Supplementary Materials for
|
|
|
- Opal Hodge
- 5 years ago
- Views:
Transcription
1 /DC1 Supplementary Materials for An Inhibitor of Mutant IDH1 Delays Growth and Promotes Differentiation of Glioma Cells Dan Rohle, Janeta Popovici-Muller, Nicolaos Palaskas, Sevin Turcan, Christian Grommes, Carl Campos, Jennifer Tsoi, Owen Clark, Barbara Oldrini, Evangelia Komisopoulou, Kaiko Kunii, Alicia Pedraza, Stefanie Schalm, Lee Silverman, Alexandra Miller, Fang Wang, Hua Yang, Yue Chen, Andrew Kernytsky, Marc K. Rosenblum, Wei Liu, Scott A. Biller, Shinsan M. Su, Cameron W. Brennan, Timothy A. Chan, Thomas G. Graeber, Katharine E. Yen,* Ingo K. Mellinghoff* *Corresponding author. (K.E.Y.); (I.K.M.) Published 4 April 2013 on Science Express DOI: /science This PDF file includes: Materials and Methods Supplementary Text Figs. S1 to S16 Tables S1 and S2 References
2 Materials and Methods Determination of compound inhibition potency against the R132H. This assay was used for both the HTS and subsequent IC50 determinations to map the SAR of evolved compounds. In the primary reaction, the reduction of α-kg acid to 2-HG is accompanied by a concomitant oxidation of NADPH to NADP. The amount of NADPH remaining at the end of the reaction time is measured in a secondary diaphorase/resazurin reaction in which the NADPH is consumed in a 1:1 molar ratio with the conversion of resazurin to the highly fluorescent resorufin. Uninhibited reactions exhibit a low fluorescence at the end of the assay, while reactions in which the consumption of NADPH by R132H IDH1 has been inhibited by a small molecule show a high fluorescence. The primary reaction was performed in a volume of 50 µl 1X Buffer (150 mm NaCl, 20 mm Tris 7.5, 10 mm MgCl2, 0.05% (w/v) bovine serum albumin), contained 2 nm R132H IDH1, 1 mm alphaketoglutarate, and 4 µm NADPH, and was conducted for sixty minutes at 25 C. To perform the secondary reaction, 25 µl of 1X buffer containing 36 µg/ml diaphorase and 30 mm resazurin was added to the primary reaction and incubated for a further 10 minutes at 25 C. Florescence was read on a Spectramax platereader at Ex 544 Em 590. Recombinant protein was expressed and purified as described in (1). Compounds or compound dilutions were prepared in 100% DMSO concentration and diluted 1:100 into the final reaction. R132C IDH1 was assayed under similar conditions, with the exception that the 1X Buffer was 50 mm K2HP04, ph 6.5; 40 mm NaHCO3; 5 mm MgCl2; 10% glycerol; 0.03% (w/v) bovine serum albumin. Assay of the IDH1 wild-type enzyme reaction for determination of inhibitor potency to this nontargeted version of the enzyme. IDH1 wild-type enzyme was assayed in a modified version of the assay used for R132H IDH1. Since this enzyme converts NADP to NADPH stoichiometrically with the conversion of isocitrate to alpha-ketoglutarate, NADPH product can be continuously assayed by direct coupling to the diaphorase/rezasurin system and reading resorufin production at Ex 544 Em 590. Assays were conducted in 50 µl of 1X Buffer (150 mm NaCl; 20 mm Tris, ph 7.5; 10 mm MgCl2; 0.05% (w/v) BSA; 2 mm B-ME) containing 50 µm NADP, 70 µm DL-isocitrate and 31.2 ng/ml IDH1wt enzyme. The direct coupling system was comprised of 20 µg/ml diaphorase and 40 µm resazurin. Assay of the IDH2 R140Q and IDH2 R172K enzyme reaction for determination of inhibitor potency. Inhibitory potency against the IDH2 R140Q and IDH2 R172K enzymes was determined in an endpoint assay in which the amount of NADPH remaining at the end of the reaction was measured by the addition of a large excess of diaphorase and resazurin. IDH2 R140Q was diluted to 0.25 µg/ml in 40 µl 1X Assay Buffer (150 mm NaCl, 50 mm potassium phosphate ph 7.5, 10 mm MgCl2, 10% glycerol, 2 mm B-ME, 0.03% BSA) and incubated for 16 hours at 25 C in the presence of 1 µl of compound in DMSO. The reaction was started with the addition of 10 µl of Substrate Mix (20 µm NADPH, 8 µm alpha-ketoglutarate, in 1X Assay Buffer) and incubated for 1 hour at 25 C. Then, remaining NADPH was measured by the addition of 25 µl of Detection Mix (36 µg/ml diaphorase, 18 µm resazurin in 1X Assay Buffer), incubated for 5 minutes at 25 C, and read as described above. IDH2 R172K was assayed as for IDH2 R140Q with the following modifications: 1.25 µg/ml of protein was used, the Substrate Mix contained 50 µm NADPH and 6.4 µm alpha-ketoglutarate, and the compound was incubated for 1 hour before starting the reaction. 2
3 Assay of the IDH2 wild-type enzyme reaction for determination of inhibitor potency to this nontargeted family member of the enzyme. Inhibitory potency of compounds against the IDH2 WT enzyme was determined in a coupled assay to diaphorase. In this assay, production of NADPH by IDH2 WT was linked to a concomitant reduction of resazurin to the highly fluorescent resorufin. Enzyme was diluted to 0.06 µg/ml in 40 µl 1X Assay Buffer (150 mm NaCl, 50 mm potassium phosphate ph 7, 10 mm MgCl2, 10% glycerol, 2 mm B-ME, 0.03% BSA), to which 1 µl of compound was added in DMSO. The mixture was incubated for 1 hour at room temperature. The reaction was started with the addition of 10 µl of Substrate Mix (200 µm isocitrate, 175 µm NADP, 60 µg/ml diaphorase, 200 µm resazurin, in 1X Assay Buffer), and run for 30 minutes at room temperature. The reaction was halted with the addition of 25 µl of 6% SDS and read on a Spectramax Plate Reader at Ex544/Em590. Glioma Cell Lines. TS603, TS676, and TS516 glioma cells were derived from glioma patients who underwent tumor resection at Memorial Sloan-Kettering Cancer Center (MSKCC). All human samples were collected after patients signed informed consent according to MSKCC IRBguidelines. HT1080 cells were obtained from ATCC. Ink4a/Arf -/- neuroprogenitor cells (NPCs) were derived as previously described (2). Generation of sublines with inducible IDH1 knockdown. TS603 cells and HT1080 cells were infected with lentiviral particles containing either ptripz empty vector (OpenBiosystems), ptripz IDH1 sh484 (targeting the CDS), or ptripz IDH1 sh1894 (targeting the 3 -UTR). To produce lentiviral particles, 293T cells were co-transfected with ptripz vectors, pspax2 (Addgene #12260), and pmd2.g (Addgene #12259) and supernatant containing viral particles was collected at 36 and 72 hours. Generation of lines with R132H-IDH1 overexpression. Immortalized Normal Human Astrocytes (NHAs) and Ink4a/Arf -/- neurospheres were infected with retroviral particles containing pqcxih (Clontech #631516) or pqcxih R132H-IDH1. NHAs were a kind gift from Russ Pieper (University of California, San Francisco.) Ink4a/ARF-/- neuroprogenitor cells were isolated from the subventricular zone of mice 6 days after birth as previously described (2). To produce retroviral particles, GP2-293 cells were co-transfected with pqcxih vectors and pvsv-g (Clontech #631512). Soft Agar assays. Cells were seeded in triplicates at 150,000 cells/well in Neurocult media (Stem Cell Technologies) containing 0.4% Noble agar and growth factor supplements (20ng/mL EGF, 10ng/mL bfgf). Cells were plated between two layers of Neurocult media and growth factors containing 0.65% Nobel agar. Colonies were stained 4 weeks after plating with either crystal violet (0.005%) or thiazolyl blue tetrazolium bromide (Sigma, M5655) and quantified using imagine software (Oxford Optronix) and an image processing algorithm (Charm algorithm, Oxford Optronix). In-vivo experiments. SCID mice were injected subcutaneously with 10 6 glioma cells, which were suspended in 100 µl of a 50:50 mixture of growth media and Matrigel (BD #356237). Once tumors had reached a measurable size, mice were randomized into the indicated treatment groups. All experiments followed Institutional Animal Care and Use Committee (IACUC)-approved procedures. 3
