Characterization of antibodies against endogenous mouse Gli2 and Gli3
|
|
|
- Linda Kelley
- 5 years ago
- Views:
Transcription
1 Supplemental Material for Chen et al. Cilium independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved Miao-Hsueh Chen 1*, Christopher W. Wilson 1*, Ya-Jun Li 1, Kelvin King Lo Law 2, Chi-Sheng Lu 1, Rhodora Gacayan 1, Xiaoyun Zhang 2, Chi-chung Hui 2, and Pao-Tien Chuang 1# 1 Cardiovascular Research Institute, University of California, San Francisco, CA Program in Developmental & Stem Cell iology, The Hospital for Sick Children and Department of Molecular Genetics, University of Toronto, Toronto, Canada * These authors contributed equally to this work. # Correspondence and requests for materials should be addressed to Pao-Tien Chuang ( paotien.chuang@ucsf.edu). Table of Contents Supplemental Figure S1 Characterization of antibodies against endogenous mouse Gli2 and Supplemental Figure S2 Gli2 and, but not Sufu, display dynamic localization on the primary cilium Supplemental Figure S3 Endogenous Gli2 and localize to the tip of the primary cilium in mouse embryonic fibroblasts Supplemental Figure S4 Gli2 and protein levels are greatly reduced in both the nuclear and cytoplasmic fractions in the absence of Sufu Supplemental Figure S5 Gli2 and transcript levels are not significantly changed in Sufu / MEFs or embryos Supplemental Figure S6 Primary cilia fail to form in the absence of Kif3a Supplemental Figure S7 Hh pathway reporters can be activated only by exogenous Gli activators in Kif3a / mouse embryonic fibroblasts Supplemental Figure S8 Sufu / and Kif3a / mouse embryonic fibroblasts have altered response to Shh, but not Wnt3a Supplemental Figure S9 Quantification of localization of overexpressed proteins Supplemental Figure S1 Efficient knockdown of primary cilium function by overexpressed dnkif3b or Kif3a shrna Supplemental Figure S11 Expression of mouse, zebrafish and Drosophila Smo in Smo-deficient mouse embryonic fibroblasts Supplemental Figure S12 Subcellular distributions of mouse Gli2 and in mouse embryonic fibroblasts in the presence of Drosophila or zebrafish Sufu i
2 Supplemental Figure S13 is broadly expressed in Hh-responsive tissues during mouse embryogenesis Supplemental Figure S14 and localization in CHO, COS7, and MEF cell lines Supplemental Figure S15 does not affect MyoD or FoxC2 protein levels Supplemental Figure S16 Gli2, but not Gli1, activity is inhibited by Supplemental Figure S17 Cul3 alone (without ) fails to promote Gli2 and ubiquitination Supplemental Figure S18 SPOP reduces Gli2 and protein levels in a proteasome-dependent manner Supplemental Figure S19 directs Gli2 and to Cul3 () foci Supplemental Figure S2 knockdown in Sufu-deficient MEFs enhances Hh pathway activity Supplemental Figure S21 Overexpressed does not localize to the primary cilium in mouse embryonic fibroblasts Supplemental Figure S22 Addition of Ptch1 to Ptch1 / MEFs decreases Hh responsiveness in the absence of exogenous Hh and fails to promote Hh pathway activation in the presence of Hh ligand Supplemental Figure S23 Mouse and zebrafish Sufu bind Gli proteins more strongly than Sufu D159A or fly Sufu Supplemental Figure S24 Summary of changes of Gli protein dynamics in Sufu and Kif3a mutants Supplemental Figure S25 SAP18 localization is unaffected by Sufu, and SAP18 does not significantly affect Gli1 or Gli2 activity Supplemental Figure S26 The Hh pathway is largely unaffected by pharmacologic modulation of protein kinase A (PKA) activity in Sufu / MEFs Supplemental Figure S27 Hh stimulation does not affect the interactions between Sufu and Gl2/3 Supplemental Figure S28 Sufu / MEFs are refractory to exogenous Smo and Ptch1 Supplemental Table S1 Quantification of endogenous Gli levels from Western blots Supplemental Materials and Methods and References ii
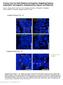
3 A anti- Gli2 MEF embryo MEF embryo wt Gli2 / wt Gli2 / anti- wt / wt / Gli2 25 Gli FL 25 FL R 75 R 75 antitubulin antitubulin / ShhN Gli2 AC AC Gli2 ShhN AC AC Supplemental Figure S1: Characterization of antibodies against endogenous mouse Gli2 and (A) Western blots of lysates derived from wild-type (wt), Gli2 /, and / mouse embryonic fibroblasts (MEFs) and embryos. ands corresponding to endogenous Gli2, full-length (FL), and repressor (R) are not detected in the corresponding null cells and embryos. Note that some nonspecific bands are apparent in / MEFs, consistent with antibodies developed by others that can detect endogenous (Wang et al., 2). Tubulin was used as the loading control and numbers on the right indicate positions of protein standards. () Gli2 / and / MEFs were stained with antibodies against either Gli2 or and acetylated tubulin (AC) (to mark primary cilia). No Gli2 or immunoreactivity was seen in the primary cilium in null cells, in the presence or absence (data not shown) of ShhN.
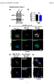
4 A 8% 7% 6% 5% 4% 3% 2% 1% % 9% 8% 7% 6% 5% 4% 3% 2% 1% % 1% 9% 8% 7% 6% 5% 4% 3% 2% 1% % % 1 hr 6 hr 24 hr 1 hr 6 hr 24 hr mock ShhN 1 hr 6 hr 24 hr Time of treatment Time of treatment Treatment Time of treatment % Sufu / Sufu / Sufu / Sufu / Sufu / Sufu / Sufu msufu msufud159a / / Sufu / zsufu dsufu Sufu msufu msufud159a zsufu 4% 35% 3% 25% 2% 15% 1% 5% ShhN ShhN % 8% 7% 6% 5% 4% 3% 2% 1% 1% 9% 8% 7% 6% 5% 4% 3% 2% 1% % Sufu / Sufu / / dsufu Sufu msufu Sufu / Sufu / msufud159a zsufu Sufu / dsufu C 8% 4% 7% 7% 35% 6% 6% 3% 5% 5% 4% 3% 2% 25% 2% 15% 1% 4% 3% 2% 1% 5% 1% % % % Ptch1 / Ptch1 / ( Sufu shrna) Ptch1 / Ptch1 / ( Sufu shrna) Ptch1 / Ptch1 / ( Sufu shrna) Supplemental Figure S2: Gli2 and, but not Sufu, display dynamic localization on the primary cilium Quantification of the number of cilia showing Gli2,, and Sufu staining in mouse embryonic fibroblasts (MEFs). For all individual data points, a minimum of two independent experiments was performed, with a minimum of 1 cilia counted in each experiment. (A) Gli2 and staining on the primary cilium in wild-type MEFs increases after stimulation with ShhN. Gli2 is not observed on the primary cilium in the absence of ShhN, but the number of Gli2 cilia increase with prolonged exposure to ligand. is typically found in approximately 4% of cilia; this number transiently increases to ~9% after brief (1 hr) exposure to ShhN, with a corresponding increase in intensity of staining along the entire length of the cilium. y contrast, mouse Sufu is detected in roughly 9% of cilia irrespective of the state of Hedgehog (Hh) pathway activation. () Retroviral introduction of FLAG-tagged mouse (m), zebrafish (z), and fly (d) Sufu into Sufu / MEFs restores Gli2 and localization to the primary cilium. Mouse and zebrafish Sufu rescue ciliary localization of Gli2 and to a greater extent than fly Sufu, consistent with the ability of the respective Sufu proteins to rescue total cellular Gli2 and levels. Expression of msufu D159A, which has reduced binding affinity for Gli proteins, does not rescue Gli2 or ciliary localization. Interestingly, epitope-tagged zsufu and dsufu were not detected on the primary cilium. (C) Knockdown of Sufu via shrna in Ptch1 / MEFs eliminates detectable Gli2, and Sufu staining from the primary cilium. Error bars are standard deviation (s.d.).
5 wt MEFs AC Gli2 DAPI merge γ tubulin Gli2 DAPI merge wt MEFs AC DAPI merge γ tubulin DAPI merge Supplemental Figure S3: Endogenous Gli2 and localize to the tip of the primary cilium in mouse embryonic fibroblasts Immunofluorescence of wild-type (wt) mouse embryonic fibroblasts (MEFs) using antibodies against acetylated tubulin (AC) (labeling primary cilia, red), γ-tubulin (labeling the basal body, red), Gli2 (green), and (green). Gli2 and staining is often detected at the end of the primary cilium, distinct from anti-γ tubulin staining. This suggests that Gli2 and localize to the tip (and not the base) of the primary cilium.
