATGTCCACGT AAGTCCATGTGCTGGCAGCACT ATCACCACTCCGAAGTCCGTCT CGGCTGCAGGAAAGTTTACA
Table S1. Primer sequences used for qrt-pcr. ACTB LCOR KLF6 CTBP1 CDKN1A CDH1 ATF3 PLAU MMP9 TFPI2 CACCATTGGCAATGAGCGGTTC AGGTCTTTGCGGATGTCCACGT AAGTCCATGTGCTGGCAGCACT ATCACCACTCCGAAGTCCGTCT CGGCTGCAGGAAAGTTTACA
FOXO Reporter Kit PI3K/AKT Pathway Cat. #60643
 Data Sheet FOXO Reporter Kit PI3K/AKT Pathway Cat. #60643 Background The PI3K/AKT signaling pathway is essential for cell growth and survival. Disruption of this pathway or its regulation has been linked
Data Sheet FOXO Reporter Kit PI3K/AKT Pathway Cat. #60643 Background The PI3K/AKT signaling pathway is essential for cell growth and survival. Disruption of this pathway or its regulation has been linked
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC
 Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplemental Information
 Supplemental Information Tobacco-specific Carcinogen Induces DNA Methyltransferases 1 Accumulation through AKT/GSK3β/βTrCP/hnRNP-U in Mice and Lung Cancer patients Ruo-Kai Lin, 1 Yi-Shuan Hsieh, 2 Pinpin
Supplemental Information Tobacco-specific Carcinogen Induces DNA Methyltransferases 1 Accumulation through AKT/GSK3β/βTrCP/hnRNP-U in Mice and Lung Cancer patients Ruo-Kai Lin, 1 Yi-Shuan Hsieh, 2 Pinpin
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Chairoungdua et al., http://www.jcb.org/cgi/content/full/jcb.201002049/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Expression of CD9 and CD82 inhibits Wnt/ -catenin
Supplemental material Chairoungdua et al., http://www.jcb.org/cgi/content/full/jcb.201002049/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Expression of CD9 and CD82 inhibits Wnt/ -catenin
UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DIVISION OF BASIC MEDICAL SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY
 1 UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DIVISION OF BASIC MEDICAL SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY GLUCOSE HOMEOSTASIS An Overview WHAT IS HOMEOSTASIS? Homeostasis
1 UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DIVISION OF BASIC MEDICAL SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY GLUCOSE HOMEOSTASIS An Overview WHAT IS HOMEOSTASIS? Homeostasis
Supplementary information. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins
 Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplemental Data. Short Article. ATF4-Mediated Induction of 4E-BP1. Contributes to Pancreatic β Cell Survival. under Endoplasmic Reticulum Stress
 Cell Metabolism, Volume 7 Supplemental Data Short Article ATF4-Mediated Induction of 4E-BP1 Contributes to Pancreatic β Cell Survival under Endoplasmic Reticulum Stress Suguru Yamaguchi, Hisamitsu Ishihara,
Cell Metabolism, Volume 7 Supplemental Data Short Article ATF4-Mediated Induction of 4E-BP1 Contributes to Pancreatic β Cell Survival under Endoplasmic Reticulum Stress Suguru Yamaguchi, Hisamitsu Ishihara,
Supplementary Materials and Methods
 Supplementary Materials and Methods Hepatocyte toxicity assay. Freshly isolated hepatocytes were incubated for overnight with varying concentrations (-25 µm) of sodium glycochenodeoxycholate (GCDC) or
Supplementary Materials and Methods Hepatocyte toxicity assay. Freshly isolated hepatocytes were incubated for overnight with varying concentrations (-25 µm) of sodium glycochenodeoxycholate (GCDC) or
Supplementary Information
 Supplementary Information GADD34-deficient mice develop obesity, nonalcoholic fatty liver disease, hepatic carcinoma and insulin resistance Naomi Nishio and Ken-ichi Isobe Department of Immunology, Nagoya
Supplementary Information GADD34-deficient mice develop obesity, nonalcoholic fatty liver disease, hepatic carcinoma and insulin resistance Naomi Nishio and Ken-ichi Isobe Department of Immunology, Nagoya
Supplementary Information. Glycogen shortage during fasting triggers liver-brain-adipose. neurocircuitry to facilitate fat utilization
 Supplementary Information Glycogen shortage during fasting triggers liver-brain-adipose neurocircuitry to facilitate fat utilization Supplementary Figure S1. Liver-Brain-Adipose neurocircuitry Starvation
Supplementary Information Glycogen shortage during fasting triggers liver-brain-adipose neurocircuitry to facilitate fat utilization Supplementary Figure S1. Liver-Brain-Adipose neurocircuitry Starvation
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
Supplementary information
 Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
