SUPPLEMENTARY INFORMATION
|
|
|
- Jacob Cook
- 5 years ago
- Views:
Transcription
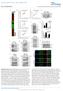
1 DOI: /ncb3073 LATS2 Binding ability to SIAH2 Degradation by SIAH / / 1-0 Supplementary Figure 1 Schematic drawing of LATS2 deletion mutants and their responses to interaction with and degradation by SIAH2. A schematic diagram of LATS2 deletion mutants is shown on the left panel. Blue represents as LATS Conserved Domain 1(LCD1) (aa.1-160), yellow as LATS Conserved Domain 2(LCD2) (aa ), red as PAPA repeat (aa ), green as Protein Binding Domain (PBD) (aa.618-0), purple as Kinase Domain. LATS2 binding activity to SIAH2 is shown in the middle panel as assayed in Fig 1d. indicates as binding, - as no binding, and -/ as less binding. The right panel shows the responses of LATS2 mutants to SIAH2-induced degradation. Cells expressing constant amounts of mutant and increasing amounts of were subjected to immunoblotting. indicates response to SIAH2-induced degradation and - indicates non-response. 1
2 a b - - RM c MG132 d BA1 e f CHX(Hours) RM CHX(Hours) Supplementary Figure 2 SIAH2 destabilizes LATS2 through proteasome. (a, b) LATS2 degradation was dosage-dependent on SIAH2 (a), but not on SIAH2 RM, an E3 ligase-activity-dead mutant of SIAH2 (b). (c, d) SIAH2-mediated LATS2 degradation was inhibited by proteasomal inhibitor MG132 (c), but not by lysosomal inhibitor BA-1 (d). (e, f) The half-life of LATS2 was shortened by SIAH2 (e) but not by SIAH2 RM (f) in cycloheximide chase assays. 2
3 a b Input IP: Flag - - Flag-SIAH1 RM Flag-SIAH1 Flag-SIAH1 RM RM Flag-SIAH1 c Input IP: Myc Input IP: Flag Myc-LATS1 RM d RM Myc-LATS1 e Input IP: Flag - - Myc-LATS1 Flag-SIAH1 RM Supplementary Figure 3 LATS1/2 is specifically regulated by SIAH2 but not by SIAH1. (a) No co-immunoprecipitation of exogenously expressed by Flag-SIAH1 RM. (b) LATS2 degradation is specifically promoted by SIAH2 but not by SIAH1. (c) Co-immunoprecipitation of exogenous expressed by Myc-LATS1 and vice versa. (d) LATS1 stability is regulated by SIAH2 but not by SIAH2 RM. (e) No co-immunoprecipitation of exogenously expressed Myc-LATS1 by Flag- SIAH1 RM. 3
4 a b DMSO MG132 c Hif1α Relative mrna level P = 0.2 P = 0.3 P = 0.5 Scramble YAP#1 YAP#2 Nor Hy Hypoxia IB:YAP IB:HIF1α IB: Hypoxia (MG132) Scramble 1# 2# YAP RNAi IB:OH-HIF1α (Pro564) IB:HIF1 α IB:YAP IB: Supplement Figure 4 YAP protects HIF1α from proteasomal degradation under hypoxia. (a) Scramble and YAP knockdown MDA-MB-231 cells were cultured under hypoxic conditions. Total RNA was extracted and subjected to RT-qPCR analysis for HIF1α mrna expression. (b) MG132 stabilized HIF1α in YAP knockdown cells under hypoxic conditions. (c) Scramble and YAP knockdown MDA-MB-231 cells were cultured under hypoxic conditions in the presence of MG132. Protein expressions were analyzed by immunoblotting with indicated antibodies. Data in a is the mean of n=3 independent experiments and error bars indicate s.d.. Two-tailed, unpaired Student s t-test. 4
5 26 17 input Figure 1a Figure 1b Figure 1c Figure 1d Figure 1f Figure 2a YAP Figure 2b Figure 2c Figure 2e Figure 2g Supplement Figure 5 continued Uncropped images of key blots/gels. 5
6 Figure 2h Figure 2i Figure 2l Figure 2k Cyr61 Hif1a LATS2 p-yap LATS2 YAP Cyr61 CTGF p-yap SIAH2 Figure 3a Figure 3g Figure 3e Supplement Figure 5 continued Uncropped images of key blots/gels. 6
7 input IgG LATS2 LATS2 YAP p-yap LATS2 Ub CTGF Cyr61 Figure 3i Figure 3j Figure 5a Figure 5i Figure 5j Supplement Figure 5 continued Uncropped images of key blots/gels. 7
8 YAP HIF1a 26 YAP-S127A-Myc HA-VHL Figure 5m Hif1a Figure 5o Hypoxia Figure 5n Normoxia Figure 5p Supplement Figure 5 continued Uncropped images of key blots/gels. 8
9 Supplementary Table 1: Identification of LATS2 as a SIAH2-associated protein. Protein Number of peptides 2-oxoglutarate dehyddrogenase 35 2-oxoglutarate dehydrogenase-like 16 Serine/threonine-protein kinase LATS2 5 Homeodomain-interacting protein kinase 4 SH3 domain-containing RING finger protein 3 (POSH2) 4 E3 ubiquitin-protein ligase SH3RF1 (POSH) 1 Supplementary Table 1 Identification of LATS2 as a SIAH2-associated protein. LATS2 and known interacting partners of SIAH2 and their related proteins are listed together with the number of peptides for each protein identified by mass spectrometry. 9
10 Supplementary Table 2: Species aligned for conservation of LATS2 lysine residues. No. Species 1 Homo sapiens 2 Mus musculus 3 Rattus norvegicus 4 Monodelphis domestica 5 Pan troglodytes 6 Pongo abelii 7 Nomascus leucogenys 8 Ailuropoda melanoleuca 9 Sus scrofa 10 Bos taurus 11 Ovis aries 12 Equus caballus 13 Loxodonta africana 14 Canis familiaris 15 Felis catus 16 Myotis lucifugus 17 Sarcophilus harrisii 18 Tasmanian devil 19 Callithrix jacchus 20 Oryctolagus cuniculus 21 Gallus gallus 22 Meleagris gallopavo 23 Anas platyrhynchos 24 Ficedula albicollis 25 Taeniopygia guttata 26 Xenopus tropicalis 27 Pelodiscus sinensis 28 Danio rerio 29 Drosophila melanogaster Supplementary Table 2 Species aligned for conservation of LATS2 lysine residues. Human LATS2 were aligned with the listed 28 species to identify those conserved lysine residues in Human LATS2 ( ). 10
11 Supplementary Table 3: LATS2 protein is downregulated and inversely correlated with SIAH2 protein levels in human breast cancer LATS2-low LATS2-high Total Breast tumor 138 (68.0%) 65 (32.0%) 203 Normal tissue 18 (25.7%) 52 (74.3%) 70 P = R = SIAH2-low SIAH2-high Total Breast tumor 105(51.5%) 99(48.5%) 204 Normal tissue 69(89.6%) 8(10.4%) 77 P = R = 0.35 LATS2-low LATS2-high Total SIAH2-low 60 (57.7%) 44(42.2%) 104 SIAH2-high 78 (78.8%) 21 (21.2%) 99 Total P = 1.3x10-3 R = Supplementary Table 3 LATS2 protein is downregulated and inversely correlated with SIAH2 protein levels in human breast cancer. LATS2 and SIAH2 protein expression status in normal breast and breast carcinoma specimens. Correlation between LATS2 and SIAH2 protein levels in human breast tumors. Statistical significance was determined by χ 2 test. R is the correlation coefficient. 11
12 Supplementary Table 4: Statistics source data Fig. 4a 0h 48h Vector SIAH2 SIAH2 RM Vector SIAH2 SIAH2 RM Vector # # # Mean S.D Fig. 4b Vector SIAH2 SIAH2 RM # # # # Mean S.D Fig. 4c Scramble SIAH2 RNAi SIAH2&LATS2 RNAi # # # Mean S.D Fig. 4d Scramble SIAH2 RNAi SIAH2&LATS2 RNAi # # # Mean S.D Supplementary Table 4 Statistics source data. Statistics source data of Figure 4a-d 12
Supplementary Figure 1. CFTR protein structure and domain architecture.