6 antitubulin anti- Gli2 Gli2 mock wt MEFs Sufu / MEFs N C N C ShhN mock ShhN antitubulin wt MEFs Sufu / MEFs wt MEFs Sufu / MEFs wt MEFs Sufu / MEFs anti- N C N C N C N C N C N C 25 FL R 75 anti- RG1 anti- RG1 Supplemental Figure S4: Gli2 and protein levels are greatly reduced in both the nuclear and cytoplasmic fractions in the absence of Sufu Western blots of nuclear (N) and cytoplasmic (C) fractions derived from wild-type (wt) and Sufu / mouse embryonic fibroblasts (MEFs) through differential salt extraction. Gli2 is present in the cytoplasmic fraction and is barely detectable in the nuclear fraction of wt MEFs. Gli2 distribution is unaffected by Shh treatment. repressor (R) is present in the nuclear fraction of untreated MEFs and disappears upon addition of Shh ligand. Full-length (FL) mainly resides in the cytoplasm and its level is enhanced by Shh stimulation. Lack of full-length nuclear Gli2 and could be due to rapid nuclear export or nuclear degradation, similar to Ci (Chen et al. 1999). Gli2 and are barely detectable in either the nucleus or cytoplasm of Sufu-deficient MEFs. RG1 and tubulin were used to assess successful isolation of the nuclear and cytoplasmic fractions respectively; numbers on the right indicate positions of protein standards.
7 A 1.2 MEF embryo 1.2 MEF embryo wt Sufu / wt Sufu / wt Sufu / wt Sufu / Supplemental Figure S5: Gli2 and transcript levels are not significantly changed in Sufu / MEFs or embryos (A) Measurement of Gli2 transcripts in Sufu / mouse embryonic fibroblasts (MEFs) or embryos. The ratio of Gli2 to β-actin transcript was calculated and normalized to the wild-type value. Results are the mean of three independent experiments. Error bar indicates standard deviation (s.d.). () Measurement of transcripts in Sufu / MEFs or embryos. The ratio of to β-actin transcript was calculated and normalized to the wild-type value. Results are the mean of three independent experiments. Error bar indicates standard deviation (s.d.).
8 A % ciliated cells 9% 8% AC DAPI merge 6% 4% 2% AC DAPI merge % wild-type MEFs Kif3a / MEFs Supplemental Figure S6: Primary cilia fail to form in the absence of Kif3a (A) Immunofluorescence of wild-type (wt) and Kif3a / mouse embryonic fibroblasts (MEFs) using antibodies against acetylated (AC) tubulin (labeling primary cilia, red, arrows). Primary cilia cannot be detected in Kif3a-deficient MEFs. () Quantification of ciliated cells in wt and Kif3a / MEFs. Error bars are standard deviation (s.d.).
9 A 25 wild-type MEFs 12 Kif3a / MEFs Supplemental Figure S7: Hh pathway reporters can be activated only by exogenous Gli activators in Kif3a / mouse embryonic fibroblasts Wild-type (A) or Kif3a / () mouse embryonic fibroblasts (MEFs) were transfected with the 8xGliSluc reporter, the prl-tk reporter (for normalization), and empty vector or expression constructs encoding Smo, SmoM2, Ptch1, Gli2, and. In wild-type MEFs, the Hh reporter is activated by exogenous ShhN ligand, the chemical agonists purmorphamine (Sinha and Chen, 26), 2-αhydroxysterol (2αOHC) (Corcoran and Scott, 26), and exogenous Smo, SmoM2, and Gli2. Kif3a / MEFs are not activated by exogenous Hh ligand or chemical agonists; activation of 8xGliS-luc is achieved only by the addition of exogenous Gli activators (represented by Gli2). Data are representative of three independent experiments; error bars are standard deviation (s.d.).
10 A C wild-type MEFs ShhN ShhN ShhN ShhN Kif3a / MEFs wild-type MEFs Sufu / MEFs wild-type MEFs Kif3a / MEFs Sufu / MEFs Wnt3a Wnt3a Supplemental Figure S8: Sufu / and Kif3a / mouse embryonic fibroblasts have altered response to Shh, but not Wnt3a Mouse embryonic fibroblasts (MEFs) were transfected with Hh reporter (8xGliS-luc) or Wnt reporter (SuperTOPflash) and treated with either ShhN- or Wnt3a-conditioned medium. Date are representative of three independent experiments; error bars are standard deviations (s.d.). (A) Wild-type MEFs exhibit a robust response to ShhN, whereas Kif3a / MEFs are unresponsive. () Sufu / MEFs show an elevated basal level of 8xGliS-luc reporter activity when compared to wildtype MEFs, but do not exhibit a significantly elevated level of reporter activity when exposed to ShhN. Data are normalized to basal reporter activity in wild-type MEFs. (C) Wild-type, Sufu /, and Kif3a / MEFs display normal responsiveness to the canonical Wnt ligand Wnt3a assayed by the SuperTOPflash reporter.
11 A 35% wild-type MEFs 45% Sufu / MEFs 3% 25% 2% 4% 35% 3% 25% 15% 1% 5% 2% 15% 1% 5% % Gli1 FLAG Gli2 FLAG FLAG % Gli1 FLAG Gli2 FLAG FLAG 1% 9% 8% 7% 6% 5% 4% 3% 2% 1% % wild-type MEFs SPOPGli1 SPOPGli2 SPOP 1% 9% 8% 7% 6% 5% 4% 3% 2% 1% % Kif3a / MEFs Gli1SPOP Gli2SPOP SPOP C 9% 8% 7% 6% 5% 4% 3% 2% 1% % Smo / MEFs msmo zsmo dsmo Supplemental Figure S9: Quantification of localization of overexpressed proteins (A) Quantification of the percentage of cilia from wild-type or Sufu / mouse embryonic fibroblasts (MEFs) with localization of overexpressed Gli1-FLAG, Gli2-FLAG, or -FLAG. All three Gli proteins were detectable on cilia of Sufu / MEFs. () Quantification of the number of transfected cells expressing both SPOP and Gli1, Gli2, or that displayed colocalization of SPOP and Glis in discrete nuclear or cytoplasmic foci. In both wild-type and Kif3a / MEFs, over 9% of doubly transfected cells exhibited colocalization of SPOP and Gli2/ in punctae. In contrast, less than 1% of doubly transfected cells displayed Gli1 and SPOP colocalization. (C) Quantification of the number of transfected, ciliated Smo / MEFs showing mouse (msmo), zebrafish (zsmo), or Drosophila (dsmo) Smo localization along the length of the primary cilium. 7-8% of transfected ciliated cells showed ciliary localization of msmo and zsmo, whereas dsmo was barely detected in cilia.
12 A 8% wild-type MEFs 6% 4% 2% anti- % 9% 8% lentiviral vector (GFP) wild-type MEFs Kif3a shrna FL % 1 4% 2% % GFP dnkif3b-gfp antitubulin C 6 5 wild-type MEFs ShhN ShhN lentiviral vector (GFP) Kif3a shrna Supplemental Figure S1: Efficient knockdown of primary cilium function by overexpressed dnkif3b or Kif3a shrna (A) Quantification of ciliated cells in wild-type (wt) mouse embryonic fibroblasts (MEFs) and wt MEFs expressing dominant negative (dn) Kif3b or Kif3a shrna. GFP cells (harboring the lentiviral vector for shrna-mediated knockdown or mammalian expression vector for dnkif3b overexpression) were scored. Expression of dnkif3b or Kif3a shrna resulted in a significant reduction of ciliated cells. () Western blots of lysates derived from wt and Kif3a / MEFs, and wt MEFs expressing dnkif3b or Kif3a shrna. The ratio of full-length (FL) to repressor (R) is altered in Kif3a / MEFs due to loss of the primary cilium. Similar changes were observed in wt MEFs expressing dnkif3b or Kif3a shrna, indicating efficient disruption of primary cilium function. Tubulin was used as the loading control and numbers on the right indicate positions of protein standards. (C) Inactivation of primary cilium function via Kif3a shrna led to reduced Hh pathway activation assayed using the 8xGliS-luc reporter.
13 msmo anti-msmo DAPI merge Smo / MEFs zsmo anti-msmo DAPI merge dsmo FLAG anti-dsmo anti-flag merge Supplemental Figure S11: Expression of mouse, zebrafish and Drosophila Smo in Smodeficient mouse embryonic fibroblasts Mouse (msmo), zebrafish (zsmo) and FLAG-tagged Drosophila Smo (dsmo FLAG ) were expressed in Smo / mouse embryonic fibroblasts (MEFs). Anti-mSmo antibodies can detect both msmo and zsmo. In transfected MEFs that expresses Smo from a given species, similar levels of expression were obtained. Identical patterns of dsmo staining were observed using either anti-flag or anti-dsmo antibodies (Developmental Studies Hybridoma ank).
