let-7-complex MicroRNAs Regulate the Temporal Identity of Drosophila Mushroom Body Neurons via chinmo
|
|
|
- Bryan Ross
- 5 years ago
- Views:
Transcription
1 Short Article let-7-complex MicroRNAs Regulate the Temporal Identity of Drosophila Mushroom Body Neurons via chinmo Yen-Chi Wu, 1 Ching-Huan Chen, 1 Adam Mercer, 1 and Nicholas S. Sokol 1, * 1 Department of Biology, Indiana University, Bloomington, IN 47405, USA *Correspondence: nsokol@indiana.edu SUMMARY Many neural lineages display a temporal pattern, but the mechanisms controlling the ordered production of neuronal subtypes remain unclear. Here, we show that Drosophila let-7 and mir-125, cotranscribed from the let-7-complex (let-7-c) locus, regulate the transcription factor chinmo to control temporal cell fate in the mushroom body (MB) lineage. We find that let-7-c is activated in postmitotic neurons born during the larval-to-pupal transition, when transitions among three MB subtypes occur. Loss or increase of let-7-c delays or accelerates these transitions, respectively, and leads to cell fate transformations. Consistent with our identification of let-7 and mir-125 sites in a recently identified 6 kb extension of the chinmo 3 0 UTR, Chinmo is elevated in let-7-c mutant MBs. In addition, we show that let-7-c acts upstream of chinmo and that let-7-c phenotypes are caused by elevated chinmo. Thus, these heterochronic mirnas, originally identified in C. elegans, underlie progenitor cell multipotency during the development of diverse bilateria. INTRODUCTION Although the ordered production of neuronal subtypes is a pervasive feature of animal nervous system development, the underlying molecular mechanism of this phenomenon is not well understood (Jacob et al., 2008; Pearson and Doe, 2004). In Drosophila, for example, the posttranscriptional regulation of the BTB-zinc finger chronologically inappropriate morphogenesis (chinmo) appears to drive the sequential generation of mushroom body (MB) neurons (Zhu et al., 2006). A center for olfactory learning and memory, the MB is comprised of four subtypes that are born in an invariant order (g / a 0 /b 0 / pioneer a/b / a/b) by multipotent progenitor cells during development (Lee et al., 1999; Zhu et al., 2003, 2006). These progenitors, or neuroblasts (Nbs), undergo self-renewing cell divisions to generate a ganglion mother cell (GMC), which divides once more to produce neurons that will differentiate into one of the MB subtypes. High Chinmo levels in postmitotic neurons specify early-born cell fates (g, a 0 /b 0 ), whereas low levels specify lateborn cell fates (pioneer a/b, a/b). The downregulation of Chinmo in progressively later-born neurons thereby determines the birth order of MB subtypes and occurs at the posttranscriptional level (Zhu et al., 2006). The mechanism responsible for generating this temporal gradient of Chinmo during development remains unknown, although it may involve the chinmo 5 0 untranslated region (UTR) because this region controls Chinmo levels in the adult brain (Zhu et al., 2006). Mechanisms controlling the ordered production of cells during C. elegans development have been identified by forward genetic analysis and involve posttranscriptional regulation by micro- RNAs (mirnas) (reviewed in Ambros, 2011). These mirnas include members of the let-7 and mir-125/lin-4 families, which promote temporal cell fate transitions during C. elegans development by binding to sequences in the 3 0 UTRs of target mrnas and repressing their expression (Lee et al., 1993; Reinhart et al., 2000). Deletion of these mirnas leads to heterochronic phenotypes in which cell fates are inappropriately reiterated during development. The sequences and temporal expression patterns of heterochronic mirnas are broadly conserved among diverse bilateria (Pasquinelli et al., 2000), suggesting a conserved function. However, there is no evidence in any system other than C. elegans that these mirnas promote one stage-specific differentiation program over another. Thus, despite their conservation, it remains unclear whether heterochronic genes represent a fundamental pathway that controls temporal cell fate during animal development. Here, we provide evidence that the switch-like function of let-7 and mir-125/lin-4 in C. elegans has been adapted to the formation of a temporal cell fate gradient of Chinmo in Drosophila. In Drosophila, let-7 and mir-125 are cotranscribed from a single locus, known as the let-7-complex (let-7-c), along with a third ancient mirna, mir-100 (Sokol et al., 2008). These let-7-c mirnas are not detected early in development, but their upregulation during the larval-to-pupal transition coincides with the downregulation of Chinmo and is partially dependent on transcriptional activation by the Ecdysone Receptor (Chawla and Sokol, 2012). let-7-c mutants appear morphologically normal but display behavioral defects, indicating a role for these mirnas during nervous system development (Caygill and Johnston, 2008; Sokol et al., 2008). The spatiotemporal expression profile of let-7-c mirnas along with their known function in C. elegans suggest that these mirnas may play a role in the temporal identity of MB neurons. 202 Developmental Cell 23, , July 17, 2012 ª2012 Elsevier Inc.
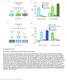
2 Figure 1. Temporal Onset of let-7-c in MB Neurons (A) let-7-c GKI crossed to UAS-mCD8::GFP (green) is detected in the MBs (indicated by dotted box) of adult brains counterstained with neuropil marker nc82 (magenta). (B) An increasing gradient of let-7 mirna is expressed in CNS tissue at the larval-to-pupal transition, along with mir-100 and mir-125 (see Figure S1). Total RNA was extracted from CNS tissues dissected from staged animals, and probed for U6 snrna as loading control. (C) let-7-cp 12.5kb ::lacz (white in top panels, green in bottom panels) is not detected in wandering L3 MBs stained for Dac (magenta, bottom panels) but is detected in increasing numbers of MB neurons in white prepupae (0), 12 hr pupae (12), and 24 hr pupae (24). Asterisks mark the location of Nbs and GMCs. (D) let-7-cp 12.5kb ::lacz (white in top panel, red in bottom panel) is detected in a white prepupae MB clone expressing GFP (green, outlined) and Dac (blue) that was induced 18 hr earlier. Scale bars, 25 mm (A) and 10 mm (C and D). See also Figure S1. RESULTS let-7-c Is Expressed in MB Neurons Born during the L3-to-Pupal Transition Expression of a let-7-c::gal4 transcriptional reporter was detected in the adult MBs (Figure 1A), suggesting a potential role for let-7-c mirnas during MB development. To correlate let-7-c expression with the sequential production of g, a 0 /b 0, pioneer a/b (p. a/b), and a/b neurons, we analyzed the temporal onset of let-7-c mirnas during central nervous system (CNS) development. Levels of let-7 (Figure 1B) as well as mir-100 and mir-125 (see Figures S1A and S1B available online) gradually increased in total CNS tissue during the larval-to-pupal transition, the time when a 0 /b 0 / p. a/b and p. a/b / a/b transitions occur (Zhu et al., 2003, 2006). To determine the timing of let-7-c onset specifically in MB neurons, we analyzed two additional transcriptional reporters, let-7-cp 12.5kb ::lacz and let-7-c D3miR ::optgal4 (Chawla and Sokol, 2012; Figure S1C), within the CNS. These were used instead of let-7-c::gal4, which displayed an 21 hr developmental lag in CNS reporter expression relative to let-7-c mirna northern blots (Y.-C.W. and N.S.S., unpublished data). let-7-cp 12.5kb ::lacz was detected in increasing numbers of MB neurons in 0, 12, and 24 hr pupae (Figure 1C), correlating with the temporal gradient of let-7-c mirnas detected by northern blot (Figures 1B, S1A, and S1B). In addition, let-7-cp 12.5kb ::lacz expression colocalized with the MB marker Dachshund (Dac), which labels postmitotic MB neurons that are at least 8 10 hr old (Martini et al., 2000), but was conspicuously absent from regions containing MB Nbs and GMCs (Figure 1C, asterisks). Although let-7-cp 12.5kb ::lacz was not consistently detected in MB neurons from wandering third-instar larvae (L3), it was expressed in almost all Dac-positive MB neurons in 24 hr pupae (Figure 1C). Expression of the let-7-c D3miR ::optgal4 reporter was similar though slightly delayed relative to let-7-cp 12.5kb ::lacz (Figures S1D S1F), presumably reflecting a Gal4/UAS system lag. To identify the age of the neurons that first expressed let-7-cp 12.5kb ::lacz in white prepupae, we used the mosaic analysis with a repressible cell marker (MARCM) technique (Lee and Luo, 1999) to analyze let-7-cp 12.5kb ::lacz expression in MB neurons born during the late L3 stage. let-7-cp 12.5kb ::lacz was detected in clones of recently born neurons in 0 hr white prepupae (Figure 1D). Taken together, these data indicated that let-7-c is activated in a few MB neurons born in late larvae, and its expression expands during early metamorphosis to include almost all postmitotic MB neurons. Developmental Cell 23, , July 17, 2012 ª2012 Elsevier Inc. 203
3 Figure 2. MB Temporal Identity Is Sensitive to let-7-c Dosage (A) Percentages of different subtypes of MB neurons (y axis) among single- and two-cell clones that were induced in wild-type and let-7-c mutant animals at different times relative to puparium formation (x axis). (B) Percentages of different subtypes of MB neurons (y axis) among single- and two-cell clones that were induced in wild-type and 4Xlet-7-C larvae at different times relative to puparium formation (x axis). (C) Average number of c305a-gal4-positive a 0 /b 0 neurons in wild-type (2X) or let-7-c KO1 and let-7-c KO2 transheterozgyotes (0X). *p = (D) Average number of c305a-gal4-positive a 0 /b 0 neurons in wild-type (2X) and 4Xlet-7-C (4X) adults. ****p < (E) Average number of c708a-gal4-positive p. a/b neurons in wild-type (2X), let-7-c KO1 and let-7-c KO2 transheterozgyotes (0X), let-7-c KO2 heterozygotes harboring a let-7-c rescuing transgene (2X-R), and wild-type harboring a let-7-c rescuing transgene (3X). ****p < comparing either 2X and 0X or 2X and 3X. (F and G) OK107-Gal4, UAS-mCD8::GFP-labeled wild-type (F) and let-7-c KO2 (G) adult MB clones generated in newly hatched larvae and stained with anti-fasii antibodies. Although the a, a 0, b 0, and g lobes appear normal in the let-7-c mutant clone (compare arrowheads in G to F), b neurons display a midline crossing defect (arrow in G). Scale bar in (F) is 50 mm and refers to (G). Numbers in parentheses represent the total number of clones (A and B) or brains (C E) analyzed for each genotype. Values are presented as mean ± SEM. See also Figure S2. let-7-c Has Dosage-Dependent Effects on MB Transitions The spatial and temporal onset of let-7-cp 12.5kb ::lacz suggested that let-7-c mirnas may promote the a 0 /b 0 / p. a/b and p. a/b / a/b transitions in MB neuron identities. To test this, we used the MARCM technique to label individual neurons born in staged wild-type and let-7-c mutant animals (Figures 2A and S2A S2D). Consistent with previous reports 204 Developmental Cell 23, , July 17, 2012 ª2012 Elsevier Inc.