Supplementary information Human Cytomegalovirus MicroRNA mir-us4-1 Inhibits CD8 + T Cell Response by Targeting ERAP1 Sungchul Kim, Sanghyun Lee, Jinwook Shin, Youngkyun Kim, Irini Evnouchidou, Donghyun
Supplementary Figure 1
 Supplementary Figure 1 Constitutive EGFR signaling does not activate canonical EGFR signals (a) U251EGFRInd cells with or without tetracycline exposure (24h, 1µg/ml) were treated with EGF for 15 minutes
Supplementary Figure 1 Constitutive EGFR signaling does not activate canonical EGFR signals (a) U251EGFRInd cells with or without tetracycline exposure (24h, 1µg/ml) were treated with EGF for 15 minutes
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1 Schematic depiction of the tandem Fc GDF15. Supplementary Figure 2 Supplementary Figure 2 Gfral mrna levels in the brains of both wild-type and knockout Gfral
Supplementary Figure 1 Supplementary Figure 1 Schematic depiction of the tandem Fc GDF15. Supplementary Figure 2 Supplementary Figure 2 Gfral mrna levels in the brains of both wild-type and knockout Gfral
Hepatic insulin signaling regulates VLDL secretion and atherogenesis in mice
 Hepatic insulin signaling regulates VLDL secretion and atherogenesis in mice Seongah Han,, Domenico Accili, Alan R. Tall J Clin Invest. 2009;119(4):1029-1041. https://doi.org/10.1172/jci36523. Research
Hepatic insulin signaling regulates VLDL secretion and atherogenesis in mice Seongah Han,, Domenico Accili, Alan R. Tall J Clin Invest. 2009;119(4):1029-1041. https://doi.org/10.1172/jci36523. Research
Oncolytic Adenovirus Complexes Coated with Lipids and Calcium Phosphate for Cancer Gene Therapy
 Oncolytic Adenovirus Complexes Coated with Lipids and Calcium Phosphate for Cancer Gene Therapy Jianhua Chen, Pei Gao, Sujing Yuan, Rongxin Li, Aimin Ni, Liang Chu, Li Ding, Ying Sun, Xin-Yuan Liu, Yourong
Oncolytic Adenovirus Complexes Coated with Lipids and Calcium Phosphate for Cancer Gene Therapy Jianhua Chen, Pei Gao, Sujing Yuan, Rongxin Li, Aimin Ni, Liang Chu, Li Ding, Ying Sun, Xin-Yuan Liu, Yourong
Supplemental material for Hernandez et al. Dicoumarol downregulates human PTTG1/Securin mrna expression. through inhibition of Hsp90
 Supplemental material for Hernandez et al. Dicoumarol downregulates human PTTG1/Securin mrna expression through inhibition of Hsp90 Dicoumarol-Sepharose co-precipitation. Hsp90 inhibitors can co-precipitate
Supplemental material for Hernandez et al. Dicoumarol downregulates human PTTG1/Securin mrna expression through inhibition of Hsp90 Dicoumarol-Sepharose co-precipitation. Hsp90 inhibitors can co-precipitate
What systems are involved in homeostatic regulation (give an example)?
 1 UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DIVISION OF BASIC MEDICAL SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY GLUCOSE HOMEOSTASIS (Diabetes Mellitus Part 1): An Overview
1 UNIVERSITY OF PNG SCHOOL OF MEDICINE AND HEALTH SCIENCES DIVISION OF BASIC MEDICAL SCIENCES DISCIPLINE OF BIOCHEMISTRY AND MOLECULAR BIOLOGY GLUCOSE HOMEOSTASIS (Diabetes Mellitus Part 1): An Overview
Synthesis of Substituted 2H-Benzo[e]indazole-9-carboxylate as Potent Antihyperglycemic Agent that May Act through IRS-1, Akt and GSK-3β Pathways
![Synthesis of Substituted 2H-Benzo[e]indazole-9-carboxylate as Potent Antihyperglycemic Agent that May Act through IRS-1, Akt and GSK-3β Pathways Synthesis of Substituted 2H-Benzo[e]indazole-9-carboxylate as Potent Antihyperglycemic Agent that May Act through IRS-1, Akt and GSK-3β Pathways](/thumbs/96/128474991.jpg) Electronic Supplementary Material (ESI) for MedChemComm. This journal is The Royal Society of Chemistry 2016 Supplementary Data Synthesis of Substituted 2H-Benzo[e]indazole-9-carboxylate as Potent Antihyperglycemic
Electronic Supplementary Material (ESI) for MedChemComm. This journal is The Royal Society of Chemistry 2016 Supplementary Data Synthesis of Substituted 2H-Benzo[e]indazole-9-carboxylate as Potent Antihyperglycemic
Data Sheet TIGIT / NFAT Reporter - Jurkat Cell Line Catalog #60538
 Data Sheet TIGIT / NFAT Reporter - Jurkat Cell Line Catalog #60538 Background: TIGIT is a co-inhibitory receptor that is highly expressed in Natural Killer (NK) cells, activated CD4+, CD8+ and regulatory
Data Sheet TIGIT / NFAT Reporter - Jurkat Cell Line Catalog #60538 Background: TIGIT is a co-inhibitory receptor that is highly expressed in Natural Killer (NK) cells, activated CD4+, CD8+ and regulatory