4 Immunohistochemistry and Image Analysis. Paraffin-embedded tumor xenografts were sectioned at a thickness of 5 μm/slide. Antigen retrieval, immunohistochemical detection, and counter staining were performed using a Ventana Discovery Ultra autostainer (Ventana). Primary antibodies were diluted as indicated: cleaved caspase-3 (Cell Signaling, #9665) 1:1000, H3K9Me3 (Abcam, #8898) 1:300, and Ki67 (DAKO, #M7240) 1:100. To determine the percentage of immunohistochemical positive cells, we assessed the total number of positively stained nuclei 100/total number of nuclei on hematoxylin counterstaining. Three separate regions were analyzed for each tumor and captured with a camera (SPOT Imaging Solutions). Digitized images were segmented with segmentation techniques such as density and size thresholding to distinguish negative from positive objects using the image analysis software (ImageJ; NIH). The segmentation process resulted in the generation of binary images from which the number of stained objects and total numbers of nuclei was determined. Results were statistically analyzed using One-way ANOVA on GraphPad Prism 5. In-vitro Differentiation Assays. Glass coverslips (Corning) were coated with PBS diluted laminin (Invitrogen, ) for 2 hours, washed with PBS and then single cell dissociated cells were plated onto the coated coverslips at a density of 30,000/cm2 and cells were allowed to attach overnight. The following day individual coverslips were placed into differentiating conditions (1%FBS with 1uM retinoic acid) along with treatments for 4 or 7 days with the media being replenished on day 4. Cells were then fixed with 4% PFA for 30 minutes at room temperature, and permeabilized with a 0.05% triton-x solution. Primary antibodies were then applied; Nestin 1:200 (Millipore), GFAP 1:1000 (Dako), and DAPI followed by secondary antibodies with fluorescent tags (Invitrogen), and then mounted on slides. Digitized Images were obtained and segmentation techniques such as density and size thresholding was done on 3 fields of view from each slide with an average of 1500 cells/view using ImageJ software (NIH). Chromatin Immunoprecipitation: TS603 cells were grown in medium containing either AGI-5198 (1.5µM) or DMSO vehicle control. One week prior to harvest cells were transferred to differentiation medium (DMEM F12; 15mM HEPES; 0.06% glucose; B27 without vitamin A; N2; Insulin/transferrin; 1% FBS) containing freshly added retinoic acid (1µM). ChIP of non-crosslinked cells was then carried out using established ChIP methods. 350µg of lysate was immunoprecipitated-using anti-h3k9me3 (ab8898, abcam), H3K27me3 (#9733S, Cell Signaling) or Rabbit Control IgG (ab46540, abcam). After washing, ChIP DNA was eluted from protein G beads and analyzed by RT-PCR using SYBR green (Applied Biosystems). Relative occupancy was calculated using the standard curve method and fold enrichment versus IgG. Enrichment in AGI treated cells was normalized to vehicle control. Means and standard deviation were calculated from 4 technical replicates. RT-PCR. RNA was isolated from mg of tumor tissue using the Qiagen RNeasy Plus protocol. RNA concentrations were then normalized and cdna generated using the high-capacity cdna reverse transcription protocol from Applied Biosciences (ABI catalog number ). Q-PCR was on a 7900HT Fast Real Time PCR machine using a standard delta relative quantitation (ΔΔRQ) QPCR protocol. Cycles length was 50C 2 minutes, 95C 10 minutes, then 40 cycles of 95C 15 seconds and 60C 1 minute. TaqMan (R) Gene Expression Assay: ATP1A2 (FAM): SMID: Hs _m1, GFAP(FAM): SMID: Hs _m1, APQ4(FAM): SMID: Hs _m1, CNP(FAM): SMID: Hs _m1, CSPG4(NG2)(FAM): SMID: Hs _m1. RNA and DNA Extraction. DNA and RNA was extracted from microdissected sections from TS603 xenografts that had been snap frozen in liquid nitrogen and stored at -80ºC. Xenograft DNA was extracted with the Qiagen DNeasy Blood and Tissue Kit protocol, and xenograft RNA was 4
5 extracted using the Qiagen RNeasy Lipid Tissue protocol. In addition, DNA from TS603 cells treated with DMSO or 1.5uM AGI-5198 at passages 0, 2, 4, 15, and 20 was extracted using the Qiagen DNeasy Blood and Tissue Kit protocol. Data Analysis. TS603 expression data was RMA normalized from the raw CEL files in Partek. Hierarchal clustering was performed on log transformed and normalized data. Analysis of variance was performed using Partek Genomic Suite and genes were filtered for p-value<0.05. Clustering was performed using Cluster 3.0 with average Pearson correlation as the similarity metric and visualized using Treeview. Functional analysis was performed using GSEA with the MolSigDB C2- pathways downloaded from the Broad Institute website ( gsea/msigdb/collections.jsp). The rank list was created by collapsing to max probe with signal to noise ratio as metric. One thousand permutations were employed to generate nominal p-values. Genes significantly (p-value <= 0.05) upregulated or downregulated in both AGI mg/kg and 450 mg/kg treatment groups with a fold change >=1.5 were selected. Enrichment analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) using all human genes as the background set and with Ingenuity IPA using Agilent Human Genome U133 Plus 2.0 probes as the background set. The methylation array beta value data was generated from intensity values using Genome Studio Software (Illumina) and imported into Partek. The β- values are given by β = (methylated probe intensity/(methylated probe intensity + unmethylated probe intensity)). TS603 xenograft delta beta values were calculated for all probes as delta = mean β treated mean β vehicle. Corresponding intermediate grade glioma delta beta values were calculated as delta = mean β mutant mean β wildtype. CIMP Analysis in TS603 Cells. The previously published intermediate grade glioma (WHO Grade II/III) whole genome methylation beta values (3) were used to calculate mean beta values for each class and their frequency distribution was plotted alongside the vehicle-treated TS603 xenograft data without further normalization. Intermediate grade glioma (WHO Grade II/III) and high grade glioma (WHO Grade IV) methylation data was limited to only probes that corresponded to the transcription start sites (140,000 probes). The most variant probes were identified to create two unique sets of ~5000 probes that would define CIMP status. TS603 methylation data was then incorporated into either the LGG or the TCGA files, and the resulting datasets where analyzed with 2 dimensional hierarchical cluster on the un-standardized beta values using Pearson s dissimilarity as the similarity metric. Integration of Methylation and Transcription Data. For our integrated analysis of DNA methylation and RNA expression changes, methylation data were logit transformed and gene expression data were log2 transformed after RMA normalization. We calculated p-values using a Student s t-test (two-sample equal variance). P-values were then negative log10 transformed for the purpose of visualization, assigned directionality based on IDH1 mutation (low grade glioma tumors) or treatment (mouse xenograft), and plotted on log-log scatter plots. Negative directionality corresponds to a decrease and positive directionality corresponds to an increase in transcription or methylation. Signed log10 p-values for transcription and methylation probes were matched by gene symbol, based on the annotations provided by respective platform manufacturers (Illumina and Affymetrix). Threshold lines on the log10 plots were drawn at -1.3 and 1.3 (p-value = 0.05). All RNA expression arrays and DNA methylation arrays were run at the Genomics Core Facility at Memorial Sloan-Kettering Cancer Center. 5
