SUPPLEMENTARY INFORMATION
|
|
|
- Sibyl Hampton
- 5 years ago
- Views:
Transcription
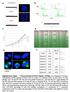
1 SUPPLMNTRY INFORMTION OI: 1.138/ncb2411 mbryoid ody Formation Pluripotent ells Multipotent Progenitor ells day ispase Formation day 7 replating s day 12 MP p1 day 17 MP p2 day 27 MP p4 mbryoid bodies d7 Replated d12 MP p3 1.2 GS Relative xpression to HPRT set to TX NNOG 1.8 lizarin red 1x Hematoxylin - osin lizarin red 2x Tolouidine blue lkaline Phosphatase HOHST 2x J RIPS pluripotent day 4 MP p MP p1 MP p3 MP p7 F ollagen II -HRP Hematoxylin counterstain Q-PR Target Forward Reverse viral PPRG2 TGGGTGTGGG TTTTGTTGGGTTG PPRG2 GGGGTTGGTTG TGTGTGTGG /P GTGTTTGG TTTTGTG IPOQ GTGGTTGTTTGGGG GTTGGTGGGTTGG FP4 TTGGGGTTT GTTGTTGGG HSL TGTGTGTTGTG GGGGGTT LPL TGTGGGTGGGGGTT GGTTTTGGTTTG HPRT TGTGGTG GGTTTTTGGT Y1 TTGGGGGTGTGTTGG GGGTTGG LOVL3 TGTGTTTGG GGTGGGTTTTT UP1 TTGGGTGG GTTGTTTGTTG PG1 GTTTTTTT TTGGTGGTTTGT TX3 GGTTTTGTGG GTTGGTGGTGT GS GGGGGTGGG TGTGGTTG NNOG TTTGTGGGTGGT GGGTGTTGTGG PRM16 (endo) GGTTTT GGGTTGGGTTT PRM16 (v+e) GGTTTTG TGGGGTTGGG Figure S1 erivation and characterization of MPs from hpss. ) xperimental scheme depicting the timeline and steps necessary to derive mesenchymal progenitor cells (MPs) from human pluripotent stem cells (hpss). hpss were differentiated as embryoid bodies (s) in suspension culture. Formed s were replated on dishes, and outgrowing differentiated cells were passaged several times and were analyzed for expression of mesenchymal surface markers and subsequently used in differentiation experiments. ) rightfield images showing different stages during the derivation of MPs. From left to right: embryoid bodies 7 days after placement of pluripotent cells into suspension culture in ultra-low-attachment dishes; replated embryoid bodies at day 12 (MP passage ) just before cells were passaged; MP passage 3, day 25 after formation, showing a homogenous population of cells with the appearance of fibroblasts. ) qrt- PR characterization of MPs over time. ells were analyzed as hpss, 4 day old s, and after various passages (passage, 1, 3, 7) for the mesendoderm marker GS (top), the mesoderm marker TX (middle) and the pluripotency marker NNOG. (n=3, relative expression to HPRT, largest expression set to 1). ) MPs were differentiated into osteoblasts. To confirm the osteoblast differentiation, the top panel shows from left to right: an alizarin red staining at 1 fold, an alizarin red staining at 2 fold magnification and immunocytochemistry for alkaline phopshatase at 2 fold magnification. ) The chondrocyte differentiation was confirmed by staining sectioned MS microspheres with hematoxylin and eosin (left), tolouidine blue was used to stain glycosaminoglycans (middle) and immunohistochemistry against chondrocyte specific ollagen II (right and bottom). F) List of Oligonucleotides used to perform quantitative RT-PR reactions in this study Macmillan Publishers Limited. ll rights reserved.
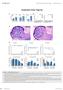
2 SUPPLMNTRY INFORMTION Stro HUS 9 J RiPS HUS 9 J RiPS Figure S2 hps-derived MPs express characteristic mesenchymal stem cell surface markers. ) Flow cytometry for the surface antigens 15, 73, 44, 19 and 4 presented as histograms. HUS 1 p39 (pink), Human Monocytes (dark green), HUS 2 MP p5 (light green), HUS 8 MP p4 (orange), J RiPS #1.1 MP p11 (light blue), SV p7 (red). The x-axis indicates the relative fluorescent intensity of the indicated antibody from 1 to 1. on a logarithmic scale. The y-axis represents the percentage of cells. ) Flow cytometry for the surface antigens Stro1, 15, 73, 44, 29, 4 and unstained controls presented as histograms. J RiPS #1.1p15 (blue), J RiPS #1.1 MP p6 (green), HUS 9 p3 (orange) and p7 (violet) The x-axis indicates the relative fluorescent intensity from 1 to 1. on a logarithmic scale. The y-axis represents the percentage of cells. ) Table showing the results of the two flow cytometry experiments. Numbers represent the percentage of positive cells. If appropriate, positive stainings were distinguished as high or low expression groups. ) Flow nalysis of MP surface antigens. The gating tree was set as follows. Left column: FS/SS (represents the distribution of cells in the light scatter based on size and intracellular composition, respectively) to right column: live gate (P, P-y5, FIT, which represents the fraction of the positive stained cells (Stro1, 29, 15, 73, 44 and 4) Macmillan Publishers Limited. ll rights reserved.
3 SUPPLMNTRY INFORMTION -3.5 log log Stro dipose derived stromal vascular cells dipose derived stromal vascular cells HUS 8 derived mesenchymal progenitor cells HUS 8 derived mesenchymal progenitor cells J RiPS derived mesenchymal progenitor cells HUS 2 derived mesenchymal progenitor cells HUS 2 derived mesenchymal progenitor cells J RiPS derived mesenchymal progenitor cells one Marrow derived MSs Study 1 one Marrow derived MSs Study 1 one Marrow derived MSs Study 1 HUV confluent HUV confluent HUV sparse HUV sparse mbryonic Stem ells ell line H9 day 1 primitive neural epithelial cells mbryonic Stem ells ell line H9 day 1 neural epithelial cells mbryonic Stem ells ell line H9 day 17 definitive neural epithelial cells mbryonic Stem ells ell line H9 day 17 definitive neural epithelial cells mbryonic Stem ells ell line H9 day 1 primitive neural epithelial cells mbryonic Stem ells ell line H9 day 1 primitive neural epithelial cells mbryonic Stem ells ell line H9 day 1 primitive neural epithelial cells mbryonic Stem ells ell line H9 day 17 definitive neural epithelial cells mbryonic Stem ells ell line H9 day 17 definitive neural epithelial cells mbryonic Stem ells ell line H9 day 17 definitive neural epithelial cells mbryonic Stem ells ell line H9 day 17 definitive neural epithelial cells mbryonic Stem ells ell line H9 day 1 neural epithelial cells mbryonic Stem ells ell line H9 day 6 embryoid body mbryonic Stem ells ell line H9 day 6 embryoid body mbryonic Stem ells ell line H9 day 6 embryoid body mbryonic Stem ells ell line H9 mbryonic Stem ells ell line H9 mbryonic Stem ells ell line H9 Stroma normal breast tissue Stroma normal breast tissue Stroma normal breast tissue Stroma normal breast tissue Stroma normal breast tissue HUS 8 derived mesenchymal progenitor cells HUS 8 derived mesenchymal progenitor cells HUS 2 derived mesenchymal progenitor cells HUS 2 derived mesenchymal progenitor cells J RiPS derived mesenchymal progenitor cells J RiPS derived mesenchymal progenitor cells dipose derived stromal vascular cells dipose derived stromal vascular cells one Marrow derived MSs Study 1 one Marrow derived MSs Study 1 one Marrow derived MSs Study 1 Figure S3 hps-derived MPs display a global transcriptional profile similar to mesenchymal stem or progenitor cells. GO entries on the ffymetrix Human Genome U133 Plus 2. platform were selected randomly from a pool of entries that contained the key words MSs and hss along with several studies focused on various tissue and cell types. ll array data was processed and normalized using the RM feature in the ioconductor affy package in R. Probesets were mapped to and median collapsed onto HUGO gene symbol identifiers and median centered by array. ) Unsupervised hierarchical clustering and heatmap across all represented genes and all arrays acquired from GO. Genes are rows; conditions are columns. Genes are colored according to probe intensity, with blue denoting lower than the median expression, and red denoting higher than the median expression. ) lose up of the array tree cluster in. ) rray clustering and heatmap of the MS subset of arrays from and with the flow cytometry markers which are represented characteristic for a mesenchymal stem cell fate Macmillan Publishers Limited. ll rights reserved.
