Supplemental Information. Integrative Transcriptome Analyses. of Metabolic Responses in Mice Define. Pivotal LncRNA Metabolic Regulators
|
|
|
- Anthony Maxwell
- 6 years ago
- Views:
Transcription
1 Cell Metabolism, Volume 24 Supplemental Information Integrative Transcriptome Analyses of Metabolic Responses in Mice Define Pivotal LncRNA Metabolic Regulators Ling Yang, Ping Li, Wenjing Yang, Xiangbo Ruan, Kurtis Kiesewetter, Jun Zhu, and Haiming Cao
2 A. B. Mouse feeding Liver RNA extracaon Ad Libitum Fast GCK AK35241 AK3369 AK539 AK79937 Gm16551 AK Ad libitum Fast Expression Profiling 18,431 non-redundant lncrnas 25,376 coding transcripts Fast Fast (285) (237) lncrnas AK45415 AK7778 AK1936 AK18645 AK85787 AK47141 AK16437 Ad libitum Fast C. RPKM Correla,on between mircoarray and RNA-seq >.5 r =.8 >.1 r =.75 > (all) r =.71 Figure S1, related to Figure 1. Genome-wide analyses of liver lncrnas and in mice during fasting and refeeding. (A) Experimental outline. MRNAs and lncrnas in the livers of mice fed ad libitum (Ad Libitum), subject to a 24-hour fast (Fast), or a 24-hour fast followed by a 4-hour refeeding () were analyzed by Mouse LncRNA Array V2. (8 x 6K, ArrayStar). P value <.5 and fold change >= 2 were used to identify differentially expressed genes (Fast vs Ad libitum or vs Fast, n=4). The numbers of genes regulated by Fast and are indicated, and the numbers of genes regulated by Fast and reversed by are indicated in brackets. (B) Validation of microarray results by quantitative real-time PCR analysis. Expression levels of one mrna, glucose kinase (GCK) and 13 lncrnas were quantified. Error bars are SEM, n=4. P<.5 (Fast vs Ad libitum), p<.5 ( vs Fast). (C) Pearson correlation of log2-transformd fold changes between microarray and RNA-seq for differentially expressed lncrnas identified by microarray.
3 A. B. Liver Adipose Muscle Group Condi+on Sample ID Group Condi+on Sample ID Group Condi+on Sample ID CH-1 FR-A1 FR-M1 Ad CH-2 Ad FR-A2 Ad FR-M2 Libitum CH-3 Libitum FR-A3 Libitum FR-M3 CH-4 FR-A4 FR-M4 CH-9 FR-A9 FR-M9 Group1 Fast CH-1 FR-A1 FR-M1 Group1 Fast Group1 Fast CH-11 FR-A11 FR-M11 CH-12 FR-A12 FR-M12 CH-17 FR-A17 FR-M17 CH-18 FR-A18 FR-M18 CH-19 FR-A19 FR-M19 CH-2 FR-A2 FR-M2 HD-L6 HD-F6 Normal HD-L9 Normal HD-F9 Normal HD-M9 diet HD-L15 diet HD-F15 diet HD-M15 control HD-L26 control HD-F26 control HD-M26 HD-L32 HD-F32 HD-M32 HD-L7 HD-F7 HD-M7 HD-L13 HD-F13 HD-M13 Group2 HFD 48h HD-L14 Group2 H48h HD-F14 Group2 H48h HD-M14 HD-L16 HD-F16 HD-M16 HD-L17 HD-F17 HD-L19 HD-F19 HD-L25 HD-F25 HD-M25 HFD 12w HD-L3 H12w HD-F3 H12w HD-M3 HD-L31 HD-F31 HD-M31 HD-L33 HD-F33 HD-M33 OB-L2 OB-F2 OB-M2 OB-L6 OB-F6 WT OB-L14 WT OB-F14 WT OB-M14 OB-L18 OB-F18 OB-M18 Group3 OB-L2 Group3 OB-F2 Group3 OB-M2 OB-L3 OB-F3 OB-L7 OB-F7 OB-M7 ob/ob OB-L9 ob/ob OB-F9 ob/ob OB-M9 OB-L13 OB-F13 OB-M13 OB-L15 OB-F15 OB-M15 C. lncrna proportion % 13.99% Exon number Relaonship of lncrnas to 3.92% 5.49% 8.95% 54.19% >14 Bidirec1onal Intergenic Intronic an1sense An1sense Sense intronic Sense overlapping D. E. F. Fast Liver Fast Adipose Fast Muscle oxidation reduction steroid biosynthetic process sterol biosynthetic process cholesterol biosynthetic process steroid metabolic process detection of abiotic stimulus response to abiotic stimulus regulation of cyclic nucleotide metabolic process regulation of nucleotide metabolic process energy reserve metabolic process..1.2 regulation of apoptosis regulation of programmed cell death regulation of cell death negative regulation of apoptosis negative regulation of programmed cell death..5.1 steroid biosynthetic process cofactor metabolic process oxidation reduction hexose metabolic process coenzyme metabolic process Liver..2.4 defense response intracellular signaling cascade positive regulation of biosynthetic process positive regulation of nitrogen compound metabolic process positive regulation of cellular biosynthetic process Adipose..2.4 positive regulation of cellular biosynthetic process positive regulation of biosynthetic process positive regulation of macromolecule metabolic process positive regulation of gene expression positive regulation of nitrogen compound metabolic process Muscle h HFD Liver oxidation reduction proteolysis involved in cellular protein catabolic process cellular protein catabolic process modification-dependent protein catabolic process modification-dependent macromolecule catabolic process..1.2 lipid biosynthetic process sterol biosynthetic process sterol metabolic process cholesterol biosynthetic process M phase of mitotic cell cycle 48h HFD Adipose fat cell differentiation acute-phase response acute inflammatory response white fat cell differentiation response to wounding 48h HFD Muscle w HFD Liver 12w HFD Adipose 12w HFD Muscle drug metabolic process positive regulation of macromolecule metabolic process regulation of multicellular organism growth positive regulation of gene expression cell surface receptor linked signal transduction..5.1 response to wounding inflammatory response cell activation chemotaxis taxis muscle cell differentiation RNA splicing mrna metabolic process mrna processing RNA splicing..5.1 ob/ob Liver ob/ob Adipose ob/ob Muscle oxidation reduction fatty acid metabolic process response to wounding lipid biosynthetic process steroid metabolic process..1.2 inflammatory response response to wounding immune response defense response phagocytosis in utero embryonic development chordate embryonic development embryonic development ending in birth or egg hatching regulation of growth cellular component morphogenesis..5.1
4 Figure S2, related to Figure 1. Sample information and biological processes regulated by various metabolic conditions. (A) Sample information. (B) Distribution of exon numbers of the lncrna transcripts covered in this study. This exon information is obtained from the NONCODE 216 mouse database. (C) Distribution of different types of lncrnas covered in this study based on their relative position to nearby coding genes. (D-F) Top 5 gene ontology (GO) biological process terms for differentially expressed in each metabolic condition in liver (D), adipose (E), and muscle tissue (F).
5 A. B. C. Liver Adipose Muscle steroid biosynthetic process oxidation reduction acyl-coa metabolic process steroid metabolic process lipid biosynthetic process cholesterol biosynthetic process cholesterol metabolic process sterol metabolic process fatty acid metabolic process sterol biosynthetic process cofactor metabolic process coenzyme metabolic process isoprenoid biosynthetic process isoprenoid metabolic process acute inflammatory response M phase of mitotic cell cycle mitosis nuclear division mitotic cell cycle organelle fission M phase cell division cell cycle phase cell cycle cell cycle process lipid biosynthetic process steroid metabolic process hexose metabolic process cholesterol metabolic process sterol metabolic process acute-phase response fatty acid metabolic process fatty acid biosynthetic process acute inflammatory response osteoblast development negative regulation of molecular function organic acid biosynthetic process carboxylic acid biosynthetic process response to cytokine stimulus negative regulation of cell differentiation D. E. F. Liver Adipose Muscle lncrnas lncrnas lncrnas G. H. I. Liver Adipose Muscle lncrnas 29 lncrnas lncrnas Figure S3, related to Figure 2. GO biological process terms for highly regulated and correlation analyses of metabolically sensitive lncrnas with coding genes. (A-C) Top GO terms for that were regulated at least three times in liver (A), adipose (B) and muscle (C). (D-F) Heat map with hierarchical clustering of the positive correlations between protein coding and the metabolically sensitive lncrnas in liver (D), adipose (E), and muscle (F) by Pearson s correlation method. (G-I) Heat map with hierarchical clustering of the negative correlations between protein coding and the metabolically sensitive lncrna genes in liver (G), adipose (H), and muscle (I) by Pearson s correlation method.