6 Beta and Delta Beta Values. β-values represent the ratio of methylated/(methylated + unmethylated) DNA and are reported for ~ 485,000 probes in the Infinium Human Methylation450 Arrays. Figure 4 B shows the distribution of β-values in vehicle versus AGI-5198 treated TS603 xenograft tumors. For the prolonged in vitro exposure of TS603 cells (Fig. S16), delta beta values were calculated from biological duplicates as delta = mean β treated mean β vehicle for each of the four passage points. Delta beta values greater than 0.2 or less than -0.2 were included in the overlap analysis. Hierarchical Clustering. For analysis of gene expression patterns from AGI-5198-treated TS603 glioma xenografts, clustering was performed on log transformed and normalized data. Analysis of variance was performed using Partek Genomic Suite and genes were filtered for p-value < Clustering was performed using Cluster 3.0 with average Pearson correlation as the similarity metric and visualized using Treeview. Probes were filtered using an ANOVA p-value threshold of 0.05 between the three treatment groups (n = 6492 probes). Supplementary Text Changes in DNA methylation during prolonged in-vitro culture with midh1 inhibitor. Treatment of leukemia cells with the DNA methyltransferase inhibitor decitabine (DAC) has been reported to result in DNA hypomethylation (i.e., a left-shift in the β-value distribution) within a timeframe of days (4). This is in contrast to our observations following acute inhibition of mutant IDH1 in glioma cells. Active DNA demethylation involves multiple steps after the initial hydroxylation of 5- methylcytosine by TET proteins (5) and may follow slower kinetics than passive demethylation occurring when maintenance DNA methylation is blocked. We therefore examined the effects of AGI-5198 on genome-wide DNA methylation during prolonged drug exposure in-vitro. We cultured TS603 glioma cells continuously in the presence of 1.5µM AGI-5198 or vehicle, collected DNA samples at multiple passage numbers (# 2, #4, #15, #20), hybridized the DNA to methylation arrays, and determined the difference in DNA methylation between AGI-5198 and vehicle treated cells (expressed as a delta-β-value for each probe). Independent biological replicates were collected for each condition. At early passage numbers (passage # 2 and 4), we noted a narrow distribution of delta β-values indicating no significant differences in DNA methylation between vehicle and AGI treated samples. The number of probes with a decrease in delta β-value (demethylation) of 0.2 or greater increased markedly between the two early passages (n=3 and n=7 probes, respectively) and the late passages (n=8012 and n=6746 probes, respectively). The majority of probes (5935/8012 = 74.1 %) with a decrease in delta β-value of 0.2 or greater at passage 15 also showed a similar or greater decreased in DNA methylation at passage 20 (Fig.S16) whereas we observed minimal overlap (1 probe) between the two early passages. While these data point to an active role of the mutant IDH1 enzyme in epigenetic remodeling, changes in DNA methylation in response to inhibition of the mutant enzyme were limited to a small fraction of the genome (< 2 %) and followed kinetics that are unlikely to drive gene expression changes within the short timeframe of our in-vivo xenograft experiments. Gene expression programs associated with in-vivo growth inhibition by mutant IDH1 inhibitor. To identify pathways that are associated with the growth-inhibitory effects of AGI-5198, we ran Affymetrix RNA expression arrays for all TS603 xenograft tumors in the AGI-5198 dose-response experiment for which we were able to isolate RNA and DNA of sufficient quality and quantity (23/30 total tumors). 41 probes showed a statistically significant (p<0.05) change in RNA expression ( 1.5-fold) in both AGI-5198 treated cohorts (i.e., 150 mg/kg and 450 mg/kg groups)(table S2). Functional annotation algorithms (Ingenuity Systems, 6
7 assign many of these genes to Cardiovascular System Development and Function (PER1, CTGF, GADD45A, LEPR, PIK3R1, ACTA2, PECAM1) and Tissue Morphology (PER1, CHRNA1, CALCRL, CTGF, LEPR, PIK3R1, ACTA2, PTGDS, TSC22D3, SERPINE2, SLITRK6, GADD45A, PROS1, ST8SIA2, PECAM1). Integrated analysis of DNA methylation and RNA expression data. An integrated analysis of the DNA methylation data and RNA expression data showed that, in contrast to human gliomas where anti-correlated DNA methylation and gene expression changes are readily appreciated in a comparison between IDH-mutant and IDH wildtype tumors (Fig.S15A), much less change and no correlation is seen in a comparison of AGI-5198 and vehicle treated R132H-IDH1 mutant glioma xenografts (Fig.S15B/C). 7
8 References 1. L. Dang et al., Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739 (2009). doi: /nature C. Lu et al., IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483, 474 (2012). doi: /nature S. Turcan et al., IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483, 479 (2012). doi: /nature H.-C. Tsai et al., Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell 21, 430 (2012). doi: /j.ccr J. S. You, P. A. Jones, Cancer genetics and epigenetics: Two sides of the same coin? Cancer Cell 22, 9 (2012). doi: /j.ccr
9 Supplementary Figures Fig. S1. Assay for determination of compound inhibition potency against the R132H-IDH1 enzyme. A Diaphorase/Resazurin coupled system reaction was used for both the High-Throughput Screen and subsequent IC50 determinations to map the SAR of compounds (see Methods Section). Fig. S2 Pharmacokinetic in-vivo profile of AGI-5198 in TS603 glioma xenografts. (A) Dosing schedule for pharmacokinetic analysis of AGI-5198 in TS603 xenograft bearing mice. (B) Concentration of AGI-5198 measured in plasma and tumor samples. Fig. S3 Treatment with AGI-5198 does not result in loss of body weight. Shown are the body weight of SCID mice treated daily with AGI-5198 (450mg/kg daily by gavage; n=20 per group). Fig. S4 midh1 inhibitor reduces proliferation of R132H-IDH1 mutant glioma cells in mice. Shown is immunohistochemistry (IHC) of TS603 xenografts treated with vehicle (top) or AGI-5198 (bottom). TS603 glioma xenografts show reduced staining for Ki-67 (left panel), but no difference in staining for cleaved caspase-3 (right panel). Tumors were harvested after 16 days of treatment with AGI Scale bar = 100µM. Fig. S5 IDH1-knockdown impairs growth of R132H-IDH1 mutant glioma cells in soft-agar. Shown are the number of colonies of R132H-IDH1 glioma cells (TS603) engineered to express two different doxycycline-inducible IDH1-shRNAs and grown in soft agar in absence or presence of doxycycline (DOX) (* p < 0.05). Fig. S6 Doxycycline does not impair the in-vivo growth of R132H-IDH1 glioma cells (TS603). 9
10 Shown are tumor volumes of TS603 cells engineered to express the empty shrna vector and treated with doxycyline. Doxycycline-containing chow was started after subcutaneous xenograft tumors were established. n = 15 mice per cohort. Fig. S7 Depletion of midh1 protein and intratumoral R-2HG in TS603-IDH1-shRNA xenograft tumors from doxycycline-treated mice. The left panel shows IHC with an R132H-IDH1 specific antibody. The right panel shows intratumoral R-2HG concentrations. Mice were treated with doxycycline, vehicle, or AGI-5198 as indicated in the text. Scale bar =100µM. n=15 mice per cohort. Fig. S8 IDH1 knockdown impairs growth of R132C-IDH1 mutant HT1080 fibrosarcoma cells. (A) Western blot of HT1080 cells that were engineered to express empty vector or two doxycyclineinducible IDH1 shrnas and treated with 2.5 µg/ml doxycycline (DOX) or vehicle for 4 days. (B) Doxycycline-induced IDH1 knockdown impairs colony formation of HT1080 cells in soft agar (* p < 0.05). (C) HT1080 xenografts that harbor the inducible IDH1 shrna show impaired growth when mice are treated with doxycycline (* p < 0.05). n = 11 mice per cohort. Doxycycline was started after subcutaneous tumors were established. Fig. S9 R132H-IDH1 mutant TS603 glioma cells and IDH1/2-mutant gliomas show an increased number of hypermethylated gene loci using Illumina Infinium Human Methylation450 Arrays. Shown is a comparison of genome wide DNA methylation (β value distribution) of IDH1- mutant TS603 xenografts (n=6), IDH1/2- mutant human gliomas (n=49), and IDH1/2-wildtype gliomas (n=31). Note the increased number of highly-methylated probes (β values > 0.7) in IDH1/2-mutant gliomas and IDH1- mutant TS603 glioma xenografts (compared to IDH1/2-wildtype gliomas). Fig. S10 R132H-IDH1 mutant TS603 glioma cells are G-CIMP positive. IDH1- mutant TS603 glioma cells cluster with IDH1/2-mutant glioblastomas (WHO grade IV) (A) and IDH1/2-mutant intermediate grade gliomas (WHO Grade II/III) (B). Unsupervised twodimensional hierarchical clustering was performed using the top ~5000 most variant (β value) transcription start site (TSS) methylation probes (see Methods Section). Fig. S11 AGI-5198 (450 mg/kg) reduces H3K9m3 levels in IDH1 mutant glioma cells. 10
