HMGB1 links chronic liver injury to progenitor responses and hepatocarcinogenesis
|
|
|
- Julius Bond
- 6 years ago
- Views:
Transcription
1 HMGB1 links chronic liver injury to progenitor responses and hepatocarcinogenesis Celine Hernandez 1, Peter Huebener 1,2, Jean-Philippe Pradere 3, Daniel J. Antoine 4, Richard A. Friedman 5, and Robert F. Schwabe 1 1 Department of Medicine, Columbia University, New York, NY 132, USA 2 Department of Medicine, University Medical Center Hamburg-Eppendorf, Hamburg, Germany 3 Institut National de la Santé et de la Recherche Médicale (INSERM) U148, Institute of Cardiovascular and Metabolic Disease, Toulouse, France 4 MRC Centre for Inflammation Research, University of Edinburgh, United Kingdom 5 Biomedical Informatics Shared Resource, Herbert Irving Comprehensive Cancer Center and Department of Biomedical Informatics, Columbia University, New York, NY 132, USA. Correspondence to: Robert F. Schwabe, 113 St.Nicholas Ave, ICRC, Room 926, New York, NY 132; phone: ; rfs212@cumc.columbia.edu The authors declare no conflict of interest. These authors contributed equally.
2 Cell death is a key driver of disease progression and carcinogenesis in chronic liver disease (CLD), highlighted by the well-established clinical correlation between hepatocellular death and risk for the development of cirrhosis and hepatocellular carcinoma (HCC). Moreover, hepatocellular death is sufficient to trigger fibrosis and HCC in mice. However, the pathways through which cell death drives CLD progression remain elusive. Here, we tested the hypothesis that high-mobility group box 1 (HMGB1), a damage-associated molecular pattern (DAMP) with key roles in acute liver injury, may link cell death to injury responses and hepatocarcinogenesis in CLD. While liver-specific HMGB1 deficiency did not significantly affect chronic injury responses such as fibrosis, regeneration and inflammation, it inhibited ductular/progenitor cell expansion and hepatocyte metaplasia. HMGB1 promoted ductular expansion independently of active secretion in a non-autonomous fashion, consistent with its role as DAMP. Liver-specific HMGB1 deficiency reduced HCC development in three models with chronic injury but not in a model lacking chronic liver injury. Similar to CLD, HMGB1 ablation reduced the expression of progenitor and oncofetal markers, a key determinant of HCC aggressiveness, in tumors. In summary, HMGB1 links hepatocyte death to ductular reaction, progenitor signature and hepatocarcinogenesis in CLD.
3 INTRODUCTION Chronic liver disease (CLD) causes approximately two million deaths per year worldwide (1). In patients with CLD, the development of liver cirrhosis and hepatocellular carcinoma (HCC) are the main contributors to morbidity and mortality and remain a significant clinical problem. HCC is the third leading cause of cancer mortality worldwide, and the fastest rising cause of cancer mortality in the US (1, 2). Importantly, cirrhosis and HCC are strongly linked to CLD and the presence of chronic hepatocellular death (3). Accordingly, a large body of well-designed clinical studies has demonstrated a profoundly increased risk for the development of cirrhosis (4, 5) and HCC (6-8) as well as liver-specific mortality (9) in patients with persistently elevated levels of serum ALT (reviewed in (3)). Interestingly, the increased risk for cirrhosis and HCC development in patients with high ALT is seen in different types of liver diseases, suggesting that cell death is a common risk factor for the progression (3). This concept is further supported by studies in mouse models with genetically-induced chronic hepatocyte death, such as mice with hepatocyte-specific deletion of Tak1, Nemo or Mcl-1, which spontaneously develop liver fibrosis and HCC (1-13), suggesting that chronic cell death is sufficient to initiate liver disease development and drive its progression. However, the mechanisms through which hepatocellular death promotes the development of liver cirrhosis and HCC remain poorly understood. Accordingly, there are currently no approved treatments that target cell death pathways to prevent the development of cirrhosis and HCC besides the treatment of the underlying disease. The liver reacts to hepatocellular death with a wide range of injury responses that include inflammation, hepatocyte regeneration, fibrosis and the appearance of ductular
4 reactions. Ductular reactions contain cell populations, termed ductular or hepatic progenitor cells, which are thought to represent an alternative cellular source for the generation of hepatocytes that becomes relevant when hepatocyte lose their capacity to proliferate. While this concept has been developed based on histopathological evidence in patients (14), the role of ductular cells in animal models remains a matter of debate (14). Despite strong expansion of ductular cells, lineage tracing studies have shown that these cells do not significantly contribute to the generation of functional hepatocytes in the most commonly used models with a progenitor response, such as DDC and CDE diet as well as Mdr2 knockout mice, most likely due to the insufficient suppression of hepatocyte proliferation (15-18). On the other hand, mouse and zebrafish models with severely suppressed hepatocyte proliferation have suggested an important role of ductular cell in hepatocyte formation (19-22). Moreover, human LGR5+ duct-derived liver stem cells differentiate into hepatocytes in vitro and in vivo (23). Furthermore, hepatocytes can undergo reversible ductular metaplasia with expression of A6, CK19, osteopontin and Sox9 (24, 25). Together, these studies suggest cellular plasticity as well as contextdependent role of specific cell types in the chronically injured liver. Although hepatic injury responses are geared toward repair and regeneration, they often become maladaptive in the long term, thereby contributing to the development of cirrhosis and HCC (3). In murine models, in which HCC arises in response to carcinogens or in the setting of chronic injury, tumors originate from hepatocytes and not from ductular cells (26, 27). In DEN-induced HCC, tumor-forming cells express progenitor markers such as A6, CK19, H19 and Epcam (28), suggesting that hepatocyte metaplasia may be a first step in hepatocarcinogenesis. Moreover, a large
5 body of clinical studies from the last decade has shown worse prognosis in HCCs with progenitor signature or expression of oncofetal markers (29-36). In summary, there are tight links between the expression of progenitor markers, HCC development and clinical outcomes. One of the first events following cell death in the liver and other organs is the release of damage-associated molecular patterns (DAMPs), which mediate sterile inflammation via specific receptors (37, 38). In addition to regulating sterile inflammation, it has been suggested that DAMPs may also be involved in repair responses, thereby linking cell death to wound healing responses such as regeneration and fibrogenesis and possibly also cancer development (38-4). Here, we tested the hypothesis that DAMPs provide a molecular link between chronic liver injury, maladaptive wound healing and HCC development, focusing on high-mobility group box 1 (HMGB1), a DAMP with key roles in sterile inflammation following acute liver injury (41). Mice with hepatic HMGB1 deficiency displayed a profound reduction of ductular reactions in multiple models of chronic liver injury. Hepatic HMGB1 deficiency reduced hepatocyte metaplasia and inhibited tumor development in three HCC models with chronic injury, but not in an HCC model that lacks chronic injury. Moreover, HMGB1 deficiency reduced the expression of progenitor markers, a key feature of aggressive HCC, within tumors. Together, our findings suggest that HMGB1 links hepatocyte injury to ductular reactions, hepatocyte metaplasia and HCC development in CLD.
6 RESULTS HMGB1 exerts no major impact on hepatic inflammation, regeneration and fibrogenesis in the chronically injured liver Previously, we demonstrated a key role for HMGB1 in sterile inflammation following acute liver injury (41). As the majority of morbidity and mortality of liver disease arises in patients with CLD, we now sought to determine whether HMGB1 may also play a role in biological processes that contribute to key features of CLD, such as the induction of inflammation, fibrosis, regeneration and ductular reactions. As such, it is conceivable that dying hepatocytes might employ DAMPs to trigger regeneration of surviving hepatocytes, or instruct cells from other compartments to respond to injury and initiate wound healing responses. To test this hypothesis, we subjected mice with a liver-specific deletion of HMGB1 (Hmgb1 hep ), and Hmgb1 fl/fl or Hmgb1 wt/wt control mice to three different and well-characterized models of chronic liver injury, including the 3,5-diethoxycarbonyl- 1,4-dihydrocollidine (DDC) diet model (42), Mdr2 knockout (Mdr2 KO ) mice (43) and Tak1 hep mice (1). Of note, we had previously shown that Hmgb1 hep mice displayed efficient reduction of HMGB1 in parenchymal cells but did not exhibit any abnormalities in hepatic architecture, injury, fibrogenesis or gene expression under baseline conditions (41, 44), suggesting that intracellular HMGB1 does not play a key role in the maintenance of adult liver homeostasis. Tak1 hep, Mdr2 KO, and DDC-treated mice displayed strong inflammation, fibrogenesis and compensatory proliferation (Suppl.Fig.1-4) as well as robust ductular reactions, with characteristic increases in the expression of progenitor markers Cd133, H19 and oncofetal marker Afp, and increased cytokeratin staining (Fig.1A-F). In contrast to our previous studies in acute liver injury, we only
7 observed a minor to moderate role for HMGB1 in the regulation of neutrophil recruitment, with a significant reduction in the DDC model, a borderline reduction in the Mdr2 KO model and no significant changes in the Tak1 hep model (Suppl.Fig.1A,C,E). Likewise, there were no significant differences in CD45+ cell recruitment in all three CLD models and no consistent alterations of inflammation with markers such as Tnf and Cd2 mrna unaffected and Il6 mrna upregulated in some models and downregulated in others (Suppl.Fig.1B,D,F). There was no difference in macrophage recruitment in the Mdr2 KO model, a trend towards reduced macrophages in the DDC model and reduced macrophages in the Tak1 model in HMGB1-deleted mice (Suppl.Fig.2). Together, these data suggest that the regulation of sterile inflammation and neutrophil recruitment by HMGB1 is mostly restricted to acute settings, when other inflammatory mediators such as chemokines and cytokines or gut-derived PAMPs are not yet released. We also did not see a major role for HMGB1 in the regulation of liver fibrosis in all three CLD models (Suppl.Fig.3A-C). Likewise, we did not observe consistent alterations in proliferation, as determined by Ki-67 or phospho-histone H3 immunohistochemistry and mki67 qpcr, with similar hepatocyte proliferation in Hmgb1 wt/wt and Hmgb1 hep mice in Tak1 hep model, increased proliferation in Hmgb1 hep mice in the Mdr2 KO model, and decreased proliferation in Hmgb1 hep mice in the DDC model (Suppl.Fig.4A-C). To further determine whether HMGB1 is required for hepatocyte proliferation, we subjected Hmgb1 fl/fl and Hmgb1 hep mice to two-thirds partial hepatectomy (Suppl.Fig.4D), or a single injection of CCl 4 (Suppl.Fig.4E). In both models, we did not observe a role for HMGB1 in the regulation of hepatocyte proliferation suggesting that the observed minor alterations in liver regeneration may be restricted to some models with ductular reactions.
8 HMGB1 is required but not sufficient for ductular reactions in the liver We therefore next examined the possible role for HMGB1 in the regulation of ductular reactions. In contrast to the minor effects on inflammation, proliferation and fibrosis, we observed a strong and consistent effect of hepatic HMGB1 deficiency on ductular reactions. Hmgb1 hep mice that were either crossed to Tak1 hep mice or Mdr2 KO mice, or treated with DDC diet displayed a significant decrease of cytokeratin-positive (Fig.1A, C, E) and A6-positive cells (Suppl.Fig.5A-B) as well as a profound reduction of Cd133, H19 and/or Afp mrna expression (Fig. 1B,D,F). Of note, HMGB1 deletion did not affect serum ALT levels (Fig.1 B,D,F) thus excluding the possibility that reduced ductular reactions in Hmgb1 hep mice might have been caused by a decrease in liver injury. Similar to above models, we also found inhibited ductular reactions in the methionecholine-deficient ethionine-supplemented diet (MCDE) model (Suppl. Fig.6). Although we found a strong release of HMGB1 as seen by increased HMGB1 serum levels following treatment with hepatoxin CCl 4, we did not observe ductular reactions in this model (Fig.1G). Together with our findings in the Tak1 hep, the Mdr2 KO, the DDC and MCDE models, our data indicate that HMGB1 is required but not sufficient for the development of ductular reactions. Passively released hepatocyte HMGB1 promotes ductular reactions via cell-extrinsic mechanisms and RAGE As the employed deletion strategy via Albumin-Cre mice not only ablates HMGB1 in hepatocytes but also in the biliary compartment (26, 41), we next wanted to exclude that the lack of HMGB1 in ductular cells impaired their ability to expand. For this purpose,
9 we deleted HMGB1 selectively in hepatocytes, employing AAV8-TBG-Cre, in which Cre expression is controlled by the hepatocyte-specific thyroxine-binding globulin (TBG) promoter, as a hepatocyte-specific deletion approach (24, 41). This strategy resulted in a significant reduction of hepatic Hmgb1 mrna and absent HMGB1 expression in hepatocytes but not in other liver cell types (Fig.2A-B), thus affecting signals from hepatocytes to other cell types including ductular cells, but leaving HMGB1 within ductular cells intact. Mdr2 KO mice with AAV8-TBG-Cre-mediated hepatocyte-specific HMGB1 deletion displayed a significant reduction of progenitor markers Cd133 and H19, and decreased cytokeratin staining in comparison to mice injected with AAV8-TBG- LacZ control virus (Fig.2C-D). These finding were confirmed in the DDC model where we observed a reduction of Cd133 and H19 mrna and cytokeratin staining in mice treated with AAV8-TBG-Cre versus mice receiving AAV8-TGB-LacZ virus (Fig.2E-F). Together, our findings in the DDC and Mdr2 KO models exclude that the intracellular loss of HMGB1 within the biliary compartment simply blocked ductular reactions, further supporting our hypothesis that hepatocellular HMGB1, acting as DAMP, is essential for the induction of ductular reactions. Extracellular HMGB1 can be released via passive leakage from dead cells (45) or via active secretion, the latter being mediated through acetylation of HMGB1 at multiple lysine residues (46). To determine whether passive release or active secretion of HMGB1 was driving ductular reactions, we inhibited HMGB1 secretion during DDC-induced liver injury using ethyl pyruvate, an established inhibitor of HMGB1 acetylation and secretion (47, 48). Although HMGB1 acetylation was completely suppressed by ethyl pyruvate
10 (Fig.3A-B), we did not observe significant effects on cytokeratin and A6 expression and Cd133 and Afp mrna and only a borderline significant reduction of H19 mrna (Fig.3C-E) Moreover, HMGB1 serum levels were not significantly decreased (Fig.3F) despite the observed loss of acetylated HMGB1. Together these findings suggest that passive HMGB1 release from dying hepatocytes rather than active secretion is the dominant driver of ductular reactions in the investigated settings. HMGB1 binds to several receptors including RAGE (encoded by Ager), TLR4 and TLR9. We next determined which one of these three HMGB1 receptors was involved in promoting ductular reactions in vivo, using Ager-deficient (Rage KO ), Tlr4-deficient (Tlr4 KO ) and Tlr9-deficient (Tlr9 KO ) mice. We found strongly reducted ductular reactions with significantly decreased cytokeratin staining and reduced H19 and Afp mrna expression as well as a trend towards decreased Cd133 mrna in livers from Rage KO mice (Fig.4A). In contrast, Tlr4 KO and Tlr9 KO mice did not show impaired ductular reactions in the DDC diet model, as demonstrated by unaltered cytokeratin staining and unaltered or increased Cd133, H19 and Afp mrna expression in Tlr4 KO and Tlr9 KO mice (Fig.4B-C). Similar to Hmgb1 hep mice, we did not find alterations of liver injury in Rage KO mice, thus excluding that reduced ductular reactions were merely a consequence of lower injury in Rage KO mice (Fig.4A). As we also observed small decreases in macrophages in HMGB1-deleted mice in some models (Suppl.Fig.2), we additionally determined whether macrophages might be a cell population through which HMGB1 might indirectly trigger progenitor responses.