14 mgli2 m wt MEFs Gli2 DAPI merge mgli2 dsufu Gli2 DAPI merge m dsufu wt MEFs Gli2 dsufu merge dsufu merge mgli2 zsufu m zsufu wt MEFs Gli2 zsufu merge zsufu merge Supplemental Figure S12: Subcellular distributions of mouse Gli2 and in mouse embryonic fibroblasts in the presence of Drosophila or zebrafish Sufu Mouse Gli2 and (mgli2 and m, green) were expressed singly or together with either Drosophila Sufu (dsufu, red) or zebrafish Sufu (zsufu, red) in wild-type (wt) mouse embryonic fibroblasts (MEFs). mgli2 expression pattern is unaffected by the presence of dsufu. y contrast, dsufu appears to sequester m in the cytoplasm. This is consistent with the observation that dsufu has a higher affinity for m than mgli2 as determined by co-immunoprecipitation. Co-expression of zsufu with mgli2 and m led to prominent cytoplasmic distributions of mgli2 and m, consistent with sequestration of mgli2 and m by zsufu via high-affinity binding.
15 sense anti-sense sense anti-sense 9.5 dpc 1.5 dpc Supplemental Figure S13: is broadly expressed in Hh-responsive tissues during mouse embryogenesis Whole mount in situ hybridization, using digoxigenin-labeled ribo-probes on wild-type mouse embryos at 9.5 and 1.5 days post coitus (dpc). All views are lateral. is broadly expressed in the developing embryos, including the neural tube, somites, branchial arches, and limbs where Hedgehog (Hh) signaling plays a key role in tissue patterning. Sufu is also broadly expressed at this stage (not shown). This is consistent with our model in which regulates Gli protein levels by antagonizing Sufu activity. Control sense probe of gave no signal.
16 A CHO COS7 DAPI merge DAPI merge DAPI merge DAPI merge merge merge MEF MEF DAPI merge DAPI merge Supplemental Figure S14: and localization in CHO, COS7, and MEF cell lines (A) -Myc and -FLAG were expressed singly or together in CHO and COS7 cells. Notably, both nuclear and cytoplasmic punctae were seen in CHO cells. is primarily nuclear and perinuclear in COS7 cells, similar to published reports in this cell line (Nagai et al., 1997) and HeLa cells (Kwon et al., 26). is concentrated in the nucleus, but is redistributed to -containing foci both in CHO and COS7 cells. () -Myc was transfected in wild-type mouse embryonic fibroblasts (MEFs), and cells were incubated overnight in the absence or presence of the nuclear export inhibitor leptomycin (LM). was largely cytosolic in the absence of LM, but was retained in the nucleus and formed some foci/punctae after LM exposure. We speculate that the subcellular distribution of varies from cell line to cell line, and may depend on the relative amounts of both nuclear import/export factors, as well as components of relevant E3 ubiquitin ligase complexes.
17 anti- FLAG FoxC2 MyoD FLAG MyoD FLAG FLAG FoxC2 FLAG FoxC2 FLAG FLAG 75 MyoD 5 37 antitubulin Supplemental Figure S15: does not affect MyoD or FoxC2 protein levels (A) Western blot of lysates derived from HEK 293T cells expressing FLAG-tagged MyoD or FoxC2 singly or in combination with FLAG-tagged probed with anti-flag antibodies. Co-expression of MyoD or FoxC2 with has no effect on protein levels of MyoD or FoxC2. This suggests that the effects of on Gli2/3 protein levels (Fig. 5) are specific. Tubulin was used as the loading control and numbers on the right indicate positions of protein standards.
18 A 12% 1% 8% 6% J J J J J SPOP EXT2 4% 2% % ng 1 ng 4 ng 12 ng Construct (ng) 12% 1% J J J J SPOP EXT2 8% J 6% 4% 2% % ng 1 ng 4 ng 12 ng Construct (ng) Supplemental Figure S16: Gli2, but not Gli1, activity is inhibited by HEK 293T cells were transfected with either Gli1 (A) or Gli2 () and increasing amounts of or EXT2 (control). significantly reduces the ability of Gli2 to transactivate the 8xGliS-luc Hh reporter, but has a much less pronounced effect on Gli1. EXT2 expression does not significantly modulate Gli1 or Gli2 transactivation. Data shown are the mean of four independent experiments; error bars are standard error of mean (s.e.m.).
19 Sufu Cul3 FLAG-Gli2 HA-Ub Sufu Cul3 FLAG- HA-Ub Supplemental Figure S17: Cul3 alone (without ) fails to promote Gli2 and ubiquitination Western blots of immunoprecipitated Gli2 and (epitope-tagged with one copy of FLAG) to detect poly-ubiquitinated Gli proteins., a T (ric-a-brac-tramtrack-road complex) and MATH (Meprin and TRAF homolgy) domain-containing protein, serves as a substrate-specific adaptor to bridge substrate and Cul3-based E3 ligase [that also contains the RING finger protein RING-box protein 1 (Rbx1), also known as Roc1 or Hrt1]. The current model suggests that Rbx1 is required for neddylation of Cul3, which activates Cu3 complexes by providing a recognition site for ubiquitinconjugating enzyme (E2), leading to ubiquitination of Gli2 and (Sumara et al. 28). Cul3 alone (without ) is not sufficient to promote Gli2 or ubiquitination. IP, immunoprecipitation; W, Western blot.
20 A DMSO MG132 DMSO MG132 Gli2 Gli2 tubulin tubulin Supplemental Figure S18: SPOP reduces Gli2 and protein levels in a proteasomedependent manner HEK 293T cells were transfected with FLAG-Gli2 or FLAG-, in the absence or presence of. A reduction in Gli2 (A) and protein levels () was seen in the presence of ; this was inhibited with the addition of the proteasome inhibitor MG132. Tubulin serves as the loading control.
21 Gli2 Cul3 Gli2 Cul3 DAPI merge Cul3 Cul3 DAPI merge Cul3 Cul3 DAPI merge Gli2 Cul3 Gli2 Cul3 DAPI merge Cul3 Cul3 DAPI merge Supplemental Figure S19: directs Gli2 and to Cul3 () foci has been shown to be an adaptor protein for Cul3 ubiquitin ligases (Kwon et al., 26). In wildtype mouse embryonic fibroblasts (MEFs), co-expressed Cul3-Myc and Gli2-FLAG or -FLAG do not extensively overlap (top two rows). y contrast (and in agreement with the above report), coexpressed -HA and Cul3-Myc show overlap in punctae in the nucleus and cytoplasm (middle row). Transfection of with Gli2/ and Cul3 caused co-localization of Gli2 and with Cul3 punctae (bottom two rows). Thus, at the subcellular level, likely directs Gli2 and to the Cul3-based E3 ubiquitin ligase complex for polyubiquitination and subsequent degradation.
22 4 ShhN ShhN mock Sufu / MEFs shrna Supplemental Figure S2: knockdown in Sufu-deficient MEFs enhances Hh pathway activity (A) Hedgehog (Hh) reporter assays using the 8xGliS-luc reporter in Sufu / mouse embryonic fibroblasts (MEFs) and Sufu / MEFs expressing shrna in the presence or absence of exogenous Shh. Hh pathway activation is enhanced in Sufu / MEFs in which is efficiently knocked down, consistent with restored Gli2/3 protein levels (Fig. 5E). Restored Gli proteins mainly reside in the cytoplasm (data not shown). Error bars are standard deviation (s.d.).
23 wt MEFs AC DAPI merge Supplemental Figure S21: Overexpressed does not localize to the primary cilium in mouse embryonic fibroblasts (A) Myc-tagged was expressed in wild-type (wt) mouse embryonic fibrolblasts (MEFs) and detected using anti-myc antibodies. (green) does not localize to the primary cilium labeled by anti-acetylated (AC) tubulin antibodies (red).
24 12% Ptch1 / MEFs ShhN ShhN 1% 8% 6% 4% 2% % mptch1 (pcdna3) (ng) Supplemental Figure S22: Addition of Ptch1 to Ptch1 / MEFs decreases Hh responsiveness in the absence of exogenous Hh and fails to promote Hh pathway activation in the presence of Hh ligand Hedgehog (Hh) activity assays using the 8xGliS-luc reporter in Ptch1 / mouse embryonic fibroblasts (MEFs) transfected with varying quantities of Ptch1. Addition of increasing amounts of Ptch1 to Ptch1 / MEFs reduces Hh responsiveness in the absence of exogenous Shh. Reduced Hh responsiveness can be restored by Shh stimulation. However, unlike a similar experiment performed on Sufu / MEFs (Fig. 6A), increasing amounts of Ptch1 in Ptch1 / MEFs does not promote an increase in Hh responsiveness in the presence of exogenous Shh. This suggests that Ptch1 does not play a positive role in Hh activation, consistent with the observation that the Hh pathway is maximally activated in Ptch1 / MEFs in a ligand-independent manner. Error bars are standard deviation (s.d.).