11 Shown are representative images of R132H-IDH1 mutant TS603 glioma xenografts treated with either vehicle (top left) or 450 mg/kg AGI-5198 (bottom left). The top right and bottom right panels show a negative (external granule cell layer of p7 murine cerebellum) and a positive (murine myocardium, E16.5) staining control. Scale bar = 50µM. Fig. S12 AGI-5198 does not affect H3K9M3 methylation in IDH1-wildtype cells. (A) Quantification of H3K9 trimethylation in IDH1-wildtype glioma xenografts (TS516) treated for two weeks with vehicle, 150 mg/kg AGI-5198, or 450 mg/kg AGI (B) Western blot of NHAs engineered to express empty vector (pqcxih) or R132H-IDH1 (pqcxih) and treated with AGI (1.5 µm) for the indicated time (in days). NHAs were a kind gift from Russ Pieper (University of California, San Francisco.) Fig. S13 AGI-5198 impairs tumor cell proliferation in R132H-IDH1 glioma xenografts. Shown are representative Ki-67 IHC images (left) and image quantification (% positive cells)(right) from mice treated with for two weeks with vehicle, 150 mg/kg AGI-5198, or 450 mg/kg AGI-5198 (* p < 0.05). White scale bars represent 200µM. n=15 mice per cohort. Fig. S14 Hierarchical clustering of genome wide RNA expression from vehicle and AGI-5198-treated TS603 glioma xenografts. Clustering was performed on log transformed and normalized data (see Methods Section). Fig. S15 AGI-5198-induced changes in RNA expression (x-axis) correlate poorly with changes in DNA methylation (y-axis) in R132H-IDH1 glioma xenografts. The relationship between IDH1-mutation associated differences in RNA expression and DNA methylation in human low grade gliomas is shown in (A) as a point of reference. Signed log10 p- values express the statistical significance of the difference. Threshold lines are drawn to represent statistical significance at p-value=0.05 (log10(0.05) = 1.3). Directionality (sign) is assigned based on increasing (positive) or decreasing (negative) methylation (y-axis) or RNA expression (xaxis)(see Methods Section). Green data points represent probes with significant decrease in methylation and increase in gene expression (methylation signed log10 p-value <-1.3, gene expression signed log10 p-value >1.3). Red data points represent probes with a significant increase in methylation and decrease in gene expression (methylation signed log10 p-value >1.3, gene expression signed log10 p-value < -1.3). While anti-correlated DNA methylation and RNA expression changes are obvious in the comparison of IDH-mutant versus IDH-wildtype human 11
12 gliomas (A), much less change and no correlation is seen in response to midh1-inhibitor treatment of the xenografts tumors (B: 150 mg/kg, C: 450 mg/kg). Fig. S16 Progressive changes in DNA methylation during continued in-vitro exposure of R132H-IDH1 TS603 cells to AGI Shown is the number of probes on the Illumina Infinium Human Methylation 450 Arrays that show a decrease in DNA-methylation (delta β < -0.2) between vehicle and AGI-5198 treated cells at each indicated passage number. Replicate arrays were run for all conditions. Note the increasing number of probes that show a consistent change in DNA methylation (Venn Diagram) between the two early passages (passage # 2 and # 4, top) versus two late passages (passage # 15 and 20, bottom). 12
13 Table S1. List of genes that are upregulated or downregulated in TS603 xenografts in response to AGI-5198 treatment (compared to vehicle). Direction Rank Fold Change Probeset ID Gene Symbol Gene Title p-value UP _at TNNC1 troponin C type 1 (slow) UP _s_at COL1A2 collagen, type I, alpha UP _at EDIL3 UP _at IGFL1 UP _s_at MFAP5 UP _x_at PTGDS UP _x_at PTGDS EGF-like repeats and discoidin I-like domains 3 insulin growth factor-like family member 1 microfibrillar associated protein 5 prostaglandin D2 synthase 21kDa (brain) prostaglandin D2 synthase 21kDa (brain) UP _at COL1A2 Collagen, type I, alpha UP _s_at COL1A2 collagen, type I, alpha UP _s_at IGFBP7 UP _at ZBTB16 UP _s_at CXCL14 UP _s_at CXCL14 UP _s_at MYL9 UP _x_at PTGDS UP _at MFAP5 UP _at NNMT UP _at ANKRD1 UP _at AQP1 UP _at ACTA2 UP _s_at S100A4 UP _s_at CES1 UP _s_at NNMT UP _at LOC insulin-like growth factor binding protein 7 zinc finger and BTB domain containing 16 chemokine (C-X-C motif) ligand 14 chemokine (C-X-C motif) ligand 14 myosin, light polypeptide 9, regulatory prostaglandin D2 synthase 21kDa (brain) microfibrillar associated protein 5 nicotinamide N- methyltransferase ankyrin repeat domain 1 (cardiac muscle) aquaporin 1 (Colton blood group) actin, alpha 2, smooth muscle, aorta S100 calcium binding protein A4 (calcium protein, calvasculin, metastasin, murine placental homolog) carboxylesterase 1 (monocyte/macrophage serine esterase 1) nicotinamide N- methyltransferase hypothetical gene supported by AJ UP _at AQP4 aquaporin UP _at EPHB1 EPH receptor B UP _s_at TIMP3 UP _at CTGF TIMP metallopeptidase inhibitor 3 (Sorsby fundus dystrophy, pseudoinflammatory) connective tissue growth factor
14 UP _s_at C19ORF33 UP _s_at ATP1A2 UP _s_at MMP11 chromosome 19 open reading frame 33 ATPase, Na+/K+ transporting, alpha 2 (+) polypeptide matrix metallopeptidase 11 (stromelysin 3) DOWN _at CREG2 DOWN _s_at RGS4 DOWN _x_at --- DOWN _at --- DOWN _at RAB27B DOWN _at PLA2G7 cellular repressor of E1Astimulated genes 2 regulator of G-protein signalling 4 Transcribed locus, weakly similar to NP_ hypothetical protein FLJ25952 [Homo sapiens] CDNA FLJ45742 fis, clone KIDNE RAB27B, member RAS oncogene family phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) /// phospholipase A2, group VII (plateletactivating factor acetylhydrolase, plasma) DOWN _at DOCK2 Dedicator of cytokinesis
15 Table S2. List of genes that are upregulated or downregulated (compared to vehicle) in TS603 glioma xenografts treated with both doses of AGI-5198 (150 mg/kg and 450 mg/kg). Direction Probeset ID Gene Symbol Gene Title UP _x_at PTGDS UP _x_at PTGDS UP _at CHRNA1 UP _x_at PTGDS UP _a_at COL5A1 UP _at FRMD4B UP _at ST8SIA2 prostaglandin D2 synthase 21kDa (brain) prostaglandin D2 synthase 21kDa (brain) cholinergic receptor, nicotinic, alpha 1 (muscle) prostaglandin D2 synthase 21kDa (brain) Collagen, type V, alpha 1 FERM domain containing 4B ST8 alpha-nacetylneuraminide alpha-2,8- sialyltransferase mg/kg p-value 150 mg/kg fold change 450 mg/kg p-value 450 mg/kg fold change UP _at CLDN11 claudin UP _at DNAJB4 UP _s_at TSC22D3 UP _at NR1D2 UP _at CRADD UP _s_at CARD16 /// CASP1 UP _at CALCRL UP _at KCNIP1 UP _at CTGF UP _at PLP2 UP _s_at KLF9 UP _at IFI27 DnaJ (Hsp40) homolog, subfamily B, member 4 TSC22 domain family, member 3 nuclear receptor subfamily 1, group D, member 2 CASP2 and RIPK1 domain containing adaptor with death domain caspase recruitment domain family, member 16 /// caspase 1, apoptosis-related cysteine calcitonin receptor-like Kv channel interacting protein 1 connective tissue growth factor proteolipid protein 2 (colonic epitheliumenriched) Kruppel-like factor 9 interferon, alphainducible protein