 A Plasma Membrane NH ₂ COOH Supplementary Figure. CFT protein structure and domain architecture. (A) Open state CFT homology model, ribbon representation from Serohijos et al. 8 PNAS 5:356. CFT domains
A Plasma Membrane NH ₂ COOH Supplementary Figure. CFT protein structure and domain architecture. (A) Open state CFT homology model, ribbon representation from Serohijos et al. 8 PNAS 5:356. CFT domains
Supporting Information
 Supporting Information McCullough et al. 10.1073/pnas.0801567105 A α10 α8 α9 N α7 α6 α5 C β2 β1 α4 α3 α2 α1 C B N C Fig. S1. ALIX Bro1 in complex with the C-terminal CHMP4A helix. (A) Ribbon diagram showing
Supporting Information McCullough et al. 10.1073/pnas.0801567105 A α10 α8 α9 N α7 α6 α5 C β2 β1 α4 α3 α2 α1 C B N C Fig. S1. ALIX Bro1 in complex with the C-terminal CHMP4A helix. (A) Ribbon diagram showing
supplementary information
 DOI: 10.1038/ncb1875 Figure S1 (a) The 79 surgical specimens from NSCLC patients were analysed by immunohistochemistry with an anti-p53 antibody and control serum (data not shown). The normal bronchi served
DOI: 10.1038/ncb1875 Figure S1 (a) The 79 surgical specimens from NSCLC patients were analysed by immunohistochemistry with an anti-p53 antibody and control serum (data not shown). The normal bronchi served
Supplementary Data to: Marco Mariotti and Roderic Guigó
 Supplementary Data to: Selenoprofiles: profile-based scanning of eukaryotic genome sequences for selenoprotein genes Marco Mariotti and Roderic Guigó Section S1: patterns used with SECISearch We report
Supplementary Data to: Selenoprofiles: profile-based scanning of eukaryotic genome sequences for selenoprotein genes Marco Mariotti and Roderic Guigó Section S1: patterns used with SECISearch We report
Supplementary Material and Methods. Cell culture. HEK293T cells were maintained in DMEM high glucose supplemented with 10% fetal
 Supplementary Material and Methods Cell culture. HEK293T cells were maintained in DMEM high glucose supplemented with 10% fetal bovine serum (Gibco), penicillin (50 U/ml, Life Technology), and streptomycin
Supplementary Material and Methods Cell culture. HEK293T cells were maintained in DMEM high glucose supplemented with 10% fetal bovine serum (Gibco), penicillin (50 U/ml, Life Technology), and streptomycin
Supplementary Figure 1
 S U P P L E M E N TA R Y I N F O R M AT I O N DOI: 10.1038/ncb2896 Supplementary Figure 1 Supplementary Figure 1. Sequence alignment of TERB1 homologs in vertebrates. M. musculus TERB1 was derived from
S U P P L E M E N TA R Y I N F O R M AT I O N DOI: 10.1038/ncb2896 Supplementary Figure 1 Supplementary Figure 1. Sequence alignment of TERB1 homologs in vertebrates. M. musculus TERB1 was derived from
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12005 a S Ψ ΨΨ ΨΨ ΨΨ Ψ Ψ Ψ Ψ Ψ ΨΨ Ψ Ψ ΨΨΨΨΨΨ S1 (a.a. 1 751) S2 (a.a. 752 1353) S1 Fc S1 (a.a. 1-747) Fc b HCoV-EMC-S1-Fc SARS-CoV-S1-Fc HCoV-EMC-S1-Fc SARS-CoV-S1-Fc kda - 170 - - 130
doi:10.1038/nature12005 a S Ψ ΨΨ ΨΨ ΨΨ Ψ Ψ Ψ Ψ Ψ ΨΨ Ψ Ψ ΨΨΨΨΨΨ S1 (a.a. 1 751) S2 (a.a. 752 1353) S1 Fc S1 (a.a. 1-747) Fc b HCoV-EMC-S1-Fc SARS-CoV-S1-Fc HCoV-EMC-S1-Fc SARS-CoV-S1-Fc kda - 170 - - 130
Supplementary Figure 1
 Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Supplementary Figure 1 YAP negatively regulates IFN- signaling. (a) Immunoblot analysis of Yap knockdown efficiency with sh-yap (#1 to #4 independent constructs) in Raw264.7 cells. (b) IFN- -Luc and PRDs
Supplementary Figure 1. MAT IIα is Acetylated at Lysine 81.