25 A Gli1 Myc msufu FLAG Gli1 Myc zsufu FLAG Gli1 Myc dsufu FLAG Gli2 msufu FLAG Gli2 zsufu FLAG Gli2 dsufu FLAG msufu FLAG zsufu FLAG dsufu FLAG in IP in IP in IP in IP in IP in IP in IP in IP in IP Gli1 Myc msufu FLAG Gli1 Myc msufu D159AFLAG Gli2 msufu FLAG Gli2 msufu D159AFLAG msufu FLAG msufu D159AFLAG in IP in IP in IP in IP in IP in IP Supplemental Figure S23: Mouse and zebrafish Sufu bind Gli proteins more strongly than Sufu D159A or fly Sufu HEK 293T cells were transfected with various Sufu-FLAG constructs and either Gli-Myc, Gli2, or constructs. Immunoprecipitations were performed with anti-flag beads as described in Materials and Methods. (A) Mouse (msufu) and zebrafish Sufu (zsufu) bind strongly to Gli1, Gli2, and, and zsufu consistently binds more Gli2 than msufu. Fly Sufu (dsufu) binds weakly to Gli1, Gli2, and, correlating with its reduced ability to restore a dynamic range of Hedgehog (Hh) response and the partial rescue of Gli levels in Sufu / mouse embryonic fibroblasts (MEFs) (Figure 4). This may be due to coevolution of the relevant interacting surfaces of dsufu and Ci. () Wild-type msufu binds Gli1, Gli2, and with greater affinity than msufu D159A mutant. As with dsufu, this weakened binding affinity correlates with a reduced rescue of Hh response in Figure 4. In, input; IP, immunopreciptate; W, Western blot.
26 genotype phenotype Gli2 and proteins wild-type production of Gli activators and repressors Kif3a / (no primary cilium) failure to generate Gli activators and repressors Sufu / reduced levels of full-length Gli proteins and Gli repressors Sufu / ; Kif3a / reduced levels of full-length Gli proteins and Gli repressors Supplemental Figure S24: Summary of changes of Gli protein dynamics in Sufu and Kif3a mutants Production of Gli activators and repressors is compromised in Kif3a mutants in which primary cilia fail to form. y contrast, loss of Sufu led to great reduction in full-length Gli2 and and subsequently Gli repressors. Sufu / ; Kif3a / double mutants resemble Sufu single mutants both in their phenotypes and Gli protein dynamics, suggesting that Sufu functions independently of the primary cilium to control Gli protein levels.
27 A Sufu Sap18 wt MEFs Sufu Sap18 DAPI merge Sufu Sap18 Sufu / MEFs Sufu Sap18 DAPI merge Sufu Sap18 Ptch1 / MEFs 14 wild-type MEFs Sufu 6 Sap18 Sufu / MEFs DAPI 8 merge Ptch1 / MEFs Supplemental Figure S25: SAP18 localization is unaffected by Sufu, and SAP18 does not significantly affect Gli1 or Gli2 activity (A) Human SAP18-FLAG and mouse Sufu-Myc were co-expressed in wild-type (top row), Sufu / (middle row), and Ptch1 / (bottom row) mouse embryonic fibroblasts (MEFs). In all cases, SAP18 was found primarily in the nucleus, and Sufu in both the cytoplasm and nucleus. SAP18 was also found in the nucleus in the absence of Sufu in all three MEF lines (data not shown). () SAP18 was transfected alone, with Gli1, Gli2, or with Sufu and Gli1 or Gli2 in wild-type, Sufu /, or Ptch1 / MEFs. R (truncated after amino acid residue 722) was used as a positive control. Notably, SAP18 alone or in combination with Gli1 or Gli2 did not affect basal or stimulated levels of the 8xGliS-luc reporter. A small amount of overexpressed Sufu had a modest effect on Gli activation, similar to what was previously reported (Cheng and ishop, 22). No synergistic repression of Gli1 or Gli2 activity with Sufu and SAP18 was observed in any of the cell lines. y contrast, R consistently inhibited basal and Gli1/Gli2-stimulated reporter activity. Data are the mean of three independent experiments.
28 Sufu / MEFs (clone #1) mock ShhN FSK IMX Firefly/Renilla Firefly/β Gal 2.5 Sufu / MEFs (clone #16) mock ShhN FSK IMX 2.5 Sufu / MEFs (clone #17) mock ShhN FSK IMX Supplemental Figure S26: The Hh pathway is largely unaffected by pharmacologic modulation of protein kinase A (PKA) activity in Sufu / MEFs Three independent clones of Sufu / mouse embryonic fibroblasts (MEFs) were transfected with 8xGliS-luc Hh reporter and either prl-tk or hsp68-lacz for normalization. Treatment with ShhNconditioned medium had no effect. Stimulation of the PKA pathway with forskolin (FSK, which activates adenylyl cyclase) or 3-isobutyl-1-methylxanthine (IMX, which inhibits camp and cgmp phosphodiesterases) had no stimulatory or inhibitory effect in clones #16 and 17. We observed an increase in prl-tk reporter activity upon FSK or IMX treatment in clone #1, and thus normalized to hsp68-lacz, which showed no increase in β-galactosidase activity after PKA stimulation. Taken together, activation of PKA through two different mechanisms in three different cell lines resulted in either no change, or a modest increase in Hh reporter activity in the absence of Sufu. Error bars are standard deviation (s.d.).
29 Gli2 msufu FLAG ShhN ShhN in IP in IP msufu FLAG ShhN ShhN in IP in IP Supplemental Figure S27: Hh stimulation does not affect the interactions between Sufu and Gl2/3 Wild-type mouse embryonic fibroblasts were co-transfected with Sufu-FLAG and either Gli2, or constructs, and treated with mock or ShhN-conditioned media. Immunoprecipitations were performed with anti-flag beads as described Materials and Methods. Mouse Sufu (msufu) binds strongly to Gli2 and, and the physical interactions between Sufu and Gli2/3 are not affected by stimulation with ShhN-conditioned media. In, input; IP, immunopreciptate; W, Western blot.
30 A wild-type MEFs mock Smo SmoM2 Ptch Sufu / MEFs mock Smo SmoM2 Ptch1 Supplemental Figure S28: Sufu / MEFs are refractory to exogenous Smo and Ptch1 Wild-type or Sufu / () mouse embryonic fibroblasts (MEFs) were transfected with Hh reporters (8xGliS-luc), empty vector, and Smo, SmoM2, or Ptch1. In wild-type MEFs, Smo weakly activates the reporter in the absence of ligand, stronger activation is seen with SmoM2, and Ptch1 inhibits reporter activity by ~5%. In Sufu / MEFs, no significant effect is seen with Smo, Smo M2, or Ptch1, suggesting that the absence of Sufu renders cell insensitive to the actions of upstream membrane components of the pathway. Data are representative of three independent experiments; error bars are standard deviations (s.d.).
31 Supplemental Table S1: Quantification of endogenous Gli levels from Western blots Figure 2A: Gli2 wild-type Gli2 / Sufu / Ptch1 / Kif3a / Gli Figure 2A: wild-type / Sufu / Ptch1 / Kif3a / FL R Figure 2C wild-type Sufu / Kif3a / Kif3a / Sufu shrna Gli FL R Figure 3 wild-type Sufu / Sufu / Ptch1 dnkif3b Ptch1 / dnkif3b Gli FL R Figure 4D wild-type Sufu / Sufu / Sufu / Sufu / msufu zsufu dsufu Gli FL R Figure 5E wild-type Sufu / wild-type shrna Sufu / shrna Gli FL R Figure 6 wild-type Sufu / Ptch1 / Ptch1 / Sufu shrna Gli FL R Gli2, R, FL and tubulin band intensities were quantified using NIH ImageJ, and Gli/tubulin ratios calculated. For Figure 2A, Gli2 and null ratios were subtracted from all other samples. Gli/tubulin ratios were then normalized to the wild-type sample on each gel.
32 Supplemental Materials and Methods Constructs Human SAP18-FLAG was a gift from Danny Reinberg (Zhang et al. 1997). Cullin3-Myc was provided by P. Renee Yew. A truncated form of in prk5 (original construct from Philip eachy) was generated through PCR; this form is truncated after amino acid 722. hsp68-lacz was a gift from rian lack. Cell lines COS-7 cells were maintained in DMEM supplemented with 1% fetal bovine serum (FS), penicillin/streptomycin, and L-glutamine (Gibco). CHO cells were maintained in Ham s F-12 (UCSF Cell Culture Facility) supplemented with 1% fetal bovine serum, penicillin/streptomycin, and L- glutamine. Cells were transfected with Fugene 6 (Roche) per the manufacturer s instructions. Leptomycin (Calbiochem) was used at 5 nm. RNA extraction and RT-PCR RNA was extracted from cells or embryos using the Rneasy Mini Kit (Qiagen). Primers used for RT- PCR were: Gli1: GACTGCCGCTGGGATGGTTGCAG (forward), GCGTGAATAGGACTTCCGACAGCCT (reverse); Gli2 forward 5 GTTCCAAGGCCTACTCTCGCCTG 3, reverse 5 CTTGAGCAGTGGAGCACGGACAT 3 (Hu et al. 26); forward 5 AGCAAGCAGGAGCCTGAAGTCAT 3, reverse 5 GTCTTGAGTAGGCTTTTGTGCAA 3 (uttitta et al. 23); Ptch1: AGCGTCTGGTAGACGCAGATGGC (forward), TAGGCCGTTGAGGTAGAAAGGGA (reverse); β- actin forward 5 TGTTACCAACTGGGACGACA 3, reverse 5 CTCTCAGCTGTGGTGGTGAA 3 (uttitta et al. 23). Nuclear-cytoplasmic fractionation Subcellular fractionation was performed essentially as described (arnfield et al. 25). riefly, harvested cell pellets were resuspended in uffer A (1 mm HEPES ph 7.5, 1 mm KCl, 1mM EDTA, 1 mm DTT,.4% NP-4) and incubated on ice for 15 min. Cell nuclei were pelleted by centrifugation at 1, rpm for 3 min. The cytoplasmic fraction was removed, and nuclear pellets were washed three times in uffer A prior to resuspension in uffer (2 mm HEPES ph 7.9, 4 mm NaCl, 1 mm EDTA, 1 mm DTT, 1% glycerol). Nuclear supernatants were collected after centrifugation at 13, rpm for 5 min. For Western blotting, anti-α tubulin (1:2, Sigma) and anti-rg1 (Imbalzano et al. 1996) (1:2, gift from Geeta Narlikar) were used to control for fraction purity. Co-immunoprecipitation Wild-type mouse embryonic fibroblasts (MEFs) were transfected with Sufu-FLAG and Gli2 or, using Fugene 6 (Roche). 48 hr post-transfection, cells were mock treated or exposed to a 1:5 dilution of ShhN-conditioned media for 1 hr. Cells were harvested and immunoprecipitated as described in Materials and Methods. Wnt responsiveness assays MEFs were transfected and treated as described in Materials and Methods, except SuperTOPflash was used in place of 8xGliS-luc. Wnt3A-conditioned medium was produced as described (Willert et al. 23).