4 SUPPLMNTRY INFORMTION SV 1 - PPRγ differentiation SV 1 control differentiation SV 2 - PPRγ differentiation SV 2 control differentiation Pluripotent ells 73+ ells Immature dipocytes Mature dipocytes d d6 d16 d35 +ox -ox rightfield HOHST PPRG2 P OIPY Figure S4 Programming with PPRG2 reduces variation of SV differentiation and allows long-term culture of hps-derived after doxycycline withdrawal. -) Two distinct SV lines were differentiated for 21 days (16 days with doxycycline followed by 5 days without doxycycline). oth lines were either untransduced (un) or transduced () with Lenti- PPRG and Lenti-rtT. xperiments were conducted in biological triplicates. Shown are representative images of Oil-Red-O stained. From top to bottom: ) cells; ) un control cells; ) cells; F) un control cells. ) In vitro programming of hpss into. PPRG2- s were differentiated with adipogenic media in the presence of doxycycline for 16 days, followed by an additional 2 days of differentiation in the absence of doxycycline. Top panel left: brightfield image showing the morphology of the differentiated mature. Top panel right: staining with HOHST dye. ottom panel left: Immunostaining for P (red). ottom panel right: Neutral lipid using OIPY dye (green) Macmillan Publishers Limited. ll rights reserved.
5 PRM16 SUPPLMNTRY INFORMTION i ii P Relative xpression to HPRT iii RN copy number: 7.5x 1 7 copies/ml ~ 1.5x1 8 copies/ml Picture Volume rtt Volume GFP i 5 μl 5 μl ii 5 μl 25 μl iii 5 μl 125 μl iv 5 μl 66 μl ells counted GFP + cells % positive cells ST. viral PPRG2 PPRG rtt control 48h PT -OX h PT +OX d PT/ 5d -OX 3.58 iv Relative xpression to HPRT normalized to UP IPONTIN I untransduced PPRG2 PPRG2-PRM16 - OX + OX P-PRM16 PPRG2- P-PRM16 PPRG2-P Figure S5 fficient transduction of hps-derived MPs with inducible lentiviral PPRG2 and P programs a brown adipocyte fate. ) In one well of a 12-well dish, approximately 5, s were transduced with a fixed volume of Lenti-rtT viral supernatant and declining volumes of Lenti-GFP viral supernatant. ells were induced with doxycyline for 48 hours, and GFP fluorescence pictures were acquired using fixedexposure settings. The viral copy number for the Lenti-GFP virus in the supernatants of virus preparations was determined using the Lenti-X qrt- PR method (S). ) Using 5 μl of Lenti-rtT and 5 μl of Lenti-GFP viral supernatants, 5, s plated in wells of a 12-well dish were transduced and exposed to doxycyline for 48 hours. Percentages of GFP-positive cells were determined by counting cells from three independent experiments (S). ) oxycycline-inducible expression of PPRG2. Quantitative RT-PR for viral PPRG2 cn expression normalized to endogenous PPRG expression, except for primary fat for which endogenous PPRG expression normalized to HPRT is shown. From left to right: J RiPS MPs transduced with Lenti-rtT only (control); s transduced with Lenti-rtT and Lenti-PPRG2 cultured in the absence ( OX) or presence (+OX) of doxycycline for 48 hours; s transduced with Lenti-rtT and Lenti-PPRG2 and differentiated for 16 days with doxycycline followed by 5 days without doxycycline; primary fat (S). ) MPs were transduced with Lenti-rtT and Lenti-PRM16 (top panel), or Lenti-rtT and Lenti-P (bottom panel) respectively. The cells were either left untreated (left panel) or exposed to 7 ng/ml of doxycycline for 48h (right panel). Immunostaining was performed using the antibodies against PRM16 or P. (1x magnification) ) ombinations of Lenti-PPRG2, lenti-p and PRM16 were screened for the potential to induce brown fat differentiation. The cells were differentiated for 21 days (14 days with doxycycline followed by 7 days without doxycycline). Shown are the result of subsequent quantitative RT-PR toward UP1 as a brown adipocyte marker, IPONTIN and I, both adipocyte markers. P values in the bar graphs represent two-tailed Student t-tests between the various conditions and un expression values. (n=3, relative expression to HPRT, normalized to 1, *P <.5; P <.1 (S)) Macmillan Publishers Limited. ll rights reserved.
6 SUPPLMNTRY INFORMTION PPRG PPRG -7.5 log2 7.5 PPRG -8 log2 1 PPRG -8 log2 1 PPRG -6 log2 6 P P P PP PP PP P P PP PP PP P PPRG PPRG-P PPRG-P-PRM16-6 log2 6 Figure S6 PPRG2-P and PPRG2-P-PRM16 exhibit global transcriptional profiles consistent with a brown adipocyte fate. ) ffymetix 1. ST microarrays: SVs, s, and s either not exposed to adipogenic media () or cultured with adipogenic media and transduced with lenti-pprg (+PPRG) were compared to primary using ffymetix 1. ST microarrays. Shown is a heatmap of hierarchical clustering performed on the 2,136 differentially expressed genes at a 5% false discovery rate. Genes are rows; conditions are columns. Genes are colored according to average expression level across all samples, with blue denoting lower expression, and red denoting higher expression. ) gilent G G3 microarray: Global transcriptional profiling of PPRG-. SVs, s, and J RiPS MPs either not exposed to adipogenic media () or cultured with adipogenic media and transduced with lenti-pprg (+PPRG) were compared to primary gilent G G3 microarrays. Shown here is unsupervised hierarchical clustering and heatmap across all probes above background on arrays. Genes are rows; conditions are columns. Genes are colored according to probe intensity, with blue denoting lower than the median expression, and red denoting higher than the median expression. ) Heatmap of hierarchical clustering performed on the 4,74 differentially expressed genes at a 5% false discovery rate. Genes are rows; conditions are columns. Genes are colored according to probe intensity with blue denoting lower than the median expression, and red denoting higher than the median expression. ) Row centered heatmap of hierarchical clustering performed on the 4,74 differentially expressed genes at a 5% false discovery rate. Genes are rows; conditions are columns. Genes are colored according to average expression level across all samples, with blue denoting lower expression, and red denoting higher expression. ) Global transcriptional profiling of (PPRG2)-, (PPRG2-P) or (PPRG2-P-PRM16). SVs, s, and s either not exposed to adipogenic media () or cultured with adipogenic media and transduced with lenti-pprg2 (+PPRG2), (PPRG2-P) or (PPRG2-P- PRM16) were compared to primary using the gilent G3 Human G microarray platform. Shown is a row centered heatmap of hierarchical clustering performed on the 2,136 differentially expressed genes at a 1% false discovery rate. Probe sets are colored according to average expression level across all samples, with blue denoting lower expression, and red denoting higher expression Macmillan Publishers Limited. ll rights reserved.
7 SUPPLMNTRY INFORMTION un 5 42: TG 44:2 TG 44:1 TG 44: TG 46:4 TG 46:3 TG 46:2 TG 46:1 TG 46: TG 48:5 TG 48:4 TG 48:3 TG 48:2 TG 48:1 TG 48: TG 5:6 TG 5:5 TG 5:4 TG 5:3 TG 5:2 TG 5:1 TG 5: TG 52:7 TG 52:6 TG 52:5 TG 52:4 TG 52:3 TG 52:2 TG 52:1 TG 52: TG 54:8 TG 54:7 TG 54:6 TG 54:5 TG 54:4 TG 54:3 TG 54:2 TG 54:1 TG 56:9 TG 56:8 TG 56:7 TG 56:6 TG 56:5 TG 56:4 TG 56:3 TG 56:2 TG 58:12 TG 58:11 TG 58:1 TG 58:9 TG 58:8 TG 58:7 TG 58:6 TG 6:12 TG 6:11 TG un 5 42: TG 44:2 TG 44:1 TG 44: TG 46:4 TG 46:3 TG 46:2 TG 46:1 TG 46: TG 48:5 TG 48:4 TG 48:3 TG 48:2 TG 48:1 TG 48: TG 5:6 TG 5:5 TG 5:4 TG 5:3 TG 5:2 TG 5:1 TG 5: TG 52:7 TG 52:6 TG 52:5 TG 52:4 TG 52:3 TG 52:2 TG 52:1 TG 52: TG 54:8 TG 54:7 TG 54:6 TG 54:5 TG 54:4 TG 54:3 TG 54:2 TG 54:1 TG 56:9 TG 56:8 TG 56:7 TG 56:6 TG 56:5 TG 56:4 TG 56:3 TG 56:2 TG 58:12 TG 58:11 TG 58:1 TG 58:9 TG 58:8 TG 58:7 TG 58:6 TG 6:12 TG 6:11 TG 25 SV un 5 42: TG 44:2 TG 44:1 TG 44: TG 46:4 TG 46:3 TG 46:2 TG 46:1 TG 46: TG 48:5 TG 48:4 TG 48:3 TG 48:2 TG 48:1 TG 48: TG 5:6 TG 5:5 TG 5:4 TG 5:3 TG 5:2 TG 5:1 TG 5: TG 52:7 TG 52:6 TG 52:5 TG 52:4 TG 52:3 TG 52:2 TG 52:1 TG 52: TG 54:8 TG 54:7 TG 54:6 TG 54:5 TG 54:4 TG 54:3 TG 54:2 TG 54:1 TG 56:9 TG 56:8 TG 56:7 TG 56:6 TG 56:5 TG 56:4 TG 56:3 TG 56:2 TG 58:12 TG 58:11 TG 58:1 TG 58:9 TG 58:8 TG 58:7 TG 58:6 TG 6:12 TG 6:11 TG un un SV un 32:2 G 32:1 G 34:2 G 34:1 G 36:3 G 36:2 G 36:1 G 32:2 G 32:1 G 34:2 G 34:1 G 36:3 G 36:2 G 36:1 G 32:2 G 32:1 G 34:2 G 34:1 G 36:3 G 36:2 G 36:1 G un un SV un :1 LP 16: LP 18:2 LP 18:1 LP 18: LP 2:4 LP 22:6 LP 16:1 LP 16: LP 18:2 LP 18:1 LP 18: LP 2:4 LP 22:6 LP 16:1 LP 16: LP 18:2 LP 18:1 LP 18: LP 2:4 LP 22:6 LP Figure S7 PPRG2- white lipid profiles resemble primary white adipose tissue. Lipidomic profiling of SV- and hpsderived. The cellular lipid content of SVs, s, and s into with PPRG2 was analyzed using a tandem mass spectroscopy lipidomics platform and compared to primary adipose tissue. Shown are the relative abundances of several long-chain triacylglyceride species in each cell type. The x-axis denotes the total number of carbon atoms in the fatty-acid chains:unsaturated bonds. The y-axis represents the relative abundance of each lipid analyte. are the most abundant triacylglycerides, of a size range between 42: to 6:11. are diacylglycerides of a size range between 32:2 to 36:1. are lysophosphatidylcholine lipids, part of the membrane lipids present in all cells. The displayed size range is between 16:1 and 22: Macmillan Publishers Limited. ll rights reserved.