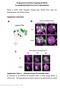
4 (Zhu et al., 2003, 2006), our fine-scale temporal mapping indicated that the a 0 /b 0 / p. a/b and p. a/b / a/b transitions began 18 to 8 hr and 4 to 0 hr before puparium formation in wild-type animals, respectively (Figures 2A and S2E). In contrast, let-7-c mutants displayed delays in both of these transitions. For example, 100% of wild-type neurons born during the 6 hr period after puparium formation displayed a/b characteristics, whereas 29% of let-7-c mutants born during this same period displayed a 0 /b 0 characteristics, and another 59% displayed p. a/b characteristics (Figure 2A). Because these results suggested that removal of let-7-c mirnas resulted in delayed transitions, we reasoned that an increase in let-7-c dosage might accelerate these transitions. To test this hypothesis, MARCM analysis was repeated in animals that carried two extra copies of the let-7-c locus and contained elevated levels of let-7 (Figure S2J). These 4Xlet-7-C animals displayed accelerated transitions: only 5% of neurons born 4 hr before puparium formation of wild-type larvae displayed late-type a/b characteristics, whereas 38% of neurons born during the same time frame did in 4Xlet-7-C larvae (Figure 2B). These data indicate that let-7-c has dosage-sensitive effects on the temporal transitions between MB identities. Such changes in the transitions between MB subtypes during neurogenesis might be reflected as alterations in the ratios of a 0 /b 0,p.a/b, and a/b neurons in the adult MBs. To test this, we used previously described MB subtype-specific Gal4 reporters (Aso et al., 2009; Lin et al., 2007; Zhu et al., 2006) to quantify the number of each class of MB neuron in animals with varying doses of let-7-c. Consistent with the MARCM analysis, we found that the number of earlier-born a 0 /b 0 neurons as detected by the c305a-gal4 driver was increased in let-7-c adults (393.8 ± 9.3, n = 6) relative to wild-type adults (363.4 ± 10.9, n = 5) (Figure 2C) and decreased in 4Xlet-7-C adults (303 ± 15.3, n = 9) relative to wild-type adults (354 ± 5.5, n = 8) (Figure 2D). Conversely, the number of later-born p. a/b neurons as detected by the c708a- Gal4 driver was decreased in let-7-c adults (23.9 ± 7.4, n = 31) and increased in 3Xlet-7-C adults (74.6 ± 8.4, n = 24) relative to wild-type adults (57.5 ± 3.3, n = 16) (Figure 2E). No evidence for changes in the size of either the g or a/b populations was identified using the H24- and c739-gal4 reporters, respectively (Figures S2G and S2H). Importantly, the total number of MB neurons generated by a single MB Nb was also not affected by removal of let-7-c (Figure S2I). Despite these penetrant temporal identity defects, the morphology of let-7-c MBs was grossly normal, although we noted some p. a/b and a/b axons that inappropriately crossed the midline (Figures 2G and S2N S2Q). Taken together, these data indicated that manipulation of let- 7-C dosage leads to cell fate transformations. In contrast to the relatively mild morphological defects associated with let-7-c loss, forced expression of let-7-c beginning in L1 MB lineages resulted in dramatic effects on adult MB morphology (Figures 3A and 3C). These phenotypes included the apparent elimination of g as well as a 0 /b 0 lobes, indicating that let-7-c mirnas promote later-born a/b cell fates at the expense of earlier-born g and a 0 /b 0 cell fates. This transformation appeared identical to the phenotypes resulting from loss of the temporal identity factor chinmo (compare Figures 3C and 3E; Zhu et al., 2006). To test whether Chinmo expression levels were affected by precocious expression of let-7-c, larval MB clones ectopically expressing let-7-c were stained for Chinmo protein. Whereas abundant Chinmo was detected in wild-type MB neurons at this stage (Figure 3B), Chinmo levels were depleted in MB neurons either expressing a UAS-let-7-C transgene (Figure 3D) or homozygous for the protein null chinmo 1 allele (Figure 3F). These data indicated that the let-7-c temporal identity phenotypes are likely due to changes in Chinmo levels, and also suggested that chinmo might be a direct target of let- 7-C mirnas. Prior analysis found that posttranscriptional control of chinmo was mediated via its 5 0 UTR, but not its 2.5 kb 3 0 UTR (Zhu et al., 2006), which is where mirna binding sites are usually found. These results are consistent with the absence of predicted let-7-c mirna binding sites in the annotated chinmo 3 0 UTR (Brennecke et al., 2005; Grün et al., 2005; Lewis et al., 2005). Recent microarray and deep sequencing analyses, however, indicate that the full-length chinmo 3 0 UTR is 8.5 kb, considerably longer than previous gene annotations suggested (Cherbas et al., 2011; Graveley et al., 2011; Smibert et al., 2012) (Figure S3A). Thus, let-7-c mirnas might regulate Chinmo expression via binding sites in the 6 kb portion of the chinmo 3 0 UTR that was previously unannotated. Chinmo Is Regulated by let-7 and mir-125 via Its 3 0 UTR To investigate whether chinmo was directly regulated by let-7-c mirnas, we used a cell culture assay to test whether ectopic let-7-c mirnas repressed expression of a luciferase reporter containing the full-length 8.5 kb chinmo 3 0 UTR (Figure 3G). Ectopic expression of all three mirnas resulted in an 8.8-fold repression of luciferase. This was predominantly due to let-7 and mir-125: ectopic let-7 alone repressed luciferase 5.5-fold, and ectopic mir-125 alone repressed luciferase 2.5-fold, whereas ectopic mir-100 alone repressed luciferase only 1.1- fold. Control assays indicated that all three mirnas were expressed in cell culture and could repress control reporters (Figures S3B S3G). Consistent with this cell culture experiment, we found that precocious expression of let-7 and mir-125 but not mir-100 in L1 resulted in reduced Chinmo levels in L3 MBs and morphological defects in adult MBs (Figure S4). We therefore searched the 8.5 kb chinmo 3 0 UTR for putative let-7 and mir-125 binding sites using the RNAhybrid program (Rehmsmeier et al., 2004) and identified six let-7 sites and four mir-125 binding sites that are conserved between Drosophila species and have a free energy of at most 18 kcal/mol (Figure S3A). Because most of these sites clustered in a 1.4 kb region of the chinmo 3 0 UTR, we prepared two new luciferase reporters containing either a wild-type or mutated version of this fragment in which seed sequences of the let-7 and mir- 125 sites were deleted (Figure S3A). Ectopic expression of all three let-7-c mirnas together or either let-7 alone or mir-125 alone repressed a luciferase reporter containing the wild-type fragment 8.7-, 8.4-, and 3.4-fold, respectively (Figure 3G). In contrast, repression of the mutated version was significantly reduced, indicating that let-7 and mir-125 binding sites mediated reporter repression. To determine whether the chinmo 3 0 UTR was regulated by endogenous let-7-c mirnas, we analyzed the effect let-7-c dosage had on the expression of a reporter transgene. In this assay, flies containing the eye pigmentation gene white fused to its own 3 0 UTR displayed dark eye color with an absorbance Developmental Cell 23, , July 17, 2012 ª2012 Elsevier Inc. 205
5 Figure 3. let-7-c mirnas Regulate Chinmo Expression (A, C, and E) OK107-Gal4, UAS-mCD8::GFP-labeled wild-type (A), UAS-let-7-C (C), and chinmo 1 (E) adult MB clones generated in newly hatched larvae and stained with anti-fasii antibodies. In wild-type, g lobes are weakly stained with FasII, whereas a 0 and b 0 are devoid of FasII staining. Note the absence of g, a 0, and b 0 lobes in UAS-let-7-C and chinmo 1 MBs (asterisks). Arrowheads indicate a, a 0, b, b 0, and g lobes in the wild-type clone (A). Both the UAS-let-7-C and chinmo 1 clones are apparently lacking the a 0 /b 0 and g lobes (arrowheads with asterisks in C and E, respectively). Scale bar in (A) is 50 mm and refers to (C) and (E). 206 Developmental Cell 23, , July 17, 2012 ª2012 Elsevier Inc.