33 β-galactosidase assays β-galactosidase activity was measured using a Luminescent β-galactosidase detection kit II (Clontech) on an LmaxII 384 luminometer (Molecular Devices) according to manufacturer s instructions. Pharmacological modulation of PKA activity Hh reporter activity assays were conducted as described in Materials and Methods. Forskolin (FSK) and 3-isobutyl-1-methylxanthine (IMX) were from Sigma, and were used at concentrations of 1 µm and 15 µm, respectively.
34 Supplemental References arnfield, P.C., Zhang, X., Thanabalasingham, V., Yoshida, M., and Hui, C.C. 25. Negative regulation of Gli1 and Gli2 activator function by Suppressor of fused through multiple mechanisms. Differentiation 73(8): uttitta, L., Mo, R., Hui, C.C., and Fan, C.M. 23. Interplays of Gli2 and and their requirement in mediating Shh-dependent sclerotome induction. Development 13(25): Chen, C.H., von Kessler, D.P., Park, W., Wang,., Ma, Y., and eachy, P.A Nuclear trafficking of Cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell 98(3): Cheng, S.Y. and ishop, J.M. 22. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18-mSin3 corepressor complex. Proc Natl Acad Sci U S A 99(8): Corcoran, R.. and Scott, M.P. 26. Oxysterols stimulate Sonic hedgehog signal transduction and proliferation of medulloblastoma cells. Proc Natl Acad Sci U S A 13(22): Hu, M.C., Mo, R., hella, S., Wilson, C.W., Chuang, P.-T., Hui, C.C., and Rosenblum, N.D. 26. GLI3-dependent transcriptional repression of Gli1, Gli2 and kidney patterning genes disrupts renal morphogenesis. Development 133(3): Imbalzano, A.N., Schnitzler, G.R., and Kingston, R.E Nucleosome disruption by human SWI/SNF is maintained in the absence of continued ATP hydrolysis. J iol Chem 271(34): Kwon, J.E., La, M., Oh, K.H., Oh, Y.M., Kim, G.R., Seol, J.H., aek, S.H., Chiba, T., Tanaka, K., ang, O.S. et al. 26. T domain-containing speckle-type POZ protein (SPOP) serves as an adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase. J iol Chem 281(18):
35 Nagai, Y., Kojima, T., Muro, Y., Hachiya, T., Nishizawa, Y., Wakabayashi, T., and Hagiwara, M Identification of a novel nuclear speckle-type protein, SPOP. FES Lett 418(1-2): Sinha, S. and Chen, J.K. 26. Purmorphamine activates the Hedgehog pathway by targeting Smoothened. Nat Chem iol 2(1): Sumara, I., Maerki, S., and Peter, M. 28. E3 ubiquitin ligases and mitosis: embracing the complexity. Trends Cell iol 18(2): Wang,., Fallon, J.F., and eachy, P.A. 2. Hedgehog-regulated processing of produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 1(4): Willert, K., rown, J.D., Danenberg, E., Duncan, A.W., Weissman, I.L., Reya, T., Yates, J.R., 3rd, and Nusse, R. 23. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423(6938): Zhang, Y., Iratni, R., Erdjument-romage, H., Tempst, P., and Reinberg, D Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89(3):
Supporting Information
 Supporting Information Kim et al. 10.1073/pnas.0912180106 SI Materials and Methods DNA Constructs. The N-terminally-tagged mouse Gli2 expression construct, pcefl/3 HA-Gli2, was constructed by ligating
Supporting Information Kim et al. 10.1073/pnas.0912180106 SI Materials and Methods DNA Constructs. The N-terminally-tagged mouse Gli2 expression construct, pcefl/3 HA-Gli2, was constructed by ligating
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Coordinated Translocation of Mammalian Gli Proteins and Suppressor of Fused to the Primary Cilium
 Coordinated Translocation of Mammalian Gli Proteins and Suppressor of Fused to the Primary Cilium Huiqing Zeng 1, Jinping Jia 1, Aimin Liu 1,2 * 1 Department of Biology, Eberly College of Science, The
Coordinated Translocation of Mammalian Gli Proteins and Suppressor of Fused to the Primary Cilium Huiqing Zeng 1, Jinping Jia 1, Aimin Liu 1,2 * 1 Department of Biology, Eberly College of Science, The
Phosphoinositides Regulate Ciliary Protein Trafficking to Modulate Hedgehog Signaling
 Developmental Cell Supplemental Information Phosphoinositides Regulate Ciliary Protein Trafficking to Modulate Hedgehog Signaling Francesc R. Garcia-Gonzalo, Siew Cheng Phua, Elle C. Roberson, Galo Garcia
Developmental Cell Supplemental Information Phosphoinositides Regulate Ciliary Protein Trafficking to Modulate Hedgehog Signaling Francesc R. Garcia-Gonzalo, Siew Cheng Phua, Elle C. Roberson, Galo Garcia
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous
 Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
293T cells were transfected with indicated expression vectors and the whole-cell extracts were subjected
 SUPPLEMENTARY INFORMATION Supplementary Figure 1. Formation of a complex between Slo1 and CRL4A CRBN E3 ligase. (a) HEK 293T cells were transfected with indicated expression vectors and the whole-cell
SUPPLEMENTARY INFORMATION Supplementary Figure 1. Formation of a complex between Slo1 and CRL4A CRBN E3 ligase. (a) HEK 293T cells were transfected with indicated expression vectors and the whole-cell
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh-
 1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Chairoungdua et al., http://www.jcb.org/cgi/content/full/jcb.201002049/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Expression of CD9 and CD82 inhibits Wnt/ -catenin
Supplemental material Chairoungdua et al., http://www.jcb.org/cgi/content/full/jcb.201002049/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Expression of CD9 and CD82 inhibits Wnt/ -catenin
p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO
 Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
Supplementary Information p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO Yuri Shibata, Masaaki Oyama, Hiroko Kozuka-Hata, Xiao Han, Yuetsu Tanaka,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure S1: Endogenous interaction between RNF2 and H2AX: Whole cell extracts from 293T were subjected to immunoprecipitation with anti-rnf2 or anti-γ-h2ax antibodies
SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure S1: Endogenous interaction between RNF2 and H2AX: Whole cell extracts from 293T were subjected to immunoprecipitation with anti-rnf2 or anti-γ-h2ax antibodies
Developmental Biology
 Developmental Biology 362 (2012) 141 153 Contents lists available at SciVerse ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology Dual function of suppressor of
Developmental Biology 362 (2012) 141 153 Contents lists available at SciVerse ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology Dual function of suppressor of
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer
 Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
Supplementary Information TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer Yabing Mu, Reshma Sundar, Noopur Thakur, Maria Ekman, Shyam Kumar Gudey, Mariya
supplementary information
 DOI: 10.1038/ncb2153 Figure S1 Ectopic expression of HAUSP up-regulates REST protein. (a) Immunoblotting showed that ectopic expression of HAUSP increased REST protein levels in ENStemA NPCs. (b) Immunofluorescent
DOI: 10.1038/ncb2153 Figure S1 Ectopic expression of HAUSP up-regulates REST protein. (a) Immunoblotting showed that ectopic expression of HAUSP increased REST protein levels in ENStemA NPCs. (b) Immunofluorescent
Supplemental Materials. STK16 regulates actin dynamics to control Golgi organization and cell cycle
 Supplemental Materials STK16 regulates actin dynamics to control Golgi organization and cell cycle Juanjuan Liu 1,2,3, Xingxing Yang 1,3, Binhua Li 1, Junjun Wang 1,2, Wenchao Wang 1, Jing Liu 1, Qingsong
Supplemental Materials STK16 regulates actin dynamics to control Golgi organization and cell cycle Juanjuan Liu 1,2,3, Xingxing Yang 1,3, Binhua Li 1, Junjun Wang 1,2, Wenchao Wang 1, Jing Liu 1, Qingsong
Supplementary Materials
 Supplementary Materials Supplementary Figure S1 Regulation of Ubl4A stability by its assembly partner A, The translation rate of Ubl4A is not affected in the absence of Bag6. Control, Bag6 and Ubl4A CRISPR
Supplementary Materials Supplementary Figure S1 Regulation of Ubl4A stability by its assembly partner A, The translation rate of Ubl4A is not affected in the absence of Bag6. Control, Bag6 and Ubl4A CRISPR
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
DOI:.38/ncb2822 a MTC02 FAO cells EEA1 b +/+ MEFs /DAPI -/- MEFs /DAPI -/- MEFs //DAPI c HEK 293 cells WCE N M C P AKT TBC1D7 Lamin A/C EEA1 VDAC d HeLa cells WCE N M C P AKT Lamin A/C EEA1 VDAC Figure
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells
 Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells Immunofluorescence staining of H3K9me3 and 53BP1 in PH and HGADFN003 (HG003) cells at 24 h after γ-irradiation. Scale bar,
Supplementary Figure S1: Defective heterochromatin repair in HGPS progeroid cells Immunofluorescence staining of H3K9me3 and 53BP1 in PH and HGADFN003 (HG003) cells at 24 h after γ-irradiation. Scale bar,
(a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable
 Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
SUPPLEMENTARY FIGURE LEGENDS
 SUPPLEMENTARY FIGURE LEGENDS Supplemental FIG. 1. Localization of myosin Vb in cultured neurons varies with maturation stage. A and B, localization of myosin Vb in cultured hippocampal neurons. A, in DIV
SUPPLEMENTARY FIGURE LEGENDS Supplemental FIG. 1. Localization of myosin Vb in cultured neurons varies with maturation stage. A and B, localization of myosin Vb in cultured hippocampal neurons. A, in DIV
Primary Cilia Can Both Mediate and Suppress Hedgehog Pathway- Dependent Tumorigenesis (Supplementary Figures and Materials)
 Primary Cilia Can Both Mediate and Suppress Hedgehog Pathway- Dependent Tumorigenesis (Supplementary Figures and Materials) Sunny Y. Wong, Allen D. Seol, Po-Lin So, Alexandre N. Ermilov, Christopher K.