8 Human Primary Fat Primary Mouse rown Fat SUPPLMNTRY INFORMTION HSL PPRG P p = 2* FP4 IPOQ ,85 p = 2*1 p = 2.2* , ,55 3,9 4 3, ,6 1, ,3 16 * 65 8 HSL LPL 14.5 p = 4.3* p = 2.2* * * * Relative xpression to HPRT d2 set to IPOQ FP * * PPRG +PPRG HUS 8 MP J RiPS [2] MP HUS 8 MP J RiPS [2] HUS 8 MP J RiPS [1] MP J RiPS [2] MP -PPRG +PPRG HUS 8 MP J RiPS [2] MP HUS 8 MP J RiPS [2] HUS 8 MP J RiPS [1] MP J RiPS [2] MP. d2 d4 d8 d12 d16 oxycyline withdrawal 3 Leptin Release + PPRG Glycerol Release untransduced (control) PPRG2-P PPRG2-P-PRM16 Lepin [pg/ml] Glycerol/Protein [control set to 1] * 5 5. HUS 8 MP J RiPS MP HUS 9 MP. -iso +iso -iso +iso -iso +iso i F Pluripotent ells MPs rown dipocytes day - d d5 d16 d25 +ox -ox Primary Mouse iirown Fat iii HOHST M1281 HOHST (Human Nuclei) M1281 (Human Nuclei) rightfield UP1 HOHST UP1 rightfield 2x 4x (1.5 digital zoom) Relative xpression to HPRT d5 (untransdcued) set to 1 4. PRM16 (endogenous transcript) PRM16 (viral+endogenous transcript) J RiPS #1.1 p1 MP p6 day - day 5 day 5 day 5 day 25 day 25 day 25 Pluripotent ells untransduced PPRG2-P PPRG2-P-PRM16 Figure S8 dditional transcriptional and functional characterization of hps-derived. ) Quantitative RT-PR analysis of the expression of adipogenic marker genes: PPRG, P, FP4, IPOQ, HSL, and LPL. The data represent three biological replicates and are shown as relative expression to the housekeeping gene HPRT. Shown in yellow are un, control cells. Shown in green are un, differentiated cell lines. Shown in red are, differentiated cell lines. Shown in blue and, in some graphs, on a separate scale are gene expression levels in primary fat obtained from the pannus of a patient who underwent elective surgery. P values in the bar graphs represent two-tailed Student t-tests between and un expression values for each cell line. n=3 *P <.5; P <.1 (S). P values shown under each gene name represent NOV analyses among all expression values for the and un cell lines. ) hps-derived MPs were transduced with PPRG2 and adipogenic differentiation was initiated through administration of doxycycline in adipogenic medium. oxycycline was withdrawn at indicated time points (X-axis). ll cells were differentiated until day 21. qrt-pr was performed for adipocyte marker genes HSL, IPOQ, FP4. xpression values represent two biological replicates and are shown as relative to HPRT expression in each sample. Relative expression was set to 1 (S). ) Leptin release assay: nzyme-linked immunosorbent assay (LIS) for leptin in the supernatant of hps-derived PPRG2 exposed to adipogenic media, ) Glycerol Release after Isopreterenol exposure: hps-derived brown respond to Isopreterenol by releasing glycerol from the cells. Glycerol was measured in J RiPS-derived MPs that were differentiated with adipogenic media alone (untransduced, white bars), with expression of a combination of (PPRG2-P) (light grey bars) or (PPRG2-P-PRM16) (dark grey bars). The cells were measured at basal level (-iso) and after exposure to Isopreterenol (+iso). The quantity of released glycerol (in μg) was normalized to the total amount of protein (in mg) for each sample (n = 6 Student s t-test P <.1 (SM)). ) Immunostaining of primary mouse brown adipose tissue and human adipose tissue: i) Mouse interscapular T were used as M1281 negative and UP1 positive control. (pictures taken using confocal,microscopy, with 4 fold magnification + confocal digital zoom). ii) rightfield picture (left), UP1 immunostain (middle) and overlay of UP1 and HOHST stain of primary mouse T (magnification 2x). iii) rightfield morphology of human primary fat. (magnification 2x) F) Quantitative RT-PR analysis for the expression of PRM16 in pluripotent cells (black), untransduced MPs (white) or with expression of a combination of (PPRG2-P) (light grey bars) or (PPRG2-P-PRM16) (dark grey bars). ells were collected at day 5 (under exposure to doxycycline) and day 25 (doxycycline withdrawn from media) and expression was measured using oligos detecting only endogenous expressed PRM16 (top) and with oligos detecting endogenous and viral expression. The data is shown as relative expression to the housekeeping gene HPRT, with the untransduced control set as Macmillan Publishers Limited. ll rights reserved.
9 SUPPLMNTRY INFORMTION Figure S9 Western blot analysis of insulin signaling of hps-derived white. The human pluripotent stem cells (hpss)-derived were incubated with insulin alone, free fatty acid mixture (FF) alone, or both. ) Western membrane was incubated with Ser-473 phosphorylated KT antibody. While insulin induced Ser-473 phosphorylation on KT, FF treatment attenuated insulin signaling. ) Western membrane was incubated with total KT. ) Lamin / was used as a loading control. Protein bands corresponding antibodies indicated in the square boxes of the original blots Macmillan Publishers Limited. ll rights reserved.
10 SUPPLMNTRY INFORMTION Supplemental ata Table 1 Worksheet 1: VI GO term analysis for genes up-regulated in PPRG2 and primary over cells in ffymetrix data. Worksheet 2: VI GO term analysis for genes down-regulated in PPRG2 and primary cells in ffymetrix data. Worksheet 3: VI GO term analysis for genes up-regulated in primary over PPRG2 samples in ffymetrix data. Worksheet 4: VI GO term analysis for genes up-regulated in PPRG2 samples over primary samples in ffymetrix data. Supplemental ata Table 2 Work sheet 1: SM output for all genes when testing PPRG2 and primary against all samples for ffymetrix data and Work sheet 2: gilent data. Worksheet 3: SM output for all genes when testing primary against PPRG2 samples for ffymetrix and Worksheet4: gilent data Macmillan Publishers Limited. ll rights reserved.
Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 )
 770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
770 771 Supplementary Table 1. The primers used for quantitative RT-PCR. Gene name Forward (5 > 3 ) Reverse (5 > 3 ) Human CXCL1 GCGCCCAAACCGAAGTCATA ATGGGGGATGCAGGATTGAG PF4 CCCCACTGCCCAACTGATAG TTCTTGTACAGCGGGGCTTG
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
DOI:.38/ncb3399 a b c d FSP DAPI 5mm mm 5mm 5mm e Correspond to melanoma in-situ Figure a DCT FSP- f MITF mm mm MlanaA melanoma in-situ DCT 5mm FSP- mm mm mm mm mm g melanoma in-situ MITF MlanaA mm mm
Males- Western Diet WT KO Age (wks) Females- Western Diet WT KO Age (wks)
 Relative Arv1 mrna Adrenal 33.48 +/- 6.2 Skeletal Muscle 22.4 +/- 4.93 Liver 6.41 +/- 1.48 Heart 5.1 +/- 2.3 Brain 4.98 +/- 2.11 Ovary 4.68 +/- 2.21 Kidney 3.98 +/-.39 Lung 2.15 +/-.6 Inguinal Subcutaneous
Relative Arv1 mrna Adrenal 33.48 +/- 6.2 Skeletal Muscle 22.4 +/- 4.93 Liver 6.41 +/- 1.48 Heart 5.1 +/- 2.3 Brain 4.98 +/- 2.11 Ovary 4.68 +/- 2.21 Kidney 3.98 +/-.39 Lung 2.15 +/-.6 Inguinal Subcutaneous
Supplementary Figures
 Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
Supplementary Figures Supplementary Figure 1 Characterization of stable expression of GlucB and sshbira in the CT26 cell line (a) Live cell imaging of stable CT26 cells expressing green fluorescent protein
IKK-dependent activation of NF-κB contributes to myeloid and lymphoid leukemogenesis by BCR-ABL1
 Supplemental Figures BLOOD/2014/547943 IKK-dependent activation of NF-κB contributes to myeloid and lymphoid leukemogenesis by BCR-ABL1 Hsieh M-Y and Van Etten RA Supplemental Figure S1. Titers of retroviral
Supplemental Figures BLOOD/2014/547943 IKK-dependent activation of NF-κB contributes to myeloid and lymphoid leukemogenesis by BCR-ABL1 Hsieh M-Y and Van Etten RA Supplemental Figure S1. Titers of retroviral
Supplementary Figure 1
 Supplementary Figure 1 how HFD how HFD Epi WT p p Hypothalamus p p Inguinal WT T Liver Lean mouse adipocytes p p p p p p Obese mouse adipocytes Kidney Muscle Spleen Heart p p p p p p p p Extracellular
Supplementary Figure 1 how HFD how HFD Epi WT p p Hypothalamus p p Inguinal WT T Liver Lean mouse adipocytes p p p p p p Obese mouse adipocytes Kidney Muscle Spleen Heart p p p p p p p p Extracellular
TEB. Id4 p63 DAPI Merge. Id4 CK8 DAPI Merge
 a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
a Duct TEB b Id4 p63 DAPI Merge Id4 CK8 DAPI Merge c d e Supplementary Figure 1. Identification of Id4-positive MECs and characterization of the Comma-D model. (a) IHC analysis of ID4 expression in the
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Supplementary Figures Supplementary Figure 1. Confirmation of Dnmt1 conditional knockout out mice. a, Representative images of sorted stem (Lin - CD49f high CD24 + ), luminal (Lin - CD49f low CD24 + )
Influenza virus exploits tunneling nanotubes for cell-to-cell spread
 Supplementary Information Influenza virus exploits tunneling nanotubes for cell-to-cell spread Amrita Kumar 1, Jin Hyang Kim 1, Priya Ranjan 1, Maureen G. Metcalfe 2, Weiping Cao 1, Margarita Mishina 1,
Supplementary Information Influenza virus exploits tunneling nanotubes for cell-to-cell spread Amrita Kumar 1, Jin Hyang Kim 1, Priya Ranjan 1, Maureen G. Metcalfe 2, Weiping Cao 1, Margarita Mishina 1,
SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer.
 Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Supplementary Figure 1 SSM signature genes are highly expressed in residual scar tissues after preoperative radiotherapy of rectal cancer. Scatter plots comparing expression profiles of matched pretreatment
Expanded View Figures
 Shao-Ming Shen et al Role of I in MT of cancers MO reports xpanded View igures igure V1. nalysis of the expression of I isoforms in cancer cells and their interaction with PTN. RT PR detection of Ish and
Shao-Ming Shen et al Role of I in MT of cancers MO reports xpanded View igures igure V1. nalysis of the expression of I isoforms in cancer cells and their interaction with PTN. RT PR detection of Ish and
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
doi:10.1038/nature12652 Supplementary Figure 1. PRDM16 interacts with endogenous EHMT1 in brown adipocytes. Immunoprecipitation of PRDM16 complex by flag antibody (M2) followed by Western blot analysis
Nature Immunology: doi: /ni.3412
 Supplementary Figure 1 Gata1 expression in heamatopoietic stem and progenitor populations. (a) Unsupervised clustering according to 100 top variable genes across single pre-gm cells. The two main cell
Supplementary Figure 1 Gata1 expression in heamatopoietic stem and progenitor populations. (a) Unsupervised clustering according to 100 top variable genes across single pre-gm cells. The two main cell
SUPPLEMENTARY INFORMATION
 DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
DOI: 1.138/ncb3355 a S1A8 + cells/ total.1.8.6.4.2 b S1A8/?-Actin c % T-cell proliferation 3 25 2 15 1 5 T cells Supplementary Figure 1 Inter-tumoral heterogeneity of MDSC accumulation in mammary tumor
Supplemental Information. Granulocyte-Monocyte Progenitors and. Monocyte-Dendritic Cell Progenitors Independently
 Immunity, Volume 47 Supplemental Information Granulocyte-Monocyte Progenitors and Monocyte-endritic ell Progenitors Independently Produce Functionally istinct Monocytes lberto Yáñez, Simon G. oetzee, ndre
Immunity, Volume 47 Supplemental Information Granulocyte-Monocyte Progenitors and Monocyte-endritic ell Progenitors Independently Produce Functionally istinct Monocytes lberto Yáñez, Simon G. oetzee, ndre
Online Appendix Material and Methods: Pancreatic RNA isolation and quantitative real-time (q)rt-pcr. Mice were fasted overnight and killed 1 hour (h)
 Online Appendix Material and Methods: Pancreatic RNA isolation and quantitative real-time (q)rt-pcr. Mice were fasted overnight and killed 1 hour (h) after feeding. A small slice (~5-1 mm 3 ) was taken
Online Appendix Material and Methods: Pancreatic RNA isolation and quantitative real-time (q)rt-pcr. Mice were fasted overnight and killed 1 hour (h) after feeding. A small slice (~5-1 mm 3 ) was taken
Acute lung injury in children : from viral infection and mechanical ventilation to inflammation and apoptosis Bern, R.A.
 UvA-DARE (Digital Academic Repository) Acute lung injury in children : from viral infection and mechanical ventilation to inflammation and apoptosis Bern, R.A. Link to publication Citation for published
UvA-DARE (Digital Academic Repository) Acute lung injury in children : from viral infection and mechanical ventilation to inflammation and apoptosis Bern, R.A. Link to publication Citation for published
ab Adipogenesis Assay Kit (Cell-Based)
 ab133102 Adipogenesis Assay Kit (Cell-Based) Instructions for Use For the study of induction and inhibition of adipogenesis in adherent cells. This product is for research use only and is not intended
ab133102 Adipogenesis Assay Kit (Cell-Based) Instructions for Use For the study of induction and inhibition of adipogenesis in adherent cells. This product is for research use only and is not intended
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2638 Figure S1 Morphological characteristics of fetal testes and ovaries from 6.5-20 developmental weeks. Representative images of Hematoxylin and Eosin staining of testes and ovaries over
DOI: 10.1038/ncb2638 Figure S1 Morphological characteristics of fetal testes and ovaries from 6.5-20 developmental weeks. Representative images of Hematoxylin and Eosin staining of testes and ovaries over
Supplemental figure 1. PDGFRα is expressed dominantly by stromal cells surrounding mammary ducts and alveoli. A) IHC staining of PDGFRα in
 Supplemental figure 1. PDGFRα is expressed dominantly by stromal cells surrounding mammary ducts and alveoli. A) IHC staining of PDGFRα in nulliparous (left panel) and InvD6 mouse mammary glands (right
Supplemental figure 1. PDGFRα is expressed dominantly by stromal cells surrounding mammary ducts and alveoli. A) IHC staining of PDGFRα in nulliparous (left panel) and InvD6 mouse mammary glands (right
Primer sequences Target Sequence F Sequence R TNF-α (Tnfa) TCAGCCGATTTGCTATCTCAT A
 Supplementary Table 1. Q- and RT-PR primers used in this study. Primer sequences Target Sequence F Sequence R TNF-α (Tnfa) TGGTTTGTTTT GTTTGGGGTTG T hemokine (- motif) ligand 5 (cl5) GTGTTTGTTT TGGTGGTG