 IP: Flag a Mascot PTM Modified Mass Error Position Gene Names Score Score Sequence m/z [ppm] 81 MAT2A;AMS2;MATA2 35.6 137.28 _AAVDYQK(ac)VVR_ 595.83-2.28 b Pre-immu After-immu Flag- WT K81R WT K81R / Flag
IP: Flag a Mascot PTM Modified Mass Error Position Gene Names Score Score Sequence m/z [ppm] 81 MAT2A;AMS2;MATA2 35.6 137.28 _AAVDYQK(ac)VVR_ 595.83-2.28 b Pre-immu After-immu Flag- WT K81R WT K81R / Flag
(A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939 (5 µm)
 Supplementary Figure Legends Figure S1. Tankyrase inhibition suppresses cell proliferation in an axin/β-catenin independent manner. (A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939
Supplementary Figure Legends Figure S1. Tankyrase inhibition suppresses cell proliferation in an axin/β-catenin independent manner. (A) SW480, DLD1, RKO and HCT116 cells were treated with DMSO or XAV939
SUPPLEMENTARY MATERIAL
 SYPPLEMENTARY FIGURE LEGENDS SUPPLEMENTARY MATERIAL Figure S1. Phylogenic studies of the mir-183/96/182 cluster and 3 -UTR of Casp2. (A) Genomic arrangement of the mir-183/96/182 cluster in vertebrates.
SYPPLEMENTARY FIGURE LEGENDS SUPPLEMENTARY MATERIAL Figure S1. Phylogenic studies of the mir-183/96/182 cluster and 3 -UTR of Casp2. (A) Genomic arrangement of the mir-183/96/182 cluster in vertebrates.
supplementary information
 DOI: 10.1038/ncb2153 Figure S1 Ectopic expression of HAUSP up-regulates REST protein. (a) Immunoblotting showed that ectopic expression of HAUSP increased REST protein levels in ENStemA NPCs. (b) Immunofluorescent
DOI: 10.1038/ncb2153 Figure S1 Ectopic expression of HAUSP up-regulates REST protein. (a) Immunoblotting showed that ectopic expression of HAUSP increased REST protein levels in ENStemA NPCs. (b) Immunofluorescent
Uninformative BRCA Tests. Rebecca Sutphen, M.D. Professor, USF College of Medicine Chief Medical Officer, InformedDNA
 Uninformative RCA Tests Rebecca Sutphen, M.D. Professor, USF College of Medicine Chief Medical Officer, InformedDNA Uninformative RCA tests 1. R/OV cancer patient with normal results 2. Cancer-free person
Uninformative RCA Tests Rebecca Sutphen, M.D. Professor, USF College of Medicine Chief Medical Officer, InformedDNA Uninformative RCA tests 1. R/OV cancer patient with normal results 2. Cancer-free person
Kirschner, 2005). Briefly, parallel MEF cultures were isolated from single littermate
 SUPPLEMENTAL MATERIALS AND METHODS Generation of MEFs, osteoblasts and cell culture Prkar1a -/- and control MEFs were generated and cultured as described (Nadella and Kirschner, 2005). Briefly, parallel
SUPPLEMENTAL MATERIALS AND METHODS Generation of MEFs, osteoblasts and cell culture Prkar1a -/- and control MEFs were generated and cultured as described (Nadella and Kirschner, 2005). Briefly, parallel
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis
 Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
Supplementary Figure 1. PD-L1 is glycosylated in cancer cells. (a) Western blot analysis of PD-L1 in breast cancer cells. (b) Western blot analysis of PD-L1 in ovarian cancer cells. (c) Western blot analysis
mirna-guided regulation at the molecular level
 molecular level Hervé Seitz IGH du CNRS, Montpellier, France March 3, 2016 microrna target prediction . microrna target prediction mirna: target: 2 7 5 N NNNNNNNNNNNNNN 3 NNNNNN the seed microrna target
molecular level Hervé Seitz IGH du CNRS, Montpellier, France March 3, 2016 microrna target prediction . microrna target prediction mirna: target: 2 7 5 N NNNNNNNNNNNNNN 3 NNNNNN the seed microrna target
Supplemental Information. NRF2 Is a Major Target of ARF. in p53-independent Tumor Suppression
 Molecular Cell, Volume 68 Supplemental Information NRF2 Is a Major Target of ARF in p53-independent Tumor Suppression Delin Chen, Omid Tavana, Bo Chu, Luke Erber, Yue Chen, Richard Baer, and Wei Gu Figure
Molecular Cell, Volume 68 Supplemental Information NRF2 Is a Major Target of ARF in p53-independent Tumor Suppression Delin Chen, Omid Tavana, Bo Chu, Luke Erber, Yue Chen, Richard Baer, and Wei Gu Figure
**! Yuan et al., Supplemental Figure 1, related to Figure 1! EYA2 modulates the transcriptional activity of ERb, but not ERa! -DPN! +DPN!
 Yuan et al., Supplemental Figure 1, related to Figure 1! EY2 modulates the transcriptional activity of ERb, but not ERa!! B! -DPN! +DPN! * ERb GPDH MCF7 MD-MB-231 Primary BC - KD - KD #1 #2 #3 Relative
Yuan et al., Supplemental Figure 1, related to Figure 1! EY2 modulates the transcriptional activity of ERb, but not ERa!! B! -DPN! +DPN! * ERb GPDH MCF7 MD-MB-231 Primary BC - KD - KD #1 #2 #3 Relative
Supplementary Materials
 Supplementary Materials Supplementary Figure S1 Regulation of Ubl4A stability by its assembly partner A, The translation rate of Ubl4A is not affected in the absence of Bag6. Control, Bag6 and Ubl4A CRISPR
Supplementary Materials Supplementary Figure S1 Regulation of Ubl4A stability by its assembly partner A, The translation rate of Ubl4A is not affected in the absence of Bag6. Control, Bag6 and Ubl4A CRISPR
Phosphoproteomics and the Origin and Operations of the Kineome
 Phosphoproteomics and the Origin and Operations of the Kineome Presented by Steven Pelech, Ph.D. Professor, Department of Medicine, University of British Columbia President & CSO, Kinexus Bioinformatics
Phosphoproteomics and the Origin and Operations of the Kineome Presented by Steven Pelech, Ph.D. Professor, Department of Medicine, University of British Columbia President & CSO, Kinexus Bioinformatics
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
Supplementary Information
 Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Supplementary Information mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis Zhang et al. a b d e g h Rel. Luc. Act. Rel. mrna Rel. mrna
Ccdc124: A novel protein involved in cytokinesis.
 Ccdc124: A novel protein involved in cytokinesis. Uygar H. Tazebay Gebze Technical University Department of Molecular Biology and Genetics Gebze, Turkey International Conference on Science and Technology
Ccdc124: A novel protein involved in cytokinesis. Uygar H. Tazebay Gebze Technical University Department of Molecular Biology and Genetics Gebze, Turkey International Conference on Science and Technology
Supplementary figure legends
 Supplementary figure legends SUPPLEMENTRY FIGURE S1. Lentiviral construct. Schematic representation of the PCR fragment encompassing the genomic locus of mir-33a that was introduced in the lentiviral construct.