6 Figure 4. Chinmo Suppresses let-7-c MB Phenotypes and Is Epistatic to let-7-c (A) Average number of c708a Gal4-positive p. a/b neurons in adults that are wild-type (+) or heterozygous ( ) for chinmo 1 (columns 3, 5, 6), let-7- C KO1 (columns 2, 3), or let-7-c KO2 (columns 4, 5). ****p < comparing either column 2 and 3 or column 4 and 5. Values are presented as mean ± SEM. (B) Percentages of different subtypes among single- and two-cell wild-type, chinmo 1, or chinmo 1, let-7-c KO2 mutant clones that were induced at different times relative to puparium formation (x axis). (C) OK107-Gal4, UAS-mCD8::GFP-labeled chinmo 1, let-7-c KO2 double-mutant adult MB clone generated in newly hatched larvae and stained with anti-fasii antibodies. Note the absence of g, a 0, and b 0 lobes (asterisks). Scale bar, 50 mm. (D) let-7 and mir-125 are upregulated at the larval-to-pupal transition and promote transitions in MB temporal identity by repressing chinmo. Numbers in parentheses represent the total number of brains (A) or clones (B) analyzed for each genotype. of absorbance units (Figures 3H and S3H). In contrast, flies carrying the white open reading frame fused to either the chinmo 3 0 UTR or the known let-7 regulated abrupt 3 0 UTR displayed a significantly lighter eye color (Figures 3H, S3J, and S3L). Flies whose eyes were comprised exclusively of let-7-c KO1 mutant cells and that carried the same reporter transgenes displayed darker eye color (Figures 3G, S3K, and S3M). Therefore, endogenous let-7-c mirnas are required for chinmo 3 0 UTR-mediated regulation of white reporter expression. Having established that endogenous let-7-c mirnas can regulate protein expression through the chinmo 3 0 UTR, we looked for aberrant Chinmo protein expression in let-7-c mutant tissue. Chinmo is ordinarily downregulated during metamorphosis and is virtually absent in adults (Figure 3I; Zhu et al., 2006). In contrast, Chinmo is abundantly expressed in adult let-7-c brains, including throughout the MB (Figure 3J). Elevated levels of Chinmo are first detected in 48 hr let-7-c pupae (data not shown). Collectively, these data indicate that chinmo is a direct target of let-7-c mirnas. let-7-c MB Phenotypes Are Due to Elevated Chinmo Given that the downregulation of Chinmo is required for the ordered production of MB neurons, we reasoned that let-7-c mutant MB phenotypes were due to elevated Chinmo. Consistently, phenotypes resulting from either reduced let-7-c or increased chinmo dosage were similar: adults with one copy of let-7-c or three copies of chinmo had a decrease in p. a/b number from 58 to between 36 and 45 (Figure 4A; Zhu et al., 2006). We therefore tested whether reduction in chinmo dosage could suppress the heterozygous let-7-c MB phenotypes, using p. a/b number as our assay to analyze genetic interaction. Removal of one copy of chinmo in flies heterozygous for two different let-7-c null alleles restored p. a/b number to wild-type (Figure 4A). We also tested whether chinmo was epistatic to let-7-c by analyzing chinmo 1 ; let-7-c KO double-mutant phenotypes. MARCM of single-/two-cell clones in staged animals indicated that chinmo 1 ; let-7-c KO double-mutant neurons aberrantly acquired late-type neurite trajectories (Figure 4B), a phenotype identical to that displayed by chinmo 1 single-mutant neurons (Figure 4B; Zhu et al., 2006) and opposite to that displayed by let-7-c single-mutant neurons (compare Figure 4B with Figure 2A). Similarly, MBs generated from chinmo 1 ; let-7- C KO double-mutant Nbs appeared identical to MBs generated from chinmo 1 single-mutant Nbs (Figure 4C). This analysis indicated that chinmo was epistatic to let-7-c and that the let-7-c MB temporal identity phenotypes were a consequence of chinmo misregulation. (B, D, and F) OK107-Gal4-labeled wild-type (B), UAS-let-7-C (D), and chinmo 1 mutant (F) L3 MB clones labeled with GFP generated in newly hatched larvae and stained with anti-chinmo antibodies. Clones are outlined in yellow in the Chinmo channel. Scale bar in (B) is 7.5 mm and refers to (D) and (F). (G) Fold repression of luciferase reporters containing the 8.5 kb full-length chinmo 3 0 UTR, a 1.4 kb chinmo 3 0 UTR fragment, a mutated 1.4 kb 3 0 UTR fragment, or the white 3 0 UTR in cell cultures in which let-7-c mirnas were ectopically expressed either together or individually. *p = ,***p = , and **p = comparing repression of chinmo 1.4kb versus chinmo 1.4kbmut by let-7-c mirnas, let-7 alone, or mir-125 alone, respectively. (H) Quantification of pigment levels from flies harboring white reporter transgenes containing the white (w), chinmo (chin), or abrupt (ab) 3 0 UTR and containing either wild-type (+) or let-7-c KO1 mutant ( ) eye cells. **p = or **p = comparing chinmo or abrupt 3 0 UTR in wild-type and let-7-c KO1 cells. Values are presented as mean ± SEM. (I and J) Wild-type (I) and let-7-c (J) adult brains harboring OK107-Gal4 and UAS-mCD8::ChRFP insertions and stained with anti-chinmo antibodies. MBs are outlined. Scale bars, 25 mm. See also Figures S3 and S4. Developmental Cell 23, , July 17, 2012 ª2012 Elsevier Inc. 207
7 DISCUSSION Drosophila let-7-c is activated throughout the developing nervous system at the larval-to-pupal transition, where it triggers cell fate transitions in MB cell lineages and likely other lineages as well. Here, we provide evidence that let-7-c mirnas participate in the formation of a temporal gradient of Chinmo in postmitotic MB neurons via sequences in a recently annotated extension of the chinmo 3 0 UTR. Posttranscriptional regulation of chinmo is complex and involves both its 5 0 as well as its 3 0 UTRs. The 5 0 UTR regulates Chinmo expression in adult MB cells (Zhu et al., 2006), although the identity of the responsible trans-acting factors and their relationship with mirna-mediated silencing pathways remain to be identified. Chinmo expression is also posttranscriptionally regulated in MB Nbs (Zhu et al., 2006) and may involve either the 5 0 and/or 3 0 UTR, although likely not the let-7-c mirnas because they appear to be expressed exclusively in postmitotic cells. The work presented here can be reconciled with a previous observation that the general disruption of mirna processing does not affect MB temporal cell fate specification (Yu and Lee, 2007) if let-7-c mirnas are either counteracted by additional MB mirnas or are processed by noncanonical mechanisms in the MBs (Yang and Lai, 2011). Chinmo is closely related to another known let-7 target, Abrupt (Caygill and Johnston, 2008). Like chinmo, abrupt appears to repress morphologies associated with adult neuronal cell fates (Li et al., 2004; Sugimura et al., 2004), suggesting that this family of BTB-zinc fingers may generally function as temporal cell fate determinants during nervous system formation. Whether let-7-c mirnas regulate abrupt and/or chinmo to control temporal identity in other adult lineages in the developing fly nervous system remains to be determined. Our finding that let-7-c mirnas promote one neural differentiation program over another suggests that these conserved mirnas may underlie the multipotency of vertebrate neural stem cells as well. Available evidence indicates that these mirnas display temporal patterns of onset during nervous system formation in a variety of vertebrates. During zebrafish development, for example, let-7 is first detected 72 hr after fertilization (Pasquinelli et al., 2000) and is found specifically in the spinal cord and brain, along with mir-100 and mir-125 (Wienholds et al., 2005). In addition, mir-125 and let-7 are enriched in human and mouse brain samples (Sempere et al., 2004) and are also sharply upregulated in pluripotent P19 mouse cells as they differentiate into neurons (Wulczyn et al., 2007). The ordered production of neurons with distinct morphologies may therefore generally involve let-7-c mirnas, and their postdevelopmental modulation may regulate the neuronal remodeling that contributes to adult function as well. P(w ORF ::w 30UTR ) (this study), P(w ORF ::ab 30UTR ) (this study), P(w ORF ::chin 30UTR ) (this study), UAS-GFP::nls (BL4775 and 4776), UAS-mCD8::GFP (BL5130), and UAS-mCD8::ChRFP (BL27391 and 27392). Strains for MARCM analysis included (1) UAS-mCD8::GFP, hsflp; FRT 3L2A ; OK107; (2) FRT 3L2A, tubp-gal80; (3)UAS-mCD8::GFP, hsflp; let-7-c KO1 /CyO ; FRT 3L2A ; OK107; (4) let-7-c KO2 ; FRT 3L2A, tubp-gal80/cyo-tm6b; (5) UAS-mCD8::GFP, hsflp ; FRT 40A, tubgal80 ; OK107; (6) FRT 40A ; (7) P(W8, let-7-c); FRT 40A ; P(W8, let-7-c); (8) FRT 40A ; UAS-let-7-C; (9) FRT 40A, chinmo 1, UASmCD8::GFP/CyO; (10) UAS-mCD8::GFP, hsflp; chinmo 1, let-7-c KO2, FRT 40A /CyO; OK107; and (11) let-7-c KO1, FRT 40 tubp-gal80/cyo. New transgenics were generated by Rainbow Transgenic Services (Camarillo, CA, USA). Stocks with multiple genetic elements were obtained by meiotic recombination and/or genetic crosses. The let-7-c KO2 allele was generated in parallel with the let-7-c GKI allele (Sokol et al., 2008) and is identical except that the endogenous let-7-c locus was replaced with white rather than white and gal4. Immunostaining and Microscopy Antibodies used for immunostaining included rat-anti-chinmo (1:500; this study), mouse anti-fasii and anti-dac (1:5 and 1:100; Developmental Studies Hybridoma Bank), rabbit anti-dsred (1:5,000; Clontech), rabbit anti-gfp (1:1,000; Invitrogen), rabbit anti-b-gal (1:500; MP Biomedicals), and chicken anti-b-gal (1:500; Abcam). Immunostaining was performed as previously described (Chawla and Sokol, 2012). Antibodies were generated in rats (Cocalico Biologicals) against a His-tagged version of full-length Chinmo-PA protein that was expressed and purified according to standard methods (Cultivation and Bioprocessing Facility, Indiana University). Images were collected on a Leica SP5 (Light Microscopy Imaging Center, Indiana University) or a TCS confocal microscope. Confocal stacks were merged using Leica LSM software or Adobe Photoshop. Samples whose staining was directly compared were prepared and imaged in parallel and under identical conditions. For Nb clone, H24, c305a, and c739 neuron counts, MBs were optically sectioned in 0.5 mm increments, and the total number of neurons was determined by manually counting the number of b-gal- or GFP-positive cells section by section, ensuring that cells present on consecutive sections were counted only once. Statistical analysis was performed and histograms generated using GraphPad Prism software. p Values were calculated using a two-tailed paired t test. Values are presented as mean ± SEM. Cell Culture Procedures Drosophila KC-167 cells were cultured in CCM3 at 23C. Cells were transfected in 48-well plates with 25 ng of tub-gal4 plasmid DNA, 100 ng UAS-miRNA plasmid DNA, and 25 ng of 3 0 UTR-containing psicheck2 (Promega) plasmid DNA using Effectene (QIAGEN). Luciferase assays were performed using the Dual-Luciferase reporter system (Promega). Transfections were performed in quadruplicate, and resulting luciferase levels were averaged. Fold repression was calculated relative to control samples transfected with UAS-let-7- C D3miR. instead of mirna-encoding plasmids. Additional molecular biology techniques as well as schemes used for plasmid construction are described in detail in Supplemental Experimental Procedures. SUPPLEMENTAL INFORMATION Supplemental Information includes four figures and Supplemental Experimental Procedures and can be found with this article online at org/ /j.devcel EXPERIMENTAL PROCEDURES Drosophila Strains Mutants included let-7-c GKI and let-7-c KO1 (Sokol et al., 2008), let-7-c KO2 (this study), and chinmo 1 (Zhu et al., 2006). Transgenic insertions included let-7-cp 12.5kb ::lacz (Chawla and Sokol, 2012), let-7-c D3miR ::optgal4 (this study), P(W8, let-7-c) (Sokol et al., 2008), c708a-gal4 (Zhu et al., 2006), c305a-gal4 (Lin et al., 2007), H24-Gal4 (Lin et al., 2007), c739-gal4 (Lin et al., 2007), OK107-Gal4 (BL854), UAS-let-7-C (Sokol et al., 2008), ACKNOWLEDGMENTS We thank Erika Bach, Ann-Shyn Chiang, Pam Geyer, Yuh-Nung Jan, Tzumin Lee, Liz Perkins, Barret Pfeiffer, Hiromu Tanimoto, Peizhang Xu, the Bloomington Drosophila Stock Center, and the Developmental Studies Hybridoma Bank for reagents; Peter and Lucy Cherbas, Thomas Kaufman, and the Light Microscopy Center for access to equipment; Victor Ambros, Chris Hammell, and Arthur Luhur for comments on the manuscript; and the National Institute of Mental Health (award R01MH087511) for support. 208 Developmental Cell 23, , July 17, 2012 ª2012 Elsevier Inc.