Supplementary Table 1. Q- and RT-PR primers used in this study. Primer sequences Target Sequence F Sequence R TNF-α (Tnfa) TGGTTTGTTTT GTTTGGGGTTG T hemokine (- motif) ligand 5 (cl5) GTGTTTGTTT TGGTGGTG
s u p p l e m e n ta ry i n f o r m at i o n
 Figure S1 Characterization of tet-off inducible cell lines expressing GFPprogerin and GFP-wt lamin A. a, Western blot analysis of GFP-progerin- or GFP-wt lamin A- expressing cells before induction (0d)
Figure S1 Characterization of tet-off inducible cell lines expressing GFPprogerin and GFP-wt lamin A. a, Western blot analysis of GFP-progerin- or GFP-wt lamin A- expressing cells before induction (0d)
Supplemental Figures Supplemental Figure 1:
 Supplemental Figures Supplemental Figure 1: Representative FACS data showing Concurrent Brain cell type Acquisition using either Percoll PLUS (top row) or myelin removal beads (bottom two rows). Debris
Supplemental Figures Supplemental Figure 1: Representative FACS data showing Concurrent Brain cell type Acquisition using either Percoll PLUS (top row) or myelin removal beads (bottom two rows). Debris
Supporting Information Table of Contents
 Supporting Information Table of Contents Supporting Information Figure 1 Page 2 Supporting Information Figure 2 Page 4 Supporting Information Figure 3 Page 5 Supporting Information Figure 4 Page 6 Supporting
Supporting Information Table of Contents Supporting Information Figure 1 Page 2 Supporting Information Figure 2 Page 4 Supporting Information Figure 3 Page 5 Supporting Information Figure 4 Page 6 Supporting
D CD8 T cell number (x10 6 )
 IFNγ Supplemental Figure 1. CD T cell number (x1 6 ) 18 15 1 9 6 3 CD CD T cells CD6L C CD5 CD T cells CD6L D CD8 T cell number (x1 6 ) 1 8 6 E CD CD8 T cells CD6L F Log(1)CFU/g Feces 1 8 6 p
IFNγ Supplemental Figure 1. CD T cell number (x1 6 ) 18 15 1 9 6 3 CD CD T cells CD6L C CD5 CD T cells CD6L D CD8 T cell number (x1 6 ) 1 8 6 E CD CD8 T cells CD6L F Log(1)CFU/g Feces 1 8 6 p
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
doi:10.1038/nature10866 a b 1 2 3 4 5 6 7 Match No Match 1 2 3 4 5 6 7 Turcan et al. Supplementary Fig.1 Concepts mapping H3K27 targets in EF CBX8 targets in EF H3K27 targets in ES SUZ12 targets in ES
activation with anti-cd3/cd28 beads and 3d following transduction. Supplemental Figure 2 shows
 Supplemental Data Supplemental Figure 1 compares CXCR4 expression in untreated CD8 + T cells, following activation with anti-cd3/cd28 beads and 3d following transduction. Supplemental Figure 2 shows the
Supplemental Data Supplemental Figure 1 compares CXCR4 expression in untreated CD8 + T cells, following activation with anti-cd3/cd28 beads and 3d following transduction. Supplemental Figure 2 shows the
SUPPLEMENTARY INFORMATION
 Supplemental Figure 1. Furin is efficiently deleted in CD4 + and CD8 + T cells. a, Western blot for furin and actin proteins in CD4cre-fur f/f and fur f/f Th1 cells. Wild-type and furin-deficient CD4 +
Supplemental Figure 1. Furin is efficiently deleted in CD4 + and CD8 + T cells. a, Western blot for furin and actin proteins in CD4cre-fur f/f and fur f/f Th1 cells. Wild-type and furin-deficient CD4 +
SUPPLEMENTAL INFORMATIONS
 1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
1 SUPPLEMENTAL INFORMATIONS Figure S1 Cumulative ZIKV production by testis explants over a 9 day-culture period. Viral titer values presented in Figure 1B (viral release over a 3 day-culture period measured
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Fig. 1: Quality assessment of formalin-fixed paraffin-embedded (FFPE)-derived DNA and nuclei. (a) Multiplex PCR analysis of unrepaired and repaired bulk FFPE gdna from
Supplementary Figure 1 Supplementary Fig. 1: Quality assessment of formalin-fixed paraffin-embedded (FFPE)-derived DNA and nuclei. (a) Multiplex PCR analysis of unrepaired and repaired bulk FFPE gdna from
Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression.
 Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Supplementary Figure 1 Lentiviral Delivery of Combinatorial mirna Expression Constructs Provides Efficient Target Gene Repression. a, Design for lentiviral combinatorial mirna expression and sensor constructs.
Expanded View Figures
 Sarah Kit Leng Lui et al USP26 stabilizes SM7 MO reports xpanded View igures igure V1. USP26 enhances SM2 phosphorylation and T-b-mediated transcription. raph representing relative luciferase values obtained
Sarah Kit Leng Lui et al USP26 stabilizes SM7 MO reports xpanded View igures igure V1. USP26 enhances SM2 phosphorylation and T-b-mediated transcription. raph representing relative luciferase values obtained
Supplemental Figure 1. Intracranial transduction of a modified ptomo lentiviral vector in the mouse
 Supplemental figure legends Supplemental Figure 1. Intracranial transduction of a modified ptomo lentiviral vector in the mouse hippocampus targets GFAP-positive but not NeuN-positive cells. (A) Stereotaxic
Supplemental figure legends Supplemental Figure 1. Intracranial transduction of a modified ptomo lentiviral vector in the mouse hippocampus targets GFAP-positive but not NeuN-positive cells. (A) Stereotaxic
a b G75 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server.
 a b G75 2 2 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server. a. Overlay of top 10 models generated by I-TASSER illustrates the potential effect of 7 amino acid insertion
a b G75 2 2 G60 Sw-2 Sw-1 Supplementary Figure 1. Structure predictions by I-TASSER Server. a. Overlay of top 10 models generated by I-TASSER illustrates the potential effect of 7 amino acid insertion
Nature Immunology: doi: /ni Supplementary Figure 1. Huwe1 has high expression in HSCs and is necessary for quiescence.
 Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplementary Figure 1 Huwe1 has high expression in HSCs and is necessary for quiescence. (a) Heat map visualizing expression of genes with a known function in ubiquitin-mediated proteolysis (KEGG: Ubiquitin
Supplemental Table 1. Primer sequences for transcript analysis
 Supplemental Table 1. Primer sequences for transcript analysis Primer Sequence (5 3 ) Primer Sequence (5 3 ) Mmp2 Forward CCCGTGTGGCCCTC Mmp15 Forward CGGGGCTGGCT Reverse GCTCTCCCGGTTTC Reverse CCTGGTGTGCCTGCTC
Supplemental Table 1. Primer sequences for transcript analysis Primer Sequence (5 3 ) Primer Sequence (5 3 ) Mmp2 Forward CCCGTGTGGCCCTC Mmp15 Forward CGGGGCTGGCT Reverse GCTCTCCCGGTTTC Reverse CCTGGTGTGCCTGCTC
Nature Genetics: doi: /ng Supplementary Figure 1. Phenotypic characterization of MES- and ADRN-type cells.
 Supplementary Figure 1 Phenotypic characterization of MES- and ADRN-type cells. (a) Bright-field images showing cellular morphology of MES-type (691-MES, 700-MES, 717-MES) and ADRN-type (691-ADRN, 700-
Supplementary Figure 1 Phenotypic characterization of MES- and ADRN-type cells. (a) Bright-field images showing cellular morphology of MES-type (691-MES, 700-MES, 717-MES) and ADRN-type (691-ADRN, 700-
Nature Immunology: doi: /ni Supplementary Figure 1. Characteristics of SEs in T reg and T conv cells.
 Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Supplementary Figure 1 Characteristics of SEs in T reg and T conv cells. (a) Patterns of indicated transcription factor-binding at SEs and surrounding regions in T reg and T conv cells. Average normalized
Figure S1. PMVs from THP-1 cells expose phosphatidylserine and carry actin. A) Flow
 SUPPLEMENTARY DATA Supplementary Figure Legends Figure S1. PMVs from THP-1 cells expose phosphatidylserine and carry actin. A) Flow cytometry analysis of PMVs labelled with annexin-v-pe (Guava technologies)
SUPPLEMENTARY DATA Supplementary Figure Legends Figure S1. PMVs from THP-1 cells expose phosphatidylserine and carry actin. A) Flow cytometry analysis of PMVs labelled with annexin-v-pe (Guava technologies)
Supplementary Figures
 Supplementary Figures Supplementary Fig. 1. Galectin-3 is present within tumors. (A) mrna expression levels of Lgals3 (galectin-3) and Lgals8 (galectin-8) in the four classes of cell lines as determined
Supplementary Figures Supplementary Fig. 1. Galectin-3 is present within tumors. (A) mrna expression levels of Lgals3 (galectin-3) and Lgals8 (galectin-8) in the four classes of cell lines as determined
Formula A: Percentage Wound Closure: % C = percentage wound closure, A0 = Area of wound at day 0, Ai = Area of wound at 7 days.