11 However, consistent with previous studies (49), we observed no decrease in ductular reactions following macrophage depletion (Suppl.Fig.7). Based on these findings, we subsequently focused on direct effect of HMGB1 on bipotential progenitor cells with the goal to identify signals through which HMGB1 affects ductular reactions. Disulfide HMGB1 mediates effects on progenitor cells via Erk HMGB1 bioactivity is dependent on post-translational modifications with fully-reduced HMGB1 promoting cell migration but not inflammation, whereas disulfide HMGB1 has cytokine-like proinflammatory activity (5). Similar to our previous studies in acute liver injury, we found that disulfide HMGB1 was the most abundant form of HMGB1 in the chronically injured liver and that this form increased with the chronicity of liver injury (Fig.5A). More importantly, stimulation with these two forms of HMGB1, which have distinct bioactivities, revealed that only the disulfide form of HMGB1 upregulated Cd133 mrna in BMOL cells (Fig.5B), a well characterized bipotential liver progenitor cell line (51). As YAP and Notch are well-established and powerful signals driving hepatocyte metaplasia towards a ductular phenotype in adult livers (24, 52) as well as hepatocarcinogenesis (53), we determined if HMGB1 could act through these pathways. However, we found no difference of Notch target gene expression after treating of BMOL cells with HMGB1, which was further confirmed by similar levels of Notch target genes in control and HMGB1-deleted mice in vivo in multiple models (Suppl.Fig.8). Following treatment of BMOL cells with HMGB1, YAP target genes as well as YAP reporter activity decreased (Suppl.Fig.9). In contrast, we observed a decrease in some YAP target genes in HMGB1-deleted mice in vivo (Suppl.Fig.9). As our experiment had
12 not shown any direct effect of HMGB1 on YAP target genes on BMOL progenitor cells in vitro, and as we did not see a major role for hepatocyte YAP in ductular reactions after three weeks of DDC diet (data not shown), we therefore reasoned that the reduction of YAP target genes in HMGB1-deleted mice is likely reflecting the decrease in ductular cells (which are enriched in YAP and YAP target genes) rather than indicating a lack of HMGB1-induced YAP activation. Accordingly, when normalizing our qpcr results to progenitor marker Cd133, one YAP target gene was significantly upregulated in HMGB1-deleted mice in one CLD model and another YAP target genes remained significantly downregulated in one CLD model where all others showed no significant differences between floxed and HMGB1-deleted mice (data not shown). Therefore, we sought to identify additional signals through which disulfide HMGB1 may affect ductular reactions. For this purpose, we performed a phosphoscreen in BMOL cells treated with disulfide HMGB1. In this screen, we found that Erk phosphorylation and phosphorylation of its target CREB, but not other pathways were strongly induced by disulfide HMGB1 (Fig.5C), which was further confirmed by western blot analysis (Fig.5D, upper panel). In contrast, fully reduced HMGB1 did not stimulate Erk phosphorylation (Fig.5D, lower panel). Inhibition of Erk phosphorylation strongly decreased the expression of Cd133 mrna in BMOL cells (Fig.5E), suggesting that this pathway mediates effects of HMGB1 on progenitor cells. This finding is supported by previous studies that have described a key role of Erk in the regulation of CD133 (54, 55). Moreover, treatment with disulfide HMGB1 moderately but significantly promoted the proliferation of BMOL cells (Fig.5F), suggesting that HMGB1 contributes to progenitor expansion via this pathway. Indeed, we found that there was a reduction of ps1-h3/krt19-double positive
13 proliferating progenitor cells in HMGB1-deleted mice after three weeks of DDC diet (Fig.5G). Consistent with our in vivo studies in knockout mice as well as a previously published study (56), we also found that inhibition of RAGE blunted HMGB1-induced Erk phosphorylation whereas TLR4 or TLR9 blockade had no major effect (Fig.5H). In summary, our findings suggest that HMGB1-induced activation of RAGE triggers the proliferation and expansion of progenitor cells. HMGB1 promotes hepatocyte metaplasia and links chronic injury to HCC development. Chronic liver injury may result in the development of HCC, the third leading cause of world-wide cancer mortality. Accordingly, chronic hepatocellular death strongly increases the risk for HCC development (6, 7). Moreover, the expression of progenitor and oncofetal genes is common in HCC and adversely affects prognosis (29-36). Consistent with previous studies (24, 25), we found via lineage tracing via AAV8-TBG- Cre that hepatocytes undergo ductular metaplasia, as seen by co-expression of Cre reporter TdTom, demonstrating hepatocyte origin, and ductular marker A6, osteopontin and Sox9 (Fig.6A-D). In mice that had received DDC diets for 3 weeks, A6-positive hepatocytes constituted less than 1% of all A6-positive cells in the liver (Fig.6B). Of note, A6-positive metaplastic hepatocytes were reduced by 68% (p<.1) in HMGB1- deleted mice (Fig.6B). Similarly, there was also a signficant reduction of TdTom- and OPN-double-positive and TdTom- and Sox9-double-positive hepatocytes (Fig.6C-D) as well as a significant reduction of Spp1 (encoding osteopontin) and Sox9 mrna (Fig.6E- F) in HMGB1-deleted mice. Together, these findings suggest that HMGB1 acts on hepatocytes to promote their ductular metaplasia, but that this largely HMGB1-dependent
14 response contributed only a small fraction to the overall ductular response at the time points that we studied. As A6-, CK19-, AFP-, H19-, Epcam- and Sox9-expressing cells may function as liver cancer progenitors (28), and HMGB1 had a major role regulating expression of these markers in the liver and the expression of A6 in hepatocytes in particular, we next tested the hypothesis that HMGB1 may provide a link between hepatocellular death and HCC development. For this purpose, we subjected Hmgb1 fl/fl and Hmgb1 hep mice to models of hepatocarcinogenesis that either incorporate or lack chronic liver injury (Fig.7A-G). To mimic the development of HCC in chronically injured, inflamed and fibrotic livers in patients, we employed the well-established combination of the carcinogen diethynitrosamine (DEN) with chronic injection of the hepatotoxin CCl 4 or with the above-described DDC diet model (57, 58). The DEN+CCl 4 model resulted in significantly increased release of HMGB1 in comparison to DEN only model (Fig.7D). In the DEN+CCl 4 model, we found a significant reduction of HCC development, as determined by tumor number, and liver body weight ratio, and a borderline significant reduction (p=.6) in tumor size (Fig.7A-C). To further confirm the contribution of HMGB1 to injury-driven hepatocarcinogenesis, we next tested its role in mice with hepatocyte-specific deletion of Tak1. In this model, mice spontaneously develop HCC (1, 59) as a result of chronic cell death without the need for the injection of carcinogens. Similar to the DEN+CCl 4 model, we observed a significant reduction of tumor number and size, and a borderline significant reduction in liver body weight ratio (Suppl.Fig.1A- C). Likewise, we also observed a reduction in tumor formation in mice treated with DEN plus DDC diet (Suppl.Fig.1D-E), which triggers the development of HCC in the presence of chronic injury, ductular reactions and progenitor marker expression (58). In
15 contrast, when we subjected Hmgb1 hep or Hmgb1 fl/fl control mice to DEN-induced hepatocarcinogenesis, a purely genotoxic model without chronic liver injury and HMGB1 release (Fig.7D), we observed abundant tumors in both groups of mice, without significant differences in tumor number, size or liver body-weight ratio (Fig.7E-G). The finding that cell death is required to reveal effects of hepatic HMGB1 strongly suggests that HMGB1 acts as DAMP and excludes the possibility that the lack of intracellular HMGB1 might have simply impaired the ability of tumor-initiating cells to form tumors. HMGB1 controls progenitor signature but not fibrosis, inflammation or proliferation in HCC To better understand mechanisms through which HMGB1 promotes hepatocarcinogenesis in the injured liver, we focused on tumor-promoting injury responses. Similar to our findings in chronic injury models, we did not observe differences in proliferation, fibrosis or inflammation, as demonstrated by similar Ki67, picrosirius, CD45+ cells and F4/8 staining, and similar inflammatory gene expression in tumors from Hmgb1 hep and Hmgb1 fl/fl mice (Fig.8A-C, Suppl.Fig.11A-C). Likewise, we did not find a role for HMGB1 in the recruitment of neutrophils (Fig.8D), a cell population that contributes role to the development of liver cancer (6), and whose numbers positively correlate with worse prognosis (61, 62), in HCC. As we observed a moderate role for HMGB1 in neutrophil recruitment in earlier stages of injury (Suppl.Fig.1A-C) and as HMGB1 might not only contribute to neutrophil recruitment but also to neutrophil activation, we additionally performed functional experiments with neutrophil elastase (encoded by Elane) deficient mice, which have defective neutrophil
16 effector functions but display normal neutrophil recruitment (41, 63). However, we found no significant contribution of neutrophil activation to HCC development, with similar tumor number, size and liver body weight ratio in wild-type and Elane-deficient littermates (Fig.8E) suggesting that neutrophil effector functions are not required for hepatocarcinogenesis. Moreover, we did not find differences in the recruitment of CD3+ T cells (Suppl.Fig.11D), a cell population able to mediate anti-tumor immune responses, or genotoxic stress as determined by H2AX staining (Suppl.Fig.11E). Additionally, we did not observe a reduction in YAP protein and Yap1 and Wwtr1 mrna levels and even an increase in TAZ protein levels in Hmgb1 hep mice (Suppl.Fig.12A). Accordingly, YAP target genes as determined by qpcr (Suppl.Fig.12B) or RNA sequencing (Table S1), were either similar in Hmgb1 fl/fl and Hmgb1 hep mice or higher in Hmgb1 hep mice. In summary, our data demonstrate that the promotion of HCC by HMGB1 is not explained by effects on inflammation, anti-tumor responses, regeneration, fibrogenesis or genotoxic stress, or by HMGB1-mediated modulation of YAP/TAZ. To better understand the influence of HMGB1 on hepatocarcinogenesis and follow up on above-described finding that HMGB1 promotes the ductular metaplasia of hepatocytes (Fig.6), we performed RNA sequencing in tumors from DEN+CCl 4 -treated Hmgb1 hep and Hmgb1 fl/fl mice. Unsupervised clustering of differentially expressed genes showed that tumors from Hmgb1 hep mice clustered closer with normal liver than tumors from Hmgb1 fl/fl mice (Fig.9A, Table S1). Accordingly, 675 out of 848 genes (79.6%) in tumors from Hmgb1 hep mice had expression levels in the direction of normal liver (p<2.2e-16). A more mature phenotype of tumors from Hmgb1 hep mice was further supported by the strong reduction of oncofetal/progenitor markers Cd133, H19 and Afp in Hmgb1 hep
17 tumors in our RNA sequencing data (Table S1), which was confirmed by qpcr and immunohistochemical AFP and A6 staining (Fig.9B-D). These findings are consistent with clinical findings in which HMGB1 serum levels were increased in HCC patients and strongly correlated with AFP levels (64). As we had previously shown that A6-positive tumor cells within HCC are derived from hepatocytes (26), our findings demonstrate that HMGB1 is a key pathway driving metaplasia and progenitor signature in hepatocytes and hepatocyte-derived tumor cells. Notably, the presence of progenitor markers is one the most characteristic features of dedifferentiated HCC and a large body of literature has demonstrated a positive correlation with worse clinical outcomes (29-36). As we had found a key role for HMGB1 receptor RAGE in driving ductular reactions in CLD, we next investigated the role of RAGE in HCC development and observed a significant reduction of tumor development in Rage KO in the DEN+CCl 4 model as demonstrated by reduction of tumor number, size and liver body weight ratio (Suppl.Fig.13). DISCUSSION Cell death is a key component of CLD and considered an important driver of progression to fibrosis, cirrhosis and HCC (3). Although DAMPs including HMGB1 contribute to sterile inflammation in the setting of acute liver injury (38, 41, 65), their role in other injury responses and contribution to CLD remain enigmatic. Our data implicate HMGB1 as an important hepatocyte DAMP that regulates specific cell death responses in the chronically injured liver. Our findings suggest that the main effect of HMGB1 in CLD is the promotion of ductular reactions, whereas other injury responses such as regeneration, inflammation or fibrosis are not significantly affected. As we observed a strong but
18 incomplete suppression of ductular reactions in Hmgb1 hep mice, HMGB1 represents a key contributor for the efficient execution of this response but is not the only driver. This is consistent with findings that pathways such as b -catenin (66, 67), CNN1 (68), and FGF signaling (69) also promote ductular reactions in the liver. Moreover, CCl 4 -induced liver injury did not trigger a significant progenitor response despite increased HMGB1 release. Together, these findings indicate that HMGB1 release is necessary but not sufficient to trigger an effective progenitor response in the liver. Our data using hepatocyte-specific HMGB1 deletion via AAV8-TBG-Cre in two injury models also exclude the possibility that the lack of intracellular HMGB1 in the ductular compartment may have impaired their ability to respond to injury. Of note, the majority of A6-positive ductular cells were not derived from hepatocytes, indicating that hepatocyte-derived HMGB1 acts in a nonautonomous fashion on other cell types to drive ductular reactions. As such, we found that HMGB1 was able to trigger Erk activation and proliferation in BMOL progenitor cells in vitro, suggesting that effects of HMGB1 on ductular reactions in the injured liver are direct. In conjunction with our finding that HMGB1-deleted mice had a lower amount of Ki67+ progenitors ductular cells and an extensive phospho-pathway screen in HMGB1-treated BMOL progenitor cells, these data suggest that HMGB1 stimulates ductular expansion predominantly via Erk activation and proliferation. Our in vivo data in Tlr4 KO, Tlr9 KO and Rage KO mice and in vitro data using RAGE, TLR4 and TLR9 inhibitors demonstrate that HMGB1 contributes to ductular reaction via RAGE. Using liposomal clodronate, we excluded macrophages as a target that may indirectly mediate progenitor responses, but we cannot completely rule out that HMGB1 may additionally affect progenitors through indirect mechanisms that involve other cell types. Our studies
19 using ethyl pyruvate also showed that active secretion of HMGB1 is not a key driver in DDC-induced ductular reactions, further emphasizing the key role of HMGB1 from dying cells in this setting. These data are further supported by our finding that secreted, i.e. acetylated form of disulfide HMGB1 was less abundant than the non-acetylated form in DDC-mediated injury. However, it is conceivable that secretion of acetylated HMGB1 is more abundant and may drive ductular reactions in other settings. Although hepatocytes are the primary source for liver regeneration in multiple mouse models including DDC and CDE diets, CCl 4 injection and partial hepatectomy (15, 16), these models do not achieve efficient suppression of hepatocyte proliferation and may not be ideal to test the contribution of ductular cells (14). Recent studies in novel mouse models (21, 22) as well as data from zebrafish (19, 2) demonstrated a key contribution of the ductular compartment to liver regeneration in settings where hepatocyte proliferation is efficiently blocked. Accordingly, Lgr5+ liver stem cells can generate hepatocytes in vitro and in vivo (23). However, it remains unclear whether HMGB1- mediated ductular cell expansion contributes to hepatocyte generation from this source and whether this may represent a regenerative mechanism in the setting of severe injury. Further studies on HMGB1 in models with efficient suppression of hepatocyte proliferation in combination with positive lineage tracing of ductular cells will be required to answer this question. The second key finding of our study was the reduction of HCC in mice with liver-specific HMGB1 deletion. Hepatocellular death is a risk factor for HCC development (3, 6, 7), and our data suggest that HMGB1 may provide a molecular link between cell death and
20 HCC development in the chronically injured liver. This hypothesis is supported by our finding that HMGB1 did not significantly affect HCC development in a mouse model that lacks chronic cell death and subsequent HMGB1 release, whereas HCC development was blocked in three different HCC models with chronic cell death. These data not only demonstrate the key role of HMGB1 as tumor-promoting DAMP in the setting chronic injury, but also exclude that reduced HCC development was caused the loss of intracellular HMGB1. Similar to our data on HMGB1 in ductular reactions, we found that RAGE was the most likely candidate receptor through which HMGB1 mediated its effect on hepatocarcinogenesis. This finding is consistent with a published study in the Mdr2 KO model, in which Rage KO mice developed less HCC (56). Together, our findings suggest that the HMGB1-RAGE signaling axis provides a molecular link between cell death and hepatocarcinogenesis and is likely a key component of maladaptive wound healing responses, which are geared towards repairing the injured liver but become maladaptive and increase risk for HCC development in the long term. Importantly, tumors from Hmgb1 hep mice were more similar to normal liver with lower expression of progenitor and oncofetal markers in comparison to tumors from Hmgb1 fl/fl mice. High expression of these markers, reflecting poorly differentiated and stem cell-enriched tumors, has been correlated with poor clinical prognosis in a wide body of literature (29-36). Notably, serum HMGB1 levels are strongly increased in HCC patients and correlate with AFP levels as well as tumor size (64). Progenitor cells do not give rise to HCC in a large number of murine models, including the DEN+CCl 4 and DEN+DDC models employed in the current study (26, 27) As CCl 4 -induced liver injury is not associated with the development of ductular reactions, it is likely that HMGB1 induces protumorigenic
21 signals (e.g. in the in the DEN+CCl 4 model) directly in preneoplastic or tumor cells. Based on a study showing that tumor-forming cells, which are hepatocyte-derived in DEN-induced HCC, express progenitor markers such as A6, CK19, H19, Epcam and Sox9 (28), it is conceivable that HMGB1-mediated ductular metaplasia could increase the tumor-initiating capacity of hepatocytes. Future studies, using tracing of metaplastic hepatocytes, are needed to determine whether these cells are more tumorigenic and how deletion of the HMGB1/RAGE signaling axis affects tumor formation from this source. In addition, HMGB1-mediated increases of progenitor markers within already established tumor could promote more aggressive tumor behavior as demonstrated by the strong clinical correlation between progenitor signature and clinical outcomes (29-36). Further investigations are required to determine these different possibilities and to determine downstream signals through which HMGB1 contributes to hepatocarcinogenesis and tumor dedifferentiation. In contrast to a recent study that reported HMGB1-regulated transcriptional activation of YAP in HCC (7), we did not observe a role for HMGB1 in regulating YAP or TAZ mrna or protein expression or the expression of YAP target genes in HCC. This is consistent with our previous studies in normal liver (44) in which microarray analysis did not reveal differences in the expression of Yap1, Wwtr1 (encoding TAZ) or YAP target genes Ankrd1, Ctgf, Cyr61 and Spp1, and most likely reflects the regulation of YAP and TAZ activity through posttranstranslational modification and protein stability rather than through transcription. Regardless of the underlying mechanism by which HMGB1 promotes hepatocarcinogenesis, our data suggest that interfering with this pathway could delay HCC development in patients with CLD. Glycyrrhizin, a phytochemical that inhibits HMGB1 (71), is widely used for
22 treatment of liver disease in Asia, and there are indications that glycyrrhizin can reduce HCC development in subsets of patients (72). Our study also demonstrates that hepatocellular HMGB1 is not a master DAMP that regulates all wound healing responses in the chronically injured liver. As such, we did not observe an important contribution of HMGB1 in cell death responses such as fibrogenesis, inflammation and proliferation. However, we observed a reduction of neutrophil recruitment in some models and a trend towards reduced neutrophil recruitment in other models in early stages of chronic liver injury. These findings consistent with our previous findings on HMGB1/RAGE-mediated neutrophil recruitment in acute injury (41), but suggest that other pathways are more potent regulators of neutrophil recruitment in CLD. In contrast to a recent study on the role of HMGB1 in liver regeneration following acetaminophen intoxication (73), we did not observe a major role for HMGB1 in liver regeneration except for reduction of proliferation in the DDC model. In addition to using a different model, Tirone et al administrated recombinant non-oxidizable HMGB1, whereas we deleted HMGB1. Thus, it is likely that the effects of HMGB1 on liver regeneration are model- and redox-dependent. Our results on the role of HMGB1 in liver fibrosis also differ from a recently published study (74). Our findings are based on three different models (Tak1 hep, Mdr2 KO, and DDC diet) and were additionally confirmed in the MCDE diet model and in CCl 4 -induced liver injury (data not shown). Moreover, liver-specific deletion of HMGB1 in mice with hepatic autophagy deficiency due to loss of ATG7 also did not display alterations of liver fibrosis (personal communication from Dr. XM Yin, see accompanying manuscript submission), further confirming our data. Together, these findings suggest that additional cell death pathways, possibly mediated
23 by other DAMPs or apoptotic bodies, must link cell death and fibrosis in the chronically injured liver. In summary, our findings suggest that DAMPs mediate cell death responses in context-specific and cell type-specific manner, and that multiple branches of the hepatic wound healing response are most likely regulated through a variety of DAMPs rather than one master DAMP. METHODS Mice. Mice were maintained on a 12-hour dark/12-hour light cycle with free access to food and water. Hmgb1 fl/fl mice (41, 44) were backcrossed to C57Bl/6 background at least five times. C57BL/6 mice, Albumin-Cre mice, Cre reporter line TdTomato (Stock # 7914), Tlr4 KO mice (Stock #7227) and Tak1 fl/fl mice (Stock #1139) were purchased from The Jackson Laboratory. Rage KO mice were a gift of Ann-Marie Schmidt (New York University, New York, New York, USA). Mdr2 KO mice (in FVB background) were obtained from Dr. Detlef Schuppan (University of Mainz). For conditional knockout of Hmgb1 in the Tak1 hep model, Tak1 fl/fl or Tak1 fl/fl and Hmgb1 fl/fl double-floxed mice expressing Albumin-Cre were bred with floxed or double-floxed mice negative for Albumin-Cre. For conditional deletion of Hmgb1 in Mdr2 KO mice, Hmgb1 fl/fl mice expressing Albumin-Cre were backcrossed three times to Mdr2 KO, followed by further interbreeding of the F3 generation. HCC induction and evaluation. HCC was induced by a combination of DEN and CCl 4 injections for the majority of experiments. Male offspring was injected with a single
24 intraperitoneal dose of diethylnitrosamine (DEN, 25 mg/kg body weight, Sigma-Aldrich, St. Louis, MO) at day 15 post partum. Four weeks later, mice were treated with 15 weekly i.p injections of CCl 4 (.5 ml/g body weight, dissolved in corn oil at a ratio of 1:3, Sigma-Aldrich) and sacrificed 8-1 weeks after the last CCl 4 injection. Some mice were fed a DDC diet for 4 months starting 4 weeks after receiving DEN and sacrificed 8 months after the initial DEN injection. As nongenotoxic model of HCC, Hmgb1 wt/wt /Tak1 hep and Hmgb1 hep /Tak1 hep mice were sacrificed at 9 months of age. As pure genotoxic model of HCC, some mice were treated with a single dose of DEN (25 mg/kg, at day 15 postpartum) and sacrificed 11 months later. For HCC quantification, tumor number and largest tumor size were determined by counting the number of visible tumors (exceeding 1 mm in diameter) and measuring the size of the largest tumor with a caliper, respectively (57). Chronic liver injury models. Chronic liver injury was induced in 8-1 week old male mice by feeding mice.1% DDC diet or MCD diet supplemented with.15% ethionine for 3 weeks and 2 weeks, respectively. For selective deletion of HMGB1 in hepatocytes, Hmgb1 fl/fl mice were injected i.v. with 1 11 genome copies of AAV8-TBG-Cre, AAV8- TBG-LacZ (58) on day 15 post partum (for the Mdr2 KO model) or 6 weeks post partum (for all other mice). For some experiments, mice fed DDC diet were treated with ethyl pyruvate (4 mg/kg, i.p. given twice weekly through the entire time mice were fed DDC diet). For some experiments, mice fed DDC diet were treated with liposomal clodronate or control liposomes (4 ul/g body weight, i.p., both from Liposoma, Amsterdam, Netherland), at day 4, day 8 and day 12 after starting the diet.
