Supplementary Figure 1 The plasmids used for the GPCR-CRISPR ChaCha and the GPCR-CRISPR Tango systems. (a) The plasmids for the GPCR-CRISPR ChaCha
|
|
|
- Garry Whitehead
- 5 years ago
- Views:
Transcription
1 Supplementary Figure 1 The plasmids used for the GPCR-CRISPR ChaCha and the GPCR-CRISPR Tango systems. (a) The plasmids for the GPCR-CRISPR ChaCha system. The dcas9-vpr effector is fused to the C-terminus of a nuclear export signal (NES)-flanked Beta-Arrestin 2 (ARRB2) with a TEV protease cleavage site (TCS), and a small linker. Embedded in the VPR activator is a nuclear localization signal (NLS), and at the C-terminus mcherry is fused for visualization. A single guide RNA (sgrna) is expressed by the murine U6 (mu6) promoter, and a shown stream cassette contains EF1ɑ-driven GPCR with the AVPR V2 tail fused to the TEV protease (TEVp). A BFP is fused to the C-terminus of GPCR-V2-TEVp via a p2a sequence for visualization. These plasmids are co-transfected into a stable reporter cell line containing GFP under control of the Doxycycline (Dox)-inducible TRE3G promoter. The size of the GPCR fusion protein is 2.7kb. (b) The plasmids for the GPCR-CRISPR ChaCha system. The size of the GPCR fusion protein is 8.4kb. 1
2 Supplementary Figure 2 Modeling to estimate the number of dcas9 molecules released per receptor. (a) The plasmid system for gene activation using free dcas9-vpr. (b) The plasmid system for gene activation using the ChaCha system. (c) Measurement and modeling EGFP reporter activation via Dox-inducible dcas9- VPR-mCherry. Left, schematic of the HEK293T cell line containing genomically integrated lentiviral ptre3gdcas9-vpr-mcherry, puas-gfp-pef1α-rtta-p2a-puror, and pu6-sguas8, see Supplementary Table 1. Dox induces expression of dcas9-vpr-mcherry, which with sguas8 binds and activates the GFP reporter. Plots demonstrate GFP fluorescence (a.u.) as a function of mcherry fluorescence (a.u., y-axis; a surrogate for Dox), and CNO concentration (M, x-axis). Both the experimental (middle) and fitted-model plots (right) demonstrate a Dox-dependent, CNO-independent graded GFP response. Model fitting using nonlinear regression indicates n < 1 with correlation coefficient r = 0.97 (right). See Methods for the detailed model. 2
3 Supplementary Figure 3 Characterizing design parameters of hm3d-crispr ChaCha. (a) Left, flexible glycine-serine (GS) linkers (variants A E) before or after TEV cleavage site (TCS) were varied as indicated. Right, comparison of GFP activation for variants A E, after 1 day of +/- CNO treatment. (b) Left, TCS variants F I with increasing proteolytic strengths. Red wedge indicates increasing proteolytic cleavage efficiency. Right, comparison of GFP activation for these variants after 1 day of +/- CNO treatment. (c) Comparison of 3
4 GFP activation by receptor promoter variants (A, J, K) as indicated, after 1 day of +/- CNO treatment. The grey bars represent GFP activation using free dcas9-vpr with or without a targeting sgrna (sgtet) for 24h or 48h. +/- indicates with or without CNO. The fold of activation displayed on top compares +/-CNO conditions. Data was normalized to the free dcas9-vpr without a targeting sgrna. (d) Quantification of receptor (GPCR-V2- TEVp) and adaptor (ARRB2-TCS-dCas9-VPR) expression levels for BFP (receptor), mcherry (adaptor), and GFP (reporter) triple positive cells from experiments in (c) for with and without CNO. The data represent three independent experiments with technical replicates. The grey shaded region shows samples with a higher ratio of adaptor to receptor expression, which also show a higher dynamic range for gene activation in (c). 4
5 Supplementary Figure 4 Additional fields of view from time-lapse microscopy of hm3d-crispr ChaCha activation in live HEK293T cells. Related to Fig. 2a and Supplementary Movies
6 Supplementary Figure 5 Independent experimental replicate of the measured CNO dose response curve for the hm3d-crispr ChaCha. Related to Fig. 2b. 6
7 Supplementary Figure 6 Characterization of sgrna efficiency for target gene activation. (a-b) Screen of sgrnas for IL2 and IFN-γ cytokine activation and secretion using ELISA. sguas8 is used as a non-targeting control. The best performing sgrnas (red) are used for experiments in the main text. The dotted line represents the detection limit of ELISA (4 pg/ml). (c-d) Screen of sgrnas for HBB and HBG activation using qpcr. sggal4 was used as a non-targeting control. The best performing sgrnas (red) were used in the main text. 7
8 Supplementary Figure 7 (a-b) Activation of HBB and HBG using the hm3d-crispr ChaCha system measured by quantitative PCR (qpcr) with or without 10 μm CNO compared to positive and negative controls using free dcas9-vpr. Related to Fig. 3b. 8
9 Supplementary Figure 8 Control experiments for the multiplexed activation of two genes using CRISPR ChaCha. (a) The control experiment for the multiplexed activation of two endogenous genes using free dcas9-vpr. The plasmids used for the experiments are shown on the top. Measurement of IL2 (orange) and IFN-! (blue) by ELISA with different combinations of targeting sgrnas is shown on the bottom. (b) The control experiment for the multiplexed activation of two endogenous genes using a non-targeting sgrna (sggal4) or a single sgrna for IL2 (sgil2) or IFN-! (sginfg). The plasmids used for the experiments are shown on the top. Measurement of IL2 and IFN-! expression and secretion by ELISA with and without CNO is shown in the bottom diagrams. Related to Fig. 3c. 9