Exendin-4-mediated cell proliferation in beta-cells: focusing on the transcription factors
 Exendin-4-mediated cell proliferation in beta-cells: focusing on the transcription factors Seo-Yoon Chang, Jae Min Cho, Yang-Hyeok Jo, Myung-Jun Kim Departments of Physiology, College of Medicine, The
Exendin-4-mediated cell proliferation in beta-cells: focusing on the transcription factors Seo-Yoon Chang, Jae Min Cho, Yang-Hyeok Jo, Myung-Jun Kim Departments of Physiology, College of Medicine, The
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
doi:10.1038/nature12215 Supplementary Figure 1. The effects of full and dissociated GR agonists in supporting BFU-E self-renewal divisions. BFU-Es were cultured in self-renewal medium with indicated GR
Regulation of glucose metabolism from a liver-centric perspective
 OPEN (2016) 48, e218; doi:10.1038/emm.2015.122 & 2016 KSBMB. All rights reserved 2092-6413/16 www.nature.com/emm REVIEW Regulation of glucose metabolism from a liver-centric perspective Hye-Sook Han, Geon
OPEN (2016) 48, e218; doi:10.1038/emm.2015.122 & 2016 KSBMB. All rights reserved 2092-6413/16 www.nature.com/emm REVIEW Regulation of glucose metabolism from a liver-centric perspective Hye-Sook Han, Geon
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11095 Supplementary Table 1. Summary of the binding between Angptls and various Igdomain containing receptors as determined by flow cytometry analysis. The results were summarized from
doi:10.1038/nature11095 Supplementary Table 1. Summary of the binding between Angptls and various Igdomain containing receptors as determined by flow cytometry analysis. The results were summarized from
Supplementary Materials and Methods
 Supplementary Materials and Methods Immunoblotting Immunoblot analysis was performed as described previously (1). Due to high-molecular weight of MUC4 (~ 950 kda) and MUC1 (~ 250 kda) proteins, electrophoresis
Supplementary Materials and Methods Immunoblotting Immunoblot analysis was performed as described previously (1). Due to high-molecular weight of MUC4 (~ 950 kda) and MUC1 (~ 250 kda) proteins, electrophoresis
m 6 A mrna methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer
 SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
NOVEMBER 2, 2012 VOLUME 287 NUMBER 45 JOURNAL OF BIOLOGICAL CHEMISTRY 38041
 THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 287, NO. 45, pp. 38041 38049, November 2, 2012 2012 by The American Society for Biochemistry and Molecular Biology, Inc. Published in the U.S.A. Activation of Cannabinoid
THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 287, NO. 45, pp. 38041 38049, November 2, 2012 2012 by The American Society for Biochemistry and Molecular Biology, Inc. Published in the U.S.A. Activation of Cannabinoid
supplementary information
 DOI: 1.138/ncb1 Control Atg7 / NAC 1 1 1 1 (mm) Control Atg7 / NAC 1 1 1 1 (mm) Lamin B Gstm1 Figure S1 Neither the translocation of into the nucleus nor the induction of antioxidant proteins in autophagydeficient
DOI: 1.138/ncb1 Control Atg7 / NAC 1 1 1 1 (mm) Control Atg7 / NAC 1 1 1 1 (mm) Lamin B Gstm1 Figure S1 Neither the translocation of into the nucleus nor the induction of antioxidant proteins in autophagydeficient
SUPPLEMENTARY INFORMATION. Supplementary Figures S1-S9. Supplementary Methods
 SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
ZL ZDF ZDF + E2 *** Visceral (g) ZDF
 Body Weight (g) 4 3 2 1 ** * ZL ZDF 6 8 1 12 14 16 Age (weeks) B * Sub-cutaneous (g) 16 12 8 4 ZL ZDF Visceral (g) 25 2 15 1 5 ZL ZDF Total fat pad weight (g) 4 3 2 1 ZDF ZL Supplemental Figure 1: Effect
Body Weight (g) 4 3 2 1 ** * ZL ZDF 6 8 1 12 14 16 Age (weeks) B * Sub-cutaneous (g) 16 12 8 4 ZL ZDF Visceral (g) 25 2 15 1 5 ZL ZDF Total fat pad weight (g) 4 3 2 1 ZDF ZL Supplemental Figure 1: Effect
Supplemental information
 Carcinoemryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation y Chiang et al. Supplemental
Carcinoemryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation y Chiang et al. Supplemental
Role of KLF15 in regulation of hepatic gluconeogenesis and metformin action
 Diabetes Publish Ahead of Print, published online April 14, 2010 KLF15 and metformin action Role of KLF15 in regulation of hepatic gluconeogenesis and metformin action Running title: KLF15 and metformin
Diabetes Publish Ahead of Print, published online April 14, 2010 KLF15 and metformin action Role of KLF15 in regulation of hepatic gluconeogenesis and metformin action Running title: KLF15 and metformin