25 Partial hepatectomy and acute CCl 4 -induced liver injury. A two-thirds partial hepatectomy performed as described (75). Acute liver injury and subsequent hepatocyte proliferation was induced by a single i.p injection of CCl 4 (.5 ml/kg body weight dissolved in corn oil at a ratio of 1:3). Hepatocyte tracing. To trace hepatocytes and determine whether HMGB1 deletion alters the presence of hepatocyte-derived ductular cells, mice that were either wild-type or Hmgb1 fl/fl and also expressed Cre reporter TdTomato were injected with 1 11 genome copies of AAV8-TBG-Cre. One week later, mice were treated with DDC diet for 3 weeks. Subsequently ductular metaplasia of hepatocytes was determined in liver sections by identification of TdTom-positive cells co-expressing A6, Sox9 or OPN. Immunohistochemistry and immunofluorescence. Paraffin-embedded or frozen liver sections were stained with primary antibodies directed against A6 (76) (a gift of Dr. Valentina Factor, NIH, Bethesda, Maryland, USA), AFP (Proteintech, catalog AP), pan-cytokeratin (Dako, catalog Z622), cytokeratin 19 (Developmental Studies Hybridoma Bank at the University of Iowa, Troma III), Ki67 (Abcam, catalog ab16667), Ly-6B.2 (AbD Serotec, clone MCA771G), anti-f4/8 (AbD Serotec, clone CI:A3-1, catalog MCA497R), CD3 (Thermo Fisher, catalog RM-917-S), CD45 (BD Pharmingen, catalog 55539), YAP (Abcam, catalog ab2527), ps1-h3 (Santa Cruz, catalog sc- 8656), Sox9 (Millipore, catalog AB5535) and OPN (R&D systems, catalog AF88). Detection was performed using either fluorescent secondary antibody with various fluorescent conjugates (all from Thermofisher; chicken anti-goat, A21467; donkey antirabbit, A2126 or A2127; donkey anti-rat, A2128) or the Vectastain ABC HRP kit (Vector Laboratories) using DAB or VIP substrate, followed by counterstaining with
26 either DAPI (ThermoFisher, D136) or hematoxylin or methyl green. Fluorescent images were taken on a Nikon A1 confocal laser microscope. Non-fluorescent images were taken on an Olympus microscope coupled to a Retiga camera (Q Imaging). Morphometric quantification of immunohistochemical staining was done using Adobe Photoshop with the exception of quantification of hepatocyte and progenitor cell proliferation, which were done using Fiji. For quantification of hepatocyte proliferation, size and circularity thresholds were set so that nuclei of non-hepatocyte cells were not counted. For some experiments, immunohistochemistry was done both by fluorescence and VIP substrate, with fluorescent images displayed as representative images and quantification using VIP staining. Western blot analysis and phospho-kinase screen. Western blot for the detection of phosphorylated Erk was performed using primary mouse antibody detecting YAP/TAZ (Cell Signaling, catalog 8418). Membranes were reprobed with HRP-conjugated GAPDH (Sigma, catalog G9295). After detection of Thr22/Tyr24 phosphorylated Erk (Cell Signaling, catalog 916), membranes were stripped and relabeled with a rabbit antibody against Erk (Cell Signaling, catalog 4695). Phospho-kinase screen was performed using a phospho-kinase array (R&D systems, ARY3B) according to the manufacturer s instructions. RNA isolation and qpcr. RNA was isolated from cells and tissues by column purification and on-column DNAse treatment (Roche Diagnostics). Following reverse transcription, qpcr was performed using primer-probe pairs (Applied Biosystems) and relative standard curves as described (57).
27 Analysis of HMGB1 by electrospray ionization liquid chromatography MS/MS. Posttranslational modifications of HMGB1 were detected in mouse plasma using liquid chromatography MS/MS using an AB Sciex QTRAP 55 or an AB Sciex TripleTOF 56 equipped with a NanoSpray II source by in-line liquid chromatography using a U3 HPLC System connected to a 18 μm 2 mm nanoacquity UPLC C18 trap column and a 75 μm 15 cm nanoacquity UPLC BEH13 C18 column (Waters) using previously described conditions (41, 77). RNA sequencing and bioinformatics. RNA (RIN>8, as determined by Bioanalyzer 21, Agilent) from normal liver HCC (n=3) or HCC from Hmgb1 fl/fl (n=6) or Hmgb1 hep (n=6) mice were used to construct libraries using the Illumina TruSeq RNA prep kit according to manufacturer s instructions. 3M 1 bp single end sequencing was performed on the Illumina 25 at the Sulzberger Columbia Genome Center. These data were deposited in the Gene Expression Omnibus (GEO) (GSE89689). Differential expression was determined between both Hmgb1 hep HCC and Hmgb1 fl/fl HCC, and between Hmgb1 fl/fl HCC and normal liver. Counts were normalized with TMM (78). Differential expression was estimated using weighted Limma-Voom, with a significance cutoff of the Benjamini- Hochberg false discovery rate (fdr).5, an absolute log 2 fold change.6 and restriction to the twelve most significant pathways, as determined by ipathwayguide, plus progenitor and oncofetal genes Afp, H19 and Prom1. Both identification and calculation were performed in R. TMM-normalized counts of these genes were transformed by log 2 (counts+.5) centered, clustered with average linkage clustering using cluster 3., and displayed with JavaTreeview. The statistical significance of genes
28 from Hmgb1 hep that show expression in the direction of normal liver versus those that did not was calculated with the binomial test. Cell lines and cell culture experiments. BMOL cells (provided by Dr. George Yeoh, University of Western Australia) (51) were seeded in 12-well or 24-well plates in William s E medium (ThermoFisher) supplemented with 1% fetal bovine serum (Gemini), recombinant mouse EGF (1 ng/ml, ThermoFisher) and human recombinant insulin (1 mg/ml, ThermoFisher). Cells were treated next day with either disulfide or fully-reduced form of HMGB1 (79) for the indicated times. Cells were then lysed for gene expression and western blot analysis. When indicated cells were pre-treated 3 minutes with MEK inhibitor UO126 (Sigma). To determine proliferation, cells were grown in media containing 1% fetal bovine serum and growth factors, followed by addition of HMGB1 and MTT assay (Invitrogen). For some experiments, cells were pretreated with anti-rage (5 μg/ml, Santa Cruz, catalog sc ) or isotype control (5 μg/ml, Santa Cruz, sc-3877), anti-tlr4 (5 μg/ml, InvivoGen, catalog mabmtlr4md2) or isotype control (5 μg/ml, InvivoGen, catalog mabg2a-ctlrt), and TLR9 antagonist (ODN 288, 1 μm, InvivoGen, catalog tlrl-288) or control nucleotide (ODN 288 control, InvivoGen, catalog tlrl-288c) for 8 hours, followed by treatment with HMGB1. Study approval. All animal procedures were performed with Columbia University IACUC approval and were in accordance with the Guide for the Care and Use of Laboratory Animals (National Academies Press. 8th edition. Revised 211).
29 Statistics. All data are expressed as the mean ± SEM. For comparison of 2 groups, an unpaired two-tailed t test was used. AUTHOR CONTRIBUTIONS CH designed and performed in vivo experiments on liver injury and hepatocarcinogenesis and in vitro experiments, analyzed data and wrote the manuscript. PH designed and performed in vivo experiments related on liver injury and hepatocarcinogenesis, and analyzed data. JPP performed in vivo experiments related to hepatocarcinogenesis. RAF analyzed RNA sequencing data and performed bioinformatics. DJA coordinated HMGB1 bioanalysis and recombinant HMGB1 production. RFS conceptualized and oversaw all aspects of the study, designed experiments, analyzed data and wrote the manuscript. ACKNOWLEDGEMENTS We would like to thank Dr. Geum Youn Gwak (Sungkyunkwan University School of Medicine, Seoul, Korea) who contributed to the generation of Hmgb1 fl/fl mice, Dr. George Yeoh (University of Western Australia, Perth, Australia) and Dr. Bin Gao (NIH, Bethesda, Maryland, USA) for providing BMOL cells, and Dr. Valentina Factor (NIH, Bethesda, Maryland, USA) for providing A6 antibody. This study was supported by funding from the National Institutes of Health (R1CA2597 and U1AA21912, both to RFS) and a fellowship from the German Research Foundation (Hu 1953/1-1).
30 FIGURE LEGENDS Figure 1. HMGB1 is required for ductular reactions. A-B. 8 week-old mice with hepatic deletion of HMGB1 (Hmgb1 Δhep, n=9) showed fewer cytokeratin-positive ductular cells (A), lower expression of Cd133, Afp and H19 mrna but similar ALT (B) compared to Hmgb1 wt/wt controls (n=9) in the Tak1 Δhep model. C-D. In the Mdr2 KO model, 8-week old Hmgb1 Δhep mice (n=13) showed fewer cytokeratin-positive ductular cells (C), lower expression of Afp and H19 mrna but similar ALT (E) compared to to Hmgb1 fl/fl controls (n=1). E-F. Following three weeks of DDC diet, Hmgb1 Δhep mice (n=9) displayed fewer cytokeratin-positive ductular cells (E), lower expression of Afp and H19 mrna but similar ALT (F) compared to Hmgb1 fl/fl controls (n=6). G. Untreated mice (n=4) and mice treated with six CCl 4 injections (n=11) displayed similar levels of cytokeratin-positive cells. HMGB1 serum levels were determined by ELISA in mice that have received (n=3) or not (n=2) six injections of CCL 4. All data are expressed as means ± SEM. qpcr data are shown as fold induction compared to normal liver. Statistical significance was determined by unpaired two-tailed t-test. * p<.5; ** p<.1; *** p<.1. Scale bar 1 µm. Figure 2. HMGB1 from hepatocellular sources drives ductular reactions. A. 2 weekold Hmgb1 fl/fl Mdr2 KO mice were infected with AAV8-TBG-Cre (i.v., 1 11 genome copies, n=11) or AAV8-TBG-LacZ (i.v., 1 11 genome copies, n=1) and sacrificed 6 weeks later. Immunohistochemical HMGB1 staining demonstrated efficient deletion of HMGB1 from hepatocytes in AAV8-TBG-Cre-, but not in AAV8-TBG-LacZ-infected mice (arrows
31 pointing at hepatocytes). B. HMGB1 deletion was confirmed by qpcr in Hmgb1 fl/fl mice infected with AAV8-TBG-Cre or AAV8-TBG-LacZ. C-D. Immunohistochemical cytokeratin staining (C) and qpcr for Cd133 and H19 (D) demonstrated reduced ductular reactions in AAV8-TBG-Cre-infected mice. E-F. 3 week-old Hmgb1 fl/fl mice were infected with AAV8-TBG-Cre (n=8) or AAV8-TBG-LacZ (n=7) as above, followed by three week-long treatment with DDC diet two weeks after AAV infection. Immunohistochemical cytokeratin staining (E) and qpcr for Cd133 and H19 (F) demonstrated reduced ductular reactions in AAV8-TBG-Cre-infected mice. All data are expressed as means ± SEM. qpcr data are shown as fold induction compared to normal liver. Statistical significance was determined by unpaired two-tailed t-test. * p<.5; **, p<.1; *** p<.1. Scale bar 1 µm. Figure 3. Active HMGB1 secretion does not drive ductular reactions. A. C57BL/6 males were pretreated with saline (n=11) or ethyl pyruvate (n=8) followed by DDC diet for 3 weeks as shown. B. HMGB1 isoforms were determined by liquid chromatography MS/MS in plasma from untreated mice as well as in plasma from DDC diet-fed mice that received saline or ethyl pyruvate. C-D. Immunohistochemistry shows similar expression of cytokeratin (C) and A6 (D) in saline and ethyl pyruvate treated mice. E. qpcr shows similar expression of Cd133, Afp and H19 mrna in mice treated with saline or ethyl pyruvate. All data are expressed as means ± SEM. qpcr data are expressed as fold induction compared to normal liver. Statistical significance was determined by unpaired two-tailed t-test. Scale bar 1 µm.