6 A. Exisng annotaon from Ensemble B. C YFP SREBP1c lacz sh Gm16551 sh2 D. mg/dl Plasma TG lacz sh Gm16551 sh2 Iden%fied mouse liver transcript by RACE E. mg/g liver lacz sh Liver TG Gm16551 sh1 Gm16551 sh2 I. J. IdenEEes: 67% E value: 4e Gm16551 sequence (Ensemble) Human homolog of Gm16551 (RP11-29K1.2, Ensemble) F. Relative gene expression lacz sh+yfp Gm16551 sh1+yfp Gm lacz sh+dnsrebp1c Gm16551 sh1+dnsrebp1c PCK Fetal Heart Fetal Kidney Fetal Liver FAS G. Gm16551 human homolog Fetal Muscle Heart Gm16551 RNA level Kidney YFP+Vector SREBP1c+Vector SREBP1c+Gm16551 Liver Muscle. Gm16551 K. H. Onecut1 mg/g liver Human Liver Gm16551 human homolog ACLY Normal liver FAS Liver TG YFP+Vector SREBP1c+Vector SREBP1c+Gm16551 Fatty liver SCD1 Figure S4, related to Figure 4. Analysis of an SREBP1c induced lncrna, Gm (A) Browser shot of Gm16551 region from Ensembl and the Gm16551 transcript is defined by RACE using mouse liver samples. (B) Expression levels of fatty acid synthase (FAS) gene in mouse liver transduced with YFP or constitutively active mouse SREBP1c adenoviruses. Error bars are SEM, n=5. P<.5. (C) Expression levels of Gm16551, Onecut1, ACLY, FAS and SCD1 in the livers of control (lacz sh) and Gm16551 knockdown (Gm16551 sh2) mice. Error bars are SEM, n=7. P<.5. (D) Plasma TG levels in control (lacz sh) and Gm16551 knockdown (Gm16551 sh2) mice. Error bars are SEM, n=7. P<.5. (E) Liver TG contents in control (lacz sh) and Gm16551 knockdown (Gm16551 sh1/sh2) mice. Error bars are SEM, n=6. P<.5. (F) Expression levels of Gm16551 and PCK1 in the livers of mice receiving lacz sh plus YFP, lacz plus dominant negative SREBP1c, Gm16551 knockdown (Gm16551 sh1) plus YFP, and dominant negative SREBP1c plus Gm16551 knockdown adenoviruses. Error bars are SEM, n=5. P<.5 (compared to control). (G) Expression levels of Gm16551 in the livers of mice receiving YFP plus vector, active SREBP1c plus vector, and active SREBP1c plus
7 Gm16551 adenoviruses. Error bars are SEM, n=7. P<.5. (H) Liver TG contents in mice receiving YFP plus vector, active SREBP1c plus vector, and active SREBP1c plus Gm16551 adenoviruses. Error bars are SEM, n=7. P<.5. (I) The alignment of mouse Gm16551 with its human homolog. (J) Quantitative real-time PCR analysis of human homolog of Gm16551 in human tissues of fetal heart, adult heart, kidney, liver and muscle. (K) Expression levels of human homolog of Gm16551 in the liver of patients with fatty liver disease. Error bars are SEM, n=5. P<.5. SUPPLEMENTAL TABLES AND LEGENDS Table S1, related to Figure 1. RNA-seq results of differentially expressed lncrnas identified by microarray. One sheet in this table, for those differentially expressed lncrnas (fasting vs ad libitum) identified by microarray, NONCODE IDs and their log2 transformed fold changes in both microarray and RNA-seq are listed. Table S2, related to Figure 2. Metabolically sensitive and lncrnas in liver, adipose and muscle tissue. Four sheets in this table, seqnames of metabolically sensitive from liver, adipose and muscle are listed in one sheet. Metabolically sensitive lncrnas for each tissue are listed in individual sheet with their chromosome, transcript start and end sites, strand, exons number, transcript length information. Table S3, related to Figure 2. Correlation values of liver, adipose, and muscle mrna and metabolically sensitive lncrna gene pairs. Three sheets in this table, the correlation coefficient of each pair of mrna and lncrna in liver, adipose or muscle was calculated by Pearson s correlation method. The first row is seqnames of lncrnas and the first column is the seqnames of coding genes. Table S4, related to Figure 2. GO terms of correlated with representative metabolically sensitive lncrnas in liver, adipose and muscle tissue. Twenty-four sheets are in this table, 8 metabolically sensitive lncrnas were selected for each tissue, and gene ontology
8 (GO) biological process terms for positively and negatively correlated with these lncrnas are listed. Table S5, related to Figure 4. Human homologs of metabolically sensitive lncrnas. One sheet in this table, including the mouse transcript information, human transcript information, and blastn results. Table S6, related to Experimental Procedures. A list of qrt-pcr primers. Mouse genes Sense primer An/-sense primer AK85787 TCTGGCTGCCTGTGGAAGA AGTGCTAGAGAGCCAAGAGTTTTACA uc9kuu.1 CCGAAAGCTGCTTAACTCCTAAGA GAGATCCAGCAATGCACTTCAC AK16911 AAGTAGGGAGCCGAGGGTAACT AACGCCAGCTCCCATACCT AK253 TCTCCAGGATCATGTGCTCC GGCTGGCTGGTTTTGTTGTT ENSMUST1617 (Gm16551) AAAATCAACGCAAAGGAAAGGA CGGCAGGTTGCTTCTGACA uc8txr.1 AGCAGCAACATTATCAGCATCTTC TGGCAGCGGCAGTATCTG AK3369 TGAAGCAGGTCTAAATGGGACAA TGCTCATGGTGACTGGTATGG AK9289 GATCCGGCCCTTACTGTGAG CCCACAGACAACTTTCCACA FGF21 CTGCTGGGGGTCTACCAAG CTGCGCCTACCACTGTTCC FAS GGAGGTGGTGATAGCCGGTAT TGGGTAATCCATAGAGCCCAG SREBP1c GGAGCCATGGATTGCACATT GGCCCGGGAAGTCACTGT G6P CGACTCGCTATCTCCAAGTGA GTTGAACCAGTCTCCGACCA PCK1 CCCAAGGCAACTTAAGGGCTAT CTGAGGTGCCAGGAGCAACT PK TCAAGGCAGGGATGAACATTG CACGGGTCTGTAGCTGAGTG RPIA AAGGCCGAGGAGGCTAAGAA CTTTCAGCTATTCGCTGCACA GLUT2 TCAGAAGACAAGATCACCGGA GTCATAGCCGAACTGGAAGGA GCK CCGTGATCCGGGAAGAGAA GGGAAACCTGACAGGGATGAG apoa4 ATCAAGAAGGAGCTGGAGGA GGTGCTCCTGCAACTTCTGC apoc2 ATGGGGTCTCGGTTCTTCCT GTCTTCTGGTACAGGTCTTTGG IGFBP1 CTGCCAAACTGCAACAAGAATG GGTCCCCTCTAGTCTCCAGA ACC1 CTTCCTGACAAACGAGTCTGG CTGCCGAAACATCTCTGGGA HNF4a AAATGTGCAGGTGTTGACCA CACGCTCCTCCTGAAGAATC FOXO1 GGGTCCCACAGCAACGATG CGAGGACGAAATGTACTCCAGTT chrebp CTGGGGACCTAAACAGGAGC GAAGCCACCCTATAGCTCCC GPAM CCTCTTTTGCCACAACATCA CGAGCCTCCGTCTTATGAAA ACLY CAGCCAAGGCAATTTCAGAGC CTCGACGTTTGATTAACTGGTCT SCD1 TTC TTG CGA TAC ACT CTG GTG C CGG GAT TGA ATG TTC TTG TCG T Ac/n GGCTGTATTCCCCTCCATCG CCAGTTGGTAACAATGCCATGT 18s AGTCCCTGCCCTTTGTACACA CGATCCGAGGGCCTCACTA Human gene Sense primer An/-sense primer H-lncLMS5 ATGGCTGCAAATGTGTGTGT CACAAGCCCAAATTCCCTCC