16 UP _at LOC UP _at SDPR UP _at ZBTB16 uncharacterized LOC serum deprivation response zinc finger and BTB domain containing UP _at UP _at PIK3R1 UP _at GADD45A phosphoinositide- 3-kinase, regulatory subunit 1 (alpha) growth arrest and DNA-damageinducible, alpha UP _at LEPR leptin receptor UP _s_at PROS1 protein S (alpha) UP _at ACTA2 UP _at PER1 UP _at PECAM1 UP _s_at HMGA2 UP _at ZNF883 UP _at CA13 /// LOC DOWN _s_at WSB1 DOWN _at SLITRK6 DOWN _at SERPINE2 DOWN _at PPFIA2 DOWN _at LYPD1 DOWN _at SLITRK6 actin, alpha 2, smooth muscle, aorta period homolog 1 (Drosophila) platelet/endothelial cell adhesion molecule 1 high mobility group AT-hook 2 zinc finger protein 883 carbonic anhydrase XIII /// uncharacterized LOC WD repeat and SOCS box containing 1 SLIT and NTRKlike family, member 6 serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), me protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein LY6/PLAUR domain containing 1 SLIT and NTRKlike family, member DOWN _s_at RSPO3 R-spondin DOWN _s_at SLITRK6 SLIT and NTRKlike family, member
17 Fig.S1
18 Fig.S2 A Dose (mpk) # of time point # of Dose Comments Time points (hr) post last dose BID 12 hr interval BID 12 hr interval Vehicle 8 3 BID 12 hr interval 1, 3, 8, 12, 24 1, 3, 8, 12, 24 0 B
19 Fig.S Body weight (g) Vehicle 450 mg/kg 0 Day -6 Day -1 Day 4 Day 9 Day 14 Day 19
20 Fig.S4 Ki-67 IHC Cl. Caspase Vehicle AGI-5198 (450 mg/kg)
21 Fig.S5 R132H-IDH1 (TS603 glioma) 6000 * * DOX IDH1 shrna#1 IDH1 shrna#2 Colony number
22 Fig.S6 R132H-IDH1 (TS603 glioma) Tumor growth (fold day 0) vector vector Dox Days
23 R-2-HG (mm) Vehicle AGI-5198 IDH1 shrna#1 IDH1 shrna#2 Fig.S7 Vector Vector IDH1 shrna IDH1 shrna IHC: R132H-IDH
24 Fig.S8 A B C Colony Number * * Tumor Volume IDH1-shRNA IDH1-shRNA Dox * DOX vector IDH1 shrna#1 IDH1 shrna# Days HT1080 Fibrosarcoma R132C-IDH1
25 Fig.S IDH1-wt glioma (n=31) IDH1-mt glioma (n=49) Number of Probes TS603 xeno (n=6) DNA methylation value
26 Fig.S10 A B Glioblastoma (WHO Grade IV) WHO Grade II/III Glioma IDH Mutant CIMP POS Tumors IDH Mutant CIMP POS Tumors TS603 (R132H-IDH1) CIMP NEG Tumors TS603 (R132H-IDH1) CIMP NEG Tumors
27 Fig.S11 R132H-IDH1 (TS603 glioma) Glioma Xenografts Vehicle Staining Control AGI-5198(450mg/kg) Negative IHC: H3K9m3 Positive
28 Fig.S12 A H3K9M3 (% positive cells) IDH1-wildtype glioma (TS516) B NHA vector NHA R132H-IDH1 IB: H3K9M3 IB: H3 [AGI-5198] (mg/kg)
29 Fig.S13 R132H-IDH1 (TS603 glioma) Vehicle AGI mg/kg AGI mg/kg IHC: Ki67 % positive cells Vehicle * * 150 NS 450 [AGI-5198] (mg/kg)
30 Fig.S Vehicle_mr1 Vehicle_mr2 Vehicle_mr4 Vehicle_mr5 Vehicle_mr6 Vehicle_mr7 Vehicle_mr9 Vehicle_mr10 150mg/kg_mr12 150mg/kg_mr13 150mg/kg_mr14 150mg/kg_mr15 150mg/kg_mr16 150mg/kg_mr17 150mg/kg_mr18 150mg/kg_mr19 450mg/kg_mr31 450mg/kg_mr32 450mg/kg_mr34 450mg/kg_mr36 450mg/kg_mr38 450mg/kg_mr39 450mg/kg_mr40
31 Fig.S15 A Gene expression Log p-values B Gene expression Log p-values C Gene expression Log p-values Methylation Log p-values Methylation Log p-values Demethylated and expressed in IDH mutant Methylated and repressed in IDH mutant Demethylated and expressed in 150 mg/kg Methylated and repressed in 150 mg/kg Demethylated and expressed in 450 mg/kg Methylated and repressed in 450 mg/kg Methylation Log p-values
32 Fig.S16 Passage 2 Probes n= Passage 4 Probes n=7 Passage 15 Probes n=8012 Passage 20 Probes n=
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH
 EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
Somatic mutations in the metabolic enzyme. An Inhibitor of Mutant IDH1 Delays Growth and Promotes Differentiation of Glioma Cells REPORTS
 process. The heterozygous IDH2/R140Q mutation was present in all of the colonies cultured with or without AGI-6780, confirming that the inhibitor promotes outgrowth and differentiation of cells harboring
process. The heterozygous IDH2/R140Q mutation was present in all of the colonies cultured with or without AGI-6780, confirming that the inhibitor promotes outgrowth and differentiation of cells harboring
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Erzsebet Kokovay, Susan Goderie, Yue Wang, Steve Lotz, Gang Lin, Yu Sun, Badrinath Roysam, Qin Shen,
 Cell Stem Cell, Volume 7 Supplemental Information Adult SVZ Lineage Cells Home to and Leave the Vascular Niche via Differential Responses to SDF1/CXCR4 Signaling Erzsebet Kokovay, Susan Goderie, Yue Wang,
Cell Stem Cell, Volume 7 Supplemental Information Adult SVZ Lineage Cells Home to and Leave the Vascular Niche via Differential Responses to SDF1/CXCR4 Signaling Erzsebet Kokovay, Susan Goderie, Yue Wang,
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the
 Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the genome-wide methylation microarray data. Mean ± s.d.; Student
Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the genome-wide methylation microarray data. Mean ± s.d.; Student
Supplementary Figure 1. IDH1 and IDH2 mutation site sequences on WHO grade III
 Supplementary Materials: Supplementary Figure 1. IDH1 and IDH2 mutation site sequences on WHO grade III patient samples. Genomic DNA samples extracted from punch biopsies from either FFPE or frozen tumor
Supplementary Materials: Supplementary Figure 1. IDH1 and IDH2 mutation site sequences on WHO grade III patient samples. Genomic DNA samples extracted from punch biopsies from either FFPE or frozen tumor
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3
 Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
Impact of hyper-o-glcnacylation on apoptosis and NF-κB activity SUPPLEMENTARY METHODS
 SUPPLEMENTARY METHODS 3D culture and cell proliferation- MiaPaCa-2 cell culture in 3D was performed as described previously (1). Briefly, 8-well glass chamber slides were evenly coated with 50 µl/well
SUPPLEMENTARY METHODS 3D culture and cell proliferation- MiaPaCa-2 cell culture in 3D was performed as described previously (1). Briefly, 8-well glass chamber slides were evenly coated with 50 µl/well
Supplementary Information Titles Journal: Nature Medicine
 Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Supplementary Information Titles Journal: Nature Medicine Article Title: Corresponding Author: Supplementary Item & Number Supplementary Fig.1 Fig.2 Fig.3 Fig.4 Fig.5 Fig.6 Fig.7 Fig.8 Fig.9 Fig. Fig.11
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Supplementary Figure 1: Neuregulin 1 increases the growth of mammary organoids compared to EGF. (a) Mammary epithelial cells were freshly isolated,
 1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1: Neuregulin 1 increases the growth of mammary organoids compared to EGF. (a) Mammary epithelial cells were freshly isolated, embedded in matrigel and exposed
1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1: Neuregulin 1 increases the growth of mammary organoids compared to EGF. (a) Mammary epithelial cells were freshly isolated, embedded in matrigel and exposed
m 6 A mrna methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer
 SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
General Laboratory methods Plasma analysis: Gene Expression Analysis: Immunoblot analysis: Immunohistochemistry:
 General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
HEK293FT cells were transiently transfected with reporters, N3-ICD construct and
 Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Information Luciferase reporter assay HEK293FT cells were transiently transfected with reporters, N3-ICD construct and increased amounts of wild type or kinase inactive EGFR. Transfections
Supplementary Figure 1: Digitoxin induces apoptosis in primary human melanoma cells but not in normal melanocytes, which express lower levels of the
 Supplementary Figure 1: Digitoxin induces apoptosis in primary human melanoma cells but not in normal melanocytes, which express lower levels of the cardiac glycoside target, ATP1A1. (a) The percentage
Supplementary Figure 1: Digitoxin induces apoptosis in primary human melanoma cells but not in normal melanocytes, which express lower levels of the cardiac glycoside target, ATP1A1. (a) The percentage
Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types.
 Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
Supplementary Figure 1 Comparison of open chromatin regions between dentate granule cells and other tissues and neural cell types. (a) Pearson correlation heatmap among open chromatin profiles of different
SUPPLEMENT. Materials and methods
 SUPPLEMENT Materials and methods Cell culture and reagents Cell media and reagents were from Invitrogen unless otherwise indicated. Antibiotics and Tet-certified serum were from Clontech. In experiments
SUPPLEMENT Materials and methods Cell culture and reagents Cell media and reagents were from Invitrogen unless otherwise indicated. Antibiotics and Tet-certified serum were from Clontech. In experiments
Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma
 Supplementary information for: Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma Rintaro Hashizume 1, Noemi Andor 2, Yuichiro Ihara 2, Robin Lerner 2, Haiyun
Supplementary information for: Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma Rintaro Hashizume 1, Noemi Andor 2, Yuichiro Ihara 2, Robin Lerner 2, Haiyun
hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This
 SUPPLEMENTAL FIGURE LEGEND Fig. S1. Generation and characterization of. (A) Coomassie staining of soluble hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This protein was expressed
SUPPLEMENTAL FIGURE LEGEND Fig. S1. Generation and characterization of. (A) Coomassie staining of soluble hexahistidine tagged GRP78 devoid of the KDEL motif (GRP78-His) on SDS-PAGE. This protein was expressed
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism SUPPLEMENTARY FIGURES, LEGENDS AND METHODS
 A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
SUPPLEMENTARY INFORMATION. Supplementary Figures S1-S9. Supplementary Methods
 SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
SUPPLEMENTARY INFORMATION
 Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Supplementary Figures Supplementary Figure S1. Binding of full-length OGT and deletion mutants to PIP strips (Echelon Biosciences). Supplementary Figure S2. Binding of the OGT (919-1036) fragments with
Data Sheet TIGIT / NFAT Reporter - Jurkat Cell Line Catalog #60538
 Data Sheet TIGIT / NFAT Reporter - Jurkat Cell Line Catalog #60538 Background: TIGIT is a co-inhibitory receptor that is highly expressed in Natural Killer (NK) cells, activated CD4+, CD8+ and regulatory
Data Sheet TIGIT / NFAT Reporter - Jurkat Cell Line Catalog #60538 Background: TIGIT is a co-inhibitory receptor that is highly expressed in Natural Killer (NK) cells, activated CD4+, CD8+ and regulatory
(A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and a
 Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION
 Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION X. Shawn Liu 1, 3, Bing Song 2, 3, Bennett D. Elzey 3, 4, Timothy L. Ratliff 3, 4, Stephen F. Konieczny
Supplementary Information POLO-LIKE KINASE 1 FACILITATES LOSS OF PTEN-INDUCED PROSTATE CANCER FORMATION X. Shawn Liu 1, 3, Bing Song 2, 3, Bennett D. Elzey 3, 4, Timothy L. Ratliff 3, 4, Stephen F. Konieczny
Supplementary Information
 Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
HCC1937 is the HCC1937-pcDNA3 cell line, which was derived from a breast cancer with a mutation
 SUPPLEMENTARY INFORMATION Materials and Methods Human cell lines and culture conditions HCC1937 is the HCC1937-pcDNA3 cell line, which was derived from a breast cancer with a mutation in exon 20 of BRCA1
SUPPLEMENTARY INFORMATION Materials and Methods Human cell lines and culture conditions HCC1937 is the HCC1937-pcDNA3 cell line, which was derived from a breast cancer with a mutation in exon 20 of BRCA1
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY INFORMATION doi:1.138/nature9814 a A SHARPIN FL B SHARPIN ΔNZF C SHARPIN T38L, F39V b His-SHARPIN FL -1xUb -2xUb -4xUb α-his c Linear 4xUb -SHARPIN FL -SHARPIN TF_LV -SHARPINΔNZF -SHARPIN
SUPPLEMENTARY DATA. Assessment of cell survival (MTT) Assessment of early apoptosis and cell death
 Overexpression of a functional calcium-sensing receptor dramatically increases osteolytic potential of MDA-MB-231 cells in a mouse model of bone metastasis through epiregulinmediated osteoprotegerin downregulation
Overexpression of a functional calcium-sensing receptor dramatically increases osteolytic potential of MDA-MB-231 cells in a mouse model of bone metastasis through epiregulinmediated osteoprotegerin downregulation
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans.
 Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure S1. Generation of LSL-EZH2 conditional transgenic mice.
 Downstream Col1A locus S P P P EP Genotyping with P1, P2 frt PGKneopA + frt hygro-pa Targeting vector Genotyping with P3, P4 P1 pcag-flpe P2 P3 P4 frt SApA CAG LSL PGKATG frt hygro-pa C. D. E. ormal KRAS
Downstream Col1A locus S P P P EP Genotyping with P1, P2 frt PGKneopA + frt hygro-pa Targeting vector Genotyping with P3, P4 P1 pcag-flpe P2 P3 P4 frt SApA CAG LSL PGKATG frt hygro-pa C. D. E. ormal KRAS
Table S1. Sequence of human and mouse primers used for RT-qPCR measurements.
 Table S1. Sequence of human and mouse primers used for RT-qPCR measurements. Ca9, carbonic anhydrase IX; Ndrg1, N-myc downstream regulated gene 1; L28, ribosomal protein L28; Hif1a, hypoxia inducible factor
Table S1. Sequence of human and mouse primers used for RT-qPCR measurements. Ca9, carbonic anhydrase IX; Ndrg1, N-myc downstream regulated gene 1; L28, ribosomal protein L28; Hif1a, hypoxia inducible factor
RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using
 Supplementary Information Materials and Methods RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using Trizol reagent (Invitrogen,Carlsbad, CA) according to the manufacturer's instructions.
Supplementary Information Materials and Methods RNA extraction, RT-PCR and real-time PCR. Total RNA were extracted using Trizol reagent (Invitrogen,Carlsbad, CA) according to the manufacturer's instructions.
(a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable
 Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
(A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC,
 Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
Supplemental Figure Legends Figure S1. Cell line characterization (A) Cells grown in monolayer were fixed and stained for surfactant protein-c (SPC, green) and co-stained with DAPI to visualize the nuclei.
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figure 1. mrna targets were found in exosomes and absent in free-floating supernatant. Serum exosomes and exosome-free supernatant were
 Supplementary Figure 1. mrna targets were found in exosomes and absent in free-floating supernatant. Serum exosomes and exosome-free supernatant were separated via ultracentrifugation and lysed to analyze
Supplementary Figure 1. mrna targets were found in exosomes and absent in free-floating supernatant. Serum exosomes and exosome-free supernatant were separated via ultracentrifugation and lysed to analyze
Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing)
 Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing) platform with laser manipulation to efficiently purify lung
Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing) platform with laser manipulation to efficiently purify lung
SUPPLEMENTARY FIGURES AND TABLES
 SUPPLEMENTARY FIGURES AND TABLES Supplementary Figure S1: CaSR expression in neuroblastoma models. A. Proteins were isolated from three neuroblastoma cell lines and from the liver metastasis of a MYCN-non
SUPPLEMENTARY FIGURES AND TABLES Supplementary Figure S1: CaSR expression in neuroblastoma models. A. Proteins were isolated from three neuroblastoma cell lines and from the liver metastasis of a MYCN-non
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation
 Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Nature Methods: doi: /nmeth.3115
 Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
Supplementary Figure 1 Analysis of DNA methylation in a cancer cohort based on Infinium 450K data. RnBeads was used to rediscover a clinically distinct subgroup of glioblastoma patients characterized by
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis
 Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
Supplementary Figure 1: STAT3 suppresses Kras-induced lung tumorigenesis (a) Immunohistochemical (IHC) analysis of tyrosine 705 phosphorylation status of STAT3 (P- STAT3) in tumors and stroma (all-time
Supplemental figure 1. PDGFRα is expressed dominantly by stromal cells surrounding mammary ducts and alveoli. A) IHC staining of PDGFRα in
 Supplemental figure 1. PDGFRα is expressed dominantly by stromal cells surrounding mammary ducts and alveoli. A) IHC staining of PDGFRα in nulliparous (left panel) and InvD6 mouse mammary glands (right
Supplemental figure 1. PDGFRα is expressed dominantly by stromal cells surrounding mammary ducts and alveoli. A) IHC staining of PDGFRα in nulliparous (left panel) and InvD6 mouse mammary glands (right
Supplementary data Supplementary Figure 1 Supplementary Figure 2
 Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array.
 Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
Supplementary Table 1. Genes analysed for expression by angiogenesis gene-array. Gene symbol Gene name TaqMan Assay ID UniGene ID 18S rrna 18S ribosomal RNA Hs99999901_s1 Actb actin, beta Mm00607939_s1
marker. DAPI labels nuclei. Flies were 20 days old. Scale bar is 5 µm. Ctrl is
 Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Supplementary Figure 1. Validation of astrocytes. Primary astrocytes were
 Supplementary Figure 1. Validation of astrocytes. Primary astrocytes were separated from the glial cultures using a mild trypsinization protocol. Anti-glial fibrillary acidic protein (GFAP) immunofluorescent
Supplementary Figure 1. Validation of astrocytes. Primary astrocytes were separated from the glial cultures using a mild trypsinization protocol. Anti-glial fibrillary acidic protein (GFAP) immunofluorescent
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Correlation between LKB1 and YAP expression in human lung cancer samples. (a) Representative photos showing LKB1 and YAP immunohistochemical staining in human
Supplementary Figures Supplementary Figure 1 Correlation between LKB1 and YAP expression in human lung cancer samples. (a) Representative photos showing LKB1 and YAP immunohistochemical staining in human
Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein
 Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein Lei Wang 1, Tian-Peng Zhang 1, Yuan Zhang 2, Hai-Lian
Protection against doxorubicin-induced myocardial dysfunction in mice by cardiac-specific expression of carboxyl terminus of hsp70-interacting protein Lei Wang 1, Tian-Peng Zhang 1, Yuan Zhang 2, Hai-Lian
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Rabbit Polyclonal antibody to NFkB p65 (v-rel reticuloendotheliosis viral oncogene homolog A (avian))
 Datasheet GeneTex, Inc : Toll Free 1-877-GeneTex (1-877-436-3839) Fax:1-949-309-2888 info@genetex.com GeneTex International Corporation : Tel:886-3-6208988 Fax:886-3-6208989 infoasia@genetex.com Date :
Datasheet GeneTex, Inc : Toll Free 1-877-GeneTex (1-877-436-3839) Fax:1-949-309-2888 info@genetex.com GeneTex International Corporation : Tel:886-3-6208988 Fax:886-3-6208989 infoasia@genetex.com Date :
A263 A352 A204. Pan CK. pstat STAT3 pstat3 STAT3 pstat3. Columns Columns 1-6 Positive control. Omentum. Rectosigmoid A195.
 pstat3 75 Pan CK A A263 A352 A24 B Columns 1-6 Positive control A195 A22 A24 A183 Rectal Nodule STAT3 pstat3 STAT3 pstat3 Columns 7-12 Omentum Rectosigmoid Left Ovary Right Ovary Omentum Uterus Uterus
pstat3 75 Pan CK A A263 A352 A24 B Columns 1-6 Positive control A195 A22 A24 A183 Rectal Nodule STAT3 pstat3 STAT3 pstat3 Columns 7-12 Omentum Rectosigmoid Left Ovary Right Ovary Omentum Uterus Uterus
Supplementary Materials for
 immunology.sciencemag.org/cgi/content/full/2/16/eaan6049/dc1 Supplementary Materials for Enzymatic synthesis of core 2 O-glycans governs the tissue-trafficking potential of memory CD8 + T cells Jossef
immunology.sciencemag.org/cgi/content/full/2/16/eaan6049/dc1 Supplementary Materials for Enzymatic synthesis of core 2 O-glycans governs the tissue-trafficking potential of memory CD8 + T cells Jossef
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung
 Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
well for 2 h at rt. Each dot represents an individual mouse and bar is the mean ±
 Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Supplementary data: Control DC Blimp-1 ko DC 8 6 4 2-2 IL-1β p=.5 medium 8 6 4 2 IL-2 Medium p=.16 8 6 4 2 IL-6 medium p=.3 5 4 3 2 1-1 medium IL-1 n.s. 25 2 15 1 5 IL-12(p7) p=.15 5 IFNγ p=.65 4 3 2 1
Supplementary Figure 1 (Related with Figure 4). Molecular consequences of Eed deletion. (a) ChIP analysis identifies 3925 genes that are associated
 Supplementary Figure 1 (Related with Figure 4). Molecular consequences of Eed deletion. (a) ChIP analysis identifies 3925 genes that are associated with the H3K27me3 mark in chondrocytes (see Table S1,
Supplementary Figure 1 (Related with Figure 4). Molecular consequences of Eed deletion. (a) ChIP analysis identifies 3925 genes that are associated with the H3K27me3 mark in chondrocytes (see Table S1,
Supplementary Figure 1
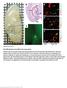 Supplementary Figure 1 AAV-GFP injection in the MEC of the mouse brain C57Bl/6 mice at 4 months of age were injected with AAV-GFP into the MEC and sacrificed at 7 days post injection (dpi). (a) Brains
Supplementary Figure 1 AAV-GFP injection in the MEC of the mouse brain C57Bl/6 mice at 4 months of age were injected with AAV-GFP into the MEC and sacrificed at 7 days post injection (dpi). (a) Brains
SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer.
 Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Supplemental information
 Carcinoemryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation y Chiang et al. Supplemental
Carcinoemryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation y Chiang et al. Supplemental
Supplementary Figure 1
 CD31 FN Supplementary Figure 1 a Multivariate Cox regression analysis of predicting factors for disease-free and overall survival in 435 HNSCC patients b FN staining in whole sections of HNSCC c FN expression
CD31 FN Supplementary Figure 1 a Multivariate Cox regression analysis of predicting factors for disease-free and overall survival in 435 HNSCC patients b FN staining in whole sections of HNSCC c FN expression
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
Gladstone Institutes, University of California (UCSF), San Francisco, USA
 Fluorescence-linked Antigen Quantification (FLAQ) Assay for Fast Quantification of HIV-1 p24 Gag Marianne Gesner, Mekhala Maiti, Robert Grant and Marielle Cavrois * Gladstone Institutes, University of
Fluorescence-linked Antigen Quantification (FLAQ) Assay for Fast Quantification of HIV-1 p24 Gag Marianne Gesner, Mekhala Maiti, Robert Grant and Marielle Cavrois * Gladstone Institutes, University of
Supplemental Table 1. Biochemical and Cellular Potency and Selectivity of PF
 Supplemental Table 1. Biochemical and Cellular Potency and Selectivity of PF- 02341066 Assay IC 50 nm Selectivity Ratio d Biochemical Activity In Vitro c-met/hgfr enzyme (Ki, nm) a 4 NA Cellular Activity
Supplemental Table 1. Biochemical and Cellular Potency and Selectivity of PF- 02341066 Assay IC 50 nm Selectivity Ratio d Biochemical Activity In Vitro c-met/hgfr enzyme (Ki, nm) a 4 NA Cellular Activity
Supplemental Figure 1. Genes showing ectopic H3K9 dimethylation in this study are DNA hypermethylated in Lister et al. study.
 mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
mc mc mc mc SUP mc mc Supplemental Figure. Genes showing ectopic HK9 dimethylation in this study are DNA hypermethylated in Lister et al. study. Representative views of genes that gain HK9m marks in their
Supplemental Figure 1. Western blot analysis indicated that MIF was detected in the fractions of
 Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
SUPPLEMENTARY FIGURES AND TABLE
 SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
SUPPLEMENTARY FIGURES AND TABLE Supplementary Figure S1: Characterization of IRE1α mutants. A. U87-LUC cells were transduced with the lentiviral vector containing the GFP sequence (U87-LUC Tet-ON GFP).
Supplementary Material
 Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
Supplementary Material Nuclear import of purified HIV-1 Integrase. Integrase remains associated to the RTC throughout the infection process until provirus integration occurs and is therefore one likely
Supplementary Information Supplementary Fig. 1. Elevated Usp9x in melanoma and NRAS mutant melanoma cells are dependent on NRAS for 3D growth.
 Supplementary Information Supplementary Fig. 1. Elevated Usp9x in melanoma and NRAS mutant melanoma cells are dependent on NRAS for 3D growth. a. Immunoblot for Usp9x protein in NRAS mutant melanoma cells
Supplementary Information Supplementary Fig. 1. Elevated Usp9x in melanoma and NRAS mutant melanoma cells are dependent on NRAS for 3D growth. a. Immunoblot for Usp9x protein in NRAS mutant melanoma cells
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at
 Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at E10.5 were double-stained for TUNEL (red) and PECAM-1 (green).