8 Received: April 9, 2012 Revised: May 15, 2012 Accepted: May 19, 2012 Published online: July 16, 2012 REFERENCES Ambros, V. (2011). MicroRNAs and developmental timing. Curr. Opin. Genet. Dev. 21, Aso, Y., Grübel, K., Busch, S., Friedrich, A.B., Siwanowicz, I., and Tanimoto, H. (2009). The mushroom body of adult Drosophila characterized by GAL4 drivers. J. Neurogenet. 23, Brennecke, J., Stark, A., Russell, R.B., and Cohen, S.M. (2005). Principles of microrna-target recognition. PLoS Biol. 3, e85. Caygill, E.E., and Johnston, L.A. (2008). Temporal regulation of metamorphic processes in Drosophila by the let-7 and mir-125 heterochronic micrornas. Curr. Biol. 18, Chawla, G., and Sokol, N.S. (2012). Hormonal activation of let-7-c micrornas via EcR is required for adult Drosophila melanogaster morphology and function. Development 139, Cherbas, L., Willingham, A., Zhang, D., Yang, L., Zou, Y., Eads, B.D., Carlson, J.W., Landolin, J.M., Kapranov, P., Dumais, J., et al. (2011). The transcriptional diversity of 25 Drosophila cell lines. Genome Res. 21, Graveley, B.R., Brooks, A.N., Carlson, J.W., Duff, M.O., Landolin, J.M., Yang, L., Artieri, C.G., van Baren, M.J., Boley, N., Booth, B.W., et al. (2011). The developmental transcriptome of Drosophila melanogaster. Nature 471, Grün, D., Wang, Y.L., Langenberger, D., Gunsalus, K.C., and Rajewsky, N. (2005). microrna target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 1, e13. Jacob, J., Maurange, C., and Gould, A.P. (2008). Temporal control of neuronal diversity: common regulatory principles in insects and vertebrates? Development 135, Lee, R.C., Feinbaum, R.L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, Lee, T., and Luo, L. (1999). Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, Lee, T., Lee, A., and Luo, L. (1999). Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development 126, Lewis, B.P., Burge, C.B., and Bartel, D.P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microrna targets. Cell 120, Li, W., Wang, F., Menut, L., and Gao, F.B. (2004). BTB/POZ-zinc finger protein abrupt suppresses dendritic branching in a neuronal subtype-specific and dosage-dependent manner. Neuron 43, Lin, H.H., Lai, J.S., Chin, A.L., Chen, Y.C., and Chiang, A.S. (2007). A map of olfactory representation in the Drosophila mushroom body. Cell 128, Martini, S.R., Roman, G., Meuser, S., Mardon, G., and Davis, R.L. (2000). The retinal determination gene, dachshund, is required for mushroom body cell differentiation. Development 127, Pasquinelli, A.E., Reinhart, B.J., Slack, F., Martindale, M.Q., Kuroda, M.I., Maller, B., Hayward, D.C., Ball, E.E., Degnan, B., Müller, P., et al. (2000). Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408, Pearson, B.J., and Doe, C.Q. (2004). Specification of temporal identity in the developing nervous system. Annu. Rev. Cell Dev. Biol. 20, Rehmsmeier, M., Steffen, P., Hochsmann, M., and Giegerich, R. (2004). Fast and effective prediction of microrna/target duplexes. RNA 10, Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, Sempere, L.F., Freemantle, S., Pitha-Rowe, I., Moss, E., Dmitrovsky, E., and Ambros, V. (2004). Expression profiling of mammalian micrornas uncovers a subset of brain-expressed micrornas with possible roles in murine and human neuronal differentiation. Genome Biol. 5, R13. Sokol, N.S., Xu, P., Jan, Y.N., and Ambros, V. (2008). Drosophila let-7 microrna is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 22, Smibert, P., Miura, P., Westholm, J.O., Shenker, S., May, G., Duff, M.O., Zhang, D., Eads, B.D., Carlsom, J., Brown, J.B., et al. (2012). Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Reports 1, Sugimura, K., Satoh, D., Estes, P., Crews, S., and Uemura, T. (2004). Development of morphological diversity of dendrites in Drosophila by the BTB-zinc finger protein abrupt. Neuron 43, Wienholds, E., Kloosterman, W.P., Miska, E., Alvarez-Saavedra, E., Berezikov, E., de Bruijn, E., Horvitz, H.R., Kauppinen, S., and Plasterk, R.H. (2005). MicroRNA expression in zebrafish embryonic development. Science 309, Wulczyn, F.G., Smirnova, L., Rybak, A., Brandt, C., Kwidzinski, E., Ninnemann, O., Strehle, M., Seiler, A., Schumacher, S., and Nitsch, R. (2007). Post-transcriptional regulation of the let-7 microrna during neural cell specification. FASEB J. 21, Yang, J.S., and Lai, E.C. (2011). Alternative mirna biogenesis pathways and the interpretation of core mirna pathway mutants. Mol. Cell 43, Yu, H.H., and Lee, T. (2007). Neuronal temporal identity in post-embryonic Drosophila brain. Trends Neurosci. 30, Zhu, S., Chiang, A.S., and Lee, T. (2003). Development of the Drosophila mushroom bodies: elaboration, remodeling and spatial organization of dendrites in the calyx. Development 130, Zhu, S., Lin, S., Kao, C.F., Awasaki, T., Chiang, A.S., and Lee, T. (2006). Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell 127, Developmental Cell 23, , July 17, 2012 ª2012 Elsevier Inc. 209
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
www.sciencemag.org/cgi/content/full/1171320/dc1 Supporting Online Material for A Frazzled/DCC-Dependent Transcriptional Switch Regulates Midline Axon Guidance Long Yang, David S. Garbe, Greg J. Bashaw*
Supplementary Figure 1 Madm is not required in GSCs and hub cells. (a,b) Act-Gal4-UAS-GFP (a), Act-Gal4-UAS- GFP.nls (b,c) is ubiquitously expressed
 Supplementary Figure 1 Madm is not required in GSCs and hub cells. (a,b) Act-Gal4-UAS-GFP (a), Act-Gal4-UAS- GFP.nls (b,c) is ubiquitously expressed in the testes. The testes were immunostained with GFP
Supplementary Figure 1 Madm is not required in GSCs and hub cells. (a,b) Act-Gal4-UAS-GFP (a), Act-Gal4-UAS- GFP.nls (b,c) is ubiquitously expressed in the testes. The testes were immunostained with GFP
mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons
 Supplemental Material mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons Antoine de Chevigny, Nathalie Coré, Philipp Follert, Marion Gaudin, Pascal
Supplemental Material mir-7a regulation of Pax6 in neural stem cells controls the spatial origin of forebrain dopaminergic neurons Antoine de Chevigny, Nathalie Coré, Philipp Follert, Marion Gaudin, Pascal
glial cells missing and gcm2 Cell-autonomously Regulate Both Glial and Neuronal
 glial cells missing and gcm2 Cell-autonomously Regulate Both Glial and Neuronal Development in the Visual System of Drosophila Carole Chotard, Wendy Leung and Iris Salecker Supplemental Data Supplemental
glial cells missing and gcm2 Cell-autonomously Regulate Both Glial and Neuronal Development in the Visual System of Drosophila Carole Chotard, Wendy Leung and Iris Salecker Supplemental Data Supplemental
Nature Neuroscience: doi: /nn Supplementary Figure 1
 Supplementary Figure 1 Expression of escargot (esg) and genetic approach for achieving IPC-specific knockdown. (a) esg MH766 -Gal4 UAS-cd8GFP (green) and esg-lacz B7-2-22 (red) show similar expression
Supplementary Figure 1 Expression of escargot (esg) and genetic approach for achieving IPC-specific knockdown. (a) esg MH766 -Gal4 UAS-cd8GFP (green) and esg-lacz B7-2-22 (red) show similar expression
Supplemental Data SUPPLEMENTAL EXPERIMENTAL PROCEDURES
 Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cédric Maurange, Louise Cheng and Alex P. Gould Supplemental Data SUPPLEMENTAL EXPERIMENTAL PROCEDURES
Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cédric Maurange, Louise Cheng and Alex P. Gould Supplemental Data SUPPLEMENTAL EXPERIMENTAL PROCEDURES
he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003)
 MicroRNAs: Genomics, Biogenesis, Mechanism, and Function (D. Bartel Cell 2004) he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003) Vertebrate MicroRNA Genes (Lim et al. Science
MicroRNAs: Genomics, Biogenesis, Mechanism, and Function (D. Bartel Cell 2004) he micrornas of Caenorhabditis elegans (Lim et al. Genes & Development 2003) Vertebrate MicroRNA Genes (Lim et al. Science
MicroRNA expression profiling and functional analysis in prostate cancer. Marco Folini s.c. Ricerca Traslazionale DOSL
 MicroRNA expression profiling and functional analysis in prostate cancer Marco Folini s.c. Ricerca Traslazionale DOSL What are micrornas? For almost three decades, the alteration of protein-coding genes
MicroRNA expression profiling and functional analysis in prostate cancer Marco Folini s.c. Ricerca Traslazionale DOSL What are micrornas? For almost three decades, the alteration of protein-coding genes
Supplementary Figure 1. Procedures to independently control fly hunger and thirst states.