Supplementary figure legends SUPPLEMENTRY FIGURE S1. Lentiviral construct. Schematic representation of the PCR fragment encompassing the genomic locus of mir-33a that was introduced in the lentiviral construct.
supplementary information
 DOI: 1.138/ncb1 Control Atg7 / NAC 1 1 1 1 (mm) Control Atg7 / NAC 1 1 1 1 (mm) Lamin B Gstm1 Figure S1 Neither the translocation of into the nucleus nor the induction of antioxidant proteins in autophagydeficient
DOI: 1.138/ncb1 Control Atg7 / NAC 1 1 1 1 (mm) Control Atg7 / NAC 1 1 1 1 (mm) Lamin B Gstm1 Figure S1 Neither the translocation of into the nucleus nor the induction of antioxidant proteins in autophagydeficient
RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh-
 1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
1 a b Supplementary Figure 1. Effects of GSK3b knockdown on poly I:C-induced cytokine production. RAW264.7 cells stably expressing control shrna (Con) or GSK3b-specific shrna (sh- GSK3b) were stimulated
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
DOI: 10.1038/ncb3076 Supplementary Figure 1 btrcp targets Cep68 for degradation during mitosis. a) Cep68 immunofluorescence in interphase and metaphase. U-2OS cells were transfected with control sirna
Mouse Clec9a ORF sequence
 Mouse Clec9a gene LOCUS NC_72 13843 bp DNA linear CON 1-JUL-27 DEFINITION Mus musculus chromosome 6, reference assembly (C57BL/6J). ACCESSION NC_72 REGION: 129358881-129372723 Mouse Clec9a ORF sequence
Mouse Clec9a gene LOCUS NC_72 13843 bp DNA linear CON 1-JUL-27 DEFINITION Mus musculus chromosome 6, reference assembly (C57BL/6J). ACCESSION NC_72 REGION: 129358881-129372723 Mouse Clec9a ORF sequence
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3445 In the format provided by the authors and unedited. a b HIF1A 14 12 Pearson R =.5419 P
DOI: 1.138/ncb3445 In the format provided by the authors and unedited. a b HIF1A 14 12 Pearson R =.5419 P
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
DOI: 10.1038/ncb2988 Supplementary Figure 1 Kif7 L130P encodes a stable protein that does not localize to cilia tips. (a) Immunoblot with KIF7 antibody in cell lysates of wild-type, Kif7 L130P and Kif7
S1a S1b S1c. S1d. S1f S1g S1h SUPPLEMENTARY FIGURE 1. - si sc Il17rd Il17ra bp. rig/s IL-17RD (ng) -100 IL-17RD
 SUPPLEMENTARY FIGURE 1 0 20 50 80 100 IL-17RD (ng) S1a S1b S1c IL-17RD β-actin kda S1d - si sc Il17rd Il17ra rig/s15-574 - 458-361 bp S1f S1g S1h S1i S1j Supplementary Figure 1. Knockdown of IL-17RD enhances
SUPPLEMENTARY FIGURE 1 0 20 50 80 100 IL-17RD (ng) S1a S1b S1c IL-17RD β-actin kda S1d - si sc Il17rd Il17ra rig/s15-574 - 458-361 bp S1f S1g S1h S1i S1j Supplementary Figure 1. Knockdown of IL-17RD enhances
Trim29 gene-targeting strategy. (a) Genotyping of wildtype mice (+/+), Trim29 heterozygous mice (+/ ) and homozygous mice ( / ).
 Supplementary Figure 1 Trim29 gene-targeting strategy. (a) Genotyping of wildtype mice (+/+), Trim29 heterozygous mice (+/ ) and homozygous mice ( / ). (b) Immunoblot analysis of TRIM29 in lung primary
Supplementary Figure 1 Trim29 gene-targeting strategy. (a) Genotyping of wildtype mice (+/+), Trim29 heterozygous mice (+/ ) and homozygous mice ( / ). (b) Immunoblot analysis of TRIM29 in lung primary
SUPPLEMENTARY INFORMATION
 Testing the accuracy of ancestral state reconstruction The accuracy of the ancestral state reconstruction with maximum likelihood methods can depend on the underlying model used in the reconstruction.
Testing the accuracy of ancestral state reconstruction The accuracy of the ancestral state reconstruction with maximum likelihood methods can depend on the underlying model used in the reconstruction.
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation
 Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna
 Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna targeted for hamster PAQR3. At 24 h after the transfection,
Supplementary Figure 1. PAQR3 knockdown inhibits SREBP-2 processing in CHO-7 cells CHO-7 cells were transfected with control sirna or a sirna targeted for hamster PAQR3. At 24 h after the transfection,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
DOI: 10.1038/ncb3021 Supplementary figure 1 Characterisation of TIMPless fibroblasts. a) Relative gene expression of TIMPs1-4 by real time quantitative PCR (RT-qPCR) in WT or ΔTimp fibroblasts (mean ±
293T cells were transfected with indicated expression vectors and the whole-cell extracts were subjected
 SUPPLEMENTARY INFORMATION Supplementary Figure 1. Formation of a complex between Slo1 and CRL4A CRBN E3 ligase. (a) HEK 293T cells were transfected with indicated expression vectors and the whole-cell
SUPPLEMENTARY INFORMATION Supplementary Figure 1. Formation of a complex between Slo1 and CRL4A CRBN E3 ligase. (a) HEK 293T cells were transfected with indicated expression vectors and the whole-cell
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
www.sciencesignaling.org/cgi/content/full/8/364/ra18/dc1 Supplementary Materials for The tyrosine phosphatase (Pez) inhibits metastasis by altering protein trafficking Leila Belle, Naveid Ali, Ana Lonic,
Predictive PP1Ca binding region in BIG3 : 1,228 1,232aa (-KAVSF-) HEK293T cells *** *** *** KPL-3C cells - E E2 treatment time (h)
 Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
Relative expression ERE-luciferase activity activity (pmole/min) activity (pmole/min) activity (pmole/min) activity (pmole/min) MCF-7 KPL-3C ZR--1 BT-474 T47D HCC15 KPL-1 HBC4 activity (pmole/min) a d
Supplemental Information. LEAP2 Is an Endogenous Antagonist. of the Ghrelin Receptor
 Cell Metabolism, Volume 27 Supplemental Information LEAP2 Is an Endogenous Antagonist of the Ghrelin Receptor Xuecai Ge, Hong Yang, Maria A. Bednarek, Hadas Galon-Tilleman, Peirong Chen, Michael Chen,
Cell Metabolism, Volume 27 Supplemental Information LEAP2 Is an Endogenous Antagonist of the Ghrelin Receptor Xuecai Ge, Hong Yang, Maria A. Bednarek, Hadas Galon-Tilleman, Peirong Chen, Michael Chen,
Supplementary Figure 1. Procedures for p38 activity imaging in living cells. (a) Schematic model of the p38 activity reporter. The reporter consists
 Supplementary Figure 1. Procedures for p38 activity imaging in living cells. (a) Schematic model of the p38 activity reporter. The reporter consists of: (i) the YPet domain (an enhanced YFP); (ii) the
Supplementary Figure 1. Procedures for p38 activity imaging in living cells. (a) Schematic model of the p38 activity reporter. The reporter consists of: (i) the YPet domain (an enhanced YFP); (ii) the
Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent Protein.