Supplementary Figure 1. EC-specific Deletion of Snail1 Does Not Affect EC Apoptosis. (a,b) Cryo-sections of WT (a) and Snail1 LOF (b) embryos at E10.5 were double-stained for TUNEL (red) and PECAM-1 (green).
Recombinant Protein Expression Retroviral system
 Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
Recombinant Protein Expression Retroviral system Viruses Contains genome DNA or RNA Genome encased in a protein coat or capsid. Some viruses have membrane covering protein coat enveloped virus Ø Essential
Supplemental Figure S1. RANK expression on human lung cancer cells.
 Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
SUPPLEMENTAL INFORMATION
 SUPPLEMENTAL INFORMATION EXPERIMENTAL PROCEDURES Tryptic digestion protection experiments - PCSK9 with Ab-3D5 (1:1 molar ratio) in 50 mm Tris, ph 8.0, 150 mm NaCl was incubated overnight at 4 o C. The
SUPPLEMENTAL INFORMATION EXPERIMENTAL PROCEDURES Tryptic digestion protection experiments - PCSK9 with Ab-3D5 (1:1 molar ratio) in 50 mm Tris, ph 8.0, 150 mm NaCl was incubated overnight at 4 o C. The
Procaspase-3. Cleaved caspase-3. actin. Cytochrome C (10 M) Z-VAD-fmk. Procaspase-3. Cleaved caspase-3. actin. Z-VAD-fmk
 A HeLa actin - + + - - + Cytochrome C (1 M) Z-VAD-fmk PMN - + + - - + actin Cytochrome C (1 M) Z-VAD-fmk Figure S1. (A) Pan-caspase inhibitor z-vad-fmk inhibits cytochrome c- mediated procaspase-3 cleavage.
A HeLa actin - + + - - + Cytochrome C (1 M) Z-VAD-fmk PMN - + + - - + actin Cytochrome C (1 M) Z-VAD-fmk Figure S1. (A) Pan-caspase inhibitor z-vad-fmk inhibits cytochrome c- mediated procaspase-3 cleavage.
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC
 Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
Supplementary Materials and Methods
 Supplementary Materials and Methods Immunoblotting Immunoblot analysis was performed as described previously (1). Due to high-molecular weight of MUC4 (~ 950 kda) and MUC1 (~ 250 kda) proteins, electrophoresis
Supplementary Materials and Methods Immunoblotting Immunoblot analysis was performed as described previously (1). Due to high-molecular weight of MUC4 (~ 950 kda) and MUC1 (~ 250 kda) proteins, electrophoresis
Islet viability assay and Glucose Stimulated Insulin Secretion assay RT-PCR and Western Blot
 Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Islet viability assay and Glucose Stimulated Insulin Secretion assay Islet cell viability was determined by colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide assay using CellTiter
Biodegradable Zwitterionic Nanogels with Long. Circulation for Antitumor Drug Delivery
 Supporting Information Biodegradable Zwitterionic Nanogels with Long Circulation for Antitumor Drug Delivery Yongzhi Men, Shaojun Peng, Peng Yang, Qin Jiang, Yanhui Zhang, Bin Shen, Pin Dong, *, Zhiqing
Supporting Information Biodegradable Zwitterionic Nanogels with Long Circulation for Antitumor Drug Delivery Yongzhi Men, Shaojun Peng, Peng Yang, Qin Jiang, Yanhui Zhang, Bin Shen, Pin Dong, *, Zhiqing
Figure S1. Sorting nexin 9 (SNX9) specifically binds psmad3 and not psmad 1/5/8. Lysates from AKR-2B cells untreated (-) or stimulated (+) for 45 min
 Figure S1. Sorting nexin 9 (SNX9) specifically binds psmad3 and not psmad 1/5/8. Lysates from AKR2B cells untreated () or stimulated () for 45 min with 5 ng/ml TGFβ or 10 ng/ml BMP4 were incubated with
Figure S1. Sorting nexin 9 (SNX9) specifically binds psmad3 and not psmad 1/5/8. Lysates from AKR2B cells untreated () or stimulated () for 45 min with 5 ng/ml TGFβ or 10 ng/ml BMP4 were incubated with
Protocol for A-549 VIM RFP (ATCC CCL-185EMT) TGFβ1 EMT Induction and Drug Screening
 Protocol for A-549 VIM RFP (ATCC CCL-185EMT) TGFβ1 EMT Induction and Drug Screening Introduction: Vimentin (VIM) intermediate filament (IF) proteins are associated with EMT in lung cancer and its metastatic
Protocol for A-549 VIM RFP (ATCC CCL-185EMT) TGFβ1 EMT Induction and Drug Screening Introduction: Vimentin (VIM) intermediate filament (IF) proteins are associated with EMT in lung cancer and its metastatic
Supplementary Figure 1. SA-β-Gal positive senescent cells in various cancer tissues. Representative frozen sections of breast, thyroid, colon and
 Supplementary Figure 1. SA-β-Gal positive senescent cells in various cancer tissues. Representative frozen sections of breast, thyroid, colon and stomach cancer were stained with SA-β-Gal and nuclear fast
Supplementary Figure 1. SA-β-Gal positive senescent cells in various cancer tissues. Representative frozen sections of breast, thyroid, colon and stomach cancer were stained with SA-β-Gal and nuclear fast
MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands)
 Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Supplemental data Materials and Methods Cell culture MTC-TT and TPC-1 cell lines were cultured in RPMI medium (Gibco, Breda, The Netherlands) supplemented with 15% or 10% (for TPC-1) fetal bovine serum
Supplementary Table 1. Primer sequences for conventional RT-PCR on mouse islets
 Supplementary Table 1. Primer sequences for conventional RT-PCR on mouse islets Gene 5 Forward 3 5 Reverse 3.T. Product (bp) ( C) mnox1 GTTCTTGGGCTGCCTTGG GCTGGGGCGGCGG 60 300 mnoxa1 GCTTTGCCGCGTGC GGTTCGGGTCCTTTGTGC
Supplementary Table 1. Primer sequences for conventional RT-PCR on mouse islets Gene 5 Forward 3 5 Reverse 3.T. Product (bp) ( C) mnox1 GTTCTTGGGCTGCCTTGG GCTGGGGCGGCGG 60 300 mnoxa1 GCTTTGCCGCGTGC GGTTCGGGTCCTTTGTGC
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein
 Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
Supplementary Figure 1. AdipoR1 silencing and overexpression controls. (a) Representative blots (upper and lower panels) showing the AdipoR1 protein content relative to GAPDH in two independent experiments.
SUPPLEMENTARY INFORMATION
 doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
Electron micrograph of phosphotungstanic acid-stained exosomes derived from murine
 1 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Physical properties of murine DC-derived exosomes. a, Electron micrograph of phosphotungstanic acid-stained exosomes derived from
1 SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURES Supplementary Figure 1. Physical properties of murine DC-derived exosomes. a, Electron micrograph of phosphotungstanic acid-stained exosomes derived from
fl/+ KRas;Atg5 fl/+ KRas;Atg5 fl/fl KRas;Atg5 fl/fl KRas;Atg5 Supplementary Figure 1. Gene set enrichment analyses. (a) (b)
 KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10860 Supplementary Discussion It remains unclear why H3K9 demethylation appeared to be more sensitive to suppression than at least some other histone methylation marks as a result of
doi:10.1038/nature10860 Supplementary Discussion It remains unclear why H3K9 demethylation appeared to be more sensitive to suppression than at least some other histone methylation marks as a result of
Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq
 Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq Philipp Bucher Wednesday January 21, 2009 SIB graduate school course EPFL, Lausanne ChIP-seq against histone variants: Biological
Computational Analysis of UHT Sequences Histone modifications, CAGE, RNA-Seq Philipp Bucher Wednesday January 21, 2009 SIB graduate school course EPFL, Lausanne ChIP-seq against histone variants: Biological
Nature Genetics: doi: /ng Supplementary Figure 1. SEER data for male and female cancer incidence from
 Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
Supplementary Figure 1 SEER data for male and female cancer incidence from 1975 2013. (a,b) Incidence rates of oral cavity and pharynx cancer (a) and leukemia (b) are plotted, grouped by males (blue),