 Formulae for Wound Closure and Stereology (15, 16, 20) Formula A: Percentage Wound Closure: % C = percentage wound closure, A0 = Area of wound at day 0, Ai = Area of wound at 7 days. A0 Ai 100 %C = x A0
Formulae for Wound Closure and Stereology (15, 16, 20) Formula A: Percentage Wound Closure: % C = percentage wound closure, A0 = Area of wound at day 0, Ai = Area of wound at 7 days. A0 Ai 100 %C = x A0
Supplemental Information. CD4 + CD25 + Foxp3 + Regulatory T Cells Promote. Th17 Cells In Vitro and Enhance Host Resistance
 Immunity, Volume 34 Supplemental Information D4 + D25 + + Regulatory T ells Promote Th17 ells In Vitro and Enhance Host Resistance in Mouse andida albicans Th17 ell Infection Model Pushpa Pandiyan, Heather
Immunity, Volume 34 Supplemental Information D4 + D25 + + Regulatory T ells Promote Th17 ells In Vitro and Enhance Host Resistance in Mouse andida albicans Th17 ell Infection Model Pushpa Pandiyan, Heather
B220 CD4 CD8. Figure 1. Confocal Image of Sensitized HLN. Representative image of a sensitized HLN
 B220 CD4 CD8 Natarajan et al., unpublished data Figure 1. Confocal Image of Sensitized HLN. Representative image of a sensitized HLN showing B cell follicles and T cell areas. 20 µm thick. Image of magnification
B220 CD4 CD8 Natarajan et al., unpublished data Figure 1. Confocal Image of Sensitized HLN. Representative image of a sensitized HLN showing B cell follicles and T cell areas. 20 µm thick. Image of magnification
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
DOI: 10.1038/ncb2607 Figure S1 Elf5 loss promotes EMT in mammary epithelium while Elf5 overexpression inhibits TGFβ induced EMT. (a, c) Different confocal slices through the Z stack image. (b, d) 3D rendering
Pathologic Stage. Lymph node Stage
 ASC ASC a c Patient ID BMI Age Gleason score Non-obese PBMC 1 22.1 81 6 (3+3) PBMC 2 21.9 6 6 (3+3) PBMC 3 22 84 8 (4+4) PBMC 4 24.6 68 7 (3+4) PBMC 24. 6 (3+3) PBMC 6 24.7 73 7 (3+4) PBMC 7 23. 67 7 (3+4)
ASC ASC a c Patient ID BMI Age Gleason score Non-obese PBMC 1 22.1 81 6 (3+3) PBMC 2 21.9 6 6 (3+3) PBMC 3 22 84 8 (4+4) PBMC 4 24.6 68 7 (3+4) PBMC 24. 6 (3+3) PBMC 6 24.7 73 7 (3+4) PBMC 7 23. 67 7 (3+4)
Supplementary Figure 1. Nature Neuroscience: doi: /nn.4547
 Supplementary Figure 1 Characterization of the Microfetti mouse model. (a) Gating strategy for 8-color flow analysis of peripheral Ly-6C + monocytes from Microfetti mice 5-7 days after TAM treatment. Living
Supplementary Figure 1 Characterization of the Microfetti mouse model. (a) Gating strategy for 8-color flow analysis of peripheral Ly-6C + monocytes from Microfetti mice 5-7 days after TAM treatment. Living
Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original
 Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original phenotypic screening was n=40. For specific tests, the
Supplementary Fig. 1 eif6 +/- mice show a reduction in white adipose tissue, blood lipids and normal glycogen synthesis. The cohort of the original phenotypic screening was n=40. For specific tests, the
Layered-IHC (L-IHC): A novel and robust approach to multiplexed immunohistochemistry So many markers and so little tissue
 Page 1 The need for multiplex detection of tissue biomarkers. There is a constant and growing demand for increased biomarker analysis in human tissue specimens. Analysis of tissue biomarkers is key to
Page 1 The need for multiplex detection of tissue biomarkers. There is a constant and growing demand for increased biomarker analysis in human tissue specimens. Analysis of tissue biomarkers is key to
Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma.
 Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
Supplementary Figure 1 Breeding scheme, transgenes, histological analysis and site distribution of SB-mutagenized osteosarcoma. (a) Breeding scheme. R26-LSL-SB11 homozygous mice were bred to Trp53 LSL-R270H/+
Supplementary Figure 1 Chemokine and chemokine receptor expression during muscle regeneration (a) Analysis of CR3CR1 mrna expression by real time-pcr
 Supplementary Figure 1 Chemokine and chemokine receptor expression during muscle regeneration (a) Analysis of CR3CR1 mrna expression by real time-pcr at day 0, 1, 4, 10 and 21 post- muscle injury. (b)
Supplementary Figure 1 Chemokine and chemokine receptor expression during muscle regeneration (a) Analysis of CR3CR1 mrna expression by real time-pcr at day 0, 1, 4, 10 and 21 post- muscle injury. (b)
Supplementary Figure 1. BMS enhances human T cell activation in vitro in a
 Supplementary Figure 1. BMS98662 enhances human T cell activation in vitro in a concentration-dependent manner. Jurkat T cells were activated with anti-cd3 and anti-cd28 antibody in the presence of titrated
Supplementary Figure 1. BMS98662 enhances human T cell activation in vitro in a concentration-dependent manner. Jurkat T cells were activated with anti-cd3 and anti-cd28 antibody in the presence of titrated
Supplementary material. Supplementary Figure legends
 Supplementary material Supplementary Figure legends Supplementary Figure 1: Senescence-associated proliferation stop in response to oncogenic N-RAS expression Proliferation of NHEM cells without (ctrl.)
Supplementary material Supplementary Figure legends Supplementary Figure 1: Senescence-associated proliferation stop in response to oncogenic N-RAS expression Proliferation of NHEM cells without (ctrl.)
Supplemental Figures Legends and Supplemental Figures. for. pirna-guided slicing of transposon transcripts enforces their transcriptional
 Supplemental Figures Legends and Supplemental Figures for pirn-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear pirn repertoire Kirsten-ndré
Supplemental Figures Legends and Supplemental Figures for pirn-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear pirn repertoire Kirsten-ndré
General Laboratory methods Plasma analysis: Gene Expression Analysis: Immunoblot analysis: Immunohistochemistry:
 General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
General Laboratory methods Plasma analysis: Plasma insulin (Mercodia, Sweden), leptin (duoset, R&D Systems Europe, Abingdon, United Kingdom), IL-6, TNFα and adiponectin levels (Quantikine kits, R&D Systems
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Binding capacity of DNA-barcoded MHC multimers and recovery of antigen specificity
 Supplementary Figure 1 Binding capacity of DNA-barcoded MHC multimers and recovery of antigen specificity (a, b) Fluorescent-based determination of the binding capacity of DNA-barcoded MHC multimers (+barcode)
Supplementary Figure 1 Binding capacity of DNA-barcoded MHC multimers and recovery of antigen specificity (a, b) Fluorescent-based determination of the binding capacity of DNA-barcoded MHC multimers (+barcode)
Supplementary Information. Supplementary Figure 1
 Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Supplementary Information Supplementary Figure 1 1 Supplementary Figure 1. Functional assay of the hcas9-2a-mcherry construct (a) Gene correction of a mutant EGFP reporter cell line mediated by hcas9 or
Nature Immunology: doi: /ni Supplementary Figure 1. Transcriptional program of the TE and MP CD8 + T cell subsets.
 Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Supplementary Figure 1 Transcriptional program of the TE and MP CD8 + T cell subsets. (a) Comparison of gene expression of TE and MP CD8 + T cell subsets by microarray. Genes that are 1.5-fold upregulated
Supplementary Table; Supplementary Figures and legends S1-S21; Supplementary Materials and Methods
 Silva et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability Supplementary Table; Supplementary Figures and legends S1-S21; Supplementary
Silva et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability Supplementary Table; Supplementary Figures and legends S1-S21; Supplementary
AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination
 AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination Supplementary Figure 1. Generation of the adult-onset, liver-specific GH receptor knock-down (alivghrkd, Kd) mouse
AAV-TBGp-Cre treatment resulted in hepatocyte-specific GH receptor gene recombination Supplementary Figure 1. Generation of the adult-onset, liver-specific GH receptor knock-down (alivghrkd, Kd) mouse
0.0 All T-lymph B-lymph Sarcomas Carcinomas Germ cell. Tumor type
 Fig S1 A B Tumors per mouse 1.2 1.0 0.8 0.6 0.4 0.2 0.0 All T-lymph B-lymph Sarcomas Carcinomas Germ cell Tumor type -/- (n = 46) Q/- (n = 76) Q/Q (n = 31) Tumors per mouse 1.2 1.0 0.8 0.6 0.4 0.2 0.0
Fig S1 A B Tumors per mouse 1.2 1.0 0.8 0.6 0.4 0.2 0.0 All T-lymph B-lymph Sarcomas Carcinomas Germ cell Tumor type -/- (n = 46) Q/- (n = 76) Q/Q (n = 31) Tumors per mouse 1.2 1.0 0.8 0.6 0.4 0.2 0.0
Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures
 Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures Supplementary Figure 1 mir-128-3p is highly expressed in chemoresistant, metastatic
Type of file: PDF Size of file: 0 KB Title of file for HTML: Supplementary Information Description: Supplementary Figures Supplementary Figure 1 mir-128-3p is highly expressed in chemoresistant, metastatic
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/3/114/ra23/dc1 Supplementary Materials for Regulation of Zap70 Expression During Thymocyte Development Enables Temporal Separation of CD4 and CD8 Repertoire Selection
www.sciencesignaling.org/cgi/content/full/3/114/ra23/dc1 Supplementary Materials for Regulation of Zap70 Expression During Thymocyte Development Enables Temporal Separation of CD4 and CD8 Repertoire Selection
a" b" 2N c" d" e" f" !!Aurora!A!!!CP110!