 prfp-vector RFP Exon1 Intron RFP Exon2 prfp-mir-124 mir-93/124 RFP Exon1 Intron RFP Exon2 Untransfected prfp-vector prfp-mir-93 prfp-mir-124 Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent
prfp-vector RFP Exon1 Intron RFP Exon2 prfp-mir-124 mir-93/124 RFP Exon1 Intron RFP Exon2 Untransfected prfp-vector prfp-mir-93 prfp-mir-124 Supplementary Fig. 1. Delivery of mirnas via Red Fluorescent
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk
 Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary Figure 1. Normal T lymphocyte populations in Dapk -/- mice. (a) Normal thymic development in Dapk -/- mice. Thymocytes from WT and Dapk -/- mice were stained for expression of CD4 and CD8.
Supplementary information. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins
 Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
Supplementary information inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins Takuya Tada, Yanzhao Zhang, Takayoshi Koyama, Minoru Tobiume, Yasuko Tsunetsugu-Yokota, Shoji
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
DOI: 1.138/ncb222 / b. WB anti- WB anti- ulin Mitotic index (%) 14 1 6 2 T (h) 32 48-1 1 2 3 4 6-1 4 16 22 28 3 33 e. 6 4 2 Time (min) 1-6- 11-1 > 1 % cells Figure S1 depletion leads to mitotic defects
Reviewers' comments: Reviewer #1 (Remarks to the Author):
 Reviewers' comments: Reviewer #1 (Remarks to the Author): In this manuscript, Song et al. identified FBXW7 as a new positive regulator for RIG-Itriggered type I IFN signaling pathway. The authors observed
Reviewers' comments: Reviewer #1 (Remarks to the Author): In this manuscript, Song et al. identified FBXW7 as a new positive regulator for RIG-Itriggered type I IFN signaling pathway. The authors observed
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC
 Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
Supplementary Figure 1. Characterization of NMuMG-ErbB2 and NIC breast cancer cells expressing shrnas targeting LPP. NMuMG-ErbB2 cells (a) and NIC cells (b) were engineered to stably express either a LucA-shRNA
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH
 EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
Boucher et al NCOMMS B
 1 Supplementary Figure 1 (linked to Figure 1). mvegfr1 constitutively internalizes in endothelial cells. (a) Immunoblot of mflt1 from undifferentiated mouse embryonic stem (ES) cells with indicated genotypes;
1 Supplementary Figure 1 (linked to Figure 1). mvegfr1 constitutively internalizes in endothelial cells. (a) Immunoblot of mflt1 from undifferentiated mouse embryonic stem (ES) cells with indicated genotypes;
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/10/471/eaah5085/dc1 Supplementary Materials for Phosphorylation of the exocyst protein Exo84 by TBK1 promotes insulin-stimulated GLUT4 trafficking Maeran Uhm,
www.sciencesignaling.org/cgi/content/full/10/471/eaah5085/dc1 Supplementary Materials for Phosphorylation of the exocyst protein Exo84 by TBK1 promotes insulin-stimulated GLUT4 trafficking Maeran Uhm,
File name: Supplementary Information Description: Supplementary figures, supplementary tables and supplementary references.
 Description Supplementary Files File name: Supplementary Information Description: Supplementary figures, supplementary tables and supplementary references. File name: Peer review file Supplementary Figure
Description Supplementary Files File name: Supplementary Information Description: Supplementary figures, supplementary tables and supplementary references. File name: Peer review file Supplementary Figure
Identification of non-ser/thr-pro consensus motifs for Cdk1 and their roles in mitotic regulation of C2H2 zinc finger proteins and Ect2
 SUPPLEMENTARY INFORMATION Identification of non-ser/thr-pro consensus motifs for Cdk1 and their roles in mitotic regulation of C2H2 zinc finger proteins and Ect2 Kazuhiro Suzuki, Kosuke Sako, Kazuhiro
SUPPLEMENTARY INFORMATION Identification of non-ser/thr-pro consensus motifs for Cdk1 and their roles in mitotic regulation of C2H2 zinc finger proteins and Ect2 Kazuhiro Suzuki, Kosuke Sako, Kazuhiro
(a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable
 Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
Supplementary Figure 1. Frameshift (FS) mutation in UVRAG. (a) Schematic diagram of the FS mutation of UVRAG in exon 8 containing the highly instable A 10 DNA repeat, generating a premature stop codon
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Díaz et al., http://www.jcb.org/cgi/content/full/jcb.201209151/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Hypoxia induces invadopodia formation in different epithelial
Supplemental material Díaz et al., http://www.jcb.org/cgi/content/full/jcb.201209151/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Hypoxia induces invadopodia formation in different epithelial
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
Supplementary Figure 1 Mutational analysis of the SA2-Scc1 interaction in vitro and in human cells. (a) Autoradiograph (top) and Coomassie stained gel (bottom) of 35 S-labeled Myc-SA2 proteins (input)
SUPPLEMENTAL FIGURE LEGENDS
 SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure S1: Endogenous interaction between RNF2 and H2AX: Whole cell extracts from 293T were subjected to immunoprecipitation with anti-rnf2 or anti-γ-h2ax antibodies
SUPPLEMENTAL FIGURE LEGENDS Supplemental Figure S1: Endogenous interaction between RNF2 and H2AX: Whole cell extracts from 293T were subjected to immunoprecipitation with anti-rnf2 or anti-γ-h2ax antibodies
Supplementary Information
 Supplementary Information A METTL3-METTL14 complex mediates mammalian nuclear RNA N 6 -adenosine methylation Jianzhao Liu, 1,2 Yanan Yue, 1,2 Dali Han, 1 Xiao Wang, 1 Ye Fu, 1 Liang Zhang, 1 Guifang Jia,
Supplementary Information A METTL3-METTL14 complex mediates mammalian nuclear RNA N 6 -adenosine methylation Jianzhao Liu, 1,2 Yanan Yue, 1,2 Dali Han, 1 Xiao Wang, 1 Ye Fu, 1 Liang Zhang, 1 Guifang Jia,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/430/ra57/dc1 Supplementary Materials for The 4E-BP eif4e axis promotes rapamycinsensitive growth and proliferation in lymphocytes Lomon So, Jongdae Lee, Miguel
www.sciencesignaling.org/cgi/content/full/9/430/ra57/dc1 Supplementary Materials for The 4E-BP eif4e axis promotes rapamycinsensitive growth and proliferation in lymphocytes Lomon So, Jongdae Lee, Miguel
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Differential expression of mirnas from the pri-mir-17-92a locus.
 Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
This PDF file includes: Supplementary Figures 1 to 6 Supplementary Tables 1 to 2 Supplementary Methods Supplementary References
 Structure of the catalytic core of p300 and implications for chromatin targeting and HAT regulation Manuela Delvecchio, Jonathan Gaucher, Carmen Aguilar-Gurrieri, Esther Ortega, Daniel Panne This PDF file
Structure of the catalytic core of p300 and implications for chromatin targeting and HAT regulation Manuela Delvecchio, Jonathan Gaucher, Carmen Aguilar-Gurrieri, Esther Ortega, Daniel Panne This PDF file
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous
 Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Supplementary Figure 1. Spatial distribution of LRP5 and β-catenin in intact cardiomyocytes. (a) and (b) Immunofluorescence staining of endogenous LRP5 in intact adult mouse ventricular myocytes (AMVMs)
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3311 A B TSC2 -/- MEFs C Rapa Hours WCL 0 6 12 24 36 pakt.s473 AKT ps6k S6K CM IGF-1 Recipient WCL - + - + - + pigf-1r IGF-1R pakt ps6 AKT D 1 st SILAC 2 nd SILAC E GAPDH FGF21 ALKPGVIQILGVK
DOI: 10.1038/ncb3311 A B TSC2 -/- MEFs C Rapa Hours WCL 0 6 12 24 36 pakt.s473 AKT ps6k S6K CM IGF-1 Recipient WCL - + - + - + pigf-1r IGF-1R pakt ps6 AKT D 1 st SILAC 2 nd SILAC E GAPDH FGF21 ALKPGVIQILGVK
Evolution of Leptin Structure and Function
 At the Cutting Edge DOI: 10.1159/000328435 Received: January 31, 2011 Accepted after revision: April 11, 2011 Published online: June 16, 2011 Evolution of Leptin Structure and Function Robert J. Denver
At the Cutting Edge DOI: 10.1159/000328435 Received: January 31, 2011 Accepted after revision: April 11, 2011 Published online: June 16, 2011 Evolution of Leptin Structure and Function Robert J. Denver
INSULIN AND ISLET AMYLOID POLYPEPTIDE GENES IN DIFFERENT SPECIES
 BIOLOGY INSULIN AND ISLET AMYLOID POLYPEPTIDE GENES IN DIFFERENT SPECIES PAUL GAGNIUC 1, CONSTANTIN IONESCU-TIRGOVISTE 2 1 Institute of Genetics, University of Bucharest, Romania 2 National Institute of
BIOLOGY INSULIN AND ISLET AMYLOID POLYPEPTIDE GENES IN DIFFERENT SPECIES PAUL GAGNIUC 1, CONSTANTIN IONESCU-TIRGOVISTE 2 1 Institute of Genetics, University of Bucharest, Romania 2 National Institute of
Supplementary Information for. A cancer-associated BRCA2 mutation reveals masked nuclear. export signals controlling localization
 Supplementary Information for A cancer-associated BRCA2 mutation reveals masked nuclear export signals controlling localization Anand D Jeyasekharan 1, Yang Liu 1, Hiroyoshi Hattori 1,3, Venkat Pisupati
Supplementary Information for A cancer-associated BRCA2 mutation reveals masked nuclear export signals controlling localization Anand D Jeyasekharan 1, Yang Liu 1, Hiroyoshi Hattori 1,3, Venkat Pisupati
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature05732 SUPPLEMENTARY INFORMATION Supplemental Data Supplement Figure Legends Figure S1. RIG-I 2CARD undergo robust ubiquitination a, (top) At 48 h posttransfection with a GST, GST-RIG-I-2CARD
doi: 10.1038/nature05732 SUPPLEMENTARY INFORMATION Supplemental Data Supplement Figure Legends Figure S1. RIG-I 2CARD undergo robust ubiquitination a, (top) At 48 h posttransfection with a GST, GST-RIG-I-2CARD
Supplemental Fig. 1. Relative mrna Expression. Relative mrna Expression WT KO WT KO RT 4 0 C
 Supplemental Fig. 1 A 1.5 1..5 Hdac11 (ibat) n=4 n=4 n=4 n=4 n=4 n=4 n=4 n=4 WT KO WT KO WT KO WT KO RT 4 C RT 4 C Supplemental Figure 1. Hdac11 mrna is undetectable in KO adipose tissue. Quantitative
Supplemental Fig. 1 A 1.5 1..5 Hdac11 (ibat) n=4 n=4 n=4 n=4 n=4 n=4 n=4 n=4 WT KO WT KO WT KO WT KO RT 4 C RT 4 C Supplemental Figure 1. Hdac11 mrna is undetectable in KO adipose tissue. Quantitative
A. List of selected proteins with high SILAC (H/L) ratios identified in mass
 Supplementary material Figure S1. Interaction between UBL5 and FANCI A. List of selected proteins with high SILAC (H/L) ratios identified in mass spectrometry (MS)-based analysis of UBL5-interacting proteins,
Supplementary material Figure S1. Interaction between UBL5 and FANCI A. List of selected proteins with high SILAC (H/L) ratios identified in mass spectrometry (MS)-based analysis of UBL5-interacting proteins,
Supplemental Table 1. List of cell lines used
 Supplemental Table 1. List of cell lines used Common species name Latin species name Source Common Marmoset Southern Lesser Bushbaby Callithrix jacchus In-house (1 line) Coriell number (if applicable)
Supplemental Table 1. List of cell lines used Common species name Latin species name Source Common Marmoset Southern Lesser Bushbaby Callithrix jacchus In-house (1 line) Coriell number (if applicable)
PREPARED FOR: U.S. Army Medical Research and Materiel Command Fort Detrick, Maryland
 AD Award Number: W81XWH-10-1-1029 TITLE: PRINCIPAL INVESTIGATOR: Mu-Shui Dai, M.D., Ph.D. CONTRACTING ORGANIZATION: Oregon Health Science niversity, Portland, Oregon 97239 REPORT DATE: October 2013 TYPE
AD Award Number: W81XWH-10-1-1029 TITLE: PRINCIPAL INVESTIGATOR: Mu-Shui Dai, M.D., Ph.D. CONTRACTING ORGANIZATION: Oregon Health Science niversity, Portland, Oregon 97239 REPORT DATE: October 2013 TYPE