 Supplementary Figure 1 Procedures to independently control fly hunger and thirst states. (a) Protocol to produce exclusively hungry or thirsty flies for 6 h water memory retrieval. (b) Consumption assays
Supplementary Figure 1 Procedures to independently control fly hunger and thirst states. (a) Protocol to produce exclusively hungry or thirsty flies for 6 h water memory retrieval. (b) Consumption assays
BIOL2005 WORKSHEET 2008
 BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
BIOL2005 WORKSHEET 2008 Answer all 6 questions in the space provided using additional sheets where necessary. Hand your completed answers in to the Biology office by 3 p.m. Friday 8th February. 1. Your
Supplementary Figures
 J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
J. Cell Sci. 128: doi:10.1242/jcs.173807: Supplementary Material Supplementary Figures Fig. S1 Fig. S1. Description and/or validation of reagents used. All panels show Drosophila tissues oriented with
100 mm Sucrose. +Berberine +Quinine
 8 mm Sucrose Probability (%) 7 6 5 4 3 Wild-type Gr32a / 2 +Caffeine +Berberine +Quinine +Denatonium Supplementary Figure 1: Detection of sucrose and bitter compounds is not affected in Gr32a / flies.
8 mm Sucrose Probability (%) 7 6 5 4 3 Wild-type Gr32a / 2 +Caffeine +Berberine +Quinine +Denatonium Supplementary Figure 1: Detection of sucrose and bitter compounds is not affected in Gr32a / flies.
Supplemental Information. Otic Mesenchyme Cells Regulate. Spiral Ganglion Axon Fasciculation. through a Pou3f4/EphA4 Signaling Pathway
 Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Neuron, Volume 73 Supplemental Information Otic Mesenchyme Cells Regulate Spiral Ganglion Axon Fasciculation through a Pou3f4/EphA4 Signaling Pathway Thomas M. Coate, Steven Raft, Xiumei Zhao, Aimee K.
Chapter 2. Investigation into mir-346 Regulation of the nachr α5 Subunit
 15 Chapter 2 Investigation into mir-346 Regulation of the nachr α5 Subunit MicroRNA s (mirnas) are small (< 25 base pairs), single stranded, non-coding RNAs that regulate gene expression at the post transcriptional
15 Chapter 2 Investigation into mir-346 Regulation of the nachr α5 Subunit MicroRNA s (mirnas) are small (< 25 base pairs), single stranded, non-coding RNAs that regulate gene expression at the post transcriptional
marker. DAPI labels nuclei. Flies were 20 days old. Scale bar is 5 µm. Ctrl is
 Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Supplementary Figure 1. (a) Nos is detected in glial cells in both control and GFAP R79H transgenic flies (arrows), but not in deletion mutant Nos Δ15 animals. Repo is a glial cell marker. DAPI labels
Cross species analysis of genomics data. Computational Prediction of mirnas and their targets
 02-716 Cross species analysis of genomics data Computational Prediction of mirnas and their targets Outline Introduction Brief history mirna Biogenesis Why Computational Methods? Computational Methods
02-716 Cross species analysis of genomics data Computational Prediction of mirnas and their targets Outline Introduction Brief history mirna Biogenesis Why Computational Methods? Computational Methods
Noncoding or non-messenger RNAs are
 The functions of animal micrornas Victor Ambros Dartmouth Medical School, Department of Genetics, Hanover, New Hampshire 03755, USA (e-mail: vra@dartmouth.edu) MicroRNAs (mirnas) are small RNAs that regulate
The functions of animal micrornas Victor Ambros Dartmouth Medical School, Department of Genetics, Hanover, New Hampshire 03755, USA (e-mail: vra@dartmouth.edu) MicroRNAs (mirnas) are small RNAs that regulate
SUPPLEMENTARY INFORMATION
 doi: 10.1038/nature07173 SUPPLEMENTARY INFORMATION Supplementary Figure Legends: Supplementary Figure 1: Model of SSC and CPC divisions a, Somatic stem cells (SSC) reside adjacent to the hub (red), self-renew
doi: 10.1038/nature07173 SUPPLEMENTARY INFORMATION Supplementary Figure Legends: Supplementary Figure 1: Model of SSC and CPC divisions a, Somatic stem cells (SSC) reside adjacent to the hub (red), self-renew
effects on organ development. a-f, Eye and wing discs with clones of ε j2b10 show no
 Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
Supplementary Figure 1. Loss of function clones of 14-3-3 or 14-3-3 show no significant effects on organ development. a-f, Eye and wing discs with clones of 14-3-3ε j2b10 show no obvious defects in Elav
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
doi:10.1038/nature10643 Supplementary Table 1. Identification of hecw-1 coding polymorphisms at amino acid positions 322 and 325 in 162 strains of C. elegans. WWW.NATURE.COM/NATURE 1 Supplementary Figure
Isoform-Specific Control of Male Neuronal Differentiation and Behavior in Drosophila by the fruitless Gene
 Current Biology 16, 1063 1076, June 6, 2006 ª2006 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2006.04.039 Isoform-Specific Control of Male Neuronal Differentiation and Behavior in Drosophila by
Current Biology 16, 1063 1076, June 6, 2006 ª2006 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2006.04.039 Isoform-Specific Control of Male Neuronal Differentiation and Behavior in Drosophila by
Supplementary Figure 1. Flies form water-reward memory only in the thirsty state
 1 2 3 4 5 6 7 Supplementary Figure 1. Flies form water-reward memory only in the thirsty state Thirsty but not sated wild-type flies form robust 3 min memory. For the thirsty group, the flies were water-deprived
1 2 3 4 5 6 7 Supplementary Figure 1. Flies form water-reward memory only in the thirsty state Thirsty but not sated wild-type flies form robust 3 min memory. For the thirsty group, the flies were water-deprived
micrornas (mirna) and Biomarkers
 micrornas (mirna) and Biomarkers Small RNAs Make Big Splash mirnas & Genome Function Biomarkers in Cancer Future Prospects Javed Khan M.D. National Cancer Institute EORTC-NCI-ASCO November 2007 The Human
micrornas (mirna) and Biomarkers Small RNAs Make Big Splash mirnas & Genome Function Biomarkers in Cancer Future Prospects Javed Khan M.D. National Cancer Institute EORTC-NCI-ASCO November 2007 The Human
50mM D-Glucose. Percentage PI. L-Glucose
 Monica Dus, Minrong Ai, Greg S.B Suh. Taste-independent nutrient selection is mediated by a brain-specific Na+/solute cotransporter in Drosophila. Control + Phlorizin mm D-Glucose 1 2mM 1 L-Glucose Gr5a;Gr64a
Monica Dus, Minrong Ai, Greg S.B Suh. Taste-independent nutrient selection is mediated by a brain-specific Na+/solute cotransporter in Drosophila. Control + Phlorizin mm D-Glucose 1 2mM 1 L-Glucose Gr5a;Gr64a
Circular RNAs (circrnas) act a stable mirna sponges
 Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Circular RNAs (circrnas) act a stable mirna sponges cernas compete for mirnas Ancestal mrna (+3 UTR) Pseudogene RNA (+3 UTR homolgy region) The model holds true for all RNAs that share a mirna binding
Senior Thesis. Presented to. The Faculty of the School of Arts and Sciences Brandeis University
 Effects of the microrna let-7 on sleep regulation in Drosophila Senior Thesis Presented to The Faculty of the School of Arts and Sciences Brandeis University Undergraduate Program in Biology Leslie Griffith,
Effects of the microrna let-7 on sleep regulation in Drosophila Senior Thesis Presented to The Faculty of the School of Arts and Sciences Brandeis University Undergraduate Program in Biology Leslie Griffith,
Studying The Role Of DNA Mismatch Repair In Brain Cancer Malignancy
 Kavya Puchhalapalli CALS Honors Project Report Spring 2017 Studying The Role Of DNA Mismatch Repair In Brain Cancer Malignancy Abstract Malignant brain tumors including medulloblastomas and primitive neuroectodermal
Kavya Puchhalapalli CALS Honors Project Report Spring 2017 Studying The Role Of DNA Mismatch Repair In Brain Cancer Malignancy Abstract Malignant brain tumors including medulloblastomas and primitive neuroectodermal
Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v)
 SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
SUPPLEMENTARY MATERIAL AND METHODS Soft Agar Assay. For each cell pool, 100,000 cells were resuspended in 0.35% (w/v) top agar (LONZA, SeaKem LE Agarose cat.5004) and plated onto 0.5% (w/v) basal agar.