32 Figure 4. HMGB1 drives ductular reactions via RAGE but not TLR4 or TLR9. Male mice were treated with DDC diet for 3 weeks. A-C. Cytokeratin expression and Cd133, Afp and H19 mrna levels were determined by IHC or qpcr in wt (n=7) or Rage KO (n=8) mice. Liver injury was assessed by serum ALT determination. D-F. Cytokeratin expression, Cd133, Afp and H19 mrna expression and liver injury were determined in wt (n=7) or Tlr4 KO (n=8) mice as above. G-I. Cytokeratin expression, Cd133, Afp and H19 mrna expression and liver injury were determined in wt (n=9) or Tlr9 KO (n=1) mice. Data are expressed as means±sem. qpcr data are shown as fold induction compared to normal liver. Statistical significance was determined by unpaired two-tailed t-test. * p<.5. ** p<.1. Scale bar 1 µm. Figure 5. Disulfide HMGB1 but not fully reduced HMGB1 upregulates CD133 and promotes progenitor/ductular proliferation. A. HMGB1 isoforms were determined by liquid chromatography MS/MS in plasma from DEN+1xCCl 4 - or DEN+6xCCl 4 -treated mice. B. BMOL cells were treated with recombinant disulfide HMGB1 (2 µg/ml) and fully reduced HMGB1 (2 µg/ml) for the indicated times, followed by analysis of Cd133 mrna by qpcr. C. Phospho-kinase screen in untreated disulfide HMGB1-treated (.5 µg/ml for 3 minutes) BMOL cells. D. BMOL cells were treated with disulfide ( Dis ) HMGB1 (.5 µg/ml) and fully reduced ( FR ) HMGB1 (.5 µg/ml) for the indicated times, followed by western blot analysis for phosphorylated and total Erk. E. BMOL cells were pretreated with MEK inhibitor UO126 (1 mm) for 3 minutes, followed by treatment with disulfide HMGB1 (.5 µg/ml) and analysis of Cd133 mrna by qpcr. F. Proliferation was determined by MTT assay in BMOL cells treated with disulfide
33 HMGB1 (.5 µg/ml) and fully reduced HMGB1 (.5 µg/ml) for the indicated times. G. Cytokeratin 19 and ps1-h3 double-positive cells were detected by confocal microscopy in Hmgb1 fl/fl and Hmgb1 Δhep mice treated with DDC diet (n=6 and n=9 mice, respectively), and quantified. H. BMOL cells were preincubated with blocking RAGE and TLR4 antibodies, TLR9 inhibitor or appropriate controls followed by treatment with disulfide HMGB1 (.5 µg/ml) for 15 minutes and p-erk and Erk western blot. Data are expressed as means ± SEM. qpcr data are expressed as fold induction compared to untreated cells, proliferation is expressed as fold increase compared to the h time point. Statistical significance was determined by unpaired two-tailed t-test. *<.5; ** p<.1; *** p<.1. Scale bar 1 µm. Figure 6. HMGB1 promotes ductular metaplasia of hepatocytes. A. Seven week-old HMGB1 wild-type mice (Hmgb1 wt/wt, n=12) or floxed HMGB1 alleles (Hmgb1 Δhep, n=8), both expressing TdTom, were infected with AAV8-TBG-Cre (i.v., 1 11 genome copies) followed by three week-long treatment with DDC diet, one week after AAV infection. B- D. Hepatocyte metaplasia was compared using dual immunofluorescent staining for A6 and TdTom (B), osteopontin (OPN) and TdTom (C), or Sox9 and TdTom (D). Hmgb1 Δhep mice showed a reduction of cells double-positive for A6 and TdTom compared to Hmgb1 wt/wt mice as well as of the percentage of A6 and TdTom double-positive cells among all A6-positive cells (B). Hmgb1 Δhep mice showed a reduction of OPN- and TdTom-double positive cells (C) as well as a reduction of Sox9- and TdTom-double positive cells (D) when compared to Hmgb1 wt/wt mice. E-F. qpcr showing decreased expression of osteopontin (encoded by Spp1) and Sox9 mrna in DDC-treated Hmgb1 Δhep
34 mice. qpcr data are shown as fold induction compared to normal liver. All data are expressed as means ± SEM. Significance was determined by unpaired two-tailed t-test. * p<.5; p<.1; *** p<.1. Scale bar 1 µm. Figure 7. HMGB1 promotes hepatocarcinogenesis in the presence but not the absence of chronic liver injury. A-C. Male Hmgb1 fl/fl (n=17) and Hmgb1 Δhep (n=19) mice were treated with DEN and CCl 4 and sacrificed as indicated (A). Livers were photographed (B) and tumor number, size and liver body weight ratio were determined (C). D. HMGB1 serum levels were determined by ELISA in mice that had either received DEN only (n=4), DEN plus a single injection of CCl 4 (n=4) or DEN plus 3 injections of CCl 4 (n=4). E-G. Male Hmgb1 fl/fl (n=11) and Hmgb1 Δhep (n=13) mice were treated with DEN and sacrificed as indicated (E). Livers were photographed (F) and tumor number, size and liver body weight ratio were determined (G). Data are expressed as means ± SEM. Statistical significance was determined by unpaired two-tailed t-test. ** p<.1; *** p<.1. Scale bar 1 cm. Figure 8. HMGB1 does not regulate fibrosis, proliferation or inflammation in HCC. A-D. Tumors from DEN+15xCCl 4 -treated Hmgb1 fl/fl (n=12) and Hmgb1 Δhep (n=12) mice were analyzed by IHC and qpcr to determine proliferation (A), fibrosis (B), infiltrating CD45+ leukocytes (C) and Ly6B-positive neutrophils (D). E. Hepatocarcinogenesis was induced in male wild-type (WT, n=1) and neutrophil elastase-deficient (Elane KO, n=11) mice, using the combination of DEN and 15 CCl 4 injections, followed by quantification
35 of tumor number, size and liver-body weight ratio. Data are expressed as means ± SEM. qpcr data are shown as fold induction compared to normal liver. Statistical significance was determined by unpaired two-tailed t-test. Scale bar 1 µm. Figure 9. HMGB1 promotes tumor dedifferentiation and progenitor signature. Tumors from DEN+15xCCl 4 -treated Hmgb1 fl/fl (n=6) and Hmgb1 Δhep (n=6) mice as well as healthy control livers (n=3) were subjected to RNA sequencing. A. Heatmap displays genes in most relevant pathways as described in Material and Methods. B. Expression of Cd133, Afp and H19 was confirmed by qpcr. C-D. AFP (C) and A6 (D) expression were determined by immunohistochemistry in Hmgb1 fl/fl (n=7-12) and Hmgb1 Δhep (n=1-12) tumors. Data are expressed as means ± SEM. qpcr data are shown as fold induction compared to normal liver. Statistical significance was determined by unpaired two-tailed t-test. * p<.5; ** p<.1. Scale bar 1 µm. REFERENCES 1. Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, et al. Global and regional mortality from 235 causes of death for 2 age groups in 199 and 21: a systematic analysis for the Global Burden of Disease Study 21. Lancet. 212;38(9859): El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 211;365(12): Luedde T, Kaplowitz N, and Schwabe RF. Cell death and cell death responses in liver disease: mechanisms and clinical relevance. Gastroenterology. 214;147(4): e4. 4. Tai DI, Lin SM, Sheen IS, Chu CM, Lin DY, and Liaw YF. Long-term outcome of hepatitis B e antigen-negative hepatitis B surface antigen carriers in relation to
36 changes of alanine aminotransferase levels over time. Hepatology. 29;49(6): Ghany MG, Kleiner DE, Alter H, Doo E, Khokar F, Promrat K, Herion D, Park Y, Liang TJ, and Hoofnagle JH. Progression of fibrosis in chronic hepatitis C. Gastroenterology. 23;124(1): Chen CF, Lee WC, Yang HI, Chang HC, Jen CL, Iloeje UH, Su J, Hsiao CK, Wang LY, You SL, et al. Changes in serum levels of HBV DNA and alanine aminotransferase determine risk for hepatocellular carcinoma. Gastroenterology. 211;141(4):124-8, 8 e Lee MH, Yang HI, Lu SN, Jen CL, Yeh SH, Liu CJ, Chen PJ, You SL, Wang LY, Chen WJ, et al. Hepatitis C virus seromarkers and subsequent risk of hepatocellular carcinoma: long-term predictors from a community-based cohort study. J Clin Oncol. 21;28(3): Boege Y, Malehmir M, Healy ME, Bettermann K, Lorentzen A, Vucur M, Ahuja AK, Bohm F, Mertens JC, Shimizu Y, et al. A Dual Role of Caspase-8 in Triggering and Sensing Proliferation-Associated DNA Damage, a Key Determinant of Liver Cancer Development. Cancer Cell. 217;32(3): e1. 9. Ruhl CE, and Everhart JE. Elevated serum alanine aminotransferase and gammaglutamyltransferase and mortality in the United States population. Gastroenterology. 29;136(2): e Bettermann K, Vucur M, Haybaeck J, Koppe C, Janssen J, Heymann F, Weber A, Weiskirchen R, Liedtke C, Gassler N, et al. TAK1 suppresses a NEMOdependent but NF-kappaB-independent pathway to liver cancer. Cancer Cell. 21;17(5): Hikita H, Kodama T, Shimizu S, Li W, Shigekawa M, Tanaka S, Hosui A, Miyagi T, Tatsumi T, Kanto T, et al. Bak deficiency inhibits liver carcinogenesis: a causal link between apoptosis and carcinogenesis. J Hepatol. 212;57(1): Luedde T, Beraza N, Kotsikoris V, van Loo G, Nenci A, De Vos R, Roskams T, Trautwein C, and Pasparakis M. Deletion of NEMO/IKKgamma in liver parenchymal cells causes steatohepatitis and hepatocellular carcinoma. Cancer Cell. 27;11(2): Weber A, Boger R, Vick B, Urbanik T, Haybaeck J, Zoller S, Teufel A, Krammer PH, Opferman JT, Galle PR, et al. Hepatocyte-specific deletion of the antiapoptotic protein myeloid cell leukemia-1 triggers proliferation and hepatocarcinogenesis in mice. Hepatology. 21;51(4): Michalopoulos GK, and Khan Z. Liver Stem Cells: Experimental Findings and Implications for Human Liver Disease. Gastroenterology. 215;149(4): Malato Y, Naqvi S, Schurmann N, Ng R, Wang B, Zape J, Kay MA, Grimm D, and Willenbring H. Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J Clin Invest. 211;121(12): Yanger K, Knigin D, Zong Y, Maggs L, Gu G, Akiyama H, Pikarsky E, and Stanger BZ. Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell. 214;15(3): Schaub JR, Malato Y, Gormond C, and Willenbring H. Evidence against a stem cell origin of new hepatocytes in a common mouse model of chronic liver injury. Cell Rep. 214;8(4):933-9.
37 18. Jors S, Jeliazkova P, Ringelhan M, Thalhammer J, Durl S, Ferrer J, Sander M, Heikenwalder M, Schmid RM, Siveke JT, et al. Lineage fate of ductular reactions in liver injury and carcinogenesis. J Clin Invest. 215;125(6): Choi TY, Ninov N, Stainier DY, and Shin D. Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology. 214;146(3): He J, Lu H, Zou Q, and Luo L. Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology. 214;146(3):789-8 e Lu WY, Bird TG, Boulter L, Tsuchiya A, Cole AM, Hay T, Guest RV, Wojtacha D, Man TY, Mackinnon A, et al. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat Cell Biol. 215;17(8): Raven A, Lu WY, Man TY, Ferreira-Gonzalez S, O'Duibhir E, Dwyer BJ, Thomson JP, Meehan RR, Bogorad R, Koteliansky V, et al. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature. 217;547(7663): Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, Ellis E, van Wenum M, Fuchs SA, de Ligt J, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 215;16(1-2): Yanger K, Zong Y, Maggs LR, Shapira SN, Maddipati R, Aiello NM, Thung SN, Wells RG, Greenbaum LE, and Stanger BZ. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 213;27(7): Tarlow BD, Pelz C, Naugler WE, Wakefield L, Wilson EM, Finegold MJ, and Grompe M. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell. 214;15(5): Mu X, Espanol-Suner R, Mederacke I, Affo S, Manco R, Sempoux C, Lemaigre FP, Adili A, Yuan D, Weber A, et al. Hepatocellular carcinoma originates from hepatocytes and not from the progenitor/biliary compartment. J Clin Invest. 215;125(1): Shin S, Wangensteen KJ, Teta-Bissett M, Wang YJ, Mosleh-Shirazi E, Buza EL, Greenbaum LE, and Kaestner KH. Genetic lineage tracing analysis of the cell of origin of hepatotoxin-induced liver tumors in mice. Hepatology. 216;64(4): He G, Dhar D, Nakagawa H, Font-Burgada J, Ogata H, Jiang Y, Shalapour S, Seki E, Yost SE, Jepsen K, et al. Identification of liver cancer progenitors whose malignant progression depends on autocrine IL-6 signaling. Cell. 213;155(2): Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, Mikaelyan A, Roberts LR, Demetris AJ, Sun Z, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 26;12(4): Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, Budhu A, Zanetti KA, Chen Y, Qin LX, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 28;68(5):
38 31. Yang XR, Xu Y, Yu B, Zhou J, Qiu SJ, Shi GM, Zhang BH, Wu WZ, Shi YH, Wu B, et al. High expression levels of putative hepatic stem/progenitor cell biomarkers related to tumour angiogenesis and poor prognosis of hepatocellular carcinoma. Gut. 21;59(7): Villanueva A, Hoshida Y, Battiston C, Tovar V, Sia D, Alsinet C, Cornella H, Liberzon A, Kobayashi M, Kumada H, et al. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 211;14(5): e Kim H, Choi GH, Na DC, Ahn EY, Kim GI, Lee JE, Cho JY, Yoo JE, Choi JS, and Park YN. Human hepatocellular carcinomas with "Stemness"-related marker expression: keratin 19 expression and a poor prognosis. Hepatology. 211;54(5): Ji J, and Wang XW. Clinical implications of cancer stem cell biology in hepatocellular carcinoma. Semin Oncol. 212;39(4): Won C, Kim BH, Yi EH, Choi KJ, Kim EK, Jeong JM, Lee JH, Jang JJ, Yoon JH, Jeong WI, et al. Signal transducer and activator of transcription 3-mediated CD133 up-regulation contributes to promotion of hepatocellular carcinoma. Hepatology. 215;62(4): Agopian VG, Harlander-Locke MP, Markovic D, Zarrinpar A, Kaldas FM, Cheng EY, Yersiz H, Farmer DG, Hiatt JR, and Busuttil RW. Evaluation of Patients With Hepatocellular Carcinomas That Do Not Produce alpha-fetoprotein. JAMA Surg. 217;152(1): Lotze MT, Zeh HJ, Rubartelli A, Sparvero LJ, Amoscato AA, Washburn NR, Devera ME, Liang X, Tor M, and Billiar T. The grateful dead: damage-associated molecular pattern molecules and reduction/oxidation regulate immunity. Immunol Rev. 27;22( Kubes P, and Mehal WZ. Sterile inflammation in the liver. Gastroenterology. 212;143(5): Hernandez C, Huebener P, and Schwabe RF. Damage-associated molecular patterns in cancer: a double-edged sword. Oncogene. 216;35 (46): Venereau E, Ceriotti C, and Bianchi ME. DAMPs from Cell Death to New Life. Front Immunol. 215;6( Huebener P, Pradere JP, Hernandez C, Gwak GY, Caviglia JM, Mu X, Loike JD, Jenkins RE, Antoine DJ, and Schwabe RF. The HMGB1/RAGE axis triggers neutrophil-mediated injury amplification following necrosis. J Clin Invest. 215;125(2): Mariotti V, Strazzabosco M, Fabris L, and Calvisi DF. Animal models of biliary injury and altered bile acid metabolism. Biochim Biophys Acta Trauner M, Fickert P, and Wagner M. MDR3 (ABCB4) defects: a paradigm for the genetics of adult cholestatic syndromes. Semin Liver Dis. 27;27(1): Huebener P, Gwak GY, Pradere JP, Quinzii CM, Friedman R, Lin CS, Trent CM, Mederacke I, Zhao E, Dapito DH, et al. High-mobility group box 1 is dispensable for autophagy, mitochondrial quality control, and organ function in vivo. Cell Metab. 214;19(3):
39 45. Bonaldi T, Talamo F, Scaffidi P, Ferrera D, Porto A, Bachi A, Rubartelli A, Agresti A, and Bianchi ME. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 23;22(2): Scaffidi P, Misteli T, and Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 22;418(6894): Dave SH, Tilstra JS, Matsuoka K, Li F, DeMarco RA, Beer-Stolz D, Sepulveda AR, Fink MP, Lotze MT, and Plevy SE. Ethyl pyruvate decreases HMGB1 release and ameliorates murine colitis. J Leukoc Biol. 29;86(3): Kim YM, Park EJ, Kim JH, Park SW, Kim HJ, and Chang KC. Ethyl pyruvate inhibits the acetylation and release of HMGB1 via effects on SIRT1/STAT signaling in LPS-activated RAW264.7 cells and peritoneal macrophages. Int Immunopharmacol. 216;41( Boulter L, Govaere O, Bird TG, Radulescu S, Ramachandran P, Pellicoro A, Ridgway RA, Seo SS, Spee B, Van Rooijen N, et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat Med. 212;18(4): Venereau E, Casalgrandi M, Schiraldi M, Antoine DJ, Cattaneo A, De Marchis F, Liu J, Antonelli A, Preti A, Raeli L, et al. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J Exp Med. 212;29(9): Tirnitz-Parker JE, Tonkin JN, Knight B, Olynyk JK, and Yeoh GC. Isolation, culture and immortalisation of hepatic oval cells from adult mice fed a cholinedeficient, ethionine-supplemented diet. Int J Biochem Cell Biol. 27;39(12): Yimlamai D, Christodoulou C, Galli GG, Yanger K, Pepe-Mooney B, Gurung B, Shrestha K, Cahan P, Stanger BZ, and Camargo FD. Hippo pathway activity influences liver cell fate. Cell. 214;157(6): Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 29;16(5): Kemper K, Versloot M, Cameron K, Colak S, de Sousa e Melo F, de Jong JH, Bleackley J, Vermeulen L, Versteeg R, Koster J, et al. Mutations in the Ras-Raf Axis underlie the prognostic value of CD133 in colorectal cancer. Clin Cancer Res. 212;18(11): Tabu K, Kimura T, Sasai K, Wang L, Bizen N, Nishihara H, Taga T, and Tanaka S. Analysis of an alternative human CD133 promoter reveals the implication of Ras/ERK pathway in tumor stem-like hallmarks. Mol Cancer. 21;9( Pusterla T, Nemeth J, Stein I, Wiechert L, Knigin D, Marhenke S, Longerich T, Kumar V, Arnold B, Vogel A, et al. Receptor for advanced glycation endproducts (RAGE) is a key regulator of oval cell activation and inflammation-associated liver carcinogenesis in mice. Hepatology. 213;58(1): Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, Caviglia JM, Khiabanian H, Adeyemi A, Bataller R, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 212;21(4):54-16.