Tumor suppressor Spred2 interaction with LC3 promotes autophagosome maturation and induces autophagy-dependent cell death
 www.impactjournals.com/oncotarget/ Oncotarget, Supplementary Materials 2016 Tumor suppressor Spred2 interaction with LC3 promotes autophagosome maturation and induces autophagy-dependent cell death Supplementary
www.impactjournals.com/oncotarget/ Oncotarget, Supplementary Materials 2016 Tumor suppressor Spred2 interaction with LC3 promotes autophagosome maturation and induces autophagy-dependent cell death Supplementary
Plasma exposure levels from individual mice 4 hours post IP administration at the
 Supplemental Figure Legends Figure S1. Plasma exposure levels of MKC-3946 in mice. Plasma exposure levels from individual mice 4 hours post IP administration at the indicated dose mg/kg. Data represent
Supplemental Figure Legends Figure S1. Plasma exposure levels of MKC-3946 in mice. Plasma exposure levels from individual mice 4 hours post IP administration at the indicated dose mg/kg. Data represent
Live cell imaging of trafficking of the chaperone complex vaccine to the ER. BMDCs were incubated with ER-Tracker Red (1 M) in staining solution for
 Live cell imaging of trafficking of the chaperone complex vaccine to the ER. BMDCs were incubated with ER-Tracker Red (1 M) in staining solution for 15 min at 37 C and replaced with fresh complete medium.
Live cell imaging of trafficking of the chaperone complex vaccine to the ER. BMDCs were incubated with ER-Tracker Red (1 M) in staining solution for 15 min at 37 C and replaced with fresh complete medium.
A Central Role of MG53 in Metabolic Syndrome. and Type-2 Diabetes
 A Central Role of MG53 in Metabolic Syndrome and Type-2 Diabetes Yan Zhang, Chunmei Cao, Rui-Ping Xiao Institute of Molecular Medicine (IMM) Peking University, Beijing, China Accelerated Aging in China
A Central Role of MG53 in Metabolic Syndrome and Type-2 Diabetes Yan Zhang, Chunmei Cao, Rui-Ping Xiao Institute of Molecular Medicine (IMM) Peking University, Beijing, China Accelerated Aging in China
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11700 Figure 1: RIP3 as a potential Sirt2 interacting protein. Transfected Flag-tagged Sirt2 was immunoprecipitated from cells and eluted from the Sepharose beads using Flag peptide.
doi:10.1038/nature11700 Figure 1: RIP3 as a potential Sirt2 interacting protein. Transfected Flag-tagged Sirt2 was immunoprecipitated from cells and eluted from the Sepharose beads using Flag peptide.
Supplementary materials for: 28-Way Vertebrate Alignment and Conservation Track in the UCSC Genome Browser
 Supplementary materials for: 28-Way Vertebrate Alignment and Conservation Track in the UCSC Genome Browser Miller, Rosenbloom, Hardison, Hou, Taylor, Raney, Burhans, King, Baertsch, Blankenberg, Kosakovsky
Supplementary materials for: 28-Way Vertebrate Alignment and Conservation Track in the UCSC Genome Browser Miller, Rosenbloom, Hardison, Hou, Taylor, Raney, Burhans, King, Baertsch, Blankenberg, Kosakovsky
MOLECULE PHYLOGENETIC ANALYSIS OF GENE ENCODING ASPARTATE AMINOTRANSFERASE
 Analele Ştiinţifice ale Universităţii Alexandru Ioan Cuza, Secţiunea Genetică şi Biologie Moleculară, TOM IX, 2008 MOLECULE PHYLOGENETIC ANALYSIS OF GENE ENCODING ASPARTATE AMINOTRANSFERASE XIAOSHENG DING
Analele Ştiinţifice ale Universităţii Alexandru Ioan Cuza, Secţiunea Genetică şi Biologie Moleculară, TOM IX, 2008 MOLECULE PHYLOGENETIC ANALYSIS OF GENE ENCODING ASPARTATE AMINOTRANSFERASE XIAOSHENG DING
Obesity is healthy for cetaceans: Evidence from pervasive positive. selection in genes related to triacylglycerol metabolism
 Obesity is healthy for cetaceans: Evidence from pervasive positive selection in genes related to triacylglycerol metabolism Zhengfei Wang 1, Zhuo Chen 1, 2, Shixia Xu 1, Wenhua Ren 1, Kaiya Zhou 1 and
Obesity is healthy for cetaceans: Evidence from pervasive positive selection in genes related to triacylglycerol metabolism Zhengfei Wang 1, Zhuo Chen 1, 2, Shixia Xu 1, Wenhua Ren 1, Kaiya Zhou 1 and
gliomas. Fetal brain expected who each low-
 Supplementary Figure S1. Grade-specificity aberrant expression of HOXA genes in gliomas. (A) Representative RT-PCR analyses of HOXA gene expression in human astrocytomas. Exemplified glioma samples include
Supplementary Figure S1. Grade-specificity aberrant expression of HOXA genes in gliomas. (A) Representative RT-PCR analyses of HOXA gene expression in human astrocytomas. Exemplified glioma samples include
p = formed with HCI-001 p = Relative # of blood vessels that formed with HCI-002 Control Bevacizumab + 17AAG Bevacizumab 17AAG
 A.. Relative # of ECs associated with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 ol b p < 0.001 Relative # of blood vessels that formed with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 l b p = 0.002 Control IHC:
A.. Relative # of ECs associated with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 ol b p < 0.001 Relative # of blood vessels that formed with HCI-001 1.4 1.2 1.0 0.8 0.6 0.4 0.2 0.0 l b p = 0.002 Control IHC:
Supplemental Table 1. Plasma NEFA and liver triglyceride levels in ap2-hif1ako and ap2-hif2ako mice under control and high fat diets.