9 SUPPLEMENTAL EXPERIMENTAL PROCEDURES RNA extraction and quantitative real-time PCR analysis Total RNA was isolated from tissue samples or cells using Trizol reagent (Invitrogen). After Turbo DNA-free DNase treatment (Ambion), reverse transcription was carried out with the SuperScript III First-Strand Synthesis system (Invitrogen) using 1 µg of RNAs. Quantitative real-time RT-PCR was performed on a ViiA 7 Real-Time PCR System (Applied Biosystems Inc.). The PCR program was: 2 min 3 s at 95 C for enzyme activation, 4 cycles of 15 s at 95 C, and 1 min at 6 C. Melting curve analysis was performed to confirm the specific real-time PCR products. Primer sequences used are provided in Table S6. RNA-seq We selected ad Libitum and fasting liver samples to run RNA-seq on an Illumina HiSeq3 sequencer and obtained ~1 million strand-specific paired 5-mer reads per sample. The raw reads were mapped to the same NONCODE 216 mouse database using BWA (bwa aln; bwa sampe with the default parameters). Proper-paired reads were then counted on each noncoding transcript to calculate the RPKM by normalizing the read count on each transcript with transcript length and total library size. For those differentially expressed lncrnas identified by microarray (fasting vs ad libtum), the correlation between Microarray and RNA-seq was calculated by Pearson Correlation using log2-transformed fold changes. Pseudogenes were excluded from this correlation analysis because differentiating them from their corresponding coding genes using an RNA-seq mapping process is challenging. Isolation and culture of mouse primary hepatocytes Primary hepatocytes were isolated from male C57BL/6 mice fed with a normal chow diet. Mice were anesthetized with Ketamine (1mg/kg) and Xylazine (1mg/kg), and the livers were perfused with Krebs Ringer buffer with glucose at a rate of 5ml/min for 8 minutes, followed by continuous perfusion with the same buffer containing collagenase (Liberase TM Research Grade, Roche) for 1 minutes. Hepatocytes were harvested and purified with Percoll. The viability of hepatocytes was examined by trypan blue exclusion. Only cell isolates with viability over 9% were used. Hepatocytes were plated onto collagen-coated plates in DMEM (no glucose)
10 supplemented with 5.5mM glucose, 2mM GlutaMAX, 1nM Dexamethasone, 1nM insulin and 1% Cosmic Calf Serum (CCS). 5-6hrs after plating, cells were switched to maintenance medium as indicated in each experiment. Immunoblotting For immunoblotting analyses, livers were lysed in RIPA buffer (Cell Signaling Technology) containing phosphatase inhibitors (Sigma) and a protease inhibitor cocktail (Roche). The lysates were subjected to SDS PAGE, transferred to polyvinylidene fluoride (PVDF) membranes, and incubated with the primary antibody followed by the fluorescence conjugated secondary antibody (LI-COR). The bound antibody was visualized using a quantitative fluorescence imaging system (LI-COR). The primary antibodies used were an ACLY antibody (4332s, Cell Signaling), FAS antibody (sc-214, Santa Cruz), SCD1 antibody (2794s, Cell Signaling), and Tubulin antibody (2146s, Cell Signaling). Triglyceride (TG) Measurement Liver and plasma TG levels were measured using a kit from Sigma following the manufacturer s protocols.
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
Defective Hepatic Autophagy in Obesity Promotes ER Stress and Causes Insulin Resistance
 Cell Metabolism, Volume 11 Supplemental Information Defective Hepatic Autophagy in Obesity Promotes ER Stress and Causes Insulin Resistance Ling Yang, Ping Li, Suneng Fu, Ediz S. Calay, and Gökhan S. Hotamisligil
Cell Metabolism, Volume 11 Supplemental Information Defective Hepatic Autophagy in Obesity Promotes ER Stress and Causes Insulin Resistance Ling Yang, Ping Li, Suneng Fu, Ediz S. Calay, and Gökhan S. Hotamisligil
Supplementary information
 Supplementary information High fat diet-induced changes of mouse hepatic transcription and enhancer activity can be reversed by subsequent weight loss Majken Siersbæk, Lyuba Varticovski, Shutong Yang,
Supplementary information High fat diet-induced changes of mouse hepatic transcription and enhancer activity can be reversed by subsequent weight loss Majken Siersbæk, Lyuba Varticovski, Shutong Yang,
AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination
 AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination Supplementary Figure 1. Generation of the adult-onset, liver-specific GH receptor knock-down (alivghrkd, Kd) mouse
AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination Supplementary Figure 1. Generation of the adult-onset, liver-specific GH receptor knock-down (alivghrkd, Kd) mouse
General Laboratory methods Plasma analysis: Gene Expression Analysis: Immunoblot analysis: Immunohistochemistry:
 General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism SUPPLEMENTARY FIGURES, LEGENDS AND METHODS
 A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
A Hepatocyte Growth Factor Receptor (Met) Insulin Receptor hybrid governs hepatic glucose metabolism Arlee Fafalios, Jihong Ma, Xinping Tan, John Stoops, Jianhua Luo, Marie C. DeFrances and Reza Zarnegar
18s AAACGGCTACCACATCCAAG CCTCCAATGGATCCTCGTTA. 36b4 GTTCTTGCCCATCAGCACC AGATGCAGCAGATCCGCAT. Acc1 AGCAGATCCGCAGCTTG ACCTCTGCTCGCTGAGTGC
 Supplementary Table 1. Quantitative PCR primer sequences Gene symbol Sequences (5 to 3 ) Forward Reverse 18s AAACGGCTACCACATCCAAG CCTCCAATGGATCCTCGTTA 36b4 GTTCTTGCCCATCAGCACC AGATGCAGCAGATCCGCAT Acc1
Supplementary Table 1. Quantitative PCR primer sequences Gene symbol Sequences (5 to 3 ) Forward Reverse 18s AAACGGCTACCACATCCAAG CCTCCAATGGATCCTCGTTA 36b4 GTTCTTGCCCATCAGCACC AGATGCAGCAGATCCGCAT Acc1
Over-expression of MKP-3 and knockdown of MKP-3 and FOXO1 in primary rat. day, cells were transduced with adenoviruses expressing GFP, MKP-3 or shgfp,
 SUPPLEMENTAL METHODS Over-expression of MKP-3 and knockdown of MKP-3 and FOXO1 in primary rat hepatocytes Primary rat hepatocytes were seeded as described in experimental procedures. The next day, cells
SUPPLEMENTAL METHODS Over-expression of MKP-3 and knockdown of MKP-3 and FOXO1 in primary rat hepatocytes Primary rat hepatocytes were seeded as described in experimental procedures. The next day, cells
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies
 Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Supplemental Table 1 Primer sequences (mouse) used for real-time qrt-pcr studies Gene symbol Forward primer Reverse primer ACC1 5'-TGAGGAGGACCGCATTTATC 5'-GCATGGAATGGCAGTAAGGT ACLY 5'-GACACCATCTGTGATCTTG
Males- Western Diet WT KO Age (wks) Females- Western Diet WT KO Age (wks)
 Relative Arv1 mrna Adrenal 33.48 +/- 6.2 Skeletal Muscle 22.4 +/- 4.93 Liver 6.41 +/- 1.48 Heart 5.1 +/- 2.3 Brain 4.98 +/- 2.11 Ovary 4.68 +/- 2.21 Kidney 3.98 +/-.39 Lung 2.15 +/-.6 Inguinal Subcutaneous
Relative Arv1 mrna Adrenal 33.48 +/- 6.2 Skeletal Muscle 22.4 +/- 4.93 Liver 6.41 +/- 1.48 Heart 5.1 +/- 2.3 Brain 4.98 +/- 2.11 Ovary 4.68 +/- 2.21 Kidney 3.98 +/-.39 Lung 2.15 +/-.6 Inguinal Subcutaneous
Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion
 Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Supplementary Figure S1. Effects of UBL5 knockdown on cell cycle distribution and sister chromatid cohesion A. Representative examples of flow cytometry profiles of HeLa cells transfected with indicated
Supplementary Materials
 Supplementary Materials 1 Supplementary Table 1. List of primers used for quantitative PCR analysis. Gene name Gene symbol Accession IDs Sequence range Product Primer sequences size (bp) β-actin Actb gi