 DLD1/Reference a" 2N 2N 2N/DLD1 2N/ /DLD1 c" d" e" f" TargetID 2N.AVG_Sig 2N.Det Pval.AVG_Sig.Det Pval Diff Pval DiffScore SYMBOL ILMN_26396 18.35238 0.0080058 44.81118 0.0021834 0.000323 34.90542 KRTHA4
DLD1/Reference a" 2N 2N 2N/DLD1 2N/ /DLD1 c" d" e" f" TargetID 2N.AVG_Sig 2N.Det Pval.AVG_Sig.Det Pval Diff Pval DiffScore SYMBOL ILMN_26396 18.35238 0.0080058 44.81118 0.0021834 0.000323 34.90542 KRTHA4
Nature Medicine: doi: /nm.2109
 HIV 1 Infects Multipotent Progenitor Cells Causing Cell Death and Establishing Latent Cellular Reservoirs Christoph C. Carter, Adewunmi Onafuwa Nuga, Lucy A. M c Namara, James Riddell IV, Dale Bixby, Michael
HIV 1 Infects Multipotent Progenitor Cells Causing Cell Death and Establishing Latent Cellular Reservoirs Christoph C. Carter, Adewunmi Onafuwa Nuga, Lucy A. M c Namara, James Riddell IV, Dale Bixby, Michael
Santulli G. et al. A microrna-based strategy to suppress restenosis while preserving endothelial function
 ONLINE DATA SUPPLEMENTS Santulli G. et al. A microrna-based strategy to suppress restenosis while preserving endothelial function Supplementary Figures Figure S1 Effect of Ad-p27-126TS on the expression
ONLINE DATA SUPPLEMENTS Santulli G. et al. A microrna-based strategy to suppress restenosis while preserving endothelial function Supplementary Figures Figure S1 Effect of Ad-p27-126TS on the expression
stem cell products Basement Membrane Matrix Products Rat Mesenchymal Stem Cell Growth and Differentiation Products
 stem cell products Basement Membrane Matrix Products Rat Mesenchymal Stem Cell Growth and Differentiation Products Stem Cell Qualified Extracellular Matrix Proteins Stem cell research requires the finest
stem cell products Basement Membrane Matrix Products Rat Mesenchymal Stem Cell Growth and Differentiation Products Stem Cell Qualified Extracellular Matrix Proteins Stem cell research requires the finest
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
www.sciencesignaling.org/cgi/content/full/8/375/ra41/dc1 Supplementary Materials for Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Supplementary Figure 1. Generation of a conditional allele of the Kindlin-2 gene. (A) A restriction map of the relevant genomic region of Kindlin-2 (top), the targeting construct
SUPPLEMENTARY INFORMATION Supplementary Figure 1. Generation of a conditional allele of the Kindlin-2 gene. (A) A restriction map of the relevant genomic region of Kindlin-2 (top), the targeting construct
Supporting Information
 Supporting Information lpek et al. 1.173/pnas.1121217 SI Materials and Methods Mice. cell knockout, inos / (Taconic arms), Rag1 /, INγR /, and IL-12p4 / mice (The Jackson Laboratory) were maintained and/or
Supporting Information lpek et al. 1.173/pnas.1121217 SI Materials and Methods Mice. cell knockout, inos / (Taconic arms), Rag1 /, INγR /, and IL-12p4 / mice (The Jackson Laboratory) were maintained and/or
Supplementary Figure 1. The CagA-dependent wound healing or transwell migration of gastric cancer cell. AGS cells transfected with vector control or
 Supplementary Figure 1. The CagA-dependent wound healing or transwell migration of gastric cancer cell. AGS cells transfected with vector control or 3xflag-CagA expression vector were wounded using a pipette
Supplementary Figure 1. The CagA-dependent wound healing or transwell migration of gastric cancer cell. AGS cells transfected with vector control or 3xflag-CagA expression vector were wounded using a pipette
SD-1 SD-1: Cathepsin B levels in TNF treated hch
 SD-1 SD-1: Cathepsin B levels in TNF treated hch. A. RNA and B. protein extracts from TNF treated and untreated human chondrocytes (hch) were analyzed via qpcr (left) and immunoblot analyses (right) for
SD-1 SD-1: Cathepsin B levels in TNF treated hch. A. RNA and B. protein extracts from TNF treated and untreated human chondrocytes (hch) were analyzed via qpcr (left) and immunoblot analyses (right) for
7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans.
 Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
Supplementary Figure 1 7SK ChIRP-seq is specifically RNA dependent and conserved between mice and humans. Regions targeted by the Even and Odd ChIRP probes mapped to a secondary structure model 56 of the
PCs Acyl chain double bonds. PEs
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2014 Male/Female ratio Male/Female ratio PCs 1.3 1.2 1.1 1 0.9 0.8 0.7 0.6 0.5 0.4 0 1 2
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2014 Male/Female ratio Male/Female ratio PCs 1.3 1.2 1.1 1 0.9 0.8 0.7 0.6 0.5 0.4 0 1 2
MALDI Activity 4 MALDI-TOF Mass Spectrometry: Data Analysis
 MALDI Activity 4 MALDI-TOF Mass Spectrometry: Data Analysis Model 1: Introduction to Triacylglycerides (TAGs) In MALDI Activity 3 you learned how to open your raw mass spectrum data and use the MMass program
MALDI Activity 4 MALDI-TOF Mass Spectrometry: Data Analysis Model 1: Introduction to Triacylglycerides (TAGs) In MALDI Activity 3 you learned how to open your raw mass spectrum data and use the MMass program
Figure 1. Dnmt3b expression in murine and human knee joint cartilage. (A) Representative images
 Figure Legends Figure. expression in murine and human knee joint cartilage. () Representative images showing that Dnmta is not expressed in chondrocytes from mo W articular cartilage [Dnmta expression
Figure Legends Figure. expression in murine and human knee joint cartilage. () Representative images showing that Dnmta is not expressed in chondrocytes from mo W articular cartilage [Dnmta expression
MicroRNAs Modulate the Noncanonical NF- B Pathway by Regulating IKK Expression During Macrophage Differentiation
 MicroRNAs Modulate the Noncanonical NF- B Pathway by Regulating IKK Expression During Macrophage Differentiation Tao Li 1 *, Michael J. Morgan 1 *, Swati Choksi 1, Yan Zhang 1, You-Sun Kim 2#, Zheng-gang
MicroRNAs Modulate the Noncanonical NF- B Pathway by Regulating IKK Expression During Macrophage Differentiation Tao Li 1 *, Michael J. Morgan 1 *, Swati Choksi 1, Yan Zhang 1, You-Sun Kim 2#, Zheng-gang
Figure S1. Sorting nexin 9 (SNX9) specifically binds psmad3 and not psmad 1/5/8. Lysates from AKR-2B cells untreated (-) or stimulated (+) for 45 min
 Figure S1. Sorting nexin 9 (SNX9) specifically binds psmad3 and not psmad 1/5/8. Lysates from AKR2B cells untreated () or stimulated () for 45 min with 5 ng/ml TGFβ or 10 ng/ml BMP4 were incubated with
Figure S1. Sorting nexin 9 (SNX9) specifically binds psmad3 and not psmad 1/5/8. Lysates from AKR2B cells untreated () or stimulated () for 45 min with 5 ng/ml TGFβ or 10 ng/ml BMP4 were incubated with
SUPPLEMENTARY MATERIAL
 SUPPLEMENTARY MATERIAL IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the T H 17 cell it reg cell balance Rajatava Basu 1,5, Sarah K.
SUPPLEMENTARY MATERIAL IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the T H 17 cell it reg cell balance Rajatava Basu 1,5, Sarah K.
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation.
 Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supplementary Figure 1: si-craf but not si-braf sensitizes tumor cells to radiation. (a) Embryonic fibroblasts isolated from wildtype (WT), BRAF -/-, or CRAF -/- mice were irradiated (6 Gy) and DNA damage
Supporting Information
 Supporting Information Kayama et al. 1.173/pnas.11193119 SI Materials and Methods Reagents. 1-methyl-L-tryptophan and nordihydroguaiaretic acid were purchased from Sigma Aldrich. N W -hydroxyl-nor-l-arginine,
Supporting Information Kayama et al. 1.173/pnas.11193119 SI Materials and Methods Reagents. 1-methyl-L-tryptophan and nordihydroguaiaretic acid were purchased from Sigma Aldrich. N W -hydroxyl-nor-l-arginine,
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells
 Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1.TRIM33 binds β-catenin in the nucleus. a & b, Co-IP of endogenous TRIM33 with β-catenin in HT-29 cells (a) and HEK 293T cells (b). TRIM33 was immunoprecipitated, and the amount of
Supplementary Figure 1. Using DNA barcode-labeled MHC multimers to generate TCR fingerprints
 Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
Supplementary Figure 1 Using DNA barcode-labeled MHC multimers to generate TCR fingerprints (a) Schematic overview of the workflow behind a TCR fingerprint. Each peptide position of the original peptide
GPR120 *** * * Liver BAT iwat ewat mwat Ileum Colon. UCP1 mrna ***
 a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
a GPR120 GPR120 mrna/ppia mrna Arbitrary Units 150 100 50 Liver BAT iwat ewat mwat Ileum Colon b UCP1 mrna Fold induction 20 15 10 5 - camp camp SB202190 - - - H89 - - - - - GW7647 Supplementary Figure
SUPPLEMENTARY INFORMATION
 doi: 1.138/nature7221 Brown fat selective genes 12 1 Control Q-RT-PCR (% of Control) 8 6 4 2 Ntrk3 Cox7a1 Cox8b Cox5b ATPase b2 ATPase f1a1 Sirt3 ERRα Elovl3/Cig3 PPARα Zic1 Supplementary Figure S1. stimulates
doi: 1.138/nature7221 Brown fat selective genes 12 1 Control Q-RT-PCR (% of Control) 8 6 4 2 Ntrk3 Cox7a1 Cox8b Cox5b ATPase b2 ATPase f1a1 Sirt3 ERRα Elovl3/Cig3 PPARα Zic1 Supplementary Figure S1. stimulates
Supplementary Figure 1
 Supplementary Figure 1 A B mir-141, human cell lines mir-2c, human cell lines mir-141, hepatocytes mir-2c, hepatocytes Relative RNA.1.8.6.4.2 Relative RNA.3.2.1 Relative RNA 1.5 1..5 Relative RNA 2. 1.5
Supplementary Figure 1 A B mir-141, human cell lines mir-2c, human cell lines mir-141, hepatocytes mir-2c, hepatocytes Relative RNA.1.8.6.4.2 Relative RNA.3.2.1 Relative RNA 1.5 1..5 Relative RNA 2. 1.5
In vitro human regulatory T cell suppression assay
 Human CD4 + CD25 + regulatory T cell isolation, in vitro suppression assay and analysis In vitro human regulatory T cell suppression assay Introduction Regulatory T (Treg) cells are a subpopulation of
Human CD4 + CD25 + regulatory T cell isolation, in vitro suppression assay and analysis In vitro human regulatory T cell suppression assay Introduction Regulatory T (Treg) cells are a subpopulation of
Nature Immunology: doi: /ni Supplementary Figure 1
 Supplementary Figure 1 Fatty acid oxidation is emphasized in 1 macrophages compared with that in macrophages. Gene expression of mitochondrial OXPHOS (Atp5j, Cox4i1, Uqcrc1/2, Ndufs1, Sdhb) and β-oxidation
Supplementary Figure 1 Fatty acid oxidation is emphasized in 1 macrophages compared with that in macrophages. Gene expression of mitochondrial OXPHOS (Atp5j, Cox4i1, Uqcrc1/2, Ndufs1, Sdhb) and β-oxidation
Nature Genetics: doi: /ng Supplementary Figure 1. HOX fusions enhance self-renewal capacity.
 Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
Supplementary Figure 1 HOX fusions enhance self-renewal capacity. Mouse bone marrow was transduced with a retrovirus carrying one of three HOX fusion genes or the empty mcherry reporter construct as described
4. Th1-related gene expression in infected versus mock-infected controls from Fig. 2 with gene annotation.
 List of supplemental information 1. Graph of mouse weight loss during course of infection- Line graphs showing mouse weight data during course of infection days 1 to 10 post-infections (p.i.). 2. Graph
List of supplemental information 1. Graph of mouse weight loss during course of infection- Line graphs showing mouse weight data during course of infection days 1 to 10 post-infections (p.i.). 2. Graph
Imtiyaz et al., Fig. S1
 . Imtiyaz et al., Fig. S1 1. 1.1 1% O.1.5 Lin/Sca-1/IL-7Rα GMPs.17 MPs.3 Days 3% O.1 MEPs.35 D3 Days 1, N, N, H, H 1 1 Days Supplemental Figure S1. Macrophage maturation, proliferation and survival are
. Imtiyaz et al., Fig. S1 1. 1.1 1% O.1.5 Lin/Sca-1/IL-7Rα GMPs.17 MPs.3 Days 3% O.1 MEPs.35 D3 Days 1, N, N, H, H 1 1 Days Supplemental Figure S1. Macrophage maturation, proliferation and survival are
SUPPLEMENTARY FIGURES AND TABLES
 SUPPLEMENTARY FIGURES AND TABLES Supplementary Figure S1: CaSR expression in neuroblastoma models. A. Proteins were isolated from three neuroblastoma cell lines and from the liver metastasis of a MYCN-non
SUPPLEMENTARY FIGURES AND TABLES Supplementary Figure S1: CaSR expression in neuroblastoma models. A. Proteins were isolated from three neuroblastoma cell lines and from the liver metastasis of a MYCN-non
Transient β-hairpin Formation in α-synuclein Monomer Revealed by Coarse-grained Molecular Dynamics Simulation
 Transient β-hairpin Formation in α-synuclein Monomer Revealed by Coarse-grained Molecular Dynamics Simulation Hang Yu, 1, 2, a) Wei Han, 1, 3, b) Wen Ma, 1, 2 1, 2, 3, c) and Klaus Schulten 1) Beckman
Transient β-hairpin Formation in α-synuclein Monomer Revealed by Coarse-grained Molecular Dynamics Simulation Hang Yu, 1, 2, a) Wei Han, 1, 3, b) Wen Ma, 1, 2 1, 2, 3, c) and Klaus Schulten 1) Beckman
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2535 Figure S1 SOX10 is expressed in human giant congenital nevi and its expression in human melanoma samples suggests that SOX10 functions in a MITF-independent manner. a, b, Representative
DOI: 10.1038/ncb2535 Figure S1 SOX10 is expressed in human giant congenital nevi and its expression in human melanoma samples suggests that SOX10 functions in a MITF-independent manner. a, b, Representative
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10188 Supplementary Figure 1. Embryonic epicardial genes are down-regulated from midgestation stages and barely detectable post-natally. Real time qrt-pcr revealed a significant down-regulation
doi:10.1038/nature10188 Supplementary Figure 1. Embryonic epicardial genes are down-regulated from midgestation stages and barely detectable post-natally. Real time qrt-pcr revealed a significant down-regulation
Supplemental Information. Increased 4E-BP1 Expression Protects. against Diet-Induced Obesity and Insulin. Resistance in Male Mice
 Cell Reports, Volume 16 Supplemental Information Increased 4E-BP1 Expression Protects against Diet-Induced Obesity and Insulin Resistance in Male Mice Shih-Yin Tsai, Ariana A. Rodriguez, Somasish G. Dastidar,
Cell Reports, Volume 16 Supplemental Information Increased 4E-BP1 Expression Protects against Diet-Induced Obesity and Insulin Resistance in Male Mice Shih-Yin Tsai, Ariana A. Rodriguez, Somasish G. Dastidar,
Expanded View Figures
 The EMO Journal Dnmt regulates translational fidelity Francesca Tuorto et al Expanded View Figures x E3 cells/µl W g/l HG x e3 cells/µl PLT % of cell type 8 day 8 month % of cell type 8 day 8 month 8 day
The EMO Journal Dnmt regulates translational fidelity Francesca Tuorto et al Expanded View Figures x E3 cells/µl W g/l HG x e3 cells/µl PLT % of cell type 8 day 8 month % of cell type 8 day 8 month 8 day
Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing)
 Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing) platform with laser manipulation to efficiently purify lung
Supplementary Figure 1. Identification of tumorous sphere-forming CSCs and CAF feeder cells. The LEAP (Laser-Enabled Analysis and Processing) platform with laser manipulation to efficiently purify lung
SUPPLEMENTARY INFORMATION
 doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
doi:.38/nature8975 SUPPLEMENTAL TEXT Unique association of HOTAIR with patient outcome To determine whether the expression of other HOX lincrnas in addition to HOTAIR can predict patient outcome, we measured
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
SUPPLEMENTARY INFORMATION doi:10.1038/nature11429 S1a 6 7 8 9 Nlrc4 allele S1b Nlrc4 +/+ Nlrc4 +/F Nlrc4 F/F 9 Targeting construct 422 bp 273 bp FRT-neo-gb-PGK-FRT 3x.STOP S1c Nlrc4 +/+ Nlrc4 F/F casp1
Supplementary Materials
 Supplementary Materials Figure S1. MTT Cell viability assay. To measure the cytotoxic potential of the oxidative treatment, the MTT [3-(4,5-dimethylthiazol- 2-yl)-2,5-diphenyl tetrazolium bromide] assay
Supplementary Materials Figure S1. MTT Cell viability assay. To measure the cytotoxic potential of the oxidative treatment, the MTT [3-(4,5-dimethylthiazol- 2-yl)-2,5-diphenyl tetrazolium bromide] assay