40 58. Mu X, Pradere JP, Affo S, Dapito DH, Friedman R, Lefkovitch JH, and Schwabe RF. Epithelial Transforming Growth Factor-beta Signaling Does Not Contribute to Liver Fibrosis but Protects Mice From Cholangiocarcinoma. Gastroenterology. 216;15(3): Inokuchi S, Aoyama T, Miura K, Osterreicher CH, Kodama Y, Miyai K, Akira S, Brenner DA, and Seki E. Disruption of TAK1 in hepatocytes causes hepatic injury, inflammation, fibrosis, and carcinogenesis. Proc Natl Acad Sci U S A. 21;17(2): Wilson CL, Jurk D, Fullard N, Banks P, Page A, Luli S, Elsharkawy AM, Gieling RG, Chakraborty JB, Fox C, et al. NFkappaB1 is a suppressor of neutrophildriven hepatocellular carcinoma. Nat Commun. 215;6( Templeton AJ, McNamara MG, Seruga B, Vera-Badillo FE, Aneja P, Ocana A, Leibowitz-Amit R, Sonpavde G, Knox JJ, Tran B, et al. Prognostic role of neutrophil-to-lymphocyte ratio in solid tumors: a systematic review and metaanalysis. J Natl Cancer Inst. 214;16(6):dju Li YW, Qiu SJ, Fan J, Zhou J, Gao Q, Xiao YS, and Xu YF. Intratumoral neutrophils: a poor prognostic factor for hepatocellular carcinoma following resection. J Hepatol. 211;54(3): Belaaouaj A, McCarthy R, Baumann M, Gao Z, Ley TJ, Abraham SN, and Shapiro SD. Mice lacking neutrophil elastase reveal impaired host defense against gram negative bacterial sepsis. Nat Med. 1998;4(5): Cheng BQ, Jia CQ, Liu CT, Lu XF, Zhong N, Zhang ZL, Fan W, and Li YQ. Serum high mobility group box chromosomal protein 1 is associated with clinicopathologic features in patients with hepatocellular carcinoma. Dig Liver Dis. 28;4(6): McDonald B, Pittman K, Menezes GB, Hirota SA, Slaba I, Waterhouse CC, Beck PL, Muruve DA, and Kubes P. Intravascular danger signals guide neutrophils to sites of sterile inflammation. Science. 21;33(62): Apte U, Thompson MD, Cui S, Liu B, Cieply B, and Monga SP. Wnt/beta-catenin signaling mediates oval cell response in rodents. Hepatology. 28;47(1): Hu M, Kurobe M, Jeong YJ, Fuerer C, Ghole S, Nusse R, and Sylvester KG. Wnt/beta-catenin signaling in murine hepatic transit amplifying progenitor cells. Gastroenterology. 27;133(5): Kim KH, Chen CC, Alpini G, and Lau LF. CCN1 induces hepatic ductular reaction through integrin alphavbeta(5)-mediated activation of NF-kappaB. J Clin Invest. 215;125(5): Utley S, James D, Mavila N, Nguyen MV, Vendryes C, Salisbury SM, Phan J, and Wang KS. Fibroblast growth factor signaling regulates the expansion of A6- expressing hepatocytes in association with AKT-dependent beta-catenin activation. J Hepatol. 214;6(5): Chen R, Zhu S, Fan XG, Wang H, Lotze MT, Zeh HJ, 3rd, Billiar TR, Kang R, and Tang D. HMGB1 Controls Liver Cancer Initiation through YAP-dependent Aerobic Glycolysis. Hepatology Mollica L, De Marchis F, Spitaleri A, Dallacosta C, Pennacchini D, Zamai M, Agresti A, Trisciuoglio L, Musco G, and Bianchi ME. Glycyrrhizin binds to high-
41 mobility group box 1 protein and inhibits its cytokine activities. Chem Biol. 27;14(4): Veldt BJ, Hansen BE, Ikeda K, Verhey E, Suzuki H, and Schalm SW. Long-term clinical outcome and effect of glycyrrhizin in 193 chronic hepatitis C patients with non-response or relapse to interferon. Scand J Gastroenterol. 26;41(9): Tirone M, Tran NL, Ceriotti C, Gorzanelli A, Canepari M, Bottinelli R, Raucci A, Di Maggio S, Santiago C, Mellado M, et al. High mobility group box 1 orchestrates tissue regeneration via CXCR4. J Exp Med. 218;215(1): Arriazu E, Ge X, Leung TM, Magdaleno F, Lopategi A, Lu Y, Kitamura N, Urtasun R, Theise N, Antoine DJ, et al. Signalling via the osteopontin and high mobility group box-1 axis drives the fibrogenic response to liver injury. Gut Mitchell C, and Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 28;3(7): Engelhardt NV, Factor VM, Medvinsky AL, Baranov VN, Lazareva MN, and Poltoranina VS. Common antigen of oval and biliary epithelial cells (A6) is a differentiation marker of epithelial and erythroid cell lineages in early development of the mouse. Differentiation. 1993;55(1): Antoine DJ, Jenkins RE, Dear JW, Williams DP, McGill MR, Sharpe MR, Craig DG, Simpson KJ, Jaeschke H, and Park BK. Molecular forms of HMGB1 and keratin-18 as mechanistic biomarkers for mode of cell death and prognosis during clinical acetaminophen hepatotoxicity. J Hepatol. 212;56(5): Robinson MD, and Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 21;11(3):R Yang H, Lundback P, Ottosson L, Erlandsson-Harris H, Venereau E, Bianchi ME, Al-Abed Y, Andersson U, Tracey KJ, and Antoine DJ. Redox modification of cysteine residues regulates the cytokine activity of high mobility group box-1 (HMGB1). Mol Med. 212;18(25-9.
42 A Mdr2 KO Tak1 Δhep C E DDC diet G Hmgb1 wt/wt Hmgb1 fl/fl Hmgb1 fl/fl Hmgb1 Δhep Hmgb1 Δhep Hmgb1 Δhep Control 6xCCl 4 CK (%) CK (%) CK (%) CK (%) * unt ctl (n=2) HMGB1 wt HMGB1 ff *** *** B D F Cd133 mrna (fold) Cd133 mrna (fold) Cd133 mrna (fold) Serum HMGB1 (ng/ml) * ctls (n=2) HMGB1 wt * 1 2 *** Afp mrna (fold) Afp mrna (fold) Afp mrna (fold) * ctls (n=2) HMGB1 wt 1 2 6, 4, 2, 8 *** *** ** Untreated 6x CCl 4 H19 mrna (fold) H19 mrna (fold) H19 mrna (fold) , 4, 3, 2, 1, WT Tak1 Δhep Hmgb1 wt/wt Tak1 Δhep Hmgb1 Δhep * WT Hmgb1 fl/fl Mdr2 KO Hmgb1 fl/fl Mdr2 KO Hmgb1 Δhep ALT [U/I] ALT [U/I] Ctrl Hmgb1 fl/fl DDC Hmgb1 fl/fl DDC Hmgb1 Δhep ** ALT [U/I] Figure 1. HMGB1 is required but not sufficient for ductular reactions. A-B. 8 week-old mice with hepatic deletion of HMGB1 (Hmgb1 Δhep, n=9) showed reduction of cytokeratin-positive ductular cells (A), lower expression of Cd133, Afp and H19 mrna but similar ALT (B) in comparison to Hmgb1 wt/wt controls (n=9) in the Tak1 Δhep model. C-D. In the Mdr2 KO model, 8-week old Hmgb1 Δhep mice (n=13) showed reduction of cytokeratin-positive ductular cells (C), lower expression of Afp and H19 mrna but similar ALT (D) in comparison to Hmgb1 fl/fl controls (n=1). E-F. Following three weeks of DDC diet, Hmgb1 Δhep mice (n=9) showed reduction of cytokeratin-positive ductular cells (E), lower expression of Afp and H19 mrna but similar ALT (F) in comparison to Hmgb1 fl/fl controls (n=6). G. Mice either untreated (n=4) or treated with six injections of CCl 4 (n=11) displayed similar level of cytokeratin-positive cells. HMGB1 serum levels were determined by ELISA in mice that have received (n=3) or not (n=2) six injections of CCL 4. All data are expressed as means ± SEM. qpcr data are shown as fold induction compared to normal liver. Statistical significance was determined by two-tailed t-test. * p<.5;; ** p<.1;; *** p<.1. Scale bar 1 µm.
43 A C DDC diet Mdr2 KO E Hmgb1 fl/fl x AAV8-TBG-LacZ Hmgb1 fl/fl x AAV8-TBG-LacZ Hmgb1 fl/fl x AAV8-TBG-LacZ Hmgb1 fl/fl x AAV8-TBG-Cre Hmgb1 fl/fl x AAV8-TBG-Cre Hmgb1 fl/fl x AAV8-TBG-Cre CK (%) CK (%) B Hmgb1 mrna (fold) *** ** ** unt ctl (n=2) HMGB1 ff HMGB1 ff D F Hmgb1fl/fl AAV8-LacZ Hmgb1 Δhep Hmgb1fl/fl AAV8-Cre WT Hmgb1 fl/fl Mdr2 KO Hmgb1 Mdr2 KO Hmgb1 Cd133 mrna (fold) Untreated 1 Hmgb1 2 fl/fl DDC Hmgb1 DDC Hmgb1 1 *** 5 * 5 *** Cd133 mrna (fold) * fl/fl AAV8-LacZ fl/fl AAV8-Cre Figure 2. HMGB1 from hepatocellular sources is required for ductular 1 2 reactions. 3 A. 1 2 week-old ctls Hmgb1 fl/fl Mdr2 KO mice were infected with AAV8-TBG-Cre (i.v., 1 11 genome copies, n=11) or AAV8-TBG- (n=2) LacZ (i.v., 1 11 genome copies, n=1) and sacrificed 6 weeks later. Immunohistochemical HMGB1 staining was performed, demonstrating efficient deletion of HMGB1 from hepatocytes in AAV8-TBG-Cre-, but not in AAV8-TBG-LacZ-infected mice (arrows pointing at hepatocytes). B. HMGB1 deletion was confirmed by qpcr in Hmgb1 fl/fl mice infected with AAV8-TBG-Creor AAV8-TBG-LacZ. C-D. Immunohistochemical cytokeratin staining (C) and qpcr for Cd133 and H19 (D) demonstrated reduced ductular reactions in AAV8-TBG-Cre-infected mice. E-F. 3 week old Hmgb1 fl/fl mice were infected with AAV8-TBG-Cre (i.v genome copies, n=8) or AAV8-TBG-LacZ (i.v., 1 11 genome copies, n=7) followed by treatment with DDC diet for 3 weeks two weeks after AAV infection. Immunohistochemical cytokeratin staining (E) and qpcr for Cd133 and H19 (F) demonstrated reduced ductular reactions in AAV8-TBG-Cre-infected mice. All data are expressed as means ± SEM. qpcr data are shown as fold induction compared to normal liver. Statistical significance was determined by two-tailed t-test. * p<.5;; **, p<.1;; *** p<.1. Scale bar 1 µm. H19 mrna (fold) H19 mrna (fold) fl/fl AAV8-LacZ fl/fl AAV8-Cre *** 1 2
44 A Ethyl Pyruvate Sac W8 Birth B W11 DDC diet Untreated Fully reduced HMGB1 Fully reduced HMGB1 DDC + Saline Disulfide HMGB1 acetylated Disulfide HMGB1 C DDC + Ethyl pyruvate Fully reduced HMGB1 acetylated Saline Fully reduced HMGB1 Disulfide HMGB1 Ethyl pyruvate Ctrl DDC + Saline DDC + Ethyl pyruvate 4.4 CK (%) D Saline Ethyl pyruvate A6 (%) F 1 1 Afp mrna (fold) H19 mrna (fold) 8 8 Cd133 mrna (fold) E P= Serum HMGB1 (ng/ml) Figure 3. Active HMGB1 secretion does not drive ductular reactions. A. C57BL/6 males were pre- treated with saline (n=11) or ethyl pyruvate (n=8) followed by treatment with DDC diet for 3 weeks as shown. B. HMGB1 isoforms were determined by liquid chromatography MS/MS in plasma from untreated mice as well as in plasma from DDC diet- fed mice that either received saline or ethyl pyruvate. C- D. Immunohistochemistry shows similar expression of cytokeratin (C) and A6 (D) in saline and ethyl pyruvate treated mice. E. qpcr shows similar expression of Cd133, Afp and H19 mrna in mice treated with saline or ethyl pyruvate. All data are expressed as means ± SEM. qpcr data are expressed as fold induction compared to normal liver. Statistical significance was determined by two- tailed t- test. Scale bar 1 µm.