Supplementary Materials 1 Supplementary Table 1. List of primers used for quantitative PCR analysis. Gene name Gene symbol Accession IDs Sequence range Product Primer sequences size (bp) β-actin Actb gi
Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified
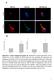 Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified for RFP-LC3 puncta (red dots) representing both autolysosomes
Suppl. Figure 1. T 3 induces autophagic flux in hepatic cells. (A) RFP-GFP-LC3 transfected HepG2/TRα cells were visualized and cells were quantified for RFP-LC3 puncta (red dots) representing both autolysosomes
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
Requires Signaling though Akt2 Independent of the. Transcription Factors FoxA2, FoxO1, and SREBP1c
 Cell Metabolism, Volume 14 Supplemental Information Postprandial Hepatic Lipid Metabolism Requires Signaling though Akt2 Independent of the Transcription Factors FoxA2, FoxO1, and SREBP1c Min Wan, Karla
Cell Metabolism, Volume 14 Supplemental Information Postprandial Hepatic Lipid Metabolism Requires Signaling though Akt2 Independent of the Transcription Factors FoxA2, FoxO1, and SREBP1c Min Wan, Karla
Supplementary Information. Deciphering the Venomic Transcriptome of Killer- Wasp Vespa velutina
 Supplementary Information Deciphering the Venomic Transcriptome of Killer- Wasp Vespa velutina Zhirui Liu, Shuanggang Chen, You Zhou, Cuihong Xie, Bifeng Zhu, Huming Zhu, Shupeng Liu, Wei Wang, Hongzhuan
Supplementary Information Deciphering the Venomic Transcriptome of Killer- Wasp Vespa velutina Zhirui Liu, Shuanggang Chen, You Zhou, Cuihong Xie, Bifeng Zhu, Huming Zhu, Shupeng Liu, Wei Wang, Hongzhuan
Supporting Information Table of content
 Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
Supporting Information Table of content Supporting Information Fig. S1 Supporting Information Fig. S2 Supporting Information Fig. S3 Supporting Information Fig. S4 Supporting Information Fig. S5 Supporting
ZL ZDF ZDF + E2 *** Visceral (g) ZDF
 Body Weight (g) 4 3 2 1 ** * ZL ZDF 6 8 1 12 14 16 Age (weeks) B * Sub-cutaneous (g) 16 12 8 4 ZL ZDF Visceral (g) 25 2 15 1 5 ZL ZDF Total fat pad weight (g) 4 3 2 1 ZDF ZL Supplemental Figure 1: Effect
Body Weight (g) 4 3 2 1 ** * ZL ZDF 6 8 1 12 14 16 Age (weeks) B * Sub-cutaneous (g) 16 12 8 4 ZL ZDF Visceral (g) 25 2 15 1 5 ZL ZDF Total fat pad weight (g) 4 3 2 1 ZDF ZL Supplemental Figure 1: Effect
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Supplementary Figure 1 U1 inhibition causes a shift of RNA-seq reads from exons to introns. (a) Evidence for the high purity of 4-shU-labeled RNAs used for RNA-seq. HeLa cells transfected with control
Figure S1. Body composition, energy homeostasis and substrate utilization in LRH-1 hep+/+ (white bars) and LRH-1 hep-/- (black bars) mice.
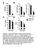 Figure S1. Body composition, energy homeostasis and substrate utilization in LRH-1 hep+/+ (white bars) and LRH-1 hep-/- (black bars) mice. (A) Lean and fat masses, determined by EchoMRI. (B) Food and water
Figure S1. Body composition, energy homeostasis and substrate utilization in LRH-1 hep+/+ (white bars) and LRH-1 hep-/- (black bars) mice. (A) Lean and fat masses, determined by EchoMRI. (B) Food and water
Supplemental Table 1. List of primers used for real time PCR.
 Supplemental Table 1. List of primers used for real time PCR. Primer Sequence Primer Sequence Mouse Pcsk9-F TTGCAGCAGCTGGGAACTT Mouse Scd1-F CATCATTCTCATGGTCCTGCT Mouse Pcsk9-R CCGACTGTGATGACCTCTGGA Mouse
Supplemental Table 1. List of primers used for real time PCR. Primer Sequence Primer Sequence Mouse Pcsk9-F TTGCAGCAGCTGGGAACTT Mouse Scd1-F CATCATTCTCATGGTCCTGCT Mouse Pcsk9-R CCGACTGTGATGACCTCTGGA Mouse
Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle.
 Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle. (a) mrna levels of Dj1 measured by quantitative RT-PCR in soleus, gastrocnemius (Gastroc.) and extensor digitorum longus
Supplementary Figure 1. DJ-1 modulates ROS concentration in mouse skeletal muscle. (a) mrna levels of Dj1 measured by quantitative RT-PCR in soleus, gastrocnemius (Gastroc.) and extensor digitorum longus
CD31 5'-AGA GAC GGT CTT GTC GCA GT-3' 5 ' -TAC TGG GCT TCG AGA GCA GT-3'
 Table S1. The primer sets used for real-time RT-PCR analysis. Gene Forward Reverse VEGF PDGFB TGF-β MCP-1 5'-GTT GCA GCA TGA ATC TGA GG-3' 5'-GGA GAC TCT TCG AGG AGC ACT T-3' 5'-GAA TCA GGC ATC GAG AGA
Table S1. The primer sets used for real-time RT-PCR analysis. Gene Forward Reverse VEGF PDGFB TGF-β MCP-1 5'-GTT GCA GCA TGA ATC TGA GG-3' 5'-GGA GAC TCT TCG AGG AGC ACT T-3' 5'-GAA TCA GGC ATC GAG AGA
Supplementary Information. MicroRNA-33b knock-in mice for an intron of sterol regulatory
 Supplementary Information MicroRNA-33b knock-in mice for an intron of sterol regulatory element-binding factor 1 (Srebf1) exhibit reduced HDL-C in vivo Takahiro Horie, Tomohiro Nishino, Osamu Baba, Yasuhide
Supplementary Information MicroRNA-33b knock-in mice for an intron of sterol regulatory element-binding factor 1 (Srebf1) exhibit reduced HDL-C in vivo Takahiro Horie, Tomohiro Nishino, Osamu Baba, Yasuhide
Supplemental Information. Genetic Regulation of Plasma Lipid Species. and Their Association with Metabolic Phenotypes
 Cell Systems, Volume 6 Supplemental Information Genetic Regulation of Plasma Lipid Species and Their Association with Metabolic Phenotypes Pooja Jha, Molly T. McDevitt, Emina Halilbasic, Evan G. Williams,
Cell Systems, Volume 6 Supplemental Information Genetic Regulation of Plasma Lipid Species and Their Association with Metabolic Phenotypes Pooja Jha, Molly T. McDevitt, Emina Halilbasic, Evan G. Williams,
Formalin-fixed paraffin-embedded sections of liver from a recipient mouse sacrificed after two rounds
 Supplementary figure legends Supplementary Figure 1 Fah + hepatocytes in a Fah -/- mouse transplanted with sorted cells. Formalin-fixed paraffin-embedded sections of liver from a recipient mouse sacrificed
Supplementary figure legends Supplementary Figure 1 Fah + hepatocytes in a Fah -/- mouse transplanted with sorted cells. Formalin-fixed paraffin-embedded sections of liver from a recipient mouse sacrificed
Supplementary Information. Glycogen shortage during fasting triggers liver-brain-adipose. neurocircuitry to facilitate fat utilization
 Supplementary Information Glycogen shortage during fasting triggers liver-brain-adipose neurocircuitry to facilitate fat utilization Supplementary Figure S1. Liver-Brain-Adipose neurocircuitry Starvation
Supplementary Information Glycogen shortage during fasting triggers liver-brain-adipose neurocircuitry to facilitate fat utilization Supplementary Figure S1. Liver-Brain-Adipose neurocircuitry Starvation
Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells.
 SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
SUPPLEMENTAL FIGURE AND TABLE LEGENDS Supplemental Figure S1. Expression of Cirbp mrna in mouse tissues and NIH3T3 cells. A) Cirbp mrna expression levels in various mouse tissues collected around the clock
For pair feeding, mice were fed 2.7g of HFD containing tofogliflozin
 Materials and Methods Pair Feeding Experiment For pair feeding, mice were fed 2.7g of HFD containing tofogliflozin (0.005%), which is average daily food intake of mice fed control HFD ad libitum at week
Materials and Methods Pair Feeding Experiment For pair feeding, mice were fed 2.7g of HFD containing tofogliflozin (0.005%), which is average daily food intake of mice fed control HFD ad libitum at week
Ambient temperature regulated flowering time
 Ambient temperature regulated flowering time Applications of RNAseq RNA- seq course: The power of RNA-seq June 7 th, 2013; Richard Immink Overview Introduction: Biological research question/hypothesis
Ambient temperature regulated flowering time Applications of RNAseq RNA- seq course: The power of RNA-seq June 7 th, 2013; Richard Immink Overview Introduction: Biological research question/hypothesis
Supplementary Figure 1
 Supplementary Figure 1 A B mir-141, human cell lines mir-2c, human cell lines mir-141, hepatocytes mir-2c, hepatocytes Relative RNA.1.8.6.4.2 Relative RNA.3.2.1 Relative RNA 1.5 1..5 Relative RNA 2. 1.5
Supplementary Figure 1 A B mir-141, human cell lines mir-2c, human cell lines mir-141, hepatocytes mir-2c, hepatocytes Relative RNA.1.8.6.4.2 Relative RNA.3.2.1 Relative RNA 1.5 1..5 Relative RNA 2. 1.5
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Supplementary Table 1. Metabolic parameters in GFP and OGT-treated mice
 Supplementary Table 1. Metabolic parameters in GFP and OGT-treated mice Fasted Refed GFP OGT GFP OGT Liver G6P (mmol/g) 0.03±0.01 0.04±0.02 0.60±0.04 0.42±0.10 A TGs (mg/g of liver) 20.08±5.17 16.29±0.8
Supplementary Table 1. Metabolic parameters in GFP and OGT-treated mice Fasted Refed GFP OGT GFP OGT Liver G6P (mmol/g) 0.03±0.01 0.04±0.02 0.60±0.04 0.42±0.10 A TGs (mg/g of liver) 20.08±5.17 16.29±0.8
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
SUPPLEMENTARY DATA. Supplementary Table 1. Primer sequences for qrt-pcr
 Supplementary Table 1. Primer sequences for qrt-pcr Gene PRDM16 UCP1 PGC1α Dio2 Elovl3 Cidea Cox8b PPARγ AP2 mttfam CyCs Nampt NRF1 16s-rRNA Hexokinase 2, intron 9 β-actin Primer Sequences 5'-CCA CCA GCG
Supplementary Table 1. Primer sequences for qrt-pcr Gene PRDM16 UCP1 PGC1α Dio2 Elovl3 Cidea Cox8b PPARγ AP2 mttfam CyCs Nampt NRF1 16s-rRNA Hexokinase 2, intron 9 β-actin Primer Sequences 5'-CCA CCA GCG
Supplementary Figures and Tables
 Supplementary Figures and Tables Supplementary Figure 1. Study design and sample collection. S.japonicum were harvested from C57 mice at 8 time points after infection. Total number of samples for RNA-Seq:
Supplementary Figures and Tables Supplementary Figure 1. Study design and sample collection. S.japonicum were harvested from C57 mice at 8 time points after infection. Total number of samples for RNA-Seq:
Online Data Supplement. Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2
 Online Data Supplement Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2 Yi Lin and Zhongjie Sun Department of physiology, college of
Online Data Supplement Anti-aging Gene Klotho Enhances Glucose-induced Insulin Secretion by Upregulating Plasma Membrane Retention of TRPV2 Yi Lin and Zhongjie Sun Department of physiology, college of
Supplementary Figure 1: Additional metabolic parameters of obesity mouse models and controls. (a) Body weight, (b) blood glucose and (c) insulin
 Supplementary Figure 1: Additional metabolic parameters of obesity mouse models and controls. (a) Body weight, (b) blood glucose and (c) insulin resistance index of homeostatic model assessment (HOMA IR)
Supplementary Figure 1: Additional metabolic parameters of obesity mouse models and controls. (a) Body weight, (b) blood glucose and (c) insulin resistance index of homeostatic model assessment (HOMA IR)
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
Figures S1-S5, Figure Legends, Table S1 List of primers used in the study
 Insulin receptor alternative splicing is regulated by insulin signaling and modulates beta cell survival Pushkar Malakar,4, Lital Chartarifsky,4, Ayat Hija, Gil Leibowitz 3, Benjamin Glaser 3, Yuval Dor,
Insulin receptor alternative splicing is regulated by insulin signaling and modulates beta cell survival Pushkar Malakar,4, Lital Chartarifsky,4, Ayat Hija, Gil Leibowitz 3, Benjamin Glaser 3, Yuval Dor,
Validation & Assay Performance Summary
 Validation & Assay Performance Summary LanthaScreen IGF-1R GripTite Cells Cat. no. K1834 Modification Detected: Phosphorylation of Multiple Tyr Residues on IGF-1R LanthaScreen Cellular Assay Validation
Validation & Assay Performance Summary LanthaScreen IGF-1R GripTite Cells Cat. no. K1834 Modification Detected: Phosphorylation of Multiple Tyr Residues on IGF-1R LanthaScreen Cellular Assay Validation
FH- FH+ DM. 52 Volunteers. Oral & IV Glucose Tolerance Test Hyperinsulinemic Euglycemic Clamp in Non-DM Subjects ACADSB MYSM1. Mouse Skeletal Muscle
 A 52 Volunteers B 6 5 4 3 2 FH- FH+ DM 1 Oral & IV Glucose Tolerance Test Hyperinsulinemic Euglycemic Clamp in Non-DM Subjects ZYX EGR2 NR4A1 SRF target TPM1 ACADSB MYSM1 Non SRF target FH- FH+ DM2 C SRF
A 52 Volunteers B 6 5 4 3 2 FH- FH+ DM 1 Oral & IV Glucose Tolerance Test Hyperinsulinemic Euglycemic Clamp in Non-DM Subjects ZYX EGR2 NR4A1 SRF target TPM1 ACADSB MYSM1 Non SRF target FH- FH+ DM2 C SRF
Supplementary Figure 1
 VO (ml kg - min - ) VCO (ml kg - min - ) Respiratory exchange ratio Energy expenditure (cal kg - min - ) Locomotor activity (x count) Body temperature ( C) Relative mrna expression TA Sol EDL PT Heart
VO (ml kg - min - ) VCO (ml kg - min - ) Respiratory exchange ratio Energy expenditure (cal kg - min - ) Locomotor activity (x count) Body temperature ( C) Relative mrna expression TA Sol EDL PT Heart
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Frequency of alternative-cassette-exon engagement with the ribosome is consistent across data from multiple human cell types and from mouse stem cells. Box plots showing AS frequency
Supplementary Figure 1 Frequency of alternative-cassette-exon engagement with the ribosome is consistent across data from multiple human cell types and from mouse stem cells. Box plots showing AS frequency
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
Supplementary Figure 1
 A B D Relative TAp73 mrna p73 Supplementary Figure 1 25 2 15 1 5 p63 _-tub. MDA-468 HCC1143 HCC38 SUM149 MDA-468 HCC1143 HCC38 SUM149 HCC-1937 MDA-MB-468 ΔNp63_ TAp73_ TAp73β E C Relative ΔNp63 mrna TAp73
A B D Relative TAp73 mrna p73 Supplementary Figure 1 25 2 15 1 5 p63 _-tub. MDA-468 HCC1143 HCC38 SUM149 MDA-468 HCC1143 HCC38 SUM149 HCC-1937 MDA-MB-468 ΔNp63_ TAp73_ TAp73β E C Relative ΔNp63 mrna TAp73
Abbreviations: P- paraffin-embedded section; C, cryosection; Bio-SA, biotin-streptavidin-conjugated fluorescein amplification.
 Supplementary Table 1. Sequence of primers for real time PCR. Gene Forward primer Reverse primer S25 5 -GTG GTC CAC ACT ACT CTC TGA GTT TC-3 5 - GAC TTT CCG GCA TCC TTC TTC-3 Mafa cds 5 -CTT CAG CAA GGA
Supplementary Table 1. Sequence of primers for real time PCR. Gene Forward primer Reverse primer S25 5 -GTG GTC CAC ACT ACT CTC TGA GTT TC-3 5 - GAC TTT CCG GCA TCC TTC TTC-3 Mafa cds 5 -CTT CAG CAA GGA
Supplementary Table 3. 3 UTR primer sequences. Primer sequences used to amplify and clone the 3 UTR of each indicated gene are listed.
 Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Supplemental Figure 1. DLKI-DIO3 mirna/mrna complementarity. Complementarity between the indicated DLK1-DIO3 cluster mirnas and the UTR of SOX2, SOX9, HIF1A, ZEB1, ZEB2, STAT3 and CDH1with mirsvr and PhastCons
Supplementary Materials and Methods
 Supplementary Materials and Methods Hepatocyte toxicity assay. Freshly isolated hepatocytes were incubated for overnight with varying concentrations (-25 µm) of sodium glycochenodeoxycholate (GCDC) or
Supplementary Materials and Methods Hepatocyte toxicity assay. Freshly isolated hepatocytes were incubated for overnight with varying concentrations (-25 µm) of sodium glycochenodeoxycholate (GCDC) or
Supplemental Information. Increased 4E-BP1 Expression Protects. against Diet-Induced Obesity and Insulin. Resistance in Male Mice
 Cell Reports, Volume 16 Supplemental Information Increased 4E-BP1 Expression Protects against Diet-Induced Obesity and Insulin Resistance in Male Mice Shih-Yin Tsai, Ariana A. Rodriguez, Somasish G. Dastidar,
Cell Reports, Volume 16 Supplemental Information Increased 4E-BP1 Expression Protects against Diet-Induced Obesity and Insulin Resistance in Male Mice Shih-Yin Tsai, Ariana A. Rodriguez, Somasish G. Dastidar,
Serum Amyloid A3 Gene Expression in Adipocytes is an Indicator. of the Interaction with Macrophages
 Serum Amyloid A3 Gene Expression in Adipocytes is an Indicator of the Interaction with Macrophages Yohei Sanada, Takafumi Yamamoto, Rika Satake, Akiko Yamashita, Sumire Kanai, Norihisa Kato, Fons AJ van
Serum Amyloid A3 Gene Expression in Adipocytes is an Indicator of the Interaction with Macrophages Yohei Sanada, Takafumi Yamamoto, Rika Satake, Akiko Yamashita, Sumire Kanai, Norihisa Kato, Fons AJ van
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
BIOL212 Biochemistry of Disease. Metabolic Disorders - Obesity
 BIOL212 Biochemistry of Disease Metabolic Disorders - Obesity Obesity Approx. 23% of adults are obese in the U.K. The number of obese children has tripled in 20 years. 10% of six year olds are obese, rising
BIOL212 Biochemistry of Disease Metabolic Disorders - Obesity Obesity Approx. 23% of adults are obese in the U.K. The number of obese children has tripled in 20 years. 10% of six year olds are obese, rising
Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63.
 Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
Supplementary Figure Legends Supplementary Figure S1. Gene expression analysis of epidermal marker genes and TP63. A. Screenshot of the UCSC genome browser from normalized RNAPII and RNA-seq ChIP-seq data
Supplementary Information
 Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
Supplementary Information Supplementary Figure 1. CD4 + T cell activation and lack of apoptosis after crosslinking with anti-cd3 + anti-cd28 + anti-cd160. (a) Flow cytometry of anti-cd160 (5D.10A11) binding
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
Supplementary Figures Supplementary Figure 1. Heatmap of GO terms for differentially expressed genes. The terms were hierarchically clustered using the GO term enrichment beta. Darker red, higher positive
ACC ELOVL MCAD. CPT1α 1.5 *** 0.5. Reverbα *** *** 0.5. Fasted. Refed
 Supplementary Figure A 8 SREBPc 6 5 FASN ELOVL6.5.5.5 ACC.5.5 CLOCK.5.5 CRY.5.5 PPARα.5.5 ACSL CPTα.5.5.5.5 MCAD.5.5 PEPCK.5.5 G6Pase 5.5.5.5 BMAL.5.5 Reverbα.5.5 Reverbβ.5.5 PER.5.5 PER B Fasted Refed
Supplementary Figure A 8 SREBPc 6 5 FASN ELOVL6.5.5.5 ACC.5.5 CLOCK.5.5 CRY.5.5 PPARα.5.5 ACSL CPTα.5.5.5.5 MCAD.5.5 PEPCK.5.5 G6Pase 5.5.5.5 BMAL.5.5 Reverbα.5.5 Reverbβ.5.5 PER.5.5 PER B Fasted Refed
Supporting Information
 Supporting Information Charalambous et al. 10.1073/pnas.1406119111 SI Experimental Procedures Serum and Tissue Biochemistry. Enzymatic assay kits were used for determination of plasma FFAs (Roche), TAGs
Supporting Information Charalambous et al. 10.1073/pnas.1406119111 SI Experimental Procedures Serum and Tissue Biochemistry. Enzymatic assay kits were used for determination of plasma FFAs (Roche), TAGs
GPR120 *** * * Liver BAT iwat ewat mwat Ileum Colon. UCP1 mrna ***
 a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
control kda ATGL ATGLi HSL 82 GAPDH * ** *** WT/cTg WT/cTg ATGLi AKO/cTg AKO/cTg ATGLi WT/cTg WT/cTg ATGLi AKO/cTg AKO/cTg ATGLi iwat gwat ibat
 body weight (g) tissue weights (mg) ATGL protein expression (relative to GAPDH) HSL protein expression (relative to GAPDH) ### # # kda ATGL 55 HSL 82 GAPDH 37 2.5 2. 1.5 1..5 2. 1.5 1..5.. Supplementary
body weight (g) tissue weights (mg) ATGL protein expression (relative to GAPDH) HSL protein expression (relative to GAPDH) ### # # kda ATGL 55 HSL 82 GAPDH 37 2.5 2. 1.5 1..5 2. 1.5 1..5.. Supplementary
RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers
 Xu et al. Molecular Cancer (2019) 18:8 https://doi.org/10.1186/s12943-018-0932-8 LETTER TO THE EDITOR RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers
Xu et al. Molecular Cancer (2019) 18:8 https://doi.org/10.1186/s12943-018-0932-8 LETTER TO THE EDITOR RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers
Supplementary Table 2. Conserved regulatory elements in the promoters of CD36.