Meigo governs dendrite targeting specificity by modulating Ephrin level and N-glycosylation
 Meigo governs dendrite targeting specificity by modulating Ephrin level and N-glycosylation Sayaka U. Sekine, Shuka Haraguchi, Kinhong Chao, Tomoko Kato, Liqun Luo, Masayuki Miura, and Takahiro Chihara
Meigo governs dendrite targeting specificity by modulating Ephrin level and N-glycosylation Sayaka U. Sekine, Shuka Haraguchi, Kinhong Chao, Tomoko Kato, Liqun Luo, Masayuki Miura, and Takahiro Chihara
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
Supplementary Figures Supplementary Figure 1. nrg1 bns101/bns101 embryos develop a functional heart and survive to adulthood (a-b) Cartoon of Talen-induced nrg1 mutation with a 14-base-pair deletion in
Strategies to determine the biological function of micrornas
 Strategies to determine the biological function of micrornas Jan Krützfeldt, Matthew N Poy & Markus Stoffel MicroRNAs (mirnas) are regulators of gene expression that control many biological processes in
Strategies to determine the biological function of micrornas Jan Krützfeldt, Matthew N Poy & Markus Stoffel MicroRNAs (mirnas) are regulators of gene expression that control many biological processes in
SUPPLEMENTARY FIGURE LEGENDS
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Negative correlation between mir-375 and its predicted target genes, as demonstrated by gene set enrichment analysis (GSEA). 1 The correlation between
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1 Negative correlation between mir-375 and its predicted target genes, as demonstrated by gene set enrichment analysis (GSEA). 1 The correlation between
a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation,
 Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
Supplementary Information Supplementary Figures Supplementary Figure 1. a) List of KMTs targeted in the shrna screen. The official symbol, KMT designation, gene ID and specifities are provided. Those highlighted
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
DOI: 10.1038/ncb2566 Figure S1 CDKL5 protein expression pattern and localization in mouse brain. (a) Multiple-tissue western blot from a postnatal day (P) 21 mouse probed with an antibody against CDKL5.
The microrna bantam Functions in Epithelial Cells to Regulate Scaling Growth of Dendrite Arbors in Drosophila Sensory Neurons
 Article The microrna bantam Functions in Epithelial Cells to Regulate Scaling Growth of Dendrite Arbors in Drosophila Sensory Neurons Jay Z. Parrish, 1,2 Peizhang Xu, 1,2 Charles C. Kim, 1 Lily Yeh Jan,
Article The microrna bantam Functions in Epithelial Cells to Regulate Scaling Growth of Dendrite Arbors in Drosophila Sensory Neurons Jay Z. Parrish, 1,2 Peizhang Xu, 1,2 Charles C. Kim, 1 Lily Yeh Jan,
(,, ) microrna(mirna) 19~25 nt RNA, RNA mirna mirna,, ;, mirna mirna. : microrna (mirna); ; ; ; : R321.1 : A : X(2015)
 35 1 Vol.35 No.1 2015 1 Jan. 2015 Reproduction & Contraception doi: 10.7669/j.issn.0253-357X.2015.01.0037 E-mail: randc_journal@163.com microrna ( 450052) microrna() 19~25 nt RNA RNA ; : microrna (); ;
35 1 Vol.35 No.1 2015 1 Jan. 2015 Reproduction & Contraception doi: 10.7669/j.issn.0253-357X.2015.01.0037 E-mail: randc_journal@163.com microrna ( 450052) microrna() 19~25 nt RNA RNA ; : microrna (); ;
MicroRNAs Modulate Hematopoietic Lineage Differentiation
 MicroRNAs Modulate Hematopoietic Lineage Differentiation Harvey F. Lodish, Chang-Zheng Chen, David P. Bartel Whitehead Institute for Biomedical Research, Department of Biology, Massachusetts Institute
MicroRNAs Modulate Hematopoietic Lineage Differentiation Harvey F. Lodish, Chang-Zheng Chen, David P. Bartel Whitehead Institute for Biomedical Research, Department of Biology, Massachusetts Institute
Supplementary Figure S1: TIPF reporter validation in the wing disc.
 Supplementary Figure S1: TIPF reporter validation in the wing disc. a,b, Test of put RNAi. a, In wildtype discs the Dpp target gene Sal (red) is expressed in a broad stripe in the centre of the ventral
Supplementary Figure S1: TIPF reporter validation in the wing disc. a,b, Test of put RNAi. a, In wildtype discs the Dpp target gene Sal (red) is expressed in a broad stripe in the centre of the ventral
LABORATÓRIUMI GYAKORLAT SILLABUSZ SYLLABUS OF A PRACTICAL DEMOSTRATION. financed by the program
 TÁMOP-4.1.1.C-13/1/KONV-2014-0001 projekt Az élettudományi-klinikai felsőoktatás gyakorlatorientált és hallgatóbarát korszerűsítése a vidéki képzőhelyek nemzetközi versenyképességének erősítésére program
TÁMOP-4.1.1.C-13/1/KONV-2014-0001 projekt Az élettudományi-klinikai felsőoktatás gyakorlatorientált és hallgatóbarát korszerűsítése a vidéki képzőhelyek nemzetközi versenyképességének erősítésére program
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Differential expression of mirnas from the pri-mir-17-92a locus.
 Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
Supplementary Figure 1 Differential expression of mirnas from the pri-mir-17-92a locus. (a) The mir-17-92a expression unit in the third intron of the host mir-17hg transcript. (b,c) Impact of knockdown
Nature Genetics: doi: /ng.3731
 Supplementary Figure 1 Circadian profiles of Adarb1 transcript and ADARB1 protein in mouse tissues. (a) Overlap of rhythmic transcripts identified in the previous transcriptome analyses. The mouse liver
Supplementary Figure 1 Circadian profiles of Adarb1 transcript and ADARB1 protein in mouse tissues. (a) Overlap of rhythmic transcripts identified in the previous transcriptome analyses. The mouse liver
Supplementary Figure 1 (Related with Figure 4). Molecular consequences of Eed deletion. (a) ChIP analysis identifies 3925 genes that are associated
 Supplementary Figure 1 (Related with Figure 4). Molecular consequences of Eed deletion. (a) ChIP analysis identifies 3925 genes that are associated with the H3K27me3 mark in chondrocytes (see Table S1,
Supplementary Figure 1 (Related with Figure 4). Molecular consequences of Eed deletion. (a) ChIP analysis identifies 3925 genes that are associated with the H3K27me3 mark in chondrocytes (see Table S1,
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Wang and Page-McCaw, http://www.jcb.org/cgi/content/full/jcb.201403084/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Extracellular anti-wg staining is specific. Note
Supplemental material Wang and Page-McCaw, http://www.jcb.org/cgi/content/full/jcb.201403084/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. Extracellular anti-wg staining is specific. Note
MicroRNA-mediated incoherent feedforward motifs are robust
 The Second International Symposium on Optimization and Systems Biology (OSB 8) Lijiang, China, October 31 November 3, 8 Copyright 8 ORSC & APORC, pp. 62 67 MicroRNA-mediated incoherent feedforward motifs
The Second International Symposium on Optimization and Systems Biology (OSB 8) Lijiang, China, October 31 November 3, 8 Copyright 8 ORSC & APORC, pp. 62 67 MicroRNA-mediated incoherent feedforward motifs
genomics for systems biology / ISB2020 RNA sequencing (RNA-seq)
 RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
RNA sequencing (RNA-seq) Module Outline MO 13-Mar-2017 RNA sequencing: Introduction 1 WE 15-Mar-2017 RNA sequencing: Introduction 2 MO 20-Mar-2017 Paper: PMID 25954002: Human genomics. The human transcriptome
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
Supplementary Figure 1 Effect of HSP90 inhibition on expression of endogenous retroviruses. (a) Inducible shrna-mediated Hsp90 silencing in mouse ESCs. Immunoblots of total cell extract expressing the
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells
 MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells Margaret S Ebert, Joel R Neilson & Phillip A Sharp Supplementary figures and text: Supplementary Figure 1. Effect of sponges on
Programmed Cell Death (apoptosis)
 Programmed Cell Death (apoptosis) Stereotypic death process includes: membrane blebbing nuclear fragmentation chromatin condensation and DNA framentation loss of mitochondrial integrity and release of
Programmed Cell Death (apoptosis) Stereotypic death process includes: membrane blebbing nuclear fragmentation chromatin condensation and DNA framentation loss of mitochondrial integrity and release of
A Genetic Program for Embryonic Development
 Concept 18.4: A program of differential gene expression leads to the different cell types in a multicellular organism During embryonic development, a fertilized egg gives rise to many different cell types
Concept 18.4: A program of differential gene expression leads to the different cell types in a multicellular organism During embryonic development, a fertilized egg gives rise to many different cell types
klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach
![klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach klp-18 (RNAi) Control. supplementary information. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach](/thumbs/91/104639484.jpg) DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
DOI: 10.1038/ncb1891 A. starting strain: AV335 [emb-27(g48); GFP::histone; GFP::tubulin] bleach embryos let hatch overnight transfer to RNAi plates; incubate 5 days at 15 C RNAi food L1 worms adult worms
Supplementary Figure 1 hlrrk2 promotes CAP dependent protein translation.
 ` Supplementary Figure 1 hlrrk2 promotes CAP dependent protein translation. (a) Overexpression of hlrrk2 in HeLa cells enhances total protein synthesis in [35S] methionine/cysteine incorporation assays.
` Supplementary Figure 1 hlrrk2 promotes CAP dependent protein translation. (a) Overexpression of hlrrk2 in HeLa cells enhances total protein synthesis in [35S] methionine/cysteine incorporation assays.
Supplementary Information. Preferential associations between co-regulated genes reveal a. transcriptional interactome in erythroid cells
 Supplementary Information Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells Stefan Schoenfelder, * Tom Sexton, * Lyubomira Chakalova, * Nathan
Supplementary Information Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells Stefan Schoenfelder, * Tom Sexton, * Lyubomira Chakalova, * Nathan
Supplemental Information. Memory-Relevant Mushroom Body. Output Synapses Are Cholinergic
 Neuron, Volume 89 Supplemental Information Memory-Relevant Mushroom Body Output Synapses Are Cholinergic Oliver Barnstedt, David Owald, Johannes Felsenberg, Ruth Brain, John-Paul Moszynski, Clifford B.