10 Supplementary Figure 9 The intracellular linker affects natural GPCR ChaCha performance. Inset: variant A is the original ChaCha architecture; variant L is a C-terminal tail replacement with that of V2, and variant M is a C-terminal tail and H8 domain replacement to V2. The data shows GFP reporter activation measured by flow cytometry for each variant with (+) or without (-) of cognate ligands for 1 day. The free dcas9-vpr with or without a targeting sgrna (sgtet) was used as controls. Dots represent biological measurements, and bars represent means from 2 independent experiments. The fold of activation displayed on top of bars compares +/- treatment for GPCR variants, or with or without a targeting sgrna for the free dcas9-vpr. 10
11 Supplementary Figure 10 Independent experimental replicate of the measured neuromedin B dose response curve for NMBR-CRISPR ChaCha. Related to Fig. 4c. 11
12 Supplementary Figure 11 Experimental data on characterizing the leaky activation of the ChaCha System. (a) Expression of GFP for free dcas9-vpr with a non-targeting sgrna (sggal4) or a targeting sgrna (sgtet) 24 h after transfection (grey, left), the full ChaCha system with or without CNO (blue, middle), and the full ChaCha system with the TEVp removed from the C-terminus of the receptor (orange, right). The system used in each group is shown on top of the bars. The GFP fluorescence is normalized to the free dcas9-vpr with a non-targeting sggal4. Bars represent the mean, and data shown is from two independent experiments with three technical replicates. 12
13 Supplementary Table 1. Plasmid constructs used in this study listed by figure. ChaCha variants are indicated in parentheses. Constructs used to generate the two reporter cell lines are described in the Methods. Figure Constructs Plasmid ID 1b pcdna3: dcas9-vpr-mcherry pslq3958 phr: U6-sgTET EF1a-hM3D-V2-TEV-p2a-BFP (A) pslq3956 phr: EF1a-hM3D-V2-TEV-p2a-BFP (A) pslq5768 pcdna3: hm3d-v2-tcs-dcas9-nls-vpr-mcherry pslq3875 phr: U6-sgTET EF1a-NES-ARRB2-NES-TEV-p2a-BFP pslq5781 1c phr: U6-sgUAS8 EF1a-hM3D-V2-TEV-p2a-BFP (A) pslq5779 phr: TRE3G-NES-ARRB2-NES-TCS-dCas9-VPR-mCherry (A) pslq5751 phr: EF1a -rtta-p2a-puro WPRE UAS -GFP bgh-pa. pslq5738 2a phr: U6-sgTET SFFV-hM3D-V2-TEV-p2a-BFP (K) pslq5777 2b phr: U6-sgTET SFFV-hM3D-V2-TEV-p2a-BFP (K) pslq5777 2c phr: U6-sgUAS8 EF1a-hM3D-V2-TEV-p2a-BFP (A) pslq5779 phr: TRE3G-NES-ARRB2-NES-TCS-dCas9-VPR-mCherry (A) pslq5751 phr: EF1a -rtta-p2a-puro WPRE UAS -GFP bgh-pa. pslq5738 3a pcdna3: dcas9-vpr-mcherry pslq3958 phr: EF1a-hM3D-V2-linker-TEV-p2a-EGFP (A) pslq5767 phr: U6-sgGal4 EF1a-puro-t2a-BFP - phr: U6-sgIL2 EF1a-puro-t2a-BFP - phr: U6-sgIFNG EF1a-puro-t2a-BFP - 3b pcdna3: dcas9-vpr-mcherry pslq3958 phr: EF1a-hM3D-V2-linker-TEV-p2a-EGFP (A) pslq5767 phr: U6-sgGal4 EF1a-puro-t2a-BFP - phr: U6-sgHBB EF1a-puro-t2a-BFP - phr: U6-sgHBG EF1a-puro-t2a-BFP - 3c pcdna3: dcas9-vpr-mcherry pslq3958 phr: EF1a-hM3D-V2-linker-TEV-p2a-EGFP (A) pslq5767 phr: hu6-sggal4 mu6-sggal4 EF1a-puro-t2a-BFP - phr: hu6-sgil2 mu6-sgifng EF1a-puro-t2a-BFP - 4b. phr: U6-sgTET EF1a-KORD-V2-TEV-p2a-BFP (A) pslq5736 phr: U6-sgTET EF1a-LPAR1-V2-TEV-p2a-BFP (L) pslq5774 phr: U6-sgTET EF1a-CXCR4-V2-TEV-p2a-BFP (M) pslq5771
14 phr: U6-sgTET EF1a-NMBR-V2-TEV-p2a-BFP (A) pslq3780 phr: U6-sgTET EF1a-ADRB2-V2-TEV-p2a-BFP (A) pslq5901 phr: U6-sgTET EF1a-AVPR2-V2-TEV-p2a-BFP (A) pslq5908 phr: U6-sgTET EF1a-TRHR-V2-TEV-p2a-BFP (A) pslq5909 4c phr: U6-sgTET EF1a-NMBR-V2-TEV-p2a-BFP (A) pslq3780 4d pcdna3: dcas9-vpr-mcherry pslq3958 phr: EF1a-NMBR-V2-linker-TEV-p2a-EGFP (A) pslq5815 phr: U6-sgGal4 EF1a-puro-t2a-BFP - phr: U6-sgIFNG EF1a-puro-t2a-BFP - Supplementary Figures S2c phr: U6-sgUAS8 EF1a-hM3D-V2-TEV-p2a-BFP (A) pslq5779 phr: TRE3G-dCas9-VPR-mCherry pslq5750 phr: EF1a -rtta-p2a-puro WPRE UAS -GFP bgh-pa. pslq5738 S3a pcdna3: dcas9-vpr-mcherry pslq3958 phr: U6-sgTET EF1a-hM3D-V2-TEV-p2a-BFP (A) pslq3956 phr: EF1a-hM3D-V2-TEV-p2a-BFP (A) pslq5768 pcdna3: NES-ARRB2-NES-TCS-dCas9-VPR-mCherry (B) pslq5740 pcdna3: NES-ARRB2-NES-TCS-dCas9-VPR-mCherry (C) pslq5741 pcdna3: NES-ARRB2-NES-TCS-dCas9-VPR-mCherry (D) pslq5744 pcdna3: NES-ARRB2-NES-TCS-dCas9-VPR-mCherry (E) pslq5745 S3b pcdna3: dcas9-vpr-mcherry pslq3958 phr: U6-sgTET EF1a-hM3D-V2-TEV-p2a-BFP (A) pslq3956 phr: EF1a-hM3D-V2-TEV-p2a-BFP (A) pslq5768 pcdna3: NES-ARRB2-NES-TCS-dCas9-VPR-mCherry (F) pslq5727 pcdna3: NES-ARRB2-NES-TCS-dCas9-VPR-mCherry (G) pslq5728 pcdna3: NES-ARRB2-NES-TCS-dCas9-VPR-mCherry (H) pslq5729 pcdna3: NES-ARRB2-NES-TCS-dCas9-VPR-mCherry (I) pslq5730 S3c pcdna3: dcas9-vpr-mcherry pslq3958 phr: U6-sgTET EF1a-hM3D-V2-TEV-p2a-BFP (A) pslq3956 phr: EF1a-hM3D-V2-TEV-p2a-BFP (A) pslq5768 phr: U6-sgTET PGK-hM3D-V2-TEV-p2a-BFP (J) pslq5776 phr: U6-sgTET SFFV-hM3D-V2-TEV-p2a-BFP (K) pslq5777 S4 phr: U6-sgTET SFFV-hM3D-V2-TEV-p2a-BFP (K) pslq5777 S5 phr: U6-sgTET SFFV-hM3D-V2-TEV-p2a-BFP (K) pslq5777