Primary Cilia Can Both Mediate and Suppress Hedgehog Pathway- Dependent Tumorigenesis (Supplementary Figures and Materials) Sunny Y. Wong, Allen D. Seol, Po-Lin So, Alexandre N. Ermilov, Christopher K.
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11700 Figure 1: RIP3 as a potential Sirt2 interacting protein. Transfected Flag-tagged Sirt2 was immunoprecipitated from cells and eluted from the Sepharose beads using Flag peptide.
doi:10.1038/nature11700 Figure 1: RIP3 as a potential Sirt2 interacting protein. Transfected Flag-tagged Sirt2 was immunoprecipitated from cells and eluted from the Sepharose beads using Flag peptide.
supplementary information
 DOI: 10.1038/ncb1875 Figure S1 (a) The 79 surgical specimens from NSCLC patients were analysed by immunohistochemistry with an anti-p53 antibody and control serum (data not shown). The normal bronchi served
DOI: 10.1038/ncb1875 Figure S1 (a) The 79 surgical specimens from NSCLC patients were analysed by immunohistochemistry with an anti-p53 antibody and control serum (data not shown). The normal bronchi served
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplemental Information. The Hedgehog Pathway Effector Smoothened. Exhibits Signaling Competency in the Absence. of Ciliary Accumulation
 Chemistry & Biology, Volume 21 Supplemental Information The edgehog Pathway Effector Exhibits Signaling Competency in the Absence of Ciliary Accumulation Chih-Wei Fan, Baozhi Chen, Irene Franco, Jianming
Chemistry & Biology, Volume 21 Supplemental Information The edgehog Pathway Effector Exhibits Signaling Competency in the Absence of Ciliary Accumulation Chih-Wei Fan, Baozhi Chen, Irene Franco, Jianming
Appendix. Table of Contents
 Appendix Table of Contents Appendix Figures Figure S1: Gp78 is not required for the degradation of mcherry-cl1 in Hela Cells. Figure S2: Indel formation in the MARCH6 sgrna targeted HeLa clones. Figure
Appendix Table of Contents Appendix Figures Figure S1: Gp78 is not required for the degradation of mcherry-cl1 in Hela Cells. Figure S2: Indel formation in the MARCH6 sgrna targeted HeLa clones. Figure
File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables. File Name: Peer Review File Description:
 File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables File Name: Peer Review File Description: Primer Name Sequence (5'-3') AT ( C) RT-PCR USP21 F 5'-TTCCCATGGCTCCTTCCACATGAT-3'
File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables File Name: Peer Review File Description: Primer Name Sequence (5'-3') AT ( C) RT-PCR USP21 F 5'-TTCCCATGGCTCCTTCCACATGAT-3'
Supplementary information. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins
 Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This
 SUPPLEMENTAL FIGURE LEGEND Fig. S1. Generation and characterization of. (A) Coomassie staining of soluble hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This protein was expressed
SUPPLEMENTAL FIGURE LEGEND Fig. S1. Generation and characterization of. (A) Coomassie staining of soluble hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This protein was expressed
MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands)
 Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
S1a S1b S1c. S1d. S1f S1g S1h SUPPLEMENTARY FIGURE 1. - si sc Il17rd Il17ra bp. rig/s IL-17RD (ng) -100 IL-17RD
 SUPPLEMENTARY FIGURE 1 0 20 50 80 100 IL-17RD (ng) S1a S1b S1c IL-17RD β-actin kda S1d - si sc Il17rd Il17ra rig/s15-574 - 458-361 bp S1f S1g S1h S1i S1j Supplementary Figure 1. Knockdown of IL-17RD enhances
SUPPLEMENTARY FIGURE 1 0 20 50 80 100 IL-17RD (ng) S1a S1b S1c IL-17RD β-actin kda S1d - si sc Il17rd Il17ra rig/s15-574 - 458-361 bp S1f S1g S1h S1i S1j Supplementary Figure 1. Knockdown of IL-17RD enhances
SUPPLEMENTARY INFORMATION. Supplementary Figures S1-S9. Supplementary Methods
 SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
Supplementary Table 1. List of primers used in this study
 Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
Supplementary Table 1. List of primers used in this study Gene Forward primer Reverse primer Rat Met 5 -aggtcgcttcatgcaggt-3 5 -tccggagacacaggatgg-3 Rat Runx1 5 -cctccttgaaccactccact-3 5 -ctggatctgcctggcatc-3
The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein
 THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
THESIS BOOK The functional investigation of the interaction between TATA-associated factor 3 (TAF3) and p53 protein Orsolya Buzás-Bereczki Supervisors: Dr. Éva Bálint Dr. Imre Miklós Boros University of
Supporting Information
 Supporting Information Palmisano et al. 10.1073/pnas.1202174109 Fig. S1. Expression of different transgenes, driven by either viral or human promoters, is up-regulated by amino acid starvation. (A) Quantification
Supporting Information Palmisano et al. 10.1073/pnas.1202174109 Fig. S1. Expression of different transgenes, driven by either viral or human promoters, is up-regulated by amino acid starvation. (A) Quantification
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Developing Molecularly Targeted Therapies for Basal Cell Carcinoma. Ivor Caro, MD, FAAD
 Developing Molecularly Targeted Therapies for Basal Cell Carcinoma Ivor Caro, MD, FAAD Disclosures Genentech, Inc Medical Director, Dermatology (employee) Stock holder Hedgehog Signaling Pathway Fundamental
Developing Molecularly Targeted Therapies for Basal Cell Carcinoma Ivor Caro, MD, FAAD Disclosures Genentech, Inc Medical Director, Dermatology (employee) Stock holder Hedgehog Signaling Pathway Fundamental
s u p p l e m e n ta ry i n f o r m at i o n
 Figure S1 Characterization of tet-off inducible cell lines expressing GFPprogerin and GFP-wt lamin A. a, Western blot analysis of GFP-progerin- or GFP-wt lamin A- expressing cells before induction (0d)
Figure S1 Characterization of tet-off inducible cell lines expressing GFPprogerin and GFP-wt lamin A. a, Western blot analysis of GFP-progerin- or GFP-wt lamin A- expressing cells before induction (0d)
SUPPLEMENTARY FIGURES AND TABLE
 SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
SUPPLEMENTARY INFORMATION
 Figure S1 Treatment with both Sema6D and Plexin-A1 sirnas induces the phenotype essentially identical to that induced by treatment with Sema6D sirna alone or Plexin-A1 sirna alone. (a,b) The cardiac tube
Figure S1 Treatment with both Sema6D and Plexin-A1 sirnas induces the phenotype essentially identical to that induced by treatment with Sema6D sirna alone or Plexin-A1 sirna alone. (a,b) The cardiac tube
Supplementary Figure 1. MAT IIα is Acetylated at Lysine 81.