Neuron, Volume 89 Supplemental Information Memory-Relevant Mushroom Body Output Synapses Are Cholinergic Oliver Barnstedt, David Owald, Johannes Felsenberg, Ruth Brain, John-Paul Moszynski, Clifford B.
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Mir-184 affects lifespan via regulating the Activin pathway in Drosophila
 1 Mir-184 affects lifespan via regulating the Activin pathway in Drosophila Senior Thesis Presented to The Faculty of the School of Arts and Sciences Brandeis University Undergraduate Program in the Department
1 Mir-184 affects lifespan via regulating the Activin pathway in Drosophila Senior Thesis Presented to The Faculty of the School of Arts and Sciences Brandeis University Undergraduate Program in the Department
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1. Formation of the AA5x. a, Camera lucida drawing of embryo at 48 hours post fertilization (hpf, modified from Kimmel et al. Dev Dyn. 1995 203:253-310). b, Confocal microangiogram
Supplementary Figure 1. Formation of the AA5x. a, Camera lucida drawing of embryo at 48 hours post fertilization (hpf, modified from Kimmel et al. Dev Dyn. 1995 203:253-310). b, Confocal microangiogram
SUPPLEMENTARY FIG. S2. Representative counting fields used in quantification of the in vitro neural differentiation of pattern of dnscs.
 Supplementary Data SUPPLEMENTARY FIG. S1. Representative counting fields used in quantification of the in vitro neural differentiation of pattern of anpcs. A panel of lineage-specific markers were used
Supplementary Data SUPPLEMENTARY FIG. S1. Representative counting fields used in quantification of the in vitro neural differentiation of pattern of anpcs. A panel of lineage-specific markers were used
Supplemental Information. Ciliary Beating Compartmentalizes. Cerebrospinal Fluid Flow in the Brain. and Regulates Ventricular Development
 Current Biology, Volume Supplemental Information Ciliary Beating Compartmentalizes Cerebrospinal Fluid Flow in the Brain and Regulates Ventricular Development Emilie W. Olstad, Christa Ringers, Jan N.
Current Biology, Volume Supplemental Information Ciliary Beating Compartmentalizes Cerebrospinal Fluid Flow in the Brain and Regulates Ventricular Development Emilie W. Olstad, Christa Ringers, Jan N.
A Drosophila model for alcohol reward. Supplemental Material
 Kaun et al. Supplemental Material 1 A Drosophila model for alcohol reward Supplemental Material Kaun, K.R. *, Azanchi, R. *, Maung, Z. *, Hirsh, J. and Heberlein, U. * * Department of Anatomy, University
Kaun et al. Supplemental Material 1 A Drosophila model for alcohol reward Supplemental Material Kaun, K.R. *, Azanchi, R. *, Maung, Z. *, Hirsh, J. and Heberlein, U. * * Department of Anatomy, University
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1. Ras V12 expression in the entire eye-antennal disc does not cause invasive tumours. a, Eye-antennal discs expressing Ras V12 in all cells (marked with GFP, green) overgrow moderately
Supplementary Figure 1. Ras V12 expression in the entire eye-antennal disc does not cause invasive tumours. a, Eye-antennal discs expressing Ras V12 in all cells (marked with GFP, green) overgrow moderately
Zhu et al, page 1. Supplementary Figures
 Zhu et al, page 1 Supplementary Figures Supplementary Figure 1: Visual behavior and avoidance behavioral response in EPM trials. (a) Measures of visual behavior that performed the light avoidance behavior
Zhu et al, page 1 Supplementary Figures Supplementary Figure 1: Visual behavior and avoidance behavioral response in EPM trials. (a) Measures of visual behavior that performed the light avoidance behavior
Nature Genetics: doi: /ng Supplementary Figure 1. Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms.
 Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
Supplementary Figure 1 Immunofluorescence (IF) confirms absence of H3K9me in met-2 set-25 worms. IF images of wild-type (wt) and met-2 set-25 worms showing the loss of H3K9me2/me3 at the indicated developmental
RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice
 SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
SUPPLEMENTARY INFORMATION RNA interference induced hepatotoxicity results from loss of the first synthesized isoform of microrna-122 in mice Paul N Valdmanis, Shuo Gu, Kirk Chu, Lan Jin, Feijie Zhang,
SUPPLEMENTARY FIGURES
 SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
SUPPLEMENTARY FIGURES 1 Supplementary Figure 1, Adult hippocampal QNPs and TAPs uniformly express REST a-b) Confocal images of adult hippocampal mouse sections showing GFAP (green), Sox2 (red), and REST
EMBO REPORT SUPPLEMENTARY SECTION. Quantitation of mitotic cells after perturbation of Notch signalling.
 EMBO REPORT SUPPLEMENTARY SECTION Quantitation of mitotic cells after perturbation of Notch signalling. Notch activation suppresses the cell cycle indistinguishably both within and outside the neural plate
EMBO REPORT SUPPLEMENTARY SECTION Quantitation of mitotic cells after perturbation of Notch signalling. Notch activation suppresses the cell cycle indistinguishably both within and outside the neural plate
PEER REVIEW FILE. Reviewers' Comments: Reviewer #1 (Remarks to the Author):
 PEER REVIEW FILE Reviewers' Comments: Reviewer #1 (Remarks to the Author): In this manuscript entitled "Neural circuits for long-term water-reward memory processing in thirsty Drosophila", the authors
PEER REVIEW FILE Reviewers' Comments: Reviewer #1 (Remarks to the Author): In this manuscript entitled "Neural circuits for long-term water-reward memory processing in thirsty Drosophila", the authors
Supplementary Figure 1
 Supplementary Figure 1 Kif1a RNAi effect on basal progenitor differentiation Related to Figure 2. Representative confocal images of the VZ and SVZ of rat cortices transfected at E16 with scrambled or Kif1a
Supplementary Figure 1 Kif1a RNAi effect on basal progenitor differentiation Related to Figure 2. Representative confocal images of the VZ and SVZ of rat cortices transfected at E16 with scrambled or Kif1a
Problem Set 5 KEY
 2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
2006 7.012 Problem Set 5 KEY ** Due before 5 PM on THURSDAY, November 9, 2006. ** Turn answers in to the box outside of 68-120. PLEASE WRITE YOUR ANSWERS ON THIS PRINTOUT. 1. You are studying the development
Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment
 Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development
Overview: Conducting the Genetic Orchestra Prokaryotes and eukaryotes alter gene expression in response to their changing environment In multicellular eukaryotes, gene expression regulates development
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS)
 Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Figure S1. Venn diagram analysis of mrna microarray data and mirna target analysis. (a) Western blot analysis of T lymphoblasts (CLS) and their exosomes (EXO) in resting (REST) and activated
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
www.sciencesignaling.org/cgi/content/full/6/283/ra57/dc1 Supplementary Materials for JNK3 Couples the Neuronal Stress Response to Inhibition of Secretory Trafficking Guang Yang,* Xun Zhou, Jingyan Zhu,
CONTRACTING ORGANIZATION: Cold Spring Harbor Laboratory Cold Spring Harbor, NY 11724
 AD Award Number: W81XWH-10-1-0450 TITLE: The role of NF1 in memory retrieval PRINCIPAL INVESTIGATOR: Yi Zhong, Ph.D. CONTRACTING ORGANIZATION: Cold Spring Harbor Laboratory Cold Spring Harbor, NY 11724
AD Award Number: W81XWH-10-1-0450 TITLE: The role of NF1 in memory retrieval PRINCIPAL INVESTIGATOR: Yi Zhong, Ph.D. CONTRACTING ORGANIZATION: Cold Spring Harbor Laboratory Cold Spring Harbor, NY 11724
Correspondence: mirna regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration
 Correspondence: mirna regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration Alison A. Staton, Holger Knaut, and Antonio J. Giraldez Supplementary Note Materials and
Correspondence: mirna regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration Alison A. Staton, Holger Knaut, and Antonio J. Giraldez Supplementary Note Materials and
The subcortical maternal complex controls symmetric division of mouse zygotes by
 The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics Xing-Jiang Yu 1,2, Zhaohong Yi 1, Zheng Gao 1,2, Dan-dan Qin 1,2, Yanhua Zhai 1, Xue Chen 1,
The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics Xing-Jiang Yu 1,2, Zhaohong Yi 1, Zheng Gao 1,2, Dan-dan Qin 1,2, Yanhua Zhai 1, Xue Chen 1,
Chapter 5 A Dose Dependent Screen for Modifiers of Kek5
 Chapter 5 A Dose Dependent Screen for Modifiers of Kek5 "#$ ABSTRACT Modifier screens in Drosophila have proven to be a powerful tool for uncovering gene interaction and elucidating molecular pathways.
Chapter 5 A Dose Dependent Screen for Modifiers of Kek5 "#$ ABSTRACT Modifier screens in Drosophila have proven to be a powerful tool for uncovering gene interaction and elucidating molecular pathways.
High AU content: a signature of upregulated mirna in cardiac diseases
 https://helda.helsinki.fi High AU content: a signature of upregulated mirna in cardiac diseases Gupta, Richa 2010-09-20 Gupta, R, Soni, N, Patnaik, P, Sood, I, Singh, R, Rawal, K & Rani, V 2010, ' High
https://helda.helsinki.fi High AU content: a signature of upregulated mirna in cardiac diseases Gupta, Richa 2010-09-20 Gupta, R, Soni, N, Patnaik, P, Sood, I, Singh, R, Rawal, K & Rani, V 2010, ' High
Neurotrophic factor GDNF and camp suppress glucocorticoid-inducible PNMT expression in a mouse pheochromocytoma model.