15 S6a pcdna3: dcas9-vpr-mcherry pslq3958 phr: U6-sgUAS8 EF1a-puro-t2a-BFP - phr: U6-sgIL2-1 EF1a-puro-t2a-BFP - phr: U6-sgIL2-2 EF1a-puro-t2a-BFP - phr: U6-sgIL2-3 EF1a-puro-t2a-BFP - phr: U6-sgIL2-4 EF1a-puro-t2a-BFP - S6b pcdna3: dcas9-vpr-mcherry pslq3958 phr: U6-sgIFNG-1 EF1a-puro-t2a-BFP - phr: U6-sgIFNG-2 EF1a-puro-t2a-BFP - phr: U6-sgIFNG-3 EF1a-puro-t2a-BFP - S7 pcdna3: dcas9-vpr-mcherry pslq3958 phr: U6-sgGAL4 EF1a-puro-t2a-BFP - phr: U6-sgHBB2 EF1a-puro-t2a-BFP phr: U6-sgHBG3 EF1a-puro-t2a-BFP phr: EF1a-hM3D-V2-linker-TEV-p2a-EGFP (A) pslq5767 S8a pcdna3: dcas9-vpr-mcherry pslq3958 phr: hu6-sggal4 mu6-sggal4 EF1a-puro-t2a-BFP phr: hu6-sgil2 mu6-sggal4 EF1a-puro-t2a-BFP phr: hu6-sggal4 mu6-sgifng EF1a-puro-t2a-BFP phr: hu6-sgil2 mu6-sgifng EF1a-puro-t2a-BFP phr: EF1a-hM3D-V2-linker-TEV-p2a-EGFP (A) pslq5767 S8b phr: hu6-sggal4 mu6-sggal4 EF1a-puro-t2a-BFP phr: hu6-sgil2 mu6-sggal4 EF1a-puro-t2a-BFP phr: hu6-sggal4 mu6-sgifng EF1a-puro-t2a-BFP phr: hu6-sgil2 mu6-sgifng EF1a-puro-t2a-BFP S9 pcdna3: dcas9-vpr-mcherry pslq3958 phr: EF1a-hM3D-V2-TEV-p2a-BFP (A) pslq5768 phr: U6-sgTET EF1a-CXCR4-V2-TEV-p2a-BFP (A) pslq3779 phr: U6-sgTET EF1a-CXCR4-V2-TEV-p2a-BFP (L) pslq5770 phr: U6-sgTET EF1a-CXCR4-V2-TEV-p2a-BFP (M) pslq5771 phr: U6-sgTET EF1a-NMBR-V2-TEV-p2a-BFP (A) pslq3780 phr: U6-sgTET EF1a-NMBR-V2-TEV-p2a-BFP (L) pslq5772 phr: U6-sgTET EF1a-NMBR-V2-TEV-p2a-BFP (M) pslq5773 phr: U6-sgTET EF1a-LPAR1-V2-TEV-p2a-BFP (A) pslq3781 phr: U6-sgTET EF1a-LPAR1-V2-TEV-p2a-BFP (L) pslq5774 phr: U6-sgTET EF1a-LPAR1-V2-TEV-p2a-BFP (M) pslq5775 S10 phr: U6-sgTET EF1a-NMBR-V2-TEV-p2a-BFP (A) pslq3780 S11 pcdna3: dcas9-vpr-mcherry pslq3958 phr: U6-sgTET EF1a-puro-t2a-BFP
16 phr: U6-sgGal4 EF1a-puro-t2a-BFP phr: U6-sgTET EF1a-hM3D-V2-TEV-p2a-BFP (A) phr: U6-sgTET EF1a-hM3D-V2-p2a-BFP (A) pslq3956 pslq5725
17 Supplementary Table 2: Sequences for S. pyogenes sgrnas used in the study. Name sgtet sggal4 sguas8 Spacer Sequence GTACGTTCTCTATCACTGATA GAGCACTGTCCTCCGAACGT GTACTCCGACCTCTAGTGT sgil2-1 sgil2-2 sgil2-3 sgil2-4 GTGATTAAAGAGAGTGATA GCAATTTATACTGTTAATTC GTTACATTAGCCCACACTT GAAAAACTGTTTCATACAGA sgifng-1 sgifng-2 sgifng-3 GAGATGGTGACAGATAGGCA GAATGGCACAGGTGGGCATAA GTACCTCCCCACTTCGCCC sghbb1 sghbb2 sghbb3 GTGGAGCCACACCCTAGGGT GAAGAGCCAAGGACAGGTA GTACCTGTCCTTGGCTCTTC HBG1 HBG2 HBG3 HBG4 GTCAAGGCAAGGCTGGCCAACCCA GCTATTGGTCAAGGCAAGGC GCTAAACTCCACCCATGGGT GATATTTGCATTGAGATAGTG
18 Supplementary Table 3. cdna resources used for receptor cloning. Receptor cdna Repository Catalogue No. hm3d Addgene KORD Addgene CXCR4 Addgene LPAR1 Addgene NMBR Addgene ADRB2 Addgene TRHR cdna Resource Center ( TRHR AVPR2 Addgene 66227
19 Supplementary Table 4. GPCR ligands used. Receptor Ligand Concentration Manufacturer Catalogue No. Media Conditions hm3d Clozapine N-Oxide (CNO) 20 μm Sigma C0832 Full Serum KORD Salvinorin B (SALB) 2 μm Tocris 5611 Full Serum CXCR4 Stromal-cell Derived Factor 1 (SDF1) 0.05 μm BioLegend Serum Free LPAR1 Lysophosphatidic acid (LPA) 10 μm Santa Cruz Biotech sc Serum Free NMBR Neuromedin B (NMB) 1 μm Tocris 1908 Serum Free ADRB2 Isoproterenol HCL (ISO) 10 μm Tocris 1747 Full Serum TRHR Thyrotropin Releasing Horomone (TRH) 10 μm Sigma P MG Full Serum AVPR2 Arg8- Vassopressin (AVP) 10 μm Tocris 2935 Full Serum
20 Supplementary Table 5. Primers used for qpcr qpcr Primer GAPDH.F GAPDH.R HBG.F HBG.R HBB.F HBB.R Sequence TGCCAAATATGATGACATCAAGAA GGAGTGGGTGTCGCTGTTGC GCTGAGTGAACTGCACTGTGA GAATTCTTTGCCGAAATGGA GCACGTGGATCCTGAGAACT ATTGGACAGCAAGAAAGCGAG
21 Supplementary Table 6. Statistical Analyses of Experimental Data See Figure Legends and Methods for descriptions of statistical tests used. Fold changes are mean values that are reflected in the figures. Statistically significant differences (P < 0.05) are bolded. Figure Condition 1 Condition 2 Fold P-value 1b dcas9-vpr (N) dcas9-vpr (T) hm3d Tango (- CNO) hm3d Tango (+ CNO) (- CNO) (+ CNO) 4.4 < a dcas9-vpr (sggal4) dcas9-vpr (sgil2) >11* <0.001 (sggal4, - CNO) (sgil2, - CNO) (sggal4, + CNO) (sgil2, + CNO) b.d.l.* <0.001 dcas9-vpr (sggal4) dcas9-vpr (sgifng) >92* <0.001 (sggal4, - CNO) (sggal4, + CNO) b.d.l.* - (sgifng, - CNO) (sgifng, + CNO) 11 < b (sggal4, - CNO) (sggal4, + CNO) (sghbb, - CNO) (sghbb, + CNO) (sggal4, - CNO) (sggal4, + CNO) (sghbg, - CNO) (sghbg, + CNO) c (sgil2/sgifng, - CNO) ( sgil2/sgifng,, + CNO) 3.3 < (sgil2/sgifng, - CNO) ( sgil2/sgifng,, + CNO) a KORD ChaCha (-SALB) KORD ChaCha (+SALB) 6.4 <0.001 LPAR1 ChaCha (- LPA) Variant L CXCR4 ChaCha (- SDF1) Variant M (+ LPA) Variant L CXCR4 ChaCha (+ SDF1) Variant M <0.001 NMBR ChaCha (- NMB) NMBR ChaCha (+ NMB)
22 Variant A Variant A ADRB2 ChaCha (-ISO) ADRB2 ChaCha (+ISO) AVPR2 ChaCha (-AVP) AVPR2 ChaCha (+AVP) 3.1 < TRHR ChaCha (-TRH) TRHR ChaCha (+TRH) 3.9 < dcas9-vpr (sggal4) dcas9-vpr (sgifng) >97 < d NMBR ChaCha (sggal4, - NMB) NMBR ChaCha (sggal4, + NMB) n.d.* - NMBR ChaCha (sgifng, - NMB) NMBR ChaCha (sgifng, + NMB) S1a dcas9-vpr (N) dcas9-vpr (T), 24 hrs 39 < dcas9-vpr (N) dcas9-vpr (T), 48 hrs Variant A (-CNO) Variant B (-CNO) Variant C (-CNO) Variant D (-CNO) Variant E (-CNO) Variant A Variant B Variant C Variant D Variant E 4.3 < S1b dcas9-vpr (N) dcas9-vpr (T), 24 hrs 39 < dcas9-vpr (N) dcas9-vpr (T), 48 hrs Variant A (-CNO) Variant F (-CNO) Variant G (-CNO) Variant A Variant F Variant G 4.1 < <
23 Variant H (-CNO) Variant I (-CNO) Variant H Variant I 1.5 < S1c dcas9-vpr (N) dcas9-vpr (T), 24 hrs 39 < dcas9-vpr (N) dcas9-vpr (T), 48 hrs Variant A (-CNO) Variant J (-CNO) Variant K (-CNO) Variant A Variant J Variant K < < S6a dcas9-vpr (sguas8) dcas9-vpr (sgil2-1) dcas9-vpr (sguas8) dcas9-vpr (sgil2-2) b.d.l.* - dcas9-vpr (sguas8) dcas9-vpr (sgil2-3) dcas9-vpr (sguas8) dcas9-vpr (sgil2-4) b.d.l.* - S6b dcas9-vpr (sguas8) dcas9-vpr (sgifng-1) 78 - dcas9-vpr (sguas8) dcas9-vpr (sgifng-2) - - dcas9-vpr (sguas8) dcas9-vpr (sgifng-3) b.d.l.* - S6c dcas9-vpr (sggal4) dcas9-vpr (sghbb-1) dcas9-vpr (sggal4) dcas9-vpr (sghbb-2) dcas9-vpr (sggal4) dcas9-vpr (sghbb-3) dcas9-vpr (sggal4) dcas9-vpr (sghbg-1) dcas9-vpr (sggal4) dcas9-vpr (sghbg-2) dcas9-vpr (sggal4) dcas9-vpr (sghbg-3) dcas9-vpr (sggal4) dcas9-vpr (sghbg-4) S7a dcas9 VPR (sggal4) dcas9 VPR (sghbb) (sggal4, - CNO) (sggal4, + CNO)