 Supplemental Table 1. Plasma NEFA and liver triglyceride levels in Hif1aKO and Hif2aKO mice under control and high fat diets. Hif1a (n=6) Hif1aK O (n=6) Hif2a Hif2aK O Hif1a (n=5) Hif1aKO (n=5) Hif2a Hif2aK
Supplemental Table 1. Plasma NEFA and liver triglyceride levels in Hif1aKO and Hif2aKO mice under control and high fat diets. Hif1a (n=6) Hif1aK O (n=6) Hif2a Hif2aK O Hif1a (n=5) Hif1aKO (n=5) Hif2a Hif2aK
Hipk2 and PP1c Cooperate to Maintain Dvl Protein Levels Required for Wnt Signal Transduction
 Cell Reports Article Hipk2 and PP1c Cooperate to Maintain Dvl Protein Levels Required for Wnt Signal Transduction Nobuyuki Shimizu, 1 Shizuka Ishitani, 1 Atsushi Sato, 2 Hiroshi Shibuya, 2 and Tohru Ishitani
Cell Reports Article Hipk2 and PP1c Cooperate to Maintain Dvl Protein Levels Required for Wnt Signal Transduction Nobuyuki Shimizu, 1 Shizuka Ishitani, 1 Atsushi Sato, 2 Hiroshi Shibuya, 2 and Tohru Ishitani
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS
 Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Supplementary Figure 1. Prevalence of U539C and G540A nucleotide and E172K amino acid substitutions among H9N2 viruses. Full-length H9N2 NS nucleotide sequences (a, b) or amino acid sequences (c) from
Nature Structural & Molecular Biology: doi: /nsmb.1529
 0.0389 0.0927 0.2641 0.4115 0.3630 0.3618 0.5153 0.3840 0.0357 1.026 0.0866 0.0423 0.1296 0.2052 0.2869 0.0948 0.6285 0.0835 0.0543 0.1135 0.1445 0.1501 0.3272 0.0418 0.4993 0.0624 0.0938 0.1189 0.0768
0.0389 0.0927 0.2641 0.4115 0.3630 0.3618 0.5153 0.3840 0.0357 1.026 0.0866 0.0423 0.1296 0.2052 0.2869 0.0948 0.6285 0.0835 0.0543 0.1135 0.1445 0.1501 0.3272 0.0418 0.4993 0.0624 0.0938 0.1189 0.0768
Bassoon and Piccolo maintain synapse integrity by regulating protein ubiquitination and degradation
 Manuscript EMBO-2012-82783 Bassoon and Piccolo maintain synapse integrity by regulating protein ubiquitination and degradation Clarissa L Waites, Sergio A Leal-Ortiz, Nathan Okerlund, Hannah Dalke, Anna
Manuscript EMBO-2012-82783 Bassoon and Piccolo maintain synapse integrity by regulating protein ubiquitination and degradation Clarissa L Waites, Sergio A Leal-Ortiz, Nathan Okerlund, Hannah Dalke, Anna
Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were
 Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were isolated from wild type (PKC-θ- WT) or PKC-θ null (PKC-θ-KO)
Supplementary Figure 1 CD4 + T cells from PKC-θ null mice are defective in NF-κB activation during T cell receptor signaling. CD4 + T cells were isolated from wild type (PKC-θ- WT) or PKC-θ null (PKC-θ-KO)
Nature Structural & Molecular Biology: doi: /nsmb.3218
 Supplementary Figure 1 Endogenous EGFR trafficking and responses depend on biased ligands. (a) Lysates from HeLa cells stimulated for 2 min. with increasing concentration of ligands were immunoblotted
Supplementary Figure 1 Endogenous EGFR trafficking and responses depend on biased ligands. (a) Lysates from HeLa cells stimulated for 2 min. with increasing concentration of ligands were immunoblotted
File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables. File Name: Peer Review File Description:
 File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables File Name: Peer Review File Description: Primer Name Sequence (5'-3') AT ( C) RT-PCR USP21 F 5'-TTCCCATGGCTCCTTCCACATGAT-3'
File Name: Supplementary Information Description: Supplementary Figures and Supplementary Tables File Name: Peer Review File Description: Primer Name Sequence (5'-3') AT ( C) RT-PCR USP21 F 5'-TTCCCATGGCTCCTTCCACATGAT-3'
m 6 A mrna methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer
 SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
Intrinsically Disordered Proteins. Alex Cioffi June 22 nd 2013
 Intrinsically Disordered Proteins Alex Cioffi June 22 nd 2013 Implications in Human Disease Adenovirus Early Region 1A (E1A) and Cancer α-synuclein and Parkinson s Disease Amyloid β and Alzheimer s Disease
Intrinsically Disordered Proteins Alex Cioffi June 22 nd 2013 Implications in Human Disease Adenovirus Early Region 1A (E1A) and Cancer α-synuclein and Parkinson s Disease Amyloid β and Alzheimer s Disease
Supporting information for: Structural basis for complement Factor I control and its disease-associated sequence polymorphisms.
 Supporting information for: Structural basis for complement Factor I control and its disease-associated sequence polymorphisms. Pietro Roversi 1a, Steven Johnson 1a, Joseph J.E. Caesar a, Florence McLean
Supporting information for: Structural basis for complement Factor I control and its disease-associated sequence polymorphisms. Pietro Roversi 1a, Steven Johnson 1a, Joseph J.E. Caesar a, Florence McLean
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2535 Figure S1 SOX10 is expressed in human giant congenital nevi and its expression in human melanoma samples suggests that SOX10 functions in a MITF-independent manner. a, b, Representative
DOI: 10.1038/ncb2535 Figure S1 SOX10 is expressed in human giant congenital nevi and its expression in human melanoma samples suggests that SOX10 functions in a MITF-independent manner. a, b, Representative
Supplemental Information Thrombopoietin induces production of nucleated thrombocytes from liver cells in Xenopus laevis
 Supplemental Information Thrombopoietin induces production of nucleated thrombocytes from liver cells in Xenopus laevis Yuta Tanizaki 1, Megumi Ichisugi 2, Miyako Shimoji-Obuchi 3, Takako Ishida-Iwata
Supplemental Information Thrombopoietin induces production of nucleated thrombocytes from liver cells in Xenopus laevis Yuta Tanizaki 1, Megumi Ichisugi 2, Miyako Shimoji-Obuchi 3, Takako Ishida-Iwata