 161 Neurotrophic factor GDNF and camp suppress glucocorticoid-inducible PNMT expression in a mouse pheochromocytoma model. Marian J. Evinger a, James F. Powers b and Arthur S. Tischler b a. Department
161 Neurotrophic factor GDNF and camp suppress glucocorticoid-inducible PNMT expression in a mouse pheochromocytoma model. Marian J. Evinger a, James F. Powers b and Arthur S. Tischler b a. Department
Head of College Scholars List Scheme. Summer Studentship Report Form
 Head of College Scholars List Scheme Summer Studentship 2019 Report Form This report should be completed by the student with his/her project supervisor. It should summarise the work undertaken during the
Head of College Scholars List Scheme Summer Studentship 2019 Report Form This report should be completed by the student with his/her project supervisor. It should summarise the work undertaken during the
mirna Dr. S Hosseini-Asl
 mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
mirna Dr. S Hosseini-Asl 1 2 MicroRNAs (mirnas) are small noncoding RNAs which enhance the cleavage or translational repression of specific mrna with recognition site(s) in the 3 - untranslated region
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2419 Figure S1 NiGFP localization in Dl mutant dividing SOPs. a-c) time-lapse analysis of NiGFP (green) in Dl mutant SOPs (H2B-RFP, red; clones were identified by the loss of nlsgfp) showing
DOI: 10.1038/ncb2419 Figure S1 NiGFP localization in Dl mutant dividing SOPs. a-c) time-lapse analysis of NiGFP (green) in Dl mutant SOPs (H2B-RFP, red; clones were identified by the loss of nlsgfp) showing
Department of Cell and Developmental Biology, Vanderbilt University, Nashville, TN
 UNC-6/Netrin mediates dendritic self-avoidance Cody J. Smith 1, Joseph D. Watson 1,4,5, Miri K. VanHoven 2, Daniel A. Colón-Ramos 3 and David M. Miller III 1,4,6 1 Department of Cell and Developmental
UNC-6/Netrin mediates dendritic self-avoidance Cody J. Smith 1, Joseph D. Watson 1,4,5, Miri K. VanHoven 2, Daniel A. Colón-Ramos 3 and David M. Miller III 1,4,6 1 Department of Cell and Developmental
A Normal Exencephaly Craniora- Spina bifida Microcephaly chischisis. Midbrain Forebrain/ Forebrain/ Hindbrain Spinal cord Hindbrain Hindbrain
 A Normal Exencephaly Craniora- Spina bifida Microcephaly chischisis NTD Number of embryos % among NTD Embryos Exencephaly 52 74.3% Craniorachischisis 6 8.6% Spina bifida 5 7.1% Microcephaly 7 1% B Normal
A Normal Exencephaly Craniora- Spina bifida Microcephaly chischisis NTD Number of embryos % among NTD Embryos Exencephaly 52 74.3% Craniorachischisis 6 8.6% Spina bifida 5 7.1% Microcephaly 7 1% B Normal
Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates
 Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates L. Cheng, A. Arata, R. Mizuguchi, Y. Qian, A. Karunaratne, P.A. Gray, S. Arata, S. Shirasawa, M. Bouchard,
Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates L. Cheng, A. Arata, R. Mizuguchi, Y. Qian, A. Karunaratne, P.A. Gray, S. Arata, S. Shirasawa, M. Bouchard,
Concentration memory-dependent synaptic plasticity of a taste circuit regulates salt chemotaxis in Caenorhabditis elegans
 Supplementary Information for: Concentration memory-dependent synaptic plasticity of a taste circuit regulates salt chemotaxis in Caenorhabditis elegans Hirofumi Kunitomo, Hirofumi Sato, Ryo Iwata, Yohsuke
Supplementary Information for: Concentration memory-dependent synaptic plasticity of a taste circuit regulates salt chemotaxis in Caenorhabditis elegans Hirofumi Kunitomo, Hirofumi Sato, Ryo Iwata, Yohsuke
Article. The Color-Vision Circuit in the Medulla of Drosophila. Javier Morante 1 and Claude Desplan 1, * 1
 Current Biology 18, 1 13, April 22, 2008 ª2008 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2008.02.075 The Color-Vision Circuit in the Medulla of Drosophila Article Javier Morante 1 and Claude Desplan
Current Biology 18, 1 13, April 22, 2008 ª2008 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2008.02.075 The Color-Vision Circuit in the Medulla of Drosophila Article Javier Morante 1 and Claude Desplan
m 6 A mrna methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer
 SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
SUPPLEMENTARY INFORMATION Articles https://doi.org/10.1038/s41556-018-0174-4 In the format provided by the authors and unedited. m 6 A mrna methylation regulates AKT activity to promote the proliferation
Nature Neuroscience: doi: /nn Supplementary Figure 1. MADM labeling of thalamic clones.
 Supplementary Figure 1 MADM labeling of thalamic clones. (a) Confocal images of an E12 Nestin-CreERT2;Ai9-tdTomato brain treated with TM at E10 and stained for BLBP (green), a radial glial progenitor-specific
Supplementary Figure 1 MADM labeling of thalamic clones. (a) Confocal images of an E12 Nestin-CreERT2;Ai9-tdTomato brain treated with TM at E10 and stained for BLBP (green), a radial glial progenitor-specific
Supplemental Information. Induction of Expansion and Folding. in Human Cerebral Organoids
 Cell Stem Cell, Volume 20 Supplemental Information Induction of Expansion and Folding in Human Cerebral Organoids Yun Li, Julien Muffat, Attya Omer, Irene Bosch, Madeline A. Lancaster, Mriganka Sur, Lee
Cell Stem Cell, Volume 20 Supplemental Information Induction of Expansion and Folding in Human Cerebral Organoids Yun Li, Julien Muffat, Attya Omer, Irene Bosch, Madeline A. Lancaster, Mriganka Sur, Lee
Conditional and reversible disruption of essential herpesvirus protein functions
 nature methods Conditional and reversible disruption of essential herpesvirus protein functions Mandy Glaß, Andreas Busche, Karen Wagner, Martin Messerle & Eva Maria Borst Supplementary figures and text:
nature methods Conditional and reversible disruption of essential herpesvirus protein functions Mandy Glaß, Andreas Busche, Karen Wagner, Martin Messerle & Eva Maria Borst Supplementary figures and text:
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature19357 Figure 1a Chd8 +/+ Chd8 +/ΔSL Chd8 +/+ Chd8 +/ΔL E10.5_Whole brain E10.5_Whole brain E10.5_Whole brain E14.5_Whole brain E14.5_Whole brain E14.5_Whole
SUPPLEMENTARY INFORMATION doi:10.1038/nature19357 Figure 1a Chd8 +/+ Chd8 +/ΔSL Chd8 +/+ Chd8 +/ΔL E10.5_Whole brain E10.5_Whole brain E10.5_Whole brain E14.5_Whole brain E14.5_Whole brain E14.5_Whole
Neuroepithelial Cells and Neural Differentiation
 Neuroepithelial Cells and Neural Differentiation Neurulation The cells of the neural tube are NEUROEPITHELIAL CELLS Neural crest cells migrate out of neural tube Neuroepithelial cells are embryonic stem
Neuroepithelial Cells and Neural Differentiation Neurulation The cells of the neural tube are NEUROEPITHELIAL CELLS Neural crest cells migrate out of neural tube Neuroepithelial cells are embryonic stem
Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid.
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Figure 1. SC35M polymerase activity in the presence of Bat or SC35M NP encoded from the phw2000 rescue plasmid. HEK293T
Alternative Splicing and Genomic Stability
 Alternative Splicing and Genomic Stability Kevin Cahill cahill@unm.edu http://dna.phys.unm.edu/ Abstract In a cell that uses alternative splicing, the total length of all the exons is far less than in
Alternative Splicing and Genomic Stability Kevin Cahill cahill@unm.edu http://dna.phys.unm.edu/ Abstract In a cell that uses alternative splicing, the total length of all the exons is far less than in
MicroRNA and Male Infertility: A Potential for Diagnosis
 Review Article MicroRNA and Male Infertility: A Potential for Diagnosis * Abstract MicroRNAs (mirnas) are small non-coding single stranded RNA molecules that are physiologically produced in eukaryotic
Review Article MicroRNA and Male Infertility: A Potential for Diagnosis * Abstract MicroRNAs (mirnas) are small non-coding single stranded RNA molecules that are physiologically produced in eukaryotic
SUPPLEMENTARY MATERIAL
 SYPPLEMENTARY FIGURE LEGENDS SUPPLEMENTARY MATERIAL Figure S1. Phylogenic studies of the mir-183/96/182 cluster and 3 -UTR of Casp2. (A) Genomic arrangement of the mir-183/96/182 cluster in vertebrates.
SYPPLEMENTARY FIGURE LEGENDS SUPPLEMENTARY MATERIAL Figure S1. Phylogenic studies of the mir-183/96/182 cluster and 3 -UTR of Casp2. (A) Genomic arrangement of the mir-183/96/182 cluster in vertebrates.
Supplementary Information
 Supplementary Information Supplementary s Supplementary 1 All three types of foods suppress subsequent feeding in both sexes when the same food is used in the pre-feeding test feeding. (a) Adjusted pre-feeding
Supplementary Information Supplementary s Supplementary 1 All three types of foods suppress subsequent feeding in both sexes when the same food is used in the pre-feeding test feeding. (a) Adjusted pre-feeding
Supplementary Figure 1
 Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Supplementary Figure 1 Asymmetrical function of 5p and 3p arms of mir-181 and mir-30 families and mir-142 and mir-154. (a) Control experiments using mirna sensor vector and empty pri-mirna overexpression
Figure S1. Molecular confirmation of the precise insertion of the AsMCRkh2 cargo into the kh w locus.
 Supporting Information Appendix Table S1. Larval and adult phenotypes of G 2 progeny of lines 10.1 and 10.2 G 1 outcrosses to wild-type mosquitoes. Table S2. List of oligonucleotide primers. Table S3.
Supporting Information Appendix Table S1. Larval and adult phenotypes of G 2 progeny of lines 10.1 and 10.2 G 1 outcrosses to wild-type mosquitoes. Table S2. List of oligonucleotide primers. Table S3.