 IP: Flag a Mascot PTM Modified Mass Error Position Gene Names Score Score Sequence m/z [ppm] 81 MAT2A;AMS2;MATA2 35.6 137.28 _AAVDYQK(ac)VVR_ 595.83-2.28 b Pre-immu After-immu Flag- WT K81R WT K81R / Flag
IP: Flag a Mascot PTM Modified Mass Error Position Gene Names Score Score Sequence m/z [ppm] 81 MAT2A;AMS2;MATA2 35.6 137.28 _AAVDYQK(ac)VVR_ 595.83-2.28 b Pre-immu After-immu Flag- WT K81R WT K81R / Flag
Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2)
 Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
Supplemental Methods Cells and reagents. Synaptopodin knockdown (1) and dynamin knockdown (2) podocytes were cultured as described previously. Staurosporine, angiotensin II and actinomycin D were all obtained
SUPPLEMENTARY INFORMATION
 Supplementary Table 1. Cell sphingolipids and S1P bound to endogenous TRAF2. Sphingolipid Cell pmol/mg TRAF2 immunoprecipitate pmol/mg Sphingomyelin 4200 ± 250 Not detected Monohexosylceramide 311 ± 18
Supplementary Table 1. Cell sphingolipids and S1P bound to endogenous TRAF2. Sphingolipid Cell pmol/mg TRAF2 immunoprecipitate pmol/mg Sphingomyelin 4200 ± 250 Not detected Monohexosylceramide 311 ± 18
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein
 Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
effects on organ development. a-f, Eye and wing discs with clones of ε j2b10 show no
 Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
Supplemental Figure 1
 Supplemental Figure 1 A S100A4: SFLGKRTDEAAFQKLMSNLDSNRDNEVDFQEYCVFLSCIAMMCNEFFEGFPDK Overlap: SF G DE KLM LD N D VDFQEY VFL I M N FF G PD S100A2: SFVGEKVDEEGLKKLMGSLDENSDQQVDFQEYAVFLALITVMCNDFFQGCPDR
Supplemental Figure 1 A S100A4: SFLGKRTDEAAFQKLMSNLDSNRDNEVDFQEYCVFLSCIAMMCNEFFEGFPDK Overlap: SF G DE KLM LD N D VDFQEY VFL I M N FF G PD S100A2: SFVGEKVDEEGLKKLMGSLDENSDQQVDFQEYAVFLALITVMCNDFFQGCPDR
Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna
 Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna targeted for hamster PAQR3. At 24 h after the transfection,
Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna targeted for hamster PAQR3. At 24 h after the transfection,
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis
 Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC
 Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
Expanded View Figures
 MO reports PR3 dephosphorylates TZ Xian-o Lv et al xpanded View igures igure V1. PR3 dephosphorylates and inactivates YP/TZ., Overexpression of tight junction proteins Pals1 () or LIN7 () has no effect
MO reports PR3 dephosphorylates TZ Xian-o Lv et al xpanded View igures igure V1. PR3 dephosphorylates and inactivates YP/TZ., Overexpression of tight junction proteins Pals1 () or LIN7 () has no effect
(A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939 (5 µm)
 Supplementary Figure Legends Figure S1. Tankyrase inhibition suppresses cell proliferation in an axin/β-catenin independent manner. (A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939
Supplementary Figure Legends Figure S1. Tankyrase inhibition suppresses cell proliferation in an axin/β-catenin independent manner. (A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939
Myf5 is a direct target of long-range Shh signaling and Gli regulation for muscle specification
 Myf5 is a direct target of long-range Shh signaling and Gli regulation for muscle specification Marcus K. Gustafsson, 1,3,4 Hua Pan, 1,3,4 Deborah F. Pinney, 1,3 Yongliang Liu, 1,3 Anna Lewandowski, 1,3
Myf5 is a direct target of long-range Shh signaling and Gli regulation for muscle specification Marcus K. Gustafsson, 1,3,4 Hua Pan, 1,3,4 Deborah F. Pinney, 1,3 Yongliang Liu, 1,3 Anna Lewandowski, 1,3
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/439/ra78/dc1 Supplementary Materials for Small heterodimer partner mediates liver X receptor (LXR) dependent suppression of inflammatory signaling by promoting
www.sciencesignaling.org/cgi/content/full/9/439/ra78/dc1 Supplementary Materials for Small heterodimer partner mediates liver X receptor (LXR) dependent suppression of inflammatory signaling by promoting
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supporting Online Material Material and Methods References Supplemental Figures S1, S2, and S3
 Supporting Online Material Material and Methods References Supplemental Figures S1, S2, and S3 Sarbassov et al. 1 Material and Methods Materials Reagents were obtained from the following sources: protein
Supporting Online Material Material and Methods References Supplemental Figures S1, S2, and S3 Sarbassov et al. 1 Material and Methods Materials Reagents were obtained from the following sources: protein
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature7221 Brown fat selective genes 12 1 Control Q-RT-PCR (% of Control) 8 6 4 2 Ntrk3 Cox7a1 Cox8b Cox5b ATPase b2 ATPase f1a1 Sirt3 ERRα Elovl3/Cig3 PPARα Zic1 Supplementary Figure S1. stimulates
doi: 1.138/nature7221 Brown fat selective genes 12 1 Control Q-RT-PCR (% of Control) 8 6 4 2 Ntrk3 Cox7a1 Cox8b Cox5b ATPase b2 ATPase f1a1 Sirt3 ERRα Elovl3/Cig3 PPARα Zic1 Supplementary Figure S1. stimulates
Supplementary Figure 1. The CagA-dependent wound healing or transwell migration of gastric cancer cell. AGS cells transfected with vector control or
 Supplementary Figure 1. The CagA-dependent wound healing or transwell migration of gastric cancer cell. AGS cells transfected with vector control or 3xflag-CagA expression vector were wounded using a pipette
Supplementary Figure 1. The CagA-dependent wound healing or transwell migration of gastric cancer cell. AGS cells transfected with vector control or 3xflag-CagA expression vector were wounded using a pipette
(A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and a
 Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Predictive PP1Ca binding region in BIG3 : 1,228 1,232aa (-KAVSF-) HEK293T cells *** *** *** KPL-3C cells - E E2 treatment time (h)
 Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature06994 A phosphatase cascade by which rewarding stimuli control nucleosomal response A. Stipanovich*, E. Valjent*, M. Matamales*, A. Nishi, J.H. Ahn, M. Maroteaux, J. Bertran-Gonzalez,
doi: 10.1038/nature06994 A phosphatase cascade by which rewarding stimuli control nucleosomal response A. Stipanovich*, E. Valjent*, M. Matamales*, A. Nishi, J.H. Ahn, M. Maroteaux, J. Bertran-Gonzalez,
Peli1 negatively regulates T-cell activation and prevents autoimmunity
 Peli1 negatively regulates T-cell activation and prevents autoimmunity Mikyoung Chang 1,*, Wei Jin 1,5,*, Jae-Hoon Chang 1, Yi-chuan Xiao 1, George Brittain 1, Jiayi Yu 1, Xiaofei Zhou 1, Yi-Hong Wang
Peli1 negatively regulates T-cell activation and prevents autoimmunity Mikyoung Chang 1,*, Wei Jin 1,5,*, Jae-Hoon Chang 1, Yi-chuan Xiao 1, George Brittain 1, Jiayi Yu 1, Xiaofei Zhou 1, Yi-Hong Wang
SUPPLEMENTARY INFORMATION
 a c e doi:10.1038/nature10407 b d f Supplementary Figure 1. SERCA2a complex analysis. (a) Two-dimensional SDS-PAGE gels of SERCA2a complexes. A silver-stained SDSPAGE gel is shown, which reveals a 12 kda
a c e doi:10.1038/nature10407 b d f Supplementary Figure 1. SERCA2a complex analysis. (a) Two-dimensional SDS-PAGE gels of SERCA2a complexes. A silver-stained SDSPAGE gel is shown, which reveals a 12 kda
Supplementary Figure 1 Induction of cellular senescence and isolation of exosome. a to c, Pre-senescent primary normal human diploid fibroblasts
 Supplementary Figure 1 Induction of cellular senescence and isolation of exosome. a to c, Pre-senescent primary normal human diploid fibroblasts (TIG-3 cells) were rendered senescent by either serial passage
Supplementary Figure 1 Induction of cellular senescence and isolation of exosome. a to c, Pre-senescent primary normal human diploid fibroblasts (TIG-3 cells) were rendered senescent by either serial passage
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature05732 SUPPLEMENTARY INFORMATION Supplemental Data Supplement Figure Legends Figure S1. RIG-I 2CARD undergo robust ubiquitination a, (top) At 48 h posttransfection with a GST, GST-RIG-I-2CARD
doi: 10.1038/nature05732 SUPPLEMENTARY INFORMATION Supplemental Data Supplement Figure Legends Figure S1. RIG-I 2CARD undergo robust ubiquitination a, (top) At 48 h posttransfection with a GST, GST-RIG-I-2CARD
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus
 Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Boucher et al NCOMMS B
 1 Supplementary Figure 1 (linked to Figure 1). mvegfr1 constitutively internalizes in endothelial cells. (a) Immunoblot of mflt1 from undifferentiated mouse embryonic stem (ES) cells with indicated genotypes;
1 Supplementary Figure 1 (linked to Figure 1). mvegfr1 constitutively internalizes in endothelial cells. (a) Immunoblot of mflt1 from undifferentiated mouse embryonic stem (ES) cells with indicated genotypes;
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. Neither the activation nor suppression of the MAPK pathway affects the ASK1/Vif interaction. (a, b) HEK293 cells were cotransfected with plasmids encoding
Supplementary Figure 1 Supplementary Figure 1. Neither the activation nor suppression of the MAPK pathway affects the ASK1/Vif interaction. (a, b) HEK293 cells were cotransfected with plasmids encoding
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures
 Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
Type of file: PDF Title of file for HTML: Supplementary Information Description: Supplementary Figures Type of file: MOV Title of file for HTML: Supplementary Movie 1 Description: NLRP3 is moving along
Figure S1, Beyer et al.