24 (sghbb, - CNO) (sghbb, + CNO) S7b dcas9 VPR (sggal4) dcas9 VPR (sghbg) (sggal4, - CNO) (sghbg, - CNO) (sggal4, + CNO) (sghbg, + CNO) S8a dcas9 VPR (sggal4/sggal4) dcas9 VPR (sgil2/sggal4) For IL2 measurements 3.3 < dcas9 VPR (sggal4/sggal4) dcas9 VPR (sggal4/sgifng) For IL2 measurements b.d.l.* - dcas9 VPR (sggal4/sggal4) dcas9 VPR (sgil2/sgifng) For IL2 measurements 3.27 < dcas9 VPR (sggal4/sggal4) dcas9 VPR (sgil2/sggal4) For IFNG measurements b.d.l.* - dcas9 VPR (sggal4/sggal4) dcas9 VPR (sggal4/sgifng) For IFNG measurements dcas9 VPR (sggal4/sggal4) dcas9 VPR (sgil2/sgifng) For IFNG measurements S8b (sggal4/sggal4, - CNO) (sggal4/sggal4, + CNO) For IL2 Measurements b.d.l.* - (sgil2/sggal4, - CNO) (sgil2/sggal4, -+CNO) For IL2 Measurements 2.8 < (sggal4/sgifng, - CNO) (sggal4/sgifng, + CNO) For IL2 Measurements b.d.l.* - (sgil2/sgifng, - CNO) (sgil2/sgifng, + CNO) For IL2 Measurements 3.3 < (sggal4/sggal4, - CNO) (sggal4/sggal4, + CNO) For IFNG Measurements b.d.l.* - (sgil2/sggal4, - CNO) (sgil2/sggal4, -+CNO) For IFNG Measurements b.d.l.* - (sggal4/sgifng, - CNO) (sggal4/sgifng, + CNO) For IFNG Measurements
25 (sgil2/sgifng, - CNO) (sgil2/sgifng, + CNO) For IFNG Measurements S9 dcas9-vpr (N) dcas9-vpr (T) 33x CXCR4 ChaCha (- SDF1) Variant A CXCR4 ChaCha (- SDF1) Variant L CXCR4 ChaCha (- SDF1) Variant M NMBR ChaCha (- NMB) Variant A NMBR ChaCha (- NMB) Variant L NMBR ChaCha (- NMB) Variant M LPAR ChaCha (- NMB) Variant A LPAR ChaCha (- NMB) Variant L LPAR ChaCha (- NMB) Variant M CXCR4 ChaCha (+ SDF1) Variant A CXCR4 ChaCha (+ SDF1) Variant L CXCR4 ChaCha (+ SDF1) Variant M NMBR ChaCha (+ NMB) Variant A NMBR ChaCha (+ NMB) Variant L NMBR ChaCha (+ NMB) Variant M LPAR ChaCha (+ NMB) Variant A LPAR ChaCha (+ NMB) Variant L LPAR ChaCha (+ NMB) Variant M S11 dcas9-vpr (sggal4) dcas9-vpr (sgtet) 39 < (- CNO) (+ CNO) 2.16 < ΔTEVp (-CNO) ΔTEVp *For ELISA values below the detection limit of 4 pg/ml, fold changes are estimates compared to that limit; b.d.l., below detectable limit. For sgrna screening, where only one experiment with technical replicates were performed, only fold change is reported. 5
Supplementary Information. Supplementary Figure 1
 Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion
 Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Supplemental Figure 1
 Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Supplemental Figure 1 1a 1c PD-1 MFI fold change 6 5 4 3 2 1 IL-1α IL-2 IL-4 IL-6 IL-1 IL-12 IL-13 IL-15 IL-17 IL-18 IL-21 IL-23 IFN-α Mut Human PD-1 promoter SBE-D 5 -GTCTG- -1.2kb SBE-P -CAGAC- -1.kb
Appendix. Table of Contents
 Appendix Table of Contents Appendix Figures Figure S1: Gp78 is not required for the degradation of mcherry-cl1 in Hela Cells. Figure S2: Indel formation in the MARCH6 sgrna targeted HeLa clones. Figure
Appendix Table of Contents Appendix Figures Figure S1: Gp78 is not required for the degradation of mcherry-cl1 in Hela Cells. Figure S2: Indel formation in the MARCH6 sgrna targeted HeLa clones. Figure
Electron micrograph of phosphotungstanic acid-stained exosomes derived from murine
 1 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Physical properties of murine DC-derived exosomes. a, Electron micrograph of phosphotungstanic acid-stained exosomes derived from
1 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Physical properties of murine DC-derived exosomes. a, Electron micrograph of phosphotungstanic acid-stained exosomes derived from
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Supplementary Table 1 Clinicopathological characteristics of 35 patients with CRCs
 Supplementary Table Clinicopathological characteristics of 35 patients with CRCs Characteristics Type-A CRC Type-B CRC P value Sex Male / Female 9 / / 8.5 Age (years) Median (range) 6. (9 86) 6.5 (9 76).95
Supplementary Table Clinicopathological characteristics of 35 patients with CRCs Characteristics Type-A CRC Type-B CRC P value Sex Male / Female 9 / / 8.5 Age (years) Median (range) 6. (9 86) 6.5 (9 76).95
Supplementary Figure 1. Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Nature Immunology: doi: /ni.
 Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
Supplementary Figure 1. Establishment of prostacyclin-secreting hmscs. (a) PCR showed the integration of the COX-1-10aa-PGIS transgene into the
 Supplementary Figure 1. Establishment of prostacyclin-secreting hmscs. (a) PCR showed the integration of the COX-1-10aa-PGIS transgene into the genomic DNA of hmscs (PGI2- hmscs). Native hmscs and plasmid
Supplementary Figure 1. Establishment of prostacyclin-secreting hmscs. (a) PCR showed the integration of the COX-1-10aa-PGIS transgene into the genomic DNA of hmscs (PGI2- hmscs). Native hmscs and plasmid
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
Supplementary material. Supplementary Figure legends
 Supplementary material Supplementary Figure legends Supplementary Figure 1: Senescence-associated proliferation stop in response to oncogenic N-RAS expression Proliferation of NHEM cells without (ctrl.)
Supplementary material Supplementary Figure legends Supplementary Figure 1: Senescence-associated proliferation stop in response to oncogenic N-RAS expression Proliferation of NHEM cells without (ctrl.)
Nature Immunology: doi: /ni Supplementary Figure 1. Huwe1 has high expression in HSCs and is necessary for quiescence.
 Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
MII. Supplement Figure 1. CapZ β2. Merge. 250ng. 500ng DIC. Merge. Journal of Cell Science Supplementary Material. GFP-CapZ β2 DNA
 A GV GVBD MI DNA CapZ β2 CapZ β2 Merge B DIC GFP-CapZ β2 Merge CapZ β2-gfp 250ng 500ng Supplement Figure 1. MII A early MI late MI Control RNAi CapZαβ DNA Actin Tubulin B Phalloidin Intensity(A.U.) n=10
A GV GVBD MI DNA CapZ β2 CapZ β2 Merge B DIC GFP-CapZ β2 Merge CapZ β2-gfp 250ng 500ng Supplement Figure 1. MII A early MI late MI Control RNAi CapZαβ DNA Actin Tubulin B Phalloidin Intensity(A.U.) n=10
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/381/ra59/dc1 Supplementary Materials for Analysis of single-cell cytokine secretion reveals a role for paracrine signaling in coordinating macrophage responses
www.sciencesignaling.org/cgi/content/full/8/381/ra59/dc1 Supplementary Materials for Analysis of single-cell cytokine secretion reveals a role for paracrine signaling in coordinating macrophage responses
ILC1 and ILC3 isolation and culture Following cell sorting, we confirmed that the recovered cells belonged to the ILC1, ILC2 and
 Supplementary Methods and isolation and culture Following cell sorting, we confirmed that the recovered cells belonged to the, ILC2 and subsets. For this purpose we performed intracellular flow cytometry
Supplementary Methods and isolation and culture Following cell sorting, we confirmed that the recovered cells belonged to the, ILC2 and subsets. For this purpose we performed intracellular flow cytometry
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) (b) (c) (d) (e) (f) (g) .
 Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) The inhibitory effects of new antibodies (Mab17 and Mab18). They were investigated in in vitro fertilization
Supplementary Figure 1. Properties of various IZUMO1 monoclonal antibodies and behavior of SPACA6. (a) The inhibitory effects of new antibodies (Mab17 and Mab18). They were investigated in in vitro fertilization
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature05732 SUPPLEMENTARY INFORMATION Supplemental Data Supplement Figure Legends Figure S1. RIG-I 2CARD undergo robust ubiquitination a, (top) At 48 h posttransfection with a GST, GST-RIG-I-2CARD
doi: 10.1038/nature05732 SUPPLEMENTARY INFORMATION Supplemental Data Supplement Figure Legends Figure S1. RIG-I 2CARD undergo robust ubiquitination a, (top) At 48 h posttransfection with a GST, GST-RIG-I-2CARD
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1
 Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Dual Targeting Nanoparticle Stimulates the Immune
 Dual Targeting Nanoparticle Stimulates the Immune System to Inhibit Tumor Growth Alyssa K. Kosmides, John-William Sidhom, Andrew Fraser, Catherine A. Bessell, Jonathan P. Schneck * Supplemental Figure
Dual Targeting Nanoparticle Stimulates the Immune System to Inhibit Tumor Growth Alyssa K. Kosmides, John-William Sidhom, Andrew Fraser, Catherine A. Bessell, Jonathan P. Schneck * Supplemental Figure
Nature Immunology: doi: /ni Supplementary Figure 1. Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice.
 Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
Supplementary Figure 1 Gene expression profile of CD4 + T cells and CTL responses in Bcl6-deficient mice. (a) Gene expression profile in the resting CD4 + T cells were analyzed by an Affymetrix microarray
Nature Medicine: doi: /nm.4322
 1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
1 2 3 4 5 6 7 8 9 10 11 Supplementary Figure 1. Predicted RNA structure of 3 UTR and sequence alignment of deleted nucleotides. (a) Predicted RNA secondary structure of ZIKV 3 UTR. The stem-loop structure
Conditional and reversible disruption of essential herpesvirus protein functions
 nature methods Conditional and reversible disruption of essential herpesvirus protein functions Mandy Glaß, Andreas Busche, Karen Wagner, Martin Messerle & Eva Maria Borst Supplementary figures and text:
nature methods Conditional and reversible disruption of essential herpesvirus protein functions Mandy Glaß, Andreas Busche, Karen Wagner, Martin Messerle & Eva Maria Borst Supplementary figures and text:
Doctoral Degree Program in Marine Biotechnology, College of Marine Sciences, Doctoral Degree Program in Marine Biotechnology, Academia Sinica, Taipei,
 Cyclooxygenase 2 facilitates dengue virus replication and serves as a potential target for developing antiviral agents Chun-Kuang Lin 1,2, Chin-Kai Tseng 3,4, Yu-Hsuan Wu 3,4, Chih-Chuang Liaw 1,5, Chun-
Cyclooxygenase 2 facilitates dengue virus replication and serves as a potential target for developing antiviral agents Chun-Kuang Lin 1,2, Chin-Kai Tseng 3,4, Yu-Hsuan Wu 3,4, Chih-Chuang Liaw 1,5, Chun-
Supplementary Figure S1 (a) (b)
 Supplementary Figure S1: IC87114 does not affect basal Ca 2+ level nor nicotineinduced Ca 2+ influx. (a) Bovine chromaffin cells were loaded with Fluo-4AM (1 μm) in buffer A containing 0.02% of pluronic
Supplementary Figure S1: IC87114 does not affect basal Ca 2+ level nor nicotineinduced Ca 2+ influx. (a) Bovine chromaffin cells were loaded with Fluo-4AM (1 μm) in buffer A containing 0.02% of pluronic
Supplementary Materials and Methods
 Supplementary Materials and Methods Reagents and antibodies was purchased from iaffin GmbH & Co KG. Cisplatin (ristol-myers Squibb Co.) and etoposide (Sandoz Pharma Ltd.) were used. Antibodies recognizing
Supplementary Materials and Methods Reagents and antibodies was purchased from iaffin GmbH & Co KG. Cisplatin (ristol-myers Squibb Co.) and etoposide (Sandoz Pharma Ltd.) were used. Antibodies recognizing
Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical
 Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical representation using all SNPs (n= 13,515,798) including the region on chromosome 1 including SORT1 which was previously
Supplementary Figure 1. Quantile-quantile (Q-Q) plots. (Panel A) Q-Q plot graphical representation using all SNPs (n= 13,515,798) including the region on chromosome 1 including SORT1 which was previously
 Supplementary Figure Legends Supplementary Figure S1. Aurora-A is essential for SAC establishment in early mitosis. (a-c) RPE cells were treated with DMSO (a), MLN8237 (b) or BI2536 (c) for Two hours.
Supplementary Figure Legends Supplementary Figure S1. Aurora-A is essential for SAC establishment in early mitosis. (a-c) RPE cells were treated with DMSO (a), MLN8237 (b) or BI2536 (c) for Two hours.
Supplementary Figure 1
 Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
The clathrin adaptor Numb regulates intestinal cholesterol. absorption through dynamic interaction with NPC1L1
 The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
The clathrin adaptor Numb regulates intestinal cholesterol absorption through dynamic interaction with NPC1L1 Pei-Shan Li 1, Zhen-Yan Fu 1,2, Ying-Yu Zhang 1, Jin-Hui Zhang 1, Chen-Qi Xu 1, Yi-Tong Ma
Supplementary Information
 Supplementary Information Supplementary Figure 1! a! b! Nfatc1!! Nfatc1"! P1! P2! pa1! pa2! ex1! ex2! exons 3-9! ex1! ex11!!" #" Nfatc1A!!" Nfatc1B! #"!" Nfatc1C! #" DN1! DN2! DN1!!A! #A!!B! #B!!C! #C!!A!
Supplementary Information Supplementary Figure 1! a! b! Nfatc1!! Nfatc1"! P1! P2! pa1! pa2! ex1! ex2! exons 3-9! ex1! ex11!!" #" Nfatc1A!!" Nfatc1B! #"!" Nfatc1C! #" DN1! DN2! DN1!!A! #A!!B! #B!!C! #C!!A!
Nature Methods: doi: /nmeth.4257
 Supplementary Figure 1 Screen for polypeptides that affect cellular actin filaments. (a) Table summarizing results from all polypeptides tested. Source shows organism, gene, and amino acid numbers used.