 Supplementary Table 1. RT-qPCR primers for CD3, PPARg and CEBP. Assay Forward Primer Reverse Primer 1A CAT TTG TGG CCT TGT GCT CTT TGA TGA GTC ACA GAA AGA ATC AAT TC 1B AGG AAA TGA ACT GAT GAG TCA CAG
Supplementary Table 1. RT-qPCR primers for CD3, PPARg and CEBP. Assay Forward Primer Reverse Primer 1A CAT TTG TGG CCT TGT GCT CTT TGA TGA GTC ACA GAA AGA ATC AAT TC 1B AGG AAA TGA ACT GAT GAG TCA CAG
Supplemental Data. Shin et al. Plant Cell. (2012) /tpc YFP N
 MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
MYC YFP N PIF5 YFP C N-TIC TIC Supplemental Data. Shin et al. Plant Cell. ()..5/tpc..95 Supplemental Figure. TIC interacts with MYC in the nucleus. Bimolecular fluorescence complementation assay using
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
SUPPLEMENTARY FIGURES Figure S1. Clinical significance of ZNF322A overexpression in Caucasian lung cancer patients. (A) Representative immunohistochemistry images of ZNF322A protein expression in tissue
An Analysis of MDM4 Alternative Splicing and Effects Across Cancer Cell Lines
 An Analysis of MDM4 Alternative Splicing and Effects Across Cancer Cell Lines Kevin Hu Mentor: Dr. Mahmoud Ghandi 7th Annual MIT PRIMES Conference May 2021, 2017 Outline Introduction MDM4 Isoforms Methodology
An Analysis of MDM4 Alternative Splicing and Effects Across Cancer Cell Lines Kevin Hu Mentor: Dr. Mahmoud Ghandi 7th Annual MIT PRIMES Conference May 2021, 2017 Outline Introduction MDM4 Isoforms Methodology
Supplementary Figure 1 a
 Supplementary Figure a Normalized expression/tbp (A.U.).6... Trip-br transcripts Trans Trans Trans b..5. Trip-br Ctrl LPS Normalized expression/tbp (A.U.) c Trip-br transcripts. adipocytes.... Trans Trans
Supplementary Figure a Normalized expression/tbp (A.U.).6... Trip-br transcripts Trans Trans Trans b..5. Trip-br Ctrl LPS Normalized expression/tbp (A.U.) c Trip-br transcripts. adipocytes.... Trans Trans
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Table 2. Plasma lipid profiles in wild type and mutant female mice submitted to a HFD for 12 weeks wt ERα -/- AF-1 0 AF-2 0
 Supplementary Table 1. List of specific primers used for gene expression analysis. Genes Primer forward Primer reverse Hprt GCAGTACAGCCCCAAAATGG AACAAAGTCTGGCCTGTATCCA Srebp-1c GGAAGCTGTCGGGGTAGCGTC CATGTCTTCAAATGTGCAATCCAT
Supplementary Table 1. List of specific primers used for gene expression analysis. Genes Primer forward Primer reverse Hprt GCAGTACAGCCCCAAAATGG AACAAAGTCTGGCCTGTATCCA Srebp-1c GGAAGCTGTCGGGGTAGCGTC CATGTCTTCAAATGTGCAATCCAT
Identification of Tissue-Specific Protein-Coding and Noncoding. Transcripts across 14 Human Tissues Using RNA-seq
 Identification of Tissue-Specific Protein-Coding and Noncoding Transcripts across 14 Human Tissues Using RNA-seq Jinhang Zhu 1, Geng Chen 1, Sibo Zhu 1,2, Suqing Li 3, Zhuo Wen 3, Bin Li 1, Yuanting Zheng
Identification of Tissue-Specific Protein-Coding and Noncoding Transcripts across 14 Human Tissues Using RNA-seq Jinhang Zhu 1, Geng Chen 1, Sibo Zhu 1,2, Suqing Li 3, Zhuo Wen 3, Bin Li 1, Yuanting Zheng
Supplementary methods:
 Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
Supplementary methods: Primers sequences used in real-time PCR analyses: β-actin F: GACCTCTATGCCAACACAGT β-actin [11] R: AGTACTTGCGCTCAGGAGGA MMP13 F: TTCTGGTCTTCTGGCACACGCTTT MMP13 R: CCAAGCTCATGGGCAGCAACAATA
SUPPLEMENTARY INFORMATION. Supp. Fig. 1. Autoimmunity. Tolerance APC APC. T cell. T cell. doi: /nature06253 ICOS ICOS TCR CD28 TCR CD28
 Supp. Fig. 1 a APC b APC ICOS ICOS TCR CD28 mir P TCR CD28 P T cell Tolerance Roquin WT SG Icos mrna T cell Autoimmunity Roquin M199R SG Icos mrna www.nature.com/nature 1 Supp. Fig. 2 CD4 + CD44 low CD4
Supp. Fig. 1 a APC b APC ICOS ICOS TCR CD28 mir P TCR CD28 P T cell Tolerance Roquin WT SG Icos mrna T cell Autoimmunity Roquin M199R SG Icos mrna www.nature.com/nature 1 Supp. Fig. 2 CD4 + CD44 low CD4
Supplementary Information
 Supplementary Information GADD34-deficient mice develop obesity, nonalcoholic fatty liver disease, hepatic carcinoma and insulin resistance Naomi Nishio and Ken-ichi Isobe Department of Immunology, Nagoya
Supplementary Information GADD34-deficient mice develop obesity, nonalcoholic fatty liver disease, hepatic carcinoma and insulin resistance Naomi Nishio and Ken-ichi Isobe Department of Immunology, Nagoya
Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most
 Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most differentially expressed between human synovial fibroblasts
Supplementary Figure 1 MicroRNA expression in human synovial fibroblasts from different locations. MicroRNA, which were identified by RNAseq as most differentially expressed between human synovial fibroblasts
Supplementary Figure 1
 Supplementary Figure 1 how HFD how HFD Epi WT p p Hypothalamus p p Inguinal WT T Liver Lean mouse adipocytes p p p p p p Obese mouse adipocytes Kidney Muscle Spleen Heart p p p p p p p p Extracellular
Supplementary Figure 1 how HFD how HFD Epi WT p p Hypothalamus p p Inguinal WT T Liver Lean mouse adipocytes p p p p p p Obese mouse adipocytes Kidney Muscle Spleen Heart p p p p p p p p Extracellular
Supplementary Figure 1. DNA methylation of the adiponectin promoter R1, Pparg2, and Tnfa promoter in adipocytes is not affected by obesity.
 Supplementary Figure 1. DNA methylation of the adiponectin promoter R1, Pparg2, and Tnfa promoter in adipocytes is not affected by obesity. (a) Relative amounts of adiponectin, Ppar 2, C/ebp, and Tnf mrna
Supplementary Figure 1. DNA methylation of the adiponectin promoter R1, Pparg2, and Tnfa promoter in adipocytes is not affected by obesity. (a) Relative amounts of adiponectin, Ppar 2, C/ebp, and Tnf mrna
Supplementary Figure S I: Effects of D4F on body weight and serum lipids in apoe -/- mice.
 Supplementary Figures: Supplementary Figure S I: Effects of D4F on body weight and serum lipids in apoe -/- mice. Male apoe -/- mice were fed a high-fat diet for 8 weeks, and given PBS (model group) or
Supplementary Figures: Supplementary Figure S I: Effects of D4F on body weight and serum lipids in apoe -/- mice. Male apoe -/- mice were fed a high-fat diet for 8 weeks, and given PBS (model group) or
Quantitative Real-Time PCR was performed as same as Materials and Methods.
 Supplemental Material Quantitative Real-Time PCR Quantitative Real-Time PCR was performed as same as Materials and Methods. Expression levels in the aorta were normalized to peptidylprolyl isomerase B
Supplemental Material Quantitative Real-Time PCR Quantitative Real-Time PCR was performed as same as Materials and Methods. Expression levels in the aorta were normalized to peptidylprolyl isomerase B
Hmgcoar AGCTTGCCCGAATTGTATGTG TCTGTTGTAACCATGTGACTTC. Cyp7α GGGATTGCTGTGGTAGTGAGC GGTATGGAATCAACCCGTTGTC
 Supplement Table I: primers for Real Time RT-PCR Gene Foward Reverse Hmgcoar AGCTTGCCCGAATTGTATGTG TCTGTTGTAACCATGTGACTTC Cyp7α GGGATTGCTGTGGTAGTGAGC GGTATGGAATCAACCCGTTGTC Cyp27a1 GTGGTCTTATTGGGTACTTGC
Supplement Table I: primers for Real Time RT-PCR Gene Foward Reverse Hmgcoar AGCTTGCCCGAATTGTATGTG TCTGTTGTAACCATGTGACTTC Cyp7α GGGATTGCTGTGGTAGTGAGC GGTATGGAATCAACCCGTTGTC Cyp27a1 GTGGTCTTATTGGGTACTTGC
Supporting Information. Supporting Tables. S-Table 1 Primer pairs for RT-PCR. Product size. Gene Primer pairs
 Supporting Information Supporting Tables S-Table 1 Primer pairs for RT-PCR. Gene Primer pairs Product size (bp) FAS F: 5 TCTTGGAAGCGATGGGTA 3 429 R: 5 GGGATGTATCATTCTTGGAC 3 SREBP-1c F: 5 CGCTACCGTTCCTCTATCA
Supporting Information Supporting Tables S-Table 1 Primer pairs for RT-PCR. Gene Primer pairs Product size (bp) FAS F: 5 TCTTGGAAGCGATGGGTA 3 429 R: 5 GGGATGTATCATTCTTGGAC 3 SREBP-1c F: 5 CGCTACCGTTCCTCTATCA
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
HSP72 HSP90. Quadriceps Muscle. MEF2c MyoD1 MyoG Myf5 Hsf1 Hsp GLUT4/GAPDH (AU)
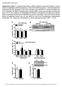 Supplementary Figure 1. Impaired insulin action in HSP72 deficient muscle and myotubes in culture cannot be explained by altered myogenesis or reduced total GLUT4 expression. Genes associated with myogenesis
Supplementary Figure 1. Impaired insulin action in HSP72 deficient muscle and myotubes in culture cannot be explained by altered myogenesis or reduced total GLUT4 expression. Genes associated with myogenesis
Supplementary Figure S1. Effect of Glucose on Energy Balance in WT and KHK A/C KO
 Supplementary Figure S1. Effect of Glucose on Energy Balance in WT and KHK A/C KO Mice. WT mice and KHK-A/C KO mice were provided drinking water containing 10% glucose or tap water with normal chow ad
Supplementary Figure S1. Effect of Glucose on Energy Balance in WT and KHK A/C KO Mice. WT mice and KHK-A/C KO mice were provided drinking water containing 10% glucose or tap water with normal chow ad
ANSC/NUTR 601 GENERAL ANIMAL NUTRITION Stearoyl-CoA desaturase, VLDL metabolism, and obesity
 ANSC/NUTR 601 GENERAL ANIMAL NUTRITION Stearoyl-CoA desaturase, VLDL metabolism, and obesity I. Stearoyl coenzyme A desaturase and obesity in rodents A. Stearoyl coenzyme A desaturase (SCD) is the 9 desaturase.