45 A DDC diet B DDC diet C DDC diet Rage WT Rage KO 66 ** p= * Tlr4 WT Tlr9 WT Tlr4 KO Tlr9 KO CK (%) CK (%) CK (%) Cd133 mrna (fold) Cd133 mrna (fold) Afp mrna (fold) Afp mrna (fold) Afp mrna (fold) * H19 mrna (fold) H19 mrna (fold) H19 mrna (fold) p= Ctrl WT DDC WT DDCRage KO ALT [U/I] ALT [U/I] ALT [U/I] Ctrl WT DDC WT DDC Tlr4 KO Ctrl WT DDC WT DDC Tlr9 KO Figure 4. HMGB1 drives ductular reactions via RAGE but not TLR4 or TLR9. A-C. Male mice were treated with DDC diet for 3 weeks. Cytokeratin expression and Cd133, Afp and H19 mrna levels were determined by IHC or qpcr, and liver injury by serum ALT in (A) wt (n=7) or Rage KO (n=8) mice, (B) in wt (n=7) or Tlr4 KO (n=8) mice, or (C) in wt (n=9) or Tlr9 KO (n=1) mice. Data are expressed as means±sem. qpcr data are shown as fold induction compared to normal liver. Statistical significance was determined by two-tailed t-test. * p<.5.** p<.1. Scale bar 1 µm. Cd133 mrna (fold) 3 2 1
46 DEN+1xCCl A 4 DEN+6xCCl 4 B 4. Intensity (%) C Dis-HMGB1 Untreated E 1% 5% UO HMGB perk - H Erk - Disulfide HMGB1 24,585.( 24,587.2( % % % % % % % % % ERK CREB a RAGE HMGB1 perk - Erk - ERK CREB Fully reduced HMGB1 Mass (kda) Cd133 mrna (fold) DMSO UO ** 1 2 (%)1 Intensity ( ( Fold induction % % F Fold inincrease Disulfide HMGB ,585.( Fully reduced HMGB1 24,587.2( Disulfide HMGB1 acetylated 25,467.1( 25,469.( % % % % % % % % % ( ( p-erk1/2 ERK1/2 CREB p-creb (S133) a TLR HMGB1 perk - Erk - D 24h 48h 48h 72h 72h Fully reduced HMGB1 acetylated Mass (kda) Untreated HMGB1 Disulfide HMGB1 Fully Reduced 4 *** 3. ** h 48h 24h 48h Disulfide Fully reduced HMGB1 HMGB1 Dis-HMGB1 (min) perk - Figure 5. Disulfide HMGB1 but not fully reduced HMGB1 upregulates CD133 and promotes progenitor/ductular proliferation. A. HMGB1 isoforms were determined by liquid chromatography MS/MS in plasma from DEN+1xCCl 4 - or DEN+6xCCl 4 -treated mice. B. BMOL cells were treated with recombinant disulfide HMGB1 (2 µg/ml) and fully reduced HMGB1 (2 µg/ml) for the indicated times, followed by analysis of Cd133 mrna by qpcr. C. Phospho screen for kinase activation in untreated disulfide HMGB1- treated (.5 µg/ml for 3 minutes) BMOL cells. D. BMOL cells were treated with disulfide ( Dis ) HMGB1 (.5 µg/ml) and fully reduced ( FR ) HMGB1 (.5 µg/ml) for the indicated times, followed by western blot analysis for phosphorylated and total Erk. E. BMOL cells were pretreated with MEK inhibitor UO126 (1 µm) for 3 minutes, followed by treatment with disulfide HMGB1 (.5 µg/ml) and analysis of Cd133 mrna by qpcr. F. Proliferation was determined by MTT assay in BMOL cells treated with disulfide HMGB1 (.5 µg/ml) and fully reduced HMGB1 (.5 µg/ml) for the indicated times. G. Cytokeratin 19 and ps1-h3 double-positive cells were detected by confocal microscopy in Hmgb1 fl/fl and Hmgb1 Δhep mice treated with DDC diet (n=6 and n=9 mice, respectively), and quantified. H. BMOL cells were preincubated with blocking RAGE and TLR4 antibodies, TLR9 inhibitor or appropriate controls followed by treatment with disulfide HMGB1 (.5 µg/ml) for 15 minutes and p-erk and Erk western blot. Data are expressed as means ± SEM. qpcr data are expressed as fold induction compared to untreated cells, proliferation is expressed as fold increase compared to the h time point. Statistical significance was determined by two-tailed t-test. *<.5;; ** p<.1;; *** p<.1. Scale bar 1 µm. * Cd133 mrna (fold) Erk - FR-HMGB1 (min) perk - Erk - ** G TLR9 inh HMGB1 perk - Erk - Hmgb1 fl/fl Hmgb1 Δhep ps1-h3+ CK19+ cells/field ** 1 2
47 A B Hmgb1 Δhep Hmgb1 wt/wt C Hmgb1 Δhep Hmgb1 wt/wt D Hmgb1 Δhep Hmgb1 wt/wt Birth TdTom TdTom TdTom TdTom TdTom TdTom AAV8-TBG-Cre W7 A6 A6 OPN OPN Sox9 Sox9 W8 DDC diet W11 Merge Merge Merge Merge Merge Merge A6 + TdTom + (Cells per field) OPN + TdTom + (Cells per field) Sox9 + TdTom + (Cells per field) *** * DDC Hmgb1 wt/wt DDC Hmgb1 Δhep Figure 6. HMGB1 promotes ductular metaplasia of hepatocytes. A. Seven week old HMGB1 wild-type mice (Hmgb1 wt/wt, n=12) expressing TdTom or mice expressing floxed HMGB1 alleles and TdTom (Hmgb1 Δhep, n=8) were infected with AAV8-TBG-Cre (i.v., 1 11 genome copies) followed by treatment with DDC diet for 3 weeks, one week after AAV infection. B-D. Hepatocyte metaplasia was compared using dual immunofluorescent staining for A6 and TdTom (B), osteopontin (OPN) and TdTom (C), or Sox9 and TdTom (D). Hmgb1 Δhep mice showed a reduction of cells double-positive for A6 and TdTom compared to Hmgb1 wt/wt mice as well as of the percentage of A6 and TdTom double-positive cells among all A6-positive cells (B). Hmgb1 Δhep mice showed a reduction of OPN- and TdTom-double positive cells (C) as well as a reduction of Sox9- an dtdtom-double positive cells (D) when compared to Hmgb1 wt/wt mice. E-F. qpcr showing decreased expression of osteopontin (encoded by Spp1) and Sox9 mrna in DDC-treated Hmgb1 Δhep mice. qpcr data are shown as fold induction compared to normal liver. All data are expressed as means ± SEM. Significance was determined by two-tailed t-test. * p<.5;; ** p<.1;; *** p<.1. Scale bar 1 µm. *** A6 + TdTom + cells (% of A6+ cells) E Spp1 mrna (fold) F Sox9 mrna (fold) *** 1 2 ** 1 2 **
48 A B Hmgb1 fl/fl C E F Hmgb1 fl/fl G Tumor number (n) Tumor number (n)1 Birth DEN W2 W6 *** 15 x CCL 4 Tumor size (mm) p=.7 Sac W28 Δh Hmgb1 fl/fl Hmgb1 Δhep Hmgb1 fl/fl Hmgb1 Δhep Hmgb1 fl/fl Hmgb1 Δhep Birth DEN W2 Tumor size (mm) 255 Sac W44 Hmgb1 fl/fl Hmgb1 Δhep Hmgb1 fl/fl Δh Hmgb1 Δhep Liver Body Ratio (%) Liver Body Ratio (%) Hmgb1 Δhep ** Hmgb1 fl/fl D Hmgb1 Δhep Serum HMGB1 (ng/ml) ** *** DEN DEN DEN xCCl 4 3xCCl 4 Figure 7. HMGB1 promotes hepatocarcinogenesis in the presence but not the absence of chronic liver injury. A-C. Male Hmgb1 fl/fl (n=17) and Hmgb1 Δhep (n=19) mice were treated with DEN and CCl 4 and sacrificed as indicated (A). Livers were photographed (B) and tumor number, size and liver body weight ratio were determined (C). D. HMGB1 serum levels were determined by ELISA in mice that had either received DEN only (n=4), DEN plus a single injection of CCl 4 (n=4) or DEN plus 3 injections of CCl 4 (n=4). E-G. Male Hmgb1 fl/fl (n=11) and Hmgb1 Δhep (n=13) mice were treated with DEN and sacrificed as indicated (E). Livers were photographed (F) and tumor number, size and liver body weight ratio were determined (G). Data are expressed as means ± SEM. Statistical significance was determined by two-tailed t-test. ** p<.1;; *** p<.1. Scale bar 1 cm. Hmgb1 Δhep
49 Ki67+ cells/field mki67 mrna (fold) p=.5 WT KO p= WT KO Liver Body Ratio (%) 3 3 Timp1 mrna (fold) E 2 Tumor size (mm) Hmgb1Δhep 44 1 O Hmgb1f/f Hmgb1Δhep KO Hmgb1fl/fl Acta2 mrna (fold).5.5 Col1a1 mrna (fold) WT Sirius red (%) Elane Elane Ctrl Hmgb1fl/fl Tumor Hmgb1fl/fl Tumor Hmgb1Δhep T HCC D Hmgb1Δhep Tumor number (n) HCC C Hmgb1fl/fl Hmgb1Δhep Leukocytes (%) HCC B Hmgb1fl/fl Neutrophils (%) HCC A 2 p= WT KO Figure 8. HMGB1 does not regulate proliferation, fibrosis or inflammation in HCC. A- D. Tumors from DEN+15xCCl4- treated Hmgb1fl/fl (n=12) and Hmgb1Δhep (n=12) mice were analyzed by IHC and qpcr to determine proliferation (A), fibrosis (B), infiltrating CD45+ leukocytes (C) and Ly6B- positive neutrophils (D). E. Hepatocarcinogenesis was induced in male wild- type (WT, n=1) and neutrophil elastase- deficient (ElaneKO, n=11) mice, using the combination of DEN and 15 CCl4 injections, followed by quantification of tumor number, size and liver- body weight ratio. Data are expressed as means ± SEM. qpcr data are shown as fold induction compared to normal liver. Statistical significance was determined by two- tailed t- test. Scale bar 1 µm.
50 A Normal HCC HCC liver Hmgb1 Δhep Hmgb1 fl/fl B Cd133 mrna (fold) Afp mrna (fold) H19 mrna (fold) * ** C D Hmgb1 fl/fl Hmgb1 Δhep Ctrl Hmgb1 fl/fl Tumor Hmgb1 fl/fl Tumor Hmgb1 Δhep **. Ctrl 1 2HCC Hmgb1 fl/fl Hmgb1 Δhep * Ctrl 1 2HCC Figure 9. HMGB1 promotes tumor dedifferentiation and progenitor signature. Tumors from DEN+15xCCl 4 -treated Hmgb1 fl/fl (n=6) and Hmgb1 Δhep (n=6) mice as well as healthy control livers (n=3) were subjected to RNA sequencing. A. Heatmap displays genes in most relevantpathways as described in Material and Methods. B. Expression of Cd133, Afp and H19 was confirmed by qpcr. C-D. AFP (C) and A6 (D) were determined by immunohistochemistry in Hmgb1 fl/fl (n=7-12) and Hmgb1 Δhep (n=1-12) tumors. Data are expressed as means ± SEM. qpcr data are shown as fold induction compared to normal liver. Statistical significance was determined by two-tailed t-test. * p<.5; ** p<.1. Scale bar 1 µm. AFP (%) A6 (%)
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.
 Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
Supplementary Figure 1: Hsp60 / IEC mice are embryonically lethal (A) Light microscopic pictures show mouse embryos at developmental stage E12.5 and E13.5 prepared from uteri of dams and subsequently genotyped.
Supplemental Figure S1. RANK expression on human lung cancer cells.
 Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
Supplemental Figure S1. RANK expression on human lung cancer cells. (A) Incidence and H-Scores of RANK expression determined from IHC in the indicated primary lung cancer subgroups. The overall expression
HMGB1 promotes ductular reaction and tumorigenesis in autophagy-deficient livers
 HMGB1 promotes ductular reaction and tumorigenesis in autophagy-deficient livers Bilon Khambu,, Zheng Dong, Xiao-Ming Yin J Clin Invest. 2018. https://doi.org/10.1172/jci91814. Research Article Cell biology
HMGB1 promotes ductular reaction and tumorigenesis in autophagy-deficient livers Bilon Khambu,, Zheng Dong, Xiao-Ming Yin J Clin Invest. 2018. https://doi.org/10.1172/jci91814. Research Article Cell biology
(a) Significant biological processes (upper panel) and disease biomarkers (lower panel)
 Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1. Functional enrichment analyses of secretomic proteins. (a) Significant biological processes (upper panel) and disease biomarkers (lower panel) 2 involved by hrab37-mediated secretory
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
fl/+ KRas;Atg5 fl/+ KRas;Atg5 fl/fl KRas;Atg5 fl/fl KRas;Atg5 Supplementary Figure 1. Gene set enrichment analyses. (a) (b)
 KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
KRas;At KRas;At KRas;At KRas;At a b Supplementary Figure 1. Gene set enrichment analyses. (a) GO gene sets (MSigDB v3. c5) enriched in KRas;Atg5 fl/+ as compared to KRas;Atg5 fl/fl tumors using gene set
Supplementary Table 1. Characterization of HNSCC PDX models established at MSKCC
 Supplementary Table 1. Characterization of HNSCC PDX models established at MSKCC Supplementary Table 2. Drug content and loading efficiency estimated with F-NMR and UV- Vis Supplementary Table 3. Complete
Supplementary Table 1. Characterization of HNSCC PDX models established at MSKCC Supplementary Table 2. Drug content and loading efficiency estimated with F-NMR and UV- Vis Supplementary Table 3. Complete
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the
 Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the genome-wide methylation microarray data. Mean ± s.d.; Student
Supplementary Figure 1. HOPX is hypermethylated in NPC. (a) Methylation levels of HOPX in Normal (n = 24) and NPC (n = 24) tissues from the genome-wide methylation microarray data. Mean ± s.d.; Student
Supplemental Table 1. Primer sequences for transcript analysis
 Supplemental Table 1. Primer sequences for transcript analysis Primer Sequence (5 3 ) Primer Sequence (5 3 ) Mmp2 Forward CCCGTGTGGCCCTC Mmp15 Forward CGGGGCTGGCT Reverse GCTCTCCCGGTTTC Reverse CCTGGTGTGCCTGCTC
Supplemental Table 1. Primer sequences for transcript analysis Primer Sequence (5 3 ) Primer Sequence (5 3 ) Mmp2 Forward CCCGTGTGGCCCTC Mmp15 Forward CGGGGCTGGCT Reverse GCTCTCCCGGTTTC Reverse CCTGGTGTGCCTGCTC
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
Supplemental figure 1. PDGFRα is expressed dominantly by stromal cells surrounding mammary ducts and alveoli. A) IHC staining of PDGFRα in
 Supplemental figure 1. PDGFRα is expressed dominantly by stromal cells surrounding mammary ducts and alveoli. A) IHC staining of PDGFRα in nulliparous (left panel) and InvD6 mouse mammary glands (right
Supplemental figure 1. PDGFRα is expressed dominantly by stromal cells surrounding mammary ducts and alveoli. A) IHC staining of PDGFRα in nulliparous (left panel) and InvD6 mouse mammary glands (right
Supplementary Figure 1: Neuregulin 1 increases the growth of mammary organoids compared to EGF. (a) Mammary epithelial cells were freshly isolated,
 1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1: Neuregulin 1 increases the growth of mammary organoids compared to EGF. (a) Mammary epithelial cells were freshly isolated, embedded in matrigel and exposed
1 2 3 4 5 6 7 8 9 10 Supplementary Figure 1: Neuregulin 1 increases the growth of mammary organoids compared to EGF. (a) Mammary epithelial cells were freshly isolated, embedded in matrigel and exposed
FOR REVIEW. BMB Reports - Manuscript Submission. Manuscript Draft. Manuscript Number: BMB
 BMB Reports - Manuscript Submission Manuscript Draft Manuscript Number: BMB-18-095 Title: Insulin Receptor Substrate 2:A Bridge between Hippo and AKT Pathways Article Type: Perspective (Invited Only) Keywords:
BMB Reports - Manuscript Submission Manuscript Draft Manuscript Number: BMB-18-095 Title: Insulin Receptor Substrate 2:A Bridge between Hippo and AKT Pathways Article Type: Perspective (Invited Only) Keywords:
Stimulating healthy tissue regeneration by targeting the 5-HT 2B receptor in chronic liver disease.
 Stimulating healthy tissue regeneration by targeting the 5-HT 2B receptor in chronic liver disease. Mohammad R Ebrahimkhani, Fiona Oakley, Lindsay B Murphy, Jelena Mann, Anna Moles, Maria J Perugorria,
Stimulating healthy tissue regeneration by targeting the 5-HT 2B receptor in chronic liver disease. Mohammad R Ebrahimkhani, Fiona Oakley, Lindsay B Murphy, Jelena Mann, Anna Moles, Maria J Perugorria,
LPS LPS P6 - + Supplementary Fig. 1.
 P6 LPS - - - + + + - LPS + + - - P6 + Supplementary Fig. 1. Pharmacological inhibition of the JAK/STAT blocks LPS-induced HMGB1 nuclear translocation. RAW 267.4 cells were stimulated with LPS in the absence
P6 LPS - - - + + + - LPS + + - - P6 + Supplementary Fig. 1. Pharmacological inhibition of the JAK/STAT blocks LPS-induced HMGB1 nuclear translocation. RAW 267.4 cells were stimulated with LPS in the absence
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
SUPPLEMENTARY INFORMATION FOR Liver X Receptor α mediates hepatic triglyceride accumulation through upregulation of G0/G1 Switch Gene 2 (G0S2) expression I: SUPPLEMENTARY METHODS II: SUPPLEMENTARY FIGURES
General Laboratory methods Plasma analysis: Gene Expression Analysis: Immunoblot analysis: Immunohistochemistry:
 General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
DOI: 10.1038/ncb3461 In the format provided by the authors and unedited. Supplementary Figure 1 (associated to Figure 1). Cpeb4 gene-targeted mice develop liver steatosis. a, Immunoblot displaying CPEB4
Exploring a Link Between Spy1 and Hepatocellular Carcinoma Progression
 University of Windsor Scholarship at UWindsor UWill Discover Undergraduate Conference UWill Discover 2016 Mar 29th, 4:00 PM - 5:00 PM Exploring a Link Between Spy1 and Hepatocellular Carcinoma Progression
University of Windsor Scholarship at UWindsor UWill Discover Undergraduate Conference UWill Discover 2016 Mar 29th, 4:00 PM - 5:00 PM Exploring a Link Between Spy1 and Hepatocellular Carcinoma Progression
Supplemental Information. Otic Mesenchyme Cells Regulate. Spiral Ganglion Axon Fasciculation. through a Pou3f4/EphA4 Signaling Pathway
 Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
(A) PCR primers (arrows) designed to distinguish wild type (P1+P2), targeted (P1+P2) and excised (P1+P3)14-
 1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
1 Supplemental Figure Legends Figure S1. Mammary tumors of ErbB2 KI mice with 14-3-3σ ablation have elevated ErbB2 transcript levels and cell proliferation (A) PCR primers (arrows) designed to distinguish
Supplementary Figure 1
 Supplementary Figure 1 Control Pancreatitis Supplementary Figure 2 A Panc Liver SI Spleen H 2 O B EZH2 fl/fl C EZH2 fl/fl 37bp EZH2 ERK2 D E 5 EZH2 fl/fl Fasting Glucose (mg/dl) 2 18 16 14 12 1 8 6 4 2
Supplementary Figure 1 Control Pancreatitis Supplementary Figure 2 A Panc Liver SI Spleen H 2 O B EZH2 fl/fl C EZH2 fl/fl 37bp EZH2 ERK2 D E 5 EZH2 fl/fl Fasting Glucose (mg/dl) 2 18 16 14 12 1 8 6 4 2
Kinase-independent functions of RIPK1 regulate hepatocyte survival and liver carcinogenesis
 The Journal of Clinical Investigation Kinase-independent functions of RIPK1 regulate hepatocyte survival and liver carcinogenesis Trieu-My Van, 1,2,3 Apostolos Polykratis, 1,2,3 Beate Katharina Straub,
The Journal of Clinical Investigation Kinase-independent functions of RIPK1 regulate hepatocyte survival and liver carcinogenesis Trieu-My Van, 1,2,3 Apostolos Polykratis, 1,2,3 Beate Katharina Straub,
Supplementary Figure 1:
 Supplementary Figure 1: (A) Whole aortic cross-sections stained with Hematoxylin and Eosin (H&E), 7 days after porcine-pancreatic-elastase (PPE)-induced AAA compared to untreated, healthy control aortas
Supplementary Figure 1: (A) Whole aortic cross-sections stained with Hematoxylin and Eosin (H&E), 7 days after porcine-pancreatic-elastase (PPE)-induced AAA compared to untreated, healthy control aortas
marker. DAPI labels nuclei. Flies were 20 days old. Scale bar is 5 µm. Ctrl is
 Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Supplemental Information. Tissue Myeloid Progenitors Differentiate. into Pericytes through TGF-b Signaling. in Developing Skin Vasculature
 Cell Reports, Volume 18 Supplemental Information Tissue Myeloid Progenitors Differentiate into Pericytes through TGF-b Signaling in Developing Skin Vasculature Tomoko Yamazaki, Ani Nalbandian, Yutaka Uchida,
Cell Reports, Volume 18 Supplemental Information Tissue Myeloid Progenitors Differentiate into Pericytes through TGF-b Signaling in Developing Skin Vasculature Tomoko Yamazaki, Ani Nalbandian, Yutaka Uchida,
(A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and a
 Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
Supplementary figure legends Supplementary Figure 1. Expression of Shh signaling components in a panel of gastric cancer. (A) RT-PCR for components of the Shh/Gli pathway in normal fetus cell (MRC-5) and
Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression
 Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Supplementary Figure 1 Supplementary Figure 1 Role of Raf-1 in TLR2-Dectin-1-mediated cytokine expression. Quantitative real-time PCR of indicated mrnas in DCs stimulated with TLR2-Dectin-1 agonist zymosan
Supplementary Figure 1. Repression of hepcidin expression in the liver of mice treated with
 Supplementary Figure 1. Repression of hepcidin expression in the liver of mice treated with DMN Immunohistochemistry for hepcidin and H&E staining (left). qrt-pcr assays for hepcidin in the liver (right).