 Figure S1, eyer et al. Pax7 Myogenin si sitrl Hoechst T = 72h 14 1.8.6.4.2 12 1 8 6 4 2 24h 48h 96h diff. sitrl siset1 212 72h diff. b1 td r t Se km MyH Vinculin Myogenin β-ctin Vinculin MW b1 ka td r
Figure S1, eyer et al. Pax7 Myogenin si sitrl Hoechst T = 72h 14 1.8.6.4.2 12 1 8 6 4 2 24h 48h 96h diff. sitrl siset1 212 72h diff. b1 td r t Se km MyH Vinculin Myogenin β-ctin Vinculin MW b1 ka td r
A. List of selected proteins with high SILAC (H/L) ratios identified in mass
 Supplementary material Figure S1. Interaction between UBL5 and FANCI A. List of selected proteins with high SILAC (H/L) ratios identified in mass spectrometry (MS)-based analysis of UBL5-interacting proteins,
Supplementary material Figure S1. Interaction between UBL5 and FANCI A. List of selected proteins with high SILAC (H/L) ratios identified in mass spectrometry (MS)-based analysis of UBL5-interacting proteins,
HCC1937 is the HCC1937-pcDNA3 cell line, which was derived from a breast cancer with a mutation
 SUPPLEMENTARY INFORMATION Materials and Methods Human cell lines and culture conditions HCC1937 is the HCC1937-pcDNA3 cell line, which was derived from a breast cancer with a mutation in exon 20 of BRCA1
SUPPLEMENTARY INFORMATION Materials and Methods Human cell lines and culture conditions HCC1937 is the HCC1937-pcDNA3 cell line, which was derived from a breast cancer with a mutation in exon 20 of BRCA1
Supplemental Figure 1. Western blot analysis indicated that MIF was detected in the fractions of
 Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
SUPPLEMENTARY INFORMATION
 Supplementary Discussion The cell cycle machinery and the DNA damage response network are highly interconnected and co-regulated in assuring faithful duplication and partition of genetic materials into
Supplementary Discussion The cell cycle machinery and the DNA damage response network are highly interconnected and co-regulated in assuring faithful duplication and partition of genetic materials into
(Stratagene, La Jolla, CA) (Supplemental Fig. 1A). A 5.4-kb EcoRI fragment
 SUPPLEMENTAL INFORMATION Supplemental Methods Generation of RyR2-S2808D Mice Murine genomic RyR2 clones were isolated from a 129/SvEvTacfBR λ-phage library (Stratagene, La Jolla, CA) (Supplemental Fig.
SUPPLEMENTAL INFORMATION Supplemental Methods Generation of RyR2-S2808D Mice Murine genomic RyR2 clones were isolated from a 129/SvEvTacfBR λ-phage library (Stratagene, La Jolla, CA) (Supplemental Fig.
Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the
 Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the genome-wide methylation microarray data. Mean ± s.d.; Student
Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the genome-wide methylation microarray data. Mean ± s.d.; Student
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
SUPPLEMENTARY FIGURES Supplementary Figure 1. (A) Left, western blot analysis of ISGylated proteins in Jurkat T cells treated with 1000U ml -1 IFN for 16h (IFN) or left untreated (CONT); right, western
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human
 Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
Fig. S1. Subcellular localization of overexpressed LPP3wt-GFP in COS-7 and HeLa cells. Cos7 (top) and HeLa (bottom) cells expressing for 24 h human LPP3wt-GFP, fixed and stained for GM130 (A) or Golgi97
Supplemental Information
 Supplemental Information Tobacco-specific Carcinogen Induces DNA Methyltransferases 1 Accumulation through AKT/GSK3β/βTrCP/hnRNP-U in Mice and Lung Cancer patients Ruo-Kai Lin, 1 Yi-Shuan Hsieh, 2 Pinpin
Supplemental Information Tobacco-specific Carcinogen Induces DNA Methyltransferases 1 Accumulation through AKT/GSK3β/βTrCP/hnRNP-U in Mice and Lung Cancer patients Ruo-Kai Lin, 1 Yi-Shuan Hsieh, 2 Pinpin
Supplementary Figure 1
 Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Supplemental Figures
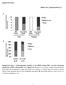 Supplemental Figures Supplemental Figure 1. Fasting-dependent regulation of the SREBP ortholog SBP-1 and lipid homeostasis mediated by the SIRT1 ortholog SIR-2.1 in C. elegans. (A) Wild-type or sir-2.1(lof)
Supplemental Figures Supplemental Figure 1. Fasting-dependent regulation of the SREBP ortholog SBP-1 and lipid homeostasis mediated by the SIRT1 ortholog SIR-2.1 in C. elegans. (A) Wild-type or sir-2.1(lof)
supplementary information
 Figure S1 Nucleotide binding status of RagA mutants. Wild type and mutant forms of MycRagA was transfected into HEK293 cells and the transfected cells were labeled with 32 Pphosphate. MycRagA was immunoprecipitated
Figure S1 Nucleotide binding status of RagA mutants. Wild type and mutant forms of MycRagA was transfected into HEK293 cells and the transfected cells were labeled with 32 Pphosphate. MycRagA was immunoprecipitated
Supplementary Figure 1. Baf60c and baf180 are induced during cardiac regeneration in zebrafish. RNA in situ hybridization was performed on paraffin
 Supplementary Figure 1. Baf60c and baf180 are induced during cardiac regeneration in zebrafish. RNA in situ hybridization was performed on paraffin sections from sham-operated adult hearts (a and i) and
Supplementary Figure 1. Baf60c and baf180 are induced during cardiac regeneration in zebrafish. RNA in situ hybridization was performed on paraffin sections from sham-operated adult hearts (a and i) and
Supplementary Figure 1 (Related with Figure 4). Molecular consequences of Eed deletion. (a) ChIP analysis identifies 3925 genes that are associated
 Supplementary Figure 1 (Related with Figure 4). Molecular consequences of Eed deletion. (a) ChIP analysis identifies 3925 genes that are associated with the H3K27me3 mark in chondrocytes (see Table S1,
Supplementary Figure 1 (Related with Figure 4). Molecular consequences of Eed deletion. (a) ChIP analysis identifies 3925 genes that are associated with the H3K27me3 mark in chondrocytes (see Table S1,
Supporting Information
 Supporting Information Gack et al. 1.173/pnas.8494715 SI Text Cell Culture, Transfection, and Reagents. HEK293T, MEF, Vero, EcoPack2-293 (D iosciences), HeLa, HCT116, Huh7, NHLF, 2fTGH wild-type, U3A,
Supporting Information Gack et al. 1.173/pnas.8494715 SI Text Cell Culture, Transfection, and Reagents. HEK293T, MEF, Vero, EcoPack2-293 (D iosciences), HeLa, HCT116, Huh7, NHLF, 2fTGH wild-type, U3A,
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
E3 ligase TRIM72 negatively regulates myogenesis by IRS-1 ubiquitination
 E3 ligase negatively regulates myogenesis by IRS-1 ubiquitination Young-Gyu Ko College of Life Science and Biotechnology Korea University MyHC immunofluorescence Lipid rafts are plasma membrane compartments
E3 ligase negatively regulates myogenesis by IRS-1 ubiquitination Young-Gyu Ko College of Life Science and Biotechnology Korea University MyHC immunofluorescence Lipid rafts are plasma membrane compartments
Luminescent platforms for monitoring changes in the solubility of amylin and huntingtin in living cells
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2016 Contents Supporting Information Luminescent platforms for monitoring changes in the
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS
 Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
TFEB-mediated increase in peripheral lysosomes regulates. Store Operated Calcium Entry
 TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
TFEB-mediated increase in peripheral lysosomes regulates Store Operated Calcium Entry Luigi Sbano, Massimo Bonora, Saverio Marchi, Federica Baldassari, Diego L. Medina, Andrea Ballabio, Carlotta Giorgi