Supplementary Figure 1 Screen for polypeptides that affect cellular actin filaments. (a) Table summarizing results from all polypeptides tested. Source shows organism, gene, and amino acid numbers used.
sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5
 sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Lu et al., http://www.jcb.org/cgi/content/full/jcb.201012063/dc1 Figure S1. Kinetics of nuclear envelope assembly, recruitment of Nup133
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Lu et al., http://www.jcb.org/cgi/content/full/jcb.201012063/dc1 Figure S1. Kinetics of nuclear envelope assembly, recruitment of Nup133
Supplementary information. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins
 Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Highly Significant Antiviral Activity of HIV-1 LTR-Specific Tre-
 Supporting Information Hauber et al. page 1 Text S1 - Supporting Information Highly Significant Antiviral Activity of HIV-1 LTR-Specific Tre- Recombinase in Humanized Mice Ilona Hauber 1, Helga Hofmann-Sieber
Supporting Information Hauber et al. page 1 Text S1 - Supporting Information Highly Significant Antiviral Activity of HIV-1 LTR-Specific Tre- Recombinase in Humanized Mice Ilona Hauber 1, Helga Hofmann-Sieber
well for 2 h at rt. Each dot represents an individual mouse and bar is the mean ±
 Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Nature Immunology: doi: /ni.3631
 Supplementary Figure 1 SPT analyses of Zap70 at the T cell plasma membrane. (a) Total internal reflection fluorescent (TIRF) excitation at 64-68 degrees limits single molecule detection to 100-150 nm above
Supplementary Figure 1 SPT analyses of Zap70 at the T cell plasma membrane. (a) Total internal reflection fluorescent (TIRF) excitation at 64-68 degrees limits single molecule detection to 100-150 nm above
Supplementary Information. Novel lentiviral vectors with mutated reverse transcriptase for mrna delivery of TALE nucleases
 Supplementary Information Novel lentiviral vectors with mutated reverse transcriptase for mrna delivery of TALE nucleases Ulrike Mock 1, Kristoffer Riecken 1, Belinda Berdien 1, Waseem Qasim 2, Emma Chan
Supplementary Information Novel lentiviral vectors with mutated reverse transcriptase for mrna delivery of TALE nucleases Ulrike Mock 1, Kristoffer Riecken 1, Belinda Berdien 1, Waseem Qasim 2, Emma Chan
Choosing Between Lentivirus and Adeno-associated Virus For DNA Delivery
 Choosing Between Lentivirus and Adeno-associated Virus For DNA Delivery Presenter: April 12, 2017 Ed Davis, Ph.D. Senior Application Scientist GeneCopoeia, Inc. Outline Introduction to GeneCopoeia Lentiviral
Choosing Between Lentivirus and Adeno-associated Virus For DNA Delivery Presenter: April 12, 2017 Ed Davis, Ph.D. Senior Application Scientist GeneCopoeia, Inc. Outline Introduction to GeneCopoeia Lentiviral
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
Supplementary Figure 1. IL-12 serum levels and frequency of subsets in FL patients. (A) IL-12
 1 Supplementary Data Figure legends Supplementary Figure 1. IL-12 serum levels and frequency of subsets in FL patients. (A) IL-12 serum levels measured by multiplex ELISA (Luminex) in FL patients before
1 Supplementary Data Figure legends Supplementary Figure 1. IL-12 serum levels and frequency of subsets in FL patients. (A) IL-12 serum levels measured by multiplex ELISA (Luminex) in FL patients before
Supplementary Information. Tissue-wide immunity against Leishmania. through collective production of nitric oxide
 Supplementary Information Tissue-wide immunity against Leishmania through collective production of nitric oxide Romain Olekhnovitch, Bernhard Ryffel, Andreas J. Müller and Philippe Bousso Supplementary
Supplementary Information Tissue-wide immunity against Leishmania through collective production of nitric oxide Romain Olekhnovitch, Bernhard Ryffel, Andreas J. Müller and Philippe Bousso Supplementary
CRISPRaTest Functional dcas9-activator Assay Kit v1 Last update: 2018/07/04 Cellecta, Inc.
 CRISPRaTest Functional dcas9-activator Assay Kit v1 Last update: 2018/07/04 Cellecta, Inc. Copyright (c) 2018 Cellecta, Inc. All Rights Reserved. Table of Contents 1. CRISPRaTest Functional dcas9-activator
CRISPRaTest Functional dcas9-activator Assay Kit v1 Last update: 2018/07/04 Cellecta, Inc. Copyright (c) 2018 Cellecta, Inc. All Rights Reserved. Table of Contents 1. CRISPRaTest Functional dcas9-activator
Rapid parallel measurements of macroautophagy and mitophagy in
 Supplemental Figures Rapid parallel measurements of macroautophagy and mitophagy in mammalian cells using a single fluorescent biosensor Sargsyan A, Cai J, Fandino LB, Labasky ME, Forostyan T, Colosimo
Supplemental Figures Rapid parallel measurements of macroautophagy and mitophagy in mammalian cells using a single fluorescent biosensor Sargsyan A, Cai J, Fandino LB, Labasky ME, Forostyan T, Colosimo
LPS LPS P6 - + Supplementary Fig. 1.
 P6 LPS - - - + + + - LPS + + - - P6 + Supplementary Fig. 1. Pharmacological inhibition of the JAK/STAT blocks LPS-induced HMGB1 nuclear translocation. RAW 267.4 cells were stimulated with LPS in the absence
P6 LPS - - - + + + - LPS + + - - P6 + Supplementary Fig. 1. Pharmacological inhibition of the JAK/STAT blocks LPS-induced HMGB1 nuclear translocation. RAW 267.4 cells were stimulated with LPS in the absence
Supplemental Materials and Methods Plasmids and viruses Quantitative Reverse Transcription PCR Generation of molecular standard for quantitative PCR
 Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
Supplemental Materials and Methods Plasmids and viruses To generate pseudotyped viruses, the previously described recombinant plasmids pnl4-3-δnef-gfp or pnl4-3-δ6-drgfp and a vector expressing HIV-1 X4
SUPPLEMENTARY LEGENDS...
 TABLE OF CONTENTS SUPPLEMENTARY LEGENDS... 2 11 MOVIE S1... 2 FIGURE S1 LEGEND... 3 FIGURE S2 LEGEND... 4 FIGURE S3 LEGEND... 5 FIGURE S4 LEGEND... 6 FIGURE S5 LEGEND... 7 FIGURE S6 LEGEND... 8 FIGURE
TABLE OF CONTENTS SUPPLEMENTARY LEGENDS... 2 11 MOVIE S1... 2 FIGURE S1 LEGEND... 3 FIGURE S2 LEGEND... 4 FIGURE S3 LEGEND... 5 FIGURE S4 LEGEND... 6 FIGURE S5 LEGEND... 7 FIGURE S6 LEGEND... 8 FIGURE
Supplementary Materials for
 www.sciencetranslationalmedicine.org/cgi/content/full/4/117/117ra8/dc1 Supplementary Materials for Notch4 Normalization Reduces Blood Vessel Size in Arteriovenous Malformations Patrick A. Murphy, Tyson
www.sciencetranslationalmedicine.org/cgi/content/full/4/117/117ra8/dc1 Supplementary Materials for Notch4 Normalization Reduces Blood Vessel Size in Arteriovenous Malformations Patrick A. Murphy, Tyson
Supplemental Information
 Cell Host & Microbe, Volume 14 Supplemental Information HIV-1 Induces the Formation of Stable Microtubules to Enhance Early Infection Yosef Sabo, Derek Walsh, Denis S. Barry, Sedef Tinaztepe, Kenia de
Cell Host & Microbe, Volume 14 Supplemental Information HIV-1 Induces the Formation of Stable Microtubules to Enhance Early Infection Yosef Sabo, Derek Walsh, Denis S. Barry, Sedef Tinaztepe, Kenia de
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.
 Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
Supplemental information contains 7 movies and 4 supplemental Figures
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplemental information contains 7 movies and 4 supplemental Figures Movies: Movie 1. Single virus tracking of A4-mCherry-WR MV
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplemental information contains 7 movies and 4 supplemental Figures Movies: Movie 1. Single virus tracking of A4-mCherry-WR MV
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis
 Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
B220 CD4 CD8. Figure 1. Confocal Image of Sensitized HLN. Representative image of a sensitized HLN
 B220 CD4 CD8 Natarajan et al., unpublished data Figure 1. Confocal Image of Sensitized HLN. Representative image of a sensitized HLN showing B cell follicles and T cell areas. 20 µm thick. Image of magnification
B220 CD4 CD8 Natarajan et al., unpublished data Figure 1. Confocal Image of Sensitized HLN. Representative image of a sensitized HLN showing B cell follicles and T cell areas. 20 µm thick. Image of magnification
Supplementary Figure 1. Deletion of Smad3 prevents B16F10 melanoma invasion and metastasis in a mouse s.c. tumor model.
 A B16F1 s.c. Lung LN Distant lymph nodes Colon B B16F1 s.c. Supplementary Figure 1. Deletion of Smad3 prevents B16F1 melanoma invasion and metastasis in a mouse s.c. tumor model. Highly invasive growth
A B16F1 s.c. Lung LN Distant lymph nodes Colon B B16F1 s.c. Supplementary Figure 1. Deletion of Smad3 prevents B16F1 melanoma invasion and metastasis in a mouse s.c. tumor model. Highly invasive growth
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
Supplementary Figure 1. Using DNA barcode-labeled MHC multimers to generate TCR fingerprints
 Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
SUPPLEMENTARY INFORMATION. Supp. Fig. 1. Autoimmunity. Tolerance APC APC. T cell. T cell. doi: /nature06253 ICOS ICOS TCR CD28 TCR CD28
 Supp. Fig. 1 a APC b APC ICOS ICOS TCR CD28 mir P TCR CD28 P T cell Tolerance Roquin WT SG Icos mrna T cell Autoimmunity Roquin M199R SG Icos mrna www.nature.com/nature 1 Supp. Fig. 2 CD4 + CD44 low CD4
Supp. Fig. 1 a APC b APC ICOS ICOS TCR CD28 mir P TCR CD28 P T cell Tolerance Roquin WT SG Icos mrna T cell Autoimmunity Roquin M199R SG Icos mrna www.nature.com/nature 1 Supp. Fig. 2 CD4 + CD44 low CD4
L1 on PyMT tumor cells but Py117 cells are more responsive to IFN-γ. (A) Flow
 A MHCI B PD-L1 Fold expression 8 6 4 2 Fold expression 3 2 1 No tx 1Gy 2Gy IFN Py117 Py117 Supplementary Figure 1. Radiation and IFN-γ enhance MHCI expression and PD- L1 on PyMT tumor cells but Py117 cells
A MHCI B PD-L1 Fold expression 8 6 4 2 Fold expression 3 2 1 No tx 1Gy 2Gy IFN Py117 Py117 Supplementary Figure 1. Radiation and IFN-γ enhance MHCI expression and PD- L1 on PyMT tumor cells but Py117 cells
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
Figure S1. HP1α localizes to centromeres in mitosis and interacts with INCENP. (A&B) HeLa
 SUPPLEMENTARY FIGURES Figure S1. HP1α localizes to centromeres in mitosis and interacts with INCENP. (A&B) HeLa tet-on cells that stably express HP1α-CFP, HP1β-CFP, or HP1γ-CFP were monitored with livecell
SUPPLEMENTARY FIGURES Figure S1. HP1α localizes to centromeres in mitosis and interacts with INCENP. (A&B) HeLa tet-on cells that stably express HP1α-CFP, HP1β-CFP, or HP1γ-CFP were monitored with livecell
Supplementary Information
 Supplementary Information Overexpression of Fto leads to increased food intake and results in obesity Chris Church, Lee Moir, Fiona McMurray, Christophe Girard, Gareth T Banks, Lydia Teboul, Sara Wells,
Supplementary Information Overexpression of Fto leads to increased food intake and results in obesity Chris Church, Lee Moir, Fiona McMurray, Christophe Girard, Gareth T Banks, Lydia Teboul, Sara Wells,
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Rapid blue-light mediated induction of protein interactions in living cells
 Nature Methods Rapid blue-light mediated induction of protein interactions in living cells Matthew J Kennedy, Robert M Hughes, Leslie A Peteya, Joel W Schwartz, Michael D Ehlers & Chandra L Tucker Supplementary
Nature Methods Rapid blue-light mediated induction of protein interactions in living cells Matthew J Kennedy, Robert M Hughes, Leslie A Peteya, Joel W Schwartz, Michael D Ehlers & Chandra L Tucker Supplementary
CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus
 0 0 CRISPR/Cas cleavage of viral DNA efficiently suppresses hepatitis B virus Authors: Vyas Ramanan,, Amir Shlomai,, David B.T. Cox,,,, Robert E. Schwartz,,, Eleftherios Michailidis, Ankit Bhatta, David
0 0 CRISPR/Cas cleavage of viral DNA efficiently suppresses hepatitis B virus Authors: Vyas Ramanan,, Amir Shlomai,, David B.T. Cox,,,, Robert E. Schwartz,,, Eleftherios Michailidis, Ankit Bhatta, David
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
<10. IL-1β IL-6 TNF + _ TGF-β + IL-23
 3 ns 25 ns 2 IL-17 (pg/ml) 15 1 ns ns 5 IL-1β IL-6 TNF
3 ns 25 ns 2 IL-17 (pg/ml) 15 1 ns ns 5 IL-1β IL-6 TNF
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplemental Data Figure S1 Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells.
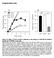 Supplemental Data Figure S1. Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells. (A) Specific lysis of IFN-γ-treated SKBR-3 cells in the absence
Supplemental Data Figure S1. Effect of TS2/4 and R6.5 antibodies on the kinetics of CD16.NK-92-mediated specific lysis of SKBR-3 target cells. (A) Specific lysis of IFN-γ-treated SKBR-3 cells in the absence
Table S1. Viral load and CD4 count of HIV-infected patient population
 Table S1. Viral load and CD4 count of HIV-infected patient population Subject ID Viral load (No. of copies per ml of plasma) CD4 count (No. of cells/µl of blood) 28 7, 14 29 7, 23 21 361,99 94 217 7, 11
Table S1. Viral load and CD4 count of HIV-infected patient population Subject ID Viral load (No. of copies per ml of plasma) CD4 count (No. of cells/µl of blood) 28 7, 14 29 7, 23 21 361,99 94 217 7, 11
Influenza virus exploits tunneling nanotubes for cell-to-cell spread
 Supplementary Information Influenza virus exploits tunneling nanotubes for cell-to-cell spread Amrita Kumar 1, Jin Hyang Kim 1, Priya Ranjan 1, Maureen G. Metcalfe 2, Weiping Cao 1, Margarita Mishina 1,
Supplementary Information Influenza virus exploits tunneling nanotubes for cell-to-cell spread Amrita Kumar 1, Jin Hyang Kim 1, Priya Ranjan 1, Maureen G. Metcalfe 2, Weiping Cao 1, Margarita Mishina 1,
CD4 + T cells recovered in Rag2 / recipient ( 10 5 ) Heart Lung Pancreas
 a CD4 + T cells recovered in Rag2 / recipient ( 1 5 ) Heart Lung Pancreas.5 1 2 4 6 2 4 6 Ctla4 +/+ Ctla4 / Ctla4 / Lung Ctla4 / Pancreas b Heart Lung Pancreas Ctla4 +/+ Ctla4 / Ctla4 / Lung Ctla4 / Pancreas
a CD4 + T cells recovered in Rag2 / recipient ( 1 5 ) Heart Lung Pancreas.5 1 2 4 6 2 4 6 Ctla4 +/+ Ctla4 / Ctla4 / Lung Ctla4 / Pancreas b Heart Lung Pancreas Ctla4 +/+ Ctla4 / Ctla4 / Lung Ctla4 / Pancreas
SD-1 SD-1: Cathepsin B levels in TNF treated hch
 SD-1 SD-1: Cathepsin B levels in TNF treated hch. A. RNA and B. protein extracts from TNF treated and untreated human chondrocytes (hch) were analyzed via qpcr (left) and immunoblot analyses (right) for
SD-1 SD-1: Cathepsin B levels in TNF treated hch. A. RNA and B. protein extracts from TNF treated and untreated human chondrocytes (hch) were analyzed via qpcr (left) and immunoblot analyses (right) for
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Krenn et al., http://www.jcb.org/cgi/content/full/jcb.201110013/dc1 Figure S1. Levels of expressed proteins and demonstration that C-terminal
Supplementary Figure 1
 Supplementary Figure 1 Expression of apoptosis-related genes in tumor T reg cells. (a) Identification of FOXP3 T reg cells by FACS. CD45 + cells were gated as enriched lymphoid cell populations with low-granularity.