Supplementary Figure 1. Repression of hepcidin expression in the liver of mice treated with DMN Immunohistochemistry for hepcidin and H&E staining (left). qrt-pcr assays for hepcidin in the liver (right).
Erzsebet Kokovay, Susan Goderie, Yue Wang, Steve Lotz, Gang Lin, Yu Sun, Badrinath Roysam, Qin Shen,
 Cell Stem Cell, Volume 7 Supplemental Information Adult SVZ Lineage Cells Home to and Leave the Vascular Niche via Differential Responses to SDF1/CXCR4 Signaling Erzsebet Kokovay, Susan Goderie, Yue Wang,
Cell Stem Cell, Volume 7 Supplemental Information Adult SVZ Lineage Cells Home to and Leave the Vascular Niche via Differential Responses to SDF1/CXCR4 Signaling Erzsebet Kokovay, Susan Goderie, Yue Wang,
Supplemental Figure 1. Western blot analysis indicated that MIF was detected in the fractions of
 Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
Supplemental Figure Legends Supplemental Figure 1. Western blot analysis indicated that was detected in the fractions of plasma membrane and cytosol but not in nuclear fraction isolated from Pkd1 null
Quantification of early stage lesions for loss of p53 should be shown in the main figures.
 Reviewer #1 (Remarks to the Author): Expert in prostate cancer The manuscript "Clonal dynamics following p53 loss of heterozygosity in Kras-driven cancers" uses a number of novel genetically engineered
Reviewer #1 (Remarks to the Author): Expert in prostate cancer The manuscript "Clonal dynamics following p53 loss of heterozygosity in Kras-driven cancers" uses a number of novel genetically engineered
AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination
 AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination Supplementary Figure 1. Generation of the adult-onset, liver-specific GH receptor knock-down (alivghrkd, Kd) mouse
AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination Supplementary Figure 1. Generation of the adult-onset, liver-specific GH receptor knock-down (alivghrkd, Kd) mouse
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/4/199/ra75/dc1 Supplementary Materials for Signaling by the Matrix Proteoglycan Decorin Controls Inflammation and Cancer Through PDCD4 and MicroRNA-21 Rosetta
www.sciencesignaling.org/cgi/content/full/4/199/ra75/dc1 Supplementary Materials for Signaling by the Matrix Proteoglycan Decorin Controls Inflammation and Cancer Through PDCD4 and MicroRNA-21 Rosetta
Supplementary fig. 1. Crystals induce necroptosis does not involve caspases, TNF receptor or NLRP3. A. Mouse tubular epithelial cells were pretreated
 Supplementary fig. 1. Crystals induce necroptosis does not involve caspases, TNF receptor or NLRP3. A. Mouse tubular epithelial cells were pretreated with zvad-fmk (10µM) and exposed to calcium oxalate
Supplementary fig. 1. Crystals induce necroptosis does not involve caspases, TNF receptor or NLRP3. A. Mouse tubular epithelial cells were pretreated with zvad-fmk (10µM) and exposed to calcium oxalate
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1. Behavioural effects of ketamine in non-stressed and stressed mice. Naive C57BL/6 adult male mice (n=10/group) were given a single dose of saline vehicle or ketamine (3.0 mg/kg,
Supplementary Figure 1. Behavioural effects of ketamine in non-stressed and stressed mice. Naive C57BL/6 adult male mice (n=10/group) were given a single dose of saline vehicle or ketamine (3.0 mg/kg,
Impact of Sox9 Dosage and Hes1-mediated Notch Signaling in Controlling the Plasticity of Adult Pancreatic Duct Cells in Mice
 Impact of Sox9 Dosage and Hes1-mediated Notch Signaling in Controlling the Plasticity of Adult Pancreatic Duct Cells in Mice Shinichi Hosokawa 1,3,Kenichiro Furuyama 1,3, Masashi Horiguchi 1,3,Yoshiki
Impact of Sox9 Dosage and Hes1-mediated Notch Signaling in Controlling the Plasticity of Adult Pancreatic Duct Cells in Mice Shinichi Hosokawa 1,3,Kenichiro Furuyama 1,3, Masashi Horiguchi 1,3,Yoshiki
Supplementary Materials and Methods
 Supplementary Materials and Methods Whole Mount X-Gal Staining Whole tissues were collected, rinsed with PBS and fixed with 4% PFA. Tissues were then rinsed in rinse buffer (100 mm Sodium Phosphate ph
Supplementary Materials and Methods Whole Mount X-Gal Staining Whole tissues were collected, rinsed with PBS and fixed with 4% PFA. Tissues were then rinsed in rinse buffer (100 mm Sodium Phosphate ph
Supplemental Figure 1. Intracranial transduction of a modified ptomo lentiviral vector in the mouse
 Supplemental figure legends Supplemental Figure 1. Intracranial transduction of a modified ptomo lentiviral vector in the mouse hippocampus targets GFAP-positive but not NeuN-positive cells. (A) Stereotaxic
Supplemental figure legends Supplemental Figure 1. Intracranial transduction of a modified ptomo lentiviral vector in the mouse hippocampus targets GFAP-positive but not NeuN-positive cells. (A) Stereotaxic
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
Supplementary Information Supplementary Fig. 1. Elevated Usp9x in melanoma and NRAS mutant melanoma cells are dependent on NRAS for 3D growth.
 Supplementary Information Supplementary Fig. 1. Elevated Usp9x in melanoma and NRAS mutant melanoma cells are dependent on NRAS for 3D growth. a. Immunoblot for Usp9x protein in NRAS mutant melanoma cells
Supplementary Information Supplementary Fig. 1. Elevated Usp9x in melanoma and NRAS mutant melanoma cells are dependent on NRAS for 3D growth. a. Immunoblot for Usp9x protein in NRAS mutant melanoma cells
SUPPLEMENTARY DATA. Supplementary Table 2. Antibodies used for Immunofluoresence. Supplementary Table 3. Real-time PCR primer sequences.
 Supplementary Table 2. Antibodies used for Immunofluoresence. Antibody Dilution Source Goat anti-pdx1 1:100 R&D Systems Rabbit anti-hnf6 1:100 Santa Cruz Biotechnology Mouse anti-nkx6.1 1:200 Developmental
Supplementary Table 2. Antibodies used for Immunofluoresence. Antibody Dilution Source Goat anti-pdx1 1:100 R&D Systems Rabbit anti-hnf6 1:100 Santa Cruz Biotechnology Mouse anti-nkx6.1 1:200 Developmental
Figure S1. Generation of inducible PTEN deficient mice and the BMMCs (A) B6.129 Pten loxp/loxp mice were mated with B6.
 Figure S1. Generation of inducible PTEN deficient mice and the BMMCs (A) B6.129 Pten loxp/loxp mice were mated with B6.129-Gt(ROSA)26Sor tm1(cre/ert2)tyj /J mice. To induce deletion of the Pten locus,
Figure S1. Generation of inducible PTEN deficient mice and the BMMCs (A) B6.129 Pten loxp/loxp mice were mated with B6.129-Gt(ROSA)26Sor tm1(cre/ert2)tyj /J mice. To induce deletion of the Pten locus,
Supplementary Figure 1. Expression of CUGBP1 in non-parenchymal liver cells treated with TGF-β
 Supplementary Figures Supplementary Figure 1. Expression of CUGBP1 in non-parenchymal liver cells treated with TGF-β and LPS. Non-parenchymal liver cells were isolated and treated with or without TGF-β
Supplementary Figures Supplementary Figure 1. Expression of CUGBP1 in non-parenchymal liver cells treated with TGF-β and LPS. Non-parenchymal liver cells were isolated and treated with or without TGF-β
Supporting Information
 Supporting Information Pang et al. 10.1073/pnas.1322009111 SI Materials and Methods ELISAs. These assays were performed as previously described (1). ELISA plates (MaxiSorp Nunc; Thermo Fisher Scientific)
Supporting Information Pang et al. 10.1073/pnas.1322009111 SI Materials and Methods ELISAs. These assays were performed as previously described (1). ELISA plates (MaxiSorp Nunc; Thermo Fisher Scientific)
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
www.sciencesignaling.org/cgi/content/full/8/407/ra127/dc1 Supplementary Materials for Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism Chao-Yung Wang,* Shian-Sen
SUPPLEMENTARY INFORMATION
 Supplemental Figure 1. Furin is efficiently deleted in CD4 + and CD8 + T cells. a, Western blot for furin and actin proteins in CD4cre-fur f/f and fur f/f Th1 cells. Wild-type and furin-deficient CD4 +
Supplemental Figure 1. Furin is efficiently deleted in CD4 + and CD8 + T cells. a, Western blot for furin and actin proteins in CD4cre-fur f/f and fur f/f Th1 cells. Wild-type and furin-deficient CD4 +
ROMANIAN ACADEMY INSTITUTE OF CELLULAR BIOLOGY AND PATHOLOGY NICOLAE SIMIONESCU. PhD THESIS Summary
 ROMANIAN ACADEMY INSTITUTE OF CELLULAR BIOLOGY AND PATHOLOGY NICOLAE SIMIONESCU PhD THESIS Summary INVOLVEMENT OF ALARMIN HMGB1 IN INFLAMMATORY PROCESSES ASSOCIATED WITH VASCULAR DYSFUNCTION IN HYPERLIPIDEMIA
ROMANIAN ACADEMY INSTITUTE OF CELLULAR BIOLOGY AND PATHOLOGY NICOLAE SIMIONESCU PhD THESIS Summary INVOLVEMENT OF ALARMIN HMGB1 IN INFLAMMATORY PROCESSES ASSOCIATED WITH VASCULAR DYSFUNCTION IN HYPERLIPIDEMIA
Supplementary Figure 1: Lgr5 expression in liver was induced by CCL4 and decreased after liver recovery. Mice were treated with single dose of CCL4
 Supplementary Figure 1: Lgr5 expression in liver was induced by CCL4 and decreased after liver recovery. Mice were treated with single dose of CCL4 or control oil, the livers were collected at various
Supplementary Figure 1: Lgr5 expression in liver was induced by CCL4 and decreased after liver recovery. Mice were treated with single dose of CCL4 or control oil, the livers were collected at various
Title: Human breast cancer associated fibroblasts exhibit subtype specific gene expression profiles
 Author's response to reviews Title: Human breast cancer associated fibroblasts exhibit subtype specific gene expression profiles Authors: Julia Tchou (julia.tchou@uphs.upenn.edu) Andrew V Kossenkov (akossenkov@wistar.org)
Author's response to reviews Title: Human breast cancer associated fibroblasts exhibit subtype specific gene expression profiles Authors: Julia Tchou (julia.tchou@uphs.upenn.edu) Andrew V Kossenkov (akossenkov@wistar.org)
Supplementary Figure 1
 Supplementary Figure 1 a Percent of body weight! (%) 4! 3! 1! Epididymal fat Subcutaneous fat Liver SD Percent of body weight! (%) ** 3! 1! SD Percent of body weight! (%) 6! 4! SD ** b Blood glucose (mg/dl)!
Supplementary Figure 1 a Percent of body weight! (%) 4! 3! 1! Epididymal fat Subcutaneous fat Liver SD Percent of body weight! (%) ** 3! 1! SD Percent of body weight! (%) 6! 4! SD ** b Blood glucose (mg/dl)!
Microarray Analysis and Liver Diseases
 Microarray Analysis and Liver Diseases Snorri S. Thorgeirsson M.D., Ph.D. Laboratory of Experimental Carcinogenesis Center for Cancer Research, NCI, NIH Application of Microarrays to Cancer Research Identifying
Microarray Analysis and Liver Diseases Snorri S. Thorgeirsson M.D., Ph.D. Laboratory of Experimental Carcinogenesis Center for Cancer Research, NCI, NIH Application of Microarrays to Cancer Research Identifying
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH
 EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
EPIGENETIC RE-EXPRESSION OF HIF-2α SUPPRESSES SOFT TISSUE SARCOMA GROWTH Supplementary Figure 1. Supplementary Figure 1. Characterization of KP and KPH2 autochthonous UPS tumors. a) Genotyping of KPH2
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation
 Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Supplementary Figure 1 ITGB1 and ITGA11 increase with evidence for heterodimers following HSC activation. (a) Time course of rat HSC activation indicated by the detection of -SMA and COL1 (log scale).
Title:Role of LPAR3, PKC and EGFR in LPA-induced cell migration in oral squamous carcinoma cells
 Author's response to reviews Title:Role of LPAR3, PKC and EGFR in LPA-induced cell migration in oral squamous carcinoma cells Authors: Ingvild J Brusevold (i.j.brusevold@medisin.uio.no) Ingun H Tveteraas
Author's response to reviews Title:Role of LPAR3, PKC and EGFR in LPA-induced cell migration in oral squamous carcinoma cells Authors: Ingvild J Brusevold (i.j.brusevold@medisin.uio.no) Ingun H Tveteraas
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer. Application Note
 Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
Analysis of small RNAs from Drosophila Schneider cells using the Small RNA assay on the Agilent 2100 bioanalyzer Application Note Odile Sismeiro, Jean-Yves Coppée, Christophe Antoniewski, and Hélène Thomassin
Supplementary data Supplementary Figure 1 Supplementary Figure 2
 Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Supplementary data Supplementary Figure 1 SPHK1 sirna increases RANKL-induced osteoclastogenesis in RAW264.7 cell culture. (A) RAW264.7 cells were transfected with oligocassettes containing SPHK1 sirna
Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 )
 770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5
 sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
sfigure 1 Styx mutant mice recapitulate the phenotype of SHIP -/- mice. (A) Analysis of the genomic sequences of a styx mutant reveals a T to A transversion in the donor splice site of intron 5 (GTAAC
SUPPLEMENTARY INFORMATION
 1. Supplementary Figures and Legends Supplementary Fig. 1. S1P-mediated transcriptional regulation of integrins expressed in OP/monocytoid cells. Real-time quantitative PCR analyses of mrna for two integrins,
1. Supplementary Figures and Legends Supplementary Fig. 1. S1P-mediated transcriptional regulation of integrins expressed in OP/monocytoid cells. Real-time quantitative PCR analyses of mrna for two integrins,
Expression of acid base transporters in the kidney collecting duct in Slc2a7 -/-
 Supplemental Material Results. Expression of acid base transporters in the kidney collecting duct in Slc2a7 -/- and Slc2a7 -/- mice. The expression of AE1 in the kidney was examined in Slc26a7 KO mice.
Supplemental Material Results. Expression of acid base transporters in the kidney collecting duct in Slc2a7 -/- and Slc2a7 -/- mice. The expression of AE1 in the kidney was examined in Slc26a7 KO mice.