ANSC/NUTR 601 GENERAL ANIMAL NUTRITION Stearoyl-CoA desaturase, VLDL metabolism, and obesity I. Stearoyl coenzyme A desaturase and obesity in rodents A. Stearoyl coenzyme A desaturase (SCD) is the 9 desaturase.
Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR
 Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) cadl CTTGGGGGCGCGTCT CTGTTCTTTTGTGCCGTTTCG cyl-coenzyme Dehydrogenase, very
Supplementary Table 1. Primer Sequences Used for Quantitative Real-Time PCR Gene Forward Primer (5-3 ) Reverse Primer (5-3 ) cadl CTTGGGGGCGCGTCT CTGTTCTTTTGTGCCGTTTCG cyl-coenzyme Dehydrogenase, very
Edge attributes. Node attributes
 HMDB ID input pane Network view Node attributes Edge attributes Supplemental Figure 1. Cytoscape App MetBridge Generator. The left pane is Cytoscape App MetBridge Generator (code name: rsmetabppi). User
HMDB ID input pane Network view Node attributes Edge attributes Supplemental Figure 1. Cytoscape App MetBridge Generator. The left pane is Cytoscape App MetBridge Generator (code name: rsmetabppi). User
SUPPLEMENTARY DATA. Supplementary Table 1. Primers used in qpcr
 Supplementary Table 1. Primers used in qpcr Gene forward primer (5'-3') reverse primer (5'-3') β-actin AGAGGGAAATCGTGCGTGAC CAATAGTGATGACCTGGCCGT Hif-p4h-2 CTGGGCAACTACAGGATAAAC GCGTCCCAGTCTTTATTTAGATA
Supplementary Table 1. Primers used in qpcr Gene forward primer (5'-3') reverse primer (5'-3') β-actin AGAGGGAAATCGTGCGTGAC CAATAGTGATGACCTGGCCGT Hif-p4h-2 CTGGGCAACTACAGGATAAAC GCGTCCCAGTCTTTATTTAGATA
Supplementary information for: Community detection for networks with. unipartite and bipartite structure. Chang Chang 1, 2, Chao Tang 2
 Supplementary information for: Community detection for networks with unipartite and bipartite structure Chang Chang 1, 2, Chao Tang 2 1 School of Life Sciences, Peking University, Beiing 100871, China
Supplementary information for: Community detection for networks with unipartite and bipartite structure Chang Chang 1, 2, Chao Tang 2 1 School of Life Sciences, Peking University, Beiing 100871, China
Supporting Information
 Supporting Information Franco et al. 10.1073/pnas.1015557108 SI Materials and Methods Drug Administration. PD352901 was dissolved in 0.5% (wt/vol) hydroxyl-propyl-methylcellulose, 0.2% (vol/vol) Tween
Supporting Information Franco et al. 10.1073/pnas.1015557108 SI Materials and Methods Drug Administration. PD352901 was dissolved in 0.5% (wt/vol) hydroxyl-propyl-methylcellulose, 0.2% (vol/vol) Tween
Supplementary Figure 1.
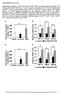 Supplementary Figure 1. FGF21 does not exert direct effects on hepatic glucose production. The liver explants from C57BL/6J mice (A, B) or primary rat hepatocytes (C, D) were incubated with rmfgf21 (2
Supplementary Figure 1. FGF21 does not exert direct effects on hepatic glucose production. The liver explants from C57BL/6J mice (A, B) or primary rat hepatocytes (C, D) were incubated with rmfgf21 (2
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Sherman SI, Wirth LJ, Droz J-P, et al. Motesanib diphosphate
Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna
 Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna targeted for hamster PAQR3. At 24 h after the transfection,
Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna targeted for hamster PAQR3. At 24 h after the transfection,
c Tuj1(-) apoptotic live 1 DIV 2 DIV 1 DIV 2 DIV Tuj1(+) Tuj1/GFP/DAPI Tuj1 DAPI GFP
 Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
Supplementary Figure 1 Establishment of the gain- and loss-of-function experiments and cell survival assays. a Relative expression of mature mir-484 30 20 10 0 **** **** NCP mir- 484P NCP mir- 484P b Relative
genomics for systems biology / ISB2020 RNA sequencing (RNA-seq)
 RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
Supplementary Figure 1 IL-27 IL
 Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Tim-3 Supplementary Figure 1 Tc0 49.5 0.6 Tc1 63.5 0.84 Un 49.8 0.16 35.5 0.16 10 4 61.2 5.53 10 3 64.5 5.66 10 2 10 1 10 0 31 2.22 10 0 10 1 10 2 10 3 10 4 IL-10 28.2 1.69 IL-27 Supplementary Figure 1.
Expanded View Figures
 Expanded View Figures A B C D E F G H I J K L Figure EV1. The dysregulated lipid metabolic phenotype of mouse models of metabolic dysfunction is most pronounced in the fasted state. A L Male 12-weeks-old
Expanded View Figures A B C D E F G H I J K L Figure EV1. The dysregulated lipid metabolic phenotype of mouse models of metabolic dysfunction is most pronounced in the fasted state. A L Male 12-weeks-old
Figure S1. IRF5 mrna expression is not expressed modulated by steatosis grade in
 9-INS-RG-TR- SUPPLEMENTRY MTERILS B IRF mrn expression 1 Control Fatty liver NSH HCV αsm IRF 1 ERG IRF < -33 33- Steatosis (%) < Figure S1. IRF mrn expression is not expressed modulated by steatosis grade
9-INS-RG-TR- SUPPLEMENTRY MTERILS B IRF mrn expression 1 Control Fatty liver NSH HCV αsm IRF 1 ERG IRF < -33 33- Steatosis (%) < Figure S1. IRF mrn expression is not expressed modulated by steatosis grade
Figure S1A. Blood glucose levels in mice after glucose injection
 ## Figure S1A. Blood glucose levels in mice after glucose injection Blood glucose (mm/l) 25 2 15 1 5 # 15 3 6 3+3 Time after glucose injection (min) # Figure S1B. α-kg levels in mouse livers after glucose
## Figure S1A. Blood glucose levels in mice after glucose injection Blood glucose (mm/l) 25 2 15 1 5 # 15 3 6 3+3 Time after glucose injection (min) # Figure S1B. α-kg levels in mouse livers after glucose
SUPPLEMENTAL DATA AGING, July 2014, Vol. 6 No. 7
 SUPPLEMENTAL DATA Figure S1. Muscle mass changes in different anatomical regions with age. (A) The TA and gastrocnemius muscle showed a significant loss of weight in aged mice (24 month old) compared to
SUPPLEMENTAL DATA Figure S1. Muscle mass changes in different anatomical regions with age. (A) The TA and gastrocnemius muscle showed a significant loss of weight in aged mice (24 month old) compared to
Mouse Meda-4 : chromosome 5G bp. EST (547bp) _at. 5 -Meda4 inner race (~1.8Kb)
 Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
Supplemental.Figure1 A: Mouse Meda-4 : chromosome 5G3 19898bp I II III III IV V a b c d 5 RACE outer primer 5 RACE inner primer 5 RACE Adaptor ORF:912bp Meda4 cdna 2846bp Meda4 specific 5 outer primer
Table S1. Quantitative RT-PCR primers
 Table S1. Quantitative RT-PCR primers Gene Forward Primer Reverse Primer Human ApoB gcaagcagaagccagaagta ccatttggagaagcagtttgg Human ApoA1 gaaagctgcggtgctgac agtggccaggtccttcact Human MTP acggccattcccattgtg
Table S1. Quantitative RT-PCR primers Gene Forward Primer Reverse Primer Human ApoB gcaagcagaagccagaagta ccatttggagaagcagtttgg Human ApoA1 gaaagctgcggtgctgac agtggccaggtccttcact Human MTP acggccattcccattgtg