Supplementary Figure 1 Expression of apoptosis-related genes in tumor T reg cells. (a) Identification of FOXP3 T reg cells by FACS. CD45 + cells were gated as enriched lymphoid cell populations with low-granularity.
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
Supplementary Figure 1. ETBF activate Stat3 in B6 and Min mice colons
 Supplementary Figure 1 ETBF activate Stat3 in B6 and Min mice colons a pstat3 controls Pos Neg ETBF 1 2 3 4 b pstat1 pstat2 pstat3 pstat4 pstat5 pstat6 Actin Figure Legend: (a) ETBF induce predominantly
Supplementary Figure 1 ETBF activate Stat3 in B6 and Min mice colons a pstat3 controls Pos Neg ETBF 1 2 3 4 b pstat1 pstat2 pstat3 pstat4 pstat5 pstat6 Actin Figure Legend: (a) ETBF induce predominantly
Supplementary Fig. S1. Schematic diagram of minigenome segments.
 open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
open reading frame 1565 (segment 5) 47 (-) 3 5 (+) 76 101 125 149 173 197 221 246 287 open reading frame 890 (segment 8) 60 (-) 3 5 (+) 172 Supplementary Fig. S1. Schematic diagram of minigenome segments.
Supplementary Material
 Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
Supplementary Material Summary: The supplementary information includes 1 table (Table S1) and 4 figures (Figure S1 to S4). Supplementary Figure Legends Figure S1 RTL-bearing nude mouse model. (A) Tumor
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
Pre-made Lentiviral Particles for Fluorescent Proteins
 Pre-made Lentiviral Particles for Fluorescent Proteins Catalog# Product Name Amounts Fluorescent proteins expressed under sucmv promoter: LVP001 LVP001-PBS LVP002 LVP002-PBS LVP011 LVP011-PBS LVP012 LVP012-PBS
Pre-made Lentiviral Particles for Fluorescent Proteins Catalog# Product Name Amounts Fluorescent proteins expressed under sucmv promoter: LVP001 LVP001-PBS LVP002 LVP002-PBS LVP011 LVP011-PBS LVP012 LVP012-PBS
Constitutive Reporter Lentiviral Vectors Expressing Fluorescent Proteins
 Constitutive Reporter Lentiviral Vectors Expressing Fluorescent Proteins www.vectalys.com/products/ Constitutive Reporter Lentiviral Vectors Catalog Number referring to this User Manual: 0008VCT; 0009VCT;
Constitutive Reporter Lentiviral Vectors Expressing Fluorescent Proteins www.vectalys.com/products/ Constitutive Reporter Lentiviral Vectors Catalog Number referring to this User Manual: 0008VCT; 0009VCT;
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3443 In the format provided by the authors and unedited. Supplementary Figure 1 TC and SC behaviour during ISV sprouting. (a) Predicted outcome of TC division and competitive Dll4-Notch-mediated
DOI: 10.1038/ncb3443 In the format provided by the authors and unedited. Supplementary Figure 1 TC and SC behaviour during ISV sprouting. (a) Predicted outcome of TC division and competitive Dll4-Notch-mediated
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein
 Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures
 Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
Waking up dormant tumor suppressor genes with zinc fingers, TALEs and the CRISPR/dCas9 system
 /, Vol. 7, No. 37 Waking up dormant tumor suppressor genes with zinc fingers, TALEs and the CRISPR/dCas9 system Benjamin Garcia-Bloj 1,2,4, Colette Moses 1,2, Agustin Sgro 1, Janice Plani-Lam 1, Mahira
/, Vol. 7, No. 37 Waking up dormant tumor suppressor genes with zinc fingers, TALEs and the CRISPR/dCas9 system Benjamin Garcia-Bloj 1,2,4, Colette Moses 1,2, Agustin Sgro 1, Janice Plani-Lam 1, Mahira
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols.
 Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
Supplementary Figure 1. Schematic diagram of o2n-seq. Double-stranded DNA was sheared, end-repaired, and underwent A-tailing by standard protocols. A-tailed DNA was ligated to T-tailed dutp adapters, circularized
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Subcellular segregation of VGluT2-IR and TH-IR within the same VGluT2-TH axon (wild type rats). (a-e) Serial sections of a dual VGluT2-TH labeled axon. This axon (blue outline) has
Supplementary Figure 1 Subcellular segregation of VGluT2-IR and TH-IR within the same VGluT2-TH axon (wild type rats). (a-e) Serial sections of a dual VGluT2-TH labeled axon. This axon (blue outline) has
and follicular helper T cells is Egr2-dependent. (a) Diagrammatic representation of the
 Supplementary Figure 1. LAG3 + Treg-mediated regulation of germinal center B cells and follicular helper T cells is Egr2-dependent. (a) Diagrammatic representation of the experimental protocol for the
Supplementary Figure 1. LAG3 + Treg-mediated regulation of germinal center B cells and follicular helper T cells is Egr2-dependent. (a) Diagrammatic representation of the experimental protocol for the
CD3 coated cover slips indicating stimulatory contact site, F-actin polymerization and
 SUPPLEMENTAL FIGURES FIGURE S1. Detection of MCs. A, Schematic representation of T cells stimulated on anti- CD3 coated cover slips indicating stimulatory contact site, F-actin polymerization and microclusters.
SUPPLEMENTAL FIGURES FIGURE S1. Detection of MCs. A, Schematic representation of T cells stimulated on anti- CD3 coated cover slips indicating stimulatory contact site, F-actin polymerization and microclusters.
Supplemental Information
 Supplemental Information Tobacco-specific Carcinogen Induces DNA Methyltransferases 1 Accumulation through AKT/GSK3β/βTrCP/hnRNP-U in Mice and Lung Cancer patients Ruo-Kai Lin, 1 Yi-Shuan Hsieh, 2 Pinpin
Supplemental Information Tobacco-specific Carcinogen Induces DNA Methyltransferases 1 Accumulation through AKT/GSK3β/βTrCP/hnRNP-U in Mice and Lung Cancer patients Ruo-Kai Lin, 1 Yi-Shuan Hsieh, 2 Pinpin
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was
 Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
Supplementary Figure 1 IMQ-Induced Mouse Model of Psoriasis. IMQ cream was painted on the shaved back skin of CBL/J and BALB/c mice for consecutive days. (a, b) Phenotypic presentation of mouse back skin
File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables. File Name: Peer Review File Description:
 File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables File Name: Peer Review File Description: Primer Name Sequence (5'-3') AT ( C) RT-PCR USP21 F 5'-TTCCCATGGCTCCTTCCACATGAT-3'
File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables File Name: Peer Review File Description: Primer Name Sequence (5'-3') AT ( C) RT-PCR USP21 F 5'-TTCCCATGGCTCCTTCCACATGAT-3'