PBMC from each patient were suspended in AIM V medium (Invitrogen) with 5% human
 Anti-CD19-CAR transduced T-cell preparation PBMC from each patient were suspended in AIM V medium (Invitrogen) with 5% human AB serum (Gemini) and 300 international units/ml IL-2 (Novartis). T cell proliferation
Anti-CD19-CAR transduced T-cell preparation PBMC from each patient were suspended in AIM V medium (Invitrogen) with 5% human AB serum (Gemini) and 300 international units/ml IL-2 (Novartis). T cell proliferation
were isolated from the freshly drawn blood of healthy donors and ACS patients using the
 Supplemental Figure 1. Quality control of CD4 + T-cell purification. CD4 + T cells were isolated from the freshly drawn blood of healthy donors and ACS patients using the RosetteSep CD4 + T Cell Enrichment
Supplemental Figure 1. Quality control of CD4 + T-cell purification. CD4 + T cells were isolated from the freshly drawn blood of healthy donors and ACS patients using the RosetteSep CD4 + T Cell Enrichment
Supplemental Experimental Procedures
 Cell Stem Cell, Volume 2 Supplemental Data A Temporal Switch from Notch to Wnt Signaling in Muscle Stem Cells Is Necessary for Normal Adult Myogenesis Andrew S. Brack, Irina M. Conboy, Michael J. Conboy,
Cell Stem Cell, Volume 2 Supplemental Data A Temporal Switch from Notch to Wnt Signaling in Muscle Stem Cells Is Necessary for Normal Adult Myogenesis Andrew S. Brack, Irina M. Conboy, Michael J. Conboy,
Canberra, Australia). CD11c-DTR-OVA-GFP (B6.CD11c-OVA), B6.luc + and. Cancer Research Center, Germany). B6 or BALB/c.FoxP3-DTR-GFP mice were
 Supplemental Materials and Methods Mice Female C57BL/6 (B6, I-E null, H-2 b ), BALB/c (H-2 d ) + ), FVB/N (H-2 q, I-E null, CD45.1 + ), and B6D2F1 (H-2 b/d ) mice were purchased from the Animal Resources
Supplemental Materials and Methods Mice Female C57BL/6 (B6, I-E null, H-2 b ), BALB/c (H-2 d ) + ), FVB/N (H-2 q, I-E null, CD45.1 + ), and B6D2F1 (H-2 b/d ) mice were purchased from the Animal Resources
Reviewers' comments: Reviewer #1 (Remarks to the Author):
 Reviewers' comments: Reviewer #1 (Remarks to the Author): In this manuscript, Song et al. identified FBXW7 as a new positive regulator for RIG-Itriggered type I IFN signaling pathway. The authors observed
Reviewers' comments: Reviewer #1 (Remarks to the Author): In this manuscript, Song et al. identified FBXW7 as a new positive regulator for RIG-Itriggered type I IFN signaling pathway. The authors observed
Supporting Information
 Supporting Information Franco et al. 10.1073/pnas.1015557108 SI Materials and Methods Drug Administration. PD352901 was dissolved in 0.5% (wt/vol) hydroxyl-propyl-methylcellulose, 0.2% (vol/vol) Tween
Supporting Information Franco et al. 10.1073/pnas.1015557108 SI Materials and Methods Drug Administration. PD352901 was dissolved in 0.5% (wt/vol) hydroxyl-propyl-methylcellulose, 0.2% (vol/vol) Tween
Supplementary Information
 Supplementary Information GADD34-deficient mice develop obesity, nonalcoholic fatty liver disease, hepatic carcinoma and insulin resistance Naomi Nishio and Ken-ichi Isobe Department of Immunology, Nagoya
Supplementary Information GADD34-deficient mice develop obesity, nonalcoholic fatty liver disease, hepatic carcinoma and insulin resistance Naomi Nishio and Ken-ichi Isobe Department of Immunology, Nagoya
Exosomes/tricalcium phosphate combination scaffolds can enhance bone regeneration by activating the PI3K/Akt signalling pathway
 Exosomes/tricalcium phosphate combination scaffolds can enhance bone regeneration by activating the PI3K/Akt signalling pathway Jieyuan Zhang, Xiaolin Liu, Haiyan Li, Chunyuan Chen, Bin Hu, Xin Niu, Qing
Exosomes/tricalcium phosphate combination scaffolds can enhance bone regeneration by activating the PI3K/Akt signalling pathway Jieyuan Zhang, Xiaolin Liu, Haiyan Li, Chunyuan Chen, Bin Hu, Xin Niu, Qing
Supplementary Materials. for Garmy-Susini, et al, Integrin 4 1 signaling is required for lymphangiogenesis and tumor metastasis
 Supplementary Materials for Garmy-Susini, et al, Integrin 4 1 signaling is required for lymphangiogenesis and tumor metastasis 1 Supplementary Figure Legends Supplementary Figure 1: Integrin expression
Supplementary Materials for Garmy-Susini, et al, Integrin 4 1 signaling is required for lymphangiogenesis and tumor metastasis 1 Supplementary Figure Legends Supplementary Figure 1: Integrin expression
Intracellular MHC class II molecules promote TLR-triggered innate. immune responses by maintaining Btk activation
 Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining Btk activation Xingguang Liu, Zhenzhen Zhan, Dong Li, Li Xu, Feng Ma, Peng Zhang, Hangping Yao and Xuetao
Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining Btk activation Xingguang Liu, Zhenzhen Zhan, Dong Li, Li Xu, Feng Ma, Peng Zhang, Hangping Yao and Xuetao
Cover Page. The handle holds various files of this Leiden University dissertation.
 Cover Page The handle http://hdl.handle.net/1887/23854 holds various files of this Leiden University dissertation. Author: Marel, Sander van der Title: Gene and cell therapy based treatment strategies
Cover Page The handle http://hdl.handle.net/1887/23854 holds various files of this Leiden University dissertation. Author: Marel, Sander van der Title: Gene and cell therapy based treatment strategies
Supplementary Materials
 Supplementary Materials Figure S1. MTT Cell viability assay. To measure the cytotoxic potential of the oxidative treatment, the MTT [3-(4,5-dimethylthiazol- 2-yl)-2,5-diphenyl tetrazolium bromide] assay
Supplementary Materials Figure S1. MTT Cell viability assay. To measure the cytotoxic potential of the oxidative treatment, the MTT [3-(4,5-dimethylthiazol- 2-yl)-2,5-diphenyl tetrazolium bromide] assay
Introduction: 年 Fas signal-mediated apoptosis. PI3K/Akt
 Fas-ligand (CD95-L; Fas-L) Fas (CD95) Fas (apoptosis) 年 了 不 度 Fas Fas-L 力 不 Fas/Fas-L T IL-10Fas/Fas-L 不 年 Fas signal-mediated apoptosis 度降 不 不 力 U-118, HeLa, A549, Huh-7 MCF-7, HepG2. PI3K/Akt FasPI3K/Akt
Fas-ligand (CD95-L; Fas-L) Fas (CD95) Fas (apoptosis) 年 了 不 度 Fas Fas-L 力 不 Fas/Fas-L T IL-10Fas/Fas-L 不 年 Fas signal-mediated apoptosis 度降 不 不 力 U-118, HeLa, A549, Huh-7 MCF-7, HepG2. PI3K/Akt FasPI3K/Akt
Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic
 Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic B cells from WT Nfat2 +/+, TCL1 Nfat2 +/+ and TCL1 Nfat2
Supplementary Figure 1: Expression of NFAT proteins in Nfat2-deleted B cells (a+b) Protein expression of NFAT2 (a) and NFAT1 (b) in isolated splenic B cells from WT Nfat2 +/+, TCL1 Nfat2 +/+ and TCL1 Nfat2
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung
 Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Supplementary Figure 1. A. Bar graph representing the expression levels of the 19 indicated genes in the microarrays analyses comparing human lung immortalized broncho-epithelial cells (AALE cells) expressing
Endocannabinoid-activated Nlrp3 inflammasome in infiltrating macrophages mediates β- cell loss in type 2 diabetes
 Endocannabinoid-activated Nlrp3 inflammasome in infiltrating macrophages mediates β- cell loss in type 2 diabetes T Jourdan, G Godlewski, R Cinar, A Bertola, G Szanda, J Liu, J Tam, T Han, B Mukhopadhyay,
Endocannabinoid-activated Nlrp3 inflammasome in infiltrating macrophages mediates β- cell loss in type 2 diabetes T Jourdan, G Godlewski, R Cinar, A Bertola, G Szanda, J Liu, J Tam, T Han, B Mukhopadhyay,
Supplementary Figure 1. Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Nature Immunology: doi: /ni.
 Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Supplementary Figure 1 Efficiency of Mll4 deletion and its effect on T cell populations in the periphery. Expression of Mll4 floxed alleles (16-19) in naive CD4 + T cells isolated from lymph nodes and
Hepatitis B Antiviral Drug Development Multi-Marker Screening Assay
 Hepatitis B Antiviral Drug Development Multi-Marker Screening Assay Background ImQuest BioSciences has developed and qualified a single-plate method to expedite the screening of antiviral agents against
Hepatitis B Antiviral Drug Development Multi-Marker Screening Assay Background ImQuest BioSciences has developed and qualified a single-plate method to expedite the screening of antiviral agents against
Boucher et al NCOMMS B
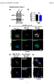 1 Supplementary Figure 1 (linked to Figure 1). mvegfr1 constitutively internalizes in endothelial cells. (a) Immunoblot of mflt1 from undifferentiated mouse embryonic stem (ES) cells with indicated genotypes;
1 Supplementary Figure 1 (linked to Figure 1). mvegfr1 constitutively internalizes in endothelial cells. (a) Immunoblot of mflt1 from undifferentiated mouse embryonic stem (ES) cells with indicated genotypes;
SUPPLEMENTARY INFORMATION. Supplementary Figures S1-S9. Supplementary Methods
 SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
SUPPLEMENTARY INFORMATION SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane Jian Huang 1,2#, Jie Yan 1,2#, Jian Zhang 3#, Shiguo Zhu 1, Yanli Wang
Supplementary Figure 1
 VO (ml kg - min - ) VCO (ml kg - min - ) Respiratory exchange ratio Energy expenditure (cal kg - min - ) Locomotor activity (x count) Body temperature ( C) Relative mrna expression TA Sol EDL PT Heart
VO (ml kg - min - ) VCO (ml kg - min - ) Respiratory exchange ratio Energy expenditure (cal kg - min - ) Locomotor activity (x count) Body temperature ( C) Relative mrna expression TA Sol EDL PT Heart
mm Distance (mm)
 b a Magnet Illumination Coverslips MPs Objective 2575 µm 1875 µm 1575 µm 1075 µm 875 µm 545 µm 20µm 2 3 0.5 0.3mm 1 1000 100 10 1 0.1 1000 100 10 1 0.1 Field Induction (Gauss) 1.5 0 5 10 15 20 Distance
b a Magnet Illumination Coverslips MPs Objective 2575 µm 1875 µm 1575 µm 1075 µm 875 µm 545 µm 20µm 2 3 0.5 0.3mm 1 1000 100 10 1 0.1 1000 100 10 1 0.1 Field Induction (Gauss) 1.5 0 5 10 15 20 Distance
Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3
 Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
Supplemental Figure Legends. Figure S1. ERBB3 mrna levels are elevated in Luminal A breast cancers harboring ERBB3 ErbB3 gene copy number gain. Supplemental Figure S1. ERBB3 mrna levels are elevated in
IL-1 receptor antagonist ameliorates inflammasome-dependent alcoholic steatohepatitis in mice
 IL-1 receptor antagonist ameliorates inflammasome-dependent alcoholic steatohepatitis in mice Jan Petrasek,, Evelyn A. Kurt-Jones, Gyongyi Szabo J Clin Invest. 2012;122(10):3476-3489. https://doi.org/10.1172/jci60777.
IL-1 receptor antagonist ameliorates inflammasome-dependent alcoholic steatohepatitis in mice Jan Petrasek,, Evelyn A. Kurt-Jones, Gyongyi Szabo J Clin Invest. 2012;122(10):3476-3489. https://doi.org/10.1172/jci60777.
RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice
 SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
Supplemental Information. Induction of Expansion and Folding. in Human Cerebral Organoids
 Cell Stem Cell, Volume 20 Supplemental Information Induction of Expansion and Folding in Human Cerebral Organoids Yun Li, Julien Muffat, Attya Omer, Irene Bosch, Madeline A. Lancaster, Mriganka Sur, Lee
Cell Stem Cell, Volume 20 Supplemental Information Induction of Expansion and Folding in Human Cerebral Organoids Yun Li, Julien Muffat, Attya Omer, Irene Bosch, Madeline A. Lancaster, Mriganka Sur, Lee
The Enhancement of Toxin-Induced Liver Fibrosis in Fatty Liver Disease. Ekihiro Seki, M.D.,Ph.D. University of California San Diego
 The Enhancement of Toxin-Induced Liver Fibrosis in Fatty Liver Disease Ekihiro Seki, M.D.,Ph.D. University of California San Diego - A manufactured chemical. - Does not exist naturally. Carbon tetrachloride
The Enhancement of Toxin-Induced Liver Fibrosis in Fatty Liver Disease Ekihiro Seki, M.D.,Ph.D. University of California San Diego - A manufactured chemical. - Does not exist naturally. Carbon tetrachloride
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Reviewers' comments: Reviewer #1 (Remarks to the Author):
 Reviewers' comments: Reviewer #1 (Remarks to the Author): In this manuscript, Hasan et al analyse the transcriptional program used by pathogenic Th17 cells raised in the presence of IL-23 as compared to
Reviewers' comments: Reviewer #1 (Remarks to the Author): In this manuscript, Hasan et al analyse the transcriptional program used by pathogenic Th17 cells raised in the presence of IL-23 as compared to
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
Patnaik SK, et al. MicroRNAs to accurately histotype NSCLC biopsies
 Patnaik SK, et al. MicroRNAs to accurately histotype NSCLC biopsies. 2014. Supplemental Digital Content 1. Appendix 1. External data-sets used for associating microrna expression with lung squamous cell
Patnaik SK, et al. MicroRNAs to accurately histotype NSCLC biopsies. 2014. Supplemental Digital Content 1. Appendix 1. External data-sets used for associating microrna expression with lung squamous cell
Functional characterisation of hepatitis B viral X protein/microrna-21 interaction in HBVassociated hepatocellular carcinoma
 RESEARCH FUND FOR THE CONTROL OF INFECTIOUS DISEASES Functional characterisation of hepatitis B viral X protein/microrna-21 interaction in HBVassociated hepatocellular carcinoma CH Li, SC Chow, DL Yin,
RESEARCH FUND FOR THE CONTROL OF INFECTIOUS DISEASES Functional characterisation of hepatitis B viral X protein/microrna-21 interaction in HBVassociated hepatocellular carcinoma CH Li, SC Chow, DL Yin,
EGFR kinase activity is required for TLR4 signaling and the septic shock response
 Manuscript EMBOR-2015-40337 EGFR kinase activity is required for TLR4 signaling and the septic shock response Saurabh Chattopadhyay, Manoj Veleeparambil, Darshana Poddar, Samar Abdulkhalek, Sudip K. Bandyopadhyay,
Manuscript EMBOR-2015-40337 EGFR kinase activity is required for TLR4 signaling and the septic shock response Saurabh Chattopadhyay, Manoj Veleeparambil, Darshana Poddar, Samar Abdulkhalek, Sudip K. Bandyopadhyay,
Pair-fed % inkt cells 0.5. EtOH 0.0
 MATERIALS AND METHODS Histopathological analysis Liver tissue was collected 9 h post-gavage, and the tissue samples were fixed in 1% formalin and paraffin-embedded following a standard procedure. The embedded
MATERIALS AND METHODS Histopathological analysis Liver tissue was collected 9 h post-gavage, and the tissue samples were fixed in 1% formalin and paraffin-embedded following a standard procedure. The embedded
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus
 Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Supplementary Fig. 1. GPRC5A post-transcriptionally down-regulates EGFR expression. (a) Plot of the changes in steady state mrna levels versus changes in corresponding proteins between wild type and Gprc5a-/-
Metabolic ER stress and inflammation in white adipose tissue (WAT) of mice with dietary obesity.
 Supplementary Figure 1 Metabolic ER stress and inflammation in white adipose tissue (WAT) of mice with dietary obesity. Male C57BL/6J mice were fed a normal chow (NC, 10% fat) or a high-fat diet (HFD,
Supplementary Figure 1 Metabolic ER stress and inflammation in white adipose tissue (WAT) of mice with dietary obesity. Male C57BL/6J mice were fed a normal chow (NC, 10% fat) or a high-fat diet (HFD,
Problem Set 8 Key 1 of 8
 7.06 2003 Problem Set 8 Key 1 of 8 7.06 2003 Problem Set 8 Key 1. As a bright MD/PhD, you are interested in questions about the control of cell number in the body. Recently, you've seen three patients
7.06 2003 Problem Set 8 Key 1 of 8 7.06 2003 Problem Set 8 Key 1. As a bright MD/PhD, you are interested in questions about the control of cell number in the body. Recently, you've seen three patients
Supplemental Information
 Supplemental Information Tobacco-specific Carcinogen Induces DNA Methyltransferases 1 Accumulation through AKT/GSK3β/βTrCP/hnRNP-U in Mice and Lung Cancer patients Ruo-Kai Lin, 1 Yi-Shuan Hsieh, 2 Pinpin
Supplemental Information Tobacco-specific Carcinogen Induces DNA Methyltransferases 1 Accumulation through AKT/GSK3β/βTrCP/hnRNP-U in Mice and Lung Cancer patients Ruo-Kai Lin, 1 Yi-Shuan Hsieh, 2 Pinpin
